- 1National Risk Assessment Laboratory for Antimicrobial Resistance of Animal Original Bacteria, South China Agricultural University, Guangzhou, China
- 2Guangdong Provincial Key Laboratory of Veterinary Pharmaceutics Development and Safety Evaluation, South China Agricultural University, Guangzhou, China
- 3Jiangsu Co-Innovation Centre for Prevention and Control of Important Animal Infectious Diseases and Zoonoses, Yangzhou, China
We screened 30 Klebsiella pneumoniae isolates from dogs and cats at a single animal hospital in Guangdong Province, China. Among them, 12 K. pneumoniae strains possessed high-level resistance to amikacin and gentamicin and these were screened for 16S rRNA methyltransferase (16S-RMTase) genes. And then the genes positive isolates were detected for ESBLs (extended spectrum β-lactamases) and analyzed by pulsed-field gel electrophoresis, multilocus sequence typing, PCR-based replicon typing and plasmid analysis. The genetic profiles of rmtB were also determined by PCR mapping. The twelve 16S-RMTase gene-positive isolates were rmtB (11/30) and armA (2/30) with one isolate carrying both genes. Extended spectrum β-lactamases genes were represented by blaCTX-M-55 (9/12), blaCTX-M-27 (2/12) and blaCTX-M-14 (1/12). The twelve 16S-RMTase containing strains were grouped into five clonal patterns and ST37 was the most prevalent sequence type. Ten rmtB-bearing plasmids conjugated successfully and all belonged to IncN and IncF (F33:A-:B-) incompatibility groups. Nine of the transconjugants carried a 97 kb plasmid and the other harbored both ∼60 and ∼200 kb plasmids. rmtB and blaCTX-M-55 were present on the same plasmid and indicated the co-transfer of these two genes, with the rmtB gene showing highly relevant relationships with IS26 and Tn3. Our findings suggested a high prevalence of 16S-RMTase genes in K. pneumonia ST37 from dogs and cats. Additional studies are needed to trace the evolutionary path of this type of resistance among the K. pneumonia isolates, and to determine whether they have been transferred to humans.
Introduction
Klebsiella pneumoniae is an opportunistic pathogen associated with a wide spectrum of community- and hospital-acquired infections (Lee et al., 2011). Increasing resistance to multiple antimicrobial agents has compromised the effectiveness of K. pneumoniae treatment options (Lee et al., 2011). Coexistence of resistance genes on the same plasmid appears to be the primary cause of the spread of resistance determinants (Luo et al., 2011). This has resulted in the appearance of multidrug resistant (MDR) or even pan drug-resistant (PDR) strains (Luo et al., 2011). Acquisition of MDR by Enterobacteriaceae members has become a global concern especially for K. pneumonia PDR strains (Zacharczuk et al., 2011a; Wasyl et al., 2015; Grevskott et al., 2017).
Aminoglycoside resistance is due primarily to aminoglycoside-modifying enzymes. Acetyltransferases, nucleotidyltransferases, and phosphotransferases inactivate commonly used aminoglycosides such as gentamicin (GEN) and tobramycin (Yang et al., 2011). Recently, plasmid-encoded 16S rRNA methyltransferases (16S-RMTase) have emerged in the Enterobacteriaceae family and in a group of glucose-non-fermentative microbes (Zacharczuk et al., 2011b). This is a new resistance mechanism to 4,6-disubstituted 2-deoxystreptamines and 4,5-disubstituted 2-deoxystreptamines. These structures encompass the majority of clinically important aminoglycosides (Zacharczuk et al., 2011b). Since the first report in 2003, ten 16S-RMTase-encoding genes, rmtA, rmtB, rmtC, rmtD, rmtE, rmtF, rmtG, rmtH, armA, and npmA, have been identified (O’Hara et al., 2013).
Reports on the prevalence of the 16S-RMTases have increased in the past years with the majority focused on the human clinical isolates including K. pneumoniae (Mezzatesta et al., 2013; Belbel et al., 2014; Bartoloni et al., 2016). Moreover, the majority of reports on dissemination of armA and rmtB to various Enterobacteriaceae species were focused on E. coli isolated from chickens and pigs (Yao et al., 2011; Yang Y. et al., 2015). Previous research has reported the occurrence of the ArmA methyltransferase in an ST11 clone of K. pneumoniae isolated from cats and dogs in Spain (Hidalgo et al., 2013a). Another research focused on the dissemination of rmtB-blaCTX-M-9 group genes and rmtB-qepA in Enterobacteriaceae isolates of dogs and cats in China mainly related to E. coli (Deng et al., 2011a). However, researches on prevalence and mechanisms of 16S-RMTase spread in pets via K. pneumonia strains in China are lacking.
Because of the common use of antimicrobial agents and the close contact with humans, companion animals may become potential sources for dissemination of antimicrobial resistance (Lloyd, 2007). Thus, we decided to identify the prevalence of 16S-RMTase genes among K. pneumonia isolates from dogs and cats in a veterinary hospital in Guangdong Province, China.
Materials and Methods
Bacterial Isolates
Thirty clinical K. pneumoniae strains were recovered from diseased dogs and cats during July, August, and September in 2010 and from July to October in 2012 at one animal hospital in Guangdong Province, China. Further information about the antimicrobial treatments of these pets were unfortunately not available. Samples were collected from the exudates of the infection areas (urinary tract infections, skin infections, or intra-abdominal infections) or feces, then seeded on MacConkey agar at 37°C. Each isolate was from a single animal. All bacterial species were identified with classical biochemical methods and confirmed using a matrix-assisted laser desorption and ionization time-of-flight mass spectrometry (MALDI-TOF MS) method (Shimadzu, Japan). As 16S-RMTase can confer high-level resistance to both GEN and amikacin (AMK), all isolates were subcultured in MacConkey agar containing 64 mg/L GEN and 64 mg/L AMK to screen for GEN/AMK-resistant isolates.
Detection of Resistance Genes and Antimicrobial Susceptibility Testing
All the strains exhibiting aminoglycoside resistance were screened for 16S-RMTase genes including rmtA, rmtB, rmtC, rmtD, rmtE, rmtF, rmtG, rmtH, armA, and npmA (Hidalgo et al., 2013b; Xia et al., 2016). Aminoglycoside-resistant isolates were further analyzed for the presence of extended spectrum β-lactamase (ESBL) genes (blaTEM, blaCTX-M, and blaSHV) using previously described primers (Liao et al., 2015).
The minimal inhibitory concentration (MIC) of ampicillin (AMP), cefotaxime (CTX), meropenem, ciprofloxacin (CIP), norfloxacin (NOR), GEN, kanamycin (KAN), AMK, streptomycin (STR), neomycin (NEO), apramycin (APR), chloramphenicol (CHL), florfenicol (FFC), tetracycline (TET), sulfamethoxazole/trimethoprim (SXT) were determined by the agar dilution method according to Clinical and Laboratory Standards Institute guidelines (Clinical and Laboratory Standards Institute, 2015). E. coli ATCC 25922 was used as a quality control strain.
Clonal Relatedness
Chromosomal DNA digested with XbaI restriction enzyme was used for pulsed-field gel electrophoresis (PFGE) to analyze the genetic relatedness of all isolates containing 16S-RMTase genes (CHEF Mapper1, Bio-Rad Laboratories, Hercules, CA, USA) as previously described (Fang et al., 2015). PFGE patterns were analyzed with the Dice coefficient and the unweighted pair group method with average linkages (UPGMA) clustering method using BioNumerics software (Applied Maths, Ghent, Belgium). PFGE types were defined with >90% similarity between clusters. Multilocus sequence typing (MLST) of 16S-RMTase genes containing K. pneumoniae was performed according to the protocols described on the K. pneumoniae database website1. Seven chromosomal genes were PCR amplified and sequenced. Then the sequences were compared with those reference sequences, submitted to the MLST database to determine allele numbers and STs (Belbel et al., 2014).
Transfer of the 16S-RMTase Genes and Plasmids Replicon Typing
Isolates positive for 16S-RMTase genes were selected for conjugation experiments by the broth-mating method using STR-resistant E. coli C600 as the recipient to determine the transferability of 16S-RMTase genes. Transconjugants were selected on MacConkey agar plates supplemented with STR (1000 μg/mL) and AMK (64 μg/mL). Transconjugants harboring 16S-RMTase genes were confirmed by PCR and antimicrobial susceptibility testing as described above (Fang et al., 2015). Antimicrobial susceptibility testing of the transconjugants and co-transfer of other resistance genes as mentioned above were also determined.
Plasmids were preliminarily classified according to their incompatibility group by using the PCR-based replicon typing (PBRT) scheme described previously (Yao et al., 2011). To better characterize IncF plasmids, replicon sequence typing of IncF plasmids was performed according to the protocol previously described (Yang Q.E. et al., 2015). The A- and B- symbols indicate the absence of the FIA and FIB replicons, respectively.
Plasmid Analysis
PFGE with S1 nuclease (TakaRa Biotechnology, Dalian, China) digestion of genomic DNA was performed for all the transconjugants as described previously (Xia et al., 2015). After Southern blotting, fix the DNA to a Hybond-N+ membrane (GE Healthcare, Little Chalfont, UK), the plasmids were probed with the 16S-RMTase genes and blaCTX-M-1G/9G gene (DIG High Prime DNA Labeling and Detection Starter Kit I, Roche Applied Science, Mannheim, Germany). All the transconjugants were subjected to restriction enzyme digestion (EcoRI) analysis to clarify whether a specific plasmid had been disseminated among the isolates (Deng et al., 2011b). EcoRI-RFLP types were defined with >80% similarity between clusters using the method described as PFGE analysis.
Genetic Profiles of 16S-RMTase Genes
Reports on the genetic contexts of the 16S-RMTase genes indicated that IS26, Tn3, ISCR1, and ISCR3 played an important role in the dissemination of these genes. These transfer events enabled horizontal spread among different bacterial lineages (Deng et al., 2011b). Here we focused on the link between 16S-RMTase genes and mobile elements by PCR mapping. The primers using in PCR mapping were listed in Supplementary Table S1.
Ethics Statement
In this study, the owners of the companion animals from which fecal swabs and the exudates of the infection areas were taken gave permission for their animals to be used in this study. Written informed consent was obtained, and the study protocol in our research was approved by the South China Agriculture University Animal ethics committee and carried out in accordance with relevant guidelines.
Results
Prevalence of 16S-RMTase Genes and Antimicrobial Susceptibility Testing
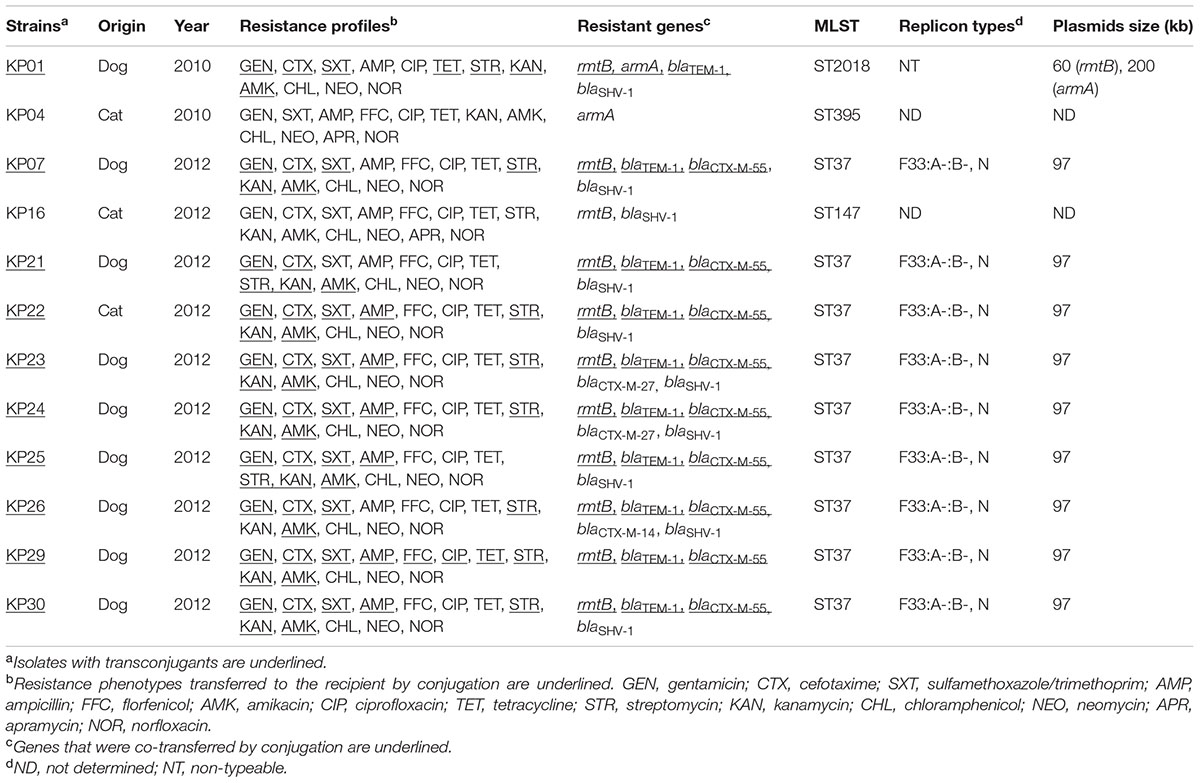
Among 30 K. pneumoniae isolates from the hospital, 12 were resistant to AMK and GEN. However, all K. pneumoniae strains contained the 16S-RMTase genes rmtB (11/30) and/or armA (2/30) and a single isolate carried both genes (1/30). The AMK/GEN-resistant K. pneumoniae isolates showed an MDR phenotype (resistant to three or more classes of antimicrobials). Apart from aminoglycoside antibiotics, there was a very high frequency (>90.0%) of resistance to AMP, CTX, SXT, TET, CIP, NOR, CHL, and FFC. The most frequently observed pattern of MDR was GEN-CTX-SXT-AMP-CIP-TET-STR-KAN-AMK-CHL-NEO-NOR (Table 1).
Characterization of Resistance Genes
We examined the prevalence of ESBL and AME genes in the 16S-RMTase gene-positive isolates. Among these, nine isolates contained blaCTX-M that were distributed as blaCTX-M-55 (6), blaCTX-M-55 and blaCTX-M-27 (2), and blaCTX-M-55 and blaCTX-M-14 (1). Two other ESBLs were also present in these isolates as blaTEM-1 (10/12) and blaSHV -1 (10/12) (Table 1).
PFGE Typing and MLST
PFGE was successfully performed in all twelve 16S-RMTase genes positive K. pneumoniae isolates and were grouped into five clonal patterns designated PFGE types A to E (Figure 1). The predominant PFGE type was type C that accounted for 67% (8/12) of the isolates, suggesting that one small clonal outbreak had occurred. The MLST results revealed that one ST was the most prevalent in this collection: ST37 (9/12). ST2018, ST395, and ST147 each had one isolate (Table 1). The results obtained by MLST were consistent with those obtained by PFGE; eight rmtB-positive K. pneumoniae strains showing similar PFGE patterns were belonged to ST37.
Transfer of the 16S-RMTase Genes and Plasmid Analysis
Plasmids carrying rmtB from 10 isolates were successfully transferred to recipients by conjugation. All the transconjugants were highly resistant to AMK, GEN, and KAN (MICs ≥ 512 μg/mL). PBRT of the plasmid incompatibility groups showed that nine rmtB-bearing plasmids from K. pneumoniae isolates carried two replicons. The two replicons were confirmed to be IncFII in combination with IncN, and the other rmtB-bearing plasmid was non-typeable. All the IncFII plasmids were further determined to be F33:A-:B-.
Nine of the 10 transconjugants carried one approximately 97 kb plasmid. The other one harbored both ∼60 kb (rmtB positive) and ∼200 kb (armA positive) plasmids at the same time. Moreover, rmtB and blaCTX-M-55 genes were located on the same 97 kb plasmid. The RFLP analysis indicated that 7 out of the 10 transconjugants had identical plasmid restriction patterns. The two remaining plasmids shared the same RFLP type (Figure 2).
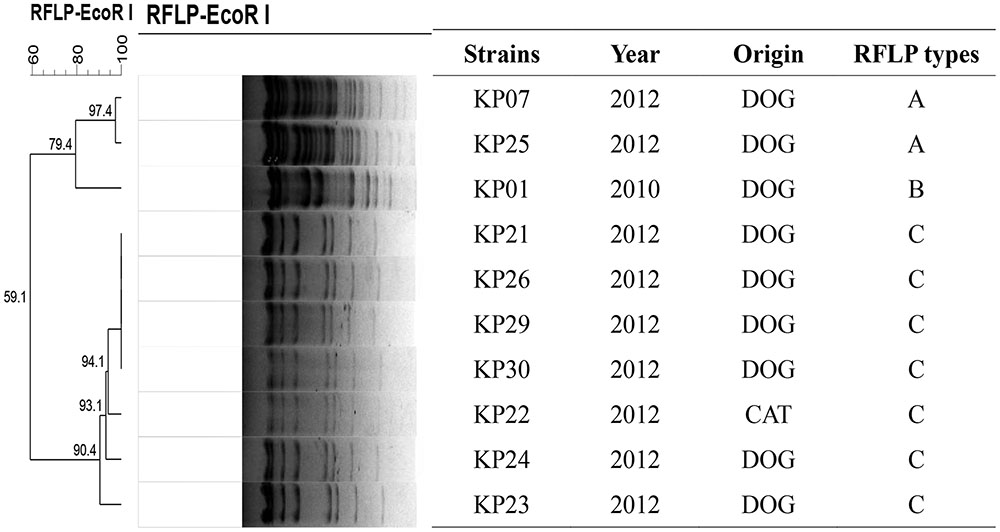
FIGURE 2. EcoRI-RFLP analysis of the 10 transconjugants in the 16S-RMTase-positive K. pneumoniae isolates.
Genetic Profile Analysis
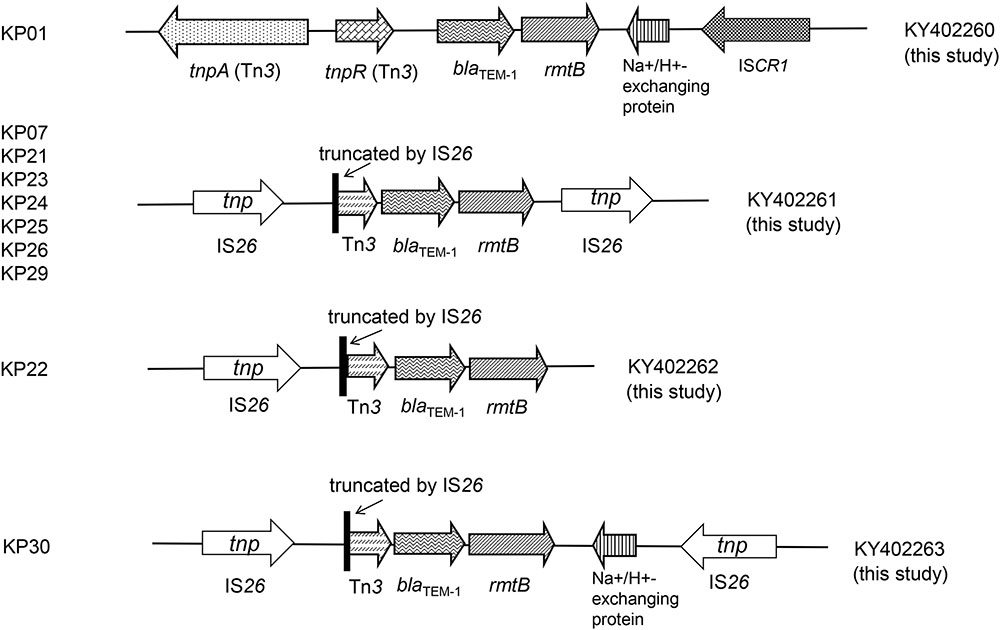
The genetic profiles were successfully obtained in the 10 rmtB-positive transconjugants. We found four new genetic profiles in the 10 rmtB-positive transconjugants and all possessed Tn3 and blaTEM-1 directly upstream of rmtB. Tn3 was truncated by IS26 in 9 of the 10 transconjugants, which were inserted at the same site (3573 of Tn3). The other isolate KP01 harbored the intact tnpA–tnpR genetic structure of Tn3, with a Na+/H+-exchanging protein gene and ISCR1 downstream rmtB. IS26 was also found directly downstream of rmtB in seven plasmids, which was located in the same orientation to the 5′ IS26 elements. Moreover, the Na+/H+-exchanging protein gene located between rmtB and IS26 (in the opposite orientation) occurred in strain KP30 plasmid (Figure 3). The four unique sequences were deposited in GenBank under accession numbers KY402260, KY402261, KY402262, and KY402263.
Discussion
The 16S-RMTase genes confer broad and high-level resistance to most clinically available aminoglycosides (Habeeb et al., 2013). This has become a worldwide concern especially when these genes are combined with other classes of antimicrobial resistance determinants. This has led to the emergence of MDR strains (Habeeb et al., 2013).
In the present study, our results highlighted a high prevalence of 16S-RMTase genes in K. pneumoniae isolates from pets compared with other reports of human infections (Yan et al., 2004; Yu et al., 2009). Similarly, these 16S-RMTase-positive K. pneumoniae isolates were all MDR strains and mostly resistant to β-lactams, aminoglycosides, TETs, amphenicols, sulfonamides, and fluoroquinolones. Fortunately, the isolates we investigated in this study were susceptible to carbapenem antibiotics, which was different from other studies (Zacharczuk et al., 2011b; Li et al., 2012).
The twelve 16S-RMTase-positive K. pneumoniae isolates were successfully typed into five different groups by PFGE. Group C predominated and contained eight isolates indicating that the rmtB-positive isolates were spread by clonal dissemination. MLST analysis indicated four STs: ST37, ST2018, ST395, and ST147. The most prevalent ST in this collection was ST37 containing eight isolates belonging to the same PFGE type C group.
Previous reports had documented that ST11 was the most prevalent 16S-RMTase-positive K. pneumoniae in human isolates. ST101 was the second frequently reported ST type, which often co-harbored KPC-2 (K. pneumoniae carbapenemase) in 16S-RMTase-positive K. pneumoniae (Li et al., 2012; Mezzatesta et al., 2013; Hu et al., 2014; Seiffert et al., 2014). However, the sharing of ST37 between isolates in our study is important because KPC-producing K. pneumoniae ST37 of human origin has been detected only sporadically in China (Yang et al., 2013; Liu et al., 2015). This observation might provide further support in concerns about transmission of resistance between companion animals and humans.
ESBL genes, especially CTX-M genes, frequently coexist and co-transfer with 16S-RMTase genes in Enterobacteriaceae members including K. pneumoniae (Yan et al., 2004; Bogaerts et al., 2007; Zacharczuk et al., 2011a). In this study, PCR and sequence analyses revealed that blaCTX-M-55 and blaTEM-1 were co-transferred with rmtB in 9 and 10 transconjugants of K. pneumoniae strains, respectively. This confirmed an intimate association between ESBLs and 16S-RMTase genes (Deng et al., 2011a; Xia et al., 2016). In addition, PCR-based plasmid replicon typing indicated that the nine transconjugants were in the FII and N incompatibility groups. All IncFII plasmids belonged to the same F33:A-:B- subgroup. This finding is similar with reports of rmtB-positive E. coli from food animals, pets and humans in China (Yu et al., 2010; Deng et al., 2011a,b; Yao et al., 2011; Xia et al., 2016).
This data indicated the horizontal transmission of the prevailing plasmids from variety bacterial species between different hosts. Moreover, our plasmid digestion profiles and S1-PFGE analysis confirmed that the prevalence of the rmtB-positive K. pneumoniae isolates was due to the dissemination of a ∼97 kb plasmid containing blaCTX-M-55.
Previous studies indicated that Tn3, IS26 and ISCR3 were associated with the transmission of rmtB (Doi and Arakawa, 2007; Deng et al., 2011b). In this study, the rmtB gene showed highly relevant relationships with IS26 and Tn3. The presence of a truncated Tn3 inserted via IS26 upstream of rmtB, and IS26 inserting in the same direction located downstream of rmtB, was the most prevalent structure in seven transconjugants. Within this group, five plasmids of the corresponding transconjugants shared similar EcoRI-RFLP patterns (Figures 2, 3). This indicated a plasmid with this particular genetic profile was the origin of rmtB in the K. pneumoniae isolates from this group of pets.
In conclusion, rmtB was the most prevalent 16S-RMTase gene of K. pneumoniae that originated in pets. The dissemination of rmtB-producing K. pneumoniae isolates in this study appeared to be clonal spread and horizontal transfer. ST37 K. pneumoniae and F33:A-:B- plasmids sized ∼97 kb with the same genetic profile mediated by IS26 transmission both contributed to the dissemination of rmtB. In this study, we investigated only a single veterinary hospital. An organized surveillance effort is required to better understand the scope of this problem and identify control measures among hospitals.
Author Contributions
Y-HL and X-PL designed and organized the study. JX and L-XF did the research. KC, G-HX, and X-RW did the assisted help. JS analyzed the data and wrote the paper.
Funding
This work was supported by the Program for Changjiang Scholars and Innovative Research Team in University of Ministry of Education of China (Grant No. IRT13063), the Special Fund for Agro-scientific Research in the Public Interest (Grant No. 201203040), and the Natural Science Foundation of Guangdong Province (Grant No. S2012030006590).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.00529/full#supplementary-material
Footnotes
References
Bartoloni, A., Sennati, S., Di Maggio, T., Mantella, A., Riccobono, E., Strohmeyer, M., et al. (2016). Antimicrobial susceptibility and emerging resistance determinants (blaCTX-M, rmtB, fosA3) in clinical isolates from urinary tract infections in the Bolivian Chaco. Int. J. Infect. Dis. 43, 1–6. doi: 10.1016/j.ijid.2015.12.008
Belbel, Z., Chettibi, H., Dekhil, M., Ladjama, A., Nedjai, S., and Rolain, J. M. (2014). Outbreak of an armA methyltransferase-producing ST39 Klebsiella pneumoniae clone in a pediatric Algerian Hospital. Microb. Drug Resist. 20, 310–315. doi: 10.1089/mdr.2013.0193
Bogaerts, P., Galimand, M., Bauraing, C., Deplano, A., Vanhoof, R., De Mendonca, R., et al. (2007). Emergence of ArmA and RmtB aminoglycoside resistance 16S rRNA methylases in Belgium. J. Antimicrob. Chemother. 59, 459–464. doi: 10.1093/jac/dkl527
Clinical and Laboratory Standards Institute (2015). Performance Standards for Antimicrobial Susceptibility Testing; Twenty-fifth Informational Supplement (M100-S25). Wayne, PA: Clinical and Laboratory Standards Institute.
Deng, Y., He, L., Chen, S., Zheng, H., Zeng, Z., Liu, Y., et al. (2011a). F33:A-:B- and F2:A-:B- plasmids mediate dissemination of rmtB-blaCTX-M-9 group genes and rmtB-qepA in Enterobacteriaceae isolates from pets in China. Antimicrob. Agents Chemother. 55, 4926–4929. doi: 10.1128/AAC.00133-11
Deng, Y., Zeng, Z., Chen, S., He, L., Liu, Y., Wu, C., et al. (2011b). Dissemination of IncFII plasmids carrying rmtB and qepA in Escherichia coli from pigs, farm workers and the environment. Clin. Microbiol. Infect. 17, 1740–1745. doi: 10.1111/j.1469-0691.2011.03472.x
Doi, Y., and Arakawa, Y. (2007). 16S ribosomal RNA methylation: emerging resistance mechanism against aminoglycosides. Clin. Infect. Dis. 45, 88–94. doi: 10.1086/518605
Fang, L. X., Sun, J., Li, L., Deng, H., Huang, T., Yang, Q. E., et al. (2015). Dissemination of the chromosomally encoded CMY-2 cephalosporinase gene in Escherichia coli isolated from animals. Int. J. Antimicrob. Agents 46, 209–213. doi: 10.1016/j.ijantimicag.2015.04.003
Grevskott, D. H., Svanevik, C. S., Sunde, M., Wester, A. L., and Lunestad, B. T. (2017). Marine bivalve mollusks as possible indicators of multidrug-resistant Escherichia coli and other species of the Enterobacteriaceae family. Front. Microbiol. 8:24. doi: 10.3389/fmicb.2017.00024
Habeeb, M. A., Haque, A., Nematzadeh, S., Iversen, A., and Giske, C. G. (2013). High prevalence of 16S rRNA methylase RmtB among CTX-M extended-spectrum beta-lactamase-producing Klebsiella pneumoniae from Islamabad, Pakistan. Int. J. Antimicrob. Agents 41, 524–526. doi: 10.1016/j.ijantimicag.2013.02.017
Hidalgo, L., Gutierrez, B., Ovejero, C. M., Carrilero, L., Matrat, S., Saba, C. K., et al. (2013a). Klebsiella pneumoniae sequence type 11 from companion animals bearing ArmA methyltransferase, DHA-1 beta-lactamase, and QnrB4. Antimicrob. Agents Chemother. 57, 4532–4534. doi: 10.1128/AAC.00491-13
Hidalgo, L., Hopkins, K. L., Gutierrez, B., Ovejero, C. M., Shukla, S., Douthwaite, S., et al. (2013b). Association of the novel aminoglycoside resistance determinant RmtF with NDM carbapenemase in Enterobacteriaceae isolated in India and the UK. J. Antimicrob. Chemother. 68, 1543–1550. doi: 10.1093/jac/dkt078
Hu, F., Munoz-Price, L. S., Depascale, D., Rivera, J. I., and Doi, Y. (2014). Klebsiella pneumoniae sequence type 11 isolate producing RmtG 16S rRNA methyltransferase from a patient in Miami, Florida. Antimicrob. Agents Chemother. 58, 4980–4981. doi: 10.1128/AAC.02632-14
Lee, C. H., Liu, J. W., Li, C. C., Chien, C. C., Tang, Y. F., and Su, L. H. (2011). Spread of ISCR1 elements containing blaDHA-(1) and multiple antimicrobial resistance genes leading to increase of flomoxef resistance in extended-spectrum-beta-lactamase-producing Klebsiella pneumoniae. Antimicrob. Agents Chemother. 55, 4058–4063. doi: 10.1128/AAC.00259-11
Li, J. J., Sheng, Z. K., Deng, M., Bi, S., Hu, F. S., Miao, H. F., et al. (2012). Epidemic of Klebsiella pneumoniae ST11 clone coproducing KPC-2 and 16S rRNA methylase RmtB in a Chinese university hospital. BMC Infect. Dis. 12:373. doi: 10.1186/1471-2334-12-373
Liao, X. P., Xia, J., Yang, L., Li, L., Sun, J., Liu, Y. H., et al. (2015). Characterization of CTX-M-14-producing Escherichia coli from food-producing animals. Front. Microbiol. 6:1136. doi: 10.3389/fmicb.2015.01136
Liu, Y., Wan, L. G., Deng, Q., Cao, X. W., Yu, Y., and Xu, Q. F. (2015). First description of NDM-1-, KPC-2-, VIM-2- and IMP-4-producing Klebsiella pneumoniae strains in a single Chinese teaching hospital. Epidemiol. Infect. 143, 376–384. doi: 10.1017/S0950268814000995
Lloyd, D. H. (2007). Reservoirs of antimicrobial resistance in pet animals. Clin. Infect. Dis. 45(Suppl. 2), S148–S152. doi: 10.1086/519254
Luo, Y., Yang, J., Zhang, Y., Ye, L., Wang, L., and Guo, L. (2011). Prevalence of beta-lactamases and 16S rRNA methylase genes amongst clinical Klebsiella pneumoniae isolates carrying plasmid-mediated quinolone resistance determinants. Int. J. Antimicrob. Agents 37, 352–355. doi: 10.1016/j.ijantimicag.2010.12.018
Mezzatesta, M. L., Gona, F., Caio, C., Adembri, C., Dell’utri, P., Santagati, M., et al. (2013). Emergence of an extensively drug-resistant ArmA- and KPC-2-producing ST101 Klebsiella pneumoniae clone in Italy. J. Antimicrob. Chemother. 68, 1932–1934. doi: 10.1093/jac/dkt116
O’Hara, J. A., Mcgann, P., Snesrud, E. C., Clifford, R. J., Waterman, P. E., Lesho, E. P., et al. (2013). Novel 16S rRNA methyltransferase RmtH produced by Klebsiella pneumoniae associated with war-related trauma. Antimicrob. Agents Chemother. 57, 2413–2416. doi: 10.1128/AAC.00266-13
Seiffert, S. N., Marschall, J., Perreten, V., Carattoli, A., Furrer, H., and Endimiani, A. (2014). Emergence of Klebsiella pneumoniae co-producing NDM-1, OXA-48, CTX-M-15, CMY-16, QnrA and ArmA in Switzerland. Int. J. Antimicrob. Agents 44, 260–262. doi: 10.1016/j.ijantimicag.2014.05.008
Wasyl, D., Kern-Zdanowicz, I., Domanska-Blicharz, K., Zajac, M., and Hoszowski, A. (2015). High-level fluoroquinolone resistant Salmonella enterica serovar Kentucky ST198 epidemic clone with IncA/C conjugative plasmid carrying bla(CTX-M-25) gene. Vet. Microbiol. 175, 85–91. doi: 10.1016/j.vetmic.2014.10.014
Xia, J., Sun, J., Cheng, K., Li, L., Fang, L. X., Zou, M. T., et al. (2016). Persistent spread of the rmtB 16S rRNA methyltransferase gene among Escherichia coli isolates from diseased food-producing animals in China. Vet. Microbiol. 188, 41–46. doi: 10.1016/j.vetmic.2016.03.018
Xia, J., Sun, J., Li, L., Fang, L. X., Deng, H., Yang, R. S., et al. (2015). First report of the IncI1/ST898 conjugative plasmid carrying rmtE2 16S rRNA methyltransferase gene in Escherichia coli. Antimicrob. Agents Chemother. 59, 7921–7922. doi: 10.1128/AAC.01235-15
Yan, J. J., Wu, J. J., Ko, W. C., Tsai, S. H., Chuang, C. L., Wu, H. M., et al. (2004). Plasmid-mediated 16S rRNA methylases conferring high-level aminoglycoside resistance in Escherichia coli and Klebsiella pneumoniae isolates from two Taiwanese hospitals. J. Antimicrob. Chemother. 54, 1007–1012. doi: 10.1093/jac/dkh455
Yang, J., Ye, L., Guo, L., Zhao, Q., Chen, R., Luo, Y., et al. (2013). A nosocomial outbreak of KPC-2-producing Klebsiella pneumoniae in a Chinese hospital: dissemination of ST11 and emergence of ST37, ST392 and ST395. Clin. Microbiol. Infect. 19, E509–E515. doi: 10.1111/1469-0691.12275
Yang, J., Ye, L., Wang, W., Luo, Y., Zhang, Y., and Han, L. (2011). Diverse prevalence of 16S rRNA methylase genes armA and rmtB amongst clinical multidrug-resistant Escherichia coli and Klebsiella pneumoniae isolates. Int. J. Antimicrob. Agents 38, 348–351. doi: 10.1016/j.ijantimicag.2011.04.021
Yang, Q. E., Sun, J., Li, L., Deng, H., Liu, B. T., Fang, L. X., et al. (2015). IncF plasmid diversity in multi-drug resistant Escherichia coli strains from animals in China. Front. Microbiol. 6:964. doi: 10.3389/fmicb.2015.00964
Yang, Y., Zhang, A., Lei, C., Wang, H., Guan, Z., Xu, C., et al. (2015). Characteristics of plasmids coharboring 16S rRNA methylases, CTX-M, and virulence factors in Escherichia coli and Klebsiella pneumoniae isolates from chickens in China. Foodborne Pathog. Dis. 12, 873–880. doi: 10.1089/fpd.2015.2025
Yao, Q., Zeng, Z., Hou, J., Deng, Y., He, L., Tian, W., et al. (2011). Dissemination of the rmtB gene carried on IncF and IncN plasmids among Enterobacteriaceae in a pig farm and its environment. J. Antimicrob. Chemother. 66, 2475–2479. doi: 10.1093/jac/dkr328
Yu, F., Wang, L., Pan, J., Yao, D., Chen, C., Zhu, T., et al. (2009). Prevalence of 16S rRNA methylase genes in Klebsiella pneumoniae isolates from a Chinese teaching hospital: coexistence of rmtB and armA genes in the same isolate. Diagn. Microbiol. Infect. Dis. 64, 57–63. doi: 10.1016/j.diagmicrobio.2009.01.020
Yu, F. Y., Yao, D., Pan, J. Y., Chen, C., Qin, Z. Q., Parsons, C., et al. (2010). High prevalence of plasmid-mediated 16S rRNA methylase gene rmtB among Escherichia coli clinical isolates from a Chinese teaching hospital. BMC Infect. Dis. 10:184. doi: 10.1186/1471-2334-10-184
Zacharczuk, K., Piekarska, K., Szych, J., Jagielski, M., Hidalgo, L., San Millan, A., et al. (2011a). Plasmid-borne 16S rRNA methylase ArmA in aminoglycoside-resistant Klebsiella pneumoniae in Poland. J. Med. Microbiol. 60, 1306–1311. doi: 10.1099/jmm.0.024026-0
Keywords: Klebsiella pneumoniae, 16S rRNA methyltransferases, companion animals, MLST
Citation: Xia J, Fang L-X, Cheng K, Xu G-H, Wang X -R, Liao X-P, Liu Y-H and Sun J (2017) Clonal Spread of 16S rRNA Methyltransferase-Producing Klebsiella pneumoniae ST37 with High Prevalence of ESBLs from Companion Animals in China. Front. Microbiol. 8:529. doi: 10.3389/fmicb.2017.00529
Received: 04 January 2017; Accepted: 14 March 2017;
Published: 29 March 2017.
Edited by:
Benoit Doublet, National Institute for Agricultural Research, FranceCopyright © 2017 Xia, Fang, Cheng, Xu, Wang, Liao, Liu and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jian Sun, jiansun@scau.edu.cn
 Jing Xia
Jing Xia Liang-Xing Fang1,2
Liang-Xing Fang1,2 Xiao-Ping Liao
Xiao-Ping Liao Jian Sun
Jian Sun