The Last Two Remaining Populations of the Critically Endangered Estuarine Pipefish Are Inbred and Not Genetically Distinct
- 1Centre for Ecological Genomics and Wildlife Conservation, Department of Zoology, University of Johannesburg, Auckland Park, South Africa
- 2Department of Ichthyology and Fisheries Science, Rhodes University, Makhanda, South Africa
- 3National Research Foundation—South African Institute for Aquatic Biodiversity, Makhanda, South Africa
The critically endangered estuarine pipefish, Syngnathus watermeyeri, is one of Africa’s rarest fish species and currently faces a significant risk of extinction. A combination of anthropogenic and natural factors threaten submerged macrophyte beds in the two South African estuaries (Bushmans and Kariega) in which the species’ only two known remaining populations reside. Here, we genotyped 34 pipefish from both populations using genome-wide data to determine whether the two estuaries harbour distinct genetic diversity, such that translocating individuals between them might improve the genetic health of both. Our results show that both populations are highly inbred, and no statistically significant genetic structure was found between them. Moreover, individuals both within and between estuaries were very closely related to each other. These results indicate that the remaining populations of the estuarine pipefish suffer from the adverse genetic effects of small population sizes. Even though recent surveys have estimated population sizes in the order of thousands of individuals, these may fluctuate considerably. Although the translocation of genetically similar individuals between habitats will not increase local genetic diversity, the creation of additional populations across the species’ historical range may be a suitable conservation strategy to prevent further loss of genetic diversity, and to minimise the overall extinction risk posed by environmental stochasticity.
Introduction
Intertidal submerged macrophyte beds dominated by seagrass are ecologically important habitats that provide foraging sites, shelter, and nursery grounds for many aquatic species, thereby supporting a greater density of animals per unit area than any other habitat type within estuaries and lagoons (Alfaro, 2006; Rasheed and Unsworth, 2011). However, seagrass habitats are declining globally, with 10 seagrass species (14% of all the known species) at risk of extinction (Short et al., 2011). With the loss of seagrass beds, a cascade in the loss of biodiversity and abundance of other aquatic species that depend on these habitats is expected (Jackson et al., 2001; Hughes et al., 2009). Threats to seagrasses include natural events such as flooding and siltation, which can temporarily result in the complete extirpation of seagrass beds (Riddin and Adams, 2012), but also anthropogenic factors such as coastal development, modification of shorelines, pollution and excessive bait collection (Duarte, 2002; Orth et al., 2006; Pillay et al., 2010; Stankovic et al., 2021), as well as climate change (Aller et al., 2019).
In southern Africa, the eelgrass Zostera capensis is a dominant keystone seagrass species (Bandeira and Gell, 2003; Adams, 2016). Listed as Vulnerable on the IUCN Red List, with a decreasing population trend (Short et al., 2010; Van der Colff, 2016), it has been reported in 62 of 290 South African estuaries, with all seagrass beds combined covering a total area of only between 7 and 20 km2 (Bandeira and Gell, 2003; Adams, 2016). Despite the ecological significance of seagrass, only two of these estuaries (Langebaan and Knysna) have some formal protection status (Adams, 2016). Their macrophyte beds harbour animal species with high conservation status, including the Endangered Knysna seahorse, Hippocampus capensis Boulenger, 1900, which is found in Knysna and two nearby estuaries (Mkare et al., 2017), and the Critically Endangered pulmonate limpet Siphonaria compressa Allanson, 1958, which occurs in both Langebaan and Knysna (Allanson and Herbert, 2005). A third species of conservation concern, the Critically Endangered estuarine pipefish, Syngnathus watermeyeri Smith, 1963, is strongly associated with macrophytes in two estuaries (Kariega and Bushmans; Figure 1) that have not been afforded any formal level of protection (Whitfield et al., 2017).
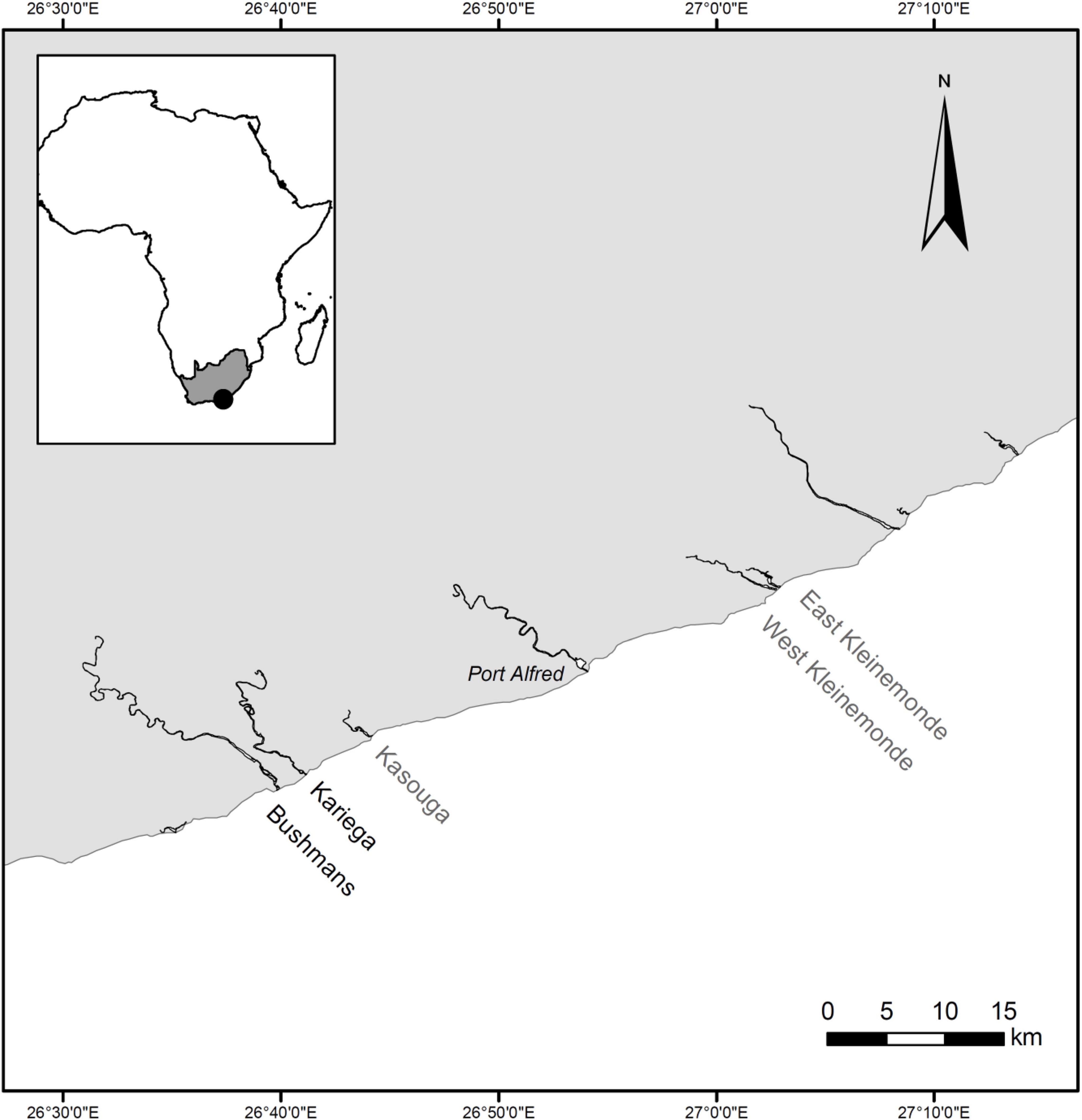
Figure 1. Present and past distribution of the estuarine pipefish, Syngnathus watermeyeri, on the eastern south coast of South Africa. The species is currently found in the two estuaries labelled in black; populations in the estuaries with grey labels are believed to be extinct. The estuarine pipefish has never been reported from any of the other estuaries shown on this map.
Pipefishes, along with seahorses, pipehorses and seadragons, are part of the family Syngnathidae, which are predominantly marine teleosts (Pollard, 1984; Dawson, 1985; Gordina, 1991). Syngnathids have a low fin surface area, which limits their distributions to environments with slow water currents (Castejón-Silvo et al., 2021). They are typically small, benthic and specialised diurnal visual feeders that occupy small home ranges (especially mated males) (Foster and Vincent, 2004; Vincent et al., 2005; Kuiter, 2010). Syngnathids are often an important component of seagrass beds, mimicking macrophytes with cryptic colours and shapes. Syngnathids that are dependent on seagrass beds are important indicator species that can be used to evaluate the conservation value and health of estuarine seagrass beds (Shokri et al., 2009). The conservation of syngnathids can thus provide benefits to other species that share the seagrass habitats, as correlations with other fish species in density and spatial variations in assemblages have been found (Shokri et al., 2009).
Syngnathus watermeyeri was originally reported from three estuaries located along the warm-temperate south coast of South Africa, namely Bushmans, Kariega, and Kasouga (Figure 1; Smith, 1963; Dawson, 1986). The estuarine pipefish was recognised as a threatened species in 1987 (Skelton, 1987) before being declared provisionally extinct in 1996 (Whitfield and Bruton, 1996) after it had not been recorded for several decades. In 1997, the species was discovered in the East Kleinemonde Estuary, where it had not previously been recorded (Figure 1; Cowley and Whitfield, 2001). Twelve pipefish from the East Kleinemonde were subsequently transferred to the adjacent West Kleinemonde Estuary (Bell et al., 2003), which may already have had a viable population since the two estuaries are sometimes linked in the mouth region. Following its localised extinction in these two estuaries after a flood event in May 2003, the estuarine pipefish was rediscovered in two of its historical habitats, namely the Bushmans (in 2005) and Kariega (in 2006) estuaries (Vorwerk et al., 2007; Whitfield et al., 2017). Both estuaries are clear, marine-dominated, permanently open systems with salinity values that are close to those of the sea along most of their lengths (Teske and Wooldridge, 2003). Although these conditions are a result of excessive freshwater abstraction in the estuaries’ catchments (Bate et al., 2002), they provide ideal conditions for the growth of extensive beds of aquatic macrophytes (Adams, 2016), which provide habitat for the pipefishes.
Recent estimates based on survey data suggest that the current population size of S. watermeyeri may be considerably larger than the 100–250 individuals reported in the IUCN Red List entry (Pollom, 2016), and may actually be in the order of tens of thousands of individuals if all known estuaries where they have occurred are occupied (Whitfield et al., 2017). However, total population size may fluctuate considerably, depending on estuarine conditions and pipefish occurrences prevailing at the time. The effects on genetic diversity resulting from severe population bottlenecks that result from either local reduction of pipefish populations, or the founder events associated with recolonisation of habitats where the species has become extinct, are currently unknown. Such information is necessary to develop conservation measures to minimise the risk of this species becoming extinct in the wild.
In the present study, we used genome-wide data from individuals sampled non-destructively to investigate genetic diversity and population structure in the last two remaining populations of this Critically Endangered species. Our work serves as the first step toward the development of a conservation plan for S. watermeyeri. Specifically, we hypothesised that each of the two remaining populations has unique genetic diversity, which would indicate that translocations of individuals between estuaries can counteract the negative consequences of inbreeding by maximising the genetic diversity of both populations.
Materials and Methods
Specimen Collection, DNA Extraction and Genotype Sequencing
The procedures for animal handling and genetic sample collection were approved by the Department of Forestry, Fisheries and the Environment (DFFE) of the Republic of South Africa (permit reference number: RES2020/101), and ethical clearance was granted by the Faculty of Science Ethics Committee at the University of Johannesburg (Ethics Reference Number: 2020-02-06/Teske_Weiss). All necessary steps were undertaken to reduce the handling time of the fish and prevent prolonged exposure to air, light and heat.
A total of 34 individuals, 11 from the Bushmans Estuary and 23 from the Kariega Estuary, were collected in seagrass beds using a 5 m seine net with a mesh size of 1 cm during incoming and low tide in July 2020. If one accepts the most recent IUCN population size estimates, these sample sizes represent a notable portion of these populations (∼10%).
A small piece of tissue (<1 mm2) from the upper edge of the dorsal fin was excised using a pair of sterile surgical scissors. The scissors were disinfected before the collection of each fin clip using a combination of flame and 70% ethanol sterilization. This non-lethal method was tested on other syngnathids by Teske et al. (2003), Woodall et al. (2012), and Mkare et al. (2017), with no evidence for long-term negative impacts on growth or survival. Fin clips were immediately placed into a digestion solution containing 180 μl of QIAGEN buffer ATL and 20 μl of Proteinase K (Qiagen GmbH, Hilden, Germany). The genomic DNA was extracted and purified from fin clips using the QIAGEN DNeasy Blood and Tissue extraction kit, following the manufacturer’s protocol. All 34 DNA extractions were of high quality based on agarose gel electrophoresis (minimal degradation), and assessment by NanoDrop 2000 (Thermo Fisher Scientific, Wilmington, United States) spectrophotometer and Qubit 2.0 (Thermo Fisher Scientific) fluorimeter (OD260/OD280 ratios of 1.8–2.0 and > 1.5 μg of total DNA). Genomic libraries were constructed using the Genotyping-by-Sequencing (GBS) method (Elshire et al., 2011). Genomic DNA was digested with NEB restriction enzymes Mse I and Msp I (New England Biolabs, Ipswitch, United States), and libraries were prepared using NEB ligation matermix and purified with Agencourt XP beads (Beckman Coulter, Indianapolis, United States). The purified libraries were then amplified using MyTaq (Bioline, Memphis, United States) and standard Illumina TrueSeq amplification primers (Illumina Inc., San Diego, United States), according to the manufacturers’ instructions. This step was followed by pooling and clean-up of amplified libraries that involved removal of PCR primers and small amplicons by means of Agencourt XP beads, and removal of PCR enzyme by additional purification on Qiagen MinElute columns. Further steps included normalization using Trimmer Kit (Evrogen, Moscow, Russia), reamplification using MyTaq and i5-Adaptor primer, size selection (removal of fragments < 250 and > 500 bp on an LMP agarose gel), and 150 bp paired-end sequencing on an Illumina NextSeq 550 (San Diego, United States) using the NextSeq 500/550 v2 sequencing kit.
Data Analysis
The sequences were assembled de novo using the ipyrad 0.9.77 pipeline (Eaton and Overcast, 2020). Default filtering and assembly parameters were specified except that the adapter trimming parameter was set to “strict mode” and the maximum fraction of heterozygous bases was set to eight.
Two SNP datasets were then generated. The first used ipyrad’s output file in ustr format and was used to assess population structure and population-level inbreeding. This file is generated by randomly including only one SNP from each putative locus in the output to minimise the magnitude of Linkage Disequilibrium (LD), but SNPs that are not in Hardy-Weinberg equilibrium (HWE) are retained. Departures from HWE due to a heterozygote deficit are expected if populations are distinct (Wahlund, 1928; Waples, 2015), and the a priori removal of loci that do not conform to Hardy-Weinberg principles would thus result in reduced power to detect population structure (Waples, 2015).
The second dataset was based on ipyrad’s Variant Call Format (VCF) output file and excluded loci out of HWE (i.e., with unusual heterozygosity levels) to ensure that the majority of the remaining sites evolve neutrally. All variant sites with more than 20% missing genotypes across all 34 individuals, with a minor allele frequency of less than 0.01, covered by less than eight reads, and those with an LD coefficient (r2) of more than 60% were filtered in Bctools version 1.9 (Danecek et al., 2021). This dataset was used to calculate allelic richness and nucleotide diversity, and to estimate effective population size (Ne), as well as inbreeding and relatedness coefficients.
The spatial distribution of genetic diversity between estuaries was investigated using three different methods. First, FST was calculated in Arlequin version 3.5.5 (Excoffier and Lischer, 2010), with the P-value based on 1,000 permutations. In the second method, a Discriminant Analysis of Principal Components (DAPC) was performed in adegenet version 2.5.1 (Jombart et al., 2020). The raw data were first transformed into lower dimensions using a Principal Component Analysis (PCA). Then, a k-means clustering method was performed on the data, and different clustering solutions (K = 1–3, each representing a putative number of distinct populations) were compared using the associated Bayesian Information Criterion (BIC; Schwarz, 1978), and individuals from the two estuarine populations were compared by means of a DAPC scatterplot with two populations (K = 2). The third method used ancestry assignment of individuals to pre-specified genetic clusters representing distinct populations (K = 1–3), which was conducted in fastSTRUCTURE (Raj et al., 2014). Estimations were computed directly from the posterior distributions of model parameters using a simple prior and a convergence criterion of 10–6. The most likely number of populations was identified based on marginal likelihoods. Each individual’s ancestry proportions (Q) assuming two populations (K = 2) were visualised by means of a barplot.
The inbreeding coefficient FIS (Wright, 1949), and observed (HO) and expected (HE) heterozygosity for each population, were calculated in the R package diveRsity version 1.9.90 (Keenan et al., 2013). The same program was used to calculate the mean allelic richness across all loci for each population using rarefaction to account for the smaller sample size from the Bushmans Estuary (n = 11), and 95% confidence intervals for both FIS and allelic richness were calculated based on 1,000 bootstrap replications. Nucleotide diversity (π; Tajima, 1983), was calculated using the sliding windows method implemented in VCFtools version 0.1 (Danecek et al., 2011).
The degree of relatedness between individuals from two estuaries was quantified based on the proportion of shared homologous alleles that are identical-by-descent within and between individuals. To this end, Jacquard’s condensed identity coefficients (Δ1−Δ9) (Jacquard, 2012) were calculated using NgsRelate version 2 (Hanghøj et al., 2019), and the pairwise inbreeding coefficient (Fa = Δ1 + Δ2 + Δ3 + Δ4, Fb = Δ1 + Δ2 + Δ5 + Δ6) (Jacquard, 1972), the coefficient of kinship (Jacquard, 2012), and the relatedness coefficient (rab = 2θab) (Hedrick and Lacy, 2015) were estimated for pairs of individuals. Combining these coefficients is suitable to describe identity-by-descent within an individual, and to reconstruct gene descent histories among individuals in a population (Wang, 2017), particularly in cases where the ancestors of the sampled individuals were themselves inbred. For all pairs of individuals within and between estuaries, the mean, 95% confidence interval and non-outlier range of the inbreeding coefficient (Fab) and the relatedness coefficient (rab) were visualised in Statistica version 14.0.0.15 (TIBCO Software Inc., Palo Alto, United States).
The effective population size (Ne) for the estuarine pipefish was estimated using the LD method implemented in NeEstimator version 2 (Do et al., 2014). In this case, a modified version of the second (neutral) dataset was used for which no filtering for LD was applied. As NeEstimator uses a method that is based on the extent of LD between adjacent loci within populations, excessive filtering of LD can affect estimates of Ne. Two estimates of Ne were reported. The first estimate was based on a minor allele frequency threshold of 0.02, and the second was based on a threshold of 0.05.
Results
The sequencing run produced an average of 1,203,529 quality filter sequences per sample. The ipyrad assembly assigned 18,001 single nucleotide polymorphic sites across 34 individuals (Supplementary Table 1). After additional filtering steps, the final dataset used to assess genetic structure (which retained SNPs that departed from Hardy-Weinberg expectations) contained 4,883 SNPs, and the selectively neutral dataset contained 3,769 SNPs (Supplementary Table 2). The percentage of missing SNPs per individual for each dataset is shown in Supplementary Table 3.
No genetic structure was found between estuaries, with FST = 0.01 (P = 0.36) and DAPC (Supplementary Figure 1A) identifying a single population cluster. A DAPC scatterplot enforcing K = 2 recovered the individuals from each estuary as a single cluster, but with three outlier individuals (Supplementary Figure 1B). Although the marginal likelihoods from the fastSTRUCTURE analysis were slightly higher for K = 2 than for K = 1 (Supplementary Figure 1C), this was not confirmed by an ancestry assignment barplot, where the same three outlier individuals as in the DAPC scatterplot were partially assigned to a second population that showed no clear affiliation with either estuary (Supplementary Figure 1D).
The population-level inbreeding coefficients (FIS) were significantly positive in both estuarine populations (Table 1). In both cases, observed (HO) heterozygosity estimates were smaller than expected (HE) heterozygosity estimates, indicating heterozygote deficiencies typical of inbred populations. The 95% confidence intervals of both FIS and mean allelic richness broadly overlapped for the two estuarine populations, indicating that they did not differ significantly. Tajima’s π, on the other hand, was greater for Kariega.
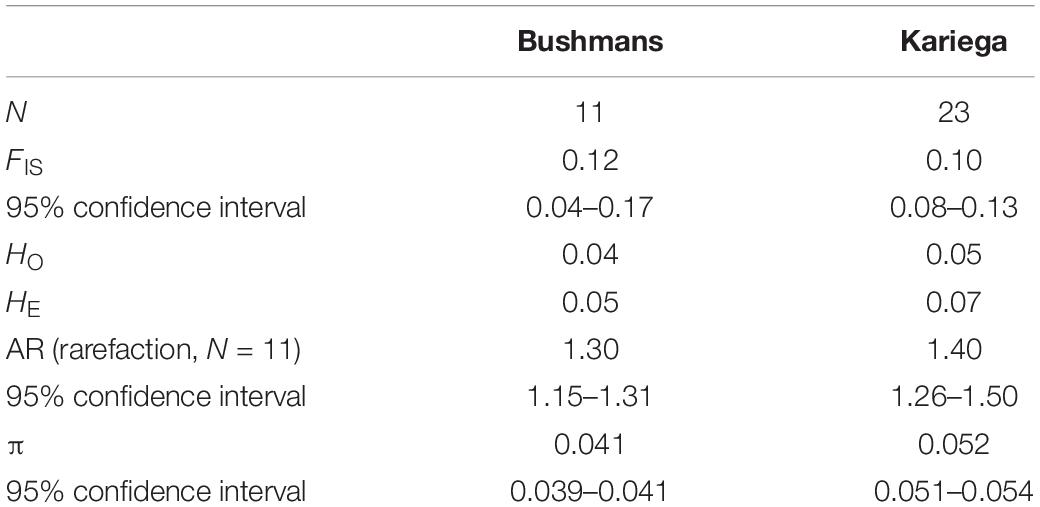
Table 1. Demographic statistics for the two estuarine populations of Syngnathus watermeyeri based on a dataset of selectively neutral SNPs.
The average inbreeding (Fab) and relatedness (rab) coefficients between pairs of individuals were strongly influenced by the estimated values for the Δ4 and Δ6 coefficients (the probability of identity-by-descent of two alleles within an individual), and point to a considerable level of heterozygote deficiency in both populations. The inbreeding coefficients were significantly positive for both populations, and this was also true when Fab was compared between individuals from different populations (Figure 2A). Moreover, many individuals were found that were closely related (Figure 2B). Within estuaries, a total of 71% (Bushmans) and 20% (Kariega) of the pairs of individuals had a relatedness coefficient that exceeded the relatedness expected between second cousins, and in 4% (Bushmans) and 1% (Kariega) of pairs of individuals, rab exceeded that expected between first cousins. High relatedness was also found between pairs of individuals from different estuaries, and thus corroborates the finding that the two populations are not distinct, with 34% having an rab that exceeded the level of relatedness between second cousins, and 2% having an rab that exceeded relatedness between first cousins.
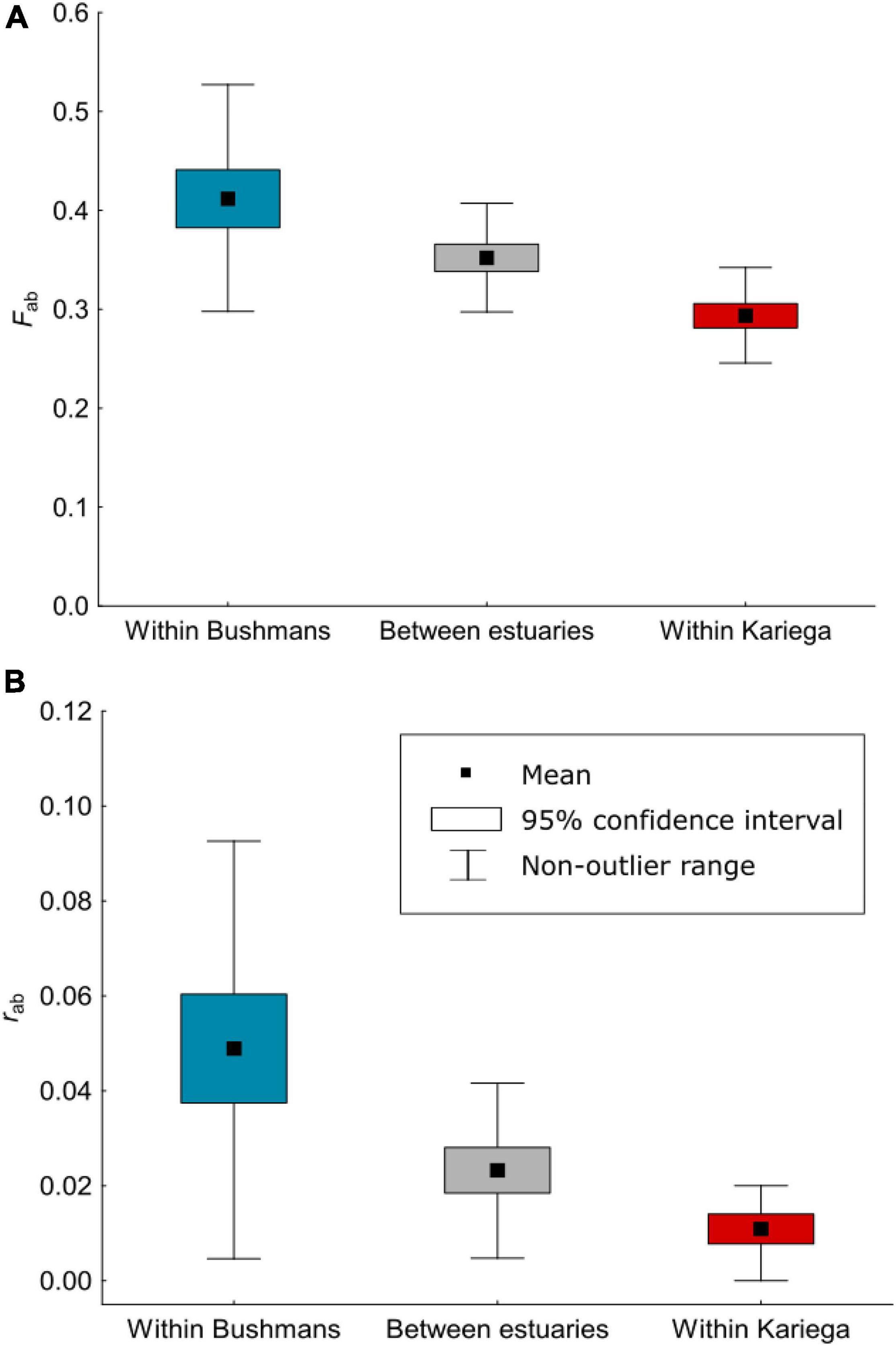
Figure 2. Mean pairwise inbreeding coefficients (A) and mean pairwise relatedness coefficients (B) between individuals of Syngnathus watermeyeri within and between the two estuaries estimated from genomic data using a model of identity-by-descent.
The overall effective population size of the remaining populations of Syngnathus watermeyeri was estimated at 199 individuals (95% CI: 153–280) using a 0.05 threshold for the lowest allele frequency, or 114 individuals (95% CI: 105–125) when a 0.02 threshold was used.
Discussion
The estuarine pipefish, Syngnathus watermeyeri, is one of the rarest fish species in Africa, and the fact that extensive recent surveys have documented it in only two estuaries highlights the urgency of comprehensive conservation initiatives to reduce its extinction risk. Previous genetic research based on maternally inherited mitochondrial DNA found that genetic diversity in S. watermeyeri is low compared to that reported for the closely related congener S. temminckii Kaup, 1856, a species that is abundant throughout temperate South Africa and that also occurs in the Bushmans and Kariega estuaries (Whitfield et al., 2017). The present study based on more informative biparentally-inherited, genome-wide markers not only confirmed low genetic diversity in S. watermeyeri, but assessments of inbreeding, pairwise relatedness, and genetic structure indicate that the species’ extinction risk is considerably greater than was previously thought.
Population-level inbreeding (FIS) and mean pairwise individual-based (Fab) coefficients of inbreeding were significantly positive in both populations of S. watermeyeri, indicating a heterozygote deficiency resulting from mating between close kin. This result not only suggests that this species likely suffers from reduced fitness because of the accumulation of deleterious alleles (i.e., inbreeding depression), but also that its evolutionary potential to adapt to future ecological and climatic challenges is diminished (Frankham, 1997, 2005).
Genome-wide measures of relatedness are considered to be suitable alternatives to traditional pedigree-based estimates in wild populations for which long-term genealogical data that are required to reconstruct a pedigree are not easily obtainable (Lopes et al., 2013; Taylor, 2015). In cases where the individuals that gave rise to the present generation were themselves inbred, they are potentially even superior because pedigree-based methods assume that the founders are unrelated (Speed and Balding, 2015; Hanghøj et al., 2019).
Population-level inbreeding coefficients found in S. watermeyeri (FIS = 0.10–0.12) were similar to those of several other threatened fish species, such as the Chinese sturgeon, Acipenser sinensis (FIS = 0.148; Liu et al., 2018) and red-tailed barb, Gonoproktopterus curmuca (FIS = 0.115; Musammilu et al., 2014), and they exceeded those of the Sichuan taimen, Hucho bleekeri (FIS = 0.003; Zhang et al., 2020). The magnitude of mean coefficients of inbreeding based on pairs of individuals (Fab) in S. watermeyeri (Fab = 0.31–0.40) was similar to that reported for the endangered Pyrenean desman, Galemys pyrenaicus (0.33; Escoda et al., 2017), and mean relatedness (rab = 0.02–0.05) approached that for the critically endangered harlequin frog Atelopus varius (0.049; Byrne et al., 2021).
Not only are estimates for inbreeding and relatedness in S. watermeyeri high, but the estimated effective population size for the species is low. This suggests that the long-term population size is closer to the conservative IUCN estimate of ∼250 individuals rather than the thousands of individuals calculated by Whitfield et al. (2017), although it is possible that high census population sizes may be attained temporarily when conditions are favourable. Repeated population bottlenecks during the recent evolutionary history of S. watermeyeri may have eroded genetic diversity to the point of creating two genetically depauperate populations.
The absence of genetic structure between the two populations may indicate that levels of gene flow are sporadically high and/or ongoing, as the two estuaries are separated by a narrow 2.5 km stretch of coastline. However, the combination of low genetic structure and low genetic diversity may point to a recent founder event. Failure to find this species in the Kariega Estuary over several decades of regular sampling suggests potential recolonisation by individuals originating from either the populations that previously resided in the East and West Kleinemonde estuaries, or from the Bushmans Estuary, which was not regularly surveyed and where a small population may have survived unnoticed for several decades. Whitfield et al. (2017) hypothesised that occasional flooding may have facilitated the dispersal of S. watermeyeri between adjacent estuaries by washing individuals into the sea. Such flood events are sporadic (Jordaan et al., 2017) and, given the poor swimming ability of pipefishes and the erratic nature of the region’s nearshore currents, are unlikely to result in large numbers of estuarine pipefishes reaching suitable habitats in nearby estuaries (Whitfield et al., 2017). For the same reason, unauthorised translocations by well-intentioned citizen scientists cannot be ruled out to explain the presence of S. watermeyeri in the Bushmans and Kariega estuaries, but this is unlikely.
Barring gene flow from potentially unreported populations of S. watermeyeri, the current low level of genetic diversity is at risk of declining even further unless concrete conservation measures are taken. In addition to the fixation of deleterious alleles and the species’ reduced adaptive potential, stochastic events like floods, as well as pollution events, the introduction of diseases, alien predators, and competitors, can singly, or in combination, drive these pipefish populations to extinction. Perhaps the greatest threat to the estuarine pipefish in both the Bushmans and Kariega estuaries is a prolonged drought, together with the impact of catchment impoundments, that results in a complete loss of river flow for an extended period such that the zooplankton food chain which supports the pipefish population collapses (Whitfield et al., 2017).
Translocation of individuals represents a suitable management strategy when genetic diversity in individual populations is low (Whiteley et al., 2015), and the risk of outbreeding depression is not a valid concern in this case because of the evident lack of genetic structure between populations (Frankham et al., 2011). However, given the fact that both estuaries are essentially inhabited by the same inbred population, conservation efforts need to focus on preventing any further loss of genetic diversity. Although both populations potentially comprise thousands of individuals (Whitfield et al., 2017), floods can result in the partial or complete loss of macrophyte beds (Riddin and Adams, 2012; Whitfield et al., 2017), while prolonged droughts result in reductions in the abundance and diversity of zooplanktonic prey (Whitfield et al., 2017). As such, both floods and droughts can potentially result in very significant reductions in population size, and even localised extinction. Preventing the occurrence of prolonged periods during which the estuarine pipefish populations remain small appears to be the only means of reducing the risk that the remaining genetic diversity will be further reduced by inbreeding and genetic drift to a degree that may be detrimental for the species’ survival.
While conservation translocations do not represent a suitable means of improving the genetic health of the two remaining pipefish populations, they need to be considered to both maximise the species’ overall effective population size, and to minimise the impact of local extinctions on the erosion of the remaining genetic diversity. These aims could be achieved by reintroducing individuals to additional estuaries within the species’ historical range, in addition to captive breeding. The success of such conservation measures requires that the cause of past population declines and their impacts on the remaining populations be investigated, and that feasible steps be undertaken to mitigate such factors (Semlitsch, 2002; George et al., 2009; Ayres et al., 2012; Grant et al., 2019). In the case of the estuarine pipefish, these causes are primarily natural phenomena that cannot be readily controlled or prevented, although impoundments in the catchment areas of both the Bushmans and Kariega estuaries accentuate the climatic issues. Additional research about the species’ ecology, behaviour and dietary preferences may provide valuable information on how to identify suitable locations and times for release that will maximise the chances of successful rehabilitation of this species throughout its historical range.
Data Availability Statement
The raw read datasets generated and analyzed in this study were submitted to the NCBI Sequence Read Archive (SRA) repository and are available under BioProject PRJNA777762 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA777762).
Ethics Statement
This animal study was reviewed and approved by the Faculty of Science Ethics Committee at the University of Johannesburg (Ethics Reference Number: 2020-02-06/Teske_Weiss).
Author Contributions
PC, HK, AW, and PT conceived the research. PC and PT generated funding and obtained permits. S-EW, AE-K, PC, NJ, and PT participated in sampling. S-EW, AE-K, HK, and PT analysed the data. S-EW, HK, and NJ created figures. HK, BJvV, and PT supervised the student. All authors contributed to the article and approved the submitted version.
Funding
This study was funded by the Mohamed bin Zayed Species Conservation Fund (project number 172516007), with additional funding provided by the University of Johannesburg (URC/FRC grant).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We acknowledge the use of infrastructure/equipment provided by the NRF-SAIAB Aquatic Genomics Research Platform and the funding channelled through the NRF-SAIAB Institutional Support System, and we thank Jody Oliver, Ofentse Ntshudisane, Laura Tensen, and Louw Claassens for helping with the sampling. The Centre for High-Performance Computing (CHPC) is thanked for granting us access to the Lengau Cluster, and the University of Johannesburg IT services are acknowledged for their support. S-EW is grateful to the University of Johannesburg for awarding him a Global Excellence and Stature (GES) 4.0 Master’s Fellowship.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.756595/full#supplementary-material
References
Adams, J. B. (2016). Distribution and status of Zostera capensis in South African estuaries — a review. S. Afr. J. Bot. 107, 63–73. doi: 10.1016/j.sajb.2016.07.007
Alfaro, A. C. (2006). Benthic macro-invertebrate community composition within a mangrove/seagrass estuary in northern New Zealand. Estuar. Coast. Shelf Sci. 66, 97–110. doi: 10.1016/j.ecss.2005.07.024
Allanson, B. R., and Herbert, D. G. (2005). A newly discovered population of the critically endangered false limpet Siphonaria compressa Allanson, 1958 (Pulmonata: Siphonariidae), with observations on its reproductive biology: research letter. S. Afr. J. Sci. 101, 95–97. doi: 10.10520/EJC96338
Aller, E. A., Eklöf, J. S., Gullström, M., Kloiber, U., Linderholm, H. W., and Nordlund, L. M. (2019). Temporal variability of a protected multispecific tropical seagrass meadow in response to environmental change. Environ. Monit. Assess. 191:774. doi: 10.1007/s10661-019-7977-z
Ayres, R., Nicol, M., and Raadik, T. A. (2012). Guidelines for the Translocation of Barred Galaxias (Galaxias fuscus) for Conservation Purposes. East Melbourne, VIC: Dept. of Sustainability and Environment.
Bandeira, S., and Gell, F. (2003). “The seagrasses of Mozambique and Southeastern Africa,” in World Atlas of Seagrasses, eds E. P. Green and F. T. Short (Berkeley, CA: University of California Press), 93–100.
Bate, G. C., Whitfield, A. K., Adams, J. B., Huizinga, P., and Wooldridge, T. H. (2002). The importance of the river-estuary interface (REI) zone in estuaries. Water SA 28, 271–279. doi: 10.4314/wsa.v28i3.4894
Bell, E. M., Lockyear, J. F., McPherson, J. M., Dale Marsden, A., and Vincent, A. C. J. (2003). First field studies of an endangered South African seahorse, Hippocampus capensis. Environ. Biol. Fish. 67, 35–46. doi: 10.1023/A:1024440717162
Byrne, A. Q., Richards-Zawacki, C. L., Voyles, J., Bi, K., Ibáñez, R., and Rosenblum, E. B. (2021). Whole exome sequencing identifies the potential for genetic rescue in iconic and critically endangered Panamanian harlequin frogs. Glob. Change Biol. 27, 50–70. doi: 10.1111/gcb.15405
Castejón-Silvo, I., Terrados, J., Nguyen, T., Jutfelt, F., and Infantes, E. (2021). Increased energy expenditure is an indirect effect of habitat structural complexity loss. Funct. Ecol. 35, 2316–2328. doi: 10.1111/1365-2435.13876
Cowley, P. D., and Whitfield, A. K. (2001). Ichthyofaunal characteristics of a typical temporarily open/closed estuary on the southeast coast of South Africa. Ichthyol. Bull. JLB Smith Inst. Ichthyol. 71, 1–19.
Danecek, P., Auton, A., Abecasis, G., Albers, C. A., Banks, E., DePristo, M. A., et al. (2011). The variant call format and VCFtools. Bioinformatics 27, 2156–2158.
Danecek, P., Bonfield, J. K., Liddle, J., Marshall, J., Ohan, V., Pollard, M. O., et al. (2021). Twelve years of SAMtools and BCFtools. Gigascience 10:giab008. doi: 10.1093/gigascience/giab008
Dawson, C. E. (1985). Indo-Pacific Pipefishes (Red Sea to the Americas). Ocean Springs, MS: Gulf Coast Research Laboratory.
Dawson, C. E. (1986). “Family no. 145: Syngnathidae,” in Smith’s Sea Fishes, eds M. M. Smith and P. C. Heemstra (Johannesburg: Macmillan South Africa), 445–458.
Do, C., Waples, R. S., Peel, D., Macbeth, G. M., Tillett, B. J., and Ovenden, J. R. (2014). NeEstimator v2: re-implementation of software for the estimation of contemporary effective population size (Ne) from genetic data. Mol. Ecol. Res. 14, 209–214. doi: 10.1111/1755-0998.12157
Eaton, D. A. R., and Overcast, I. (2020). ipyrad: interactive assembly and analysis of RADseq datasets. Bioinformatics 36, 2592–2594. doi: 10.1093/bioinformatics/btz966
Elshire, R. J., Glaubitz, J. C., Sun, Q., Poland, J. A., Kawamoto, K., Buckler, E. S., et al. (2011). A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS One 6:e19379. doi: 10.1371/journal.pone.0019379
Escoda, L., González-Esteban, J., Gómez, A., and Castresana, J. (2017). Using relatedness networks to infer contemporary dispersal: application to the endangered mammal Galemys pyrenaicus. Mol. Ecol. 26, 3343–3357.
Excoffier, L., and Lischer, H. E. (2010). Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Res. 10, 564–567. doi: 10.1111/j.1755-0998.2010.02847.x
Foster, S. J., and Vincent, A. C. J. (2004). Life history and ecology of seahorses: implications for conservation and management. Fish Biol. 65, 1–61. doi: 10.1111/j.0022-1112.2004.00429.x
Frankham, R. (1997). Do island populations have less genetic variation than mainland populations? Heredity 78, 311–327. doi: 10.1038/hdy.1997.46
Frankham, R. (2005). Genetics and extinction. Biol. Conserv. 126, 131–140. doi: 10.1016/j.biocon.2005.05.002
Frankham, R., Ballou, J. D., Eldridge, M. D. B., Lacy, R. C., Ralls, K., Dudash, M. R., et al. (2011). Predicting the probability of outbreeding depression. Conserv. Biol. 25, 465–475. doi: 10.1111/j.1523-1739.2011.01662.x
George, A. L., Kuhajda, B. R., Williams, J. D., Cantrell, M. A., Rakes, P. L., and Shute, J. R. (2009). Guidelines for propagation and translocation for freshwater fish conservation. Fisheries 34, 529–545. doi: 10.1577/1548-8446-34.11.529
Gordina, A. (1991). Distribution, reproduction, and diet of the pelagic pipefish, Syngnathus schmidti, in the Black Sea. J. Ichthyol. 31, 110–119.
Grant, L., Johnson, C., and Thiessen, C. (2019). Evaluating the efficacy of translocation: maintaining habitat key to long-term success for an imperiled population of an at-risk species. Biodivers. Conserv. 28, 2727–2743. doi: 10.1007/s10531-019-01789-6
Hanghøj, K., Moltke, I., Andersen, P. A., Manica, A., and Korneliussen, T. S. (2019). Fast and accurate relatedness estimation from high-throughput sequencing data in the presence of inbreeding. Gigascience 8:giz034. doi: 10.1093/gigascience/giz034
Hedrick, P. W., and Lacy, R. C. (2015). Measuring relatedness between inbred individuals. J. Hered. 106, 20–25. doi: 10.1093/jhered/esu072
Hughes, A. R., Williams, S. L., Duarte, C. M., Heck, K. L., and Waycott, M. (2009). Associations of concern: declining seagrasses and threatened dependent species. Front. Ecol. Environ. 7:242–246. doi: 10.1890/080041
Jackson, E. L., Rowden, A. A., Attrill, M. J., Simon, J. B., and Jones, M. B. (2001). The Importance of Seagrass Beds as a Habitat for Fishery Species. London: CRC Press.
Jacquard, A. (2012). The Genetic Structure of Populations, Vol. 5. Berlin: Springer Science & Business Media.
Jombart, T., Kamvar, Z. N., Collins, C., Luštrik, R., Beugin, M. P., Knaus, B. J., et al. (2020). Adegenet: Exploratory Analysis of Genetic and Genomic Data. R Package Version 2.
Jordaan, A. J., Sakulski, D. M., and Muyambo, F. (2017). Vulnerability, Adaptation to and Coping with Drought: The Case of Commercial and Subsistence Rain fed Farming in the Eastern Cape. Report to the Water Research Commission and Department of Agriculture, Forestry and Fisheries, Vol. 1. Gezina: Water Research Commission.
Keenan, K., McGinnity, P., Cross, T. F., Crozier, W. W., and Prodöhl, P. A. (2013). diveRsity: an R package for the estimation and exploration of population genetics parameters and their associated errors. Methods Ecol. Evol. 4, 782–788. doi: 10.1111/2041-210X.12067
Kuiter, R. H. (2010). Seahorses and their relatives. J. Fish Biol. 77, 308–309. doi: 10.1111/j.1095-8649.2010.02678.x
Liu, J., You, X., Xu, P., Zhuang, P., Zheng, Y., Zhang, K., et al. (2018). Assessing the genetic diversity of the critically endangered Chinese sturgeon Acipenser sinensis using mitochondrial markers and genome-wide single-nucleotide polymorphisms from RAD-seq. Sci. China Life Sci. 61, 1090–1098. doi: 10.1007/s11427-017-9254-6
Lopes, M. S., Silva, F. F., Harlizius, B., Duijvesteijn, N., Lopes, P. S., Guimarães, S. E., et al. (2013). Improved estimation of inbreeding and kinship in pigs using optimized SNP panels. BMC Genet. 14:92. doi: 10.1186/1471-2156-14-92
Mkare, T. K., Vuuren, B. J., and Teske, P. R. (2017). Conservation implications of significant population differentiation in an endangered estuarine seahorse. Biodivers. Conserv. 26, 1275–1293. doi: 10.1007/s10531-017-1300-5
Musammilu, K. K., Abdul-Muneer, P. M., Gopalakrishnan, A., Basheer, V. S., Gupta, H., Mohindra, V., et al. (2014). Identification and characterization of microsatellite markers for the population genetic structure in endemic red-tailed barb, Gonoproktopterus curmuca. Mol. Biol. Rep. 41, 3051–3062. doi: 10.1007/s11033-014-3164-z
Orth, R. J., Carruthers, T. J. B., Dennison, W. C., Duarte, C. M., Fourqurean, J. W., Heck, K. L., et al. (2006). A global crisis for seagrass ecosystems. Bioscience 56, 987–996.
Pillay, D., Branch, G. M., Griffiths, C. L., Williams, C., and Prinsloo, A. (2010). Ecosystem change in a South African marine reserve (1960–2009): role of seagrass loss and anthropogenic disturbance. Mar. Ecol. Prog. Ser. 415, 35–48. doi: 10.3354/meps08733
Pollard, D. A. (1984). A review of ecological studies on seagrass—fish communities, with particular reference to recent studies in Australia. Aquat. Bot. 18, 3–42. doi: 10.1016/0304-3770(84)90079-2
Pollom, R. (2016). Syngnathus watermeyeri. The IUCN Red List of Threatened Species. Gland: International Union for Conservation of Nature. doi: 10.2305/IUCN.UK.2017-3.RLTS.T41030A67621860.en
Raj, A., Stephens, M., and Pritchard, J. K. (2014). fastSTRUCTURE: variational inference of population structure in large SNP data sets. Genetics 197, 573–589.
Rasheed, M. A., and Unsworth, R. K. F. (2011). Long-term climate-associated dynamics of a tropical seagrass meadow: implications for the future. Mar. Ecol. Prog. Ser. 422, 93–103. doi: 10.3354/meps08925
Riddin, T., and Adams, J. B. (2012). Predicting macrophyte states in a small temporarily open/closed estuary. Mar. Freshw. Res. 63, 616–623. doi: 10.1071/MF11224
Semlitsch, R. D. (2002). Critical elements for biologically based recovery plans of aquatic-breeding amphibians. Conserv. Biol. 16, 619–629. doi: 10.1046/j.1523-1739.2002.00512.x
Shokri, M. R., Gladstone, W., and Jelbart, J. (2009). The effectiveness of seahorses and pipefish (Pisces: Syngnathidae) as a flagship group to evaluate the conservation value of estuarine seagrass beds. Aquat. Conserv. Mar. Freshw. Ecosyst. 19, 588–595. doi: 10.1002/aqc.1009
Short, F. T., Coles, R., Waycott, M., Bujang, J., Fortes, M., Prathep, A., et al. (2010). Zostera capensis. The IUCN Red List of Threatened Species. Gland: International Union for Conservation of Nature. doi: 10.2305/IUCN.UK.2010-3.RLTS.T173370A7001305.en
Short, F. T., Polidoro, B., Livingstone, S. R., Carpenter, K. E., Bandeira, S., Bujang, J. S., et al. (2011). Extinction risk assessment of the world’s seagrass species. Biol. Conserv. 144, 1961–1971. doi: 10.1016/j.biocon.2011.04.010
Skelton, P. H. (1987). South African Red Data Book – Fishes. National Scientific Programmes Unit. Pretoria: CSIR.
Smith, J. L. B. (1963). Fishes of the Family Syngnathidae from the Red Sea and the Western Indian Ocean. Ichthyological Bulletin No. 28. Grahamstown: Department of Ichthyology, Rhodes University.
Speed, D., and Balding, D. J. (2015). Relatedness in the post-genomic era: is it still useful? Nat. Rev. Genet. 16, 33–44. doi: 10.1038/nrg3821
Stankovic, M., Hayashizaki, K.-I., Tuntiprapas, P., Rattanachot, E., and Prathep, A. (2021). Two decades of seagrass area change: organic carbon sources and stock. Mar. Pollut. Bull. 163:111913. doi: 10.1016/j.marpolbul.2020.111913
Tajima, F. (1983). Evolutionary relationship of DNA sequences in finite populations. Genetics 105, 437–460.
Taylor, H. R. (2015). The use and abuse of genetic marker-based estimates of relatedness and inbreeding. Ecol. Evol. 5, 3140–3150. doi: 10.1002/ece3.1541
Teske, P. R., Cherry, M. I., and Matthee, C. A. (2003). Population genetics of the endangered Knysna seahorse, Hippocampus capensis. Mol. Ecol. 12, 1703–1715. doi: 10.1046/j.1365-294X.2003.01852.x
Teske, P. R., and Wooldridge, T. H. (2003). What limits the distribution of subtidal macrobenthos in permanently open and temporarily open/closed South African estuaries? Salinity vs. sediment particle size. Estuar. Coast. Shelf Sci. 57, 225–238. doi: 10.1016/S0272-7714(02)00347-5
Van der Colff, A. (2016). Zostera capensis Setchell. National Assessment: Red List of South African Plants Version 2020.1. Available online at: http://redlist.sanbi.org/species.php?species=4111-1 (accessed May 29, 2021)
Vincent, A. C. J., Evans, K. L., and Marsden, A. D. (2005). Home range behaviour of the monogamous Australian seahorse, Hippocampus whitei. Environ. Biol. Fish. 72, 1–12. doi: 10.1007/s10641-004-4192-7
Vorwerk, P. D., Froneman, P. W., and Paterson, A. W. (2007). Recovery of the critically endangered river pipefish, Syngnathus watermeyeri, in the Kariega Estuary, Eastern Cape province. S. Afr. J. Sci. 103, 199–201.
Wahlund, S. (1928). Zusammensetzung von Populationen und Korrelationserscheinungen vom Standpunkt der Vererbungslehre aus betrachtet. Hereditas 11, 65–106.
Wang, J. (2017). Estimating pairwise relatedness in a small sample of individuals. Heredity 119, 302–313.
Waples, R. S. (2015). Testing for Hardy–Weinberg proportions: have we lost the plot? J. Hered. 106, 1–19. doi: 10.1093/jhered/esu062
Whiteley, A. R., Fitzpatrick, S. W., Funk, W. C., and Tallmon, D. A. (2015). Genetic rescue to the rescue. Trends Ecol. Evol. 30, 42–49. doi: 10.1016/j.tree.2014.10.009
Whitfield, A. K., and Bruton, M. N. (1996). Extinction of the river pipefish Syngnathus watermeyeri in the Eastern Cape Province, South Africa. S. Afr. J. Sci. 92, 59–60.
Whitfield, A. K., Mkare, T. M., Teske, P. R., James, N. C., and Cowley, P. D. (2017). Life-histories explain the conservation status of two estuary-associated pipefishes. Biol. Conserv. 212, 256–264. doi: 10.1016/j.biocon.2017.06.024
Woodall, L. C., Jones, R., Zimmerman, B., Guillaume, S., Stubbington, T., Shaw, P., et al. (2012). Partial fin-clipping as an effective tool for tissue sampling seahorses, Hippocampus spp. J. Mar. Biol. Assoc. U. K. 92, 1427–1432. doi: 10.1017/S0025315411001810
Wright, S. (1949). The genetical structure of populations. Ann. Eugen. 15, 323–354. doi: 10.1111/j.1469-1809.1949.tb02451.x
Keywords: conservation translocation, Critically Endangered species, estuarine pipefish, inbreeding, next-generation sequencing, population genomics, seagrass fauna, Syngnathus watermeyeri
Citation: Weiss S-E, Emami-Khoyi A, Kaiser H, Cowley PD, James NC, Jansen van Vuuren B, Whitfield AK and Teske PR (2022) The Last Two Remaining Populations of the Critically Endangered Estuarine Pipefish Are Inbred and Not Genetically Distinct. Front. Mar. Sci. 8:756595. doi: 10.3389/fmars.2021.756595
Received: 10 August 2021; Accepted: 25 November 2021;
Published: 05 January 2022.
Edited by:
Christian Marcelo Ibáñez, Andres Bello University, ChileReviewed by:
Mariano González-Castro, Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), ArgentinaJuan Andrés López, University of Alaska Fairbanks, United States
Nicolás I. Segovia, University of Chile, Chile
Copyright © 2022 Weiss, Emami-Khoyi, Kaiser, Cowley, James, Jansen van Vuuren, Whitfield and Teske. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Arsalan Emami-Khoyi, ekarsalan@gmail.com; Peter R. Teske, pteske101@gmail.com
 Sven-Erick Weiss
Sven-Erick Weiss Arsalan Emami-Khoyi
Arsalan Emami-Khoyi Horst Kaiser2
Horst Kaiser2  Paul D. Cowley
Paul D. Cowley Nicola C. James
Nicola C. James Bettine Jansen van Vuuren
Bettine Jansen van Vuuren Alan K. Whitfield
Alan K. Whitfield Peter R. Teske
Peter R. Teske