Factors Determining the Susceptibility of Bacteria to Antibacterial Photodynamic Inactivation
- 1Department of Pharmaceutical Microbiology, Faculty of Pharmacy Medical University of Gdańsk, Gdańsk, Poland
- 2Laboratory of Molecular Diagnostics, Intercollegiate Faculty of Biotechnology, University of Gdańsk and Medical University of Gdańsk, Gdańsk, Poland
Photodynamic inactivation of microorganisms (aPDI) is an excellent method to destroy antibiotic-resistant microbial isolates. The use of an exogenous photosensitizer or irradiation of microbial cells already equipped with endogenous photosensitizers makes aPDI a convenient tool for treating the infections whenever technical light delivery is possible. Currently, aPDI research carried out on a vast repertoire of depending on the photosensitizer used, the target microorganism, and the light delivery system shows efficacy mostly on in vitro models. The search for mechanisms underlying different responses to photodynamic inactivation of microorganisms is an essential issue in aPDI because one niche (e.g., infection site in a human body) may have bacterial subpopulations that will exhibit different susceptibility. Rapidly growing bacteria are probably more susceptible to aPDI than persister cells. Some subpopulations can produce more antioxidant enzymes or have better performance due to efficient efflux pumps. The ultimate goal was and still is to identify and characterize molecular features that drive the efficacy of antimicrobial photodynamic inactivation. To this end, we examined several genetic and biochemical characteristics, including the presence of individual genetic elements, protein activity, cell membrane content and its physical properties, the localization of the photosensitizer, with the result that some of them are important and others do not appear to play a crucial role in the process of aPDI. In the review, we would like to provide an overview of the factors studied so far in our group and others that contributed to the aPDI process at the cellular level. We want to challenge the question, is there a general pattern of molecular characterization of aPDI effectiveness? Or is it more likely that a photosensitizer-specific pattern of molecular characteristics of aPDI efficacy will occur?
Introduction
Antibacterial photodynamic inactivation (aPDI) is a therapeutic option used in the treatment of infectious diseases. It is based on a combination of a photosensitizer (PS), light and oxygen to remove highly metabolically active cells. These cells may be microorganisms, such as fungi (1), viruses (2), or bacteria (3). The main element in aPDI is a triplet excited photosensitizer, whose action can lead to the formation of singlet oxygen and radicals, known as reactive oxygen species (ROS) (4). These reactive species cause damage to biological molecules, which promotes cell death, the desired effect of aPDI.
In comparison with other methods of treatment, aPDI has several advantages. Photoactivation allows local treatment, which reduces the side effects of photodynamic therapy. In addition, aPDI has several cellular targets, and therefore, it is not considered to lead to the development of resistance to the treatment. However, despite these advantages and increasing knowledge about the effectiveness of photodynamic inactivation, the clinical application of aPDI is still not widespread and mainly concerns anticancer applications (5). Thus, researching the molecular mechanisms involved in the photoinactivation of microorganisms warrants serious attention. Many researchers have focused exclusively on finding new photosensitizers or modifying existing ones to maximize the quantum yield of singlet oxygen in in vitro tests, while ignoring molecular aspects within microbial cells and the factors that affect differences in the susceptibility of microbes to photoinactivation. Notably, the essential biological targets in photodynamic reactions for achieving effective eradication of microorganisms are still unclear, as are the genetic or phenotypic features of microorganisms that determine their response to photoinactivation (6). In our opinion, understanding the mechanism and phenomena occurring in microbial cells in response to photodynamic reactions is indispensable for increasing their effective clinical use. The primary motivation for writing this review was to identify the molecular phenomena occurring in the cells of microorganisms during and after photosensitization that may influence the effectiveness of photodynamic inactivation and to identify the elements that determine the existence of microbial phenotypes with differences in vulnerability to aPDI.
Photodynamic inactivation includes aPDI employing exogenously administered photosensitizers (PSs) and antimicrobial blue light (aBL), leading to the excitation of endogenously produced photosensitizing compounds, i.e., intracellular porphyrins and flavins.
There are two types of reactions that occur in aPDI (Figure 1); a type I reaction, which generates ROS such as superoxide (·), hydroxyl radical (·OH), and hydrogen peroxide (H2O2), or a type II reaction, in which mainly singlet oxygen (1) is produced. Type I and type II reaction products can be produced simultaneously in PDI, with the proportion of each being dependent on the type of photosensitizer used and the ionic strength of its solvent (7). Recently, a type III reaction has been added to the well-known mechanisms of type I and II, in which, regardless of the presence of oxygen, free radicals of inorganic compounds are formed, which can also be involved in the photoinactivation of microorganisms (8).
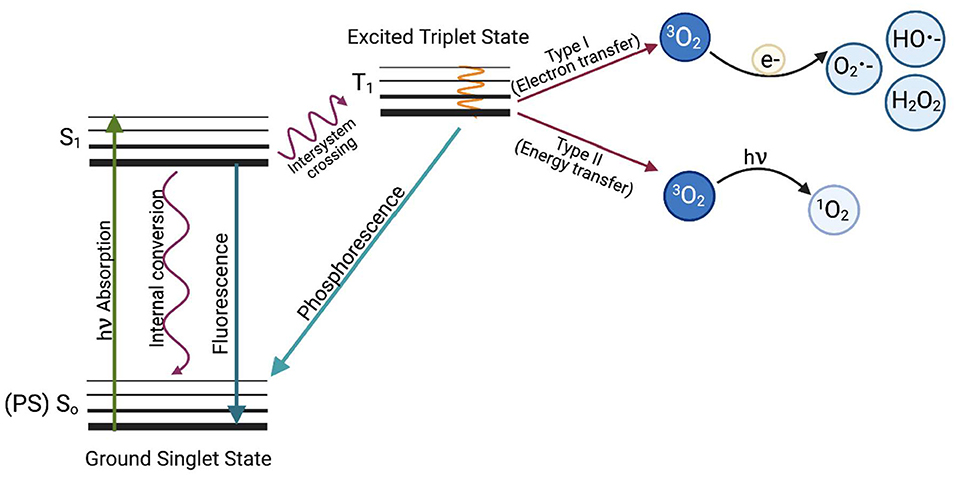
Figure 1. The Jabłoński diagram illustrates the mechanism of photodynamic inactivation. aPDI can be divided into two types of reactions. The type I (electron transfer) reaction occurs when a photon is absorbed by the PS, which causes PS excitation and transition from the ground PS singlet state (So) to the excited PS singlet state (S1) and later to the excited triplet state via the inter-system crossing (T1). When excited, the photosensitizer can transfer an electron (e−) to molecular oxygen (3O2) to produce oxygen radicals and other reactive oxygen species such as superoxide anion (O2 •−), hydroxyl radical (HO•−) or hydrogen peroxide (H2O2) or can react with biomolecules such as membrane lipids. The first stage of the Type II reaction (energy transfer) is similar, but the photosensitizer in the triplet excited state (T1) transfers energy directly to molecular oxygen. This produces highly reactive singlet oxygen (1O2). Based on Oruba and Chomyszyn-Gajewska (9).
ROS generated during aPDI and aBL target several biomolecules within a cell (Figure 2): membrane lipids, proteins, nucleic acids, carbohydrates, and thiols. Inside a cell (Figure 3), proteins seem to be the main target, as they are the most abundant. Singlet oxygen, which is produced by most porphyrin derivatives, reacts with aromatic amino acids as well as those containing sulfur, which results in the accumulation of toxic products (10). In nucleic acids, the formation of 8-oxo,7,8-dihydro-2'-deoxyguanosine (8-oxo-G) may lead to frameshift mutations (11). ROS cause lipid peroxidation, disturbing the cell membrane integrity and energy production and transport processes (12). Cell envelopes are considered significant photoinactivation targets, which may be supported by the fact that, in general, gram-positive [G (+)] bacteria display higher susceptibility to photoinactivation than gram-negative [G (–)] bacteria. This probably results from differences in the structure of cell envelopes between these two microbial groups (Figure 2). G (+) and G (–) bacteria have a similar cytoplasmic membrane, however, they differ significantly in peptidoglycan thickness and the presence of the outer membrane. The outer membrane in G (–) species constitutes an additional block of the physical interaction of PS with intracellular ROS-sensitive molecules. The thick but quite porous structure of the peptidoglycan in G (+) species filled with teichoic and teichuronic acids is not a very difficult barrier for PS to overcome. The situation is different in G (–) where the outer membrane containing lipopolysaccharides is tight and difficult to penetrate (Figure 2).
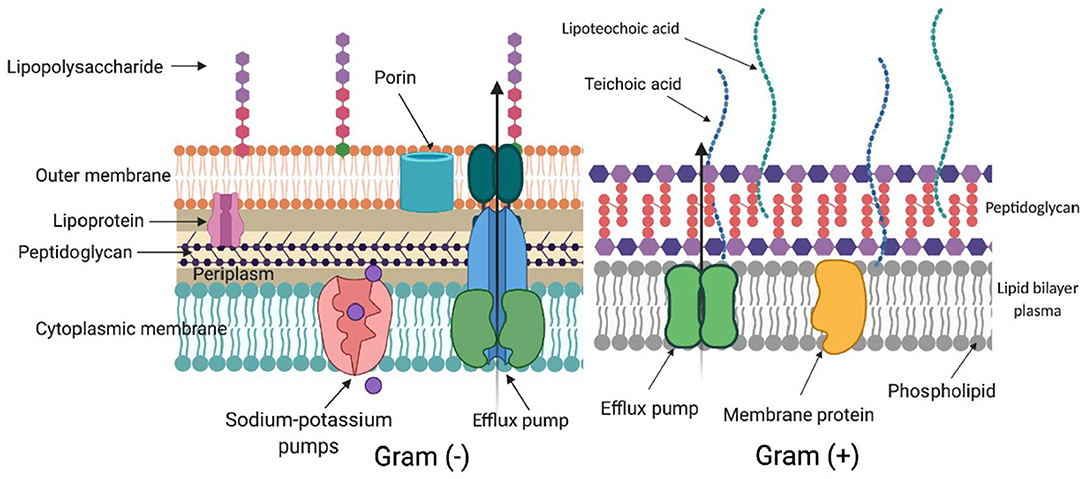
Figure 2. Differences in cell wall structure in gram-negative and gram-positive bacteria. Schematic representation of a cell wall in gram-negative [Gram (–)] and gram-positive [Gram (+)] species. The complex structure of two lipid membranes: cytoplasmic and an outer membrane are responsible for generally lower aPDI effectiveness against gram-negative bacteria. On the contrary, gram-positive species with their thick although relatively porous peptidoglycan are more easily photoinactivated. The main elements in the cell wall that can be involved in photodynamic inactivation are depicted. (Created with BioRender).
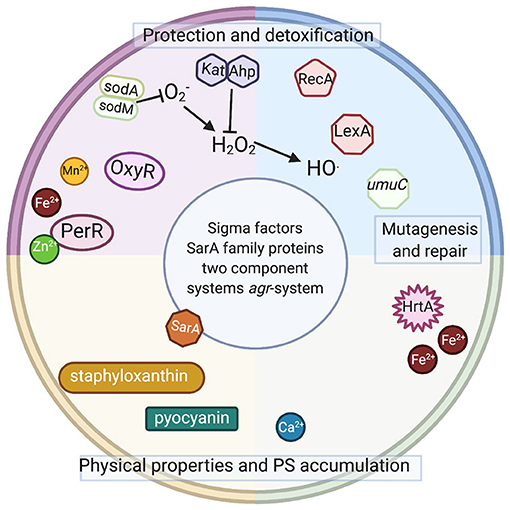
Figure 3. Intracellular elements involved or potentially involved in photodynamic inactivation. Schematic representation of intracellular elements that were studied with respect of photodynamic inactivation of bacteria. Sod, superoxide dismutase; OxyR, PerR, peroxide-responsive regulators in gram-positive and gram-negative species; Kat, catalase in gram-positive and gram-negative species; Ahp, alkyl hydroperoxide reductase in gram-positive and gram-negative species; RecA, recombinase A; LexA, SOS system repressor, umuC, gene coding for an error-prone polymerase in gram-positive and gram-negative species; HrtA, heme-regulated transporter A in S. aureus; SarA, transcriptional regulator in S. aureus; agr, virulence accessory gene regulator in S. aureus. Staphyloxanthin (membrane embedded) and pyocyanin (secreted) are dyes produced by S. aureus and P. aeruginosa. Please see the text for detailed explanations (Created with BioRender).
It should be remembered that in the case of bacterial cells it is extremely difficult to analyze individual structural elements of the cell, for example by flow cytometry techniques, where the bacterial cells are often at the limit of the resolution of the method. Obtaining information on the role or behavior of individual cell structures in the process of photoinactivation of bacteria requires the use of sophisticated techniques to which access is often limited, such as atomic force microscopy. Gram-negative species have a more complex cell envelope structure, which may explain their higher tolerance to photoinactivation (13). Despite the crucial role of the cell envelope in the bactericidal activity of photoinactivation, the actual contribution of DNA damage to the outcome of phototreatment should not be underestimated.
Phototreatment Tolerance
Due to their non-selective, multitargeting and ROS-dependent mechanisms of action, aPDI and aBL are considered unlikely to induce bacterial resistance and/or tolerance. Resistance/tolerance development has been extensively studied for both phototreatments (aPDI and aBL) over the past decade (14–29). Kashef and Hamblin reviewed many of these works in depth (30). However, the phenomenon of resistance/tolerance was not observed in any of the publications included in this review (30). Guffey et al. suggested that S. aureus may be capable of developing adaptation to blue light irradiation. Subsequent applications of blue light (405 nm) to subcultured generations of S. aureus (ATCC 25923) were increasingly useful through four cycles; however, beginning with the 5th cycle, the effectiveness of phototreatment decreased (24). In turn, in studies performed by Amin et al., Pseudomonas aeruginosa exhibited reduced susceptibility to sublethal aBL treatment after nine cycles of photoinactivation. The fraction of the surviving cells was increased by approximately two log10 units compared to the first cycle (28). Although the authors did not consider these results to reflect resistance, in our opinion, this observation could indicate possible tolerance development. Our studies indicated the development of S. aureus tolerance to RB-mediated aPDI and aBL (405 nm) when the S. aureus USA300 JE2 strain was subjected to 15 cycles of both sublethal treatments. Potential reductions in susceptibility to aPDI and aBL were examined after the 5th, 10th, and 15th consecutive cycles. The developed adaptation was stable after five cycles of subculturing without aPDI/aBL exposure. The development of aPDI/aBL tolerance was also observed in clinical MRSA and MSSA strains as well as in other representatives of gram-positive species, i.e., Enterococcus faecium and Streptococcus agalactiae. A key point of the obtained results was the lack of cross-tolerance between RB-aPDI and aPDI mediated by other PSs, i.e., [7-(dimethylamino)phenothiazin-3-ylidene]-dimethylazanium chloride (New Methylene Blue, NMB) and 5,10,15,20-tetrakis(1-methyl-4-pyridinio)porphyrin tetra(p-toluenesulfonate (TMPyP), along with a lack of cross-tolerance between aPDI and aBL. It should be highlighted that the developed aPDI/aBL tolerance cannot be considered resistance since the administration of more rigorous conditions, i.e., increased PS concentrations and higher light doses, caused bacterial eradication (20). In our most recent study, we also demonstrated the development of aPDI tolerance (21). The application of 10 cycles of sublethal aPDI resulted in significant tolerance development for all tested S. agalactiae strains, including both the reference ATCC 27956 strain and clinical isolates (2306/06 and 2974/07). The developed tolerance decreased aPDI efficacy by up to 3 log10 units in viable counts. Moreover, we observed the phenotypic stability of the developed tolerance after passaging the cells for the next five cycles with no selection pressure. The obtained results indicated increased tolerance after passaging that reduced the aPDI efficacy by 5 log10 units (21).
The studies mentioned above indicate that the developed adaptation results from genetic alterations and is sustained in subsequent generations without selective pressure. Currently, no data indicate whether the observed tolerance has any particular pattern, i.e., which particular genetic elements underlie the observed phenomenon of increased tolerance to the studied aPDI or aBL applications. Several elements (Figure 3) discussed below might be expected to be involved in the response to the phototreatments.
Oxidative Stress Sensing and Detoxifying Mechanisms
Since bacterial cells are exposed to ROS produced both internally (aerobic growth, endogenous PSs, e.g., porphyrins) and externally (oxidative burst of macrophages, drugs, environment), there are several elements involved in oxidative stress sensing and ROS detoxification. Most of them are present in both gram-negative and gram-positive bacteria that live in aerobic conditions.
The General View on Oxidative Stress Sensing and Detoxifying Mechanisms in Gram-Positive and Gram-Negative Bacteria
The main transcription factor involved in sensing oxidative stress in bacterial cells sensing oxidative stress is OxyR. It can detect an increased level of peroxide by thiol-disulfide exchange. Its analog, PerR, acts by a metal-mediated peroxide-sensing mechanism. PerR, as a metalloprotein, contains Zn2+ and a second metal ion—Fe2+ or Mn2+. These proteins have a regulatory function; PerR containing Fe2+ responds to low levels of H2O2, and PerR containing Mn2+ responds poorly to elevated H2O2 levels (32). Furthermore, PerR is one of the ferric uptake regulator (Fur) homologs. PerR is a manganese-dependent transcriptional repressor that controls the transcription of catalase (Kat) and alkyl hydroperoxide reductase (Ahp). In S. aureus, PerR can repress Fur transcription and act as an autoregulator. It is induced by elevated iron and H2O2 concentrations (33). To avoid ROS intoxication due to the Fenton reaction, strict regulation of the free iron concentration is necessary. Fur, active as a dimer, binds structural Zn2+ and regulatory Fe2+. Fur can actively repress transcription by blocking the polymerase binding site of the promoter or control target gene expression via secondary regulators. Fur also positively regulates the expression of iron co-factored superoxide dismutase (sodM) and downregulates Mn-containing SodA (superoxide dismutase A). Fur transcription is activated by OxyR, which also activates the transcription of KatG and AhpCF in E. coli (34). Another important element activated by OxyR is Mnt, a metal ion-activated transcriptional repressor of manganese transporter genes. Manganese ions are actively imported by MntH mostly after OxyR activation; they are crucial for the activation of SodA and can also substitute iron in some metalloenzymes (35). Furthermore, PerR and OxyR can regulate mnt genes in different species. Therefore, Fur, PerR, and MntR form an integrated network controlling peroxide stress resistance as well as Fe2+ and Mn2+ homeostasis (33). In addition to iron and manganese, zinc, and copper also play important roles in oxidative stress. All of these metals can be cofactors for superoxide dismutases: iron for SodB, manganese for SodA and SodM, and copper/zinc for SodC (36). The main mechanism used by E. coli to sense superoxide anions functions via SoxRS. SoxR activates the transcription of SoxS, which in turn activates the production of several enzymes, including superoxide dismutase and DNA repair enzymes (37). Interestingly, the SoxRS system was shown to partially protect cells from singlet oxygen after treatment with (dipotassium;2,3,4,5-tetrachloro-6-(2,4,5,7-tetraiodo-3-oxido-6-oxoxanthen-9-yl)benzoate (rose bengal, RB) and illumination (38).
Oxidative Stress Elements Studied With Respect to aPDI
During aPDI, the excited state PS can generate ROS, including the superoxide anion (O2·-) and, in the presence of divalent ions (Fe2+), which enable the Fenton reaction, hydroxyl radical (HO·) or hydrogen peroxide (H2O2) (type I mechanism). It can also produce highly reactive singlet oxygen, 1O2 (type II mechanism). Superoxide dismutases and catalases can catalyze the reaction of O2·-, HO· and H2O2 to H2O and molecular oxygen, which can allow bacteria to avoid eradication when treated with PSs acting based on type I mechanism. On the other hand, there is no known enzyme specific for the detoxification of singlet oxygen, which makes mechanism type II PSs more promising as antibacterial agents (39).
One of the singlet oxygen producers, [7-(dimethylamino)phenothiazin-3-ylidene]-dimethylazanium chloride (methylene blue, MB), was used to study the E. coli response to oxidative damage. Kim et al. observed that the oxyR overexpression mutant was much more resistant to singlet oxygen-mediated cellular damage than the oxyR deletion mutant (28). The oxyR overexpression mutant also exhibited significantly higher activities of antioxidant enzymes such as Sod and Kat, which suggested that the oxyR regulon plays an important protective role in singlet oxygen-mediated cellular damage (40). Among many oxidative stress-induced compounds, OxyR can partially control the production of pyocyanin. Hendiani et al. observed pyocyanin gene upregulation after MB photoinactivation (41). Another transcription regulator, Fur, was investigated in an H. pylori mutant in which the fur gene was replaced with a kanamycin resistance marker. After MB-aPDI, the H. pylori mutant showed a 10,000-fold decreased viable cell number compared with wild-type H. pylori, which indicates a role of Fur in the response to aPDI-induced oxidative stress. Interestingly, a 3-fold higher increase in the level of 8-oxo-G indicated DNA damage (42).
The high importance of SodA and SodB in the survival of aPDI by E. coli was observed by Herbig et al. (32). A mutant that was not able to produce SodA and SodB was more susceptible to MB-PDI than the wild-type strain. However, the addition of external quenching agents such as Sod and Kat was sufficient to protect both strains from MB-aPDI. The addition of these enzymes was not sufficient when both strains were treated with TMPyP-aPDI. In addition, with TMPyP, the differences in survival between strains were not significant. Researchers did not observe changes in the expression of sodA and sodB for any of the treatments (43). On the other hand, in earlier studies, superoxide dismutase activation was observed after 6 h of growth in the presence of manganese and azure C, MB, thionine, or (7-amino-8-methylphenothiazin-3-ylidene)-dimethylammonium chloride (toluidine blue O, TBO) under room light (33). Additionally, E. coli B, with induced levels of superoxide dismutase and catalase, exhibited marked resistance to azure C phototoxicity (44). Misba and Khan performed the analysis of gene expression in Streptococcus mutans biofilms after azure A or NMB aPDI (34). They observed a 2-fold increase in expression of the sod gene (45). Buchovec et al. also studied the gene expression of oxyR, fur, sodA, ahpC, ahpF, katG, katE, and sodC using chlorophyllin-based photosensitization against Salmonella enterica. They observed significantly increased expression of ahpC and a slight increase in oxyR. The rest of the genes had expression changes <1.5-fold (46). On the other hand, when Dosselli et al. performed proteomic analysis of S. aureus after T4 porphyrin-aPDI, they observed that AhpC showed the most pronounced decrease in the intensity level of proteins and an 8-fold increase in catalase (KatA) protein levels (47). KatA was also investigated in P. aeruginosa after treatment with blue light or TBO-aPDI. The isogenic mutant katA− was significantly more sensitive to both treatments than the wild-type strain (48). The activity of Sod and Kat was investigated in E. coli after treatment with illuminated riboflavin. The activity of both enzymes was significantly reduced in comparison to treatment with light or riboflavin only (49). The activity of Sod was also checked after visible light illumination of S. aureus and E. coli in the presence of TA/Fe3+/AgNP nanofilm (38). The illumination of bacteria in the presence of the nanofilm caused a decrease in the activity of Sod in both species (50). Finally, our research team investigated S. aureus Sod isogenic mutants deprived of either sodA or sodM or both genes. We did not observe differences in the effectiveness of protoporphyrin IX (PPIX)-aPDI against all strains tested. However, in the medium without Mn2+ ions, the double sodAM mutant was highly susceptible to aPDI (39). We also examined 8 clinical isolates of S. aureus (4 MRSA and 4 MSSA), including strains that were highly resistant and strains that were highly vulnerable to photodynamic inactivation. We observed that Sod activity as well as sodA and sodM transcript levels increased after PPIX-based aPDI but only in aPDI-sensitive strains. The fact that an increase in Sod activity is observed only in aPDI-susceptible cells emphasizes that this is probably not a direct factor affecting S. aureus vulnerability to PPIX-based aPDI (51).
On the other hand, in our recent research concerning S. agalactiae tolerance to RB-aPDI, we observed that immediately after sublethal treatment, sod expression increased in both the control and tolerant strains, while ahpC expression decreased in both (21).
In conclusion, few studies have investigated the role of oxidative stress sensing and detoxifying mechanisms in photoinactivation. Different factors were examined in different bacterial species and after treatment with different photosensitizing compounds. We still lack profound knowledge of the aPDI mechanism of action and therefore possible bacterial actions that allow them to escape applied treatment. Does the complex regulatory antioxidant system in bacteria confer protection against aPDI? The answer seems to be positive, as many studies indicate that antioxidant enzymes are necessary to protect microbial cells from aPDI-induced ROS. Another issue remains unsolved, however: whether aPDI-generated ROS can specifically induce the production of enzymatic detoxifying enzymes, which might further lead to adaptation to elevated levels of ROS.
Key Master Regulator of Stress Response
Agr
One of the most important and best-studied regulatory systems in S. aureus is the accessory gene regulator (Agr). Agr was first described in 1986 and plays a crucial role in the pathogenesis of S. aureus, responding to various signals from the environment—availability of energy, bacterial cell density (quorum sensing) or nutritional status (52, 53). Park et al. studied the S. aureus response to aPDI at the transcription level (54). Transcriptional profiling indicated that sublethal aPDI combined with chlorin e6 (Ce6) and laser light (λmax = 664 nm) generated a bacterial stress response and activated Agr-dependent gene regulation. This result suggested the role of the Agr regulator in the S. aureus response to photooxidative stress generated during aPDI. Additionally, lethal aPDI conditions using two different combinations—Ce6 with laser light and pheophorbide a (Pa) with red LED light (λmax = 635 nm)—produced similar results. Unlike wild-type S. aureus, which survived the treatment conditions, the agr mutant strain demonstrated hypersusceptibility to aPDI. These observations proved that S. aureus requires an Agr regulator for protection from oxidative stress when exposed to aPDI (54). Grinholc et al. published similar conclusions (55). Clinical S. aureus strains lacking a functional agr system treated with protoporphyrin IX (PPIX) and irradiated with polarized light (620–780 nm) revealed increased susceptibility to aPDI. Moreover, the same group studied two isogenic S. aureus strains characterized by different agr statuses (agr-positive and agr, agrA-, agrB-, agrC-negative strains) and indicated a significant difference between agr-positive and agr-negative strains in response to PPIX-aPDI with 620–780 nm light (55). In contrast, Gándara et al. indicated no association between aPDI treatment using TBO with non-coherent light and agr gene status in S. aureus strains: RN6390 (rsbU−, agr+), SH1000 (rsbU+, agr+), and RN6911 (rsbU−, agr). However, in this case, the genetic background of the studied strains also differed with respect to another important transcription regulator, namely, rsbU (56). These significant variations in strain responses to aPDI may be because different PSs induce different mechanisms of photodamage.
Sar
Another regulatory network that plays a role in the bacterial stress response is staphylococcal accessory regulator A (SarA) (57). SarA is a positive regulator of agr and alternative sigma factor σB activity (58, 59). It can activate the expression of various virulence factors, including enterotoxin B, toxic shock syndrome toxin (TSST-1) and most cell-bound proteins (extracellular protein A, fibrinogen and fibronectin-binding proteins). SarA downregulates several proteases, lipases and nucleases (60, 61). The involvement of SarA in photooxidative stress is not fully understood and has not been examined to date. However, Ballal and Manna published the first report revealing the role of SarA in modulating oxidative stress resistance in S. aureus (62). Under both aerobic and microaerophilic conditions, the levels of sodM and sodA transcripts were markedly elevated in the sarA mutant compared to the wild-type S. aureus strain. Studies of various oxidative stress-inducing chemicals indicated that in the presence of diamide, a significant increase in sodM transcription was observed in the isogenic sarA mutant strain. Additionally, a small increase in sodA transcription was observed in the sarA mutant strain in the presence of tert-butyl hydroperoxide (t-BOOH). However, exposure to hydrogen peroxide (H2O2) and cumene hydroperoxide (CuOOH) did not affect sodM and sodA expression. It was suggested that SarA plays a role in the regulation of Sod transcription in S. aureus and oxidative stress resistance (62). Research has shown, however, that the cysteine in the structure of the SarA protein that might be a redox sensing element is sensitive to alkylation rather than oxidation (63). A more in-depth analysis of the direct impact of the SarA regulatory network on the bacterial response to aPDI needs to be conducted.
Sigma B
σB (encoded by the sigB gene) is an alternative sigma factor activated in the bacterial response to environmental conditions that regulates the expression of over 150 genes involved in the stress response (64, 65). The SigB operon also regulates the transcription of virulence genes, biofilm formation, membrane transport, cell internalization, persistence and antibiotic resistance (66, 67). Kossakowska-Zwierucho et al. revealed the significant role of the sigB operon of the S. aureus response to aPDI (68). We observed a marked decrease in the bacterial survival of sigB operon mutants compared to the wild-type S. aureus USA300 strain. This aPDI efficacy was noticed with the use of protoporphyrin IX diarginate (PPArg2), zinc phthalocyanine (ZnPC) and RB. The group obtained the same effect for other pairs of strains – RN6390 (deletion in rsbU gene) and SH1000 (restored deletion in rsbU gene) (68). Moreover, sequencing of the genes from the SigB operon (rsbU, rsbV, rsbW, sigB) of clinical S. aureus isolates highly susceptible to aPDI treatment revealed mutations in the rsbU gene, a positive regulator of SigB activity. Additionally, RsbU-defective S. aureus strains indicated a decrease in σB activity (measured by asp23 transcript level, which is exclusively controlled by σB factor). Therefore, the SigB operon and rsbU gene, in particular, were proven to be essential elements for the S. aureus response to aPDI (68). However, similar to Kossakowska-Zwierucho et al. and Gándara et al. demonstrated that aPDI treatment was ineffective when TBO was used on RsbU-defective mutants. This ineffectiveness might be attributed to TBO being a different type of PS from PPArg2, ZnPC, RB. TBO is characterized as a Type I PS, acting largely via radical species, whereas the PSs involved in the study conducted by Kossakowksa-Zwierucho et al. (PPArg2, ZnPC, RB) are strong singlet oxygen producers (Type II PSs). It was suggested that S. aureus strains defective in σB are easily eradicated by aPDI treatment using photosensitizers, producing a high yield of singlet oxygen (56, 68).
DNA Repair Enzymes
The Role of the recA Gene and RecA Protein in the Phototreatment Outcome
recA is an element of the SOS system, a bacterial response to genotoxic stress, during which bacteria induce several phenotypic changes through the differential regulation of genes and through rearranging and mutating their genome, sometimes acquiring characteristics that enable bacterial survival and adaptation to stress conditions. SOS induction is linked with the network of other essential stress responses and can be modulated under various circumstances to fit bacterial needs (69). The mechanism of the SOS response is well-known and well-described (70, 71) (Figure 4). It is induced by a single-stranded DNA (ssDNA), and two other key composites are involved in the SOS response: LexA, which is a transcriptional repressor, and RecA, which is an inducer (72). RecA binds ssDNA in the form of a nucleofilament, which is crucial for catalysis of the self-cleavage of the LexA repressor and enables expression of the SOS genes (73).
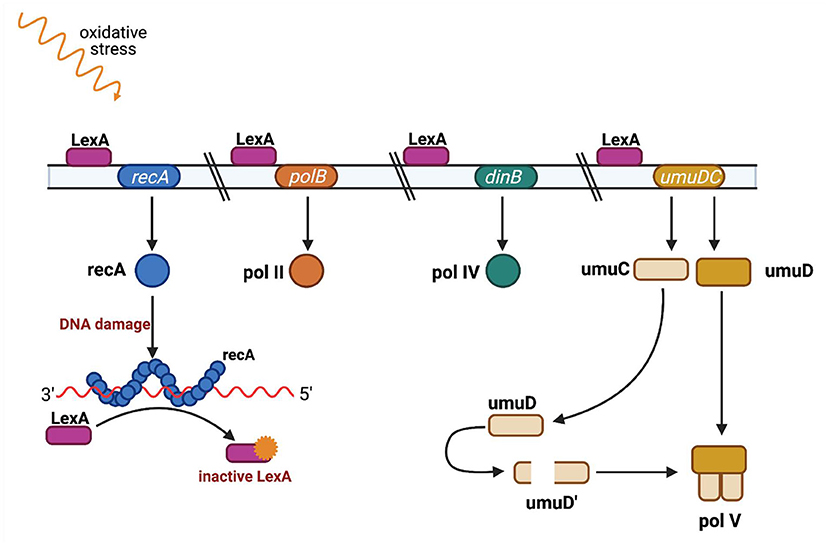
Figure 4. SOS regulation. The LexA protein is attached to an operator upstream of the SOS response genes and blocks their transcription. Single-stranded DNA fragments (ssDNA) accumulate when DNA is damaged (eg. as a result of UV radiation or ciprofloxacin). RecA protein binds to single-stranded DNA fragments to form nucleoprotein fibers that stimulate the SOS response. The active form of RecA protein interacts with the LexA repressor and causes LexA to dissociate from the operator. When LexA is not attached to an operator, genes responsible for DNA repair are transcribed, such as polB gene encoding polymerase II, dinB gene encoding polymerase IV, and the umuC and umuD genes encoding proteins UmuC and UmuD, which are subunits of polymerase V. Polymerases IV and V replicate damaged DNA. Adapted from Tropp (31). (Created with BioRender).
Membrane proteins and other components of the cell envelope are considered to be the major targets of oxidative stress caused by antimicrobial photoinactivation; however, the ROS produced during antimicrobial photoinactivation have the potential to cause DNA damage. Photocleavage of bacterial DNA is believed to occur as a secondary effect in extensively photoinactivated cells (6). It has been critically discussed within our recent study in which we examined the correlation between DNA damage and aPDI (74). The research investigated whether DNA damage occurs during sublethal doses of phototreatment (resulting in a 0.1-1 log10 unit reduction in cell viability) and demonstrated that the administration of exogenous PSs, i.e., TMPyP, RB, zinc phthalocyanine (ZnPc), NMB and TBO, or antimicrobial blue light (aBL, 405 nm), led to the excitation of endogenously produced photosensitizing chromophores, indicating that both aPDI and aBL exert (i) increased DNA damage (analyzed by gel electrophoresis), (ii) increased recA expression (luminescence assay conducted in the recA-lux strain), and (iii) increased levels of RecA protein (examined by Western blot analysis).
The same study aimed to examine whether the presence of RecA protein or inhibition of the recA gene could influence the effectiveness of photoinactivation; thus, wild-type USA300 JE2 and its isogenic derivative strain JE2 recA (recA-negative, NE805) were subjected to sublethal aPDI and aBL inactivation. Moreover, the wild-type USA300 JE2 strain was photoinactivated in the presence of novobiocin, is a well-known recA-downregulating agent, at MIC concentrations. As a result, the wild-type strain with inhibited RecA (by novobiocin) or strain NE805 with a disrupted recA gene demonstrated increased susceptibility to TMPyP-, RB-, ZnPc-, NMP-, and TBO-mediated aPDI as well as susceptibility to aBL. This result indicates that aPDI or aBL treatment results in DNA damage and subsequent cell death. There is also an important question of whether aPDI/aBL treatment results in increased expression of recA, which would indicate DNA photodamage upon aPDI or aBL. Indeed, sublethal doses of aPDI/aBL resulted in increased levels of recA gene expression, as measured by the increase in bioluminescent signal of recA promoter-lux constructs (75). Accordingly, RecA protein levels increased upon photoinactivation. This reveals that DNA damage occurs not only in the case of bacterial cells that are intensively photoinactivated (more than 2–3 log10 unit reduction), as was suggested previously (76–78), but could also be detected at lower doses of photoinactivation, when the majority of bacteria are still viable (confirmed with the cell membrane integrity assay). Within the same study, the results of the luminescence assay that enabled tracking of the activity of the recA promoter were subjected to Western blot analysis, demonstrating the induction of recA expression upon phototreatment.
In our most recent studies (20, 21) concerning aPDI and aBL, we confirmed that after consecutive sublethal phototreatments, increased recA expression occurs. The studies mentioned above demonstrate that aPDI/aBL treatment efficacy could be improved by controlling the DNA repair system, e.g., by inhibiting or eliminating the expression of the recA gene, and that recA is a critical element in the bacterial response to photoinactivation.
Role of LexA in Phototreatment Outcome
LexA is a late-induced gene and can inhibit SOS induction when the genotoxic signal is stopped (LexA cleavage is no longer promoted) (69).
Our previous study examined, using a luminescence assay in lexA-lux and recA-lux strains, whether RecA determines the response of S. aureus to phototreatment independently of LexA (repressor of SOS regulon). The obtained results indicated that phototreatments induced both lexA and recA expression. To investigate the role of RecA alone (without LexA interference), the S. aureus strain with uncleavable LexA (HG001 lexAG94E) was used. However, the response of the mutant strain was not significantly different from that of the wild-type strain.
Role of the umuC Gene in Increased Mutation Frequency and aPDI/aBL Tolerance Development
The expression of umuCD, like that of several other loci, is induced by DNA damage, and it is regulated by products of the recA and lexA genes (79). Similar to LexA, UmuCD production depends on proteolytic cleavage catalyzed by RecA nucleofilament (80). umuCD codes PolV, a stress-responsive error-prone DNA polymerase. PolV proceeds with DNA replication on damaged DNA by incorporating any base across the DNA lesion. PolV can incorporate correct or incorrect bases across the lesion on the template strand and has no proofreading activity, which may have a highly mutagenic effect due to an increased frequency of spontaneous mutation (81, 82). The outcome of such mutagenesis could lead to a phenomenon broadly defined as an adaptation (69).
Our previously published study investigated potential acceleration in the mutation frequency due to phototreatment. The rifampicin-resistant mutant selection assay is considered an adequate methodology for mutation rate estimation because, in the case of S. aureus, resistance to rifampicin may result from a single spontaneous mutation in rpoB. The group examined the development of spontaneous resistance to an antibiotic after plating on agar plates containing 4 × MIC (minimal inhibitory concentration) rifampicin to recover spontaneous antibiotic-resistant mutants. S. aureus USA300 JE2 did not demonstrate significant dissimilarities in response to sublethal phototreatment (0.1–1 log10 reduction in CFU) in comparison to basal mutation frequencies (74). We studied potential increases in the mutation frequency after the 5th, 10th, and 15th consecutive cycles of exposure to RB-mediated aPDI or aBL in the S. aureus USA300 JE2 strain. The results indicated that sublethal doses of aPDI or aBL induced genetic alterations in S. aureus. An increased mutation rate could result directly from induced DNA damage or from the activity of error-prone DNA Pol V. We employed aPDI or aBL to investigate whether phototreatments led to SOS response activation and increased expression levels of recA and umuC. In the case of both sublethal phototreatments (aPDI and aBL), we observed increased expression levels of recA and umuC. To confirm the possible SOS-dependent mechanism, we employed two transposon mutants derived from the S. aureus USA300 JE2 strain: with disrupted recA (NE805) and umuC gene (NE445). The obtained results indicated that aPDI, as well as aBL tolerance development, might be considered recA-dependent processes since no tolerance to either phototreatment upon five consecutive sublethal aPDI or aBL cycles was observed in the case of S. aureus NE805 lacking the functional recA gene. In contrast, studies revealed that only aBL tolerance acquisition is considered an umuC-dependent process: S. aureus lacking the functional umuC gene developed significant aPDI tolerance but expressed no aBL tolerance after five consecutive cycles of sublethal aPDI or aBL.
The described findings confirm that aPDI/aBL may result in DNA lesions that could lead to increased SOS- and umuC-dependent and umuC-independent mutagenesis.
Heterogenous Response to APDI and Genetic Background
Our several reports indicated that susceptibility across S. aureus isolates to aPDI is strain dependent (55, 83–86). We examined several 100 strains in total. Under the same experimental conditions (protoporphyrin IX-mediated aPDI, wavelength range of 620–780 nm, fluence of 50 J/cm2), the different strains expressed a heterogeneous response to photoinactivation, ranging from 0 to 5.1 log10-unit reductions in viable counts.
Determination of the molecular markers underlying the mechanism of the heterogeneous bacterial response to PDI treatment would have substantial clinical importance. We performed AluI and RsaI digestion of the agr gene PCR product, which revealed existing correlations between the determined digestion profiles and PDI susceptibility (55). Furthermore, the functionality of the agr system affected the S. aureus response to PDI. Based on our results, we concluded that the agr gene may be a genetic factor affecting the strain-dependent response to PDI. It is also known that MRSA and MSSA have different responses to aPDI. Data obtained by our group demonstrated that MRSA strains are significantly more resistant to photoinactivation than MSSA strains. However, the difference observed did not result from the presence of mec, antimicrobial susceptibility or resistance mechanisms, or the ability to form biofilms in vitro (85). We compared the genetic backgrounds of the MRSA strains [SCCmec types, spa types and main clonal complexes, (CC)] with their susceptibility to protoporphyrin IX-mediated aPDI. SCCmec typing revealed no differences in response to photoinactivation. However, the detection of spa types and clonal complexes clustered the studied population of MRSA strains according to their response to photodynamic oxidation. Strains of CC30 (ST36) demonstrated susceptibility to photoinactivation, and CC1 manifested a decreased response to PpIX-aPDI. Moreover, spa typing identified isolates that were more tolerant (t032) and more susceptible to phototreatment (t051, t015) (86). The association between the response to aPDI and clonal lineages displays the important role of genetic background in aPDI effectiveness. The described results make a case for the development of a diagnostic tool with the prognostic value of aPDI efficacy according to an identified genetic background of S. aureus isolates; nevertheless, the mechanism underlying this phenomenon is still poorly understood.
Physical Membrane Properties
Lipid Composition of Bacterial Membranes
The different responses of gram-negative and gram-positive microorganisms to photoinactivation with anionic PSs are obviously due to the presence in gram-negative species (e.g., K. pneumoniae and H. influenzae) of phospholipids, lipoproteins and polysaccharides in the additional outer envelope (87). In gram-negative bacteria, the prevalence of neutral lipids is represented by phosphatidylethanolamine (PE) (70–75% of total phospholipids), and the other 25% of the lipid composition consists of anionic phosphatidylglycerols (PGs). In gram-positive bacteria, the PE content is lower. Moreover, the membranes of bacterial cells contain cardiolipin (CL), another anionic lipid. All bacterial cells contain at least 15% anionic lipids, such as CL or PG, and exposure to such anionic lipids with lipoteichoic acids (LTA) or lipopolysaccharide (LPS) reduces the diffusion of PS into the cytosol (30, 88). Differences in photoinactivation efficiency may also be related to gram-negative species having structure A in LPS and its polysaccharide composition.
Photochemical reactions that involve the type I mechanism often result in the degradation of unsaturated phospholipids due to the abstraction of allylic hydrogens from these compounds (89). The radical species thus formed may undergo reaction with oxygen to yield lipid hydroperoxides. Lipid peroxidation is detrimental to membrane integrity, leading to loss of fluidity and increased ion permeability. Other cell wall/membrane targets include aminolipids and peptides. Thus, the inactivation of membrane enzymes and receptors is also possible.
Proteins in the cell membrane are significant photoinactivation targets; thus, singlet oxygen causes protein oxidation and the formation of carbonyls and aggregates and contributes to protein folding or unfolding (90, 91). The reaction of OH• with amino acids, e.g., proline, generates protein carbonyls (92). Overall, the data presented by Dosselli et al. suggest that phototreatment leads to specific damage that depends on the localization of a PS in a cell. The presence of proteins in the cell membrane is very closely related to the dynamics of the lipid bilayer. For example, when the homeostasis of the environment is disturbed via oxidative stress conditions, the stabilization of membrane proteins by electrostatic interactions among the lipid head groups, charged amino acid residues, and fatty acids is disrupted (93). Such disturbance of membrane fluidity can lead to lethal effects. The membrane fluidity of bacterial and mammalian cells can be interrupted as a result of membrane lipid oxidation via the action of reactive oxygen species (e.g., singlet oxygen) (93). The disruption of membrane functions associated with a decrease in membrane fluidity may contribute to the bactericidal effect of photoinactivation with TBO (93). Another observation of changes in the fluidity of the cytoplasmic membrane was investigated with TBO and red light (632.8 nm) treatment of Porphyromonas gingivalis. To evaluate the changes in membrane fluidity, a trimethyl-ammonium diphenyl hexatriene probe (TMA-DPH) was applied. This compound contains a positively charged trimethylammonium substituent that incorporates the molecule into the plasma membrane; therefore, it binds in proportion to the free membrane surface. When P. gingivalis cells were treated with TBO (82 μM) and 0.88 J of helium/neon laser red light, the reduction in viable cell count was estimated as 7 log10. This lethal treatment and even exposure to PS alone (without irradiation) led to a significant reduction in cytoplasmic membrane fluidity. Fluorescence measurement with the TMA-DPH compound confirmed these results. The decrease in membrane fluidity after the exposure of cells to PS can be explained by the occurrence of membrane damage after exposure to visible white light. On the other hand, increased membrane fluidity is a result of TBO aggregates, which can “bind” with membrane proteins and are therefore influenced by their properties. The explanation of the increased membrane fluidity after exposure to light and TBO is not clear. The authors suggest that the obtained effect could result from peroxidation of membrane lipids or from protein-protein crosslinking (93). In another experiment, E. coli cells were irradiated with white light for 1.5 h in the presence of 5 μM Tri-Py+-Me-PF-tricationic porphyrin [(5,10,15-tris(1-methylpyridinium-4-yl)-20-(pentafluorophenyl)porphyrin. The application of this PS and white light led to a decrease of approximately 6 log10 CFU/ml. As a part of the experiment, the photodynamic oxidation of E. coli membrane phospholipids was examined with a lipidomic approach. The fatty acid composition and lipid hydroperoxide level in E. coli extracts were investigated before and after aPDI treatment. The experimental outcome revealed that upon aPDI treatment, the concentration of lipid hydroperoxides was 93.6% higher than in a non-phototreated cell. The fatty acid composition revealed that aPDI induced the formation of a high amount of lipid hydroperoxides in the E. coli lipid extract and decreased the unsaturated C16:1 and C18:1 fatty acids. Lipid hydroperoxide derivatives of phosphatidylethanolamines with C16:1, C18:1, and C18:2 fatty acyl chains were also detected after photoinactivation (12).
The Role of Bacterial Pigments in aPDI
Many bacterial species, including pathogens, produce, and accumulate pigments within cell membranes. The primary function of those pigments is neutralization of oxidative stress produced by light exposure. Within microbial cells pigments additionally serve to stabilize membrane and protect lipids to assure cell envelop integrity.
Gram-Positive Bacteria
One of the most abundant bacterial pigments produced by S. aureus is staphyloxanthin. The gold C30 triterpenoid [α-D-glucopyranosyl-1-O-(4,4'-diaponeurosporen-4-oate)-6-O-(12-methyltetradecanoate)] is localized in the staphylococcal membrane. This compound alternates between single and double bonds, which can quench oxidation by reactive oxygen species (94). This carotenoid compound is synthesized by six enzymes in the biosynthetic pathway encoded in the crtOPQMN operon and is regulated by two-component signal transduction systems (TGSs) (95). The pigmentation of S. aureus cells is not the main factor responsible for the resistance of this pathogen to phototreatment. This staphylococcal pigment can interact with proteins and DNA (96). The effect of S. aureus pigmentation on the survival rate of this pathogen was investigated with the use of chlorin-ce6 PS and red light (97). The experiment was performed on the wild-type strain S. aureus Newman and the non-pigmented isogenic mutant ΔcrtM (without the gene for dehydrosqualene synthase CrtM). The application of chlorin-ce 6 at a concentration ≥128 μM led to 5 log10 reductions in the bacterial population of the Newman wild-type strain, and ΔcrtM exhibited the same reduction in bacterial viability when PS was applied at concentrations ≥64 μM. These results suggest that pigmentation can influence the effectivity of photoinactivation; thus, pigmented S. aureus was more resistant to singlet oxygen action than its mutant without pigment (97). It was shown that the presence of carotenoids is crucial for the cell membrane integrity of S. aureus and influences protection from oxidative stress (94). It is also worth mentioning that polar carotenoids can modulate the fluidity properties of lipid membranes (94). Experiments performed by Kossakowska-Zwierucho et al. suggested that the enhanced membrane fluidity could be responsible for the increased effectiveness of aPDI. Membrane fluidity, which is a temperature-dependent parameter, was estimated for S. aureus strains (SA144, SA145, and SA147) by measuring the fluorescence polarization anisotropy (r) of 1.6-diphenyl-1.3.5-hexatriane (DPH). DPH is a lipophilic fluorescent probe that can localize in the hydrophobic regions of the cell membrane. Analysis of fluorescence anisotropy for the tested S. aureus strains SA144, SA145, and SA147 (which differ in the level of pigmentation) revealed that low membrane fluidity was observed for highly pigmented cells (SA144 SA147). However, for weakly pigmented cells (e.g., strain SA145), the fluorescence anisotropy value was estimated to be low, thus confirming the high level of membrane fluidity. The exposure of these three strains to photoinactivation conditions (5 nM ZnPC (zinc phthalocyanine) and red light 627 nm) revealed that strains exhibiting wild-type pigmentation and lack of pigmentation (SA 144 and SA 145) were much more sensitive to aPDI than the strain with high pigmentation (SA 147). On the other hand, the irradiation of cells with green light in the presence of RB (0.1 μM) showed a reduction in bacterial viability (by ~4 log10 CFU/ml), which indicates that the lack of protection against photooxidative stress was detected only for the strain lacking pigmentation (68). Enterococcus species are very often linked with resistance to vancomycin and human infections; however, there are many interesting species associated with soil that contain yellow pigment (98). For example, E. casseliflavus, E. mundtii, or E. sulfureus can produce carotenoids similar to those found in S. aureus. Experiments performed on S. aureus mutants deficient in the production of yellow pigment, as mentioned above, are more susceptible to photooxidative stress. For similar experimental purposes, the E. faecalis reference non-pigmented strain, E. casseliflavus (pigmented) and Enterococcus species environmental isolates from Avalon Beach (lightly pigmented) were used for tests. Bacteria were exposed to conditions simulating seawater at pH 8.1 and to a broad range of the visible light spectrum, 290–800 nm, without the addition of any photosensitizing agent. The results of these experiments revealed that non-pigmented E. faecalis strains were significantly reduced faster than pigmented strains (E. casseliflavus and pigmented E. faecalis environmental isolates) (98). The results of this research suggest that pigments of Enterococcus spp. could quench reactive oxygen species and be responsible for the photooxidation process.
Gram-Negative Bacteria
Literature data on the photoinactivation and pigmentation of microorganisms were presented by Orlandi et al., who investigated the gram-negative species P. aeruginosa. This pathogen is known as a producer of various pigments: pyocyanin, phenazine, pyoverdine, pyorubin, and pyomelanin. These pigments are crucial in the context of photoinactivation due to their ability to survive under oxidative stress conditions. Experiments performed by Orlandi et al. aimed to identify whether P. aeruginosa pigments contribute to their relative tolerance to photooxidative stress. For this purpose, differently pigmented wild-type and transposon mutants of the P. aeruginosa PAO1 strain were used: RE- pyomelanine overproducer; BL- pyocyanin producer; and YE- pyoverdine overproducer. All four strains cultivated in LB medium or M9 with glucose were subjected to photoinactivation with TBO or TMPyP. Each photosensitizing agent has a different mechanism of action: TBO acts mainly via a type I mechanism, whereas TMPyP acts via a type II mechanism. To determine the influence of pyomelanin or pyoverdine on the oxidative stress response, mutants were treated with 25 μM TBO and irradiance at 140 J/cm2 or with 5 μM TMPyP and 210 J/cm2 of light irradiance, respectively. BL cells cultivated in M9 and glucose medium were not successfully inactivated with either PS, whereas YE, RE and wild-type BL cells were sensitive to both phototreatments (99). Other literature data showed that P. aeruginosa pyoverdine blue pigment can absorb visible light at 415 nm. This property suggests that this pyoverdine can participate in photodynamic reactions because it can absorb visible light (100). Colorful pigments are also found in Prevotella intermedia and Prevotella nigriscens, which are microorganisms able to colonize the oral cavity that are capable of producing black pigments. P. gingivalis is known as an aetiological agent of periodontal diseases. It is also easily recognized as a black pigment producer (101). Experiments performed with visible light (in the range 380 to 520 nm) revealed that the exposure of P. intermedia and P. nigrescens to visible light doses of 4.2 J/cm2 and of P. melaninogenica at a dose of 21 J/cm2 led to a reduction of 5 log10 (0.001% of survival fraction) in comparison to the control. Chromatographic analysis identified 267 ng/mg, 47 ng/mg and 41 ng/mg endogenous porphyrin in bacterial cells of P. intermedia, P. nigrescens, and P. melaninogenica, respectively (102). Another study also confirmed photoinactivation with aBL against species producing the black iron pigment (103). The pigment produced by P. gingivalis, P. intermedia and P. nigriscens is reported in the literature as μ-oxo bishaem ([Fe(III)PPIX]2O) (104). A bacterial cell-surface layer of μ-oxo bishaem can act defensively by protecting bacterial cells against hydrogen peroxide (104). The abovementioned species use haem as an iron source for their growth; thus, they can accumulate black pigment that mainly consists of μ-oxobishaem at the cell surface. Despite the predomination of iron-protoporphyrin pigment, these species can also accumulate iron-free PpIX, which can be easily excited with visible light. This process leads to energy transfer from the porphyrin triplet state to molecular oxygen to produce singlet oxygen (type II mechanism of action) (102, 103). The effectiveness of photoinactivation with the engagement of free PpIX has been proven many times in the literature, and the use of exogenously administered PSs, e.g., TBO, highlights another interesting fact related to the response of cells to photoinactivation treatment (93, 101).
Photosensitizer Accumulation in Cells
The accumulation of a PS is one of the crucial factors to establish an efficient photodynamic approach for bacterial eradication. However, according to the literature data, this intracellular accumulation is not a necessary element for aPDI. Accumulation in cells is generally described by three main mechanisms: (I) external action, (II) intracellular action (including self-promoted uptake), and (III) active transport (Figure 5). All of them have provided efficient photokilling of both gram-positive and gram-negative bacteria (6). However, there are additional factors that affect the accumulation of PSs.
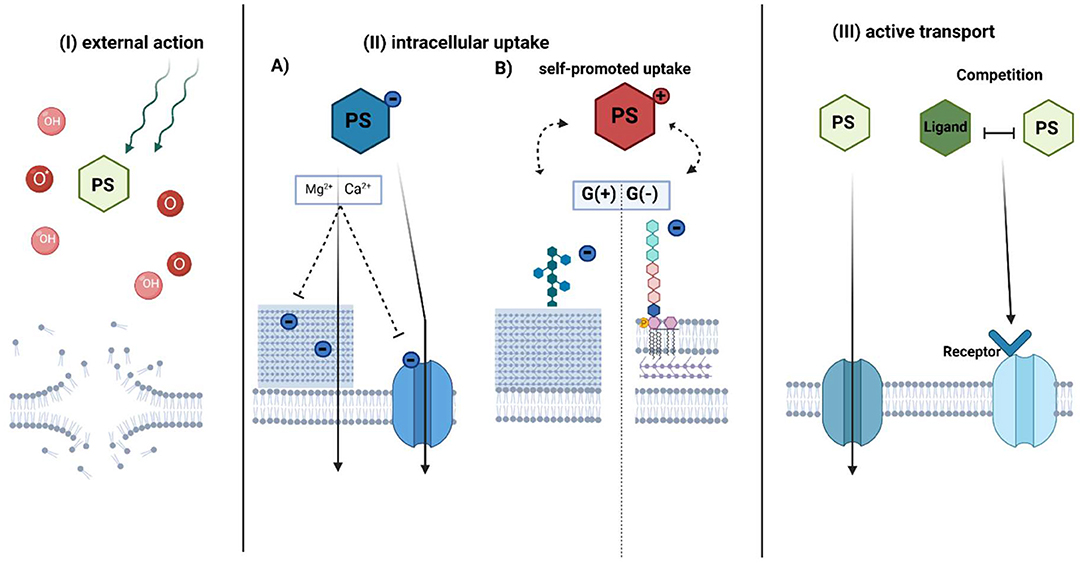
Figure 5. Proposed mechanisms of photosensitizer accumulation in aPDI treatment. The photosensitizer uptake mechanism can be divided into three general pathways: (I) external action, (II) intracellular action, and III) active transport-dependent accumulation. In the external action (I), PS does not accumulate inside the bacterial cell, however, due to the diffusion length of generated ROS (OH) or singlet oxygen (O*) upon irradiation, it causes the membrane disruption. The intracellular action (II) depends on the PS charge. (A) Anionic PS is accumulated by electrostatic interactions or active transport. The addition of divalent cations neutralizes negative charge on the bacterial envelope and promotes uptake of negatively charged PS through membranes. (B) Cationic PSs use mechanism known as “self-promoted uptake.” The positively charged compound is bound to the negatively charged teichoic acids on the peptidoglycan of the G(+) or lipopolysaccharides on the outer membrane of G(-) bacteria. The photosensitizer is associated with the surface layer, while after photosensitization it promotes cell disruption. In active transport (III), PS is taken up inward through a membrane protein channel such as porins or by mimicking receptor binding due to structure similarity to its natural ligand. PS-photosensitizer, O*- singlet oxygen, OH- radicals referred in general as ROS. (Created with BioRender).
(I) External Action
Photodynamic targets (such as cellular components and nucleic acids) are dependent on the localization of PS and the maximal diffusion length of generated ROS and singlet oxygen (105). There is no requirement for a PS to be intracellularly taken up to promote oxidative damage; thus, the proximity of PS to bacteria might promote efficient photokilling. According to recent studies, cationic charged TMPyP does not accumulate inside the cells of E. coli. While remaining outside, however, it could provide significant phototoxicity by singlet oxygen generation (106). Additionally, Gollmer et al. showed that TMPyP accumulates minimally in the cytoplasm and the cell wall of S. aureus but only after cell wall damage (107). Nevertheless, these results are not in accordance with previous studies on the localization of TMPyP (108), so further investigation of the external action of PSs is important.
(II) Intracellular Uptake
Simple diffusion of PSs through physical barriers, such as membranes, is restricted to either gram-positive or gram-negative bacteria due to the cell wall structure (109). The primary issue for uptake is the structural organization and charge of the PS. Anionic PSs accumulate in bacterial cells by electrostatic interactions or via active transport (110). The addition of divalent ions such as Mg2+ or Ca2+ promotes the accumulation of such PSs through higher activation of protein transporters or neutralization of the negative charges of bacterial envelopes, thus reducing the repulsion between anionic PS and the bacterial cell wall (110). In general, gram-negative microorganisms are more aPDI-resistant with applied anionic or neutral PS than gram-positive bacteria (111). Only by adding membrane-disrupting agents such as EDTA (112) or polycationic peptide, for example, polymixin B (113–115), can these PSs accumulate in gram-negative cells (110).
Cationic PSs could be intracellularly accumulated by electrostatic bonds in a self-promoted uptake mechanism (110). The peptidoglycans of gram-positive bacteria include negatively charged teichoic acids that constitute binding sites for PSs. Cationic PSs such as MB or TBO could also be taken up into gram-negative cells by a self-promoted mechanism. MB binds as a dimer to negatively charged LPS, causing an association with the surface layer and cell disruption after photosensitization (116). Negatively charged lipopolysaccharides are stabilized by electrostatic bonds with surrounding divalent ions such as Mg2+ or Ca2+, which act compatibly with PS. Divalent ions promote the inhibitory effect on cationic PS uptake. Saji et al. showed that the accumulation of MB by a self-promoted mechanism was reflected by the effectiveness of aPDI. A reduction of E. faecalis cells was achieved with 6.25 μM MB in the absence of divalent ions upon irradiation. In the presence of divalent ions, a PS concentration of up to 100 μM was achieved. Additive divalent ions inhibit MB binding to LPS (117) and accumulation, which was also confirmed in uptake assays (110).
Compared to MB, the cationic PS TBO, displayed a higher reduction in bacterial burden after photodynamic treatment (118, 119). Spectrophotometric studies revealed that TBO has a greater affinity for different types of LPS from MB (116). Contrary to expectations, MB-mediated photoinactivation was significantly more inhibited by CaCl2 than TBO-mediated photoinactivation. It was suggested that MB could also be electrostatically bound to charged amino acids in proteins such as siderophores (120). The complex of MB and the protein receptor on siderophores translocates PS through the membrane. However, the mechanism involving LPS is much more efficient in photokilling. The two mechanisms could coexist and influence aPDI effectiveness (120).
The self-promoted uptake pathway was also considered a mechanism of action for cationic pyridinium zinc phthalocyanine in E. coli (121) or cationic meso-substituted porphyrins in P. aeruginosa (122), Vibro anguillarum and E. coli cells (123). However, the number of cationic charges associated with the neutral PS might influence its hydrophobicity (124, 125). The increased amphiphilic character of porphyrins seems to enhance their affinity to the envelope of gram-negative bacteria, which improves accumulation. This might be accomplished by the asymmetric charge distribution or by adding cationic charges (6). Different numbers of positive charges of meso-porphyrins result in changes in the log10 reduction of aPDI (125–127).
(III) Active Transport
In active transport, PSs might be bound to the bacterial cell wall and then transported into the plasma membrane. In general, hydrophilic PSs are better able to penetrate through protein channels such as porins. Porins transport low molecular weight (up to 700 kDa) hydrophilic compounds (128, 129) and could mediate the active transport of PS. Divalent ions and trypsin may influence the uptake of PSs (110, 122). Trypsin promotes the functional impairment of cell wall-associated proteins, while divalent ions might have an impact on protein activity. The mechanism of RB uptake was proposed to be mediated by proteins based on the inhibitory aPDI effect after treatment of cells with trypsin (110).
The mechanism of exogenous porphyrin accumulation is still not fully understood. One of the proposed hypotheses is recognition by the haem transport machinery due to its similar structure to Fe3+-protoporphyrin IX (known as a heme). The well-characterized transport system is the staphylococcal iron-regulated surface determinant system (Isd), described in detail in many research studies by the Skaar group (130–133). Briefly, in a low-iron environment, heme-acquisition machinery recognizes and binds free haem or haemoproteins using an Isd protein system (130, 133). Haem is transferred to the cytoplasm using ABC-like membrane transporters, then degraded by an oxygenase to release free iron from the porphyrin structure (134). Paradoxically, too high intracellular concentration could be a challenge for S. aureus due to the toxicity of free iron (131). The HssRS (two-component system required for haem sensing)/HrtAB (haem-regulated transporter) system senses and pumps out an excess haem (135). Based on our investigation, mutants with impaired HrtA protein function accumulated PpIX in the largest amount and were the most efficiently eradicated upon PpIX-based photoinactivation in comparison to other haem transporter mutants, ΔIsdD, ΔHtsA, and wild-type S. aureus. However, the observed results do not correlate solely to protein function but also to a physical membrane modification that lacks the HrtA protein. It cannot be ruled out that PpIX may be a substrate for the HrtAB efflux pump (136). Metalloporphyrins, haem analogs, could be a potential group for targeting haem machinery (137). Metals in the oxidation state (III) have an affinity to the haem receptor on the Isd protein and mimic the properties of the natural ligand (138). Studies on the accumulation of metalloporphyrins showed that S. aureus cannot detoxify most of the toxic analogs through pump efflux in the same manner as heme. Most of them accumulate in the staphylococcal cell membrane (135). Recent studies of aPDI with Ga3+ PPIX showed rapid, diffusion-limited uptake of PS correlated with the appearance of cell-surface hemin receptors. Additionally, photoinactivation was more potent than in the case of PpIX (139). These indications might open a novel pathway for metalloporphyrins in the aPDI approach.
Microbial efflux pumps (MEPs) are transport proteins localized in the bacterial membrane that are involved in the detoxification process of toxic substances in bacteria. Some efflux pumps selectively pump out specific antibiotics, while multidrug resistance pumps (MDRs) remove structurally diverse compounds with a different mechanism of action. A well-known major facilitator-type MEP is NorA in S. aureus, while in gram-negative bacteria, a three-component RND (resistance nodulation division) efflux pump is typified by MexAB-OprM in P. aeruginosa. It has been suggested that amphipathic cations are natural substrates of MEP. Tegos et al. published studies on the accumulation of cationic phenothiazinium salts in knock-out and overexpressed MDR pump mutants of S. aureus, P. aeruginosa, and E. coli. The MDR-aberrant phenotype indicated an increased accumulation of all phenothiazinium dyes in each MDR knockout mutant in the studied species. The uptake of non-phenothiazinium dyes remained unchanged in NorA(+), NorA(–), and wild-type S. aureus, which indicated that these compounds are not recognized by MDRs (140). Adding inhibitors specific to the type of efflux pump promoted the cellular uptake of phenothiazinium salts in S. aureus and P. aeruginosa and was correlated with a higher aPDI efficiency (141). NorA inhibitors covalently attached to the structure of MB demonstrated the efficiency of aPDI and higher accumulation in MRSA (methicillin-resistant S. aureus) than MB alone (142). Additionally, those hybrids showed enhanced E. coli and Acinetobacter baumannii cell killing upon irradiation under both in vitro and in vivo conditions, despite neither species expressing this type of efflux pump (143). In contrast, Grinholc et al. observed that amphiphilic cationic PPArg2 was not a substrate for the NorA efflux pump even when an efflux inhibitor was used (84). Efflux pumps could be potential targets for increasing the intracellular level of suitable PSs and improving the activity of aPDI toward gram-positive and gram-negative bacteria.
(IV) Additive Factors
Photosensitizer affinity to the bacterial cell depends on more than the cell structure or physicochemical properties (Figure 1). Factors such as the composition of the culture medium and its additive components, PS concentration, solvent or cell density might also have a significant impact on the effectiveness of accumulation.
As referred previously, monovalent and divalent ions had a concentration-dependent effect on PS accumulation in gram-positive and gram-negative bacteria (122). Divalent cations such as Ca2+ have a stronger effect on ionic strength than monovalent cations such as Na+. The ionic strength of a solution has a strong impact on the activity of polyvalent ions (114, 122). Additionally, the presence of EDTA might exhibit increased or decreased uptake, depending on the charge of the PS, which is also reflected in aPDI efficiency (110). The protein composition in the medium also influences the effectiveness of accumulation. A 7-fold reduction in the protein content in the medium resulted in a greater reduction in A. baumannii cultures after photoinactivation with TMPyP (144). For instance, proteins such as human albumin or lipoproteins interact with PPIX (136). However, different proteins have limited affinity for certain PSs (144).
Another approach for a higher uptake of PS is preincubation with antimicrobial peptides (AMPs). AMPs are amphipathic amino acids with up to 50 amino acids. Positively charged peptides are bound to anionic groups on the bacterial membrane and promote the creation of pores (145). A study by our group revealed a 20-fold decrease in RB concentration and fluence in the photoinactivation of P. aeruginosa after supplementation with AMPs. Combined treatments with RB and PEX (pexiganan) or CAM (camel) resulted in a doubled uptake rate of RB in comparison to RB alone (146). The uptake of MB in the presence of AMP aurein 1.2 was also double rated. However, studies on chlorin-e6 combined with AMPs showed a decrease in uptake of PS with a synergistic effect in aPDI. This synergy was observed only in photokilling with MB and chlorin-e6, which indicated PS-dependent action (147).
Effective uptake also relies on the PS concentration. Cationic PSs are more efficient for aPDI and accumulate at a lower concentration than neutral or anionic PSs. The uptake of all types of PS was inversely dependent on cell density (13). Notably, the solvent might also contribute to the accumulation process, resulting in different aPDI effects. Studies on MB uptake in E. faecalis revealed higher accumulation in water, while photokilling was reduced in comparison to other solvents (148). Water as a solvent promotes the aggregation of MB and TBO at the cell wall of bacteria (119). Aggregated PS exhibits a reduction in the quantum yield of singlet oxygen, resulting in diminished bacterial eradication upon light illumination (76). The most appropriate solvent for PS should be taken into consideration while optimizing the aPDI approach.
The uptake rate of PS may be strain dependent. In the work of both Gad et al. and Tegos et al., there was a significant difference in uptake and aPDI efficiency in various S. aureus strains; however, there was no relevant correlation (149). Investigations by our team suggested that the level of PPArg2 uptake was linked to the level of aPDI susceptibility of MRSA and MSSA (84, 150). Interestingly, aPDI tolerance might be a result of the decreased accumulation of PS. Recent studies by Pierański et al. showed decreased accumulation of RB inside an aPDI-tolerant S. agalactiae strain after 10 cycles of aPDI treatment. Without selective pressure, uptake remained at the same level. Multiple aPDI treatments promote cell envelope-related modifications, resulting in decreased PS uptake. Lower PS availability results in less effective photokilling of bacteria. This study is the first investigation of decreased PS uptake as a possible microbial strategy to gain aPDI tolerance (21).
Conclusions
In this review, we discuss several issues contributing to the various responses of bacteria to aPDI or aBL. All the issues addressed contribute to the aPDI or aBL outcome to a greater or lesser extent. The mechanisms of aPDI or aBL are non-specific; the toxic effects of ROS are directed at many cellular targets. Thus, various bacteria for photoinactivation depend on a particular photosensitizer that accumulates in the cell to produce photoinduced ROS, which ultimately causes cytotoxicity and cell death. The potential problem with aPDI or aBL lies in the potential to introduce damage to DNA, which, in the case of SOS induction, may lead to the mutation and fixation of a particular genetic feature in the population that equips the bacteria better to resist aPDI or aBL. Another very important aspect of aPDI should be taken into account, namely photoinactivation based on photosensitizing compounds bound to a solid support. The direction of research on solid-based photosensitizing compounds focuses primarily on the production of materials with specific properties: efficient ROS generation, resistance to self-quenching, high photooxidation efficiency. These properties may differ for the same photosensitizing compound depending on whether it will be free or bound to a solid material (151).
For solid-based PS, two aspects must be considered: (i) the first situation where the photosensitizing compound cannot enter the bacterial cell due to its immobilization on an insoluble carrier. In this situation, the cell is affected through the extracellular structures associated with the cell wall and the cell membrane. In this case, the signal generated by aPDI is strong enough to cause the desired phototoxic effect and cell death. Potentially, one could also consider the situation (ii), in which the signal generated at the level of the external cell structures is sent inside the cell and cytotoxic effects are induced inside the cell. However, this is a purely hypothetical situation, and so far no experimental data has shown such a situation. In the light of the current state of knowledge, the action of photosensitizing compounds related to the solid medium consists in destroying external cell structures, especially the cytoplasmic membrane (gram-positive bacteria) and the cytoplasmic and outer membrane (gram-negative bacteria) that lead to leakage of cellular content or disruption of critical enzymes (152). This type of in-depth analysis to explain the mechanism underlying the effects of aPDI or aBL on specific photosensitizers either in solution or based on solid material requires further study.
Author Contributions
AR-Z wrote a fragment related to the influence of aPDI on DNA. AW prepared a fragment on the properties of cell membranes. KM prepared a fragment on accumulation. MP described oxidative stress. PO described stress cell regulators. MG prepared an introduction and edited manuscript. JN did literature search, prepared an outline of the manuscript, edited the entire text, made a substantive, stylistic, and linguistic correction. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by NCN grant no. NCN 2017/27/B/NZ7/02323.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Javed F, Samaranayake LP, Romanos GE. Treatment of oral fungal infections using antimicrobial photodynamic therapy: a systematic review of currently available evidence. Photochem Photobiol Sci. (2014) 13:726–34. doi: 10.1039/C3PP50426C
2. Wiehe A, O'brien JM, Senge MO. Trends and targets in antiviral phototherapy. Photochem Photobiol Sci. (2019) 18:2565–612. doi: 10.1039/c9pp00211a
3. Cieplik F, Deng D, Crielaard W, Buchalla W, Hellwig E, Al-Ahmad A, et al. Antimicrobial photodynamic therapy – what we know and what we don't. Crit Rev Microbiol. (2018) 44:571–89. doi: 10.1080/1040841X.2018.1467876
4. Vatansever F, de Melo WCMA, Avci P, Vecchio D, Sadasivam M, Gupta A, et al. Antimicrobial strategies centered around reactive oxygen species - bactericidal antibiotics, photodynamic therapy, and beyond. FEMS Microbiol Rev. (2013) 37:955–89. doi: 10.1111/1574-6976.12026
5. Hamblin MR. Photodynamic therapy for cancer: what's past is prologue. Photochem Photobiol. (2020) 96:506–16. doi: 10.1111/php.13190
6. Alves E, Faustino MA, Neves MG, Cunha A, Tome J, Almeida A. An insight on bacterial cellular targets of photodynamic inactivation. Future Med Chem. (2014) 6:141–64. doi: 10.4155/fmc.13.211
7. Huang L, Xuan Y, Koide Y, Zhiyentayev T, Tanaka M, Hamblin MR. Type i and Type II mechanisms of antimicrobial photodynamic therapy: an in vitro study on gram-negative and gram-positive bacteria. Lasers Surg Med. (2012) 44:490–9. doi: 10.1002/lsm.22045
8. Hamblin MR, Abrahamse H. Oxygen-independent antimicrobial photoinactivation: type iii photochemical mechanism? Antibiotics. (2020) 9:53. doi: 10.3390/antibiotics9020053
9. Oruba Z, Chomyszyn-Gajewska M. [Application of photodynamic therapy in dentistry – literature review]. Przegl Lek. (2016) 73:857–61.
10. Michaeli A, Feitelson J. Reactivity of singlet oxygen toward amino acids and peptides. Photochem Photobiol. (1994) 59:284–9. doi: 10.1111/j.1751-1097.1994.tb05035.x
11. Cadet J, Douki T, Ravanat J-L. Oxidatively generated base damage to cellular DNA. Free Radic Biol Med. (2010) 49:9–21. doi: 10.1016/j.freeradbiomed.2010.03.025
12. Alves E, Santos N, Melo T, Maciel E, Dõria ML, Faustino MAF, et al. Photodynamic oxidation of Escherichia coli membrane phospholipids: new insights based on lipidomics. Rapid Commun Mass Spectrom. (2013) 27:2717–28. doi: 10.1002/rcm.6739
13. Demidova TN, Hamblin MR. Effect of cell-photosensitizer binding and cell density on microbial photoinactivation. Antimicrob Agents Chemother. (2005) 49:2329–35. doi: 10.1128/AAC.49.6.2329-2335.2005
14. Pedigo LA, Gibbs AJ, Scott RJ, Street CN. Absence of bacterial resistance following repeat exposure to photodynamic therapy. In: Kessel DH, editor. Proceedings of SPIE 7380, Photodynamic Therapy: Back to the Future. Seattle, WA. doi: 10.1117/12.822834
15. Tavares A, Carvalho CMB, Faustino MA, Neves MGPMS, Tomé JPC, Tomé AC, et al. Antimicrobial photodynamic therapy: study of bacterial recovery viability and potential development of resistance after treatment. Mar Drugs. (2010) 8:91–105. doi: 10.3390/md8010091
16. Al-Mutairi R, Tovmasyan A, Batinic-Haberle I, Benov L. Sublethal photodynamic treatment does not lead to development of resistance. Front Microbiol. (2018) 9:01699. doi: 10.3389/fmicb.2018.01699
17. Leanse LG, Harrington OD, Fang Y, Ahmed I, Goh XS, Dai T. Evaluating the potential for resistance development to antimicrobial blue light (at 405 nm) in gram-negative bacteria: And in vivo studies. Front Microbiol. (2018) 9:2403. doi: 10.3389/fmicb.2018.02403
18. de Annunzio SR, de Freitas LM, Blanco AL, da Costa MM, Carmona-Vargas CC, de Oliveira KT, et al. Susceptibility of Enterococcus faecalis and Propionibacterium acnes to antimicrobial photodynamic therapy. J Photochem Photobiol B Biol. (2018) 178:545–50. doi: 10.1016/j.jphotobiol.2017.11.035
19. de Freitas LM, Blanco AL, Fontana CR. Antimicrobial photodynamic therapy proved not to induce bacterial resistance. In: Light-Based Diagnosis and Treatment of Infectious Diseases, Vol. 10479. Bellingham, WA: SPIE (2018).
20. Rapacka-Zdonczyk A, Wozniak A, Pieranski M, Woziwodzka A, Bielawski KP, Grinholc M. Development of Staphylococcus aureus tolerance to antimicrobial photodynamic inactivation and antimicrobial blue light upon sub-lethal treatment. Sci Rep. (2019) 9:1–18. doi: 10.1038/s41598-019-45962-x
21. Pieranski M, Sitkiewicz I, Grinholc M. Increased photoinactivation stress tolerance of Streptococcus agalactiae upon consecutive sublethal phototreatments. Free Radic Biol Med. (2020) 160:657–69. doi: 10.1016/j.freeradbiomed.2020.09.003
22. Giuliani F, Martinelli M, Cocchi A, Arbia D, Fantetti L, Roncucci G. In vitro resistance selection studies of rlp068/cl, a new zn(ii) phthalocyanine suitable for antimicrobial photodynamic therapy. Antimicrob Agents Chemother. (2010) 54:637–42. doi: 10.1128/AAC.00603-09
23. Costa L, Tomé JPC, Neves MGPMS, Tomé AC, Cavaleiro JAS, Faustino MAF, et al. Evaluation of resistance development and viability recovery by a non-enveloped virus after repeated cycles of aPDT. Antiviral Res. (2011) 91:278–282. doi: 10.1016/j.antiviral.2011.06.007
24. Guffey JS, Payne W, Jones T, Martin K. Evidence of resistance development by Staphylococcus aureus to an in vitro, multiple stage application of 405 nm light from a supraluminous diode array. Photomed Laser Surg. (2013) 31:179–82. doi: 10.1089/pho.2012.3450
25. Guffey JS, Payne W, Martin K, Dodson C. Delaying the onset of resistance formation: effect of manipulating dose, wavelength, and rate of energy delivery of 405-, 464-, and 850-nanometer light for Staphylococcus aureus. Wounds a Compend Clin Res Pract. (2014) 26:95–100.
26. Zhang Y, Zhu Y, Gupta A, Huang Y, Murray CK, Vrahas MS, et al. Antimicrobial blue light therapy for multidrug-resistant acinetobacter baumannii infection in a mouse burn model: implications for prophylaxis and treatment of combat-related wound infections. J Infect Dis. (2014) 209:1963–71. doi: 10.1093/infdis/jit842
27. Zhang Y, Zhu Y, Chen J, Wang Y, Sherwood ME, Murray CK, et al. Antimicrobial blue light inactivation of Candida albicans: In vitro and in vivo studies. Virulence. (2016) 7:536–45. doi: 10.1080/21505594.2016.1155015
28. Amin RM, Bhayana B, Hamblin MR, Dai T. Antimicrobial blue light inactivation of Pseudomonas aeruginosa by photo-excitation of endogenous porphyrins: In vitro and in vivo studies. Lasers Surg Med. (2016) 48:562–8. doi: 10.1002/lsm.22474
29. Tomb RM, Maclean M, Coia JE, MacGregor SJ, Anderson JG. Assessment of the potential for resistance to antimicrobial violet-blue light in Staphylococcus aureus. Antimicrob Resist Infect Control. (2017) 6:100. doi: 10.1186/s13756-017-0261-5
30. Kashef N, Hamblin MR. Can microbial cells develop resistance to oxidative stress in antimicrobial photodynamic inactivation? Drug Resist Updat. (2017) 31:31–42. doi: 10.1016/j.drup.2017.07.003
31. Tropp BE. Principles of Molecular Biology. Jones & Bartlett Learning. (2012). Available online at: https://books.google.pl/books?id=2-IlMqaQyE4C
32. Herbig AF, Helmann JD. Roles of metal ions and hydrogen peroxide in modulating the interaction of the Bacillus subtilis PerR peroxide regulon repressor with operator DNA. Mol Microbiol. (2002) 41:849–859. doi: 10.1046/j.1365-2958.2001.02543.x
33. Horsburgh MJ, Clements MO, Crossley H, Ingham E, Foster SJ. PerR controls oxidative stress resistance and iron storage proteins and is required for virulence in Staphylococcus aureus. Infect Immun. (2001) 69:3744–54. doi: 10.1128/IAI.69.6.3744-3754.2001
34. Cornelis P, Wei Q, Andrews SC, Vinckx T. Iron homeostasis and management of oxidative stress response in bacteria. Metallomics. (2011) 3:540. doi: 10.1039/c1mt00022e
35. Anjem A, Varghese S, Imlay JA. Manganese import is a key element of the OxyR response to hydrogen peroxide in Escherichia coli. Mol Microbiol. (2009) 72:844–58. doi: 10.1111/j.1365-2958.2009.06699.x
36. Strohmeier Gort A, Ferber DM, Imlay JA. The regulation and role of the periplasmic copper, zinc superoxide dismutase of Escherichia coli. Mol Microbiol. (1999) 32:179–91. doi: 10.1046/j.1365-2958.1999.01343.x
37. Pomposiello PJ, Demple B. Global adjustment of microbial physiology during free radical stress. Adv Microb Physiol. (2002) 46:319–41. doi: 10.1016/S0065-2911(02)46007-9
38. Saenkham P, Utamapongchai S, Vattanaviboon P, Mongkolsuk S. Agrobacterium tumefaciens iron superoxide dismutases have protective roles against singlet oxygen toxicity generated from illuminated Rose Bengal. FEMS Microbiol Lett. (2008) 289:97–103. doi: 10.1111/j.1574-6968.2008.01382.x
39. Liu T, Hu X, Wang Y, Meng L, Zhou Y, Zhang J, et al. Triazine-based covalent organic frameworks for photodynamic inactivation of bacteria as type-II photosensitizers. J Photochem Photobiol B Biol. (2017) 175:156–162. doi: 10.1016/j.jphotobiol.2017.07.013
40. Sun YK, Eun JK, Park JW. Control of singlet oxygen-induced oxidative damage in Escherichia coli. J Biochem Mol Biol. (2002) 35:353–7. doi: 10.5483/bmbrep.2002.35.4.353
41. Hendiani S, Pornour M, Kashef N. Quorum-sensing-regulated virulence factors in Pseudomonas aeruginosa are affected by sub-lethal photodynamic inactivation. Photodiagnosis Photodyn Ther. (2019) doi: 10.1016/j.pdpdt.2019.02.010
42. Park Y, Kim H, Choi S. The role of helicobacter pylori's fur protein in the oxidative stress induced by photodynamic therapy. Korean J Microbiol. (2011) 47:124–9.
43. Faraj Tabrizi P, Wennige S, Berneburg M, Maisch T. Susceptibility of sodA- and sodB-deficient: Escherichia coli mutant towards antimicrobial photodynamic inactivation via the type I-mechanism of action. Photochem Photobiol Sci. (2018) 17:352–62. doi: 10.1039/c7pp00370f
44. Martin JP, Logsdon N. Oxygen radicals are generated by dye-mediated intracellular photooxidations: a role for superoxide in photodynamic effects. Arch Biochem Biophys. (1987) 256:39–49. doi: 10.1016/0003-9861(87)90423-1
45. Misba L, Khan AU. Enhanced photodynamic therapy using light fractionation against Streptococcus mutans biofilm: type I and type II mechanism. Future Microbiol. (2018) 13:437–454. doi: 10.2217/fmb-2017-0207
46. Buchovec I, Lukseviciute V, Kokstaite R, Labeikyte D, Kaziukonyte L, Luksiene Z. Inactivation of Gram (–) bacteria Salmonella enterica by chlorophyllin-based photosensitization: mechanism of action and new strategies to enhance the inactivation efficiency. J Photochem Photobiol B Biol. (2017) 172:1–10. doi: 10.1016/j.jphotobiol.2017.05.008
47. Dosselli R, Millioni R, Puricelli L, Tessari P, Arrigoni G, Franchin C, et al. Molecular targets of antimicrobial photodynamic therapy identified by a proteomic approach. J Proteomics. (2012) 77:329–43. doi: 10.1016/j.jprot.2012.09.007
48. Orlandi VT, Martegani E, Bolognese F. Catalase A is involved in the response to photooxidative stress in Pseudomonas aeruginosa. Photodiagnosis Photodyn Ther. (2018) 22:233–40. doi: 10.1016/j.pdpdt.2018.04.016
49. Khan S, P MR, Rizvi A, Alam MM, Rizvi M, Naseem I. ROS mediated antibacterial activity of photoilluminated riboflavin: a photodynamic mechanism against nosocomial infections. Toxicol Reports. (2019) 6:136–42. doi: 10.1016/j.toxrep.2019.01.003
50. Xu Z, Wang X, Liu X, Cui Z, Yang X, Yeung KWK, et al. Tannic Acid/Fe3+/Ag nanofilm exhibiting superior photodynamic and physical antibacterial activity. ACS Appl Mater Interfaces. (2017) 9:39657–71. doi: 10.1021/acsami.7b10818
51. Nakonieczna J, Michta E, Rybicka M, Grinholc M, Gwizdek-Wiśniewska A, Bielawski KP. Superoxide dismutase is upregulated in Staphylococcus aureus following protoporphyrin-mediated photodynamic inactivation and does not directly influence the response to photodynamic treatment. BMC Microbiol. (2010) 10:323. doi: 10.1186/1471-2180-10-323
52. Recsei P, Kreiswirth B, O'Reilly M, Schlievert P, Gruss A, Novick RP. Regulation of exoprotein gene expression in Staphylococcus aureus by agr. Mol Gen Genet MGG. (1986) 202:58–61. doi: 10.1007/BF00330517
53. Roux A, Todd DA, Velazquez JV, Cech NB, Sonenshein AL. CodY-Mediated regulation of the Staphylococcus aureus Agr system integrates nutritional and population density signals. J Bacteriol. (2014) 196:1184–96. doi: 10.1128/JB.00128-13
54. Park HJ Moon YH Yoon HE Park YM Yoon JH Bang IS. Agr function is upregulated by photodynamic therapy for Staphylococcus aureus and is related to resistance to photodynamic therapy. Microbiol Immunol. (2013) 57:547–52. doi: 10.1111/1348-0421.12070
55. Grinholc M, Nakonieczna J, Negri A, Rapacka-Zdonczyk A, Motyka A, Fila G, et al. The agr function and polymorphism: impact on Staphylococcus aureus susceptibility to photoinactivation. J Photochem Photobiol B Biol. (2013) 129:100–7. doi: 10.1016/j.jphotobiol.2013.10.006
56. Gándara L, Mamone L, Dotto C, Buzzola F, Casas A. Sae regulator factor impairs the response to photodynamic inactivation mediated by Toluidine blue in Staphylococcus aureus. Photodiagnosis Photodyn Ther. (2016) 16:136–41. doi: 10.1016/j.pdpdt.2016.09.005
57. Cheung AL, Koomey JM, Butler CA, Projan SJ, Fischetti VA. Regulation of exoprotein expression in Staphylococcus aureus by a locus (sar) distinct from agr. Proc Natl Acad Sci USA. (1992) 89:6462–66. doi: 10.1073/pnas.89.14.6462
58. Chien Y, Manna AC, Cheung AL. SarA level is a determinant of agr activation in Staphylococcus aureus. Mol Microbiol. (1998) 30:991–1001. doi: 10.1046/j.1365-2958.1998.01126.x
59. Manna AC, Bayer MG, Cheung AL. Transcriptional analysis of different promoters in the sar locus in Staphylococcus aureus. J Bacteriol. (1998) 180:3828–36. doi: 10.1128/JB.180.15.3828-3836.1998
60. Chan PF, Foster SJ. Role of SarA in virulence determinant production and environmental signal transduction in Staphylococcus aureus. J Bacteriol. (1998) 180:6232–41. doi: 10.1128/JB.180.23.6232-6241.1998
61. Dunman PM, Murphy E, Haney S, Palacios D, Tucker-Kellogg G, Wu S, et al. Transcription profiling-based identification ofStaphylococcus aureus genes regulated by the agrand/or sarA loci. J Bacteriol. (2001) 183:7341–53. doi: 10.1128/JB.183.24.7341-7353.2001
62. Ballal A, Manna AC. Regulation of superoxide dismutase (sod) genes by sara in Staphylococcus aureus. J Bacteriol. (2009) 191:3301–10. doi: 10.1128/JB.01496-08
63. Chen PR, Brugarolas P, He C. Redox signaling in human pathogens. Antioxidants Redox Signal. (2011) 14:1107–18. doi: 10.1089/ars.2010.3374
64. Gertz S, Engelmann S, Schmid R, Ziebandt A-K, Tischer K, Scharf C, et al. Characterization of the ςB Regulon inStaphylococcus aureus. J Bacteriol. (2000) 182:6983–6991. doi: 10.1128/JB.182.24.6983-6991.2000
65. Senn MM, Giachino P, Homerova D, Steinhuber A, Strassner J, Kormanec J, et al. Molecular analysis and organization of the σB operon in Staphylococcus aureus. J Bacteriol. (2005) 187:8006–19. doi: 10.1128/JB.187.23.8006-8019.2005
66. Bischoff M, Dunman P, Kormanec J, Macapagal D, Murphy E, Mounts W, et al. Microarray-based analysis of the Staphylococcus aureus σB regulon. J Bacteriol. (2004) 186:4085–99. doi: 10.1128/JB.186.13.4085-4099.2004
67. Guldimann C, Boor KJ, Wiedmann M, Guariglia-Oropeza V. Resilience in the face of uncertainty: sigma factor b fine-tunes gene expression to support homeostasis in gram-positive bacteria. Appl Environ Microbiol. (2016) 82:4456–69. doi: 10.1128/AEM.00714-16
68. Kossakowska-Zwierucho M, Kazmierkiewicz R, Bielawski KP, Nakonieczna J. Factors Determining Staphylococcus aureus Susceptibility to photoantimicrobial chemotherapy: rsbu activity, staphyloxanthin level, and membrane fluidity. Front Microbiol. (2016) 7:1–14. doi: 10.3389/fmicb.2016.01141
69. Baharoglu Z, Mazel D. SOS, the formidable strategy of bacteria against aggressions. FEMS Microbiol Rev. (2014) 38:1126–45. doi: 10.1111/1574-6976.12077
71. Michel B. After 30 years of study, the bacterial sos response still surprises us. PLoS Biol. (2005) 3:e255. doi: 10.1371/journal.pbio.0030255
72. Cirz RT, Jones MB, Gingles NA, Minogue TD, Jarrahi B, Peterson SN, et al. Complete and SOS-mediated response of Staphylococcus aureus to the antibiotic ciprofloxacin. J Bacteriol. (2007) 189:531–9. doi: 10.1128/JB.01464-06
73. Courcelle J, Khodursky A, Peter B, Brown PO, Hanawalt PC. Comparative gene expression profiles following UV exposure in wild-type and SOS-deficient Escherichia coli. Genetics. (2001) 158:41–64.
74. Grinholc M, Rodziewicz A, Forys K, Rapacka-Zdonczyk A, Kawiak A, Domachowska A, et al. Fine-tuning recA expression in Staphylococcus aureus for antimicrobial photoinactivation: importance of photo-induced DNA damage in the photoinactivation mechanism. Appl Microbiol Biotechnol. (2015) 99:9161–76. doi: 10.1007/s00253-015-6863-z
75. Mesak LR, Miao V, Davies J. Effects of subinhibitory concentrations of antibiotics on SOS and DNA repair gene expression in Staphylococcus aureus. Antimicrob Agents Chemother. (2008) 52:3394–7. doi: 10.1128/AAC.01599-07
76. George S, Kishen A. Influence of photosensitizer solvent on the mechanisms of photoactivated killing of Enterococcus faecalis. Photochem Photobiol. (2008) 84:734–40. doi: 10.1111/j.1751-1097.2007.00244.x
77. Caminos DA, Spesia MB, Pons P, Durantini EN. Mechanisms of Escherichia coli photodynamic inactivation by an amphiphilic tricationic porphyrin and 5,10,15,20-tetra(4-N,N,N-trimethylammoniumphenyl) porphyrin. Photochem Photobiol Sci. (2008) 7:1071. doi: 10.1039/b804965c
78. Spesia MB, Caminos DA, Pons P, Durantini EN. Mechanistic insight of the photodynamic inactivation of Escherichia coli by a tetracationic zinc(II) phthalocyanine derivative. Photodiagnosis Photodyn Ther. (2009) 6:52–61. doi: 10.1016/j.pdpdt.2009.01.003
79. Walker GC. Mutagenesis and inducible responses to deoxyribonucleic acid damage in Escherichia coli. Microbiol Rev. (1984) 48:60–93. doi: 10.1128/MMBR.48.1.60-93.1984
80. Nohmi T, Battista JR, Dodson LA, Walker GC. RecA-mediated cleavage activates UmuD for mutagenesis: mechanistic relationship between transcriptional derepression and posttranslational activation. Proc Natl Acad Sci USA. (1988) 85:1816–20. doi: 10.1073/pnas.85.6.1816
81. Pagès V, Fuchs RP. Uncoupling of leading- and lagging-strand DNA replication during lesion bypass in vivo. Science (80-). (2003) 300:1300–3. doi: 10.1126/science.1083964
82. Friedberg EC, Wagner R, Radman M. Specialized DNA polymerases, cellular survival, and the genesis of mutations. Science (80-). (2002) 296:1627–30. doi: 10.1126/science.1070236
83. Grinholc M, Szramka B, Kurlenda J, Graczyk A, Bielawski KP. Bactericidal effect of photodynamic inactivation against methicillin-resistant and methicillin-susceptible Staphylococcus aureus is strain-dependent. J Photochem Photobiol B Biol. (2008) 90:57–63. doi: 10.1016/j.jphotobiol.2007.11.002
84. Grinholc M, Zawacka-Pankau J, Gwizdek-Wiśniewska A, Bielawski KP. Evaluation of the role of the pharmacological inhibition of Staphylococcus aureus multidrug resistance pumps and the variable levels of the uptake of the sensitizer in the strain-dependent response of Staphylococcus aureus to PPArg2-based photodynamic Ina. Photochem Photobiol. (2010) 86:1118–26. doi: 10.1111/j.1751-1097.2010.00772.x
85. Grinholc M, Rapacka-Zdonczyk A, Rybak B, Szabados F, Bielawski KP. Multiresistant strains are as susceptible to photodynamic inactivation as their naïve counterparts: protoporphyrin ix-mediated photoinactivation reveals differences between methicillin-resistant and methicillin-sensitive Staphylococcus aureus Strai. Photomed Laser Surg. (2014) 32:121–9. doi: 10.1089/pho.2013.3663
86. Rapacka-Zdonczyk A, Larsen AR, Empel J, Patel A, Grinholc M. Association between susceptibility to photodynamic oxidation and the genetic background of Staphylococcus aureus. Eur J Clin Microbiol Infect Dis. (2014) 33:577–86. doi: 10.1007/s10096-013-1987-5
87. Nakonechny F, Nisnevitch M., Nitzan Y, Firer MA. New techniques in antimicrobial photodynamic therapy: scope of application and overcoming drug resistance in nosocomial infections. In: Méndez-Vilas A, editor. Science Against Microbial Pathogens: Communicating Current Research and Technological Advances, Vol. 1. Formatex Research Center (2011). p. 684–91. Available online at: http://www.formatex.info/microbiology3/book/684-691.pdf
88. Epand RM, Epand RF. Lipid domains in bacterial membranes and the action of antimicrobial agents. Biochim Biophys Acta - Biomembr. (2009) 1788:289–94. doi: 10.1016/j.bbamem.2008.08.023
89. Wainwright M. Photodynamic antimicrobial chemotherapy (PACT). J Antimicrob Chemother. (1998) 42:13–28. doi: 10.1093/jac/42.1.13
90. Cabiscol E, Tamarit J, Ros J. Oxidative stress in bacteria and protein damage by reactive oxygen species. Int Microbiol. (2000) 3:3–8. doi: 10.2436/im.v3i1.9235
91. Davies MJ. Singlet oxygen-mediated damage to proteins and its consequences. Biochem Biophys Res Commun. (2003) 305:761–70. doi: 10.1016/S0006-291X(03)00817-9
92. Dharmaraja AT. Role of Reactive Oxygen Species (ROS) in therapeutics and drug resistance in cancer and bacteria. J Med Chem. (2017) 60:3221–40. doi: 10.1021/acs.jmedchem.6b01243
93. Bhatti M, MacRobert A, Henderson B, Wilson M. Exposure of Porphyromonas gingivalis to Red Light in the Presence of the Light-Activated Antimicrobial Agent Toluidine Blue Decreases Membrane Fluidity. Curr Microbiol. (2002) 45:118–122. doi: 10.1007/s00284-001-0095-4
94. Mishra NN, Liu GY, Yeaman MR, Nast CC, Proctor RA, McKinnell J, et al. Carotenoid-related alteration of cell membrane fluidity impacts Staphylococcus aureus susceptibility to host defense peptides. Antimicrob Agents Chemother. (2011) 55:526–31. doi: 10.1128/AAC.00680-10
95. Hall JW, Yang J, Guo H, Ji Y. The Staphylococcus aureus AirSR two-component system mediates reactive oxygen species resistance via transcriptional regulation of staphyloxanthin production. Infect Immun. (2017) 85:1–12. doi: 10.1128/IAI.00838-16
96. Clauditz A, Resch A, Wieland K-P, Peschel A, Götz F. Staphyloxanthin plays a role in the fitness of Staphylococcus aureus and its ability to cope with oxidative stress. Infect Immun. (2006) 74:4950–3. doi: 10.1128/IAI.00204-06
97. Winkler K, Simon C, Finke M, Bleses K, Birke M, Szentmáry N, et al. Photodynamic inactivation of multidrug-resistant Staphylococcus aureus by chlorin e6 and red light (λ = 670 nm). J Photochem Photobiol B Biol. (2016) 162:340–7. doi: 10.1016/j.jphotobiol.2016.07.007
98. Maraccini PA, Ferguson DM, Boehm AB. Diurnal variation in enterococcus species composition in polluted ocean water and a potential role for the enterococcal carotenoid in protection against photoinactivation. Appl Environ Microbiol. (2012) 78:305–10. doi: 10.1128/AEM.06821-11
99. Chiodaroli L, Tolker-Nielsen T, Orlandi VT, Bolognese F, Barbieri P. Pigments influence the tolerance of Pseudomonas aeruginosa PAO1 to photodynamically induced oxidative stress. Microbiology. (2015) 161:2298–309. doi: 10.1099/mic.0.000193
100. Sabino CP, Wainwright M, dos Anjos C, Sellera FP, Baptista MS, Lincopan N, et al. Inactivation kinetics and lethal dose analysis of antimicrobial blue light and photodynamic therapy. Photodiagnosis Photodyn Ther. (2019) 28:186–91. doi: 10.1016/j.pdpdt.2019.08.022
101. Yoshida A, Sasaki H, Toyama T, Araki M, Fujioka J, Tsukiyama K, et al. Antimicrobial effect of blue light using Porphyromonas gingivalis pigment. Sci Rep. (2017) 7:5225. doi: 10.1038/s41598-017-05706-1
102. Soukos NS, Som S, Abernethy AD, Ruggiero K, Dunham J, Lee C, et al. Phototargeting Oral Black-Pigmented Bacteria. Antimicrob Agents Chemother. (2005) 49:1391–96. doi: 10.1128/AAC.49.4.1391-1396.2005
103. Fontana CR, Song X, Polymeri A, Goodson JM, Wang X, Soukos NS. The effect of blue light on periodontal biofilm growth in vitro. Lasers Med Sci. (2015) 30:2077–86. doi: 10.1007/s10103-015-1724-7
104. Smalley JW, Silver J, Birss AJ, Withnall R, Titler PJ. The haem pigment of the oral anaerobes Prevotella nigrescens and Prevotella intermedia is composed of iron(III) protoporphyrin IX in the monomeric form. Microbiology. (2003) 149:1711–18. doi: 10.1099/mic.0.26258-0
105. Maisch T, Baier J, Franz B, Maier M, Landthaler M, Szeimies R-M, et al. The role of singlet oxygen and oxygen concentration in photodynamic inactivation of bacteria. Proc Natl Acad Sci USA. (2007) 104:7223–28. doi: 10.1073/pnas.0611328104
106. Preuß A, Zeugner L, Hackbarth S, Faustino MAF, Neves MGPMS, Cavaleiro JAS, et al. Photoinactivation of Escherichia coli (SURE2) without intracellular uptake of the photosensitizer. J Appl Microbiol. (2013) 114:36–43. doi: 10.1111/jam.12018
107. Gollmer A, Felgentraeger A, Maisch T, Flors C. Real-time imaging of photodynamic action in bacteria. J Biophotonics. (2017) 10:264–70. doi: 10.1002/jbio.201500259
108. Ragàs X, Agut M, Nonell S. Singlet oxygen in Escherichia coli: New insights for antimicrobial photodynamic therapy. Free Radic Biol Med. (2010) 49:770–6. doi: 10.1016/j.freeradbiomed.2010.05.027
109. Denyer SP, Maillard J. Cellular impermeability and uptake of biocides and antibiotics in Gram-negative bacteria. Soc Appl Microbiol. (2002) 92:35S−45S. doi: 10.1046/j.1365-2672.92.5s1.19.x
110. George S, Hamblin MR, Kishen A. Uptake pathways of anionic and cationic photosensitizers into bacteria. Photochem Photobiol Sci. (2009) 8:788. doi: 10.1039/b809624d
111. Malik Z, Ladan H. Photodynamic Inactivation of Gram-Negative Problems and Possible Solutions Bacteria. (1990). p. 262–6.
112. Bertoloni G, Rossi F, Valduga G, Jori G, Ali H, van Lier JE. Photosensitizing activity of water- and lipid-soluble phthalocyanines on prokaryotic and eukaryotic microbial cells. Microbios. (1992) 71:33–46.
113. Nitzan Y, Gutterman M, Malik Z, Ehrenberg B. Inactivation of gram-negative bacteria by photosensitized porphyrins. Photochem Photobiol. (1992) 55:89–96. doi: 10.1111/j.1751-1097.1992.tb04213.x
114. Vaara M. Agents that increase the permeability of the outer membrane. Microbiol Rev. (1992) 56:395–411.
115. Wiese A, Mu¨nstermann M, Gutsmann T, Lindner B, Kawahara K, Za¨hringer U. Molecular mechanisms of polymyxin b-membrane interactions: direct correlation between surface charge density and self-promoted transport. J Membr Biol. (1998) 138:127–38.
116. Usacheva MN, Teichert MC, Biel MA. The interaction of lipopolysaccharides with phenothiazine dyes. Lasers Surg Med. (2003) 33:311–9. doi: 10.1002/lsm.10226
117. Usacheva MN, Teichert MC, Sievert CE, Biel MA. Effect of Ca+ on the photobactericidal efficacy of methylene blue and toluidine blue against gram-negative bacteria and the dye affinity for lipopolysaccharides. Lasers Surg Med. (2006) 38:946–54. doi: 10.1002/lsm.20400
118. Rolim JPML, De-Melo MAS, Guedes SF, Albuquerque-Filho FB, de Souza JR, Nogueira NAP, et al. The antimicrobial activity of photodynamic therapy against Streptococcus mutans using different photosensitizers. J Photochem Photobiol B Biol. (2012) 106:40–6. doi: 10.1016/j.jphotobiol.2011.10.001
119. Usacheva MN, Teichert MC, Biel MA. The role of the methylene blue and toluidine blue monomers and dimers in the photoinactivation of bacteria. J Photochem Photobiol B Biol. (2003) 71:87–98. doi: 10.1016/j.jphotobiol.2003.06.002
120. Usacheva MN, Teichert MC, Usachev YM, Sievert CE, Biel MA. Interaction of the photobactericides methylene blue and toluidine blue with a fluorophore inPseudomonas aeruginosa cells. Lasers Surg Med. (2008) 40:55–61. doi: 10.1002/lsm.20593
121. Minnock A, Vernon DI, Schofield J, Griffiths J, Parish JH, Brown SB. Mechanism of uptake of a cationic water-soluble pyridinium zinc phthalocyanine across the outer membrane ofEscherichia coli. Antimicrob Agents Chemother. (2000) 44:522–7. doi: 10.1128/AAC.44.3.522-527.2000
122. Lambrechts SAG, Aalders MCG, Langeveld-Klerks DH, Khayali Y, Lagerberg JWM. Effect of monovalent and divalent cations on the photoinactivation of bacteria with meso-substituted cationic porphyrins. Photochem Photobiol. (2004) 79:297. doi: 10.1562/SA-03-15.1
123. Merchat M, Bertolini G, Giacomini P, Villanueva A, Jori G. Meso-substituted cationic porphyrins as efficient photosensitizers of Gram-positive and Gram-negative bacteria. J Photochem Photobiol. (1996) 32:153–157.
124. Merchat M, Spikes JD, Bertoloni G, Jori G. Studies on the mechanism of bacteria photosensitization by meso-substituted cationic porphyrins. J Photochem Photobiol. (1996) 35:149–57.
125. Alves E, Costa L, Carvalho CM, Tomé JP, Faustino MA, Neves MG, et al. Charge effect on the photoinactivation of Gram-negative and Gram-positive bacteria by cationic meso-substituted porphyrins. BMC Microbiol. (2009) 9:70. doi: 10.1186/1471-2180-9-70
126. Caminos DA, Spesia MB, Durantini EN. Photodynamic inactivation of Escherichia coli by novel meso-substituted porphyrins by 4-(3-N,N,N-trimethylammoniumpropoxy)phenyl and 4-(trifluoromethyl)phenyl groups. Photochem Photobiol Sci. (2006) 5:56–65. doi: 10.1039/B513511G
127. Banfi S, Caruso E, Buccafurni L, Battini V, Zazzaron S, Barbieri P, et al. Antibacterial activity of tetraaryl-porphyrin photosensitizers: an in vitro study on Gram negative and Gram positive bacteria. J Photochem Photobiol B Biol. (2006) 85:28–38. doi: 10.1016/j.jphotobiol.2006.04.003
128. Costa-Riu N, Burkovski A, Krämer R, Benz R. PorA represents the major cell wall channel of the gram-positive bacterium Corynebacterium glutamicum. J Bacteriol. (2003) 185:4779–86. doi: 10.1128/JB.185.16.4779-4786.2003
129. Rieβ FG, Elflein M, Benk M, Schiffler B, Benz R, Garton N, et al. The cell wall of the pathogenic bacterium rhodococcus equi contains two channel-forming proteins with different properties. J Bacteriol. (2003) 185:2952–60. doi: 10.1128/JB.185.9.2952-2960.2003
130. Choby JE, Skaar EP. Heme synthesis and acquisition in bacterial pathogens. J Mol Biol. (2016) 428:3408–28. doi: 10.1016/j.jmb.2016.03.018
131. Anzaldi LL, Skaar EP. Overcoming the heme paradox: heme toxicity and tolerance in bacterial pathogens. Infect Immun. (2010) 78:4977–89. doi: 10.1128/IAI.00613-10
132. Cassat JE, Skaar EP. Iron in infection and immunity. Cell Host Microbe. (2013) 13:509–19. doi: 10.1016/j.chom.2013.04.010
133. Hammer ND, Skaar EP. Molecular mechanisms of Staphylococcus aureus Iron acquisition. Annu Rev Microbiol. (2011) 65:129–47. doi: 10.1146/annurev-micro-090110-102851
134. Hammer ND, Reniere ML, Cassat JE, Zhang Y, Hirsch AO, Indriati Hood M, et al. Two heme-dependent terminal oxidases power Staphylococcus aureus organ-specific colonization of the vertebrate host. MBio. (2013) 4:1–9. doi: 10.1128/mBio.00241-13
135. Wakeman CA, Stauff DL, Zhang Y, Skaar EP. Differential activation of Staphylococcus aureus heme detoxification machinery by heme analogues. J Bacteriol. (2014) 196:1335–42. doi: 10.1128/JB.01067-13
136. Nakonieczna J, Kossakowska-Zwierucho M, Filipiak M, Hewelt-Belka W, Grinholc M, Bielawski KP. Photoinactivation of Staphylococcus aureus using protoporphyrin IX: the role of haem-regulated transporter HrtA. Appl Microbiol Biotechnol. (2016) 100:1393–405. doi: 10.1007/s00253-015-7145-5
137. Stojiljkovic I, Kumar V, Srinivasan N. Non-iron metalloporphyrins: potent antibacterial compounds that exploit haem/Hb uptake systems of pathogenic bacteria. Mol Microbiol. (1999) 31:429–42. doi: 10.1046/j.1365-2958.1999.01175.x
138. Moriwaki Y, Caaveiro J, Tanaka Y, Tsutumi H, Hamachi I, Tsumoto K. Molecular basis of recognition of antibacterial porphyrins by heme- transporter IsdH-NEAT3 of Staphylococcus aureus. Acs Biochem. (2011) 50:7311–20. doi: 10.1021/bi200493h
139. Morales-de-Echegaray AV, Maltais TR, Lin L, Younis W, Kadasala NR, Seleem MN, et al. Rapid uptake and photodynamic inactivation of staphylococci by Ga(III)-protoporphyrin IX. ACS Infect Dis. (2018) 4:1564–73. doi: 10.1021/acsinfecdis.8b00125
140. Tegos GP, Hamblin MR, Tegos GP, Hamblin MR. Phenothiazinium antimicrobial photosensitizers are substrates of bacterial multidrug resistance pumps phenothiazinium antimicrobial photosensitizers are substrates of bacterial multidrug resistance pumps. Antimicrob Agents Chemother. (2006) 50:196–203. doi: 10.1128/AAC.50.1.196-203.2006
141. Tegos GP, Masago K, Aziz F, Higginbotham A, Stermitz FR, Hamblin MR. Inhibitors of bacterial multidrug efflux pumps potentiate. Antimicrob Agents Chemother. (2008) 52:3202–9. doi: 10.1128/AAC.00006-08
142. Rineh A, Dolla NK, Ball AR, Magana M, Bremner JB, Hamblin MR, et al. Attaching the NorA Efflux Pump Inhibitor INF55 to methylene blue enhances antimicrobial photodynamic inactivation of methicillin-resistant Staphylococcus aureus in vitro and in vivo. ACS Infect Dis. (2018) 3:756–6. doi: 10.1021/acsinfecdis.7b00095.Details
143. Rineh A, Bremner JB, Hamblin MR, Ball AR. Attaching NorA efflux pump inhibitors to methylene blue enhances antimicrobial photodynamic inactivation of Escherichia coli and Acinetobacter baumannii in vitro and in vivo. Bioorganic Med Chem Lett. (2018) 28:2736–2740. doi: 10.1016/j.bmcl.2018.02.041
144. Nitzan Y, Balzam-sudakevitz A, Ashkenazi H. Photochemistry eradication of acinetobacter baumannii by photosensitized agents in vitro. J Photochem Photobiol B. (1998) 42:211–8.
145. Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. (2002) 415:389–95. doi: 10.1038/415389a
146. Nakonieczna J, Wolnikowska K, Ogonowska P, Neubauer D, Bernat A, Kamysz W. Rose bengal-mediated photoinactivation of multidrug resistant pseudomonas aeruginosa is enhanced in the presence of antimicrobial peptides. Front Microbiol. (2018) 9:1–15. doi: 10.3389/fmicb.2018.01949
147. Freitas LM De, Lorenzón EN, Santos-filho NA, Henrique L, Zago DP, Uliana MP, et al. Antimicrobial Photodynamic therapy enhanced by the peptide. Sci Rep. (2018) 8:1–15. doi: 10.1038/s41598-018-22687-x
148. George S, Kishen A. Photophysical, photochemical, and photobiological characterization of methylene blue formulations for light-activated root canal disinfection. J Biomed Opt. (2007) 12:034029. doi: 10.1117/1.2745982
149. Gad F, Zahra T, Hasan T, Hamblin MR. Effects of growth phase and extracellular slime on photodynamic inactivation of gram-positive pathogenic bacteria. Antimicrob Agents Chemother. (2004) 48:2173–8. doi: 10.1128/AAC.48.6.2173-2178.2004
150. Kossakowska M, Nakonieczna J, Kawiak A, Kurlenda J, Bielawski KP, Grinholc M. Discovering the mechanisms of strain-dependent response of Staphylococcus aureus to photoinactivation: oxidative stress toleration, endogenous porphyrin level and strain's virulence. Photodiagnosis Photodyn Ther. (2013) 10:348–55. doi: 10.1016/j.pdpdt.2013.02.004
151. Castro KADF, Moura NMM, Figueira F, Ferreira RI, Simões MMQ, Cavaleiro JAS, et al. New materials based on cationic porphyrins conjugated to chitosan or titanium dioxide: Synthesis, characterization and antimicrobial efficacy. Int J Mol Sci. (2019) 20:2522. doi: 10.3390/ijms20102522
Keywords: membrane fluidity, staphyloxanthin, antioxidant enzymes, transcription regulators, reactive oxygen species, DNA photodamage
Citation: Rapacka-Zdończyk A, Woźniak A, Michalska K, Pierański M, Ogonowska P, Grinholc M and Nakonieczna J (2021) Factors Determining the Susceptibility of Bacteria to Antibacterial Photodynamic Inactivation. Front. Med. 8:642609. doi: 10.3389/fmed.2021.642609
Received: 16 December 2020; Accepted: 12 April 2021;
Published: 12 May 2021.
Edited by:
Yolanda Gilaberte, Hospital Universitario Miguel Servet, SpainReviewed by:
Natalia Mayumi Inada, University of São Paulo, BrazilReza Ghiladi, North Carolina State University, United States
Copyright © 2021 Rapacka-Zdończyk, Woźniak, Michalska, Pierański, Ogonowska, Grinholc and Nakonieczna. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Joanna Nakonieczna, joanna.nakonieczna@biotech.ug.edu.pl
 Aleksandra Rapacka-Zdończyk
Aleksandra Rapacka-Zdończyk Agata Woźniak
Agata Woźniak Klaudia Michalska
Klaudia Michalska Michał Pierański2
Michał Pierański2  Patrycja Ogonowska
Patrycja Ogonowska Mariusz Grinholc
Mariusz Grinholc Joanna Nakonieczna
Joanna Nakonieczna