Chaotic Genetic Patchiness in the Highly Valued Atlantic Stalked Barnacle Pollicipes pollicipes From the Iberian Peninsula: Implications for Fisheries Management
- 1Departamento de Biología Funcional, Universidad de Oviedo, Oviedo, Spain
- 2Departamento de Bioquímica, Genética e Inmunología, Centro de Investigación Mariña (CIM-UVIGO), Universidade de Vigo, Vigo, Spain
- 3Adaptation et Diversité en Milieu Marin (AD2M) UMR 7144, Station Biologique de Roscoff, Sorbonne Université - CNRS, Roscoff, France
- 4Departamento de Biología de Organismos y Sistemas, Universidad de Oviedo, Oviedo, Spain
- 5Future Oceans Lab, Centro de Investigación Mariña, Universidade de Vigo, Vigo, Spain
- 6Laboratório de Ciências do Mar, MARE – Marine and Environmental Sciences Centre, Universidade de Évora, Évora, Portugal
- 7Departamento de Biologia, Escola de Ciências e Tecnologia, Universidade de Évora, Évora, Portugal
- 8Centro de Experimentación Pesquera, Consejería de Desarrollo Rural y Recursos Naturales del Principado de Asturias, Gijón, Spain
- 9Estación de Ciencias Mariñas de Toralla (ECIMAT), Universidade de Vigo, Vigo, Spain
- 10Department of Biological Sciences, University of South Carolina, Columbia, SC, United States
- 11Coastal Ecology Group, Faculty of Marine Sciences, Universidade de Vigo, Vigo, Spain
The stalked barnacle Pollicipes pollicipes inhabits rocky shores from the Atlantic coasts of Brittany (France) to Senegal. Because of the culinary traditions of southern Europe, stalked barnacles represent an important target species for local fisheries on the Iberian Peninsula. To manage this fishery sustainably, it is therefore important to assess the dynamics of local populations over the Iberian coast, and how they are interconnected at a wider scale using finely tuned genetic markers. In this work, a new enriched library of GT microsatellites for P. pollicipes was prepared and sequenced using Ion Torrent™ Next Gen-Sequencing Technology. 1,423 adults and juveniles were sampled in 15 localities of three geographic regions: southern Portugal, Galicia and Asturias (both in northern Spain). Twenty polymorphic loci arranged in five multiplex PCRs were then tested and validated as new molecular tools to address the spatial and temporal genetic patterns of P. pollicipes. Our results revealed high genetic diversity among adults. However, juveniles were genetically more structured than their adult counterparts, which alternatively displayed much more connectivity among the three studied regions. The lack of spatial genetic heterogeneity in adults may be due to the overlapping of several generations of settlers coming from different geographic origins, which mainly depends on the orientation of residual currents along the coast during reproduction. The genetic differentiation of juveniles may indeed be congruent with Iberian Peninsula hydrodynamics, which can produce chaotic genetic patchiness (CGP) at small temporal scales due to sweepstake reproductive success, collective dispersal and/or self-recruitment. Remarkably, most of the genetic heterogeneity of juveniles found in this work was located in Galicia, which could represent an admixture between distinct metapopulations or an old refuge for the most northern populations. To conclude, high genetic variation in P. pollicipes can lead to the false impression of population panmixia at the Iberian scale by masking more restricted and current-driven larval exchanges between regions. This possibility should be taken into consideration for further specific management and conservation plans for the species over the Iberian Peninsula.
Introduction
The percentage of stocks exploited at biologically unsustainable levels increased from 10% in 1974 to 34.2% in 2017 (FAO, 2020) after decades of management strategies based on catch-rate limitations (i.e., the EU Common Fisheries Policy). As an alternative or complementary approach, management practices are increasingly incorporating the spatial allocation of fishing intensity through marine protected areas, marine zoning, or spatial user rights, particularly for sessile or low-motility species (Lorenzen et al., 2010; Rassweiler et al., 2012). Optimization of these processes depends on the accurate estimation of the connectivity among management units, mediated by the dispersal of the planktonic larval stages (Silva et al., 2019). In this regard, a fundamental issue concerns whether the dispersal scales are consistent with the management scales (Ouréns et al., 2015). Although advection by ocean currents should lead to long dispersal distances exceeding the scale of management, there is increasing evidence that long-distance dispersal may be rare on ecological time scales (Palumbi, 2003; Selkoe et al., 2010; D’Aloia et al., 2015). This phenomenon can be explained by a combination of seascape characteristics such as eddies, gyres or upwellings of deep water bodies and specific larval behavior that would favor local retention and reduced dispersal (Morgan et al., 2009, 2018; Barshis et al., 2011; Kough et al., 2013). An additional line of evidence reveals surprising patterns of spatial and temporal genetic structure observed in some marine species at a scale where genetic variation should be efficiently homogenized by gene flow via larval dispersal, collectively coined chaotic genetic patchiness (CGP) (Johnson and Black, 1982; Hedgecock and Pudovkin, 2011; Eldon et al., 2016).
The stalked barnacle (Pollicipes pollicipes) is a pollicipedomorph cirriped (Chan et al., 2021) inhabiting rocky coasts that are highly exposed to waves in the northeast Atlantic. Its range extends from southwestern England through the coasts of Brittany (France), Spain, Portugal, and West Africa to Dakar (Senegal) (Barnes, 1996, 2008; Southward, 2008; Fernandes et al., 2010). In the Iberian Peninsula, it represents a highly valued resource that reaches very high market prices due to an old gastronomic tradition (Molares and Freire, 2003; Jacinto et al., 2011; Rivera et al., 2014). Remains of its consumption have been found in early Holocene archeological sites, mainly associated with Mesolithic and Neolithic shell-middens on both the Atlantic and Mediterranean coasts (Álvarez-Fernández et al., 2010, 2013; Álvarez-Fernández, 2011). Between 2013 and 2016, the European stalked barnacle fisheries have an annual economic value of EUR 10 million, involving approximately 500 t of landings and 2,100 professional fishers (Aguión et al., 2021). At some localities, the pressure exerted by poachers can be extremely high (more than 60% of their catches), especially in banned areas or periods (Jacinto et al., 2010; Rivera et al., 2014; Ruiz-Díaz et al., 2020).
Management of the stalked barnacle fishery in the Iberian Peninsula is highly heterogeneous (Aguión et al., 2021). In Galicia (NW Spain) since 1992, the regional government has developed a co-management system between fishers’ guilds (“cofradías”) and the fisheries authority through territorial user rights for fishing (TURFs) (Molares and Freire, 2003; Macho et al., 2013), where exclusive right of access are granted to fishing communities (Costello et al., 2010; Rivera et al., 2014). Similarly, in the West coast of Asturias (N Spain), the barnacle fishery has been managed through a co-management system with TURFs since 1994 (Rivera et al., 2014, 2017). Both Galicia and western Asturias present adaptive spatial management with nested scales at regional, local and patch/rock levels; recognized to promote fisheries sustainability (Aguión et al., 2021). However, on the eastern coast of Cape Peñes (eastern Asturias) and Portugal, the fishery is managed at a regional scale through general regulations without management plans (Aguión et al., 2021). In Portugal, however, there are two protected areas subjected to specific regulations for harvesting P. pollicipes: the Reserva Natural das Berlengas (RNB) and the Parque Natural do Sudoeste Alentejano e Costa Vicentina (PNSACV) (Sousa et al., 2013; Cruz et al., 2015; Carvalho et al., 2017). The first one (RNB) is subjected to local management, resembling a TURF in many aspects (Aguión et al., 2021). Currently, there is interest and potential for developing co-management systems similar to the one in Galicia and western Asturias in both Portuguese protected areas (Cruz et al., 2015; Sousa et al., 2020). Among different management approaches, TURFs represent the best option for the sustainable management of small-scale sessile fisheries (Gutiérrez et al., 2011; Rivera et al., 2017; Aguión et al., 2021). However, the design of management areas mandates a good understanding of population renewals for which estimates of connectivity are crucial (Aceves-Bueno et al., 2017; Silva et al., 2019). Dispersal, settlement, and subsequent recruitment are decisive processes in the population dynamics of marine invertebrates with planktonic larval stages, allowing the connection between remote populations and leading to meta-populations that are globally viable (Cowen and Sponaugle, 2009).
Pollicipes pollicipes larvae go through six planktotrophic nauplius stages before turning into a lecithotrophic stage, called cypris. According to Molares et al. (1994) and Franco et al. (2016, 2017), the pelagic larval development is finalized after 15 days to 1 month under optimal conditions in the laboratory, whereas in the natural environment, the total pelagic larval duration is estimated to last 2 months (Cruz, 2000; Macho, 2006). The presence of stalked barnacles on the shore might favor the settlement of cyprids, because recruitment is intense on conspecifics (e.g., Cruz et al., 2010; Fernandes et al., 2021). The dynamics of ocean circulation are recognized as important aspects in shaping connectivity patterns among marine populations (Treml et al., 2008). In this situation, significant effort is required to study population dynamics locally to adequately manage the resource (Molares and Freire, 2003). For P. pollicipes, a minimum potential passive migration distance of 600 km during the planktonic stage has been suggested (Quinteiro et al., 2007); nevertheless, reanalysis of genetic data and basic biophysical modeling point to modest dispersal distances in the range of tens of kilometers in the Asturian region (Rivera et al., 2013). At a large spatial scale, it has been suggested that P. pollicipes displays a metapopulation structure, where disconnected adult populations share a common larval pool (the n-islands model hypothesis) (Molares and Freire, 2003). However, the metapopulation structure has not yet been addressed. Alternatively, species with long larval dispersal potential, such as P. pollicipes, may exhibit surprising patterns of spatial and temporal genetic structure. CGP (Johnson and Black, 1982; Hedgecock and Pudovkin, 2011; Eldon et al., 2016) has been consistently reported in marine species that broadcast larvae at a scale where genetic variation should be efficiently homogenized by gene flow via larval dispersal. Eldon et al. (2016) reviewed and discussed how selection, sweepstake reproductive success, collective dispersal, and temporal shifts in local population dynamics may play a crucial role in generating such unexpected patterns. Moreover, Pineda et al. (2006) reported the existence of “recruitment windows” in a close barnacle species (Semibalanus balanoides), in which after a recruitment period of approximately 3 months, only recruits able to settle in just a couple of weeks survive after settlement and mature into adults. In spite of its interest for the management of this species, the processes that shape the genetic structure of P. pollicipes in the Atlantic Ocean have not been studied.
Genetic markers are a powerful tool for fisheries management because they present an array of very useful applications: they can address the correct identification of species, delimit distinct fish stocks (Borrell et al., 2012; Papa et al., 2020), assess relatedness levels within populations (Veliz et al., 2006; Plough et al., 2014), expose population connectivity (Pascual et al., 2017; Muñoz-Ramírez et al., 2020), estimate larval dispersal (Van Wyngaarden et al., 2017) and larval diversity (Chen et al., 2013; Wong et al., 2014; Alshari et al., 2021) or the source-sink dynamics within the population structure (Pineda et al., 2007; Brault et al., 2013; Lindegren et al., 2014). Genetic data, however, integrate information on the past demographic history of populations and are not always easily applicable for the present-days management for marine species with high fecundity and dispersal capabilities (Gagnaire et al., 2015). Estimating some of the population parameters that are crucial for stock management imposes the need to develop numerous highly polymorphic markers. These will help to discriminate between past and present-day processes that shape populations of species with highly effective population sizes (e.g., Hongjamrassilp et al., 2020). Despite the economic relevance of the P. pollicipes fishery, only a few articles have been published on the genetics of the stalked barnacles, most of which are based on mitochondrial markers (Quinteiro et al., 2007; Campo et al., 2010; Rivera et al., 2013). According to Quinteiro et al. (2007), the panmixia hypothesis is rejected, and 5 population groups are established: (1) Brittany; (2) Asturias-East; (3) Galicia, Portugal and Morocco; (4) Canary Islands; and (5) Cape Verde Islands, with the latter being extremely divergent. The Cape Verde population was later considered a new species (Van Syoc et al., 2010) and described as Pollicipes caboverdensis (Fernandes et al., 2010). Campo et al. (2010) revealed genetic differences among populations between Brittany (France) and the rest of the species distribution range, while Rivera et al. (2013) described small-scale, asymmetric connectivity in gooseneck barnacle populations, when reanalyzing data from Campo et al. (2010) for the Cantabrian coast. Microsatellites usually display high levels of genetic variation and can detect subtle genetic differentiation among populations separated by only a few hundred kilometers (Borrell et al., 2012). Moreover, they seem to be very useful to detect parentage/familial structures, when assessing the origin of recruits (St-Onge et al., 2015; Couvray and Coupé, 2018; Dubé et al., 2020). Microsatellite markers have been previously developed and, in some cases, used to infer the population genetic structure for several closely related acorn barnacles, such as S. balanoides (Dufresne et al., 1999; Flight et al., 2012); Chthamalus montagui (Pannacciulli et al., 2005; Fontani, 2009); Tetraclita spp. (Dawson et al., 2010; Chen et al., 2015); Megabalanus coccopoma (Reigel et al., 2015); Chelonibia testudinaria (Ewers-Saucedo et al., 2016, 2017); Notochthamalus scabrosus (Barahona et al., 2019) and two stalked barnacle species: Pollicipes elegans (Plough and Marko, 2014) and P. pollicipes (Seoane-Miraz et al., 2015; Fernandes et al., in prep.). The latter appear to have shown positive results with specific cross-amplifications in the congeners P. elegans, Pollicipes polymerus, and P. caboverdensis.
The aim of the present study was to revisit and test the previously described genetic homogeneity of P. pollicipes at the scale of the Iberian Peninsula with highly polymorphic microsatellite markers. The final goal is to provide support for the design of adequate and sustainable fishery management plans, using an in-depth analysis of genetic patterns inferred from a hierarchical geographic sampling of the barnacle populations along the Iberian coastline. However, preliminary tests using published microsatellite markers have provided inconsistent and non-reproducible PCR results in two different and independent genetic labs, necessitating the development of new highly variable genetic markers for the species P. pollicipes (this study).
Materials and Methods
Study Area and Sampling
A total of 1,423 individuals from 15 different localities belonging to three Atlantic regions of the Iberian Peninsula covering the most important spots of the barnacle fishery were sampled. These three regions were SW Portugal, Galicia (NW Spain), and W Asturias (N Spain). Thus, a hierarchical sampling of populations was performed in which five distinct localities were sampled within each region (Figure 1). The five localities belonging to Portugal are Aljezur (AL), Azenha do Mar (AZ), Cabo Sardão (CO), Malhão (MA) and Sines (CS). The five localities belonging to Galicia are Baiona (BA), Cabo Home (CH), Aguiño (AG), Camelle (CA) and A Coruña (AC). The five localities belonging to Asturias are La Cruz (PC), El Cuerno (CU), Las Llanas (LM), La Erbosa (ER), and El Corviru (EC) (Figure 1). Samples were transferred to the laboratory and frozen on the same day of collection until further individualization and labeling.
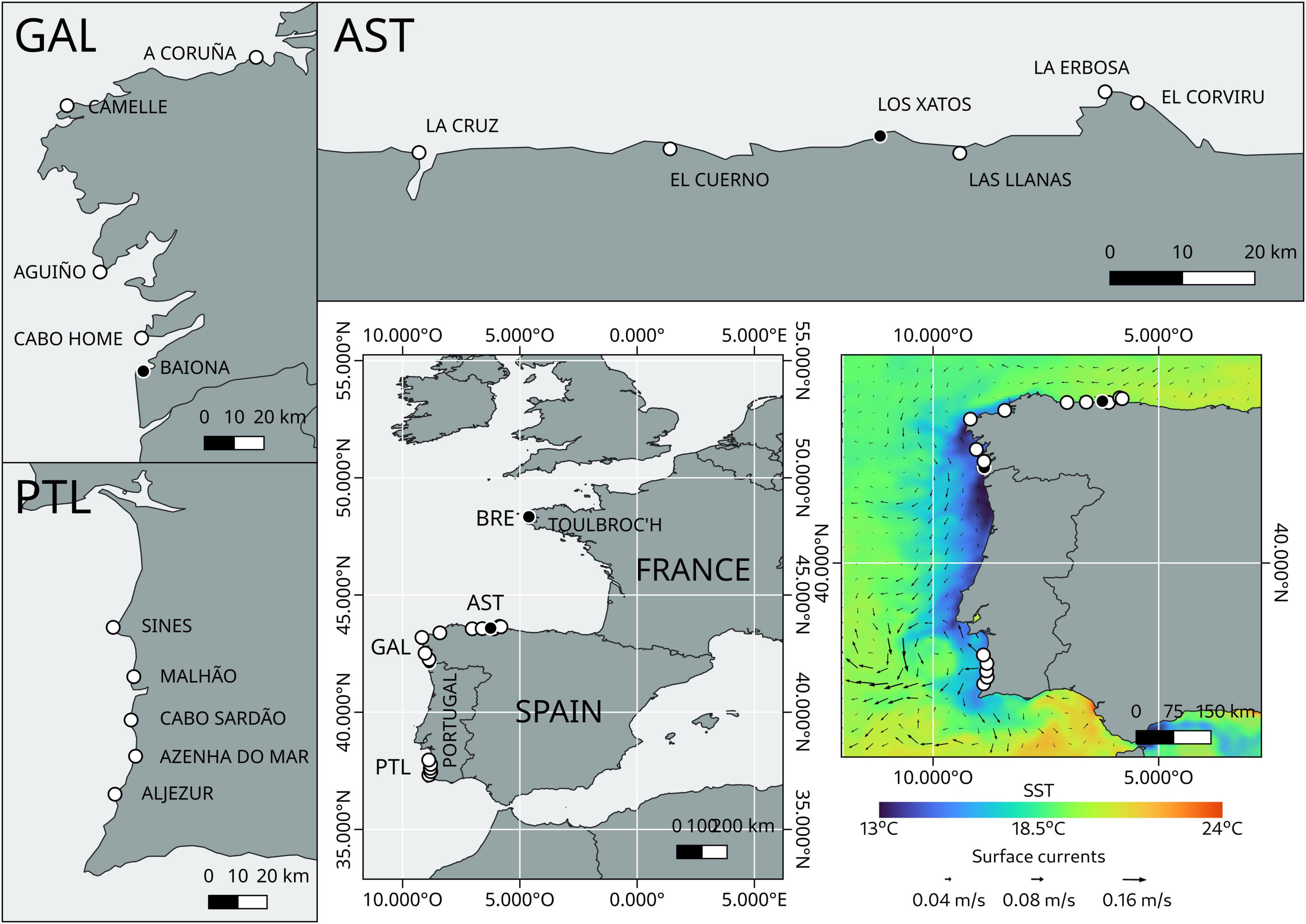
Figure 1. Study regions of the Iberian Peninsula for genetic analyses of Pollicipes pollicipes using microsatellites: Galicia (GAL); Asturias (AST), and SW Portugal (PTL). A total of 15 localities were sampled (white dots) for population genetic analyses. Three localities from the Bay of Biscay [including one from the French Brittany (BRE)] were initially sampled for microsatellites developing procedures (black dots). Also depicted is the upwelling circulation characteristic of the summer along the Atlantic coast of the Iberian Peninsula.
Within each of the targeted localities, one hundred individuals collected in September and October 2017 were randomly sampled within two distinct developmental cohorts according to their rostrum-carinal (RC) length [see Figure 4 in Parada et al. (2013)] (50 adults of commercial size greater than >18 mm; 50 juveniles between 2 and 4 mm). As barnacles are usually found in groups of sessile individuals, fixed on primary rocky substrates with small juveniles attached to adult peduncles (Cruz et al., 2010), juveniles were first removed from the adults, avoiding the collection of more than one juvenile by adult and then treated secondarily. Each barnacle was put individually in a tube previously labeled and preserved in absolute ethanol at room temperature. In the laboratory, a small portion of the peduncle muscle was dissected from each individual for genomic DNA extraction. In the case of adults, special care was taken to dissect the tissue from the inner part of the peduncle to avoid possible contamination by attached post-larvae (cyprids) and juveniles.
To characterize the typical upwelling circulation during the stalked barnacle larval season in summer/autumn 2017 along the coasts of northern and western Iberia, sea surface temperature (SST) along with modeled sea surface currents datasets were retrieved during the peak of meridional Ekman transport at central Portugal on 11-08-2017. Daily 4 km SST data were obtained from the all-satellites combined Copernicus’ product1. The 5-days averaged meridional and zonal components of the surface currents were obtained from the OSCAR model with a spatial resolution2 of 0.33°.
Microsatellite Markers and Multiplex PCR Development
Genomic DNA from five adult individuals was extracted using the EZNA® Mollusk Kit (Omega Bio-Tek Inc., Norcross, GA, United States). An enriched biotin-labeled CT/GT library for dinucleotides was obtained using the methodology described by Bloor et al. (2001) and Sotelo et al. (2007), where DNA was digested with HaeIII (NEB). Digestions were run in 1.5% agarose gels stained with ethidium bromide. Fragments between 400 and 800 bp were excised from gels and purified using a QIAquick Gel Extraction Kit (Qiagen). Fragments were ligated to a double-stranded adaptor using ligase (NEB) and enriched by PCR using oligoA. Purified PCR products were denatured and incubated with 200 pmol of 5′ biotinylated (CT)12 and (GT)12 probes (Invitrogen) attached to streptavidin-coated magnetic beads (Streptavidin MagneSphere Paramagnetic Particles, Promega). Hybridization was carried out in 6 SSC for 30 min at 60°C in a thermocycler. Specific fragments were recovered after washing the bead suspension with solutions progressively desalted at 60°C, and subsequently amplified using Oligo A. A DNA library was prepared using an Ion Plus Fragment Library Kit (Thermo Fisher Scientific, Austin, TX, United States) according to the manufacturer’s protocol. Next-generation Ion Torrent sequencing of the library was conducted using the Ion Torrent platform on an Ion PGM System (Life Technologies, Foster City, CA, United States) using Ion PGM 400 sequencing reagents and Ion 318v2 chips following the manufacturer’s instructions at the University of Vigo Central Services (CACTI). Quality control procedures and filtering of the resulting reads were afforded using PRINSEQ software (Schmieder and Edwards, 2011). Tag Sequence Check and Sequence Duplication routines were used to trim adapters and eliminate duplicates. Sequences shorter than 100 bp with a mean quality Phred score lower than 20 were removed. Tandem Repeats Finder (Benson, 1999) was used with all the parameters by default for locating and displaying tandem repetitions in DNA sequences. Forward and reverse primers were designed for effective microsatellite amplifications using FastPCR 6.5 software following Kalendar et al.’s (2009) recommendations. Finally, primers were proposed and tested by individual PCR on 30 individuals of P. pollicipes from 3 distinct geographic populations: 10 individuals from Baiona (Galicia, Spain), 10 individuals from Los Xatos (Asturias, Spain) and 10 individuals from Toulbroc’h (Brittany, France) (Figure 1). PCR tests were equally subdivided between different research laboratories with 41 primers tested per laboratory at the University of Vigo, the University of Oviedo and the Roscoff Marine Station. In this way, microsatellite markers were calibrated between geographic regions and the three institutes involved in the project.
Individual PCRs were conducted in a 20 μL total volume with Green GoTaq® Flexi Buffer (1×) (Promega Corporation, Madison, WI, United States), MgCl2 (2.5 mM), dNTPs (0.5 mM), 0.2 μM of each primer, 0.1 U of GoTaq® G2 Flexi Polymerase (Promega Corporation, Madison, WI, United States), and 50 ng of DNA in sterile distilled water. The PCR program included an initial 5 min denaturation step at 95°C, 35 cycles of denaturation at 95°C for 30 s, annealing at 57°C for 30 s and elongation at 72°C for 30 s. PCR products were visualized using electrophoresis on a 2% agarose gel stained with SimplySafe™ (EURx, Gdañsk, Poland). Primer pairs without amplification, leading to a multiband pattern or a band size differing from its expected size, were discarded. Twelve microsatellite loci were amplified reliably and arranged in three multiplex PCRs (M1, M2, and M3 with four microsatellite markers per multiplex each) using Multiplex Manager 1.2 software (Holleley and Geerts, 2009) according to the dye colors and expected amplicon sizes. In addition to the twelve microsatellite markers retained with this screening, eight microsatellite markers previously developed in a parallel study (Fernandes et al., in prep) were tested, calibrated, and added in two supplementary multiplexes (M4 and M5) following the previously detailed methodology. This process resulted in a total of 5 multiplex PCRs. Forward primers were labeled using fluorescent dyes: 6-FAM™, NED™, VIC™, and PET® (Applied Biosystems, Foster City, CA, United States) (Table 1). PCR products were sequenced at the Genomer platform of the Roscoff Marine Station and at Servicios Científico-Técnicos of the University of Oviedo. Allele sizes were manually scored using GeneMapper v.4.0 (Applied Biosystems, Foster City, CA, United States).
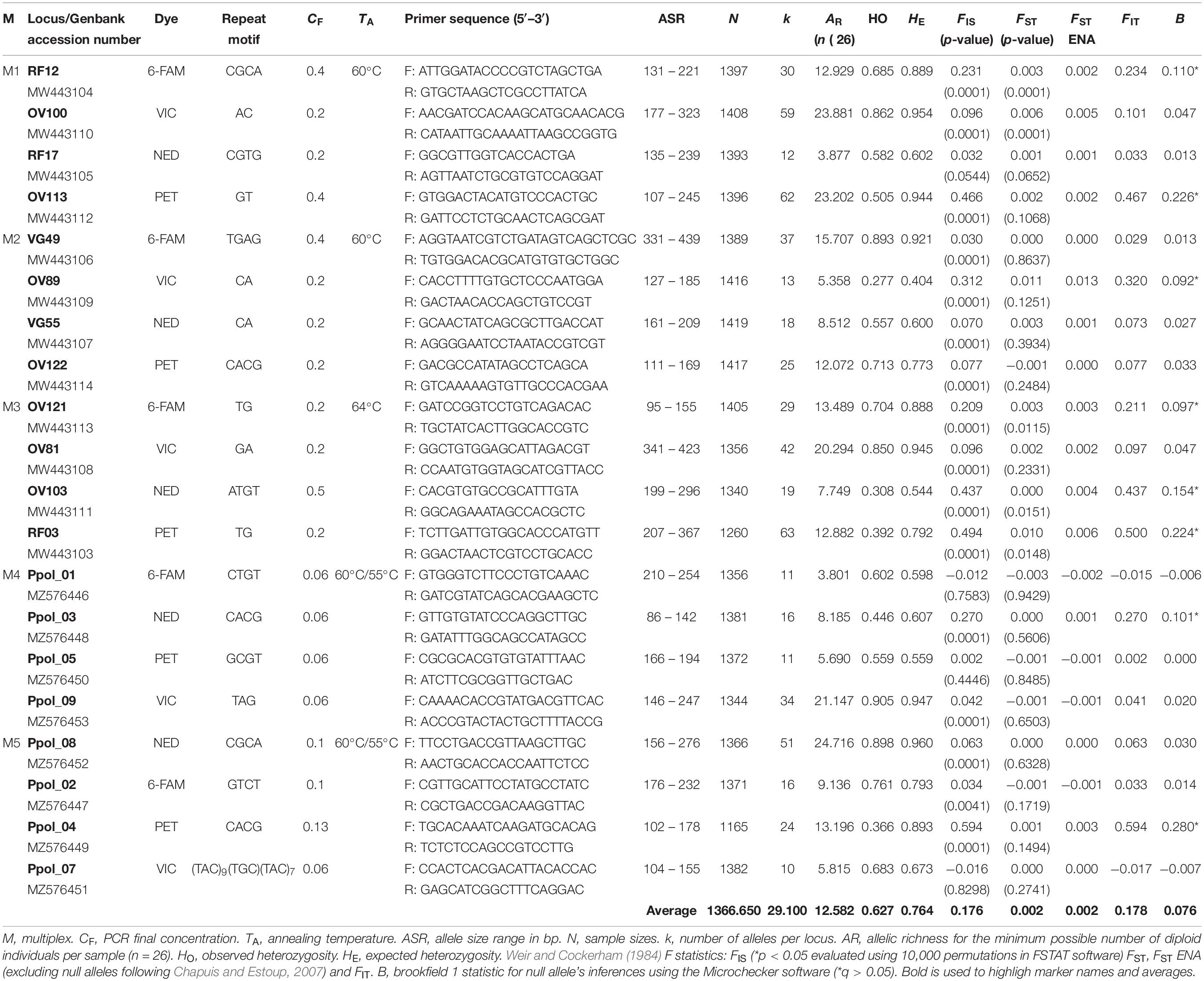
Table 1. Overall microsatellites information based on multiplex PCRs typifying P. pollicipes populations coming from 15 different localities of the Iberian Peninsula.
Multiplex PCR and Microsatellite Genotyping
As explained above, all adult DNA was extracted with the EZNA® Mollusk Kit (Omega Bio-Tek Inc., Norcross, GA, United States). Juvenile DNA was extracted using the Chelex® 100 (Bio-Rad Laboratories Inc., Hercules, CA, United States) method (Estoup et al., 1996). PCRs were carried out following a unidirectional workflow that started in a pre-PCR room to prepare PCR plates. Amplification by PCR and processing of the subsequent PCR products always took place in a post-PCR area to avoid any possible contamination. M1, M2, and M3 multiplex PCRs were conducted using the QIAGEN Multiplex PCR Kit (QIAGEN Inc., Venlo, Netherlands) in a final reaction volume of 13 μL with the following components: 1× QIAGEN Multiplex PCR Master Mix, 1× Q-Solution, 50 ng of DNA template and 0.2–0.5 μM of each primer (Table 1). PCR conditions consisted of an initial denaturation step at 95°C for 15 min, followed by 40 cycles at 94°C for 30 s, an annealing temperature of 60°C (M1 and M2) or 64°C (M3) for 1:30 min and 72°C for 1 min, with a final extension at 60°C for 30 min. M4 and M5 multiplex PCRs were incorporated and tested in a later stage and they were conducted using the TouchDown PCR technique (Hecker and Roux, 1996). TouchDown PCRs were conducted in a 15 μL total volume with Colorless GoTaq® Flexi Buffer (1×) (Promega Corporation, Madison, WI, United States), MgCl2 (1.5 mM for M4 and 1.16 for M5), dNTPs (0.1 mM), 0.06–0.13 μM of each primer (Table 1), 0.4 U of GoTaq® G2 Flexi Polymerase (Promega Corporation, Madison, WI, United States), 200 ng/μL bovine serum albumin (BSA), and 5–10 ng of DNA in distilled water. The samples were initially heated at 95°C for 5 min, followed by 10 cycles consisting of 95°C for 30 s, 60°C (decreasing incrementally by 0.5°C per cycle) for 40 s, and 72°C for 40 s, followed by 25 cycles at 95°C for 30 s, 55°C for 40 s, and 72°C for 40 s, culminating in a final cycle at 72°C for 10 min. PCR results were checked on a 2% agarose gel. For each multiplex amplification, 2 μL of reaction product (diluted 1/40 with Milli-Q water for M1, M2, and M3) was mixed with 9.5 μL of Hi-Di formamide (Applied Biosystems, Foster City, CA, United States) and 0.50 μL of SM594 molecular weight marker (Mauger et al., 2012). The mixture was heated at 94°C for 5 min, immediately chilled on ice for 2 min, loaded in an ABI Prism® 3130XL automatic sequencer (Applied Biosystems, Foster City, CA, United States) of 16 capillaries using POP-7 polymer and run at 60°C, 15 kV, 1,200 s using the sequencing platform Plateforme Genomer (Station Biologique de Roscoff). To ensure that the allele spread calibration held between the set of samples analyzed, controls were included in each plate to be genotyped as reference genotypes. Each genotype was then scored after analyzing the amplification products with Genemapper 4.0 (Applied Biosystems, Foster City, CA, United States).
Population Genetic Analyses
The allele frequencies, number of alleles per locus (k), observed heterozygosity (HO) and unbiased expected heterozygosity (HE) were calculated with GENETIX 4.05 (Belkhir et al., 2004). Moreover, possible genotyping errors and null allele frequency estimation were conducted using MICRO-CHECKER 2.2.3 (Van Oosterhout et al., 2004) and FreeNa (Chapuis and Estoup, 2007) with a number of replicates fixed to 10,000. Moreover, to explore the influence of null alleles on data we assessed FIS and FST correlation, FIS and the number of missing data (putative null homozygotes) correlation and estimated the StrdErrFIS and StrdErrFST values following the De Meeûs (2018) and Manangwa et al. (2019) [but see Waples (2018)]. The significance of correlations was tested with a unilateral (ρ > 0) Spearman’s rank correlation test with Rcmdr package (Fox, 2005, 2007) for R. Furthermore, for each population, the number of private alleles was calculated with GENALEX 6.5.03 (Peakall and Smouse, 2012).
Possible deviations from expected proportions in Hardy Weinberg’s equilibrium and linkage disequilibrium for each locus and population were assessed using FSTAT 2.94 software (Goudet, 1995). FSTAT 2.94 software (Goudet, 1995) was used to calculate the allelic richness (AR) and to determine the fixation indices (F-statistics) within and across populations using the method described by Weir and Cockerham (1984). Significance levels of FIS were estimated by permutating alleles between genotypes within samples 2,000 times and adjusted following Bonferroni correction (Rice, 1989) from all tested juvenile and adult samples. To test self-recruitment, the relatedness between individuals (RXY) was estimated with the “related” package in R (Pew et al., 2015). The relative performance of seven different relatedness estimators was examined (dyadml, lynchli, lynchrd, quellergt, ritland, trioml, and wang) through comparison of the observed values to expected values generated from a simulated sample set of 400 individuals of known relatedness [with one hundred individuals from 4 categories: parent-offspring (RXY = 0.500), full-sib (RXY = 0.500), half-sib (RXY = 0.250), and unrelated pairs (RXY = 0.000)]. The results showed that the dyadic likelihood relatedness estimator (dyadml) provided the most consistent estimates through all possible levels of kinship; therefore, it was performed with 500 iterations. The bottleneck hypothesis was tested using the software BOTTLENECK 1.2.02 (Piry et al., 1999) under the two-phased model of mutation (TPM), taking into account 90% single stepwise mutations with a variance of 12.
Comparisons between regions and between cohorts (adults and juveniles) were conducted using a two-sided statistical analysis included in the FSTAT software for several statistics [AR, HO, HE, FIS, FST, relatedness (R), and corrected relatedness]. In addition, FST values were estimated using FreeNa, which estimates unbiased FST following the ENA method (Chapuis and Estoup, 2007). The FST values and associated p-values between cohorts and within and between regions were also calculated using FSTAT 2.94 (Goudet, 1995) to test for the regional and local structure. To assess the significance levels of FST, multilocus genotypes were permutated 2,000 times between pairs of samples, and the significance threshold was obtained by applying a false discovery rate (FDR) over samples (Benjamini and Hochberg, 1995). Partial Mantel tests to estimate the correlation between genetic and geographical distance were performed with FSTAT 2.94 (Goudet, 1995) using the INA correction method for the chord distance (Cavalli-Sforza and Edwards, 1967) (DCSE) provided by FreeNA (Chapuis and Estoup, 2007) and combining a ln transformation of Haversine geographic distances following Séré et al. (2017) and Rousset’s θ/(1-θ) and a log transformation of Haversine geographic distances with 10,000 permutations (Rousset, 1997).
The software BayeScan v2.1. (Foll and Gaggiotti, 2008) was used to identify candidate loci deviating from neutral expectations from genetic data using differences in allele frequencies between populations. Twenty pilot runs of 5,000 iterations each, followed by an additional burn-in of 50,000 iterations and then 5,000 samplings with a thinning interval of 10, were conducted. To correct for multiple testing, the program computes q-values based on the posterior probability for each locus. Loci with α-values significantly > 0 and q-values < 0.05 were defined as “outliers” –, i.e., loci putatively under directional selection. Loci with α-values significantly < 0 were considered putatively under balancing selection. The remaining loci were classified as neutral.
An analysis of molecular variance (AMOVA) implemented in Arlequin 3.5.1.3 (Excoffier et al., 2005) to partition genetic variation across nested levels, regions and sites within regions was used. For the AMOVA, the number of different alleles was used as a measure of genetic variation (FST-like option in Arlequin), and 10 000 permutations were used to test for statistical significance. Moreover, the “adegenet” package in R was used to estimate the genetic differentiation and visualize individual clustering with principal component analysis (DAPC, Jombart, 2008; Jolliffe, 2011) among adults and juveniles from each of the three regions separately and both among adults and among juveniles for all three regions pooled together. A neighbor-joining (NJ) tree based on the pairwise Nei’s genetic distance DA (Nei and Takezaki, 1983) for all microsatellites and localities (15 localities; adults and juveniles grouped together) and then adding temporal cohorts as independent samples (i.e., 15 localities and 2 cohorts, 30 samples) was constructed with the software POPTREEW (Takezaki et al., 2014) using 10 000 bootstraps and visualized in The Interactive Tree of Life3 (Letunic and Bork, 2019). Finally, STRUCTURE 2.3.4 (Pritchard et al., 2000) was also run to explore the population structure with Bayesian clustering. STRUCTURE was run using the 15 localities and 30 samples using admixture (Gilbert et al., 2012; Novembre, 2016) and also using adults and juveniles taken separately from the three regions (Portugal, Galicia and Asturias) in the same conditions to explore putative genetic units. The settings used were an admixture model from K = 1 to K = 30 in 20 runs following Evanno et al. (2005) and (Gilbert et al., 2012). Assignment clusters were made with burn-in periods of 20,000 and 200,000 Markov chain Monte Carlo repetitions. The most likely value of K was chosen using the delta K statistic (Evanno et al., 2005) using STRUCTURE HARVESTER software (Earl and VonHoldt, 2012), and visualization and grouping of the individual STRUCTURE runs was performed using CLUMPAK (Kopelman et al., 2015).
Results
The typical upwelling circulation during the stalked barnacle larval season in summer/autumn 2017 along the coasts of northern and western Iberia, sea surface temperature (SST) along with modeled sea surface currents datasets revealed that the SST patterns showed strong onshore advection of cold waters (13–15°C) on the Galician and Portuguese shelves with upwelling filaments extending further offshore especially at the upwelling centers of Fisterra, A Guarda, and Cape da Roca (Figure 1). Slightly onshore cooling indicative of upwelling was also observed along the western Cantabrian coast. Westward and southward currents in the order of few cm/s off the Cantabrian and Atlantic shores, respectively, clearly pointed to upwelling circulation (Figure 1). These flows are weaker close to the coast probably due to friction with the coastal boundary layer. Off southern/central Portugal in between Cape da Roca and Cape San Vincente, an anticlockwise cyclonic eddy was apparent with strong southward currents (>10 cm/s) along its western side. The dynamic structure of this feature matched SST patterns remarkably well, with a warm core (18°C) surrounded by colder upwelled waters (14°C).
The microsatellites markers development process produced libraries with a total amount of 42,860 reads showing a mean sequence length of 91.61 ± 103.29 bp (minimum length: 25 bp – maximum length: 517 bp) and a mean GC content of 63.66 ± 18.90%. A total of 10 781 sequences with a mean sequence length of 244.48 ± 97.61 bp, a length range of 418 bp and a mean GC content of 50.30 ± 5.61%, resulted after quality control procedures. A total of 1,140 sequences containing di, tri, tetra, and pentanucleotides were selected after locating and displaying tandem repetitions in DNA sequences. Finally, 123 pairs of primers were proposed and tested in three different research laboratories (University of Vigo, University of Oviedo, and the Roscoff Marine Station). A new set of twelve microsatellite loci currently arranged into three multiplex PCRs (M1, M2, and M3) was developed for the stalked barnacle P. pollicipes in this work (Genbank accession numbers: MW443103–MW443114). Moreover, eight previously developed microsatellite loci by Fernandes et al. (in prep) were also tested and included in another two multiplexes (M4 and M5, Genbank accession numbers: MZ576446–MZ576456). This procedure resulted in a total of 5 multiplex PCRs (Table 1) leading to scorable and reproducible genotypes for all 20 microsatellite loci. None of these loci showed evidence of linkage disequilibrium between alleles (p > 0.05). These loci were highly polymorphic and exhibited approximately 15% private alleles (n = 87) only present at one locality (Tables 1, 2).
The number of alleles per locus (k) varied greatly from 10 to 63 between loci, with an average of 29.10, and yielded an average (min–max) allelic richness (AR) of 12.170 (11.074–12.918) per locality. The observed and expected heterozygosities across loci ranged from HO = 0.277 (M2; OV89) and HE = 0.404 (M2; OV89) to HO = 0.905 (M4; Ppol_09) and HE = 0.960 (M5; Ppol_08), with observed and expected multilocus mean heterozygosities equal to 0.627 (0.561–0.667) and 0.764 (0.746–0.785), respectively (Table 1). All markers and all populations showed significant deviations from Hardy–Weinberg equilibrium (mean FIS = 0.179) due to heterozygote deficiencies (Table 2). When testing these markers for null alleles with MICRO-CHECKER 2.2.3. (Van Oosterhout et al., 2004) and FreeNA (Chapuis and Estoup, 2007), we found that heterozygote deficiency could be due to null alleles for at least 8 highly polymorphic loci: RF12 locus (Brookfield 1 Statistic = 0.110); OV113 locus (B = 0.226); OV89 locus (B = 0.092); OV121 locus (B = 0.097); OV103 locus (B = 0.154); RF03 locus (B = 0.224); Ppol_03 locus (B = 0.101); and Ppol_04 locus (B = 0.280) (Table 1). The correlation between FIS and FST appeared to be significant (Spearman’s ρ = 0.606, p-value = 0.005). However, FIS and the number of missing data (putative null homozygotes) were not correlated (Spearman’s ρ = 0.098, p-value = 0.3402) and the standard error for FIS (StrdErrFIS = 0.044) was higher than for FST (StrdErrFST = 0.001). The mean overall FST value for the 20 microsatellites was FST = 0.002 (P = 0.0001), and three loci clearly showed higher FST values [OV100 (FST = 0.006), OV89 (FST = 0.011), and RF03 (FST = 0.010)] (Table 1).
The comparative analysis for levels of genetic variation between regions [Portugal (PTL), Galicia (GAL), and Asturias (AST)] revealed no significant differences for expected heterozygosities (HS) (Table 2). Slight differences in genetic diversities were, however, observed depending on the population parameter estimated. Galicia showed the highest values for allelic richness and observed and expected heterozygosity in adults and juvenile populations (Table 2). Portugal showed the highest number of private alleles (mean APPTL = 3.3), which was mainly attributable to adults (mean AP = 4.4) (Table 2). Significant differences in allelic richness were also observed for juveniles (ARPTL: 11.569; ARGAL: 12.418; ARAST: 11.900; p < 0.01) and in observed heterozygosity for adults (HOPTL: 0.640; HOGAL: 0.656; HOAST: 0.615; p < 0.05) and juveniles (HOPTL: 0.605; HOGAL: 0.648; HOAST: 0.601; p < 0.01) (Table 2). This phenomenon was especially obvious in Portuguese samples, where the average number of private alleles decreased by 50% (APAD: 4.4 to APJV: 2.2) (Table 2). In this later region, significant differences were found between adults and juveniles both in terms of allele richness (ARAD: 12.318; ARJV: 11.569; p < 0.01) and observed heterozygosity (HOAD: 0.640; HOJV: 0.605; p < 0.05) or expected heterozygosity (HSAD: 0.773; HSJV: 0.757; p < 0.01) (Table 2). Globally, juveniles were also more related to each other (RXY = 0.067) than their adult (RXY = 0.058) counterparts, as indicated by relatedness analyses. Juveniles from Portugal (RXY value = 0.073, p < 0.002) and Asturias (RXY = 0.070, p < 0.002) were much more related than expected from panmixia. Bottleneck software showed that none of the 30 samples tested (15 localities × 2 cohorts) exhibited a significant excess of predicted heterozygotes under the TPM model and could not be considered to have experienced a recent genetic bottleneck (Table 2). When the bottleneck hypothesis was tested with all juveniles and adults together (15 samples) and at the regional scale (15 samples grouped in 3 regions, for 2 cohorts), the statistics remained non-significant (Table 2).
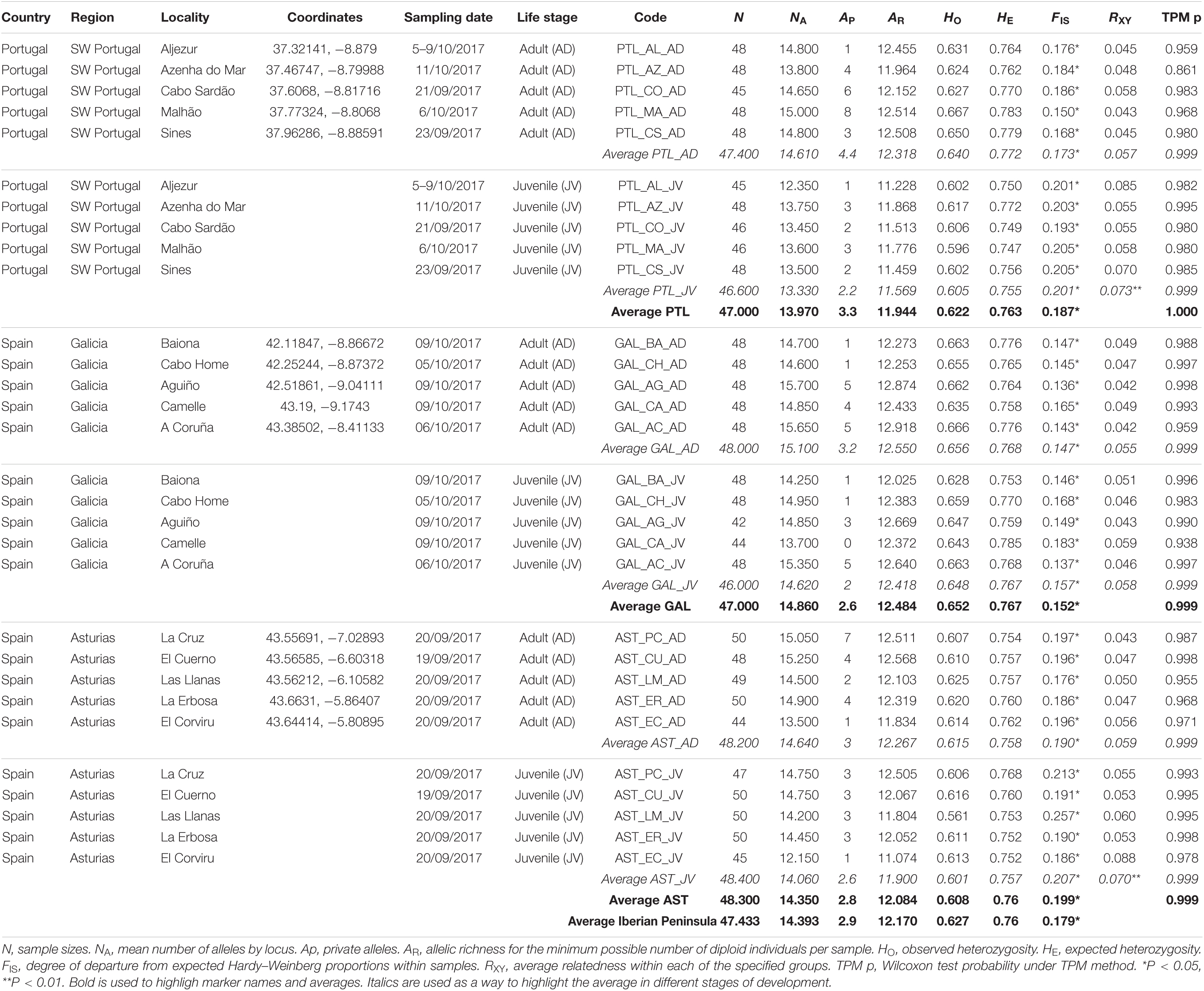
Table 2. Genetic variability of P. pollicipes populations (adults vs. juveniles) coming from 15 distinct localities along the Atlantic Iberian Peninsula coastline.
According to the overall FST, there was no significant genetic differentiation of adults between and within regions (Figure 2A). Only 20 out of the 75 possible pairwise FST values between adult samples from different regions (25.3%) showed p-values lower than the 0.05 cutoff value, and these critical values were more often encountered between Galicia and Asturias (12/25 = 48%) (Figure 2A). However, no p-values remained significant after FDR correction (Figure 2A). In contrast, the overall FST statistics estimated for the juveniles between and within regions indicated notable regional and spatial structuring (Figure 2B). Pairwise FST estimated between juvenile samples from Galicia and Portugal (12/25 = 48% before and 6/25 = 24% after FDR) and between Galicia and Asturias (13/25 = 52% before and 6/25 = 24% after FDR) confirmed this trend and showed clear regional structuring (Figure 2B). Asturias and Portugal were, however, less differentiated from each other, with fewer significant pairwise FST values (3/25 = 12% before and 2/25 = 8% after FDR) (Figure 2B). Some spatial structuring within regions was detected for juveniles using pairwise FST analyses but only in the case of Portugal (2/15 = 13% before and 1/15 = 6% after FDR) (Figure 2B). The pairwise FST analyses between adults and juveniles within regions revealed that only 8 out of the 75 possible comparisons (11%) had p-values lower than the 0.05 cutoff threshold for Portugal and Asturias (but not in Galicia), which, however, did not remain significant after FDR correction (Figures 2C–E).
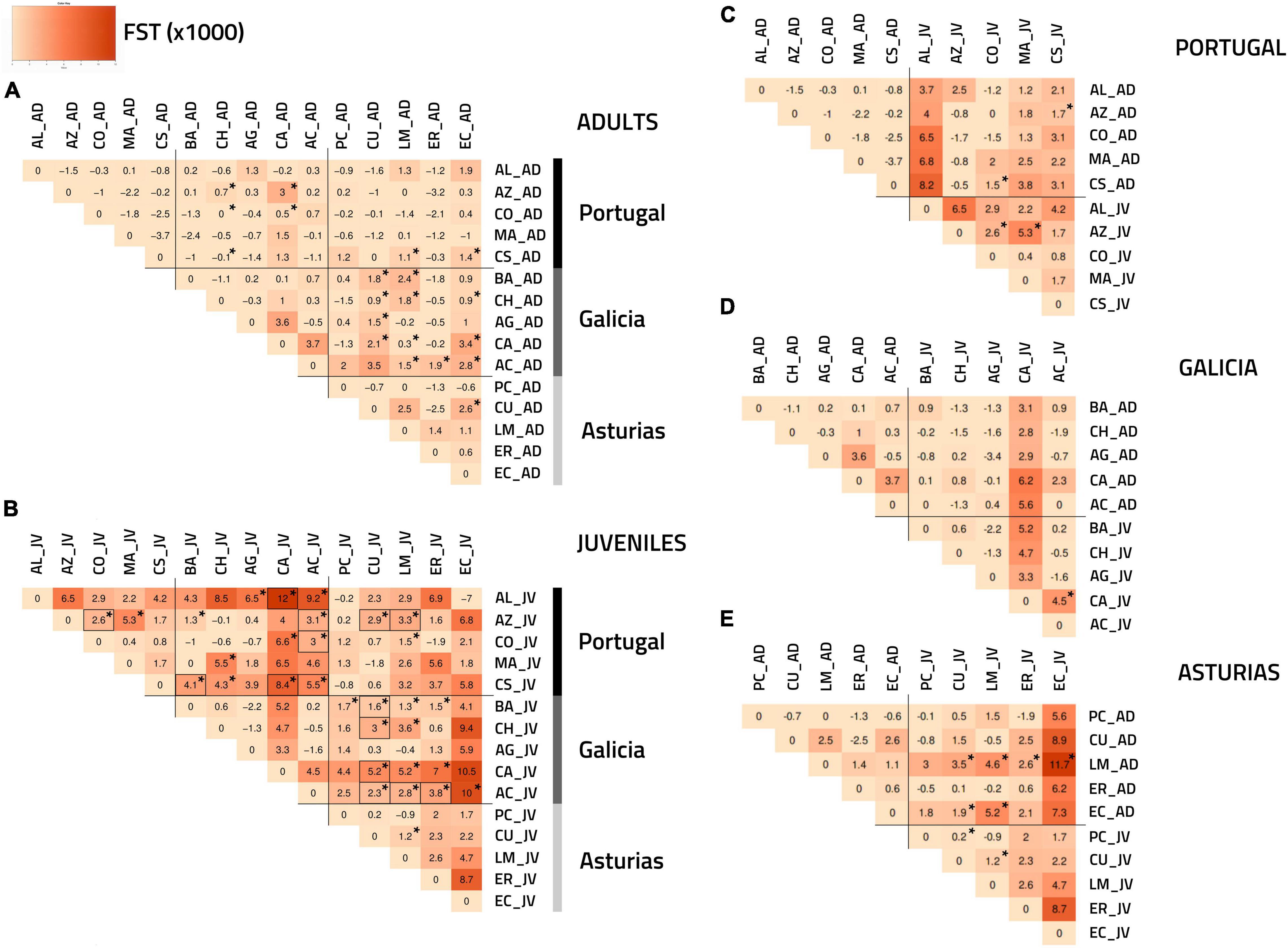
Figure 2. FST heatmaps [based on Weir and Cockerham (1984)] following genetic analyses of P. pollicipes using microsatellites along the Iberian Peninsula. The darker the color, the higher the FST value. Asterisks (*) indicate significant p-values (p < 0.05) while significant values after a FDR correction (Benjamini and Hochberg, 1995) are highlighted by black rectangles: (A) adults (AD) and (B) juvenile (JV) analyses among regions. (C) Portugal, (D) Galicia, and (E) Asturias analyses between the two developmental stages (adults vs. juvenile) within each of the three regions.
The analysis conducted with BayeScan v2.1 for outlier detection resulted in no loci under selection or biased by species admixture and hybridization which have the same expectations in terms of outliers; the twenty loci showed signatures of balanced or purifying selection with negative alpha values. The results of the partial Mantel tests indicated no correlation between genetic and geographic distances, with R2 = 1.61 and p-value = 0.1916 for adults and R2 = 3.16 and p-value = 0.0698 for juveniles using the INA correction method for DCSE, and R2 = 0.06 and p-value = 0.8002 for adults and R2 = 0.22 and p-value = 0.6241 for juveniles using the Rousset method. The population structure was therefore closer to an n-island model than a stepping stone model, and the pairwise FST between adjacent sites often exceeded those obtained between geographically distant locations.
The DAPC analyses and the hierarchical analysis of molecular variance (AMOVA) using ϕST statistics showed no significant genetic differentiation of adults among and within regions [AD: ϕCT(among) = 0.00032, p > 0.05; ϕSC(within) = 0.00013, p > 0.05] (Figure 3A). However, a globally significant genetic differentiation for juveniles among and within regions was found [JUV: ϕCT(among) = 0.00093, p < 0.05; ϕSC(within) = 0.00217 p < 0.001] (Figure 3B). The analyses also revealed significant genetic heterogeneity between P. pollicipes generations in Portugal [ϕCT(among) = 0.00127, p < 0.01] and Asturias [ϕCT(among) = 0.00120, p < 0.01], but not in Galicia (Figures 3C–E). The neighbor-joining tree using adults and juveniles grouped together by localities clearly separated Galicia with high bootstrapping values (i.e., 90%), where Camelle and Baiona fall apart from the rest of the Galician localities, after which two other different Portuguese and Asturian clades appeared (Figure 4A). When all populations (15 localities and 2 cohorts, 30 samples) were analyzed, the neighbor-joining tree again showed Galicia samples falling apart and becoming heterogeneous, whereas the Portuguese and Asturian samples were mixed together, with aggregations showing low bootstrap values (Figure 4B). The STRUCTURE runs using admixture suggested 3 genetic clusters [Evanno’s k = 3, L(K) = −117589.9100] when all populations (30 samples) were analyzed (Figure 5A). The STRUCTURE results also indicated the co-occurrence of 2 genetic clusters [Evanno’s k = 2, L(K) = −60822.8150] for adults (Figure 5B) and 3 clusters [Evanno’s k = 3, L(K) = −56241.9750] for juveniles when run separately (Figure 5C).
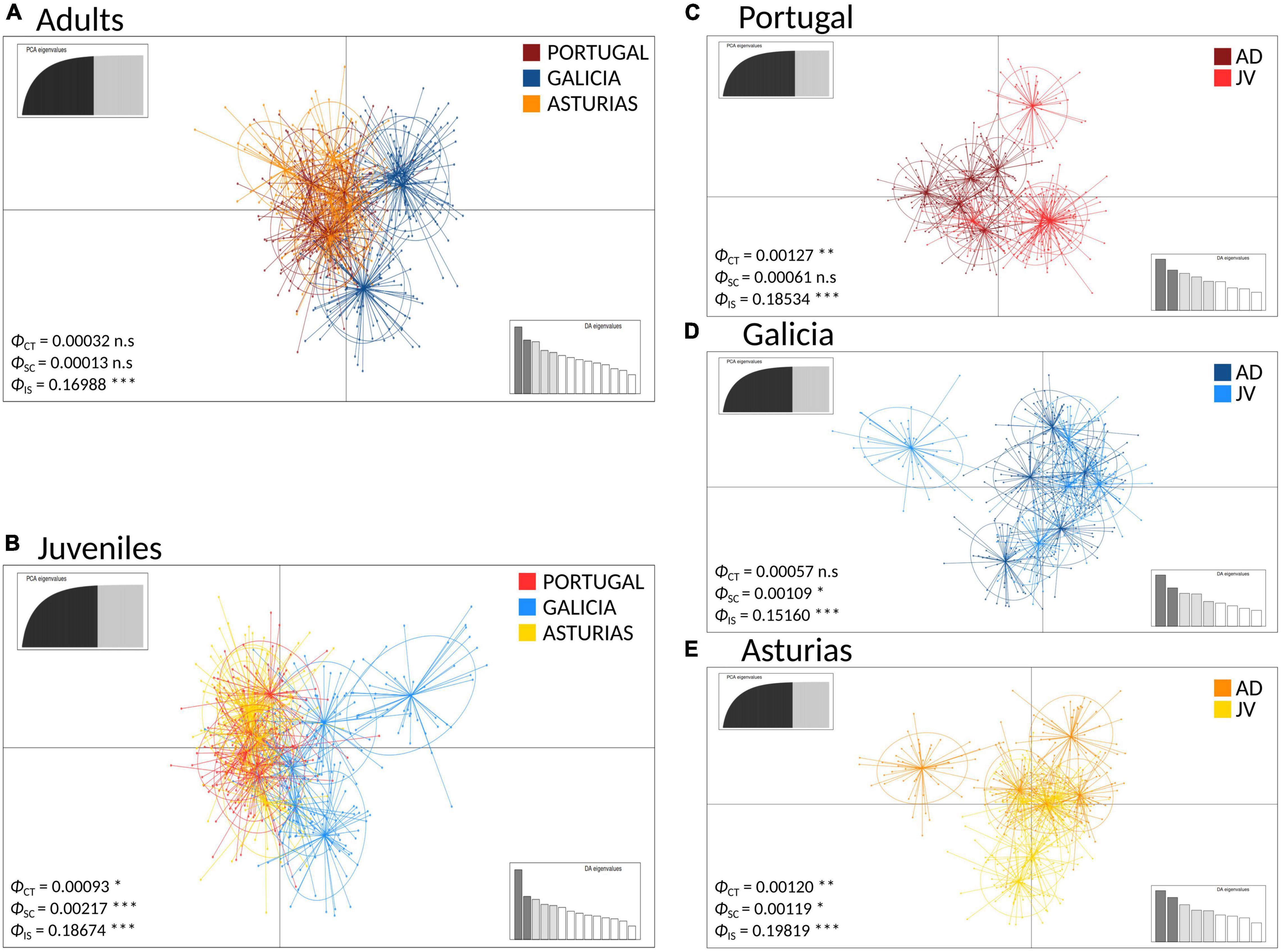
Figure 3. Genetic clustering using Discriminant Analysis of Principal Components (DAPC) of P. pollicipes populations using microsatellites along the Iberian Peninsula. In each case the ϕ statistics are shown together with their p-values after AMOVA analyses (ϕCT: among groups, ϕSC: among populations within groups, ϕIS: among individuals within populations): (A) adults (AD) and (B) juvenile (JV) analyses among regions. (C) Portugal, (D) Galicia, and (E) Asturias analyses between the two developmental stages (adults vs. juvenile) within each of the three regions. *P < 0.05, **P < 0.01, ***P < 0.001.
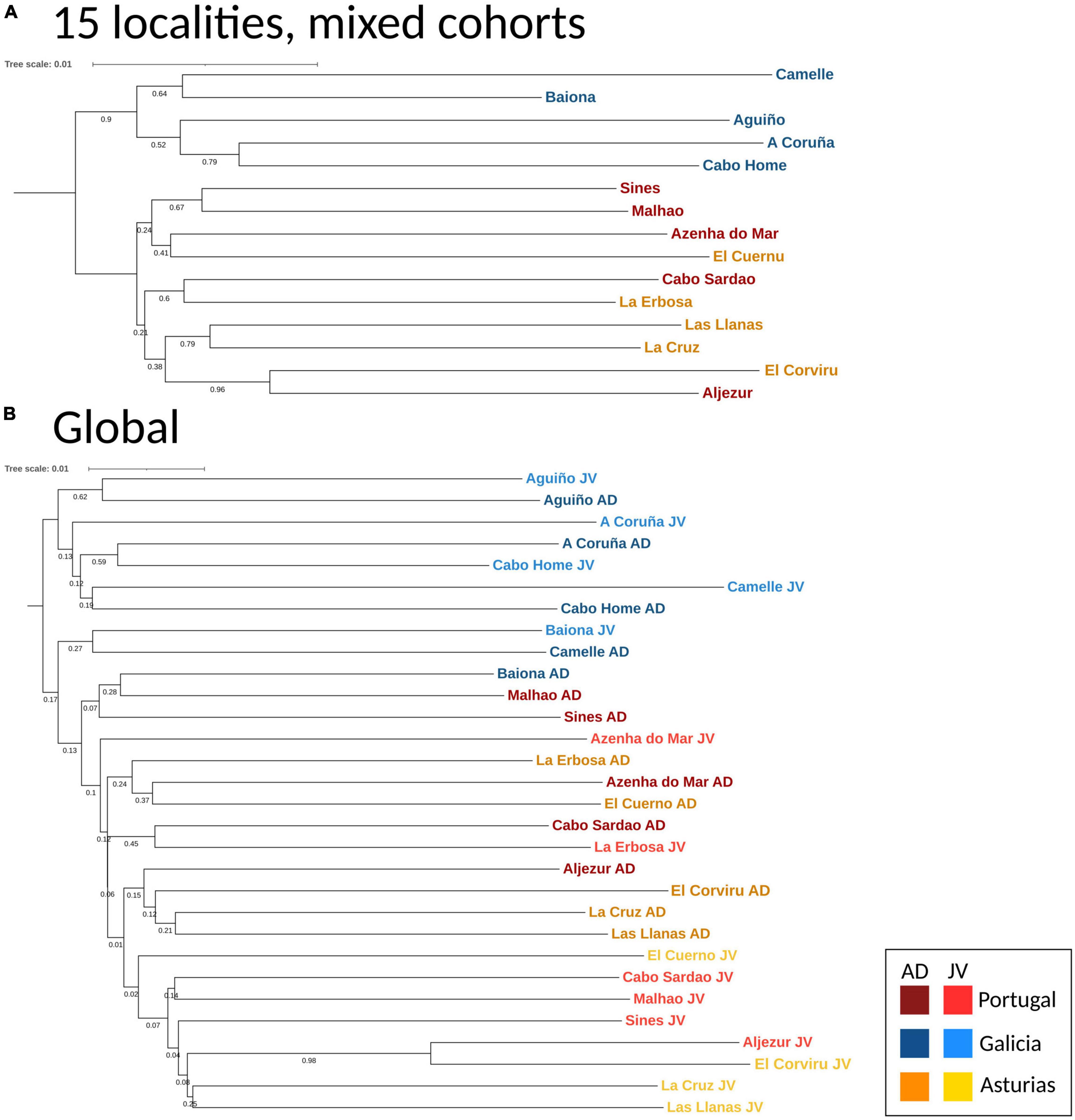
Figure 4. Neighbor Joining trees using DA distance (Nei and Takezaki, 1983) of P. pollicipes populations using microsatellites along the Iberian Peninsula. (A) Global analysis for 15 localities (adults and juveniles mixed together), (B) global analysis in 30 samples (localities + developmental stages, i.e., adults and juveniles).
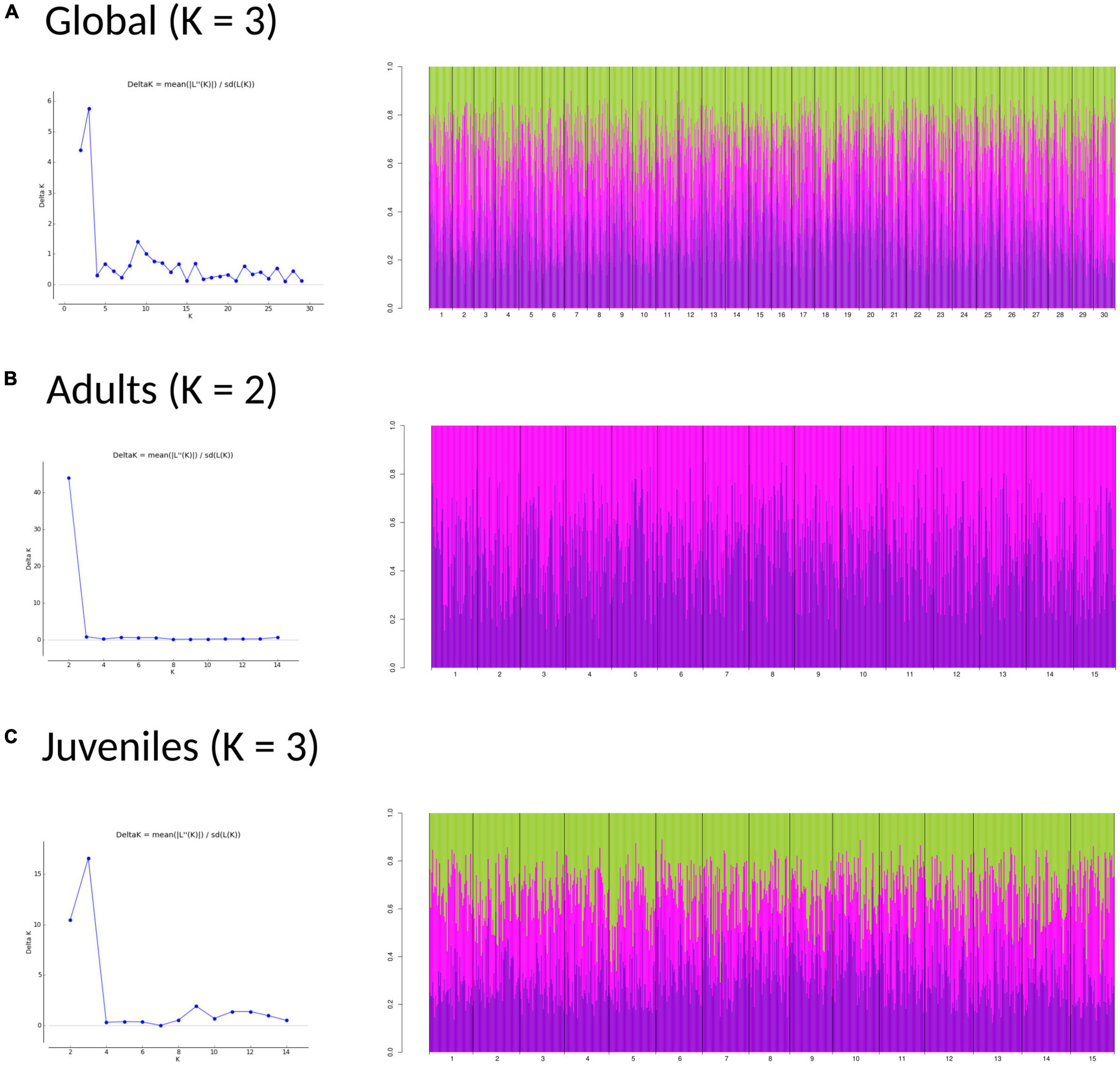
Figure 5. Structure bar-plot showing the assignment probabilities for each genotyped individual under admixture model. Global (A), adults (B), and juveniles (C) analysis. Each bar correspond with one individual.
Discussion
The analyses using twenty new microsatellite loci aimed to define, more accurately, the temporal and spatial evolution of the genetic structure of stalked barnacle P. pollicipes. This species is highly appreciated in the Spanish and Portuguese markets, and its management must be based on reliable scientific data. Previous studies have suggested that larval dispersal driven by ocean currents, in particular, the Iberian Poleward Current have played a crucial role in determining the population structure, and two distinct regional configurations have been established using mitochondrial DNA for P. pollicipes within its distribution range along the northeastern Atlantic. Quinteiro et al. (2007) suggested that P. pollicipes is structured into four genetically differentiated groups: French populations, eastern Asturian populations, Galician-Portuguese populations, and Canarian populations. Conversely, Campo et al. (2010) suggested the presence of only two groups, among which French populations were highlighted as a peculiar and differentiated genetic entity, as a result of a past population fragmentation during Pleistocene glacial/interglacial periods. Regardless, later studies based on estimates of population migration rates have suggested that barnacle population connectivity occurred on a small scale and in an asymmetric manner in the Cantabrian coast (Rivera et al., 2013). Information based on highly variable nuclear molecular markers can provide crucial information on both population connectivity and stock renewal for this species within the Iberian Peninsula. This information is needed for the delimitation of conservation/management units in this fishery and the improvement of the management plans and the performance of TURFs.
Genetic diversity contributes to the ability of a species to respond to environmental changes, and highly fecund species that release high numbers of small eggs into the environment (the so−called r−strategists) are much more polymorphic than species that produce a small number of relatively large offspring and provide parental care (called K−strategists) (Ellegren and Galtier, 2016). Recent studies in S. balanoides have confirmed that barnacles harbor high levels of genome-wide genetic variation (Nunez et al., 2021). The level of genetic diversity of P. pollicipes found in this work was particularly high. We observed higher levels of genetic variation in P. pollicipes than in other barnacles of the same genus, such as P. elegans (Plough and Marko, 2014). Our results showed that Galicia exhibited the highest values for allelic richness and observed and expected heterozygosity in adult and juvenile populations. Conversely, newly settled cohorts (juvenile) had a lower genetic diversity than adults across all the studied regions, particularly when examining both allelic richness and private alleles.
The main principal assumption of the Hardy–Weinberg principle is that the sample comes from a single, randomly mating population where perturbing forces (such as selection, genetic drift, mutation, and migration) are absent or balanced (Waples, 2014). All loci and populations showed significant deviations from Hardy-Weinberg equilibrium in this work due to, more or less pronounced, heterozygote deficiencies. This phenomenon could be the consequence of local admixtures of genetically differentiated populations (Wahlund effect), assortative mating, inbreeding, selection (Palumbi, 2003) and finally null alleles. The presence of null alleles has been reported in the vast majority of previous microsatellite studies in barnacles (Dufresne et al., 1999; Pannacciulli et al., 2005; Plough and Marko, 2014; Reigel et al., 2015; Abreu et al., 2016; Ewers-Saucedo et al., 2016) as well as in other marine invertebrate species such as clams (Borrell et al., 2014; Chiesa et al., 2016; Rico et al., 2017), octopus (Greatorex et al., 2000; De Luca et al., 2016), sea urchins (Mccartney et al., 2004; Carlon and Lippé, 2007), jellyfish (Aglieri et al., 2014), and polychetes (Jolly et al., 2003, 2009, 2014). The presence of null alleles is an inherent trait of microsatellite loci and is caused by mutations in the primer sequences, leading to the lack of amplification and the dropout of alleles (Selkoe and Toonen, 2006). In addition, an increase of the null allele frequency would be expected with the increase of alleles per locus and previous studies have indicated that the presence of null alleles seems to be particularly common in populations with high effective population sizes (Chapuis and Estoup, 2007). Although the presence of null alleles leads to an overestimation of both FST and genetic distances in cases of significant population differentiation (Chapuis and Estoup, 2007), our results showed no differences worth considering for both the FST or FST ENA values. It has been argued that the conservative approach of discarding loci deviating from Hardy-Weinberg equilibrium expectations could rob us of our most informative markers, weakening our ability to interpret biological phenomena (Dharmarajan et al., 2013). Moreover, De Meeûs (2018) stated that in case of null alleles, FIS and FST are augmented and a positive correlation is expected between FIS and FST as is expected a positive correlation between FIS and the number of missing data (putative null homozygotes), and StrdErrFIS being at least twice StrdErrFST. If such correlations do not exist and if StrdErrFIS > StrdErrFST, then a Wahlund effect better explains the data (De Meeûs, 2018; Manangwa et al., 2019). Waples (2018) had also argued about this and simulated 10% of null alleles suggesting that caution in interpreting FIS × FST correlations under conditions where null alleles might be common it is indeed necessary and more efforts will be needed for a comprehensive evaluation of this complex topic. In this work, panmixia is rarely met for any locus (Table 1), we found positive FIS × FST correlations, StrdErrFIS > StrdErrFST and we did not find positive correlations between FIS and the number of missing data (putative null homozygotes) pointing out to the fact that, even when null alleles are present, other biological factors also play a fundamental role to explain significant heterozygote deficits in our data.
Heterozygote deficiencies can as well be the result of local admixtures of genetically differentiated cohorts in populations, or due to sweepstake reproductive effort (Waples, 1998; Hedgecock and Pudovkin, 2011). Growth of individuals in P. pollicipes populations is highly heterogeneous (Cruz et al., 2010; Jacinto et al., 2015), so that individuals of similar size may differ greatly in age. Our adult samples likely contained a mixture of cohorts from different reproductive and dispersal events, potentially leading to significant departures from Hardy-Weinberg equilibrium, locally. Genetic heterogeneity of cohorts can potentially blur the genetic signal in adults and may decrease the genetic differences over time, given that the geographic origin of migrants might change throughout the breeding/dispersal seasons depending on prevailing local hydrodynamics during these periods. However, it should be noted that a special care was taken in this work to sample only one cohort of juveniles with a specific size (2–4 mm RC). If the deficiencies of heterozygotes were due the superimposition of cohorts, juveniles should not show such deficiencies. This was clearly not the case here as our results demonstrated that juvenile mean FIS values were higher than those for adults in all the three regions (Table 2). It has been stated that the surf zone and its surrounding nearshore waters are known to act as selective barriers to the onshore transport of many larval invertebrates on the local scale (Porri et al., 2006; Rilov et al., 2008). The permeability of such barrier is modulated by small scale topography that generates retentive oceanographic features like coastal fronts (Pineda, 1999; Shanks et al., 2003). In fact, the larvae of P. pollicipes and other barnacles have been shown to accumulate in great numbers at internal waves and river plume fronts off the Cantabrian coast only at some specific locations (Weidberg et al., 2014; Höfer et al., 2017). In this topic the available evidence are indeed scarce, however, genetic data seems to confirm it.
Pollicipes pollicipes has asynchronous broods during the reproductive season which usually occurs from March to September (e.g., Cardoso and Yule, 1995; Cruz and Hawkins, 1998; Pavón, 2003; Macho, 2006), where several batches of larvae are produced, and potentially lead to the co-occurrence of different settlement events. Juveniles sampled in this study might, however, come from one to few settlement events. Despite the possibility of several discrete settlement events, post larval mortality might favor one specific batch of survivors, and in the end, the 2–4 mm RC juveniles might become more related than what would have been expected from the mixing of several reproductive events. Pineda et al. (2006) found that recruitment to the reproductive stage of acorn barnacles (S. balanoides) was composed of survivors that settled in a recruitment window. The recruitment window (to reproduction in the case of the Pineda study, to 2–4 mm in our study) might be narrower than the recruitment season. If by some reason these survivors correspond to larvae that are genetically more related, then a pattern of genetic differentiation could occur among recruits. The concept of a “recruitment window” proposed by Pineda et al. (2006) matches quite well with Hedgecock’s “sweepstakes-chance matching hypothesis” also known as “sweepstakes reproductive success hypothesis,” which is based in part on the observation of reduced genetic variability in young-of-the-year populations relative to adult populations. This reduced genetic variability among recruits suggests that the surviving young of the year are the products of spawning by only a small fraction of the adult population, which, according to Hedgecock’s hypothesis, happened to produce their offspring at a place and time that was suitable for survival (Hedgecock, 1994). Moreover, barnacles rear embryos in bags before hatching and there is also the possibility that the larval release is only efficient for a small proportion of the reproductive adults depending on the local hydrodynamics. In this work, we found evidence indicative of reproductive sweepstakes in adult and juvenile samples. Although globally, the relatedness coefficients estimated for P. pollicipes were in the same range as those from other studies previously conducted with barnacles (Veliz et al., 2006; Plough et al., 2014), they were significantly slightly greater in juveniles (i.e., Asturias and Portugal) compared with adults. Juveniles were significantly more related to each other than expected from random mixing despite their larval entrainment in the water column during the planktonic phase.
Unexpected genetic differentiation in marine invertebrates can occur due to three neutral processes: sweepstake reproductive success (Hedgecock, 1994), collective dispersal (Johnson et al., 1993; Li and Hedgecock, 1998) and asynchronous population dynamics (Eldon et al., 2016), but also selective processes during the settlement process. According to the Hedgecock’s “sweepstakes-chance matching hypothesis” or selective sweepstakes (Hedgecock, 1994), only a fortunate combination (hence sweepstakes) of reproductive traits and oceanographic conditions would allow an individual to complete the long mobile phase from spawning and fertilization through larval survival to recruitment back to the adult habitat. In a highly fecund species and a locally heterogeneous oceanographic setting, this would involve strong selection favoring just a handful of genotypes at each locality, leading to a local-scale genetic mosaic but a relatively large-scale uniformity. Post-larval settlement selection under different environmental conditions has been argued to create CGP in coastal areas of temperate regions over a mosaic of contrasting habitats able to impose a strong differential selective sieve or a target for habitat choice in larvae (Eldon et al., 2016). We detected significant genetic differentiation for juveniles among and within regions (but not for adults), together with significant genetic heterogeneity between P. pollicipes generations. However, we did not find evidence of such selective processes for the assayed microsatellites. There seemed to be a genome-wide pattern that was more parsimoniously explained by neutral processes such as sweepstake reproductive success, which may greatly reduce the genetic diversity of a given cohort while provoking unexpected heterozygote deficiencies, as seen previously, by mimicking local bottlenecks (genetic diversity drawn from a small subset of parents). In addition to this phenomenon, genetic differentiation may persist in recruits when dispersal is limited in space, when larvae from different cohorts do not mix completely during dispersal (collective dispersal), or when local conditions may promote self-recruitment (Eldon et al., 2016).
The genetic data obtained in this work, after applying dissimilar approaches (F and ϕST statistics, Discriminant Principal Component and Bayesian analyses), pointed all out to the existence of significant genetic heterogeneity in the Iberian coasts rejecting previous findings using mitochondrial DNA. The results herein highlighted Galicia as a peculiar genetic entity possibly representing a superimposition of two distinct metapopulations or potentially an old refuge for the most northern populations from France (not sampled in this study). Among Galician northernmost populations, Camelle (CA) and A Coruña (AC) are also the most differentiated from Portugal and Asturias and may have a specific demographic history. The sampled P. pollicipes populations are located along the Atlantic Iberian coast, whose hydrodynamic patterns have been well studied. The western peninsular coast (SW Portugal and Galicia) is characterized by a complex current system subject to strong seasonality and mesoscale variability, showing inverse patterns between summer and winter in the upper layers of the shelf and slope. During spring and summer (coinciding with P. pollicipes breeding season), northerly winds along the coast are dominant, causing coastal upwelling and producing a southward current on the surface and a northward undercurrent on the slope. In the Cantabrian Sea (Asturias) the surface currents flow generally eastward in winter and early spring and shift westward in late spring and summer following the wind force with intermittent summer upwelling events west of Cape Peñes (ICES, 2021). Different aspects of the oceanographic circulation in Iberia were reviewed by Relvas et al. (2007). Casabella et al. (2014) divided the upwelling affecting the coasts of Galicia into three regions: Rías Baixas, Fisterra-Bares and Cantabrian. These two locations (CA and AC) would be found in the Fisterra-Bares region, which is the region with a greater intensity of upwelling, although the period favorable for upwelling is longer in the region of Rias Baixas (sampled here i.e., Baiona). Galician juveniles showed clear genetical differences from those of Portugal and Asturias. The main explanation for this distinction is that the Biscay Bay Current, characterized by a wide gyre, can trap larvae, and thus should favor self-recruitment and perhaps local importations from the French and Cantabrian populations. This ocean circulation could also be responsible for the differentiation between juveniles from Asturian and Galicia. Previous studies on adult barnacles have found significant differences between the Asturian and Galician localities (Quinteiro et al., 2007). However, it should be noted that most of the Asturian sites sampled in this study are located to the West of Cape Peñes, while the site sampled by Quinteiro et al. (2007) was located to the East of the same cape, which has been described as a biogeographic barrier (Anadón and Niell, 1980). Rivera et al. (2013) showed that during a year of high upwelling activity (2009), the theoretical P. pollicipes recruitment success was 94%, with a recruitment peak predicted 56 km west of the emission point. Consistently, migration rates derived from genetic analyses showed that westward dispersal was much more likely along the Cantabrian coast, which matches the upwelling driven circulation typical of the stalked barnacle larval season in summer/autumn (Figure 1). Thus, the recurrence of upwelling may not only define the spatial scale and direction of the dispersal process but also the genetic structure of the barnacle metapopulation.
The Western Iberian upwelling system represents an important crossroad between Lusitanian and boreal temperate species (Jolly et al., 2006; Maggs et al., 2008). Upwelling/downwelling wind-driven circulation and tides are recurrent physical processes along the Atlantic Iberian coastlines and are among the most energetic phenomena that can affect near-shore circulation during the spring and summer periods when reproduction occurs and, during the summer and beginning of autumn in the case of recruitment (Queiroga et al., 2007). However, when studying a strong upwelling region in the northeastern Pacific coast, Morgan et al. (2009) observed that the larvae of most invertebrate species remain close to the shore even during strong upwelling, where high local retention and limited connectivity have been evidenced in populations of several species, such as Petrolisthes cinctipes (Hameed et al., 2016) or in the red rock lobster Panulirus interruptus (Iacchei et al., 2013). Despite this phenomenon, upwelling areas have been pointed out as probable climate change refuges for the distribution of Fucus guiryi, other barnacles such as S. balanoides and other sessile marine species (Gómez and Lunt, 2007 for a review; Hoarau et al., 2007; Provan and Bennett, 2008; Lourenço et al., 2016; Herrera, 2019). In addition, Campo et al. (2010) suggested the existence of a Pleistocene refuge area off the coast of North Africa and two additional northern glacial refuges for P. pollicipes, in the English Channel/Brittany region and in the northwestern Iberian Peninsula.
Previous studies have mentioned that the southern region of Portugal also represents a well-known upwelling area (Lourenço et al., 2016) with a high level of barnacle larval settlement (Queiroga et al., 2007) and recruitment (Aguión et al., in prep.). Remarkably, the number of private alleles was significantly higher in adults there when compared with those from Galicia and in Asturias. Portuguese juveniles were, however, significantly less genetically diversified and more related to each other than expected based on random mating. Moreover, Nolasco et al. (in prep.) show that connectivity matrices integrated over the period of the observations (July 2017 to July 2019) indicate high levels of larval retention. Such retention is probably caused by the recurrent eddies driven by upwelling circulation observed off southern-central Portugal in between Cape Roca and Cape San Vicente (Figure 1; Haynes et al., 1993; Batteen et al., 2000; Sánchez and Relvas, 2003; Peliz et al., 2004). These findings suggest that Portuguese populations are likely to export more migrants than they receive. As Queiroga et al. (2007) hypothesized, regular exchanges of larvae over the distance separating the southern and northern parts of Portugal are unlikely. Conversely, the Portugal Current, which shows a north- or southward direction, depending on the season, could be an important factor in promoting gene flow between our sampling locations in southern Portugal and other, unsampled, P. pollicipes southernmost areas such as the Canarian and North African coasts. Nevertheless, microsatellite markers have recently shown a genetic differentiation between European and African P. pollicipes populations (Fernandes et al., in prep).
The correct management of marine ecosystems relies on understanding the scale and magnitude of connectivity among populations through the identification of adaptive genetic differences (Almany et al., 2009; Aceves-Bueno et al., 2017), because locally adapted populations should be considered poorly-connected, separate management units (Waples, 1998). Our results suggested that P. pollicipes populations in the Iberian Peninsula possibly exhibit a “CGP” structure, which extends from a few kilometers apart to as much as hundreds of kilometers apart. This phenomenon has clear consequences for the sustainable management of resources. Currently, an increasing number of small-scale fisheries have successfully implemented co-managed TURFs; a governance arrangement that enables the collaboration across diverse stakeholders, develops new knowledge and increases the capacity of the system to deal with new drivers (Rivera et al., 2014). However, the design of TURFs does not usually account for the spatial configuration of resources (Aceves-Bueno et al., 2017) due to the multi-species nature of fisheries. This mismatch between management and biological scales can compromise the sustainability of sessile stocks (Ouréns et al., 2015), like barnacles. However, a better understanding of the spatial structure and larval dynamics of the population, permits the redefinition of management units according to population boundaries. In addition to these management measures, it would be interesting to implement networks of protected areas at detailed scales to ensure that propagules are available when and where conditions are favorable for their survival (Larson and Julian, 1999; Ouréns et al., 2015).
Conclusion
New molecular markers have been developed in the highly valued species P. pollicipes and offer useful tools to provide a better fine-tuning assessment of its population dynamics along the Iberian Peninsula. P. pollicipes displays high genetic diversity, which is attributable to large effective population sizes representing a well-connected network of local populations. However, temporal and spatial genetic differentiation of populations over regional scales, on one hand, and a significant reduction in genetic diversity in juveniles, on the other hand, clearly indicate that patterns of exchanges together with seasonal wind-induced upwelling may induce genetic differences between settlers throughout generations. Such patterns of CGP are likely due to sweepstake reproductive success with possible collective dispersal or episodic self-recruitment events. Therefore, our P. pollicipes genetic dataset suggests that recruitment may be stochastic and highly dependent on climatic conditions with multiple sources of emissions. These phenomena may have strong implications in terms of management plans over the whole Iberian Peninsula with the need to protect a series of putative sources within each region. Future research should combine genetic information at broader spatiotemporal scales with larval dispersal models based on ecological and biological characteristics of P. pollicipes. This means, among others, mapping the complete species distribution and tracking the genetic structure of age groups over time and space. It also means applying new sequencing technologies to fully understand the dynamics of larval exchanges and the post-larval settlement of the stalked barnacle but also to better apprehend how environmental variations shape genomic variation in this species.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Author Contributions
MP: methodology, formal analysis, investigation, writing—original draft, and writing—review and editing. PM: conceptualization, supervision, methodology, and writing—review and editing. MB and NW: methodology and writing—review and editing. JLA: conceptualization, supervision, project administration, funding acquisition, and writing—review and editing. AA, JA, JNF, LG-F, and KJG: writing—review and editing. JC: methodology. TC and GM: conceptualization and writing—review and editing. EG-V: conceptualization, funding acquisition, and writing—review and editing. ET and DJ: conceptualization, supervision, and writing—review and editing. YJB: conceptualization, supervision, funding acquisition, and writing—review and editing. All authors contributed to manuscript revision, read, and approved the submitted version.
Funding
This research was funded by the project PERCEBES (BiodivERsA) (PCIN-2016-120) and (PCIN-2016-063), the projects AYUD/2021/50967 and ECOSIFOOD (MCI-20- PID2019-108481RB-I00/AEI/10.13039/501100011033). This work also had the support of Fundação para a Ciência e Tecnologia (FCT), through the strategic project UIDB/04292/2020-MARE granted to MARE and Xunta de Galicia (GRC, ED431C 2020-05). AA was supported by a FPU fellowship (Ministerio de Ciencia, Innovacion y Universidades de España, Grant No. FPU2016-04258) and KJG was supported through the Severo Ochoa Ph.D. program (Principado de Asturias, PA-18-PF-BP17-184). GM was supported by post-doctoral contracts from project PERCEBES (PCIN-2016-063) and MARISCO (CTM2014-51935-R, Ministerio de Economía y Competitividad). NW was funded by NASA grant 80NSSC20K0074 during data processing and manuscript elaboration. This is a contribution of the Marine Observatory of Asturias (OMA) and the Biotechnology Institute of Asturias (IUBA).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank the barnacle harvesters at the Brittany, Asturias, Galicia, Alentejo, and Algarve coasts, who provided part of the samples for this study. A. Roca, G. Machado, F. Ayllón, J. L. Hórreo, and C. García made all useful comments about the initial manuscript in a Ph.D. dissertation evaluation.
Abbreviations
CGP, chaotic genetic patchiness; TURF, territorial use rights for fishing.
Footnotes
- ^ https://cds.climate.copernicus.eu/cdsapp#!/dataset/satellite-sea-surface-temperature?tab=overview
- ^ https://coastwatch.pfeg.noaa.gov/erddap/griddap/jplOscar_LonPM180.html
- ^ https://itol.embl.de
References
Abreu, N. M. N., Marçal, I., Borges Duarte, A., Bettini Pitombo, F., Vilasboa, A., and Gusmão, J. (2016). Microsatellite markers for barnacle studies: isolation and characterization of polymorphic microsatellite loci from the invasive barnacle Megabalanus coccopoma (Darwin, 1854) and its cross-amplification in the Southern Atlantic endemic species Megabalanus. Biochem. Syst. Ecol. 66, 224–228. doi: 10.1016/j.bse.2016.04.006
Aceves-Bueno, E., Cornejo-Donoso, J., Miller, S. J., and Gaines, S. D. (2017). Are territorial use rights in fisheries (TURFs) sufficiently large? Mar. Policy. 78, 189–195. doi: 10.1016/j.marpol.2017.01.024
Aglieri, G., Papetti, C., Zane, L., Milisenda, G., Boero, F., and Piraino, S. (2014). First evidence of inbreeding, relatedness and chaotic genetic patchiness in the holoplanktonic jellyfish Pelagia noctiluca (Scyphozoa, Cnidaria). PLoS One 9:e99647. doi: 10.1371/journal.pone.0099647
Aguión, A., Ojea, E., García-Flórez, L., Cruz, T., Garmendia, J. M., Davoult, D., et al. (2021). Establishing a governance threshold in small-scale fisheries to achieve sustainability. Ambio 51, 652–665. doi: 10.1007/s13280-021-01606-x
Almany, G. R., Connolly, S. R., Heath, D. D., Hogan, J. D., Jones, G. P., McCook, L. J., et al. (2009). Connectivity, biodiversity conservation and the design of marine reserve networks for coral reefs. Coral Reefs 28, 339–351. doi: 10.1007/s00338-009-0484-x
Alshari, N. F. M. A. H., Ahmad, S. Z., Azlan, A., Lee, Y. H., Azzam, G., and Nor, S. A. M. (2021). Metabarcoding of fish larvae in the Merbok River reveals species diversity and distribution along its mangrove environment. Zool. Stud. 60:76. doi: 10.6620/ZS.2021.60-76
Álvarez-Fernández, E. (2011). Humans and marine resource interaction reappraised: archaeofauna remains during the late Pleistocene and Holocene in Cantabrian Spain. J. Anthropol. Archaeol. 30, 327–343. doi: 10.1016/j.jaa.2011.05.005
Álvarez-Fernández, E., Barrera, I., Borja, A., Fernández, M., Iriarte, M., and Arrizabalaga, A. (2013). Biometric analysis of the stalked barnacle Pollicipes pollicipes, at a Holocene archaeological site in Jaizkibel (Basque Country, northern Spain). Holocene 23, 1373–1380. doi: 10.1177/0959683613489584
Álvarez-Fernández, E., Ontañón-Peredo, R., and Molares-Vila, J. (2010). Archaeological data on the exploitation of the goose barnacle Pollicipes pollicipes (Gmelin, 1790) in Europe. J. Archaeol. Sci. 37, 402–408. doi: 10.1016/j.jas.2009.10.003
Anadón, R., and Niell, F. X. (1980). Distribución longitudinal de macrófitos en la costa asturiana (N de España). Investig. Pesq. 45, 143–156.
Barahona, M., Broitman, B. R., Faugeron, S., Jaugeon, L., Ospina-Alvarez, A., Véliz, D., et al. (2019). Environmental and demographic factors influence the spatial genetic structure of an intertidal barnacle in central-northern Chile. Mar. Ecol. Prog. Ser. 612, 151–165. doi: 10.3354/meps12855
Barnes, M. (1996). Pedunculate cirripedes of the genus Pollicipes. Oceanogr. Mar. Biol. Annu. Rev. 34, 303–394.
Barnes, M. (2008). Pollicipes pollicipes A Goose Barnacle. Available online at: https://www.marlin.ac.uk/species/detail/43 (accessed March 15, 2021).
Barshis, D. J., Sotka, E. E., Kelly, R. P., Sivasundar, A., Menge, B. A., Barth, J. A., et al. (2011). Coastal upwelling is linked to temporal genetic variability in the acorn barnacle Balanus glandula. Mar. Ecol. Prog. Ser. 439, 139–150. doi: 10.3354/meps09339
Batteen, M. L., Martinez, J. R., Bryan, D. W., and Buch, E. J. (2000). A modeling study of the coastal eastern boundary current system off Iberia and Morocco. J. Geophys. Res. Oceans 105, 14173–14195. doi: 10.1029/2000JC900026
Belkhir, K., Borsa, P., Chikhi, L., Raufaste, N., and Bonhomme, F. (2004). GENETIX 4.05, Logiciel Sous Windows TM Pour la Génétique des Populations. Montpellier: Université de Montpellier II. Laboratoire Génome, Populations, Interactions, CNRS UMR 5000.
Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 57, 289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x
Benson, G. (1999). Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27, 573–580. doi: 10.1093/nar/27.2.573
Bloor, P. A., Barker, F. S., Watts, P. C., Noyes, H. A., and Kemp, S. J. (2001). Microsatellite Libraries by Enrichment, Version 1.0. Animal Genomics Laboratory, School of Biological Sciencies. Liverpool: University of Liverpool.
Borrell, Y. J., Arias-Pérez, A., Freire, R., Valdés, A., Sánchez, J. A., Méndez, J., et al. (2014). Microsatellites and multiplex PCRs for assessing aquaculture practices of the grooved carpet shell Ruditapes decussatus in Spain. Aquaculture 426–427, 49–59. doi: 10.1016/j.aquaculture.2014.01.010
Borrell, Y. J., Piñera, J. A., Sánchez Prado, J. A., and Blanco, G. (2012). Mitochondrial DNA and microsatellite genetic differentiation in the European anchovy Engraulis encrasicolus L. ICES J. Mar. Sci. 69, 1357–1371. doi: 10.1093/icesjms/fss129
Brault, S., Stuart, C. T., Wagstaff, M. C., and Rex, M. A. (2013). Geographic evidence for source-sink dynamics in deep-sea neogastropods of the eastern North Atlantic: an approach using nested analysis. Glob. Ecol. Biogeogr. 22, 433–439. doi: 10.1111/geb.12005
Campo, D., Molares, J., Garcia, L., Fernandez-Rueda, P., Garcia-Gonzalez, C., and Garcia-Vazquez, E. (2010). Phylogeography of the European stalked barnacle (Pollicipes pollicipes): identification of glacial refugia. Mar. Biol. 157, 147–156. doi: 10.1007/s00227-009-1305-z
Cardoso, A. C., and Yule, A. B. (1995). Aspects of the reproductive biology of Pollicipes pollicipes (Cirripedia; Lepadomorpha) from the southwest coast of Portugal. Neth. J. Aquat. Ecol. 29, 391–396. doi: 10.1007/BF02084238
Carlon, D. B., and Lippé, C. (2007). Eleven new microsatellite markers for the tropical sea urchin Tripneustes gratilla and cross-amplification in Tripneustes ventricosa. Mol. Ecol. Notes 7, 1002–1004. doi: 10.1111/j.1471-8286.2007.01755.x
Carvalho, A. N., Vasconcelos, P., Piló, D., Pereira, F., and Gaspar, M. B. (2017). Socio-economic, operational and technical characterization of the harvesting of gooseneck barnacle (Pollicipes pollicipes) in SW Portugal: insights towards fishery co-management. Mar. Policy 78, 34–44. doi: 10.1016/j.marpol.2017.01.008
Casabella, N., Lorenzo, M. N., and Taboada, J. J. (2014). Trends of the galician upwelling in the context of climate change. J. Sea Res. 93, 23–27. doi: 10.1016/j.seares.2014.01.013
Cavalli-Sforza, L. L., and Edwards, A. W. (1967). Phylogenetic analysis. Models and estimation procedures. Am. J. Hum. Genet. 19(3 Pt 1):233. doi: 10.1111/j.1558-5646.1967.tb03411.x
Chan, B. K. K., Dreyer, N., Glenner, H., Pérez-Losada, M., Ewers-Saucedo, C., Gale, A. S., et al. (2021). The evolutionary diversity of barnacles and a classification of fossil and living forms. Zool. J. Linn. Soc. 193, 789–846. doi: 10.1093/zoolinnean/zlaa160
Chapuis, M. P., and Estoup, A. (2007). Microsatellite null alleles and estimation of population differentiation. Mol. Biol. Evol. 24, 621–631. doi: 10.1093/molbev/msl191
Chen, H. N., Chan, B. K., and Tsang, L. M. (2015). Transcriptome derived microsatellite markers of Tetraclita kuroshioensis and cross amplification among Tetraclita spp. Biochem. Syst. Ecol. 63, 13–16. doi: 10.1016/j.bse.2015.09.015
Chen, H. N., Hoeg, J. T., and Chan, B. K. K. (2013). Morphometric and molecular identification of individual barnacle cyprids from wild plankton: an approach to detecting fouling and invasive barnacle species. Biofouling 29, 133–145. doi: 10.1080/08927014.2012.753061
Chiesa, S., Lucentini, L., Freitas, R., Nonnis Marzano, F., Ferrari, C., Filonzi, L., et al. (2016). Null alleles of microsatellites for Manila clam Ruditapes philippinarum. Anim. Genet. 47, 135–136. doi: 10.1111/age.12382
Costello, C. J., Lynham, J., Lester, S. E., and Gaines, S. D. (2010). Economic incentives and global fisheries sustainability. Annu. Rev. Resour. Econ. 2, 299–318. doi: 10.1146/annurev.resource.012809.103923
Couvray, S., and Coupé, S. (2018). Three-year monitoring of genetic diversity reveals a micro-connectivity pattern and local recruitment in the broadcast marine species Paracentrotus lividus. Heredity 120, 110–124. doi: 10.1038/s41437-017-0013-6
Cowen, R. K., and Sponaugle, S. (2009). Larval dispersal and marine population connectivity. Annu. Rev. Mar. Sci. 1, 443–466. doi: 10.1146/annurev.marine.010908.163757
Cruz, T., Castro, J. J., and Hawkins, S. J. (2010). Recruitment, growth and population size structure of Pollicipes pollicipes in SW Portugal. J. Exp. Mar. Biol. Ecol. 392, 200–209. doi: 10.1016/j.jembe.2010.04.020
Cruz, T., and Hawkins, S. J. (1998). Reproductive cycle of Pollicipes pollicipes at Cabo de Sines, south-west coast of Portugal. J. Mar. Biol. Assoc. U. K. 78, 483–496. doi: 10.1017/S0025315400041576
Cruz, T., Jacinto, D., Sousa, A., Penteado, N., Pereira, D., Fernandes, J. N., et al. (2015). The state of the fishery, conservation and management of the stalked barnacle Pollicipes pollicipes in Portugal. Mar. Environ. Res. 112, 73–80. doi: 10.1016/j.marenvres.2015.10.005
Cruz, T. P. G. (2000). Biologia e Ecologia do Percebe Pollicipes pollicipes (Gmelin, 1790), no Litoral Sudoeste Portugês. Évora: Universidade de Évora.
D’Aloia, C. C., Bogdanowicz, S. M., Francis, R. K., Majoris, J. E., Harrison, R. G., and Buston, P. M. (2015). Patterns, causes, and consequences of marine larval dispersal. Proc. Natl. Acad. Sci. U.S.A. 112, 13940–13945. doi: 10.1073/pnas.1513754112
Dawson, M. N., Grosberg, R. K., Stuart, Y. E., and Sanford, E. (2010). Population genetic analysis of a recent range expansion: mechanisms regulating the poleward range limit in the volcano barnacle Tetraclita rubescens. Mol. Ecol. 19, 1585–1605. doi: 10.1111/j.1365-294X.2010.04588.x
De Luca, D., Catanese, G., Procaccini, G., and Fiorito, G. (2016). Octopus vulgaris (Cuvier, 1797) in the mediterranean sea: genetic diversity and population structure. PLoS One 11:e0149496. doi: 10.1371/journal.pone.0149496
De Meeûs, T. (2018). Revisiting FIS, FST, Wahlund effects, and null alleles. J. Hered. 109, 446–456. doi: 10.1093/jhered/esx106
Dharmarajan, G., Beatty, W., and Rohodes, O. E. (2013). Heterozygote deficiencies caused by a wahlund effect: dispelling unfounded expectations. J. Wildl. Manag. 77, 226–234. doi: 10.1002/jwmg.458
Dubé, C. E., Boissin, E., Mercière, A., and Planes, S. (2020). Parentage analyses identify local dispersal events and sibling aggregations in a natural population of Millepora hydrocorals, a free-spawning marine invertebrate. Mol. Ecol. 29, 1508–1522. doi: 10.1111/mec.15418
Dufresne, F., Parent, M., and Bernatchez, L. (1999). Isolation and characterization of microsatellite markers in the acorn barnacle Semibalanus balanoides. Mol. Ecol. 8, 1558–1559. doi: 10.1046/j.1365-294X.1999.07225.x
Earl, D. A., and VonHoldt, B. M. (2012). Structure harvester: a website and program for visualizing STRUCTURE output and implementing the evanno method. Conserv. Genet. Resour. 4, 359–361. doi: 10.1007/s12686-011-9548-7
Eldon, B., Riquet, F., Yearsley, J., Jollivet, D., and Broquet, T. (2016). Current hypotheses to explain genetic chaos under the sea. Curr. Zool. 62, 551–566. doi: 10.1093/cz/zow094
Ellegren, H., and Galtier, N. (2016). Determinants of genetic diversity. Nat. Rev. Genet. 17, 422–433. doi: 10.1038/nrg.2016.58
Estoup, A., Largiadèr, C. R., Perrot, E., and Chourrout, D. (1996). Rapid one-tube DNA extraction for reliable PCR detection of fish polymorphic markers and transgenes. Mol. Mar. Biol. Biot 5, 295–298.
Evanno, G., Regnaut, S., and Goudet, J. (2005). Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 14, 2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x
Ewers-Saucedo, C., Chan, B. K. K., Zardus, J. D., and Wares, J. P. (2017). Parallel patterns of host-specific morphology and genetic admixture in sister lineages of a commensal barnacle. Biol. Bull. 232, 171–185. doi: 10.1086/693356
Ewers-Saucedo, C., Zardus, J. D., and Wares, J. P. (2016). Microsatellite loci discovery from nextgeneration sequencing data and loci characterization in the epizoic barnacle Chelonibia testudinaria (Linnaeus, 1758). PeerJ 2016:e2019. doi: 10.7717/peerj.2019
Excoffier, L., Laval, G., and Schneider, S. (2005). Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol. Bioinforma. 1:117693430500100. doi: 10.1177/117693430500100003
Fernandes, J. N., Cruz, T., and Van Syoc, R. (2010). Pollicipes caboverdensis sp. nov. (Crustacea: Cirripedia: Scalpelliformes), an intertidal barnacle from the cape verde Islands. Zootaxa 2557, 29–38. doi: 10.11646/zootaxa.2557.1.3
Fernandes, J. N., Jacinto, D., Penteado, N., Sousa, A., Mateus, D., Seabra, M. I., et al. (2021). Ten years of monitoring recruitment of the edible stalked barnacle Pollicipes pollicipes: linking to oceanographic variability. Limnol. Oceanogr. 66, 2309–2318. doi: 10.1002/lno.11755
Flight, P. A., O’Brien, M. A., Schmidt, P. S., and Rand, D. M. (2012). Genetic structure and the North American postglacial expansion of the barnacle. Semibalanus balanoides. J. Hered. 103, 153–165. doi: 10.1093/jhered/esr083
Foll, M., and Gaggiotti, O. (2008). A genome-scan method to identify selected loci appropriate for both dominant and codominant markers: a bayesian perspective. Genetics 180, 977–993. doi: 10.1534/genetics.108.092221
Fontani, S. (2009). Genetic Biodiversity of the European Barnacle Chthamalus Montagui. Doctoral dissertation. Plymouth PL: University of Plymouth. Doctoral dissertation
Fox, J. (2005). Getting started with the R commander: a basic-statistics graphical user interface to R. J. Stat. Softw. 14, 1–42. doi: 10.1007/0-387-22769-5_1
Franco, S. C., Aldred, N., Cruz, T., and Clare, A. S. (2017). Effects of culture conditions on larval growth and survival of stalked barnacles (Pollicipes pollicipes). Aquac. Res. 48, 2920–2933. doi: 10.1111/are.13125
Franco, S. C., Aldred, N., Cruz, T., Clare, A. S., and Clare, A. S. (2016). Modulation of gregarious settlement of the stalked barnacle, Pollicipes pollicipes: a laboratory study. Sci. Mar. 80, 217–228. doi: 10.3989/scimar.04342.01A
Gagnaire, P. A., Broquet, T., Aurelle, D., Viard, F., Souissi, A., Bonhomme, F., et al. (2015). Using neutral, selected, and hitchhiker loci to assess connectivity of marine populations in the genomic era. Evol. Appl. 8, 769–786. doi: 10.1111/eva.12288
Gilbert, K. J., Andrew, R. L., Bock, D. G., Franklin, M. T., Kane, N. C., Moore, J. S., et al. (2012). Recommendations for Utilizing and Reporting Population Genetic Analyses: The Reproducibility of Genetic Clustering Using the Program Structure. Hoboken, NJ: John Wiley & Sons, Ltd, doi: 10.1111/j.1365-294X.2012.05754.x
Gómez, A., and Lunt, D. H. (2007). “Refugia within refugia: patterns of phylogeographic concordance in the Iberian Peninsula,” in Phylogeography of Southern European Refugia, eds S. Weiss and N. Ferrand (Dordrecht: Springer), 155–188. doi: 10.1007/1-4020-4904-8_5
Goudet, J. (1995). FSTAT (Version 1.2): a computer program to calculate f-statistics. J. Hered. 86, 485–486. doi: 10.1093/oxfordjournals.jhered.a111627
Greatorex, E. C., Jones, C. S., Murphy, J., Key, L. N., Emery, A. M., and Boyle, P. R. (2000). Microsatellite markers for investigating population structure in Octopus vulgaris (Mollusca: Cephalopoda). Mol. Ecol. 9, 641–642. doi: 10.1046/j.1365-294x.2000.00882-7.x
Gutiérrez, N. L., Hilborn, R., and Defeo, O. (2011). Leadership, social capital and incentives promote successful fisheries. Nature 470, 386–389. doi: 10.1038/nature09689
Hameed, S. O., Wilson White, J., Miller, S. H., Nickols, K. J., and Morgan, S. G. (2016). Inverse approach to estimating larval dispersal reveals limited population connectivity along 700 km of wave-swept open coast. Proc. R. Soc. B Biol. Sci. 283:20160370. doi: 10.1098/rspb.2016.0370
Haynes, R., Barton, E. D., and Pilling, I. (1993). Development, persistence, and variability of upwelling filaments off the Atlantic coast of the Iberian Peninsula. J. Geophys. Res. Oceans 98, 22681–22692. doi: 10.1029/93JC02016
Hecker, K. H., and Roux, K. H. (1996). High and low annealing temperatures increase both specificity and yield in touchdown and stepdown PCR. BioTechniques 20, 478–485. doi: 10.2144/19962003478
Hedgecock, D. (1994). Does variance in reproductive success limit effective population sizes of marine organisms. Genet. Evol. Aquat. Organ. 122, 122–134.
Hedgecock, D., and Pudovkin, A. I. (2011). Sweepstakes reproductive success in highly fecund marine fish and shellfish: a review and commentary. Bull. Mar. Sci. 87, 971–1002. doi: 10.5343/bms.2010.1051
Herrera, M. (2019). The Effects of Climate Change on Reproduction and Recruitment Success of the Acorn Barnacle Semibalanus Balanoides at its Southernmost European Distribution limit (Galicia, Spain). Doctoral dissertation. Pontevedra: Universidade de Vigo. Doctoral dissertation
Hoarau, G., Coyer, J. A., Veldsink, J. H., Stam, W. T., and Olsen, J. L. (2007). Glacial refugia and recolonization pathways in the brown seaweed Fucus serratus. Mol. Ecol. 16, 3606–3616. doi: 10.1111/j.1365-294X.2007.03408.x
Höfer, J., Muñiz, C., Weidberg, N., García-Flórez, L., and Acuña, J. L. (2017). High densities of stalked barnacle larvae (Pollicipes pollicipes) inside a river plume. J. Plankton Res. 39, 316–329. doi: 10.1093/plankt/fbw093
Holleley, C. E., and Geerts, P. G. (2009). Multiplex manager 1.0: a cross-platform computer program that plans and optimizes multiplex PCR. BioTechniques 46, 511–517. doi: 10.2144/000113156
Hongjamrassilp, W., Murase, A., Miki, R., and Hastings, P. A. (2020). Journey to the west: trans-pacific historical biogeography of fringehead blennies in the genus Neoclinus (Teleostei: Blenniiformes). Zool. Stud. 59, 1–12. doi: 10.6620/ZS.2020.59-09
Iacchei, M., Ben-Horin, T., Selkoe, K. A., Bird, C. E., García-Rodríguez, F. J., and Toonen, R. J. (2013). Combined analyses of kinship and FST suggest potential drivers of chaotic genetic patchiness in high gene-flow populations. Mol. Ecol. 22, 3476–3494. doi: 10.1111/mec.12341
ICES (2021). “Bay of biscay and the iberian coast ecoregion – ecosystem overview,” in Report of the ICES Advisory Committee, 2021. ICES Advice 2021, Section 6.1, (Toronto, ON: ICES), doi: 10.17895/ices.advice.9436
Jacinto, D., Cruz, T., Silva, T., and Castro, J. J. (2010). Stalked barnacle (Pollicipes pollicipes) harvesting in the berlengas nature reserve, portugal: temporal variation and validation of logbook data. ICES J. Mar. Sci. 67, 19–25. doi: 10.1093/icesjms/fsp226
Jacinto, D., Cruz, T., Silva, T., and Castro, J. J. (2011). Gestión de la explotación de percebe (Pollicipes pollicipes) en la reserva natural de berlengas (Portugal): evaluación del tope de capturas y talla mínima. Sci. Mar. 75, 439–445. doi: 10.3989/scimar.2011.75n3439
Jacinto, D., Penteado, N., Pereira, D., Sousa, A., and Cruz, T. (2015). Growth rate variation of the stalked barnacle Pollicipes pollicipes (Crustacea: Cirripedia) using calcein as a chemical marker. Sci. Mar. 79, 117–123. doi: 10.3989/scimar.04135.08B
Johnson, M. S., and Black, R. (1982). Chaotic genetic patchiness in an intertidal limpet. Siphonaria sp. Mar. Biol. 70, 157–164. doi: 10.1007/BF00397680
Johnson, M. S., Holborn, K., and Black, R. (1993). Fine-scale patchiness and genetic heterogeneity of recruits of the corallivorous gastropod Drupella cornus. Mar. Biol. 117, 91–96. doi: 10.1007/BF00346429
Jolliffe, I. (2011). Principal Component Analysis. Berlin: Springer Berlin Heidelberg, doi: 10.1007/978-3-642-04898-2
Jolly, M., Viard, F., Weinmayr, G., Gentil, F., Thiébaut, E., and Jollivet, D. (2003). Does the genetic structure of Pectinaria koreni (Polychaeta: Pectinariidae) conform to a source–sink metapopulation model at the scale of the Baie de Seine? Helgoland Mar. Res. 56, 238–246. doi: 10.1007/s10152-002-0123-1
Jolly, M. T., Guyard, P., Ellien, C., Gentil, F., Viard, F., Thiébaut, E., et al. (2009). Population genetics and hydrodynamic modeling of larval dispersal dissociate contemporary patterns of connectivity from historical expansion into European shelf seas in the polychaete Pectinaria koreni. Limnol. Oceanogr. 54, 2089–2106. doi: 10.4319/lo.2009.54.6.2089
Jolly, M. T., Thiébaut, E., Guyard, P., Gentil, F., and Jollivet, D. (2014). Meso-scale hydrodynamic and reproductive asynchrony affects the source–sink metapopulation structure of the coastal polychaete Pectinaria koreni. Mar. Biol. 161, 367–382. doi: 10.1007/s00227-013-2342-1
Jolly, M. T., Viard, F., Gentil, F., and Thiébaut, É, and Jollivet, D. (2006). Comparative phylogeography of two coastal polychaete tubeworms in the Northeast Atlantic supports shared history and vicariant events. Mol. Ecol. 15, 1841–1855. doi: 10.1111/j.1365-294X.2006.02910.x
Jombart, T. (2008). adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24, 1403–1405. doi: 10.1093/bioinformatics/btn129
Kalendar, R., Lee, D., and Schulman, A. H. (2009). FastPCR software for PCR primer and probe design and repeat search. Genes Genomes Genomics 3, 1–14.
Kopelman, N. M., Mayzel, J., Jakobsson, M., Rosenberg, N. A., and Mayrose, I. (2015). Clumpak: a program for identifying clustering modes and packaging population structure inferences across K. Mol. Ecol. Resour. 15, 1179–1191. doi: 10.1111/1755-0998.12387
Kough, A. S., Paris, C. B., and Butler, M. J. IV (2013). larval connectivity and the international management of fisheries. PLoS One 8:e0064970. doi: 10.1371/journal.pone.0064970
Larson, R. J., and Julian, R. M. (1999). Spatial and temporal genetic patchiness in marine populations and their implications for fisheries management. Cal. Coop. Ocean. Fish. 1999, 94–99.
Letunic, I., and Bork, P. (2019). Interactive Tree of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47, W256–W259. doi: 10.1093/nar/gkz239
Li, G., and Hedgecock, D. (1998). Genetic heterogeneity, detected by PCR-SSCP, among samples of larval pacific oysters (Crassostrea gigas) supports the hypothesis of large variance in reproductive success. Can. J. Fish. Aquat. Sci. 55, 1025–1033. doi: 10.1139/f97-312
Lindegren, M., Andersen, K. H., Casini, M., and Neuenfeldt, S. (2014). A metacommunity perspective on source-sink dynamics and management: the Baltic Sea as a case study. Ecol. Appl. 24, 1820–1832. doi: 10.1890/13-0566.1
Lorenzen, K., Steneck, R. S., Warner, R. R., Parma, M., Coleman, F. C., and Leber, K. M. (2010). The spatial dimensions of fisheries: putting it all in place. Bull. Mar. Sci. 86, 169–177.
Lourenço, C. R., Zardi, G. I., McQuaid, C. D., Serrão, E. A., Pearson, G. A., Jacinto, R., et al. (2016). Upwelling areas as climate change refugia for the distribution and genetic diversity of a marine macroalga. J. Biogeogr. 43, 1595–1607. doi: 10.1111/jbi.12744
Macho, G. (2006). Ecología Reproductiva y Larvaria del Percebe y Otros cirrípedos en Galicia. Doctoral dissertation. Pontevedra: Universidade de Vigo. Doctoral dissertation
Macho, G., Naya, I., Freire, J., Villasante, S., and Molares, J. (2013). The key role of the barefoot fisheries advisors in the Co-managed TURF system of Galicia (NW Spain). Ambio 42, 1057–1069. doi: 10.1007/s13280-013-0460-0
Maggs, C. A., Castilho, R., Foltz, D., Henzler, C., Jolly, M. T., Kelly, J., et al. (2008). Evaluating signatures of glacial refugia for North Atlantic benthic marine taxa. Ecology 89, S108–S122. doi: 10.1890/08-0257.1
Manangwa, O., De Meeûs, T., Grébaut, P., Ségard, A., Byamungu, M., and Ravel, S. (2019). Detecting Wahlund effects together with amplification problems: Cryptic species, null alleles and short allele dominance in Glossina pallidipes populations from Tanzania. Mol. Ecol. Resour. 19, 757–772. doi: 10.1111/1755-0998.12989
Mauger, S., Couceiro, L., and Valero, M. (2012). A simple and cost-effective method to synthesize an internal size standard amenable to use with a 5-dye system. Prime Res. Biotgechnol. 2, 2315–5299.
Mccartney, M. A., Brayer, K., and Levitan, D. R. (2004). Polymorphic microsatellite loci from the red urchin, Strongylocentrotus franciscanus, with comments on heterozygote deficit. Mol. Ecol. Notes 4, 226–228. doi: 10.1111/j.1471-8286.2004.00624.x
Molares, J., and Freire, J. (2003). Development and perspectives for community-based management of the goose barnacle (Pollicipes pollicipes) fisheries in Galicia (NW Spain). Fish. Res. 65, 485–492. doi: 10.1016/j.fishres.2003.09.034
Molares, J., Tilves, F., and Pascual, C. (1994). Larval development of the pedunculate barnacle Pollicipes cornucopia (Cirripedia: Scalpellomorpha) reared in the laboratory. Mar. Biol. 120, 261–264. doi: 10.1007/BF00349686
Morgan, S. G., Fisher, J. L., Miller, S. H., McAfee, S. T., and Largier, J. L. (2009). Nearshore larval retention in a region of strong upwelling and recruitment limitation. Ecology 90, 3489–3502. doi: 10.1890/08-1550.1
Morgan, S. G., Miller, S. H., Robart, M. J., and Largier, J. L. (2018). Nearshore larval retention and cross-shelf migration of benthic crustaceans at an upwelling center. Front. Mar. Sci. 5:161. doi: 10.3389/fmars.2018.00161
Muñoz-Ramírez, C. P., Barnes, D. K. A., Cárdenas, L., Meredith, M. P., Morley, S. A., Roman-Gonzalez, A., et al. (2020). Gene flow in the antarctic bivalve Aequiyoldia eightsii (Jay, 1839) suggests a role for the antarctic peninsula coastal current in larval dispersal: gene flow patterns in a. eightsii. R. Soc. Open Sci. 7:200603. doi: 10.1098/rsos.200603
Nei, M., and Takezaki, N. (1983). Estimation of genetic distances and phylogenetic trees from DNA analysis. Proc. World Cong. Genet. Appl. Livstock. Prod. 21, 405–412.
Novembre, J. (2016). Pritchard, stephens, and donnelly on population structure. Genetics 204, 391–393. doi: 10.1534/genetics.116.195164
Nunez, J. C. B., Rong, S., Damian-Serrano, A., Burley, J. T., Elyanow, R. G., Ferranti, D. A., et al. (2021). Ecological load and balancing selection in circumboreal barnacles. Mol. Biol. Evol. 38, 676–685. doi: 10.1093/molbev/msaa227
Ouréns, R., Naya, I., and Freire, J. (2015). Mismatch between biological, exploitation, and governance scales and ineffective management of sea urchin (Paracentrotus lividus) fisheries in Galicia. Mar. Policy 51, 13–20. doi: 10.1016/j.marpol.2014.07.015
Palumbi, S. R. (2003). Population genetics, demographic connectivity, and the design of marine reserves. Ecol. Appl. 13, 146–115. doi: 10.1890/1051-0761(2003)013[0146:pgdcat]2.0.co;2
Pannacciulli, F. G., Piyapattanakorn, S., Bishop, J. D. D., Hawkins, S. J., and Maclean, N. (2005). Isolation of highly polymorphic microsatellite markers from the intertidal barnacle Chthamalus montagui Southward. Mol. Ecol. Notes 5, 641–643. doi: 10.1111/j.1471-8286.2005.01027.x
Papa, Y., Oosting, T., Valenza-Troubat, N., Wellenreuther, M., and Ritchie, P. A. (2020). Genetic Stock Structure of New Zealand Fish and the Use of Genomics in Fisheries Management: An Overview and Outlook. Singapore: Taylor and Francis Asia Pacific, doi: 10.1080/03014223.2020.1788612
Parada, J. M. M., Iglesias, E., Outeiral, R., and Molares, J. (2013). Diameter of the base of the capitulum as a biometric variable of the goose barnacle Pollicipes pollicipes (Gmelin, 1789) (Cirripedia. Lepadomorpha). Crustaceana 86, 1527–1538. doi: 10.1163/15685403-00003251
Pascual, M., Rives, B., Schunter, C., and MaCpherson, E. (2017). Impact of life history traits on gene flow: a multispecies systematic review across oceanographic barriers in the Mediterranean Sea. PLoS One 12:e0176419. doi: 10.1371/journal.pone.0176419
Pavón, M. C. (2003). Biología y Variables Poblacionales del Percebe, Pollicipes pollicipes (Gmelin, 1790) en Asturias. Doctoral dissertation. Pontevedra: Universidad de Oviedo. Doctoral dissertation
Peakall, R., and Smouse, P. E. (2012). GenALEx 6.5: genetic analysis in excel. Population genetic software for teaching and research-an update. Bioinformatics 28, 2537–2539. doi: 10.1093/bioinformatics/bts460
Peliz, A., Santos, A. M. P., Oliveira, P. B., and Dubert, J. (2004). Extreme cross-shelf transport induced by eddy interactions southwest of Iberia in winter 2001. Geophys. Res. Lett. 31:L08301. doi: 10.1029/2004GL019618
Pew, J., Muir, P. H., Wang, J., and Frasier, T. R. (2015). related: an R package for analysing pairwise relatedness from codominant molecular markers. Mol. Ecol. Resour. 15, 557–561. doi: 10.1111/1755-0998.12323
Pineda, J. (1999). Circulation and larval distribution in internal tidal bore warm fronts. Limnol. Oceanogr. 44, 1400–1414. doi: 10.4319/lo.1999.44.6.1400
Pineda, J., Hare, J. A., and Sponaugle, S. (2007). Larval transport and dispersal in the coastal ocean and consequences for population connectivity. Oceanography 20, 22–39. doi: 10.5670/oceanog.2007.27
Pineda, J., Starczak, V., and Stueckle, T. A. (2006). Timing of successful settlement: demonstration of a recruitment window in the barnacle Semibalanus balanoides. Mar. Ecol. Prog. Ser. 320, 233–237. doi: 10.3354/meps320233
Piry, S., Luikart, G., and Cornuet, J. M. M. (1999). Bottleneck: a computer program for detecting recent reductions in the effective population size using allele frequency data. J. Hered. 90, 502–503. doi: 10.1002/anie.198209122
Plough, L. V., and Marko, P. B. (2014). Characterization of microsatellite loci and repeat density in the gooseneck barnacle, Pollicipes elegans, using next generation sequencing. J. Hered. 105, 136–142. doi: 10.1093/jhered/est064
Plough, L. V. L., Moran, A., Marko, P. P., Andersson, M., Jennions, M., Petrie, M., et al. (2014). Density drives polyandry and relatedness influences paternal success in the Pacific gooseneck barnacle, Pollicipes elegans. BMC Evol. Biol. 14:81. doi: 10.1186/1471-2148-14-81
Porri, F., McQuaid, C. D., and Radloff, S. (2006). Spatio-temporal variability of larval abundance and settlement of Perna perna: differential delivery of mussels. Mar. Ecol. Prog. Ser. 315, 141–150. doi: 10.3354/meps315141
Pritchard, J. K., Stephens, M., and Donnelly, P. (2000). Inference of population structure using multilocus genotype data. Genetics 155, 945–959. doi: 10.1093/genetics/155.2.945
Provan, J., and Bennett, K. D. (2008). Phylogeographic insights into cryptic glacial refugia. Trends Ecol. Evol. 23, 564–571. doi: 10.1016/j.tree.2008.06.010
Queiroga, H., Cruz, T., dos Santos, A., Dubert, J., González-Gordillo, J. I., Paula, J., et al. (2007). Oceanographic and behavioural processes affecting invertebrate larval dispersal and supply in the western Iberia upwelling ecosystem. Prog. Oceanogr. 74, 174–191. doi: 10.1016/j.pocean.2007.04.007
Quinteiro, J., Rodríguez-Castro, J., and Rey-Méndez, M. (2007). Population genetic structure of the stalked barnacle Pollicipes pollicipes (Gmelin, 1789) in the northeastern Atlantic: Influence of coastal currents and mesoscale hydrographic structures. Mar. Biol. 153, 47–60. doi: 10.1007/s00227-007-0783-0
Rassweiler, A., Costello, C., and Siegel, D. A. (2012). Marine protected areas and the value of spatially optimized fishery management. P. Natl. Acad. Sci. U.S.A. 109, 11884–11889. doi: 10.1073/pnas.1116193109
Reigel, A. M., Harrison, J. S., and Gleason, D. F. (2015). Tetranucleotide microsatellites for the barnacle Megabalanus coccopoma (Darwin, 1854). Biochem. Syst. Ecol. 62, 159–163. doi: 10.1016/j.bse.2015.08.010
Relvas, P., Barton, E. D., Dubert, J., Oliveira, P. B., Peliz, A., Da Silva, J. C. B., et al. (2007). Physical oceanography of the western Iberia ecosystem: latest views and challenges. Prog. Oceanogr. 74, 149–173. doi: 10.1016/j.pocean.2007.04.021
Rice, W. R. (1989). Analyzing tables of statistical tests. Evolution 43, 223–225. doi: 10.2307/2409177
Rico, C., Cuesta, J. A., Drake, P., Macpherson, E., Bernatchez, L., and Marie, A. D. (2017). Null alleles are ubiquitous at microsatellite loci in the Wedge Clam (Donax trunculus). PeerJ 5:e3188. doi: 10.7717/peerj.3188
Rilov, G., Dudas, S. E., Menge, B. A., Grantham, B. A., Lubchenco, J., and Schiel, D. R. (2008). The surf zone: a semi-permeable barrier to onshore recruitment of invertebrate larvae? J. Exp. Mar. Biol. Ecol. 361, 59–74. doi: 10.1016/j.jembe.2008.04.008
Rivera, A., Gelcich, S., García-Flórez, L., and Acuña, J. L. (2017). Trends, drivers, and lessons from a long-term data series of the Asturian (northern Spain) gooseneck barnacle territorial use rights system. Bull. Mar. Sci. 93, 35–51. doi: 10.5343/bms.2015.1080
Rivera, A., Gelcich, S., García-Florez, L., Alcázar, J. L., and Acuña, J. L. (2014). Co-management in Europe: Insights from the gooseneck barnacle fishery in Asturias. Spain. Mar. Policy 50, 300–308. doi: 10.1016/j.marpol.2014.07.011
Rivera, A., Weidberg, N., Pardiñas, A. F., González-Gil, R., García-Flórez, L., and Acuña, J. L. (2013). Role of upwelling on larval dispersal and productivity of gooseneck barnacle populations in the Cantabrian sea: management implications. PLoS One 8:e78482. doi: 10.1371/journal.pone.0078482
Rousset, F. (1997). Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics 145, 1219–1228. doi: 10.1093/genetics/145.4.1219
Ruiz-Díaz, R., Liu, X., Aguión, A., Macho, G., deCastro, M., Gómez-Gesteira, M., et al. (2020). Social-ecological vulnerability to climate change in small-scale fisheries managed under spatial property rights systems. Mar. Policy 121:104192. doi: 10.1016/j.marpol.2020.104192
Sánchez, R. F., and Relvas, P. (2003). Spring–summer climatological circulation in the upper layer in the region of Cape St. Vincent, Southwest Portugal. ICES J. Mar. Sci. 60, 1232–1250. doi: 10.1016/S1054-3139(03)00137-1
Schmieder, R., and Edwards, R. (2011). Quality control and preprocessing of metagenomic datasets. Bioinformatics 27, 863–864. doi: 10.1093/bioinformatics/btr026
Selkoe, K. A., and Toonen, R. J. (2006). Microsatellites for ecologists: a practical guide to using and evaluating microsatellite markers. Ecol. Lett. 9, 615–629. doi: 10.1111/j.1461-0248.2006.00889.x
Selkoe, K. A., Watson, J. R., White, C., Horin, T. B., Iacchei, M., Mitarai, S., et al. (2010). Taking the chaos out of genetic patchiness: seascape genetics reveals ecological and oceanographic drivers of genetic patterns in three temperate reef species. Mol. Ecol. 19, 3708–3726. doi: 10.1111/j.1365-294X.2010.04658.x
Seoane-Miraz, D., Martínez-Lage, A., and González-Tizón, A. M. (2015). Characterization of 15 polymorphic microsatellite loci in gooseneck barnacle Pollicipes pollicipes (Gmelin, 1789), and cross-amplification in other Pollicipes species. Conserv. Genet. Resour. 7, 591–593. doi: 10.1007/s12686-015-0436-4
Séré, M., Thévenon, S., Belem, A. M. G., and De Meeûs, T. (2017). Comparison of different genetic distances to test isolation by distance between populations. Heredity 119, 55–63. doi: 10.1038/hdy.2017.26
Shanks, A. L., McCulloch, A., and Miller, J. (2003). Topographically generated fronts, very nearshore oceanography and the distribution of larval invertebrates and holoplankters. J. Plankton Res. 25, 1251–1277. doi: 10.1093/plankt/fbg090
Silva, C. N. S., MacDonald, H. S., Hadfield, M. G., Cryer, M., and Gardner, J. P. A. (2019). Ocean currents predict fine-scale genetic structure and source-sink dynamics in a marine invertebrate coastal fishery. ICES J. Mar. Sci. 76, 1007–1018. doi: 10.1093/icesjms/fsy201
Sotelo, G., Morán, P., and Posada, D. (2007). Identification and characterization of microsatellite loci in the spiny spider crab Maja brachydactyla. Conserv. Genet. 8, 245–247. doi: 10.1007/s10592-006-9141-x
Sousa, A., Jacinto, D., Penteado, N., Martins, P., Fernandes, J., Silva, T., et al. (2013). Patterns of distribution and abundance of the stalked barnacle (Pollicipes pollicipes) in the central and southwest coast of continental Portugal. J. Sea Res. 83, 187–194. doi: 10.1016/j.seares.2013.04.005
Sousa, A., Jacinto, D., Penteado, N., Pereira, D., Silva, T., Castro, J. J., et al. (2020). Temporal variation of the fishers’ perception about the stalked barnacle (Pollicipes pollicipes) fishery at the Berlengas Nature Reserve (Portugal). Reg. Stud. Mar. Sci. 38:101378. doi: 10.1016/j.rsma.2020.101378
Southward, A. J. (2008). “Barnacles: keys and notes for the identification of British species,” in Synopses of the British fauna (New Series), eds J. H. Crothers and P. J. Hayward (London: The Linnean Society of London and the Estuarine and Coastal Sciences Association by Field Studies Council Shrewsbury), viii+1–144.
St-Onge, P., Tremblay, R., and Sévigny, J. M. (2015). Tracking larvae with molecular markers reveals high relatedness and early seasonal recruitment success in a partially spawning marine bivalve. Oecologia 178, 733–746. doi: 10.1007/s00442-015-3245-2
Takezaki, N., Nei, M., and Tamura, K. (2014). POPTREEW: web version of POPTREE for constructing population trees from allele frequency data and computing some other quantities. Mol. Biol. Evol. 31, 1622–1624. doi: 10.1093/molbev/msu093
Treml, E. A., Halpin, P. N., Urban, D. L., and Pratson, L. F. (2008). Modeling population connectivity by ocean currents, a graph-theoretic approach for marine conservation. Landsc. Ecol. 23, 19–36. doi: 10.1007/s10980-007-9138-y
Van Oosterhout, C., Hutchinson, W. F., Wills, D. P. M., and Shipley, P. (2004). MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes 4, 535–538. doi: 10.1111/j.1471-8286.2004.00684.x
Van Syoc, R. J., Fernandes, J. N., Carrison, D. A., and Grosberg, R. K. (2010). Molecular phylogenetics and biogeography of Pollicipes (Crustacea: Cirripedia), a Tethyan relict. J. Exp. Mar. Biol. Ecol. 392, 193–199. doi: 10.1016/j.jembe.2010.04.024
Van Wyngaarden, M., Snelgrove, P. V. R., DiBacco, C., Hamilton, L. C., Rodríguez-Ezpeleta, N., Jeffery, N. W., et al. (2017). Identifying patterns of dispersal, connectivity and selection in the sea scallop. Placopecten magellanicus, using RADseq-derived SNPs. Evol. Appl. 10, 102–117. doi: 10.1111/eva.12432
Veliz, D., Duchesne, P., Bourget, E., and Bernatchez, L. (2006). Genetic evidence for kin aggregation in the intertidal acorn barnacle (Semibalanus balanoides). Mol. Ecol. 15, 4193–4202. doi: 10.1111/j.1365-294X.2006.03078.x
Waples, R. S. (1998). Separating the wheat from the chaff: patterns of genetic differentiation in high gene flow species. J. Hered. 89, 438–450. doi: 10.1093/jhered/89.5.438
Waples, R. S. (2014). Testing for hardy–weinberg proportions: have we lost the plot? J. Hered. 106, 1–19. doi: 10.1093/jhered/esu062
Waples, R. S. (2018). Null alleles and FIS x FST correlations. J. Hered. 109, 457–461. doi: 10.1093/jhered/esy013
Weidberg, N., Lobón, C., López, E., Flórez, L. G., Rueda, M. D. P. F., Largier, J. L., et al. (2014). Effect of nearshore surface slicks on meroplankton distribution: role of larval behaviour. Mar. Ecol. Prog. Ser. 506, 15–30. doi: 10.3354/meps10777
Weir, B. S., and Cockerham, C. C. (1984). Estimating F-statistics for the analysis of population structure. Evolution 38, 1358–1370. doi: 10.2307/2408641
Wong, J. Y., Chen, H. N., Chan, B. K. K., Tan, I. K. P., and Chong, V. C. (2014). A combined morphological and molecular approach in identifying barnacle cyprids from the matang mangrove forest reserve in malaysia: essentials for larval ecology studies. Raffles B. Zool. 62, 317–329. doi: 10.5281/zenodo.5756768
Keywords: stalked barnacle, multiplex PCR, microsatellite, small-scale fisheries, recruitment, stock management, connectivity
Citation: Parrondo M, Morán P, Ballenghien M, Acuña JL, Aguión A, Arrontes J, Chiss J, Cruz T, Fernandes JN, García-Flórez L, García-Vázquez E, Geiger KJ, Macho G, Thiébaut E, Weidberg N, Jollivet D and Borrell YJ (2022) Chaotic Genetic Patchiness in the Highly Valued Atlantic Stalked Barnacle Pollicipes pollicipes From the Iberian Peninsula: Implications for Fisheries Management. Front. Mar. Sci. 9:801780. doi: 10.3389/fmars.2022.801780
Received: 25 October 2021; Accepted: 09 February 2022;
Published: 15 March 2022.
Edited by:
Simone Libralato, Istituto Nazionale di Oceanografia e di Geofisica Sperimentale, ItalyCopyright © 2022 Parrondo, Morán, Ballenghien, Acuña, Aguión, Arrontes, Chiss, Cruz, Fernandes, García-Flórez, García-Vázquez, Geiger, Macho, Thiébaut, Weidberg, Jollivet and Borrell. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yaisel J. Borrell, borrellyaisel@uniovi.es
†Present address: Gonzalo Macho, Fisheries Consultant, Victoria, Seychelles
 Marina Parrondo
Marina Parrondo Paloma Morán
Paloma Morán Marion Ballenghien3
Marion Ballenghien3  Jose L. Acuña
Jose L. Acuña Alba Aguión
Alba Aguión Teresa Cruz
Teresa Cruz Joana N. Fernandes
Joana N. Fernandes Eva García-Vázquez
Eva García-Vázquez Katja J. Geiger
Katja J. Geiger Gonzalo Macho
Gonzalo Macho Didier Jollivet
Didier Jollivet Yaisel J. Borrell
Yaisel J. Borrell