Targeted surveillance detected novel beaked whale circovirus in ten new host cetacean species across the Pacific basin
- 1Human Nutrition, Food, and Animal Sciences, College of Tropical Agriculture and Human Resources, University of Hawaíi at Mānoa, Honolulu, HI, United States
- 2Health and Stranding Lab, Hawaíi Institute of Marine Biology, University of Hawaíi at Mānoa, Kānéohe, HI, United States
- 3National Marine Fisheries Service Pacific Islands Regional Office, National Oceanic and Atmospheric Administration, Pago Pago, American Samoa
The first cetacean circovirus, beaked whale circovirus (BWCV), was recently reported in a Longman’s beaked whale (Indopacetus pacificus) stranded in Hawai‘i and represents an emergent disease with unknown population impacts. In other species, circovirus infection may cause mortality or opportunistic co-infection by other pathogens. We report on a targeted surveillance of stranded cetaceans in the Pacific basin, including archived beaked whale species, strandings where pathological findings suggested disease presence, mass stranded animals, and additional individuals to represent a broad range of Hawaiian cetacean species. Archived tissues primarily from the brain, kidney, liver, lung, spleen, and lymph nodes of individuals stranded between 2000 and 2020 (n=30) were tested by PCR for the presence of BWCV. Suspect positive tissue amplicons were confirmed as BWCV through sequencing. Of the screened individuals, 15 animals tested positive in one or more tissues, with a single striped dolphin (Stenella coeruleoalba) testing positive in all six tissues. The highest rate of detection among positive cases was found in the brain (69%), followed by lymph (67%) and lung tissues (64%). Additionally, co-infections of cetacean morbillivirus (n=3), Brucella ceti (n=1), and Toxoplasma gondii (n=1) were found among the positive cases. These results expand the potential host range for BWCV into ten additional odontocete species. New host species include a dwarf sperm whale (Kogia sima) that stranded on O‘ahu in 2000, predating the initial case of BWCV. The results broaden the known geographic range of BWCV to Saipan in the Western Pacific, and American Samoa in the South Pacific, where stranded Cuvier’s beaked whales (Ziphius cavirostris) tested positive. Although the clinical significance is currently unknown, this study demonstrates that BWCV has a high prevalence within targeted cetacean screening efforts. Infectious diseases pose a major threat to cetaceans and BWCV may represent an important emerging disease within populations spanning the central, Western, and South Pacific.
Introduction
Circoviruses are small, non-enveloped viruses with a circular genomic structure of single-stranded DNA (Breitbart et al., 2017). Circoviruses have long been recognized for their impact on mortality in the pet trade and agricultural industries, causing psittacine beak and feather disease in parrots, as well as porcine respiratory disease complex and post-weaning multi-systemic wasting syndrome in pigs (Crowther et al., 2003; Rose et al., 2012; Fogell et al., 2018). New viral species are recognized when nucleotide similarity to existing species falls below 80% (Breitbart et al., 2017) and in recent years many novel species and strains of circoviruses have been discovered, although their pathological significance can be difficult to discern. Many of these new circoviruses have been found in mammals, with recently discovered novel species being present in pandas, elk, wolverines, bats, and the first marine mammal (Fisher et al., 2020; Landrau-Giovannetti et al., 2020; Lecis et al., 2020; Bandoo et al., 2021; Dai et al., 2021). Additionally, novel avian strains have been discovered within species of duck and penguin, the latter of which was connected to a feather-loss disorder, demonstrating that pathogenicity is present in these emergent viruses (Morandini et al., 2019; Levy et al., 2020; Wang et al., 2021).
Circovirus infections do not always result in a pathogenic response, though many of the strains across various species can cause negative health impacts (Gavier-Widén et al., 2012). Disease due to pathogenic circovirus strains includes necrosis and inflammation in the brain, lung, liver, heart, spleen, intestine, and lymph tissues (Woods and Latimer, 2000; Rampin et al., 2006; Seo et al., 2014; Bexton et al., 2015; Yang et al., 2015). These viruses are frequently associated with respiratory illnesses (Lin et al., 2011; Seo et al., 2014; Chen et al., 2021) and several wasting diseases (Gavier-Widén et al., 2012; Seo et al., 2014; Yang et al., 2015). Circoviruses have been directly linked to reproductive failure and mortality in fish, birds, and swine, often in newly hatched or young offspring, although these outcomes can be found in infected juveniles and adults as well (Woods et al., 1993; Lőrincz et al., 2011; Grasland et al., 2013; Yang et al., 2015). There is also the potential for indirect negative impacts to infected hosts, with lymphoid depletion observed in cases of chronic circovirus infections that may indicate immune suppression (Yang et al., 2015; Mao et al., 2017; Palinski et al., 2017). Co-infections by other viruses and bacteria have been documented in both mammals and birds found to be infected with circoviruses (Zaccaria et al., 2016; Dal Santo et al., 2020; Lagan Tregaskis et al., 2020; Zhen et al., 2021).
Circovirus transmission can occur through multiple vectors and between different species. Horizontal transmission has been documented through direct contact with infected secretions such as feces, urine, and airborne respiratory aerosols (Patterson, 2010; Verreault et al., 2010; Rose et al., 2012). Vertical transmission from parent to offspring in mammals has been documented in transplacental investigations, dietary supplementation of pregnant test subjects, as well as embryonic survival studies (Mateusen et al., 2007; Ren et al., 2013; Dong et al., 2016). It has also been proposed as a likely route of transmission for avian circovirus strains (Li et al., 2014; Wang et al., 2016). Circoviruses have been shown to cross from one species to another (Firth et al., 2009; Zhai et al., 2017; Li et al., 2019). Porcine circovirus has been demonstrated to cross between species at the family level with a strain of porcine circovirus 2 being able to pass back and forth between swine and buffalo (Zhai et al., 2017). At the Order level, an avian strain of circovirus was recently found in the rainbow bee-eater (Merops ornatus), a Coraciiform species, when previously it had been thought to be restricted to Psittaciform and Columbiform birds, and it is presumed that circovirus transmitted from one Order of birds to another (Peters et al., 2014; Sarker et al., 2015). It is currently unknown if and how marine mammal circoviruses may transmit in terms of vectors and/or among and between different species.
The full genome of the first known marine mammal circovirus was characterized in a Longman’s beaked whale (Indopacetus pacificus) that stranded in Maui, Hawai‘i in 2010, which is now recognized as beaked whale circovirus (BWCV) (GenBank Acc. No. MN103538.1) (Landrau-Giovannetti et al., 2020). Longman’s beaked whales have been described as the second most poorly known of all whale species, with fewer than 20 strandings having occurred world-wide (Kobayashi et al., 2021). The potential threats that this species may face are poorly understood, including the population level impact of infectious disease. The stranded Maui, Hawai‘i individual was co-infected with both morbillivirus and herpesvirus at time of death, confounding the ability to determine if the pathology observed can be attributed to the presence of circovirus that was later discovered in archived tissues from this same individual (West et al., 2013; Landrau-Giovannetti et al., 2020). Necropsy and histopathology findings from the Longman’s beaked whale infected with BWCV included pathology that is common to pathogenic circoviruses in other species, including lymphoid depletion and pulmonary edema, but these may also be associated with the co-infections described in this individual and/or a result of the immunosuppressive effect of circovirus infections documented in other species (West et al., 2013; Seo et al., 2014; Pendl and Tizard, 2016).
Sequencing of the full BWCV genome allowed for the development of DNA primers and subsequent polymerase chain reaction (PCR) amplification of the viral DNA to test a suite of tissues for BWCV presence in the infected Longman’s beaked whale which were all positive by PCR (Landrau-Giovannetti et al., 2020). To our knowledge, the Longman’s beaked whale represents the only marine mammal tested by PCR to date for the presence of a circovirus. This investigation aimed to determine if BWCV was limited to a single Longman’s beaked whale stranding, or if the virus may be present among other cetacean species. We conducted PCR screening for this virus in archived tissues from previously stranded individuals that represent a wide range of cetacean species (13 total species). Other objectives involved investigating the potential spatial and temporal scope of BWCV presence across the Pacific basin. In order to address this, we included previously stranded cetaceans from the main Hawaiian Islands, American Samoa, Guam, and the Commonwealth of the Northern Mariana Islands in our screening, as well as archived samples dating back to 2000 to better understand the emergence of beaked whale circovirus in the Pacific.
Materials and methods
Animal selection
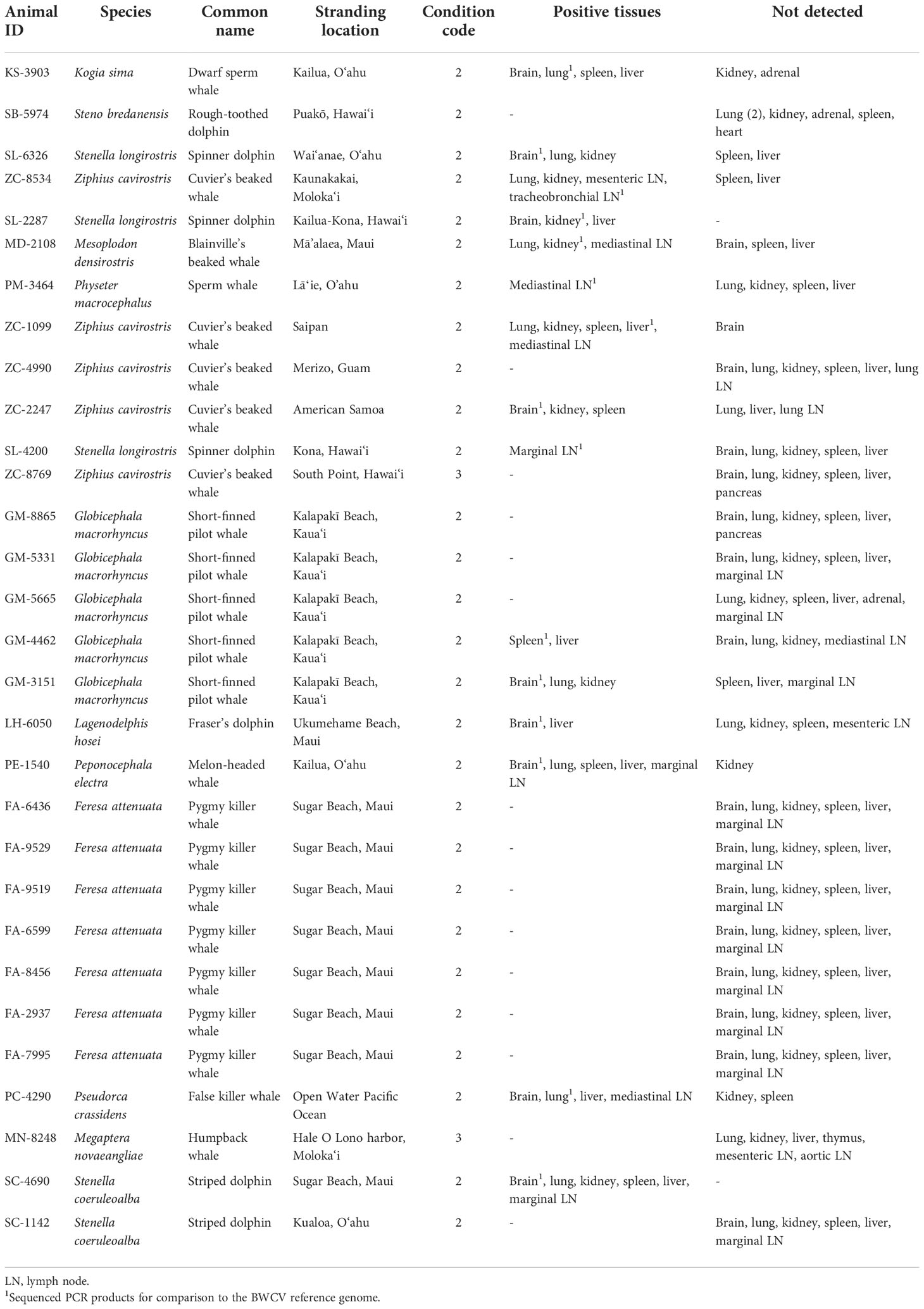
To investigate the presence of circovirus among cetacean species throughout the Pacific Ocean basin, 30 previously stranded individuals were targeted for screening of BWCV presence (Table 1). Selected specimens for screening included the following: five Cuvier’s beaked whales (Ziphius cavirostris), a Blainville’s beaked whale (Mesoplodon densirostris), five short-finned pilot whales (Globicephala macrorhynchus) that mass stranded in 2017, and seven pygmy killer whales (Feresa attenuata) that stranded during a prolonged mass stranding event in 2019, as well as a humpback whale (Megaptera novaeangliae), a sperm whale (Physeter macrocephalus), a dwarf sperm whale (Kogia sima), a false killer whale (Pseudorca crassidens), a melon-headed whale (Peponocephala electra), and several species of dolphin. Tissues tested were obtained from the University of Hawai‘i Health and Stranding Laboratory’s tissue archive of cetaceans that have stranded in Hawai‘i and other regions of the Pacific since 1998, from animals with carcass condition codes of 2 and 3 (Geraci and Lounsbury, 2005). Field and laboratory necropsies of stranded animals included detailed gross examination of organs and tissues and collection of tissue samples. Initial cases selected for this study were archived beaked whale species, being that the virus was initially discovered in a Longman’s beaked whale. Stranded animals of other species with pathological findings associated with infectious disease, as well as those with confirmed presence of other diseases, were also selected for testing. Finally, representatives from mass stranding events that have occurred in Hawai‘i were investigated, as disease has been linked to mass strandings in other regions (Garrigue et al., 2016; Mazzariol et al., 2017; Vargas-Castro et al., 2020). Additional individuals were selected for screening to represent a broad assessment of the various cetacean species that regularly inhabit the region (Baird, 2016).
BWCV detection by PCR
Tissues selected for testing from each individual targeted samples of brain, lung, kidney, spleen, liver, and lymph tissue (Table 1). When these specific tissues were unavailable, other available tissue types were substituted in order to test at least 6 different tissues from each individual when possible. Two cases had less than a full suite of tissues available, SL-2287 and PM-3464, which had three and five tissues available respectively for analysis. Brain and lung tissues from the original Longman’s beaked whale BWCV case were used as a positive control throughout the screening process (Table 2). DNA was extracted from tissue samples using DNeasy Blood and Tissue kits (Qiagen, Germantown, Maryland) and quantified using a Qubit 4 fluorometer (Thermo Fisher Scientific, Waltham, Massachusetts). Utilizing traditional polymerase chain reaction (PCR), each tissue extract was run in triplicate to test for the presence of BWCV. The PCR protocol used primers and thermocycler settings adapted from Landrau-Giovannetti et al., 2020, that were based on gene sequences obtained from both capsid and replication-associated proteins. The BWCV forward primer 5’ CTTCAGATTCCCCGTCAAGA 3’ and BWCV reverse primer 5’ GTCTCCCCACAATGGTTCAC 3’ were used with an initial denaturation at 94°C for five minutes, 40 cycles of denaturing at 94°C for 30 seconds, annealing at 56°C for 30 seconds, and extension at 72°C for 30 seconds, with a final extension step at 72°C for five minutes. A no template control was utilized in all runs to check for primer dimerization and ensure the quality of results.
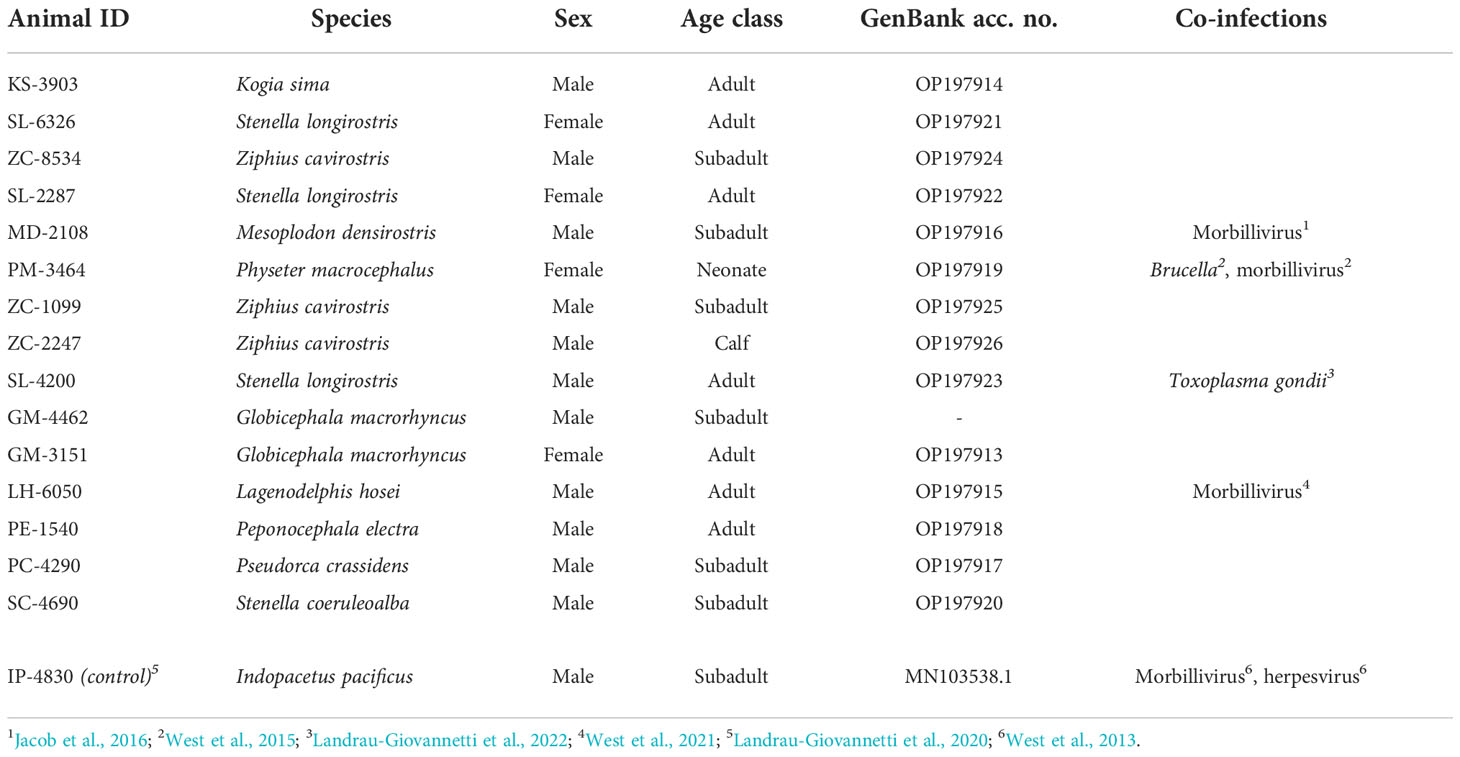
Table 2 Animal cases that tested positive for beaked whale circovirus, their demographics, and detected presence of other infections.
Amplified products were visualized through gel electrophoresis, with bands of 400bp in size indicating the likely presence of BWCV. Bands from the positive control and a minimum of one band from each positive case were excised from the gels and the PCR product was cleaned using Qiaquick PCR and Gel Cleanup Kits (Qiagen, Germantown, Maryland). Amplified DNA was sequenced at the Advanced Studies in Genomics, Proteomics, and Bioinformatics lab at the University of Hawai‘i.
Phylogenetic analyses
Phylogenetic analyses were performed on the resultant sequences for comparison to the BWCV reference genome (GenBank Acc. No. MN103538.1), as well as to other recognized and recently discovered circovirus genomes, including those closest in genetic similarity to BWCV (Landrau-Giovannetti et al., 2020). Geneious Prime v.2022.2.1 software and the NCBI BLAST database were then used to annotate, align, and perform analyses on the study sequences. The MAFFT Alignment v.1.5.0 plugin was used under default parameters to align the sequences and the MrBayes v.3.2.6 plugin was used to produce phylogenetic trees based on Bayesian parameters with a General Time-Reversible substitution model using a 1,100,000 chain length and 100,000 burn-in length (Huelsenbeck and Ronquist, 2001; Ronquist and Huelsenbeck, 2003; Katoh and Standley, 2013).
Results
Of the thirty animals that were tested in this study, fifteen animals were positive for BWCV in one or more tissues (Table 1). Among these cases, brain tissue was the most consistently positive tissue type (69%), followed by lymph tissue (67%) and lung tissue (64%). BWCV was detected in lower frequencies in kidney, liver, and spleen tissue of positive cases.
Positive cases were detected among ten different species that have stranded or were by-caught in the Pacific Islands region since 2000, bringing the total to eleven known cetacean species with BWCV when including the initial Longman’s beaked whale (Figure 1). New species where BWCV was detected as part of this screening effort included Cuvier’s beaked whales (n=3), Blainville’s beaked whales (n=1), sperm whales (n=1), dwarf sperm whales (n=1), spinner dolphins (Stenella longirostris) (n=3), striped dolphins (Stenella coeruleoalba) (n=1), Fraser’s dolphins (Lagenodelphis hosei) (n=1), melon-headed whales (n=1), false killer whales (n=1), and short-finned pilot whales (n=2, both from a single mass stranding event) (Table 2). Positive cases were found in seven adults (out of 15 tested), six subadults (out of 11 tested), one calf (out of three tested), and one neonate (only individual representing this age class). Both males and females were positive for the virus. The majority of these positive cases occurred geographically within the main Hawaiian Islands (n=12), on the islands of Hawai‘i, Kaua‘i, Maui, Moloka‘i, and O‘ahu. Two of the BWCV positive Cuvier’s beaked whales stranded in the Western and South Pacific. Stranding locations of positive Cuvier’s beaked whales included American Samoa and Saipan in the Commonwealth of the Northern Mariana Islands. A single positive individual was by-caught approximately 700 km south of the main Hawaiian Islands.
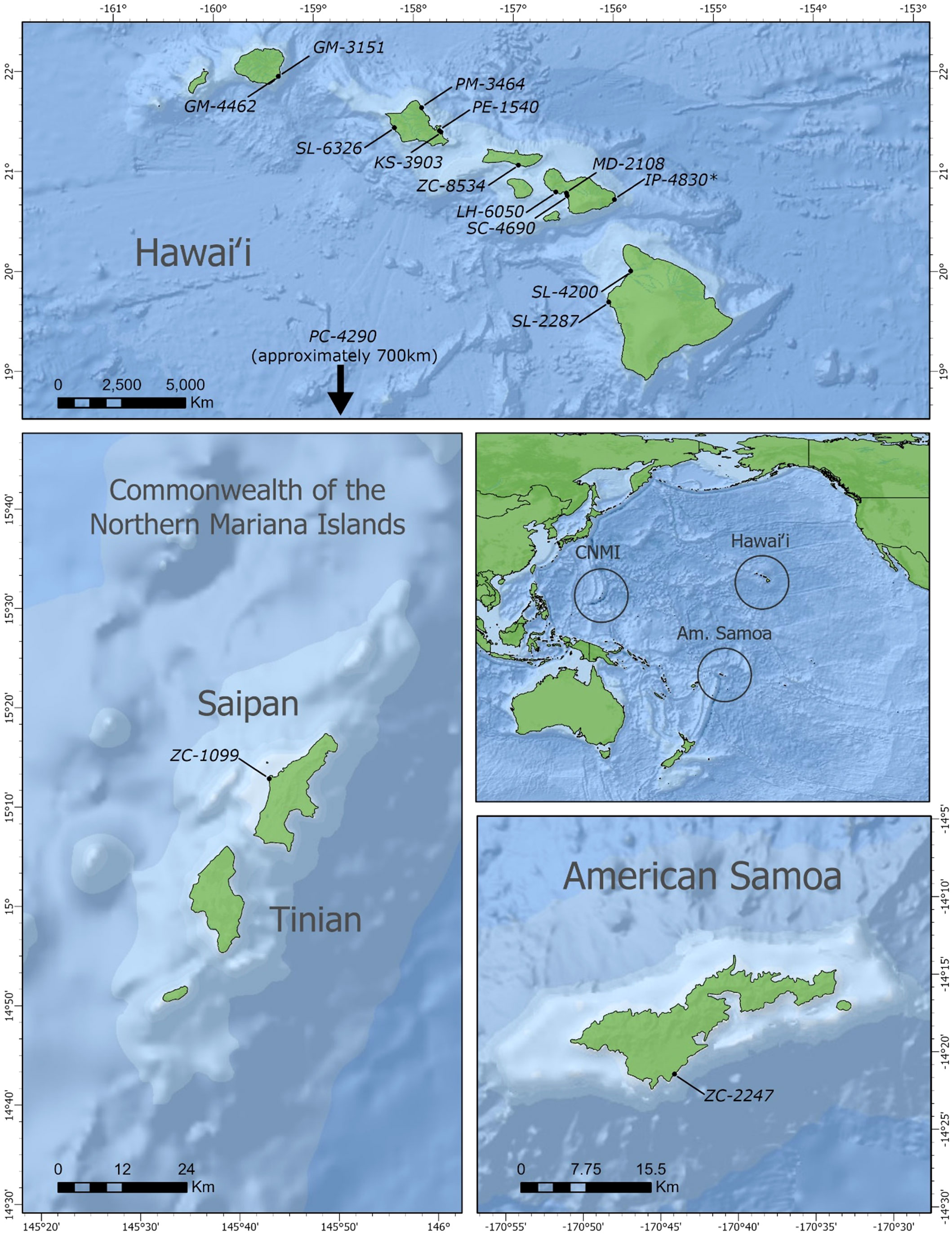
Figure 1 Stranding locations of beaked whale circovirus positive animals. Hawaiian Islands (n=13, *initial Longman's beaked whale); Saipan (n=1); American Samoa (n=1). Location of open ocean by-caught false killer whale not pictured. Base map source: ESRI, GEBCO, NOAA, National Geographic, DeLorme, NaturalVue.
The average percent similarity of all study sequences to the BWCV reference genome was 96.7%. The average query cover was 93.3%, with 13 of the individuals greater than 97.8%. Two outliers were apparent, SL-4200 (spinner dolphin) and GM-4462 (short-finned pilot whale), having a query cover of 71.5% and 33.0% respectively. Because of the low query cover and short length of the final sequence of GM-4462, it was not included in the phylogenetic analyses performed, although it was still deemed positive based on its comparison to GenBank reference genomes. Phylogenetic analyses of these sequences produced results that support high similarity between infected individuals and significant difference from other species of circovirus (Figure 2). The posterior probability (PP) of the analyzed study sequences and the BWCV reference genome was 1, indicating high confidence in the consensus tree. Within this group there was an additional division including sequences from a striped dolphin (SC-4690), spinner dolphin (SL-2287), and a Cuvier’s beaked whale (ZC-1099) (PP=0.65). The reference genomes with the next closest similarity to the BWCV grouping included bat circovirus (GenBank Acc. No. KX756986), canine circovirus (GenBank Acc. No. JQ821392) and bamboo rat circovirus (GenBank Acc. No. MF497827).
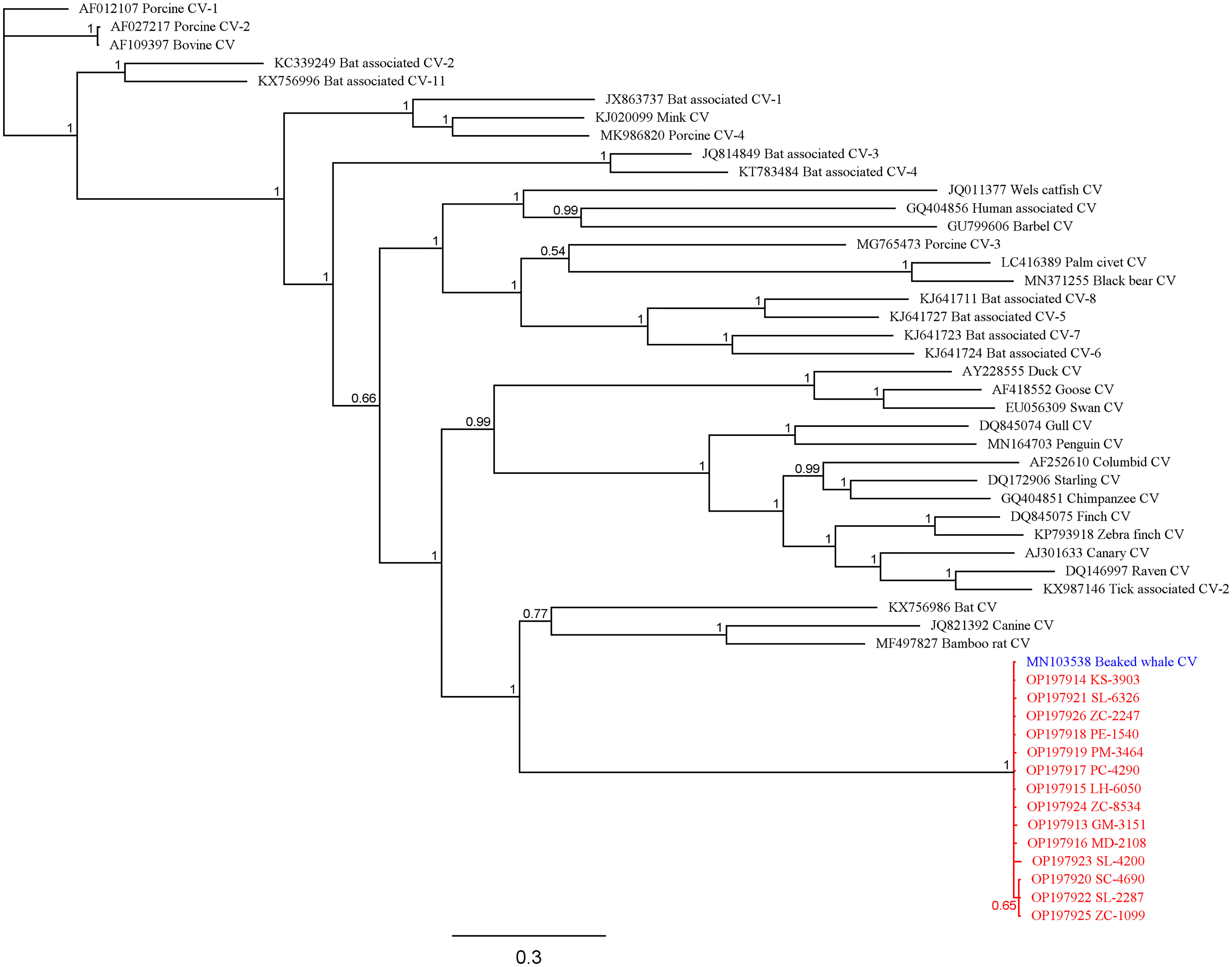
Figure 2 Bayesian phylogenetic tree showing relationships between study sequences (red), BWCV reference genome (blue), and other circoviruses (black). Posterior probabilities are denoted at branching nodes.
Co-infections were present among some of the BWCV positive individuals (Table 2). The initial Longman’s beaked whale, a sperm whale, a Blainville’s beaked whale, and a Fraser’s dolphin were co-infected with cetacean morbillivirus (West et al., 2013; West et al., 2015; Jacob et al., 2016; West et al., 2021). In addition to morbillivirus, an alpha herpesvirus was present in the Longman’s beaked whale and Brucella ceti in the neonate sperm whale (West et al., 2013; West et al., 2015). Disseminated toxoplasmosis was diagnosed in an adult male spinner dolphin (Landrau-Giovannetti et al., 2022). A presumed healthy pelagic false killer whale in robust body condition that was by-caught outside of the Exclusive Economic Zone off Hawai‘i tested positive for BWCV, but did not show signs of systemic disease.
This study detected positive cases across the entire time span of the archived samples tested, from 2000 to 2020. The majority of the positive cases were found in animals that stranded in the latter half of this time frame (10 of the 15 positive cases occurring after 2010), which is consistent with increased carcass recovery and necropsy efforts during this time.
Discussion
Findings from this study have significantly increased our understanding of BWCV in marine mammals, including the expansion of known host species, the geographic range of BWCV presence, and further insight into when this infectious agent may have emerged in the central Pacific. Our results indicate 50% prevalence of BWCV among the 30 individual cetaceans tested, and demonstrates that despite being named beaked whale circovirus, this recently identified circovirus species can infect a number of other host cetacean species. The current study confirms the discovery of BWCV in ten new cetacean species in addition to the initial Longman’s beaked whale case and includes individuals of diverse age and sex classes. BWCV was found in two additional species from family Ziphiidae (beaked whales), as well as in animals from the families Delphinidae (oceanic dolphins), Kogiidae (dwarf and pygmy sperm whales) and Physeteridae (sperm whales). This suggests that this virus is wide-spread across odontocete species in the central Pacific, with infections likely in a greater number of cetacean species than we report, due to the fact that more than 20 cetacean species have been documented in the waters around Hawai‘i (Baird, 2016). Not all species were investigated in this study, and only a very limited number of individuals were screened for the species that were examined. Of the 13 cetacean species where a small number of representative individuals were screened in the current study, only 3 species were found to be negative for BWCV: rough-toothed dolphins (Steno bredanensis) (n=1), humpback whales (n=1), and pygmy killer whales (n=7), the last of which only tested individuals that were part of a prolonged mass stranding event, and thus did not vary significantly by time or location. It would be valuable to test additional solitary stranding cases of species where BWCV was not detected. Additionally, this study focused predominantly on odontocetes, with the screening of only a single mysticete. Future work in the Pacific should increase screening of individuals representing each species tested to date and investigate presence in other cetacean species that inhabit the region but have not yet been assessed for BWCV.
In terms of geographic scope, our findings indicate that BWCV is a much more wide-spread virus than suggested by the isolated case of the Longman's beaked whale stranding on Maui. Our results show that BWCV is present in cetacean hosts across the Pacific basin, including the central, Western and South Pacific (Figure 1). Infected individuals were identified across the Hawaiian archipelago, on the islands of Hawai‘i, Kaua‘i, Maui, Moloka‘i, and O‘ahu. Our results also indicated positive circovirus findings in Cuvier’s beaked whales that stranded in Saipan in 2011 and American Samoa in 2015. The high genetic similarities between BWCV positive cases from the central, Western, and South Pacific and the BWCV reference genome suggests that these infections represent the same strain of BWCV described in the Longman’s beaked whale stranded on Maui (Figure 2). This high similarity is somewhat surprising considering distances exceeding 6,000 km between locations in the Pacific basin where BWCV positive animals were identified. With such a high prevalence rate, combined with the greatly expanded geographic scope identified, future work should include testing of marine mammals from other locations in both the Pacific and globally. It is currently unknown if BWCV is present in cetaceans from other ocean basins of the world.
The current study provided a unique opportunity to investigate the emergence of BWCV in the central Pacific. The initial discovery of BWCV occurred 10 years after the stranding of the Longman’s beaked whale in 2010, where the stranding cause was attributed to morbillivirus (West et al., 2013; Landrau-Giovannetti et al., 2020). Our screening of archived tissues detected BWCV in 4 of 6 tested tissues in a dwarf sperm whale that stranded in 2000 on the island of O‘ahu, which indicates that BWCV had emerged in the central Pacific at least 10 years prior to the initial case (Landrau-Giovannetti et al., 2020). Other infectious diseases recently described from the Pacific Islands region have indicated a date discrepancy between their discovery and evidence of earlier emergence. In the case of beaked whale morbillivirus, this strain of morbillivirus was first identified from a stranding in 2010 but was later found in a humpback whale that stranded in 1998, indicating that it had been present in the region for at least 12 years prior to its discovery (West et al., 2013; Jacob et al., 2016). It is currently unknown when BWCV emerged in the central Pacific but this study demonstrates its persistence for over a 20-year period that dates back to the year 2000.
Circoviruses have a broad range of effects on their hosts, many times associated with severe impacts to the brain and lungs (Seo et al., 2014; Bexton et al., 2015). Lymphoid depletion commonly occurs with infections of several strains of circoviruses, which is suggestive of immunosuppression (Yang et al., 2015; Mao et al., 2017; Palinski et al., 2017). Canine circovirus (GenBank Acc. No. JQ821392), determined to be of relatively close relation to BWCV, is known to cause pathologic lesions of internal tissues (Li et al., 2013; Kubiski, 2017). The most frequently positive tissues examined in this study were the brain, lung, and lymph tissues which are commonly infected by pathogenic circoviruses (Yang et al., 2015; Palinski et al., 2017; Opriessnig et al., 2020). Co-infection by other pathogens is common with other circoviruses (Yang et al., 2015). Cetacean morbillivirus, a potentially fatal disease, was found in four of the BWCV cases from the Pacific Islands, including the initial Longman’s beaked whale case, as well as a neonate sperm whale, a subadult Blainville’s beaked whale, and a Fraser’s dolphin infected with a novel morbillivirus strain (West et al., 2013; West et al., 2015; Jacob et al., 2016; West et al., 2021). Fatally disseminated toxoplasmosis was found in an adult spinner dolphin that was positive for BWCV (Landrau-Giovannetti et al., 2022). Brucella was present in several of the tissues of the neonate sperm whale and likely caused the pathology observed, and an alpha herpesvirus was also described in the initial Longman’s beaked whale, making both of these cases tri-infections (West et al., 2013; West et al., 2015). The individuals with BWCV and confirmed co-infections had respiratory disease, lymphoid depletion and poor body condition indicative of wasting (Landrau-Giovannetti et al., 2020; Landrau-Giovannetti et al., 2022; West et al., 2021). Conversely, other positive cases had no apparent pathology observed. In particular, the pelagic false killer whale that was by-caught by a fishing vessel was found to be infected with BWCV in several tissues, including brain, lung, liver, and mediastinal lymph node. At the time of necropsy, the animal was assessed as in robust body condition and gross pathology was not observed. However, this animal was frozen on board the fishing vessel where it was by-caught prior to coming to port which limited histopathological examination in this case.
Through this study, BWCV has been shown to have a range of host species, yet transmission vectors of this virus remain unclear. In other cetacean diseases, vertical transmission is believed to occur in utero, as well as through infected milk ingestion during nursing, as has been observed in both Brucella and morbillivirus infections (Guzmán-Verri et al., 2012; Van Bressem et al., 2014). The discovery of BWCV in both a neonate sperm whale and a Cuvier’s beaked whale calf suggest these are likely routes of infection. Horizontal transmission of porcine circovirus 2 has been documented through direct contact with infected secretions such as feces, urine, and airborne respiratory aerosols (Patterson, 2010; Verreault et al., 2010; Rose et al., 2012). Horizontal transmission of cetacean viruses, such as morbillivirus, papillomavirus, and herpesvirus are assumed to occur by physical contact and the transfer of bodily fluids, via inhalation of aerosols exhaled from infected individuals (blow), reproductive activity, or other social behaviors. This is further supported by the presence of unique viral strains in multiple cetacean species that inhabit Pacific waters (Gottschling et al., 2011; Jo et al., 2018; Weiss et al., 2020; Sierra et al., 2022), and this study similarly contributes to the identification of BWCV in 11 of these species. Considering the highly social nature of cetacean groups and that different cetacean species interact, it is probable that cross species infections of BWCV and other diseases occur. Several of the cetacean species with observed interspecies interactions in Hawaiian waters were positive for BWCV in this study, including melon-headed whales, pilot whales, spinner dolphins, Cuvier’s beaked whales, Blainville’s beaked whales, and false killer whales (Baird, 2016).
Future work should focus on investigating the clinical significance of such a high prevalence rate in a targeted screening and examine the pathologies observed in co-infected and non co-infected individuals in order to identify BWCV infection associated disease. The current study focused on six tissues per animal, but an increased sample size would aid in establishing the breadth of infection in archived and newly stranded animals, and in evaluating the extent of systemic infection. This would allow for a more detailed analysis of the distribution of the pathogen among tissues. Investigations into the potential pathogenicity of BWCV are important in light of the high prevalence rate. While traditional PCR is effective at detecting viral sequences and ideal for a broad screening effort, it provides limited insight into clinical manifestations. The current study did not address the pathological significance of BWCV but instead focused on a PCR screening effort to investigate prevalence. Additional methods, such as testing that focuses on cellular effects would aid in describing the pathological significance of BWCV. In situ hybridization (ISH) binds viral DNA specific probes to infected tissues, labeling them by chemical or radioactive means, which allows for visualization of viral inclusion bodies, aiding in determination of the level of infection and impact to the infected cells. Similar to ISH, immunohistochemistry (IHC) provides another potential method to better understand BWCV by visualizing viral infection through histological staining and microscopy, although IHC procedures focus on labeling the antigens within infected cells, rather than the DNA labeled in ISH. This will likely require the development of BWCV associated antibodies through in vivo injection and antibody isolation from the tissues of test animals. Commercially available antibodies for other strains of circovirus are unlikely to exhibit enough genetic similarity for cross reactivity to be successful in BWCV IHC staining. Implementation of such methodology would allow for a detailed analysis of BWCV infections and its cellular effects. This study demonstrated the wide-spread presence of BWCV among cetacean species and across geographic locations throughout the Pacific over the past 20 years, yet much still remains to be learned about the clinical significance of this emerging cetacean disease.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The animal study was reviewed and approved by Institutional Animal Care and Use Committee.
Author contributions
CC, KW, and IS-K wrote the main manuscript text and CC prepared the tables and figures. MM conducted necropsy and sample preservation for the stranded Cuvier’s beaked whale from American Samoa that was positive for BWCV. CC performed laboratory and phylogenetic analyses and interpretation was carried out by CC, KW, and IS-K. All authors contributed to the manuscript and approved the submitted version.
Funding
This work was supported by Naval Sea Systems (Cooperative Agreement #N62742-20-2-0001) and the National Marine Fisheries John H. Prescott Marine Mammal Rescue Assistance Grant Program.
Acknowledgments
We are grateful to the Pacific Islands Marine Mammal Stranding Response Network of volunteers for conducting stranding response, recovery, and transport of cetacean carcasses for necropsy across the Pacific basin. We also thank Jamie Marchetti for coordinating recovery of the by-caught false killer whale and Brent Tibbatts for leading cetacean necropsies in Guam. We would like to acknowledge Nicholas Hofmann and the necropsy team at the Health and Stranding Lab. We are grateful to Conner Humann for assistance in extraction of DNA samples for PCR and sequencing work. All work was conducted under NOAA National Marine Fisheries Service permit #18786-06.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Baird (2016). The lives of Hawa‘i's dolphins and whales: Natural history and conservation (University of Hawai‘i Press, Honolulu, HI).
Bandoo R. A., Bautista J., Lund M., Newkirk E., Squires J., Varsani A., et al. (2021). Identification of novel circovirus and anelloviruses from wolverines using a non-invasive faecal sampling approach. Infect Genet. Evol. 93, 104914. doi: 10.1016/j.meegid.2021.104914
Bexton S., Wiersma L. C., Getu S., Run P. R., van, Verjans G. M. G. M., Schipper D., et al. (2015). Detection of circovirus in foxes with meningoencephalitis. Emerg. Infect. Dis. J. Volume 21, Number 7, 1205–1208. doi: 10.3201/eid2107.150228
Breitbart M., Delwart E., Rosario K., Segalés J., Varsani A., ICTV Report Consortium (2017). ICTV virus taxonomy: Circoviridae. J. Gen. Virol. 98 (8), 1997–1998. doi: 10.1099/jgv.0.000871
Chen S., Zhang L., Li X., Niu G., Ren L. (2021). Recent progress on epidemiology and pathobiology of porcine circovirus 3. Viruses 13 (10), 1944–. doi: 10.3390/v13101944
Crowther R. A., Berriman J. A., Curran W. L., Allan G. M., Todd D. (2003). Comparison of the structures of three circoviruses: Chicken anemia virus, porcine circovirus type 2, and beak and feather disease virus. J. Virol. 77 (24), 13036–13041. doi: 10.1128/JVI.77.24.13036-13041.2003
Dai Z., Wang H., Feng Z., Ma L., Yang S., Shen Q., et al. (2021). Identification of a novel circovirus in blood sample of giant pandas (Ailuropoda melanoleuca). Infect Genet. Evol. 95, 105077. doi: 10.1016/j.meegid.2021.105077
Dal Santo A. C., Cezario K. C., Bennemann P. E., Machado S. A., Martins M. (2020). Full-genome sequences of porcine circovirus 3 (PCV3) and high prevalence in mummified fetuses from commercial farms in Brazil. Microbial Pathogenesis 141, 104027. doi: 10.1016/j.micpath.2020.104027
Dong W. N., Jun L. J., Bing W. A., Zhe Z., Yan C., Yu M., et al. (2016). Vertical transmission of PCV2b to fetuses in sows intramuscularly infected with PCV2b. Polish J. Vet. Sci. 19 (3), 471–476. doi: 10.1515/pjvs-2016-0059
Firth C., Charleston M. A., Duffy S., Shapiro B., Holmes E. C. (2009). Insights into the evolutionary history of an emerging livestock pathogen: Porcine circovirus 2. J. Virol. 83 (24), 12813–12821. doi: 10.1128/JVI.01719-09
Fisher M., Harrison T. M. R., Nebroski M., Kruczkiewicz P., Rothenburger J. L., Ambagala A., et al. (2020). Discovery and comparative genomic analysis of elk circovirus (ElkCV), a novel circovirus species and the first reported from a cervid host. Sci. Rep. 10 (1), 19548. doi: 10.1038/s41598-020-75577-6
Fogell D. J., Martin R. O., Bunbury N., Lawson B., Sells J., McKeand A. M., et al. (2018). Trade and conservation implications of new beak and feather disease virus detection in native and introduced parrots: BFDV in native and introduced parrots. Conserv. Biol. 32 (6), 1325–1335. doi: 10.1111/cobi.13214
Garrigue C., Oremus M., Dodémont R., Bustamante P., Kwiatek O., Libeau G., et al. (2016). A mass stranding of seven Longman’s beaked whales (Indopacetus pacificus) in New Caledonia, South Pacific. Mar. Mammal Sci. 32 (3), 884–910. doi: 10.1111/mms.12304
Gavier-Widén D., Duff J. P., Meredith A. (2012). Infectious diseases of wild mammals and birds in Europe (Wiley-Blackwell, United Kingdom).
Geraci J. R., Lounsbury V. J. (2005). Marine mammals ashore: A field guide for strandings (Second Edition). National Aquarium in Baltimore, Inc. Baltimore, MD.
Gottschling M., Bravo I. G., Schulz E., Bracho M. A., Deaville R., Jepson P. D., et al. (2011). Modular organizations of novel cetacean papillomaviruses. Mol. Phylogenet. Evol. 59 (1), 34–42. doi: 10.1016/j.ympev.2010.12.013
Grasland B., Blanchard P., Kéranflec’h A., Bigault L., Oger A., Rose N., et al. (2013). Evaluation of the transmission of porcine circovirus type 2 (PCV-2) genogroups a and b with semen from infected specific-pathogen-free boars. Vet. Microbiol. 162 (2), 381–387. doi: 10.1016/j.vetmic.2012.10.011
Guzmán-Verri C., González-Barrientos R., Hernández-Mora G., Morales J.-A., Baquero-Calvo E., Chaves-Olarte E., et al. (2012). Brucella ceti and brucellosis in cetaceans. Front. Cell. Infect Microbiol. 2. doi: 10.3389/fcimb.2012.00003
Huelsenbeck J. P., Ronquist F. (2001). MRBAYES: Bayesian inference of phylogeny. Bioinformatics 17, 754–755. doi: 10.1093/bioinformatics/17.8.754
Jacob J. M., West K. L., Levine. G., Sanchez S., Jensen B. A. (2016). Initial characterization of the novel beaked whale morbillivirus in Hawaiian cetaceans. Dis. Aquat. Organisms. 117, 215–227. doi: 10.3354/dao02941
Jo W. K., Kruppa J., Habierski A., van de Bildt M., Mazzariol S., Di Guardo G., et al. (2018). Evolutionary evidence for multi-host transmission of cetacean morbillivirus. Emerg Microbes Infect 7 (1), 1–15. doi: 10.1038/s41426-018-0207-x
Katoh K., Standley D. M. (2013). MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30 (4), 772–780. doi: 10.1093/molbev/mst010
Kobayashi N., Tokutake K., Yoshida H., Okabe H., Miyamoto K., Ito H., et al. (2021). The first stranding record of Longman's beaked whale (Indopacetus pacificus) in Okinawa, Japan. Aquat. Mammals 47 (2), 153–174. doi: 10.1578/AM.47.2.2021.153
Kubiski S. (2017). Initial studies on the prevalence and pathogenesis of canine circovirus (ProQuest Dissertations Publishing) Ann Arbor, MI.
Lagan Tregaskis P., Staines A., Gordon A., Sheridan P., McMenamy M., Duffy C., et al. (2020). Co-Infection status of novel parvovirus’s (PPV2 to 4) with porcine circovirus 2 in porcine respiratory disease complex and porcine circovirus-associated disease from 1997 to 2012. Transbound. Emerg. Dis. 68 (4), 1979–1994. doi: 10.1111/tbed.13846
Landrau-Giovannetti N., Subramaniam K., Brown M. A., Ng T. F. F., Rotstein D. S., West K., et al. (2020). Genomic characterization of a novel circovirus from a stranded Longman’s beaked whale (Indopacetus pacificus). Virus Res. 277, 197826. doi: 10.1016/j.virusres.2019.197826
Landrau-Giovannetti N., Waltzek T. B., López-Orozco N., Su C., Rotstein D., Levine G., et al. (2022). Prevalence and genotype of Toxoplasma gondii in stranded Hawaiian cetaceans. Dis. Aquat. Org. 152, 27–36. doi: 10.3354/dao03699
Lecis R., Mucedda M., Pidinchedda E., Zobba R., Pittau M., Alberti A. (2020). Genomic characterization of a novel bat-associated circovirus detected in European Miniopterus schreibersii bats. Virus Genes 56 (3), 325–328. doi: 10.1007/s11262-020-01747-3
Levy H., Fiddaman S. R., Djurhuus A., Black C. E., Kraberger S., Smith A. L., et al. (2020). Identification of circovirus genome in a chinstrap penguin (Pygoscelis antarcticus) and Adélie penguin (Pygoscelis adeliae) on the Antarctic peninsula. Viruses 12 (8), 858. doi: 10.3390/v12080858
Li J., Xing G., Zhang C., Yang H., Li G., Wang N., et al. (2019). Cross-species transmission resulted in the emergence and establishment of circovirus in pig. Infect Genet. Evol. 75, 103973. doi: 10.1016/j.meegid.2019.103973
Li L., McGraw S., Zhu K., Leutenegger C. M., Marks S. L., Kubiski S., et al. (2013). Circovirus in tissues of dogs with vasculitis and hemorrhage. Emerg Infect. Dis. 19 (4), 534–541. doi: 10.3201/eid1904.121390
Li Z., Wang X., Zhang R., Chen J., Xia L., Lin S., et al. (2014). Evidence of possible vertical transmission of duck circovirus. Vet. Microbiol. 174 (1–2), 229–232. doi: 10.1016/j.vetmic.2014.09.001
Lin K., Wang C., Murtaugh M. P., Ramamoorthy S. (2011). Multiplex method for simultaneous serological detection of porcine reproductive and respiratory syndrome virus and porcine circovirus type 2. J. Clin. Microbiol. 49 (9), 3184–3190. doi: 10.1128/JCM.00557-11
Lőrincz M., Cságola A., Farkas S. L., Székely C., Tuboly T. (2011). First detection and analysis of a fish circovirus. J. Gen. Virol. 92 (8), 1817–1821. doi: 10.1099/vir.0.031344-0
Mao Y., Li J. J., Liu Y., Dong W., Pang P., Deng Z. B. (2017). Imbalance of intestinal immune function in piglets infected by porcine circovirus type 2 during the fetal period. Acta Veterinaria Hungarica 65 (1), 135–146. doi: 10.1556/004.2017.014
Mateusen B., Maes D. G. D., Van Soom A., Lefebvre D., Nauwynck H. J. (2007). Effect of a porcine circovirus type 2 infection on embryos during early pregnancy. Theriogenology 68 (6), 896–901. doi: 10.1016/j.theriogenology.2007.07.014
Mazzariol S., Centelleghe C., Di Provvido A., Di Renzo L., Cardeti G., Cersini A., et al. (2017). Dolphin morbillivirus associated with a mass stranding of sperm whales, Italy. Emerg Infect. Dis. 23 (1), 144–146. doi: 10.3201/eid2301.160239
Morandini V., Dugger K. M., Ballard G., Elrod M., Schmidt A., Ruoppolo V., et al. (2019). Identification of a novel Adélie penguin circovirus at Cape Crozier (Ross island, Antarctica). Viruses 11 (12), 1088. doi: 10.3390/v11121088
Opriessnig T., Karuppannan A. K., Castro A. M. M. G., Xiao C.-T. (2020). Porcine circoviruses: Current status, knowledge gaps and challenges. Virus Res. 286, 198044. doi: 10.1016/j.virusres.2020.198044
Palinski R., Piñeyro P., Shang P., Yuan F., Guo R., Fang Y., et al. (2017). A novel porcine circovirus distantly related to known circoviruses is associated with porcine dermatitis and nephropathy syndrome and reproductive failure. J. Virol. 91 (1), e01879–16. doi: 10.1128/JVI.01879-16
Patterson A. (2010). Horizontal transmission of porcine circovirus type 2 (PCV2) (ProQuest Dissertation Publishing). Ann Arbor, MI. doi: 10.31274/etd-180810-1202. [Doctor of Philosophy, Iowa State University, Digital Repository].
Pendl H., Tizard I. (2016). “Chapter 11–immunology,” in Current therapy in avian medicine and surgery. Eds. Speer B. L., Saunders W. B., 400–432, Saunders, Philadelphia, PA. doi: 10.1016/B978-1-4557-4671-2.00020-3
Peters A., Patterson E. I., Baker B. G. B., Holdsworth M., Sarker S., Ghorashi S. A., et al. (2014). Evidence of psittacine beak and feather disease virus spillover into wild critically endangered orange-bellied parrots (Neophema chrysogaster). J. Wildlife Dis. 50 (2), 288–296. doi: 10.7589/2013-05-121
Rampin T., Manarolla G., Pisoni G., Recordati C., Sironi G. (2006). Circovirus inclusion bodies in intestinal muscle cells of a canary. Avian Pathol. 35 (4), 277–279. doi: 10.1080/03079450600817057
Ren W., Wu M., Luo W., Huang R., Yin Y., Li Y., et al. (2013). Dietary supplementation with proline confers a positive effect in both porcine circovirus-infected pregnant and non-pregnant mice. Br. J. Nutr. 110 (8), 1492–1499. doi: 10.1017/S0007114513000652
Ronquist F., Huelsenbeck J. P. (2003). MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574. doi: 10.1093/bioinformatics/btg180
Rose N., Opriessnig T., Grasland B., Jestin A. (2012). Epidemiology and transmission of porcine circovirus type 2 (PCV2). Virus Res. 164 (1), 78–89. doi: 10.1016/j.virusres.2011.12.002
Sarker S., Moylan K. G., Ghorashi S. A., Forwood J. K., Peters A., Raidal S. R. (2015). Evidence of a deep viral host switch event with beak and feather disease virus infection in rainbow bee-eaters (Merops ornatus). Sci. Rep. 5 (1), 14511–14511. doi: 10.1038/srep14511
Seo H. W., Park S.-J., Park C., Chae C. (2014). Interaction of porcine circovirus type 2 and Mycoplasma hyopneumoniae vaccines on dually infected pigs. Vaccine 32 (21), 2480–2486. doi: 10.1016/j.vaccine.2014.02.088
Sierra E., Fernández A., Fernández-Maldonado C., Sacchini S., Felipe-Jiménez I., Segura-Göthlin S., et al. (2022). Molecular characterization of herpesviral encephalitis in cetaceans: Correlation with histopathological and immunohistochemical findings. Animals: an Open Access J. MDPI 12 (9), 1149. doi: 10.3390/ani12091149
Van Bressem M.-F., Duignan P., Banyard A., Barbieri M., Colegrove K., De Guise S., et al. (2014). Cetacean morbillivirus: Current knowledge and future directions. Viruses 6 (12), 5145–5181. doi: 10.3390/v6125145
Vargas-Castro I., Crespo-Picazo J. L., Rivera-Arroyo B., Sánchez R., Marco-Cabedo V., Jiménez-Martínez M.Á., et al. (2020). Alpha- and gammaherpesviruses in stranded striped dolphins (Stenella coeruleoalba) from Spain: first molecular detection of gammaherpesvirus infection in central nervous system of odontocetes. BMC Vet. Res. 16 (1), 288–288. doi: 10.1186/s12917-020-02511-3
Verreault D., Létourneau V., Gendron L., Massé D., Gagnon C. A., Duchaine C. (2010). Airborne porcine circovirus in Canadian swine confinement buildings. Vet. Microbiol. 141 (3–4), 224–230. doi: 10.1016/j.vetmic.2009.09.013
Wang J., Liu L., Li R., Yuan W. (2016). Rapid detection of porcine circovirus 2 by recombinase polymerase amplification. J. Vet. Diagn. Invest. 28 (5), 574–578. doi: 10.1177/1040638716654201
Wang Y., Zhang D., Bai C., Guo X., Gao W., Li M., et al. (2021). Molecular characteristics of a novel duck circovirus subtype 1d emerging in Anhui, China. Virus Res. 295, 198216. doi: 10.1016/j.virusres.2020.198216
Weiss M. N., Franks D. W., Balcomb K. C., Ellifrit D. K., Silk M. J., Cant M. A., et al. (2020). Modelling cetacean morbillivirus outbreaks in an endangered killer whale population. Biol. Conserv. 242, 108398. doi: 10.1016/j.biocon.2019.108398
West K. L., Levine G. A., Jacob J., Jensen B. A., Sanchez S., Colegrove K., et al. (2015). Co-Infection and vertical transmission of Brucella and Morbillivirus in a neonatal sperm whale. J. Wildlife Dis 51 (1), 227–232. doi: 10.7589/2014-04-092
West K. L., Sanchez S., Rotstein D., Robertson K. M., Dennison S., Levine G., et al. (2013). A Longman’s beaked whale (Indopacetus pacificus) strands in Maui, Hawai‘i, with first case of morbillivirus in the central Pacific. Mar. Mammal Sci. 29 (4), 767–776. doi: 10.1111/j.1748-7692.2012.00616.x
West K. L., Silva-Krott I., Landrau-Giovannetti N., Rotstein D., Saliki J., Raverty S., et al. (2021). Novel cetacean morbillivirus in a rare Fraser’s dolphin (Lagenodelphis hosei) stranding from Maui, Hawaiʻi. Sci. Rep. 11 (1), 15986. doi: 10.1038/s41598-021-94460-6
Woods L. W., Latimer K. S. (2000). Circovirus infection of nonpsittacine birds. J. Avian Med. Surg. 14 (3), 154–163. doi: 10.1647/1082-6742(2000)014[0154:CIONB]2.0.CO;2
Woods L. W., Latimer K. S., Barr B. C., Niagro F. D., Campagnoli R. P., Nordhausen R. W., et al. (1993). Circovirus-like infection in a pigeon. J. Vet. Diagn. Invest. 5 (4), 609–612. doi: 10.1177/104063879300500417
Yang Y., Shi R., She R., Mao J., Zhao Y., Du F., et al. (2015). Fatal disease associated with swine hepatitis E virus and porcine circovirus 2 co-infection in four weaned pigs in China. BMC Veterinary Res. 11 (1), 77. doi: 10.1186/s12917-015-0375-z
Zaccaria G., Malatesta D., Scipioni G., Di Felice E., Campolo M., Casaccia C., et al. (2016). Circovirus in domestic and wild carnivores: An important opportunistic agent? Virol. (New York N.Y.) 490, 69–74. doi: 10.1016/j.virol.2016.01.007
Zhai Z., Lin T., Zhang H., Wen X.-H., Zhou X.-R., Jia C.-L., et al. (2017). Reappearance of buffalo-origin-like porcine circovirus type 2 strains in swine herds in southern China. New Microbes New Infect 17 (C), 98–100. doi: 10.1016/j.nmni.2017.02.009
Keywords: circovirus, cetacean, odontocete, disease, Pacific, beaked whale, PCR, phylogenetics
Citation: Clifton CW, Silva-Krott I, Marsik MG and West KL (2023) Targeted surveillance detected novel beaked whale circovirus in ten new host cetacean species across the Pacific basin. Front. Mar. Sci. 9:945289. doi: 10.3389/fmars.2022.945289
Received: 16 May 2022; Accepted: 03 November 2022;
Published: 26 January 2023.
Edited by:
Peter H. Dutton, Southwest Fisheries Science Center (NOAA), United StatesReviewed by:
Idaira Felipe-Jiménez, University of Las Palmas de Gran Canaria, SpainCristiane Kolesnikovas, Animal R3 Association, Brazil
Copyright © 2023 Clifton, Silva-Krott, Marsik and West. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cody W. Clifton, cclifton@hawaii.edu
 Cody W. Clifton
Cody W. Clifton Ilse Silva-Krott
Ilse Silva-Krott Michael G. Marsik3
Michael G. Marsik3