Alkaline Peptone Water-Based Enrichment Method for mcr-3 From Acute Diarrheic Outpatient Gut Samples
- 1Department of Clinical Laboratory, Second Affiliated Hospital of Zhejiang University, School of Medicine, Hangzhou, China
- 2University of Connecticut, Storrs, CT, United States
- 3The Affiliated High School of Hangzhou Normal University, Hangzhou, China
A third plasmid-mediated colistin resistance gene, mcr-3, is increasingly being reported in Enterobacteriaceae and Aeromonas spp. from animals and humans. To investigate the molecular epidemiology of mcr in the gut flora of Chinese outpatients, 152 stool specimens were randomly collected from outpatients in our hospital from May to June, 2017. Stool specimens enriched in alkaline peptone water or Luria-Bertani (LB) broth were screened for mcr-1, mcr-2, and mcr-3 using polymerase chain reaction (PCR)-based assays. Overall, 19.1% (29/152) and 5.3% (8/152) of the stool samples enriched in alkaline peptone water were PCR-positive for mcr-1 and mcr-3, respectively, while 2.7% (4/152) of samples were positive for both mcr-1 and mcr-3. Strains isolated from the samples that were both mcr-1- and mcr-3-positive were subjected to antimicrobial susceptibility testing by broth microdilution. They were also screened for the presence of other resistance genes by PCR, while multilocus sequence typing and whole-genome sequencing were used to investigate the molecular epidemiology and genetic environment, respectively, of the resistance genes. mcr-3-positive Aeromonas veronii strain 126-14, containing a mcr-3. 8-mcr-3-like2 segment, and mcr-1-positive Escherichia coli strain 126-1, belonging to sequence type 1485, were isolated from the sample from a diarrheic butcher with no history of colistin treatment. A. veronii 126-14 had a colistin minimum inhibitory concentration (MIC) of 2 µg/mL and was susceptible to antibiotics in common use, while E. coli 126-1 produced TEM-1, CTX-M-55, and CTX-M-14 β-lactamases and was resistant to colistin, ceftazidime, and cefotaxime. Overall, there was a higher detection rate of mcr-3-carrying strains with low colistin MICs from the samples enriched in alkaline peptone water than from samples grown in LB broth.
Introduction
Since the identification of a third plasmid-mediated colistin resistance gene, mcr-3, in a porcine Escherichia coli isolate from China in 2017 (1), several mcr-3 variants have been detected in clinical E. coli and Salmonella isolates from Denmark, Spain, and China (2–5). The amino acid sequence of MCR-3 is highly similar to that of phosphoethanolamine transferases from various Aeromonas and Enterobacteriaceae species (1). Ling et al. reported that chromosomally located mcr-3 variants, including mcr-3.3 and mcr-3-like, which were identified in Aeromonas veronii from chicken meat, showed 95.2 and 84.2% nucleotide sequence identity, respectively, to mcr-3 from E. coli of porcine origin (6). Interestingly, the reported minimum inhibitory concentration (MIC) of colistin for the mcr-3-carrying A. veronii isolate from chicken meat was 2 µg/mL while colistin MICs for the mcr-3-positive Enterobacteriaceae were in the range of 4–8 µg/mL. Thus, mcr-3-positive Aeromonas spp. strains are likely to go undetected by routine clinical tests. Our previous studies have established an optimized enrichment method for the screening of mcr-1 from human gut and environmental water sources (7, 8), in which the mcr-1-carrying strains demonstrated MICs for colistin of 1–32 µg/mL. As Aeromonas spp. generally prefer an alkaline pH, we improved the enrichment method using alkaline peptone water. In this study, we used the newly developed enrichment method to investigate the epidemiology of mcr in the gut flora of outpatients treated in our hospital.
Materials and Methods
Stool Specimens and Microbial Enrichment
A total of 152 stool specimens were randomly collected from outpatients suffering from acute diarrhea at the Intestinal Clinic of the Second Affiliated Hospital of the Zhejiang University School of Medicine from May to June 2017. Aliquots (~1 g) of each stool sample were individually inoculated into 5 mL of alkaline peptone water (Binhe, Hangzhou, China) and 5 mL of Luria-Bertani (LB) broth for enrichment overnight at 35°C. The alkaline peptone water was adjusted to a pH of 8.4–9.2 and contained 15.0 g/L of tryptone, 4.0 g/L of beef extract, and 10.0 g/L of NaCl.
Detection of mcr-Positive Isolates by Enrichment Culture
Following incubation, each enrichment culture tube was inverted 10 times to resuspend the cells and a 1-mL aliquot of suspension was transferred to a fresh 1.5-mL tube. The suspension was centrifuged for 3 min at 8,000 rpm, after which the supernatant was discarded and 1 mL of 0.9% (w/v) saline was added to wash and resuspend the cell pellet. The centrifugation step was then repeated, and 70 µL of ultra-pure water was added to the pellet, which was then boiled for 5 min. Following centrifugation, a 3-µL aliquot of the supernatant was used as template for polymerase chain reaction (PCR) amplification of mcr-1, mcr-2, and mcr-3 as described previously (1, 9, 10).
Following initial PCR-based screening, four of the alkaline peptone water enrichment cultures tested positive for both mcr-1 and mcr-3 and were therefore selected for colony isolation. A 10-µL aliquot of suspension from the enrichment cultures was inoculated onto Salmonella–Shigella agar plates and incubated at 37°C overnight. Resultant colonies were selected for further purification and confirmation of the presence of the mcr genes using the PCR-based method described above. Final identification of the mcr-positive colonies was performed by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (Bruker Daltonik GmbH, Bremen, Germany) analysis. As this method cannot distinguish between E. coli and Shigella spp., Kligler Iron Agar and Motility-indole-Urea medium were added to help identify E. coli strains. An mcr-3-positive A. veronii isolate and a mcr-1-positive E. coli isolate were identified from one of the four mcr-1- and mcr-3-positive alkaline peptone water enrichment cultures, and were tested for antimicrobial susceptibility and screened for the presence of other common β-lactamase-encoding genes using further PCR.
Antimicrobial Susceptibility Testing
The MICs of eight antibiotics against mcr-positive isolates were determined using a broth microdilution procedure. The susceptibilities of each of the isolates to meropenem, ceftazidime, cefotaxime, cefoperazone–sulbactam, amikacin, and ciprofloxacin were determined according to the Clinical and Laboratory Standards Institute guidelines (11). The breakpoints for colistin and tigecycline against E. coli were obtained from the European Committee on Antimicrobial Susceptibility Testing breakpoint tables (12). E. coli ATCC25922 was used as a quality control strain for broth microdilution assays.
Detection of Other Common β-Lactamase-Encoding Genes
Additional β-lactamase-encoding genes, including blaTEM, blaSHV, blaCTX-M-1-group, and blaCTX-M-9-group were detected by PCR using previously described primers and conditions (13).
Multilocus Sequence Typing (MLST)
Molecular typing of the mcr-1-positive E. coli isolate was performed by MLST using conditions and primers described on the MLST website (http://mlst.warwick.ac.uk/mlst/dbs/Ecoli). The sequences of the seven housekeeping genes were compared with those in the E. coli MLST database.
Whole-Genome Sequencing
The selected mcr-3-positive A. veronii isolate was submitted for 300-bp paired-end whole-genome sequencing using the Illumina Hiseq 2500 system (Annoroad, Beijing, China). The raw Illumina reads were assembled into a draft genome sequence using CLC Genomics Workbench 9.0 (CLC Bio, Aarhus, Denmark). Antibiotic resistance genes were analyzed using SRST2 (14), with reference sequences for the antibiotic resistance genes obtained from the ARG-ANNOT database (15).
Results
Detection of mcr-Positive Isolates Following Enrichment Culture
Following enrichment in alkaline peptone water, 19.1% (29/152) and 5.3% (8/152) of the stool samples were PCR-positive for mcr-1 and mcr-3, respectively, while 18.4% (28/152) of the samples enriched in LB broth were positive for mcr-1. None of the LB enrichment samples tested positive for mcr-3 (Table 1), and none of the samples from either enrichment method were positive for mcr-2.
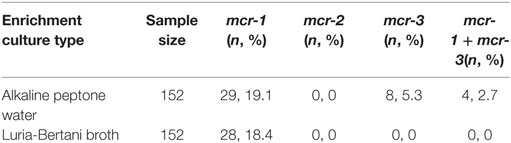
Table 1. Initial polymerase chain reaction screening results for the presence of mcr genes in enrichment stool cultures.
An mcr-3-positive A. veronii isolate (strain 126-14) and a mcr-1-positive E. coli isolate (strain 126-1) were simultaneously isolated from the same alkaline peptone water-enriched stool sample. The sample was collected from a 42-year-old male pork butcher with no medication history of colistin. He was admitted to the gastroenterology clinic for 2 days suffering from acute abdominal pain and diarrhea following ingestion of watermelon. He developed a fever (38.4°C), and stool analysis showed the yellow loose stool did not contain any leukocytes or erythrocytes. Levofloxacin and viable Lactobacillus acidophilus tablets were administrated, and the patient attained complete remission.
Antimicrobial Susceptibility Testing
Identified as sequence type 1485 by MLST, mcr-1-positive E. coli isolate 126-1 showed resistance to colistin, ceftazidime, and cefotaxime, and additional PCR analyses confirmed the co-existence of blaTEM-1, blaCTX-M-55, and blaCTX-M-14 in this strain. mcr-3-positive A. veronii isolate 126-14 was susceptible to all tested antibiotics, and had MICs for colistin and tigecycline of 2 and 1 µg/mL, respectively (Table 2).
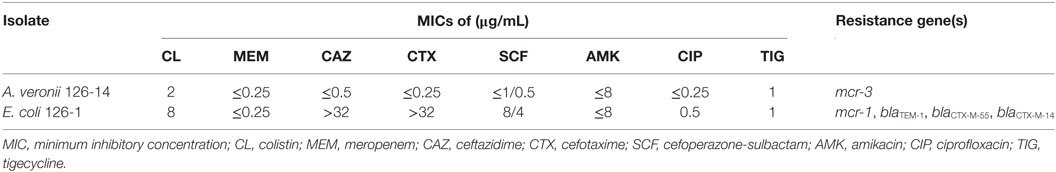
Table 2. MICs and resistance gene profiles of mcr-3-positive Aeromonas veronii 126-14 and mcr-1-positive Escherichia coli 126-1.
Whole-Genome Sequencing
Whole-genome sequencing of mcr-3 positive A. veronii isolate 126-14 produced 146 contigs. Two adjacent mcr-3 variants, the novel upstream variant termed mcr-3.8 and the downstream variant termed mcr-3-like2, were located on 5,338-bp contig 85 and were separated by only 66 bp. The sequences of these two variants have been deposited in GenBank under accession no. PPTE01000085.1. Both variants showed >99.0% nucleotide and amino acid sequence identity to the mcr-3.3 and mcr-3-like genes in A. veronii isolated from chicken meat, and 95.9 and 87.2% nucleotide sequence identity and 95.8 and 84.8% amino acid sequence identity, respectively, to the original mcr-3 gene (Figures 1 and 2). Similar to the mcr-3.3-mcr-3-like segment in a previously identified A. veronii isolate (GenBank accession no. MF495680), the mcr-3.8-mcr-3-like2 segment in A. veronii 126-14 was surrounded by both hypothetical genes and diacylglycerol kinase alpha-encoding gene dgkA but lacked transfer elements (Figure 1).
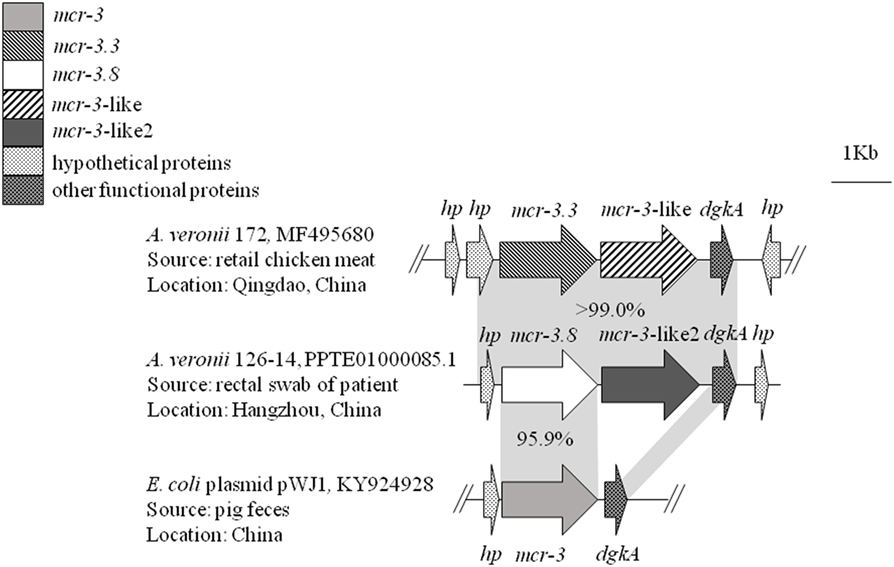
Figure 1. The genetic environment of the mcr-3.8-mcr-3-like2 segment in the Aeromonas veronii isolate identified in this study. Arrows represent the directions of the genes. Gray shading indicates two areas with significant similarity.
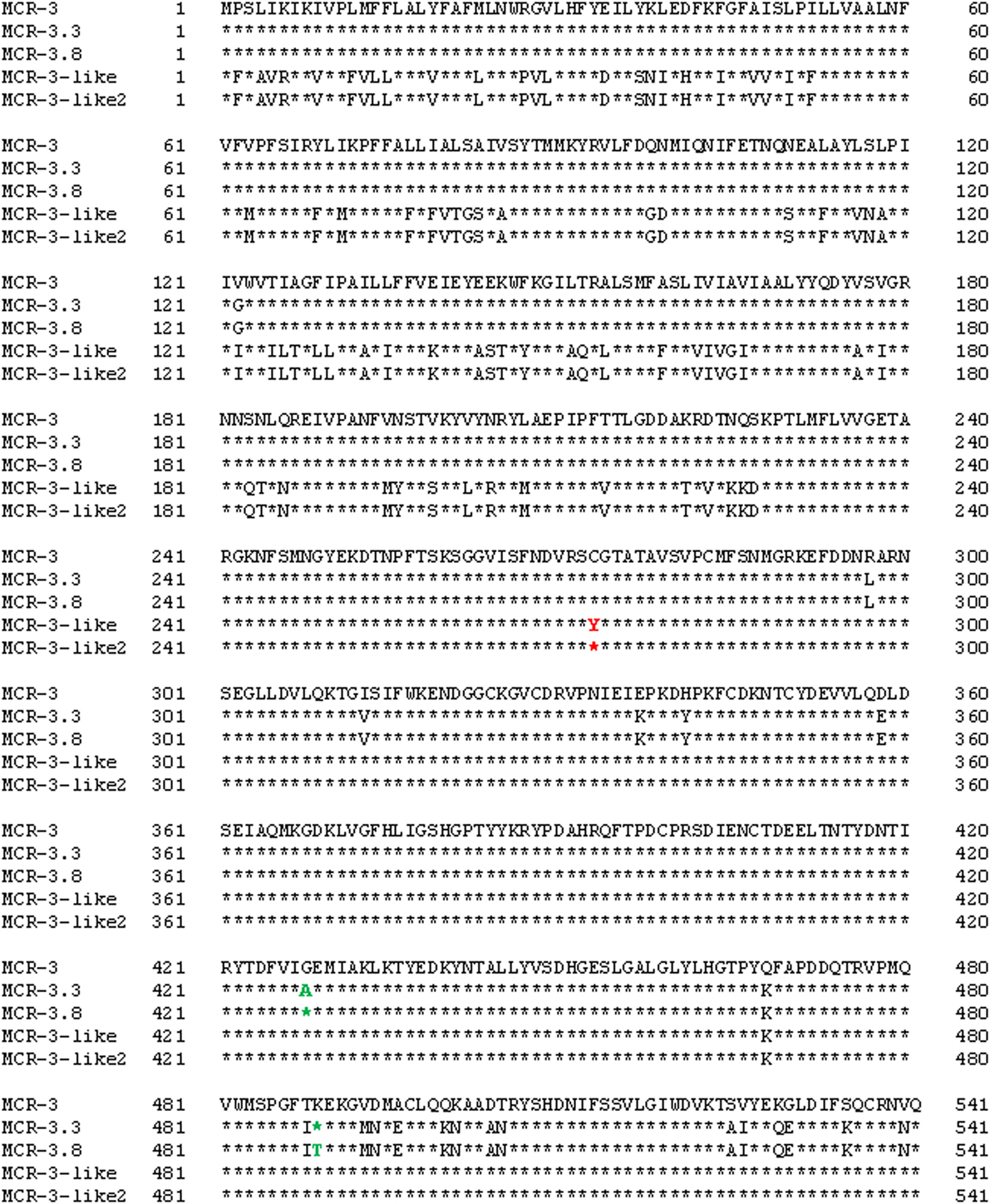
Figure 2. Alignment of the MCR-3, MCR-3.3, MCR-3-like, MCR-3.8, and MCR-3-like2 sequences from Escherichia coli (GenBank accession no. KY924928) and Aeromonas isolates (GenBank accession no. MF495680 and PPTE01000085.1).
Discussion
To the best of our knowledge, this study is the first report of the co-occurrence of a mcr-3-positive A. veronii isolate and a mcr-1-positive E. coli isolate from the gut of a diarrheic outpatient. Gram-negative Aeromonas spp. cause various infections in both humans and animals, with Aeromonas-associated diarrhea and gastroenteritis the most frequent manifestations of infection in humans. Aeromonas infections affect all age groups, including both healthy and immunocompromised individuals (16). The transferable colistin resistance gene mcr-1 has been reported in Enterobacteriaceae isolated both from food-producing animals and humans, with carriage rates of 5.1 and 6.2%, respectively (17, 18). These carriage rates indicate that the emergence and spread of mcr-1 probably occurred first in isolates of animal origin, which then spread to humans. In this study, the mcr-3-positive A. veronii strain isolated from the fecal sample might have been the cause of the acute diarrhea. The patient was employed as a pork butcher and had no history of colistin use, which indicated that the mcr-3 and the mcr-1 genes likely originated from isolates of food–animal origin. This suggests that foodstuffs of farm animal origin may act as a critical transmission vehicle in the dissemination of mobile colistin resistance genes from animal-associated with human-associated bacteria.
In this study, 19.1% (29/152) and 5.3% (8/152) of the stool samples enriched in alkaline peptone water were PCR-positive for mcr-1 and mcr-3, respectively, while only 18.4% (28/152) of the LB broth-enriched samples tested positive for mcr-1. This suggests that the alkaline peptone water enrichment step before direct sample testing resulted in higher sensitivity of the mcr screening from human stool samples compared with LB broth cultivation. In addition, the detection rate of mcr-1 in this study (approximately 19%) was much higher than that reported previously (about 6%) (18), probably due to the enrichment step and direct enriched sample testing by PCR. Furthermore, mcr-3-positive A. veronii isolate 126-14 had a colistin MIC of 2 µg/mL, which was in agreement with the previously reported colistin MIC for a mcr-3-carrying A. veronii isolate from chicken meat (6). Therefore, these results suggest that unsupplemented Salmonella–Shigella agar rather than medium supplemented with colistin is better for selection of mcr-3-carrying Aeromonas spp. strains. Overall, only one mcr-3-positive A. veronii isolate was recovered from the four PCR-positive alkaline peptone water-enriched stool samples. Although Aeromonas spp. are widely distributed in soil (19), foodstuffs (20), and aquatic environments (21), they usually constitute a small percentage of the human gut flora (22). As such, the number of Aeromonas spp. present in an enrichment culture may still be too low to detect via direct culture plating methods.
In conclusion, the alkaline peptone water enrichment method was optimal for detection of mcr-3-carrying strains with low colistin MICs from the human gut microbiota. The method is simple to perform and can be used in any laboratory that is equipped to perform PCR assays and can obtain alkaline peptone water at the proper pH. In addition, as the human intestine may serve as a reservoir for antibiotic resistance genes, including mcr-3, and play an important role in horizontal gene transfer, the rapid horizontal spread of mobile colistin resistance genes between and within Enterobacteriaceae and Aeromonas spp. in the human gut should be closely monitored.
Ethics Statement
Written informed consent was obtained from the patient for the publication of this research. The Ethics Committee of the Second Affiliated Hospital of Zhejiang University, School of Medicine, already approved this work.
Author Contributions
RZ and HZ designed and supervised the study. HW participated in stool sample collecting. QS, LS, HW, and ZH conducted the microbial enrichment, PCR, antimicrobial susceptibility testing, and strain characterization. YH analyzed the whole-genome sequencing. RZ, QS, YH, HZ, and LS contributed to the data interpretation and manuscript writing. All the authors have approved the final version and have agreed to be accountable for all aspects of the work.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding
The study was supported by grants from the National Natural Science Foundation of China (81661138002 and 81501805).
References
1. Yin WJ, Li H, Shen YB, Liu ZH, Wang SL, Shen ZQ, et al. Novel plasmid-mediated colistin resistance gene mcr-3 in Escherichia coli. mBio (2017) 8(3):e543–517. doi:10.1128/mBio.00543-17
2. Roer L, Hansen F, Stegger M, Sönksen UW, Hasman H, Hammerum AM. Novel mcr-3 variant, encoding mobile colistin resistance, in an ST131 Escherichia coli isolate from bloodstream infection, Denmark, 2014. Euro Surveill (2017) 22(31):30584. doi:10.2807/1560-7917.ES.2017.22.31.30584
3. Hernández M, Iglesias MR, Rodríguez-Lázaro D, Gallardo A, Quijada NM, Miguela-Villoldo P, et al. Co-occurrence of colistin-resistance genes mcr-1 and mcr-3 among multidrug-resistant Escherichia coli isolated from cattle, Spain, September 2015. Euro Surveill (2017) 22(31):30586. doi:10.2807/1560-7917.ES.2017.22.31.30586
4. Litrup E, Kiil K, Hammerum AM, Roer L, Nielsen EM, Torpdahl M. Plasmid-borne colistin resistance gene mcr-3 in salmonella isolates from human infections, Denmark, 2009–17. Euro Surveill (2017) 22(31):30587. doi:10.2807/1560-7917.ES.2017.22.31.30587
5. Liu L, Feng Y, Zhang XX, McNally A, Zong ZY. A new variant of mcr-3 in an extensively drug-resistant Escherichia coli clinical isolate carrying mcr-1 and blaNDM-5. Antimicrob Agents Chemother (2017) 61(12):1757–1717. doi:10.1128/AAC.01757-17
6. Ling ZR, Yin WJ, Li H, Zhang QD, Wang XM, Wang Z, et al. Chromosome-mediated mcr-3 variants in Aeromonas veronii from chicken meat. Antimicrob Agents Chemother (2017) 61:1272–1217. doi:10.1128/AAC.01272-17
7. Hu YY, Wang YL, Sun QL, Huang ZX, Wang HY, Zhang R, et al. Colistin resistance gene mcr-1 in gut flora of children. Int J Antimicrob Agents (2017) 50:593–7. doi:10.1016/j.ijantimicag.2017.06.011
8. Zhou HW, Zhang T, Ma JH, Fang Y, Wang HY, Huang ZX, et al. Occurrence of plasmid- and chromosome-carried mcr-1 in waterborne Enterobacteriaceae in China. Antimicrob Agents Chemother (2017) 61:e00017–17. doi:10.1128/AAC.00017-17
9. Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis (2016) 16(2):161–8. doi:10.1016/S1473-3099(15)00424-7
10. Xavier BB, Lammens C, Ruhal R, Kumar-Singh S, Butaye P, Goossens H, et al. Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia coli, Belgium, June 2016. Euro Surveill (2016) 21(27). doi:10.2807/1560-7917.ES.2016.21.27.30280
11. Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing. M100. 27th ed. Wayne, PA: (2017). Available online at: https://clsi.org/standards/products/microbiology/documents/m100/ (Accessed: July, 2017).
12. European Committee on Antimicrobial Susceptibility Testing (EUCAST). Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 7.1. Växjö (2017). Available online at: http://www.eucast.org/clinical_breakpoints/ (Accessed: July, 2017).
13. Yu YS, Ji SJ, Chen YG, Zhou WL, Wei ZQ, Li LJ, et al. Resistance of strains producing extended-spectrum beta-lactamases and genotype distribution in China. J Infect (2007) 54(1):53–7. doi:10.1016/j.jinf.2006.01.014
14. Inouye M, Dashnow H, Raven LA, Schultz MB, Pope BJ, Tomita T, et al. SRST2: rapid genomic surveillance for public health and hospital microbiology labs. Genome Med (2014) 6(11):90. doi:10.1186/s13073-014-0090-6
15. Gupta SK, Padmanabhan BR, Diene SM, Lopez-Rojas R, Kempf M, Landraud L, et al. ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob Agents Chemother (2014) 58(1):212–20. doi:10.1128/AAC.01310-13
16. Silva LCAD, Leal-Balbino TC, Melo BST, Mendes-Marques CL, Rezende AM, Almeida AMP, et al. Genetic diversity and virulence potential of clinical and environmental Aeromonas spp. isolates from a diarrhea outbreak. BMC Microbiol (2017) 17(1):179. doi:10.1186/s12866-017-1089-0
17. Yang YQ, Li YX, Song T, Yang YX, Jiang W, Zhang AY, et al. Colistin resistance gene mcr-1 and its variant in Escherichia coli isolates from chickens in China. Antimicrob Agents Chemother (2017) 61(5):e1204–16. doi:10.1128/AAC.01204-16
18. Zhong LL, Phan HTT, Shen C, Doris-Vihta K, Sheppard AE, Huang X, et al. High rates of human fecal carriage of mcr-1-positive multi-drug resistant Enterobacteriaceae isolates emerge in China in association with successful plasmid families. Clin Infect Dis (2018) 66(5):676–85. doi:10.1093/cid/cix885
19. Goswami G, Deka P, Das P, Bora SS, Samanta R, Boro RC, et al. Diversity and functional properties of acid-tolerant bacteria isolated from tea plantation soil of Assam. 3 Biotech (2017) 7(3):229. doi:10.1007/s13205-017-0864-9
20. Gowda TK, Reddy VR, Devleesschauwer B, Zade NN, Chaudhari SP, Khan WA, et al. Isolation and seroprevalence of Aeromonas spp. Among common food animals slaughtered in Nagpur, Central India. Foodborne Pathog Dis (2015) 12(7):626–30. doi:10.1089/fpd.2014.1922
21. Chenia HY, Duma S. Characterization of virulence, cell surface characteristics and biofilm-forming ability of Aeromonas spp. isolates from fish and sea water. J Fish Dis (2017) 40(3):339–50. doi:10.1111/jfd.12516
Keywords: mcr-3, Aeromonas veronii, diarrheic outpatient, alkaline peptone water, enrichment method
Citation: Sun Q, Hu Y, Zhou H, Shu L, Wang H, Huang Z and Zhang R (2018) Alkaline Peptone Water-Based Enrichment Method for mcr-3 From Acute Diarrheic Outpatient Gut Samples. Front. Med. 5:99. doi: 10.3389/fmed.2018.00099
Received: 29 November 2017; Accepted: 26 March 2018;
Published: 03 May 2018
Edited by:
Margaret Ip, The Chinese University of Hong Kong, ChinaReviewed by:
Ákos Tóth, National Public Health Institute (OKI), HungaryAna Afonso, Universidade de São Paulo, Brazil
Copyright: © 2018 Sun, Hu, Zhou, Shu, Wang, Huang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rong Zhang, zhang-rong@zju.edu.cn
 Qiaoling Sun1
Qiaoling Sun1
 Rong Zhang
Rong Zhang