In silico characterization of Cnidarian’s antimicrobial peptides
- 1Grupo de Investigación en Biología Molecular y Genética (BIOMOGEN), Facultad de Ciencias Básicas, Departamento de Biología, Universidad de Pamplona, Pamplona, Colombia
- 2Grupo de Investigación en Medicina (GINUMED), Facultad de Salud, Corporación Universitaria Rafael Núñez, Cartagena, Colombia
- 3Grupo de Investigación en Microbiología, Industria y Ambiente (GIMIA), Facultad de Ciencias Básicas, Departamento de Microbiología, Universidad Santiago de Cali, Cali, Colombia
The characterization of active compounds in Cnidaria sheds light on a large bank of substances against multiresistant bacteria related to diseases in humans, which makes it a cutting edge with a repertoire of antimicrobial molecules worthy of bioprospecting analysis. Thus, the main nabof this research was to characterize antimicrobial peptides (AMP) belonging to the defensin family in different species of Cnidarians through bioinformatic approaches. To this, an exhaustive search was carried out for sequences homologous to antimicrobial peptides belonging to the defensin family in genomes availables for Cnidarians. Also, 3D models of AMP were obtained by modeling based on homology, functional characterization of peptides found was performed with machine learning approaches. Characterization of twelve peptides derived from 11 Cnidarian species was possible due to 3D modeling, which showed structural similarity with defensins reported in several species such as Nasonia vitripennis, Pisum sativum, Solanum lycopersicum, and Aurelia aurita. Also, different physicochemical properties such as hydrophobic moment, hydrophobicity, net charge, amphiphilic index, and isoelectric point were evaluated. These peptides showed values that are ideal for AMP. Further, functional characterization showed a bactericidal potential of 20 peptides against multiresistant bacteria Escherichia coli, Pseudomonas aeruginosa, and Klebsiella pneumoniae. These peptides with action potential were found in 17 species from Cnidarians and obtained by homology through the defensin Aurelin, described in the Cnidarian Aurelia aurita, and Mus musculus’ Beta-defensin 7. Finally, a phylogenetic tree was performed, it showed that defensins are distributed in all Cnidarians regardless of the taxonomic group. Thus, the origin of defensins in the Phylum Cnidaria is not monophyletic. Our results show that Cnidaria has AMP with structural and physicochemical characteristics similar to those described in defensins of insects, mammals, and plants. The structural characteristics of these peptides, their physicochemical properties, and their functional potential outline them as promising templates for the discovery of new antibiotics.
Introduction
Cnidarians is a primitive phylum of invertebrates that emerged 700 million years ago. They are organisms with simple anatomy and a dysmorphic life cycle that allow them to use varied environments and resources (Calcagno, 2014). This phylum is represented by pelagic species, both freshwater (Technau and Steele, 2011) and marine (Cheng, 2021), and sessile benthic organisms such as hermatypic corals (Veron, 2001). Among its representatives are corals, hydras, jellyfish, sea anemones, Portuguese warships, sea pens, sea whips and sea fans (Veron, 2001).
The genomes of Cnidarians have a great complexity that is reflected in the repertoire of innate immune genes. Several researches have allowed the description of a variety of molecules involved in the immunological processes of innate immunity (Leal et al., 2022). One of these processes is immunological recognition, in which molecular patterns in a pathogen are recognized to induce a response that leads to its destruction. Among the receptors that have been studied the most are: Toll-like receptors (TLR), associated with the activation of transcription factors of the NF-κB family; NOD-like receptors, which have been attributed the ability to activate several immune pathways, such as the NF-κB pathway, interferon, mitogen-activated protein kinase (MAPK), and inflammasome (Yap and Hwang, 2018; Emery et al., 2021); lectins, carbohydrate-binding proteins involved in complement cascade activation and opsonization (Kimura et al., 2009; Poole et al., 2016); and RIG-I-like receptors (RLR), associated with the activation of the interferon system (Lewandowska et al., 2021). Also, signal translation mediators have been identified that activated immune responses such as phagocytosis, opsonization, lysis, and production of antimicrobial peptides (Lewandowska et al., 2021).
Among the innate defense systems, antimicrobial peptides (AMPs) are considered conserved mechanisms, since they are found in marine organisms, plants, and mammals, among others (Haney et al., 2017). Its mechanisms of action include the inhibition of organelles such as ribosomes, blockage of RNA synthesis or of important compounds for the cell membrane of pathogens (Bechinger and Gorr, 2017; Moravej et al., 2018). Organisms belonging to the phylum Cnidaria are exposed to various groups of infections, and although pathways of the innate immune system have been described (Miller et al., 2007; Augustin and Bosch, 2010), little is known about the role of AMPs in this group of marine organisms. Cnidarians are exposed to interactions with bacterial communities and viruses (Stabili et al., 2018; Lewandowska et al., 2020). This, added to climate change, means that the immune system of these organisms is subject to biological pressure, however, some components of the immune system of these organisms have been studied very little.
Defensins represent one of the largest families of AMPs (Rivas et al., 2006). They are low molecular weight cysteine-rich cationic peptides subdivided into three subfamilies (Dong et al., 2016): α-defensins, they have been widely identified in humans, monkeys, and several rodent species; β-defensins, a widely studied subfamily with the widest taxonomic distribution, including invertebrates and plants; and finally, θ-defensins, which have a circular structure and, this family is the less numerous in mammals. Currently, in Cnidaria only one antimicrobial peptide has been reported with characteristics similar to defensins (Ovchinnikova et al., 2006). Aurelin, is a peptide described in the jellyfish Aurelia aurita composed of 40 amino acid residues active against Gram-positive and Gram-negative bacteria (Ovchinnikova et al., 2006). Given the gap of knowledge about these AMPs belonging to the defensin family in Cnidaria, this research aimed to advance the understanding of the antimicrobial peptide system of this Phylum. Thus, in silico strategies were used to characterized peptides with functional and structural features of AMP and their possible antibiotic properties.
Methods
To achieve the aims proposed in this research, the methods were developed in four phases. First, an exhaustive search was carried out for sequences homologous to antimicrobial peptides belonging to the defensin family from genomes database for Cnidarians. Second, three-dimensional (3D) modeling was performed by sequence homology, which allowed the structural characterization of peptides. Third, a physicochemical characterization of the structurally modeled peptides was carried out. Also, 3D modeling of hydrophobic and hydrophilic surfaces was performed. Fourth, a functional characterization was addressed by computational analysis. Finally, the phylogenetic tree from structurally modeled peptides and peptides that showed putative bactericidal activity, was built by using the Maximum Likelihood method.
Local database
Several Antimicrobial peptide sequences from different vertebrates (Homo sapiens, Mus musculus), plants, insects and jellyfish Aurelia aurita (specifically, defensin Aurelin, already structurally and functionally characterized PDB: 2LG4_A) from the public Protein Data Bank (https://www.rcsb.org/) and NCBI (HTTPS://www.ncbi.nlm.nih.gov/) were used to search into the Cnidarian’s genomes database such as REEFGENOMICS (http://reefgenomics.org/), OIST Marine Genomics Unit (https://marinegenomics.oist.jp/gallery), National Human Genome Research Institute (NHGRI) (https://research.nhgri.nih.gov/hydra/), CassiopeaBase (https://cassiopeabase.org/resources/cxamachana-genome/), UniProt (https://www.uniprot.org/), via protein blast searches. We also use the following sequences from Nematostella vectensis and Hydra magnipapillata as a query: 1) jgi_Nemve1_206031 sequence, obtained from a first search using AMP Aurelin described in Aurelia aurita. 2) Hma2.232062 sequence, obtained from a first search for Beta-defensin 7 (mBD-7) described in Mus musculus. A total of 40 defensins with their secondary structure described, and with proven bactericidal action (Data Sheet 1. FASTA) were used to the search and built an AMP local database.
Peptides characterization
The 3D structure of the putatives peptides in Cnidarian was performed by homology-based modeling using the Swiss model server (Schwede et al., 2003). These models were optimized using the UCF Chimera software (Pettersen et al., 2004). In addition, the evaluation of the quality of the models was adressed through a Ramachandran plot analysis and the QMEANDisCo Global and GMQE estimators.
Also, the AMPs were characterized physicochemically. Physicochemical traits such as hydrophobicity, hydrophobic moment, net charge, amphiphilic index, isoelectric point, and weight were tested by the Property Calculation tool of the DBAASP database (Pirtskhalava et al., 2020). In addition, the antimicrobial potential of the AMPs obtained to Cnidaria was predicted using the PAASS tool in the DBAASP database (https://dbaasp.org/).
Analysis of phylogenetic relationships
For the phylogenetic analysis, a file in Fasta format (Data Sheet 2. FASTA) was created to perform multiple alignments with the MUSCLE tool in the MEGA X program (Tamura et al., 2021). The most likely evolutionary model was found, and a phylogenetic tree was built using the Maximum Likelihood (ML) method, with 1000 starting replicates and the Dayhoff model (Schwarz and Dayhoff, 1979). A discrete gamma distribution, calculated with the local base sequences constructed in this work, was used to model the different evolutionary rates between sites (+G, parameter = 5.0564).
Results
Fifty sequences were obtained from different Cnidarian species that showed a similarity above 25% with AMP belonging to the family of defensins described in higher organisms (Supplementary Table 1) (Data Sheet 3. FASTA). These 50 sequences were subjected to different analyzes to find putative antimicrobial peptides structurally similar to defensins, with putative action against particular microbial species.
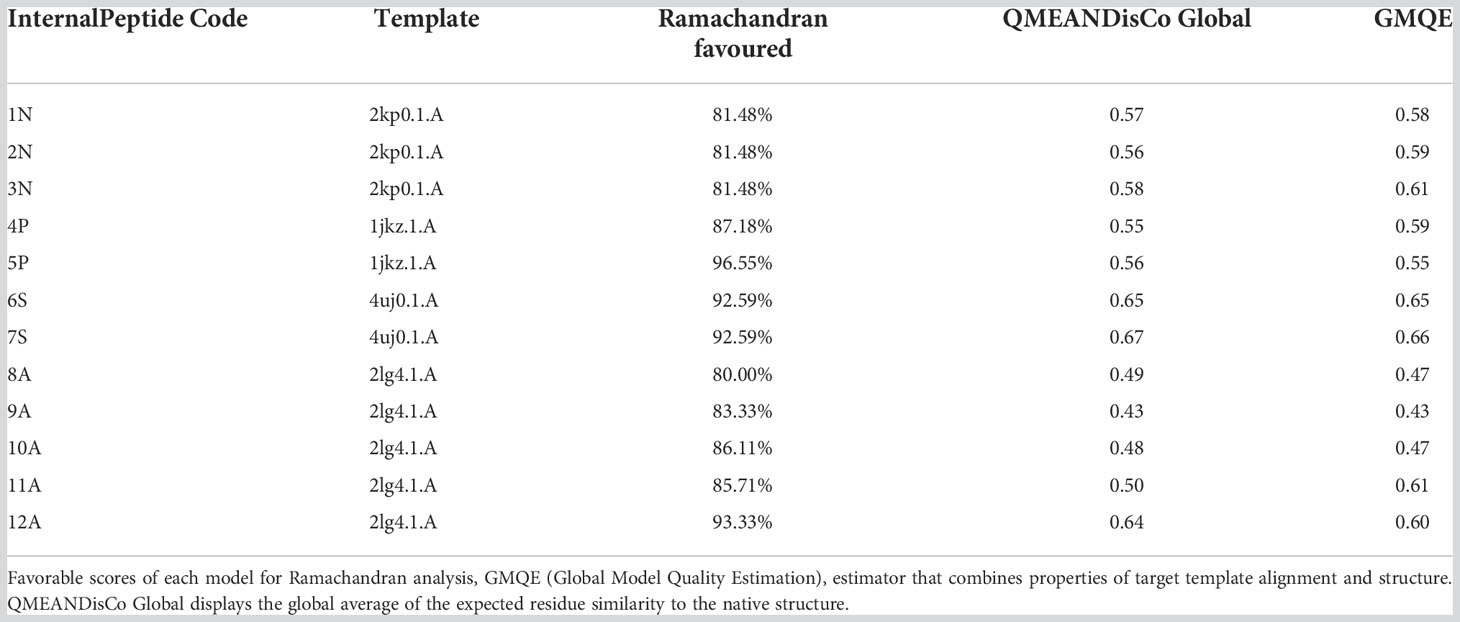
Structural modeling
Of fifty putative antimicrobial peptide sequences, only twelve sequences (from 11 species of Cnidarians) met the minimum criteria to solve their 3D structure. Five peptides presented α-helical conformations, and seven antiparallel α-helical and β-sheet conformations (Figure 1; Table 1). Three peptides (1N, 2N, 3N) were modeled through the defensin nasonin-1 (PDB ID: 2KP0) from Nasonia vitripennis. Two other peptides (4P, 5P) were modeled through Defensin-1 (Psd1; PDB ID: 1JKZ) described from Pisum sativum. While defensin gamma-thionin (TPP3; PDB ID: 4uj0), described from Solanum lycopersicum, served as a template to obtain 2 more peptides (6S, 7S). Finally, five peptide structures (8A, 9A, 10A, 11A, 12A) were built using as a template the defensin Aurelin (PDB ID: 2LG4_A) from Aurelia aurita. The alignment of the sequences of the different structural models obtained from Cnidaria, with the defensin sequences used as templates (Supplementary Figure 2).
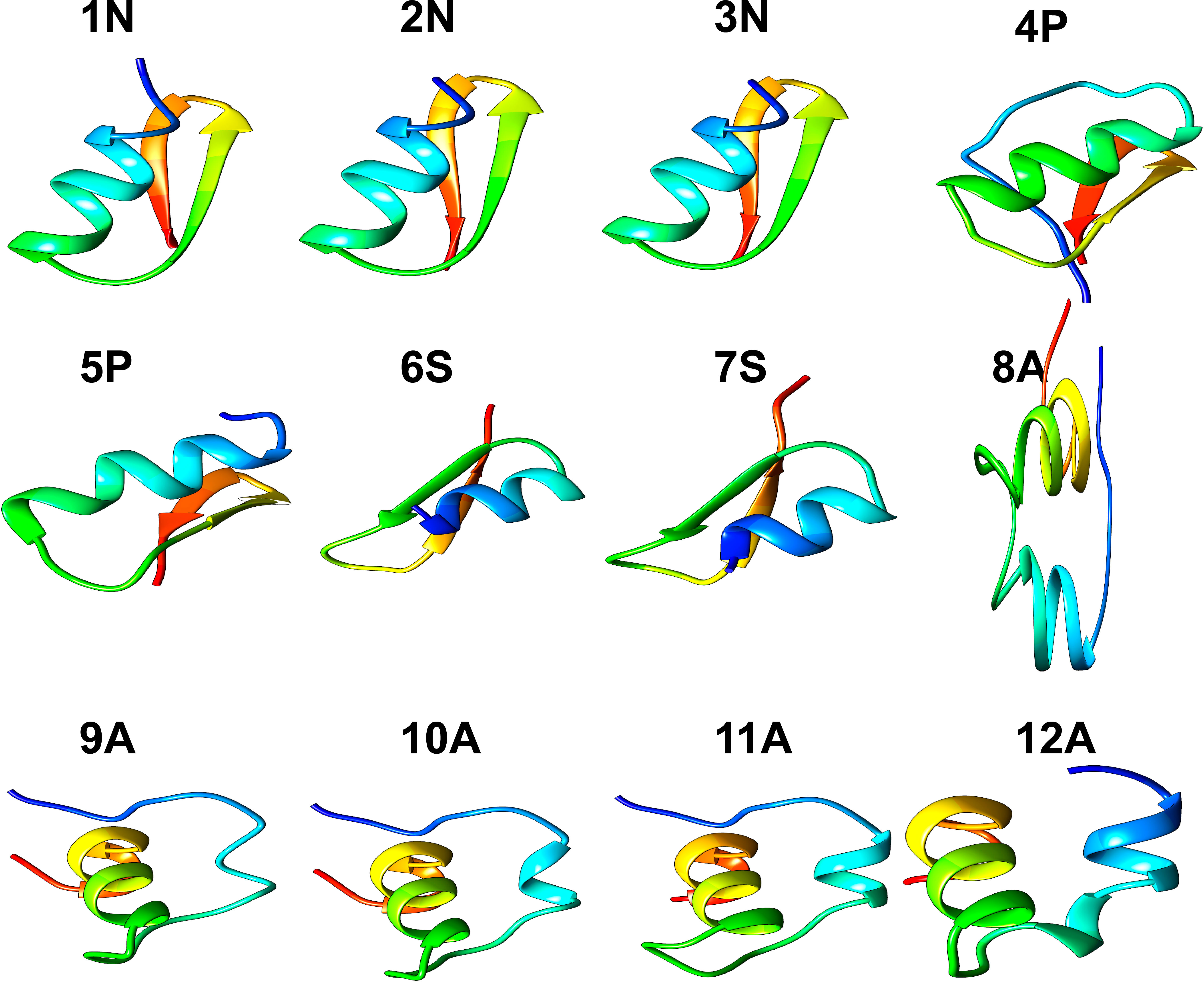
Figure 1 3D structure of peptides from 11 species of Cnidarians (shown in Table 1) with putative antimicrobial action.
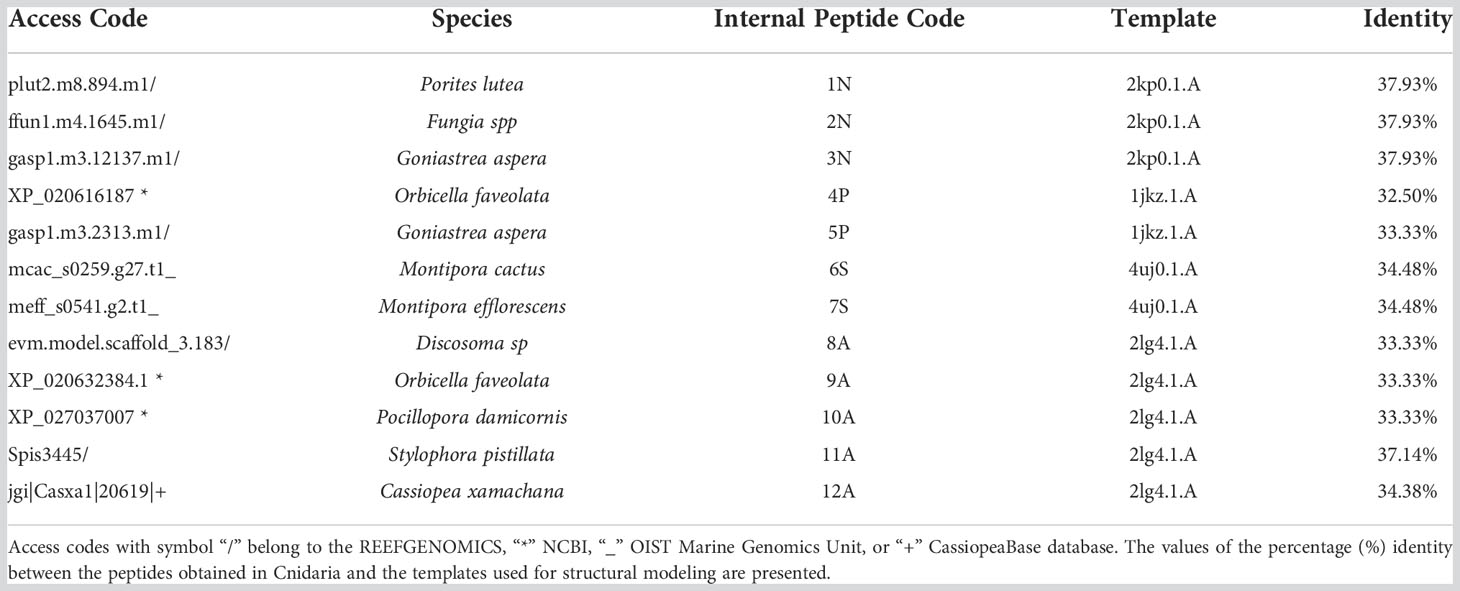
Table 1 Putative antimicrobial peptides from Cnidaria with minimum criteria to obtain a 3D structure.
AMP’s models quality
Ramachandran plots allowed visualized both amino acids present in energetically favored regions for dihedral angles, as well as amino acids in questionable regions for each of the 3D models (Supplementary Figure 1). The structure that presented the largest number of amino acids (Leal et al., 2022) with dihedral angles “not allowed” was that of peptide 8A described in the species Discosoma sp. On the other hand, the 5P peptide described in Goniastrea aspera showed that all its amino acids have dihedral angle conformations for forming α-helix and β-sheet structures. The peptides that showed the most favorable values for this analysis were those described in the species Goniastrea aspera (5P), Montipora cactus (6S), Montipora efflorescens (7S) and Cassiopea xamachana (12A), with Ramachandran values of 96.55%, 92.59%, 92.59%, and 93.33%, respectively (Table 2).
Using the GMQE and QMEANDisCo estimators, we were found that the models generally had a quality that ranges between 0 and 1. These estimators are based on different geometric properties where the expected similarity of the model with the native structure is evaluated. In general, scores below 0.5 indicate a low-quality model (Studer et al., 2020). The peptides with the most favorable values for this analysis were 3N (Goniastrea aspera), 6s (Montipora cactus), 7S (Montipora efflorescens), and 12A (Cassiopea xamachana). On the other hand, the peptides that showed low-quality values were: 8A (Discosoma sp), 9A (Orbicella faveolata), and 10A (Pocillopora damicornis) (Table 2).
Physicochemical characterization of peptides
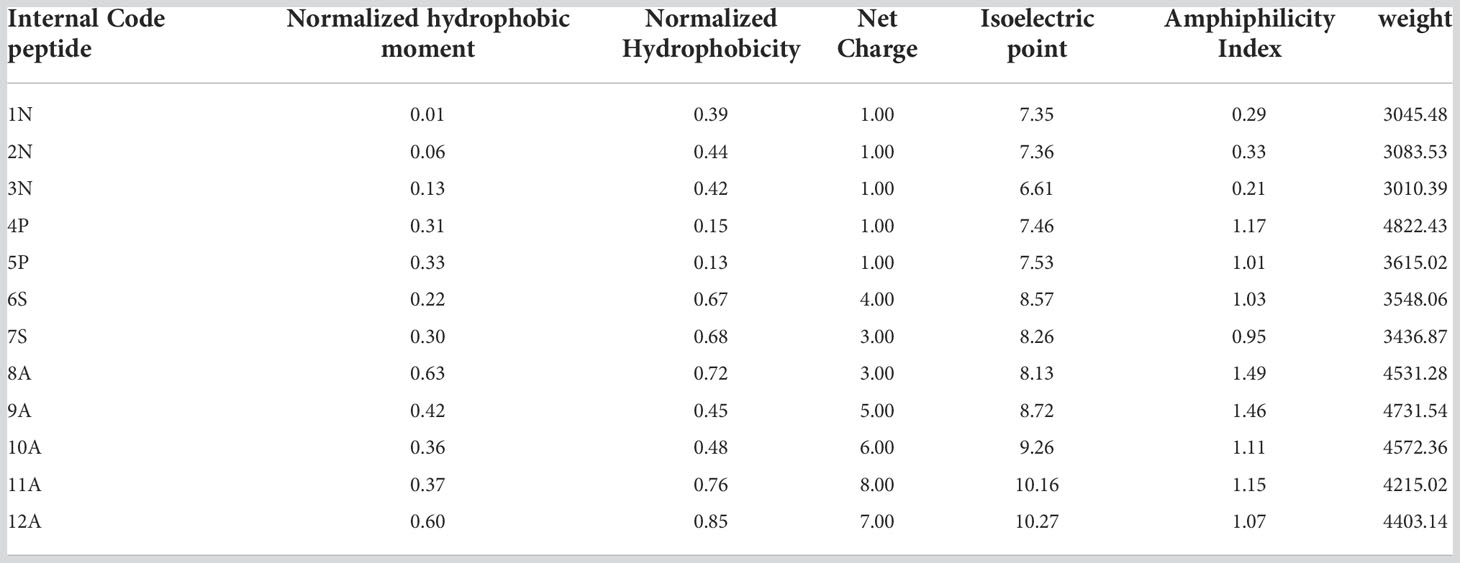
Different physicochemical features from the 12 structurally modeled peptides, were obtained (Table 3). Hydrophobicity profiles between -3 and 5 in the Wimley-White scale were obtained; where positive values represent greater hydrophobicity (White and Wimley, 1998). Also, hydrophobic moment values ranging between 0.01 and 0.63 were obtained, with higher values in peptides 12A, 9A, and 8A. Regarding the general hydrophobicity of the peptide, peptides with values between 0.39 and 0.83 were obtained. The peptides 7S, 8A, 11A, and 12A were the most favored for this trait. The net charge for all peptides was positive, ranging from +1 to +8. Isoelectric point values ranged from 6.61 to 9.89, with higher values for peptides 9A, 10A, 11A, and 12A. Moreover, the amphiphilic index values ranged between 0.21 and 1.49. Values greater than zero indicate amphiphilic (Mitaku et al., 2002), peptides 4P, 11A, 9A, and 8A showed the highest values for this feature. Finally, the 3D structure of hydrophobic and hydrophilic surfaces of the 12 structurally characterized peptides are represented (Figure 2).
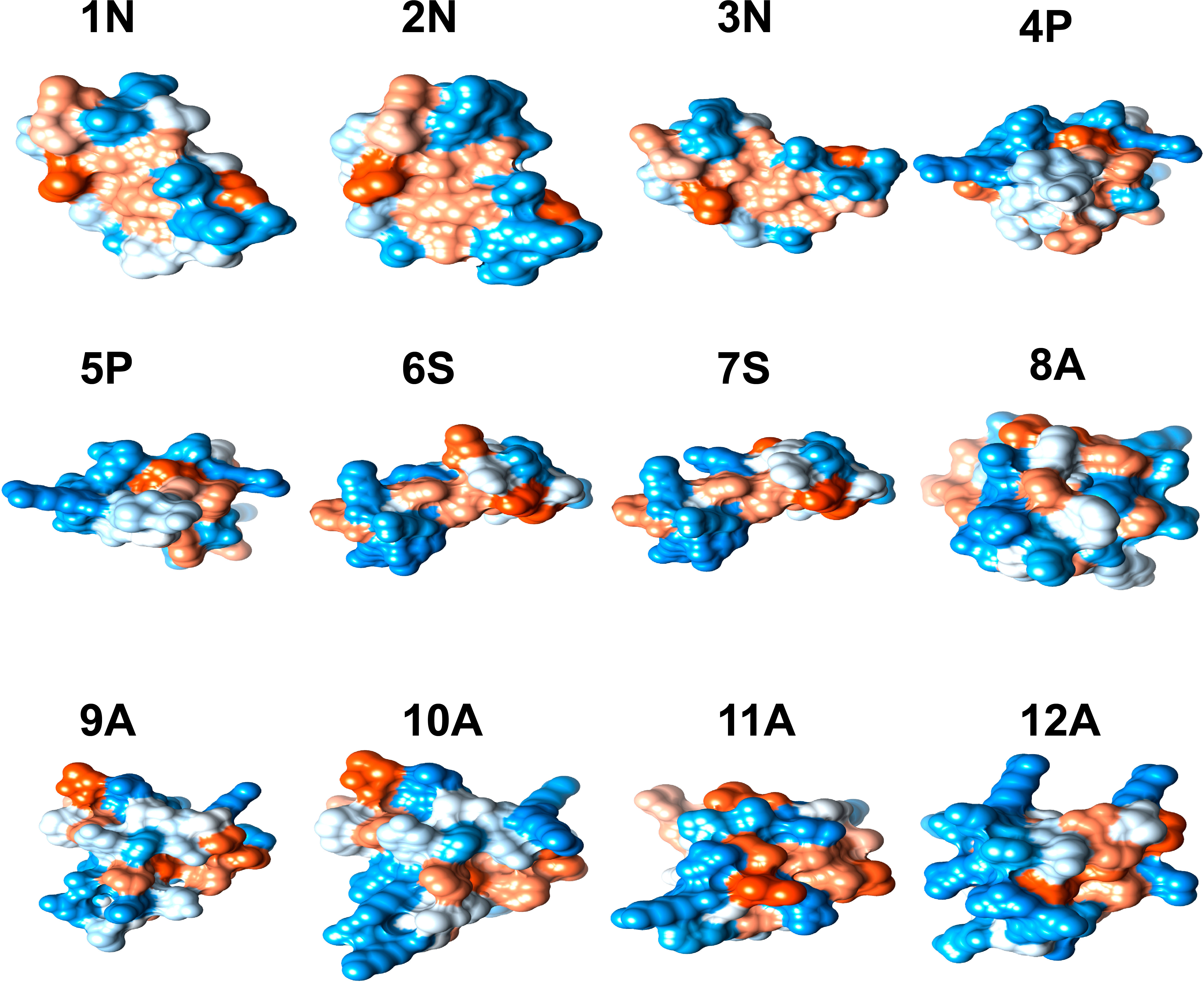
Figure 2 3D structure of hydrophobic and hydrophilic regions of peptides with antimicrobial potential modeled in 11 Cnidarian species (Table 3). The blue color represents hydrophilic regions and the orange hydrophobic regions.
Prediction of activity antimicrobial against specific species
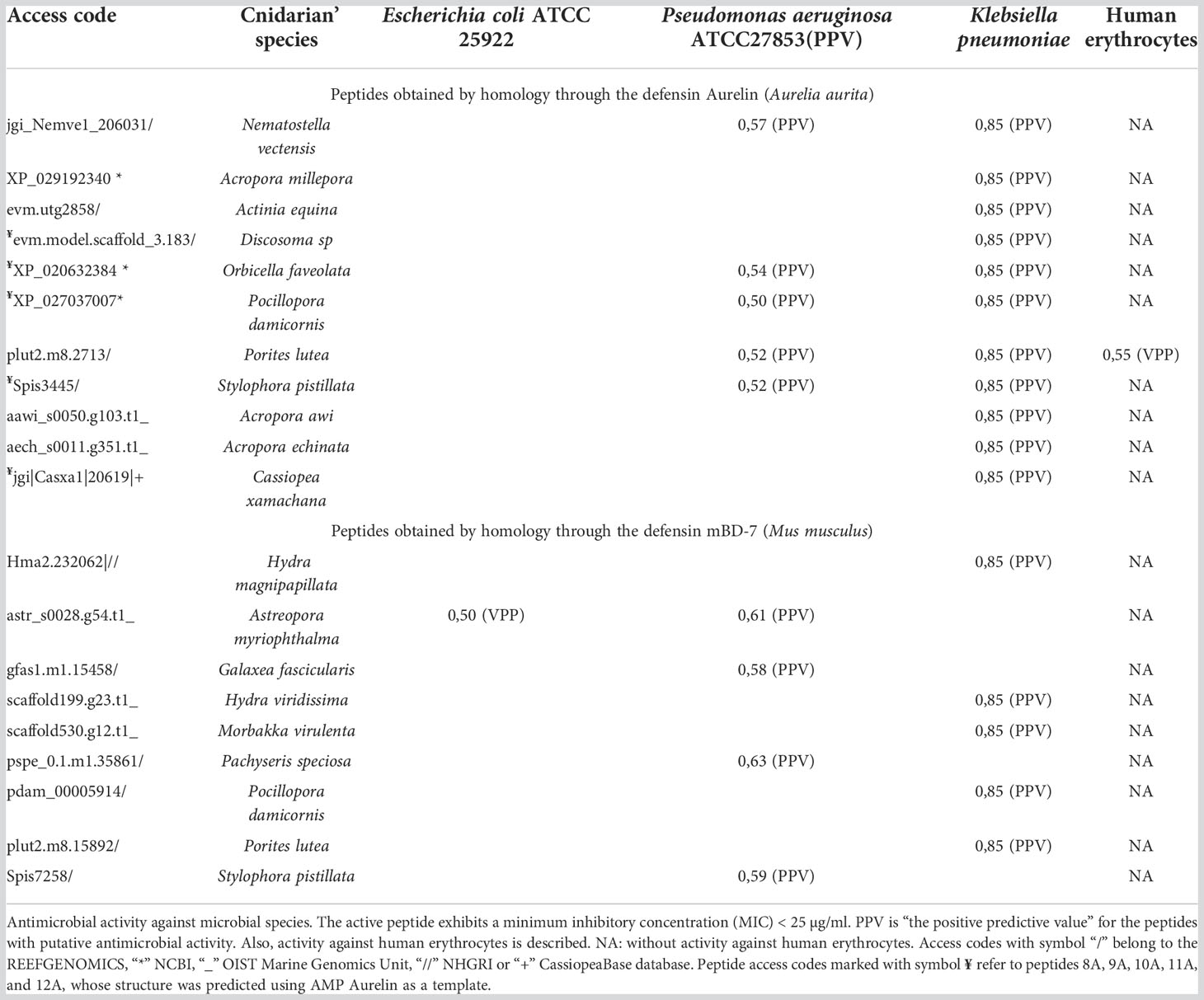
Using the PASS tool, the putative antimicrobial action of all 50 peptides (Supplementary Table 1) against specific microbial species was analyzed. After analysis, we found that 20 peptides showed a potential antimicrobial activity against species such as Escherichia coli, Pseudomonas aeruginosa, and Klebsiella pneumoniae (Table 4). Eleven peptides showed putative antimicrobial action against Klebsiella pneumoniae; three peptides against Pseudomonas aeruginosa; one peptide against Escherichia coli and Pseudomonas aeruginosa; and five peptides against Pseudomonas aeruginosa and Klebsiella pneumoniae. Only a peptide from the species Porites lutea showed putative antimicrobial activity against human erythrocytes. These peptides were obtained by homology through the defensin Aurelin described in the Cnidarian Aurelia aurita, and Beta-defensin 7 (mBD-7) described in Mus musculus.
Analysis of phylogenetic relationships
A total of 33 sequences, including peptides structurally and functionally characterized as AMP, and different defensins used as a template for their characterization, in addition to a Homo sapiens defensin, were analyzed to find a phylogenetic relationship between the peptides from the taxonomic groups different.
The phylogenetic tree showed that some proteins with putative antimicrobial activity characterized in Cnidaria are closely related to AMPs described in other organisms (Figure 3). A node with resampling support of 43% can be seen, which groups different peptides characterized in Cnidarians with the defensins Aurelin and Nuetrophil. This random resampling percentage is close to the ideal (≥50) for validating nodes in the tree. Also, a close relationship is show between the peptide characterized in Nematostella vectensis (Table 4) and the Neutrophil AMP described in Homo sapiens with resampling support of 29%. In another node, with resampling support of 31%, are the defensin Psd1 described in Pisum Sativum, TPP3 described in Solanum lycopersicum, and Beta-defensin 7 (mBD-7) described in Mus musculus. Also, it is show that the peptides described in Montipora cactus and Montipora efflorescens share a close common ancestor with the defensins Psd1 and TPP3.
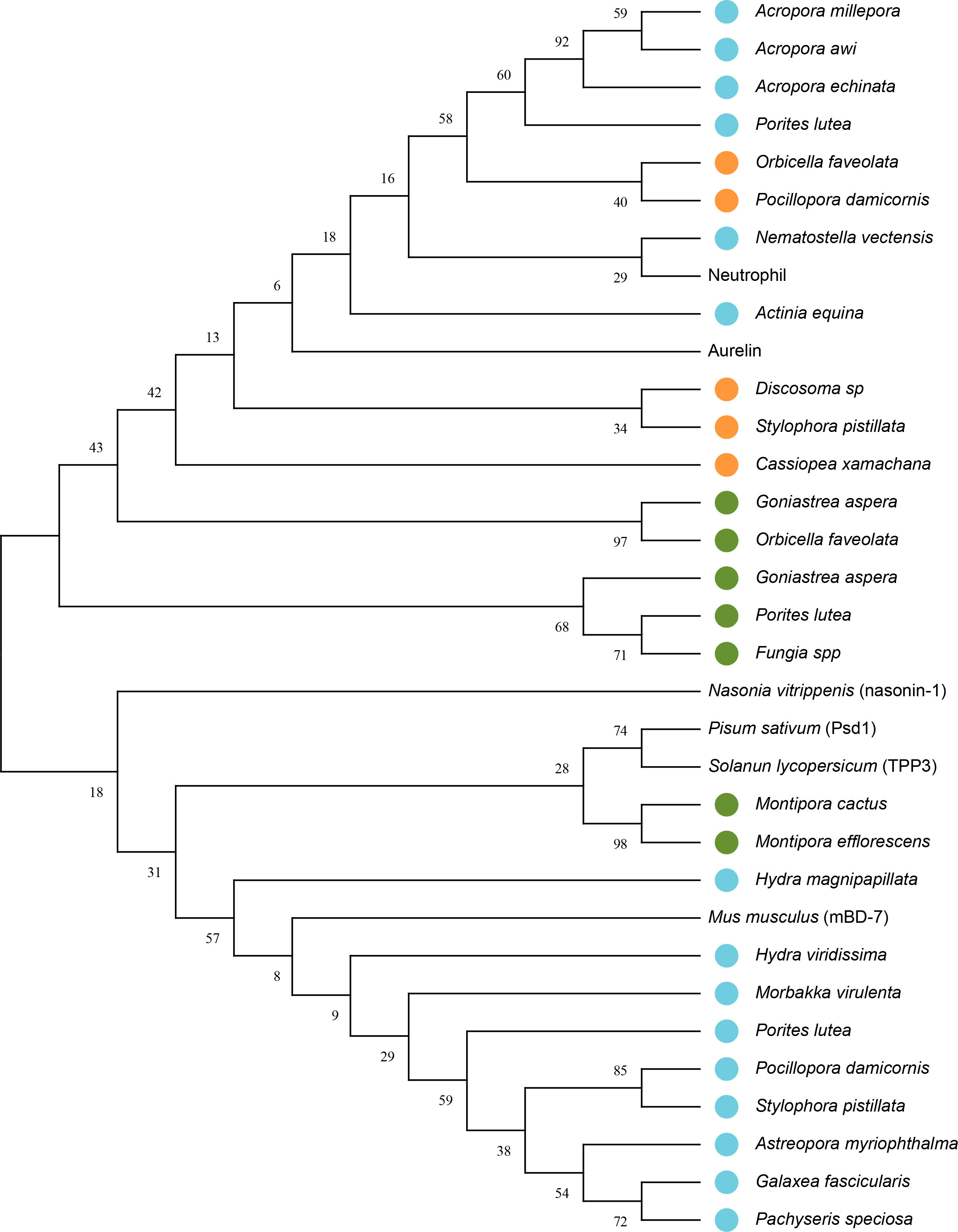
Figure 3 Phylogenetic tree with putative sequences for AMPs from Cnidaria. The bootstrap consensus tree unrooted inferred from 1000 replicates using the Maximum Likelihood method and the model based on the Dayhoff matrix (Schwarz and Dayhoff, 1979). This analysis involved 33 amino acid sequences. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches. Initial tree(s) for the heuristic search were obtained automatically by applying the Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the Dayhoff model and then selecting the topology with a higher log-likelihood value. A discrete Gamma distribution was used to model the evolutionary rate differences between sites (5 categories (+G, parameter = 5.6968)). There was a total of 56 positions in the final data set. Blue circles represent peptides that showed putative antimicrobial activity against multiresistant bacteria, orange circles represent peptides that showed putative antimicrobial activity and were structurally characterized; and green circles represent peptides that were only structurally characterized. Evolutionary analyzes were performed in MEGA11 (Tamura et al., 2021).
Discussion
Structural modeling of peptides
Three dimensional models of twelve peptides with antimicrobial potential were obtained by comparative modeling. The three models obtained from the defensin nasonin-1 showed values for the Ramachandran analysis close to the ideal percentage for this measure (>98%) (Table 2). The GMQE and QMEANDisCo estimators showed favorable values that indicate structural similarity between the modeled peptide and the native structure, which allows an approximation of the possible secondary structure for these peptides. After modeling, the typical CSαβ fold (cysteine-stabilized αβ fold) was confirmed in the three peptides (Figure 1), characteristic of insect defensins (Kovaleva et al., 2020), made up of an α-helix and a two-chain β-sheet. Also, the alignment of 6 cysteine residues in the conserved domain in nasonin-1 was evidenced (Supplementary Figure 2). These residues are related to the formation of disulfide bonds that give stability to the α-helix and β-sheet structure (Rivas et al., 2006). The similarity in sequence and the structural similarity of these peptides with the defensin nasonin-1, together with the presence of conserved cysteine residues, are indications that these peptides possibly have antimicrobial properties similar to those displayed by this defensin. Studies have shown that nasonin-1 is an AMP with bactericidal activity against two species of Gram-negative bacteria with lethal concentrations of 1.52 μM for Stenotrophomonus sp. LZ-1 and 15.4 μM for Escherichia coli ATCC 25922 (Tian et al., 2010).
Models from defensins described in plants, specifically in the species Pisum sativum (Psd1) and Solanum lycopersicum (TPP3), showed the following: All peptides showed a structure with a CSαβ fold (Figure 1), and GMQE and QMEANDisCo values indicating structural similarity between the modeled peptide and the native structure. For peptide 5P, the most favorable value was obtained for the Ramachandran analysis, with 96.55%, thus being the model that came closest to the ideal score for its validation. In terms of sequence similarity, unlike the defensins described in plants, which have 8 cysteine residues that stabilize the CSαβ structure (Kovaleva et al., 2020), the peptides modeled from these defensins showed 5 to 6 residues. However, alignment of some of these residues within the α-helical and β-sheet structures is observed (Supplementary Figure 2).
Defensins with the CSαβ structure are widely distributed in different insect species and show activity mainly against Gram-positive bacteria (Manniello et al., 2021). For example, the AMP Sapecin described in Sarcophaga peregrina, has the ability to inhibit the growth of the Gram-positive bacteria Staphylococcus aureus, through the permeabilization of the plasma membrane (Takeuchi et al., 2004). In the case of plants, defensins with this folding have been reported to be active against fungal species such as Fusarium Oxysporum, Fusarium Graminearum, and Botrytis cinerea (Kovaleva et al., 2020). This type of structure is also present in serine protease inhibitor peptides, amylase inhibitors, and scorpion toxins, suggesting that the CSαβ motif has emerged during evolution as a polyvalent structure used in peptides with different functions (Dimarcq et al., 1998).
All peptides modeled after the defensin Aurelin showed the typical 6-cysteine domain and a structure composed of α-helix regions (Figure 1). Peptide 12A described in Cassiopea xamachana was the one that showed the greatest structural similarity with this AMP. It has been determined that Aurelin is an AMP with defensin characteristics since it has a secondary structure composed of two α-helix regions crossed by three disulfide bonds, formed from 6 cysteine residues (Ovchinnikova et al., 2006). This antimicrobial peptide has shown activity against Gram-positive and Gram-negative bacteria such as Listeria monocytogenes, with a minimum inhibitory concentration (MIC) of 22.6 µg*mL-1, and Escherichia coli with a MIC of 7.6 µg*mL-1 (Ovchinnikova et al., 2006). It should be noted that all Cnidarian peptides obtained from AMP Aurelin show putative action against 3 microbial species: Escherichia coli, Pseudomonas aeruginosa, and Klebsiella pneumoniae (Table 4). This allows us to propose these peptides as possible bactericidal agents and supports that AMP belonging to the defensin family could be widely distributed in this Phylum.
Physicochemical properties of peptides
Physicochemical characteristics such as hydrophobic moment, hydrophobicity, net charge, amphiphilic index, and isoelectric point were determined for all structurally modeled peptides in different Cnidaria species. These properties are specific to AMPs and are critical to their mechanisms of action (Kumar et al., 2018; Vélez et al., 2021). Regarding the net charge, all peptides showed a positive charge (Table 3). It has been determined that the initial binding of AMP with bacterial and fungal membranes is based on the electrostatic interaction between the cationic charge of the peptides and different negatively charged components of the cell membrane of the pathogen (Zhang et al., 2021). Also, variation in peptide charge has been reported to correlate with antimicrobial activity (Kumar et al., 2018). This was evidenced in a study carried out on AMP magainin II, where increasing the load from +3 to +5 improved activity and selectivity against Gram-positive and Gram-negative bacteria, and decreased hemolytic activity. Likewise, a load greater than +5 led to an increase in hemolytic activity and loss of antimicrobial activity (Dathe et al., 2001). In this investigation, peptides 6S, 7S, 8A, and 9A presented charges that ranged between +3 and +5, which could indicate greater selectivity and antimicrobial activity. Normalized hydrophobicity (Overall Peptide Hydrophobicity) showed values above zero for all peptides (Table 3). This scale represents the free energy transfer of amino acids from water to the interface of a lipid bilayer (White and Wimley, 1998). It has been suggested that the increase in the hydrophobicity of the peptide favors its insertion into the membrane. However, excessive levels of hydrophobicity can cause a loss of microbial selectivity (Chen et al., 2007). For this property, values were obtained that ranged between 0.13 and 0.83, optimal values for antimicrobial activity. Favorable values were also obtained for the hydrophobic moment, this property refers to the hydrophobicity of the peptide in the α helix approximation (Pirtskhalava et al., 2020), and is indicative of the formation of an amphipathic α helix structure (Vélez et al., 2021). Similarly, positive values were obtained for the amphiphilic index, this characteristic is common in all AMPs with α helix regions since it allows the formation of two faces, the polar and non-polar face that facilitate interaction with the bacterial membrane. (3. 4).
The values obtained for these physicochemical characteristics and the structure of the peptides found here reflect contact capacity and adherence to the cell membrane of bacteria, a crucial process in the mechanisms of action of AMPs. Also, the isoelectric point values were between 8 and 9 feature which guarantees that these peptides can bind to the cell membrane of bacteria in physiological pH conditions. In this investigation, 7 peptides were obtained with values that oscillated in this range (Table 3).
Prediction of antimicrobial activity against specific microbial species
Fifty sequences obtained from Cnidaria showing similarity with defensins described in different organisms, their specific antimicrobial action was evaluated. In total, 20 peptides obtained from the defensins Aurelin and β-defensin 7 (mBD-7) showed potential action against Gram-negative bacteria Escherichia coli, Pseudomonas aeruginosa, and Klebsiella pneumoniae (Table 4). Several peptides showed putative bactericidal activity against the bacterium Klebsiella pneumoniae, a microorganism of pharmaceutical interest that causes a wide range of infections, including pneumonia, urinary tract infections, bacteremia, and liver abscesses (Paczosa and Mecsas, 2016). Similarly, a significant number of peptides showed potential activity against the bacterium Pseudomonas aeruginosa, which is responsible of several infections in immunocompromised patients (Paz-Zarza et al., 2019). Only one peptide showed a putative activity against Escherichia coli, a microorganism that can be pathogenic and cause damage, producing different clinical pictures (Rodríguez-Angeles, 2002). These microorganisms are opportunistic and are related to nosocomial infections, with one characteristic in common, their high resistance to antibiotics, a circumstance that is worrying due to the increase in serious infections caused by these pathogens (Rodríguez-Angeles, 2002; Paczosa and Mecsas, 2016; Paz-Zarza et al., 2019). In addition to the putative bactericidal activity of these peptides, they showed inactivity against human erythrocytes, a property that increases their value for bioprospecting studies. Other types of peptides found in Cnidaria have shown these characteristics through in vitro tests; for example, the AMP Arminin 1a described in Hydra magnipapillata exhibits potent activity against the multidrug-resistant bacterium Staphylococcus aureus without affecting human erythrocyte membranes (Augustin et al., 2009). Likewise, synthetic peptides predicted from the transcriptome of Hydractinia symbiolongicarpus showed activity against Staphylococcus aureus, Staphylococcus epidermidis, Pseudomonas aeruginosa, Acinetobacter baumannii, and Enterobacter aerogenes bacteria, without producing hemolytic effects on animal cells (Martínez Mondragón, 2019).
In vitro characterization studies have demonstrated the effectiveness of the tool (PAASS) used for this analysis. The characterization of AMPs through this algorithm has allowed the prediction of active peptides against strains of Escherichia coli ATCC 25922, with a prediction of biological activity in 93% of the 15 cases analyzed. In this same study, all peptides showed antimicrobial activity against 8 strains out of 16 tested (Vishnepolsky et al., 2019).
Peptide sequences obtained from AMP β-defensin 7 (mBD-7) only had two cysteine residues. On the other hand, all the peptides obtained from AMP Aurelin showed the 6 typical cysteine residues characteristic of mammalian and insect defensins. Aditionally, 5 of the 11 peptides with action potential showed structural homology, which allowed their structure to be predicted.
Phylogenetic relationships between AMPs defensin-type from Cnidaria
Phylogenetic analyzes performed between the putative AMPs found in Cnidaria indicate a close relationship with different defensins (Figure 3). Peptide described in Nematostella vectensis, which showed activity against Pseudomonas aeruginosa and Klebsiella pneumoniae (Table 4), is closely related to the Neutrophil AMP from Homo sapiens. The common ancestry between these two peptides, the six conserved cysteine residues, and the functional similarity, support the homology between these two proteins. Also, defensins Aurelin and Nuetrophil share a close common ancestor with different peptides described in Cnidarian species. In addition, many of the sequences share a high similarity, which indicates that the diversification of these peptides is possibly due to gene duplication (Vargas and Zardoya, 2012) or convergence events (Gallardo, 2017). The defensin nasonin-1 did not show a close phylogenetic relationship with the peptides described in Cnidaria. The peptides modeled from this defensin (1N, 2N, 3N) clustered at distant nodes, which could indicate a case of convergent evolution (Gallardo, 2017). Also, a close phylogenetic relationship was found between defensins Psd1 and TPP3 with structurally characterized peptides from Montipora cactus (6S) and Montipora efflorescens (7S). Finally, the peptides predicted from the defensin mBD-7 (Table 4) are grouped with this AMP, without sharing a close common ancestor, which may be due to the absence of the 6 typical cysteine residues present in defensins from mammals. All together, we can conclude that the AMPs described for Cnidaria do not describe a monophyletic relationship at the taxonomic level.
Advances and limitations
Although we performed an intensive search for peptides using structurally and functionally characterized defensin-type AMPs from different organisms as queries, perhaps we lost information about potential genes encoding this type of peptide because we relied on protein blast searches. However, the databases such as REEFGENOMICS and OIST Marine Genomics Unit used in this study support their information in genetic models with gene annotation updated.
Peptides with characteristics of defensins from different Cnidarian species with desirable physicochemical and structural features were obtained. Twenty of these peptides showed putative antimicrobial activity against different multiresistant pathogenic bacteria of pharmaceutical interest, and inactivity against human erythrocytes. The above, outlines them as candidates for therapeutic applications. These peptides with putative antibiotic properties could be evaluated to be used to treat corals that face threats of an infectious nature, such as the white band disease that affects acroporid corals (Aronson and Precht, 2001) or the devastating Stony Coral Tissue Loss Disease (SCTLD) that currently affecting the integrity of the Mesoamerican reef (Camacho-Vite et al., 2022). This type of method was tested on the species Montastraea cavernosa, where different antibiotics were used to treat diseased corals (Shilling et al., 2021). In this way, it was possible to show that the treatment had a success rate of 95% in the healing of individual lesions caused by the disease (Shilling et al., 2021). It is possible that the treatment of diseases in corals with AMPs obtained from different Cnidarians shows greater efficiency since cytotoxicity problems would be avoided.
Given the importance of the results obtained, it is pertinent to carry out future research through in vitro tests that allow them to be validated. In this sense, here we present an efficient methodology for the in-silico characterization of antimicrobial peptides for therapeutic purposes, because we were able to reduce the bank of possible defensin-type AMPs that could be characterized experimentally. However, it will have to be ratified by experimental methods.
On the other hand, phylogenetic analyzes indicate that defensins are a primitive family of AMPs that have been conserved throughout evolution, which outlines these molecules as important candidates for studies on innate immune evolution.
Conclusions
One of the main effector mechanisms of innate immunity is AMPs. In this research, we found several putative AMPs with structural characteristics of defensins from different species of Cnidarians. Also, many of these peptides showed putative antimicrobial activity against multiresistant pathogenic bacteria, which allows us to propose them to be profiled as promising templates for the discovery of new antibiotics. Moreover, these putative defensins are a family of AMPs with a high degree of conservation, which prevents them from being differentiated by taxonomic level.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
Conceived and designed the experiments: LS-B and MM. Performed the experiments: EL, MM and LS-B. Analyzed the data: EL, MM, and LS-B. Contributed materials/analysis tools: LS-B and MM. Wrote the article: EL, LS-B, and MM. All authors contributed to the article and approved the submitted version.
Funding
This research has been funded by Dirección General de Investigaciones of Universidad Santiago de Cali under call No. DGI-01-2022.
Acknowledgments
We thank to the Dirección General de Investigaciones from the Universidad Santiago de Cali for supporting this publication through call No. DGI-01-2022. To the Vicerectoría de Investigaciones from Universidad de Pamplona for its support of this research and the reviewers for their comments, which helped to improve this work. Also, we thank to Ilya Geraldine Palacios González for her support with the computing power to carry out some of the analyzes performed in this investigation.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.1065717/full#supplementary-material
References
Aronson R. B., Precht W. F. (2001). White-band disease and the changing face of Caribbean coral reefs. Ecol. Etiol. Newly emerging Mar. Dis. 159, 25–38. doi: 10.1007/978-94-017-3284-0_2
Augustin R., Anton-Erxleben F., Jungnickel S., Hemmrich G., Spudy B., Podschun R., et al. (2009). Activity of the novel peptide arminin against multiresistant human pathogens shows the considerable potential of phylogenetically ancient organisms as drug sources. Antimicrob. Agents Chemother. 53 (12), 5245–5250. doi: 10.1128/AAC.00826-09
Augustin R., Bosch T. C. (2010). Cnidarian immunity: A tale of two barriers. Adv. Exp. Med. Biol. 708, 1–16. doi: 10.1007/978-1-4419-8059-5_1
Bechinger B., Gorr S. U. (2017). Antimicrobial peptides: Mechanisms of action and resistance. J. Dent. Res. 96 (3), 254–260. doi: 10.1177/0022034516679973
Calcagno J. A. (2014). “Los Invertebrados marinos,” in Fundación de historia natural félix de azara. (Ciudad autónoma de Buenos Aires: Fundación de historia natural Félix de azara).
Camacho-Vite C., Estrada-Saldívar N., Pérez-Cervantes E. (2022). Alvarez-Filip l differences in the progression rate of SCTLD in pseudodiploria strigosa are related to colony size and morphology. Front. Mar. Sci. 9, 1328. doi: 10.3389/fmars.2022.790818
Cheng K. (2021). Learning in cnidaria: A systematic review. Learn Behav. 49 (2), 175–189. doi: 10.3758/s13420-020-00452-3
Chen Y., Guarnieri M. T., Vasil A. I., Vasil M. L., Mant C. T., Hodges R. S. (2007). Role of peptide hydrophobicity in the mechanism of action of alpha-helical antimicrobial peptides. Antimicrob. Agents Chemother. 51 (4), 1398–1406. doi: 10.1128/AAC.00925-06
Dathe M., Nikolenko H., Meyer J., Beyermann M., Bienert M. (2001). Optimization of the antimicrobial activity of magainin peptides by modification of charge. FEBS Lett. 501 (2-3), 146–150. doi: 10.1016/s0014-5793(01)02648-5
Dimarcq J. L., Bulet P., Hetru C., Hoffmann J. (1998). Cysteine-rich antimicrobial peptides in invertebrates. Biopolymers 47 (6), 465–477. doi: 10.1002/(SICI)1097-0282(1998)47:6<465
Dong H., Lv Y., Zhao D., Barrow P., Zhou X. (2016). Defensins: The case for their use against mycobacterial infections. J. Immunol. Res. 2016, 7515687. doi: 10.1155/2016/7515687
Emery M. A., Dimos B. A., Mydlarz L. D. (2021). Cnidarian pattern recognition receptor repertoires reflect both phylogeny and life history traits. Front. Immunol. 12. doi: 10.3389/fimmu.2021.689463
Gallardo M. H. (2017) Evolución. El curso de la vida. primera edición electrónica. Available at: http://sitiosciencias.uach.cl/EvolucionElCursodelaVida2017.pdf.
Haney E. F., Mansour S. C., Hancock R. E. (2017). Antimicrobial peptides: An introduction. Methods Mol. Biol. 1548, 3–22. doi: 10.1007/978-1-4939-6737-7_1
Kimura A., Sakaguchi E., Nonaka M. (2009). Multi-component complement system of cnidaria: C3, bf, and MASP genes expressed in the endodermal tissues of a sea anemone, nematostella vectensis. Immunobiology 214 (3), 165–178. doi: 10.1016/j.imbio.2009.01.003
Kovaleva V., Bukhteeva I., Kit O. Y., Nesmelova I. V. (2020). Plant defensins from a structural perspective. Int. J. Mol. Sci. 21 (15), 5307. doi: 10.3390/ijms21155307
Kumar P., Kizhakkedathu J. N., Straus S. K. (2018). Antimicrobial peptides: Diversity, mechanism of action and strategies to improve the activity and biocompatibility In vivo. Biomolecules 8 (1), 4. doi: 10.3390/biom8010004
Leal E., Suescún-Bolívar L. P., Múnera M. (2022). Mecanismos inmunológicos en cnidaria: un sistema inmune basal de gran complejidad e interés bioprospectivo. Rev. Biología Trop. 70, e49798. doi: 10.15517/rev.biol.trop.v70i1.49798
Lewandowska M., Hazan Y., Moran Y. (2020). Initial virome characterization of the common Cnidarian Lab model nematostella vectensis. Viruses 12 (2), 218. doi: 10.3390/v12020218
Lewandowska M., Sharoni T., Admoni Y., Aharoni R., Moran Y. (2021). Functional characterization of the Cnidarian antiviral immune response reveals ancestral complexity. Mol. Biol. Evol. 38 (10), 4546–4561. doi: 10.1093/molbev/msab197
Manniello M. D., Moretta A., Salvia R., Scieuzo C., Lucchetti D., Vogel H., et al. (2021). Insect antimicrobial peptides: potential weapons to counteract the antibiotic resistance. Cell. Mol. Life Sci. 78 (9), 4259–4282. doi: 10.1007/s00018-021-03784-z
Martínez Mondragón S. (2019). Actividad bactericida de péptidos antimicrobianos sintetizados a partir del transcriptoma del cnidario hydractinia symbiolongicarpus (Colombia: Universidad de los Andes). Available at: http://repositorio.uniandes.edu.co/bitstream/handle/1992/45807/u828114.pdf?sequence=1&isAllowed=y.
Miller D. J., Hemmrich G., Ball E. E., Hayward D. C., Khalturin K., Funayama N., et al. (2007). The innate immune repertoire in Cnidaria–ancestral complexity and stochastic gene loss. Genome Biol. 8 (4), R59. doi: 10.1186/gb-2007-8-4-r59
Mitaku S., Hirokawa T., Tsuji T. (2002). Amphiphilicity index of polar amino acids as an aid in the characterization of amino acid preference at membrane-water interfaces. Bioinformatics 18 (4), 608–616. doi: 10.1093/bioinformatics/18.4.608
Moravej H., Moravej Z., Yazdanparast M., Heiat M., Mirhosseini A., Moosazadeh Moghaddam M., et al. (2018). Antimicrobial peptides: Features, action, and their resistance mechanisms in bacteria. Microb. Drug Resist. 24 (6), 747–767. doi: 10.1089/mdr.2017.0392
Ovchinnikova T. V., Balandin S. V., Aleshina G. M., Tagaev A. A., Leonova Y. F., Krasnodembsky E. D., et al. (2006). Aurelin, a novel antimicrobial peptide from jellyfish aurelia aurita with structural features of defensins and channel-blocking toxins. Biochem. Biophys. Res. Commun. 348 (2), 514–523. doi: 10.1016/j.bbrc.2006.07.078
Paczosa M. K., Mecsas J. (2016). Klebsiella pneumoniae: Going on the offense with a strong defense. Microbiol. Mol. Biol. Rev. 80 (3), 629–661. doi: 10.1128/MMBR.00078-15
Paz-Zarza V. M., Mangwani-Mordani S., Martínez-Maldonado A., Álvarez-Hernández D., Solano-Gálvez S. G., Vázquez-López R. (2019). Pseudomonas aeruginosa: patogenicidad y resistencia antimicrobiana en la infección urinaria. Rev. Chil. infectología. 36 (2), 180–189. doi: 10.4067/S0716-10182019000200180
Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., et al. (2004). UCSF chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25 (13), 1605–1612. doi: 10.1002/jcc.20084
Pirtskhalava M., Amstrong A. A., Grigolava M., Chubinidze M., Alimbarashvili E., Vishnepolsky B., et al. (2020). DBAASP v3: database of antimicrobial/cytotoxic activity and structure of peptides as a resource for development of new therapeutics. Nucleic Acids Res. 49 (D1), D288–D297. doi: 10.1093/nar/gkaa991
Poole A. Z., Kitchen S. A., Weis V. M. (2016). The role of complement in Cnidarian-dinoflagellate symbiosis and immune challenge in the Sea anemone aiptasia pallida. Front. Microbiol. 7, 519. doi: 10.3389/fmicb.2016.00519
Rivas S. B., Sada E., Hernández P. R., Tsutsumi V. (2006). Péptidos antimicrobianos en la inmunidad innata de enfermedades infecciosas. Salud pública Méx. 48, 62–71. doi: 10.1590/S0036-36342006000100010
Rodríguez-Angeles G. (2002). Principales características y diagnóstico de los grupos patogenos de escherichia coli. Salud Publica Mex. 44 (5), 464–475. doi: 10.1590/S0036-36342002000500011
Schwarz R., Dayhoff M. (1979). Matrices for detecting distant relationships (Washington: National Biomedical Research Foundation), 353–358.
Schwede T., Kopp J., Guex N., Peitsch M. C. (2003). SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 31 (13), 3381–3385. doi: 10.1093/nar/gkg520
Shilling E. N., Combs I. R., Voss J. D. (2021). Assessing the effectiveness of two intervention methods for stony coral tissue loss disease on montastraea cavernosa. Sci. Rep. 11 (1), 8566. doi: 10.1038/s41598-021-86926-4
Stabili L., Parisi M. G., Parrinello D., Cammarata M. (2018). Cnidarian interaction with microbial communities: From aid to animal's health to rejection responses. Mar. Drugs 16 (9), 296. doi: 10.3390/md16090296
Studer G., Rempfer C., Waterhouse A. M., Gumienny R., Haas J., Schwede T. (2020). QMEANDisCo - distance constraints applied on model quality estimation. Bioinformatics 36, 1765–1771. doi: 10.1093/bioinformatics/btz828
Takeuchi K., Takahashi H., Sugai M., Iwai H., Kohno T., Sekimizu K., et al. (2004). Channel-forming membrane permeabilization by an antibacterial protein, sapecin: determination of membrane-buried and oligomerization surfaces by NMR. J. Biol. Chem. 279 (6), 4981–4987. doi: 10.1074/jbc.M307815200
Tamura K., Stecher G., Kumar S. (2021). MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38 (7), 3022–3027. doi: 10.1093/molbev/msab120
Technau U., Steele R. E. (2011). Evolutionary crossroads in developmental biology: Cnidaria. Development 138 (8), 1447–1458. doi: 10.1242/dev.048959
Tian C., Gao B., Fang Q., Ye G., Zhu S. (2010). Antimicrobial peptide-like genes in nasonia vitripennis: a genomic perspective. BMC Genomics 11, 187. doi: 10.1186/1471-2164-11-187
Vargas P., Zardoya R. (2012). El árbol de la vida: sistemática y evolución de los seres vivos (Vargas, Pablo).
Vélez A., Mera C., Orduz S., Branch J. W. (2021). Synthetic antimicrobial peptides generation using recurrent neural networks. Dyna 88 (216), 210–219. doi: 10.15446/dyna.v88n216.88799
Veron J. E. (2001). The state of coral reef science. Science 293 (5537), 1996–1997. doi: 10.1126/science.293.5537.1996b
Vishnepolsky B., Zaalishvili G., Karapetian M., Nasrashvili T., Kuljanishvili N., Gabrielian A., et al. (2019). De novo design and In vitro testing of antimicrobial peptides against gram-negative bacteria. Pharm. (Basel) 12 (2), 82. doi: 10.3390/ph12020082
White S. H., Wimley W. C. (1998). Hydrophobic interactions of peptides with membrane interfaces. Biochim. Biophys. Acta 1376 (3), 339–352. doi: 10.1016/S0304-4157(98)00021-5
Yap W. Y., Hwang J. S. (2018). Response of cellular innate immunity to Cnidarian pore-forming toxins. Molecules 23 (10), 2537. doi: 10.3390/molecules23102537
Keywords: defensins-like peptides, peptides’ three-dimensional structure, peptides’ physicochemical properties, antimicrobial activity, invertebrates
Citation: Leal E, Múnera M and Suescún-Bolívar LP (2022) In silico characterization of Cnidarian’s antimicrobial peptides. Front. Mar. Sci. 9:1065717. doi: 10.3389/fmars.2022.1065717
Received: 10 October 2022; Accepted: 29 November 2022;
Published: 19 December 2022.
Edited by:
Qiong Shi, BGI Academy of Marine Sciences, ChinaCopyright © 2022 Leal, Múnera and Suescún-Bolívar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Luis Parmenio Suescún-Bolívar, biomolgen@hotmail.com
†These authors have contributed equally to this work
 Edwar Leal
Edwar Leal Marlon Múnera
Marlon Múnera Luis Parmenio Suescún-Bolívar
Luis Parmenio Suescún-Bolívar