- 1Agricultural Bioinformatics Key Laboratory of Hubei Province, College of Informatics, Huazhong Agricultural University, Wuhan, China
- 2State Key Laboratory of Agricultural Microbiology, College of Life Science and Technology, Huazhong Agricultural University, Wuhan, China
As an industrial bacterium, Bacillus licheniformis DW2 produces bacitracin which is an important antibiotic for many pathogenic microorganisms. Our previous study showed AbrB-knockout could significantly increase the production of bacitracin. Accordingly, it was meaningful to understand its genome features, expression differences between wild and AbrB-knockout (ΔAbrB) strains, and the regulation of bacitracin biosynthesis. Here, we sequenced, de novo assembled and annotated its genome, and also sequenced the transcriptomes in three growth phases. The genome of DW2 contained a DNA molecule of 4,468,952 bp with 45.93% GC content and 4,717 protein coding genes. The transcriptome reads were mapped to the assembled genome, and obtained 4,102∼4,536 expressed genes from different samples. We investigated transcription changes in B. licheniformis DW2 and showed that ΔAbrB caused hundreds of genes up-regulation and down-regulation in different growth phases. We identified a complete bacitracin synthetase gene cluster, including the location and length of bacABC, bcrABC, and bacT, as well as their arrangement. The gene cluster bcrABC were significantly up-regulated in ΔAbrB strain, which supported the hypothesis in previous study of bcrABC transporting bacitracin out of the cell to avoid self-intoxication, and was consistent with the previous experimental result that ΔAbrB could yield more bacitracin. This study provided a high quality reference genome for B. licheniformis DW2, and the transcriptome data depicted global alterations across two strains and three phases offered an understanding of AbrB regulation and bacitracin biosynthesis through gene expression.
Introduction
Bacillus licheniformis (B. licheniformis) is a Gram-positive bacterium that is widely used in multiple fields: in agriculture as a probiotic and microbial fertilizer (Pötter et al., 2001), and in biotechnology industry for the production of enzymes, acetoin and poly-γ-glutamic acid (γ-PGA) (Veith et al., 2004; Liu et al., 2011; Konglom et al., 2012). This facultative anaerobic organism can also produce a variety of antibiotics such as bacitracin, which is an important peptide antibiotic for many pathogenic microorganisms secreted by certain strains of B. licheniformis and B. subtilis (Johnson et al., 1945; Mcinerney et al., 1990). The branched cyclic dodecyl peptide bacitracin is the most active against other Gram-positive and certain Gram-negative microorganisms, which is synthesized by a large non-ribosomal multi-enzyme complex bacABC (Konz et al., 1997). The AbrB gene of B. licheniformis and B. subtilis is known as an important global transcription repressor, but also act as an activator for some genes by binding to promoters (Kim et al., 2003). AbrB has a wide range of regulatory functions, including cell wall and membrane synthesis, bacterial chemotaxis, antibiotic synthesis, amino acid synthesis and transport, protein modification and so on (Qian et al., 2002; Strauch et al., 2007). AbrB directly regulates more than 100 genes and indirectly influence hundreds genes. The evidence suggests that expression level of AbrB is inhibited by another regulatory gene Spo0A-P, leading to an opposite tendency of AbrB gene expression compares with that of Spo0A-P, i.e., high expression during the logarithmic phase and low expression during stationary phase (Strauch et al., 1990; Shafikhani and Leighton, 2004; Banse et al., 2008; Chumsakul et al., 2011).
Since the project of B. subtilis genome was completed as the first gram-positive bacterium (Kunst et al., 1997), more and more Bacillus strains have been sequenced including B. licheniformis ATCC14580, 9945A, WX-02 and DSM13; B. subtilis CGMCC 12426, BSD-2, and so on (Veith et al., 2004; Guo et al., 2015; Li et al., 2015; Liu et al., 2016). In contrast, some other studies are limited to single or several proteins and metabolites, for example, the production of increased isopentenyl pyrophosphate (IPP) (Cain et al., 1993; Chalker et al., 2000), the secretion of exopolysaccharides (Pollock et al., 1994) and bcrABC protein transporting bacitracin in B. subtilis (Podlesek et al., 1995, 2000). Although these studies provide great advances, there has never been a comprehensive research focused on the effects of AbrB-knockout in term of whole genome and transcriptome, especially the effect on the bacitracin gene cluster.
Our previous study showed that AbrB-knockout could significantly increase the production of bacitracin, and the yields were 17.5% higher than that of the wild-type strain (Wang et al., 2017). As an industrial B. licheniformis strain, DW2 can produce an extra yield using the improvement way of AbrB-knockout in industrial application. In the present study, we sequenced, annotated the genome and transcriptome of an important industrial B. licheniformis strain DW2, and obtained the following results. (1) The genome of B. licheniformis DW2 is 4,468,952 bp with 45.93% GC content and 4,717 predicted coding sequences (CDSs). (2) B. licheniformis DW2 has good collinearity with other B. licheniformis strains, and a unique ∼100 kb genomic sequences. (3) N-6-methylated adenines (6mA) and 4-methylated cytosines (4mC) methylation of B. licheniformis DW2 was analyzed, and two 6mA motifs were identified. (4) There are 369∼1,517 differentially expressed genes (DEGs) between the transcriptome of wild and ΔAbrB strains in logarithmic growth phase, transitional phase and stationary growth phase. (5) We identified the location and length of bacitracin synthetase gene cluster in B. licheniformis DW2. In this gene cluster, the transporter genes bcrABC were significantly highly expressed in ΔAbrB strain, which supported the assumption that bcrABC genes could pump bacitracin out to avoid self-intoxication. The study produced a reference genome of B. licheniformis, also enabled us to highlight global changes in gene expression and provided theory and data support for industrial process of bacitracin production.
Materials and Methods
Genomic DNA Preparation
The DW2 strain used in this study was provided by China Center for Type Culture Collection (CCTCC), whose number was CCTCC M2011344. B. licheniformis DW2 was grown on Luria Bertani (LB) agar plates or in LB broth at 37°C, supplemented with antibiotics (tetracycline, 20 mg/mL; ampicillin, 50 mg/mL; kanamycin, 20 mg/mL) when necessary. The seed culture (1 mL) was inoculated into a 250 mL Erlenmeyer flask containing 20 mL bacitracin fermentation medium [6% soybean meal, 4.5% starch, 0.1% (NH4)2SO4, 0.6% CaCO3, pH 6.5∼7.0], followed by 40 h incubation on a rotary shaker (180 rpm) at 37°C. Genomic DNA was extracted using a standard SDS-Phenol procedure (Sambrook et al., 1982) according to the manufacturer’s instructions, combined with phenol-chloroform extraction and RNase A treatment.
PacBio SMRT Sequencing
We determined the complete genome sequence of B. licheniformis DW2 using PacBio single molecule real-time (SMRT) technology. We prepared a 3 to 20 kb genomic DNA library suitable for P6/C4 chemistry. Using two SMRT cell on PacBio RSII sequencing platform, 116,486 polymerase reads with mean read length of 4,268 bp were obtained. The polymerase reads were assembled de novo with Hierarchical Genome Assembly Process 3 (HGAP3) within the SMRT Analysis version 2.3.0 software (Chin et al., 2013). Subsequently, the best assembly was selected, and Minimus 2 was used for trimming the circular contig (Sommer et al., 2007). The replication origin was determined by aligning the DW2 genomic sequence with B. licheniformis DSM13.
For identification of methylated bases and modification motifs, the “RS_Modification_and_Motif_Analysis.1” protocol in SMRT Portal under default parameter settings was used, with the assembled genome. Putative restriction modification systems have been identified using the Restriction-ModificationFinder-1.0 server1 based on the Restriction Enzyme database (REBASE) (Roberts et al., 2015).
The Annotation of B. licheniformis DW2 Genome
After the complete sequence was obtained, the genome was annotated using the rapid annotation tool RAST (Ogata et al., 1999; Aziz et al., 2008). In addition, the unigenes were annotated by the public protein databases of Kyoto Encyclopedia of Genes and Genomes (KEGG) (Galperin et al., 2015), Cluster of Orthologous Groups of Proteins (COG) (Gene Ontology Consortium, 2015), Swiss-Prot protein2, NCBI non-redundant (NR3), and Gene Ontology (GO) (Lagesen et al., 2007) using BLASTP to get corresponding functional annotation information. As each sequence had many alignments, we retain the optimal comparison results as the annotation of the gene to ensure its accuracy. The predictions of rRNA and tRNA were performed using RNAmmer (Lowe and Eddy, 1997) and tRNAscan-SE (Zhang et al., 2000), respectively. Insertion sequence (IS) elements were predicted with IS Finder (Benson, 1999).
Whole-Genome Alignment and Identification of Operon Structures
We constructed and visualized the multiple genome alignment of four complete genomes of B. licheniformis strains DW2, ATCC14580, 9945A and WX-02 using Mauve v2.4.0 (Darling et al., 2010). Pairwise collinear comparisons of the four genome sequences were performed using Mummer3 (Kurtz et al., 2004). Operon of B. licheniformis DW2 was identified using DOOR 2.0 algorithm for prokaryotic operon analysis (Mao et al., 2014).
RNA Isolation and Library Preparation for ssRNA-seq
RNA samples were isolated at three time points for two bacterial samples (wild strain and ΔAbrB strain): 14 h (logarithmic growth phase), 22 h (transitional phase) and 25 h (stationary phase). Cells were collected by centrifugation at 12,000 rpm for 5 min, and then transferred to a 10-ml centrifuge tube after grinding in liquid nitrogen. Cells were then lysed in 1 mL of TRIzol for 30–60 s. To the lysate, 200 μL of chloroform was added, and the sample was then mixed by inversion and incubated at room temperature for 15 min. The sample was then centrifuged at 12,000 rpm for 15 min at 4°C, and supernatant was precipitated with an equal volume of isopropanol at room temperature for 10 min. After centrifugation, the supernatant was discarded, and the pellet was air dried and dissolved in 20–40 μL of RNase-free water. Total RNA was treated with RNase-free DNase I for 30 min at 37°C to remove genomic DNA. RNA concentration was measured using Qubit® RNA Assay Kit in Qubit® 2.0 Fluorometer (Life Technologies, Carlsbad, CA, United States). At the same time, RNA integrity was assessed using the RNA Nano 6000 Assay Kit of the Bioanalyzer 2100 system (Agilent Technologies, Santa Clara, CA, United States). Each sample had three replicates, and all biological replicates were processed in separate batches from each other.
Libraries for ssRNA sequencing (Novogene Experimental Department) were constructed at three time points (14, 22, and 25 h). A total amount of 3 μg RNA per sample was used as input material for the RNA sample preparations. Sequencing libraries were generated using NEBNext® UltraTM Directional RNA Library Prep Kit for Illumina® (NEB, United States) following manufacturer’s recommendation and index codes were added to attribute sequences to each sample. Briefly, (epicentre Ribo-ZeroTM) fragmentation was carried out using divalent cations under elevated temperature in NEBNext First Strand Synthesis Reaction Buffer (5X). First strand cDNA was synthesized using random hexamer primer and M-MuLV Reverse Transcriptase (RNaseH-). Second strand cDNA synthesis was subsequently performed using DNA Polymerase I and RNase H. In the reaction buffer, dNTPs with dTTP were replaced by dUTP. Remaining overhangs were converted into blunt ends via exonuclease/polymerase activities. After adenylation of 3′ ends of DNA fragments, NEBNext Adaptor with hairpin loop structure were ligated to prepare for hybridization. In order to select cDNA fragments of preferentially 150∼200 bp in length, the library fragments were purified with AMPure XP system (Beckman Coulter, Beverly, MA, United States). Then 3 μl USER Enzyme (NEB, United States) was used with size-selected, adaptor-ligated cDNA at 37°C for 15 min followed by 5 min at 95°C before PCR. Then PCR was performed with Phusion High-Fidelity DNA polymerase, Universal PCR primers and Index (X) Primer. At last, products were purified (AMPure XP system) and library quality was assessed on the Agilent Bioanalyzer 2100 system.
Mapping Reads to B. licheniformis DW2 Genome and Analyzing Differentially Expressed Genes
All the raw reads were initially processed to obtain clean reads by the following steps: (1) reads with adaptor were discarded; (2) reads with ambiguous bases (undetermined bases, N) larger than 10% were removed; (3) low-quality reads that contained more than 50% Q-value < 5 bases were discarded. After filtering, all clean reads were aligned to the assembled genome using Bowtie2 (Langmead and Salzberg, 2012), and reads with ambiguous alignments or more than three mismatches were discarded. Only uniquely mapped reads were used for further analysis. The per-base–format coverage depth and read counts were calculated using BEDTools (Quinlan and Hall, 2010). Because different samples may have different total read counts, sequencing depth, and biases, the normalized transcription level of genes was expressed in reads per kilobase of ORF per million mapped reads (RPKM) (Mortazavi et al., 2008). The transcriptome raw reads have been deposited in NCBI Sequence Read Archive database under accession number SRP045205. In addition, we generated a distance matrix for expression data based on RPKM values by using Pearson correlation as dissimilarity metric (El-Sharnouby et al., 2013). Clustering was performed and deprograms were generated using the cluster package in R.
Differentially expressed genes between wild strain and ΔAbrB strain were identified using the DESeq R package with the MARS (MA-plot-based method with Random Sampling) model (Anders and Huber, 2010). The DEGs were screened with the false discovery rate (FDR) threshold of ≤0.01 and an absolute value of fold change (FC) ≥2.
Results and Discussion
Genomic Features and Comparative Genomics of B. licheniformis DW2
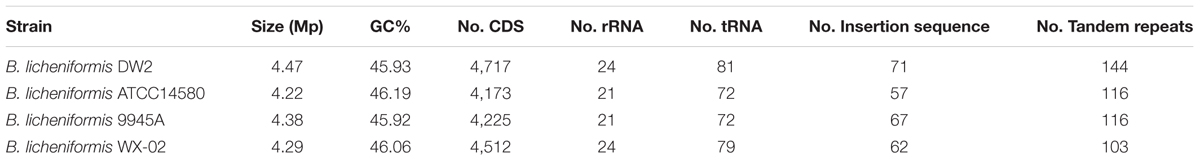
Bacillus licheniformis DW2 genome has one circular DNA with 4,468,952 bp length and 45.93% GC content. The DNA molecule (Figure 1) contained 4,717 predicted CDSs accounting for ∼87% of the genome. Among the predicted CDSs, 3,896 were assigned biological functions or putative functions and 821 could be annotated as hypothetical protein. In addition, 24 rRNA genes and 81 tRNA-coding genes were identified, and 71 IS elements were found. Additional information about the genome statistics is shown in Table 1.
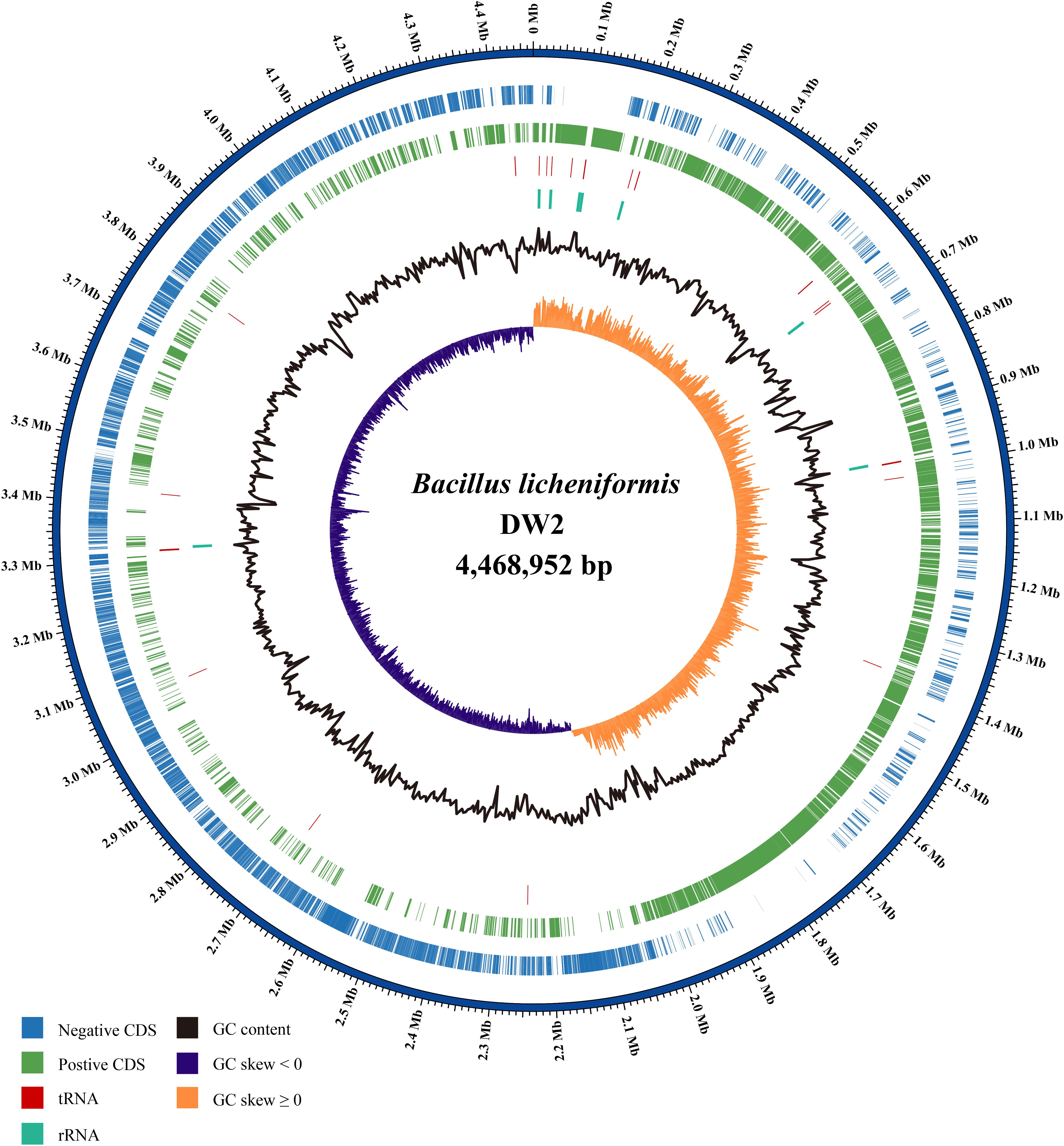
FIGURE 1. The circular plot of B. licheniformis DW2 chromosome. Circles are numbered from 1 (outermost) to 7 (innermost). Circle 1 represents the whole chromosome; Circles 2 and 3 show the locations of predicted CDSs on the positive and negative strands, respectively; Circle 4, tRNA genes; Circle 5, rRNA genes; Circle 6, %G + C; Circle 7, GC skew ((G - C)/(G + C)).
To obtain further insights into B. licheniformis DW2 genome, we performed comparative genomics analysis with other three available B. licheniformis genomes: DW2, ATCC14580, 9945A and WX-02 (Table 2). B. licheniformis DW2 had the largest genome size and number of predicted CDSs, IS and tandem repeat sequence. However, the number of rRNAs and tRNAs was consistent with the others and the number of rRNAs was almost same. The multi-genome alignments showed the four genomes shared homologous blocks in order (Figure 2A). We found that B. licheniformis DW2 genome contained a specific fragment at 3.3∼3.4 Mb with 113 CDSs, most of them were hypothetical proteins and phage-associated proteins (such as phage-like protein, phage protein, phage capsid, and scaffold). Considering that only B. licheniformis DW2 can highly yield peptide antibiotics in the above four strains, we hypothesized that this region might be associated with the yield of peptide antibiotics and inhibition of pathogens. The B. licheniformis DW2 genome was collinear with the other three B. licheniformis strains (Supplementary Figure S1), and it has the best consistency with B. licheniformis 9945A.
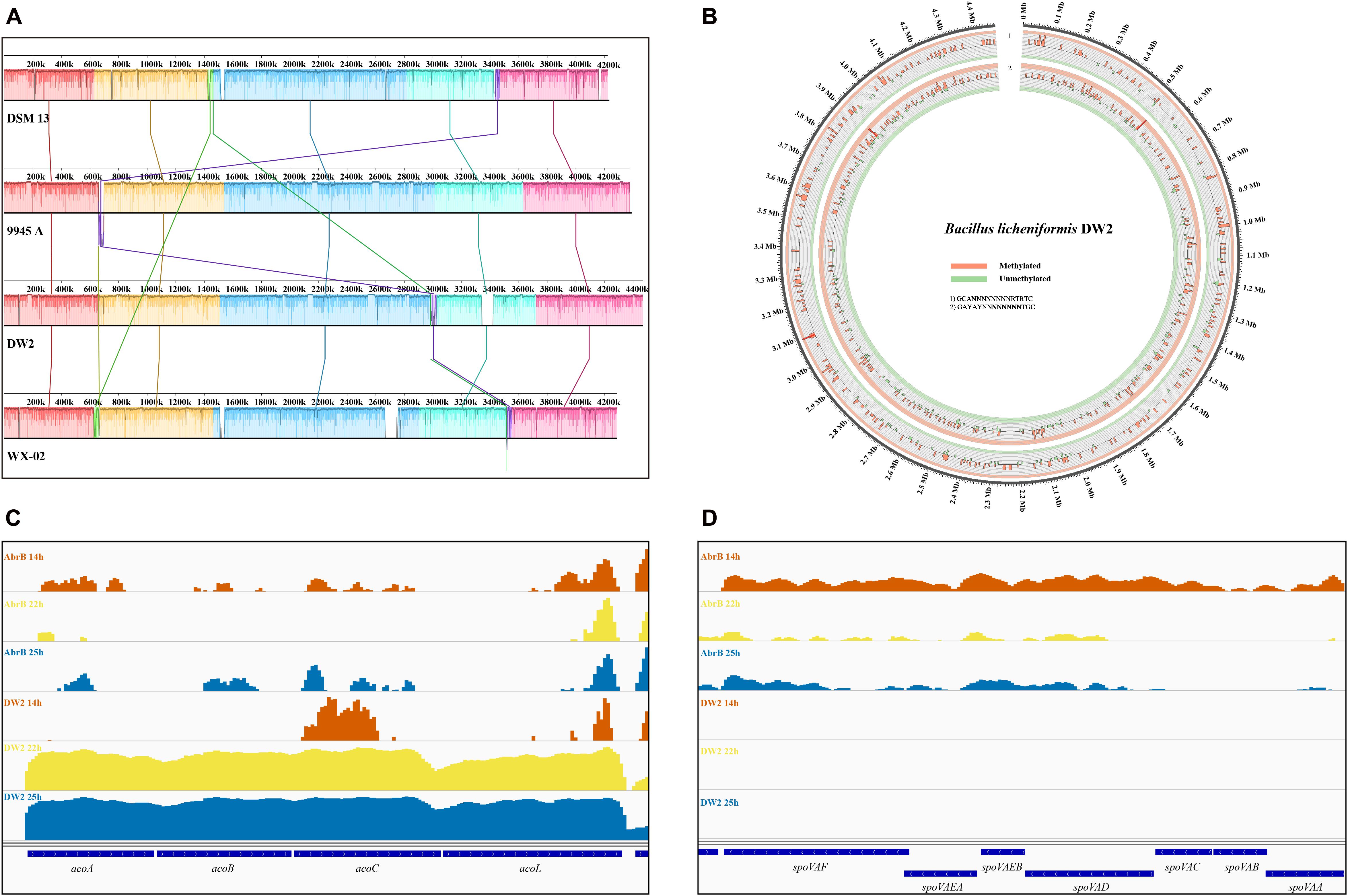
FIGURE 2. Comparative genomics of four B. licheniformis strains and other genomic features of B. licheniformis DW2. (A) Multiple genomic alignments of four B. licheniformis strains. (B) Distribution of 6mA motifs in B. licheniformis DW2 genome. (C) The operon acoABCL associated with acetoin catabolism in B. licheniformis DW2. (D) The stage V sporulation protein in B. licheniformis DW2.
DNA Methylation Features in B. licheniformis DW2
DNA methylation is an essential epigenetic modification that can change the activity of a DNA segment without changing the sequence. It typically represses gene transcription if it locates in a gene promoter (Laird, 2010). In bacteria DNA methylation often acts as a cellular defense system against phage infection that confers a selective advantage to the host bacterium (Sitaraman, 2016). If a foreign DNA is introduced into the cell, it will be degraded by sequence-specific restriction enzymes while the methylated DNA of the bacteria is not recognized by the restriction enzymes. Therefore, DNA methylation can be viewed as a primitive immune system to protect bacteria from bacteriophage attack (Sitaraman, 2016). DNA methylation transfers methyl groups from adenosylmethionine to adenine or cytosine by DNA methyltransferase to form 6-methyladenine (6mA), 4-methyl cell (4-Methylcytosine, 4mC) and 5-Methylcytosine (5mC), of which 6mA and 4mC are mainly found in prokaryotes (Kumar et al., 1994; Sánchezromero et al., 2015). PacBio SMRT technology has been used to simultaneously detect DNA methylation modification during single-molecule genome sequencing (Flusberg et al., 2010). In order to characterize the methylation of B. licheniformis DW2, using the SMRT Analysis Modification and Motif detection, we identified 2,939 6 mA, 2,784 4 mC and 88,319 unspecific “modified bases” where the type of modification was not recognized by the software.
We then detected candidate methylation motifs using a sliding window of 5 kb, and identified two dominant motifs (GCANNNNNNNNRTRTC and GAYAYNNNNNNNNTGC), both of them were recognized by N-6 adenine-specific methyltransferases. Figure 2B shows the distribution of two motifs in B. licheniformis DW2 genome, approximately 40% of 6 mA bases are clustered into the two motifs. However, there is no consensus motif for 4mC-methylated bases or other unspecific modified bases. We aligned the two motifs to a comprehensive restriction-modification (RM) system database REBASE (Roberts et al., 2015) to make sure whether the two motifs have been identified in other species, and found that both motifs could not match existing recognition sequences of the restriction systems.
Prediction and Analysis of Operons in B. licheniformis DW2
We predicted 1021 operons based on RNA-seq data of B. licheniformis DW2 using software DOOR. Among these operons, 446 (43.7%) of them are composed of two genes and 19 of them are composed of more than 10 genes. Figure 2C shows the operon acoABCL associated with acetoin catabolism, where acoR is a transcriptional regulator, acoL is dihydrolipoamide dehydrogenase, and acoABC is a gene related to acetoin dehydrogenase (Ali et al., 2001). Many microorganisms can convert carbohydrates into acetone during glycolysis to avoid over-acidification. As shown in the figure, these genes had high expression at 22 and 25 h in DW2 strain, while they were almost not expressed at 14 h in DW2 and all the three time points in ΔAbrB strain. These results indicate that AbrB may have a great effect on the expression of this operon, and this effect is time-specific, which has a significant effect in stationary phase and no obvious effect in logarithmic phase.
Figure 2D shows the stage V sporulation protein. The spoVA operon of B. subtilis is expressed only in the developing spore during sporulation and at least five SpoVA proteins, SpoVAA, -B, -C, -D, and -Eb, are necessary for normal B. subtilis spore formation (Perez-Valdespino et al., 2014). Many genes of ΔAbrB strain showed much higher expression than DW2 strain, which fitted the basic fact that AbrB was a global regulatory gene and had inhibition effect on many genes. The expression level significantly increased when AbrB gene was knocked out. The ΔAbrB genes at 14 h were highly expressed compared with that at 22 and 25 h. In general, B. licheniformis would not sporulate during logarithmic growth phase. As for spoVAC and spoVAB, we observed that the expression at 22 and 25 h were significantly lower than those at 14 h, which indicated they were activated during logarithmic growth phase. These results suggest that knockout of AbrB is favorable for sporulation. Then the knockout of AbrB may be able to directly relieve the inhibition of AbrB on other genes related to spore synthesis, thereby increasing the transcription level of spore-related genes and promote the synthesis of spores.
Differentially Expressed Genes (DEGs) of Wild and ΔAbrB B. licheniformis DW2 Strains under Different Conditions
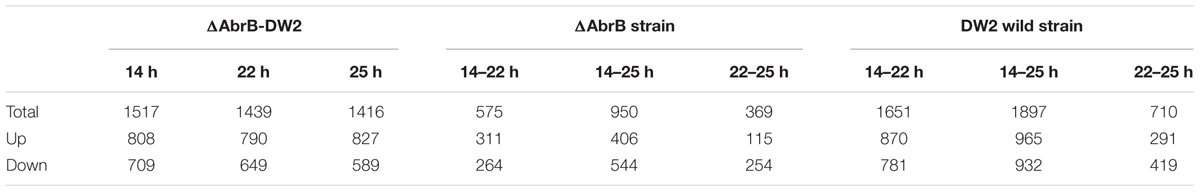
In the experiment of RNA isolation and preparation, we performed three biological replicates. Consequently, we obtained RNA-seq data at three time points from two strains and three biological replicates, 18 sets of RNA-seq data in total. It is important to examine the correlation of the three biological replicates. We calculated the Spearman coefficient of correlation among the data sets. The results in Figure 3A show that the RPKM of biological replicates are highly correlated. The AbrB gene has been completely knocked out according to the expression levels (Figure 3B).
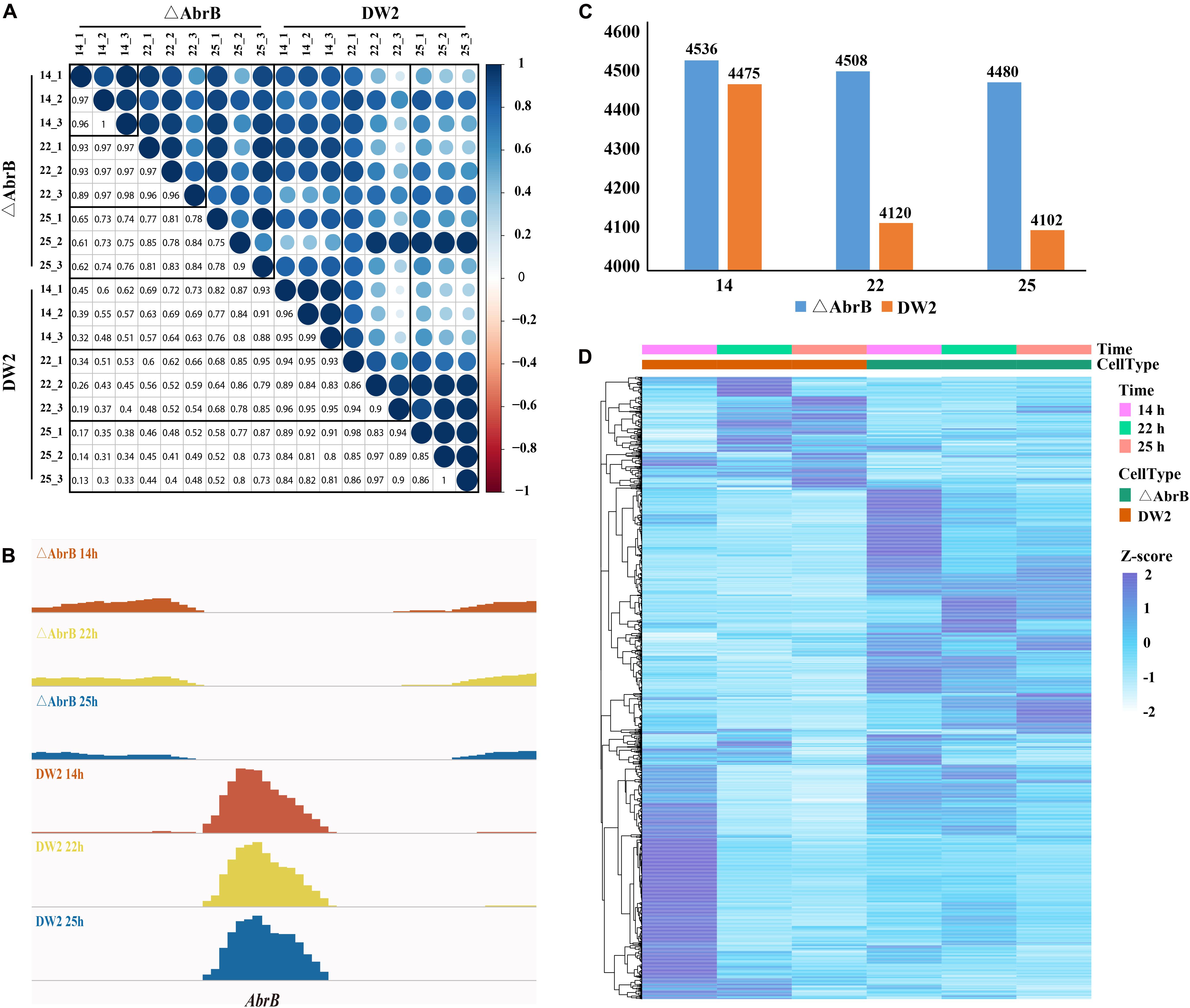
FIGURE 3. Gene expression of wild B. licheniformis DW2 and AbrB-knockout strains. (A) The correlation of three biological replicates in three time points of wild and AbrB-knockout strains. (B) The AbrB gene was knocked out and not expressed in AbrB-knockout strain. (C) The number of expressed genes in three different time points of wild and AbrB-knockout strains. (D) The different gene expression pattern between wild and AbrB-knockout strains.
We screened the expressed genes of wild-type strain and ΔAbrB strain at different time points with the threshold of RPKM > 1, and the number of expressed genes in different time points were in the range of 4,102∼4,536 (Figure 3C). DEGs were obtained using DEseq package between the two data sets of both strains and three growth phases (Table 3). As shown in Figure 3D, the number of expressed genes of ΔAbrB increased significantly than that of wild type in all three time points, indicating many inhibited genes expressed after AbrB gene was knocked out. In Table 3, more than a few hundred genes were up-regulated and down-regulated between wild type and ΔAbrB strains in all time points, which supported the above conclusion that AbrB can regulate numerous genes directly and indirectly. The number of expressed genes of DW2 wild type at 14 h is greater than that at 22 and 25 h. It is shown that more genes are activated in logarithmic growth phase, while quite a few of them are absent or very lowly expressed in stationary and transitional phases. Both up-regulated and down-regulated genes in ΔAbrB are far fewer than that of wild type, as shown in Table 3, may be caused by the inhabitation absence of AbrB. Many regulated genes expressed at all time points after AbrB was knocked out, and no longer were differentially genes. We also note that the number of DEGs correlate with time length. Among three columns either in wild type or ΔAbrB type, the middle column represents the DEGs numbers between 25 and 14 h, which are significantly greater the others. On the contrary, the DEGs between 22 and 25 h with the shortest time length correspond to the smallest numbers. It suggested that more and more genes begin to transcript or stop transcription and become DEGs with the development process of organism. To gain insight into the differential expression of these DEGs, complete linkage hierarchical cluster analysis based on RPKM values was performed using R. The heat map showed significant differential expression between strains, we identified six broad clusters of genes that exhibited expression changes over time. Genes in cluster found that about 30% of the DEGs in DW2 strains show high expression at logarithmic growth phase (14 h) and low expression at transitional phase (22 h) and stationary phase (25 h).
To understand the function of these DEGs, they were grouped according to their COG functional categories. As shown in Figure 4A, a majority of the functional categories in B. licheniformis DW2 contained both induce and repressed genes, indicating that this bacterium needs to balance many biological pathways under different conditions. We found that the pathways had a very unequal distribution, and some of them were significantly affected, such as ‘Carbohydrate transport and metabolism (G),’ ‘Amino Acid transport and metabolism (E),’ ‘Cell wall/membrane/envelope biogenesis (M),’ ‘Energy production and conversion (C)’ and ‘Lipid transport and metabolism (I).’ Notably, almost all DEGs in the functional categories of ‘cell motility (N)’ were down-regulated in logarithmic growth phase (14 h) and up-regulated in transitional phase (22 h) and stationary phase (25 h). Previous observations suggested one of the major functions of AbrB in repressing biofilm formation by repressing signal peptidase sipW, and sipW-processed protein may have a role in a motility structure in B. subtilis (Hamon et al., 2004). We speculated that AbrB repressed some genes related to cell motility in transitional phase (22 h) and stationary phase (25 h).
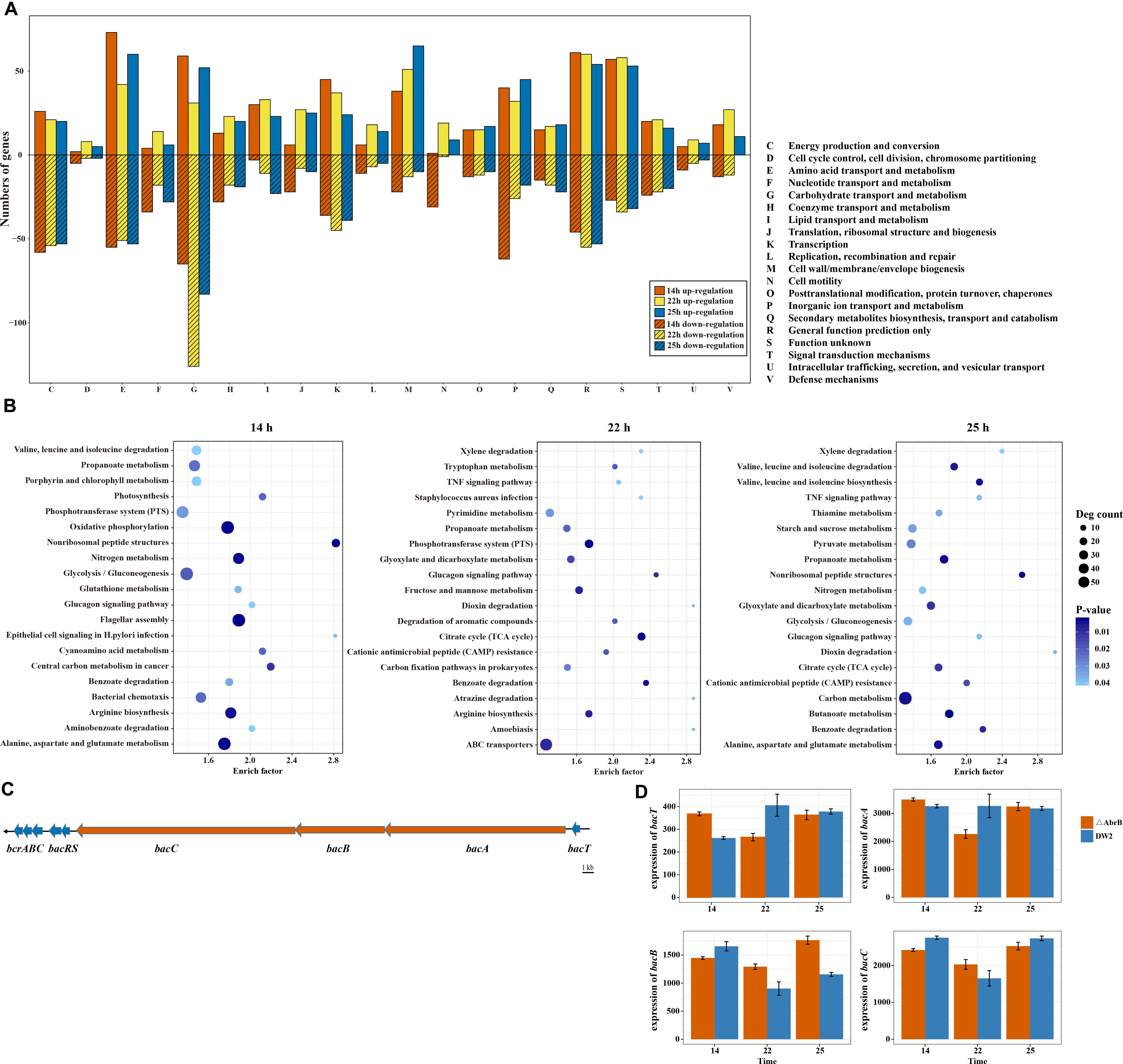
FIGURE 4. Functional enrichment of DEGs and different expression of bacitracin biosynthesis related genes. (A) The functional categories of DEGs in three different time points of wild type and AbrB-knockout strains. (B) The top 20 significantly enriched pathways of DEGs in three time points. (C) The cluster of bacitracin biosynthesis related genes in B. licheniformis DW2. (D) The different expression of bacitracin biosynthesis related genes bacA, bacB, bacC and bacT between wild and AbrB-knockout strains.
In addition, we mapped all DEGs to KEGG terms to identify significantly enriched metabolic pathways and got 190 pathways, which contained 1,271 DEGs. To obtain a better visual display of the results, top 20 significantly enriched pathways of each time points were shown in Figure 4B. The pathway flagellar assembly was significantly at logarithmic growth phase (14 h), which was consistent with the results of COG function enrichment. We also found that the number of genes in significantly enriched pathways was different among three time points, 14 h is much more than 22 and 25 h. These results also coincide with the variations of DEGs RPKM in ΔAbrB strain.
The Expression of Bacitracin Biosynthesis Related Genes
Bacitracin, comprised of 12 cyclic polypeptides, is an important non-ribosomal peptide antibiotic. It is produced by several Bacillus strains that are usually active against other Gram-positive and some strains of Gram-negative microorganisms, but not against the strain itself. B. subtilis C126 was reported as a bacitracin producer (Azevedo et al., 1993). B. subtilis 168, which has no bacitracin synthetase, but is more sensitive to bacitracin than B. licheniformis, has several homologs of bcrABC (Ohki et al., 2003). In previous studies, the bacitracin synthetase gene cluster was clearly described (Konz et al., 1997; Neumüller et al., 2001; Mascher et al., 2003). A thioesterase bacT and a non-ribosomal peptide synthetase (NPRS) bacABC operon compose the bacitracin synthetase gene cluster in B. licheniformis. The bcrABC genes of transporter system which are hypothesized to pump out bacitracin from the cells localize downstream of bacABC, between the bacABC operon and transporter system bcrABC is two-component system bacRS. Using antiSMASH 4.0.0 tool (Blin et al., 2017), we identified a complete bacitracin synthetase gene cluster in B. licheniformis DW2, and all genes of this cluster shows high similarity (identity > 90%) to bacitracin synthetase operon in B. licheniformis strain ATCC 10716 (Figure 4C). The bacT gene is 705 bp and encodes a protein of 235 amino acids. The bacABC gene cluster starts from downstream of the bacT gene and encoding bacitracin synthetase, it consists of the gene bacA (15,771 bp), bacB (7,809 bp), and bacC (19,080 bp). Gene bacR (717 bp) and bacS (1,035 bp) encode two-component regulatory system bacRS. The bcrABC gene cluster locates after 80 bp downstream of the bacRS genes, which is composed of bcrA (921 bp), bcrB (735 bp), and bcrC (612 bp) and encoding ABC transporter. These findings provided gene level information of bacitracin biosynthesis for B. licheniformis DW2.
Previous study suggested that AbrB might repress the expression of both bacT and bacA through binding to the promoter regions in B. licheniformis (Wang et al., 2017). Although bacABC and bacT are in the same bacitracin synthetase gene cluster, their transcription levels tremendously vary in both wild type and ΔAbrB (Figure 4D). The highest expressed gene bacA has about 10-fold RPKM comparing with bacT, the lowest expressed one; while bacB and bacC are between the two. In addition, the genes of bacitracin synthetase cluster did not share an identical differentiated expression manner when AbrB was knocked out, the up/down regulations of bacT and bacABC did not show an obvious regularity. However, at 22 h their expression level of ΔAbrB were lower than those of 14 and 25 h, which was found in all the above four genes, especially in bacT and bacA.
Notably, the bcrABC were significantly up-regulated in ΔAbrB strain at all three time points in contrast to wild type (Supplementary Figure S3). In previous study, B. licheniformis was hypothesized to pump out bacitracin for self-resistance by bcrABC (Neumüller et al., 2001). Besides, experiments showed that ΔAbrB strains would increase bacitracin yield (Supplementary Figure S2; Wang et al., 2017). Our findings provided evidence for bacitracin up-regulation and transport, which was accordance with the above two studies and linked them together. Comparing with wild type, AbrB-knockout strains secreted more bacitracin, thus they had to increase resistance to the extra bacitracin in order to avoid self-intoxication. The up-regulation of bcrABC enabled ΔAbrB strain produce more transporters to remove the bacitracin out of the cell. So it is reasonable to view the differential expression of bcrABC as a response to the change of bacitracin production.
Conclusion
In this study, we successfully assemble the complete genome of B. licheniformis DW2 using PacBio SMRT sequencing system and de novo assembly based on the HGAP method. The genomic information obtained in this study would help subsequent comparative genomic analysis with other B. licheniformis strains. Meanwhile, we conducted comprehensive analyses of B. licheniformis DW2 by ssRNA-seq in wild and ΔAbrB strains. The ssRNA-seq data enabled us to comparatively analyze DEGs between different strains and growth phases and provided us information on bacitracin synthesis. In previous studies, B. licheniformis was considered to transport bacitracin for self-resistance by bcrABC (Neumüller et al., 2001). The global transcription regulator AbrB represses the transcription of the bacitracin synthase operon by directly binding to the bacitracin synthase operon promoter region (Wang et al., 2017). In our study, we identified a complete bacitracin synthetase gene cluster in the genome including the location and length of each gene. The expressions of bcrABC were significantly upregulated in ΔAbrB strain. These findings provided evidence for bacitracin up-regulation and removing out of the cell, which were consistent with the above two studies and linked them together. This study would be helpful for improving the industrial process of bacitracin production through AbrB-knockout and further understanding of bacitracin transport in B. licheniformis.
Accession Numbers
The genomic and transcriptomic sequencing data of B. licheniformis DW2 generated in this study have been submitted to BIG Data Center (http://bigd.big.ac.cn) under accession numbers GWHAAAE00000000 and CRA000557.
Author Contributions
L-LC and J-XG conceived the study. C-CS, JG, and J-MS analyzed the data. S-WC and DW performed the experiments. C-CS, L-LC, and J-XG wrote the paper.
Funding
This work was supported by National Natural Science Foundation of China (31271406, 31571351) and the Natural Science Foundation of Hubei Province (2015CFA044).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are grateful to Dr. Jack Chen at Simon Fraser University for valuable suggestions in data analysis.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.00307/full#supplementary-material
Footnotes
- ^ https://cge.cbs.dtu.dk/services/Restriction-ModificationFinder-1.0/
- ^ http://www.expasy.ch/sprot
- ^ http://www.ncbi.nlm.nih.gov
References
Ali, N. O., Bignon, J., Rapoport, G., and Debarbouille, M. (2001). Regulation of the acetoin catabolic pathway is controlled by sigma L in Bacillus subtilis. J. Bacteriol. 183, 2497–2504. doi: 10.1128/JB.183.8.2497-2504.2001
Anders, S., and Huber, W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11:R106. doi: 10.1186/gb-2010-11-10-r106
Aziz, R. K., and Bartels, D., Best, A. A., DeJongh, M., Disz, T., and Edwards, R. A. (2008). The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75
Azevedo, E. C., Rios, E. M., Fukushima, K., and Campos-Takaki, G. M. (1993). Bacitracin production by a new strain of bacillus subtilis. Appl. Biochem. Biotechnol. 42, 1–7. doi: 10.1007/BF02788897
Banse, A. V., Chastanet, A., Rahn-Lee, L., Hobbs, E. C., and Losick, R. (2008). Parallel pathways of repression and antirepression governing the transition to stationary phase in Bacillus subtilis. Proc. Natl. Acad. Sci. U.S.A. 105, 15547–15552. doi: 10.1073/pnas.0805203105
Benson, G. (1999). Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27, 573–580. doi: 10.1093/nar/27.2.573
Blin, K., Wolf, T., Chevrette, M. G., Lu, X., Schwalen, C. J., Kautsar, S. A., et al. (2017). antiSMASH 4.0-improvements in chemistry prediction and gene cluster boundary identification. Nucleic Acids Res. 45, W36-W41. doi: 10.1093/nar/gkx319
Cain, B. D., Norton, P. J., Eubanks, W., and Nick, H. S. Allen, C. M. (1993). Amplification of the bacA gene confers bacitracin resistance to Escherichia coli. J. Bacteriol. 175, 3784–3789. doi: 10.1128/jb.175.12.3784-3789.1993
Chalker, A. F., Ingraham, K. A., Lunsford, R. D., Bryant, A. P., Bryant, J., Wallis, N. G., et al. (2000). The bacA gene, which determines bacitracin susceptibility in Streptococcus pneumoniae and Staphylococcus aureus, is also required for virulence. Microbiology 146, 1547–1553. doi: 10.1099/00221287-146-7-1547
Chin, C. S., Alexander, D. H., Marks, P., Klammer, A. A., Drake, J., Heiner, C., et al. (2013). Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 10, 563–569. doi: 10.1038/nmeth.2474
Chumsakul, O., Takahashi, H., Oshima, T., Hishimoto, T., Kanaya, S., Ogasawara, N., et al. (2011). Genome-wide binding profiles of the Bacillus subtilis transition state regulator AbrB and its homolog Abh reveals their interactive role in transcriptional regulation. Nucleic Acids Res. 39, 414–428. doi: 10.1093/nar/gkq780
Darling, A. E., Mau, B., and Perna, N. T. (2010). Progressive Mauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147. doi: 10.1371/journal.pone.0011147
El-Sharnouby, S., Redhouse, J., and White, R. A. (2013). Genome-wide and cell-specific epigenetic analysis challenges the role of polycomb in Drosophila spermatogenesis. PLoS Genet. 9:e1003842. doi: 10.1371/journal.pgen.1003842
Flusberg, B. A., Webster, D. R., Lee, J. H., Travers, K. J., Olivares, E. C., Clark, T. A., et al. (2010). Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat. Methods 7, 461–465. doi: 10.1038/nmeth.1459
Galperin, M. Y., Makarova, K. S., Wolf, Y. I., and Koonin, E. V. (2015). Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res. 43, 261–269. doi: 10.1093/nar/gku1223
Gene Ontology Consortium (2015). Gene ontology consortium: going forward. Nucleic Acids Res. 43, D1049–1056. doi: 10.1093/nar/gku1179
Guo, J., Cheng, G., Gou, X. Y., Xing F., Li S., Han, Y. C., et al. (2015). Comprehensive transcriptome and improved genome annotation of Bacillus licheniformis WX-02. FEBS Lett. 589, 2372–2381. doi: 10.1016/j.febslet.2015.07.029
Hamon, M. A., Stanley, N. R., Britton, R. A., Grossman, A. D., and Lazazzera, B. A. (2004). Identification of AbrB-regulated genes involved in biofilm formation by Bacillus subtilis. Mol. Microbiol. 52, 847–860. doi: 10.1111/j.1365-2958.2004.04023.x
Johnson, B. A., and Anker, H. Meleney, F. L. (1945). Bacitracin: a new antibiotic produced by a member of the B. subtilis group. Science 102, 376–377. doi: 10.1126/science.102.2650.376
Kim, H. J., Kim, S. I., Ratnayake-lecamwasam, M., Tachikawa, K., Sonenshein, A. L., and Strauch, M. (2003). Complex regulation of the Bacillus subtilis aconitase gene. J. Bacteriol. 185, 1672–1680. doi: 10.1128/JB.185.5.1672-1680.2003
Konglom, N., Chuensangjun, C., Pechyen, C., and Sirisansaneeyakul, S. (2012). Production of poly-γ-glutamic acid by Bacillus licheniformis, synthesis and characterization. J. Met. Mater. Miner. 22, 7–11.
Konz, D., Klens, A., Schörgendorfer, K., and Marahiel, M. A. (1997). The bacitracin biosynthesis operon of Bacillus licheniformis ATCC 10716: molecular characterization of three multi-modular peptide synthetases. Chem. Biol. 4, 927–937. doi: 10.1016/S1074-5521(97)90301-X
Kumar, S., Cheng, X., Klimasauskas, S., Mi, S., Posfai, J., Roberts, R. J., et al. (1994). The DNA (cytosine-5) methyltransferases. Nucleic Acids Res. 22, 1–10. doi: 10.1093/nar/22.1.1
Kunst, F., Ogasawara, N., Moszer, I., Albertini, A. M., Alloni, G., Azevedo, V., et al. (1997). The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390, 249–256. doi: 10.1038/36786
Kurtz, S., Phillippy, A., Delcher, A. L., Smoot, M., Shumway, M., Antonescu, C., et al. (2004). Versatile and open software for comparing large genomes. Genome Biol. 5:R12. doi: 10.1186/gb-2004-5-2-r12
Lagesen, K., Hallin, P., Rødland, E. A., Staerfeldt, H. H., Rognes, T., and Ussery, D. W. (2007). RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 35, 3100–3108. doi: 10.1093/nar/gkm160
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Laird, P. W. (2010). Principles and challenges of genome-wide DNA methylation analysis. Nat. Rev. Genet. 11, 191–203. doi: 10.1038/nrg2732
Li, E., Yang, H., Wang, X., Wan, L. Pan, H., and Zhu, D. (2015). Whole-genome sequencing and analysis of inosine-producing strain Bacillus subtilis ATCC 13952. Acta Microbiol. Sin. 55, 1560–1567.
Liu, H., Yin, S., An, L., Zhang, G., Cheng, H., Xi, Y., et al. (2016). Complete genome sequence of Bacillus subtilis BSD-2, a microbial germicide isolated from cultivated cotton. J. Biotechnol. 230, 26–27. doi: 10.1016/j.jbiotec.2016.05.019
Liu, Y., Zhang, S., Yong, Y.-C., Ji, Z., Ma, X., Xu, Z., et al. (2011). Efficient production of acetoin by the newly isolated Bacillus licheniformis strain MEL09. Process Biochem. 46, 390–394. doi: 10.1016/j.procbio.2010.07.024
Lowe, T. M., and Eddy, S. R. (1997). tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25, 955–964. doi: 10.1093/nar/25.5.0955
Mao, X., Ma, Q., Zhou, C., Chen, X., Zhang, H., Yang, J., et al. (2014). Door 2.0: presenting operons and their functions through dynamic and integrated views. Nucleic Acids Res. 42, 654–659. doi: 10.1093/nar/gkt1048
Mascher, T., Margulis, N. G., Wang, T., Ye, R. W., and Helmann, J. D. (2003). Cell wall stress responses in Bacillus subtilis: the regulatory network of the bacitracin stimulon. Mol. Microbiol. 50, 1591–1604. doi: 10.1046/j.1365-2958.2003.03786.x
Mcinerney, M. J., Javaheri, M., and Nagle, D. P. (1990). Properties of the biosurfactant produced by bacillus licheniformis strain JF-2. J. Ind. Microbiol. 5, 95–101. doi: 10.1007/BF01573858
Mortazavi, A., Williams, B. A., McCue, K., Schaeffer, L., and Wold, B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628. doi: 10.1038/nmeth.1226
Neumüller, A. M., Konz, D., and Marahiel, M. A. (2001). The two-component regulatory system BacRS is associated with bacitracin ’self-resistance’ of Bacillus licheniformis ATCC 10716. Eur. J. Biochem. 268, 3180–3189. doi: 10.1046/j.1432-1327.2001.02203.x
Ogata, H., Goto, S., Sato, K., Fujibuchi, W., Bono, H., and Kanehisa, M. (1999). KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 27, 29–34. doi: 10.1093/nar/27.1.29
Ohki, R., Giyanto, Tateno, K., Masuyama, W., Moriya, S., and Kobayashi, K. (2003). The BceRS two-component regulatory system induces expression of the bacitracin transporter, BceAB, in Bacillus subtilis. Mol. Microbiol. 49, 1135–1144. doi: 10.1046/j.1365-2958.2003.03653.x
Perez-Valdespino, A., Li, Y., Setlow, B., Ghosh, S., Pan, D., and Korza, G. (2014). Function of the SpoVAEa and SpoVAF proteins of Bacillus subtilis spores. J. Bacteriol. 196, 2077–2088. doi: 10.1128/JB.01546-14
Podlesek, Z., Comino, A., Herzog-Velikonja, B., Zgur-Bertok, D., Komel, R., and Grabnar, M. (1995). Bacillus licheniformis bacitracin resistance ABC transporter: relationship to mammalian multidrug resistance. Mol. Microbiol. 16, 969–976. doi: 10.1111/j.1365-2958.1995.tb02322.x
Podlesek, Z., Comino, A., Herzog-Velikonja, B., and Grabnar, M. (2000). The role of the bacitracin ABC transporter in bacitracin resistance and collateral detergent sensitivity. FEMS Microbiol. Lett. 188, 103–106. doi: 10.1111/j.1574-6968.2000.tb09176.x
Pollock, T. J., Thorne, L., Yamazaki, M., and Mikolajczak, M. J., and Armentrout, R. W. (1994). Mechanism of bacitracin resistance in Gram-negative bacteria that synthesize exopolysaccharides. J. Bacteriol. 176, 6229–6237. doi: 10.1128/jb.176.20.6229-6237.1994
Pötter, M., Oppermann-Sanio, F. B., and Steinbüchel, A. (2001). Cultivation of bacteria producing polyamino acids with liquid manure as carbon and nitrogen source. Appl. Environ. Microbiol. 67, 617–622. doi: 10.1128/AEM.67.2.617-622.2001
Qian, Q., Lee, C. Y., Helmann, J. D., and Strauch, M. A. (2002). AbrB is a regulator of the sigma(w) regulon in Bacillus subtilis. FEMS Microbiol. Lett. 211, 219–223.
Quinlan, A. R., and Hall, I. M. (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. doi: 10.1093/bioinformatics/btq033
Sitaraman, R. (2016). The role of DNA restriction-modification systems in the biology of Bacillus anthracis. Front. Microbiol. 7, 1–6. doi: 10.3389/fmicb.2016.00011
Roberts, R. J., Vincze, T., Posfai, J., and Macelis, D. (2015). Rebase-a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res. 43, D298-299. doi: 10.1093/nar/gku1046
Sambrook, J., Fritsch, E. F., and Maniatis, T. (1982). Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor laboratory press.
Sánchezromero, M. A., Cota I., and Casadesús, J. (2015). DNA methylation in bacteria: from the methyl group to the methylome. Curr. Opin. Microbiol. 25, 9–16. doi: 10.1016/j.mib.2015.03.004
Shafikhani, S. H., and Leighton, T. (2004). AbrB and Spo0E control the proper timing of sporulation in Bacillus subtilis. Curr. Microbiol. 48, 262–269. doi: 10.1007/s00284-003-4186-2
Sommer, D. D., Delcher, A. L., Salzberg, S. L., and Pop, M. (2007). Minimus: a fast, lightweight genome assembler. BMC Bioinformatics 8:64. doi: 10.1186/1471-2105-8-64
Strauch, M., Webb, V., Spiegelman, G., and Hoch, J. A. (1990). The SpoOA protein of Bacillus subtilis is a repressor of the abrB gene. Proc. Natl. Acad. Sci. U.S.A. 87, 1801–1805. doi: 10.1073/pnas.87.5.1801
Strauch, M. A., Bobay, B. G., Cavanagh, J., Yao, F., Wilson, A., and Le, B. Y. (2007). Abh and AbrB control of Bacillus subtilis antimicrobial gene expression. J. Bacteriol. 189, 7720–7732. doi: 10.1128/JB.01081-07
Veith, B., Herzberg, C., Steckel, S., Feesche, J., Maurer, K. H., Ehrenreich, P., et al. (2004). The complete genome sequence of Bacillus licheniformis DSM13, an organism with great industrial potential. J. Mol. Microbiol. Biotechnol. 7, 204–211. doi: 10.1159/000079829
Wang, D., Wang, Q., Qiu, Y., Nomura, C. T., Li, J., and Chen, S. (2017). Untangling the transcription regulatory network of the bacitracin synthase operon in Bacillus licheniformis DW2. Res. Microbiol. 168, 515–523. doi: 10.1016/j.resmic.2017.02.010
Keywords: Bacillus licheniformis, reference genome, transcriptome, AbrB-knockout, differentially expressed genes, bacitracin biosynthesis
Citation: Shu C-C, Wang D, Guo J, Song J-M, Chen S-W, Chen L-L and Gao J-X (2018) Analyzing AbrB-Knockout Effects through Genome and Transcriptome Sequencing of Bacillus licheniformis DW2. Front. Microbiol. 9:307. doi: 10.3389/fmicb.2018.00307
Received: 30 November 2017; Accepted: 09 February 2018;
Published: 26 February 2018.
Edited by:
Feng Gao, Tianjin University, ChinaReviewed by:
Mingfeng Cao, Iowa State University, United StatesOzlem Ates Duru, Nişantaşı University, Turkey
Copyright © 2018 Shu, Wang, Guo, Song, Chen, Chen and Gao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jun-Xiang Gao, gao200@mail.hzau.edu.cn
 Cheng-Cheng Shu1
Cheng-Cheng Shu1 Shou-Wen Chen
Shou-Wen Chen Ling-Ling Chen
Ling-Ling Chen Jun-Xiang Gao
Jun-Xiang Gao