- 1Department of Veterinary Preventive Medicine, College of Veterinary Medicine, Nanjing Agricultural University, Nanjing, China
- 2State Key Laboratory of Agricultural Microbiology, College of Veterinary Medicine, Huazhong Agricultural University, Wuhan, China
- 3School of Bioengineering, Shandong Provincial Key Laboratory of Microbial Engineering, Qilu University of Technology, Jinan, China
- 4Department of Clinical Veterinary Science, College of Veterinary Medicine, Nanjing Agricultural University, Nanjing, China
Uropathogenic Escherichia coli (UPEC) are the primary causative agents of urinary tract infections. Some UPEC isolates are able to infect renal proximal tubule cells, and can potentially cause pyelonephritis. We have previously shown that to fulfill their physiological roles renal proximal tubule cells accumulate high concentrations of α-ketoglutarate (KG) and that gene cluster c5032–c5039 contribute to anaerobic utilization of KG by UPEC str. CFT073, thereby promoting its in vivo fitness. Given the importance of utilizing KG for UPEC, this study is designed to investigate the roles of two transporters KgtP and C5038 in KG utilization, their transcriptional regulation, and their contributions to UPEC fitness in vivo. Our phylogenetic analyses support that kgtP is a widely conserved locus in commensal and pathogenic E. coli, while UPEC-associated c5038 was acquired through horizontal gene transfer. Global anaerobic transcriptional regulators Fumarate and nitrate reduction (FNR) and ArcA induced c5038 expression in anaerobiosis, and C5038 played a major role in anaerobic growth on KG. KgtP was required for aerobic growth on KG, and its expression was repressed by FNR and ArcA under anaerobic conditions. Analyses of FNR and ArcA binding sites and results of EMS assays suggest that FNR and ArcA likely inhibit kgtP expression through binding to the –35 region of kgtP promoter and occluding the occupancy of RNA polymerases. Gene c5038 can be specifically induced by KG, whereas the expression of kgtP does not respond to KG, yet can be stimulated during growth on glycerol. In addition, c5038 and kgtP expression were further shown to be controlled by different alternative sigma factors RpoN and RpoS, respectively. Furthermore, dual-strain competition assays in a murine model showed that c5038 mutant but not kgtP mutant was outcompeted by the wild-type strain during the colonization of murine bladders and kidneys, highlighting the importance of C5038 under in vivo conditions. Therefore, different transcriptional regulation led to distinct roles played by C5038 and KgtP in KG utilization and fitness in vivo. This study thus potentially expanded our understanding of UPEC pathobiology.
Introduction
Urinary tract infection (UTI) is one of the most common bacterial infections in humans, and represents a significant clinical issue worldwide (Foxman, 2010). The primary causative agent of UTIs is uropathogenic Escherichia coli (UPEC), which is responsible for ∼80–90% of community-acquired UTIs (Russo and Johnson, 2003; Spurbeck and Mobley, 2013). Despite that most previous studies of UPEC pathogenesis have been focused on traditional virulence factors, increasing attention is being paid to UPEC’s metabolic adaptive mechanisms, which impact colonization and survival in host’s urinary tract (Alteri and Mobley, 2015; Conover et al., 2016). UPECs have evolved multiple systems and strategies to snatch needed iron, combatting extremely low iron availability within host (Opal et al., 1990; Russo et al., 2001, 2002; Torres et al., 2001). Possession of a complete dsdCXA locus responsible for the detoxification of D-serine allows UPEC to use D-serine as the sole carbon and nitrogen source in urine (Roesch et al., 2003). In addition, most UPEC strains carry a genomic island involved in utilizing α-ketoglutarate (KG) under anaerobic conditions, which significantly increased UPEC’s fitness in a mouse model for UTI (Cai et al., 2013). KG is an abundant metabolite in the UPEC’s infection site – renal proximal tubule cells (Mobley et al., 1990; Chassin et al., 2008; Melican et al., 2008; Pichon et al., 2009), with an intracellular concentration of up to 400 μM (Boyd and Goldstein, 1979; Martin et al., 1989; Pritchard, 1995). KG lies at the intersection between the carbon and nitrogen metabolic pathways, and has long been recognized to coordinate carbon and nitrogen metabolism (Kim and Gadd, 2008).
A BLAST search revealed that the great majority of UPEC strains encode at least two transporters possibly associated with utilization of KG (Cai et al., 2013). A genomic island-encoded gene c5038 was predicted to encode a putative dicarboxylate transporter with 13 transmembrane alpha-helices, showing 49% similarity to citrate/succinate antiporter CitT (Pos et al., 1998) at the amino acid level. Its expression is activated by two-component regulatory system (TCS) KguSR in response to KG under anaerobic conditions (Cai et al., 2013). Fumarate and nitrate reduction (FNR) positively regulates c5038 expression by directly affecting the expression of KguSR (KG utilization sensor and regulator) (Barbieri et al., 2014). The other gene, kgtP (KG permease), encodes a KG:H+ symporter in E. coli K-12 strain (Seol and Shatkin, 1991, 1992). Alkaline phosphatase fusion study showed that KgtP contains 12 transmembrane segments (Seol and Shatkin, 1993). Genome-wide analysis of ArcA binding sites and its modulon in E. coli K-12 revealed that kgtP is negatively affected by ArcA (aerobic respiratory control), which binds to the upstream region of kgtP in vivo under anaerobic conditions (Liu and De Wulf, 2004; Park et al., 2013). Despite the knowledge gained of KgtP in K-12 strain and C5038, the individual roles and transcriptional regulation of C5038 and KgtP in UPEC during KG utilization remain unknown.
In this study, we describe KgtP and C5038 contribute differentially to growth on KG under aerobic and anaerobic conditions. Further analyses of their expression levels in various growth conditions and of roles played by regulators ArcA, FNR, RpoN, RpoS, and CRP (catabolite receptor protein) can account for the findings of different contributions to growth on KG and to colonization of murine urinary tracts. Overall, our results suggest that C5038 is a specialized, anaerobic KG importer, but KgtP is a generalized, aerobic KG importer. This report, therefore, should substantially improve our understanding of UPEC physiology.
Materials and Methods
Ethics Statement
All animal experimental procedures were conducted according to the guidelines of Experimental Animal Management Measures of Jiangsu Province and were approved by the Laboratory Animal Monitoring Committee of Jiangsu Province (China).
Bacterial Strains and Culture Conditions
Strains and plasmids used in this study are listed in Supplementary Table S1. Aerobic growth was achieved by shaking in air at 160 rpm, and anaerobic growth by incubating in a BugBox chamber (Ruskinn, UK) filled with a gas mixture (N2, 85%; CO2, 5%; H2, 10%). For genetic manipulations, all E. coli strains were grown routinely in lysogenic broth (LB) medium (OXOID). For growth and gene expression studies, bacteria were generally grown aerobically or anaerobically in M9 minimal salts with certain carbon sources indicated, supplemented with 2 mM MgSO4, 0.1 mM CaCl2, and 1 μg ml-1 vitamin B1 (Cai et al., 2013). Glucose (0.4% m/v) or glycerol (0.4% v/v) (Sinopharm, China) was added as energy substrates, as indicated. When used, electron acceptor trimethylamine N-oxide (TMAO) and dicarboxylates were present at 40 mM. Selective antibiotics and IPTG (Isopropyl β-D-1-thiogalactopyranoside) were added when necessary at the following concentrations: ampicillin (Amp), 100 μg ml-1; kanamycin (Kan), 50 μg ml-1;chloramphenicol (Chl), 25 μg ml-1; IPTG, 1 mM. All reagents were purchased from Sigma-Aldrich unless otherwise noted.
For growth studies, fresh colonies of wild-type (WT) CFT073 and its derivative mutant strains were picked and inoculated in 3 ml LB medium. After OD600 of bacterial culture reached about 1.0, bacteria were washed once with phosphate-buffered saline (PBS) and normalized, followed by being subcultured 1:100 into M9 medium. OD600 value at different time points was measured and recorded using spectrophotometer (Eppendorf, basic model).
Recombinant DNA Techniques
Polymerase chain reaction (PCR), DNA ligation, electroporation and DNA gel electrophoresis were performed according to Sambrook and Russell (2001) unless otherwise indicated. All oligonucleotide primers were purchased from BGI Technology Solutions Co., Ltd. (BGI, Guangzhou, China) and are listed in Supplementary Table S2. All restriction and DNA-modifying enzymes were purchased from New England Biolabs and were used based on the suppliers’ recommendations. Recombinant plasmids, PCR products, and restriction fragments were purified using TaKaRa PCR purification kit or gel extraction kit (TaKaRa, a Clontech company) as recommended by the supplier. DNA sequencing was performed at Shanghai Sunny Biotech Co., Ltd. DNA and amino acid sequence analyses were performed using Clone Manager software (Scientific & Educational Software, Morrisville, NC, USA) and the search engine1 was used to find homologous sequences. ClustalW program was used to perform multiple sequences alignments. Phylogenetic trees were made using MEGA software (Tamura et al., 2011; Hall, 2013).
Deletion mutants were constructed using the lambda red recombinase system described by Datsenko and Wanner (2000). For complementation studies, the coding sequences of genes plus their promoter regions (400 bp upstream of start codons) were amplified from the CFT073 genome and independently cloned into pGEN-MCS (Lane et al., 2007) using EcoRI and SalI restriction sites. Plasmid pCJ112 for plasmid-borne LacZ fusion studies was constructed by replacing the R6K origin in pVIK112 [this plasmid was created for making chromosomal transcriptional fusions of lacZ reporter gene (Kalogeraki and Winans, 1997)] with p15A origin from pBAD30 (Guzman et al., 1995) plasmid using EcoRI and BamHI restriction sites (See Supplementary Figure S1 for detail). The resulting plasmid was tested to show the functionality and an undetectable basal level without inserting any promoters upstream of promoterless lacZ. Pbla constitutively expressed promoter was cloned from pGEN-MCS plasmid by amplifying 400 bp upstream of bla gene as previous described (Sperandio et al., 1999). All constructs were verified by Sanger Sequencing (Shanghai Sunny Biotech Co., Ltd).
β-Galactosidase Assays
Overnight LB cultures of CFT073 and its derivative strains containing the gene of interest-lacZ fusions were washed with PBS once and then were diluted 1:100 in LB or M9 medium with the carbon sources indicated. Empty vector pCJ112 and pCJ112 containing Pbla-lacZ were used under all conditions as controls. Cultures were grown at 37°C to log phase or stationary phase. Samples were diluted 1:1 in Z buffer and assayed for β-galactosidase activity using ortho-Nitrophenyl-β-galactoside (ONPG) as a substrate as described previously (Miller, 1972).
5’-RACE (Rapid Amplification of cDNA Ends) PCR to Identify Transcriptional Start Site (TSS)
The transcriptional start site (TSS) site of c5038 was identified by using ExactSTART 5′- and 3′ -RACE Kit (Epicentre) with some modifications. Briefly, bacteria were grown in M9 media supplemented with glycerol and KG to log phase. RNA was stabilized by RNAprotect Bacterial Reagent (QIAGEN) and extracted using an RNeasy Mini Kit (QIAGEN) with a 1-h in-tube DNase digestion (QIAGEN) to remove possible DNA contamination according to the manufacturer’s instructions. Complete removal of DNA contamination was confirmed by using reverse transcription PCR. Three biological replicates of each sample were prepared. The concentration of RNA was determined using a Spectrophotometer (Eppendorf, basic model). The Steps A and B were omitted as they were designed for treating Eukaryotic RNA. About 5 μg of RNA were used for the Step C in the manual: 5’-RACE Acceptor Oligo Ligation step. Control reactions were included according to the manual. The primers which yielded reliable PCR products were listed in Supplementary Table S2.
Electrophoretic Mobility Shift Assays
To study the binding of proteins of interest to DNA probes, electrophoretic mobility shift assays (EMSAs) were performed using the commercialized EMSA kit (Invitrogen, Carlsbad, CA, USA) (Shen et al., 2011; Cai et al., 2013). pET28a prokaryotic expression system (Novagen) was used to over-produce recombinant proteins. His6-ArcA (Jiang et al., 2015), His6-(FNR-FNR)2 (Shan et al., 2012) and His6-RpoN fusion proteins were individually purified to homogeneity using Ni-NTA Spin Columns (QIAGEN) and dialyzed against the binding buffer. Protein concentrations were measured using BCA protein assay kit (Pierce). DNA probes (230 bp upstream of the start codon of kgtP or c5038) were PCR amplified using specific primers and gel-purified. The negative control probe was amplified from the coding region of Chloramphenicol resistance gene-cat. The concentrations of DNA fragments were determined by Biospectrometer (Eppendorf). EMSAs were performed by adding increasing amounts of purified fusion proteins (0240 nM) to DNA probes (10 nM) in binding buffer [10 mM Tris (pH 7.5), 1 mM EDTA, 1 mM dithiothreitol, 50 mM KCl, 50 mM MgCl2, 1 μg ml-1 bovine serum albumin (NEB)) and incubating for 30 min at room temperature (His6-ArcA and His6-RpoN), or 20 min at 37°C (His6-(FNR-FNR)2]. Reaction mixtures were then mixed with the loading dye (bromophenol blue, 0.25%; xylene cyanol FF, 0.25%; Ficoll 400, 15%), followed by electrophoresis on a 6% polyacrylamide gel in 0.5 × TBE buffer (44.5 mM Tris, 44.5 mM boric acid, 1 mM EDTA, pH 8.0) at 200V for 45 min in an ice-bath. The gel was stained in 0.5 × TBE buffer containing 1 × SYBR Gold nucleic acid staining solution (Life technology, USA) for 15 min. Gels were then visualized and photographed using GelDoc-it Imager (UVP).
Experimental UTIs
Mouse infection studies were performed according to the methods of Johnson et al. (2005). Female Balb/C mice (about 6 to 7 weeks of age) were anesthetized by ketamine/xylazine/Midazolam and inoculated via transurethral catheterization with a 50 μl (∼109 CFU) challenge inoculum per mouse. Overnight statically grown LB cultures of CFT073 and the mutant strains were pelleted and resuspended in sterile PBS, mixed in equal number, and adjusted to make challenge inocula. To determine the initial CFU/ml, dilutions of each inoculum were plated onto LB plates with and without chloramphenicol (kgtP::ChlR mutant) or kanamycin (c5038::KanR mutant). After 48 h, the mice were sacrificed and the bladder and kidneys were aseptically removed and homogenized in tubes containing PBS using a Bioprep-24 bullet homogenizer (Allsheng, Hangzhou in China). Dilutions (10 μl in each droplet) of the homogenized tissue were then spotted onto quadruplicate LB plates with and without antibiotics to determine the bacterial load. The numbers of colonies on selective plates were subtracted from those on LB plates to obtain the number of WT bacteria. A group of 10 or 6 (for complementation studies) mice for each dual-strain challenge were used to determine alterations in fitness, and assays were performed at least twice. Competitive index is defined as the ratio of mutant to the WT isolated divided by the ratio of mutant to the WT in the inoculums. the single-gene deletion mutants
Statistical Analyses
Statistical tests were performed using GraphPad Prism (version 5.0 for Windows, San Diego, CA, USA). For animal experiments data, Wilcoxon signed rank test was used to determine significance of competitive indices with a hypothetical median of 1 (numberWT = numbermutant). For other data, unpaired Student t-test was used to estimate differences between samples. The threshold for statistical significance was a P-value < 0.05.
Results
C5038 and KgtP Played Differential Roles in KG Utilization under Different Oxygen Tension Conditions
Gene c5038 is encoded on a metabolic island implicated in the anaerobic utilization of KG in UPEC (Cai et al., 2013). Bioinformatic analysis revealed that C5038 belongs to the Divalent Anion:Na+ Symporter (DASS) Family (Saier et al., 2006). Characterized DASS members in bacteria include succinate transporter VcINDY, citrate transporter CitT, L-tartrate/succinate transporter TtdT, C4-dicarboxylates uptake system DccT, YbhI, and SdcS (Markovich, 2012). C5038 on average shares 29% homology with these proteins. Sequence alignment of these DASS members (Supplementary Figure S2A) revealed that C5038 contains a SAT (serine–alanine–threonine) signature, which is similar to the relatively conserved SNT (serine–asparagine–threonine) motif involved in substrates-binding (Mancusso et al., 2012). The kgtP locus in K-12 is 96.4% identical to kgtP in UPEC str. CFT073 at DNA level, with 47 differences between them. At the amino acid level, only three differences exist between them, all of which are non-charged amino acids. KgtP shares no homology with C5038 at both DNA and amino acid levels. The promoter regions of kgtP in K-12 and CFT073 have an 88% sequence identity to each other (Supplementary Figure S2B).
To determine the roles of C5038 and KgtP in growth of CFT073 in various media, we compared the growth of the WT, the single-gene deletion mutants (Δc5038 and ΔkgtP, respectively), and the double-gene deletion mutant (Δc5038ΔkgtP) in different media under aerobic conditions. We observed no significant difference in growth kinetics among these strains in LB or M9 medium supplemented with glucose (Gluc) or glycerol (Glyc) as the sole carbon source (data not shown).
When cultured in M9 using KG as the sole carbon source under aerobic conditions, the Δc5038 mutant showed similar growth kinetics as WT, indicating that c5038 was not important for growth under these conditions (Figure 1A). In contrast, deletion of kgtP abolished CFT073’s growth (P < 0.0001) and kgtP mutant displayed no detectable growth within 40 h. To rule out the possible masking effect of kgtP on c5038’s role, we compared the growth of two mutants, ΔkgtP mutant and Δc5038ΔkgtP mutant. The results showed no significant difference in growth between ΔkgtP mutant and Δc5038ΔkgtP mutant (Figure 1A), further supporting that c5038 played a negligible role in KG utilization in aerobiosis. Additionally, the introduction of a complementation plasmid (pkgtP) carrying kgtP controlled by its native promoter restored the growth of kgtP mutant (Figure 1B). These results suggest that kgtP is required whereas c5038 is dispensable for growth on KG under aerobic conditions.
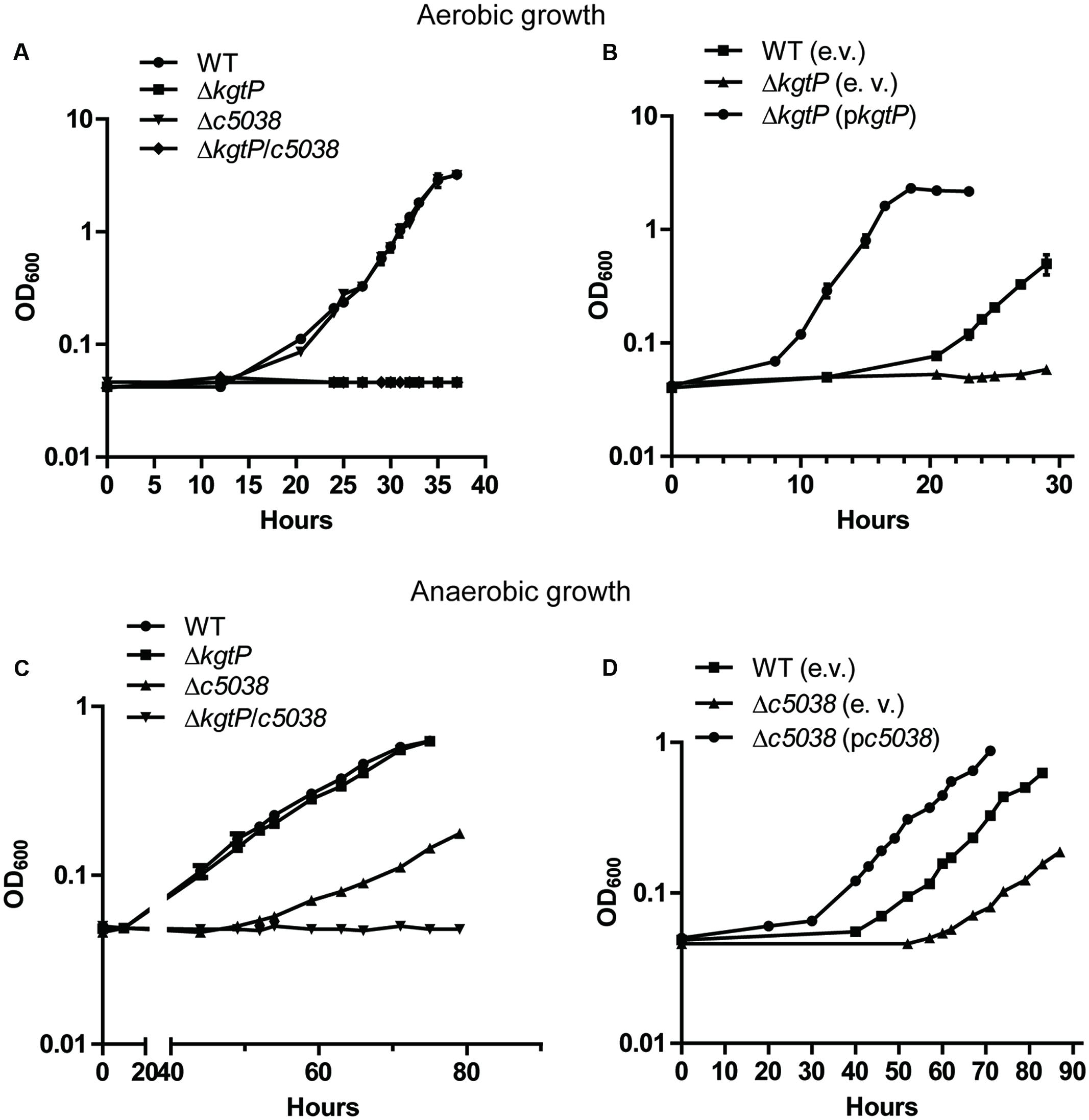
FIGURE 1. Contributions of c5038 and kgtP to growth on KG under aerobic (A,B) and anaerobic (C,D) conditions. Various strains were grown in M9 medium containing KG as the sole carbon and energy source. Trimethylamine N-oxide (TMAO) was added as electron acceptor under anaerobic conditions. Optical density at different time points was determined. Growth curves represent the average measurement at each time point from triplicate experiments. Student t-test was used for statistical analysis. e. v., empty vector; pc5038, complementation plasmid for c5038; pkgtP, complementation plasmid for kgtP.
Under anaerobic conditions, deletion of kgtP did not affect CFT073’s growth, whereas loss of c5038 dramatically reduced its growth in M9 with KG as the sole carbon source, as compared to WT (Figure 1C). Transformation of the c5038 mutant with a complementation plasmid (pc5038) carrying c5038 controlled by its native promoter significantly improved c5038 mutant’s growth (Figure 1D). To rule out the possible masking effect of c5038 on kgtP’s role under these conditions, we compared the growth of Δc5038 mutant and Δc5038ΔkgtP mutant. We observed a difference in growth between Δc5038 mutant and Δc5038ΔkgtP mutant at 60 h time point and thereafter (P < 0.05) (Figure 1C). These data indicate that under anaerobic conditions, c5038 played a major role and kgtP a minor role in growth on KG. Of note, in both aerobiosis and anaerobiosis, the complementation strains apparently had much shorter lag phases and higher growth rates than the WT strain carrying the empty vector. This was likely due to the fact that the complementation plasmid vector is a multi-copy plasmid (∼15 copies per cell), which can cause overexpression of kgtP or c5038.
Anaerobiosis Induced c5038 but Repressed kgtP Expression
Given that kgtP and c5038 contributed differentially to growth on KG under aerobic and anaerobic conditions, we sought to unravel their transcriptional levels under these conditions. The TSS of c5038 was obtained using 5′-RACE PCR (Figure 2B). The –10 and –35 sites of kgtP promoter were predicted by Virtual Footprint program (Munch et al., 2005) and Bprom program (Solovyev and Salamov, 2011), which presented consistent output with high reliability. The putative ribosome binding sites were also indicated. The predicted TSS of kgtP in CFT073 was highly similar to that in K-12 identified by 5′-RACE and deep sequencing (Cho et al., 2009). The promoter regions of kgtP and c5038 were then cloned to drive promoterless lacZ gene on a plasmid, respectively, and the resulting plasmid constructs were individually introduced into LMP10 strain (CFT073 ΔlacZYA). In parallel, a control plasmid carrying constitutively expressed promoter Pbla-lacZ fusion was also transformed into LMP10 strain. β-galactosidase activities were measured to indicate expression levels. As shown in Figure 2C, c5038 expression was not detectable in aerobiosis, but was induced in anaerobiosis. The expression of kgtP was moderate under aerobic conditions, but was greatly repressed under anaerobic conditions, with a 40-fold reduction. It is also evident that anaerobic expression level of c5038 was significantly higher than aerobic expression level of kgtP (about 10-fold difference). In the control strain, Pbla-lacZ expression did not exhibit difference between under aerobic and anaerobic conditions. These results clearly showed that under anaerobic conditions, c5038 was highly expressed whereas kgtP was severely repressed, thereby at least partially explaining the phenotypic difference in growth.
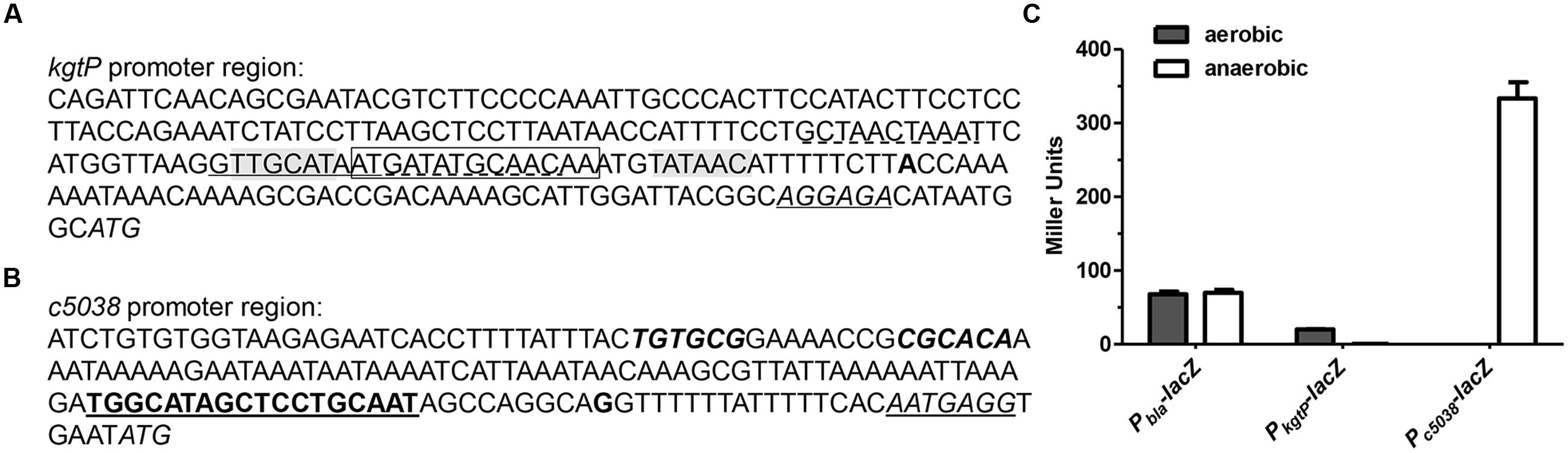
FIGURE 2. c5038 transcription was induced whereas kgtP repressed in anaerobiosis. (A) Promoter region of kgtP. (B) Promoter region of c5038. Italic, start codon; italic and underlined, putative ribosome binding site; bold, transcription start site; bold and underlined, RpoN binding site/σ54 motif (–12 and –24 regions); italic and bold, inverted repeats for KguR binding; shaded, –10 and –35 regions; Underlined (solid), perfect ArcA binding site; underlined (dashed), imperfect ArcA binding site; boxed, FNR binding site. (C) kgtP and c5038 expression under aerobic and anaerobic conditions. Bacteria were grown aerobically or anaerobically in M9 medium containing KG as the sole carbon source. TMAO was used as an electron acceptor for anaerobic tests. Expression of c5038-lacZ or kgtP-lacZ was indicated by β-galactosidase activity. Pbla-lacZ constitutive expression was used as a control. Bars represent means ± SEM from three independent experiments performed in triplicate.
FNR and ArcA Induced c5038 But Repressed kgtP Expression
Fumarate and nitrate reduction and ArcA are two master regulators mediating bacterial adaptation to environmental oxygen availability (Constantinidou et al., 2006; Park et al., 2013; Jiang et al., 2015). To probe the regulatory roles of arcA and fnr in expression of c5038 and kgtP, we examined the effects of single deletion and double deletion of fnr and arcA on expression levels of c5038 and kgtP under aerobic and anaerobic conditions. Under aerobic conditions, deletion of fnr did not affect kgtP expression while loss of arcA upregulated kgtP expression about fivefold. Under anaerobic conditions, deletion of either fnr or arcA caused upregulation of kgtP expression (4-fold and 700-fold increase, respectively). Deletion of both fnr and arcA led to a greater increase in kgtP expression than individual deletions of fnr and arcA. These results indicate that both fnr and arcA negatively modulate kgtP expression. Further, in LMP10 strain, anaerobiosis repressed kgtP expression about 40-fold, as compared to aerobic expression, whereas in LMP10Δfnr and LMP10ΔarcA mutants, anaerobiosis repressed kgtP expression 10-fold and 4-fold, respectively (Figure 3A). These results clearly demonstrate that in either fnr or arcA mutant, anaerobic repression of kgtP was partially relieved compared with that in their parental strain LMP10, suggesting both fnr and arcA are involved in the anaerobic repression, with ArcA being a major player.
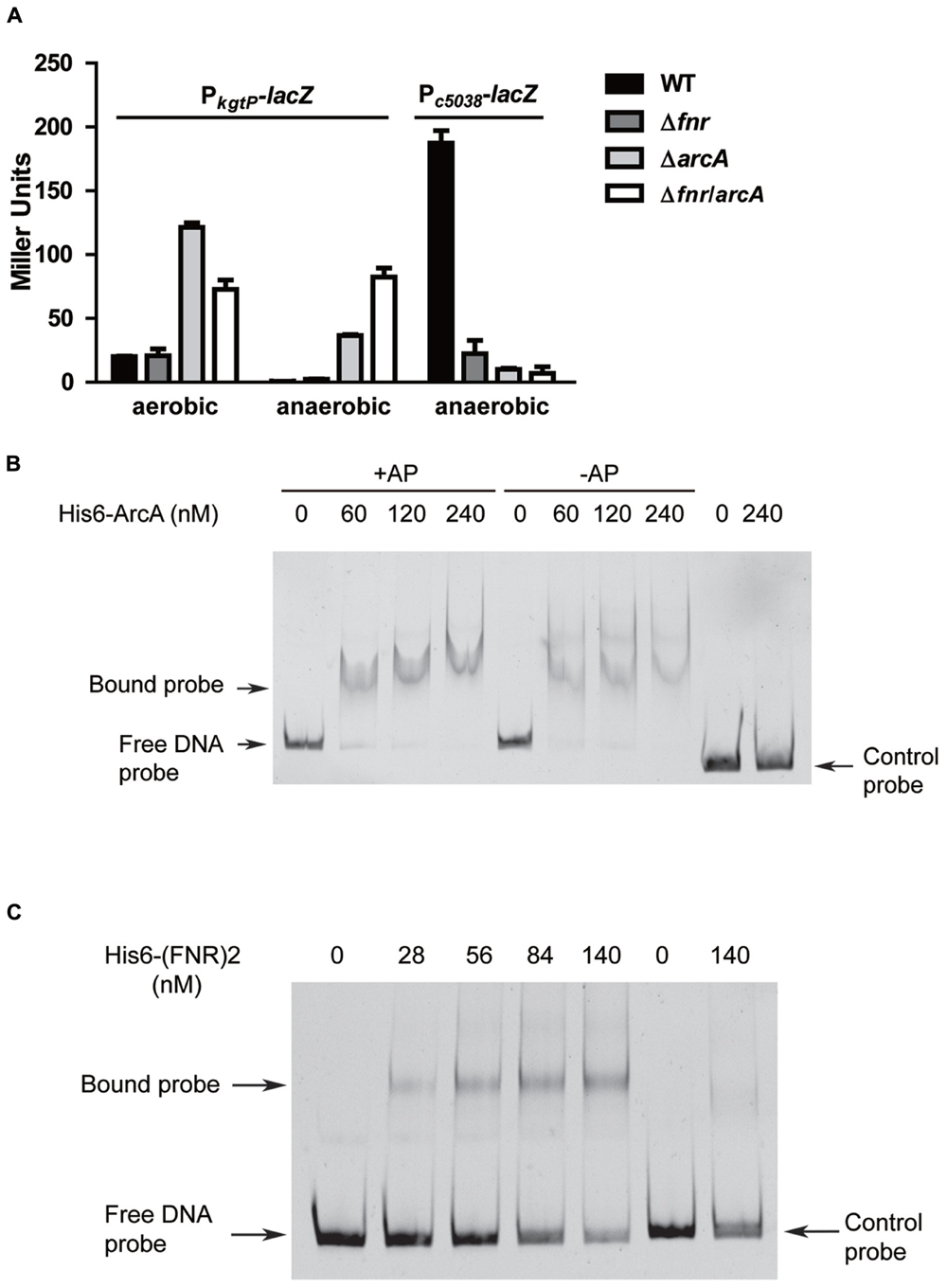
FIGURE 3. Roles of fnr and arcA in regulating kgtP and c5038 expression in response to varying oxygen tension. (A) Loss of fnr or arcA leads to different effects on kgtP and c5038 expression. Various strains were grown aerobically or anaerobically in M9 medium containing KG as the sole carbon source. TMAO was used as electron acceptor anaerobically. Expression of c5038-lacZ or kgtP-lacZ in different strains was represented by β-galactosidase activity. (B) Non-radioactive EMSA studying the binding of ArcA to kgtP promoter regions. (C) Non-radioactive EMSA studying the binding of FNR to kgtP promoter regions. Gel-extracted PCR products of kgtP promoter region and cat (ChlR gene) coding region were used as probes. Purified His6-ArcA or His6-(FNR)2 fusion protein was added in different concentrations in each reaction mixture as indicated. DNA fragments were stained with SYBR green. AP, acetyl phosphate.
We next tried to understand the mechanisms by which ArcA and FNR repress the expression of kgtP. Sequence analyses of promoters repressed by ArcA or FNR suggest that ArcA and FNR inhibit gene expression through competitive binding to –10 or/and –35 region, thereby blocking occupancy of RNA polymerases (Myers et al., 2013; Park et al., 2013). Using a recently developed binding consensus sequence of ArcA ([GT][TA][TA][AG][AC][AT][AT][AT][AT][AT], each pair of letters in each bracket indicate two most frequent bases in that position, with the first being the most frequent) (Park et al., 2013), we were able to identify one perfect match (GTTGCATAAT) that overlaps the putative –35 region and two imperfect matches close to the perfect one (Figure 2A). Similarly, using a recently developed binding consensus sequence of FNR ([TA][TC][GA][AC][TC]nnnn[AG][TA][CT][AG][AC]) (Myers et al., 2013), we were able to identify a perfect match (ATGATatgcAACAA) that lies between –10 and –35 region. To test whether ArcA and FNR directly binds to the promoter region of kgtP, EMS assays were carried out. DNA fragments with sizes around 230 bp containing the predicted sites were prepared. Purified His6-tagged ArcA and FNR variant (FNRD154A) proteins (Shan et al., 2012) were obtained by nickel affinity chromatography. As shown in Figure 3B, the DNA probe of kgtP promoter region can be shifted by ArcA protein, but the negative control probe cannot be shifted. With the same amounts of protein, the presence of acetyl phosphate appears to not affect binding of ArcA to DNA under our testing conditions. Also, FNR recombinant protein can shift the DNA probe of kgtP, but not the control probe (Figure 3C). These results indicate that ArcA and FNR can bind to the promoter region of kgtP directly. Altogether, these data suggest that ArcA and FNR likely repress kgtP expression through binding to promoter region and occluding the occupancy of RNAP.
For c5038, deletion of either fnr or arcA or both resulted in a significant reduction in c5038-lacZ expression in comparison to that in the LMP10 strain (P < 0.01), suggesting both fnr and arcA positively modulate c5038 expression. We next used EMSA to test if ArcA directly associates with DNA probe of c5038 promoter region. The results show that ArcA protein cannot shift the DNA probes (Supplementary Figure S3), raising the possibility that ArcA may not regulate c5038 expression directly. Taken together, these data indicate that FNR and ArcA stimulated c5038 but repressed kgtP expression.
Response of c5038 and kgtP Expression to Different Stimuli and the Role of CRP
In contrast to c5038, what stimuli induce kgtP’s expression is still unclear. To test if the expression of kgtP is influenced by the addition of KG, we measured and compared the expression levels of kgtP during bacterial growth in M9 (Glyc), M9 (Glyc and KG), and M9 (KG), respectively. Surprisingly, kgtP expression in all three media is comparable (Figure 4A), indicating that kgtP expression does not respond to the presence of KG. We next tested and compared the expression of kgtP in LB rich medium, M9 (Gluc), and M9 (Glyc). The results showed that the order of kgtP expression levels is as follows: M9 (Glyc) > M9 (Gluc) > LB.
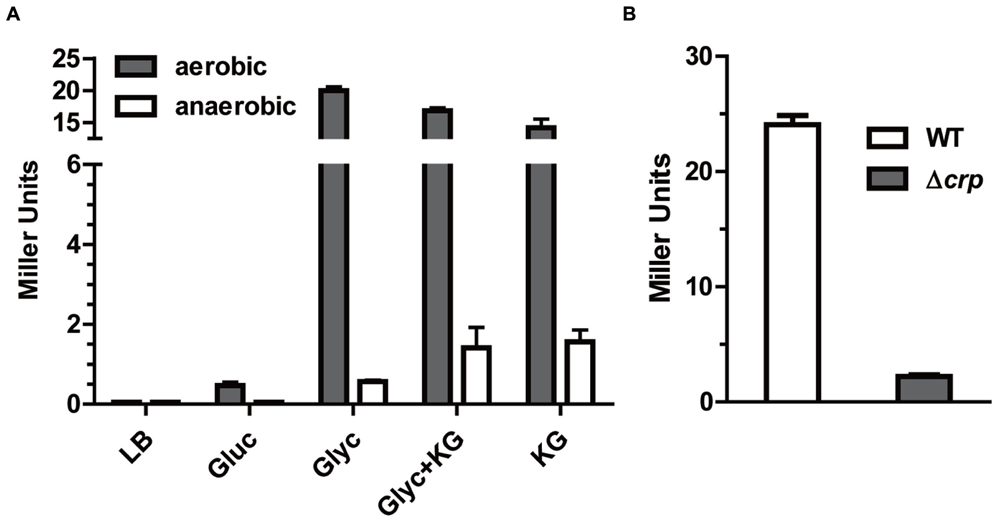
FIGURE 4. Response of kgtP expression to growth in different media and contribution of global regulator CRP. (A) kgtP expression does not respond to the presence of KG. Bacteria were grown aerobically or anaerobically in different media containing the indicated carbon and energy source. TMAO was used as an electron acceptor for anaerobic tests. Gluc, glucose; Glyc, glycerol. (B) Role of CRP in kgtP expression. Bacteria were grown in M9 containing glycerol as sole carbon source. Expression of kgtP-lacZ was indicated by β-galactosidase activity. Bars represent means ± SEM from three independent experiments performed in triplicate.
The cAMP-CRP complex is a global regulatory element that induces a number of genes involved in the utilization of carbons in the absence of glucose (Botsford and Harman, 1992; Kolb et al., 1993), including glycerol catabolic regulon (Iuchi et al., 1990). We therefore examined whether CRP contributes to the regulation of kgtP expression. As shown in Figure 4B, kgtP expression in the strain lacking crp was downregulated, as compared to the parental strain LMP10. This result indicates that CRP positively regulates kgtP expression. Collectively, these data suggest that kgtP might be responsive to the carbon state of the cell, and be used for carbon scavenging.
Roles of Different Alternative Sigma Factors in the Expression of c5038 and kgtP
KguR involved in direct activation of c5038 was predicted to belong to the category of bacterial enhancer-binding proteins (bEBPs) which interact with σ54 factor (Bush and Dixon, 2012), raising the possibility that c5038 is a new member of the σ54 regulon. The “A,” –30 site relative to the start codon was identified to be the TSS of c5038 (Figure 2B). Further analysis of the promoter region of c5038 did reveal a motif that is highly similar to the σ54 binding consensus “–24 to –12” region (Figure 2B), suggesting that c5038 transcription might depend on sigma54. To assess the contribution of rpoN (the gene coding for sigma54) in c5038 expression, we compared the expression levels of c5038 in LMP10 with that in LMP10ΔrpoN. As shown in Figure 5A, deletion of rpoN abolished the KG induction of c5038 expression, as compared to parental strain LMP10. To ensure that no unwanted mutation was responsible for such phenotype, a complementation study was performed by introducing a copy of plasmid-borne rpoN controlled by its putative native promoter. The result showed that with rpoN reintroduced, the expression level of c5038 was rescued.
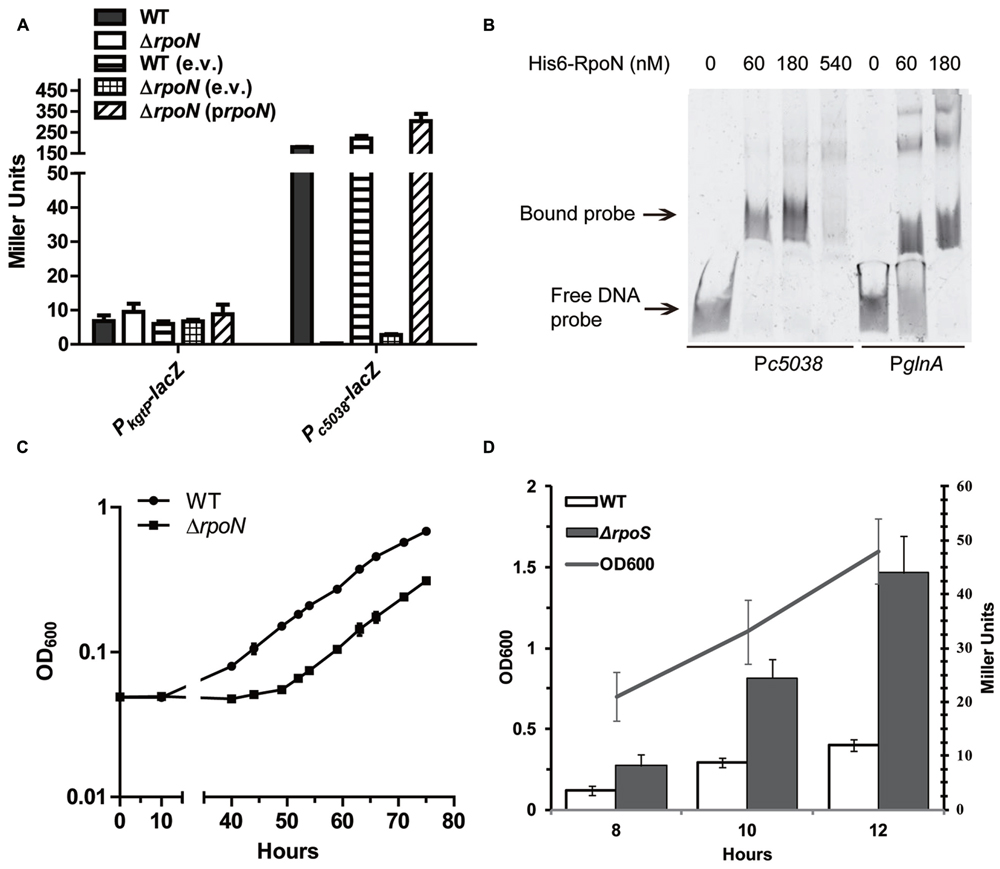
FIGURE 5. Role of alternative σ54 encoded by rpoN in kgtP and c5038 expression. (A) Loss of RpoN affects kgtP and c5038 expression differently. Various strains were grown aerobically (kgtP) or anaerobically (c5038) in M9 medium containing glycerol (kgtP) or KG (c5038) as the sole carbon source. TMAO was used as electron acceptor anaerobically. Expression of c5038-lacZ or kgtP-lacZ in different strains was represented by the β-galactosidase activity. e.v., empty vector; prpoN, complementation plasmid carrying rpoN. (B) Non-radioactive EMSA studying the binding of RpoN to promoter regions. PCR products of glnA (right three lanes) and c5038 (left four lanes) promoter regions were used as probes. Purified His6-RpoN fusion protein was added in different concentration in each reaction mixture as indicated. DNA fragments were stained with SYBR green. (C) Growth of the wild-type (WT) and rpoN deletion mutant. Bacteria were cultured in M9 minimal medium supplemented with KG as the sole carbon source and TMAO as an electron acceptor. Samples were taken at the indicated time points, and their OD600 was measured. Each point in the growth curve represents the average measurement from triplicate experiments. (D) The role of rpoS in kgtP expression in different growth phases. Bacteria were grown aerobically in M9 medium containing glycerol as the sole carbon source. Expression of kgtP-lacZ was represented by the β-galactosidase activity.
Sigma54 protein was shown to be capable of binding to target promoter regions independent of core RNA polymerase (Shingler, 2011). To evaluate if σ54 protein directly associates with c5038 promoter region, EMSA was used to study the binding of σ54 proteins to DNA molecules. σ54 was over-produced in E. coli as His6-RpoN fusion protein, which was then purified to homogeneity against nickel column. glnA promoter region, which was previously shown to be directly associated with RpoN (Huo et al., 2006), was included as a positive control; on the other hand, xylA promoter region, a member of σ70 regulon (Song and Park, 1997; Desai and Rao, 2010), was included as a negative control. As we expected, His6-RpoN fusion protein can shift PglnA and Pc5038 DNA probes, but not the PxylA (Figure 5B; Supplementary Figure S4) probe. Altogether, these data indicate that c5038 is a new member in the RpoN regulon.
Because RpoN is required for c5038 expression which is important for growth of CFT073 on KG, we then examined the growth kinetics of the WT and ΔrpoN mutant in M9 (KG) containing abundant nitrogen source NH4Cl under anaerobic conditions. The results demonstrate that ΔrpoN mutants had a longer lag phase and grew significantly slower than their parental WT strain (P < 0.001) (Figure 5C). These data suggest that RpoN contributes to anaerobic utilization of KG, possibly through regulating c5038 expression.
In contrast to c5038, deletion of rpoN had no effect on kgtP expression (Figure 5A). We have shown earlier that kgtP expression remains high in M9 (Glyc), which is considered to be a carbon- and amino acid-limited environment. RpoS, also known as sigma38 factor, is a stress-responsive global regulator, which is induced under stress conditions such as carbon, phosphorus, nitrogen, or amino acid scarcity (Notley and Ferenci, 1996). Thus, rpoS was deleted from LMP10 strain, and its effects on kgtP expression were determined. Unexpectedly, loss of RpoS increased kgtP expression by about twofold, irrespective of growth phase (Figure 5D). Therefore, RpoS is a negative modulator of kgtP expression.
c5038 But Not kgtP Contributed to UPEC Fitness In vivo
Now that c5038 and kgtP were demonstrated to play differential roles in vitro, we next tried to investigate if loss of c5038 or kgtP affects UPEC fitness in vivo. Mice were challenged transurethrally with a 1:1 mixture of WT and a mutant, followed by recovering bacteria from tissues 48 h post inoculation and calculating competitive indices. Note that when inoculated in LB medium with 1:1 ratio, any two strains contained in the inoculums were recovered equally in numbers during exponential growth phase, indicating that they were equally fit in vitro. Figure 6A showed that the c5038 mutant was significantly outcompeted by the WT in both bladder and kidneys (P-value equals 0.0005 and 0.0458, respectively). In vivo complementation experiments were performed to confirm that the mutation is not polar and the disadvantage is not caused by a secondary mutation (Figure 6B). The stable low-copy pGEN plasmid was used as it was shown to be well maintained in CFT073 up to 48 h even in the absence of antibiotic pressure. The Δc5038 mutant carrying empty vector displayed the expected competitive disadvantage in bladder and kidneys when co-challenging mice with the WT containing pGEN (P-value equals 0.0161 and 0.0342, respectively) whereas Δc5038 harboring a copy of c5038 on pGEN plasmid was equally fit as WT (pGEN). Therefore, reintroduction the c5038 gene into the c5038 mutant can potentially complement the negative effect on colonization caused by mutation of c5038.
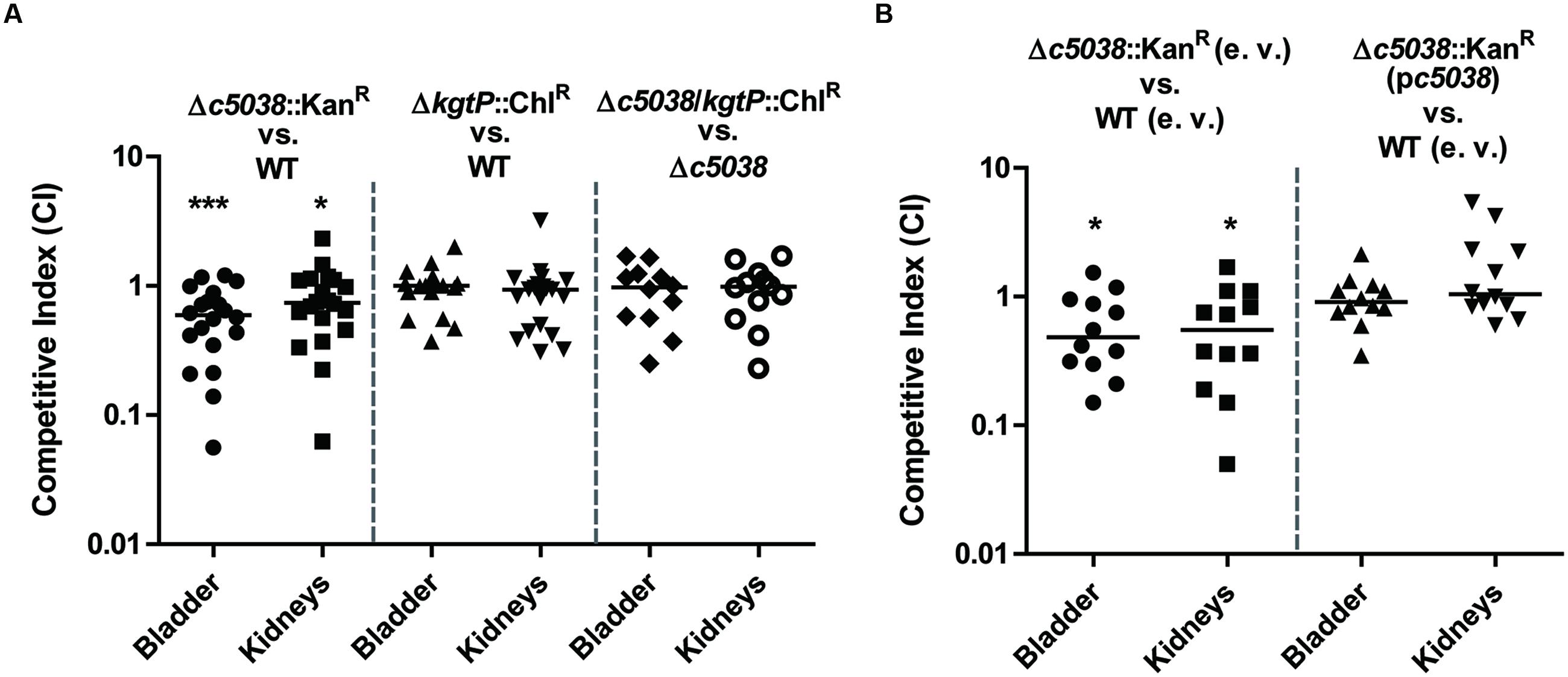
FIGURE 6. Fitness of c5038 and kgtP mutants during co-infections with the WT. Bacteria were recovered from mice transurethrally challenged with a 1:1 mixture of two strains. Colonies on LB plates and LB plates containing the indicated antibiotics were enumerated. Competitive indices at 48 hpi were determined by dividing the ratio of two strains recovered by the ratio of the same two strains in the inoculum. Each group contains 10 or 6 mice, and experiments were performed at least twice. Bars represent the medians, and a Wilcoxon signed rank test was used to determine the significance of CI against a hypothetical median of 1. The threshold for statistical significance was a P-value < 0.05. ∗P < 0.05; ∗∗∗P < 0.001. (A) In vivo competition of Δc5038::KanR/WT, ΔkgtP::ChlR/WT, and Δc5038/kgtP::ChlR/Δc5038. (B) In vivo complementation study of c5038.
In contrast to the c5038 mutant, the kgtP mutant colonized murine urinary tracts as well as the WT (Figure 6A, CI was not significantly different from the hypothetical value 1). To rule out the possible masking effect of c5038 on kgtP, we tested the effects of mutating kgtP in Δc5038 genetic background on fitness in vivo. Deletion of kgtP in Δc5038 background had no impact on colonization of mice by CFT073, further corroborating that kgtP is not important for colonization of mice by UPEC. Taken together, these results suggest that c5038 but not kgtP is important for UPEC fitness in vivo.
Phylogenetic Analysis of c5038 and kgtP
In light of distinct roles of C5038 and KgtP in KG utilization and in vivo fitness of CFT073, their phylogenetic trees were constructed in order to understand their evolutionary routes (Escherichia spp. strains were excluded). Comparing the two phylogenetic trees (Supplementary Figures S5A,B), it is evident that the two transporters have different patterns: KgtP is clustered with many E. coli close relatives whereas C5038 is clustered with only two of them, with others being Pseudomonads and Burkholderiales. These data strongly indicate that C5038 and KgtP have distinct evolutionary trajectories, supporting our suggestion that c5038 was acquired through horizontal gene transfer.
Discussion
Bacterial pathogens usually possess more than one system/factor to execute certain cellular function critical for pathogenicity (Brussow et al., 2004). In UPEC, multiple fimbrial genes and iron acquisition systems exist and contribute to UPEC’s fitness in different host niches and environments (Garcia et al., 2011; Spurbeck et al., 2011). In this study, we described multiple aspects of transcriptional regulation of two transporters, C5038 and KgtP, and effects of their regulation on physiological roles played by the two transporters. The expression of c5038 was induced solely by KG under anaerobic conditions, which was mediated by ArcA and FNR regulators responsible for adaptation to low-oxygen environments. Thus, it can be expected that possession of c5038 can significantly increase UPEC’s growth on KG anaerobically (Figures 1C,D). Phylogenetic analysis supports that c5038 was acquired through horizontal gene transfer. Further, c5038 is highly prevalent in UPEC strains (78%), but very lowly in diarrheagenic E. coli strains (0%) (Cai et al., 2013). These findings prompted us to reason that C5038 is a specialized protein that carries out important functions for UPEC under in vivo conditions. During infection by UPEC, host sites such as renal proximal tubule cells could provide abundant KG (Martin et al., 1989; Pritchard, 1995) while oxygen tension is low (Melican et al., 2008). Indeed, loss of c5038 led to reduced fitness during colonization of murine urinary tracts by UPEC (Figure 6A). Although the c5038 mutant was outcompeted less than twofold, it is plausible that under our testing conditions, C5038 did not exert effects to its full potential. Acquisition of genes or factors that promote fitness of bacterial pathogens is a recurring theme. For example, in response to tetrathionate, genomic island-encoded loci ttrABC provide a growth advantage for Salmonella by utilizing tetrathionate anaerobically as an electron receptor produced by inflamed guts (Price-Carter et al., 2001; Winter et al., 2010).
The expression of kgtP is not inducible by KG and appears to be constitutively expressed in K-12 strain, implying that kgtP is expressed at similar levels in different environments (Seol and Shatkin, 1992). We did show here that kgtP expression in CFT073 does not respond to the addition of KG, but its expression was high in M9 (Glyc) and low in M9 (Gluc) and LB. This was consistent with previous KG accumulation assays demonstrating that maximal intracellular concentration of KG was achieved for bacteria grown on glycerol, followed by those on Gluc and LB (Seol and Shatkin, 1992). Because one of the major stresses E. coli face during growth on glycerol is limited carbon availability, it is tempting to hypothesize that kgtP senses carbon availability of bacterial cells and that importing KG could scavenge carbons (Yan et al., 2011) and help improve carbon status, considering the role of KG in providing carbon skeletons for both TCA cycle (tricarboxylic acid cycle) and amino acid metabolism. Supporting data is that the expression level of kgtP was higher in stationary phase when carbon sources are mostly depleted (Supplementary Figure S6), and was regulated by CRP and stress sigma factor σS. However, more biochemical evidence is needed to further address that hypothesis. Similar to KgtP in K-12, KgtP in UPEC str. CFT073 is also required for aerobic growth using KG as the sole carbon source. On the contrary, KgtP only played a minor role in bacterial growth on KG under anaerobic conditions (Figure 1C). Expectedly, loss of KgtP did not result in any competitive disadvantage in the animal model. Overall, we for the first time revealed kgtP is subject to regulation by oxygen tension, which was mediated by ArcA and FNR.
The TCA cycle is a bi-functional pathway that generates ATP during the catabolic process and supplies skeletons in the anabolic process of biosynthesis (Kim and Gadd, 2008). As a result, transport, assimilation, and conversion of intermediates in TCA cycle are subject to regulation in response to oxygen availability (Nakano et al., 1998; Kim and Gadd, 2008). For four-carbon (C4)-dicarboxylates in TCA cycle including succinate, fumarate, and malate, E. coli possess both aerobic and anaerobic transporters, namely, DctA (Davies et al., 1999) and DcuABC (Zientz et al., 1996; Golby et al., 1998). Similar to DctA, KgtP requires protons to import its substrate-KG (Seol and Shatkin, 1992). Similar to DcuABC which exchanges their substrates with succinate, we also found that overexpression of c5038 led to decreased succinate assimilation, suggesting that C5038 likely exported succinate (data not shown). To ensure optimal assimilation of KG, KgtP as a proton symporter needs to be inhibited in low-oxygen environments in order to save energy, whereas C5038 can function in a substrate-product exchange fashion which is less energy-consuming and thus highly desirable. Indeed, our data show that FNR and ArcA co-repress kgtP but co-activate c5038 expression, emphasizing roles of FNR and ArcA in promoting a coordinated response to KG in anaerobic environments. Likewise, for C4-dicarboxylate transporters, DctA is repressed by ArcA under anaerobic conditions (Davies et al., 1999) while Dcu system is activated by FNR under anaerobic conditions (Engel et al., 1992). Conceivably, possessing functional alternatives with optimized regulatory patterns for importing KG should greatly promote UPEC’s fitness in changing environments. To our knowledge, for the first time, we reveal bacteria carry both aerobic and anaerobic transporter systems for KG. The conformity between C4- and C5-dicarboxylates transporters makes us speculate that such transport systems and regulatory patterns might be widely existed for other important metabolites in bacteria.
Since FNR activates arcA expression under anaerobic conditions (Compan and Touati, 1994), it raises a question of whether FNR can affect kgtP expression through ArcA. Our data showed that in the absence of arcA, mutation of fnr can still increase the expression of kgtP by about twofold, indicating that FNR can regulate kgtP independent of ArcA. ArcA represses kgtP expression to a higher degree than FNR does, this can be at least partially attributable to the fact that ArcA can potentially bind to three putative binding sites, whereas FNR can only bind to one site. By binding to more than one site, ArcA has a potential to form multimer (Pellicer et al., 1999; Jeon et al., 2001); in contrast, FNR usually works as a dimer (Lazazzera et al., 1996; Jervis and Green, 2007). In the arcA mutant, kgtP expression in aerobiosis was higher than that in anaerobiosis, this may be due to that (1) fnr can still repress gene expression in anaerobiosis when arcA is absent, (2) there are other activating elements (probably cAMP-CRP) that function better under aerobic conditions, or (3) the metabolic changes caused by arcA deletion.
Like TCS response regulator KguR and inducer KG, sigma54 factor is also essential in activating c5038 expression, thereby adding one more level of control in the existing regulatory network for orchestrating c5038 expression. It is well established that members of sigma54 regulon are primarily involved in nitrogen metabolism, although exceptions exist (Shingler, 2011). The inclusion of sigma54 in c5038 regulatory network is expected to have great biological significance as nitrogen availability and KG accumulation are inversely correlated (Reyes-Ramirez et al., 2001; Yan et al., 2011). Recent studies have suggested that UPEC can encounter environments with limited nitrogen source during infection (Snyder et al., 2004). Therefore, it is likely that expression of c5038 helps accumulate KG and further to adapt to nitrogen-limited urinary tracts (Snyder et al., 2004; Hagan and Mobley, 2007). Notably, RpoN coding for sigma54 factor and a bEBP regulator MifR were both required for the expression of KG transporter PA5530 (Lundgren et al., 2014) in Pseudomonas aeruginosa, highlighting the possibility of commonly using a similar mechanism to link KG and nitrogen metabolism. By contrast, sigma54 factor does not contribute to kgtP expression, further distinguishing it from c5038.
In summary, we employed bioinformatic, biochemical, and genetic tools to study two transporters involved in KG utilization, demonstrating that C5038 is likely an anaerobic KG importer but KgtP is an aerobic KG importer. Their distinct roles can be at least in part attributed to distinctive transcriptional regulatory patterns. The development of these two systems should provide adaptive advantages for UPEC in various environments.
Author Contributions
WC, SY, and HZ conceived and designed the experiments; WC, YY, and XC performed the experiments; WC and HZ analyzed the data; WC, SY, and HZ contributed reagents/materials/analysis tools; WC, SY, and HZ wrote the paper. All authors contributed, read and approved the final manuscript.
Funding
This work was funded by the Natural Science Foundation of China (31470243), the Fundamental Research Funds for the Central Universities, and Science and Technology Development Program of Shandong Province (Grant: 2014GSF120006).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Huochun Yao for providing an anaerobic chamber, and Mr. Haobo Zhang and Ms. Xuhua Chen for assistance in animal experiments.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.00275/full#supplementary-material
Footnotes
References
Alteri, C. J., and Mobley, H. L. (2015). Metabolism and fitness of urinary tract pathogens. Microbiol. Spectr. 3. doi: 10.1128/microbiolspec.MBP-0016-2015
Barbieri, N. L., Nicholson, B., Hussein, A., Cai, W., Wannemuehler, Y. M., Dell’anna, G., et al. (2014). FNR regulates expression of important virulence factors contributing to pathogenicity of uropathogenic Escherichia coli. Infect. Immun. 82, 5086–5098. doi: 10.1128/IAI.02315-14
Boyd, T. A., and Goldstein, L. (1979). Kidney metabolite levels and ammonia production in acute acid-base alterations in the rat. Am. J. Physiol. 236, E289–E295.
Brussow, H., Canchaya, C., and Hardt, W. D. (2004). Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol. Mol. Biol. Rev. 68, 560–602. doi: 10.1128/MMBR.68.3.560-602.2004
Bush, M., and Dixon, R. (2012). The role of bacterial enhancer binding proteins as specialized activators of sigma(54)-dependent transcription. Microbiol. Mol. Biol. Rev. 76, 497–529. doi: 10.1128/MMBR.00006-12
Cai, W., Wannemuehler, Y., Dell’anna, G., Nicholson, B., Barbieri, N. L., Kariyawasam, S., et al. (2013). A novel two-component signaling system facilitates uropathogenic Escherichia coli’s ability to exploit abundant host metabolites. PLoS Pathog. 9:e1003428. doi: 10.1371/journal.ppat.1003428
Chassin, C., Vimont, S., Cluzeaud, F., Bens, M., Goujon, J. M., Fernandez, B., et al. (2008). TLR4 facilitates translocation of bacteria across renal collecting duct cells. J. Am. Soc. Nephrol. 19, 2364–2374. doi: 10.1681/ASN.2007121273
Cho, B. K., Zengler, K., Qiu, Y., Park, Y. S., Knight, E. M., Barrett, C. L., et al. (2009). The transcription unit architecture of the Escherichia coli genome. Nat. Biotechnol. 27, 1043–1049. doi: 10.1038/nbt.1582
Compan, I., and Touati, D. (1994). Anaerobic activation of arca transcription in Escherichia coli – roles of Fnr and Arca. Mol. Microbiol. 11, 955–964. doi: 10.1111/j.1365-2958.1994.tb00374.x
Conover, M. S., Hadjifrangiskou, M., Palermo, J. J., Hibbing, M. E., Dodson, K. W., and Hultgren, S. J. (2016). Metabolic requirements of Escherichia coli in intracellular bacterial communities during urinary tract infection pathogenesis. MBio 7:e00104-16. doi: 10.1128/mBio.00104-16
Constantinidou, C., Hobman, J. L., Griffiths, L., Patel, M. D., Penn, C. W., Cole, J. A., et al. (2006). A reassessment of the FNR regulon and transcriptomic analysis of the effects of nitrate, nitrite, NarXL, and NarQP as Escherichia coli K12 adapts from aerobic to anaerobic growth. J. Biol. Chem. 281, 4802–4815. doi: 10.1074/jbc.M512312200
Datsenko, K. A., and Wanner, B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645. doi: 10.1073/pnas.120163297
Davies, S. J., Golby, P., Omrani, D., Broad, S. A., Harrington, V. L., Guest, J. R., et al. (1999). Inactivation and regulation of the aerobic C(4)-dicarboxylate transport (dctA) gene of Escherichia coli. J. Bacteriol. 181, 5624–5635.
Desai, T. A., and Rao, C. V. (2010). Regulation of arabinose and xylose metabolism in Escherichia coli. Appl. Environ. Microbiol. 76, 1524–1532. doi: 10.1128/AEM.01970-09
Engel, P., Kramer, R., and Unden, G. (1992). Anaerobic fumarate transport in Escherichia coli by an fnr-dependent dicarboxylate uptake system which is different from the aerobic dicarboxylate uptake system. J. Bacteriol. 174, 5533–5539. doi: 10.1128/jb.174.17.5533-5539.1992
Foxman, B. (2010). The epidemiology of urinary tract infection. Nat. Rev. Urol. 7, 653–660. doi: 10.1038/nrurol.2010.190
Garcia, E. C., Brumbaugh, A. R., and Mobley, H. L. (2011). Redundancy and specificity of Escherichia coli iron acquisition systems during urinary tract infection. Infect. Immun. 79, 1225–1235. doi: 10.1128/IAI.01222-10
Golby, P., Kelly, D. J., Guest, J. R., and Andrews, S. C. (1998). Transcriptional regulation and organization of the dcuA and dcuB genes, encoding homologous anaerobic C4-dicarboxylate transporters in Escherichia coli. J. Bacteriol. 180, 6586–6596.
Guzman, L. M., Belin, D., Carson, M. J., and Beckwith, J. (1995). Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177, 4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995
Hagan, E. C., and Mobley, H. L. (2007). Uropathogenic Escherichia coli outer membrane antigens expressed during urinary tract infection. Infect. Immun. 75, 3941–3949. doi: 10.1128/IAI.00337-07
Hall, B. G. (2013). Building phylogenetic trees from molecular data with MEGA. Mol. Biol. Evol. 30, 1229–1235. doi: 10.1093/molbev/mst012
Huo, Y. X., Tian, Z. X., Rappas, M., Wen, J., Chen, Y. C., You, C. H., et al. (2006). Protein-induced DNA bending clarifies the architectural organization of the sigma54-dependent glnAp2 promoter. Mol. Microbiol. 59, 168–180. doi: 10.1111/j.1365-2958.2005.04943.x
Iuchi, S., Cole, S. T., and Lin, E. C. (1990). Multiple regulatory elements for the glpA operon encoding anaerobic glycerol-3-phosphate dehydrogenase and the glpD operon encoding aerobic glycerol-3-phosphate dehydrogenase in Escherichia coli: further characterization of respiratory control. J. Bacteriol. 172, 179–184. doi: 10.1128/jb.172.1.179-184.1990
Jeon, Y., Lee, Y. S., Han, J. S., Kim, J. B., and Hwang, D. S. (2001). Multimerization of phosphorylated and non-phosphorylated ArcA is necessary for the response regulator function of the Arc two-component signal transduction system. J. Biol. Chem. 276, 40873–40879. doi: 10.1074/jbc.M104855200
Jervis, A. J., and Green, J. (2007). In vivo demonstration of FNR dimers in response to lower O-2 availability. J. Bacteriol. 189, 2930–2932. doi: 10.1128/JB.01921-06
Jiang, F., An, C., Bao, Y., Zhao, X., Jernigan, R. L., Lithio, A., et al. (2015). ArcA controls metabolism, chemotaxis, and motility contributing to the pathogenicity of avian pathogenic Escherichia coli. Infect. Immun. 83, 3545–3554. doi: 10.1128/IAI.00312-15
Johnson, J. R., Jelacic, S., Schoening, L. M., Clabots, C., Shaikh, N., Mobley, H. L., et al. (2005). The IrgA homologue adhesin Iha is an Escherichia coli virulence factor in murine urinary tract infection. Infect. Immun. 73, 965–971. doi: 10.1128/IAI.73.2.965-971.2005
Kalogeraki, V. S., and Winans, S. C. (1997). Suicide plasmids containing promoterless reporter genes can simultaneously disrupt and create fusions to target genes of diverse bacteria. Gene 188, 69–75. doi: 10.1016/S0378-1119(96)00778-0
Kim, B. H., and Gadd, G. M. (2008). Bacterial Physiology and Metabolism. Cambridge: Cambridge University Press. doi: 10.1017/CBO9780511790461
Kolb, A., Busby, S., Buc, H., Garges, S., and Adhya, S. (1993). Transcriptional regulation by camp and its receptor protein. Annu. Rev. Biochem. 62, 749–795. doi: 10.1146/annurev.bi.62.070193.003533
Lane, M. C., Alteri, C. J., Smith, S. N., and Mobley, H. L. (2007). Expression of flagella is coincident with uropathogenic Escherichia coli ascension to the upper urinary tract. Proc. Natl. Acad. Sci. U.S.A. 104, 16669–16674. doi: 10.1073/pnas.0607898104
Lazazzera, B. A., Beinert, H., Khoroshilova, N., Kennedy, M. C., and Kiley, P. J. (1996). DNA binding and dimerization of the Fe-S-containing FNR protein from Escherichia coli are regulated by oxygen. J. Biol. Chem. 271, 2762–2768. doi: 10.1074/jbc.271.5.2762
Liu, X., and De Wulf, P. (2004). Probing the ArcA-P modulon of Escherichia coli by whole genome transcriptional analysis and sequence recognition profiling. J. Biol. Chem. 279, 12588–12597. doi: 10.1074/jbc.M313454200
Lundgren, B. R., Villegas-Penaranda, L. R., Harris, J. R., Mottern, A. M., Dunn, D. M., Boddy, C. N., et al. (2014). Genetic analysis of the assimilation of C5-dicarboxylic acids in Pseudomonas aeruginosa PAO1. J. Bacteriol. 196, 2543–2551. doi: 10.1128/JB.01615-14
Mancusso, R., Gregorio, G. G., Liu, Q., and Wang, D. N. (2012). Structure and mechanism of a bacterial sodium-dependent dicarboxylate transporter. Nature 491, 622–626. doi: 10.1038/nature11542
Markovich, D. (2012). Sodium-sulfate/carboxylate cotransporters (SLC13). Curr. Top. Membr. 70, 239–256. doi: 10.1016/B978-0-12-394316-3.00007-7
Martin, M., Ferrier, B., and Baverel, G. (1989). Transport and utilization of alpha-ketoglutarate by the rat kidney in vivo. Pflugers Archiv. 413, 217–224. doi: 10.1007/BF00583533
Melican, K., Boekel, J., Mansson, L. E., Sandoval, R. M., Tanner, G. A., Kallskog, O., et al. (2008). Bacterial infection-mediated mucosal signalling induces local renal ischaemia as a defence against sepsis. Cell. Microbiol. 10, 1987–1998. doi: 10.1111/j.1462-5822.2008.01182.x
Miller, J. H. (1972). Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory.
Mobley, H. L., Green, D. M., Trifillis, A. L., Johnson, D. E., Chippendale, G. R., Lockatell, C. V., et al. (1990). Pyelonephritogenic Escherichia coli and killing of cultured human renal proximal tubular epithelial cells: role of hemolysin in some strains. Infect. Immun. 58, 1281–1289.
Munch, R., Hiller, K., Grote, A., Scheer, M., Klein, J., Schobert, M., et al. (2005). Virtual footprint and PRODORIC: an integrative framework for regulon prediction in prokaryotes. Bioinformatics 21, 4187–4189. doi: 10.1093/bioinformatics/bti635
Myers, K. S., Yan, H., Ong, I. M., Chung, D., Liang, K., Tran, F., et al. (2013). Genome-scale analysis of Escherichia coli FNR reveals complex features of transcription factor binding. PLoS Genet. 9:e1003565. doi: 10.1371/journal.pgen.1003565
Nakano, M. M., Zuber, P., and Sonenshein, A. L. (1998). Anaerobic regulation of Bacillus subtilis Krebs cycle genes. J. Bacteriol. 180, 3304–3311.
Notley, L., and Ferenci, T. (1996). Induction of RpoS-dependent functions in glucose-limited continuous culture: what level of nutrient limitation induces the stationary phase of Escherichia coli? J. Bacteriol. 178, 1465–1468. doi: 10.1128/jb.178.5.1465-1468.1996
Opal, S. M., Cross, A. S., Gemski, P., and Lyhte, L. W. (1990). Aerobactin and alpha-hemolysin as virulence determinants in Escherichia coli isolated from human blood, urine, and stool. J. Infect. Dis. 161, 794–796. doi: 10.1093/infdis/161.4.794
Park, D. M., Akhtar, M. S., Ansari, A. Z., Landick, R., and Kiley, P. J. (2013). The bacterial response regulator ArcA uses a diverse binding site architecture to regulate carbon oxidation globally. PLoS Genet. 9:e1003839. doi: 10.1371/journal.pgen.1003839
Pellicer, M. T., Lynch, A. S., De Wulf, P., Boyd, D., Aguilar, J., and Lin, E. C. C. (1999). A mutational study of the ArcA-P binding sequences in the aldA promoter of Escherichia coli. Mol. Gen. Genet. 261, 170–176. doi: 10.1007/s004380050954
Pichon, C., Hechard, C., Du Merle, L., Chaudray, C., Bonne, I., Guadagnini, S., et al. (2009). Uropathogenic Escherichia coli AL511 requires flagellum to enter renal collecting duct cells. Cell. Microbiol. 11, 616–628. doi: 10.1111/j.1462-5822.2008.01278.x
Pos, K. M., Dimroth, P., and Bott, M. (1998). The Escherichia coli citrate carrier CitT: a member of a novel eubacterial transporter family related to the 2-oxoglutarate/malate translocator from spinach chloroplasts. J. Bacteriol. 180, 4160–4165.
Price-Carter, M., Tingey, J., Bobik, T. A., and Roth, J. R. (2001). The alternative electron acceptor tetrathionate supports B12-dependent anaerobic growth of Salmonella enterica serovar typhimurium on ethanolamine or 1,2-propanediol. J. Bacteriol. 183, 2463–2475. doi: 10.1128/JB.183.8.2463-2475.2001
Pritchard, J. B. (1995). Intracellular alpha-ketoglutarate controls the efficacy of renal organic anion transport. J. Pharmacol. Exp. Ther. 274, 1278–1284.
Reyes-Ramirez, F., Little, R., and Dixon, R. (2001). Role of Escherichia coli nitrogen regulatory genes in the nitrogen response of the Azotobacter vinelandii NifL-NifA complex. J. Bacteriol. 183, 3076–3082. doi: 10.1128/JB.183.10.3076-3082.2001
Roesch, P. L., Redford, P., Batchelet, S., Moritz, R. L., Pellett, S., Haugen, B. J., et al. (2003). Uropathogenic Escherichia coli use d-serine deaminase to modulate infection of the murine urinary tract. Mol. Microbiol. 49, 55–67. doi: 10.1046/j.1365-2958.2003.03543.x
Russo, T. A., Carlino, U. B., and Johnson, J. R. (2001). Identification of a new iron-regulated virulence gene, ireA, in an extraintestinal pathogenic isolate of Escherichia coli. Infect. Immun. 69, 6209–6216. doi: 10.1128/IAI.69.10.6209-6216.2001
Russo, T. A., and Johnson, J. R. (2003). Medical and economic impact of extraintestinal infections due to Escherichia coli: focus on an increasingly important endemic problem. Microbes Infect. 5, 449–456. doi: 10.1016/S1286-4579(03)00049-2
Russo, T. A., Mcfadden, C. D., Carlino-Macdonald, U. B., Beanan, J. M., Barnard, T. J., and Johnson, J. R. (2002). IroN functions as a siderophore receptor and is a urovirulence factor in an extraintestinal pathogenic isolate of Escherichia coli. Infect. Immun. 70, 7156–7160. doi: 10.1128/IAI.70.12.7156-7160.2002
Saier, M. H. Jr., Tran, C. V., and Barabote, R. D. (2006). TCDB: the Transporter Classification Database for membrane transport protein analyses and information. Nucleic Acids Res. 34, D181–D186. doi: 10.1093/nar/gkj001
Sambrook, J., and Russell, D. W. (2001). Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
Seol, W., and Shatkin, A. J. (1991). Escherichia coli kgtP encodes an alpha-ketoglutarate transporter. Proc. Natl. Acad. Sci. U.S.A. 88, 3802–3806. doi: 10.1073/pnas.88.9.3802
Seol, W., and Shatkin, A. J. (1992). Escherichia coli alpha-ketoglutarate permease is a constitutively expressed proton symporter. J. Biol. Chem. 267, 6409–6413.
Seol, W. G., and Shatkin, A. J. (1993). Membrane topology model of Escherichia coli alpha-ketoglutarate permease by phoa fusion analysis. J. Bacteriol. 175, 565–567. doi: 10.1128/jb.175.2.565-567.1993
Shan, Y., Pan, Q., Liu, J., Huang, F., Sun, H., Nishino, K., et al. (2012). Covalently linking the Escherichia coli global anaerobic regulator FNR in tandem allows it to function as an oxygen stable dimer. Biochem. Biophys. Res. Commun. 419, 43–48. doi: 10.1016/j.bbrc.2012.01.121
Shen, Z., Pu, X. Y., and Zhang, Q. (2011). Salicylate functions as an efflux pump inducer and promotes the emergence of fluoroquinolone-resistant Campylobacter jejuni mutants. Appl. Environ. Microbiol. 77, 7128–7133. doi: 10.1128/AEM.00763-11
Shingler, V. (2011). Signal sensory systems that impact Sigma 54-dependent transcription. FEMS Microbiol. Rev. 35, 425–440. doi: 10.1111/j.1574-6976.2010.00255.x
Snyder, J. A., Haugen, B. J., Buckles, E. L., Lockatell, C. V., Johnson, D. E., Donnenberg, M. S., et al. (2004). Transcriptome of uropathogenic Escherichia coli during urinary tract infection. Infect. Immun. 72, 6373–6381. doi: 10.1128/IAI.72.11.6373-6381.2004
Solovyev, V., and Salamov, A. (2011). “Automatic annotation of microbial genomes and metagenomic sequences,” in Metagenomics and Its Applications in Agriculture, Biomedicine and Environmental Studies, ed. R. W. Li (Hauppauge, NY: Nova Science Publishers), 61–78.
Song, S., and Park, C. (1997). Organization and regulation of the D-xylose operons in Escherichia coli K-12: XylR acts as a transcriptional activator. J. Bacteriol. 179, 7025–7032. doi: 10.1128/jb.179.22.7025-7032.1997
Sperandio, V., Mellies, J. L., Nguyen, W., Shin, S., and Kaper, J. B. (1999). Quorum sensing controls expression of the type III secretion gene transcription and protein secretion in enterohemorrhagic and enteropathogenic Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 96, 15196–15201. doi: 10.1073/pnas.96.26.15196
Spurbeck, R. R., Stapleton, A. E., Johnson, J. R., Walk, S. T., Hooton, T. M., and Mobley, H. L. (2011). Fimbrial profiles predict virulence of uropathogenic Escherichia coli strains: contribution of ygi and yad fimbriae. Infect. Immun. 79, 4753–4763. doi: 10.1128/IAI.05621-11
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Torres, A. G., Redford, P., Welch, R. A., and Payne, S. M. (2001). TonB-dependent systems of uropathogenic Escherichia coli: aerobactin and heme transport and TonB are required for virulence in the mouse. Infect. Immun. 69, 6179–6185. doi: 10.1128/IAI.69.10.6179-6185.2001
Winter, S. E., Thiennimitr, P., Winter, M. G., Butler, B. P., Huseby, D. L., Crawford, R. W., et al. (2010). Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 467, 426–429. doi: 10.1038/nature09415
Yan, D., Lenz, P., and Hwa, T. (2011). Overcoming fluctuation and leakage problems in the quantification of intracellular 2-oxoglutarate levels in Escherichia coli. Appl. Environ. Microbiol. 77, 6763–6771. doi: 10.1128/AEM.05257-11
Keywords: uropathogenic Escherichia coli, transcriptional regulation, α-ketoglutarate, transporter, sigma factor
Citation: Cai W, Cai X, Yang Y, Yan S and Zhang H (2017) Transcriptional Control of Dual Transporters Involved in α-Ketoglutarate Utilization Reveals Their Distinct Roles in Uropathogenic Escherichia coli. Front. Microbiol. 8:275. doi: 10.3389/fmicb.2017.00275
Received: 27 October 2016; Accepted: 09 February 2017;
Published: 21 February 2017.
Edited by:
Yuji Morita, Aichi Gakuin University, JapanReviewed by:
William Schwan, University of Wisconsin–La Crosse, USAJuan Xicohtencatl-Cortes, Hospital Infantil de Mexico Federico Gomez, Mexico
Copyright © 2017 Cai, Cai, Yang, Yan and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shigan Yan, yanshigan@qlu.edu.cn Haibin Zhang, haibinzh@njau.edu.cn
 Wentong Cai
Wentong Cai Xuwang Cai
Xuwang Cai Yongwu Yang
Yongwu Yang Shigan Yan
Shigan Yan Haibin Zhang
Haibin Zhang