Investigation of the molecular mechanisms which contribute to the survival of the polychaete Platynereis spp. under ocean acidification conditions in the CO2 vent system of Ischia Island (Italy)
- 1Department of Biosciences University of Milan, Milan, Italy
- 2Department of Integrative Marine Ecology, Ischia Marine Centre, Stazione Zoologica Anton Dohrn, Ischia (Naples), Italy
- 3Unitech OMICs, Mass Spectrometry Platform, University of Milan, Milan, Italy
- 4Department of Pharmacological and Molecular Sciences, University of Milan, Milan, Italy
- 5National Institute of Oceanography and Applied Geophysics, OGS, Trieste, Italy
The continuous increase of CO2 emissions in the atmosphere due to anthropogenic activities is one of the most important factors that contribute to Climate Change and generates the phenomenon known as Ocean Acidification (OA). Research conducted at the CO2 vents of Castello Aragonese (Ischia, Italy), which represents a natural laboratory for the study of OA, demonstrated that some organisms, such as polychaetes, thrive under acidified conditions through different adaptation mechanisms. Some functional and ecological traits promoting tolerance to acidification in these organisms have been identified, while the molecular and physiological mechanisms underlying acclimatisation or genetic adaptation are still largely unknown. Therefore, in this study we investigated epigenetic traits, as histone acetylation and methylation, in Platynereis spp. individuals coming from the Castello vent, and from a nearby control site, in two different periods of the year (November-June). Untargeted metabolomics analysis was also carried out in specimens from the two sites. We found a different profile of acetylation of H2B histone in the control site compared to the vent as a function of the sampling period. Metabolomic analysis showed clear separation in the pattern of metabolites in polychaetes from the control site with respect to those from the Castello vent. Specifically, a significant reduction of lipid/sterols and nucleosides was measured in polychaetes from the vent. Overall results contribute to better understand the potential metabolic pathways involved in the tolerance to OA.
1. Introduction
Anthropic pressures on marine environments are dramatically reducing biodiversity and ecosystem functionality. Currently, there is still uncertainty concerning the actual impacts of environmental disturbances at different hierarchical level of biological organization, from single species up to the community composition. Therefore, within this scenario the challenge is to identify mechanisms by which organisms respond to changes which lead some species to become “winners” and other “loosers” in future ocean ecosystems (Watson, 2018). Ocean acidification (OA) is one of the most relevant environmental threat to marine biodiversity. Therefore, understanding how organisms cope with low pH is extremely important to foster conservation and restoration of marine ecosystems under such global stressor. Although studies on OA have seen an important increase in the last 10 years, the great majority of them have been short- or middle-term experiments conducted in laboratory/mesocosms conditions, usually on a single organism per time and on a single stage of their life cycle. If in a hand they were useful to understand the physiological effects of OA and the tolerance window of a certain organism, in the other one they do not give enough information about the adaptation potential of a natural population or of a community of organisms to OA. In this context, the study of populations naturally subjected to acidified conditions- such as those living in CO2 vent systems- can provide essential information to understand resistance and resilience of organisms and so to make comprehensive predictions of ecological processes which can occur in future acidified oceans (Foo et al., 2018; González-Delgado and Hernández, 2018).
Naturally acidified systems, such as CO2 vents, although cannot be considered perfect analogues to study the ocean acidification problem at large and global scale, due to their limited extension and depth and to variability in space and time in pH/carbonate chemistry conditions, give useful information on long term effects to exposure to high pCO2/low pH and carbonate chemistry alterations, on natural populations of various organisms. Therefore they represent a natural laboratory and model system for experimental studies, which allow to integrate effects on biogeochemical cycles over hundreds years or millennia contributing in identifying winner and looser organisms, test model species and scale-up laboratory and mesocosm experiments to test ecophysiological limits and performances, show what major ecological features and species interactions occur along gradients of increasing pCO2/low pH conditions.
The CO2 vent systems off the Castello Aragonese at the Ischia island represents one of the best studied vent systems in the world (Hall-Spencer et al., 2008; Foo et al., 2018). The vents, distributed on both sides of the Castello islet, show a well-defined gradient of OA, due to different abundance and intensity of gas bubbling from the bottom, and going from mean values <7.4 units into intermediate values of 7.8-7.9 and finally normal pH conditions (8.1) far from the bubbling. The vents host a rich cover of seaweeds of the shallow rocky reefs and the seagrass Posidonia oceanica habitat (Foo et al., 2018). Associated to these macrophytes, there are many invertebrates, some of which occur also in the most acidified areas of the vents and which have been object of many ecological studies (see Foo et al., 2018 for a review). Among the invertebrates, polychaetes are one of the dominant groups, with up to more than 150 species recorded in the whole Castello area (Kroeker et al., 2011; Gambi et al., 2016; Gambi et al., 2019; unpublished data).
A first reciprocal transplant experiment carried out in the Castello vents showed that some polychaete species are genetically adapted to pCO2/low pH, while other employ physiological plasticity to persist under OA conditions (Calosi et al., 2013a). Conversely, a study by Lucey and co-authors (2016) pointed out that for the calcifying spirorbid polychaete Simplaria sp. neither local adaptations nor phenotypic plasticity could support the abundance of the species in the Castello vents, but that instead the species might be favored by some life traits and likely exploit the buffering capacity of Posidonia oceanica to persist to pCO2/low pH. Finally, some ecological and life history traits emerged as be favorable to thrive under low pH conditions, such as a filter feeding and herbivore feeding habit, small size and brooding of offspring (Gambi et al., 2016).
However, only a few of these species occur in relatively high abundances all along the gradient, including the most acidified zones, and are present all year around (Ricevuto et al., 2014). Among such taxa, two sibling species representatives of the genus Platynereis, P. cf dumerilii and P. cf massiliensis, are particularly interesting since both species have been intensively utilised as model species (mainly Platynereis dumerilii) (see the review by Özpolat et al., 2021). At the Ischia vents both taxa have been investigated as regards their distribution (Ricevuto et al., 2014), reproductive biology and genetic differentiation around the vents (Lucey et al., 2015; Wäge et al., 2017). The two sibling species which are morphologically indistinguishable, show a striking differences in their reproductive biology, with P. cf dumerilii as a free spawner, semelparous form with epitokous transformation at maturation, while P. cf massiliensis is a brooding (inside the tube), iteroparous and without epitoke’ transformation in mature worms (Wäge et al., 2017; Özpolat et al., 2021). Both species occur in the vents, although P. cf massiliensis seems to be more abundant, and in some vent systems restricted, to the acidified areas, while P. cf dumerilii occurs both inside and outside the vents (Vizzini et al., 2017; Wäge et al., 2017; Valvassori et al., 2019).
Besides, some ecophysiological responses to OA stress by mean of reciprocal transplant experiments have been investigated. These studies have included investigations on metabolic rates (Calosi et al., 2013a), antioxidant capacity (Ricevuto et al., 2015b), gene expression, and metabolic machinery (Wäge et al., 2018), and antioxidant efficiency (Valvassori et al., 2019). Specifically, the study by Wäge et al. (2018) showed that individuals of Platynereis collected from the vent showed a lower transcription of the genes sodium- hydrogen antiporter (NHE) involved in acid-base regulation and in paramyosin, involved in cytoskeleton processes, while the NADH dehydrogenase gene, involved in energy metabolism, was significantly up-regulated. Moreover, Platynereis inhabiting the Castello vent displayed enhanced basal antioxidant efficiency compared to organisms living under ambient pH conditions (Ricevuto et al., 2015a), even though the activity of several antioxidant enzymes was not increased (Valvassori et al., 2019).
Up to now, the potential molecular mechanisms underpinning the tolerance of the species to OA remain mostly unknown. To overcome this gap of knowledge, this study aimed to investigate some molecular and metabolic endpoints that could underlie the adaptive response to OA in the polychaete Platynereis spp. in the CO2 vent site of Ischia Island. In particular, we evaluated the potential involvement of epigenetic mechanisms in the adaptive response of these polychaetes to OA, through the analysis of histone modifications. Environmental epigenetics is at its infancy stage, but there is evidence that epigenetic modifications play a central role in the organisms’ adaptation to several environmental stressors (Eirin-Lopez and Putnam, 2019). Epigenetic modifications can contribute to physiological plasticity, but can also promote genetic adaptation (Jeremias et al., 2018). Two recent studies carried out on tropical corals showed that DNA methylation enhanced the ability to cope with OA (Putnam et al., 2016; Liew et al., 2018). In this view, the analysis of epigenetic changes, could provide relevant insight into the molecular mechanisms which promote the development of adapted phenotypes. Besides, we assessed the potential changes occurring in the metabolome of these polychaetes. Some studies carried out in population of ectotherms living in the CO2 vent system of Ischia showed that adaptation to OA influences specific metabolic pathways such as the energetic and antioxidant and immune metabolism (Calosi et al., 2013a; Ricevuto et al., 2015a; Migliaccio et al., 2019). From these evidences, through the application of untargeted metabolomics analysis we aimed to achieve a broader picture of the metabolic changes occurring in the polychaetes under OA conditions. In particular, this analysis allows to understand two different aspects such as: the metabolic state and performance of organisms under acidic stress and the potential metabolic pathways involved in the acclimatization to OA.
2. Methods
2.1. Polychaete collection
Individuals of Platynereis spp. were collected in November 2019 and June 2020 for epigenetic analysis and a further collection has been carried out in September 2021 for metabolomics analysis. Organisms were collected from sites of the Castello Aragonese CO2 vent system, named S2/S3 in previous studies (e.g., Calosi et al., 2013a; Ricevuto et al., 2015a; Ricevuto et al., 2015b) centered in 40°43’57.9” N, 13°57’51.8” E on the south side of the Castello (from now on named Vent). According to physico-chemical characterization carried out or summarized in previous studies (e.g., Hall-Spencer et al., 2008; Kroeker et al., 2011; Foo et al., 2018) the mean pH at this sites, is around 7.8, representing the realistic condition that could be reached by the end of this century. As reference, polychaetes were also collected from a non-acidified area (pH ~ 8.1) located at San Pietro promontory 40°44’47.6” N, 13°56’40.42” E, about 4 km from the vents and having hydrodynamic conditions, temperature and salinity similar to that of the vent site (Ricevuto et al., 2015a; Migliaccio et al., 2019) (from now on named Ctrl) (Figure 1).
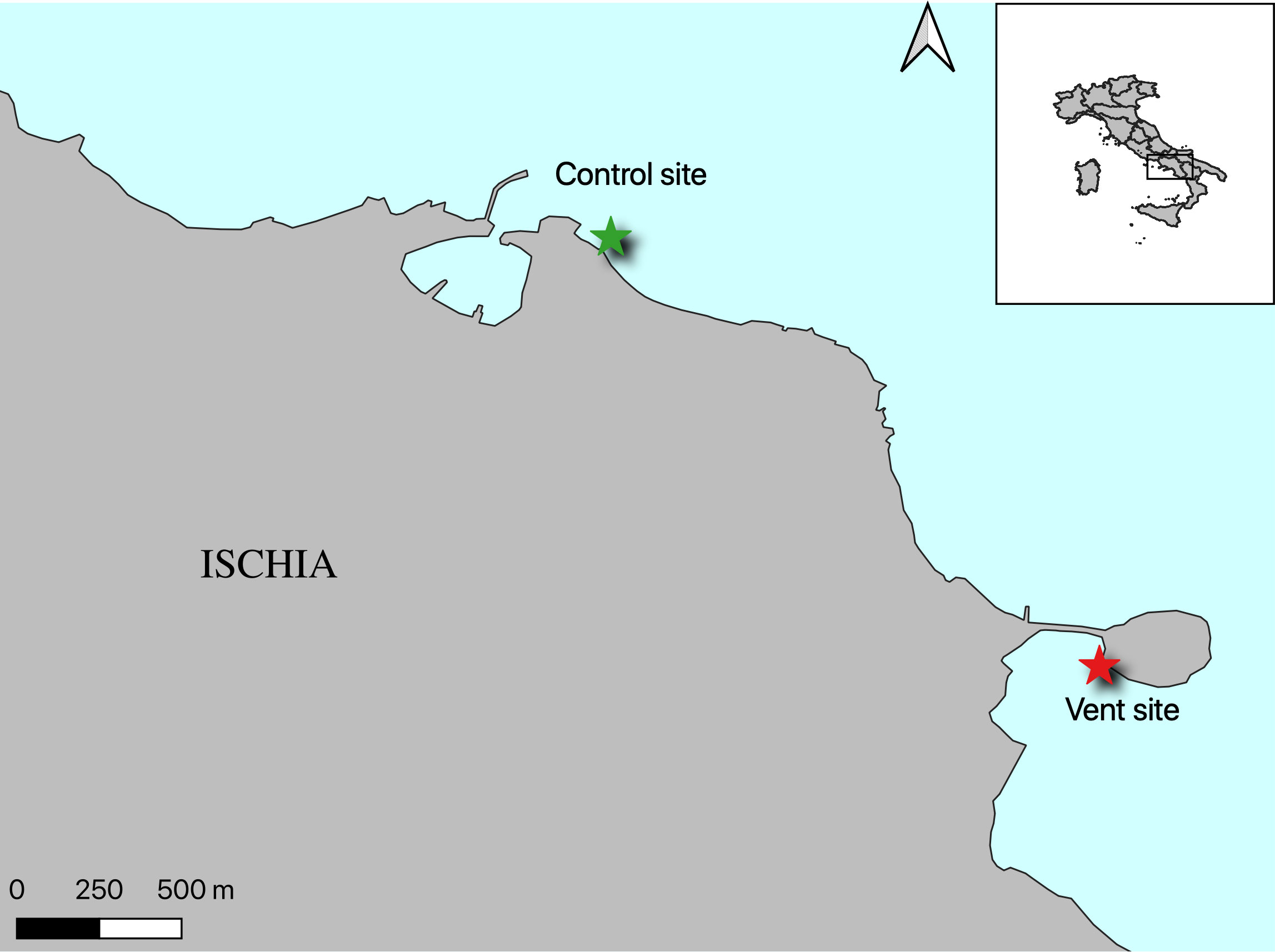
Figure 1 Location of the two sampling sites: Vent site at the Castello Aragonese (40°43’57.9”N - 13°57’51.8”E), Control site at San Pietro Point (40°44’47.6’’ N; 13°56’40.42’’E).
Since these polychaetes live in association with macroalgae species, mainly Halopteris scoparia and Dictyota spp., the macroalgae were collected by hand by scuba divers at 1-2 m depth in all collection sites and put in fabric bags subsequently inserted in larger plastic bags to avoid the animals to escape.
The collected material was transported to the lab within one hour from sampling and each fabric bag was placed on a 10 l bucket under current sea water flow so that the material was kept well oxygenated. Worms were sorted in the following 3-4 hours, and identified at the stereomicroscope. Individuals of Platynereis spp. were pooled in various sample replicates, and stored at -80°C prior to epigenetics and metabolomics analysis.
2.2. Western blot analysis for histone modifications
Three pools of 10 individuals from each site were homogenized in 100 µL of RIPA buffer (10 mM Tris HCl pH 8.0.1 mM EDTA, 0.5 mM EGTA, 0.1% SDS, 0.1% sodium deoxycholate, 140 mM NaCl, 1% Triton X-100, 1 mM protease inhibitor cocktail, phenilmethylsulfonyl fluoride) with TissueLyserII QIAGEN® set at a frequency of 30/s for 90 sec each and then centrifuged at 12,000 x g at 4°C for 10 min. The supernatants were stored at -80°C. The protein content was determined following the method described by Bradford (1976), using bovine serum albumin (BSA) as standard (0.1-0.5 mg mL-1 r2 > 0.99).
Western blot analysis has been carried out according to standard (Libetti et al., 2020) method properly adapted for Platynereis. Five or ten µg (for histone modifications) of extracts were loaded on a 5–14% SDS-polyacrylamide gel with 10 µL of loading buffer (200 mM Tris HCl pH 6.8, 8% SDS, 40% glycerol, 20% β mercaptoethanol). Proteins were transferred to a nitrocellulose membranes, which were incubated in milk 3% dissolved in TBST for 45 minutes at room temperature, to avoid non-specific binding, and with primary rabbit antibodies diluted (diluted 1:2000 in TBST and BSA 4%) overnight at 4°C on a shaker. The primary antibodies used for this analysis were specific for the detection of the following histones: αH2B and its modification H2BK16Ac (Active motif 39121), H3 and its respective modifications H3K27Ac (Active motif 39135) and H3K4Me2 (Abcam 32356). After three washes with TBST, membranes were incubated with the secondary horseradish peroxidase-conjugated anti-rabbit antibodies (diluted 1:2000 in TBST and Milk 3%) for 1 h at room temperature. Band detection was carried out using a Protein Detection System (Genespin) and a ChemiDoc (Bio-Rad). Gel images are shown as SI (Figure S1). Images were exported using Image-Lab software (Bio-Rad) and were analyzed through ImageJ software, in order to calculate the ratio between the histone modification and the histone in each sample.
The differences on histone modifications were tested using a non-parametric PERMutational multivariate Analysis Of Variance (PERMANOVA, Anderson, 2001) applied on the Euclidean distance matrix of square root transformed data, including two crossed factors: site fixed with two levels (vents, control) and season fixed with two levels (November, June). In cases where results were significant, PERMANOVA was used to test for the interactive effect of site and season. The software package PRIMER 6 PERMANOVA Plus (PRIMER-E Ltd, Plymouth, UK was used for statistical analyses.
2.3. Untargeted metabolomics
The analysis was carried out on 4 pools of 30 individuals from each site. Metabolites were extracted from lyophilized polychaete pooled tissues with 500 µL of methanol:ethanol mixture (1:1, v:v). Samples were vortexed for 30 seconds, shaked at 37°C for 90 minutes, centrifuged at 14,000 x g for 10 min to 4°C and the organic phase was evaporated under a stream of nitrogen. The residues were dissolved in 50 μL of methanol and five microliters of each sample extracted were analysed in LC-MS/MS.
All analysis were performed at the Unitech OMICs platform (University of Milano, Italy) using an ExionLC™ AD system connected to TripleTOF™ 6600 System equipped with Turbo V™ Ion Source with ESI Probe (SCIEX, MA, USA. Samples were separated on CORTECS UPLC T3 C18 - 2.1 x 150 mm x 1.6 µm (Waters®). The temperature was set at 40°C.The analytes were eluted with the following gradient: from 99% buffer A (0.1% formic acid in water) to 95% buffer B (0.1% formic acid in acetonitrile) in 8 min. Constant flow rate: 400 µL min−1. Total run: 20 min. MS spectra were collected, in both positive and negative polarities, in Full-Mass Scan from 50 to 1500 Da (100 ms accumulation time) and in IDA® mode (Information Dependent Acquisition) from 50 to 1500 Da (40 ms accumulation time, top 20 spectra per cycle 0.95 s). Nitrogen was used as a nebulizing gas (GS1, 40 psi), turbo spray gas (GS2, 55 psi), and curtain gas (CUR, 35 psi). Spray Voltage was fixed at 4.5 kV (-4.5 kV in negative mode), declustering potential (DP) was 60 eV, the collision energy was 30 eV with a collision energy spread (CES) of 15 eV and source temperature was 550°C.
The data were analyzed using SCIEX OS 1.4 software (SCIEX™), together with two processing functions: 1) FormulaFinder: to identify possible compound formula based on TOF-MS spectra (compound molecular weight); 2) LibraryView™ (ver 1.0): to search MS/MS spectra with built-in accurate mass spectral libraries (Natural Products and Metabolite High Resolution MS/MS Spectral Library) which contain over 1000 compounds. Each sample was injected in triplicate. The average value of the corresponding areas is reported, then normalized for the mg of extracted sample. The identifications (ID) were obtained based on the value m/z (parent ion) achieved and determined in high resolution. Statistical analysis was carried out by MetaboAnalyst ver. 5.0.
3. Results
3.1. Histone post translational modifications (Histone PTMs)
Regarding histone H2B and in particular the lysine K16, the H2BAc/H2B ratio did not show significant effects related to the period or related to the site, while the interaction of both factors resulted in significant differences (Table S1; PERMANOVA). Specifically, a significant decrease in the ratio H2BAc/H2B has been observed in organisms from the control site collected in June with respect to November (p(MC)= 0.007, t= 4.954 Figure 2A); whereas no differences were detected in the individuals from the CO2 vent site with respect to the different sampling time. A significant difference in H2BAc/H2B ratio was also observed between polychaetes from the control and the vent site in June (p(MC)= 0.0078, t= 4.9196), with lower levels in individuals from the control site with respect to those from the vent.
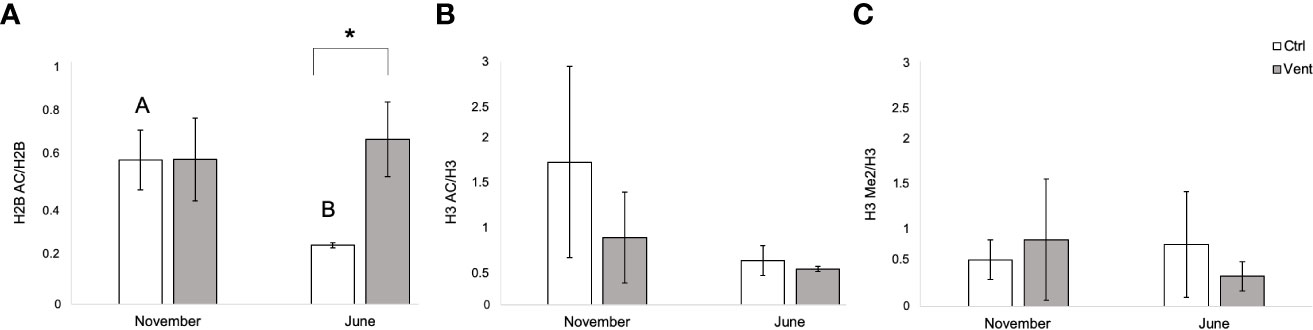
Figure 2 Ratio of histone modifications H2BK16Ac/H2B (A), H3K27Ac/H3 (B) and H3K4Me2/H3 (C) measured through western blotting in in Platynereis spp. collected in November and in June at the non-acidified control site and in the CO2 vents of Castello Aragonese. Different letter means statistically significant differences among polychaetes from the two sites (p ≤ 0.05). The asterisk (*) means statistically difference among polychaetes from the same site at different collecting periods.
A similar trend related to the sampling period has been observed also for the acetylation of the lysine K27 of histone H3, with a decrease of the ratio in the month of June in the individuals sampled in the control site with respect to the ratio measured in November, albeit the difference was not statistically significant (Figure 2B). Concerning the methylation of the lysine K4 of histone H3 there were no significant differences of the ratio, neither correlated with the period nor with the sampling site (Figure 2C).
3.2. Effects on the metabolome
A clear separation in the profile of metabolites in polychaetes from the control site with respect to those from the Castello vent has been observed (Figure 3A). PLS-DA analysis showed 50 metabolites with Variable Importance in Projection (VIP) >1, belonging mostly to nucleosides, amino acids and osmolytes, and lipids/sterols (Figure 3B).
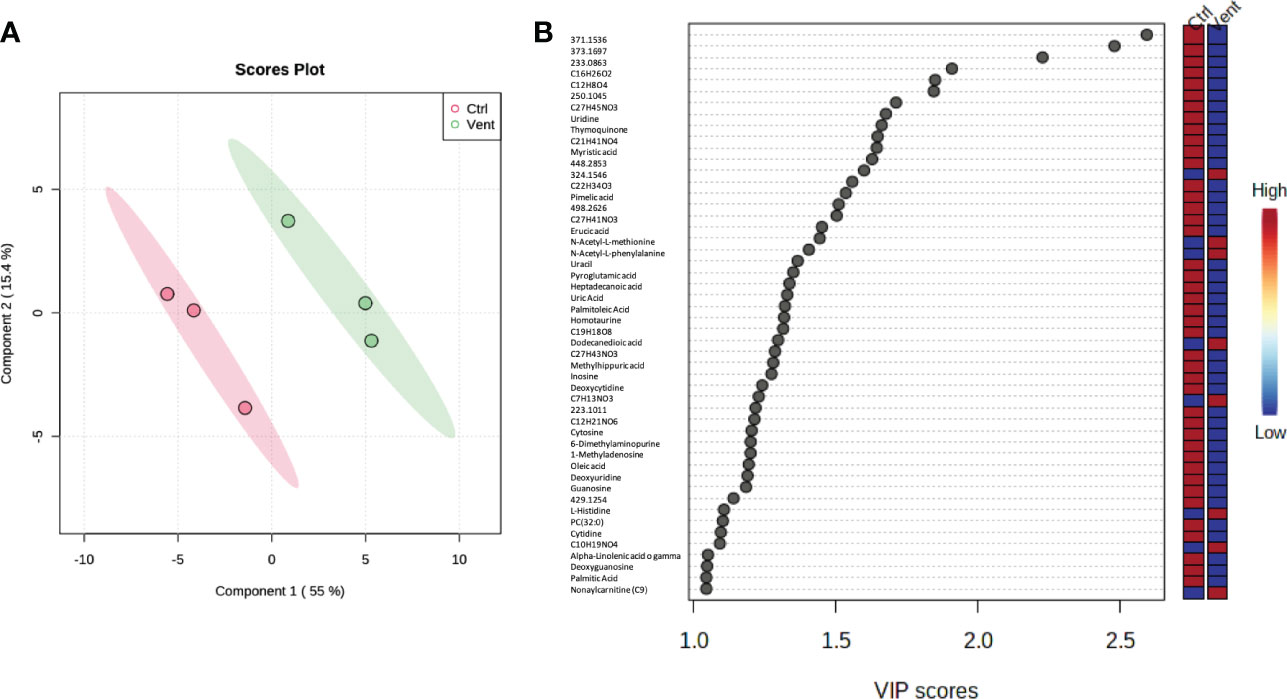
Figure 3 Scores plot between the selected PCs (A), the explained variances are shown in brackets. Important features identified by PLS-DA (B). The colored boxes on the right indicate the relative concentrations of the corresponding metabolite in each group.
The statistical analysis showed a significant modification in the level of 11 ID known and 8 unidentified metabolites (Table 1). Concerning the known metabolites, lipids/sterols, nucleosides and amino acids were significantly different among the two populations (control vs vent). Some fatty acids resulted significantly down-regulated in organisms from the vent with respect to the control. Several purines and pyrimidines were less abundant in organisms from the Castello vent respect to control. A lower content of homotaurine, thymoxiquinone and uric acid was also observed in polychaetes of the vent, with respect to organisms from control. Conversely the amino acids L-hystidine showed a higher content in organisms from the CO2 vent.
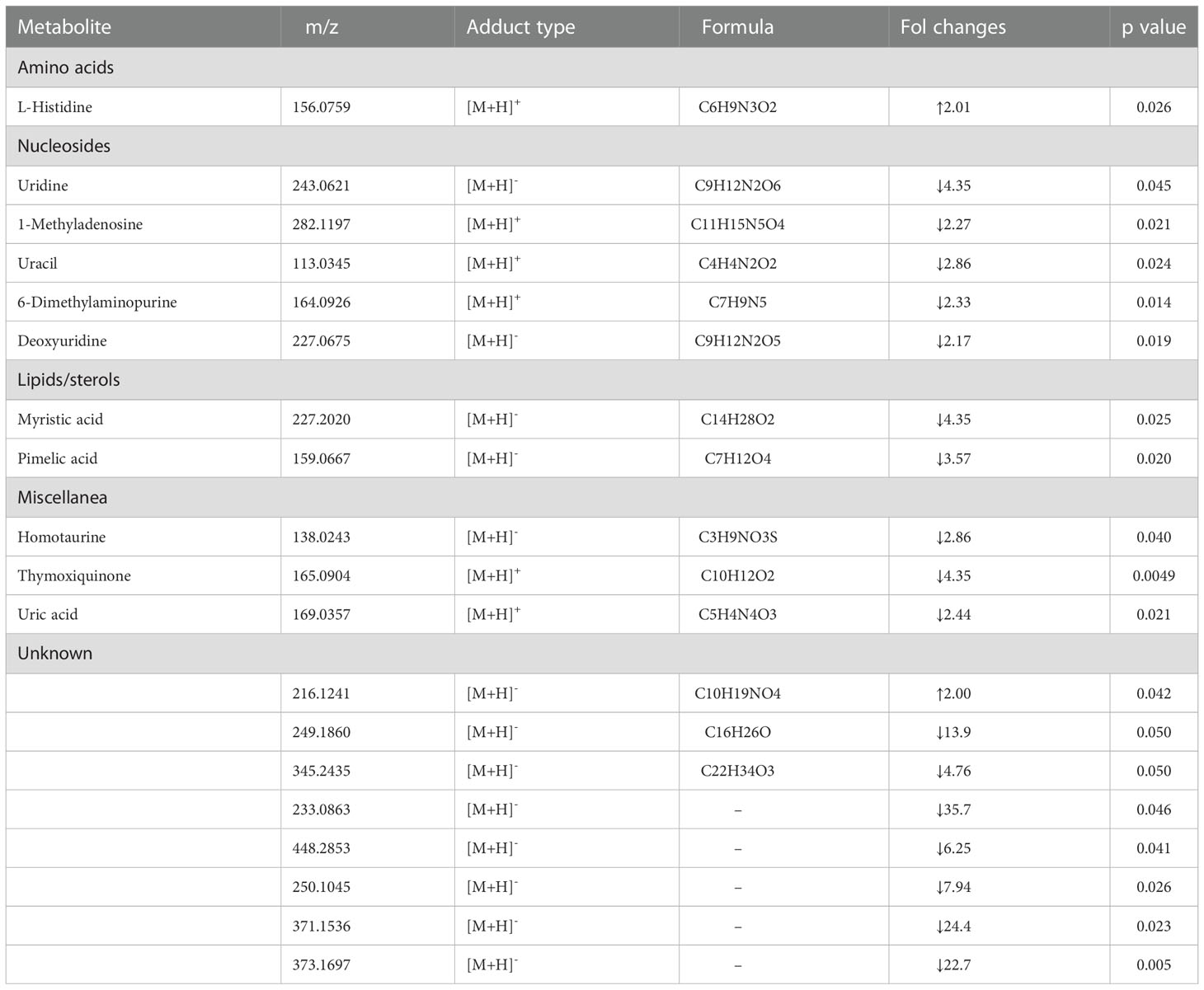
Table 1 Changes in the abundance of metabolites measured in individuals of Platynereis spp. collected in the Castello vent site with respect to the non-acidified San Pietro site.
4. Discussion
The tolerance/adaptation of organisms to OA is a complex puzzle made by several pieces concerning mechanisms and processes, which remain mostly unknown. Therefore, this study aims to improve the current understanding of molecular and metabolic mechanisms that could underlie the adaptive response to OA in the polychaetes Platynereis spp. from the Castello CO2 vent site of the Ischia Island.
Upon exposure to environmental stress, epigenetic mechanisms are identified to play a key role to maintain physiological and biological functions of the organisms (Turner, 2000; Jeremias et al., 2018). So far, the majority of the studies carried out on epigenetics in marine species focused on mollusks, cnidarians, crustaceans, echinoderms and on some fish species (reviewed by Eirin-Lopez and Putnam, 2019). In particular, recent studies carried out on the tropical coral Stylophora pistillata and on the pteropod Limacina helicina antarctica, pointed out that DNA methylation might enhance the ability to cope with OA (Liew et al., 2018; Bogan et al., 2020). As far as polychaetes, a study carried out on the Antarctic Spiophanes tcherniai assessed a correlation between the metabolic rate and the pattern of DNA methylation, linked to an increase of temperature (Marsh and Pasqualone, 2014). In all these studies, DNA methylation was the main epigenetic modification investigated. However, histone post-translational modifications (PTMs) may also play a relevant role in organisms’ adaptation to environmental stressors (Fallet et al., 2020). Indeed, the increase of histone acetylation in response to environmental stressors have been documented in different plants species. For instance, it has been observed that drought and heat stress increase the acetylation of histone H3 lysine 9 and histone H4 lysine 5 levels in maize and rice (Li et al., 2014; Zhao et al., 2014a; Zhao et al., 2014b; Wang et al., 2014). The involvement of histone acetylation in behavioral plasticity, including stress adaptation has been also observed in adult rats (Tsankova et al., 2006; Weaver et al., 2004; Chandramohan et al., 2007) and mice (Fischer et al., 2007) and in Cobb chicks (Kisliouk and Meiri, 2009). Besides, Hu et al. (2019) identified the histone acetyltransferase CBP-1 as one of the epigenetic regulators in heath stress in the nematode Caenorabditis elegans. Conversely, very few studies investigated this epigenetic mechanism in marine organisms. Gibson et al. (2011) focused on the potential presence of characteristic changes in these modifications throughout ontogeny (at representative stages from oocyte to adult) in the marine annelid Polydora cornuta. In particular, three modifications were detected throughout development (H3K14ac, H3K9me, and H3K4me2), indicating that histone modifications are present in a marine invertebrate and that they showed peculiar changes correlated with tissue differentiation and with life stages. In the Eastern oysters Crassostrea virginica exposed to the dinoflagellate Karenia brevis an increase in histone H2A.X phosphorylation (γH2A.X) has been measured likely related to the increase of oxidative damage induced by the brevetoxins produced by K. brevis (González-Romero et al., 2017). In another study, Fellous and co-authors (2015) investigated methylation of H3 histone (H3K4, H3K9 and H3K27 residues) during larval stages of Crassostrea gigas upon exposure to thermal stress (both cold and heath). A significant hypermetilation was the most frequent modification observed, although also hypomethylation was also recorded at the lower temperature.
Based on the findings from previous studies, one would expect an increase of acetylation in summer. Nevertheless, during the summer season there are intense fluctuations of several environmental factors other than temperature, such as solar radiation, oxygen, food availability, algal coverage etc. which might contribute to change the profile of histone PTMs. To the best of our knowledge, only one study focused on the effects of seasonal fluctuation on these epigenetic end-points (Ishihara et al., 2019), where an increase in acetylation level of the histones H3 and H4 in the bullfrog Lithobates catesbeianus was observed during summer. Therefore, our study represents the first attempt to investigate PTMs in a marine species related to OA, and also contribute to improve current knowledge on the potential mechanisms underpinning histone acetylation in marine invertebrates.
Among the different histone PTMs, acetylation and methylation of key lysins were investigated in the present research. These modifications affect the inter-nucleosomal interactions and consequently they alter the overall chromatin structure (Bannister and Kouzarides, 2011).
Our results highlighted a significant reduction of the H2BAc/H2B ratio in the individuals coming from the control site and sampled in the month of June, while no differences were observed between the two different periods of the year for the organisms thriving at the CO2 vent. A similar result was assessed also for the acetylation of the histone H3. Histone acetylation is an epigenetic modification that generally leads to an open chromatin environment and consequently to gene activation (Turner, 2000; Bannister and Kouzarides, 2011; Guida, 2013; Kumar et al., 2021). In fact, the enzyme histone acetyltransferase (HATs) catalyzes the addition of acetyl groups to the lysine residues of the N-terminal tails of histones, which neutralizes the positive charge of this aminoacid and weakens the interactions with the DNA, making chromatin more accessible to enzymes.
Individuals coming from the control site showed a reduction of acetylation of both histone H2B and H3 in the month of June, with respect to November.
Besides, our result is in contrast with what has been observed in the bullfrog Lithobates catesbeianus by Ishihara et al. (2019). Nonetheless, the modification analyzed in the bullfrog referred to different residues of lysine, specifically: the acetylation and methylation of K9 residue and the methylation of K36, which might explain the difference in respect to our results. Moreover, the discrepancy might be also due to the different ecological and physiological traits of polychaetes with respect to amphibians. Indeed, a species-specificity in epigenetic mechanisms related to environmental fluctuations, such as thermal variation, has already been pointed out (McCaw et al., 2022).
Conversely, the seasonal trend of H2BAc observed in organisms coming from the CO2 vent suggests that these individuals seem to be forced in maintaining higher level of acetylation also in the summer season. Hence, this result suggests the potential involvement of this mechanism as an adaptive strategy toward OA in Platynereis spp. This higher level of acetylation of histone H2B may be indicative of a major accessibility of the DNA to enzymes involved in transcription. In fact, the acetylated lysine residues in histone H2B are necessary for the transcriptional activation of some genes, particularly those that play a role in NAD biosynthesis or vitamin metabolism (Parra et al., 2006; Kupkova et al., 2021). But this modification may also affect other important DNA processes, such as replication, recombination and repair of DNA damages (Turner, 2000; Bannister and Kouzarides, 2011; Hofmann, 2017; Eirin-Lopez and Putnam, 2019).
The lack of modulation of H3 methylation, neither correlated to the period nor to the sampling site, suggests that this specific histone modification might play a minor role in seasonal acclimatization, as already described in bullfrogs (Ishihara et al., 2019). Otherwise, this result might be due to the fact that other residues of lysine may be involved, through other PTMs that might counterbalance the effect.
Since histones are highly conserved across eukaryotic species (Mariño-Ramirez et al., 2006), our result highlights the importance of increasing the current knowledge concerning histone PTMs in marine wild populations naturally subjected to extreme environmental conditions, to contribute to the current understanding of the role of this epigenetic mechanism in shaping the tolerance/adaptation of organisms to future global changes. Indeed, it would also be important to understand whether these alterations occur at the expense of key physiological functions or whether such mechanisms might entail tolerance towards environmental disturbances.
The application of untargeted metabolomics in polychaetes has been carried out in few studies so far, focused mostly on the evaluation of the effects on the metabolic profile of different environmental pollutants such as hydrocarbons, microplastics (Missawi et al., 2022) and metals (Sinclair et al., 2019). On the contrary, no information is currently available regarding the effects of environmental stressors related to climate changes on the metabolome of polychaetes.
Concerning our results, the classes of metabolites that contributed most to distinguish the metabolic profile of vent organisms compared to controls are amino acids and osmolytes, fatty acids and nucleosides.
An imbalance of purine and pyrimidine metabolism seems to occur in polychaetes from the vent, also confirmed by the reduction in these organisms of uric acid, which is the end product of purine catabolism (Maiuolo et al., 2016). A similar down regulation of purine and pyrimidine metabolism has been observed in juveniles of the Dungeness crab subjected to low pH (7.45) for 32 days (Trigg et al., 2019) suggesting that this cellular mechanism could be a target of OA, which could lead to an alteration of the synthesis of RNA and DNA and of the DNA repair mechanism. This result agrees with our observations concerning acetylation of histone H2B.
The content of some fatty acids was reduced in polychaetes from the vent site compared to controls. This suggests an increase of fatty acid beta oxidation to boost energy necessary to meet the energetic demand requested to survive under OA conditions. Our findings are consistent with what has been observed for populations of purple sea urchin (Strongylocentrotus purpuratus) locally adapted to upwelling phenomenon, leading to low pH, which showed an up-regulation of genes involved in fatty acids beta oxidation (Evans et al., 2017). In line with our results, a strong decrease of fatty acid content has been measured also in larvae of the American lobster (Homarus americanus) subjected to OA (Noisette et al., 2021). These findings are in agreement also with observations made in a previous study by Calosi et al. (2013a). The authors observed that Platynereis and other polychaete species living in the Castello vent displayed a higher metabolic rate with respect to organisms living under non-acidified conditions; at the same time, Platynereis worms had significantly lower size than those of the control site, likely due to allocation of energy reserve to maintain basic metabolism penalizing growth. In the present study, however, a measurement of the size of the worms was not made, to confirm this.
The modulation of amino acids observed in organisms from the vent with respect to controls might be correlated with the need of organisms to keep osmoregulation and acid-base regulation under hypercapnia, as already suggested for the Dungeness crab by Trigg et al. (2019) and could be responsible for the non-bicarbonate buffering capacities, which protect extracellular pH during acute CO2 exposure (Melzner et al., 2009). Indeed, the ability of an organism to maintain the acid-base balance and the ionic regulation is considered a key feature that allow to persist under high pCO2/low pH conditions (Calosi et al., 2013b; Lewis et al., 2016) and our results suggest that this condition could occur also in Platynereis. Moreover, histidine has an antioxidant role, being a scavenger of reactive oxygen species (Wade and Tucker, 1998). Since OA is considered a prooxidant stressor, based on in vivo and in situ studies (Ricevuto et al., 2016 and citation therein), the observed increase in histidine suggests that the organisms activated a homeostatic response to counteract the prooxidant condition generated by OA, which could contribute to boost their ability to survive in the vent system.
A significant decrease of homotaurine was also observed in Platynereis spp. of the CO2 vent system, with respect to those of the control. Homotaurine is a homolog of taurine naturally present in several seaweeds (Terriente-Palacios and Castellari, 2022), hence, a low content of this metabolite might suggest a different food availability or different assimilation/consumption processes for the two populations. To this respect, food availability, represented by Platynereis spp. by various macroalgae and epiphytes of Posidonia oceanica where they live as mesograzer (Gambi et al., 2000), in the vents show a higher coverage and higher seasonal persistence, respect to control (Ricevuto et al., 2015b). In addition, nitrogen content in plant tissues under OA conditions showed higher values respect to control (Ricevuto et al., 2015b). Therefore, the pattern of homotaurine, observed in worms from the vents, could be somehow related to this feature.
Given the key role of this osmolyte in osmoregulation and protection against oxidative stress (Yancey, 2005), further researches are warranted to clarify the potential mechanisms underlying the observation.
The changes in the metabolome observed in Platynereis spp. from the Castello vent are not totally consistent with the previous observations made on polychaetes and other invertebrates of the vent, which showed that antioxidant and immune metabolism were involved in the adaptation to OA (Ricevuto et al., 2015a; Migliaccio et al., 2019). In particular, in our study we did not detect metabolites with immunomodulation role, furthermore only few metabolites involved in the antioxidant response were modulated in organisms from the Castello vent with respect to controls in the present study. These discrepancies could be related to the different analytical approach used, namely metabolomics vs biochemical measurements and proteomics (Ricevuto et al., 2015a; Migliaccio et al., 2019). In addition, among the 19 significantly modified metabolites, 8 metabolites were unidentified. In this view, the application of other omic analyses might improve the outcomes of this research. Another explanation for the differences is likely due to the fact that the response to OA stress is known to be species-specific (Wilson-McNeal et al., 2020), thus it is possible that different organisms, such as sea urchins, show a different mechanism of tolerance to OA.
5. Conclusions
Overall results from the analysis of histone modifications and metabolomics suggested that OA interfere in energy-yelding metabolic reactions. Based on the first evidences provided from this study future investigation should look at evaluating whether the physiological adaptation mechanism to OA occur at the expense of life-history traits such as for instance body growth, feeding or reproduction, that could have repercussions at higher hierarchical levels. Our study confirmed the importance of CO2 vent systems as natural analogues to study the effects of OA, since they provide realistic information on the impacts of this phenomenon on marine organisms and contribute to understand the potential of some species to thrive under future-predicted acidified oceans.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author contributions
MM, CDT, and MCG conceptualization; MN, ACh, MM, and AC performed the field samplings; SS, performed epigenetics analysis; MF, FF, and DC performed metabolomics analysis; MF and ACh, data elaboration; SS, MCG, and CDT paper writing; CDT and MM provided resources; all authors contributed to writing and revision of the text and figures. All authors contributed to the article and approved the submitted version.
Funding
The project was financed by the grant Line 2 of the Research Support Plan from the University of Milan. During this study MN was supported by a fellowship from the Stazione Zoologica Anton Dohrn, ACh was supported by a fellowship from the Stazione Zoologica Anton Dohrn in collaboration with the University of Naples Federico II – AFRIMED. During the initial period of this research MCG was senior investigator at the Ischia Marine Center of the Stazione Zoologica Anton Dohrn, Naples (Italy).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.1067900/full#supplementary-material
References
Anderson M. J. (2001). A new method for non-parametric multivariate analysis of variance. Austral Ecol. 26, 32–46. doi: 10.1111/j.1442-9993.2001.01070.pp.x
Bannister A. J., Kouzarides T. (2011). Regulation of chromatin by histone modifications. Cell. Res. 21, 381–395. doi: 10.1038/cr.2011.22
Bogan S. N., Johnson K. M., Hofmann G. E. (2020). Changes in genome-wide methylation and gene expression in response to future pCO2 extremes in the Antarctic pteropod Limacina helicina antarctica. Front. Mar. Sci. 6. doi: 10.3389/fmars.2019.00788
Bradford M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. doi: 10.1006/abio.1976.9999
Calosi P., Rastrick S. P. S., Graziano M., Thomas S. C., Baggini C., Carter H. A., et al. (2013b). Distribution of sea urchins living near shallow water CO2 vents is dependent upon species acid-base and ion-regulatory abilities. Mar. Pollut. Bull. 73, 470–484. doi: 10.1016/j.marpolbul.2012.11.040
Calosi P., Rastrick S. P. S., Lombardi C., de Guzman H. J., Davidson L., Jahnke M., et al. (2013a). Adaptation and acclimatization to ocean acidification in marine ectotherms: an in situ transplant experiment with polychaetes at a shallow CO2 vent system. Phil. Trans. R. Soc B. 368, 20120444. doi: 10.1098/rstb.2012.0444
Chandramohan Y., Droste S. K., Reul J. M. (2007). Novelty stress induces phospho-acetylation of histone H3 in rat dentate gyrus granule neurons through coincident signalling via the n-methyl-D-aspartate receptor and the glucocorticoid receptor: relevance for c-fos induction. J. Neurochem. 101, 815–828. doi: 10.1111/j.1471-4159.2006.04396.x
Eirin-Lopez J. M., Putnam H. M. (2019). Editorial: Marine environmental epigenetics. Front. Mar. Sci. 8. doi: 10.3389/fmars.2021.685075
Evans T. G., Pespeni M. H., Hofmann G. E., Palumbi S. R., Sanford E. (2017). Transcriptomic responses to seawater acidification among sea urchin populations inhabiting a natural pH mosaic. Mol. Ecol. 26, 2257–2275. doi: 10.1111/mec.14038
Fallet M., Luquet E., David P., Cosseau C. (2020). Epigenetic inheritance and intergenerational effects in mollusks. Gene. 729, 144166. doi: 10.1016/j.gene.2019.144166
Fischer A., Sananbenesi F., Wang X., Dobbin M., Tsai L. H. (2007). Recovery of learning and memory is associated with chromatin remodelling. Nature 447, 178–182. doi: 10.1038/nature05772
Foo S. A., Byrne M., Ricevuto E., Gambi M. C. (2018). The carbon dioxide vents of ischia, Italy, a natural system to assess impacts of ocean acidification on marine ecosystems: An overview of research and comparisons with other vent systems. Oceanogr. Mar. Biol. ann. review 56, 237–310. doi: 10.1098/rstb.2012.0444
Gambi M. C., Gaglioti M., Del Pasqua M., Giangrande A. (2019). Polychaetes and ocean acidification: a synthesis on taxonomic and functional diversity at the CO2 vents of Ischia (Italy). Biol. Mar. Mediter. 26, 271–272.
Gambi M. C., Musco L., Giangrande A., Badalamenti F., Micheli F., Kroeker K. J. (2016). Distribution and functional traits of polychaetes in a CO2 vent system: winners and losers among closely related species. Mar. Ecol. Prog. Ser. 550, 121–134. doi: 10.3354/meps11727
Gambi M. C., Zupo V., Buia M. C., Mazzella L. (2000). Feeding ecology of the polychaete Platynereis dumerilii (Audouin & Milne edwards) (Nereididae) in the seagrass Posidonia oceanica system: role of the epiphytic flora. Ophelia 53, 189–202. doi: 10.1080/00785326.2000.10409449
Gibson G., Hart C., Pierce R., Lloyd V. (2011). Ontogenetic survey of histone modification in an annelid. Genet. Res. Int. 2012, 392903. doi: 10.1155/2012/392903
González-Delgado S., Hernández J. C. (2018). The importance of natural acidified systems in the study of ocean acidification: What have we learned? Adv. Mar. Biol. 80, 57–99. doi: 10.1098/rstb.2012.0439
González-Romero R., Suarez-Ulloa V., Rodriguez-Casariego J., Garcia-Souto D., Diaz G., Smith A., et al. (2017). Effects of Florida red tides on histone variant expression and DNA methylation in the Eastern oyster Crassostrea virginica. Aquat. Toxicol. 186, 196–204. doi: 10.1016/j.aquatox.2017.03.006
Guida S. M. (2013). Environmental epigenetics in polychaetes (University of Delaware ProQuest Dissertations Publishing). Available at: https://www.proquest.com/openview/bfffcda14e34b4351b6a63b509fb1129/1?pq-origsite=gscholar&cbl=18750.
Hall-Spencer J. M., Rodolfo-Metalpa R., Martìn S., Ransome E., Fine M., Turner S. M., et al. (2008). Volcanic carbon dioxide vents show ecosystem effects of ocean acidification. Nature 454, 96–99. doi: 10.1038/nature07051
Hofmann G. E. (2017). Ecological epigenetics in marine metazoans. Front. Mar. Sci. 4. doi: 10.3389/fmars.2017.00004
Hu Y., Lu Y., Zhao Y., Zhou D. X. (2019). Histone acetylation dynamics integrates metabolic activity to regulate plant response to stress. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.01236
Ishihara A., Sapon M. A., Yamauchi K. (2019). Seasonal acclimatization and thermal acclimation induce global histone epigenetic changes in liver of bullfrog (Lithobates catesbeianus) tadpole. Comp. Biochem. Physiol. A. 230, 39–48. doi: 10.1016/j.cbpa.2018.12.014
Jeremias G., Barbosa J., Marques S. M., Asselman J., Gonçalves F. J. M., Pereira J. L. (2018). Synthesizing the role of epigenetics in the response and adaptation of species to climate change in freshwater ecosystems. Mol. Ecol. 27, 2790–2806. doi: 10.1111/mec.14727
Kisliouk T., Meiri N. (2009). A critical role for dynamic changes in histone H3 methylation at the Bdnf promoter during postnatal thermotolerance acquisition. Eur. J. Neurosci. 30, 1909–1922. doi: 10.1111/j.1460-9568.2009.06957.x
Kroeker K. J., Micheli F., Gambi M. C., Martz T. R. (2011). Divergent ecosystem responses within a benthic marine community to ocean acidification. Proc. Natl. Acad. Sci. 108, 14515–14520. doi: 10.1073/pnas.1107789108
Kumar V., Thakur J. K., Prasad M. (2021). Histone acetylation dynamics regulating plant development and stress responses. Cell. Mol. Life Sci. 78, 4467–4486. doi: 10.1007/s00018-021-03794-x
Kupkova K., Shetty S. J., Haque R., Petri J., Auble D. T. (2021). Histone H3 lysine 27 acetylation profile undergoes two global shifts in undernourished children and suggests altered one−carbon metabolism. Clin. Epigenet. 13, 182. doi: 10.1186/s13148-021-01173-8
Lewis C., Ellis R. P., Vernon E., Elliot K., Newbatt S., Wilson R. W. (2016). Ocean acidification increases copper toxicity differentially in two key marine invertebrates with distinct acid-base responses. Sci. Rep. 6, 21554. doi: 10.1038/srep21554
Libetti D., Bernardini A., Sertic S., Messina G., Dolfini D., Mantovani R. (2020). The switch from NF-YA1 to NF-YAs isoform impairs myotubes formation. Cells 9, 789. doi: 10.3390/cells9030789
Liew Y. L., Zoccola D., Li Y., Tambutté E., Venn A. A., Michell C. T., et al. (2018). Epigenome-associated phenotypic acclimatization to ocean acidification in a reef-building coral. Sci. Adv. 4. doi: 10.1126/sciadv.aar8028
Li H., Yan S., Zhao L., Tan J., Zhang Q., Gao F., et al. (2014). Histone acetylation associated up-regulation of the cell wall related genes is involved in salt stress induced maize root swelling. BMC Plant Biol. 14, 105. doi: 10.1186/1471-2229-14-105
Lucey N. M., Lombardi C., DeMarchi L., Schulze A., Gambi M. C., Calosi P. (2015). To brood or not to brood: Are marine invertebrates that protect their offspring more resilient to ocean acidification? Sci. Rep. 5, 12009. doi: 10.1038/srep12009
Maiuolo J., Oppedisano F., Gratteri S., Muscoli C., Mollace V. (2016). Regulation of uric acid metabolism and excretion. Int. J. Cardiol. 213, 8–14. doi: 10.1016/j.ijcard.2015.08.109
Mariño-Ramirez L., Jordan I. K., Landsman D. (2006). Multiple independent evolutionary solutions to core histone gene regulation. Gen. Biol. 2, R122. doi: 10.1186/gb-2006-7-12-r122
Marsh A. G., Pasqualone A. A. (2014). DNA Methylation and temperature stress in an Antarctic polychaete, Spiophanes tcherniai. Fron. Physiol. 5. doi: 10.3389/fphys.2014.00173
McCaw B. A., Stevenson T. J., Lancaster L. T. (2022). Epigenetic responses to temperature and climate. Integr. Comp. Biol. 60, 1469–1480. doi: 10.1093/icb/icaa049
Melzner F., Gutowska M. A., Langenbuch M., Dupont S., Lucassen M., Thorndyke M. C., et al. (2009). Physiological basis for high CO2 tolerance in marine ectothermic animals: pre-adaptation through lifestyle and ontogeny? Biogeosciences 6, 2313–2331. doi: 10.5194/bg-6-2313-2009
Migliaccio O., Pinsino A., Maffioli E., Smith A. M., Agnisola C., Matranga V., et al. (2019). Living in future ocean acidification, physiological adaptive responses of the immune system of sea urchins resident at a CO2 vent system. Sci. Total Environ. 672, 938–950. doi: 10.1016/j.scitotenv.2019.04.005
Missawi O., Venditti M., Cappello T., Zitouni N., De Marco G., Boughattas I., et al. (2022). Autophagic event and metabolomic disorders unveil cellular toxicity of environmental microplastics on marine polychaete Hediste diversicolor. Environ. Pollut. 302, 119106. doi: 10.1016/j.envpol.2022.119106
Noisette F., Calosi P., Madeira D., Chemel M., Menu-Courey K., Piedalue S., et al. (2021). Tolerant larvae and sensitive juveniles: Integrating metabolomics and whole-organism responses to define life-stage specific sensitivity to ocean acidification in the American lobster. Metabolites 11, 584. doi: 10.3390/metabo11090584
Özpolat B. D., Randel N., Williams E. A., Bezares−Calderón L. A., Andreatta G., Balavoine G., et al. (2021). The nereid on the rise: Platynereis as a model system. EvoDevo 12, 10. doi: 10.1186/s13227-021-00180-3
Parra M. A., Kerr D., Fahy D., Pouchnik D. J., Wyrick J. J. (2006). Deciphering the roles of the histone H2B n-terminal domain in genome-wide transcription. Mol. Cell. Biol. 26, 3842–3852. doi: 10.1128/MCB.26.10.3842–3852.2006
Putnam H. M., Davidson J. M., Gates R. D. (2016). Ocean acidification influences host DNA methylation and phenotypic plasticity in environmentally susceptible corals. Evol. Appl. 9, 1165–1178. doi: 10.1111/eva.12408
Ricevuto E., Benedetti M., Regoli F., Spicer J. I., Gambi M. C. (2015a). Antioxidant capacity of polychaetes occurring at a natural CO2 vent system: Results of an in situ reciprocal transplant experiment. Mar. Environ. Res. 112, 44–51. doi: 10.1016/j.marenvres.2015.09.005
Ricevuto E., Kroeker K. J., Ferrigno F., Micheli F., Gambi M. C. (2014). Spatio-temporal variability of polychaete colonization at volcanic CO2 vents (Italy) indicates high tolerance to ocean acidification. Mar. Biol. 161, 2909–2919. doi: 10.1007/s00227-014-2555-y
Ricevuto E., Lanzoni I., Fattorini D., Regoli F., Gambi M. C. (2016). Arsenic speciation and susceptibility to oxidative stress in the fanworm Sabella spallanzanii (Gmelin) (Annelida, Sabellidae) under naturally acidified conditions: An in situ transplant experiment in a Mediterranean CO2 vent system. Sci. Tot. Environ. 544, 765–773. doi: 10.1016/j.scitotenv.2015.11.154
Ricevuto E., Vizzini S., Gambi M. C. (2015b). Ocean acidification effects on stable isotope signatures and trophic interactions of polychaete consumers and organic matter sources at a CO2 shallow vent system. J. Exp. Mar. Biol. 468, 105–117. doi: 10.1016/j.jembe.2015.03.016
Sinclair G. M., O’Brien A. L., Keough M., De Souza D. P., Dayalan S., Kanojia K., et al. (2019). Using metabolomics to assess the sub−lethal effects of zinc and boscalid on an estuarine polychaete worm over time. Metabolomics 15, 108. doi: 10.1007/s11306-019-1570-x
Terriente-Palacios C., Castellari M. (2022). Levels of taurine, hypotaurine and homotaurine, and amino acids profiles in selected commercial seaweeds, microalgae, and algae-enriched food products. Food Chem. 368, 130770. doi: 10.1016/j.foodchem.2021.130770
Trigg S. A., McElhany P., Maher M., Perez D., Shallin Busch D., Nichols K. M. (2019). Uncovering mechanisms of global ocean change effects on the dungeness crab (Cancer magister) through metabolomics analysis. Sci. Rep. 9, 10717. doi: 10.1038/s41598-019-46947-6
Tsankova N. M., Berton O., Renthal W., Kumar A., Neve R. L., Nestler E. J. (2006). Sustained hippocampal chromatin regulation in a mouse model of depression and antidepressant action. Nat. Neurosci. 9, 519–525. doi: 10.1038/nn1659
Turner B. M. (2000). Histone acetylation and an epigenetic code. BioEssays 22, 836–845. doi: 10.1002/1521-1878(200009)22:9<836::AID-BIES9>3.0.CO;2-X
Valvassori G., Benedetti M., Regoli F., Gambi M. C. (2019). Antioxidant efficiency of Platynereis spp. (Annelida, Nereididae) under different pH conditions at a CO2 vent’s system. J. Mar. Sci. 2019, 8415916. doi: 10.1155/2019/8415916
Vizzini S., Martínez-Crego B., Andolina C., Massa-Gallucci A., Connell S. D., Gambi M. C. (2017). Ocean acidification as a driver of community simplification via the collapse of higher-order and rise of lower-order consumers. Sci. Rep. 7, 4018. doi: 10.1038/s41598-017-03802-w
Wade A. M., Tucker H. N. (1998). Antioxidant characteristics of l-histidine. J. Nutr. Biochem. 9, 308–315. doi: 10.1016/S0955-2863(98)00022-9
Wäge J., Rotchell J. M., Gambi M. C., Hardege J. D. (2018). Target gene expression studies on Platynereis dumerilii and Platynereis cfr massiliensis at the shallow CO2 vents off ischia, Italy. Estuar. Coast. Shelf. Sci. 207, 351–358. doi: 10.1016/j.ecss.2017.11.012
Wäge J., Valvassori G., Hardege J. D., Schulze A., Gambi M. C. (2017). The sibling polychaetes Platynereis dumerilii and Platynereis massiliensis in the Mediterranean Sea: are phylogeographic patterns related to exposure to ocean acidification? Mar. Biol. 164, 199. doi: 10.1007/s00227-017-3222-x
Wang L., Zhu W., Fang L., Sun X., Su L., Liang Z., et al. (2014). Genome-wide identification of WRKY family genes and their response to cold stress in Vitis vinifera. BMC Plant Biol. 14, 103. doi: 10.1186/1471-2229-14-103
Watson G. J. (2018). “Winners” and “losers” in the anthropocene: Understanding adaptation through phenotypic plasticity. Funct. Ecol. 32, 1906–1907. doi: 10.1111/1365-2435.13108
Weaver I. C., Cervoni N., Champagne F. A., D’Alessio A. C., Sharma S., Seckl J. R., et al. (2004). Epigenetic programming by maternal behavior. Nat. Neurosci. 7:847–854. doi: 10.1038/nn1276
Wilson-McNeal A., Hird C., Hobbs C., Nielson C., Smith C. E., Wilson R. W., et al. (2020). Fluctuating seawater pCO2/pH induces opposing interactions with copper toxicity for two intertidal invertebrates. Sci. Tot. Environ. 748, 141370. doi: 10.1016/j.scitotenv.2020.141370
Yancey P. H. (2005). Organic osmolytes as compatible, metabolic and counteracting cytoprotectants in high osmolarity and other stresses. J. Exper. Biol. 208, 2819–2830. doi: 10.1242/jeb.01730
Zhao L., Wang P., Hou H., Zhang H., Wang Y., Yan S., et al. (2014a). Transcriptional regulation of cell cycle genes in response to abiotic stresses correlates with dynamic changes in histone modifications in maize. PloS One 9 (8), e106070. doi: 10.1371/journal.pone.0106070
Keywords: ocean acidification, polychaetes, metabolomics, histone modifications, adaptation, CO2 vents, Mediterranean Sea
Citation: Signorini SG, Munari M, Cannavacciuolo A, Nannini M, Dolfini D, Chiarore A, Farè F, Fontana M, Caruso D, Gambi MC and Della Torre C (2023) Investigation of the molecular mechanisms which contribute to the survival of the polychaete Platynereis spp. under ocean acidification conditions in the CO2 vent system of Ischia Island (Italy). Front. Mar. Sci. 9:1067900. doi: 10.3389/fmars.2022.1067900
Received: 12 October 2022; Accepted: 30 December 2022;
Published: 18 January 2023.
Edited by:
Peng Jin, University of Guangzhou, ChinaCopyright © 2023 Signorini, Munari, Cannavacciuolo, Nannini, Dolfini, Chiarore, Farè, Fontana, Caruso, Gambi and Della Torre. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Camilla Della Torre, camilla.dellatorre@unimi.it; Marco Munari, marco.munari@szn.it
 Silvia Giorgia Signorini
Silvia Giorgia Signorini Marco Munari
Marco Munari Antonio Cannavacciuolo
Antonio Cannavacciuolo Matteo Nannini
Matteo Nannini Diletta Dolfini
Diletta Dolfini Antonia Chiarore
Antonia Chiarore Fiorenza Farè
Fiorenza Farè Manuela Fontana3
Manuela Fontana3  Donatella Caruso
Donatella Caruso Maria Cristina Gambi
Maria Cristina Gambi Camilla Della Torre
Camilla Della Torre