Understanding the effects of salinity and Vibrio harveyi on the gut microbiota profiles of Litopenaeus vannamei
- 1Institute of Marine Biotechnology, Universiti Malaysia Terengganu, Kuala Nerus, Malaysia
- 2Faculty of Fisheries and Food Science, Universiti Malaysia Terengganu, Kuala Nerus, Malaysia
- 3GeneSEQ Sdn Bhd, Selangor Darul Ehsan, Malaysia
- 4Institute of Tropical Aquaculture and Fisheries Research, Universiti Malaysia Terengganu, Kuala Nerus, Malaysia
- 5Department of Marine Bioresource Science, Faculty of Fisheries, Chattogram Veterinary and Animal Sciences University, Chattogram, Bangladesh
- 6College of Marine Life Sciences, Frontiers Science Center for Deep Ocean Multispheres and Earth System, Institute of Evolution and Marine Biodiversity, Ocean University of China, Qingdao, China
- 7UMT-OUC Joint Academic Center for Marine Studies, Universiti Malaysia Terengganu, Kuala Nerus, Malaysia
Increasing evidence have revealed a positive correlation between gut microbiota and shrimp health, in which a healthy shrimp gut consists of a complex and stable microbial community. Given that both abiotic and biotic factors constantly regulate shrimp gut microbiota, any changes affecting the levels of these factors could cause modification to the gut microbiota assemblage. The goal of this study was to explore the effects of salinity levels and pathogenic Vibrio harveyi infection on the diversity, structure, composition, interspecies interaction, and functional pathways of Litopenaeus vannamei gut microbiota. Juvenile shrimp were cultured at 5 ppt, 20 ppt, and 30 ppt for two months prior to Vibrio harveyi infection. After pathogenic V. harveyi challenge test, genomic DNA was isolated from the shrimp gut, and subjected to the 16S rRNA metagenomic sequencing analysis. We observed that gut microbiota diversity of shrimp cultured at 5 ppt and 30 ppt were lower than those cultured at 20 ppt after exposure to V. harveyi infection, suggesting that shrimp cultured at the two former salinity levels were more susceptible to V. harveyi infection. Network analysis also showed that shrimp cultured at 20 ppt exhibit a more stable bacterial network with complex interspecies interaction, even after induced by V. harveyi. Moreover, the presence of a high number of beneficial bacteria such as Pseudoruegeria, Rhodovulum, Ruegeria, Shimia and Lactobacillus in shrimp cultured at 20 ppt might have played a role in inhibiting the growth of V. harveyi and other potentially pathogenic bacteria. Besides, bacterial functional pathway prediction has also shown that metabolic pathways such as phenylalanine metabolism, glycine, serine and threonine metabolism, starch and sucrose metabolism, glyoxylate and dicarboxylate metabolism, carbon metabolism and biofilm formation process were significantly higher in shrimp cultured at 20 ppt. Collectively, our results suggested that 20 ppt is an optimal salinity that suppresses the growth of V. harveyi and potential pathogenic bacteria in the shrimp gut, which could possibly minimize the risk of pathogenic infection for sustainable production of healthy shrimp.
Introduction
The Pacific white shrimp Litopenaeus vannamei, an endemic to the intertropical Pacific coast of South America, is amongst the most extensively cultured crustacean species in the world due to its fast growth, high yield, euryhaline nature and high tolerance to high-density farming condition (Panigrahi et al., 2018; Araneda et al., 2020; Soares et al., 2021). Like other aquaculture species, L. vannamei is also vulnerable to the global climate change impacts, which include the more frequent occurrence of extreme weather events resulting in drastic fluctuations of abiotic factors levels such as pH, carbon dioxide (CO2), dissolved oxygen (DO), water temperature and salinity (Kathyayani et al., 2019; Liang et al., 2019; AghaKouchak et al., 2020). The rapid alterations to these abiotic factors may result in variations in viral and bacterial loads, causing severe physiological stress to cultured species, leading to disease outbreaks and devastating economic losses in the shrimp farming industry (Huang et al., 2020; Millard et al., 2021). Although L. vannamei is adaptable to a wide range of salinity levels (0.5 ppt to 40 ppt), climatic effects such as heavy rainfall and large culture water changes during farming activities can cause rapid changes to the salinity levels, leading to osmotic imbalance, metabolic disorders, and suppressed immunity of the shrimp (Saoud et al., 2003; Li et al., 2008; Cheng and Chen, 2000; Bauer et al., 2020).
Gut microbiota is known as a metabolically active “organ”, since bacterial metabolism in the gut is responsible for most of the gut functions such as improving immune response, nutrient absorption, intestinal tract homeostasis, physiological regulation, and pathogen defense (Hou et al., 2020; Qu et al., 2021; Huang et al., 2022). Extensive research has revealed that abiotic variables, food composition, trophic levels, and developmental stages have a profound effect on the gut microbiota of aquatic animals (Xiong et al., 2017; Cornejo-Granados et al., 2018; Huang et al., 2018; Lim et al., 2019; Wilkes-Walburn et al., 2019; Hou et al., 2020; Xue et al., 2020). Collectively, salinity was observed to be the principal abiotic factor influencing the structure and function of ambient and host gut microbiota in the shrimp aquaculture system (Hou et al., 2017). Salinity stress is thought to have a deleterious effect on the stability of the environmental microbiome by enhancing potential pathogens and modifying the ambient microbial populations (Xiong et al., 2017). Undeniably, dysbiosis or shifts in the host gut microbiota are often closely linked with the occurrence of various bacterial diseases in L. vannamei (Dai et al., 2020; Huang et al., 2020; Zeng et al., 2020). An environmentally abundant pathogen may reach a distressed shrimp gut through the oral route, resulting in indirect alteration of shrimp gut microbiota (Rungrassamee et al., 2016; Dong et al., 2021; Prathiviraj et al., 2021). Notably, maintaining the population balance and resilience of gut microbiota against pathogenic organisms is crucial to reduce the severity of the disease outbreak within the host.
Recently, high-throughput and cost-effective 16S rRNA metagenomic sequence analysis has been widely employed to examine the bacterial communities of various environments and biological hosts to provide a deeper insight into the microflora profiles of the targeted samples (Deng et al., 2020; Chaiyapechara et al., 2022). In fact, the outbreaks of Vibrio-associated disease in aquatic species are closely linked with the presence of opportunistic Vibrio pathogens, dysbiosis of host gut microbiota, and abrupt changes in the salinity levels (Deng et al., 2020; Hou et al., 2020; Xue et al., 2020; Dong et al., 2021). Understanding the relationship between salinity level and shrimp gut microbiota assemblages will extend our knowledge of shrimp immune-physiological responses toward pathogenic bacterial infection. In the present study, we used 16S rRNA metagenomic analysis to comprehensively compare the structure and function of core bacterial signatures in the L. vannamei guts cultured under different salinity levels (5 ppt, 20 ppt, and 30 ppt) and challenged with pathogenic Vibrio harveyi, with the following questions: i) What are the differences in core bacterial communities in shrimp gut cultured at different salinity levels, with or without exposure to the pathogenic V. harveyi? (ii) What ecological processes that shape the microbiota assemblage of shrimp gut cultured at different salinity levels? (iii) How does interspecies interaction and functions of the shrimp gut microbiota affect shrimp’s susceptibility towards V. harveyi infection? Despite the fact that actual climatic change would not cause rapid and extreme fluctuations in salinity levels of more than 25 ppt, the vast salinity gradients (5 ppt, 20 ppt, 30 ppt) in the present study were selected to determine shrimp physiological tolerance in environment of different salinity levels, via gut microbiota assemblage. Being the most common salinity level used in shrimp farms within the Southeast Asian regions, 30 ppt was selected as the control salinity in the present study. The other two salinity levels experimented in this study were 5 ppt (oligohaline/low salinity) and 20 ppt (lower polyhaline/medium salinity), which were reported to modulate shrimp growth and innate immunity (Lin et al., 2012; Chen et al., 2015; Gao et al., 2016; Huang et al., 2022). We also provide the first effort to determine the combined effects of both abiotic (salinity) and biotic (V. harveyi) factors on the L. vannamei gut microbiota signatures. Our findings will serve as a starting point for the study of other abiotic and biotic factors affecting shrimp gut microbiota, as well as a stepping-stone towards developing microbe-based therapeutics for pre-emptive control of shrimp disease.
Methodology
Animal and experimental design
Juveniles of Litopenaeus vannamei with an average of 2 g of body weight were obtained from a commercial farm and were acclimatized for a week in six tanks (500 L) at a density of 240 shrimp per tank in the hatchery of the Institute of Aquaculture Tropical and Fisheries Sciences (AKUATROP), Universiti Malaysia Terengganu (UMT). The shrimp were acclimated to 5 ppt, 20 ppt and 30 ppt (as a control salinity level similar to those maintained in the commercial farm) through a daily decrement of 1.5 ppt prior to a two-month cultivation period. The shrimp were fed a commercial diet (Protein: 45% and Fat: 6%; Cargill, USA) for three times daily based on 10% of their body weight. To maintain good water quality, the feed leftover were removed once in a day using sterile syphoning method, while 50% of the shrimp culture water were changed once in every three days. Water quality parameters were recorded twice a week and maintained at a temperature of 26-28°C, pH 7.5-7.9, temperature of 26–28°C, dissolved oxygen of 4.8–6.4 mg L-1, and total ammonia nitrogen of < 0.02 mg L-1.
Pathogenic Vibrio harveyi challenge test
Vibrio harveyi strain (BB120) which was isolated from diseased L. vannamei was obtained from the Institute of Marine Biotechnology, UMT and cultured for 24 hours (h) at 35°C on tryptic soy agar (TSA; Miller) with 1.5% sodium chloride (NaCl). Bacterial colonies were then grown to stationary phase at 35°C for 24 h in tryptic soy broth (TSB) containing 1.5 percent NaCl. After being harvested by centrifugation at 10,000 x g for 15 minutes, Vibrio harveyi cells were rinsed with phosphate-buffered saline (PBS) and then resuspended with the rearing water of L. vannamei. Prior to pathogenic challenge test, the growth curve, pathogenicity test and species confirmation were conducted on V. harveyi. The density of V. harveyi was determined using spectrophotometer (Shimadzu, Japan). Each salinity treatments were divided into two groups, notably the control/non-infected (C) and V. harveyi infected groups (VH), each consisting of three replicates (N=3). The pathogenic challenge test used approximately 2.7 x 107 CFU mL-1 pure culture of V. harveyi. The immersion method was applied to mimic the natural bacterial infection route (Soto-Rodriguez et al., 2015), where L. vannamei individuals (VH group) were immersed in 500 L of culture water containing 50 mL of 2.7 x 107 CFU mL-1 of the bacterial suspension for 1.5 h before they were transferred back to experimental tanks. Shrimp survivability was recorded, and shrimp individuals were collected in triplicates following pathogenic challenge test prior to 16S rRNA metagenomic analysis. The shrimp gut was aseptically dissected and stored at -80°C freezer in a sterile tube until microbial genomic DNA extraction. All the experimental procedures followed the guidelines approved by the Biosecurity and Ethic Committee of Universiti Malaysia Terengganu.
Microbial genomic DNA extraction, library preparation and 16S rRNA metagenomic sequencing
Following the manufacturer’s protocols, Wizard Genomic DNA Purification Kit (Promega, USA) was used to extract the total genomic DNA of L. vannamei from whole gut tissues, and their quality and quantity were accessed using BioDrop Fluorometer (BioDrop, UK) and 1.5% agarose gel. Then, a Qubit 2.0 Fluorometer (Invitrogen, USA) was used to quantify the integrity of DNA samples. The high-quality DNA was used for library construction using the MetaVx™ Library Preparation kit (GENEWIZ, USA). A specific primer set (Forward: 5’-CCTACGGRRBGCASCAGKVRVGAAT-3’ and Reverse: 5’-GGACTACNVGGGTWTCTAATCC-3’) was used to amplify the hypervariable V3-V4 region of the bacterial 16S rRNA gene (Wang et al., 2020). The total PCR mixture volume was 25μl, containing TransStart Taq DNA polymerase (0.5μL), TransStart buffer (2.5μL), dNTPs (2μL), primers (1μL each primer) and template DNA (20ng). PCR was programmed into an initial denaturation for 3 min at 94°C; 24 cycles of denaturation for 5 s at 95°C, annealing 90 s at 57°C, and extension for 10 s at 72°C; and final extension for 10 min at 72°C. Indexed adapters were added to the ends of the amplicons, and the libraries were purified using magnetic beads. Clusters were generated using purified libraries, and the Illumina Miseq platform (Illumina, USA) was used to generate 250 bp paired-end. The sequencing data obtained from this study were submitted to the National Center for Biotechnology Information (NCBI) database (Accession number: PRJNA815689).
16S rRNA metagenomic data analysis
The paired-end sequences were merged and overlapped using fastp v0.21 (Chen et al., 2018). Demultiplexing, sequence primers, barcodes and adaptor were trimmed using cutadapt v1.18 (Martin, 2011). The demultiplexed and trimmed reads were processed with QIIME2 v.2021.4 (Bolyen et al., 2019). Briefly, the sequences were denoised using dada2 (Callahan et al., 2016) within QIIME2 v.2021.4. Taxonomic assignments of the amplicon sequence variances (ASV) using q2-feature-classifier (Bokulich et al., 2018) was mapped on the latest Genome Taxonomy Database (GTDB) (Parks et al., 2020). ASVs with taxonomic assignments were selected for subsequent analyses. Both ASV tables and taxonomic classification tables were imported into MicrobiomeAnalyst (Chong et al., 2020) to generate α- and β-diversity, taxonomic bar plot, LEfSe (Linear Discriminant Analysis (LDA >2.0) Effect size), and correlation network analysis.
The α-diversity indices (Shannon index, Chao1 index, Good’s coverage, Rarefaction curve, and Observe OTUs) were calculated. β-diversity based on Bray-Curtis dissimilarity were statistically calculated using permutational multivariate analysis of variance (PERMANOVA) with a significant value of 0.05 in MicrobiomeAnalyst (Chong et al., 2020). Bray-Curtis dissimilarity was used to compare the similarities of bacterial community between control (C) and VH groups. Principal Coordinates Analysis (PCoA) based on Bray-Curtis dissimilarity was applied to visualize the variation and dissimilarity in bacterial structure between i) the control (C) and Vibrio harveyi infected (VH) groups and ii) three different salinity levels (5 ppt, 20 ppt and 30 ppt). Correlation network analysis was conducted using Sparse Correlation Compositional (SparCC) at significant value of P <0.05 to identify potential interactions between the bacterial community in C and VH group (Faust et al., 2012; Friedman and Alm, 2012). Heatmap for core bacterial community within the group was generated based on clusters with an accumulated relative abundance of more than 1% and sample prevalence of more than 20% was included. Further, Canonical Correspondence Analysis (CCA) was conducted to measure the relationship between environmental factor (salinity), shrimp growth increment and bacterial community in shrimp gut using RStudio 4.2.1 (Mantel test; P<0.05) (Oksanen et al., 2018). t-test was performed to compare the growth increment of shrimp cultured at each different salinity level (5 ppt, 20 ppt, 30 ppt).
The functional pathways of L. vannamei gut microbiota were predicted from 16S rRNA metagenomic sequences using PICRUSt. Similar set of paired-end sequences which were analyzed using MicrobiomeAnalyst, were used to determine the functional pathways in the EzBioCloud pipeline (ChunLab. Inc., Seoul, Republic of Korea; Yoon et al., 2017; EzBioCloud Database Update 2021.07.07). The functional prediction pathways were then annotated to the Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway (www.genome.jp/kegg/pathway; Kanehisa et al., 2014) and was compared between C and VH groups.
Result
Microbiota diversity of Litopenaeus vannamei cultured at different salinity levels followed by exposure to Vibrio harveyi infection
A total of 201,405 high-quality sequences were retained after quality check and removal of chimeric sequences. The rarefaction curve based on Good coverage (Supplementary Figure S1) indicated that the number of sequences per sample with a range of 6,000 to 12,500 sequence samples size was sufficient to capture the α-diversity of microbial communities in both treatment groups [Vibrio harveyi infected (VH) and control (C)] and a total of 344 OTUs were identified in this study. α-diversity indices, including Shannon’s index, Chao 1’s index and observed OTUs showed higher diversity in 20VH than 5VH and 30VH respectively, following V. harveyi infection (Figure 1). However, the α-diversity was found to be higher in shrimp individuals cultured at 30 ppt, than 5 ppt and 20 ppt (Supplementary Figure S2). The PCoA plot relying on Bray-Curtis dissimilarity displayed a clear discrimination in gut bacterial community composition of L. vannamei between the C and VH groups, and also at different salinity levels (F-value: 2.49; R-squared: 0.51; P <0.001) (Figure 2A. PERMANOVA indicated that the bacterial community composition of the gut L. vannamei was significantly different between the VH and C groups (F-value: 2.46; R-squared: 0.13; P <0.015) (Figure 2B, suggesting that the presence of pathogenic V. harveyi has altered the gut bacterial community composition of L. vannamei regardless of different salinity levels.
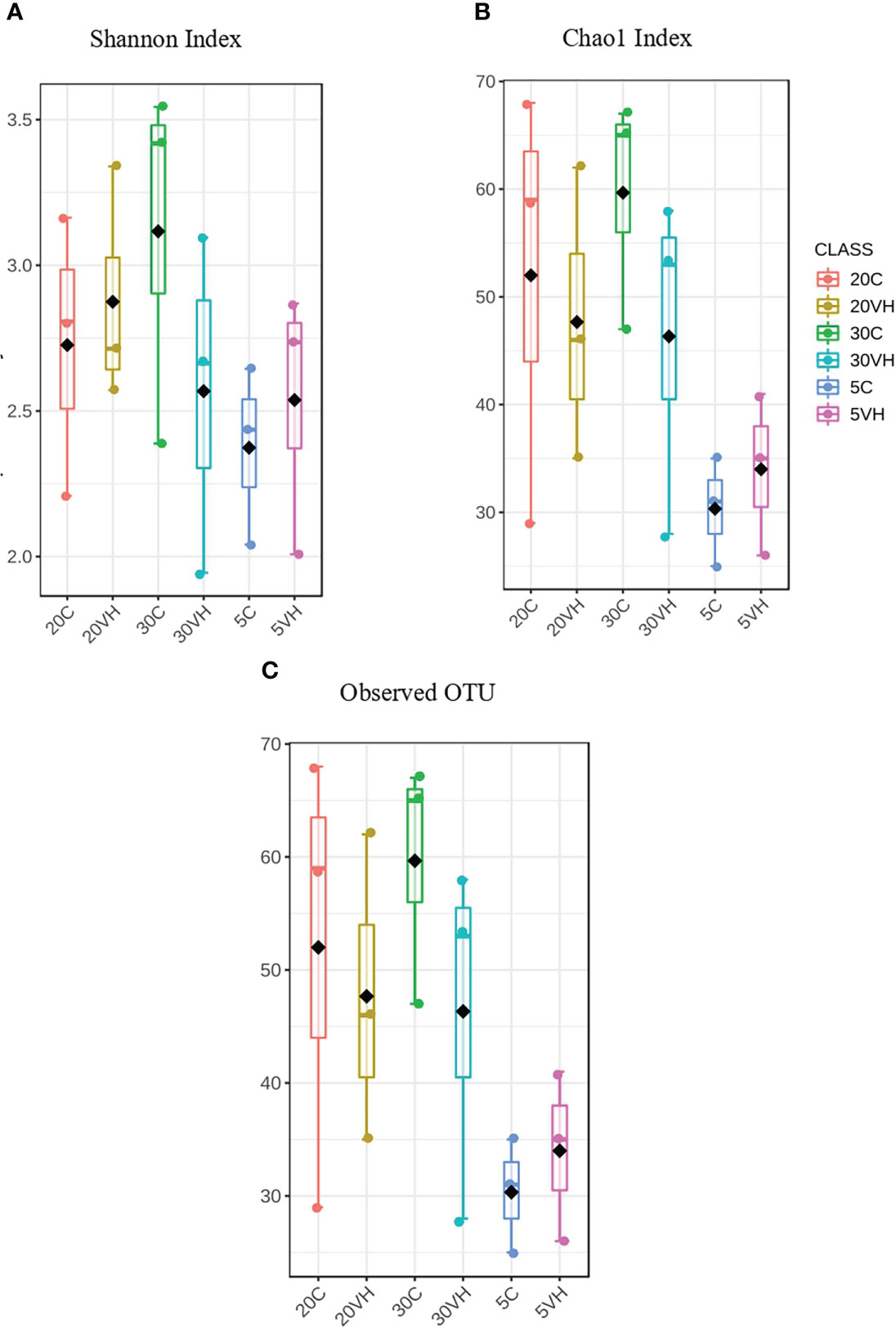
Figure 1 The α-diversity analysis of each group treatments of Litopenaues vannamei cultured at different salinity level and challenged with Vibrio harveyi (VH, Vibrio harveyi infected group and C, Control group; 5, 5 ppt; 20, 20 ppt and 30, 30 ppt). (A) Shannon index box plot and (B) Chao 1 index box plot and (B) Observed OTUs box plot.
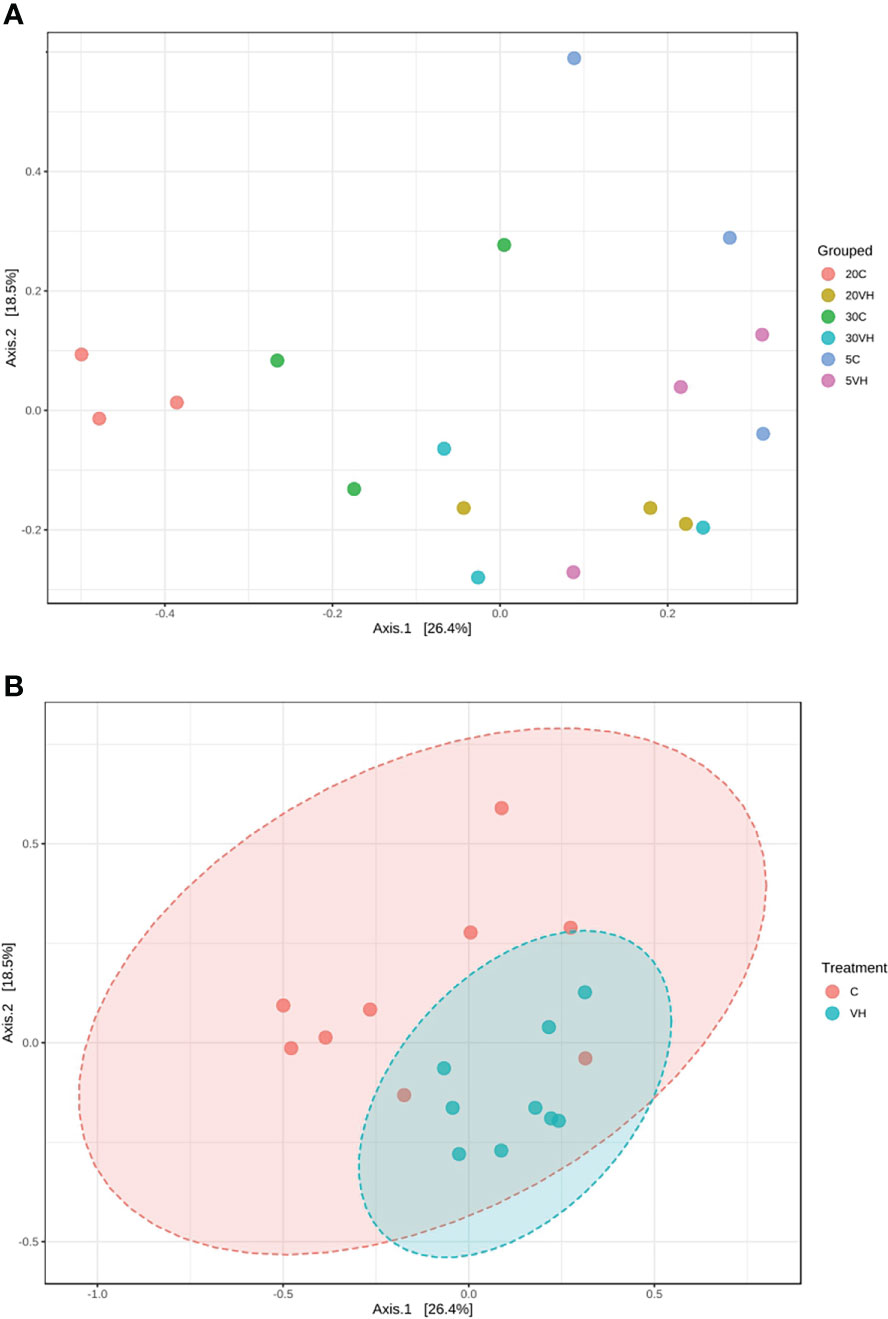
Figure 2 Principal co-ordinates analysis (PCoA) plot based on the Bray-Curtis dissimilarities of bacterial community structures of the Litopenaues vannamei cultured at different salinity levels and challenged with Vibrio harveyi, (A) Comparison of bacterial community between all treatment groups and (B) Comparison of bacterial community between C and VH group. (VH, Vibrio harveyi infected group and C, Control group; 5, 5 ppt; 20, 20 ppt and 30, 30 ppt).
Comparison of gut microbiota composition between two groups of L. vannamei (C and VH) cultured at different salinity levels
To compare the gut microbiota composition of L. vannamei (C and VH), we assessed their bacterial composition profiles at the phylum, family and genus levels. At phylum level, Proteobacteria (5C: 78.98%, 20C: 78.9% and 30C: 78.38%), Firmicutes (5C: 9.85%, 20C: 5.08% and 30C: 3.57%), and Bacterioidota (5C: 5.19%, 20C: 11.53%, and 30C: 14.54%) were the dominant phyla in all C groups cultured at different salinity levels (Figure 3). In the presence of pathogenic V. harveyi, Fusobacteriota (5VH: 4.53% and 20VH: 6.95%) emerged as the third most dominant phylum in 5VH and 20VH after Proteobacteria (5VH: 72.87% and 20VH: 62.51%) and Firmicutes (5VH: 18.28% and 20VH: 26.86%), while 30VH showed dominance profile (Proteobacteria 79.56%, Firmicutes 14.22% and Bacteriodata 5.67%) similar to those of all C groups. At the family level, Vibrionaceae (5C: 64.71% and 5VH: 68.13%), Rhodobacteraceae (5C: 5.92%), Fusobacteriaceae (5VH: 4.53%) and UBA3375 (5C: 19.16% and 5VH: 17.56%) (Figure 4) dominated the gut microbiota at 5 ppt. At 20 ppt, the dominant families were Vibrionaceae (20C: 17.09% and 20VH: 49.23%), Rhodobacteraceae (20C: 55.85% and 20VH: 7.26%), Marinilabiliaceae (20C: 8.7%) and UBA3375 (20VH: 26.79%). The 30 ppt dominance profile was similar to that of 20 ppt, notably Vibrionaceae (30C: 56.39% and 30VH: 66.83%), Rhodobacteraceae (30C: 11.42% and 30VH: 7.89%), Marinilabiliaceae (30C: 7.15%) and UBA3375 (30VH: 14.22%). The relative abundance of Vibrionaceae was higher in all VH groups than C groups.
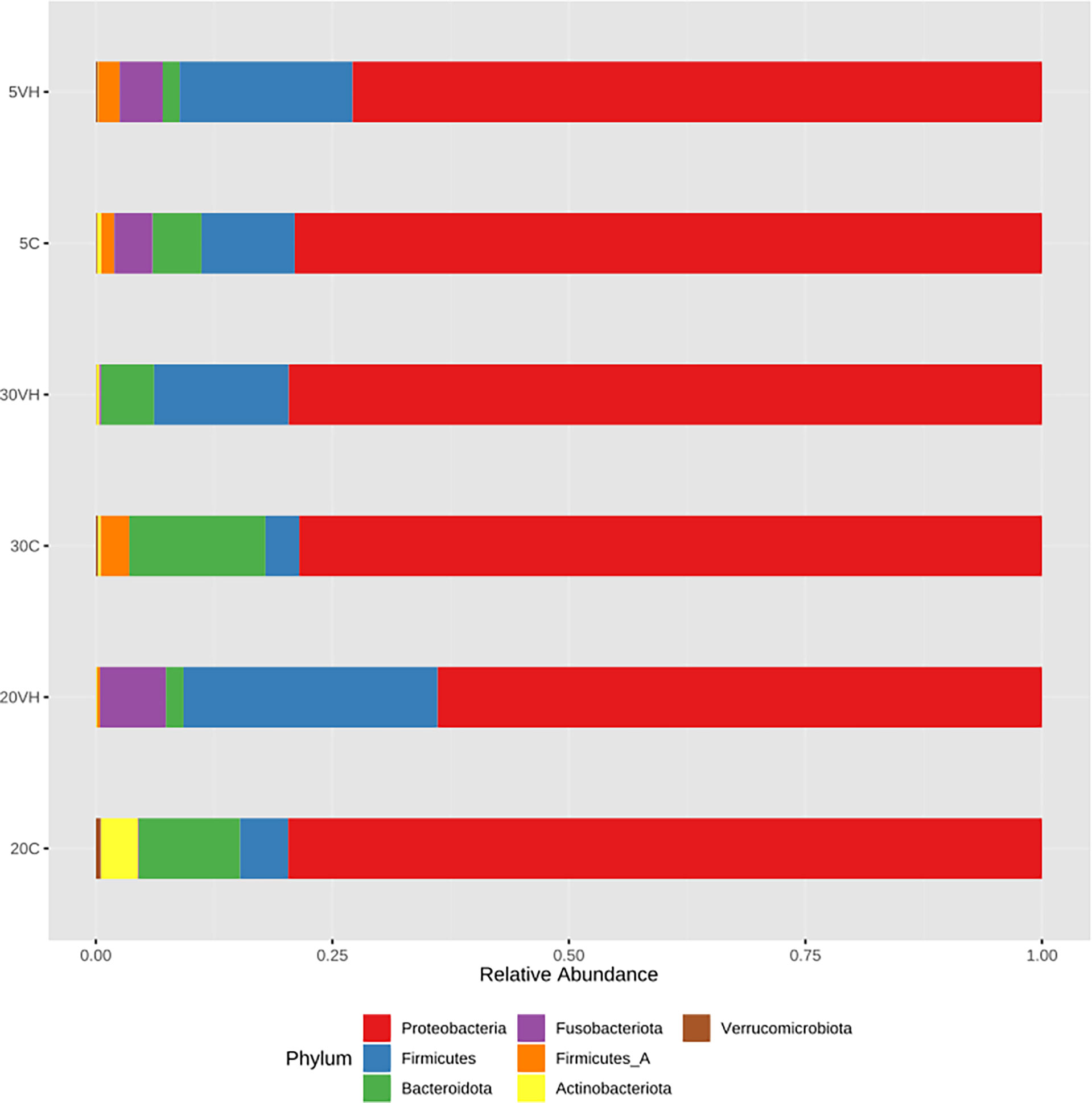
Figure 3 Phylum level gut bacterial profiles of Litopenaues vannamei cultured at different salinity levels and exposed to Vibrio harveyi infection (VH: Vibrio harveyi infected group and C, Control group; 5, 5 ppt; 20, 20 ppt and 30: 30 ppt).
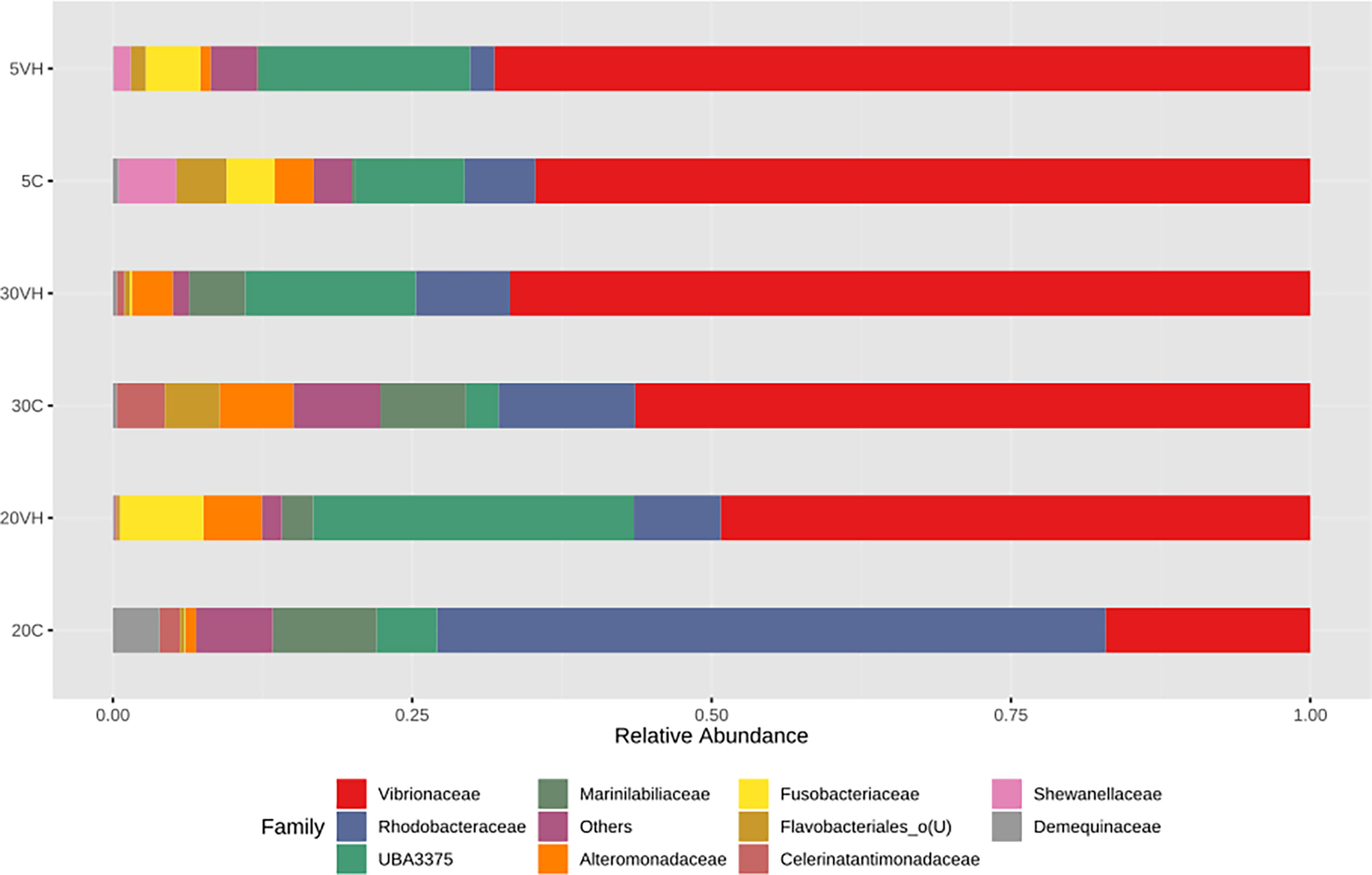
Figure 4 Family level gut bacterial profiles of Litopenaues vannamei cultured at different salinity levels and exposed to Vibrio harveyi infection (VH: Vibrio harveyi infected group and C, Control group; 5, 5 ppt, 20, 20 ppt and 30, 30 ppt).
At the genus level, the composition of the bacterial community composition was very diverse (Figure 5). In L. vannamei cultured at 5 ppt, Vibrio (5C: 63.50% and 5VH: 35.65%), unclassified genus from Rhodobacteraceae family (5C: 5.56%), unclassified genus from Vibrionaceae family (5VH: 29.16%) and UBA3375 (5C: 9.16% and 5VH: 17.56%) were the most prevalent genera. The most abundant genera in 20C consist of unclassified genus from Rhodobacteraceae family (40.06%), Vibrio (13.18%), and Salipiger (7.66%). Notably, Vibrio, unclassified genus from Fusobacteriaceae family, and UBA3375 were the most dominant genera accounting for 42.11%, 6.94%, and 26.79% of the total gut microbiota composition in 20VH. While for L. vannamei cultured at 30 ppt, the dominant genera were Vibrio (30C: 48.28% and 30VH: 47.11%), unclassified genus from Rhodobacteraceae family (30C: 5.62%), unclassified genus from Vibrionaceae family (30C: 7.51% and 30VH: 17.69%) and UBA3375 (30VH: 14.22%). LEfSe’s result showed that 11 core bacterial genera were observed in the gut microbiota of L. vannamei (LDA >2.0; P <0.05). Shewanella was found to be significantly most abundant genus in 5C, while Salipiger, Shimia, Pseudoruegeria, Amaricoccus and Rubritalea were core bacteria in 20C. In 20VH, the core genera were identified from the genus Marivivens, unclassified Fusobacteriaceae and unclassified Legionellaceae. The main genus in 30C was Rhodovulum, while in 30VH, Oceanospirillum (Figure 6).
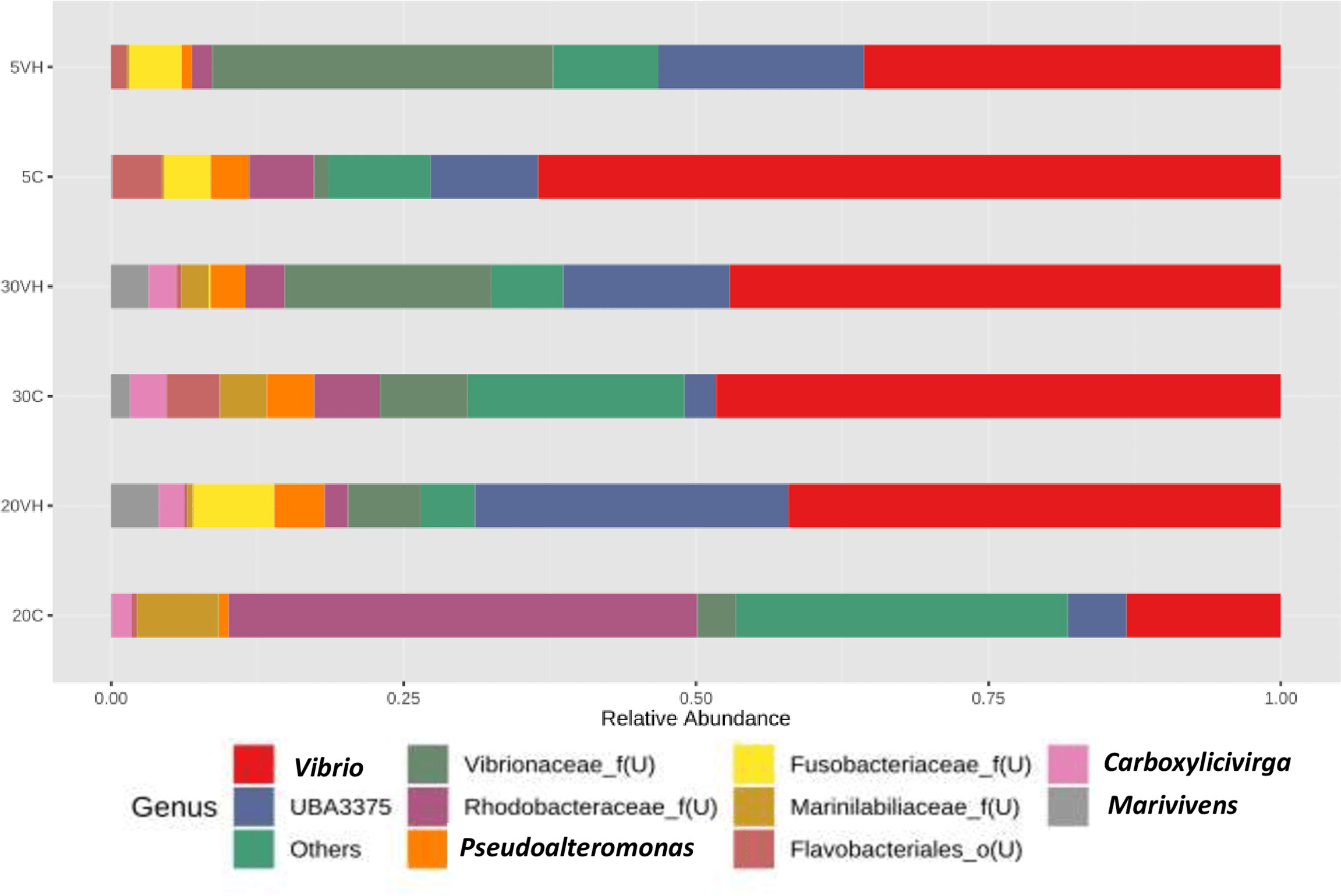
Figure 5 Genus level gut bacterial profiles of Litopenaues vannamei cultured at different salinity levels and exposed to Vibrio harveyi (VH: Vibrio harveyi infected group and C, Control group; 5, 5 ppt, 20, 20 ppt and 30, 30 ppt).
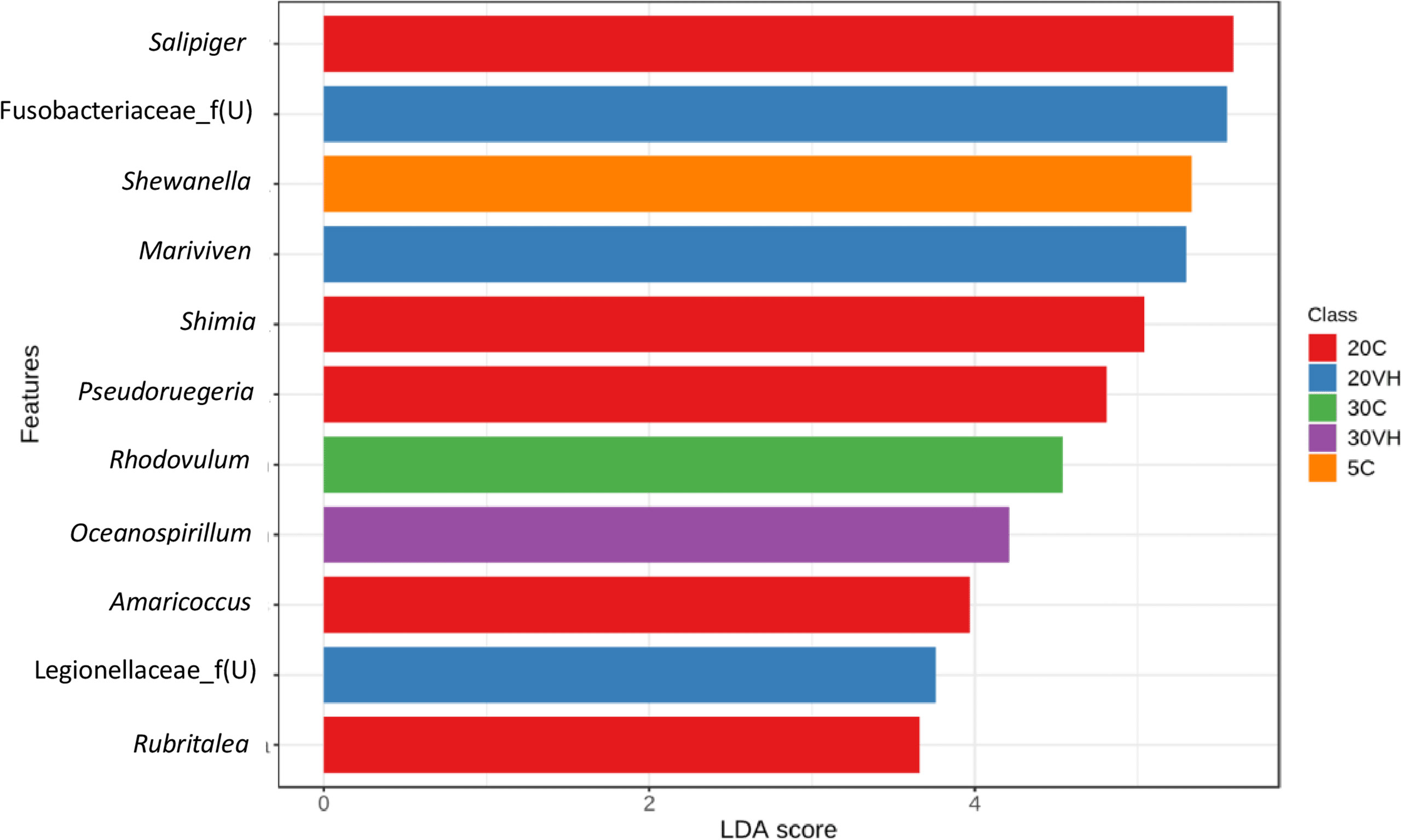
Figure 6 The core genera of the gut microbiota in Litopenaues vannamei cultured at different salinity levels and exposed to Vibrio harveyi (VH: Vibrio harveyi infected group and C, Control group; 5, 5 ppt, 20, 20 ppt and 30, 30 ppt).
Interestingly, the abundance of beneficial bacterial under the genera of Pseudoruegeria, Rubritalea, Roseibium, Demequina, unclassified Rhodobacteraceae, Amaricoccus, Salipiger, Ruegeria, Shimia, and Lactobacillus were lower in all VH groups compared to their corresponding C groups. Notably, the pathogenic bacterial genera under the genera of Vibrio, Photobacterium and unclassified genus from Vibrionaceae family were increased in the VH group (Figure 7).
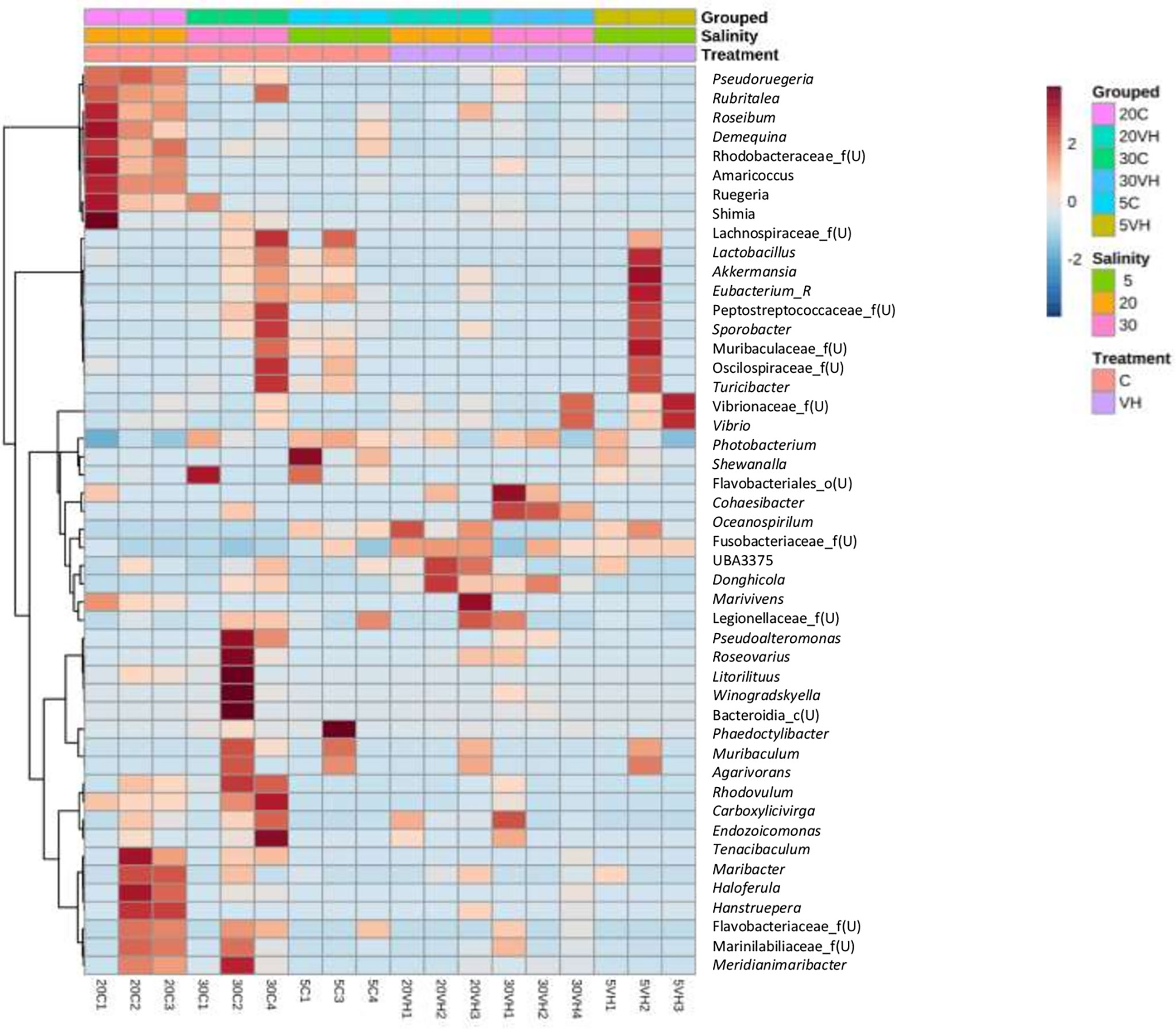
Figure 7 The heatmap analysis of top 50 genera on gut microbiota of Litopenaues vannamei cultured at different salinity levels and exposed to Vibrio harveyi. The red colour show the higher abundance of the genera, and the blue colour is the lower abundance (VH, Vibrio harveyi infected group and C, Control group; 5, 5 ppt, 20, 20 ppt and 30, 30 ppt).
Correlation between bacteria community and genus Vibrio
The abundance of genus Vibrio was positively correlated with those of 11 genera (unclassified genus from Flavobacteria family, unclassified genus from Fusobacteriaceae family, UBA3375, Phaeodactylibac, Turicibacter, Cohaesibacter, unclassified genus from Vibrionaceae family, unclassified genus from Muribaculacea family, Sporobacter, Oceanospirillum, and Pseudoalteromon). However, 14 bacterial genera were found to be negatively correlated with the presence of genus Vibrio, namely the Carboxylicivirga, unclassified genus from Legionellacea family, Roseibium, Litorilituus, Meridianimariba, Rubritalea, unclassified genus from Flavobacteria family, Agarivorans, Haloferula, Tenacibaculum, Ruegeria, Shimia, Rhodovulum, and Pseudoruegeria, which reduced or suppressed the presence of the genus Vibrio (Figure 8). However, based on the bacterial co-occurrence network, only three genera were negatively correlated with genus Vibrio, namely Shimia, Pseudoreugeria and Haloferula for both C and VH groups (Supplementary Figure S4).
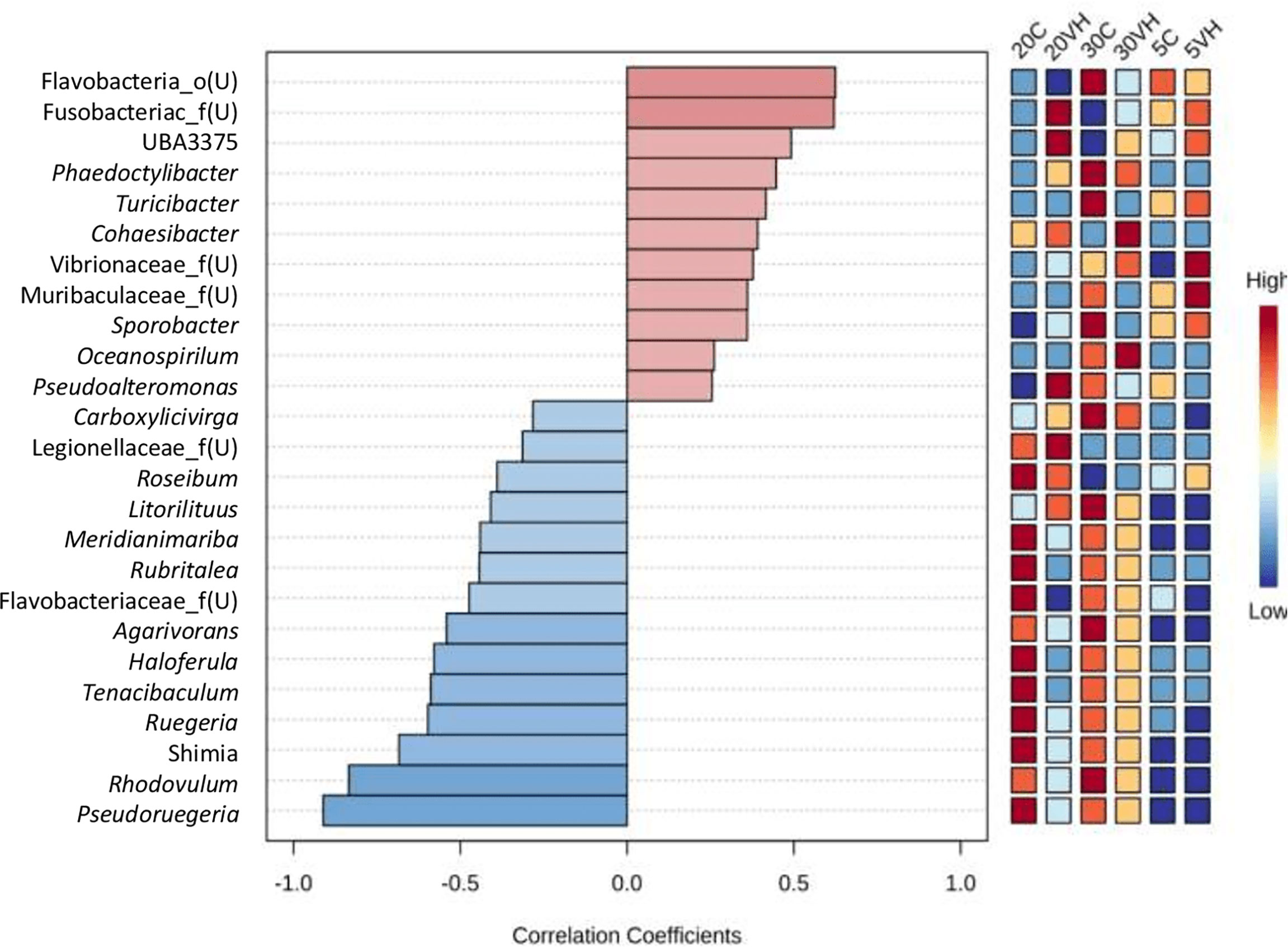
Figure 8 Top genus of gut microbiota in all Litopenaues vannamei correlated with the abundance of genus Vibrio. The blue bars indicated negative correlation between selected genus with genus Vibrio, whereas the pale red bars show positive interactions (VH, Vibrio harveyi infected group and C, Control group; 5, 5 ppt, 20, 20 ppt and 30, 30 ppt).
To visualize the relationship between microbe and environment and operation parameters of the processes in the shrimp gut, Canonical Correspondent Analysis (CCA) was performed based on the top 10 most abundant genera (Figure 10). Notably, unclassified genus from Rhodobacteraceae family was identified as the most dominant genus in the gut of shrimp cultured at 20 ppt, and was observed to be positively correlated with 20 ppt and shrimp growth (Figure 10). Besides, genus Vibrio was found to be predominant in the gut of shrimp cultured at 5 ppt and 30 ppt. The abundance of unclassified genus from Vibrionaceae family, unclassified genus from Fusobacteriaceae family, Marivivens and UBA3375 was strongly associated with the V. harveyi infected shrimp groups (5VH, 20VH and 30VH). Particularly, these genera dominated the shrimp gut following V. harveyi infection.
Functional prediction analysis by PICRUSt
Predicted microbial functional profiles were generated in PICRUSt and LEfSe (P <0.05, LDA > 2.0). Figure 9 revealed the significant difference of KEGG pathways based on metagenomics data between the C and VH groups (Figure 9). In the 5 ppt and 30 ppt treatment groups, only one pathway was significantly different between the C and VH groups, notably the Phosphotransferase system (PTS) (2.97) in 5 ppt and the oxidative phosphorylation pathway (2.59) in 30 ppt (Figures 9A, C). On the other hand, the 20 ppt group showed significant difference in several metabolic pathways including phenylalanine metabolism, glycine, serine and threonine metabolism, starch and sucrose metabolism, glyoxylate and dicarboxylate metabolism, and carbon metabolism (Figure 9B). 20VH exhibited a significantly higher abundance for eight different KEGG pathways, which are related to cellular processes (bacteria chemotaxis, LDA score of 3.0; biofilm formation, LDA score of 3.32; flagellar assembly, LDA score of 3.03), environmental information processing, bacterial secretion system (LDA score of 3.04), phosphotransferase system, PTS (LDA score of 3.38), starch and sucrose metabolism (LDA score of 3.09), carbon metabolism (LDA score of 3.19), and the genetic information processing pathway (LDA score of 3.38). The six KEGG pathways which were significantly enriched in 20C were related to metabolism (Phenylalanine metabolism, LDA score of 2.98; glycine, serine and threonine metabolism, LDA score of 3.04; metabolic pathway, LDA score of 3.09; glyoxylate and dicarboxylate metabolism, LDA score of 3.18; microbial metabolism in diverse environments, LDA score of 3.52; biosynthesis of antibiotics, LDA score of 3.62) (Figure 9B).
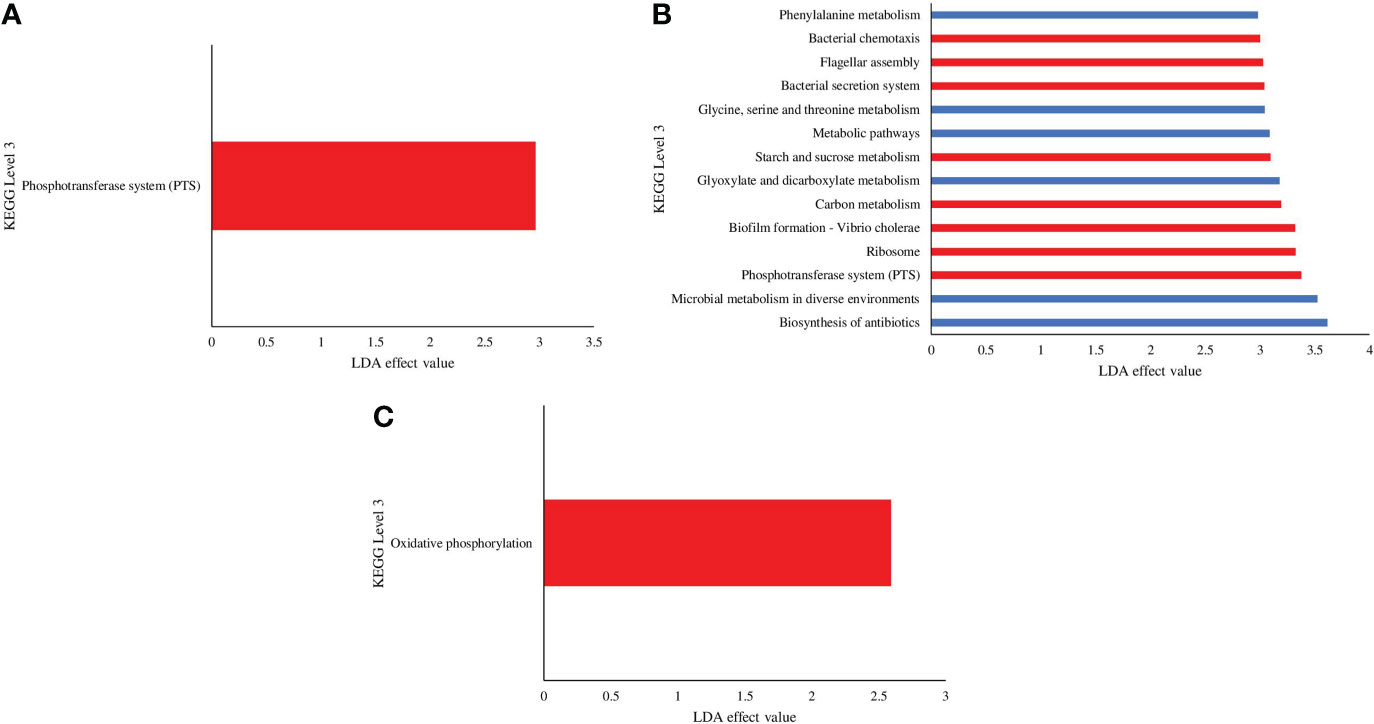
Figure 9 KEGG pathways found at significantly different abundances in the gut microbiota metagenomics profiles of Litopenaues vannamei cultured at (A) 5 ppt, (B) 20 ppt and (C) 30 ppt followed by exposure to Vibrio harveyi infection. The significant value is P < 0.05. All the P-value was less than 0.05. Red bars represent VH group, while blue bars represents C group (VH, Vibrio harveyi infected group and C, Control group; 5, 5 ppt, 20, 20 ppt and 30, 30 ppt).
Discussion
The shrimp gut microbiota composition is often associated with the host health in which the alteration of the gut microbiota community and functions, may reduce host immunity, leading to shrimp diseases outbreak (Huang et al., 2020; Zeng et al., 2020). Evidently, biotic and abiotic factors are observed to play a central role in shaping gut microbiota (Chen et al., 2017; Huang et al., 2018; Md Zoqratt et al., 2018; Deng et al., 2020; Hou et al., 2020; Xue et al., 2020). The present study reveals that water salinity (abiotic factor) and pathogenic V. harveyi (biotic factor) have greatly influenced the shrimp gut microbiota, with observed marked variations in the bacterial taxonomic composition, core taxa, interspecies interactions, correlation and prediction functions between experimented shrimp groups (5C, 5VH, 20C, 20VH, 30C, 30VH).
Our 16S rRNA gene amplicon sequence analysis suggested that gut microbiota α-diversity was lower in the VH groups than the C groups (Figure 1). Previous research has found that gut microbial diversity is positively correlated to the host health and is significantly reduced when the host is infected with pathogens (Xiong et al., 2015; Dai et al., 2017; Xiong et al., 2017). The presence of pathogenic V. harveyi may have likely stimulated the rapid growth of bacteria from Vibrionaceae family that have further reduced the shrimp gut diversity. Indeed, gut microbiota diversity has long been regarded as a key indicator of bacterial community stability, implicating that higher bacteria diversity signifies a more stable community with greater resilience to pathogenic infection (Ives and Carpenter, 2007; Shade et al., 2012; De Schryver and Vadstein, 2014). In this study, the diversity of the bacterial community in 20VH was higher than 5VH and 30VH, suggesting that L. vannamei cultured at 20 ppt were more resistance towards pathogenic V. harveyi infection (Figure 1). The PCoA biplot also showed that the bacterial community in all VH groups of varying salinity levels were clustered together (Figure 2). Interestingly, 5C and 30C were clustered closer to the VH groups (Figure 2A, indicating that shrimp that were cultured at 5 ppt and 30 ppt were more susceptible to V. harveyi infection than those that were cultured in 20 ppt. Similar observation have also been reported where the gut bacterial community of the diseased shrimp were usually clustered together, apart from those of non-infected shrimp (Rungrassamee et al., 2016; Chen et al., 2017; Yang et al., 2017; Deng et al., 2020).
Interspecies interaction in the shrimp gut is closely associated with the niche processes which lead to microbial co-occurrence within biological communities (Hou et al., 2020; Huang et al., 2022). In addition, the network complexity is closely linked to network stability (Ruan et al., 2020; Yuan et al., 2021). Previous research has revealed that the health of aquatic hosts is positively connected with the intricacy of interspecies interactions (Zhu et al., 2016; Nie et al., 2017). It is also reported that diseased or infected shrimp reduced the complexity of species-to-species interactions as well as the cooperative activities that take place between species (Zhu et al., 2016; Chen et al., 2017). L. vannamei infected with V. parahaemolyticus had a lower gut bacterial network complexity than those that were non-infected (Chen et al., 2017; Yao et al., 2018). Consistent with these findings, C groups possessed a more complex gut bacterial network than VH groups (Supplementary Figure S4). This study also demonstrated that 20 ppt could enhance the gut bacterial network’s complexity (Supplementary Figure S4), implying that this salinity could improve the microbial ecological network’s stability in the shrimp gut.
Vibrio, Photobacterium, and unclassified genus from Vibrionaceae family predominated the gut of L. vannamei following V. harveyi infection (Figure 5). Our finding corroborates with those of previous studies, highlighting that the abundance of Vibrio in the host gut that has been exposed to Vibrio sp. infection could trigger the growth of another potential pathogenic bacteria such as Photobacterium, Sphingomonas, Atopostipes, Staphylococcus, and Acinetobacter (Deng et al., 2020; Joe et al., 2021). Our results observed that pathogenic Vibrio, Photobacterium and unclassified genus from Vibrionaceae family were found to be more dominant than beneficial bacteria with immunomodulatory properties, indicating that these pathogenic bacteria outcompete them for nutrients and habitat niches within the shrimp gut. The higher abundance of pathogenic bacteria could also increase the occurrence of both primary and secondary disease infection in infected shrimp with reduced immunity levels (Selvin and Lipton, 2003). It has been proven that the overabundance of certain bacterial community may have impacted the disease structure of the host and trigger the changes in the health status of the gut microbiota (Xiong et al., 2015). Furthermore, Vibrio species, such as V. harveyi, V. parahaemolyticus, V. alginolyticus, V. owensii and V. anguillarum were reported as common pathogenic bacteria in various marine animals (Lee et al., 2015; Chen et al., 2017; Deng et al., 2020; Deris et al., 2020; Zhang et al., 2020; Liao et al., 2022; Muthukrishnan et al., 2022). In fact, an overabundance of Vibrio species was found to be intimately related to the progression of shrimp, lobster, and crab diseases such as AHPND, Vibrio-caused bacteraemia, shell disease, luminescent vibriosis and septic hepatopancreatic necrosis disease (Diggles et al., 2000; Vogan et al., 2002; Jayasree et al., 2006; Nie et al., 2017; Morales-Covarrulbias et al., 2018; Xia et al., 2018; Prachumwat et al., 2020; Nurhafizah et al., 2021; Lioa et al., 2022).
Contradictorily, contrasting abundance patterns between pathogenic Vibrio species and beneficial Pseudoruegeria, and Ruegeria species in 20C and 20VH may indicate that the salinity of 20 ppt could suppress the growth of Vibrio while enhancing the growth of those beneficial bacteria. This observation was supported by the significantly strong negative correlation between pathogenic Vibrio and beneficial bacteria (Figures 7, 8), signifying that beneficial bacterium from genera Pseudoruegeria, Rhodovulum, Ruegeria, Shimia and Lactobacillus in the present study could inhibit the growth or colonization of pathogenic bacteria from genera Vibrio, unclassified genus from Vibrionaceae family and Photobacterium. In general, Lactobacillus and Rhodovulum are probiotics that are colonized within the digestive tracts and are equipped with anti-microbial properties that inhibit the growth of pathogenic bacteria such as Vibrio sp. in fish and shrimp (Slover and Danziger, 2008; Liu et al., 2013; Chang et al., 2019; Koga et al., 2022). On the other hand, Shimia, Reugeria and Pseudoruegeria are beneficial bacteria with metabolic capability that could synthesize various bioactive molecules which are involved in the host adaptation and defence mechanism (Kanukollu et al., 2016; Abisado et al., 2018; Cho et al., 2020). In fact, Ruegeria was also documented to possess Vibrio sp. growth suppressing ability (Miura et al., 2019).
CCA was conducted to reveal the correlationship between gut microbiota community, environmental factor and shrimp growth performance. While shrimps cultured at 20 ppt showed positive correlationship with the shrimp growth increment, the abundance of unclassified genus from Rhodobacteraceae was also observed to be associated with the shrimp growth (Figure 10 and Supplementary Figure S5). In fact, L. vannamei cultured at 20 ppt was reported to have higher growth increment compared to those that were cultured at 2 ppt and 10 ppt respectively (Gao et al., 2016; Ponce-Palafox et al., 2019). As previously mentioned, Ruegeria, a genus from Rhodobacteraceae family may have established an antagonistic beneficial bacterial community which inhibit the growth of other pathogenic bacteria in the rearing environment (Miura et al., 2019). Furthermore, potential pathogenic bacteria such as Photobacterium and unclassified genus from Vibrionaceae family were significantly higher at 5 ppt and 30 ppt after exposure to pathogenic V. harveyi infection (Figures 4, 7), suggesting that these salinity levels are optimal in promoting the growth of these pathogenic bacteria (Figure 10). Our core taxa analysis also shows that potential pathogenic bacteria have dominated the gut microbiota in shrimp cultured at 5 ppt and 30 ppt (Supplementary Figure S3). In fact, pathogenic bacteria such as Vibrio, Propiogenium, Spongiimonas and Synchiccus were reported as core taxa in the gut of shrimp cultured at higher salinity levels (Hou et al., 2020). Both drastic fluctuations in salinity level and/or extreme salinity levels could trigger a disease outbreak by adversely affecting the shrimp’s defense mechanism and increasing their susceptibility to bacterial infection (Wang and Chen, 2006).
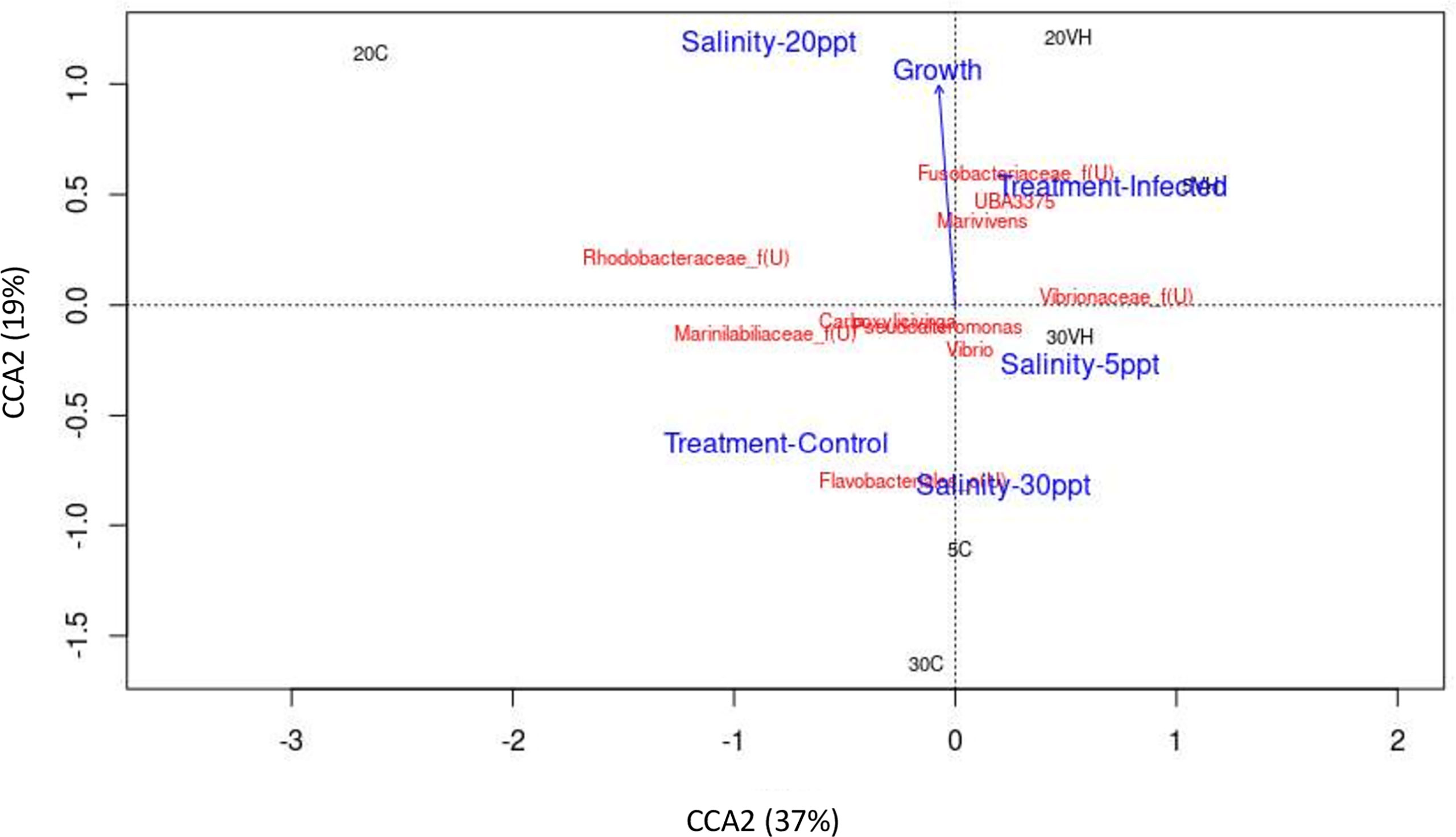
Figure 10 The correlation between gut microbiota of Litopenaeus vannamei cultured at different salinity levels and exposed to Vibrio harveryi infection with environmental or operational variables using Canonical correspondence analysis (CCA). The arrow length represents the strength of the correlation between the environmental variables and the microbes. The longer the arrow length, the stronger the correlation. The perpendicular distance between microbes and environmental variable axes in the plot reflects their correlations. The smaller the distance, the stronger the correlation. Treatment-Control, Control group. Treatment-Infected, Vibrio harveyi infected group, Salinity-5ppt, 5 ppt; Salinity-20ppt, 20 ppt; and Salinity-30ppt, 30 ppt. Growth (Growth increment of shrimp).
Functional prediction of gut microbiota unveils the potential roles and associated pathways of gut bacteria in shrimp cultured under different salinity levels and exposed to pathogenic V. harveyi infection. Interestingly, energy metabolism and oxidative phosphorylation pathways were significantly induced in 30VH (Figure 9C). Enrichment of energy metabolism pathway indicated that more free energy was produced to promote reproduction and growth of shrimp gut bacteria (Huang et al., 2019). On the other hand, the oxidative phosphorylation pathway is a crucial adhesion regulator for Vibrio species that increase their adhesiveness to the host gut (Paola et al., 2003; Huang et al., 2019; Xu et al., 2021). The phosphotransferase system (PTS) was found to be significantly enriched in the shrimp gut at 5VH and 20VH (Figures 9A, B). Besides regulating the virulence of gut bacteria, PTS is also linked to numerous regulatory functions related to metabolism pathways, chemotaxis, potassium transportation, and catabolite repression (Beth, 2010; Milohanic et al., 2014; Lim et al., 2019). In addition, the colonization process of the pathogenic V. harveyi in the shrimp gut may have triggered the significant increase of bacteria chemotaxis, bacterial secretion system, flagellar assembly, and biofilm formation mechanism in the gut microbiome of 20VH (Figure 9B). The flagellar assembly (motility organelle) is often associated with the chemical information signalled by the bacteria chemotaxis mechanism that enable bacteria to efficiently and rapidly respond to chemically favorable environments while avoiding repellents. Vibrio alginolyticus have been reported to use these mechanisms to adhere to host tissue, under salinity stress condition (Wang et al., 2015). The bacterial secretion system is involved in the adhesion process by controlling the secretion of intercellular protein complexes as virulence factors to invade host gut cells (Millet et al., 2014; Wang et al., 2015; Matilla and Krell, 2018). Furthermore, biofilm formation is not only involved in bacterial colonization and adhesion in the host tissues, but also act as a protective shield for microbes to thrive in harsh environments (Chavez de Paz et al., 2007; Koerdt et al., 2010; Amires et al., 2015; Charles et al., 2017; Cihan et al., 2017; Mallick et al., 2018; Yin et al., 2019). Besides, the formation of biofilm was considered as one of the crucial stress response of a particular bacteria towards the changes within the host environments (Fang et al., 2016; Rossi et al., 2018). Previous studies have also reported that pathogenesis related mechanisms such as bacterial secretion, quorum sensing and biofilm formation were significantly induced in the gut of host infected by V. harveyi (Khan et al., 2020; Deng et al., 2020). Evidently, Vibrio infection in L. vannamei have been shown to cause minor to drastic alterations in various shrimp metabolic functions such as cell cycle, glycine, serine, threonine, cysteine, methionine, purine, pyrimidine, pyruvate, quorum sensing and lipid metabolism (Kumar et al., 2021; Alvariz-Ruiz et al., 2022). Therefore, based on this PICRUSt analysis, we hypothesize that 20 ppt may likely be one of the environmental stressors to V. harveyi, given that biofilm formation process was significantly higher in shrimp gut microbiota of 20VH. In fact, our functional prediction analysis have also proven that salinity level could be the key regulator for host-gut bacterial osmotic adaptation, energy metabolism and metabolic functions. Our findings are consistent with results from previous studies, suggesting that salinity gradients may affect the function of bacterial communities and the survivability of bacterial communities (Oren, 2011; Sunagawa et al., 2015; Hou et al., 2017). Accordingly, genes associated with gut microbial metabolism pathways (phenylalanine metabolism, glycine, serine and threonine metabolism, metabolic pathway, glyoxylate and dicarboxylate metabolism, microbial metabolism in diverse environments, and biosynthesis of antibiotics) were significantly higher in the gut microbiota of shrimp cultured at 20 ppt. Our results provide insight into the importance of metabolic functions of shrimp gut microbiota in responding to salinity changes.
Conclusion
Varying salinity levels and the presence of pathogenic V. harveyi altered the diversity, structure, composition, interspecies interaction and functional pathways of L. vannamei gut microbiota. The gut microbiota diversity of shrimp cultured at 5 ppt and 30 ppt were lower than 20 ppt after exposure to pathogenic V. harveyi infection. Clustering of shrimp gut bacteria from 5C and 30C with the VH groups suggested that both shrimp cultured at these salinity levels were more susceptible to pathogenic V. harveyi infection. On the other hand, shrimp cultured at 20 ppt (20C and 20VH) appeared to exhibit a more stable bacterial network with complex interspecies interaction. Moreover, higher abundance of beneficial bacteria such as Pseudoruegeria, Rhodovulum, Ruegeria, Shimia and Lactobacillus in the gut of shrimp cultured at 20 ppt might have inhibited the growth of pathogenic bacterial despite the presence of pathogenic V. harveyi compared to shrimp cultured at 5 ppt and 30 ppt. The biofilm formation process was observed to be significantly higher in the gut microbiota of 20VH, suggesting that 20 ppt may be a stressor that impedes the growth of V. harveyi, which further protects shrimp from primary or secondary pathogenic infection. Additionally, pathogenesis-related microbial functions such as oxidative phosphorylation pathways, phosphotransferase system (PTS), bacteria chemotaxis, bacterial secretion system, flagellar assembly, and biofilm formation were altered due to the proliferation of V. harveyi in the shrimp gut. Besides, CCA has also supported that shrimp cultured at 20 ppt have the highest growth increment compared to those cultured at 5 ppt and 30 ppt. The basis understanding of the microbiota profiles from this study provide insights into regulating water salinity range into 20 ± 2 ppt to improve the overall immune health of shrimp to defend against pathogenic infection. However, study on the host-microbe-environmental interactions incorporating data of host responses towards pathogenic infection and salinity stress may be promising future work in strategizing shrimp disease prevention measures. Full-length sequencing of 16S rRNA or even 16S-ITS using long read sequencing technology will be instructive to improve the classification, resolution and detection of microbial communities down to species level. Quorum sensing of the gut microbiota and the transcriptomic analysis of shrimp may reveal the regulatory mechanism of both microbe and host towards external stressors.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/ - PRJNA815689.
Author contributions
ZMD, WLL, SI designed the study. ZMD carried out the experiments. ZMD, SI and HG performed bioinformatics analysis. ZMD and WLL wrote the manuscript. SI, HG, MI, MN, MA, WM, LY, MD-D, and YYS reviewed and give critical comments on the manuscript. All the authors have contributed to the completion of this manuscript and have approved the submitted version.
Funding
The Ministry of Higher Education, Malaysia funded this study under the Fundamental Research Grant Scheme (FRGS) (Reference number: FRGS/1/2018/WAB01/UMT/02/6).
Acknowledgments
This research was conducted at the Institute of Marine Biotechnology (IMB), Institute of Tropical Aquaculture and Fisheries Research (AKUATROP) and Faculty of Fisheries and Food Science, Universiti Malaysia Terengganu. We would like to thank the science officers in these institutions for their technical assistance.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.974217/full#supplementary-material
Supplementary Figure 1 | The rarefaction curves analysis based on Good coverage of each group treatments of Litopenaues vannamei cultured at different salinity level and challenged with Vibrio harveyi (VH: Vibrio harveyi infected group and C: Control group; 5: 5 ppt, 20: 20 ppt and 30: 30 ppt).
Supplementary Figure 2 | α-diversity analysis of each group treatments of Litopenaues vannamei cultured at different salinity level and challenged with Vibrio harveyi (A) Shannon index box plot and (B) Chao 1 index box plot (5: 5 ppt, 20: 20 ppt and 30: 30 ppt).
Supplementary Figure 3 | The core genera in the gut microbiota of Litopenaeus vannamei cultured at different salinity levels (5: 5 ppt, 20: 20 ppt and 30: 30 ppt).
Supplementary Figure 4 | Bacterial correlation network (SparCC) of Litopenaues vannamei gut, cultured at different salinity levels and exposed to Vibrio harveyi. Each node represents a bacterial genus and the colours of nodes affiliated to different treatment group.
Supplementary Figure 5 | Growth increment of Litopenaues vannamei gut, cultured at different salinity levels (N=3; t-test P<0.05).
References
Abisado R. G., Benomar S., Klaus J. R., Dandekar A. A., Chandler J. R. (2018). Bacterial quorum sensing and microbial community interactions. mBio 9, 02331–02317. doi: 10.1128/mBio.01749-18
AghaKouchak A., Chiang F., Huning L. S., Love C. A., Mallakpour I., Mazdiyasni O., et al. (2020). Climate extremes and compound hazards in a warming world. Ann. Rev. Earth Planetary Sci. 48 (1), 519–548. doi: 10.1146/annurev-earth-071719-055228
Alvarez-Ruiz S. A. P., Luna-González A., Escamilla-Montes R., Fierro-Coronado A., Diarte-Plata G., García-Gutiérrez C., et al. (2022). Gut bacterial profile associated with healthy and diseased (AHPND) shrimp Penaeus vannamei. Lat. Am. J. Aquat. Res. 50 (2), 197–211. doi: 10.3856/vol50-issue2-fulltext-2839
Amjres H., Bejar V., Quesada E., Carranza D., Abrini J., Sinquin C., et al. (2015). Characterization of haloglycan, an exopolysaccharide produced by Halomonas stenophila HK30. Int. J. Biol. Macromol 72, 117–124. doi: 10.1016/j.ijbiomac.2014.07.052
Araneda M., Gasca-Leyva E., Vela M. A., Domínguez-May R. (2020). Effects of temperature and stocking density on intensive culture of pacific white shrimp in freshwater. J. Thermal Biol. 94, 102756. doi: 10.1016/j.jtherbio.2020.102756
Bauer J., Teitge F., Neffe L., Adamek M., Jung A., Peppler C., et al. (2020). Impact of a reduced water salinity on the composition of vibrio spp. in recirculating aquaculture systems for pacific white shrimp (Litopenaeus vannamei) and its possible risks for shrimp health and food safety. J. Fish. Dis. 00, 1–17. doi: 10.1111/jfd.13270
Beth A. L. (2010). The phosphoenolpyruvate phosphotransferase system: as important for biofilm formation by Vibrio cholerae as it is for metabolism in Escherichia coli. J. Bacteriol. 192 (16), 4083–4085. doi: 10.1128/JB.00641-10
Bokulich N. A., Benjamin D. K., Jai R. R., Matthew D., Evan B., Rob K., et al. (2018). Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s Q2-Feature-Classifier plugin. Microbiome 6 (1), 90. doi: 10.1186/s40168-018-0470-z
Bolyen E., Jai R. R., Matthew R. D., Nicholas A. B., Christian C. A., Gabriel A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37 (8), 852–857. doi: 10.1038/s41587-019-0209-9
Callahan B. J., McMurdie P. J., Rosen M. J., Han A. W., Johnson A. J. A., Holmes S. P. (2016). DADA2: High-resolution sample inference from illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Chaiyapecharaa S., Uengwetwanit T., Arayamethakornb S., Bunphimpaphaa P., Phromson M., Jangsuttivorawat W., et al. (2022). Understanding the host-Microbe-Environment interactions: Intestinal microbiota and transcriptomes of black tiger shrimp Penaeus monodon at different salinity levels. Aquaculture 546, 737371. doi: 10.1016/j.aquaculture.2021.737371
Chang B. V., Liao C. S., Chang Y. T., Chao W. L., Yeh S. L., Kuo D. L., et al. (2019). Investigation of a farm-scale multitrophic recirculating aquaculture system with the addition of Rhodovulum sulfidophilum for milkfish (Chanos chanos) coastal aquaculture. Sustainability 11 (7), 1880. doi: 10.3390/su11071880
Charles C. J., Rout S. P., Patel K. A., Akbar S., Laws A. P., Jackson B. R., et al. (2017). Floc formation reduces the pH stress experienced by microorganisms living in alkaline environments. Appl. Environ. Microbiol. 83, 02985–02916. doi: 10.1128/AEM.02985-16
Chavez de Paz L. E., Bergenholtz G., Dahlén G., Svensäter G. (2007). Response to alkaline stress by root canal bacteria in biofilms. Int. Endod. J. 40, 344–355. doi: 10.1111/j.1365-2591.2006.01226.x
Cheng W., Chen J. C. (2000). Effects of pH, temperature and salinity on immune parameters of the freshwater prawn. Fish Shellfish Immunol. 10, 387–391. doi: 10.1006/fsim.2000.0264
Chen Y. Y., Blanc M., Lin Y. C., Huang C. L., Chen J. C. (2015). Growth and immune parameters in white shrimp Litopenaeus vannamei reared at different salinities. J of the Fish Socie. of Taiwan 42, 63–72. doi: 10.29822/JFST.201503_42(1).0006
Chen W. Y., Ng T. H., Wu J. H., Chen J. W., Wang H. C. (2017). Microbiome dynamics in a shrimp grow-out pond with possible outbreak of acute hepatopancreatic necrosis disease. Sci. Rep. 7, 9395. doi: 10.1038/s41598-017-09923-6
Chen S., Yanqing Z., Yaru C., Jia G. (2018). Fastp: an ultra-fast all-in-One FASTQ preprocessor. Bioinformatics 34 (17), i884–i890. doi: 10.1093/bioinformatics/bty560
Cho S. H., Lee E., Ko S. R., Jin S., Song Y., Ahn C. Y., et al. (2020). Elucidation of the biosynthetic pathway of vitamin b groups and potential secondary metabolite gene clusters via genome analysis of a marine bacterium pseudoruegeria sp. M32A2M. J. Microbiol. Biotechnol. 30, 505–514. doi: 10.4014/jmb.1911.11006
Chong J., Peng L., Guangyan Z., Jianguo X. (2020). Using MicrobiomeAnalyst for comprehensive statistical, functional, and meta-analysis of microbiome data. Nat. Protoc. 15 (3), 799–821. doi: 10.1038/s41596-019-0264-1
Cihan A. C., Karaca B., Ozel B. P., Kilic T. (2017). Determination of the biofilm production capacities and characteristics of members belonging to bacillaceae family. World J. Microbiol. Biotechnol. 33, 118. doi: 10.1007/s11274-017-2271-0
Cornejo-Granados F., Gallardo-Becerra L., Leonardo-Reza M., Ochoa-Romo J. P., Ochoa-Leyva A. (2018). A meta-analysis reveals the environmental and host factors shaping the structure and function of the shrimp microbiota. PeerJ 10 6, 5382. doi: 10.7717/peerj.5382
Dai W., Sheng Z., Chen J., Xiong J. (2020). Shrimp disease progression increases the gut bacterial network complexity and abundances of keystone taxa. Aquaculture 517, 734802. doi: 10.1016/j.aquaculture.2019.734802
Dai W. F., Yu W. N., Zhang J. J., Zhu J. Y., Tao Z., Xiong J. B. (2017). The gut eukaryotic microbiota influences the growth performance among cohabitating shrimp. Appl. Microbiol. Biotechnol. 01, 6447–6457. doi: 10.1007/s00253-017-8388-0
Deng Y., Zhang Y., Chen H., Xu L., Wang Q., Feng J. (2020). Gut–liver immune response and gut microbiota profiling reveal the pathogenic mechanisms of Vibrio harveyi in pearl gentian grouper (Epinephelus lanceolatus ♂ × e. fuscoguttatus). Front. Immunol. 11. doi: 10.3389/fimmu.2020.607754
Deris Z. M., Iehata S., Sahimi M. B. M. K., Do T. D., Sorgeloos P., Sung Y. Y., et al. (2020). Immune and bacterial toxin genes expression in different giant tiger prawn, Penaeus monodon post-larvae stages following AHPND-causing strain of Vibrio parahaemolyticus challenge. Aqua. Rep. 16, 248. doi: 10.1016/j.aqrep.2019.100248
De Schryver P., Vadstein O. (2014). Ecological theory as a foundation to control pathogenic invasion in aquaculture. ISME J. 8, 2360–2368. doi: 10.1038/ismej.2014.84
Diggles B. K., Moss G. A., Carson J., Anderson C. D. (2000). Luminous vibriosis in rock lobster Jasus verreauxi (Decapoda: Palinuridae) phyllosoma larvae associated with infection by Vibrio harveyi. Dis. Aquat. Organ. 43 (2), 127–121. doi: 10.3354/dao043127
Dong P., Guo H., Wang Y., Wang R., Chen H., Zhao Y., et al. (2021). Gastrointestinal microbiota imbalance is triggered by the enrichment of Vibrio in subadult Litopenaeus vannamei with acute hepatopancreatic necrosis disease. Aquaculture 533, 736199. doi: 10.1016/j.aquaculture.2020.736199
Dong X., Wang H., Xie G., Zou P., Guo C., Liang Y., et al. (2017). An isolate of Vibrio campbellii carrying the pirVP gene causes acute hepatopancreatic necrosis disease. Emerging Microbes Infect. 6 (1), 1–3. doi: 10.1038/emi.2016.131
Fang F. C., Frawley E. R., Tapscott T., Vazquez-Torres A. (2016). Bacterial stress responses during host infection. Cell Host Microbe 10 20 (2), 133–143. doi: 10.1016/j.chom.2016.07.009
Faust K., Sathirapongsasuti J. F., Izard J., Segata N., Gevers D., Raes J., et al. (2012). Microbial Co-occurrence relationships in the human microbiome. PloS Comput. Biol. 8 (7), e1002606. doi: 10.1371/journal.pcbi.1002606
Friedman J., Alm E. J. (2012). Inferring correlation networks from genomic survey data. PloS Comput. Biol. 8 (9), e1002687. doi: 10.1371/journal.pcbi.1002687
Gao W., Tian L., Huang T., Yao M., Hu W., Xu Q. (2016). Effect of salinity on the growth performance, osmolarity and metabolism-related gene expression in white shrimp Litopenaeus vannamei. Aqua. Rep. 4, 125–129. doi: 10.1016/j.aqrep.2016.09.001
Hou D., Huang Z., Zeng S., Liu J., Wei D., Deng X., et al. (2017). Environmental factors shape water microbial community structure and function in shrimp cultural enclosure ecosystems. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.02359
Hou D., Zhou R., Zeng S., Wei D., Deng X., Xing C., et al. (2020). Intestine bacterial community composition of shrimp varies under low- and high-salinity culture conditions. Front. Microbiol. 16, 11. doi: 10.3389/fmicb.2020.589164
Huang L., Guo H., Liu Z., Chen C., Wang K., Huang X., et al. (2022). Contrasting patterns of bacterial communities in the rearing water and gut of Penaeus vannamei in response to exogenous glucose addition. Mar. Life Sci. Technol. 4, 222–236. doi: 10.1007/s42995-021-00124-9
Huang L., Huang L., Zhao L., Qin Y., Su Y., Yan Q. (2019). The regulation of oxidative phosphorylation pathway on Vibrio alginolyticus adhesion under adversities. Microbiol. Open 8 (8), e00805. doi: 10.1002/mbo3.805
Huang W., Li H., Cheng C., Ren C., Chen T., Jiang X., et al. (2018). Analysis of the transcriptome data in Litopenaeus vannamei reveals the immune basis and predicts the hub regulation-genes in response to high pH stress. PLoS One 13 (12), e0207771. doi: 10.1371/journal.pone.0207771
Huang F., Pan L., Song M., Tian C., Gao S. (2018). Microbiota assemblages of water, sediment, and intestine and their associations with environmental factors and shrimp physiological health. Appl. Microbiol. Biotechnol. 102, 8585–8598. doi: 10.1007/s00253-018-9229-5
Huang Z., Zeng S., Xiong J., Hou D., Zhou R., Xing C., et al. (2020). Microecological ‘Koch’s postulates reveal that intestinal microbiota dysbiosis contributes to shrimp white feces syndrome. Microbiome 8 (1), 1–13. doi: 10.1186/s40168-020-00802-3
Ives A. R., Carpenter S. R. (2007). Stability and diversity of ecosystems. Science 317, 58–62. doi: 10.1126/science.1133258
Jayasree L., Janakiram P., Madhavi R. (2006). Characterization of associated with diseased shrimp from culture ponds of andhra pradesh (India). J. World Aqua. Soc. 37 (4), 523–532. doi: 10.1111/j.1749-7345.2006.00066.x
Joe X. J. T., Tseng Y. C., Wu J. L., Lu M. W. (2021). The alteration of intestinal microbiota profile and immune response in Epinephelus coioides during pathogen infection. Life 11 (2), 99. doi: 10.3390/life11020099
Kanehisa M., Goto S., Sato Y., Kawashima M., Furumichi M., Tanabe M. (2014). Data, information, knowledge and principle: Back to metabolism in KEGG. Nuclei Acid Res. 42, D199–D205. doi: 10.1093/nar/gkt1076
Kanukollu S., Voget S., Pohlner M., Vandieken V., Petersen J., Kyrpides N. C., et al. (2016). Genome sequence of shimia str. SK013, a representative of the roseobacter group isolated from marine sediment. Genom. Scien. 11, 25. doi: 10.1186/s40793-016-0143-0
Kathyayani S. A., Poornima M., Sukumaran S., Nagavel A., Muralidhar M. (2019). Effect of ammonia stress on immune variables of pacific white shrimp Penaeus vannamei under varying levels of pH and susceptibility to white spot syndrome virus. Ecotoxicol. Environ. Saf. 184, 109626. doi: 10.1016/j.ecoenv.2019.109626
Khan F., Tabassum N., Pham D. T. N., Oloketuyi S. F., Kim Y. M. (2020). Molecules involved in motility regulation in Escherichia coli cells: a review. Biofouling 36 (8), 889–908. doi: 10.1080/08927014.2020.1826939
Koerdt A., Godeke J., Berger J., Thormann K. M., Albers S. V. (2010). Crenarchaeal biofilm formation under extreme conditions. PloS One 5 (11), e14104. doi: 10.1371/journal.pone.0014104
Koga A., Goto M., Hayashi S., Yamamoto S., Miyasaka H. (2022). Probiotic effects of a marine purple non-sulfur bacterium, Rhodovulum sulfidophilum KKMI01, on kuruma shrimp (Marsupenaeus japonicus). Microorganisms 10 (2), 244. doi: 10.3390/microorganisms10020244
Kumar R., Tung T. C., Ng T. H., Chang C. C., Chen Y. L., Chen Y. M., et al. (2021). Metabolic alterations in shrimp stomach during acute hepatopancreatic necrosis disease and effects of taurocholate on Vibrio parahaemolyticus. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.631468
Lee C. T., Chen I. T., Yang Y. T., Ko T. P., Huang Y. T., Huang J. Y., et al. (2015). The opportunistic marine pathogen Vibrio parahaemolyticus becomes virulent by acquiring a plasmid that expresses a deadly toxin. PNAS 112 (34), 10798–10803. doi: 10.1073/pnas.1503129112
Liang Q., Wu X., Yang P., Kong J., Wei W., Qiao X., et al. (2019). The role of delta-1-Pyrroline-5-Carboxylate dehydrogenase (P5cdh) in the pacific white shrimp (Litopenaeus vannamei) during biotic and abiotic stress. Aquat. Toxicol. 208, 1–11. doi: 10.1016/j.aquatox.2018.12.016
Liao G., Wu Q., Mo B., Zhou J., Li J., Zou J., Fan L. (2022). Intestinal morphology and microflora to Vibrio alginolyticus in pacific white shrimp (Litopenaeus vannamei). Fish Shellfish Immunol. 121, 437–445. doi: 10.1016/j.fsi.2022.01.026
Li E., Chen L., Zeng C., Yu N., Xiong Z., Chen X., et al. (2008). Comparison of digestive and antioxidant enzymes activities, haemolymph oxyhemocyanin contents and hepatopancreas histology of white shrimp, Litopenaeus vannamei, at various salinities. Aquaculture 274 (1), 80–86. doi: 10.1016/j.aquaculture.2007.11.001
Lim S., Seo H. S., Jeong J., Yoon H. (2019). Understanding the multifaceted roles of the phosphoenolpyruvate: Phosphotransferase system in regulation of Salmonella virulence using a mutant defective in ptsi and crr expression. Microbiol. Res. 223, 63 71. doi: 10.1016/j.micres.2019.04.002
Lin Y. C., Chen J. C., Morni W. Z. W., Suhaili A. S. N. A., Kuo Y. H., Chang Y. H., et al. (2012). Modulation of the innate immune system in white shrimp Litopenaeus vannamei following long-term low salinity exposure. Fish Shellfish Immunol. 33, 324–331. doi: 10.1016/j.fsi.2012.05.006
Lioa X., Su Y., Wang B., Qin H., Hu S., et al. (2022). The effect of water temperature on the pathogenicity of decapod iridescent virus 1 (DIV1) in Litopenaeus vannamei. Israeli J. Aqua-Bamidgeh. 74, 1–9. doi: 10.46989/001c.33012
Li H., Ren C., Jiang X., Cheng C., Ruan Y., Zhang X., et al. (2019). Na+/H+ exchanger (NHE) in pacific white shrimp (Litopenaeus vannamei): Molecular cloning, transcriptional response to acidity stress, and physiological roles in pH homeostasis. PloS One 14 (2), e0212887. doi: 10.1371/journal.pone.0212887
Liu W. S., Ren P. F., He S. X., Xu L., Yang Y. L., Gu Z. M., et al. (2013). Comparison of adhesive gut bacteria composition, immunity, and disease resistance in juvenile hybrid tilapia fed two different Lactobacillus strains. Fish Shellfish Immunol. 35 (1), 54–62. doi: 10.1016/j.fsi.2013.04.010
Mallick I., Bhattacharyya C., Mukherji S., Dey D., Sarkar S. C., Mukhopadhyay U. K., et al. (2018). Effective rhizo inoculation and biofilm formation by arsenic immobilizing halophilic plant growth promoting bacteria (PGPB) isolated from mangrove rhizosphere: a step towards arsenic rhizoremediation. Sci. Total Environ. 610–611, 1239–1250. doi: 10.1016/j.scitotenv.2017.07.234
Martin M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17 (1), 10–12. doi: 10.14806/ej.17.1.200
Matilla M. A., Krell T. (2018). The effect of bacterial chemotaxis on host infection and pathogenicity. FEMS Microbiol. Rev.1 42(1), 40–46. doi: 10.1093/femsre/fux052
Md Zoqratt M. Z. H., Eng W. W. H., Thai B. T., Austin C. M., Gan H. M. (2018). Microbiome analysis of pacific white shrimp gut and rearing water from Malaysia and Vietnam: Implications for aquaculture research and management. PeerJ 6, e5826. doi: 10.7717/peerj.5826
Millard R. S., Ellis R. P., Bateman K. S., Bickley L. K., Tyler C. R., van Aerle R., et al. (2021). How do abiotic environmental conditions influence shrimp susceptibility to disease? a critical analysis focussed on white spot disease. J. Invert. Pathol. 186, 107369. doi: 10.1016/j.jip.2020.107369
Millet Y. A., Alvarez D., Ringgaard S., von Andrian U. H., Davis B. M., Waldor M. K. (2014). Insights into Vibrio cholerae intestinal colonization from monitoring fluorescently labeled bacteria. PloS Pathog. 10, e1004405. doi: 10.1371/journal.ppat.1004405
Milohanic E., Bouraoui H., Derkaoui M., Zébré A. C., Cao T. N., Kentache T., et al. (2014). The bacterial phosphoenolpyruvate: Carbohydrate phosphotransferase system: Regulation by protein phosphorylation and phosphorylation-dependent protein-protein interactions. Microbiol. Mol. Biol. Rev. 78, 231–256. doi: 10.1128/mmbr.00001-14
Miura N., Motone K., Takagi T., Aburaya S., Watanabe S., Aoki W., et al. (2019). Ruegeria sp. strains isolated from the reef-building coral Galaxea fascicularis inhibit growth of the temperature-dependent pathogen Vibrio coralliilyticus. Mar. Biotechnol. 21 (1), 1–8. doi: 10.1007/s10126-018-9853-1
Morales-Covarrubias M. S., Cu íellar-Anjel J., Varela-Mejías A., Elizondo-Ovares C. (2018). Shrimp bacterial infections in Latin America: A review. Asian Fisheries Sci. 31, 76–87. doi: 10.33997/j.afs.2018.31.S1.005
Muthukrishnan S., Shariff M., Ina-Salwany M. Y., Yusoff F., Natrah I. (2022). Expression of AHPND toxin genes (pirAB), quorum sensing master regulator gene (luxR) and transmembrane transcriptional regulator gene (toxR) in vibrio parahaemolyticus and Vibrio harveyi during infection of Penaeus vannamei (Bonne 1931). Aquaculture 551, 737895. doi: 10.1016/j.aquaculture.2022.737895
Nie L., Zhou Q., Qiao Y., Chen J. (2017). Interplay between the gut microbiota and immune responses of ayu (Plecoglossus altivelis) during Vibrio anguillarum infection. Fish Shellfish Immunol. 68, 479–487. doi: 10.1016/j.fsi.2017.07.054
Nurhafizah W. W. I., Kok Leong L., Abdul Razzak L., Musa N., Danish-Daniel M., Catherine Zainathan S., et al. (2021). Virulence properties and pathogenicity of multidrug-resistant Vibrio harveyi associated with luminescent vibriosis in pacific white shrimp, Penaeus vannamei. J. Invert. Patho. 186, 107594 doi: 10.1016/j.jip.2021.107594
Oksanen J., Guillaume Blanchet F., Friendly M., Kindt R., Legendre P., McGlinn D., et al. (2018). “Vegan: Community ecology package,” in R package version 2.5-1. Available at: https://CRAN.R-project.org/package=vegan.
Oren A. (2011). Thermodynamic limits to microbial life at high salt concentration. Environ. Microbiol. 13, 1908–1923. doi: 10.1111/j.1462-2920.2010.02365.x
Panigrahi A., Saranya C., Sundaram M., Kannan S. V., Das R. R., Kumar R. S., et al. (2018). Carbon: Nitrogen (C:N) ratio level variation influences microbial community of the system and growth as well as immunity of shrimp (Litopenaeus vannamei) in biofloc based culture system. Fish Shellfish Immunol. 81, 329–337. doi: 10.1016/j.fsi.2018.07.035
Paola C., Giovambattista P., Elisa G., Letizia T., Renata C., Giovanni R., et al. (2003). Reactive oxygen species as essential mediators of cell adhesion: The oxidative inhibition of a FAK tyrosine phosphatase is required for cell adhesion. J. Cell Biol. 161 (5), 933–944. doi: 10.1083/jcb.200211118
Parks D. H., Maria C., Pierre-Alain C., Christian R., Aaron J. M., Philip H. (2020). A complete domain-to-Species taxonomy for bacteria and archaea. Nat. Biotechnol. 9, 1079–1086. doi: 10.1038/s41587-020-0501-8
Ponce-Palafox J. T., Pavia Á., López D., Arredondo-Figueroa J., Lango-Reynoso F., Castañeda-Chávez M., et al. (2019). Response surface analysis of temperature-salinity interaction effects on water quality, growth and survival of shrimp Penaeus vannamei postlarvae raised in biofloc intensive nursery production. Aquaculture 503, 312–321. doi: 10.1016/j.aquaculture.2019.01.020
Prachumwat A., Wechprasit P., Srisala J., Kriangsaksri R., Flegel T. W., Thitamadee S., et al. (2020). Shewanella khirikhana sp. nov. - a shrimp pathogen isolated from a cultivation pond exhibiting early mortality syndrome. Microbial Biotechnol. 13 (3), 781–795. doi: 10.1111/1751-7915.13538
Prathiviraj R., Rajeev R., Fernandes H., Rathna K., Lipton A. N., Selvin J., et al. (2021). A gelatinized lipopeptide diet effectively modulates immune response, disease resistance and gut microbiome in Penaeus vannamei challenged with Vibrio parahaemolyticus. Fish Shellfish Immunol. 112, 92–107. doi: 10.1016/j.fsi.2021.02.018
Qu B., Zhang S., Ma Z., Gao Z. (2021). Hepatic cecum: a key integrator of immunity in amphioxus. Mar. Life Sci. Technol. 3, 279–292. doi: 10.1007/s42995-020-00080-w
Rossi E., Paroni M., Landini P. (2018). Biofilm and motility in response to environmental and host-related signals in gram negative opportunistic pathogens. J. Appl. Microbiol. 125, 1587–1602. doi: 10.1111/jam.14089
Ruan Y., Wang T., Guo S., Ling N., Shen Q. (2020). Plant grafting shapes complexity and Co-occurrence of rhizobacterial assemblages. Microb. Ecol. 80, 643–655. doi: 10.1007/s00248-020-01532-7
Rungrassamee W., Klanchui A., Maibunkaew S., Karoonuthaisiri N. (2016). Bacterial dynamics in intestines of the black tiger shrimp and the pacific white shrimp during Vibrio harveyi exposure. J. Invert. Patho. 133, 12–19. doi: 10.1016/j.jip.2015.11.004
Saoud I. P., Davis D. A., Rouse D. B. (2003). Sustainability studies of inland well waters for Litopenaeus vannamei culture. Aquaculture 217 (1), 373–383. doi: 10.1016/S0044-8486(02)00418-0
Selvin J., Lipton A. P. (2003). Vibrio alginolyticus associated with white spot disease of Penaeus monodon. Dis. Aquat. Org. 57, 147–150. doi: 10.3354/dao057147
Shade A., Peter H., Allison S. D., Baho D., Berga M., Bürgmann H., et al. (2012). Fundamentals of microbial community resistance and resilience. Front. Microbiol. 3. doi: 10.3389/fmicb.2012.00417
Slover C. M., Danziger L. (2008). Lactobacillus: a review. Clin. Microbiol. Newslett. 30 (4), 23–27. doi: 10.1016/j.clinmicnews.2008.01.006
Soares P. E. T., Dantas M. D. A., Silva-Portela R. D. C. B., Agnez-Lima L. F., Lanza D. C. F. (2021). Characterization of Penaeus vannamei mitogenome focusing on genetic diversity. PloS One 16 (7), 0255291. doi: 10.1371/journal.pone.025529
Soto-Rodriguez S. A., Gomez-Gil B., Lozano-Olvera R., Betancourt-Lozano M., Morales-Covarrubias M. S. (2015). Field and experimental evidence of Vibrio parahaemolyticus as the causative agent of acute hepatopancreatic necrosis disease of cultured shrimp (Litopenaeus vannamei) in northwestern Mexico. Appl. Environ. Microbiol. 81 (5), 1689–1699. doi: 10.1128/AEM.03610-14
Sunagawa S., Coelho L. P., Chaffron S., Kultima J. R., Labadie K., Salazar G., et al. (2015). Structure and function of the global ocean microbiome. Science 348, 1261359. doi: 10.1126/science.1261359
Vogan C. L., Costa-Ramos C., Rowley A. F. (2002). Shell disease syndrome in the edible crab, Cancer pagurus isolation, characterization and pathogenicity of chitinolytic bacteria. Microbiol. 148 (3), 743–754. doi: 10.1099/00221287-148-3-743
Wang F. I., Chen J. C. (2006). Effect of salinity on the immune response of tiger shrimp Penaeus monodon and its susceptibility to Photobacterium damselae subsp. damselae. Fish Shellfish Immunol. 20, 671–681. doi: 10.1016/j.fsi.2005.08.003
Wang R., Guo Z., Tang Y., Kuang J., Duan Y., Lin H., et al. (2020). Effects on development and microbial community of shrimp Litopenaeus vannamei larvae with probiotics treatment. AMB Expr. 10, 109. doi: 10.1186/s13568-020-01041-3
Wang L., Huang L., Su Y., Qin Y., Kong W., Ma Y., et al. (2015). Involvement of the flagellar assembly pathway in Vibrio alginolyticus adhesion under environmental stresses. Front. Cell Infect. Microbiol. 5. doi: 10.3389/fcimb.2015.00059
Wilkes-Walburn J., Wemheuer B., Thomas T., Copeland E., O’Connor W., Booth M., et al. (2019). Diet and diet-associated bacteria shape early microbiome development in yellowtail kingfish (Seriola lalandi). Microb. Biotech. 12 (2), 275–288. doi: 10.1111/1751-7915.13323
Xia M., Pei F., Mu C., Ye Y., Wang C. (2018). Disruption of bacterial balance in the gut of Portunus trituberculatus induced by Vibrio alginolyticus infection. J. Ocean. Limnol. 36 (5), 1891–1898. doi: 10.1007/s00343-018-7121-x
Xiong J., Wang K., Wu J. F., Lin L., Qian Q., Jie, et al. (2015). Changes in intestinal bacterial communities are closely associated with shrimp disease severity. Appl. Microbiol. Biotech. 99 (16), 6911–6919. doi: 10.1007/s00253-015-6632-z
Xiong J., Zhu J., Dai W., Dong C., Qiu Q., Li C. (2017). Integrating gut microbiota immaturity and disease-discriminatory taxa to diagnose the initiation and severity of shrimp disease. Environ. Microbiol. 19 (4), 1490–1501. doi: 10.1111/1462-2920.13701
Xue M., He Y., Chen D., Wang L., Liang H., Liu J., et al. (2020). Temporal dynamics of aquatic microbiota and their correlation with environmental factors during larviculture of the shrimp Litopenaues vannamei. Aquaculture 529,529. doi: 10.1016/j.aquaculture.2020.735605
Xu X., Li H., Qi X., Chen Y., Qin Y., Zheng J., et al. (2021). cheA, cheB, cheR, cheV, and cheY are involved in regulating the adhesion of Vibrio harveyi. Front. Cell Infect. Microbiol. 10. doi: 10.3389/fcimb.2020.591751
Yang H. T., Zou S. S., Zhai L. J., Wang Y., Zhang F. M., An L. G., et al. (2017). Pathogen invasion changes the intestinal microbiota composition and induces innate immune responses in the zebrafish intestine. Fish Shellfish Immunol. 71, 35–42. doi: 10.1016/j.fsi.2017.09.075S1050464817305946
Yao Z., Yang K., Huang L., Huang X., Qiuqian L., Wang K., et al. (2018). Disease outbreak accompanies the dispersive structure of shrimp gut bacterial community with a simple core microbiota. AMB Express 8, 120. doi: 10.1186/s13568-018-0644-x
Yin W., Wang Y., Liu L., He J. (2019). Biofilms: The microbial “Protective clothing” in Extreme Environments. Int. J. Mol. Sci. 20, 3423. doi: 10.3390/ijms20143423
Yoon S. H., Ha S. M., Kwon S., Lim J., Kim Y., Seo H., et al. (2017). Introducing EzBioCloud: A taxonomically united database on 16S rRNA gene sequences and whole genome assemblies. J. Syst. Evol. Microbiol. 67, 1613–1617. doi: 10.1099/ijsem.0.001755
Yuan M., Guo X., Wu L., Zhang Y., Xiao N., Ning D., et al. (2021). Climate warming enhances microbial network complexity and stability. Nat. Clim. Change 11, 343–348. doi: 10.1038/s41558-021-00989-9
Zeng S., Zhou R., Bao S., Li X., Deng Z., Hou D., et al. (2020). Identification of multigene biomarker for shrimp white feces syndrome by full-length transcriptome sequencing. Front. Genet. 11. doi: 10.3389/fgene.2020.00071
Zhang X. H., He X., Austin B. (2020). Vibrio harveyi: A serious pathogen of fish and invertebrates in mariculture. Mar. Life Sci. Techno. l2, 231–245. doi: 10.1007/s42995-020-00037-z
Keywords: Pacific white shrimp, climate change, 16S rRNA metagenomic sequencing, shrimp immunity, abiotic factors
Citation: Deris ZM, Iehata S, Gan HM, Ikhwanuddin M, Najiah M, Asaduzzaman M, Wang M, Liang Y, Danish-Daniel M, Sung YY and Wong LL (2022) Understanding the effects of salinity and Vibrio harveyi on the gut microbiota profiles of Litopenaeus vannamei. Front. Mar. Sci. 9:974217. doi: 10.3389/fmars.2022.974217
Received: 21 June 2022; Accepted: 22 August 2022;
Published: 04 October 2022.
Edited by:
Yu Zhang, Shanghai Jiao Tong University, ChinaCopyright © 2022 Deris, Iehata, Gan, Ikhwanuddin, Najiah, Asaduzzaman, Wang, Liang, Danish-Daniel, Sung and Wong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Li Lian Wong, lilian@umt.edu.my
†These authors have contributed equally to this work
 Zulaikha Mat Deris
Zulaikha Mat Deris Shumpei Iehata2†
Shumpei Iehata2†  Mhd Ikhwanuddin
Mhd Ikhwanuddin Musa Najiah
Musa Najiah Md Asaduzzaman
Md Asaduzzaman Min Wang
Min Wang Yantao Liang
Yantao Liang Muhd Danish-Daniel
Muhd Danish-Daniel Yeong Yik Sung
Yeong Yik Sung Li Lian Wong
Li Lian Wong