Morphological and Genetic Recovery of Coral Polyps After Bail-Out
- Marine Biophysics Unit, Okinawa Institute of Science and Technology Graduate University, Okinawa, Japan
Contemporary advances in microfluidic and molecular techniques have enabled coral studies to shift from reef and colony scales to polyp- and molecular-level investigations. Polyp bail-out provides an alternative approach to acquire solitary polyps for studies at finer scales. Although induction of polyp bail-out has been reported in several studies, polyp health after bail-out has not been investigated. In this study, we monitored morphological and genetic changes in Pocillopora acuta polyps after bail-out induced by hyperosmosis. In isosmotic conditions, over 80% of bailed-out polyps survived, of which half regenerated normal polyp morphology within 5 days, including a polarized polyp body, extended tentacles, and a distinguishable oral disk. In contrast, the remaining polyps degenerated into tissue ball-like structures that resemble multicellular aggregates reported in earlier studies. In morphologically recovered polyps, transcriptomic analysis showed that ∼87% of genes altered during bail-out induction recovered from stress status, suggesting resumption of metabolism, cell division, and immunity, while in degenerated polyps, only ∼71% of genes recovered. Quantitative polymerase chain reaction data further demonstrated that genetic recovery of energy production, cell proliferation, and immune response was achieved in morphologically recovered polyps within 3 days after bail-out, but was not fully accomplished in degenerated polyps even after 5 days. Our findings indicate that solitary polyps generated by hyperosmosis-induced bail-out can recover rapidly from physiological stress under laboratory conditions, suggesting that bailed-out polyps could be used as new models for coral research.
Introduction
Scleractinian corals (Cnidaria: Anthozoa) are the focus of considerable research, due to their pivotal ecological roles in coral reef ecosystems and their economic significance (Cesar et al., 2003; Wilkinson, 2008; Osinga et al., 2011). Because coral colonies secrete the structural framework of coral reefs, early research was conducted mostly on individual colonies or at population and community scales that consider entire coral reefs as single entities. Since the mid-20th century, such pioneering work has broadened our knowledge of the influences of many biotic and abiotic factors on coral survival and reef-building capacity (Buddemeier and Kinzie, 1976; Dennison and Barnes, 1988; Erez et al., 2011), enabling predictions of changes in reef ecosystems in response to on-going global climate change.
In recent decades, advances in microfluidic and molecular techniques have made it possible for coral research at polyp scale, the basic building blocks of coral colonies (Bromage et al., 2009; Shapiro et al., 2016; Luo et al., 2020; Pang et al., 2020). Individual polyps enable detailed visualization of cellular and molecular responses in corals, which is typically constrained at colony scale by the calcareous skeletons. In addition, studies at the scale of single polyps allow more precise characterization of biological factors that can vary within a colony, such as polyp senescence and microbiome composition in individual polyps. Conventionally, individual coral polyps are acquired from newly hatched larvae or planulae (Titlyanov et al., 1998; Schwarz et al., 1999; Hirose et al., 2008). Since reproduction of most stony corals occurs mainly in summer, sample acquisition has necessarily been seasonal. Also, like many animals, scleractinian corals show pronounced ontogenetic changes during development. Planktonic coral larvae transform into primary polyps through metamorphosis, which involves dramatic histological transformations and transcriptomic shifts (Vandermeulen, 1975; Grasso et al., 2008, 2011). Post-settlement, establishment of symbiosis, initiation of calcification, and sexual maturation also alter coral biochemistry and physiology (Reyes-Bermudez et al., 2009; Inoue et al., 2012; Tanaka et al., 2013). Biological parameters revealed in juvenile polyps therefore reflect a transient stage and do not fully represent those of adult corals.
To acquire individual polyps or cells from adult corals, mechanical or enzymatic treatments are usually applied to extract coral soft tissues from their calcareous skeletons (Vizel et al., 2011; Lecointe et al., 2013; Feuillassier et al., 2014). These methods, however, exert great stress on the corals, from which a period of weeks is typically required for isolated polyps to recover. Recently, polyp bail-out, a novel coral stress response, has captured the attention of coral researchers. Distinct from the abovementioned methods, polyp bail-out represents an intrinsic mechanism in some stony corals to disrupt coloniality and dissociate into isolated polyps in response to environmental stresses (Sammarco, 1982). To date, a variety of stimuli, such as insecticides, acidification, and thermal stress, have been shown to induce polyp bail-out in scleractinian corals (Kvitt et al., 2015; Fordyce et al., 2017; Wecker et al., 2018). Using hyperosmotic treatments, previous studies further demonstrated that the bail-out response in pocilloporid corals can be induced within 24 h in a predictable manner (Shapiro et al., 2016; Chuang and Mitarai, 2020; Liu et al., 2020). In contrast to mechanical or enzymatic separation, isolated polyps from polyp bail-out are mostly intact in gross morphology and are able to resettle and resume skeletogenesis within days (Sammarco, 1982; Kvitt et al., 2015; Shapiro et al., 2016). This stress response therefore provides an alternative micropropagation method to acquire adult coral polyps.
Given that calcification is an important index in assessing ecological functions of corals (De’ath et al., 2009; Carricart-Ganivet et al., 2012), resumption of skeletogenesis after resettlement is generally considered a sign of polyp recovery from bail-out (Sammarco, 1982; Kvitt et al., 2015; Shapiro et al., 2016). However, detached polyps from bail-out have also been reported to survive and begin feeding in free-living form, although such polyps seem to conduct no calcification (Goreau and Goreau, 1959; Serrano et al., 2018). Appraising the health of bailed-out polyps thus requires assessment of parameters other than calcification, such as their metabolism or energy reserves (Rodrigues and Grottoli, 2007; Lesser, 2013). Recently, genomic and transcriptomic data of several pocilloporid corals have been released (Cunning et al., 2018; Wecker et al., 2018; Chuang and Mitarai, 2020), shedding light on polyp health determination from a molecular perspective. In this study, we examined morphological and genetic dynamics in coral polyps after hyperosmosis-induced polyp bail-out. To our knowledge, this study is the first to monitor post-bail-out polyp recovery from both morphological and molecular perspectives. The results provide a basis for future studies on more specific aspects of physiology in bailed-out polyps and long-term, for their use as new models for coral research.
Materials and Methods
Coral Sampling and Maintenance
In 2018–2020 we purchased ten Pocillopora acuta corals from the Onna Village Fishery Cooperative in Okinawa, Japan. Prior to experiments, all coral colonies were acclimated outdoors for more than 1 month in flowing, sand-filtered seawater (FSW) at the OIST Marine Science Section at Seragaki (OMSSS).
Polyp Bail-Out Induction and Polyp Recovery Setup
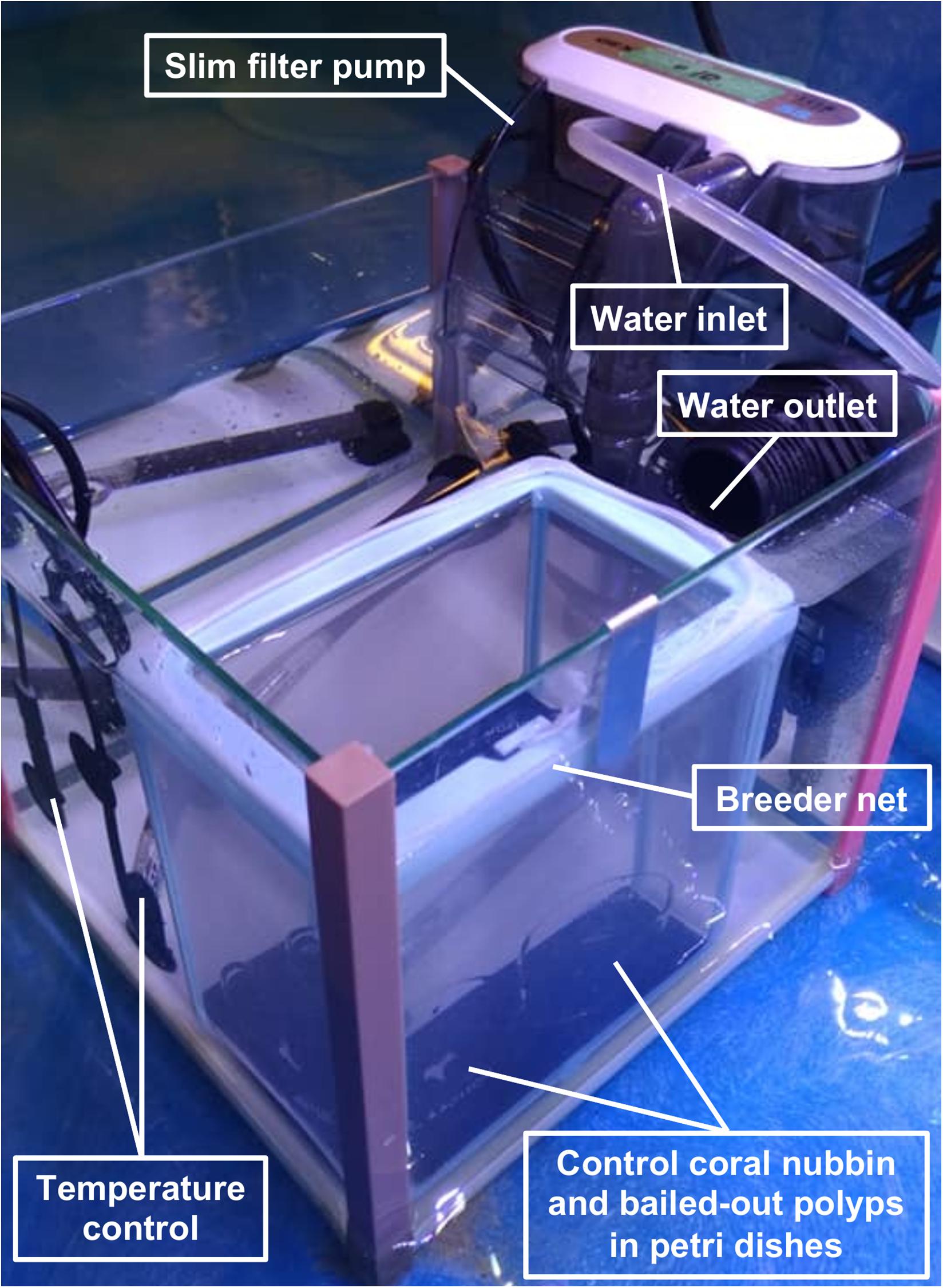
In the present study, we induced polyp bail-out in P. acuta using the hyperosmotic method described in our previous report (Chuang and Mitarai, 2020). For each P. acuta colony in the experiment, two coral nubbins (∼1 cm in length) were broken off with a bone cutter 1 day before the experiment and were placed separately in two 5-L indoor tanks filled with FSW (33–35‰; one for polyp bail-out induction and one as a control group). To induce polyp bail-out, high-salinity seawater (prepared at a salinity 13–15‰ higher than ambient) was pumped into the bail-out induction tank at a fixed rate of 4 mL/min. While for the control group, FSW was slowly supplied (rate not measured) to establish a semi-closed system, designed to compensate for evaporation during the experiment. Each tank was equipped with a 3.4 W slim filter pump to facilitate mixing/circulation of seawater in the tank. In both tanks, seawater temperature was controlled at 25°C (± 0.3°C) and light was provided at ∼150 μmol photons m-2 s-1 with a 12-h light-dark cycle. After 24–25 h of hyperosmotic treatment, with salinity ∼11‰ higher than ambient, polyp bail-out was observed. Detached polyps were collected with a plastic pipette and transferred immediately to the control tank, at where they were placed in a glass petri dish and were allowed to recover for 5 days in isosmotic conditions (33–35‰). As solitary polyps were easily suspended by water movement and were prone to be captured by the slim filter pump, an aquarium breeder net (mesh size: 200–300 μm) was set up in the control tank to host bailed-out polyps and the coral nubbin in the control group (Figure 1). The same experiment was repeated in two batches in 2019, with three (first batch; spring season) and five (second batch; winter season; one individual was included in both batches) P. acuta colonies, respectively.
Whole Transcriptome RNA Sequencing
Five days after bail-out, we observed two morphotypes among bailed-out polyps. Some polyps displayed typical polyp morphological features (recovered polyps) while others degenerated into globular structures (degenerated polyps; Figure 2). In the first batch of experiments, we collected recovered polyps (sample labeled RP_day5; 7–8 polyps/individual coral; N = 3) and healthy coral nubbins in the control group (HC_day5; N = 3) at day5 post-bail-out. In the second batch of experiments, most bailed-out polyps degenerated after 5 days in the control tank. This might be due to seasonal fluctuation of quality in incoming seawater, which was supplied as a semi-closed system in the control tank, or a delay of polyp collection after the bail-out response, which happened slightly earlier than expected in the second batch of experiments. We therefore collected only degenerated polyps (DP_day5; 10 polyps/individual coral; N = 5) in the second batch of experiments. Sampled coral tissues were immediately preserved in 1 mL TRIZOL reagent and RNA was extracted following the process described in our previous report (Chuang and Mitarai, 2020). RNA samples with RIN > 7 (RNA integrity number, checked with an Agilent Bioanalyzer 2100) were used to construct libraries (strand-specific, polyA-RNA purified). Illumina Novaseq 6000 150 × 150-bp paired-end RNA sequencing was then performed by the DNA Sequencing Section (SQC) at the Okinawa Institute of Science and Technology (OIST).
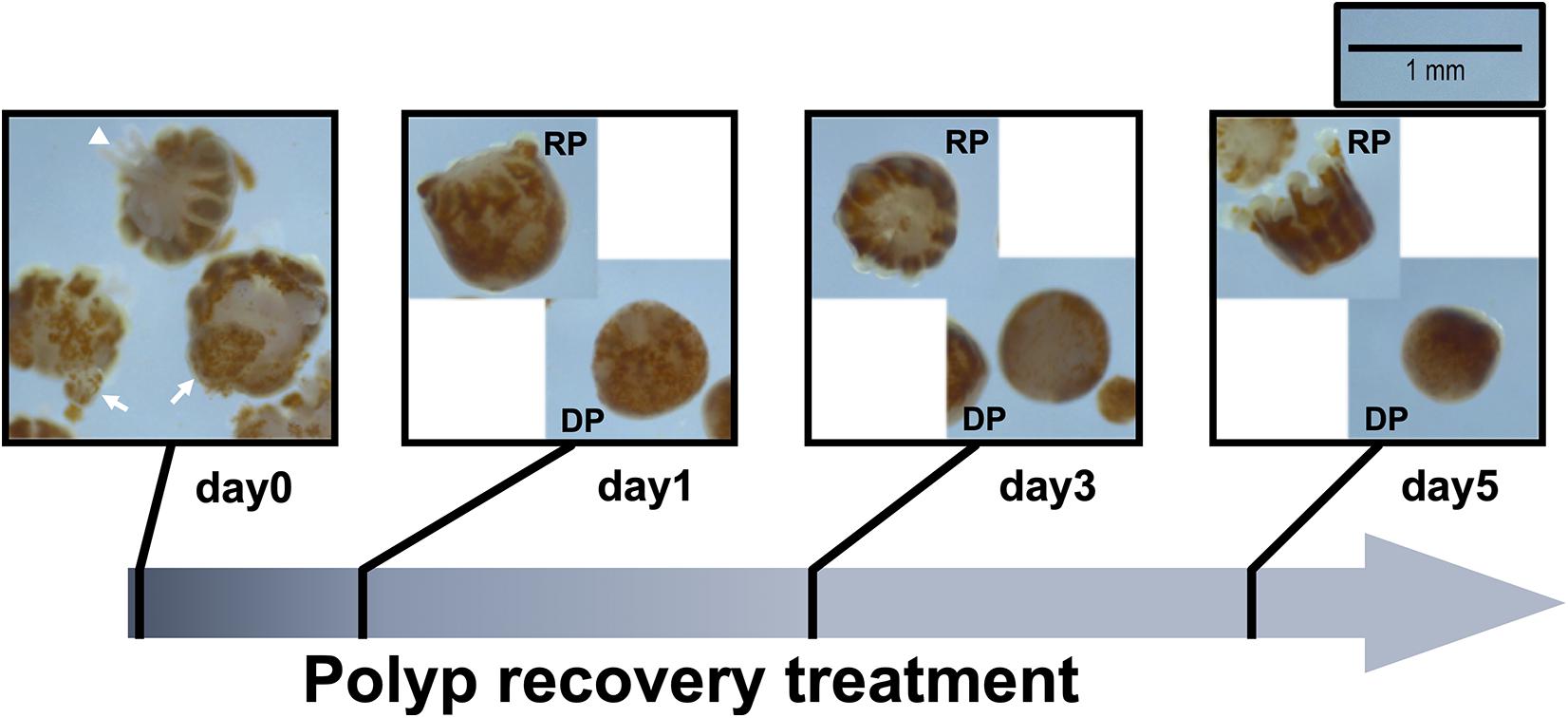
Figure 2. Morphological changes of Pocillopora acuta polyps post-bail-out. Residues of coenosarc tissues (arrow) and extruding mesentery filaments (arrowhead) are observable in some polyps right after bail-out (day0). After recovery for 1 day in isosmotic conditions, polyp bodies become smooth and morphological differentiation can be seen among bailed-out polyps, with some polyps recovering typical morphological features such as tentacles and oral-aboral polarization (RP), while other polyps have degenerated into tissue balls (DP).
Transcriptomic Data Analysis
To assess genetic recovery from the hyperosmosis-induced bail-out response, RNA-seq data of healthy P. acuta colonies (SRR10696827, SRR10708186, and SRR10708226; herein labeled HC_day0) and bailed-out polyps (SRR10696823, SRR10708182, and SRR10708229; labeled BP_day0) collected in our previous study were downloaded from the NCBI GenBank database to construct a reference of stressed status. Raw RNA-seq reads sequenced in the present study and those downloaded from the GenBank database were trimmed with Trimmomatic v0.36 (Bolger et al., 2014) and quantified using RSEM v1.3.2 (Li and Dewey, 2011) based on the P. acuta transcriptome assembly constructed in our previous report (GenBank accession: GIDI00000000). Gene functional annotation was conducted by searching the SwissProt eukaryotic database (downloaded on 3 Jul 2019), with a criterion of E < 10–5 for a valid BLASTX match. Annotated transcripts (N = 17,908) were subsequently subjected to differential gene expression (DE) analysis using edgeR (Robinson et al., 2010). Considering that experimental conditions differed between the present study (filtered natural seawater, 150 μmol photons m-2 s-1) and our previous study (artificial seawater, 300 μmol photons m-2 s-1), DE analysis was conducted separately for HC_day0 vs. BP_day0 and for pairwise comparisons among the three conditions in this study (HC_day5, RP_day5, and DP_day5), based on the following criteria: false discovery rate (FDR) < 0.001, absolute change > 2x, and > 5 counts per million (CPM) in at least 3 libraries in a given pairwise comparison. To identify alteration of cellular functions during and after the bail-out response, Gene Ontology (GO) enrichment analysis was conducted using DAVID bioinformatics resources v6.8 (Huang et al., 2008, 2009), with all annotated transcripts as a background reference and GOTERM_BP_DIRECT as analytical output.
Examination of Polyp Recovery Dynamics
To further examine polyp recovery dynamics, in 2020 we repeated the experiment on three P. acuta specimens (N = 3; non-overlapping with the first or second batch) and monitored dynamics of morphological and genetic changes of coral polyps after bail-out. At day0 (right after bail-out), day1, day3, and day5 post-bail-out, morphological changes in bailed-out polyps were recorded using a stereo microscope (Leica, Germany). Morphological recovery/degeneration was determined by the presence of recognizable tentacles and oral-aboral polarization. Polyp size was measured as the longest axis of the polyp body using imageJ (Schneider et al., 2012). For genetic analysis, five nubbins from each colony were employed in the experiment (one nubbin for bail-out induction and four nubbins in the control group). At day0, day1, day3, and day5 post-bail-out, bailed-out polyps and healthy coral nubbins were collected and preserved in TRIZOL reagent. Since morphological differentiation of bailed-out polyps was distinguishable 1 day after bail-out, recovered and degenerated polyps were collected separately at day1, day3, and day5 post-bail-out.
Quantitative Polymerase Chain Reaction Analysis of Gene Expression
For samples collected in the recovery dynamics experiments, RNA was extracted following the same protocol as above and reverse-transcriptase polymerase chain reaction was conducted using SuperScript IV VILO Master Mix (Invitrogen, United States). Ten genes related to energy reserves, cell proliferation, adhesion, and immunity (Table 1) were selected from the P. acuta transcriptome assembly as molecular markers to assess functional recovery in bailed-out polyps using a quantitative polymerase chain reaction (qPCR) assay. The β-tubulin gene mentioned in our previous study was adopted as an internal control gene in this study (Chuang and Mitarai, 2020). For each qPCR reaction, 200 μM of primer pair and 1 μL cDNA (concentration not determined) were prepared in iQ SYBR Green Supermix (Bio-Rad, United States) to a final volume of 10 μL. qPCR was performed on a StepOnePlus Real-Time PCR system (Thermo Fisher Scientific, United States) under the conditions: 3 min at 95°C, 40 cycles of 15 s at 95°C, and 30 s at 60°C. Each qPCR reaction was performed in two technical replicates, of which the averaged cycle threshold (CT) value was employed to estimate the relative expression of a given target gene to the internal control gene (ΔCT = CTcontrol gene – CTtarget gene). Statistical significance between gene expression in bailed-out polyps (recovered or degenerated) and that in healthy corals at each time point was tested using a paired sample t-test.
Results
Survival and Morphological Changes of Polyps After Bail-Out
In the present study we monitored morphological changes of P. acuta polyps after hyperosmosis-induced bail-out. Right after bail-out, morphological vestiges of the bail-out response could be observed in detached polyps, including rough aboral surfaces, protruding mesentery filaments, and in some polyps, residues of coenosarc tissues (day0; Figure 2). These morphological abnormalities mostly vanished after 1 day in isosmotic conditions, resulting in isolated polyps with smooth polyp bodies (day1; Figure 2). Differentiation of polyp morphotypes could be seen at this stage, with some polyps having typical polyp features such as tentacles and oral-aboral polarization, while others degenerated into spheres of tissue. The morphological divergence became more distinguishable after 3 days in isosmotic conditions (day3; Figure 2) and some degree of tentacle extension was observable in recovered polyps at day5 post-bail-out (day5; Figure 2). During the whole experimental period, bailed-out polyps tended to sink in seawater, but floated slightly above the bottoms of their glass petri dishes, with no signs of resettlement or calcification in either recovered or degenerated polyps. Zooxanthellae were retained in polyps throughout the experimental period and in some bailed-out polyps, cytoplasmic streaming-like zooxanthella movement was observed in polyp bodies during the period from day1 to day5 post-bail-out (Supplementary Video 1). In total, ∼84% of polyps survived 5 days after bail-out, among which 52% recovered normal polyp morphology and 48% degenerated. Compared to polyp sizes at day0 post-bail-out, recovered polyps showed a conspicuous size decrease at day1 post-bail-out (∼19%) and remained relatively stable afterward, with a roughly 24% decrease in mean size by day5 post-bail-out. In contrast, degenerated polyps decreased in size about 45% by day5 post-bail-out (Figure 3 and Table 2).
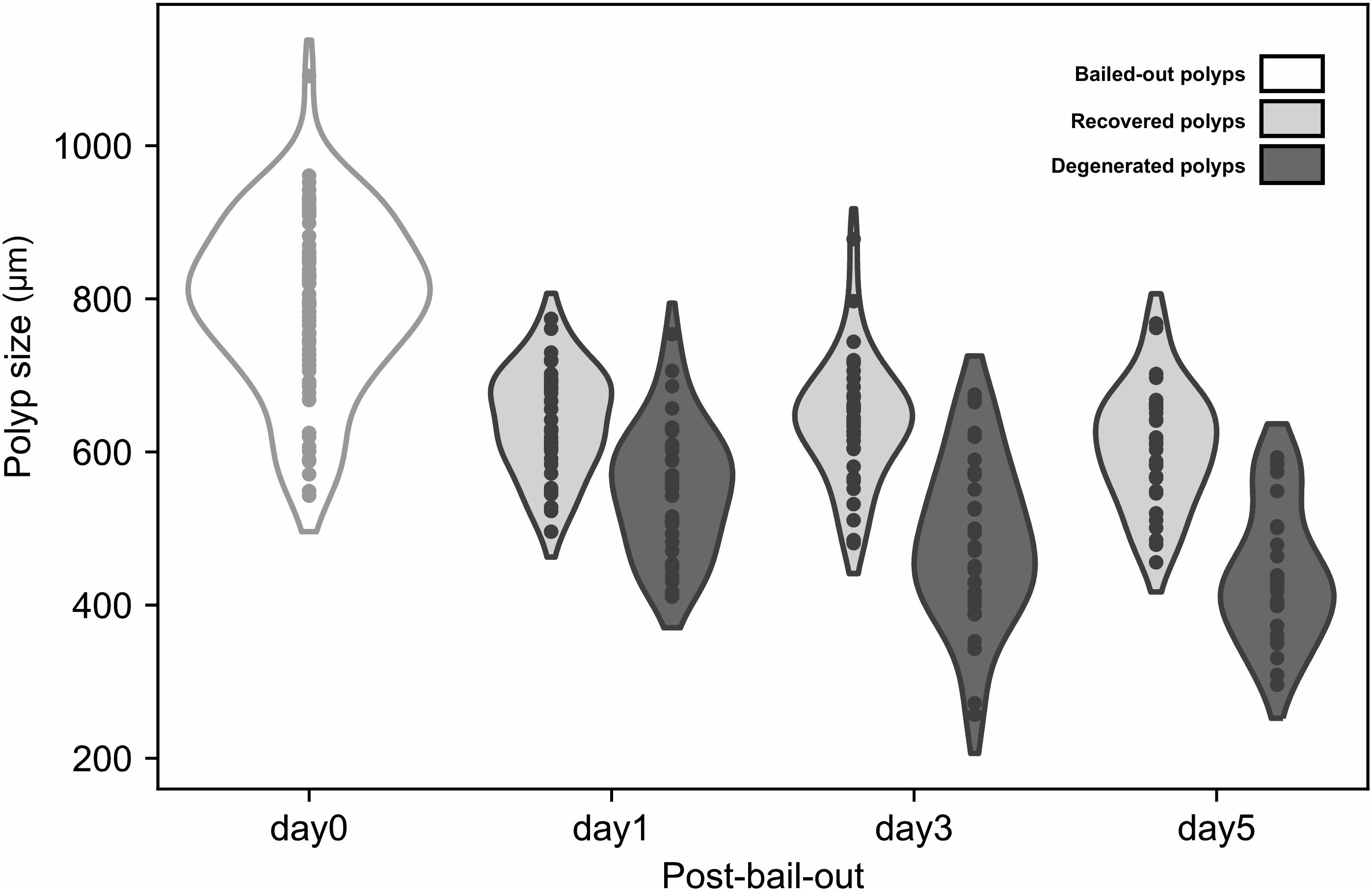
Figure 3. Size reduction of bailed-out polyps. Bailed-out polyps of different morphotypes (recovered or degenerated) are displayed for day1, day3, and day5 post-bail-out.
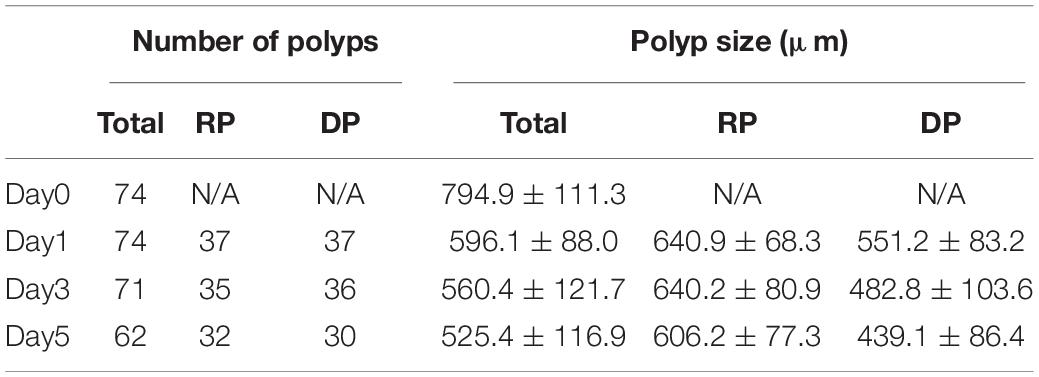
Table 2. Numbers and sizes (presented as means ± standard deviations) of polyps with recovered (RP) or degenerate morphology (DP) post-bail-out.
Transcriptomic Changes in Polyps at Day5 Post-Bail-Out
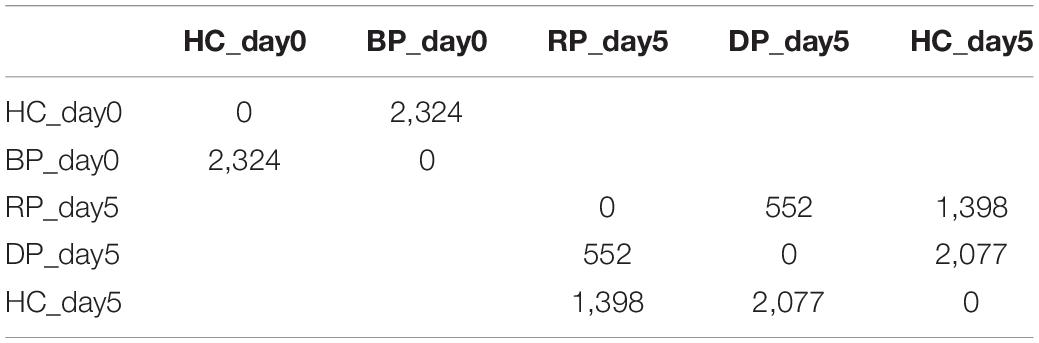
Between healthy corals (HC_day0) and bailed-out polyps (BP_day0) sequenced in our previous study (Chuang and Mitarai, 2020), significant expression changes were identified in 2,324 functionally annotated transcripts. For transcriptomes constructed in this study, 1,398 and 2,077 differentially expressed genes (DEG) were identified in recovered polyps (RP_day5) and degenerated polyps (DP_day5), compared to healthy corals collected at day5 post-bail-out (HC_day5), respectively. While for the comparison between RP_day5 and DP_day5, 552 DEGs were identified (numbers of DEGs in pairwise comparisons are summarized in Table 3). To examine molecular recovery of P. acuta polyps from bail-out, DEGs identified in the comparison BP_day0 vs. HC_day0 were cross-referenced to those identified in RP_day5 vs. HC_day5 and those in DP_day5 vs. HC_day5. In RP_day5, 1,063 bail-out-activated and 954 bail-out-suppressed DEGs showed recovery from bail-out (recovered DEGs), while 143 and 164 DEGs remained up-regulated and down-regulated, compared to HC_day5, respectively (non-recovered DEGs; Figure 4). GO enrichment analysis showed that recovered DEGs in RP_day5 overrepresented for biological processes related to metabolism, apoptosis, cell division, immune system, and intracellular signal transduction (Figure 4). Among non-recovered DEGs in RP_day5, cell adhesion, collagen fibril organization, and multicellular organism development were overrepresented (Figure 4). In DP_day5, 840 bail-out-activated DEGs and 816 bail-out-suppressed DEGs showed recovery from stressed status, while 366 and 302 DEGs stayed up-regulated and down-regulated, compared to HC_day5, respectively (Figure 5). GO analysis of recovered DEGs in DP_day5 showed significant enrichment in biological processes related to metabolism, DNA repair, immune system, and intracellular signal transduction. For non-recovered DEGs in DP_day5, GO terms such as cell adhesion, multicellular organism development, proteolysis, cell differentiation, and those related to epithelium or skeleton development were significantly overrepresented (Figure 5). For DEGs in the comparison RP_day5 vs. DP_day5, significant GO enrichment was identified in translation, cell cycle, DNA replication, cell differentiation, and Notch signaling pathway (Figure 6).
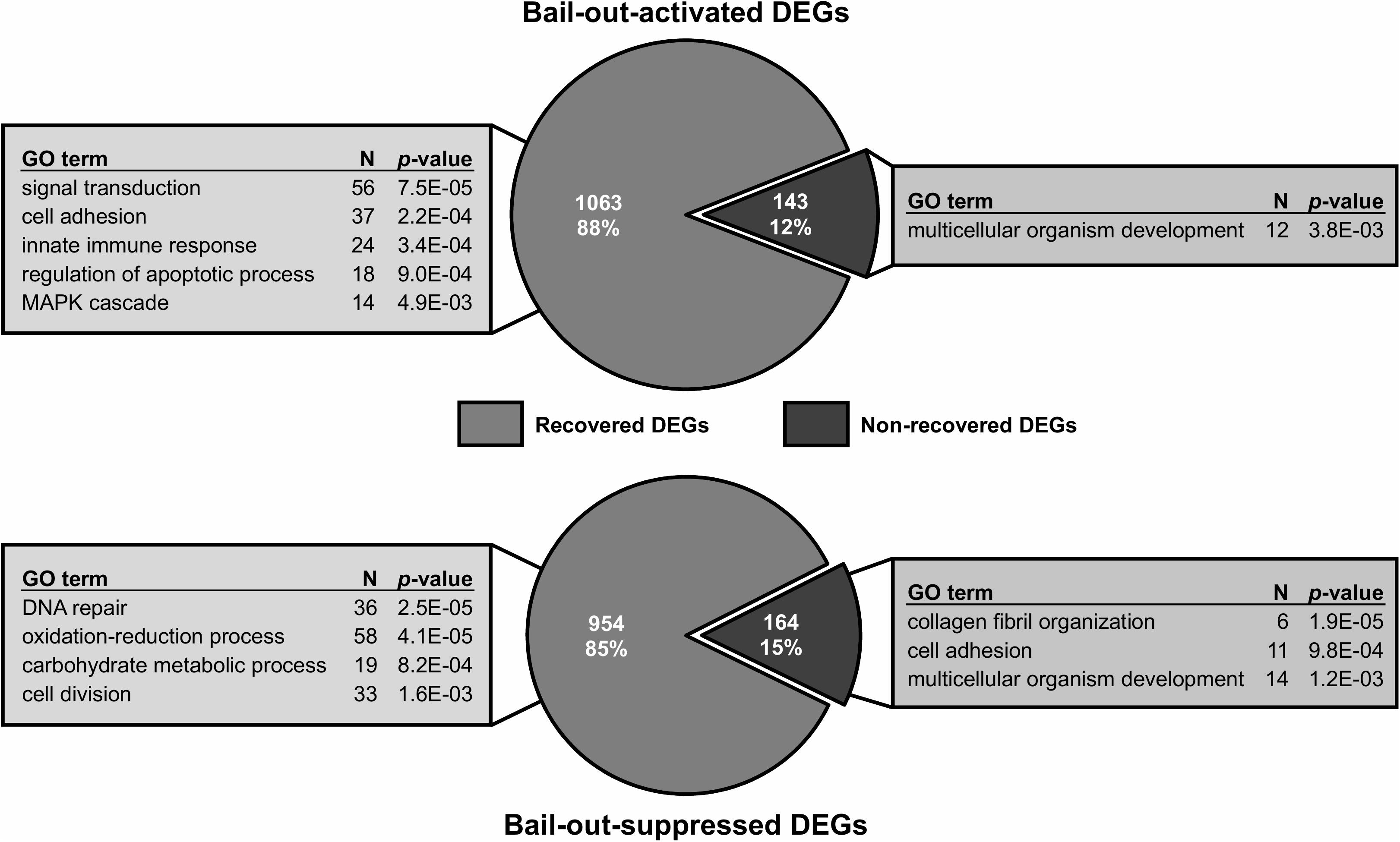
Figure 4. Recovery of bail-out-induced genetic changes in recovered polyps at day5 post-bail-out. Numbers of differentially expressed genes (DEGs) up- or down-regulated during polyp bail-out and significantly overrepresented gene ontology (GO) categories (p < 0.005) are displayed. The number of unique genes in each GO category is indicated.
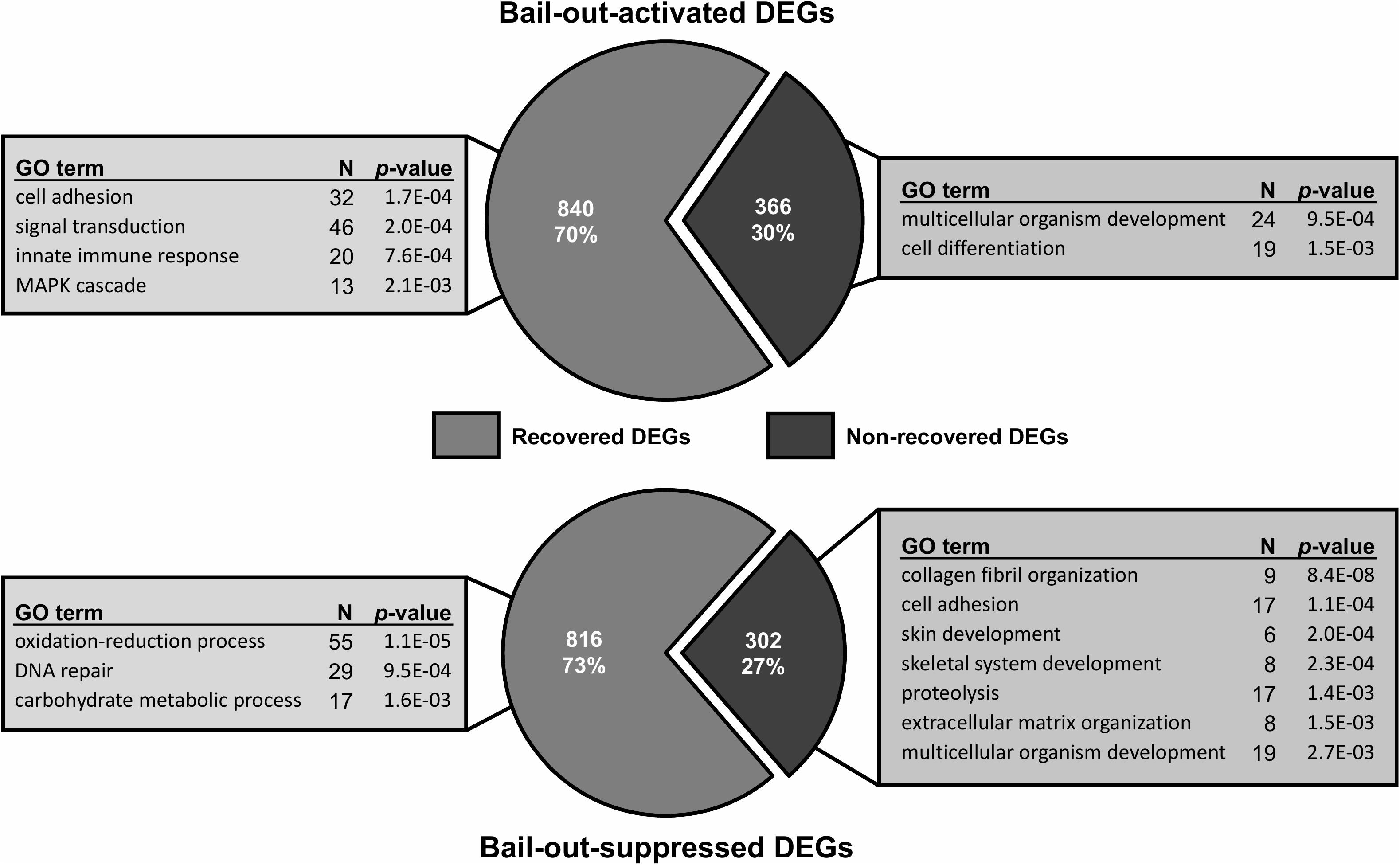
Figure 5. Recovery of bail-out-induced genetic changes in degenerated polyps at day5 post-bail-out. Numbers of differentially expressed genes (DEGs) up- or down-regulated during polyp bail-out and significantly overrepresented gene ontology (GO) categories (p < 0.005) are displayed. The number of unique genes in each GO category is indicated.
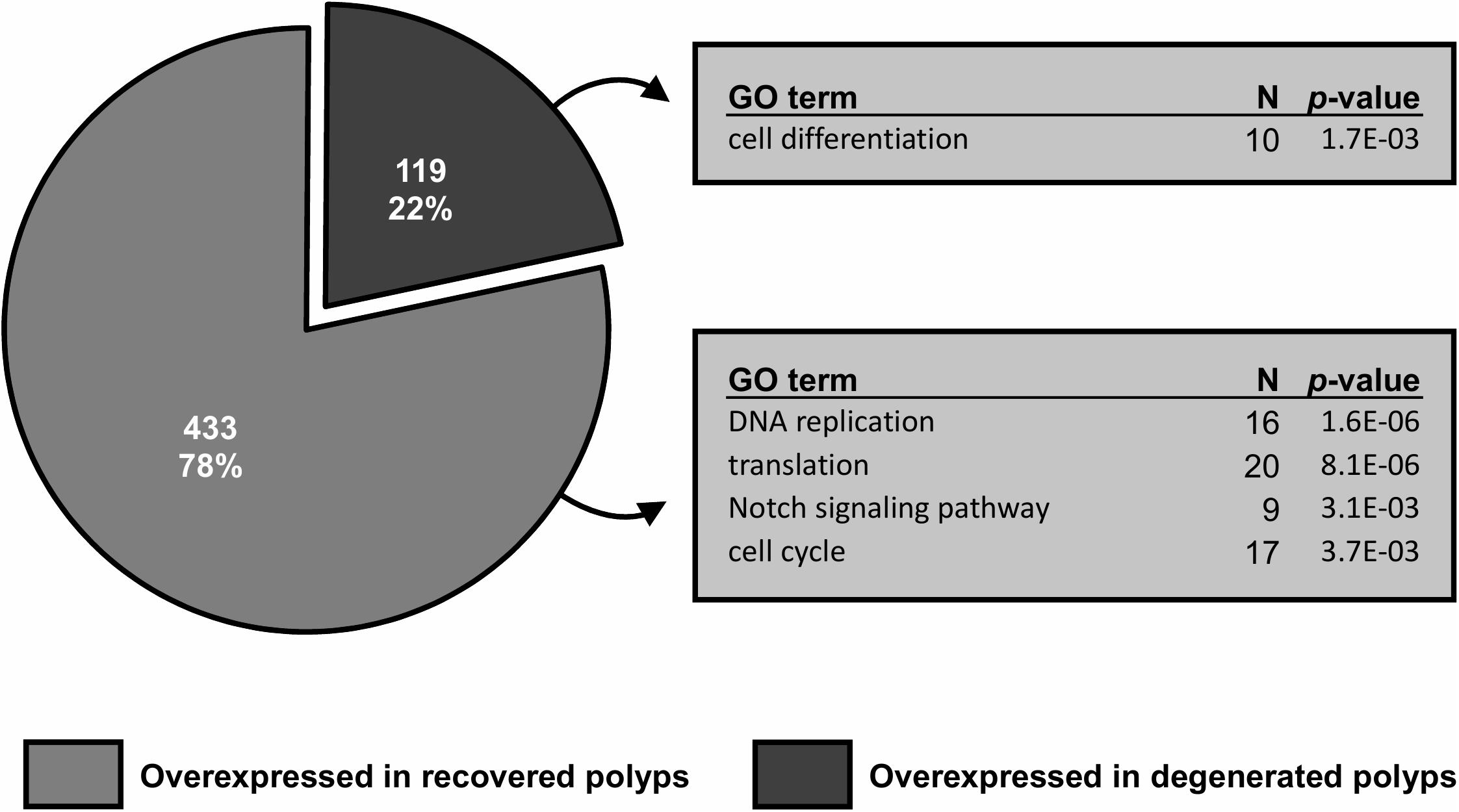
Figure 6. Gene differential expression (DE) analysis of recovered and degenerated polyps at day5 post-bail-out. Numbers of differentially expressed genes (DEGs) overexpressed in each polyp morphotype are displayed. Selected biological processes significantly overrepresented (p < 0.005) in gene ontology (GO) enrichment analysis and the number of unique genes in each GO category are indicated.
Post-Bail-Out Genetic Dynamics
To further examine dynamics of genetic changes in bailed-out polyps, we checked expression profiles of ten genes related to metabolism, cell proliferation, immunity, and cell adhesion during the experimental period using a qPCR assay. Compared to healthy corals, polyps right after bail-out showed significant expression changes in all selected genes except gene for complement C3 (p < 0.075; Figure 7). For ATP synthase, NADH-ubiquinone oxidoreductase, succinate dehydrogenase flavoprotein subunit, proliferation marker protein Ki-67, proliferating cell nuclear antigen, and DNA replication licensing factor Mcm6 genes, recovered polyps showed expression recovery to the levels of healthy corals within 3 to 5 days after bail-out. In contrast, in degenerated polyps, expression of these genes remained significantly suppressed during the experimental period except for day3 post-bail-out, at which great variation resulted in statistical insignificance. For toll-like receptor 6 and nuclear factor NF-kappa-B p105 subunit genes, expression recovery from bail-out-induced overexpression was seen in both polyp morphotypes 1 day after bail-out, with the former gene gradually returning to the expression level of healthy corals within 3 days post-bail-out, while the latter remained slightly under-expressed throughout the experiment. For complement C3 in both polyp morphotypes, significant upregulation was only seen at day1 post-bail-out, after which expression returned to the level of healthy corals. In contrast to all the above-mentioned genes, the integrin alpha-4 subunit gene showed expression recovery at day1 post-bail-out. However, at day3 and day5 post-bail-out, the integrin alpha-4 gene was re-up-regulated in both recovered and degenerated polyps, compared to its expression level in healthy corals.
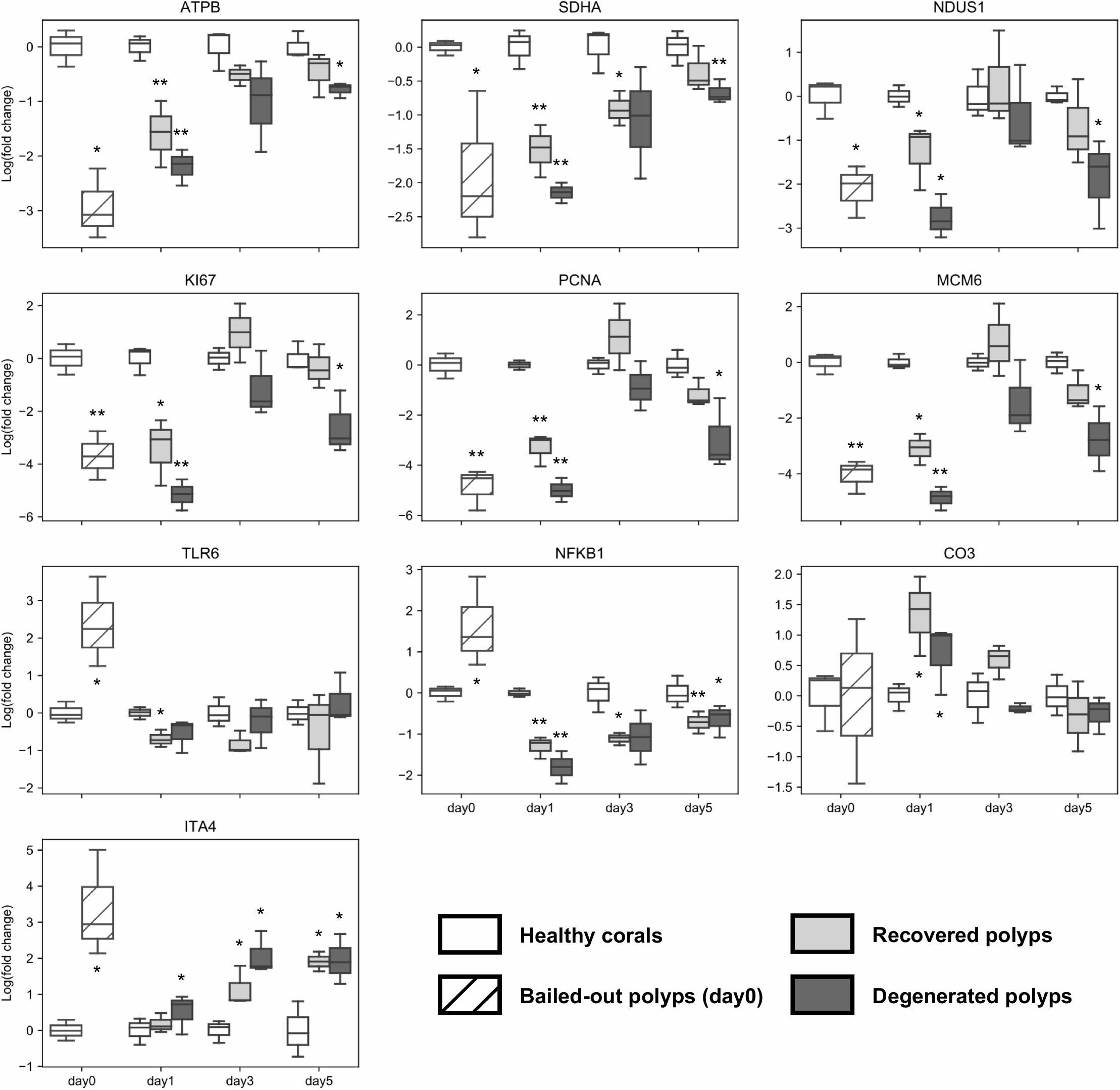
Figure 7. Post-bail-out expression dynamics of ten selected genes. Gene expression is normalized to an internal control gene (Tubulin beta chain) and to mean expression levels of healthy corals at each given time point. Statistical differences between bailed-out polyps and healthy corals at each time point were examined using paired t-tests. Groups showing significant differences compared to healthy corals are indicated (*p < 0.075; **p < 0.01). Gene name abbreviations: ATPB, ATP synthase subunit beta; SDHA, Succinate dehydrogenase flavoprotein subunit; NDUS1, NADH-ubiquinone oxidoreductase 75 kDa subunit; KI67, Proliferation marker protein Ki-67; PCNA, Proliferating cell nuclear antigen; MCM6, DNA replication licensing factor Mcm6; TLR6, Toll-like receptor 6; NFKB1, Nuclear factor NF-kappa-B p105 subunit; CO3, Complement C3; and ITA4, Integrin alpha-4.
Discussion
Recovery of Coral Polyps After Bail-Out
Polyp bail-out has been proposed as a micropropagation method for some scleractinian corals (Shapiro et al., 2016; Liu et al., 2020). However, the health status of polyps after bail-out has rarely been examined. In the present study, we monitored morphological and genetic changes of P. acuta polyps after hyperosmosis-induced bail-out. Among bailed-out polyps collected in this study, about 43% regenerated clearly distinguishable morphological features, such as tentacles and body polarity. Survival and morphological recovery of bailed-out polyps is possibly determined by a suite of extrinsic and intrinsic factors, such as the initial health condition of a coral colony, the rapidity of polyp escape from stressful environments, and seawater quality in recovery environments, as suggested in previous studies (Shapiro et al., 2016; Pang et al., 2020). Deviations of these factors from their optima may significantly impair polyp survival and recovery, which likely contributed to the low polyp recovery rate in our second batch of experiments, as well as in some of our preliminary experiments (0%; data not shown). Although in the present study, experiments were conducted only until 5 days post-bail-out, recovered polyps survived for over 2 weeks in our preliminary experiments, without strict monitoring of recovery conditions (data not shown). This lifespan is comparable to those reported in previous studies applying microfluidic techniques for post-bail-out polyp cultivation, in which recommencement of skeletogenesis was even observed (Shapiro et al., 2016; Luo et al., 2020; Pang et al., 2020). Based on morphological observations and computational modeling, Pang et al. (2020) suggested that continuous supply of DO (dissolved oxygen) and DIC (dissolved inorganic carbon) is essential for long-term cultivation of isolated polyps. Longer survival is thus expected for recovered polyps acquired using the method described in this study, if better control of seawater chemistry is maintained.
In other cnidarian models, tentacle morphogenesis has been demonstrated, mediated by a common mechanism that underlies proliferation of progenitor cells around the oral pole and subsequent migration toward developing tentacles (Fritz et al., 2013; Fujita et al., 2019). Consistent with this developmental pattern, our results suggest resumption of cell proliferation in recovered polyps within 3 days post-bail-out, a time span coincident with observed tentacle regeneration. This morphological regeneration implies recovery of heterotrophy in detached polyps after bail-out. Indeed, it has been reported that detached coral polyps can feed normally in the free-living stage and can survive for weeks (Goreau and Goreau, 1959). In bleached corals, heterotrophy has been proposed to supply up to 150% of coral metabolic demands and is attributable to the survival and recovery of coral animals after bleaching (Grottoli et al., 2006; Houlbrèque and Ferrier-Pagès, 2009; Levas et al., 2016). Given that bailed-out polyps sink in seawater (either hypersaline or of ambient salinity), rapid resumption of heterotrophy might help polyps to survive in deeper environments, where photosynthesis is compromised. However, since well-regenerated tentacles and oral disks were observed only at later stages of our experimental period, heterotrophy is believed to have contributed little to survival of bailed-out polyps in this study. Estimating the significance of heterotrophy relative to the energy budget of bailed-out polyps, especially at longer time scales, would be a fruitful area of research and should better illustrate the ecological role of polyp bail-out in scleractinian corals.
In addition to resumption of heterotrophy, bailed-out polyps retained symbiotic zooxanthellae throughout the experimental period, suggesting preservation of photosynthetic ability in coral polyps after bail-out. Interestingly, in some bailed-out polyps, we observed cytoplasmic streaming-like movement of zooxanthellae within polyp bodies during their recovery. Coincident with this observation, the complement C3-encoding gene in our qPCR assay showed significant upregulation in bailed-out polyps at day1 post-bail-out and gradually returned to the expression level of healthy corals thereafter. In other cnidarian models, the complement system has been thought to function in establishment and maintenance of host-dinoflagellate symbiosis (Kvennefors et al., 2010; van de Water et al., 2015; Poole et al., 2016). Fluctuating expression of the complement system therefore implies alteration of the endosymbiotic status between Symbiodinium and host coral cells in bailed-out polyps. Given that complete resumption of metabolism in bailed-out polyps takes a few days, as suggested by our qPCR results, disruption of endosymbiosis probably enables coral cells to avoid overloading with photosynthates as well as related byproducts during the metabolic readjustment period. Retention of de-partnered zooxanthellae within the polyp body likely builds up a dinoflagellate pool in proximity to coral cells, which enables rapid reestablishment of symbiont populations, as well as a photosynthetic energy supply chain in bailed-out polyps, once their metabolism resumes. It has been generally acknowledged that photosynthates from Symbiodinium afford the majority of daily carbon requirements in host coral animals (Muscatine and Porter, 1977; Muscatine et al., 1981; Lesser et al., 2000). Securing this main energy source, therefore, may be credited for the rapid recovery of bailed-out polyps observed in this study, which is remarkably shorter than that for bleaching recovery (Toller et al., 2001; Nakamura et al., 2003; Thomas and Palumbi, 2017). Together, our results show that coral polyps can rebuild their energy reserves and resume fundamental biological functions rapidly after hyperosmosis-induced bail-out. Considering that resettlement and skeletogenesis can ensue after polyp bail-out (Sammarco, 1982; Shapiro et al., 2016; Liu et al., 2020), this coral micropropagation method enables acquisition of live, healthy coral polyps, facilitating subsequent applications, such as coral culturing and in vivo coral physiological and molecular studies.
Alteration of Notch Signaling Leads to Cell De-Differentiation in Degenerated Polyps
While in the present study 43% of bailed-out polyps showed morphological regeneration, 41% degenerated instead, yielding polyps morphologically similar to the “tissue balls” reported in Lecointe et al. (2013). Based on histological evidence, Lecointe et al. (2013) suggested involvement of tissue reorganization and possible arrest of cell division in the formation of tissue balls. Consistent with these findings, our genetic data showed sustained alteration of cell differentiation, skin development, and incomplete recovery of cell proliferation in degenerated polyps. Furthermore, when compared to recovered polyps, degenerated polyps showed suppression of several fundamental cellular functions, including translation, DNA replication, and cell cycle. This morphological transformation therefore may represent a dormant stage that allows corals to survive stressful events by reducing energy demands. Interestingly, the Notch signaling pathway was significantly down-regulated in degenerated polyps. In vertebrate models, Notch signaling functions in neural cell de-differentiation (Mirsky et al., 2008; Jopling et al., 2011) as well as in trans-differentiation of various cell types (Sawey et al., 2007; He et al., 2014; Vega et al., 2014). With its evolutionary conservation in metazoans (Bi et al., 2016), it can therefore be hypothesized that the Notch signaling pathway helps to determine polyp fates after bail-out: regeneration or degeneration.
Post-Bail-Out Genetic Dynamics Shed Light on Induction of Polyp Resettlement
Resettlement of bailed-out polyps was proposed as a sequential process consisting of: (1) initial attachment of polyps to suitable substrates, (2) flattening of polyp bodies while strengthening connections between polyps and substrates, and (3) re-commencement of skeletogenesis (Shapiro et al., 2016). In the present study, expression changes of several cell adhesion genes were identified in polyps both during bail-out induction and after 5 days of recovery, which may reflect the isolated nature of bailed-out polyps. Interestingly, our qPCR analysis showed that an integrin α4 gene, up-regulated during polyp bail-out induction, recovered to levels typical of healthy corals after 1 day in isosmotic conditions, but was then up-regulated again at day3 and day5 post-bail-out. Integrins α4β1 (VLA-4) and α4β7 are cell surface receptors that recognize the extracellular matrix glycoprotein fibronectin (Mould et al., 1990; Postigo et al., 1993), which reportedly facilitates attachment of coral polyps to substrates (Domart-Coulon et al., 2004; Puverel et al., 2005). Thus, re-upregulation of α4 integrins seen in our experiments may represent molecular preparation for “programmed” resettlement. The “sinking” nature of bailed-out polyps in this scenario can then be considered as a behavioral adaptation that facilitates searching for suitable resettlement sites. Nevertheless, no resettlement was seen in the present study, either in recovered or degenerated polyps. Future studies on the inducibility of polyp resettlement are expected to illuminate the ecological significance of polyp bail-out in shaping coral distributions, with an eye to using polyp bail-out for coral propagation and restoration.
Data Availability Statement
Datasets generated for this study can be found in the NCBI GenBank database with the following accession numbers: RNA-Seq raw data: NCBI SRA: SRR12639762–SRR12639772.
Author Contributions
P-SC conducted the majority of this project, including project design, the main experiments, data analysis, and manuscript writing. KI participated in parts of the experiments and contributed to manuscript writing. SM contributed to the design of this project and manuscript writing. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by JSPS KAKENHI Grant Number JP18J20226. The authors gratefully acknowledge generous support to the Marine Biophysics Unit from Okinawa Institute of Science and Technology Graduate University (OIST).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the OIST DNA Sequencing Section for constructing RNA-Seq libraries and performing sequencing and the OIST Scientific Computing and Data Analysis Section for providing the high-performance computing service for transcriptome assembly. We thank Dr. Steven D. Aird for editing and commenting on the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.609287/full#supplementary-material
References
Bi, P., Yue, F., Sato, Y., Wirbisky, S., Liu, W., Shan, T., et al. (2016). Stage-specific effects of Notch activation during skeletal myogenesis. eLife 5:e17355.
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bromage, E., Carpenter, L., Kaattari, S., and Patterson, M. (2009). Quantification of coral heat shock proteins from individual coral polyps. Mar. Ecol. Prog. Ser. 376, 123–132. doi: 10.3354/meps07812
Carricart-Ganivet, J. P., Cabanillas-Teran, N., Cruz-Ortega, I., and Blanchon, P. (2012). Sensitivity of calcification to thermal stress varies among genera of massive reef-building corals. PLoS One 7:e32859. doi: 10.1371/journal.pone.0032859
Cesar, H., Burke, L., and Pet-Soede, L. (2003). The Economics of Worldwide Coral Reef Degradation. Arnhem: Cesar Environmental Economics Consulting.
Chuang, P.-S., and Mitarai, S. (2020). Signaling pathways in the coral polyp bail-out response. Coral Reefs 39, 1535–1548. doi: 10.1007/s00338-020-01983-x
Cunning, R., Bay, R., Gillette, P., Baker, A., and Traylor-Knowles, N. (2018). Comparative analysis of the Pocillopora damicornis genome highlights role of immune system in coral evolution. Sci. Rep. 8:16134.
De’ath, G., Lough, J. M., and Fabricius, K. E. (2009). Declining coral calcification on the Great Barrier Reef. Science 323, 116–119. doi: 10.1126/science.1165283
Dennison, W. C., and Barnes, D. J. (1988). Effect of water motion on coral photosynthesis and calcification. J. Exp. Mar. Biol. Ecol. 115, 67–77. doi: 10.1016/0022-0981(88)90190-6
Domart-Coulon, I., Tambutté, S., Tambutté, E., and Allemand, D. (2004). Short term viability of soft tissue detached from the skeleton of reef-building corals. J. Exp. Mar. Biol. Ecol. 309, 199–217. doi: 10.1016/j.jembe.2004.03.021
Erez, J., Reynaud, S., Silverman, J., Schneider, K., and Allemand, D. (2011). “Coral calcification under ocean acidification and global change,” in Coral Reefs: An Ecosystem in Transition, eds Z. Dubinsky and N. Stambler (Dordrecht: Springer), 151–176. doi: 10.1007/978-94-007-0114-4_10
Feuillassier, L., Martinez, L., Romans, P., Engelmann-Sylvestre, I., Masanet, P., Barthélémy, D., et al. (2014). Survival of tissue balls from the coral Pocillopora damicornis L. exposed to cryoprotectant solutions. Cryobiology 69, 376–385. doi: 10.1016/j.cryobiol.2014.08.009
Fordyce, A. J., Camp, E. F., and Ainsworth, T. D. (2017). Polyp bailout in Pocillopora damicornis following thermal stress. F1000Res. 6:687. doi: 10.12688/f1000research.11522.2
Fritz, A. E., Ikmi, A., Seidel, C., Paulson, A., and Gibson, M. C. (2013). Mechanisms of tentacle morphogenesis in the sea anemone Nematostella vectensis. Development 140, 2212–2223. doi: 10.1242/dev.088260
Fujita, S., Kuranaga, E., and Nakajima, Y.-I. (2019). Cell proliferation controls body size growth, tentacle morphogenesis, and regeneration in hydrozoan jellyfish Cladonema pacificum. PeerJ 7:e7579. doi: 10.7717/peerj.7579
Goreau, T. F., and Goreau, N. I. (1959). The physiology of skeleton formation in corals. II. Calcium deposition by hermatypic corals under various conditions in the reef. Biol. Bull. 117, 239–250. doi: 10.2307/1538903
Grasso, L., Negri, A., Foret, S., Saint, R., Hayward, D., Miller, D. J., et al. (2011). The biology of coral metamorphosis: molecular responses of larvae to inducers of settlement and metamorphosis. Dev. Biol. 353, 411–419. doi: 10.1016/j.ydbio.2011.02.010
Grasso, L. C., Maindonald, J., Rudd, S., Hayward, D. C., Saint, R., Miller, D. J., et al. (2008). Microarray analysis identifies candidate genes for key roles in coral development. BMC Genomics 9:540. doi: 10.1186/1471-2164-9-540
Grottoli, A. G., Rodrigues, L. J., and Palardy, J. E. (2006). Heterotrophic plasticity and resilience in bleached corals. Nature 440, 1186–1189. doi: 10.1038/nature04565
He, J., Lu, H., Zou, Q., and Luo, L. (2014). Regeneration of liver after extreme hepatocyte loss occurs mainly via biliary transdifferentiation in zebrafish. Gastroenterology 146, 789–800.e8.
Hirose, M., Yamamoto, H., and Nonaka, M. (2008). Metamorphosis and acquisition of symbiotic algae in planula larvae and primary polyps of Acropora spp. Coral Reefs 27, 247–254. doi: 10.1007/s00338-007-0330-y
Houlbrèque, F., and Ferrier-Pagès, C. (2009). Heterotrophy in tropical scleractinian corals. Biol. Rev. Camb. Philos. Soc. 84, 1–17. doi: 10.1111/j.1469-185x.2008.00058.x
Huang, D. W., Sherman, B. T., and Lempicki, R. A. (2008). Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13. doi: 10.1093/nar/gkn923
Huang, D. W., Sherman, B. T., and Lempicki, R. A. (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57. doi: 10.1038/nprot.2008.211
Inoue, M., Shinmen, K., Kawahata, H., Nakamura, T., Tanaka, Y., Kato, A., et al. (2012). Estimate of calcification responses to thermal and freshening stresses based on culture experiments with symbiotic and aposymbiotic primary polyps of a coral, Acropora digitifera. Glob. Planet. Change 92-93, 1–7. doi: 10.1016/j.gloplacha.2012.05.001
Jopling, C., Boue, S., and Belmonte, J. C. I. (2011). Dedifferentiation, transdifferentiation and reprogramming: three routes to regeneration. Nat. Rev. Mol. Cell Biol. 12, 79–89. doi: 10.1038/nrm3043
Kvennefors, E. C. E., Leggat, W., Kerr, C. C., Ainsworth, T. D., Hoegh-Guldberg, O., and Barnes, A. C. (2010). Analysis of evolutionarily conserved innate immune components in coral links immunity and symbiosis. Dev. Comp. Immunol. 34, 1219–1229. doi: 10.1016/j.dci.2010.06.016
Kvitt, H., Kramarsky-Winter, E., Maor-Landaw, K., Zandbank, K., Kushmaro, A., Rosenfeld, H., et al. (2015). Breakdown of coral colonial form under reduced pH conditions is initiated in polyps and mediated through apoptosis. Proc. Natl. Acad. Sci. U.S.A. 112, 2082–2086. doi: 10.1073/pnas.1419621112
Lecointe, A., Cohen, S., Gèze, M., Djediat, C., Meibom, A., and Domart-Coulon, I. (2013). Scleractinian coral cell proliferation is reduced in primary culture of suspended multicellular aggregates compared to polyps. Cytotechnology 65, 705–724. doi: 10.1007/s10616-013-9562-6
Lesser, M. (2013). Using energetic budgets to assess the effects of environmental stress on corals: are we measuring the right things? Coral Reefs 32, 25–33. doi: 10.1007/s00338-012-0993-x
Lesser, M. P. L., Mazel, C., Phinney, D., and Yentsch, C. S. (2000). Light absorption and utilization by colonies of the congeneric hermatypic corals Montastraea faveolata and Montastraea cavernosa. Limnol. Oceanogr. 45, 76–86. doi: 10.4319/lo.2000.45.1.0076
Levas, S., Grottoli, A. G., Schoepf, V., Aschaffenburg, M., Baumann, J., Bauer, J. E., et al. (2016). Can heterotrophic uptake of dissolved organic carbon and zooplankton mitigate carbon budget deficits in annually bleached corals? Coral Reefs 35, 495–506. doi: 10.1007/s00338-015-1390-z
Li, B., and Dewey, C. N. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323. doi: 10.1186/1471-2105-12-323
Liu, C., Cheng, S. H., and Lin, S. (2020). Illuminating the dark depths inside coral. Cell. Microbiol. 22:e13122.
Luo, Y., Zhao, J., He, C., Lu, Z., and Lu, X. (2020). Miniaturized platform for individual coral polyps culture and monitoring. Micromachines 11:127. doi: 10.3390/mi11020127
Mirsky, R., Woodhoo, A., Parkinson, D. B., Arthur-Farraj, P., Bhaskaran, A., and Jessen, K. R. (2008). Novel signals controlling embryonic Schwann cell development, myelination and dedifferentiation. J. Peripher. Nerv. Syst. 13, 122–135. doi: 10.1111/j.1529-8027.2008.00168.x
Mould, A., Wheldon, L., Komoriya, A., Wayner, E., Yamada, K., and Humphries, M. (1990). Affinity chromatographic isolation of the melanoma adhesion receptor for the IIICS region of fibronectin and its identification as the integrin alpha 4 beta 1. J. Biol. Chem. 265, 4020–4024. doi: 10.1016/s0021-9258(19)39696-6
Muscatine, L., Mccloskey, L. R., and Marian, R. E. (1981). Estimating the daily contribution of carbon from zooxanthellae to coral animal respiration 1. Limnol. Oceanogr. 26, 601–611. doi: 10.4319/lo.1981.26.4.0601
Muscatine, L., and Porter, J. W. (1977). Reef corals: mutualistic symbioses adapted to nutrient-poor environments. Bioscience 27, 454–460. doi: 10.2307/1297526
Nakamura, T., Yamasaki, H., and Van Woesik, R. (2003). Water flow facilitates recovery from bleaching in the coral Stylophora pistillata. Mar. Ecol. Prog. Ser. 256, 287–291. doi: 10.3354/meps256287
Osinga, R., Schutter, M., Griffioen, B., Wijffels, R. H., Verreth, J. A., Shafir, S., et al. (2011). The biology and economics of coral growth. Mar. Biotechnol. 13, 658–671.
Pang, A.-P., Luo, Y., He, C., Lu, Z., and Lu, X. (2020). A polyp-on-chip for coral long-term culture. Sci. Rep. 10:6964.
Poole, A. Z., Kitchen, S. A., and Weis, V. M. (2016). The role of complement in cnidarian-dinoflagellate symbiosis and immune challenge in the sea anemone Aiptasia pallida. Front. Microbiol. 7:519. doi: 10.3389/fmicb.2016.00519
Postigo, A. A., Sanchez-Mateos, P., Lazarovits, A., Sanchez-Madrid, F., and De Landazuri, M. (1993). Alpha 4 beta 7 integrin mediates B cell binding to fibronectin and vascular cell adhesion molecule-1. Expression and function of alpha 4 integrins on human B lymphocytes. J. Immunol. 151, 2471–2483.
Puverel, S., Tambutte, E., Zoccola, D., Domart-Coulon, I., Bouchot, A., Lotto, S., et al. (2005). Antibodies against the organic matrix in scleractinians: a new tool to study coral biomineralization. Coral Reefs 24, 149–156. doi: 10.1007/s00338-004-0456-0
Reyes-Bermudez, A., Desalvo, M. K., Voolstra, C. R., Sunagawa, S., Szmant, A. M., Iglesias-Prieto, R., et al. (2009). Gene expression microarray analysis encompassing metamorphosis and the onset of calcification in the scleractinian coral Montastraea faveolata. Mar. Genomics 2, 149–159. doi: 10.1016/j.margen.2009.07.002
Robinson, M. D., Mccarthy, D. J., and Smyth, G. K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Rodrigues, L. J., and Grottoli, A. G. (2007). Energy reserves and metabolism as indicators of coral recovery from bleaching. Limnol. Oceanogr. 52, 1874–1882. doi: 10.4319/lo.2007.52.5.1874
Sammarco, P. W. (1982). Polyp bail-out: an escape response to environmental stress and a new means of reproduction in corals. Mar. Ecol. Prog. Ser. 10, 57–65. doi: 10.3354/meps010057
Sawey, E. T., Johnson, J. A., and Crawford, H. C. (2007). Matrix metalloproteinase 7 controls pancreatic acinar cell transdifferentiation by activating the Notch signaling pathway. Proc. Natl. Acad. Sci. U.S.A. 104, 19327–19332. doi: 10.1073/pnas.0705953104
Schneider, C. A., Rasband, W. S., and Eliceiri, K. W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. doi: 10.1038/nmeth.2089
Schwarz, J. A., Krupp, D. A., and Weis, V. M. (1999). Late larval development and onset of symbiosis in the scleractinian coral Fungia scutaria. Biol. Bull. 196, 70–79. doi: 10.2307/1543169
Serrano, E., Coma, R., Inostroza, K., and Serrano, O. (2018). Polyp bail-out by the coral Astroides calycularis (Scleractinia, Dendrophylliidae). Mar. Biodivers. 48, 1661–1665. doi: 10.1007/s12526-017-0647-x
Shapiro, O. H., Kramarsky-Winter, E., Gavish, A. R., Stocker, R., and Vardi, A. (2016). A coral-on-a-chip microfluidic platform enabling live-imaging microscopy of reef-building corals. Nat. Commun. 7:10860.
Tanaka, Y., Iguchi, A., Inoue, M., Mori, C., Sakai, K., Suzuki, A., et al. (2013). Microscopic observation of symbiotic and aposymbiotic juvenile corals in nutrient-enriched seawater. Mar. Pollut. Bull. 68, 93–98. doi: 10.1016/j.marpolbul.2012.12.017
Thomas, L., and Palumbi, S. R. (2017). The genomics of recovery from coral bleaching. Proc. Biol. Sci. 284:20171790. doi: 10.1098/rspb.2017.1790
Titlyanov, E., Titlyanova, T., Loya, Y., and Yamazato, K. (1998). Degradation and proliferation of zooxanthellae in planulae of the hermatypic coral Stylophora pistillata. Mar. Biol. 130, 471–477. doi: 10.1007/s002270050267
Toller, W. W., Rowan, R., and Knowlton, N. (2001). Repopulation of zooxanthellae in the Caribbean corals Montastraea annularis and M. faveolata following experimental and disease-associated bleaching. Biol. Bull. 201, 360–373.
van de Water, J. A., Ainsworth, T. D., Leggat, W., Bourne, D. G., Willis, B. L., and Van Oppen, M. J. (2015). The coral immune response facilitates protection against microbes during tissue regeneration. Mol. Ecol. 24, 3390–3404.
Vandermeulen, J. (1975). Studies on reef corals. III. Fine structural changes of calicoblast cells in Pocillopora damicornis during settling and calcification. Mar. Biol. 31, 69–77.
Vega, M. E., Giroux, V., Natsuizaka, M., Liu, M., Klein-Szanto, A. J., Stairs, D. B., et al. (2014). Inhibition of Notch signaling enhances transdifferentiation of the esophageal squamous epithelium towards a Barrett’s-like metaplasia via KLF4. Cell Cycle 13, 3857–3866.
Vizel, M., Loya, Y., Downs, C. A., and Kramarsky-Winter, E. (2011). A novel method for coral explant culture and micropropagation. Mar. Biotechnol. 13, 423–432.
Wecker, P., Lecellier, G., Guibert, I., Zhou, Y., Bonnard, I., and Berteaux-Lecellier, V. (2018). Exposure to the environmentally-persistent insecticide chlordecone induces detoxification genes and causes polyp bail-out in the coral P. damicornis. Chemosphere 195, 190–200.
Keywords: polyp bail-out, micropropagation, polyp recovery, genetics, metabolism
Citation: Chuang P-S, Ishikawa K and Mitarai S (2021) Morphological and Genetic Recovery of Coral Polyps After Bail-Out. Front. Mar. Sci. 8:609287. doi: 10.3389/fmars.2021.609287
Received: 23 September 2020; Accepted: 04 March 2021;
Published: 25 March 2021.
Edited by:
Daniel Wangpraseurt, University of Cambridge, United KingdomCopyright © 2021 Chuang, Ishikawa and Mitarai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Po-Shun Chuang, ps.chuang@oist.jp
 Po-Shun Chuang
Po-Shun Chuang Kota Ishikawa
Kota Ishikawa Satoshi Mitarai
Satoshi Mitarai