Detection of the Largest Deep-Sea-Endemic Teleost Fish at Depths of Over 2,000 m Through a Combination of eDNA Metabarcoding and Baited Camera Observations
- 1Research Institute for Global Change (RIGC), Japan Agency for Marine-Earth Science and Technology (JAMSTEC), Yokosuka, Japan
- 2Institute of Industrial Science, The University of Tokyo, Meguro-Ku, Japan
- 3Department of Collection Management, Natural History Museum and Institute, Chiba, Chiba, Japan
The Yokozuna Slickhead Narcetes shonanmaruae is a recently described deep-sea fish species and an active-swimming, relatively large top predator in Suruga Bay, Japan. Its only known habitat is the deepest part of the bay (>2,000 m); six individuals have been collected thus far (up to 138 cm in total length). During our monitoring survey of faunal diversity on seamounts within marine protected areas in Japanese waters, environmental DNA (eDNA) metabarcoding revealed the Yokozuna Slickhead 12S ribosomal RNA gene sequence on/around three seamounts belonging to the Nishi-Shichito Ridge (at depths of around 2,000 m) located 400–600 km south of the known locality. A baited camera system deployed at the foot of one of the three seamounts at a depth of 2,091 m captured a Yokozuna Slickhead individual that was over 250 cm in total length, threatening Pacific Grenadiers Coryphaenoides acrolepis around the bait and attacking the bait cage. A combination of eDNA metabarcoding and baited camera observation represents a powerful tool for the detection of rare predatory fish species and the study of their ecology even in the deep sea, thus helping to better understand vulnerable marine ecosystems and reveal the impact of the rapidly changing global ocean.
Introduction
Top predators play an important role in maintaining species diversity and ecosystem functions (Sergio et al., 2005; Baum and Worm, 2009; Estes et al., 2011; Wallach et al., 2015). The extinction and reintroduction of gray wolves in Yellowstone National Park are examples of drastic changes in top predator populations that affect the entire ecosystem (Ripple and Beschta, 2012). Similar instances can be enumerated not only in terrestrial but also in marine ecosystems (Myers et al., 2007; Baum and Worm, 2009; Roff et al., 2016).
Marine environments are increasingly affected by global climate change and anthropogenic activities, leading to oceanic warming, acidification, and deoxygenation even in deep-sea regions (Levin and Le Bris, 2015). Such global changes are assumed to initially affect large, predatory consumers and have subsequent repercussions for organisms at lower trophic levels (Zarnetske et al., 2012). Thus, there is an urgent need to elucidate the present biodiversity and abundance of predatory fishes inhabiting deep-sea sites (Schmitz, 2007; Tecchio et al., 2013). However, due to the relatively small population sizes, the probability of detecting top predators is usually low in all environments, especially in the hydrosphere where organisms are hidden beneath water layers (Jerde et al., 2011).
Environmental DNA (eDNA) detection represents a powerful tool for assessing aquatic biodiversity (Miya et al., 2015; Yamamoto et al., 2017) and is also used for the efficient detection of “hard-to-find species,” including top predators (Jerde et al., 2011; Wilcox et al., 2013; Fukumoto et al., 2015; Thomsen and Willerslev, 2015; Bakker et al., 2017; Weltz et al., 2017; Boussarie et al., 2018; Lafferty et al., 2018; Postaire et al., 2020; Ip et al., 2021). Detection of a species-specific eDNA sequence implies the presence of this species in a certain area.
However, further ecological information, such as body size, population, sex, maturity, coloration, and behavior, is not obtained in most cases through eDNA metabarcoding alone. Baited camera observation is one of the complementary methods for detecting predatory/scavenging species, which not only indicates the diversity of species but also provides insight into population size, abundance, behavior, and relationship with conspecific or heterospecific individuals (Stoner et al., 2008; Brooks et al., 2011; Devine et al., 2018; Fujiwara et al., 2021b; Sakaue et al., 2021; Aoki et al., 2022).
The Yokozuna Slickhead Narcetes shonanmaruae is a recently discovered large predator in Suruga Bay, Japan (Fujiwara et al., 2021a). Compound-specific isotope analyses of specific amino acids and DNA metabarcoding analyses of the stomach contents revealed that the slickhead consumes relatively large fish species and is a top predator at depths greater than 2,000 m in Suruga Bay (Fujiwara et al., 2021a). Only six individuals have been collected so far, with their distribution limited to the Suruga Bay mouth, and the extent of their habitat remains elusive (Fujiwara et al., 2021a).
In 2020 and 2021, two research cruises were conducted to develop biodiversity monitoring methods for the management of deep-sea marine protected areas (MPAs) and monitoring the deep-sea MPAs in Japanese waters (Hookabe et al., 2021; Koeda et al., 2021; Hookabe et al., 2022; Jimi et al., 2022; Komai et al., 2022). To understand fish diversity including large predatory consumers, metabarcoding analyses of fish and baited camera observations were conducted on/around six seamounts on the Nishi-Shichito Ridge as well as the Central and Western Mariana ridges, which are designated as MPAs. During the research cruises, we coincidently discovered novel habitats of the Yokozuna Slickhead, which were distantly separated from the type locality. Unexpected threatening behavior and colossal body size were also reported.
Materials and Methods
Water Sampling and Metabarcoding of Fish
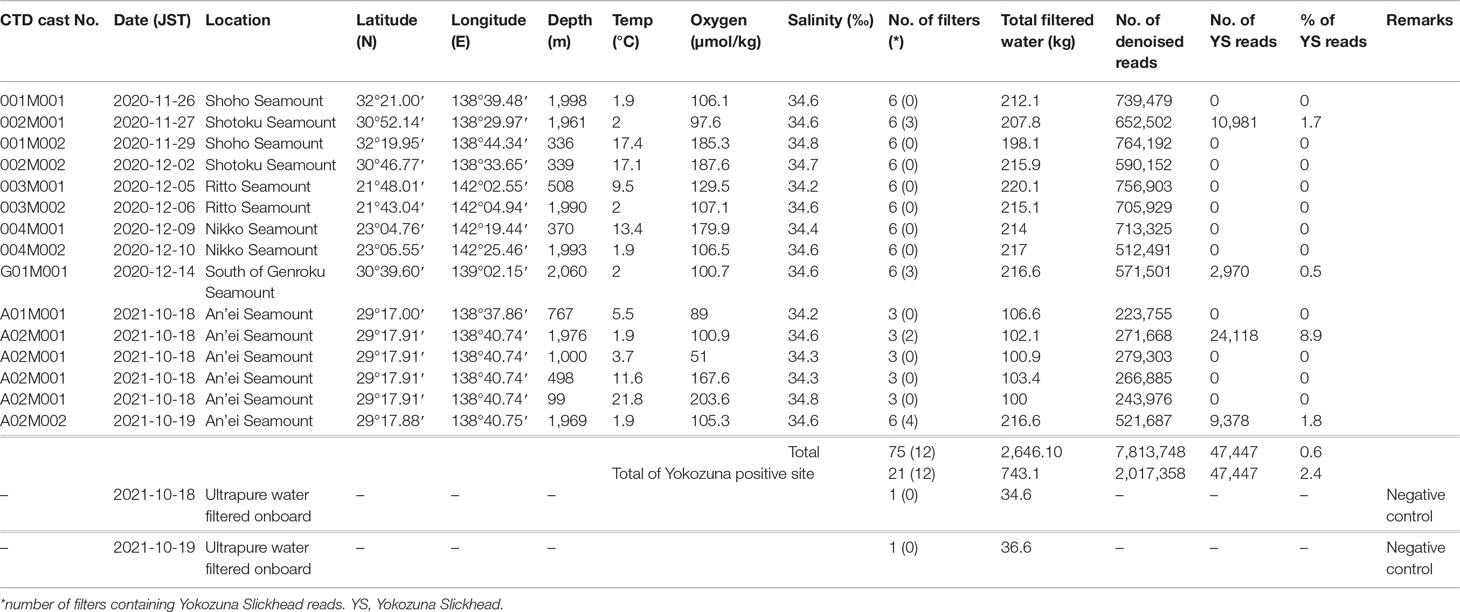
During cruises KM20-10C and KM21-E04C, seawater sampling and physico-chemical measurements were conducted using an SBE 32 Carousel Water Sampler (Sea-Bird Electronics, Bellevue, USA), which was composed of 36 bottles of 12-L Niskin water samplers with an SBE 9plus CTD (Sea-Bird Electronics) installed on R/V Kaimei belonging to the Japan Agency for Marine-Earth Science and Technology (JAMSTEC). Sampling locations are shown in Table 1 and Figure 1. Approximately 30 L of the collected seawater was immediately filtered onboard using an enclosed type filter, Sterivex-HV Pressure Filter Unit (0.45 µm pore size, PVDF membrane, gamma-irradiated, sterile) (Merck KGaA, Darmstadt, Germany) according to previous studies (Miya et al., 2016; Kawato et al., 2021), and the filters were stored at −30°C in freezers onboard. To monitor contamination during the filtration process, blank samples (negative control) were prepared onboard by filtering approximately 30 L ultrapure water during the KM21-E04C cruise. Subsequent experiments were conducted in land-based laboratories after the cruises. eDNA extraction, first-round PCR amplification for eDNA metabarcoding of fish using MiFish primers, paired-end library preparation through second-round PCR, and MiSeq sequencing were conducted according to previous studies (Miya et al., 2015; Kawato et al., 2021; Oka et al., 2021). Data preprocessing and taxon assignment were also conducted according to the previous study (Oka et al., 2021). The sequences reported here have been deposited in the DNA Data Bank of Japan (DDBJ) database under accession numbers LC710825–LC710836. All physico-chemical data were acquired using a CTD in real-time via a coaxial cable.
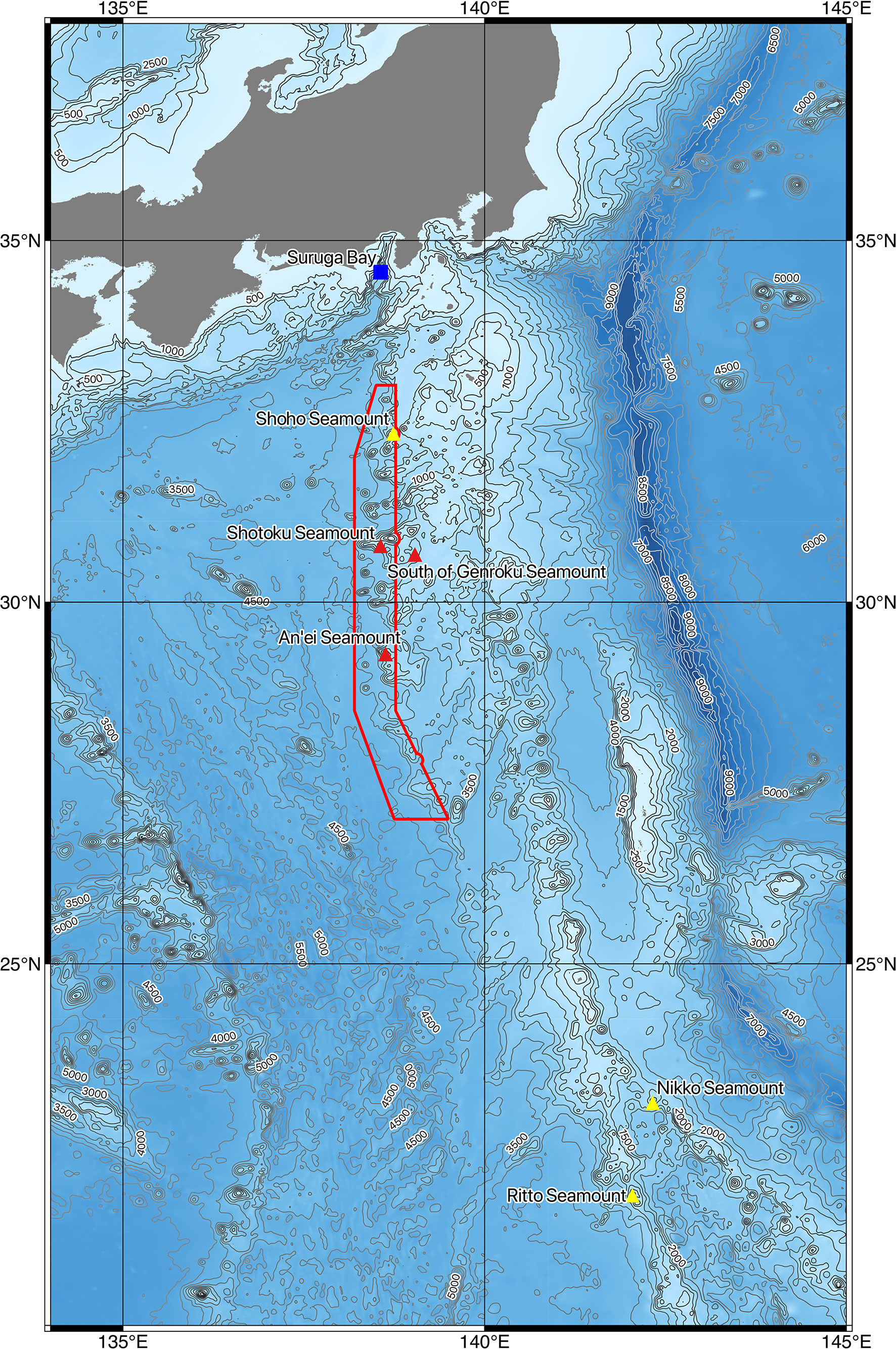
Figure 1 Research area and location of CTD casts and baited camera deployments. This map was created using QGIS software version 3.10 (https://qgis.org/), and bathymetric data etopo1 (doi:10.7289/V5C8276M) was supplied by the NOAA National Geophysical Data Center. Triangle: research locations; red triangle: Yokozuna Slickhead (Narcetes shonanmaruae)-positive sites; yellow triangle: Yokozuna Slickhead-negative sites; blue square: type locality of Yokozuna Slickhead; red box: the Nishi-Shichito Ridge Marine Protected Area.
Baited Camera Observation
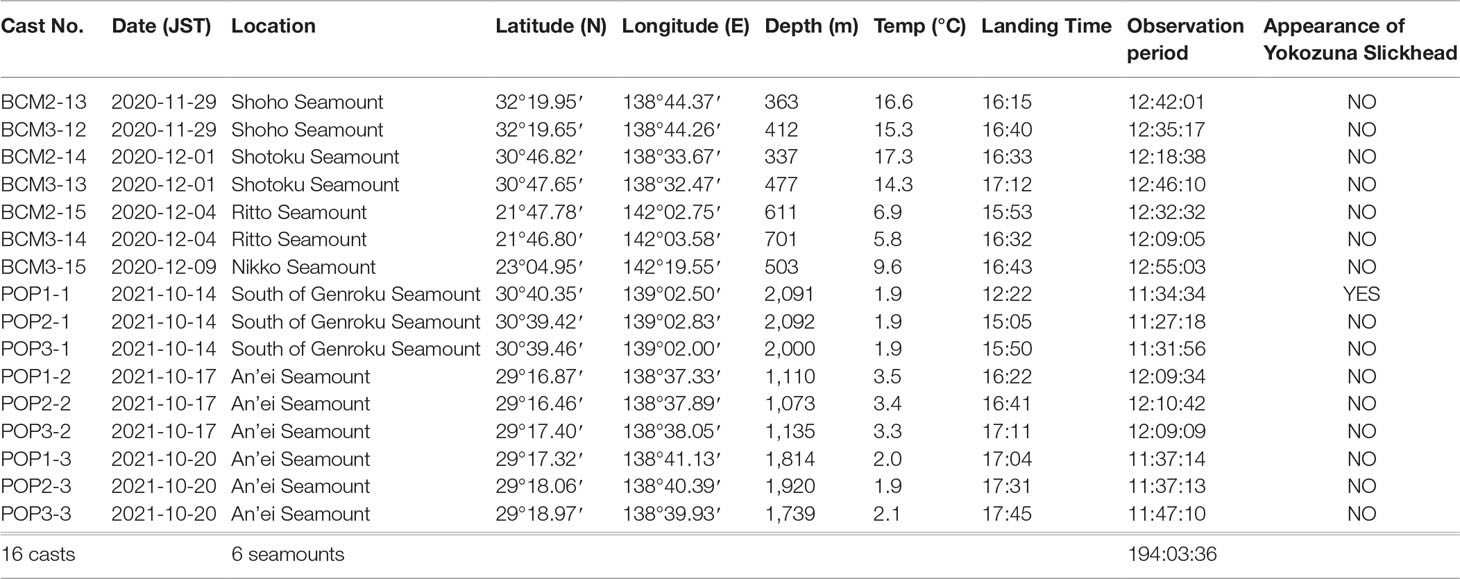
Baited camera systems were deployed to estimate the diversity and population density of predators/scavengers during the KM20-10C and KM21-E04C cruises. Detailed information on the casts is shown in Table 2. Two types of baited camera systems were used. Both were deployed from R/V Kaimei in the free-fall mode; one, named “BCM,” was retrieved by a remotely operated vehicle KM-ROV installed on R/V Kaimei, and the other, named “POP,” released a sinker through an acoustic release command and rose to the surface by itself. Both BCM and POP were composed of the following: a digital still camera DSC-RX0 (Sony, Tokyo, Japan) equipped with a Beastgrip × Kenko Pro Series 0.75× wide-angle lens (Kenko Tokina, Tokyo, Japan) (horizontal angle: 97.6°; vertical angle: 74.6° both in air); two LED flashlights (model no. OL0183) (KC Fire, Guangdong, China); three aluminum alloy housings for the camera and flashlights; an electro-magnetic current profiler Infinity-EM (JFE Advantech, Nishinomiya, Japan) for BCM or an electro-magnetic current profiler Infinity-Deep (JFE Advantech) for POP; a miniature conductivity, temperature, and depth (CTD) logger DST-CTD (Star-Oddi, Garðabær, Iceland); an acoustic release transponder STE (Kaiyo Denshi, Tsurugashima, Japan); an ROV Homer (Sonardyne International, Yateley, UK); a stainless steel bait cage containing approximately 0.7 kg of fresh mackerel; a syntactic foam TG2000 (Trelleborg AB, Trelleborg, Sweden) for BCM or a syntactic foam TG3000 (Trelleborg AB) for POP; and a stainless steel frame for BCM or a vinyl chloride frame for POP. Additionally, POP was equipped with a miniature flasher MMF-7500 (Novatech, Montreal, Canada), a miniature VHF radio beacon MMB-7500 (Novatech), an internal manufacturing sinker-release device, internal manufacturing backup release timer, a 20-kg steel sinker, and a customized plastic buoy 10AS-30 (Plastech Industrial, Tainan, Taiwan). Video recording was started approximately 20 min before free fall and yielded 13-h video footage. All data, including video footage, CTD, and current profiles, were recovered when the system was retrieved onboard.
Size Estimate of the Yokozuna Slickhead
The size of Yokozuna Slickhead was estimated from the video images. The postorbital head width and the interorbital distance were measured using video grabs and corrected using the bait cage width (260 mm). The standard length (SL) was first estimated using the average ratio of the postorbital head width (9.08% in standard length) and the interorbital distance (7.33% in standard length) (Fujiwara et al., 2021a). The total length (TL) was calculated using the mean ratio of SL/TL (87.79%) (Fujiwara et al., 2021a).
Results
Fish eDNA Metabarcoding
Eight CTD casts were conducted during KM20-10C and seven during KM21-E04C (Table 1). More than 2,600 kg of seawater was filtered using 75 cartridge filters at 15 sites on/around six seamounts (35 ± 2 kg of seawater filtered/cartridge) (Table 1). Approximately 7.8 million denoised reads were acquired using MiSeq sequencing. The reads of Yokozuna Slickhead were detected in seawater samples collected at a depth of 1,961 m at Shotoku Seamount, at a depth of 2,060 m south of Genroku Seamount, and at depths of 1,969 and 1,976 m at An’ei Seamount. No Yokozuna Slickhead reads were detected at depths shallower than 1,900 m, and detection was limited to the Nishi-Shichito Ridge (Table 1 and Figure 1). At all sampling sites where Yokozuna Slickhead reads were detected, more than half of the filters contained the reads (50–67% appearance shown in Table 1). Of these positive sites, the ratio of the Yokozuna Slickhead reads ranged from 0.5 to 8.9% (2.4% of the total reads in the positive samples) (Table 1). Even at the same location (CTD cast no. A02M001), Yokozuna Slickhead reads were detected near the seafloor but not in a midwater column (Table 1). No PCR product was detected from the blank samples.
Baited Camera Observation
Seven baited camera casts were conducted during KM20-10C and nine during KM21-E04C, at depths from 337 to 2,092 m on/around six seamounts (Table 2). More than 11 h of video sequences were recorded during each cast, and a total of 194 h of video footage was recorded by baited cameras (Table 2).
A single individual of Yokozuna Slickhead was observed at a depth of 2,091 m south of Genroku Seamount during cast no. POP1-1 at 17:52 (Japan Standard time; JST) on October 14, 2021 (Table 2; Figures 1, 2, and Supplementary Video 1). The Yokozuna Slickhead individual came around the bait 5 h 29 min 50 s after the baited camera landed (Figure 2A). Two Pacific Grenadiers Coryphaenoides acrolepis and two Snubnosed eels Simenchelys parasitica had already arrived around the bait before the arrival of the slickhead (Supplementary Video 1). The slickhead swam into the video frame from the right side against the current (Supplementary Video 1).

Figure 2 Yokozuna Slickhead Narcetes shonanmaruae. In situ video grabs recorded using a baited camera system at a depth of 2,091 m south of Genroku Seamount during cast no. POP1-1 on October 14, 2021. (A) First arrival of a Yokozuna Slickhead individual (on the right side) at the baited camera system; (B) First threatening behavior toward Pacific Grenadiers Coryphaenoides acrolepis; (C) Second threatening behavior to the Pacific Grenadiers; (D) Second arrival of the Yokozuna Slickhead at the baited camera system.
After first contacting one of the Pacific Grenadiers, the slickhead opened its mouth wide twice in a display of threatening behavior, and all the grenadiers around the bait cage quickly disappeared (Figures 2B, C and Supplementary Video 1). The slickhead attacked the bait cage and disappeared to the far right of the baited camera (Supplementary Video 1). Its eyes were deep blue, and its skin was bumpy, damaged, and ridden with parasites (Figure 2 and Supplementary Video 1). Approximately 4 min later, the same individual re-entered the video frame from the right side at 17:56 (JST) and came closer to the bait cage (Figure 2D). However, this time, the fish suddenly turned around and disappeared to the far left without attacking the bait cage. The recording duration of the first and second arrival of the slickhead was approximately 1.5 and 1 min, respectively.
The TL of the Yokozuna Slickhead was estimated from the video images. Five measurements of the postorbital head width and four measurements of the interorbital distance were conducted from five video grabs, and the average SL and TL were estimated to be 222 cm and 253 cm, respectively.
Environmental Factors at the Time of Yokozuna Slickhead Appearance
The presence of Yokozuna Slickhead was confirmed a total of five times during this study (four recorded through eDNA metabarcoding and one through baited camera observation), as shown in Tables 1, 2. The depth, temperature, salinity, and oxygen concentration were between 1,961 and 2,091 m, 1.9 and 2.0°C, 34.6‰, and 97.6 and 105.3 µmol·kg-1, respectively, at the time of appearance of the Yokozuna Slickhead (Tables 1, 2). The salinity data acquired during the baited camera observations were not used because the accuracy of the salinity sensor installed on the miniature CTD was low during deployment.
Discussion
In this study, we discovered novel habitats of the Yokozuna Slickhead and observed its unexpected threatening behavior through the use of eDNA metabarcoding and baited camera observation. Despite its large size, the Yokozuna Slickhead had not been recognized until 2021, and only four individuals have been collected from Suruga Bay thus far (Fujiwara et al., 2021a), with two additional individuals collected from the same location for television show projects. Although the population density in Suruga Bay was not estimated due to an insufficient number of baited camera casts (Fujiwara et al., 2021a; Aoki et al., 2022), the extremely high trophic position implied its rarity (Fujiwara et al., 2021a). In fact, Yokozuna Slickhead reads were not detected from all the filters for each Yokozuna Slickhead-positive site, and even at the Yokozuna Slickhead-positive sites, the average detection rate of Yokozuna Slickhead reads was only 2.4% (Table 1). Despite the rarity, eDNA metabarcoding was able to reliably detect the “hard-to-find species” even in low-biomass offshore areas. Therefore, eDNA metabarcoding would be an effective method to monitor fish biodiversity in deep-sea MPAs. To increase the likelihood of detecting the “hard-to-find species,” the use of as many sample replicates as possible is recommended.
Many attempts have been made to uncover the diversity of aquatic organisms, including rare species, through the use of eDNA metabarcoding, thereby expanding the knowledge of fauna diversity in various environments (reviewed in Miya, 2022). However, it is not easy to infer ecological aspects such as body size, population, sex, maturity, coloration, and behavior through eDNA metabarcoding alone. The baited camera survey is a complementary method for non-invasively studying these ecological characteristics of predators/scavengers, especially in the deep sea (Stoner et al., 2008; Brooks et al., 2011; Devine et al., 2018; Fujiwara et al., 2021b). The target area of a baited camera survey is confined to the light irradiation range and is considered to be much smaller than that of eDNA analysis (Murakami et al., 2019). Therefore, a combination of a wide-range survey through eDNA metabarcoding and a spot survey using the baited camera represents a powerful tool for elucidating the ecology of “hard-to-find species,” such as predators in the deep sea.
The novel habitats of the Yokozuna Slickhead were found on Nishi-Shichito Ridge, more than 400 km south of the type locality. The type specimens of slickhead were collected from Suruga Bay at depths between 2,171 m and 2,572 m, and the video sequence was recorded at a depth of 2,572 m in the bay (Fujiwara et al., 2021a). Therefore, the present study expanded the geographic range and depth (1,961–2,091 m) of the habitat. The water temperature range of Yokozuna Slickhead appearance in this study (1.9–2.0°C) was comparable to that determined in our previous study (1.6–2.0°C) (Fujiwara et al., 2021a). The Yokozuna Slickhead did not appear on the Shoho Seamount, which is located between Suruga Bay and other seamounts where its distribution was confirmed (Figure 1 and Tables 1, 2). The environmental factors on Shoho Seamount were similar to those in Suruga Bay and the other seamounts. It is difficult to conclude whether the distribution of Yokozuna Slickhead is divided near the Shoho Seamount, as only one CTD cast was performed, and no baited camera cast was conducted around a depth of 2,000 m on the seamount. There was no evidence of Yokozuna Slickhead distribution on the Nikko and Ritto seamounts located approximately 800 km south of the An’ei Seamount, the southernmost known habitat of the slickhead. Again, the environmental factors were similar between Yokozuna Slickhead habitats and the Nikko and Ritto seamounts. Notably, CTD cast was performed only once per seamount, and no baited camera was deployed around a depth of 2,000 m on the seamounts. Thus, further research is needed to reveal the distinct distribution of this species.
In general, slickheads are mesopelagic or benthopelagic fish, and their behavioral ecology is largely unknown (Jones and Breen, 2013). Although slickheads have been caught in demersal trawls, images and video footages taken by baited cameras and submersibles are rare (Trenkel et al., 2004; Priede et al., 2010; Pakhorukov and Parin, 2012; Zintzen et al., 2012). Trenkel et al. (2004) suggested that the lack of video footage was due to the slickheads swimming a few meters above the seafloor and being out of the field of view. Pakhorukov and Parin (2012) suggested that the slickheads avoid the submersible’s light. The Yokozuna Slickhead was observed via baited cameras deployed on the seafloor, equipped with a red LED light (Fujiwara et al., 2021a) or with two white LED lights in the present study. Therefore, the Yokozuna Slickhead is possibly benthopelagic and swims closer to the seafloor. Further, red and white lights did not seem to have a significant negative effect on its foraging behavior.
The threatening behavior of the Yokozuna Slickhead against Pacific Grenadiers was unexpected (Figures 2B, C). The slickhead vigorously opened its mouth wide toward a grenadier, and the grenadier quickly fled (Figure 2B and Supplementary Video 1). Nevertheless, the slickhead widened its mouth even further (Figure 2C and Supplementary Video 1). As such threatening behavior would not be seen by grenadiers in the darkness at 2,000 m of depth, it was probably threatening other individuals via sound or pressure waves. The Yokozuna Slickhead is presumed to be a top predator in the deepest part of Suruga Bay owing to its high trophic position (TP = 4.9), estimated from compound-specific isotope analyses, and its predation of relatively large fish (Bassozetus sp.), as inferred via stomach content analysis (Fujiwara et al., 2021a). Although slickheads are generally considered fragile with watery flesh (Childress and Nygaard, 1973), this aggressive, threatening behavior may indicate that the Yokozuna Slickhead is a top predator not only in Suruga Bay but also in this area.
The Yokozuna Slickhead body size estimated from video footage (253 cm in TL) was much larger than that from the original description (122–138 cm in TL) (Fujiwara et al., 2021a). The skin surface was bumpy, damaged, and parasite-infested, indicating that the individual was quite old (Figure 2 and Supplementary Video 1). To the best of our knowledge, there are only seven species from six families of teleost fish with a body length of 2 m or more that may be distributed deeper than 2,000 m (Figure 3) (Froese and Pauly, 2022). Among these, only two species, i.e., the Yokozuna Slickhead and the Giant Grenadier Albatrossia pectoralis, are endemic to the deep sea as adults (>98% of records of appearance were deeper than 200 m according to the Ocean Biodiversity Information System [OBIS]) (OBIS, 2022). Currently, the Yokozuna Slickhead is the largest deep-sea-endemic teleost fish at depths of over 2,000 m (Figure 3). The scavenging ability and broad gape of the Yokozuna Slickhead are thought to be correlated with its colossal body size and relatively high TP (Fujiwara et al., 2021a). The Giant Grenadier usually lives at depths greater than 400 m (Figure 4), and the trophic positions of large adult individuals are relatively high (TP up to 4.44 ± 0.12) (Chuchukalo and Napazakov, 2012). The presence of these large species implies the existence of an ecological niche of colossal predators with high trophic status even at depths greater than 2,000 m. The distributions of these two species are limited within the North Pacific, which may suggest that the deep waters of the North Pacific provide sufficient energy to meet the nutritional requirements of such predators. Apparently, there is a spatial partitioning between the two species in the North Pacific (Figure 4). The Yokozuna Slickhead is distributed at greater depths and lower latitudes than the Giant Grenadier (Figure 4). Considering this result, the Yokozuna Slickhead may be distributed further south at depths greater than 2,000 m in the North Pacific.
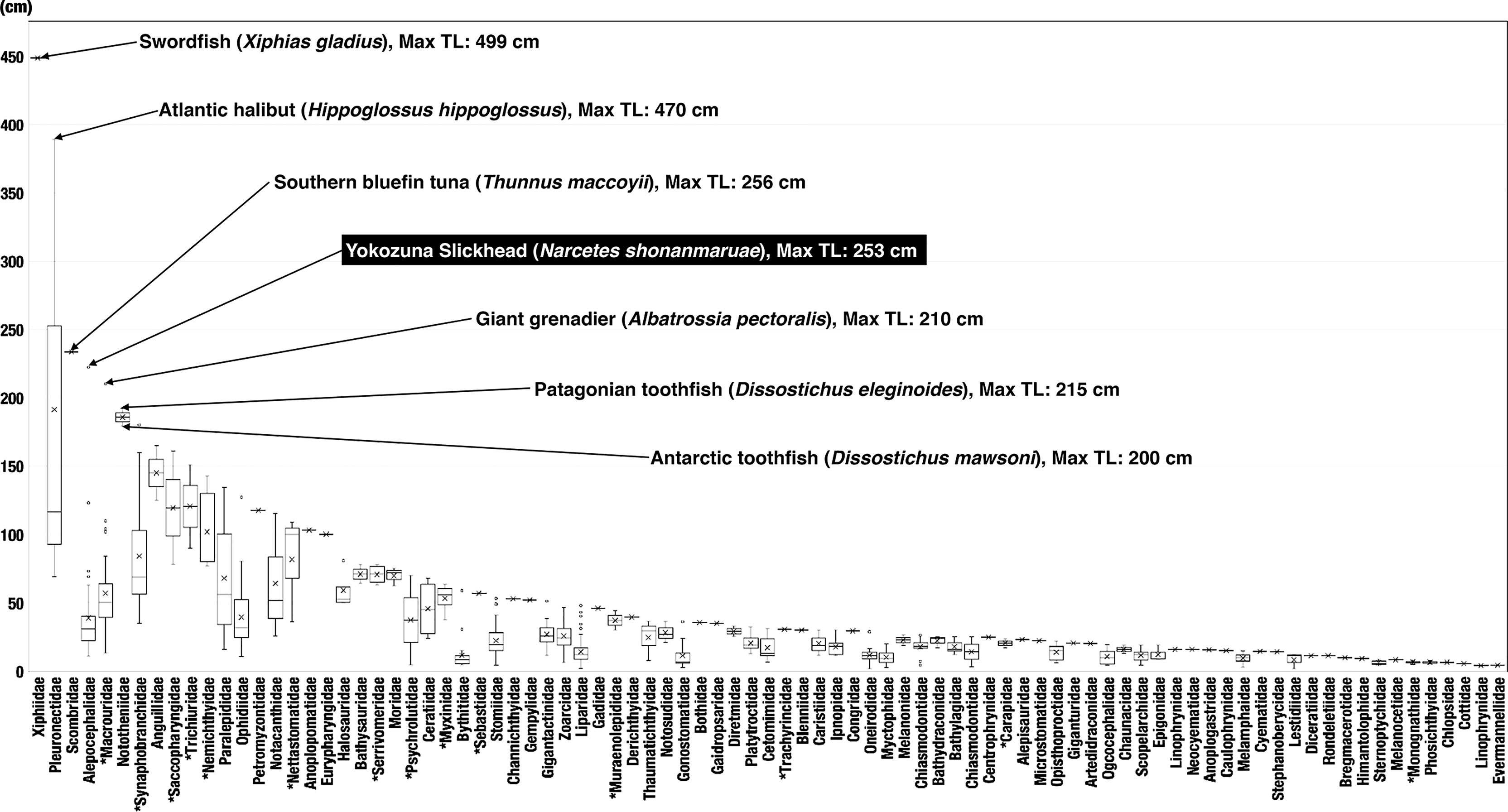
Figure 3 Box plot of maximum standard length (SL) of teleost species that live deeper than 2,000 m from each family. Species with total length (TL) over 2 m are labeled. Size information is primarily based on FishBase (www.fishbase.org). The Yokozuna Slickhead is highlighted. *families displayed in TL.
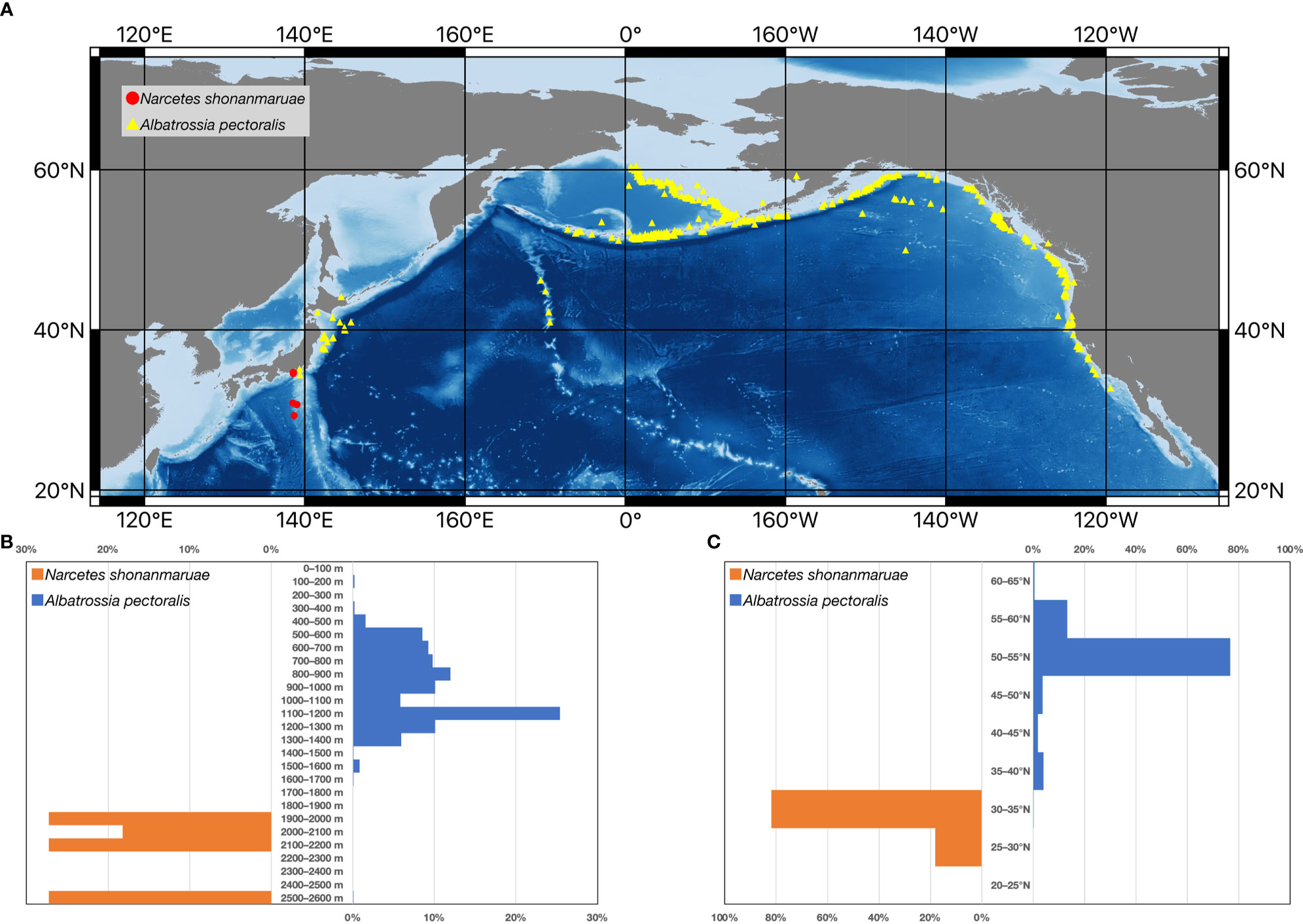
Figure 4 Distribution patterns of the Yokozuna Slickhead Narcetes shonanmaruae and the Giant Grenadier Albatrossia pectoralis. (A) Geographic distribution of the Yokozuna Slickhead and the Giant Grenadier. (B) Depth distribution of the Yokozuna Slickhead and the Giant Grenadier. (C) Latitudinal distribution of the Yokozuna Slickhead and the Giant Grenadier. The distribution information of the Giant Grenadier is derived from the Ocean Biodiversity Information System (OBIS) (OBIS, 2022).
Our understanding of deep-sea predators is limited. The Giant Grenadier exhibits a typical form of “rattail,” i.e., the caudal area is narrowed to the posterior point. The swimming speed of the Giant Grenadier was presumed to be slow (Rodgveller et al., 2017), and individuals could be caught by trawling (Napazakov and Chuchukalo, 2011; Hutchinson and Anderl, 2012). In contrast, the Yokozuna Slickhead is fusiform and has a narrowed but robust caudal peduncle with a relatively large emarginate caudal fin (Fujiwara et al., 2021a). A Yokozuna Slickhead turned vigorously with a single tail stroke and rapidly disappeared from the video frame recorded using a baited camera (Fujiwara et al., 2021a). All Yokozuna Slickheads were captured using longlines, and none have been caught in trawl nets thus far, probably owing to their high swimming ability. According to information from OBIS, there were less than 10 longline records deeper than 2,000 m and more than 1,000 trawl records at the same depth range (OBIS, 2022). Most deep-sea surveys are likely to have failed to collect large, active swimmers due to incompatibility with collection methods. Further metabarcoding and baited camera surveys, in addition to longline research, will reveal active-swimming predators lurking in the deep waters of global oceans.
In 2010, “Aichi Target 11” declared that by 2020, at least 17% of terrestrial and inland water and 10% of coastal and marine areas, especially areas of particular importance for biodiversity and ecosystem services, will be conserved through effective and equitable management, in addition to ecologically representative and well-connected systems of protected areas and other effective area-based conservation measures (OECMs). Further, these would be integrated into the wider landscapes and seascapes (Conference of the parties to the convention on biological diversity, 2010). Since 2010, over 21 million km2 have been placed within protected and conserved areas, meaning that 42% of these areas have been added within the last decade (UNEP-WCMC and IUCN, 2021). As a result, the May 2021 WDPA and WD-OECM showed that at least 22.5 million km2 (16.64%) of land and inland water ecosystems, as well as 28.1 million km2 (7.74%) of coastal waters and the ocean, are within protected areas and OECMs (UNEP-WCMC and IUCN, 2021). However, the understanding of the biodiversity in protected and conserved areas remains relatively poor, especially in offshore environments (Vad et al., 2017; Ferrari et al., 2018; Hookabe et al., 2021; Koeda et al., 2021; Hookabe et al., 2022; Jimi et al., 2022; Komai et al., 2022). In fact, all newly discovered habitats of the Yokozuna Slickhead, with the exception of the Genroku Seamount, are in the MPAs established in 2020 in Japanese waters. It is suggested that climate change should have a stronger impact on top predators, disrupting vertical interactions, and thereby affecting many species across trophic levels (Zarnetske et al., 2012). Understanding the ecology of top predators, especially in areas of particular importance for biodiversity and ecosystem services such as MPAs, is urgently needed to assess and determine the impact of rapidly changing global oceans.
Data Availability Statement
The gene sequences can be obtained from the DNA Data Bank of Japan (DDBJ) database under accession numbers LC710825–LC710836. The datasets generated and analyzed during this study are available from the corresponding author upon request.
Ethics Statement
The animal study was reviewed and approved by Research Safety Committee of the Japan Agency for Marine-Earth Science and Technology.
Author Contributions
YF and ST conceived the concept of the study. MK and TY collected DNA samples. ST and YF conducted the baited camera experiment. ST, YF, and KM developed the baited camera system. SS, TY, TS, MM, and MK conducted DNA sequencing and taxon assignment. YF wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was partly supported by the Environment Research and Technology Development Fund (JPMEERF20S20700, 20S20710, and 20S20720) of the Environmental Restoration and Conservation Agency of Japan. This cruise was partly funded by an MPA monitoring project outsourced by the Ministry of the Environment of Japan. This work was partly supported by JSPS KAKENHI Grant Number JP16H04611 for YF.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Drs. Katsunori Fujikura; Akinori Yabuki; and Yuriko Nagano (JAMSTEC), Mr. Tetsuji Maki (JAMSTEC), the captain and the crew of R/V Kaimei, the operation team of KM-ROV, and all the other participants of KM20-10C and KM21-E04C cruises for research support, Dr. Rachel Atanacio (FishBase) for providing a FishBase data set, Ms. Seiko Sano for laboratory assistance, Mr. Yoshikazu Saito for sampling information on a Yokozuna Slickhead, and Dr. Aya Yamazaki for data mining and collection. We also thank Editage (www.editage.jp) for English language editing.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.945758/full#supplementary-material
Supplementary Video 1 | Video footage of the Yokozuna Slickhead Narcetes shonanmaruae taken by a baited camera system deployed at a depth of 2,091 m south of Genroku Seamount on October 14, 2021.
References
Aoki K., Fujiwara Y., Tsuchida S. (2022). Estimating Deep-Sea Fish Population Density From the Odour Extension Area: A Theoretical Basis and Comparison With the Conventional Methods. Front. Mar. Sci. 9, 854958. doi: 10.3389/fmars.2022.854958
Bakker J., Wangensteen O. S., Chapman D. D., Boussarie G., Buddo D., Guttridge T. L., et al. (2017). Environmental DNA Reveals Tropical Shark Diversity in Contrasting Levels of Anthropogenic Impact. Sci. Rep. 7 (1), 16886. doi: 10.1038/s41598-017-17150-2
Baum J. K., Worm B. (2009). Cascading Top-Down Effects of Changing Oceanic Predator Abundances. J. Anim. Ecol. 78 (4), 699–714. doi: 10.1111/j.1365-2656.2009.01531.x
Boussarie G., Bakker J., Wangensteen O. S., Mariani S., Bonnin L., Juhel J. B., et al. (2018). Environmental DNA Illuminates the Dark Diversity of Sharks. Sci. Adv. 4 (5), eaap9661. doi: 10.1126/sciadv.aap9661
Brooks E. J., Sloman K. A., Sims D. W., Danylchuk A. J. (2011). Validating the Use of Baited Remote Underwater Video Surveys for Assessing the Diversity, Distribution and Abundance of Sharks in the Bahamas. Endanger. Species. Res. 13 (3), 231–243. doi: 10.3354/esr00331
Childress J. J., Nygaard M. H. (1973). The Chemical Composition of Midwater Fishes as a Function of Depth of Occurrence Off Southern California. Deep-Sea Res. Oceanogr. Abstr. 20 (12), 1093–1109. doi: 10.1016/0011-7471(73)90023-5
Chuchukalo V. I., Napazakov V. V. (2012). Specific Features of Feeding and Trophic Status of Mass Species of the Family Macrouridae in the Northwestern Part of the Pacific Ocean. J. Ichthyol. 52 (10), 756–781. doi: 10.1134/S0032945212100013
Conference of the parties to the convention on biological diversityDecision adopted by the conference of the parties to the convention on biological diversity at its tenth meeting: X/2 (2010) The Strategic Plan for Biodiversity 2011-2020 and the Aichi Biodiversity Targets. Available at: https://www.cbd.int/doc/decisions/cop-10/cop-10-dec-02-en.pdf.
Devine B. M., Wheeland L. J., Fisher J. A. D. (2018). First Estimates of Greenland Shark (Somniosus Microcephalus) Local Abundances in Arctic Waters. Sci. Rep. 8 (1), 974. doi: 10.1038/s41598-017-19115-x
Estes J. A., Terborgh J., Brashares J. S., Power M. E., Berger J., Bond W. J., et al. (2011). Trophic Downgrading of Planet Earth. Science 333 (6040), 301–306. doi: 10.1126/science.1205106
Ferrari R., Marzinelli E. M., Ayroza C. R., Jordan A., Figueira W. F., Byrne M., et al. (2018). Large-Scale Assessment of Benthic Communities Across Multiple Marine Protected Areas Using an Autonomous Underwater Vehicle. PLoS One 13 (3), e0193711. doi: 10.1371/journal.pone.0193711
Froese R., Pauly D. (2022) FishBase. Available at: www.fishbase.org (Accessed April 7 2022).
Fujiwara Y., Kawato M., Poulsen J. Y., Ida H., Chikaraishi Y., Ohkouchi N., et al. (2021a). Discovery of a Colossal Slickhead (Alepocephaliformes: Alepocephalidae): An Active-Swimming Top Predator in the Deep Waters of Suruga Bay, Japan. Sci. Rep. 11 (1) 2490. doi: 10.1038/s41598-020-80203-6
Fujiwara Y., Matsumoto Y., Sato T., Kawato M., Tsuchida S. (2021b). First Record of Swimming Speed of the Pacific Sleeper Shark Somniosus Pacificus Using a Baited Camera Array. J. Mar. Biol. Assoc. U. K. 101 (2), 457–464. doi: 10.1017/S0025315421000321
Fukumoto S., Ushimaru A., Minamoto T. (2015). A Basin-Scale Application of Environmental DNA Assessment for Rare Endemic Species and Closely Related Exotic Species in Rivers: A Case Study of Giant Salamanders in Japan. J. Appl. Ecol. 52 (2), 358–365. doi: 10.1111/1365-2664.12392
Hookabe N., Jimi N., Yokooka H., Tsuchida S., Fujiwara Y. (2021). Lacydonia Shohoensis (Annelida, Lacydoniidae) Sp. Nov. – a New Lacydonid Species From Deep-Sea Sunken Wood Discovered at the Nishi-Shichito Ridge, North-Western Pacific Ocean. J. Mar. Biol. Assoc. U. K. 101 (6), 927–933. doi: 10.1017/S0025315421000862
Hookabe N., Kajihara H., Chernyshev A. V., Jimi N., Hasegawa N., Kohtsuka H., et al. (2022). Molecular Phylogeny of the Genus Nipponnemertes (Nemertea: Monostilifera: Cratenemertidae) and Descriptions of 10 New Species, With Notes on Small Body Size in a Newly Discovered Clade. Front. Mar. Sci. doi: 10.3389/fmars.2022.906383
Hutchinson C. E., Anderl D. M. (2012). Giant Grenadier (Albatrossia Pectoralis) (Seattle: noaa fisheries). Available at: http://www.afsc.noaa.gov/REFM/Age/default.htm (Accessed April 29 2022).
Ip Y. C. A., Chang J. J. M., Lim K. K. P., Jaafar Z., Wainwright B. J., Huang D. (2021). Seeing Through Sedimented Waters: Environmental DNA Reduces the Phantom Diversity of Sharks and Rays in Turbid Marine Habitats. BMC Ecol. Evol. 21 (1), 166. doi: 10.1186/s12862-021-01895-6
Jerde C. L., Mahon A. R., Chadderton W. L., Lodge D. M. (2011). Sight-Unseen” Detection of Rare Aquatic Species Using Environmental DNA. Conserv. Lett. 4 (2), 150–157. doi: 10.1111/j.1755-263X.2010.00158.x
Jimi N., Chen C., Fujiwara Y. (2022). Two New Species of Branchinotogluma (Polynoidae: Annelida) From Chemosynthesis-Based Ecosystems in Japan. Zootaxa 5138 (1), 17–30. doi: 10.11646/zootaxa.5138.1.2
Jones M. R. L., Breen B. B. (2013). Food and Feeding Relationships of Three Sympatric Slickhead Species (Pisces: Alepocephalidae) From Northeastern Chatham Rise, New Zealand. Deep-Sea. Res. I.: Oceanogr. Res. Pap. 79, 1–9. doi: 10.1016/j.dsr.2013.04.011
Kawato M., Yoshida T., Miya M., Tsuchida S., Nagano Y., Nomura M., et al. (2021). Optimization of Environmental DNA Extraction and Amplification Methods for Metabarcoding of Deep-Sea Fish. MethodsX 8, 101238. doi: 10.1016/j.mex.2021.101238
Koeda K., Takashima S., Yamakita T., Tsuchida S., Fujiwara Y. (2021). Deep-Sea Fish Fauna on the Seamounts of Southern Japan With Taxonomic Notes on the Observed Species. J. Mar. Sci. Eng. 9 (11), 1294. doi: 10.3390/jmse9111294
Komai T., Tsuchida S., Fujiwara Y. (2022). New Record of a Rarely Collected Caridean Shrimp Bathypalaemonella Pandaloides (Rathbun 1906) (Decapoda: Bathypalaemonellidae) From the West Mariana Ridge, Northwestern Pacific. Zootaxa 5129 (2), 272–284. doi: 10.11646/zootaxa.5129.2.7
Lafferty K. D., Benesh K. C., Mahon A. R., Jerde C. L., Lowe C. G. (2018). Detecting Southern California’s White Sharks With Environmental DNA. Front. Mar. Sci. 5. doi: 10.3389/fmars.2018.00355
Levin L. A., Le Bris N. (2015). The Deep Ocean Under Climate Change. Science 350 (6262), 766–768. doi: 10.1126/science.aad0126
Miya M. (2022). Environmental DNA Metabarcoding: A Novel Method for Biodiversity Monitoring of Marine Fish Communities. Ann. Rev. Mar. Sci. 14 (1), 161–185. doi: 10.1146/annurev-marine-041421-082251
Miya M., Minamoto T., Yamanaka H., Oka S.-i., Sato K., AYamamoto S., et al. (2016). Use of a Filter Cartridge for Filtration of Water Samples and Extraction of Environmental DNA. JoVE (117), e54741. doi: 10.3791/54741
Miya M., Sato Y., Fukunaga T., Sado T., Poulsen J. Y., Sato K., et al. (2015). MiFish, a Set of Universal PCR Primers for Metabarcoding Environmental DNA From Fishes: Detection of More Than 230 Subtropical Marine Species. R. Soc Open Sci. 2 (7), 150088. doi: 10.1098/rsos.150088
Murakami H., Yoon S., Kasai A., Minamoto T., Yamamoto S., Sakata M. K., et al. (2019). Dispersion and Degradation of Environmental DNA From Caged Fish in a Marine Environment. Fish. Sci. 85 (2), 327–337. doi: 10.1007/s12562-018-1282-6
Myers R. A., Baum J. K., Shepherd T. D., Powers S. P., Peterson C. H. (2007). Cascading Effects of the Loss of Apex Predatory Sharks From a Coastal Ocean. Science 315 (5820), 1846–1850. doi: 10.1126/science.1138657
Napazakov V. V., Chuchukalo V. I. (2011). Feeding of Albatrossia Pectoralis (Macrouridae) on the Continental Slope of Eastern Kamchatka and the Kurils. J. Ichthyol. 51 (4), 343–351. doi: 10.1134/S0032945211030052
OBIS (2022) Ocean Biodiversity Information System (Intergovernmental Oceanographic Commission of UNESCO). Available at: www.obis.org (Accessed March 28 2022).
Oka S.-I., Doi H., Miyamoto K., Hanahara N., Sado T., Miya M. (2021). Environmental DNA Metabarcoding for Biodiversity Monitoring of a Highly Diverse Tropical Fish Community in a Coral Reef Lagoon: Estimation of Species Richness and Detection of Habitat Segregation. Environ. DNA 3 (1), 55–69. doi: 10.1002/edn3.132
Pakhorukov N. P., Parin N. V. (2012). Visual Observations of Fish at the Whale Ridge (Atlantic Ocean) From the Sever-2 Manned Underwater Vehicle. J. Ichthyol. 52 (9), 579–591. doi: 10.1134/S0032945212050050
Postaire B. D., Bakker J., Gardiner J., Wiley T. R., Chapman D. D. (2020). Environmental DNA Detection Tracks Established Seasonal Occurrence of Blacktip Sharks (Carcharhinus Limbatus) in a Semi-Enclosed Subtropical Bay. Sci. Rep. 10 (1), 11847. doi: 10.1038/s41598-020-68843-0
Priede I. G., Godbold J. A., King N. J., Collins M. A., Bailey D. M., Gordon J. D. M. (2010). Deep-Sea Demersal Fish Species Richness in the Porcupine Seabight, NE Atlantic Ocean: Global and Regional Patterns. Mar. Ecol. 31 (1), 247–260. doi: 10.1111/j.1439-0485.2009.00330.x
Ripple W. J., Beschta R. L. (2012). Trophic Cascades in Yellowstone: The First 15 Years After Wolf Reintroduction. Biol. Conserv. 145 (1), 205–213. doi: 10.1016/j.biocon.2011.11.005
Rodgveller C. J., Hutchinson C. E., Harris J. P., Vulstek S. C., Guthrie C. M. (2017). Otolith Shape Variability and Associated Body Growth Differences in Giant Grenadier, Albatrossia Pectoralis. PLoS One 12 (6), e0180020. doi: 10.1371/journal.pone.0180020
Roff G., Doropoulos C., Rogers A., Bozec Y. M., Krueck N. C., Aurellado E., et al. (2016). The Ecological Role of Sharks on Coral Reefs. Trends Ecol. Evol. 31 (5), 395–407. doi: 10.1016/j.tree.2016.02.014
Sakaue J., Maeda K., Miller M. J., Sakai R., Tahara K.-i., Abe H., et al. (2021). New Insights About the Behavioral Ecology of the Coelacanth Latimeria Chalumnae Video Recorded in the Absence of Humans Off South Africa. Front. Mar. Sci. 8. doi: 10.3389/fmars.2021.755275
Schmitz O. J. (2007). Predator Diversity and Trophic Interactions. Ecology 88 (10), 2415–2426. doi: 10.1890/06-0937.1
Sergio F., Newton I., Marchesi L. (2005). Conservation: Top Predators and Biodiversity. Nature 436 (7048), 192. doi: 10.1038/436192a
Stoner A. W., Laurel B. J., Hurst T. P. (2008). Using a Baited Camera to Assess Relative Abundance of Juvenile Pacific Cod: Field and Laboratory Trials. J. Exp. Mar. Biol. Ecol. 354 (2), 202–211. doi: 10.1016/j.jembe.2007.11.008
Tecchio S., Coll M., Christensen V., Company J. B., Ramírez-Llodra E., Sardà F. (2013). Food Web Structure and Vulnerability of a Deep-Sea Ecosystem in the NW Mediterranean Sea. Deep-Sea. Res. Pt. I. 75, 1–15. doi: 10.1016/j.dsr.2013.01.003
Thomsen P. F., Willerslev E. (2015). Environmental DNA – an Emerging Tool in Conservation for Monitoring Past and Present Biodiversity. Biol. Conserv. 183, 4–18. doi: 10.1016/j.biocon.2014.11.019
Trenkel V. M., Francis R. I. C. C., Lorance P., Mahévas S., Rochet M.-J., Tracey D. M. (2004). Availability of Deep-Water Fish to Trawling and Visual Observation From a Remotely Operated Vehicle (ROV). Mar. Ecol. Prog. Ser. 284, 293–303. doi: 10.3354/meps284293
UNEP-WCMC, and IUCN (2021) Protected Planet Report 2020 (Cambridge UK; Gland, Switzerland: UNEP-WCMC and IUCN). Available at: https://livereport.protectedplanet.net (Accessed April 20 2022).
Vad J., Orejas C., Moreno-Navas J., Findlay H. S., Roberts J. M. (2017). Assessing the Living and Dead Proportions of Cold-Water Coral Colonies: Implications for Deep-Water Marine Protected Area Monitoring in a Changing Ocean. PeerJ 5, e3705. doi: 10.7717/peerj.3705
Wallach A. D., Ripple W. J., Carroll S. P. (2015). Novel Trophic Cascades: Apex Predators Enable Coexistence. Trends Ecol. Evol. 30 (3), 146–153. doi: 10.1016/j.tree.2015.01.003
Weltz K., Lyle J. M., Ovenden J., Morgan J. A. T., Moreno D. A., Semmens J. M. (2017). Application of Environmental DNA to Detect an Endangered Marine Skate Species in the Wild. PLoS One 12 (6), e0178124. doi: 10.1371/journal.pone.0178124
Wilcox T. M., McKelvey K. S., Young M. K., Jane S. F., Lowe W. H., Whiteley A. R., et al. (2013). Robust Detection of Rare Species Using Environmental DNA: The Importance of Primer Specificity. PLoS One 8 (3), e59520. doi: 10.1371/journal.pone.0059520
Yamamoto S., Masuda R., Sato Y., Sado T., Araki H., Kondoh M., et al. (2017). Environmental DNA Metabarcoding Reveals Local Fish Communities in a Species-Rich Coastal Sea. Sci. Rep. 7, 40368. doi: 10.1038/srep40368
Zarnetske P. L., Skelly D. K., Urban M. C. (2012). Biotic Multipliers of Climate Change. Science 336 (6088), 1516–1518. doi: 10.1126/science.1222732
Keywords: eDNA metabarcoding, baited camera, rare species, top predator, Yokozuna Slickhead, marine protected area, Nishi-Shichito Ridge, MiFish primer
Citation: Fujiwara Y, Tsuchida S, Kawato M, Masuda K, Sakaguchi SO, Sado T, Miya M and Yoshida T (2022) Detection of the Largest Deep-Sea-Endemic Teleost Fish at Depths of Over 2,000 m Through a Combination of eDNA Metabarcoding and Baited Camera Observations. Front. Mar. Sci. 9:945758. doi: 10.3389/fmars.2022.945758
Received: 16 May 2022; Accepted: 01 June 2022;
Published: 01 July 2022.
Edited by:
Clara F. Rodrigues, University of Aveiro, PortugalReviewed by:
Jon Thomassen Hestetun, Norwegian Research Institute (NORCE), NorwayHiroshi Yamasaki, Kyushu University, Japan
Copyright © 2022 Fujiwara, Tsuchida, Kawato, Masuda, Sakaguchi, Sado, Miya and Yoshida. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yoshihiro Fujiwara, fujiwara@jamstec.go.jp
 Yoshihiro Fujiwara
Yoshihiro Fujiwara Shinji Tsuchida
Shinji Tsuchida Masaru Kawato
Masaru Kawato Kotohiro Masuda
Kotohiro Masuda Sakiko Orui Sakaguchi
Sakiko Orui Sakaguchi Tetsuya Sado
Tetsuya Sado Masaki Miya
Masaki Miya Takao Yoshida
Takao Yoshida