Reactivation of a Vaccine Escape Hepatitis B Virus Mutant in a Cambodian Patient During Anti-Hepatitis C Virus Therapy
- 1Medicine/General Internal Medicine and Infectious Diseases, Massachusetts General Hospital, Harvard Medical School, Boston, MA, United States
- 2Laboratory of Systems Pharmacology, Harvard Medical School, Boston, MA, United States
- 3Molecular Epidemiology and Bioinformatics Team, DVH/NCHHSTP/CDC, Centers for Disease Control and Prevention (CDC), Atlanta, GA, United States
- 4Cooper Medical School of Rowan University, Camden, NJ, United States
- 5Wellesley College, Wellesley, MA, United States
- 6Gastrointestinal Division, Massachusetts General Hospital, Harvard Medical School, Boston, MA, United States
- 7Institut de recherche pour le développement(IRD), Marseille, France
- 8Chiang Mai University, Chiang Mai, Thailand
- 9Harvard School of Public Health, Boston, MA, United States
- 10Institut National de la Tranfusion Sanguine INSER U1134, Paris, France
A 76-year-old Cambodian man co-infected with hepatitis B virus (HBV) and hepatitis C virus (HCV) 6c-1 presented for care. HBV DNA was intermittently detectable despite anti-HBs levels being above the protective threshold. During treatment for HCV, HBV DNA levels increased. Sequencing revealed multiple mutations including vaccine escape mutation and mutations predicted to enhance fitness. This case represents exacerbation of an HBV vaccine escape mutant during a direct-acting antiviral therapy.
Background
Hepatitis B virus (HBV) reactivation has been reported during hepatitis C virus (HCV) treatment, leading to a need for close surveillance of HBV-infected patients undergoing HCV therapy. We here report a case of HBV reactivation during HCV treatment, with the finding that the reactivated HBV was a vaccine escape mutant.
Case Report
A 76-year-old man from Cambodia presented for viral hepatitis care. Institutional Review Board approval was waived for this case report. The patient had a medical history of benign prostatic hyperplasia, supraventricular tachycardia, adenomatous polyp of the colon, spinal stenosis, essential thrombocythemia, and hemoglobin E trait. The patient was co-infected with HBV and HCV and had never received treatment for either virus. The patient did not recall any past HBV vaccination. Risk factors for HBV and HCV exposure were unknown. The patient emigrated from Cambodia, where he served in the military. He had no history of blood transfusion, or surgery in Cambodia. He had no tattoos and no history of unsafe injection practices. The patient’s family had no history of liver cancer, liver disease, or cirrhosis. The patient had no history of jaundice or hospitalization for liver disease. On presentation, he had a complaint only of constant generalized weakness. He did not have nausea, vomiting, diarrhea, anorexia, change in bowel pattern, hematochezia, hematemesis, or blood when brushing teeth, rash, arthralgias, unexpected change in weight, abdominal pain, or swelling. Physical examination revealed normal vital signs and no evidence of end-stage liver disease, including no ascites or edema, no appreciable hepatosplenomegaly, no asterixis, no spider angiomata, no jaundice, and clear mentation. Laboratory values revealed creatinine of 1.01 mg/dl, alkaline phosphatase of 43I U/l, ALT of 22 U/l, and AST of 29 U/l. AFP was 3.2 ng/ml. Complete blood count revealed white blood cell count of 5, hemoglobin of 13, and platelet count of 436. HBV DNA 1 month prior to presentation had been 120 IU/ml. The patient had detectable total HBV core antibody (anti-HBc). HBV e antibody and e antigen were not detected. The HBV surface antigen antibody (anti-HBs) titer was at 106 mIU/ml and surface antigen (HBsAg) was undetectable. HBV DNA testing was repeated upon presentation and was reported as <20 IU/ml (HBV DNA not quantifiable). HIV 1,2 Ag/Ab test was negative. HCV genotype was found to be 6c-1, and HCV viral load was 14,500,000 IU/ml. HDV RNA was undetectable. Abdominal ultrasound revealed no suspicious liver lesions and cholelithiasis without cholecystitis. A biopsy was not performed but FibroSure® (LabCorp) blood test indicated F3 fibrosis with bridging. Initial counseling related to HCV and HBV was performed. HCV genotype and HBV viral load were identified for all specimens collected during HCV treatment.
One month later, the patient was seen in follow-up. HCV genotype had been identified as HCV 6c-1.
Based on AASLD guidelines, accessed on September 13, 2016, both sofosbuvir (400 mg)/velpatasvir (100 mg) and ledipasvir (90 mg)/sofosbuvir (400 mg) are acceptable treatments for HCV genotype 6, although the evidence rating was slightly stronger for sofosbuvir (400 mg)/velpatasvir (100 mg). A prior authorization was submitted for sofosbuvir (400 mg)/velpatasvir (100 mg), but rejected by the patient’s insurance with a recommendation to substitute sofosbuvir (400 mg)/velpatasvir (100 mg) with ledipasvir (90 mg)/sofosbuvir (400 mg) 1 pill once daily for 12 weeks and was approved 3 weeks later. Prior to treatment initiation, HBV viral load was found to be <20 IU/ml (Figure 1). One week following treatment initiation, the patient was seen in follow-up. He was tolerating ledipasvir (90 mg)/sofosbuvir (400 mg) well, with improvement of his generalized weakness, and complained only of mild dry mouth. HCV viral load was 19 IU/ml. One week later, the patient was called back to clinic for HBV counseling, as a new black box warning had been added to the ledipasvir (90 mg)/sofosbuvir (400 mg) package insert recommending close HBV monitoring of patients on direct-acting antiviral therapy (DAA). HBV viral load was checked and found to be 287 IU/ml (up from <20 IU/ml 1.5 and 3 months earlier) (Figure 1). HCV viral load was <15 IU/ml. A plan was made to monitor the patient every 2 weeks for clinical symptoms and virologic evidence of worsening HBV, with agreement to present to a clinic earlier if any concerning symptoms developed in the interim (e.g., nausea, vomiting, jaundice, anorexia, or extreme fatigue).
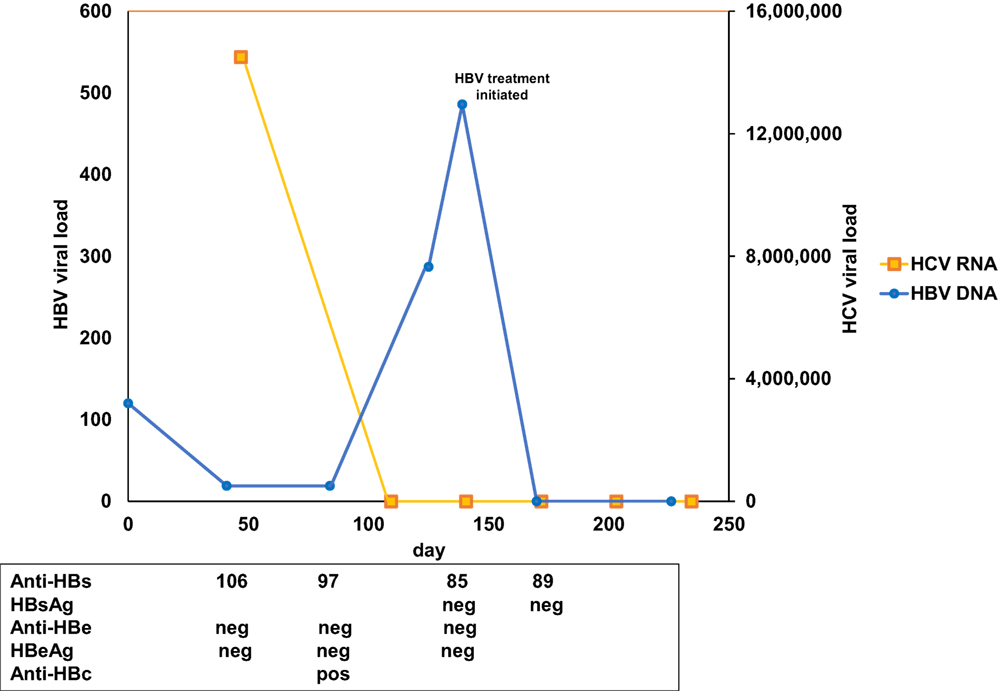
Figure 1. Chronological results for hepatitis B virus (HBV) and hepatitis C virus (HCV) studies. Y-axis indicates HBV (left) and HCV (right) viral load over time (X-axis). HCV RNA is indicated in yellow, HBV DNA in blue. HBV surface antibody (Anti-HBs), surface antigen (HBsAg), E antibody (anti-HBe), E antigen (HBeAg), and core antibody (anti-HBc) results are presented corresponding to the day of collection.
Two weeks later, the patient returned for follow-up and was found to have undetectable HCV RNA and HBV viral load of 486 IU/ml (up from 287 IU/ml) (Figure 1). Due to concern for potential fulminant HBV reactivation while on DAA therapy (1) (Table S1 in Supplementary Material), the patient was initiated on tenofovir therapy 300 mg once daily, with a plan for an indefinite course due to advanced fibrosis. The patient was seen 2 weeks following tenofovir initiation, was doing well without complaints, and was found to have undetectable HBV DNA as well as undetectable HCV RNA. The patient completed a 12-week course of HCV treatment without further event and maintained undetectable HBV DNA and HCV RNA through 12 weeks following the end of HCV treatment at the time of this writing.
Because the patient had exhibited detectable HBV DNA in the presence of HBV surface antibody at the level above the protective threshold, there was concern for the presence of an HBV vaccine escape mutant. Serum from day 139, with HBV DNA 486 IU/ml, was sent for sequencing analysis to the Center for Disease Control, Division of Viral Hepatitis Laboratory. Complete sequence results are presented (Figure S1 in Supplementary Material). The HBV genotype was identified to be C1 (98% identical to published isolate SEA-01, GenBank KM999990). The serological subtype was identified to be adrq+. Eleven mutations were identified (Table 1), including (1) S gene/polymerase: a 6-nucleotide (nt) deletion at nt 45–50 resulting in the removal of amino acids LY from pre-S2 and PI from polymerase, (2) S gene: G145A, a known immune escape S Ag mutation, (3–5) T118M, P120T, P142T: S Ag mutations affecting the a-determinant, (6) polymerase: V173A, (7) nt 1380: frameshift mutation in 50% of reads, (8) X gene: frameshift mutation at 1,581 in 52% of reads, (9) premature stop codon at 1,619, truncating X from 154 to 71 aa, (10) core: basal core promoter mutation A1762T, and (11) core: basal core promoter mutation G1764A. A summary of the mutations and prior literature describing these mutations (Table 1; Table S2 in Supplementary Material) and a schematic of the mutations (Figure 2) are presented.
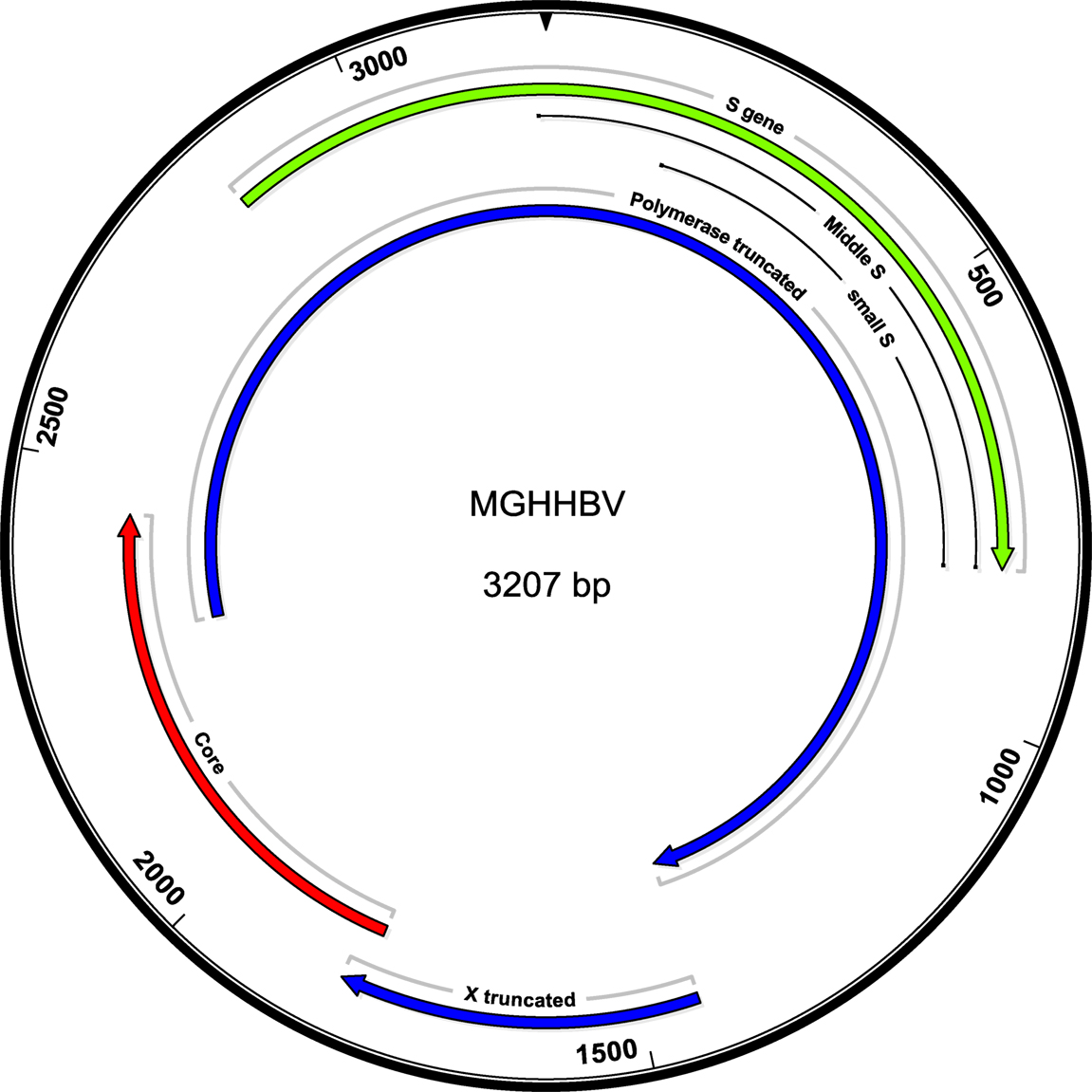
Figure 2. Schematic of the mutant genome. The in-frame deletion is not affecting the ORFS, but shortens pre-S2 and Pol by the corresponding two amino acids. A total of 1,380 truncates the polymerase to 82 amino acids.
Consent
Written informed consent was obtained from the participant for the publication of this case report.
Serologic Assays
Hepatitis B virus e Ag/Ab testing was performed using VITROS Immunodiagnostic Products for anti-HBe, HBeAg (http://www.mayomedicallaboratories.com/test-catalog/Performance/8311). For the anti-HBs test, the ARCHITECT AUSAB assay (Abbott Diagnostics) was used. ARCHITECT AUSAB is a chemiluminescent microparticle immunoassay (CMIA) for the quantitative determination of anti-HBs in human adult and pediatric serum and plasma (dipotassium EDTA, lithium heparin, and sodium heparin) and neonatal serum. For the HBsAg test, the ARCHITECT HBsAg qualitative assay (Abbott diagnostics) was used. The ARCHITECT HBsAg qualitative assay is a CMIA for the qualitative detection of HBsAg in human adult and pediatric serum and plasma and neonate serum. For the HBV core Ab test, the ARCHITECT Core assay (Abbott diagnostics) was used. The ARCHITECT Core assay is a CMIA for the qualitative detection of IgG and IgM antibodies to hepatitis B core antigen (anti-HBc) in human adult and pediatric serum and plasma (dipotassium EDTA, lithium heparin, and sodium heparin) and neonatal serum.
HBV DNA Quantification
Hepatitis B virus DNA quantitative assay was performed using Roche Diagnostics (Cobas AmpliPrep/Cobas TaqMan v2.0).
HBV DNA Sequencing
Hepatitis B virus DNA was amplified using modified primers (19) that produce a linearized product of the complete genome, followed by random enzymatic shearing, barcoding, and size selection to generate a shotgun next-generation sequencing (NGS) library for the Illumina platform. NGS was done using Illumina v2 150 cycle kit. Reads (150 nt) were assembled by CLC Genomics Workbench (v 10.03, Qiagen, Aarhus) into a complete HBV genome of 3,207 nt at an average coverage of 12,414.78x.
Discussion
This case represents reactivation of an HBV immune escape mutant containing potential fitness-enhancing mutations during HCV clearance in a non-vaccinated, non-transplant patient. This case is unique for several reasons. First, anti-HBs was detectable simultaneously with HBV DNA, while the patient was negative for HBsAg, HBeAg, and anti-HBe, which is consistent with anti-HBs escape and occult infection. Second, the HBV DNA levels, though detectable within the 3 months prior to treatment, increased from <20 to 400 IU/ml during HCV 6c-1 clearance with DAA, consistent with HBV reactivation due to HCV clearance, as has been reported by others during both DAA and IFN—mediated clearance of HCV (Table S1 in Supplementary Material). Finally, although HBV immune escape mutants have been reported to be of lower or equal fitness compared to wild type (20, 21), the current sequence revealed concurrent immune escape and fitness-enhancing mutations as listed in Table 1.
There are limitations of this case report. The consensus sequence extracted from the NGS data may not correspond to any actual intra-host HBV variant existing in the patient. As for instance, one mutation, or deletion in preS2, may not coexist on the same HBV DNA molecule in the patient with, for example, a frameshift in Pol or X. Deletions 1,380 and 1,581 are found in 50 and 52% of the reads. However, the remaining reads exhibit a wild-type sequence. These data do not allow establishing the cis-coexistence of the described mutations. The distance between mutations prevents their identification in one fragment. For the frameshift mutation at nt 1,380 detected in 50% of reads, it is not possible to guarantee that this was not due to PCR error. However, PCR errors of this sort are 100 times less frequent than substitution errors (which happen with about 1.2 × E^4 frequency) and 10 times less frequent than recombination errors. They also tend to occur in homopolymer or short repeat regions and the location nt 1,375 does not contain either.
It is important to note that the sequence described here includes both immune escape and two fitness-enhancing mutations (Table 1). HBV immune escape mutants have been reported to be of lower, equal, and higher infectivity compared to wild type (4, 20–22). While these mutations are frequently described among intra-host HBV variants (23–25), and we have no direct evidence that immune escape and fitness-enhancement mutations occur on the same DNA molecule, their coexistence is nonetheless concerning, given the potential public health consequences of vaccine escape mutants (21). While mutant forms of HBV in cases of occult HBV may theoretically escape detection and could present a risk to blood safety, current methods for blood screening include HBsAg, anti-HBc, and HBV DNA. Although negative for HBsAg, the case described here was positive for anti-HBc and intermittently positive for HBV DNA; therefore, screening would have detected this case through the inclusion of anti-HBc. The greater safety concern for cases such as the one here described relates to the risk of transmission to persons who have been vaccinated (21) and would not be aware of risk related to exposure in the absence of full sequencing. It is not clear how common occult HBV (HBsAg negative/DNA positive) and anti-HBs escape mutations (anti-HBs positive/DNA positive) are among HBV-infected patients in the United States. Prior studies in China and France have noted between 3.4 (26), 4.9 (27), and 8.9% (14) of HBV-infected patients to have coexistent HBsAg and anti-HBs. Anti-HBs escape mutations have been described in the USA in infants born to HBV-infected mothers following postnatal HBV vaccine and hepatitis B immune globulin (HBIG) prophylaxis and in many liver-transplant recipients who develop HBV re-infection despite HBIG prophylaxis (28–30) as well as one HBV vaccinated patient post lung transplant on hemodialysis (31). To the best of our knowledge, this is the first report of a mutation in the “a”-determinant region (G145A) in a patient in the USA with chronic HBV infection in the absence of receiving HBV vaccine or HBIG (28, 31, 32). G–A substitution leads to a 50% loss of a-determinant and infectivity in vitro, while the more common G–R substitution leads to a loss of a-determinant but a gain in infectivity in vitro, explaining why this G145R preferentially emerges (20, 33). The positive charge of R enhances attachment to cell surface heparan sulfate and hence infectivity.
This case is significant because it highlights the potential need for a greater sequence surveillance of populations outside those previously known to be high risk for escape mutations (transplant patients and infants born to HBV-infected mothers).
In addition to immune escape mutation, the HBV sequence analysis in this case revealed HBV genotype C1 with a small deletion in preS2. PreS2 deletion mutations have been previously described especially in Asian patients with HBV genotype C (2) and are associated with a higher likelihood of hepatocellular carcinoma (HCC) in both adults and children, as well as the coexistence of HBsAg and anti-HBs (34–36). Mutations in the HBV pre-S2 region have been demonstrated in vitro to cause transactivation of reporter genes, which is thought to contribute to the development of HCC (37, 38). Subsequent studies have determined that preS2 deletion mutations are common in patients with HCC, compared to HBV-infected patients without HCC (39) and that preS2 deletions are associated with the histological progression of ground-glass hepatocytes (40). Ground-glass hepatocytes have been associated with HBV-related fibrosis and HCC (41, 42). Proposed models through which preS2 deletion might cause hepatic pathology include (1) the loss of an HBV epitope of cytotoxic T lymphocytes, favoring viral escape from host immune attack; (2) an increased endoplasmic reticulum (ER) stress, due to accumulation of intracellular envelope proteins (43), contributing to carcinogenesis; and/or (3) the upregulation of cyclin A and cyclooxygenase which decreases the distance between S promoter and transcription initiation site of S mRNA, changing the ratios between the 3 HBV envelope proteins (L-/M-/S-HBsAg) and also increasing ER stress (44). It is notable that the previously described pre-S2 deletions are larger than the two amino acid deletions described here. However, the LY dipeptide is well conserved in all HBV genotypes; therefore, the mutation observed here may point to a crucial importance of LY. There are currently no guidelines for enhanced HCC monitoring among patients with preS2 deletions (of any size).
It is important to note that the combination of “protective” anti-HBs and negative HBsAg observed in this case could have provided false reassurance of HBV immune control and had anti-HBc and subsequent HBV DNA not been checked. The presence of positive anti-HBc prompted HBV DNA testing and sequencing, which revealed important mutations and HBV reactivation during treatment. Had this case progressed to fulminant HBV reactivation, the risk of transmission to vaccinated persons, such as health-care workers, was potentially high, given the potential coexistence of vaccine escape mutations and enhanced replication mutations described. This case underscores the importance of checking anti-HBc in all at-risk patients and subsequently monitoring HBV viral load, especially during HCV treatment. The frequency at which HBV viral load should be monitored, especially during and after HCV clearance and other immune-disruptive events, is not clear. Our current practice is to check HBV DNA every 2–4 weeks during HCV treatment of anti-HBc-positive patients. Finally, this case raises the question of when HBV sequence analysis should be performed and how those data could be centrally managed. HBsAg-negative anti-HBs escape mutants, if transmitted, could undermine vaccine efforts and also go unnoticed without appropriate evolution of diagnostic tests (45, 46).
Furthermore, there is evolving evidence that HBsAg variants may influence HBV treatment response (47).
The potential for this problem should be anticipated, a highly curated central repository for HBV-mutant strains be initiated, and strategy for vaccine adaptation be in place. Roque-Afonso et al. point out that “understanding the prevalence of potential antigenic variation of the HBsAg is fundamental for assay design and to future changes in vaccine formulation” (48). Similarly, Kim et al. suggest that the monitoring of mutation types in GTc with nested PCR (especially in occult cases) “should be imperative for appropriate control of HBV…because…certain mutations in HBV pre-S/-S region might alter (1) HBsAg antigenicity or (2) secretion capacity leading to immune escape infections and eventually emergence of HBV variants in the vaccinated population” (44). Overall, this case underscores the message of Roque-Afonso and Kim and highlights the importance of anti-HBc testing in all at-risk patients, the need for an active surveillance program for HBV sequences, and the importance of ongoing interpretation and anticipation of potential HBV mutation effects on vaccine and antiviral efficacy.
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Ethics Statement
This study was carried out in accordance with the recommendations of the Partners Institutional Review Board with written informed consent from the subject. The subject gave written informed consent in accordance with the Declaration of Helsinki. The protocol was approved by the Partners Institutional Review Board.
Author Contributions
DF, GJ, CS, KA, RC, and GL evaluated the clinical case and contributed to the description of the case evolution. DF, YK, LP, LG, AM, SC, PK, GJ, and CS evaluated the HBV sequence results and contributed to HBV sequence literature review. All authors contributed to the manuscript preparation.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding
Support for this work was provided by the Massachusetts General Hospital Scholarly Writing Award (DF), NIH NIAID (K08AI108629, DF), and the NIH P50 GM107618 (DF).
Supplementary Material
The Supplementary Material for this article can be found online at https://www.frontiersin.org/articles/10.3389/fmed.2018.00097/full#supplementary-material.
Table S1. Literature review of hepatitis B virus reactivation during hepatitis C virus treatment.
Table S2. Literature review of hepatitis B virus vaccine escape mutants.
References
1. Bersoff-Matcha SJ, Cao K, Jason M, Ajao A, Jones SC, Meyer T, et al. Hepatitis B virus reactivation associated with direct-acting antiviral therapy for chronic hepatitis C virus: a review of cases reported to the U.S. Food and Drug Administration adverse event reporting system. Ann Intern Med (2017) 166(11):792–8. doi:10.7326/M17-0377
2. Sa-Nguanmoo P, Tangkijvanich P, Thawornsuk N, Vichaiwattana P, Prianantathavorn K, Theamboonlers A, et al. Molecular epidemiological study of hepatitis B virus among migrant workers from Cambodia, Laos, and Myanmar to Thailand. J Med Virol (2010) 82(8):1341–9. doi:10.1002/jmv.21828
3. Zanetti AR, Tanzi E, Manzillo G, Maio G, Sbreglia C, Caporaso N, et al. Hepatitis B variant in Europe. Lancet (1988) 2(8620):1132–3.
4. Tong S, Revill P. Overview of hepatitis B viral replication and genetic variability. J Hepatol (2016) 64(1 Suppl):S4–16. doi:10.1016/j.jhep.2016.01.027
5. Sticchi L, Caligiuri P, Cacciani R, Alicino C, Bruzzone B. Epidemiology of HBV S-gene mutants in the Liguria Region, Italy: implications for surveillance and detection of new escape variants. Hum Vaccin Immunother (2013) 9(3):568–71.
6. Rodríguez Lay LA, Corredor MB, Villalba MC, Frómeta SS, Wong MS, Valdes L, et al. Genetic diversity of the hepatitis B virus strains in Cuba: absence of West-African genotypes despite the transatlantic slave trade. PLoS One (2015) 10(5):e0125052.
7. Salpini R, Fokam J, Ceccarelli L, Santoro MM, Nanfack A, Sosso SM, et al. High burden of HBV-infection and atypical HBV strains among HIV-infected cameroonians. Curr HIV Res (2016) 14(2):165–71.
8. Aghakhani A, Mohraz M, Aghasadeghi MR, Banifazl M, Vahabpour R, Karami A, et al. Occult hepatitis B virus infection and S gene escape mutants in HIV-infected patients after hepatitis B virus vaccination. Int J STD AIDS (2016) 27(11):967–72. doi:10.1177/0956462415602419
9. Huy TT, Ishikawa K, Ampofo W, Izumi T, Nakajima A, Ansah J, et al. Characteristics of hepatitis B virus in Ghana: full length genome sequences indicate the endemicity of genotype E in West Africa. J Med Virol (2006) 78(2):178–84. doi:10.1002/jmv.20525
10. Wang XY, Harrison TJ, He X, Chen QY, Li GJ, Liu MH, et al. The prevalence of mutations in the major hydrophilic region of the surface antigen of hepatitis B virus varies with subgenotype. Epidemiol Infect (2015) 143(16):3572–82. doi:10.1017/S0950268815000242
11. Amini-Bavil-Olyaee S, Vucur M, Luedde T, Trautwein C, Tacke F. Differential impact of immune escape mutations G145R and P120T on the replication of lamivudine-resistant hepatitis B virus e antigen-positive and -negative strains. J Virol (2010) 84(2):1026–33. doi:10.1128/JVI.01796-09
12. Avellón A, Echevarria JM. Frequency of hepatitis B virus ‘a’ determinant variants in unselected Spanish chronic carriers. J Med Virol (2006) 78(1):24–36.
13. Weber B. Genetic variability of the S gene of hepatitis B virus: clinical and diagnostic impact. J Clin Virol (2005) 32(2):102–12.
14. Lada O, Benhamou Y, Poynard T, Thibault V. Coexistence of hepatitis B surface antigen (HBs Ag) and anti-HBs antibodies in chronic hepatitis B virus carriers: influence of “a” determinant variants. J Virol (2006) 80(6):2968–75. doi:10.1128/JVI.80.6.2968-2975.2006
15. Ye Q, Shang SQ, Li W. A new vaccine escape mutant of hepatitis B virus causes occult infection. Hum Vaccin Immunother (2015) 11(2):407–10. doi:10.4161/21645515.2014.994461
16. Ishigami M, Ogura Y, Hirooka Y, Goto H. Change of strategies and future perspectives against hepatitis B virus recurrence after liver transplantation. World J Gastroenterol (2015) 21(36):10290–8. doi:10.3748/wjg.v21.i36.10290
17. Locarnini S, McMillan J, Bartholomeusz A. The hepatitis B virus and common mutants. Semin Liver Dis (2003) 23(1):5–20.
18. Hung CH, Chen CH, Lu SN, Wang JH, Hu TH, Huang CM, et al. Precore/core promoter mutations and hepatitis B virus genotype in hepatitis B and C dually infected patients treated with interferon-based therapy. Antiviral Res (2012) 93(1):55–63. doi:10.1016/j.antiviral.2011.10.018
19. Günther S, Li BC, Miska S, Krüger DH, Meisel H, Will H. A novel method for efficient amplification of whole hepatitis B virus genomes permits rapid functional analysis and reveals deletion mutants in immunosuppressed patients. J Virol (1995) 69(9):5437–44.
20. Salisse J, Sureau C. A function essential to viral entry underlies the hepatitis B virus “a” determinant. J Virol (2009) 83(18):9321–8. doi:10.1128/JVI.00678-09
21. Teo CG, Locarnini SA. Potential threat of drug-resistant and vaccine-escape HBV mutants to public health. Antivir Ther (2010) 15(3 Pt B):445–9. doi:10.3851/IMP1556
22. Kwei K, Tang X, Lok AS, Sureau C, Garcia T, Li J, et al. Impaired virion secretion by hepatitis B virus immune escape mutants and its rescue by wild-type envelope proteins or a second-site mutation. J Virol (2013) 87(4):2352–7. doi:10.1128/JVI.02701-12
23. Kaneko S, Miller RH. Heterogeneity of the core gene sequence in a patient chronically infected with hepatitis B virus. J Infect Dis (1989) 160(5):903–4. doi:10.1093/infdis/160.5.903
24. Okamoto H, Imai M, Kametani M, Nakamura T, Mayumi M. Genomic heterogeneity of hepatitis B virus in a 54-year-old woman who contracted the infection through materno-fetal transmission. Jpn J Exp Med (1987) 57(4):231–6.
25. Gerken G, Kremsdorf D, Capel F, Petit MA, Dauguet C, Manns MP, et al. Hepatitis B defective virus with rearrangements in the preS gene during chronic HBV infection. Virology (1991) 183(2):555–65. doi:10.1016/0042-6822(91)90984-J
26. Huang X, Qin Y, Zhang P, Tang G, Shi Q, Xu J, et al. PreS deletion mutations of hepatitis B virus in chronically infected patients with simultaneous seropositivity for hepatitis-B surface antigen and anti-HBS antibodies. J Med Virol (2010) 82(1):23–31. doi:10.1002/jmv.21669
27. Zhang JM, Xu Y, Wang XY, Yin YK, Wu XH, Weng XH, et al. Coexistence of hepatitis B surface antigen (HBsAg) and heterologous subtype-specific antibodies to HBsAg among patients with chronic hepatitis B virus infection. Clin Infect Dis (2007) 44(9):1161–9. doi:10.1086/513200
28. Tabor E. Infections by hepatitis B surface antigen gene mutants in Europe and North America. J Med Virol (2006) 78(Suppl 1):S43–7. doi:10.1002/jmv.20606
29. Moraleda G, Barcena R, Del Campo S, Oton E, Moreno J, Foruni JR. De novo HBV infection caused by an anti-HBc positive donor in a vaccinated liver transplant recipient in spite of anti-HBs response. Am J Transplant (2006) 6(2):438–40. doi:10.1111/j.1600-6143.2005.01189.x
30. Ghany MG, Ayola B, Villamil FG, Gish RG, Rojter S, Vierling JM, et al. Hepatitis B virus S mutants in liver transplant recipients who were reinfected despite hepatitis B immune globulin prophylaxis. Hepatology (1998) 27(1):213–22. doi:10.1002/hep.510270133
31. Foy MC, Thio CL, Hwang HS, Saulynas M, Hamilton JP, Fine DM, et al. False-negative hepatitis B virus (HBV) surface antigen in a vaccinated dialysis patient with a high level of HBV DNA in the United States. Clin Vaccine Immunol (2012) 19(5):820–2. doi:10.1128/CVI.05696-11
32. Nainan OV, Khristova ML, Byun K, Xia G, Taylor PE, Stevens CE, et al. Genetic variation of hepatitis B surface antigen coding region among infants with chronic hepatitis B virus infection. J Med Virol (2002) 68(3):319–27. doi:10.1002/jmv.10206
33. Sureau C, Salisse J. A conformational heparan sulfate binding site essential to infectivity overlaps with the conserved hepatitis B virus a-determinant. Hepatology (2013) 57(3):985–94. doi:10.1002/hep.26125
34. Su IJ, Wang LH, Hsieh WC, Wu HC, Teng CF, Tsai HW, et al. The emerging role of hepatitis B virus pre-S2 deletion mutant proteins in HBV tumorigenesis. J Biomed Sci (2014) 21:98. doi:10.1186/s12929-014-0098-7
35. Huang HP, Hsu HY, Chen CL, Ni YH, Wang HY, Tsuei DJ, et al. Pre-S2 deletions of hepatitis B virus and hepatocellular carcinoma in children. Pediatr Res (2010) 67(1):90–4. doi:10.1203/PDR.0b013e3181c1b0b7
36. Abe K, Thung SN, Wu HC, Tran TT, Le Hoang P, Truong KD, et al. Pre-S2 deletion mutants of hepatitis B virus could have an important role in hepatocarcinogenesis in Asian children. Cancer Sci (2009) 100(12):2249–54. doi:10.1111/j.1349-7006.2009.01309.x
37. Caselmann WH, Meyer M, Kekulé AS, Lauer U, Hofschneider PH, Koshy R. A trans-activator function is generated by integration of hepatitis B virus preS/S sequences in human hepatocellular carcinoma DNA. Proc Natl Acad Sci U S A (1990) 87(8):2970–4. doi:10.1073/pnas.87.8.2970
38. Kekulé AS, Lauer U, Meyer M, Caselmann WH, Hofschneider PH, Koshy R. The preS2/S region of integrated hepatitis B virus DNA encodes a transcriptional transactivator. Nature (1990) 343(6257):457–61. doi:10.1038/343457a0
39. Li X, Qin Y, Liu Y, Li F, Liao H, Lu S, et al. PreS deletion profiles of hepatitis B virus (HBV) are associated with clinical presentations of chronic HBV infection. J Clin Virol (2016) 82:27–32. doi:10.1016/j.jcv.2016.06.018
40. Wang HC, Huang W, Lai MD, Su IJ. Hepatitis B virus pre-S mutants, endoplasmic reticulum stress and hepatocarcinogenesis. Cancer Sci (2006) 97(8):683–8. doi:10.1111/j.1349-7006.2006.00235.x
41. Winckler K, Junge U, Creutzfeldt W. Ground-glass hepatocytes in unselected liver biopsies. Ultrastructure and relationship to hepatitis B surface antigen. Scand J Gastroenterol (1976) 11(2):167–70.
42. Mathai AM, Alexander J, Kuo FY, Torbenson M, Swanson PE, Yeh MM. Type II ground-glass hepatocytes as a marker of hepatocellular carcinoma in chronic hepatitis B. Hum Pathol (2013) 44(8):1665–71. doi:10.1016/j.humpath.2013.01.020
43. Tai PC, Suk FM, Gerlich WH, Neurath AR, Shih C. Hypermodification and immune escape of an internally deleted middle-envelope (M) protein of frequent and predominant hepatitis B virus variants. Virology (2002) 292(1):44–58. doi:10.1006/viro.2001.1239
44. Kim H, Lee SA, Kim DW, Lee SH, Kim BJ. Naturally occurring mutations in large surface genes related to occult infection of hepatitis B virus genotype C. PLoS One (2013) 8(1):e54486. doi:10.1371/journal.pone.0054486
45. Thibault V, Servant-Delmas A, Ly TD, Roque-Afonso AM, Laperche S. Performance of HBsAg quantification assays for detection of hepatitis B virus genotypes and diagnostic escape-variants in clinical samples. J Clin Virol (2017) 89:14–21. doi:10.1016/j.jcv.2017.02.001
46. Servant-Delmas A, Mercier-Darty M, Ly TD, Wind F, Alloui C, Sureau C, et al. Variable capacity of 13 hepatitis B virus surface antigen assays for the detection of HBsAg mutants in blood samples. J Clin Virol (2012) 53(4):338–45. doi:10.1016/j.jcv.2012.01.003
47. Velay A, Jeulin H, Eschlimann M, Malvé B, Goehringer F, Bensenane M, et al. Characterization of hepatitis B virus surface antigen variability and impact on HBs antigen clearance under nucleos(t)ide analogue therapy. J Viral Hepat (2016) 23(5):387–98. doi:10.1111/jvh.12498
Keywords: hepatitis B virus, preS2 deletion, surface antibody escape mutant, HBV vaccine escape mutant, HCV
Citation: Fusco DN, Ganova-Raeva L, Khudyakov Y, Punkova L, Mohamed A, Cheon SSY, Koirala P, Andersson KL, Jourdain G, Sureau C, Chung RT and Lauer G (2018) Reactivation of a Vaccine Escape Hepatitis B Virus Mutant in a Cambodian Patient During Anti-Hepatitis C Virus Therapy. Front. Med. 5:97. doi: 10.3389/fmed.2018.00097
Received: 04 December 2017; Accepted: 26 March 2018;
Published: 30 April 2018
Edited by:
Anne-Mieke Vandamme, KU Leuven, BelgiumReviewed by:
Ana Afonso, Universidade de São Paulo, BrazilAleksandra Barac, University of Belgrade, Serbia
Stephen Locarnini, Victorian Infectious Diseases Reference Laboratory, Australia
Copyright: © 2018 Fusco, Ganova-Raeva, Khudyakov, Punkova, Mohamed, Cheon, Koirala, Andersson, Jourdain, Sureau, Chung and Lauer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dahlene N. Fusco, dnfusco@mgh.harvard.edu
 Dahlene N. Fusco
Dahlene N. Fusco Lilia Ganova-Raeva3
Lilia Ganova-Raeva3
 Scarlett Se Yun Cheon
Scarlett Se Yun Cheon Gonzague Jourdain
Gonzague Jourdain Camille Sureau
Camille Sureau Georg Lauer
Georg Lauer