DNA repair-related genes are the key for stony coral ancestors to survive under elevated levels of UVR
- 1Laboratory of Marine Organism Taxonomy and Phylogeny, Qingdao Key Laboratory of Marine Biodiversity and Conservation, Institute of Oceanology, Chinese Academy of Sciences, Qingdao, China
- 2State Key Laboratory of Marine Geology, Tongji University, Shanghai, China
- 3Southern Marine Science and Engineering Guangdong Laboratory (Zhuhai), Zhuhai, China
- 4College of Marine Science, University of Chinese Academy of Sciences, Beijing, China
DNA damage response (DDR) is a complicated network to defend against physical or chemical changes in DNA in all animals. Elevated levels of ultraviolet radiation (UVR) caused DNA damage, which was a reason for the mass extinction that occurred at the Devonian/Carboniferous (D/C) boundary approximately 359 million years ago (Ma). However, the molecular adaptation of the stony coral ancestors that strangely survived the D/C boundary mass extinction is not well understood. In the present study, the molecular clock analysis using fourfold degenerate sites of 1,463 homologous genes of different stony coral species (a representative group of marine organisms with calcareous skeletons) suggested that their common ancestors originated 384.24 Ma, i.e., slightly earlier than the D/C transition. We identified 21 rapidly evolving genes (REGs) and 49 positive selection genes (PSGs) that were significantly enriched in diverse pathways, including the mitotic cell cycle process, intracellular protein transport, and DNA synthesis involved in DNA repair. Interestingly, four REGs and 21 PSGs were significantly enriched in DDR pathways, including the mitotic cell cycle process, DNA synthesis involved in DNA repair, and cellular response to DNA damage stimulus pathways. We hypothesize that enriched DDR genes are likely involved in the enhanced ability of ancient stony corals to detect and repair DNA damage. For example, the DNA polymerase epsilon catalytic subunit (POLE) gene, which encodes the DNA polymerase epsilon catalytic subunit A, mediates interaction with the other three catalytic subunits through its nonenzymatic carboxy-terminal domain. POLE may potentially enhance the binding ability to other subunits to strengthen its function because eight positive selection sites were distributed in the C-terminal of POLE and on the surface of the simulated 3D protein model. Therefore, our results demonstrated for the first time that the precise transfer of DNA information may help stony coral ancestors survive in elevated levels of ultraviolet radiation, suggesting that DDR levels may be critical to the environmental adaptation of calcareous skeletal organisms during climate change.
Introduction
Genomic integrity is essential for normal cell function and ensures faithful DNA transmission during cell division, a process that is key under stressful environmental conditions (Giglia-Mari et al., 2011). For example, ultraviolet radiation (UVR) is one of the environmental health hazards that causes DNA damage in organisms. DDR is a fundamental mechanism activated in response to DNA lesions. This mechanism consists of pathways that include DNA damage recognition and repair (e.g., mismatch repair, base excision repair, nucleotide excision repair, double-strand break repair), checkpoint activation, and cell cycle arrest (Friedberg, 2003). The DNA repair pathway is essential to maintain genome integrity by acting as an adaptive system to face extreme environmental conditions such as high UVR levels (Qu et al., 2021; Zhang Z.H. et al., 2021).
A mass extinction event occurred at the Devonian/Carboniferous (D/C) boundary at 359 Ma. It has been proposed that high UVR levels were one of the primary causes leading to the D/C mass extinction, an observation supported by the discovery of malformed land plant spores in east Greenland (Marshall et al., 2020; Voosen, 2020). In marine organisms, Rugosa constitutes a clade of corals that underwent extinction during the D/C boundary. However, stony corals (Scleractinia) survived in shallow seas facing high UVR levels (Fagerstrom, 1994; Quattrini et al., 2020; Mcfadden et al., 2021). Stony corals are typical marine species whose ancestors evolved to acquire beneficial features such as symbiotic relationships with algae and aragonite calcareous skeletons (Quattrini et al., 2020; Mcfadden et al., 2021). According to molecular phylogenetic analysis, the stony coral ancestors originated during the Middle Devonian, even earlier (Stolarski et al., 2011; Campoy et al., 2020; Mcfadden et al., 2021; Stolarski et al., 2021), or later than Devonian (Park et al., 2012). The estimated origin of ancient stony corals is crucial in determining whether this taxon was exposed to high UVR. Coincidentally, living stony corals are also facing environmental challenges, making molecular mechanism research on the resistance of ancient stony corals to UVR urgent.
Evidence mounts that UVR may cause coral bleaching (Lesser et al., 1990; Gleason and Wellington, 1993; Lesser, 1997). Oxidative stress is an underlying stress response of corals to high temperatures and is exacerbated under high UVR conditions (Lesser, 2011). Oxidative stress induces DNA, protein, and lipid damage and the accumulation of reduced oxygen intermediates (i.e., superoxide radicals, singlet oxygen, hydrogen peroxide, and hydroxyl radicals), which results in an abnormal cell cycle and the expulsion of symbiont algae from stony coral hosts (Inoue and Kawanishi, 1995; Keyer et al., 1995; Keyer and Imlay, 1996). Moreover, high intensity and long periods of exposure to UV can directly cause DNA lesions, generating cyclobutane pyrimidine dimers (CPD) and pyrimidine (6-4) pyrimidone photoproducts (6-4PP). We aimed to explore whether the resistance to UVR of ancient stony corals is associated with DDR pathways.
In the present study, the fourfold degenerate sites of protein-coding genes of 11 Anthozoa and Hydra vulgaris were used to reconstruct phylogenetic relationships, estimate their divergence times, and conduction selection analyses. We aimed to shed light on the underlying molecular mechanism of tolerance of ancient stony corals to high UVR levels. Our results provide new insights into the pathways by which these marine organisms with calcareous skeletons face environmental challenges.
Materials and methods
Identification of orthologous genes
In this study, genome annotations of a set of 12 species were selected as the dataset after eliminating the low assembly quality, including five stony coral species (Acropora digitifera, Acropora millepora, Orbicella faveolate, Pocillopora damicornis, and Stylophora pistillata), six anemones or soft coral species (Actinia tenebrosa, Exaiptasia diaphana, Nematostella vectensis, Xenia sp. Carnegie-2017, Dendronephthya gigantea, and Renilla reniformis), and H. vulgaris, which was selected as an outgroup (Supplementary Table S1). Genome data were downloaded from the National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov) and the Reef Genomics database (http://reefgenomics.org) (Sherry et al., 2001; Liew et al., 2016). The divergence time estimates were similar to those reported by Mcfadden et al. (2021), which suggests that our dataset and the number of species used in this study were reasonable.
Those transcripts that were shorter than 150 bp or those with a length that was not multiple of three bases were eliminated using our in-house Perl scripts. Single-copy orthologous genes were identified by OrthoFinder (Emms and Kelly, 2019) using the all-against-all BLASTP algorithm.
Phylogenetic tree construction
A total of 1,463 single-copy orthologous genes were aligned using default parameter values of the “alignSequences” and “exportAlignment” modules in the Macse software (Ranwez et al., 2018). Poorly aligned regions with gaps and nonhomologous fragments were removed using the Gblock server (Castresana, 2000). Next, we concatenated the aligned sequences to produce single reads in Fasta format. The fourfold degenerate sites were identified using our Perl script for phylogenetic tree construction. The phylogenetic tree was inferred by IQ-TREE (Nguyen et al., 2015) using the maximum-likelihood (ML) method with 1,000 bootstrap pseudo-replicates and default parameter values. The “-m MFP” parameter that could cause the ModelFinder program to select the best model was used.
Divergence time evaluation
Based on molecular clock theory, divergence dating analyses were conducted using the MCMCTree program of the PAML package (Bromham and Penny, 2003; Yang, 2007). The dates of the three nodes that were constrained by fossil records (Hydra–Anthozoa: 512–741 Ma, Pennatulacea–Octocorallia: 218–419 Ma, and Scleractinia–Hexacorallia: 436.7–541 Ma) were based on the Timetree website (Kumar et al., 2017) (http://www.timetree.org). Two independent divergence time analyses were run using a relaxed molecular clock (clock = 2). The posterior distribution of parameters was estimated by the Markov chain Monte Carlo sampling with 20,000 iterations, a burn-in of 2,000, and a sampling frequency of 10. The results of the analyses were separately examined using Tracer (Rambaut et al., 2018) to determine the convergence between parameters, node ages, and likelihood values. The estimated sample size for each parameter was above 200.
Selection pressure analysis
A total of 1,225 single-copy orthologous genes were recovered after removing sequences containing premature stop codons. The reads were aligned using PRANK with the “-codon” parameter (Löytynoja, 2014) after removing ambiguously aligned regions by Gblocks with the “−t = c, −b5 = h” parameters (Castresana, 2000).
To assess past selection in genes of ancient stony corals, we estimated the nonsynonymous (dN) to synonymous substitution (dS) rate ratio (ω = dN/dS), where ω > 1, = 1, and < 1 indicate positive, neutral, and purifying selection values, respectively. The ω value of each protein-coding gene was estimated by the ML model using the Codeml program of the PAML package (Yang, 2007). The results of the phylogenetic tree were used as input for both models, and we defined the ancestral branch of stony corals as the foreground.
In the branch model, the null model assumed that all branches in the tree had the same evolutionary rate by setting the parameter “model” to zero, and the alternative model allowed ancient stony corals (foreground) and other (background) branches to evolve at two different rates by setting the parameter “model” to two. The likelihood ratio test (LRT) was used to evaluate the significance of the difference between null and alternative models. A significant LRT value leads to the acceptance of the alternative hypothesis. The false discovery rate (FDR) method was used to correct for multiple testing in the R software (Benjamini and Hochberg, 1995). The genes showing high ω and significant p-adjusted values in the ancestral branch of stony corals were identified as rapidly evolving genes (REGs).
In the branch-site model, the null model assumed that all codons and branches evolved neutrally by setting parameters to “model = 2, NSsites = 2, fix_omega = 1, omega = 1,” and the alternative model allowed positive selection at individual codons along the ancestral branch of stony corals (model = 2, NSsites = 2, fix_omega = 0). The LTR method was used to identify positive selection genes (PSGs) with a BEB posterior probability > 0.9. Finally, we identified whether these positive selection sites are shared by extant stony corals using the “Segregating Site Identification” tool of the FasParser software package (Sun, 2017).
Enrichment analysis
To determine the processes or pathways the REGs and PSGs were related to, we combined the two gene sets for enrichment analyses. Gene Ontology (GO, including molecular functions, biological processes, and cellular components) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses were performed using the Metascape website (http://metascape.org), setting a p-value cutoff of 0.01. The protein–protein interaction network was obtained using the String database (https://cn.string-db.org/) with default parameter values.
Tridimensional modeling of the POLE protein
We then selected the most critical gene, the DNA polymerase epsilon catalytic subunit (POLE), for in-depth analysis. The POLE gene sequence of A. millepora with the best assembly quality was translated and used to model the tridimensional (3D) protein structure by the Swiss-Model website (https://swissmodel.expasy.org), and the quality of the prediction model was evaluated. Amino acids under positive selection were then labeled on the 3D structure by PyMol (Delano, 2002). For the positive selection sites of POLE shared by stony corals, we checked if they were radical amino acid substitutions according to the classification of Zhang (2000).
Results
Phylogenetic tree and divergence times
A total of 1,463 one-to-one orthologous genes were identified in the genomes of 11 Anthozoa species and H. vulgaris (Figure 1; Supplementary Table S1) using Orthofinder and our in-house Perl scripts. The bootstrap values for all nodes of the ML tree inferred from fourfold degenerate sites (37.5k bp) were 100%. As shown in Figure 1, Hexacorallia and Octocorallia were monophyletic within the Anthozoa class. Scleractinia and its two subgroups (complex and robust) were also monophyletic. The branch topologies were consistent with the phylogenetic relationships reported by Mcfadden et al. (2021) and inferred from hundreds of ultraconserved elements and exon loci.
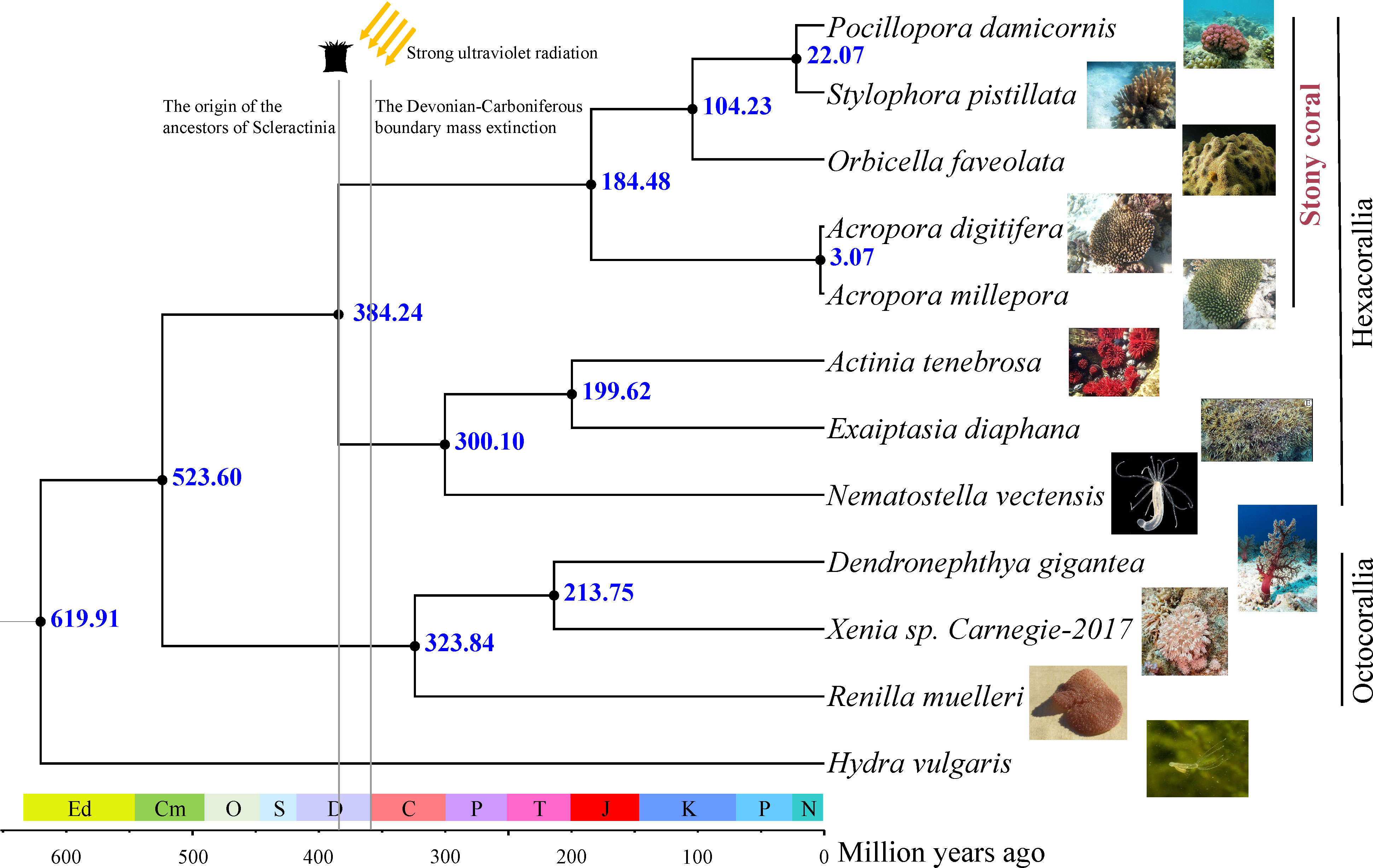
Figure 1 Phylogenetic tree and divergence times based on the fourfold degenerate sites of 1,463 one-to-one orthologous genes. The images of each species are from the Internet.
The ML tree and sequence data were used as the input for MCMCTree to estimate divergence times. Figure 1 shows the divergence time estimates for all nodes in the phylogenetic tree. The mean age for the Anthozoa class was 619.91 Ma (95% highest posterior density (HPD): 399.95–774.69 Ma), a younger estimation than the divergence age of 771 Ma reported by Quattrini et al. (2020) and similar to the suggested by Erwin et al. (2011), who estimated the anthozoan clade originated during the Ediacaran period. The divergence time of Scleractinia was estimated to occur during the Middle Devonian around 384.24 Ma (95% HPD: 235.72–540.80 Ma), a value consistent with the age estimation of 383 Ma reported by Quattrini et al. (2020). The split between complex and robust groups of stony corals was estimated to occur during the Jurassic period (184.48 Ma, HPD: 87.65–284.39 Ma).
The selection of genes related to DDR
To test whether divergent selective pressures acted on stony coral ancestors, the branch model and branch-site model were used to identify REGs and PSGs, respectively. After FDR correction, 21 REGs (Figure 2A; Supplementary Table S2) and 49 PSGs (Figure 2B; Supplementary Table S3) in the ancestral branch of stony corals were identified. A total of 2,321 amino acid changes shared by living stony corals were identified in 49 PSGs (Supplementary Table S4), parts of which were also identified as positive selection sites (Supplementary Table S5).
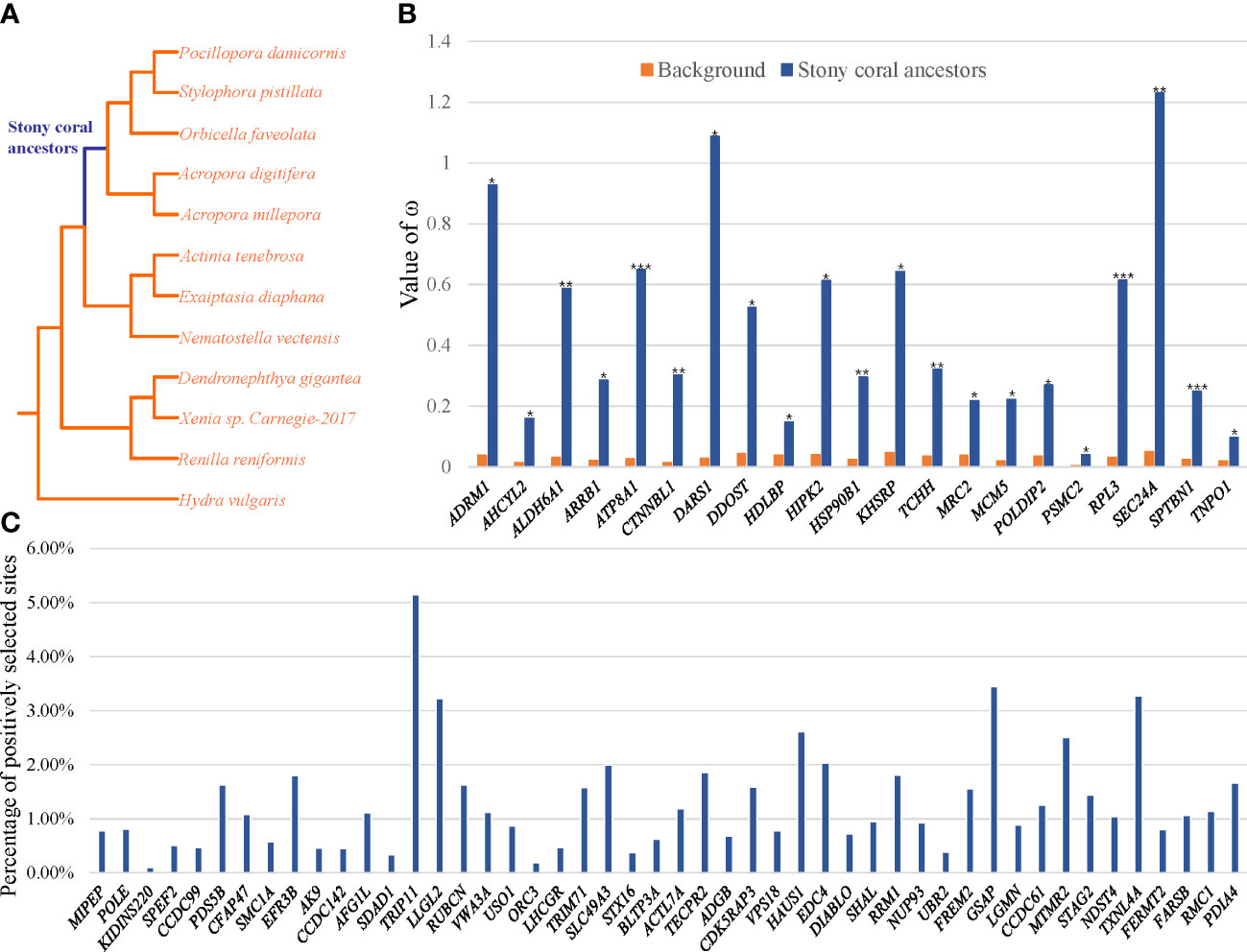
Figure 2 (A) The phylogenetic tree used in the selection analysis, with stony coral ancestors as the foreground branch. (B) The REGs of stony coral ancestors. (C) The PSGs of stony coral ancestors. The vertical axis represents the percentage of the number of positively selected sites for each gene.
For the 70 genes under rapidly evolving or positive selection, the functional enrichment analyses revealed that they were significantly enriched (p < 0.01) in 20 GO terms and KEGG pathways (Figure 3A; Supplementary Table S6). The top three GO terms were mitotic cell cycle process (p = 3.40e−7), intracellular protein transport (p = 3.56e−06), and DNA synthesis involved in DNA repair (p = 9.86e−5). Moreover, identified genes were involved in KEGG pathways related to the cell cycle (p = 2.03e−4) and protein processing in the endoplasmic reticulum (p = 6.47e−4) (Figure 3A; Supplementary Table S6).
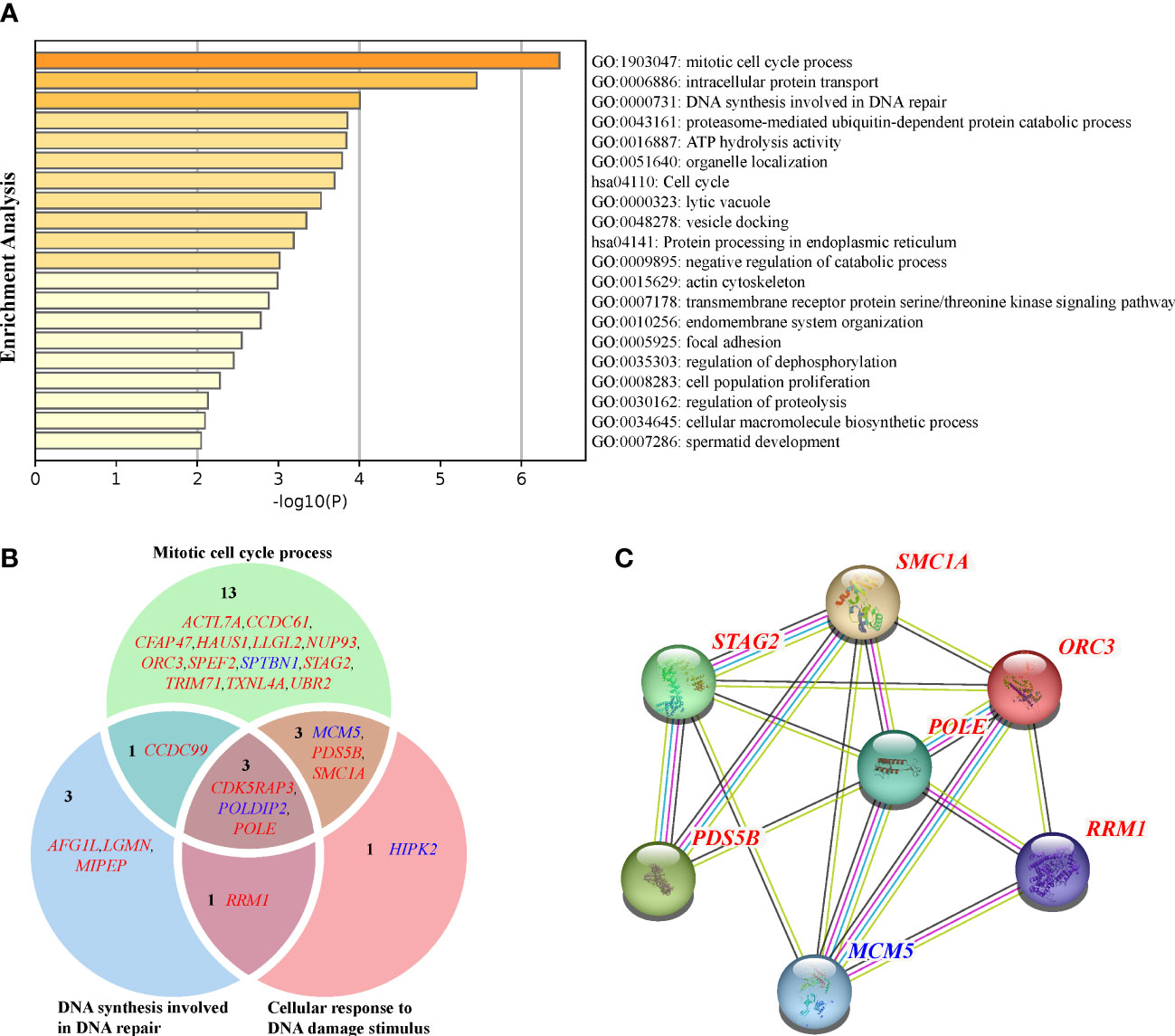
Figure 3 (A) GO and KEGG enrichment of the union of the REGs and PSGs. (B) Venn diagram of the GO terms (mitotic cell cycle process, DNA synthesis involved in DNA repair, and cellular response to DNA damage stimulus) related to DDR. REGs with blue fonts, and PSGs with red fonts. (C) Protein–protein interaction network of the REGs and PSGs. The edges represent protein–protein associations, blue represents “from curated databases,” red represents “experimentally determined,” yellow represents “text mining,” and black represents “co-expression.”.
The enrichment analyses revealed a total of 21 of 49 PSGs (42.86%) and four of 21 REGs (19.05%) related to the faithful DNA transmission (Figure 3B). There were 17 PSGs and three REGs related to the mitotic cell cycle process, seven PSGs and one REG related to the DNA synthesis involved in DNA repair, and five PSGs and three REGs related to the Cellular response to DNA damage stimulus (p = 2.87e-4). The Venn diagram illustrated the intersection of genes between the above three GO terms (Figure 3B). The POLDIP2 gene codes the DNA polymerase delta-interacting protein 2, with ω = 0.27 of the stony coral ancestors, significantly higher than the ω = 0.04 of background branches (FDR = 1.36e−2; Supplementary Table S2). CDK5 regulatory subunit-associated protein 3 (CDK5RAP3) and POLE genes were found to have seven (p = 1.83e−3) and 16 (p = 6.88e−4) sites under positive selection, respectively. The protein–protein interaction network of genes involved in faithful transmission of the DNA information showed that the average node degree was the value of 1.28 for all 25 input genes, and only seven genes formed a network (p = 1.68e−7; Figure 3C). Notably, POLE was the only gene linked to the others by co-expression or text mining.
We explored positive selection sites of the POLE gene due to the high number of connections of POLE in the protein–protein interaction network. The analysis revealed that half of the POLE-positive selection sites in stony coral species were fixed and distributed in the region encoding the C-terminal portion of the protein (Figure 4A). Compared with the properties of the homologous amino acids of sea anemones or soft corals, these amino acid changes of the stony coral ancestors all belonged to radical changes, according to Zhang (2000). The high-quality 3D protein structure of POLE showed that most of the positive selection sites were located on the surface of the protein (Figure 4B, C).
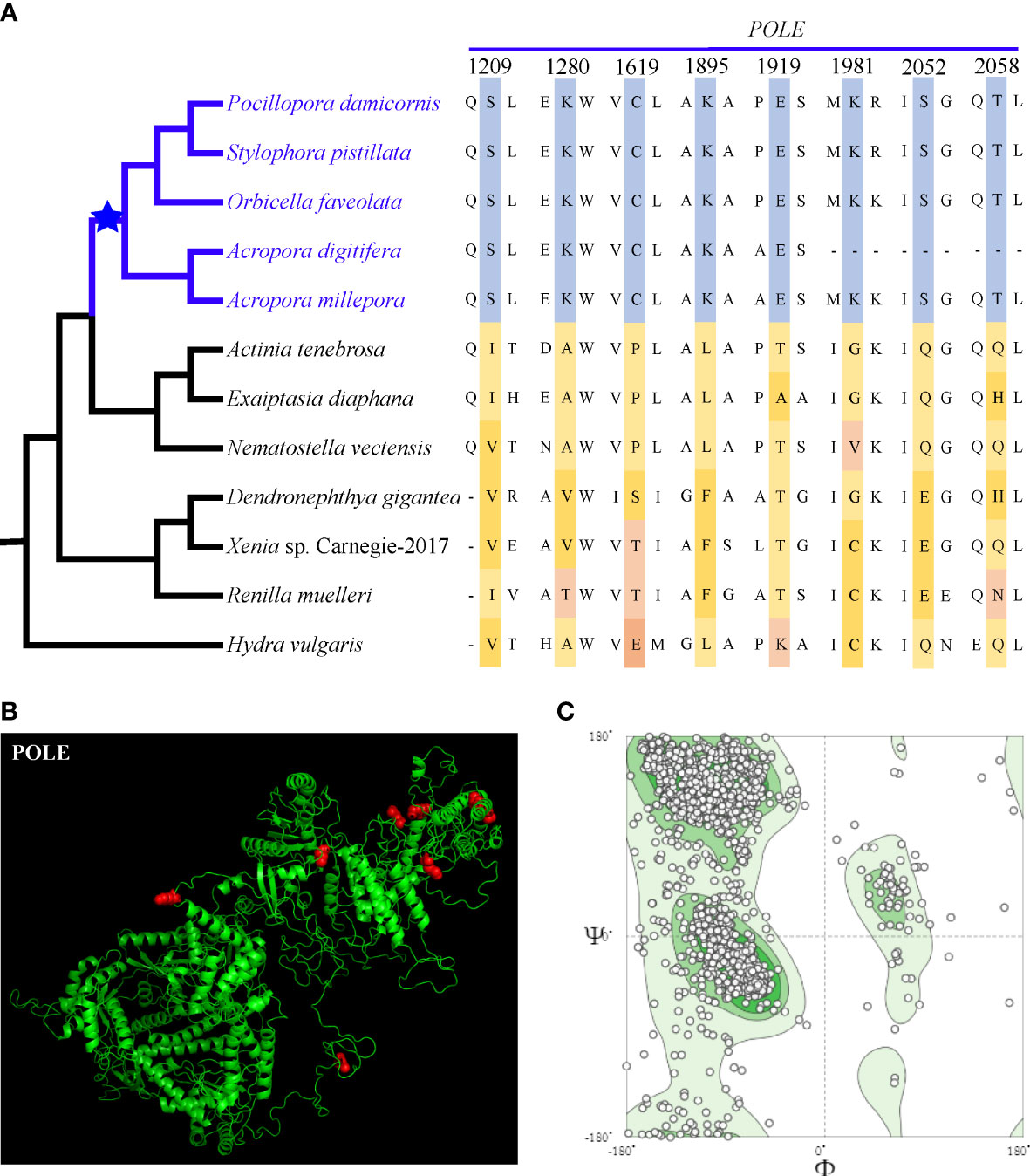
Figure 4 (A) Positive selection sites of POLE shared by extant stony corals, which are marked in red in (B) 3D structural modeling. (C) The quality evaluation of the 3D structural modeling. Most of the points are in the dark green area, indicating that our predictions are of high quality.
Discussion
Stony corals are a group of widely distributed marine organisms with calcareous skeletons that survived mass extinction events. Nowadays, coral reef ecosystems are seriously threatened due to the increased frequency and intensity of stressors such as water warming and ocean acidification. The elucidation of the mechanisms associated with the adaptive molecular evolution of ancient stony coral groups during the Devonian can shed light on how living coral species face global environmental challenges. In this study, the results of the time divergence analysis suggested that the origin of stony corals was slightly earlier than the mass extinction at the D/C boundary, an estimation consistent with those reported for a wide range of species within the taxonomic group (Mcfadden et al., 2021). Furthermore, our results suggest that DDR genes were essential to survive the high UVR levels that characterized the D/C transition.
Origin of ancient stony corals
The time-calibrated analysis of the class Anthozoa reported by Mcfadden et al. (2021) is considered the most reliable in the field because the authors analyzed hundreds of conserved noncoding regions and exons in more than 200 anthozoan species. The consistency between our divergence age estimates and those previously estimated by Mcfadden et al. (2021) suggests that protein-coding gene sequences contain enough phylogenetic information to understand the evolutionary history of stony corals. However, Park et al. (2012) found that stony coral ancestors arose during the Triassic, i.e., later than reported by Mcfadden et al. (2021), probably due to a small sample size or mitogenomes without enough phylogenetic information. Incomplete taxon sampling may bias divergence age estimates toward younger ages. Therefore, it is feasible that ancient stony corals originated much earlier, an observation supported by the estimation of nuclear and mitochondrial rDNA sequences (Stolarski et al., 2011; Campoy et al., 2020) and complete mitogenomes (Stolarski et al., 2021). Regardless, these analyses suggested that stony coral ancestors originated during the Middle Devonian or even earlier than this period. These observations suggest that stony corals were exposed to high UVR levels soon after the origin of the group, i.e., at the D/C boundary (Marshall et al., 2020).
Potential adaptive strategies of stony corals to high UVR levels
Ultraviolet-B (UV-B) radiation levels in the low latitudes have been projected to be higher by 2%–3% at the end of the twenty-first century than in 1980 due to the increased atmospheric concentration of ozone-depleting substances (Bais et al., 2015). Stony corals inhabiting tropical shallow-water habitats cannot swim or migrate like other marine animals, being constantly exposed to environmental conditions such as high temperatures and UVR that accelerate DNA damage (Hoegh-Guldberg et al., 2007; Spalding and Brown, 2015).
Corals display a series of adaptive strategies to face UVR, including mycosporine-like amino acids that provide a wide-band absorption response to UVR (310–360 nm), fluorescent pigments to absorb and dissipate photons from the solar radiation, and antioxidant enzymes to protect against reactive oxygen species damage (Drollet et al., 1993; Lesser, 2000; Torregiani and Lesser, 2007; Baird et al., 2009). Moreover, DNA repair pathways during replication and faithful DNA transmission also play roles in the adaptation to high UVR levels (Mitchell and Karentz, 1993; Van De Poll et al., 2001; Liñán-Cabello et al., 2010). It has been shown that animals and plants on the Qinghai–Tibet Plateau and tropical marine animals exhibit enhanced DNA damage repair responses (Svanfeldt et al., 2014; Qiao et al., 2016; Qu et al., 2021). Cells of the coral Seriatopora hystrix showed efficient DNA repair pathways under exposure to experimental UVR conditions, suggesting that other coral species may also exhibit stringent DNA repair mechanisms (Svanfeldt et al., 2014). Signatures of positive selection delimit genomic regions that are or have been functionally important. We identified stony coral DDR genes under significantly positive selection. Our results suggest that ancient stony corals survived the D/C mass extinction via the evolution of genes related to DDR pathways as part of an adaptive strategy to face high UVR levels.
PSGs of stony coral ancestors related to DDR
Several studies have shown that phenotypic plasticity contributes to the ability of corals to adapt to environmental stress (Kenkel and Matz, 2016; Putnam et al., 2016), but the detection of adaptive variation in genomes is slightly insufficient. Focusing on the stony coral ancestors originating in the Devonian, our study explored the adaptive variation in protein-coding genes in the genome to adapt to environmental stress. Genes under positive selection were related to DDR pathways that ensure faithful DNA transmission. Therefore, our results suggest that natural selection may have favored the fixation of adaptive mutations in the DDR genes of ancient stony corals.
Three main pathways were identified, including the mitotic cell cycle process, DNA synthesis involved in DNA repair, and the cellular response to DNA damage stimulus. Furthermore, seven genes (POLE, RRM1, MCMC5, PDS5B, SMC1A, ORC3, and STAG2) may play a central role in DDR due to their co-expression in protein–protein interaction networks. The DNA polymerase ϵ complex, which consists of four subunits and includes the largest catalytic subunit coded by POLE, is believed to play an essential role during eukaryotic DNA replication and is thought to participate in recombination and DNA repair (Kesti et al., 1999). Surprisingly, it has been shown that an essential region of the DNA polymerase ϵ complex is the POLE nonenzymatic carboxy-terminal domain, which not only mediates the interaction of the catalytic subunit with the three smaller regulatory subunits but also links the replication machinery to the S-phase checkpoint (Pospiech and Syväoja, 2003). Besides its function during DNA replication, POLE has also been shown to be involved in UV-induced DNA damage repair (Burgers, 1998; Pospiech and Syväoja, 2003). We found that positive selection sites for POLE are shared by all stony coral species and distributed in the encoding C-terminal portion of the protein. The 3D structure modeling of POLE revealed that amino acid substitutions are mainly located on the surface of the predicted protein structure. We hypothesize that amino acid substitutions in the POLE protein of the DNA polymerase ϵ complex enhanced its DNA replication fidelity and repair after UV-induced damage by modifying the mechanism of action and the interaction among subunits. More experiments are needed to test the DNA repair activity of the DNA polymerase ϵ complex in stony coral species, particularly those associated with CPD and 6-4PP repair after UV-induced damage. The catalytic subunit M1 of the ribonucleotide reductase protein encoded by the RRM1 gene plays a pivotal role in DNA repair by providing dNTPs during this process, which accumulate rapidly in damaged sites (Niida et al., 2010). PDS5 is a protein that plays roles during sister chromatid cohesion, DNA damage repair, gene transcription, and DNA replication (Zhang N. et al., 2021). SMC1A is a component of the recombination protein complex involved in DNA recombination repair pathways, and mutations in the SMC1A gene have been identified in several human cancers (Musio, 2020). Our results show that the PSGs identified in the present study are essential during normal cellular processes and DDR pathways. Therefore, we speculate that DDR genes were critical for the adaptation of ancient stony corals to high UVR levels during the D/C mass extinction event.
Conclusions
The present study is the first comparative analysis identifying genes in stony corals under positive selection and involved in DDR pathways. Our results suggest that mutations in DDR genes may have contributed to their survival after exposure to high UVR levels during the D/C mass extinction event. Therefore, the faithful transmission of the DNA information to progeny by DNA repair is one of the mechanisms for adapting to UVR, and calcareous skeletal organisms are no exception. Recently, anthropogenic activities have threatened the survival of marine organisms with calcareous skeletons, including stony corals. Our study also suggests that the adaptation of marine organisms with calcareous skeletons involves basic cellular processes such as the faithful transmission of genetic material. The adaptive mechanism driven by the accumulation of beneficial variants over many generations is extremely slow, constituting a challenge for living organisms and determining responses to the current climate change scenario.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author contributions
TW, YL, and ZJ conceived and designed the study. TW performed the bioinformatics analysis. TW, YL, and ZJ interpreted the data and wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Key Program of National Natural Science Foundation of China (No. 41930533), the National Key R&D Program of China (Grant No. 2021YFF0502500), and the Strategic Priority Research Program of the Chinese Academy of Sciences (No. XDB42000000).
Acknowledgments
Special thanks are due to Dr. Xin Huang for technical support. We also thank referee for helpful comments on the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1170565/full#supplementary-material
Supplementary Table 1 | Genome information for the twelve species used in Genome-wide comparative genomic analysis.
Supplementary Table 2 | The results of the REGs of stony coral ancestors.
Supplementary Table 3 | The results of the PSGs of stony coral ancestors.
Supplementary Table 4 | Amino acid changes shared by living stony corals for PSGs.
Supplementary Table 5 | The overlap between the specific amino acid changes for living stony corals and positive selection sites.
Supplementary Table 6 | The results of GO and KEGG enrichments.
References
Baird A. H., Bhagooli R., Ralph P. J., Takahashi S. (2009). Coral bleaching: the role of the host. Trends Ecol. Evol. 24, 16–20. doi: 10.1016/j.tree.2008.09.005
Bais A., Mckenzie R., Bernhard G., Aucamp P., Ilyas M., Madronich S., et al. (2015). Ozone depletion and climate change: impacts on UV radiation. Photochemical Photobiological Sci. 14, 19–52. doi: 10.1039/C0PP90034F
Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. society: Ser. B (Methodological) 57, 289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x
Bromham L., Penny D. (2003). The modern molecular clock. Nat. Rev. Genet. 4, 216–224. doi: 10.1038/nrg1020
Burgers P. M. (1998). Eukaryotic DNA polymerases in DNA replication and DNA repair. Chromosoma 107, 218–227. doi: 10.1007/s004120050300
Campoy A. N., Addamo A. M., Machordom A., Meade A., Rivadeneira M. M., Hernández C. E., et al. (2020). The origin and correlated evolution of symbiosis and coloniality in scleractinian corals. Front. Mar. Sci. 7. doi: 10.3389/fmars.2020.00461
Castresana J. (2000). Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17, 540–552. doi: 10.1093/oxfordjournals.molbev.a026334
Delano W. L. (2002). Pymol: an open-source molecular graphics tool. CCP4 Newsl. Protein Crystallogr. 40, 82–92.
Drollet J., Glaziou P., Martin P. (1993). A study of mucus from the solitary coral fungia fungites (Scleractinia: fungiidae) in relation to photobiological UV adaptation. Mar. Biol. 115, 263–266. doi: 10.1007/BF00346343
Emms D. M., Kelly S. (2019). OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20, 1–14. doi: 10.1186/s13059-019-1832-y
Erwin D. H., Laflamme M., Tweedt S. M., Sperling E. A., Pisani D., Peterson K. J. (2011). The Cambrian conundrum: early divergence and later ecological success in the early history of animals. science 334, 1091–1097. doi: 10.1126/science.1206375
Fagerstrom J. A. (1994). The history of Devonian-carboniferous reef communities: extinctions, effects, recovery. Facies 30, 177–191. doi: 10.1007/BF02536896
Giglia-Mari G., Zotter A., Vermeulen W. (2011). DNA Damage response. Cold Spring Harbor Perspect. Biol. 3, a000745. doi: 10.1101/cshperspect.a000745
Gleason D. F., Wellington G. M. (1993). Ultraviolet radiation and coral bleaching. Nature 365, 836–838. doi: 10.1038/365836a0
Hoegh-Guldberg O., Mumby P. J., Hooten A. J., Steneck R. S., Greenfield P., Gomez E., et al. (2007). Coral reefs under rapid climate change and ocean acidification. science 318, 1737–1742. doi: 10.1038/365836a0
Inoue S., Kawanishi S. (1995). Oxidative DNA damage induced by simultaneous generation of nitric oxide and superoxide. FEBS Lett. 371, 86–88. doi: 10.1016/0014-5793(95)00873-8
Kenkel C. D., Matz M. V. (2016). Gene expression plasticity as a mechanism of coral adaptation to a variable environment. Nat. Ecol. Evol. 1, 14. doi: 10.1038/s41559-016-0014
Kesti T., Flick K., Keränen S., Syväoja J. E., Wittenberg C. (1999). DNA Polymerase ϵ catalytic domains are dispensable for DNA replication, DNA repair, and cell viability. Mol. Cell 3, 679–685. doi: 10.1016/S1097-2765(00)80361-5
Keyer K., Gort A. S., Imlay J. A. (1995). Superoxide and the production of oxidative DNA damage. J. bacteriology 177, 6782–6790. doi: 10.1128/jb.177.23.6782-6790.1995
Keyer K., Imlay J. A. (1996). Superoxide accelerates DNA damage by elevating free-iron levels. Proc. Natl. Acad. Sci. 93, 13635–13640. doi: 10.1073/pnas.93.24.13635
Kumar S., Stecher G., Suleski M., Hedges S. B. (2017). TimeTree: a resource for timelines, timetrees, and divergence times. Mol. Biol. Evol. 34, 1812–1819. doi: 10.1093/molbev/msx116
Lesser M. P. (1997). Oxidative stress causes coral bleaching during exposure to elevated temperatures. Coral reefs 16, 187–192. doi: 10.1007/s003380050073
Lesser M. P. (2000). Depth-dependent photoacclimatization to solar ultraviolet radiation in the Caribbean coral montastraea faveolata. Mar. Ecol. Prog. Ser. 192, 137–151. doi: 10.3354/meps192137
Lesser M. P. (2011). Coral bleaching: causes and mechanisms. In: Dubinsky Z., Stambler N. (eds) Coral reefs: an ecosystem transition (Dordrecht: Springert), 405–419. doi: 10.1007/978-94-007-0114-4_23
Lesser M. P., Stochaj W., Tapley D., Shick J. (1990). Bleaching in coral reef anthozoans: effects of irradiance, ultraviolet radiation, and temperature on the activities of protective enzymes against active oxygen. Coral reefs 8, 225–232. doi: 10.1007/BF00265015
Liew Y. J., Aranda M., Voolstra C. R. (2016). Reefgenomics. org-a repository for marine genomics data. Database (Oxford) 2016, baw152. doi: 10.1093/database/baw152
Liñán-Cabello M. A., Flores-Ramírez L. A., Cobo-Díaz J. F., Zenteno-Savin T., Olguín-Monroy N. O., Olivos-Ortiz A., et al. (2010). Response to short term ultraviolet stress in the reef-building coral pocillopora capitata (Anthozoa: scleractinia). Rev. Biología Trop. 58, 103–118. doi: 10.15517/rbt.v58i1.5197
Löytynoja A. (2014). Phylogeny-aware alignment with PRANK. Multiple sequence alignment Methods, 1079. doi: 10.1007/978-1-62703-646-7_10
Marshall J. E., Lakin J., Troth I., Wallace-Johnson S. M. (2020). UV-B radiation was the Devonian-carboniferous boundary terrestrial extinction kill mechanism. Sci. Adv. 6, eaba0768. doi: 10.1126/sciadv.aba0768
Mcfadden C. S., Quattrini A. M., Brugler M. R., Cowman P. F., Dueñas L. F., Kitahara M. V., et al. (2021). Phylogenomics, origin, and diversification of anthozoans (Phylum cnidaria). Systematic Biol. 70, 635–647. doi: 10.1093/sysbio/syaa103
Mitchell D. L., Karentz D. (1993). The induction and repair of DNA photodamage in the environment. In: Young A. R., Moan J., Björn L. O., Nultsch W. (eds) Environ. UV Photobiol. (Boston, MA: Springer), 345–377. doi: 10.1007/978-1-4899-2406-3_12
Musio A. (2020). The multiple facets of the SMC1A gene. Gene 743, 144612. doi: 10.1016/j.gene.2020.144612
Nguyen L.-T., Schmidt H. A., Von Haeseler A., Minh B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Niida H., Katsuno Y., Sengoku M., Shimada M., Yukawa M., Ikura M., et al. (2010). Essential role of Tip60-dependent recruitment of ribonucleotide reductase at DNA damage sites in DNA repair during G1 phase. Genes Dev. 24, 333–338. doi: 10.1101/gad.1863810
Park E., Hwang D.-S., Lee J.-S., Song J.-I., Seo T.-K., Won Y.-J. (2012). Estimation of divergence times in cnidarian evolution based on mitochondrial protein-coding genes and the fossil record. Mol. Phylogenet. Evol. 62, 329–345. doi: 10.1016/j.ympev.2011.10.008
Pospiech H., Syväoja J. E. (2003). DNA Polymerase e-more than a polymerase. TheScientificWorldJournal 3, 87–104. doi: 10.1100/tsw.2003.08
Putnam H. M., Davidson J. M., Gates R. D. (2016). Ocean acidification influences host DNA methylation and phenotypic plasticity in environmentally susceptible corals. Evolutionary Appl. 9, 1165–1178. doi: 10.1111/eva.12408
Qiao Q., Wang Q., Han X., Guan Y., Sun H., Zhong Y., et al. (2016). Transcriptome sequencing of crucihimalaya himalaica (Brassicaceae) reveals how arabidopsis close relative adapt to the qinghai-Tibet plateau. Sci. Rep. 6, 1–8. doi: 10.1038/srep21729
Qu Y., Chen C., Chen X., Hao Y., She H., Wang M., et al. (2021). The evolution of ancestral and species-specific adaptations in snowfinches at the qinghai–Tibet plateau. Proc. Natl. Acad. Sci. 118, e2012398118. doi: 10.1073/pnas.2012398118
Quattrini A. M., Rodríguez E., Faircloth B. C., Cowman P. F., Brugler M. R., Farfan G. A., et al. (2020). Palaeoclimate ocean conditions shaped the evolution of corals and their skeletons through deep time. Nat. Ecol. Evol. 4, 1531–1538. doi: 10.1038/s41559-020-01291-1
Rambaut A., Drummond A. J., Xie D., Baele G., Suchard M. A. (2018). Posterior summarization in Bayesian phylogenetics using tracer 1.7. Systematic Biol. 67, 901–904. doi: 10.1093/sysbio/syy032
Ranwez V., Douzery E. J., Cambon C., Chantret N., Delsuc F. (2018). MACSE v2: toolkit for the alignment of coding sequences accounting for frameshifts and stop codons. Mol. Biol. Evol. 35, 2582–2584. doi: 10.1093/molbev/msy159
Sherry S. T., Ward M.-H., Kholodov M., Baker J., Phan L., Smigielski E. M., et al. (2001). dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 29, 308–311. doi: 10.1093/nar/29.1.308
Spalding M. D., Brown B. E. (2015). Warm-water coral reefs and climate change. Science 350, 769–771. doi: 10.1126/science.aad0349
Stolarski J., Coronado I., Murphy J. G., Kitahara M. V., Janiszewska K., Mazur M., et al. (2021). A modern scleractinian coral with a two-component calcite–aragonite skeleton. Proc. Natl. Acad. Sci. 118, e2013316117. doi: 10.1073/pnas.2013316117
Stolarski J., Kitahara M. V., Miller D. J., Cairns S. D., Mazur M., Meibom A. (2011). The ancient evolutionary origins of scleractinia revealed by azooxanthellate corals. BMC evolutionary Biol. 11, 1–11. doi: 10.1186/1471-2148-11-316
Sun Y.-B. (2017). FasParser: a package for manipulating sequence data. Zoological Res. 38, 110. doi: 10.24272/j.issn.2095-8137.2017.017
Svanfeldt K., Lundqvist L., Rabinowitz C., Sköld H. N., Rinkevich B. (2014). Repair of UV-induced DNA damage in shallow water colonial marine species. J. Exp. Mar. Biol. Ecol. 452, 40–46. doi: 10.1016/j.jembe.2013.12.003
Torregiani J. H., Lesser M. P. (2007). The effects of short-term exposures to ultraviolet radiation in the Hawaiian coral montipora verrucosa. J. Exp. Mar. Biol. Ecol. 340, 194–203. doi: 10.1016/j.jembe.2006.09.004
Van De Poll W. H., Eggert A., Buma A. G., Breeman A. M. (2001). Effects of UV-b-induced DNA damage and photoinhibition on growth of temperate marine red macrophytes: habitat-related differences in UV-b tolerance. J. Phycology 37, 30–38. doi: 10.1046/j.1529-8817.2001.037001030.x
Voosen P. (2020). UV Radiation blamed in ancient mass extinction. Am. Assoc. Advancement Science 368, 926. doi: 10.1126/science.368.6494.926
Yang Z. (2007). PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591. doi: 10.1093/molbev/msm088
Zhang J. (2000). Rates of conservative and radical nonsynonymous nucleotide substitutions in mammalian nuclear genes. J. Mol. Evol. 50, 56–68. doi: 10.1007/s002399910007
Zhang Z. H., Chang X., Su D. Y., Yao R., Liu X. D., Zhu H., et al. (2021). Comprehensive transcriptome analyses of two oocystis algae provide insights into the adaptation to qinghai–Tibet plateau. J. Systematics Evol. 59, 1209–1219. doi: 10.1111/jse.12589
Keywords: stony coral ancestors, ultraviolet radiation, DNA damage repair, positive selection, mass extinction, calcareous skeleton organisms
Citation: Wu T, Lei Y and Jian Z (2023) DNA repair-related genes are the key for stony coral ancestors to survive under elevated levels of UVR. Front. Mar. Sci. 10:1170565. doi: 10.3389/fmars.2023.1170565
Received: 21 February 2023; Accepted: 17 April 2023;
Published: 02 May 2023.
Edited by:
Haifeng Gu, State Oceanic Administration, ChinaReviewed by:
Nanjing Ji, Jiangsu Ocean Universiity, ChinaLeo Lai Chan, City University of Hong Kong, Hong Kong SAR, China
Copyright © 2023 Wu, Lei and Jian. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yanli Lei, leiyanli@qdio.ac.cn; Zhimin Jian, jian@tongji.edu.cn
 Tianzhen Wu
Tianzhen Wu Yanli Lei
Yanli Lei Zhimin Jian2*
Zhimin Jian2*