A Systematic Review of Deep Learning Techniques for Tuberculosis Detection From Chest Radiograph
- Computer Science Discipline, School of Mathematics, Statistics and Computer Science, University of KwaZulu-Natal, Durban, South Africa
The high mortality rate in Tuberculosis (TB) burden regions has increased significantly in the last decades. Despite the possibility of treatment for TB, high burden regions still suffer inadequate screening tools, which result in diagnostic delay and misdiagnosis. These challenges have led to the development of Computer-Aided Diagnostic (CAD) system to detect TB automatically. There are several ways of screening for TB, but Chest X-Ray (CXR) is more prominent and recommended due to its high sensitivity in detecting lung abnormalities. This paper presents the results of a systematic review based on PRISMA procedures that investigate state-of-the-art Deep Learning techniques for screening pulmonary abnormalities related to TB. The systematic review was conducted using an extensive selection of scientific databases as reference sources that grant access to distinctive articles in the field. Four scientific databases were searched to retrieve related articles. Inclusion and exclusion criteria were defined and applied to each article to determine those included in the study. Out of the 489 articles retrieved, 62 were included. Based on the findings in this review, we conclude that CAD systems are promising in tackling the challenges of the TB epidemic and made recommendations for improvement in future studies.
Introduction
Tuberculosis (TB) is ranked among the leading causes of death. About 10 million persons fell ill globally from TB infections in 2019 (1). TB is triggered by the Mycobacterium bacteria that usually affect the lungs (pulmonary) but sometimes affect other parts of the body (extrapulmonary) (2). Many TB patients lose their lives yearly due to diagnostic delay, misdiagnosis, and lack of appropriate treatments (3, 4). Although TB is a global challenge, the mortality rate is more prevalent in low and middle-income nations (5).
TB is certainly treatable if diagnosed early for appropriate treatment. Early diagnosis is essential for successful treatment, preventing further spread, and significantly reducing the mortality rate in line with the World Health Organization (WHO) End TB Strategy (1). The gold standard for TB screening is Sputum culture. However, posterior-anterior chest radiographs (CXR) are an effective technique with low-cost and moderately low radiation doses for screening lung abnormalities to achieve prompt results (6). CXR has been adequately employed in developed countries to analyze individuals exhibiting active TB symptoms. At the same time, its application is limited in developing countries where TB is most prevalent (7, 8). High TB burden regions lack the skilled and radiological expertise required to interpret CXR images adequately (9, 10).
In the last decades, several efforts have been made using Artificial Intelligence (AI) to develop a Computer-Aided Detection (CAD) system to advance automatic object/image recognition tasks and overcome the challenges of a skilled workforce. Machine Learning (ML) and Deep Learning (DL) are the predominant AI techniques employed to develop CAD systems for analyzing CXR images. Both techniques have had a significant impact, but the DL approach, such as Convolutional Neural Network (CNN), has become more prominent for analyzing different pulmonary abnormalities in the medical domain, most importantly in diagnosing TB. The application of an efficient classification tool is vital for improving the quality of diagnosis while reducing the time taken to analyze a large volume of CXRs (11). This endeavor is to achieve the global decline in TB incidence to about 5% annually compared to the current 2% yearly as part of the World Health Organization strategy to end TB (1).
The contribution of this systematic review is to present an extensive summary of the various state-of-the-art CAD system proposed in the literature for the classification of TB. Ultimately, only the CAD system developed using Deep Learning models is considered in this study detailing the diagnostic accuracy between 2017 and 2021. The rest of the paper is structured as follows: section Methodology presents the study methodology. The results are presented in section Results. Section Discussion presents the discussion, while the conclusion is expressed in section Conclusion and Recommendations.
Methodology
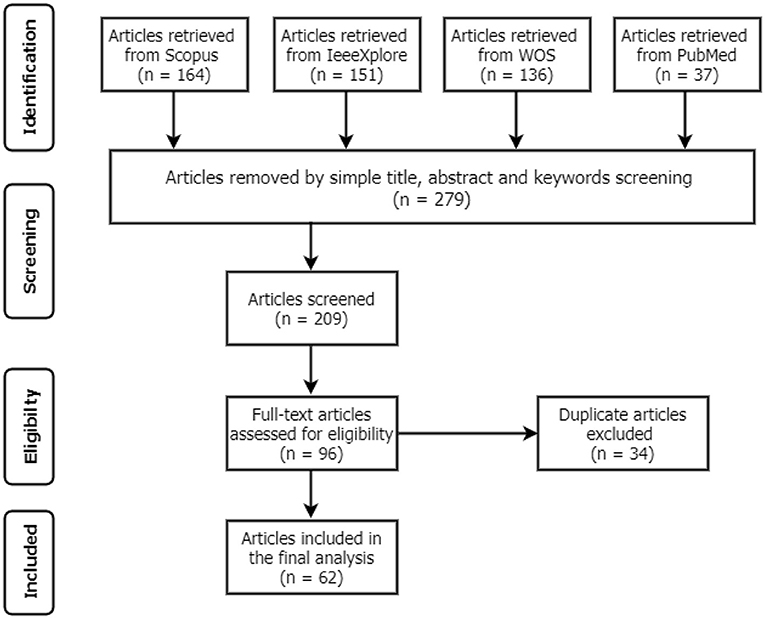
This systematic review aims to establish various CAD systems related to Tuberculosis diagnosis from CXR using DL techniques. The study followed the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) procedures (12) to identify the standards for inclusion and exclusion, as shown in Figure 1. These standards were formulated based on the present study objectives and the research questions. All articles that satisfied the following conditions were included and excluded if otherwise:
• Articles that considered only pulmonary tuberculosis disease.
• Employed at least one deep learning technique as a classifier
• CXR is the only medical imagine for screening tuberculosis.
• Articles published between January 2017 and September 2021.
• Articles are entirely written in English.
• Articles must be full text. All others, such as abstract, preprints are excluded.
Search Strategy
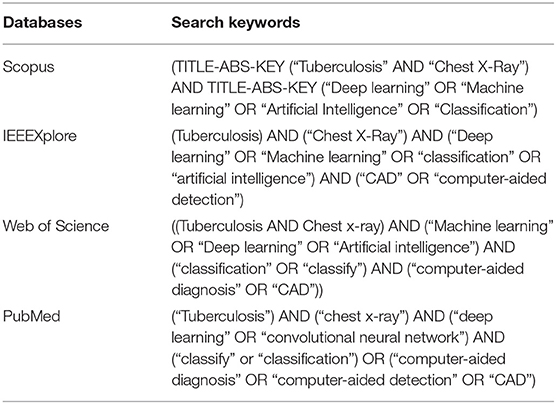
The search strategy was developed to identify relevant published articles in Scopus (https://www.scopus.com/), IEEEXplore (https://ieeexplore.ieee.org/), Web of Science (www.webofscience.com), and PubMed (https://www.ncbi.nlm.nih.gov/pubmed/). Searches were performed across the four databases using the following search keywords: “tuberculosis,” “chest x-ray,” “classification,” “artificial intelligence,” “computer-aided diagnosis,” “deep learning.” These keywords were used to form Boolean search strings according to searching standards on different databases. The configuration of keywords used to retrieve all relevant articles from each database search engine is presented in Table 1.
Study Selection
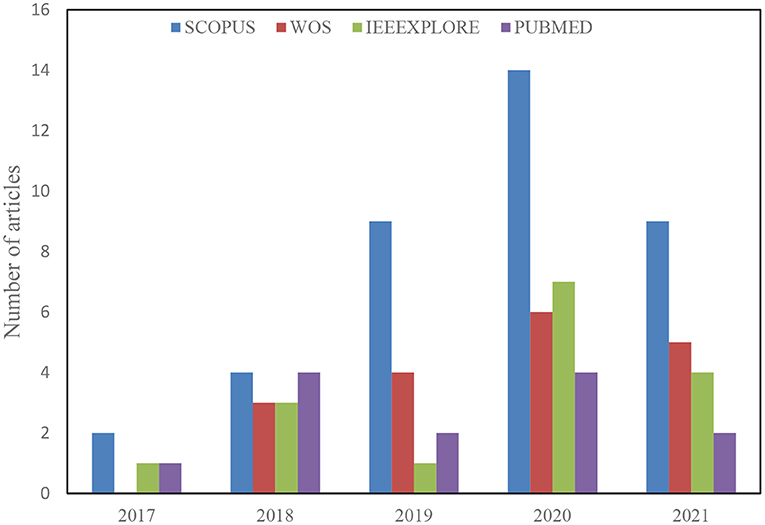
A total of 488 articles were retrieved from the initial search on all the databases. The next step then scans the article topics and keywords to identify highly related papers from the irrelevant ones. This step is followed by the overview reading of the abstract, methods, and Conclusion to further screen for relevant papers for full-text reading and better understanding. Thus, 209 articles were considered for the title and abstract screening, out of which 96 papers were found suitable for full-text reading, and a total of 62 were finally included in the analysis after removing 34 duplicates. The detailed structure for the study selection is presented in Figures 1, 2 shows the numbers of articles included per year and databases. It is necessary to note that some relevant articles might have been unintentionally omitted.
Data Extraction
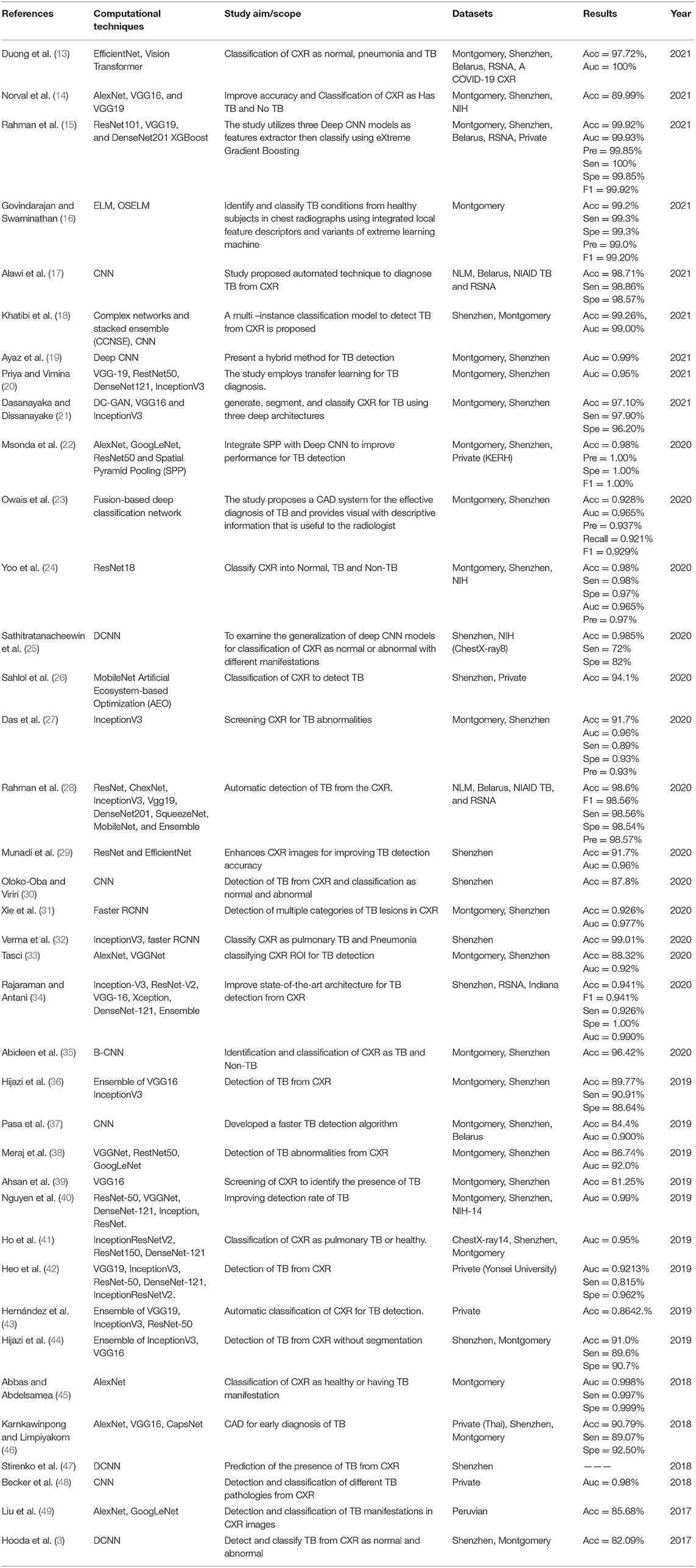
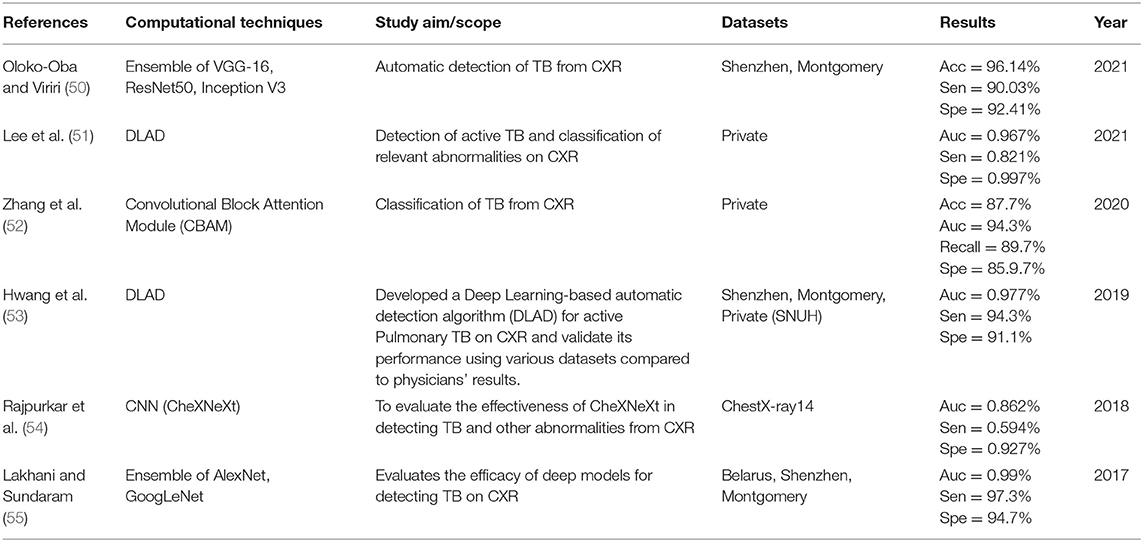
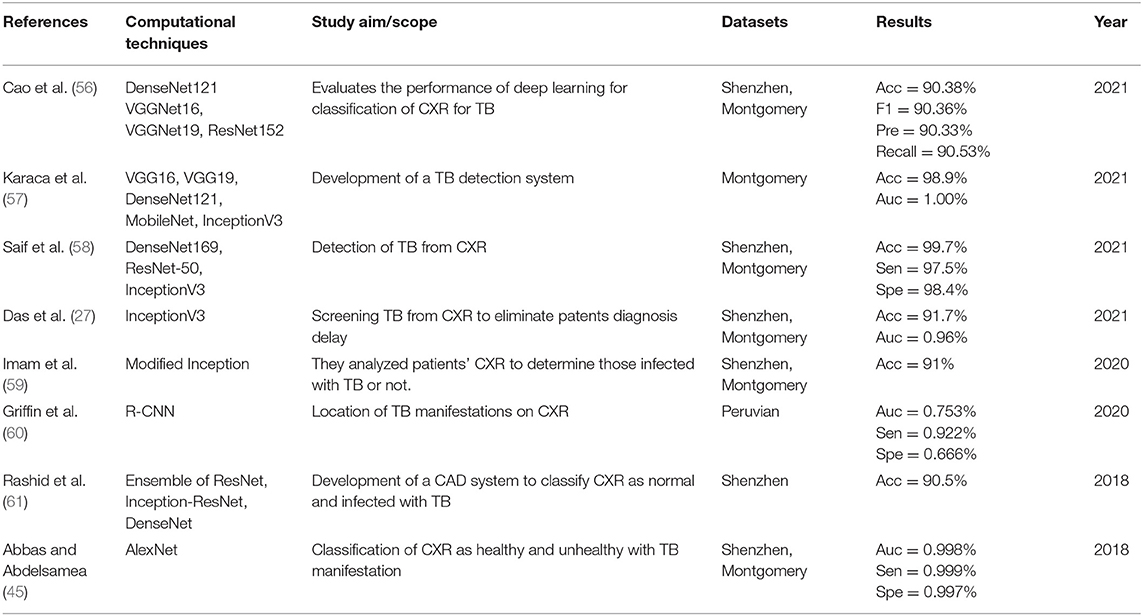
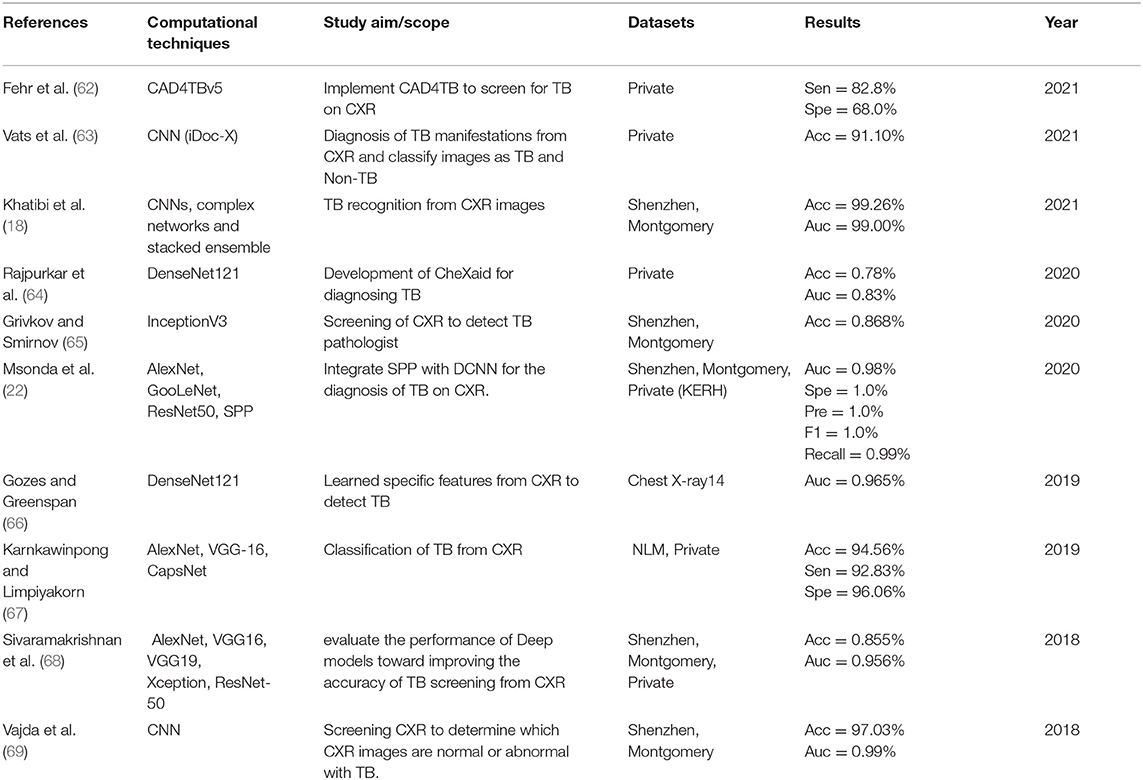
The details extracted from each article are presented in Tables 2–5. Each table represents the search results from the Scopus, IEEE Xplore, PubMed, and Web of Science databases. Data extracted include study aim and scope, computational techniques (DL models), CXR datasets, evaluation metrics/results achieved, and publication year. The database search shows that most IEEEXplore articles are conference proceedings and included in this systematic review, along with a few conference articles from springer and ACM. This is due to the articles' quality and contribution to the subject matter.
Results
This study reviewed computer-aided diagnosis systems articles to detect pulmonary TB from January 2017 until September 2021. It was observed from the articles that the development of CAD follows a standard framework involving four steps as follows: “pre-processing” is the first step which deals with cleaning up the CXR images by eliminating noise and enhancing for clarity. The second step is “segmentation” of a region of interest from the entire image, which is the lung field region in the case of CXR. The third step is the “Feature extraction,” where discriminative features are identified and selected for further analysis in the “classification” step, where the various images are categorized as normal or abnormal (infected) with TB. Several techniques such as handcraft, machine learning, and deep learning have been employed to diagnose TB, but DL has recorded more success in this regard; hence our interest was to analyze the CAD system based on one or more DL techniques as the classifier for TB detection.
The descriptive analysis of the results is presented in Tables 2–5. These tables show the computational technique, study scope, datasets, evaluation criteria, and results. Articles that utilized CXR as the only imaging modality and employed DL as the only computational technique for developing CAD are considered.
Imaging Modalities
Different screening procedures are used to confirm the presence of TB. Still, Chest Radiograph, otherwise referred to as Chest X-ray (CXR), is a radiograph tool used to detect abnormalities in the lungs and nearby structures. In recent years, CXR has remained a vital method for screening TB and other lung diseases and hence recommended by WHO due to its high sensitivity, wide availability, and relatively less expensive (70, 71).
Deep Learning Techniques
Many DL techniques have been used for screening, predicting, and diagnosing TB. In most classification algorithms, a dataset is required for training and testing with many samples of inputs and outputs to learn from. Model is developed using the training set to calculate how best to map examples of input data to a specific class label; then, the model validation is accomplished using the test set (72).
In Tables 2–5, only the results obtained from the test set are extracted. Some studies employed more the one DL technique to find the optimal results for diagnosing TB diseases. Only the best results are documented in this study if more than one accuracy is reported in an article.
Datasets
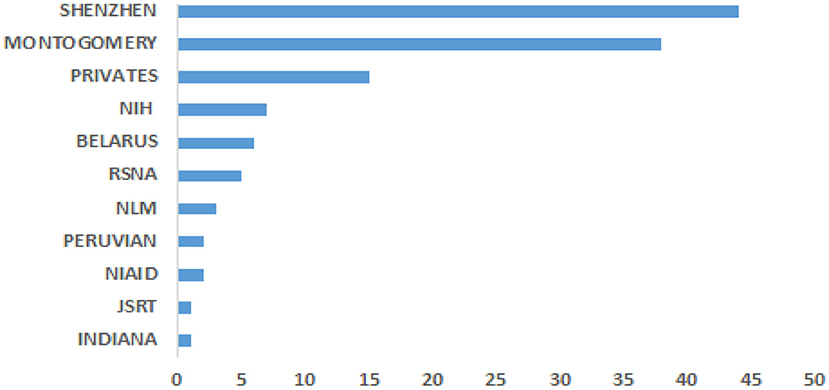
Data is crucial for developing CAD required to solve life-threatening diseases, including TB, as one of the leading causes of worldwide death. The various popular datasets that have been used in developing TB detection algorithms contain de-identified CXR images to protect the privacy of the patients. In other words, the identity of the patients is not disclosed. Most of the datasets are accompanied by radiological interpretation of the observed manifestation that can serve as groundtruth. These datasets are made available to the research communities to foster state-of-the-art research into finding lasting solutions to the early diagnosis of TB manifestations. Some of these public datasets include Montgomery County (73), Shenzhen (73), Peruvian (49), KIT (74), MIMIC-CXR (75), Belarus, NIH (chest x-ray 8, 14) (76), JSRT. Figure 3 shows the datasets frequency of use.
Evaluation Metrics
Once a model is trained using some training images, the test dataset is then employed to assess the quality of the model. It is evident from the data extraction process that different evaluation metrics exist for assessing model performance. Most of the popular evaluation metrics applied to the development of CAD system includes accuracy, sensitivity, specificity, and AUC. These evaluation metrics are briefly explained as follows:
Accuracy is the rate of the correct samples from the total number of samples examined (77). The equation gives the accuracy:
Sensitivity is the proportion of the actual positive samples that are correctly identified as positive (78). This metric is given as:
Specificity is the ratio of the actual negative samples that are correctly identified as negative (78). This metric is given as:
Precision, otherwise known as a positive predictive value, is the ratio of positive samples that are accurately predicted (77). This is given as:
AUC-ROC Curve: sometimes written as AUROC, specify the rate at which a model distinguishes between classes [84]. It tells the probability of separating negative samples from positive samples. Thresholds are set to determine the ROC curve, which is the plot of True Positive Rate (TPR) against False Positive Rate (FPR) given as:
Where TP, TN, FP, FN, TPR, and FPR are true positive, true negative, false positive, false negative, true positive rate, and false positive rate.
Discussion
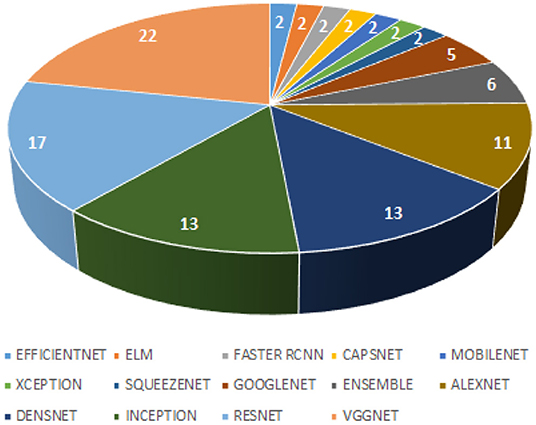
This systematic review extensively searched the various Deep Learning classifiers for diagnosing TB from CXR. Automatic detection of TB has received mammoth attention in the last decades resulting in many publications with state-of-the-art techniques. Figure 4 presents a hierarchical chart of computational techniques categorized according to the frequency of usage in CAD systems for TB based on the included articles. Despite some good accuracies reported in some studies, we found some limitations in the existing studies concerning methods and reported accuracy, which should be a point of consideration for developing CAD systems in the future. Many studies measured diagnostic accuracy without evaluating the risk of bias emerging from the datasets that were used. It is essential that the accuracy of CAD systems is evaluated using a different set of datasets (CXR images) from the set used for training. In other words, avoid
• Using the same set of CXR images for training and testing.
• Testing with CXR images that were not used for training but originated from the same image subset.
• Using images with class imbalance, and
• Using unannotated CXR images.
Otherwise, the diagnostic accuracy evaluation is likely to be exaggerated and could impact the overall generalization of the system. In general, about 80% of the studies used the same CXR images for training and testing or did not report on it.
Conclusion and Recommendations
This systematic review intends to inform researchers of the existing Deep Learning classifiers and assist them in developing a CAD system for the efficient diagnosis of TB. Different state-of-the-art Deep Learning techniques that have been used to detect TB have been explored and presented in Tables 2–5. Generally, it is challenging to compare the methods with each other extensively. Factors like the types of datasets used for evaluation, number of image samples used, evaluation metrics approach, and model parameters tunning make the comparison complex.
As evident from the literature, these pre-trained models (VggNet, ResNet, AlexNet, DenseNet, and Inception) are the most popular and have been extensively explored for the classification of TB, as shown in Figure 4. Despite the effectiveness of Deep Learning models in detection and classification tasks, CAD systems for clinical diagnosis are still challenging in a real-world scenario. The physicians and radiologists see CAD intervention as a threat to their jobs rather than a supporting system to improve physicians' performance in terms of time, effort, efficiency, and affordability, especially in developing countries. This review found that most existing works focused on development studies rather than clinical studies.
One of the likely weaknesses of this review is that it is limited to only the studies written in English because there might be other high-quality studies written in other languages. Also, we restricted the computational techniques to only Deep Learning classifiers, which could increase the risk of classifiers bias where studies that employed a hybrid of both Machine Learning and Deep Learning could have achieved better performance. Furthermore, this study did not undertake meta-analyses due to variations of algorithms used. Also, the raw data required to meta-analyze the diagnostic accuracy for most studies were unavailable.
However, it is recommended that studies be carried out using standardized public datasets that contain additional masks of the images that can be used as groundtruth to detect the infected aspect of the pulmonary images. It is highly recommended that models are trained with a set of images and evaluated on a different set of images. For instance, training a model with the Shenzhen datasets and evaluating it on the Montgomery dataset will validate better generalization. It is also recommended that future CAD systems focus more on clinical evaluation and should be able to identify foreign objects such as buttons and rings that look like a nodule on the CXR images, which may lead to misclassification.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
Author Contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. World Health Organization. Global Tuberculosis Report 2020: Executive Summary. Available online at: https://apps.who.int/iris/handle/10665/337538 (accessed November 29, 2021). License: CC BY-NC-SA 3.0 IGO.
2. Baral SC, Karki DK, Newell JN. Causes of stigma and discrimination associated with tuberculosis in Nepal: a qualitative study. BMC Public Health. (2007) 7:1–0. doi: 10.1186/1471-2458-7-211
3. Hooda R, Sofat S, Kaur S, Mittal A, Meriaudeau F. Deep-learning: a potential method for tuberculosis detection using chest radiography. In: 2017 IEEE International Conference on Signal and Image Processing Applications (ICSIPA) (IEEE) (2017), 497–502.
4. Zumla A, George A, Sharma V, Herbert RH, Oxley A, Oliver M. The WHO 2014 global tuberculosis report—further to go. Lancet Global Health. (2015) 3:e10–2. doi: 10.1016/S2214-109X(14)70361-4
5. Sathitratanacheewin S, Pongpirul K. Deep learning for automated classification of tuberculosis-related chest x-ray: dataset specificity limits diagnostic performance generalizability. arXiv[Prepint].arXiv:1811.07985. (2018).
6. World Health Organization. Chest Radiography in Tuberculosis Detection: Summary of Current WHO Recommendations and Guidance on Programmatic Approaches. (2016). World Health Organization. Available online at: https://apps.who.int/iris/handle/10665/252424 (accessed November 29, 2021).
7. Williams FH. The use of X-ray examinations in pulmonary tuberculosis. Boston Med Surg J. (1907) 157:850–3. doi: 10.1056/NEJM190712261572602
8. Pande T, Pai M, Khan FA, Denkinger CM. Use of chest radiography in the 22 highest tuberculosis burden countries. Eur Respir J. (2015) 46:1816–9. doi: 10.1183/13993003.01064-2015
9. Noor NM, Rijal OM, Yunus A, Mahayiddin AA, Peng GC, Abu-Bakar SA. A statistical interpretation of the chest radiograph for the detection of pulmonary tuberculosis. In: 2010 IEEE EMBS Conference on Biomedical Engineering and Sciences (IECBES) (IEEE). (2010), 47–51.
10. Pedrazzoli D, Lalli M, Boccia D, Houben R, Kranzer K. Can tuberculosis patients in resource-constrained settings afford chest radiography? Eur Respir J. (2017) 49:1601877. doi: 10.1183/13993003.01877-2016
11. Schmidhuber J. Deep learning in neural networks: an overview. Neural Netw. (2015) 61:85–117. doi: 10.1016/j.neunet.2014.09.003
12. Moher D, Liberati A, Tetzlaff J, Altman DG, Prisma Group. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. (2009) 6:e1000097. doi: 10.1371/journal.pmed.1000097
13. Duong LT, Le NH, Tran TB, Ngo VM, Nguyen PT. Detection of tuberculosis from chest X-ray images: boosting the performance with vision transformer and transfer learning. Expert Syst Appl. (2021) 184:115519. doi: 10.1016/j.eswa.2021.115519
14. Norval M, Wang Z, Sun Y. Evaluation of image processing technologies for pulmonary tuberculosis detection based on deep learning convolutional neural networks. J Adv Inform Technol. (2019) 12:253–9. doi: 10.12720/jait.12.3.253-259
15. Rahman M, Cao Y, Sun X, Li B, Hao Y. Deep pre-trained networks as a feature extractor with XGBoost to detect tuberculosis from chest X-ray. Comput Elect Eng. (2021) 93:107252. doi: 10.1016/j.compeleceng.2021.107252
16. Govindarajan S, Swaminathan R. Extreme learning machine based differentiation of pulmonary tuberculosis in chest radiographs using integrated local feature descriptors. Comput Methods Progr Biomed. (2021) 204:106058. doi: 10.1016/j.cmpb.2021.106058
17. Alawi AE, Al-basser A, Sallam A, Al-sabaeei A, Al-khateeb H. Convolutional neural networks model for screening tuberculosis disease. In: 2021 International Conference of Technology, Science and Administration (ICTSA) (IEEE). (2021), 1–5.
18. Khatibi T, Shahsavari A, Farahani A. Proposing a novel multi-instance learning model for tuberculosis recognition from chest X-ray images based on CNNs, complex networks and stacked ensemble. Phys Eng Sci Med. (2021) 44:291–311. doi: 10.1007/s13246-021-00980-w
19. Ayaz M, Shaukat F, Raja G. Ensemble learning based automatic detection of tuberculosis in chest X-ray images using hybrid feature descriptors. Phys Eng Sci Med. (2021) 44:183–94. doi: 10.1007/s13246-020-00966-0
20. Priya PA, Vimina ER. Tuberculosis detection from CXR: an approach using transfer learning with various CNN architectures. In: International Conference on Communication, Computing and Electronics Systems: Proceedings of ICCCES 2020. Vol. 733 (Springer Nature). (2021), 407p.
21. Dasanayaka C, Dissanayake MB. Deep learning methods for screening pulmonary tuberculosis using chest X-rays. Comput Methods Biomech Biomed Eng Imaging Vis. (2021) 9:39–49. doi: 10.1080/21681163.2020.1808532
22. Msonda P, Uymaz SA, Karaagaç SS. Spatial pyramid pooling in deep convolutional networks for automatic tuberculosis diagnosis. Traitement du Signal. (2020) 37:1075–1084. doi: 10.18280/ts.370620
23. Owais M, Arsalan M, Mahmood T, Kim YH, Park KR. Comprehensive computer-aided decision support framework to diagnose tuberculosis from chest X-ray images: data mining study. JMIR Med Inform. (2020) 8:e21790. doi: 10.2196/21790
24. Yoo SH, Geng H, Chiu TL, Yu SK, Cho DC, Heo J, et al. Study on the TB and non-TB diagnosis using two-step deep learning-based binary classifier. J Instrument. (2020) 15:P10011. doi: 10.1088/1748-0221/15/10/P10011
25. Sathitratanacheewin S, Sunanta P, Pongpirul K. Deep learning for automated classification of tuberculosis-related chest X-Ray: dataset distribution shift limits diagnostic performance generalizability. Heliyon. (2020) 6:e04614. doi: 10.1016/j.heliyon.2020.e04614
26. Sahlol AT, Abd Elaziz M, Tariq Jamal A, Damaševičius R, Farouk Hassan O. A novel method for detection of tuberculosis in chest radiographs using artificial ecosystem-based optimization of deep neural network features. Symmetry. (2020) 12:1146. doi: 10.3390/sym12071146
27. Das D, Santosh KC, Pal U. Inception-based deep learning architecture for tuberculosis screening using chest X-rays. In: 2020 25th International Conference on Pattern Recognition (ICPR). Vol. 10 (IEEE) (2021), 3612–9.
28. Rahman T, Khandakar A, Kadir MA, Islam KR, Islam KF, Mazhar R, et al. Reliable tuberculosis detection using chest X-ray with deep learning, segmentation and visualization. IEEE Access. (2020) 8:191586–601. doi: 10.1109/ACCESS.2020.3031384
29. Munadi K, Muchtar K, Maulina N, Pradhan B. Image enhancement for tuberculosis detection using deep learning. IEEE Access. (2020) 8:217897–907. doi: 10.1109/ACCESS.2020.3041867
30. Oloko-Oba M, Viriri S. Tuberculosis abnormality detection in chest X-rays: a deep learning approach. In: International Conference on Computer Vision and Graphics. Vol. 14 (Cham: Springer) (2020), 121–32.
31. Xie Y, Wu Z, Han X, Wang H, Wu Y, Cui L, et al. Computer-aided system for the detection of multicategory pulmonary tuberculosis in radiographs. Journal of Healthcare Engineering. (2020) 2020:1–12. doi: 10.1155/2020/9205082
32. Verma D, Bose C, Tufchi N, Pant K, Tripathi V, Thapliyal A. An efficient framework for identification of tuberculosis and pneumonia in chest X-ray images using Neural Network. Procedia Computer Science. (2020) 171:217–24. doi: 10.1016/j.procs.2020.04.023
33. Tasci E. Pre-processing effects of the tuberculosis chest x-ray images on pre-trained cnns: an investigation. In The International Conference on Artificial Intelligence and Applied Mathematics in Engineering. Vol. 20 (Cham: Springer). (2019), 589–596. doi: 10.1007/978-3-030-36178-5_48
34. Rajaraman S, Antani SK. Modality-specific deep learning model ensembles toward improving TB detection in chest radiographs. IEEE Access. (2020) 8:27318–26. doi: 10.1109/ACCESS.2020.2971257
35. Abideen ZU, Ghafoor M, Munir K, Saqib M, Ullah A, Zia T, et al. Uncertainty assisted robust tuberculosis identification with bayesian convolutional neural networks. IEEE Access. (2020) 8:22812–25. doi: 10.1109/ACCESS.2020.2970023
36. Hijazi MH, Hwa SK, Bade A, Yaakob R, Jeffree MS. Ensemble deep learning for tuberculosis detection using chest X-Ray and canny edge detected images. IAES Int J Artif Intel. (2019) 8:429. doi: 10.11591/ijai.v8.i4.pp429-435
37. Pasa F, Golkov V, Pfeiffer F, Cremers D, Pfeiffer D. Efficient deep network architectures for fast chest X-ray tuberculosis screening and visualization. Sci Rep. (2019) 9:1–9. doi: 10.1038/s41598-019-42557-4
38. Meraj SS, Yaakob R, Azman A, Rum SN, Shahrel A, Nazri A, et al. Detection of pulmonary tuberculosis manifestation in chest x-rays using different convolutional neural network (CNN) models. Int J Eng Adv Technol. (2019) 9:2270–5. doi: 10.35940/ijeat.A2632.109119
39. Ahsan M, Gomes R, Denton A. Application of a convolutional neural network using transfer learning for tuberculosis detection. In: 2019 IEEE International Conference on Electro Information Technology (EIT) (IEEE) (2019), 427–33.
40. Nguyen QH, Nguyen BP, Dao SD, Unnikrishnan B, Dhingra R, Ravichandran SR, et al. Deep learning models for tuberculosis detection from chest x-ray images. In: 2019 26th International Conference on Telecommunications (ICT) (IEEE) (2019), 381–5.
41. Ho TK, Gwak J, Prakash O, Song JI, Park CM. Utilizing pretrained deep learning models for automated pulmonary tuberculosis detection using chest radiography. In: 11th Asian Conference on Intelligent Information and Database Systems, ACIIDS (Springer Verlag) (2019), 395–403.
42. Heo SJ, Kim Y, Yun S, Lim SS, Kim J, Nam CM, et al. Deep learning algorithms with demographic information help to detect tuberculosis in chest radiographs in annual workers' health examination data. Int J Environ Res Public Health. (2019) 16:250. doi: 10.3390/ijerph16020250
43. Hernández A, Panizo Á, Camacho D. An ensemble algorithm based on deep learning for tuberculosis classification. In: International Conference on Intelligent Data Engineering and Automated Learning (Cham: Springer). (2019), 145–54.
44. Hijazi M, Yang L, Alfred R, Mahdin H, Yaakob R. Ensemble deep learning for tuberculosis detection. Indonesian J Elect Eng Comput Sci. (2020) 8:429. doi: 10.11591/ijeecs.v17.i2.pp1014-1020
45. Abbas A, Abdelsamea MM. Learning transformations for automated classification of manifestation of tuberculosis using convolutional neural network. In: 2018 13th International Conference on Computer Engineering and Systems (ICCES) (IEEE). (2018), 122–6.
46. Karnkawinpong T, Limpiyakorn Y. Chest X-ray analysis of tuberculosis by convolutional neural networks with affine transforms. In: Proceedings of the 2018 2nd International Conference on Computer Science and Artificial Intelligence. (2018), 90–3.
47. Stirenko S, Kochura Y, Alienin O, Rokovyi O, Gordienko Y, Gang P, et al. Chest X-ray analysis of tuberculosis by deep learning with segmentation and augmentation. In: 2018 IEEE 38th International Conference on Electronics and Nanotechnology (ELNANO) (IEEE) (2018), 422–8.
48. Becker AS, Blüthgen C, Sekaggya-Wiltshire C, Castelnuovo B, Kambugu A, Fehr J, et al. Detection of tuberculosis patterns in digital photographs of chest X-ray images using deep learning: feasibility study. Int J Tubercul Lung Dis. (2018) 22:328–35. doi: 10.5588/ijtld.17.0520
49. Liu C, Cao Y, Alcantara M, Liu B, Brunette M, Peinado J, et al. TX-CNN: Detecting tuberculosis in chest X-ray images using convolutional neural network. In: 2017 IEEE International Conference on Image Processing (ICIP) (IEEE). (2017), 2314–8.
50. Oloko-Oba M, Viriri S. Ensemble of convolution neural networks for automatic tuberculosis classification. In: International Conference on Computational Collective Intelligence (Cham: Springer) (2021), 549–59. doi: 10.1007/978-3-030-88081-1_41
51. Lee JH, Park S, Hwang EJ, Goo JM, Lee WY, Lee S, et al. Deep learning–based automated detection algorithm for active pulmonary tuberculosis on chest radiographs: diagnostic performance in systematic screening of asymptomatic individuals. Eur Radiol. (2021) 31:1069–80. doi: 10.1007/s00330-020-07219-4
52. Zhang R, Duan H, Cheng J, Zheng Y. A study on tuberculosis classification in chest X-ray using deep residual attention networks. In: 2020 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC) (IEEE) (2020), 1552–5.
53. Hwang EJ, Park S, Jin KN, Kim JI, Choi SY, Lee JH, et al. Development and validation of a deep learning–based automatic detection algorithm for active pulmonary tuberculosis on chest radiographs. Clin Infect Dis. (2019) 69:739–47. doi: 10.1093/cid/ciy967
54. Rajpurkar P, Irvin J, Ball RL, Zhu K, Yang B, Mehta H, et al. Deep learning for chest radiograph diagnosis: a retrospective comparison of the CheXNeXt algorithm to practicing radiologists. PLoS Med. (2018) 15:e1002686. doi: 10.1371/journal.pmed.1002686
55. Lakhani P, Sundaram B. Deep learning at chest radiography: automated classification of pulmonary tuberculosis by using convolutional neural networks. Radiology. (2017) 4:574–82. doi: 10.1148/radiol.2017162326
56. Cao K, Zhang J, Huang M, Deng T. X-ray classification of tuberculosis based on convolutional networks. In: 2021 IEEE International Conference on Artificial Intelligence and Industrial Design (AIID) (IEEE). (2021), 125–9. doi: 10.1109/AIID51893.2021.9456476
57. Karaca BK, Güney S, Dengiz B, Agildere M. Comparative study for tuberculosis detection by using deep learning. In: 2021 44th International Conference on Telecommunications and Signal Processing (TSP) (IEEE) (2021), 88–91.
58. Saif AF, Imtiaz T, Shahnaz C, Zhu WP, Ahmad MO. Exploiting cascaded ensemble of features for the detection of tuberculosis using chest radiographs. IEEE Access. (2021) 9:112388–99. doi: 10.1109/ACCESS.2021.3102077
59. Imam OT, Haque M, Shahnaz C, Imran SA, Islam MT, Islam MT. Detection of tuberculosis from chest X-ray images based on modified inception deep neural network model. In: 2020 IEEE International Women in Engineering (WIE) Conference on Electrical and Computer Engineering (WIECON-ECE) (IEEE). (2020), 360–3.
60. Griffin T, Cao Y, Liu B, Brunette MJ. Object detection and segmentation in chest X-rays for tuberculosis screening. In: 2020 Second International Conference on Transdisciplinary AI (TransAI) (IEEE) (2020), 34–42.
61. Rashid R, Khawaja SG, Akram MU, Khan AM. Hybrid RID network for efficient diagnosis of tuberculosis from chest X-rays. In: 2018 9th Cairo International Biomedical Engineering Conference (CIBEC) (IEEE) (2018), 167–70.
62. Fehr J, Konigorski S, Olivier S, Gunda R, Surujdeen A, Gareta D, et al. Computer-aided interpretation of chest radiography reveals the spectrum of tuberculosis in rural South Africa. NPJ Dig Med. (2021) 4:1–10. doi: 10.1101/2020.09.04.20188045
63. Vats S, Singh S, Kala G, Tarar R, Dhawan S. iDoc-X: an artificial intelligence model for tuberculosis diagnosis and localization. J Discrete Math Sci Cryptogr. (2021) 24:1257–72. doi: 10.1080/09720529.2021.1932910
64. Rajpurkar P, O'Connell C, Schechter A, Asnani N, Li J, Kiani A, et al. CheXaid: deep learning assistance for physician diagnosis of tuberculosis using chest x-rays in patients with HIV. NPJ Dig Med. (2020) 3:1–8. doi: 10.1038/s41746-020-00322-2
65. Grivkov AV, Smirnov AA. Application of convolutional neural networks for diagnostics of tuberculosis. AIP Conf Proc. (2020) 2313:080011.
66. Gozes O, Greenspan H. Deep feature learning from a hospital-scale chest x-ray dataset with application to TB detection on a small-scale dataset. In: 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC) (IEEE) (2019), 4076–9.
67. Karnkawinpong T, Limpiyakorn Y. Classification of pulmonary tuberculosis lesion with convolutional neural networks. J Phys Conf Series. (2019) 1195:012007. doi: 10.1088/1742-6596/1195/1/012007
68. Sivaramakrishnan R, Antani S, Candemir S, Xue Z, Abuya J, Kohli M, et al. Comparing deep learning models for population screening using chest radiography. Med Imaging 2018 Comput Aided Diag. (2018) 10575:105751E. doi: 10.1117/12.2293140
69. Vajda S, Karargyris A, Jaeger S, Santosh KC, Candemir S, Xue Z, et al. Feature selection for automatic tuberculosis screening in frontal chest radiographs. J Med Syst. (2018) 42:1–11. doi: 10.1007/s10916-018-0991-9
70. Puddy E, Hill C. Interpretation of the chest radiograph. Continuing education in anaesthesia. Crit Care Pain. (2007) 7:71–5. doi: 10.1093/bjaceaccp/mkm014
71. Gilpin C, Korobitsyn A, Migliori GB, Raviglione MC, Weyer K. The world health organization standards for tuberculosis care and management. Eur Respir J. (2018) 51:1800098. doi: 10.1183/13993003.00098-2018
72. Brownlee J. 4 Types of Classification Tasks in Machine Learning. Machine Learning Mastery, 4p. Available online at: https://machinelearningmastery.com/types-of-classification-in~machine-learning (accessed November 25, 2021).
73. Jaeger S, Candemir S, Antani S, Wáng YX, Lu PX, Thoma G. Two public chest X-ray datasets for computer-aided screening of pulmonary diseases. Quant Imaging Med Surg. (2014) 4:475. doi: 10.3978/j.issn.2223-4292.2014.11.20
74. Irvin J, Rajpurkar P, Ko M, Yu Y, Ciurea-Ilcus S, Chute C, et al. Chexpert: a large chest radiograph dataset with uncertainty labels and expert comparison. Proc AAAI conf Artif Intell. (2019) 33:590–7. doi: 10.1609/aaai.v33i01.3301590
75. Johnson AE, Pollard TJ, Greenbaum NR, Lungren MP, Deng CY, Peng Y, et al. MIMIC-CXR-JPG, a large publicly available database of labeled chest radiographs. arXiv[Prepint].arXiv:1901.07042. (2019).
76. Hwang S, Kim HE, Jeong J, Kim HJ. A novel approach for tuberculosis screening based on deep convolutional neural networks. Med Imaging Comput Aided Diag. (2016) 9785:97852W. doi: 10.1117/12.2216198
77. Lalkhen AG, McCluskey A. Clinical tests: sensitivity and specificity. Continuing Educ Anesth Crit Care Pain. (2008) 8:221–3. doi: 10.1093/bjaceaccp/mkn041
Keywords: tuberculosis, chest radiograph, computer-aided diagnosis, deep learning, systematic review
Citation: Oloko-Oba M and Viriri S (2022) A Systematic Review of Deep Learning Techniques for Tuberculosis Detection From Chest Radiograph. Front. Med. 9:830515. doi: 10.3389/fmed.2022.830515
Received: 07 December 2021; Accepted: 14 February 2022;
Published: 10 March 2022.
Edited by:
Emmanuel Twumasi Osei, University of British Columbia Okanagan, CanadaReviewed by:
Anjali Agrawal, Teleradiology Solutions, IndiaMuhammad Usman Akram, National University of Sciences and Technology (NUST), Pakistan
Copyright © 2022 Oloko-Oba and Viriri. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Serestina Viriri, viriris@ukzn.ac.za
 Mustapha Oloko-Oba
Mustapha Oloko-Oba Serestina Viriri
Serestina Viriri