Effects of temperature and microbial disruption on juvenile kelp Ecklonia radiata and its associated bacterial community
- 1School of Life and Environmental Sciences, The University of Sydney, Sydney, NSW, Australia
- 2Sydney Institute of Marine Science, Mosman, NSW, Australia
- 3School of Biological, Earth and Environmental Sciences, University of New South Wales, Sydney, NSW, Australia
- 4Institute of Marine and Antarctic Studies (IMAS), University of Tasmania, Hobart, TAS, Australia
- 5Singapore Centre for Environmental Life Sciences Engineering, Nanyang Technological University, Singapore, Singapore
Ocean warming can affect the development and physiological responses of kelps, and under future climate change scenarios, increasing seawater temperatures pose a major threat to these habitat-forming species. However, little is known about the effects of warming on epiphytic bacterial communities and how an altered microbiome may interact with temperature stress, affecting the condition and survival of kelp, particularly of the potentially more vulnerable early life stages. Here, we tested the effects of thermal stress on the growth and physiological responses of juvenile kelp Ecklonia radiata in which their epiphytic bacterial community was experimentally disrupted using antimicrobials, simulating dysbiosis. We hypothesized that, under thermal stress (23°C, simulating a extreme scenario of ocean warming in Tasmania), kelp with a disrupted bacterial community would be more strongly affected than kelp with an undisrupted microbiome or kelp under ambient temperature (14°C) but with a disrupted microbiota. Thermal stress reduced growth, increased tissue bleaching and negatively affected net photosynthesis of kelp. In addition, a substantial change in the epiphytic bacterial community structure was also found under thermal stress conditions, with an increase in the abundance of potentially pathogenic bacterial groups. However, microbial disruption did not act synergistically with thermal stress to affect kelp juveniles. These results suggest that effects of elevated temperature on juvenile kelps is not microbially-mediated and that juveniles may be less susceptible to disruptions of their microbiome.
Introduction
Microbial communities associated with host macro-organisms are a crucial component of many coastal ecosystems and hold an important role in the survival, performance and reproduction of many aquatic organisms (Abreu et al., 2022). Host-microbiome relationships have been studied in many marine biological systems (Apprill, 2017) providing evidence that hosts and their associated microbiomes constitute functional ecological units, i.e. “holobionts” (Dittami et al., 2021).
In temperate coastal environments, kelps are often dominant organisms, playing an essential role as habitat-forming species (i.e., shelter or nursery effects; Hollants et al., 2013) and strongly contributing to local primary productivity (Dolliver and O’Connor, 2022). Kelp-associated epiphytic bacterial communities are composed of diverse generalist and specialist groups that are prevalent across multiple macroalgal species (e.g. Weigel et al., 2022) and form important interactions that can be both beneficial and detrimental to the host (Hollants et al., 2013). For example, bacterial communities may provide their host with nutrients (e.g. vitamin synthesis and nitrogen cycling; Croft et al., 2005 and Pfister et al., 2019), biosynthesis of antibacterial compounds (including terpenes and polyphenols; Goecke et al., 2010) used for pathogenic defence and antifouling (Kanagasabhapathy et al., 2006), and can influence zoospore settlement (e.g. bacterial biofilm produce chemical signalling to guide zoospore settlement on host surfaces; Joint et al., 2002), recruitment (e.g. facilitate gametophyte growth; Morris et al., 2016) and growth (e.g. enhances sporophyte development through nutrient exchange; Singh and Reddy, 2014). Disruption to host-microbiota interactions can therefore have detrimental effects on the kelp-holobiont, with potential consequences to the ecosystems they underpin including increases in pathogenic invasions (e.g. Qiu et al., 2019 with ocean warming), changes in kelp population dynamics (i.e. impacts on growth, survival, and reproduction) and disruption of biogeochemical cycles, and food webs (Wilkins et al., 2019). Importantly, such disruptions may exacerbate the effects of environmental stressors on the host because the importance of beneficial interactions between hosts and their microbiota appears to be strongest under environmental stress (Fuggle et al., 2023).
Habitat-forming kelps are presently under increasing threat due to climate change and ocean warming which are direct factors that have severely decreased kelp populations, affected their associated microbiome (Qiu et al., 2019) and in turn impacted the ecosystem services and processes they regulate (Wernberg et al., 2011; Monserrat et al., 2022). Temperature is one of the main drivers that can pose a direct risk to habitat forming species (Ji and Gao, 2021) but can also become a long lasting stressor under current scenarios of increasing temperatures due to climate change. Changes on the overall density, diversity, composition and specific taxa of host-associated bacterial communities occur as a consequence of increasing temperatures (Stratil et al., 2013; Minich et al., 2018) and has been seen to lead to detrimental effects on kelp development and their defence mechanisms against pathogens (Minich et al., 2018; Qiu et al., 2019). However, other studies have also determined that temperature variability across sites (Weigel and Pfister, 2019) and seasonal effects (e.g. Nereocystis luetkeana with a persisting reservoir of microbes across years in this annual species, Weigel et al., 2022), may not severely affect kelp-associated microbiomes and their functions which can persist through time even with marked seasonal changes, suggesting that kelp-associated microbiomes may be resilient to temperature variation, as seen in other systems (Philippot et al., 2021). Differences in the temperature response of kelps across studies, may not only be a reflection of direct differences driven by kelp species specific thermal tolerances, life stage, genotype or the magnitude of warming events; but also other indirect factors that involve changes in their habitat (e.g. shift in ecological interactions) or synergic effects with other stressors (Smale, 2020) Thus, if kelp already living within a stressful scenario (e.g., increased temperature) are affected by a discrete pulse disturbance (e.g. nutrients, xenobiotic effluents or salinity changes) that creates an altered state of the bacterial communities (i.e. dysbiosis, Plante, 2017) , these factors may interact, strongly affecting host survival and performance.
Here, we experimentally examined the interactions between temperature stress and microbiota disruption on a key habitat-forming kelp, Ecklonia radiata (Ecklonia hereafter). Ecklonia is a dominant subtidal laminarian kelp that is broadly distributed along ~8,000 km of subtropical and temperate rocky reefs in Australia (Wernberg et al., 2019). Ecklonia, forms large underwater forests that provide important local ecosystem services (Bennett et al., 2016) but are highly threatened by ocean warming (Wernberg et al., 2011). We exposed juvenile kelp to ambient and stress-inducing temperatures (simulating a relevant ocean warming scenario; Britton et al. in review) and experimentally disrupted their epiphytic bacterial community using antibiotic and antiseptic agents. It was hypothesised that Ecklonia with a disrupted microbiome would have lower growth, poor condition (i.e., higher erosion and bleached tissue) and physiology (e.g. lower oxygen production through photosynthesis) than Ecklonia with an undisrupted microbiome and that such effects would be strongest on kelp under thermal stress. Finally, we hypothesised that changes in host performance would be related with changes in their associated bacterial community structure (and predicted function) throughout the experiment.
Materials and methods
Field collection and experimental setup
Juvenile Ecklonia were haphazardly collected from subtidal rocky reefs at a depth of 7 m at Coal Point, Bruny Island, in southeast Tasmania (-43.33430 S, 147.32493 W) in the austral autumn (May 2022). Immediately after collection, juveniles were placed inside plastic bags with water from the site, stored in cold containers (~10°C) and transported to the lab at the Institute of Marine and Antarctic Studies (IMAS, University of Tasmania). Juvenile Ecklonia (defined as sporophytes with a length <15 cm and with a linear oblong blade with low spinousity; Stage I, Kirkman, 1981) were selected for this study as they are predicted to be more sensitive than adults to disturbances and stressors such as temperature (Coelho et al., 2000).
Upon arrival to the lab, 64 juveniles were selected based on i) low abundance of visible epibionts and ii) no visible signs of obvious stress (i.e., bleaching or blade erosion). Autoclaved filtered seawater (AFSW, filtered at 0.2 µm and sterilized for 2h at 121°C) was used to remove any visible epibionts from the juveniles and then placed individually into an experimental setup with temperature monitored and kept constant (± 0.2°C) with a specific light regime (see details of this setup and light regime in Figure 1A based on, Britton et al. in review). A low concentration of nutrients was provided within each chamber to avoid any stress on the juveniles due to nutrient limitation (nutrient source: Sodium nitrate, NaNO3 CAS: 7631-99-4, Merck; to an initial concentration of 5 µM per chamber). Juveniles were left in this setup overnight and after this period they were attached via the holdfast to autoclaved stones using UV sterilized rubber bands (see Figure 1A).
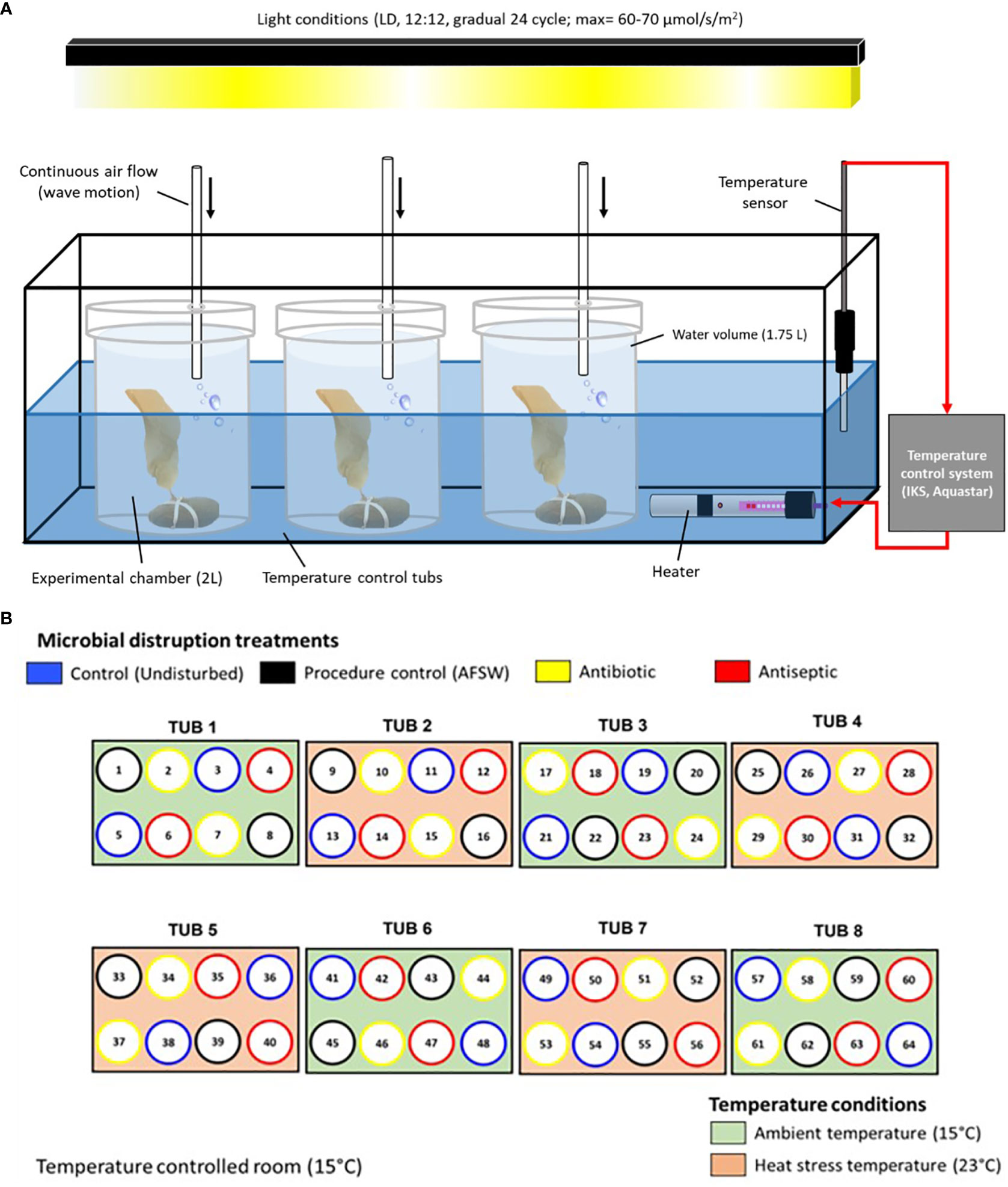
Figure 1 Experimental setup showing (A) placement of Ecklonia juveniles within experimental chambers, temperature control tubs and light regulation. Light conditions in the experimental setup were kept under a pre-programmed regime in which light intensity was gradually varied throughout a 24 h cycle (i.e., Dark period: 0 µmol photons m-2 s-1; light period: morning/afternoon average intensity = 25-35 µmol photons m-2 s-1 and midday maximum = 60-70 photons m-2 s-1, LED lights, Zeus-70, Ledzeal, Hong Kong). (B) bird eye view of the arrangement of the 8 temperature control tubs each holding 8 experimental chambers. Color coding shows the random assignment of microbial disruptions treatments and temperature conditions within the temperature-controlled room. All juveniles allocated to microbial disruption treatments were exposed every 2-3 days to simulate a press disturbance.
Experimental design and treatment allocation
After being placed in the experimental setup, juveniles were randomly allocated to 2 temperature conditions: 1) ambient (15°C, simulating their natural thermal optimum, Visch et al., 2023; n=32) and 2) a temperature designed to induce thermal stress (23°C, simulating an extreme scenario of ocean warming in Tasmania, CSIRO and Bureau of Meteorology, 2020 n=32). Temperature was gradually increased (2°C per day for four days) until reaching the expected value for the heat condition (23°C). Experimental start time for the heat condition was staggered due to this gradual temperature increase. No heating was applied to ambient conditions and were left to reach the expected temperature (15°C) through exposure to the temperature set for the room.
Juveniles were left for 5 days to acclimatise to their corresponding temperature conditions before randomly allocating them to 4 microbial disruption treatments: 1) Controls (undisrupted juveniles, n=8), 2) Procedural controls (PC, disturbed with AFSW for 30 mins, n=8), 3) Antibiotics (Antibiotic Antimycotic Solution, Sigma-Aldrich®, 2% in AFSW for 30 mins, n=8) and 4) Antiseptic (Povidone-Iodine solution, Betadine®, 2% in AFSW, 1 min, n=8). Details of treatment allocation, the chemical composition and details on application of these solutions are given in Figure 1B and Supplementary Table 1, based on pilot work with Ecklonia juveniles in these same systems. Pilot work showed that these treatments successfully disrupted the microbiota but did not have direct effects on the host (see additional Supplementary Figure 1 for more details on the pilot work). Juveniles were returned to new experimental chambers after exposure to the microbial disruption treatments. Water changes were conducted every two days in which juveniles were transferred to new sterile chambers with clean filtered sea water and new nutrient availability (5 µm NaNO3), as well as exposed again to their corresponding microbial disruption treatment. This was done to simulate a press disruption (Bender et al., 1984) which is thought to maintain the predicted bacterial disruption or dysbiosis (see pilot work, Supplementary Figure 1). The experiment lasted 16 days after 5 days of acclimatisation, at which juveniles were sampled for morphological, physiological and microbial analysis.
Host responses: absolute growth and condition indicators
Photographs of all the juveniles were taken at the start and at the end of the experiment to obtain different morphological values that were then used to calculate absolute growth rate (cm/day), blade erosion (cm/d) and blade bleaching (%). These morphological values include total blade length, stipe length and distance to a visible mark (used to determine blade growth). Blade surface area was also calculated but only used for microbial analysis (see below). Details of these calculations can be found in Supplementary Figure 2 and all morphological measurements were processed using the image processing software Fiji (Image J, Schindelin et al., 2012).
Host responses: physiological measurements
Net photosynthesis and respiration (µmol O2/h/g ww) were measured independently for each juvenile at the end of the experiment and similarly to methods detailed in Britton et al. (2020). Briefly, juveniles were placed within respirometry chambers (310 mL) with constant water motion, appropriate levels of irradiance and adjusted to the corresponding temperature condition (i.e., ambient vs heat). Measurements of dissolved oxygen were taken at the start and after 3 h of incubation with a portable oxygen sensor (i.e., DO, µmol O2/L, Fibox 4, PreSen). A similar procedure was done for dark respiration, with the difference that incubation was done in complete darkness and left overnight (i.e., 11 h). Water used within all chambers for both physiological measurements was AFSW and was gassed with N2 to ensure that all initial DO measurements started at ~70% saturation to avoid any oxygen oversaturation. Blanks with no juveniles in the chambers were included and used to correct values of oxygen concentration. Details of the calculations used for final values of the physiological responses are included in Supplementary Table 2.
Microbial responses: DNA processing and bioinformatics
At the end of the experiment, juveniles were sampled for microbial analysis by first removing loose epiphytic bacteria from the lamina using AFSW and then passing a sterilised swab across the whole surface of one side of the lamina for 30 s (as per Marzinelli et al., 2015). An unused swab was left in the experimental area for 30 s and was included as a blank to determine possible ambient contamination. Swabs were immediately frozen at -80°C until DNA extraction. DNA was extracted randomly from a subset of the swabs (N=40, considering n=5 in each microbial disruption treatment at each temperature condition) using a DNA extraction kit following manufacturer’s protocols (PowerSoil DNA Isolation kit, Qiagen ®). Procedural blanks were included by performing the DNA extraction protocol with sterile water. All samples were quantified (Nanodrop 1000, Thermo Fisher Scientific ®) and stored at -20°C. All DNA extracts were then sent to the Ramaciotti Centre for Genomics (University of New South Wales, Sydney) where PCR and sequencing were performed. An amplicon library was constructed by using universal primer pairs targeting the V3-V4 hypervariable region of the bacterial 16S rRNA gene (Klindworth et al., 2013; 314F, 5’- CCTACGGGNGGCWGCAG-3’; and 805R, 5’-GACTACHVGGGTATCTAATCC-’3) and a standard single-step PCR procedure. A second PCR procedure was performed to include index and adapter sequences for sample identification and multiplexing for the dual-indexed sequenciation. Sequencing was performed on an Illumina MiSeq v3 platform (2x300bp) using a reagent Kit v3 600-cycle and approximately 10% PhiX control spike-in.
Raw demultiplexed fastQ sequences were quality trimmed based on the maximum truncation lengths obtained upon inspection of quality error plots and expected base pair median Q scores (truncation lengths, Forward: 290 and reverse: 260; maximum expected errors= 0; and median Q scores < 20). An error-based model was constructed and used to optimize the denoising algorithm (i.e., DADA2) for sample inferences. Subsequently, demultiplexed sequences were merged, an ASV table constructed, and chimeric sequences removed. An initial taxonomic assignment was conducted using a Naive Bayes classifier trained from the SILVA v.138 database (Quast et al., 2013) and aimed at identifying sequences from non-bacterial sources (e.g. eukaryotes), chloroplast and mitochondrial DNA. To optimize the taxonomic assignment, a second classifier was applied, Genome Taxonomy Database (GTDB v.202, Parks et al., 2022), which provides a higher taxonomic resolution but poor chloroplast/mitochondrial identification. For both taxonomic assignments, a minimum bootstrap confidence = 50 was used for taxonomic identification to avoid any incongruencies during classification (ASV < 50 minBoot, were treated as unclassified taxa).The resulting ASV table was processed further by removing singletons, contaminating sequences (based on unused swabs and DNA extraction controls, R package decontam, Davis et al., 2018) and low abundant taxa (i.e. abundance < 0.001% across the whole dataset). A total of 98.52% of the original sequences remained in the processed ASV table. To effectively account for compositional, sparsity and inter sample heterogeneity biases, the ASV table was normalized using a Wrench method (Kumar et al., 2018; as a suggested best method of normalization, Swift et al., 2023). All the bioinformatic pipeline was based on the DADA2 workflow (R package dada, Callahan et al., 2016) and performed using R v.4.2.2.
Statistical analyses
Initially, the effects of temperature (fixed factor, 2 levels), microbial disruption treatments (fixed factor, 4 levels) and their interaction on host morphological and physiological responses, were evaluated through general linear mixed models including tub as a random factor (random terms evaluated through likelihood ratio test, package lmerTest, Kuznetsova et al., 2017). However, individual tub effects were not found to influence results at any level (all p values > 0.072) so analyses were simplified to include only fixed terms, their interaction and evaluated through general linear models (LM). All model assumptions (i.e., linearity, homogeneity of variance and normality) were validated (R package performance, Lüdecke et al., 2021) and any outlier or influential values were identified and removed (R packages, car, Fox and Weisberg, 2019; and EnvStats, Millard, 2013). Model inferences were calculated through an ANOVA (Type III, alpha<0.05, R package car, Fox and Weisberg, 2019) and if any significant differences were found within the levels of microbial disruption treatments, post hoc contrasts were performed (Tukey HSD; R package emmeans, Lenth, 2022) .
Alpha diversity indices (Richness and Shannon-Wiener diversity index) for the bacterial communities were calculated using the R package phyloseq (McMurdie and Holmes, 2013) and the influence of temperature and microbial disruption analysed as above. To further analyse the influence of host morphological and physiological responses on the associated bacterial community, linear Pearson correlations were calculated (R package corrplot, Wei et al., 2013). A multivariate analysis was also conducted to determine the effects of the fixed factors on the bacterial community. For this, the normalized ASV table was square-root transformed and a resemblance table was calculated using the Bray-Curtis dissimilarity index. An ASV accumulation curve was constructed with the transformed dataset to evaluate sampling effort across the whole community (R package vegan, Oksanen et al., 2013). A Permutational Analysis of Variance using Bray-Curtis dissimilarities was performed to determine compositional differences across the fixed factors and the interaction therein (Type III, PERMANOVA+ v.1.0.5 and Primer-e v.6.1.15). Estimates of variance components (EVC) were extracted from this analysis to further explore the magnitude of effect that each fixed factor or interaction had on the overall bacterial community. If compositional differences were found within the microbial disruption treatments, multivariate pairwise post hoc tests were done. To examine assumptions of the above analysis and also test hypotheses about variation in bacterial community structure, a multivariate homogeneity of group dispersion and a permutation test for each fixed factor was performed (PERMDISP, R package vegan, Oksanen et al., 2013 based on model validation proposed by Anderson, 2014). Multivariate structural results were visualized through a non-metric multidimensional scaling ordination (nMDS; R package vegan, Oksanen et al., 2013).
A distance-based redundancy analysis (dbRDA; R package vegan, Oksanen et al., 2013) was used to determine the influence of host morphological and physiological responses on the bacterial community composition. For this, a stepwise model selection was done using the Akaike information criteria (AIC) to determine the most optimum model using only the best host response predictors. Marginal tests (i.e., permutation tests) and estimation of linear trends (i.e., envfit, R package vegan, Oksanen et al., 2013) were used to determine the importance of each predictor to overall bacterial community composition. All host responses were not found to have strong multicollinearity (Variance Inflation factor <4) and were transformed and normalized to increase data homogeneity and reduce skewness.
To provide further evidence of specific bacterial taxa that could explain the observed compositional differences above, a multivariate generalised linear model assuming a negative binomial distribution was performed including all bacterial abundance data (R package mvabund, Wang et al., 2012). Overall significant differences between main fixed factors were determined by an analysis of deviance (alpha <0.05). Subsequently, for each ASV, univariate tests were used to identify the ASVs that presented significant differences in abundance across fixed factors (R package mvabund, Wang et al., 2012). As the package mvabund is unable to provide multiple pairwise comparisons between factor sublevels, separate linear models and post hoc tests similar to the above analyses were performed. For this, only the ten most abundant and prevalent ASVs across all samples were included.
An additional analysis was attempted to predict the potential functional metabolic capacity of the bacterial communities and how are these affected by the temperature conditions and microbial disruption (R package Tax4Fun2, Wemheuer et al., 2020). Briefly, a user specific genomic reference database for Ecklonia was constructed based on the default reference database in Tax4Fun2 (i.e., Ref100NR) and user provided metagenome-assembled genomes (MAGs; Song et al., 2022). For this, we incorporated user-defined data as metagenome-assembled genomes (MAGs) from Ecklonia radiata into Tax4Fun2 to increase its predictive power and accuracy (Song et al., 2022). Raw sequences from the present study were aligned against our user-designed reference database (i.e., BLAST) and ASVs abundances were compared to functional information to create a sample-specific functional profile. Finally this profiles were summarized to relevant metabolic pathways (KEGG pathways, Kanehisa and Goto, 2011). Final functional predictions only included ASV that passed a similarity threshold (>97%). A similar multivariate analysis (i.e., PERMANOVA) and dbRDA were done as above to detect if these predicted functional profiles differed across the fixed factors. Unfortunately, it was impossible to provide reliable results of functional predictions using this dataset (see details of the results for this analysis on Box S1).
Results
Host responses
Increased temperature significantly affected the growth, bleaching and physiology of Ecklonia juveniles. Juveniles in the thermal stress condition had reduced absolute growth rates (32% less; F1,53 = 4.11 and p<0.05) and a higher proportion of bleached tissue (70% increase; F1,53 = 5.88 and p= 0.02) compared to juveniles in ambient temperatures (Figures 2A, B; Table 1). Erosion rates over the experiment did not differ between the temperature conditions (Table 1, F1,53 = 2.61 and p=0.111). Thermal stress also decreased net photosynthesis (60% reduction, F1,53 = 7.48 and p<0.001) and respiration (68% reduction, F1,53 = 8.32 and p=0.006; Figures 2C, D and Table 1). No effects of microbial disruption treatments, nor their interaction with temperature, were detected for any of the host morphological or physiological responses (Table 1; overall p>0.06).
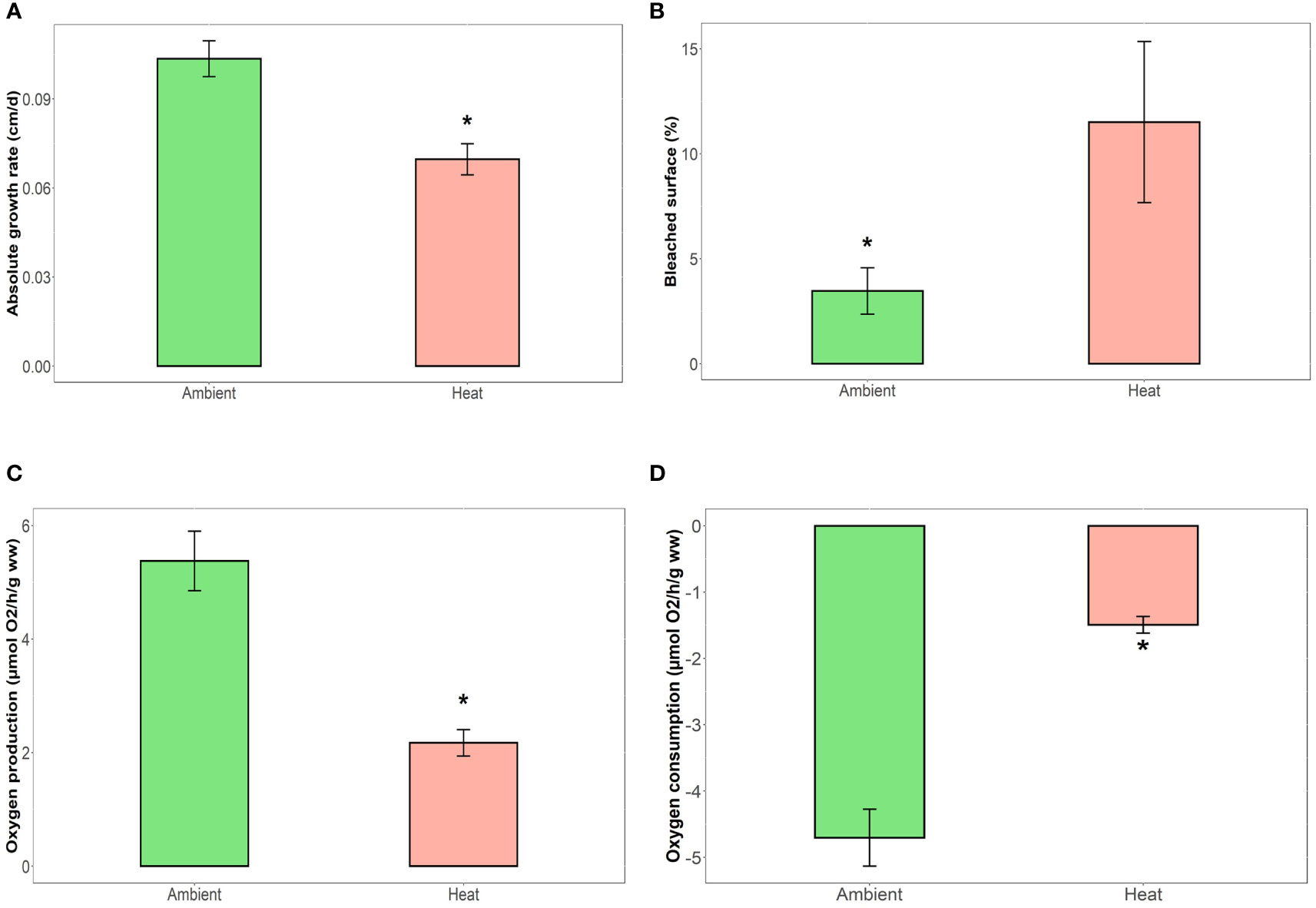
Figure 2 Effect of temperature condition (2 levels, Ambient [n=31] vs. Heat stress [n=30]) on host morphological and physiological responses including juvenile (A) absolute growth rate (cm/day), (B) laminar bleaching (%), (C) net photosynthesis (µmol O2/h/g ww) and (D) aerobic respiration (µmol O2/h/g ww, lower values indicate higher oxygen consumption). Values shown are mean ± SE and differences between temperature conditions are shown with an asterisk (*).
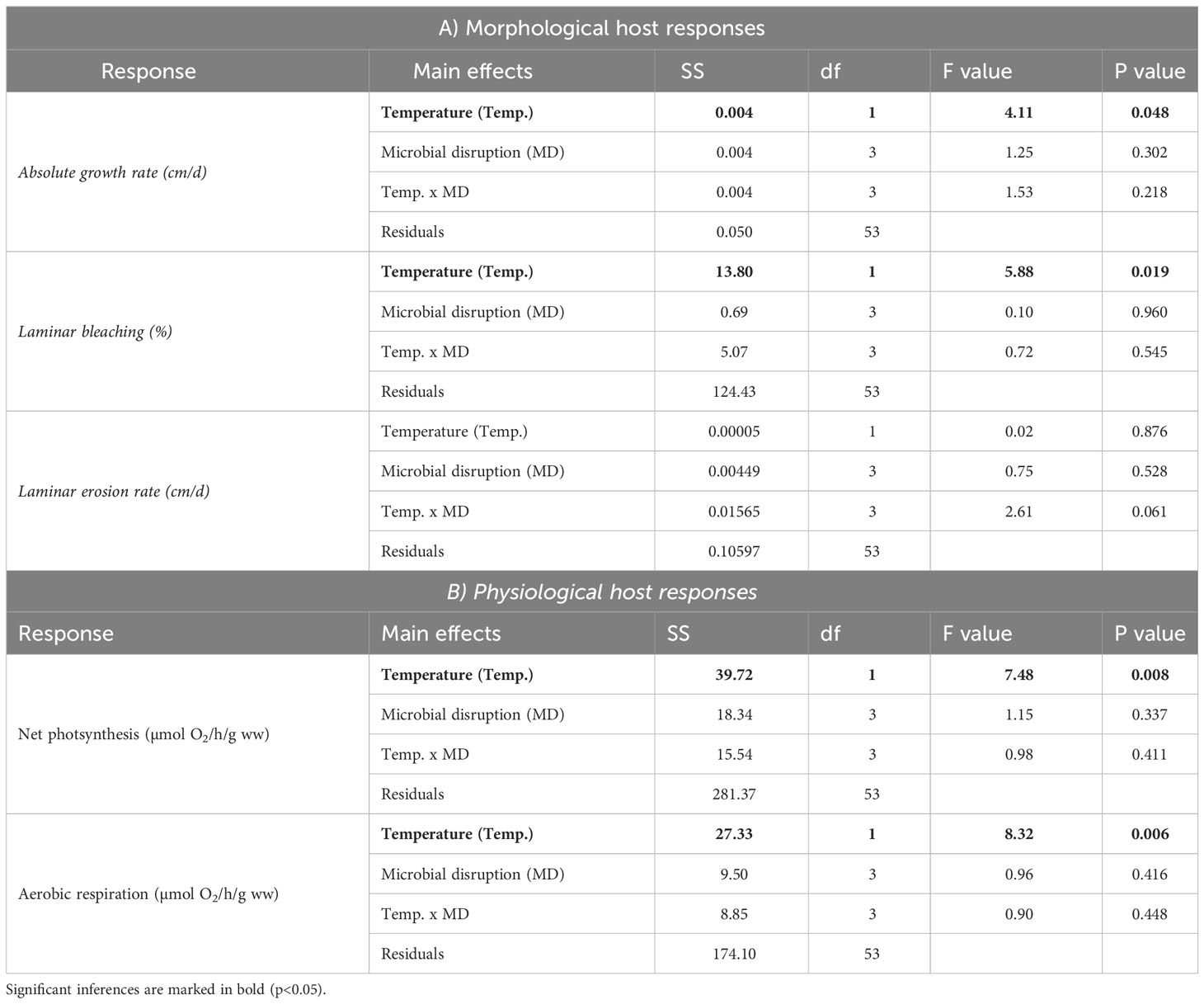
Table 1 Summary of the One-way ANOVA to determine differences in the (A) morphological and (B) physiological host responses caused by the experimental temperature conditions (2 levels, Ambient vs. Heat stress), Microbial disruption treatments (4 levels, Undisturbed, procedure control, antibiotics and antiseptic) and interactions between these main effects.
Responses of host associated bacterial communities
Following bioinformatic processing, 2256 ASVs were identified and used in subsequent analyses. Temperature had no effect on bacterial alpha diversity, but microbial disruption with antibacterials significantly affected bacterial diversity (Shannon-Wiener, Table 2 and Supplementary Figure 3), with bacterial diversity lower in treatments exposed to the antiseptic solution compared to undisrupted juveniles (Supplementary Figure 5, t ratio,3,32 = 3.57 and p=0.006). Bacterial richness and diversity were not correlated with morphological or physiological responses of the host (Supplementary Figure 4, Pearson correlations, -0.23 > R< 0.2 and p>0.26).
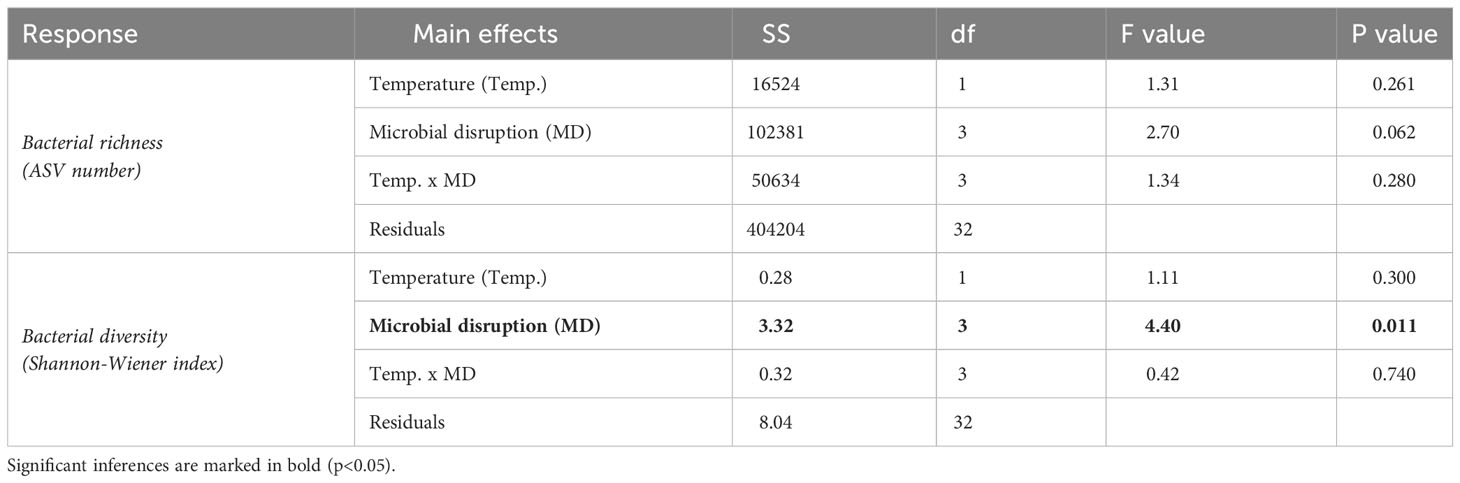
Table 2 Summary of the ANOVA performed to determine significant differences in bacterial richness and diversity across temperature conditions (2 levels, Ambient [n=20] vs. Heat stress [n=20]) and microbial disruption treatments (4 levels, Control [n=10], procedural control [n=10], antibiotics [n=10] and antiseptics [n=10]).
Bacterial community structure was significantly influenced by the interaction of both temperature and microbial disruption treatments (Figure 3, Supplementary Table 3 and Supplementary Figures 7 and 8; PERMANOVA: pseudo-F1,32 = 1.52 and p=0.004). Further multivariate pairwise contrasts determined that bacterial community structure in ambient conditions differed between the antibiotic and antiseptic treatments (t1,8 = 2.43 and p=0.009) as well as the antibiotic and antiseptic treatments with the control groups (t1,8<2.32 and p<0.034; Figure 3 and Supplementary Table 4). In thermal stress treatments, the bacterial structure from the antibiotic treatments was only different from the antiseptic treatment (t1,8 = 2.04 and p=0.008) but not controls. In contrast, antiseptic treatments in the thermal stress conditions did show differences in bacterial structure compared to controls (Control: t1,8 = 1.88 and p=0.009; PC: t1,8 = 1.82 and p=0.008). Bacterial community structure was always different between temperature conditions across the different microbial disruption treatments (Figure 3 and Supplementary Table 4). Differences in bacterial structure across the evaluated groups were not affected by intra-group structural variability (Supplementary Table 3; PERMIDSP).
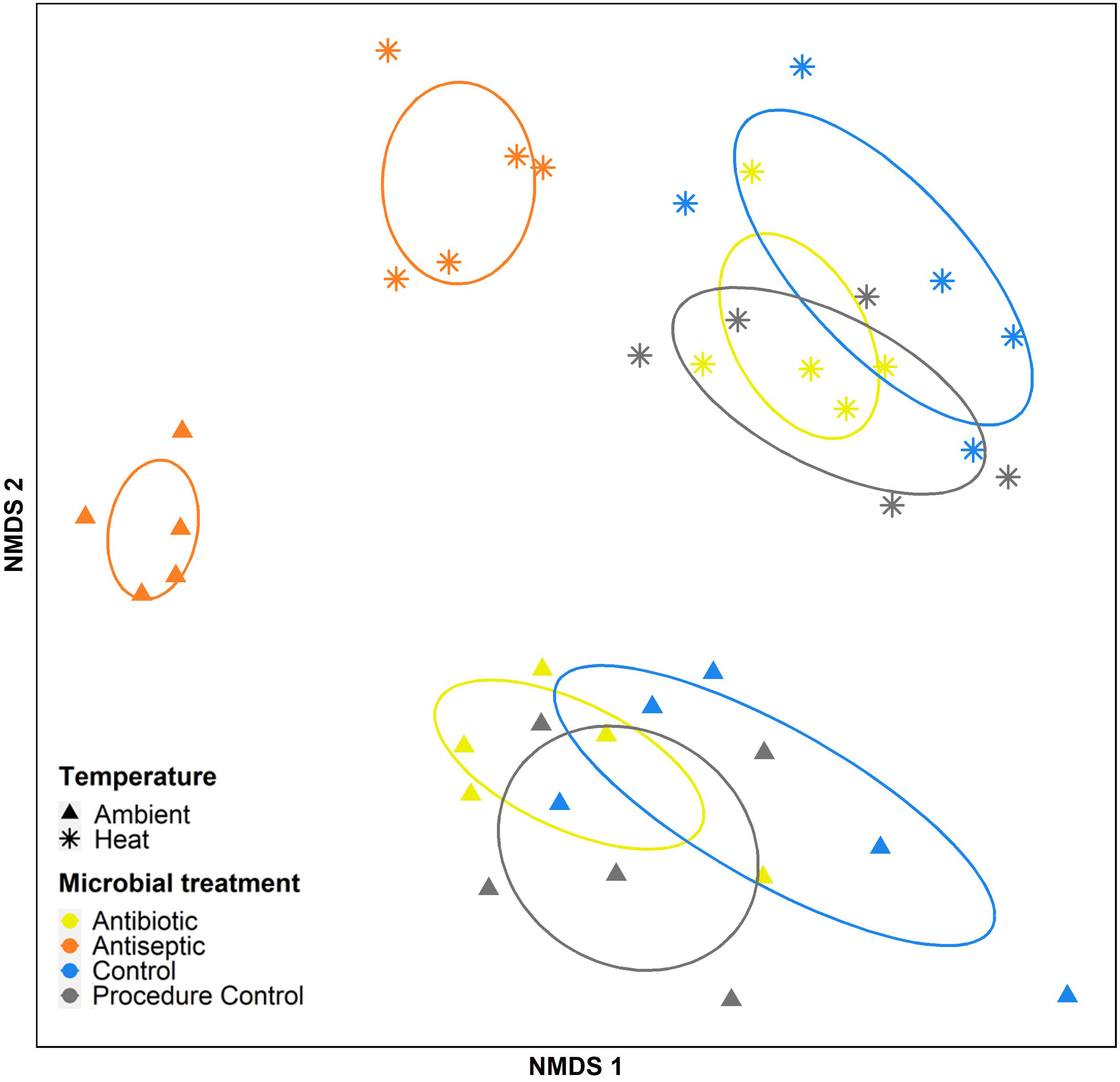
Figure 3 Non-metric multidimensional scaling ordination (NMDS, stress=0.16) based on Bray-Curtis dissimilarity matrix showing the effects of temperature conditions (2 levels, Ambient [n=20] vs. Heat stress [n=20]) and microbial disturbance treatments (4 levels, Control [n=10], procedural control [n=10], antibiotics [n=10] and antiseptics [n=10]) on lamina-associated bacterial communities in Ecklonia juveniles. Pairwise contrasts are shown in Supplementary Table 6.
A distance-based redundancy analysis (dbRDA) was done to further assess variables that could predict differences in bacterial community composition. In this analysis all host morphological and physiological responses were included but only absolute algal growth rate, and respiration, were significantly related with bacterial community structure (Supplementary Figure 6 and marginal effects, Supplementary Table 5, absolute growth rate: F1,37 = 1.94 and p=0.01; respiration: F1,37 = 3 and p=0.001).
A differential abundance analysis (mvabund) was then done to explore which specific bacterial ASVs changed significantly as communities shifted overall (see Supplementary Figures 7, 8 for an overview of dominant taxa across all samples). More than 452 ASVs changed, but only the 10 ASVs with the highest average relative abundance (0.38-7.23%) and a high frequency of occurrence (>95% prevalence across all individual hosts in the experiment) were analysed further (see details of this selection in Supplementary Table 6). Overall, results on the selected ASVs show that specific taxa were affected by both the antibiotic and antiseptic treatments compared to control, but this effect varied between temperature conditions in some cases. Additionally, some ASVs were also affected by temperature conditions independently of microbial disruption.
The abundance of several ASVs of the genus Alteromonas increased in the antiseptic disruption treatment compared to other microbial disruption treatments in both temperature conditions (Supplementary Table 8). An ASV assigned to the genus Sulfitobacter also presented an increase in the antiseptic treatment but only for ambient conditions (Supplementary Table 9). Additionally, some ASVs showed an increase in abundance in heat conditions independently of microbial disruption treatments (Supplementary Table 7, assigned to the genera Hyphomonas, Thalassospira, Wenyingzhuangia and Mariniblastus) although one ASV showed this pattern exclusively in the control groups (Supplementary Table 9, unidentified genus of the class Gammaproteobacteria). In contrast, one ASV decreased its relative abundance across all disruption treatments in heat conditions for the exception of the antiseptic group were it remained lower in abundance for both temperature conditions (Supplementary Table 9, genus Paraglaciecola).
Results of the functional prediction analysis (i.e., Tax4Fun2) provided low prediction and demonstrated that accuracy and reliability of functional predictions may be highly affected by specific treatments (e.g. antiseptic disruption treatment) and the user-defined data. Data repositories for an accurate functional prediction (i.e. default or user-defined) need to be more context-specific and with a higher representation of local areas in which kelp associated microbes may be driven by specific environmental conditions (for further details of results and discussion of this analysis see Box S1, Supplementary Table 10 and Supplementary Figure 9).
Discussion
Environmental stressors such as ocean warming and disruptions to host-microbiota interactions can affect habitat-forming species such as kelps; however we have little understanding of how these two factors interact. (Egan et al., 2013; although see some increased temperature effects on macroalgal growth, Roth-Schultze et al., 2018 and Minich et al., 2018; and disease progression, Case et al., 2011). We found strong effects of thermal stress on both components of the kelp holobiont, including strong morphological and physiological effects on host performance and a structural shift in the associated bacterial communities. However, and contrary to our hypothesis, disruption to the kelp microbiota were not directly correlated with negative effects on the host and did not exacerbate temperature effects on juvenile kelp, although clear signs of a bacterial community shifts were observed. So, while early stages of Ecklonia are vulnerable to thermal stress, the microbiota may not play a substantive role in resistance to that stress. Alternatively, it may be that even though the bacterial community changed, these retain key functions that permitted the maintenance of essential interactions with the host even under high thermal stress.
Strong effects of temperature on the macroalgal holobiont
Our results highlight the strong effect that thermal stress had on decreasing growth, net photosynthesis and respiration. Net photosynthesis and respiration were severely affected by higher temperatures, possibly through the alteration of heat-sensitive structures (e.g. components of photosynthesis such as electron transporters, Davison, 1991) and enzymatic biochemical processes linked to nutrient acquisition and assimilation (Ji and Gao, 2021). For example, photosystem II, ATP synthase and other important metabolic enzymes in the photosynthetic pathway are highly sensitive to increased temperatures (Allakhverdiev et al., 2008). A similar effect is expected for other metabolic pathways such as aerobic respiration, in which high temperature may not only impair enzymatic activities and structural cell integrity but also increment the production of reactive oxygen species that may further damage the metabolic machinery (Eggert, 2012). Ecklonia has considerable phenotypic plasticity, enabling a broad thermal tolerance and adaptive responses to a range of different temperatures (Wernberg et al., 2019). Such thermal traits include biochemical changes at the cellular level (e.g., gene expression, protein identity and cell membrane fluidity; Wang et al., 2018; Barkina et al., 2020) or physiological responses like photosynthesis, respiration and growth that were analysed in the present study (Eggert, 2012). However, such thermal adaptability in Ecklonia would be compromised at temperatures higher than their optimal thermal tolerance range where performance will decrease or be permanently damaged (Harley et al., 2012).
Thermal adaptability of phenotypic traits and negative effects of temperature as a stressor to macroalgae are not always comparable across species (e.g. eurythermic or stenothermic species, Eggert, 2012) or present high intra-specific variability (i.e. larger vulnerability on earlier life stages, Qiu et al., 2019). For Ecklonia juveniles, significant damage to their metabolic system could also be influenced by other factors that increases their susceptibility to thermal stress including their specific natural thermal conditions in the field (i.e., sampled Ecklonia inhabit a cold environment in southeast Tasmania) and inherent vulnerability of early life stages. Under thermal stress, Ecklonia juveniles are likely to primarily focus resource allocation for the repair of damaged metabolic pathways and further protection to thermal stress (as seen in other brown algae, Davison, 1991).This would undoubtedly hinder their development, as seen here with absolute growth rate of Ecklonia juveniles significantly reduced at higher temperatures, but also potentially limiting their capacity to defend against pathogenic invasions and diseases as seen in other studies with kelp (Minich et al., 2018; Qiu et al., 2019).
A substantial level of bleaching (i.e. laminar depigmentation) and alteration of the bacterial communities in the laminar surface occurred in Ecklonia juveniles exposed to high temperatures. Bleaching in macroalgae is a natural physiological mechanism in response to thermal stress in which photosynthetic pigments are lost and nutrient availability is reduced (Rishworth et al., 2018) but which can be related with changes to epiphytic bacteria under high temperatures (Marzinelli et al., 2015; Qiu et al., 2019). As described above, a reallocation of resources to counteract the effects of thermal stress are likely in Ecklonia juveniles that present both bleaching, reduced photosynthesis and low biomass. However, bleaching has also been associated to macroalgal diseases in thermal stress, in some cases resulting in the increase abundance of multiple opportunistic pathogens that degrade algal tissues and can disrupt photosynthetic efficiency (Campbell et al., 2011; Case et al., 2011 and Zozaya-Valdés et al., 2016).
Shifts in bacterial community structure, association with lower absolute growth rates (as shown by the dbRDA) and changes in the abundance of specific bacterial taxa in Ecklonia juveniles exposed to higher temperatures, suggests that there may have been an increase of pathogenic bacteria in high temperature treatments in this study. Several identified bacterial genera that increased their abundance in heat conditions are known as opportunistic pathogens in red and brown algae that can cause bleaching (i.e. Thalassospira, Azizi et al., 2018; de Mesquita et al., 2019) or have strong tissue degradation capacities (i.e. Wenyingzhuangia, Masasa et al., 2021). In contrast, other bacterial taxa were severely affected by higher temperatures which include genera such as Sulfitobacter and Mariniblastus that are known to play an important role in morphogenesis and nutrient degradation in macroalgae (Weiss et al., 2017; Alsufyani et al., 2020; Weigel et al., 2022). It is likely that many of the identified bacterial taxa had a higher thermal tolerance and contributed to the poor state of Ecklonia juveniles. Interestingly however, other bacterial taxa that are known to provide beneficial effects to macroalgae also increased their abundance at higher temperatures. This is especially noticeable by the increase of the genus Hyphomonas that plays a role in inducing tissue morphogenesis and nutrient exchange (Fukui et al., 2014). The prevalence of this group may show evidence that even under a long lasting severe thermal stress (16 days), Ecklonia juveniles may still retain a functional microbiome that potentially support their survival although further research is needed to test this.
Tolerance and synergic effects of temperature and microbial dysbiosis in Ecklonia juveniles
The antiseptic solution was an effective press disturbance that changed the associated bacterial community of Ecklonia juveniles, however its effect to create a microbial dysbiosis varied between temperature conditions and was only directly linked with two host phenotypic traits. Lower bacterial diversity and shifts in bacterial structure were observed in both temperature conditions (Supplementary Figure 2) however for the latter, the additional effect of these bacterial structural shifts on Ecklonia growth rates and respiration, could evidence a synergic effect of microbial disruption and thermal stress (i.e. dbRDA and Supplementary Table 5). Additionally, in both temperature conditions, the antiseptic treatment substantially favoured the proliferation of the genus Alteromonas that could potentially play a role in these synergic effects.
The genus Alteromonas is a cosmopolitan taxa that appears as an epiphyte in many tropical and temperate macroalgal species (Kumar et al., 2016) and is known to participate in algal nutrient cycling and exchange through polysaccharide degradation (i.e. alginate, Mitulla et al., 2016; Minich et al., 2018; Koch et al., 2019). In healthy algae, Alteromonas appear normally in low abundance (Fernandes et al., 2012) however they are known to be a highly opportunistic bacteria with a broad metabolic capacity to degrade algal cell walls (Kumar et al., 2016). Under an unhealthy or stressed state, macroalgae may have a higher abundance of this group and can potentially become a pathogenic taxon to the host (Egan et al., 2013). In this study, the increase of this potentially pathogenic taxa under both temperature conditions and when disturbed, shows the possibility that Ecklonia may retain a level of protection to avoid decay or disease progression under ambient conditions, but this interaction could change under thermal stress. A level of protection in ambient conditions under an altered microbiome may be regulated by the innate immune system of the algae (e.g. receptor mediated immunity, production of reactive oxygen species or secondary metabolites such as aromatic compounds, Weinberger, 2007; Kloareg et al., 2021) or/and by the support of a perduring microbiome that is still able to provide a protective role. The prevalence and high abundance of the genus Sulfitobacter in the antiseptic treatment and under ambient temperatures, could evidence the above finding as this taxa is known to be involved in algal growth and development and could benefit the host even under microbial dysbiosis (Alsufyani et al., 2020). In contrast, under thermal stress, Sulfitobacter decreases its abundance while the Alteromonas remain mostly unchanged and could potentially contribute to the decreased in growth and respiration of Ecklonia although further experimental assessments are needed to verify this. However, the capacity of this bacterial group to adapt to a broad range of environmental conditions including high temperatures (Koch et al., 2019), makes it a good candidate to be associated with detrimental effect on the host brough by the synergic relationship between thermal stress and microbial dysbiosis. Further functional assessments under thermal stress could provide a broader understanding of the positive or negative interactions of this and other bacterial groups with Ecklonia juveniles.
Antiseptic solution more effective to achieve bacterial community shifts
A microbial shift was achieved in this study by exposing Ecklonia juveniles to press disturbances using two different disruption approaches intended to both 1) decrease the overall abundance of bacterial taxa (antiseptic solution) or 2) selectively remove specific taxa to change the overall community structure (antibiotic solution). Similar manipulative experiments with macroalgae-associated microbiomes have been attempted to untangle the ecological importance of these microbial communities on host performance (e.g. McGrath et al., 2023 in review) and trophic interactions with other epifauna (e.g. Singh et al., 2021). Results here show that the antiseptic solution decrease bacterial abundance compared to control groups, as well as producing strong shifts in the bacterial community structure and an increase in the abundance of specific taxa (i.e., several ASVs of the genus Alteromonas and one Sulfitobacter). Comparatively, the antibiotic solution presented lessened effects with only noticeable differences to controls in community structure in the ambient temperature and one prevalent bacterial ASV that decreased in abundance (i.e., Unidentified genus from the class Gammaproteobacteria). Effectiveness of the antiseptic compared to the antibiotic solution as a stronger agent to achieve microbial shifts and a potential dysbiosis, is further evidenced by the largely different bacterial structure between both treatments that is also driven by the decrease of some bacterial groups exclusively in the antiseptic solution (e.g. genus Paraglaciecola and the multiple Alteromonas).
Similar manipulative experiments using Iodine as an oxidant agent to remove bacterial communities in macrophyte tissues have been successfully used in the past without strong negative effects on the host (Kientz et al., 2011; Borburema et al., 2021; Fuggle et al., 2023). Here, no noticeable morphological or physiological effects were seen in Ecklonia juveniles as a consequence of the antiseptic solution (i.e. Povidone-Iodine solution 2%, as seen in other studies with similar concentrations and evaluated through changes in macroalgal biomass; Bostrychia binderi, Borburema et al., 2021) although effects due to the accumulation and assimilation of Iodine by Ecklonia or associated bacteria cannot be discarded (i.e. iodine is a key metabolic compound in algae, Küpper and Carrano, 2019; and for bacterial groups, Gupta et al., 2017). From the results obtained here, it is clear that both microbial dysbiosis approaches worked as intended, but it was the antiseptic solution that showed highest impact to create an alteration of the microbiome independently from the temperature conditions. However, it is acknowledged that assessment of the potential direct chemical effects on the host of both solutions (i.e. antiseptic and antibiotic) did not include histological assessments (e.g. to detect cell integrity, Kientz et al., 2011) and controls which involve the re-inoculation/introduction of the microbiota (“conventionalized” controls, Callens et al., 2018).
Conclusion
Temperature is a strong abiotic driver that shapes coastal communities through its direct effect on the performance, reproduction and interaction between many species (Kordas et al., 2011). In recent years, climate change and human-derived ocean warming have affected macroalgal communities to a level that has rapidly affected their survival and distribution (Takolander et al., 2017) and is expected to continue in coming decades with possible stronger effects in colder waters where many algal species have a narrow thermal range (Piñeiro-Corbeira et al., 2018). Further, evidence exists that a dysbiosis of their associated microbiome could lead to sublethal effects (i.e., altered metabolism, lower innate immunity responses or disease) that can ultimately lead to high mortality in more severe scenarios of future ocean warming (Qiu et al., 2019).
Here, we provide further evidence of the negative effects of thermal stress in Ecklonia juveniles and bring valuable information of the effects on epiphytic bacterial communities. Results here highlight the strong influence of temperature on kelps, and clearly show an effect on the associated bacterial communities (i.e., structural changes and shifts in the abundance of prevalent bacterial ASVs) that could be linked to the overall condition of Ecklonia. This study also shows that an experimentally applied press disturbances (i.e., antiseptic solution) can cause a clear shifts of these bacterial communities without causing observable toxic effects on the host development or physiology. However, there is little evidence to suggest a strong synergic effect with temperature. It is possible that the duration of the experiment was not long enough to be able to assess the impacts of the microbial removal press disturbance to host condition and physiological responses. However, alternative explanations may point to Ecklonia juveniles having enough innate immunological responses and adaptations that may permit them to survive even with a microbial dysbiosis. Additionally, kelp could also have a resilient epiphytic bacterial community that may change under microbial dysbiosis (i.e., structural changes or shift in the abundance of specific bacterial taxa) but yet retain a consistent and highly redundant functional profile (Burke et al., 2011) that enables the maintenance of key interactions that benefit the development and performance of the host.
Data availability statement
Raw sequence reads for the 16S amplicon sequencing is now publicly available through the NCBI SRA with the BiProject code: https://www.ncbi.nlm.nih.gov/, PRJNA1033398.
Author contributions
SVG: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. CH: Conceptualization, Methodology, Resources, Supervision, Validation, Visualization, Writing – review & editing. DB: Conceptualization, Investigation, Methodology, Resources, Validation, Visualization, Writing – review & editing. EB: Investigation, Methodology, Validation, Visualization, Writing – review & editing. PS: Conceptualization, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Visualization, Writing – review & editing. EM: Conceptualization, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Visualization, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by funds from the Australian Research Council to EM and PS.
Acknowledgments
We thank all the staff and students from the Institute of Marine and Antarctic Studies (IMAS, UTAS Tasmania) including Allyson Nardelli, Wouter Visch, Cayne Layton, Jakop Schwoerbel, Olivia Wynn, Pam Quayle and Eva Smid for field and laboratory assistance, specially at stages conducted during the COVID-19 pandemic.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1332501/full#supplementary-material
References
Abreu A., Bourgois E., Gristwood A., Troublé R., Acinas S. G., Bork P., et al. (2022). European marine biological resource centre - European research infrastructure consortium (EMBRC-ERIC). Priorities ocean microbiome Res. Nat. Microbiol. 7 (7), 937–947. doi: 10.1038/s41564-022-01145-5
Allakhverdiev S. I., Kreslavski V. D., Klimov V. V., Los D. A., Carpentier R., Mohanty P. (2008). Heat stress: An overview of molecular responses in photosynthesis. Photosynthesis Res. 98 (1), 541–550. doi: 10.1007/s11120-008-9331-0
Alsufyani T., Califano G., Deicke M., Grueneberg J., Weiss A., Engelen A. H., et al. (2020). Macroalgal–bacterial interactions: Identification and role of thallusin in morphogenesis of the seaweed Ulva (Chlorophyta). J. Exp. Bot. 71 (11), 3340–3349. doi: 10.1093/jxb/eraa066
Anderson M. J. (2014). Permutational multivariate analysis of variance (PERMANOVA). Wiley statsref: statistics reference online, 1–15. doi: 10.1002/9781118445112.stat07841
Apprill A. (2017). Marine animal microbiomes: toward understanding host–microbiome interactions in a changing ocean. Front. Mar. Sci. 4. doi: 10.3389/fmars.2017.00222
Azizi A., Mohd Hanafi N., Basiran M. N., Teo C. H. (2018). Evaluation of disease resistance and tolerance to elevated temperature stress of the selected tissue-cultured Kappaphycus alvarezii Doty 1985 under optimized laboratory conditions. 3 Biotech. 8 (8), 321. doi: 10.1007/s13205-018-1354-4
Barkina M., Pomazenkova L. A., Chopenko N. S., Velansky P. V., Ya. E., Sanina N. M. (2020). Influence of warm-acclimation rate on polar lipids of ulva lactuca. Russian J. Plant Physiol. 67 (1), 111–121. doi: 10.1134/S1021443720010021
Bender E. A., Case T. J., Gilpin M. E. (1984). Perturbation experiments in community ecology: theory and practice. Ecology 65 (1), 1–13. doi: 10.2307/1939452
Bennett S., Wernberg T., Connell S. D., Hobday A. J., Johnson C. R., Poloczanska E. S. (2016). The “Great Southern Reef”: Social, ecological and economic value of Australia’s neglected kelp forests. Mar. Freshw. Res. 67 (1), 47. doi: 10.1071/MF15232
Borburema H. D. S., Barbosa Ê.N.A., Miranda G. E. C. (2021). Decontamination protocol of the macroalga Bostrychia binderi Harvey (Rhodophyta) for unialgal cultures and laboratory studies. Hoehnea 48, e582020. doi: 10.1590/2236-8906-58/2020
Britton D., Schmid M., Noisette F., Havenhand J. N., Paine E. R., McGraw C. M., et al. (2020). Adjustments in fatty acid composition is a mechanism that can explain resilience to marine heatwaves and future ocean conditions in the habitat-forming seaweed Phyllospora comosa (Labillardière) C. Agardh. Global Change Biol. 26 (6), 3512–3524. doi: 10.1111/gcb.15052
Burke C., Steinberg P., Rusch D., Kjelleberg S., Thomas T. (2011). Bacterial community assembly based on functional genes rather than species. Proc. Natl. Acad. Sci. 108 (34), 14288–14293. doi: 10.1073/pnas.1101591108
Callahan B. J., McMurdie P. J., Rosen M. J., Han A. W., Johnson A. J. A., Holmes S. P. (2016). DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13 (7), 581–583. doi: 10.1038/nmeth.3869
Callens M., Watanabe H., Kato Y., Miura J., Decaestecker E. (2018). Microbiota inoculum composition affects holobiont assembly and host growth in Daphnia. Microbiome 6, 1–12. doi: 10.1186/s40168-018-0444-1
Campbell A. H., Harder T., Nielsen S., Kjelleberg S., Steinberg P. D. (2011). Climate change and disease: Bleaching of a chemically defended seaweed. Global Change Biol. 17 (9), 2958–2970. doi: 10.1111/j.1365-2486.2011.02456.x
Case R. J., Longford S. R., Campbell A. H., Low A., Tujula N., Steinberg P. D., et al. (2011). Temperature induced bacterial virulence and bleaching disease in a chemically defended marine macroalga. Environ. Microbiol. 13 (2), 529–537. doi: 10.1111/j.1462-2920.2010.02356.x
Coelho S. M., Rijstenbil J. W., Brown M. T. (2000). Impacts of anthropogenic stresses on the early development stages of seaweeds. J. Aquat. Ecosystem Stress Recovery 7 (4), 317–333. doi: 10.1023/A:1009916129009
Croft M. T., Lawrence A. D., Raux-Deery E., Warren M. J., Smith A. G. (2005). Algae acquire vitamin B12 through a symbiotic relationship with bacteria. Nature 438 (7064), 90–93. doi: 10.1038/nature04056
Davis N. M., Proctor D. M., Holmes S. P., Relman D. A., Callahan B. J. (2018). Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome 6 (1), 226. doi: 10.1186/s40168-018-0605-2
Davison I. R. (1991). Environmental effects on algal photosynthesis: temperature. J. Phycology 27 (1), 2–8. doi: 10.1111/j.0022-3646.1991.00002.x
de Mesquita M. M. F., Crapez M. A. C., Teixeira V. L., Cavalcanti D. N. (2019). Potential interactions bacteria-brown algae. J. Appl. Phycology 31 (2), 867–883. doi: 10.1007/s10811-018-1573-4
Dittami S. M., Arboleda E., Auguet J.-C., Bigalke A., Briand E., Cárdenas P., et al. (2021). A community perspective on the concept of marine holobionts: Current status, challenges, and future directions. PeerJ 9, e10911. doi: 10.7717/peerj.10911
Dolliver J., O’Connor N. (2022). Whole system analysis is required to determine the fate of macroalgal carbon: A systematic review. J. Phycology 58 (3), 364–376. doi: 10.1111/jpy.13251
Egan S., Harder T., Burke C., Steinberg P., Kjelleberg S., Thomas T. (2013). The seaweed holobiont: Understanding seaweed–bacteria interactions. FEMS Microbiol. Rev. 37 (3), 462–476. doi: 10.1111/1574-6976.12011
Eggert A. (2012). “Seaweed responses to temperature,” in Seaweed biology: novel insights into ecophysiology, ecology and utilization. Eds. Wiencke C., Bischof K. (Berlin, Heidelberg: Springer), 47–66. doi: 10.1007/978-3-642-28451-9_3
Fernandes N., Steinberg P., Rusch D., Kjelleberg S., Thomas T. (2012). Community structure and functional gene profile of bacteria on healthy and diseased thalli of the red seaweed delisea pulchra. PloS One 7 (12), e50854. doi: 10.1371/journal.pone.0050854
Fox J., Weisberg S. (2019). An R Companion to Applied Regression, Third edition. Sage, Thousand Oaks CA. Available at: https://socialsciences.mcmaster.ca/jfox/Books/Companion/.
Fuggle R. E., Gribben P. E., Marzinelli E. M. (2023). Experimental evidence root-associated microbes mediate seagrass response to environmental stress. J. Ecol. 111 (5), 1079–1093. doi: 10.1111/1365-2745.14081
Fukui Y., Abe M., Kobayashi M., Yano Y., Satomi M. (2014). Isolation of hyphomonas strains that induce normal morphogenesis in protoplasts of the marine red alga pyropia yezoensis. Microbial Ecol. 68 (3), 556–566. doi: 10.1007/s00248-014-0423-4
Goecke F., Labes A., Wiese J., Imhoff J. F. (2010). Chemical interactions between marine macroalgae and bacteria. Mar. Ecol. Prog. Ser. 409, 267–299. doi: 10.3354/meps08607
Gupta V., Jain M., Reddy C. R. K. (2017). “Macroalgal functional genomics: A missing area,” in Systems biology of marine ecosystems. Eds. Kumar M., Ralph P. (Edinburgh, Scotland: Springer International Publishing), 3–12. doi: 10.1007/978-3-319-62094-7_1
Harley C. D., Anderson K. M., Demes K. W., Jorve J. P., Kordas R. L., Coyle T. A., et al. (2012). Effects of climate change on global seaweed communities. J. Phycology 48 (5), 1064–1078. doi: 10.1111/j.1529-8817.2012.01224.x
Hollants J., Leliaert F., De Clerck O., Willems A. (2013). What we can learn from sushi: A review on seaweed-bacterial associations. FEMS Microbiol. Ecol. 83 (1), 1–16. doi: 10.1111/j.1574-6941.2012.01446.x
Ji Y., Gao K. (2021). Chapter Two - Effects of climate change factors on marine macroalgae: A review. Adv. Mar. Biol. 88, 91–136. doi: 10.1016/bs.amb.2020.11.001
Joint I., Tait K., Callow M. E., Callow J. A., Milton D., Williams P., et al. (2002). Cell-to-cell communication across the prokaryote-eukaryote boundary. Science 298 (5596), 1207–1207. doi: 10.1126/science.1077075
Kanagasabhapathy M., Sasaki H., Haldar S., Yamasaki S., Nagata S. (2006). Antibacterial activities of marine epibiotic bacteria isolated from brown algae of Japan. Ann. Microbiol. 56, 167–173. doi: 10.1007/BF03175000
Kientz B., Thabard M., Cragg S. M., Pope J., Hellio C. (2011). A new method for removing microflora from macroalgal surfaces: An important step for natural product discovery. Botm 54 (5), 457–469. doi: 10.1515/BOT.2011.053
Kirkman H. (1981). The first year in the life history and the survival of the juvenile marine macrophyte, Ecklonia radiata (Turn.) J. Agardh. J. Exp. Mar. Biol. Ecol. 55 (2), 243–254. doi: 10.1016/0022-0981(81)90115-5
Klindworth A., Pruesse E., Schweer T., Peplies J., Quast C., Horn M., et al. (2013). Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 41 (1), e1. doi: 10.1093/nar/gks808
Kloareg B., Badis Y., Cock J. M., Michel G. (2021). Role and evolution of the extracellular matrix in the acquisition of complex multicellularity in eukaryotes: A macroalgal perspective. Genes 12 (7), 1059. doi: 10.3390/genes12071059
Koch H., Dürwald A., Schweder T., Noriega-Ortega B., Vidal-Melgosa S., Hehemann J.-H., et al. (2019). Biphasic cellular adaptations and ecological implications of Alteromonas macleodii degrading a mixture of algal polysaccharides. ISME J. 13 (1), 92–103. doi: 10.1038/s41396-018-0252-4
Kordas R. L., Harley C. D. G., O’Connor M. I. (2011). Community ecology in a warming world: The influence of temperature on interspecific interactions in marine systems. J. Exp. Mar. Biol. Ecol. 400 (1), 218–226. doi: 10.1016/j.jembe.2011.02.029
Kumar M. S., Slud E. V., Okrah K., Hicks S. C., Hannenhalli S., Corrada Bravo H. (2018). Analysis and correction of compositional bias in sparse sequencing count data. BMC Genomics 19 (1), 799. doi: 10.1186/s12864-018-5160-5
Kumar V., Zozaya-Valdes E., Kjelleberg S., Thomas T., Egan S. (2016). Multiple opportunistic pathogens can cause a bleaching disease in the red seaweed Delisea pulchra. Environ. Microbiol. 18 (11), 3962–3975. doi: 10.1111/1462-2920.13403
Küpper F. C., Carrano C. J. (2019). Key aspects of the iodine metabolism in brown algae: a brief critical review. Metallomics 11 (4), 756–764. doi: 10.1039/c8mt00327k
Kuznetsova A., Brockhoff P. B., Christensen R. H. B. (2017). lmerTest package: tests in linear mixed effects models. J. Stat. Softw. 82 (13), 1–26. doi: 10.18637/jss.v082.i13
Lenth R. (2022). emmeans: Estimated Marginal Means, aka Least-Squares Means. R package version 1.7.4-1. Available at: https://CRAN.R-project.org/package=emmeans.
Lüdecke D., Ben-Shachar M. S., Patil I., Waggoner P., Makowski D. (2021). performance: An R package for assessment, comparison and testing of statistical models. J. Open Source Software 6 (60), 3139.
Marzinelli E. M., Campbell A. H., Zozaya Valdes E., Vergés A., Nielsen S., Wernberg T., et al. (2015). Continental-scale variation in seaweed host-associated bacterial communities is a function of host condition, not geography. Environ. Microbiol. 17 (10), 4078–4088. doi: 10.1111/1462-2920.12972
Masasa M., Kushmaro A., Kramarsky-Winter E., Shpigel M., Barkan R., Golberg A., et al. (2021). Mono-specific algal diets shape microbial networking in the gut of the sea urchin Tripneustes gratilla elatensis. Anim. Microbiome 3 (1), 79. doi: 10.1186/s42523-021-00140-1
McGrath A. H., Lema K., Egan S., Wood G., Vadillo Gonzalez S., Kjelleberg S., et al. (2023). Disentangling direct vs indirect effects of microbiome manipulations in a habitat-forming marine holobiont. doi: 10.21203/rs.3.rs-3012963/v1
McMurdie P. J., Holmes S. (2013). phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PloS One 8 (4), e61217. doi: 10.1371/journal.pone.0061217
Millard S. P. (2013). EnvStats: An R Package for Environmental Statistics. Springer, New York. Available at: https://www.springer.com.
Minich J. J., Morris M. M., Brown M., Doane M., Edwards M. S., Michael T. P., et al. (2018). Elevated temperature drives kelp microbiome dysbiosis, while elevated carbon dioxide induces water microbiome disruption. PloS One 13 (2), e0192772. doi: 10.1371/journal.pone.0192772
Mitulla M., Dinasquet J., Guillemette R., Simon M., Azam F., Wietz M. (2016). Response of bacterial communities from California coastal waters to alginate particles and an alginolytic Alteromonas macleodii strain. Environ. Microbiol. 18 (12), 4369–4377. doi: 10.1111/1462-2920.13314
Monserrat M., Comeau S., Verdura J., Alliouane S., Spennato G., Priouzeau F., et al. (2022). Climate change and species facilitation affect the recruitment of macroalgal marine forests. Sci. Rep. 12 (1), 18103. doi: 10.1038/s41598-022-22845-2
Morris M. M., Haggerty J. M., Papudeshi B. N., Vega A. A., Edwards M. S., Dinsdale E. A. (2016). Nearshore pelagic microbial community abundance affects recruitment success of giant kelp, Macrocystis pyrifera. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.01800
Oksanen J., Blanchet F. G., Kindt R., Legendre P., Minchin P. R., O’hara R. B., et al. (2013). Package ‘vegan’. Community Ecol. Package 2 (9), 1–295.
Parks D. H., ChuvoChina M., Rinke C., Mussig A. J., Chaumeil P.-A., Hugenholtz P. (2022). GTDB: An ongoing census of bacterial and archaeal diversity through a phylogenetically consistent, rank normalized and complete genome-based taxonomy. Nucleic Acids Res. 50 (D1), D785–D794. doi: 10.1093/nar/gkab776
Pfister C. A., Altabet M. A., Weigel B. L. (2019). Kelp beds and their local effects on seawater chemistry, productivity, and microbial communities. Ecology 100 (10), e02798. doi: 10.1002/ecy.2798
Philippot L., Griffiths B. S., Silke L. (2021). Microbial community resilience across ecosystems and multiple disturbances. Microbiol. Mol. Biol. Rev. 85.2, 10–1128. doi: 10.1128/mmbr.00026-20
Piñeiro-Corbeira C., Barreiro R., Cremades J., Arenas F. (2018). Seaweed assemblages under a climate change scenario: Functional responses to temperature of eight intertidal seaweeds match recent abundance shifts. Sci. Rep. 8 (1), 12978. doi: 10.1038/s41598-018-31357-x
Plante C. J. (2017). Defining disturbance for microbial ecology. Microbial Ecol. 74 (2), 259–263. doi: 10.1007/s00248-017-0956-4
Qiu Z., Coleman M. A., Provost E., Campbell A. H., Kelaher B. P., Dalton S. J., et al. (2019). Future climate change is predicted to affect the microbiome and condition of habitat-forming kelp. Proc. R. Soc. B: Biol. Sci. 286 (1896), 20181887. doi: 10.1098/rspb.2018.1887
Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., et al. (2013). The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 41 (D1), D590–D596. doi: 10.1093/nar/gks1219
Rishworth G. M., Perissinotto R., Bird M. S., Pelletier N. (2018). Grazer responses to variable macroalgal resource conditions facilitate habitat structuring. R. Soc. Open Sci. 5 (1), 171428. doi: 10.1098/rsos.171428
Roth-Schultze A. J., Thomas T., Steinberg P., Deveney M. R., Tanner J. E., Wiltshire K. H., et al. (2018). The effects of warming and ocean acidification on growth, photosynthesis, and bacterial communities for the marine invasive macroalga Caulerpa taxifolia. Limnology Oceanography 63 (1), 459–471. doi: 10.1002/lno.10739
Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., et al. (2012). Fiji: An open-source platform for biological-image analysis. Nat. Methods 9 (7), 676–682. doi: 10.1038/nmeth.2019
Singh C. L., Huggett M. J., Lavery P. S., Säwström C., Hyndes G. A. (2021). Kelp-associated microbes facilitate spatial subsidy in a detrital-based food web in a shoreline ecosystem. Front. Mar. Sci. 8. doi: 10.3389/fmars.2021.678222
Singh R. P., Reddy C. R. K. (2014). Seaweed–microbial interactions: key functions of seaweed-associated bacteria. FEMS Microbiol. Ecol. 88 (2), 213–230. doi: 10.1111/1574-6941.12297
Smale D. A. (2020). Impacts of ocean warming on kelp forest ecosystems. New Phytol. 225 (4), 1447–1454. doi: 10.1111/nph.16107
Song W., Zhang S., Thomas T. (2022). MarkerMAG: Linking metagenome-assembled genomes (MAGs) with 16S rRNA marker genes using paired-end short reads. Bioinformatics 38 (15), 3684–3688. doi: 10.1093/bioinformatics/btac398
Stratil S. B., Neulinger S. C., Knecht H., Friedrichs A. K., Wahl M. (2013). Temperature-driven shifts in the epibiotic bacterial community composition of the brown macroalga Fucus vesiculosus. Microbiol. Open 2 (2), 338–349. doi: 10.1002/mbo3.79
Swift D., Cresswell K., Johnson R., Stilianoudakis S., Wei X. (2023). A review of normalization and differential abundance methods for microbiome counts data. Wiley Interdisciplinary Reviews: Computational Statistics 15 (1), e1586. doi: 10.1002/wics.1586
Takolander A., Cabeza M., Leskinen E. (2017). Climate change can cause complex responses in Baltic Sea macroalgae: A systematic review. J. Sea Res. 123, 16–29. doi: 10.1016/j.seares.2017.03.007
Visch W., Lush H., Schwoerbel J., Hurd C. L. (2023). Nursery optimization for kelp aquaculture in the Southern Hemisphere: The interactive effects of temperature and light on growth and contaminants. Appl. Phycology 4 (1), 44–53. doi: 10.1080/26388081.2023.2174903
Wang Y. I., Naumann U., Wright S. T., Warton D. I. (2012). mvabund–an R package for model-based analysis of multivariate abundance data. Methods Ecol. Evol. 3 (3), 471–474. doi: 10.1111/j.2041-210X.2012.00190.x
Wang W., Teng F., Lin Y., Ji D., Xu Y., Chen C., et al. (2018). Transcriptomic study to understand thermal adaptation in a high temperature-tolerant strain of Pyropia haitanensis. PloS One 13 (4), e0195842. doi: 10.1371/journal.pone.0195842
Wei T., Simko V., Levy M., Xie Y., Jin Y., Zemla J. (2013). corrplot: Visualization of a correlation matrix. R Package version 0.73, 230(11).
Weigel B. L., Miranda K. K., Fogarty E. C., Watson A. R., Pfister C. A. (2022). Functional insights into the kelp microbiome from metagenome-assembled genomes. MSystems 7 (3), e01422–e01421. doi: 10.1128/msystems.01422-21
Weigel B. L., Pfister C. A. (2019). Successional dynamics and seascape-level patterns of microbial communities on the canopy-forming kelps Nereocystis luetkeana and Macrocystis pyrifera. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.00346
Weinberger F. (2007). Pathogen-induced defense and innate immunity in macroalgae. Biol. Bull. 213 (3), 290–302. doi: 10.2307/25066646
Weiss A., Costa R., Wichard T. (2017). Morphogenesis of Ulva mutabilis (Chlorophyta) induced by Maribacter species (Bacteroidetes, Flavobacteriaceae). Botanica Marina 60 (2), 197–206. doi: 10.1515/bot-2016-0083
Wemheuer F., Taylor J. A., Daniel R., Johnston E., Meinicke P., Thomas T., et al. (2020). Tax4Fun2: Prediction of habitat-specific functional profiles and functional redundancy based on 16S rRNA gene sequences. Environ. Microbiome 15 (1), 11. doi: 10.1186/s40793-020-00358-7
Wernberg T., Coleman M. A., Babcock R. C., Bell S. Y., Bolton J. J., Connell S. D., et al. (2019). Biology and ecology of the globally significant kelp Ecklonia radiata. Oceanography Mar. Biol. 57, 265–324. doi: 10.1201/9780429026379-6
Wernberg T., Russell B. D., Moore P. J., Ling S. D., Smale D. A., Campbell A., et al. (2011). Impacts of climate change in a global hotspot for temperate marine biodiversity and ocean warming. J. Exp. Mar. Biol. Ecol. 400 (1-2), 7–16. doi: 10.1016/j.jembe.2011.02.021
Wilkins L. G., Leray M., O’Dea A., Yuen B., Peixoto R. S., Pereira T. J., et al. (2019). Host-associated microbiomes drive structure and function of marine ecosystems. PloS Biol. 17 (11), e3000533. doi: 10.1371/journal.pbio.3000533
Keywords: microbiome, thermal stress, dysbiosis, kelp, disturbances
Citation: Vadillo Gonzalez S, Hurd CL, Britton D, Bennett E, Steinberg PD and Marzinelli EM (2024) Effects of temperature and microbial disruption on juvenile kelp Ecklonia radiata and its associated bacterial community. Front. Mar. Sci. 10:1332501. doi: 10.3389/fmars.2023.1332501
Received: 03 November 2023; Accepted: 27 December 2023;
Published: 18 January 2024.
Edited by:
Taewoo Ryu, Okinawa Institute of Science and Technology Graduate University, JapanReviewed by:
Raquel Xavier, Centro de Investigacao em Biodiversidade e Recursos Geneticos (CIBIO-InBIO), PortugalBrooke Weigel, University of Washington, United States
Copyright © 2024 Vadillo Gonzalez, Hurd, Britton, Bennett, Steinberg and Marzinelli. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sebastian Vadillo Gonzalez, sebastian.vadillo@sydney.edu.au
 Sebastian Vadillo Gonzalez
Sebastian Vadillo Gonzalez Catriona L. Hurd
Catriona L. Hurd Damon Britton
Damon Britton Eloise Bennett4
Eloise Bennett4  Peter D. Steinberg
Peter D. Steinberg Ezequiel M. Marzinelli
Ezequiel M. Marzinelli