The natural products discovered in marine sponge-associated microorganisms: structures, activities, and mining strategy
- 1Li Dak Sum Marine Biopharmaceutical Research Center, Department of Marine Pharmacy, College of Food and Pharmaceutical Sciences, Ningbo University, Ningbo, Zhejiang, China
- 2National-Regional Joint Engineering Research Center for Soil Pollution Control and Remediation in South China, Guangdong Key Laboratory of Integrated Agro-Environmental Pollution Control and Management, Institute of Eco-Environmental and Soil Sciences, Guangdong Academy of Sciences, Guangzhou, China
- 3Department of Food Science and Engineering, College of Food and Pharmaceutical Sciences, Ningbo University, Ningbo, China
- 4School of Biotechnology and Health Sciences, Wuyi University, Jiangmen, China
It is well known that microorganisms are abundant in sponges, which make up 50%-60% of the host biomass. Moreover, mounting evidence suggests that bacteria, fungi, and cyanobacteria, all associated with sponges, are the true creators of the bioactive compounds identified from sponges. The discovery of 774 structurally active compounds from 1998 to 2017 presents a good overview of natural product resources in sponge-associated microorganisms. During the last 5 years, many new molecules, including peptides, polyketides, alkaloids, and terpenes, have been identified from sponge-associated microorganisms through various mining strategies, exhibiting a wide range of biological activities, such as anti-microbial, anti-cancer, enzyme inhibition, and antioxidant properties. In this paper, 140 compounds produced by sponge-associated microorganisms from 2017 to 2022 are systematically discussed in terms of their structures, biological activities, and strain sources, as well as the mining strategies, which not only further updates the natural product library of sponge-associated microorganisms but also provides a new guideline for exploring the “dark matter” in sponges.
1 Introduction
Natural products, also known as secondary metabolites, are small molecules produced by living organisms including plants, invertebrates, and microorganisms. They have been frequently utilized in clinical treatment and are crucial for the development of contemporary medicine (Newman and Cragg, 2018; Liu et al., 2020b). The ocean covers more than 70% of the earth’s surface and is the home for a lot of living organisms, making it an excellent target to explore natural products (Martins et al., 2019). The statistics data show that 2659 compounds were identified from sponges between 2010 and 2019, occupying 47.2% of the marine natural products discovered from all invertebrates (Calado et al., 2022). As a result, sponges are regarded as one good source of bioactive compounds and are frequently referred to as “chemical factories” or “gold mines” (Gavriilidou et al., 2021).
The sponges (phylum Porifera) are the simplest and likely the most ancient group of animals with a fossil record dating over 580 million years (Li et al., 1998; Love et al., 2009). They attach to the solid substrate in the sea and feed on bacteria and bacteria-sized particles in the filtered water. Therefore, sponges are also responsible for the purification of water indirectly (Taylor et al., 2007). Sponges are important parts of the benthic zone with a worldwide distribution, from freshwater to marine and from intertidal to the deep sea (Taylor et al., 2007). So far, as many as 15,000 species have been identified, including mainly are Calcarea, Hexactinellida, Homoscleromorpha, and Demospongiae (Kiran et al., 2018), but their diversity may be richer (Abdelmohsen et al., 2014). Another reason for the ongoing interest in sponges is their close association with microorganisms, including bacteria, fungi, and viruses (Buttachon et al., 2018; Kiran et al., 2018). Microorganisms have been known to form a variety of distribution relationships with sponges, including extracellular exosymbiosis, extracellular endo-symbiosis, intracellular symbiosis, and intranuclear symbiosis, accounting for 50%-60% of the host biomass (Lee et al., 2001; Zhang et al., 2017). Indeed, the relationship between sponges and microorganisms is very complicated, because the microbial population can be selectively enriched by sponge-active metabolites (Sathiyanarayanan et al., 2017).
Under the ecological pressure of intense competition, overgrowth, poisoning, infection, and predation, sponges have evolved defensively chemical weapons (secondary metabolites) to protect themselves from threats (Proksch, 1994), including amino acids, nucleosides, polyethers, alkaloids, macrolides, porphyrins, terpenes, and sterols. Most of them exhibit anti-cancer, anti-inflammatory, anti-fungal, and anti-viral activities (Shukla, 2016). However, mounting evidence shows that these weapons actually originate from microorganisms rather than sponges themselves. The surface or inner space of the sponge is richer in nutrients than seawater, thus providing a perfect and safe habitat for microorganisms (Lee et al., 2001). In return, microorganisms participate in the construction of the sponge’s chemical defense system. For example, Bovio et al. discovered cyclo-L-Trp-L-Ala, echinulin, neoechinulin A, dihydroauroglaucin, and flavoglaucin from the sponge-associated fungus Eurotium chevalieri MUT 2316, showing the inhibition on the adhesion and growth of marine bacteria and microalgae (Bovio et al., 2019).
Sponge-associated microorganisms, as important treasures for natural product discovery, have received a lot of attention. Of 774 natural products were identified from sponge-associated microorganisms between 1998 and 2017 (Cheng et al., 2020). Meanwhile, a series of review articles systematically make a systematic discussion the natural products from sponge-associated microbes (Abdelaleem et al., 2022; Alahmari et al., 2022; Amelia et al., 2022; Esposito et al., 2022; Bibi et al., 2017). Therefore, with the technology being updated, many new research methods have been applied to explore natural products from sponges and their associated microorganisms, which accelerate the discovery of new compounds. To update the knowledge in this field, we summarize the most recent progress on the origins, structures, and bioactivities of active natural products from sponge-associated microorganisms during the last 5 years (2017-2022). Moreover, a thorough discussion of the mining strategy is also included.
2 New bioactive compounds from sponge-associated microorganisms
There are 45 publications released between 2017 and 2022, reporting 140 new structures identified from sponge-associated microorganisms, such as sesquiterpenes, polyketides, peptides, alkaloids, etc. The new compounds exhibit a variety of biological activities, including anti-microbial, anti-cancer, enzyme inhibition, and so on.
2 1 Antimicrobial activity
Antibiotics are a great discovery of modern medicine. The application of antibiotics in clinical treatment has greatly improved the survival rate of human beings (Abdelmohsen et al., 2014). However, many antibiotics no longer have the clinical therapeutic impact as before due to the prevalence of drug resistance. Searching for new antibiotics is therefore an important way to resist the shortage of new medicine. Previous studies have shown that sponge-associated microorganisms contain a variety of antibacterial compounds, showing a good inhibitory effect on Staphylococcus aureus, Escherichia coli, Bacillus subtilis, and other bacteria. Some compounds could even inhibit drug-resistant bacteria, such as Methicillin-resistant Staphylococcus aureus (MRSA), Methicillin-resistant Streptococcus epidermidis (MRSE), and vancomycin-resistant Enterococci (VRE) (Cheng et al., 2020). Besides, some newly discovered compounds from sponge-associated microorganisms exhibit potent anti-fungal activity (Liang et al., 2023).
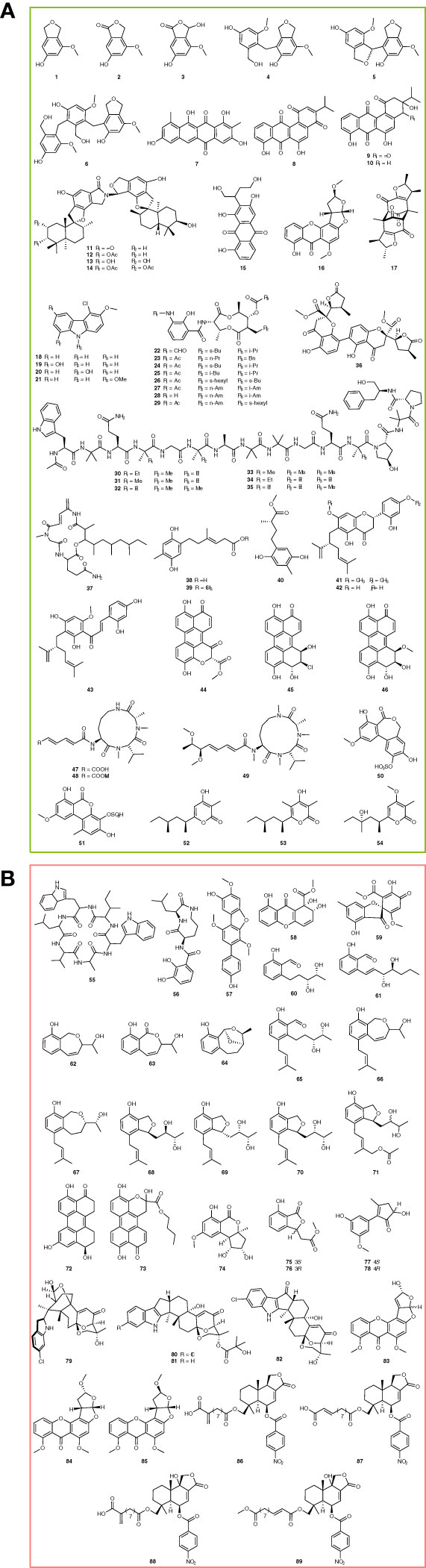
To explore new antibiotics, Wang et al. found cymopolyphenols A-F (Figure 1A, 1-6) from sponge-associated Cymostachys sp. NBUF082 by using the “One Strain, Many Compounds” (OSMAC) strategy combined with LC-MS/MS-based molecular networking. Antibacterial tests showed that cymopolyphenols A and C-F were effective against several Gram-positive and Gram-negative bacteria (MIC, 16-64 μg/ml), including Pseudoalteromonas carrageenovora, Vibrio shilanii, V. scophthalmi, V. alginolyticus, Salmonella typhi, Pseudomonas aeruginosa, S. aureus, and B. pumilus (Wang et al., 2021). Persiamycin A (Figure 1A, 7) is a new linear tetracycline compound acquired from sponge-associated Streptomonospora sp. PA3. It displayed an inhibitory effect on S. aureus, Klebsiella pneumoniae, and P. aeruginosa, and the diameters of the inhibition zone are 9.2-9.7 mm (Matroodi et al., 2020). Different from persiamycin A, nocardiopsistins A-C (Figure 1A, 8-10) are a group of new angular tetracyclines, which were discovered from the sponge-associated Nocardiopsis sp. HB-J378 and exhibited antibacterial activity against MRSA (MIC, 3.12-12.5 μg/mL) (Xu et al., 2018).
Chartarlactams are new phenylspirodrimane-type dimers with antibacterial activity. Liu et al. identified chartarlactams Q-T (Figure 1A, 11-14) from a sponge-associated Stachybotrys chartarum WGC-25. The bioassay showed that chartarlactams Q-S inhibited S. aureus with MIC values of 8, 16, and 4 μg/mL, respectively. Besides, it was found that chartarlactam T can inhibit the ZIKV virus by blocking virus entry and RNA replication in the antiviral experiment, indicating a multi-target feature (Liu et al., 2020a). In searching for new natural products, versiconol B and oxygensterigmatocystin I (Figure 1A, 15-16) were discovered from sponge-associated Aspergillus sp. F40. According to the structures, versiconol B belongs to the anthraquinone family, while oxygensterigmatocystin I is a member of xanthone. The antibacterial test showed that versiconol B inhibited S. aureus and V. parahaemolyticus with MIC values of 48 and 24 μg/mL, respectively (Tian et al., 2018). Penicitrinone G (Figure 1A, 17), a new citrinin derivative, was identified from the sponge-associated Penicillium citrinum WK-P9 through a bioassay-guided strategy. It could inhibit 8 pathogenic bacteria at 64 μg/mL, including B. megaterium DSM32, B. subtilis JH642, B. subtilis DSM10, Micrococcus luteus ATCC 4698, M. smegmatis ATCC 607, Listeria monocytogenes DSM20600, S. aureus ATCC25923, and E. coli K12 (Sabdaningsih et al., 2020).
Chlocarbazomycins A-D (Figure 1A, 18-21) are a group of new chlorinated carbazole alkaloids, which were identified from the sponge-associated Streptomyces diacarni LHW51701. The bioassay showed chlocarbazomycin C could inhibit MRSA, M. smegmatis, B. mycoides, and C. albicans with a MIC value of 32 μg/mL, while the other three compounds are against MRSA, M. smegmatis, B. mycoides, and C. albicans at 128 μg/mL (Cheng et al., 2021). Li et al. found 8 novel antimycins I-P (Figure 1A, 22-29) from the sponge-associated Streptomyces sp. NBU3104 by metabolomics and genomics. Among them, antimycin I was effective against phytopathogenic fungi, such as C. albicans, P. expanses, P. lemon, and Botrytis cinerea, with MIC values of 2, 4, 4, and 1 μg/mL, respectively. Moreover, antimycin I could effectively inhibit the gray mold of apples in vivo (MIC, 8 μg/mL), showing good potential for agricultural applications (Li et al., 2022). In addition, Hao et al. identified 6 novel 16-residue peptaibols, acremopeptaibols A-F (Figure 1A, 30-35), from sponge-associated Acremonium sp. IMB18-086. Compared with other members of the SF3 protein peptide family, acremopeptoils A-F lacked the highly conserved Thr6 and Hyp10 residues. Among them, acremopeptoils A and E showed antibacterial activity against S. aureus, MRSA, B. subtilis, and C. albicans (MIC, 16-64 μg/mL) (Hao et al., 2021).
Kumla et al. discovered a new dihydrochromone dimer, paecilin E (Figure 1A, 36), from the sponge-associated Neosartorya fennelliae KUFA 0811. It displayed an inhibition activity against S. aureus and Enterococcus faecalis with MIC values of 32 μg/mL and 16 μg/mL, respectively (Kumla et al., 2017). Rakicidin F (Figure 1A, 37) was identified from the sponge-associated Streptomyces sp. GKU 220 by chromatography, showing inhibition against B. subtilis and E. coli in the disk diffusion test at a dosage of 25 μg (Kitani et al., 2018). Ketidocillinones A–C (Figure 1A, 38-40) are three new polyketides, which discovered from the sponge-associated fungus Penicillium sp. HDN151272, possessing antibacterial activity against P. aeurigenosa, M. phlei, and Methicillin-resistant coagulase-negative staphylococci (MRCNS) with MIC values ranging from 1.56 to 25 µg/mL (Shah et al., 2020). Flavonoids are a group of natural substances with variable phenolic structures. Cao et al. discovered three new lavandulylated flavonoids, (2S,2″S)-6-lavandulyl-7,4′-dimethoxy-5,2′-dihydroxylflavanone, (2S,2″S)-6-lavandulyl-5,7,2′,4′-tetrahydroxylflavanone, and (2″S)-5′-lavandulyl-2′-methoxy-2,4,4′,6′-tetrahydroxylchalcone (Figure 1A, 41-43), from sponge-associated Streptomyces sp. G248. They displayed inhibitory effects against 5 pathogenic microorganisms, including E. faecalis, S. aureus, B. cereus, P. aeruginosa, and C. albicans, and their MIC values range is 1-8 μg/mL (Cao et al., 2019). Three new perylenequinone derivatives, xanalterate A, altertoxin VIII, and IX (Figure 1A, 44-46), were identified from the sponge-associated Alternaria sp. HDN19-690. However, only xanalterate A showed inhibition activity against 6 bacterial strains (MRCNS, B. subtilis, Proteus mirabilis, Bacillus cereus, E. coli, and M. phlei) with MIC values ranging from 3.13 to 12.5 µM (Hou et al., 2022).
Sclerotiotides M–O (Figure 1A, 47-49) are a class of aspochracin-type cyclic tripeptides with a side chain. They were identified from sponge-associated Aspergillus insulicola HDN151418, and sclerotiotides M and N displayed antimicrobial activity against a panel of pathogenic strains, including B. cereus, Proteus species, M. phlei, B. subtilis, Vibrio parahemolyticus, Edwardsiella tarda, MRCNS, and MRSA, with MIC values ranging from 1.56 to 25.0 µM (Sun et al., 2020). Alterlactone 5′-O-sulfate and 3′-hydroxyalternariol 5-O-methyl ether-3′-O-sulfate are two new dibenzopyrones identified from sponge-associated Alternaria sp. SCSIOS02F49 (Figure 1A, 50-51). Among them, alterlactone 5′-O-sulfate was shown to alter the external structure of S. aureus and caused the rupture or deformation of the cell membranes (Chen et al., 2022b). In addition, Ding et al. found three new polyketides, nipyrones A–C (Figure 1A, 52-54), from a marine sponge-associated Aspergillus niger, but only nipyrone C inhibited S. aureus and B. subtilis with MIC values of 8 μg/mL and 16 μg/mL, respectively (Ding et al., 2019).
2.2 Cytotoxicity
Natural products are rich resources for anti-cancer compounds. Moreover, some of them or their derivatives have been developed into clinical-used medicines. During the last 5 years, a lot of new compounds with potent anticancer properties have been identified from sponge-associated microorganisms, providing more drug leads for pharmaceutical research. To explore new natural products, Ibrahim et al. identified Nocardiopsis sp. UR67 strain from the red sea sponge Callyspongia sp. and identified a new cyclic hexapeptide, nocardiotide A (Figure 1B, 55). It exhibited cytotoxicity against the murine CT26 colon carcinoma, human HeLa cervix carcinoma, and human MM.1S multiple myeloma cell lines with IC50 values of 12, 11, and 8 μM/mL, respectively (Ibrahim et al., 2018). Petrocidin A (Figure 1B, 56) is a new cyclic dipeptide identified in Streptomyces sp. SBT348, which was identified from the Mediterranean sponge Petrosia ficiformis. It showed antiproliferative activity against human promyelocytic leukemia cells HL-60 and human colon cancer cell line HT-29 with IC50 values of 3.9 and 5.3 μg/mL (Cheng et al., 2017).
Wang et al. identified a new p-terphenyl alcohol, 4-O-methylcandidusin A (Figure 1B, 57), from the sponge-associated Aspergillus candidus OUCMDZ-1051. It selectively inhibited cancer cell lines MDA-MB-468, BT474, and A431 with IC50 values of 1.84, 6.05, and 0.98 μmol/L, respectively (Wang et al., 2020). Globosuxanthone F and 2’-hydroxy bisdechlorogeodin (Figure 1B, 58-59) are two polyketides discovered from the sponge-associated Pleosporales sp. NBUF144. Among them, globosuxanthone F was found an antiproliferative effect against CCRF-CEM human acute lymphatic leukemia cells with an IC50 value of 0.46 μM, without showing any cytotoxicity at 20 μM (Zhou et al., 2021b). Lei et al. discovered 12 new polyketide derivatives, heterocornols A-L (Figure 1B, 60-71) from the sponge-associated Pestalotiopsis heterocornis. The heterocornols A-C and F-H possessed cytotoxicity against 4 human cancer cell lines, including the human gastric carcinoma cell line, human large-cell lung carcinoma cell line, human prostate cancer cell line, and human hepatocellular carcinoma cell line. Their IC50 values ranged from 15.0 to 83.5 μM. Interestingly, these polyketide derivatives also exhibited antibacterial activity against S. aureus and B. subtilis (IC50, 25-100 μg/mL) (Lei et al., 2017).
In addition, Pang et al. identified 7 new compounds from the sponge-associated Alternaria sp. SCSIO41014, including altertoxin VII, butyl xanalterate, nordihydroaltenuenes A, (S)-isoochracinate A1, (R)-isoochracinate A2, (S)-alternariphent A1 and (R)-alternariphent A2 (Figure 1B, 72-78). However, only altertoxin VII displayed cytotoxicity against human erythroleukemia, human gastric carcinoma cells, and hepatocellular carcinoma cells with IC50 values of 26.58, 8.75, and 13.11 μg/mL, respectively (Pang et al., 2018b). Ascandinines A-D (Figure 1B, 79-82) are classified as a new type of indole diterpenoid, which were identified from sponge-associated Aspergillus candidus HDN15-152. Among them, ascandinine D showed cytotoxicity against HL-60 cells with an IC50 value of 7.8 μM (Zhou et al., 2021a). Aspergillus nomius NC06 was identified from the marine sponge Neopetrosia chaliniformis and proved to synthesize three new oxisterigmatocystins J-L (Figure 1B, 83-85). In the bioassay test, oxisterigmatocystins J and K induced apoptosis and cell death in HT 29 colon cancer cells with IC50 values of 6.28 and 15.14 µM (Artasasta et al., 2021). Insulicolides D–G (Figure 1B, 86-89) belong to two new nitrobenzoyl sesquiterpenoids, which are identified from a sponge-associated Aspergillus insulicola HDN151418. Further studies indicated that insulicolide D could significantly suppress cell proliferation to induce apoptosis and blocked migration and invasion of pancreatic ductal adenocarcinoma (PDAC) cell lines with IC50 values of 2.3-22.9 µM (Sun et al., 2022).
2.3 Enzyme inhibitors
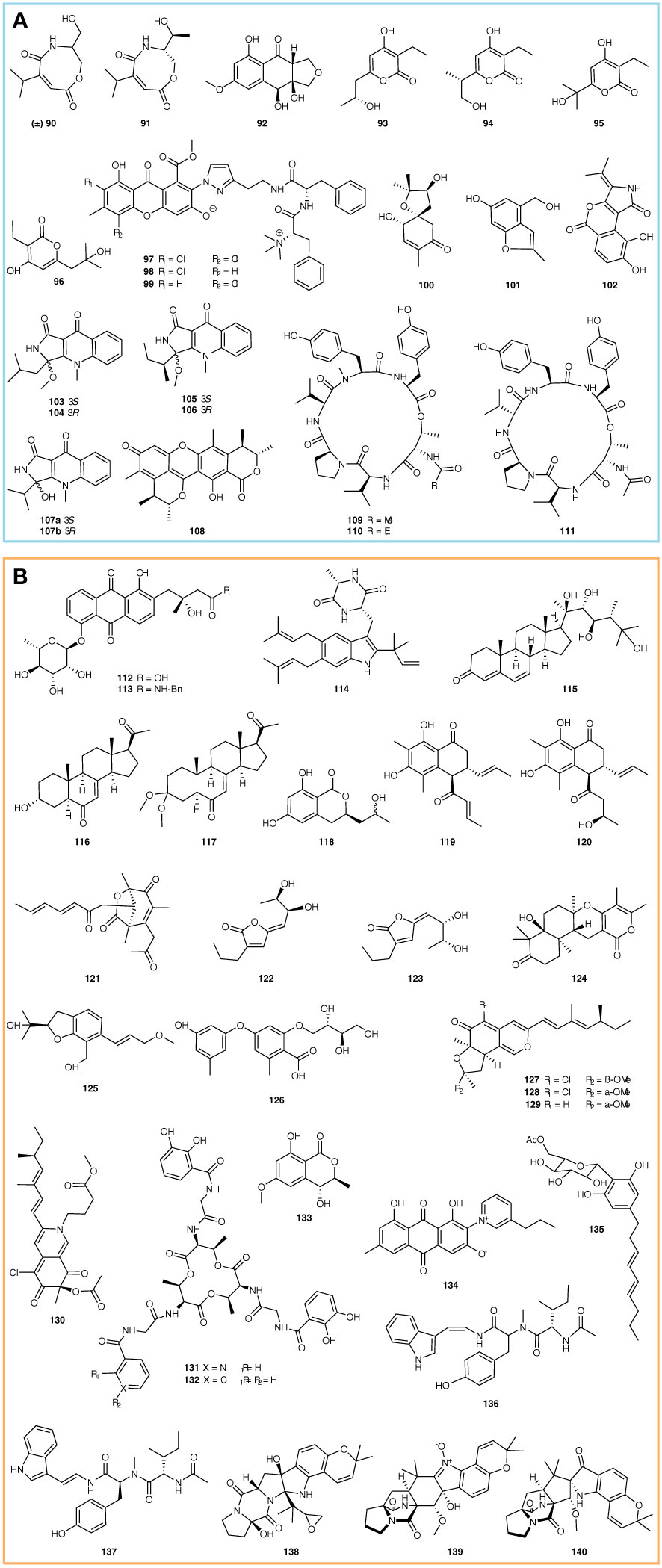
Enzyme inhibitors are an important type of medicine, which have been widely used in clinical treatment against various diseases. For example, the well-known antivirus drugs (Paxlovid and Molnupiravir), which belong to protease inhibitors, saved millions of lives in the battle against Coronavirus (COVID-19) (Wen et al., 2022). It is known that many new compounds with enzyme inhibition activity have been found in sponge-associated microorganisms during the past 5 years. For example, two new carboxamides ((±)-vochysiamide C and (+)-vochysiamide B) and a new polyketide (4S,3aS,9aR-3a,9a-deoxy-3a hydroxy-1-dehydroxyarthrinone) (Figure 2A, 90-92) were identified from the sponge-associated Arthrinium sp. SCSIO 41421 using chromatography. The (±)-vochysiamide C and (+)-vochysiamide B showed inhibition on 79.38% and 84.22% acetylcholinesterase, respectively, at a concentration of 50 μg/mL, while 86% of acetylcholinesterase was blocked by 4S,3aS,9aR-3a,9a-deoxy-3a hydroxy-1-dehydroxyarthrinone at 50 μg/mL (She et al., 2022). Bai et al. discovered 4 new α-pyrone derivatives, germicidins P-S from Streptomyces sp. 18A01 (Figure 2A, 93-96), through a combination of the Global Natural Products Social Molecular Networking (GNPS), the LC-DAD-MS profile, and hexokinase II (HK2) inhibitory activity screening. The bioassay showed that germicidins P-S inhibit HK2 with IC50 values ranging from 5.1 to 11.0 μM. Moreover, the molecular docking simulations indicated that the ligand has a strong affinity to Arg91 of HK2 instead of Asn89, resulting in higher hexokinase inhibitory action (Bai et al., 2021). Pancreatic lipase is the most important enzyme for hydrolyzing dietary fat, which can promote digestion and appetite. With the pancreatic lipase as the screening model, Jiao et al. identified 3 new chlorinated PKS-NRPS hybrid metabolites, flavipesides A-C (Figure 2A, 97-99), from the sponge-associated A. flavipes 164013. They exhibited a strong inhibition on pancreatic lipase at IC50 values of 0.23, 0.07, and 0.14 μM, respectively (Jiao et al., 2020).
In addition, two new compounds, acremine S and acremine T (Figure 2A, 100-101) were identified from sponge-associated Acremonium persicinum KUF1007. The bioassay showed that acremine S and acremine T exhibited inhibition on 10.42% and 14.08% of acetylcholinesterase at 6.6 µM, respectively, while 30.71% and 10.53% of butyrylcholinesterase were blocked by acremine S and acremine T at the concentration 6.25 µM (Alves et al., 2019). Rotinsulu et al. discovered a new protein tyrosine phosphatase (PTP) 1B inhibitor, cladosporamide A (Figure 2A, 102), from sponge-associated Cladosporium sp. TPU1507. It displayed inhibition activity against PTP1B and T-cell PTP with IC50 values of 48 and 54 μM, respectively (Rotinsulu et al., 2018). Five new N-methyl-4-quinolones, including quinolactacins E1 and E2, quinolactacins F1 and F2, quinolactacin G, and one new citrinin dimer derivative, dicitrinol D (Figure 2A, 103-108) were identified from the sponge-associated Penicillium sp. SCSIO41303. The enzyme inhibition test showed that quinolactacin F1 inhibited pancreatic lipase with an IC50 value of 24.6 μg/mL (Guo et al., 2021). Tang et al. discovered three new cyclohexapeptides, petrosamides A–C (Figure 2A, 109-111), from the sponge-associated fungus Aspergillus sp. 151304. These peptides displayed significant and dose-dependent pancreatic lipase inhibition activities with IC50 values of 7.6, 1.8, and 0.5 μM, respectively (Tang et al., 2020).
2.4 Others
Besides the anti-microbial, cytotoxic, and enzyme inhibition, some compounds from sponge-associated microorganisms display other types of bioactivities or no activities. In this study, we group them into ‘others’ for the compound discussion at a convenience. For example, fridamycins H and fridamycins I (Figure 2B, 112-113) are two new types II polyketides synthesized by the sponge-associated bacteria Actinokineospora spheciospongiae sp. DSM 45935T. In the experiment against Trypanosoma brucei TC221, fridamycin H showed anti-trypanosomal activity at 48 and 72 h (IC50, 7.18 and 3.35 μM) (Tawfike et al., 2019). Fintiamin (Figure 2B, 114) is a diketopiperazine-indole alkaloid identified from the Eurotium sp. identified from the marine sponge Ircinia variabilis. As one terpenoid-dipeptide derivative, fintiamin was made up of amino acids and terpenoid moieties and showed an affinity to the cannabinoid CB1 receptor (Elsebai et al., 2021). Cladosporisteroids A-C and (3R)-3-(2-hydroxypropyl)-6,8-dihydroxy-3,4-dihydroiso-coumarin (Figure 2B, 115-118) were identified from sponge-associated Cladosporium sp. SCSIO41007, but only cladosporisteroid B exhibited inhibition activity against H3N2 with the IC50 value of 16.2 μM (Pang et al., 2018a). Chen et al. discovered 3 new sorbicillinoid pigments, stasorbicillinoids A-C (Figure 2B, 119-121), from the sponge-associated Stagonospora sp. SYSU-MS7888 through phenotype combined with metabolomic and genomic (PMG) strategies. These compounds showed potent anti-inflammatory activity (IC50, 7.1-14.0 μM) (Chen et al., 2022a). Two new γ-lactones, aspergilactones A-B (Figure 2B, 122-123), were discovered from sponge-associated Aspergillus sp. LS45 and inhibited root growth of Arabidopsis thaliana Columbia-0 at a concentration of 100 μM (Huang et al., 2019).
In addition, a series of new compounds identified from sponge-associated microorganisms exhibit no bioactivities. For example, Shin et al. identified a new α-pyrone sesquiterpene with an angular tetracycline, ochraceopone F (Figure 2B, 124), from sponge-associated A. flocculosus. Ochraceopone F had no antiproliferative effect on human cancer cell lines HCT 15, NUGC-3, NCI-H23, ACHN, PC-3, and MDA-MB-231 at a concentration of 30 μg/mL (Shin et al., 2018). Asperspin A and asperther A (Figure 2B, 125-126) are two new molecules discovered from the sponge-associated Aspergillus sp. LS34 through the OSMAC culture strategy (Li et al., 2019). Sclerotiorins A-D (Figure 2B, 127-130) belong to one group of azaphilone compounds, which were identified from the sponge-associated P. sclerotiorum OUCMDZ-3839 (Jia et al., 2019). Wu et al. identified two siderophores, bacillibactins E-F (Figure 2B, 131-132) from the sponge-associated Bacillus sp., which are characterized by nicotinic and benzoic acid moieties in their structures (Wu et al., 2020). (3S,4R)-4-hydroxy-6-methoxymellein, anthraquinone, and acetyl carnemycin E (Figure 2B, 133-135) were identified from sponge-associated Aspergillus stellatus KUFA 2017 (Machado et al., 2022). Luo et al. discovered 2 new isomeric-modified tripeptides, aspergillamides C-D (Figure 2B, 136-137) from the sponge-associated fungus Aspergillus terreus SCSIO 41008 (Luo et al., 2019). Speramide C, 3,21-epi-taichunamide F, and 2-epi-amoenamide C (Figure 2B, 138-140) are three new 2,5-diketopiperazines identified from the sponge-associated Aspergillus sclerotiorum GDST-2013-0501. Speramide C represents the first prenylated indole alkaloid with an ethylene oxide ring at the isopentenyl side chain (Wang et al., 2022). Even though these compounds failed to exhibit biological activity, their new structures still enrich the natural product library of sponge-associated microorganisms.
During the last 5 years (2017-2022), 140 new compounds were identified from sponge-associated microorganisms (Supplementary, Table S1). The statistical data show that most of them come from actinomycetes (bacteria) and ascomycota (fungi). However, it is found that 76% of compounds are of fungal origin. Among those different natural product categories, polyketides occupy 47.85% followed by peptides (12.14%) and alkaloids (4.28%). Even though newly discovered compounds exhibit a variety of biological activities, the antimicrobial and anti-cancer compounds occupy 38.57% and 25%, respectively, providing a lot of lead compounds for drug development.
3 Methodological options for target natural products
3.1 Genome mining
Natural product discovery is developing at a faster speed than before due to technological developments. Genome mining is a technology that uses computer programs to extract useful information from genome data, including gene structure, sequence, quantity, and other molecular features. It examines the conserved regions of enzymes and gene clusters on the genome to guide the discovery of new enzymes and the identification of novel natural products (Zhao and Yang, 2011). This approach has been widely applied in natural product discovery due to a large amount of sequencing data in the database (Blin et al., 2019; Russell and Truman, 2020), and many new compounds have been discovered (Albarano et al., 2020; Kenshole et al., 2021).
Because of the update in sequencing technology, the cost and time consumed to generate genome sequences have significantly decreased over the past decade, providing a good basis for the application of genome mining strategy. Meanwhile, the bioinformatics software for biosynthetic gene cluster (BGC) prediction with high accuracy makes it possible to mine BGCs from massive sequence data. Anti-SMASH (antibiotics & Secondary Metabolite Analysis Shell) is currently the most widely used tool for predicting biosynthetic gene clusters (Medema et al., 2011). The new version (antiSMASH 6.0) could effectively detect splicing enzymes in ribosomally synthesized and post-translationally modified peptides (RiPPs) clusters and support the prediction of 71 types of gene cluster types. Moreover, it could highlight the modular structure of multi-module BGC and integrate the BGC comparison function. PRISM (Prediction Informatics for Secondary Metabolomes) is another commonly used biosynthetic gene cluster prediction tool, and it was updated for the fourth iteration in 2020. The latest version, PRISM 4, can not only generate accurate structure predictions for known BGCs but also perform structural predictions for cryptic BGCs. It could even predict chemical structures from metagenomic data and structure-activity relationships of compounds (Skinnider et al., 2020). Nowadays, it is not a time-costing mission to obtain ten thousand BGC in a limited time with the aid of large genome data and powerful tools. However, it does matter to group BGC into gene cluster families to decrease the analytical difficulty and increase the data process capacity. To this end, Navarro-Muñoz et al. developed the BiG-SCAPE (Biosynthetic Gene Similarity Clustering and Prospecting Engine) and CORASON (core analysis of syntenic orthologues to prioritize natural product gene clusters) automatically construct BGC similarity network and analyze the BGC phylogeny, realizing the high-throughput analysis of BGC (Navarro-Muñoz et al., 2020). Overall, the development and use of increasingly sophisticated bioinformatics tools make a great advance in the mining of natural products.
Numerous studies have demonstrated the advantages of genome mining in discovering novel compounds from sponge-associated microorganisms. In search of the natural products from sponge-associated microorganisms, Matsuda et al. used genome mining to select the terpene synthase bifunctional terpene synthases gene from Emericella variecolor NBRC 32302 for heterologous expression. Finally, a new terpene hydrocarbon, astellifadiene, was identified and identified (Matsuda et al., 2016). Besides, Alex and Antunes detected the presence of several biosynthetic gene clusters-polyketide synthases, nonribosomal peptide synthetase, and siderophore from sponge-associated Pseudovibrio sp. POLY-S9 with genome mining (Alex and Antunes, 2015). Overall, the development and use of increasingly sophisticated bioinformatics tools make a great advance in the mining of natural products.
3.2 Metabolomics analysis
Metabolomics is a discipline that studies the chemical process of metabolites. Its basic goal is to conduct a comprehensive qualitative and/or quantitative analysis of all metabolites present in living systems to reflect changes in their metabolites after stimulation with the characteristics of the simple operation (Liu and Locasale, 2017; Zhao et al., 2018). At present, metabolomics is widely used in natural product mining research, and the most common-used technology is liquid chromatography-mass spectrometry (LC-MS) (Salem et al., 2020). The liquid chromatography separation system can greatly reduce the complexity of the sample entering the mass spectrometer. Meanwhile, mass spectrometry is very sensitive to capturing a lot of ions from trace amounts of samples (Zhang et al., 2018). Therefore, the emergence of LC-MS/MS provides a powerful tool for compound structure elucidation.
In terms of metabolomics data analysis, the emergence of many new tools has made it possible to realize the automation and high throughput of data analysis (Perez De Souza et al., 2020). Yang et al. demonstrated that molecular networking, an approach that organizes MS/MS data based on chemical similarity, is a powerful complement to traditional dereplication strategies. It visualizes the output data in the network and dereplicates new chemical entities by comparing it to the MS/MS data of known compounds (Yang et al., 2013). In addition, Principal Component Analysis (PCA) is another common-used option for metabolomics analysis, which is used to analyze multivariate data by transforming many correlated variables into smaller linear data sets to define a data set (Kemsley et al., 2007). The PCA analysis could simplify the complexity of high-dimensional data while preserving trends and patterns (Lever et al., 2017).
The PCA and molecular networks are widely used as powerful tools in separating metabolites, avoiding tedious separation work for compounds. Based on the chemical uniqueness of PCA, Cheng et al. used liquid chromatography/high-resolution mass spectrometry (LC-HRMS) and nuclear magnetic resonance (NMR) to analyze the bacteria identified from the Mediterranean sponge Petrosia ficiformis. Finally, a new compound, named petrocidin A (Cheng et al., 2017), was identified from Streptomyces sp. SBT348. In addition, Li et al. discovered 8 new antibacterial compounds (antimycins I-P) by analyzing the secondary metabolites of the sponge-associated actinomycete Streptomyces sp. NBU3104 through integrating genome mining and molecular networking (Li et al., 2022), indicating a potent application of metabolomics in natural product discovery from sponge-associated microorganisms.
3.3 Bioactivity-guided approaches
Bioassay is one traditional strategy for bioactive compound discovery. The bioassay on crude extract not only confirmed the existence of bioactive compounds but also track the functional molecule in the crude extracts. However, if some biological activity is due to a mixture or synergistic effects from more components, it is not easy to find real clues. Although this strategy is believed to lead to the repeated discovery of known compounds, it still plays an important role in exploring new natural products from a variety of organisms, such as sponge-associated microorganisms. Devi et al. screened the crude extracts of marine bacteria identified from the surface of the sponge Halichondria sp. against 16 clinical pathogenic bacteria. With the bioassay guiding, three compounds, including indole, 3-phenylpropionic acid, and a dimer 4,4’-oxybis[3-phenylpropionic acid] were identified with a combination of ESI-MS and NMR (Devi et al., 2010). Gavriilidou et al. screened the extracts of 21 sponge-associated bacteria for antibacterial and anticancer compounds. Finally, one extract was found against MRSA, and more than 70% of extracts exhibited cytotoxicity (Gavriilidou et al., 2021).
4 Discussion and prospect
Sponges have been proven a rich reservoir of bioactive compounds. With a lot of new natural products being identified from sponge-associated microorganisms, it is believed that the real producers of bioactive compounds from sponges might be the microorganisms rather than the sponges themselves. Therefore, discovering natural products from sponge-associated microorganisms not only provides more leading compounds for drug development but also is of great significance to understand the interaction between sponges and their associated microorganisms (Pita et al., 2018). In the past 5 years, 140 new compounds were identified from sponge-associated microorganisms, exhibiting great chemical diversity. Although most of them exhibit bioactivity in the in vitro tests, their biological targets and mechanism of action are still elusive, which needs further study in the future.
Nowadays, the discovery of new natural products is much faster than ever due to technological development. However, there are still a lot of new challenges that deserved to pay more attention. For example, the study of the biological function and ecological roles of microbial secondary metabolites usually requires the combination of multiple technologies to resolve the research difficulties, such as genomics, metabolomics, synthetic biology, and so on. With the technology being updated at a high frequency, we believe more compounds will be uncovered from sponge-associated microorganisms, and their diverse biological functions will provide us with a new perspective on drug development and ecological protection.
Author contributions
PL, HL, YZ, MW, and LWL conceived, designed, and wrote this manuscript. XZ and LLL improved the quality of the manuscript by compiling information in the form of tables and figures. All authors read and approved the manuscript. All authors contributed to the article.
Funding
This research was funded by the Startup Foundation of Ningbo University (422010882, 422110473, 422207513), Ningbo Natural Science Foundation (2021S129, 2021Z046).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1191858/full#supplementary-material
References
Abdelaleem E. R., Samy M. N., Abdelmohsen U. R., Desoukey S. Y. (2022). Natural products potential of Dictyoceratida sponges-associated micro-organisms. Lett. Appl. Microbiol. 74 (1), 8–16. doi: 10.1111/lam.13559
Abdelmohsen U. R., Bayer K., Hentschel U. (2014). Diversity, abundance, and natural products of marine sponge-derived actinomycetes. Nat. Prod. Rep. 31 (3), 381–399. doi: 10.1039/c3np70111e
Alahmari N. A., Hassoubah S. A., Alaidaroos B. A. (2022). Sponges-associated marine bacteria as sources of antimicrobial compounds. Nov. Res. Microbiol. J. 6 (5), 1742–1767. doi: 10.21608/nrmj.2022.267424
Albarano L., Esposito R., Ruocco N., Costantini M. (2020). Genome mining as new challenge in natural products discovery. Mar. Drugs 18 (4), 199. doi: 10.3390/md18040199
Alex A., Antunes A. (2015). Whole genome sequencing of the symbiont Pseudovibrio sp. from the intertidal marine sponge Polymastia penicillus revealed a gene repertoire for host-switching permissive lifestyle. Genome. Biol. Evol. 7 (11), 3022–3032. doi: 10.1093/gbe/evv199
Alves A. J. S., Pereira J. A., Dethoup T., Cravo S., Mistry S., Silva A. M. S., et al. (2019). A new meroterpene, a new benzofuran derivative and other constituents from cultures of the marine sponge-derived fungus Acremonium persicinum KUFA 1007 and their anticholinesterase activities. Mar. Drugs 17 (6), 379. doi: 10.3390/md17060379
Amelia T. S. M., Suaberon F. A. C., Vad J., Fahmi A. D. M., Saludes J. P., Bhubalan K. (2022). Recent advances of marine sponge-derived microorganisms as a source of commercially viable natural products. Mar. Biotechnol. 24 (3), 492–512. doi: 10.1007/s10126-022-10130-2
Artasasta M. A., Yanwirasti Y., Taher M., Djamaan A., Ariantari N. P., Edrada-Ebel R. A., et al. (2021). Apoptotic activity of new oxisterigmatocystin derivatives from the marine-derived fungus Aspergillus nomius NC06. Mar. Drugs 19 (11), 631. doi: 10.3390/md19110631
Bai Y., Yi P., Zhang Y., Hu J., Wang Y., Ju J., et al. (2021). Structure-based molecular networking for the target discovery of novel germicidin derivatives from the sponge-derived Streptomyces sp. 18A01. J. Antibiot. 74 (11), 799–806. doi: 10.1038/s41429-021-00447-w
Bibi F., Faheem M., Azhar E. I., Yasir M., Alvi S. A., Kamal M. A., et al. (2017). Bacteria from marine sponges: a source of new drugs. Curr. Drug Metab. 18 (1), 11–15. doi: 10.2174/1389200217666161013090610
Blin K., Shaw S., Steinke K., Villebro R., Ziemert N., Lee S. Y., et al. (2019). antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic. Acids Res. 47 (W1), W81–W87. doi: 10.1093/nar/gkz310
Bovio E., Fauchon M., Toueix Y., Mehiri M., Varese G. C., Hellio C. (2019). The sponge-derived fungus Eurotium chevalieri MUT 2316 and its bioactive molecules: potential applications in the field of antifouling. Mar. Biotechnol. 21 (6), 743–752. doi: 10.1007/s10126-019-09920-y
Buttachon S., Ramos A. A., Inácio A., Dethoup T., Gales L., Lee M., et al. (2018). Bis-indolyl benzenoids, hydroxypyrrolidine derivatives and other constituents from cultures of the marine sponge-derived fungus Aspergillus candidus KUFA0062. Mar. Drugs 16 (4), 119. doi: 10.3390/md16040119
Calado R., Mamede R., Cruz S., Leal M. C. (2022). Updated trends on the biodiscovery of new marine natural products from invertebrates. Mar. Drugs 20 (6), 389. doi: 10.3390/md20060389
Cao D. D., Trinh T. T. V., Mai H. D. T., Vu V. N., Le H. M., Vu Thi Q., et al. (2019). Antimicrobial lavandulylated flavonoids from a sponge-derived Streptomyces sp. G248 in east vietnam sea. Mar. Drugs 17 (9), 529. doi: 10.3390/md17090529
Chen S., Guo H., Wu Z., Wu Q., Jiang M., Li H., et al. (2022a). Targeted discovery of sorbicillinoid pigments with anti-inflammatory activity from the sponge-derived fungus Stagonospora sp. SYSU-MS7888 using the PMG strategy. J. Agric. Food. Chem. 70 (48), 15116–15125. doi: 10.1021/acs.jafc.2c05940
Chen Y., Liu C., Kumaravel K., Nan L., Tian Y. (2022b). Two new sulfate-modified dibenzopyrones with anti-foodborne bacteria activity from sponge-derived fungus Alternaria sp. SCSIOS02F49. Front. Microbiol. 13, 879674. doi: 10.3389/fmicb.2022.879674
Cheng Y. J., Chen N. N., Li J., Su J. C., Yang J. Y., Zhang C. X., et al. (2021). Antimicrobial chlorinated carbazole alkaloids from the sponge-derived actinomycete Streptomyces diacarni LHW51701. Chin. J. Chem. 39 (5), 1188–1192. doi: 10.1002/cjoc.202000736
Cheng C., Othman E. M., Stopper H., Edrada-Ebel R., Hentschel U., Abdelmohsen U. R. (2017). Isolation of petrocidin a, a new cytotoxic cyclic dipeptide from the marine sponge-derived bacterium Streptomyces sp. SBT348. Mar. Drugs 15 (12), 383. doi: 10.3390/md15120383
Cheng M. M., Tang X. L., Sun Y. T., Song D. Y., Cheng Y. J., Liu H., et al. (2020). Biological and chemical diversity of marine sponge-derived microorganisms over the last two decades from 1998 to 2017. Molecules. 25 (4), 853. doi: 10.3390/molecules25040853
Devi P., Wahidullah S., Rodrigues C., Souza L. D. (2010). The sponge-derived bacterium Bacillus licheniformis SAB1: a source of antimicrobial compounds. Mar. Drugs 8 (4), 1203–1212. doi: 10.3390/md8041203
Ding L., Ren L., Li S., Song J., Han Z., He S., et al. (2019). Production of new antibacterial 4-hydroxy-α-pyrones by a marine fungus Aspergillus niger cultivated in solid medium. Mar. Drugs 17 (6), 344. doi: 10.3390/md17060344
Elsebai M. F., Schoeder C. T., Müller C. E. (2021). Fintiamin: a diketopiperazine from the marine sponge-derived fungus Eurotium sp. Arch. Pharm. (Weinheim) 354 (11), e2100206. doi: 10.1002/ardp.202100206
Esposito R., Federico S., Bertolino M., Zupo V., Costantini M. (2022). Marine demospongiae: a challenging treasure of bioactive compounds. Mar. Drugs 20 (4), 244. doi: 10.3390/md20040244
Gavriilidou A., Mackenzie T. A., Sánchez P., Tormo J. R., Ingham C., Smidt H., et al. (2021). Bioactivity screening and gene-trait matching across marine sponge-derived bacteria. Mar. Drugs 19 (2), 75. doi: 10.3390/md19020075
Guo T. T., Song M. M., Han W. R., Zhu J. H., Liu Q. C., Wang J. F. (2021). New N-methyl-4-quinolone alkaloid and citrinin dimer derivatives from the sponge-derived fungus Penicillium sp. SCSIO 41303. Phytochem. Lett. 46, 29–35. doi: 10.1016/j.phytol.2021.09.012
Hao X. M., Li S. S., Ni J., Wang G. Y., Li F., Li Q., et al. (2021). Acremopeptaibols A-F, 16-residue peptaibols from the sponge-derived Acremonium sp. IMB18-086 cultivated with heat-killed Pseudomonas aeruginosa. J. Nat. Prod. 84 (11), 2990–3000. doi: 10.1021/acs.jnatprod.1c00834
Hou Z. X., Sun X. C., Chen X. D., Zhang G. J., Che Q., Li D. H., et al. (2022). Xanalterate a, altertoxin VIII and IX, perylenequinone derivatives from antarctica-sponge-derived fungus Alternaria sp. HDN19-690. Tetrahedron. Lett. 96, 153778. doi: 10.1016/j.tetlet.2022.153778
Huang L. M., Ding L. J., Li X. H., Wang N., Yan Y. S., Yang M. X., et al. (2019). A new lateral root growth inhibitor from the sponge-derived fungus Aspergillus sp. LS45. Bioorg. Med. Chem. Lett. 29 (13), 1593–1596. doi: 10.1016/j.bmcl.2019.04.051
Ibrahim A. H., Attia E. Z., Hajjar D., Anany M. A., Desoukey S. Y., Fouad M. A., et al. (2018). New cytotoxic cyclic peptide from the marine sponge-derived Nocardiopsis sp. UR67. Mar. Drugs 16 (9), 290. doi: 10.3390/md16090290
Jia Q., Du Y. Q., Wang C., Wang Y., Zhu T. H., Zhu W. M. (2019). Azaphilones from the marine sponge-derived fungus Penicillium sclerotiorum OUCMDZ-3839. Mar. Drugs 17 (5), 260. doi: 10.3390/md17050260
Jiao W. H., Xu Q. H., Ge G. B., Shang R. Y., Zhu H. R., Liu H. Y., et al. (2020). Flavipesides A–C, PKS-NRPS hybrids as pancreatic lipase inhibitors from a marine sponge symbiotic fungus Aspergillus flavipes 164013. Org. Lett. 22 (5), 1825–1829. doi: 10.1021/acs.orglett.0c00150
Kemsley E. K., Le Gall G., Dainty J. R., Watson A. D., Harvey L. J., Tapp H. S., et al. (2007). Multivariate techniques and their application in nutrition: a metabolomics case study. Br. J. Nutr. 98 (1), 1–14. doi: 10.1017/S0007114507685365
Kenshole E., Herisse M., Michael M., Pidot S. J. (2021). Natural product discovery through microbial genome mining. Curr. Opin. Chem. Biol. 60, 47–54. doi: 10.1016/j.cbpa.2020.07.010
Kiran G. S., Sekar S., Ramasamy P., Thinesh T., Hassan S., Lipton A. N., et al. (2018). Marine sponge microbial association: towards disclosing unique symbiotic interactions. Mar. Environ. Res. 140, 169–179. doi: 10.1016/j.marenvres.2018.04.017
Kitani S., Ueguchi T., Igarashi Y., Leetanasaksakul K., Thamchaipenet A., Nihira T. (2018). Rakicidin f, a new antibacterial cyclic depsipeptide from a marine sponge-derived Streptomyces sp. J. Antibiot. 71 (1), 139–141. doi: 10.1038/ja.2017.92
Kumla D., Aung T. S., Buttachon S., Dethoup T., Gales L., Pereira J. A., et al. (2017). A new dihydrochromone dimer and other secondary metabolites from cultures of the marine sponge-derived fungi Neosartorya fennelliae KUFA 0811 and Neosartorya tsunodae KUFC 9213. Mar. Drugs 15 (12), 375. doi: 10.3390/md15120375
Lee Y. K., Lee J. H., Lee H. K. (2001). Microbial symbiosis in marine sponges. J. Microbiol. 39 (4), 254–264.
Lei H., Lin X. P., Han L., Ma J., Dong K. L., Wang X. B., et al. (2017). Polyketide derivatives from a marine-sponge-derived fungus Pestalotiopsis heterocornis. Phytochemistry 142, 51–59. doi: 10.1016/j.phytochem.2017.06.009
Lever J., Krzywinski M., Altman N. (2017). Points of significance: principal component analysis. Nat. Methods 14 (7), 641–642. doi: 10.1038/nmeth.4346
Li C. W., Chen J. Y., Hua T. E. (1998). Precambrian sponges with cellular structures. Science 279 (5352), 879–882. doi: 10.1126/science.279.5352.879
Li W. H., Ding L. J., Li J., Wen H. M., Liu Y., Tan S. L., et al. (2022). Novel antimycin analogues with agricultural antifungal activities from the sponge-derived actinomycete Streptomyces sp. NBU3104. J. Agric. Food. Chem. 70 (27), 8309–8316. doi: 10.1021/acs.jafc.2c02626
Li W., Ding L. J., Wang N., Xu J. Z., Zhang W. Y., Zhang B., et al. (2019). Isolation and characterization of two new metabolites from the sponge-derived fungus Aspergillus sp. LS34 by OSMAC approach. Mar. Drugs 17 (5), 283. doi: 10.3390/md17050283
Liang J. Q., She J. L., Fu J., Wang J. M., Ye Y. X., Yang B., et al. (2023). Advances in natural products from the marine-sponge-derived microorganisms with antimicrobial activity in the last decade. Mar. Drugs 21 (4), 236. doi: 10.3390/md21040236
Liu Z., Frank M., Yu X. Q., Yu H. Q., Tran-Cong N. M., Gao Y., et al. (2020b). Secondary metabolites from marine-derived fungi from China. Prog. Chem. Organic Natural Products 111, 81–153. doi: 10.1007/978-3-030-37865-3_2
Liu D., Li Y., Guo X. C., Ji W., Lin W. H. (2020a). Chartarlactams q-t, dimeric phenylspirodrimanes with antibacterial and antiviral activities. Chem. Biodiversity 17 (6), e2000170. doi: 10.1002/cbdv.202000170
Liu X., Locasale J. W. (2017). Metabolomics: a primer. Trends. Biochem. Sci. 42 (4), 274–284. doi: 10.1016/j.tibs.2017.01.004
Love G. D., Grosjean E., Stalvies C., Fike D. A., Grotzinger J. P., Bradley A. S., et al. (2009). Fossil steroids record the appearance of demospongiae during the cryogenian period. Nature. 457 (7230), 718–721. doi: 10.1038/nature07673
Luo X. W., Lin Y., Lu Y. J., Zhou X. F., Liu Y. H. (2019). Peptides and polyketides isolated from the marine sponge-derived fungus Aspergillus terreus SCSIO 41008. Chin. J. Nat. Med. 17 (2), 149–154. doi: 10.1016/s1875-5364(19)30017-2
Machado F. P., Rodrigues I. C., Gales L., Pereira J. A., Costa P. M., Dethoup T., et al. (2022). New alkylpyridinium anthraquinone, isocoumarin, c-glucosyl resorcinol derivative and prenylated pyranoxanthones from the culture of a marine sponge-derived fungus, Aspergillus stellatus KUFA 2017. Mar. Drugs 20 (11), 672. doi: 10.3390/md20110672
Martins B. T., da Silva M. C., Pinto M., Cidade H., Kijjoa A. (2019). Marine natural flavonoids: chemistry and biological activities. Nat. Prod. Res. 33 (22), 3260–3272. doi: 10.1080/14786419.2018.1470514
Matroodi S., Siitonen V., Baral B., Yamada K., Akhgari A., Metsa-Ketela M. (2020). Genotyping-guided discovery of persiamycin a from sponge-derived halophilic Streptomonospora sp. PA3. Front. Microbiol. 11, 1237. doi: 10.3389/fmicb.2020.01237
Matsuda Y., Mitsuhashi T., Lee S., Hoshino M., Mori T., Okada M., et al. (2016). Astellifadiene: structure determination by NMR spectroscopy and crystalline sponge method, and elucidation of its biosynthesis. ChemInform 55 (19), 5785–5788. doi: 10.1002/anie.201601448
Medema M. H., Blin K., Cimermancic P., de Jager V., Zakrzewski P., Fischbach M. A., et al. (2011). antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic. Acids Res. 39 (suppl_2), W339–W346. doi: 10.1093/nar/gkr466
Navarro-Muñoz J. C., Selem-Mojica N., Mullowney M. W., Kautsar S. A., Tryon J. H., Parkinson E. I., et al. (2020). A computational framework to explore large-scale biosynthetic diversity. Nat. Chem. Biol. 16 (1), 60–68. doi: 10.1038/s41589-019-0400-9
Newman D. J., Cragg G. M. (2018). Natural products as sources of new drugs from 1981 to 2014. J. Nat. Prod. 79 (3), 629–661. doi: 10.1021/acs.jnatprod.5b01055
Pang X. Y., Lin X. P., Wang J. F., Liang R., Tian Y. Q., Salendra L., et al. (2018a). Three new highly oxygenated sterols and one new dihydroisocoumarin from the marine sponge-derived fungus Cladosporium sp. SCSIO41007. Steroids. 129, 41–46. doi: 10.1016/j.steroids.2017.12.001
Pang X. P., Lin X. P., Wang P., Zhou X. F., Yang B., Wang J. F., et al. (2018b). Perylenequione derivatives with anticancer activities isolated from the marine sponge-derived fungus, Alternaria sp. SCSIO41014. Mar. Drugs 16 (8), 280. doi: 10.3390/md16080280
Perez De Souza L., Alseekh S., Brotman Y., Fernie A. R. (2020). Network-based strategies in metabolomics data analysis and interpretation: from molecular networking to biological interpretation. Expert. Rev. Proteomics. 17 (4), 243–255. doi: 10.1080/14789450.2020.1766975
Pita L., Rix L., Slaby B. M., Franke A., Hentschel U. (2018). The sponge holobiont in a changing ocean: from microbes to ecosystems. Microbiome 6 (1), 1–18. doi: 10.1186/s40168-018-0428-1
Proksch P. (1994). Defensive roles for secondary metabolites from marine sponges and sponge-feeding nudibranchs. Toxicon. 32 (6), 639–655. doi: 10.1016/0041-0101(94)90334-4
Rotinsulu H., Yamazaki H., Sugai S., Iwakura N., Wewengkang D. S., Sumilat D. A., et al. (2018). Cladosporamide a, a new protein tyrosine phosphatase 1B inhibitor, produced by an indonesian marine sponge-derived Cladosporium sp. J. Nat. Med. 72 (3), 779–783. doi: 10.1007/s11418-018-1193-y
Russell A. H., Truman A. W. (2020). Genome mining strategies for ribosomally synthesised and post-translationally modified peptides. Comput. Struct. Biotechnol. J. 18, 1838–1851. doi: 10.1016/j.csbj.2020.06.032
Sabdaningsih A., Liu Y., Mettal U., Heep J., Wang L., Cristianawati O., et al. (2020). A new citrinin derivative from the Indonesian marine sponge-derived fungus Penicillium citrinum. Mar. Drugs 18 (4), 227. doi: 10.3390/md18040227
Salem M. A., de Souza L. P., Serag A., Fernie A. R., Farag M. A., Ezzat S. M., et al. (2020). Metabolomics in the context of plant natural products research: from sample preparation to metabolite analysis. Metabolites 10 (1), 37. doi: 10.3390/metabo10010037
Sathiyanarayanan G., Saibaba G., Kiran G. S., Yang Y. H., Selvin J. (2017). Marine sponge-derived bacteria as a potential source for polyhydroxyalkanoates. Crit. Rev. Microbiol. 43 (3), 294–312. doi: 10.1080/1040841X.2016.1206060
Shah M., Sun C. X., Sun Z. C., Zhang G. J., Che Q., Gu Q. Q., et al. (2020). Antibacterial polyketides from antarctica sponge-derived fungus Penicillium sp. HDN151272. Mar. Drugs 18 (2), 71. doi: 10.3390/md18020071
She J. L., Chen Y., Ye Y. X., Lin X. P., Yang B., Xiao J., et al. (2022). New carboxamides and a new polyketide from the sponge-derived fungus Arthrinium sp. SCSIO 41421. Mar. Drugs 20 (8), 475. doi: 10.3390/md20080475
Shin H. J., Choi B.-K., Trinh P. T. H., Lee H.-S., Kang J. S., Van T. T. T., et al. (2018). Suppression of RANKL-induced osteoclastogenesis by the metabolites from the marine fungus Aspergillus flocculosus isolated from a sponge Stylissa sp. Mar. Drugs 16 (1), 14. doi: 10.3390/md16010014
Shukla S. (2016). Secondary metabolites from marine microorganisms and therapeutic efficacy: a mini review. Indian J. Mar. Sci. 45 (10), 1245–1254.
Skinnider M. A., Johnston C. W., Gunabalasingam M., Merwin N. J., Kieliszek A. M., MacLellan R. J., et al. (2020). Comprehensive prediction of secondary metabolite structure and biological activity from microbial genome sequences. Nat. Commun. 11 (1), 6058. doi: 10.1038/s41467-020-19986-1
Sun C. X., Liu X. Y., Sun N., Zhang X. M., Shah M., Zhang G. J., et al. (2022). Cytotoxic nitrobenzoyl sesquiterpenoids from an antarctica sponge-derived Aspergillus insulicola. J. Nat. Prod. 85 (4), 987–996. doi: 10.1021/acs.jnatprod.1c01118
Sun C. X., Zhang Z. P., Ren Z. L., Yu L., Zhou H., Han Y. X., et al. (2020). Antibacterial cyclic tripeptides from antarctica-sponge-derived fungus Aspergillus insulicola HDN151418. Mar. Drugs 18 (11), 532. doi: 10.3390/md18110532
Tang W. Z., Liu J. T., Hu Q., He R. J., Guan X. Q., Ge G. B., et al. (2020). Pancreatic lipase inhibitory cyclohexapeptides from the marine sponge-derived fungus aspergillus sp. J. Nat. Prod. 83 (7), 2287–2293. doi: 10.1021/acs.jnatprod.0c00549
Tawfike A., Attia E. Z., Desoukey S. Y., Hajjar D., Makki A. A., Schupp P. J., et al. (2019). New bioactive metabolites from the elicited marine sponge-derived bacterium Actinokineospora spheciospongiae sp. nov. AMB. Express. 9 (1), 12. doi: 10.1186/s13568-018-0730-0
Taylor M. W., Radax R., Steger D., Wagner M. (2007). sponge-derived microorganisms: evolution, ecology, and biotechnological potential. Microbiol. Mol. Biol. Rev. 71 (2), 295–347. doi: 10.1128/MMBR.00040-06
Tian Y. Q., Lin S. T., Kumaravel K., Zhou H., Wang S. Y., Liu Y. H. (2018). Polyketide-derived metabolites from the sponge-derived fungus Aspergillus sp. F40. Phytochem. Lett. 27, 74–77. doi: 10.1016/j.phytol.2018.06.009
Wang C. Y., Liu X. H., Zheng Y. Y., Ning X. Y., Zhang Y. H., Fu X. M., et al. (2022). 2,5-diketopiperazines from a sponge-derived fungus Aspergillus sclerotiorum. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.808532
Wang D. Y., Qu P., Zhou J. Y., Wang Y., Wang L. P., Zhu W. M. (2020). p-terphenyl alcohols from a marine sponge-derived fungus, Aspergillus candidus OUCMDZ-1051. Mar. Life. Sci. Tech. 2 (3), 262–267. doi: 10.1007/s42995-020-00039-x
Wang T. T., Zhou J., Zou J. B., Shi Y. T., Zhou W. L., Shao P., et al. (2021). Discovery of cymopolyphenols a-f from a marine mesophotic zone aaptos sponge-derived fungus Cymostachys sp. NBUF082. Front. Microbiol. 12, 638610. doi: 10.3389/fmicb.2021.638610
Wen W., Chen C., Tang J. K., Wang C. Y., Zhou M. Y., Cheng Y. R., et al. (2022). Efficacy and safety of three new oral antiviral treatment (molnupiravir, fluvoxamine and paxlovid) for COVID-19: a meta-analysis. Ann. Med. 54 (1), 516–523. doi: 10.1080/07853890.2022.2034936
Wu Q. H., Throckmorton K., Maity M., Chevrette M. G., Braun D. R., Rajski S. R., et al. (2020). Bacillibactins e and f from a marine sponge-derived Bacillus sp. J. Nat. Prod. 84 (1), 136–141. doi: 10.1021/acs.jnatprod.0c01170
Xu D. B., Nepal K. K., Chen J., Harmody D., Zhu H. N., McCarthy P. J., et al. (2018). Nocardiopsistins a-c: new angucyclines with anti-MRSA activity isolated from a marine sponge-derived Nocardiopsis sp. HB-J378. Synth. Syst. Biotechnol. 3 (4), 246–251. doi: 10.1016/j.synbio.2018.10.008
Yang J. Y., Sanchez L. M., Rath C. M., Liu X., Boudreau P. D., Bruns N., et al. (2013). Molecular networking as a dereplication strategy. J. Nat. Prod. 76 (9), 1686–1699. doi: 10.1021/np400413s
Zhang P. X., Zhang W. G. L., Lang Y., Qu Y., Chu F. N., Chen J. F., et al. (2018). Mass spectrometry-based metabolomics for tuberculosis meningitis. Clin. Chim. Acta 483, 57–63. doi: 10.1016/j.cca.2018.04.022
Zhang H. W., Zhao Z. P., Wang H. (2017). Cytotoxic natural products from marine sponge-derived microorganisms. Mar. Drugs 15 (3), 68. doi: 10.3390/md15030068
Zhao X. Q., Yang T. H. (2011). Draft genome sequence of the marine sediment-derived actinomycete Streptomyces xinghaiensis NRRL B24674T. Am. Soc Microbiol. 193 (19), 5543–5543. doi: 10.1128/JB.05689-11
Zhao Q., Zhang J. L., Li F. (2018). Application of metabolomics in the study of natural products. Nat. Prod. Bioprospect. 8 (4), 321–334. doi: 10.1007/s13659-018-0175-9
Zhou G. L., Sun C. X., Hou X. W., Che Q., Zhang G. J., Gu Q. Q., et al. (2021a). Ascandinines a-d, indole diterpenoids, from the sponge-derived fungus Aspergillus candidus HDN15-152. J. Org. Chem. 86 (3), 2431–2436. doi: 10.1021/acs.joc.0c02575
Keywords: sponge, sponge-associated microorganism, natural product, biological activity, mining strategy
Citation: Li P, Lu H, Zhang Y, Zhang X, Liu L, Wang M and Liu L (2023) The natural products discovered in marine sponge-associated microorganisms: structures, activities, and mining strategy. Front. Mar. Sci. 10:1191858. doi: 10.3389/fmars.2023.1191858
Received: 22 March 2023; Accepted: 09 May 2023;
Published: 19 May 2023.
Edited by:
Bo Wang, Shandong University of Science and Technology, ChinaReviewed by:
Carlos Jimenez, University of A Coruña, SpainCopyright © 2023 Li, Lu, Zhang, Zhang, Liu, Wang and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liwei Liu, liuliwei@nbu.edu.cn; Min Wang, wangm@wyu.edu.cn
†These authors have contributed equally to this work
 Peishan Li
Peishan Li Hongmei Lu1†
Hongmei Lu1†  Min Wang
Min Wang Liwei Liu
Liwei Liu