Ecological Fitness of Vibrio cholerae, Vibrio parahaemolyticus, and Vibrio vulnificus in a Small-Scale Population Dynamics Study
- Department of Life Sciences and Chemistry, Jacobs University Bremen, Bremen, Germany
The potential spread of infectious diseases in response to climate change and rising sea surface temperatures in temperate regions has been a growing concern for the past several decades. Extreme heat waves in the North Atlantic and North Sea regions have been correlated with an increase in human Vibrio infections; of particular concern to human health are Vibrio cholerae, Vibrio parahaemolyticus, and Vibrio vulnificus. While these species are well-known to cause disease in humans, most environmental strains are not pathogenic. Studying not only the behavior of the pathogenic strains, but that of non-pathogenic environmental isolates, may better elucidate their ecological relationship in their native microbiome and the dispersal of these species in coastal regions. Using red fluorescent protein-tagged and gentamycin-resistant V. cholerae, V. parahaemolyticus, and V. vulnificus strains, we investigated whether increasing temperatures confer greater competitive fitness to these species when incubated within a natural North Sea water sample still containing its microbiome in a small-scale niche investigation. Increased incubation temperatures alone did not confer a competitive advantage to V. cholerae, V. parahaemolyticus, and V. vulnificus. The microbial community could limit Vibrio growth at all temperatures. To the best of our knowledge, we also demonstrate the first (albeit unintentional) genetic modification of multiple species of marine bacteria through the introduction of a genetically modified V. vulnificus strain into a natural water sample in a contained system.
Introduction
Rising sea surface temperatures (SST) have been studied for their pronounced impact on the abundance, distribution, and composition of marine ecosystems (Poloczanska et al., 2013). Due to their significance to global element and nutrient cycling, and a lack of historical data to compare with, studies on the potential effects of rising temperatures on marine prokaryotes, in particular, have been in high demand (Vezzulli et al., 2016). The potential spread of infectious diseases in response to climate change has been well expressed, as reviewed by Harvell et al. (2002). Vibrio cholerae has been used to illustrate the idea of the “pathogen advantage theory,” which describes how pathogens of warm-blooded animals may hold a competitive advantage over non-pathogenic organisms in their native environment under rising temperatures, leading to a rise in abundance in the pathogen over time (Colwell, 2004). The relative abundance of Vibrios was found to be correlated with rising SST in the North Atlantic and North Sea (Vezzulli et al., 2016). In temperate regions, such as in the North Sea, extreme heat waves (SST over 18°C) in coastal areas are correlated with an increase in human Vibrio infections (Baker-Austin et al., 2017) manifesting primarily in the form of gastroenteritis, but also wound infections and primary septicemia in rarer cases. Therefore, understanding not only the prevalence of these pathogenic Vibrios in the environment, but also their relationship to the greater microbial community, is crucial to the understanding of the dispersal of these organisms and the potential risk they pose to aquaculture and human health.
Members of the genus Vibrio are found ubiquitously in marine and freshwater environments. Genetically diverse, these heterotrophic bacteria have been found to inhabit a wide variety of niches, both as free-living cells and symbionts. From copepods to mollusks, and from marine aggregates to fish, Vibrio bacteria have been at the heart of many marine symbiotic and ecological studies. With over 110 species described, the genus Vibrio represents one of the largest fractions of culturable marine bacteria (Farmer and Janda, 2015; Logue and Nde, 2017), many of which are pathogenic to animals or humans. These pathogenic Vibrio species are of particular interest to the aquaculture industry and to human health in coastal areas. Twelve Vibrio species are known to be pathogenic to humans, of these, V. cholerae, Vibrio parahaemolyticus, and Vibrio vulnificus are amongst the best described and most prominent in their ability to cause disease.
Laboratory studies investigating the growth dynamics of V. cholerae, V. parahaemolyticus, and V. vulnificus are often restricted to well characterized laboratory strains or known pathogenic variants but neglect the environmental isolates. Each species represents a genetically diverse group, and relatively few strains cause disease in humans (Nishibuchi and Kaper, 1995; Faruque et al., 1998; Strom and Paranjpye, 2000; Alam et al., 2007; Vieira et al., 2011; Islam et al., 2013). Ecological studies regarding the overall abundance of these organisms and characterizing their environment have become crucial for informing our knowledge for how and where Vibrio interact with other organisms in their niches affected by changing SST. Traditional in vitro studies on the interactions of environmental V. cholerae, V. parahaemolyticus, and V. vulnificus isolates are uncommon, largely because they are often laborious and require labeled bacteria that can be tracked within a sample of mixed organisms. Despite these challenges, such in vitro approaches can provide great insight into the growth behavior of bacteria under changing conditions.
Herein, we investigated whether increasing temperatures confer greater competitive fitness to V. cholerae, V. parahaemolyticus, or V. vulnificus when co-incubated with a natural water sample taken from the North Sea. We were particularly interested in whether a SST increase alone leads to higher abundances, or if the surrounding microbial community will limit the growth of these Vibrio species regardless of temperature change. Using red fluorescent protein (RFP)-tagged and gentamycin-resistant (GmR) V. cholerae, V. parahaemolyticus, and V. vulnificus strains, we could monitor their abundance over time in a native North Sea water sample when exposed to three distinct temperatures. Our test hypothesis was that increased incubation temperatures would lead to a slower decline in our RFP-tagged Vibrios in the mixed sample than the coldest test temperature. We report that in our summer water sample, increasing temperatures alone did not confer a competitive advantage to V. cholerae, V. parahaemolyticus, and V. vulnificus. To the best of our knowledge, we also demonstrate the first RFP-labeling of multiple different species of marine bacteria through the introduction of a genetically modified clone into a contained natural water sample.
Materials and Methods
Water Sample Description
A two-liter water sample was taken from surface waters near Helgoland, Germany, at the long-term sampling station “Kabeltonne” at Helgoland Roads (54°11.3′N; 7°54.0′E) in early August 2019. The water was collected into two sterile one-liter Schott flasks and stored on ice in the dark until arrival in the lab. The in situ temperature at the time of sampling was 17°C. Upon arrival in the lab, the water sample was split into a Natural Sea Water (NaSW) sample, which was used to simulate the native or natural microbial community of the water column in the North Sea, and a Sterile Sea Water (SSW) control sample, which was filter-sterilized through a 0.22 μM pore-size filter.
Media and Antibiotics
Labeled Vibrio strains were routinely grown on Marine Broth (MB) agar (Sonnenschein et al., 2011) or MB liquid medium supplemented with either gentamycin (Gm) or chloramphenicol (Cm). The final concentration of each antibiotic in the media was always 50 μg mL–1 and 10 μg mL–1, respectively. V. parahaemolyticus strains were grown on MB agar plates prepared with a higher concentration of agar (3%), to prevent the swarming of the colonies and to obtain clear CFU counts. Dilutions were routinely made in sterile-filtered North Sea Water (NSW), previously stored in bulk at 4°C and autoclaved before use.
Experimental Niche Analyses
Bacterial strains used in this study, and their relevant characteristics, are shown in Table 1. Previously, multiple genetically modified Vibrio clones were obtained via bi-parental mating with an Escherichia coli strain carrying a suicide vector with a Tn5 transposon coding for the red fluorescent protein (RFP) gene (dsRedExpress), the gentamycin resistance gene (GmR), and the chloramphenicol resistance gene (CmR) (Thorstenson and Ullrich, 2020). These RFP-labeled clones behave comparably to the wild-type (WT) strain when grown in MB medium and sterilized NSW.
All labeled Vibrio strains were reactivated from 15% glycerol stocks (stored at −80°C), onto MB agar plates supplemented with Gm, and grown for 14 h at 37°C. A single colony was then used to inoculate a 10 mL pre-culture of MB broth supplemented with Gm, and grown for 12 h at 37°C. The pre-culture (1 mL) was used to inoculate a 100 mL culture of MB broth supplemented with Gm and allowed to grow at 28°C for 20–22 h with shaking at 180 rpm. Bacterial cultures were demonstrated to be free of contaminants via light microscopy and plating on non-selective MB agar. Each RFP-tagged Vibrio strain was confirmed as their supposed species via species-specific multiplex PCR (Neogi et al., 2010) (data not shown) before use in the following niche experiments.
The overnight cultures of Vibrio were serially diluted to obtain a concentration of approximately 1 × 106 CFU mL–1 in sterile NSW. Then, 0.1 mL of this dilution was inoculated, in triplicate, into 10 mL of either NaSW or SSW, in borosilicate test tubes, to have a starting concentration of 105 CFU mL–1. The set-up was such that each Vibrio strain had a triplicate NaSW and SSW incubation for each growth temperature of 17°C, 20°C, and 22°C. A negative control series of NaSW and SSW (each in triplicate per temperature), was inoculated with the sterile NSW used for dilution of the Vibrio cultures. A diagram of the experimental set-up is found in Supplementary Figure 1. All tubes were incubated at 180 rpm in the dark for up to 72 h. A sub-sample was taken every 24 h and serially diluted onto MB agar supplemented with Gm for CFU enumeration. All agar plates were incubated at 25°C for up to 48 h.
For CFU enumeration, all colonies were counted per plate after 24 and after 48 h of incubation at 25°C. This was to ensure that all plates were counted at least twice. The corresponding control plates were also kept for the 48 h and repeatedly checked for contamination or growth. No growth occurred during this time on the controls, consequently we can assume all bacteria that did grow on our experimental plates are a result of the experiment, and not from contamination or natural bacterial resistance to Gm in the water samples. For CFU enumeration, the detection limit was set at the least diluted sample containing at least 30 colonies or more, otherwise the number was noted but not considered statistically relevant.
Double Agar Overlay Assay
To detect the presence of any possible phages in our sea water sample, a standard double agar overlay assay (DAOA) was performed after the completion of the niche analyses experiment. The host strain, VpR-DSMZ, was grown in 10 mL MB broth at 17°C and 20°C. An aliquot (3 mL) of soft agar (2 g peptone, 0.4 g yeast extract, 0.04 g FePO4, 300 mL NSW, 100 mL dH2O, 2.4 g agar, pH of 7.6) was melted at 95°C in a water bath. Then the aliquots were transferred to a heat block of 42°C. The remaining NaSW and SSW water from the 17°C and 20°C VpR-DSMZ niche analysis were pooled together and split into two fractions, the first was sterile-filtered again via a 0.2 μM syringe filter and the second fraction was sterile-filtered and autoclaved. The resulting samples were serially diluted 1: 10 and 1: 100 and used as so-called “phage eluate” samples. The phage eluate (0.1 mL) was added directly to the center of a 10 mL MB agar plate. The host strain was briefly vortexed and then 0.5 mL was added to an aliquot of soft agar, this mix was quickly poured onto the center of the plate and spread by swirling the plate gently. The plates were incubated for 24 and 48 h, both at 17°C and 20°C, respectively.
A second DAOA was performed, except the original NaSW water was used as the starting source for the phage enumeration. This sample was again split into a non-autoclaved and an autoclaved sterile-filtered subsample. From then on, the experiment was performed identically as detailed above.
Species Identification and Transposon Detection via Polymerase Chain Reaction
All primers pairs and annealing temperatures used for these experiments are detailed in Table 2. All Vibrio strains were routinely re-identified via species-specific multiplex PCR published by Neogi et al. (2010). Additional isolates from the VvR-21 niche analysis were characterized with 16S rRNA gene sequencing. In most cases, an identification down to the genus level could be reliably made.
To see if we could amplify the RFP gene found in the transposon originally used in the construction of VvR-21 (Thorstenson and Ullrich, 2020), in the new bacterial isolates from the VvR-21 niche analysis, the rfp6 primers were used. All isolate DNA was tested alongside the DNA of VvR-21 as a positive control. All PCR products were analyzed on a 1% agarose gel and scored as either a presence or absence (±) of the 600 bp rfp6 amplification product.
All PCR reagents were performed as recommended from the manufacturer New England Biolabs (NEB) (DNA Taq Polymerase, NEB #M0273L) for a 25 μL volume reaction for the 16S rRNA sequencing. The rfp6 PCR used OneTaq DNA Polymerase (NEB#M0480L) for a 25 μL volume reaction. The thermocycling conditions were as follows; for 16S: initial denaturation for 2 min at 95°C, followed by 35 cycles of 0.5 min denaturation at 95°C, primer annealing of 0.5 min, primer extension of 1.5 min at 72°C, followed by a final extension time of 3 min at 72°C; for all other PCRs: initial denaturation for 3 min at 95°C, followed by 35 cycles of 0.5 min denaturation at 95°C, primer annealing of 0.5 min, primer extension of 0.5 min at 72°C, followed by a final extension time of 3 min at 72°C. All PCR products were analyzed via gel electrophoresis on a 1% agarose/TBE gel run for 30 min at 120 V. The 16S PCR products were cleaned via PCR Clean-up and Gel Extraction Kit (NEB #T1030S) and sent for sequencing via Eurofins Genomics, per the manufacturers’ protocols.
Results
In order to test whether increased SST alone led to an increased abundance or elevated competitive-fitness of V. cholerae, V. parahaemolyticus, and V. vulnificus, we inoculated filter-sterilized Sea Water (SSW) and non-sterile Native Sea Water (NaSW) samples with four different strains of RFP-labeled, GmR -Vibrio strains (VcR-7-2, VpR-DSMZ, VpR-NSI, and VvR-21) in triplicate (Table 1). An additional set of negative controls (SSW and NaSW without an inoculation), were made in triplicate per temperature and sampled along with the experimental incubations. The experiment was done at three temperatures, the in situ temperature at the time of sampling (17°C), 20°C, and 22°C to reflect probable and extreme increased SST in the North Sea in the future. The abundance of our labeled Vibrio strains was observed over a period of 72 h, via subsequent serial dilutions and plating on MB agar supplemented with Gm every 24 h. The negative controls were sampled and plated on the same selective media.
Throughout the course of the experiment, all selective plates from the NaSW and SSW negative controls remained free of contamination and growth. Since no bacterial growth was observed from the NaSW and SSW negative controls, we conclude that all growth on the selective plates from the experimental plates comes from the added labeled Vibrio strains and do not arise from a natural genetic resistance to Gm in the native microbial sample.
Vibrio cholerae
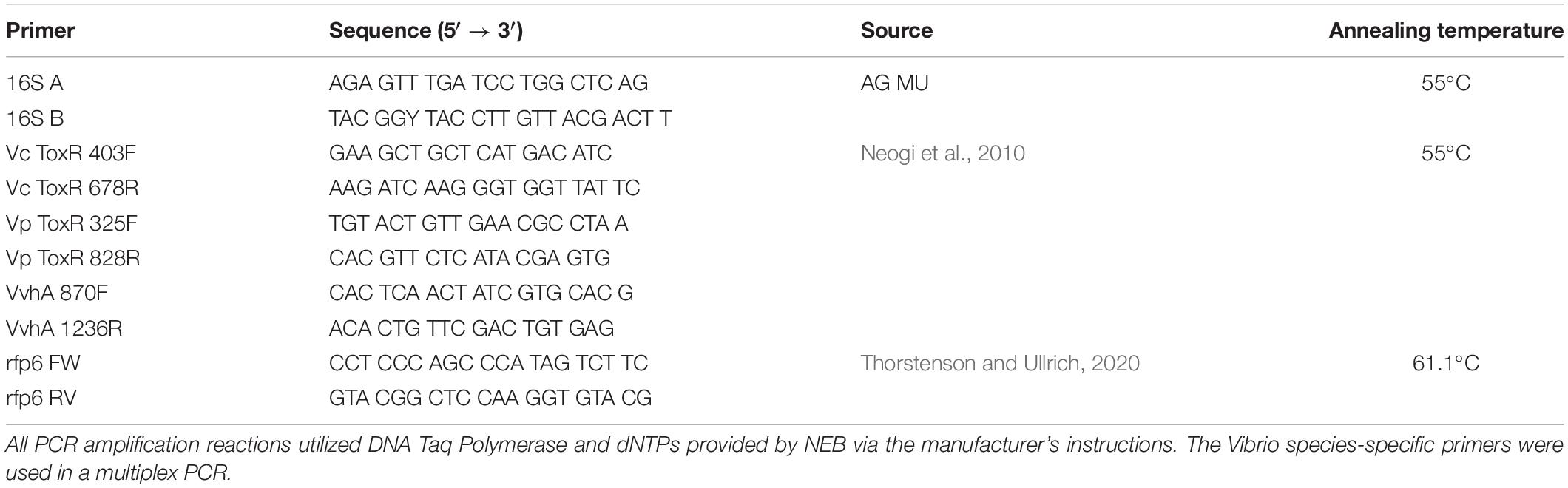
When RFP-tagged V. cholerae clone VcR-7-2 was incubated at 17°C, we did not see any significant changes in abundance in the SSW incubations and observed a steady decrease in abundance in the NaSW incubations (Figure 1). With the increased temperatures of 20°C and 22°C there was a slight increase in abundance (2.2- and 1.8- fold, respectively) in the SSW incubations from 0 to 24 h, after which there was no significant change up through 72 h. Meanwhile, in the NaSW, VcR-7-2 abundance steadily declined in the 20°C and 22°C incubations. Temperature was not significantly correlated with the rate of decline across the NaSW incubations (Figure 2). While we did see that temperatures above 20°C led to a slightly higher V. cholerae abundance in the SSW incubations, higher temperatures did not confer any competitive advantage to VcR-7-2 in the NaSW incubations over the 17°C incubations.
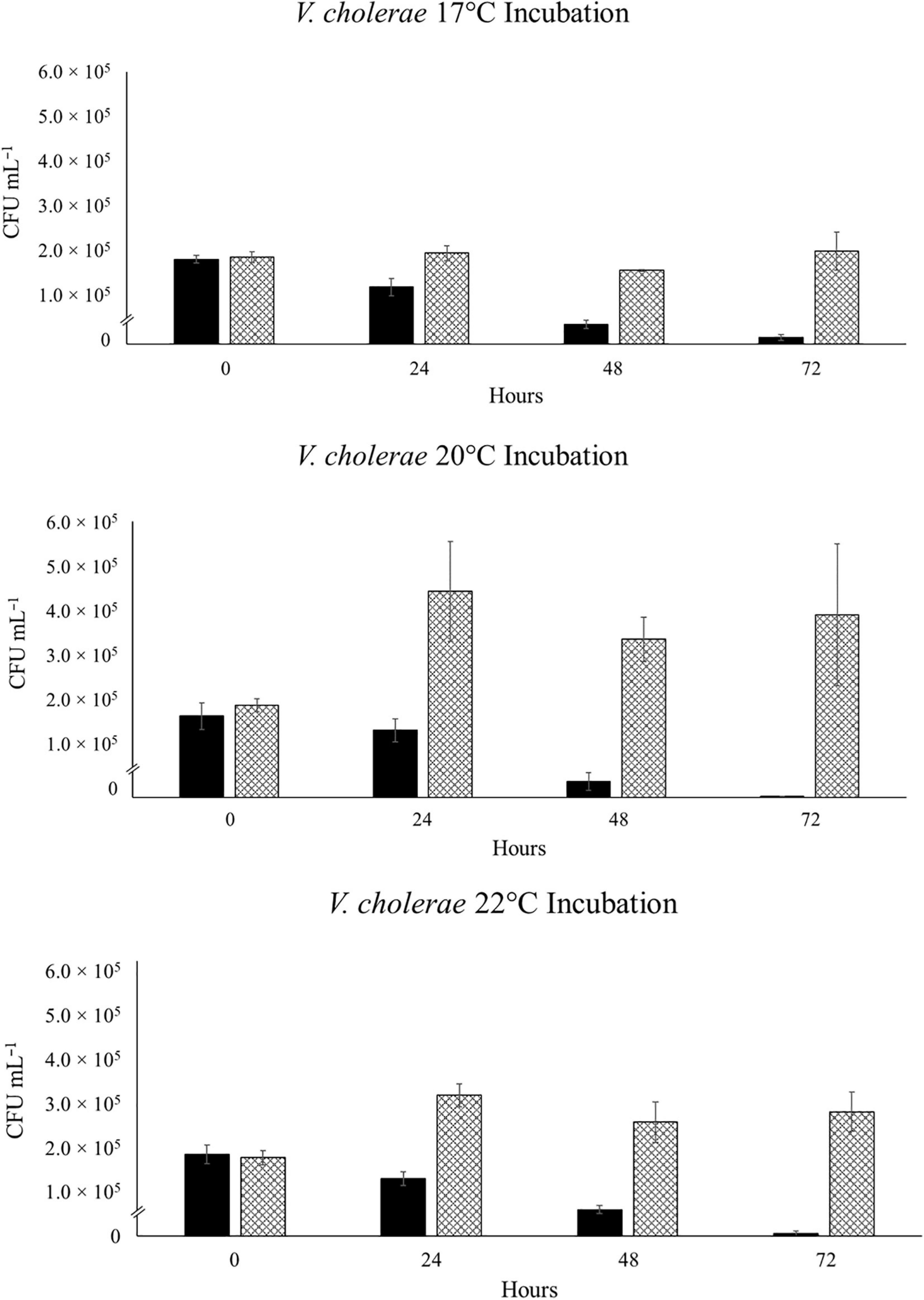
Figure 1. Abundance of RFP-labeled V. cholerae in NaSW and SSW over 72 h. Strain VcR-7-2 was inoculated into a non-sterile native sea water sample (NaSW, black bars) and a sterile-filtered sea water sample (SSW, hatched bars) and incubated for 72 h at three discrete temperatures: 17°C (Top), 20°C (Middle), and 22°C (Bottom). Error bars represent the standard deviation between triplicate samples.
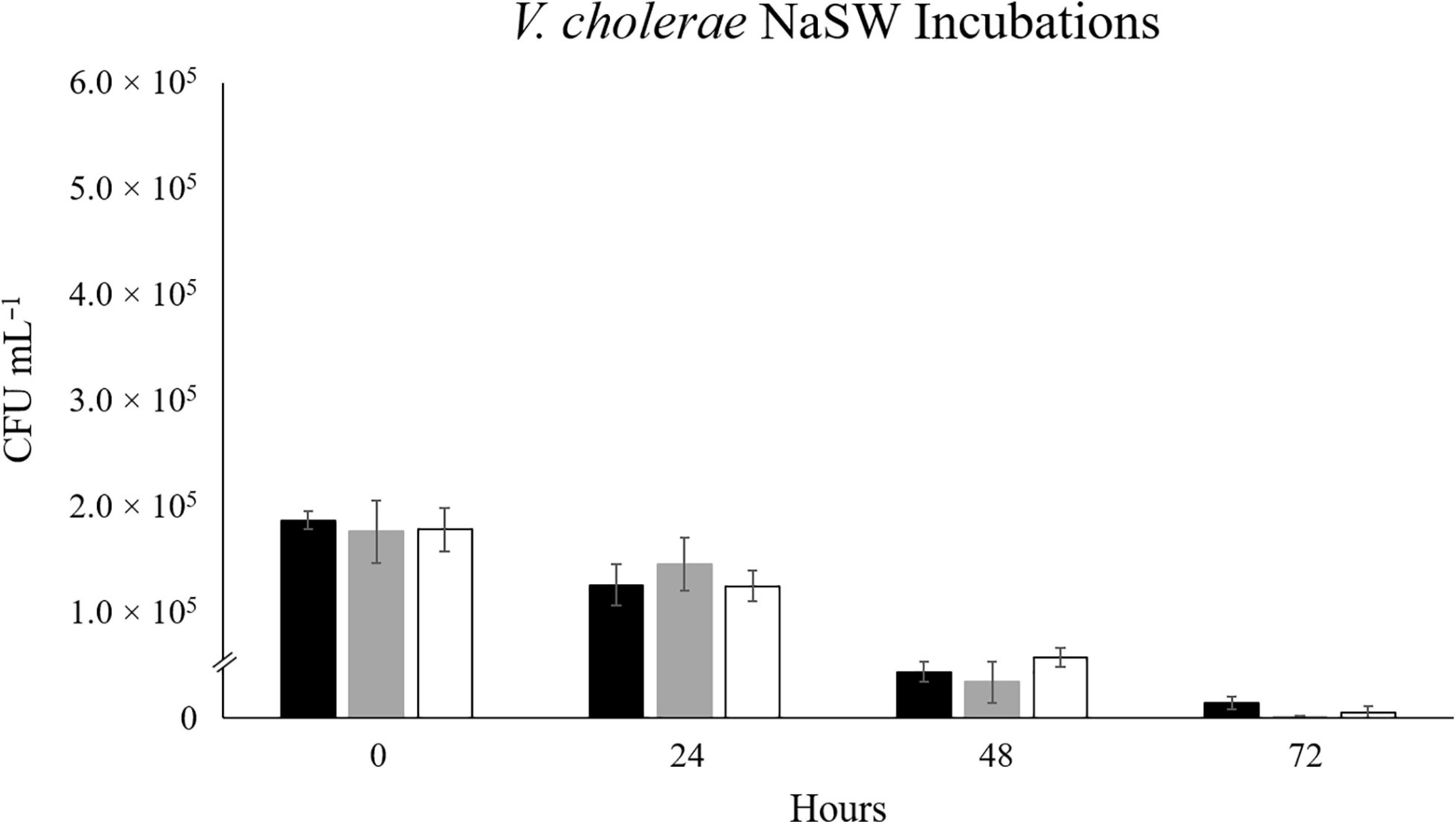
Figure 2. The Effect of Temperature on the Abundance of RFP-tagged V. cholerae in NaSW. Strain VcR-7-2 was inoculated into a non-sterile native sea water sample (NaSW) and incubated for 72 h at three discrete temperatures: 17°C (black), 20°C (gray), and 22°C (white). Error bars represent the standard deviation between triplicates.
Vibrio parahaemolyticus
Two strains of RFP-tagged V. parahaemolyticus (VpR) were tested in the niche experiments. One strain (VpR-DSMZ) was originally isolated from cockles which caused food poisoning in humans and had been in use in the laboratory for some time. VpR-DSMZ has been relatively well characterized and is known to contain the Kanagawa virulence factors, tdh and trh, associated with human disease (Nishibuchi and Kaper, 1995). The other strain is a North Sea isolate (VpR-NSI) which does not contain these factors and, to our knowledge, is not pathogenic to humans. Two strains of V. parahaemolyticus were chosen for the niche analysis to not only compare the behavior of two different strains of the same species, but also to test one strain presumably more adapted to the North Sea than the other.
In the VpR-DSMZ incubation at 17°C, we see a slow, 20.7-fold, increase in abundance over 72 h in SSW. A smaller, initial increase (6.3-fold) after 24 h, was observed in the NaSW incubations, followed by a rapid decrease by 72 h (Figure 3). After 24 h in the 20°C incubation, a 5.7-fold and 13.9-fold increase in abundance were observed in the NaSW and SSW incubations, respectively. Fold increases were determined with the average CFU mL–1, although if we consider the larger standard deviation for this time point, we see there was perhaps not as large an increase in abundance between the NaSW and SSW incubation as the numbers would suggest. In contrast to the incubations with VcR-7-2, after 24 h the VpR-DSMZ abundances did not remain constant in the SSW incubations, but rather, rapidly declined. Similarly, in the 22°C incubations, after a 6.7-fold increase after 24 h in both NaSW and SSW, VpR-DSMZ abundance rapidly declines in both incubations. To summarize, temperatures above 20°C featured an initial increase in abundance in both the NaSW and SSW incubations, which was followed by a decline nearly to the detection limit by 72 h in both NaSW and SSW. Nonetheless, the final abundance in the SSW was always higher than in the NaSW across the three temperatures.
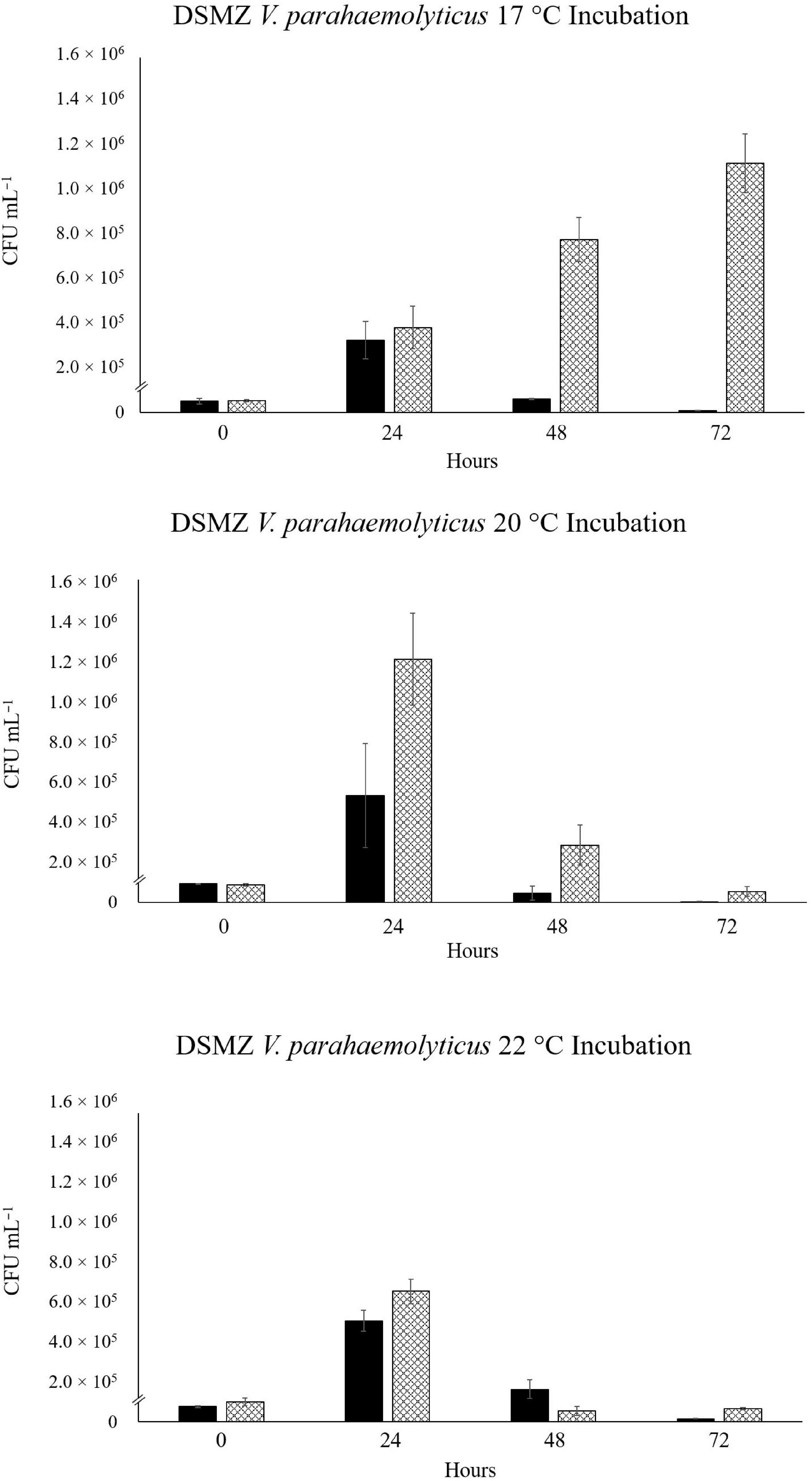
Figure 3. Abundance of RFP-labeled V. parahaemolyticus in NaSW and SSW over 72 h. Strain VpR-DSMZ was inoculated into a non-sterile native sea water sample (NaSW, black bars) and a sterile-filtered sea water sample (SSW, hatched bars) and incubated for 72 h at three discrete temperatures: 17°C (Top), 20°C (Middle), and 22°C (Bottom). Error bars represent the standard deviation between triplicates.
For the North Sea V. parahaemolyticus, VpR-NSI, a steady and comparable decline in abundance was observed in the NaSW and SSW incubations across all three temperatures (Figure 4). VpR-NSI abundance was still enumerable after 72 h in the 17°C NaSW incubation, but not in the 20°C and 22°C NaSW incubations. Neither the total abundance nor the rate of decline of VpR-NSI in SSW strongly correlate with temperature. Across all three temperatures, the abundance of VpR-NSI in the SSW incubations was greater than that of NaSW incubations after 72 h. Both NaSW and SSW incubations featured similar decline rates in VpR-NSI abundance.
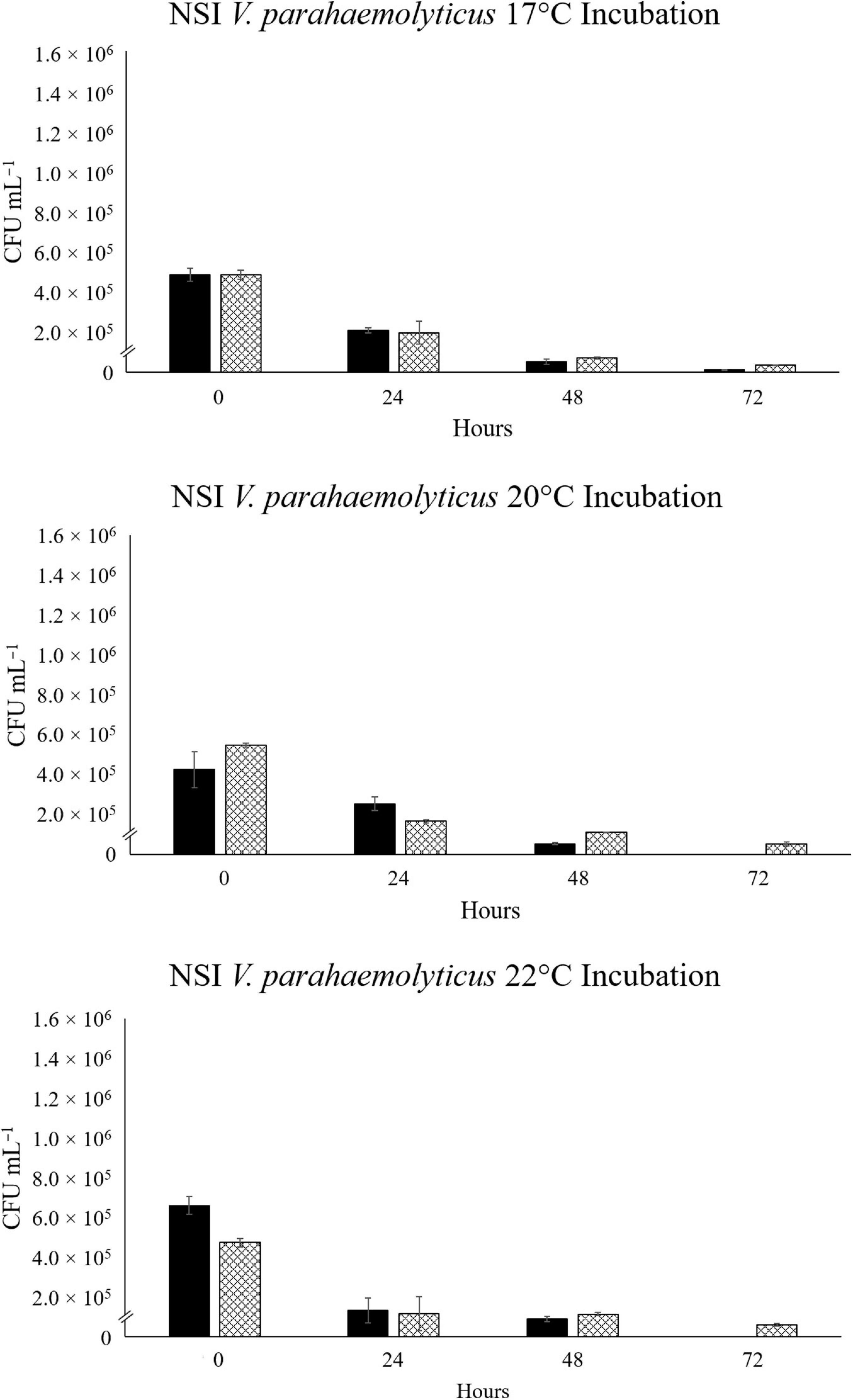
Figure 4. Abundance of RFP-labeled V. parahaemolyticus strain VpR-NSI in NaSW and SSW over 72 h. Strain VpR-NSI was inoculated into a non-sterile native sea water sample (NaSW, black bars) and a sterile-filtered sea water sample (SSW, hatched bars) and incubated for 72 h at three discrete temperatures: 17°C (Top), 20°C (Middle), and 22°C (Bottom). Error bars represent the standard deviation between triplicates.
This unexpected decline in V. parahaemolyticus abundance in SSW, led us to theorize on the various possibilities for cell death of the VpR strains in “sterile” sea water. Both strains had been previously incubated in autoclaved NSW at 28°C, and no significant fluctuations in abundance was recorded for either strain over a period of 72 h (data not shown). We repeated the NSW incubations at 20°C for VpR-DSMZ to see if we could receive a similar result to the 20°C SSW experiment (Figure 5). This was not the case, and like in the 28°C incubations, there were no major changes in VpR-DSMZ abundance over 72 h incubation.
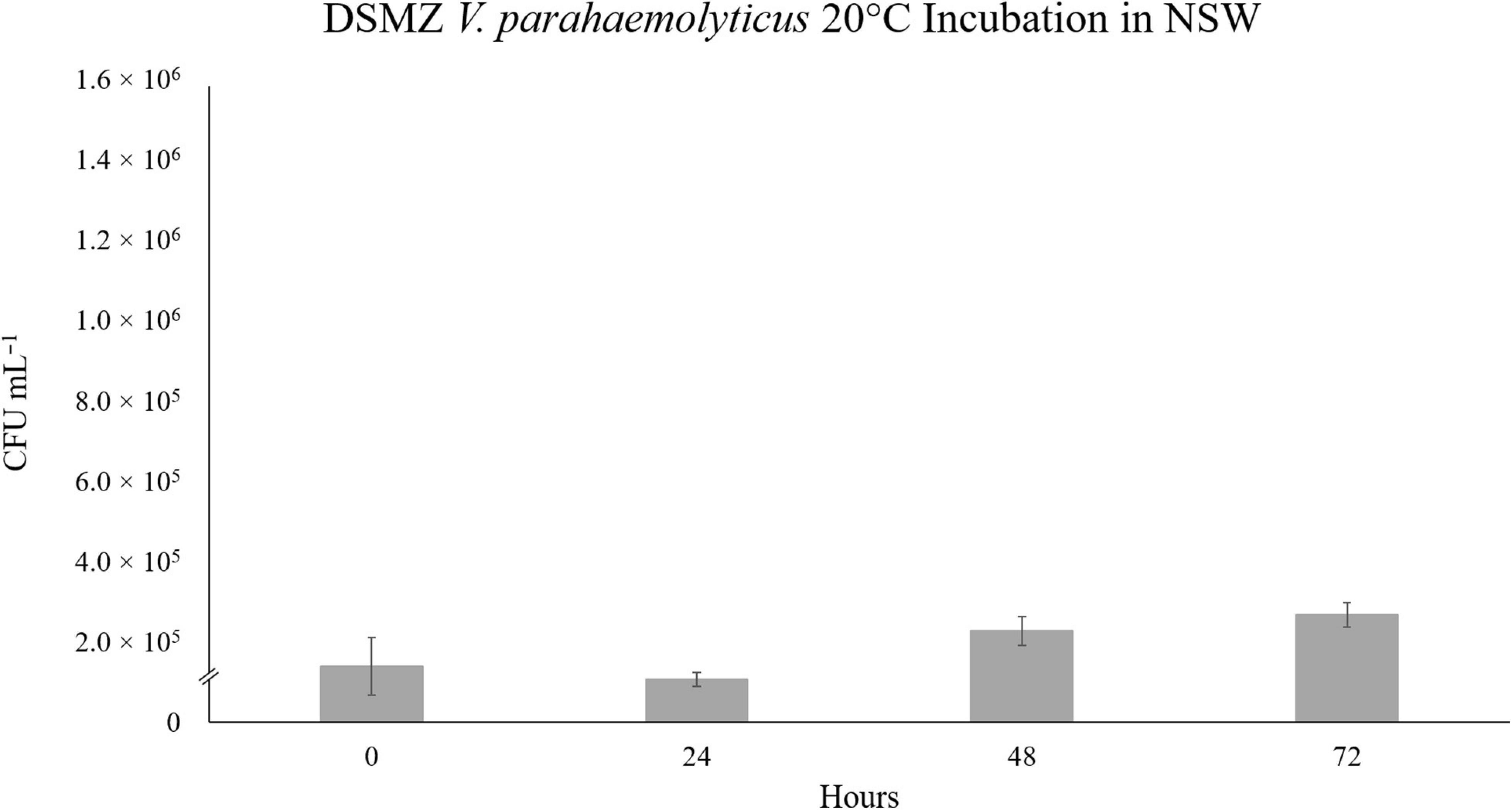
Figure 5. Strain VpR-DSMZ Abundance in Sterile NSW at 20°C. The starting culture and inoculation at t0 were prepared as described for the niche analyses experiments, except sterile-filtered and autoclaved North Sea Water (NSW) was used in place of the NaSW or SSW. Error bars represent the standard deviation between triplicate samples.
A standard DAOA protocol was used to assess if we could cultivate any phages or other microorganisms that could have escaped the first filtration and be contributing to the decline in V. parahaemolyticus in the SSW. No plaque forming units (PFUs) were observed across the two different temperatures (17°C and 20°C) used to cultivate a lawn of VpR-DSMZ.
Vibrio vulnificus
Introduction of RFP-tagged V. vulnificus clone VvR-21 into NaSW and SSW yielded entirely unexpected and surprising results. While the plates from the SSW incubations contained only V. vulnificus-like colonies and were free from contaminations, many morphologically different bacterial colonies formed on the NaSW plates in addition to the VvR-21 colonies (Figure 6). This non-Vibrio growth was seen in all three temperature incubations and was observed only after 24 h co-incubation with VvR-21 in NaSW. There was no obvious correlation between the type of bacterial contamination on the plate and temperature.
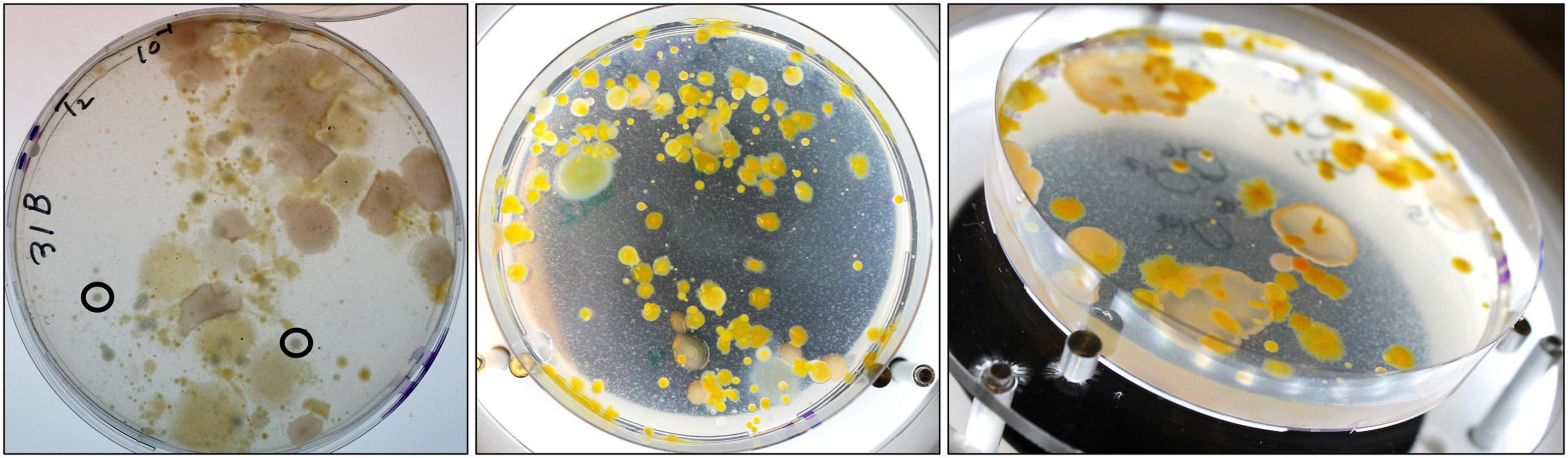
Figure 6. Non-V. vulnificus growth on MB agar supplemented with Gentamycin. Chosen plates are representative of the bacterial growth from all temperature incubations. (Left) NaSW incubation, 48 h, 22°C. Image taken from the bottom of the agar plate, circled colonies are representative of “V. vulnificus -like colonies”. (Middle) NaSW incubation, 72 h, 17°C. Image taken from top of plate without lid. (Right) NaSW incubation, 72 h, 17°C. Image taken at an angle with the lid off to better visualize the different morphologies of the non-Vibrio bacteria.
The t0 plates (samples taken at inoculation) had only V. vulnificus-like colonies. CFU enumeration was attempted based on the number of V. vulnificus-like colonies still countable on the plates at later time points, but since the non-Vibrio bacteria grew densely, it would be misleading to over-interpret colony numbers and draw conclusions from such estimated CFUs. We instead summarize the results of the niche analyses with VvR-21 based on the simple presence or absence of V. vulnificus-like colonies on the plates in Table 3.
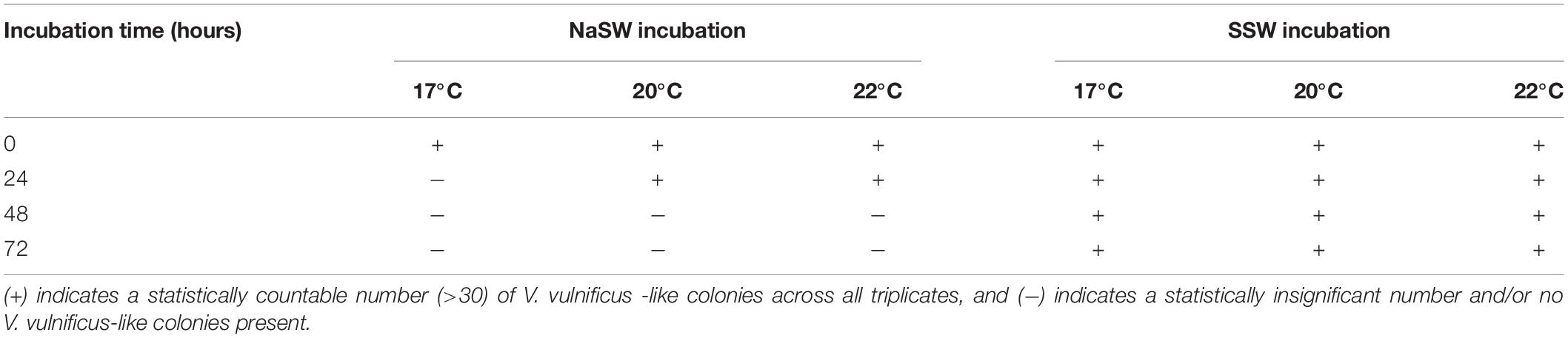
Table 3. Presence or absence of V. vulnificus -like colonies on selective media throughout the course of the niche analyses.
At 17°C, there were not enough colonies on the plates (>30) to be statistically significant in the NaSW incubations after 24 h. For 20°C and 22°C, the number of statistically significant colonies lasted up through 24 h before disappearing completely by the 48 h. In all three temperatures, statistically countable colonies were counted on all SSW plates throughout 72 h. However, like in the V. parahaemolyticus incubations, a noticeable decline in abundance was observed in the SSW at all three temperatures (Figure 7).
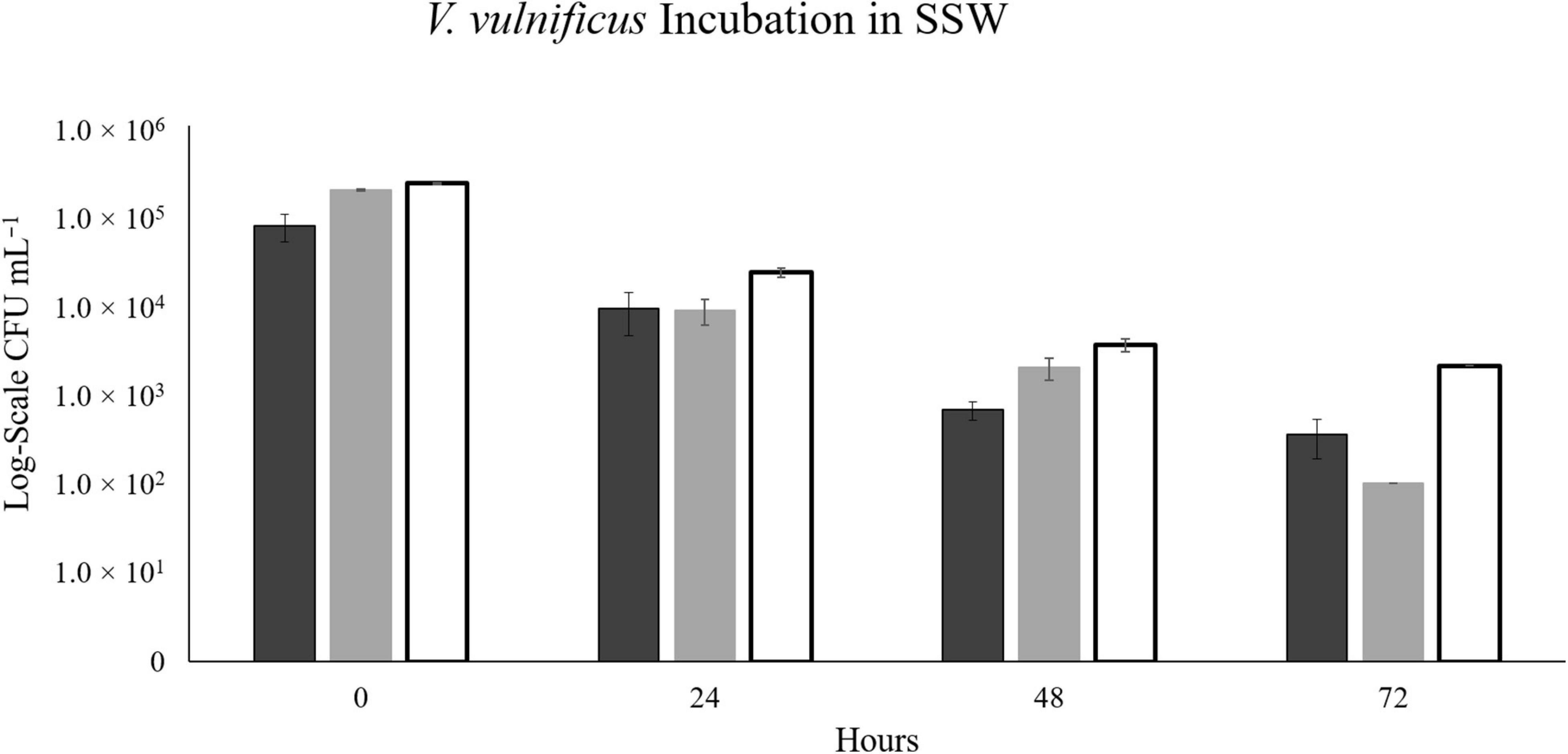
Figure 7. The abundance of V. vulnificus strain VvR-21 in SSW over 72 h. Strain VvR-21 was inoculated into a sterile-filtered sea water sample (SSW) and incubated for 72 h at three discrete temperatures: 17°C (black), 20°C (gray), and 22°C (white). Error bars represent the standard deviation between triplicates.
Non-Vibrio colonies were only observed on the NaSW plates after 24 h incubation, or in other words, the t0 NaSW plates only contained V. vulnificus-like colonies similar to the SSW plates. Since no bacteria grew on the NaSW negative control plates throughout the experiment, and a similar contamination did not occur in the VcR or VpR experiments, we surmise that the GmR was likely conferred from VvR-21 to these other non-Vibrio bacteria during the initial 24 h co-incubation, rather than natural GmR occurring in the water sample.
It is interesting to note that VvR-21 consistently conferred GmR to the same types of bacteria over the course of the experiment: at least 10 morphologically different colonies were consistently observed on the NaSW plates (Figure 6). To identify the bacteria which grew on the selective media, thirty colonies were chosen from the first niche analysis for continued propagation and isolation on selective media. VvR-21 was originally constructed with a transposon carrying the genes for RFP, GmR, and CmR (Thorstenson and Ullrich, 2020). In short, this transposon was delivered into VvR-21 via a bi-parental mating procedure and a suicide vector. To ensure that the isolates obtained their GmR from VvR-21 and did not pick up the resistance from another source, we also screened for the RFP expression via fluorescence microscopy (FM) and PCR. In addition, isolates were plated on MB agar containing Gm and Cm. Twenty-five isolates were able to be stably maintained on MB agar supplemented with Gm and Cm. These isolates represent nine different genera as identified by 16S rRNA gene sequencing (Table 4). Eighteen of the isolates were positive for the RFP gene by PCR amplification. Curiously, only ten isolates actively expressed RFP under our standard fluorescence microscopy methods.
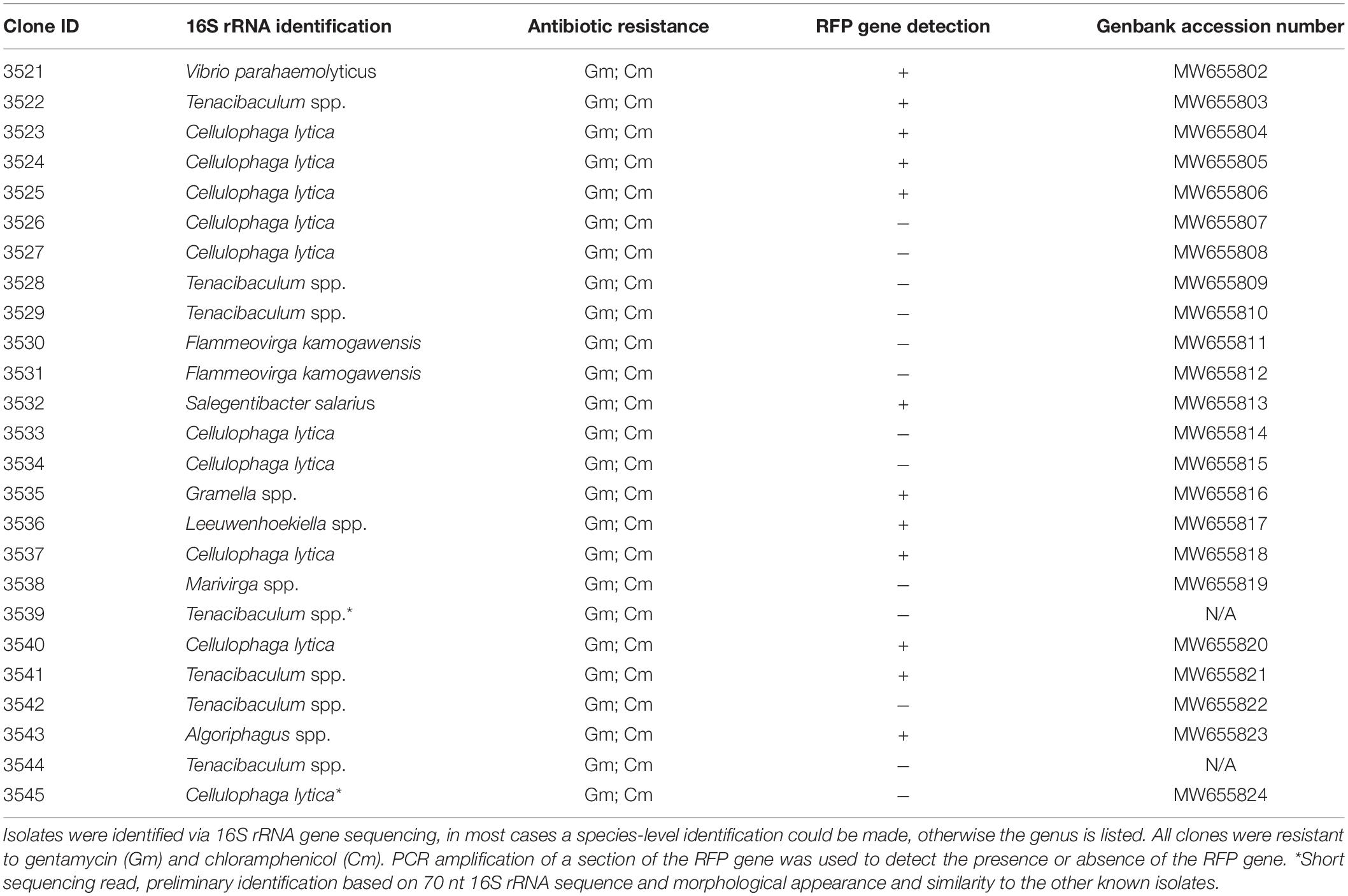
Table 4. Identification and characterization of non-V. vulnificus bacteria from the NaSW incubations with VvR-21.
To see if the non-Vibrio growth phenomenon was repeatable, the niche analysis with VvR-21 was repeated a second time with the same source of water for the NaSW and SSW, which had been stored for five days in the dark at 4°C. At the same time, we repeated the niche analysis with a separate strain of RFP-labeled V. vulnificus, VvR-25. This strain had been constructed in the same manner as VvR-21 and carries the same transposon (Thorstenson and Ullrich, 2020). This second experiment was to confirm that the transfer of the transposon was unique to the VvR-21 strain and not to V. vulnificus as a species. All incubations for the repeat VvR-21 and VvR-25 experiments were done at 20°C. Again, NaSW and SSW inoculated with sterile NSW served as the negative controls. The second VvR-21 niche analysis (Supplementary Figure 2) had comparable results to the original analysis with non-V. vulnificus bacterial growth occurring after 24 h co-incubation in NaSW. In this repeated experiment, V. vulnificus-like colonies were enumerable for both the NaSW and SSW after 72 h, at similar amounts. A lower morphological diversity in bacterial colonies on the selective plates was observed (data not shown), presumably due to the storage and subsequent bottle effect in the water sample.
For the VvR-25 niche analysis, the abundance remained stable in the SSW incubations through 48 h at 2.5 × 105 CFU mL–1 ± 1.0 × 105 (Supplementary Figure 3). At 72 h, the abundance had dropped to 5.2 × 103 CFU mL–1 ± 1.6 × 103. For the NaSW incubations, the abundance decreased to below the detection limit by 72 h. While the repeat “mock-niche” VvR-21 and VvR-25 experiments cannot be directly compared to the original niche experiments, these results suggest that the V. vulnificus strains behaved similarly to the V. parahaemolyticus incubations in SSW.
Discussion
Herein, a small-scale niche analysis was conducted to look at three selected Vibrio species’ competitive fitness when co-incubated with a living and active native microbial community sampled from the North Sea. It has long been proposed that an increase in SST by a few degrees as a result of climate change may lead to a selective advantage for pathogens (Colwell, 1996). The abundance of human pathogenic Vibrios has been increasing in the North Atlantic in correlation to rising SST (Oberbeckmann et al., 2011; Vezzulli et al., 2016). Due to their prevalence and impact on human health (Huehn et al., 2014; Bier et al., 2015), we investigated the link between the behavioral patterns of V. cholerae, V. parahaemolyticus, and V. vulnificus in response to temperature changes in a controlled, in vitro, setting. We selected a small temperature range, 17°C (in situ at the time of sampling), 20°C, and 22°C, to reflect probable increases in summer SST. In contrast to the pathogenic advantage theory, our results suggest that at least in terms of “competitive fitness,” increased temperatures alone do not lead to better survival rates for V. cholerae, V. parahaemolyticus, and V. vulnificus when incubated with a native microbial community. In contrast, the native microbiome of the North Sea sample seems to limit the spread of these Vibrio species irrespective of elevated temperatures.
Vibrio cholerae
Vibrio cholerae upheld the null hypothesis in the niche analysis experiments. While we saw a small increase (approximately 2-fold) in the SSW incubations above 20°C versus the 17°C incubations, temperature did not have an impact on the abundance and survival rate in the NaSW incubations. The abundances of V. cholerae declined at comparable rates when co-incubated with the NaSW community across all three temperatures indicating that VcR-7-2 could not successfully outcompete the natural microflora of the water samples or avoid predation. Previously V. cholerae was grown in 0.22 μm pore-sized filtered and autoclaved freshwater samples taken from a river, lake, and wastewater treatment plant effluent (Vital et al., 2007). The concentration of apparent assimilable organic carbon (AOC) correlated positively with final cell concentration of V. cholerae, suggesting the concentration and type of AOC was important to the growth of V. cholerae. When V. cholerae was grown with native lake water communities, a positive correlation between increased temperature and the specific growth rate achieved by V. cholerae was found, however, this was done with a much larger temperature range of 20–30°C. Ultimately, like in our study, the authors concluded that the ability of V. cholerae to compete with freshwater bacteria for nutrients was not impacted when the temperature was raised (Vital et al., 2007).
Kirschner et al. (2008) found that, apart from chitinase, enzymatic changes measured before and after exponential growth in V. cholerae pure cultures did not correlate with increased temperature. Instead, they observed a positive correlation between chitinase activity and temperature, which suggests an indirect correlation between temperature and V. cholerae abundances via zooplankton abundances. Changes in zooplankton and phytoplankton community composition in response to changing SST have been well documented in the North Sea (Richardson, 2008; Alvarez-Fernandez et al., 2012).
Copepod exoskeletons have been shown to support large populations of Vibrio species, including pathogenic V. cholerae (Huq et al., 1983; Tamplin et al., 1990; Constantin de Magny et al., 2014). Long et al. (2005) found that V. cholerae were less sensitive to antagonistic interactions on agarose particles (meant to simulate zooplankton) at temperatures above 30°C in vitro. In this study, changes in V. cholerae abundances in response to temperature within a random water sample were observed without further investigating potential changes in V. cholerae abundances in the attached and planktonic fractions. In future studies, we believe it is important to investigate possible changes in V. cholerae behavior and abundances in separate size fractions of North Sea water. It is likely that temperature influences V. cholerae abundances and behavior indirectly by influencing the zooplankton and phytoplankton communities.
Vibrio parahaemolyticus
Two separate RFP -tagged strains of V. parahaemolyticus were tested in separate niche experiments using the same water from the North Sea. Both strains exhibited different behaviors from the experiments using V. cholerae and V. vulnificus. Neither strain behaved as expected in reference to our null and experimental hypotheses.
When grown at 17°C, VpR-DSMZ showed a relatively “slow” increase throughout 72 h in the SSW incubations, and a short initial increase in the NaSW incubations, which was quickly followed by a rapid decline to below the detection limit. Indicating that, at least within the first 24 h, this strain could grow (comparatively) uninhibited by the microbial community. What is particularly interesting is the behavior of VpR-DSMZ at the two elevated temperatures (20 and 22°C). For the initial 24 h growth, we again see a small increase in abundance in both the NaSW and SSW, but then in both the SSW and NaSW, the abundance rapidly declines by 72 h, suggesting that at elevated temperatures the initial boom in growth resulted in a rapid consumption of available nutrients. This strain also may have become vulnerable to either a build-up of toxic metabolic by-product or antimicrobial activities in the water. It is important to note that the 20°C and 22°C SSW incubations still had a detectable amount of V. parahaemolyticus after 72 h, whereas in the NaSW incubations V. parahaemolyticus was no longer detectable after 48 h indicating that grazing in the NaSW still contributed notably to the decline of this strain.
Our North Sea isolate, VpR-NSI, behaved similarly across all three temperatures. In contrast to VpR-DSMZ, there was no initial increase in abundance after 24 h, but there was a similar decline in both the SSW and NaSW incubations by 72 h suggesting that this strain, unlike VpR-DSMZ, employs a different strategy than rapid growth and resource consumption, in the same environment. Across the NaSW incubations there was no significant difference in decline rates, again leading us to conclude that temperature alone does not confer better fitness amongst V. parahaemolyticus to avoid grazing. VpR-NSI was no longer detectable by 72 h in the NaSW, but was still detectable in SSW, much like VpR-DSMZ, further indicating that for the VpR strains, predator activity in the NaSW played an important role.
Vibriosis (disease caused by Vibrio species) and cholera outbreaks are strongly correlated with SST greater than 18°C around the world, as reviewed by Baker-Austin et al. (2017). It is possible that temperatures greater than 17°C, allow for a much faster rate of growth in V. parahaemolyticus (particularly the VpR-DSMZ strain). This rapid growth could have led to a faster depletion in nutrients in the culture or a faster accumulation of toxic metabolic by-products, which, in-turn, led to a faster collapse in the 20°C and 22°C incubations. VpR-DSMZ was expected to be less adapted to the North Sea environment. As disease in humans has been attributed to this strain, it may be more naturally suited to “boom-and-then-bust” growth strategies (Lockwood et al., 2014; Strayer et al., 2017) as opposed to the non-pathogenic VpR-NSI strain.
With the exception that we did not observe an initial increase in abundance in either SSW or NaSW incubations after 24 h, the VpR-NSI strain behaved very similarly to VpR-DSMZ in the niche experiments. Both strains grew similarly across all temperatures, with a slower decline in abundances noted in SSW than in the NaSW incubations. The faster extinction observed in the NaSW can be explained by direct interactions with the community, such as predation or the competition for nutrients. Ostensibly, a number of protozoan grazers and other such predators of Vibrio would have been in our NaSW samples, we did not see any direct evidence of these larger grazers in our SSW samples. At this time, we hypothesize the decline in SSW abundances is due to an overconsumption of nutrients early at the beginning of the experiment, or there was a strong antimicrobial inhibitor in the original water source, such as phage activity or the presence of bacteriocins. The decline in the SSW incubations, which was not observed in the V. cholerae incubations, implicates that either something was left in the water after filtration through a 0.22 μM filter or that the SSW itself was not suitable for longer-term maintenance of V. parahaemolyticus. Additionally, temperatures above 17°C could have induced phage-like activity or other anti-bacterial compounds to become more active, leading to the observed decline in V. parahaemolyticus abundances.
Filtration through a 0.22 μM pore-size filter does not lead to perfect sterilization of the water sample (Hahn et al., 2004), and the viral component of the community could easily pass through this filter. While we did investigate via light microscopy for larger microbial contamination (data not shown), we did not perform viral enumeration. To see if we could detect or isolate V. parahaemolyticus phages in the NSW sample after the completion of the niche analyses, we performed a standard DAOA with autoclaved and non-autoclaved SSW and NaSW samples, as well as with autoclaved water from the original sampling, which was stored at 4°C in the interim. We did not observe the formation of any plaques on our VpR-DSMZ lawns after incubation. We acknowledge that this was not an ideal set-up to detect Vibrio phages, and we cannot completely rule out phage activity at this time. Ideally, future experiments should analyze the viral abundance throughout the experimental timeframe via the methods discussed in John et al. (2011) and Cunningham et al. (2015).
Previously, VpR-DSMZ and VpR-NSI were grown at 28°C in a separate sea water source, filtered through a 0.22 μM filter and then autoclaved (NSW) prior to the niche experiments. In those trial tests, we did not see any changes in V. parahaemolyticus abundance (Thorstenson and Ullrich, 2020). After the niche experiments, we tested VpR-DSMZ in the same NSW, with an incubation at 20°C for 72 h. Again, the concentration of V. parahaemolyticus remained stable at the starting concentration at inoculation over 72 h. Based on these experiments in the NSW, it is unlikely the decline in V. parahaemolyticus abundance observed in the SSW incubations is a specific attribute of our genetically modified bacteria, but rather is attributed to the state of the SSW used for the incubation.
Vital et al. (2010) studied the impact of assimilable organic carbon (AOC) concentration on the growth of V. cholerae. They report that AOC concentration alone does not describe V. cholerae growth, but that the composition of the AOC available seems to greatly impact V. cholerae abundance as well. Perhaps these V. parahaemolyticus strains consumed their preferred AOC (or another limited nutrient) quickly in the experiment, which ultimately led to their decline in the SSW.
The original purpose of this experiment was to determine if increasing temperatures influence the competitive fitness of V. parahaemolyticus in its natural habitat. While our SSW results do implicate that the water sample itself likely played a substantial role in the decline of V. parahaemolyticus abundance, the finding that VpR declined in abundance more in the NaSW incubation than in the SSW demonstrated that predation and competition for resources still played an important role. Similar to V. cholerae, there was no significant difference in the NaSW incubations across the three temperatures for both V. parahaemolyticus strains. The interactions of V. parahaemolyticus and V. vulnificus with a natural seawater community has been studied before within the context of bioconcentration by the oyster, Crassostrea virginica (Richards et al., 2012). In NaSW, the oysters bioconcentrated both V. parahaemolyticus and V. vulnificus species for 24 h when incubated at 22°C, and the counts rapidly declined to negligible levels after 72 h resembling the results of our study. Several Vibrio -predating species were identified as the cause of the decline of V. parahaemolyticus and V. vulnificus overtime in the seawater from Richards et al. (2012), namely Bdellovibrio species, Bdellovibrio-like, and Micavibrio aeruginosavorus-like bacteria. Whether any such bacterial organisms were presented in the NaSW sample in the current study remains unknown.
Vibrio vulnificus
When V. vulnificus strain VvR-21 was incubated in NaSW, an entirely unexpected outcome was observed. Non-V. vulnificus bacteria could grow on the same selective medium used to cultivate VcR, VpR, and the negative controls after 24 h co-incubation with VvR-21. At t0, only VvR-21 colonies were observed on the selective plates whereas from 24 to 72 h, non-V. vulnificus colonies occurred and were abundant. This phenomenon was observed in all three temperature incubations. It is important to remember that the negative controls, in which no Vibrio inoculum was added to the applied NaSW and SSW samples, did not result in the growth of any contaminating bacterial colonies throughout the course of the experiment, indicating that an appropriate concentration of Gm was used on the selective plates to prevent growth of the native microbiota.
We believe that the results of the VvR-21 niche experiments provide good preliminary evidence that a genetic element was transferred from the modified V. vulnificus strain to the non-Vibrio bacteria. To recapitulate: The VcR-7-2, VpR-DSMZ, VpR-NSI, VvR-21, and (later on) VvR-25 strains, as well as the NaSW and SSW blanks (negative controls) all used the same source of water sample. The only time non-Vibrio growth occurred on the selective (Gm-containing) medium used for enumeration, was when VvR-21 was inoculated into the sample, providing evidence that the Gm resistance found in the non-Vibrio growth is somehow related to the VvR-21 strain and not due to natural resistance in the water sample. The GmR cassette carried by VvR-21 is flanked by the RFP gene and a CmR cassette (Thorstenson and Ullrich, 2020). Accordingly, non-Vibrio bacterial growth were streaked for isolation on Gm- and Cm-containing media. Twenty-five of these isolates were able to be stably maintained on this media. RFP expression was directly observed via fluorescent microscopy in 10 isolates. A PCR amplification of a section of the RFP gene contained in VvR-21 was able to be amplified in 13 of isolates, providing further evidence that a genetic element encoding the RFP, GmR, and CmR genes was transferred from VvR-21 to these isolates. The mechanism of this transfer is currently unknown. Only ten isolates expressed RFP when visualized via FM microscopy, suggesting either growth conditions were not optimal to induce expression, or possibly that expression of RFP was toxic in these isolates.
The transposon insertion in VvR-21 was initially described as occurring in a so-far uncharacterized region of V. vulnificus chromosome II (Clone no. 150, Thorstenson and Ullrich, 2020). Chromosome II in Vibrio is known to be highly variable even within the same species, houses primarily niche adaptation genes, and strongly contributes to horizontal gene transfer (HGT) within the genus (Okada et al., 2005; Eiler et al., 2006). Multiple studies have found native Vibrio plasmids and “super-plasmids” which were conjugable to other bacteria (Chen et al., 2003; Miller et al., 2007; Valiente et al., 2008; Erauso et al., 2011), we hypothesize that VvR-21 may also host a native conjugable plasmid of a similar nature.
The 25 non-Vibrio isolates from the original VvR-21 incubation in NaSW were identified via 16S rRNA gene sequencing as belonging to nine separate genera. The majority of those isolates were identified as Cellulophaga lytica and Tenacibaculum species. The remaining isolates were identified as V. parahaemolyticus, Flammeovirga kamogawensis, Salegentibacter salarius, Gramella species, Leeuwenhoekiella species, Marivirga species, and Algoriphagus species. Apart from the one V. parahaemolyticus isolate (a gammaproteobacteria), the remaining isolates are all members of the Bacteroidetes phylum. Many of these Bacteroidetes are flavobacteria, a group well characterized and previously isolated from not only the North Sea, but also specifically from our sampling area near Helgoland, Germany (Hahnke and Harder, 2013). Flavobacteria often account for a relatively high percentage of the bacterial populations in the water column (Eilers et al., 2000). These bacteria were likely to be in high abundance in our water sample, increasing their chances for possible HGT with VvR-21 during the incubation.
In terms of testing the impact of increasing temperatures on the competitive fitness of V. vulnificus, the growth of these non-V. vulnificus bacteria obfuscated the CFU enumeration. Attempts were made to count only the Vibrio-like colonies, but since one V. parahaemolyticus isolate was recovered from the plates in addition to the other Bacteroidetes isolates masking the V. vulnificus-like colonies, we are reluctant to draw conclusions from the CFU estimates. Since the VvR-25 experiment was performed after the original set of experiments, it would be misleading to compare the VvR-25 results directly to those of V. cholerae and the V. parahaemolyticus without acknowledging the caveats in this approach. Broadly speaking, VvR-25 was observed on the SSW plates from all three temperatures, while in the NaSW incubations, only those incubations above 20°C had growth after t0 hours. When incubated at 17°C, VvR-25 was only observed on the t0 plates. This provides preliminary evidence that temperatures above 20°C provided some benefit to V. vulnificus. When we look exclusively at the SSW incubations, across all three temperatures we see a similar decrease in V. vulnificus abundance after 48 h, much like in the V. parahaemolyticus incubations.
To summarize, this study did not reveal evidence supporting the hypothesis that increased temperature alone confers increased competitive fitness in the survival of V. cholerae, V. parahaemolyticus, or V. vulnificus. This contrasts with, but is not entirely in contradiction with, the multiple ecological studies wherein increasing Vibrio abundances (and human Vibrio infections) were strongly correlated with rising sea surface temperatures (Vezzulli et al., 2016; Baker-Austin et al., 2017). Colwell proposed that increased temperatures may provide benefits for pathogens capable of inhabiting warm-blooded animals (2004). Perhaps to expand on this idea more broadly, increased temperatures may benefit those bacteria capable of inhabiting a wide range of ecological niches (and not just warm-blooded animals), and the three Vibrio species studied here certainly match that description. From the sea and freshwater environments to the human gut and skin, V. cholerae, V. parahaemolyticus, and V. vulnificus inhabit a wide range of habitats (Vital et al., 2007; Kirschner et al., 2008). In addition, while increasing temperatures have been thought to lead to a selective advantage for pathogens, it also effects the growth of grazers and other members of the microbial consortium. Vibrios have been found attached to marine particles and zooplankton, such as copepods, in addition to living planktonically (Huq et al., 1983; Colwell and Grimes, 1984; Tamplin et al., 1990; Kirschner et al., 2008). In future studies, a focus on how Vibrio interacts with marine particles and zooplankton specifically under increasing temperatures would greatly benefit our understanding of the behavior of these bacteria.
While temperature may not confer a special competitive advantage to these bacteria directly and when free-living, temperature is likely influencing the observed increasing Vibrio abundances in coastal waters via indirect pathways, such as: promoting algal growth, excretion of exploitable metabolites, grazing or viral lysis of heterotrophic predators. V. cholerae’s correlation with temporal and seasonal changes (as reviewed by Constantin de Magny et al., 2014) suggests that a specific microbial consortium allows for the rapid growth of Vibrio. A better understanding of how changing SST affects the entire planktonic community will aid our understanding of survival and proliferation of pathogenic Vibrios in the ocean.
For future studies, we suggest an emphasis on the joint need for understanding zooplankton community composition shifts under changing temperatures in vivo, and controlled in vitro experimentation using fractionated water samples and labeled Vibrio bacteria. Through this combined approach, we could better elucidate how the behavior of Vibrio changes within separate micro-niches undergoing environmental changes. It would be interesting to see if, for example, V. cholerae is more likely to attach in algal groups or assume a free-living lifestyle in response to changing temperatures due to increasing or decreasing assimilable nutrient concentrations. Such an approach could also better elucidate the role of temperature on the protozoan grazing of bacteria.
To our knowledge, this study presents the first evidence that an RFP-tagged organism was used to, albeit inadvertently, label multiple other bacterial species when inoculated into a natural water sample. This outcome has the potential for a wide range of applications and promotes future study of the mechanism of transfer from the labeled VvR-21 clone to these multiple other species.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
CT: conceptualization, methodology, validation, formal analysis, and writing – original draft preparation. MU: supervision. Both authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Deutsche Forschungsgemeinschaft (DFG), grant number UL169/7-1.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Gunnar Gerdts for the consultation on the design of the niche analyses and the Biological Institute Helgoland for the water sample. In addition, a special thank you to Nina Bartlau for the delivery of the water sample and to Tara Helmi Turkki for the assistance in the preparation of the niche experiments.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.623988/full#supplementary-material
References
Alam, M., Sultana, M., Nair, G. B., Siddique, A. K., Hasan, N. A., Sack, R. B., et al. (2007). Viable but nonculturable Vibrio cholerae O1 in biofilms in the aquatic environment and their role in cholera transmission. Proc. Natl. Acad. Sci. U.S.A. 104, 17801–17806. doi: 10.1073/pnas.0705599104
Alvarez-Fernandez, S., Lindeboom, H., and Meesters, E. (2012). Temporal changes in plankton of the North Sea: community shifts and environmental drivers. Mar. Ecol. Prog. Ser. 462, 21–38. doi: 10.3354/meps09817
Baker-Austin, C., Trinanes, J., Gonzalez-Escalona, N., and Martinez-Urtaza, J. (2017). Non-Cholera Vibrios: the microbial barometer of climate change. Trends Microbiol. 25, 76–84. doi: 10.1016/j.tim.2016.09.008
Bier, N., Diescher, S., and Strauch, E. (2015). Multiplex PCR for detection of virulence markers of Vibrio vulnificus. Lett. Appl. Microbiol. 60, 414–420. doi: 10.1111/lam.12394
Chen, C.-Y., Wu, K.-M., Chang, Y.-C., Chang, C.-H., Tsai, H.-C., Liao, T.-L., et al. (2003). Comparative genome analysis of Vibrio vulnificus, a marine pathogen. Genome Res. 13, 2577–2587. doi: 10.1101/gr.1295503
Colwell, R. R. (1996). Global climate and infectious disease: the cholera paradigm. Science 274, 2025–2031. doi: 10.1126/science.274.5295.2025
Colwell, R. R. (2004). Infectious disease and environment: cholera as a paradigm for waterborne disease. Int. Microbiol. 7, 285–289.
Colwell, R. R., and Grimes, D. J. (1984). Vibrio diseases of marine fish populations. Helgoländer Meeresunt. 37, 265–287. doi: 10.1007/BF01989311
Constantin de Magny, G., Hasan, N. A., and Roche, B. (2014). How community ecology can improve our understanding of cholera dynamics. Front. Microbiol. 5:137. doi: 10.3389/fmicb.2014.00137
Cunningham, B. R., Brum, J. R., Schwenck, S. M., Sullivan, M. B., and John, S. G. (2015). An inexpensive, accurate, and precise wet-mount method for enumerating aquatic viruses. Appl. Environ. Microbiol. 81, 2995–3000.
Eiler, A., Johansson, M., and Bertilsson, S. (2006). Environmental influences on Vibrio populations in Northern temperate and boreal coastal waters (Baltic and Skagerrak Seas). Appl. Environ. Microbiol. 72, 6004–6011. doi: 10.1128/AEM.00917-06
Eilers, H., Pernthaler, J., Glöckner, F. O., and Amann, R. (2000). Culturability and in situ abundance of pelagic bacteria from the North Sea. Appl. Environ. Microbiol. 66, 3044–3051. doi: 10.1128/AEM.66.7.3044-3051.2000
Erauso, G., Lakhal, F., Bidault-Toffin, A., Le Chevalier, P., Bouloc, P., Paillard, C., et al. (2011). Evidence for the role of horizontal transfer in generating pVT1, a large mosaic conjugative plasmid from the clam pathogen, Vibrio tapetis. PLoS One 6:e016759. doi: 10.1371/journal.pone.0016759
Farmer, J. J., and Janda, J. M. (2015). “Vibrionaceae,” in Bergey’s Manual of Systematics of Archaea and Bacteria, eds M. E. Trujillo, S. Dedysh, P. DeVos, B. Hedlund, P. Kämpfer, F. A. Rainey, and W. B. Whitman (Atlanta, GE: American Cancer Society), 1–7. doi: 10.1002/9781118960608.fbm00212
Faruque, S. M., Albert, M. J., and Mekalanos, J. J. (1998). Epidemiology, genetics, and ecology of toxigenic Vibrio cholerae. Microbiol. Mol. Biol. Rev. 62, 1301–1314. doi: 10.1128/MMBR.62.4.1301-1314.1998
Fujino, T., Okuno, Y., Nakada, D., Aoyama, A., Fukai, K., Mukai, T., et al. (1953). On the bacteriological examination of shirasu-food poisoning. Med. J. Osaka Univ. 4, 299–304.
Hahn, M. W., Stadler, P., Wu, Q. L., and Pöckl, M. (2004). The filtration-acclimatization method for isolation of an important fraction of the not readily cultivable bacteria. J. Microbiol. Methods 57, 379–390. doi: 10.1016/j.mimet.2004.02.004
Hahnke, R. L., and Harder, J. (2013). Phylogenetic diversity of Flavobacteria isolated from the North Sea on solid media. Syst. Appl. Microbiol. 36, 497–504. doi: 10.1016/j.syapm.2013.06.006
Harvell, C. D., Mitchell, C. E., Ward, J. R., Altizer, S., Dobson, A. P., Ostfeld, R. S., et al. (2002). Climate warming and disease risks for terrestrial and marine biota. Science 296, 2158–2162. doi: 10.1126/science.1063699
Huehn, S., Eichhorn, C., Urmersbach, S., Breidenbach, J., Bechlars, S., Bier, N., et al. (2014). Pathogenic vibrios in environmental, seafood and clinical sources in Germany. Int. J. Med. Microbiol. 304, 843–850. doi: 10.1016/j.ijmm.2014.07.010
Huq, A., Small, E. B., West, P. A., Huq, M. I., Rahman, R., and Colwell, R. R. (1983). Ecological relationships between Vibrio cholerae and planktonic crustacean copepods. Appl. Environ. Microbiol. 45, 275–283. doi: 10.1128/AEM.45.1.275-283.1983
Islam, A., Labbate, M., Djordjevic, S. P., Alam, M., Darling, A., Melvold, J., et al. (2013). Indigenous Vibrio cholerae strains from a non-endemic region are pathogenic. Open Biol. 3, 120181. doi: 10.1098/rsob.120181
John, S. G., Mendez, C. B., Deng, L., Poulos, B., Kauffman, A. K. M., Kern, S., et al. (2011). A simple and efficient method for concentration of ocean viruses by chemical flocculation. Environ. Microbiol. Rep. 3, 195–202. doi: 10.1111/j.1758-2229.2010.00208.x
Kirschner, A. K. T., Schlesinger, J., Farnleitner, A. H., Hornek, R., Süß, B., Golda, B., et al. (2008). Rapid growth of Planktonic Vibrio cholerae Non-O1/Non-O139 Strains IN a large Alkaline Lake in Austria: dependence on temperature and dissolved organic carbon quality. Appl. Environ. Microbiol. 74, 2004–2015. doi: 10.1128/AEM.01739-07
Logue, C. M., and Nde, C. W. (2017). “Rapid microbiological methods in food diagnostics,” in Advances in Food Diagnostics, eds F. Toldrá and L. M. L. Nollet (Hoboken, NJ: Wiley), 153–185.
Long, R. A., Rowley, D. C., Zamora, E., Liu, J., Bartlett, D. H., and Azam, F. (2005). Antagonistic interactions among marine bacteria impede the proliferation of Vibrio cholerae. Appl. Environ. Microbiol. 71, 8531–8536. doi: 10.1128/AEM.71.12.8531-8536.2005
Miller, M. C., Keymer, D. P., Avelar, A., Boehm, A. B., and Schoolnik, G. K. (2007). Detection and transformation of genome segments that differ within a coastal population of Vibrio cholerae strains. Appl. Environ. Microbiol. 73, 3695–3704. doi: 10.1128/AEM.02735-06
Neogi, S. B., Chowdhury, N., Asakura, M., Hinenoya, A., Haldar, S., Saidi, S. M., et al. (2010). A highly sensitive and specific multiplex PCR assay for simultaneous detection of Vibrio cholerae, Vibrio parahaemolyticus and Vibrio vulnificus. Lett. Appl. Microbiol. 51, 293–300. doi: 10.1111/j.1472-765X.2010.02895.x
Nishibuchi, M., and Kaper, J. B. (1995). Thermostable direct hemolysin gene of Vibrio parahaemolyticus: a virulence gene acquired by a marine bacterium. Infect. Immun. 63, 2093–2099.
Oberbeckmann, S., Wichels, A., Wiltshire, K. H., and Gerdts, G. (2011). Occurrence of Vibrio parahaemolyticus and Vibrio alginolyticus in the German Bight over a seasonal cycle. Anton. Van Leeuwenhoek 100, 291–307. doi: 10.1007/s10482-011-9586-x
Okada, K., Iida, T., Kita-Tsukamoto, K., and Honda, T. (2005). Vibrios commonly possess two chromosomes. J. Bacteriol. 187, 752–757. doi: 10.1128/JB.187.2.752-757.2005
Poloczanska, E. S., Brown, C. J., Sydeman, W. J., Kiessling, W., Schoeman, D. S., Moore, P. J., et al. (2013). Global imprint of climate change on marine life. Nat. Clim. Chang. 3, 919–925. doi: 10.1038/nclimate1958
Richards, G. P., Fay, J. P., Dickens, K. A., Parent, M. A., Soroka, D. S., and Boyd, E. F. (2012). Predatory bacteria as natural modulators of Vibrio parahaemolyticus and Vibrio vulnificus in seawater and oysters. Appl. Environ. Microbiol. 78, 7455–7466. doi: 10.1128/AEM.01594-12
Richardson, A. J. (2008). In hot water: zooplankton and climate change. ICES J. Mar. Sci. 65, 279–295. doi: 10.1093/icesjms/fsn028
Sonnenschein, E. C., Gärdes, A., Seebah, S., Torres-Monroy, I., Grossart, H.-P., and Ullrich, M. S. (2011). Development of a genetic system for Marinobacter adhaerens HP15 involved in marine aggregate formation by interacting with diatom cells. J. Microbiol. Methods 87, 176–183. doi: 10.1016/j.mimet.2011.08.008
Strayer, D. L., D’Antonio, C. M., Essl, F., Fowler, M. S., Geist, J., Hilt, S., et al. (2017). Boom-bust dynamics in biological invasions: towards an improved application of the concept. Ecol. Lett. 20, 1337–1350. doi: 10.1111/ele.12822
Strom, M. S., and Paranjpye, R. N. (2000). Epidemiology and pathogenesis of Vibrio vulnificus. Microb. Infect. 2, 177–188. doi: 10.1016/S1286-4579(00)00270-7
Tamplin, M. L., Gauzens, A. L., Huq, A., Sack, D. A., and Colwell, R. R. (1990). Attachment of Vibrio cholerae serogroup O1 to zooplankton and phytoplankton of Bangladesh waters. Appl. Environ. Microbiol. 56, 1977–1980.
Thorstenson, C. A., and Ullrich, M. S. (2020). Developing a universal and efficient method for the rapid selection of stable fluorescent protein-tagged pathogenic Vibrio species. J. Mar. Sci. Eng. 8:804. doi: 10.3390/jmse8100804
Valiente, E., Lee, C. T., Lamas, J., Hor, L., and Amaro, C. (2008). Role of the virulence plasmid pR99 and the metalloprotease Vvp in resistance of Vibrio vulnificus serovar E to eel innate immunity. Fish Shellf. Immunol. 24, 134–141. doi: 10.1016/j.fsi.2007.10.007
Vezzulli, L., Grande, C., Reid, P. C., Hélaouët, P., Edwards, M., Höfle, M. G., et al. (2016). Climate influence on Vibrio and associated human diseases during the past half-century in the coastal North Atlantic. Proc. Natl. Acad. Sci. U.S.A. 113, E5062–E5071. doi: 10.1073/pnas.1609157113
Vieira, R. H. S. F., Costa, R. A., Menezes, F. G. R., Silva, G. C., Theophilo, G. N. D., Rodrigues, D. P., et al. (2011). Kanagawa-Negative, tdh- and trh-Positive Vibrio parahaemolyticus isolated from fresh oysters marketed in Fortaleza, Brazil. Curr. Microbiol. 63, 126–130. doi: 10.1007/s00284-011-9945-x
Vital, M., Füchslin, H. P., Hammes, F., and Egli, T. (2007). Growth of Vibrio cholerae O1 Ogawa Eltor in freshwater. Microbiology 153, 1993–2001. doi: 10.1099/mic.0.2006/005173-0
Keywords: Vibrio, ecological niche, competitive fitness, microbiome interactions, pathogen
Citation: Thorstenson CA and Ullrich MS (2021) Ecological Fitness of Vibrio cholerae, Vibrio parahaemolyticus, and Vibrio vulnificus in a Small-Scale Population Dynamics Study. Front. Mar. Sci. 8:623988. doi: 10.3389/fmars.2021.623988
Received: 30 October 2020; Accepted: 05 March 2021;
Published: 06 April 2021.
Edited by:
Marcelino T. Suzuki, Sorbonne Université, FranceReviewed by:
Michele Kiyoko Nishiguchi, University of California, Merced, United StatesMarie-Agnès Travers, Institut Français de Recherche pour l’Exploitation de la Mer (IFREMER), France
Copyright © 2021 Thorstenson and Ullrich. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Candice A. Thorstenson, c.thorstenson@jacobs-university.de
 Candice A. Thorstenson
Candice A. Thorstenson Matthias S. Ullrich
Matthias S. Ullrich