Biogeography and Connectivity Across Habitat Types and Geographical Scales in Pacific Abyssal Scavenging Amphipods
- 1Life Sciences Department, Natural History Museum, London, United Kingdom
- 2Department of Marine Sciences, University of Gothenburg, Gothenburg, Sweden
- 3NORCE Norwegian Research Centre, Bergen, Norway
- 4National Oceanography Centre, Southampton, United Kingdom
- 5Department of Oceanography, University of Hawai’i at Mānoa, Honolulu, HI, United States
- 6Monterey Bay Aquarium Research Institute, Moss Landing, CA, United States
- 7School of Biological and Marine Sciences, University of Plymouth, Plymouth, United Kingdom
- 8Departamento de Biodiversidad, Ecología y Evolución, Universidad Complutense de Madrid, Madrid, Spain
- 9Departamento de Ciencias de la Vida, EU-US Marine Biodiversity Group, Universidad de Alcalá, Alcalá de Henares, Spain
Recently, there has been a resurgent interest in the exploration of deep-sea mineral deposits, particularly polymetallic nodules in the Clarion-Clipperton Zone (CCZ), central Pacific. Accurate environmental impact assessment is critical to the effective management of a new industry and depends on a sound understanding of species taxonomy, biogeography, and connectivity across a range of scales. Connectivity is a particularly important parameter in determining ecosystem resilience, as it helps to define the ability of a system to recover post-impact. Scavenging amphipods in the superfamilies Alicelloidea Lowry and De Broyer, 2008 and Lysianassoidea Dana, 1849 contribute to a unique and abundant scavenging community in abyssal ecosystems. They are relatively easy to sample and in recent years have become the target of several molecular and taxonomic studies, but are poorly studied in the CCZ. Here, a molecular approach is used to identify and delimit species, and to investigate evolutionary relationships of scavenging amphipods from both abyssal plain and deep (>3000 m) seamount habitats in three APEIs (Areas of Particular Environmental Interest, i.e., designated conservation areas) in the western CCZ. A total of 17 different morphospecies of scavenging amphipods were identified, which include at least 30 genetic species delimited by a fragment of the cytochrome c oxidase subunit I (COI) barcode gene. The scavenging communities sampled in the western CCZ included the most common species (Abyssorchomene gerulicorbis (Shulenberger and Barnard, 1976), A. chevreuxi (Stebbing, 1906), Paralicella caperesca Shulenberger and Barnard, 1976, and P. tenuipes Chevreux, 1908) reported for other ocean basins. Only four morphospecies, representing five genetic species, were shared between APEIs 1, 4, and 7. The two abyssal plain sites at APEIs 4 and 7 were dominated by two and three of the most common scavenging species, respectively, while the APEI 1 seamount site was dominated by two species potentially new to science that appeared to be endemic to the site. The presence of common species in all sites and high genetic diversity, yet little geographic structuring, indicate connectivity over evolutionary time scales between the areas, which span about 1500 km. Similar to recent studies, the differences in amphipod assemblages found between the seamount and abyssal sites suggest that ecological conditions on seamounts generate distinct community compositions.
Introduction
For decades, it has been known that large deposits of metals such as nickel, iron, copper, cobalt, and manganese, in the form of polymetallic nodules exist in the abyssal deep sea (Mewes et al., 2014). However, with an increasing demand for these metals to fuel high-technology industry, deep-sea mining is now being more actively considered (Hein et al., 2020). The Clarion-Clipperton Zone (CCZ), located in the central Pacific, has one of the largest known deposits of polymetallic nodules and deep-sea mining exploration in the area has greatly increased in the last decade. The seafloor minerals in the CCZ lie in areas beyond national jurisdiction and are managed by the International Seabed Authority (ISA), which has granted 18 exploration contracts to date (ISA, 2021). In addition, the ISA has implemented a regional environmental management plan, provisionally designating nine representative areas where mining activities will be prohibited (Areas of Particular Environmental Interest, APEI) to preserve the biodiversity and ecosystem functions across the region (Wedding et al., 2013). Yet, their effectiveness depends on several key ecological criteria, particularly the degree to which APEI community compositions are representative of mining contract areas and whether they could maintain regional connectivity in the event of mining-related habitat loss.
Connectivity is a particularly important parameter in determining ecosystem resilience, as it helps to define the ability of a system to recover post-impact (Gollner et al., 2017). Thus, connectivity studies are critical for the effective management of nodule fields targeted for deep-sea mining. Many biodiversity baseline surveys have been conducted in the CCZ (Glover et al., 2015). However, most of these have focused on surveying contract areas, which is an ISA regulatory requirement, with the APEIs remaining largely unexplored. Moreover, while most surveys have aimed to describe general patterns of biodiversity and community structure, our understanding of species ranges and genetic connectivity is still very limited. To date, only two studies have investigated genetic connectivity patterns in the CCZ (Taboada et al., 2018; Janssen et al., 2019), with data restricted to the eastern CCZ. However, larger scale studies spanning the entire CCZ and adjacent areas are obviously required to fully understand species ranges and connectivity.
Necrophagous amphipods in the superfamiles Alicelloidea Lowry and De Broyer, 2008 and Lysianassoidea Dana, 1849, could provide insight into connectivity patterns for highly mobile, demersal species in the CCZ given their ubiquity in bathyal and abyssal ecosystems (Lacey et al., 2016). Scavenging amphipods can be easily collected in large numbers using baited traps, which have been in use since 1888 (Horton and Thurston, 2014). They are highly motile, with some deep-sea species within the genus Paralicella Chevreux, 1908 showing remarkably high and reciprocal pan-Pacific migration between hadal trench populations (Ritchie et al., 2017). Many deep-sea scavenging amphipod species were previously considered to be cosmopolitan (Christiansen et al., 1990), with large distributional and bathymetric ranges, occurring in bathyal, abyssal, and hadal zones. More recently, genetic diversity studies have uncovered complexes of cryptic species within these “cosmopolitan” taxa (Havermans, 2016), revealing biogeographic patterns and bathymetric structuring within the deep sea (Havermans et al., 2013; Lacey et al., 2016).
Although scavenging amphipods are generally well studied at shallower depths (Sainte-Marie, 1992), they remain little studied in the abyssal CCZ. Two recent studies have looked into the diversity of this group, using morphological and molecular data, in contractor areas and APEIs in the eastern CCZ (Patel et al., 2020; Mohrbeck et al., 2021). In our study, the DeepCCZ expedition targeted APEIs in the western CCZ, which remains largely unexplored. During the expedition in May–June 2018, on board the R/V Kilo Moana, baited traps were deployed in APEIs 1, 4, and 7, collecting more than 5,000 specimens of scavenging amphipods. Here, the biodiversity and connectivity patterns of necrophagous amphipods within these areas, spanning approximately 1500 km, are explored. Species delimitation is performed using both morphological and genetic data, and phylogenetic relationships are inferred. Additionally, the biodiversity of assemblages from the three sites is described, and connectivity between these sites, across the CCZ, and amongst other abyssal regions is inferred.
Materials and Methods
Study Sites, Specimen Sampling and Processing
APEIs 1, 4, and 7 are the westernmost APEIs (Figure 1), and were surveyed during the DeepCCZ expedition on board the R/V Kilo Moana in May–June 2018. Scavenging amphipods were collected using a large baited trap (∼1.2 m wide, 1 m high, 2 m long), with two PVC tube traps (10” diameter with funneled entrances; Leitner et al., 2017) inside, specifically designed to trap necrophagous amphipods. The PVC tube traps were baited with ∼1 kg of chopped Pacific mackerel (Scomber japonicus). Baited traps were deployed five times for a minimum of 22 h (22.7–47.1 h) on benthic landers at less than 50 cm above the seafloor at four different sites: APEI 1 seamount (deployment KM1808-074: 4175 m depth, 22.7 h), APEI 4 abyssal plain (deployment KM1808-052: 4872 m depth, 43.4 h), twice on APEI 7 abyssal plain (deployment KM1808-007: 4871 m depth, 30.7 h; and deployment KM1808-018: 4871 m depth, 47.1 h), and APEI 7 seamount (deployment KM1808-032: 3203 m, 25.1 h) (Leitner et al., 2017). More than 5700 specimens were collected during the four deployments (see Figure 1), but unfortunately only a single specimen was recovered from the deployment on the seamount on APEI 7. This was likely due to the large numbers of synaphobranchid eels observed at this site, with 12 individuals recovered from this trap (Leitner et al., 2021). Amphipods from each deployment were processed separately, and a total of about 850 specimens from all traps were sorted and grouped into morphospecies at sea. These sorted specimens were photographed and preserved in 80%, non-denatured ethanol. The remaining unsorted amphipods were also preserved in 80%, non-denatured ethanol. Preserved specimens were shipped to the Natural History Museum, London for further analyses.
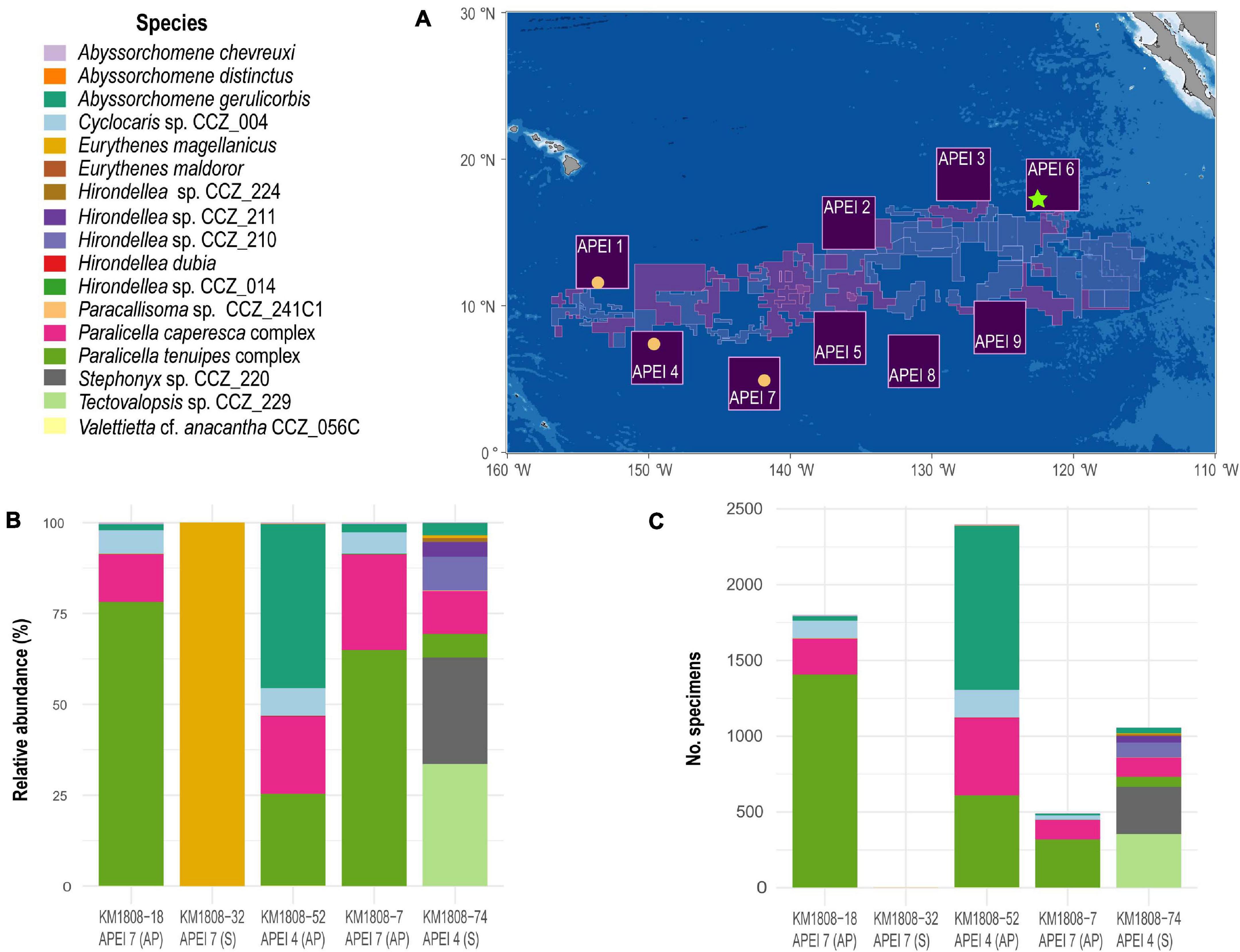
Figure 1. Map of the Clarion-Clipperton Zone indicating the Areas of Particular Environmental Interest [APEIs; (A)]. Yellow circles indicate the sites where baited-traps were deployed during the DeepCCZ expedition, with bar plots indicating the relative abundance (B) and total number of specimens collected (C) of each of the different morphospecies of scavenging amphipod recovered at each site. During deployment KM1808-32 on APEI 7, only a single specimen was recovered. The green star indicates the site where the baited trap was deployed during the JC120 expedition, and from which only a few specimens were included in this study. S, seamount, AP, abyssal plain.
An additional 36 amphipods from the APEI 6 (Figure 1), collected during the JC120 cruise on the Royal Research Ship James Cook in 2015 (Jones, 2015), were included in our genetic analyses.
Alpha-Diversity
All specimens were morphologically examined and sorted into different morphotypes using external morphological characters. Specimens were identified to the lowest taxonomic level possible, considering both morphological characters and genetic sequences. Additionally, representatives of the different morphotypes identified were morphologically examined in more detail by a taxonomic expert (TH). Taxonomic classification and authorities follow WoRMS (Horton et al., 2020b) and can be checked by following the hyperlinks in Table 1.
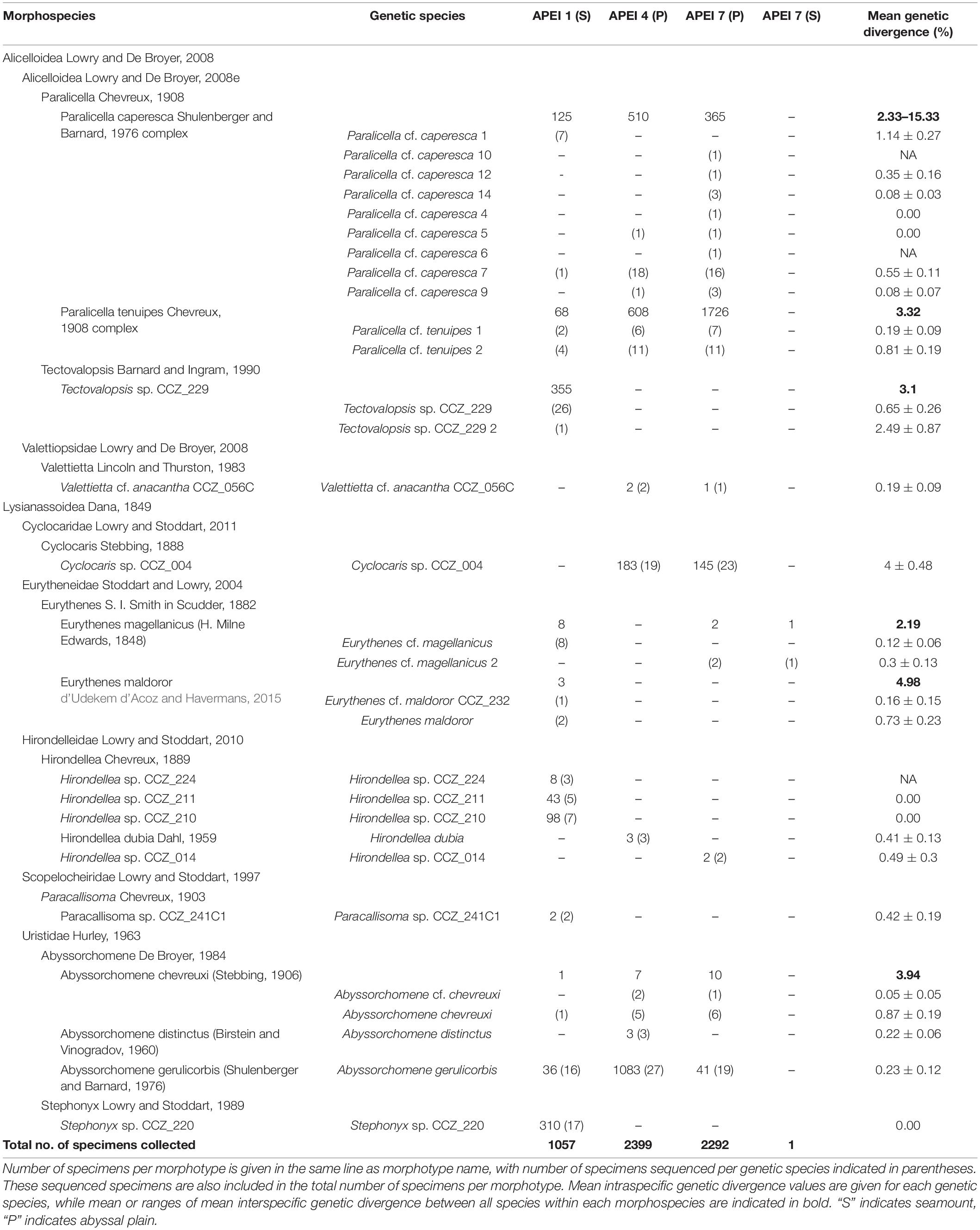
Table 1. Morphospecies and genetic species of scavenging amphipods found in the Areas of Particular Environmental Interest (APEIs) 1, 4, and 7.
Analyses were performed in R version 4.0.2 (R Development Core Team, 2017), using several packages, and all plots generated using ggplot2 (Wickham, 2009). Individual-based abundance data were used to describe alpha-diversity of morphospecies in the three APEIs. The seamount site on APEI 7 was excluded from these analyses, as only a single specimen was recovered here. Rarefaction curves were used to visualize sampling effort, and were generated using the R package vegan (Oksanen et al., 2018). Diversity was estimated as effective number of species (i.e., Hill numbers; Jost, 2006) for different orders of diversity (q), that indicate sensitivity to common and rare species, using the R package iNEXT (Hsieh et al., 2020). Diversity profiles were generated using a continuum of q values (from 0 to 5 every 0.10) to visualize the contribution of rare and abundant species in the community (See Supplementary S1). Order zero diversity (q = 0) only takes into account presence, thus representing species richness. Order one diversity (q = 1) weighs all species frequencies equally without favoring rare or common species and is equivalent to the inverse of the Shannon-Wiener entropy index. The order two diversity (q = 2) places more weight on the frequencies of abundant species and discounts rare species, equivalent to the inverse of Simpson’s concentration index.
DNA Extraction, Amplification, and Sequencing
A reverse taxonomic approach was used to improve and speed up taxonomic assignments, as well as to allow comparisons between studies for which sequences of indeterminate species have been provided. Several representatives of each morphotype were thus photographed and one or several pleopods were used for DNA extraction. DNA extraction was performed using the DNeasy Blood and Tissue Kit (Qiagen). A fragment of the barcoding gene cytochrome c oxidase subunit I (COI) was amplified for all selected specimens in order to perform single-locus species delimitation. A fragment of the genes 16S rRNA (16S) and 18S rRNA (18S) were additionally amplified for at least a single representative of each morphotype for phylogenetic inference. The PCR mix for each reaction contained 10.5 μl of Red Taq DNA Polymerase 1.1X MasterMix (VWR), 0.5 μl of each primer (10 μM) and 1 μl of DNA template. The mitochondrial COI was amplified using the LCO1490 (GGTCAACAAATCATAAAGATATTGG) and HCO2198 (TAAACTTCAGGGTGACCAAAAAATCA) primers (Folmer et al., 1994) with an initial denaturation at 95°C for 5 min, followed by 35 cycles of denaturation at 95°C for 45 s, annealing at 51°C for 45 s, and extension at 72°C for 1 min, with a final extension of 72°C for 10 min. The mitochondrial 16S was successfully amplified with the universal primers 16sar-L (CGCCTGTTTATCAAAAACAT) and 16sbr-H (CCGGTCTGAACTCAGATCACGT) (Palumbi, 1996) using an initial denaturation at 94°C for 2 min, followed by 30 cycles of denaturation at 94°C for 15 s, annealing at 55°C for 30 s, extension at 68°C for 30 s, and a final extension at 68°C for 4 min. The nuclear 18S was amplified using the primers 18SA (AYCTGGTTGATCCTGCCAGT; Medlin et al., 1988) and 18SB (ACCTTGTTACGACTTTTACTTCCTC; Nygren and Sundberg, 2003) with an initial denaturation at 95°C for 5 min, followed by 30 cycles of denaturation at 95°C for 30 s, annealing at 59°C for 30 sec, and extension at 72°C for 1 min, with a final extension at 72°C for 2 min. The primers used for sequencing were the same as those for amplifications, with an additional set of internal primers for 18S, 620F (TAAAGYTGYTGCAGTTAAA; Nygren and Sundberg, 2003) and 1324R (CGGCCATGCACCACC; Cohen et al., 1998). PCR products were purified using a Millipore Multiscreen 96-well PCR Purification System and sequenced using the forward and reverse primers mentioned above using an ABI 3730XL DNA Analyzer (Applied Biosystems) at The Natural History Museum Sequencing Facilities. For each gene fragment, contigs were assembled by aligning both forward and reverse sequences using Geneious 7.0.61; chromatograms were visually inspected and ambiguous base calls were corrected manually. Sequences were deposited on GenBank with accession numbers MZ443996–MZ444014 (16S), MZ444015–MZ444046 (18S), and MZ474200–MZ474475 (COI).
Phylogenetic Analyses
In addition to the sequences generated for this study, sequences from Genbank were included in downstream analyses (see Supplementary Table 1). Sequences for COI (276 sequences generated for this study and 482 sequences from public databases; Supplementary Table 1) were aligned using MUSCLE in MEGA X (Kumar et al., 2018); with nucleotides translated into amino acids to identify pseudogenes based on the presence of stop codons. Sequences of 16S (19 generated for this study and 55 from public databases) and 18S (32 generated for this study and 32 from public databases) were aligned using MAFFT version 7 (Katoh et al., 2019) with the iterative refinement method FFT-NS-i, and unalignable regions were filtered in Gblocks server http://molevol.cmima.csic.es/castresana/Gblocks_server.html (Castresana, 2000), allowing gap positions within final blocks and less strict flanking positions. These alignments were used in three different datasets: (1) concatenated COI, 16S, and 18S alignment of a few representatives per morphotype, (2) a reduced dataset: COI alignment from a single representative per unique haplotype, and (3) complete dataset: all COI sequences. For the concatenated dataset, individual gene-alignments were concatenated in Geneious. Also, best substitution models for each partition (each marker, and each codon position for COI) were determined using PartitionFinder (Lanfear et al., 2017).
Phylogenetic trees were estimated using partitioned maximum-likelihood (RAxML v8.2.10; Stamatakis, 2006) and Bayesian inference (BEAST v2.4.7; Bouckaert et al., 2014), with the best inferred substitution model for each partition. In RAxML, the most common substitution model for each taxon was selected, with the best maximum-likelihood (ML) tree and support values estimated from 1000 rapid Bootstrap and 20 ML searches. BEAST analyses were performed with trees and clock models linked, a Yule tree model, and relaxed clock log normal. Two independent runs of a maximum 100M steps were combined after discarding 20% as burn-in. Runs were checked for convergence and a median consensus tree was estimated from the combined post-burn-in samples.
Species Delimitation
In order to estimate the number of species of scavenging amphipods collected during the DeepCCZ expedition, single-locus species delimitation was carried out (Supplementary S3). Three different methods were considered, including both distance- and tree-based approaches, because different methods are known to yield different results on the same dataset (Dellicour and Flot, 2018). Two different datasets for species delimitation were also used, the complete COI-only dataset and the reduced COI-only dataset that included a single representative per haplotype.
The barcode gap method ABGD (Automatic Barcode Gap Discovery; Puillandre et al., 2012) is a distance-based approach that separates Molecular Operational Taxonomic Units (MOTUs), hereby referred to as species, by identifying a threshold pairwise distance known as the barcode gap. ABGD was run on both full and reduced datasets, with K80 distances, default parameters for P (P = 0.001000, 0.001668, 0.002783, 0.004642, 0.007743, 0.012915, 0.021544, 0.035938, 0.059948, and 0.100000), and different values of X (X = 0.5, 0.8, 1.0, and 1.5).
Also, two different tree-based delimitation approaches were used: single-threshold GMYC (General Mixed Yule Coalescent; Pons et al., 2006) and PTP (Poisson tree processes; Zhang et al., 2013). Both tree-based approaches require a phylogenetic tree, with GMYC requiring an ultrametric (i.e., BEAST) tree to fit models of inter- and intraspecific processes and defining the limits of these. PTP does not require an ultrametric tree, as the speciation rate is modeled directly from the number of substitutions which is reflected in the branch lengths. Herein, the most conservative approach was considered, hence basing species delimitation on results from ABGD on the full dataset. However, some clusters/singletons identified by ABGD as separate putative species were nested in well-supported clades, from both ML and BI trees. These were combined to ensure all species were monophyletic. The full dataset of 758 barcodes, including GenBank sequences, was reduced to a CCZ dataset with 585 barcodes, excluding putative species delimited that were not found in the CCZ, to facilitate visualization of results.
Genetic Diversity, Population Differentiation and Connectivity
In order to investigate genetic diversity, as well as genetic differentiation, and qualitatively assess connectivity between localities, only the six species for which there were at least 30 COI sequences available were considered. mtDNA diversity metrics including haplotype number (h), haplotype diversity (Hd), nucleotide diversity (pi), and number of segregating sites (S), were estimated for each species using the R packages pegas (Paradis, 2010) and ape (Paradis and Schliep, 2019) (See Supplementary S4). Basic statistics of genetic diversity, including the analysis of molecular variance (AMOVA; Excoffier et al., 1992) and pairwise Fixation index (Fst; Weir and Cockerham, 1984), were estimated between different localities (e.g., APEIs 1, 4, 6, and 7, UK-1, OMS) using the R packages poppr (Kamvar et al., 2014) and hierfstat (Goudet and Jombart, 2020). Fst values were not estimated for Paralicella cf. caperesca 14, as two of the haplotypes differed in a single base and the other two haplotypes defined had a missing base. Genetic connectivity was visualized using minimum-spanning tree haplotype networks built using pegas (Paradis, 2010).
Results
A total of 5749 specimens were captured during five 22 + hour deployments of baited traps, of which 1057 individual amphipods were captured on the APEI 1 seamount, 2399 on the APEI 4 abyssal plain site, 2292 (1801 plus 491 individuals from two different deployments) on the APEI 7 abyssal plain site, and a single specimen on the APEI 7 seamount site (Table 1). Thus, samples were collected from three different APEIs (APEI 1, APEI 4, and APEI 7) and from two different habitats (seamount and abyssal plain).
Alpha-Diversity and Compositional Assemblages
The 5749 amphipods recovered were morphologically identified as 17 morphospecies, belonging to seven families and nine genera (Table 1). Seven of these species are likely new to science. Four morphospecies were found in all three investigated western APEIs (Abyssorchomene gerulicorbis (Shulenberger and Barnard, 1976), A. chevreuxi (Stebbing, 1906), Paralicella caperesca Shulenberger and Barnard, 1976, and P. tenuipes Chevreux, 1908); three were found only in two APEIs, and 10 were found in a single APEI (Table 1; Figure 2). APEI 1 was the most species-rich area with 12 morphotypes recorded, and both APEIs 4 and 7 had eight. The assemblage composition of the three areas was highly dominated by a few species (Figure 1), with common deep-sea amphipod scavenger species dominating the communities in APEI 4 (Abyssorchomene gerulicorbis: 45%, Paralicella tenuipes complex: 25%, and P. caperesca complex: 21%) and APEI 7 (P. tenuipes complex: 75%, and P. caperesca complex: 16%). However, the seamount in APEI 1, the only seamount site with a large sample, was dominated by two new species that were also restricted to this site (Tectovalopsis sp. CCZ_229: 34%, Stephonyx sp. CCZ_220: 29%). The APEI 1 seamount site was the most species-rich, as well as the most diverse when accounting for the contribution of common and rare species using the inverse of Simpsons concentration (4.7 spp.), followed by APEI 4 (3.6 spp.), and APEI 7 (1.7 spp.). Diversity profiles for the three sites showed highly uneven communities (Figure 3).
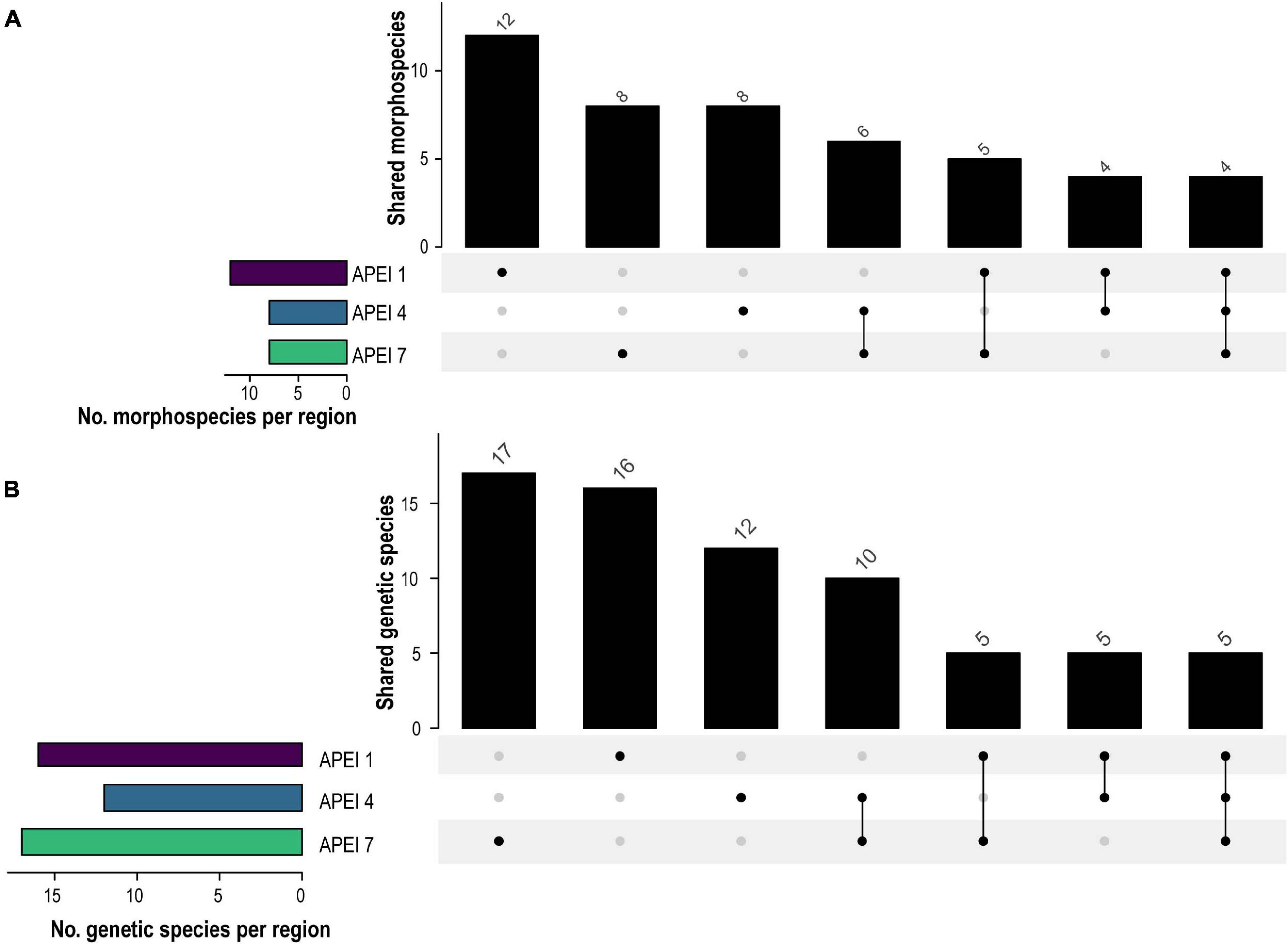
Figure 2. UpSet plots showing shared number of morphospecies (A) and genetic species (B) species between western CCZ APEIs (APEI 1, 4, and 7) in vertical bars and total number of species per site indicated by horizontal bars.
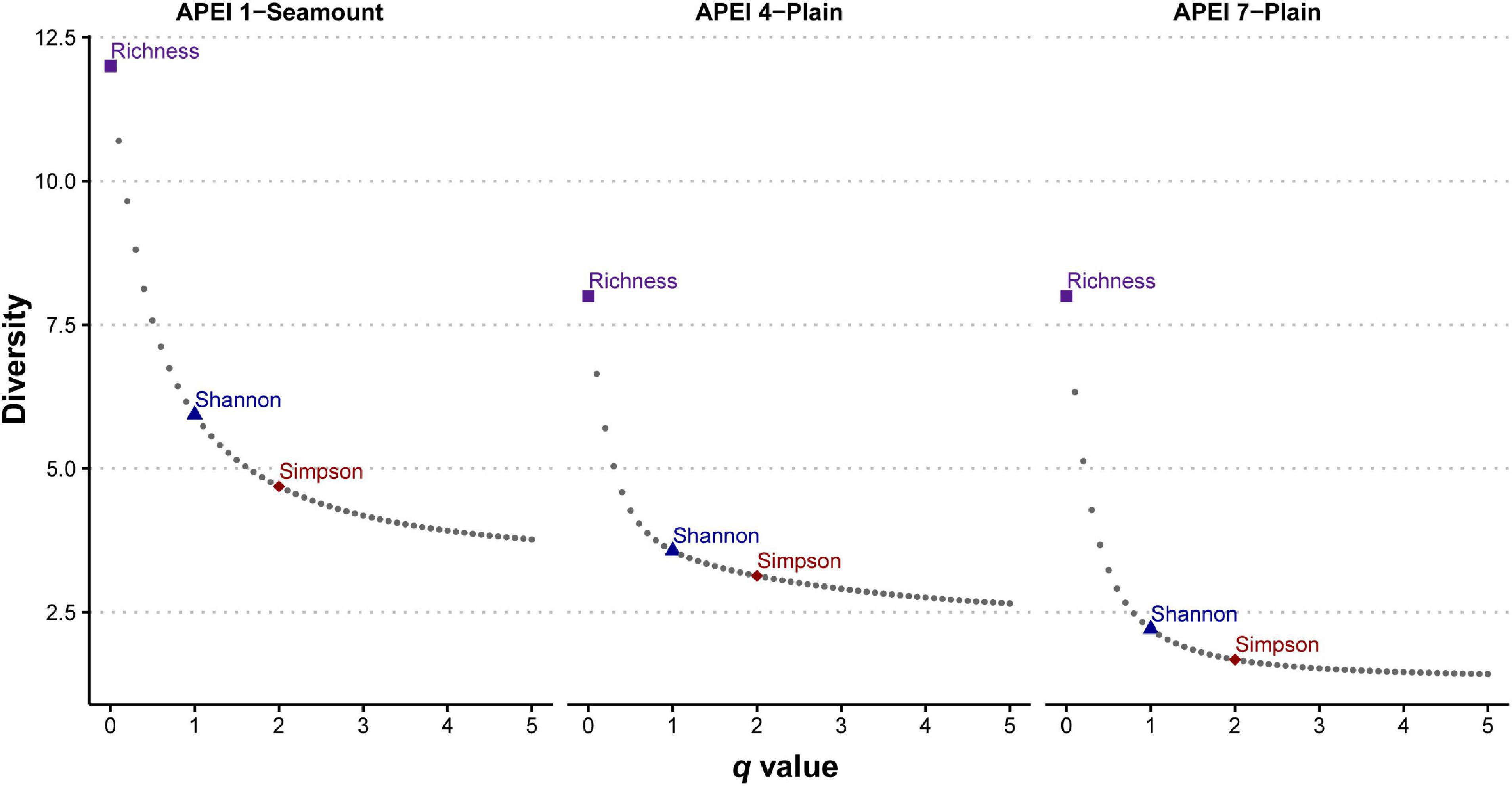
Figure 3. Diversity profile plots for scavenging amphipods in western APEIs (Area of Particular Environmental Interest) 1, 4, and 7 showing higher diversity in APEI 1 and a strong dominance of species in each community. X-axis indicates different values of q (i.e., diversity order), with y-axis indicating diversity as effective number of species (i.e., Hill numbers). Values of q = 0, q = 1, and q = 2 are indicated as this correspond to values of species richness, the inverse of Shannon-Wiener entropy index, and the inverse of Simpson’s concentration index, respectively.
There were two deployments on the abyssal plain in APEI 7, KM1808-007, and KM1808-018, with different bottom times of 30.7 and 47.1 h, respectively. The number of morphospecies recovered was higher (8) in the deployment with the longest bottom time than in deployment KM1808-007 with shorter bottom time (6 morphospecies; Figure 1, Supplementary S1). The number of specimens was also higher in the trap with longest bottom time (1801 vs. 491), and this general trend was also observed for the other deployments, except KM1808-032 (Supplementary S1) which recovered a single specimen. Relative abundance of species was very similar for both deployments on the abyssal plain in APEI 7, but P. caperesca relative abundance declined (from 26.27 to 13.1%) with increased bottom time while P. tenuipes relative abundance increased (from 64.9 to 78.1%) with increased bottom time.
Phylogenetic Analyses
In this study, a total of 239 sequences of the barcoding gene COI, with average length of 612 bp, were generated from samples from the western CCZ and 36 from samples from the eastern CCZ. Nucleotide composition was T: 36.6%, C: 18.2%, A: 26.7%, and G: 18.5. In addition, 22 and 32 sequences for the 16S and 18S markers, respectively, were generated from specimens belonging to different morphotypes to infer phylogenetic relationships (Figure 4).
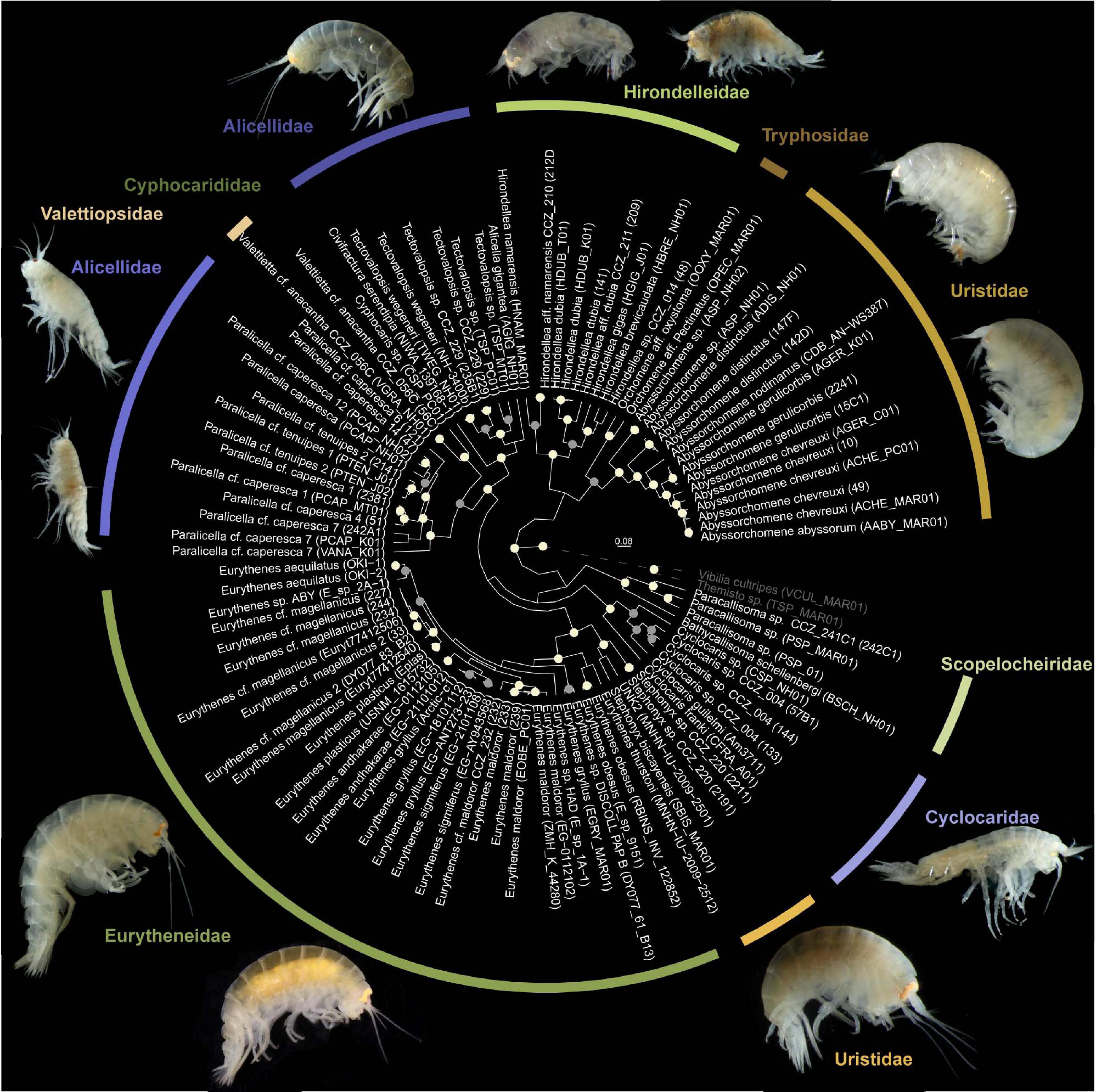
Figure 4. Bayesian phylogenetic tree from a concatenated alignment of the COI, 16S, and 18S genes of the superfamilies Alicelloidea and Lysianassoidea. Only highly supported nodes are highlighted, with posterior support values of 1.0 colored in white and ≥0.95 in gray.
The Bayesian phylogenetic tree showed a distinct clade belonging to the superfamily Alicelloidea, represented in the phylogeny by the families Alicellidae and Valettiopsidae (Figure 4). However, the family Cyphocarididae Lowry and Stoddart, 1997, belonging to the Lysianassoidea, was recovered within the Alicelloidea. Members of the superfamily Lysianassoidea were not recovered in the same clade. The family Alicellidae was not recovered as monophyletic, with Paralicella forming a separate clade from Tectovalopsis, Civifractura Weston, Peart and Jamieson, 2020 and Alicella Chevreux, 1899. The family Uristidae appeared also as paraphyletic, with Stephonyx and Abyssorchomene forming two separate clades. The phylogeny showed that Stephonyx is more closely related to Eurythenes (Eurytheneidae) than to Abyssorchomene. The latter appeared as a sister taxon to Orchomene Boeck, 1871 in the Tryphosidae Lowry and Stoddart, 1997 family.
Species Delimitation
Different species delimitation methods identified a different number of putative species within the 758 barcodes, being assigned to 35–131 putative species (ABGD: 35–41, PTP: 44–131, GMYC: 43–45; Figure 5, Table 2). The 239 COI sequences from the western CCZ were assigned to 29–80 putative species (ABGD: 29–33, PTP: 34–85, GMYC: 34–36). PTP performed poorly on the complete dataset, greatly over-splitting taxa. Although there was no clear barcode gap identified when plotting pairwise divergence distances (Supplementary S3), both ABGD and GMYC species hypotheses on the full dataset were similar. Hence, species delimitation was based on the output from ABGD as it was more conservative, and because a few species delimited by GMYC were recovered with low posterior values. Some clusters/singletons recovered as separate putative species were considered the same to ensure monophyly of all species.
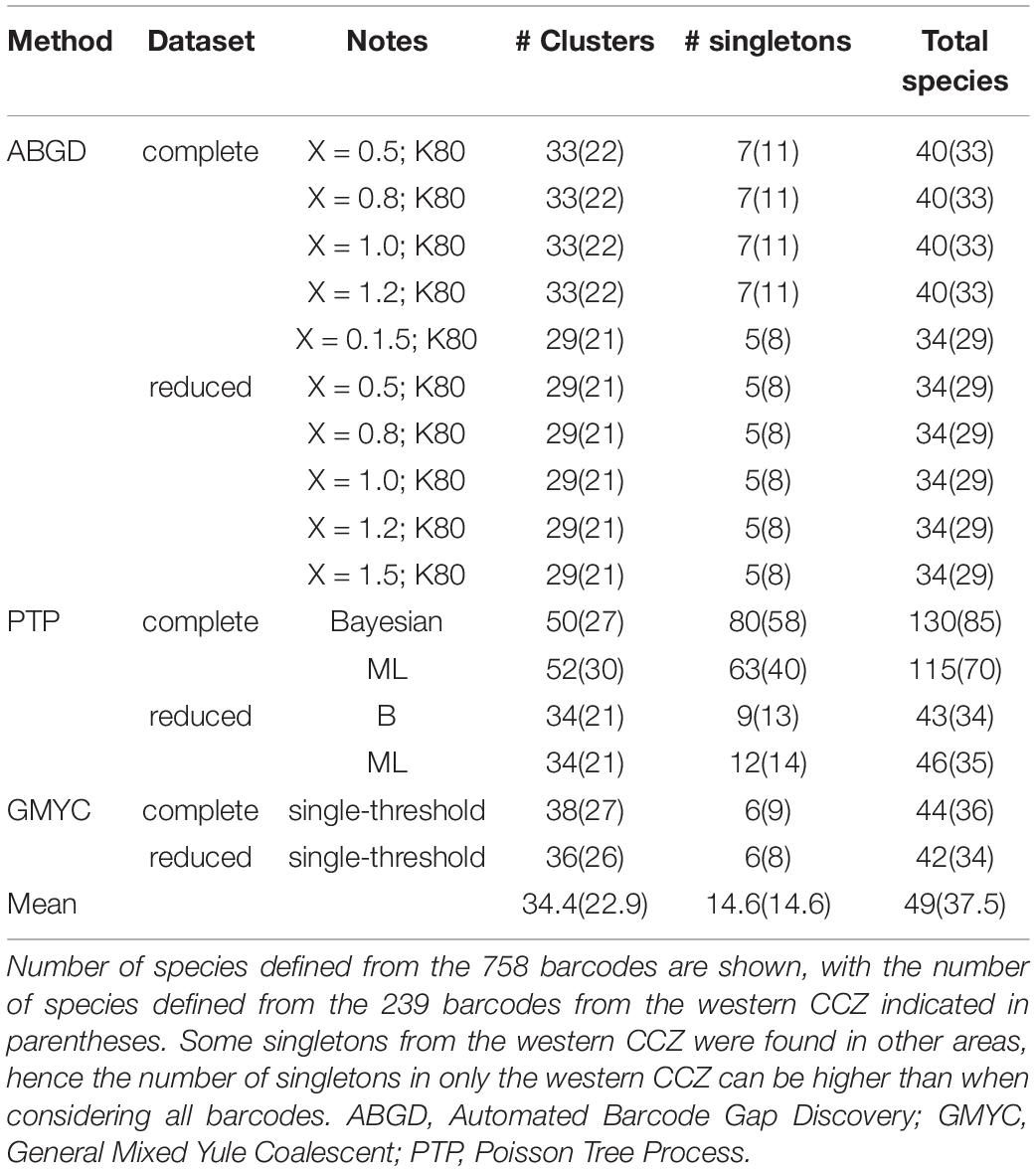
Table 2. Table indicating the 16 different approaches for species delimitation considered in this study.
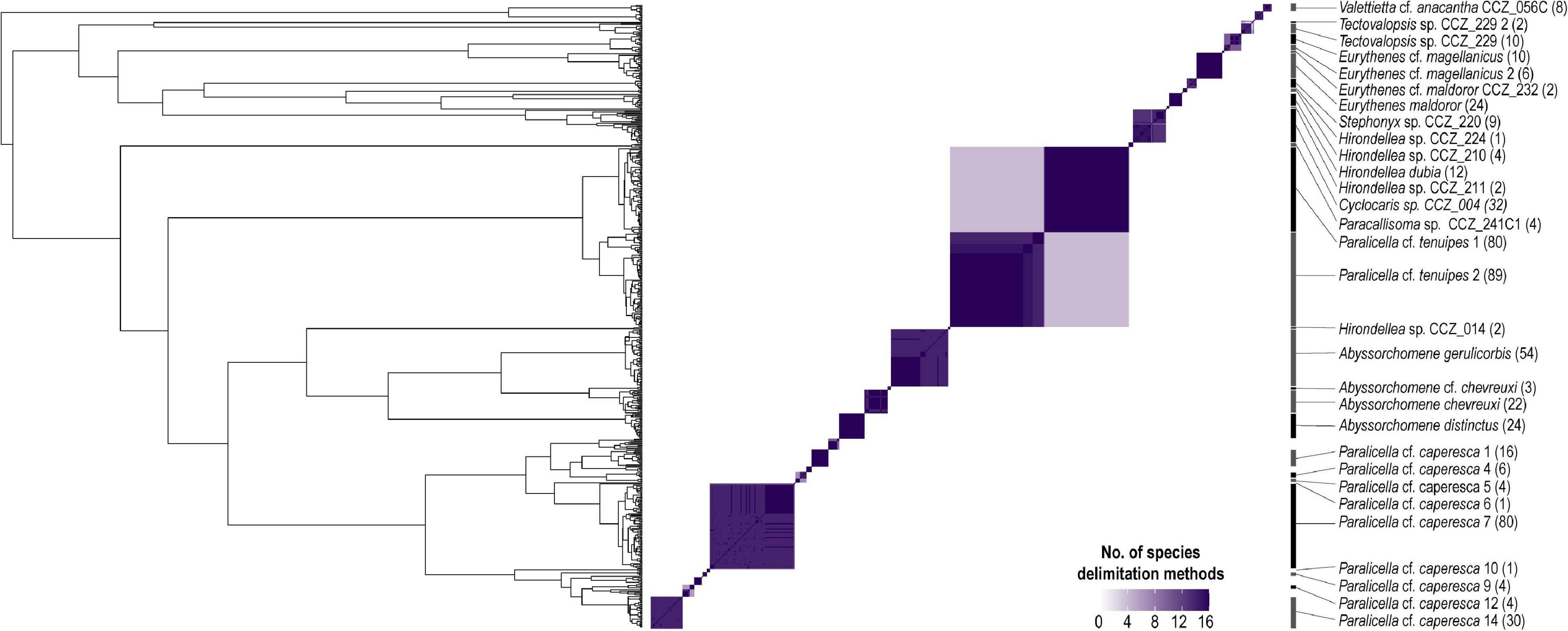
Figure 5. Bayesian phylogenetic tree from COI-only data (CCZ dataset; i.e., 585 sequences) indicating species delimitation for the 30 genetic species found to occur in the western CCZ. Tip labels representing specimens distributed in the western CCZ are highlighted. The heatmap indicates the number of species delimitation methods that recovered each different cluster, with species boundaries considered for downstream analyses indicated on the right. Number in brackets indicates number of specimens.
Mean intraspecific divergence (K2P distance) varied between putative species, ranging from 0% (e.g., Hirondellea sp. CCZ_210) to 4% (Cyclocaris sp. CCZ_004) (Table 1). Mean interspecific divergence between species within the same morphotypes ranged from 2.19% (Eurythenes magellanicus) to 15.33% (Paralicella caperesca complex) (Table 1 and Supplementary Table 2).
In total, 30 putative species of scavenging amphipods were found in the western CCZ. Considering previously published sequences from the eastern CCZ (Mohrbeck et al., 2021), and sequences generated for this study from some specimens collected during the MIDAS expedition, 11 of the species delimited were found only in the western CCZ. A total of 36 species were found to occur in the CCZ, with 20 distributed in the eastern CCZ, 14 of those spanning the entire CCZ and 3 being restricted to the eastern CCZ (the remaining two are shared with areas adjacent to the CCZ). APEI 7 had the highest species richness (17), followed by APEI 1 (16), and APEI 4 (12) (Figures 2, 5). APEI 1, the seamount site, had the highest number of unique species (6), as well as fewest species shared with other sites. The most shared species (10) occurred between both abyssal plain sites, APEIs 4 and 7.
Several morphospecies were separated into multiple genetic species (Table 1, Supplementary S5), for which further morphological analyses are needed in order to determine diagnostic characters for each group. A few of these genetic species are consistent with previous studies. Valettietta cf. anacantha was recovered as two separate clades (Figure 6), Va2 and Va1 in Mohrbeck et al. (2021). Both lineages were also recovered in our samples from the eastern CCZ, but only Va2 (V. cf. anacantha CCZ_056C) was found in the western CCZ.
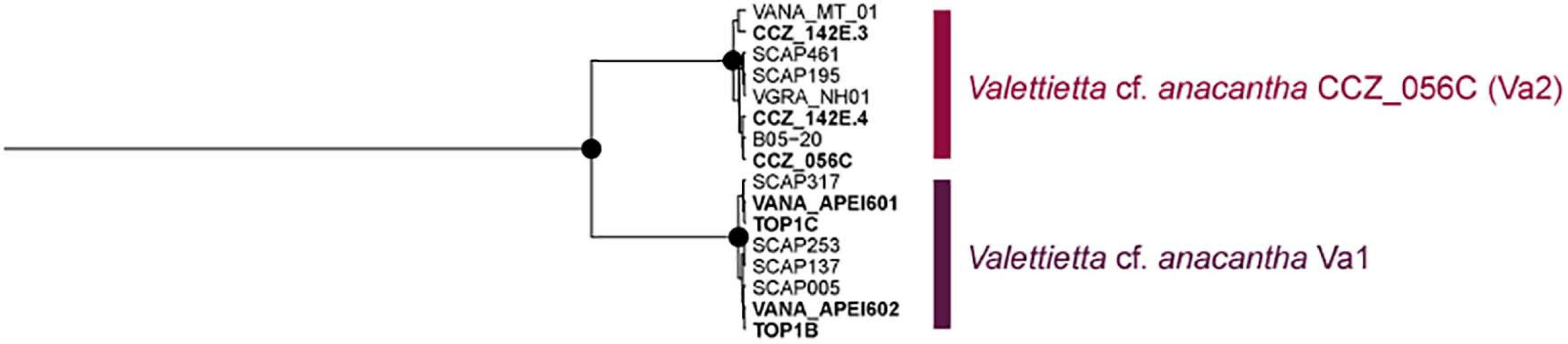
Figure 6. Bayesian COI-only phylogenetic tree for the morphospecies Valettietta cf. anacantha. The two genetic clades are indicated on the right. Node posterior support values of 1.0 are indicated as black circles. Sequences generated in this study are highlighted in bold, with western CCZ labels starting with “CCZ_”.
Eurythenes magellanicus was also recovered as two separate genetic species, both found in the western CCZ (Figure 7, Supplementary S5). Havermans (2016) identified two clades, Eg4 and Eg5, within the species, plus an additional sequence (TAI-1; Havermans et al., 2013) recovered outside of both clades. In the western CCZ, representatives of Eg4 were found, along with a clade including the sequence TAI-1, but did not recover representatives of Eg5. The clade including the sequence TAI-1 includes specimens collected in the Pacific Ocean (Sea of Okhotsk: OKI-3 (Narahara-Nakano et al., 2018), and off Taiwan: TAI-1, and the western CCZ), but also a specimen collected in the Porcupine Abyssal Plain (DY077_83_B2; Horton et al., 2020a). In a recent revision of the genus Eurythenes (d’Udekem d’Acoz and Havermans, 2015), specimens from both clades Eg4 and Eg5 were included as voucher sequences for E. magellanicus, and a detailed morphological description was provided based on a specimen from Eg5. Eurythenes maldoror was also recovered as two separate clades, both collected in the western CCZ. Specimens from the clade including the holotype sequence (ArgB-7, GenBank JX887151) were thus listed as E. maldoror, and the remaining ones as E. cf. maldoror CCZ_232.
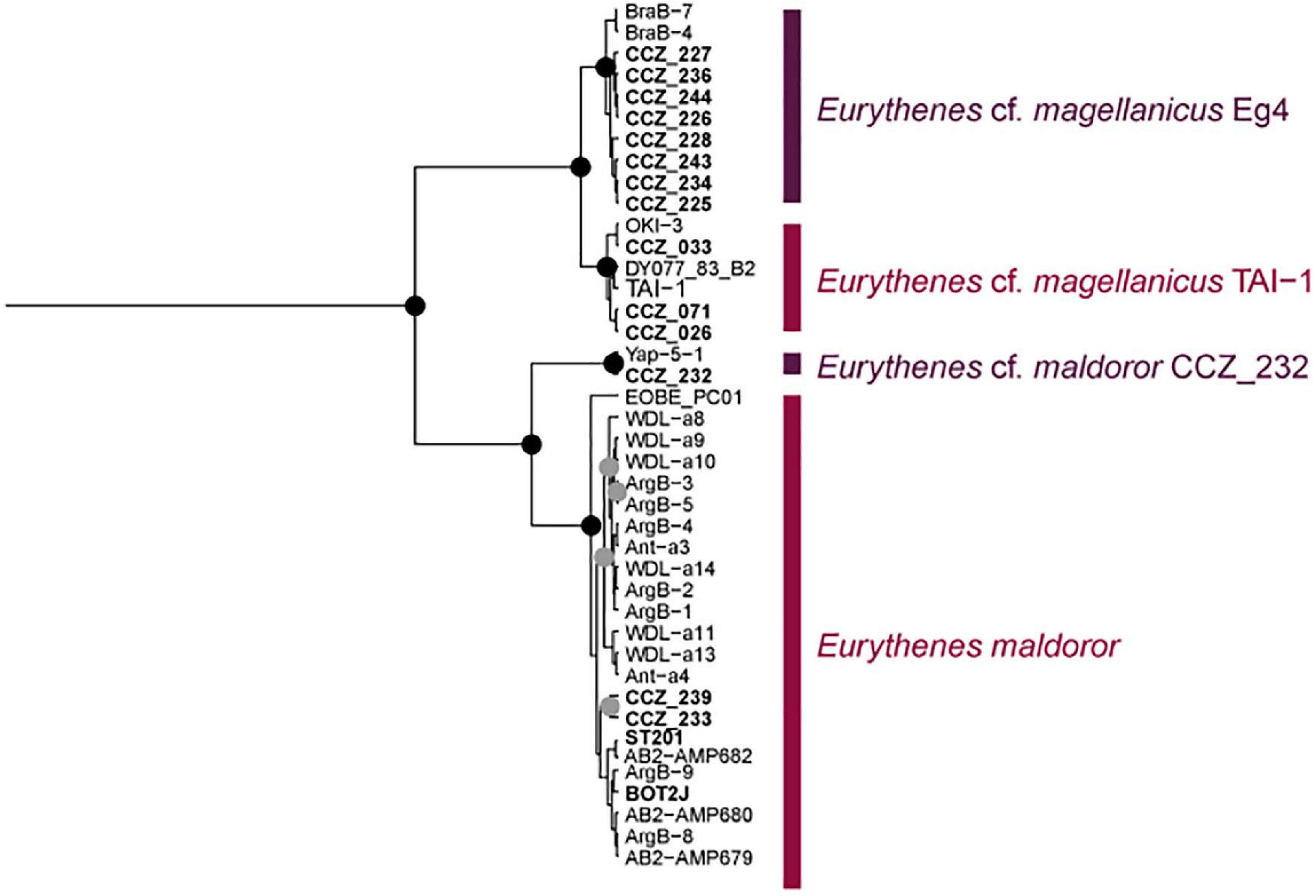
Figure 7. Bayesian COI-only phylogenetic tree for species of Eurythenes distributed in the western CCZ. Genetic species are indicated on the right. Node posterior support values of 1.0 are indicated as black circles, and values ≥ 0.95 in gray circles. Sequences generated in this study are highlighted in bold, with western CCZ labels starting with “CCZ_”.
Genetic Structuring of Scavenging Amphipods
Six species, for which there were at least 30 COI sequences available, were chosen for connectivity analyses –two species within the Paralicella caperesca complex, two species within the P. tenuipes complex, Abyssorchomene gerulicorbis, and Cyclocaris sp. CCZ_004. A total of 363 individuals were included in the analyses (154 from the western CCZ), with 77 unique haplotypes found (Table 3). These species are also distributed in the eastern CCZ, and in other regions except for a single species for which the only samples were from the CCZ (Paralicella cf. caperesca 14). Several haplotypes were found in all species, with haplotype diversity (Hd) being high for all except P. cf. caperesca 14. Cyclocaris sp. CCZ_004 had the highest Hd (0.92), with 20 different haplotypes (h) found across 31 samples.
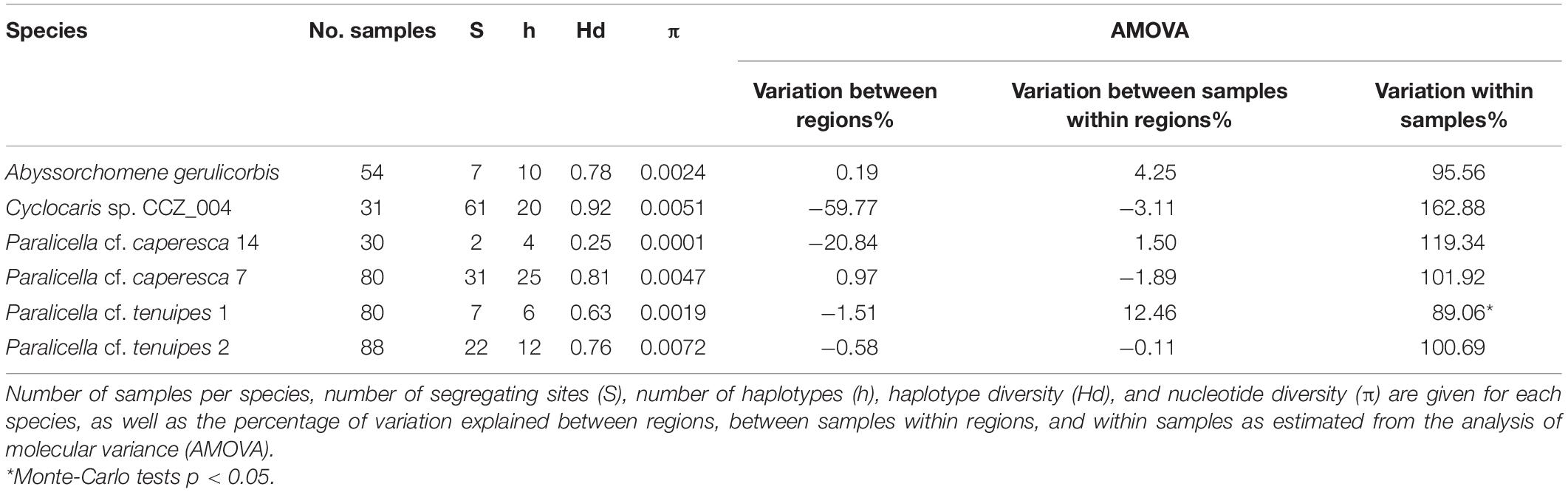
Table 3. Population genetic statistics for six genetic species of scavenging amphipods occurring in the western CCZ.
Minimum-spanning tree haplotype networks were constructed to visualize genetic differentiation between regions (Figure 8). No genetic differentiation was observed, with haplotypes being shared across different localities. This is also consistent with the results obtained from the AMOVA, with most of the variation being found within localities (>89.06%; Table 3 and Supplementary S4). Pairwise fixation index (Fst) also suggested there is no divergence between areas in the CCZ (Table 4, Supplementary S4), with very low values estimated for all species but P. cf. tenuipes 1. Fst values considered as significant for differentiating population (>0.15) were found between APEI 1-APEI 7, APEI 1-OMS, APEI 4-APEI 6, APEI 4-OMS, APEI 6-APEI 7, APEI 6-OMS, and APEI 6-UK-1.
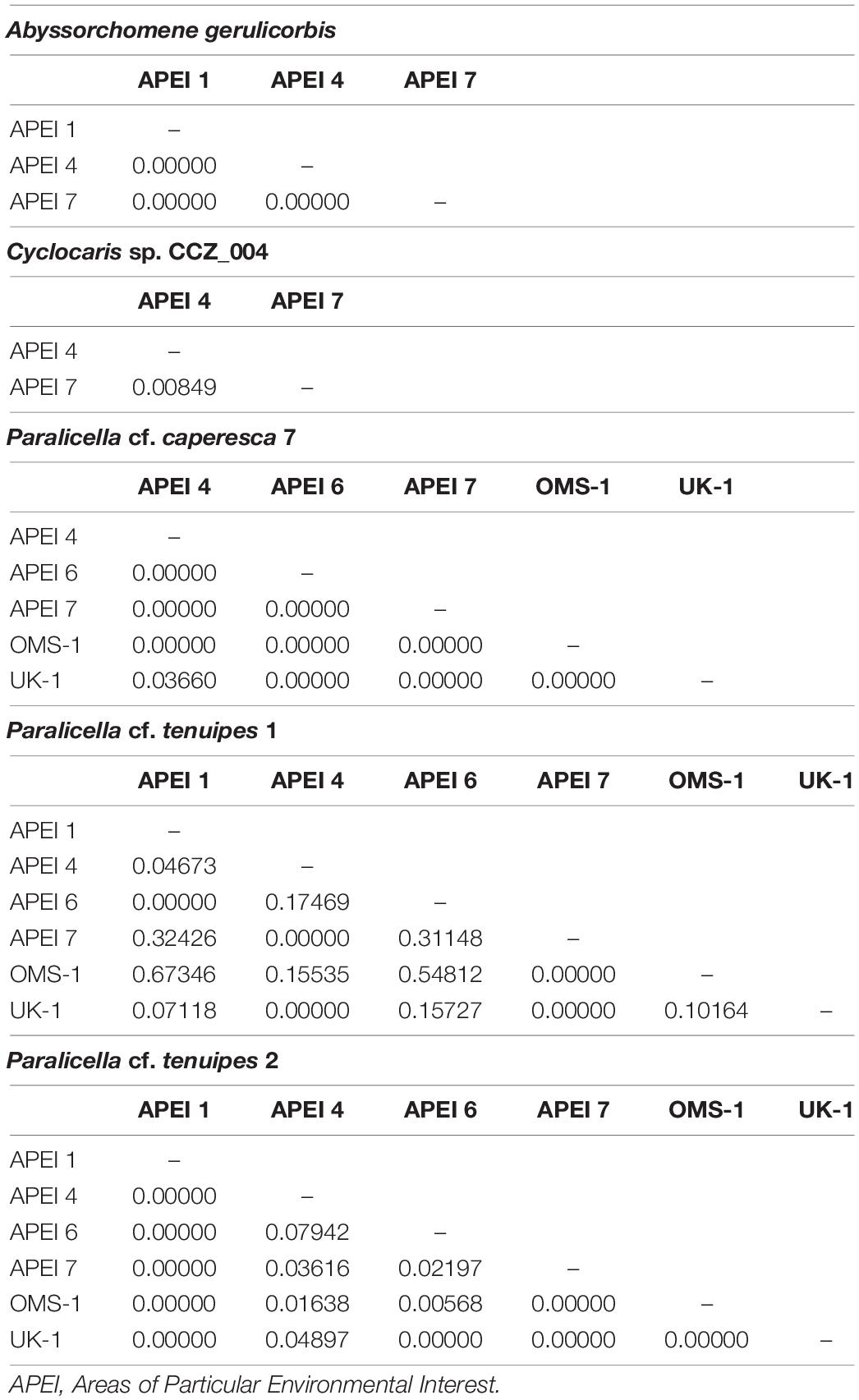
Table 4. Pairwise Fixation index (FST) for genetic species of scavenging amphipods collected in different areas within the Clarion-Clipperton Zone (CCZ).
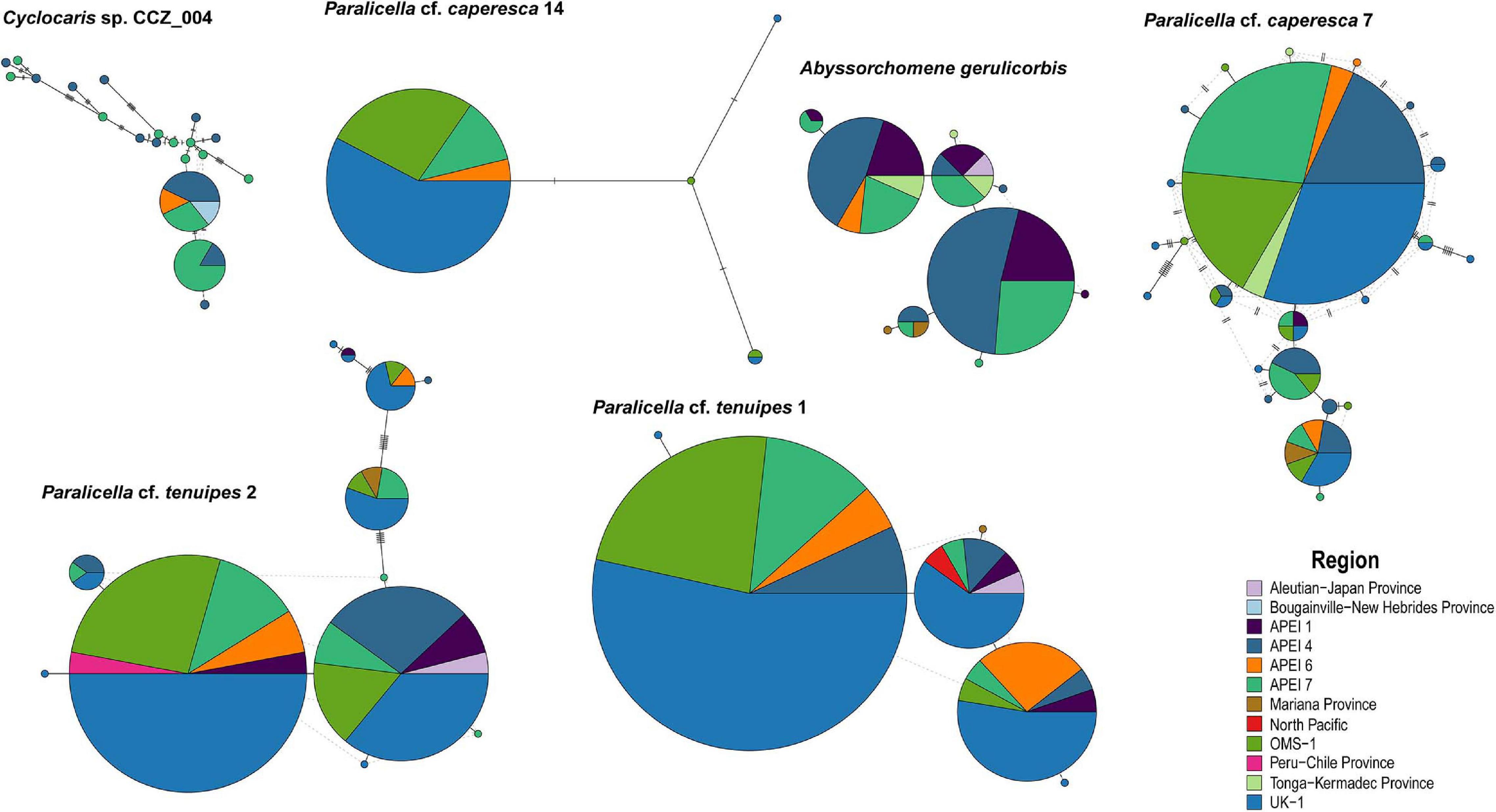
Figure 8. Minimum-spanning tree haplotype networks for COI for six species of scavenging amphipods. Circles are proportional to the number of samples and all regions are color-coded.
Discussion
With increasing interest in deep-sea mining, there is an urgent need for accurate estimates of species diversity, as well as understanding the processes generating and maintaining it, in order to inform environmental management plans. Despite the large number of scientific expeditions that have been carried out in the CCZ, only two studies have focused on necrophagous amphipods (Patel et al., 2020; Mohrbeck et al., 2021), which are important components of abyssal ecosystems. This study contributes to our understanding of the diversity, biogeography, and connectivity of bait-attending necrophagous amphipods in the CCZ and is the first carried out in the western CCZ.
Morphological Diversity of Scavenging Amphipods
The western CCZ, with 17 different morphospecies, appears to be much more species rich than the eastern CCZ. However, this apparent higher diversity could be an artifact of sampling. Although the western CCZ has sustained considerably less sampling effort, with only five baited trap deployments, sampling has encompassed a wider range of habitats (abyssal plains and seamounts), as well as larger depth ranges (3203–4872 m). Nonetheless, sampling adequacy has been evaluated using rarefaction plots for a previous study in the eastern CCZ (Patel et al., 2020), as well as herein (see Supplementary S1), with all species accumulation curves plateauing except for one CCZ station included in Patel et al. (2020). The recent study by Mohrbeck et al. (2021) included a subsample of 645 specimens and found the same species richness (10 morphospecies) for the eastern CCZ (Patel et al., 2020). Because one study was strictly morphology-based, it is not possible to determine if some of the species identified to genus level only are shared between the eastern CCZ studies. Nonetheless, both reported the same genera (Eurythenes, Paralicella, Abyssorchomene, Paracallisoma, and Valettietta), with the addition of a single species of Hirondellea only reported by Patel et al. (2020). Based on the species overlap found in the present study, it is very likely that most of the species reported in Patel et al. (2020) represent the same species reported in Mohrbeck et al. (2021).
In the western CCZ, species richness was highest in APEI 1, the seamount site, with 12 morphospecies. The abyssal APEI 4 and 7 sites had eight morphospecies each, although composition differed between the assemblages. APEI 7 was highly dominated (91%) by species of the Paralicella tenuipes and P. caperesca complexes, as reported for the eastern areas in 2015 (UK-1 Stratum B, OMS and APEI 6; Mohrbeck et al., 2021) and previous studies north of Hawaii (Ingram and Hessler, 1983). APEI 4 was dominated by Abyssorchomene gerulicorbis, P. tenuipes complex, and P. caperesca complex, differing from APEI 7 and previous findings for the eastern CCZ. Both Mohrbeck et al. (2021) and Patel et al. (2020) reported another species of Abyssorchomene, A. distinctus, to be dominant in several sites, with a westward decrease in abundance. In our study, A. distinctus only occurred in APEI 4, with 3 specimens identified. Interestingly, the most common abyssal species (Abyssorchomene gerulicorbis, A. chevreuxi, Paralicella caperesca, and P. tenuipes) were not found in large numbers in APEI 1, the seamount site, with the community dominated (∼63%) instead by two new species restricted to this site (Tectovalopsis sp. CCZ_229 and Stephonyx sp. CCZ_220).
While scavenging amphipods are easily collected using baited-traps, and trapping is the only method to collect them in large numbers, this sampling strategy is always biased (Kaïm-Malka, 2005). Different species have different chemosensory abilities, as well as different swimming speeds, which affect distance of attraction to the bait (Horton et al., 2020c). Stronger odor plumes can also be more attractive, and while these depend on currents, no correlation has been found between current speeds and relative abundance of trapped amphipods (Leitner et al., 2017). Relative abundances of different species attracted to baited traps can also be biased because some developmental stages might be fasting (e.g., brooding females) or have different diets (e.g., juveniles), therefore not attend bait (Kaïm-Malka, 2005). Very few replicated bait-trap studies have been carried out, but differences in species composition between these traps, with different deployment times and at different heights above the seabed, have suggested a possible succession of scavenging species (Horton et al., 2020c). Two baited traps were deployed in the abyssal plains at APEI 7. Morphospecies diversity was higher in the deployment with the longest bottom time, where the “rare” species Eurythenes magellanicus and Valettietta sp. were recovered in low numbers (two and one individuals, respectively). This possibly indicates a later arrival to the trap, as previously suggested (Horton et al., 2020a), indicating that collection of “rare” species necessitates increased time on the seabed. Despite the limited data with few deployments, our results suggest that abundance increases with deployment times (Supplementary S1). However, the overall relative abundance of the common species seem to be similar between the only replicated deployments on the abyssal plain on APEI 7, but relative abundances of P. caperesca decreased while P. tenuipes increased with longer bottom times. While longer deployment times seem to better recover “rare” species (Horton et al., 2020a), the dominance of new or “rare” species (Stephonyx sp. and Tectovalopsis sp.) in the seamount site is unlikely to be explained by sampling bias.
Temporal and spatial variability in scavenging communities have been linked to environmental conditions. A recent study found that compositional changes in amphipod scavenging communities in the Atlantic were related to changes in Particulate Organic Carbon (POC) flux to the seafloor, shifting between communities dominated by specialist scavengers of food-falls (Paralicella) (Duffy et al., 2016; Havermans and Smetacek, 2018) to opportunistic scavengers (Eurythenes and Abyssorchomene) that include organic matter in their diets (Horton et al., 2020c). While the differences in community composition observed in the three western CCZ sites may be driven by different environmental conditions across the three sites, the difference in species composition between the seamount and abyssal plain sites is striking, with the seamount site dominated by genera not commonly reported in the literature such as Paralicella, Eurythenes and Abyssorchomene (e.g., Fujii et al., 2013; Lacey et al., 2016).
Net primary production and POC data suggest there is a southeast to northwest gradient of declining food availability and productivity in the western CCZ (Washburn et al., 2021b); for example, APEI 1 site is considered oligotrophic (0.06 mg m-3; McQuaid et al., 2020; Washburn et al., 2021b). Despite the low food availability, the seamount site in APEI 1 had the highest diversity of scavenging amphipods. For this site, very low abundances of predators, with an overall maximum of 3 individuals of a single predator species observed in either of the two baited camera deployments, were reported (Leitner et al., 2021). The high diversity could be driven by the low abundance of predators, but could also be driven by the seamount higher habitat heterogeneity (Washburn et al., 2021a) or by its oligotrophic condition (McQuaid et al., 2020). Oligotrophy has been associated with lower abundances but higher species diversity of scavenging amphipods compared to eutrophic sites (Horton et al., 2020c), and higher habitat heterogeneity has been also linked to higher diversity. Seamounts have been suggested to be biodiversity hotspots (Rowden et al., 2010). Additionally, seamounts have been posited to have endemic faunas, while also acting as faunal reservoirs for surrounding areas (Clark et al., 2010). However, very little overlap has been reported for megafauna between seamount and abyssal plain sites in the CCZ (Cuvelier et al., 2020), with large differences in the composition of scavenging fish communities. This may indicate seamount faunas in the CCZ are distinct from those of abyssal plains. The species composition of necrophagous amphipods between the seamount and abyssal plain sites in this study differed drastically, with the seamount appearing to have a distinct scavenging fauna. Five out of the seven potentially new species identified in this study were only found in APEI 1 on the seamount. The seamount site was also the shallowest (4084 m; abyssal plain sites > 4800 m depth). While depth has been identified to structure scavenging communities (Lacey et al., 2016), it is very unlikely this explains the difference in composition since these bathymetric changes in species assemblages occur at the abyssal-hadal transition zone around 6000 m depth (Jamieson et al., 2011; Lacey et al., 2016), and the eastern CCZ sites are also shallower (∼4200 m depth). Owing to the lack of sampling in abyssal plains in APEI 1, and in seamounts in APEIs 4 and 7, it is not possible to determine if the difference in species composition and diversity is attributed to the geoform, or to different environmental conditions in APEI 1 leading to general differences in species composition from other CCZ sites (Patel et al., 2020; Mohrbeck et al., 2021).
Commonly reported species (i.e., Abyssorchomene gerulicorbis, A. chevreuxi, Eurythenes maldoror, and E. magellanicus) were found in very low numbers in our samples, in at least one of the three sites. This is in accordance with previous findings in amphipods (Mohrbeck et al., 2021) and gastropods (McClain and Hardy, 2010; McClain, 2021), suggesting that local rarity is not necessarily linked to small geographic ranges. There was also overlap of species with narrow ranges in adjacent areas. Hirondellea dubia had only been reported for southwest Pacific trenches, where it is a dominant component of the hadal scavenging amphipod community (Lacey et al., 2016). This species was found in very low numbers in APEI 4, the southwestern most APEI, closer to the likely core area of distribution of the species. Moreover, four putative new species of Hirondellea were also found. These also appear to be restricted to a single area, with three of them only occurring at the seamount site in APEI 1.
Genetic Diversity, Population Differentiation and Connectivity
Single-locus species delimitation is being more commonly used for uncovering biodiversity in remote and poorly studied systems, as it can provide a fast and accurate assessment (Glover et al., 2015, 2016; Dahlgren et al., 2016; Wiklund et al., 2017). Species delimitation was performed using a fragment of the barcoding cytochrome c oxidase subunit I (COI), which has been used in scavenging amphipods previously (e.g., Havermans, 2016; Horton et al., 2020a). In our study, it revealed a diversity greater than that estimated from morphological characters, with at least 30 species identified from barcode sequences compared to 17 morphospecies identified from external morphological characters, similar to a previous study for the CCZ (Mohrbeck et al., 2021). However, there was disagreement between the different methods for species delimitation with regard to the inferred species numbers. This has been previously reported (Rubinoff et al., 2006), and it has been suggested that estimates of species boundaries should be treated with caution and different approaches should be compared (Dellicour and Flot, 2018).
The distance-based approach ABGD is known to lump species (Dellicour and Flot, 2018) and, as expected, it inferred the lowest number of species in our datasets (35–41 putative species for the complete dataset; Table 2). In contrast, the GMYC approach (Pons et al., 2006) tends to oversplit lineages (Correa et al., 2017; da Silva et al., 2017), yet it yielded only a slightly larger number of species than ABGD (43–45 putative species for the complete dataset; Table 2). bPTP (bayesian Poisson tree processes; Zhang et al., 2013) has been found to be sensitive to different mutation rates and has been reported to produce the least accurate results in empirical datasets (Hofmann et al., 2019). In this study, the results produced using the reduced dataset, excluding redundant haplotypes, were very similar to those from GMYC (44–47 species for the complete dataset; Table 2). However, when all haplotypes were considered, bPTP greatly oversplit lineages (116–131 species for the complete dataset; Table 2), even for those in which genetic divergences were low (e.g., Abyssorchomene gerulicorbis; mean intraspecific divergence: 0.23%).
Several widespread lineages of scavenging amphipods have been identified as complexes of species using COI. While several differ morphologically and have subsequently been described as new species (e.g., Havermans et al., 2011; d’Udekem d’Acoz and Havermans, 2015; Havermans, 2016; Weston et al., 2020), cryptic lineages have yet to be subjected to more detailed morphological analyses, despite having been identified within common species such as Paralicella caperesca (Ritchie et al., 2017). The morphological characters commonly used to separate P. caperesca from P. tenuipes have been reported as not being robust enough to accurately discriminate the species (Ritchie et al., 2015; 2017), but these two species were easily separated by the conspicuous red eyes in P. tenuipes, as reported by Mohrbeck et al. (2021). However, both species were found to represent complexes of cryptic species. Mohrbeck et al. (2021) identified at least four genetically distinct clusters within P. caperesca, whereas nine (mean interspecific divergence ranging from 2.33–15.33%) were identified in this study, five of which were found in both the western and eastern CCZ and belong to three of the clades identified by Mohrbeck et al. (2021) (Pc1, Pc3, and Pc4), with Pc2 being absent from the western CCZ. It is important to highlight that some of the P. caperesca MOTUs hypothesized by Mohrbeck et al. (2021) differ from the ones in this study, with some considered in here as multiple MOTUs. This reflects differences between methods for species delimitation and highlights the need to carry out further studies targeting a larger number of genes, as well as detailed morphological studies that will provide evidence to support one of the different species hypotheses available. For instance, several thresholds for species discrimination in amphipods have been proposed, ranging from the widely used 3% (Hebert et al., 2003) to 16% (e.g., Fiser et al., 2015). Although a more recent study proposed a threshold of 7% to discriminate amphipod species (Tempestini et al., 2018), and was also used for scavenging amphipods in the CCZ (Mohrbeck et al., 2021), no clear barcode gap was found in this study (Supplementary S3), and interspecific distances were estimated >2%. There was also support for this as the different approaches considered inferred a similar number of species.
There was no evidence of geographic genetic differentiation, with haplotypes being shared across different sites in the CCZ and with other adjacent areas (e.g., Kermadec, Peru-Chile, South Fiji, Mariana, and New Hebrides trenches; Figure 8). Additionally, none of the haplotype networks showed a strong star-like pattern, with only P. cf. caperesca 14 and P. cf. caperesca 7 having a somewhat predominant central haplotype. However, the starburst pattern of P. cf. caperesca 7 is indicative of low geographic structure and might suggest rapid population expansion (Neubaum et al., 2007). Although we acknowledge that the use of COI alone could prevent us from detecting genetic geographic structuring, COI has shown similar patterns to nuclear microsatellite markers in squat lobsters (Yan et al., 2020). Thus, the lack of geographic structuring observed in the widespread scavenging amphipods included in this study might not necessarily be an artifact of COI lacking resolution, but rather the result of them being highly motile, with high levels of gene flow across the Pacific Ocean, as has been shown for a study on Paralicella spp. using microsatellites (Ritchie et al., 2017).
Available data thus currently support the hypothesis that many mobile necrophagous amphipods are wide ranging and thus could be relatively resistant to localized mining impacts, although apparent endemism (e.g., the species found in APEI 1) and/or reduced motility in select species could place them under greater threat (Havermans et al., 2011). There are very few comparable studies of other CCZ taxa, but in the small nodule-dwelling sponge, Plenaster craigi Lim and Wiklund, 2017, evidence of geographic genetic structure was found using microsatellite DNA, suggesting recolonization of this sessile species into disturbed areas may be much slower (Taboada et al., 2018). These two groups of taxa, sessile nodule dwelling P. craigi and highly motile scavenging amphipods may make useful contrasts in future monitoring studies, being functionally very different, relatively easy to collect and with potentially differing patterns of resilience to mining impacts.
Phylogenetic Relationships of Scavenging Amphipods
Our phylogenetic analyses of scavenging amphipods recovered all genera included as monophyletic. However, higher taxa were recovered as polyphyletic, differing from the current classification (Horton et al., 2020b). A finding consistent with other published phylogenies (Corrigan et al., 2014; Ritchie et al., 2015; Copilas-Ciocianu et al., 2020) was the recovery of the superfamilies Alicelloidea and Lysianassoidea as non-monophyletic. Those published phylogenies also recovered the families Alicellidae and Uristidae as polyphyletic, and while our results are concordant, they all differ in phylogenetic relationships at the family level. Most families were recovered with high support values, but there was no resolution for deeper nodes in the phylogeny, most likely due to targeting only three genetic markers (COI, 16S, and 18S), as these were the most abundant in the datasets considered for this study. In contrast, both phylogenies from Ritchie et al. (2015) and Copilas-Ciocianu et al. (2020) showed high support values in deeper nodes in the phylogeny, but showed contrasting hypotheses of phylogenetic relationships between families, possibly also as a result of choice of genetic markers and taxa representativeness. Further studies to elucidate phylogenetic relationships of scavenging amphipods should therefore consider a wider sample of scavenging genera and a much larger number of genetic markers.
Conclusion
This study has again demonstrated the advantages of molecular taxonomy over morphology-based taxonomy in taxa where cryptic speciation is pervasive, allowing for fast but reliable diversity estimates in poorly studied groups. However, different approaches for species delimitation yielded different results, making it necessary to contrast results from different approaches and datasets. The finding of potentially cryptic species within described, widely distributed morphospecies such as Paralicella caperesca highlights the need for detailed, integrative studies that aim to identify and describe cryptic diversity. The study also contributes to our understanding of the diversity and connectivity of necrophagous amphipods in the western Clarion-Clipperton Zone (CCZ), in particular within the poorly studied APEI regions. A higher diversity, of both morphospecies and genetic species was found from a single expedition to the western CCZ than from multiple expeditions in eastern CCZ spanning several years. Within the western CCZ, differences in composition and diversity of scavenging communities between sites on seamounts and those on abyssal plains were observed, possibly driven by habitat heterogeneity and different environmental conditions between the sites, but further studies are required to understand the drivers of the diversity patterns observed. It is critical to understand the drivers of diversity, and the potential for biodiversity reservoirs within different topographic features in the CCZ, and in the currently conserved APEI regions, to allow sustainable management of any future deep-sea mining industry.
Data Availability Statement
The original contributions presented in the study are publicly available. This data can be found here: Sequences were deposited in GenBank with accession numbers MZ443996–MZ444014 (16S), MZ444015–MZ444046 (18S), and MZ474200–MZ474475 (COI). Code used for all analyses has been made available as Supplementary Material.
Author Contributions
JD, RD, AL, KM, HW, ST, DJ, and CS collected and processed samples. GB-C and ST generated genetic sequences. TH carried out detailed morphological examination. AG, TD, and GB-C designed the study, with GB-C carrying out the analyses. All authors contributed to drafting and reviewing the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank the masters, crew and technical support staff on the R/V Kilo Moana and ROV Luukai for their expertise in collecting samples. We thank the primary funder, the Gordon and Betty Moore Foundation grant no. 5596 and NOAA Office of Ocean Exploration (grant #NA17OAR0110209). We also acknowledge funding from UK Seabed Resources, the UK Natural Environment Research Council grant numbers NE/T003537/1 and NE/T002913/1, and The Norwegian Research Council (JPIO Mining Impact 2). We also acknowledge Dr. Lauren Hughes and Muriel Rabone (NHM) for curatorial support, and Elena Luigli (NHM) and Claire Griffin (NHM) for lab support. This is a contribution from SOEST, University of Hawai‘i at Mānoa.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.705237/full#supplementary-material
Footnotes
References
Bouckaert, R., Heled, J., Kuhnert, D., Vaughan, T., Wu, C. H., Xie, D., et al. (2014). BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 10:e1003537. doi: 10.1371/journal.pcbi.1003537
Castresana, J. (2000). Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17, 540–552. doi: 10.1093/oxfordjournals.molbev.a026334
Christiansen, B., Pfannkuche, O., and Thiel, H. (1990). Vertical distribution and population structure of the necrophagous amphipod Eurythenes gryllus in the west European basin. Mar. Ecol. Prog. Ser. 66, 35–45. doi: 10.3354/meps066035
Clark, M. R., Rowden, A. A., Schlacher, T., Williams, A., Consalvey, M., Stocks, K. I., et al. (2010). The ecology of seamounts: structure, function, and human impacts. Ann. Rev. Mar. Sci. 2, 253–278. doi: 10.1146/annurev-marine-120308-081109
Cohen, B. L., Gawthrop, A., and Cavalier-Smith, T. (1998). Molecular phylogeny of brachiopods and phoronids based on nuclear-encoded small subunit ribosomal RNA gene sequences. Philos. Trans. R. Soc. Lond. B Biol. Sci. 353, 2039-2061.
Copilas-Ciocianu, D., Borko, S., and Fiser, C. (2020). The late blooming amphipods: global change promoted post-Jurassic ecological radiation despite Palaeozoic origin. Mol. Phylogenet. Evol. 143:106664. doi: 10.1016/j.ympev.2019.106664
Correa, C., Vasquez, D., Castro-Carrasco, C., Zuniga-Reinoso, A., Ortiz, J. C., and Palma, R. E. (2017). Species delimitation in frogs from South American temperate forests: the case of Eupsophus, a taxonomically complex genus with high phenotypic variation. PLoS One 12:e0181026. doi: 10.1371/journal.pone.0181026
Corrigan, L. J., Horton, T., Fotherby, H., White, T. A., and Hoelzel, A. R. (2014). Adaptive evolution of deep-sea amphipods from the superfamily lysiassanoidea in the North Atlantic. Evol. Biol. 41, 154–165. doi: 10.1007/s11692-013-9255-2
Cuvelier, D., Ribeiro, P. A., Ramalho, S. P., Kersken, D., Martinez Arbizu, P., and Colaço, A. (2020). Are seamounts refuge areas for fauna from polymetallic nodule fields? Biogeosciences 17, 2657–2680. doi: 10.5194/bg-17-2657-2020
da Silva, R., Peloso, P. L. V., Sturaro, M. J., Veneza, I., Sampaio, I., Schneider, H., et al. (2017). Comparative analyses of species delimitation methods with molecular data in snappers (Perciformes: Lutjaninae). Mitochondrial DNA A DNA Mapp. Seq. Anal. 29, 1108–1114. doi: 10.1080/24701394.2017.1413364
Dahlgren, T. G., Wiklund, H., Rabone, M., Amon, D. J., Ikebe, C., Watling, L., et al. (2016). Abyssal fauna of the UK-1 polymetallic nodule exploration area, Clarion-Clipperton zone, central Pacific Ocean: Cnidaria. Biodivers. Data J. 4:e9277. doi: 10.3897/bdj.4.e9277
Dellicour, S., and Flot, J. F. (2018). The hitchhiker’s guide to single-locus species delimitation. Mol. Ecol. Resour. 18, 1234–1246. doi: 10.1111/1755-0998.12908
d’Udekem d’Acoz, C., and Havermans, C. (2015). Contribution to the systematics of the genus Eurythenes S.I. Smith in Scudder, 1882 (Crustacea: Amphipoda: Lysianassoidea: Eurytheneidae). Zootaxa 3971, 1–80. doi: 10.11646/zootaxa.3971.1.1
Duffy, G. A., Lawler, S. F., and Horton, T. (2016). Scavenging amphipods of the Angolan deep-sea habitat, with a focus on Abyssorchomene distinctus (Birstein and Vinogradov, 1960) (Amphipoda: Lysianassoidea). J. Crustac. Biol. 36, 417–426. doi: 10.1163/1937240x-00002448
Excoffier, L., Smouse, P. E., and Quattro, J. M. (1992). Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondria DNA restriction sites. Genetics 131, 479–491. doi: 10.1093/genetics/131.2.479
Fiser, Z., Altermatt, F., Zaksek, V., Knapic, T., and Fiser, C. (2015). Morphologically cryptic amphipod species are “ecological clones” at regional but not at local scale: a case study of four Niphargus species. PLoS One 10:e0134384. doi: 10.1371/journal.pone.0134384
Folmer, O., Black, M., Hoeh, W., Lutz, R., and Vrijenhoek, R. C. (1994). DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 3, 294–299.
Fujii, T., Kilgallen, N. M., Rowden, A. A., and Jamieson, A. J. (2013). Deep-sea amphipod community structure across abyssal to hadal depths in the Peru-Chile and Kermadec trenches. Mar. Ecol. Prog. Ser. 492, 125–138. doi: 10.3354/meps10489
Glover, A., Dahlgren, T., Wiklund, H., Mohrbeck, I., and Smith, C. (2015). An end-to-end DNA taxonomy methodology for benthic biodiversity survey in the Clarion-Clipperton zone, central Pacific Abyss. J. Mar. Sci. Eng. 4:2. doi: 10.3390/jmse4010002
Glover, A. G., Wiklund, H., Rabone, M., Amon, D. J., Smith, C. R., O’hara, T., et al. (2016). Abyssal fauna of the UK-1 polymetallic nodule exploration claim, Clarion-Clipperton Zone, central Pacific Ocean: echinodermata. Biodivers. Data J. 4:e7251.
Gollner, S., Kaiser, S., Menzel, L., Jones, D. O. B., Brown, A., Mestre, N. C., et al. (2017). Resilience of benthic deep-sea fauna to mining activities. Mar. Environ. Res. 129, 76–101.
Goudet, J., and Jombart, T. (2020). hierfstat: Estimation and Tests of Hierarchical F-Statistics. R package version 0.5-7. Available online at: https://CRAN.R-project.org/package=hierfstat. (accessed April 7, 2021)
Havermans, C. (2016). Have we so far only seen the tip of the iceberg? Exploring species diversity and distribution of the giant amphipod Eurythenes. Biodiversity 17, 12–25. doi: 10.1080/14888386.2016.1172257
Havermans, C., Nagy, Z. T., Sonet, G., De Broyer, C., and Martin, P. (2011). DNA barcoding reveals new insights into the diversity of Antarctic species of orchomene sensu lato (Crustacea: Amphipoda: Lysianassoidea). Deep Sea Res. 2 Top Stud. Oceanogr. 58, 230–241. doi: 10.1016/j.dsr2.2010.09.028
Havermans, C., and Smetacek, V. (2018). Bottom-up and top-down triggers of diversification: a new look at the evolutionary ecology of scavenging amphipods in the deep sea. Prog. Oceanogr. 164, 37–51. doi: 10.1016/j.pocean.2018.04.008
Havermans, C., Sonet, G., D’udekem D’acoz, C., Nagy, Z. T., Martin, P., Brix, S., et al. (2013). Genetic and morphological divergences in the cosmopolitan deep-sea amphipod Eurythenes gryllus reveal a diverse abyss and a bipolar species. PLoS One 8:e74218. doi: 10.1371/journal.pone.0074218
Hebert, P. D., Ratnasingham, S., and Dewaard, J. R. (2003). Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proc. R. Soc. B Biol. Sci. 270(Suppl 1), S96–S99.
Hein, J. R., Koschinsky, A., and Kuhn, T. (2020). Deep-ocean polymetallic nodules as a resource for critical materials. Nat. Rev. Earth Environ. 1, 158–169. doi: 10.1038/s43017-020-0027-0
Hofmann, E. P., Nicholson, K. E., Luque-Montes, I. R., Kohler, G., Cerrato-Mendoza, C. A., Medina-Flores, M., et al. (2019). Cryptic diversity, but to what extent? discordance between single-locus species delimitation methods within mainland anoles (Squamata: Dactyloidae) of Northern Central America. Front. Genet. 10:11. doi: 10.3389/fgene.2019.00011
Horton, T., Cooper, H., Vlierboom, R., Thurston, M., Hauton, C., and Young, C. R. (2020a). Molecular phylogenetics of deep-sea amphipods (Eurythenes) reveal a new undescribed species at the Porcupine Abyssal plain, north east Atlantic ocean. Prog. Oceanogr. 183:102292. doi: 10.1016/j.pocean.2020.102292
Horton, T., Lowry, J., De Broyer, C., Bellan-Santini, D., Coleman, C. O., Corbari, L., et al. (2020b). World Amphipoda Database. Lysianassidira [Online]. Available online at: http://www.marinespecies.org/aphia.php?p=taxdetails&id=1055696 (accessed April 7, 2021)
Horton, T., and Thurston, M. H. (2014). A revision of the bathyal and abyssal necrophage genus Cyclocaris Stebbing, 1888 (Crustacea: Amphipoda: Cyclocaridae) with the addition of two new species from the Atlantic Ocean. Zootaxa 3796, 507–527. doi: 10.11646/zootaxa.3796.3.6
Horton, T., Thurston, M. H., Vlierboom, R., Gutteridge, Z., Pebody, C. A., Gates, A. R., et al. (2020c). Are abyssal scavenging amphipod assemblages linked to climate cycles? Prog. Oceanogr. 184:102318. doi: 10.1016/j.pocean.2020.102318
Hsieh, T. C., Ma, K. H., and Chao, A. (2020). iNEXT: iNterpolation and EXTrapolation for Species Diversity. Available online at: http://chao.stat.nthu.edu.tw/wordpress/software-download/. (accessed April 7, 2021)
ISA (2021). Exploration Contracts [Online]. Available online at: https://isa.org.jm/exploration-contracts (accessed April 29, 2021).
Jamieson, A. J., Kilgallen, N. M., Rowden, A. A., Fujii, T., Horton, T., Lörz, A. N., et al. (2011). Bait-attending fauna of the kermadec trench, SW Pacific Ocean: evidence for an ecotone across the abyssal–hadal transition zone. Deep Sea Res. 1 Oceanogr. Res. Pap. 58, 49–62. doi: 10.1016/j.dsr.2010.11.003
Janssen, A., Stuckas, H., Vink, A., and Arbizu, P. M. (2019). Biogeography and population structure of predominant macrofaunal taxa (Annelida and Isopoda) in abyssal polymetallic nodule fields: implications for conservation and management. Mar. Biodivers. 49, 2641–2658. doi: 10.1007/s12526-019-00997-1
Jones, D. O. B. (2015). RRS James Cook Cruise JC120 15 Apr–19 May 2015. Manzanillo to Manzanillo, Mexico. Managing Impacts of Deep-Sea Resource Exploitation (MIDAS): Clarion-Clipperton Zone North Eastern Area of Particular Environmental Interest. (Southampton, National Oceanography Centre, Southampton)
Kaïm-Malka, R. A. (2005). Biology and life cycle of Tmetonyx similis (G. O. Sars, 1891) (Amphipoda, Lysianassidae), a scavenging amphipod from the continental slope of the Mediterranean. J. Nat. Hist. 39, 3163–3186. doi: 10.1080/00222930500240502
Kamvar, Z. N., Tabima, J. F., and Grunwald, N. J. (2014). Poppr: an R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 2:e281. doi: 10.7717/peerj.281
Katoh, K., Rozewicki, J., and Yamada, K. D. (2019). MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 20, 1160–1166. doi: 10.1093/bib/bbx108
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Lacey, N. C., Rowden, A. A., Clark, M. R., Kilgallen, N. M., Linley, T., Mayor, D. J., et al. (2016). Community structure and diversity of scavenging amphipods from bathyal to hadal depths in three South Pacific Trenches. Deep Sea Res. 1 Oceanogr. Res. Pap. 111, 121–137. doi: 10.1016/j.dsr.2016.02.014
Lanfear, R., Frandsen, P. B., Wright, A. M., Senfeld, T., and Calcott, B. (2017). PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological hylogenetic analyses. Mol. Biol. Evol. 34, 772–773.
Leitner, A. B., Durden, J. M., Smith, C. R., Klingberg, E. D., and Drazen, J. C. (2021). Synaphobranchid eel swarms on abyssal seamounts: largest aggregation of fishes ever observed at abyssal depths. Deep Sea Res. 1 Oceanogr. Res. Pap. 167:103423. doi: 10.1016/j.dsr.2020.103423
Leitner, A. B., Neuheimer, A. B., Donlon, E., Smith, C. R., and Drazen, J. C. (2017). Environmental and bathymetric influences on abyssal bait-attending communities of the Clarion Clipperton Zone. Deep Sea Res. 1 125, 65–80. doi: 10.1016/j.dsr.2017.04.017
McClain, C. R. (2021). The commonness of rarity in a deep-sea taxon. Oikos 130, 863–878. doi: 10.1111/oik.07602
McClain, C. R., and Hardy, S. M. (2010). The dynamics of biogeographic ranges in the deep sea. Proc. Biol. Sci. 277, 3533–3546. doi: 10.1098/rspb.2010.1057
McQuaid, K. A., Attrill, M. J., Clark, M. R., Cobley, A., Glover, A. G., Smith, C. R., et al. (2020). Using habitat classification to assess representativity of a protected area network in a large, data-poor area targeted for deep-sea mining. Front. Mar. Sci. 7:558860. doi: 10.3389/fmars.2020.558860
Medlin, L., Elwood, H., Stickel, S., and Sogin, M. (1988). The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions. Gene 71, 491–499. doi: 10.1016/0378-1119(88)90066-2
Mewes, K., Mogollón, J. M., Picard, A., Rühlemann, C., Kuhn, T., Nöthen, K., et al. (2014). Impact of depositional and biogeochemical processes on small scale variations in nodule abundance in the Clarion-Clipperton fracture zone. Deep Sea Res. 1 Oceanogr. Res. Pap. 91, 125–141. doi: 10.1016/j.dsr.2014.06.001
Mohrbeck, I., Horton, T., Jażdżewska, A. M., and Martínez Arbizu, P. (2021). DNA barcoding and cryptic diversity of deep-sea scavenging amphipods in the Clarion-Clipperton zone (Eastern Equatorial Pacific). Mar. Biodivers. 51:26.
Narahara-Nakano, Y., Nakano, T., and Tomikawa, K. (2018). Deep-sea amphipod genus Eurythenes from Japan, with a description of a new Eurythenes species from off Hokkaido (Crustacea: Amphipoda: Lysianassoidea). Mar. Biodivers. 48, 603–620. doi: 10.1007/s12526-017-0758-4
Neubaum, M. A., Douglas, M. R., Douglas, M. E., and O’shea, T. J. (2007). Molecular ecology of the big brown bat (Eptesicus fuscus): genetic and natural history variation in a hybrid zone. J. Mammal. 88, 1230–1238. doi: 10.1644/06-mamm-a-228r1.1
Nygren, A., and Sundberg, P. (2003). Phylogeny and evolution of reproductive modes in Autolytinae (Syllidae, Annelida). Mol. Phylogenet. Evol. 29, 235–249.
Oksanen, J., Blanchet, G., Friendly, M., Kindt, R., Legendre, P., Mcglinn, D., et al. (2018). vegan: Community Ecology Package. R package Version 2.4-6. Available online at: http://CRAN.Rproject.org/package=vegan. (accessed April 7, 2021).
Palumbi, S. R. (1996). “Nucleic acid II: the polymerase chain reaction,” in Molecular Systematics, eds D. M. Hillis, G. Moritz, and B. K. Mable (Sunderland, MA: Sinauer Associates).
Paradis, E. (2010). pegas: an R package for population genetics with an integrated-modular approach. Bioinformatics 26, 419–420. doi: 10.1093/bioinformatics/btp696
Paradis, E., and Schliep, K. (2019). ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 35, 526–528. doi: 10.1093/bioinformatics/bty633
Patel, T., Robert, H., D’udekem D’acoz, C., Martens, K., De Mesel, I., Degraer, S., et al. (2020). Biogeography and community structure of abyssal scavenging Amphipoda (Crustacea) in the Pacific Ocean. Biogeosciences 17, 2731–2744. doi: 10.5194/bg-17-2731-2020
Pons, J., Barraclough, T., Gomez-Zurita, J., Cardoso, A., Duran, D., Hazell, S., et al. (2006). Sequence-based species delimitation for the DNA taxonomy of undescribed insects. Syst. Biol. 55, 595–609. doi: 10.1080/10635150600852011
Puillandre, N., Lambert, A., Brouillet, S., and Achaz, G. (2012). ABGD, automatic barcode gap discovery for primary species delimitation. Mol. Ecol. 21, 1864–1877. doi: 10.1111/j.1365-294x.2011.05239.x
R Development Core Team (2017). R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. Vienna: R Foundation for Statistical Computing.
Ritchie, H., Jamieson, A. J., and Piertney, S. B. (2015). Phylogenetic relationships among hadal amphipods of the Superfamily lysianassoidea: implications for taxonomy and biogeography. Deep Sea Res. 1 Oceanogr. Res. Pap. 105, 119–131.
Ritchie, H., Jamieson, A. J., and Piertney, S. B. (2017). Population genetic structure of two congeneric deep-sea amphipod species from geographically isolated hadal trenches in the Pacific Ocean. Deep Sea Res. 1 Oceanogr. Res. Pap. 119, 50–57. doi: 10.1016/j.dsr.2016.11.006
Rowden, A. A., Dower, J. F., Schlacher, T. A., Consalvey, M., and Clark, M. R. (2010). Paradigms in seamount ecology: fact, fiction and future. Mar. Ecol. 31, 226–241. doi: 10.1111/j.1439-0485.2010.00400.x
Rubinoff, D., Cameron, S., and Will, K. (2006). A genomic perspective on the shortcomings of mitochondrial DNA for “barcoding” identification. J. Hered. 97, 581–594. doi: 10.1093/jhered/esl036
Sainte-Marie, B. (1992). “Foraging of scavenging deep-sea lysianassoid amphipods,” in Deep-Sea Food Chains and the Global Carbon Cycle, eds G. T. Rowe and V. Pariente (Dordrecht: Springer).
Stamatakis, A. (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690. doi: 10.1093/bioinformatics/btl446
Taboada, S., Riesgo, A., Wiklund, H., Paterson, G. L. J., Koutsouveli, V., Santodomingo, N., et al. (2018). Implications of population connectivity studies for the design of marine protected areas in the deep sea: an example of a demosponge from the Clarion-Clipperton Zone. Mol. Ecol. 27, 4657–4679. doi: 10.1111/mec.14888
Tempestini, A., Rysgaard, S., and Dufresne, F. (2018). Species identification and connectivity of marine amphipods in Canada’s three oceans. PLoS One 13:e0197174. doi: 10.1371/journal.pone.0197174
Washburn, T. W., Jones, D. O. B., Wei, C.-L., and Smith, C. R. (2021a). Environmental heterogeneity throughout the Clarion-Clipperton zone and the potential representativity of the APEI Network. Front. Mar. Sci. 8:661685. doi: 10.3389/fmars.2021.661685
Washburn, T. W., Menot, L., Bonifácio, P., Pape, E., Błaz_Ewicz, M., Bribiesca-Contreras, G., et al. (2021b). Patterns of macrofaunal biodiversity across the Clarion-Clipperton zone: an area targeted for deep-sea mining. Front. Mar. Sci. 8:626571. doi: 10.3389/fmars.2021.626571
Wedding, L. M., Friedlander, A. M., Kittinger, J. N., Watling, L., Gaines, S. D., Bennett, M., et al. (2013). From principles to practice: a spatial approach to systematic conservation planning in the deep sea. Proc. R. Soc. Lond. B Biol. Sci. 280:20131684. doi: 10.1098/rspb.2013.1684
Weir, B. S., and Cockerham, C. C. (1984). Estimating F-statistics for the analysis of population structure. Evolution 38, 1358–1370. doi: 10.2307/2408641
Weston, J. N. J., Carrillo-Barragan, P., Linley, T. D., Reid, W. D. K., and Jamieson, A. J. (2020). New species of Eurythenes from hadal depths of the Mariana Trench, Pacific Ocean (Crustacea: Amphipoda). Zootaxa 4748, 4741–4749.
Wiklund, H., Taylor, J. D., Dahlgren, T. G., Todt, C., Ikebe, C., Rabone, M., et al. (2017). Abyssal fauna of the UK-1 polymetallic nodule exploration area, Clarion-Clipperton Zone, central Pacific Ocean: Mollusca. Zookeys 707, 1–46. doi: 10.3897/zookeys.707.13042
Yan, R.-J., Schnabel, K. E., Rowden, A. A., Guo, X.-Z., and Gardner, J. P. A. (2020). Population structure and genetic connectivity of squat lobsters (Munida Leach, 1820) associated with vulnerable marine ecosystems in the Southwest Pacific Ocean. Front. Mar. Sci. 6:791. doi: 10.3389/fmars.2019.00791
Keywords: scavenging amphipods, connectivity, biodiversity, Clarion-Clipperton Zone, deep-sea mining, seamount
Citation: Bribiesca-Contreras G, Dahlgren TG, Horton T, Drazen JC, Drennan R, Jones DOB, Leitner AB, McQuaid KA, Smith CR, Taboada S, Wiklund H and Glover AG (2021) Biogeography and Connectivity Across Habitat Types and Geographical Scales in Pacific Abyssal Scavenging Amphipods. Front. Mar. Sci. 8:705237. doi: 10.3389/fmars.2021.705237
Received: 04 May 2021; Accepted: 07 July 2021;
Published: 27 July 2021.
Edited by:
Sylvie Marylène Gaudron, Sorbonne Université, FranceReviewed by:
Sven Richard Laming, University of Aveiro, PortugalJorge Paramo, University of Magdalena, Colombia
Copyright © 2021 Bribiesca-Contreras, Dahlgren, Horton, Drazen, Drennan, Jones, Leitner, McQuaid, Smith, Taboada, Wiklund and Glover. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guadalupe Bribiesca-Contreras, l.bribiesca-contreras@nhm.ac.uk
 Guadalupe Bribiesca-Contreras
Guadalupe Bribiesca-Contreras Thomas G. Dahlgren
Thomas G. Dahlgren Tammy Horton
Tammy Horton Jeffrey C. Drazen
Jeffrey C. Drazen Regan Drennan
Regan Drennan Daniel O.B. Jones
Daniel O.B. Jones Astrid B. Leitner
Astrid B. Leitner Kirsty A. McQuaid
Kirsty A. McQuaid Craig R. Smith
Craig R. Smith Sergio Taboada1,8,9
Sergio Taboada1,8,9  Helena Wiklund
Helena Wiklund Adrian G. Glover
Adrian G. Glover