Application of artificial intelligence in diagnosis and treatment of colorectal cancer: A novel Prospect
- 1Department of General Surgery, The First Affiliated Hospital of Dalian Medical University, Dalian, China
- 2Department of Respiratory, The First Affiliated Hospital of Dalian Medical University, Dalian, China
- 3Institute of Laboratory Animal Science, Chinese Academy of Medical Sciences and Comparative Medicine Center, Peking Union Medical College, Beijing, China
In the past few decades, according to the rapid development of information technology, artificial intelligence (AI) has also made significant progress in the medical field. Colorectal cancer (CRC) is the third most diagnosed cancer worldwide, and its incidence and mortality rates are increasing yearly, especially in developing countries. This article reviews the latest progress in AI in diagnosing and treating CRC based on a systematic collection of previous literature. Most CRCs transform from polyp mutations. The computer-aided detection systems can significantly improve the polyp and adenoma detection rate by early colonoscopy screening, thereby lowering the possibility of mutating into CRC. Machine learning and bioinformatics analysis can help screen and identify more CRC biomarkers to provide the basis for non-invasive screening. The Convolutional neural networks can assist in reading histopathologic tissue images, reducing the experience difference among doctors. Various studies have shown that AI-based high-level auxiliary diagnostic systems can significantly improve the readability of medical images and help clinicians make more accurate diagnostic and therapeutic decisions. Moreover, Robotic surgery systems such as da Vinci have been more and more commonly used to treat CRC patients, according to their precise operating performance. The application of AI in neoadjuvant chemoradiotherapy has further improved the treatment and efficacy evaluation of CRC. In addition, AI represented by deep learning in gene sequencing research offers a new treatment option. All of these things have seen that AI has a promising prospect in the era of precision medicine.
1. Introduction
Colorectal cancer (CRC) is a common disease that threatens the public health. According to the International Agency for Research on Cancer, there were an estimated 1.93 million new cases of CRC worldwide in 2020, making itself in the third place in the most common cancer list (Figure 1A). The incidence of CRC is particularly significant in countries undergoing social and economic transition. In China, there were about 560 thousand newly diagnosed cases of CRC in 2020, second only to lung cancer in terms of morbidity (1) (Figure 1B). Based on the GLOBOCAN 2020 cancer assessment and population data from the World Health Organization (WHO), the number of new cases of CRC in China is estimated to reach 590 thousand in 2022, more than any other countries in the world (2). Besides smoking, obesity, and unhealthy lifestyle, the incidence of CRC is also related to gender, genetic cause, and family factors (3–6). Currently, the main diagnostic methods for CRC include laboratory tests, endoscopy, imaging and histopathology examination, etc. Traditional ways of treating CRC entail surgery, radiotherapy, and post-metastasis therapy, among others (7–10). Despite all these tools, the rise in CRC incidence and mortality is alarming. With the recent attention of early screening and the rapid development of precision medicine (4, 11), a new diagnosis and treatment model for CRC is on the horizon.
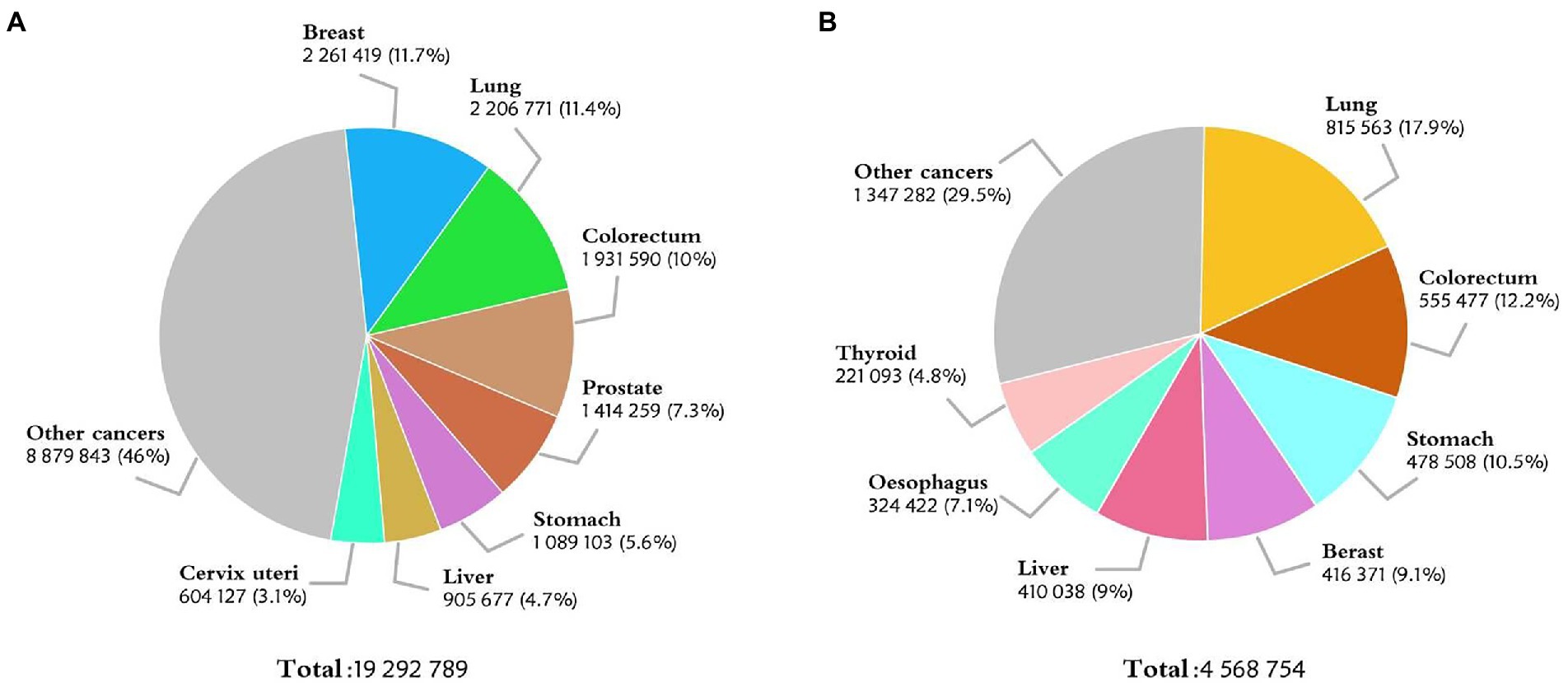
Figure 1. (A) Estimated number of new cases in 2020, World, both sexes, all ages. (B) Estimated number of new cases in 2020, China, both sexes, all ages. (Data source: GLOBOCAN 2020).
Artificial intelligence (AI) can be understood as studying the principle of human intelligence activities, constructing an artificial system with certain intelligence, and studying how to let computers complete the work that requires human intelligence in the past. Nowadays, with the rapid development of computer technology and the vigorous promotion of precision medicine, the application of AI in medicine is also in full swing (12, 13). AI applications in medicine are now divided into virtual and physical branches (12). Machine learning (ML) is an essential subbranch of AI. It also can be divided into subsets such as deep learning (DL), supervised learning (SL), semi-supervised learning (SSL), support vector machine (SVM), random forest (RF), and convolutional neural network (CNN) (14–17) (Figure 2). Among them, DL and CNN are representatives of the most successful algorithms used in medicine in recent years (17, 18). They play a very broad role in data management (19), information control (20), diagnosis prediction (21), and drug delivery (22). The branches of physics mainly include medical equipment (23) and robot applications, such as da Vinci robot system, which is widely used today (24, 25). This article mainly discusses the development of AI and its application in the diagnosis and treatment of CRC.
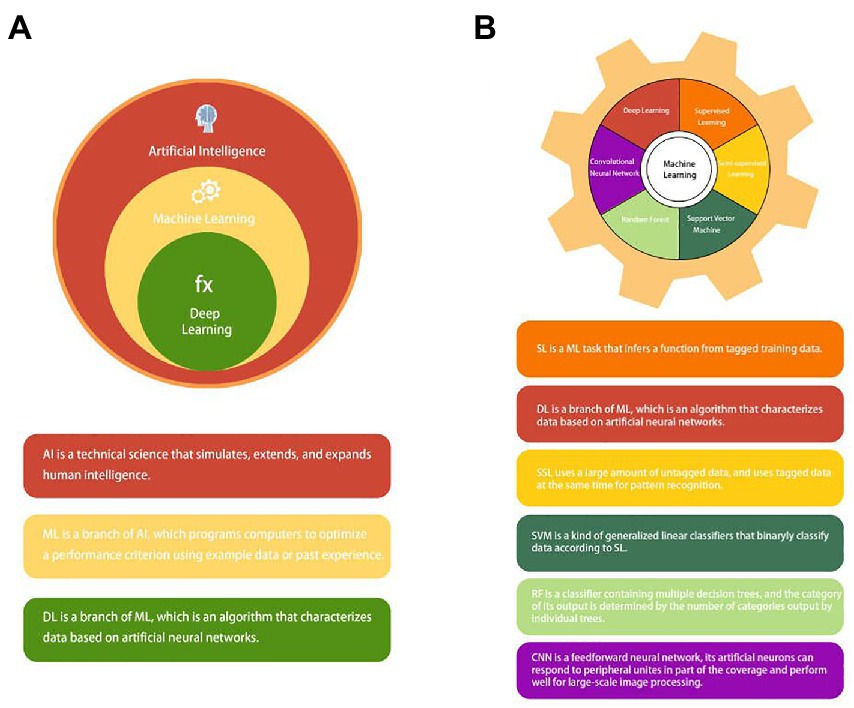
Figure 2. (A) The concept and relationship of Artificial intelligence (AI), machine learning (ML), and deep learning (DL). (B) Common types of machine learning (ML): supervised learning (SL); deep learning (DL); semi-supervised learning (SSL); support vector machine (SVM); random forest (RF); and convolutional neural network (CNN).
2. The development of AI
It is generally believed that the development of AI evolved from robots. The word “robot” first appeared in the works of the Czech dramatist Karel Capek in the early 20th century, referring to forced labor or compulsory work (26). However, the topic about robots has been popular in Chinese and Western cultures for a long time. Hephaestus and his robotic dogs constantly featured in ancient Greek and Roman myths. Aristotle’s genius prediction for robots in his Politics also reflected people’s beautiful vision of robots at that time. There are also many stories about automata in ancient China. More than 3,000 years ago, a mechanic named Yan Shi presented King Mu of Zhou to a human-size mechanical device. To drink and recite poems with Liu every day, Emperor Yang of the Sui Dynasty ordered the craftsman to create a wooden mechanical man according to Liu’s figure. This wooden man could kneel and toast like Liu (12, 26, 27). Leonardo da Vinci also made a major contributor to medical physics. He presented his invention of automata robots, a mobile knight, in 1495. In the da Vinci notebook, we found details and sketches of manufacturing robots. With the approval of the FDA, the first generation da Vinci surgical system was manufactured and put on the market in 1999. So far, robot-assisted surgery has been widely used (12, 26–28).
In 1950, Alan Turing experimentally detected some machines that showed intelligent behavior like human, which we call the “Turing test.” At the Dartmouth conference in 1956, John McCarthy and his team officially proposed the concept of AI (13, 29–31). Over the past 60 years, AI has made great progress in all walks of life. AI has apparent advantages in solving complex nonlinear parameter problems, and its accurate prediction ability has made it widely used in waste generation, collection, management, and conversion processes (32). AI is also considered to play an important role in addressing rising demand, road forecasting, planning, and management, self-driving and safety (33, 34). The development of educational AI can reduce the expenditure budget and the burden on teachers and provide more personalized teaching services for each student. On the other hand, it can also help realize educational opportunities for more students (35). Over the past few years, the AI player represented by AlphaGo has defeated the world Go champions, including Jie Ke, indicating that we have made exciting progress in the computer Go. These results are primarily based on the creative combination of deep convolutional neural networks (DCNN) and Monte Carlo tree search (MCTS). We still need to further understand the working mechanism of the model through the visual operation (36). AI can also have an impact on monitoring and controlling the spread of the virus. Vaishya et al. have obtained a result-driving technology through experiments, which can play a role in the early screening of COVID-19, the detection and tracking of infected patients, the formulation of adjuvant treatment plans, as well as the development of vaccines (37).
With the development of ML and DL, AI has become more and more widely used in medicine and has bright prospects in disease prediction, treatment, and prevention (29, 38, 39). This article mainly reviews the diagnosis and treatment of AI in CRC.
3. The application of AI in CRC diagnosis
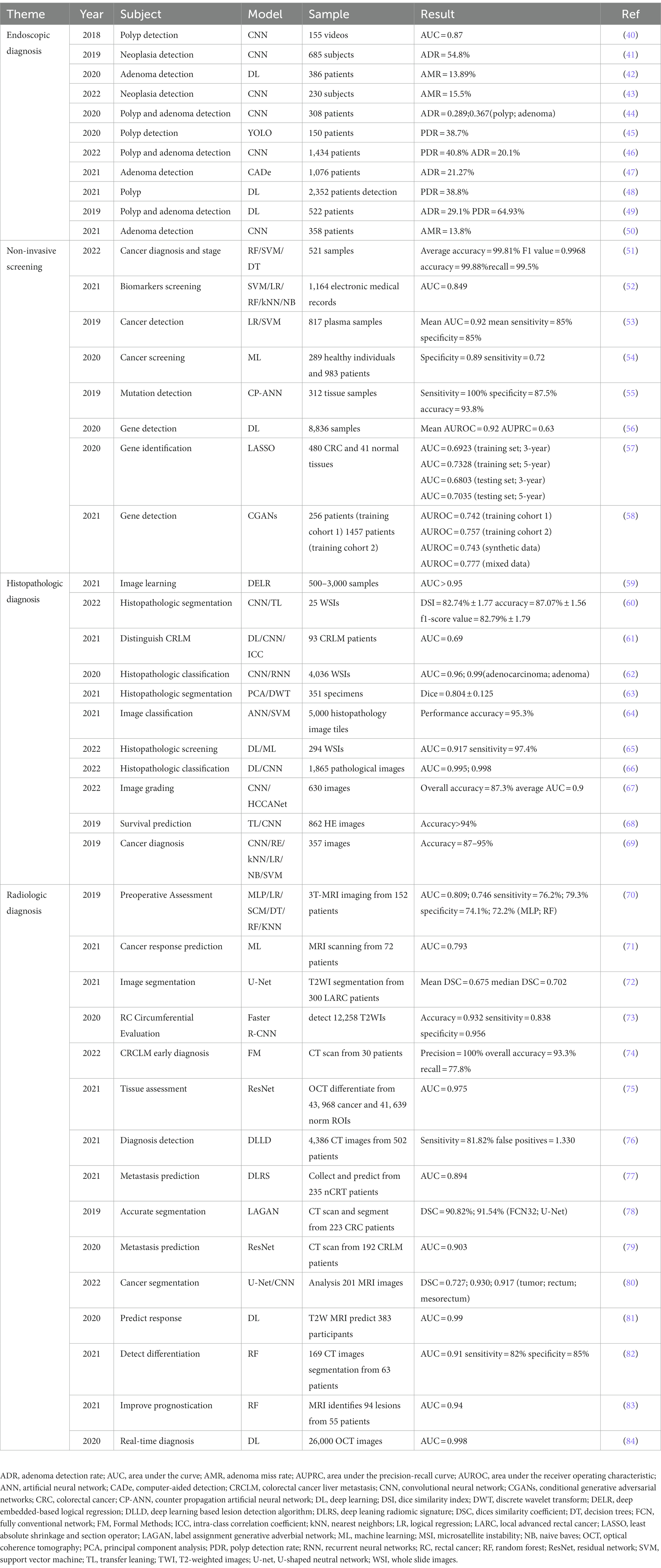
The application of AI in CRC screening can improve the early screening rate and thus significantly reduce the incidence and mortality of CRC patients. The bioinformatics tools embedded in AI can help screen and identify more CRC biomarkers. AI assisted pathology recognition technology can help pathologists improve efficiency, reduce workload, and lower the rate of misdiagnosis and missed diagnosis. ML, widely used in AI image recognition, can greatly improve the readability of medical images, reduce empirical errors, objectively provide reliable and comprehensive reference opinions, and help doctors make more accurate clinical decisions (Figure 3). Common AI models for CRC diagnosis are summarized in Table 1.
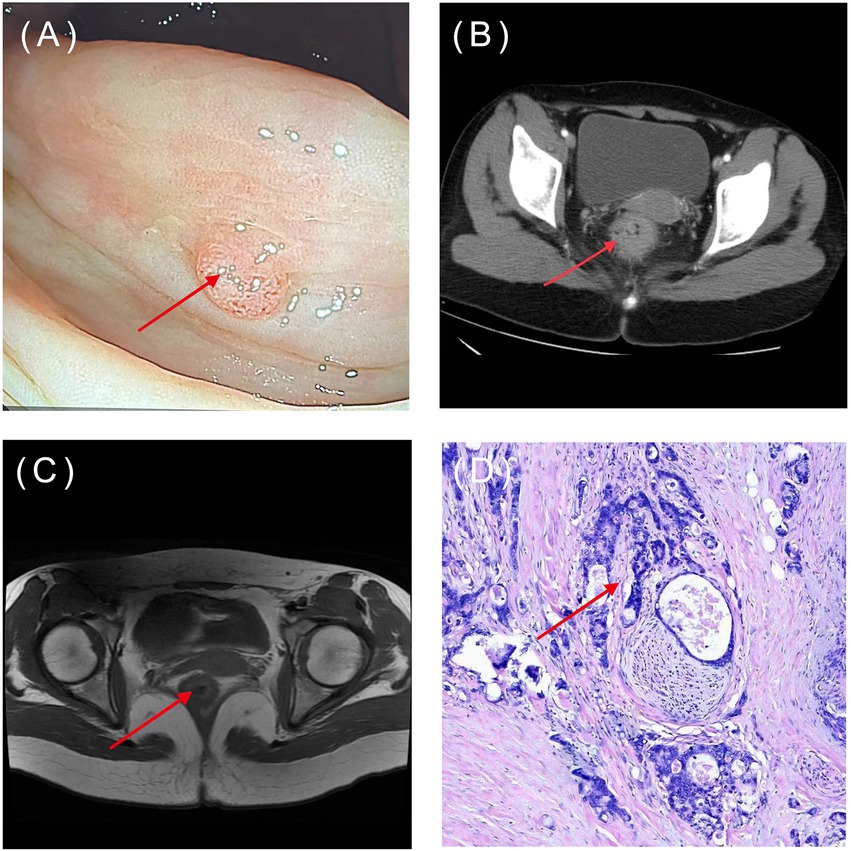
Figure 3. Common types of CRC diagnostic images: (A) Endoscopy; (B) CT; (C) MRI; (D) pathology image (HE×100). The arrow indicates the location of the lesions. (Image source: The First Affiliated Hospital of Dalian Medical University.)
3.1. Endoscopic diagnosis
Colonoscopy has long been regarded as the gold standard procedure for diagnosing colorectal diseases, and it is strongly recommended as an early screening criterion by national associations (85). Due to the high operator variability of quality, challenging and frequently inadequate preparation, high loss of work productivity, and so on, the detection rate of polyps and adenomas in early colorectal screening often varies greatly (86).
Researchers have used computer-aided detection (CADe) systems and AI, based primarily on DL algorithms, to improve the speed and accuracy of clinical detection of CRC and reduce the detection of missed lesions (50). The adenoma detection rate (ADR) is a reliable indicator for CRC detection. A higher ADR is often linked to a lower incidence and mortality in CRC patients (87). The combination of AI and colonoscopy can effectively improve ADR. It not only reduces the risk of CRC but also achieves the purpose of accurate resection, avoiding excessive burden on clinical work caused by the resection of many non-neoplastic polyps. A YOLOV3 AI algorithm was utilized to detect real-time polyps via media. It could accomplish an excellent effect in a short time. It was economical and affordable, making it suited for large-scale promotions in underdeveloped areas (88). Some adenomas and polyps detected by the real-time CADe system are small and low risk. Such adenomas and polyps are also easily ignored by endoscopists using traditional colonoscopy. Therefore this CADe system increases the incidence of CRC to a certain extent (49). A large number of studies are supporting AI-assisted colonoscopy in the diagnosis of CRC. Some optical biopsy techniques, which can capture real-time images manually, have also been proved to have a lot of potential in medical applications (45, 89). Another widely recognized colonoscopy index is adenoma miss rate (AMR), which refers to the difference between the lesions detected by consecutive endoscopy. Kamba et al. established a CADe system that uses the CNN algorithm to aid in the detection of AMR. The result demonstrated that the AMR of the CADe-assisted group was 22.9%, which was lower than that of the standard colonoscopy group. At the same time, the difference in ADR between the two groups was also only 10.9% (Figure 4). These results showed that AMR has greatly decreased with the assistance of AI. The AMR was more sensitive in detecting lesions than ADR indicators (50).
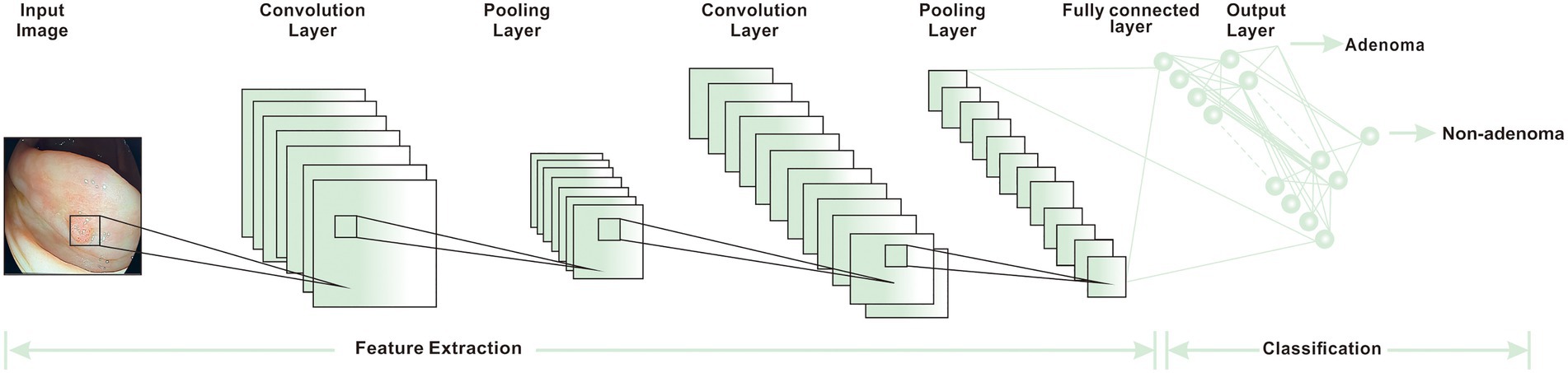
Figure 4. The basic workflow of CNN (15).
In addition, after the colorectal polyps are screened out, it is also very necessary to identify benign and malignant polyps. As endoscopic tools, including magnifying endoscopy, chromoendoscopy, confocal laser endomicroscopy, and autofluorescence endoscopy, continue to advance, the combination of AI and colorectal endoscopy has injected new impetus for future diagnosis (90). Japanese experts previously used narrow-band imaging (NBI) to classify magnifying endoscopes. They were earlier to apply NBI imaging technology and magnifying endoscopes in the clinical application (91). By combining NBI with magnification endoscopy, Gonai et al. calculated the difference between microvessel density in colorectal lesions, which was used as the differential point for the diagnosis of CRC and adenoma (92). In the same year, some experts in the United States developed CADe algorithms to distinguish tumor polyps and non-tumor polyps based on probe confocal laser endoscopy. The sensitivity, specificity and sensitivity are more than 90% (93). It is expected that the use of AI can greatly improve the accuracy of endoscopic diagnosis, and reduce the misdiagnosis rate and overtreatment.
3.2. Non-invasive screening
The main form of non-invasive screening is the detection of various relatively specific tumor markers from ascites, feces, blood, and other samples. Compared with colonoscopy screening, the non-invasive screening is not only lower risk, but it’s preparation time is shorter (94). However, there are few effective tumor markers for the detection of early CRC. Common non-invasive screening methods such as fecal occult blood test (FOBT) and carcinoembryonic antigen (CEA) have low sensitivity and specificity (95–97). In recent years, ML has been widely used in medical data analysis, which can help us improve the accuracy of existing biomarkers and screen out more potential marker genes (51).
CEA is one of the most studied colorectal tumor markers, but the screening of serum CEA has limited sensitivity in asymptomatic people (97). Li et al. extracted some of the most common markers from the laboratory blood tests and used five ML models to identify CRC patients from healthy people. The results showed that the logistic regression model could greatly improve the sensitivity and specificity of CEA. The model was an effective, economical, and non-invasive method for CRC identification (52). Common genes used for CRC DNA mutation detection include BRAF and KRAS, but these tumor markers are not sensitive or specific enough to show CRC (98). In 2019, Zhang et al. used near-infrared (NIR) spectroscopy combined with counter propagation artificial neural network (CP-ANN) to distinguish a BRAF V600E mutant from wild-type samples in CRC tissues. This method had a sensitivity of 100% and specificity of 87.5% for detecting the BRAF V600E mutant in CRC. It could be used for the auxiliary diagnosis of the BRAF mutation in CRC (55). The detection of abnormal DNA methylation markers in plasma or feces is a promising approach for the non-invasive early diagnosis of CRC. The SEPT9 gene methylation test has been used commercially as an alternative for CRC screening (94, 99, 100). However, the specificity of detection methods based on single DNA methylation sites is limited (101). In 2019, Kel et al. used a method called “Walking away” to collect data samples from 300 CRC patients, analyzing the data by ML and bioinformatics methods. They ultimately selected six DNA methylation epigenetic biomarkers for optimal cancer detection potential (102). The combined detection of abnormal DNA methylation markers can improve the detection rate of CRC.
Bioinformatics tools are increasingly used to analyze the pathogenesis of cancer. The effective combination of bioinformatics analysis and ML can screen and identify early CRC biomarkers, playing a role in evaluating the prognosis of CRC patients (51, 103). Hammad et al. recently identified 105 differentially expressed genes (DEGs) and 10 hub genes through bioinformatics analysis of gene expression microarray data in the Gene Expression Omnibus (GEO) database. The researchers used these tools, including SVM, Receiver operating characteristic curve (ROC), and survival analyses, to predict the diagnostic value of hub genes as CRC biomarkers. The results showed that the area under ROC curve (AUC) values of all genes were more than 0.92, confirming that these genes are expected to be biomarkers for CRC (98) With the development of sequencing technology, many non-coding RNAs (ncRNAs) have been discovered. The ncRNAs include messenger RNAs (mRNAs), microRNA (miRNAs), long non-coding RNAs (lncRNAs) and so on. Their extracellular properties are stable, easy to extract and preserve, and thus they can be studied in different body fluids (54, 104–106). MicroRNAs (miRNAs) are a class of endogenous ncRNAs with a length of about 22 nucleotides. They have a variety of important regulatory functions in the cell and are associated with the development and metastasis of cancer. There is increasing evidence that miRNAs are potential biomarkers of CRC (105, 106). In 2019, Zhang et al. identified whether miR-31 could be used as a biomarker for the diagnosis of CRC lymph node metastasis (LNM) through bioinformatics analysis techniques, including Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis, correlation analysis, survival analysis, validation of expression levels, protein–protein interactive (PPI) network construction, and Gene ontology (GO). These researchers found that miR-31 was significantly increased in the plasma and tissue of CRC patients with LNM. They predicted that TNS1 might be a targeted protein for miR-31, which has an important prognostic value for patients (105). Wang et al. also found that the expression level of miR-1-3p was down-regulated in CRC by the bioinformatics analysis, suggesting that the miR-1-3p may have the potential to diagnose CRC and inhibit tumor cells progression (106).
3.3. Histopathologic diagnosis
Pathology is the gold standard for tumor diagnosis. It can identify the tumor cell types, stage the tumor and guide the treatment plan of patients. Meanwhile, it can also be used as a prognosis and tumor recurrence predictor. Now, a lot of diagnostic work still needs to be completed by pathologists alone. With the continuous breakthrough of digital pathology (DP) technology, it is expected to become the development direction of pathology in the future. DP can be used in image retrieval, pattern recognition, ML, and DL. By extracting corresponding quantitative features or identifying specific regions of interest (ROI), DP can build AI computer-aided diagnostic system with automatic recognition functions. Many studies have proved that the application of AI technology can help pathologists improve diagnosis efficiency, reduce workload, and improve the working environment, increasing diagnosis rate and reducing misdiagnosis rate (107). Kasahara et al. used AI to collect 146 T1 CRC cases. They analyzed the nuclear morphological characteristics in hematoxylin and eosin (HE)-stained slide images. The results suggested that the model could increase the accuracy of preoperative lymph node metastasis prediction, which needs to be further verified in clinical practice (108).
One of the most successful examples of DL’s widespread application in medicine is CNN, which has virtually reinvented image analysis technology. In a retrospective study, researchers obtained more than 100,000 HE image patches from 86 CRC tissue slides, and they used these images to train CNN based on transfer learning (TL). The results showed a nine-class accuracy of over 94% in 7180 independent data sets of 25 CRC patients. It does confirm that CNN can separate histological images and predict the survival rate of CRC patients after treatment (68). High-intensive workload and accuracy requirement for reading pathological images sometimes lead to the misdiagnosis of pathologists. Wang et al. proposed a CNN-based method to classify a large number of histopathological images. The AUC of this model was up to 0.988, with the ability to distinguish CRC from other benign tissues (18). Furthermore, by using CNN, some researchers proposed a clinically comparable technology. This model could be used to stage tumor and classify HE-stained colon histopathological images. They confirmed that the model’s classification could reach more than 90% equally by processing four different data sets (109).
Nowadays, most AI-assisted pathology recognition technologies are achieved by SL. In order to address the problem of massive data markers in SL, an SSL based on the average teacher structure was proposed. A series of expansion experiments also confirmed that SSL significantly reduces the amount of impractical labeled data and expands the fundamental reality of AI in medical work. SSL achieved the comparable effect as SL with less labeled data (110). More interestingly, an monogram model combined with ML-pathomics, immunoscore, radiomics, and clinical factors, was proved to effectively predict the postoperative prognosis of patients with CRC lung metastasis (111). These indicate significant progress in AI-assisted pathological reading. The AI-assisted pathology recognition is expected to break the current limitations that are only applied to the primary screening stage, providing more guidance and decision-making for treatment and prognosis of the CRC patients.
3.4. Radiologic diagnosis
Radiomics refer to a technology that converts medical images into high-dimensional available data for cancer diagnosis and prognosis (112). At present, Methods commonly used for CRC imaging evaluation include MRI, CT, and ultrasonography. Conventional imaging evaluation methods have certain shortcomings, such as limited local tumor evaluation, low tumor staging accuracy, and excessive reliance on the clinical diagnose of imaging physicians (113). Although functional magnetic resonance including diffusion-weighted imaging (DWI), T2-weighted imaging (T2WI) shows high predictive performance (112, 114). AI systems will learn to extract and integrate a large amount of imaging information to achieve a more accurate diagnosis.
The combination of AI and radiomics can extract information from various imaging data. It can be used for tumor segmentation, feature extraction, and the model establishment, eventually achieving the purpose of tumor quantitative evaluation. It has gradually become a crucial component of CRC precision diagnosis and treatment. In 2018, Liu et al. proposed a label assignment generative adversarial network (Lagan), and accurately segmented ROI in the analysis and diagnosis of CRC with CT. The application of this computer-aided segmentation can save time and labor costs. It has been proved that Lagan is a robust model that can be applied to more medical physics network tasks (21). Not long ago, Hamabe et al. developed a rectal cancer segmentation software based on a U-Net deep neural network. They used this software for MRI image simulation analysis of 201 preoperative patients. It segmented the rectum, mesorectum, and tumors respectively, and eventually obtained DICE similarity coefficients (DSC) to artificial segmentation (rectum: DSC = 0.930, mesorectum: DSC = 0.917, tumor: DSC = 0.727) (80). Compared with MRI radiomics, which has been studied more in CRC, ultrasound is mainly used for early diagnosis of CRC. Song et al. designed a deep multi-view fusion network system based on endorectal ultrasonography (ERUS) to identify benign and malignant colorectal tumors, which could effectively reduce the workload of ultrasonic experts and the misdiagnosis rate (115).
Radiomics prediction of local advanced rectal cancer (LARC) after treatment is another crucial application. The ERUS can be used to identify early CRC, however, the diagnostic value for metastasis of advanced CRC is limited (113). The MRI examination after neoadjuvant treatment may be unreliable for pathologic complete response (PCR) identification (116). Interestingly, we can predict and stratify the risk of patients after neoadjuvant chemoradiotherapy (NCRT) by establishing a nomogram. This nomogram combines multi-parametric MRI information and clinicopathological factors, eliminating the effects of PCR intervention and helping to choose more accurate treatment options (77). Similarly, Farri et al. established an AI model based on the MRI image texture features to evaluate the PCR of 55 LARC patients after NCRT. The results showed that the AUC of 0.86, confirming that it is valuable in speculating PCR patients after NCRT (117). Radiomics is still widely used in the study of distal metastasis. Last year, Rocca et al. proposed to use the formal methods (FMS) combined with CT to monitor the liver metastasis of CRC. The overall accuracy rate reached 93.3%. The FMS appears to be trustworthy and valuable (74). There were also the combined models of radiomics, immunomics, and pathomics. They were beneficial in predicting lung metastasis of CRC (111).
4. The application of AI in CRC treatment
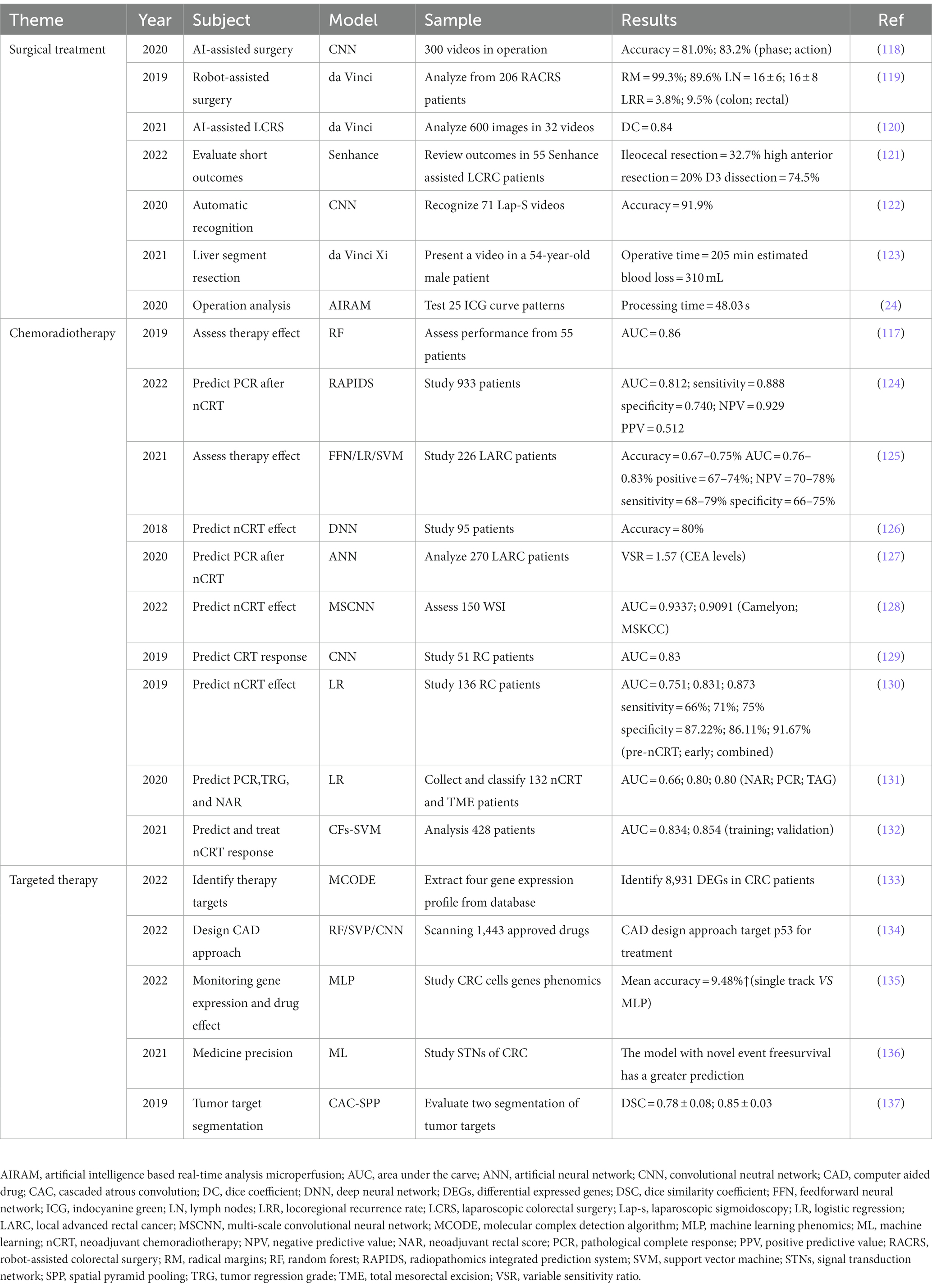
The treatments for CRC include surgical therapy, chemotherapy, targeted therapy, and other combined therapies. The application of AI in the treatment of CRC can design appropriate therapeutic plans for patients, providing patients with more personalized and precise medical decisions, and improving the prognosis. Common AI models for CRC treatment are summarized in Table 2.
4.1. Surgical treatment
In recent years, with the development of minimally invasive surgery, the application of AI in the field of surgery has also been gradually valued. Compared with more complicated CRC surgery, AI was applied earlier in lung cancer and breast cancer surgery (138). Notably, people have begun to study the application of AI in colorectal surgery, and gradually realize that AI can provide a new direction for the development of colorectal surgery. Much research data on CRC is pouring out. Computer vision (CV) is a subfield of AI that can be used to analyze and evaluate video data (138). Some researchers in Japan have collected and analyzed 300 videos of laparoscopic colorectal surgery, hoping that these data sets may be employed to optimize the CNN performance in CV (118). South Korean scholars also conducted AI-based research on the perfusion of the indocyanine green (ICG) angiography system under laparoscopic colorectal surgery in the same year. They collected 200 ROIs from every 50 patients, a total of 10,000 ICG curves. And then they classified these data sets into 25 curve modes, confirming that the virtual microcirculation analysis system is more accurate with the assistance of AI (24).
Now, robotic surgery also achieves more remarkable development in the field of colorectal surgery, especially in rectal surgery. Even in the relatively difficult and complex transanal total mesorectal excision (taTME), which requires rich experience in laparoscopic operation, the robotic surgery has also been proved feasible (139). Compared with open surgery and laparoscopic surgery, robotic surgery has many advantages, such as shorter hospital stay, less perioperative bleeding, fewer complications, and improved postoperative quality of life. At the same time, it also can reduce the difficulty of the surgeon’s operation and relieve fatigue (140–142). In terms of long-term effects, the robotic surgery’s recurrence rate and mortality rate are comparable to that of laparoscopic surgery (143, 144). Recently, Igaki et al. successfully developed a flat image navigation system, which could be used to help surgeons identify anatomical tissue during TME. This system needs more image data to improve the accuracy of its recognition for future evaluation (120).
At present, the most widely used robot on the market is the da Vinci robot system, which has developed to the fourth generation (Figure 5). The fourth-generation robot has been dramatically improved in the cantilever system, with more subtle vision and precise operation, caring more about personalized needs. However, some problems remain, such as extended operation time, limited movement range, and poor sensory system (25, 145). The da Vinci SP system is a single-hole robot. It has only one robotic arm and three surgical instrument arms, which can be connected to three machines through one port. Experiments have confirmed that the da Vinci SP system is an excellent model for taTME and natural orifice specimen extraction (139, 145, 146). One of the biggest problems of robotic surgery is the high cost (145, 147). It indicates that the promotion of robotic surgery requires government financial support. With the progress of science and technology and establishment of a unified market, the cost of robotic surgery will also be reduced.
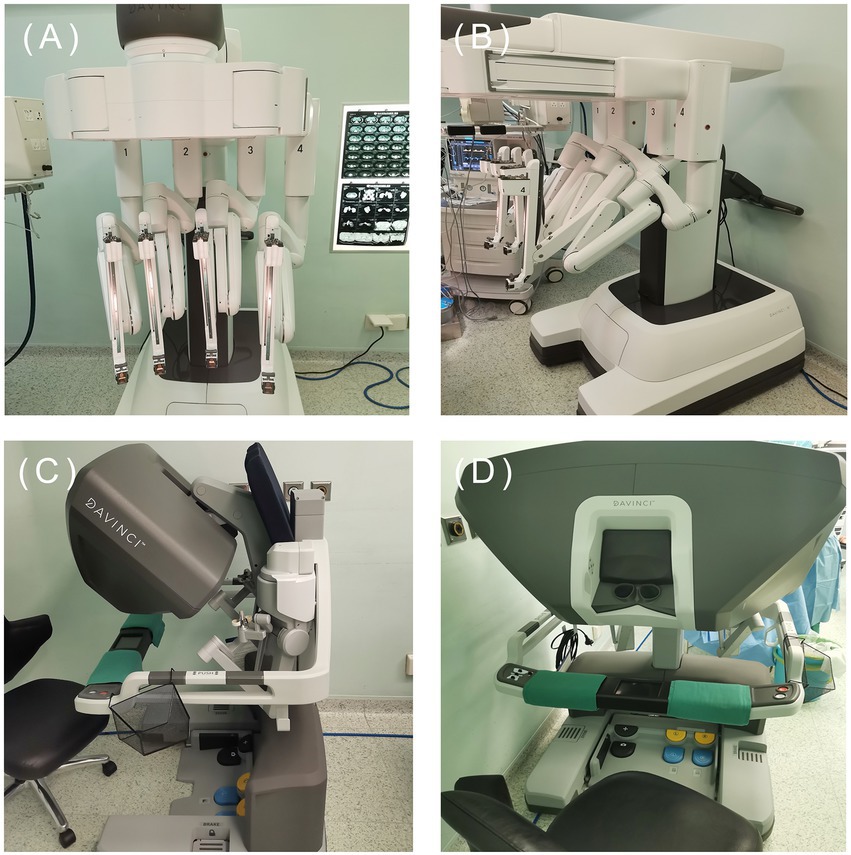
Figure 5. The fourth generation da Vinci. (Image source: The First Affiliated Hospital of Dalian Medical University.)
4.2. Chemoradiotherapy
NCRT is of great significance for CRC treatment, especially for patients with rectal cancer. Adjuvant chemotherapy is primarily used in the patients who are classified as intermediate risk (148). However, most patients do not need additional chemotherapy, so an accurate clinical decision-making is particularly important. The addition of AI is helpful for the treatment decision and efficacy evaluation of NCRT patients. In recent years, as one of the tools that can effectively reduce medical malpractice, AI-based clinical decision support systems (CDSSs) have attracted widespread attention. Experts in South Korea processed a chemotherapy recommender for CRC. This is the first CDSS in the country to reflect real data. It has satisfactory accuracy (AUC > 0.95), but the drawback is that the data source is relatively specific and single (149). Recently, kleppe et al. invented a DoMore-v1-CRC marker, developing the CDSSs based on DL to make a new risk division for patients after colectomy. When patients were classified as low risk, they could be exempted from NCRT. As a result, the survival rate of these patients improved significantly (148).
The prognosis evaluation of CRC patients is an important part for clinical doctors to choose appropriate treatment plans. DL-based assisted MRI can predict the metastasis of LARC patients receiving NCRT, which is a hot topic of current research (77, 124). Notably, Farrando et al. developed a classifier to predict the response of LARC patients who underwent NCRT. By evaluating the expression of lncRNAs, the researchers achieved satisfactory results (AUC = 0.93) (150). The researchers used biomarkers with significant stability, such as lncRNAs, in combination with available massive computational power to accurately predict drug resistance (151).
4.3. Targeted therapy
Targeted therapy is one of the effective methods for the treatment of CRC. Epithelial growth factor receptor (EGFR) is one of the vital drug targets. KRAS gene is highly sensitive to EGFR (152). However, the non-invasive prediction of the KRAS mutation state inCRC is considered as a significant challenge. Recently, some scholars have used the DL method based on a residual neural network to achieve this goal, attaining high predictive performance (AUC = 0.90) on the axis. This is helpful for further targeted treatment of CRC (153). The mutation rate of the BRAF gene in CRC can be as high as 10%. In another study, Beal et al. used a simpler RF data model to predict the V600E mutation in the BRAF (153). Many studies have supported that using AI to detect genetic mutations in CRC is a reliable way (151). These simple and cheap models will be the right choice for patients.
Abnormal mutations in genes and chromosomes can also cause drug resistance, which brings many obstacles to the treatment of CRC. In such an environment, the targeted therapy through drug delivery platform will contribute to precision medicine in the future (22, 151). Russo et al. used AI-based prediction model to analyze the patients who may have drug resistance before and after treatment. They achieved a sound effect (average AUC = 0.90) in the classification and targeted precision treatment (154). Interestingly, with the help of artificial algorithms, Hu et al. studied the competitive endogenous RNA (ceRNA) network about lncRNA and proposed 144 core genes for the first time, which could be used as target drugs for the treatment of CRC (155). The CRC research at the genetic scale can help us to understand the pathogenesis of tumors at the molecular level well, thus providing theoretical support for the diagnosis and treatment of CRC.
5. Discussion
Recently, the application of AI in CRC surgery has dabbled in various fields, and it has also achieved satisfactory results. There are indeed some challenges that we need to face and overcome. The development of AI needs to be supported by three elements: big data, computing power, and algorithm model. Today, the development of big data in China is still in the primary stage. More high-quality data is needed, and the interaction between data centers should be stronger. Without a large amount of high-quality data as the basis, even the most advanced algorithm model will not help. Therefore, we urgently need to standardize big medical data and increase interoperability among multiple centers (90, 147, 156). Moreover, there are specific problems in various imaging omics and AI models. For example, most models are based on retrospective data. The cases have strict admission and exclusion criteria. But there are more or fewer differences in the imaging standards of each center. Therefore, the repeatability of these models and the effectiveness of their application in the real world have yet to be fully evaluated (156).
Furthermore, the results obtained through the DL still lack interpretability. It cannot correctly judge the cause and give a reasonable explanation of the process and internal information of the algorithm. This phenomenon is called the “black box” problem in DL. Although there are still different opinions on the application of the black box in medicine, the transparency and interpretability of the algorithm have been taken as the core principles. It ensures that medical staff, patients, and other relevant personnel can fully understand clinical decisions, avoiding infringement of patients’ privacy, unclear responsibilities, and other ethical issues (156, 157). Now scholars have begun to study how the DL model makes decisions based on images and additional information, analyzing the causal relationship in its black box. Shao et al. used the DNN model to predict the survival rate of more than 20,000 patients within a year after major cardiovascular surgery. An impact score was defined innovatively to explain the results of the model prediction. The research on an interpretable DL is becoming more and more popular (158).
The development of AI in medicine still faces several limitations, and its application in colorectal surgery is also in its infancy. AI has indeed provided broad prospects for development in this field. This does not mean that AI can replace the role of clinicians. There is a need to strengthen cooperation between clinicians and computer experts to break through various transformation barriers. We should also evaluate clinicians’ acceptance of different AI systems and minimize the interference of AI in diagnosis and treatment.
6. Conclusion
With the continuous improvement of big clinical data, AI will develop rapidly in medicine. AI-based on various algorithms that combine with multiple medical imaging big data help to improve the early detection rate and diagnosis of CRC, conducting the early and systematic evaluation of patients. Moreover, it enhances the effect of adjuvant therapy, such as NCRT and targeted treatment, strengthening patients’ prognosis monitoring. Through continuous optimization and development, AI will make greater contributions to the diagnosis and treatment of CRC in the era of precision medicine.
Author contributions
ZY wrote the paper. CY and LZ performed the revision and approval of the final version. LZ and ZY performed literature research. SQ corrected the writing of the paper. All authors contributed to the article and approved the submitted version.
Acknowledgments
We would acknowledge the support of the First Affiliated Hospital of Dalian Medical University (grant no. 20Z12020).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Sung, H, Ferlay, J, Siegel, RL, Laversanne, M, Soerjomataram, I, Jemal, A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2021) 71:209–49. doi: 10.3322/caac.21660
2. Xia, C, Dong, X, Li, H, Cao, M, Sun, D, He, S, et al. Cancer statistics in China and United States, 2022: profiles, trends, and determinants. Chinese Med J-Peking. (2022) 135:584–90. doi: 10.1097/CM9.0000000000002108
3. Dekker, E, Tanis, PJ, Vleugels, JLA, Kasi, PM, and Wallace, MB. Colorectal cancer. Lancet. (2019):394. doi: 10.1016/S0140-6736(19)32319-0
4. Buccafusca, G, Proserpio, I, Tralongo, AC, Rametta Giuliano, S, and Tralongo, P. Early colorectal cancer: diagnosis, treatment and survivorship care. Crit Rev Oncol Hematol. (2019) 136:20–30. doi: 10.1016/j.critrevonc.2019.01.023
5. Kuipers, EJ, Grady, WM, Lieberman, D, TJJS, S, PGB, C, Seufferlein, T, et al. Colorectal cancer. Nat Rev Dis Primers. (2015) 1:15065. doi: 10.1038/nrdp.2015.65
6. Thanikachalam, K, and Khan, G. Colorectal cancer and nutrition. Nutrients. (2019) 11:164. doi: 10.3390/nu11010164
7. Ciardiello, F, Ciardiello, D, Martini, G, Napolitano, S, Tabernero, J, and Cervantes, A. Clinical management of metastatic colorectal cancer in the era of precision medicine. CA Cancer J Clin. (2022) 72:372–401. doi: 10.3322/caac.21728
8. Ganesh, K, Stadler, ZK, Cercek, A, Mendelsohn, RB, Shia, J, Segal, NH, et al. Immunotherapy in colorectal cancer: rationale, challenges and potential. Nat Rev Gastro Hepat. (2019) 16:361–75. doi: 10.1038/s41575-019-0126-x
9. Kijima, S. Preoperative evaluation of colorectal cancer using CT colonography, MRI, and PET/CT. World J Gastroentero. (2014) 20:16964. doi: 10.3748/wjg.v20.i45.16964
10. Zhang, Y, Chen, Z, and Li, J. The current status of treatment for colorectal cancer in China. Medicine. (2017) 96:e 8242. doi: 10.1097/MD.0000000000008242
11. Onyoh, EF, Hsu, W, Chang, L, Lee, Y, Wu, M, and Chiu, H. The rise of colorectal cancer in Asia: epidemiology, screening, and management. Curr Gastroenterol Rep. (2019) 21:36. doi: 10.1007/s11894-019-0703-8
12. Hamet, P, and Tremblay, J. Artificial intelligence in medicine. Metabolism. (2017) 69:S36–40. doi: 10.1016/j.metabol.2017.01.011
13. Ramesh, AN, Kambhampati, C, Monson, J, and Drew, PJ. Artificial intelligence in medicine. Ann Roy Coll Surg. (2004) 86:334–8. doi: 10.1308/147870804290
14. Ranka, S, Reddy, M, and Noheria, A. Artificial intelligence in cardiovascular medicine. Curr Opin Cardiol. (2021) 36:26–35. doi: 10.1097/HCO.0000000000000812
15. Tsigelny, IF. Artificial intelligence in drug combination therapy. Brief Bioinform. (2019) 20:1434–48. doi: 10.1093/bib/bby004
16. Howard, J. Artificial intelligence: implications for the future of work. Am J Ind Med. (2019) 62:917–26. doi: 10.1002/ajim.23037
17. Sultan, AS, Elgharib, MA, Tavares, T, Jessri, M, and Basile, JR. The use of artificial intelligence, machine learning and deep learning in oncologic histopathology. J Oral Pathol Med. (2020) 49:849–56. doi: 10.1111/jop.13042
18. Wang, KS, Yu, G, Xu, C, Meng, XH, Zhou, J, Zheng, C, et al. Accurate diagnosis of colorectal cancer based on histopathology images using artificial intelligence. BMC Med. (2021) 19:76. doi: 10.1186/s12916-021-01942-5
19. Sheikh, A, Anderson, M, Albala, S, Casadei, B, Franklin, BD, Richards, M, et al. Health information technology and digital innovation for national learning health and care systems. Lancet Digit Health. (2021) 3:e383–96. doi: 10.1016/S2589-7500(21)00005-4
20. Hassan, C, Spadaccini, M, Iannone, A, Maselli, R, Jovani, M, Chandrasekar, VT, et al. Performance of artificial intelligence in colonoscopy for adenoma and polyp detection: a systematic review and meta-analysis. Gastrointest Endosc. (2021) 93:77–85.e6. doi: 10.1016/j.gie.2020.06.059
21. Liu, X, Guo, S, Zhang, H, He, K, Mu, S, Guo, Y, et al. Accurate colorectal tumor segmentation for CT scans based on the label assignment generative. Med Phys. (2019) 46:3532–42. doi: 10.1002/mp.13584
22. Azad, MS, Fathi, M, Cho, WC, Barzegari, A, Dadashi, H, Dadashpour, M, et al. Recent advances in targeted drug delivery systems for resistant colorectal cancer. Cancer Cell Int. (2022) 22:196. doi: 10.1186/s12935-022-02605-y
23. Muehlematter, UJ, Daniore, P, and Vokinger, KN. Approval of artificial intelligence and machine learning-based medical devices in the USA and Europe (2015–20): a comparative analysis. Lancet Digital Health. (2021) 3:e195–203. doi: 10.1016/S2589-7500(20)30292-2
24. Park, S, Park, H, Baek, K, Ahn, H, Lee, IY, and Son, GM. Artificial intelligence based real-time microcirculation analysis system for laparoscopic colorectal surgery. World J Gastroentero. (2020) 26:6945–62. doi: 10.3748/wjg.v26.i44.6945
25. Ngu, JC, Sim, S, Yusof, S, Ng, C, and Wong, AS. Insight into the da Vinci® xi – technical notes for single-docking left-sided colorectal procedures. Int J Med Robot. (2017) 13:e1798. doi: 10.1002/rcs.1798
26. Yates, DR, Vaessen, C, and Roupret, M. From Leonardo to da Vinci: the history of robot-assisted surgery in urology. BJU Int. (2011):108. doi: 10.1111/j.1464-410X.2011.10576,10600.x
27. Iavazzo, C, GkegkE, XD, Iavazzo, P, and Gkegkes, ID RAZVOJ ROBOTA KROZ POVIJEST DO DA VINCIJEVOG ROBOTA: Hrvatsko znanstveno društvo za povijest zdravstvene kulture. Acta med-hist Adriat. (2014). 247.
28. Kron, T, and Krishnan, P. Leonardo DaVinci's contributions to medical physics and biomedical engineering: celebrating the life of a ‘polymath’. Australas Phys Eng Sci Med. (2019) 42:403–5. doi: 10.1007/s13246-019-00757-2
29. Kaul, V, Enslin, S, and Gross, SA. History of artificial intelligence in medicine. Gastrointest Endosc. (2020) 92:807–12. doi: 10.1016/j.gie.2020.06.040
30. Muthukrishnan, N, Maleki, F, Ovens, K, Reinhold, C, Forghani, B, and Forghani, R. Brief history of artificial intelligence. Neuroimag Clin N Am. (2020) 30:393–9. doi: 10.1016/j.nic.2020.07.004
31. A Turing test for artificial intelligence in cancer. Nat Cancer. (2020) 1:137–8. doi: 10.1038/s43018-020-0041-7
32. Abdallah, M, Abu Talib, M, Feroz, S, Nasir, Q, Abdalla, H, and Mahfood, B. Artificial intelligence applications in solid waste management: a systematic research review. Waste Manag. (2020) 109:231–46. doi: 10.1016/j.wasman.2020.04.057
33. Sadek, AW. Artificial intelligence in transportation. Transportation research circular E-C113. (2007).
34. Abduljabbar, R, Dia, H, Liyanage, S, and Bagloee, SA. Applications of artificial intelligence in transport: an overview. Sustainability-Basel. (2019) 11:189. doi: 10.3390/su11010189
35. Zawacki-Richter, O, Marín, VI, Bond, M, and Gouverneur, F. Systematic review of research on artificial intelligence applications in higher education – where are the educators? Int J Educ Technol High Educ. (2019) 16:39. doi: 10.1186/s41239-019-0171-0
36. Pang, Y, and Ito, T. Visualization techniques to give insight into the operation of the go policy network. TAAI. (2020). 35–40.doi: 10.1109/TAAI51410.2020.00015
37. Vaishya, R, Javaid, M, Khan, IH, and Haleem, A. Artificial intelligence (AI) applications for COVID-19 pandemic. Diabetes Metab Syndr Clin Res Rev. (2020) 14:337–9. doi: 10.1016/j.dsx.2020.04.012
38. Malycha, J, Bacchi, S, and Redfern, O. Artificial intelligence and clinical deterioration. Curr Opin Crit Care. (2022) 28:315–21. doi: 10.1097/MCC.0000000000000945
39. Yin, Z, and Wong, STC. Artificial intelligence unifies knowledge and actions in drug repositioning. Emerg Top Life Sci. (2021) 5:803–13. doi: 10.1042/ETLS20210223
40. Misawa, M, Kudo, S, Mori, Y, Cho, T, Kataoka, S, Yamauchi, A, et al. Artificial intelligence-assisted polyp detection for colonoscopy: initial experience. Gastroenterology. (2018) 154:2027–9. doi: 10.1053/j.gastro.2018.04.003
41. Repici, A, Badalamenti, M, Maselli, R, Correale, L, Radaelli, F, Rondonotti, E, et al. Efficacy of real-time computer-aided detection of colorectal neoplasia in a randomized trial. Gastroenterology. (2020) 159:512–20. doi: 10.1053/j.gastro.2020.04.062
42. Wang, P, Liu, P, Glissen Brown, JR, Berzin, TM, Zhou, G, Lei, S, et al. Lower adenoma miss rate of computer-aided detection-assisted colonoscopy vs routine white-light colonoscopy in a prospective tandem study. Gastroenterology. (2020) 159:1252–61. doi: 10.1053/j.gastro.2020.06.023
43. Wallace, MB, Sharma, P, Bhandari, P, East, J, Antonelli, G, Lorenzetti, R, et al. Impact of artificial intelligence on miss rate of colorectal neoplasia. Gastroenterology. (2022) 163:295–304. doi: 10.1053/j.gastro.2022.03.007
44. Su, JR, Li, Z, Shao, XJ, Ji, CR, Ji, R, Zhou, RC, et al. Impact of a real-time automatic quality control system on colorectal polyp and adenoma detection: A prospective randomized controlled study (with videos). Gastrointest Endosc. (2020) 91:415–24. doi: 10.1016/j.gie.2019.08.026
45. Luo, Y, Zhang, Y, Liu, M, Lai, Y, Liu, P, Wang, Z, et al. Artificial intelligence-assisted colonoscopy for detection of colon polyps: a prospective, randomized cohort study. J Gastrointest Surg. (2021) 25:2011–8. doi: 10.1007/s11605-020-04802-4
46. Lu, Y, Lu, S, Huang, Y, Cai, S, Le, P, Hsu, F, et al. A novel convolutional neural network model as an alternative approach to bowel preparation evaluation before colonoscopy in the COVID-19 era: a multicenter, single-blinded, randomized study. Am J Gastroenterol. (2022) 117:1437–43. doi: 10.14309/ajg.0000000000001900
47. Yao, L, Zhang, L, Liu, J, Zhou, W, He, C, Zhang, J, et al. Effect of an artificial intelligence-based quality improvement system on efficacy of a computer-aided detection system in colonoscopy: a four-group parallel study. Endoscopy. (2022) 54:757–68. doi: 10.1055/a-1706-6174
48. Xu, L, He, X, Zhou, J, Zhang, J, Mao, X, Ye, G, et al. Artificial intelligence-assisted colonoscopy: a prospective, multicenter, randomized controlled trial of polyp detection. Cancer Med. (2021) 10:7184–93. doi: 10.1002/cam4.4261
49. Wang, P, Berzin, TM, Glissen Brown, JR, Bharadwaj, S, Becq, A, Xiao, X, et al. Real-time automatic detection system increases colonoscopic polyp and adenoma detection rates: a prospective randomised controlled study. Gut. (2019) 68:1813–9. doi: 10.1136/gutjnl-2018-317500
50. Kamba, S, Tamai, N, Saitoh, I, Matsui, H, Horiuchi, H, Kobayashi, M, et al. Reducing adenoma miss rate of colonoscopy assisted by artificial intelligence: a multicenter randomized controlled trial. J Gastroenterol. (2021) 56:746–57. doi: 10.1007/s00535-021-01808-w
51. Su, Y, Tian, X, Gao, R, Guo, W, Chen, C, Chen, C, et al. Colon cancer diagnosis and staging classification based on machine learning and bioinformatics analysis. Comput Biol Med. (2022) 145:105409. doi: 10.1016/j.compbiomed.2022.105409
52. Li, H, Lin, J, Xiao, Y, Zheng, W, Zhao, L, Yang, X, et al. Colorectal cancer detected by machine learning models using conventional laboratory test data. Technol Cancer Res T. (2021) 20:1180552841. doi: 10.1177/15330338211058352
53. Wan, N, Weinberg, D, Liu, T, Niehaus, K, Ariazi, EA, Delubac, D, et al. Machine learning enables detection of early-stage colorectal cancer by whole-genome sequencing of plasma cell-free DNA. BMC Cancer. (2019) 19:832. doi: 10.1186/s12885-019-6003-8
54. Tanos, R, Tosato, G, Otandault, A, Al Amir Dache, Z, Pique Lasorsa, L, Tousch, G, et al. Machine learning-assisted evaluation of circulating DNA quantitative analysis for cancer screening. Adv Sci. (2020) 7:2000486. doi: 10.1002/advs.202000486
55. Zhang, X, Yang, Y, Wang, Y, and Fan, Q. Detection of the BRAF V600E mutation in colorectal cancer by NIR spectroscopy in conjunction with counter propagation artificial neural network. Molecules. (2019) 24:2238. doi: 10.3390/molecules24122238
56. Echle, A, Grabsch, HI, Quirke, P, van den Brandt, PA, West, NP, Hutchins, GGA, et al. Clinical-grade detection of microsatellite instability in colorectal tumors by deep learning. Gastroenterology. (2020) 159:1406–16. doi: 10.1053/j.gastro.2020.06.021
57. Huang, X, Cai, W, Yuan, W, and Peng, S. Identification of key lncRNAs as prognostic prediction models for colorectal cancer based on LASSO. Int J Clin Exp Pathol. (2020) 13:675–84.
58. Krause, J, Grabsch, HI, Kloor, M, Jendrusch, M, Echle, A, Buelow, RD, et al. Deep learning detects genetic alterations in cancer histology generated by adversarial networks. J Pathol. (2021) 254:70–9. doi: 10.1002/path.5638
59. Jiao, Y, Yuan, J, Qiang, Y, and Fei, S. Deep embeddings and logistic regression for rapid active learning in histopathological images. Comput Meth Prog Bio. (2021) 212:106464. doi: 10.1016/j.cmpb.2021.106464
60. Hosseinzadeh Kassani, S, Hosseinzadeh Kassani, P, Wesolowski, MJ, Schneider, KA, and Deters, R. Deep transfer learning based model for colorectal cancer histopathology segmentation: a comparative study of deep pre-trained models. Int J Med Inform. (2022) 159:104669. doi: 10.1016/j.ijmedinf.2021.104669
61. Starmans, MPA, Buisman, FE, Renckens, M, Willemssen, FEJA, van der Voort, SR, Groot Koerkamp, B, et al. Distinguishing pure histopathological growth patterns of colorectal liver metastases on CT using deep learning and radiomics: a pilot study. Clin Exp Metastas. (2021) 38:483–94. doi: 10.1007/s10585-021-10119-6
62. Iizuka, O, Kanavati, F, Kato, K, Rambeau, M, Arihiro, K, and Tsuneki, M. Deep learning models for histopathological classification of gastric and colonic epithelial Tumours. Sci Rep. (2020) 10:1504. doi: 10.1038/s41598-020-58467-9
63. Kim, H, Yoon, H, Thakur, N, Hwang, G, Lee, EJ, Kim, C, et al. Deep learning-based histopathological segmentation for whole slide images of colorectal cancer in a compressed domain. Sci Rep. (2021) 11:22520. doi: 10.1038/s41598-021-01905-z
64. Trivizakis, E, Ioannidis, GS, Souglakos, I, Karantanas, AH, Tzardi, M, and Marias, K. A neural pathomics framework for classifying colorectal cancer histopathology images based on wavelet multi-scale texture analysis. Sci Rep. (2021) 11:15546. doi: 10.1038/s41598-021-94781-6
65. Ho, C, Zhao, Z, Chen, XF, Sauer, J, Saraf, SA, Jialdasani, R, et al. A promising deep learning-assistive algorithm for histopathological screening of colorectal cancer. Sci Rep. (2022) 12:2222. doi: 10.1038/s41598-022-06264-x
66. Byeon, S, Park, J, Cho, YA, and Cho, B. Automated histological classification for digital pathology images of colonoscopy specimen via deep learning. Sci Rep. (2022) 12:12804. doi: 10.1038/s41598-022-16885-x
67. Zhou, P, Cao, Y, Li, M, Ma, Y, Chen, C, Gan, X, et al. HCCANet: histopathological image grading of colorectal cancer using CNN based on multichannel fusion attention mechanism. Sci Rep. (2022) 12:15103. doi: 10.1038/s41598-022-18879-1
68. Kather, JN, Krisam, J, Charoentong, P, Luedde, T, Herpel, E, Weis, C, et al. Predicting survival from colorectal cancer histology slides using deep learning: a retrospective multicenter study. PLoS Med. (2019) 16:e1002730. doi: 10.1371/journal.pmed.1002730
69. Lichtblau, D, and Stoean, C. Cancer diagnosis through a tandem of classifiers for digitized histopathological slides. PLoS One. (2019) 14:e209274. doi: 10.1371/journal.pone.0209274
70. Ma, X, Shen, F, Jia, Y, Xia, Y, Li, Q, and Lu, J. MRI-based radiomics of rectal cancer: preoperative assessment of the pathological features. BMC Med Imaging. (2019) 19:86. doi: 10.1186/s12880-019-0392-7
71. Delli Pizzi, A, Chiarelli, AM, Chiacchiaretta, P, d'Annibale, M, Croce, P, Rosa, C, et al. MRI-based clinical-radiomics model predicts tumor response before treatment in locally advanced rectal cancer. Sci Rep. (2021) 11:5379. doi: 10.1038/s41598-021-84816-3
72. Zhu, HT, Zhang, XY, Shi, YJ, Li, XT, and Sun, YS. Automatic segmentation of rectal tumor on diffusion-weighted images by deep learning with U-net. J Appl Clin Med Phys. (2021) 22:324–31. doi: 10.1002/acm2.13381
73. Wang, D, Xu, J, Zhang, Z, Li, S, Zhang, X, Zhou, Y, et al. Evaluation of rectal cancer circumferential resection margin using faster region-based convolutional neural network in high-resolution magnetic resonance images. Dis Colon Rectum. (2020) 63:143–51. doi: 10.1097/DCR.0000000000001519
74. Rocca, A, Brunese, MC, Santone, A, Avella, P, Bianco, P, Scacchi, A, et al. Early diagnosis of liver metastases from colorectal cancer through CT Radiomics and formal methods: a pilot study. J Clin Med. (2022) 11:31. doi: 10.3390/jcm11010031
75. Luo, H, Li, S, Zeng, Y, Cheema, H, Otegbeye, E, Ahmed, S, et al. Human colorectal cancer tissue assessment using optical coherence tomography catheter and deep learning. J Biophotonics. (2022) 15:e202100349. doi: 10.1002/jbio.202100349
76. Kim, K, Kim, S, Han, K, Bae, H, Shin, J, and Lim, JS. Diagnostic performance of deep learning-based lesion detection algorithm in CT for detecting hepatic metastasis from colorectal cancer. Korean J Radiol. (2021) 22:912. doi: 10.3348/kjr.2020.0447
77. Liu, X, Zhang, D, Liu, Z, Li, Z, Xie, P, Sun, K, et al. Deep learning radiomics-based prediction of distant metastasis in patients with locally advanced rectal cancer after neoadjuvant chemoradiotherapy: a multicentre study. EBioMedicine. (2021) 69:103442. doi: 10.1016/j.ebiom.2021.103442
78. Liu, X, Guo, S, Zhang, H, He, K, Mu, S, Guo, Y, et al. Accurate colorectal tumor segmentation for CT scans based on the label assignment generative adversarial network. Med Phys. (2019) 46:3532–42. doi: 10.1002/mp.13584
79. Wei, J, Cheng, J, Gu, D, Chai, F, Hong, N, Wang, Y, et al. Deep learning-based radiomics predicts response to chemotherapy in colorectal liver metastases. Med Phys. (2020) 48:513–22. doi: 10.1002/mp.14563
80. Hamabe, A, Ishii, M, Kamoda, R, Sasuga, S, Okuya, K, Okita, K, et al. Artificial intelligence–based technology for semi-automated segmentation of rectal cancer using high-resolution MRI. PLoS One. (2022) 17:e269931. doi: 10.1371/journal.pone.0269931
81. Zhang, XY, Wang, L, Zhu, HT, Li, ZW, Ye, M, Li, XT, et al. Predicting rectal cancer response to neoadjuvant Chemoradiotherapy using deep learning of diffusion kurtosis MRI. Radiology. (2020) 296:56–64. doi: 10.1148/radiol.2020190936
82. Grosu, S, Wesp, P, Graser, A, Maurus, S, Schulz, C, Knösel, T, et al. Machine learning-based differentiation of benign and premalignant colorectal polyps detected with CT Colonography in an asymptomatic screening population: a proof-of-concept study. Radiology. (2021) 299:326–35. doi: 10.1148/radiol.2021202363
83. Daye, D, Tabari, A, Kim, H, Chang, K, Kamran, SC, Hong, TS, et al. Quantitative tumor heterogeneity MRI profiling improves machine learning–based prognostication in patients with metastatic colon cancer. Eur Radiol. (2021) 31:5759–67. doi: 10.1007/s00330-020-07673-0
84. Zeng, Y, Xu, S, Chapman, WC, Li, S, Alipour, Z, Abdelal, H, et al. Real-time colorectal cancer diagnosis using PR-OCT with deep learning. Theranostics. (2020) 10:2587–96. doi: 10.7150/thno.40099
85. Viscaino, M, Torres Bustos, J, Muñoz, P, Auat Cheein, C, and Cheein, FA. Artificial intelligence for the early detection of colorectal cancer: a comprehensive review of its advantages and misconceptions. World J Gastroentero. (2021) 27:6399–414. doi: 10.3748/wjg.v27.i38.6399
86. JB, WEISS, NS, CETEL, and DE, WEISS. Colorectal cancer screening: colonoscopy has disadvantages. Letters The Editor. (2019) 110:774–6. doi: 10.1038/ajg.2015.365
87. Kaminski, MF, Wieszczy, P, Rupinski, M, Wojciechowska, U, Didkowska, J, Kraszewska, E, et al. Increased rate of adenoma detection associates with reduced risk of colorectal cancer and death. Gastroenterology. (2017) 153:98–105. doi: 10.1053/j.gastro.2017.04.006
88. Li, JW, Chia, T, Fock, KM, Chong, KDW, Wong, YJ, and Ang, TL. Artificial intelligence and polyp detection in colonoscopy: use of a single neural network to achieve rapid polyp localization for clinical use. J Gastroen Hepatol. (2021) 36:3298–307. doi: 10.1111/jgh.15642
89. Byrne, MF, Chapados, N, Soudan, F, Oertel, C, Linares Pérez, M, Kelly, R, et al. Real-time differentiation of adenomatous and hyperplastic diminutive colorectal polyps during analysis of unaltered videos of standard colonoscopy using a deep learning model. Gut. (2018) 68:94–100. doi: 10.1136/gutjnl-2017-314547
90. Mitsala, A, Tsalikidis, C, Pitiakoudis, M, Simopoulos, C, and Tsaroucha, AK. Artificial intelligence in colorectal cancer screening, diagnosis and treatment. A new era. Curr Oncol. (2021) 28:1581–607. doi: 10.3390/curroncol28030149
91. Iwatate, M, Sano, Y, Tanaka, S, Kudo, SE, Saito, S, Matsuda, T, et al. Validation study for development of the Japan NBI expert team classification of colorectal lesions. Digest Endosc. (2018) 30:642–51. doi: 10.1111/den.13065
92. Gonai, T, Kawasaki, K, Nakamura, S, Yanai, S, Akasaka, R, Sato, K, et al. Microvascular density under magnifying narrow-band imaging endoscopy in colorectal epithelial neoplasms. Intestinal. Research. (2020) 18:107–14. doi: 10.5217/ir.2019.00061
93. Taunk, P, Atkinson, CD, Lichtenstein, D, Rodriguez-Diaz, E, and Singh, SK. Computer-assisted assessment of colonic polyp histopathology using probe-based confocal laser endomicroscopy. Int J Color Dis. (2019) 34:2043–51. doi: 10.1007/s00384-019-03406-y
94. Ferrari, A, Neefs, I, Hoeck, S, Peeters, M, and Van Hal, G. Towards novel non-invasive colorectal cancer screening methods: a comprehensive review. Cancers. (2021) 13:1820. doi: 10.3390/cancers13081820
95. Li, JN, and Yuan, SY. Fecal occult blood test in colorectal cancer screening. J Digest Dis. (2019) 20:62–4. doi: 10.1111/1751-2980.12712
96. Kanth, P, and Inadomi, JM. Screening and prevention of colorectal cancer. BMJ. (2021) 374:1855. doi: 10.1136/bmj.n1855
97. Thomas, DS, Fourkala, E, Apostolidou, S, Gunu, R, Ryan, A, Jacobs, I, et al. Evaluation of serum CEA, CYFRA21-1 and CA125 for the early detection of colorectal cancer using longitudinal preclinical samples. Brit J Cancer. (2015) 113:268–74. doi: 10.1038/bjc.2015.202
98. Hammad, A, Elshaer, M, and Tang, X. Identification of potential biomarkers with colorectal cancer based on bioinformatics analysis and machine learning. Math Biosci Eng. (2021) 18:8997–9015. doi: 10.3934/mbe.2021443
99. Bresalier, RS. Colorectal cancer screening in a changing world. Gastroenterol Clin N. (2022) 51:577–91. doi: 10.1016/j.gtc.2022.05.002
100. Tepus, M, and Yau, TO. Non-invasive colorectal cancer screening: an overview. Gastrointestinal Tumors. (2020) 7:62–73. doi: 10.1159/000507701
101. Mikeska, T, and Craig, J. DNA methylation biomarkers: cancer and beyond. Genes-Basel. (2014) 5:821–64. doi: 10.3390/genes5030821
102. Kel, A, Boyarskikh, U, Stegmaier, P, Leskov, LS, Sokolov, AV, Yevshin, I, et al. Walking pathways with positive feedback loops reveal DNA methylation biomarkers of colorectal cancer. BMC Bioinformatics. (2019) 20:119. doi: 10.1186/s12859-019-2687-7
103. Zhou, H, Yang, Z, Yue, J, Chen, Y, Chen, T, Mu, T, et al. Identification of potential hub genes via bioinformatics analysis combined with experimental verification in colorectal cancer. Mol Carcinogen. (2020) 59:425–38. doi: 10.1002/mc.23165
104. Liu, Z, Liu, L, Weng, S, Guo, C, Dang, Q, Xu, H, et al. Machine learning-based integration develops an immune-derived lncRNA signature for improving outcomes in colorectal cancer. Nat Commun. (2022) 13:816. doi: 10.1038/s41467-022-28421-6
105. Zhang, W, Ming, X, Rong, Y, Huang, C, Weng, H, Chen, H, et al. Diagnostic value investigation and bioinformatics analysis of mi R-31 in patients with lymph node metastasis of colorectal cancer. Anal Cell Pathol. (2019). doi: 10.1155/2019/9740475
106. Wang, JY, Huang, JC, Chen, G, and Wei, DM. Expression level and potential target pathways of miR-1-3p in colorectal carcinoma based on 645 cases from 9 microarray datasets. Mol Med Rep. (2018) 17:5013–20. doi: 10.3892/mmr.2018.8532
107. Acs, B, Rantalainen, M, and Hartman, J. Artificial intelligence as the next step towards precision pathology. J Intern Med. (2020) 288:62–81. doi: 10.1111/joim.13030
108. Kasahara, K, Katsumata, K, Saito, A, Ishizaki, T, Enomoto, M, Mazaki, J, et al. Artificial intelligence predicts lymph node metastasis or risk of lymph node metastasis in T1 colorectal cancer. Int J Clin Oncol. (2022) 27:1570–9. doi: 10.1007/s10147-022-02209-6
109. Dabass, M, Vashisth, S, and Vig, R. A convolution neural network with multi-level convolutional and attention learning for classification of cancer grades and tissue structures in colon histopathological images. Comput Biol Med. (2022) 147:105680. doi: 10.1016/j.compbiomed.2022.105680
110. Yu, G, Sun, K, Xu, C, Shi, X, Wu, C, Xie, T, et al. Accurate recognition of colorectal cancer with semi-supervised deep learning on pathological images. Nat Commun. (2021) 12:6311. doi: 10.1038/s41467-021-26643-8
111. Wang, R, Dai, W, Gong, J, Huang, M, Hu, T, Li, H, et al. Development of a novel combined nomogram model integrating deep learning-pathomics, radiomics and immunoscore to predict postoperative outcome of colorectal cancer lung metastasis patients. J Hematol Oncol. (2022) 15:11. doi: 10.1186/s13045-022-01225-3
112. Li, P, Song, G, Wu, R, Li, H, Zhang, R, Zuo, P, et al. Multiparametric MRI-based machine learning models for preoperatively predicting rectal adenoma with canceration. MAGMA. (2021) 34:707–16. doi: 10.1007/s10334-021-00915-2
113. Wong, C, Fu, Y, Li, M, Mu, S, Chu, X, Fu, J, et al. MRI-based artificial intelligence in rectal cancer. J Magn Reson Imaging. (2022) 57:45–56. doi: 10.1002/jmri.28381
114. Horvat, N, Veeraraghavan, H, Nahas, C, Bates, D, Ferreira, FR, Zheng, J, et al. Combined artificial intelligence and radiologist model for predicting rectal cancer treatment response from magnetic resonance imaging: an external validation study. Abdom Radiol (NY). (2022) 47:2770–82. doi: 10.1007/s00261-022-03572-8
115. Song, D, Zhang, Z, Li, W, Yuan, L, and Zhang, W. Judgment of benign and early malignant colorectal tumors from ultrasound images with deep multi-view fusion. Comput Meth Prog Bio. (2022) 215:106634. doi: 10.1016/j.cmpb.2022.106634
116. Koh, D. Using deep learning for MRI to identify responders to Chemoradiotherapy in rectal cancer. Radiology. (2020) 296:65–6. doi: 10.1148/radiol.2020200417
117. Ferrari, R, Mancini-Terracciano, C, Voena, C, Rengo, M, Zerunian, M, Ciardiello, A, et al. MR-based artificial intelligence model to assess response to therapy in locally advanced rectal cancer. Eur J Radiol. (2019) 118:1–9. doi: 10.1016/j.ejrad.2019.06.013
118. Kitaguchi, D, Takeshita, N, Matsuzaki, H, Oda, T, Watanabe, M, Mori, K, et al. Automated laparoscopic colorectal surgery workflow recognition using artificial intelligence: experimental research. Int J Surg. (2020) 79:88–94. doi: 10.1016/j.ijsu.2020.05.015
119. Polat, F, Willems, LH, Dogan, K, and Rosman, C. The oncological and surgical safety of robot-assisted surgery in colorectal cancer: outcomes of a longitudinal prospective cohort study. Surg Endosc. (2019) 33:3644–55. doi: 10.1007/s00464-018-06653-2
120. Igaki, T, Kitaguchi, D, Kojima, S, Hasegawa, H, Takeshita, N, Mori, K, et al. Artificial intelligence-based Total Mesorectal excision plane navigation in laparoscopic colorectal surgery. Dis Colon Rectum. (2022) 65:e329–33. doi: 10.1097/DCR.0000000000002393
121. Sasaki, M, Hirano, Y, Yonezawa, H, Shimamura, S, Kataoka, A, Fujii, T, et al. Short-term results of robot-assisted colorectal cancer surgery using Senhance digital laparoscopy system. Asian J Endosc Surg. (2022) 15:613–8. doi: 10.1111/ases.13064
122. Kitaguchi, D, Takeshita, N, Matsuzaki, H, Takano, H, Owada, Y, Enomoto, T, et al. Real-time automatic surgical phase recognition in laparoscopic sigmoidectomy using the convolutional neural network-based deep learning approach. Surg Endosc. (2020) 34:4924–31. doi: 10.1007/s00464-019-07281-0
123. Machado, MAC, Mattos, BH, Lobo Filho, MM, and Makdissi, FF. Robotic resection of Postero-superior liver segments (7, 8) (with video). J Gastrointest Surg. (2021) 25:574–5. doi: 10.1007/s11605-020-04799-w
124. Feng, L, Liu, Z, Li, C, Li, Z, Lou, X, Shao, L, et al. Development and validation of a radiopathomics model to predict pathological complete response to neoadjuvant chemoradiotherapy in locally advanced rectal cancer: a multicentre observational study. Lancet Digit Health. (2022) 4:e8–e17. doi: 10.1016/S2589-7500(21)00215-6
125. Haak, HE, Gao, X, Maas, M, Waktola, S, Benson, S, Beets-Tan, RGH, et al. The use of deep learning on endoscopic images to assess the response of rectal cancer after chemoradiation. Surg Endosc. (2022) 36:3592–600. doi: 10.1007/s00464-021-08685-7
126. Bibault, J, Giraud, P, Housset, M, Durdux, C, Taieb, J, Berger, A, et al. Deep learning and Radiomics predict complete response after neo-adjuvant chemoradiation for locally advanced rectal cancer. Sci Rep. (2018) 8:12611. doi: 10.1038/s41598-018-30657-6
127. Huang, C, Huang, M, Huang, C, Tsai, H, Su, W, Chang, W, et al. Machine learning for predicting pathological complete response in patients with locally advanced rectal cancer after neoadjuvant chemoradiotherapy. Sci Rep. (2020) 10:12555. doi: 10.1038/s41598-020-69345-9
128. Zhang, D, Duan, Y, Guo, J, Wang, Y, Yang, Y, Li, Z, et al. Using multi-scale convolutional neural network based on multi-instance learning to predict the efficacy of neoadjuvant Chemoradiotherapy for rectal cancer. IEEE J Transl Eng Health Med. (2022) 10:1–8. doi: 10.1109/JTEHM.2022.3156851
129. Shi, L, Zhang, Y, Nie, K, Sun, X, Niu, T, Yue, N, et al. Machine learning for prediction of chemoradiation therapy response in rectal cancer using pre-treatment and mid-radiation multi-parametric MRI. Magn Reson Imaging. (2019) 61:33–40. doi: 10.1016/j.mri.2019.05.003
130. Shu, Z, Fang, S, Ye, Q, Mao, D, Cao, H, Pang, P, et al. Prediction of efficacy of neoadjuvant chemoradiotherapy for rectal cancer: the value of texture analysis of magnetic resonance images. Abdom Radiol. (2019) 44:3775–84. doi: 10.1007/s00261-019-01971-y
131. Shaish, H, Aukerman, A, Vanguri, R, Spinelli, A, Armenta, P, Jambawalikar, S, et al. Radiomics of MRI for pretreatment prediction of pathologic complete response, tumor regression grade, and neoadjuvant rectal score in patients with locally advanced rectal cancer undergoing neoadjuvant chemoradiation: an international multicenter study. Eur Radiol. (2020) 30:6263–73. doi: 10.1007/s00330-020-06968-6
132. Jiang, W, Li, M, Tan, J, Feng, M, Zheng, J, Chen, D, et al. A nomogram based on a collagen feature support vector machine for predicting the treatment response to neoadjuvant Chemoradiotherapy in rectal cancer patients. Ann Surg Oncol. (2021) 28:6408–21. doi: 10.1245/s10434-021-10218-4
133. Sharma, A, Yadav, D, Rao, P, Sinha, S, Goswami, D, Rawal, RM, et al. Identification of potential therapeutic targets associated with diagnosis and prognosis of colorectal cancer patients based on integrated bioinformatics analysis. Comput Biol Med. (2022) 146:105688. doi: 10.1016/j.compbiomed.2022.105688
134. Patrício, RPS, Videira, PA, and Pereira, F. A computer-aided drug design approach to discover tumour suppressor p 53 protein activators for colorectal cancer therapy. Bioorgan Med Chem. (2022) 53:116530. doi: 10.1016/j.bmc.2021.116530
135. Orazio M, D, Murdocca, M, Mencattini, A, Casti, P, Filippi, J, Antonelli, G, et al. Machine learning phenomics (MLP) combining deep learning with time-lapse-microscopy for monitoring colorectal adenocarcinoma cells gene expression and drug-response. Sci Rep. (2022) 12:8545. doi: 10.1038/s41598-022-12364-5
136. Nwaokorie, A, and Fey, D. Personalised medicine for colorectal cancer using mechanism-based machine learning models. Int J Mol Sci. (2021) 22:9970. doi: 10.3390/ijms22189970
137. Men, K, Boimel, P, Janopaul-Naylor, J, Zhong, H, Huang, M, Geng, H, et al. Cascaded atrous convolution and spatial pyramid pooling for more accurate tumor target segmentation for rectal cancer radiotherapy. Phys Med Biol. (2018) 63:185016. doi: 10.1088/1361-6560/aada6c
138. Quero, G, Mascagni, P, Kolbinger, FR, Fiorillo, C, De Sio, D, Longo, F, et al. Artificial intelligence in colorectal cancer surgery: present and future perspectives. Cancers. (2022) 14:3803. doi: 10.3390/cancers14153803
139. Ribero, D, Baldassarri, D, and Spinoglio, G. Robotic ta TME using the da Vinci SP: technical notes in a cadaveric model. Updat Surg. (2021) 73:1125–9. doi: 10.1007/s13304-021-01002-w
140. Sheng, S, Zhao, T, and Wang, X. Comparison of robot-assisted surgery, laparoscopic-assisted surgery, and open surgery for the treatment of colorectal cancer. Medicine. (2018) 97:e11817. doi: 10.1097/MD.0000000000011817
141. Grosek, J, Ales Kosir, J, Sever, P, Erculj, V, and Tomazic, A. Robotic versus laparoscopic surgery for colorectal cancer: a case-control study. Radiol Oncol. (2021) 55:433–8. doi: 10.2478/raon-2021-0026
142. Kim, MJ, Park, SC, Park, JW, Chang, HJ, Kim, DY, Nam, B, et al. Robot-assisted versus laparoscopic surgery for rectal cancer. Ann Surg. (2018) 267:243–51. doi: 10.1097/SLA.0000000000002321
143. Flynn, J, Larach, JT, Warrier, S, and Heriot, A. Whither robotic colorectal surgery? ANZ J Surg. (2020) 90:1230–2. doi: 10.1111/ans.16067
144. Pinar, I, Fransgaard, T, Thygesen, LC, and Gögenur, I. Long-term outcomes of robot-assisted surgery in patients with colorectal cancer. Ann Surg Oncol. (2018) 25:3906–12. doi: 10.1245/s10434-018-6862-2
145. Baek, S, Piozzi, GN, and Kim, S. Optimizing outcomes of colorectal cancer surgery with robotic platforms. Surg Oncol. (2021) 37:101559. doi: 10.1016/j.suronc.2021.101559
146. Luo, R, Zheng, F, Zhang, H, Zhu, W, He, P, and Liu, D. Robotic natural orifice specimen extraction surgery versus traditional robotic-assisted surgery (NOTR) for patients with colorectal cancer: a study protocol for a randomized controlled trial. Trials. (2021) 22:121. doi: 10.1186/s13063-021-05077-z
147. Liang, F, Wang, S, Zhang, K, Liu, T, and Li, J. Development of artificial intelligence technology in diagnosis, treatment, and prognosis of colorectal cancer. World J Gastro Oncol. (2022) 14:124–52. doi: 10.4251/wjgo.v14.i1.124
148. Kleppe, A, Skrede, OJ, De Raedt, S, Hveem, TS, Askautrud, HA, Jacobsen, JE, et al. A clinical decision support system optimising adjuvant chemotherapy for colorectal cancers by integrating deep learning and pathological staging markers: a development and validation study. Lancet Oncol. (2022) 23:1221–32. doi: 10.1016/S1470-2045(22)00391-6
149. Park, J, Baek, J, Sym, SJ, Lee, KY, and Lee, Y. A data-driven approach to a chemotherapy recommendation model based on deep learning for patients with colorectal cancer in Korea. BMC Med Inform Decis. (2020) 20:241. doi: 10.1186/s12911-020-01265-0
150. Ferrando, L, Cirmena, G, Garuti, A, Scabini, S, Grillo, F, Mastracci, L, et al. Development of a long non-coding RNA signature for prediction of response to neoadjuvant chemoradiotherapy in locally advanced rectal adenocarcinoma. PLoS One. (2020) 15:e226595. doi: 10.1371/journal.pone.0226595
151. Nussinov, R, Tsai, C, and Jang, H. Anticancer drug resistance: an update and perspective. Drug Resist Update. (2021) 59:100796. doi: 10.1016/j.drup.2021.100796
152. Sanchez-Ibarra, HE, Jiang, X, Gallegos-Gonzalez, EY, Cavazos-González, AC, Chen, Y, Morcos, F, et al. NRAS, and BRAF mutation prevalence, clinicopathological association, and their application in a predictive model in Mexican patients with metastatic colorectal cancer: a retrospective cohort study. PLoS One. (2020) 15:e235490. doi: 10.1371/journal.pone.0235490
153. He, K, Liu, X, Li, M, Li, X, Yang, H, and Zhang, H. Noninvasive KRAS mutation estimation in colorectal cancer using a deep learning method based on CT imaging. BMC Med Imaging. (2020) 20:59. doi: 10.1186/s12880-020-00457-4
154. Russo, V, Lallo, E, Munnia, A, Spedicato, M, Messerini, L, AurizioR, D, et al. Artificial intelligence predictive models of response to cytotoxic chemotherapy alone or combined to targeted therapy for metastatic colorectal cancer patients: a systematic review and meta-analysis. Cancers. (2022) 14:4012. doi: 10.3390/cancers14164012
155. Hu, D, Zhang, B, Yu, M, Shi, W, and Zhang, L. Identification of prognostic biomarkers and drug target prediction for colon cancer according to a competitive endogenous RNA network. Mol Med Rep. (2020) 22:620–32. doi: 10.3892/mmr.2020.11171
156. Qiu, H, Ding, S, Liu, J, Wang, L, and Wang, X. Applications of artificial intelligence in screening, diagnosis, treatment, and prognosis of colorectal cancer. Curr Oncol. (2022) 29:1773–95. doi: 10.3390/curroncol29030146
157. Beltramin, D, Lamas, E, and Bousquet, C. Ethical issues in the utilization of black. Stud Health Technol Inform. (2022) 2022:249–52. doi: 10.3233/SHTI220709
Keywords: artificial intelligence, colorectal cancer, machine learning, deep learning, bioinformatics analysis, screening, diagnosis, therapy
Citation: Yin Z, Yao C, Zhang L and Qi S (2023) Application of artificial intelligence in diagnosis and treatment of colorectal cancer: A novel Prospect. Front. Med. 10:1128084. doi: 10.3389/fmed.2023.1128084
Edited by:
Ming-zhe Ma, Fudan University, ChinaReviewed by:
Hui Yang, First Affiliated Hospital of Wannan Medical College, ChinaXiang Kong, Wannan Medical College, China
Copyright © 2023 Yin, Yao, Zhang and Qi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chenhui Yao, gavinyaoch@126.com
†These authors have contributed equally to this work
 Zugang Yin1
Zugang Yin1  Chenhui Yao
Chenhui Yao Limin Zhang
Limin Zhang