Bacterial etiologies, antimicrobial susceptibility pattern and associated factors among patients suspected sterile body site infections at Debre Markos Comprehensive Specialized Hospital, Northwest Ethiopia
- Department of Medical Laboratory Science, College of Health Sciences, Debre Markos University, Debre Markos, Ethiopia
Background: Sterile body locations are usually associated with clinical urgency and life-threatening illnesses, and they are typically contaminated with diverse bacterial etiologies. If the bacteria acquire resistance to antimicrobial drugs, the public health crisis will only worsen. In developing countries, drug-resistant bacteria are common because of poor surveillance, diagnostic capacity, and control measures. Early diagnosis, and assessing the drug resistance and factors associated with infection are important to combat the drug resistance and treatment. This study aimed to assess the bacterial etiologies, antimicrobial susceptibility pattern, and possible associated factors among patients suspected of sterile body sites.
Methods: A hospital-based cross-sectional study was conducted from June 2022 to August 2022 at Debre Markos Comprehensive Specialized Hospital in Amhara regional state, Ethiopia. One hundred seven study participants were selected using consecutive convenient sampling techniques. A structured questionnaire was used to collect socio-demographic and clinical data. Gram stain was done for a preliminary report and inoculated into blood agar, MacConkey agar, and chocolate agar and incubated aerobically and micro aerobically at 37°C for 24 h. Antimicrobial susceptibility testing was done by the modified Kirby Bauer’s disk diffusion method. Data were analyzed using bivariate and multivariate logistic regression was used. A p-value less than 0.05 is considered as statistically significant.
Results: The overall magnitude of sterile body site infection among study participants was 7.5% (14/187). The majority of the isolates were Gram-negative bacteria with the predominant species Enterobacter cloacae accounting for 28.57% (4/14). Among isolates 78.57%(11/14) of them were multidrug-resistant isolates. Being inpatient, co-morbidity, and alcohol consumption were significantly associated with sterile body site infection.
Conclusion: In our study, Gram-negative bacteria were the predominant bacteria that infects sterile body fluid. The prevalence of multi-drug resistance bacteria isolates was significantly high. Therefore, before prescribing an empirical treatment, a medical professional should identify the bacterial etiology of sterile body fluids and the susceptibility of microbes to the drug.
Introduction
Sterile body fluids (SBF), which include cerebrospinal fluid (CSF), peritoneal fluid, pleural fluid, and synovial fluid, are fluids that have no normal flora of bacteria. Sterile body infection and drug resistance bacteria is life threatening ceases. One of the most urgent public health problems facing the world today is antibiotic resistance. It is typically associated with higher medical costs, longer hospital stays, and more mortality (1, 2). Because of insufficient policy implementation for the prevention and control of drug resistance and laboratory infrastructure inadequacy for obtaining samples and identifying the particular agent, the situation simply gets worse. It was more challenging in those settings since it was necessary to collect samples from invasive locations and confirm the agents for diseases including meningitis, peritonitis, septic arthritis, and pleural empyema (3–8). These circumstances force medical professional to treat patients empirically, which exacerbates the problem of drug resistance already facing public health (1, 2).
Meningitis, peritonitis, septic arthritis, and pleural empyema are only a few of the potentially fatal diseases caused by bacterial infections in sterile body sites (3–8). A worldwide study found that there were 2.5 million cases of meningitis and 236,000 deaths from the disease (9). Also Wunrow et al. (9), bacterial meningitis affects approximately 1.2 million people each year and causes almost 170,000 deaths globally (10). In developing nations, bacterial meningitis continues to pose a serious threat to public health (11). The highest burden of bacterial meningitis occurs in sub-Saharan Africa, known as the meningitis belt (12). Despite the availability of effective antimicrobials, mortality rates in developing nations are higher, ranging from 16 to 32% (13).
Furthermore, spontaneous bacterial peritonitis (SBP) contributes 10 to 30% prevalence and its prevalence even increased among hospitalized patients (25–30%), of which 15% of the infection rate is caused by post-operative procedures (4, 14, 15). This disease is significantly associated with a high level of mortality (90%) (4). Gram-negative flora is the major cause of SBP and mortality rates reach up to 10 to 46% (16). Moreover, bacterial-related pleural infection has increased from 7.6 to 14.9% globally with a 20% mortality rate (15, 17). Also, Septic arthritis is a serious orthopedic issue and the incidence of septic arthritis of native joints has been reported to be around two cases per 100,000 people per year (18, 19). In this regard, the prevalence of bacterial isolates from sterile body fluids in Ethiopia is 11.5–20.2%. Of this, 10.6–40.7% of the isolates were Gram-positive bacteria whereas the rest 9.6–74.6% were Gram-negative bacteria (5–8).
Currently, the emergency of multidrug-resistant (MDR) bacterial infection challenges treatment success and leads to increased mortality (20, 21). MDR bacteria are bacteria that are resistant to three or more classes of antimicrobial drugs. Ethiopia, a developing country in Sub-Saharan Africa, further aggravates the problem with its inadequate laboratory infrastructure, poor antimicrobial surveillance and control measures, and high rates of antimicrobial misuse from drug stores even without a medical prescription (22, 23).
For instance, a study conducted by Frehiwot and colleagues in Ethiopia found a high level of MDR (75.9%) in the bacterial isolates. According to this study 100% and 90% of MDR was from CoNS isolates and K. pneumoniae isolates, respectively (6). The main reason for the high prevalence of drug resistance in this setting could be related to a habit of self-medication of antimicrobials from drug stores, poor microbiology facilities, and diagnostic capacity (24). Studies have also shown that surgical procedures, trauma, age, hospitalization, chronic illnesses (HIV/AIDS, hepatitis, cirrhosis, cancer, and diabetes mellitus), as well as other behavioral factors (alcohol misuse and cigarette smoking), are significantly associated to bacterial invasion into the sterile body site (25–27).
Therefore, assessing the prevalence of bacterial etiology, and antimicrobial susceptibility patterns, and addressing potential associated factors of bacterial infection of sterile body sites in settings where poor surveillance and control measures, inadequate laboratory infrastructure, medical professionals treat patients empirically, as well as a lack of published data on this matter, could contribute to better management of bacterial diseases and address factors for the development of drug resistance.
Materials and methods
Study setting, design, and population
A hospital-based cross-sectional study was conducted among patients suspected of bacterial sterile body site infections at Debre Markos Comprehensive Specialized Hospital (DMCSH), Northwest Ethiopia, from June 2022 to August 2022 from patients suspected of sterile body fluid infections and had no a history of antimicrobials intake within the last two weeks. Socio-demographic (age, gender, marital status, residence, occupation, income, and educational status), clinical data (history of hospitalization, co-morbidities, cigarette smoking), and other possible associated factors were collected by using a pretested structured questionnaire. DMCSH is found in Debre Markos town and provides specialized health care services through its medical, clinical and diagnostic departments for approximately 255,248 people per year. In the present study, the prevalence of bacterial growth was dependent variable whereas the Socio-demographic data (age, gender, residence, occupation, educational status, and monthly income), Clinical-related data (Patient setting (inpatient, outpatient), history of Hospital admission, history of invasive procedures& surgery, and clinical features), and Co-morbidity (DM, HIV, TB, chronic liver disease, cancer &others) and Behavioral factors (Alcoholism, Smoking cigarettes) were independent variables.
Sample size and sampling technique
The sample size was determined using a single population proportion formula by considering a prevalence (p) of 14.1% from the Tikur Anbesa Hospital, a 95% confidence interval of Z = (1.96), and a margin of error (d) of 5% (6).
Consecutive convenient sampling technique was used to recruit study participants.
Specimen collection, transportation and bacteriological analysis
One to five mL of sterile body fluid samples (lumbar puncture, paracentesis, thoracentesis, and arthrocentesis) were collected aseptically in sterile glass tubes from each participant by experienced physicians. The sample collection tube was labeled and closed tightly. Then specimens were transported to the hospital laboratory within 10 min and laboratory analyses were started within 30 min of specimen delivery.
Inoculation, incubation and isolation of bacterial agent
A drop of the suspension was inoculated into blood agar, chocolate agar, and MacConkey agar plates (Oxoid Ltd., Basingstoke, and Hampshire, United Kingdom). Blood and chocolate agar plates were incubated at 35–37°C in a candle jar with a capnophilic (5–10% CO2) environment and MacConkey agar was incubated aerobically for the isolation of Gram-negative aerobic bacteria and examined after 24 h. Mannitol salt agar (Oxoid Ltd., Basingstoke, and Hampshire, United Kingdom) was used to isolate S. aureus when beta-hemolysis was observed from the blood agar plate. For mixed colonies, a sub-culture of blood agar and chocolate agar was performed to get pure colonies (28).
Identification of bacterial isolates
Isolates were identified by colonial morphology such as hemolytic reaction on blood agar plate, pigment production or color changes surrounding carbohydrate fermenting colonies on MacConkey agar plate. After obtaining pure colonies, further identification was conducted using standard bacteriological techniques including Gram reaction, colony morphology, and biochemical tests. A battery of biochemical test such as catalase test, coagulase test, Optochin sensitivity test, bacitracin sensitivity test, mannitol test, carbohydrate utilization, indole production, mannitol fermentation, citrate utilization, lysine decarboxylation, H2S production, triple sugar iron utilization, and motility testing used to identify Gram positive and Gram negative bacteria responsible for sterile body fluid infections (28).
Antimicrobial susceptibility testing
Using a sterile wire loop, up to 3–5 pure colonies from culture plate were packed and emulsified with sterile normal saline in a test tube and compare with turbidity of the 0.5 McFarland standards. The suspension was inoculated and uniformly distributed on Mueller Hinton agar plates using a sterile cotton bud and incubated at 37°C for 15 min. The appropriate disks impregnated with antimicrobial agents were placed on the surface of agar plates (29). For the AST various antibiotic disks such as gentamicin (GN, 10 μg), erythromycin (ERY, 15 μg), clindamycin (CLD, 2 μg) trimethoprim/sulfamethoxazole (SXT, 1.25/23.75 μg), penicillin (P, 10 units), vancomycin (VA, 30 μg), cefoxitin (FOX, 30 μg), tetracycline (TCY, 30 μg), doxycycline (30 μg), and oxacillin (OXA, 1 μg) (Oxoid, LTD, UK) were used for Gram-positive isolates. Ampicillin (AMP, 10 μg), amoxicillin-clavulanic acid (AMC, 30 μg), ceftriaxone (CRO, 30 μg), ceftazidime (CAZ, 30 μg), amikacin (AMK, 30 μg), gentamicin (GN, 10 μg), meropenem (MEM, 10 μg), imipenem (IMP, 10 μg), ciprofloxacin (CIP, 5 μg), trimethoprim/sulfamethoxazole (SXT,1.25/23.75 μg), piperacillin (10 μg), piperacillin /tazobactam (TZB,100/10 μg), tobramycin (TOB 30 μg) (Oxoid, LTD, UK) were used for Gram-negative isolates (30). To assure the quality of laboratory work all aseptic measures were strictly followed. For instance the sterility of prepared culture media was checked by overnight incubating 5% of the batch at 35–37°C and observed for bacterial growth. The prepared culture media, biochemical test, and antimicrobial susceptibility tests were checked by inoculating the American Type Culture Collection (ATCC) reference strains of Escherichia coli (ATCC-25922), Staphylococcus aureus (ATCC-25923), Streptococcus pneumoniae (ATCC 49619), and Pseudomonas aeruginosa (ATCC-27853) which were used as positive quality control throughout the study.
Data analysis and interpretation
Data were entered into Epi-data version 4.6 software and exported to SPSS version 25 for analysis. Antimicrobial susceptibility pattern was analyzed by World Health Organization (WHO) NET 2022 software. Binary logistic regression models were used to predict the relationship between dependent and independent variables. Variables with a p-value <0.25 in the bivariate logistic regression analysis were moved to a multivariate logistic regression analysis. Adjusted odds ratios with 95% confidence interval (CI) were used and variables with a p-value less than 0.05 in multivariate analysis were considered as statistically significant.
Ethical considerations
Ethical clearance was obtained from the Ethical Review Committee of the College of Health Sciences, Debre Markos University (Hsc/R/C/Ser/PG/Co/197/11/14), and Amhara Public Health Institute (APHI). Written informed consent and assent were obtained from each participant and parents or legal guardians. Information obtained from this study used only for the purpose of the study.
Results
Sociodemographic characteristics
In this study, a total of 187 sterile body fluid samples were collected from patients suspected of bacterial infections. Among enrolled participants, the majority 103/187 (55.1%) of study participants were males, 72/187 (38.5%) were unable to read and write, and 105/187 (56.1%) were rural residents. The median and interquartile range of the age of the study participants were 35 and 26, respectively (Table 1).
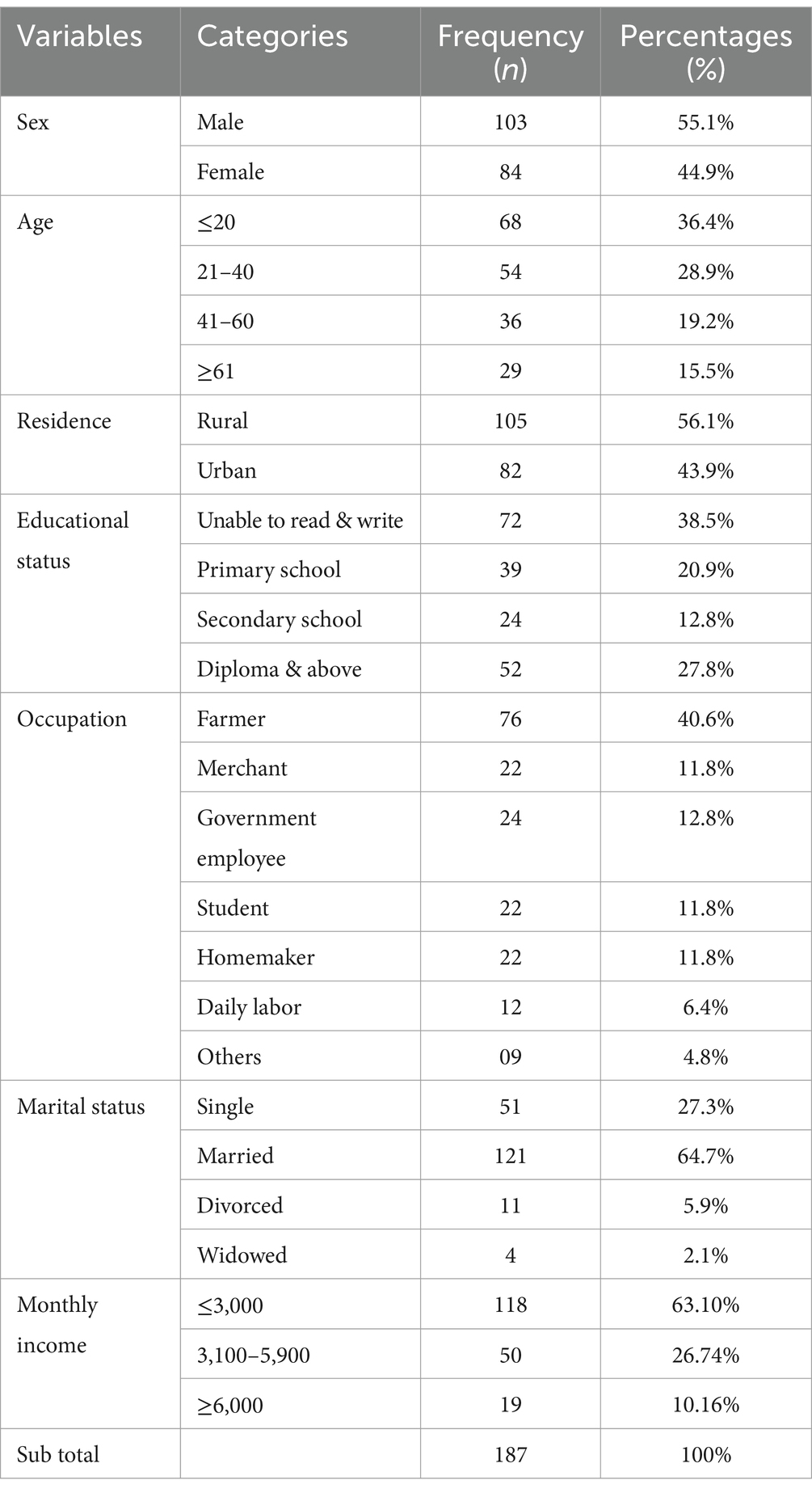
Table 1. Socio-demographic characteristics of patients suspected of sterile body site infections at DMCSH, from June 2022 to August 2022.
Clinical and sample-related characteristics
Out of 187 body fluid samples, CSF was the most frequently encountered sterile body site accounting for 48.7% (91/187) followed by a peritoneal fluid at 27.2%(51/187). The majority of body fluids were clear 78.1% (146/187) while 8.6% (16/187) of body fluids were turbid.
Prevalence of bacterial isolates
From a total of 187 specimens, 14 were culture-positive with overall prevalence of 7.5% (14/187) (95% CI: 3.72 to 11.27%). Of these 35.71% (5/14) were from age group below 20 years old. Most of the isolated bacteria were Gram-negative with 64.28% (9/14). Among these, E. cloacae 28.57% (4/14) and P. aeruginosa 21.42% (3/14) were the most predominant, while the remaining cases were Gram-positive cocci and account 37.72% (5/14). Of these, 21.42% (3/14), 14.28% (2/14) were S. pneumoniae and S. aureus, respectively (Figure 1). The most common bacterial isolates found in peritoneal fluid were E. cloacae, S. aureus, S. pneumoniae, and E. coli. Of these bacterial isolates, E. cloacae followed by S. aureus accounted for the highest percentage. P. aeruginosa was the most common bacterial isolated in CSF, followed by E. coli and E. cloacae. S. pneumoniae was the most common isolated from pleural fluid, followed by P. aeruginosa. No bacteria were isolated from synovial fluid.

Figure 1. Bacterial isolates from patients diagnosed with sterile body site infections at DMCSH, from June 2022 to August 2022.
Antimicrobial susceptibility pattern for Gram-negative isolates
Most effective antimicrobials against E. cloacae isolates (n = 4) were amikacin (100%) and chloramphenicol (75%), imipenem (75%), and piperacillin/tazobactam (75%). In contrast, the highest resistance to cefuroxime, cefepime, and trimethoprim-sulfamethoxazole (75%) was observed against these isolates. The most effective antimicrobials against P. aeruginosa isolates (n = 3) were amikacin (100%), tobramycin (100%), and piperacillin/tazobactam (100%). Although, the highest resistance to gentamycin (66.7%) was observed against these isolates. All the E. coli isolates (n = 2) were susceptible to ceftazidime (100%), ceftriaxone (100%), tobramycin (100%), and chloramphenicol (100%). But they were 50%% resistant to cefuroxime, ciprofloxacillin, meropenem, imipenem, amikacin, and gentamycin (Table 2).
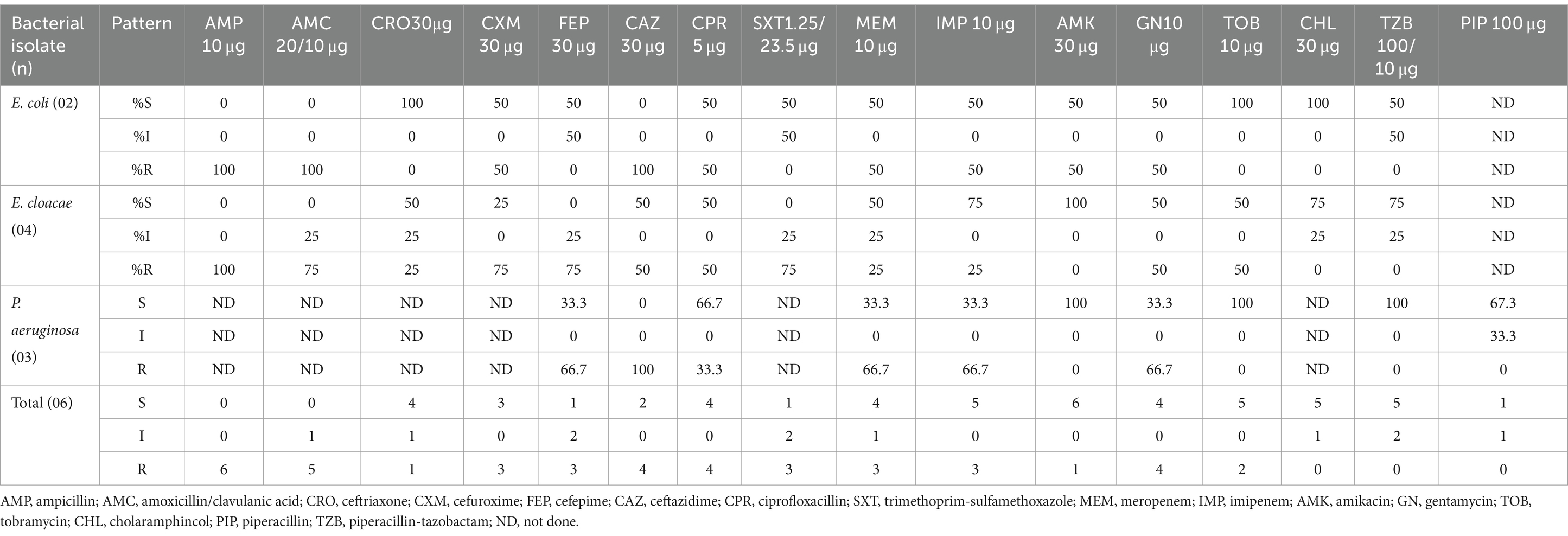
Table 2. Antimicrobial susceptibility patterns of Gram-negative isolates from patients suspected of sterile body site infections at DMCSH, from June 2022 to August 2022.
Antimicrobial susceptibility pattern for Gram-positive isolates
S. aureus (n = 2) isolates were highly susceptible to gentamycin, ciprofloxacin, and clindamycin (100%). But, all S. aureus isolates were 100% resistant to cefoxitin (methicillin resistance staphylococcus aureus), doxycycline, and erythromycins. S. pneumoniae (n = 3) was 100% susceptible to clindamycin and chloramphenicol. On contrary, all S. pneumoniae 100% resistant to trimethoprim-sulfamethoxazole (Table 3).
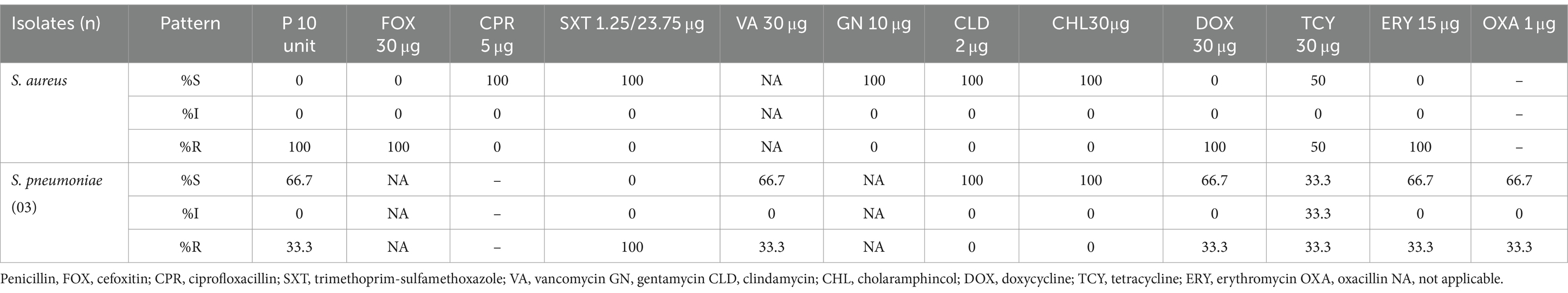
Table 3. Antimicrobial susceptibility patterns of Gram-positive bacterial isolate from patients suspected of sterile body site infections at DMCSH, from June 2022 to August 2022.
Multi-drug resistance of bacterial isolate from sterile body fluids
Out of 14 isolated pathogenic bacteria, 78.57% (11/14) (95% CI: 56.30–99.70%) of them were found to have multidrug resistance (MDR). Among these isolated bacteria, Gram-positive and Gram-negative had shown multiple drug resistance of 60% (3/5) and 88.89% (8/9) respectively. Among Gram-negative bacteria isolates, all (36.36%) E. cloacae had multi-drug resistance. Similarly, S. aureus had shown 100% (2/2) multi-drug resistance among Gram-positive bacteria isolates.
Factors associated with bacterial isolates of sterile body site infections
Patients in the inpatient department were 10.267 times more likely to be infected than outpatients [AOR =10.267, 95% CI: (2.37, 44.50), p value =0.002]. Similarly, patients with comorbidity were 6.31 times more likely to develop bacterial infections than those without comorbidity [AOR = 6.31, 95% CI: (1.487–26.771), p value =0.012]. Patients who consumed alcohol regularly were 5.075 times more likely to develop bacterial infections [AOR = 5.075, 95%CI:(1.157–22.258)], p value =0.03 (Table 4).

Table 4. Bivariate and multivariate analysis of factors associated with patients suspected of sterile body site infections at DMCSH, from June 2022 to August 2022.
Discussion
In the present study, the overall prevalence of bacterial pathogens was 7.5% (14/187) (95% CI: 3.72 to 11.27%). This finding is comparable with previous studies from India (10.81%) (31), and Nepal (10.7%) (32). In contrast, it is higher than reports from India 2.8% (33). But lower than other studies in Mekelle, Ethiopia (20.2%), Harar, Ethiopia (16.7%) (8), Tikur Anbesa, Ethiopia (14.1%) (6), India (14.78%) (34), and Brazil (21.3%) (35). These variations can be attributed to differences in sample processing, seasonal variation, and the practice of infection control.
In this study, the most frequently isolated microorganisms were Gram-negative bacteria (64.3%). A similar finding is reported from previous studies in EPHI, Ethiopia (74.6) (36), and India (67.8%) (37). In contrast, studies from Mekelle, Ethiopia (52.3%) (5), Turkey (61.7%) (38), and India (85.2%) (34) reported that Gram-positive bacteria were found to be the most common isolates. This difference could be due to different study areas, hospital-acquired infections, and different standard infection prevention control.
In this study, E. cloacae was the most frequent bacterial etiologies causing sterile bodies infection, in contrast. E. coli was the most prevalent isolated bacteria in a previous study conducted in EPHI, Ethiopia (36), Harar, Ethiopia (8), India (33), and Nepal (32). The detection of E. cloacae may be an indication of an opportunistic pathogen associated with hospital-acquired infections, due to poor infection prevention practices such as overcrowding, lack of standard facilities with controlled ventilation, and poor sterilization of all gowns and surgical equipment.
In our study, amikacin (88.9%), cholaramphincol (83.3%), piperacillin/tazobactam (77.8%), and imipenem (75%) were found to be the most effective against Gram-negative isolates. Whereas ampicillin (0%), cefepime (11.3%), and trimethoprim-sulfamethoxazole (16.7%) were the least effective antimicrobials against those Gram-negative isolates. This finding is comparable to previous studies in EPHI, Ethiopia (36), and India (31). This may be due to frequent use of these antimicrobials, the practice of self-medication, limited diagnostic facilities, and inappropriate antibiotic use.
In our study, all Gram-positive isolates were resistant to penicillin and cefoxitin, but most of the isolates were susceptible to erythromycins, doxycycline, and tetracycline. Similarly, high levels of resistance to beta-lactam agents were reported in previous studies in Ethiopia (8, 36) and India (39). This may be due to the presence of mobile-resistant factors, widely usage of these drugs in the treatment of different infections, and irrational use of antimicrobials.
All S. aureus isolates were methicillin-resistant (MRSA), similar to the study conducted in Harar, Ethiopia (8). But this finding is higher than study conducted in India, 36.6% (34), Nepal (30%) (32), and Addis Ababa, Ethiopia (50%) (6). This may be due to geographic variation, differences in infection control practice, and differences in treatments used. This may be related to various factors. As evidenced, for example, by this study, patients in the inpatient department had an infection rate that was 10.267 times higher than that of outpatients [AOR =10.267, 95% CI: (2.37, 44.50), p value =0.002]. Similarly, individuals with co-morbidity had a 6.31-fold increased risk of bacterial infections [AOR = 6.31, 95% CI: (1.487–26.771), p value =0.012] compared to those without co-morbidity.
In this study, high rates of multi-drug resistance (MDR) (78.57%) (95% CI: 56.30–99.70%) were reported, which is comparable to the findings in Addis Ababa, Ethiopia (75%) (6, 36), Harar, Ethiopia (76.4%) (8), Mekelle, Ethiopia (90%) (5) and India (62.9%) (40). But higher than study conducted in Nepal (40%) (32). These variations may be due to different bacterial isolates, patients’ awareness of the use of the antimicrobials, the difference in antibiotic prescribing policies, easy availability of some drugs without a prescription, isolated strain difference, and indiscriminate use of common antimicrobials. In addition, poor infection control practice (the use of alcohol, bleach, disposable gloves, sanitizer), along with poor hospital sanitation conditions, waste management guidelines, and inadequate infrastructure, raise serious concerns in Ethiopia since they may contribute to the emergence of drug resistance.
Being an inpatient, comorbidity, and the habit of drinking alcohol had a statistically significant association with bacterial infection of sterile body fluids. This finding is consistent with a study from Harar, Ethiopia (8). This could be due to the hospital’s level of environmental hygiene and surgical invasive procedures. Another significant factor associated with bacterial infection of sterile body fluids was having comorbidities. This might be due to low immune status due to any underlying medical conditions.
Drinking alcohol was also statistically significant to bacterial infection of sterile body fluids. This result agrees with studies in Arba Minch, Ethiopia (25). The reason might be due to common alcohol consumption influences the contribution of the intestinal microbial populations by way of disturbing the stability of intestinal homeostasis and impairs the phagocytic function of macrophages and neutrophils recruitment, as well as minimizes bacterial clearance inside the lungs.
In the present study, the main shortcomings were the limited sample size and inadequate hospital infrastructure. The microbiology laboratory, for example, lacks anaerobic culture, which is expensive and requires specialized equipment. Hence, the identification was limited to anaerobic bacteria because only aerobic cultures were carried out. This might be one of the reasons for the low culture-positive yield in this study.
Conclusion
E. cloacae, P. aeruginosa, and S. pneumoniae were the most frequently isolated bacteria from various sterile bodily fluids. Antibiotics resistance was high among Gram-negative isolates. The majority of the isolates have shown multidrug resistance (MDR. Being an inpatient, comorbidity, and drinking alcohol were significant predictors for bacterial infection of sterile body fluids. Thus, there should be; regular monitoring of antimicrobial resistance patterns of infected body fluids, periodic antimicrobial surveillance, effective antibiotic policy with good medical practices, and proper infection prevention practices to control the disease transmission. Furthermore, as this study was done in a single study area, a multicenter study should be done to improve the clinical utility of the study.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Ethics statement
The studies involving humans were approved by Ethical Review Committee of the College of Health Science, Debre Markos University. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
DA: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Resources, Software, Writing – original draft, Writing – review & editing. GD: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Software, Supervision, Validation, Visualization, Writing – review & editing. AA: Conceptualization, Formal analysis, Investigation, Methodology, Visualization, Writing – review & editing. AE: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. Debre Markos University funded only for material supports.
Acknowledgments
We acknowledge Amhara Public Health Institute, Debre Markos Comprehensive Specialized Hospital, the staff members of Debre Markos Comprehensive Specialized Hospital microbiology laboratory, and study participants.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Ayukekbong, JA, Ntemgwa, M, and Atabe, AN. The threat of antimicrobial resistance in developing countries: causes and control strategies. Antimicrob Resist Infect Control. (2017) 6:47. doi: 10.1186/s13756-017-0208-x
2. Cegielski, J, Tudor, C, Volchenkov, G, and Jensen, P. Antimicrobial drug resistance and infection prevention/control: lessons from tuberculosis. Int J Infect Control. (2021) 17:1. doi: 10.3396/ijic.v17.20840
3. van de Beek, D, Brouwer, M, Hasbun, R, Koedel, U, Whitney, CG, and Wijdicks, E. Community-acquired bacterial meningitis. Nat Rev Dis Prim. (2016) 2:1–20. doi: 10.1038/nrdp.2016.74
4. De, A, Bankar, S, and Baveja, S. Comparison of three culture methods for diagnosis of spontaneous bacterial peritonitis (SBP) in adult patients with cirrhosis. Int J Curr Microbiol App Sci. (2014) 3:156–60.
5. Tsegay, E, Hailesilassie, A, Hailekiros, H, Niguse, S, Saravanan, M, and Abdulkader, M. Bacterial isolates and drug susceptibility pattern of sterile body fluids from tertiary hospital, northern Ethiopia: a four-year retrospective study. J Pathogens. (2019) 2019:1–6. doi: 10.1155/2019/5456067
6. Teklehymanot, F, Legese, M, and Desta, K. Bacterial profile and their antimicrobial resistance patterns from body fluids at Tikur Anbesa specialized hospital, Addis Ababa, Ethiopia. Biol Med (Aligarh). (2017) 9:408. doi: 10.4172/0974-8369.1000408
7. Ebrahim, S . Bacterial profile and antimicrobial susceptibility pattern of isolates recovered from sterile body fluids referred to the national reference laboratory. Ethiop J Public Health Nutr. (2020) 4:32–36.
8. Shume, T, Tesfa, T, Mekonnen, S, Asmerom, H, Tebeje, F, and Weldegebreal, F. Aerobic bacterial profile and their antibiotic susceptibility patterns of sterile body fluids among patients at Hiwot Fana specialized university hospital, Harar, eastern Ethiopia. Infect Drug Resist. (2022) 15:581–93. doi: 10.2147/IDR.S351961
9. Wunrow, HY, Bender, RG, Vongpradith, A, Sirota, SB, Swetschinski, LR, Novotney, A, et al. Global, regional, and national burden of meningitis and its aetiologies, 1990– 2019: a systematic analysis for the global burden of disease study 2019. Lancet Neurol. (2023) 22:685–711. doi: 10.1016/S1474-4422(23)00195-3
10. Owusu, M, Nguah, SB, Boaitey, YA, Badu-Boateng, E, Abubakr, A-R, Lartey, RA, et al. Aetiological agents of cerebrospinal meningitis: a retrospective study from a teaching hospital in Ghana. Ann Clin Microbiol Antimicrob. (2012) 11:28–8. doi: 10.1186/1476-0711-11-28
11. Brouwer, MC, Tunkel, AR, and van de Beek, D. Epidemiology, diagnosis, and antimicrobial treatment of acute bacterial meningitis. Clin Microbiol Rev. (2010) 23:467–92. doi: 10.1128/CMR.00070-09
12. Gudina, EK, Tesfaye, M, Adane, A, Lemma, K, Shibiru, T, Pfister, HW, et al. Challenges of bacterial meningitis case management in low income settings: an experience from Ethiopia. Trop Med Int Health. (2016) 21:870–8. doi: 10.1111/tmi.12720
13. Nwadioha, S, Nwokedi, E, Onwuezube, I, Egesie, J, and Kashibu, E. Bacterial isolates from cerebrospinal fluid of children with suspected acute meningitis in a Nigerian tertiary hospital. Niger Postgrad Med J. (2013) 20:9–13. doi: 10.4103/1117-1936.165489
14. Goldstein, EJ, and Snydman, DR. Intra-abdominal infections: review of the bacteriology, antimicrobial susceptibility and the role of ertapenem in their therapy. J Antimicrob Chemother. (2004) 53:ii29–36. doi: 10.1093/jac/dkh201
15. Nwagboso, CI, and Ekeng, BE. Microbiological profile and antibiotic resistance pattern of empyema thoracis in Calabar, Nigeria. Trop Doct. (2021) 51:523–6. doi: 10.1177/00494755211032844
16. Dever, JB, and Sheikh, MY. Review article: spontaneous bacterial peritonitis--bacteriology, diagnosis, treatment, risk factors and prevention. Aliment Pharmacol Ther. (2015) 41:1116–31. doi: 10.1111/apt.13172
17. Rosenstengel, A . Pleural infection-current diagnosis and management. J Thorac Dis. (2012) 4:186–93. doi: 10.3978/j.issn.2072-1439.2012.01.12
18. Emamifar, A, Andreasen, RA, Andersen, NS, and Hansen, IMJ. Septic arthritis and subsequent fatal septic shock caused by Vibrio vulnificus infection. Case Rep. (2015) 2015:bcr2015212014. doi: 10.1136/bcr-2015-212014
19. Kaandorp, CJ, Dinant, HJ, van de Laar, MA, Moens, HJB, Prins, APA, and Dijkmans, BA. Incidence and sources of native and prosthetic joint infection: a community based prospective survey. Ann Rheum Dis. (1997) 56:470–5. doi: 10.1136/ard.56.8.470
20. Solomkin, JS, Mazuski, JE, Baron, EJ, Sawyer, RG, Nathens, AB, DiPiro, JT, et al. Guidelines for the selection of anti-infective agents for complicated intra-abdominal infections. Clin Infect Dis. (2003) 37:997–1005. doi: 10.1086/378702
21. Sodani, S, and Hawaldar, R. Bacteriological profile and antibiotic sensitivity pattern in various body fluids–a retrospective study. Indian J Microbiol Res. (2020) 7:51–8. doi: 10.18231/j.ijmr.2020.012
22. Iskandar, K, Molinier, L, Hallit, S, Sartelli, M, Catena, F, Coccolini, F, et al. Drivers of antibiotic resistance transmission in low-and middle-income countries from a “one health” perspective—a review. Antibiotics. (2020) 9:372. doi: 10.3390/antibiotics9070372
23. Dugassa, J, and Shukuri, N. Review on antibiotic resistance and its mechanism of development. J Health Med Nurs. (2017) 1:1–17.
24. Chokshi, A, Sifri, Z, Cennimo, D, and Horng, H. Global contributors to antibiotic resistance. J Global Infect Dis. (2019) 11:36–42. doi: 10.4103/jgid.jgid_110_18
25. Alelign, D, Ameya, G, and Siraj, M. Bacterial pathogens, drug-resistance profile and its associated factors from patients with suspected peritonitis in southern Ethiopia. Infect Drug Resist. (2021) 14:4107–17. doi: 10.2147/IDR.S335103
26. Alelign, D, Ameya, G, Siraj, M, and Fenta, F. Pleural infections: antimicrobial susceptibility patterns of bacterial isolates and associated factors in suspected hospitalized patients at Arba Minch general hospital, southern Ethiopia. Open Microbiol J. (2022) 16. doi: 10.2174/18742858-v16-e2208050
27. Assegu Fenta, D, Lemma, K, Tadele, H, Tadesse, BT, and Derese, B. Antimicrobial sensitivity profile and bacterial isolates among suspected pyogenic meningitis patients attending at Hawassa university hospital: cross-sectional study. BMC Microbiol. (2020) 20:1–10. doi: 10.1186/s12866-020-01808-5
28. Cheesbrough, M . District laboratory practice in tropical countries, part 2. Cambridge, England: Cambridge university press (2005).
29. Murray, PR, Rosenthal, KS, and Pfaller, MA. Medical microbiology E-book. Amsterdam, Netherlands: Elsevier Health Sciences (2020).
30. Humphries, R, Bobenchik, AM, Hindler, JA, and Schuetz, AN. Overview of changes to the clinical and laboratory standards institute performance standards for antimicrobial susceptibility testing, M100. J Clin Microbiol. (2021) 59:e00213–21. doi: 10.1128/JCM.00213-21
31. Rouf, M, and Nazir, A. Aerobic bacteriological profile and antimicrobial sensitivity pattern of bacteria isolated from sterile body fluids: a study from a tertiary Care Hospital in North India. J Med Microbiol. (2019) 28:1–10. doi: 10.9734/mrji/2019/v28i130123
32. Shrestha, LB, Bhattarai, NR, and Khanal, B. Bacteriological profile and antimicrobial susceptibility pattern among isolates obtained from body fluids. J Nepal Health Res Counc. (2019) 17:173–7. doi: 10.33314/jnhrc.v0i0.1656
33. Sultana, S, Palvai, S, Lakshmi, GJ, and Reddy, PS. Bacteriological profile and antimicrobial susceptibility among isolates obtained from sterile body fluids at a tertiary care Centre. J Dent Med Sci. (2021) 20:47–50.
34. Vishalakshi, B, Hanumanthappa, P, and Krishna, S. A study on aerobic bacteriological profile of sterile body fluids. In J Curr Microbiol App Sci. (2016) 5:120–6. doi: 10.20546/ijcmas.2016.505.013
35. Daur, AV, Klimak, F, Cogo, LL, Botão, GD, Monteiro, CLB, and Dalla Costa, LM. Enrichment methodology to increase the positivity of cultures from body fluids. Braz J Infect Dis. (2006) 10:372–3. doi: 10.1590/S1413-86702006000600002
36. Ebrahim, S, Abera, A, Terfe, K, Tsiga, E, Ayenew, Z, Addis, T, et al. Bacterial profile and antimicrobial susceptibility pattern of isolates recovered from sterile body fluids referred to the National Reference Laboratory. Ethiopia. J Public Health Nutr. (2020) 4:32–36.
37. Vijaya Durga, S, and Anuradha, B. An aerobic bacteriological profile and antibio gram of various body fluids from a tertiary care hospital in Telangana, India–a 5 year study. Int J Curr Microbiol App Sci. (2019) 8:592–601. doi: 10.20546/ijcmas.2019.808.071
38. Çetin, ES, Kaya, S, Demirci, M, and Aridogan, BC. Comparison of the BACTEC blood culture system versus conventional methods for culture of normally sterile body fluids. Adv Ther. (2007) 24:1271–7. doi: 10.1007/BF02877773
39. Sandhya, EMASR, Dutta, A, Das, B, Sharma, T, and Hazarika, M. A study of bacteriological profile of sterile body fluids in a tertiary care hospital. Int J Sci Res. (2019) 8:41–5.
Keywords: antimicrobial susceptibility pattern, bacterial etiologies, associated factors, sterile body fluids, Ethiopia
Citation: Admas D, Demeke G, Adugna A and Esmael A (2024) Bacterial etiologies, antimicrobial susceptibility pattern and associated factors among patients suspected sterile body site infections at Debre Markos Comprehensive Specialized Hospital, Northwest Ethiopia. Front. Med. 11:1260841. doi: 10.3389/fmed.2024.1260841
Edited by:
Amir Sasan Mozaffari Nejad, Jiroft University of Medical Sciences, IranReviewed by:
Mohammad Reza Arabestani, Hamadan University of Medical Sciences, IranFaham Khamesipour, Tehran University of Medical Sciences, Iran
Copyright © 2024 Admas, Demeke, Adugna and Esmael. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gebreselassie Demeke, gebredemeke@yahoo.com
 Dires Admas
Dires Admas Gebreselassie Demeke
Gebreselassie Demeke Adane Adugna
Adane Adugna Ahmed Esmael
Ahmed Esmael