An exome-wide study of renal operational tolerance
- 1Human Genetics Unit, Institut de Recherche Interdisciplinaire en Biologie Humaine et Moléculaire (IRIBHM), Université Libre de Bruxelles (ULB), Brussels, Belgium
- 2Interuniversity Institute of Bioinformatics in Brussels (IB2), Université Libre de Bruxelles - Vrije Universiteit Brussel (ULB-VUB), Brussels, Belgium
- 3Department of Nephrology, Antwerp University Hospital and Laboratory of Experimental Medicine, University of Antwerp, Antwerp, Belgium
- 4CHU Nantes, Nantes Université, INSERM, Center for Research in Transplantation and Translational Immunology, CR2TI, UMR 1064, ITUN, Nantes, France
- 5Brussels Interuniversity Genomics High Throughput Core (BRIGHTcore), VUB-ULB, Brussels, Belgium
- 6Center for Medical Genetics, Reproduction and Genetics, Reproduction Genetics and Regenerative Medicine, Vrije Universiteit Brussel, UZ Brussel, Brussels, Belgium
- 7Department of Biostatistics, University of Washington, Seattle, WA, United States
- 8Transplant Laboratory, Institute for Clinical and Experimental Medicine, Prague, Czechia
- 9Department of Genetics, Hôpital Erasme, ULB Center of Human Genetics, Université Libre de Bruxelles, Brussels, Belgium
- 10Department of Genetics, Hôpital Universitaire des Enfants Reine Fabiola, ULB Center of Human Genetics, Université Libre de Bruxelles, Brussels, Belgium
- 11Center for Human Genetics, Clinique Universitaires Saint Luc, Brussels, Belgium
- 12Istanbul Tip Fakültesi, Istanbul School of Medicine, Internal Medicine, Nephrology, Istanbul, Türkiye
- 13Department of Nephrology, Hospital del Mar, Institute Mar for Medical Research, Barcelona, Spain
- 14Department of Nephrology, Hospital Universitario 12 de Octubre, Madrid, Spain
- 15Nephrology Center, Santaros Klinikos, Medical Faculty, Vilnius University, Vilnius, Lithuania
- 16Department of Nephrology, Hospital Centre EpiCURA, Baudour, Belgium
- 17CHU Nantes, Centre d'Investigation Clinique en Biothérapie, Centre de Ressources Biologiques (CRB), Nantes, France
- 18LabEx IGO “Immunotherapy, Graft, Oncology”, Nantes, France
- 19Department of Genetic Medicine and Development, Faculty of Medicine, Université de Geneve, Geneva, Switzerland
Background: Renal operational tolerance is a rare and beneficial state of prolonged renal allograft function in the absence of immunosuppression. The underlying mechanisms are unknown. We hypothesized that tolerance might be driven by inherited protein coding genetic variants with large effect, at least in some patients.
Methods: We set up a European survey of over 218,000 renal transplant recipients and collected DNAs from 40 transplant recipients who maintained good allograft function without immunosuppression for at least 1 year. We performed an exome-wide association study comparing the distribution of moderate to high impact variants in 36 tolerant patients, selected for genetic homogeneity using principal component analysis, and 192 controls, using an optimal sequence-kernel association test adjusted for small samples.
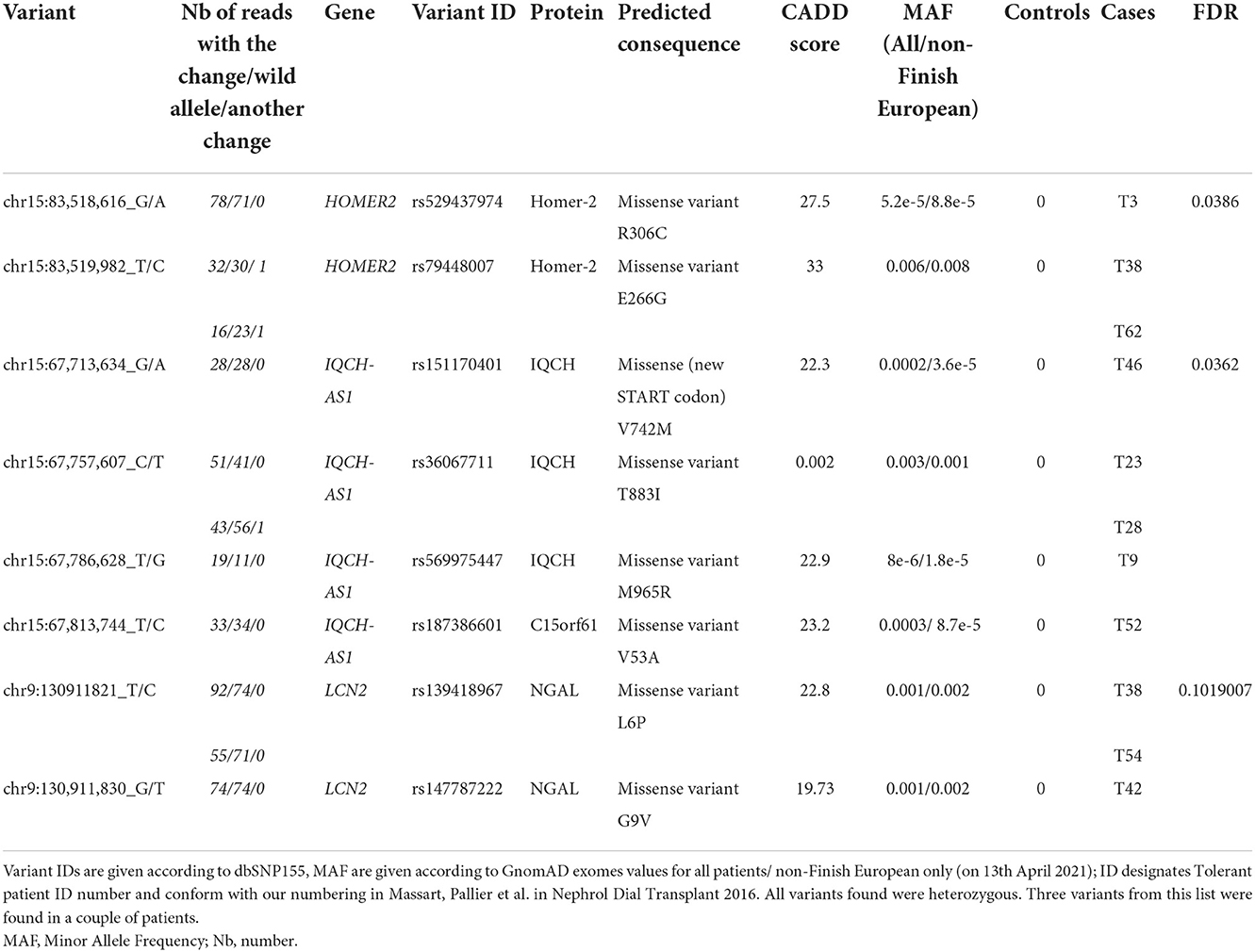
Results: We identified rare variants of HOMER2 (3/36, FDR 0.0387), IQCH (5/36, FDR 0.0362), and LCN2 (3/36, FDR 0.102) in 10 tolerant patients vs. 0 controls. One patient carried a variant in both HOMER2 and LCN2. Furthermore, the three genes showed an identical variant in two patients each. The three genes are expressed at the primary cilium, a key structure in immune responses.
Conclusion: Rare protein coding variants are associated with operational tolerance in a sizable portion of patients. Our findings have important implications for a better understanding of immune tolerance in transplantation and other fields of medicine.
ClinicalTrials.gov, identifier: NCT05124444.
Introduction
Renal transplantation is considered the best treatment option for end-stage renal disease. However, graft survival comes at the price of life-long immunosuppression that may cause cardiovascular disease, cancer and infection. Exceptional patients will however maintain long-lasting allograft function, in spite of complete discontinuation of immunosuppression (1–3). The absence of destructive immunological response toward the allograft, contrasting with global immunocompetence and a normal response against other immunological challenges is referred to as 'operational tolerance'. Renal operational tolerance is associated with prolonged allograft and patient survival (1, 2, 4, 5). The discontinuation of immunosuppression in tolerant patients generally results from patient non-compliance or from medical decisions grounded on a life-threatening infection or cancer. However, under current transplantation protocols, and despite non-adherence being so common (6) spontaneous tolerance is very rarely observed. In a recent European survey, clinicians identified tolerance in 3/10,000 transplanted patients (2). Because non-compliance is usually undisclosed and in the absence of reliable biomarkers, most tolerant kidney recipients are discovered by chance, suggesting that the incidence of tolerance may be underestimated. Inducing renal tolerance in human recipients is possible through strategies of combined kidney and bone marrow transplantation, resulting in transient chimerism (7). Unfortunately, these strategies are challenging and far from routine clinical implementation. Thus, a better understanding of the molecular pathways and mechanisms involved in spontaneous tolerance might help design innovative tolerance induction protocols and medications.
Most studies of renal operational tolerance to date aimed at identifying reliable biomarkers for the detection of patients with reduced immunosuppression needs or for monitoring tolerance induction protocols [for review, see (3)]. Such studies included antigen-specific functional assays, circulating cell immuno-phenotyping, transcriptomic analyses and mechanistic studies. All together they showed that operational tolerance relies on the expansion of diverse alloreactive immune cells with inhibitory phenotypes and suppressive properties that maintain graft protection against inflammation injury. It remains unclear however whether the changes observed were causes or consequences of tolerance and currently, nothing explains why a kidney recipient initially receiving standard immunosuppression will eventually develop a form of immunoquiescence toward an alloreactive graft.
Because various extreme phenotypes are enriched in trait-causing alleles (8–14), we hypothesized that some tolerant patients may carry inherited, high impact gene variants allowing tolerance to develop. We reasoned that even Mendelian or near-Mendelian inheritance of tolerance would go unnoticed, because the phenotype is only revealed by transplantation, which is never performed experimentally in asymptomatic relatives of transplanted patients. As most high impact variants are found in protein-coding sequences of the genome (15), we sampled one of the world's largest DNA collections of tolerant patients (n = 40) and performed an exome-wide association study.
Materials and methods
Recruitment and data analysis
Patients were recruited from August 22, 2015 through October 29, 2019 and originated from transplantation centers in 12 different European countries (Austria, Belgium, Czech Republic, France, Germany, Holland, Italy, Lithuania, Romania, Spain, Switzerland, Turkey). Data analysis was performed from February 7, 2018 until September 29, 2021. The study protocol was approved at Erasme-ULB Ethics Committee under the refence 2015/379) and all participants signed a written informed consent form approved by their local institutional review board. This paper adheres to the Helsinki Declaration of 1975, as revised in 2013. The trial was registered in ClinicalTrials.gov under the access number NCT05124444.
Eligibility criteria
Tolerant patients (i) were transplanted with an allogenic kidney; (ii) were 18 y or older; (iii) maintained good allograft function (serum creatinine < 1.7 mg/dL and proteinuria ≤ 1 g/day or /g creatinine; or values above these limits but stable with a maximum variability of 20%) in the absence of immunosuppression for at least 1 year. Patients with a history of bone marrow/other solid organ transplantation or who had been included in a protocol of tolerance induction were not eligible. Control patients were (i) healthy persons or persons with a condition unrelated to kidney or immunological disease; (ii) of European ancestry according to the investigator. All participants signed a written informed consent form approved by their local institutional review board.
DNA samples
Following a large European survey involving 256 renal transplantation centers in 28 countries (2), we collected DNA samples and clinical data from 40 tolerant patients originating from 12 countries. Twenty-two patients' DNA samples were available at Nantes University, FR (IRB protocol number: RC14_0431) and 18 other patients were prospectively sampled as part as the TOlerance MOlecular and Genome-wide studies with Renal Allograft recipient Material [TOMOGRAM study, launched by the ERA-EDTA DESCARTES working group (http://wwwa.era-edtaworkinggroups.org/en-US/group/descartes)]. The samples originating from the Nantes University biocollection were sequenced over 100 base pairs from both ends on an Illumina HiSEq2000 (Aros Applied Biotechnology, DK) with a mean depth of 68 ± 7. The remaining 18 patients DNAs were paired-end sequenced over 125 base pairs on an Illumina HiSeq 1500 platform (Brightcore, BE) with a mean depth of 121 ± 28. The exome sequencing data from the 40 tolerant patients were compared with the dataset of 197 in house controls previously sequenced the same way as the TOMOGRAM cases. Details on DNA sequencing and variant analysis are available in the Supplementary material. Characteristics of tolerant patients are given in Supplementary Table 1.
Principal component analysis
PCA of exome data is a commonly used multivariate analysis that reduces the data's dimensionality while their covariance remains preserved. This analysis reduced the high dimensional dataset to a small number of dimensions termed principal components. Here we performed a principal component decomposition of the whole set of exome data (n = 85,899 variants after applying the filters described above), without LD pruning, to identify outliers due to ancestry using the function snpgds PCA of the R Bioconductor package SNPRelate v1.12.2.2.
Case-control analysis
The rarity of operational tolerance—our cohort of 40 patients is one of the world largest—(3) represents a major obstacle to the exhaustive study of its underlying genetics. However, extreme phenotypes are often driven by highly penetrant genetic variants of large effect making small (9, 13) or very small samples (16–18) sufficient to identify the causative gene. We reasoned that our limited number of patients would allow for discovering such variants, if present, and may encourage building larger cohorts in the future. To demonstrate our hypothesis, we built a pilot study comparing 40 tolerant patients to 197 controls from the general population with the objective to demonstrate enrichment of variants with large effect in the tolerant patients. This design called “single extreme vs. control” has proven to be an effective and reasonable method to increase the study power without resorting to a high number of, less accessible, exome-sequenced patients with the opposite phenotype (19). Healthy controls are specially relevant when we study a rare trait because the contamination in the control population will remain low with little or no impact on the study power (18). Finally, given the high number of genes/variants tested, we reported both false discovery rates (FDRs) and p-values following Bonferroni correction as measures of statistical significance after correction for multiple hypothesis testing (20).
In practice, we filtered exome data for moderate to high-impact variants affecting exonic, gene promoter or splice-site regions (for details see Supplementary material) and performed a principal component analysis (PCA) to identify outliers by ancestry. We studied exome-wide association using an optimal sequence kernel association test with small sample adjustment (aSKAT-O) which behaves well over a wide range of allele frequencies (21). This test can detect differences in both rare and common variants but upweights the contribution of rare variants that are likely trait-causing. The aSKAT-O can furthermore be used as a burden test or a variance component test by changing the kernel weight (Burden configuration when Rho = 1; SKAT configuration when Rho = 0) (Supplementary Table 2). We introduced principal components (PCs) 1 to 3 as covariables into our aSKAT-O model in order to correct for a possible population stratification artifact and an additional PC issued from the comparison of patients according to the sequencing machine in order to correct for a possible batch effect. We performed both by-gene analysis, with the aim to capture allelic heterogeneity, and by-variant analysis.
Control of the variants
All genes and variants arising from aSKAT-O analyses had to pass three control steps to ensure they were true and not resulting from a batch effect due to the use of two different sequencers. Firstly, all variants were carefully examined (at the level of the BAM files) for any irregularity in the alignment processes. All variants but one (rs529437974—lack of material) were confirmed by Sanger sequencing. Secondly, a new aSKAT-O was performed with the sequencer as an outcome (instead of the phenotype) and the genes or variants predicting the sequencer were excluded (Supplementary Figure 1). Thirdly, we performed a new SKAT analysis comparing our controls (n = 192) to the tolerant patients after exclusion of the 9 non-European (3 Maghrebin, 6 Turkish) (n = 31) in order to rule out a spurious genetic association due to population stratification. Of note, three variants shared by a pair of tolerant patients each were further investigated to rule out a family relationship between the carriers that could bias the genotype-phenotype association. Patients carrying the same variants were compared for the proportion of single nucleotide polymorphisms (SNPs) with a minor allele frequency (MAF) > 5%) (Supplementary Table 2).
Results
Principal component analysis
The filtration steps (see Study Flow Chart in Figure 1) yielded 85,899 moderate to high impact variants in 16,390 genes. PCA of exome data excluded 5 outliers by ancestry out of 197 controls and 4 (T26, T43, T53, and T63) out of 40 tolerant patients (Figure 2).
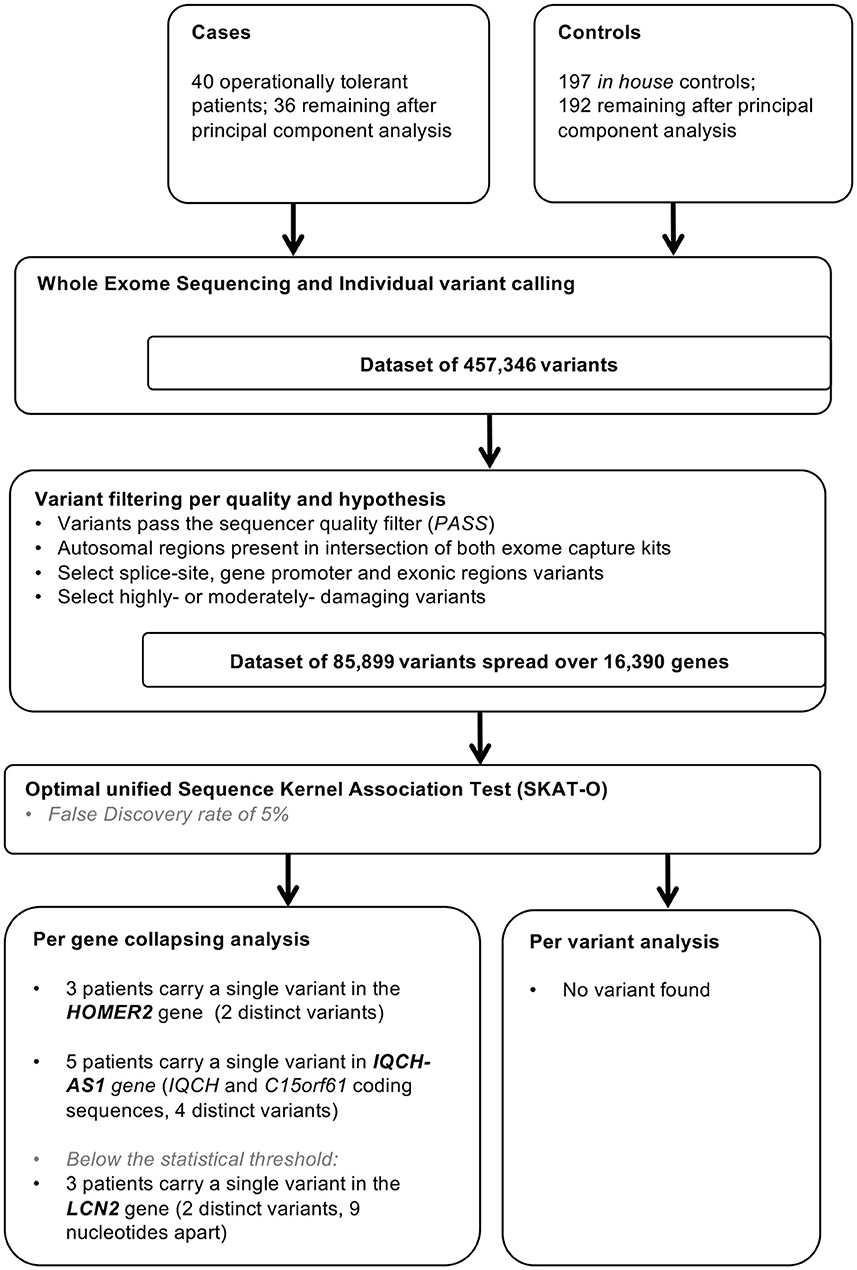
Figure 1. Study flow chart. In house controls were selected for absence of history of renal dysfunction and for homogeneity in principal component analysis.
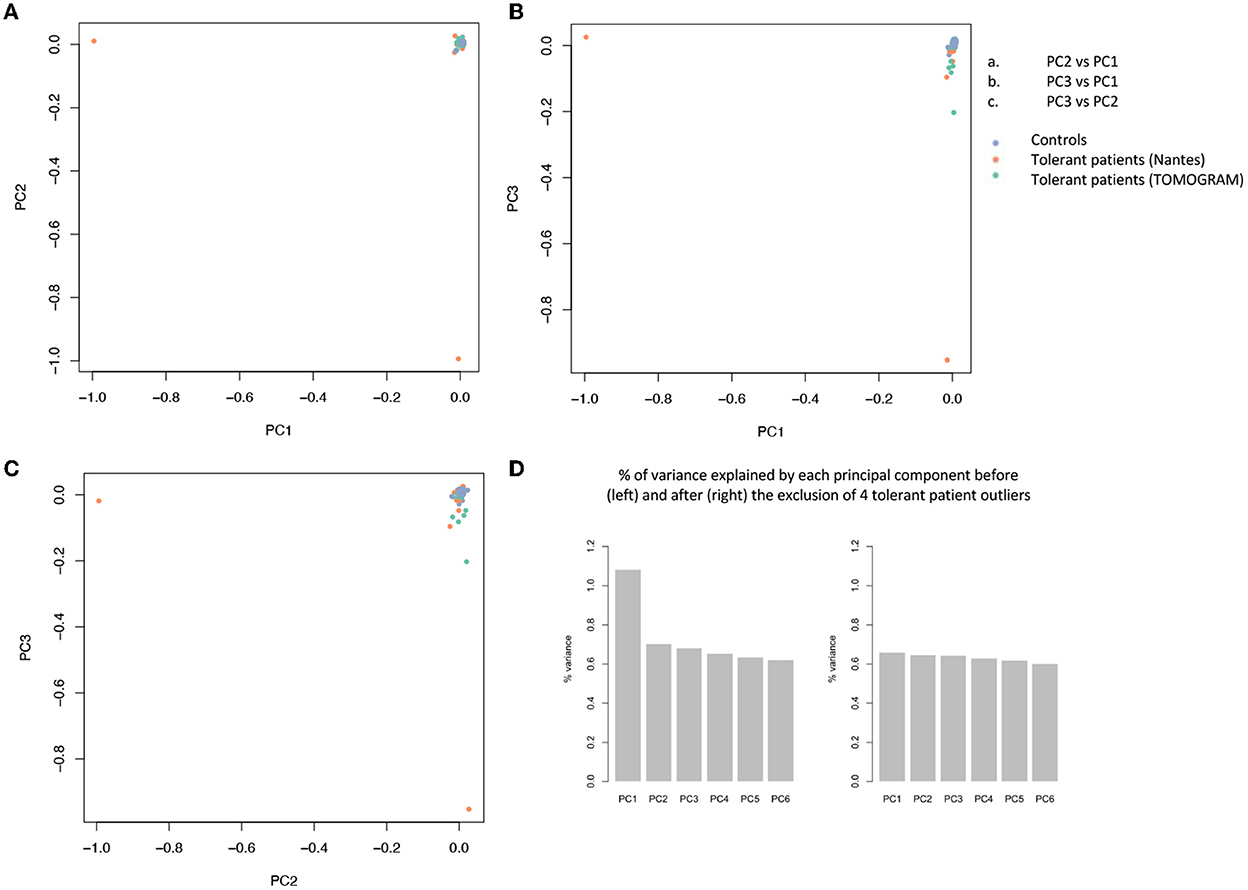
Figure 2. Principal component analysis (PCA) of 85.899 variants. PCA is used to reduce the dimensionality of a data set with many variables/features while minimizing the information loss and thus capturing as much of the data's variability as possible. The principal components are linear functions of the variables in the original data set. Principal components are identified by solving an eigenvalue/eigen vector problem. (A–C) Each dot represents a patient and is distributed in function of the 2 first principal components. Blue dots represent controls, orange dots represent tolerant patients sequenced on the AROS platform (Nantes cohort), green dots represent patients sequenced on the BIGRE platform (TOMOGRAM cohort). (D) % of variance explained by each principal component before (left) and after (right) the removal of 4 tolerant patient outliers (T26, T43, T53, and T63).
Per gene and per variant comparisons
We compared the distribution of variants within each gene (n = 16,390) in the 36 tolerant patients vs. 192 controls (Table 1, Figure 3). We identified two genes HOMER2 (entrez ID 9455) and IQCH-AS1 (entrez ID 100506686); FDR = 0.0387 and 0.0362, respectively; p = 0.1160 and 0.1086, respectively. Three patients (8%) carried a variant in HOMER2. The same variant (rs79448007, p.Glu266Gly) was found in two patients (T38 and T62, Table 1). Five patients (14%) carried a IQCH-AS1 gene variant, two of whom shared the same variant (p = 0.03 for the null hypothesis of random selection among the tolerant group, Supplementary methods) (T23 and T28, Table 1). No control subject carried a moderate or highly damaging variant in these two genes. IQCH-AS1 encodes a long non-coding RNA, and variants were filtered in because the complementary DNA strand encompasses two protein-coding genes: IQCH (most of IQCH-AS1 length) and C15orf61. These two genes remain significant after excluding the 9 tolerant patients of foreign origin (data not shown). A third gene, LCN2 (entrez ID 3934) showed a weaker association (FDR = 0.102; p = 0.4847), with three patients (8%) carrying a variant vs. 0 controls. Again, two patients shared the same LCN2 variant (T38 and T54, Table 1). Both LCN2 variants were located 9 base pairs apart, in the 20 amino-acid long signal peptide of the encoded NGAL protein, suggesting the possibility of a shared functional effect. Of note, one patient (T38) carried both a LCN2 and a HOMER2 variant. The 8 variants identified in the 10 patients were rare [i.e., allele frequency < 1% (22)] single nucleotide substitutions. All three genes encode proteins that affect primary cilia function (p = 0.01; Supplementary methods). We ruled out close ancestry in patients sharing a same variant by quantifying their SNPs homology (Supplementary Table 3). The clinical characteristics of the tolerant patients with genetic variants are given in Table 2. Four out of the 10 tolerant patients had received a fully-matched graft for loci HLA A, B and DR. Interestingly, patient T38 who carried variants in both HOMER2 and LCN2 had remained tolerant over an especially long time (173 months at the time of this analysis). SNP array analysis in a subgroup of 19 tolerant patients (from Nantes) detected no copy number variants (CNVs) over any of the three loci (Supplementary methods).
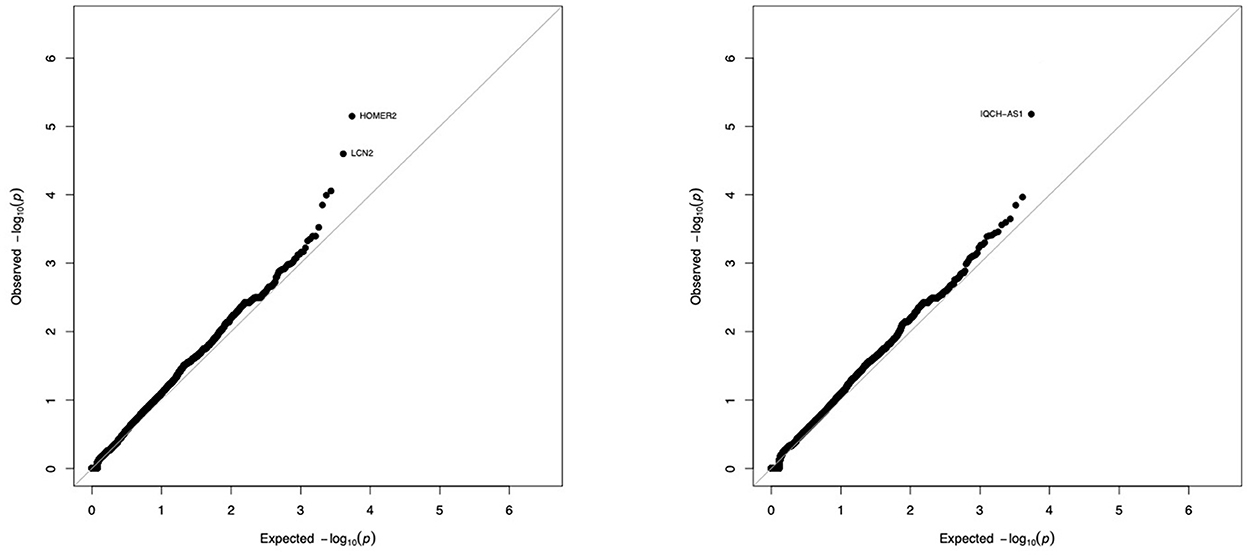
Figure 3. Gene-by-gene comparison of tolerant patients and controls. The distribution of exome variants with predicted moderate or high impact was compared in 36 kidney allograft tolerant recipients and 192 unrelated controls of European ancestry, using SKAT-O adjusted for small sample size. Results are presented as –log10 quantile-quantile plots of observed P-values (Y axis) vs. expected P-values (X axis) if variants were equally distributed in cases and controls, when Rho is set to 0 (SKAT-like test, left panel) or Rho is set to 1 (Burden–like test, right panel). Five genes (3 on the left panel, 2 on the right panel) were not considered significant because of an artefactual signal produced by the sequencing machine (see Supplementary Figure 1).
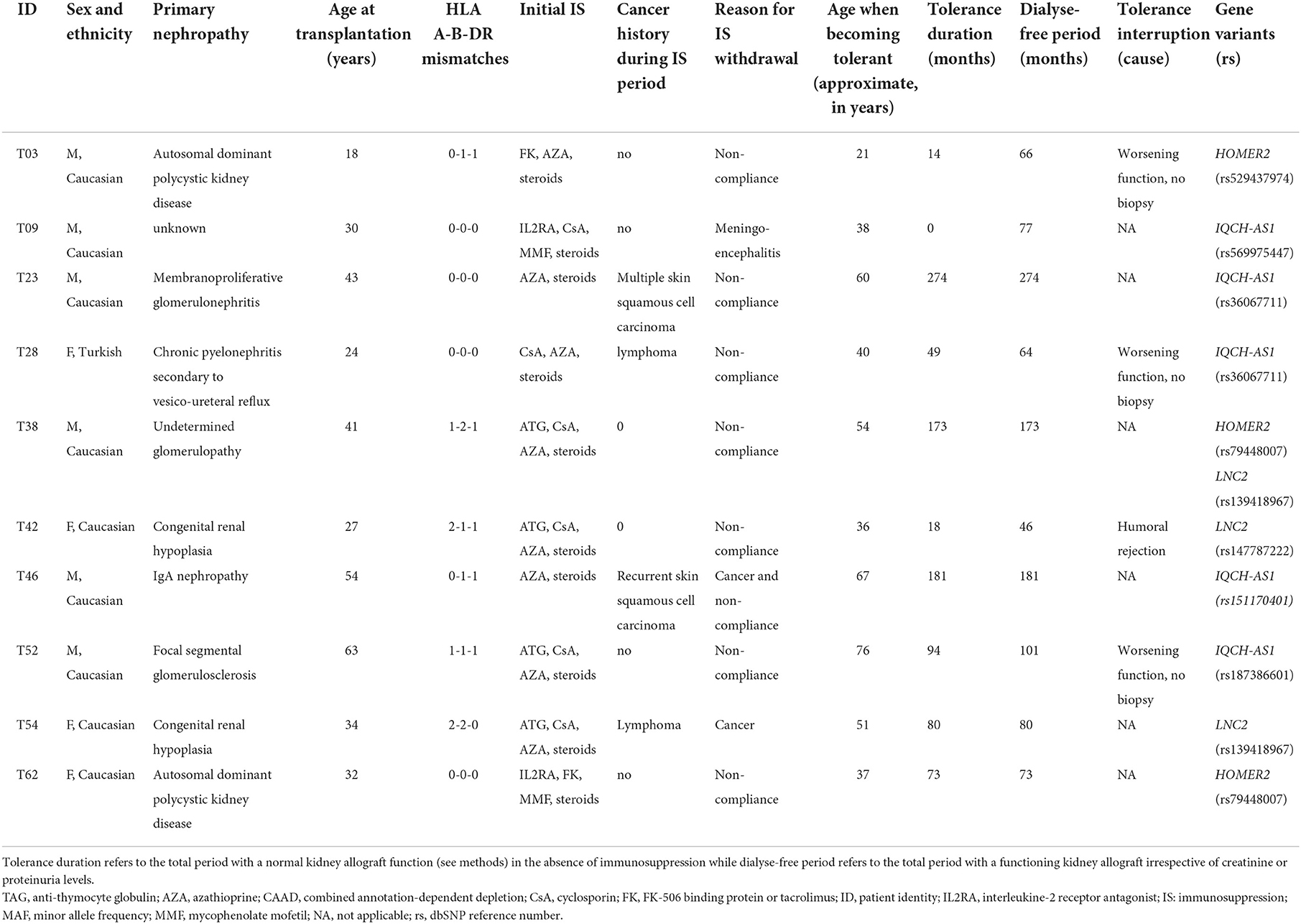
Table 2. Characteristics of the variant-bearing tolerant patients identified through per-gene aSKAT-O.
We then used the aSKAT-O to compare the frequency of each variant (n = 85,899) in the 36 tolerant vs. 192 controls, and did not identify any variant with statistical significance.
Discussion
Overall, our results show a statistically significant association of spontaneous renal tolerance with moderate to high impact coding variants in HOMER2 and IQCH/C15orf61, and a likely association with LCN2. HOMER2 is an actin-binding, scaffolding protein expressed at various specialized primary cilia (23), and a critical regulator of intracellular Ca2+ signaling (24). Homer2 and 3 compete with calcineurin, resulting in a negative regulation of T-cell activation and Homer2,3-deficient mice develop an autoimmune-like pathology (25). IQCH encodes the IQ motif-containing protein H also known as SPATA17 (spermatogenesis associated 17), a cilia and flagella-associated protein involved in spermatogenesis (26). IQ motif containing proteins bind calmodulin (27), the key transducer of intracellular calcium signals in T cells, and alter its binding to calcium. C15orf61 (chromosome 15 open reading frame 61) is an uncharacterized coding gene located immediately downstream of IQCH in a non-imprinted portion of chromosome 15. LCN2 is expressed at the primary cilium (28, 29). It encodes NGAL (neutrophil gelatinase-associated lipocalin), an iron-trafficking protein involved in multiple immunological processes (30–32), including common variable immunodeficiency (33), but its role in transplantation remains obscure (34, 35). Furthermore, HOMER2, IQCH, and NGAL are all cilia-associated proteins. The T cell cilium is viewed as a specialized compartment that assembles at the plasma membrane upon TCR binding and interaction with the antigen-presenting cell or target cell, producing the so-called “immune synapse” (36, 37). The immune synapse is a place of intense TCR/CD3 recycling which directly impacts on the duration and intensity of TCR signaling. This process is dependent upon actin polymerization and intracellular calcium release and is critical for the development of T cell full effector potential (38, 39). A spurious association between renal tolerance and the primary cilium resulting from a cluster of ciliopathies in our cohort appears unlikely. While 2/10 variant-carrying tolerant patients suffered from a recognized ciliopathy (Table 2), none of the genes above are associated with a renal disease, either in the OMIM database or in a large whole-exome study (40). Furthermore, our results are consistent with the reported association of tolerance with a coding variant in PARVG, encoding an actin-binding protein (41), and also with a GWAS of the mirror phenotype, acute rejection of renal allograft, that showed a signal close to the CCDC67 locus (42), encoding a key protein for ciliogenesis (43). Finally, accumulating reports of immunological disorders linked to dysfunctional actin dynamics at the immune synapse further support the causality of the association (44, 45). It might seem contradictory that patients carrying putative tolerance variants would still be immunocompetent. It is however well-known that transplantation tolerance in primates cannot be achieved by blocking a single mechanism (46, 47). It is therefore likely that renal allograft tolerance results from a combination of the hereditary predisposition that we showed here, and the use of multiple immunosuppressive drugs at the time of transplantation. To our knowledge, this is the first unbiased, exome-wide study of tolerance in solid organ transplantation. While we identified genetic variants in only a subset of our patients (10/36, 27%), indicating causal heterogeneity, other mechanisms, e.g., epigenetic changes, might possibly involve the same molecules, or other molecules in the same pathway, like Homer1 as suggested elsewhere (48). The unusually high proportion (25%) of donor-recipient pairs that were fully matched for the HLA A, B and DR loci, also argues for a possible contribution of donor-recipient genetic interactions (49).
Our study has several limitations. First, because documented cases of allograft tolerance in renal transplantation are extremely rare, we could not replicate our results in an independent cohort. Our study stems from a near-complete survey of more than 218,000 renal transplant recipients in 28 countries, where tolerance was reported in 3 per 10,000 renal recipients (2). Contrasting with other successful genetic projects on common phenotypes (diabetes, chronic kidney disease, and many more), our recruitment was necessarily limited. We designed our study specifically to overcome this issue (SKAT adjusted for small samples, FDR, enlarged control group), resulting in several significant rare variants in biologically relevant genes. While a plethora of GWA studies for many kinds of traits has called for stringent guidelines on p-values and replication cohorts, the number of tolerant patients needed to meet these standards is simply not feasible. Nevertheless, the FDRs of ~0.03 are highly convincing, indicating only a 3% chance that the results are false discoveries (20). Second, while our study allowed to unmask a few cases of possibly Mendelian tolerance—akin to studies of rare Mendelian diseases where only a few patients can uncover the underlying gene (16, 17)—it was underpowered to detect heterogeneous and/or polygenic factors in the remaining patients. We may also have missed variants of large effect as we did not cover every exon of the genome, partly because of the limits of exome capture kits (50). Finally, in the context of a rare trait and a posteriori identification we must acknowledge that data collection could not be exhaustively controlled. Nevertheless, this is the first exome-wide study of tolerance and provides a springboard for candidate gene studies and meta-analyses.
In conclusion, our work strongly suggests that spontaneous renal allograft tolerance stems from moderate to high impact, coding variants of primary cilium genes in at least a sizeable subgroup of patients (10/36, 27%). Further research should confirm our results and investigate the mechanisms involved.
Data availability statement
The bam files of the 38 tolerant patients who consented to further use of their data by all bona fide researchers are deposited in the European Genome-Phenome Archive (EGAS00001007154 and 155).
Ethics statement
The studies involving human participants were reviewed and approved by Erasme-ULB Ethics Committee, Université Libre de Bruxelles, Brussels, Belgium. The patients/participants provided their written informed consent to participate in this study.
Author contributions
DA and MA conceived the study. AM and OV implemented the biocollection of the TOMOGRAM study with the support of the DESCARTES ERA-EDTA board members and MG the biocollection of Nantes University with the support of the DIVAT Consortium. AM, OV, MG, MS, JP, MM, and the RENAL TOLERANCE INVESTIGATORS managed IRB approval process and collected patient data and samples. AM and MG reviewed patients' phenotypes. JD, RBS, ET, SVD, and CV obtained IRB-approved and consent from control patients. AM, CP, and VJ performed DNA extraction, preparation of the samples, and Sanger validation of the variants. BC, DC, and DD created patient libraries, sequenced the samples, and prepared them for CNV analysis. JS performed variant calling and annotation. LG, JS, DC, GS, SD, VJ, IP, OV, and PH contributed significant intellectual content. CO performed the principal component analysis and the case-control study under the guidance of ME. AM, CO, RD, ME, DA, SB, and MA analyzed the data and wrote the manuscript. AM, MA, SB, and DA supervised all aspects of the project. All authors revised the manuscript. All authors contributed to the article and approved the submitted version.
Group members of the Renal Tolerance Investigators
Maria Aguilar Rodríguez1, Friederike Bachmann2, Rajendra Bahadur Shahi3, Frederike Bemelman4, Luboslav Bena5, Luigi Biancone3, Laura Braun7, Klemens Budde8, Alejandro Camargo-Salamanca9, Katia Clemente10, Hulya Colak11, Adrian Covic12, Jacques Degreve3, Philippe Gatault13, François Glowacki14, Karine Hadaya15,16, Marc Hazzan14, Bénédicte Janbon15,16, Christophe Legendre17, Umberto Maggiore18, Marius Miglinas19, Anja Mühlfeld20, Maarten Naesens21, Christian Noël14, Rainer Oberbauer22, Evangeline Pillebout23, Gian Benedetto Piredda24, Francesco Pisani10, Ana Ramírez Puga25, Tomas Reischig26, Francisco González-Roncero1, Søren Schwartz Sørensen27, Daniel Seron Micas28, Nurhan Seyahi29, Dimitrie Siriopol12, Goce Spasovski30, Jean-François Subra31, Erik Teugels3, Serhan Tuǧlular32, Sonia Van Dooren33, Catheline Vilain34,35, Florence Villemain31, Xavier Warling36, Bruno Watschinger22 and Laurent Weekers37
1Hospital Universitario Virgen del Rocío, Unidad de Gestión Clínica de Urología y Nefrología, Sevilla, Spain
2Charité University Medicine Berlin, Campus Virchow Klinikum, Nierentransplantationsambulanz, Berlin, Germany
3Laboratory of Medical and Molecular Oncology (LMMO), Vrije Universiteit Brussel (VUB), Brussels, Belgium
4Renal Transplant Unit, Department of Nephrology, Academic Medical Center, Amsterdam, Netherlands
5Transplant Centre, University Hospital Louis Pasteur, Košice, Slovakia
6Department of Medical Sciences, University of Torino, Torino, Italy
7Hôpital de jour de Néphrologie, Service de Néphrologie et Transplantation Rénale, Nouvel Hôpital Civil, CHU, Strasbourg, France
8Charité - Universitätsmedizin Berlin, Medizinische Klinik mit Schwerpunkt Nephrologie und Internistische Intensivmedizin, Berlin, Germany
9Colombiana de Trasplantes, Bogotá, Colombia
10U.O.C. Trapianti D'Organo, L'Aquila, Italy
11Department of Nephtology, Tepecik Training and Research Hospital, Izmir, Turkey
12Dr. CI Parhon University Hospital, Lasi, Romania
13Service Néphrologie - Immunoclinique, CHRU, Hôpital Bretonneau, Tours, France
14Nephrology Department, University Hospital of Lille, Lille, France
15Department of Nephrology and Hypertension, Geneva University Hospitals, Geneva, Switzerland
16Transplantation rénale, CHU Grenoble, Grenoble, France
17Service de Néphrologie-Transplantation, Hôpital Necker, Université de Paris, Paris, France
18Nephrology Unit, Department of Medicine and Surgery, University of Parma, Parma, Italy
19Nephrology Center, Medical Faculty, Vilnius University, Hospital Santaros Klinikos, Vilnius, Lithuania
20Division of Nephrology, Uniklinik RWTH Aachen, Aachen, Germany
21Department of Microbiology, Immunology and Transplantation, KU Leuven, Leuven, Belgium
22Department of Internal Medicine III, Nephrology, Medical University Vienna, AKH Wien, Vienna, Austria
23Service de Néphrologie, Hôpital Saint-Louis, Paris, France
24Kidney Transplant Azienda Ospedaliera G. Brotzu, Kidney Diseases, Cagliari, Italy
25Universitario Insular de Gran Canaria, Servicio de nefrología, Las Palmas, Spain
26Department of Internal Medicine I, Charles University Teaching Hospital Pilsen, Pilsen, Czechia
27Department of Nephrology, Rigshospitalet, University of Copenhagen, Copenhagen, Denmark
28Servicio de Nefrología, Vall d'Hebron Hospital Universitari, Vall d'Hebron Barcelona Hospital Campus, Barcelona, Spain
29Department of Nephrology, İstanbul University-Cerrahpaşa, Cerrahpaşa Medical Faculty, Istanbul, Turkey
30Department of Nephrology, Medical Faculty, University of St. Cyril and Methodius, Skopje, Macedonia
31Service de Néphrologie-Dialyse-Transplantation, CHU, Angers, France
32Marmara School of Medicine Hastanesi (Nephrology), S.B. Marmara Üniversitesi EAH Mimar Sinan Cd., Istanbul, Turkey
33Center for Medical Genetics, Reproduction and Genetics, Reproduction Genetics and Regenerative Medicine, Vrije Universiteit Brussel, UZ Brussel, Brussels, Belgium
34ULB Center of Human Genetics, Department of Genetics, Hôpital Erasme, Université Libre de Bruxelles, Brussels, Belgium
35Department of Genetics, Hôpital Universitaire des Enfants Reine Fabiola, ULB Center of Human Genetics, Université Libre de Bruxelles, Brussels, Belgium
36Department of Nephrology, Centre Hospitalier Régional de la Citadelle, Liège, Belgium
37Nephrology Department, Avenue de l'Hôpital 1, CHU of Liege, Liège, Belgium
Funding
AM was supported by the Fonds Erasme; the Fonds de la Recherche Scientifique Médicale (FRSM, PDR 23670170); the Fonds Carine VYGHEN; and the Fonds Horlait-Dapsens). MA was supported by the Belgian FNRS (T.0174.15); and Fonds Erasme. RD was supported by a Marie Skłodowska-Curie fellowship (IF-EF) from the European Union's Horizon 2020 research and innovation program under Grant Agreement No. 706296 and the ANR project KTD-innov (ANR-17-RHUS-0010) thanks to the French government financial support managed by the French National Research Agency (ANR) within investments into the future program. This work was performed in the context of the IHU-Cesti project (ANR-10-IBHU-005), the DHU Oncogreffe, the LabEx IGO program (no. ANR-11-LABX-0016), the ANR project PRELUD (ANR-18-CE17-0019) and the ANR project BIKET (ANR-17-CE17-0008). The IHU-Cesti project was also supported by Nantes Métropole and Région Pays de la Loire. The laboratory received funding from the European Union's Horizon 2020 Research and Innovation Program under Grant Agreement No. 754995. OV was supported by the Ministry of Health of the Czech Republic (NV19-06-00031). TOMOGRAM was also supported logistically and financially by the ERA-EDTA – DESCARTES working group, the Department of Nephrology at Antwerp University Hospital, Belgium and the Institute for Clinical and Experimental Medicine (IKEM) in Prague, Czech Republic. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Open access funding provided by University of Geneva.
Acknowledgments
We are in debt to the patients, clinicians, and study nurses, who took part in this study and especially to the members of the DESCARTES ERA-EDTA working group and the DIVAT consortium for mounting the biocollection. We thank Ms. Prokopova and Mr. Snelders for administrative support, Eric Trepo for interesting discussion, the cytogenetics unit of Erasme Hospital, Brussels, Belgium for preparing cell lines, as well as the biological resource centre for biobanking from Nantes (Nantes Université, CHU Nantes, Centre de resources biologiques (CRB), F-44000 Nantes, France) (BRIF: BB-0033-00040).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmed.2022.976248/full#supplementary-material
References
1. Brouard S, Pallier A, Renaudin K, Foucher Y, Danger R, Devys A, et al. The natural history of clinical operational tolerance after kidney transplantation through twenty-seven cases. Am J Transplant. (2012) 12:3296–307. doi: 10.1111/j.1600-6143.2012.04249.x
2. Massart A, Pallier A, Pascual J, Viklicky O, Budde K, Spasovski G, et al. The DESCARTES-Nantes survey of kidney transplant recipients displaying clinical operational tolerance identifies 35 new tolerant patients and 34 almost tolerant patients. Nephrol Dial Transplant. (2016) 31:1002–13. doi: 10.1093/ndt/gfv437
3. Massart A, Ghisdal L, Abramowicz M, Abramowicz D. Operational tolerance in kidney transplantation and associated biomarkers. Clin Exp Immunol. (2017) 189:138–57. doi: 10.1111/cei.12981
4. Starzl TE, Murase N, Demetris AJ, Trucco M, Abu-Elmagd K, Gray EA, et al. Lessons of organ-induced tolerance learned from historical clinical experience. Transplantation. (2004) 77:926–9. doi: 10.1097/01.TP.0000117780.74133.74
5. Scandling JD, Busque S, Shizuru JA, Lowsky R, Hoppe R, Dejbakhsh-Jones S, et al. Chimerism, graft survival, and withdrawal of immunosuppressive drugs in HLA matched and mismatched patients after living donor kidney and hematopoietic cell transplantation. Am J Transplant. (2015) 15:695–704. doi: 10.1111/ajt.13091
6. Takemoto SK, Pinsky BW, Schnitzler MA, Lentine KL, Willoughby LM, Burroughs TE, et al. A retrospective analysis of immunosuppression compliance, dose reduction and discontinuation in kidney transplant recipients. Am J Transplant. (2007) 7:2704–11. doi: 10.1111/j.1600-6143.2007.01966.x
7. Oura T, Cosimi AB, Kawai T. Chimerism-based tolerance in organ transplantation: preclinical and clinical studies. Clin Exp Immunol. (2017) 189:190–6. doi: 10.1111/cei.12969
8. Cohen JC, Kiss RS, Pertsemlidis A, Marcel YL, McPherson R, Hobbs HH. Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science. (2004) 305:869–72. doi: 10.1126/science.1099870
9. Emond MJ, Louie T, Emerson J, Zhao W, Mathias RA, Knowles MR, et al. Exome sequencing of extreme phenotypes identifies DCTN4 as a modifier of chronic Pseudomonas aeruginosa infection in cystic fibrosis. Nat Genet. (2012) 44:886–9. doi: 10.1038/ng.2344
10. Johansen CT, Wang J, Lanktree MB, Cao H, McIntyre AD, Ban MR, et al. Excess of rare variants in genes identified by genome-wide association study of hypertriglyceridemia. Nat Genet. (2010) 42:684–7. doi: 10.1038/ng.628
11. Tsai CT, Hsieh CS, Chang SN, Chuang EY, Juang JM, Lin LY, et al. Next-generation sequencing of nine atrial fibrillation candidate genes identified novel de novo mutations in patients with extreme trait of atrial fibrillation. J Med Genet. (2015) 52:28–36. doi: 10.1136/jmedgenet-2014-102618
12. Do R, Stitziel NO, Won HH, Jorgensen AB, Duga S, Angelica Merlini P, et al. Exome sequencing identifies rare LDLR and APOA5 alleles conferring risk for myocardial infarction. Nature. (2015) 518:102–6. doi: 10.1038/nature13917
13. McLaren CE, Emond MJ, Subramaniam VN, Phatak PD, Barton JC, Adams PC, et al. Exome sequencing in HFE C282Y homozygous men with extreme phenotypes identifies a GNPAT variant associated with severe iron overload. Hepatology. (2015) 62:429–39. doi: 10.1002/hep.27711
14. Mackelprang RD, Bamshad MJ, Chong JX, Hou X, Buckingham KJ, Shively K, et al. Whole genome sequencing of extreme phenotypes identifies variants in CD101 and UBE2V1 associated with increased risk of sexually acquired HIV-1. PLoS Pathog. (2017) 13:e1006703. doi: 10.1371/journal.ppat.1006703
15. Botstein D, Risch N. Discovering genotypes underlying human phenotypes: past successes for mendelian disease, future approaches for complex disease. Nat Genet. (2003) 33(Suppl.):228–37. doi: 10.1038/ng1090
16. Ng SB, Bigham AW, Buckingham KJ, Hannibal MC, McMillin MJ, Gildersleeve HI, et al. Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat Genet. (2010) 42:790–3. doi: 10.1038/ng.646
17. Ng SB, Turner EH, Robertson PD, Flygare SD, Bigham AW, Lee C, et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. (2009) 461:272–6. doi: 10.1038/nature08250
18. Guo MH, Dauber A, Lippincott MF, Chan YM, Salem RM, Hirschhorn JN. Determinants of power in gene-based burden testing for monogenic disorders. Am J Hum Genet. (2016) 99:527–39. doi: 10.1016/j.ajhg.2016.06.031
19. Emond MJ, Louie T, Emerson J, Chong JX, Mathias RA, Knowles MR, et al. Exome sequencing of phenotypic extremes identifies CAV2 and TMC6 as interacting modifiers of chronic pseudomonas aeruginosa infection in cystic fibrosis. PLoS Genet. (2015) 11:e1005273. doi: 10.1371/journal.pgen.1005273
20. He Z, Liu L, Wang C, Le Guen Y, Lee J, Gogarten S, et al. Identification of putative causal loci in whole-genome sequencing data via knockoff statistics. Nat Commun. (2021) 12:3152. doi: 10.1038/s41467-021-22889-4
21. Lee S, Emond MJ, Bamshad MJ, Barnes KC, Rieder MJ, Nickerson DA, et al. Optimal unified approach for rare-variant association testing with application to small-sample case-control whole-exome sequencing studies. Am J Hum Genet. (2012) 91:224–37. doi: 10.1016/j.ajhg.2012.06.007
22. Cirulli ET, Goldstein DB. Uncovering the roles of rare variants in common disease through whole-genome sequencing. Nat Rev Genet. (2010) 11:415–25. doi: 10.1038/nrg2779
23. Martin E, Girardello R, Dittmar G, Ludwig A. New insights into the organization and regulation of the apical polarity network in mammalian epithelial cells. FEBS J. (2021) 288:7073–95. doi: 10.1111/febs.15710
24. Worley PF, Zeng W, Huang G, Kim JY, Shin DM, Kim MS, et al. Homer proteins in Ca2+ signaling by excitable and non-excitable cells. Cell Calcium. (2007) 42:363–71. doi: 10.1016/j.ceca.2007.05.007
25. Huang GN, Huso DL, Bouyain S, Tu J, McCorkell KA, May MJ, et al. NFAT binding and regulation of T cell activation by the cytoplasmic scaffolding Homer proteins. Science. (2008) 319:476–81. doi: 10.1126/science.1151227
26. Nie DS, Liu Y, Juan H, Yang X. Overexpression of human SPATA17 protein induces germ cell apoptosis in transgenic male mice. Mol Biol Rep. (2013) 40:1905–10. doi: 10.1007/s11033-012-2246-z
27. Putkey JA, Kleerekoper Q, Gaertner TR, Waxham MN. A new role for IQ motif proteins in regulating calmodulin function. J Biol Chem. (2003) 278:49667–70. doi: 10.1074/jbc.C300372200
28. Wei F, Karihaloo A, Yu Z, Marlier A, Seth P, Shibazaki S, et al. Neutrophil gelatinase-associated lipocalin suppresses cyst growth by Pkd1 null cells in vitro and in vivo. Kidney Int. (2008) 74:1310–8. doi: 10.1038/ki.2008.395
29. Cassidy H, Slyne J, Higgins M, Radford R, Conlon PJ, Watson AJ, et al. Neutrophil gelatinase-associated lipocalin (NGAL) is localised to the primary cilium in renal tubular epithelial cells - a novel source of urinary biomarkers of renal injury. Biochim Biophys Acta Mol Basis Dis. (2019) 1865:165532. doi: 10.1016/j.bbadis.2019.165532
30. Flo TH, Smith KD, Sato S, Rodriguez DJ, Holmes MA, Strong RK, et al. Lipocalin 2 mediates an innate immune response to bacterial infection by sequestrating iron. Nature. (2004) 432:917–21. doi: 10.1038/nature03104
31. Schreiber A, Rousselle A, Klocke J, Bachmann S, Popovic S, Bontscho J, et al. Neutrophil gelatinase-associated lipocalin protects from ANCA-induced GN by inhibiting TH17 immunity. J Am Soc Nephrol. (2020) 31:1569–84. doi: 10.1681/ASN.2019090879
32. Chen W, Li W, Zhang Z, Tang X, Wu S, Yao G, et al. Lipocalin-2 exacerbates lupus nephritis by promoting Th1 cell differentiation. J Am Soc Nephrol. (2020) 31:2263–77. doi: 10.1681/ASN.2019090937
33. Vlkova M, Chovancova Z, Nechvatalova J, Connelly AN, Davis MD, Slanina P, et al. Neutrophil and granulocytic myeloid-derived suppressor cell-mediated T cell suppression significantly contributes to immune dysregulation in common variable immunodeficiency disorders. J Immunol. (2019) 202:93–104. doi: 10.4049/jimmunol.1800102
34. Ashraf MI, Schwelberger HG, Brendel KA, Feurle J, Andrassy J, Kotsch K, et al. Exogenous lipocalin 2 ameliorates acute rejection in a mouse model of renal transplantation. Am J Transplant. (2016) 16:808–20. doi: 10.1111/ajt.13521
35. Weng YC, Huang YT, Chiang IC, Tsai PJ, Su YW, Chou WH. Lipocalin-2 mediates the rejection of neural transplants. FASEB J. (2021) 35:e21317. doi: 10.1096/fj.202001018R
36. Le Borgne M, Shaw AS. Immunology. Do T cells have a cilium? Science. (2013) 342:1177–8. doi: 10.1126/science.1248078
37. Finetti F, Baldari CT. Compartmentalization of signaling by vesicular trafficking: a shared building design for the immune synapse and the primary cilium. Immunol Rev. (2013) 251:97–112. doi: 10.1111/imr.12018
38. Huppa JB, Gleimer M, Sumen C, Davis MM. Continuous T cell receptor signaling required for synapse maintenance and full effector potential. Nat Immunol. (2003) 4:749–55. doi: 10.1038/ni951
39. Hartzell CA, Jankowska KI, Burkhardt JK, Lewis RS. Calcium influx through CRAC channels controls actin organization and dynamics at the immune synapse. Elife. (2016) 5:e14850. doi: 10.7554/eLife.14850
40. Groopman EE, Marasa M, Cameron-Christie S, Petrovski S, Aggarwal VS, Milo-Rasouly H, et al. Diagnostic utility of exome sequencing for kidney disease. N Engl J Med. (2019) 380:142–51. doi: 10.1056/NEJMoa1806891
41. Danger R, Thervet E, Grisoni ML, Puig PL, Pallier A, Tregouet D, et al. PARVG gene polymorphism and operational renal allograft tolerance. Transplant Proc. (2012) 44:2845–8. doi: 10.1016/j.transproceed.2012.09.034
42. Ghisdal L, Baron C, Lebranchu Y, Viklicky O, Konarikova A, Naesens M, et al. Genome-wide association study of acute renal graft rejection. Am J Transplant. (2017) 17:201–9. doi: 10.1111/ajt.13912
43. Zhao H, Zhu L, Zhu Y, Cao J, Li S, Huang Q, et al. The Cep63 paralogue Deup1 enables massive de novo centriole biogenesis for vertebrate multiciliogenesis. Nat Cell Biol. (2013) 15:1434–44. doi: 10.1038/ncb2880
44. Tsyklauri O, Niederlova V, Forsythe E, Prasai A, Drobek A, Kasparek P, et al. Bardet-Biedl Syndrome ciliopathy is linked to altered hematopoiesis and dysregulated self-tolerance. EMBO Rep. (2021) 22:e50785. doi: 10.15252/embr.202050785
45. Sprenkeler EGG, Webbers SDS, Kuijpers TW. When actin is not actin' like it should: a new category of distinct primary immunodeficiency disorders. J Innate Immun. (2021) 13:3–25. doi: 10.1159/000509717
47. Miller ML, Chong AS, Alegre ML. Fifty shades of tolerance: beyond a binary tolerant/non-tolerant paradigm. Curr Transplant Rep. (2017) 4:262–9. doi: 10.1007/s40472-017-0166-5
48. Rodriguez RM, Hernandez-Fuentes MP, Corte-Iglesias V, Saiz ML, Lozano JJ, Cortazar AR, et al. Defining a methylation signature associated with operational tolerance in kidney transplant recipients. Front Immunol. (2021) 12:709164. doi: 10.3389/fimmu.2021.709164
49. Rodriguez DS, Jankowska-Gan E, Haynes LD, Leverson G, Munoz A, Heisey D, et al. Immune regulation and graft survival in kidney transplant recipients are both enhanced by human leukocyte antigen matching. Am J Transplant. (2004) 4:537–43. doi: 10.1111/j.1600-6143.2004.00385.x
Keywords: exome sequencing, renal transplantation, operational tolerance, NGAL, LCN2, Homer2, IQCH, primary cilium
Citation: Massart A, Danger R, Olsen C, Emond MJ, Viklicky O, Jacquemin V, Soblet J, Duerinckx S, Croes D, Perazzolo C, Hruba P, Daneels D, Caljon B, Sever MS, Pascual J, Miglinas M, the Renal Tolerance Investigators, Pirson I, Ghisdal L, Smits G, Giral M, Abramowicz D, Abramowicz M and Brouard S (2023) An exome-wide study of renal operational tolerance. Front. Med. 9:976248. doi: 10.3389/fmed.2022.976248
Received: 23 June 2022; Accepted: 31 October 2022;
Published: 17 May 2023.
Edited by:
Xiaopeng Hu, Beijing Chaoyang Hospital, Capital Medical University, ChinaReviewed by:
Swastika Sur, University of California, San Francisco, United StatesXin Geng, Eastern Hepatobiliary Surgery Hospital, China
Copyright © 2023 Massart, Danger, Olsen, Emond, Viklicky, Jacquemin, Soblet, Duerinckx, Croes, Perazzolo, Hruba, Daneels, Caljon, Sever, Pascual, Miglinas, the Renal Tolerance Investigators, Pirson, Ghisdal, Smits, Giral, Abramowicz, Abramowicz and Brouard. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marc Abramowicz, marc.abramowicz@unige.ch
†These authors have contributed equally to this work and share first authorship
‡These authors have contributed equally to this work and share last authorship
§The names of the authors directly contributing to the consortium and the building of the dataset are listed in Group members of the Renal Tolerance Investigators section
 Annick Massart
Annick Massart Richard Danger
Richard Danger Catharina Olsen2,5,6†
Catharina Olsen2,5,6†  Ondrej Viklicky
Ondrej Viklicky Petra Hruba
Petra Hruba Julio Pascual
Julio Pascual Daniel Abramowicz
Daniel Abramowicz Marc Abramowicz
Marc Abramowicz Sophie Brouard
Sophie Brouard