Propidium Monoazide based selective iDNA monitoring method improves eDNA monitoring for harmful algal bloom Alexandrium species
- Department of Marine Environmental Engineering, Gyeongsang National University, Tongyeong, Gyeongnam, Republic of Korea
eDNA, also known as environmental DNA, has garnered significant attention due to its potential applications in various fields such as biodiversity assessment, species distribution monitoring, ecological interaction analysis, and quantitative analysis. However, the presence of non-selective DNA signals in eDNA samples poses challenges in accurately detecting species, assessing biodiversity, and conducting quantitative analysis. To address these limitations, this study developed a novel method for selectively detecting iDNA from specific species in eDNA samples. The method involved the application of PMA treatment to Alexandrium spp. effectively preventing the detection of non-selective exDNA signals. Additionally, by optimizing the filter size used in the sampling process, the researchers were able to selectively collect and analyze iDNA from species of interest, particularly Alexandrium spp. Furthermore, the study successfully demonstrated the selective collection and analysis of iDNA from Alexandrium spp. cysts present in the sediment layer, further strengthening the findings. The results indicated that the combined use of PMA treatment and filter size optimization significantly enhanced the selective detection capability of iDNA. The successful selective detection of iDNA from eDNA in the sediment layer highlights the practical applicability of the developed method. This study holds promise for advancing eDNA monitoring technology by providing a selective iDNA detection method utilizing PMA. Moreover, these findings lay the foundation for effectively utilizing iDNA in environmental conservation, monitoring, and ecological research.
1 Introduction
Harmful algal blooms (HABs) are widespread, with severe implications for public health and aquatic ecosystems. They can significantly negatively impact fisheries, leading to reduced productivity and economic losses. Alexandrium spp. is a species known to cause HABs and is one of the organisms responsible for paralytic shellfish poisoning (PSP), a serious threat to human health when contaminated shellfish are consumed (Samsur et al., 2006). Alexandrium spp. produce saxitoxins (Whedon, 1936), which can accumulate in shellfish such as mussels, clams, and oysters and have harmful or fatal effects on humans or animals that consume them. Impacts of PSP outbreaks include poisoning and death from contaminated shellfish or fish, loss of wild and farmed seafood resources, damage to tourism and recreational activities, changes in ocean trophic structure, and deaths of marine mammals, fish, and seabirds (Anderson et al., 2012). This distribution of Alexandrium spp. has expanded to regional and global scales (Hallegraeff and Bolch, 1992).
Alexandrium spp. reproduce by both asexual and sexual reproductive cycles (Anderson, 1980), with trophozoites developing into blooms through cell division. Sexual reproduction results in the formation of swimming zygotes (planozygotes) by sexual induction, which develop into dormant spores in subsequent stages (Anderson, 1998). This sexual reproduction may contribute to maintaining genetic heterogeneity and maintaining and enhancing adaptability to the environment. Moreover, spores are more resistant to unfavorable environments compared to trophozoites and play an important role as a seed population for subsequent seasonal flowering the following year (Anderson and Wall, 1978). Therefore, it is very important to monitor the density distribution of cells and dormant spores in order to minimize PSP. However, it is difficult to identify microalgae by morphological features and dormant spores do not have species-specific morphological features that can be distinguished (Fukuyo, 1985). Furthermore, these methods require specialized expertise (Hopkins and Freckleton, 2002).
Environmental DNA (eDNA) refers to genetic material acquired from environmental samples, encompassing DNA originating from a wide range of sources including unicellular or small multicellular organisms, tissue particles (such as shed cells and feces), as well as gametes from multicellular organisms. Furthermore, eDNA also includes distinct fractions such as extracellular DNA (exDNA) and intracellular DNA (iDNA) (Pietramellara et al., 2009; Nagler et al., 2022). eDNA has emerged as a valuable tool for monitoring the presence and distribution of aquatic organisms (Lawson Handley, 2015). eDNA-based monitoring allows for qualitative detection using metabarcoding (Grzebyk et al., 2017) and quantitative analysis of single species using quantitative polymerase chain reaction (qPCR) with species-specific primers and probes (Salter et al., 2019). In comparison to conventional monitoring methods, eDNA monitoring offers advantages. Traditional monitoring relies on the ability to discern morphological characteristics, a task that can be challenging or even unfeasible for elusive organisms, such as microbes, which are difficult to identify. However, eDNA-based approaches overcome these limitations by relying solely on the organism’s DNA for identification. Additionally, while conventional monitoring involves time-consuming field sampling, sample sorting, and consulting identification guidebooks, eDNA monitoring offers a relatively easy and rapid process of water sampling and species identification. Quantifying biomass directly at the eDNA level in seawater samples is difficult (Günther et al., 2018). Presumably, this is because eDNA decays at different rates depending on environmental variables such as temperature, UV, pH, etc. (Strickler et al., 2015) and varies with organism distribution and habitat. Nevertheless, assessment of eDNA diversity and species can provide a more accurate approximation than traditional monitoring, and quantitative analysis of eDNA using qPCR is showing promise (Park et al., 2014; Valentini et al., 2016; Pont et al., 2023). While a cell is in a viable state, iDNA can remain relatively stable and contains the genetic information of that cell. On the other hand, exDNA is the DNA of an organism that has been released into the external environment. It can be derived from cellular decay, cellular effluents, feces, or dead organisms. Detection of exDNA is likely to overestimate microbial abundance by up to 55% (Carini et al., 2016) and detect organisms that have already died or moved on to other locations. If only iDNA can be selectively collected, quantitative changes in iDNA can indicate when a cell or organism is surviving or when cells are actively reproducing. Also, since iDNA is related to cell number, it can be correlated with cell density or biomass.
Propidium monoazide (PMA) is a photoreactive binding reagent (Nocker and Camper, 2006) that is impermeable to cell membranes. This means that PMA is permeable to the DNA of damaged or dead cells, but does not bind to the DNA of normally viable cells. Because of this property, PMA can be utilized to reduce false positives by binding to exDNA. PMA tends not to bind effectively to iDNA. This is because PMA is only permeable to the damaged cell membrane, whereas iDNA located inside the cell is more reliably protected. Therefore, the application of PMA to eDNA samples enables selective binding to exDNA, predominantly situated outside the cell. This approach facilitates the exclusive detection of iDNA, significantly enhancing the precision of eDNA monitoring. By detecting only DNA present within intact cells, this approach reduces potential false positives due to exDNA and enables sensitive detection of eDNA signals associated with microbial activity or the presence of biological species. In conclusion, PMA can reduce false positives in eDNA monitoring by selectively binding to exDNA as a photo-reactive DNA insertion agent. This can help minimize the influence of iDNA and obtain accurate biological information.
In this study, the applicability of PMA to cells and dormant spores of the marine harmful alga Alexandrium spp. was evaluated, and the feasibility of iDNA monitoring was confirmed by comparing live and dead cells at known concentrations. In addition, only iDNA was selectively collected from field samples by filtering seawater using pore size-specific filters, and the eDNA present in sediments was compared by the PMA-iDNA method. Thus, the PMA-iDNA monitoring method is expected to contribute to quantitative eDNA monitoring techniques by reducing the possibility of false positives. In addition, given the characteristics of iDNA, it is expected to provide a more accurate estimate of the presence or activity of living cells. The introduction of these PMA-iDNA monitoring techniques is expected to improve the reliability and usefulness of eDNA monitoring in various fields such as environmental monitoring, ecological research, and aquatic life management.
2 Materials and methods
2.1 Target species and controls
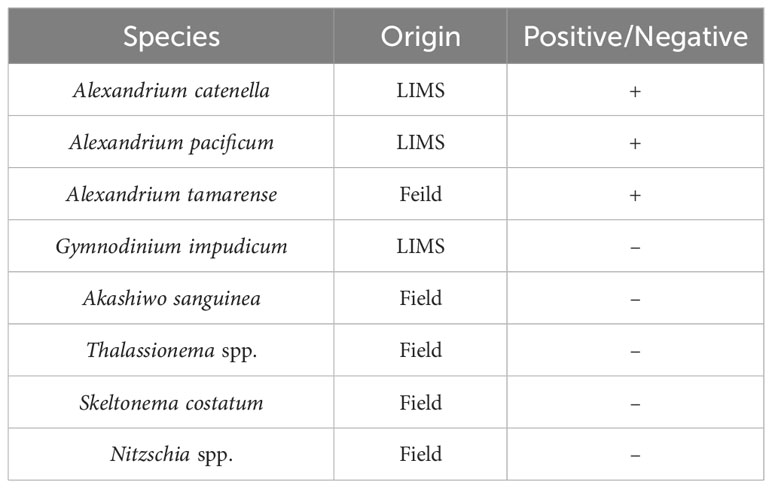
The target species Alexandrium catenella (LIMS-PS-3427), A. Pacificum (LIMS-PS-2611), and Gymnodinium impudicum, (LIMS-PS-3373) were obtained from the Library of Marine Samples (LIMS, Korea) in May 2022. In addition, Thalassionema spp., Skeletonema spp., Nitzschia spp., and Akashiwo sanguinea were collected, isolated, and cultured under a microscope. In addition, the dominant species Alexandrium tamarense was collected from Masan Bay, Korea in April 2023 and isolated and cultured. They were amplified using Euk-A (5′-AACCTGGTTGATCCTGCCAGT-3′) and Euk-B (5′-GATCCTTCTGCAGGTTCACCTAC-3′) primers (Medlin et al, 1988). Species identification was made by sequencing, and all were used as controls for false positive tests. All organisms were cultured for at least 6 weeks in f/2 medium under fluorescent lights with a 24 hr photoperiod of 3,000-10,000 Lux at a temperature of 20 ± 2°C. A. catenella cells were cultured for 4 weeks at 15°C without f/2 medium to induce cyst formation. A. catenella cells, cysts, and A. tamarense cells were diluted to concentrations ranging from 10,000-1, 6,000-1, and 50,000-5 cells/mL, respectively, in order to create a standard curve for qPCR quantification. It’s important to acknowledge that variations in cell size can lead to varying amounts of DNA when using DNA dilution methods (Behrenfeld et al., 2021). These standard curves provide a means to accurately determine the concentration of A. catenella cells, cysts, and A. tamarense based on their corresponding Ct values, enabling precise quantitative analysis (Lee et al., 2017).
2.2 Cell disruption and PMA treatment
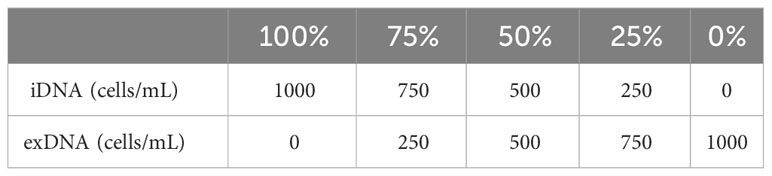
To selectively detect only iDNA using PMA, exDNA was generated using two methods: heat treatment of A. catenella cells (1,000 cells/mL) and cyst cultures (1,000 cells/mL) at 60°C for 30 min and exposure to 1% Triton x-100 (Junsei, Japan) for 10 min to selectively disrupt cell membranes. Dead cells were assumed to be exDNA and non-dead cells were assumed to be iDNA. The iDNA and exDNA were mixed at ratios of 100, 75, 50, 25, and 0% (Table 1) and the mixed DNA was defined as eDNA. Samples for each concentration were treated with 10 mM of PMAxx™ dye (Biotium, USA). The samples were incubated in the dark at room temperature for 10 min and then exposed to blue LED on ice for 30 min. Furthermore, to add credibility to, we utilized A. tamarense at a concentration of 1,000 cells/mL. The sample was partitioned into two distinct groups representing 100% and 0% viable cell counts, achieved through treatment with Triton X-100. The evaluation was organized into the following experimental groups: A) PMA untreated, DNase untreated, B) PMA untreated, DNase treated, C) PMA treated, DNase treated, D) PMA treated, DNase untreated. The DNase treatment was administered, with a 5 μL volume used instead of the manufacturer-recommended 1 μL, as per their protocol, employing DNase I, RNase-free (Thermo Scientific, USA). Additionally, Group C involved treating with DNase first, followed by the application of PMA. Each sample was centrifuged at 13,000 x g for 5 min and extracted using the AccuPrep® Plant Genomic DNA Extraction Kit (Bioneer, Korea) according to the manufacturer’s instructions.
2.3 DNA extraction and primer design for quantitative analysis
Primer was designed to specifically detect the LSU region of the A. catenella 18s rRNA gene (DQ785887.1) from the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/blast) using Primer 3 (Whitehead Institute and Howard Hughes Medical Institute, MD) and Oligo Calc (Oligonucleotide Properties Calculator software). Multiple sequence alignments were performed using the NCBI, and data were visualized using NCBI MSA Viewer to confirm species specificity (Figure S1).
The designed primer sets Alx2 Forward (5′ATTTTCCTGCGGGGTGTGGA′3), and Alx2 Reverse (5′TCCGTGTTTCAAGACGGGGTCA′3) were used to adjust the qPCR template final concentration to 20 μL. qPCR template was 10 μL Prime Q-Master Mix with UDG (Genetbio, Korea), 7 μL tertiary distilled water, 1 μL Alx2 Foward, 1 μL Alx2 Reverse, and 1 μL DNA template. SYBR Green qPCR was performed using a CFX Opus 96 (BioRad, USA) at 95°C for 5 min with 35 cycles (95°C for 30 sec, 59°C for 30 sec, 72°C for 30 sec), and all qPCR runs were performed in triplicate for reliability.
2.4 Field sampling methods and marine samples preparation
Masan Bay, a representative semi-enclosed sea in South Korea, is well known for its contaminated waters in the past. In April 2023, samples were collected at 3 Stations in Masan Bay (Figure 1). All filters used in the study have a diameter of 47 mm. The filter pore size was set to 30 μm in order to selectively collect iDNA, considering the typical size range of Alexandrium spp. (30-40 μm) (Anderson et al., 2005). To compare PMA-iDNA monitoring, samples from the field were taken to the lab and subjected to comparison using filters commonly utilized in many eDNA studies, specifically 0.2, and 1.2 μm filters (Lacoursière-Roussel et al., 2016). To selectively collect and compare iDNA, seawater filtration in the field was improvised using a filter with 30 μm pores and a custom 3D-printed filter holder connected to a pump (Table 2; Figure 2). The filter was stored in 5 ml tubes on ice and transported to the laboratory. In order to verify primer specificity, sampling was conducted in August 2023, a period when Alexandrium spp. were not anticipated to appear due to high water temperatures. Water samples were collected from the coastal waters of Tongyeong using a 0.2 μm filter to capture eDNA (Figure 1).
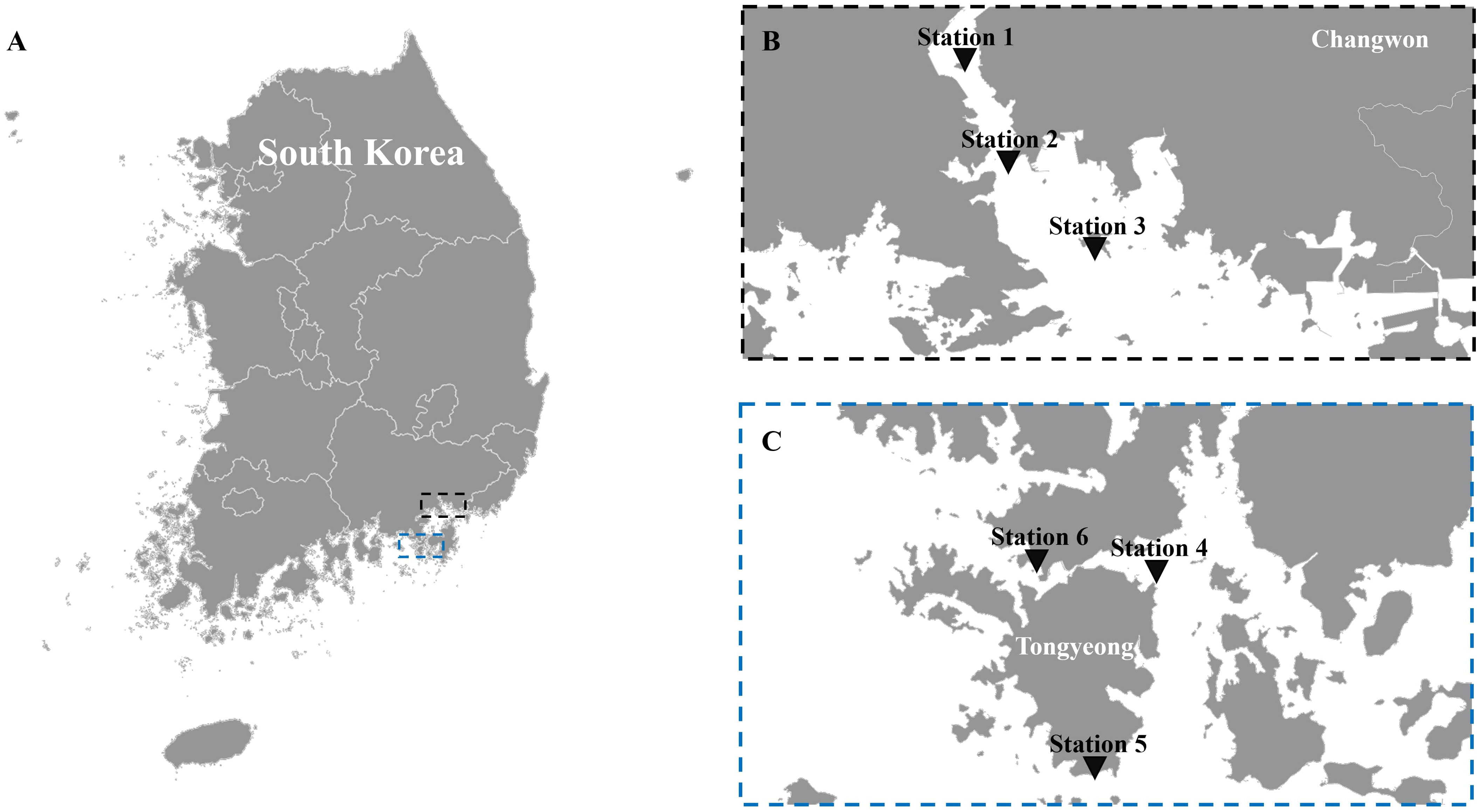
Figure 1 The sampling locations. (A) map of Korea, (B) stations 1, 2, and 3 in Masan Bay for field sampling in April 2023 to compare filter pore sizes, and (C) stations 4, 5, and 6 off the coast of Tongyeong in August 2023, when Alexandrium spp. are not present.
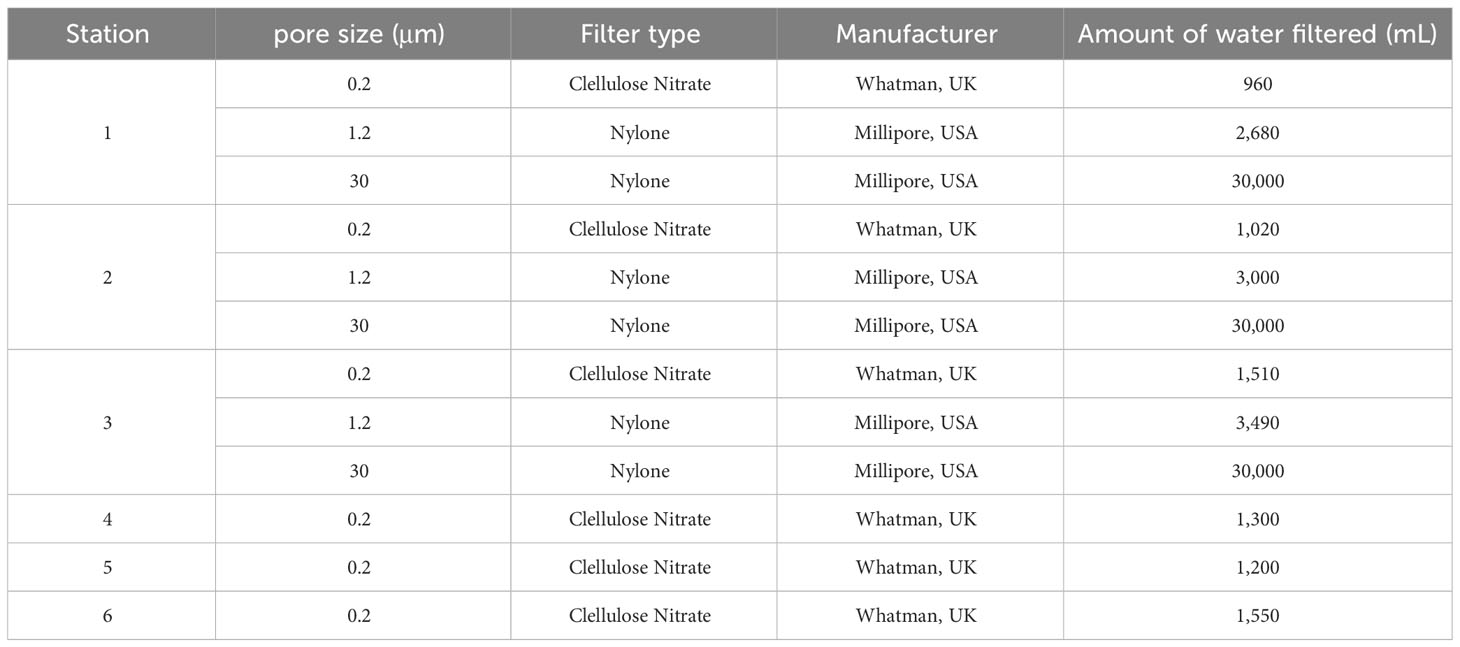
Table 2 Filters and seawater filtration volumes are used to collect eDNA (exDNA and iDNA) in the field.
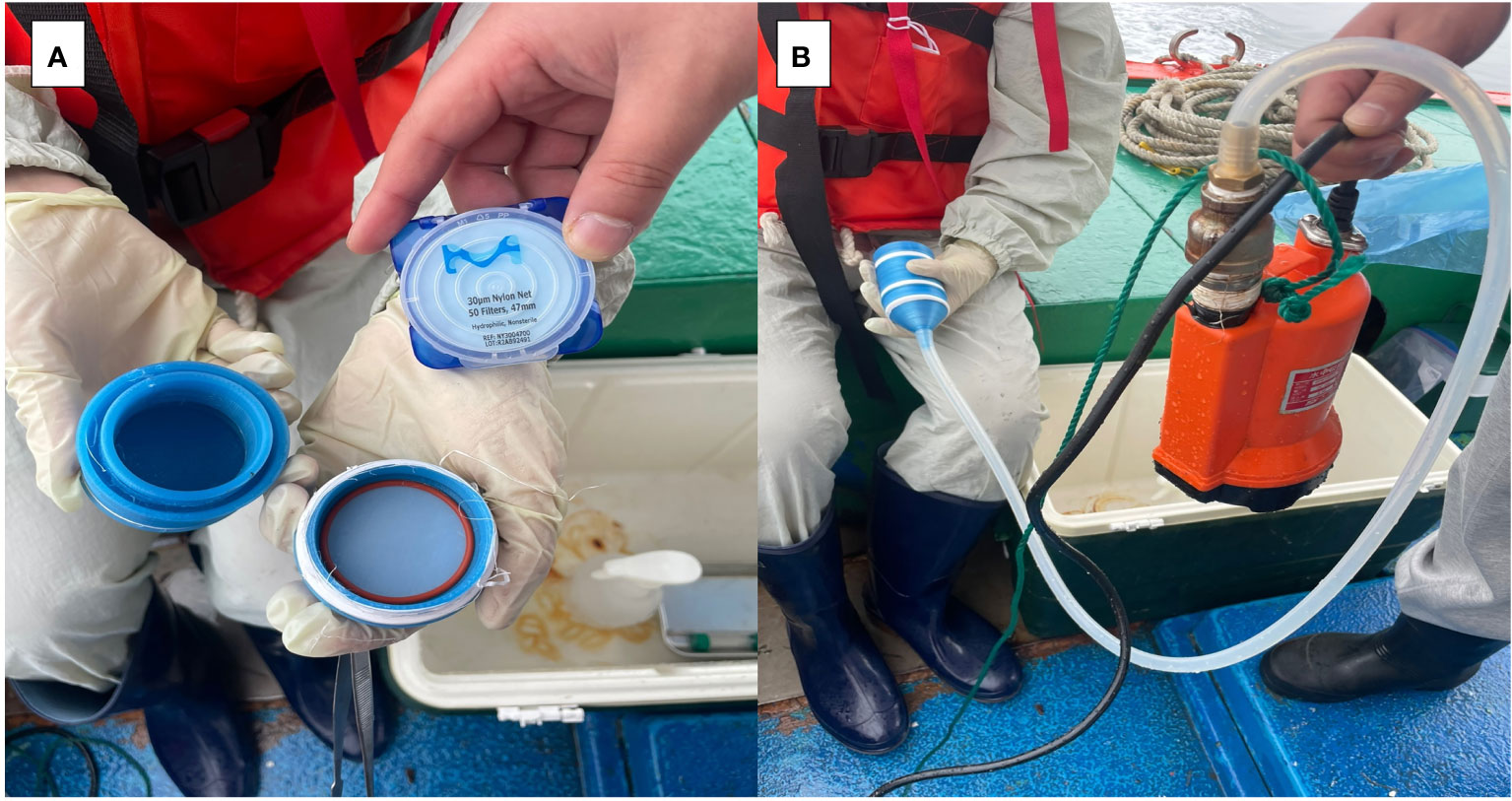
Figure 2 Filtration of eDNA using filters with 30um pore size in the field. (A) Filter holder fabricated with a 3D printer, (B) Filter holder connected to a pump.
To fabricate the filter holder, 132D Design software (http://www.123dapp.com/) was utilized to design the model, and subsequently, the design file was converted using Cubicreator4 V4.4.0. (Cubicon, Korea). 3D printing was performed on a Cubicorn Single Plus printer (Cubicorn, Korea) using PLA filament (3DBUYER, UK). The filter holder has an 800 μm pore-size plate in the inlet for primary filtration of other suspended solids.
In addition, 4 L of seawater was collected at each vertex in sterilized collecting bottles, stored on ice, transported to the laboratory, and immediately filtered using filters with 0.2 and 1.2 μm pores until the filter clogged (Table 2). To collect sediment samples, a van Veen Grab with a surface area of 0.1 m2 was used to collect sediment from each vertex. Sediment was transported to the laboratory on ice in sterile collection bags, stored at -4°C for 24 hr, and DNA was extracted. Water quality survey data (water temperature, pH, DO, turbidity, salinity, Chl-a) for Masan Bay in April 2023 were obtained from the Korea Marine Environment Management Cooperation MEIS (Marine Environmental Information System) (KOEM, https://www.meis.go.kr/mei/observe/port.do) (Table S1). In addition, Suspended Solids affecting sample collection using filters are described in the Supplementary Material (Table S2). Seawater was collected from all sampling sites, fixed with Lugol’s solution, and concentrated for analysis. A Sedgwick-Rafter chamber under a phase contrast microscope counting in triplicate, and data are shown in Supplementary Tables 3, 4. Qualitative analysis was performed at 400-1,000x magnification.
2.5 Filter and sediment eDNA extraction
The sample filter from the field and the filter treated in the lab were transferred to a 60 mm diameter Petri dish to which 3 mL of PBS was added. Then, it was shaken at 150 rpm for 2 hr in a shaker at 20°C. Using sterilized forceps and a scalpel, collect the filtered sample and divide it equally between two 1.5 mL tubes. The PMA-Untreated eDNA group was composed of one sample, while the PMA-Treated eDNA group consisted of another sample. The PMA-Untreated eDNA group was extracted by spin-down of 1.5 mL tubes at 13,000 RPM for 5 min, discarding the supernatant and using the AccuPrep® Plant Genomic DNA Extraction Kit (Bioneer, Korea) according to the manufacturer’s instructions. For PMA treatment, the PMA-treated eDNA group was centrifuged at 13,000 RPM for 5 min, the supernatant discarded, and the DNA was extracted as above after exposure to 400 μL of PBS and 1 μL of PMAxx™ dye in blue light (450-490 nm) for 30 min.
To extract DNA from sediment samples, 20 g of sediment from each vertex was transferred to a 50 mL tube. Subsequently, 10 mL of PBS was added, and the mixture was centrifuged at 2,000 RPM for 20 min. The supernatant was carefully transferred to a new tube and further centrifuged at 13,000 RPM for 5 min. Following this step, the supernatant was discarded, and 800 μL of PBS was added to the sediment pellet. The resulting mixture was divided equally, with half of it transferred to one 1.5 mL tube for the PMA Untreated eDNA group and the other half transferred to another 1.5 mL tube for the PMA Treated eDNA group. DNA extraction was then performed on both groups following the same procedure as described above.
2.6 Statistical analysis
The correlation between cell concentration and Cycle of Threshold (Ct) value was determined using a standard curve. Statistical analysis comparing the results PMA Untreated eDNA group and PMA Treated eDNA group was performed using a t-test after checking for normal distribution. All statistical analyses and graphs were visualized using R Studio and Excel.
3 Results
Species specificity testing was conducted using primers specifically designed to detect only Alexandrium spp. The tested species included Alexandrium catenella, Alexandrium pacificum, and Alexandrium tamarense. Additionally, the Dinoflagellata Gymnodinium impudicum and Akashiwo sanguinea, as well as the Diatoms Thalassionema species, Skeletonema costatum, and Nitzschia spp., which coexist in the same Marine region, were included in the testing (Table 3). The results of the species specificity test for the primer indicate that no amplification was observed in any of the species tested, except for Alexandrium spp. This confirms the specific detection of Alexandrium spp. by the Alx2 primer set, as evidenced by the positive amplification observed exclusively in the tested Alexandrium spp. samples. Morphological analysis using traditional microscopy showed that Alexandrium spp. were the priority species at Stations 1, 2, and 3 in Masan Bay in April 2023 (Table S3), and Alexandrium spp. were not detected at Stations 4, 5, and 6 in August 2023 (Table S4).
In August 2023, the eDNA detection results for stations 4, 5, and 6 showed a significant contrast between the untreated and treated PMA groups. Specifically, the untreated PMA group showed a Ct value indicating DNA amplification, while the PMA-treated group showed no Ct value, suggesting that DNA amplification was successfully inhibited. This indicates that the Alx2 primer set specifically detects Alexandrium spp. and that exDNA is a potential source of error in eDNA monitoring (Figure 3).
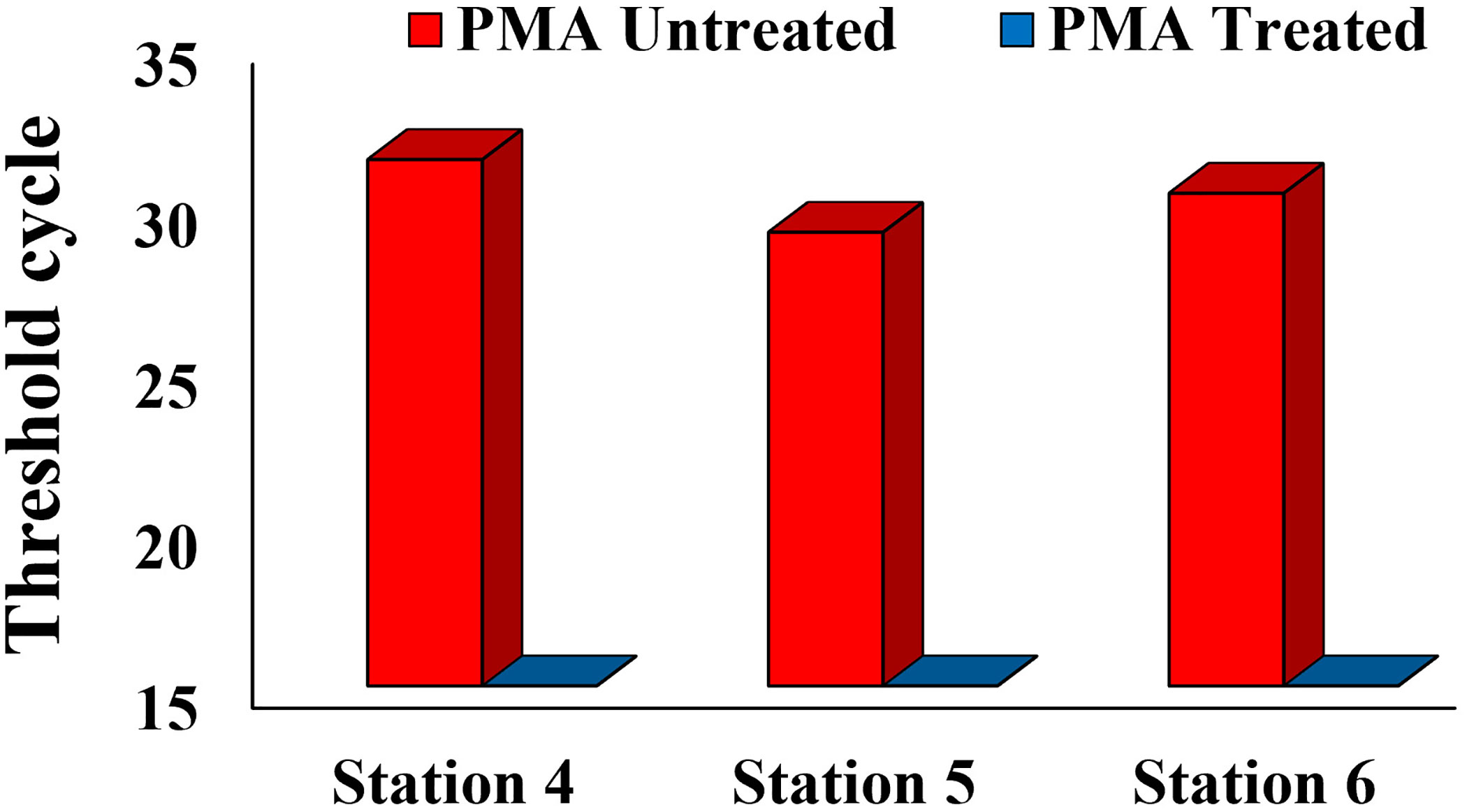
Figure 3 Comparison of eDNA Detection between PMA Untreated Group and PMA Treated Group in the Tongyeong Coastal Area in August 2023.
Quantitative analysis is significantly facilitated by the utilization of standard curves. In this study, we formulated standard curves to establish a correlation between the concentration of A. catenella cells, cysts, and A. tamarense cells and their corresponding Ct values (Figure 4). The generation of Standard curves using the dilution series of A. catenella cells and cysts yielded high R2 values of 0.9963 and 0.9965, which are close to 1, while A. tamarense cells also showed a value of 0.9919, close to 1. These R2 values indicate a strong correlation between the concentrations of A. catenella cells and cysts and their corresponding Ct values. The high R2 values suggest that the standard curves are highly reliable and accurate in quantifying the target species. Finally, to obtain log cell counts for field samples, the standard curve for cells of A. catenella and A. tamarense (Figure 4D) showed an R2 value of 0.9911.
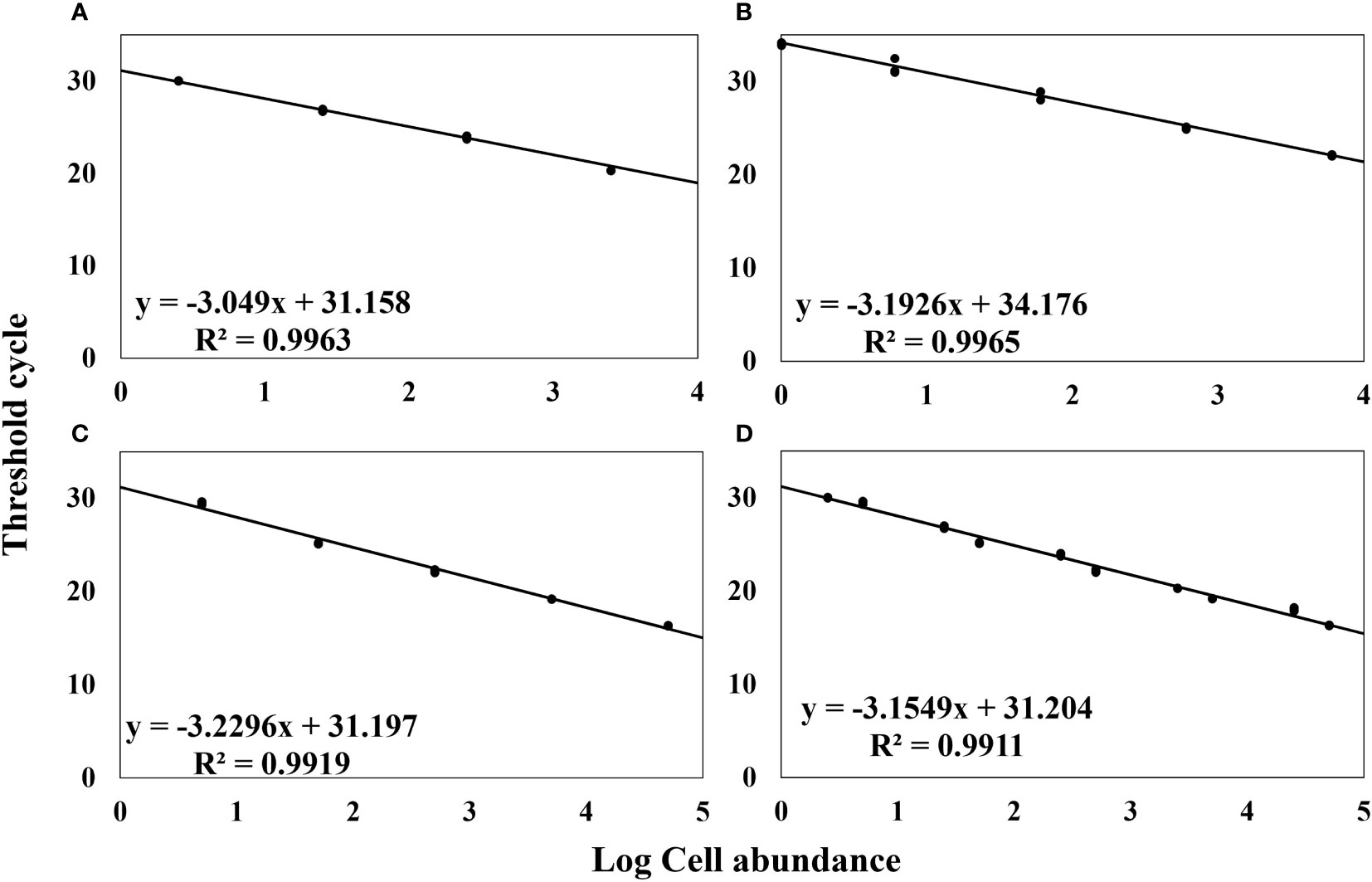
Figure 4 Standard Curve of Cell Concentration (log) and Ct Values. (A) Alexandrium catenella Cells Concentration, (B) Alexandrium catenella Cysts Concentration, (C) Alexandrium tamarense Cells Concentration, (D) Alexandrium catenella and Alexandrium tamarense Cells Concentration.
A standard curve enables the mathematical modeling of the relationship between concentration and Ct value, providing a means to accurately estimate the concentration in various samples. This makes standard curves valuable for comparative analysis across different samples or experimental conditions. By analyzing multiple samples on a consistent basis, it becomes possible to compare relative concentration differences or determine statistical variances between groups. These capabilities make standard curves an important tool in quantifying and comparing target concentrations in a study.
To assess the effectiveness of the PMA-iDNA monitoring method, laboratory-cultured A. catenella cells and cysts were divided into two groups: PMA Untreated and PMA Treated. This division allowed for a comparison of the performance of the monitoring method with and without the use of PMA. Furthermore, the slopes of the standard curves for Cells and Cysts presented in Figures 4A, B were used to convert the Ct values into logarithmic values. The results of PMA treatment for each percentage of the sample heat-treated at 60°C for 30 min (Figure 5) showed no significant difference between PMA Untreated and PMA Treated groups at 100% for both cells and cysts. However, as the percentage of exDNA increased, the Log cells abundance also decreased, indicating that PMA selectively detects only iDNA. However, even in the PMA Untreated group, the Log value decreased as the percentage of exDNA increased. It is worth noting that the heat treatment method used could potentially affect the eDNA.
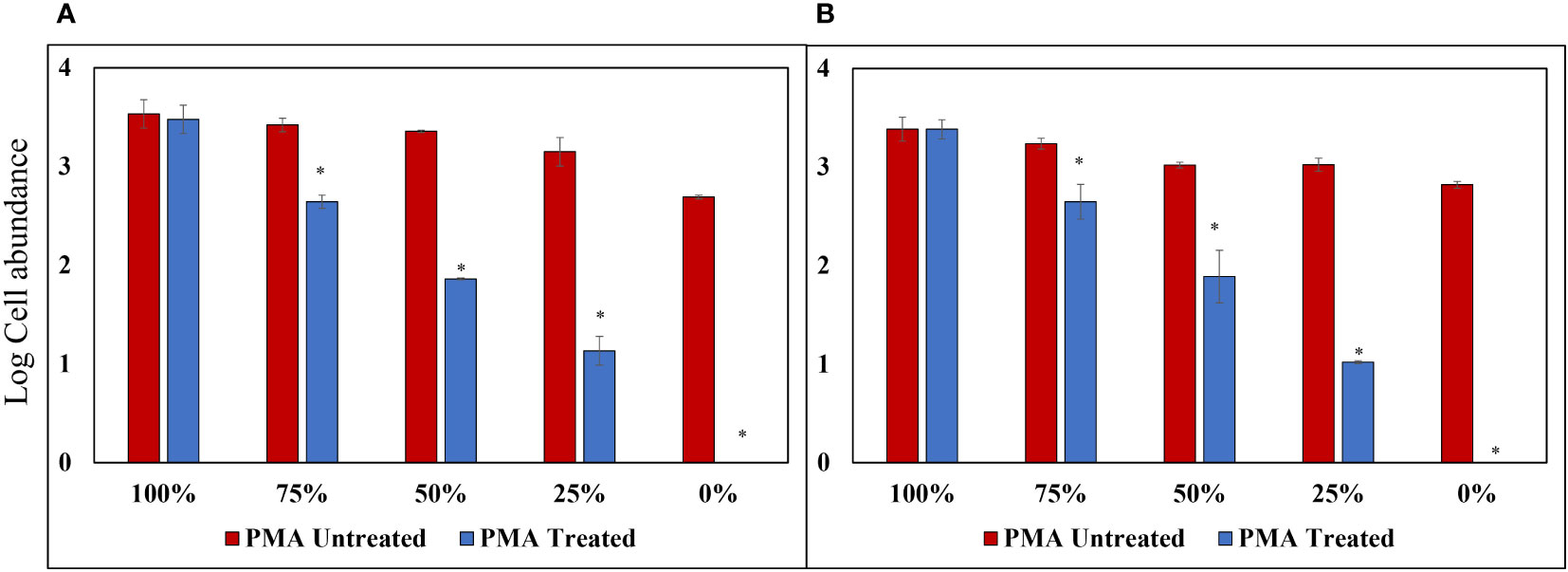
Figure 5 PMA Treatment Comparison for Samples Heat-Treated at 60°C for 30 min. (A) Alexandrium catenella cells (1,000 cells/mL). (B) Alexandrium catenella cysts (1,000 cells/mL). *P<0.05.
Triton X-100 (1%), a non-ionic surfactant, was used to selectively disrupt cells without causing DNA damage; this method showed no decrease in log values for the PMA Untreated group, contrast the heat-treated method, for both cells and cysts, and both cells and cysts showed P-values above 0.05 at 100% concentration. Similarly, the log cell abundance of the PMA-treated group decreased as the percentage of exDNA increased (Figure 6). The results obtained from the study suggest that Triton X-100 (1%) effectively disrupts cells without causing eDNA damage. The findings also confirm the ability of PMA treatment to selectively detect iDNA. Notably, no significant differences were observed between the PMA Untreated and PMA Treated groups at a concentration of 100% for both cells and cysts, regardless of PMA treatment. However, as the percentage decreased, the Log cells abundance decreased, indicating that PMA selectively detects iDNA. However, both cells and cysts showed a log cell count in the 0% group. The findings of this study have the potential to make a significant contribution to the enhancement and standardization of eDNA quantification methods utilizing qPCR. The accurate analysis of iDNA and exDNA through the application of PMA treatment shows promise for advancing eDNA monitoring techniques. By effectively differentiating between iDNA and exDNA, researchers can improve the accuracy and reliability of eDNA-based assessments, thereby enhancing our understanding of ecosystems and facilitating more robust environmental monitoring and management strategies. Furthermore, to add credibility to the above experiment, the results of each of the four groups treated with DNase and PMA showed that log cell abundance remained consistent in Group A regardless of the percentage of viable cells at 100% and 0%. In contrast, Groups B, C, and D showed higher values of log cell abundance at 100% viable cells and lower values at 0% (Figure 7).
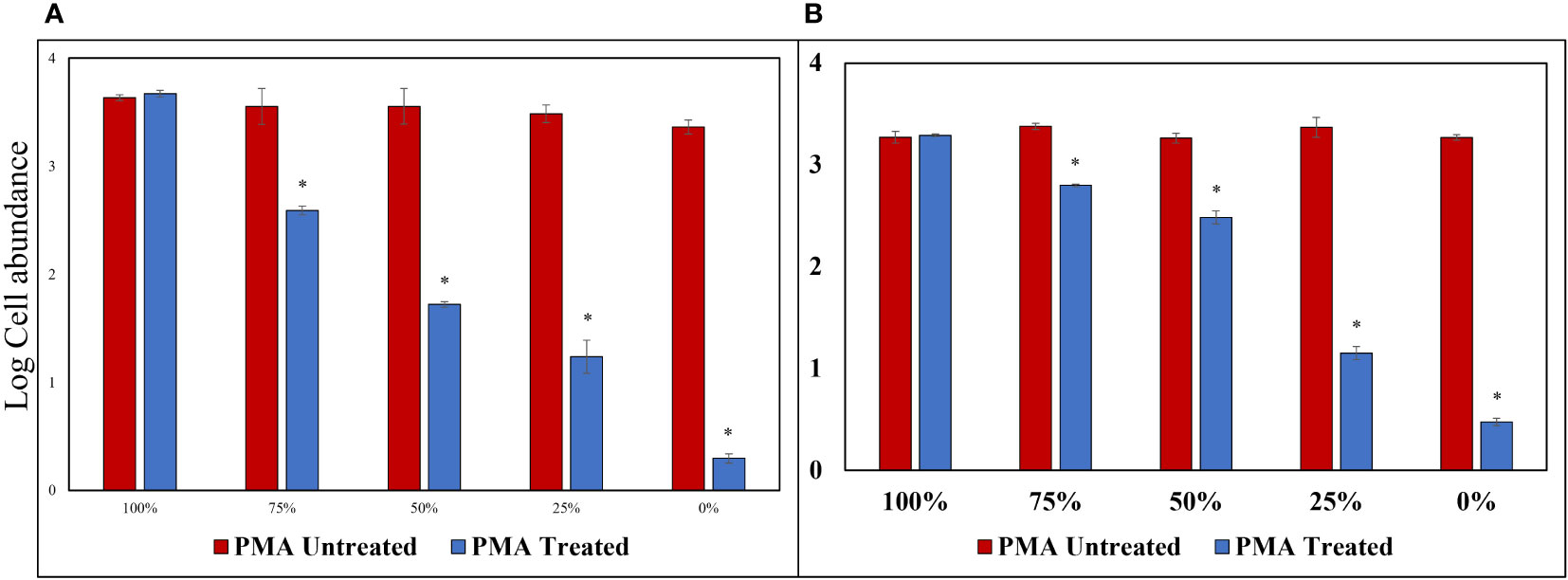
Figure 6 PMA Treatment Comparison for Samples Treated with Triton X-100 (1%). (A) Alexandrium catenella cells (1,000 cells/mL). (B) Alexandrium catenella cysts (1,000 cells/mL). *P<0.05.
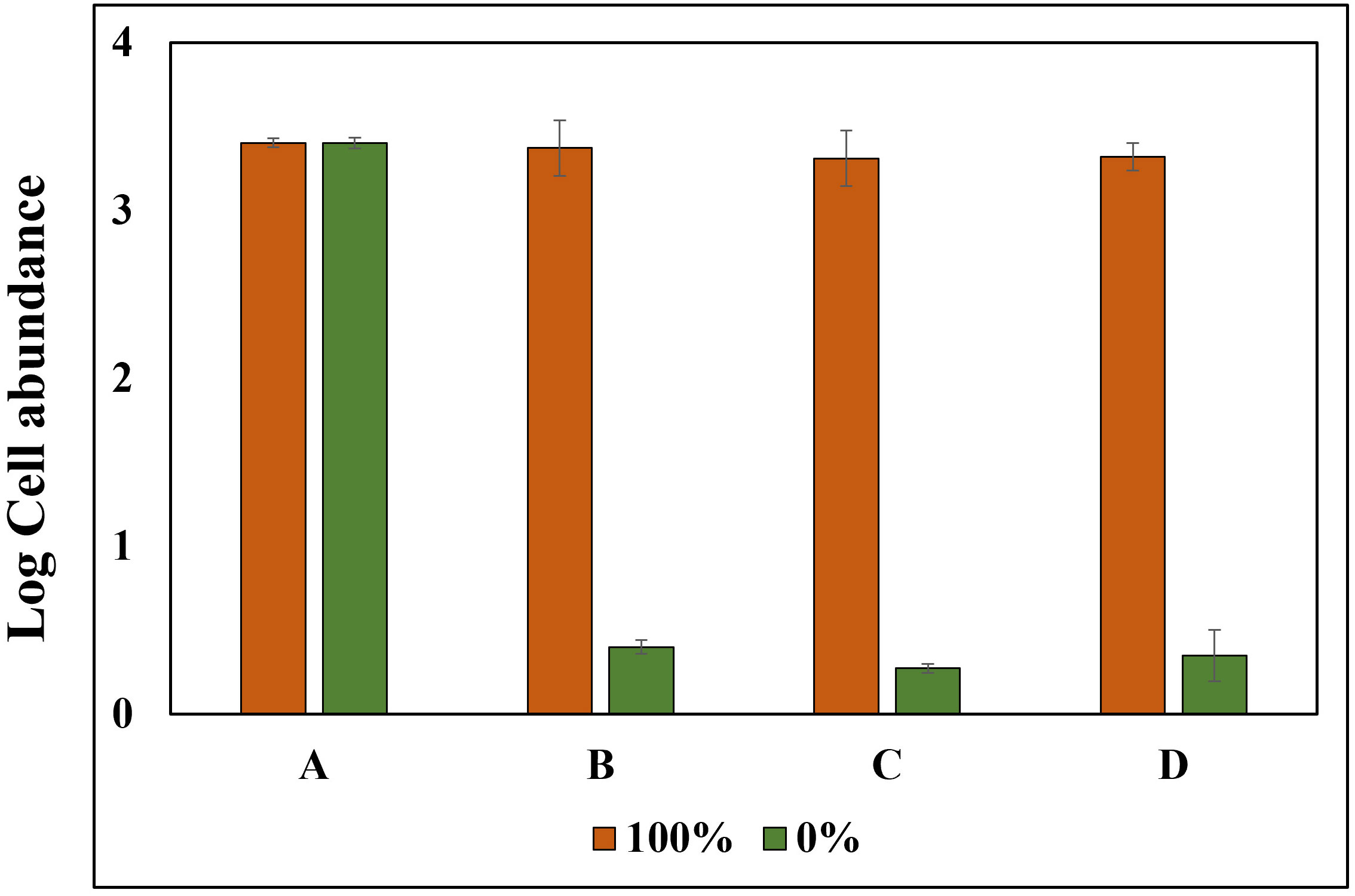
Figure 7 Experimental Setup and Treatment Categories for PMA Verification using Alexandrium tamarense. Experiments were performed involving Alexandrium tamarense with live cell ratios of 100% and 0%. The samples were systematically divided into four distinct treatment groups: (A) Untreated with PMA, Untreated with DNase. (B) Untreated with PMA, Treated with DNase. (C) Treated with PMA, Treated with DNase. (D) Treated with PMA, Untreated with DNase.
PMA-treated and untreated groups were compared with pore size filters of 0.2 μm and 1.2 μm (Turner et al., 2014; Kumar et al., 2022), which are commonly used pore sizes in conventional eDNA sampling methods, as well as a relatively large filter pore of 30 μm, in order to selectively collect only iDNA from field sampling (Figure 8). In the absence of PMA treatment, the log cell abundance of Alexandrium spp. at all vertices was observed to be in the order of 1.2 μm, 0.2 μm, and 30 μm. However, upon treating the filter samples with PMA and extracting DNA, it was found that the abundance order shifted to 30 μm, 1.2 μm, and 0.2 μm. This observation suggests that the smaller pore size filter successfully captures a wide range of eDNA types, including both exDNA and iDNA. Pore size filters of 30 μm selectively capture iDNA and relatively large microorganisms larger than 30 μm, while allowing exDNA and other microorganisms to pass through. As a consequence, the yield of exDNA is reduced, but the filter effectively increases the yield of iDNA. The results presented highlight the distinction between conventional eDNA sampling methods utilizing small pore sizes and iDNA selective collection methods employing filters with relatively large pores and PMA treatment. By selectively collecting only iDNA through PMA processing, errors in eDNA analysis, such as quantitative overestimation and the inability to differentiate between living and dead organisms, can be minimized. The combination of PMA treatment and an appropriate filter size can indeed be a useful strategy for effectively collecting iDNA during field sampling. However, it is important to note that the size of the DNA can result in variations in the relative amount of iDNA generated by the method used in this study, particularly for certain microbial species, cells of higher organisms, and small microorganisms. This consideration should be taken into account when conducting comparative studies of iDNA from different organisms in the environment.
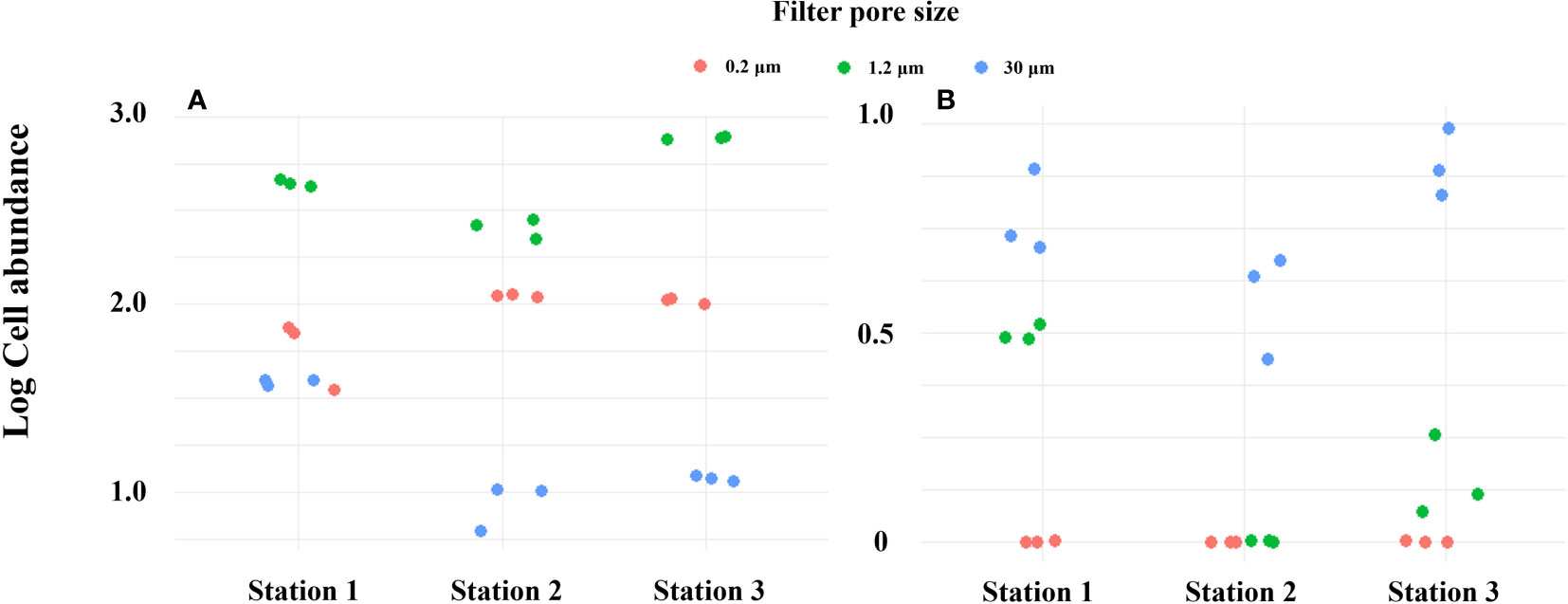
Figure 8 DNA yield by filter pore size in Masan Bay in April 2023. (A) PMA Untreated group. (B) PMA Treated group.
It is suggested that sediment layers have the ability to store DNA for long periods of time in biological monitoring (Corinaldesi et al., 2008). Deposits act as reservoirs for exDNA and iDNA, with exDNA predominantly representing the majority rather than iDNA (Corinaldesi et al., 2005). Therefore, the method can distinguish between living and dead organisms and has the capability to detect not only organisms that currently exist but also those that existed in the past. Given the low abundance of Alexandrium spp. at lower depths (Martin et al., 2005), it can be inferred that the DNA detected in the sediment layer originated from cysts or that exDNA was present within the sediment layer. In April 2023, the PMA untreated in Masan Bay sediments exhibited a high abundance of Alexandrium spp., whereas the treated samples showed a low abundance (Figure 9). In marine sediments, eDNA can originate from living cells or organisms, as well as natural or anthropogenic processes (Dell'Anno and Danovaro, 2005). Therefore, DNA extraction from sediment samples cannot exclude DNA derived from deceased organisms. Nevertheless, by selectively detecting only iDNA, the PMA-treated method appears to overcome the limitations from a sediment biomonitoring perspective. The application of selective iDNA targeted analysis can potentially prove valuable in biomonitoring studies, aiding in the differentiation between living and deceased organisms (Corinaldesi et al., 2018).
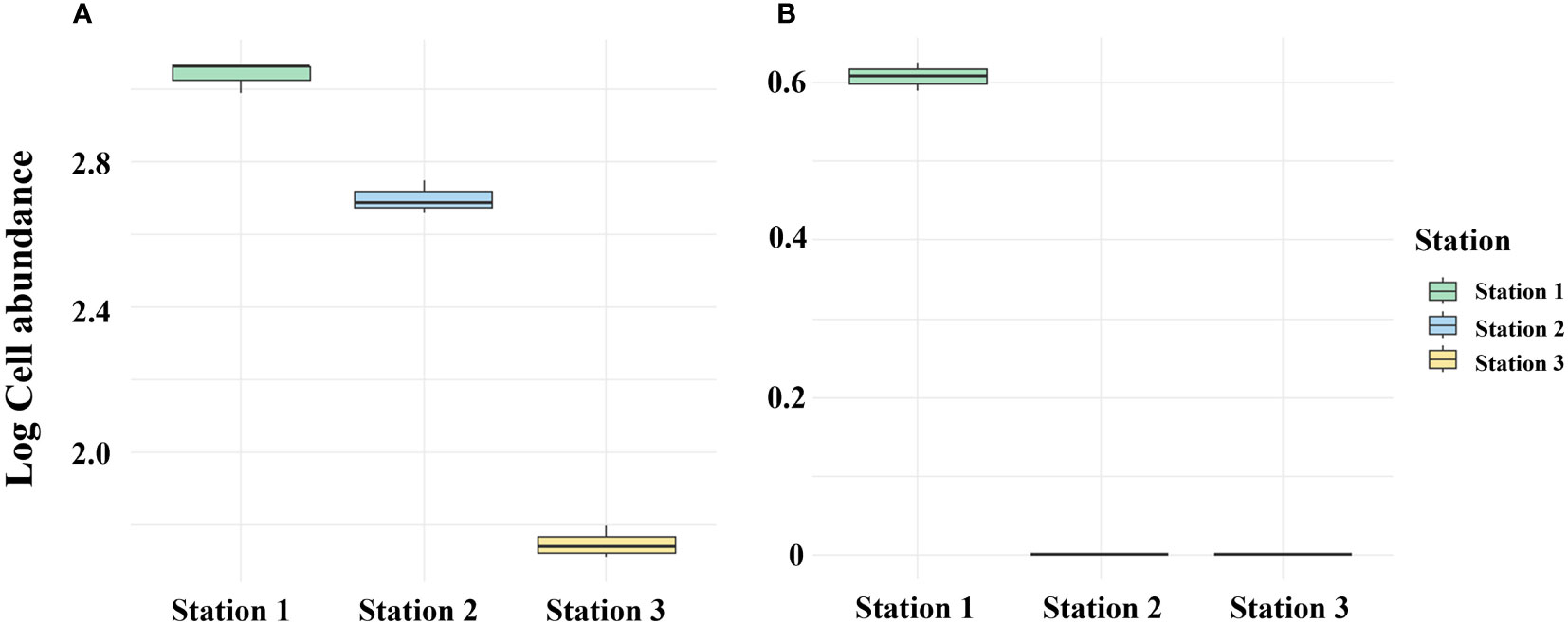
Figure 9 Comparison between PMA-treated and untreated eDNA groups of April 2023 Massan Bay sediment. (A) PMA Untreated group. (B) PMA Treated group.
4 Discussion
Alexandrium spp. are recognized as the main contributor to red tide blooms in various marine regions worldwide (Shikata et al., 2020). These organisms are also significant harmful algae, particularly in coastal or shelf waters, capable of disrupting marine ecosystems (Anderson et al., 2012). Red tide events, which can have adverse ecological and economic impacts, are often linked to Alexandrium spp. and can be influenced by environmental factors such as oceanic conditions, temperature, and nutrient levels (Min and Kim, 2023). As a result, monitoring Alexandrium spp. in the environment becomes essential to understanding the effects of human activities on marine ecosystems. For this purpose, targeted primers were developed. 3 Alexandrium species were cultivated in the laboratory for a minimum of 6 weeks, with an additional species from the same group used as a control for comparison. Significantly, no amplification was observed in species other than Alexandrium spp., confirming the specificity of this primer design. Such precision in primer design has the potential to enhance environmental monitoring and early warning systems. This primer configuration, tailored specifically for Alexandrium spp., demonstrates the promising potential for practical application in environmental assessment and protection.
In this study, for quantitative analysis using eDNA in sediment layers and seawater, standard curves were generated to investigate the relationship between A. catenella cells, cysts, and A. tamarense concentrations and Ct values. This analysis confirmed the feasibility of modeling the relationship between concentrations and Ct values, allowing us to estimate concentrations in other samples. In addition, a high R2 value indicates that the standard curve fits the data well and contributes to the reliability and accuracy of the measurement results.
If protected from physical degradation, exDNA can persist for years (Agnelli et al., 2007). While iDNA is protected from external degradation factors in the environment and has a longer half-life, internal degradation factors associated with cell death can result in DNA fragmentation (Hotchkiss et al., 2009). In conclusion, the findings suggest that different forms of DNA, including extracellular eDNA, damaged intracellular eDNA, and intact intracellular eDNA, may undergo distinct degradation processes (Hirohara et al., 2021). These processes can be influenced by various factors such as environmental conditions, enzymatic activity, and the physiological state of the cells or organisms. Understanding these separate degradation processes is crucial for interpreting eDNA data accurately and assessing its persistence and reliability as a biomonitoring tool. Therefore, it is considered more reliable to selectively detect iDNA in seawater than to detect total eDNA. By specifically targeting and analyzing iDNA, the focus is shifted to intact cell-derived DNA, which provides a clearer indication of the presence and abundance of living organisms. This approach helps to minimize potential errors and biases associated with the detection of degraded or extracellular DNA, contributing to a more accurate assessment of biodiversity, ecological dynamics, and environmental monitoring in marine ecosystems.
The results presented in Figure 5 demonstrate that the heat treatment method leads to the absence of log cell values in both the cell and cyst categories within the PMA 0% treatment group. Additionally, there is a gradual decrease in log cell counts for eDNA (iDNA, exDNA) in both cell and cyst categories in the absence of PMA treatment. These observations strongly suggest that the heat treatment method affects both iDNA and exDNA. However, the PMA-iDNA experiment, which generates randomized exDNA, highlights that only sufficient iDNA is detected. Contrastingly, Triton X-100 treatment did not impact the log cell count of eDNA (iDNA, exDNA) in the no PMA group, offering a distinct approach from the heat treatment. Nevertheless, DNA amplification occurred in the 0% PMA treatment group, implying that Triton X-100 might not completely degrade all iDNA. Furthermore, the higher log cell count observed in cysts in the PMA-treated group, as opposed to cells, aligns with the well-established understanding that cyst membranes are more resilient than cells. In summary, the heat treatment method’s effect primarily pertained to exDNA without detecting iDNA in the 0% live cell group. On the other hand, the Triton X-100 treatment method identified iDNA while not affecting exDNA within the same group. Both methodologies collectively highlight PMA’s exclusive covalent binding to exDNA, enabling the selective detection of iDNA while excluding exDNA. The potential of iDNA-PMA monitoring is further supported by the insights presented in Figure 7. In this fig, the results of the four groups subjected to DNase and PMA treatment lend credibility to the aforementioned experiments. Specifically, the outcomes demonstrate that log cell abundance remained stable in Group A regardless of the percentage of viable cells, spanning from 100% down to 0%. In contrast, Groups B, C, and D exhibited higher log cell abundance values at 100% viable cells, which progressively decreased to lower values at 0% (Figure 7). This reinforcement underscores the reliability and effectiveness of the iDNA-PMA monitoring approach.
In our study, we observed that the eDNA yields were high when using relatively small pore sizes of 0.2 and 1.2 μm filters in seawater. However, when it came to the selective detection of iDNA treated with PMA, which is designed to target DNA from living organisms, the yields with 0.2 and 1.2 μm filters were lower compared to the 30 μm filter. This suggests that employing a combination of PMA treatment and a larger pore size filter, such as 30 μm, may be a useful strategy for effectively collecting iDNA during field sampling. By selectively capturing iDNA while minimizing the capture of other particles and exDNA, this approach can enhance the accuracy and reliability of iDNA analysis, providing valuable insights into the presence and activity of living organisms in aquatic environments. In August 2023, eDNA detection at stations 4, 5, and 6 exhibited a notable contrast between untreated and PMA-treated groups (Figure 3). exDNA is generated from cell breakdown post-mortem or active shedding by organisms in the environment (Ibanez de Aldecoa et al., 2017). Particularly, exDNA introduced into soil or water displays significant resistance to rapid degradation, persisting for hours to days (Nielsen et al., 2007). Therefore, DNA amplification in the PMA-untreated group likely indicates exDNA detection, while the absence of amplification in the PMA-treated group implies specificity of the Alx2 primer set for Alexandrium spp. These outcomes enable us to anticipate errors in eDNA monitoring due to the specificity of primers and the persistence of exDNA.
Alexandrium spp. cysts exist in the sediment layer depending on environmental conditions and give rise to HABs (Dale, 1983). The eDNA derived from these cysts serves as a crucial indicator for detecting the distribution and occurrence of Alexandrium spp. in the ocean. However, DNA extraction from sediments carries the risk of overestimation or false positives. To tackle these challenges, sediment samples can be treated with PMA to assess the potential of environmental monitoring for Alexandrium spp. cysts. PMA-treated eDNA samples selectively detect only iDNA. Consequently, the separation of PMA-treated eDNA samples into exDNA and iDNA improves the accuracy of eDNA detection.
While eDNA monitoring serves various purposes in environmental monitoring, such as biodiversity surveys and species detection, it has limitations in quantitative detection and epidemiological surveys. However, PMA-iDNA monitoring selectively detects intact DNA and can overcome these limitations. However, in the future, there is a need to improve the capability of PMA-iDNA to investigate a wide range of biological groups by optimizing the target species’ iDNA size and the filter size used. It is essential to understand the variations in iDNA quantity that may arise based on the DNA size of specific microbial species, higher organisms, cells, and tiny microorganisms. This understanding will serve as the foundation for accurate iDNA monitoring of diverse biological groups.
5 Conclusion
To the best of our knowledge, the detection of iDNA in Seawater and sediments using PMA treatment and filter size optimization for improved selective detection of iDNA has been pioneered in this study. A method has been developed to prevent non-selective detection of eDNA through PMA treatment and minimize potential errors in eDNA sampling by optimizing the filter size. This approach reduces the risk of quantitative overestimation in eDNA studies and enhances the accuracy of species identification. The method is not only applicable to the analysis of Alexandrium spp. eDNA but also holds promise for the assessment of other harmful algal blooms (HABs) or phytoplankton, as well as for higher organism PMA-iDNA monitoring assays. Furthermore, it can serve as a rapid and reliable approach to mitigate errors in eDNA analysis.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
K-WY: Conceptualization, Data curation, Investigation, Methodology, Writing- original draft. H-SS: Data curation, Formal analysis, Investigation, Writing- review & editing. M-JS: Formal analysis, Investigation, Data curation, Writing- review & editing. MC-K: Conceptualization, Data curation, Funding acquisition, Methodology, Project administration, Resources, Supervision, Validation, Visualization, Writing- review & editing, Writing- original draft.
Funding
The authors declare financial support was received for the research, authorship, and/or publication of this article. This research was supported by Korea Institute of Marine Science & Technology Promotion (KIMST) funded by the Ministry of Oceans and Fisheries, (20220252).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1257343/full#supplementary-material
References
Agnelli A., Ascher J., Corti G., Ceccherini M. T., Pietramellara G., Nannipieri P. (2007). Purification and isotopic signatures (Δ 13 C, Δ 15 N, Δ 14 C) of soil extracellular DNA. Biol. Fertility Soils 44, 353–361. doi: 10.1007/s00374-007-0213-y
Anderson D. M. (1980). Effects of temperature conditioning on development and germination of gonyaulax tamarensis (Dinophyceae) hypnozygotes 1. J. Phycology 16 (2), 166–172. doi: 10.1111/j.1529-8817.1980.tb03013.x
Anderson D. M. (1998). Physiology and bloom dynamics of toxic alexandrium species, with emphasis on life cycle transitions. Nato Asi Ser. G Ecol. Sci. 41, 29–48.
Anderson D. M., Alpermann T. J., Cembella A. D., Collos Y., Masseret E., Montresor M. (2012). The globally distributed genus alexandrium: multifaceted roles in marine ecosystems and impacts on human health. Harmful Algae 14, 10–35. doi: 10.1016/j.hal.2011.10.012
Anderson D. M., Kulis D. M., Keafer B. A., Gribble K. E., Marin R., Scholin C. A. (2005). Identification and enumeration of alexandrium spp. from the gulf of maine using molecular probes. Deep Sea Res. Part II: Topical Stud. Oceanography 52 (19), 2467–2490. doi: 10.1016/j.dsr2.2005.06.015
Anderson D. M., Wall D. (1978). Potential importance of benthic cysts of Gonyaulax Tamarensis and G. excavata in initiating toxic dinoflagellate blooms 1, 2, 3. J. Phycol. 14, 224–234.
Behrenfeld M. J., O’Malley R., Boss E., Karp-Boss L., Mundt C. (2021). Phytoplankton biodiversity and the inverted paradox. ISME Commun. 1 (1), 52. doi: 10.1038/s43705-021-00056-6
Carini P., Marsden P. J., Leff J. W., Morgan E. E., Strickland M. S., Fierer N. (2016). Relic DNA is abundant in soil and obscures estimates of soil microbial diversity. Nat. Microbiol. 2 (3), 1–6. doi: 10.1038/nmicrobiol.2016.242
Corinaldesi C., Beolchini F., Dell’Anno A. (2008). Damage and degradation rates of extracellular DNA in marine sediments: implications for the preservation of gene sequences. Mol. Ecol. 17 (17), 3939–3951. doi: 10.1111/j.1365-294X.2008.03880.x
Corinaldesi C., Danovaro R., Dell'Anno A. (2005). Simultaneous recovery of extracellular and intracellular DNA suitable for molecular studies from marine sediments. Appl. Environ. Microbiol. 71 (1), 46–50. doi: 10.1128/AEM.71.1.46-50.2005
Corinaldesi C., Tangherlini M., Manea E., Dell’Anno A. (2018). Extracellular DNA as a genetic recorder of microbial diversity in benthic deep-sea ecosystems. Sci. Rep. 8 (1), 1–9. doi: 10.1038/s41598-018-20302-7
Dale B. (1983). “Dinoflagellate resting cysts: benthic plankton,” in Survival strategies of the algae, 69–136.
Dell'Anno A., Danovaro R. (2005). Extracellular DNA plays a key role in deep-sea ecosystem functioning. Science 309 (5744), 2179. doi: 10.1126/science.1117475
Fukuyo Y. (1985). Morphology of protogonyaulax tamarensis (Lebour) taylor and protogonyaulax catenella (Whedon and kofoid) taylor from Japanese coastal waters. Bull. Mar. Sci. 37 (2), 529–537.
Grzebyk D., Audic S., Lasserre B., Abadie E., de Vargas C., Bec B. (2017). Insights into the Harmful Algal Flora in Northwestern Mediterranean Coastal Lagoons Revealed by Pyrosequencing Metabarcodes of the 28S rRNA Gene. Harmful Algae 68, 1–16. doi: 10.1016/j.hal.2017.06.003
Günther B., Knebelsberger T., Neumann H., Laakmann S., Martínez Arbizu P. (2018). Metabarcoding of marine environmental DNA based on mitochondrial and nuclear genes. Sci. Rep. 8 (1), 14822. doi: 10.1038/s41598-018-32917-x
Hallegraeff G. M., Bolch C. J. (1992). Transport of diatom and dinoflagellate resting spores in ships' Ballast water: implications for plankton biogeography and aquaculture. J. Plankton Res. 14 (8), 1067–1084. doi: 10.1093/plankt/14.8.1067
Hirohara T., Tsuri K., Miyagawa K., Paine R. T. R., Yamanaka H. (2021). The application of PMA (Propidium monoazide) to different target sequence lengths of zebrafish eDNA: A new approach aimed toward improving environmental DNA ecology and biological surveillance. Front. Ecol. Evol. 9, 632973. doi: 10.3389/fevo.2021.632973
Hopkins G. W., Freckleton R. P. (2002). Declines in the numbers of amateur and professional taxonomists: implications for conservation. Anim. Conserv. 5, 245–249.
Hotchkiss R. S., Strasser A., McDunn J. E., Swanson P. E. (2009). Cell death. New Engl. J. Med. 361 (16), 1570–1583. doi: 10.1056/NEJMra0901217
Ibanez de Aldecoa A. L., Zafra O., González-Pastor JoséE. (2017). Mechanisms and regulation of extracellular DNA release and its biological roles in microbial communities. Front. Microbiol. 8, 1390. doi: 10.3389/fmicb.2017.01390
Kumar G., Farrell E., Reaume A. M., Eble J. A., Gaither M. R. (2022). One size does not fit all: tuning eDNA protocols for high-and low-turbidity water sampling. Environ. DNA 4 (1), 167–180. doi: 10.1002/edn3.235
Lacoursière-Roussel A., Rosabal M., Bernatchez L. (2016). Estimating Fish Abundance and Biomass from eDNA Concentrations: Variability among Capture Methods and Environmental Conditions. Mol. Ecol. Resour. 16 (6), 1401–1414. doi: 10.1111/1755-0998.12522
Lawson Handley L. (2015). How will the ‘molecular revolution’contribute to biological recording? Biol. J. Linn. Soc. 115 (3), 750–766. doi: 10.1111/bij.12516
Lee S. Y., Jeong H. J., Seong K. A., Lim A. S., Kim J. H., Lee K. H., et al. (2017). Improved real-time PCR method for quantification of the abundance of all known ribotypes of the ichthyotoxic dinoflagellate cochlodinium polykrikoides by comparing 4 different preparation methods. Harmful Algae 63, 23–31. doi: 10.1016/j.hal.2017.01.006
Martin J. L., Page F. H., Hanke A., Strain P. M., LeGresley M. M. (2005). Alexandrium fundyense vertical distribution patterns during 1982, 2001 and 2002 in the offshore bay of fundy, eastern Canada. Deep Sea Res. Part II: Topical Stud. Oceanography 52 (19), 2569–2592. doi: 10.1016/j.dsr2.2005.06.010
Medlin L., Elwood H. J., Stickel S., Sogin M. L. (1988). The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions. Gene 71 (2), 491–499. doi: 10.1016/0378-1119(88)90066-2
MEIS (Marine Environmental Information System). Marine environment observation & Survey. Available at: https://www.meis.go.kr/mei/observe/port.do.
Min J., Kim K. Y. (2023). Seasonal change and subniche dynamics of three alexandrium species in the korea strait. Harmful Algae 125, 102420. doi: 10.1016/j.hal.2023.102420.
Nagler M., Podmirseg S. M., Ascher-Jenull J., Sint D., Traugott M. (2022). Why eDNA fractions need consideration in biomonitoring. Mol. Ecol. Resour. 22 (7), 2458–2705. doi: 10.1111/1755-0998.13658.
Nielsen K. M., Johnsen PålJ., Bensasson D., Daffonchio D. (2007). Release and persistence of extracellular DNA in the environment. Environ. Biosafety Res. 6 (1-2), 37–53. doi: 10.1051/ebr:2007031
Nocker A., Camper A. K. (2006). Selective removal of DNA from dead cells of mixed bacterial communities by use of ethidium monoazide. Appl. Environ. Microbiol. 72 (3), 1997–2004. doi: 10.1128/AEM.72.3.1997-2004.2006
Park B. S., Wang P., Kim J. H., Kim J.-H., Gobler C. J., Han M.-S. (2014). Resolving the Intra-Specific Succession within Cochlodinium Polykrikoides Populations in Southern Korean Coastal Waters Via use of Quantitative PCR Assays. Harmful Algae 37, 133–141. doi: 10.1016/j.hal.2014.04.019
Pietramellara G., Ascher J., Borgogni F., Ceccherini M. T., Guerri G., Nannipieri P. (2009). Extracellular DNA in soil and sediment: fate and ecological relevance. Biol. Fertility Soils 45, 219–235. doi: 10.1007/s00374-008-0345-8
.Pont D., Meulenbroek P., Bammer V., Dejean T., Erős T., Jean P., et al. (2023). Quantitative Monitoring of Diverse Fish Communities on a Large Scale Combining eDNA Metabarcoding and qPCR. Mol. Ecol. Resour. 23 (2), 396–409. doi: 10.1111/1755-0998.13715
Salter I., Joensen M., Kristiansen R., Steingrund P., Vestergaard P. (2019). Environmental DNA concentrations are correlated with regional biomass of atlantic cod in oceanic waters. Commun. Biol. 2 (1), 461. doi: 10.1038/s42003-019-0696-8
Samsur M., Yamaguchi Y., Sagara T., Takatani T., Arakawa O., Noguchi T. (2006). Accumulation and depuration profiles of PSP toxins in the short-necked clam tapes japonica fed with the toxic dinoflagellate alexandrium catenella. Toxicon 48 (3), 323–330. doi: 10.1016/j.toxicon.2006.06.002
Shikata T., Taniguchi E., Sakamoto S., Kitatsuji S., Yamasaki Y., Yoshida M., et al. (2020). Phylogeny, growth and toxicity of the noxious red-tide dinoflagellate alexandrium leei in Japan. Regional Stud. Mar. Sci. 36, 101265. doi: 10.1016/j.rsma.2020.101265
Strickler K. M., Fremier A. K., Goldberg C. S. (2015). Quantifying Effects of UV-B, Temperature, and pH on eDNA Degradation in Aquatic Microcosms. Biol. Conserv. 183, 85–92. doi: 10.1016/j.biocon.2014.11.038
Turner C. R., Barnes M. A., Xu C. C. Y., Jones S. E., Jerde C. L., Lodge D. M. (2014). Particle size distribution and optimal capture of aqueous macrobial eDNA. Methods Ecol. Evol. 5 (7), 676–684. doi: 10.1111/2041-210X.12206
Valentini A., Taberlet P., Miaud C., Civade R., Herder J., Thomsen P. F., et al. (2016). Next-generation monitoring of aquatic biodiversity using environmental DNA metabarcoding. Mol. Ecol. 25 (4), 929–942. doi: 10.1111/mec.13428
Keywords: eDNA, IDNA, exDNA, Alexandrium spp., filter size optimization, selective detection, quantitative analysis
Citation: Yun K-W, Son H-S, Seong M-J and Kim M-C (2023) Propidium Monoazide based selective iDNA monitoring method improves eDNA monitoring for harmful algal bloom Alexandrium species. Front. Mar. Sci. 10:1257343. doi: 10.3389/fmars.2023.1257343
Received: 12 July 2023; Accepted: 29 August 2023;
Published: 15 September 2023.
Edited by:
Conghui Liu, Chinese Academy of Agricultural Sciences, ChinaReviewed by:
Feng Zhao, Chinese Academy of Sciences (CAS), ChinaHyun Woo Kim, Pukyong National University, Republic of Korea
Copyright © 2023 Yun, Son, Seong and Kim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mu-Chan Kim, kmc81@gnu.ac.kr
 Kun-Woo Yun
Kun-Woo Yun Hwa-Seong Son
Hwa-Seong Son Min-Jun Seong
Min-Jun Seong Mu-Chan Kim
Mu-Chan Kim