Full-Length Transcriptome Sequencing and Comparative Transcriptomic Analysis Provide Insights Into the Ovarian Maturation of Exopalaemon carinicauda
- 1Key Laboratory for Sustainable Utilization of Marine Fisheries Resources, Ministry of Agriculture and Rural, Yellow Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences, Qingdao, China
- 2Function Laboratory for Marine Fisheries Science and Food Production Processes, Qingdao National Laboratory for Marine Science and Technology, Qingdao, China
The ridgetail white shrimp Exopalaemon carinicauda has the potential to be used as a useful experimental organism in the field of crustacean research due to its transparent body, large egg, and short reproductive cycle. However, little is known about the regulatory mechanisms of ovarian maturation in E. carinicauda. In this study, we applied PacBio single-molecule long-read sequencing technology to unveil the whole transcriptome landscape of Exopalaemon carinicauda to better understand the molecular mechanisms of ovarian development. A total of 49.05 G polymerase read bases were generated, finally, 15, 151 unigenes were successfully identified with a mean length of 4, 257 bp and an N50 of 4, 884 bp from ovary tissues (five different ovarian developmental stages). A total of 14, 007 unigenes were successfully annotated in at least one public database. In addition, 8, 861 coding sequences, 4, 594 lncRNAs and 895 transcription factors were identified. Furthermore, we performed RNA-Seq analysis combined with Iso-Seq results to investigate ovarian maturation regulation mechanism and validated the results by quantitative real-time PCR. We annotated five vitellogenin isoforms in the full-length transcriptome, meanwhile these five genes and vitellogenin receptor suggested an important role during previtellogenesis of E. carinicauda. Genes were involved in PI3K-Akt signaling pathway, retinol metabolism, cell cycle and hedgehog signaling pathway, which showed obvious associations with ovarian maturation of E. carinicauda. We identified 40 DEGs, such as forkhead box protein L2, which may be related to ovarian development in the comparisons. The expression profiles of genes such as ecdysone receptor, ecdysone-induced protein 74EF, ecdysone inducible gene E75 and post-molt protein suggest that they were significantly associated with reproductive molting in the ovarian mature stage. In conclusion, the present study identified important genes and pathways involved in ovarian maturation, which might be useful for studying the reproductive regulation and mechanisms of ovarian maturation in E. carinicauda.
Introduction
The ridgetail white shrimp Exopalaemon carinicauda, belonging to the Palaemonidae family of crustaceans, is a major commercial mariculture species that is naturally distributed on the coasts of the Yellow Sea and Bohai Sea (Xu et al., 2010). Owing to its rapid growth and high tolerance to environmental stress, the scale at which E. carinicauda is cultured has expanded in recent years (Zhang et al., 2015a). However, aquaculture production and large-scale cultivation are still restricted by the supply of high-quality broodstock. In addition to its economic value, E. carinicauda is a useful experimental organism in the field of crustacean developmental biology due to its transparent body, large egg, and high reproductive capacity, and short reproductive cycle of only 2–3 months (Yuan et al., 2017; Liang et al., 2020; Wang et al., 2020). The female shrimps generate at least three consecutive ovarian development during reproductive stage, but the ovaries of some shrimps gradually atrophy into cystic tubes after first ovarian mature. It is important to provide valuable information for understanding the vitellogenesis and ovarian development and clarify the regulatory mechanism of ovarian development of E. carinicauda.
Previous studies have reported the different ovarian maturation stages of E. carinicauda (Wang, 1987); the ovary size and color undergo visible changes during ovarian maturation. Researchers have cloned, expressed, and studied the functions of ovarian maturation-related genes in E. carinicauda, including those that encode farnesoic acid O-methyltransferase (FAMeT), vitellogenin (Vtg) and heat shock protein 90 (Duan et al., 2014; Li et al., 2016; Liang et al., 2020). These genes have also been studied in the Palaemonidae family, including Macrobrachium nipponense (Bai et al., 2016) and Macrobrachium rosenbergii (Guo et al., 2019a; Qian and Liu, 2019). However, the regulatory genes identified using these methods are incomplete, and the regulation of ovarian maturation remains unclear. It is, therefore, essential to investigate the molecular mechanisms that regulate ovarian maturation. Next-generation RNA-seq is one method in which to do this.
Illumina RNA-Seq is a powerful tool for the identification of differentially expressed genes (DEGs) (Santos et al., 2021). However, low-quality transcripts obtained through Illumina RNA sequencing result in incorrect annotation information and limit the scope of analysis of alternative splicing variants (Zhang et al., 2020). It is difficult to identify full-length transcript in the absence of a reference sequence in most crustaceans (Bankar et al., 2015). Additionally, genome resources have been identified in only a few crustaceans because of their large genome size and complexity (Rotllant et al., 2018). Fortunately, full-length transcriptome sequencing has been employed as an effective approach to obtain high-quality transcript sequences in the absence of reference genomes. Single-molecule real-time (SMRT) sequencing developed by Pacific Biosciences (PacBio) can obtain full-length sequences without post-sequencing assembly (Holmes et al., 2021), which has been used for whole-transcriptome profiling in many crustaceans (Cao et al., 2020; Ren et al., 2020; Zheng et al., 2020).
In the present study, a high-quality full-length transcriptome of the ovaries of E. carinicauda was generated by a combination of PacBio SMRT sequencing and Illumina RNA-Seq, and was further used for comparative transcriptomic analysis of the different ovarian developmental stages. Transcript functional annotation, long noncoding RNA (lncRNA) prediction, transcription factor and coding sequence were performed based on the data. Putative genes involved in ovarian development and vitellogenesis were identified according to the characteristics of the different ovarian developmental stages. This study provides a better understanding of the molecular mechanism of ovarian development and may be a valuable resource for further investigation of E. carinicauda.
Materials and Methods
Animals and Sample Preparation
Female E. carinicauda adults (body length 52.29 ± 2.09 mm, body weight 2.37 ± 0.18 g) were collected from Haichen Aquatic Products Co. LTD in Rizhao, China. The shrimps were cultured in filtered aerated seawater at 25.0–26.0°C with an initial salinity of 31 ± 0.5, for one week. Aeration was used to maintain an adequate dissolved oxygen level. During the experimental periods, the shrimps were fed with commercial shrimp feed (~1.0 mm in diameter) twice daily (8:00 and 18:00), with food quantities equaling 10% of their body mass.
The females were assigned to one of five ovarian developmental stages using the criteria of Wang (1987) defined as: proliferative phase (stage I), the ovary is small and completely transparent when anatomically observed and its morphology and color cannot be distinguished by in vitro observation; minor growth phase (stage II), the ovary is enlarged, translucent with small black spots over its outer membrane, and positioned above the heart; major growth phase (stage III), ovarian volume continues to increase and its length has reached the 1/2 cephalothorax length, and the ovary is faintly yellow with small black dots over its ovarian membrane; mature phase (stage IV), the ovary covers almost the entire stomach, hepatopancreas, and heart, is bright yellow with dark green or tan spots, and the lateral shell of the first to fourth ventral segments is bluish; postpartum recovery (stage V), the ovary is very small and not obvious in vitro, and anatomical observation shows that it is transparent or translucent (Wang, 1987).According to the criteria, ovarian development was assessed daily by observing the size and color of gonads, and the five different ovarian developmental females were obtained in three weeks. A total of one hundred and five shrimps were collected, ten shrimps (2 individuals×5 stages) were used for histological examination, other for transcriptomic analysis. Among them, five shrimps (different ovarian development stage) were used for single-molecule full-length transcriptome sequencing, and ninety shrimps (6 individuals×5 stages×3 three replicates) were used for Illumina RNA-seq, they were separately flash-frozen in liquid nitrogen and then preserved at -80°C.
Histological Procedure
Ten samples of E. carinicauda from each developmental stage were used for histological examination. Ovarian tissue slices, taken from the middle of each ovary, were fixed in 4% paraformaldehyde for 24 h. After dehydration in a gradient of ethanol solutions, the fixed tissues were embedded in paraffin wax. Paraffin-embedded tissues were sectioned to 4 μm and stained with Mayer’s hematoxylin and eosin. Tissue sections were observed microscopically to assess the stage of ovarian development.
RNA Extraction and Quality Evaluation
Total RNA was isolated using TransZol Up Plus RNA Kit (Trans, China) according to the manufacturer’s instructions, respectively. The integrity of RNA was assessed with the Agilent 2100 Bioanalyzer (Agilent Technologies, USA) and agarose gel electrophoresis. The purity and concentration of RNA were determined using a Nanodrop 2000 microspectrophotometer (Thermo Scientific).
Library Construction and Sequencing
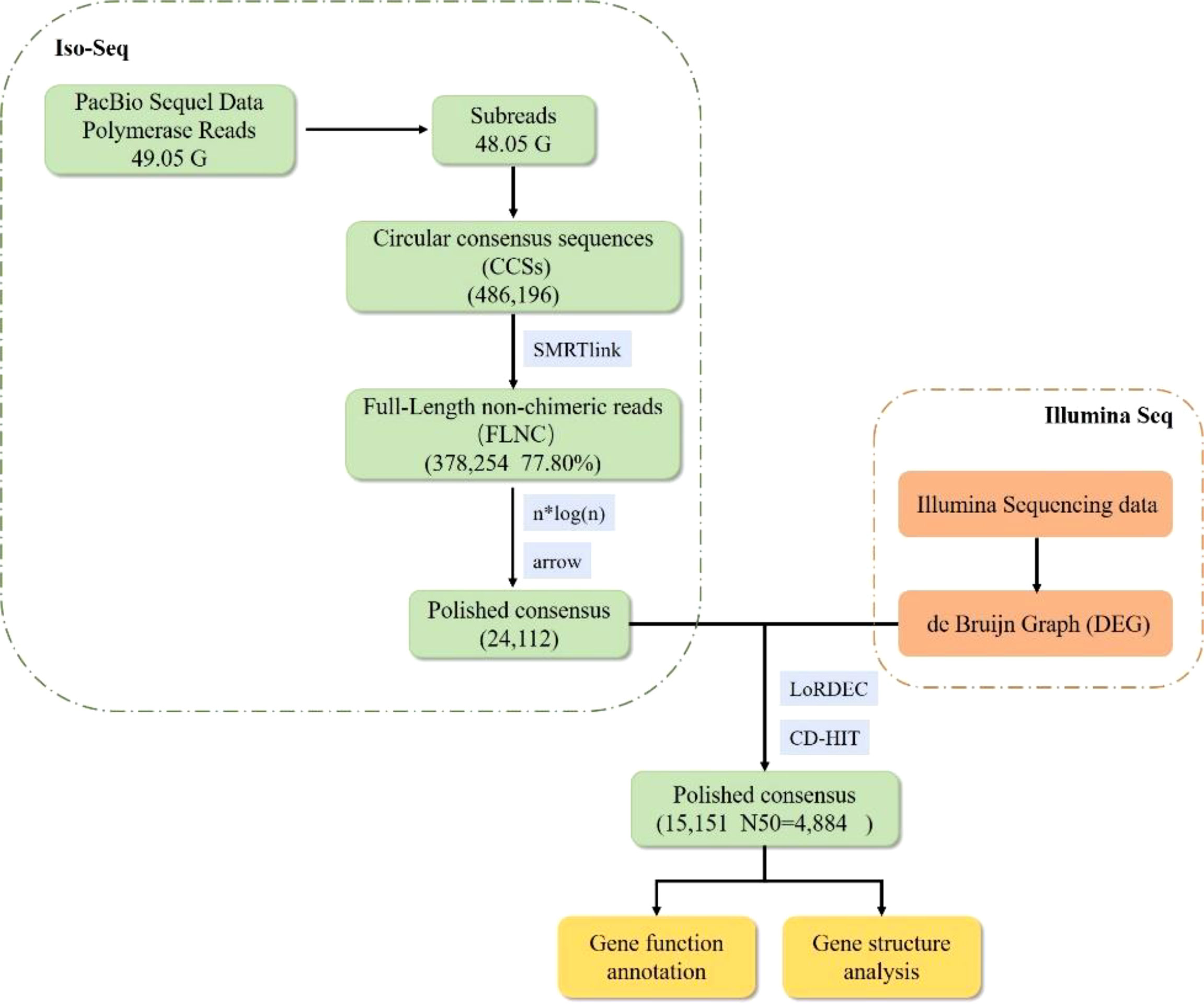
To construct the full-length transcriptome sequencing library for PacBio sequencing, qualified RNA from five ovaries of the different ovarian development stage, were pooled in equal amounts. RNA sequencing and analysis were performed using Novogene (Beijing, China). The full-length cDNA library was sequenced on a SMRT Cell of PacBio platform, and the Blue Pippin Size Selection System protocol was used as described by PacBio (PN 100-092-800-03). Briefly, mRNA was enriched using Oligo (dT) magnetic beads, and subsequently reverse transcribed into full length 1st strand cDNA. Full-length cDNAs were subjected to restoration of DNA damage, end repair, ligation to sequencing adapters, and digestion with exonuclease. Qualified libraries were sequenced on the PacBio Sequel platform according to their effective concentration and data output requirements.
PacBio Long-Read Processing
According to the PacBio protocol, the sequence data were first processed using the SMRTlink 7.0 software to remove the SMRTbell™ adapter and low-quality data. The circular consensus sequences, also known as the reads of insert, were generated from subreads with BAM files. All the reads of inserts were further classified into full-length and non full-length transcript sequences based on whether the polyA tail signal and 5′ and 3′ cDNA primers could be simultaneously observed. The full-length consensus isoforms were obtained using the ICE Quiver algorithm and subsequently polished. These polished consensus sequences were further subjected to correction using the Illumina short reads. The final transcriptome sequences were filtered by removing redundancy using the CD-HIT program with a threshold identity of 0.99.
Functional Annotation
Gene function was annotated using BLAST software (version 2.2.26) (Altschul et al., 1997) based on the following seven databases: NR (NCBI non-redundant protein sequences), NT (NCBI non-redundant nucleotide sequences), Pfam (Protein family) (Finn et al., 2013), KOG/COG (Clusters of Orthologous Groups of proteins) (Tatusov et al., 2000), Swiss-Prot (A manually annotated and reviewed protein sequence database) (Apweiler et al., 2004), KO (KEGG Ortholog database) (Kanehisa et al., 2004) and GO (Gene Ontology) (Ashburner et al., 2000).
Structure Analysis of the Transcriptome
Candidate coding regions within the transcript sequences were identified by ANGEL software (Shimizu et al., 2006). We used E. carinicauda or closely related species confident protein sequences for ANGEL training and then run the ANGEL prediction for given sequences. Transcripts with a length of more than 200 nucleotides and having more than two exons were selected as lncRNA candidates. Four computational approaches, coding-noncoding-Index (CNCI) (Sun et al., 2013), coding potential calculator (CPC) (Kang et al., 2017), PLEK (Li et al., 2014) and Pfam-scan (Bateman et al., 2000), were employed to further screen the protein-coding unigenes from the non-coding unigenes. Transcripts predicted with coding potential were filtered out, and those without coding potential formed our candidate set of lncRNAs. Transcription factor related unigenes were predicted using the AnimalTFDB 2.0 database.
RNA-Seq Library Construction and Sequencing
Total RNA was extracted using same method mentioned above. A total of 15 libraries (three replicates from five groups) were generated using the NEBNext® Ultra™ RNA Library Prep Kit (Illumina, CA, USA). The libraries were sequenced on an Illumina HiSeq 4000 platform and 150 bp paired-end raw reads were generated. Clean data were obtained by removing low-quality reads and reads containing adapters and ploy-N, and used for subsequent analysis.
Differentially Expressed Transcript Analysis
The clean reads of each RNA-seq library were aligned to the full-length reference transcriptome to obtain unique mapped reads using STAR (Dobin et al., 2013) with default parameters. The mapped reads were counted, and the expression value was normalized to fragments per kilobase of transcript per million fragments mapped. Differential expression analysis of the two groups was performed using the DEseq R package. P-values were adjusted using Benjamini and Hochberg’s approach for controlling the false discovery rate. We set the conditions of the false discovery rate <0.05 and |log2(foldchange)|≥1 as the thresholds for significantly differentially expressed transcripts.
Validation Experiments
Differentially expressed transcripts were verified using quantitative real-time PCR (qPCR) analysis. cDNA was synthesized using the PrimeScript™ Real-time PCR Kit (TaKaRa) for qPCR analysis. The specific primers of transcripts for qPCR were designed using Primer 5.0 software and listed in Supplementary Table 1. The qPCR assay was performed using the ChamQ SYBR Color qPCR Master Mix (Vazyme, China). Each qPCR reaction consisted of 10 μl 2×ChamQTM SYBR Color qPCR Master Mix, 50-100 ng of cDNA, 0.8 μl each of forward and reverse primer, 0.4 μl 50×ROX Reference Dye 2, and added sterile ddH2O to total volume of 20 μl. The PCR was performed on the ABI PRISM 7500 Sequence Detection System (Applied Biosystems, USA) under the following conditions: one cycle at 95°C for 30 s, then 40 cycles of 95°C for 10 s, 60°C for 30 s followed by 1 cycle of 95°C 15 s, 60°C for 1 min and 95°C for 15 s. The β-actin of E. carinicauda (GenBank accession number: JQ045354.1) was used as an internal control for expression analysis of E. carinicauda, which was abundant and stable in cells, and hardly affected by the external regulation (Liang et al., 2017). The primers of all assays for the real-time PCR perform were conducted in triplicate. All analyzed were based on the CT values of the PCR products. The expression was calculated with 2-ΔΔCT methods. Differences in gene expression were considered statistically significant at P<0.05 using one-way ANOVA and Tukey’s multiple comparison test.
Results
Histological Analysis of Different Ovarian Developmental Stages in E. carinicauda
According to Wang’s study, the of E. carinicauda ovarian development can be divided into five stages. Histological examination of stage I showed that basophilic oogonia (OG) were located in the ovarian middle zone, and their mean long and short diameters were 9.45 ± 2.06 μm and 7.74 ± 1.64 μm, respectively. Basophilic previtellogenic oocytes (PR) were found on the periphery of OG, and their mean long and short diameters were 33.8 ± 5.41 μm and 26.65 ± 5.23 μm, respectively (Figure 1A).
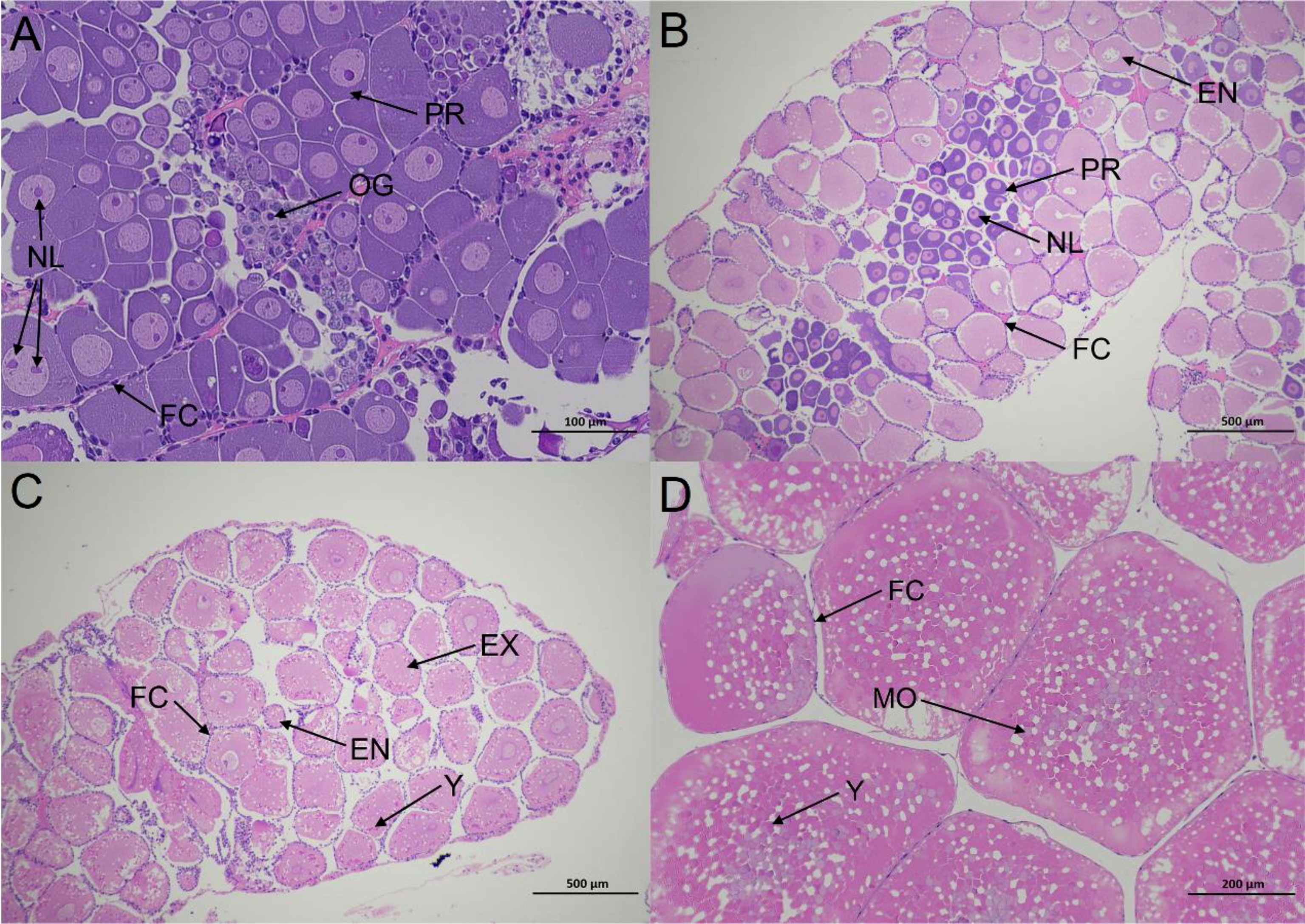
Figure 1 Histological sections illustrating the ovarian status at different stages in E.carinicauda (A) proliferative phase (Stage I), ×200. (B) minor growth phase (Stage II), ×40. (C) major growth phase (Stage III), ×40. (D) mature phase (Stage IV), ×100. EN, endogenous vitellogenic oocytes. EX, exogenous vitellogenic oocytes. FC, follicle cell; MO, mature oocytes; NL, nucleolus; OG, oogonium; PR, previtellogenic oocytes; Y, yolk granule.
In stage II, the PR were located in the ovarian middle zone, and were surrounded by endogenous vitellogenic oocytes (EN). The mean long and short diameters of EN were 153.88 ± 18.59 μm and 120.45 ± 20.59 μm, respectively (Figure 1B).In stage III, the main cell type was exogenous vitellogenic oocytes (EX) with mean long and short diameters of 313.12 ± 35.14 μm and 254.64 ± 16.13 μm, respectively. The yolk granules (YG) appeared in the EX, and the follicular cells (FC) migrated to the periphery of EX (Figure 1C).Mature oocytes (MO) were the main cell type in stage IV, with mean long and short diameters of 562.75 ± 34.51 μm and 446.8 ± 24.26 μm, respectively. The oocytes were squeezed into each other in a polygonal shape, and most cells in the nucellus were invisible (Figure 1D).
A Global Description of Full-Length Transcriptome
The full-length transcriptome of E. carinicauda was generated from pooled ovarian RNA using the PacBio Sequel platform (Figure 2). A total of 49.05 G polymerase read bases were generated. After filtering out adaptors and low-quality reads (less than 50bp), 48.05 G subreads with a mean length of 3, 472 bp were obtained. Using SMRT Link 4.0 software, 486, 196 circular consensus sequences were generated, and approximately 77.80% of them were full-length non-chimeric reads, containing 5′ and 3′ primers, and the poly (A) tail. After clustering and polishing, a total of 24, 112 consensus reads were obtained with a mean length of 4, 076 bp. These consensus reads were subsequently corrected using LoRDEC and CD-HIT to eliminate up to 99.99% sequencing errors. Finally, 15, 151 unigenes with an average length of 4, 257bp and an N50 of 4, 884 bp were successfully identified. Of these, 10, 510 were longer than 3, 000bp.
Functional Annotation of Transcript
All 15, 151 unigenes were aligned with seven different databases, including NR, GO, KEGG, KOG, NT, Pfam, and SwissProt. A total of 14, 007 (92.45%) transcripts were successfully annotated in at least one public database, and 3, 578 transcripts annotated in all the databases (Figure 3). The remaining unannotated unigenes (1, 144 unigenes) may represent novel E. carinicauda species-specific genes.
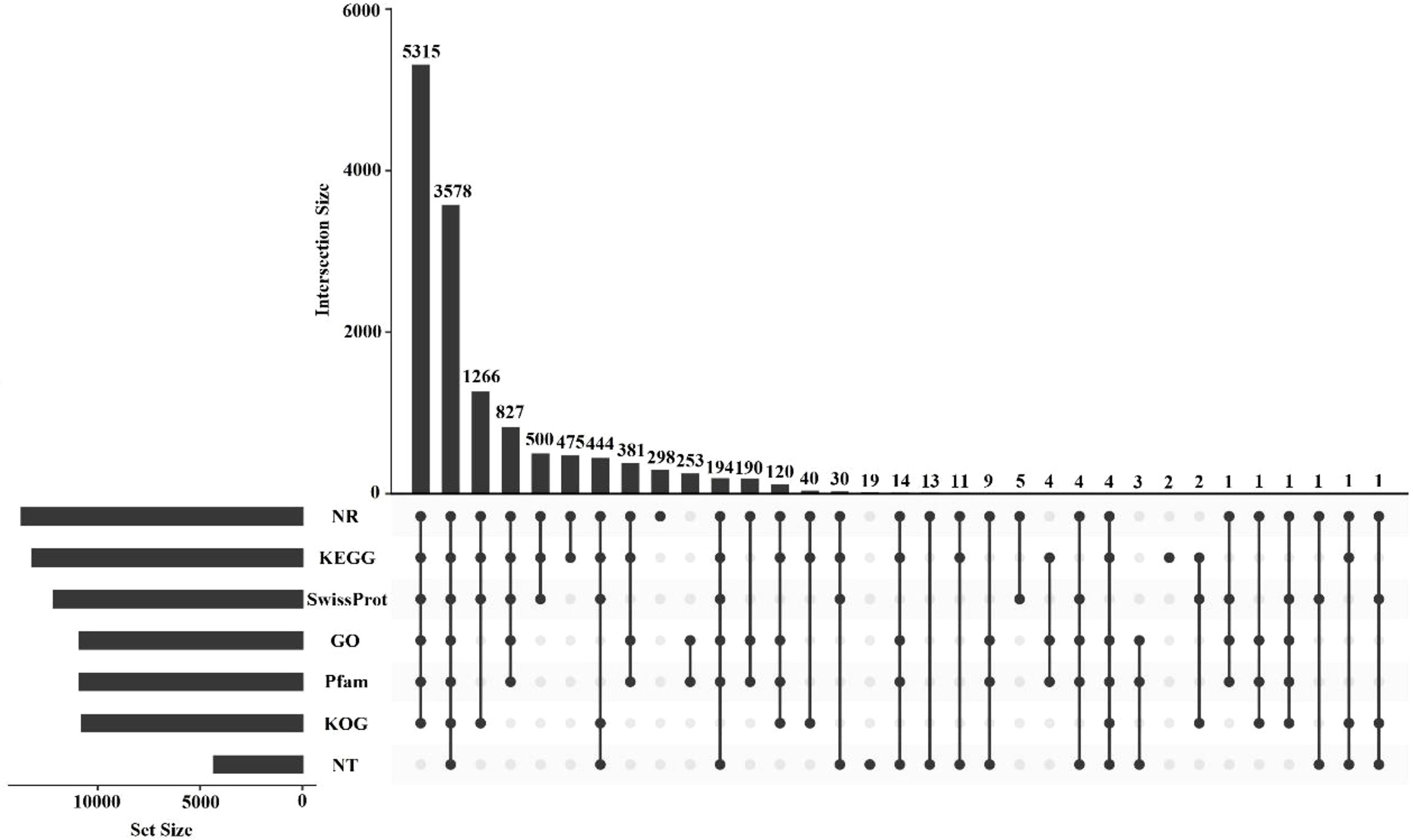
Figure 3 UpSet plot showing the results of the annotated transcripts and the number of unique annotated transcripts for each database.
GO analysis revealed that 10, 899 transcripts were assigned to 58 level-2 GO terms. Metabolic processes (4, 256), binding (7, 106) and cell parts (2, 531) were the most enriched subcategories within the biological process, molecular function, and cellular component level-2 categories, respectively (Supplementary Figure 1A).
A total of 13, 208 transcripts were mapped to KEGG Ortholog categories and grouped into 353 signaling pathways. The annotated pathways were grouped into six level-1 KEGG Ortholog terms, and signal transduction, transport and catabolism, and cancers were the top three most annotated level-2 KEGG Ortholog terms (Supplementary Figure 1B).
A total of 12, 140 transcripts were classified into 26 KOG classifications. The largest classification was general function prediction (2, 108 unigenes), followed by signal transduction mechanisms (1, 891 unigenes) then posttranslational modification, protein turnover and chaperones (1, 060 unigenes). (Supplementary Figure 1C)
Transcription Factor, Coding Sequence, and lncRNA Predictions
Using AnimalTFDB 2.0, a total of 895 transcripts were predicted to be transcription factors, whereby zf-C2H2 and ZBTB were the main ones identified (Supplementary Figure 2A). There were 8, 861 coding sequences identified by ANGEL, the distribution of the coding sequence lengths of complete open reading frames is shown in Supplementary Figure 2B. A total of 4, 594 lncRNAs were identified using the CNCI, CPC, PLEK, and Pfam approaches (Supplementary Figure 2C).
Differentially Expressed Transcript Analysis
We calculated and compared the expression levels of DEGs among the five groups (Stage I, Stage II, Stage III, Stage IV and Stage V) to identify reproduction-related genes. A total of 867 DEGs (204 up-regulated, 663 down-regulated) were detected between the Stage II and Stage I. For the Stage III and Stage II, 80 DEGs (41 up-regulated, 39 down-regulated) were detected, and for the Stage IV and Stage III, 333 DEGs (132 up-regulated, 201 down-regulated) were detected, and for the Stage IV and Stage IV, the number were 734 (389 up-regulated, 345 down-regulated) (Figure 4).
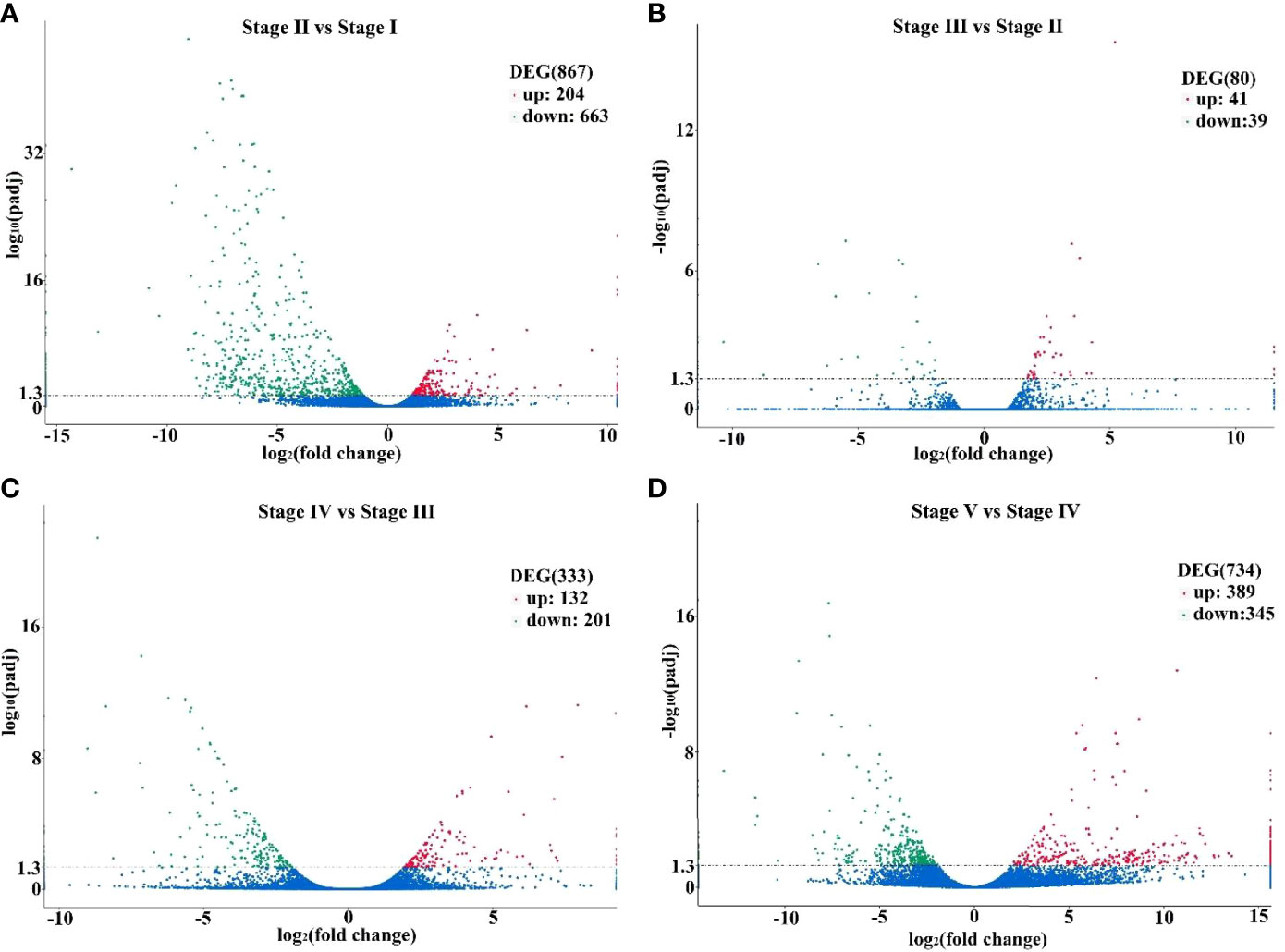
Figure 4 Volcano plots displaying differentially expressed genes in Stage II vs Stage I (A), Stage III vs Stage II (B), Stage IV vs Stage III (C) and Stage V vs Stage IV (D). Those that are significantly upregulated and downregulated are indicated in red and green, respectively, and those are not significantly different are in blue.
We recorded the most important pathways associated to ovarian development by comparing the DEGs with the KEGG pathway. Six pathways were involved in the regulation of ovarian development of E. carinicauda. There were three pathways (folate biosynthesis, PI3K-Akt signaling pathway and retinol metabolism) related to ovarian development in the comparison of Stage II and Stage I. Only folate biosynthesis pathway was significantly different between the Stage III and Stage II. There were three pathways (cell cycle, hedgehog signaling pathway and PI3K-Akt signaling pathway) were significantly different between the Stage IV and Stage III. The folate biosynthesis, phototransduction-fly and Hedgehog signaling pathway were significantly different between the Stage V and Stage IV. (Table 1)
Screening and Expression of Genes Related to Ovarian Development
Based on the analysis of DEGs, we manually screened 40 genes related to ovarian development by combining the q-value of the DEG enrichment pathways in the comparison groups Stage I vs Stage II, Stage II vs Stage III, Stage III vs Stage IV and Stage IV vs Stage V (Figure 5). The expressions of six genes were higher in the Stage II than the Stage I indicated that these genes participated in the regulation of the initiation of vitellogenesis. Seven genes and nineteen genes were highest expressed in the Stage III and Stage IV, respectively. Among them, ecdysone receptor, ecdysone-induced protein, 74EF, ecdysone inducible gene E75 and post-molt protein 1 expressed higher in the Stage IV for reproductive molting. The RYamide receptor like and dopamine receptor 2 were expressed higher in the Stage I than in the Stage II indicated that they are related to the inhibition of vitellogenesis.
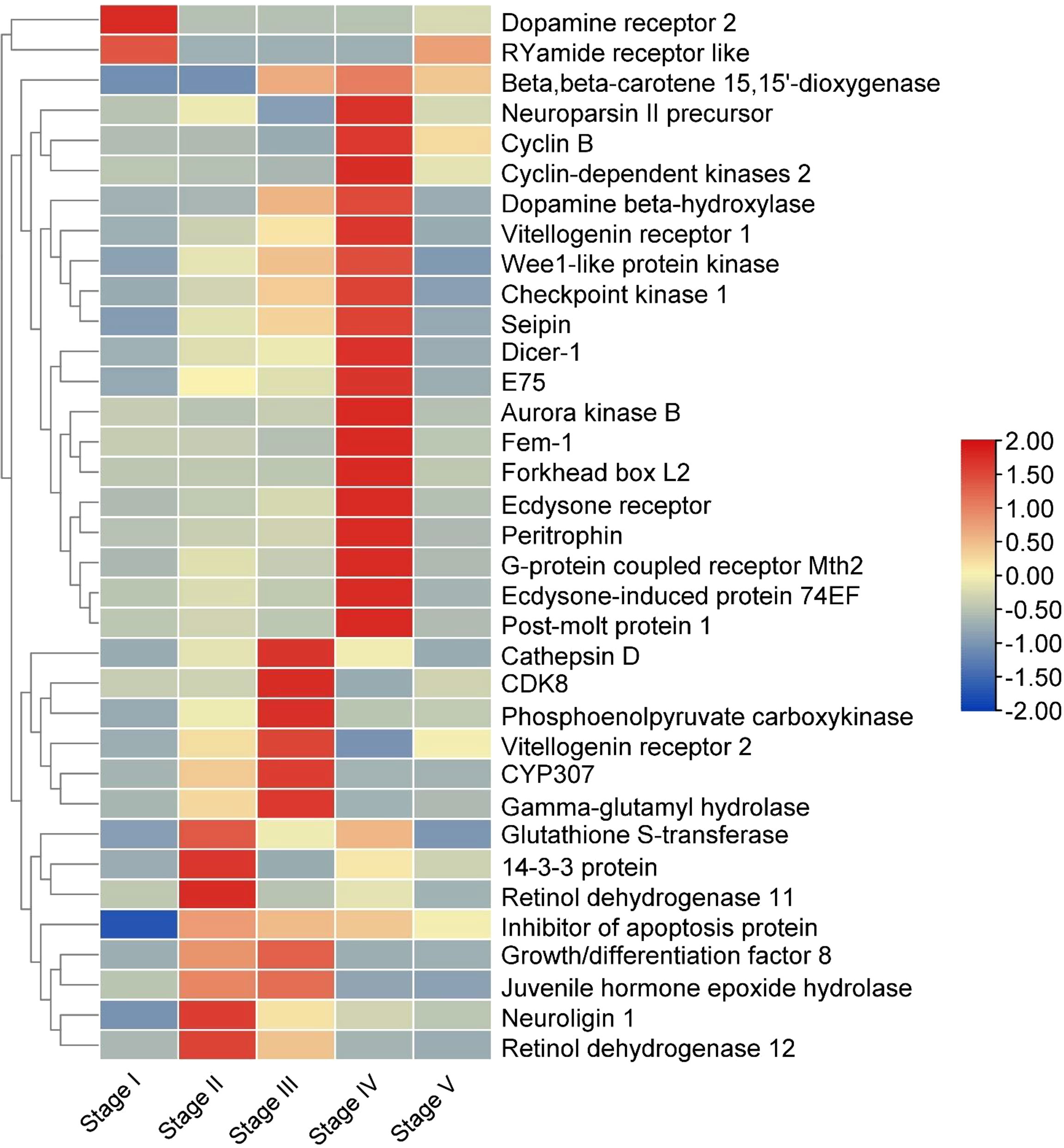
Figure 5 Heat map of genes related to ovarian development. The columns and rows indicate individuals and genes, respectively. The color scale represents FPKM after standard normalization.
Validation of DEGs by qRT-PCR
Expression levels of upregulated and downregulated unigenes were determined using quantitative real-time PCR (qRT-PCR). Between Stage II and Stage I, the expression levels were determined for five upregulated unigenes coding for CYP307, adenylyl cyclase-associated protein, checkpoint kinase 1, glutathione S-transferase and adiponectin receptor; and five downregulated unigenes coding for serine/threonine-protein kinase Doa, cyclophilin A, lipoprotein receptor 2A, guanylate cyclase PcGC-M2 precursor and ficolin-like protein 2. Similarly, between Stage IV and Stage III, the expression levels were measured for five upregulated unigenes coding for aurora kinase B, calcified cuticle protein, cyclin-dependent kinases 2, fork-head box L2, and serine/threonine-protein kinase D3, and five downregulated unigenes encoding alkaline phosphatase, growth/differentiation factor 8, cathepsin L, cyclin-dependent kinase inhibitor, and fatty acid synthase. The fold-changes determined by qRT-PCR and RNA-seq were compared for each DEG (Figure 6).
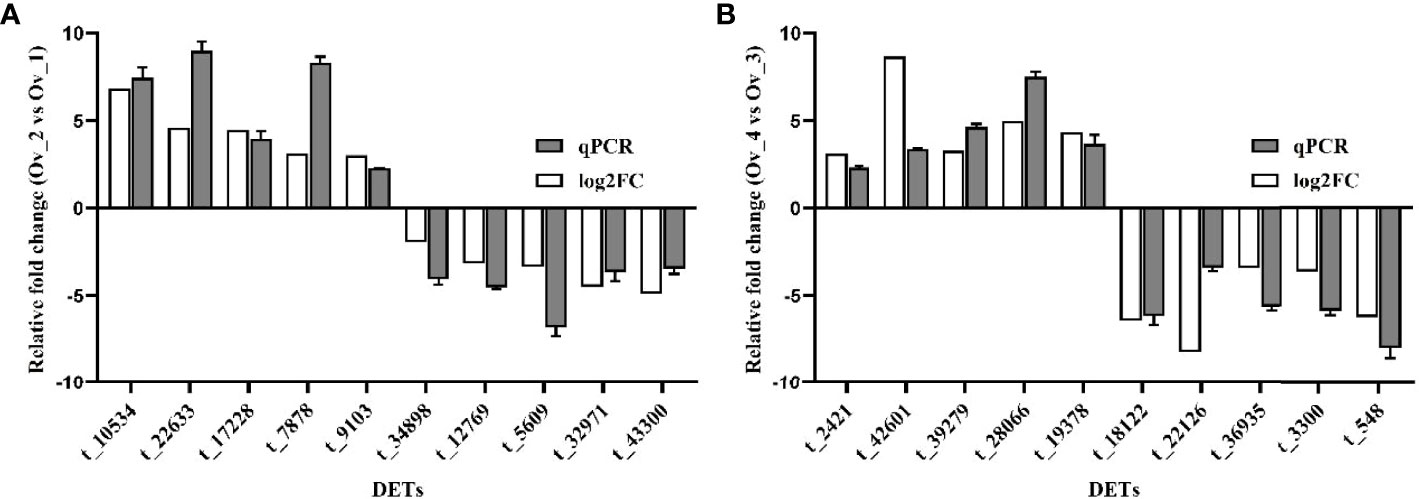
Figure 6 Validation of the expression levels of five upregulated and five downregulated DEGs in Stage II vs Stage I (A) and Stage IV vs Stage III (B) RNA-Seq and qRT-PCR data are indicated in grey and white, respectively t_10534, CYP307; t_22633, Adenylyl cyclase-associated protein; t_17228, checkpoint kinase 1; t_7878, glutathione S-transferase; t_9103, adiponectin receptor; t_34898, serine/threonine-protein kinase Doa; t_12769, cyclophilin A; t_5609, lipoprotein receptor 2A; t_32971, guanylate cyclase PcGC-M2 precursor; t_43300, ficolin-like protein 2; t_2421, aurora kinase B; t_42601, calcified cuticle protein; t_39279, cyclin-dependent kinases 2; t_28066, forkhead box L2; t_19378, Serine/threonine-protein kinase D3; t_18122, alkaline phosphatase; t_22126, growth/differentiation factor 8; t_36935, cathepsin L; t_3300, cyclin-dependent kinase inhibitor; t_548, fatty acid synthase.
Analysis of Vitellogenin in E. carinicauda
Vitellogenin (Vtg) is an important source of nutrients for the development of ovaries and embryos in almost all oviparous organisms. We annotated five members of vitellogenin in the full-length transcriptome, including t_9427, t_7810, t_7815, t_7987 and t_8253, which were subsequently named EcVtg1 to EcVtg5, respectively. EcVtg1 and EcVtg2 had four functional domains: Lipoprotein N-terminal domain, DUF1943, DUF 1081 and VWD structure. EcVtg3, EcVtg4 and EcVtg5 have the same functional domains, except for DUF 1081. All the EcVgs contained signal peptide that start at position 1 and ended at positions 18–22. The expression patterns of EcVtg1, EcVtg2, EcVtg3 and EcVtg5 were similar, and their expression was highest in the Stage III stage, whereas EcVtg4 was highest in the Stage II stage (P < 0.05) (Figure 7).
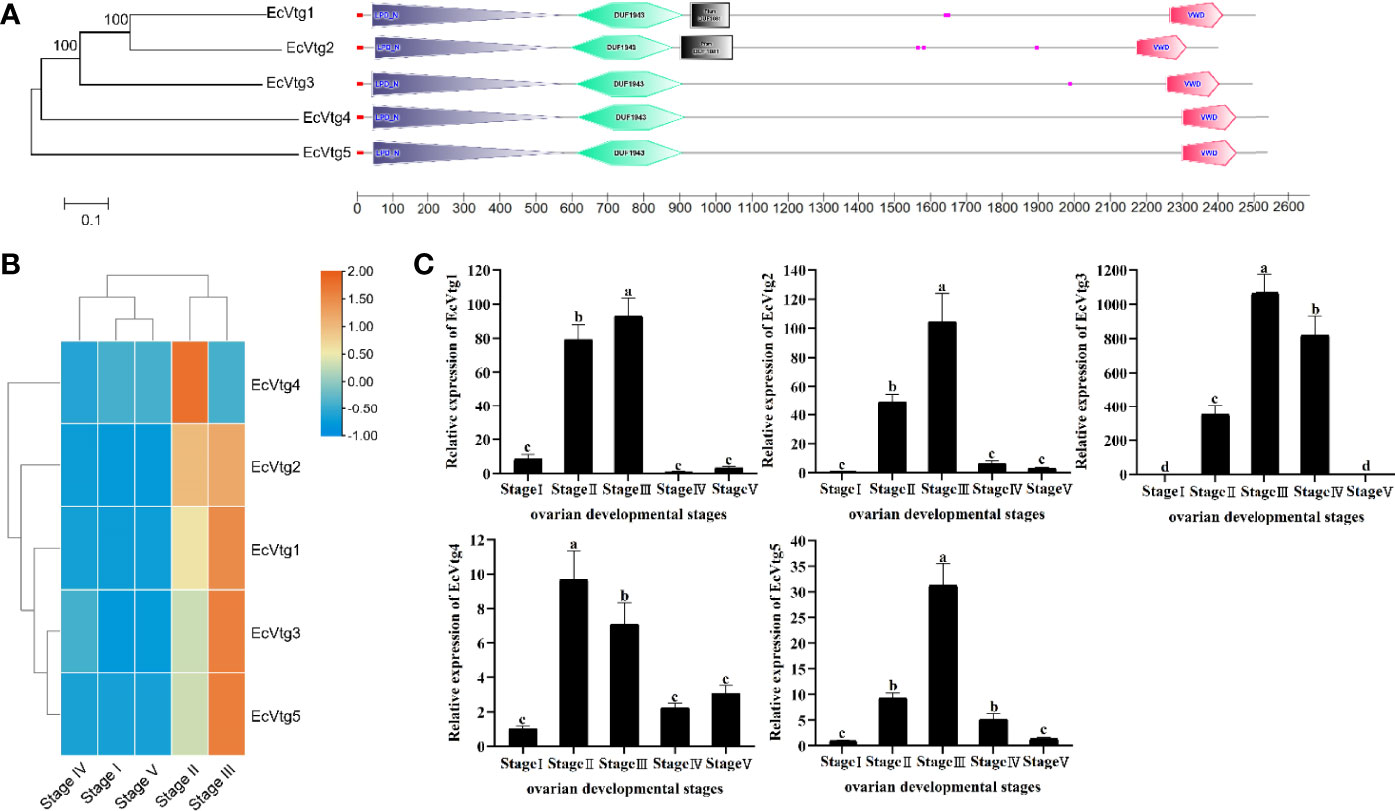
Figure 7 The five types of vitellogenin genes in E. carinicauda (A) Schematic representation of gene structures of Vtg genes in E. carinicauda. The phylogenetic relationship was shown on the left. The functional domains were marked with different colors. The signal peptide is red line, the LPD_N domain is gray boxed, the DUF1943 is green boxed, the DUF 1081 is black boxed and the VWD is red boxed. (B) Visualizing the genes using a heatmap plot. The columns and rows indicate individuals and genes, respectively. The color scale represents FPKM after standard normalization. (C) The mRNA expression of EcVtgs in five ovarian developmental stages. Data with different letters significantly differ (P < 0.05) among stages. Each bar represents the mean ± SE (n = 3). The β-actin serves as internal reference gene.
Discussion
Ovarian development is a very important physiological process for crustacean reproduction and is closely related to the reproductive capacity of the female broodstock. However, there is limited knowledge of the regulatory genes and pathways involved in crustacean ovarian development because of the limited transcriptomic information on target organs. We identified several key genes and pathways are potentially involved in the regulation of ovarian maturation within the ovary using PacBio and RNA-seq technology. The PacBio platform can achieve the full-length sequence of transcripts and identify full coding sequences and multiple encoded isoforms (Weirather et al., 2017), which is necessary for gene discovery of some species without a published genome. In this study, we first produced high confidence and full-length transcriptome data from ovary tissue (five different ovarian developmental stages) using the PacBio Sequel sequencing approach. A large amount of transcriptome data was generated, including 15, 151 high-quality full-length transcripts with a mean length of 4, 257bp, and N50 length were higher than those in a previous study that did not use full-length transcript information of the ovary (Shi et al., 2020). This dataset may provide new information for (i) studying the molecular regulatory mechanisms involved in female reproduction, (ii) developing new technology for genetic control within the aquaculture industry, and (iii) helping industry proactively solve various production problems caused by ovary maturation in E. carinicauda.
Ovarian development is a complex physiological process, which involves activation or inhibition of many genes. The cell cycle and oocyte meiosis pathways play important roles during the ovarian developmental stage in E. carinicauda. The cyclin B and cyclin-dependent kinase play an important role in the development and maturation of oocytes/ovaries in crustacean (Visudtiphole et al., 2009; Han et al., 2012; Phinyo et al., 2013; Phinyo et al., 2014). Serotonin promotes vitellogenesis of Penaeus indicus, and shrimps treated with serotonin exhibited higher transcript levels of vitellogenin and cyclin B (Tomy et al., 2016). The mRNA level of Cyclin B in mature stage was greater than that of oogonia stage in Penaeus monodon (Visudtiphole et al., 2009), Scylla paramamosain (Han et al., 2012) and Tachypleus tridentatus (Li et al., 2015c). In our study, cyclin-dependent kinases 8 was highest expressed in the major growth phase, and the cyclin B and cyclin-dependent kinases 2 were highest expressed in the mature growth phase. Techa et al. suggested that genes related to growth and molt play a critical role in the early embryogenesis of arthropods (Techa et al., 2015). The expression of cell division cycle 2 mRNA is relatively stable between oogenesis and early embryogenesis with the exception of a drop in early vitellogenesis ovary in Oncorhynchus mykiss (Qiu et al., 2008). These data indicated that four genes for cell cycle pathways not only possibly related to ovarian development of E. carinicauda, but also are maternal-mRNAs stored for newly fertilized embryos to presume translation.
Vitellogenin is an important precursor of egg yolk in nearly all oviparous animals (Harwood and Amdam, 2021). The accumulation of vitellogenin acts as a pivotal regulator of nutritional accumulation within oocytes, which drives gonadal maturation in females. Vitellogenesis originates in both the hepatopancreas and ovary in most crustacean (Zmora et al., 2007). Liang et al., found that vitellogenin of E. carinicauda is synthesized both in the ovary and hepatopancreas (Liang et al., 2020). In this study, the mRNA expression level of Vtgs and vitellogenin receptor (Vtgr) in the stage II were significantly higher than that in the stage I. The vitellogenin mRNAs are translated and stored in the stage II and stage III for the highly expressed level of five Vtgs at stage II and stage III. Most studies on the function of the vitellogenin gene in crustaceans mainly involve one vitellogenin gene, but many oviparous animals, such as fish (Williams et al., 2014) and insects (Boldbaatar et al., 2010), have multiple vitellogenin genes. Previous studies on Metapenaeus ensis (Kung et al., 2004) and Pandalopsis japonica (Jeon et al., 2011) have indicated the presence of multiple Vtg genes. In this study, five EcVtgs were identified in the full-length transcripts, which were expressed in the ovaries at different ovarian developmental stages. It is the first time that five vitellogenin isoforms has been identified in crustaceans. The relative abundance of the EcVtgs mRNA transcripts changed with ovarian development, and EcVtg1, EcVtg2, EcVtg3 and EcVtg5 were expressed similarly during the ovarian developmental stage. These results indicated that these five vitellogenin isoforms play important roles in ovarian developmental stages.
Retinol and its derivatives play key roles in the initiating meiosis in germ cells of mammalian fetal ovaries (Jiang et al., 2017), follicular development (Kawai et al., 2016), ovarian steroidogenesis (Damdimopoulou et al., 2019), and oocyte maturation (Tahaei et al., 2011). Full-length transcriptomic analysis was performed to identify 22 transcripts in retinol metabolism, and five of them were confirmed to be upregulated from the stage I to the growth phase, with four genes coding for retinol dehydrogenase (RDH) and β, β-carotene 15,15’-dioxygenase. Three RDHs, RDH11, RDH12 and RDH13, were detected in the transcriptome sequences. RDH12 is involved in steroid metabolism (Keller and Adamski, 2007), and RDH13 in zebrafish shows significantly higher expression levels in vitellogenic ovaries than in non-vitellogenic ovaries (Levi et al., 2011). Knockdown of RDH11 resulted in decreased transcription of vitellogenin and vitellogenin receptor in Procambarus clarkii (Kang et al., 2019). Our results revealed that retinol metabolism is critical for ovarian development in E. carinicauda. An appropriate feed proportion with adequate vitamin A levels in the stage I may be beneficial for the reproduction of E. carinicauda.
Juvenile hormones (JH) play important roles in the regulation of molting, development, metamorphosis and reproductive maturation in arthropods (Li et al., 2019; Santos et al., 2019). An increase in JH initiates vitellogenesis and egg development in adult insects. JH degradation in insects occurs by JH esterase and JH epoxide hydrolase (JHEH) (Lü et al., 2015). Knocking down JHEH in Colaphellus bowringi elevated JH signaling and increased expressions of vitellogenin genes (Guo et al., 2019b). In crustaceans, the JHEH-like genes have been identified in Daphnia magna (Toyota et al., 2014), M. rosenbergii (Chen et al., 2021) and Penaeus vannamei (Liu et al., 2022). JHEH-like is indispensable for the regulation of larval development and ecdysteroidogenesis in M. rosenbergii (Chen et al., 2021) and plays an essential role in shrimp survival during bacterial infection in P. vannamei (Liu et al., 2022). In this study, the expression of JHEH-like was significantly highest in the major growth stage among the five ovarian developmental stages in E. carinicauda. However, there was no evidence that JHEH plays an important role in the regulation of ovarian development in other crustacean. The JHEH-like proteins may exist in crustaceans, but compared to insects with probably different functions. We will properly investigate the function of JHEH-like gene in E. carinicauda. Reproductive development in crustaceans requires neuropeptides, ecdysone and methyl farnesoate (Nagaraju, 2011). Methyl farnesoate is the crustacean analogue of JH III. Similar to the pleiotropic functions of JH in insects, methyl farnesoate participates in numerous physiological processes in crustaceans, including molting (Nagaraju et al., 2006), sex determination (Suppa et al., 2021) and vitellogenesis (Li et al., 2021). The FAMeT catalyzes the conversion of farnesoic acid to methyl farnesoate, which is a rate-limiting step in the synthesis of methyl farnesoate. In our previous study, we have identified the expression profile of FAMeT in E. carinicauda (Li et al., 2016), which indicated that FAMeT was widely distributed but with higher expression levels in the mandibular organ. During the first ovarian development of E. carinicauda, the expression trend of FAMeT in the mandibular organ was similar to that in the hepatopancreas, which showed that the expression of FAMeT decreased gradually from stage I to stage IV, and decreased to the lowest in the stage IV, then rapidly recovered to a higher state in the stage V. Different from the expression of FAMeT in the mandibular organ and hepatopancreas, the expression of FAMeT in the stage IV of ovary was lowest among five stages, but there were no significant differences among the stage I, stage II, stage III and stage V. In this study, a full-length transcript of FAMeT was identified, but there was no significant difference between any two different stages of ovarian development in ovary. According to these results and the previous study, we supposed that the mandibular organ is the main tissue for FAMeT synthesis, which plays an important role in the regulation of ovarian development.
Neuropeptides play a critical role in regulating animal reproduction (Webster et al., 2012; Bao et al., 2015). A number of neuropeptide families were identified in crustaceans. The crustacean hyperglycemic hormone family including crustacean hyperglycemic hormone, molt inhibiting hormone, vitellogenesis inhibiting hormone and mandibular organ-inhibiting hormone, are known to regulate several important functions in decapod crustaceans such as energetic metabolism, molting and reproduction (Montagné et al., 2010; Webster et al., 2012). Crustacean hyperglycemic hormone has been shown to inhibit protein and mRNA synthesis in vitro in ovarian fragments of crustacean (Khayat et al., 1998), and might inhibit the synthesis of ecdysteroids by Y-organs and be involved in promoting vitellogenesis in Scylla paramamosain (Fu et al., 2016). Molt inhibiting hormone is a key endocrine regulator in the coordination of molting and likely reproduction in crustaceans, which simultaneously inhibits molt and induces ovarian maturation (Huang et al., 2015). The vitellogenesis-inhibiting hormone plays a major regulatory role in vitellogenesis and the reproductive cycle (Cohen et al., 2021). In our study, only crustacean hyperglycemic hormone was identified in ovary by Illumina RNA-Seq. The crustacean hyperglycemic hormone family members are best known for being secreted from the X-organ sinus gland complex of the eyestalk. In this study, crustacean hyperglycemic hormone gene was identified in ovary tissue by Illumina RNA-Seq, which is identical in amino acid sequence to that of eyestalk in our previous study (Li et al., 2015a). A total of 22 neuropeptides and related transcripts were predicted from the transcriptome data of female E. carinicauda (Table 2 and Supplementary Table 2). Of these transcripts, the neuroparsin II precursor, RYamide receptor like, dopamine receptor 2, and dopamine beta-hydroxylase were identified as DEGs in the four comparisons.
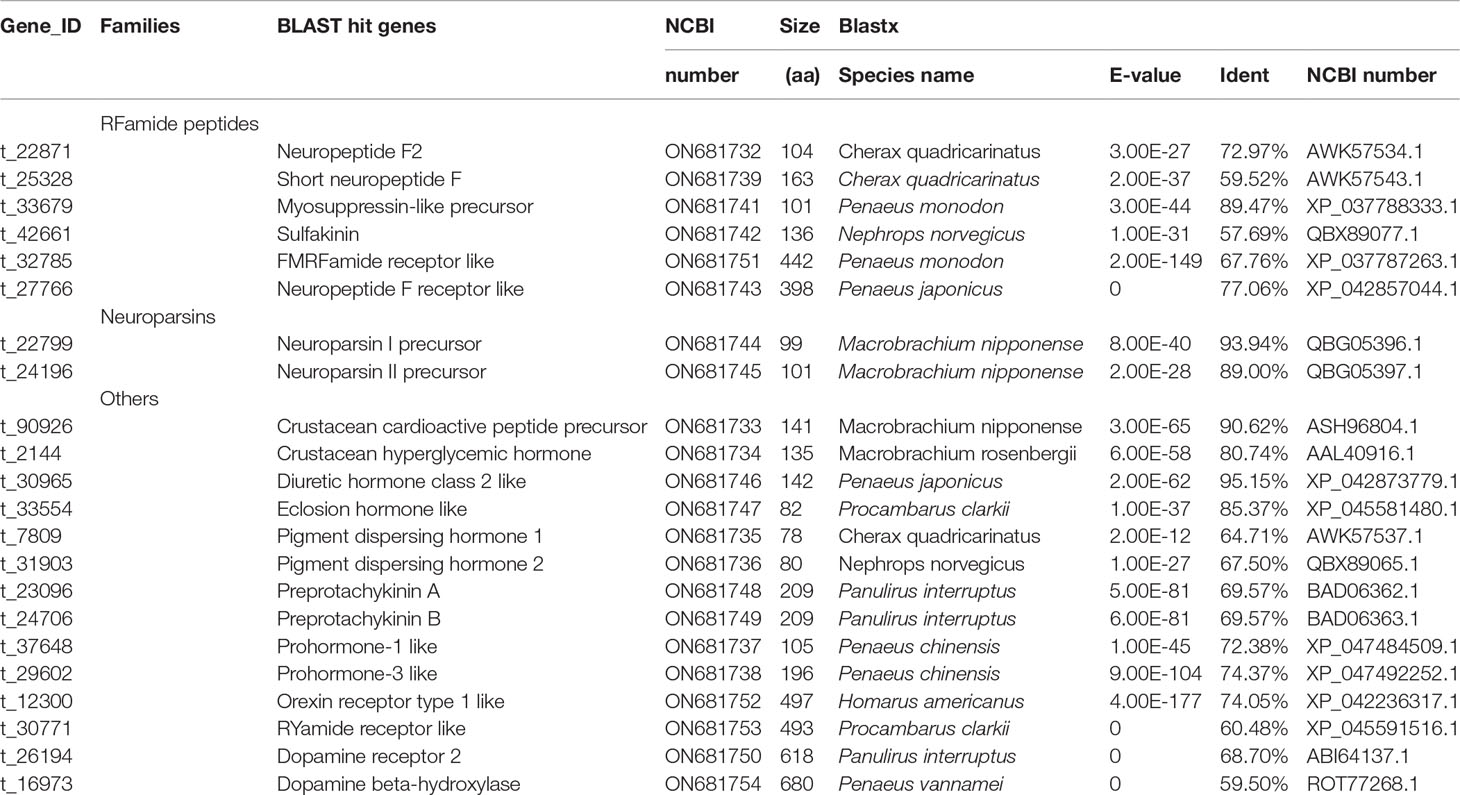
Table 2 Putative neuropeptides and related transcripts in the full-length transcriptome of E. carinicauda.
It is well-known that neuroparsin displays multiple biological activities, including antidiuretic and inhibition of vitellogenesis in insects. Also, in the crab Scylla paramamosain, the neuroparsin possibly can inhibit the production of vitellogenin in the hepatopancreas (Liu et al., 2020). In this study, the expression levels of Neuroparsin II precursor in the major growth phase was lowest in five ovarian development stages, which also indicated that the neuroparsin possibly inhibit vitellogenesis in E. carinicauda. The RYamide was thought to be another counterpart of neuropeptide Y, and may play a role in the regulation of feeding and digestion in insects (Roller et al., 2016). The RYamides exhibited changes in its relative abundance for salinity stress in Carcinus maenas (Zhang et al., 2015b). However, the function of RYamide to reproduction have not been determined in crustaceans. In this study, the expression of RYamide receptor like in stage I was higher than the growth and mature stages, which indicated that the RYamide receptor was possibly related inhibition of vitellogenesis.
The synthesis and release of neurohormones in crustaceans are believed to be regulated by biogenic amines (Richardson et al., 1991). The dopamine was shown to inhibit ovarian maturation in Procambarus clarkii (Sarojini et al., 1995). Furthermore, dopamine depressed vitellogenin synthesis through inhibition of vitellogenesis-stimulating hormone release at the thoracic ganglia in M. rosenbergii (Chen et al., 2003). In this study, the expression of dopamine receptor 2 was higher in the Stage I than the growth and mature stages, which indicated that the dopamine receptor 2 may inhibit ovarian development in E. carinicauda. Dopamine beta-hydroxylase converts dopamine to noradrenaline and plays an crucial role in catecholamine synthesis of the neuroendocrine regulatory network (Cheng et al., 2016). Cheng et al., suggested that it played a crucial role in the neuroendocrine-immune regulation of L. vannamei (Cheng et al., 2017). Due to the lack of related research, it is not clear whether dopamine beta-hydroxylase has a function in crustacean ovarian development, but our results indicated that dopamine beta-hydroxylase possibly related to vitellogenesis in E. carinicauda.The forkhead box protein L2 (foxl2) is a critical transcriptional regulator factor and has been extensively studied in crustacean and confirmed its critical roles in gonad differentiation and development (Farhadi et al., 2021; Wan et al., 2022). The knockdown of foxl2 mediated by RNAi technology repressed the expression of vtg in Eriocheir sinensis (Li et al., 2015b) and Scylla paramamosain (Wan et al., 2021). In our study, a total of a total of 895 transcripts were predicted to be transcription factors, and only 22 transcripts were the DEGs (Supplementary Table 3). Furthermore, foxl2, as transcriptional regulator factor, were highest expressed in the mature stage than that in other four stages. We speculated that foxl2 closely related to ovarian maturation in E. carinicauda.After the ovaries mature, females must molt to mate and lay eggs. Copulation occurs immediately after molting of the female, while the exoskeleton is still soft. The pre-mating molt, also known as reproductive molting, is a significant indicator of the complete maturation of the ovary (Asmat-Ullah et al., 2021). Ecdysteroids play major roles in regulating vitellogenesis, ovarian maturation and protein synthesis in decapods (Nagaraju, 2011). Crustacean ecdysteroids trigger the specific regulation of several genes in different tissues and developmental stages through interaction with the ecdysteroid receptor (EcR) (Hopkins, 2009). Low EcR expression levels have been shown to cause defective ovarian differentiation in Drosophila, egg degeneration and increased resistance to starvation (Schwedes and Carney, 2012). Higher EcR expression level was observed in the ovaries of breeding Metopograpsus messor than in the ovaries of non-breeding crabs (Shyamal et al., 2015). Additionally, EcR is necessary for ovarian development during the mid-late vitellogenesis stage in Eriocheir sinensis (Su et al., 2020). In this study, the expression of four genes (ecdysone receptor, ecdysone-induced protein 74EF, ecdysone inducible gene E75 and post-molt protein 1) which associated to molting in crustacean in the stage IV was higher than that in the other ovarian developmental stages. Techa et al., suggested that the ecdysteroid-responsive factors Callinectes sapidus, including ecdysone receptor, retinoid X receptor and molt-inhibiting hormone may be utilized at the initiation of early embryogenesis (Techa et al., 2015). Therefore, we speculate the four genes regulate the reproductive molting of E. carinicauda and plays an important role in the late vitellogenesis stage and early embryogenesis.
Conclusion
To conclude, a comprehensive full-length transcriptome ovarian tissue from five different ovarian developmental stages was obtained for the first time through SMRT sequencing in E. carinicauda. By using the full-length transcriptome as reference, we performed Illumina sequencing to analyze the gene expression difference among different ovarian developmental stages of the ovary and validated the results by quantitative real time-PCR. The PI3K-Akt signaling pathway, retinol metabolism, cell cycle and ovarian steroidogenesis pathways play important roles in ovarian development. We identified 37 DEGs that may be related to ovary maturation in significant pathways identified in the comparisons. Five vitellogenin isoforms and vitellogenin receptor suggested an important role during previtellogenesis of E. carinicauda. We can suggest that supplementation of Vitamin A to E. carinicauda feed could enhance female reproductive success. This study provides valuable information about the regulation of ovarian development in E. carinicauda and enhances the understanding of reproductive regulatory mechanisms in shrimp. The relationship between regulatory genes and ovarian development requires further study.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI accession, PRJNA746617, https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA746617.
Ethics Statement
All animal experiments were performed according to the protocols approved by The Animal Research and Ethics Committees of Yellow Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences.
Author Contributions
JW and JL conceived the study. WL collected samples and extracted RNA samples. JW and JTL participated in analyzing the sequencing data. QG verified the sequencing results. All authors contributed to the article and approved the submitted version.
Funding
This project was financially supported by National Key R & D Program of China (No. 2019YFD0900403), National Natural Science Foundation of China (No. 31902368), China Agriculture Research System of MOF and MARA (No. CARS-48), Central Public-interest Scientific Institution Basal Research Fund, CAFS (No. 2020TD46).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.906730/full#supplementary-material
References
Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., et al. (1997). Gapped BLAST and PSI-BLAST: A New Generation of Protein Database Search Programs. Nucleic Acids Res. 25 (17), 3389–3402. doi: 10.1093/nar/25.17.3389
Apweiler R., Bairoch A., Wu C. H., Barker W. C., Boeckmann B., Ferro S., et al. (2004). UniProt: The Universal Protein Knowledgebase. Nucleic Acids Res. 32, 115–119. doi: 10.1093/nar/gkh131
Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., Cherry J. M., et al. (2000). Gene Ontology: Tool for the Unification of Biology. Nat. Genet. 25 (1), 25–29. doi: 10.1038/75556
Asmat-Ullah M., Waiho K., Azra M. N., Norainy M. H., Syafaat M. N., Nahid S. A. A., et al. (2021). Induced Mating of Newly Molted Females of Orange Mud Crab, Scylla Olivacea (Herbst 1796), in Captivity. Aquaculture 533, 736159. doi: 10.1016/j.aquaculture.2020.736159
Bai H., Qiao H., Li F., Fu H., Jiang S., Zhang W., et al. (2016). Molecular and Functional Characterization of the Vitellogenin Receptor in Oriental River Prawn, Macrobrachium Nipponense. Comp. Biochem. Physiol. Part A: Mol. Integr. Physiol. 194, 45–55. doi: 10.1016/j.cbpa.2015.12.008
Bankar K. G., Todur V. N., Shukla R. N., Vasudevan M. (2015). Ameliorated De Novo Transcriptome Assembly Using Illumina Paired End Sequence Data With Trinity Assembler. Genomics Data 5, 352–359. doi: 10.1016/j.gdata.2015.07.012
Bao C., Yang Y., Huang H., Ye H. (2015). Neuropeptides in the Cerebral Ganglia of the Mud Crab, Scylla Paramamosain: Transcriptomic Analysis and Expression Profiles During Vitellogenesis. Sci. Rep. 5 (1), 17055. doi: 10.1038/srep17055
Bateman A., Birney E., Durbin R., Eddy S. R., Howe K. L., Sonnhammer E. L. L. (2000). The Pfam Protein Families Database. Nucleic Acids Res. 28 (1), 263–266. doi: 10.1093/nar/28.1.263
Boldbaatar D., Umemiya-Shirafuji R., Liao M., Tanaka T., Xuan X., Fujisaki K. (2010). Multiple Vitellogenins From the Haemaphysalis Longicornis Tick are Crucial for Ovarian Development. J. Insect Physiol. 56 (11), 1587–1598. doi: 10.1016/j.jinsphys.2010.05.019
Cao M., Zhang M., Yang N., Fu Q., Su B., Zhang X., et al. (2020). Full Length Transcriptome Profiling Reveals Novel Immune-Related Genes in Black Rockfish (Sebastes Schlegelii). Fish. Shellfish. Immunol. 106, 1078–1086. doi: 10.1016/j.fsi.2020.09.015
Chen Y. N., Fan H. F., Hsieh S. L., Kuo C. M. (2003). Physiological Involvement of DA in Ovarian Development of the Freshwater Giant Prawn, Macrobrachium Rosenbergii. Aquaculture 228 (1), 383–395. doi: 10.1016/S0044-8486(03)00324-7
Chen X., Gao Q., Cheng H., Peng F., Wang C., Xu B. (2021). Molecular Cloning and Expression Pattern of the Juvenile Hormone Epoxide Hydrolase Gene From the Giant Freshwater Prawn Macrobrachium Rosenbergii During Larval Development and the Moult Cycle. Aquacult. Res. 52 (8), 3890–3899. doi: 10.1111/are.15233
Cheng W., Ka Y.-W., Chang C.-C. (2016). Dopamine Beta-Hydroxylase Participate in the Immunoendocrine Responses of Hypothermal Stressed White Shrimp, Litopenaeus Vannamei. Fish. Shellfish. Immunol. 59, 166–178. doi: 10.1016/j.fsi.2016.10.036
Cheng W., Ka Y.-W., Chang C.-C. (2017). Involvement of Dopamine Beta-Hydroxylase in the Neuroendocrine-Immune Regulatory Network of White Shrimp, Litopenaeus Vannamei. Fish. Shellfish. Immunol. 68, 92–101. doi: 10.1016/j.fsi.2017.07.028
Cohen S., Ilouz O., Manor R., Sagi A., Khalaila I. (2021). Transcriptional Silencing of Vitellogenesis-Inhibiting and Molt-Inhibiting Hormones in the Giant Freshwater Prawn, Macrobrachium Rosenbergii, and Evaluation of the Associated Effects on Ovarian Development. Aquaculture 538, 736540. doi: 10.1016/j.aquaculture.2021.736540
Damdimopoulou P., Chiang C., Flaws J. A. (2019). Retinoic Acid Signaling in Ovarian Folliculogenesis and Steroidogenesis. Reprod. Toxicol. 87, 32–41. doi: 10.1016/j.reprotox.2019.04.007
Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., et al. (2013). STAR: Ultrafast Universal RNA-Seq Aligner. Bioinformatics 29 (1), 15–21. doi: 10.1093/bioinformatics/bts635
Duan Y., Liu P., Li J., Wang Y., Li J., Chen P. (2014). A Farnesoic Acid O-Methyltransferase (FAMeT) From Exopalaemon Carinicauda is Responsive to Vibrio Anguillarum and WSSV Challenge. Cell Stress Chaperones 19 (3), 367–377. doi: 10.1007/s12192-013-0464-5
Farhadi A., Cui W., Zheng H., Li S., Zhang Y., Ikhwanuddin M., et al. (2021). The Regulatory Mechanism of Sexual Development in Decapod Crustaceans. Front. Mar. Sci. 8. doi: 10.3389/fmars.2021.679687
Finn R. D., Bateman A., Clements J., Coggill P., Eberhardt R. Y., Eddy S. R., et al. (2013). Pfam: The Protein Families Database. Nucleic Acids Res. 42 (1), 222–230. doi: 10.1093/nar/gkt1223
Fu C., Huang X., Gong J., Chen X., Huang H., Ye H. (2016). Crustacean Hyperglycaemic Hormone Gene From the Mud Crab, Scylla Paramamosain: Cloning, Distribution and Expression Profiles During the Moulting Cycle and Ovarian Development. Aquacult. Res. 47 (7), 2183–2194. doi: 10.1111/are.12671
Guo H., Chen L.-L., Li G.-L., Deng S.-P., Zhu C.-H. (2019a). Accumulation and Depuration of Nonylphenol and Its Effect on the Expressions of Vitellogenin and Vitellogenin Receptor in Freshwater Prawn Macrobrachium Rosenbergii. Bull. Environ. Contamination Toxicol. 103 (5), 729–733. doi: 10.1007/s00128-019-02714-x
Guo S., Sun D., Tian Z., Liu W., Zhu F., Wang X.-P. (2019b). The Limited Regulatory Roles of Juvenile Hormone Degradation Pathways in Reproductive Diapause Preparation of the Cabbage Beetle, Colaphellus Bowringi. J. Insect Physiol. 119, 103967. doi: 10.1016/j.jinsphys.2019.103967
Han K., Dai Y., Zou Z., Fu M., Wang Y., Zhang Z. (2012). Molecular Characterization and Expression Profiles of Cdc2 and Cyclin B During Oogenesis and Spermatogenesis in Green Mud Crab (Scylla Paramamosain). Comp. Biochem. Physiol. Part B: Biochem. Mol. Biol. 163 (3), 292–302. doi: 10.1016/j.cbpb.2012.07.001
Harwood G., Amdam G. (2021). Vitellogenin in the Honey Bee Midgut. Apidologie 52 (4), 837–847. doi: 10.1007/s13592-021-00869-3
Holmes J. C., Scholl E. H., Dickey A. N., Hess P. R. (2021). High-Resolution Characterization of the Structural Features and Genetic Variation of Six Feline Leukocyte Antigen Class I Loci via Single Molecule, Real-Time (SMRT) Sequencing. Immunogenetics 73 (5), 381–393. doi: 10.1007/s00251-021-01221-w
Hopkins P. M., (2009)Crustacean Ecdysteroids and Their ReceptorsEcdysone: Structures and Functions,Smagghe G.Springer Verlag, Netherlands Press73–9710.1007/978-1-4020-9112-4_3
Huang H., Fu C., Chen X., Gong J., Huang X., Ye H. (2015). Molt-Inhibiting Hormone (MIH) Gene From the Green Mud Crab Scylla Paramamosain and its Expression During the Molting and Ovarian Cycle. Aquacult. Res. 46 (11), 2665–2675. doi: 10.1111/are.12421
Jeon J. M., Kim B. K., Kim Y. J., Kim H. W. (2011). Structural Similarity and Expression Differences of Two Pj-Vg Genes From the Pandalus Shrimp Pandalopsis Japonica. Fisher. Aquat. Sci. 14 (1), 22−30. doi: 10.5657/fas.2011.14.1.022
Jiang Y., Li C., Chen L., Wang F., Zhou X. (2017). Potential Role of Retinoids in Ovarian Physiology and Pathogenesis of Polycystic Ovary Syndrome. Clinica. Chimica. Acta 469, 87–93. doi: 10.1016/j.cca.2017.03.025
Kanehisa M., Goto S., Kawashima S., Okuno Y., Hattori M. (2004). The KEGG Resource for Deciphering the Genome. Nucleic Acids Res. 32, 277–280. doi: 10.1093/nar/gkh063
Kang P.-F., Mao B., Fan C., Wang Y.-F. (2019). Transcriptomic Information From the Ovaries of Red Swamp Crayfish (Procambarus Clarkii) Provides New Insights Into Development of Ovaries and Embryos. Aquaculture 505, 333–343. doi: 10.1016/j.aquaculture.2019.02.074
Kang Y., Yang D., Kong L., Hou M., Meng Y., Wei L., et al. (2017). CPC2: A Fast and Accurate Coding Potential Calculator Based on Sequence Intrinsic Features. Nucleic Acids Res. 45 (1), 12–16. doi: 10.1093/nar/gkx428
Kawai T., Yanaka N., Richards J. S., Shimada M. (2016). De Novo-Synthesized Retinoic Acid in Ovarian Antral Follicles Enhances FSH-Mediated Ovarian Follicular Cell Differentiation and Female Fertility. Endocrinology 157 (5), 2160–2172. doi: 10.1210/en.2015-2064
Keller B., Adamski J. (2007). RDH12, a Retinol Dehydrogenase Causing Leber’s Congenital Amaurosis, is Also Involved in Steroid Metabolism. J. Steroid Biochem. Mol. Biol. 104 (3), 190–194. doi: 10.1016/j.jsbmb.2007.03.015
Khayat M., Yang W. J., Aida K., Nagasawa H., Tietz A., Funkenstein B., et al. (1998). Hyperglycaemic Hormones Inhibit Protein and mRNA Synthesis Inin Vitro-Incubated Ovarian Fragments of the Marine Shrimp Penaeus Semisulcatus. Gen. Comp. Endocrinol. 110 (3), 307–318. doi: 10.1006/gcen.1998.7078
Kung S. Y., Chan S.-M., Hui J. H. L., Tsang W. S., Mak A., He J. G. (2004). Vitellogenesis in the Sand Shrimp, Metapenaeus Ensis: The Contribution From the Hepatopancreas-Specific Vitellogenin Gene (Mevg2)1. Biol. Reprod. 71 (3), 863–870. doi: 10.1095/biolreprod.103.022905
Levi L., Ziv T., Admon A., Levavi-Sivan B., Lubzens E. (2011). Insight Into Molecular Pathways of Retinal Metabolism, Associated With Vitellogenesis in Zebrafish. Am. J. Physiol.-Endocrinol. Metab. 302 (6), E626–E644. doi: 10.1152/ajpendo.00310.2011
Liang J., Zhang W., Li H., Li J.-T., Li J. (2020). Abundances of Vitellogenin and Heat Shock Protein 90 During Ovarian and Embryonic Development of Exopalaemon Carinicauda. Anim. Reprod. Sci. 223, 106633. doi: 10.1016/j.anireprosci.2020.106633
Liang B., Zhang P., Li Z., Zhao L., Lai X., Gao H., et al. (2017). Cloning, Expression and Stability Analysis of the Reference Gene Glyceraldehyde-3-Phosphate Dehydrogenase (GAPDH) in Exopalaemon Carinicauda. J. Fish. Sci. China 24 (5), 1003–1012. doi: 10.3724/SP.J.1118.2017.17018
Li X., Chen T., Jiang H., Huang J., Huang M., Xu R., et al. (2021). Effects of Methyl Farnesoate on Krüppel Homolog 1 (Kr-H1) During Vitellogenesis in the Chinese Mitten Crab (Eriocheir Sinensis). Anim. Reprod. Sci. 224, 106653. doi: 10.1016/j.anireprosci.2020.106653
Li W., Huang H., Huang J., Ye H. (2015c). Evidence for Cyclin A and Cyclin B Genes in the Ovary of the Horseshoe Crab, Tachypleus Tridentatus: Molecular Cloning and Expression Profiles. Mar. Freshw. Behav. Physiol. 48 (3), 163–176. doi: 10.1080/10236244.2015.1027540
Li J., Li J., Chen P., Liu P., He Y. (2015a). Transcriptome Analysis of Eyestalk and Hemocytes in the Ridgetail White Prawn Exopalaemon Carinicauda: Assembly, Annotation and Marker Discovery. Mol. Biol. Rep. 42 (1), 135–147. doi: 10.1007/s11033-014-3749-6
Li Z., Li J., Li J., He Y., Ge Q., Suo S. (2016). The Expression Analysis of Farnesoic Acid O-Methyl Transferase (FAMeT) Gene During the Ovarian Development Cycles of Exopalaemon Carinicauda. Prog. Fish. Sci. 37 (1), 46–51. doi: 10.11758/yykxjz.20141217001
Li G., Sun Q.-Z., Liu X.-Y., Zhang J., Dou W., Niu J.-Z., et al. (2019). Expression Dynamics of Key Ecdysteroid and Juvenile Hormone Biosynthesis Genes Imply a Coordinated Regulation Pattern in the Molting Process of a Spider Mite, Tetranychus Urticae. Exp. Appl. Acarol. 78 (3), 361–372. doi: 10.1007/s10493-019-00396-y
Liu Z., Huang Z., Zheng X., Zheng Z., Yao D., Zhang Y., et al. (2022). The Juvenile Hormone Epoxide Hydrolase Homolog in Penaeus Vannamei Plays Immune-Related Functions. Dev. Comp. Immunol. 132, 104410. doi: 10.1016/j.dci.2022.104410
Liu J., Liu A., Liu F., Huang H., Ye H. (2020). Role of Neuroparsin 1 in Vitellogenesis in the Mud Crab, Scylla Paramamosain. Gen. Comp. Endocrinol. 285, 113248. doi: 10.1016/j.ygcen.2019.113248
Li Q., Xie J., He L., Wang Y., Yang H., Duan Z., et al. (2015b). FOXL2 Down-Regulates Vitellogenin Expression at Mature Stage in Eriocheir Sinensis. Biosci. Rep. 35 (6), e00278. doi: 10.1042/bsr20150151
Li A., Zhang J., Zhou Z. (2014). PLEK: A Tool for Predicting Long Non-Coding RNAs and Messenger RNAs Based on an Improved K-Mer Scheme. BMC Bioinf. 15 (1), 311. doi: 10.1186/1471-2105-15-311
Lü F.-G., Fu K.-Y., Guo W.-C., Li G.-Q. (2015). Characterization of Two Juvenile Hormone Epoxide Hydrolases by RNA Interference in the Colorado Potato Beetle. Gene 570 (2), 264–271. doi: 10.1016/j.gene.2015.06.032
Montagné N., Desdevises Y., Soyez D., Toullec J.-Y. (2010). Molecular Evolution of the Crustacean Hyperglycemic Hormone Family in Ecdysozoans. BMC Evolution. Biol. 10 (1), 62. doi: 10.1186/1471-2148-10-62
Nagaraju G. P. C. (2011). Reproductive Regulators in Decapod Crustaceans: An Overview. J. Exp. Biol. 214 (1), 3–16. doi: 10.1242/jeb.047183
Nagaraju G. P. C., Reddy P. R., Reddy P. S. (2006). In Vitro Methyl Farnesoate Secretion by Mandibular Organs Isolated From Different Molt and Reproductive Stages of the Crab Oziotelphusa Senex Senex. Fisher. Sci. 72 (2), 410–414. doi: 10.1111/j.1444-2906.2006.01164.x
Phinyo M., Nounurai P., Hiransuchalert R., Jarayabhand P., Klinbunga S. (2014). Characterization and Expression Analysis of Cyclin-Dependent Kinase 7 Gene and Protein in Ovaries of the Giant Tiger Shrimp Penaeus Monodon. Aquaculture 432, 286–294. doi: 10.1016/j.aquaculture.2014.05.022
Phinyo M., Visudtiphole V., Roytrakul S., Phaonakrop N., Jarayabhand P., Klinbunga S. (2013). Characterization and Expression of Cell Division Cycle 2 (Cdc2) mRNA and Protein During Ovarian Development of the Giant Tiger Shrimp Penaeus Monodon. Gen. Comp. Endocrinol. 193, 103–111. doi: 10.1016/j.ygcen.2013.07.012
Qian Z., Liu X. (2019). Elucidation of the Role of Farnesoic Acid O-Methyltransferase (FAMeT) in the Giant Freshwater Prawn, Macrobrachium Rosenbergii: Possible Functional Correlation With Ecdysteroid Signaling. Comp. Biochem. Physiol. Part A: Mol. Integr. Physiol. 232, 1–12. doi: 10.1016/j.cbpa.2019.03.003
Qiu G.-F., Ramachandra R. K., Rexroad C. E., Yao J. (2008). Molecular Characterization and Expression Profiles of Cyclin B1, B2 and Cdc2 Kinase During Oogenesis and Spermatogenesis in Rainbow Trout (Oncorhynchus Mykiss). Anim. Reprod. Sci. 105 (3), 209–225. doi: 10.1016/j.anireprosci.2007.03.005
Ren Y., Li J., Guo L., Liu J. N., Wan H., Meng Q., et al. (2020). Full-Length Transcriptome and Long non-Coding RNA Profiling of Whiteleg Shrimp Penaeus Vannamei Hemocytes in Response to Spiroplasma Eriocheiris Infection. Fish. Shellfish. Immunol. 106, 876–886. doi: 10.1016/j.fsi.2020.06.057
Richardson H. G., Deecaraman M., Fingerman M. (1991). The Effect of Biogenic Amines on Ovarian Development in the Fiddler Crab, Uca Pugilator. Comp. Biochem. Physiol. Part C: Comp. Pharmacol. 99 (1), 53–56. doi: 10.1016/0742-8413(91)90074-4
Roller L., Čižmár D., Bednár B., Žitňan D. (2016). Expression of RYamide in the Nervous and Endocrine System of Bombyx Mori. Peptides 80, 72–79. doi: 10.1016/j.peptides.2016.02.003
Rotllant G., Palero F., Mather P. B., Bracken-Grissom H. D., Santos M. B. (2018). Preface: Recent Advances in Crustacean Genomics. Hydrobiologia 825 (1), 1–4. doi: 10.1007/s10750-018-3773-y
Santos C. A., Andrade S. C. S., Teixeira A. K., Farias F., Guerrelhas A. C., Rocha J. L., et al. (2021). Transcriptome Differential Expression Analysis Reveals the Activated Genes in Litopenaeus Vannamei Shrimp Families of Superior Growth Performance. Aquaculture 531, 735871. doi: 10.1016/j.aquaculture.2020.735871
Santos C. G., Humann F. C., Hartfelder K. (2019). Juvenile Hormone Signaling in Insect Oogenesis. Curr. Opin. Insect Sci. 31, 43–48. doi: 10.1016/j.cois.2018.07.010
Sarojini R., Nagabhushanam R., Fingerman M. (1995). In Vivo Effects of Dopamine and Dopaminergic Antagonists on Testicular Maturation in the Red Swamp Crayfish, Procambarus Clarkii. Biol. Bull. 189 (3), 340–346. doi: 10.2307/1542151
Schwedes C. C., Carney G. E. (2012). Ecdysone Signaling in Adult Drosophila Melanogaster. J. Insect Physiol. 58 (3), 293–302. doi: 10.1016/j.jinsphys.2012.01.013
Shi K., Li J., Lv J., Liu P., Li J., Li S. (2020). Full-Length Transcriptome Sequences of Ridgetail White Prawn Exopalaemon Carinicauda Provide Insight Into Gene Expression Dynamics During Thermal Stress. Sci. Total Environ. 747, 141238. doi: 10.1016/j.scitotenv.2020.141238
Shimizu K., Adachi J., Muraoka Y. (2006). ANGLE: A Sequencing Errors Resistant Program for Predicting Protein Coding Regions in Unfinished cDNA. J. Bioinf. Comput. Biol. 4 (3), 649–664. doi: 10.1142/s0219720006002260
Shyamal S., Anilkumar G., Bhaskaran R., Doss G. P., Durica D. S. (2015). Significant Fluctuations in Ecdysteroid Receptor Gene (EcR) Expression in Relation to Seasons of Molt and Reproduction in the Grapsid Crab, Metopograpsus Messor (Brachyura: Decapoda). Gen. Comp. Endocrinol. 211, 39–51. doi: 10.1016/j.ygcen.2014.11.006
Su Y., Guo Q., Gong J., Cheng Y., Wu X. (2020). Functional Expression Patterns of Four Ecdysteroid Receptor Isoforms Indicate Their Different Functions During Vitellogenesis of Chinese Mitten Crab, Eriocheir Sinensis. Comp. Biochem. Physiol. Part A: Mol. Integr. Physiol. 248, 110754. doi: 10.1016/j.cbpa.2020.110754
Sun L., Luo H., Bu D., Zhao G., Yu K., Zhang C., et al. (2013). Utilizing Sequence Intrinsic Composition to Classify Protein-Coding and Long Non-Coding Transcripts. Nucleic Acids Res. 41 (17), 166. doi: 10.1093/nar/gkt646
Suppa A., Gorbi G., Marková S., Buschini A., Rossi V. (2021). Transgenerational Effects of Methyl Farnesoate on Daphnia Pulex Clones: Male and Ephippia Production and Expression of Genes Involved in Sex Determination. Freshw. Biol. 66 (2), 374–390. doi: 10.1111/fwb.13644
Tahaei L. S., Eimani H., Yazdi P. E., Ebrahimi B., Fathi R. (2011). Effects of Retinoic Acid on Maturation of Immature Mouse Oocytes in the Presence and Absence of a Granulosa Cell Co-Culture System. J. Assisted Reprod. Genet. 28 (6), 553–558. doi: 10.1007/s10815-011-9579-8
Tatusov R. L., Galperin M. Y., Natale D. A., Koonin E. V. (2000). The COG Database: A Tool for Genome-Scale Analysis of Protein Functions and Evolution. Nucleic Acids Res. 28 (1), 33–36. doi: 10.1093/nar/28.1.33
Techa S., Alvarez J. V., Sook Chung J. (2015). Changes in Ecdysteroid Levels and Expression Patterns of Ecdysteroid-Responsive Factors and Neuropeptide Hormones During the Embryogenesis of the Blue Crab, Callinectes Sapidus. Gen. Comp. Endocrinol. 214, 157–166. doi: 10.1016/j.ygcen.2014.07.017
Tomy S., Saikrithi P., James N., Balasubramanian C. P., Panigrahi A., Otta S. K., et al. (2016). Serotonin Induced Changes in the Expression of Ovarian Gene Network in the Indian White Shrimp, Penaeus Indicus. Aquaculture 452, 239–246. doi: 10.1016/j.aquaculture.2015.11.003
Toyota K., Kato Y., Miyakawa H., Yatsu R., Mizutani T., Ogino Y., et al. (2014). Molecular Impact of Juvenile Hormone Agonists on Neonatal Daphnia Magna. J. Appl. Toxicol. 34 (5), 537–544. doi: 10.1002/jat.2922
Visudtiphole V., Klinbunga S., Kirtikara K. (2009). Molecular Characterization and Expression Profiles of Cyclin A and Cyclin B During Ovarian Development of the Giant Tiger Shrimp Penaeus Monodon. Comp. Biochem. Physiol. Part A: Mol. Integr. Physiol. 152 (4), 535–543. doi: 10.1016/j.cbpa.2008.12.011
Wang X. E. (1987). A Preliminary Observation on the Reproductive Biology of Exopalaemon Carinicauda. J. Zool. 22 (1), 6–10. doi: 10.13859/j.cjz.1987.01.003
Wang J., Ge Q., Li J., Li J. (2020). Effects of Inbreeding on Growth and Survival Rates, and Immune Responses of Ridgetail White Prawn Exopalaemon Carinicauda Against the Infection of Vibrio Parahaemolyticus. Aquaculture 519, 734755. doi: 10.1016/j.aquaculture.2019.734755
Wan H., Zhong J., Zhang Z., Xie Y., Wang Y. (2021). Characterization of the Foxl2 Gene Involved in the Vtg Expression in Mud Crab (Scylla Paramamosain). Gene 798, 145807. doi: 10.1016/j.gene.2021.145807
Wan H., Zhong J., Zhang Z., Zou P., Wang Y. (2022). Comparative Transcriptome Reveals the Potential Modulation Mechanisms of Spfoxl-2 Affecting Ovarian Development of Scylla Paramamosain. Mar. Biotechnol. 24 (1), 125–135. doi: 10.1007/s10126-022-10091-6
Webster S. G., Keller R., Dircksen H. (2012). The CHH-Superfamily of Multifunctional Peptide Hormones Controlling Crustacean Metabolism, Osmoregulation, Moulting, and Reproduction. Gen. Comp. Endocrinol. 175 (2), 217–233. doi: 10.1016/j.ygcen.2011.11.035
Weirather J., de Cesare M., Wang Y., Piazza P., Sebastiano V., Wang X., et al. (2017). Comprehensive Comparison of Pacific Biosciences and Oxford Nanopore Technologies and Their Applications to Transcriptome Analysis. F1000Research 6 (100), 1–32. doi: 10.12688/f1000research.10571.1
Williams V. N., Reading B. J., Hiramatsu N., Amano H., Glassbrook N., Hara A., et al. (2014). Multiple Vitellogenins and Product Yolk Proteins in Striped Bass, Morone Saxatilis: Molecular Characterization and Processing During Oocyte Growth and Maturation. Fish. Physiol. Biochem. 40 (2), 395–415. doi: 10.1007/s10695-013-9852-0
Xu W., Xie J., Shi H., Li C. (2010). Hematodinium Infections in Cultured Ridgetail White Prawns, Exopalaemon Carinicauda, in Eastern China. Aquaculture 300 (1), 25–31. doi: 10.1016/j.aquaculture.2009.12.024
Yuan J., Gao Y., Zhang X., Wei J., Liu C., Li F., et al. (2017). Genome Sequences of Marine Shrimp Exopalaemon Carinicauda Holthuis Provide Insights Into Genome Size Evolution of Caridea. Mar. Drugs 15 (7), 213. doi: 10.3390/md15070213
Zhang Y., Buchberger A., Muthuvel G., Li L. (2015b). Expression and Distribution of Neuropeptides in the Nervous System of the Crab Carcinus Maenas and Their Roles in Environmental Stress. Proteomics 15 (23-24), 3969–3979. doi: 10.1002/pmic.201500256
Zhang J., Gao S., Shi Y., Yan Y., Liu Q. (2020). Full-Length Transcriptome of Anadromous Coilia Nasus Using Single Molecule Real-Time (SMRT) Sequencing. Aquacult. Fisher. 7(6), 420−426. doi: 10.1016/j.aaf.2020.08.006
Zhang C., Li Z., Li F., Xiang J. (2015a). Effects of Starvation on Survival, Growth and Development of Exopalaemon Carinicauda Larvae. Aquacult. Res. 46 (9), 2289–2299. doi: 10.1111/are.12386
Zheng J., Wang P., Mao Y., Su Y., Wang J. (2020). Full-Length Transcriptome Analysis Provides New Insights Into the Innate Immune System of Marsupenaeus Japonicus. Fish. Shellfish. Immunol. 106, 283–295. doi: 10.1016/j.fsi.2020.07.018
Keywords: iso-seq, full-length transcripts, shrimp, vitellogenin, ovarian maturation, vitellogenesis
Citation: Wang J, Li J, Ge Q, Li W and Li J (2022) Full-Length Transcriptome Sequencing and Comparative Transcriptomic Analysis Provide Insights Into the Ovarian Maturation of Exopalaemon carinicauda. Front. Mar. Sci. 9:906730. doi: 10.3389/fmars.2022.906730
Received: 29 March 2022; Accepted: 16 June 2022;
Published: 20 July 2022.
Edited by:
Heinrich Dircksen, Stockholm University, SwedenReviewed by:
Khor Waiho, University of Malaysia Terengganu, MalaysiaSirinart Techa, National Science and Technology Development Agency (NSTDA), Thailand
Copyright © 2022 Wang, Li, Ge, Li and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jian Li, bigbird@ysfri.ac.cn
 Jiajia Wang
Jiajia Wang Jitao Li
Jitao Li Qianqian Ge2
Qianqian Ge2  Wenyang Li
Wenyang Li Jian Li
Jian Li