Characterization and Function Analysis of β, β-carotene-9′, 10′-oxygenase 2 (BCDO2) Gene in Carotenoid Metabolism of the Red Shell Hard Clam (Meretrix meretrix)
- 1College of Fisheries, Henan Normal University, Xinxiang, China
- 2Zhejiang Key Laboratory of Aquatic Germplasm Resources, College of Biological & Environmental Sciences, Zhejiang Wanli University, Ningbo, China
- 3Ninghai Institute of Mariculture Breeding and Seed Industry, Zhejiang Wanli University, Ningbo, China
The relationship between carotenoid and shellfish shell color has gained increasing attention. β, β-carotene-9′,10′-oxygenase 2 (BCDO2) is a key enzyme in animal carotenoid metabolism, and its accumulation affects the change in body color, as demonstrated in mammals, birds, and fish. However, it is unclear whether BCDO2 is involved in the formation of the red shell color of clam. To explore the molecular structure and biological function of BCDO2 gene in the process of carotenoids accumulation, in this study, the BCDO2 from hard clam Meretrix meretrix (designated as Mm-BCDO2) was cloned and characterized, and the single-nucleotide polymorphisms (SNPs) associated with shell color were detected. The results of qRT-PCR indicated that Mm-BCDO2 gene was expressed in all six tested tissues, and the expression of mantle was significantly higher than other tissues (P < 0.05). The association analysis identified 20 SNPs in the exons of Mm-BCDO2, among which three loci (i.e., c.984A > C, c.1148C > T, and c.1187A > T) were remarkably related (P < 0.05) to the shell color of clam. The western blot analysis revealed that the expression level of Mm-BCDO2 in the mantle of red shell clams was stronger than that of white shell clams (P < 0.05). Further, the immunofluorescence analysis indicated that the single-layer columnar cells at the edge of the mantle were the major sites for the Mm-BCDO2 secretion. This study explored the potential impacts of BCDO2 gene on the shell color of M. meretrix, which provided a theoretical basis for a better understanding of the important role of BCDO2 in carotenoid metabolism.
Introduction
Carotenoids are a class of fat-soluble pigments, which can be classified into two groups: the xanthophylls (oxygenated-contained group) and the carotenes (hydrocarbons without oxygenated group) (Wu et al., 2016). It has been reported that carotenoids play an important role in various aspects of animal life. For example, it acts as precursors of vitamin A and antioxidants to involve in host physiological response (Svensson and Wong, 2011; Milani et al., 2017). The zeaxanthin, lutein, and astaxanthin have a positive effect on antioxidant and light filtering by preventing vision loss and eye diseases (Li et al., 2017), hence protecting the eyesight to a certain extent (Felix et al., 2011). Moreover, carotenoids and their derivatives are involved in the coloration of feathers, skin, and shells, which play a signal role in mate selection, social competition, and species recognition (Blount and Mcgraw, 2008).
Notably, animals that are unable to de novo synthesize carotenoids can get them from the diet (Zheng et al., 2010). The metabolic process of carotenoids in animals includes a series of metabolic reactions. Enzyme degradation is the key step for the utilization of carotenoids. So far, two types of carotene oxygenase, β-carotene 15, 15′-monooxygenase (BCMO1) and β-carotene 9′, 10′-dioxygenase (BCDO2), have been identified in vertebrates (Wu et al., 2016). BCMO1 is a key enzyme that converts β-carotene into vitamin A. Most β-carotene is symmetrically cleaved to form metabolites under the action of BCMO1 (Georg et al., 2010). BCDO2 displays broad substrate specificity and can cleave β-carotene, generating beta-10′-apocarotenal and beta-ionone; contrarily, beta-10′-apocarotenal can be cleaved by BCMO1 to produce retinaldehyde and downstream derivatives including retinol and retinoic acid (Seña et al., 2016; Jin et al., 2020). In addition, BCMO1 catalyzes the asymmetric cleavage of xanthophylls to generate rosafluene and ionone (Seña et al., 2016). Studies have found that the expression level of BCDO2 affects the body color of animals (Eriksson et al., 2008). Similarly, researches on the body surface color of wall lizard have showed that the content of lutein and zeatin is remarkably higher in yellow skin lizard compared with that in white skin lizard, which corresponds to the significantly lower expression level of BCDO2 in yellow skin lizard (Andrade et al., 2019). Genetic studies on the effect of BCDO2 mutations on “yellow fat” phenotype in cattle, sheep, and chicken have showed that BCDO2 is implicated in the carotenoid homeostasis of milk color, fat color, and skin color (Eriksson et al., 2008; Berry et al., 2009; Tian et al., 2009; Vge and Boman, 2010). Genetic disruption (low-temperature expression and detergent choice) of BCDO2 can significantly affect binding and turnover rates of the recombinant enzymes with various xanthophyll substrates and then increase the concentration of dietary pigment in plasma and blood of the mice (Babino et al., 2015).
Currently, researches on the role of carotenoids on the coloring of tissues and shells have been reported in Meretrix meretrix (Lin and Dong, 2015; Cui et al., 2018), Crassostrea gigas (Ge et al., 2016; Wang et al., 2016, 2018), Pinctada fucata (Zhang et al., 2019), Hyriopsis cumingii (Li et al., 2014b), Paphia textile (Deng et al., 2018), Chlamys nobilis (Zheng et al., 2010), and Exopalaemon carinicauda (Jin et al., 2020). The hard clam (M. meretrix) mainly distributed in the coastal area of Asia is one of the most important commercially cultured bivalves in China (Lin, 2015). Notably, clam shell displays two kinds of typical and differentiated colors, including white and red (Lin, 2015). The Raman spectra of the shell layer of M. meretrix showed that the characteristic peaks of the carotenoids of the red shell clams are stronger than those of the white shell clams (Zhan, 2015). And the total carotenoid content in the marginal membrane of the red clam mantle is about 3.4 times that of the white clam, which is a very significant difference (Qi, 2014). Especially, the ability of individual clams to ingest, absorb, and metabolize carotenoids is different, which may be one of the main reasons for the difference in shell color (Meilisza et al., 2017). However, it is unclear whether this difference attributes to the expression of the BCDO2 gene in M. meretrix. To address this gap, the expression level of the BCDO2 gene was compared between red and white shell color M. meretrix. Further, we investigated the distribution and SNPs of the BCDO2 gene in the two groups. Our findings will provide valuable information for further studies on the mechanism of clam shell color formation.
Materials and Methods
Ethics Statement
The adult hard clams (M. meretrix) at the age of 2 years were collected from the genetic breeding research center of Zhejiang Wanli University, China. All experimental procedures were approved by the Institutional Animal Care and Use Committee (IACUC) of Zhejiang Wanli University, China.
Sample Collection and Preparation
Six tissues including gill, siphon, digestive gland, adductor muscle, mantle, and foot were dissected, frozen immediately in liquid nitrogen, and then stored at −80°C.
RNA Extraction and cDNA Synthesis
Total RNA was extracted from the foot, mantle, digestive gland, siphon, gill, and adductor muscle by using the Trizol method. The first-strand cDNA was synthesized using the prime script RT reagent kit with gDNA Eraser (TAKARA, Japan) according to the instructions of the manufacturer. In brief, the amplification reactions were performed in a 20 μL volume containing 7 μL tissue RNA (1,500 μg/μL), 1 μL gDNA eraser, 2 μL 5 × gDNA eraser buffer, 1 μL prime script RT enzyme mix 1, 4 μL 5 × prime script buffer 2, 1 μL RT prime mix, and 4 μL RNase free dH2O. The first-strand cDNA was synthesized by incubating the mixture at 42°C for 2 min, 37°C for 15 min, and finally, heating at 85°C for 5 s.
Cloning of Full-Length cDNA
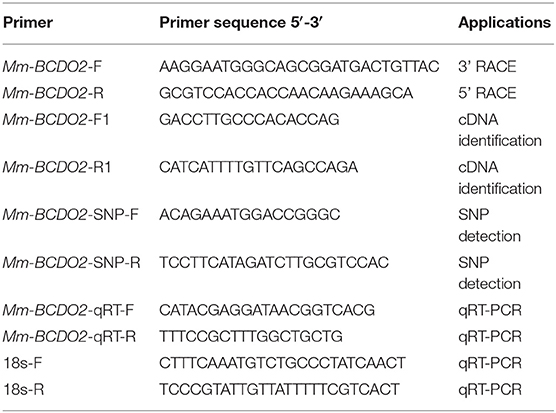
The expressed sequence tag (EST) homologous to Mm-BCDO2 was detected through the cDNA library of M. meretrix (GenBank accession no. SRX023927). A pair of gene-specific primers, Mm-BCDO2-F and Mm-BCDO2-R (Table 1), was designed on the basis of EST sequence of Mm-BCDO2 gene. The PCR amplification was synthesized following the instructions of SMARTerTM RACE cDNA amplification kit (Clontech, San Francisco, USA). The RACE PCR was performed in a 25 μL volume, which contained DEPC water, 10 × advantage 2 PCR buffer, 10 mM of dNTPs, 10 μM of gene-specific primer, 10 × universal primer A mix (UPM), 1 μL of diluted RACE cDNA, and 50 × advantage 2 polymerase mix. The amplification was carried out as follows: 5 cycles of 94°C for 30 s, 72°C for 3 min; 5 cycles of 94°C for 30 s, 70°C for 30 s, 72°C for 3 min; and 25 cycles of 94°C for 30 s, 68°C for 30 s, 72°C 3 min. PCR products were examined on 1.0% agarose gels and purified by gel extraction kit (Tiangen, Beijing, China). The purified PCR products were cloned into pEasy-T1 (Trans, Beijing, China), and T1 vector was transformed into E. coli DH5a, and the positive plasmids were selected and sequenced. A pair of gene-specific primers, Mm-BCDO2-F1 and Mm-BCDO2-R1 (Table 1), was designed based on the full-length cDNA sequence and used to confirm the accuracy of cloning and sequencing of Mm-BCDO2.
Sequence and Phylogenetic Analysis
The cDNA sequence was assembled using the BLASTX search program of the National Center for Biotechnology Information (NCBI). The open reading frame (ORF) of Mm-BCDO2 cDNA was identified using the ORF Finder program from NCBI. The theoretical isoelectric point (pI) and molecular weight (Mw) of Mm-BCDO2 protein were calculated using Compute pI/Mw Tool. Conserved domain of the deduced Mm-BCDO2 was predicted by simple modular architecture research tool (SMART). Tertiary structure of the amino acid sequence of deduced Mm-BCDO2 was analyzed using the SWISSMODEL tool. Multiple alignments of BCDO2 proteins between M. meretrix and other species were performed using ClustalW2 program. A phylogenetic tree was constructed by the neighbor-joining (NJ) method with MEGA X software.
Quantitative Expression Analysis
The mRNA expression levels of Mm-BCDO2 at different adult tissues (n = 4, four sets of samples per tissue) were analyzed by real-time quantitative reverse transcription PCR (qRT-PCR). A pair of gene-specific primers, Mm-BCDO2 qRT-F/R (Table 1), was used to quantitatively detect the expression level of Mm-BCDO2 with 18S rRNA as an internal reference. The qRT-PCR was performed in 12 μL of iTaq universal SYBR green super mix (Bio-Rad, Beijing, China), 7.2 μL deionized water, 0.8 μL of the first-strand cDNA, and 1 μL of each primer. The amplification was carried out at the following condition: incubation of 20 s at 94°C; 40 cycles of 3 s at 94°C, 15 s at 60°C, and 10 s at 72°C; and a final extension of 7 min at 72°C. All amplifications were performed in triplicate as biological replicates, and negative controls were run in the absence of cDNA templates. The 2−ΔΔCT method was used to analyze the expression level of Mm-BCDO2. The expression levels of Mm-BCDO2 among different tissues and between red and white shell clam groups were compared using the one-way ANOVA and Student's t-tests, respectively.
SNPs of Mm-BCDO2 and Their Association With Clam Shell Color
A pair of primers, Mm-BCDO2-SNP-F/R (Table 1), was designed based on the cDNA sequence of Mm-BCDO2 gene; 180 adult hard clams at 2 years of age were used for the SNP detection, collected from one population in Ningbo, Zhejiang Province, China. Total RNA was extracted from the mantle of clams, and then, the cDNA was synthetized using the methods described above. SNPs were detected by directly sequencing the PCR products using Sanger technology and the ABI3730 platform (Applied Biosystems, Foster City, CA, USA). The SNP genotypes were analyzed by using the Mutation Surveyor software version 4.0.8. The software PopGen32 was used to identify deviations from Hardy–Weinberg equilibrium and polymorphism information content (PIC) of significantly different SNP sites. The associated loci linkage disequilibrium (LD) was analyzed by the SHEsis online (http://analysis.bio-x.cn/myAnalysis.php). The difference in the genotype frequency of SNP locus between red and white shell clam populations was examined using the chi-square (χ2) text. The correlation between the SNPs of Mm-BCDO2 and clam shell color traits was analyzed by using the SPSS software version 20.0. The one-way ANOVA was adopted to compare the difference among different genotypes.
Immunofluorescence Analysis
The marginal tissues of the mantle and digestive gland were chosen from red and white shell clams, respectively. These samples were fixed with 4% paraformaldehyde, then dehydrated by gradient ethanol and xylene/n-butanol transparent, and paraffin-embedded sections (thickness 4 μm) were prepared. After baking, the tissue slices were deparaffinized with xylene, rehydrated with gradient ethanol, washed with PBS, and retrieved with EDTA antigen. Sections were then blocked for 1 h in blocking solution (5% albumin from bovine serum) at room temperature, followed by primary antibody (antibody rabbit anti-Mm-BCDO2, produced privately by HuaBio, Zhejiang, 1:200) incubation (overnight, 4°C). Primary antibody was detected by secondary antibodies Alexa Fluor 488 donkey anti-rabbit immunoglobulin G (Invitrogen, Carlsbad, California, U.S.) and diluted at 1:150. Nuclei were stained with DAPI (Beyotime, Shanghai, China). The cells were observed and photographed under a fluorescence microscope (Nikon Eclipse 80i, Tokyo, Japan).
Western Blot Detection
The mantle and digestive gland tissues were dissected from red and white shell clams, respectively. The samples were added with an appropriate amount of RIPA lysate and then homogenized. The suspension was collected to measure protein concentration by BCA method, configured SDS-PAGE gel with appropriate concentration and loaded the same amount. The protein glue with the target protein was transferred to PVDF membrane and then blocked for 1 h in blocking solution (5% skimmed milk powder solution) at room temperature, followed by primary antibody (antibody rabbit anti-Mm-BCDO2, produced privately by HuaBio, Zhejiang, 1:1,000) incubation (overnight, 4°C). Primary antibody was bound by secondary antibodies [anti-rabbit labeled with biotin HRP (1:8,000)] for 1 h. ECL luminescent substrate mixture was dropped on the PVDF membrane. The proteins were observed and photographed under gel imagers (Bio-Rad, Beijing, China).
Results
Molecular Characterization of Mm-BCDO2
The full-length cDNA of Mm-BCDO2 gene was 2,040 bp, including a 5' untranslated region (UTR) of 113 bp, an open reading frame (ORF) of 1,566 bp that encoded 521 amino acids, and a 3' UTR of 361 bp existing a tail signal (AATAAA) after terminator codon (Figure 1A). Based on the amino acid sequence encoded by the BCDO2 gene, the protein molecular weight was predicted to be 60.06 kDa, corresponding isoelectric point (pI) = 5.83, including a RPE65 domain (11–308, 356–517 aa). Mm-BCDO2 was a hydrophilic protein with a large proportion of polar amino acids. Protein structure prediction showed that Mm-BCDO2 protein composed of 9 α helices, 38 β-sheets, and a large number of random coils and had the typical characteristics of RPE65 protein family with a cleft and a right-handed twisted anti-parallel β-barrel (Figure 1B). The multiple comparisons between the BCDO2 of mollusks and model animals showed that Mm-BCDO2 had a relatively low similarity with other species (15−44%). Among them, Mm-BCDO2 had the highest homology with Tauraco erythrolophus (43.36%), the lowest homology with Trichinella spiralis (15.15%), and the homology with shellfish Mizuhopecten yessoensis (27.34%) (Figure 1C).
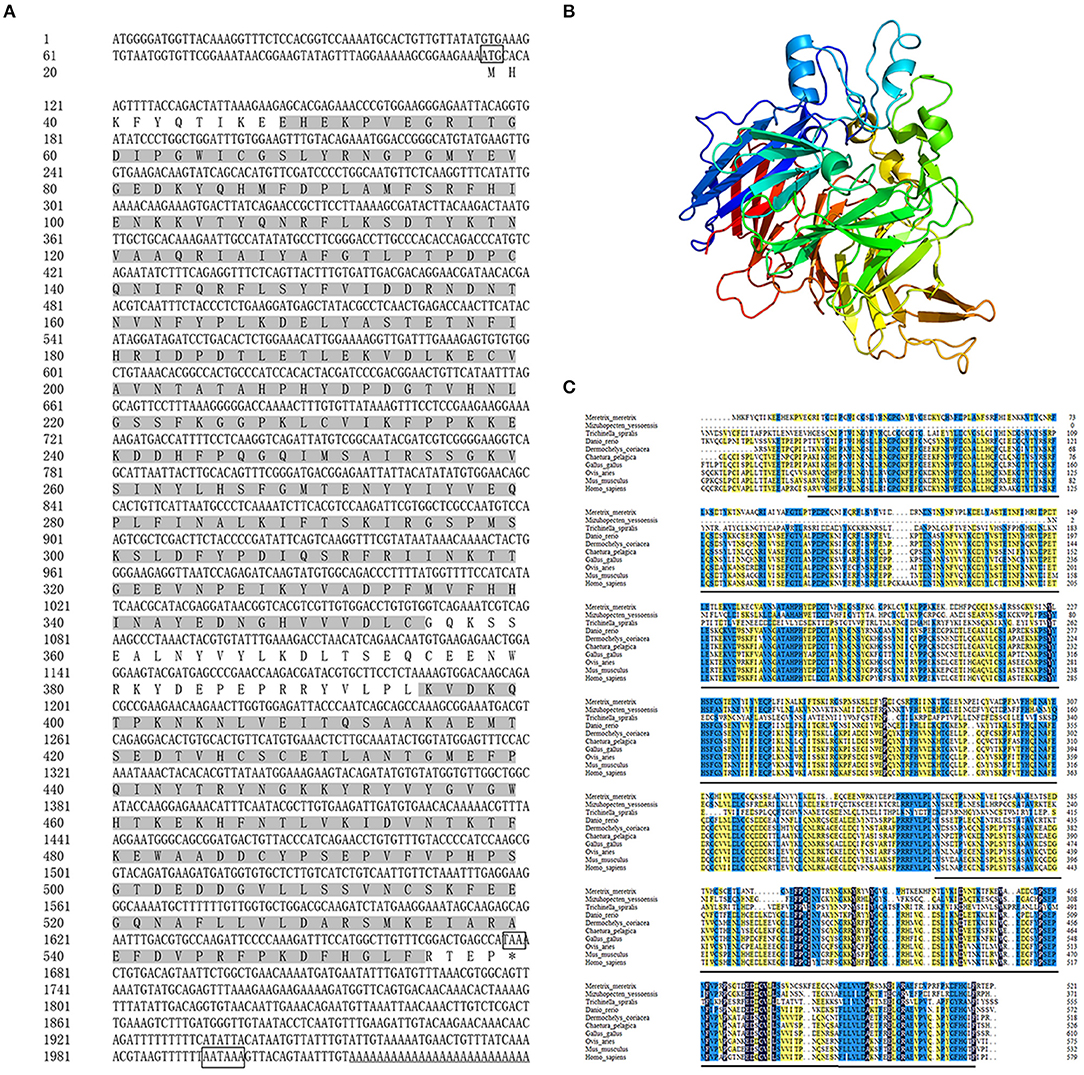
Figure 1. The information of the Mm-BCDO2 gene. (A) The full length of cDNA and deduced amino acid sequence of Mm-BCDO2. The frames represented the start codon, the stop codon, and polyadenylation signal. The gray background fill is the functional domain of RPE65. (B) The structure of Mm-BCDO2 protein. The arrow structure elements are β-strand, the helical structure elements are α-helix, and the others are random coils and turns. (C) Multiple alignments of amino acid sequences of BCDO2 from M. meretrix and other model species. Among these, the completely (=100%), strongly (≥75%), and weakly (≥50%) conserved residues are shaded in black, blue, and yellow, respectively. The black line represents the domain RPE65.
The phylogenetic tree showed that the BCDO2 proteins from vertebrates were clustered together as one branch, while that from M. meretrix and other invertebrates were gathered into one branch (Figure 2A). Eighteen BCDO2 protein sequences were analyzed and identified 10 conserved motifs, ranging in length from 6 to 50 amino acid residues. Most of the BCDO2 proteins contained motif 1–10 with the exception of M. yessoensis, Stylophora pistillata, and T. spiralis. The domain prediction analysis showed that there was a PRE65 functional domain in all tested vertebrates and invertebrates except T. spiralis (Figure 2B), indicating that the functions between them might be extremely similar.
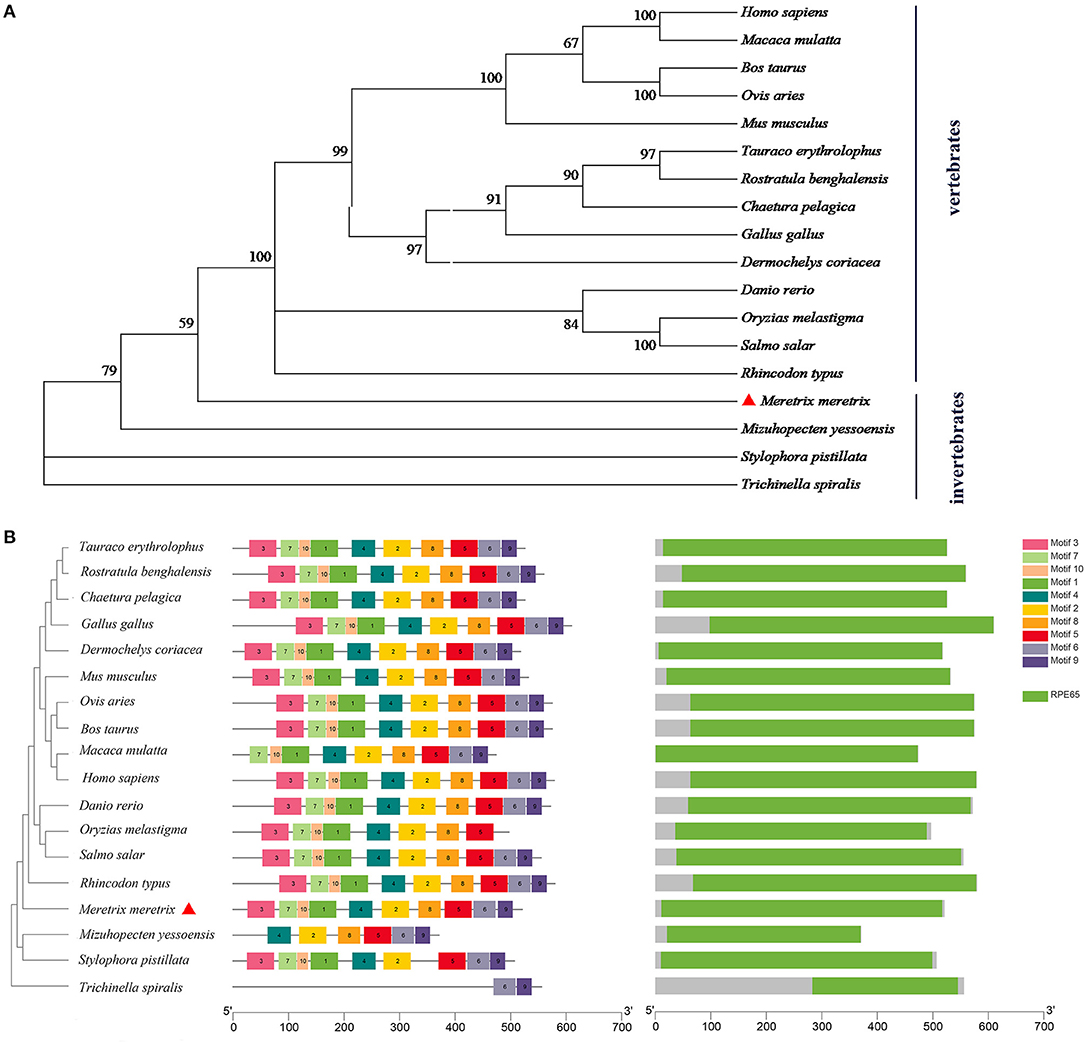
Figure 2. Analysis of evolutionary relationship among species. (A) Phylogenetic tree analysis of BCDO2 protein between M. meretrix and other species. (B) The structure of BCDO2 of M. meretrix, other bivalves and vertebrates.
High mRNA expression levels of Mm-BCDO2 were observed in the mantle and foot, followed by the digestive gland, siphon, adductor muscle, and gill (Figure 3). Notably, the expression level of Mm-BCDO2 was significantly higher (P < 0.05) in the mantle of red shell clams than that of white shell clams (Figure 3).
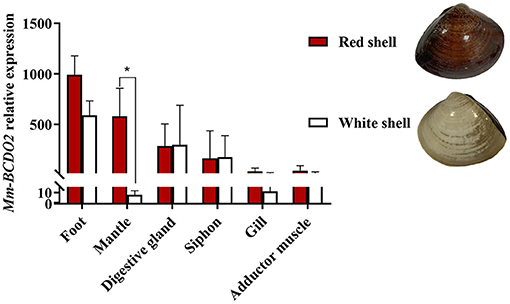
Figure 3. Relative expression levels of Mm-BCDO2 in different tissues from M. meretrix. Significant differences between red shell and white shell are indicated with an asterisk for P < 0.05.
Associations Between the SNP of Mm-BCDO2 and Shell Color of Clam
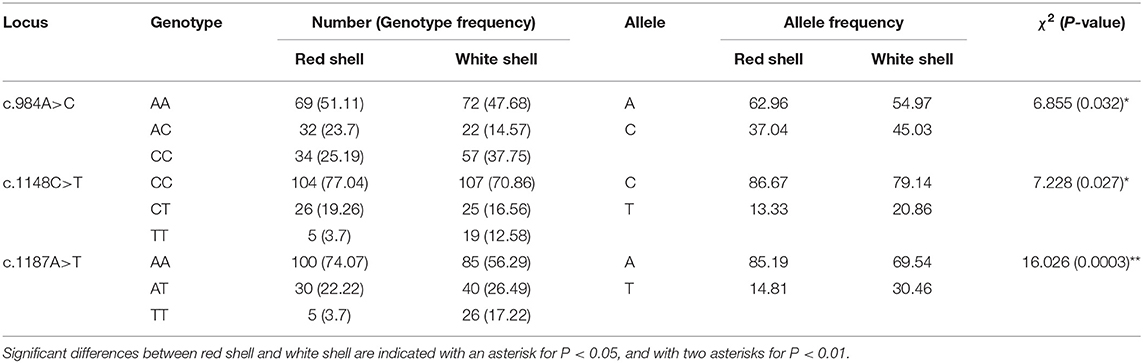
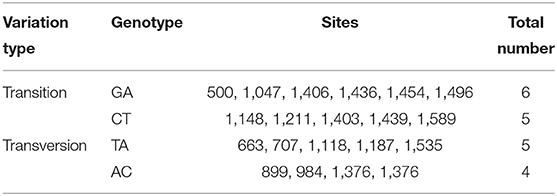
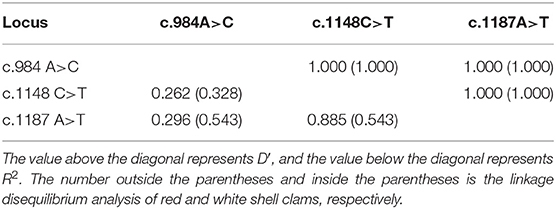
A total of 20 polymorphic loci were screened in the coding sequence of Mm-BCDO2 (Supplementary Table 1), of which three SNPs (c.984A > C, c.1148C > T, and c.1187A > T) were significantly associated with clam shell color formation (Table 2). The c.984A>C and c.1187A>T sites were non-synonymous mutations, which led to the mutation of amino acids from Lys to Gln and from Lys to Asn, respectively. The c.1148C>T site was a synonymous mutation. Statistical analysis of the mutation types of SNP sites found that there existed conversion and transversion types in the SNP of Mm-BCDO2 exons (Table 3).
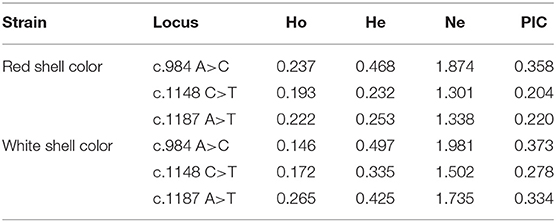
Both c.1148 C>T and c.1187 A>T were low polymorphism sites, while c.984 A>C was a moderate polymorphism site in the red shell clams. On the contrary, all the three SNPs were moderately polymorphic sites in the white shell clams (Table 4). The range of observed heterozygosity (Ho) and expected heterozygosity (He) in the red shell clams was 0.193–0.237 and 0.232–0.468, respectively, whereas the range of Ho and He in the white shell clams was 0.146–0.265 and 0.335–0.497, respectively. Further, the linkage disequilibrium results showed that the three sites were strongly linked (D′ > 0.75; Table 5).
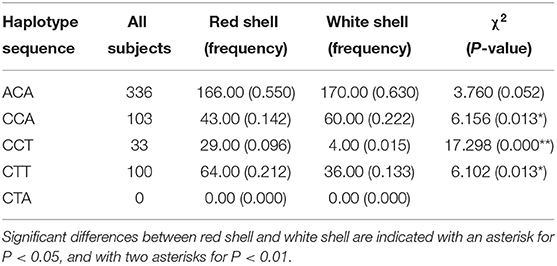
Three-locus haplotypes consisting of three SNPs were constructed to estimate the haplotype frequencies using SHEsis software. The analytical results showed that ACA was a common haplotype. CCA, CCT, and CTT haplotype frequencies for these three polymorphisms showed significant differences between the red shell and white shell (P < 0.05; Table 6).
Distribution of Mm-BCDO2 Protein in the Mantle and Digestive Gland
The immunofluorescence was used to assess the subcellular distribution of Mm-BCDO2 in the mantle and digestive gland of red and white shell clams. The result revealed that the Mm-BCDO2 was an extranuclear enzyme (Figure 4). According to the green fluorescence signal, the expression level of Mm-BCDO2 protein in single-layer columnar cells at the edge of the mantle of red clam was higher than that of white clam, but there was no difference in the muscle fibers scattered in the middle of the mantle (Figure 4A). Mm-BCDO2 was expressed on the glandular epithelium of digestive gland, while there was no difference in the expression level of Mm-BCDO2 between red and white shell clams (Figure 4B).
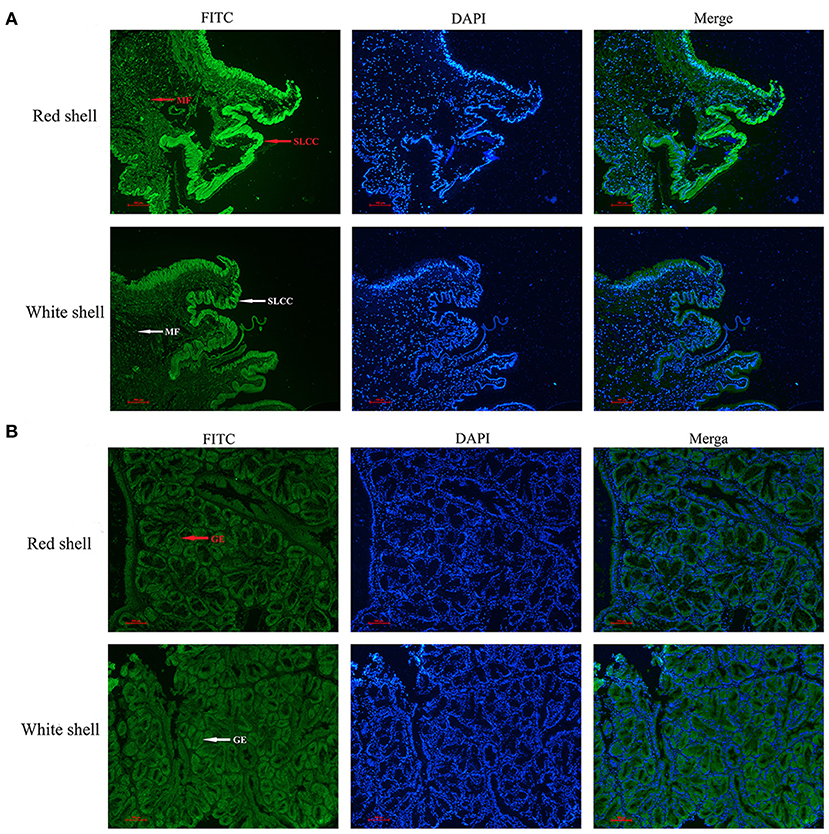
Figure 4. Distribution of BCDO2 in the mantle and digestive gland from red and white shell M. meretrix, respectively. (A) Localization of BCDO2 in single-layer columnar cells (SLCC) and muscle fibers (MF) of the mantle. (B) Localization of BCDO2 in glandular epithelium (GE) of digestive gland. Green fluorescence signal, FITC; blue fluorescence signal, DAPI.
Expression of Mm-BCDO2 Protein in the Mantle and Digestive Gland
Western blot analysis of the Mm-BCDO2 protein in the mantle and digestive gland showed that the target band appeared at the predicted molecular weight. There was no significant difference in the expression level of BCDO2 in the digestive gland between red and white shell clams (Figure 5A). Conversely, Mm-BCDO2 was highly expressed in the mantle of the red shell clams compared with that of the white shell clams (P < 0.05; Figure 5B).
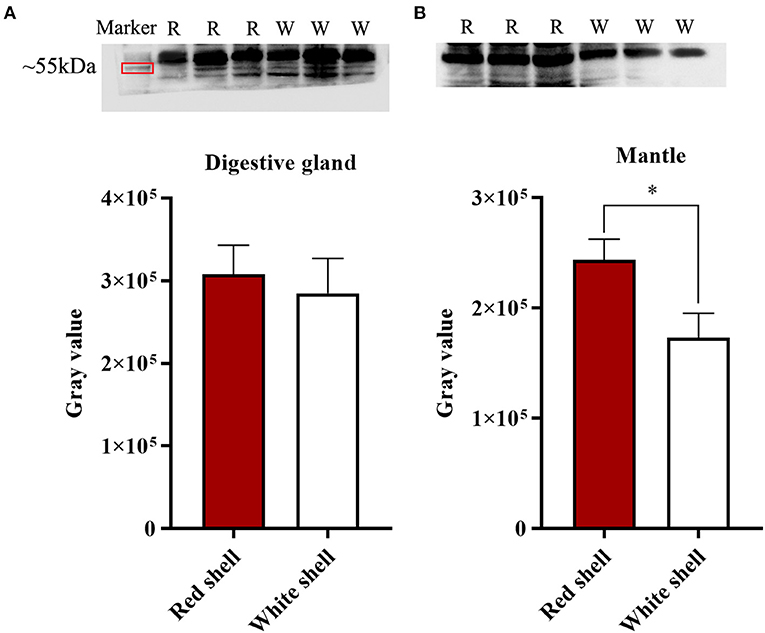
Figure 5. Western blotting analysis of Mm-BCDO2 protein in the digestive gland (A) and mantle (B) of red and white shell clams. Significant differences between red shell and white shell are indicated with an asterisk for P < 0.05.
Discussion
Studies on mammals, fish, and birds have revealed that BCDO2 can control carotenoid homeostasis, thereby affecting changes in body surface color or tissue color (Amengual et al., 2013; Costabile et al., 2016). In this study, the full-length cDNA sequence of Mm-BCDO2 was successfully cloned. Further, Mm-BCDO2 displayed low identity in sequence size with other species such as mammal and shellfish, but all of them contained the RPE65 domain. It is still controversial whether the function of BCOD2 gene is conserved in evolution. For example, RPE65 was molecularly identified as the first member of the vertebrate carotenoid cleavage dioxygenase family member (Hamel et al., 1993) and initially characterized as retinoid-binding protein rather than an enzyme catalyst (Gollapalli et al., 2003; Mata et al., 2004). Some studies question whether there exist the BCDO2 enzyme that produces vitamin A (Hansen and Maret, 1988). It has been reported that BCDO2 gene encodes an enzymatically active protein, but it showed no carotenoid cleavage activity in human (Li et al., 2014a; Seña et al., 2016). Indeed, studies have proved that BCDO2 is a catalytically competent enzyme and displays broad substrate specificity by transferring human BCDO2 splice variants into ARPE-19 cells and E. coli (Thomas et al., 2020). Furthermore, primate BCDO2 displayed a conserved structural fold and enzymatic function (Babino et al., 2015). Consistently, the BCDO2 gene in vertebrate was conserved in evolution, while that in invertebrate was quite different here. Therefore, whether the function of BCDO2 gene is conserved in evolution needs to be further explored.
Increasing studies have shown that the polymorphism of BCDO2 gene is related to color traits (Wu et al., 2020). For instance, the nonsense mutations of BCDO2 gene in the cattle lead to increased carotenoids in milk (Berry et al., 2009), while the yellow fat traits of sheep and rabbit are correlated with mutations in the coding region of BCDO2 gene (Vge and Boman, 2010; Strychalski et al., 2015, 2019; Niu et al., 2016). In addition, studies have found that some polymorphic loci of BCDO2 gene are completely linked to the skin color traits of chickens (Eriksson et al., 2008; Xu et al., 2017). These findings reveal that BCDO2 mutations cause the alterations in its function on mediating carotenoids cleavage. Non-synonymous single-nucleotide polymorphisms (SNPs) result in changes in the encoded amino acids, which in turn may affect the function of corresponding proteins (Katsonis et al., 2015; Cao et al., 2016). This is considered to be an important cause that leads to changes in animal phenotypes. In this study, the positions c.984 A>C and c.1187 A>T were non-synonymous mutations, and the position Lys984Gln caused the amino acid coded to be mutated from positively charged to uncharged, which was located near the conservative site. Similarly, the site Lys1187Asn led to the amino acid mutation from positively charged to negatively charged, which was located in the starting region of the functional domain. Three haplotype frequencies for these three polymorphisms showed significant differences between red shell clams and white shell clams (P < 0.05). We think the different genotypes of the three SNPs may associate with mRNA stability or affect Mm-BCDO2 gene expression through interaction with other transcription factors that would disturb carotenoids metabolism. It has been shown that changes in the charge of amino acids can lead to changes in redox intensity (Jiao et al., 2016). Consistent with the notion, a non-synonymous mutation in the BCDO2 gene was found to disrupt the carotenoid cleavage activity of the enzyme in a study of the coloration mechanism on birds, thereby causing a significant increase in the concentration of intact carotenoids in tissues (Anna et al., 2020). These findings suggest that BCDO2 gene plays an important role in the accumulation of carotenoids in animal tissues. Therefore, we speculate that changes of amino acid at multiple sites may affect the spatial collision between the side chain of corresponding site of the clam BCDO2 protein and the surrounding side chain, thereby changing the protein structure or causing protein folding defects, which in turn altered enzyme activities and resulted in different metabolic capabilities between red and white shell clams. Intriguingly, there was a strong linkage between c.984 A>C and c.1187 A>T, which could be inherited stably and could be an important target of selection in the evolution of color traits.
Changes in gene expression are the basis for differential phenotypes between the same species (Fallahshahroudi et al., 2018). In this study, the Mm-BCDO2 gene was expressed in all six tested tissues, with significantly higher expression in the mantle of red shell clams than white shell clams, and there was no difference in the digestive gland. These findings were further verified by western blot and immunofluorescence analysis. Consistently, the expression level of BCDO2 gene in the red feathers and beaks of Quelea quelea was significantly higher than that in the pale yellow feathers and beaks of the population (Walsh et al., 2012). The expression level of BCDO2 decreased in the skin, muscle, and adipose tissue of the white leghorn compared with that of the Red Junglefowl, but no change was observed in the liver and other internal organs. It has been proposed that the expression level of BCDO2 gene in the livers did not decrease because BCDO2 was not sensitive to the toxicity of excess carotenoids (Fallahshahroudi et al., 2019). On the contrary, the transcript levels of BCDO2-l gene was elevated in the white Chinook salmon compared with that in the red Chinook salmon (Lehnert et al., 2019). The protection of cells against apoptosis induced by carotenoid is strictly dependent on BCDO2 enzymatic function (Lobo et al., 2012). Our previous study on the clam shell color found that the genes and proteins that are related to carotenoid transport were significantly higher in the red shell clams than that in the white shell clams (Zhan, 2015). Therefore, we speculate that there are two explanations for this phenomenon. On the one hand, it is to prevent excessive carotenoid accumulation in the body from causing cellular oxidative stress, which will have side effects on the body, so more carotene oxygenases are needed to decompose carotenoids. Previous studies in mice have shown that high concentration of carotenoids can force the expression of the BCDO2 gene, so the high-level expression of carotene oxygenase in the body can balance the carotenoid concentration without causing oxidative stress (Babino et al., 2015). On the other hand, it may be that the body needs to metabolize carotenoids to show different colors (red, yellow, etc.). “Red” pigments are instead often metabolically derived from the yellow precursors, typically by oxygenation (also known as allylic oxidation), adding double-bonded oxygen atoms (“keto-groups”) to the end rings, thereby shifting the absorptance λ max toward longer wavelengths (Britton, 1995). For example, researchers have found that some birds produce red feathers by metabolizing carotenoids into derived keto carotenoids, while other birds produce red feathers by depositing high concentrations of yellow dietary carotenoids (Andersson et al., 2006). And the method of depositing carotenoids in clams needs further research.
In summary, we characterized the cDNA of Mm-BCDO2 and analyzed the relationship between its sequence characteristics and phylogenetic trait. The expression level of Mm-BCDO2 gene was significantly higher in the mantle of the red shell clams than that in the white shell clams. Moreover, the association analysis identified three shell color-related SNPs in the exons of Mm-BCDO2. Among them, the non-synonymous mutations of c.984 A>C and c.1187 A>T sites may be correlated with the accumulation of carotenoids. Overall, these findings contribute to understanding the role of Mm-BCDO2 gene in the formation of shell color via regulating carotenoid levels and provide molecular marker for shell color breeding in M. meretrix.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
This animal study was reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) of Zhejiang Wanli University, China.
Author Contributions
YD, HY, and ZL designed the study. LF performed the experiments under the support of YB. LF, HS, WD, and YD analyzed the data and wrote the manuscript. All authors read and approved the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (31772846), National Key Research and Development Program of China (2018YFD0901404), Ningbo Major Project of Science and Technology (2019B10005), Zhejiang Provincial First-Class Discipline of Bioengineering-A (ZS2019001), and National Marine Genetic Resource Center Program.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.746026/full#supplementary-material
References
Amengual, J., Widjaja Adhi, M., Rodriguez Santiago, S., Hessel, S., Golczak, M., Palczewski, K., et al. (2013). Two carotenoid oxygenases contribute to mammalian provitamin a metabolism. J. Biol. Chem. 288, 34081–34096. doi: 10.1074/jbc.M113.501049
Andersson, S., Prager, M., and Johansson, E. (2006). Carotenoid content and reflectance of yellow and red nuptial plumages in widowbirds (Euplectes spp.). Func. Ecol. 21, 272–281. doi: 10.1111/j.1365-2435.2007.01233.x
Andrade, P., Pinho, C., Pérezide Lanuza, G., Afonso, S., Brejcha, J., Rubin, C. J., et al. (2019). Regulatory changes in pterin and carotenoid genes underlie balanced color polymorphisms in the wall lizard. Proc. Natl. Acad. Sci. U.S.A. 116, 5633–5642. doi: 10.1073/pnas.1820320116
Anna, G. M., Toomey, M., Araújo, P., Lopes, R. J., Sandra, A., Myers, C. A., et al. (2020). Genetic basis of de novo appearance of carotenoid ornamentation in bare parts of canaries. Mol. Biol. Evol. 37, 1317–1328. doi: 10.1093/molbev/msaa006
Babino, D., Palczewski, G., Widjaja-Adhi, M., Kiser, P. D., and Lintig, J. V. (2015). Characterization of the role of β-carotene 9,10-dioxygenase in macular pigment metabolism. J. Biol. Chem. 290, 24844–24857. doi: 10.1074/jbc.M115.668822
Berry, S. D., Davis, S. R., Beattie, E. M., Thomas, N. L., Burrett, A. K., Ward, H. E., et al. (2009). Mutation in bovine β-carotene oxygenase 2 affects milk color. Genetics 182, 923–926. doi: 10.1534/genetics.109.101741
Blount, J. D., and Mcgraw, K. J. (2008). “Signal functions of carotenoid colouration,” in Carotenoids, ed B. George (Basel: Birkhäuser), 213–236. doi: 10.1007/978-3-7643-7499-0_11
Britton, G. (1995). Structure and properties of carotenoids in relation to function. FASEB. J. 9,1551–1559. doi: 10.1096/fasebj.9.15.8529834
Cao, R., Shi, Y., Chen, S., Ma, Y., Chen, J., Yang, J., et al. (2016). DbSAP: Single amino-acid polymorphism database for protein variation detection. Nucleic Acids Res. 45, 827–832. doi: 10.1093/nar/gkw1096
Costabile, B. K., Kim, Y. K., Iqbal, J., Zuccaro, M. V., Wassef, L., Narayanasamy, S., et al. (2016). Beta-apo-10′-carotenoids modulate placental microsomal triglyceride transfer protein expression and function to optimize transport of intact beta-carotene to the embryo. J. Biol. Chem. 291, 18525–18535. doi: 10.1074/jbc.M116.738336
Cui, B., Dong, Y., Zhao, J., Hu, L., LI, X., Yao, H., et al. (2018). Cloning and expression analysis of SRBI gene in different shell-color strains of Meretrix meretrix. Acta Hydrobiol. Sin. 42, 488–493. doi: 10.7541/2018.061
Deng, S. Z., Li, Q. C., Han, F., Zhang, X. L., Cai, M. Y., Zhang, J., et al. (2018). Analysis of total carotenoid content with different tissues or genders in Paphia textzle. Mar. Bull. 37, 165–168, 208. doi: 10.11840/j.issn.1001-6392.2018.02.006
Eriksson, J., Larson, G., Gunnarsson, U., Bed'Hom, B., Tixier-Boichard, M., Strömstedt, L., et al. (2008). Identification of the yellow skin gene reveals a hybrid origin of the domestic chicken. PLoS Genet. 4:e1000010. doi: 10.1371/journal.pgen.1000010
Fallahshahroudi, A., Løtvedt, P., Bélteky, J., Altimiras, J., and Jensen, P. (2018). Changes in pituitary gene expression may underlie multiple domesticated traits in chickens. Heredity 122, 195–204. doi: 10.1038/s41437-018-0092-z
Fallahshahroudi, A., Sorato, E., Altimiras, J., and Jensen, P. (2019). The domestic BCO2 allele buffers low-carotenoid diets in chickens: possible fitness increase through species hybridization. Genetics 212, 1445–1452. doi: 10.1534/genetics.119.302258
Felix, M. B., Snodderly, D. M., Johnson, E. J., Schalch, W., Korepcke, W., Gerss, J., et al. (2011). Nutritional manipulation of primate retinas, V: effects of lutein, zeaxanthin, and n-3 fatty acids on retinal sensitivity to blue-light-induced damage. Invest. Ophthalmol. Vis. Sci. 52, 3934–3942. doi: 10.1167/iovs.10-5898
Ge, J. L., Li, Q., and Yu, H. (2016). Selection response in mass selection of golden shell Pacific oyster (Crassostrea gigas). J. Fish. China 40, 95–100. doi: 10.11964/jfc.20150409829
Georg, L., Jennifer, L., and Gerald, R. (2010). Molecular and dietary regulation of β,β-carotene 15,15′-monooxygenase 1 (BCMO1). Arch. Insect Biochem. 502, 8–16. doi: 10.1016/j.abb.2010.06.032
Gollapalli, D. R., Maiti, P., and Rando, R. R. (2003). RPE65 operates in the vertebrate visual cycle by stereospecifically binding all-trans-retinyl esters. Biochemistry 42:11824–30. doi: 10.1021/bi035227w
Hamel, C. P., Tsilou, E., Pfeffer, B. A., Hooks, J. J., Detrick, B., and Redmond, T. M. (1993). Molecular cloning and expression of RPE65, a novel retinal pigment epithelium-specific microsomal protein that is post-transcriptionally regulated in vitro. J. Biol. Chem. 268, 15751–15757. doi: 10.1016/S0021-9258(18)82319-5
Hansen, S., and Maret, W. (1988). Retinal is not formed in vitro by enzymatic central cleavage of beta-carotene. Biochemistry 27, 200–206. doi: 10.1021/bi00401a030
Jiao, J., Yang, X., Jin, L., Gao, J., Yang, Z., Xiao, Y., et al. (2016). Conservative and variability of the important functional sites in a laccase from bacillus subtilis. Chem. J. Chin. Univer. 37, 1320–1327. doi: 10.7503/cjcu20160092
Jin, Y., Yu, Y., Zhang, C., Li, S., Zhang, X., and Li, F. (2020). Characterization and function analysis of the beta-carotene oxygenase-like genes in carotenoids metabolism of the ridgetail white prawn exopalaemon carinicauda. Front. Physiol. 11:745–760. doi: 10.3389/fphys.2020.00745
Katsonis, P., Koire, A., Wilson, S. J., Hsu, T. K., Lua, R. C., Wilkins, A. D., et al. (2015). Single nucleotide variations: biological impact and theoretical interpretation. Protein Sci. 23, 1650–1666. doi: 10.1002/pro.2552
Lehnert, S. J., Christensen, K. A., Vandersteen, W. E., Sakhrani, D., and Devlin, R. H. (2019). Carotenoid pigmentation in salmon: variation in expression at BCO2-l locus controls a key fitness trait affecting red coloration. Proc. R. Soc. B. 286, 20191588–20191596. doi: 10.1098/rspb.2019.1588
Li, B., Vachali, P. P., Gorusupudi, A., Shen, Z., Sharifzadeh, H., Besch, B. M., et al. (2014a). Inactivity of human β,β-carotene-9',10'-dioxygenase (BCO2) underlies retinal accumulation of the human macular carotenoid pigment. Proc. Natl. Acad. Sci. U.S.A. 111, 10173–10178. doi: 10.1073/pnas.1402526111
Li, B., Vachali, P. P., Shen, Z., Gorusupudi, A., Nelson, K., Besch, B. M., et al. (2017). Retinal accumulation of zeaxanthin, lutein, and β-carotene in mice deficient in carotenoid cleavage enzymes. Exp. Eye. Res. 159, 123–131. doi: 10.1016/j.exer.2017.02.016
Li, X., Bai, Z., Luo, H., Wang, G., and Li, J. (2014b). Comparative analysis of total carotenoid content in tissues of purple and white inner-shell color pearl mussel, Hyriopsis cumingii. Aquacult. Int. 22, 1577–1585. doi: 10.1007/s10499-014-9766-6
Lin, Z. (2015). Biology and Culture Technology of Meretrix meretrix. Beijing: Science Press Beijing.
Lobo, G. P., Isken, A., Hoff, S., Babino, D., and Lintig, J. V. (2012). BCDO2 acts as a carotenoid scavenger and gatekeeper for the mitochondrial apoptotic pathway. Development 139, 2966–2977. doi: 10.1242/dev.079632
Mata, N. L., Moghrabi, W. N., Lee, J. S., Bui, T. V., Radu, R. A., Horwitz, J., et al. (2004). RPE65 is a retinyl ester binding protein that presents insoluble substrate to the isomerase in retinal pigment epithelial cells. J. Biol. Chem. 279, 635–643. doi: 10.1074/jbc.M310042200
Meilisza, N., Jusadi, D., Zairin, M., Artika, I. M., Utomo, N. P., Kadarini, T., et al. (2017). Digestibility, growth and pigmentation of astaxanthin, canthaxanthin or lutein diets in Lake Kurumoi rainbowfish, Melanotaenia parva (Allen) cultured species. Aquac. Res. 48, 1–9. doi: 10.1111/are.13372
Milani, A., Basirnejad, M., Shahbazi, S., and Bolhassani, A. (2017). Carotenoids: biochemistry, pharmacology and treatment. Br. J. Pharmacol. 174, 1290–1324. doi: 10.1111/bph.13625
Niu, Y., Jin, M., Li, Y., Li, P., and Chen, Y. (2016). Biallelic b-carotene oxygenase 2 knockout results in yellow fat in sheep via CRISPR/Cas9. Anim. Genet. 48, 242–244. doi: 10.1111/age.12515
Qi, X. Y. (2014). The analysis of molecular mechanisms of shell color formation for Meretrix meretrix (dissertation). Ningbo University, Ningbo, Zhejiang.
Seña, C. D., Jian, S., Narayanasamy, S., Riedl, K. M., Yan, Y., Curley, R. W., et al. (2016). Substrate specificity of purified recombinant chicken β-carotene 9′,10′-oxygenase (bco2). J. Biol. Chem. 291, 14609–14619. doi: 10.1074/jbc.M116.723684
Strychalski, J., Brym, P., Czarnik, U., and Gugoek, A. (2015). A novel AAT-deletion mutation in the coding sequence of the BCO2 gene in yellow-fat rabbits. J. Appl. Genet. 56, 535–537. doi: 10.1007/s13353-015-0290-9
Strychalski, J., Gugoek, A., Brym, P., Antoszkiewicz, Z., and Chwastowska-Siwiecka, I. (2019). Polymorphism of the BCO2 gene and the content of carotenoids, retinol, and α-tocopherol in the liver and fat of rabbits. Rev. Bras. Zool. 48:e20180243. doi: 10.1590/rbz4820180243
Svensson, P. A., and Wong, B. B. M. (2011). Carotenoid-based signals in behavioural ecology: a review. Behaviour 148, 131–189. doi: 10.1163/000579510X548673
Thomas, L. D., Bandara, S., Parmar, V. M., Srinivasagan, R., Khadka, N., Golczak, M., et al. (2020). The human mitochondrial enzyme BCO2 exhibits catalytic activity toward carotenoids and apocarotenoids. J. Biol. Chem. 295, 15553–15565. doi: 10.1074/jbc.RA120.015515
Tian, R., Pitchford, W. S., Morris, C. A., Cullen, N. G., and Bottema, C. D. K. (2009). Genetic variation in the β, β-carotene-9′, 10′-dioxygenase gene and association with fat colour in bovine adipose tissue and milk. Anim. Genet. 41, 253–259. doi: 10.1111/j.1365-2052.2009.01990.x
Vge, D. I., and Boman, I. A. (2010). A nonsense mutation in the beta-carotene oxygenase 2 (BCO2) gene is tightly associated with accumulation of carotenoids in adipose tissue in sheep (Ovis aries). BMC Genet. 11:10–16. doi: 10.1186/1471-2156-11-10
Walsh, N., Dale, J., Mcgraw, K. J., Pointer, M. A., and Mundy, N. I. (2012). Candidate genes for carotenoid coloration in vertebrates and their expression profiles in the carotenoid-containing plumage and bill of a wild bird. Proc. Soc. B. 279, 58–66. doi: 10.1098/rspb.2011.0765
Wang, J., Li, Q., Zhong, X., Song, J., Kong, L., and Yu, H. (2018). An integrated genetic map based on EST-SNPs and QTL analysis of shell color traits in Pacific oyster Crassostrea gigas. Aquaculture 492, 226–236. doi: 10.1016/j.aquaculture.2018.04.018
Wang, X. L., Li, Q., and Kong, L. F. (2016). Construction and comparison of growth and survival among different golden shell families of Pacific oyster (Crassostrea gigas). J. Fish. China 40, 1683–1693. doi: 10.11964/jfc.20150810050
Wu, J., Chen, G., Lin, Z., Yin, Y., Chen, H., Chen, J., et al. (2020). Polymorphism of BCO2 gene and its correlation with skin yellowness in partridge chickens. China Poult. 42, 12–18. doi: 10.3389/fphys.2021.585089
Wu, L., Guo, X., Wang, W., Medeiros, D. M., Clarke, S. L., Lucas, E. A., et al. (2016). Molecular aspects of β, β-carotene-9′, 10′-oxygenase 2 in carotenoid metabolism and diseases. Exp. Biol. Med. 241, 1879–1887. doi: 10.1177/1535370216657900
Xu, J., Lin, S., Gao, X., Nie, Q., and Zhang, X. (2017). Mapping of id locus for dermal shank melanin in a Chinese indigenous chicken breed. J. Genet. 96, 977–983. doi: 10.1007/s12041-017-0862-z
Zhan, Y. L. (2015). Identification of shell color and the related gene and microRNA research of Meretrix meretrix (dissertation). Zhejiang Wanli University, Ningbo, Zhejiang.
Zhang, B., Zhu, C., Meng, Z., Liu, B., and Yu, D. (2019). Carotenoids in pearl oyster Pinctada fucata: the tissue distribution and correlation to color parameters. Pak. J. Zool. 51, 1655–1661. doi: 10.17582/journal.pjz/2019.51.5.1655.1661
Keywords: Meretrix meretrix, BCDO2, carotenoids, shell color, SNP
Citation: Fu L, Shi H, Dai W, Yao H, Bao Y, Lin Z and Dong Y (2021) Characterization and Function Analysis of β, β-carotene-9′, 10′-oxygenase 2 (BCDO2) Gene in Carotenoid Metabolism of the Red Shell Hard Clam (Meretrix meretrix). Front. Mar. Sci. 8:746026. doi: 10.3389/fmars.2021.746026
Received: 23 July 2021; Accepted: 19 August 2021;
Published: 16 September 2021.
Edited by:
Liqiang Zhao, Guangdong Ocean University (GDOU), ChinaReviewed by:
Zhe Zheng, Guangdong Ocean University (GDOU), ChinaHao Song, Institute of Oceanology, Chinese Academy of Sciences (CAS), China
Copyright © 2021 Fu, Shi, Dai, Yao, Bao, Lin and Dong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hanhan Yao, yaohanhan1020@126.com; Yinghui Dong, dongyinghui118@126.com
 Lulu Fu1,2
Lulu Fu1,2  Hanhan Yao
Hanhan Yao Yinghui Dong
Yinghui Dong