Feeding and water temperature modulate the potential activity of gut microbiota in greater amberjack juveniles (Seriola dumerili)
- 1Departamento de Biología Marina y Acuicultura. Instituto de Ciencias Marinas de Andalucía (ICMAN-CSIC), Puerto Real, Spain
- 2Department of Biotechnology and Food Science, Norwegian University of Science and Technology (NTNU), Trondheim, Norway
Gut microbial communities are highly dynamic and respond to factors such as diet and host metabolism. However, there is limited knowledge on changes in gut microbiota during the daily cycle. To investigate how interactions between feeding and water temperature affected the diurnal dynamics and activity of the microbiota, we analyzed the qualitative and quantitative composition in feces of the potentially active bacterial community along a day cycle in greater amberjack juveniles (Seriola dumerili). Bacterial density (based on qPCR of 16S rDNA) in feces was relatively stable along the day. In contrast bacterial activity (based on qPCR of 16S rRNA) increased during the period of active feeding, and was reduced to pre-feeding levels four hours after the last meal. The relative potential activity of the bacterial community in fish feces varied with postprandial time. At the phylum level there was a shift from a Spirochaetes-dominated community in the morning to increased potential activity of Proteobacteria after feeding. At lower taxonomic level, i.e. order, the bacterial community was dominated by Mycoplasmoidales, which relative potential activity was maintained throughout the day in all treatments. By contrast, for absolute potential bacterial activity at the order level there was a peak in potential activity of several bacterial orders at 6h after first feeding, with the most noticeable increase observed for Mycoplasmoidales. Results from the present work also indicated a close relation between the effect of water temperature and microbial dynamics. This is the first study assessing the quantitative and qualitative effects of water temperature and daily feeding rhythm on the gut microbiota dynamics in fish. Feeding was the main driver modulating bacterial activity, promoting a peak in bacterial activity 6h after first feeding. However, water temperature also modulated gut microbiota dynamics, with a marked effect on the time scale. In addition, results indicate that analysis of absolute bacterial potential activity provides a better resolution of metabolically active gut microbial community, since a change in the absolute activity of a single taxon can alter the relative activity of all taxa.
1 Introduction
The digestive tract houses a complex and metabolically active consortium of microorganisms that plays critical roles in the nutrition and health of the host. During the last years, many studies have focused on microbial taxonomy to describe the bacterial community composition in different animal species and how it is affected by different variables. However, the interaction between the microbiome and the host is under-explored. Gut microbial communities are highly dynamic and respond to factors such as diet and host metabolism (Schlomann and Parthasarathy, 2019). These responses can generate community dynamics at different time scales, such as circadian rhythms stimulated by light-dark cycles or feeding time. In aquaculture research, one of the horizons is to establish knowledge to optimize growth and proper development, in a way that assure animal welfare. Thus, characterizing of gut microbiome dynamics is necessary to identify and better understand the relationship it has for host physiology and fitness (Zarrinpar et al., 2014; Risely et al., 2021).
During the last years, several studies have provided a good description of relative (percent) changes in microbiota composition of different fish species and its variability as a result of manipulation of specific conditions, e.g. dietary modulation, development, rearing system and environmental microbiota (e.g. Fossmark et al., 2021; Moroni et al., 2021; Navarro-Guillén et al., 2021; Naya-Català et al., 2022). However, there is limited knowledge on gut microbiota changes during the daily cycle and on variation in absolute microbiota abundance. Absolute abundances can provide information on the extent and directionality of changes in abundance of taxa that may be masked by their relative abundances (Korem et al., 2015; Vandeputte et al., 2021). In laboratory mice, a peak in bacterial density at dusk characterized microbial circadian rhythms, coinciding with the period when mice become active and start feeding (Thaiss et al., 2016). Also meerkats (Suricata suricatta) show strong diurnal oscillations in bacterial density and composition (Risely et al., 2021). These oscillations were associated with temperature constrained foraging schedules. In fish, information on gut microbiota daily dynamics is scarce. In the leopard coral grouper (Plectropomus leopardus) the relative composition of microbiota changed between fasting and feeding conditions, and it took approximately 12 hours for the dominant phyla to change (Mekuchi et al., 2018). Comparison of microbiome composition with transcriptome and metabolome data indicated that microbial fluctuation followed nutritional input. The studies above indicated a link between feeding schedule/habits and the gut microbiome composition and functionality. However, the underlying mechanisms of this relationship and the role of intestine microbiota remain unknown.
Given the multiple impacts of the gut microbiome on the host physiology and metabolism, it is important to understand how environmental factors may influence it. A practical application of such knowledge is to improve the efficiency of aquaculture production systems. Water temperature is a crucial factor in fish production, influencing among others feed intake and gut transit time (i.e. Das et al., 2018; Yúfera et al., 2019; Mazumder et al., 2020; Navarro-Guillén et al., 2023). However, in addition to affecting the digestive capacity of the host, it can also modulate the composition of its bacterial community. Because of the ectothermic nature of fish, the bacterial community is exposed to variations in environmental temperature. Increase in water temperature resulted in increased richness (Margalef index) and diversity (Shannon-Weaver index) of the gut microbiota in turbot juveniles (Scophthalmus maximus L.) (Guerreiro et al., 2016), and in gut species richness in yellowtail kingfish juveniles (Seriola lalandi) (Soriano et al., 2018). In Chinook salmon (Oncorhynchus tshawytscha) gut relative microbiome composition changed with increasing temperature (Steiner et al., 2021). Furthermore, gut transit time, the time it takes the chyme to travel through the gastrointestinal tract, has been shown to alter the composition of the microbial community in humans. A slow transit in the colon has been associated with a high microbial richness and a tendency to substitute carbohydrate catabolism for protein catabolism. This affect the global digestive efficiency in the colon in humans (Oliphant and Allen-Vercoe, 2019). Thus, from a sustainable aquaculture perspective, understanding how water temperature affect growth, food consumption, feed conversion efficiency, and host-microbe interaction is crucial, especially in the context of climate change that can negatively affect aquaculture performance.
Greater amberjack (Seriola dumerili) is a marine fish species of great interest for the aquaculture industry worldwide due to its fast growth and excellent flesh quality (Sicuro and Luzzana, 2016). Greater amberjack exhibits a voracious appetite, and to maintain high growth rates during the juvenile stage it has to be fed several times a day. Studies of the digestive function of this species have been focused on digestive enzymes at early and juvenile stages (Navarro-Guillén et al., 2019; Pérez et al., 2020; Gamberoni et al., 2021; Navarro-Guillén et al., 2022), and gut transit time (Fernández-Montero et al., 2018; Navarro-Guillén et al., 2023). Despite the increasing interest in this species, information about gut microbiota composition is only available for related species, such as longfin yellowtail (Seriola rivoliana) and yellowtail kingfish. The intestine of S. rivoliana is dominated by the phyla Proteobacteria, Firmicutes and Bacteroidetes (Salas-Leiva et al., 2020). For yellowtail kingfish juveniles the most abundant phyla are Proteobacteria, Actinobacteria, Bacteroidetes and Firmicutes, but the relative composition of the microbiota varies with fish size (Aguilera et al., 2013; Soriano et al., 2018).
To investigate how interactions between feeding and water temperature affected the diurnal dynamics and activity of the gut microbiota, we did an experiment with pulsed feeding for six hours, followed by fasting. The experiment was run at three different water temperatures. We analyzed the protein synthesis potential of the bacterial community along a day cycle in feces of greater amberjack juveniles by RNA-based amplicon sequencing. For quantitative abundance and activity, we quantified copy numbers of total 16S DNA and RNA from 16S rRNA genes by quantitative PCR (qPCR), and used these data as a proxy of absolute bacterial density and activity, respectively.
2 Materials and methods
2.1 Fish rearing and sampling
Greater amberjack juveniles were supplied by Futuna Blue España S.L. (El Puerto de Santa María, Cádiz, Spain) and transferred to the Institute of Marine Sciences of Andalusia facilities (Puerto Real, Cádiz, Spain). Juveniles were randomly distributed in nine 1 m3 cylindroconical tanks (16 fish tank-1, with three replicate tanks per temperature treatment), and acclimated to the experimental temperatures for one week. The three replicate tanks belonged to an independent recirculating aquaculture system (RAS). During acclimation water temperature in two of the RAS was changed by 0.5°C per day to 18 and 26°C, whereas in the other RAS was kept constant at 22°C. Acclimated juveniles with 53.4 ± 19.8 g wet body weight, were reared under a light/dark cycle (12 h light: 12 h dark) at the three temperatures, with the light period starting at 08:00 h. Water salinity was 33.2 ± 0.4 g L-1, oxygen was maintained above 85% saturation, pH was 8.0 ± 0.1 and NH4< 0.25 mg NH3+NH4 L-1. The fish were fed a commercial diet (Skretting, Burgos, Spain) to apparent satiation three times a day (08:00, 12:00 and 16:00 h). After 17 days under these conditions, nine fish per temperature (3 fish per tank) were sampled every 6 h for a 24 h cycle. Sampling points were set at 08:00h (before first feeding), 14:00 h, 20:00 h and 02:00 h. For sampling, fish were euthanized with an overdose of phenoxyethanol (77699, Sigma-Aldrich) and rinsed with ethanol to avoid contamination of the intestinal samples with skin microbiota. The rectum of each fish was aseptically excised and feces were collected by stripping. At each sampling, 50 mL of water from each tank were sampled and filtered through 0.22 µm sterile filters Sterivex™ (Millipore) to collect bacteria in the water. Feces and filter samples were maintained at -80°C until analysis.
The voluntary feed intake (VFI), expressed as percentage of initial body weight per day (% IBW d-1) was calculated as:
and fish weight gain (WG, %IBW) as:
All experimental procedures complied with the Guidelines of the European Union Council (2010/63/EU) for the use and experimentation of laboratory animals, and were reviewed and approved by the Spanish National Research Council (CSIC) bioethical committee and Spanish National Veterinary Authority for project THERMODIGEST- RTI2018-096134-B-I00 (REF: 02/07/2019/107).
2.2 DNA and RNA extraction
Feces samples and STERIVEX-filters containing the water samples were freeze-dried before analyses. The feces samples were weighed and both, feces and water samples were transferred to sterile Precellys-tubes containing an approximately 2 mm thick layer of 1.4 mm zirconium oxide beads (Bertin Technologies), and 800 or 600 µl DNA/RNA shield (Zymo Research), for feces and water samples, respectively. Samples were homogenized using a Precellys 24 (Bertin Technologies) at 5500 rpm, 2 pulses, 30 seconds. DNA and RNA were extracted by using the Quick-DNA/RNA™ Magbead kit (Zymo Research), and the Kingfisher™ Flex (Thermo Fisher), following the protocol and Kingfisher script provided by Zymo Research. The eluted DNA and RNA concentrations were determined with QUBIT™ 3 (Invitrogen), using QUBIT™ dsDNA HS and QUBIT™ RNA HS assay kits, respectively, according to producer’s protocol. cDNA was made from the RNA by using the iScript™ cDNA synthesis kit (BioRad) following the manufacturer’s protocol.
2.3 Amplicon Library preparation
PCR was conducted to amplify the variable regions 3 and 4 of the 16S rRNA genes by using universal bacterial primers (Gómez de la Torre Canny et al., 2023), cDNA was used as template. PCR reactions (25 µl volume, 0.3 µM of both of the primers ill341F_Kl/805R, 0.2 mM of each dNTP, 1 µl template) were conducted using 0.4 U Phusion Hot Start polymerase (Thermo Scientific). The cycles for the PCR reaction were performed in a T100™ Thermal Cycler (BioRad), using the following cycling conditions: an initial denaturation step at 98°C for 60 s; 38 cycles of 98°C for 15 s, 55°C for 20 s, and 72°C for 20 s; and a final elongation step of 5 minutes at 72°C (Gómez de la Torre Canny et al., 2023). The resulting amplicons were purified and normalized using the Sequal Prep™ Normalization plates (96-well plates, Invitrogen) following the producer’s protocol. The amplicons were indexed in a second PCR using the Nextera® XT Index Kit v2. The PCR conditions were as described above, except that 2.5 µl of the purified normalized PCR products together with 2.5 µl of each indexing primer were used. The same cycling program as described above was used, except for only 10 cycles. After the indexing, the samples were purified and normalized using the Sequal Prep™ Normalization plates. Samples were pooled and concentrated using an Amicon® Ultra 0.5 ml centrifugal filter (30K membrane, Merck Millipore), and analyzed with a NanoDrop™ One Microvolume Spectrophotometer (Thermo Scientific™). The amplicon library was sequenced in a MiSeq run (Illumina, San Diego, CA) with v3 reagents (Illumina) and 300 paired-end at the Norwegian Sequencing Centre (NSC), University of Oslo.
2.4 Processing of sequencing data
The returned sequence data were processed by using the USEARCH V11 and UNOISE 3 pipelines (Edgar, 2010; Edgar, 2016a). Chimera removal and clustering at the 97% similarity level were performed using the UPARSE-OTU algorithm (Edgar, 2013). Taxonomy assignment was done based on the SINTAX script (Edgar, 2016b), using a confidence value threshold of 0.8 and included the RDP reference data set (version 15). The resulting amplicon sequence variant (ASV) data was processed in Excel. All chloroplasts, eukaryotes, and abundant ASVs from kit blanks and non-template controls were removed. After quality filtering and chimera removal, a total of 6,278,976 sequence reads were obtained with an average of 48,707 ± 18,395 and 35,949 ± 18,779 for feces and water samples, respectively. Finally, the ASV table was normalized to 20,000 reads per sample (the lowest read count observed for a sample) prior to multivariate analyses to avoid bias due to sequencing depth.
Since the sequences were obtained from 16S rRNA genes amplified from RNA extracts (using reverse transcribed cDNA), the present study targets taxa featuring high protein synthesis potential and does not consider quiescent/inactive or dead bacteria nor couple to cell number within the community (Rosselli et al., 2016). Therefore, this work does not describe bacterial community composition in terms of taxa abundance (as DNA-based amplicon sequencing does), but describes the metabolic activity potential of each taxon within the bacterial ecosystem, hereafter referred to as potential activity.
2.5 qPCR
To compare the amount of 16S DNA and 16S rRNA from 16S rRNA genes in the samples, qPCR was conducted. Standards for qPCR were prepared by performing PCR on some of the samples with the RT.966F (GCAACGCGMRGAACCTTACCTA) and RT.1089R (SGGACTTAACCSAACATYTCA) primers (Skjermo et al., 2015). Then the PCR products were purified using the QIAquick® PCR Purification Kit (Qiagen), following the producers’ protocol. The DNA concentration of the purified PCR products was determined with the NanoDrop™ One Microvolume Spectrophotometer (Thermo Scientific). A serial dilution of one of the purified PCR products was used as a standard for the determination of copy numbers by qPCR. Triplicate reactions of each cDNA and total DNA sample were run (4 µl of template per reaction), using a master mix (16 µl per reaction) containing SYBR® Green master mix (Thermo Scientific), DNA free water and the two primers at a final concentration of 0.3 mM each. For every run, the standard program for standard curves of the QuantStudio™ 5 Real-Time PCR System (Applied Biosystems™) was used. To estimate total copy-number of the samples, CT-values were plotted against log copy number/µl of diluted internal standards. The resulting equation was used to calculate copy-numbers for cDNA and DNA by using CT-values from the qPCR.
2.6 Statistical analysis
Statistical differences in fish growth, voluntary feed intake and weight gain were tested by one-way ANOVA and, whenever significant differences were identified, means were compared by the Post hoc multiple comparisons Tukey’s test. Before analyses, the ANOVA assumptions of normality and homogeneity of variance were tested using the Shapiro-Wilk and Levene’s tests, respectively (SPSS version 27). Percentage data (feed intake and weight wain) was arcsine-transformed before statistical analysis. Statistical analyses of potentially active bacterial community composition, based on the ASV table, were performed using the program package PAST version 3 (Hammer et al., 2001). Principal coordinate analysis (PCoA) (Davis, 1986) was based on Bray-Curtis and Sørensen similarities (Bray and Curtis, 1957; Wolda, 1981). To test for differences in the diversity of the potentially active ecosystem between sample groups, we applied one- and two-way PERMANOVA based on Bray-Curtis and Sørensen similarities (Anderson, 2001). For analysis of the oscillations on relative and absolute potential activity of specific taxa, two-way ANOVA was used with water temperature and sampling time as fixed factors (SPSS). All statistical analyses were considered significant at p<0.05. Absolute potential activity quantification was performed by multiplying relative potential activity of ASVs (% of reads) by the total amount of bacterial RNA copies (from 16S rRNA genes) in each sample quantified by qPCR (Barlow et al., 2020).
3 Results
3.1 Fish performance and voluntary feed intake
Initial body weight was, on average, 53.4 ± 19.8 g (mean ± SD, n = 140) with no statistical differences between groups. At the end of the experiment, fish reared at 26°C showed statistically higher body weight than fish reared at 18 and 22°C (Figure 1). Final average body weight was 63% higher when comparing the higher temperature with the lowest (76.0 ± 26.5, 92.9 ± 40.6 and 123.9 ± 44.9 g, for 18, 22 and 26°C, respectively. Mean ± SD, n = 36 per treatment). Weight dispersion increased with water temperature, but the coefficient of variance was similar. Daily feed consumption increased significantly with water temperature, ranging between 2.3 and 4.2% of IBW for fish reared at 18 and 26°C, respectively. Likewise, weight gain during the 17-days experimental period was significantly higher for fish reared at 26°C compared to the other two temperatures (Table 1).
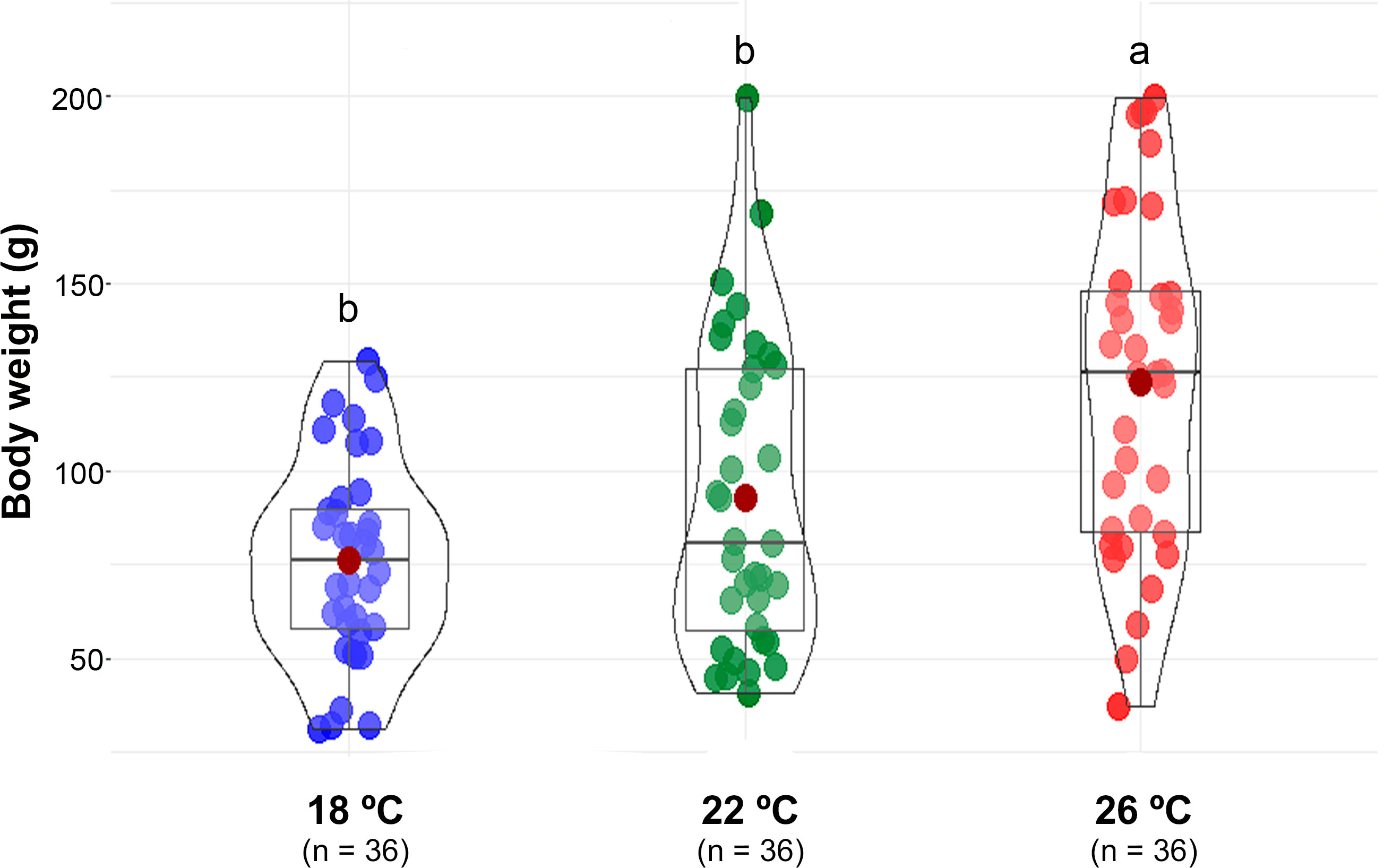
Figure 1 Box-and-violin plot of greater amberjack juveniles body weight (g) at the end of the experiment. Boxplots show the median (bold line) and the mean (dark red spot), while the violinplots represent the distribution of the data. Different letters indicate significant differences between rearing temperatures (p<0.05).
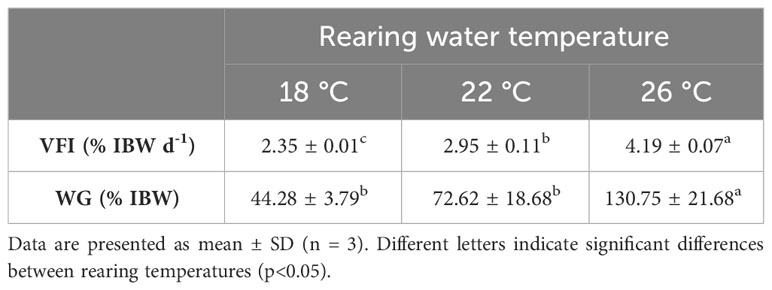
Table 1 Voluntary feed intake (VFI, % IBW d-1) and weight gain (WG, % IBW) of S. dumerili juveniles reared at 18, 22 and 26°C for 17 days.
3.2 Quantitative bacterial density and activity in the fish feces and rearing water
We used 16S rDNA and 16S rRNA copies from 16S rRNA genes as proxies for bacterial density and bacterial activity, respectively. Bacterial density in feces was relatively stable along the day, ranging, on average, from 5.43×103 to 2.92×106 16S rDNA copies per mg DW feces (Figure 2A). A two-way ANOVA revealed no effect of temperature, sampling time or the interaction between both factors on bacterial abundance per mg of gut content (p-value = 0.352, 0.439 and 0.432, respectively). By contrast, the quantitative sum of bacterial activity per sample (protein synthesis capacity, based on qPCR of 16S rRNA) differed with respect to sampling time (p-value< 0.0001), but not to temperature (p-value = 0.202) or the interaction between both factors (p-value = 0.876). The bacterial activity increased 1.5 order of magnitude during the period of active feeding, and was reduced to pre-feeding levels four hours after the last meal (Figure 2B). This pattern was also observed for the specific bacterial activity (16S rRNA:16S rDNA ratio), with almost two orders of magnitude increase during the period of active feeding. The ratio was reduced to the morning levels four hours after the last meal (Figure 2C). A statistically significant effect of the interaction between water temperature and sampling time was detected for the specific bacterial activity. The highest specific activity was reached in feces of fish reared at 26°C at 6 h post-first meal (p-values = 0.094, 0.000 and 0.045 for temperature, sampling time and the interaction between both factors, respectively).
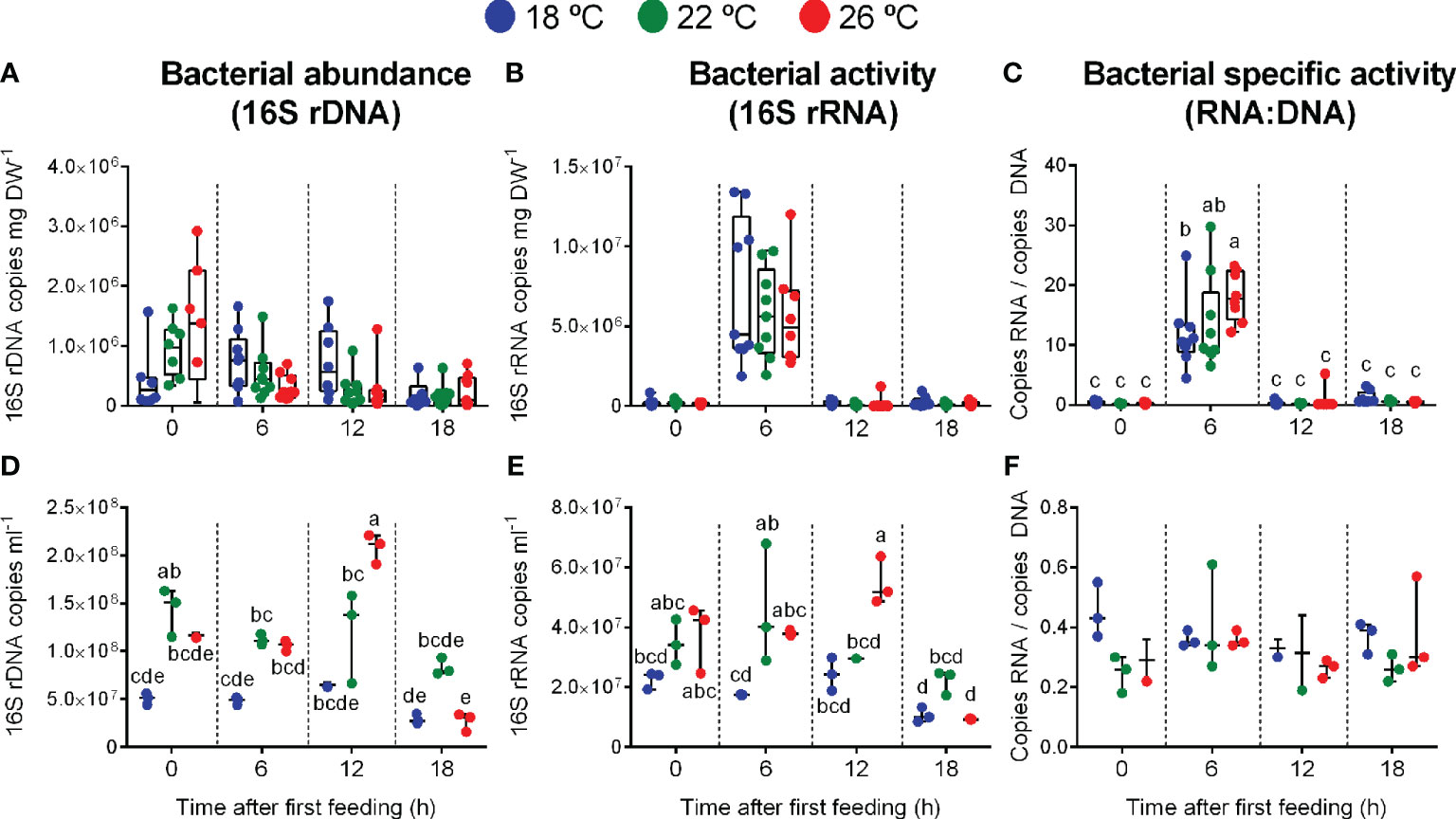
Figure 2 Bacterial density (16S rDNA copies from 16S rRNA genes), activity (16S rRNA copies from 16S rRNA genes) and specific activity (16S rRNA:16S rDNA ratio) in greater amberjack feces [(A–C); copies per mg DW feces] and rearing water [(D–F); copies per ml] for each treatment (18, 22 and 26 °C water temperature) at the different sampling points (n = 9 per temperature). Different letters mean statistical differences between sampling points for each treatment.
For bacterial abundance and activity in the water, two-way ANOVA based on the qPCR data revealed a statistically significant effect of water temperature, sampling point, and the interaction between both variables. This was observed for both bacterial abundance (16S rDNA; p-value = 0.000 for temperature, sampling time and the interaction between both factors) and bacterial activity (16S rRNA; p-values = 0.000, 0.000 and 0.007). For both variables, the highest copy numbers were observed at 26°C-water at 12 h after first feeding, while the lowest was reached 18 h after first feeding at the same water temperature (Figures 2D, E). By contrast, the specific activity of the bacteria in the water was low and stable (Figure 2F), with no effect of feeding time or water temperature (p-values = 0.245, 0.514 and 0.275 for temperature, sampling time and the interaction between both factors, respectively).
3.3 Taxonomic assessment of potentially active bacteria in the fish feces
The Illumina sequencing based on 16S rRNA yielded a total of 1858 ASVs. Two fish samples and two water samples were removed due to low number of reads (<100 reads). Globally, the relative potential activity of the bacterial community in fish feces varied in relation to postprandial time. The daily variation was more noticeable in ASVs belonging to the phyla Proteobacteria and Spirochaetes (Table 2). This fluctuation was mostly explained by changes in the relative potential activity of ASVs belonging to the orders Spirochaetes and Vibrionales. For the absolute bacterial potential activity (relative activity multiplied by rRNA copies number), a peak was observed at first postprandial sampling (6 h postprandial). The order Mycoplasmoidales was the main order responsible for this increase in absolute potential activity (Figure 3).
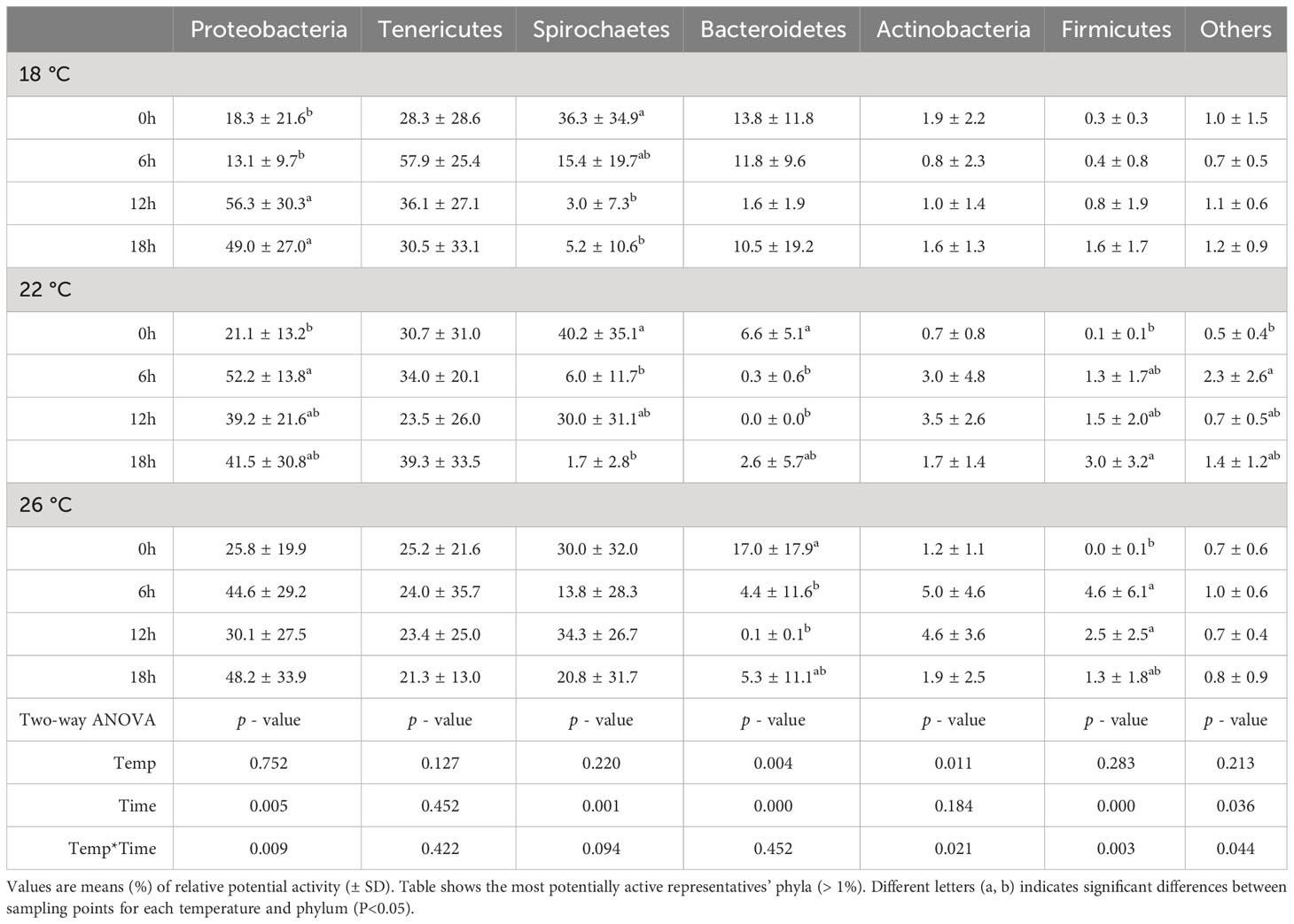
Table 2 Relative potential activity of the ASV at phylum level (expressed as percentage of the total number of reads) in feces of greater amberjack juveniles reared at 18, 22 and 26 °C and sampled at 0, 6, 12 and 18 hours post-first feeding.
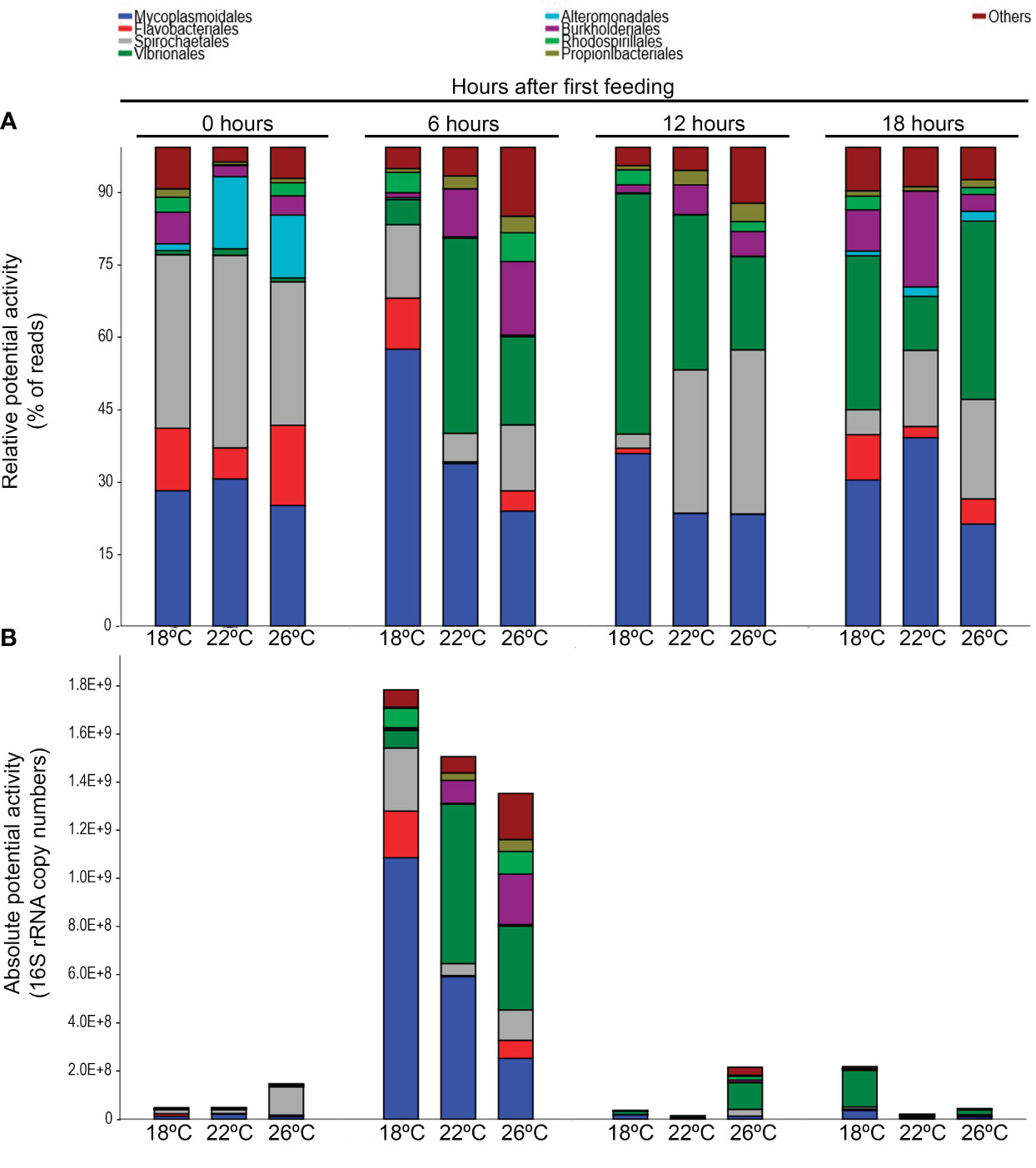
Figure 3 Structure of the potentially active bacterial community at the order level expressed as relative potential activity (A) and absolute potential activity (B) in feces of greater amberjack juveniles reared at 18, 22 and 26 °C and sampled at 0 hours (pre-feeding) and 6, 12 and 18 hours post-first meal (n = 9). The plots represent the most potentially active orders (> 1% of total reads).
Relative potential activity expressed as percent of the total number of reads at the phylum level is shown in Table 2. Postprandial time significantly affected the relative potential activity of the phyla Proteobacteria, Spirochaetes, Bacteroidetes and Firmicutes. While the relative potential activity of Proteobacteria and Firmicutes increased after feeding, it decreased for Spirochaetes and Bacteroidetes. Moreover, this effect tended to be faster with temperature increase, e.g. Firmicutes potential activity increase was statistically significant 6 h after first meal at 26°C, by contrast, at 22°C it was 18 h after first meal. Delays in the temporal oscillation in potential activity were also observed for Proteobacteria and Spirochaetes between 18 and 22°C. Overall, differences in relative potential activity between fish reared at different temperatures were found for the phyla Bacteroidetes and Actinobacteria. The relative potential activity of Bacteroidetes was higher in 18°C-fish compared to 22°C-fish, while the relative potential activity of Actinobacteria was higher in 26°C-fish compared to 18°C-fish.
Under fasting conditions (postprandial time 0h), the most potentially active orders were Spirochaetales, Mycoplasmoidales and Flavobacteriales (Figure 3A). The relative potential activity of Mycoplasmoidales was maintained throughout the sampling period, and not affected by feeding time. The same trend was observed for the order Spirochaetales in fish reared at 22 and 26°C, while at the lowest temperature the relative potential activity of this order decreased significantly along the postprandial period. By contrast, feeding promoted an increase in the relative potential activity of the order Vibrionales, which reached its maximum at 6h post first meal in fish reared at 22°C. For fish reared at 18 and 26°C the maximum was at 12 and 18h post first meal, respectively. Relative potential activity of Vibrionales in fish reared at 22°C was similar to the pre-feeding situation 18h after first meal, while feces from fish reared at 18 and 26°C still showed higher proportion. The relative potential activity of Alteromonadales decreased after feeding for all treatments, while the inverse pattern was observed for Burkholderiales.
Differences in the global potentially active bacteria between treatments, in relative terms, were only found for the orders Flavobacteriales, which was higher in fish reared at 18°C compared to 22°C, Rhodospirillales, for which the lowest relative activity was found in 22°C-fish and, and in Others which were higher in 26 than in 22°C-fish.
For absolute bacterial potential activity at the order level, there was a peak in absolute activity in several bacterial orders 6h postprandial (Figure 3B). The most noticeable increase was observed for Mycoplasmoidales, which potential activity statistically rose in all treatments. To a lesser extent, an increase in Flavobacteriales and Spirochaetales absolute activities was recorded in 18°C-reared fish, while for 22 and 26°C-fish it was for Burkholderiales absolute potential activity. An increase in Vibrionales absolute potential activity was also observed in 22 °C-reared fish 6h postprandial.
Clear statistical differences were also recorded for the alpha diversity indices based on rRNA for the feces potentially active bacterial community (richness and evenness). Sampling time did not significantly affect diversity, but water temperature did. Richness, which take into account the total number of ASVs and is insensitive to ASV activity, showed considerable variation between individuals. It was statistically higher at 18°C (p-value< 0.001). Whereas the evenness, which quantifies how similar the relative 16S RNA signal is for each ASV, was significantly lower for fish reared at 18°C (p-value=0.0003) (Figure 4).
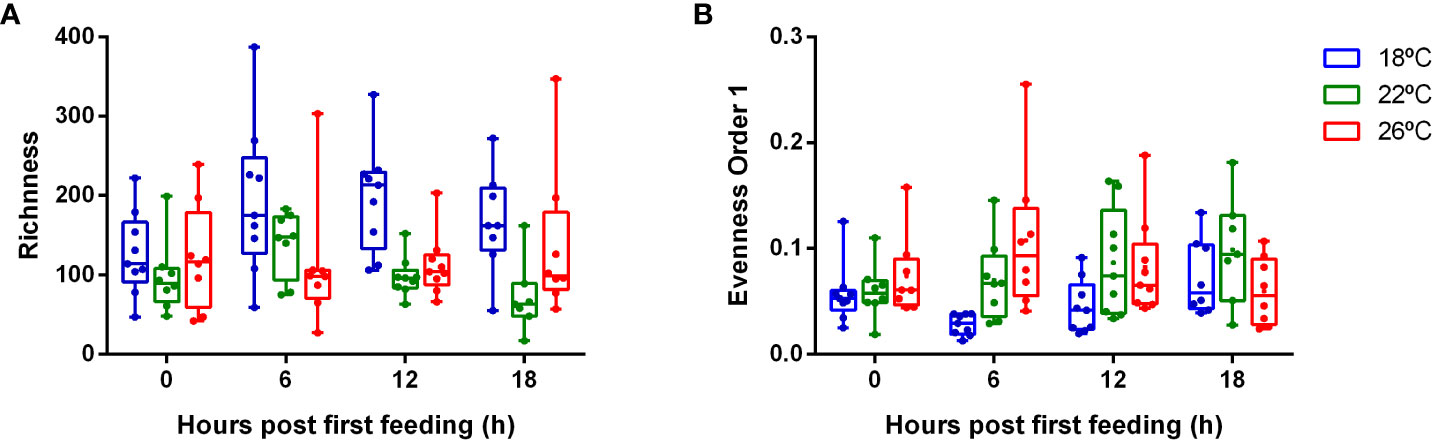
Figure 4 Box-and-wishers plots of the alpha diversity indices based on 16S sRNA sequencing: richness (A) and evenness (B) in feces of greater amberjack juveniles reared at 18 (blue), 22 (green) and 26 °C (red) and sampled at 0, 6, 12 and 18 hours post-first meal.
Two-way PERMANOVA based on Sørensen (Figure 5A) and Bray-Curtis (Figure 5B) similarities revealed significantly different fish microbiota due to temperature and sampling time. A statistical interaction between both factors was also observed for both beta diversity indices (Figure 5C). These results are reflected in the Principal Coordinate Analysis (PCoA). For the Sørensen similarity 31.64% of the variance in the dataset was explained by the first two coordinates. Coordinate 1 mainly separates samples from pre-feeding from the remaining sampling points, being more noticeable in 22 and 26°C-samples. Coordinate 2 separates samples from 18°C-fish from 22 and 26°C-fish (Figure 5A). For the Bray-Curtis similarity, the first two coordinates in PCoA explained 41.74% of the variance (Figure 5B). However, the separation of the samples according to treatment was not as clear as for the Sørensen similarity. This indicate that loss of ASVs with low relative potential activity was the main reason for the differences in the data.
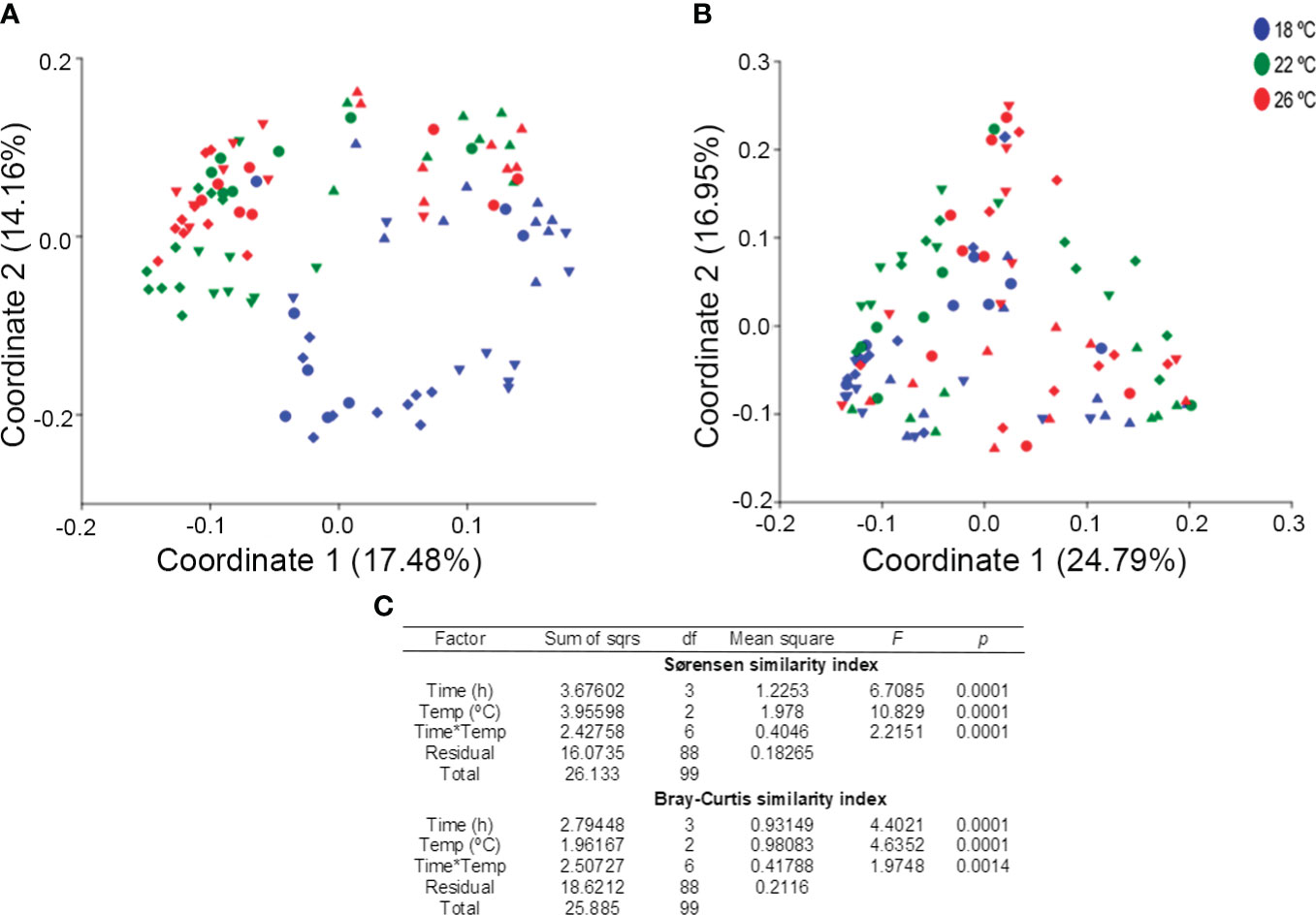
Figure 5 PCoA plots based on Sørensen (A) and Bray–Curtis (B) similarities and two-way PERMANOVA results (C) for feces of greater amberjack juveniles reared at 18 (blue), 22 (green) and 26°C (red) and sampled at ▲ pre-feeding time, ▼ 6h post-feeding, ♦ 12h post-feeding and, ● 18h post-feeding.
When the samples are separated according to water temperature, the composition of ASVs based on Sørensen similarity was different between all sampling points for fish reared at 18 and 22°C (Figure 6A). By contrast at 26°C, only the potentially active bacterial community at pre-feeding time was statistically different from the remaining sampling times (one way-PERMANOVA p-value< 0.0001 for all water temperatures; Figure 6A). By contrast, when filtered by sampling time as factor (Figure 6B), the composition of ASVs based on 16S rRNA from the different temperatures were different at all sampling points. However, feces potentially active microbiota was more similar between fish from 22 and 26°C than fish at 18°C (one way-PERMANOVA p-value<0.0002 for all sampling points).
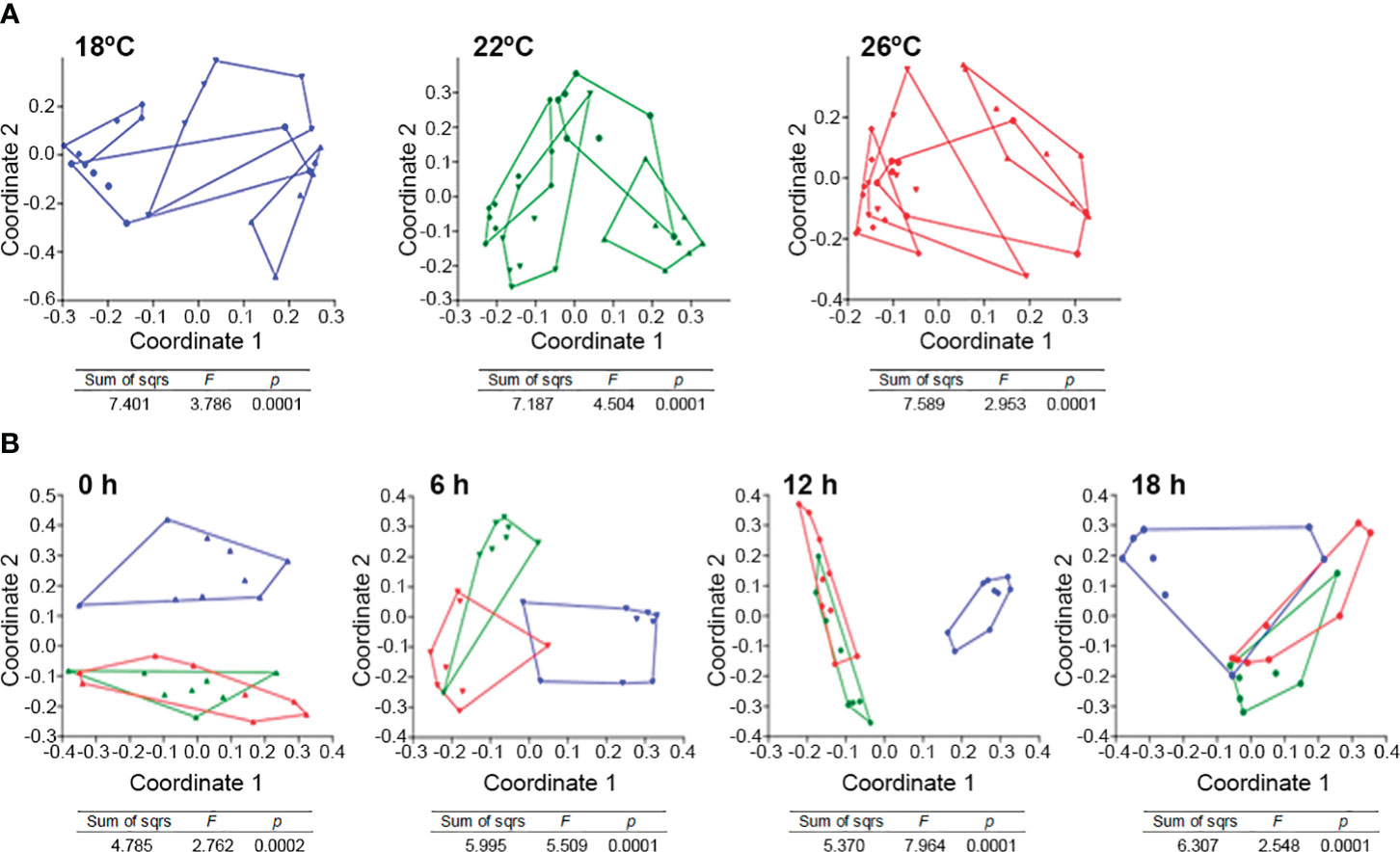
Figure 6 Principal Coordinates Analysis (PCoA) based on Sørensen similarities for greater amberjack feces reared at different water temperatures (18 °C blue, 22 °C green, 26° C red), and sampled at different experimental times (▲ pre-feeding time, ▼ 6h post-feeding, ♦ 12h post-feeding and, ● 18h post-feeding) filtered by (A) sampling time as factor and (B) water temperature as factor. One-way PERMANOVA results for each graph are included. Lines represent convex hulls for the points representing the feces microbiome for each variable.
3.4 Taxonomic assessment of potentially active bacterial community in the RAS water
Water bacterial community potential activity in all treatments was dominated by the phyla Proteobacteria and Bacteroidetes (Table 3). The relative potential activity of the 18°C-water bacterial community at the phylum level was stable along the day cycle, with no variation in the potential activity of the most abundant phyla. By contrast, in 22°C-water the relative potential activity of Proteobacteria and Planctomycetes changed during the day, reaching their maximum at 12 and 6h after first meal, respectively. In 26°C-water, all phyla tended to increase potential activity along the sampling period except for Proteobacteria, which decreased.
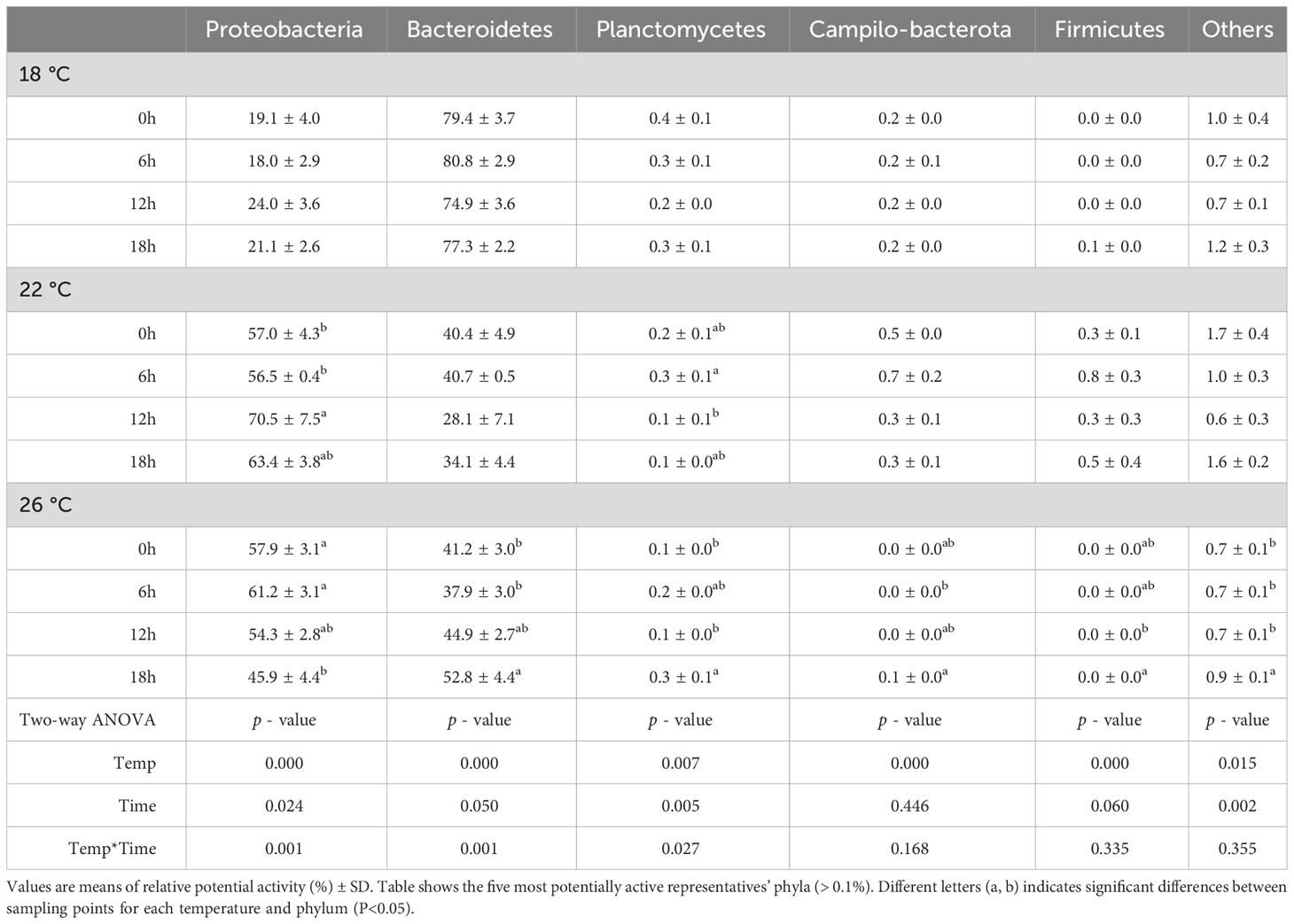
Table 3 Relative potential activity of the ASV at phylum level (expressed as percentage of the total number of reads) in water sampled at 18, 22 and 26 °C and samples at 0, 6, 12 and 18 hours post-first feeding.
Overall, the most potentially active phylum for 22 and 26°C water was Proteobacteria, with an average of 63 and 55%, respectively. The most potentially active phylum for 18°C was Bacteroidetes, accounting for 78% of the community potential activity (Table 3). Ordination by Principal Coordinate Analysis (PCoA) based on Bray-Curtis similarities explained 86% of the variance (Figure 7). 71.9% of the variance was explained by coordinate 1, while 14.1% was explained by coordinate 2. PCoA indicated that coordinate 1 clustered the potentially active bacterial structure according to water temperature, with more similarity in active water bacteria at 22 and 26°C-waters than at 18°C. In fact, a two-way PERMANOVA test revealed a significant effect of water temperature on the ordination of the samples (p = 0.001), while neither sampling time nor the interaction between both factors had significant effect (p = 0.139; 0.0546).
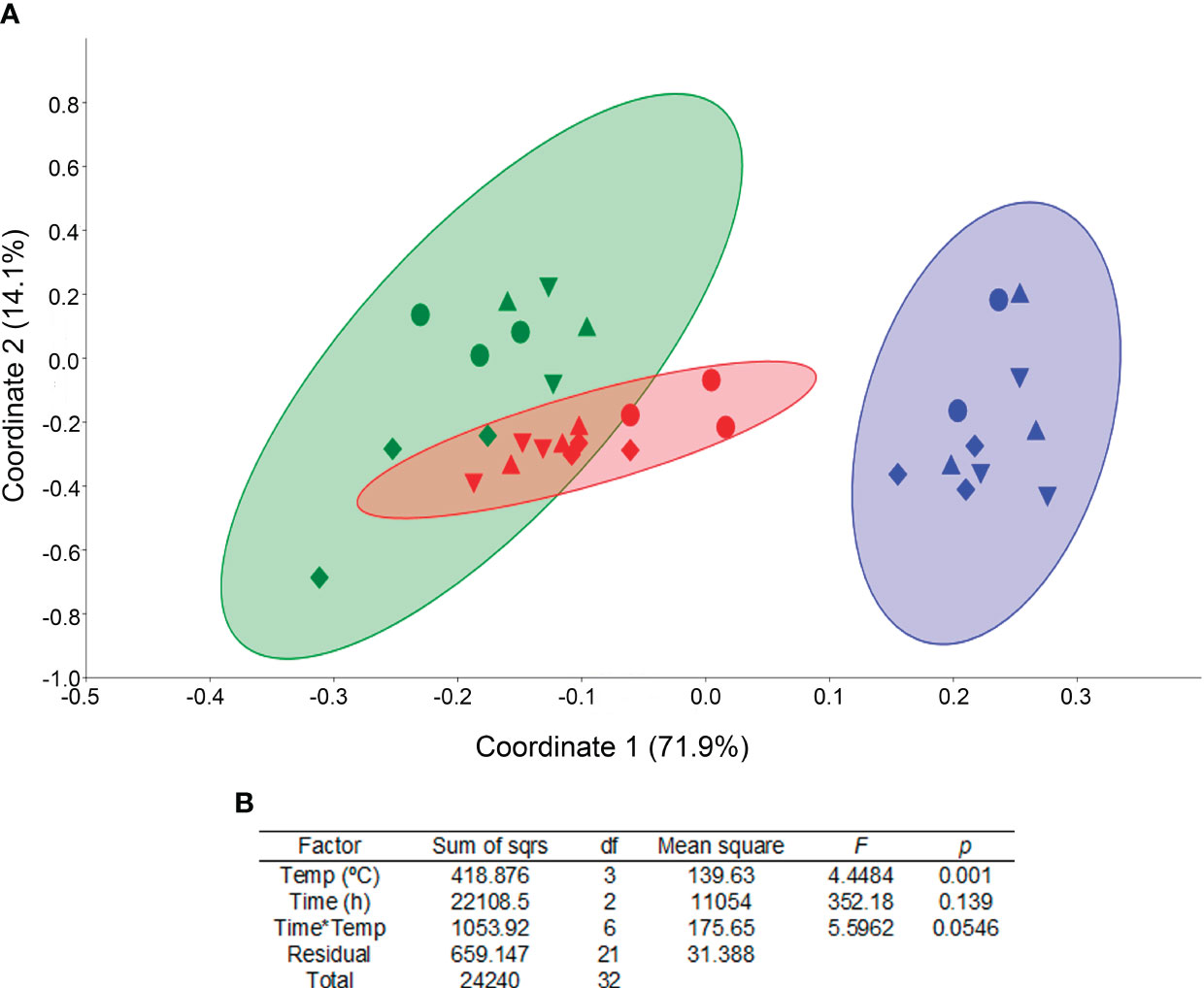
Figure 7 PCoA of composition of potentially active water microbiota community at phylum level; 18 °C blue, 22 °C green, 26 °C red and sampled at different experimental times (▲ pre-feeding time, ▼ 6h post-feeding, ♦ 12h post-feeding and, ● 18h post-feeding) (A) and two-way PERMANOVA results (B). 95% confidence ellipses are represented.
Composition of the potentially active bacteria community at order level is shown in Figure 8. The most potentially active bacterial order in rearing water at 18 and 26°C was Flavobacteriales (61.7 and 43.1%, respectively), while for 22°C water it was Alteromonadales (48.4%). A two-way ANOVA test confirmed that the potentially active water microbiota at the order level was significantly different between temperatures, sampling times and the interaction between both factors (p = 0.0001, 0.0166 and 0.0006, respectively). For 18°C-water, bacteria activity at the order level was stable during the sampling period, with significant variations only for the order Rhodobacterales. Its maximum relative potential activity was at 12h after the first feed supply. The minimum was registered at 6h after first meal. For 22°C-water, only the orders Vibrionales and Alteromonadales showed fluctuations during the day, while Vibrionales potential activity decreased significantly after first feed supply (reaching the minimum relative potential activity at 6h post first meal) Alteromonadales increased, reaching its maximum at 12h after first feed supply. By contrast, for 26°C water, the relative activity of almost all the most potentially active orders was affected by feed supply. Feed supply promoted a decrease of both orders, Vibrionales and Rhodospirillales, reaching their minimum 6h after first meal. On the order hand, Flavobacteriales, Alteromonadales, Cellvibrionales and Rhodobacteriales relative activities were increased. Alteromonadales was the most responsive order, reaching its maximum relative potential activity 6h after the first feed supply.
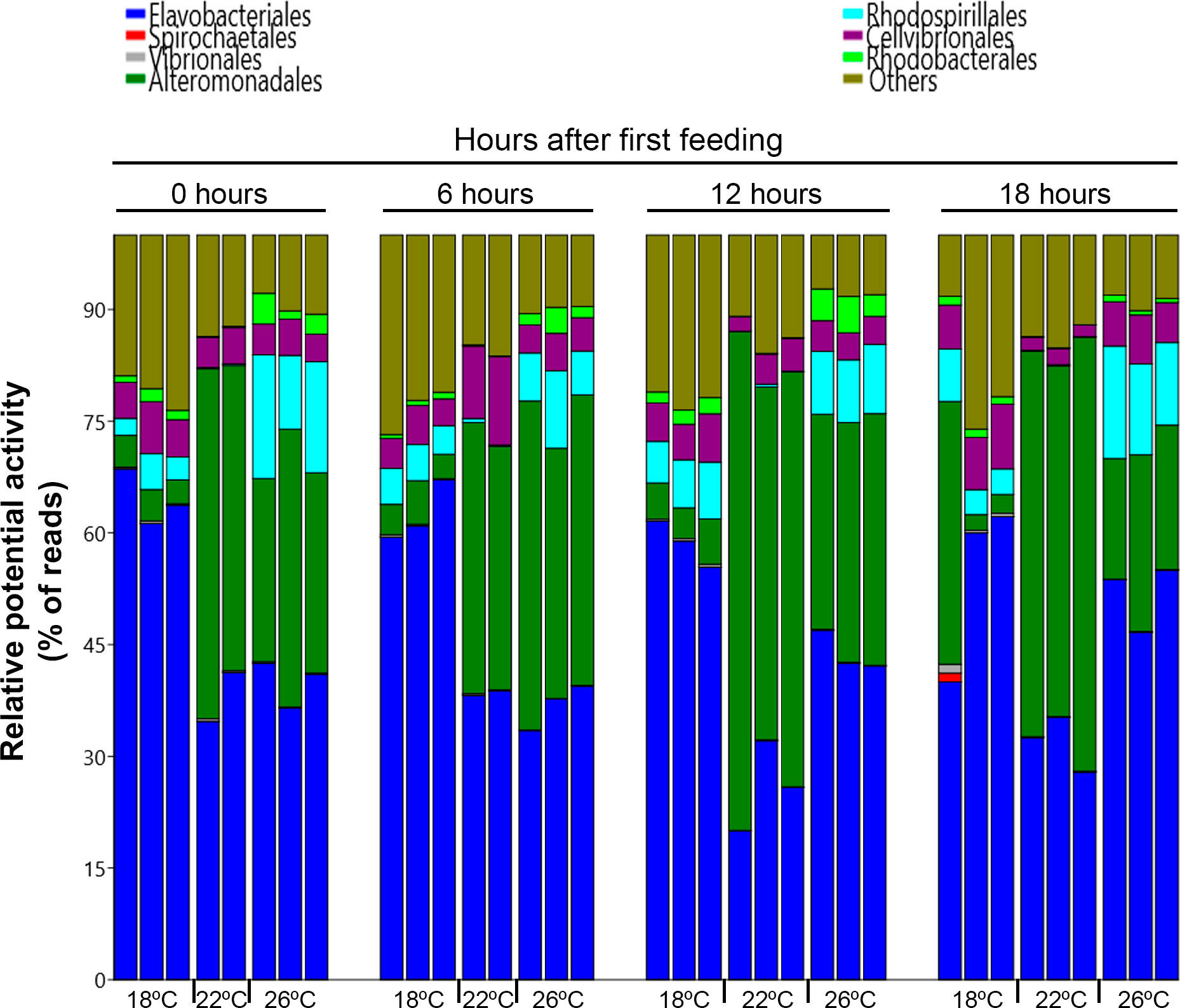
Figure 8 Structure of the potentially active bacterial community at order level (expressed as percentage of total reads) in water samples from the different treatments (18, 22 and 26 °C water temperature) at the different sampling times. The plots represent the most potentially active representatives’ orders (> 1%).
4 Discussion
To be able to identify the mechanisms through which gut microbial communities affect host health and fitness it is crucial to understand gut microbial ecosystem dynamics. To date, there is lack of information on diel fluctuations in gut microbiota potential metabolic activity. The present study is the first assessing the quantitative and qualitative effects of water temperature and daily feeding rhythm on the gut microbiota in fish, i.e. S. dumerili juveniles.
In fish, as poikilothermic animals, water temperature has a significant influence on voluntary feed intake and fish growth. Both variables increase with the increase in temperature up to an optimum, and decrease with higher temperatures (Björnsson et al., 2007; Kounna et al., 2021). In our study, and concordantly with a previous of this species (Fernández-Montero et al., 2018), fish ingested almost 2-fold more feed and grew 63% more at 26 °C than at 18°C after two weeks. In spite of this, the body weights at the three temperatures presented notable populations overlap. Therefore, we assumed that differences in body weight did not affect the observed results in the microbiota dynamics.
The 16S rDNA qPCR from 16S r RNA genes can be used for quantification of bacterial density (Wang et al., 2021). In addition, if the same procedure is performed with cDNA, obtained by reverse transcription of 16S rRNA, it can be interpreted as a proxy for activity (protein synthesis capacity) of the bacterial community (Blazewicz et al., 2013). Thus, by multiplying qPCR-based abundance or potential activity from 16S rRNA genes by relative occurrence based on amplicon sequencing, it is possible to get an estimation of absolute abundance or potential activity for each taxon, respectively.
In the present study, water temperature did not have any clear effect on bacterial abundance and activity in feces. Moreover, results from 16S rDNA qPCR revealed no statistical significant effect of sampling time on bacterial abundance per mg of gut content along the day. However, there was a tendency for higher bacterial densities early in the morning before the first meal, especially in fish reared at 22 and 26°C. These results agree with previous studies in mammalians, reporting increase in gut microbial density when individuals become active. This was observed at dawn in meerkats (Suricata suricatta) (Risely et al., 2021), and at dusk for mice (Thaiss et al., 2016; Wu et al., 2018). By contrast, we observed an increase in bacteria activity 6 h after the first meal, providing evidence that activity is driven by the arrival of the ingested feed into the intestine (Navarro-Guillén et al., 2023). In animals, physiological processes are controlled by the central and peripherals molecular clock machineries that are synchronized by internal and external signals. This is particularly relevant for farmed animals, whose wellness may rely on this metabolic coordination between endogenous rhythms and external clues imposed by production practices. Feeding time has been described as the dominant driver in the temporal orchestration of intestinal microbiome activity over the course of a day, but a functional circadian clock in the host driven by the light-dark alternation is also necessary to maintain the rhythmic variation of the microbiota (Thaiss et al., 2014; Thaiss et al., 2015). In the cited study, these fluctuations during the feeding-active phase of the host resulted in specific functional profiles of the microbiome such as energy harvest, DNA repair, and cell growth. On the other hand, detoxification and chemotaxis were more abundant during the resting phase of the host (Thaiss et al., 2014; Thaiss et al., 2015). In fish, the orchestration of the general functions of an organism is also linked by the light-dark daily cycle (Yúfera et al., 2017; Eilertsen et al., 2022). However, the digestive-intestinal molecular clock machinery is also depending on the feeding schedule (Vera et al., 2013; Gilannejad et al., 2021). Thus, a similar interaction between the feeding phases and the intestinal microbiota fluctuations to that found in mammalians is expected in the present study, although it would be necessary metatranscriptomics to be conclusive.
Temporal dynamics in the composition of water column bacteria in RAS systems have been described in some studies (Rojas-Tirado et al., 2019; Lorgen-Ritchie et al., 2021; Schoina et al., 2022). However, to the best of our knowledge, our study is the first to assess water bacterial dynamics on a daily basis. Both the highest bacterial density and activity were registered in 26°C-water 12h after first feed supply. This probably coincided with a peak in fish defecation. Gut transit time of fish, that is the time it takes the chyme to travel through the gastrointestinal tract, is faster at higher temperatures (Miegel et al., 2010; Yúfera et al., 2019; Navarro-Guillén et al., 2023). In greater amberjack juveniles reared with the same feeding protocol and temperatures as in the present study, at 26°C feed transited so fast through the gastrointestinal tract that the first and second meals were completely defecated 12 h after first feeding and the third meal reached the posterior intestine at this time. Thus, results suggest that bacterial load to the water column from fish feces is faster at the highest temperature (Navarro-Guillén et al., 2023). The trend to a decrease in bacterial abundance and activity at the end of the sampling period (18 h after first feeding) might be representative of a double pressure on the bacteria. The fish are digesting them and lack of feed load is starving them.
Feed is the main driving forces of the bacterial composition within the gut, and the microbes best able to utilize dietary nutrients have a competitive advantage (Tajima et al., 2001). Thus, the feeding schedule may influence microbiome composition and diversity, resulting in a strong link between feeding behavior and the microbiome. Our results demonstrate that gut potentially active bacterial communities exhibit 24-hour oscillations, as reported in mammalians (Thaiss et al., 2014), and provide evidence that these cyclical fluctuations are explained by feeding. However, the temporal differences in the patterns between temperatures (mainly between 18 and 22 and 26°C-fish) suggest an equally important role of the gut transit time. At the phylum level there was a shift from a Spirochaetes-dominated community in the morning to Proteobacteria dominance after feeding. At a lower taxonomic level, order, the potential activity of the bacterial community was dominated by Mycoplasmoidales, whose relative activity was maintained throughout the day in all treatments. The same was observed for the order Spirochaetales in 22 and 26°C-fish, while in 18°C-fish its relative potential activity decreased after feeding. A Spirochaetes-dominated bacterial community during fasting has been described also for Angus steers, suggesting that this order may increase in abundance when the dietary selection pressure on the microbial population is removed completely, acting in this situation as opportunistic pathogens (Welch et al., 2021). These results are in line with Navarro-Guillén et al. (2023), who described that at the end of the day cycle the gastrointestinal tract of greater amberjack was empty at the three rearing temperatures.
Assessing community changes entails interest in identifying taxa that are different in abundance or potential activity between treatments or sampling points. For that objective, it is important to distinguish the differences in the results and/or conclusions that can be obtained from relative or absolute bacteria data. Although relative abundance measurement has been widely applied to study community shifts for many biological questions, the absolute bacterial density is more suitable for analyzing bacterial dynamics in a community. The same is the case for potentially active bacteria-related (rRNA based) studies. Analysis of absolute bacterial activity provides a better resolution of the metabolic activity potential of gut bacterial community, because changes in the relative activity of a single taxon alter the relative activity of all taxa, and thus, it may overlook important biological findings (Wang et al., 2021). As an example, although the relative potential activity of Mycoplasmoidales was stable along the day cycle, the increase in bacterial activity (16S rRNA copies number from 16S rRNA genes) observed at 6h post-first feeding in the three treatments was mainly explained by an increase in absolute activity for this order. It is more likely that the changes in absolute activity of this individual taxa drove the proportion changes within the potentially active community. Despite this, information on daily dynamics in absolute bacterial abundance or activity is scarce for all types of animals. By using also absolute data, this study is presenting unbiased results of the diel dynamics of the potentially functional bacterial community in greater amberjack juveniles.
Mycoplasmoidales, belonging to the phylum Tenericutes, is a common order in the gut microbial community of aquatic animals. It seems to act as potential gut symbiont, and is shown to improve survival on a low quality diet in crustaceans (Clarke et al., 2019). One of this phylum’s more significant genus, Mycoplasma, is abundant in the gut of healthy Atlantic salmon (Salmo salar) (Bozzi et al., 2021). These bacteria are detected more frequently in carnivorous fish species than in fish with other feeding habits (Sellyei et al., 2021; Cheaib et al., 2021; Dias et al., 2021; García-Márquez et al., 2022). They are protease- and amylase-producing bacteria, thus, with the capacity to participate in protein and carbohydrate metabolism (Staats et al., 2007; Ray et al., 2012). Therefore, the increase in Mycoplasmoidales absolute potential activity in the present study suggests a potential role in nutrient provisioning, supporting the host in digestion and metabolization of food, and suggest a close relationship between the bacterial community and the digestion. A study on humans described 24-hour oscillatory patterns in the absolute abundance of some bacteria, and revealed that this diel pattern was linked to host susceptibility to obesity and glucose intolerance (Thaiss et al., 2014; Korem et al., 2015).
As mentioned above, gut transit time appears to be a major driver of gut microbiota dynamics, and it seems to influence gut microbial diversity, composition and metabolism (Procházková et al., 2023). However, studies investigating the gut microbiome in relation to transit time are limited. Water temperature is probably one of the key factors inducing changes in the gut transit time, but also in gut environmental conditions, such as luminal pH, which is also linked to gut microbial composition and activity. Navarro-Guillén et al. (2023) described in greater amberjack juveniles that each meal progresses in a different manner through the gastrointestinal tract, presenting each one of the three daily meals different gut transit and residence times. This affects to some degree two factors, the daily pattern of environmental pH within the gut and the transit of the chyme throughout the digestive tract. The food transit time decreased with increasing temperature from 18 to 26°C. Likewise, water temperature also affected the luminal pH. In fish reared at 22°C the alkaline pH in the gut was observed earlier and was maintained for a longer time than at 18 and 26°C (Navarro-Guillén et al., 2023). Microbial species richness has been positively correlated with transit time in the posterior intestine in humans (Procházková et al., 2023), which is in accordance with the higher richness found in 18°C-fish in the present study. Analysis of beta diversity based on PCoA and Sørensen similarity, revealed that 18°C-fish clustered distinctly from 22 and 26°C-fish, indicating that differences are partly driven by potentially active/inactive infrequent OTUs. In addition to the effect of water temperature on the bacterial community, it was also a clear effect of sampling time when using Sørensen similarity (Figure 8). The largest differences were observed between 18°C-fish and the other two groups at 0 and 12 h post-first feeding. These differences at the beginning of the day might be explained by the fact that the total transit time is slower at 18°C, so the intestine is empty for a shorter time. At 22 and 26°C the ingesta was almost completely evacuated at 20h after first feeding, while at 18°C it was evacuated at 24 h after first feeding (Navarro-Guillén et al., 2023). Therefore, at 18°C the fasting pressure on the gut microbiome is shorter. On the other hand, at 12 h post-first feeding, the load of ingesta into the posterior intestine is lower in 18°C-fish, due to the fact that the third meal had not yet reached this part of the intestine (Navarro-Guillén et al., 2023). Thus, results from the present work confirm the close relation between gut transit time and bacterial dynamics. Including gut transit time as a variable in gut microbiome-related studies might help to advance the knowledge of the links between gut microbiome, dietary composition of the feed and fish health.
The aquatic environment in RAS is a complex system, modulated by biotic and abiotic variables such as surface-associated and suspended bacteria, feed composition and digestibility, and feed loading (Rojas-Tirado et al., 2018). The present study indicates that water temperature also influences the potential activity of the water bacterial community, which is closely related to the fish gut bacterial community. The overall taxonomic analyses showed that Proteobacteria and Bacteroidetes were the most potentially active bacteria phyla in water samples, in accordance with previous studies in seawater RAS systems (Duarte et al., 2019). However, whereas the community activity was dominated by Bacteroidetes at the lowest temperature, Proteobacteria was the dominant phylum at 22 and 26°C. This is in agreement with fish gut potentially active bacterial community composition at the phylum level and with the assumption that the strains from the rearing water play a significant role in the assembly of the fish gut microbiome. The analysis of beta-diversity (Bray–Curtis similarity) of the potentially active bacterial communities observed in water samples revealed a clear clustering pattern driven by two main bacterial orders, Flavobacteriales and Alteromonadales. Gómez-Pereira et al. (2010) described that Flavobacteriales abundance in seawater is negatively correlated with temperature, indicating a preference for colder water. In addition, Flavobacteriales potential activity was also statistically higher in the feces of fish reared at 18°C compared to the other two treatments. On the other hand, the Alteromonadales order has an important environmental role in the uptake of nitrate in marine environments (Wawrik et al., 2012; Duarte et al., 2019). Thus, members of this order might be more metabolically active in 22 and 26°C-RAS water due to higher fish feed intake and feed loading at these temperatures, and therefore higher nutrient inputs due to fish excretion. The potential activity of Alteromonadales was also higher in feces of fish reared at the two higher temperatures. Results reveal a high degree of shared bacterial between water and fish, since the rearing water microbiota in the RAS systems is one of the primary sources of microbiota in the fish, but the fish microbiota will also settle in the water (Zhang et al., 2022).
In conclusion, this is the first study assessing the quantitative and qualitative effects of water temperature and daily feeding rhythm on the gut microbiota dynamics in a poikilothermic animal. Results demonstrated that the potentially active bacterial community in the fish gut exhibit daily oscillations. Feeding was the main driver modulating bacterial activity, promoting a peak in bacterial potential activity 6 h after first feeding, and suggesting resource limitation of the gut microbiota during non-fed periods. When analyzing relative potential activity, it differently varied with postprandial time. By contrast, when assessing absolute activity, there was only a peak in potential activity at 6 hours after first feeding, mainly due to an increase in Mycoplasmoidales absolute activity. At the phylum level it was observed a shift from a Spirochaetes-dominated community in the morning to an increased potential activity of Proteobacteria after feeding. Also water temperature modulated gut microbiota dynamics, with a marked effect on the dynamics along the timeline. At the lowest water temperature, the patterns of bacterial activity oscillation were slowed down, probably due to a slower, temperature limited digestive process and bacterial metabolic rates. Results from this study provide a starting point for developing insights into the mechanisms synchronizing daily dynamics in the structure of the potentially active bacterial community and its impact on host biological function.
Data availability statement
The data presented in the study are deposited in the NCBI repository, accession number PRJNA1001609.
Ethics statement
The animal study was approved by Spanish National Research Council (CSIC) bioethical committee and Spanish National Veterinary Authority (REF: 02/07/2019/107). The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
CN-G contributed to the conception and design of the work, acquisition, analysis and interpretation of the data and was a major contributor in writing the manuscript. EDL contributed to the acquisition and analysis of the data. EP contributed to the conception and design of the work and to the interpretation of the data. MY contributed to the funding acquisition, conception and design of the work and interpretation of the data. OV contributed to the conception and design of the work, acquisition, analysis and interpretation of the data and was a major contributor in reviewing the manuscript. All authors read and approved the final manuscript.
Funding
This research was funded by the project RTI2018-096134-B-I00 from the Spanish Ministry of Science, Innovation and Universities (MCIU), the State Research Agency (AEI) and the European Regional Development Fund (FEDER/ERDF) granted to MY. CN-G was supported by the researcher contract DOC_01203 funded by the Andalusian Plan for Research Development and Innovation (PAIDI 2020).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Aguilera E., Yany G., Romero J. (2013). Cultivable intestinal microbiota of yellowtail juveniles (Seriola lalandi) in an aquaculture system. Lat. Am. J. Aquat. Res. 41, 395–403. doi: 10.3856/vol41-issue3-fulltext-3
Anderson M. J. (2001). A new method for non-parametric multivariate analysis of variance. Aust. J. Ecol. 26, 32–46. doi: 10.1111/j.1442-9993.2001.01070.pp.x
Barlow J. T., Bogatyrev S. R., Ismagilov R. F. (2020). A quantitative sequencing framework for absolute abundance measurements of mucosal and lumenal microbial communities. Nat. Commun. 11, 2590. doi: 10.1038/s41467-020-16224-6
Björnsson B., Steinarsson A., Árnason T. (2007). Growth model for Atlantic cod (Gadus morhua): effects of temperature and body weight on growth rate. Aquaculture 271, 216–226. doi: 10.1016/j.aquaculture.2007.06.026
Blazewicz S., Barnard R., Daly R., Firestone M. K. (2013). Evaluating rRNA as an indicator of microbial activity in environmental communities: limitations and uses. ISME J. 7, 2061–2068. doi: 10.1038/ismej.2013.102
Bozzi D., Rasmussen J. A., Carøe C., Sveier H., Nordøy K., Gilbert M. T. P., et al. (2021). Salmon gut microbiota correlates with disease infection status: potential for monitoring health in farmed animals. Anim. Microbiome 3, 30. doi: 10.1186/s42523-021-00096-2
Bray J. R., Curtis J. T. (1957). An ordination of the upland forest Ccmmunities of Wisconsin. Ecol. Monogr. 27, 326–349. doi: 10.2307/1942268
Cheaib B., Yang P., Kazlauskaite R., Lindsay E., Heys C., Dwyer T., et al. (2021). Genome erosion and evidence for an intracellular niche – exploring the biology of mycoplasmas in Atlantic salmon. Aquaculture 541, 736772. doi: 10.1016/j.aquaculture.2021.736772
Clarke L. J., Suter L., King R., Bissett A., Deagle B. E. (2019). Antarctic krill are reservoirs for distinct southern ocean microbial communities. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.03226
Das S. K., Noor N. M., Kai K. S., Juan Q. Z., Iskandar N. S., De M. (2018). Effects of temperature on the growth, gastric emptying time, and oxygen consumption rate of mahseer (Tor tambroides) under laboratory conditions. Aquac. Rep. 12, 20–24. doi: 10.1016/j.aqrep.2018.08.004
Dias D., Dardengo G. M., Engrola S., Navarro-Guillén C. (2021). Characterization and comparison of the digestive physiology of two scombrids, Katsuwonus pelamis and Sarda sarda, in the Gulf of Cádiz. PloS One 16, e0249541. doi: 10.1371/journal.pone.0249541
Duarte L. N., Coelho F. J. R. C., Oliveira V., Cleary D. F. R., Martins P., Gomes N. C. M. (2019). Characterization of bacterioplankton communities from a hatchery recirculating aquaculture system (RAS) for juvenile sole (Solea Senegalensis) production. PloS One 14, e0211209. doi: 10.1371/journal.pone.0211209
Edgar R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. doi: 10.1093/bioinformatics/btq461
Edgar R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Edgar R. C. (2016a). UNOISE2: improved error-correction for Illumina 16S and ITS amplicon sequencing. bioRxiv, 081257. doi: 10.1101/081257
Edgar R. C. (2016b). SINTAX: a simple non-Bayesian taxonomy classifier for 16S and ITS sequences. bioRxiv, 074161. doi: 10.1101/074161
Eilertsen M., Dolan D. W. P., Bolton C. M., Karlsen R., Davies W. I. L., Edvardsen R. B., et al. (2022). Photoreception and transcriptomic response to light during early development of a teleost with a life cycle tightly controlled by seasonal changes in photoperiod. PloS Genet. 18, e1010529. doi: 10.1371/journal.pgen.1010529
Fernández-Montero A., Caballero M. J., Torrecillas S., Tuset V. M., Lombarte A., Ruiz Ginés R., et al. (2018). Effect of temperature on growth performance of greater amberjack (Seriola dumerili Risso 1810) juveniles. Aquac. Res. 49, 908–918. doi: 10.1111/are.13537
Fossmark R. O., Attramadal K. J. K., Nordøy K., Østerhus S. W., Vadstein O. (2021). A comparison of two seawater adaptation strategies for Atlantic salmon post-smolt (Salmo salar) grown in recirculating aquaculture systems (RAS): nitrification, water and gut microbiota, and performance of fish. Aquaculture 532, 735973. doi: 10.1016/j.aquaculture.2020.735973
Gamberoni P., Yúfera M., de las Heras V., Siguero I., Gilannejad N., Martínez-Rodríguez G., et al. (2021). Ontogeny and diurnal patterns of molecular gene expression and activity of digestive enzymes in developing greater amberjack. Aquaculture 534, 736330. doi: 10.1016/j.aquaculture.2020.736330
García-Márquez J., Cerezo I. M., Figueroa F. L., Abdala-Díaz R. T., Arijo S. (2022). First evaluation of associated gut microbiota in wild thick-lipped grey mullets (Chelon labrosus, Risso 1827). Fishes 7, 209. doi: 10.3390/fishes7040209
Gilannejad N., Moyano F. J., Martínez-Rodríguez G., Yúfera M. (2021). The digestive function of gilthead seabream juveniles in relation to feeding frequency. Aquaculture 531, 735867. doi: 10.1016/j.aquaculture.2020.735867
Gómez de la Torre Canny S., Nordgård C. T., Mathisen A. J. H., Degré Lorentsen E., Vadstein O., Bakke I. (2023). A novel gnotobiotic experimental system for Atlantic salmon (Salmo salar L.) reveals a microbial influence on mucosal barrier function and adipose tissue accumulation during the yolk sac stage. Front. Cell. Infect. Microbiol. 12. doi: 10.3389/fcimb.2022.1068302
Gómez-Pereira P., Fuchs B., Alonso C., Oliver M. J., van Beusekom J. E. E., Amann R. (2010). Distinct flavobacterial communities in contrasting water masses of the North Atlantic Ocean. ISME J. 4, 472–487. doi: 10.1038/ismej.2009.142
Guerreiro I., Enes P., Rodiles A., Merrifield D., Oliva-Teles A. (2016). Effects of rearing temperature and dietary short-chain fructooligosaccharides supplementation on allochthonous gut microbiota, digestive enzymes activities and intestine health of turbot (Scophthalmus maximus L.) juveniles. Aquacult. Nutr. 22, 631–642. doi: 10.1111/anu.12277
Hammer Ø., Harper D., Ryan P. (2001). Past: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 4, 1–9. Available at: https://palaeo-electronica.org/2001_1/past/past.pdf.
Korem T., Zeevi D., Suez J., Weinberger A., Avnit-Sagi T., Pompan-Lotan M., et al. (2015). Growth dynamics of gut microbiota in health and disease inferred from single metagenomic samples. Science 349, 1101 —1106. doi: 10.1126/science.aac4812
Kounna C., Fountoulaki E., Miliou H., Chatzifotis S. (2021). Water temperature effects on growth performance, proximate body and tissue composition, morphometric characteristics and gastrointestinal evacuation processes of juvenile meagre, Argyrosomus regius (Asso 1801). Aquaculture 540, 736683. doi: 10.1016/j.aquaculture.2021.736683
Lorgen-Ritchie M., Clarkson M., Chalmers L., Taylor J. F., Migaud H., Martin S. A. M. (2021). A temporally dynamic gut microbiome in Atlantic salmon during freshwater recirculating aquaculture system (RAS) production and post-seawater transfer. Front. Mar. Sci. 8. doi: 10.3389/fmars.2021.711797
Mazumder S. K., Ghaffar M. A., Das S. K. (2020). Effect of temperature and diet on gastrointestinal evacuation of juvenile malabar blood snapper (Lutjanus malabaricus Bloch & Schneider 1801). Aquaculture 522, 735114. doi: 10.1016/j.aquaculture.2020.735114
Mekuchi M., Asakura T., Sakata K., Yamaguchi T., Teruya K., Kikuchi J. (2018). Intestinal microbiota composition is altered according to nutritional biorhythms in the leopard coral grouper (Plectropomus leopardus). PloS One 13, e0197256. doi: 10.1371/journal.pone.0197256
Miegel R. P., Pain S. J., Wettere W. H. E. J., Howarth G. S., Stone D. A. J. (2010). Effect of water temperature on gut transit time, digestive enzyme activity and nutrient digestibility in yellowtail kingfish (Seriola lalandi). Aquaculture 308, 145–151. doi: 10.1016/j.aquaculture.2010.07.036
Moroni F., Naya-Català F., Piazzon M. C., Rimoldi S., Calduch-Giner J., Giardini A., et al. (2021). The effects of nisin-producing Lactococcus lactis strain used as probiotic on gilthead sea bream (Sparus aurata) growth, gut microbiota, and transcriptional response. Front. Mar. Sci. 8. doi: 10.3389/fmars.2021.659519
Navarro-Guillén C., Conceição L. E. C., Pinto W., Siguero I., Urrutia P., Moyano F. J., et al. (2019). Fast growing greater amberjack post-larvae require a high energy-high protein weaning diet. Aquaculture 499, 195–202. doi: 10.1016/j.aquaculture.2018.09.037
Navarro-Guillén C., do Vale Pereira G., Lopes A., Colen R., Engrola S. (2021). Egg nutritional modulation with amino acids improved performance in zebrafish larvae. PloS One 16, e0248356. doi: 10.1371/journal.pone.0248356
Navarro-Guillén C., Gilannejad N., Pérez-Hilario D., Martínez-Rodríguez G., Yúfera M. (2023). Gut transit of daily consecutive meals in greater amberjack juveniles reared at different temperatures. Aquaculture 567, 739244. doi: 10.1016/j.aquaculture.2023.739244
Navarro-Guillén C., Yúfera M., Perera E. (2022). Biochemical features and modulation of digestive enzymes by environmental temperature in the greater amberjack, Seriola dumerili. Front. Mar. Sci. 9. doi: 10.3389/fmars.2022.960746
Naya-Català F., Piazzon M. C., Torrecillas S., Toxqui-Rodríguez S., Calduch-Giner J., Fontanillas R., et al. (2022). Genetics and nutrition drive the gut microbiota succession and host-transcriptome interactions through the gilthead sea bream (Sparus aurata) production cycle. Biol. (Basel) 11, 1744. doi: 10.3390/biology11121744
Oliphant K., Allen-Vercoe E. (2019). Macronutrient metabolism by the human gut microbiome: major fermentation by-products and their impact on host health. Microbiome 7, 91. doi: 10.1186/s40168-019-0704-8
Pérez J. A., Papadakis I. E., Papandroulakis N., Cruces L., Cotou E., Gisbert E., et al. (2020). The ontogeny of greater amberjack digestive and antioxidant defence systems under different rearing conditions: A histological and enzymatic approach. Aquac. Nutr. 26, 1908–1925. doi: 10.1111/anu.13128
Procházková N., Falony G., Dragsted L. O., Licht T. R., Raes J., Roager H. M. (2023). Advancing human gut microbiota research by considering gut transit time. Gut 2, 180–191. doi: 10.1136/gutjnl-2022-328166
Ray A., Ghosh K., Ringø E. (2012). Enzyme-producing bacteria isolated from fish gut: A review. Aquac. Nutr. 18, 465–492. doi: 10.1111/j.1365-2095.2012.00943.x
Risely A., Wilhelm K., Clutton-Brock T., Manser M. B., Sommer S. (2021). Diurnal oscillations in gut bacterial load and composition eclipse seasonal and lifetime dynamics in wild meerkats. Nat. Commun. 12, 6017. doi: 10.1038/s41467-021-26298-5
Rojas-Tirado P., Pedersen P. B., Vadstein O., Pedersen L. F. (2018). Changes in microbial water quality in RAS following altered feed loading. Aquac. Eng. 81, 80–88. doi: 10.1016/j.aquaeng.2018.03.002
Rojas-Tirado P., Pedersen P. B., Vadstein O., Pedersen L. F. (2019). Microbial dynamics in RAS water: Effects of adding acetate as a biodegradable carbon-source. Aquac. Eng. 84, 106–116. doi: 10.1016/j.aquaeng.2018.12.010
Rosselli R., Romoli O., Vitulo N., Vezzi A., Campanaro S., de Pascale F., et al. (2016). Direct 16S rRNA-seq from bacterial communities: a PCR-independent approach to simultaneously assess microbial diversity and functional activity potential of each taxon. Sci. Rep. 6, 32165. doi: 10.1038/srep32165
Salas-Leiva J., Mazón-Suástegui J. M., Teles A., Tovar-Ramírez D. (2020). Structure and predictive metabolic contribution of intestinal microbiota of longfin yellowtail (Seriola rivoliana) juveniles in aquaculture systems. Mol. Biol. Rep. 47, 9627–9636. doi: 10.1007/s11033-020-05970-x
Schlomann B. H., Parthasarathy R. (2019). Timescales of gut microbiome dynamics. Curr. Opin. Microbiol. 50, 56–63. doi: 10.1016/j.mib.2019.09.011
Schoina E., Doulgeraki A. I., Miliou H., Nychas G. J. E. (2022). Dynamics of water and biofilm bacterial community composition in a Mediterranean recirculation aquaculture system. Aquac. J. 2, 164–179. doi: 10.3390/aquacj2020008
Sellyei B., Varga Z., Cech G., Varga Á., Székely C. (2021). Mycoplasma infections in freshwater carnivorous fishes in Hungary. J. Fish Dis. 44, 297–304. doi: 10.1111/jfd.13283
Sicuro B., Luzzana U. (2016). The state of Seriola spp. other than yellowtail (S. quinqueradiata) farming in the world. Rev. Fish. Sci. Aquac. 24, 314–325. doi: 10.1080/23308249.2016.1187583
Skjermo J., Bakke I., Wiborg Dahle S., Vadstein O. (2015). Probiotic strains introduced through live feed and rearing water have low colonizing success in developing Atlantic cod larvae. Aquaculture 438, 17–23. doi: 10.1016/j.aquaculture.2014.12.027
Soriano E. L., Ramírez D. T., Araujo D. R., Gómez-Gil B., Castro L. I., Sánchez C. G. (2018). Effect of temperature and dietary lipid proportion on gut microbiota in yellowtail kingfish Seriola lalandi juveniles. Aquaculture 497, 269–277. doi: 10.1016/j.aquaculture.2018.07.065
Staats C. C., Boldo J., Broetto L., Vainstein M., Schrank A. (2007). Comparative genome analysis of proteases, oligopeptide uptake and secretion systems in Mycoplasma spp. Genet. Mol. Biol. 30, 225–229. doi: 10.1590/S1415-47572007000200009
Steiner K., Laroche O., Walker S. P., Symonds J. E. (2022). Effects of water temperature on the gut microbiome and physiology of Chinook salmon (Oncorhynchus tshawytscha) reared in a freshwater recirculating system. Aquaculture 560, 738529. doi: 10.1016/j.aquaculture.2022.738529
Tajima K., Aminov R. I., Nagamine T., Matsui H., Nakamura M., Benno Y. (2001). Diet-dependent shifts in the bacterial population of the rumen revealed with real-time PCR. Appl. Environ. Microbiol. 67, 2766–2774. doi: 10.1128/AEM.67.6.2766-2774.2001
Thaiss C. A., Levy M., Korem T., Dohnalová L., Shapiro H., Jaitin D. A., et al. (2016). Microbiota diurnal rhythmicity programs host transcriptome oscillations. Cell 167, 1495–1510. doi: 10.1016/j.cell.2016.11.003
Thaiss C. A., Zeevi D., Levy M., Segal E., Elinav E. (2015). A day in the life of the meta-organism: diurnal rhythms of the intestinal microbiome and its host. Gut Microbes 6, 137–142. doi: 10.1080/19490976.2015.1016690
Thaiss C. A., Zeevi D., Levy M., Zilberman-Schapira G., Suez J., Tengeler A. C., et al. (2014). Transkingdom control of microbiota diurnal oscillations promotes metabolic homeostasis. Cell 159, 514–529. doi: 10.1016/j.cell.2014.09.048
Vandeputte D., De Commer L., Tito R. Y., Kathagen G., Sabino J., Vermeire S., et al. (2021). Temporal variability in quantitative human gut microbiome profiles and implications for clinical research. Nat. Commun. 12, 6740. doi: 10.1038/s41467-021-27098-7
Vera L. M., Negrini P., Zagatti C., Frigato E., Sánchez-Vázquez F. J., Bertolucci C. (2013). Light and feeding entrainment of the molecular circadian clock in a marine teleost (Sparus aurata). Chronobiol. Int. 30, 649–661. doi: 10.3109/07420528.2013.775143
Wang X., Howe S., Deng F., Zhao J. (2021). Current applications of absolute bacterial quantification in microbiome studies and decision-making regarding different biological questions. Microorganisms 9, 1797. doi: 10.3390/microorganisms9091797
Wawrik B., Boling W. B., Van Nostrand J. D., Xie J., Zhou J., Bronk D. A. (2012). Assimilatory nitrate utilization by bacteria on the west Florida shelf as determined by stable isotope probing and functional microarray analysis. FEMS Microbiol. Ecol. 79, 400–411. doi: 10.1111/j.1574-6941.2011.01226.x
Welch C. B., Lourenco J. M., Seidel D. S., Krause T. R., Rothrock M. J., Pringle T. D., et al. (2021). The impact of pre-slaughter fasting on the ruminal microbial population of commercial angus steers. Microorganisms 9, 2625. doi: 10.3390/microorganisms9122625
Wolda H. (1981). Similarity Indices, sample size and diversity. Oecologia 50, 296–302. doi: 10.1007/BF00344966
Wu G., Tang W., He Y., Hu J., Gong S., He Z., et al. (2018). Light exposure influences the diurnal oscillation of gut microbiota in mice. Biochem. Biophys. Res. Commun. 501, 16–23. doi: 10.1016/j.bbrc.2018.04.095
Yúfera M., Nguyen M. V., Navarro-Guillén C., Moyano F. J., Jordal A. E. O., Espe M., et al. (2019). Effect of increased rearing temperature on digestive function in cobia early juvenile. Comp. Biochem. Physiol. A 230, 71–80. doi: 10.1016/j.cbpa.2019.01.007
Yúfera M., Perera E., Mata-Sotres J. A., Calduch-Giner J., Martínez-Rodríguez G., Pérez-Sánchez J. (2017). The circadian transcriptome of marine fish (Sparus aurata) larvae reveals highly synchronized biological processes at the whole organism level. Sci. Rep. 7, 12943. doi: 10.1038/s41598-017-13514-w
Zarrinpar A., Chaix A., Yooseph S., Panda S. (2014). Diet and feeding pattern affect the diurnal dynamics of the gut microbiome. Cell Metab. 20, 1006–1017. doi: 10.1016/j.cmet.2014.11.008
Keywords: bacteria potential activity, diel oscillations, feeding, fish physiology, gut microbiota, quantitative bacteria composition, water temperature
Citation: Navarro-Guillén C, Degré Lorentsen E, Perera E, Yúfera M and Vadstein O (2023) Feeding and water temperature modulate the potential activity of gut microbiota in greater amberjack juveniles (Seriola dumerili). Front. Mar. Sci. 10:1236305. doi: 10.3389/fmars.2023.1236305
Received: 07 June 2023; Accepted: 21 August 2023;
Published: 12 September 2023.
Edited by:
Enric Gisbert, Institute of Agrifood Research and Technology (IRTA), SpainReviewed by:
Ana Teresa Gonçalves, GreenCoLab, PortugalFrancisco Vargas-Albores, National Council of Science and Technology (CONACYT), Mexico
Alberto Ruiz, IRTA Sant Carles de la Ràpita, Spain
Copyright © 2023 Navarro-Guillén, Degré Lorentsen, Perera, Yúfera and Vadstein. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Carmen Navarro-Guillén, carmen.navarro@csic.es
 Carmen Navarro-Guillén
Carmen Navarro-Guillén Eirik Degré Lorentsen
Eirik Degré Lorentsen Erick Perera
Erick Perera Manuel Yúfera
Manuel Yúfera Olav Vadstein
Olav Vadstein