Phylogeny and Pathology of Anisakids Parasitizing Stranded California Sea Lions (Zalophus californianus) in Southern California
- 1Laboratory of Aquaculture, Institute of Oceanography and Fisheries, Split, Croatia
- 2College of Veterinary Medicine, Western University of Health Sciences, Pomona, CA, United States
- 3Department of Botany, Faculty of Science, University of South Bohemia, České Budějovice, Czechia
- 4University Department of Marine Studies, University of Split, Split, Croatia
- 5Marine Mammal Care Center Los Angeles, San Pedro, CA, United States
- 6Institute of Parasitology, Biological Centre of Czech Academy of Sciences, České Budějovice, Czechia
In marine mammals, nematode-inflicted pathological lesions combined with other pathogens and factors (i.e., pollution, climate change, domoic acid poisoning events, and seasonal El Nino starvation events) negatively impact pinnipeds’ health and may cause mortality. Five California sea lions (Zalophus californianus)—a female pup, three male yearlings, and an adult female—suffered mortalities during rehabilitation at the Marine Mammal Care Center Los Angeles (San Pedro, CA). According to the necropsy reports, animals developed multisystemic parasitism as a leading cause of death, combined with malnutrition and hypoglycemia. In order to reveal host–parasite dynamics that may play a role in pinniped health and recovery, we examined the type and level of histopathological stomach lesions in California sea lions caused by anisakid nematodes. All isolated anisakids were morphologically and molecularly identified, and their phylogenetic relationships were reconstructed using the sequence of the mitochondrial COII gene. Co-parasitation of different Anisakidae spp. within the same host or lesions presented the opportunity to evaluate the existence of recombinant haplotypes and their eventual pathological pressure exerted onto host. The lesions were presented as chronic granulomatous gastritis, with moderate edema and hyperemia of the tunica submucosa and lamina propria, followed by mild, focal fibrosis of the gastric wall. Ulcerative changes with mixed leukocytic infiltrate showed to be localized, shallow, and non-perforative and with no apparent bacterial coinfection, mostly accompanied by healing granulation tissue. Isolated anisakids are grouped into three distinctively separated monophyletic clades corresponding to genera Anisakis, Contracaecum, and Pseudoterranova. Most abundant were representatives of Contracaecum ogmorhini sensu lato (55.36%), followed by Anisakis pegreffii (23.21%), Pseudoterranova azarasi (17.86%), Pseudoterranova decipiens sensu lato (1.79%), and Anisakis simplex (1.79%). Phylogenetic trees revealed no differentiation at intra-species level. Our analysis of divergence revealed Contracaecum separated from other lineages in the Jurassic period at the 176.2 Mya and Anisakis diverging from Pseudoterranova in Cenozoic period at 85.9 Mya.
Introduction
Existing data on marine mammal diversity, spatial and temporal distribution, abundance, and population structure within the marine ecosystem worldwide indicate mammals’ extreme vulnerability to exposure to significant anthropogenic impacts, such as disturbance, bycatch, and hunting (van der Elst and Everett, 2015). In addition, these animals are also gravely affected by various infectious diseases (Aznar et al., 2001), among which parasitic diseases represent a tipping over factor responsible for the regulation of marine mammal populations, on both the ecological and evolutionary scales (Raga et al., 1998).
A pinniped, California sea lion (Zalophus californianus), has occupied the Gulf of California since 3 million years ago (Late Pleistocene or earlier), being attracted to this geographic area by high primary production and large abundance of its typical prey, Monterrey sardine (Sardinops sagax). Today, three genetic groups divided into 13 breeding colonies that inhabit the North, Central, and South Gulf of California, Mexico, exhibit temporal fluctuations in population mainly influenced by Gulf’s physical environmental conditions (Masper et al., 2019). Two other populations, the US population (US or Pacific Temperate) and the western Baja California population, show population sizes above maximum net productivity level (MNPL) and within its optimum sustainable population (OSP) range but are still vulnerable to increasing sea surface temperature associated with El Nino events or similar regional ocean temperature anomalies (Laake et al., 2018).
For effective conservation of the California sea lion, it is crucial to understand the health, immune, and infectious diseases status of the population to predict and mitigate threats that could result in population bottlenecks (Mancia et al., 2012).
In respect to parasites, 24 helminth species, of which six are nematodes, have been reported to cause detrimental infections in the Z. californianus (Kuzmina et al., 2018). The lesions caused by gastrointestinal nematodes are usually characterized as mild. However, severe cases are characterized by complete perforations of the gastric wall (Sweeney and Gilmartin, 1974), which can lead to peritonitis, anorexia, weight loss, and mortality.
The most frequently described nematode family in marine mammals is Anisakidae, encompassing three predominant genera Anisakis, Contracaecum, and Pseudoterranova (Dailey, 2005). Anisakidae family is traditionally separated into two subfamilies, i.e., Anisakinae (including Anisakis and Pseudoterranova) and Contracaecinae (including Contracaecum). Nevertheless, phylogenetic relationships among Anisakidae, as well as the whole Ascaridoidea superfamily are still not fully resolved. Molecular approaches based primarily on rDNA sequence analyses (Nadler and Hudspeth, 1998, 2000; Nadler et al., 2005) have in part confirmed the traditional systematics of Ascaridoidea sensu Fagerholm (1991). In their pivotal work, Nadler and Hudspeth (1998) recovered Anisakidae as traditionally constituted using maximum likelihood (ML). However, in the maximum parsimony (MP) tree, Contracaecum was recovered as a sister taxon of Toxocara, although this relationship was not observed in the bootstrap MP consensus tree. Further advancement was made also by Nadler and Hudspeth (2000) by combining rDNA and cox2 sequence data with morphological characteristics. In the combined evidence tree, traditional Anisakidae clade was again recovered by ML, but not in bootstrap MP analysis. Interestingly, clade encompassing Anisakis and Pseudoterranova was only moderately supported in combined evidence tree. The most comprehensive molecular phylogeny analysis was recently performed by Li et al. (2018). Using sequence data from five nuclear and three mitochondrial genes, the authors recovered the traditionally constituted Anisakidae as monophyletic with strong support from both ML and Bayesian inference (BI). Moreover, the two subfamilies within the Anisakidae sensu Fagerholm (1991) received strong support from ML and BI.
Contracaecum spp. have been recorded to cause ulcerative changes in the stomach, described as lesions with raised edges, eosinophilic cellular infiltration that extends beyond the submucosa into the tunica muscularis, accompanied by hemorrhage and evident healing (Liu and Edward, 1971; Migaki et al., 1971). Parasites are usually observed entangled within granulation tissues that show a plethora of dynamic destruction and healing processes concurrently. Spraker et al. (2003) and Kuzmina et al. (2014) observed similar dynamic changes in Northern fur seal (Callorhinus ursinus) stomach ulcers affected by Anisakis, Contracaecum, and Pseudoterranova spp. In addition to the appearance of the giant multinucleated cells in the lesions, Kuzmina et al. (2014) frequently observed fibroplasia (fibrosis), although the authors could not attribute the etiology of lesions solely to anisakids, since animals harbored very diverse parasite communities (Kuzmina et al., 2018). Among these, several taxa, most notably hookworms and lungworms, can inflict more detrimental effects to their hosts, causing high morbidity or mortality rates. Hookworms of the genus Uncinaria are especially pathogenic for pups of several pinniped species, causing severe enteritis with hemorrhage and multiple systemic organ infections following penetration of the intestinal wall, a condition termed hookworm enteritis with bacteremia (HEB) (Spraker et al., 2004, 2007; Seguel et al., 2017). Lungworms, primarily belonging to the genus Parafilaroides, cause verminous pneumonia and bronchitis in several pinniped species, including California sea lion (Fleischman and Squire, 1970; Reisfeld et al., 2019). The condition is characterized by lung edema, alveolar capillary congestion, atelectasis, focal alveolar and interlobular hemorrhages, and mixed inflammatory infiltrate in the tracheal and bronchial mucosa and submucosa (Fleischman and Squire, 1970; Reisfeld et al., 2019). Moreover, Parafilaroides spp. have been reported to harbor Brucella spp. in various nematode organs and larvae, suggesting the role of these parasites in transmission of Brucella spp. among pinnipeds (Garner et al., 1997; Rhyan et al., 2018). Occasionally, besides well-documented host–parasite associations, unusual associations or aberrant migration within the host can occur, as is the case with another metastrongyloid lungworm Otostrongylus circumlitus and liver fluke Zalophotrema hepaticum. O. circumlitus normally infects seals but has been reported to cause pneumonia and severe suppurative and necrotizing arteritis in California sea lions (Kelly et al., 2005). Z. hepaticum is a common parasite of California sea lions, which resides in the liver, gallbladder, and pancreas, causing portal and biliary fibrosis and bile duct hyperplasia; but aberrant migration leading to meningoencephalitis with parenchymal necrosis, hemorrhage, and fibrosis has been documented (Stroud and Dailey, 1978; Fauquier et al., 2004).
Anisakidae nematodes have also been documented to cause stomach ulcers in stranded cetaceans (Abollo et al., 1998; Blažeković et al., 2015; Hrabar et al., 2017), although these ulcers were deemed non-fatal lesions. In general, most of the references describing nematode stomach lesions were obsolete and lacked a comprehensive, multidisciplinary approach that would allow accurate etiology and description of the lesions. Therefore, the aim of this study was to determine (i) the type and level of histopathological stomach lesions caused by anisakids in stranded California sea lions suffering mortalities during rehabilitation at the Marine Mammal Care Center Los Angeles, San Pedro, CA, United States; (ii) the species of anisakid nematodes involved in observed lesions; and (iii) their phylogenic relationships. Having a rare opportunity to observe anisakids contemporary and chronic infection within the same animal, we employed genetic population tools to evaluate potential occurrence of recombinant haplotypes that are known in anisakids (Cavallero et al., 2014; Mladineo et al., 2017a, b) and to test their potential pathological pressure exerted onto the host. In coupling histopathological observations with the phylogenetic structure of nematode communities, we aim to draw conclusions on host–parasite dynamics that may play a role in pinniped health and in the recovery of rescued California sea lions.
Materials and Methods
Collection of Samples for Histopathology
Five stranded or injured California sea lions (Zalophus californianus) were rescued from beaches in Los Angeles County (Figure 1) and admitted for rehabilitation at Marine Mammal Care Center Los Angeles, San Pedro CA. These included one female pup (ZC18-6), estimated to be 7 months of age, and three male yearlings, one estimated to be 13 months of age (ZC17-335) and the other two estimated to be 17 months of age (ZC17-359 and ZC17-361). The adult female (ZC17-340) was estimated to be more than 5 years of age. Age is based on the time of year, length, and weight of the animal and dental development and is a subjective assessment. The pup was 1–12 months of age, and the yearlings were 13–24 months of age. The pup and yearlings died during rehabilitation, and the adult female was humanely euthanized due to traumatic limb amputation associated with a shark bite. No animals were euthanized for the purpose of this study.
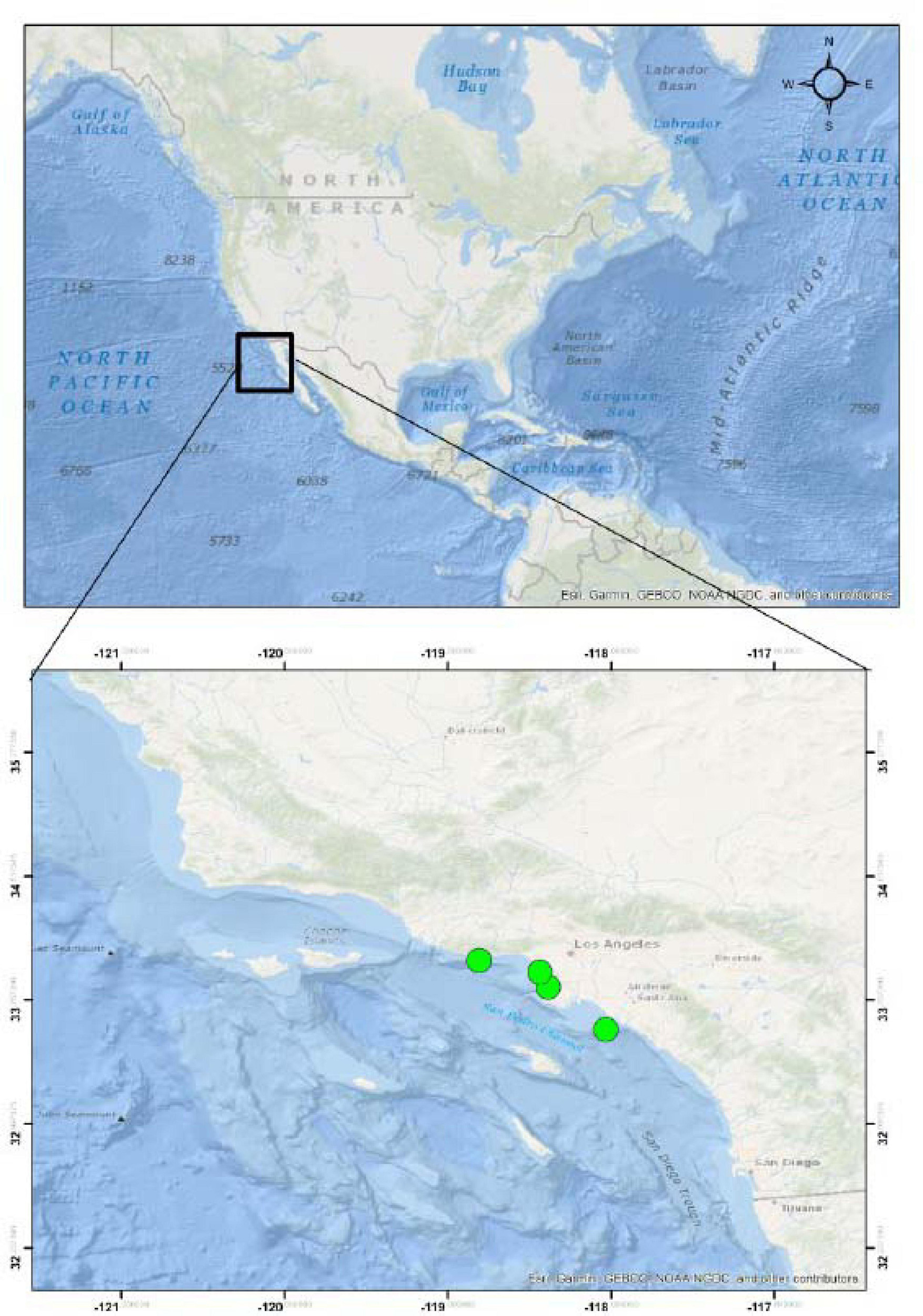
Figure 1. Stranding localities of the California sea lions (Zalophus californianus) acquired for this study.
The animals were necropsied at the Marine Mammal Care Center during spring and summer of 2017 and 2018. All research activities were approved by the IACUC of the Western University of Health Sciences (R16IACUC025), and the National Oceanic and Atmospheric Administration (NOAA) approved the acquisition of the specimens (permit 151401WCR2015PR00034). Export of these specimens to the European Union, Croatia, was also authorized by the NOAA, which provided a special letter of authorization.
Upon routine necropsies, gastric walls with stomach ulcers were excised (in total n = 10; 1–3 lesions per animal, each lesion sampled in triplicate), trimmed to the appropriate size and shape, and fixed in 10% neutral buffered formalin (NBF) solution. The specimens were paraffin-embedded using standardized histological procedures, sectioned at 5 μm thickness, and floated on gelatinized slides. The tissue sections were stained by hematoxylin and eosin stain (H&E) and Mallory’s trichrome stain (MTS), the latter using both aniline blue and light green as a collagen stain. Specimen identifiers were designated as follows: host name (i.e., ZC for Z. californianus), year of collection (i.e., 17 or 18), field number (i.e., 335), and alphabet letter (i.e., A, B, or C) that differentiates separate lesions from the same individual animal.
Scoring System for Characterization of Lesion Severity
The severity of lesions has been evaluated by the scoring system following these criteria: number of anisakid species, number of isolated nematodes, lesion depth, type of process, duration, and the number of parasitic individuals and species within the lesion. The number of anisakid species present in the lesion was scored as 1 (single sp.), 2 (two spp.), and 3 (>2 spp.). The number of individual nematodes was scored as 1 (1–5 nematodes), 2 (6–15), and 3 (>16). Lesion depth, characterized by the depth of nematode penetration into the stomach wall, was designated as shallow (in lamina epithelialis and/or l. propria), medium (tunica submucosa) and deep (t. muscularis), and scored 1, 2, and 3, respectively. Type of process, characterized by the observed pathological process, was designated as inflammatory, inflammatory and granulomatous, necrotic, or mixed process (combination of all three types of pathology) and scored 1, 2, 3, or 4, respectively. Duration of process was evaluated based on the type of inflammatory cellular infiltrate, and scored as 1 (acute), 2 (acute to chronic), and 3 (chronic) following Hrabar et al. (2017). The authors observed neutrophilic granulocytes (hereafter neutrophils) and macrophages as the predominant cell types in acute infections, and eosinophilic granulocytes (hereafter eosinophils), lymphocytes, and plasma cells in chronic infections. Finally, the severity of lesions was evaluated as the average of all individual scores of five identified criteria per lesion and animal, with the theoretical maximum value reaching 3.2. These data were presented graphically via a line and point plot using ggplot2 (Wickham, 2016) and RColorBrewer (Neuwirth, 2014) packages for R.
Nematode Identification
Nematode specimens were isolated from gastric ulcers and fixed in absolute ethanol, labeled according to the California sea lion specimen identification number, and shipped to the Institute of Oceanography and Fisheries, Split, Croatia, for morphological and molecular identification assessments. Morphological identification was performed following Gibbons (2010).
Nematode genomic DNA was extracted using DNeasy Blood and Tissue Kit (Qiagen) according to manufacturer’s instructions. PCR amplification of the mitochondrial cytochrome oxidase 2 (cox2) locus (∼600 bp) and sequencing were performed as previously described in detail (Blažeković et al., 2015). All nematodes isolated from each sea lion were molecularly identified and assigned to a specific lesion in the stomach.
Sequence Analyses
In total, 111 cox2 sequences obtained from sequencing service (Macrogene, The Netherlands) were edited by Seqscape version 2.6 (Applied Biosystems). These new sequences were added to an alignment that included 24 anisakid GenBank referent sequences, chosen based on the blast homology, geographic site of isolation, and use in authors’ previous studies. The latter included Pseudoterranova azarasi (KM853036), Pseudoterranova decipiens (AF179920 and KU558723), Pseudoterranova bulbosa (KU558720), Anisakis brevispiculata (DQ116433), Anisakis physeteris (DQ116432), Anisakis paggiae (DQ116434), Anisakis ziphidarum (DQ116430), Anisakis nascettii (DQ116431), Anisakis typica (DQ116427), Anisakis pegreffii (MF399132, JQ900762, KC809997, DQ116428, KY418057, KY418062, KY909256, and LC222461), Anisakis simplex (DQ116426 and JN786326), Anisakis berlandi (DQ116429), and Contracaecum ogmorhini (NC031649, KU558727, and KU558726) (Valentini et al., 2006; Mattiucci et al., 2013; Marigo et al., 2015; Liu et al., 2016; Bernardi et al., 2017). Two additional sequences, Parascaris equorum (AF179918) and Ascaris lumbricoides (AF179907) were used as outgroups. All new sequences were added to GenBank with the following accession numbers MN624172–MN624201 and MT912390–MT912471.
Genetic diversity (nucleotide and haplotype diversities), polymorphic sites, and haplotype numbers were measured using DnaSP version 6.0 (Librado and Rozas, 2009). Genetic distances and composition of nucleotides were examined based on the uncorrected pairwise genetic distances with 1,000 bootstraps using MEGA version 6 (Tamura et al., 2013). The substitution saturation test was conducted using DAMBE version 6.0.4 (Xia, 2013). Pairwise F-statistics (FST) was executed among lineages through Arlequin version 3.5 (Excoffier and Lischer, 2010) with populations grouped into the clades identified in the phylogenetic analysis.
Phylogeny Analysis and Molecular Dating
Phylogenetic tree reconstructions were done based on the BI and ML approaches. The appropriate model (GTR+I+G) of sequence evolution was identified using the Bayesian information criterion (BIC) for BI and the Akaike information criterion (AIC) for ML tests in JModelTest 2.1.10 (Darriba et al., 2012). A Bayesian phylogenetic analysis was carried out using the selected model in Mr.Bayes 3.2.7a (Ronquist et al., 2012). Four Monte Carlo Markov chain (MCMC) methods were simultaneously run for 40 million generations. As trees were sampled every 1,000th generation, the first 25% of the steps were discarded as burn-in. The remaining trees were combined in a 50% majority rule consensus tree. Bayesian posterior probabilities (PPs) were used to assess branch support of the BI tree. IQ-Tree web server1 with 1,000 ultrafast bootstrap pseudo-replications was applied to construct an ML tree. The topology results from ML and BI methods applied in PAUP 4.0a147 (Swofford, 2002) were compared through using the Shimodaira–Hasegawa test with 1,000 bootstrap replicates.
Additionally, to scrutinize the potential existence of recombinant haplotypes as a result of mitochondrial introgression occurring during mixed infections, we employed haplotype networks. Two haplotype networks were drawn to reveal haplotype relationships among the Anisakidae taxa. To reconstruct the phylogenetic relationships among the Anisakidae taxa, SplitsTree version 4.6 (Huson and Bryant, 2006) was used, and a neighbor-net network was generated using the uncorrected patristic distances with 1,000 bootstrap replications. To plot haplotype relationships within Anisakidae populations (populations with sample size >3), TCS algorithm was used in PopArt (Leigh and Bryant, 2015).
To evaluate whether particular lineages that emerged earlier or later in past exerted any difference in pathological pressure on the host (i.e., lesion characteristics), we estimated divergence dates using BEAST 1.8.2 (Drummond et al., 2012), which was performed based on one calibration point, i.e., the initial divergence of Anisakidae and Ascaridae, dated at 198 million years (Myr), as suggested by Vanfleteren et al. (1994) and Blaxter (2009), modeled with a normal prior with a standard deviation of 42 Myr. The best data partition and evolutionary models were selected for the timing dataset using PartitionFinder 1.1.1 (Lanfear et al., 2012). We used a Birth-Death Process, which is more appropriate when considering sequences from different species (Rambaut and Drummond, 2007). Fitness of the molecular clocks (strict, exponential relaxed, and lognormal relaxed) was assessed based on the Bayes factor support value (2LnBF; see Brandley et al., 2005), estimated by Tracer version 1.5 (Rambaut and Drummond, 2007). For the dating analyses, two independent Markov chains for 400 million generations were run, sampling every 1,000 generations and removing the first 25% burn-in trees. Tracer version 1.5 was used to check for convergence of MCMC runs, stationary distribution of the chains, adequate chain mixing, and effective sample sizes of parameters.
Demographic History of Anisakidae
To evaluate whether demographic history of the particular lineage exerted any difference in pathological pressure on the host (i.e., lesion characteristics), we used the Extended Bayesian Skyline Plot (EBSP) created in Beast version 2.4.7 (Bouckaert et al., 2014) to estimate the demographic history of Anisakidae using the cox2 dataset. Strict molecular clocks were used with a mutation rate of 1.6 × 10–7 per site per generation (Denver et al., 2000; Søe et al., 2016; Nejsum et al., 2017). The MCMC was set to a total of 400 million steps, and the Markov chain was sampled every 10,000 steps. Tracer version 1.5 was utilized to check for convergence of MCMC runs and examine effective sample sizes (>200). Bayesian Skyline Plots were created using the EBSP R function (Heled and Drummond, 2008).
Results
Histopathological Findings
One pup, three yearlings, and one adult California sea lions (Zalophus californianus) with gastric ulcerative lesions were necropsied (Supplementary Table 1). Three of five specimens had multiple ulcerative lesions in their stomach mucosa caused by members of the family Anisakidae. Multiple nematode species concurrently causing gastric ulcerations in yearlings included members of two subfamilies: Anisakinae (Anisakis pegreffii, Anisakis simplex, Pseudoterranova azarasi, and Pseudoterranova decipiens sensu lato) and Contracaecinae (Contracaecum ogmorhini sensu lato). Most animals were infected by three parasitic spp., while the rest harbored one, two, and four parasitic species (Figure 2).
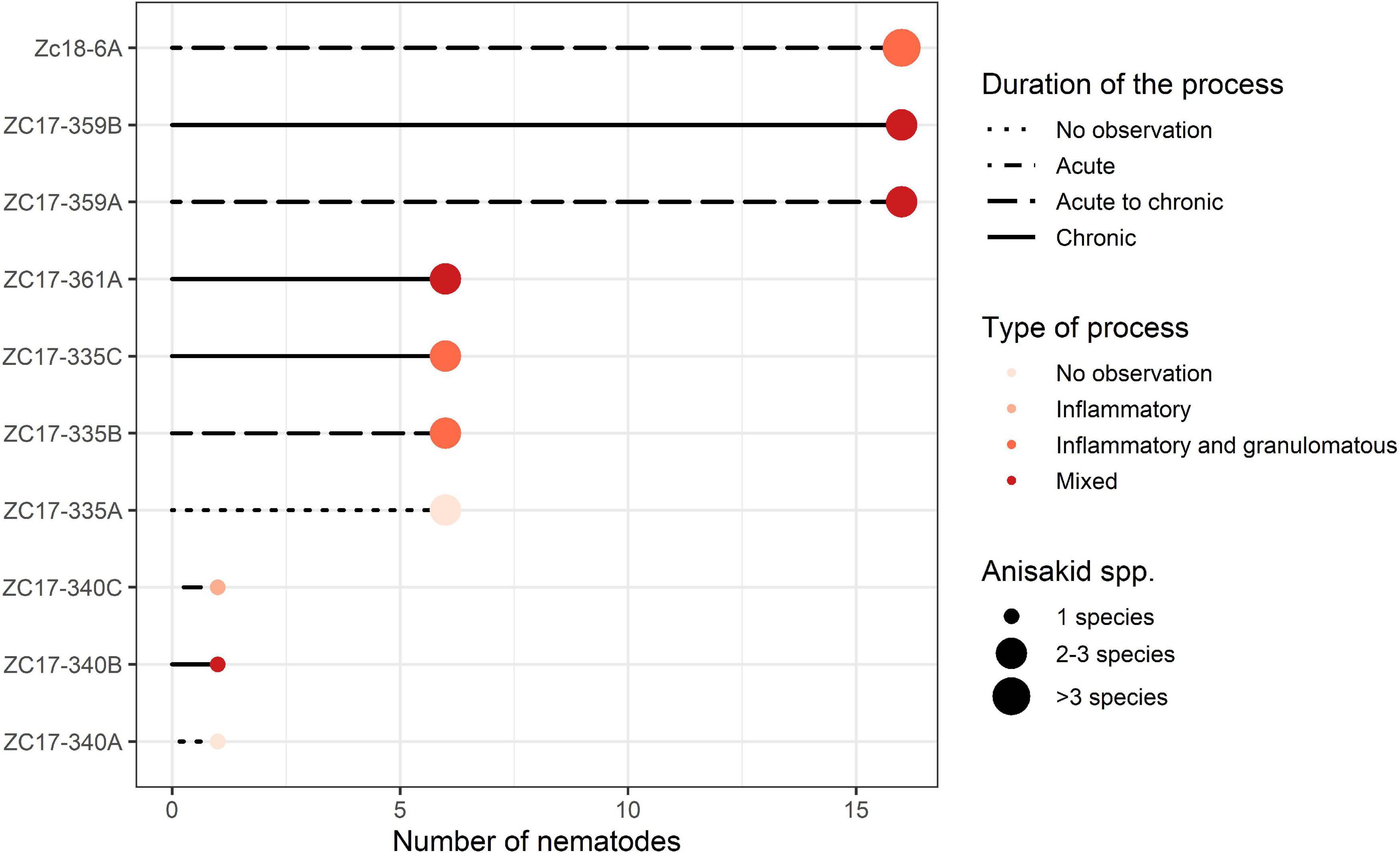
Figure 2. Graphical representation of the severity of lesions inflicted by members of the Anisakidae family per sampled California sea lion (Zalophus californianus). Animals and lesions are depicted on the y-axis and abundance of nematodes coded on the x-axis: 1 (1–5), 6 (6–15), and 16 (>16) nematodes found in the lesion. Other criteria chosen to score severity (duration and type of process and number of species found) are depicted according to the legend in the graph.
According to necropsy reports, pup and yearling sea lions died due to multisystemic parasitism, hypoglycemia, and malnutrition. In contrast, the adult female (ZC17-340) was in apparent good health but was humanely euthanized due to a traumatic limb amputation resulting from a shark bite. However, the latter also had severe gastric lesions according to our scoring chart (Supplementary Table 1 and Figure 2), with a single nematode species causing gastric ulcerations (i.e., C. ogmorhini sensu lato).
The ulcers grossly appeared as distinct protrusions into the lumen of the stomach; the edges were elevated and thickened; and centrally located depressions were of varying depths (see insets in Figures 3, 4). Nematodes were usually observed protruding from the center of the ulcers.
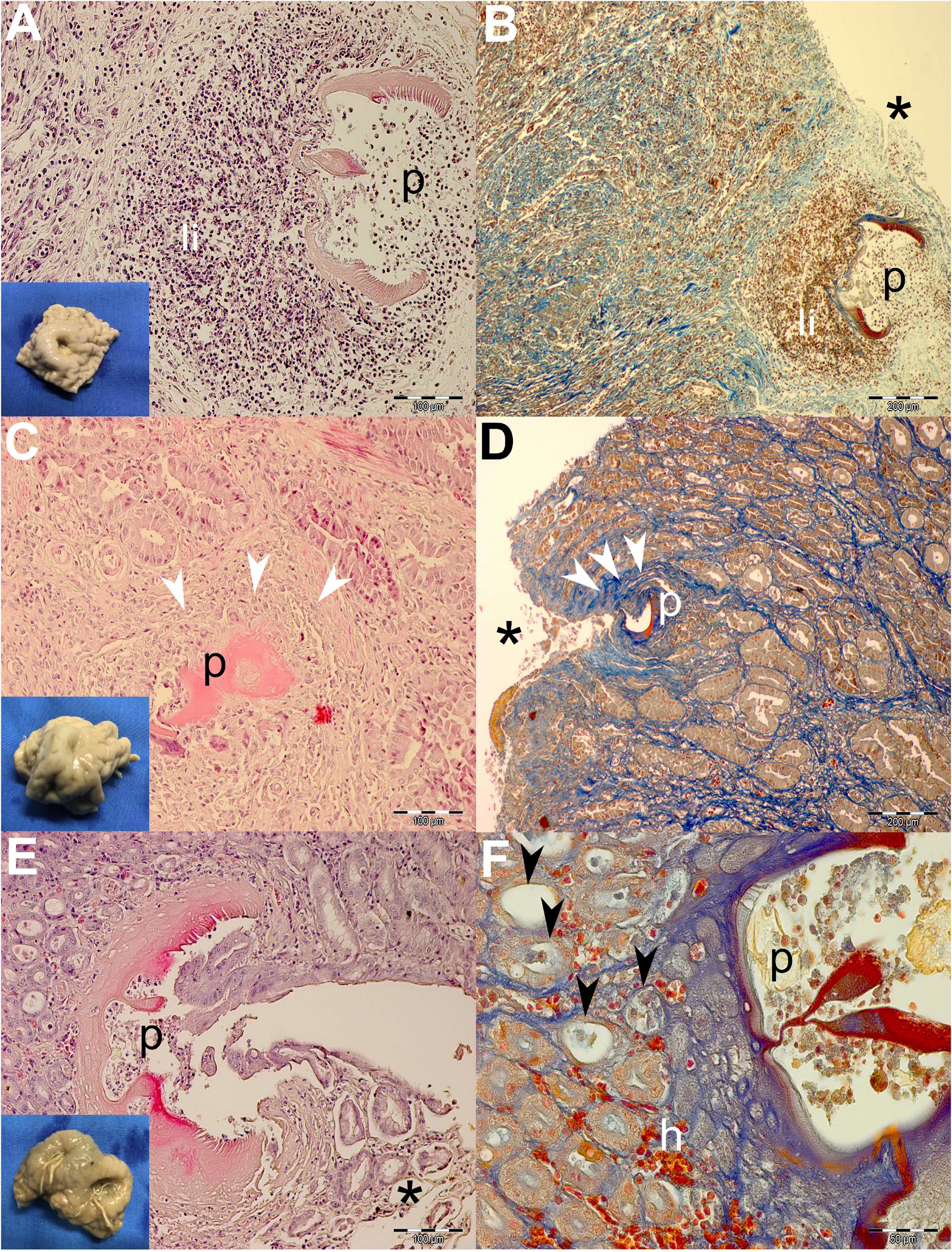
Figure 3. Gross pathological (insets) and histopathological findings of gastric lesions in California sea lions (Zalophus californianus) necropsied at the Marine Mammal Care Center in San Pedro, California, United States (spring and summer 2017 and 2018), affected by parasites of Anisakidae family. (A) Mixed leukocytic infiltrate in the lamina propria of gastric mucosa surrounding transversal section of the anisakids, stained by H&E and Mallory’s trichrome stain (MTS) (B). (C) Deposition of collagen fibers around anisakid body consistent with the formation of granulation tissue and the beginning of the healing process in deeper lamina propria and superficially among gastric acini (D). (E) Superficial penetration of the anisakid among gastric acini and consequent epithelial necrosis. (F) Miniscule hemorrhaging and ectasis of gastric acini after sloughing of the cuboidal epithelia, forming at the anisakid attachment site. h, hemorrhage; li, mixed leukocytic infiltrate; p, parasite; *, necrosis; white arrowheads, connective tissue fibrosis; black arrowheads, ectasis of gastric acini. Scale bars: 50 (F), 100 (A,C,E), and 200 μm (B,D). Staining: H&E (A,C,E) and MTS (B,D,F).
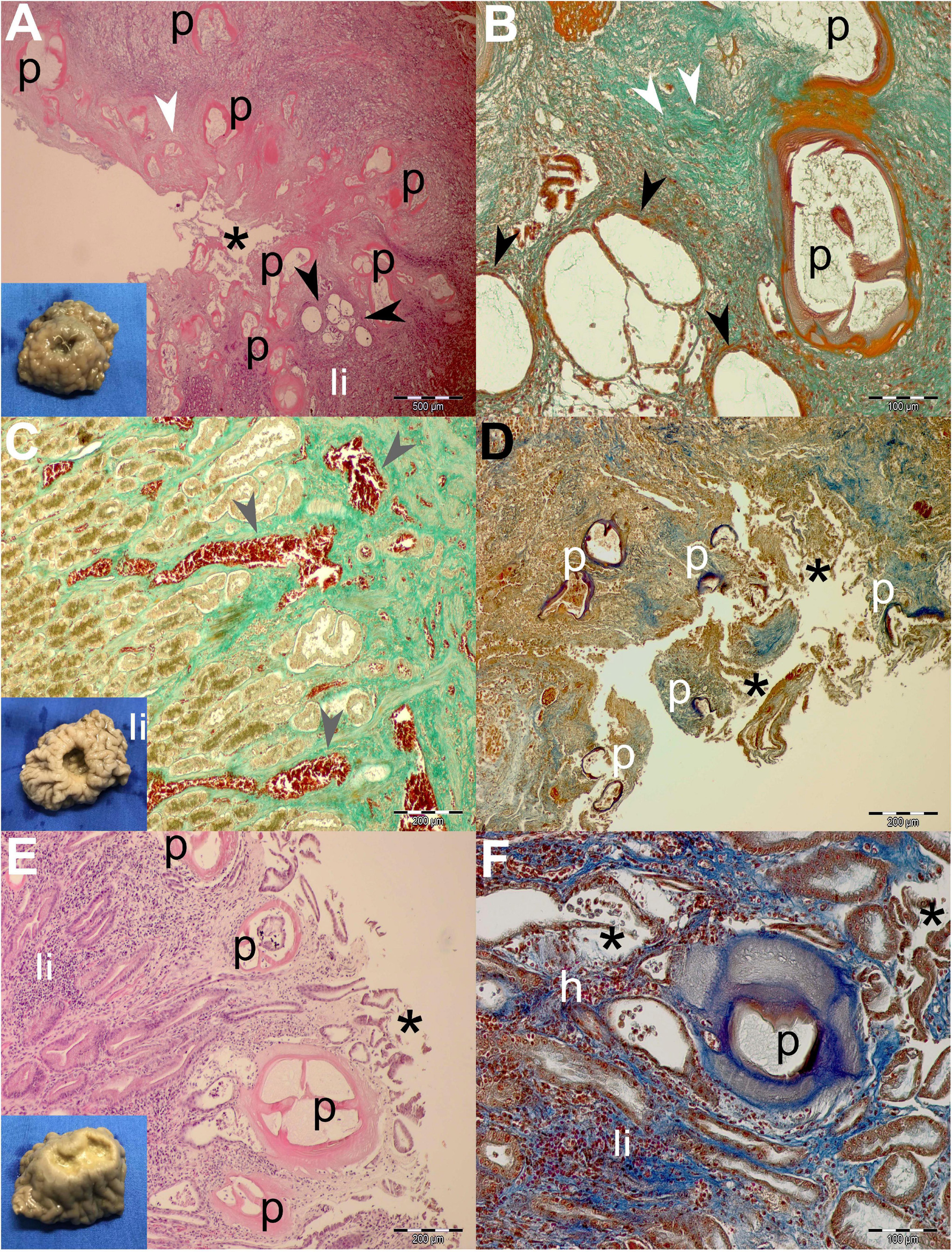
Figure 4. Gross pathological (insets) and histopathological findings of gastric lesions in California sea lions (Zalophus californianus) necropsied at the Marine Mammal Care Center in San Pedro, California, United States (spring and summer 2017 and 2018), affected by parasites of Anisakidae family. (A) Multiple anisakid parasitation site showing transversal sections of the parasites, necrotic epithelium, granulation, mixed leukocytic infiltration, and coalescence of cystic gastric acini. (B) Higher magnification of the previous section showing coalesced gastric acini forming large cysts, which consist of a single atypical cell layer and intraluminal net-like secretion and few cells, stained by Mallory trichrome stain (MTS). (C) Hemostasis in the lamina propria beneath multiple anisakid attachment in a deep ulcer. (D) Severe necrosis in a multiple anisakid ulceration affecting the tunica submucosa. (E) Mild necrosis in a shallow gastric ulcer parasitized by multiple anisakids affecting the tunica mucosa. (F) Higher magnification of the same specimen surrounded by mixed leukocyte infiltration, mild hemorrhaging, and coalescing acini. h, hemorrhage; li, mixed leukocytic infiltrate; p, parasite; *, necrosis; white arrowheads, connective tissue fibrosis; black arrowheads, ectasis of gastric acini; gray arrowhead, hemostasis. Scale bars: 100 (B,F), 200 (C–E), and 500 μm (A). Staining: H&E (A,E), MTS (D,F), and MTS (B,C,D,F).
Generally speaking, deeper lesions reaching the lamina propria and/or tunica submucosa have elicited a more robust inflammatory cellular infiltration consistent with the granulomatous processes (Figures 3A,B, 4A,B), and more shallow lesions (e.g., in the lamina epithelialis) have provoked lesser inflammatory cellular infiltration/response (Figures 3C–F, 4E,F).
Often, more superficial regions of the epithelium and the lamina propria were affected by the necrotic process (Figures 3B,D,E, 4A,D,E). Necrosis was observed in gastric glands (Figures 3E,F), prompting the sloughing of the cuboidal epithelium, accumulation of cellular debris in the acini, and its eventual ectasis (Figure 3F). Cystic acini showed a negligible intraluminal material (net-like secretion with few cellular elements), being lined by an atypical single cell layer that lost its basoapical polarity (Figures 4A,B). In advanced processes, acini coalesced forming wide cysts. Medium-intensity leukocytic infiltration was observed surrounding enlarged acini.
In addition to the inflammatory granulomatous and necrotic processes, nearly every specimen was also exhibiting progressive healing characterized by granulation tissue formation and subsequent fibrosis (Figures 3C,D). Negligible hemorrhage was observed in close contact with the nematode (Figures 3F, 4F), while on a single occasion, hemostasis was observed in the lamina propria (Figure 4C).
All ulcerative lesions had one to up to 50 nematodes embedded in the stomach mucosa (see inset in Figure 3E). The heads of the nematodes were often embedded deep into the center of the ulcer, often reaching as deep as the lamina propria and submucosa and occasionally into the tunica muscularis (Figures 3, 4). The eosinophilic to pink amorphous, hyaline-like, vacuolar substance surrounded the heads of the adults and larval stages of the parasites (Figures 5A–D). Within the substance, entrapped macrophages (Figure 5A) and granulocytes (neutrophils) were often observed (Figure 5D) showing signs of karyorrhexis and pyknosis.
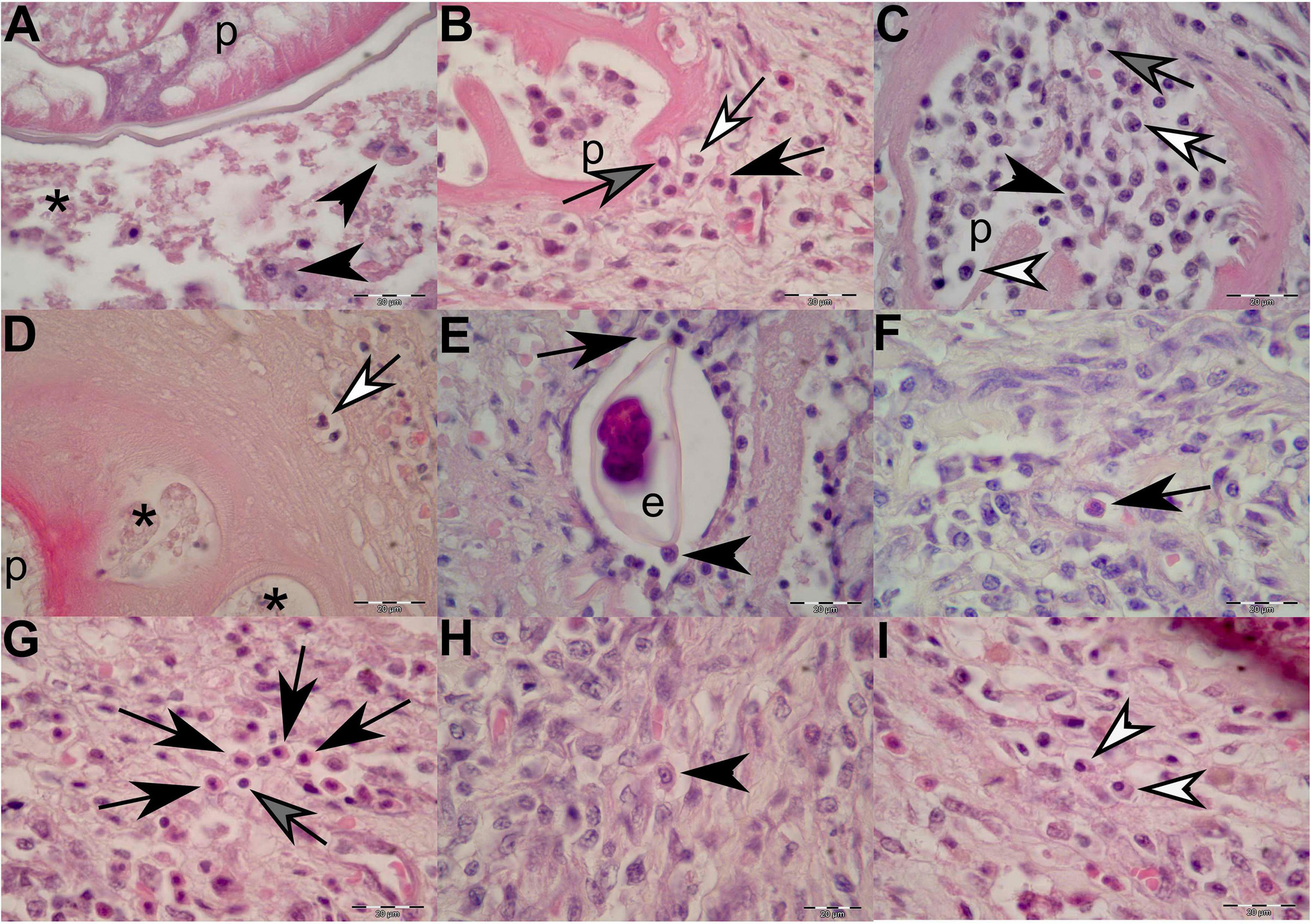
Figure 5. Histopathological findings of gastric lesions in California sea lions (Zalophus californianus) necropsied at the Marine Mammal Care Center in San Pedro, California, United States (spring and summer 2017 and 2018), affected by parasites of Anisakidae family. (A) Interface of the parasite and necrotic tissue composed of cellular debris and macrophages. (B) Lymphocytes, neutrophils, and eosinophils among connective-tissue fibers in close contact with the anisakid. (C) Anisakid pseudocoelom infiltrated by lymphocytes, neutrophils, eosinophils, and plasma cells. (D) Hyaline-like substance enveloping an anisakid, showing vacuolations filled with necrotic debris and neutrophils in early infection stage. (E) Anisakid egg enveloped by a simple squamous epithelium in the tunica submucosa and in close contact with macrophages and eosinophils. (F) A solitary eosinophil and a cluster of eosinophils (G) in the tunica submucosa. (H) A solitary macrophage with visible pseudopodia in the tunica submucosa. (I) Two plasma cell present in the tunica submucosa. p, parasite; *, necrosis; black arrowheads, macrophages; black arrows, eosinophils; gray arrows, lymphocytes; white arrows, neutrophils; white arrowheads, plasma cells. Scale bars: 20 μm. Staining: H&E.
In cases of more recent parasite attachment, hyaline-like substance was not evident; however, an inflammatory cellular infiltrate was always present, even in the early stage of the infection (Figure 5B). The body portions of the nematodes were often immersed in the necrotic debris of the lesion or freely suspended in the lumen of the stomach. In specimens in which nematodes just started penetrating through the lamina epithelialis and lamina propria, the inflammatory cellular infiltrate was mostly composed of few neutrophils and random lymphocytes and accompanied by negligible (or minimal) bleeding (Figure 3F). As the inflammatory process progressed, a more cohesive, concentric, and stronger cellular inflammatory infiltrate developed, closely resembling the granulomatous inflammatory process in which eosinophils, macrophages, lymphocytes, and plasma cells become more predominant (Figures 5B, 5F–I). Many of the macrophages (histiocytes) were activated and resembled “epithelioid” cells; thus, it was sometimes difficult to identify them (Figure 5H).
Occasionally, few nematode eggs were observed entrapped in the lamina propria and tunica submucosa, enveloped by a simple squamous epithelium and in close contact with macrophages and eosinophils (Figure 5E). On many occasions, a mixed leukocytic infiltrate composed of macrophages, neutrophils, eosinophils, lymphocytes, and plasma cells invaded the nematode pseudocoelom (Figures 5B,C).
Profuse bleeding into the tissue was also often observed, and the extent varied from ulcer to ulcer. The tunica submucosa presented prominent neovascularization, fibroblastic proliferation, and the random deposition of collagen fibers, which made this process consistent with the formation of granulation tissue and the beginning of the healing process. Deeper than that, tissue edema and hyperemia were evident. In some older ulcers, granulation tissue underwent metaplasia into more organized fibrotic tissue, indicating the chronicity of the process.
The lesions in a majority of specimens presented as chronic, ulcerative, granulomatous gastritis, with moderate edema and hyperemia of the tunica submucosa and lamina propria, followed by a mild, focal fibrosis of the gastric wall when the process took more prolonged chronic course.
Sequence Analysis
BLAST analysis revealed the presence of three genera belonging to the Anisakidae family in sampled California sea lions, i.e., Anisakis, Contracaecum, and Pseudoterranova. Most abundant were representatives of C. ogmorhini sensu lato (N = 62, 55.36%), found in all except one necropsied animal, in descending order: A. pegreffii (N = 26, 23.21%), P. azarasi (N = 20, 17.86%), P. decipiens sensu lato (N = 2, 1.79%), and A. simplex (N = 2, 1.79%).
Based on the phylogenetic reconstruction analyses, out of a total of 135 [111 new sequences and 24 sequences from the National Center for Biotechnology Information (NCBI)] aligned positions of the cox2 dataset (601 bp), 346 invariable, 53 singleton, and 180 parsimony-informative sites were identified. Also, 118 haplotypes were detected. No indels (insertions/deletions) or stop codons were detected in the alignment. The sequences had the following base compositions: T = 44.6%, C = 11.2%, A = 21.3%, and G = 22.9%. Additionally, number of individuals (N), number of haplotypes (h), nucleotide diversity (Pi), haplotype diversity (Hd), and polymorphic sites (P) are shown in Table 1.
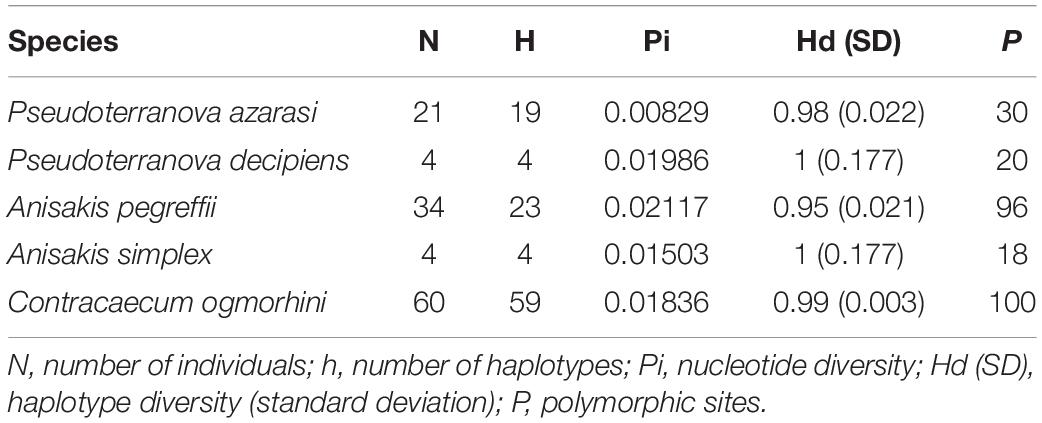
Table 1. Genetic statistics based on cox2 for populations of the Anisakidae nematodes with more than three sequences (sequences from the previous studies were considered).
The genetic distance among five different species of Anisakidae is represented in Table 2. The smallest genetic distance 0.023 was noted between P. decipiens and P. azarasi, while the distances between A. pegreffii and other species ranged from 0.049 to 0.177. The genetic distance between the C. ogmorhini and other lineages ranged from 0.177 (A. pegreffii) to 0.217 (P. decipiens). Pairwise FST evidenced significant genetic differentiation (p < 0.05) among five isolated species: the highest (0.85) and lowest (0.44) genetic differentiations were expectedly found between C. ogmorhini and A. simplex, and P. azarasi, P. decipiens, and P. azarasi, respectively (Table 3).
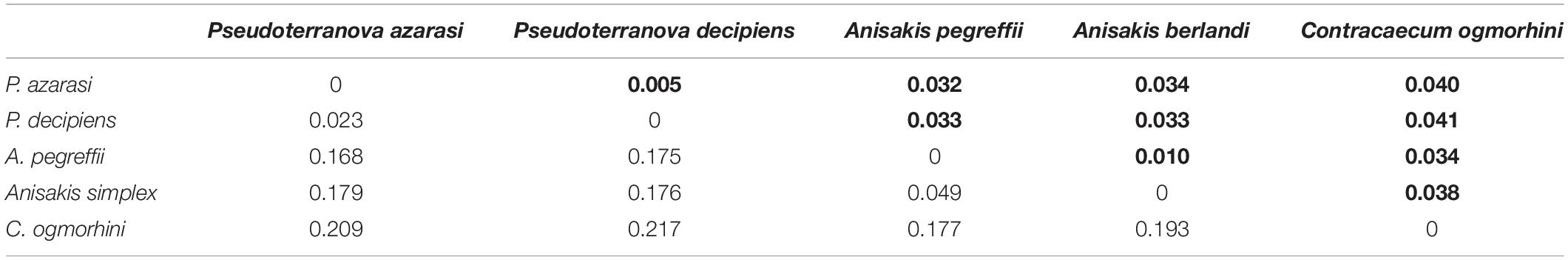
Table 2. Genetic distance based on 601-bp cox2 sequence of five species from Anisakidae (bold numbers show the standard deviation, in populations with more than three sequences).

Table 3. Matrix of pairwise FST values between populations of five species from Anisakidae isolated from California sea lions.
Saturation analyses showed that the mtDNA cox2 locus was suitable for phylogenetic analyses, with index values of the observed substitution saturation (ISS) being significantly smaller than the critical (ISSc) values, signifying minimal substitution saturation (Supplementary Figure 1).
Phylogeny Analysis and Molecular Dating
The phylogenetic relationships among 13 species of Anisakidae (herein isolated and referent species) are given in Figure 6. Both ML and BI phylogenetic trees generated congruent branching patterns, and the Shimodaira–Hasegawa test did not depict significant differences between them (p > 0.05); therefore, only the BI tree is represented. The tree revealed three distinct clades (herein represented by species or species complexes). Clade 1 includes C. ogmorhini that separated from other species with high statistical support (1 PP and 100 BS support). Clade 2 includes representatives of A. simplex sensu lato (A. simplex sensu stricto, A. pegreffii, and Anisakis berlandi) that diverged from the rest of anisakid taxa by high PP (1 PP) and bootstrap support (100 BS). Within clade 2, A. berlandi diverged from the sister clade encompassing A. simplex s.s. and A. pegreffii, showing a high support values, i.e., 1 PP and 99 BS, while the latter two differed from each other by 0.94 PP and 89 BS. Clade 3, a sister clade of A. simplex complex species, splits between Anisakis typica and the rest of the polyphyletic sister subclades formed by Anisakis spp. (Anisakis nascettii, Anisakis ziphidarum, Anisakis paggiae, Anisakis brevispiculata, and Anisakis physeteris (1 PP and 97 BS). A. nascettii splits from A. ziphidarum and its sister subclade that includes representatives of morphological type II larvae (A. paggiae, A. physeteris, and A. brevispiculata) and Pseudoterranova sister subclade (supported by 0.96 PP and 75 BS). The latter subclade is formed by Pseudoterranova bulbosa, parting from P. decipiens/P. azarasi subclade (supported by 1 PP and 91 BS). Interestingly, P. decipiens population from Western Atlantic diverged from California sea lions’ P. decipiens and P. azarasi also with high support (1 PP and 91 BS), while the latter subclade (sea lions P. decipiens/P. azarasi) diverged by 0.93 PP and 86 BS.
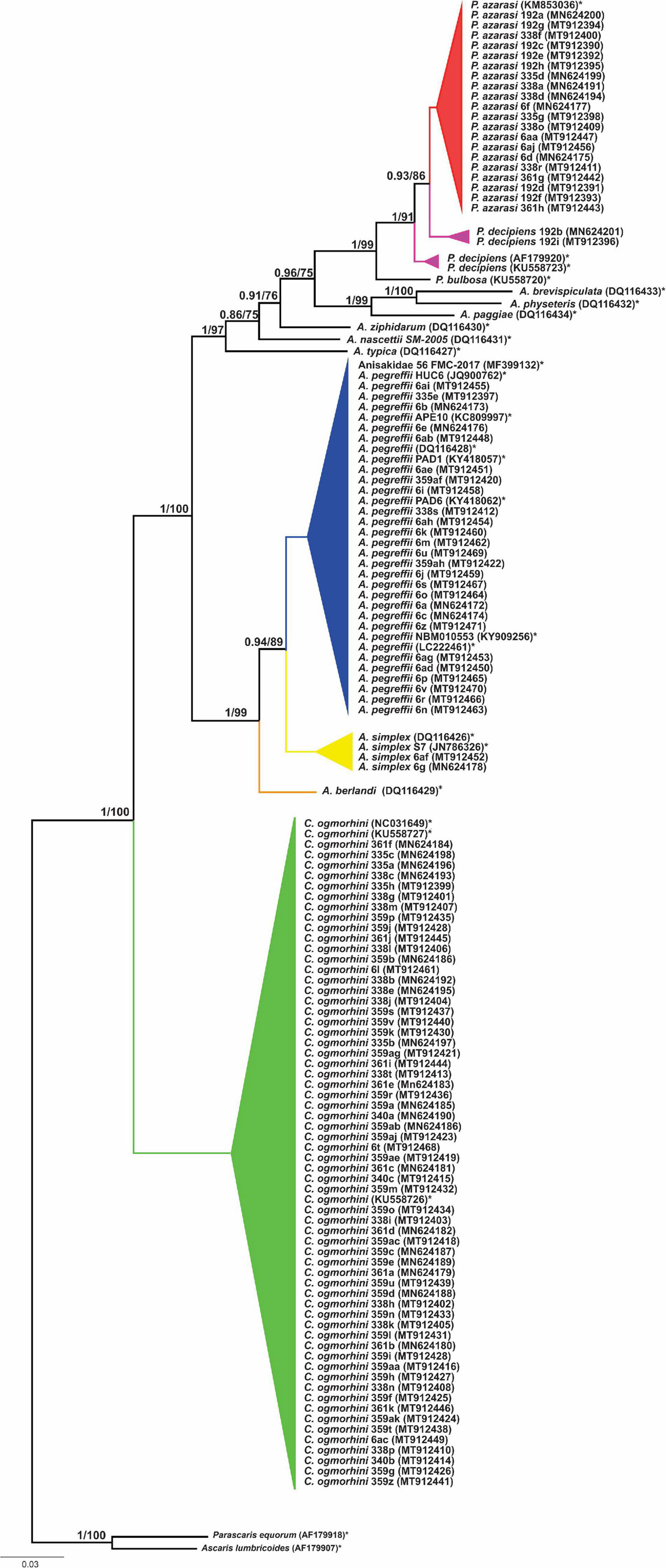
Figure 6. Phylogenetic tree reconstructed from 601 bp sequence cox2 of Anisakidae. The values on the top branches correspond to posterior probabilities and bootstrap support.
The haplotype network (Figure 7A) plotted by PopArt using anisakids cox2 (populations with sample size >3) detected five distinct species within Anisakidae group in California sea lions dataset. The similar number of mutational steps, 70 and 71, was observed between Anisakis and Pseudoterranova, and Anisakis and Contracaecum, respectively. A. simplex and A. pegreffii diverged by 20 mutational steps. Referent West Atlantic P. decipiens diverged from sea lions’ isolated P. azarasi by eight mutational steps, although the same difference (i.e., eight mutational steps) was observed between these two particular Atlantic haplotypes.
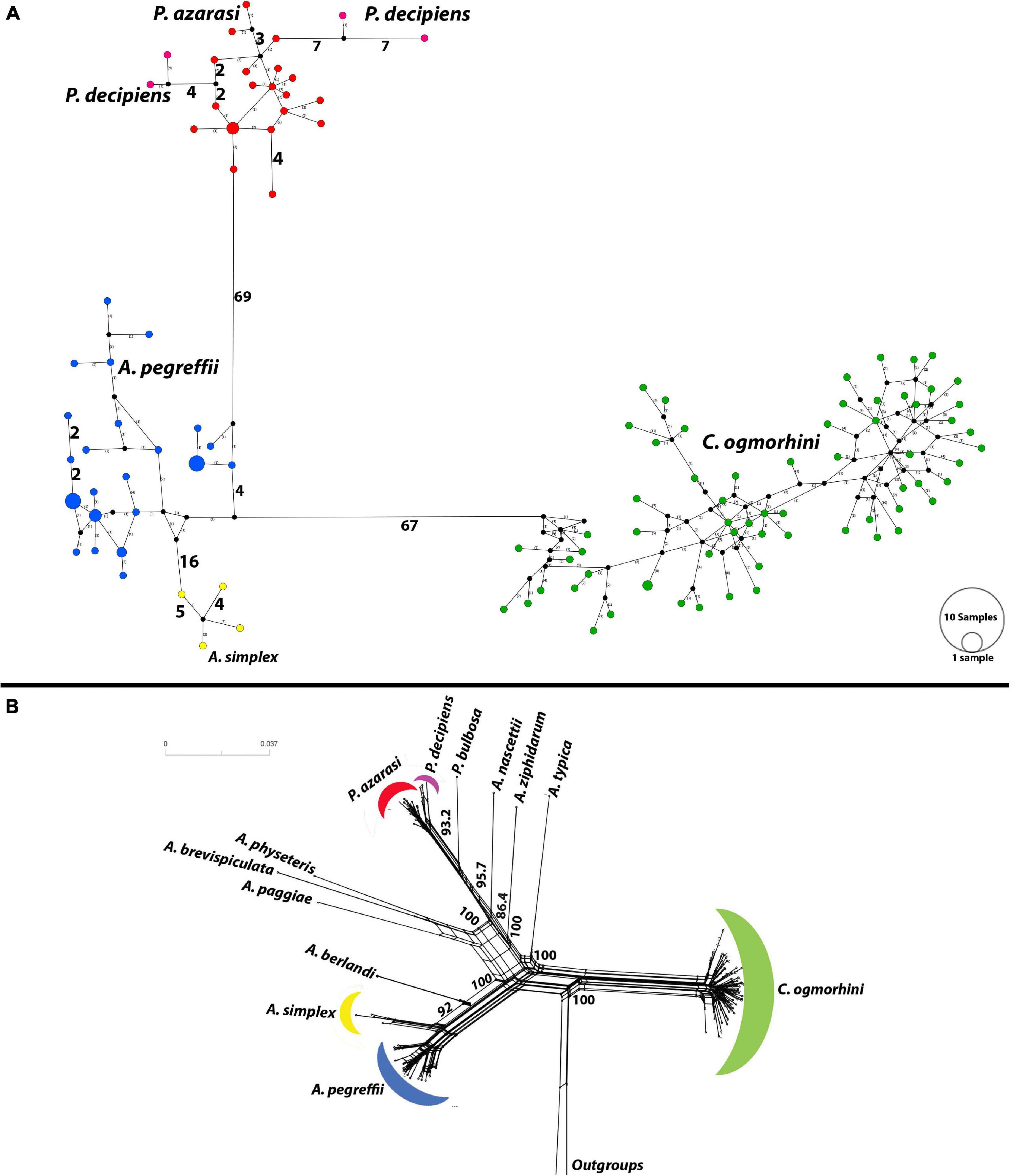
Figure 7. Haplotype network using the 601 bp of cox2 dataset. (A) Median-joining network for anisakid populations with more than three samples. Symbol size and branch lengths are proportional to the number of shared individuals per haplotype and the number of mutational steps among haplotypes, respectively. Black circles indicate unsampled or extinct haplotypes. Sizes of circles are proportional to haplotype frequencies. (B) Split tree for 13 species of anisakids with uncorrected p-distances.
Furthermore, split tree network, developed incorporating all population sizes, was consistent with phylogeny tree and split tree. C. ogmorhini diverged from other anisakids with 100% bootstrap. P. azarasi, P. decipiens, and P. bulbosa separated from each other and diverged from A. nascettii, A. ziphidarum, and A. typica with the high support value (95.7). Moreover, A. physeteris, A. brevispiculata, and A. paggiae diverged from A. berlandi, A. simplex, and A. pegreffii with 100% bootstrap (Figure 7B).
The analyses of divergence placed the basal cladogenesis among Anisakidae in the Late Mesozoic (Jurassic Age), approximately at 176.2 Mya [95% highest posterior density (HPD) interval; HPD = 167.4–180.5 Mya], separating the C. ogmorhini as the basal clade of Anisakidae. Subsequent cladogenic events dated to the Late Mesozoic (Cretaceous Age) at approximately 134.8 Mya (95% HPD = 127.4–140.1 Mya), separating A. pegreffii and A. berlandi from other species. The recent species age of Anisakidae is related to Cenozoic (Neogene Age) (Figure 8).
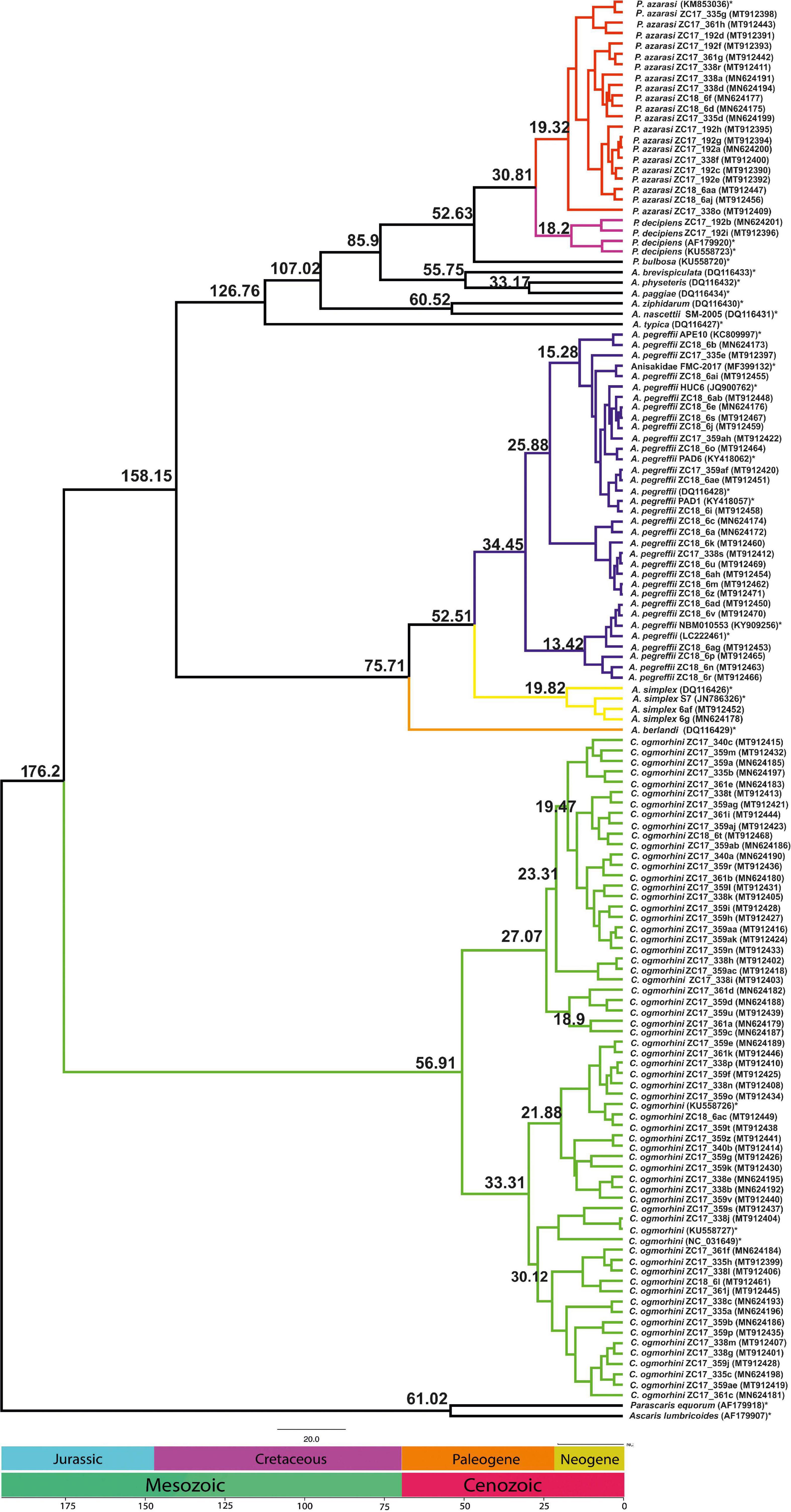
Figure 8. Chronogram resulting from divergence time analyses using the concatenated cox2 dataset with 137 sequences using two outgroups (AF179918 and AF179907), generated by BEAST v1.8.2. Branch numbers display times of divergence.
EBSP indicated a population growth curve for Anisakidae since ∼2–4 Kya. This plot revealed a gradual expansion since ∼2–4 Kya and a considerable increase in population size since 0.5–2 Kya up to the present day, with the exception of C. ogmorhini. Also, the demographic events estimated based on analysis of pairwise mismatch distribution of cox2 haplotypes found a multimodal mismatch distribution for all Anisakidae clades (Figure 9).
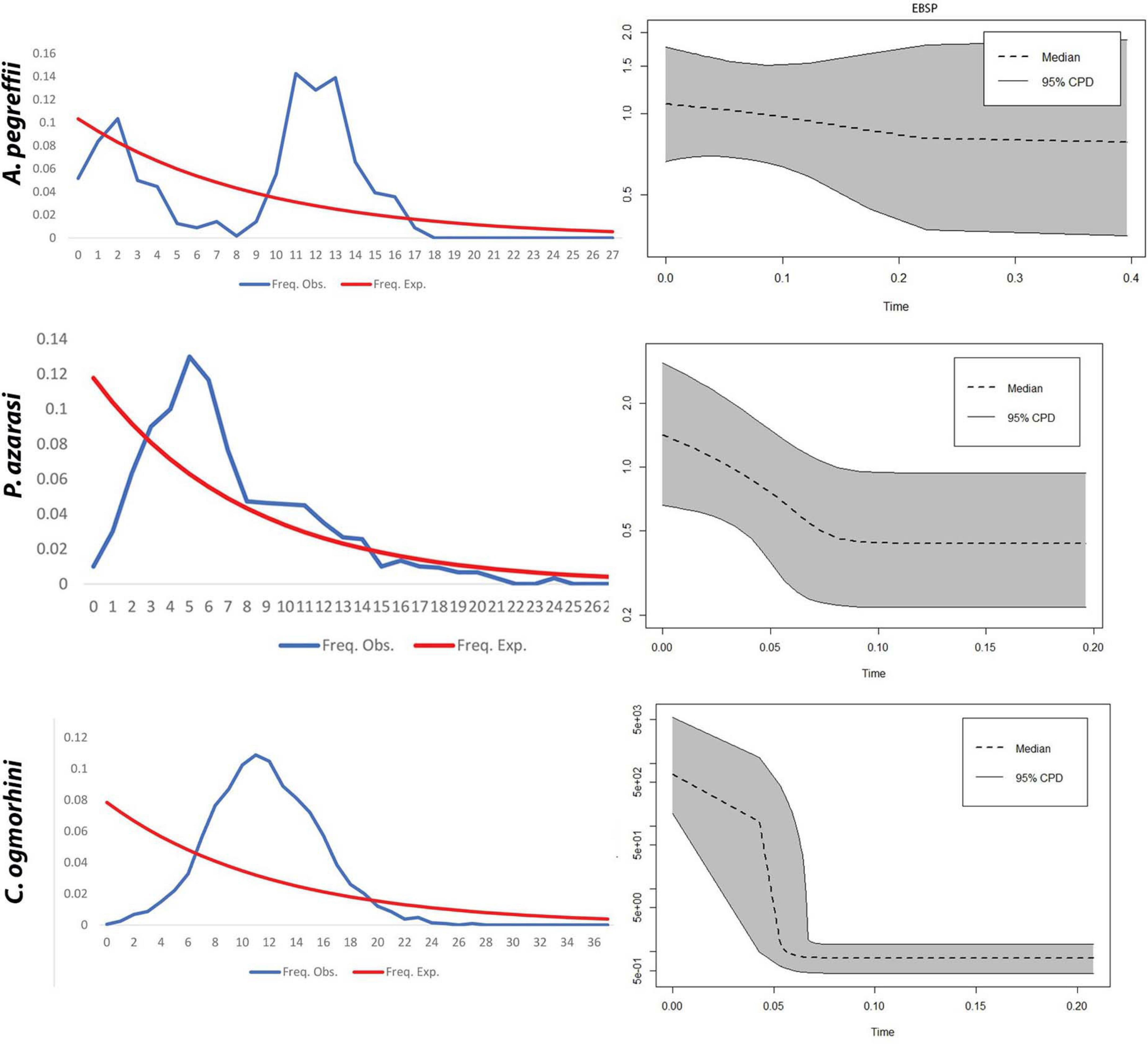
Figure 9. Extended Bayesian Skyline Plot. (A) The x-axis visualizes time before present (Mya), and the y-axis represents effective population sizes (Ne). Gray regions depict confidence intervals [95% highest posterior density (HPD) limits], and the dashed line expresses median value for the log10 of Ne (effective population size). (B) Mismatch distribution demonstrates the demographic history of Pseudoterranova azarasi, Anisakis pegreffii, and Contracaecum ogmorhini. The dotted line represents the observed distributions, and the solid black curve indicates distributions under a constant-sized population assumption.
Discussion
Nematode-inflicted pathological lesions described in marine mammals range from shallow ulcerations to full-thickness perforations of the gastric wall (Sweeney and Gilmartin, 1974), even though these infections rarely cause mortality in pinnipeds. However, combined with the pollution, climate change, domoic acid poisoning events, seasonal El Nino starvation events, and other pathogens, nematode infections may negatively impact pinnipeds’ health and cause mortality. According to the necropsy reports, four of five specimens developed multisystemic parasitism as a leading cause of death, combined with malnutrition and hypoglycemia. Only one adult female specimen, infected by Contracaecum ogmorhini sensu lato, did not succumb to multisystemic parasitism. Raga et al. (1998) previously observed that parasitic infection can strongly affect and shape pinnipeds communities, although the evidence available is limited and should be interpreted with caution. In the northern fur seals (Callorhinus ursinus), hookworm Uncinaria lucasi transmitted from mother to pup during nursing, which causes high mortalities from severe intestinal hemorrhages. U. lucasi apparently acts in a density-dependent manner, restricting the pup population when it reaches a considerable size. Whether a similar pattern is followed by other nematode species, more specifically anisakids, should be evaluated in a broader population study of the California sea lions. However, this might prove difficult to achieve, as feral animals usually harbor diverse parasite communities with cumulative effect on the host. In contrast to maternally transmitted U. lucasi, anisakids are trophically transmitted to sea lions, therefore occurring well after the pup’s weaning period, which might warrant a better level of innate and acquired immunity in those animals and consequently better prognosis for their survival, if not affected by other comorbid factors. In toothed whales, such as bottlenose (Tursiops truncatus) and striped dolphins (Stenella coeruleoalba), anisakid numbers expressed as mean intensity on average reach 4,000 specimens without causing apparent mortalities, even though infections significantly increase with the age (Blažeković et al., 2015). The host–Anisakis interaction is characterized by a convex curve, where the nematode number does not reach an asymptotic value but falls after an initial increase (Hudson and Dobson, 1995). This suggests a long and efficient coevolution of the anisakids with their final hosts and mostly mild and tolerable pathological effects within the population. However, the focus of this study in the California sea lions was exclusively on the gastric lesions and parasites, not accounting for other comorbidities or parasites (e.g., lung worms, hookworms, or acanthocephalans) that likely influenced to the greater extent the survival prognosis in these animals. Therefore, anisakids, rather than regulating the sea lion population, likely take part with other disease and environmental factors, such as predation, hunting, shortage of resources, or severe weather, expressing a compensatory effect (Aznar et al., 2001). Moreover, the animals in this study came from the population that appears to be approximately 40% above its MNPL, therefore being considered within the range of its OSP size (NOAA, 2018).
Depending on the quantitative and qualitative severity of the infection, nematode-caused ulcerations can express as destructive (necrosis), defensive (inflammation), or healing (granulation formation) processes (Liu and Edward, 1971). At the site of nematodes’ burrowing, some authors (Liu and Edward, 1971) described hyaline bluish to pink substance surrounding the head of the nematodes, which is consistent with our observations. This hyaline substance often entrapped granulocytes that subsequently undergo necrosis. Liu and Edward (1971) speculated that in the Steller sea lion (Eumetopias jubatus), this substance could in fact be either secretion of the parasites mixed with necrotic debris or a deposition of antigen–antibody complexes. Spraker et al. (2003) and Kuzmina et al. (2014) described multinucleated giant cells in the tissue immediately surrounding the nematode in the northern fur seal; however, our observations in California sea lions did not confirm the presence of those cells. The processes observed in the California sea lions seemed to be less destructive from those observed by Hrabar et al. (2017). In the three species of stranded toothed whales—namely, bottlenose dolphins, striped dolphins, and Risso’s dolphins (Grampus griseus)—Anisakis-induced alterations included severe ulcerative gastritis with mixed inflammatory infiltrate often associated with colonies of bacteria, and mild-to-moderate granulomatous gastritis with eosinophilic infiltrate. In our study, ulcerative changes with mixed leukocytic infiltrate showed to be localized, shallow, and non-perforative and with no apparent bacterial coinfection, mostly accompanied by healing granulation tissue. The difference in observed pathologies could at least partly be explained by the fact that in the study by Hrabar et al. (2017), stranded cetaceans were of much older age than the animals from the present study, allowing enough time for anisakid infections to reach high intensities and in turn cause more significant pathologies. A lack of potential secondary bacterial infection in the lesions, or perforative peritonitis and consequent septicemia, suggests a mild and self-limiting course of these infections. However, on a few occasions, necrosis of stomach glandular epithelium with ectasis and cystic gastric acini possibly caused malabsorption, which although localized in already starving pups might have influenced the survival prognosis. Namely, the nutritional status of the host can influence the pathogenesis of parasitic infection, and well-nourished animals generally withstand parasitism better than those less adequately fed (Roy et al., 2003), which is likely the case in our specimens. In some other host–parasite associations, it was shown that the effect of parasitism is highly context-dependent, and parasitized animals can sometimes perform better and show lower metabolic demand than the non-parasitized ones (Aldana et al., 2020 and references therein). Anisakid infections are generally not considered fatal except in rare cases when intestinal wall perforation can occur leading to peritonitis. However, gastric ulcers inflicted by these nematodes might be an important debilitating factor in feral animals (Dailey et al., 1985). It is therefore tempting to speculate whether the observed pathologies in the present study are a result of an already poor nutritional status caused by other parasites and whether the same effects would be observed in well-fed animals not affected by other parasite taxa.
Interestingly, some anisakids embedded in tissue lesions showed the pseudocoelom infiltrated by mixed leukocyte lineages. These lacked a well-defined cuticle and the normal inner architecture typically observed in parasite’s cross sections (i.e., cross section through the esophagus/gastrointestinal tube, excretory cell, somatic muscles, and reproductive organs), being reduced to a hyaline-like substance lined with remnants of hypodermis, eventually presenting a single lateral chord. To our knowledge, only in the experimental mice infection with the human filarial nematode, Brugia malayi and Brugia pahangi, were adherent macrophages observed to be attached to the infective larvae and eosinophils within the pseudocoel 4–6 weeks post infection (Rajan et al., 2002). Whether in the sea lions these damaged anisakids represent specimens that died naturally or were affected by host immunity still needs to be evaluated, although the finding further confers the self-limiting character of the infection.
Anisakids from California sea lions are grouped in three distinctively separated clades corresponding to genera Anisakis, Contracaecum, and Pseudoterranova, all monophyletic except the former, with strong support from both ML (BS = 100) and BI (PP = 1). This is in accordance with separation of Anisakidae into two subfamilies; the Anisakinae (including Anisakis, Pseudoterranova, Pulchascaris, and Terranova) and the Contracaecinae (including Contracaecum and Phocascaris) (Li et al., 2018). Scrutinizing potential intra- and inter-species differences of anisakids from California sea lions in relation to previously characterized taxa (Mattiucci et al., 2014; Cipriani et al., 2018), our phylogenetic trees revealed no differentiation at intra-species level. This is congruent with the observed lack of structure for A. pegreffii, Anisakis berlandi, and Anisakis simplex populations in the Mediterranean, Pacific Ocean, and Norwegian coast assessed through a multi-locus approach (Mattiucci et al., 2014), explainable by a high gene flow among the parasite populations. Interestingly, the exceptions were two California sea lions Pseudoterranova decipiens (sample ZC17-92b, GenBank # MN624201; and ZC17-192i, GenBank # MT912396) that diverged from the respective Western Atlantic population (KM853036, AF179920, and KU558723) and formed a sister group with Pseudoterranova azarasi sea lions population. Because of the scarce number of P. decipiens isolates in the sea lions, this further needs to be corroborated in a larger dataset.
Tested dataset of anisakids parasitizing California sea lion, however, did not show the existence of potential recombinant haplotypes, which was previously observed for A. simplex sensu lato species (Cavallero et al., 2014; Mladineo et al., 2017a, b). Long-term parasitation (reflected from chronic character of lesions) of mature adults within the same final host and present in robust numbers offered a rare opportunity to test the hypothesis; however, we did not find evidence to support such.
The highest-abundant anisakid lineage included the only representative of the genus Contracaecum, C. ogmorhini sensu lato (N = 60), which showed mostly unique but closely related haplotypes. This confirms the lack of cryptic species in the California sea lions from the studied geographic area and also complies with the recent observation that C. ogmorhini does not represent a complex of cryptic species, at least not supported by mitochondrial genome data (Liu et al., 2016). Namely, Mattiucci et al. (2003) differentiated C. ogmorhini sensu stricto and Contracaecum margolisi (isolated from Zalophus californianus in the Boreal region) within the C. ogmorhini species complex, based on 260 bp mitochondrial cytochrome b locus and two diagnostic enzyme loci. While mitochondrial genome phylogeny failed to differentiate cryptic species (Liu et al., 2016), further research should focus on analyses of multiple nuclear markers to improve resolution.
In contrast, haplotype networks of sea lions’ Anisakis and Pseudoterranova show that within each lineage, haplotypes are more mutated and show higher gene flow, based on the number of haplotype interconnections and mutational steps but interestingly form more common haplotypes in respect to Contracaecum. A. simplex sensu stricto and Anisakis pegreffii were clearly differentiated, as well as P. decipiens sensu lato from more abundant P. azarasi. No cryptic species within P. decipiens sensu lato have been observed from the studied sea lions (Timi et al., 2014), although a weak number of isolated specimens (N = 3) most likely affected the output.
The haplotype network for each lineage differed from a star-like phylogeny with a central, most abundant haplotype and the rest of diverging unique haplotypes, as mainly observed for Anisakis in the Adriatic, indicative of a recent rapid range expansion (Mladineo and Poljak, 2014). In contrast, lineages showed rare and unique haplotypes, especially Contracaecum where most of the missing haplotypes have been noticed (non-sampled or extinct). This was further corroborated by demographic history based on EBSP and mismatch distribution, according to which P. azarasi and C. ogmorhini ranges expanded between 60 and 80 Kya during the upper Pleistocene period (Walker et al., 2018). Both populations showed a unimodal distribution, usually associated with a panmictic population that has undergone sudden demographic expansion (Slatkin and Hudson, 1991). In contrast, sea lions A. pegreffii showed a multimodal distribution that indicates a widespread and stable population (Schneider and Excoffier, 1999). We can only speculate whether different historical events occurring in that period and directly affecting one or more anisakids’ host types (i.e., intermediate, paratenic, or final) also indirectly affected the shape and size of parasites’ population. To further explore this, a wider sampling effort of both parasite and host taxa should be undertaken.
Our dated tree revealed Contracaecum separated from other lineages in the Jurassic period at the 176.2 Mya (95% HPD = 167.4–180.5 Mya), which might be linked to host switching to fish-eating birds. Moreover, Anisakis diverged from Pseudoterranova in Cenozoic period at 85.9 Mya (95% HPD = 92.5–76.3 Mya), likely a result of lateral host switches in which host ranges increased and their parasitological pattern varied. Consequently, one of the hypotheses for host shifting could date to global sea level water during the Early Cretaceous (Li et al., 2018).
The estimated time of divergence of California sea lions’ anisakids for the most part is congruent to ascaridoid divergence as in Li et al. (2018). The exception is the separation between Anisakis/Pseudoterranova sister clades from Contracaecum, which comes later in the Jurassic in contrast to the Triassic observed in Li et al. (2018). The reason is likely the extent of analyses performed by Li et al. (2018) that was based on a wide number of ascaridoid families, eight phylogenetic markers, and subsequent calibration involving time priors from hosts rather than ascaridoids.
In conclusion, anisakids from California sea lions show recent and diverse populations, divided among Anisakis, Pseudoterranova, and Contracaecum lineages, the latter being the most abundant. No evidence for recombinant haplotypes was observed, neither as for particular lineages exerting more pressure on the host (i.e., histopathological changes) in respect to other, as the majority of animals was parasitized by 2–3 anisakid species, showing similar lesions types. Inflicted pathology is expressed as mostly a mild inflammatory reaction with self-limiting and healing processes, suggesting that nematodes can present a minor cofactor rather than a cause in mortalities suffered at the Marine Mammal Care Center. However, to elucidate the actual effect of anisakid nematodes on California sea lions a broader, population-wide study should be conducted.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
The animal study was reviewed and approved and all research activities were approved by the IACUC of the Western University of Health Sciences (R16IACUC025), and the National Oceanic Atmospheric Administration (NOAA) approved the acquisition of the specimens (permit 151401WCR2015PR00034). Export of these specimens to the European Union, Croatia, was also authorized by the NOAA that provided a special letter of authorization.
Author Contributions
HS, IM, JH, and SR-D: conceptualization. IM: data curation, funding acquisition, and project administration. HS, IM, JH, SR-D, TP, and ŽT: formal analysis. HS, JH, and SR-D: investigation. KS and LP: sample collection. IM, JH, MP, and SR-D: methodology. HS, IM, SR-D, and ŽT: visualization. HS, IM, and SR-D: writing—original draft. HS, IM, JH, LP, MP, SR-D, TP, and ŽT: writing—review and editing. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding
This research was funded by the Croatian Science Foundation (project AnisCar: Anisakis as a carcinogen: Daring to bust Lancet’s myth or revealing its true colors, grant no. IP-2018-01-8490 to IM).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.636626/full#supplementary-material
Footnotes
References
Abollo, E., López, A., Gestal, C., Benavente, P., and Pascual, S. (1998). Long-term recording of gastric ulcers in cetaceans stranded on the Galician (NW Spain) coast. Dis. Aquat. Organ. 32, 71–73. doi: 10.3354/dao032071
Aldana, M., Pulgar, J., Hernandez, B., George-Nascimento, M., Lagos, N. A., and Garcia-Huidobro, M. R. (2020). Context-dependence in parasite effects on keyhole limpets. Mar. Environ. Res. 157:104923. doi: 10.1016/j.marenvres.2020.104923
Aznar, F. J., Balbuena, J. A., Fernández, F., and Raga, J. A. (2001). ““Living together: the parasites of marine mammals,” in Marine Mammals: Biology and Conservation, eds P. G. H. Evans and J. A. Raga (New York, NY: Kluwer Academic/Plenum Publishers), 385–421. doi: 10.1007/978-1-4615-0529-7_11
Bernardi, C. E. M., Stella, S., Anastasio, A., Cattaneo, P., and Tirloni, E. (2017). Prevalence and Mean Intensity of Anisakis spp. in Two Anglerfish Species (Lophius piscatorius, Lophius budegassa) Caught in Mediterranean Sea. in Convegno SISVET (Napels: SISVET). Milano: IRIS-AIR, 199–1999.
Blaxter, M. (2009). ““Nematodes (nematoda),” in The Tree of Life, eds S. B. Hedges and S. Kumar (New York, NY: Oxford University Press), 247–250.
Blažeković, K., Lepen Pleić, I., Duras, M., Gomerčić, T., and Mladineo, I. (2015). Three Anisakis spp. isolated from toothed whales stranded along the eastern Adriatic Sea coast. Int. J. Parasitol. 45, 17–31. doi: 10.1016/j.ijpara.2014.07.012
Bouckaert, R., Heled, J., Kühnert, D., Vaughan, T., Wu, C. H., Xie, D., et al. (2014). BEAST 2: a software platform for bayesian evolutionary analysis. PLoS Comput. Biol. 10:e1003537. doi: 10.1371/journal.pcbi.1003537
Brandley, M. C., Schmitz, A., and Reeder, T. W. (2005). Partitioned Bayesian analyses, partition choice, and the phylogenetic relationships of scincid lizards. Syst. Biol. 54, 373–390. doi: 10.1080/10635150590946808
Cavallero, S., Costa, A., Caracappa, S., Gambetta, B., and D’Amelio, S. (2014). Putative hybrids between two Anisakis cryptic species: molecular genotyping using high resolution melting. Exp. Parasitol. 146, 87–93. doi: 10.1016/j.exppara.2014.08.017
Cipriani, P., Sbaraglia, G. L., Palomba, M., Giulietti, L., Bellisario, B., Bušelić, I., et al. (2018). Anisakis pegreffii (Nematoda: Anisakidae) in European anchovy Engraulis encrasicolus from the Mediterranean Sea: Fishing ground as a predictor of parasite distribution. Fish. Res. 202, 59–68. doi: 10.1016/j.fishres.2017.03.020
Dailey, M. D. (2005). “Parasites of marine mammals,” in Marine Parasitology, ed. K. Rhode (Clayton, VIC: CSIRO Publishing), 408–414.
Dailey, M. D., Kinne, O., and Lauckner, G. (1985). “Volume IV, part 2: reptilia, aves, mammalia,” in Diseases of Marine Animals, ed. O. Kinne (Helgoland: Biologishe Anstalt Helgoland), 543–884.
Darriba, D., Taboada, G. L., Doallo, R., and Posada, D. (2012). JModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 9:772. doi: 10.1038/nmeth.2109
Denver, D. R., Morris, K., Lynch, M., Vassilieva, L. L., and Thomas, W. K. (2000). High direct estimate of the mutation rate in the mitochondrial genome of Caenorhabditis elegans. Science (80-) 289, 2342–2344. doi: 10.1126/science.289.5488.2342
Drummond, A. J., Suchard, M. A., Xie, D., and Rambaut, A. (2012). Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29, 1969–1973. doi: 10.1093/molbev/mss075
Excoffier, L., and Lischer, H. E. L. (2010). Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 10, 564–567. doi: 10.1111/j.1755-0998.2010.02847.x
Fagerholm, H.-P. (1991). Systematic implications of male caudal morphology in ascaridoid nematode parasites. Syst. Parasitol. 19, 215–228. doi: 10.1007/bf00011888
Fauquier, D., Gulland, F., Haulena, M., Dailey, M., Rietcheck, R. L., and Lipscomb, T. P. (2004). Meningoencephalitis in two stranded California sea lions (Zalophus californianus) caused by aberrant trematode migration. J. Wildl. Dis. 40, 816–819. doi: 10.7589/0090-3558-40.4.816
Fleischman, R. W., and Squire, R. A. (1970). Verminous pneumonia in the California sea lion (Zalophus californianus). Vet. Pathol. 7, 89–101. doi: 10.1177/030098587000700201
Garner, M. M., Lambourn, D. M., Jeffries, S. J., Briggs Hall, P., Rhyan, J. C., Ewalt, D. R., et al. (1997). Evidence of Brucella infection in Parafilaroides lungworms in a Pacific harbor seal (Phoca vituline richardsi). J. Vet. Diagn. Invest. 9, 298–303. doi: 10.1177/104063879700900311
Gibbons, L. M. (2010). Keys to the Nematode Parasites of Vertebrates. Supplementary Volume. Wallingford: CABI.
Heled, J., and Drummond, A. J. (2008). Bayesian inference of population size history from multiple loci. BMC Evol. Biol. 8:289. doi: 10.1186/1471-2148-8-289
Hrabar, J., Bočina, I., Gudan Kurilj, A., Đuras, M., and Mladineo, I. (2017). Gastric lesions in dolphins stranded along the Eastern Adriatic coast. Dis. Aquat. Organ. 125, 125–139. doi: 10.3354/dao03137
Hudson, P. J., and Dobson, A. P. (1995). “Macroparasites: observed patterns in naturally fluctuating animal populations,” in Ecology of Infectious Diseases IN Natural Populations, eds B. T. Grenfell and A. P. Dobson (Cambridge: Cambridge University Press), 144–176. doi: 10.1017/cbo9780511629396.006
Huson, D. H., and Bryant, D. (2006). Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23, 254–267. doi: 10.1093/molbev/msj030
Kelly, T. R., Greig, D., Colegrove, K. M., Lowenstine, L. J., Dailey, M., Gulland, F. M., et al. (2005). Metastrongyloid nematode (Otostrongylus circumlitus) infection in stranded California sea lion (Zalophus californianus)-a new host-parasite association. J. Wildl. Dis. 41, 593–598. doi: 10.7589/0090-3558-41.3.593
Kuzmina, T. A., Lyons, E. T., and Spraker, T. R. (2014). Anisakids (Nematoda: Anisakidae) from stomachs of northern fur seals (Callorhinus ursinus) on St. Paul Island, Alaska: parasitological an pathological analysis. Parasitol. Res. 113, 4463–4470. doi: 10.1007/s00436-014-4131-2
Kuzmina, T. A., Spraker, T. R., Kudlai, O., Lisitsyna, O. I., Zabludovskaja, S. O., Karbowiak, G., et al. (2018). Metazoan parasites of California sea lions (Zalophus californianus): a new data and review. Int. J. Parasitol. Parasites Wildl. 7, 326–334. doi: 10.1016/j.ijppaw.2018.09.001
Laake, J. J., Lowry, M. S., DeLong, R. L., Melin, S. R., and Carretta, J. V. (2018). Population growth and status of California sea lions. J. Wildl. Managm. 82, 583–596. doi: 10.1002/jwmg.21405
Lanfear, R., Calcott, B., Ho, S. Y. W., and Guindon, S. (2012). PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol. Biol. Evol. 29, 1695–1701. doi: 10.1093/molbev/mss020
Leigh, J. W., and Bryant, D. (2015). POPART: full-feature software for haplotype network construction. Methods Ecol. Evol. 6, 1110–1116. doi: 10.1111/2041-210X.12410
Li, L., Lü, L., Nadler, S. A., Gibson, D. I., Zhang, L. P., Chen, H. X., et al. (2018). Molecular phylogeny and dating reveal a terrestrial origin in the early carboniferous for ascaridoid nematodes. Syst. Biol. 67, 888–900. doi: 10.1093/sysbio/syy018
Librado, P., and Rozas, J. (2009). DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452. doi: 10.1093/bioinformatics/btp187
Liu, G.-H., Nadler, S. A., Liu, S.-S., Podolska, M., D’Amelio, S., Shao, R., et al. (2016). Mitochondrial phylogenomics yields strongly supported hypotheses for ascaridomorph nematodes. Sci. Rep. 6:32948.
Liu, S. K., and Edward, A. G. (1971). Gastric ulcers associated with Contracaecum spp. (Nematoda: Ascaroidea) in a Steller sea lion and a white pelican. J. Wildl. Dis. 7, 266–271. doi: 10.7589/0090-3558-7.4.266
Mancia, A., Ryan, J. C., Chapman, R. W., Wu, Q., Warr, G. W., Gulland, F. M. D., et al. (2012). Health status, infection and disease in California sea lions (Zalophus californianus) studied using a canine microarray platform and machine-learning approaches. Dev. Comp. Immunol. 36, 629–637. doi: 10.1016/j.dci.2011.10.011
Marigo, J., Taniwaki, S. A., Pinto, P. L. S., Soares, R. M., and Catao-Dias, J. L. (2015). Molecular identification of Pseudoterranova azarasi larvae in cod (Gadus sp.) sold for human consumption in Brazil. Rev. Inst. Med. Trop. Sao Paulo 57, 537–539. doi: 10.1590/s0036-46652015000600015
Masper, A., Gallo-Reynoso-Reynoso, J. P., and Cisneros-Mata, M. Á, and García-Hernández, J. (2019). Review of California sea lion (Zalophus californianus) abundance, and population dynamics in the Gulf of California. Rev. Biol. Trop. 67, 833–849. doi: 10.15517/rbt.v67i4.35965
Mattiucci, S., Cianchi, R., Nascetti, G., Paggi, L., Sardella, N., Timi, J., et al. (2003). Genetic evidence for two sibling species within Contracaecum ogmorhini Johnston & Mawson, 1941 (Nematoda: Anisakidae) from otariid seals of boreal and austral regions. Syst. Parasitol. 54, 13–23. doi: 10.1023/A:1022145926409
Mattiucci, S., Cipriani, P., Webb, S. C., Paoletti, M., Marcer, F., Bellisario, B., et al. (2014). Genetic and morphological approaches distinguish the three sibling species of the Anisakis simplex species complex, with a species designation as Anisakis berlandi n. sp. for A. simplex sp. C (Nematoda: Anisakidae). J. Parasitol. 100, 199–214. doi: 10.1645/12-120.1
Mattiucci, S., Fazii, P., De Rosa, A., Paoletti, M., Megna, A. S., Glielmo, A., et al. (2013). Anisakiasis and gastroallergic reactions associated with Anisakis pegreffii infection. Italy. Emerg. Infect. Dis. 19, 496–499. doi: 10.3201/eid1903.121017
Migaki, G., van Dyke, D., and Hubbardei, R. C. (1971). Some histopathological lesions caused by helminths in marine mammals. J. Wildl. Dis. 7, 281–289. doi: 10.7589/0090-3558-7.4.281
Mladineo, I., Bušelić, I., Hrabar, J., Vrbatović, A., and Radonić, I. (2017a). Population parameters and mito-nuclear mosaicism of Anisakis spp. in the Adriatic Sea. Mol. Biochem. Parasitol. 212, 46–54. doi: 10.1016/j.molbiopara.2017.01.003
Mladineo, I., and Poljak, V. (2014). Ecology and genetic structure of zoonotic Anisakis spp. from adriatic commercial fish species. Appl. Environ. Microbiol. 80, 1281–1290. doi: 10.1128/AEM.03561-13
Mladineo, I., Trumbić, Ž, Radonić, I., Vrbatović, A., Hrabar, J., and Bušelić, I. (2017b). Anisakis simplex complex: ecological significance of recombinant genotypes in an allopatric area of the Adriatic Sea inferred by genome-derived simple sequence repeats. Int. J. Parasitol. 47, 215–223. doi: 10.1016/j.ijpara.2016.11.003
Nadler, S. A., D’Amelio, S., Dailey, M. D., Paggi, L., Siu, S., and Sakanari, J. A. (2005). Molecular phylogenetics and diagnosis of Anisakis, Pseudoterranova, and Contracaecum from Northern Pacific marine mammals. J. Parasitol. 91, 1413–1429. doi: 10.1645/ge-522r.1
Nadler, S. A., and Hudspeth, D. S. S. (1998). Ribosomal DNA and phylogeny of the Ascaridoidea (Nemata: Secernentea): implications for morphological evolution and classification. Mol. Phylogenet. Evol. 10, 221–236. doi: 10.1006/mpev.1998.0514
Nadler, S. A., and Hudspeth, D. S. S. (2000). Phylogeny of the Ascaridoidea (Nematoda: Ascaridida) based on three genes and morphology: hypotheses of structural and sequence evolution. J. Parasitol. 86, 380–393. doi: 10.2307/3284785
Nejsum, P., Hawash, M. B. F., Betson, M., Stothard, J. R., Gasser, R. B., and Andersen, L. O. (2017). Ascaris phylogeny based on multiple whole mtDNA genomes. Infect. Genet. Evol. 48, 4–9. doi: 10.1016/j.meegid.2016.12.003
Neuwirth, E. (2014). RColorBrewer: ColorBrewer Palettes. R Package Version 1.1-2. Available online at: https://cran.r-project.org/package=RColorBrewer.
NOAA (2018). California sea lion (Zalophus californianus). Available online at: https://www.fisheries.noaa.gov/species/california-sea-lion
Raga, J. A., Balbuena, J. A., Aznar, J., and Fernández, M. (1998). The impact of parasites on marine mammals: a review. Parassitologia 39, 293–296.
Rajan, T. V., Ganley, L., Paciorkowski, N., Spencer, L., Klei, T. R., and Shultz, L. D. (2002). Brugian infections in the peritoneal cavities of laboratory mice: kinetics of infection and cellular responses. Exp. Parasitol. 100, 235–247. doi: 10.1016/S0014-4894(02)00015-2
Rambaut, A., and Drummond, A. (2007). Tracer v1.4. Available online at: http://beast.bio.ed.ac.uk/Tracer
Reisfeld, L., Sacristan, C., Sanchez-Sarmiento, A. M., Costa-Silva, S., Diaz-Delgado, J., Groch, K. R., et al. (2019). Fatal pulmonary parafilaroidiasis in a free-ranging subantarctic fur seal (Arctocephalus tropicalis) coinfected with two gammaherpesviruses and Sarcocystis sp. Braz. J. Vet. Parasitol. 28, 499–503. doi: 10.1590/s1984-29612019029
Rhyan, J., Garner, M., Spraker, T., Lambourn, D., and Cheville, N. (2018). Brucella pinnipedialis in lungworms Parafilaroides sp. And Pacific harbor seals Phoca vituline Richards: proposed pathogenesis. Dis. Aquat. Org. 131, 87–94. doi: 10.3354/dao03291
Ronquist, F., Teslenko, M., Van Der Mark, P., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). Mrbayes 3.2: efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Roy, N. C., Bermingham, E. N., Sutherland, I. A., and McNabb, W. C. (2003). Nematodes and nutrient partitioning. Aust. J. Exp. Agric. 43, 1419–1426. doi: 10.1071/ea02224
Schneider, S., and Excoffier, L. (1999). Estimation of past demographic parameters from the distribution of pairwise differences when the mutation rates vary among sites: application to human mitochondrial DNA. Genetics 152, 1079–1089.
Seguel, M., Munoz, F., Navarrete, M. J., Paredes, E., Howerth, E., and Gottdenker, N. (2017). Hookworm infection in South American fur seal (Arctocephalus australis) pups: pathology and factors associated with host tissue damage and mortality. Vet. Pathol. 54, 288–297. doi: 10.1177/0300985816677151
Slatkin, M., and Hudson, R. R. (1991). Pairwise comparisons of mitochondrial DNA sequences in stable and exponentially growing populations. Genetics 129, 555–562. doi: 10.1093/genetics/129.2.555
Søe, M. J., Kapel, C. M. O., and Nejsum, P. (2016). Ascaris from humans and pigs appear to be reproductively isolated species. PLoS Negl. Trop. Dis. 10:e0004855. doi: 10.1371/journal.pntd.0004855
Spraker, T. R., DeLong, R. L., Lyons, E. T., and Melin, S. R. (2007). Hookworm enteritis with bacteremia in California sea lion pups on San Miguel Island. J. Wildl. Dis. 43, 179–188. doi: 10.7589/0090-3558-43.2.179
Spraker, T. R., Lyons, E. T., DeLong, E. T., and Zink, R. R. (2004). Penetration of the small intestine of a California sea lion (Zalophus californianus) pup by adult hookworms (Uncinaria spp.) (2004). Parasitol. Res. 92, 436–438. doi: 10.1007/s00436-003-1050-z
Spraker, T. R., Lyons, E. T., Tolliver, S. C., and Bair, H. D. (2003). Ascaridoid nematodes and associated lesions in stomachs of subadult male northern fur seals (Callorhinus ursinus) on St. Paul Island, Alaska: (1987-1999). J. Vet. Diagnostic Investig. 15, 432–437. doi: 10.1177/104063870301500505
Stroud, R. K., and Dailey, M. D. (1978). Parasites and associated pathology observed in pinnipeds stranded along the Oregon coast. J. Wildl. Dis. 14, 292–298. doi: 10.7589/0090-3558-14.3.292
Sweeney, J. C., and Gilmartin, W. G. (1974). Survey of diseases in free-living California sea lions. J. Wildl. Dis. 10, 370–376. doi: 10.7589/0090-3558-10.4.370
Swofford, D. L. (2002). PAUP∗: Phylogenetic Analysis Using Parsimony (*and other methods). Sunderland, MA: Sinauer Associates.
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Timi, J. T., Paoletti, M., Cimmaruta, R., Lanfranchi, A. L., Alarcos, A. J., Garbin, L., et al. (2014). Molecular identification, morphological characterization and new insights into the ecology of larval Pseudoterranova cattani in fishes from the Argentine coast with its differentiation from the Antarctic species, P. decipiens sp. E (NematodaAnisakidae). Vet. Parasitol. 199, 59–72. doi: 10.1016/j.vetpar.2013.09.033
Valentini, A., Mattiucci, S., Bondanelli, P., Webb, S. C., Mignucci-Giannone, A. A., Colom-Llavina, M. M., et al. (2006). Genetic relationships among Anisakis species (Nematoda: Anisakidae) inferred from mitochondrial cox2 sequences, and comparison with allozyme data. J. Parasitol. 92, 156–166. doi: 10.1645/GE-3504.1
van der Elst, R., and Everett, B. (eds) (2015). Offshore Fisheries of the Southwest Indian Ocean: Their Status and the Impact on Vulnerable Species. Durban: Oceanographic Research Institute.
Vanfleteren, J. R., Van De Peer, Y., Blaxter, M. L., Tweedie, S. A. R., Trotman, C., Lu, L., et al. (1994). Molecular genealogy of some nematode taxa as based on cytochrome c and globin amino acid sequences. Mol. Phylogenet. Evol. 3, 92–101. doi: 10.1006/mpev.1994.1012
Walker, M., Head, M. J., Berkelhammer, M., Björck, S., Cheng, H., Cwynar, L., et al. (2018). Formal ratification of the subdivision of the Holocene Series/Epoch (Quaternary System/Period): two new Global Boundary Stratotype Sections and Points (GSSPs) and three new stages/subseries. Episodes 41, 213–223. doi: 10.18814/epiiugs/2018/018016
Wickham, H. (2016). ggplot2: Elegant Graphics for Data Analysis, 2nd Edn. New York, NY: Springer-Verlag New York.
Keywords: Anisakis, Contracaecum, California sea lion, phylogenetics, pathology, Pseudoterranova, Zalophus californianus
Citation: Hrabar J, Smodlaka H, Rasouli-Dogaheh S, Petrić M, Trumbić Ž, Palmer L, Sakamaki K, Pavelin T and Mladineo I (2021) Phylogeny and Pathology of Anisakids Parasitizing Stranded California Sea Lions (Zalophus californianus) in Southern California. Front. Mar. Sci. 8:636626. doi: 10.3389/fmars.2021.636626
Received: 03 December 2020; Accepted: 08 February 2021;
Published: 08 March 2021.
Edited by:
Christian Marcelo Ibáñez, Andres Bello University, ChileReviewed by:
Serena Cavallero, Sapienza University of Rome, ItalyFrances Gulland, University of California, Davis, United States
M. Roberto García-Huidobro, Santo Tomás University, Chile
Copyright © 2021 Hrabar, Smodlaka, Rasouli-Dogaheh, Petrić, Trumbić, Palmer, Sakamaki, Pavelin and Mladineo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ivona Mladineo, ivona.mladineo@paru.cas.cz
†These authors have contributed equally to this work and share first authorship
 Jerko Hrabar
Jerko Hrabar Hrvoje Smodlaka
Hrvoje Smodlaka Somayeh Rasouli-Dogaheh
Somayeh Rasouli-Dogaheh Mirela Petrić4
Mirela Petrić4  Željka Trumbić
Željka Trumbić Ivona Mladineo
Ivona Mladineo