Gracilaria gracilis and Nannochloropsis oceanica, singly or in combination, in diets alter the intestinal microbiota of European seabass (Dicentrarchus labrax)
- 1CIIMAR/CIMAR-LA, Centro Interdisciplinar de Investigação Marinha e Ambiental, Matosinhos, Portugal
- 2ICBAS, Instituto de Ciências Biomédicas Abel Salazar, Universidade do Porto, Porto, Portugal
- 3Faculty of Biosciences and Aquaculture, Nord University, Bodø, Norway
- 4ALGAplus, Production and Trading of Seaweed and Derived Products Ltd., Iílhavo, Portugal
- 5Allmicroalgae, Microalgae Production Plant, Pataias, Portugal
Algae feeds and fish gut microbiota have been given importance in the past few years because of the necessity to rely on sustainable ingredients in aquafeeds and the link of host-associated microbes to organismal health. But little is known about the potential of algae, particularly of micro- and macroalgae combination, to shape the intestinal bacterial communities. Hence, in the present work, the 16S rRNA gene sequencing technique was employed to unravel the effects of the seaweed Gracilaria gracilis and the microalga Nannochloropsis oceanica - included either singly or in combination in the diets of European seabass - on the diversities and composition of the gut bacterial communities. Results indicated that 8% inclusion of either G. gracilis (GRA) or N. oceanica (NAN) led to a reduction in the gut microbial diversity. On the other hand, inclusion of the micro- and macroalga in a blend (NANGRA) mitigated these plausible effects on the intestinal bacterial communities. The core microbiota of European seabass was composed of both beneficial (Lactobacillus and Cetobacterium) and potentially pathogenic (Flavobacterium) bacteria. The GRA diet was associated with a lower abundance of carbohydrate degraders and also promoted the growth of bacteria capable of outcompeting fish pathogens (Sulfitobacter and Methylobacterium). On the other hand, the NAN diet led to a higher representation of the genus Bacillus, with probiotic potential, accompanied by a decrease in Vibrio, a genus encompassing several fish pathogenic species. These findings demonstrate the ability of micro- and macroalgae to modulate the gut microbiota of European seabass, with plausible implications to host gut homeostasis.
Introduction
Fish intestinal microbiota has become one of the most studied topics in the field of aquaculture, by following the trends in human and mammalian research (Diwan et al., 2022). The complex and dynamic assemblage of microorganisms in the gut microbiota is known to have crucial implications on key physiological functions (Cerf-Bensussan and Gaboriau-Routhiau, 2010; Egerton et al., 2018). Since the intestinal bacterial communities of aquatic animals are vastly shaped by diet, it is expected that nutritional manipulation of fish gut microbiota using added-value products can produce intended benefits on fish welfare and nutrition (Yukgehnaish et al., 2020). In this context, it is worth mentioning that algae are natural products and are rich in biologically active substances capable of promoting fish growth and improving the immune status and disease resistance (Wan et al., 2019; Valente et al., 2021). Specifically, two algae that have been recently explored by the feed industry are the macroalga Gracilaria gracilis and the microalga Nannochloropsis oceanica. Although there are some reports on the effect of these algae on fish growth performance, immunity, oxidative status and intestine histomorphology (Batista et al., 2020; Passos et al., 2021; Sørensen et al., 2021), little is known about their impact on the intestinal microbiota of farmed fish.
The core research on dietary modulation of fish gut microbiota has been undertaken to reveal the effect of either probiotics and prebiotics or dietary proteins and lipids (Egerton et al., 2018; Pérez-Pascual et al., 2020; Vargas-Albores et al., 2021). Studies that evaluated the changes in intestinal microbiota of farmed fish fed algae-supplemented diets have also emerged more recently (Keating et al., 2021; Sagaram et al., 2021). Other studies have focused on the impact of algae as feed ingredients (replaced fish meal/fish oil) (Rico et al., 2016; Lyons et al., 2017; Li et al., 2022) or feed additives (Jorge et al., 2019; Cerezo et al., 2022). Replacing fish meal (partially) with the macroalgae G. cornea or Ulva rigida altered the diversity of bacteria in gilthead seabream (Sparus aurata) intestine: while 15% substitution of both the seaweeds increased the diversity, 25% substitution of U. rigida decreased it (Rico et al., 2016). Li et al. (2022) reported the effects of total fish meal replacement with the microalga Chlorella vulgaris on the intestinal microbiota of largemouth bass (Micropterus salmoides); the abundance of beneficial taxa (e.g., Cetobacterium that are known as vitamin B12 producers) was increased without any differences in alpha diversity compared to the fish meal group. On the other hand, in rainbow trout (Oncorhynchus mykiss), 5% dietary Schizochytrium limacinum as partial fish oil substitute significantly increased the intestinal bacterial diversity without changing the overall microbial community structure. Nevertheless, some microbial groups including members of the lactic acid bacteria, which are commonly considered beneficial, had an increased representation in the S. limacinum group (Lyons et al., 2017). Short-term feeding of gilthead seabream with diets supplemented with N. gaditana at 7.5 g kg-1 led to an increased richness of the gut microbiota, but not significant when compared with fish fed the control diet (Jorge et al., 2019). Similarly, the diversity of gut microbiota was not altered in Atlantic cod (Gadus morhua) fed diets supplemented with U. rigida (10%) compared to the control group (Keating et al., 2021).
Based on the results reported in the available literature, micro- and macroalgae, either as ingredients or supplements, seem to modulate the gut microbiota of farmed fish, but the effects are species-specific and vastly dependent on the type of the algae and their inclusion levels. Moreover, most studies have reported the effects of individual algae. A mix of micro- and macroalgae in aquafeeds can impart synergistic modulatory effects on the gut microbiota. However, this topic is underexplored and deserves more attention because both micro- and macroalgae can be future feed ingredients/additives. Here we report for the first time the impact of the seaweed G. gracilis and the microalga N. oceanica, incorporated singly or in combination, on the composition of the microbial communities in the posterior intestine of European seabass (Dicentrarchus labrax), which is one of the most important farmed fish species of the Mediterranean region.
Material and methods
Ethical statement
The trial was performed by accredited scientists, and the animal handling and sampling procedures were in compliance with the guidelines of the European Union (directive 2010/63/EU) and Portuguese law (Decreto-Lei no. 113/2013, de 7 de Agosto) on the protection of animals used for scientific purposes. Review of the ethical process concerning all animal handling and sampling procedures was performed by CIIMAR animal welfare body (ORBEA-CIIMAR) and approved by national competent authorities.
Experimental diets, feeding trial, and sampling
Details of the experimental design and diets for the feeding trial, as well as zootechnical data can be found in Batista et al. (2020). Briefly, European seabass juveniles (approximately 6 months of age) from a commercial fish farm (SONRIONANSA S.L., Cantabria, Spain) were transported to CIIMAR facilities in Matosinhos, Portugal. Following a 2-week quarantine, twelve homogeneous groups (average body weight of 29.7 ± 0.02 g and total length of 13.7 ± 0.08 cm) of nineteen fish were transferred into 50-L fiberglass tanks of a saltwater recirculation system (RAS, density of 11.3 kg m−3). The RAS conditions were as follows: water temperature of 21 °C, salinity of 35‰, flow rate at 4 L min−1 and 12 h light/12 h dark photoperiod regime.
The experimental diets consisted of four isoproteic (53% dry matter, DM) and isolipidic (17% DM) diets supplied by Sparos Lda.: a commercial feed-based diet (CTRL) and three experimental diets with either 8% G. gracilis (GRA) or 8% N. oceanica (NAN) or a blend of 4% of each alga (NANGRA), included at the expense of fish meal and wheat meal. Ingredients and proximate composition of the experimental diets are summarized in Figure 1. To unravel the impact of the macroalga G. gracilis and the microalga N. oceanica, added singly or in combination in diets, on European seabass intestinal microbiota, fish were fed the experimental diets for 106 days. Fish in triplicate tanks per dietary treatment were fed three times a day until apparent satiation. The biotic conditions (e.g., temperature and salinity), known to have a greater impact on the gut microbiota, were kept constant throughout the feeding trial.
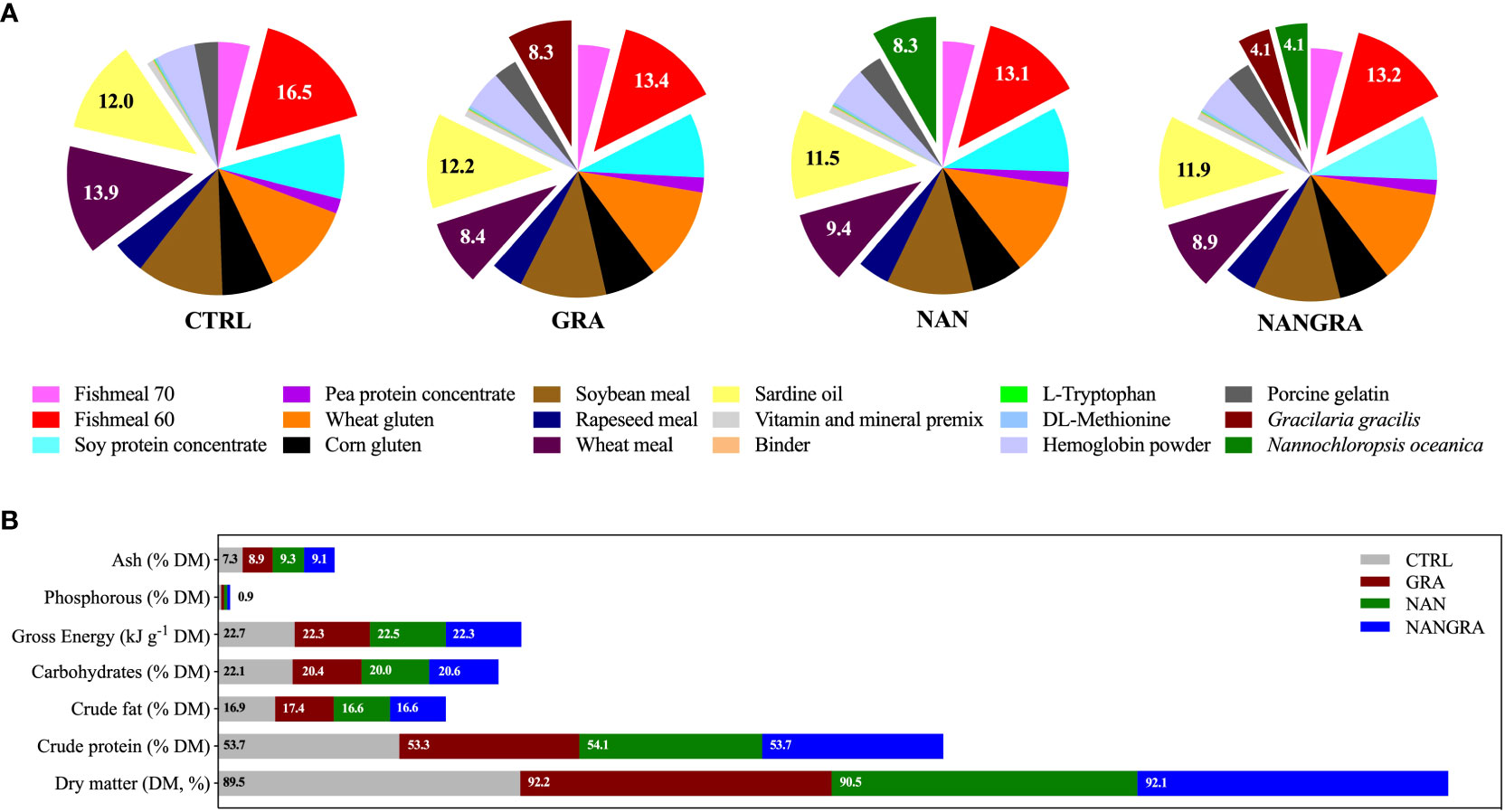
Figure 1 Ingredients in % (A) and proximate composition in % DM (B) of the experimental diets. Adapted from Batista et al. (2020).
At the end of the 15-week feeding trial, fish were starved for 24 h and sacrificed by a sharp blow on the head before sampling the tissues. The gastrointestinal tract of four fish per tank (12 fish per treatment) was removed under sterile conditions and the posterior intestine (section preceding the ileorectal valve) was opened using sterile scissors. A sterile swab (Copan Italia, Italy) was used to collect the mucus from the posterior portion. The swab was then placed in a sterile tube and immediately frozen in liquid nitrogen. Samples were stored at -80 °C until further analysis.
DNA extraction for 16S rRNA sequencing
All the procedures mentioned here were performed under sterile conditions. For DNA extraction, three samples per treatment were randomly selected to divide samples into 4 batches. Genomic DNA was extracted from all samples using QIAamp DNA stool Mini Kit (Qiagen, Germany) according to the manufacturer’s protocol, with some modifications. Briefly, swabs with mucus from the posterior intestine were first transferred to 5 mL tubes containing 1.4- and 0.1-mm Zirconium oxide beads (Cayman chemical, USA) and 2 mL of InhibitEX buffer (Qiagen). Afterwards, samples were homogenized using Precellys® Evolution tissue homogenizer at 6,000 RPM (3 cycles of 30 s, with a 30 s pause in-between) following centrifugation for 5 min at 12,000 g. The obtained pellet was then resuspended, transferred to new 2 mL tubes, and incubated at 70 °C for 20 min. Following incubation, samples were mixed with a vortex for 1 min and centrifuged for 1 min at 12,000 g. The supernatant (600 µL) was transferred into a 2 mL tube with 25 µL of proteinase K, and the following steps of the DNA extraction proceeded according to the manufacturer’s protocol (QIAamp DNA Stool Mini Kit). Finally, DNA was eluted using 75 µL of ATE buffer and DNA quantity was checked using Qubit™ dsDNA Assay (Thermo Fisher Scientific Inc., USA).
Amplicon library preparation and sequencing
Amplicon libraries were prepared under sterile conditions. The first PCR reaction targeted the hypervariable V3-V4 region of the 16S rRNA gene, using specific bacterial primers 341F (5’ CCTACGGGNGGCWGCAG 3’) and 805R (5’ GACTACNVGGGTWTCTAATCC 3’) (Klindworth et al., 2013) flanked by Illumina adapters (~ 460 bp; Illumina, USA). The PCR reactions were performed in triplicate for each sample in a 25 µL final volume, with 1 µL of each primer (10 µM), 2.5 µl of DNA template (5 ng/µL), 8 µL of water, and using 12.5 µL of Amplitaq Gold Q5® (Thermo Fisher Scientific Inc.). Thermal cycling conditions were as follows: initial denaturation step at 95°C for 10 min, 37 cycles at 95°C for 30 s, 57°C for 30 s, 72°C for 1 min, and the final extension step at 72°C for 7 min. Negative PCR controls, without DNA template, were also included. At the end of the amplification, the replicates for each sample were pooled together for the subsequent procedures. The amplified products were visualized on an agarose gel (1.5%), and the CleanNGS system (CleanNA, Netherlands) was used to purify the PCR products, according to the manufacturer’s recommendations.
A second PCR (index PCR) was performed using the purified products, using Nextera XT Index primers (Illumina) and with the following thermal cycling conditions: 95 °C for 3 min, 8 cycles at 95°C for 30 s, 55°C for 30 s, 72°C for 30 min, and a final step at 72 °C for 5 min (16S Metagenomic Sequencing Library Preparation, Illumina). The CleanNGS system (CleanNA) was then used to purify the amplicon libraries. Quality of the obtained libraries was evaluated on a Tapestation 2200 platform (Agilent Technologies, USA), and the libraries were subsequently quantified using the Quant-IT PicoGreen dsDNA assay kit (Thermo Fisher Scientific Inc.) and the Synergy2 microplate reader (Biotek, USA). Thereafter, the pooled library was quantified by Real-Time qPCR LightCycler 480 (Roche, Switzerland), using the KAPA Library quantification kit (Roche). The libraries were sequenced on an Illumina® MiSeq (PE300) platform (MiSeq Control Software 2.5.0.5 and Real-Time Analysis software 1.18.54.0).
Sequence data analysis
Raw sequence data is deposited in the Sequence Read Archive (SRA) and the accession number is PRJNA867546. The obtained reads were first truncated at 270 bp by VSEARCH (Rognes et al., 2016). Next, MICCA pipeline (v1.7.2) (Albanese et al., 2015) was used to further process the reads. Employing MICCA, the sequences were merged at a minimum overlap length of 60 bp and a maximum mismatch of 20 bp. For further analysis, reads without primers were discarded and the forward and reverse primers were trimmed off from the merged reads. Next, sequences with an expected error rate of > 0.75 (Edgar and Flyvbjerg, 2015) were filtered and sequences shorter than 400 bp were removed. The “de novo UNOISE” method implemented in MICCA was employed to perform denoising on the filtered reads. The MICCA pipeline uses UNOISE3 algorithm (Edgar, 2016) that corrects sequencing errors and determines true biological sequences at single-nucleotide resolution. Chimera and mitochondrial sequences were removed, and the amplicon sequence variants (ASVs) were generated for the downstream analyses. The RDP classifier was used to assign taxonomies to the bacterial ASVs. The alignment of the sequences was then performed using the NAST multiple sequence aligner (DeSantis et al., 2006), and a phylogenetic tree was generated using the FastTree software within the MICCA pipeline.
Phylogenetic analysis
Blastn (Chen et al., 2015) of the Basic Local Alignment Search Tool (BLAST) was employed to investigate the similarities of the sequences of the identified ASVs and bacterial sequences from the NCBI Reference Sequence Database. Bacterial sequences with 100% match/similarity were selected for the phylogenetic analysis. Thereafter, these sequences were imported into Molecular Evolutionary Genetics Analysis (MEGAX) software (Kumar et al., 2018) and multiple sequence alignment of the ASVs and sequences with high hits in BLASTn was performed using ClustalW aligner (Version 2.0) (Larkin et al., 2007). Phylogenetic analysis of the sequences was performed using the unweighted average binding among clusters (Unweighted Pair Group Method with Arithmetic mean, UPGMA) approach (Rédei (Ed.), 2008), by applying default parameters and 500 bootstrap replicates. The generated phylogenetic trees (using maximum likelihood inference) were visualized using MEGAX software.
Statistical analysis
All statistical analysis were performed on R studio version 1.4.1103. The packages “iNEXT” and “phyloseq” were used to calculate alpha diversity: overall species richness (function “ChaoRichnes”), Shannon diversity (function “ChaoShannon”) and Simpson diversity (function “ChaoSimpson”). Faith’s phylogenetic diversity was calculated using the function “pd” in package “picante” to reveal the losses and gains in species. Kruskal-Wallis’ test followed by Dunn’s test were employed to understand the differences between the experimental groups. Then employing the functions in “ggplot2”, plots were generated for overall species richness, Shannon diversity, Simpson diversity and Faith’s phylogenetic diversity (Hsieh et al., 2016). For the bacterial beta diversity analysis, weighted UniFrac distances were employed (Lozupone and Knight, 2005), and beta diversity was visualized on a principal coordinates analysis (PCoA) plot. Dispersions of the communities were first analyzed using “betadisper” and significant dissimilarities between the communities were then determined using Permutational Multivariate Analysis of Variance Using Distance Matrices (PERMANOVA with 9,999 permutations) (Anderson, 2001), implemented in “adonis” function of the vegan R-package (Oksanen et al., 2013), followed by pairwise comparisons. The package “microbiome” was employed to determine relative abundance of core taxa (Lahti et al., 2017). To identify the differentially abundant ASVs, the non-rarefied data (McMurdie and Holmes, 2014) was analyzed using the “DESeq2” package (Love et al., 2014) as rarefied data is reported to reduce the statistical power of the analysis (Weiss et al., 2017).
Results
Composition of the intestinal microbiota of European seabass fed the algae diets
All samples were sequenced in a single MiSeq run, generating a total of 4,674,383 high-quality reads with an average of 101,617 reads per sample. To account for the read count variation in the different samples, the reads were rarefied to 18,000 sequences per sample; to obtain a uniform sampling depth for comparing the different groups. Out of the 45 samples, two libraries with several reads below the cut off were discarded. A total of 43 samples were used for the downstream analysis – 11 samples for the CTRL group, 11 samples for the GRA group, 12 samples for the NAN group and 9 samples for the NANGRA group. Across all samples, 4371 ASVs were identified after denoising, belonging to 26 phyla and 306 genera.
Concerning the abundances of the intestinal bacteria of European seabass, the 26 bacterial phyla identified across all samples are displayed in Figure 2A (relative abundance per sample) and Supplementary Figure 1A (abundance per dietary treatment). The most abundant phyla were Proteobacteria, Firmicutes, Actinobacteria, Bacteroidetes, Fusobacteria, Nitrospirae and Parcubacteria (Figure 2B). Across all experimental groups, Proteobacteria was the most abundant phylum, with an average relative abundance of 35.25% in the CTRL group, 53.29% in the GRA group, 34.93% in the NAN group and 40.45% in the NANGRA group. The second most abundant phylum for the CTRL group was Bacteroidetes (12.06%), Actinobacteria for the GRA group (11.83%), and Firmicutes for the NAN and NANGRA groups (20.98 and 14.66%, respectively). The third most abundant phyla were Nitrospirae for the CTRL group (10.21%), Bacteroidetes for both GRA and NANGRA groups (9.61 and 11.46%, respectively), and Actinobacteria for the NAN group (12.65%) (Table 1).
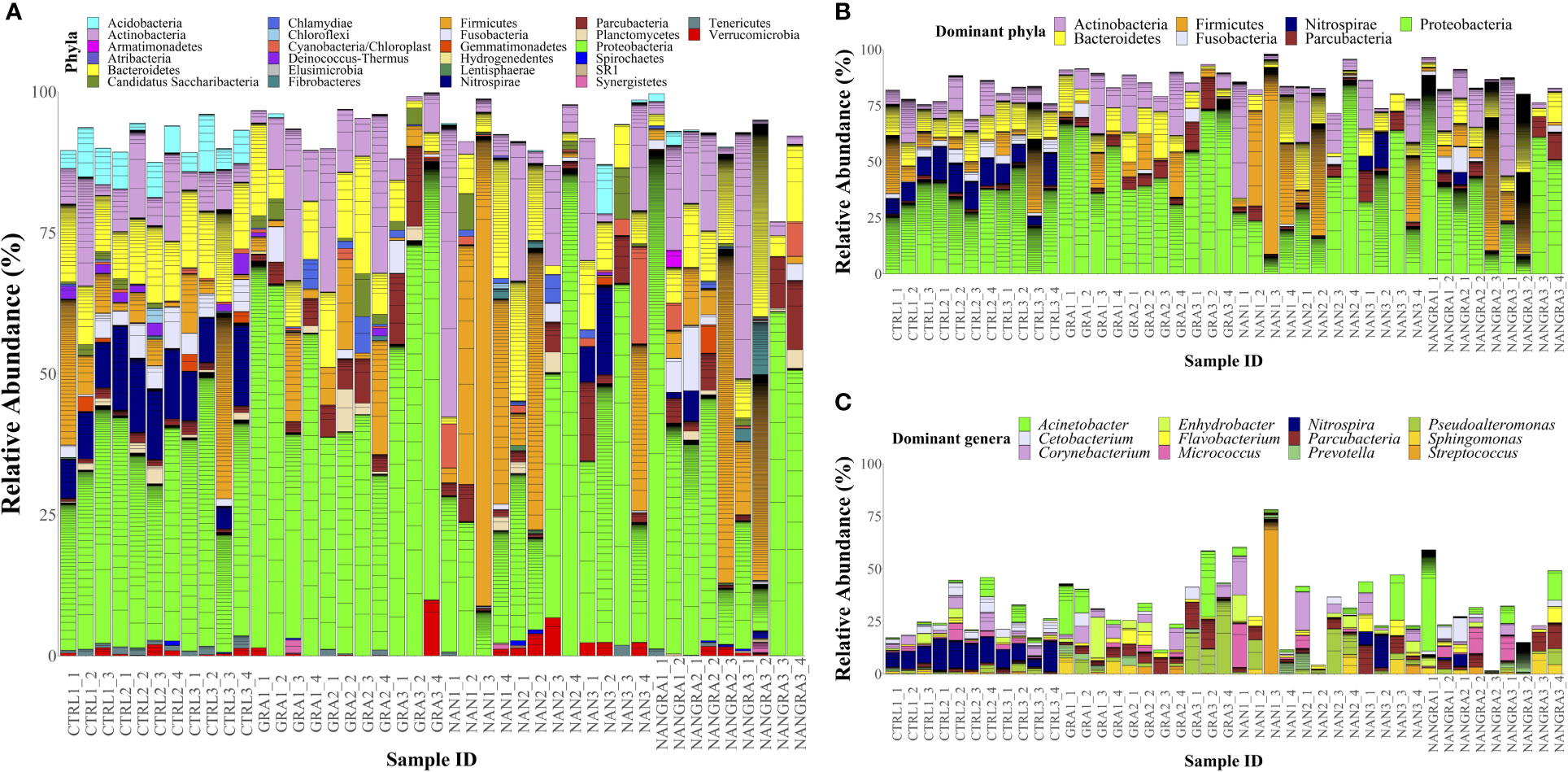
Figure 2 Relative abundance of the bacterial phyla (A), dominant phyla (B) and dominant genera (C) in the posterior intestine of European seabass fed the experimental diets. Sample ID: CTRL, control group; GRA, Gracilaria gracilis group; NAN, Nannochloropsis oceanica group; NANGRA, G gracilis and N. oceanica blend group. Color codes: Actinobacteria – pink, Bacteroidetes – yellow, Firmicutes – orange, Fusobacteria – lavender, Nitrospirae – dark blue, Parcubacteria – dark red, Proteobacteria – green. Unclassified bacteria are not shown in the figures.
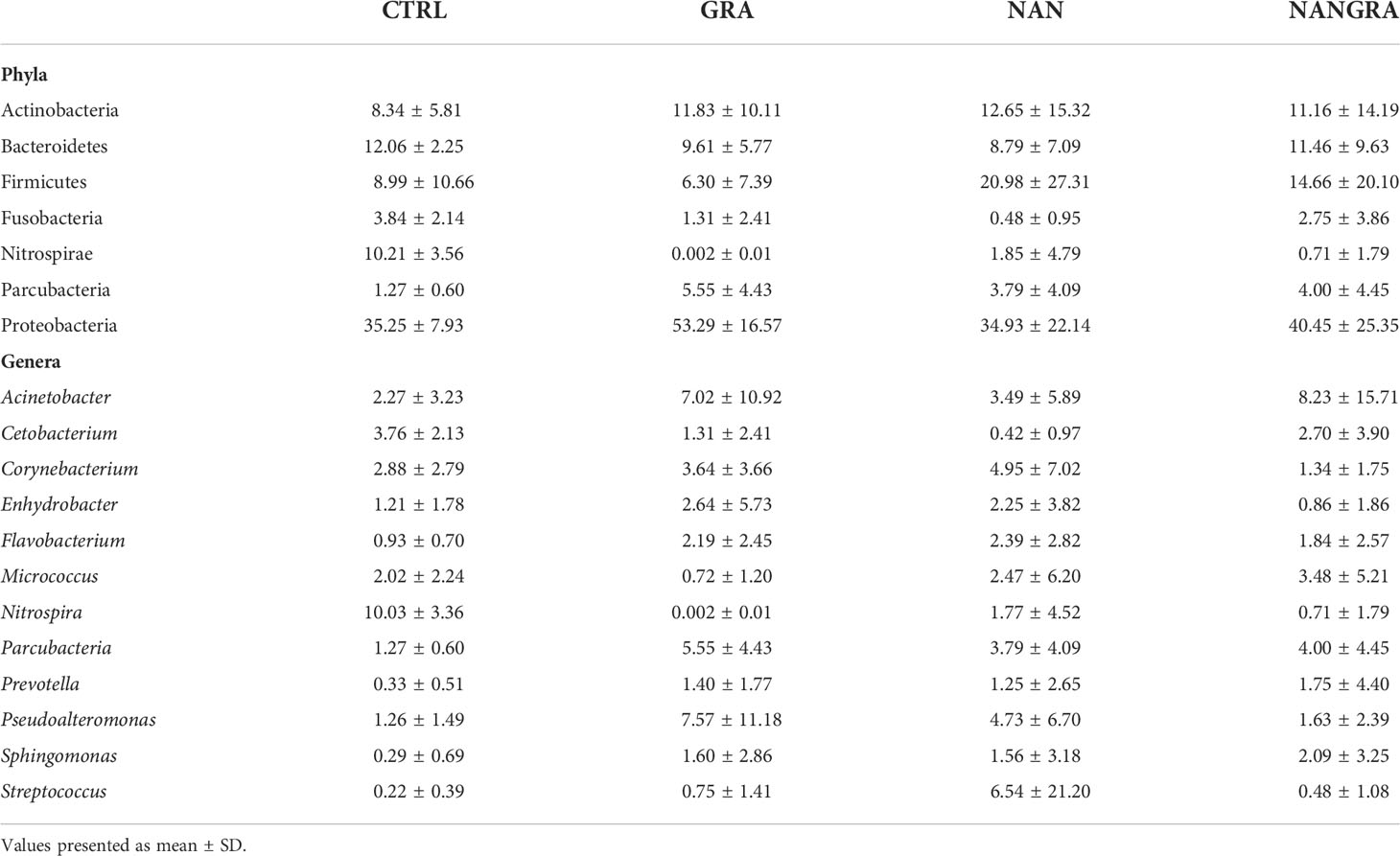
Table 1 Average relative abundance (%) of the dominant bacterial phyla and genera in the posterior intestine of European seabass fed the experimental diets.
The abundance of the dominant genera per dietary treatment are presented in Supplementary Figure 1B. The dominant genera included Acinetobacter, Cetobacterium, Corynebacterium, Enhydrobacter, Flavobacterium, Micrococcus, Nitrospira, Parcubacteria, Prevotella, Pseudoalteromonas, Sphingomonas and Streptococcus, but we did not find a clear dominance of specific genera across all intestinal samples (Figure 2C). The proportions of the dominant genera varied according to dietary treatment: the most abundant genus for the CTRL group was Nitrospira (10.03%); for the GRA group the most abundant taxa were Pseudoalteromonas (7.57%) and Acinetobacter (7.02%); for the NAN group Streptococcus had the highest proportion (6.54%); and for the NANGRA group Acinetobacter had a higher representation (8.23%) (Table 1).
Impact of the algae diets on the diversity of the intestinal microbiota
The alpha diversity analysis, displayed in Figure 3A, was evaluated using the ecological diversity measures Chao richness (measure of species richness), Shannon diversity (effective number of common species that indicates the even distribution of microbes) and Simpson diversity (effective number of dominant species). The overall species richness was significantly lower in fish that consumed the GRA diet compared to CTRL (P = 0.003), but not compared to fish fed the NAN (P = 0.06) and NANGRA (P = 0.36) diets (Figure 3A1; Kruskal-Wallis’ chi-squared = 13.467; P = 0.0037). The microbial evenness of the populations (Shannon diversity) in single algae groups (GRA and NAN) was significantly lower than in CTRL-fed fish (P = 0.009), but a similar difference was not observed for fish that consumed the algae blend (NANGRA; P = 0.14) (Figure 3A2; Kruskal-Wallis’ chi-squared = 12.593; P = 0.0056). Similarly, fish that consumed both GRA and NAN diets presented a significant reduction in the effective number of dominant species (Simpson diversity) compared to the CTRL fish (P = 0.02 and P = 0.01, respectively), while such differences were not observed in NANGRA fish (P = 0.15) (Figure 3A3; Kruskal-Wallis’ chi-squared = 11.442; P = 0.0096).
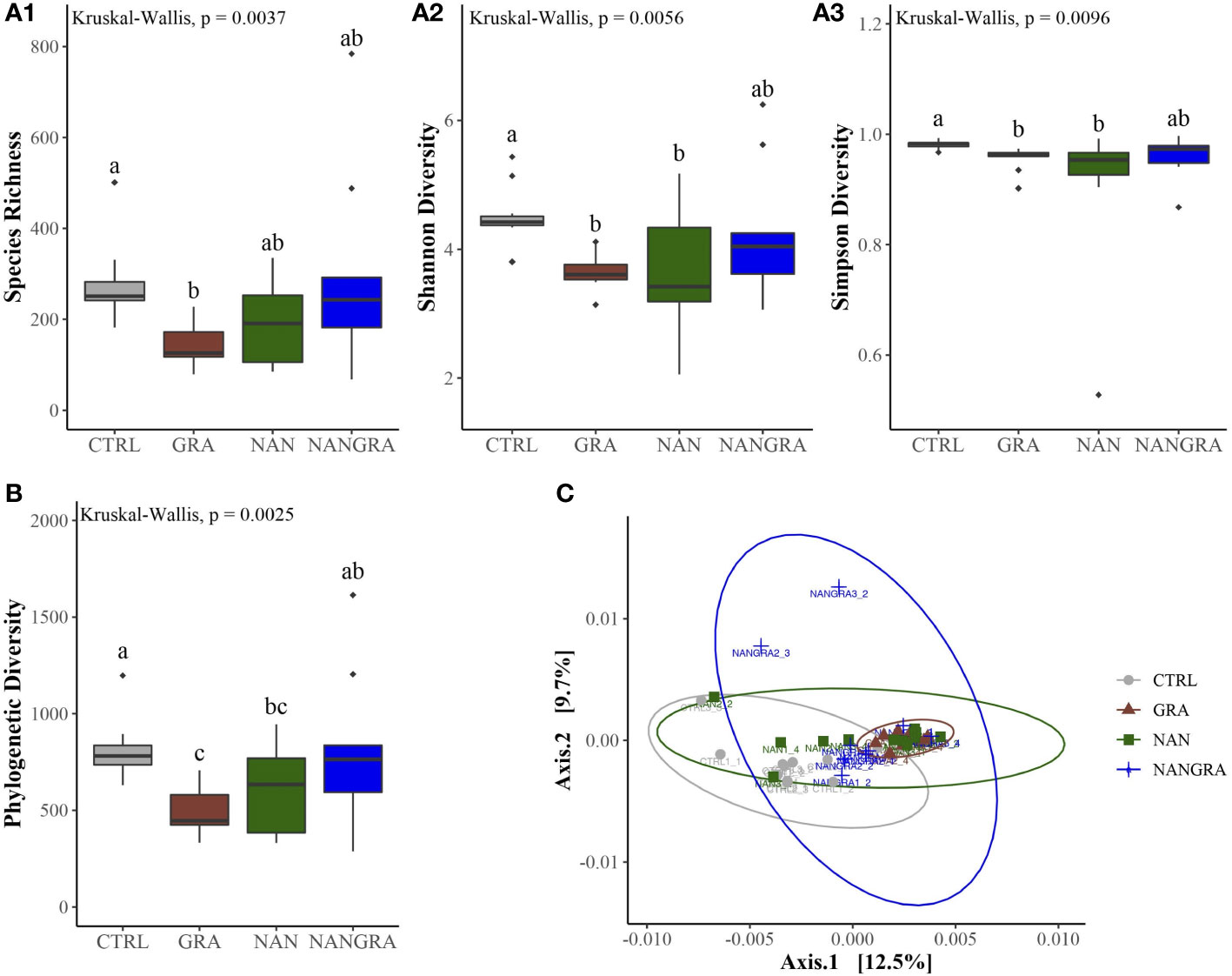
Figure 3 Diversity of the bacterial communities of the posterior intestine of European seabass fed the experimental diets. (A) Alpha diversity: species richness - A1, Shannon diversity - A2, Simpson diversity - A3; (B) Faith’s phylogenetic diversity; (C) Beta diversity visualized by a principial coordinate analysis (PCoA) plot, by employing weighted UniFrac distances. Different letters denote statistically significant differences (P < 0.05) between the dietary treatments (CTRL, control group; GRA, Gracilaria gracilis group; NAN, Nannochloropsis oceanica group; NANGRA, G gracilis and N. oceanica blend group).
Faith’s phylogenetic diversity was also lower in fish fed the GRA (P = 0.002) and NAN (P = 0.04) diets compared to CTRL, but once again not in fish that consumed the blend diet (P = 0.29). Fish fed the GRA diet had also a significantly lower Faith’s phylogenetic diversity compared to the NANGRA fish (P = 0.04) (Figure 3B; Kruskal-Wallis’ chi-squared = 13.323; P = 0.0025). For the beta diversity analysis, we incorporated weighted UniFrac distance metric to understand the dissimilarities of the community structure. The principal coordinates analysis (PCoA) plot indicated that the first two components explained 12.5 and 9.7% of the variance within the dataset. Results from the PERMANOVA test revealed that the bacterial communities of fish fed the GRA diet were significantly different from those of the CTRL fish (P = 0.01) (Figure 3C; F-statistic = 1.932, R2 = 0.129, P = 0.001).
Core bacterial taxa of the intestinal microbiota of European seabass
For the core microbial analysis, a prevalence and detection thresholds of 90 and 0.2%, respectively, were concomitantly applied. Results indicated that the core microbial taxa of the posterior intestine of European seabass were composed of three ASVs belonging to genera Flavobacterium, Parcubacteria and Lactobacillus (Figure 4A and Supplementary Figure 2A). When a less restrictive prevalence threshold was applied (i.e., 80%), 7 ASVs belonging to the genera Flavobacterium (2), Parcubacteria (1), Lactobacillus (2), Cetobacterium (1) and Propionibacterium (1) could be identified as belonging to the common core (Figure 4B and Supplementary Figure 2B).
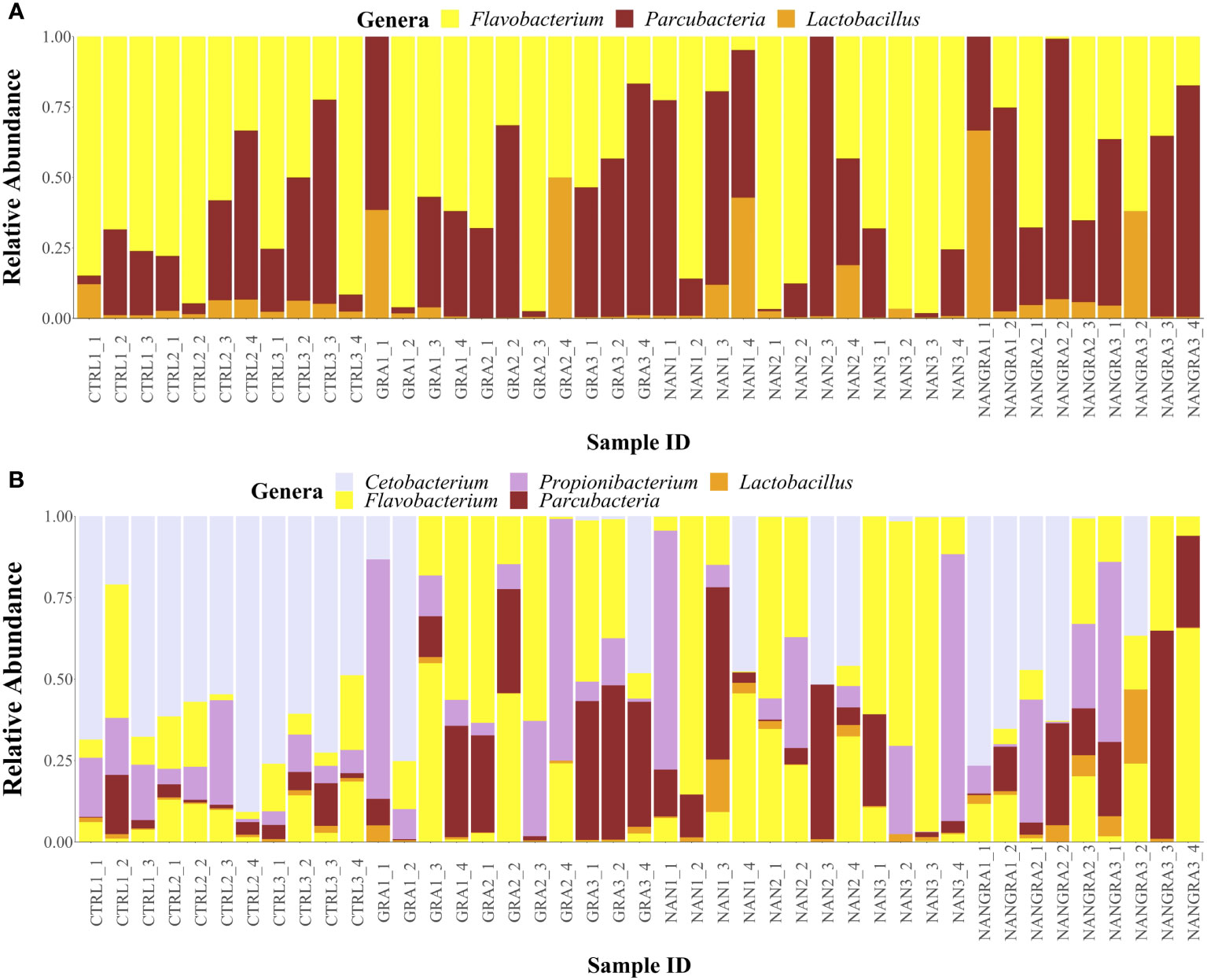
Figure 4 Relative abundance of the core bacterial taxa in the posterior intestine of European seabass fed the experimental diets. (A) Prevalence threshold of 90%; (B) prevalence threshold of 80%. Sample ID: CTRL, control group; GRA, Gracilaria gracilis group; NAN, Nannochloropsis oceanica group; NANGRA, G gracilis and N. oceanica blend group.
Modulation of the bacterial communities by the algae diets
The inclusion of macro- and microalgae, included either singly or in combination, in diets for European seabass led to significant alterations on the abundance of several groups of intestinal bacteria compared to the CTRL-fed fish, as evaluated by DESeq2 (Figure 5). The results indicate that the GRA diet, compared to CTRL, led to an increase (10 to 30-fold changes) in the abundance of Acinetobacter, Corynebacterium, Ilumatobacter, Kordia, Methylobacterium, Polaribacter, Pseudomonas and Sulfitobacter; and a decrease (10 to 30-fold changes) in the abundance of Bacillus, Exiguobacterium, Gp4, Gp6, Nitrospira, Opitutus, Rhizobium, Roseateles and Vampirovibrio (Figure 5A). Concerning the NAN diet, there was an increase in abundance of ASVs of Acinetobacter, Bacillus, Kordia, Pseudomonas, Rothia, Streptococcus and Thermicanus when compared to CTRL (20-fold change); while the abundance of some members of the Acinetobacter, Clostridium sensu strictu, Exiguobacterium, Guardnerella, Muricauda, Ruminobacter, Vampirovibrio and Vibrio was decreased, and the fold changes ranged between -20 and -30 (Figure 5B). The inclusion of the blend (NANGRA diet) led to a proliferation (15 to 25-fold changes) of Acinetobacter, Kordia, Massilia, Parcubacteria and Pseudomonas, and a decreased representation of ASVs of Clostridium sensu strictu, Guardnerella, Muricauda, Nitrospira, Opitutus, Pseudomonas and Tepidimonas (-15 to -25-fold changes) (Figure 5C).
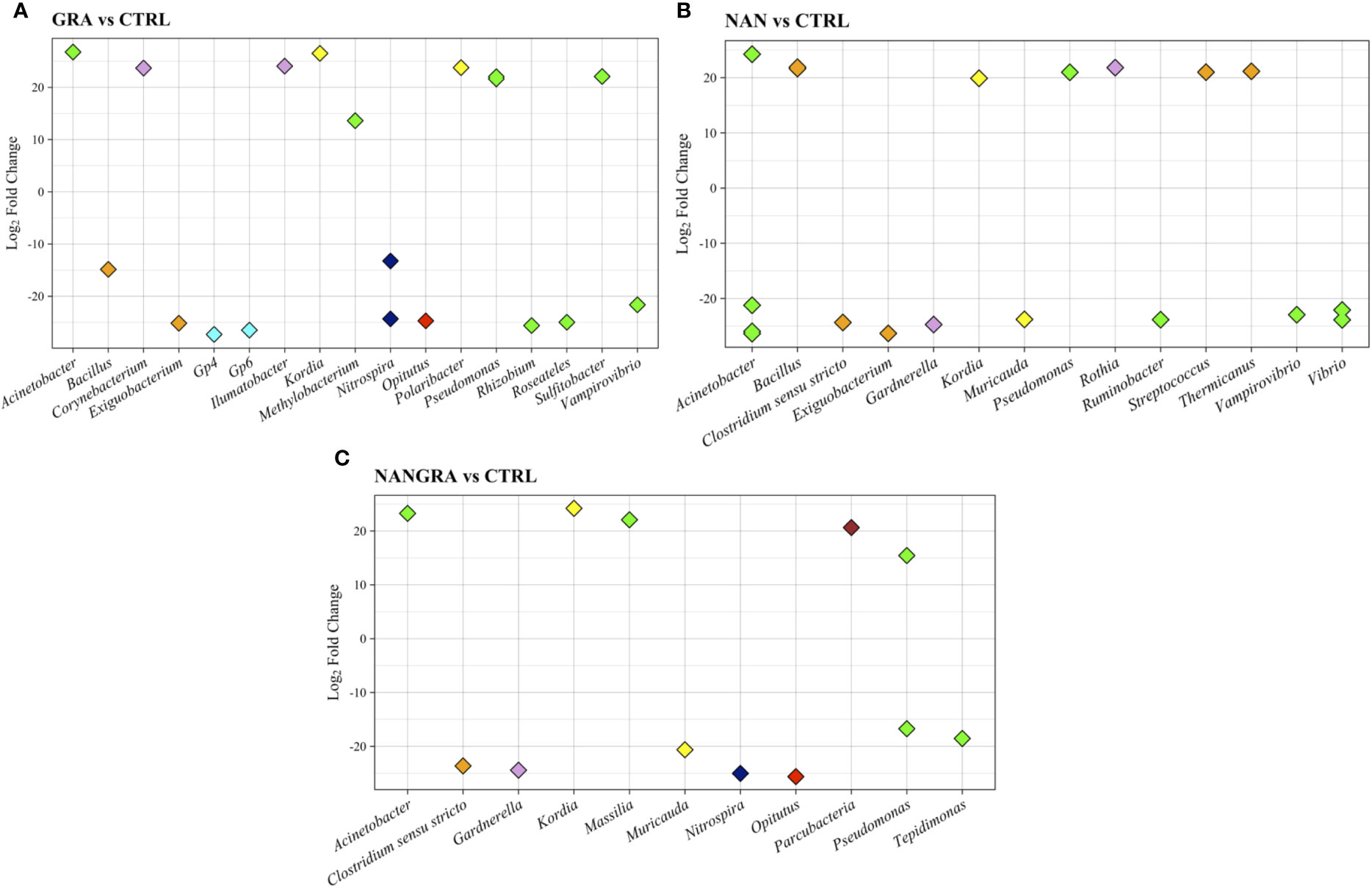
Figure 5 Differently abundant genera in the study groups. (A) GRA group vs CTRL group. (B) NAN group vs CTRL group. (C) NANGRA group vs CTRL group. Color codes: green – Proteobacteria (Acinetobacter, Methylobacterium, Pseudomonas, Rhizobium, Roseateles, Sulfitobacter, Vampirovibrio, Ruminobacter, Vibrio, Massilia and Tepidimonas); orange – Firmicutes (Bacillus, Exiguobacterium, Clostridium sensu stricto); pink – Actinobacteria (Corynebacterium, Ilumatobacter, Gardnerella and Rothia); light blue – Acidobacteria (Gp4 and Gp6); yellow – Bacteroidetes (Kordia and Muricauda); dark blue – Nitrospirae (Nitrospira); light red – Verrucomicrobia (Opitutus); dark red – Parcubacteria.
Phylogenetic analysis of the ASVs identified as core and those differently modulated by the algae diets
To understand the phylogenetic relationship of certain bacteria which are known to be beneficial and/or pathogenic we first extracted the sequences of the ASVs and performed a blast in NCBI. Thereafter, a phylogenetic tree was constructed to illustrate the genetic relationship, which indirectly indicates functional diversity if there are traits that are retained through evolution.
The phylogenetic tree that includes the ASVs identified as part of the core microbiome in the current study is presented in Figure 6A. The ASVs identified as Lactobacillus (ASVs 1679 and 1313) were found to be related to strains of Lactobacillus delbrueckii subsp. lactis, Lactobacillus acidophilus, Lactobacillus helveticus and Lactiplantibacillus plantarum. The ASV10 Cetobacterium clustered with several strains of Cetobacterium somerae, while the ASV16 Propionibacterium clustered with strains of Cutibacterium acnes. The ASVs 11 and 22, identified as Flavobacterium, are likely related to a strain of Flavobacterium succinicans, whereas the ASV19 Parcubacteria clustered with the uncultured candidate division OD1 bacterium.
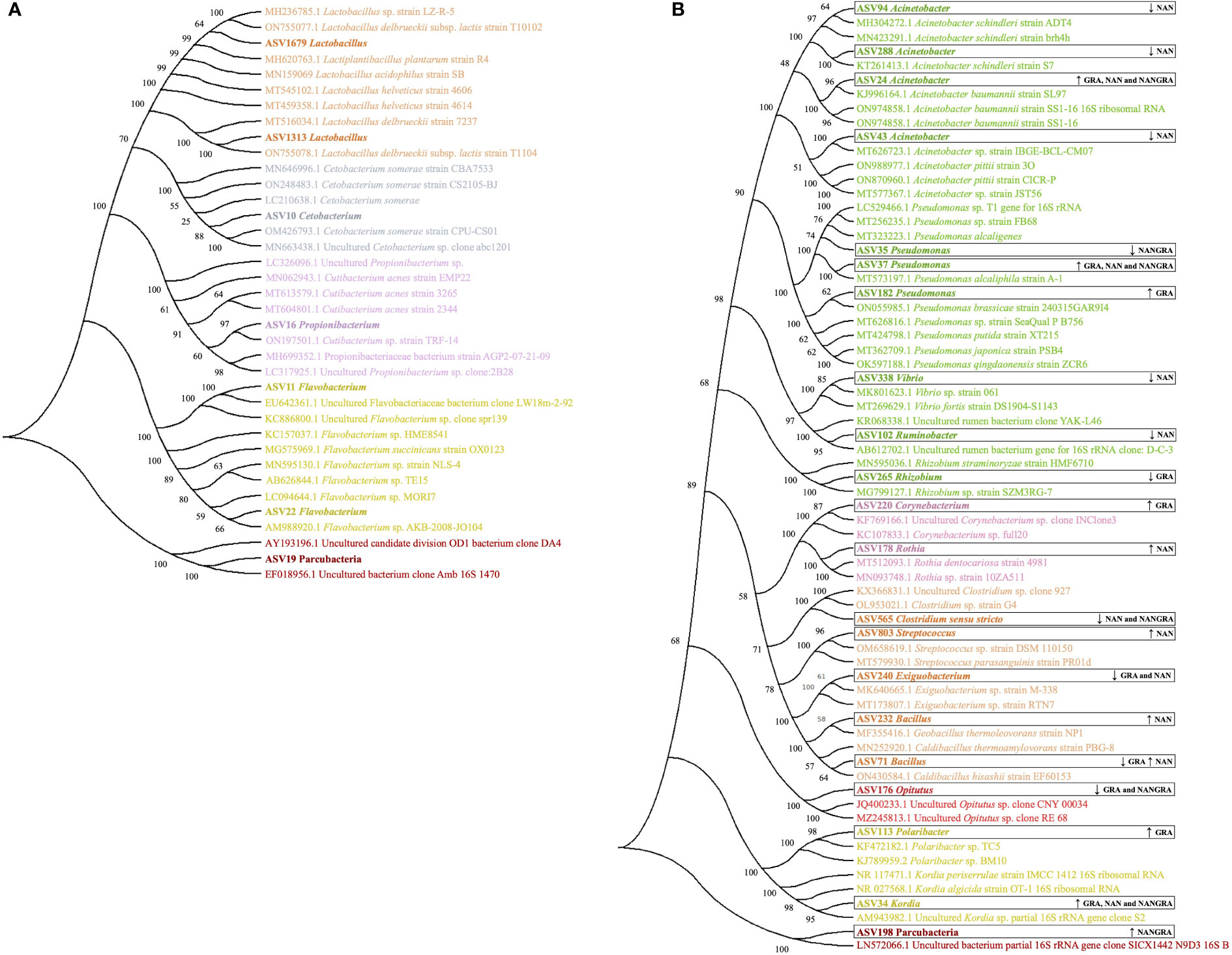
Figure 6 Phylogenetic trees generated for the ASVs of interest identified in the study. (A) ASVs identified as part of the core microbiome. (B) ASVs identified as differently abundant in the study groups. ↑ – increase in abundance; ↓ – decrease in abundance; GRA, Gracilaria gracilis group; NAN, Nannochloropsis oceanica group; NANGRA, G gracilis and N. oceanica blend group.
The phylogenetic relationships of another set of ASVs − i.e., those that were significantly modulated by the dietary treatments − with the highly similar sequences from NCBI are presented in Figure 6B. The Acinetobacter ASVs 94 and 288 clustered with strains of Acinetobacter schindleri; ASV24 appeared in a cluster with strains of Acinetobacter baumannii; and ASV43 clustered with strains of Acinetobacter pittii. The ASVs identified as Pseudomonas clustered together with species of this genus; Pseudomonas alcaligenes (ASV35), Pseudomonas alcaliphila (ASV37), Pseudomonas brassicae, Pseudomonas putida, Pseudomonas japonica and Pseudomonas qingdaonensis (ASV182). Vibrio ASV338 clustered with a strain of Vibrio fortis; Ruminobacter (ASV102) appeared in a cluster with uncultured rumen bacteria; and Rhizobium (ASV265) clustered with a strain of Rhizobium straminoryzae. Rothia (ASV178) is likely related to the strain of Rothia dentocariosa; Streptococcus (ASV803) clustered with a strain of Streptococcus parasanguinis; and the Bacillus ASVs clustered with strains of Geobacillus thermoleovorans and Caldibacillus thermoamylovorans (ASV232) and a strain of Caldibacillus hisashii (ASV71). ASV34, annotated as Kordia, was phylogenetically close to strains of Kordia periserrulae and Kordia algicida. Corynebacterium (ASV220), Clostridium sensu stricto (ASV565), Exiguobacterium (ASV240), Opitutus (ASV176), Polaribacter (ASV113), and Parcubacteria (ASV198) clustered with strains of bacteria belonging to the same genera.
Discussion
The gut microbiota of farmed fish has recently become one of the most studied topics in aquaculture research due to the recognized importance of the intestinal bacterial communities to fish health and physiology (Egerton et al., 2018). Although nutrients are known to play a crucial role in modulating the intestine microbiota and algae have been extensively studied in recent years as novel feed ingredients and additives for farmed fish (Hua et al., 2019), the knowledge about the impact of macro- and microalgae on the composition of the intestinal bacteria of fish is still limited (Cerezuela et al., 2012; Rico et al., 2016; Jorge et al., 2019; Keating et al., 2021; Sagaram et al., 2021; Cerezo et al., 2022). In the present study we evaluated the ability of the macroalga G. gracilis and the microalga N. oceanica, and a mix of these two algae, to modulate the gut bacterial profile of European seabass, a widely farmed fish species in Europe.
Gut bacterial community composition and diversity in European seabass is affected by the algae diets
The dominant bacterial phyla found in the posterior intestine of European seabass were Proteobacteria, Firmicutes, Actinobacteria, Bacteroidetes, Fusobacteria, Nitrospirae and Parcubacteria. Our findings are in line with previous studies on this fish species that have also reported the dominance of Proteobacteria, Firmicutes, Actinobacteria, Bacteroidetes and Fusobacteria in the gut (Pérez-Pascual et al., 2020; Serra et al., 2021; Liu et al., 2022). The phylum Nitrospirae has a relatively high representation in our samples from the CTRL group (10.21%), and although it has been previously reported as part of the intestinal microbiota of farmed fish, it was not found among the most abundant phyla in other fish microbiome studies (Li et al., 2019; Serra et al., 2021). As sulfate reducing bacteria, Nitrospirae are usually associated with the water and biofilters of RAS (Schmidt et al., 2016; Minich et al., 2020; Fossmark et al., 2021). Therefore, the presence in the intestine of cultured fish is most likely the result of a transfer event, as also reported by Minich et al. (2020) in Atlantic salmon reared in RAS systems. We did not observe a clear dominance of certain genera across all experimental samples, but Acinetobacter, Nitrospira, Pseudoalteromonas and Streptococcus were among the most abundant genera. The genera Acinetobacter and Streptococcus spp. have strains with probiotic effects (Swain et al., 2009; Bunnoy et al., 2019), but some species are opportunistic bacterial pathogens that cause diseases in immunocompromised fish or hosts that encounter environmental stressors (Dawood, 2020). On the other hand, Pseudoalteromonas species can provide protection against harmful bacteria present in the gut microbiota, namely Vibrio (Richards et al., 2017; Rimoldi et al., 2020). The relatively high abundance of both potentially pathogenic and beneficial bacteria found in the fish intestine denotes the importance of the commensal microbiota in the maintenance of host health, through a tight regulation of competing microorganisms.
The bacterial alpha diversity analysis revealed that the 8% inclusion of G. gracilis or N. oceanica reduced the species richness (significant only for G. gracilis), evenness of the microbial populations and the effective number of dominant species, compared to the CTRL group. Similarly, Faith’s phylogenetic diversity, that is a measure of biodiversity based on phylogeny, was also significantly lower in single algae groups. The gut microbial community of European seabass that consumed the seaweed supplemented diet (GRA) also differed significantly compared to the CTRL-fed fish. Although a decreased bacterial diversity may not always imply an unstable community, a reduction in the microbial diversity and changes in the bacterial profiles in the intestine have been associated with unhealthy fish (Li et al., 2016). Interestingly, inclusion of the algae blend eliminated these negative effects on the intestinal bacterial diversity, most probably due to the lower inclusion (4%) of each alga. In a complementary study (Batista et al., 2020), we evaluated the effects of G. gracilis and N. oceanica on growth performance, nutrient digestibility and intestinal histomorphology; a lower protein digestibility was observed for fish fed the macroalga supplemented diet, but without differences in terms of growth. The fish on this diet had the lowest number of intestinal neutral goblet cells associated with the digestive and absorptive processes. Similar to the ability bestowed by the microalga in the G. gracilis-N. oceanica blend to regain the diversity and structure of the intestinal microbiota, our previous study also indicated the positive association of the microalga with the digestion process (Batista et al., 2020). Together, these results may indicate that inclusion of different micro- and macroalgae in a blend is likely a good strategy to lessen the negative impacts of added-value natural compounds from macroalgae species.
Core microbiome of European seabass
The core microbiota (autochthonous microbiome) is composed of resident microorganisms that colonize the intestinal mucosa (Egerton et al., 2018). In the present study, three and five genera were identified as part of the core microbiota at 90 and 80% prevalence thresholds, respectively. The core bacteria were Flavobacterium, Lactobacillus, Parcubacteria, Cetobacterium and Propionibacterium. Although part of normal fish microbiota, the genus Flavobacterium is well recognized for their opportunistic nature in fish (Derome et al., 2016). Some Flavobacterium spp. are responsible for systemic infections that are difficult to control and prevent, and hence, are associated with devastating economic losses that affect the worldwide fish production (Loch and Faisal, 2015). In the present study, the Flavobacterium ASVs identified as part of the core microbiome showed a high similarity with a potentially pathogenic bacteria, F. succinicans, the likely agent of bacterial gill disease in rainbow trout cultivated in RAS (Good et al., 2015). Interestingly, the genus Lactobacillus, belonging to the lactic acid bacteria group, was also identified as part of the core microbiota of European seabass in our study. Lactobacillus, well recognized as part of the beneficial gut bacteria, provides protective effects against bacterial infections (He et al., 2017; Dawood, 2020). The core ASVs belonging to Lactobacillus clustered with L. delbrueckii subsp. lactis, L. acidophilus, L. helveticus, and Lactiplantibacillus plantarum, that were reported to have in vitro and in vivo probiotic activity (Rurangwa et al., 2009; Hosseini et al., 2016; Ahire et al., 2019; Iorizzo et al., 2022).
ASVs assigned to Parcubacteria were detected among both core microbiome and the top ASVs in our samples (between 1.3 and 5.6% average relative abundance). Genes encoding for amylases and capacities for degrading cellulose and mannose were associated with Parcubacteria isolated from sporadic permafrost zone of subarctic Quebec, and this bacteria has the capacity to produce acetate as the major end product of its metabolism (Vigneron et al., 2019). Nonetheless, the role of Parcubacteria as part as the gut microbiota of European seabass remains unclear and further studies should delve into the functionality of this bacteria.
As part of the commensal microbiota, Cetobacterium and Propionibacterium are also often described as beneficial bacteria that are commonly found in healthy fish. Here, the ASVs identified as core and belonging to Cetobacterium and Propionibacterium were closely related to the strains of C. somerae and Cutibacterium acnes, respectively. Cetobacterium is reported as protease and vitamin B12 producing bacteria (Legrand et al., 2019), and previous studies have demonstrated the ability of the fermentation products of C. somerae to improve gut health of different farmed fishes (Xie et al., 2021; Xie et al., 2022; Zhou et al., 2022). The genus Propionibacterium - producer of propionate, linolenic acid, vitamins, and antimicrobials - is known to enhance host robustness by boosting the immune response and promoting the growth of other probiotic bacteria (Zárate, 2012; Boutin et al., 2013). In gilthead seabream Propionibacterium has also been reported as an important genus of the core intestinal microbiota, producing metabolites with an important role in gut health (Piazzon et al., 2017; Piazzon et al., 2019).
The presence of taxa, namely Lactobacillus and Cetobacterium, with probiotic potential among the highly prevalent taxa in majority of the studied population indicate that these bacteria play a key role in the maintenance of biological functions, health, and disease resistance in European seabass.
Algae, singly or in combination, modulate both beneficial and potentially pathogenic groups of bacteria
The intestinal microbiota of farmed fish is a complex assemblage of microorganisms that interact and compete for nutrient acquisition and space. Both beneficial and potentially pathogenic groups of bacteria coexist in the gut, with diet playing a leading role in modulating these bacterial communities (Tarnecki et al., 2017; Dawood, 2020). In the present study, macro- and microalgae feeds affected the abundance of several groups of bacteria present in the posterior intestine of seabass when compared to fish fed the CTRL diet.
The inclusion of G. gracilis and N. oceanica, fed both singly and blended, led to a significant increase in the representation of ASVs belonging to potentially pathogenic bacteria. Notably there was an increased representation of an ASV that clustered with strains of Acinetobacter baumannii; a strain of this bacterium was previously recovered from diseased channel catfish (Ictalurus punctatus) (Xia et al., 2008). The GRA diet led to an increase in a Pseudomonas ASV that clustered together with P. putida, a bacteria known to cause disease in both humans and fish (Mao et al., 2013). Rothia, that proliferated in the NAN group, has only been reported as an opportunistic pathogen in humans (Fatahi-Bafghi, 2021). As fish may act as a vehicle for the transmission of drug-resistant human pathogens, it is of crucial importance to understand the impact of novel formulations on the fish microbial communities. Other groups of potentially pathogenic bacteria that were significantly increased only in the gut of GRA-fed fish included Corynebacterium (Tarnecki et al., 2018) and Polaribacter, a potential RAS-associated pathogen (Rud et al., 2017). It is noteworthy that some Polaribacter species are associated with important antioxidant functions (Sehnal et al., 2021). A pro-oxidative or bacterial challenge could give further insights into the impact of these potentially pathogenic genera on fish health.
The Gracilaria diets (GRA and NANGRA) were associated with a significant decrease of the genus Nitrospira. During the 15-weeks feeding trial, all abiotic conditions, including the nitrogenous compounds in the RAS system, were carefully monitored, and optimum water quality parameters were maintained in the rearing system of the fish. Therefore, the lower proportion of the genus Nitrospira (the phylum Nitrospirae) found in macroalgae groups is most likely a response to the dietary treatment. Suo et al. (2017) studied the impact of sulfide exposure on gut microbiota of Pacific white shrimp (Litopenaeus vannamei) and reported that although present in the intestine of non-exposed shrimp, Nitrospirae could not be found in sulfide-exposed groups. The authors argued that the disappearance of this bacterial phylum upon sulfide exposure could make shrimp more susceptible to nitrite toxicity. Nitrospira spp. are nitrite-oxidizing bacteria that convert nitrite into the less toxic nitrate (Philips et al., 2002). Therefore, the reduction of Nitrospira in intestine of European seabass fed G. gracilis diets might negatively impact the ability of the fish to cope with a nitrite exposure situation. Nitrite uptake in marine fish is thought to occur through the gills and intestine, and although it is less toxic for marine fish, previous studies have found negative impacts of elevated nitrite levels on growth (Ciji and Akhtar, 2019).
The algae-rich diets modulated certain groups of bacteria involved in nutrient degradation and utilization. The genus Opitutus, that are cellulose degraders commonly found on surfaces of green periphytic algae (Knack et al., 2015), was less abundant in seaweed-rich diets. Likewise, a decreased abundance of bacteria belonging to the genus Rhizobium that are known to have cellulolytic and pectolytic activity (Xia et al., 2018), was observed in the GRA group. Macroalgae, much like plants, have cellulose-based walls (Kumar et al., 2013). Therefore, these bacterial groups may aid in the degradation of the algae and plant products present in the experimental diets, and a decrease in its abundance may partially explain the lower nutrient digestibility associated with the Gracilaria diets, as reported by Batista et al. (2020). Indeed, the authors attributed the reduced protein and energy digestibility in fish fed the GRA and NANGRA diets to the presence of indigestible fibers in the macroalga cell wall that compromised the action of the digestive enzymes. The involvement of the gut microbiota in the digestive/absorptive processes in fish has been previously recognized (Butt and Volkoff, 2019), and the above-mentioned results further support these interactions. Although the genera Ruminobacter, described as starch utilizer (Darabighane et al., 2021), and Clostridium sensu stricto, previously characterized as a carbohydrate degrader (Abdelhamed et al., 2019), had a lower abundance in the intestine of fish fed the Nannochloropsis diet, in our associated study (Batista et al., 2020) we did not find a difference in nutrient or energy digestibility in the NAN group compared to the CTRL group. Moreover, fish fed the algae supplemented diets had an increased representation of an ASV that was annotated as Kordia, that is likely closely related to K. periserrulae and K. algicida. Members of K. algicida can exhibit algicidal activity and produce extracellular proteases responsible for the cell lysis of diatoms in a species-specific way (Demuez et al., 2015). Therefore, a higher abundance of Kordia in the intestine of European seabass may aid in the digestion of microalgae enriched diets. On the other hand, the genus Exiguobacterium was found to be less abundant in not only the NAN-fed fish but also the GRA-fed fish. A previous study on zebrafish has found that Exiguobacterium sp. play a role in lipid droplet formation in enterocytes to positively impact the fatty acid absorption (Semova et al., 2012). Further studies on the function of this bacterial genus on fish gut microbiota are required, but the reduced abundance of Exiguobacterium in single alga diets may indicate an impact of these algae on the metabolism of dietary fat. Nonetheless, there were no differences between the lipid digestibility and whole-body and muscle fat contents of fish that consumed algae-rich diets and those fed the CTRL diet (Batista et al., 2020).
A modulation of both beneficial (i.e., increase) and potentially pathogenic (i.e., decrease) bacteria was observed in the intestine of European seabass fed the macro- or microalgae diets. Inclusion of seaweed G. gracilis in feeds led to a higher abundance of Sulfitobacter and Methylobacterium (producer of poly-β-hydroxybutyrate that degrades short-chain fatty acids), two genera that comprise bacteria capable of inhibiting the growth of fish pathogens (Boutin et al., 2013; Wilczynski et al., 2022). Similarly, the N. oceanica feeds led to an increase in beneficial bacteria such as Bacillus in fish intestine. Spore-forming species such as Bacillus spp. are probiotics with wide application in the aquaculture industry. The Bacillus ASVs that were enriched in the gut of NAN-fed fish clustered together in the phylogenetic tree with bacterial strains such as Geobacillus thermoleovorans, Caldibacillus thermoamylovorans and C. hisashii that exhibit antimicrobial and probiotic activities (Inabu et al., 2022; Zebrowska et al., 2022). Spore-forming bacterial species belonging to the genus Bacillus have been previously isolated from the intestine of European seabass, with some isolates presenting carbohydrase activity and probiotic potential (Serra et al., 2019). From an industrial perspective, it would be interesting to further explore the ability of the microalga N. oceanica to enrich the gut of European seabass with Bacillus spp. with promising probiotic activity. Although limited information on the topic is available, some studies have reported that species belonging to the genus Acinetobacter, namely Acinetobacter lwoffii, Acinetobacter junii and A. pittii are emerging fish pathogens and their virulence against fish of the genus Schizothorax was also revealed previously (Cao et al., 2018; Malick et al., 2020). Interestingly, the NAN diet led to a decrease in several Acinetobacter ASVs, including an ASV that likely has a close phylogenetic relationship with the pathogen A. pittii. Although the genus Pseudomonas is often reported as part of the normal microbiota and certain bacteria belonging to this taxa are probiotic strains (Qi et al., 2020), there are also opportunistic fish pathogens among them that are responsible for disease outbreaks and high mortality in farms (Oh et al., 2019). NANGRA-fed fish presented a reduced abundance of an ASV belonging to Pseudomonas that is probably related to P. alcaligenes, a rare but potentially opportunistic fish pathogen (Bai et al., 2021). This microalga also decreased the abundance of the genus Vibrio, one of the most important fish pathogens responsible for devastating economic losses in fish farms. In the present study, the ASV of the genus Vibrio clustered with a strain of Vibrio fortis, a bacterium with reported pathogenicity against rainbow trout (Austin et al., 2005), and that has been found in association with enteritis in cultured seahorses (Hippocampus erectus) (Wang et al., 2016). In vitro studies have recently demonstrated the antimicrobial activity of G. gracilis and N. oceanica against some Vibrio species, namely V. harveyi and V. parahaemolyticus (Ferreira et al., 2021). Disease outbreaks caused by Vibrio are bottlenecks for the long-term sustainability of the aquaculture sector. In this context, microbiota modulation using added-value products such as those employed in this study may be a promising strategy to improve disease resistance in fishes.
Conclusion
In the present study it was demonstrated for the first time the potential of the seaweed G. gracilis and the microalga N. oceanica, incorporated singly or blended in European seabass diets, to modulate the intestinal microbiota of the fish. It was observed that 8% inclusion of the two algae (singly) led to a reduction of the gut microbial diversity, which is often associated with a negative impact. Nonetheless, inclusion of a lower percentage of each alga in a blend (4% each) was able to mitigate these plausible effects on the intestinal bacterial communities. The core microbiome of European seabass was composed of both beneficial (Lactobacillus and Cetobacterium) and potentially pathogenic (Flavobacterium) bacteria, which might suggest that the host gut homeostasis and disease resistance is dependent on a tight interaction between competing microorganisms. The algae-rich feeds modulated some groups of bacteria that are known carbohydrate degraders, with the Gracilaria-fed fish showing a decreased abundance of the genera Opitutus and Rhizobium, which may partially explain the lower nutrient digestibility observed in fish that consumed the macroalgae diet. Gracilaria promoted the growth of bacteria capable of outcompeting fish pathogens (Sulfitobacter and Methylobacterium). Nannochloropsis, on the other hand, led to a higher representation of Bacillus bacteria, widely recognized for their probiotic potential, and a decreased abundance of the potentially pathogenic bacteria belonging to the genus Acinetobacter. Nannochloropsis-fed fish also presented a lower abundance of ASVs identified as Vibrio, a highly pathogenic bacteria that affect the culture of several farmed aquatic animals of high economic value. Such results are indicative of the potential of algae to modulate the bacterial communities present in the intestine of European seabass, with possible implications to host health, nutrition, and disease resistance.
Data availability statement
The datasets presented in this study are deposited in the NCBI repository (https://www.ncbi.nlm.nih.gov), accession number PRJNA867546.
Ethics statement
This study was reviewed and approved by Review of the ethical process concerning all animal handling and sampling procedures performed by CIIMAR (Centro Interdisciplinar de Investigação Marinha e Ambiental, Matosinhos, Portugal) animal welfare body (ORBEA-CIIMAR) and approved by national competent authorities.
Author contributions
LV and VK were responsible for funding acquisition and the conceptualization of the study. MF and YA performed the formal analysis and investigation. HA and JS provided the resources for the study. MF wrote the original draft under the supervision of LV and VK, who reviewed and edited it. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by the structured R&D&I project ATLANTIDA (NORTE-01-0145-FEDER-000040), supported by the North Portugal Regional Operational Programme (NORTE2020), under the PORTUGAL 2020 Partnership Agreement and through the European Regional Development Fund (ERDF). MF acknowledges Fundação para a Ciência e a Tecnologia (FCT) for Grant SFRH/BD/144843/2019 (FCT/FSE). Financial support to CIIMAR within the scope of UIDB/04423/2020 and UIDP/04423/2020 from FCT is also acknowledged.
Acknowledgments
The support received from Bisa Saraswathy, Researcher, Nord University, for data analyses and preparation of the manuscript is acknowledged.
Conflict of interest
HA and JS were employed by the companies ALGAplus and Allmicroalgae, respectively.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.1001942/full#supplementary-material
References
Abdelhamed H., Ozdemir O., Waldbieser G., Perkins A., Lawrence M., Karsi A. (2019). Effects of florfenicol feeding on diversity and composition of the intestinal microbiota of channel catfish (Ictalurus punctatus). Aquac. Res. 50, 3663–3672. doi: 10.1111/are.14325
Ahire J. J., Mokashe N. U., Chaudhari B. L. (2019). Effect of dietary probiotic Lactobacillus helveticus on growth performance, antioxidant levels, and absorption of essential trace elements in goldfish (Carassius auratus). Probiot. Antimicrob. Proteins 11, 559–568. doi: 10.1007/s12602-018-9428-5
Albanese D., Fontana P., De Filippo C., Cavalieri D., Donati C. (2015). MICCA: a complete and accurate software for taxonomic profiling of metagenomic data. Sci. Rep. 5, 9743. doi: 10.1038/srep09743
Anderson M. (2001). A new method for non-parametric multivariate analysis of variance. Austral Ecol. 26, 32–46. doi: 10.1111/j.1442-9993.2001.01070.pp.x
Austin B., Austin D., Sutherland R., Thompson F., Swings J. (2005). Pathogenicity of vibrios to rainbow trout (Oncorhynchus mykiss, Walbaum) and Artemia nauplii. environ. Microbiol. 7, 1488–1495. doi: 10.1111/j.1462-2920.2005.00847.x
Bai J., Huo Y., Hu X., Lü A., Sun J. (2021). Characterization of pathogenic Pseudomonas alcaligenes isolated from koi carp in China. J. Aquat. Anim. Health 33, 243–251. doi: 10.1002/aah.10141
Batista S., Pereira R., Oliveira B., Baião L. F., Jessen F., Tulli F., et al. (2020). Exploring the potential of seaweed Gracilaria gracilis and microalga Nannochloropsis oceanica, single or blended, as natural dietary ingredients for European seabass Dicentrarchus labrax. J. Appl. Phycol. 32, 2041–2059. doi: 10.1007/s10811-020-02118-z
Boutin S., Bernatchez L., Audet C., Derôme N. (2013). Network analysis highlights complex interactions between pathogen, host and commensal microbiota. PloS One 8, e84772. doi: 10.1371/journal.pone.0084772
Bunnoy A., Na-Nakorn U., Srisapoome P. (2019). Probiotic effects of a novel strain, Acinetobacter KU011TH, on the growth performance, immune responses, and resistance against Aeromonas hydrophila of bighead catfish (Clarias macrocephalus Günther 1864). Microorganisms 7, 613. doi: 10.3390/microorganisms7120613
Butt R. L., Volkoff H. (2019). Gut microbiota and energy homeostasis in fish. Front. Endocrinol. 10. doi: 10.3389/fendo.2019.00009
Cao S., Geng Y., Yu Z., Deng L., Gan W., Wang K., et al. (2018). Acinetobacter lwoffii, an emerging pathogen for fish in Schizothorax genus in China. Transbound Emerg. Dis. 65, 1816–1822. doi: 10.1111/tbed.12957
Cerezo I. M., Fumanal M., Tapia-Paniagua S. T., Bautista R., Anguís V., Fernández-Díaz C., et al. (2022). Solea senegalensis bacterial intestinal microbiota is affected by low dietary inclusion of Ulva ohnoi. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.801744
Cerezuela R., Fumanal M., Tapia-Paniagua S. T., Meseguer J., Moriñigo M.Á., Esteban M.Á. (2012). Histological alterations and microbial ecology of the intestine in gilthead seabream (Sparus aurata l.) fed dietary probiotics and microalgae. Cell Tissue Res. 350, 477–489. doi: 10.1007/s00441-012-1495-4
Cerf-Bensussan N., Gaboriau-Routhiau V. (2010). The immune system and the gut microbiota: friends or foes? Nat. Rev. Immunol. 10, 735–744. doi: 10.1038/nri2850
Chen Y., Ye W., Zhang Y., Xu Y. (2015). High speed BLASTN: an accelerated MegaBLAST search tool. Nucleic Acids Res. 43, 7762–7768. doi: 10.1093/nar/gkv784
Ciji A., Akhtar M. S. (2019). Nitrite implications and its management strategies in aquaculture: a review. Rev. Aquac. 12, 878–908. doi: 10.1111/raq.12354
Darabighane B., Tapio I., Ventto L., Kairenius P., Stefański T., Leskinen H., et al. (2021). Effects of starch level and a mixture of sunflower and fish oils on nutrient intake and digestibility, rumen fermentation, and ruminal methane emissions in dairy cows. Animals 11, 1310. doi: 10.3390/ani11051310
Dawood M. A. O. (2020). Nutritional immunity of fish intestines: important insights for sustainable aquaculture. Rev. Aquac. 13, 642–663. doi: 10.1111/raq.12492
Demuez M., González-Fernández C., Ballesteros M. (2015). Algicidal microorganisms and secreted algicides: New tools to induce microalgal cell disruption. Biotechnol. Adv. 33, 1615–1625. doi: 10.1016/j.biotechadv.2015.08.003
Derome N., Gauthier J., Boutin S., Llewellyn M. (2016). “Bacterial opportunistic pathogens of fish,” in The rasputin effect: when commensals and symbionts become parasitic. Ed. Hurst C. J. (Cham: Springer International Publishing), 81–108. Available at: 10.1007/978-3-319-28170-4_4.
DeSantis T. Z. Jr., Hugenholtz P., Keller K., Brodie E. L., Larsen N., Piceno Y. M., et al. (2006). NAST: a multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res. 34, W394–W399. doi: 10.1093/nar/gkl244
Diwan A. D., Harke S. N., Gopalkrishna, Panche A. N. (2022). Aquaculture industry prospective from gut microbiome of fish and shellfish: An overview. J. Anim. Physiol. Anim. Nutr. 106, 441–469. doi: 10.1111/jpn.13619
Edgar R. C. (2016). UNOISE2: improved error-correction for illumina 16S and ITS amplicon sequencing. bioRxiv, 081257. doi: 10.1101/081257
Edgar R. C., Flyvbjerg H. (2015). Error filtering, pair assembly and error correction for next-generation sequencing reads. Bioinformatics 31, 3476–3482. doi: 10.1093/bioinformatics/btv401
Egerton S., Culloty S., Whooley J., Stanton C., Ross R. P. (2018). The gut microbiota of marine fish. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.00873
Fatahi-Bafghi M. (2021). Characterization of the Rothia spp. and their role in human clinical infections. Infect. Genet. Evol. 93, 104877. doi: 10.1016/j.meegid.2021.104877
Ferreira M., Teixeira C., Abreu H., Silva J., Costas B., Kiron V., et al. (2021). Nutritional value, antimicrobial and antioxidant activities of micro- and macroalgae, single or blended, unravel their potential use for aquafeeds. J. Appl. Phycol. 33, 3507–3518. doi: 10.1007/s10811-021-02549-2
Fossmark R. O., Attramadal K. J. K., Nordøy K., Østerhus S. W., Vadstein O. (2021). A comparison of two seawater adaptation strategies for Atlantic salmon post-smolt (Salmo salar) grown in recirculating aquaculture systems (RAS): Nitrification, water and gut microbiota, and performance of fish. Aquaculture 532, 735973. doi: 10.1016/j.aquaculture.2020.735973
Good C., Davidson J., Wiens G. D., Welch T. J., Summerfelt S. (2015). Flavobacterium branchiophilum and F. succinicans associated with bacterial gill disease in rainbow trout Oncorhynchus mykiss (Walbaum) in water recirculation aquaculture systems. J. Fish Dis. 38, 409–413. doi: 10.1111/jfd.12249
He S., Ran C., Qin C., Li S., Zhang H., de Vos W. M., et al. (2017). Anti-infective effect of adhesive probiotic Lactobacillus in fish is correlated with their spatial distribution in the intestinal tissue. Sci. Rep. 7, 13195. doi: 10.1038/s41598-017-13466-1
Hosseini M., Kolangi Miandare H., Hoseinifar S. H., Yarahmadi P. (2016). Dietary Lactobacillus acidophilus modulated skin mucus protein profile, immune and appetite genes expression in gold fish (Carassius auratus gibelio). Fish Shellfish Immunol. 59, 149–154. doi: 10.1016/j.fsi.2016.10.026
Hsieh T. C., Ma K., Chao A. (2016). iNEXT: An r package for rarefaction and extrapolation of species diversity (Hill numbers). Methods Ecol. Evol. 7, 1451–1456. doi: 10.1111/2041-210X.12613
Hua K., Cobcroft J. M., Cole A., Condon K., Jerry D. R., Mangott A., et al. (2019). The future of aquatic protein: implications for protein sources in aquaculture diets. One Earth 1, 316–329. doi: 10.1016/j.oneear.2019.10.018
Inabu Y., Taguchi Y., Miyamoto H., Etoh T., Shiotsuka Y., Fujino R., et al. (2022). Development of a novel feeding method for Japanese black calves with thermophile probiotics at postweaning. J. Appl. Microbiol. 132, 3870–3882. doi: 10.1111/jam.15519
Iorizzo M., Albanese G., Letizia F., Testa B., Tremonte P., Vergalito F., et al. (2022). Probiotic potentiality from versatile Lactiplantibacillus plantarum strains as resource to enhance freshwater fish health. Microorganisms 10, 463. doi: 10.3390/microorganisms10020463
Jorge S. S., Enes P., Serra C. R., Castro C., Iglesias P., Oliva Teles A., et al. (2019). Short-term supplementation of gilthead seabream (Sparus aurata) diets with Nannochloropsis gaditana modulates intestinal microbiota without affecting intestinal morphology and function. Aquac. Nutr. 25, 1388–1398. doi: 10.1111/anu.12959
Keating C., Bolton-Warberg M., Hinchcliffe J., Davies R., Whelan S., Wan A. H. L., et al. (2021). Temporal changes in the gut microbiota in farmed Atlantic cod (Gadus morhua) outweigh the response to diet supplementation with macroalgae. Anim. Microbiome 3, 7. doi: 10.1186/s42523-020-00065-1
Klindworth A., Pruesse E., Schweer T., Peplies J., Quast C., Horn M., et al. (2013). Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 41, e1. doi: 10.1093/nar/gks808
Knack J. J., Wilcox L. W., Delaux P.-M., Ané J.-M., Piotrowski M. J., Cook M. E., et al. (2015). Microbiomes of streptophyte algae and bryophytes suggest that a functional suite of microbiota fostered plant colonization of land. Int. J. Plant Sci. 176, 405–420. doi: 10.1086/681161
Kumar S., Gupta R., Kumar G., Sahoo D., Kuhad R. C. (2013). Bioethanol production from Gracilaria verrucosa, a red alga, in a biorefinery approach. Bioresour. Technol. 135, 150–156. doi: 10.1016/j.biortech.2012.10.120
Kumar S., Stecher G., Li M., Knyaz C., Tamura K. (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Lahti L., Shetty S., et al. (2017) Tools for microbiome analysis in r. version 1.17.2. Available at: http://microbiome.github.com/microbiome.
Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., et al. (2007). Clustal W and clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Legrand T., Wynne J., Weyrich L., Oxley A. (2019). A microbial sea of possibilities: current knowledge and prospects for an improved understanding of the fish microbiome. Rev. Aquac. 12, 1101–1134. doi: 10.1111/raq.12375
Li L., Liu X., Wang Y., Huang Y., Wang C. (2022). Effects of alternate feeding between fish meal and novel protein diets on the intestinal health of juvenile largemouth bass (Micropterus salmoides). Aquac. Rep. 23, 101023. doi: 10.1016/j.aqrep.2022.101023
Li T., Long M., Ji C., Shen Z., Gatesoupe F.-J., Zhang X., et al. (2016). Alterations of the gut microbiome of largemouth bronze gudgeon (Coreius guichenoti) suffering from furunculosis. Sci. Rep. 6, 30606. doi: 10.1038/srep30606
Liu Y., Cheng J., Xia Y., Li X., Liu Y., Liu P.-f. (2022). Response mechanism of gut microbiome and metabolism of European seabass (Dicentrarchus labrax) to temperature stress. Sci. Total Environ. 813, 151786. doi: 10.1016/j.scitotenv.2021.151786
Li H., Zhou Y., Ling H., Luo L., Qi D., Feng L. (2019). The effect of dietary supplementation with Clostridium butyricum on the growth performance, immunity, intestinal microbiota and disease resistance of tilapia (Oreochromis niloticus). PloS One 14, e0223428. doi: 10.1371/journal.pone.0223428
Loch T. P., Faisal M. (2015). Emerging flavobacterial infections in fish: A review. J. Adv. Res. 6, 283–300. doi: 10.1016/j.jare.2014.10.009
Love M. I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. doi: 10.1186/s13059-014-0550-8
Lozupone C., Knight R. (2005). UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71, 8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005
Lyons P. P., Turnbull J. F., Dawson K. A., Crumlish M. (2017). Effects of low-level dietary microalgae supplementation on the distal intestinal microbiome of farmed rainbow trout Oncorhynchus mykiss (Walbaum). Aquac. Res. 48, 2438–2452. doi: 10.1111/are.13080
Malick R. C., Bera A., Chowdhury H., Bhattacharya M., Abdulla T., Swain H. S., et al. (2020). Identification and pathogenicity study of emerging fish pathogens Acinetobacter junii and Acinetobacter pittii recovered from a disease outbreak in Labeo catla (Hamilton 1822) and Hypophthalmichthys molitrix (Valenciennes 1844) of freshwater wetland in West Bengal, India. Aquac. Res. 51, 2410–2420. doi: 10.1111/are.14584
Mao Z., Ye J., Li M., Xu H., Chen J. (2013). Vaccination efficiency of surface antigens and killed whole cell of Pseudomonas putida in large yellow croaker (Pseudosciaena crocea). Fish Shellfish Immunol. 35, 375–381. doi: 10.1016/j.fsi.2013.04.030
McMurdie P. J., Holmes S. (2014). Waste not, want not: why rarefying microbiome data is inadmissible. PloS Comput. Biol. 10, e1003531. doi: 10.1371/journal.pcbi.1003531
Minich J. J., Poore G. D., Jantawongsri K., Johnston C., Bowie K., Bowman J., et al. (2020). Microbial ecology of Atlantic salmon (Salmo salar) hatcheries: impacts of the built environment on fish mucosal microbiota. Appl. Environ. Microbiol. 86, e00411–e00420. doi: 10.1128/AEM.00411-20
Oh W. T., Kim J. H., Jun J. W., Giri S. S., Yun S., Kim H. J., et al. (2019). Genetic characterization and pathological analysis of a novel bacterial pathogen, Pseudomonas tructae, in rainbow trout (Oncorhynchus mykiss). Microorganisms 7, 432. doi: 10.3390/microorganisms7100432
Oksanen J., Blanchet F. G., Friendly M., Kindt R., Legendre P., McGlinn D., et al. (2013) Vegan: Community ecology package. r package version, 2.5-7. Available at: https://cran.rproject.org/web/packages/vegan/index.html.
Passos R., Correia A. P., Pires D., Pires P., Ferreira I., Simões M., et al. (2021). Potential use of macroalgae Gracilaria gracilis in diets for European seabass (Dicentrarchus labrax): Health benefits from a sustainable source. Fish Shellfish Immunol. 119, 105–113. doi: 10.1016/j.fsi.2021.09.033
Pérez-Pascual D., Estellé J., Dutto G., Rodde C., Bernardet J.-F., Marchand Y., et al. (2020). Growth performance and adaptability of European sea bass (Dicentrarchus labrax) gut microbiota to alternative diets free of fish products. Microorganisms 8, 1346. doi: 10.3390/microorganisms8091346
Philips S., Laanbroek H. J., Verstraete W. (2002). Origin, causes and effects of increased nitrite concentrations in aquatic environments. Rev. Environ. Sci. Biotechnol. 1, 115–141. doi: 10.1023/A:1020892826575
Piazzon M. C., Calduch-Giner J. A., Fouz B., Estensoro I., Simó-Mirabet P., Puyalto M., et al. (2017). Under control: how a dietary additive can restore the gut microbiome and proteomic profile, and improve disease resilience in a marine teleostean fish fed vegetable diets. Microbiome 5, 164. doi: 10.1186/s40168-017-0390-3
Piazzon M. C., Naya-Català F., Simó-Mirabet P., Picard-Sánchez A., Roig F. J., Calduch-Giner J. A., et al. (2019). Sex, age, and bacteria: how the intestinal microbiota is modulated in a protandrous hermaphrodite fish. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.02512
Qi X., Xue M., Cui H., Yang K., Song K., Zha J., et al. (2020). Antimicrobial activity of Pseudomonas monteilii JK-1 isolated from fish gut and its major metabolite, 1-hydroxyphenazine, against Aeromonas hydrophila. Aquaculture 526, 735366. doi: 10.1016/j.aquaculture.2020.735366
Rédei G. P. (2008). “UPGMA (unweighted pair group method with arithmetic means),” in Encyclopedia of genetics, genomics, proteomics and informatics. Ed. Rédei G. P. (Dordrecht: Springer Netherlands), 2068–2068. doi: 10.1007/978-1-4020-6754-9_17806
Richards G. P., Watson M. A., Needleman D. S., Uknalis J., Boyd E. F., Fay J. P., et al. (2017). Mechanisms for Pseudoalteromonas piscicida-induced killing of Vibrios and other bacterial pathogens. Appl. Environ. Microbiol. 83, e00175–e00117. doi: 10.1128/AEM.00175-17
Rico R. M., Tejedor-Junco M. T., Tapia-Paniagua S. T., Alarcón F. J., Mancera J. M., López-Figueroa F., et al. (2016). Influence of the dietary inclusion of Gracilaria cornea and Ulva rigida on the biodiversity of the intestinal microbiota of Sparus aurata juveniles. Aquac. Int. 24, 965–984. doi: 10.1007/s10499-015-9964-x
Rimoldi S., Gini E., Koch J. F. A., Iannini F., Brambilla F., Terova G. (2020). Effects of hydrolyzed fish protein and autolyzed yeast as substitutes of fishmeal in the gilthead sea bream (Sparus aurata) diet, on fish intestinal microbiome. BMC Vet. Res. 16, 118. doi: 10.1186/s12917-020-02335-1
Rognes T., Flouri T., Nichols B., Quince C., Mahé F. (2016). VSEARCH: a versatile open source tool for metagenomics. PeerJ 4, e2584. doi: 10.7717/peerj.2584
Rud I., Kolarevic J., Holan A. B., Berget I., Calabrese S., Terjesen B. F. (2017). Deep-sequencing of the bacterial microbiota in commercial-scale recirculating and semi-closed aquaculture systems for Atlantic salmon post-smolt production. Aquac. Eng. 78, 50–62. doi: 10.1016/j.aquaeng.2016.10.003
Rurangwa E., Laranja J. L., Van Houdt R., Delaedt Y., Geraylou Z., Van de Wiele T., et al. (2009). Selected nondigestible carbohydrates and prebiotics support the growth of probiotic fish bacteria mono-cultures in vitro. J. Appl. Microbiol. 106, 932–940. doi: 10.1111/j.1365-2672.2008.04034.x
Sørensen S. L., Ghirmay A., Gong Y., Dahle D., Vasanth G., Sørensen M., et al. (2021). Growth, chemical composition, histology and antioxidant genes of Atlantic salmon (Salmo salar) fed whole or pre-processed Nannochloropsis oceanica and Tetraselmis sp. Fishes 6, 23. doi: 10.3390/fishes6030023
Sagaram U. S., Gaikwad M. S., Nandru R., Dasgupta S. (2021). Microalgae as feed ingredients: recent developments on their role in immunomodulation and gut microbiota of aquaculture species. FEMS Microbiol. Lett. 368, fnab071. doi: 10.1093/femsle/fnab071
Schmidt V., Amaral-Zettler L., Davidson J., Summerfelt S., Good C., Schloss P. D. (2016). Influence of fishmeal-free diets on microbial communities in Atlantic salmon (Salmo salar) recirculation aquaculture systems. Appl. Environ. Microbiol. 82, 4470–4481. doi: 10.1128/AEM.00902-16
Sehnal L., Brammer-Robbins E., Wormington A. M., Blaha L., Bisesi J., Larkin I., et al. (2021). Microbiome composition and function in aquatic vertebrates: small organisms making big impacts on aquatic animal health. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.567408
Semova I., Carten J. D., Stombaugh J., Mackey L. C., Knight R., Farber S. A., et al. (2012). Microbiota regulate intestinal absorption and metabolism of fatty acids in the zebrafish. Cell Host Microbe 12, 277–288. doi: 10.1016/j.chom.2012.08.003
Serra C. R., Almeida E. M., Guerreiro I., Santos R., Merrifield D. L., Tavares F., et al. (2019). Selection of carbohydrate-active probiotics from the gut of carnivorous fish fed plant-based diets. Sci. Rep. 9, 6384. doi: 10.1038/s41598-019-42716-7
Serra C. R., Oliva-Teles A., Enes P., Tavares F. (2021). Gut microbiota dynamics in carnivorous European seabass (Dicentrarchus labrax) fed plant-based diets. Sci. Rep. 11, 447. doi: 10.1038/s41598-020-80138-y
Suo Y., Li E., Li T., Jia Y., Qin J. G., Gu Z., et al. (2017). Response of gut health and microbiota to sulfide exposure in pacific white shrimp Litopenaeus vannamei. Fish Shellfish Immunol. 63, 87–96. doi: 10.1016/j.fsi.2017.02.008
Swain S. M., Singh C., Arul V. (2009). Inhibitory activity of probiotics Streptococcus phocae PI80 and Enterococcus faecium MC13 against Vibriosis in shrimp Penaeus monodon. world J. Microbiol. Biotechnol. 25, 697–703. doi: 10.1007/s11274-008-9939-4
Tarnecki A. M., Burgos F. A., Ray C. L., Arias C. R. (2017). Fish intestinal microbiome: diversity and symbiosis unravelled by metagenomics. J. Appl. Microbiol. 123, 2–17. doi: 10.1111/jam.13415
Tarnecki A. M., Rhody N. R., Walsh C. J. (2018). Health characteristics and blood bacterial assemblages of healthy captive red drum: implications for aquaculture and fish health management. J. Aquat. Anim. Health 30, 339–353. doi: 10.1002/aah.10047
Valente L. M. P., Cabrita A. R. J., Maia M. R. G., Valente I. M., Engrola S., Fonseca A. J. M., et al. (2021). “Microalgae as feed ingredients for livestock production and aquaculture,” in Microalgae. Ed. Galanakis C. M. (Academic Press, Cambridge, Massachusetts, USA: Academic Press), 239–312. Available at: 10.1016/B978-0-12-821218-9.00009-8.
Vargas-Albores F., Martínez-Córdova L. R., Hernández-Mendoza A., Cicala F., Lago-Lestón A., Martínez-Porchas M. (2021). Therapeutic modulation of fish gut microbiota, a feasible strategy for aquaculture? Aquaculture 544, 737050. doi: 10.1016/j.aquaculture.2021.737050
Vigneron A., Cruaud P., Langlois V., Lovejoy C., Culley A., Vincent W. (2019). Ultra-small and abundant: Candidate phyla radiation bacteria are potential catalysts of carbon transformation in a thermokarst lake ecosystem. Limnol. Oceanogr. Lett. 5, 212–220. doi: 10.1002/lol2.10132
Wan A. H. L., Davies S. J., Soler-Vila A., Fitzgerald R., Johnson M. P. (2019). Macroalgae as a sustainable aquafeed ingredient. Rev. Aquac. 11, 458–492. doi: 10.1111/raq.12241
Wang X., Zhang Y., Qin G., Luo W., Lin Q. (2016). A novel pathogenic bacteria (Vibrio fortis) causing enteritis in cultured seahorses, Hippocampus erectus perry 1810. J. Fish Dis. 39 6, 765–769. doi: 10.1111/jfd.12411
Weiss S., Xu Z. Z., Peddada S., Amir A., Bittinger K., Gonzalez A., et al. (2017). Normalization and microbial differential abundance strategies depend upon data characteristics. Microbiome 5, 27. doi: 10.1186/s40168-017-0237-y
Wilczynski W., Radlinska M., Wysujack K., Czub M., Brzeziński T., Kowalczyk G., et al. (2022). Metagenomic analysis of the gastrointestinal microbiota of Gadus morhua callarias L. originating from a chemical munition dump site. Toxics 10, 206. doi: 10.3390/toxics10050206
Xia Y., Lu M., Chen G., Cao J., Gao F., Wang M., et al. (2018). Effects of dietary Lactobacillus rhamnosus JCM1136 and Lactococcus lactis subsp. lactis JCM5805 on the growth, intestinal microbiota, morphology, immune response and disease resistance of juvenile Nile tilapia, Oreochromis niloticus. Fish Shellfish Immunol. 76, 368–379. doi: 10.1016/j.fsi.2018.03.020
Xia L., Xiong D., Gu Z., Xu Z., Chen C., Xie J., et al. (2008). Recovery of Acinetobacter baumannii from diseased channel catfish (Ictalurus punctatus) in China. Aquaculture 284, 285–288. doi: 10.1016/j.aquaculture.2008.07.038
Xie M., Xie Y., Li Y., Zhou W., Zhang Z., Yang Y., et al. (2022). Stabilized fermentation product of Cetobacterium somerae improves gut and liver health and antiviral immunity of zebrafish. Fish Shellfish Immunol. 120, 56–66. doi: 10.1016/j.fsi.2021.11.017
Xie M., Zhou W., Xie Y., Li Y., Zhang Z., Yang Y., et al. (2021). Effects of Cetobacterium somerae fermentation product on gut and liver health of common carp (Cyprinus carpio) fed diet supplemented with ultra-micro ground mixed plant proteins. Aquaculture 543, 736943. doi: 10.1016/j.aquaculture.2021.736943
Yukgehnaish K., Issac P., Parimannan S., Marimuthu K., Arshad A. B., Paray B., et al. (2020). Gut microbiota metagenomics in aquaculture: factors influencing gut microbiome and its physiological role in fish. Rev. Aquac. 12, 1903–1927. doi: 10.1111/raq.12416
Zárate G. (2012). “Dairy propionibacteria: less conventional probiotics to improve the human and animal health,” in Probiotic in animals (IntechOpen, London, UK: IntechOpen), 153–202. Available at: 10.5772/50320.
Zebrowska J., Witkowska M., Struck A., Laszuk P. E., Raczuk E., Ponikowska M., et al. (2022). Antimicrobial potential of the genera Geobacillus and Parageobacillus, as well as endolysins biosynthesized by their bacteriophages. Antibiotics 11, 242. doi: 10.3390/antibiotics11020242
Zhou W., Xie M., Xie Y., Liang H., Li M., Ran C., et al. (2022). Effect of dietary supplementation of Cetobacterium somerae XMX-1 fermentation product on gut and liver health and resistance against bacterial infection of the genetically improved farmed tilapia (GIFT, Oreochromis niloticus). Fish Shellfish Immunol. 124, 332–342. doi: 10.1016/j.fsi.2022.04.019
Keywords: seaweed, microalga, algae blend, European seabass, intestinal microbiota
Citation: Ferreira M, Abdelhafiz Y, Abreu H, Silva J, Valente LMP and Kiron V (2022) Gracilaria gracilis and Nannochloropsis oceanica, singly or in combination, in diets alter the intestinal microbiota of European seabass (Dicentrarchus labrax). Front. Mar. Sci. 9:1001942. doi: 10.3389/fmars.2022.1001942
Received: 24 July 2022; Accepted: 15 September 2022;
Published: 11 October 2022.
Edited by:
Andrea Tarnecki, Auburn University Shellfish Laboratory, United StatesReviewed by:
Raquel Xavier, Centro de Investigacao em Biodiversidade e Recursos Geneticos (CIBIO-InBIO), PortugalFederico Moroni, University of Insubria, Italy
Copyright © 2022 Ferreira, Abdelhafiz, Abreu, Silva, Valente and Kiron. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Viswanath Kiron, kiron.viswanath@nord.no
 Mariana Ferreira
Mariana Ferreira Yousri Abdelhafiz
Yousri Abdelhafiz Helena Abreu4
Helena Abreu4  Luisa M. P. Valente
Luisa M. P. Valente Viswanath Kiron
Viswanath Kiron