Transcriptome analysis reveals ABA involved in the detoxification mechanism of macroalga Gracilariopsis lemaneiformis to cadmium toxicity
- Key Laboratory of Marine Biotechnology of Zhejiang Province, School of Marine Sciences, Ningbo University, Ningbo, Zhejiang, China
Introduction: Cadmium (Cd) is a significant threat environmental pollutant in the marine ecological environment offshore. The macroalga, Gracilariopsis lemaneiformis, of significant economic value, is widely cultivated along China’s coastline. Yet, little is known about the molecular mechanisms underlying Cd tolerance in macroalga.
Methods: Here, we examined the transcriptome of G. lemaneiformis exposed to Cd to identify the responses to Cd stress.
Results and discussion: Our findings revealed that Cd led to the retardation of growth rate in G. lemaneiformis, accompanied by a notable reduction in the content of photosynthetic pigments and a decrease in the expression of genes associated with the photosynthetic system and nitrogen metabolism. When exposed to Cd, there was a rapid increase in Cd levels through the upregulation of genes encoding GlZIP6 and GlIRT1. Additionally, the expression of Cd efflux transporters, GlZIP1 and GlABCG22, and the ABCC7 transporter for compartmentation to the vacuole, was induced to mitigate Cd toxicity. Cd also activated crucial genes involved in the ABA biosynthesis and enhanced ABA content, thereby inducing ABA signaling pathway. Furthermore, exogenous ABA reduced the growth inhibition of G. lemaneiformis under Cd stress. Redox homeostasis was adjusted to adapt to Cd toxicity, with thioredoxin, glutaredoxin cycle and ascorbate-glutathione cycle identified as playing significant in maintaining reactive oxygen species homeostasis. Moreover, transcription factors such as several MYBs, signal transmission factors G protein and heat shock proteins (sHSPs, HSP 40, HSP 90, HSP101) were involved in the detoxification of Cd. Collectively, this study provided a comprehensive understanding of the molecular mechanisms underpinning the of responses of macroalga G. lemaneiformis to Cd exposure.
1 Introduction
Cadmium (Cd), an inherently toxic heavy metal and non-essential element, engenders deleterious repercussions on plant development (Haider et al., 2021), as well as animal and human health (Kumar and Sharma, 2019). In recent years, there has been a marked increase in industrial discharges and marine aquaculture operations, leading to heightened concentrations of Cd, a persistent environmental contaminant. Numerous studies have indicated that Cd is the primary polluted metal with high hazard indices to aquatic ecosystem along China’s coast (Yu et al., 2022; Zhang et al., 2022a). For example, elevated levels of Cd have been detected in the water environment and sediments in electronic waste recycling areas in South China (Houessionon et al., 2021). Additionally, sedimentary deposits in the East China Sea coastal area (Zhang et al., 2022a), South China Sea (Xiao et al., 2022), and Bohai Bay (Yu et al., 2022) have all been contaminated by Cd.
The red alga, Gracilariopsis lemaneiformis (G. lemaneiformis), is a prominent economic macroalga cultivated in China’s coastal region. It serves as the primary source of agar and is also a cornerstone resource for pharmaceuticals (Zou et al., 2004). Additionally, it is widely used as an effective bait for various piscine species and abalones (Zhu et al., 2017). G. lemaneiformis has been found to absorb Cd and significantly reduce Cd concentration in sediment (Wang et al., 2014; Luo et al., 2020; Luo et al., 2021). The bioconcentration factors of Cd in G. lemaneiformis can reach to 1000 times (Wang et al., 2014). Consequently, G. lemaneiformis holds considerable potential for in-situ bioremediation of Cd in coastal areas. To date, research has primarily focused on the impact of Cd on the growth, absorption, and accumulation of Cd in G. lemaneiformis. Beyond the evident growth effects, recent studies on the Cd toxicity have highlighted complex cellular and molecular variations in higher plants and microalgae (Rahman et al., 2022; Zhang et al., 2023a). A majority of these studies have explored changes using OMICS strategies such as transcriptomics, metabolomics, ionomics, proteomics, and genomics when plants and microalgae are exposed to Cd stress (Rahman et al., 2022). Transcriptomic strategies are frequently employed to determine the significance of transcripts under environmental stressors in plant species (Rahman et al., 2022). The molecular study on the transcriptome regulatory profile was performed under Cd toxicities and results revealed the stress-responsive genes, key metabolic pathways under Cd stress, and crucial genes involved in these metabolic reactions (Chen et al., 2021; Yu et al., 2023). Furthermore, the transcriptome analysis uncovered the detoxification mechanism and genes involved in detoxification, such as hormones, transcription factors, antioxidant, heavy metal transporters, plant hormones and calcium signals to Cd toxicity (Ramegowda et al., 2020; Chen et al., 2021).
Previous research has demonstrated that Cd has negatively impacts the on growth and development of G. lemaneiformis by reducing pigment content (Xia et al., 2004). To gain a deeper understanding of the molecular regulatory networks and tolerance mechanisms in macroalgae to Cd, we examined these processes at the transcriptional level. The study aimed to investigate G. lemaneiformis’s stress response to Cd and identify the key genes involved in the detoxification processes. In the current study, we screened for a Cd concentration that caused moderate stress, measured Cd accumulation at various time points, and conducted a comparative transcriptome to investigate the genes responsive to Cd.
2 Materials and methods
2.1 Culture conditions
In this study, G. lemaneiformis strain 981 was collected from Ningde aquaculture base (Fujian, China (26°65′N, 119°66′E). Upon arrival at the laboratory, the attached sediment and coexisting algae adhering to G. lemaneiformis were eliminated thorough washing with sterile seawater. Subsequently, the robust G. lemaneiformis were cultivated under controlled conditions in an incubator for a duration of one week. The rhizoids with similar branching and length (13 ± 1 cm with the rhizome tip) were used for experiment and cultured at 23°C with a photoperiod of 12 h light: 12 h darkness and a light intensity of 50 μmol photons m-2s-1 light intensity.
2.2 Determination of relative growth rate, pigments, and glutathione content
The relative growth rate (RGR) was detected at 1, 2, 4 and 6 d post-Cd treatment with utilizing concentration of 50, 100, and 200 µM. The RGR was calculated using the formula established by Zou and Gao (2014): RGR (%) = Ln (Wt/W0) ×100. Here, Wt represents the fresh weight at the corresponding time point, while W0 represents the initial time point. Each treatment had three biological replicates.
The content of photosynthetic pigments and glutathione (GSH) content was determined at 6 d after Cd treatment in G. lemaneiformis. Photosynthetic pigments, phycoerythrin (PE) and phycocyanin (PC), were extracted with 50 mM PBS (pH 6.8). The extraction solution was centrifuged, and the absorbance was measured at 455, 564, 592, 618, and 645 nm by a spectrophotometer according to Sampath-Wiley and Neefus (2007) and Chen et al. (2022a). Chlorophyll a (Chl a) and carotenoids (Car) were extracted using methanol (5 mL) with 0.1 g G. lemaneiformis overnight in dark at 4°C. Then, the extracts were centrifuged at 10,000 rpm for 15 min at 4°C and measured at 480, 510, 652, 665, and 750 nm using full wavelength spectrophotometer according to the procedures described by Ji et al. (2019). GSH content quantified using colorimetric assay kits from Comin Bio. (Suzhou, China). In detail, approximately 0.1 g fresh sample extracted with 1 mL 50 mM PBS (pH 6.8) at 4°C and centrifuged at 8,000 rpm for 10 min at 4°C. Then, the supernatant reacted with 5,5’-dithiobis-2-nitrobenoicacid (DTNB) and measured at 412 nm by a spectrophotometer. Every physiological indicator each treatment had three biological replicates.
2.3 Determination of Cd content and bioconcentration factors
Cd level was quantified at 1, 3, 6, 12, 24 h, 3 d and 6 d after 75 µM of Cd treatment. Each time point had three biological replicates. In detail, the collected samples were rinsed thoroughly for 6 cycles with deionized water and dried in an oven at 80°C for 24 h. Then, the desiccated sample (0.1 g) was ground for nitric acid digestion. The quantification of Cd content was executed via an inductively coupled plasma mass spectrometry (ICP-MS, iCAP RQ, Thermo Fisher Scientific, CA, USA) (Zhao et al., 2022).
The bioconcentration factor (BCF) is determined by the ratio q/c, which signifies degree of enrichment level of a specific chemical in living organisms (Foster, 1976). The term “q (mg/kg DW)” represents the concentration of Cd content found in G. lemaneiformis, while “c (mg/L)” signifies the dissolved Cd concentration within a solution.
2.4 ABA quantification
Abscisic acid (ABA) measurement was performed using high-performance liquid chromatography-electrospray ionization-tandem mass spectrometry (HPLC-ESI-MS/MS) according to Shao et al. (2019) with some modifications. G. lemaneiformis samples (1.0 g fresh weight) were ground in liquid nitrogen and extracted at 4°C overnight with methanol containing 0.2% formic acid (FA) that had been spiked with internal standards. The extracts were centrifuged 1,4000 rpm at 4°C, and the resulting supernatants were dried under N2. The obtained pellet was then resuspended with 200 μL of 50% methanol (1:1, v/v), centrifuged before HPLC-ESI-MS/MS analysis. The mobile phases consisted of solvent A (0.04% [v/v] formic acid (FA) in water) and solvent B (0.04% [v/v] FA in acetonitrile). Samples were placed in an automatic sampler at 4°C with an injection volume of 4 μL. The column temperature was maintained at 45°C, and a flow rate of 400 μL/min was employed. The linear gradient was programmed as follows: 0-10 min, transitioning from 2 to 98% solvent B (v/v), 10-11.1 min, transitioning from 98 to 2% solvent B (v/v), and 11.1-13 min, hold at 2% solvent B. A Qtrap 5500 System (AB Sciex) was used for mass spectrometry in negative ion mode and multiple reaction monitoring modes. Data was analyzed using Multiquant 3.03 software.
2.5 RNA sequencing and analysis of differentially expressed genes
Total RNA was extracted from G. lemaneiformis samples (0.1 g fresh weight) using the RNeasy Plant Mini Kit (Qiagen, Hilden, Germany). Each treatment had three biological replicates. RNA-seq was performed by BGI (Shenzhen, China). SOAPnuke (https://github.com/BGI-flexlab/SOAPnuke) was used to remove reads containing adapter sequences or more than 10% unidentified nucleotides and low-quality reads to obtain clean read databases. The clean data were mapped to the reference transcripts, which combined the transcripts from genomic annotation and full-length transcripts obtained by SMRT-Seq (Chen et al., 2022b), by Bowtie2 (https://bowtie-bio.sourceforge.net/bowtie2/index.shtml). Expectation Maximization (RSEM) tool was applied to calculate the transcript abundance of each gene in each sample based on fragments per kilobase per million reads (FPKM) (Li and Dewey, 2011).
The differentially expressed genes (DEGs) was identified based on the criterion of |log2 [fold-change (FC)]| ≥ 1 and an adjusted P value (Q value) ≤ 0.001. The Gene Ontology (GO) (https://www.geneontology.org/) term and Kyoto Encyclopedia of Genes and Genomes (KEGG) (https://www.genome.jp/kegg/) pathway significantly enriched in DEGs were obtained by performing GO and KEGG enrichment analysis, respectively. A Q value ≤ 0.01 was used to determine if a GO term was significantly enriched, while a P value ≤ 0.05 was used to determine if a KEGG pathway was significantly enriched.
The Swiss-Prot (http://web.expasy.org/compute_pi/), Cell-PLoc 2.0 (http://www.csbio.sjtu.edu.cn/bioinf/Cell-PLoc-2/), and InterPro (http://prosite.expast.org/) databases were utilized to predict the molecular weight, subcellular localization, and domains of genes, respectively.
2.6 Quantitative real-time RT-PCR analysis
For the validation of transcriptome results, six DEGs were randomly selected for qRT-PCR analysis. The method of qRT-PCR was according to our previous study (Chen et al., 2022a). Gene-specific primer pairs are listed in Supplementary Table S1. As the internal reference gene, actin was employed, and the relative expression of the target gene was calculated by the 2-ΔΔCt method (Livak and Schmittgen, 2001).
2.7 Statistical analysis
The presentation of data was achieved as mean ± standard deviation (SD), with n = 3 replicates. The Univariate General Linear Models procedure of SPSS Statistics 22.0 was applied to analyze the statistical significance of the data. One-way ANOVA and Duncan’s multiple range tests were used to separate treatment averages at P <0.05 and P <0.01.
3 Results and discussion
3.1 Impact of Cd stress on growth and pigments content of G. lemaneiformis
With the escalation of Cd concentration ranging from 50 to 500 μM, a substantial reduction in the RGR of G. lemaneiformis was observed (Supplementary Figures 1A, B). The algal biomass exhibited complete discoloration and lethality at a Cd concentration of 500 μM (Supplementary Figure 1A). At 50 μM of Cd, the RGR exhibited a decline from 12.8% on the 1st day to 47.0% on the 6th day. Furthermore, the branching pattern of G. lemaneiformis displayed shorter and fewer branches compared to the control (Supplementary Figure 1). At a Cd concentration of 100 μM, the RGR registered a decrease of 63.5% on the 6th day, accompanied by the emergence of white branches. Notably, the growth was significantly suppressed at 200 μM Cd concentration, resulting in a 93.5% decline in RGR by the 6th day. This phenomenon was similar to previous studies by Xia et al. (2004), the RGR in 50, 100, 200 μM Cd-treatments decreased by 7.0%, 46.3% and 89.4% for 96 h.
Moreover, at the same Cd concentrations of 50-200 μM, the content of photosynthetic pigments, PE, PC and Chl a, exhibited marked diminishment by 24.6-38.4%, 32.3-53.4% and 6.5-13.7% after 6 d of Cd treatment, respectively (Supplementary Figures 1C–E). In contrast, the content of GSH significantly increased in a dose-dependent manner within the range of 50–200 μM Cd by 11.7–37 folds (Supplementary Figure 1F). Glutathione, the heavy metal chelating agent, chelates with Cd already located inside cells for detoxification (Fang et al., 2020; Das et al., 2021). In many studies, Cd has proved to induce an increase in GSH content (Gao et al., 2022), in consistent with our result.
To avoid a severe growth inhibition of G. lemaneiformis, the IC50 value corresponding to a 50% reduction in RGR was selected for further studies at 6 d after stress. By virtue of linear interpolation, the determined IC50 value of Cd was 75 μM.
3.2 Cd accumulation in G. lemaneiformis
In this section, we investigate the accumulation of Cd in G. lemaneiformis. The accumulation process is crucial to understanding the impact of environmental pollutants on aquatic ecosystems and the potential for bioaccumulation. To clarify the dynamics of Cd uptake in G. lemaneiformis, the accumulation of Cd in the algal was determined (Table 1). The Cd content increased rapidly within 12 h and reached a maximum at day 3, suggesting that G. lemaneiformis can quickly absorb Cd. The BCF of G. lemaneiformis ranged from 4.86 to 28.46 during the experimental period. In particular, the accumulation of Cd accounted for 54% and 93% of its maximum capacity at 1 and 3 d after Cd stress, respectively. Wang et al. (2014) reported that the maximum absorption of low concentration Cd (1, 5, 10, and 20 μg/L) in G. lemaneiformis was between 20-30 days and the BCF was 463- 1113. Our results revealed that G. lemaneiformis can quickly absorb Cd at high concentration for bioremediation.
3.3 Transcriptome analysis and gene functional annotation
To clarify the responsive gene to Cd, we performed transcriptomes analysis of G. lemaneiformis at 1 and 3 d after Cd stress. After removing adaptors, N>10 and low-quality reads, the clean reads were aligned to reference transcripts with an overall mapping rate of 91-92%. In a result, 22,806 transcripts were expressed. Among these, 13,800, 12,088, 18,856, 320 and 125 transcripts were annotated to GO, KEGG, RefSeq non-redundant proteins (NR), Transcription factors (TFs) and Plant Resistance Gene (PRG) public databases, respectively. Approximately 82.9% of these transcripts were annotated in at least one database.
3.4 Identification and validation of differentially expressed genes
For comparison, 1967 (998 up and 969 down) and 3369 (1850 up and 1519 down) DEGs were identified at 1 and 3 d after Cd stress, respectively (Supplementary Figures 2A, B). This result showed that the gene response was enhanced in G. lemaneiformis with the increase of Cd accumulation. Moreover, 1192 (592 up and 474 down) of the DEGs were co-regulated at 1 and 3 d after treatment (Supplementary Figure 2C). The qRT-PCR results were consistent with those of RNA-Seq (Supplementary Table S1), which verified the reliability of RNA-Seq.
3.5 GO, KEGG enrichment of DEGs
GO enrichment analysis of DEGs revealed that numerous GO terms were influenced by Cd. The molecular function, including “pigment binding”, “antioxidant activity”, “transmembrane transporter activity”, biological regulation such as “response to far red light” and “photosynthesis”, “ROS biosynthetic process”, “cellular oxidant detoxification” were observed at 1 and 3 d in response to Cd stress. Furthermore, cellular component including “plastoglobule”, “plastid thylakoid membrane” and “photosynthetic membrane” were also implicated (Figure 1). The GO terms “detoxification”, “antioxidant activity”, and “signal transducer activity”, which may play key roles in abiotic stress and “membrane” were also found in kenaf in the response to Cd exposure (Chen et al., 2021).
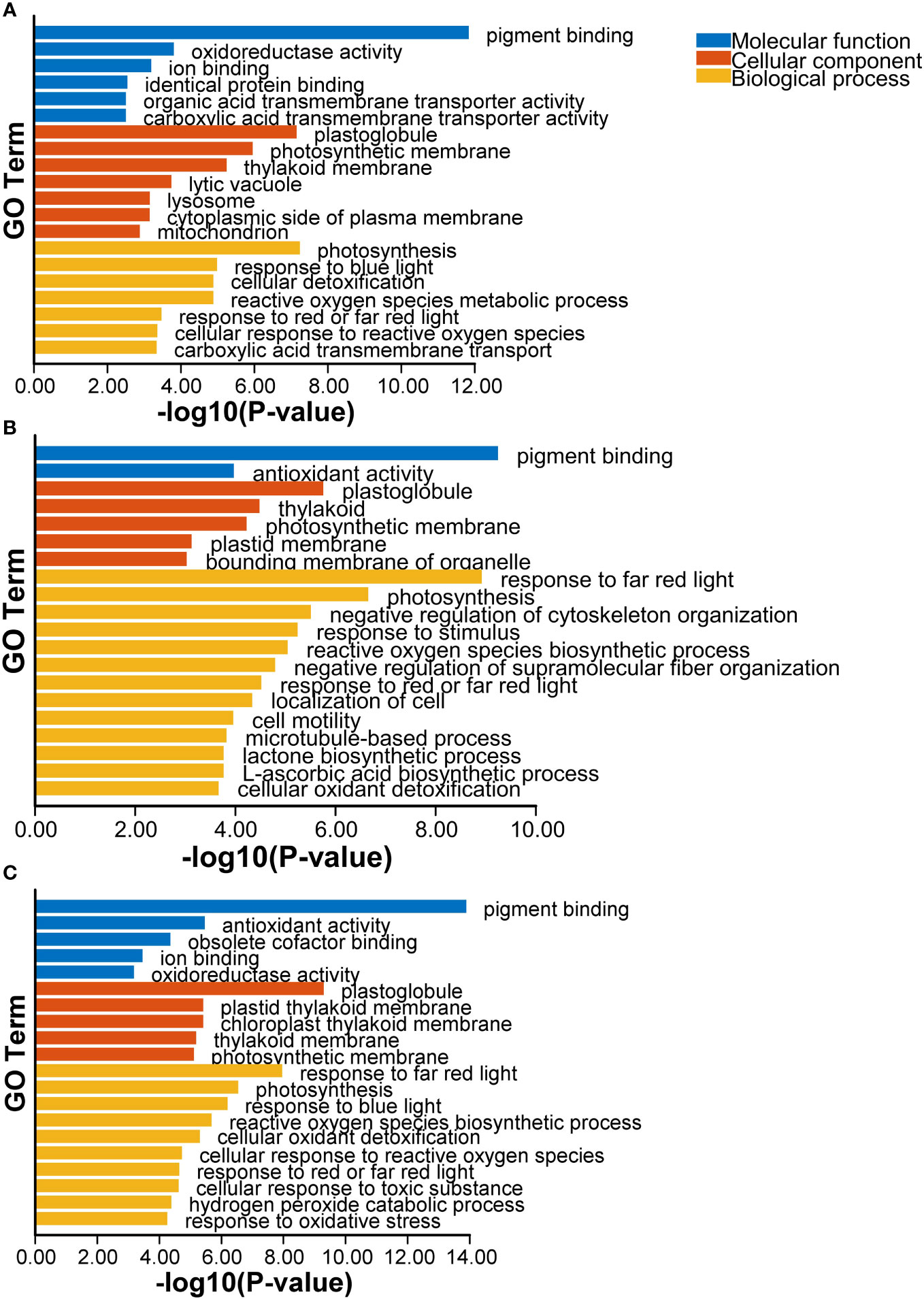
Figure 1 The top20 of GO enrichment terms of DEGs after Cd treatment at 1 d (A), 3d (B) and co-expressed at 1 and 3 d (C).
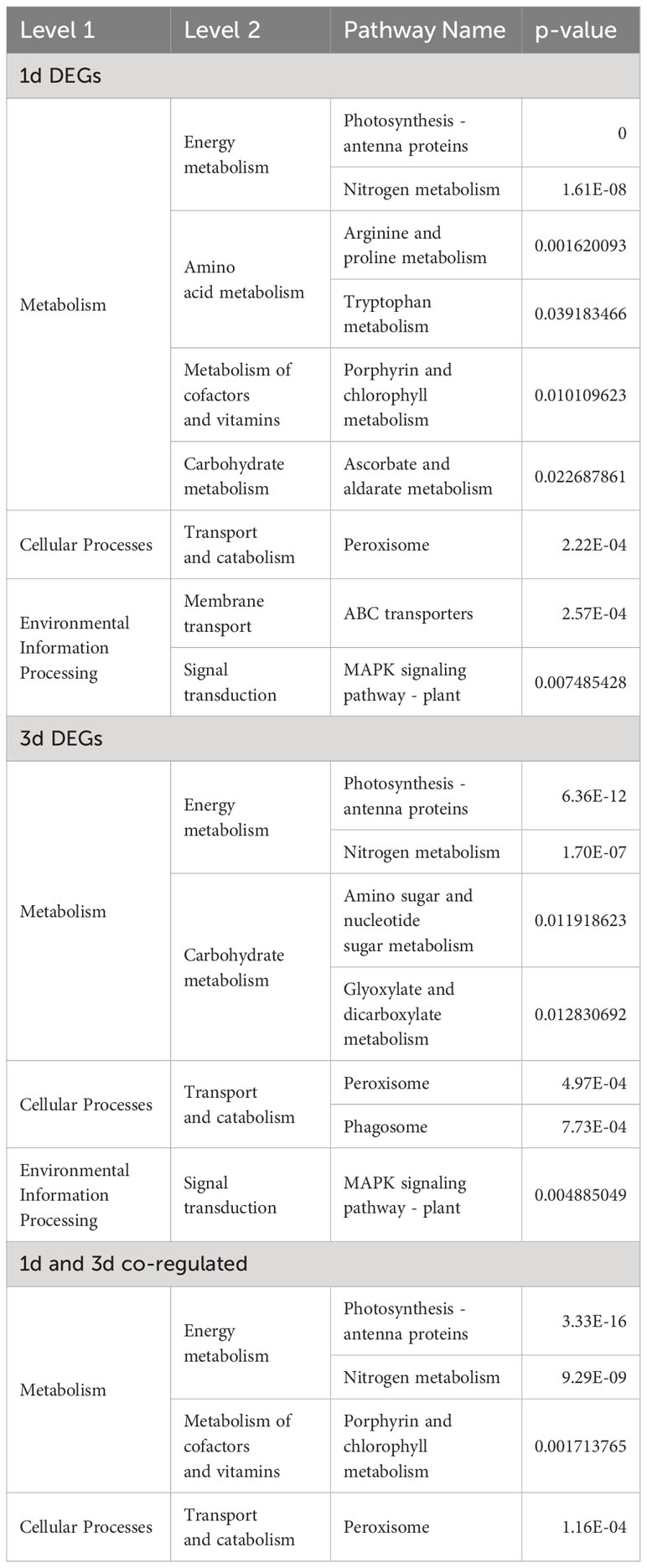
KEGG pathway analysis revealed that DEGs at 1 and 3 d were predominantly associated with metabolism processes, including energy metabolism and carbohydrate metabolism, as well as cellular processes such as “peroxisome” and “transporter” and “MAPK signaling pathway–plant” (Table 2). These data suggested that genes related to photosynthesis system, antioxidant activity, transmembrane transporter activity, energy metabolism, carbohydrate metabolism, and MAPK signaling pathway – plant play a crucial role in the response to Cd exposure in G. lemaneiformis.
3.6 Specific differentially expressed genes in response to Cd
3.6.1 DEGs involved in signal transduction and ABA biosynthesis pathway
In MAPK signal-plant pathway, we found that many pathways generated by signaling molecules and stress-induced were affected by Cd stress. However, most of genes involved in these pathways were down-regulated by Cd, for example, FLS2 involved in fg22-mediated disease resistance, NDPK2 mediated by H2O2, CAM4 mediated by wounding (Figures 2A, B). Whereas, Cd activated genes encoding serine/threonine-protein kinase SRK2 (SnRK2) involved in ABA signaling pathway (Figures 2A, B). Moreover, the expression of downstream genes encoding ABA responsive element binding factor (ABF) and MKK3 was enhanced by 1.1-1.4 folds and 1.2-1.5 folds, respectively by Cd (Supplementary Table S3). Furthermore, expression of key genes encoding 15-cis-phytoene desaturase (PDS), prolycopene isomerase (crtISO) and zeaxanthin epoxidase (ZEP) involved in ABA biosynthesis pathway was also induced by Cd (Figure 2C; Supplementary Table S3).
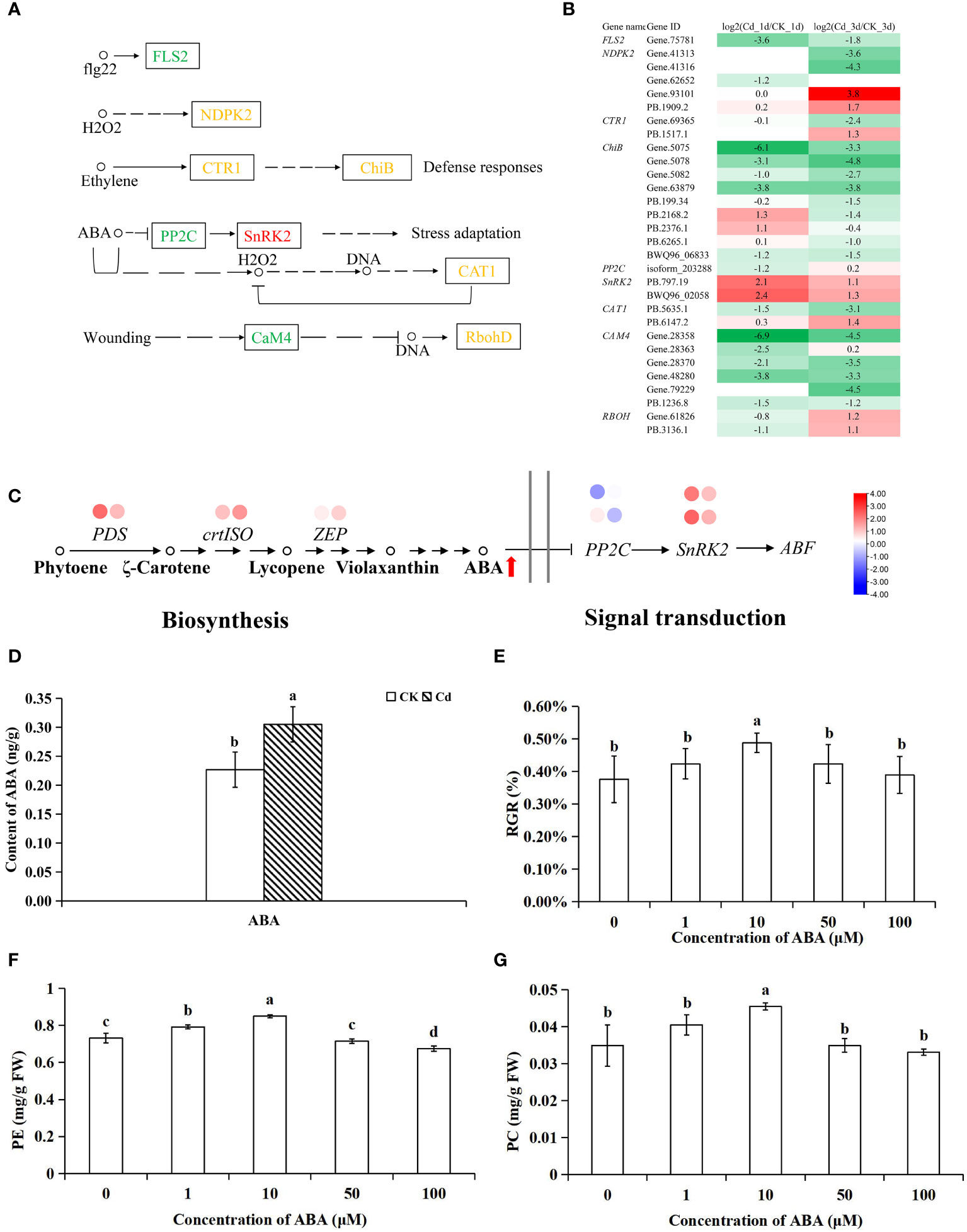
Figure 2 The expression profiles of Cd-responsive genes involved in MAPK signaling pathway-plant (A, B), ABA pathway (C) and the effect of Cd on ABA content (D), and the effect of ABA on the RGR (E), PE (F), PC (G) in G. lemaneiformis under Cd stress. Each circle beside the name of genes represents the abundance change of corresponding DEGs at 1 d (left) and 3 d (right), respectively. The red arrows next to the metabolite represent a significant increase in the content of metabolite. The different letters on the 2 columns indicate significant differences between different concentrations of treatments (P < 0.05).
As the enhanced expression of genes related to the ABA synthesis pathway was observed in G. lemaneiformis after Cd stress, ABA content quantified 1 day post-stress using UPLC-MS. The MS data and retention times of ABA are detailed in Supplementary Table S2. Notably, the presence of Cd led to a 34.5% remarkable increase in ABA content (Figure 2D).
Considering the content of hormones and expression profiles, we conducted that ABA might be play a positive role in response to Cd stress in G. lemaneiformis. To further verify the effect of ABA on growth of G. lemaneiformis under Cd stress, different concentrations of ABA were applied to G. lemaneiformis under 75 µM Cd stress. At 10 µM, ABA reduced the inhibition of Cd on the growth of G. lemaneiformis and increased the contents of PE and PC (Figures 2E–G). A number of reports have demonstrated that ABA is closely related to perception and mediation of Cd stress (Rasafi et al., 2020). For instance, in rice seedlings and lettuce, there is a rapid enhancement of ABA content under Cd stress (Hsu and Kao, 2003; Tang et al., 2020), and in Arabidopsis thaliana exogenous application of ABA has been shown to significantly decreased Cd content and alleviated Cd toxicity (Meng et al., 2022). Wang et al. (2022) reported that gene involved in ABA signaling pathway can enhance Cd resistance ability of T. hispida by regulating the production of ROS under Cd stress. Our results revealed that ABA and ABA signal may be involved in the induction of protective mechanisms against Cd toxicity in G. lemaneiformis.
3.6.2 DEGs involved in antioxidant system
GO enrichment analysis revealed that “antioxidant activity” and “cellular oxidant detoxification” was involved in the response to Cd. The antioxidant system may play key roles in Cd stress in plants (Zhang and Reynolds, 2019). In plants, the antioxidant defense system mainly consists of superoxide mutase (SOD), which converts oxygen radicals (O2−) into hydrogen peroxide (H2O2), and L-ascorbate peroxidase (APX), peroxidase (POD), and catalase (CAT), these three enzymes remove toxic H2O2 (Zhang and Reynolds, 2019). Furthermore, the sulfur-containing antioxidant including thioredoxin (Trx), ascorbate (AsA) -GSH cycle and glutaredoxin cycle, etc., may also involve in the detoxification of Cd (Smiri et al., 2011; Zhang et al., 2020; Zhang et al., 2023a). A total of 6 (3 upregulated and 3 downregulated) and 12 (8 upregulated and 4 downregulated) DEGs associated with antioxidant activity were identified at 1 and 3d after Cd stress, respectively (Figure 3). Among them, expression of 2 SOD encoding genes (SOD3 and EC-SOD) and 2 of 3 CAT encoding genes (CAT-B, CAT) was down-regulated, while APX encoding gene (APX4) and two thioredoxin (Trx) encoding genes (TRX2.1 and TRX2.2) were up-regulated (Figures 3A, C). Trxs are disulfide reductases containing a conserved active redox center thioredoxin domain (WCGPC), which also exist in GlTRX2.1 and GlTRX2.2 (Supplementary Table S4). Moreover, expression of three genes encoding thioredoxin-like disulfide reductases (TRLs) were up-regulated at 3 d after Cd stress (Figures 3A, C). GlTRLs possess a thioredoxin domain and TPR-like_helical domain (Supplementary Table S4), which catalyze the transfer of electrons from a reduced substrate to the disulfide bridge of Trx (Figure 3A). Trxs distribute the reductant to selected targets via a dithiol-disulfide exchange, resulting in the structural and functional modification of a target protein (Balsera and Buchanan, 2019). Zhang et al. reported that the thioredoxin-like protein CDSP32 improves cadmium tolerance by alleviating Cd-induced photosynthetic inhibition (Zhang et al., 2020) and modulating antioxidant system in tobacco leaves (Zhang et al., 2023b).
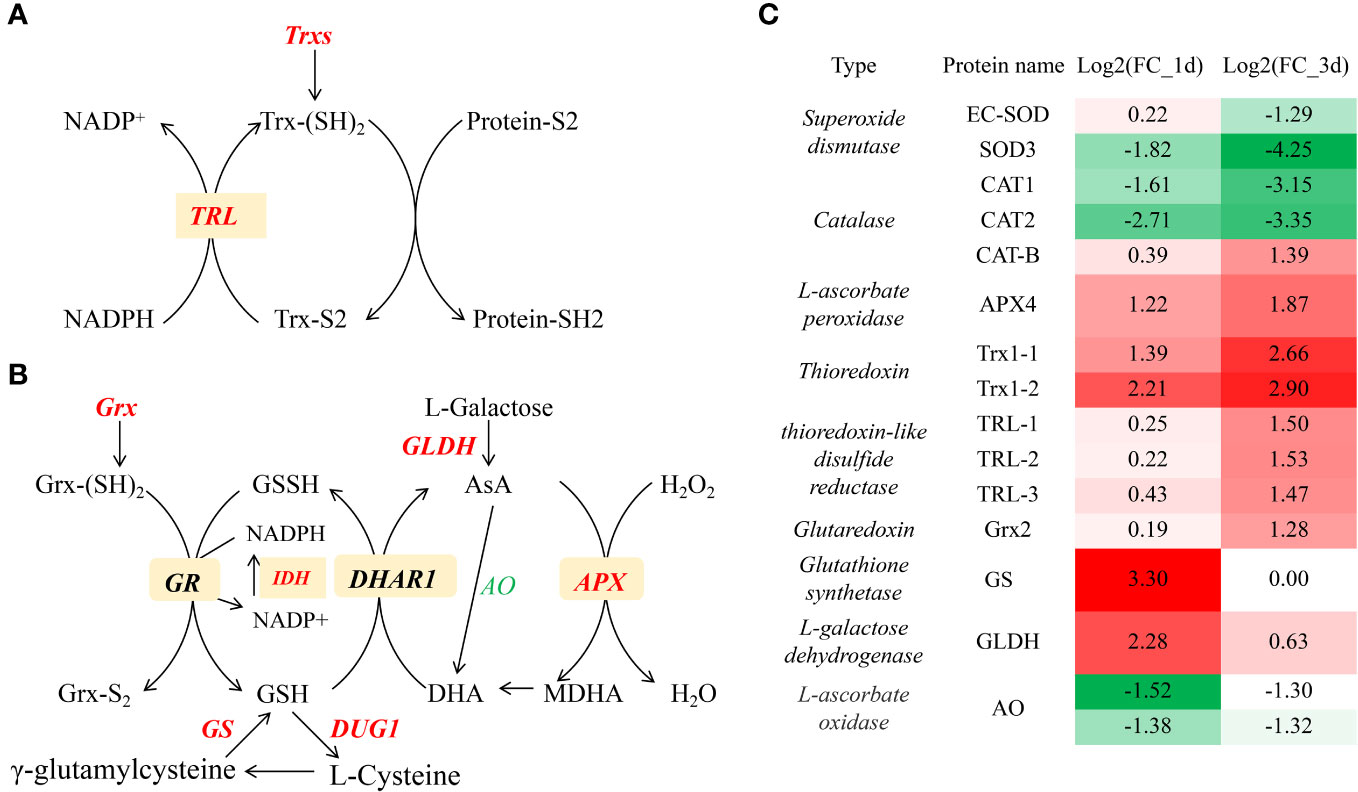
Figure 3 The expression profiles of Cd-responsive genes involved in thioredoxin pathway (A, C) and Glutaredoxin cycle, ascorbate-glutathione cycle (B, C). Italics represent genes, red and green represents up-regulation and down- regulation, respectively.
The Glutaredoxin (Grx) cycle, comprised of Grxs, GSH, and glutathione reductase (GR), is crucial for maintaining intracellular redox homeostasis (Ogata et al., 2021). Grxs are a multifunctional family of glutathione-dependent disulfide oxidoreductases. In G. lemaneiformis, expression of Grx encoding gene (GLX2) was upregulated at 3 d after Cd stress (Figures 3B, C). The ascorbate-glutathione cycle is acknowledged as a cellular redox buffer which donates electrons to various oxidized compounds in plants (Decros et al., 2023). Expression of genes encoding L-galactose dehydrogenase (GLDH) and glutathione synthetase (GS) was upregulated; these enzymes catalyze the conversion of L-galactose to ascorbate and γ-glutamylcysteine to GSH, respectively. Expression levels of APX, which detoxifies excess toxic ROS H2O2 to H2O, were also upregulated (Figure 3B). These data suggested that the Grx cycle and ascorbate-glutathione cycle alleviates Cd toxicity by scavenging ROS. Glutathione have been proposed as Cd-binding chelator (Sears, 2013). The upregulation of these genes encoding Trxs, Grxs and GSS showed that sulfur antioxidants may play important roles in controlling oxidative stress in G. lemaneiformis exposed to Cd.
3.6.3 DEGs involved in Cd transports
In plants, Cd transporters mainly include natural resistance-associated macrophage protein (Nramp), heavy metal-transporting ATPase (HMA), iron regulated transporter protein (IRT), zinc transporter protein (ZIP), ATP-binding cassette (ABC) and yellow stripe-like (YSL) families (Hu et al., 2022; Tao and Lu, 2022). Up to now, Nramps, ZIP5/6/9 and IRT1/2 are found to be involved in the uptake of Cd from the soil to roots in plants (Tao and Lu, 2022). In G. lemaneiformis, YSL transporters do not exist, and the expression of GlNramps and GlHMA was not altered by Cd. Similarly, Nramps were not differentially expressed under Cd stress in algae C. reinhardti (Zhang et al., 2023a). In G. lemaneiformis, the expression of transporter genes GlIRT1 and GlZIP6 was significantly enhanced by Cd (Figure 3). This suggested that G. lemaneiformis actively engages in the uptake and transportation of Cd upon exposure.
Plants possess several detoxification mechanisms to mitigate Cd toxicity including efflux, chelation binding to metallothioneins/PCs and compartmentation to vacuole. In rice, ZIP1, ABCG36 and HMA9 mediate Cd efflux, and ABCG8 mediates Cd efflux in Arabidopsis thaliana (Tao and Lu, 2022). In G. lemaneiformis, Cd induced the expression of genes encoding GlZIP1 and GlABCG22 for Cd efflux (Figure 4). Particularly, expression of GlZIP1 was upregulated more than 2-fold at both 1 and 3 d after Cd stress (Figure 4), and that in C. reinhardti increased by more than 5 times (Zhang et al., 2023a). However, expression of genes coding for phytochelatins or metallothioneins did not elevated by Cd. HMA3 and ABCC subfamily transported Cd into vacuole for sequestration in plants (Tao and Lu, 2022). In G. lemaneiformis, the expression of multidrug resistance protein 7 (MRP 7, ABCC subfamily) was significantly upregulated at 3 d after Cd stress (Figure 4). Cd is also can bind the dimer of GSH (Zhang et al., 2022b), which is consistent with the increase of GSH content in our result, and then transported to vacuole in form of chelates. These results indicated that G. lemaneiformis through synthesis of GSH, then combined Cd transport into the vacuole and improve Cd efflux to reduce the Cd damage.
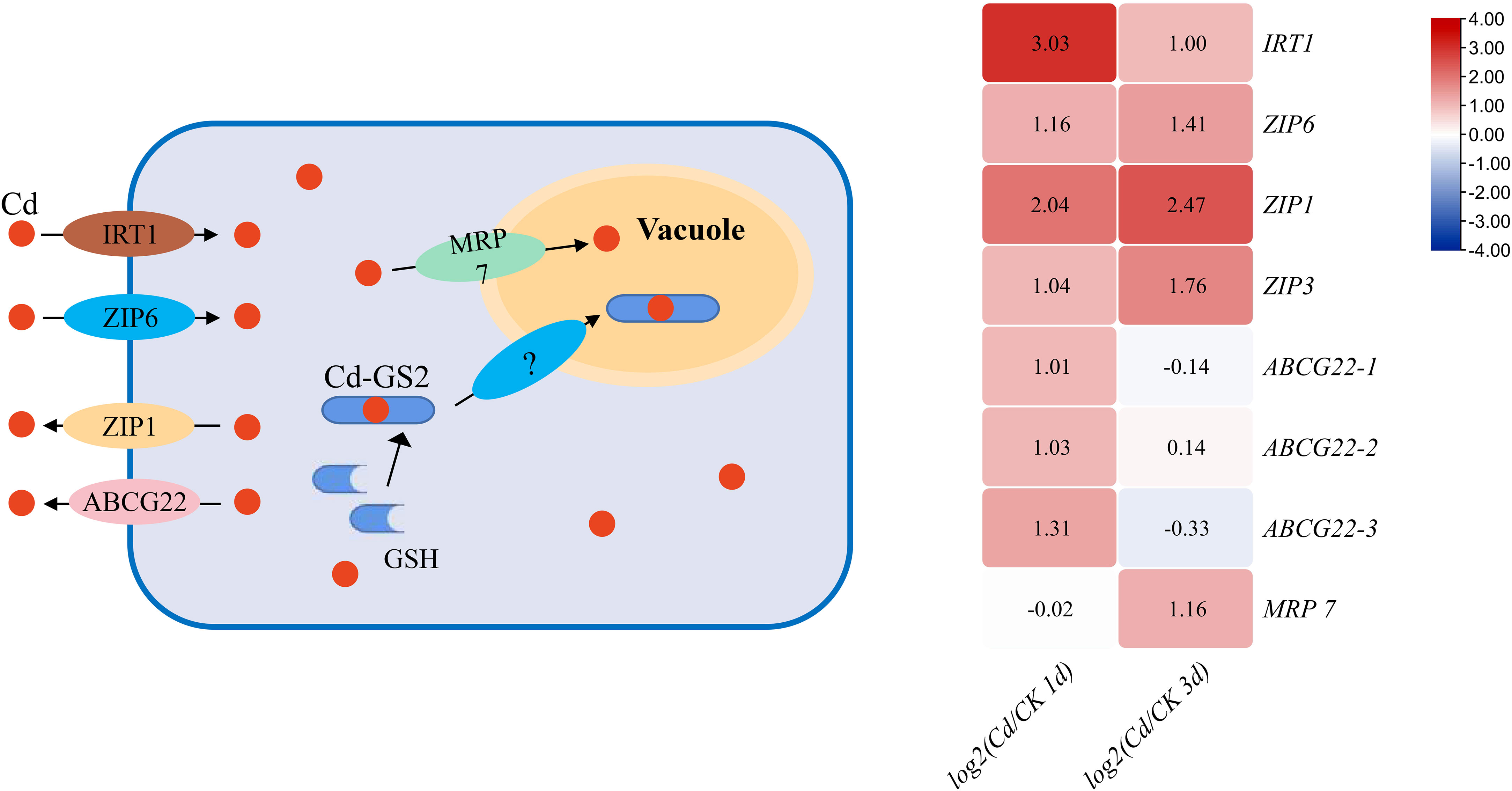
Figure 4 The DEGs of Cd uptake, efflux and vacuole sequestration genes in response to Cd at 1 and 3 d in G. lemaneiformis.
3.6.4 Cd negatively affected energy metabolism
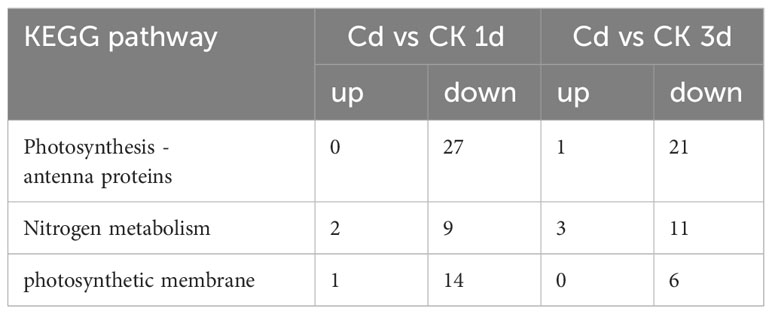
KEGG analysis revealed an enrichment of energy metabolism processes, including photosynthesis - antenna proteins and nitrogen metabolism under Cd stress. In the process of photosynthesis - antenna proteins, the genes encoding allophycocyanin (APC), phycocyanin (PC) and light-harvesting chlorophyll protein complex (LHC) were affected under Cd stress (Supplementary Table S5). In total, 27 and 22 transcripts were differentially expressed at 1 and 3 d, separately. Of these, 27 and 21 transcripts encoding ApcB-C, CpcA-C, Lhca1 and Lhca4 were down-regulated at 1 and 3 d after Cd stress, respectively (Table 3; Supplementary Table S5). This is consistent with a decrease in photosynthetic pigments PE, PC and Chl a content. In the nitrogen metabolism pathway, 9 (1 up and 8 down) and 13 (4 up and 9 down) transcripts encoding nitrate/nitrite transporter (NRT), nitrate reductase (NAD(P)H), ferredoxin-nitrite reductase, glutamine synthetase, glutamate synthase (ferredoxin) and glutamate dehydrogenase (NAD(P)+) were differentially expressed at 1 and 3 d, respectively (Supplementary Table S5). In addition, Cd destroys chloroplast structures, such as the photosynthetic membrane (Table 3; Supplementary Table S5), which is consistent with results reported in plants (Parmar et al., 2013). These findings indicated that Cd downregulated the expression of genes involved in the energy metabolism pathway to inhibit the growth of G. lemaneiformis.
3.6.5 Transcription factors, G-proteins and heat shock proteins are involved in the response to Cd stress
Many studies have shown that transcription factors are involved in the response to Cd in plants. In G. lemaneiformis, most DEGs related to the “transcription factor” including MYB, TBP-binding protein (TBP) and SNF7 were significantly upregulated under Cd stress, suggesting that TFs participate in response to Cd (Figure 5A; Supplementary Table S6). Among them, 3 transcripts encoding MYB transcription factors were significantly upregulated by Cd at both 1 and 3 d. The transcription factor MYBs is one of the most widely distributed families in plants and participate in responses to stresses by binding to the cis-elements of MYB in the promoter of target genes (Wang et al., 2021). Overexpression of AtMYB4 and SbMYB15 resulted in increased tolerance to Cd (Sapara et al., 2019; Agarwal et al., 2020). In addition, “G-Protein” was also significantly upregulated under Cd stress (Supplementary Table S7). G proteins function as molecular switches to participate in the transmission of various extracellular stimulus signals and responses to abiotic stresses in plants (Zhang et al., 2021). Studies have shown that overexpression of AGG3 can increase tolerance to Cd (Alvarez et al., 2015).
HSPs act as molecular chaperones for a variety of target proteins in response to heavy metal stress and play a crucial role in maintenance of protein homeostasis (Fragkostefanakis et al., 2015). In our results, expression of transcripts encoding HSPs including 6 sHsp, 4 Hsp 40, 1 Hsp 90-5 and 1 Hsp 101 was upregulated by Cd (Figure 5B). Particularly, sHSP was greatly induced by Cd, and the multiple was generally between 3-6 times. SHSPs, the first line of stress defense (Haslbeck and Vierling, 2015), can bind denatured proteins, stabilize them, and prevent an aggregating of denatured proteins without depending on ATP, then the denatured proteins are refolded by either the HSP70 system or the HSP40/HSP70 system (Figure 5B). The highly conserved C-terminal region of sHsp is the α-crystallin domain (ACD domain) (Haslbeck and Vierling, 2015), which present in all these DEGs of sHsp (Supplementary Table S8). In Closterium ehrenbergii, expression of sHSP10 and sHSP17.1 was significantly upregulated under heavy metal stress (Abassi et al., 2019). Expression of small heat shock protein OsMSR3 in Arabidopsis enhanced tolerance to Cd stress (Cui et al., 2013). HSP40s together with HSP70s constitute a versatile chaperone machinery for stress responses (Qiu et al., 2006). However, HSP70s were not induced by Cd in G. lemaneiformis. Hsp40 proteins contain the J domain through which they bind to Hsp70s (Qiu et al., 2006), and the J domain exist in the DEGs of GlHsp40s (Supplementary Table S8). In Venerupis philippinarum, the expression of VpHSP40 induced by Cd (Li and Dewey, 2011) and the Hsp40-like protein provided tolerance to Cd toxicity (Thakur et al., 2021). Together, these results indicated that HSPs might improve the tolerance to Cd in G. lemaneiformis.
4 Conclusions
In summary, our work revealed the response of macroalgae to Cd at the molecular level. Both transcriptome and physiological results showed that Cd was toxic to macroalgae and triggered growth and photosynthesis inhibition. Our results showed that ABA levels and the expression of genes related to ABA biosynthesis and signaling pathway was induced in the presence of Cd. And application of ABA alleviated Cd toxicity, suggesting that ABA played a key role against Cd stress in G. lemaneiformis. In addition, several ABC and ZIP transporters responding to Cd were identified. Specifically, thioredoxin, glutaredoxin cycle and ascorbate-glutathione cycle played important roles in maintaining ROS homeostasis. Furthermore, MYBs transcription factors, signal transduction factor G-protein and HSPs were involved in detoxification to Cd stress, these genes could be good Cd-responsive marker genes. Taken together, our results identified novel genes and pathways for improving Cd accumulation in algae and provided new insights into the detoxification mechanisms of macroalgae under Cd stress.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author contributions
XC: Data curation, Visualization, Writing – original draft. YT: Methodology, Writing – review & editing. XS: Supervision, Writing – review & editing. HZ: Software, Writing – review & editing. NX: Writing – review & editing, Supervision.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was financially supported by the Natural Science Foundation of Zhejiang Province (LY22C190005), the Natural Science Foundation of China (31901465), and the open fund of the State Key Laboratory of Plant Physiology and Biochemistry (SKLPPBKF2117).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2024.1352529/full#supplementary-material
References
Abassi S., Wang H., Ponmani T., Ki J. S. (2019). Small heat shock protein genes of the green algae Closterium ehrenbergii: Cloning and differential expression under heat and heavy metal stresses. Environ. Toxicol. 34 (9), 1013–1024. doi: 10.1002/tox.22772
Agarwal P., Mitra M., Banerjee S., Roy S. (2020). MYB4 transcription factor, a member of R2R3-subfamily of MYB domain protein, regulates cadmium tolerance via enhanced protection against oxidative damage and increases expression of PCS1 and MT1C in Arabidopsis. Plant Sci. 297, 110501. doi: 10.1016/j.plantsci.2020.110501
Alvarez S., Roy Choudhury S., Sivagnanam K., Hicks L. M., Pandey S. (2015). Quantitative proteomics analysis of Camelina sativa seeds overexpressing the AGG3 gene to identify the proteomic basis of increased yield and stress tolerance. J. Proteome Res. 14 (6), 2606–2616. doi: 10.1021/acs.jproteome.5b00150
Balsera M., Buchanan B. B. (2019). Evolution of the thioredoxin system as a step enabling adaptation to oxidative stress. Free Radical Biol. Med. 140, 28–35. doi: 10.1016/j.freeradbiomed.2019.03.003
Chen P., Li Z., Luo D., Jia R., Lu H., Tang M., et al. (2021). Comparative transcriptomic analysis reveals key genes and pathways in two different cadmium tolerance kenaf (Hibiscus cannabinus L.) cultivars. Chemosphere 263, 128211. doi: 10.1016/j.chemosphere.2020.128211
Chen X., Tang Y., Sun X., Zhang X., Xu N. (2022a). Comparative transcriptome analysis reveals the promoting effects of IAA on biomass production and branching of Gracilariopsis lemaneiformis. Aquaculture 548, 737678. doi: 10.1016/j.aquaculture.2021.737678
Chen X., Tang Y. Y., Yin H., Sun X., Zhang X., Xu N. (2022b). A survey of the full-length transcriptome of Gracilariopsis lemaneiformis using single-molecule long-read sequencing. BMC Plant Biol. 22 (1), 597. doi: 10.1186/s12870-022-03992-0
Cui Y. C., Xu G. Y., Wang M. L., Yu Y., Li M. J., da Rocha P. S. C. F., et al. (2013). Expression of small heat shock protein OsMSR3 in Arabidopsis enhances tolerance to cadmium stress. Plant Cell Tissue Organ Cult 113, 331–340. doi: 10.1007/s11240-012-0275-x
Das U., Rahman M. A., Ela E. J., Lee K. W., Kabir A. H. (2021). Sulfur triggers glutathione and phytochelatin accumulation causing excess Cd bound to the cell wall of roots in alleviating Cd-toxicity in alfalfa. Chemosphere 262, 128361. doi: 10.1016/j.chemosphere.2020.128361
Decros G., Dussarrat T., Baldet P., Cassan C., Cabasson C., Dieuaide-Noubhani M., et al. (2023). Enzyme-based kinetic modelling of ASC-GSH cycle during tomato fruit development reveals the importance of reducing power and ROS availability. New Phytol. 240 (1), 242–257. doi: 10.1111/nph.19160
Fang Z., Hu Z., Yin X., Song G., Cai Q. (2020). Exogenous glutathione alleviation of cd toxicity in italian ryegrass (Lolium multiflorum) by modulation of the cd absorption, subcellular distribution, and chemical form. Int. J. Environ. Res. Public Health 17 (21), 8143. doi: 10.3390/ijerph17218143
Foster P. (1976). Concentrations and concentration factors of heavy metals in brown algae. Environ. Pollut. 10 (1), 45–53. doi: 10.1016/0013-9327(76)90094-x
Fragkostefanakis S., Röth S., Schleiff E., Scharf K. D. (2015). Prospects of engineering thermotolerance in crops through modulation of heat stress transcription factor and heat shock protein networks. Plant Cell Environ. 38 (9), 1881–1895. doi: 10.1111/pce.12396
Gao Y., Li H., Song Y., Zhang F., Yang Z., Yang Y., et al. (2022). Response of glutathione pools to cadmium stress and the strategy to translocate cadmium from roots to leaves (Daucus carota L.). Sci. Total Environ. 823, 153575. doi: 10.1016/j.scitotenv.2022.153575
Haider F. U., Liqun C., Coulter J. A., Cheema S. A., Wu J., Zhang R., et al. (2021). Cadmium toxicity in plants: Impacts and remediation strategies. Ecotoxicol. Environ. Saf. 211, 111887. doi: 10.1016/j.ecoenv.2020.111887
Haslbeck M., Vierling E. (2015). A first line of stress defense: small heat shock proteins and their function in protein homeostasis. J. Mol. Biol. 427 (7), 1537–1548. doi: 10.1016/j.jmb.2015.02.002
Houessionon M. G. K., Ouendo E. D., Bouland C., Takyi S. A., Kedote N. M., Fayomi B. (2021). Environmental heavy metal contamination from electronic waste (E-waste) recycling activities worldwide: A systematic review from 2005 to 2017. Int. J. Environ. Res. Public Health 18 (7), 3517. doi: 10.3390/ijerph18073517
Hsu Y. T., Kao C. H. (2003). Role of abscisic acid in cadmium tolerance of rice (Oryza sativa L.) seedlings. Plant Cell Environ. 26 (6), 867–874. doi: 10.1046/j.1365-3040.2003.01018.x
Hu J., Chen G., Xu K., Wang J. (2022). Cadmium in cereal crops: uptake and transport mechanisms and minimizing strategies. J. Agric. Food Chem. 70 (20), 5961–5974. doi: 10.1021/acs.jafc.1c07896
Ji Z., Zou D., Gong J., Liu C., Ye C., Chen Y. (2019). The different responses of growth and photosynthesis to NH4+ enrichments between Gracilariopsis lemaneiformis and its epiphytic alga Ulva lactuca grown at elevated atmospheric CO2. Mar. Pollut. Bull. 144, 173–180. doi: 10.1016/j.marpolbul.2019.04.049
Kumar S., Sharma A. (2019). Cadmium toxicity: effects on human reproduction and fertility. Rev. Environ. Health 34 (4), 327–338. doi: 10.1515/reveh-2019-0016
Li B., Dewey C. N. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf. 12, 323. doi: 10.1186/1471-2105-12-323
Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-ΔΔ C(T)) Method. Methods 25 (4), 402–408. doi: 10.1006/meth.2001.1262
Luo H., Wang Q., Liu Z., Wang S., Long A., Yang Y. (2020). Potential bioremediation effects of seaweed Gracilaria lemaneiformis on heavy metals in coastal sediment from a typical mariculture zone. Chemosphere 245, 125636. doi: 10.1016/j.chemosphere.2019.125636
Luo H., Wang Q., Zhang C., Zhang L., Yang Y. (2021). Bioaccumulation and release of heavy metals during growth and decomposition of cultivated Gracilaria lemaneiformis. Mar. Pollut. Bull. 173 (Pt B), 113130. doi: 10.1016/j.marpolbul.2021.113130
Meng Y., Huang J., Jing H., Wu Q., Shen R., Zhu X. (2022). Exogenous abscisic acid alleviates Cd toxicity in Arabidopsis thaliana by inhibiting Cd uptake, translocation and accumulation, and promoting Cd chelation and efflux. Plant Sci. 325, 111464. doi: 10.1016/j.plantsci.2022.111464
Ogata F. T., Branco V., Vale F. F., Coppo L. (2021). Glutaredoxin: Discovery, redox defense and much more. Redox Biol. 43, 101975. doi: 10.1016/j.redox.2021.101975
Parmar P., Kumari N., Sharma V. (2013). Structural and functional alterations in photosynthetic apparatus of plants under cadmium stress. Botanical Stud. 54 (1), 45. doi: 10.1186/1999-3110-54-45
Qiu X. B., Shao Y. M., Miao S., Wang L. (2006). The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell. Mol. Life Sci. 63 (22), 2560–2570. doi: 10.1007/s00018-006-6192-6
Rahman S. U., Nawaz M. F., Gul S., Yasin G., Hussain B., Li Y., et al. (2022). State-of-the-art OMICS strategies against toxic effects of heavy metals in plants: A review. Ecotoxicol. Environ. Saf. 242, 113952. doi: 10.1016/j.ecoenv.2022.113952
Ramegowda V., Da Costa M. V. J., Harihar S., Karaba N. N., Sreeman S. M. (2020). “Abiotic and biotic stress interactions in plants: a cross-tolerance perspective,” in Priming-Mediated Stress and Cross-stress Tolerance in Crop Plants (Amsterdam: Elsevier), 267–302.
Rasafi T. E., Oukarroum A., Haddioui A., Song H., Rinklebe J. (2020). Cadmium stress in plants: a critical review of the effects, mechanisms, and tolerance strategies. Environ. Sci. Technol. 52 (5), 675–726. doi: 10.1080/10643389.2020.1835435
Sampath-Wiley P., Neefus C. D. (2007). An improved method for estimating R-phycoerythrin and R-phycocyanin contents from crude aqueous extracts of Porphyra (Bangiales, Rhodophyta). J. Appl. Phycol. 19 (2), 123–129. doi: 10.1007/s10811-006-9118-7
Sapara K. K., Khedia J., Agarwal P., Gangapur D. R., Agarwal P. K. (2019). SbMYB15 transcription factor mitigates cadmium and nickel stress in transgenic tobacco by limiting uptake and modulating antioxidative defence system. Funct. Plant Biol. 46 (8), 702–714. doi: 10.1071/FP18234
Sears M. E. (2013). Chelation: harnessing and enhancing heavy metal detoxification–a review. Sci. World J. 219840, 1–13. doi: 10.1155/2013/219840
Shao Y., Zhou H. Z., Wu Y., Zhang H., Lin J., Jiang X., et al. (2019). OsSPL3, an SBP-domain protein, regulates crown root development in rice. Plant Cell 31 (6), 1257–1275. doi: 10.1105/tpc.19.00038
Smiri M., Chaoui A., Rouhier N., Gelhaye E., Jacquot J. P., El Ferjani E. (2011). Cadmium affects the glutathione/glutaredoxin system in germinating pea seeds. Biol. Trace element Res. 142 (1), 93–105. doi: 10.1007/s12011-010-8749-3
Tang Y., Wang L., Xie Y., Yu X., Li H., Lin L., et al. (2020). Effects of exogenous abscisic acid on the growth and cadmium accumulation of lettuce under cadmium-stress conditions. Int. J. Environ. Anal. Chem. 100 (6), 720–731. doi: 10.1080/03067319.2019.1639686
Tao J., Lu L. (2022). Advances in genes-encoding transporters for cadmium uptake, translocation, and accumulation in plants. Toxics 10 (8), 411. doi: 10.3390/toxics10080411
Thakur B., Yadav R., Mukherjee A., Melayah D., Marmeisse R., Fraissinet-Tachet L., et al. (2021). Protection from metal toxicity by Hsp40-like protein isolated from contaminated soil using functional metagenomic approach. Environ. Sci. Pollut. Res. Int. 28 (14), 17132–17145. doi: 10.1007/s11356-020-12152-6
Wang P. L., Lei X. J., Wang Y. Y., Liu B. C., Wang D. N., Liu Z. Y., et al. (2022). Transcriptomic analysis of cadmium stressed tamarix hispida revealed novel transcripts and the importance of abscisic acid network. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.843725
Wang X., Niu Y., Zheng Y. (2021). Multiple functions of MYB transcription factors in abiotic stress responses. Int. J. Mol. Sci. 22 (11), 6125. doi: 10.3390/ijms22116125
Wang Z., Wang X., Ke C. (2014). Bioaccumulation of trace metals by the live macroalga Gracilaria lemaneiformis. J. Appl. Phycol. 26 (4), 1889–1897. doi: 10.1007/s10811-013-0222-1
Xia J. R., Li Y. J., Lu J., Chen B. (2004). Effects of copper and cadmium on growth, photosynthesis, and pigment content in Gracilaria lemaneiformis. Bull. Environ. Contamination Toxicol. 73 (6), 979–986. doi: 10.1007/s00128-004-0522-x
Xiao H., Shahab A., Ye F., Wei G., Li J., Deng L. (2022). Source-specific ecological risk assessment and quantitative source apportionment of heavy metals in surface sediments of Pearl River Estuary, China. Mar. Pollut. Bull. 179, 113726. doi: 10.1016/j.marpolbul.2022.113726
Yu C., Sui W., Liang B., Bao C., Ma M. (2022). Temporal trends of heavy metals in the sediments of Bohai Bay in China. Environ. Monit. Assess. 194 (2), 79. doi: 10.1007/s10661-021-09747-y
Yu M., Zhuo R., Lu Z., Li S., Chen J., Wang Y., et al. (2023). Molecular insights into lignin biosynthesis on cadmium tolerance: Morphology, transcriptome and proteome profiling in Salix matSudana. J. Hazard. Mater. 441, 129909. doi: 10.1016/j.jhazmat.2022.129909
Zhang B., Tang Y., Yu F., Peng Z., Yao S., Deng X., et al. (2023a). Translatomics and physiological analyses of the detoxification mechanism of green alga Chlamydomonas reinhardtii to cadmium toxicity. J. Hazard. Mater. 448, 130990. doi: 10.1016/j.jhazmat.2023.130990
Zhang H., Reynolds M. (2019). Cadmium exposure in living organisms: A short review. Sci. Total Environ. 678, 761–767. doi: 10.1016/j.scitotenv.2019.04.395
Zhang H., Xie P., Xu X., Xie Q., Yu F. (2021). Heterotrimeric G protein signalling in plant biotic and abiotic stress response. Plant Biol. (Stuttgart Germany) 23 (Suppl 1), 20–30. doi: 10.1111/plb.13241
Zhang H., Xu Z., Huo Y., Guo K., Wang Y., He G., et al. (2020). Overexpression of Trx CDSP32 gene promotes chlorophyll synthesis and photosynthetic electron transfer and alleviates cadmium-induced photoinhibition of PSII and PSI in tobacco leaves. J. Hazard. Mater. 398, 122899. doi: 10.1016/j.jhazmat.2020.122899
Zhang H., Yao T., Wang Y., Wang J., Song J., Cui C., et al. (2023b). Trx CDSP32-overexpressing tobacco plants improves cadmium tolerance by modulating antioxidant mechanism. Plant Physiol. Biochem. 194, 524–532. doi: 10.1016/j.plaphy.2022.11.036
Zhang J., Zhu Y., Yu L., Yang M., Zou X., Yin C., et al. (2022b). Research advances in cadmium uptake, transport and resistance in rice (Oryza sativa L.). Cells 11 (3), 569. doi: 10.3390/cells11030569
Zhang M., Sun X., Hu Y., Chen G., Xu J. (2022a). The influence of anthropogenic activities on heavy metal pollution of estuary sediment from the coastal East China Sea in the past nearly 50 years. Mar. Pollut. Bull. 181, 113872. doi: 10.1016/j.marpolbul.2022.113872
Zhao K., Bao K., Yan Y., Neupane B., Gao C. (2022). Spatial distribution of potentially harmful trace elements and ecological risk assessment in Zhanjiang mangrove wetland, South China. Mar. Pollut. Bull. 182, 114033. doi: 10.1016/j.marpolbul.2022.114033
Zhu D., Wen X., Li S., Xuan X., Li Y. (2017). Evaluation of the red alga Gracilaria lemaneiformis and brown alga Sargassum horneri as ingredients in diets for white spotted snapper Lutjanus stellatus Akazaki juveniles. J. Appl. Phycol. 29, 3211–3219. doi: 10.1007/s10811-017-1187-2
Zou D., Gao K. (2014). Temperature response of photosynthetic light- and carbon-use characteristics in the red seaweed Gracilariopsis lemaneiformis (Gracilariales, Rhodophyta). J. Phycol. 50 (2), 366–375. doi: 10.1111/jpy.12171
Keywords: Cd stress, macroalga, ABA, RNA-Seq, Gracilariopsis lemaneiformis
Citation: Chen X, Tang Y, Sun X, Zhang H and Xu N (2024) Transcriptome analysis reveals ABA involved in the detoxification mechanism of macroalga Gracilariopsis lemaneiformis to cadmium toxicity. Front. Mar. Sci. 11:1352529. doi: 10.3389/fmars.2024.1352529
Received: 11 December 2023; Accepted: 25 January 2024;
Published: 09 February 2024.
Edited by:
Satheesh Sathianeson, King Abdulaziz University, Saudi ArabiaReviewed by:
Wenjun Wang, Chinese Academy of Fishery Sciences (CAFS), ChinaShaharior Hossen, Chonnam National University, Republic of Korea
Copyright © 2024 Chen, Tang, Sun, Zhang and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nianjun Xu, xunianjun@nbu.edu.cn
 Xiaojiao Chen
Xiaojiao Chen Yueyao Tang
Yueyao Tang  Xue Sun
Xue Sun Hao Zhang
Hao Zhang Nianjun Xu
Nianjun Xu