Mitogenomes Provide Insights Into the Evolution of Thoracotremata (Brachyura: Eubrachyura)
- 1Department of Marine Organism Taxonomy and Phylogeny, Institute of Oceanology, Chinese Academy of Sciences, Qingdao, China
- 2Laboratory for Marine Biology and Biotechnology, Qingdao National Laboratory for Marine Science and Technology, Qingdao, China
- 3Shandong Province Key Laboratory of Experimental Marine Biology, Institute of Oceanology, Chinese Academy of Sciences, Qingdao, China
- 4College of Biological Sciences, University of Chinese Academy of Sciences, Beijing, China
Thoracotremata is a group of Brachyura, with 1,248 extant species. To date, parts of the thoracotreme phylogeny are not yet resolved and require further investigation. In this study, 12 new mitogenomes from the four thoracotreme superfamilies were sequenced. They contain a standard set of 37 genes, and vary in size from 15,422 (Hapalocarcinus marsupialis Stimpson, 1858 sensu lato) to 16,490 bp [Arcotheres sinensis (Shen, 1932)]. Combined with 58 thoracotreme mitochondrial genomes (mitogenomes) from GenBank, we described the evolution of gene rearrangement and the internal phylogenetic relationships of Thoracotremata, and evaluated the phylogenetic position of Cryptochiroidea and Pinnotheroidea. Nine distinct patterns of mitochondrial gene order (MGO) among thoracotreme mitogenomes are identified, with four MGOs newly found in Thoracotremata. All other gene orders are the result of transformational pathways originating from brachyuran gene order (BraGO). The different gene orders have variable levels of gene rearrangements, which involve both tRNAs and protein-coding genes. No link between variable gene arrangements (breakpoint distances) and nucleotide substitution rates (branch lengths) is found in thoracotreme crabs. The symbiotic groups, the cryptochiroid and pinnotheroid crabs, display variable MGOs (CryGO, Pin1GO, and Pin2GO), providing evidence for possible correlations of rearranged MGOs to the adaptations to specialized lifestyles. In our phylogenetic analyses, Cryptochiridae (Cryptochiroidea) show close relationship with an Ocypodoidea lineage (Camptandriidae/Xenophthalmidae/Dotillidae). Pinnotheridae (Pinnotheroidea) form the basal monophyletic clade.
Introduction
Brachyura (true crabs) is one of the most diverse groups among the extant crustaceans with over 7,250 described species, which occurred in almost any ecosystem, from aquatic to terrestrial habitats, from freshwater to marine, as well as the deep-sea and hydrothermal vents (Ma et al., 2019). Based on the gonopore positions, Guinot (1978; 1979; 1977) divided Brachyura into three sections, Podotremata, Heterotremata, and Thoracotremata. This classification was widely accepted. Podotremata was deemed as the most primitive group with various ancestral characteristics, whereas Thoracotremata featured with sternal male sexual apertures is the most derived crab group (Tsang et al., 2014; Wang et al., 2021). Later, Heterotremata and Thoracotremata were reassigned to the subsection level and together formed the section Eubrachyura, the biggest and most differentiated group of crabs (Guinot, 2008; Tsang et al., 2014). Eubrachyura is accepted as monophyletic; debates and controversies involving its internal classification still exist (Von Sternberg and Cumberlidge, 2001; Brösing et al., 2007; Ahyong et al., 2007; Ji et al., 2014; Xing et al., 2016; Xing et al., 2017; Wang et al., 2021).
The Thoracotremata encompasses 1,248 extant species (Ng et al., 2008; WoRMS, 2021). It is currently understood to comprise four superfamilies, Grapsoidea MacLeay, 1838; Ocypodoidea Rafinesque, 1815; Cryptochiroidea Paulson, 1875; and Pinnotheroidea De Haan, 1833 (Ng et al., 2008; WoRMS, 2021). Crabs from Grapsoidea and Ocypodoidea are predominant in intertidal and supratidal marine habitats. They are most diverse in the tropics, where they invade terrestrial and freshwater habitats repeatedly (Schubart et al., 2006). Within Grapsoidea, Ng et al. (2008) and Davie et al. (2015a) recognized eight families, including the Gecarcinidae, Glyptograpsidae, Grapsidae, Percnidae, Plagusiidae, Sesarmidae, Varunidae, and Xenograpsidae. Subsequently, a new family, Leptograpsodidae, was established (Guinot et al., 2018). Therefore, the Grapsoidea provisionally retained nine families (WoRMS, 2021). For Ocypodoidea, the family Ucididae was reassigned to the genus level and merged into the family Ocypodidae (Shih et al., 2016). Therefore, the current Ocypodoidea includes seven families, Camptandriidae, Dotillidae, Heloeciidae, Macrophthalmidae, Mictyridae, Ocypodidae, and Xenophthalmidae (WoRMS, 2021). The monophyly of the superfamilies Ocypodoidea and Grapsoidea is one of the most contentious issues (reviewed in Davie et al., 2015b), representing conundrums in crab systematics. Cryptochiroidea crabs are coral-dwelling crabs, which live in obligate association with their coral hosts (Wetzer et al., 2009; van der Meij, 2015; van der Meij et al., 2015). The superfamily Pinnotheroidea comprises a group of symbiotic crabs, mainly living in association with mollusks and tubeworms (Campos, 2016; Theil et al., 2016). The two thoracotreme superfamilies, Cryptochiroidea and Pinnotheroidea, are represented today by one (Cryptochiridae) and two families (Aphanodactylidae and Pinnotheridae), respectively (Ng et al., 2008; WoRMS, 2021).
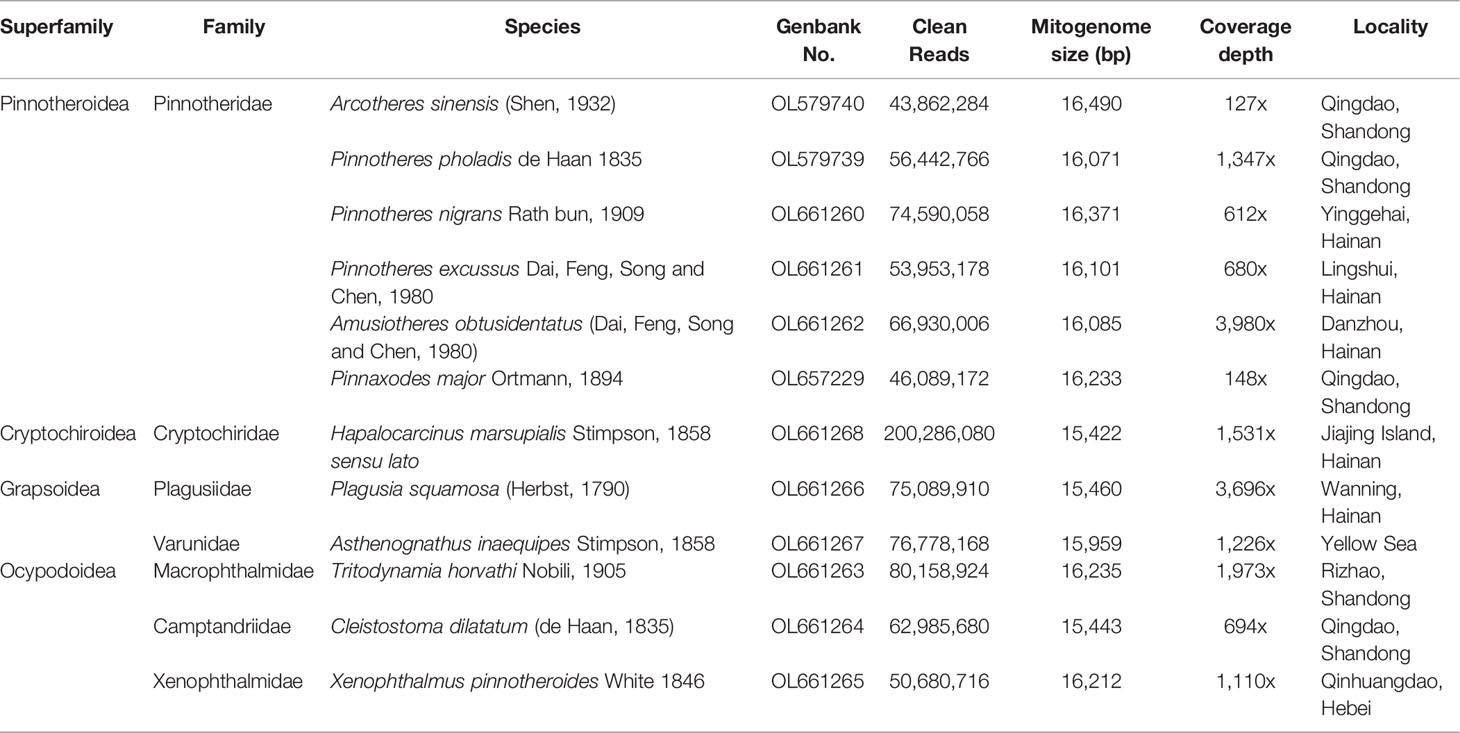
The hypotheses of phylogenetic relationships among the thoracotreme superfamilies based on molecular data are summarized in Figure 1. The polyphyly of the two major thoracotreme superfamilies, Grapsoidea and Ocypodoidea, was confirmed in these molecular studies (Schubart et al., 2006; Tsang et al., 2014; Chen et al., 2018; Wang et al., 2021). The superfamilies of Cryptochiroidea and Pinnotheroidea are nested within the grapsoid and ocypodoid clades (Tsang et al., 2014; van der Meij and Schubart, 2014; Tsang et al., 2018). However, the current analyses either contained limited thoracotreme samples or applied short gene fragments. Parts of the thoracotreme phylogeny are not yet resolved, which underscore the need for a well-supported molecular phylogeny of Thoracotremata, to solve the relationships of the families and superfamilies.
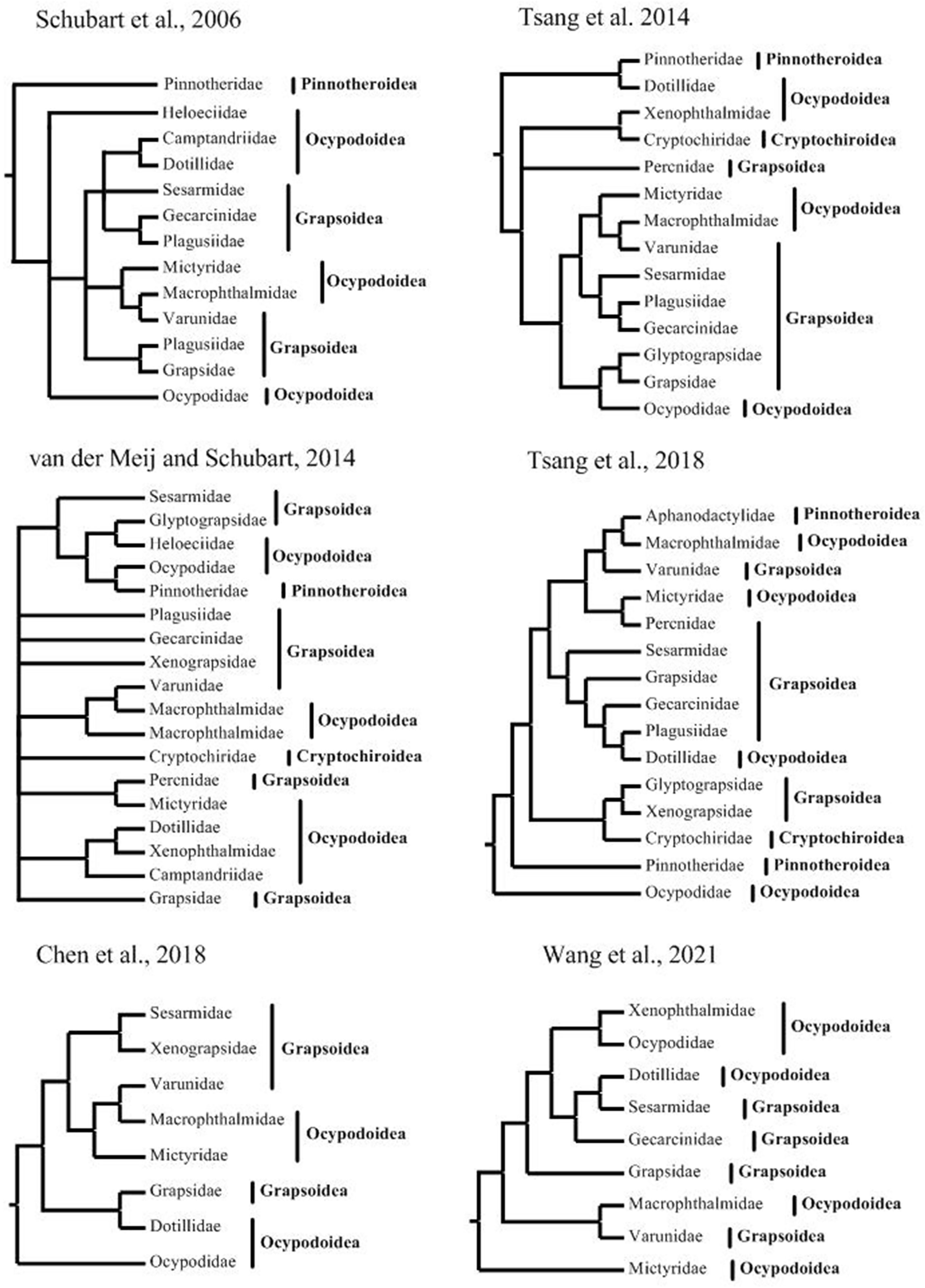
Figure 1 Recent hypotheses of relationships at families and superfamilies of Thoracotremata based on molecular data. The studies summarized here reached different conclusions.
The mitochondrial genomes (mitogenomes) provide rich phylogenetic information not only in the sequences, but also in the genomic rearrangements (Boore, 1999; Sun et al., 2003; Tan et al., 2017). In the Decapoda, novel mitochondrial gene orders have been reported across a range of lineages, such as brachyurans (Zhang et al., 2020; Wang et al., 2021), anomurans (Sun et al., 2019; Gong et al., 2020), and caridean shrimps (Ye et al., 2021). By analyzing 246 decapod mitogenomes, Tan et al. (2019) found that a large number of mitochondrial gene orders in decapods deviated from the ancestral arthropod ground pattern and unevenly distributed among infraorders, but there was limited evidence for correlations between gene rearrangement events and species ecology or lineage-specific nucleotide substitution rates. Also, within a phylogenetic context, the gene order rearrangements that may act as synapomorphies for specific lineages have provided support for the hypotheses for phylogenetic reconstruction in Decapoda (Tan et al., 2017; Tan et al., 2018; Tan et al., 2019).
Mitogenomes have been used to reconstruct the phylogenetic tree of Brachyura (Basso et al., 2017; Chen et al., 2018; Tan et al., 2018; Chen et al., 2019; Ma et al., 2019; Wang et al., 2021), but the mitogenome of Thoracotremata was usually poorly sampled. Prior to our study, only 58 thoracotreme mitogenomes were available. They were all from superfamilies Ocypodoidea and Grapsoidea, with no representation from Cryptochiroidea and Pinnotheroidea. In the context of phylogenetics, inadequate taxon sampling and taxon biases can result in topological distortions owing to the artifactual sources of error, which can impede the resolution of phylogenetic positions (Timm and Bracken-Grissom, 2015). Therefore, there is a need to improve the representation of mitogenomes for Thoracotremata, especially for the poorly represented lineages to acquire more exhaustive sampling, which is necessary to comprehensively understand some fundamental evolutionary, genomic, and phylogenetic questions (Smith, 2016).
In this study, we greatly enhanced taxonomic coverage of Thoracotremata mitogenomic data by contributing 12 new mitogenomes from all the four thoracotreme superfamilies, of which seven are the first from their superfamilies (one from Cryptochiroidea and six from Pinnotheroidea), three are first from their families (Camptandriidae, Xenophthalmidae, and Plagusiidae), and one ocypodoid and one grapsoid are supplement to their families. In this study, we aim to (1) reveal the evolution of mitochondrial gene order (MGO) among thoracotremes, (2) estimate phylogenetic relationships within Thoracotremata using mitogenomic data, and (3) examine the phylogenetic position of Cryptochiroidea and Pinnotheroidea within Thoracotremata by inclusion of representatives of all available thoracotreme superfamilies.
Materials and Methods
Specimen Collection and Mitochondrial Genome Sequencing
A total of twelve species from four thoracotreme superfamilies (Table 1) were sampled for sequencing. All specimens were preserved in 95% ethanol following collection and deposited at Marine Biological Museum, Chinese Academy of Sciences, Qingdao, China. Identification was performed morphologically according to the morphological information on crabs of the China Seas (Dai and Yang, 1991). Total genomic DNA was extracted from the samples using the E.Z.N.A® Tissue DNA kit (OMEGA, Wuhan, China) following the manufacturer’s protocols. The paired-end libraries were constructed using TruSeq™ Nano DNA Sample Prep Kit (Illumina, San Diego, CA, USA) with an insert size of 450 bp. The above libraries were sequenced by an Illumina (San Diego, CA, USA) HiSeq 4000 platform (2 × 150 bp paired-end reads).
Mitochondrial Genome Assemblies and Annotation
The paired-end raw sequences for the twelve samples were trimmed using Trimmomatic 0.39 (Bolger et al., 2014) with the following parameters: ILLUMINACLIP : TruSeq3-PE.fa:2:30:10 LEADING:3 TRAILING:3 SLIDINGWINDOW:4:15 MINLEN:75. Resulting clean reads (Table 1) were assembled de novo by NOVOPlasty software (Dierckxsens et al., 2016) with default parameters. In the seed extension algorithm achieved via NOVOPlasty, the cox1 or 16S rDNA gene fragments of the species or their closed related species were used as seed sequences (Supplementary Table 1). NOVOPlasty does not try to assemble every read, but will extend the given seed until the circular mitogenome is formed. The mitogenomes will be circularized when the length is in the expected range and both ends overlap by at least 200 bp. If circularized mitogenome was not obtained, MITObim v.1. (Hahn et al., 2013) was used to achieve assemblies using short mtDNA sequences from related species as “baits”. The clean reads can be accessed from the SRA database with the accession numbers SRR18217430-SRR18217435 and SRR18217456-SRR18217461.
MITOS web server (Bernt et al., 2013) was preliminarily used to annotate the protein-coding genes (PCGs), 2 ribosomal RNA (rRNA) and transfer RNA (tRNA) genes under default settings, and the invertebrate genetic code for mitochondria. The PCGs and rRNA genes boundaries were edited manually based on similarity with the homologous genes of other reported thoracotreme mitochondrial gene sequences. The tRNA genes and their secondary structures were further identified with ARWEN 1.2.3.c (Laslett and Canbäck, 2007). Correlations between mitochondrial genome size and length of non-coding regions were measured by the Spearman correlation coefficient carried out with IBM SPSS Statistics, release 19.0.0.1. The complete mitochondrial DNA (mtDNA) sequences can be accessed from the GenBank database with accession numbers OL661260-OL661268, OL579739-OL579740, and OL657229.
Mitogenomic Characterization and Gene Order Divergence
The nucleotide composition was calculated in MEGA 6.0 (Tamura et al., 2013), based on the invertebrate mitochondrial genetic code (genetic code = 5). AT and GC skews were determined for complete mitogenomes (plus strand) according to the following formula: AT-skew = (A-T)/(A+T) and GC-skew = (G-C)/(G+C) (Perna and Kocher, 1995). Mitochondrial gene arrangements were compared using the newest version of MitoPhAST v2.0 (Tan et al., 2015) and the same gene orders were clustered into groups. Genomic rearrangement analyses were performed with the CREx program (http://pacosy.informatik.uni-leipzig.de/crex/form) (Bernt et al., 2007). The rearrangement events involve transpositions, inversions, inverse transpositions, and tandem duplication random loss (TDRL) (Boore, 2000; Bernt et al., 2007; Bernt et al., 2008; Bernt and Middendorf, 2011). We include all the mitochondrial genes (PCGs, tRNAs, and rRNAs); thus, 37 genes were considered. We explored the relationship between gene rearrangements and branch length of each taxon. Gene order changes were measured using the breakpoint distance (Blanchette et al., 1999; Bernt et al., 2013). The breakpoint distance was quantified as the minimal number of breakpoints needed to get from the gene order under view to the Brachyura ground pattern. The branch length of each taxon was determined as root-to-leaf distance in the phylogenetic tree. Correlations between different gene orders and branch length were measured by the Spearman correlation coefficient carried out with IBM SPSS Statistics, release 19.0.0.1.
Phylogenetic Analysis
The phylogenetic trees were reconstructed with 75 species, including the newly determined sequences, of which 5 Heterotremata species were used as outgroups (Supplementary Table 2). The relationships were reconstructed based on nucleotide sequences from 13 PCGs, which were aligned separately using MAFFT (Katoh and Standley, 2013) with default settings, then concatenated into a single supermatrix with FASconCAT (Kück and Meusemann, 2010). PartitionFinder 1.1.1 (Lanfear et al., 2012) was used to select the best-fit partitioning schemes and substitution models (Supplementary Table 3). Maximum Likelihood (ML) analysis was conducted with the IQ-TREE web server (Trifinopoulos et al., 2016) with the best-fit partition schemes and models. The node reliability was assessed using 5,000 ultrafast bootstrap replicates (Minh et al., 2013). The Bayesian inference (BI) was conducted using MrBayes 3 (Ronquist and Huelsenbeck, 2003) with partition models. Runs of 10,000,000 generations (Markov chain Monte Carlo, MCMC) were conducted, with a sampling frequency of 1,000 generations to allow adequate time for convergence. In our study, the average standard deviation of split frequencies was less than 0.01 (0.000771), the estimated sample size was >200, and the potential scale reduction factor approached 1.0 already after 10 million generations, and then the run was stopped. All trees generated prior to the achievement of stationarity of the log likelihood values were discarded as burn-in (2,500 trees). The 50% majority rule consensus tree was constructed using the remaining sampled trees to estimate the Bayesian posterior probabilities. The software Tracer v1.6 (Rambaut and Drummond, 2013) was used to check all the parameters.
Results and Discussion
Mitochondrial Genome Organization
Our work successfully obtained complete mitogenomes of 12 thoracotreme species from 4 superfamilies, substantially increasing the mitogenome resources for superfamilies, which were poorly represented in previous mitogenomic studies. They contained a standard set of 37 genes, and varied in size from 15,422 [Hapalocarcinus marsupialis Stimpson, 1858 sensu lato (Bahr et al., 2021)] to 16,490 bp [Arcotheres sinensis (Shen, 1932)] (Table 1 and Supplementary Table 4). The average coverage of mitogenomes varied from 694-fold to >3,000-fold coverage. Although the mitogenome sizes of thoracotreme were similar to that of the most metazoans (Gissi et al., 2008), the length of thoracotreme mitogenomes seemed to be family-specific, instead of superfamily-specific. The size of mtDNAs from five ocypodoid families (Camptandriidae, Xenograpsidae, Dotillidae, Mictyridae, and Ocypodidae), five grapsoid families (Grapsidae, Gecarcinidae, Sesarmidae, Plagusiidae, and Varunidae), and a cryptochiroid family (Cryptochiridae) were usually less than 16,000 bp, while the two families, Macrophthalmidae and Xenophthalmidae, from Ocypodoidea, Varunidae from Grapsoidea, and Pinnotheridae from Pinnotheroidea had mtDNAs larger than 16,000 bp (Figure 2A), forming their own cluster. Two species, Chasmagnathus convexus (De Haan, 1835 [in De Haan, 1833-1850]) from Varunidae (yellow dot 1 in Figure 2A) and Macrophthalmus pacificus Dana, 1851 from Macrophthalmidae (yellow dot 2 in Figure 2A), were 15,107 and 17,226 bp in length, respectively, which were the outliers. The length variations in thoracotreme mitogenomes is mainly due to the length heterogeneity of the non-coding regions (Supplementary Table 5). A significant and strong positive correlation was observed between mitochondrial genome size and length of non-coding regions (Spearman’s rho = 0.859, p < 0.001).
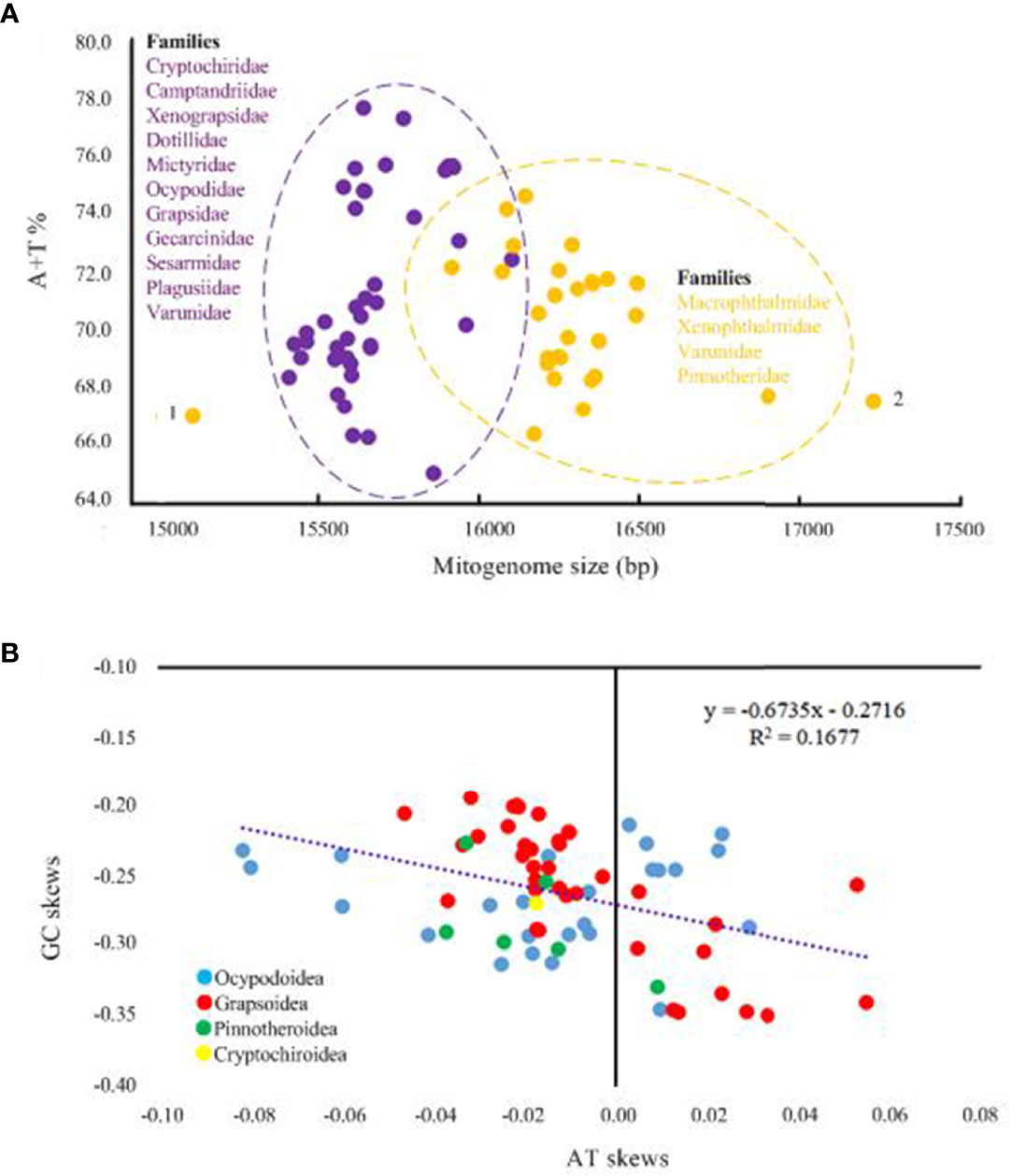
Figure 2 (A) Genome size versus A+T content of the thoracotreme mitogenomes. The yellow dots represented the families Macrophthalmidae, Xenophthalmidae, Varunidae, and Pinnotheridae. The purple dots represented the families Cryptochiridae, Camptandriidae, Xenograpsidae, Dotillidae, Mictyridae, Ocypodidae, Grapsidae, Gecarcinidae, Sesarmidae, Plagusiidae, and Varunidae. The yellow dots 1 (15,107 bp) and 2 (17,226 bp) represented Chasmagnathus convexus from Varunidae and Macrophthalmus pacificus from Macrophthalmidae, respectively, which were the outliers. (B) AT-skew versus GC-skew in the thoracotreme mitogenomes. Ocypodoidea, Grapsoidea, Cryptochiroidea, and Pinnotheroidea were represented by blue, red, yellow, and green dots, respectively.
Nucleotide composition of the thoracotreme mitogenomes was biased toward A and T (Figure 2A and Supplementary Table 5). The A+T content ranged from 65.0% in Grapsus tenuicrustatus (Herbst, 1783 [in Herbst, 1782-1790]) to 77.7% in Nanosesarma minutum (de Man, 1887). The values of the AT-skews were mostly negative, indicating more Ts than As, while the GC-skews for all thoracotreme mitogenomes were negative, suggesting that Cs were more abundant than Gs (Figure 2B). We found a strong negative relationship between the AT and GC skew (Pearson = −0.409, p < 0.0001) (Figure 2B). Two clusters can be found, one with negative GC-skews and AT-skews, the other with negative GC-skews and positive AT-skews. Note that the skewness of the thoracotreme mitogenomes were neither superfamily-specific nor family-specific. Species from four superfamilies were mixed in an irregular skewness.
The Evolution of Mitochondrial Gene Order in Thoracotremata
A total of nine distinct patterns of mitochondrial gene order (MGO) are known for the 70 available thoracotreme mtDNAs (Figure 3A). The most widespread gene order pattern is brachyuran gene order (BraGO). For some thoracotreme species, MGO patterns are shared at the family level. For example, the BraGO pattern was shared by the crabs belonging to eight families (Dotillidae, Mictyridae, Ocypodidae, Plagusiidae, Grapsidae, Gecarcinidae, Camptandriidae, and Xenophthalmidae). The mitochondrial DNA of Sesarmidae and Cryptochiridae rearranged with respect to SesGO and CryGO, respectively. The rearrangements of the two families Macrophthalmidae and Varunidae were described by the MaVaGO pattern, whereas for other species, MGO patterns are distinct among species within the same family (e.g., Varunidae and Pinnotheridae) or even genus (e.g., Xenograpsus). The Pinnotheridae species exhibit two gene orders (hereafter named Pin1GO and Pin2GO, respectively). Asthenognathus inaequipes Stimpson, 1858 was first found to have a novel gene order (AinGO) relative to other Varunidae species.
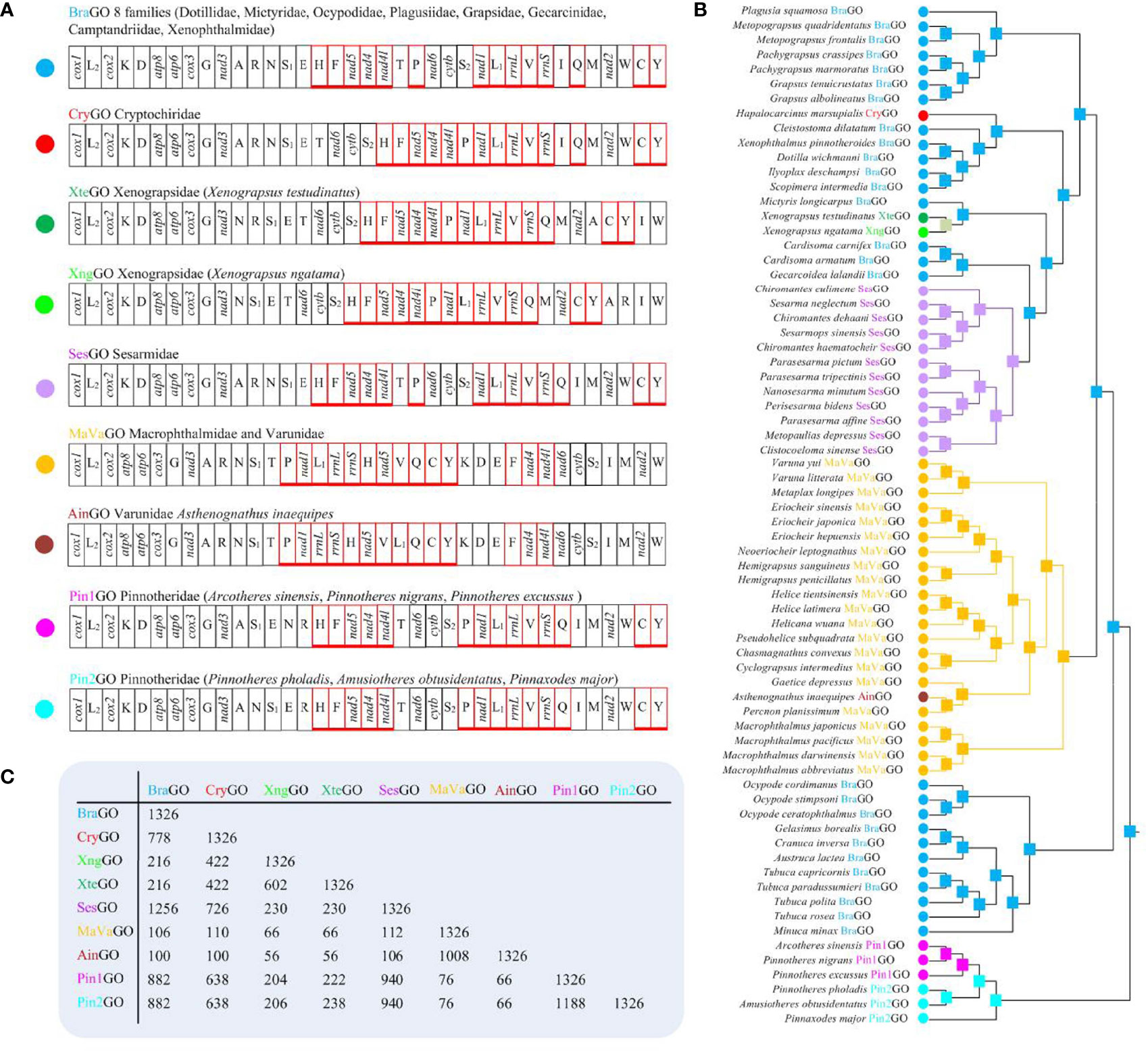
Figure 3 (A) The nine mitochondrial gene order patterns of Thoracotremata, which were linearized starting from cox1. The genes encoded on the minus strand were marked by red boxes. (B) The assignment of the gene order on the phylogenetic nodes. The solid dots and squares represented the extant and ancestral gene order patterns, respectively. (C) The number of shared common intervals values computed with the CREx program through pairwise comparisons of complete gene orders.
The ancestral gene order reconstruction shows that the BraGO pattern was the ancestral MGO of Thoracotremata, which occurred at the onset of thoracotreme clade (Figure 3B). The other Thoracotremata GOs evolved from BraGO can be separated into three groups: (i) high rearranged GO (SesGO), which shares the most common intervals (1,256) with BraGO; (ii) moderate rearranged GOs (CryGO, Pin1GO, and Pin2GO), which share 778, 882, and 882 common intervals with BraGO, respectively; and (iii) low rearranged GOs (XngGO, XteGO, MaVaGO, and AinGO), which share 216 or less common intervals with BraGO (Figure 3C). Through mechanism analysis, we found that MaVaGO, AinGO, CryGO, and XngGO (Figures 4A–D) could be produced through TDRL events from BraGO. XteGO derived from BraGO through a very complex mechanism requiring two transposition events and two TDRL events (Figure 4D). In Pin1GO, Pin2GO, and SesGO, only some tRNA genes have changed their location, implying the transposition events (Figures 4E, F).
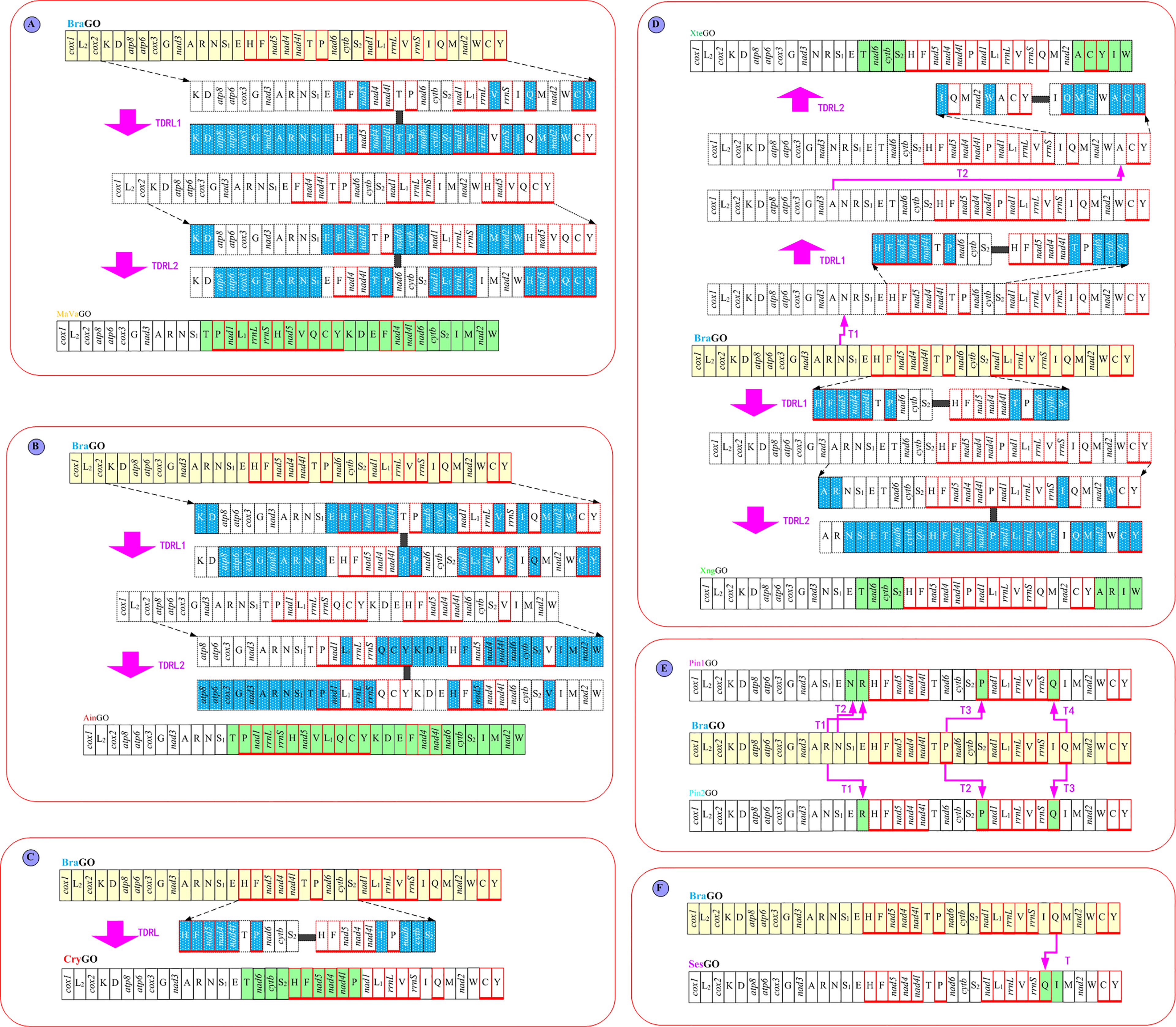
Figure 4 The evolutionary pathways generating (A) MaVaGO, (B) AinGO, (C) CryGO, (D) XteGO and XngGO, (E) Pin1GO and Pin2GO, and (F) SesGO arrangements from BraGO. tdrl, tandem duplication random loss mechanism. T, transposition events. The genes involved in rearrangement events relative to BraGO were shown with a green background.
In the present study, we newly described four MGOs, AinGO, CryGO, Pin1GO, and Pin2GO, for the subsection Thoracotremata. The evolutionary pathways generating the new arrangements are depicted in Figures 4B, C, E. The AinGO has been produced through two TDRL events that involved the genomic portion included between trnT and trnW (Figure 4B). In the CryGO pattern, a single TDRL event is necessary to explain the final rearrangement, and the genomic portion involved in the process contains 10 genes (Figure 4C). Pin1GO differs from BraGO for the transposition of trnN, trnR, trnP, and trnQ (T1–T4), which were located between trnE and trnH, trnN and trnH, trnS2 and nad1, and rrnS and trnI, instead of its placement downstream to trnA, trnT, and trnI in BraGO, respectively (Figure 4E). The transformational pathway producing Pin2GO implies three transpositions (T1–T3). Only trnR, trnP, and trnQ are transposed with respect to BraGO.
Some studies have mentioned that higher variation in mitochondrial gene order may be correlated with accelerated substitution rates, resulting in long branches and problems in nucleotide sequence-based analysis. Strong positive correlations between gene orders and elevated nucleotide substitution rates were found in arthropod and bivalve mitogenomes (Shao et al., 2003; Xu et al., 2006; Plazzi et al., 2016). In contrast to these findings, our Spearman correlation analysis indicated no link between variable gene arrangements (breakpoint distances) and nucleotide substitution rates (branch lengths) in thoracotreme crabs (Supplementary Table 6). The value of Spearman’s rho was 0.15 (p = 0.217). However, the branch lengths of taxa with rearranged GO (SesGO, CryGO, Pin1GO, Pin2GO, XngGO, XteGO, MaVaGO, and AinGO) were significantly higher than that of BraGO (Mann–Whitney U, p = 0.031). This result is consistent with the observation of Tan et al. (2019), who found that the Spearman correlation analysis does not suggest a link between variable MGOs and nucleotide/amino acid substitution rates in Decapoda. Although our correlation analysis indicated no such association for the Thoracotremata, we found that the symbiotic groups, the cryptochiroid and pinnotheroid crabs, display variable MGOs (CryGO, Pin1GO, and Pin2GO). The crabs of Cryptochiroidea and Pinnotheroidea shared a similar lifestyle. Pea crabs from the superfamily Pinnotheroidea are obligate symbionts with some marine invertebrates, such as the mollusks, tubeworms, and echinoderms (Campos, 2016; Theil et al., 2016). Gall crabs (Cryptochiroidea) are coral-dwelling crabs, which are obligate symbionts of living scleractinian coral reefs (Wetzer et al., 2009; van der Meij, 2015; van der Meij et al., 2015). Some studies have suggested possible correlations of rearranged MGOs to the adaptations to specialized lifestyles and extreme ecological niches (also see Tan et al., 2019), such as burrowing crayfish (Gan et al., 2018), freshwater crabs (Tan et al., 2018), and the species adapted to extreme deep-sea environments (Nakajima et al., 2016). This seems not absolute. Some deep-sea vent crab and shrimp species and burrowing axiid mud shrimps exhibited only a few rearranged MGOs (Tan et al., 2019). Therefore, more mitogenomes of representation from major groups with varied lifestyles and extreme ecological niches were needed to clarify exact drivers of mitochondrial gene rearrangements.
Phylogeny of Thoracotremata Based on Mitogenomes
Previous mitogenomic thoracotreme phylogenies were limited by including only Grapsoidea and Ocypodoidea taxa in studies (Chen et al., 2018; Wang et al., 2021). In this study, we presented the first expanded Thoracotremata mitochondrial phylogenetic relationship, including all the four superfamilies (Grapsoidea, Ocypodoidea, Pinnotheroidea, and Cryptochiroidea).
The ML phylogeny shared most similarities in their topologies with that obtained from Bayesian inference (Figure 5), specifically in the recovery of Pinnotheroidea as basal clade. However, several nodes were still in contention. In the BI tree, the soldier crab Mictyris longicarpus Latreille, 1806 closely grouped with the ocypodoid family Macrophthalmidae with strong nodal support (BPP = 0.96). However, in the ML tree, M. longicarpus and the grapsoid family Xenograpsidae formed a sister lineage with low support value (BS = 56). The monophyly of the grapsoid family Gecarcinidae was not supported in the BI tree (BPP = 1.00). Gecarcoidea lalandii H. Milne Edwards, 1837 was more closely related to the family Sesarmidae than its Gecarcinidae relatives Cardisoma carnifex (Herbst, 1796 [in Herbst, 1791-1796]) and Cardisoma armatum (Herklots, 1851), while the three Gecarcinidae species formed a monophyletic group in the ML tree, but with low support value (BS = 65). Although there is some discrepancy in the internal relationships among Varunidae between the ML and BI trees, the position of the newly presented grapsid species Tritodynamia horvathi Nobili, 1905 was identical. It is apparent that T. horvathi, a member of Macrophthalmidae, was shown to be more closely related to the varunid crab A. inaequipes (BS = 100, BPP = 1.00), than to other macrophthalmid crabs. This seems not surprising. The genera Tritodynamia and Asthenognathus were once placed in the family Pinnotheridae based on the morphological characteristics (Schmitt et al., 1973). Števčić (2005) raised them to family level and placed Tritodynamia in the Macrophthalmidae as a separate subfamily, the Tritodynamiinae. Cuesta et al. (2005) suggested a close relationship with the Varunidae after looking at the molecular data for several species. Later, Ng et al. (2008) placed the Tritodynamia and Asthenognathus into the family Macrophthalmidae and Varunidae, respectively. Our results questioned the validity of the subfamily Tritodynamiinae and support the inclusion of Tritodynamica in the family Varunidae.
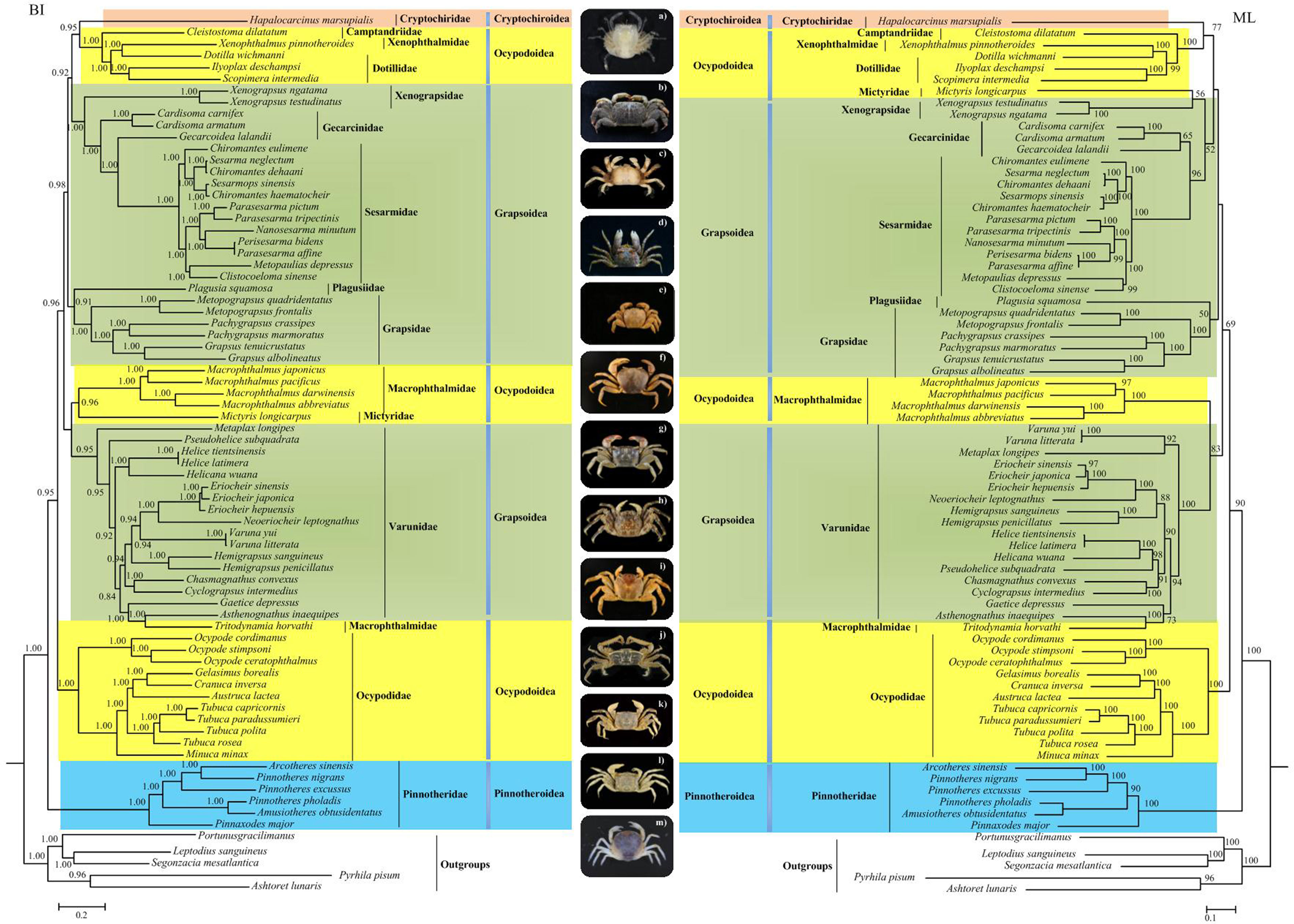
Figure 5 The phylogenetic tree of Thoracotremata derived from Bayesian inference (BI) and Maximum Likelihood (ML) analyses using 13 PCGs. The number at each node is Bayesian posterior probability (BPP) for BI and bootstrap values (BP) for ML. Ocypodoidea, Grapsoidea, Cryptochiroidea, and Pinnotheroidea were also indicated by yellow, green, red, and blue background. Photographs on the medial position show the extent of morphological diversity of the Thoracotremata. Photographs: (A) Hapalocarcinus marsupialis Stimpson, 1858 sensu lato (Cryptochiridae), (B) Cleistostoma dilatatum De Haan, 1833 (Camptandriidae), (C) Xenophthalmus pinnotheroides White, 1846 (Xenophthalmidae), (D) Dotilla wichmannide Man, 1892 (Dotillidae), (E) Xenograpsus testudinatus N.K. Ng, J.-F. Huang & P.-H. Ho, 2000 (Xenograpsidae), (F) Cardisoma carnifex (Herbst, 1796) (Gecarcinidae), (G) Parasesarma affine (De Haan, 1837) (Sesarmidae), (H) Plagusia squamosa (Herbst, 1790) (Plagusiidae), (I) Grapsus albolineatus Latreille in Milbert, 1812 (Grapsidae), (J) Macrophthalmus (Macrophthalmus) abbreviatus R.B. Manning & Holthuis, 1981 (Macrophthalmidae), (K) Helicana wuana (Rathbun, 1931) (Varunidae), (L) Ocypode stimpsoni Ortmann, 1897 (Ocypodidae), and (M) Pinnotheres pholadis De Haan 1835 (Pinnotheridae).
The crabs of Pinnotheroidea are usually symbiotic with mollusks, tubeworms, echinoids, and other invertebrates. Its phylogeny was poorly understood, with the monophyly of this group still under question (Theil et al., 2016). In our study, the Pinnotheroidea, including only one family Pinnotheridae, formed a monophyletic clade. However, previous studies have pointed out the non-monophyly of the pinnotheroid crabs (Palacios-Theil et al., 2009; Theil et al., 2016). This discrepancy may due to the limited pinnotheroid sample in our study and the heterogeneity of data. Pinnotheridae was the earliest branching in Thoracotremata, located at the base of the phylogenetic tree, which was consistent with the result of Schubart et al. (2006). In the work of Schubart et al. (2006) (using 16S and 12S), only one Pinnotheridae species, Pinnotheres pisum (Linnaeus, 1767), was sampled, which was shown to hold a basal position compared to the clade of “Grapsoidea & Ocypodoidea”. However, this presents an opposite view compared with some other previous studies (Wetzer et al., 2009; Tsang et al., 2014; van der Meij and Schubart, 2014; Tsang et al., 2018). Using a fragment of the 16S gene, Wetzer et al. (2009) found that the three Pinnotheridae species constituted a monophyletic clade, nested within a clade that includes the ocypodoid families Mictyridae, Camptandridae, and Ocypodidae. An analysis of more than 140 brachyuran species (only two Pinnotheridae species) using six nuclear protein-coding genes and two mitochondrial rRNA genes suggested that the two pea crabs clustered with Dotillidae from Ocypodoidea, forming a basal lineage of Thoracotremata (Tsang et al., 2014). Using molecular data from three markers (mitochondrial 12S and 16S rRNAs, and nuclear Histone H3), Tsang et al. (2018) revealed Pinnotheridae to be monophyletic, but widely distant from its pinnotheroid relatives (Aphanodactylidae) and instead clustered with Ocypodidae (Ocypodoidea), forming two basal thoracotreme clades. Based on the analysis of the 16S gene of 82 thoracotreme crabs, van der Meij and Schubart (2014) found that the monophyletic Pinnotheridae clade was closely related to the family Ocypodidae from Ocypodoidea.
Cryptochiroidea was represented in the present study by the species H. marsupialis from family Cryptochiridae, which was grouped with an ocypodoid lineage (Dotillidae/Xenophthalmidae/Camptandriidae). Previous studies have suggested that Cryptochiridae was a sister lineage to Xenograpsidae (Grapsoidea) (Tsang et al., 2014; Tsang et al., 2018). Close phylogenetic relationships between the Cryptochiridae and Grapsidae were proposed by Wetzer et al. (2009), who, based on 16SmtDNA, considered H. marsupialis as a “highly modified Grapsidae”, because H. marsupialis evolved within the family Grapsidae (Grapsoidea), while van der Meij and Schubart (2014) recommended that gall crabs should not be considered “highly modified Grapsidae”, but an independent lineage of grapsid crabs. As the Cryptochiroidea was represented by single species in the present study and the ML analysis only has 77 as a support value in this node, no overall conclusions about its phylogenetic position in the Thoracotremata should be made.
Although the results on the polyphyly of Grapsoidea and Ocypodoidea are consistent between the present and previous studies (Tsang et al., 2014; Basso et al., 2017; Tsang et al., 2018; Chen et al., 2019; Kobayashi et al., 2020; Zhang et al., 2020; Wang et al., 2021), the internal relationships of the families within these two groups were different. Our research provides some new insights into the phylogenetic relationships of some families with a wider taxonomic coverage. For example, Dotillidae (Ocypodoidea) and the ocypodoid families Xenophthalmidae and Camptandriidae formed a sister lineage, and Sesarmidae (Grapsoidea) grouped with its grapsoid relatives Xenograpsidae and Gecarcinidae. In contrast, in the studies of Basso et al. (2017); Tan et al. (2018), and Wang et al. (2021), which were also based on mitogenomes, Dotillidae (Ocypodoidea) and Sesarmidae (Grapsoidea) formed a sister group. Certainly, when the material from more species were obtained and the mitogenomic data were substantial, the relationships of families from Grapsoidea and Ocypodoidea can be well interpreted.
Conclusion
In the present study, we contributed twelve new mitogenomes from all the four thoracotreme superfamilies, greatly enhancing the taxonomic coverage of Thoracotremata mitogenomic data. Four new MGOs (CryGO, AinGO, Pin1GO, and Pin2GO) were described for the subsection Thoracotremata. We inferred the ancestral mitochondrial gene orders of Thoracotremata and demonstrated how the distinct patterns of gene orders could be obtained by evolutionary rearrangement events. No link between variable gene arrangements and nucleotide substitution rates was found in thoracotreme crabs. Pinnotheridae (Pinnotheroidea) formed a basal and monophyletic group. Cryptochiridae (Cryptochiroidea) was closely related to an Ocypodoidea lineage (Camptandriidae/Xenophthalmidae/Dotillidae).
Data Availability Statement
The complete mitochondrial DNA (mtDNA) sequences can be accessed from the GenBank database with the accession number OL661260-OL661268, OL579739-OL579740, OL657229.
Author Contributions
Data curation: SS. Funding acquisition: ZS and WJ. Software: SS. Writing—original draft: SS. Writing—review and editing: SS, ZS, WJ, and ZY. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by research grants from the National Science Foundation for Distinguished Young Scholars (42025603), the Key Research Program of Frontier Sciences, Chinese Academy of Sciences (QYZDB-SSWDQC036), the National Natural Science Foundation of China (42176138), and the Strategic Priority Research Program of the Chinese Academy of Sciences (XDA22050302).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.848203/full#supplementary-material
Supplementary Table 1 | The seed sequences used in mitochondrial genome assemblies using NOVOPlasty software.
Supplementary Table 2 | The species used in the phylogenetic analysis.
Supplementary Table 3 | The best-fit partitioning schemes and substitution models selected by PartitionFinder.
Supplementary Table 4 | The characteristics of the newly determined 12 thoracotreme mitogenomes.
Supplementary Table 5 | The size and nucleotide composition of the 70 thoracotreme mitogenomes.
Supplementary Table 6 | The breakpoint distances and branch lengths of thoracotremes.
References
Ahyong S. T., Lai J. C., Sharkey D., Colgan D., Ng P. K. L. (2007). Phylogenetics of the Brachyuran Crabs (Crustacea: Decapoda): The Status of Podotremata Based on Small Subunit Nuclear Ribosomal RNA. Mol. Phylogenet. Evol. 45, 576–586. doi: 10.1016/j.ympev
Bahr S., Johnson M. L., Berumen M. L., Hardenstine R. S., Rich W. A., van der Meij S. E. T. (2021). Morphology and Reproduction in the Hapalocarcinus Marsupialis Stimpson 1859 Species Complex (Decapoda: Brachyura: Cryptochiridae). J. Crustac. Biol. 41, 1–15. doi: 10.1093/jcbiol/ruab052
Basso A., Babbucci M., Pauletto M., Riginella E., Patarnello T., Negrisolo E. (2017). The Highly Rearranged Mitochondrial Genomes of the Crabs Maja Crispata and Maja Squinado (Majidae) and Gene Order Evolution in Brachyura. Sci. Rep. 7, 4096. doi: 10.1038/s41598-017-04168-9
Bernt M., Bleidorn C., Braband A., Dambach J., Donath A., Fritzsch G., et al. (2013). A Comprehensive Analysis of Bilaterian Mitochondrial Genomes and Phylogeny. Mol. Phylogenet. Evol. 69, 352–364. doi: 10.1016/j.ympev.2013.05.002
Bernt M., Merkle D., Middendorf M. (2008). An Algorithm for Inferring Mitogenome Rearrangements in a Phylogenetic Tree, in Comparative Genomics. Eds. Setubal J. C., Stoye J., Stadler P. F. (Berlin, Heidelberg: Springer), 143–157.
Bernt M., Merkle D., Ramsch K., Fritzsch G., Perseke M., Bernhard D., et al. (2007). CREx: Inferring Genomic Rearrangements Based on Common Intervals. Bioinformatics 23, 2957–2958. doi: 10.1093/bioinformatics/btm468
Bernt M., Middendorf M. A. (2011). A Method for Computing an Inventory of Metazoan Mitochondrial Gene Order Rearrangements. BMC Bioinform. 12, S6. doi: 10.1186/1471-2105-12-S9-S6
Blanchette M., Kunisawa T., Sankoff D. (1999). Gene Order Breakpoint Evidence in Animal Mitochondrial Phylogeny. J. Mol. Evol. 49, 193–203. doi: 10.1007/PL00006542
Bolger A. M., Lohse M., Usadel B. (2014). Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Boore J. L. (1999). Animal Mitochondrial Genomes. Nucleic Acids Res. 27, 1767–1780. doi: 10.1093/nar/27.8.1767
Boore J. L. (2000). “The Duplication/Random Loss Model for Gene Rearrangement Exemplified by Mitochondrial Genomes of Deuterostome Animals,” in Comparative Genomics, Computational Biology. Eds. Sankoff D., Nadeau J. H. (Dordrecht: Springer), 133–147. doi: 10.1007/978-94-011-4309-7_13
Brösing B., Richter S., Scholtz G. (2007). Phylogenetic Analysis of the Brachyura (Crustacea, Decapoda) Based on Characters of the Foregut With Establishment of a New Taxon. J. Zool. Syst. Evol. Res. 45, 20–32. doi: 10.1111/j.1439-0469.2006.00367.x
Campos E. (2016). The Pinnotheridae of the Northeastern Pacific (Alaska to Mexico): Zoogeographical Remarks and New Bivalve Hosts (Crustacea, Brachyura, Pinnotheridae). Zootaxa 4170, 311. doi: 10.11646/zootaxa.4170.2.5
Chen J. Q., Xing Y. H., Yao W. J., Xu X., Zhang C. L., Zhang Z. H., et al. (2019). Phylomitogenomics Reconfirm the Phylogenetic Position of the Genus Metaplax Inferred From the Two Grapsid Crabs (Decapoda: Brachyura: Grapsoidea). PloS One 14, e0210763. doi: 10.1371/journal.pone.0210763
Chen J. Q., Xing Y. H., Yao W. J., Zhang C. L., Zhang Z. H., Jiang G. C., et al. (2018). Characterization of Four New Mitogenomes From Ocypodoidea & Grapsoidea, and Phylomitogenomic Insights Into Thoracotreme Evolution. Gene 675, 27–35. doi: 10.1016/j.gene.2018.06.088
Cuesta J. A., Schubart C. D., Felder D. L. (2005). “Systematic Position of the Asthenognathinae Stimpson 1858 and Pseudopinnixa Carinata Ortmann (Decapoda, Brachyura): New Findings From Larval and DNA Comparisons,” in Abstracts of the Sixth International Crustacean Congress, (Glasgow, Scotland, UK), July 2005. 127.
Davie P. J. F., Guinot D., Ng P. K. L. (2015a). “Systematics and Classification of Brachyura,” in The Crustacea, Complementary to the Traité De Zoologie, vol. 9, Part C-II . Eds. Castro P., Davie P. J. F., Guinot D., Schram F. R., von Vaupe Klei J. C. (Leiden(NL) and Boston(US): Koninklijke NV, Brill), 1049–1130.
Davie P. J. F., Guinot D., Ng P. K. L. (2015b). “Phylogeny of Brachyura,” in The Crustacea, Complementary to the Traité De Zoologie, vol. 9 Part C-II . Eds. Castro P., Davie P. J. F., Guinot D., Schram F. R., von Vaupe Klei J. C. (Leiden(NL) and Boston(US): Koninklijke NV, Brill), 921–979.
Dierckxsens N., Mardulyn P., Smits G. (2016). NOVOPlasty: De Novo Assembly of Organelle Genomes From Whole Genome Data. Nucleic Acids Res. 45, e18. doi: 10.1093/nar/gkw955
Gan H. M., Tan M. H., Lee Y. P., Schultz M. B., Horwitz P., Burnham Q., et al. (2018). More Evolution Underground: Accelerated Mitochondrial Substitution Rate in Australian Burrowing Freshwater Crayfishes (Decapoda: Parastacidae). Mol. Phylogenet. Evol. 118, 88–98. doi: 10.1016/j.ympev.2017.09.022
Gissi C., Iannelli F., Pesole G. (2008). Evolution of the Mitochondrial Genome of Metazoa as Exemplified by Comparison of Congeneric Species. Heredity 101, 301–320. doi: 10.1038/hdy.2008.62
Gong L., Lu X., Wang Z., Zhu K., Liu L., Jiang L., et al. (2020). Novel Gene Rearrangement in the Mitochondrial Genome of Coenobita Brevimanus (Anomura: Coenobitidae) and Phylogenetic Implications for Anomura. Genomics 112, 1804–1812. doi: 10.1016/j.ygeno.2019.10.012
Guinot D. (1977). Propositions Pour Une Nouvelle Classification Des Crustacés Décapodes Brachyoures. C. R. Hebd. Des. Séances Acad. Des. Sci. 285, 1049–1052.
Guinot D. (1978). Principesd’une Classification Évolutive Des Crustacés Décapodes Brachyoures. Bull. Biol. Fr. Belg. 112, 211–292.
Guinot D. (1979). Données Nouvelles sur la Morphologie, la Phylogenèseet et La Taxonomie des Crustacés Décapodes Brachyoures, Mém. Mus. Natl. Hist. Nat., Sér. A, Zool. 112, 3–354.
Guinot D. (2008). Propositions Pour Une Nouvelle Classification Des Crustaces Decapodes Brachyoures. C. R. Acad. Sci. Paris D 285, 1049–1052.
Guinot D., Ng N. K., Moreno P. A. R. (2018). Review of Grapsoid Families for the Establishment of a New Family for Leptograpsodes Montgomery 1931, and a New Genus of Gecarcinidae H. Milne Edwards 1837 (Crustacea, Decapoda, Brachyura, Grapsoidea MacLeay 1838). Zoosystema 40, 547–604. doi: 10.5252/zoosystema2018v40a26
Hahn C., Bachmann L., Chevreux B. (2013). Reconstructing Mitochondrial Genomes Directly From Genomic Next-Generation Sequencing Reads-a Baiting and Iterative Mapping Approach. Nucleic Acids Res. 41, e129. doi: 10.1093/nar/gkt371
Ji Y. K., Wang A., Lu J. J., Lu X. L., Jin Y. H., Song D. H., et al. (2014). Mitochondrial Genomes of Two Brachyuran Crabs (Crustacea: Decapoda) and Phylogenetic Analysis. J. Crustac. Biol. 34, 494–503. doi: 10.1163/1937240X-00002252
Katoh K., Standley D. M. (2013). MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kobayashi G., Itoh H., Fukuda H., Kojima S. (2020). The Complete Mitochondrial Genome of the Sand Bubbler Crab Scopimera Globosa and its Phylogenetic Position. Genomics 113, 831–839. doi: 10.1016/j.ygeno.2020.10.014
Kück P., Meusemann K. (2010). FASconCAT: Convenient Handling of Data Matrices. Mol. Phylogenet. Evol. 56, 1115–1118. doi: 10.1016/j.ympev.2010.04.024
Lanfear R., Calcott B., Ho S. Y. W., Guindon S. (2012). PartitionFinder: Combined Selection of Partitioning Schemes and Substitution Models for Phylogenetic Analyses. Mol. Biol. Evol. 29, 1695–1701. doi: 10.1093/molbev/mss020
Laslett D., Canbäck B. (2007). ARWEN: A Program to Detect tRNA Genes in Metazoan Mitochondrial Nucleotide Sequences. Bioinformatics 24, 172–175. doi: 10.1093/bioinformatics/btm573
Ma K. Y., Qin J., Lin C. W., Chan T. Y., Ng P. K. L., Chu K. H., et al. (2019). Phylogenomic Analyses of Brachyuran Crabs Support Early Divergence of Primary Freshwater Crabs. Mol. Phylogenet. Evol. 135, 62–66. doi: 10.1016/j.ympev.2019.02.001
Minh B. Q., Nguyen M. A. T., von Haeseler A. (2013). Ultrafast Approximation for Phylogenetic Bootstrap. Mol. Biol. Evol. 30, 1188–1195. doi: 10.1093/molbev/mst024
Nakajima Y., Shinzato C., Khalturina M., Nakamura M., Watanabe H., Satoh N., et al. (2016). The Mitochondrial Genome Sequence of a Deep-Sea, Hydrothermal Vent Limpet, Lepetodrilus Nux, Presents a Novel Vetigastropod Gene Arrangement. Mar. Genomics 28, 121–126. doi: 10.1016/j.margen.2016.04.005
Ng P. K. L., Guinot D., Davie P. J. F. (2008). Systema Brachyurorum: Part I. An Annotated Checklist of Extant Brachyuran Crabs of the World. Raffles Bull. Zool. 17, 1–286.
Palacios-Theil E., Cuesta J. A., Campos E., Felder D. L. (2009). “Molecular Genetic Re-Examination of Subfamilies and Polyphyly in the Family Pinnotheridae,” in Crustacean Issues 18: Decapod Crustacean Phylogenetics. Eds. Martin J. W., Crandall K. A., Felder D. L. (Boca Raton (FL: CRC Press), 457–474.
Perna N. T., Kocher T. D. (1995). Patterns of Nucleotide Composition at Fourfold Degenerate Sites of Animal Mitochondrial Genomes. J. Mol. Evol. 41, 353–358. doi: 10.1007/BF01215182
Plazzi F., Puccio G., Passamonti M. (2016). Comparative Large-Scale Mitogenomics Evidences Clade-Specific Evolutionary Trends in Mitochondrial DNAs of Bivalvia. Genome Biol. Evol. 8, 2544–2564. doi: 10.1093/gbe/evw187
Rambaut A., Drummond A. (2013) Tracer V1.6. Available at: http://tree.bio.ed.ac.uk/software/tracer/.
Ronquist F., Huelsenbeck J. P. (2003). MrBayes 3: Bayesian Phylogenetic Inference Under Mixed Models. Bioinformatics 19, 1572–1574. doi: 10.1093/bioinformatics/btg180
Schmitt W. L., McCain J. C., Davidson E. (1973). “Decapoda I. Brachyura I. Fam. Pinnotheridae,” in Crustaceorum Catalogus, vol. 3. (W. Junk B.V., The Hague, NL), 1–160.
Schubart C. D., Cannicci S., Vannini M., Fratini S. (2006). Molecular Phylogeny of Grapsoid Crabs (Decapoda, Brachyura) and Allies Based on Two Mitochondrial Genes and a Proposal for Refraining From Current Superfamily Classification. J. Zool. Syst. Evol. Res. 44, 193–199. doi: 10.1111/j.1439-0469.2006.00354.x
Shao R. F., Dowton M., Murrell A., Barker S. C. (2003). Rates of Gene Rearrangement and Nucleotide Substitution Are Correlated in the Mitochondrial Genomes of Insects. Mol. Biol. Evol. 20, 1612–1619. doi: 10.1093/molbev/msg176
Shih H. T., Ng P. K. L., Davie P. J. F., Schubart C. D., Türkay M., Naderloo R., et al. (2016). Systematics of the Family Ocypodidae Rafinesque 1815 (Crustacea: Brachyura), Based on Phylogenetic Relationships, With a Reorganization of Subfamily Rankings and a Review of the Taxonomic Status of Uca Leach 1814, Sensu Lato and its Subgenera. Raffles Bull. Zool. 64, 139–175. doi: 10.5281/zenodo.5355087
Smith D. R. (2016). The Past, Present and Future of Mitochondrial Genomics: Have We Sequenced Enough mtDNAs? Brief. Funct. Genomics 15, 47–54. doi: 10.1093/bfgp/elv027
Števčić Z. (2005). The Reclassification of Brachyuran Crabs (Crustacea: Decapoda: Brachyura). Nat. Croat. 14, 1–159.
Sun S., Sha Z., Wang Y. (2019). The Complete Mitochondrial Genomes of Two Vent Squat Lobsters, Munidopsis Lauensis and M. Verrilli: Novel Gene Arrangements and Phylogenetic Implications. Ecol. Evol. 9, 12390–12407. doi: 10.1002/ece3.5542
Sun H. Y., Zhou K. Y., Song D. X. (2003). Mitochondrial Genome and Phylogenetic Reconstruction of Arthropods. Zool. Res. 24, 467–479. doi: 10.3321/j.issn:0254-5853.2003.06.012
Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Tan M. H., Gan H. M., Lee Y. P., Lintona S., Grandjeand F., Bartholomei-Santos M. L., et al. (2018). ORDER Within the Chaos: Insights Into Phylogenetic Relationships Within the Anomura (Crustacea: Decapoda) From Mitochondrial Sequences and Gene Order Rearrangements. Mol. Phylogenet. Evol. 127, 320–331. doi: 10.1016/j.ympev.2018.05.015
Tan M. H., Gan H. M., Lee Y. P., Poore G. C. B., Austin C. M. (2017). Digging Deeper: New Gene Order Rearrangements and Distinct Patterns of Codons Usage in Mitochondrial Genomes Among Shrimps From the Axiidea, Gebiidea and Caridea (Crustacea: Decapoda). PeerJ 5, e2982. doi: 10.7717/peerj.2982
Tan M. H., Gan H. M., Schultz M. B., Austin C. M. (2015). MitoPhAST, a New Automated Mitogenomic Phylogeny Tool in the Post-Genomic Era With a Case Study of 89 Decapod Mitogenomes Including Eight New Freshwater Crayfish Mitogenomes. Mol. Phylogenet. Evol. 85, 180–188. doi: 10.1016/j.ympev.2015.03.017
Tan M. H., Gan H. M., Yin P. L., Bracken-Grissom H., Chan T. Y., Miller A. D., et al. (2019). Comparative Mitogenomics of the Decapoda Reveals Evolutionary Heterogeneity in Architecture and Composition. Sci. Rep. 9, 10756. doi: 10.1038/s41598-019-47145-0
Theil E. P., Cuesta J. A., Felder D. L. (2016). Molecular Evidence for non-Monophyly of the Pinnotheroid Crabs (Crustacea: Brachyura: Pinnotheroidea), Warranting Taxonomic Reappraisal. Invertebr. Syst. 30, 1–27. doi: 10.1071/IS15023
Timm L., Bracken-Grissom H. D. (2015). The Forest for the Trees: Evaluating Molecular Phylogenies With an Emphasis on Higher-Level Decapoda. J. Crust. Biol. 35, 577–592. doi: 10.1163/1937240X-00002371
Trifinopoulos J., Nguyen L. T., von Haeseler A., Minh B. Q. (2016). W-IQ-TREE: A Fast Online Phylogenetic Tool for Maximum Likelihood Analysis. Nucleic Acids Res. 44, W232–W235. doi: 10.1093/nar/gkw256
Tsang L. M., Ahyong S. T., Shih H. T., Ng P. K. L. (2018). Further Polyphyly of Pinnotheroid Crabs: The Molecular Phylogenetic Position of the Polychaete-Associated Aphanodactylidae. Invertebr. Syst. 32, 92–99. doi: 10.1071/IS17038
Tsang L. M., Schubart S. T., Ahyong J. C., Lai E. Y., Au T., Chan P. K., et al. (2014). Evolutionary History of True Crabs (Crustacea: Decapoda: Brachyura) and the Origin of Freshwater Crabs. Mol. Biol. Evol. 31, 1173–1187. doi: 10.1093/molbev/msu068
van der Meij S. E. T. (2015). Evolutionary Diversification of Coral-Dwelling Gall Crabs (Cryptochiridae). [Dissertation/Doctor Thesis] (Rapenburg: Leiden University).
van der Meij S. E. T., Fransen C. H. J. M., Pasman L. R., Hoeksema B. W. (2015). Phylogenetic Ecology of Gall Crabs (Cryptochiridae) as Associates of Mushroom Corals (Fungiidae). Ecol. Evol. 5, 5770–5780. doi: 10.1002/ece3.1808
van der Meij S. E. T., Schubart C. D. (2014). Monophyly and Phylogenetic Origin of the Gall Crab Family Cryptochiridae (Decapoda: Brachyura). Invertebr. Syst. 28, 491–500. doi: 10.1071/IS13064
Von Sternberg R., Cumberlidge N. (2001). On the Heterotreme-Thoracotreme Distinction in the Eubrachyura De Saint Laurent 1980 (Decapoda, Brachyura). Crustaceana 74, 321–338. doi: 10.1163/156854001300104417
Wang Q., Wang J., Wu Q., Xu X. Y., Wang P., Wang Z. F. (2021). Insights Into the Evolution of Brachyura (Crustacea: Decapoda) From Mitochondrial Sequences and Gene Order Rearrangements. Int. J. Biol. Macromol. 170, 717–727. doi: 10.1016/j.ijbiomac.2020.12.210
Wetzer R., Martin J. W., Boyce S. L. (2009). “Evolutionary Origin of the Gall Crabs (Family Cryptochiridae) Based on 16S rDNA Sequence Data,” in Crustacean Issues 18: Decapod Crustacean Phylogenetics. Eds. Martin J. W., Crandall K. A., Felder D. L.. (Boca Raton, US: CRC Press), 475–490.
WoRMS (2021) Thoracotremata. Available at: http://www.marinespecies.org/aphia.php?p=taxdetails&id=106682 (Accessed October 12, 2021).
Xing Y. H., Ma X. P., Wei Y. Q., Pan D., Liu W. L., Sun H. Y. (2016). The Complete Mitochondrial Genome of the Semiterrestrial Crab, Chiromantes Neglectum (Eubrachyura: Grapsoidea: Sesarmidae). Mitochondrial DNA Part B: Resour. 1, 461–463. doi: 10.1080/23802359.2016.1186509
Xing Y. H., Zhou L. J., Hou Y., Wang X. Q., Zhang C., Zhang H. L., et al. (2017). Complete Mitochondrial Genomes From Two Species of Chinese Freshwater Crabs of the Genus Sinopotamon Recovered Using Next-Generation Sequencing Reveal a Novel Gene Order (Brachyura, Potamidae). ZooKeys 705, 41–60. doi: 10.3897/zookeys.705.11852
Xu W., Jameson D., Tang B., Higgs P. G. (2006). The Relationship Between the Rate of Molecular Evolution and the Rate of Genome Rearrangement in Animal Mitochondrial Genomes. J. Mol. Evol. 63, 375–392. doi: 10.1007/s00239-005-0246-5
Ye Y., Miao J., Guo Y. H., Gong L., Jiang L., Lü Z., et al. (2021). The First Mitochondrial Genome of the Genus Exhippolysmata (Decapoda: Caridea: Lysmatidae), With Gene Rearrangements and Phylogenetic Associations in Caridea. Sci. Rep. 11, 14446. doi: 10.1038/s41598-021-93946-7
Keywords: thoracotremata, mitogenome, evolution, gene rearrangement, phylogenetic relationship
Citation: Sun S, Jiang W, Yuan Z and Sha Z (2022) Mitogenomes Provide Insights Into the Evolution of Thoracotremata (Brachyura: Eubrachyura). Front. Mar. Sci. 9:848203. doi: 10.3389/fmars.2022.848203
Received: 04 January 2022; Accepted: 17 March 2022;
Published: 14 April 2022.
Edited by:
Qi-Lin Zhang, Kunming University of Science and Technology, ChinaReviewed by:
Christopher Mervyn Austin, Deakin University, AustraliaSancia Van Der Meij, University of Groningen, Netherlands
Li Gong, Zhejiang Ocean University, China
Copyright © 2022 Sun, Jiang, Yuan and Sha. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhongli Sha, shazl@qdio.ac.cn
 Shao’e Sun1,2,3,4
Shao’e Sun1,2,3,4  Wei Jiang
Wei Jiang Zhongli Sha
Zhongli Sha