Identification, classification and expression analysis of the Ras superfamily genes in the Pacific white shrimp, Litopenaeus vannamei
- 1School of Life and Sciences, Qingdao Agricultural University, Qingdao, China
- 2Chinese Academy of Sciences (CAS) and Shandong Province Key Laboratory of Experimental Marine Biology, Institute of Oceanology, Chinese Academy of Sciences, Qingdao, China
- 3Laboratory for Marine Biology and Biotechnology, Qingdao National Laboratory for Marine Science and Technology, Qingdao, China
- 4Center for Ocean Mega-Science, Chinese Academy of Sciences, Qingdao, China
The Ras superfamily of small guanosine triphosphatases (GTPases) are a large group of small GTP-binding proteins, which play crucial roles in basic cellular processes in all eukaryotes. In this study, by analyzing the gene structure, temporal and spatial expression patterns, a total of 108 Ras superfamily genes were identified in the genome of the Pacific white shrimp Litopenaeus vannamei. We found these genes included not only the classical Ras GTPase superfamily members, but also some unconventional and novel Ras GTPase proteins, which have unknown functions and unique expression patterns. All Ras superfamily genes of L. vannamei were highly conserved within the core G domain and closely related in phylogeny, but they might have two different evolutionary origins. In addition, different Ras GTPase genes exhibited distinct expression patterns in different tissues, development/molting stages and WSSV infection samples of L. vannamei, suggesting that they may have a high functional specialization, and play important roles in regulating the biological processes of cell differentiation, growth and development, immune response, etc. This study provides important clues for the structure, classification, evolution and function of Ras superfamily in shrimp.
1 Introduction
The Ras superfamily, also known as small guanosine triphosphatases (GTPases), or small G proteins, is a group of monomeric protein family with GTPase hydrolytic activity. They are low molecular weight (20–30 kDa) and similar to the α-subunit of G-proteins and regulate many biological processes as molecular switches, alternating between an active GTP-bound state and an inactive GDP-bound state (Phillips et al., 2008). Ras was the first members of the superfamily to be discovered, and then Ran, Rho, Rab, Arf and other families (Marcus and Mattos, 2020). They control different signal transduction pathways in cells, including proliferation, differentiation, morphology, polarity, adhesion, migration, survival, apoptosis, etc. (Goitre et al., 2014).
Ras superfamily proteins are universal components of signaling pathways in eukaryotic organisms, including vertebrates, invertebrates, yeasts and plants (Cetkovic et al., 2007). In human, using a somewhat broader definition of sequence similarity reveals an extended superfamily of more than 170 Ras superfamily members (Colicelli, 2004). Drosophila melanogaster has 68 members: 13 Ras proteins, 7 Rhos, 30 Rabs, 17 Arfs, and 1 Ran (Rojas et al., 2012). The budding yeast Saccharomyces cerevisiae contains 29 members: 1 Sar and 1 SRβ, 6 Arfs, 10 Rabs, 6 Rhos, 4 Ras, and 2 Rans (Garcia-Ranea and Valencia, 1998). Studies have shown that the proteins of each family appeared very early in the evolution of eukaryotes, and then expanded to varying degrees in various species (Colicelli, 2004; Jiang and Ramachandran, 2006).
At present, there are limited studies on Ras GTPases in crustaceans, which mainly focused on their function in immunity of shrimp. In Kuruma shrimp Marsupenaeus japonicus, Ras, Ran and Rab genes have been found, they all play important roles in resistance to virus (Ménasché et al., 2000). In Chinese shrimp Fenneropenaeus chinensis, the expression of Rap gene was up-regulated in both Vibrio harveyi and white spot syndrome virus (WSSV) infection (Ren et al., 2012). Another study showed that after WSSV infection, the expression of FcRas was significantly up-regulated in muscle of F. chinensis, while it was significantly down-regulated in hepatopancreas (Li et al., 2020). These results suggest that different Ras superfamily members may participate in the process of anti-bacterial and viral immunity in different ways in shrimp.
Ras GTPases participate in various biological processes, and many studies have confirmed that they play an important role in growth (Sato et al., 2008; Geng et al., 2016; Liu et al., 2021). In the Pacific white shrimp Litopenaeus vannamei, the most economically valuable aquaculture shrimp in the world, through genome wide association study (GWAS) analysis, we found several genomic markers related to body weight and body length were mapped to Rap-2a, which is a member of the Ras superfamily (Yu et al., 2019). Rap-2a, as part of several signaling cascades, may regulate cytoskeletal rearrangement, cell migration, and cell diffusion (Taira et al., 2004). Through RT-qPCR analysis, Rap-2a was found to have high expression in lymphatic organ, hepatopancrea, intestine and stomach, it was negatively regulated by NF-κB and contributed to growth (Yu et al., 2019; Wang et al., 2022). Additionally, another shrimp growth trait candidate gene, MMD2, was identified by our group (Wang et al., 2020); and studies showed that MMD2 can enhance the retention and activation of Ras protein in Golgi complex, and subsequently lead to the enhancement of ERK (extracellular signal-regulated kinase) signal (Huang et al., 2012). These studies suggest that the Ras superfamily may play key roles in shrimp growth.
In this study, through the analyses of genome and transcriptome data, we found that L. vannamei has a large number of Ras superfamily genes. In this study, we identified 108 Ras superfamily genes, and analyzed their gene structure, classification and expression patterns. These studies have provided an important basis for further explore the structure, evolution and function of the Ras superfamily in shrimp.
2 Materials and methods
2.1 Experimental animals
The experimental shrimp were cultured in the laboratory of the Institute of Oceanology, Chinese Academy of Sciences (Qingdao, Shandong, China), at a temperature of 25 ± 1°C, salinity of 30%, and pH of 7.5 ± 0.1, the photoperiod was maintained at 12L:12D. The aquaculture seawater was filtered, sterilized, and continuously oxygenated. All shrimp were fed three times per day at 9:00 a.m., 2:00 p.m., and 7:00 p.m. with equal weights of commercial food pellets (Dale Feed Company, Yantai, China). The average weight of the shrimp was 4.0 ± 0.8g (7.5 ± 0.5cm). The animal study was reviewed and approved by the ethics committee of the Institute of Oceanology, Chinese Academy of Sciences. We declare that all animal experiments in this study were conducted in accordance with the guidelines of UK Animals Act, 1986 and EU Directive 2010/63/EU. In these experiments, no any endangered or protected species were used.
2.2 Identification of Ras superfamily gene members
We collected all genes annotated as Ras superfamily or small G protein genes from the L. vannamei genome database (http://www.shrimpbase.net). At the same time, we screened all Ras superfamily genes in the previous RNA-Seq data from different developmental stages, molting stages, different adult tissues, and WSSV infection of the shrimp (Wei et al., 2014; Gao et al., 2015; Wang et al., 2019; Zhang et al., 2019). After comparing all the obtained sequences, eliminating redundant sequences, merging overlapping fragments and connecting broken genes, all non-redundant candidate sequences were initially identified and compared by blastx (https://blast.ncbi.nlm.nih.gov/Blast.cgi) and SMART (https://smart.embl.de/) to confirm the gene members of Ras superfamily.
2.3 Phylogenetic analysis of Ras superfamily of L. vannamei
In order to determine evolutionary relationships of the Ras superfamily members in L. vannamei, all identified Ras GTPase homologous sequences of this species and a number of representative sequences of other animals, fungi, protists and plants were aligned by ClustalW in MEGA X (www.megasoftware.net/), and phylogenetic trees were constructed using the maximum likelihood (ML) method of MEGA X with 1000 bootstrap repeats (Whelan and Goldman, 2001; Kumar et al., 2018). The phylogenetic trees were then visualized using iTOL (Letunic and Bork, 2007).
2.4 Gene structure and conservative motif
In order to illustrate the gene structure of the Ras GTPases of L. vannamei, the gene location, gene length, open reading frame (ORF), exon number and deduced amino acid number of all the obtained genes were analyzed in detail. ExPASy (http://web.expasy.org/protparam/) was used for protein molecular weight, isoelectric point analysis. Using the gene structure display server (GSDS) program (Hu et al., 2015), the coding sequence (CDS) of each gene was compared with the genome sequence of L. vannamei, and the intron and exon arrangement diagram of each gene was obtained. The gene structure of Ras GTPase proteins was drawn by TBools software (Chen et al., 2020), and the conservative domains analysis was carried out according to the amino acid sequences. In order to better analyze the conserved motifs in Ras superfamily, a conserved motif map using the Multiple EM for Motif Elicitation (MEME, https://meme-suite.org/meme/tools/meme) program was constructed to provide more detailed evidence for clarifying the structural characteristics between different categories of Ras superfamily numbers.
2.5 Gene expression analyses
According to the previous RNA-Seq data at different developmental stages, molting stages, different adult tissues and WSSV infection of L. vannamei (Wei et al., 2014; Gao et al., 2015; Zhang et al., 2019), the RPKM (Reads per kilo base per million mapped reads) value of each unigene or transcript were obtained. The expression heatmaps of Ras GTPase genes in different transcriptomes were constructed and a part of representative genes were verified and analyzed using RT-qPCR subsequently.
2.6 RNA isolation and cDNA synthesis
The healthy WSSV-free shrimp L. vannamei (9 ± 1g) were collected from laboratory culture tanks for WSSV infection study, each shrimp was injected into 1000 copies of live WSSV particles suspended in 10μl sterile phosphate-buffered saline (PBS) in vivo WSSV challenge group, the control group shrimps were injected into the same volume of PBS. A total of 15 individuals were randomly assigned in each group and equally divided into three parallel subgroups as biological replicates. At 6 hpi, same size shrimp were picked, sacrificed and dissected. A total of 3 tissues, hemocyte, lymphoid (Oka) organ, hepatopancreas were sampled and frozen in liquid nitrogen and stored at -80 °C for total RNA extraction.
About 4.0 ± 0.8g (7.5 ± 0.5cm) untreated shrimp were picked, sacrificed and dissected. A total of 12 tissues, hemocyte, muscle, intestines, stomach, lymphoid organ, gill, hepatopancreas, eye stalk, brain, ventral nerve, epidermis and heart, were sampled and frozen in liquid nitrogen and stored at -80 °C for total RNA extraction. According to the manufacturer’s instructions, RNAiso Plusreagent (TaKaRa, Japan) was used to isolate total RNA from these tissues. Then, the quality and the concentration of RNA were detected using 1% agarose gel electrophoresis and Nanodrop 2000 (Thermo Fisher Scientific, United States). The first-strand cDNA was synthesized with the PrimeScript First Strand cDNA Synthesis Kit (TaKaRa, Japan) using 1.5 µg RNA as template. The specific cDNA synthesis were as follows: the first step was to remove genomic DNA, 5×genomic DNA eraser buffer was added to the template at 42°C for 5 min, and then moving on to step 2 immediately, at 37°C for 1 h and 85°C for 5 s. Finally, the cDNA was stored at -80°C until use.
2.7 Real-time quantitative PCR
The 18S rRNA gene was selected as the internal reference gene. Primer3Plus (http://www.primer3plus) was used to design thirteen pairs of primers of the selected Ras superfamily genes. All primer sequences used in this study are shown in Supplementary Table S1. Then, an Eppendorf Mastercycler ep realplex (Eppendorf, Hamburg, Germany) was used to perform the RT-qPCR. The SuperReal PreMix Plus (SYBR Green) (TIANGEN, Beijing, China), template, primers, and DEPC-treated water were mixed in a certain proportion (Supplementary Table S2); each sample includes four technical replicates. After qPCR, melting temperature (Tm) analysis showed a single peak and a single PCR band was identified, indicating that both primers were suitable for further experiments. The qPCR steps were as follows: 94°C for 2 min, 40 cycles of 94°C for 20 s, 62°C or 55°C for 20 s (the annealing temperature of rtRas superfamily genes-F/R and rt18S-F/R were 62 and 55°C, respectively), and 72°C for 20 s. Eventually, relative expression levels were analyzed by the 2-△△Ct method (Livak and Schmittgen, 2001).
2.8 Statistical analyses
The different groups were subjected to one-way ANOVA tests using SPSS (https://www.ibm.com/cn-zh/analytics/spss-statistics-software) (version 20).
3 Results
3.1 Identification and classification of the Ras superfamily genes
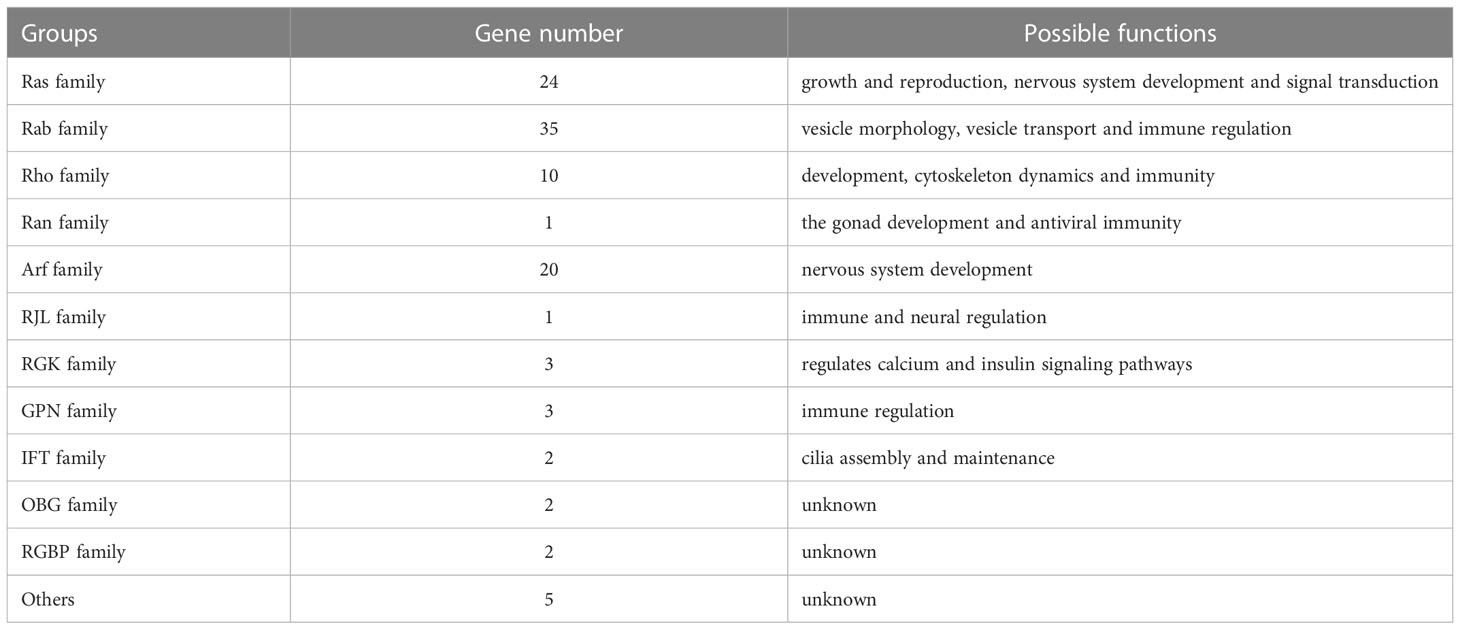
In this study, a total of 108 Ras superfamily genes of L. vannamei were screened and identified from genome and transcriptome data. The identified genes were classified and analyzed according to gene homology, conserved domain, gene annotation and evolutionary relationship. Using the SMART tool, the domains of 50 sequences were identified as the classical domains of the Ras superfamily: Ras, Rab, Arf (Sar), Rho respectively, and 49 sequences contained a small-GTPase domain, which have similar GTP binding conserved motifs to the classical Ras superfamily domains (Table 1), all them shared a set of conserved G box GDP/GTP-binding motif elements (Figure 1). Among the small-GTPase domain containing sequences, five have large molecular weight, they have not only a small-GTPase domain, but also other domains, such as BTB, FF, EFh, and RPT1 (Table 1). The remaining 9 sequences do not have the classical Ras superfamily domains or the small-GTPase domain, but they all have GTPase-related domains, and all the residues are perfectly conserved with Ras GTPases, mainly G1 and G3 motif (Figure 2A, corresponding to motif symbol 1 and 2).
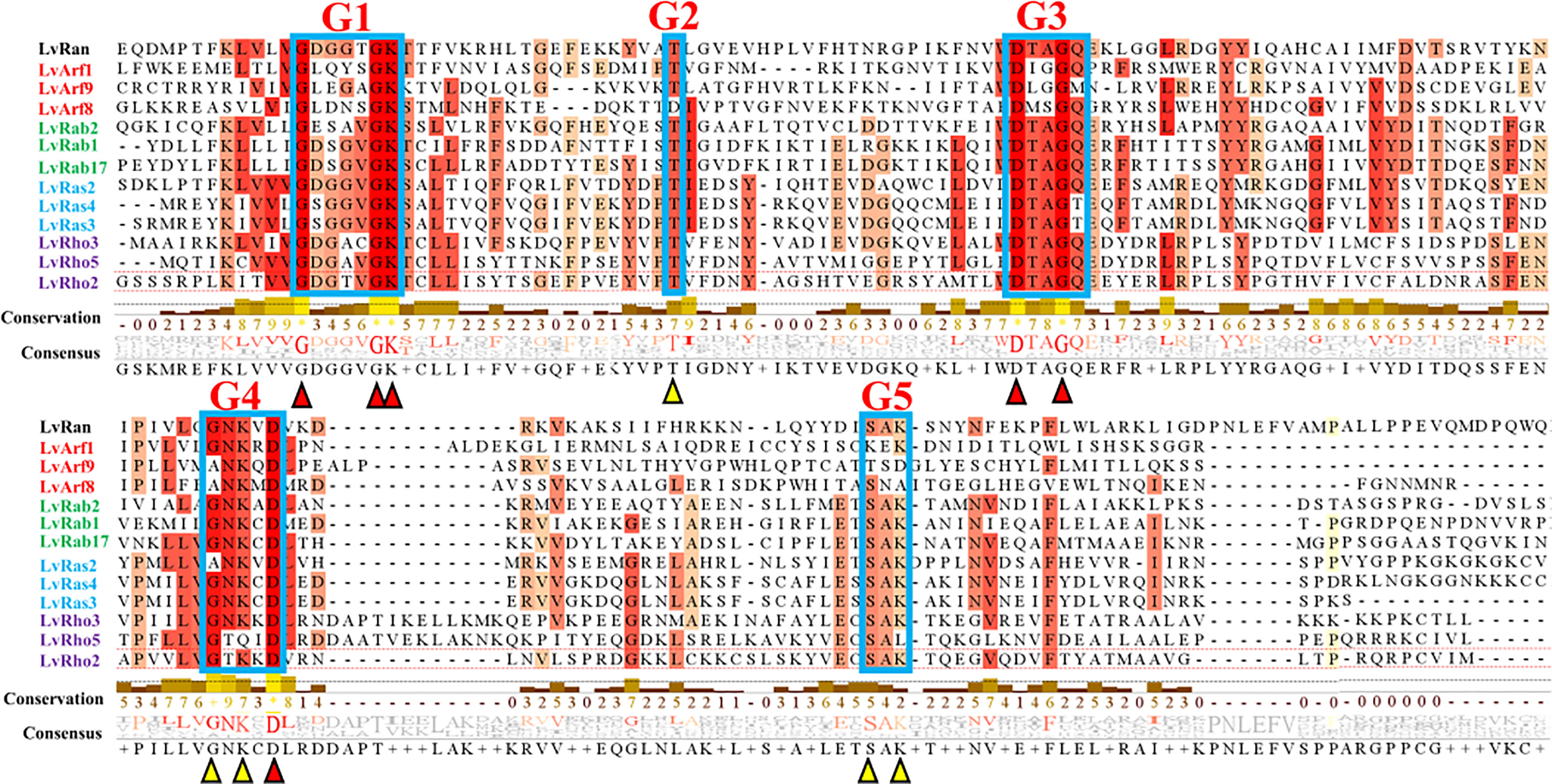
Figure 1 Multiple sequence alignment of some classical Ras superfamily protein sequences of L. vannamei. Red indicates complete agreement and orange indicates 75% ~ 99% homology. Conserved residues: G1 (GXXXXGKS/T), G2 (T), G3 (DXXGQ/H/T), G4 (T/NKXD) and G5 (C/SAK/L/T) (X stands for any amino acid). Red and yellow triangles represent major sites of conserved motifs.
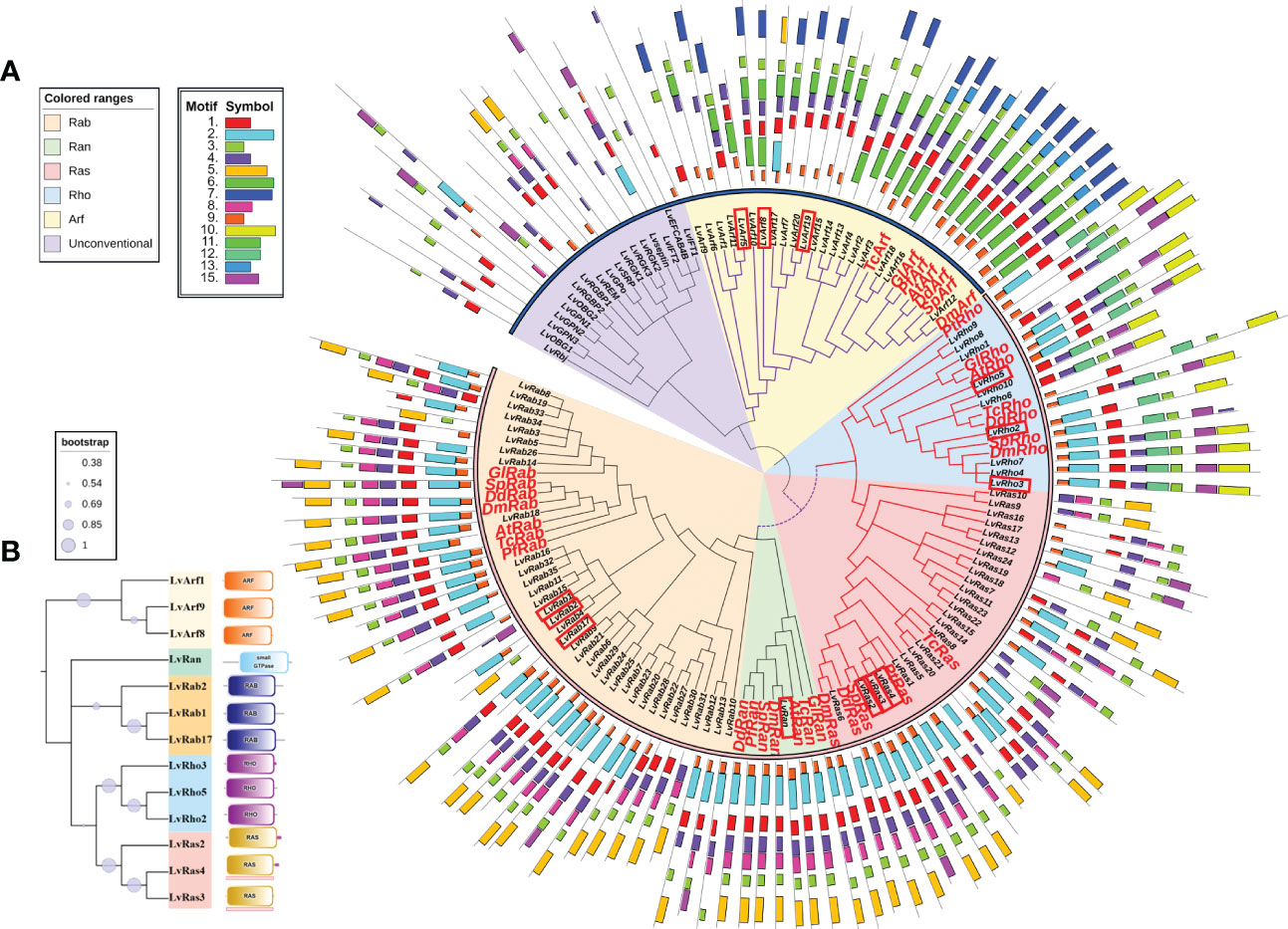
Figure 2 (A) Phylogenetic tree of Ras superfamily genes of L. vannamei and other species. The accession numbers of amino acid sequences used in phylogenetic trees were shown in Table 1 and Supplementary Table S3. The red box marks the most typical GTPases with high expression. The orange shadow represents Rab family; The green shadow represents Ran family; The red shadow represents Ras family; The blue shadow represents Rho family; The yellow shadow represents Arf family. The purple shadow represents Unconventional Ras superfamily. The phylogenetic tree periphery represents motifs of different families of L. vannamei and other species, the colored boxes indicate the conserved motifs. (B) Domains of classical Ras GTPases of different families of L. vannamei. The tree on the left illustrates the phylogenetic relationships of various families of Ras GTPases. On the right are the different domains of each family.
From the perspective of gene location, these Ras superfamily genes were distributed in 98 scaffolds, Ras, Rho, Rab and Arf families each had 1-2 two-gene clusters, and Arfs also had a three-gene cluster (LvArf4, LvArf14, and LvArf15) (Table 1).
In order to distinguish these Ras superfamily members, we compared identity as assayed by smartblast and blastp, and found that most of them were classical Ras GTPases, such as Ras, Rab, Arf, Rho, Ran, named LvRas1-24, LvRab1-35, LvArf1-20, LvRho1-10, LvRan. A small number (18) are unconventional or newly discovered members of the Ras superfamily, named LvRbj, LvRGK1-3, LvGPN1-3, LvREM, LvIFT1-2, LvRGBP1-2, LvOBG1-2, Lvseptin, LvEFCAB4B, LvSRP, LvGPo (Table 1).
3.2 Phylogenetic analysis of the Ras superfamily numbers
In order to clarify the phylogenetic relationship among Ras superfamily members of L. vannamei, we constructed phylogenetic trees using Ras superfamily genes from L. vannamei and those identified from other species (Table 1 and Supplementary Table S3). Phylogenetic analysis showed that the Ras GTPase proteins were clustered in the clades of each subfamily, although they come from different species (Figure 2A). The result suggested that the Ras superfamily members of L. vannamei could be divided into two large monophyletic groups. The Ras family and Rho family could be clustered into a clade, the number of Rab family is the largest (35 members) and form a clade with Ran family, then clustered with Ras family and Rho family into a larger clade. Arf family members were clustered together and far away from other four families, other unconventional Ras superfamily numbers were clustered together, and then clustered with the Arf family clade formed another large clade (Figure 2A).
3.3 Conserved motifs and gene structure of the Ras superfamily numbers
According to the analyses of gene structure and conserved motifs, most Ras superfamily genes share a set of conserved G box GDP/GTP-binding motif elements: GXXXXGKS/T, T, DXXGQ/TE, NXXD and SXK (X stands for any amino acid), a few genes contain only G1 box, G3 box, and G4 box (Figures 1, 2B). However, different Ras GTPases have many specific motifs, resulting in different domains (Figure 2), and the differences of their amino acid sequences mainly occur in the amino terminal and carboxyl terminal, which are considered as major protein modification sites (Figure 1).
3.4 Gene expression patterns of the Ras superfamily of L. vannamei
In order to clarify the expression patterns of Ras superfamily genes of L. vannamei, we summarized four different transcriptional profiles of 108 Ras GTPase genes into heatmaps of different tissues, development stages, molting stages and WSSV infection status (Figures 3–6; Supplementary Figures S1-6).
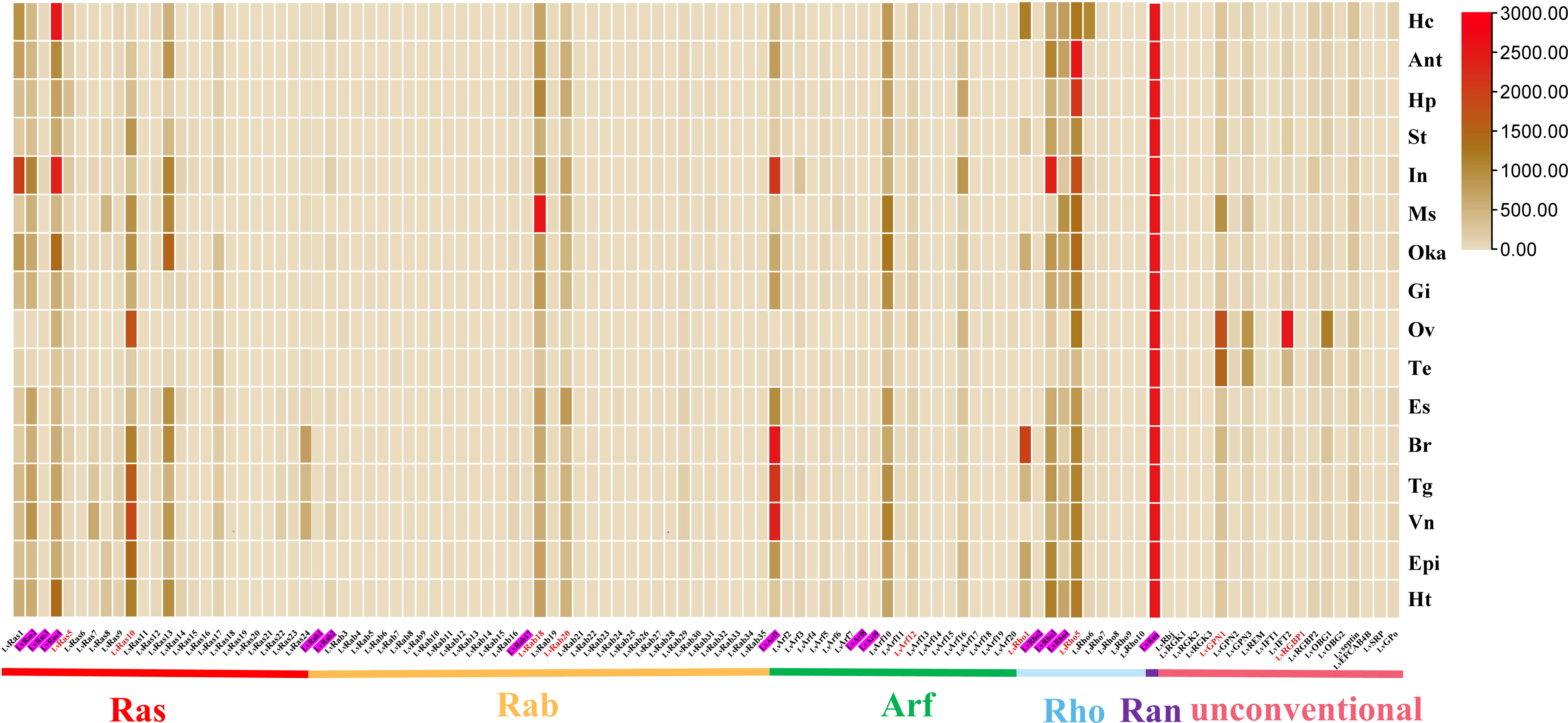
Figure 3 Ras superfamily gene expression profiles in different tissues of L. vannamei. Adult tissues: Hc, hemocyte; Ant, antenna; Ms, muscle; In, intestines; Ov, ovary; St, stomach; Oka, lymphoid organ; Gi, gill; Hp, hepatopancreas; Te, testis; Es, eye stalk; Br, brain; Tg, thoracic ganglion; Vn, ventral nerve; Epi, epidermis; and Ht, heart. Pink shading represents members of the classical Ras superfamily in each classification, red fonts represent members of the Ras superfamily with validated expression levels in each classification.
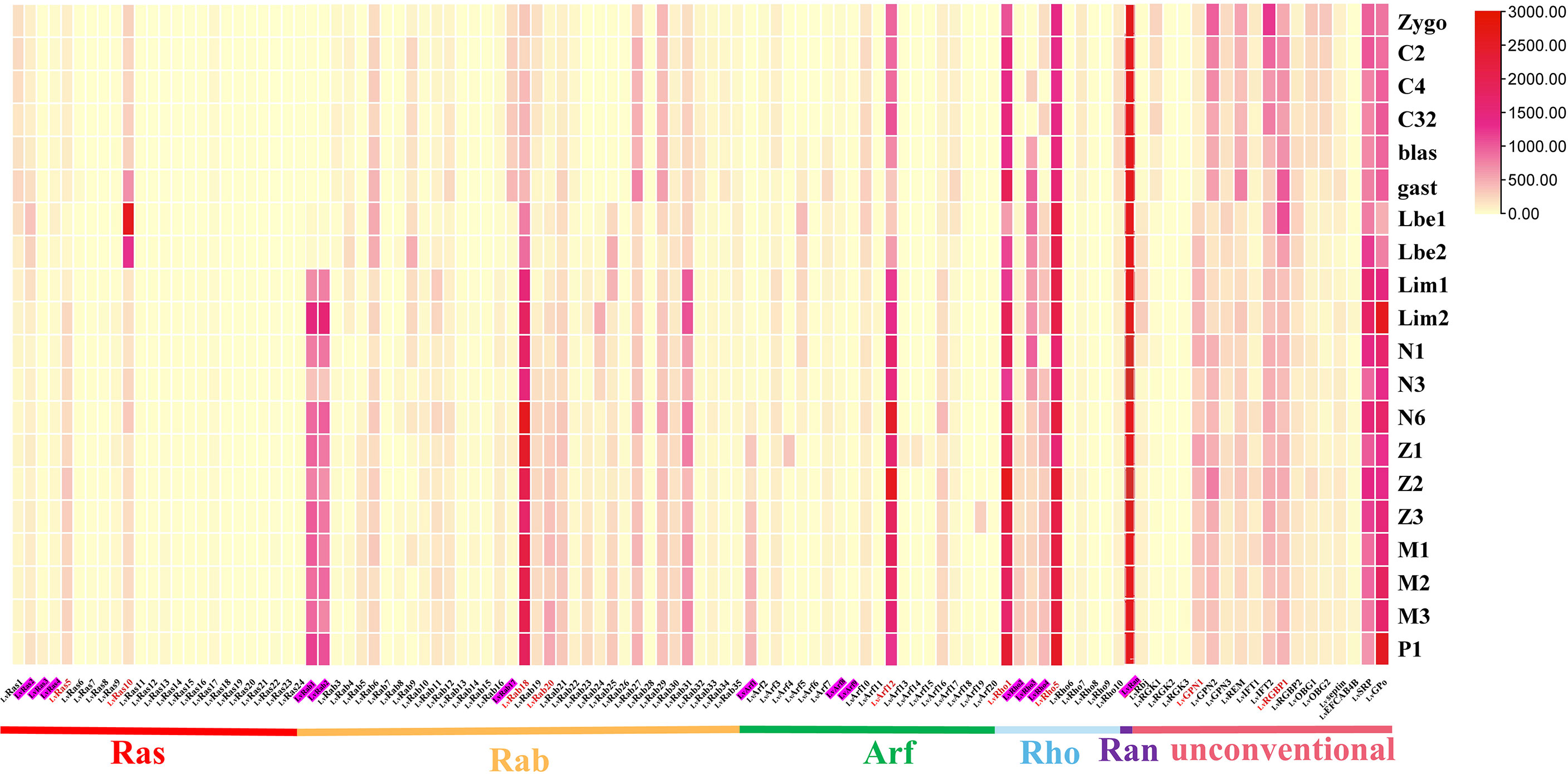
Figure 4 Ras superfamily gene expression profiles at different early developmental stages of L. vannamei. Early development stages: zygote (zygo), 2 cells (C2), 4 cells (C4), 32 cells (C32), blastula (blast), gastrula (gast), limb bud embryo I (Lbe1), limb bud embryo II (Lbe2), larva in membrane I (Lim1), larva in membrane II (Lim2), nauplius I (N1), nauplius III (N3), nauplius VI (N6), zoea I (Z1), zoea II (Z2), zoea III (Z3), mysis I (M1), mysis II (M2), mysis III (M3), and post larvae 1 (P1). Pink shading represents members of the classical Ras superfamily in each classification, red fonts represent members of the Ras superfamily with validated expression levels in each classification.
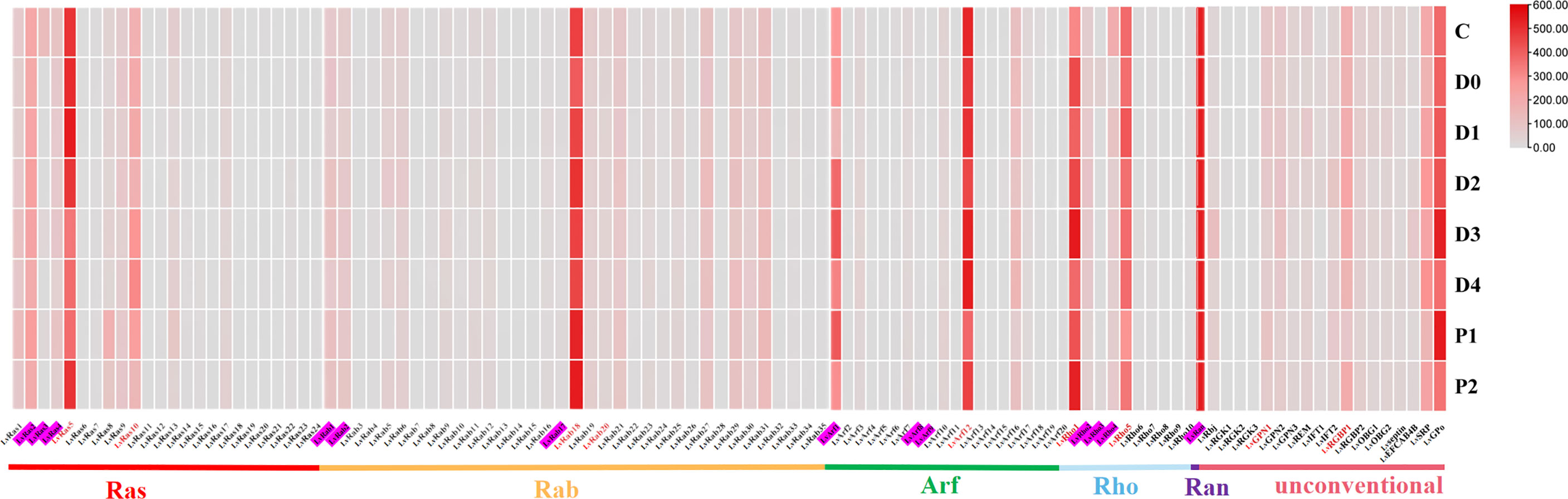
Figure 5 Ras superfamily gene expression profiles at different molting stages of L. vannamei. Molting stages: intermolting phase (C), premolting phase (D0, D1, D2, D3, and D4), and postmolting phase (P1 and P2). Pink shading represents members of the classical Ras superfamily in each classification, red fonts represent members of the Ras superfamily with validated expression levels in each classification.
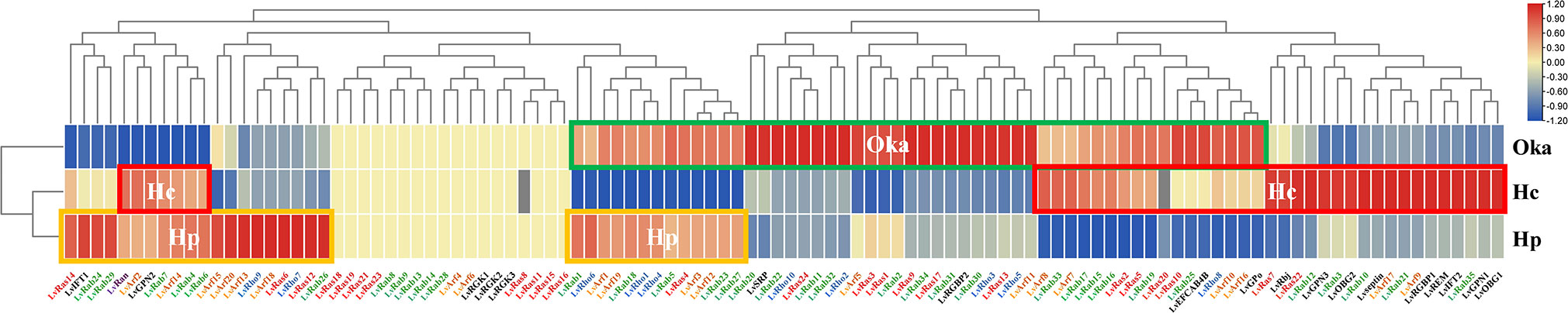
Figure 6 Changes of Ras superfamily gene expressions after WSSV infection of L. vannamei. The orange line (Hp) shows high expression in hepatopancreas tissue, the red line (Hc) shows high expression in hemocyte, the green line (Oka) shows high expression in lymphoid organ. Sterile phosphate-buffered saline (PBS) as control group, the infection lasts for 6 hours.
In different tissues of adult shrimp, most members of Ras superfamily showed low expression, a few genes were highly expressed in their respective families, such as LvRab18, LvRab20, LvRho3, LvRho5, LvRas4, LvRas10, LvArf1, LvArf10, and LvRan. (Figure 3). LvRan had the highest expression level in all the examined tissues, and its expression level is more than 3-5 times that of other members (Figure 3; Supplementary Figures S1, S2A). Other highly expressed Ras superfamily genes usually presented in some specific tissues: LvRas4 was highly expressed in hemocyte and intestines; LvRab18 was highly expressed in muscle; LvArf1 was highly expressed in brain, thoracic ganglion, ventral nerve and intestines; and LvRho5 was highly expressed in antenna, hepatopancreas and intestines. Some unconventional Ras superfamily members were highly expressed in testis and ovary tissues, such as LvGPN1, LvGPN3, LvIFT2 and LvOBG1.
At early development, most members of Ras superfamily were expressed at a higher level. LvRas1, LvRas2, LvRas5 and LvRas10 are the main expressed Ras family genes, in addition to LvRas10 at gastrula (gast) and limb bud embryo (Lbe) stages, the expression of Ras genes was not very obvious in early development as a whole. The Rab family showed two expression patterns, some genes were highly expressed mainly during the zygote to gast stages, while others were highly expressed after limb bud embryo stages. Arf12, Rho1 and Rho5 were the most expressed genes in their respective families, and their expression trends remained relatively stable during different stages (Figure 4; Supplementary Figure S3). Ran was the gene with the highest expression of all Ras superfamily numbers in the early developmental stages of L. vannamei, but with little fluctuations (Supplementary Figure S4A). Interestingly, most of the unconventional Ras superfamily genes had high expression levels at early development stages. LvSRP and LvGPo were highly expressed at the whole stages, showing that play an important role in early development. LvREM, LvGPN2, LvIFT2, and LvRGBP1 had similar expression patterns, they were all highly expressed after gastrula stage (gast). In contrast, LvRbj and LvGPN1 were highly expressed before gastrula stage (gast) (Supplementary Figures S4B-H).
In different molting stages of L. vannamei, most members of Ras superfamily showed lower expression (Figure 5; Supplementary Figure S5), and only a few genes were highly expressed in respective different families, such as LvRab18, LvRho1, LvArf12, LvRan, LvRas5. Similarly, a few unconventional members were highly expressed at different molting stages, such as LvRGBP1, LvSRP and LvGPo (Supplementary Figures S6B-H).
The hemocyte, hepatopancreas, lymphoid (Oka) organs are the three main immune related tissues. The expression level of many Ras superfamily genes was up-regulated in different organs after WSSV infection. In lymphoid organs, 19 Rab genes, 8 Rho genes, 11 Ras genes, 10 Arf genes, LvRGBP2, LvSRP, LvEFCAB4B and LvGPo were significantly up-regulated. In hepatopancreas, 11 Rab genes, 5 Rho genes, 4 Ras genes, 10 Arf genes, LvIFT1 and LvGPN2 genes were significantly up-regulated. In hematocyte, 6 Rab genes, 1 Rho gene, 7 Ras genes and 8 Arf genes were up-regulated, most unconventional Ras superfamily genes were also significantly up-regulated (except for LvRGKs, LvSRP and LvRGBP2) (Figure 6; Supplementary Table S2). The types of up-regulated Ras superfamily genes were different in different tissues, and it is possible that these genes play different roles in the immune process. For example, LvRan was up-regulated in hemocyte and hepatopancreas, but down-regulated in lymphoid organs after WSSV infection. In lymphoid organs, most of the Ras superfamily genes were up-regulated and a few genes were down-regulated, while was opposite in hepatopancreas. Among them, some Ras and Arf genes and LvRGK showed no changes in their expression levels in the three organs after WSSV infection, indicating that these genes are not affected by immune response.
3.5 Expression verification of some Ras superfamily genes
In order to verify the accuracy of the expression level of different tissues, the expression of twelve Ras superfamily genes were verified by qRT-PCR in twelve different adult tissues (Figure 7). LvRas5 and LvRas10 were highly expressed in hepatopancreas and ventral nerve respectively, and LvRas6 was highly expressed in ventral nerve, lymphoid organ and brain. LvRab18, LvRab20, LvRho1 and LvRho5 were relatively expressed in most tissues except eyestalk, epidermis and muscle, among the expression tissues, these genes were mainly highly expressed in ventral nerve, brain, lymphoid organ and hepatopancreas, which is similar to the expressed pattern of verified Ras genes above. LvArf1 and LvArf12 were expressed in most tissues, and the three tissues with the highest expression were brain, ventral nerve and gill. LvRan was highly expressed in ventral nerve, and is about 2-4 times higher than other tissues. In the unconventional Ras superfamily members, LvGPN1 and LvRGBP1 had similar expression patterns with the classical Ras superfamily members, which were mainly highly expressed in hepatopancreas, brain, heart and other tissues. In sum, the most expression levels of these genes were consistent with respective RNA-Seq results, and some differences might be caused by individual differences. We found that almost all verified members of Ras superfamily were expressed in almost all detected tissues, and showed the lowest expression level in eyestalk, epidermis and muscle tissues. It is possible that Ras GTPases play a weak role in these tissues.
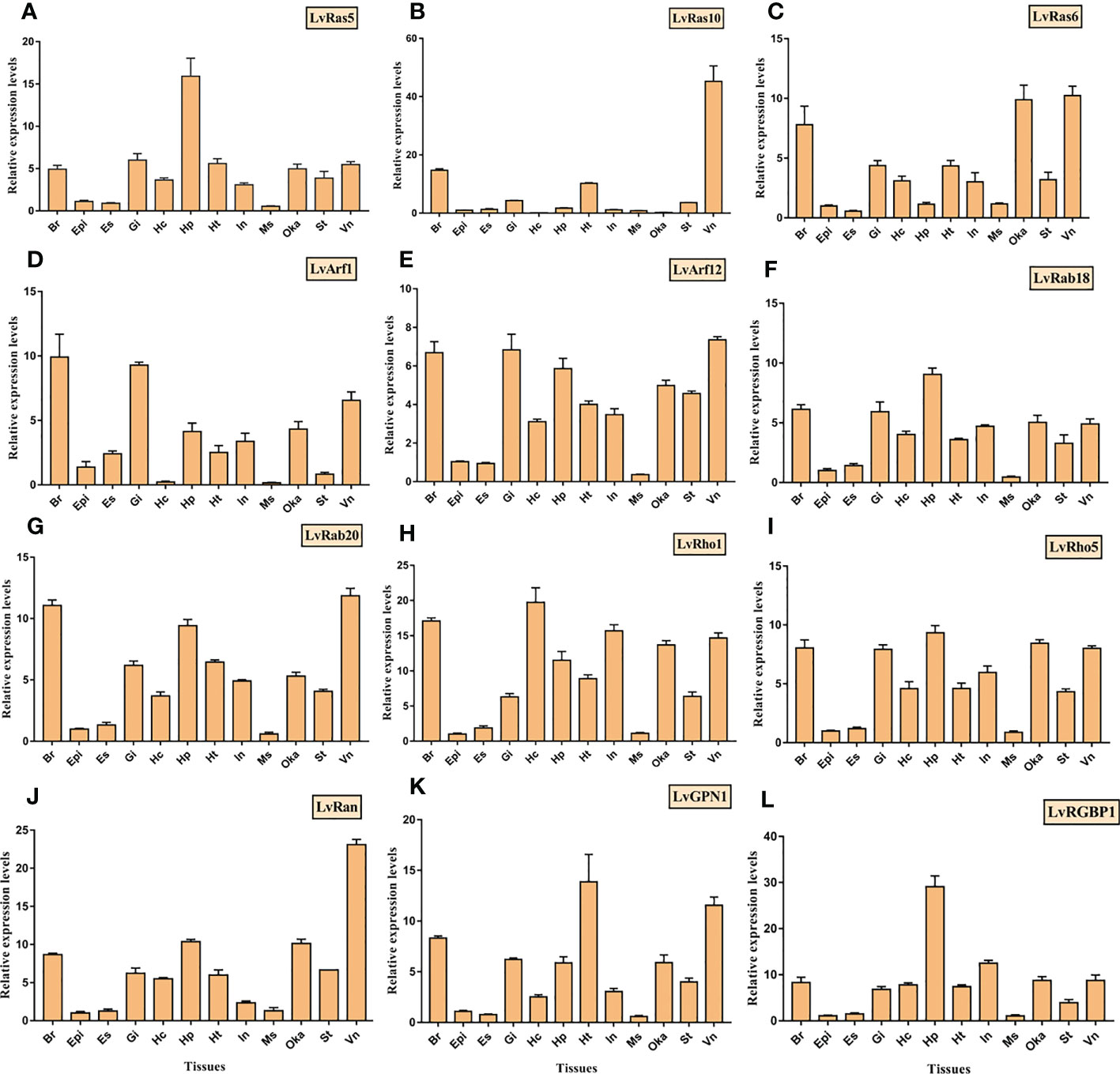
Figure 7 Tissue distributions of 12 Ras superfamily genes were detected by RT-qPCR, and (A–L) was LvRas5, LvRas10, LvRas6, LvArf1, LvArf12, LvRab18, LvRab20, LvRho1, LvRho5, LvRan, LvGPN1, LvRGBP1, respectively. Adult tissues: Hc, hemocyte; Ant, antenna; Ms, muscle; In, intestines; Ov, ovary; St, stomach; Oka, lymphoid organ; Gi, gill; Hp, hepatopancreas; Te, testis; Es, eye stalk; Br, brain; Tg, thoracic ganglion; Vn, ventral nerve; Epi, epidermis; and Ht, heart." . Please note that "LvRas5, LvRas10, LvRas6, LvArf1, LvArf12, LvRab18, LvRab20, LvRho1, LvRho5, LvRan, LvGPN1, LvRGBP1.
4 Discussion
The Ras superfamily is a large gene family, which generally contains fewer members in invertebrates and more in vertebrates, such as 68 in D. melanogaster and 46 in C. elegans, correspondingly, 170 in human and 137 in Xenopus tropicalis (Rojas et al., 2012). In metazoans, although the total number of Ras superfamily varies in different species, the Ras superfamily has traditionally been divided into five major branches: Ras, Rho, Rab, Ran, and Arf/Sar. Among them, the Rab family has the most members, and the Ran family has the least members (usually one). In this study, shrimp obviously has an expanded Ras superfamily (108 members), in which, Contains five classic Ras families and with the most Rabs and the least Rans, implying a high degree of conservation of the structure and pattern of this superfamily.
From the perspective of gene structure, most these Ras GTPase have a close genetic relationship and very similar binding GTP related motifs, indicating that these genes have the same ancestor, and the gene structure differentiation originated from different functional requirements in the process of evolution. Phylogenetic analysis showed that among these different families, Ras and Rho families are closely related, clustered on the same branch and have high homology; Rab family cluster with Ran, then combine with Ras and Rho into a larger branch. However, Arf family is a relatively separate branch, and the unconventional Ras superfamily numbers are clustered into larger branch with the Arf family. The result indicated that the members of Ras superfamily might come from two different origins, which is consistent with previous studies: Ras, Rho, Rab and Ran were probably derived from Cyanobacteria or proteobacteria, or the common ancestor of both; and Arf was probably derived from Methanogenus (Dong et al., 2007).
Comprehensive snapshots of the patterns of gene expression can provide a path toward a global and dynamic understanding of gene functions and their roles in particular biological processes or events. In this study, the expression analysis of Ras superfamily genes showed that the expression of different Ras GTPases was different under different conditions (different tissues, development/molting stages and WSSV infection). In general, from the expression pattern, it can be concluded that the functions of the Ras superfamily of shrimp are similar to other animals, which are very diverse and complex (Table 2). Only a few members of each family are highly expressed, suggesting that these genes are critical in the corresponding biological processes, while other members may correspond to other specific conditions, allowing for more fine-grained regulation. During early development, unconventional Ras family members are generally expressed more actively than the classical Ras superfamily members, which suggested that the unconventional members play a vital role in early developmental stages. In addition, after WSSV infection, almost all of Ras superfamily genes are up-regulated in different tissue, which at least suggests that these genes play an important role in development and pathogenesis.
4.1 The Ras family
Ras family is the most typical and common class of Ras superfamily. In L. vannamei, multiple Ras family members such as Ras, Rap, Ral, Rheb, RHEs and RIT were found. Gene expression analysis showed that these Ras genes were expressed in different tissues, indicating their respective functions. Most members of the Ras family in mammals are called oncogenes and well-studied. Ras proteins receive signals from cell surface receptors, these signals are transmitted among proteins through different pathways, and finally affect a variety of biological functions, such as development, proliferation, differentiation, and survival (Goitre et al., 2014). If the Ras gene was mutated, these signal pathways will be destroyed, which will lead to a variety of tumors and cancers (Bos, 1989; Goodsell, 1999; Murugan et al., 2019). In addition, some Ras family genes are considered to be involved in animal growth and reproduction. For example, the Ras gene is related to the size of catfish head (Geng et al., 2016), it is involved in the regulation of insulin pathway during oocyte vitellogenesis in female oysters (Jouaux et al., 2012). During vitellogenesis of the marine flounder Solea senegalensis, the expression of Ras homologous gene was up-regulated (Tingaud-Sequeira et al., 2009). In L. vannamei, Ras family genes also were highly expressed in ovary, such as LvRas4 and LvRas10. In previous study, we conducted growth candidate gene association analysis in two independent populations of L. vannamei, the results showed that the SNP of Ras related protein gene Rap-2a (LvRas1) was significantly correlated with growth traits (Yu et al., 2019). The above research proves that Ras family may play an important role in growth and reproduction of shrimp.
There are also other classic Ras family genes in L. vannamei, such as M-Ras and Ras related protein R-Ras2. M-Ras alone constitutes a small Ras subfamily in mammals, in L. vannamei, M-Ras gene was named as LvRas2, and showed high relative expression under most conditions, indicating that M-Ras plays an important role in shrimp. Classical Ras binds to Raf and activates ERK pathway (Endo, 2020), while M-Ras mediated cell transformation related to the weak activation of RAF/MEK/ERK pathway, other downstream effectors are also involved in the pathway, which can induce neuronal differentiation (Castro et al., 2012). In mice, both classical Ras and M-Ras are highly expressed in the central nervous system (Sun et al., 2006). In addition, neuronal differentiation of p12 cells in rats also requires the induction of classical Ras and M-Ras (Sassone-Corsi et al., 1989). R-Ras2 has also been shown to be essential for correct axonal myelination and accurate neurotransmission in mouse (Gutierrez-Erlandsson et al., 2013; Sanz-Rodriguez et al., 2018). In L. vannamei, both M-Ras (LvRas2) and R-Ras2 (LvRas6) gene were highly expressed in the thoracic ganglion, which further showed that Ras family genes may play an important role in nervous system development and signal transduction.
At present, there are few studies on Ras family of L. vannamei, but as these typical Ras superfamily proteins may play an important role in metabolism, growth, reproduction and neural development, the related mechanisms need to be further studied.
4.2 The Rab family
Rab is a kind of regulatory small molecule GTPase protein, found on eukaryotic cell and organelle membrane (Seabra et al., 2002), and it also is a group with the largest number of genes of the Ras superfamily in L. vannamei. For example, there are 57 Rab genes in Arabidopsis, 30 in D. melanogaster, and 61 in Mus musculus (Rojas et al., 2012). The main members of Rab family include Rab1, Rab3A and Rab5c.
Rab, called Ypt in yeast, plays an important role in vesicle transport (Takai et al., 2001). Rab8 and Rab5 in human have the same function as YPT in yeast, and they share high sequence similarity (Molendijk et al., 2004). It has also been reported that Rab protein plays a role in vesicle transport in plants. For example, Rab1, Rab2, Rab4, Rab5 and Rab6 in Arabidopsis have been proven to play an important role in regulating the morphology of endoplasmic reticulum, Golgi apparatus and plasma membrane vesicles (Stenmark and Olkkonen, 2001). In the squid Loligo pealei, it was verified that Myo5a and Rab3A directly bind and interact with synaptic vesicles (SVs) and participate in the transport of neuronal vesicles (Wöllert et al., 2011). In L. vannamei, studies showed that Rab might be related to the resistance mechanism of shrimp induced by environmental stress (Wang et al., 2015). In addition, Rab gene also plays an important role in shrimp immunity, Rab like protein had been proved to interact with immune related membrane proteins to regulate the phagocytosis of shrimp hemolymph cells against WSSV, and Rab27 mutation leads to immune deficiency (Wu et al., 2008; Han et al., 2011; Chen et al., 2021). Similarly, Rab gene (Rab6a) in M. japonicus was up-regulated during WSSV infection (Wu and Zhang, 2007). In this study of WSSV-infected L. vannamei, several Rab family genes were significantly up-regulated in immune organs such as Oka organ, hepatopancreas and hemocyte. Therefore, Rab family may affect vesicle morphology, vesicle transport and immune regulation in shrimp.
4.3 The Rho family
In 1985, Madaule found a Ras superfamily member Rho (RAS homolog) in the marine gastropod molluscs Aplysia (Kawasaki et al., 2004). The study confirmed that Ras has highly sequence homology with Rho, with 35% consistency in amino acid sequence. Moreover, they have the same C-terminal required for membrane attachment. In L. vannamei, Rho family were found in the same clade of the phylogenetic tree with Ras family. These results suggest that Rho family is closely related to Ras family. In vertebrates, the Rho family has undergone considerable expansion and differentiated into more than 10 subfamilies, including Rho subtypes (A, B, C, D, G, E), Rac subtypes (1, 2, 3), Cdc42, Rnd (1, 2, 3), TCL, Rho H/TTF, Chp, Wrch-1, Rif, Rho BTB1, Rho BTB2, Miro-1 and Miro-2 (Burridge and Wennerberg, 2004; Wennerberg and Der, 2004). Five Rho subfamilies, including Rhol, Cdc42, Rac2, MIG-2, Rho BTB1, were identified in L. vannamei.
As a representative and well-studied member of Rho family, Cdc42 plays a significant role in a variety of cellular processes that are dependent on the actin cytoskeleton, such as cytokinesis, cell migration, phagocytosis, morphogenesis, axon myelination, intracellular trafficking, and tumor occurrence (Etienne-Manneville and Hall, 2002; Sahai and Marshall, 2002; Moon and Zheng, 2003). Rac is another member of the Rho family with more research. In mammals, Rac is mainly involved in promoting the malignant proliferation and migration of tumor cells (Chan et al., 2007). Compared with normal striated muscle tissue, the expression of Rac1 and Cdc42 was significantly high (P < 0.05) in rhabdomyosarcoma (RMS) tissue (Li et al., 2021). In addition, Rac also plays a role in immunity, the expression of Rac2 gene in the large yellow croaker Pseudosciaena crocea was significantly up-regulated after challenge by Vibrio parahaemolyticus (Liu et al., 2017). Injection of Vibrio alginolyticus into L. vannamei induced up-regulated expression of LvRac1 (LvRho1) in hepatopancreas, then after knocking down LvRac1 and stimulating V. alginolyticus, the mortality of L. vannamei was significantly increased relative to that of the control group (Cha et al., 2015). Rho1 is another important Rho GTPase protein, and previous research has shown a role for this in immunity, cytoskeleton dynamics and embryonic development. For example, Rho1 affected the development of eggs by regulating the hormone level of the braconid wasp Microplitis mediator (Magie and Parkhurst, 2005). Rho1 also regulated the rearrangement of cytoskeleton, and then affected the cellular immunity of the cotton bollworm Helicoverpa armigera (Li et al., 2010). In the purple sea urchin Strongylocentrotus purpuratus, Rho could affect SpROCK expression by the Rho dependent signal pathway, which is essential for early embryonic development (Aguirre-Armenta et al., 2011). In this study, we suggest Rho family may play an important role in development, cytoskeleton dynamics and immunity in L. vannamei.
4.4 The Ran family
Although the Ran is small family in number, it plays an important role in many species. In H. armigera, it was reported that Ran participated in the 20-hydroxyecdysone (20E) signal transduction pathway by regulating the location of ecdysone receptor-B1 (EcR-B1) (He et al., 2010). In the brown planthopper Nilaparvata lugens, NlRan knockdown significantly delayed development and affected reproduction (Liao et al., 2019). Only one Ran gene exists in L. vannamei, and its expression is higher than that of other Ras superfamily genes. It was previously reported that the highest expression of Ran is the black tiger shrimp, Penaeus monodon is in the ovary (Zhou et al., 2012). In the expression profile of testis maturation stages of scallop, Ran expression increased dramatically during meiosis and spermatogenesis (Hino et al., 2012). In addition, there is a similar situation in mammals. Ran had a high level of expression from the late pachytene spermatocytes to early round spermatocytes in mice (López-Casas et al., 2003). Moreover, the cellular localization of Ran also changed during spermatogenesis (Kierszenbaum et al., 2002). On the other hand, Ran may also involve in the immune process. In the Kuruma shrimp M. japonicus, Ran played a vital role in antiviral immunity (Han and Zhang, 2007). Another study found that Ran interacts with myosin in M. japonicus, which can regulate blood cell phagocytosis. RNAi knockdown led to a significant increase in virus copy number in M. japonicus, and overexpression of Ran resulted in a significant decrease in virus copy number (Liu et al., 2009). When IL-4 and lysophosphatidylcholine were respectively injected into shrimp, the results indicated that the two molecules could enhance the Ran GTPase activity and improve hemocytic phagocytosis against WSSV (Zhao et al., 2011). In our study, LvRan was significantly up-regulated in hemocytes and hepatopancreas, but down-regulated in Oka after WSSV infection. The above research suggests that Ran may participate in the 20E signaling pathway, regulate the gonad development and antiviral immunity, and then affect the growth and reproduction in shrimp.
4.5 The Arf family
The Arf family is mainly divided into three subfamilies: Arf, Arf like proteins (ARLs) and Sar1. Relevant studies have confirmed that Arf and the components that promote Arf function played an important role in mediating the transport of endoplasmic reticulum to the Golgi (Balch et al., 1992; Dong et al., 2010). For example, Arf1 regulated vesicle formation, Golgi assembly and promoting vesicle division, and Arf6 promoted membrane invagination on the cell surface during endocytosis (D’Souza-Schorey and Chavrier, 2006). Arf and Rab have similar functions in vesicle formation and transportation, indicating that the connection between GTPase-mediated signaling pathways requires different Ras superfamily proteins to fulfill a common task by a cooperation (Mitin et al., 2005). In D. melanogaster, Arf6 was only involved in spermatogenesis (Lambaerts et al., 2009), it was not required for early development in mice (Doherty and McMahon, 2009). In L. vannamei, Arf4 was almost not expressed in the early development stages, however, it had a higher expression in adults. There is almost no research on Arf in crustaceans, we found most Arf genes highly expressed in brain and ventral nerve of L. vannamei, so it is speculated that Arf should have a certain effect on the nervous system in shrimp.
4.6 The unconventional Ras GTPase families in L. vannamei
The researches of the five classical Ras GTPase families have been very in-depth. Furthermore, many genes have regions predicted as or similar to the small GTPase domain, but they’re unconventional, for example, RJL family, RGK family, GPN family, IFT family, OBG family and Septin family. They have the same motif as classical Ras superfamily members, and the prediction region acts as a signal converter or molecular switch, called GTP-binding protein and has GTPase activity. Unlike the other members of the classical Ras GTPase, most these non-classical members showed extremely low levels of expression in adult tissues of L. vannamei, but most of them showed high levels of expression during early development and WSSV infection, this indicates that their functions are relatively specific and may be related to early development and immunity.
4.6.1 RJL family
RJL is a new member of Ras superfamily reported in recent years (Gao et al., 2019). Besides the GTP binding domain, members of this family also contain an additional DnaJ domain, so they are also named the DnaJ family. The family is divided into two subfamilies: Rjl and Rbj. Different from other Ras superfamily members, The RJL family lacks membrane targeting signal and their hydrolysis ability of GTP is impaired. The RJL family exists in many protozoa and deuterostome metazoans, but is obviously missing in some intermediate phyla, indicating an interesting possibility of horizontal gene transfer (HGT) between lower and higher eukaryotes (Nepomuceno-Silva et al., 2004). In human gastrointestinal cancers, Rbj was dysregulated and could promote tumor progression, the activation of MEK and ERK by Rbj indicated that RJL family might have a role in MEK/ERK signaling pathway (Gao et al., 2019; Chen et al., 2021). No any RJL family member has been reported in shrimp so far. In this study, the Rjl gene with GTP binding domain and DnaJ domain was found in L. vannamei, and called LvRbj. This gene was little highly expressed in the limb bud embryo (Lbe) and Lim stages, while in the adult, it only had low expression in eyestalk and blood cells, and only significantly highly expressed in hemolymph after WSSV interference, so LvRbj might involve in immune and neural regulation of shrimp.
4.6.2 RGK family
In this study, we identified three unconventional Ras superfamily genes of L. vannamei, LvRGK1-3. The RGK family includes Rad, Rem, Rem2 and Gem/Kir, which are called “distant cousins” of the typical G proteins and constitute the first unconventional Ras subfamily with a novel effector binding mechanism distinct from that of other Ras GTPases (Miranda et al., 2021). Rad (Ras associated with diabetes) is mainly in skeletal muscle and cardiac muscle, and its expression increase by an average of 8.6 times in muscle of type II diabetes. It is speculated that Rad may be an inhibitor of Ras, interfering with the function of normal Ras, Rab or Rap (Reynet and Kahn, 1993). In addition, Rad can be phosphorylated by PKA, but it does not affect GTP binding and GTPase activity like classical Ras superfamily members, indicating it may have a specific GAP-like activity regulation mechanism (Zhu et al., 1995). Rem is the first Ras-related GTP-binding protein whose mRNA levels are regulated by repression after stimulation (Finlin and Andres, 1997). In human, RGK GTPase family genes bind directly to Ca2+ channel β-subunits (CaVβ), serve as regulators of Ca2+ channel activity (Finlin et al., 2003). In conclusion, RGK family is mainly highly expressed in skeletal muscle, cardiac muscle and other tissues, and plays a major role in the calcium and insulin signaling pathway. In L. vannamei, the overall expression level of RGK family was low, but they were higher expressed in skeletal muscle and cardiac muscle than most other tissues.
4.6.3 GPN family
GPN-loop GTPase (GPN) is a member of P-loop NTPase, which has a GTP binding domain similar to the classical Ras superfamily (Forget et al., 2010). However, this family is rare and there are few relevant studies. In the yeast S. cerevisiae, the deletion of Gpn1 or its homologous genes Gpn2 and Gpn3 was fatal (Giaever et al., 2002), Gpn1, Gpn2, and Gpn3 were all essential proteins for cell growth, and deletion of each one resulted in cell death, suggesting that these GPN-like GTPases may be necessary for survival and their functions are not redundant (Liu et al., 2020). Three GPN subfamily genes were found in L. vannamei: LvGPN1, LvGPN2 and LvGPN3, their expression patterns were similar, mainly in gonads and muscles, and significantly increased in hemocyte after WSSV infection, but the specific role is unknown.
4.6.4 IFT family
The intraflagellar transport (IFT) family contains at least 20 different proteins and can be resolved into two smaller subunits, complexes A and B (Cole et al., 1998), complexe A contains 6 protein subunits (IFT43, 121, 122, 139, 140, 144), and complexe B contains 14 protein subunits (IFT20, 22, 25, 27, 46, 52, 54, 57, 70, 72, 74, 80, 81, 88, 172) (Fan et al., 2010). Only IFT-B complexes were found in L. vannamei: IFT22 and IFT27. As core subunits of IFT-B complexes, IFT22 and IFT27 have significant sequence homology with members of Ras superfamily. IFT27 was predicted to be Rab-like GTPase and proved to binding to GTP (Qin et al., 2007). IFT complex has a function similar to small GTPase, but its GTPase activity is very low, due to the lack of conserved catalytic Gln sites, so some GTP-activating proteins (GAPs) are needed to exchange between GTP and GDP (Bhogaraju et al., 2011). Therefore, the IFT complex, as an important structure in cilia assembly and maintenance, may be required for ciliogenesis by ferrying ciliary components using IFT complexes as cargo adaptors and may be necessary for normal life activities.
4.6.5 OBG family
OBG like-GTPase is a subfamily of P-loop GTPase, which was originally found downstream of Spo0b in the Gram-positive bacteria Bacillus subtilis (Trach and Hoch, 1989). Although these proteins contain GTP binding domains and conserved from bacteria to human, their sequence homology with other GTP-binding proteins is low (Kukimoto-Niino et al., 2004), so they are divided into a new group: OBG family, which contains three domains: OBG folding, G domain and OBG c-terminal region (OCT). In this study, an OBG-like GTPases protein of L. vannamei was identified, which has three domains, GTP1_OBG (OBG folding), FeOB_N, MMR_HSR1, among them MMR_HSR1 interacts with 50S ribosome and is necessary for binding adenine and guanine nucleotides to have complete activity. At present, research on OBG-like GTPases mainly focused on bacteria, yeast and plant chloroplasts (Chigri et al., 2009; Lin et al., 2018), and there are few reports in animals.
4.6.6 Septin family
Septin, a unique polymeric Ras superfamily protein with GTPase activity, was found in L. vannamei. Septin was described mainly as a spatial regulator of protein localization and interaction in the budding yeast, it is the key to their asymmetric cell shape and division (Spiliotis and McMurray, 2020). In human, septin2 is a cancer promoting gene, its overexpression can promote the proliferation of gastric cancer cells and inhibit apoptosis (Li et al., 2018). Septin2 gene was highly expressed in liver cancer tissues and corresponding adjacent tissues (Xu et al., 2019). Septin is classified as a member of the Ras superfamily because it has the same GTP binding motif as most Ras GTPases, and it is closely related to cell proliferation and oncogenesis. In crustaceans, the function of Septin is unclear.
5 Conclusion
In this study, based on genome and transcriptome data, we have conducted comprehensive analyses of gene structure, protein domain, and expression patterns of the Ras superfamily members in the economically important shrimp, L. vannamei. The results showed that the Ras superfamily is relatively complete in shrimp, a total of 108 Ras superfamily genes were identified. We found that shrimp contained not only all classical Ras superfamily members, but also some unconventional and novel Ras superfamily genes, these genes shared common conserved domain and motifs. From a phylogenetic point of view, Ras superfamily of L. vannamei are divided into two clades, They had different expression patterns and might have diversified functions in development, growth and immune response. These works provide important clues for future research on the function of Ras superfamily genes in crustaceans, which is of great significance for understanding growth development and immunity mechanism and promoting genetic breeding of shrimp.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
XJZ, SS, SY, and FL conceived and designed the experiments. SS and XJZ performed the experiments and data analyses. SS wrote the manuscript and prepared all the figures. XJZ, JY, and XXZ reviewed the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (Grant No. XDA24030105), the National Key R&D Program of China (2018YFD0900103, 2022YFF1000304), and the National Natural Sciences Foundation of China (31972782 and 32273102).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1063857/full#supplementary-material
References
Aguirre-Armenta B., López-Godínez J., Martínez-Cadena G., García-Soto J. (2011). Rho-kinase in sea urchin eggs and embryos. Dev. Growth Differentiation 53 (5), 704–714. doi: 10.1111/j.1440-169X.2011.01280.x
Balch W. E., Kahn R. A., Schwaninger R. (1992). ADP-ribosylation factor is required for vesicular trafficking between the endoplasmic reticulum and the cis-golgi compartment. J. Biol. Chem. 267 (18), 13053–13061. doi: 10.1016/S0021-9258(18)42380-0
Bhogaraju S., Taschner M., Morawetz M., Basquin C., Lorentzen E. (2011). Crystal structure of the intraflagellar transport complex 25/27. EMBO J. 30 (10), 1907–1918. doi: 10.1038/emboj.2011.110
Burridge K., Wennerberg K. (2004). Rho and rac take center stage. Cell 116 (2), 167–179. doi: 10.1016/s0092-8674(04)00003-0
Castro A. F., Campos T., Babcock J. T., Armijo M. E., Martínez-Conde A., Pincheira R., et al. (2012). M-ras induces ral and JNK activation to regulate MEK/ERK-independent gene expression in MCF-7 breast cancer cells. J. Cell. Biochem. 113 (4), 1253–1264. doi: 10.1002/jcb.23458
Cetkovic H., Mikoc A., Mueller W. E. G., Gamulin V. (2007). Ras-like small GTPases form a large family of proteins in the marine sponge suberites domuncula. J. Mol. Evol. 64 (3), 332–341. doi: 10.1007/s00239-006-0081-3
Cha G. H., Wang W. N., Peng T., Huang M. Z., Liu Y. (2015). A Rac1 GTPase is a critical factor in the immune response of shrimp (Litopenaeus vannamei) to vibrio alginolyticus infection. Dev. Comp. Immunol. 51 (2), 226–237. doi: 10.1016/j.dci.2015.04.004
Chan A. A. M., Brenner M., Zheng Y., Gulko P. S., Symons M. (2007). The GTPase rac regulates the proliferation and invasion of fibroblast-like synoviocytes from rheumatoid arthritis patients. Mol. Med. 13 (15-16), 297–304. doi: 10.2119/2007-00025.Chan
Chen C., Chen H., Zhang Y., Thomas H. R., Frank M. H., He Y., et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13 (8), 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen T., Yang M., Yu Z., Tang S., Wang C., Zhu X., et al. (2021). Small GTPase RBJ mediates nuclear entrapment of MEK1/MEK2 in tumor progression. Cancer Cell 39 (3), 438–440. doi: 10.1016/j.ccell.2021.02.010
Chigri F., Sippel C., Kolb M., Vothknecht U. C. (2009). Arabidopsis OBG-like GTPase (AtOBGL) is localized in chloroplasts and has an essential function in embryo development. Mol. Plant 2 (6), 1373–1383. doi: 10.1093/mp/ssp073
Cole D. G., Diener D. R., Himelblau A. L., Beech P. L., Fuster J. C., Rosenbaum J. L. (1998). Chlamydomonas kinesin-II-dependent intraflagellar transport (IFT): IFT particles contain proteins required for ciliary assembly in Caenorhabditis elegans sensory neurons. J. Cell Biol. 141 (4), 993–1008. doi: 10.1083/jcb.141.4.993
Colicelli J. (2004). Human RAS superfamily proteins and related GTPases. Sci. STKE 2004(250), Re13. doi: 10.1126/stke.2502004re13
Doherty G. J., McMahon H. T. (2009). Mechanisms of endocytosis. Annu. Rev. Biochem. 78, 857–902. doi: 10.1146/annurev.biochem.78.081307.110540
Dong J.-H., Wen J.-F., Tian H.-F. (2007). Homologs of eukaryotic ras superfamily proteins in prokaryotes and their novel phylogenetic correlation with their eukaryotic analogs. Gene 396 (1), 116–124. doi: 10.1016/j.gene.2007.03.001
Dong C., Zhang X., Zhou F., Dou H., Duvernay M. T., Zhang P., et al. (2010). ADP-ribosylation factors modulate the cell surface transport of G protein-coupled receptors. J. Pharmacol. Exp. Ther. 333 (1), 174–183. doi: 10.1124/jpet.109.161489
D’Souza-Schorey C., Chavrier P. (2006). ARF proteins: roles in membrane traffic and beyond. Nat. Rev. Mol. Cell Biol. 7 (5), 347–358. doi: 10.1038/nrm1910
Endo T. (2020). M-ras is muscle-ras, moderate-ras, mineral-ras, migration-ras, and many more-ras. Exp. Cell Res. 397 (1), 112342. doi: 10.1016/j.yexcr.2020.112342
Etienne-Manneville S., Hall A. (2002). Rho GTPases in cell biology. Nature 420 (6916), 629–635. doi: 10.1038/nature01148
Fan Z. C., Behal R. H., Geimer S., Wang Z., Williamson S. M., Zhang H., et al. (2010). Chlamydomonas IFT70/CrDYF-1 is a core component of IFT particle complex b and is required for flagellar assembly. Mol. Biol. Cell 21 (15), 2696–2706. doi: 10.1091/mbc.e10-03-0191
Finlin B. S., Andres D. A. (1997). Rem is a new member of the rad- and Gem/Kir ras-related GTP-binding protein family repressed by lipopolysaccharide stimulation. J. Biol. Chem. 272 (35), 21982–21988. doi: 10.1074/jbc.272.35.21982
Finlin B. S., Crump S. M., Satin J., Andres D. A. (2003). Regulation of voltage-gated calcium channel activity by the rem and rad GTPases. Proc. Natl. Acad. Sci. United States America 100 (24), 14469–14474. doi: 10.1073/pnas.2437756100
Forget D., Lacombe A.-A., Cloutier P., Al-Khoury R., Bouchard A., Lavallée-Adam M., et al. (2010). The protein interaction network of the human transcription machinery reveals a role for the conserved GTPase RPAP4/GPN1 and microtubule assembly in nuclear import and biogenesis of RNA polymerase II*. Mol. Cell. Proteomics 9 (12), 2827–2839. doi: 10.1074/mcp.M110.003616
Gao Y., Zhang X., Wei J., Sun X., Yuan J., Li F., et al. (2015). Whole transcriptome analysis provides insights into molecular mechanisms for molting in Litopenaeus vannamei. PloS One 10 (12), e0144350. doi: 10.1371/journal.pone.0144350
Gao Z., Xing K., Zhang C., Qi J., Wang L., Gao S., et al. (2019). Crystal structure and function of rbj: A constitutively GTP-bound small G protein with an extra DnaJ domain. Protein Cell 10 (10), 760–763. doi: 10.1007/s13238-019-0622-3
Garcia-Ranea J. A., Valencia A. (1998). Distribution and functional diversification of the ras superfamily in Saccharomyces cerevisiae. FEBS Lett. 434 (3), 219–225. doi: 10.1016/s0014-5793(98)00967-3
Geng X., Liu S., Yao J., Bao L., Zhang J., Li C., et al. (2016). A genome-wide association study identifies multiple regions associated with head size in catfish. G3 Genes Genomes Genet. 6 (10), 3389–3398. doi: 10.1534/g3.116.032201
Giaever G., Chu A. M., Ni L., Connelly C., Riles L., Véronneau S., et al. (2002). Functional profiling of the Saccharomyces cerevisiae genome. Nature 418 (6896), 387–391. doi: 10.1038/nature00935
Goitre L., Trapani E., Trabalzini L., Retta S. F. (2014). The ras superfamily of small GTPases: The unlocked secrets. Methods Mol. Biol. 1120, 1–18. doi: 10.1007/978-1-62703-791-4_1
Goodsell D. S. (1999). The molecular perspective: the ras oncogene. Oncologist 4 (3), 263–264. doi: 10.1634/theoncologist.4-3-263
Gutierrez-Erlandsson S., Herrero-Vidal P., Fernandez-Alfara M., Hernandez-Garcia S., Gonzalo-Flores S., Mudarra-Rubio A., et al. (2013). R-RAS2 overexpression in tumors of the human central nervous system. Mol. Cancer 12 (1), 127. doi: 10.1186/1476-4598-12-127
Han F., Wang X., Yang Q., Cai M., Wang Z. Y. (2011). Characterization of a RacGTPase up-regulated in the large yellow croaker Pseudosciaena crocea immunity. Fish Shellfish Immunol. 30 (2), 501–508. doi: 10.1016/j.fsi.2010.11.020
Han F., Zhang X. (2007). Characterization of a ras-related nuclear protein (Ran protein) up-regulated in shrimp antiviral immunity. Fish Shellfish Immunol. 23 (5), 937–944. doi: 10.1016/j.fsi.2007.01.022
He H. J., Wang Q., Zheng W. W., Wang J. X., Song Q. S., Zhao X. F. (2010). Function of nuclear transport factor 2 and ran in the 20E signal transduction pathway in the cotton bollworm, Helicoverpa armigera. BMC Mol. Cell Biol. 11, 1. doi: 10.1186/1471-2121-11-1
Hino H., Arimoto K., Yazawa M., Murakami Y., Nakatomi A. (2012). Ran and calcineurin can participate collaboratively in the regulation of spermatogenesis in scallop. Mar. Biotechnol. 14 (4), 479–490. doi: 10.1007/s10126-011-9429-9
Hu B., Jin J., Guo A. Y., Zhang H., Luo J., Gao G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31 (8), 1296–1297. doi: 10.1093/bioinformatics/btu817
Huang H., Jin T., He J., Ding Q., Xu D., Wang L., et al. (2012). Progesterone and AdipoQ receptor 11 links ras signaling to cardiac development in zebrafish. Arteriosc. Thromb. Vasc. Biol. 32 (9), 2158–2170. doi: 10.1161/ATVBAHA.112.252775
Jiang S.-Y., Ramachandran S. (2006). Comparative and evolutionary analysis of genes encoding small GTPases and their activating proteins in eukaryotic genomes. Physiol. Genomics 24 (3), 235–251. doi: 10.1152/physiolgenomics.00210.2005
Jouaux A., Franco A., Heude-Berthelin C., Sourdaine P., Blin J. L., Mathieu M., et al. (2012). Identification of ras, pten and p70S6K homologs in the Pacific oyster crassostrea gigas and diet control of insulin pathway. Gen. Comp. Endocrinol. 176 (1), 28–38. doi: 10.1016/j.ygcen.2011.12.008
Kawasaki S., Kimura S., Fujita R., Sasaki K. (2004). The small GTP-binding protein RhoA regulates serotonin-induced na+-current response in the neurons of Aplysia. Neurosci. Res. 48 (1), 33–43. doi: 10.1016/j.neures.2003.09.007
Kierszenbaum A. L., Gil M., Rivkin E., Tres L. L. (2002). Ran, a GTP-binding protein involved in nucleocytoplasmic transport and microtubule nucleation, relocates from the manchette to the centrosome region during rat spermiogenesis. Mol. Reprod. Dev. 63 (1), 131–140. doi: 10.1002/mrd.10164
Kukimoto-Niino M., Murayama K., Inoue M., Terada T., Tame J. R. H., Kuramitsu S., et al. (2004). Crystal structure of the GTP-binding protein obg from Thermus thermophilus HB8. J. Mol. Biol. 337 (3), 761–770. doi: 10.1016/j.jmb.2004.01.047
Kumar S., Stecher G., Li M., Knyaz C., Tamura K. (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35 (6), 1547–1549. doi: 10.1093/molbev/msy096
Lambaerts K., Wilcox-Adelman S. A., Zimmermann P. (2009). The signaling mechanisms of syndecan heparan sulfate proteoglycans. Curr. Opin. Cell Biol. 21 (5), 662–669. doi: 10.1016/j.ceb.2009.05.002
Letunic I., Bork P. (2007). Interactive tree of life (iTOL): An online tool for phylogenetic tree display and annotation. Bioinformatics 23 (1), 127–128. doi: 10.1093/bioinformatics/btl529
Li C., Li Z., Song L., Meng L., Xu G., Zhang H., et al. (2021). GEFT inhibits autophagy and apoptosis in rhabdomyosarcoma via activation of the Rac1/Cdc42-mTOR signaling pathway. Front. Oncol. 11. doi: 10.3389/fonc.2021.656608
Li J. F. L., Gao Y., Shi S., He H., Liu X., Yuan C. (2018). Expression of Septin2 and its effect on cell proliferation and apoptosis in gastric cancer. J-Global 53 (03), 332–337. doi: 10.13705/j.issn.1671-6825.2017.11.012
Li J. L. Z., Qu Z., Liu W. (2010). Research progress of Microplitis mediator and its application. J. Hebei Agric. Sci. 14 (08), 4–11. doi: 10.16318/j.cnki.hbnykx.2010.08.004
Li X., Meng X., Luan S., Luo K., Cao B., Chen B., et al. (2020). Effect of white spot syndrome virus infection on a ras gene in the Chinese shrimp Fenneropenaeus chinensis. Aquaculture 516, 734604. doi: 10.1016/j.aquaculture.2019.734604
Liao X., Ali E., Li W., He B., Gong P., Xu P., et al. (2019). Sublethal effects of sulfoxaflor on the development and reproduction of the brown planthopper, Nilaparvata lugens (Stål). Crop Prot. 118, 6–14. doi: 10.1016/j.cropro.2018.12.005
Lin D., Jiang Q., Ma X., Zheng K., Gong X., Teng S., et al. (2018). Rice TSV3 encoding obg-like GTPase protein is essential for chloroplast development during the early leaf stage under cold stress. G3 Genes Genomes Genet. 8 (1), 253–263. doi: 10.1534/g3.117.300249
Liu L. H. F., Zhang Y., Wang X., Wang Z. (2017). Cloning and analysis of Rac2 gene in disease-resistant in the Large yellow croaker (Larimichthys crocea). J. Jimei Univ. Natural Sci. 22 (04), 1–11. doi: 10.19715/j.jmuzr.2017.04.001
Liu W., Han F., Zhang X. (2009). Ran GTPase regulates hemocytic phagocytosis of shrimp by interaction with myosin. J. Proteome Res. 8 (3), 1198–1206. doi: 10.1021/pr800840x
Liu X., Xie D., Hua Y., Zeng P., Ma L., Zeng F. (2020). Npa3 interacts with Gpn3 and assembly factor Rba50 for RNA polymerase II biogenesis. FASEB J. 34 (11), 15547–15558. doi: 10.1096/fj.202001523R
Liu X., Zhang Z., Zhang M., Zhao X., Zhang T., Liu W., et al. (2021). A ras-related nuclear protein ran participates in the 20E signaling pathway and is essential for the growth and development of Locusta migratoria. Pesticide Biochem. Physiol. 178, 104945. doi: 10.1016/j.pestbp.2021.104945
Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 25 (4), 402–408. doi: 10.1006/meth.2001.1262
López-Casas P. P., López-Fernández L. A., Párraga M., Krimer D. B., del Mazo J. (2003). Developmental regulation of expression of Ran/M1 and Ran/M2 isoforms of ran-GTPase in mouse testis. Int. J. Dev. Biol. 47 (4), 307–310.
Magie C. R., Parkhurst S. M. (2005). Rho1 regulates signaling events required for proper Drosophila embryonic development. Dev. Biol. 278 (1), 144–154. doi: 10.1016/j.ydbio.2004.10.022
Marcus K., Mattos C. (2020). Water in ras superfamily evolution. J. Comput. Chem. 41 (5), 402–414. doi: 10.1002/jcc.26060
Ménasché G., Pastural E., Feldmann J., Certain S., Ersoy F., Dupuis S., et al. (2000). Mutations in RAB27A cause griscelli syndrome associated with haemophagocytic syndrome. Nat. Genet. 25 (2), 173–176. doi: 10.1038/76024
Miranda D. R., Voss A. A., Bannister R. A. (2021). Into the spotlight: RGK proteins in skeletal muscle. Cell Calcium 98, 102439. doi: 10.1016/j.ceca.2021.102439
Mitin N., Rossman K. L., Der C. J. (2005). Signaling interplay in ras superfamily function. Curr. Biol. 15 (14), R563–R574. doi: 10.1016/j.cub.2005.07.010
Molendijk A. J., Ruperti B., Palme K. (2004). Small GTPases in vesicle trafficking. Curr. Opin. Plant Biol. 7 (6), 694–700. doi: 10.1016/j.pbi.2004.09.014
Moon S. Y., Zheng Y. (2003). Rho GTPase-activating proteins in cell regulation. Trends Cell Biol. 13 (1), 13–22. doi: 10.1016/s0962-8924(02)00004-1
Murugan A. K., Grieco M., Tsuchida N. (2019). RAS mutations in human cancers: Roles in precision medicine. Semin. Cancer Biol. 59, 23–35. doi: 10.1016/j.semcancer.2019.06.007
Nepomuceno-Silva J. L., de Melo L. D., Mendonçã S. M., Paixão J. C., Lopes U. G. (2004). RJLs: a new family of ras-related GTP-binding proteins. Gene 327 (2), 221–232. doi: 10.1016/j.gene.2003.11.010
Phillips M. J., Calero G., Chan B., Ramachandran S., Cerione R. A. (2008). Effector proteins exert an important influence on the signaling-active state of the small GTPase Cdc42. J. Biol. Chem. 283 (20), 14153–14164. doi: 10.1074/jbc.M706271200
Qin H., Wang Z., Diener D., Rosenbaum J. (2007). Intraflagellar transport protein 27 is a small G protein involved in cell-cycle control. Curr. Biol. 17 (3), 193–202. doi: 10.1016/j.cub.2006.12.040
Ren Q., Zhou J., Jia Y.-P., Wang X.-W., Zhao X.-F., Wang J.-X. (2012). Cloning and characterization of rap GTPase from the Chinese white shrimp Fenneropenaeus chinensis. Dev. Comp. Immunol. 36 (1), 247–252. doi: 10.1016/j.dci.2011.07.004
Reynet C., Kahn C. R. (1993). Rad: a member of the ras family overexpressed in muscle of type II diabetic humans. Science 262 (5138), 1441–1444. doi: 10.1126/science.8248782
Rojas A. M., Fuentes G., Rausell A., Valencia A. (2012). The ras protein superfamily: Evolutionary tree and role of conserved amino acids. J. Cell Biol. 196 (2), 189–201. doi: 10.1083/jcb.201103008%
Sahai E., Marshall C. J. (2002). RHO-GTPases and cancer. Nat. Rev. Cancer 2 (2), 133–142. doi: 10.1038/nrc725
Sanz-Rodriguez M., Gruart A., Escudero-Ramirez J., de Castro F., Delgado-García J. M., Wandosell F., et al. (2018). R-Ras1 and r-Ras2 are essential for oligodendrocyte differentiation and survival for correct myelination in the central nervous system. J. Neurosci. 38 (22), 5096–5110. doi: 10.1523/jneurosci.3364-17.2018
Sassone-Corsi P., Der C. J., Verma I. M. (1989). Ras-induced neuronal differentiation of PC12 cells: possible involvement of fos and jun. Mol. Cell. Biol. 9 (8), 3174–3183. doi: 10.1128/mcb.9.8.3174-3183.1989
Sato T., Umetsu A., Tamanoi F. (2008). Characterization of the rheb-mTOR signaling pathway in mammalian cells: constitutive active mutants of rheb and mTOR. Methods Enzymol. 438, 307–320. doi: 10.1016/s0076-6879(07)38021-x
Seabra M. C., Mules E. H., Hume A. N. (2002). Rab GTPases, intracellular traffic and disease. Trends Mol. Med. 8 (1), 23–30. doi: 10.1016/s1471-4914(01)02227-4
Spiliotis E. T., McMurray M. A. (2020). Masters of asymmetry - lessons and perspectives from 50 years of septins. Mol. Biol. Cell 31 (21), 2289–2297. doi: 10.1091/mbc.E19-11-0648
Stenmark H., Olkkonen V. M. (2001). The rab GTPase family. Genome Biol. 2 (5), 3007. doi: 10.1186/gb-2001-2-5-reviews3007
Sun P., Watanabe H., Takano K., Yokoyama T., Fujisawa J., Endo T. (2006). Sustained activation of m-ras induced by nerve growth factor is essential for neuronal differentiation of PC12 cells. Genes Cells 11 (9), 1097–1113. doi: 10.1111/j.1365-2443.2006.01002.x
Taira K., Umikawa M., Takei K., Myagmar B.-E., Shinzato M., Machida N., et al. (2004). The Traf2- and nck-interacting kinase as a putative effector of Rap2 to regulate actin cytoskeleton*. J. Biol. Chem. 279 (47), 49488–49496. doi: 10.1074/jbc.M406370200
Takai Y., Sasaki T., Matozaki T. (2001). Small GTP-binding proteins. Physiol. Rev. 81 (1), 153–208. doi: 10.1152/physrev.2001.81.1.153
Tingaud-Sequeira A., Chauvigné F., Lozano J., Agulleiro M. J., Asensio E., Cerdà J. (2009). New insights into molecular pathways associated with flatfish ovarian development and atresia revealed by transcriptional analysis. BMC Genomics 10, 434. doi: 10.1186/1471-2164-10-434
Trach K., Hoch J. A. (1989). The Bacillus subtilis spo0B stage 0 sporulation operon encodes an essential GTP-binding protein. J. Bacteriol. 171 (3), 1362–1371. doi: 10.1128/jb.171.3.1362-1371.1989
Wang F., Li S., Xiang J., Li F. (2019). Transcriptome analysis reveals the activation of neuroendocrine-immune system in shrimp hemocytes at the early stage of WSSV infection. BMC Genomics 20 (1), 247. doi: 10.1186/s12864-019-5614-4
Wang L., Wang X. R., Liu J., Chen C. X., Liu Y., Wang W. N. (2015). Rab from the white shrimp Litopenaeus vannamei: characterization and its regulation upon environmental stress. Ecotoxicology 24 (7-8), 1765–1774. doi: 10.1007/s10646-015-1481-1
Wang Q., Yu Y., Zhang Q., Luo Z., Zhang X., Xiang J., et al. (2020). The polymorphism of LvMMD2 and its association with growth traits in Litopenaeus vannamei. Mar. Biotechnol. 22 (4), 564–571. doi: 10.1007/s10126-020-09977-0
Wang S., He M., Li Q., Li H., Yin B., He J. (2022). Rap2a is negatively regulated by NF-κB and contributes to growth via wnt pathway in shrimp. Front. Mar. Sci. 9. doi: 10.3389/fmars.2022.942009
Wei J., Zhang X., Yu Y., Huang H., Li F., Xiang J. (2014). Comparative transcriptomic characterization of the early development in pacific white shrimp Litopenaeus vannamei. PloS One 9 (9), e106201. doi: 10.1371/journal.pone.0106201
Wennerberg K., Der C. J. (2004). Rho-family GTPases: it’s not only rac and rho (and I like it). J. Cell Sci. 117 (Pt 8), 1301–1312. doi: 10.1242/jcs.01118
Whelan S., Goldman N. (2001). A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 18 (5), 691–699. doi: 10.1093/oxfordjournals.molbev.a003851
Wöllert T., Patel A., Lee Y. L., Provance D. W. Jr., Vought V. E., Cosgrove M. S., et al. (2011). Myosin5a tail associates directly with Rab3A-containing compartments in neurons. J. Biol. Chem. 286 (16), 14352–14361. doi: 10.1074/jbc.M110.187286
Wu W., Zhang X. (2007). Characterization of a rab GTPase up-regulated in the shrimp Peneaus japonicus by virus infection. Fish Shellfish Immunol. 23 (2), 438–445. doi: 10.1016/j.fsi.2007.01.001
Wu W., Zong R., Xu J., Zhang X. (2008). Antiviral phagocytosis is regulated by a novel rab-dependent complex in shrimp Penaeus japonicus. J. Proteome Res. 7 (1), 424–431. doi: 10.1021/pr700639t
Xu C., Zhang W., Zhang X., Zhou D., Qu L., Liu J., et al. (2019). Coupling function of cyclin-dependent kinase 2 and Septin2 in the promotion of hepatocellular carcinoma. Cancer Sci. 110 (2), 540–549. doi: 10.1111/cas.13882
Yu Y., Wang Q., Zhang Q., Luo Z., Wang Y., Zhang X., et al. (2019). Genome scan for genomic regions and genes associated with growth trait in pacific white shrimp Litopeneaus vannamei. Mar. Biotechnol. 21 (3), 374–383. doi: 10.1007/s10126-019-09887-w
Zhang X., Yuan J., Sun Y., Li S., Gao Y., Yu Y., et al. (2019). Penaeid shrimp genome provides insights into benthic adaptation and frequent molting. Nat. Commun. 10 (1), 356. doi: 10.1038/s41467-018-08197-4
Zhao Z., Jiang C., Zhang X. (2011). Effects of immunostimulants targeting ran GTPase on phagocytosis against virus infection in shrimp. Fish Shellfish Immunol. 31 (6), 1013–1018. doi: 10.1016/j.fsi.2011.08.022
Zhou F., Zheng L., Yang Q., Qiu L., Huang J., Su T., et al. (2012). Molecular analysis of a ras-like nuclear (Ran) gene from Penaeus monodon and its expression at the different ovarian stages of development. Mol. Biol. Rep. 39 (4), 3821–3827. doi: 10.1007/s11033-011-1160-0
Keywords: Ras superfamily, GTPase proteins, Litopenaeus vannamei, gene structure and function, gene expression, immunity, growth
Citation: Si S, Zhang X, Yuan J, Zhang X, Yu Y, Yang S and Li F (2023) Identification, classification and expression analysis of the Ras superfamily genes in the Pacific white shrimp, Litopenaeus vannamei. Front. Mar. Sci. 10:1063857. doi: 10.3389/fmars.2023.1063857
Received: 07 October 2022; Accepted: 04 January 2023;
Published: 23 January 2023.
Edited by:
Ana Riesgo, Museo Nacional de Ciencias Naturales CSIC, SpainReviewed by:
Nathan Kenny, University of Otago, New ZealandAida Verdes, Department of Biodiversity and Evolutionary Biology (CSIC), Spain
Copyright © 2023 Si, Zhang, Yuan, Zhang, Yu, Yang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaojun Zhang, xjzhang@qdio.ac.cn; Song Yang, yangsong1209@163.com
 Shuqing Si1,2
Shuqing Si1,2  Xiaojun Zhang
Xiaojun Zhang Jianbo Yuan
Jianbo Yuan Xiaoxi Zhang
Xiaoxi Zhang Yang Yu
Yang Yu Song Yang
Song Yang Fuhua Li
Fuhua Li