Research and prospects of environmental DNA (eDNA) for detection of invasive aquatic species in East Asia
- 1Guangdong Laboratory for Lingnan Modern Agriculture, Genome Analysis Laboratory of the Ministry of Agriculture and Rural Affairs, Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Shenzhen, China
- 2School of Agricultural Sciences, School of Agricultural Sciences, Zhengzhou University, Zhengzhou, China
The issue of biological invasions in aquatic ecosystems is becoming increasingly severe in the contemporary world. Due to the lack of monitoring and management systems for aquatic invasive species, the difficulty in identifying aquatic invasive species, and the limited effectiveness of conventional control methods in aquatic environments, biological control in water bodies is comparatively more challenging than other types of interventions. In recent years, environmental DNA (eDNA) survey methods have rapidly developed in various fields, such as biological monitoring, community ecology, paleoenvironmental research, conservation biology, and invasion ecology, due to their unique advantages of being rapid, sensitive, efficient, and non-invasive. Because of these characteristics, this innovative molecular approach has gained wider acceptance and is being increasingly utilized for the detection of biological diversity in aquatic environments. Furthermore, it has emerged as a novel technology to address the pressing and significant issue of aquatic invasive species in the vast freshwater and marine resources of the East Asian region. This paper summarizes a variety of literature sources to summarize the major aquatic invasive species in East Asian countries and the current application status of eDNA technology in their survey processes. Using China as a case study, it expounds on the prospective incorporation of the 4E strategy with eDNA technology for the surveillance of biological invasions. Furthermore, it explores the potential prospects of eDNA technology in species diversity management and policy formulation, offering theoretical guidance for establishing aquatic invasive species monitoring systems. From a technological standpoint, the integration of eDNA technology with the 4E strategy holds significant potential for application, thereby offering a promising reference for the formulation of policies related to the management of aquatic biological invasions and biodiversity.
1 Introduction
With the influence of factors such as global warming and economic globalization, biological invasion has become one of the hot topics in the field of global ecology. Biological invasion refers to the process by which organisms invade a new environment from their native habitats through natural or human-mediated pathways, causing economic losses or ecological disasters to biodiversity, agricultural and forestry production, and human health (Wan et al., 2002). Human activities have led to the efficient global transportation of thousands of species, and as trade has accelerated, the frequency of introductions has risen over time (Havel et al., 2015). For example, contaminant, stowaway, corridor and unaided pathways and other diverse routes arises the risk of alien species introduction (Hulme, 2009). Due to these intricate introduction pathways, aquatic ecosystems appear to be particularly susceptible to the threat of invasive species (Havel et al., 2015). The East Asian region encompasses vast aquatic expanses, where invasive species disrupt the dynamic equilibrium of native species and existing ecosystems (Sing and Tan, 2018). These invasions lead to shifts in the trophic levels of aquatic ecosystems, diminishing the self-purification capacity of the ecological environment and impacting the ecological system quality of aquatic habitats for aquatic organisms. Similar to other ecosystems, biological invasions in aquatic environments also consist of three consecutive stages: introduction, establishment of self-sustaining populations, and spread within recipient ecosystems. (Hulme, 2006; Davis, 2009). Early detection, diagnosis, and monitoring of invasive species are particularly important because of the benefits for rapid and effective response and the formulation of corresponding prevention and control decisions to curb the spread and expansion of invasive species (Hulme, 2006; Woodell et al., 2021).
Accurate identification of invasive species is pivotal to conservation strategies, enabling early detection and swift implementation of control measures through monitoring and surveillance activities (Mehta et al., 2007). To address these difficulties in detection at very low population densities, environmental DNA technology has emerged (MacKenzie et al., 2005; Ficetola et al., 2008). Environmental DNA (eDNA) refers to the DNA directly obtained from environmental samples, encompassing the DNA released by organisms such as animals, plants, and microorganisms into the air, soil, water, and other surroundings (Taberlet et al., 2012; Thomsen and Willerslev, 2015). Compared to traditional detection methods, aquatic eDNA technology can enhance the detectability of invasive species (Ficetola et al., 2008). The technological advantages of aquatic eDNA lie in its potential as a rapid and cost-effective tool for applied conservation biology. This encompasses the early detection of invasive species and the monitoring of species that are challenging to detect using conventional methods (Bohmann et al., 2014). In general, eDNA technology can facilitate early detection of invasive species and monitoring of inconspicuous organisms in aquatic ecosystem management, offering a more effective approach to biodiversity surveys.
2 Investigation of invasive aquatic species in East Asia
East Asia is endowed with abundant aquatic resources, including rivers, lakes, reservoirs, and oceans. However, due to globalization, international trade, tourism, and increased human activities, numerous non-native aquatic species have been introduced into the region’s water bodies. Some invasive species are intentionally imported and escape from captivity or are carelessly released into the environment. Unintentionally importing is another main path of introduction, arriving through livestock and produce, or by transport equipment such as packing material or a ship’s ballast water and hull (Lovell et al., 2006). These introduced species often possess strong reproductive capabilities and adaptability, enabling them to quickly establish and dominate new ecosystems (Knight, 2010). On the other hand, trade provides a major pathway for the introduction of invasive species (Ruiz and Carlton, 2003). As part of global business, the pet trade has become a significant channel with a great proportion of invasive mammals, birds, reptiles, amphibians, and fish. These non-native pets are sometimes released or escape from their owners and may be introduced beyond their native ranges, posing threats to biodiversity, agriculture, and public health. The pet trade particularly facilitates the spread of invasive species because commercially successful exotic pets often have larger spatial distributions and broader habitat requirements (Gippet and Bertelsmeier, 2021). By exploring diverse literature sources, this study has identified that aquatic environments in East Asia are encountering substantial risks posed by a wide array of invasive aquatic species, encompassing categories such as fish, reptiles, arthropods, mollusks, and more.
This article will focus on highlighting some of the highly invasive aquatic species in East Asia (Figure 1).
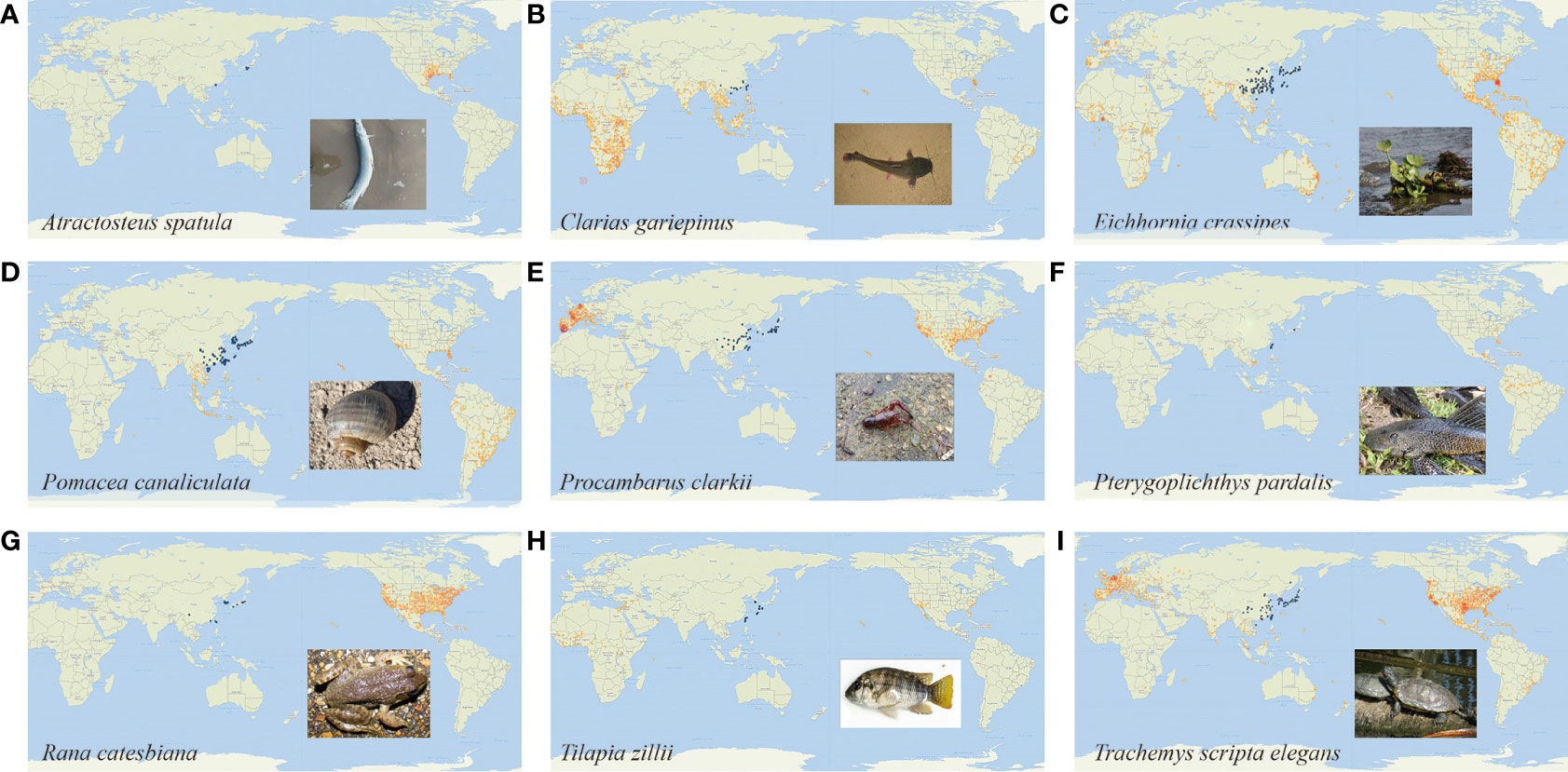
Figure 1 Morphology and distribution of typical aquatic invasive species The points on the map indicate the distribution location of the invasive species, the blue points indicate the distribution location of the invasive species in East Asia, and the yellow points indicate the distribution of the invasive species in other areas. The picture at the bottom right is a picture of the morphology of each species. (A). Atractosteus spatula (B). Clarias gariepinus (C). Eichhornia crassipes (D). Pomacea canaliculata (E). Procambarus clarkii (F). Pterygoplichthys pardalis (G). Rana catesbiana (H). Tilapia zillii (I). Trachemys scripta elegans. These images are quoted from the GBIF.org (Blackburn and Brown, 2023; de Vries and Lemmens, 2023; iNaturalist Contributor, 2023).
2.1 Pomacea canaliculata
Pomacea canaliculata, also known as the giant snail, apple snail, or mystery snail, is a freshwater gastropod mollusk belonging to the family Ampullariidae and genus Pomacea, native to the Amazon River basin in South America. It is one of the top 100 most threatening invasive species in the world (Lowe et al., 2000). In the 1980s, the P. canaliculata was introduced as a high-protein food source and for aquaculture purposes in regions of the United States and Southeast Asia (Halwart, 1994). Since its introduction to Guangdong, China in 1981, the P. canaliculata has had a significant impact on rice production in the southern region due to its strong environmental adaptability and rapid reproduction (Cai and Chen, 1990; Teo, 2001). Currently, in East Asia, P. canaliculata is distributed in China, Japan, and South Korea. It has also been found in Cambodia, Malaysia, Laos, Myanmar, the Philippines, Thailand, Vietnam, and Indonesia (Hayes et al., 2008; Matsukura et al., 2008; Xu et al., 2012).
2.2 Clarias leather
Clarias leather, also known as the Clarias catfish or African catfish, belongs to the order Siluriformes and the family Clariidae. It is native to the Nile River basin in Africa. Due to its high reproductive capacity, tolerance to low oxygen levels, and resistance to low temperatures, the C. leather was introduced to China as an aquaculture species from Egypt in 1981 (Li et al., 1984). Escapes and releases from aquaculture operations have led to its entry into natural water bodies. With its strong survival and reproductive abilities, it has spread to multiple rivers in the southern region of China, exerting significant pressure on native species (Radhakrishnan et al., 2011). In the East Asian region, the C. leathers is primarily distributed in freshwater environments such as lakes, rivers, reservoirs, and agricultural irrigation channels in China.
2.3 Rana catesbiana
The bullfrog belongs to the phylum Chordata, subphylum Vertebrata, class Amphibia, order Anura, family Ranidae, and genus Rana. It is considered one of the world’s top 100 most aggressive invasive species. Native to the eastern United States, it has been widely introduced to various regions worldwide over the past two centuries (Lever, 2003). The bullfrog was initially introduced as a food source from Cuba and Japan to mainland China in 1959, and subsequent introductions occurred multiple times during the 20th century (Wu et al., 2004). However, due to the lack of scientific management, escape and release incidents have been common, leading to the widespread proliferation of bullfrogs in wetland areas of Zhejiang, Hunan, Hubei, Guizhou, Sichuan, and Yunnan provinces in China. This proliferation poses a significant threat to the survival of native species (Wang et al., 2007; Zhan et al., 2017). In East Asia, bullfrogs are found mainly in rivers, lakes, reservoirs, and agricultural irrigation channels in China and Japan.
2.4 Procambarus clarkii
The red swamp crayfish, also known as Procambarus clarkii, belongs to the subphylum Crustacea, order Decapoda, and family Cambaridae. It is native to the northeastern part of Mexico and the central and southern regions of the United States, and has become one of the world’s notorious invasive species (Huner, 1988). It exerts harmful impacts on invaded freshwater ecosystems through predation, competition with native species, alteration of habitat characteristics, water quality, and other ecosystem services (Lodge et al., 2012). In Asia, this crayfish is widely distributed in China and Japan (Loureiro et al., 2015). It was initially introduced to Japan from New Orleans, USA, in 1927 for bullfrog aquaculture and pet trade purposes, and it can now be found throughout the country, including the Ryukyu Islands (Mito and Uesugi, 2004; Kawai and Kobayashi, 2005). In East Asia, it has a relatively wide distribution and is mainly found in rivers, lakes, reservoirs, and agricultural irrigation channels in China, Japan, and South Korea.
2.5 Tilapia zillii
The tilapia, belonging to the order Perciformes, family Cichlidae, is native to Africa and has now become widely distributed in the Americas, Europe, Australia, and China in Asia (http://www.fishbase.org) (Khaefi et al., 2014). It was introduced as a cultivated species in China in 1978 (Lou, 2000; Li et al., 2007; Deng et al., 2013). In China, although the tilapia aquaculture industry has rapidly developed nationwide, there is still a lack of comprehensive comparison regarding the ecological risks associated with tilapia introduction and aquaculture, which remains a priority in assessing the significant role of tilapia introduction (Xiong et al., 2023). In East Asia, the tilapia is relatively widely distributed, mainly in China, Japan, South Korea, and other regions, inhabiting freshwater environments such as rivers, lakes, reservoirs, and agricultural irrigation channels.
2.6 Trachemys scripta elegans
The Trachemys scripta elegans, belonging to the family Emydidae and genus Trachemys, is a subspecies of the Trachemys scripta. It is native to the area surrounding the Mississippi River to the Gulf of Mexico in the United States (Ernst, 1990). In the 1980s, the T. s. elegans was introduced to East Asia as an ornamental turtle (Shi et al., 2009). Initially brought in as pets, the T. s. elegans gained popularity due to its attractive appearance and ease of care, becoming a common choice for many people. However, with the growth of the pet trade and irresponsible ownership practices, many T. s. elegans were released into natural water bodies or escaped. These released or escaped individuals rapidly reproduced and established their populations in suitable environments (Burger, 2009). They are capable of adapting to various aquatic habitats and have become widely distributed in lakes, rivers, reservoirs, agricultural irrigation channels, and urban parks in East Asian countries. This invasive species poses a threat to native species’ survival in the region (Ma and Shi, 2017).
2.7 Atractosteus spatula
Atractosteus spatula, belonging to the class Actinopterygii, family Lepisosteidae, and genus Atractosteus, is the largest species in the gar family. It is native to North America but has recently been introduced to China, where it is considered an invasive fish species. Due to its predatory nature and consumption of many aquatic organisms in non-native habitats, it can cause a loss of native species diversity and abundance, as well as a reduction in fishery yields (Liu et al., 2023). The A. spatula is primarily distributed in freshwater habitats in countries such as China, Japan, and South Korea, including rivers, lakes, reservoirs, and agricultural irrigation channels (Xie et al., 2023).
2.8 Pterygoplichthys pardalis
Pterygoplichthys pardalis, commonly known as the leopard sailfin catfish or bulldog pleco, belongs to the order Siluriformes, family Loricariidae, and genus Pterygoplichthys. It is native to the Amazon River basin in South America and has become an invasive species in countries and regions including the United States, Mexico, South Africa, Southeast Asia, Japan (Jones et al., 2013; Ishikawa and Tachihara, 2014). In 1990, the P. pardalis was introduced to Guangdong Province, China, for its ornamental value and subsequently spread to the eastern and southern parts of the country through aquaculture trade (Li et al., 2007). Studies have found that the leopard pleco feeds on algae, zooplankton, secretion on tank walls, food remnants left by fish, and even fish eggs, posing a significant threat to the population and diversity of other fish species and disrupting the ecological balance (Tuten et al., 2009; Pound et al., 2011). In East Asia, the P. pardalis is primarily distributed in rivers, lakes, reservoirs, and agricultural irrigation channels in China, Japan, and South Korea.
2.9 Eichhornia crassipes
Eichhornia crassipes, a floating aquatic plant belonging to the Pontederiaceae family in the Eichhornia genus, is also known as “ Water Hyacinth” or “ Water Gourd.” It is native to South America and has often been introduced as an ornamental plant in countries across Asia, Africa, Europe, and North America, causing significant ecological harm (Villamagna and Murphy, 2010). Introduced to Southeast Asia during the 19th century, it was later introduced to China as an ornamental flower in 1901. In the 1930s, it was introduced to various provinces in mainland China as animal feed and promoted for its use in water purification and as an ornamental plant. Subsequently, it escaped cultivation and became a wild species. Currently, water hyacinth is widely distributed in the Yangtze River and Yellow River basins, as well as in various regions of South China (Yan et al., 2017). In East Asia, E. crassipes, with its strong environmental adaptability, is widely distributed in ponds, ditches, and waterlogged fields in China, Japan, and South Korea.
2.10 Prymnesium parvum
Prymnesium parvum is a microscopic, single-celled algae with four morphologically distinct forms. Three of the forms are scaled, bi-flagellated, and have a flexible, non-coiling, needle-like filament called a haptonema. The fourth form is a scaled, non-motile, siliceous cyst (Manton, 1966; Genitsaris et al., 2009). P. parvum is an algal species that forms harmful blooms in inland and coastal aquatic environments and is responsible for devastating fish kills causing ecological and economic damage. While blooms of P. parvum were documented in the eastern hemisphere since the early 1900s, the species has now spread widely, with blooms occurring in all southern regions of the USA and some northern regions. P. parvum is not on an alert list or listed as a regulated pest (https://www.cabidigitallibrary.org/).
3 Applications of eDNA in the detection of aquatic invasive species
In the early stages of aquatic species detection, field surveys were primarily conducted using methods such as fishing nets, boats, and complementary techniques like electrofishing for sample collection. However, these approaches were often time-consuming and labor-intensive, particularly when surveying rare species, low-density invasive species, or species that were challenging to sample. Moreover, these methods could potentially harm the target species or disrupt the ecological systems of survey sites (Snyder, 2003). Due to these factors, the scientific community sought a more efficient detection method, leading to the emergence of eDNA analysis. The eDNA technology initially emerged in the field of environmental microbiology, where it was used for the isolation and purification of microbial DNA from sediments (Ogram et al., 1987). It was not until the year 2000 that eDNA analysis gained recognition and wider application (Rondon et al., 2000). However, the first application of eDNA analysis in the field of aquatic biology occurred in 2008 when Ficetola and his colleagues used eDNA fragments extracted from water samples to monitor an invasive species, the R. catesbeiana (Ficetola et al., 2008). Since then, with the advancement of technology, eDNA techniques have been extensively applied in various aquatic ecosystems for studying fish (Doi et al., 2017), benthic organisms (Pawlowski et al., 2022), planktonic organisms (Suter et al., 2020), planktonic bacteria (Tessler et al., 2017), viruses (Mohiuddin and Schellhorn, 2015), and more. These applications include, but are not limited to, studies on aquatic community diversity (Bohmann et al., 2014), species identification (Ficetola et al., 2008), and biomonitoring (Takahara et al., 2012).
The number of studies using eDNA in different environments and species worldwide was highest for freshwater, followed by seawater, and soil and sediment (Thomaz et al., 2015). Among different species, fish were the most studied, followed by invertebrates, amphibians, mammals, plants, reptiles, and birds. The use of eDNA in research has shown a positive correlation with time, with an increasing number of studies conducted each year since 2008 (Sahu et al., 2023). This demonstrates the rising significance of eDNA technology in the field of aquatic organism detection.
As of May 2023, a search was conducted on the Web of Science database (https://www.webofscience.com/) covering the entire time period from 1998 to 2023 using the search terms “(environmental DNA OR eDNA) AND (Aquatic invasion OR Aquatic invasive species)”. Based on the publication status of articles, the research on eDNA in aquatic invasive species within the East Asian region was categorized into two main classes: those with established research methods and those currently lacking research (Table 1). The main objective was to survey and summary the use of eDNA for aquatic invasive species detection in East Asia, demonstrate the current research status of eDNA in aquatic invasions in the region, and provide reference for future studies using eDNA for monitoring aquatic invasions.
3.1 Research on species that have developed study methods in East Asia
During the research process, it was found that the use of eDNA detection technology to monitor aquatic invasive species is increasingly recognized in East Asia. It can not only detect low-abundance invasive species but also provide information about species presence and distribution. Since the pioneering use of eDNA technology for detecting R. catesbeiana in 2008 (Ficetola et al., 2008), subsequent studies in the East Asian region have successfully utilized eDNA technology to detect common aquatic invasive species and have even developed innovative methodologies. In Japan, several studies have focused on the potential of eDNA technology in ecological monitoring and conservation, particularly for invasive species detection. Successful eDNA surveys have been conducted to assess the distribution of Pacifastacus leniusculus in streams of Hokkaido (Ikeda et al., 2019). Further applications of eDNA methods have facilitated the detection of Lithobates catesbeiana and P. clarkii (Ogata et al., 2022), followed by an in-depth analysis of the influence of water quality and the abundance of T. s. elegans on the eDNA concentration of T. s. elegans in ponds (Kakuda et al., 2019). In China, DNA metabarcoding techniques have been employed to monitor the invasion of P. canaliculata in the Suzhou area. This approach demonstrated that environmental DNA metabarcoding had a much higher detection rate for P. canaliculata compared to traditional observation methods (Chen et al, 2021). Moreover, eDNA technology has been applied to distinguish between two invasive apple snail species, P. canaliculata and P. maculata, revealing their distribution patterns and enhancing tracking methods for these highly invasive and economically damaging species (Banerjee et al., 2022). The optimization of eDNA monitoring techniques is also continually advancing. Research has targeted the eDNA detection method for Trachemys scripta elegans, testing species specificity and refining reaction conditions (Lam et al., 2020). Similarly, species-specific primers and probes for frog DNA were designed, and a comparison of the extraction efficiency and cost indicated that the CTAB method was superior to the PCI method or DNeasy kits (Lin et al., 2019). In South Korea, studies have demonstrated that environmental DNA analysis is more accurate than traditional surveys in detecting the presence of Bugulina californica (Kim et al., 2021). These examples collectively highlight the effective performance of eDNA technology in aquatic invasive species detection. This innovative detection method continues to play an increasingly prominent role in the field of species monitoring.
For some species that are only distributed in East Asia, innovative methods have been innovated. For example, some studies have used Grandidierella japonica as a model species to experimentally investigate key factors such as methodology, decay characteristics, abundance estimation, and environmental monitoring using surface sediment eDNA. A novel eDNA-based method was devised to monitor benthic species through sediment samples (Wei, 2018). In another study, environmental DNA methods were employed to design a set of primers and probes specific to the Sciaenops ocellatus, investigating its distribution and biomass in the East China Sea. This study detected the distribution region of the S. ocellatus in the East China Sea (Wang et al., 2022). Furthermore, a specific eDNA detection method was established for Bufo species to detect the invasive and toxic land toad species, Bufo japonicus formosus, in Hokkaido, Japan (Mizumoto et al., 2022). These examples serve as templates from independent research conducted by East Asian scientists, serving as references for future scientific endeavors. Advancements and developments in research can be achieved through strategies such as formulating research plans, expanding collaborative networks, enhancing technical capabilities, strengthening data analysis and model construction, advocating for policy and funding support, and elevating the capacity of research teams.
During the process of conducting searches on the Web of Science database, it was observed that while East Asian scientists have been researching the application of eDNA technology for monitoring aquatic invasive species, many have made improvements to the research methods. However, they still encounter challenges and limitations. These challenges include the design and optimization of primers specific to particular species, interactions with environmental factors, and the standardization of sampling and analysis methods. Therefore, further research and collaborative efforts are still needed to promote the application of eDNA technology for monitoring aquatic invasive species in the East Asian region. The eDNA technology is continuously advancing in the East Asian region, leading to a plethora of research outcomes. For example, some studies analyzed the technical challenges and potential solutions of utilizing high-throughput sequencing technology to monitor early invasive species in marine ecosystems (Xiong et al., 2016). Other studies have investigated the effects of different filters and filtration methods on DNA capture efficiency. They found that a 0.8μm filter is the optimal pore size for membrane filtration of turbid, eutrophic, and high-density fish ponds (Li et al., 2018). Furthermore, research has demonstrated that eDNA metabarcoding can provide complementary insights into the biodiversity monitoring of zooplankton in polluted freshwater ecosystems. This involves cross-referencing traditional morphology-based methods with DNA-based approaches to ensure accurate and rapid identification of zooplankton species (Xiong et al., 2020). In summary, the East Asian region should enhance research and application efforts in the field of eDNA, driving innovation and development of relevant technologies and methodologies. This proactive approach addresses the demands of biodiversity conservation, environmental monitoring, disease diagnosis, and contributes to sustainable development and the establishment of an ecological civilization.
3.2 Species investigation to be explored in East Asia
So far, there is a lack of research in the East Asian region regarding the use of eDNA technology for monitoring certain high-risk invasive species. The Potamopyrgus antipodarum, known for its strong adaptability and reproductive capacity, has become a serious invasive species widely distributed around the world (Nentwig et al., 2018; Geist et al., 2022). The species has also shown invasive traces in Japan (Shimada and Urabe, 2003). Since there is no prior eDNA research for this species in the East Asian region, it is advisable to reference best practices and establish a comprehensive monitoring system. For example, Goldberg et al. used dose-response experiments to study the relationship between the density of P. antipodarum and the detection of eDNA over time, demonstrating the high potential of eDNA technology in aiding the early detection of widely distributed invasive aquatic invertebrates (Goldberg et al., 2013). The A. spatula has also invaded the East Asian region (Han, 2022). In order to accurately assess the extent of its invasion in East Asia, further research and monitoring are needed. For example, Ulayya et al. utilized eDNA methods to detect A. spatula as a supplementary approach to traditional monitoring methods, where eDNA and traditional detection methods were used in combination (Ulayya et al., 2020). Furthermore, during the investigative process, this study identified several invasive species with significant levels of invasion in the East Asian region, such as T. zillii (Mito and Uesugi, 2004) and P. pardalis (Li et al., 2007), however, there is a lack of eDNA-based research on these species in East Asia.
Although eDNA research in East Asia has made significant progress in recent years and achieved some important results, there is still a gap compared to the cutting-edge research level internationally. To enhance the research capacity of eDNA in East Asia, it is necessary to strengthen collaboration among research institutions, improve technical equipment and talent training, and enhance policy and funding support for eDNA research. It is important to pay attention to these invasive aquatic species, conduct early warning checks, learn from experiences, and establish a detection system for high-risk invasive species.
4 Advantages and disadvantages of eDNA technology in the study of aquatic invasions
Environment DNA technology enhances the sensitivity and efficiency of species-level identification. It can achieve early warning and monitoring of aquatic invasions, providing timely insights into species population distribution patterns and sizes. This aspect is crucial for species conservation and biodiversity preservation, forming an essential component and research foundation. Compared to traditional detection methods, eDNA technology presents significant developmental advantages in aquatic invasive species research. Firstly, there is a growing body of research aimed at optimizing operational processes and result analysis, providing comprehensive analytical procedures and operational guidelines for eDNA analysis (Deiner et al., 2015; Takahara et al., 2015). Secondly, eDNA analysis requires relatively small water samples, causing minimal disturbance to the habitats. Thirdly, eDNA analysis technology exhibits higher sensitivity in aquatic biological detection compared to traditional methods, particularly in the survey of endangered species (Dejean et al., 2012). Fourthly, the cost of eDNA surveys is relatively economical and time-efficient in comparison to conventional methods (e.g., electrofishing). Since its initial application in 2008, eDNA analysis technology has undergone over a decade of development and has matured into a pivotal tool in aquatic biological resource investigation. It is extensively applied in monitoring invasive species in specific regions, tracking the distribution of endangered species, and investigating the relationship between species distribution and climate change under the backdrop of global climate change.
The eDNA technology can achieve early warning and monitoring, and provide technical and decision-making references for eradication and management strategies. However, solely relying on comparative literature studies makes it challenging to assess the efficiency of each approach or establish a definitive eDNA protocol (Xing et al., 2022). As a result, even though eDNA methods have matured, decision-makers have been hesitant to treat eDNA-positive results as independent evidence of new invasions (i.e., lacking evidence from non-molecular methods). They perceive eDNA detection to be highly uncertain and lacking guidance on integrating this uncertainty into decisions on how and when to take action (Jerde, 2021). This is mainly due to several shortcomings of eDNA technology. Firstly, monitoring outcomes can only confirm the presence of target species in water bodies, without providing insights into the physiological status, growth, and developmental stages of those species. Secondly, further research is needed to establish the consistency and relationships between eDNA survey results and the spatiotemporal distribution of target species. The eDNA detection outcomes are significantly influenced by eDNA generation, transport, and degradation processes, with degradation playing a decisive role. Quantifying and presenting the reliability of eDNA results is a pivotal challenge that the development of eDNA technology must address. A notable area of focus for researchers is the quantification of these factors. For instance, this involves investigating eDNA drift in water, as well as examining the impact of various environmental factors on eDNA degradation (Barnes et al., 2014; Jane et al., 2014). Thirdly, the results of eDNA technology are highly dependent on existing databases, but existing databases may suffer from incomplete comparative data, and thus it may appear that some species in the results are difficult to identify. Moreover, the accuracy of different target genes varies widely across taxa, and some selected target genes may not have matching databases for comparison nowadays. Furthermore, challenges stem from potential cross-contamination during sample collection, transportation, and preservation processes. Contamination issues persist during laboratory analysis due to sample-reagent interactions, along with potential PCR inhibitors present in the samples, all of which could introduce biases and distort the experimental outcomes. In summary, the limitations of eDNA technology encompass its inability to provide comprehensive physiological insights, the need for further research on the consistency of survey results, the challenge of quantifying reliability, and the potential for contamination throughout the process. Overcoming these limitations is integral to enhancing the accuracy and applicability of eDNA technology.
5 Prospects of eDNA technology in the study of aquatic invasions
Biological invasions have resulted in significant declines in biodiversity and incurred substantial socio-economic losses and monetary expenses (Havel et al., 2015). These costs are severely underestimated and show no signs of abating, increasing threefold every decade, with damage costs estimated to be one order of magnitude higher than management expenditures (Diagne et al., 2021). Reasonable management actions and governance policies can alleviate the burdens imposed by invasive alien species. This article takes China’s 4E strategy for biological invasions as an example, analyzing the prospective application of eDNA technology in the research of aquatic biological invasions (Wan, 2022). This analysis aims to provide decision-making references and theoretical foundations for monitoring aquatic invasions.
E1 Action refers to Prevention and Early Warning, including four stages: Data-Driven Predictions, Quantitative Risk Alerts, Colonization Area Assessment, and Early Expansion Anticipation. For high-risk invasive species not yet present in East Asia, which possess strong invasiveness and potential harm, it is crucial to conduct a scientific analysis to evaluate their invasion risk and potential risk areas. eDNA technology is valuable for early prevention and warning of such high-risk species. By referencing published eDNA analysis models for high-risk invasive species, insights can be gained into their invasion patterns, trajectories, and more, allowing for the adjustment of targeted strategies for dealing with these high-risk species in East Asia. For instance, a new eDNA detection method has been developed for Pacifastacus leniusculus, Faxonius limosus, and Faxonius immunis, enabling year-round monitoring with high sensitivity. This method can also be used to specifically search for populations that have not been previously recorded or have newly emerged. Additionally, with established spatiotemporal elements considered, the use of quantitative ddPCR can further estimate population size roughly. Therefore, experimental results indicate that eDNA detection serves as a supplementary survey tool, especially for extensive screening or year-round monitoring in watersheds with limited data (Chucholl et al., 2021).
E2 Action entails Detection and Monitoring, which includes four stages: Molecular Identification Detection, Image Recognition Diagnosis, Remote Smart Monitoring, and Regional Tracking Detection. eDNA technology is primarily used for Molecular Identification Detection, enabling the early detection of the presence of a species in a new environment. This aids in early-stage management and control of the species, preventing more significant harm. For example, research has utilized eDNA technology to detect the invasive European species Rangia cuneata. It demonstrated that even in cases of sparse populations, R. cuneata could be detected in environmental DNA, allowing for rapid management responses and the tracking of invasive dynamics (Ardura et al., 2015).
E3 Action involves Eradication and Interception, including four stages: Early Eradication and Extinction, Corridor Node Interception, Ecological Barrier Segmentation, and Source Control in Quarantined Areas. eDNA technology can rapidly and extensively detect the distribution range of invasive species. It can serve as a reference for analyzing the invasion stage and path of invasive species, allowing for early eradication and interception, effectively suppressing rapid spread and reducing damage. For instance, optimization and validation of a qPCR detection method based on the H2B histone gene was conducted to quantify the co-infection levels of zebra and quagga mussels in environmental DNA samples. It was demonstrated that a highly specific qPCR detection method for environmental DNA can be an important tool for monitoring the locations of numerous invasive mussel species, with a focus on preventing the establishment of mussels in new locations (Peñarrubia et al., 2016).
E4 Action refers to Joint Control and Disaster Reduction, encompassing four stages: Traditional Biological Control, Ecological Substitute Restoration, Regional Joint Prevention and Control, and Cross-Border Collaborative Governance. eDNA technology can be used to detect the effectiveness of invasive species management efforts by comparing the concentration of residual DNA before and after control measures. This can help determine whether control methods are effective and which methods are more efficient. For instance, ddPCR technology was used to quantify the abundance of L. catesbeianus. The study showed that tadpole abundance and biomass explained 99% of the variation in eDNA concentration. Therefore, eDNA concentration can serve as an approximate value for the local bullfrog abundance in natural populations. This demonstrates that eDNA technology can be a robust and reliable tool for detecting the early stages of bullfrog invasion and quantifying changes in abundance over time, aiding in coordinating large-scale bullfrog eradication plans and evaluating their efficiency (Everts et al., 2021).
The integration of the 4E China Program with eDNA technology demonstrates the potential of eDNA technology to provide efficient and reliable reference data for early monitoring and warning of aquatic invasive species and the management of invasive species. This combination can offer strong support for the effective prevention and control of aquatic invasive species and the monitoring and protection of ecosystems. Therefore, we should acknowledge that with scientific evidence support, the eDNA method aligns with the legal standards accepted as evidence in most courts. This indicates that eDNA technology has matured into a sufficiently reliable technique. However, the question of whether eDNA methods have reached a level of maturity for transitioning from research to widespread application and integration into the management of aquatic invasive species is a critical issue to address. This is because decision-makers rarely incorporate uncertain outcomes into their decisions; false positive results may lead to unnecessary costs and inconveniences, and in more severe cases, it might trigger politically motivated reactions. Recognizing the doubts of managers regarding eDNA results, researchers have proposed solutions such as decision-support trees based on molecular best practices. These trees integrate the temporal and spatial trends of positive eDNA results relevant to human risk tolerance, thus narrowing the interface between results and management (Sepulveda et al., 2020). Furthermore, traditional morphology-based methods cannot be discarded. Despite their existing technical challenges, traditional morphological methods and eDNA technology must complement each other to ensure accurate and swift identification of aquatic invasive species. This collaboration also facilitates the analysis of the causes and consequences of biodiversity loss in ecosystems (Xiong et al., 2020). In general, routine monitoring of high-risk and high-frequency aquatic invasive species has become a management trend. For eDNA technology to better contribute to the 4E strategy, we must establish a robust biosafety risk prevention and control framework. This involves implementing a decentralized departmental management system under a coordinated mechanism, clarifying the biological security responsibilities of all stakeholders, and efficiently addressing the challenges posed by alien species invasions. Furthermore, it is essential to raise the awareness of research personnel and establish regular training programs. Continuous innovation is needed to address the existing limitations of eDNA technology. This includes the formulation of more standardized management protocols and the utilization of decision support frameworks to enhance the alignment between eDNA findings and management actions.
Author contributions
LW: Writing – original draft. WQ: Writing – review & editing, Supervision. FW: Writing – review & editing, Conceptualization, Supervision.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was funded by the National Key R&D Program of China (2022YFC2601500 & 2022YFC2601503) and the Natural Science Foundation of Shenzhen (JCYJ20190813120401662).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ardura A., Zaiko A., Martinez J. L., Samulioviene A., Semenova A., Garcia-Vazquez E. (2015). eDNA and specific primers for early detection of invasive species-A case study on the bivalve Rangia cuneata, currently spreading in Europe. Mar. Environ. Res. 112, 48–55. doi: 10.1016/j.marenvres.2015.09.013
Banerjee P., Stewart K. A., Dey G., Sharma R. K., Maity J. P., Chan M. W., et al. (2022). When conventional methods fall short: identification of invasive cryptic Golden Apple Snails (Pomacea canaliculata; P. maculata) using environmental DNA. Hydrobiologia 849 (19), 4241–4257. doi: 10.1007/s10750-022-04979-6
Barnes M. A., Turner C. R., Jerde C. L., Renshaw M. A., Chadderton W. L., Lodge D. M. (2014). Environmental conditions influence eDNA persistence in aquatic systems. Environ. Sci. Technol. 48 (3), 1819–1827. doi: 10.1021/es404734p
Blackburn D., Brown W. (2023). “University of florida herpetology,” in Occurrence dataset (Florida: Florida Museum of Natural History). doi: 10.15468/vw3dvj
Bohmann K., Evans A., Gilbert M. T. P., Carvalho G. R., Creer S., Knapp M., et al. (2014). Environmental DNA for wildlife biology and biodiversity monitoring. Trends Ecol. Evol. 29 (6), 358–367. doi: 10.1016/j.tree.2014.04.003
Boivin-Delisle D., Laporte M., Burton F., Dion R., Normandeau E., Bernatchez L. (2021). Using environmental DNA for biomonitoring of freshwater fish communities: Comparison with established gillnet surveys in a boreal hydroelectric impoundment. Environ. DNA 3 (1), 105–120. doi: 10.1002/edn3.135
Burger J. (2009). Red-eared slider turtles (Trachemys scripta elegans). Freshwater ecology and conservation lab (Freshwater Ecology and Conservation Lab, Washington: University of Washington).
Cai H., Chen R. (1990). A new harmful species-the giant apple snail. Guangdong Agric. Sci. 05, 36–38. doi: 10.16768/j.issn.1004-874x.1990.05.015
Chen J., Chen Z., Liu S., Guo W., Li D., Minamoto T., et al. (2021). Revealing an invasion risk of fish species in Qingdao underwater world by environmental DNA metabarcoding. J. Ocean U. China 20 (1), 124–136. doi: 10.1007/s11802-021-4448-2
Chen X., Fang J., Wang M., Shen Q., Sun Z., Wang B. (2021). Monitoring the invasive of Pomacea canaliculata via environmental DNA metabarcoding in Suzhou city. Plant Prot. 06), 58–65. doi: 10.16688/j.zwbh.2020389
Chucholl F., Fiolka F., Segelbacher G., Epp L. S. (2021). eDNA detection of native and invasive crayfish species allows for year-round monitoring and large-scale screening of lotic systems. Front. Environ. Sci. 9. doi: 10.3389/fenvs.2021.639380
Davies C. E., Batista F. M., Malkin S. H., Thomas J. E., Bryan C. C., Crocombe P., et al. (2019). Spatial and temporal disease dynamics of the parasite Hematodinium sp. in shore crabs, Carcinus maenas. Parasit. Vectors 12 (1), 472. doi: 10.1186/s13071-019-3727-x
Deiner K., Renshaw M. A., Li Y., Olds B. P., Lodge D. M., Pfrender M. E. (2017). Long-range PCR allows sequencing of mitochondrial genomes from environmental DNA. Methods Ecol. Evol. 8 (12), 1888–1898. doi: 10.1111/2041-210x.12836
Deiner K., Walser J. C., Mächler E., Altermatt F. (2015). Choice of capture and extraction methods affect detection of freshwater biodiversity from environmental DNA. Biol. Conserv. 183, 53–63. doi: 10.1016/j.biocon.2014.11.018
Dejean T., Valentini A., Miquel C., Taberlet P., Bellemain E., Miaud C. (2012). Improved detection of an alien invasive species through environmental DNA barcoding: the example of the American bullfrog Lithobates catesbeianus. J. Appl. Ecol. 49 (4), 953–959. doi: 10.1111/j.1365-2664.2012.02171.x
Deng F., Zhang C., Zhao Y., Zhou Q., Zhang J. (2013). Diversity and community structure of the fishes in the headstream region of the Dongjiang River. Chin. J. Zool. 48 (2), 161–173. Available at: https://www.cabdirect.org/cabdirect/abstract/20133188485.
de Vries H., Lemmens M. (2023). “Observation.org, Nature data from around the World,” in Occurrence dataset (Observation.org). doi: 10.15468/5nilie
Diagne C., Leroy B., Vaissière A. C., Gozlan R. E., Roiz D., Jarić I., et al. (2021). High and rising economic costs of biological invasions worldwide. Nature 592 (7855), 571–576. doi: 10.1038/s41586-021-03405-6
Doi H., Inui R., Akamatsu Y., Kanno K., Yamanaka H., Takahara T., et al. (2017). Environmental DNA analysis for estimating the abundance and biomass of stream fish. Freshw. Biol. 62 (1), 30–39. doi: 10.1111/fwb.12846
Egan S. P., Barnes M. A., Hwang C. T., Mahon A. R., Feder J. L., Ruggiero S. T., et al. (2013). Rapid invasive species detection by combining environmental DNA with light transmission spectroscopy. Conserv. Lett. 6 (6), 402–409. doi: 10.1111/conl.12017
Ernst C. H. (1990). “Systematics, taxonomy, variation, and geographic distribution of the slider turtle,” in Life history and ecology of the slider turtle. Ed. Gibbons J.W. (Washington: Smithsonian Institution Press), 1, 56–67.
Everts T., Halfmaerten D., Neyrinck S., De Regge N., Jacquemyn H., Brys R. (2021). Accurate detection and quantification of seasonal abundance of American bullfrog (Lithobates catesbeianus) using ddPCR eDNA assays. Sci. Rep. 11 (1), 11282. doi: 10.1038/s41598-021-90771-w
Ficetola G. F., Miaud C., Pompanon F., Taberlet P. (2008). Species detection using environmental DNA from water samples. Biol. Lett. 4 (4), 423–425. doi: 10.1098/rsbl.2008.0118
Fornillos R. J. C., Sato M. O., Tabios I. K. B., Sato M., Leonardo L. R., Chigusa Y., et al. (2019). Detection of Schistosoma japonicum and Oncomelania hupensis quadrasi environmental DNA and its potential utility to schistosomiasis japonica surveillance in the Philippines. PloS One 14 (11), e0224617. doi: 10.1371/journal.pone.0224617
Geist J. A., Mancuso J. L., Morin M. M., Bommarito K. P., Bovee E. N., Wendell D., et al. (2022). The New Zealand mud snail (Potamopyrgus antipodarum): autecology and management of a global invader. Biol. Invasions 24, 905–938. doi: 10.1007/s10530-021-02681-7
Genitsaris S., Kormas K. A., Moustaka-Gouni M. (2009). Microscopic eukaryotes living in a dying lake (Lake Koronia, Greece). FEMS Microbiol. Ecol. 69 (1), 75–83. doi: 10.1111/j.1574-6941.2009.00686.x
Gippet J. M., Bertelsmeier C. (2021). Invasiveness is linked to greater commercial success in the global pet trade. Proc. Natl. Acad. Sci. 118 (14), e2016337118. doi: 10.1073/pnas.2016337118
Goldberg C. S., Sepulveda A., Ray A., Baumgardt J., Waits L. P. (2013). Environmental DNA as a new method for early detection of New Zealand mudsnails (Potamopyrgus antipodarum). Freshw. Sci. 32 (3), 792–800. doi: 10.1899/13-046.1
Gustavson M. S., Collins P. C., Finarelli J. A., Egan D., Conchúir R.Ó., Wightman G. D., et al. (2015). An eDNA assay for Irish Petromyzon marinus and Salmo trutta and field validation in running water. J. Fish Biol. 87 (5), 1254–1262. doi: 10.1111/jfb.12781
Halwart M. (1994). The golden apple snail Pomacea canaliculata in Asian rice farming systems: present impact and future threat. Int. J. Pest Manage. 40 (2), 199–206. doi: 10.1080/09670879409371882
Han Y. (2022). The invasion of the alien species alligator gar (Atractosteus spatula) all over China. Int. J. Aquacult. 12 (2), 1–6. doi: 10.5376/ija.2022.12.0002
Havel J. E., Kovalenko K. E., Thomaz S. M., Amalfitano S., Kats L. B. (2015). Aquatic invasive species: challenges for the future. Hydrobiologia 750, 147–170. doi: 10.1007/s10750-014-2166-0
Hayes K. A., Joshi R. C., Thiengo S. C., Cowie R. H. (2008). Out of South America: multiple origins of non-native apple snails in Asia. Divers. Distrib. 14 (4), 701–712. doi: 10.1111/j.1472-4642.2008.00483.x
Hulme P. E. (2006). Beyond control: wider implications for the management of biological invasions. J. Appl. Ecol. 43 (5), 835–847. doi: 10.1111/j.1365-2664.2006.01227.x
Hulme P. E. (2009). Trade, transport and trouble: managing invasive species pathways in an era of globalization. J. Appl. Ecol. 46 (1), 10–18. doi: 10.1111/j.1365-2664.2008.01600.x
Huner J. V. (1988). “Procambarus in north america and elsewhere,” in Freshwater crayfish: biology, management and exploitation. Eds. Holdich D. M., Lowery R. S. (London: Croom Helm Publisher), 239–261.
Igawa T., Takahara T., Lau Q., Komaki S. (2019). An application of PCR-RFLP species identification assay for environmental DNA detection. PeerJ 7, e7597. doi: 10.7717/peerj.7597
Ikeda K., Doi H., Terui S., Kato A., Mitsuzuka T., Kawai T., et al. (2019). Estimating native and invasive crayfish distributions in relation to culvert barriers with environmental DNA. Freshw. Sci. 38 (3), 629–641. doi: 10.1086/704998
iNaturalist contributors. (2023). “iNaturalist research-grade observations,” in Occurrence dataset https (iNaturalist.org). doi: 10.15468/ab3s5x
Ishikawa T., Tachihara K. (2014). Introduction history of non-native freshwater fish in Okinawa-jima Island: ornamental aquarium fish pose the greatest risk for future invasions. Ichthyol. Res. 61 (1), 17–26. doi: 10.1007/s10228-013-0367-6
Jane S. F., Wilcox T. M., McKelvey K. S., Young M. K., Schwartz M. K., Lowe W. H., et al. (2014). Distance, flow and PCR inhibition: eDNA dynamics in two headwater streams. Mol. Ecol. Resour. 15 (1), 216–227. doi: 10.1111/1755-0998.12285
Jerde C. L. (2021). Can we manage fisheries with the inherent uncertainty from eDNA? J. Fish Biol. 98 (2), 341–353. doi: 10.1111/jfb.14218
Jones R. W., Weyl O. L., Swartz E. R., Hill M. P. (2013). Using a unified invasion framework to characterize Africa’s first loricariid catfish invasion. Biol. Invasions 15, 2139–2145. doi: 10.1007/s10530-013-0438-7
Kakuda A., Doi H., Souma R., Nagano M., Minamoto T., Katano I. (2019). ). Environmental DNA detection and quantification of invasive red-eared sliders, Trachemy scripta elegans, in ponds and the influence of water quality. PeerJ 7, e8155. doi: 10.7717/peerj.8155
Kawai T., Kobayashi Y. (2005). Origin and current distribution of the alien crayfish, Procambarus clarkii (Girar) in Japan. Crustaceana 78 (9), 1143–1149. doi: 10.1163/156854005775361034
Keskin E. (2014). Detection of invasive freshwater fish species using environmental DNA survey. Biochem. Syst. Ecol. 56, 68–74. doi: 10.1016/j.bse.2014.05.003
Khaefi R., Esmaeili H. R., Zareian H., Babaei S. (2014). The first record of the redbelly tilapia, Tilapia zillii (Gervai), in freshwaters of Iran. Turk. J. Zool. 38 (1), 96–98. doi: 10.3906/zoo-1304-3
Kim P., Yoon T. J., Shin S. (2021). Monthly eDNA monitoring of an invasive Bryozoan, Bugulina californica, in seawater using species-specific markers. Animals 11 (7), 1966. doi: 10.3390/ani11071966
Knight J. M. (2010). Invasive ornamental fish: a potential threat to aquatic biodiversity in peninsular India. J. Threat. Taxa 2 (2), 700–704. doi: 10.11609/JoTT.o2179.700-4
Lam I. P., Sung Y. H., Lin L., Fong J. J. (2020). Developing quantitative PCR assays to detect threatened and invasive freshwater turtles in Hong Kong using environmental DNA. Conserv. Genet. Resour. 12, 293–300. doi: 10.1007/s12686-019-01103-0
Lever C. (2003). Naturalized amphibians and reptiles of the world. (New York: Oxford University Press).
Li H., Deng X., Ye W., Ruan S. (1984). Introduction of a good strain of freshwater fish-the leather catfish. Freshw. Fish. 01, 7–12.
Li J. L., Dong Z. G., Li Y. S., Wang C. H. (2007). Alien aquatic plants and animals in China. (Shanghai: Shanghai Scientific and Technical Publishers).
Li J., Hatton-Ellis T. W., Lawson Handley L. J., Kimbell H. S., Benucci M., Peirson G., et al. (2019). Ground-truthing of a fish-based environmental DNA metabarcoding method for assessing the quality of lakes. J. Appl. Ecol. 56 (5), 1232–1244. doi: 10.1111/1365-2664.13352
Li J., Lawson Handley L.-J., Read D. S., Hänfling B. (2018). The effect of filtration method on the efficiency of environmental DNA capture and quantification via metabarcoding. Mol. Ecol. Resour. 18 (5), 1102–1114. doi: 10.1111/1755-0998.12899
Lin M., Zhang S., Yao M. (2019). Effective detection of environmental DNA from the invasive American bullfrog. Biol. Invasions 21, 2255–2268. doi: 10.1007/s10530-019-01974-2
Liu D., Xie C., Jim C. Y., Liu Y., Hou S. (2023). Predicting the potential distribution of the alien invasive alligator garspatula in China. Sustainability 15 (8), 6419. doi: 10.3390/su15086419
Lodge D. M., Deines A., Gherardi F., Yeo D. C. J., Arcella T., Baldridge A. K., et al. (2012). Global introductions of crayfishes: evaluating the impact of species invasions on ecosystem services. Annu. Rev. Ecol. Evol. Syst. 43 (1), 449–472. doi: 10.1146/annurev-ecolsys-111511-103919
Loureiro T. G., Anastácio P. M. S. G., Araujo P. B., Souty-Grosset C., Almerão M. P. (2015). Red swamp crayfish: biology, ecology and invasion-an overview. Nauplius 23, 1–19. doi: 10.1590/s0104-64972014002214
Lovell S. J., Stone S. F., Fernandez L. (2006). The economic impacts of aquatic invasive species: a review of the literature. Agric. Resour. Econ. Rev. 35 (1), 195–208. doi: 10.1017/s1068280500010157
Lowe S., Browne M., Boudjelas S., De Poorter M. (2000). 100 of the world’s worst invasive alien species: a selection from the global invasive species database Vol. Vol. 12) (Auckland: Invasive Species Specialist Group).
Ma K., Shi H. (2017). “Red-Eared Slider Trachemys scripta elegans (Wied-Neuwied),” in Biological Invasions and Its Management in China. Ed. Wan F. H. (Singapore: Springer). 13, 49–76. doi: 10.1007/978-981-10-3427-5_4
MacKenzie D. I., Nichols J. D., Sutton N., Kawanishi K., Bailey L. L. (2005). Improving inferences in population studies of rare species that are detected imperfectly. Ecology 86 (5), 1101–1113. doi: 10.1890/04-1060
Manton I. (1966). Observations on scale production in Prymnesium parvum. J. Cell Sci. 1 (3), 375–380. doi: 10.1242/jcs.1.3.375
Matsukura K., Okuda M., Kubota K., Wada T. (2008). Genetic divergence of the genus Pomacea (Gastropoda: Ampullariidae) distributed in Japan, and a simple molecular method to distinguish P. canaliculata and P. insularum. Appl. Entomol. Zool. 43 (4), 535–540. doi: 10.1303/aez.2008.535
Mehta S. V., Haight R. G., Homans F. R., Polasky S., venette R. C. (2007). Optimal detection and control strategies for invasive species management. Ecol. Econ. 61 (2-3), 237–245. doi: 10.1016/j.ecolecon.2006.10.024
Minamoto T., Hayami K., Sakata M. K., Imamura A. (2019). Real-time polymerase chain reaction assays for environmental DNA detection of three salmonid fish in Hokkaido, Japan: Application to winter surveys. Ecol. Res. 34 (1), 237–242. doi: 10.1111/1440-1703.1018
Minamoto T., Uchii K., Takahara T., Kitayoshi T., Tsuji S., Yamanaka H., et al. (2017). Nuclear internal transcribed spacer-1 as a sensitive genetic marker for environmental DNA studies in common carp Cyprinus carpio. Mol. Ecol. Resour. 17 (2), 324–333. doi: 10.1111/1755-0998.12586
Mito T., Uesugi T. (2004). Invasive alien species in Japan: the status quo and the new regulation for prevention of their adverse effects. Global Environ. Res. 8 (2), 171–193.
Mizumoto H., Kishida O., Takai K., Matsuura N., Araki H. (2022). Utilizing environmental DNA for wide-range distributions of reproductive area of an invasive terrestrial toad in Ishikari river basin in Japan. Biol. Invasions 24 (4), 1199–1211. doi: 10.21203/rs.3.rs-246449/v1
Mohiuddin M., Schellhorn H. E. (2015). Spatial and temporal dynamics of virus occurrence in two freshwater lakes captured through metagenomic analysis. Front. Microbiol. 6. doi: 10.3389/fmicb.2015.00960
Nakao R., Miyata R., Nakamura N., Muramatsu M., Okamura H., Imamura F., et al. (2023). Development of environmental DNA chip for monitoring the invasive alien fishes in dam reservoirs. Landsc. Ecol. Eng. 19 (1), 33–41. doi: 10.1007/s11355-022-00513-x
Nathan L. R., Jerde C. L., Budny M. L., Mahon A. R. (2015). The use of environmental DNA in invasive species surveillance of the Great Lakes commercial bait trade. Conserv. Biol. 29 (2), 430–439. doi: 10.1111/cobi.12381
Nentwig W., Bacher S., Kumschick S., Pyšek P., Vilà M. (2018). More than “100 worst” alien species in Europe. Biol. Invasions 20 (6), 1611–1621. doi: 10.1007/s10530-017-1651-6
Nur M. N., Ulayya N., Azis M., Maryanto A. E., Andayani N. (2020). Methods to maximize environmental DNA (eDNA) for detection the presence of Alligator Gar (Atractosteus spatula). IOP Conf. Series: Earth Environ. Sci. 538, 12018. doi: 10.1088/1755-1315/538/1/012018
Ogata S., Doi H., Igawa T., Komaki S., Takahara T. (2022). Environmental DNA methods for detecting two invasive alien species (American bullfrog and red swamp crayfish) in Japanese ponds. Ecol. Res. 37 (6), 701–710. doi: 10.1111/1440-1703.12341
Ogram A., Sayler G. S., Barkay T. (1987). The extraction and purification of microbial DNA from sediments. J. Microbiol. Meth. 7 (2-3), 57–66. doi: 10.1016/0167-7012(87)90025-x
Pawlowski J., Bruce K., Panksep K., Aguirre F. I., Amalfitano S., Apothéloz-Perret-Gentil L., et al. (2022). Environmental DNA metabarcoding for benthic monitoring: A review of sediment sampling and DNA extraction methods. Sci. Total Environ. 818, 151783. doi: 10.1016/j.scitotenv.2021.151783
Peñarrubia L., Alcaraz C., Vaate A. B. D., Sanz N., Pla C., Vidal O., et al. (2016). Validated methodology for quantifying infestation levels of dreissenid mussels in environmental DNA (eDNA) samples. Sci. Rep. 6 (1), 39067. doi: 10.1038/srep39067
Pound K. L., Nowlin W. H., Huffman D. G., Bonner T. H. (2011). Trophic ecology of a nonnative population of suckermouth catfish (Hypostomus plecostomus) in a central Texas spring-fed stream. Environ. Biol. Fish. 90 (3), 277–285. doi: 10.1007/s10641-010-9741-7
Radhakrishnan K. V., Lan Z. J., Zhao J., Qing N., Huang X. L. (2011). Invasion of the African sharp-tooth catfish Clarias gariepinus (Burchel) in South China. Biol. Invasions 13, 1723–1727. doi: 10.1007/s10530-011-0004-0
Rondon M. R., August P. R., Bettermann A. D., Brady S. F., Grossman T. H., Liles M. R., et al. (2000). Cloning the soil metagenome: a strategy for accessing the genetic and functional diversity of uncultured microorganisms. Appl. Environ. Microb. 66 (6), 2541–2547. doi: 10.1128/aem.66.6.2541-2547.2000
Ruiz G. M., Carlton J. T. (2003). Invasive species: vectors and management strategies. (Australia: Island Press).
Sahu A., Kumar N., Singh C., Singh M. (2023). Environmental DNA (eDNA): Powerful technique for biodiversity conservation. J. Nat. Conserv. 71, 126325. doi: 10.1016/j.jnc.2022.126325
Scriver M., Marinich A., Wilson C., Freeland J. (2015). Development of species-specific environmental DNA (eDNA) markers for invasive aquatic plants. Aquat. Bot. 122, 27–31. doi: 10.1016/j.aquabot.2015.01.003
Secondi J., Dejean T., Valentini A., Miaud C., Audebaud B. (2016). Detection of a global aquatic invasive amphibian, Xenopus laevis, using environmental DNA. Amphibia-Reptilia 37 (1), 131–136. doi: 10.1163/15685381-00003036
Sepulveda A. J., Nelson N. M., Jerde C. L., Luikart G. (2020). Are environmental DNA methods ready for aquatic invasive species management? Trends Ecol. Evol. 35 (8), 668–678. doi: 10.1016/j.tree.2020.03.011
Shi H., Gong S., Liang W., Hong M., Fu L., Wang J. (2009). The situation of controlling the spread of the invasive species red-eared slider (Trachemys scripta elegans) in the wild environment of China. Bull. Biol. 04, 1–3.
Shimada K., Urabe M. (2003). Comparative ecology of the alien freshwater snail Potamopyrgus antipodarum and the indigenous snail. Semisulcospira spp. Japanese J. Malacology (Japan). 62 (1-2), 39–53, 2003. doi: 10.18941/venus.62.1-2_39
Sing L. C., Tan K. S. (2018). Challenges in managing marine bio-invasions via shipping in Singapore. ASEAN J. Sci. Technol. Dev. 35 (1-2), 125–132. doi: 10.29037/ajstd.482
Skelton J., Cauvin A., Hunter M. E. (2022). Environmental DNA metabarcoding read numbers and their variability predict species abundance, but weakly in non-dominant species. Environ. DNA 00, 1–13. doi: 10.1002/edn3.355
Skinner M., Murdoch M., Loeza-Quintana T., Crookes S., Hanner R. (2020). A mesocosm comparison of laboratory-based and on-site eDNA solutions for detection and quantification of striped bass (Morone saxatilis) in marine ecosystems. Environ. DNA 2 (3), 298–308. doi: 10.1002/edn3.61
Snyder D. E. (2003). Invited overview: conclusions from a review of electrofishing and its harmful effects on fish. Rev. Fish Biol. Fisher. 13 (4), 445–453. doi: 10.1007/s11160-004-1095-9
Suter L., Polanowski A. M., Clarke L. J., Kitchener J. A., Deagle B. E. (2020). Capturing open ocean biodiversity: comparing environmental DNA metabarcoding to the continuous plankton recorder. Mol. Ecol. 30 (13), 3140–3157. doi: 10.1111/mec.15587
Taberlet P., Coissac E., Hajibabaei M., Rieseberg L. H. (2012). Environmental DNA. Mol. Ecol. 21 (8), 1789–1793. doi: 10.1111/j.1365-294X.2012.05542.x
Takahara T., Minamoto T., Doi H. (2013). Using environmental DNA to estimate the distribution of an invasive fish species in ponds. PloS One 8 (2), e56584. doi: 10.1371/journal.pone.0056584
Takahara T., Minamoto T., Doi H. (2015). Effects of sample processing on the detection rate of environmental DNA from the Common Carp (Cyprinus carpio). Biol. Conserv. 183, 64–69. doi: 10.1016/j.biocon.2014.11.014
Takahara T., Minamoto T., Yamanaka H., Doi H., Kawabata Z. I. (2012). Estimation of fish biomass using environmental DNA. PloS One 7 (4), e35868. doi: 10.1371/journal.pone.0035868
Teo S. S. (2001). Evaluation of different duck varieties for the control of the golden apple snail (Pomacea canaliculata) in transplanted and direct seeded rice. Crop Prot. 20 (7), 599–604. doi: 10.1016/s0261-2194(01)00029-1
Tessler M., Brugler M. R., DeSalle R., Hersch R., Velho L. F. M., Segovia B. T., et al. (2017). A global eDNA comparison of freshwater bacterioplankton assemblages focusing on large-river floodplain lakes of Brazil. Microb. Ecol. 73, 61–74. doi: 10.1007/s00248-016-0834-5
Thomaz S. M., Kovalenko K. E., Havel J. E., Kats L. B. (2015). Aquatic invasive species: general trends in the literature and introduction to the special issue. Hydrobiologia 746, 1–12. doi: 10.1007/s10750-014-2150-8
Thomsen P. F., Willerslev E. (2015). Environmental DNA-An emerging tool in conservation for monitoring past and present biodiversity. Biol. Conserv. 183, 4–18. doi: 10.1016/j.biocon.2014.11.019
Tréguier A., Paillisson J. M., Dejean T., Valentini A., Schlaepfer M. A., Roussel J. M. (2014). Environmental DNA surveillance for invertebrate species: Advantages and technical limitations to detect invasive crayfish Procambarus clarkii in freshwater ponds. J. Appl. Ecol. 51 (4), 871–879. doi: 10.1111/1365-2664.12262
Tsuji S., Iguchi Y., Shibata N., Teramura I., Kitagawa T., Yamanaka H. (2018). Real-time multiplex PCR for simultaneous detection of multiple species from environmental DNA: An application on two Japanese medaka species. Sci. Rep. 8 (1), 9138. doi: 10.1038/s41598-018-27434-w
Tsuji S., Shibata N. (2021). Identifying spawning events in fish by observing a spike in environmental DNA concentration after spawning. Environ. DNA 3 (1), 190–199. doi: 10.1002/edn3.153
Tuten T., Nico L. G., Jelks H. L. (2009). Non-native suckermouth armored catfishes in Florida: description of nest burrows and burrow colonies with assessment of shoreline conditions. ANSRP Bulletin, Vol-09-1. Available at: https://www.researchgate.net/publication/235024259.
Uchii K., Doi H., Minamoto T. (2016). A novel environmental DNA approach to quantify the cryptic invasion of non-native genotypes. Mol. Ecol. Resour. 16 (2), 415–422. doi: 10.1111/1755-0998.12460
Ulayya N., Andayani N., Maryanto A. E. (2020). Development of environmental DNA approaches to detect alligator gar (Atractosteus spatula) from water samples. IOP Conf. Series: Earth Environ. Sci. 481, 12013. doi: 10.1088/1755-1315/481/1/012013
Villamagna A. M., Murphy B. R. (2010). Ecological and socio-economic impacts of invasive water hyacinth (Eichhornia crassipes): a review. Freshw. Biol. 55 (2), 282–298. doi: 10.1111/j.1365-2427.2009.02294.x
Wan F. H. (2022). Biological invasion: Chinese solution. Available at: https://news.gmw.cn/2022-09/20/content_36034919.htm [Accessed August 25, 2023].
Wan F. H., Guo J. Y., Wang D. H. (2002). Alien invasive species in China: their damages and management strategies. Biodiv. Sci. 10 (1), 119–125. doi: 10.17520/biods.2002014
Wang Y., Guo Z., Pearl C. A., Li Y. (2007). Body size affects the predatory interactions between introduced American Bullfrogs (Rana catesbeiana) and native anurans in China: an experimental study. J. Herpetol. 41 (3), 514–520. doi: 10.1670/0022-1511(2007)41[514:bsatpi]2.0.co;2
Wang X., Zhang H., Lu G., Gao T. (2022). Detection of an invasive species through an environmental DNA approach: The example of the red drum Sciaenops ocellatus in the East China Sea. Sci. total Environ. 815, 152865. doi: 10.1016/j.scitotenv.2021.152865
Wei N. (2018). Analysis of sediment eDNA variation in microcosms and urban water environment for development of a tool to monitor amphipod Grandidierella japonica. [doctoral thesis]. (Tokyo (JPN): University of Tokyo).
Wilcox T. M., Carim K. J., McKelvey K. S., Young M. K., Schwartz M. K. (2015). The dual challenges of generality and specificity when developing environmental DNA markers for species and subspecies of Oncorhynchus. PloS One 10 (11), e0142008. doi: 10.1371/journal.pone.0142008
Woodell J. D., Neiman M., Levri E. P. (2021). Matching a snail’s pace: successful use of environmental DNA techniques to detect early stages of invasion by the destructive New Zealand mud snail. Biol. Invasions 23 (10), 3263–3274. doi: 10.1007/s10530-021-02576-7
Wu Z. J., Wang Y. P., Li Y. M. (2004). Natural populations of bullfrog (Rana catesbeiana) and their potential threat in the east of Zhejiang Province. Biodiv. Sci. 12 (4), 441. doi: 10.17520/biods.2004054
Wu L., Wu Q., Inagawa T., Okitsu J., Sakamoto S., Minamoto T. (2023). Estimating the spawning activity of fish species using nuclear and mitochondrial environmental DNA concentrations and their ratios. Freshw. Biol. 68 (1), 103–114. doi: 10.1111/fwb.14012
Xie W. D., Wen Z. X., Song K., Guo B. C., Fang Y. (2023). and sun, YGlobal freshwater assessment of establishment risk of invasive Alligator gar (Atractosteus spatula) and risks to freshwater ecosystems in China. H.Zool. Res. 44 (1), 90. doi: 10.24272/j.issn.2095-8137.2022.368
Xing Y., Gao W., Shen Z., Zhang Y., Bai J., Cai X., et al. (2022). A review of environmental DNA field and laboratory protocols applied in fish ecology and environmental health. Front. Environ. Sci. 10. doi: 10.3389/fenvs.2022.725360
Xiong W., Guo C., Gozlan R. E., Liu J. (2023). Tilapia introduction in China: Economic boom in aquaculture versus ecological threats to ecosystems. Rev. Aquacult. 15 (1), 179–197. doi: 10.1111/raq.12710
Xiong W., Huang X., Chen Y., Fu R., Du X., Chen X., et al. (2020). Zooplankton biodiversity monitoring in polluted freshwater ecosystems: A technical review. Environ. Sci. Ecotechnol. 1, 100008. doi: 10.1016/j.ese.2019.100008
Xiong W., Li H., Zhan A. (2016). Early detection of invasive species in marine ecosystems using high-throughput sequencing: technical challenges and possible solutions. Mar. Biol. 163, 1–12. doi: 10.1007/s00227-016-2911-1
Xu H., Qiang S., Genovesi P., Ding H., Wu J., Meng L., et al. (2012). An inventory of invasive alien species in China. NeoBiota 15, 1–26. doi: 10.3897/neobiota.15.3575
Xu M., Yang Y., Zhang J., Fu X. (2023). Challenges in invasive mussel monitoring using environmental DNA technology. Acta Ecol. Sin. 11, 4423–4433. doi: 10.5846/stxb202111163208
Yan S. H., Song W., Guo J. Y. (2017). Advances in management and utilization of invasive water hyacinth (Eichhornia crassipes) in aquatic ecosystems-a review. Crit. Rev. Biotechnol. 37 (2), 218–228. doi: 10.3109/07388551.2015.1132406
Keywords: environmental DNA, aquatic invasion, East Asia, aquatic species monitoring, risk management, policy formulation
Citation: Wang L, Wan F and Qian W (2023) Research and prospects of environmental DNA (eDNA) for detection of invasive aquatic species in East Asia. Front. Mar. Sci. 10:1284953. doi: 10.3389/fmars.2023.1284953
Received: 29 August 2023; Accepted: 06 November 2023;
Published: 23 November 2023.
Edited by:
Meng Yan, City University of Hong Kong, Hong Kong SAR, ChinaReviewed by:
Li-Kun Yang, Ocean University of China, ChinaFeng Zhao, Chinese Academy of Sciences (CAS), China
Copyright © 2023 Wang, Wan and Qian. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fanghao Wan, wanfanghao@caas.cn; Wanqiang Qian, qianwanqiang@caas.cn
 Linjing Wang
Linjing Wang Fanghao Wan
Fanghao Wan Wanqiang Qian
Wanqiang Qian