- 1National Marine Environmental Monitoring Center, Ministry of Ecology and Environment, Dalian, China
- 2State Key Lab of Estuarine and Coastal Research, East China Normal University, Shanghai, China
- 3Yantai Institute of Coastal Zone Research, Chinese Academy of Sciences, Yantai, China
In this study, the release effects of the buried clam Meretrix meretrix in a northern estuary of China (Shuangtaizi Estuary) was assessed in terms of population ecology and genetic variability by using longer-term monitoring data from 2011 (used as the baseline data) to 2018. A total of 2.4 × 108 hatchery juveniles were released from 2012 to 2014, and follow-up monitoring survey of the growth, distribution, density, and biomass of M. meretrix population was carried out. Results showed a temporary spike in the population of M. meretrix, and the population underwent considerable year-to-year fluctuations in total abundance and biomass. Meanwhile, the age structure of the populations from 2011 to 2018 is still an unstable expanding type. Genetic analysis based on 10 polymorphic microsatellite markers revealed no significant genetic differentiation and weak genetic structures between years but higher genetic diversities in 2014–2018 than in the initial period (2013) of stock restoration. Our study suggests that effective policies for management and conservation to restore M. meretrix population by local governments are still needed in the long run.
Introduction
Stock enhancement has been considered as one of the most popular tools in fishery management approach involving the release of hatchery-reared juveniles (Laikre et al., 2010; Kitada et al., 2019). More than 180 marine species were released into the wild each year worldwide (Kitada, 2018, 2020). Starting from the 1860s, enhancement programs of Salmonidae were implemented in America, Canada, Russia, Australia, New Zealand, and Japan (Hamasaki and Kitada, 2006; Chen H. et al., 2019; Tang, 2019). In addition, from the 1930s, marine stock enhancement programs were initiated in China (Zhou, 2004). In 1951, artificial release activities with tagging method were conducted (Fei and Zhang, 1990). The marine stock enhancement programs gradually formed a scale in China from the 1980s and rapidly grew into a large scale from 2000 (Luo and Zhang, 2014; Lee and Zhang, 2018; Wang et al., 2018; Chen P. M. et al., 2019; Zhou et al., 2019).
Stock enhancement activities may influence wild populations through the ecological or genetic variability and composition (Yang et al., 2014; Li et al., 2016; Grant et al., 2017; Qin et al., 2020). Evaluating effectiveness is an integral part of stock enhancement (Chen H. et al., 2019). Consequently, the global concern is how to assess the effects of marine stock enhancement and deeply understand the ecological and genetic effects of hatchery releases on wild populations while focusing on progress towards ecological goals rather than the “yes or no” alternative of success versus failure. To date, relatively few studies could be found on the effect assessment method of marine stock enhancement (Bell et al., 2008). Most studies were at experimental stages, and empirical studies (studies with data-points covering a long period of time) on assessment of the potential and effectiveness of hatchery release on wild populations are heavily sparse.
In recent years, the tagging-recapture experiment, classical fishery models (Kitada and Kishino, 2006; Xu et al., 2018), and the molecule marker technology (Gonzalez et al., 2008; Wei et al., 2016; Kitada et al., 2019) were always applied in the assessment of stock enhancement. Exploitation efficiency and resource restore effects could be estimated by the tagging methods, but the effects of the whole population, including the population structure and genetic biodiversity, are difficult to be assessed by these methods. In addition, the tagging methods could not be easily applied to most clams but only to a few large individual species (Villella and Weller, 2000; Hamasaki and Kitada, 2006). Fortunately, the molecule marker technology could make up the disadvantages in mollusk enhancement assessment (Yamakawa and Imai, 2012; Xu et al., 2018). Most clams belong to long-lived species. Therefore, long-time continuous monitoring could ensure the stated specific conditions and is necessary for the assessment of clam stock enhancement. However, in most cases, monitoring efforts were insufficient to properly assess the program effectiveness due to the difficulty of long-term monitoring. Most studies were conducted with short-time (such as 1 or 2 years) monitoring, and only a few with several years and discontinuous monitoring, which did not accurately assess the effect of marine stock enhancement. Therefore, population and genetics monitoring of breed stocks and their offspring must be carried out (Blankenship and Leber, 1995).
The clam Meretrix meretrix is widely distributed along the coastal and estuarine areas of the western Indian Ocean and Western Pacific, including China, Korea, Vietnam, Japan, and India (Wang et al., 1993; Zhang et al., 2012). The annual production of M. meretrix is approximately 3.5 × 105–4.0 × 105 t in China, accounting for more than 90% of the world (Chen et al., 2004). Unfortunately, the natural stocks of M. meretrix declined during the past 20 years because of overharvesting, disease breakout, and habitat degradation (Chen et al., 2012; Zhang et al., 2014). Therefore, taking measures to restore M. meretrix resources is urgent. To the knowledge of authors, a large-scale stock enhancement (a total of 11.8 × 108 individuals) has been carried out in the coast of China to protect and recover the collapsed M. meretrix resources. Approximately 3.7 × 108, 5.7 × 108 and 3.8 × 108 individuals of M. meretrix were artificially released in Shandong, Jiangsu, and Liaoning Provinces since 1998 (Zhang et al., 2009; Zhang, 2015). Although large scales of M. meretrix stock enhancement activities effectively mitigate the recession of resources in short term to some extent, the benefits of hatcheries for long-term clams and the conservation objectives remain unclear.
Like other invertebrates, M. meretrix stock enhancement is mainly on artificial propagation, which may change the population structure and composition. Meanwhile, stock enhancement activities may change the genetic variability and genetic composition of the recipient wild populations (Yang et al., 2014; Li et al., 2016). A rigorous evaluation requires simultaneous monitoring data for time series of population abundance, incorporated with stocking effects and genetic diversity (Kitada et al., 2019). Therefore, follow-up population ecological and genetic monitoring of M. meretrix brood stocks and their offspring is necessary for the long-term sustainability of wild population. However, research and monitoring about the ecological and genetic effects of enhanced production on wild populations of Meretrix species are generally lacking (Yamakawa and Imai, 2012).
In the present study, the survival, growth rate, and migratory pattern of the hatchery released clams in Shuangtaizi Estuary was monitored through field sampling monitoring to evaluate the potential effectiveness of the stock enhancement program. Biometric and gravimetric data were processed to evaluate the distribution of size class, abundance and biomass, and age composition of the clams. Meanwhile, the genetic variation of M. meretrix populations in Shuangtaizi Estuary from 2013 to 2018 was assessed using microsatellite markers. The population genetic structure and genetic differentiation were also analyzed, and the genetic variation trend of M. meretrix was evaluated. Comparison between the current and bibliographical data enabled an estimation of the health condition of clam stocks in the area. Understanding of M. meretrix stock enhancement from the perspectives of population ecology and genetic diversity may provide a scientific basis for stock maintenance and rational exploitation of clams and help develop strategies for the corresponding enhancement and release management.
Materials and Methods
General Situation of Artificial Release of Meretrix meretrix
A large number of hatchery juveniles M. meretrix (0.8 × 108 individuals with shell length of approximately 1.1 cm) was released annually from 2012 to 2014 (Yuan et al., 2021). The released juveniles were hatched from brood stocks of local origin, collecting from the intertidal flats of Shuangtaizi Estuary. The designated area (N 40.79590°–40.81016°, E 121.61145°–121.64759°) for clam stock enhancement in Shuangtaizi Estuary is approximately 300 hm2 (Figure 1). This area is one of the few remaining intertidal sand flats in Bohai Sea and a most important spawning and breeding ground for clams. The clam M. meretrix used to be the dominant species of these intertidal flats, but its population extensively declined starting from the 1990s due to overfishing, habitat deterioration, and diseases (Chen et al., 2012; Zhang, 2015).
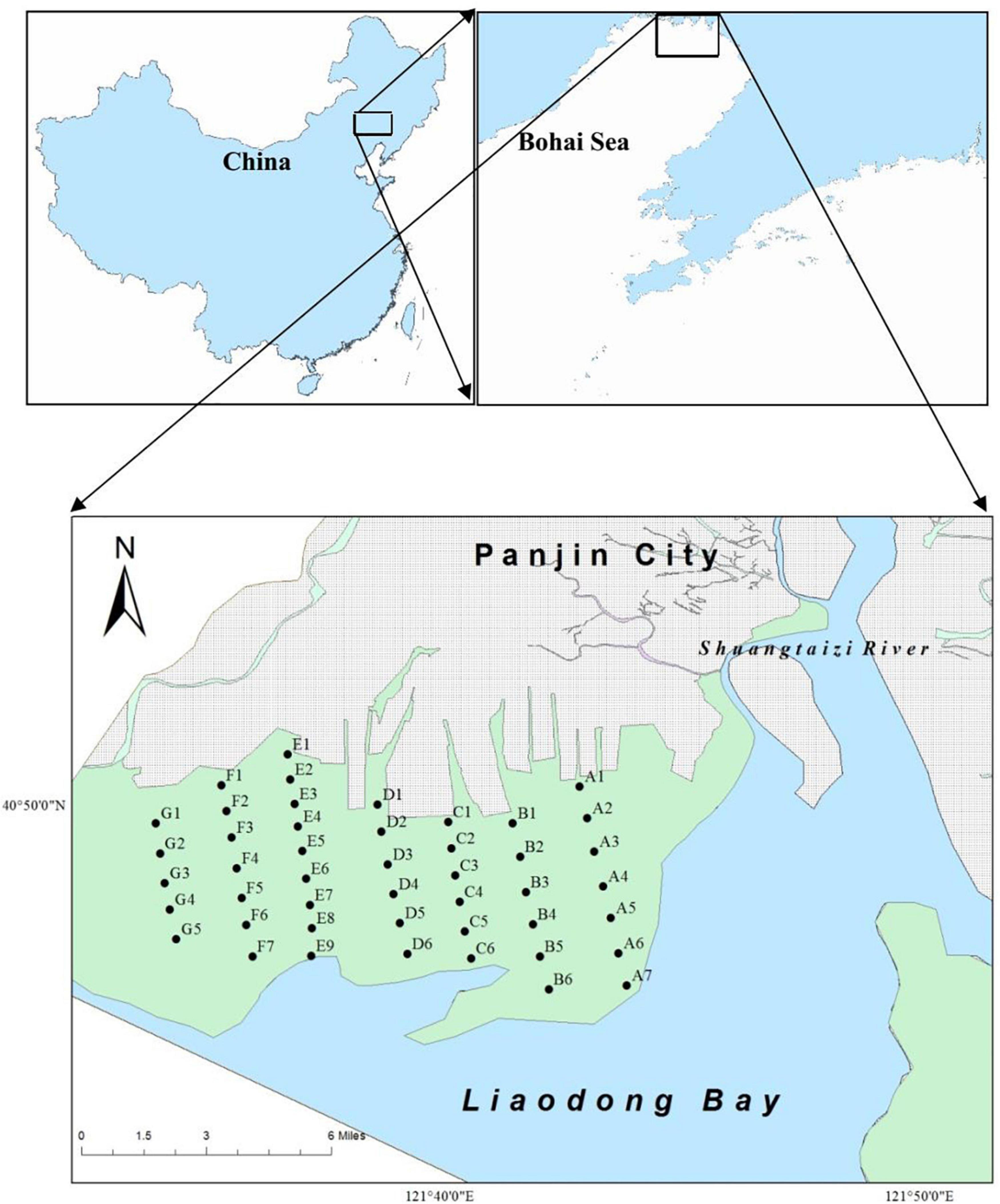
Figure 1. Artificial juvenile releasing area and monitoring sites of Meretrix meretrix in Shuangtaizi Estuary.
Field Sampling and Monitoring
Before the artificial release of M. meretrix individuals, clams were sampled at 46 sites in seven sections (A–G, perpendicular to the Shuangtaizi River current) in spring, summer, and autumn of 2011. Subsequently, the clams were also collected at these corresponding sites after the juveniles artificial release in spring, summer, and autumn of 2013–2018 (Figure 1). Sampling could not be conducted during winter because of ice cover (Gu et al., 2013). Treatment, preservation, counting, and weighing protocols were conducted in accordance with “The Specification for Marine Monitoring Part 3: Sampling Collection, Storage and Transportation” (State Standard of the People’s Republic of China, 2007). In brief, eight subsamples were randomly collected at each sampling site by a Van Veen Grab sampler (25 cm × 25 cm) to a depth of 20 cm and sieved through a 1 mm iron mesh screen with seawater. The collected individuals of M. meretrix in the sampling sites were initially stored in an icebox until delivery to the laboratory. The shell length of the clams (considered as the maximal length parallel to the hinge) was measured using a vernier caliper (accuracy of 0.1 mm), and the fresh weight was weighed using an electronic balance (accuracy of 0.1 g). The ages of the sampled M. meretrix were determined in accordance with the shell length (He and Chen, 1997).
Microsatellite DNA Genotyping
After the artificial release of clams, each summer from 2013 to 2018, additional 50 individuals were recaptured from the area and used for genetic analysis. Adductor muscle tissue was dissected from fresh specimens and maintained in 80% alcohol buffer for DNA extraction. Total genomic DNA was extracted using a phenol-chloroform-isoamyl alcohol method. Polymorphic microsatellite markers were selected according to the references (Lu et al., 2011; Dong et al., 2013; Li et al., 2016) which published microsatellite primers about M. meretrix. Furthermore, 10 polymorphic microsatellite markers with good amplification results and high polymorphism were selected for the microsatellite DNA genotyping analysis. In total, 300 individuals were genotyped with 10 polymorphic microsatellite markers (MM12295, MM1031, MM2016, MM21723, MM11, MM14, MM15, MM16, MM5358, and MM12736) (Table 1). PCR was carried out in 25 μL volume containing 1× PCR buffer, 2.5 mM of MgCl2, 0.2 mM of each dNTP, 0.04 μM of forward and reverse primers, 1 U of Taq DNA polymerase (Takara), and ∼50 ng of genomic DNA. The forward primers were 5′ end fluorescently labeled with one of the following four fluorescent dyes: 6-FAM, HEX, ROX, and TMR. The conditions of PCR amplifications were as follows: 95°C for 5 min, followed by 35 cycles of 95°C for 30 s, locus-specific annealing temperature for 30 s, 72°C for 30 s, and a final extension at 72°C for 10 min. Amplified fragments were separated on an ABI 3730xl automated sequencer with the GeneScan 500 LIZ internal size standard. Alleles were scored with GeneMapper software v.4.0 (Applied Biosystems).
Statistical Analysis
The samples collected before restocking in 2011 were used as a baseline against to compare that after restocking for population variation analysis of M. meretrix. Prior to any calculations, data were standardized by transforming the clam abundances to the number of individuals per m2 (ind/m2; density) and the biomass to the weight of gram per m2 (g/m2; biomass). The mean values of abundance and biomass among seasons were expressed as the annual average of every year. ArcGIS 10.0 was used to illustrate the sampling sites of M. meretrix across the study area. The boxplot graph of variation in the mean shell length of commercial clam was established on SPSS (version 19.0).
The samples collected from 2013 to 2018 were used for genetic variability analysis of M. meretrix population. Microsatellite-based population genetic diversity and distance within each-year population were evaluated by the number of alleles (NA), observed heterozygosity (Ho), inbreeding coefficient (FIS), pairwise FST values with 106 permutations (Markov chain length), and significance levels of exact p-values adjusted using sequential Bonferroni corrections in Arlequin version 3.5.2.2 (Excoffier and Lischer, 2010). The genetic relationships among years were estimated using GENETIX version 4 (Belkhir et al., 2004). In addition, Bayesian clustering analysis implemented in STRUCTURE version 2.3.4 (Pritchard et al., 2000) was carried out to infer the population structure. The program was run with a period burn-in length of 20,000, followed by 30,000 Markov chain Monte Carlo steps under the admixture model with no prior information on the sampling location provided. K was set from 1 to 6, with 10 replications for each K-value; the optimal K was assessed on the basis of deltaK method (Evanno et al., 2005), which was calculated using STRUCTURE HARVESTER version 0.6.94 (Earl and VonHoldt, 2012). The population structure was visualized using DISTRUCT version 1.1 (Rosenberg, 2004).
Results
Population Variations of Meretrix meretrix
Abundance and Biomass
The population of clam M. meretrix underwent considerable year-to-year fluctuations (Figure 2). Before enhancing the clam stock, the abundance of M. meretrix was 0.7 ind/m2 in Shuangtaizi Estuary in 2011. The abundance of clams began to increase and stabilized gradually after the clam artificial release over 3 consecutive years. The abundance of M. meretrix was 2.3 ind/m2 in 2013, and it showed an upward trend in 2014 and 2015, with values of 4.1 and 5.0 ind/m2, respectively. The abundance of clams dropped to 1.7 and 1.3 ind/m2 in 2016 and 2017, respectively, and then lightly increases to 2.7 ind/m2 in 2018.
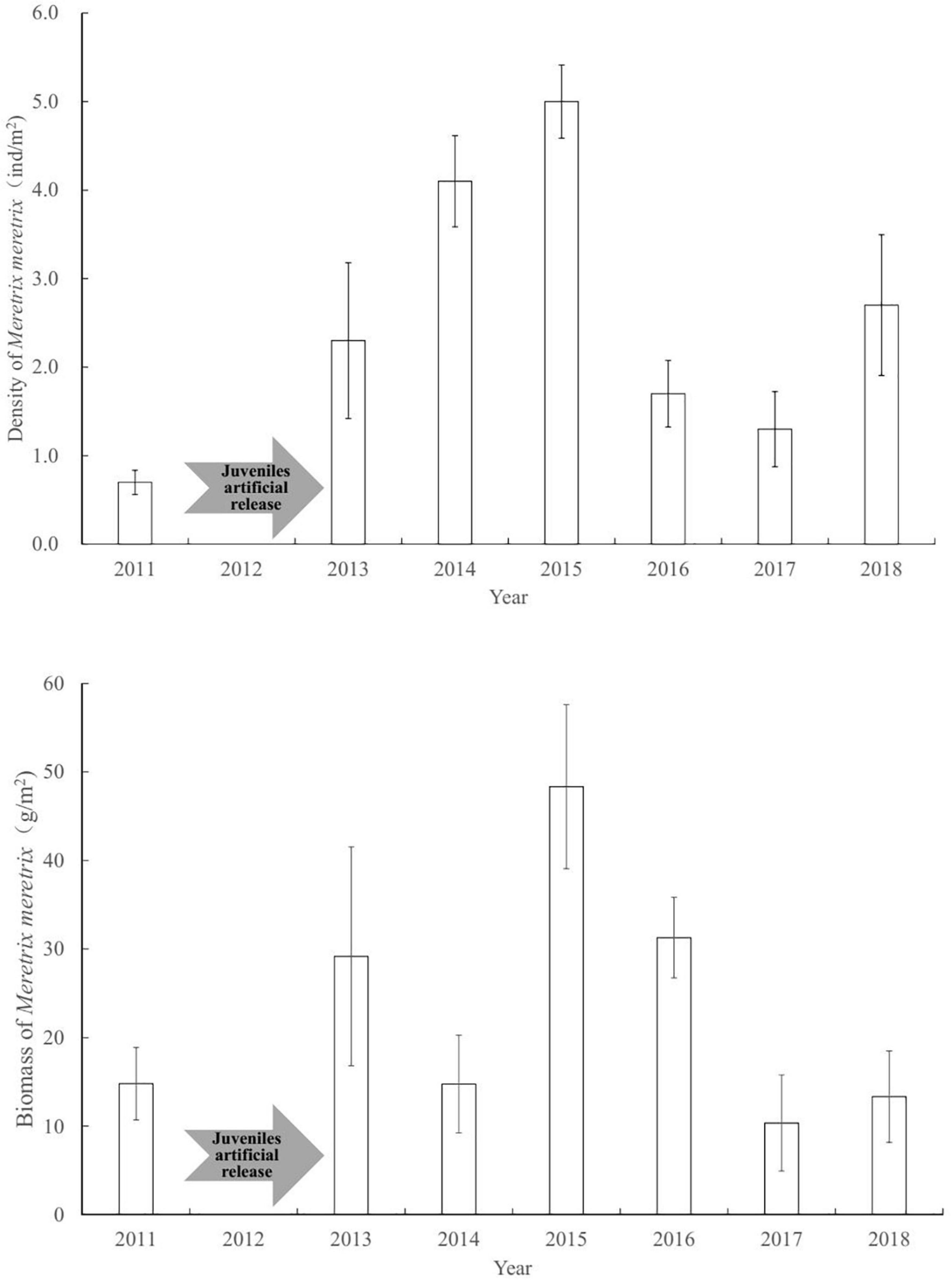
Figure 2. Density and biomass fluctuations of Meretrix meretrix population before and after clam stock enhancement. Bars indicate mean values. Error bars represent standard errors.
Similar to abundance variation, the biomass of M. meretrix revealed considerable year-to-year fluctuations (Figure 2). Before the juveniles artificial release, the biomass of M. meretrix was 14.8 g/m2 in Shuangtaizi Estuary in 2011. The mean annual biomasses of M. meretrix were 29.2 and 14.8 g/m2 in 2013 and 2014, respectively. Subsequently, the biomass reached the maximum post-release program of 48.3 g/m2 in 2015 but then decreased up to 31.3 g/m2 in 2016 (Figure 2). It finally stabilized between 10.4 and 13.3 g/m2 from 2017 to 2018.
Age Structure
Prior to the artificial release of M. meretrix individuals, the 1 year cohorts of clams were poorly represented, and accounted for only 13% of the total sampled clams found in 2011 (Figure 3). The percentages of 1- and 2-year-old clams in 2013, 2014, and 2015 approached or exceeded 50% but decreased from 2016 to 2018, with values of 16, 42, and 29%, respectively, which are close to or even more than the percentage of 4-year-old clams. Moreover, more than 3-year-old clams accounted for 84% in 2016 (Figure 3).
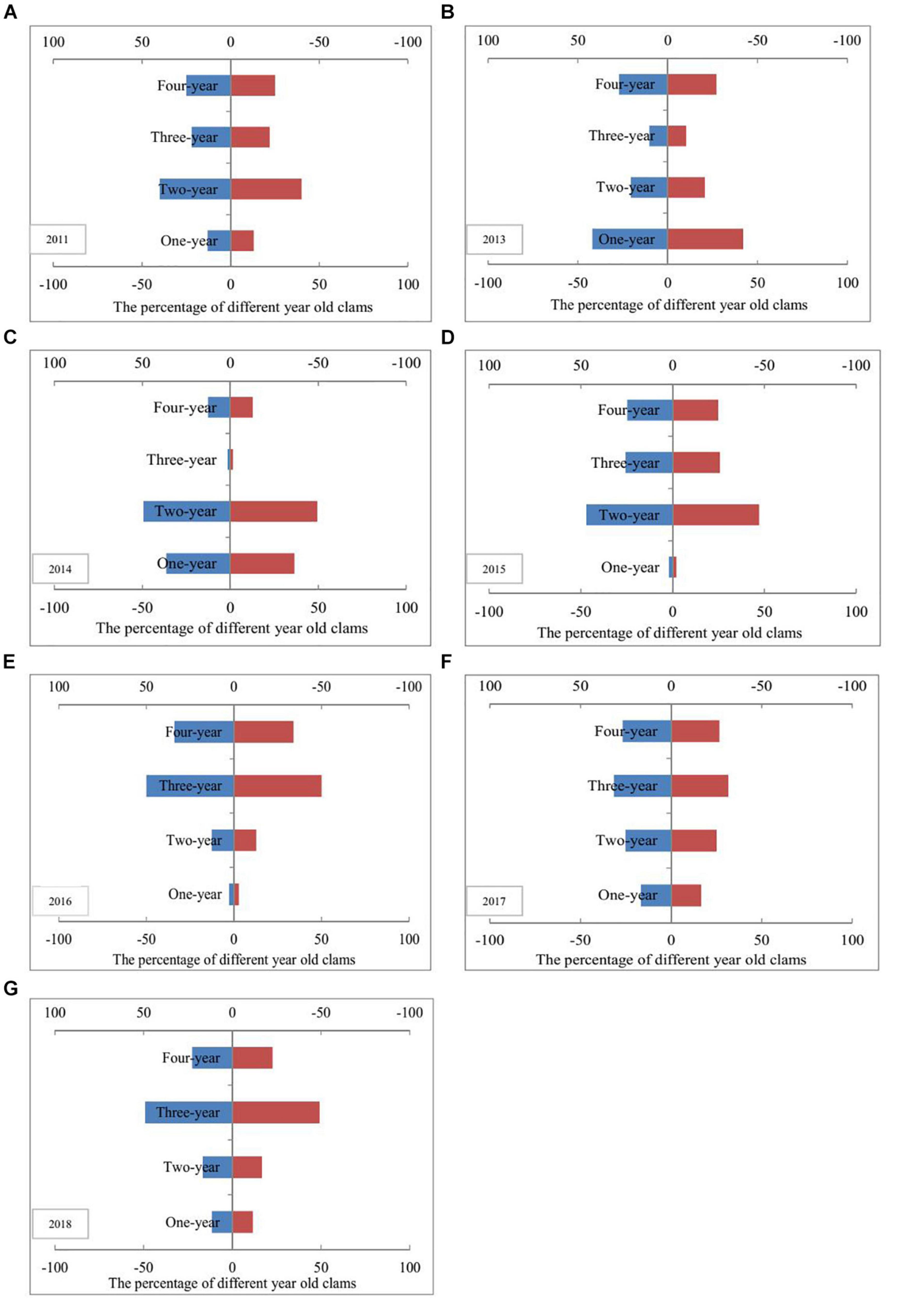
Figure 3. Age structure fluctuations of Meretrix meretrix population before and after clam stock enhancement. Panels (A–G) indicates the percentage of different year old clams in the year of 2011, 2013, 2014, 2015, 2016, 2017, and 2018, respectively.
Clam Distribution
In the aspect of distribution, the clams were found in eight sampling sites in the middle and western parts (sections C, D, F, and G) of the survey area in 2013. However, individuals were not found in sections A, B, and E of the survey area. The appearance frequency of clam individuals increased from 2014. The clams were found in two sampling sites of section A, and one sampling site in sections B and E.
Genetic Variability of Meretrix meretrix Population
Microsatellite Polymorphism Analyses
The number of average alleles showed no large variation as 11–14 alleles for per microsatellite loci were discerned between 2013 and 2018. A total of 75 alleles were amplified from 300 individuals by using 10 microsatellite markers (Table 2), with an average of 12.5 alleles for per microsatellite loci per year and the range of 11–14 alleles. The HO by populations in each year was 0.71–0.85, with a mean value of 0.79. Furthermore, the HO of population showed lower levels (0.71–0.81) in 2014–2018 than at the initial period (2013, 0.85) of stock restoration.
Population Genetic Structure
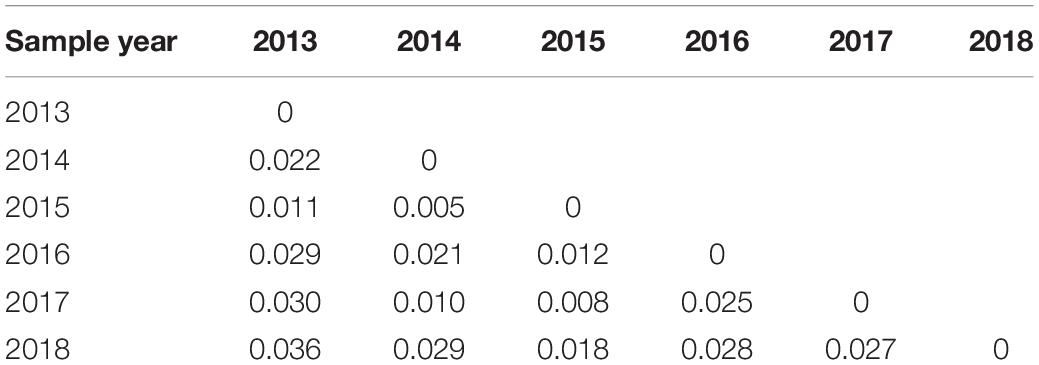
The results of pairwise population genetic differentiation indexes (FST) based on multi-locus SSR were shown in Table 3. The FST between each temporal sample showed no significant difference (P > 0.05) with low FST values (0.005–0.036), which indicated weak genetic differentiation among the different temporal samples (Table 3). Meanwhile, the FCA analysis revealed weak clustering of genotypes on the basis of sampling years (Figure 4), as also supported by low pairwise FST values among sampling years (Table 3).
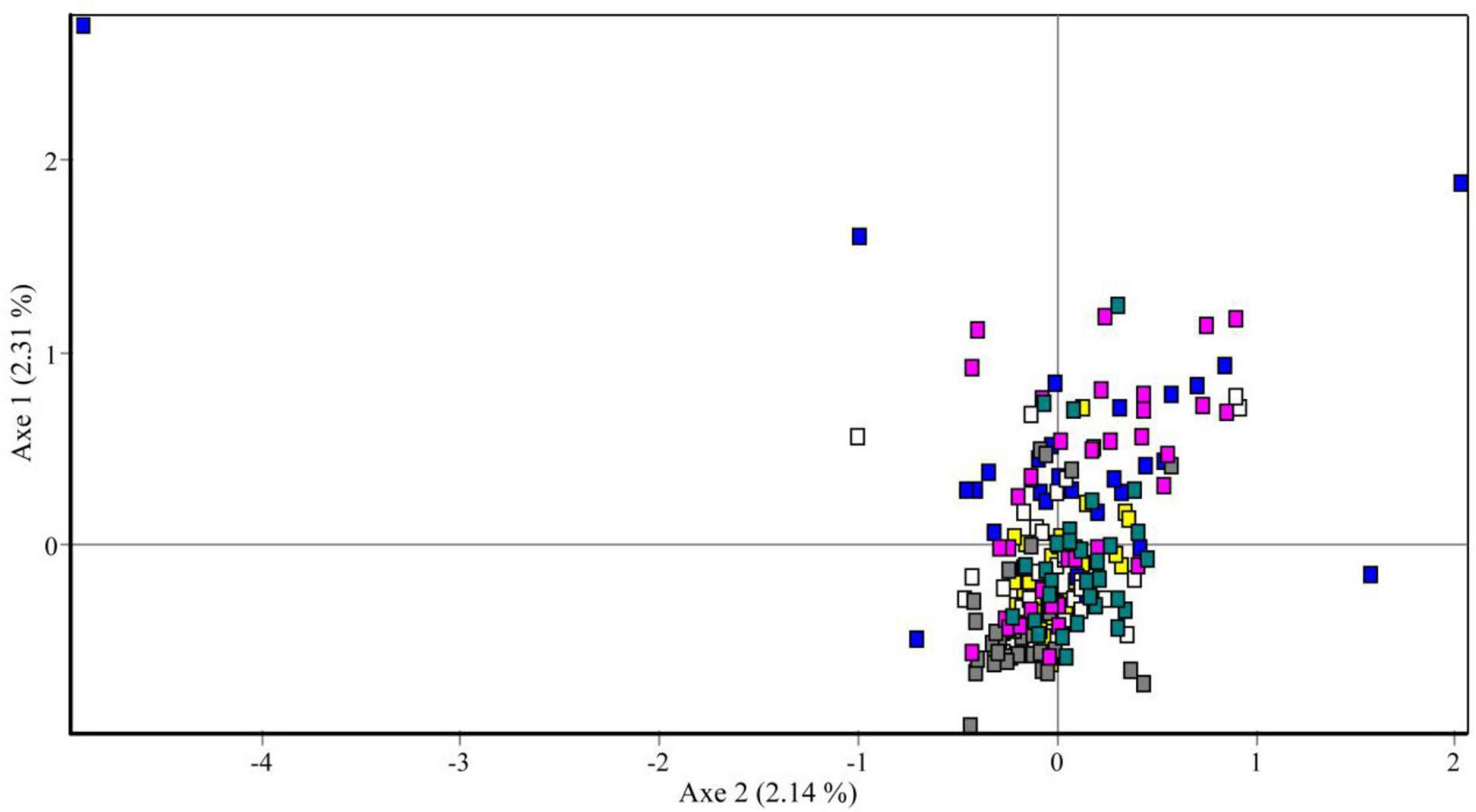
Figure 4. Factorial correspondence analysis of microsatellite data showing genetic similarities of sampled individuals. Yellow square, 2013; blue square, 2014; white square, 2015; gray square, 2016; pink square, 2017; and green square, 2018.
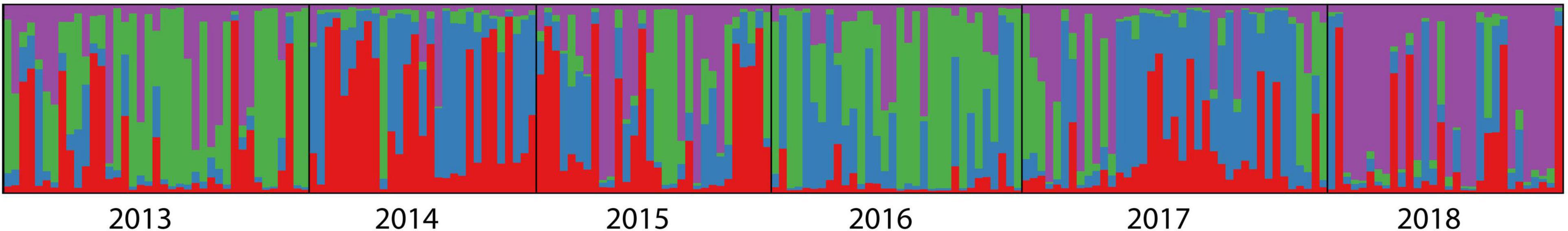
Figure 5 showed the results of the Bayesian clustering analysis for six different temporal samples of clusters (K). In STRUCTURE analysis, the delta K result showed that four clusters were the most likely model. However, the four clusters were all detected at each year (Figure 5), suggesting high genetic similarly among years.
Discussion
Population Dynamics of Clam
The results showed that the abundance of M. meretrix was low with the value of 0.7 ind/m2 in 2011. In contrast to the lower abundance of M. meretrix before the clam stock enhancement, the abundance and biomass extensively increased in 2013. The abundance increase could most probably be attributed to the recruitment events that occurred in 3 consecutive years from 2012 to 2014 in Shuangtaizi Estuary. The average abundance and biomass after the clam stock enhancement were approximately four and two times than those before the clam stock enhancement, respectively. These results suggested that the self-reproduction ability of M. meretrix population was gradually restored to a certain extent after successive artificial releasing of M. meretrix juveniles. In addition, M. meretrix is a rather long-lived species reported to be 4–5 years old (He and Chen, 1997). Under ideal conditions, the species does not need to produce excessive numbers of recruits every year, especially because growth and natural mortality were reported to be density dependent (Morello et al., 2011). The results of the present study revealed that like other invertebrate species, the natural population dynamics of M. meretrix is determined by not only density-dependent factors (Kitada and Kishino, 2006; Hamasaki and Kitada, 2008) but also environmental factors, such as more stable substrates for bivalve settlement. The decrease in environmental influences possibly intensifies the intraspecific modulation, although this needs further confirmation. Overall, the results pointed out that substantial clam recruitment events could yield a cohort so conspicuous to withstand and survive the fishing pressure to which it is subjected. Meanwhile, the source of M. meretrix may be plentiful now, but it does not mean it could sustain heavy and prolonged exploitation.
The results indicated that prior to the artificial release of M. meretrix individuals, the 1-year cohorts of clams were poorly represented, accounting for only 13% of the total sampled clams found in 2011. This age structure is probably a consequence of low recruitment success. The age structure of M. meretrix population is unreasonable and has been seriously destroyed to a critical situation. Consequently, the recovery ability of natural resources is weak. In the current study, the age structure of clams changed after a large number of M. meretrix individuals was released annually from 2012 to 2014. The results indicated that the percentage of 1-year old clam in 2013 increased by 29% compared with that in 2011. In accordance with the shell length and shell stripes of clams, the sampled 1-year old clams were presumed to come from the individuals released artificially in 2013, although the clams released were not tagged. Meanwhile, the 2-year-old clams in 2014 were presumed to come from the juveniles released artificially in 2013. The high proportion of 1-year-old clams from 2013 to 2014 contributed to the continuous artificial release activities from 2102 to 2014. The results also suggested that the high proportion of 3-year-old clams in 2016 were presumed to come from the M. meretrix individuals released between 2012 and 2014. However, the results suggested that the age structure of the populations from 2013 to 2018 is still an unstable expanding type. Two reasons possibly contributed to this phenomenon. First, the habitat was destroyed and has not been restored, and is still not conducive to clam growth. Second, the number of breeding population is still limited. Only a temporary spike in population individuals was observed after the artificial release of clam juveniles for 3 consecutive years, and the population was unstably growth-oriented. Therefore, the artificial release of clam juveniles and the habitat improvement are still needed to restore clam resources. In addition, if management measures are not taken to control overfishing, especially after the artificial release, the artificial releasing activities merely conducted for sustainable growth of the fishery resources could fail (Fang, 2014).
In addition, in terms of distribution variation, the distribution range of M. meretrix gradually expanded with the artificial propagation and artificial releasing of clam juveniles. The achievements of releasing demonstrated the feasibility to place them into intertidal flats under the existing conditions.
Genetic Variability of Meretrix meretrix
In the stock enhancement program, studies have shown that the stock enhancement activities not only restore the fishery resources but also pose potential risks of reducing the genetic diversity and changing the genetic structure of wild populations (Gonzalez et al., 2015). While the ecological effects of enhanced production on wild populations have been recognized, the genetic effects of supplementation have received much less attention. Large-scale releases of hatchery-reared juveniles have a potential to affect the genetic diversity of wild populations (Bell et al., 2008). Genetic influences on the viabilities of wild populations are more difficult to be observed and evaluated than ecological influences because genetic effects may not be as apparent as short-term shifts in abundance or age structure (Grant et al., 2017). Genetic principles have to be considered in stock supplementation, because numerous studies showed that natural disturbances and human manipulations of populations led to microevolutionary changes on short-time scales that may be irreversible (Law, 2000; Ashley et al., 2003; Hendry et al., 2011).
Sustaining genetic diversity within the populations is an essential goal in the stock enhancement programs for further adaptation to a changing environment (Allendorf and Luikart, 2007). Long-term monitoring of the possibility of genetic drift and other genetic changes are necessary for stock enhancement programs (Chen H. et al., 2019). It is regret that the clams were not sampled prior to clam release so that we do not have a genetic baseline to thorough compare against. However, the purpose of the genetic variability we analyzed is to show the effects of hatchery juveniles from local origin, but limited numbers of parents on the wild genetic structure of clams. Consequently, we analyzed and assessed the genetic diversity and genetic structure of M. meretrix population from the clam stock enhancement primary stage (2013–2014) to a later stage (2015–2018). In the present study, the obtained molecular genetic parameters (HO and FIS) showed a high level of genetic diversity of M. meretrix population and low pairwise FST values among six different temporal samples. Two reasons could be attributed to these results. First, recurrent supplementation in different years with juveniles from different batches of breeders that collected from the intertidal flat of Shuangtaizi Estuary could erase previous patterns (Arias-Pérez et al., 2016), and large numbers of native clams were used during the larvae hatchery process every year. Second, wild non-captured clams should greatly outnumber the clams released for restocking due to clam captures. Meanwhile, the results showed that the clustering of genotypes on the basis of sampling years was weak. The possible reason is that the samples were collected from different year, but individuals of all the temporal samples were overlapped to some extent. Overall, the results suggested that large-scale hatchery-reared enhancement program had no significant effect on the genetic diversity and genetic structure of the M. meretrix population. Moreover, no significant genetic differentiation was established after large-scale the stock enhancement, which is a good news for the resource protection of M. meretrix population in this case.
Yamakawa and Imai (2012) found that Meretrix petechialis populations replaced the local Meretrix lusoria and caused genetic disturbance via hybridization in Japan due to the artificial release activities of M. petechialis calms. In the present study, the structure analysis results suggested that the differentiated genetic structure of M. meretrix population to some degree and the potential risk of genetic contamination, which should not be neglected, due to the genetic diversity of hatchery-released buried clams. In addition, similar to Meretrix lamarckii, M. meretrix clams would more likely affect the genetic diversity of wild population because they are infaunal bivalves and have large fecundity (Qi et al., 2016). Therefore, native clams are encouraged to be artificially released to conserve the endangered local M. meretrix resource, thereby avoiding gene contamination by alien clams. The analysis of genetic population structure provides only a small amount of power to detect the effects of supplementation because the effects of environmental change, random drift, and migration could override small genetic signals of supplementation (Grant et al., 2017).
Depleted populations often showed heritable changes in demography with shifts to younger ages at maturity and to smaller sizes for a given age (Longhurst, 2010; Audzijonyte et al., 2013). Overharvested populations could lose genetic diversity (McCusker and Bentzen, 2010), and they showed shifts in genotypic frequencies, as noted by drops in growth rates and early maturity (Longhurst, 2010; Grant et al., 2017). Therefore, the most ecological friendly method for preserving the genetic diversity of M. meretrix resources is to limit harvests of clam stock.
Implications of Restoration Strategy for Meretrix meretrix Resources
Besides the effects of artificial releasing recruitment events reported in this study, broadened ecological consequences could be expected as bivalves play key roles in the intertidal ecosystem functioning (Zhang et al., 2014). Therefore, the results of the present study suggested that three management strategies and conservation policies could be used to protect the decreased local M. meretrix resources. First, sufficient numbers of native clam brood stocks are encouraged to be reproduced for the fewest number of generations as possible to minimize genetic changes, and large brood stock sizes could be used to reduce the loss of genetic diversity. Meanwhile, any future releases of M. meretrix must be controlled to avoid gene contamination. Second, the coastal provinces and municipalities should seek funding for the artificial releasing and recoup a percentage of funds from reselling the clams that they harvest. Meanwhile, local qualified nursery companies should be selected to assure adequate juvenile supply. Third, strict fishing regulations for M. meretrix stocks after the artificial release should be critically enforced under the supervision and management of local fishery departments. For instance, fishing regulations, including fishing methods (such as dredge pump and rakes), minimum legal size limit, seasonal and/or area closures, and total allowable catch of clams, should be implemented to provide the M. meretrix population with the chance to rejuvenate.
Conclusion
Approximately 2.4 × 108 hatchery juveniles of M. meretrix were artificially released into Shuangtaizi Estuary from 2012 to 2014. The results of ecological effect assessment indicated a temporary spike in the population numbers of M. meretrix, and the clams were not in a stable population growth condition. The genetic effects showed no significant genetic differentiation and weak genetic structures among years but higher genetic diversities in 2014–2018 than in the initial period (2013) of stock restoration. However, our study is limited to lack of genetic baseline prior to juvenile releasing. Adopting measures for releasing M. meretrix juveniles into Shuangtaizi Estuary to enhance clam stock is not only workable but also necessary. Meanwhile, management strategies and conservation policies to protect the nursery habitat recovery and fishing pressure reduction could outperform hatcheries in the future.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
AZ: conceptualization, funding acquisition, and writing – original draft preparation. HL: writing – original draft preparation, methodology, and formal analysis. XYa and LW: field investigation and formal analysis. YG and MS: formal analysis and data curation. XYu: conceptualization, funding acquisition, and writing – reviewing and editing. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Marine Public Welfare Research Project (201305043), the Open Research Fund of State Key Laboratory of Estuarine and Coastal Research (SKLEC-KF201911), the Doctoral Scientific Research Foundation of National Marine Environmental Monitoring Center (2018-A-21), and the Open Research Fund of Key Laboratory of Marine Eco-Environmental Science and Engineering, State Oceanic Administration (MESE-2018-03).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors wish to thank several referees for their professional comments on this manuscript, and the authors express their gratitude to all the volunteers who participated in the field and laboratory work.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.725238/full#supplementary-material
References
Allendorf, F. W., and Luikart, G. H. (2007). Conservation and the Genetics of Populations. Hoboken: Blackwell Publishing.
Arias-Pérez, A., Cordero, D., Borrell, Y., Sánchez, J. A., Blanco, G., Freire, R., et al. (2016). Assessing the geographic scale of genetic population management with microsatellites and introns in the clam Ruditapes decussatus. Ecol. Evol. 6, 3380–3404. doi: 10.1002/ece3.2052
Ashley, M. V., Willson, M. F., and Pergams. O. R. W., O’Dowd, D. J. (2003). Evolutionarily enlightened management. Biol. Conserv. 111, 115–123. doi: 10.1016/S0006-3207(02)00279-3
Audzijonyte, A., Kuparinen, A., Gorton, R., and Fulton, E. A. (2013). Ecological consequences of body size decline in harvested fish species: positive feedback loops in trophic interactions amplify human impact. Biol. Lett. 9:20121103. doi: 10.1098/rsbl.2012.1103
Belkhir, K., Borsa, P., and Chikhi, L. (2004). GENETIX 4.05, Logiciel Sous Windows Pour la Génétique des Populations. Laboratoire Génome, Populations, Interactions, CNRS UMR 5000. Montpellier: Université de Montpellier II.
Bell, J. D., Leber, K. M., Blankenship, H. L., Loneragan, N. R., and Masuda, R. (2008). A new era for restocking, stock enhancement and sea ranching of coastal fisheries resources. Rev. Fish. Sci. 16, 1–9. doi: 10.1080/10641260701776951
Blankenship, H. L., and Leber, K. M. (1995). A responsible approach to marine stock enhancement. Am. Fish. Soc. Symp. 15, 167–175.
Chen, H., Wang, D., Duan, X., Liu, S., Chen, D., and Li, Y. (2019). Hatchery-reared enhancement program for silver carp (Hypophthalmichthys molitrix) in the middle Yangtze River: monitoring the effectiveness based on parentage analysis. Peer J. 7:6836.
Chen, L. S., Wang, X. B., and Chen, D. Q. (2004). Research on the development of hard clam market in China mainland. J. Fish. Univ. Shanghai. 13, 283–287.
Chen, P. M., Shu, L. M., Yuan, H. R., Feng, X., Tong, F., Chen, Q., et al. (2019). Review on development, definition and classification of marine ranching in domestic and overseas. J. Fish. China 43, 1851–1869.
Chen, Y., Jiang, J. Y., Li, S. L., Li, K. W., Wang, Z. S., Liu, X. F., et al. (2012). The biological resources survey of mudflat clams and other major creatures in Panjin Geligang and Xiao He. Hebei. Fish. 1, 46–49.
Dong, Y. H., Yao, H. H., Lin, Z. H., Sun, C. S., and You, Z. J. (2013). Development of 53 novel polymorphic EST-SSR markers for the hard clam Meretrix meretrix and cross-species amplification. Conserv. Genet. Resour. 5, 811–816. doi: 10.1007/s12686-013-9914-8
Earl, D. A., and VonHoldt, B. M. (2012). Structure harvester: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 4, 359–361. doi: 10.1007/s12686-011-9548-7
Evanno, G., Madec, L., and Arnaud, J. F. (2005). Multiple paternity and postcopulatory sexual selection in a hermaphrodite: what influences sperm precedence in the garden snail Helix aspersa. Mol. Ecol. 14, 805–812. doi: 10.1111/j.1365-294x.2005.02449.x
Excoffier, L., and Lischer, H. E. L. (2010). Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 10, 564–567. doi: 10.1111/j.1755-0998.2010.02847.x
Fang, K. Q. (2014). Research on China’s Ecosystem-Based Fisheries Management. Qingdao: Qingdao Ocean University Press.
Fei, H. N., and Zhang, S. Q. (1990). Aquatic Resource Science. Beijing: China Science and Technology Press, 182–194.
Gonzalez, E. B., Aritaki, M., Knutsen, H., and Taniguchi, N. (2015). Effects of large-scale releases on the genetic structure of Red Sea Bream (Pagrus major, Temminck et Schlegel) populations in Japan. PLoS One 10:125743.
Gonzalez, E. B., Nagasawa, K., and Umino, T. (2008). Stock enhancement program for black sea bream (Acanthopagrus schlegelii) in Hiroshima bay: monitoring the genetic effects. Aquaculture 276, 36–43. doi: 10.1016/j.aquaculture.2008.02.004
Grant, W. S., Jasper, J., Bekkevold, D., and Adkison, M. (2017). Responsible genetic approach to stock restoration, sea ranching and stock enhancement of marine fishes and invertebrates. Rev. Fish. Biol. Fish. 27, 615–649. doi: 10.1007/s11160-017-9489-7
Gu, W., Liu, C. Y., Yuan, S., Li, N., Chao, J. L., Li, L. T., et al. (2013). Spatial distribution characteristics of sea-ice-hazard risk in Bohai. China Ann. Glaciol. 54, 73–79. doi: 10.3189/2013aog62a303
Hamasaki, K., and Kitada, S. (2006). A review of kuruma prawn Penoeus japonicas stock enhancement in Japan. Fish. Res. 80, 80–90. doi: 10.1016/j.fishres.2006.03.018
Hamasaki, K., and Kitada, S. (2008). The enhancement of abalone stocks: lessons from Japanese case studies. Fish. Fish. 9, 243–260. doi: 10.1111/j.1467-2979.2008.00280.x
He, C. B., and Chen, H. D. (1997). Study on the growth and ecological characteristics of Meretrix meretrix cultivated on tidal flat. J. Fish. Sci. 16, 17–20.
Hendry, A. P., Kinnison, M. T., Heino. M., Day, T., Smith, T. B., Fitt, G., et al. (2011). Evolutionary principles and their practical application. Evol. Appl. 4, 159–183. doi: 10.1111/j.1752-4571.2010.00165.x
Kitada, S. (2018). Economic, ecological and genetic impacts of marine stock enhancement and sea ranching: a systematic review. Fish. Fish. 19, 511–532. doi: 10.1111/faf.12271
Kitada, S. (2020). Lessons from Japan marine stock enhancement and sea ranching programmes over 100 years. Rev. Aquacult. 12, 1944–1969.
Kitada, S., and Kishino, H. (2006). Lessons learned from Japanese marine finfish stock enhancement programmes. Fish. Res. 80, 101–112. doi: 10.1016/j.fishres.2006.03.019
Kitada, S., Nakajima, K., Hamasaki, K., Shishidou, H., Waples, R. S., and Kishino, H. (2019). Rigorous monitoring of a largescale marine stock enhancement program demonstrates the need for comprehensive management of fisheries and nursery habitat. Sci. Rep. 9:5290.
Laikre, L., Schwartz, M. K., Waples, R. S., and Ryman, N. (2010). Compromising genetic diversity in the wild: unmonitored large-scale release of plant and animals. Trends Ecol. Evol. 25, 520–529. doi: 10.1016/j.tree.2010.06.013
Law, R. (2000). Fishing, selection, and phenotypic evolution. ICES J. Mar. Sci. 57, 659–668. doi: 10.1006/jmsc.2000.0731
Lee, S. I., and Zhang, C. I (2018). Evaluation of the effect of marine ranching activities on the Tongyeong marine ecosystem. Ocean Sci. J. 53, 557–582. doi: 10.1007/s12601-018-0045-8
Li, H. J., Zhang, J. J., Yuan, X. T., Zhang, A. G., Liu, G. Z., Shao, K. S., et al. (2016). Genetic diversity and differentiation of seven geographical populations of hard clam (Meretrix meretrix) assessed by COI and microsatellite markers. Acta. Ecol. Sin. 36, 499–507.
Lu, X., Wang, H. X., Dai, P., and Liu, B. Z. (2011). Characterization of EST-SSR and genomic-SSR markers in the clam, Meretrix meretrix. Conserv. Genet. Resour. 3, 655–658. doi: 10.1007/s12686-011-9426-3
Luo, G., and Zhang, Z. D. (2014). The development status of aquatic organisms in China. China Fish. 12, 37–39.
McCusker, M. R., and Bentzen, P. (2010). Positive relationships between genetic diversity and abundance in fishes. Mol. Ecol. 19, 4852–4862. doi: 10.1111/j.1365-294x.2010.04822.x
Morello, E. B., Martinelli, M., Antolini, B., Gramitto, M. E., Arneri, E., and Froglia, C. (2011). Population Dynamics of the Clam, Chamelea gallina, in the Adriatic Sea (Italy). Department of Earth and Environment, Marine Research at CNR, National Research Council of Italy, Roma. Italy: National Research Council, 1907–1921.
Pritchard, J. K., Stephens, M., and Donnelly, P. (2000). Inference of population structure using multilocus genotype data. Genetics 155, 945–959. doi: 10.1093/genetics/155.2.945
Qi, J. F., Zeng, N. M., Ning, Y., and Wu, Q. S. (2016). Construction of ecological risk assessment system for stock enhancement in zoobenthos. J. Fish. China. 40, 1099–1105.
Qin, C. X., Chen, P. M., Sara, G., Mo, B. L., Zhang, A. K., and Li, X. G. (2020). Ecological implications of purple sea urchin (Heliocidaris crassispina, Agassiz, 1864) enhancement on the coastal benthic food web: evidence from stable isotope analysis. Mar. Environ. Res. 158:104957. doi: 10.1016/j.marenvres.2020.104957
Rosenberg, N. A. (2004). DISTRUCT: a program for the graphical display of population structure. Mol. Ecol. Notes. 4, 137–138. doi: 10.1046/j.1471-8286.2003.00566.x
State Standard of the People’s Republic of China (2007). The Specification for Marine Monitoring Part 3: Sampling Collection, Storage and Transportation. Beijing: China Standard Press.
Tang, Q. S. (2019). Fishery resources multiplication, marine ranching, multiplication fishery and its development orientation. China Fish. 28–29.
Villella, D. R., and Weller, D. A. (2000). Evaluation of tag types and adhesives for marking freshwater mussels. J. Shellfish. Res. 1, 247–250.
Wang, R. C., Wang, Z. P., and Zhang, J. Z. (1993). Marine Shellfish Aquaculture. Qingdao: Qingdao Ocean University Press, 322–324.
Wang, Y., Yu, J., and Chen, P. (2018). Remote sensing assessment of ecological effects of marine ranching in the eastern Guangdong waters. China J. Geosci. Environ. Prot. 6, 101–113.
Wei, H. Q., Zhang, F. Y., Jiang, K. J., Jiang, Y. Z., Meng, Y. Y., Song, W., et al. (2016). Genetic diversity analysis of Marsupenaeus japonicus population for reproduction and release. J. Fish. Sci. China. 23, 530–537.
Xu, K. D., Zhou, Y. D., and Wand, Y. (2018). Effect and assessment of enhancement release of Sepiella maindroni in the northern coastal water of Zhejiang. J. Fish. Sci. China 25, 654–662. doi: 10.3724/sp.j.1118.2018.17412
Yamakawa, A. Y., and Imai, H. (2012). Hybridization between Meretrix lusoria and the alien congeneric species M. petechialis in Japan as demonstrated using DNA markers. Aquat. Invasions 7, 327–336. doi: 10.3391/ai.2012.7.3.004
Yang, S., Song, N., Zhang, X. M., Wang, Y. Z., Wang, S. J., and Gao, T. X. (2014). Genetic diversity of swimming crab (Portunus triberculatus) from four broodstock populations in stock enhancement inferred from mitochondrial control region. J. Fish. China 38, 1089–1096.
Yuan, X. T., Zhao, Q., and Zhang, A. G. (2021). The Environmental Capacity And Tidal-Flat Clams Resources Restoration in Liaodong Bay. Beijing: Science Press.
Zhang, A. G. (2015). Meretrix Meretrix Resource Restoration and its Interactions With Estuarine in Shuangtaizi estuary. Ningbo: Ningbo University, 44–49.
Zhang, A. G., Yuan, X. T., Hou, W. J., Chen, W. X., Zhao, K., Ba, F. Y., et al. (2014). Influence of biodeposition, respiration, and excretion of the buried clam Meretrix meretrix on the pelagic-benthic coupling in Shuangtaizi Estuary. Acta. Ecol. Sin. 34, 6573–6582.
Zhang, S. P., Wang, H. X., and Xu, F. S. (2012). Taxonomic study on Meretrix (Bivalvia, veneridae) from China seas. Acta. Zootax. Sin. 37, 473–479.
Zhang, X. M., Wang, X. J., Tu, Z., Zhang, P. D., Wang, Y. Z., Gao, T. X., et al. (2009). Current status and prospect of fisheries resource enhancement in Shandong Province. Chin. Fish. Econ. 27, 51–58.
Zhou, X., Zhao, X., Zhang, S. Y., and Lin, J. (2019). Marine ranching construction and management in East China Sea: programs for sustainable fishery and aquaculture. Water 11:1237. doi: 10.3390/w11061237
Keywords: Meretrix meretrix, stock enhancement, stock assessment, population dynamics, genetic effect
Citation: Zhang A, Li H, Yang X, Wang L, Gao Y, Song M and Yuan X (2021) Stock Assessment of Hatchery-Released Clam Meretrix meretrix in an Estuary of China From the Perspectives of Population Ecology and Genetic Diversity. Front. Mar. Sci. 8:725238. doi: 10.3389/fmars.2021.725238
Received: 15 June 2021; Accepted: 01 September 2021;
Published: 30 September 2021.
Edited by:
Huang Wei, Second Institute of Oceanography, Ministry of Natural Resources, ChinaReviewed by:
Jin-Xian Liu, Institute of Oceanology, Chinese Academy of Sciences, ChinaYunwei Dong, Ocean University of China, China
Copyright © 2021 Zhang, Li, Yang, Wang, Gao, Song and Yuan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiutang Yuan, eHR5dWFuQHlpYy5hYy5jbg==
 Anguo Zhang
Anguo Zhang Hongjun Li
Hongjun Li Xiaolong Yang
Xiaolong Yang Lili Wang1
Lili Wang1 Xiutang Yuan
Xiutang Yuan