Dispersal patterns of sandfish (Holothuria scabra) larvae in Manus Province, Papua New Guinea
- 1Honiara Office, The Nature Conservancy, Rove, Solomon Islands
- 2General Organization for Conservation of Coral Reefs and Turtles in the Red Sea, Jeddah, Saudi Arabia
- 3School of Mathematical Sciences, Queensland University of Technology, Brisbane, QLD, Australia
- 4Mwanus Endras Asi Resource Development Network, Lorengau, Papua New Guinea
- 5School of Biological Sciences, University of Bristol, Bristol, United Kingdom
- 6Red Sea Research Center, Division of Biological and Environmental Science and Engineering, King Abdullah University of Science and Technology, Thuwal, Saudi Arabia
- 7National Fisheries Authority, Port Moresby, Papua New Guinea
- 8University of Papua New Guinea, Port Moresby, Papua New Guinea
- 9Asia Pacific Resource Centre, The Nature Conservancy, South Brisbane, QLD, Australia
- 10Coastal People Southern Skies Centre of Research Excellence, The Nature Conservancy, South Brisbane, QLD, Australia
In many tropical nations, coastal communities seek to manage their sea cucumber fisheries by establishing locally managed marine areas on their traditional fishing grounds. These managed areas can protect spawning stocks, however the extent to which they help to replenish nearby sea cucumber fisheries is debated, as nothing is known about the scales at which sea cucumber larvae disperse. In this study, we used genetic parentage analyses and statistical modelling to provide the first empirical measure of larval dispersal patterns for a sea cucumber species. We analysed tissue samples from 765 adult and 827 juvenile sandfish (Holothuria scabra) collected from five traditional fishing grounds of the Titan tribe, from Manus Province, Papua New Guinea. All adults were sampled from the Pere fishing grounds (the larval source area), with juveniles sampled from the Pere, Mbunai, Tawi, Timonai and Mbuke fishing grounds. Parentage analysis identified 15 juveniles that were the offspring of parents sampled from Pere fishing grounds, with six of these juveniles located in the same fishing grounds as their parents. The best-fit larval dispersal kernel predicted that the average H. scabra larvae travelled 15 km from its spawning location, with 50% of larvae settling within 6.7 km of their parents and 95% of larvae settling within 59 km of their parents. These results suggest that the Titan tribes’ desire to protect their H. scabra spawning stocks by establishing a network of locally managed marine areas across 65 km of continuous coastline is a culturally appropriate strategy, which has merit from both an ecological and fisheries standpoint.
Introduction
Understanding the larval dispersal patterns of exploited marine species is important to fisheries managers as it informs the spatial scales at which these harvested populations should be managed (e.g. Sale et al., 2005; Pelc et al., 2010; Jones, 2015). From an ecological perspective, when mean larval dispersal distances are short (i.e. tens of kilometers, e.g. Almany et al., 2007) and a large proportion of larvae are locally retained, local marine reserves can be self-sustaining, and nearby fisheries can receive larval subsidy. From the perspective of economic and social theory, local larval dispersal makes small-scale management more viable (Almany et al., 2007; Hamilton et al., 2021), and creates the conditions for self-sustaining territorial user-rights fisheries – in particular, it means that the community who manage their fisheries are the primary beneficiary of sustainable behavior (Costello et al., 2015). Such management becomes much more challenging where mean larvae dispersal distances are large (i.e. in the hundreds of kilometres, e.g. Nanninga et al., 2015; Herrera et al., 2016; Williamson et al., 2016). Where larval dispersal scales substantially exceed management scales, connectivity creates economic and social externalities.
Our understanding of the scales and patterns of population connectivity in marine species has increased markedly in recent decades. Empirical field studies based on new methodologies such as genetic parentage analysis have allowed direct observations of larval dispersal events (Jones et al., 2005). These studies have assisted in the validation and calibration of increasingly sophisticated biophysical dispersal simulations (Bode et al., 2019). However, most experimental studies on larvae dispersal have focused on small fish species that are not harvested (e.g., Planes et al., 2009; Pusack et al., 2014; Shulzitski et al., 2016). Literature pertaining to large commercial species (i.e., reef fish) is gradually increasingly (e.g., Harrison et al., 2012; Almany et al., 2013; Hamilton et al., 2021). Yet despite the global interest in managing sea cucumber fisheries, we know of only one published study that has attempted to quantify sea cucumber larval dispersal (Brown et al., 2022). The study by Brown et al. (2022) involved conducting a high-resolution genomic audit of sandfish (Holothuria scabra) populations at six sites in Fiji that were separated by a maximum of 300 km. Brown et al. (2022) could not infer larval dispersal patterns of sandfish in Fiji, as they did not identify any parent-offspring relationships, however they did detect some full sibling and half sibling relationships, and concluded that sandfish in Fiji are made up of three genetically distinct populations.
Sea cucumbers, or bêche-de-mer, as they are known in their dried and processed form, have been a luxury sea food in Asia for centuries (Toral-Granda et al., 2008). Many commercially valuable sea cucumbers occur in shallow tropical waters, where they are typically exploited by small-scale fisheries that operate in low income and low governance contexts (Barclay et al., 2019). Consequently, many sea cucumber fisheries are severely overfished (Toral-Granda et al., 2008; Friedman et al., 2011), with patterns of boom and bust inherent in sea cucumber fisheries globally. In the Pacific Islands region both bottom-up and top-down management efforts have been utilized to manage these high value fisheries. Coastal communities in Fiji, Papua New Guinea and Solomon Islands have established locally managed marine areas (LMMAs) on their customarily owned reefs to allow populations of sea cucumbers to recover (e.g. PNG National Fisheries Authority, 2007; Hamilton and Lokani, 2011; Hamilton et al., 2015; Lalavanua et al., 2017). This type of community-based fisheries management is made possible by the existence of customary marine tenure systems that are common throughout the Pacific, often involving communities closing an area to some or all types of fishing for a certain period (Ruddle, 1996; Johannes, 2002; Almany et al., 2015). National fishery agencies also routinely place moratoriums on these fisheries once they are considered over-exploited at a regional scale (e.g. Christensen, 2011; Hair et al., 2019; Brown et al., 2022).
For the past two decades, the Titan communities, located along the south coast of Manus Island in Papua New Guinea, have established LMMAs within their traditional fishing grounds as a fisheries management tool for a variety of harvested species (e.g. Hamilton et al., 2005). To evaluate the effectiveness of these LMMAs, Titan fishers and staff from James Cook University and The Nature Conservancy investigated the dispersal of squaretail grouper (Plectropomus areolatus) larvae that were spawned at a fish spawning aggregation that is protected by one of these LMMAs (Almany et al., 2010, Almany et al., 2013, Almany et al., 2015). This work showed that P. areolatus had a mean larval dispersal distance of 14 km, with 20% of larvae retained within the fishing grounds of where they spawned (Almany et al., 2013).
When the findings on P. areolatus connectivity were shared with Titan fishers in 2011, communities realized that while they would obtain some direct larval subsidies from protecting fish spawning aggregations located within their fishing grounds, the scale of larval connectivity meant that this fishery would be best managed at a larger spatial scale. This information provided Titan leaders with the ecological rationale for establishing the Mwanus Endras Asi Resource Development Network (MEARDN) tribal network. MEARDN encompasses a sea area of approximately 24,000 km2 and 10,000 people; the Titan communities that make up MEARDN share a common language, religion, and ethnic identity. Most MEARDN inhabitants live in communities that are located along a continuous 65 km stretch of the south coast of Manus Island, with a smaller number of people residing on outer islands that lie up to 100 km south or east of Manus Island (Almany et al., 2015). MEARDN was established in 2013, with the goal of improving the livelihoods of the Titan people, by maintaining healthy marine ecosystems and strengthening the governance and financial capacity of the Titan people.
One of the most valuable marine resources that is found within MEARDN is H. scabra. Often called “sandfish”, this is the most valuable species in the Papua New Guinea sea cucumber fishery (Purcell et al., 2014; Hair et al., 2019). Sandfish have been commercially exported from Papua New Guinea since the 1870s (few local people traditionally consumed sea cucumber), but the trade accelerated dramatically in the 1980s, by which time Papua New Guinea had become the third largest supplier of bêche-de-mer to Hong Kong (Kinch et al., 2008). Historically sandfish were very abundant in nearshore seagrass habitats in Papua New Guinea, with reported densities in the 1980s-1990s often exceeding 1000 per ha (i.e. Lokani, 2001; Hamilton and Lokani, 2011). Because of its shallow distribution children, women, men, and the elderly participate in the sandfish fishery (Barclay et al., 2019). Its shallow distribution and high value have resulted in sandfish populations being depleted throughout Papua New Guinea (e.g. Dalzell, 1990; Hamilton and Lokani, 2011; Hair et al., 2019) and much of its range (Hamel et al., 2022). Given this vulnerability to overexploitation, H. scabra was listed as Endangered on the IUCN Red List in 2010 (Hamel et al., 2013). At the time of this study, 2014, a Papua New Guinea wide moratorium on sea cucumber fisheries had been in place since 2009, following low stock assessments for all commercially valuable sea cucumber species (Hair et al., 2016).
Sexes are separate in H. scabra and this species is a broadcast spawner. Their larvae are planktonic, settling in shallow seagrass habitats, approximately 14 days post-spawning (see review in Hamel et al., 2022). Adults inhabit seagrass areas and deeper sand and silt habitats. The species has a narrow habitat range, and is only found in shallow, low-energy environments that have muddy or sand substrates (see review in Hamel et al., 2022). Uthicke and Benzie (2001) found restricted gene flow between H. scabra populations along the north-east coast of Australia, suggesting a low dispersal ability in this species. Because of their broadcast spawning behavior many closely related congeneric species of sea cucumbers can hybridize, particularly in areas where both populations are abundant (e.g. Gkafas et al., 2023). In Papua New Guinea the golden sandfish (H. lessoni) occurs in similar habits to H. scabra, although in Manus this species occurs in very low abundances (authors, personal observations). The two species are readily distinguishable based on markedly different color patterns and presence/absence of wrinkles in the body wall (Uthicke et al., 2005). H. scabra and H. lesoni have been shown to hybridize, although the genetic integrity of either holothurian species remains intact through an unknown postzygotic mechanism that may be hybrid sterility (Uthicke et al., 2005).
During a MEARDN tribal meeting in 2013, Titan leaders expressed interest in establishing a business arm within their tribal network, as a means of enhancing the financial security of their people. Titan leaders were particularly interested in sustainably harvesting, processing, and exporting H. scabra directly to Hong Kong once the national moratorium on sea cucumber harvesting was lifted. By running all aspects of the bêche-de-mer business themselves, MEARDN and its fishers hoped to capture a greater share of the profits in this fishery. To ensure the sustainability of their H. scabra fisheries, Titan leaders also discussed the merits of establishing a network of nearshore LMMAs within MEARDN, to protect local spawning stocks of H. scabra once the fishery was opened. Since nothing was known about the larval dispersal patterns of H. scabra, TNC staff who were present at the 2013 tribal meeting were asked if they would conduct research on their larval dispersal patterns within MEARDN. This request formed the premise for the research that is reported on here.
Methods
Environmental setting
The Titan communities of Pere, Mbunai, Locha, Tawi, Timonai and M’buke participated in this study These communities are all situated near the mainland of Manus Island, and their traditional fishing grounds are shown in Figure 1. At the time of this study (2014) the national moratorium on sea cucumber harvest had been in place for five years, and Titan fishers reported that their sandfish populations were showing signs of recovery (Feary et al., 2014). In water surveys that we conducted in 2014 revealed that the average densities of sandfish in shallow seagrass habitats in Titan fishing grounds ranged from 192 to 422 individuals per hectare, with H. scabra populations dominated by juvenile size classes (Feary et al., 2014). These 2014 densities were on par with areas that have undertaken experimental, small-scale fishing of sandfish (i.e., Warrior Reefs within Torres Strait, Australia, Murphy et al., 2012).
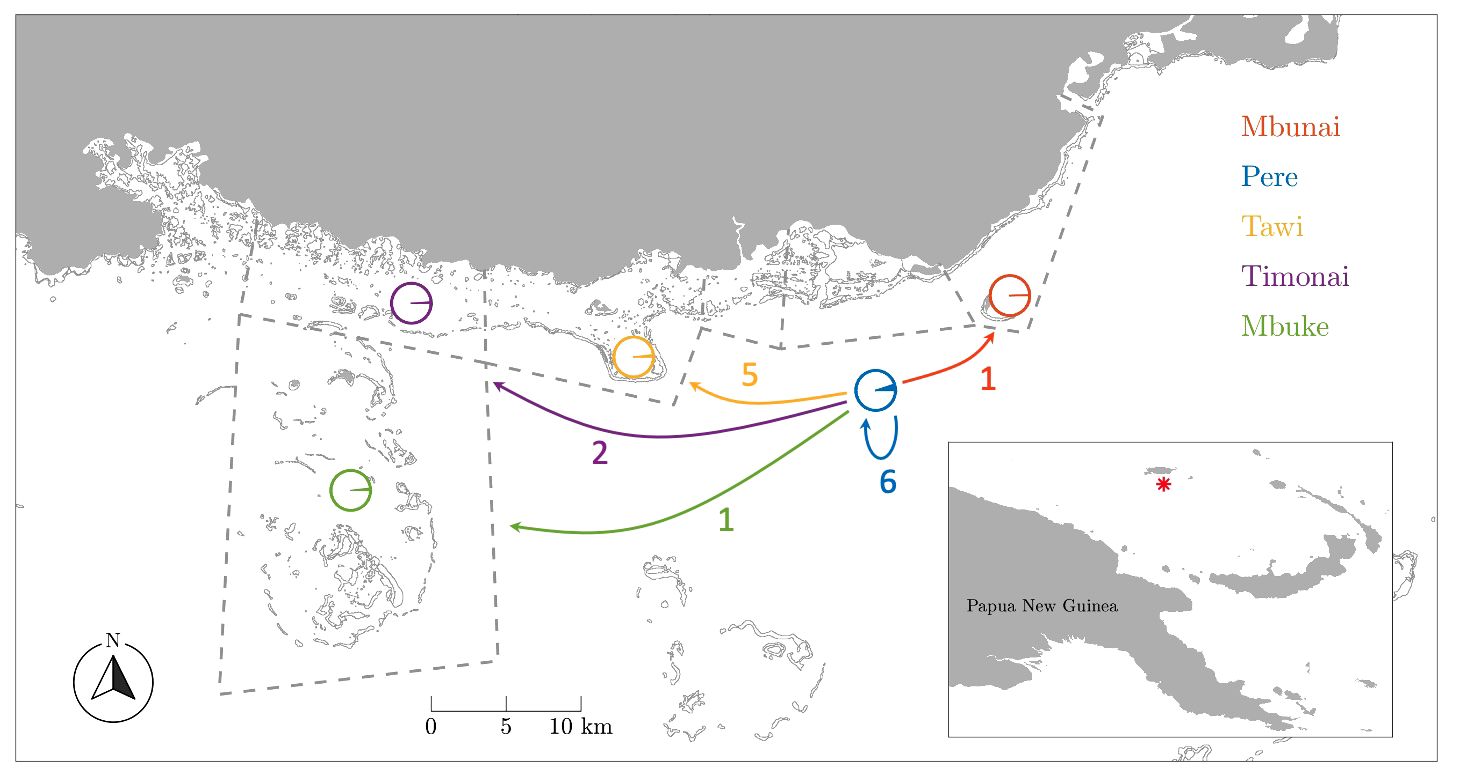
Figure 1 Dispersal map by fishing grounds. Dashed lines delineate customary marine tenure boundaries between communities. Circles indicate the proportion of juveniles sampled in each fishing ground that were assigned to parents from among the Pere samples. Arrows indicate the number of juveniles assigned to parents from Pere. Note the high number (6) and proportion (5.1%) of self-recruits from Pere.
Collection of tissue samples
Although juvenile H. scabra in Papua New Guinea have been categorized as being ≤ 21cm total length (TL) (Lokani, 1990), maturity can occur from 17cm TL to 21cm TL (Hamel et al., 2022). To be conservative, for our connectivity study we considered adults ≥ 22cm TL and juveniles to be ≤ 15 cm TL. Between 19th May to the 22nd June 2014 we worked with over one hundred Titan fishers to sample 6,465 H. scabra from 57 sites located in the Pere, Mbunai, Locha, Tawi, Timonai and Mbuke fishing grounds (Table 1). Juveniles were sampled from all six communities’ fishing grounds, whereas adults were only sampled from the Pere fishing grounds (i.e. the ‘natal site’: see Feary et al., 2014 for further details). Once collected, each H. scabra was allowed to rest for 5 mins, as individuals are prone to constrict when moved. Following this, each individual was measured (mouth to anus), a small sample of body wall tissue from the dorsal side of the animal collected and this sample immediately preserved in 98% ethanol. Between individual sample collection, all sampling instruments were placed in 98% ethanol, to reduce contamination. All individual sandfish were then replaced in the habitat they were collected. A subsample of 765 adults and 827 juveniles were analysed for parent – offspring relationships (Table 1).
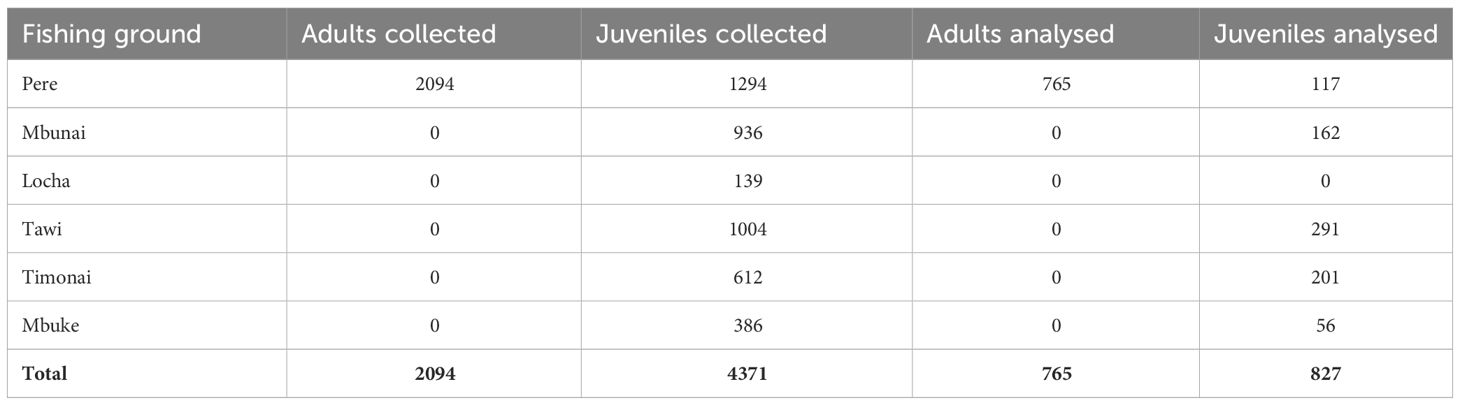
Table 1 Total number of adult and juvenile H. scabra collected from each fishing ground and the subsample of these that were genetically analysed.
Estimating the amount of H. scabra habitat within the Pere fishing grounds
To estimate the amount of H. scabra habitat within the Pere fishing grounds, we asked knowledgeable sea cucumber fishers from Pere to demarcate sandfish habitats on satellite images of their fishing grounds (Feary et al., 2014), a process known as participatory mapping. Local fishers identified three readily distinguishable H. scabra habitats in the Pere fishing grounds, that being dense and sparse seagrass habitat that occurred in shallow (1-2 m) water depths, and sand/silt habitat that occurred in water depths of 5-10 m. H. scabra habitats that were identified through participatory mapping were digitized in GIS, enabling us to calculate the total area of each H. scabra habitat in the Pere fishing grounds.
Underwater visual census surveys
For the purposes of dispersal kernel estimation, we needed to estimate the adult abundance of H. scabra in the Pere fishing grounds (the adult source area). This abundance estimate can be used to determine what proportion of the larvae were genetically marked. Between the 25th to 30th of May 2014, 180 100m2 (50m * 2m) underwater visual census (UVC) surveys were undertaken within the Pere fishing grounds (Supplementary Table S1). We surveyed dense seagrass, sparse seagrass, and sand/silt H. scabra habitats in the Pere fishing grounds. Shallow seagrass habitats were surveyed on snorkel while deeper sand/silt habitat was surveyed using SCUBA (Supplementary Table S1). All UVC surveys were conducted at night using underwater torches to coincide with the highest density of H. scabra, since this species will burrow during the day and emerge during the night (Hamel et al., 2022). On each UVC survey all H. scabra encountered were counted, with each individual’s length visually estimated and placed within 50 mm size classes (total length, TL). To determine the density of sandfish in juvenile versus adult phases, all individuals were split into ≤ 21cm TL (juvenile) and ≥ 22cm TL (mature [adult] phase) (following Lokani, 1990).
Microsatellite development and genotyping
Microsatellites were identified from a de-novo assembly of Illumina 2 x 150 bp paired-end sequencing from genomic DNA of a single H. scabra. The genomic library was prepared and sequenced on an Illumnia MiSeq platform according to the manufacturer’s instructions, using a Nextera DNA sample prep kit (Illumina). Reads were first trimmed of low-quality bases in TRIMMOMATIC v0.36 before forward and reverse reads were merged in Pear v0.9.10 (Zhang et al., 2014). Candidate tri- and tetra-nucleotide microsatellite repeats were detected using MSATCOMMANDER v 1.0.8 (Faircloth, 2008). A total of 6,473 suitable loci were identified, of which 48 were selected for multiplex PCR amplification. Primers were designed in msatcommander with optimum lengths of 26 bp, optimum annealing temperature of 60°C, and a 3’ GC clamp for higher specificity in multiplex PCR. Forward primers were labelled with 5’-modifications (6-FAM, VIC, NED and PET) for fragment analysis on an ABI 3370xl DNA Analyzer (Applied Biosystems) with the GeneScan 500 LIZ internal size standard (Applied Biosystems).
All loci were first amplified in simplex PCRs on three individuals and later combined in multiplex PCRs in eight individuals to assess the quality of amplification and adjust primer concentrations. A total of 26 loci were selected for amplification in four multiplex reactions. All four multiplex reactions were performed in a total volume of 10 µL containing 5 µL of Qiagen Multiplex Master Mix (Qiagen, Germany), 3 µL of distilled water, 1 µL of primer premix, and 1 µL template DNA. Multiplex PCRs were performed on Veriti thermocyclers following a ‘touch-down’ sequence to increase the binding specificity of primers and reduce artifacts: 15 min initial denaturation at 95°C, 5 cycles of 30 s at 95°C, 90 s at 62°C, and 60 s at 72°C, then 5 cycles of 30 s at 95°C, 90 s at 60°C, and 60 s at 72°C, then 20 cycles of 30 s at 95°C, 90 s at 58°C, and 60 s at 72°C, followed by 30 min at 60°C. All adult and juvenile samples were genotyped at all 26 loci following DNA extraction using a Nucleospin tissue extraction kit (Macherey-Nagel). Individual genotypes were scored in GENEMAPPER v4.0, and unique alleles were distinguished using marker specific binsets adapted from MSATALLELE (Alberto, 2009).
Parentage analysis and accuracy tests
All collected juveniles were screened against the total pool of adults to reveal parent–offspring relationships using a maximum-likelihood approach implemented in FAMOZ (Marshall et al., 1998; Gerber et al., 2003). The program computes log of the odds ratio (LOD) scores for assigning individuals to candidate parents based on the observed allelic frequencies at each locus. Minimum LOD score thresholds for accepting assignments to single parents and parent pairs were determined from simulation adapted from Harrison et al. (2014). Briefly, we generated five simulated datasets of individual genotypes of known and unknown decent, simulating incomplete sampling of the adult populations. The proportion of known parents in the sample was fixed at 20% and genotyping error was introduced at a rate of 1% for each locus. Simulated datasets were analysed using the pairwise likelihood score method implemented in FAMOZ and the results analysed to identify a LOD threshold resulting in the highest overall accuracy of assignments. Accuracy was measured as the proportion of correct assignments to single parents or parent pairs and the number of correct exclusions over all possible assignments (Harrison et al., 2013).
Fitting dispersal kernels to genetic parentage data
We fit several isotropic dispersal kernels to the parentage data, using the maximum likelihood methods described in Bode et al. (2018), implemented in Matlab (see Data availability statement). We considered four kernel shapes from the family of generalized normal distributions: two were thin-tailed distributions (the Gaussian and the Ribbens), and two were heavy-tailed (the Laplacian and the Cauchy). We represented the sampling locations of individuals as single points in each of the fishing grounds, creating distance and parentage matrices (the final row accounting for unassigned juveniles). We did not consider the Locha fishing grounds in this analysis as none of the juveniles from Locha were genetically analysed.
The best-fit kernel functions and parameter values were estimated using both those juveniles which could be assigned to parents, and also unassigned juveniles, since these also offer information about the scale of dispersal.
Results
Microsatellite genotyping and locus characteristics
In total, 765 adults from Pere and 827 juveniles from five fishing grounds (Pere, Mbunai; Tawi; Timonai; Mbuke) were successfully genotyped at 26 microsatellites for parentage analysis. Of the 26 loci amplified, seven were excluded from further analyses due to null alleles or poor amplification. All remaining loci were highly polymorphic (Supplementary Table S3) ranging from 7 to 51 alleles with a mean of 24.5 (SE: 2.8) alleles per locus and average heterozygosity of 0.730 (SE: 0.04). The high level of polymorphism provided a strong exclusion power (Supplementary Table S3) with an overall accuracy of 98.6% across simulated datasets with LOD scores above 6, providing very strong confidence in assigned parent-offspring pairs. The probability of assigning a juvenile to a parent that was not its true parent, knowing that the true parent was not sampled was 1.3% (false positive – type I error). Conversely, the probability of a true parent–offspring pair not being identified knowing that the true parent was sampled was 0.1% (false negative – type II error). Fifteen juveniles were genetically assigned to a single sampled parents in our samples (mean LOD score: 8.6). All fifteen individuals were assigned to adults from Pere (Table 2, Figure 1).
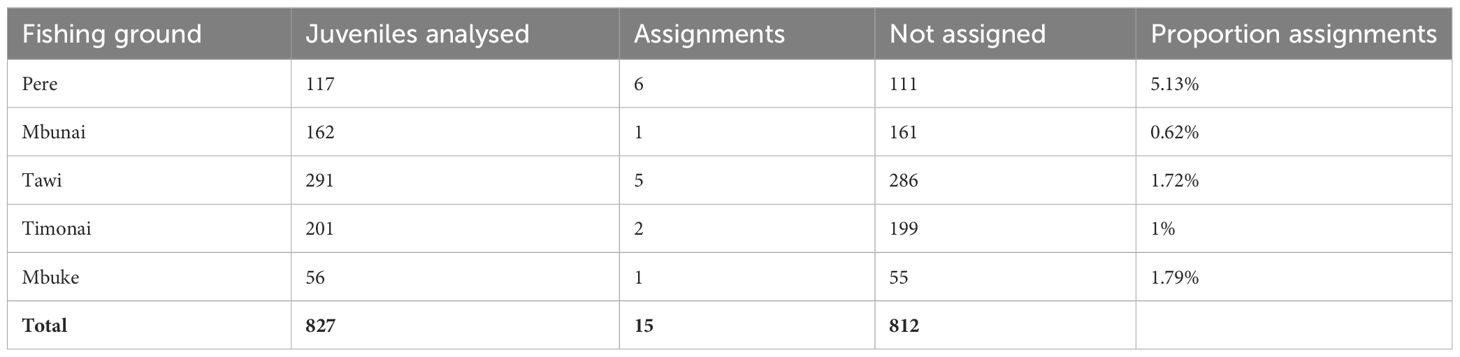
Table 2 Number and proportion of juvenile H. scabra in each fishing ground that were assigned back to a genetically analysed adult in Pere.
Larval dispersal within the MEARDN customary fishing grounds
The participatory mapping of H. scabra habitats (Supplementary Figure S1) and UVC data (Supplementary Tables S1 and S2) indicated that the 765 surveyed adults represented 3.1% of the total adult population in Pere, across all three habitat types. Using this proportion, and the number of assignments across the five fishing grounds along the coastline, we found that the best fit was provided by the Cauchy dispersal kernel function:
with . The fit implies that the mean dispersal distance in the system was 15 km, and that half of the larvae are settling within 6.7 km of their natal population. The kernel predicts that the overwhelming majority of settlement (95%) happens within 59 km of the larval source. The Cauchy kernel is particularly steep at the origin (Figure 2), suggesting that the strength of dispersal drops off very rapidly with distance.
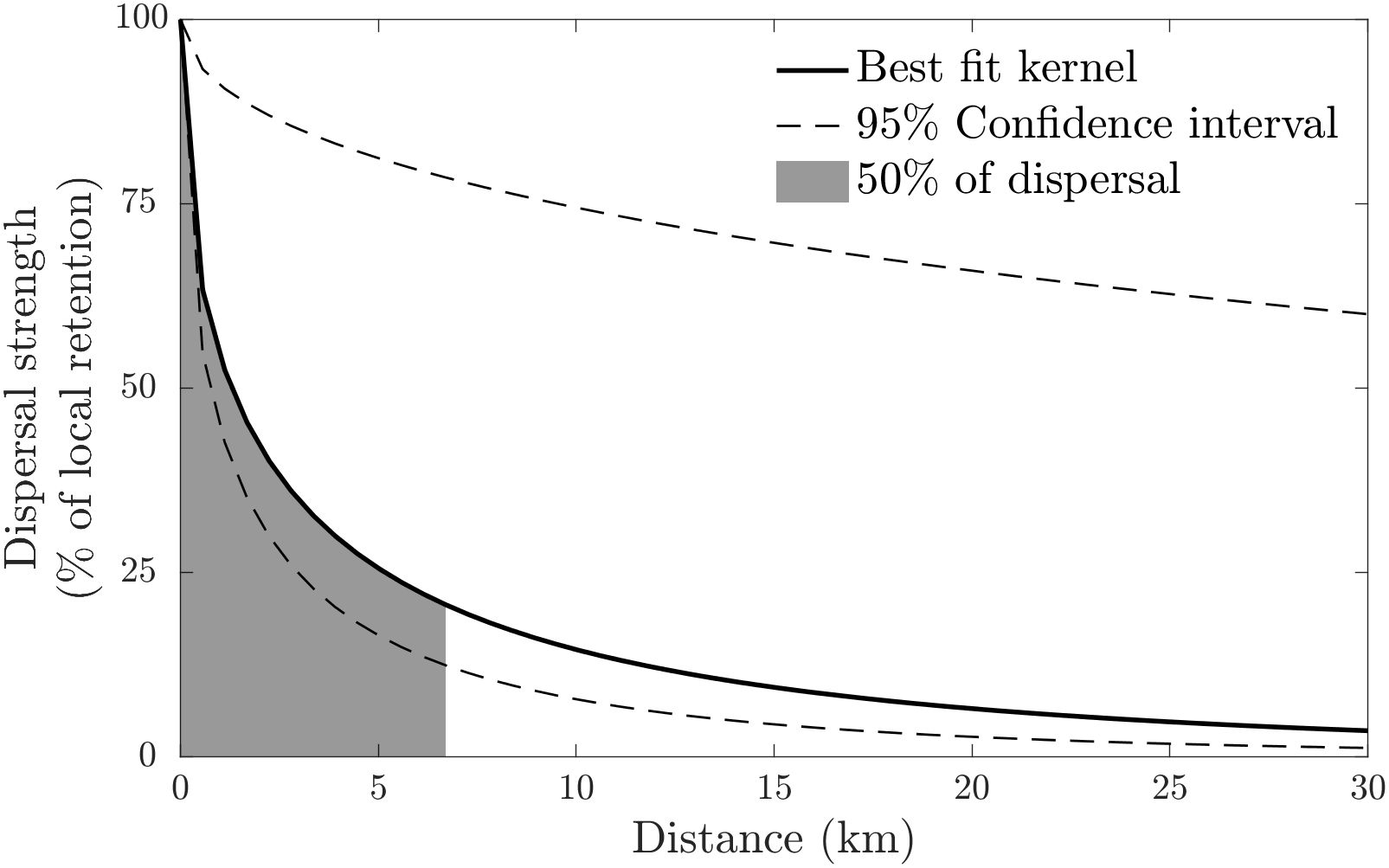
Figure 2 Best-fit larval dispersal kernel (solid black line) – a Cauchy kernel – estimated from the parentage data. Dashed lines enclose ± 1 SD of the fits to the bootstrap resampled data. Dispersal strength is given relative to the strength of local retention. The best-fit kernel estimates that 50% of larvae settle within 6.7 km of the natal reef. The estimated mean dispersal distance is 15 km.
Confidence bounds around this fit were generated using bootstrap resampling at the scale of the 5 sampling locations, which showed that 95% of the fits had values in the range (Figure 2). This choice of bootstrap resampling addresses the uncertainty produced by the choice of sample locations. Along with the relatively small number of assignments, this choice produces wide confidence bounds, since the dataset changes substantially between resamples.
Discussion
In this study, we combined genetic parentage analysis and modelling to provide the first description of larval dispersal patterns in a commercially valuable sea cucumber. We identified 15 juvenile H. scabra in the Mbuke, Mbunai, Pere, Tawi and Timonai fishing grounds that were the offspring of parents sampled from the Pere fishing grounds. A fitted dispersal kernel based on the sampling location of adult and juvenile H. scabra predicted that the mean distance of larval dispersal is 15 km. These findings imply high levels of self-recruitment within the Pere fishing grounds, as well as larval connectivity between the five MEARDN fishing grounds investigated in this study – particularly between adjacent communities (Figure 2).
We found that 50% of larvae settle within 6.7 km of their parents and 95% of larvae settle within 59 km of their parents, indicating that the studied H. scabra population is reasonably demographically closed (Sale et al., 2005). This implies that the spatial scale of MEARDN, which extends across 65 km of the Manus coastline, is likely adequate for sustainably managing the stock. Establishing a network of nearshore LMMAs that protect adult spawning stock of H. scabra would be a culturally appropriate strategy for MEARDN that has merit from both an ecological and fisheries standpoint. Although demographically closed populations can be managed at restricted spatial scales that match the size of tribal networks such as MEARDN, they are more vulnerable to recruitment overfishing (Begg et al., 1999; Walters and Martell, 2004) and localized extinction at that scale. Once depleted, demographically closed populations are unlikely to be rescued by the arrival of larvae from distant intact populations (van der Meer et al., 2015; Bonin et al., 2016). Indeed, some sea cucumber fisheries that have been overfished have failed to recover, despite many decades of complete closure (Friedman et al., 2011). The limited dispersal observed in this study confers with findings from Brown et al. (2022), who found three genetically distinct populations of H. scabra across sites that were separated by a maximum of 300 km in Fiji.
Aside from limited larval dispersal, there are several additional reasons why H. scabra stocks are well-suited to being managed through the establishment of a network of LMMAs in southern Manus. Firstly, adult H. scabra habitat is common near MEARDN communities. Establishing LMMAs for sandfish that were in line-of-sight of communities makes social enforcement more feasible (McClanahan et al., 2006), and was a topic of discussion and the 2013 MEARDN meeting. Secondly, in Papua New Guinea and across the Pacific, most LMMAs are usually< 0.5 km2 (Mills et al., 2010; Weeks et al., 2010). Because sea cucumbers are relatively sedentary, small LMMAs are likely to be sufficient for protecting their spawning stocks. For example, some large-bodied species such as the elephant trunkfish (Holothuria fuscopunctata) move less than 10 m on average over annual timescales (Purcell et al., 2023). Movement studies on H. scabra indicate that H. scabra will cover a total distance of 2–8 m per day and have home ranges of<0.05 km2 (Lee et al., 2018).
For LMMA networks to serve their fisheries management objectives individual reserves must be large enough to protect adult spawning stocks that export larval subsidies to fished areas and nearby LMMAs (Green et al., 2015). For sea cucumbers, the configuration (size, spacing and location) of LMMAs within a network should be informed by the home ranges of benthic life stages and the dispersal patterns of pelagic larvae (Green et al., 2015). We suggest that a LMMA network for H. scabra should be made up of LMMAs ≥ 40 ha (Purcell and Kirby, 2006) and separated by no more than 15 km (the approximate distance that the majority of larvae travel from their natal sites). It is noteworthy that a study by Hamel et al. (2019) challenges the notion of that H. scabra adults have restricted home ranges. Hamel et al. (2019) describes how juveniles and adult H. scabra in aquaculture enclosures in Madagascar were able to modify their buoyancy, leading them to tumble or float at speeds much faster than they can obtain through benthic crawling. Hamel et al. (2019) termed this behavior active buoyancy adjustment, and found that in experimental trials this behavior was triggered by high conspecific density, increasing water turbidity, and decreasing salinity, with active buoyancy adjustment only observed in enclosures that were located near land. If active buoyancy adjustment is common in wild populations of H. scabra, then LMMAs would need to be large to be effective once protection leads to high densities.
The findings of this study demonstrate that H. scabra, and possibly other species of the genus Holothuria, are well suited to parentage analysis. In this study, 19 H. scabra microsatellites were successfully genotyped. Each of these microsatellites had high levels of polymorphism, which provided strong exclusion power in the parentage analysis and high confidence in assigned parent-offspring pairs. The H. scabra dispersal kernel presented here is based on a low number of assignments, 15 out of out of 827 total juvenile samples. This 2% assignment rate is relatively low in dispersal analyses based on parentage assignments (Bode et al., 2018), most of which assign between 1% and 15% of juveniles. This low juvenile assignment is a result of only sampling a small proportion of a single source population, and is reflected in the large uncertainty around the best-fit larval dispersal kernel. UVC surveys indicate that there were 24,653 adult sandfish in the Pere fishing grounds when this fieldwork was conducted (Supplementary Table S2), of which only 3.1% (n = 765) were analysed for the purpose of this study.
In this study, the shallow distribution of abundant and sedentary H. scabra, combined with the assistance of knowledge local fishers, made it possible to sample 6,465 sandfish in a month. However, the human resources required to amplify all the samples were unavailable. – In particular, the costs of the technical specialists’ time to extract and genotype the samples, the cost of the necessary extraction kits, and the time to process the output from the genotyping. In detail, to extract batches of samples (e.g., 2 x 96 samples) and prepare them for fragment analysis would take 1–3 days, while processing of genotyped samples would take 3–4 days. As a consequence, we were only able to process 36.5% (n = 765) of the adult and 18.9% (n = 827) of the juvenile samples. Advances in genetic technologies may allow genotyping of a very large number of tissue samples in a small number of sequencing runs, saving months of data processing and making large-scale parentage studies on sea cucumbers more feasible.
The limited larval dispersal of H. scabra that we observed in this study provides compelling evidence that coastal communities who establish LMMAs to protect spawning stocks of sea cucumbers can directly benefit from their efforts. These LMMAs can replenish stocks within the fishing grounds they were established in, complementing national management measures such as closed seasons and size limits. Our study has also shown that although H. scabra appear to have limited larval dispersal, the animals nevertheless move across spatial scales that are larger than most communities’ traditional fishing grounds. Networks of LMMAs that span multiple fishing grounds – at the scale of MEARDN, for example – are needed to generate the greatest benefits for these fisheries. LMMAs that are protect prime adult habitat of H. scabra, are in line-of-sight of communities, are ≥ 40 ha, and are separated by other LMMAs in a network by a maximum of 15 km are likely to provide the greatest fisheries management benefits for sandfish.
Data availability statement
The code for fitting dispersal kernels to parentage data are available at https://github.com/MikeBode/Parentage_kernel_fitting/. The cleaned datasets used for the Manus example, including the code for creating Figure 2, can be found in this repository in the archive “Manus_Holothuria_scabra.zip”. The GenBank accession number for the sequences reported in this paper are PP548095-PP548120. Additional markers are available upon request to the authors.
Ethics statement
Ethical approval was not required for the study involving animals in accordance with the local legislation and institutional requirements because Research clearance, which included ethics clearance, was provided by the Mwanus Endras Asi Resource Development Network and the Papua New Guinea National Fisheries Authority.
Author contributions
PW: Project administration, Data curation, Formal analysis, Software, Validation, Writing – review & editing. DF: Data curation, Writing – review & editing, Project administration, Investigation, Methodology. MB: Formal analysis, Methodology, Software, Writing – original draft. MM: Conceptualization, Data curation, Investigation, Project administration, Writing – review & editing. HH: Data curation, Writing – review & editing, Formal analysis, Methodology. MLB: Writing – review & editing, Project administration, Resources. CM: Writing – review & editing, Data curation, Investigation. MK: Data curation, Writing – review & editing, Investigation. RH: Investigation, Conceptualization, Funding acquisition, Methodology, Project administration, Supervision, Writing – original draft.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Acknowledgments
We would like to acknowledge the following members of the sandfish survey team: Polin Chapokat, Paul Karamen, Rabbie Kanamon, Robert Maiah, Mendennis Kanawi, Pondros Lokes, Polin Manuai, Cholia Pochalon, Pokakes Pondraken, Ron Ponowan, Gai Samol and Paul Tapas. We also thank Dr. Pongie Kichawen and Luanah Yaman for logistical support. We express our thanks to the community members of Mbunai, Pere, Locha, Tawi, Timoenai and Mbuke and the Manus Provincial Government for supporting this work. Funding for this work was provided by the David and Lucile Packard Foundation, the Papua New Guinea National Fisheries Authority, and the King Abdullah University of Science and Technology.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2024.1380235/full#supplementary-material
References
Alberto F. (2009). MsatAllele_1.0: an R package to visualize the binning of microsatellite alleles. J. Hered. 100, 394–397. doi: 10.1093/jhered/esn110
Almany G. R., Berumen M. L., Thorrold S. R., Planes S., Jones G. P. (2007). Local replenishment of coral reef fish populations in a marine reserve. Science 316, 742–744. doi: 10.1126/science.1140597
Almany G. R., Hamilton R. J., Bode M., Matawai M., Potuku T., Saenz-Agudelo P., et al. (2013). Dispersal of grouper larvae drives local resource sharing in a coral reef fishery. Curr. Biol. 23, 626–630. doi: 10.1016/j.cub.2013.03.006
Almany G. R., Hamilton R. J., Matawai M., Kichawen P. (2015). Local benefits of community-based management: Using small managed areas to rebuild and sustain some coastal fisheries. SPC Traditional Mar. Resource Manage. Knowledge Inf. Bull. 35, 3–17.
Almany G. R., Hamilton R. J., Williamson D. H., Evans R. D., Jones G. P., Matawai M., et al. (2010). Research partnerships with local communities: two case studies from Papua New Guinea and Australia. Coral Reefs 29, 567–576. doi: 10.1007/s00338-010-0624-3
Barclay K., Fabinyi M., Kinch J., Foale S. (2019). Governability of high-value fisheries in low-income contexts: a case study of the sea cucumber fishery in Papua New Guinea. Hum. Ecol. 47, 381–396. doi: 10.1007/s10745-019-00078-8
Begg G. A., Friedland K. D., Pearce J. B. (1999). Stock identification and its role in stock assessment and fisheries management: An overview. Fisheries Res. 43, 1–8. doi: 10.1016/S0165-7836(99)00062-4
Bode M., Leis J. M., Mason L. B., Williamson D. H., Harrison H. B., Choukroun S., et al. (2019). Successful validation of a larval dispersal model using genetic parentage data. PLoS Biol. 17, e3000380. doi: 10.1371/journal.pbio.3000380
Bode M., Williamson D. H., Harrison H. B., Outram N., Jones G. P. (2018). Estimating dispersal kernels using genetic parentage data. Methods Ecol. Evol. 9, 490–501. doi: 10.1111/2041-210X.12922
Bonin M. C., Harrison H. B., Williamson D. H., Frisch A. J., Saenz-Agudelo P., Berumen M. L., et al. (2016). The role of marine reserves in the replenishment of a locally impacted population of anemonefish on the Great Barrier Reef. Mol. Ecol. 25, 487–499. doi: 10.1111/mec.13484
Brown K. T., Southgate P. C., Hewavitharane C. A., Lal M. M. (2022). Saving the sea cucumbers: Using population genomic tools to inform fishery and conservation management of the Fijian sandfish Holothuria (Metriatyla) scabra. PLoS One 17, e0274245. doi: 10.1371/journal.pone.0274245
Christensen A. E. (2011). Marine gold and atoll livelihoods: The rise and fall of the bêche-de-mer trade on Ontong Java, Solomon Islands. Natural Resour. Forum 35, 9–20. doi: 10.1111/narf.2011.35.issue-1
Costello C., Quérou N., Tomini A. (2015). Partial enclosure of the commons. J. Public Economics 121, 69–78. doi: 10.1016/j.jpubeco.2014.11.011
Dalzell P. (1990). Beche-de-mer production from three Papua New Guinean atolls between 1982 and 1983. SPC Beche-de-mer Inf. Bull. 1, 6–7.
Faircloth B. C. (2008). Msatcommander: detection of microsatellite repeat arrays and automated, locus-specific primer design. Mol. Ecol. Resour. 8, 92–94. doi: 10.1111/j.1471-8286.2007.01884.x
Feary D. A., Hamilton R., Matawai M., Molai C., Karo M., Almany G. (2014). Assessing sandfish population stocks within the south coast of Manus, and a summary report of sandfish connectivity field research (Queensland Australia: The Nature Conservancy Asia Pacific Division).
Friedman K., Eriksson H., Tardy E., Pakoa K. (2011). Management of sea cucumber stocks: patterns of vulnerability and recovery of sea cucumber stocks impacted by fishing. Fish Fisheries 12, 75–93. doi: 10.1111/j.1467-2979.2010.00384.x
Gerber S., Chabrier P., Kremer A. (2003). FAMOZ: a software for parentage analysis using dominant, codominant and uniparentally inherited markers. Mol. Ecol. Notes 3, 479–481. doi: 10.1046/j.1471-8286.2003.00439.x
Gkafas G. A., Sarantopoulou J., Apostologamvrou C., Antoniadou C., Exadactylos A., Fleris G., et al. (2023). Admixture of holothurian species in the hellenic seas (Eastern mediterranean) as revealed by RADseq. Sustainability 15, 11493. doi: 10.3390/su151511493
Green A. L., Maypa A. P., Almany G. R., Rhodes K. L., Weeks R., Abesamis R. A., et al. (2015). Larval dispersal and movement patterns of coral reef fishes, and implications for marine reserve network design. Biol. Rev. 90, 1215–1247. doi: 10.1111/brv.12155
Hair C., Foale S., Kinch J., Frijlink S., Lindsay D., Southgate P. C. (2019). Socioeconomic impacts of a sea cucumber fishery in Papua New Guinea: Is there an opportunity for mariculture? Ocean Coast. Manage. 179, 104826. doi: 10.1016/j.ocecoaman.2019.104826
Hair C., Foale S., Kinch J., Yaman L., Southgate P. C. (2016). Beyond boom, bust and ban: The sandfish (Holothuria scabra) fishery in the Tigak Islands, Papua New Guinea. Regional Stud. Mar. Sci. 5, 69–79. doi: 10.1016/j.rsma.2016.02.001
Hamel J. F., Eeckhaut I., Conand C., Sun J., Caulier G., Mercier A. (2022). Global knowledge on the commercial sea cucumber Holothuria scabra Vol. 91 (Elsevier Ltd: Academic Press), 1–286. doi: 10.1016/bs.amb.2022.04.001
Hamel J.-F., Mercier A., Conand C., Purcell S., Toral-Granda T.-G., Gamboa R. (2013). Holothuria Scabra (The IUCN Red List of Threatened Species 2013). e.T180257A1606648. doi: 10.2305/IUCN.UK.2013-1.RLTS.T180257A1606648.en
Hamel J. F., Sun J., Gianasi B. L., Montgomery E. M., Kenchington E. L., Burel B., et al. (2019). Active buoyancy adjustment increases dispersal potential in benthic marine animals. J. Anim. Ecol. 88, 820–832. doi: 10.1111/1365-2656.12943
Hamilton R., Gereniu G., Kaniki H., Kereseka J. (2015). A report on periodic harvests at the Paspasbarego and Parama Locally Marine Managed Areas, Choiseul, Solomon Islands (Solomon Islands: The Nature Conservancy Pacific Division). 16 pp.
Hamilton R., Lokani P. (2011). Severely overfished sea cucumbers in the autonomous region of Bougainville. SPC Beche-de-mer Inf. Bull. 31, 12–16.
Hamilton R. J., Lozano-Cortés D., Bode M., Almany G. R., Harrison H. B., Pita J., et al. (2021). Larval dispersal and fishing pressure influence recruitment in a coral reef fishery. J. Appl. Ecol. 58, 2924–2935. doi: 10.1111/1365-2664.14027
Hamilton R. J., Matawai M., Potuku T., Kama W., Lahui P., Warku J., et al. (2005). Applying local knowledge and science to the management of grouper aggregation sites in Melanesia. SPC Live Reef Fish Inf. Bull. 14, 7–19.
Harrison H. B., Feldheim K. A., Jones G. P., Ma K., Mansour H., Perumal , et al. (2014). Validation of microsatellite multiplexes for parentage analysis and species discrimination in two hybridizing species of coral reef fish (Plectropomus spp., Serranidae). Ecol. Evol. 4, 2046–2057. doi: 10.1002/ece3.1002
Harrison H. B., Saenz-Agudelo P., Planes S., Jones G. P., Berumen. M. L. (2013). Relative accuracy of three common methods of parentage analysis in natural populations. Mol. Ecol. 22, 1158–1170. doi: 10.1111/mec.12138
Harrison H. B., Williamson D. H., Evans R. D., Almany G. R., Thorrold S. R., Russ G. R., et al. (2012). Larval export from marine reserves and the recruitment benefit for fish and fisheries. Curr. Biol. 22, 1023–1028. doi: 10.1016/j.cub.2012.04.008
Herrera M., Nanninga G. B., Planes S., Jones G. P., Thorrold S. R., Saenz-Agudelo P., et al. (2016). Seascape and life-history traits do not predict self-recruitment in a coral reef fish. Biol. Lett. 12, 20160309. doi: 10.1098/rsbl.2016.0309
Johannes R. E. (2002). The renaissance of community- based marine resource management in Oceania. Annu. Rev. Ecol. Systematics 33, 317–340. doi: 10.1146/annurev.ecolsys.33.010802.150524
Jones G. P., Planes S., Thorrold S. R. (2005). Coral reef fish larvae settle close to home. Curr. Biol. 15, 1314–1318.
Jones G. P. (2015). “Mission impossible: Unlocking the secrets of coral reef fish dispersal,” in Ecology of Fishes on Coral Reefs, ed. Mora C. (Cambridge, UK: Cambridge University) 16–27.
Kinch J., Purcell S., Uthicke S., Friedman K. (2008). “Papua New Guinea: a hot spot of sea cucumber fisheries in the Western Central Pacific,” in Sea Cucumbers: A Global Review of Fisheries and Trade. Eds. Toral-Granda V., Lovatelli A., Vasconcellos M. (FAO Fisheries and Aquaculture Technical Paper No. 516, FAO, Rome), 57–77.
Lalavanua W., Mangubhai S., Vandervord C., Dulunaqio S., Fox M., Naisilisili W., et al. (2017). “Sea cucumber species richness and densities within locally managed marine areas.,” in Fiji’s Sea Cucumber Fishery: Advances in Science for Improved Management (Suva, Fiji: Wildlife Conservation Society), 4–15.
Lee S., Ford A. K., Mangubhai S., Wild C., Ferse S. C. A. (2018). Length-weight relationship, movement rates, and in situ spawning observations of Holothuria scabra (sandfish) in Fiji. SPC Beche-de-mer Inf. Bull. 38, 11–14.
Lokani P. (1990). Bêche-de-mer research and development in Papua New Guinea. SPC Beche mer Inf. Bull. 2, 8–11.
Lokani P. (2001). Survey of bêche-de-mer resource in Manus. Natl. Fisheries Authority (Port Moresby).
Marshall T. C., Slate J. B. K. E., Kruuk L. E. B., Pemberton J. M. (1998). Statistical confidence for likelihood-based paternity inference in natural populations. Mol. Ecol. 7 (5), 639–655. doi: 10.1046/j.1365-294x.1998.00374.x
McClanahan T. R., Marnane M. J., Cinner J. E., Kiene W. E. (2006). A comparison of marine protected areas and alternative approaches to coral-reef management. Curr. Biol. 16, 1408–1413. doi: 10.1016/j.cub.2006.05.062
Mills M., Weeks R., Pressey R. L., Foale S., Ban N. C. (2010). A mismatch of scales: challenges in planning for implementation of marine protected areas in the Coral Triangle. Conserv. Lett. 3, 291–303. doi: 10.1111/j.1755-263X.2010.00134.x
Murphy N. E., Skewes T. D., Dovers E., Leatherbarrow A. (2012). Assessing stock recovery using experimental fishing for sandfish on Warrior Reef, Torres Strait (Dutton Park: Final Report. CSIRO). 34 pp.
Nanninga G. B., Saenz-Agudelo P., Zhan P., Hoteit I., Berumen M. L. (2015). Not finding Nemo: Limited reef-scale retention in a coral reef fish. Coral Reefs 34, 383–392. doi: 10.1007/s00338-015-1266-2
Pelc R. A., Warner R. R., Gaines S. D., Paris C. B. (2010). Detecting larval export from marine reserves. Proc. Natl. Acad. Sci. United States America 107, 18266–18271. doi: 10.1073/pnas.0907368107
Planes S., Jones G. P., Thorrold S. R. (2009). Larval dispersal connects fish populations in a network of marine protected areas. Proc. Natl. Acad. Sci. 106 (14), 5693–5697.
PNG National Fisheries Authority (2007). “Sea cucumber survey, New Ireland Province,” in Kaveing, National Fisheries Authority and the Coastal Fisheries Management and Development Project (Port Moresby: Papua New Guinea National Fisheries Authority).
Purcell S. W., Kirby D. S. (2006). Restocking the sea cucumber Holothuria scabra: sizing no-take zones through individual-based movement modelling. Fisheries Res. 80, 53–61. doi: 10.1016/j.fishres.2006.03.020
Purcell S. W., Lovatelli A., Pakoa K. (2014). Constraints and solutions for managing Pacific Island sea cucumber fisheries with an ecosystem approach. Mar. Policy 45, 240–250. doi: 10.1016/j.marpol.2013.11.005
Purcell S. W., Rallings S. L., Hammond A. R. (2023). Long-term home ranging in the large sea cucumber, Holothuria. fuscopunctata. Coral reefs 42 (5), 1059–1066. doi: 10.1007/s00338-023-02413-4
Pusack T. J., Christie M. R., Johnson D. W., Stallings C. D., Hixon M. A. (2014). Spatial and temporal patterns of larval dispersal in a coral-reef fish metapopulation: Evidence of variable reproductive success. Mol. Ecol. 23, 3396–3408. doi: 10.1111/mec.12824
Ruddle K. (1996). “Traditional management of reef fishing,” in Reef fisheries. Eds. Polunln N. V. C., Roberts C. M. (Chapman and Hall, London), p 315–p 335.
Sale P. F., Cowen R. K., Danilowicz B. S., Jones G. P., Kritzer J. P., Lindeman K. C., et al. (2005). Critical science gaps impede use of no-take fishery reserves. Trends Ecol. Evol. 20, 74–80. doi: 10.1016/j.tree.2004.11.007
Shulzitski K., Sponaugle S., Hauff M., Walter K. D., Cowen R. K. (2016). Encounter with mesoscale eddies enhances survival to settlement in larval coral reef fishes. Proc. Natl. Acad. Sci. United States America 113, 6928–6933. doi: 10.1073/pnas.1601606113
Toral-Granda V., Lovatelli A., Vasconcellos M. (2008). “Sea cucumbers. A global review of fisheries and trade,” in FAO Fisheries and Aquaculture Technical Paper. Eds. Toral-Granda V., Lovatelli A., Vasconcellos M. (FAO, Rome), 1–4. OverviewNo. 516.
Uthicke S., Benzie J. A. H. (2001). Restricted gene flow between Holothuria scabra (Echinodermata: Holothuroidea) populations along the north-east coast of Australia and the Solomon Islands. Mar. Ecol. Prog. Ser. 216, 109–117. doi: 10.3354/meps216109
Uthicke S., Purcell S., Blockmans B. (2005). Natural hybridization does not dissolve species boundaries in commercially important sea cucumbers. Biol. J. Linn. Soc. 85, 261–270. doi: 10.1111/bij.2005.85.issue-3
van der Meer M. H., Berumen M. L., Hobbs J. P., van Herwerden L. (2015). Population connectivity and the effectiveness of marine protected areas to protect vulnerable, exploited and endemic coral reef fishes at an endemic hotspot. Coral Reefs 34 (2), 393–402. doi: 10.1007/s00338-014-1242-2
Walters C. J., Martell S. J. (2004). Fisheries ecology and management (New Jersey: Princeton University Press). doi: 10.1515/9780691214634
Weeks R., Russ G. R., Alcala A. C., White A. T. (2010). Effectiveness of marine protected areas in the Philippines for biodiversity conservation. Conserv. Biol. 24, 531–540. doi: 10.1111/j.1523-1739.2009.01340.x
Williamson D. H., Harrison H. B., Almany G. R., Berumen M. L., Bode M., Bonin M. C., et al. (2016). Large-scale, multidirectional larval connectivity among coral reef fish populations in the Great Barrier Reef Marine Park. Mol. Ecol. 25, 6039–6054. doi: 10.1111/mec.13908
Keywords: customary marine tenure, connectivity, locally managed marine areas, fisheries management, genetic parentage analyses, larval dispersal
Citation: Waldie P, Feary DA, Bode M, Matawai M, Harrison HB, Berumen ML, Molai C, Karo M and Hamilton RJ (2024) Dispersal patterns of sandfish (Holothuria scabra) larvae in Manus Province, Papua New Guinea. Front. Mar. Sci. 11:1380235. doi: 10.3389/fmars.2024.1380235
Received: 01 February 2024; Accepted: 08 April 2024;
Published: 18 April 2024.
Edited by:
Pedro M. Félix, Center for Marine and Environmental Sciences (MARE), PortugalReviewed by:
Chryssanthi Antoniadou, Aristotle University of Thessaloniki, GreeceVincent Raoult, Deakin University, Australia
Copyright © 2024 Waldie, Feary, Bode, Matawai, Harrison, Berumen, Molai, Karo and Hamilton. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Richard Hamilton, rhamilton@tnc.org
 Peter Waldie
Peter Waldie David A. Feary
David A. Feary Michael Bode3
Michael Bode3  Michael L. Berumen
Michael L. Berumen Richard J. Hamilton
Richard J. Hamilton