- 1State Key Laboratory of Applied Microbiology Southern China, Guangdong Institute of Microbiology, Guangzhou, China
- 2Guangdong Provincial Key Laboratory of Microbial Culture Collection and Application, Guangdong Open Laboratory of Applied Microbiology, Guangzhou, China
- 3School of Bioscience and Bioengineering, South China University of Technology, Guangzhou, China
- 4College of Food Science, South China Agricultural University, Guangzhou, China
Methicillin-resistant Staphylococcus aureus (MRSA) is an emerging pathogen that is difficult to treat due to the multiresistance of the bacteria upon infection. From 2011 to 2016, 1581 S. aureus strains were isolated from 4300 samples from retail foods covering most provincial capitals in China. To determine the prevalence of food-related MRSA and its genetic background in China, antibiotic resistance, staphylococcal toxin genes, staphylococcal cassette chromosome mec (SCCmec) typing, spa-typing and MLST were carried out in this study. In total, 108 (7.4%) isolates were confirmed for MRSA by phenotyping (cefoxitin) and genotyping (mecA/mecC gene). A total of 52.8% (57/108) of the MRSA isolates belonged to clonal complex 59 (CC59) (ST59, ST338, and ST3355), which was the predominant clone in this study. These CC59 isolates carried SCCmec elements of type IV, V, or III and exhibited spa type t437, t441, t543, t163, t1785, or t3485, and half of them carried major virulence genes, such as the Panton-Valentine leucocidin (PVL) gene. The secondary clones belonged to ST9 (15.7%, 17/108) with a type of t899-SCCmec III and showed a broader range of antimicrobial resistance. The remaining MRSA isolates (31.5%, 34/108) were distributed in 12 different STs and 18 different spa types. All isolates harbored at least one of the enterotoxin genes, whereas only 4 isolates (3.70%) were positive for the toxic shock syndrome toxin tsst alleles. For antibiotic susceptibility testing, all isolates were resistant to more than three antibiotics, and 79.6% of the isolates were resistant to more than 10 antibiotics. Amoxycillin/clavulanic acid, ampicillin, cefoxitin, penicillin, ceftazidime, kanamycin, streptomycin, clindamycin, and telithromycin was the most common antibiotic resistance profile (55.6%, 60/108) in the study. In summary, the results of this study implied that the major food-related MRSA isolate in China was closer to community-associated MRSA, and some of the remaining isolates (ST9-t899-SCCmec III) were supposed to livestock-associated MRSA. In addition, most MRSA isolates showed resistance to multiple drugs and harbored staphylococcal toxin genes. Thus, the pathogenic potential of these isolates cannot be ignored. In addition, further studies are needed to elucidate the transmission routes of MRSA in relation to retail foods and to determine how to prevent the spread of MRSA.
Introduction
Methicillin-resistant Staphylococcus aureus (MRSA) is a pathogen of increasing importance in hospitals as well as in the community and livestock. It can be resistant to several antibiotics and quickly disseminates worldwide. In recent years, MRSA has been attributed to an estimated 5400 extra deaths and over one million extra days of hospitalization (Kobayashi et al., 2015). S. aureus became MRSA because of the acquisition of the gene mecA or mecC, which encodes the low-affinity penicillin-binding protein 2a (PBP2a), which, unlike other PBPs, remains active and allows for cell wall biosynthesis at otherwise lethal β-lactam concentrations. The mec operon is carried by the staphylococcal cassette chromosome mec (SCCmec) and most likely originated from horizontal transfer from coagulase-negative staphylococcal species (Banerjee et al., 2008; Garcíaálvarez et al., 2011).
MRSA strains have been reported from various sources. To distinguish epidemiological groups of MRSA, it was divided into hospital-associated MRSA (HA-MRSA), community-associated MRSA (CA-MRSA) and livestock-associated MRSA (LA-MRSA) (Petinaki and Spiliopoulou, 2012). MRSA was first recognized as HA-MRSA in 1961 and then spread into the community and, later on, into healthcare facilities; in 1990, this type became recognized as CA-MRSA. LA-MRSA has always been associated with animals and is linked to a jump from animals to humans (Petinaki and Spiliopoulou, 2012). In general, the genetic backgrounds differ among different types of MRSA. In previous research, CA-MRSA strains frequently harbor SCCmec type IV or V and often produce potent toxins/virulence factors, such as Panton-Valentine leucocidin (PVL), arginine catabolic mobile element (ACME) and phenol-soluble modulins, while HA-MRSA strains typically possess larger-size SCCmec type I-III and are more resistant to other classes of antibiotics (An and Otto, 2008; Chuang and Huang, 2013). For multilocus sequence typing (MLST), sequence type 5 (ST5), ST8, ST22, ST36, and ST45 spread successfully to different regions of the world and caused substantial nosocomial disease (Deleo et al., 2010), whereas CA-MRSA showed five lineages worldwide: ST1-IV (USA400), ST8-IV (USA300), ST30-IV (Pacific/Oceania), ST59-IV and V (USA1000, Taiwan) and ST80-IV (European) (Skov, 2009). In contrast, LA-MRSA strains exhibit co-resistance to many non-β-lactam antimicrobials (e.g., antibiotics and metals), including those commonly used in animal production, and many of them belong to CC398 or CC9, as determined by MLST (Bens et al., 2006; Cui and Li, 2009; Neela et al., 2009). The SCCmec elements of LA-MRSA are different from those carried by other MRSA genotypes commonly found in the community and healthcare settings (Li et al., 2011). In addition, the majority of LA-MRSA isolates lack toxins, such as PVL and other enterotoxins (Hallin et al., 2011).
Nowadays, MRSA strains have been reported from various foods sources, such as poultry, pork, beef, milk and vegetables, suggesting that foods may serve as reservoirs (Wang et al., 2014; Wu et al., 2018). As is commonly known, foods have many different origins, and different types of MRSA are present in foods of different origin in different countries. In China, large-scale studies of the prevalence of S. aureus in food are scarce. From July 2011 to June 2016, we collected 4300 retail food samples from supermarkets, fairs and farmers’ markets that covered most of the provincial capitals of China (Supplementary Figure 1) and found 1063 (24.7%) S. aureus-positive samples from all sampling sites. To better understand the genetic background among food-related MRSA isolates in China, this study aimed to identify the MRSA isolates from our previous study and to characterize these MRSA isolates for their antimicrobial resistance profiles, their virulence genes and their genotypic types (SCCmec-, MLST, and spa types).
Materials and Methods
Bacterial Isolates
A total of 1581 S. aureus isolates were collected from 4300 retail food samples in 39 Chinese cities (Supplementary Figure 1), comprising 469 isolates from meat and meat products (bacon/sausage, poultry, pork, mutton and beef), 511 isolates from aquatic products (freshwater fish, shrimp and seafood), 368 isolates from quick-frozen products (frozen dumplings/steamed stuffed buns and frozen meat), 148 isolates from ready-to-eat food (cold vegetable/noodle dishes in sauce, fried rice/sushi, roast meat, sausage, and ham), 42 isolates from edible mushrooms, 30 isolates from vegetables and 13 isolates from pasteurized milk. These isolates were obtained between July 2011 and June 2016 according to the GB 4789.10-2010 food microbiological examination of S. aureus (National Food Safety Standards of China) and the most probable number (MPN) method (Gombas et al., 2003). They were identified by Gram stain, catalase and oxidase tests and API STAPH test strips (BioMerieux, Marcy-1’Etoile, France). Each isolate was incubated at 37°C overnight in brain heart infusion (BHI) broth. Genomic DNA was extracted using a genomic DNA extraction kit (Magen Biotech, Guangzhou, China) according to the manufacturer’s instructions. The concentration of genomic DNA was determined at 260 nm using a NanoDrop-ND-1000 UV-Vis spectrophotometer (Thermo Fisher Scientific, MA, United States).
MRSA Confirmation
Cefoxitin disks (30 μg) were used for detecting methicillin-resistant isolates. S. aureus ATCC 25923 was used as a control. The mecA/mecC gene, which has been shown to confer methicillin resistance to S. aureus (MRSA), was also detected by PCR using primers as described previously (Perez-Roth et al., 2001; Stegger et al., 2012).
Antimicrobial Susceptibility Testing
A total of 25 antimicrobial agents were tested for antimicrobial susceptibility in all MRSA isolates. Amoxycillin/clavulanic acid (AMC, 30 μg), ampicillin (AMP, 10 μg), cefepime (FEP, 10 μg), penicillin G (P, 10 U), ceftazidime (CAZ, 30 μg), amikacin (AK, 30 μg), gentamicin (CN, 10 μg), kanamycin (K, 30 μg), streptomycin (S, 25 μg), chloramphenicol (C, 30 μg), clindamycin (DA, 2 μg), erythromycin (E, 15 μg), telithromycin (TEL, 15 μg), ciprofloxacin (CIP, 5 μg), norfloxacin (NOR, 10 μg), tetracycline (TE, 30 μg), linezolid (LZD, 30 μg), trimethoprim/sulphamethoxazole 1:19 (SXT, 25 μg), rifampicin (RD, 5 μg), quinupristin/dalfopristin (QD, 15 μg), teicoplanin (TEC, 30 μg), nitrofurantoin (F, 300 μg) and fusidic acid (FD, 10 μg) were tested by the disk diffusion method using Mueller–Hinton agar and commercially available discs (Oxoid, United Kingdom). Vancomycin and daptomycin were tested by broth microdilution according to the recommendations of the Clinical and Laboratory Standards Institute (CLSI) (CLSI, 2015). Staphylococcus aureus ATCC25923 and Escherichia coli ATCC25922 were used as quality control organisms (CLSI, 2015). The isolates with linezolid resistance determined by disk diffusion were also confirmed using a microdilution test according to the CLSI method for minimum inhibitory concentrations (MICs) (CLSI, 2015). The results of the antimicrobial susceptibilities of the analyzed strains were scored according to the guidelines of the CLSI (CLSI, 2015).
Detection of Staphylococcal Toxin Genes
All MRSA isolates were tested by PCR for the presence of 20 genes coding for staphylococcal enterotoxins (sea, seb, sec, sed, see, seg, seh, sei, sej, sek, sel, sem, sen, seo, sep, seq, ser, and seu), and the tsst-1 gene encoding the TSST (Varshney et al., 2009) and the lukSF-PV (pvl) genes were determined by PCR according to a previously published method (Jarraud et al., 2002). The primers and PCR conditions are presented in Supplementary Table 1. The amplicons were stained with Goldview, loaded, and electrophoresed in 1.5% agarose at 120 V for 0.5 h and visualized under a UV transilluminator gel imaging system (GE Healthcare, WI, United States). The images were saved as TIFF files for analysis.
Molecular Typing of MRSA Isolates
All MRSA isolates were subjected to SCCmec typing, spa-typing and MLST. The SCCmec typing method was performed on the isolates by multiplex PCR as previously described (Zhang et al., 2005). The S. aureus protein A (spa) repeat region was amplified according to a published protocol (Shopsin et al., 1999). The MLST scheme used to characterize MRSA isolates is based on the sequence analysis of the following seven housekeeping genes: arcC, aroE, glpF, gmk, pta, tpi, and yqil (Enright et al., 2013).
The DNA fragments were purified using a PCR purification kit (Qiagen, Genman) and sequenced in each direction with Big Dye fluorescent terminators on an ABI 3730XL sequencer (Applied Biosystems). The spa types were randomly assigned using the SpaServer website1. For each MLST locus, an allele number was given to each distinct sequence variant, and a distinct ST number was attributed to each distinct combination of alleles at the seven genes. STs were determined using the S. aureus MLST database2. Clonal complex (CC) analysis was performed in eBURST v.3 as previously described (Feil et al., 2004). The minimum spanning tree (MST) was constructed with Bionumerics 7.6 software (Applied Maths, Sint-Martens-Latem, Belgium).
Results
Prevalence of MRSA in Food
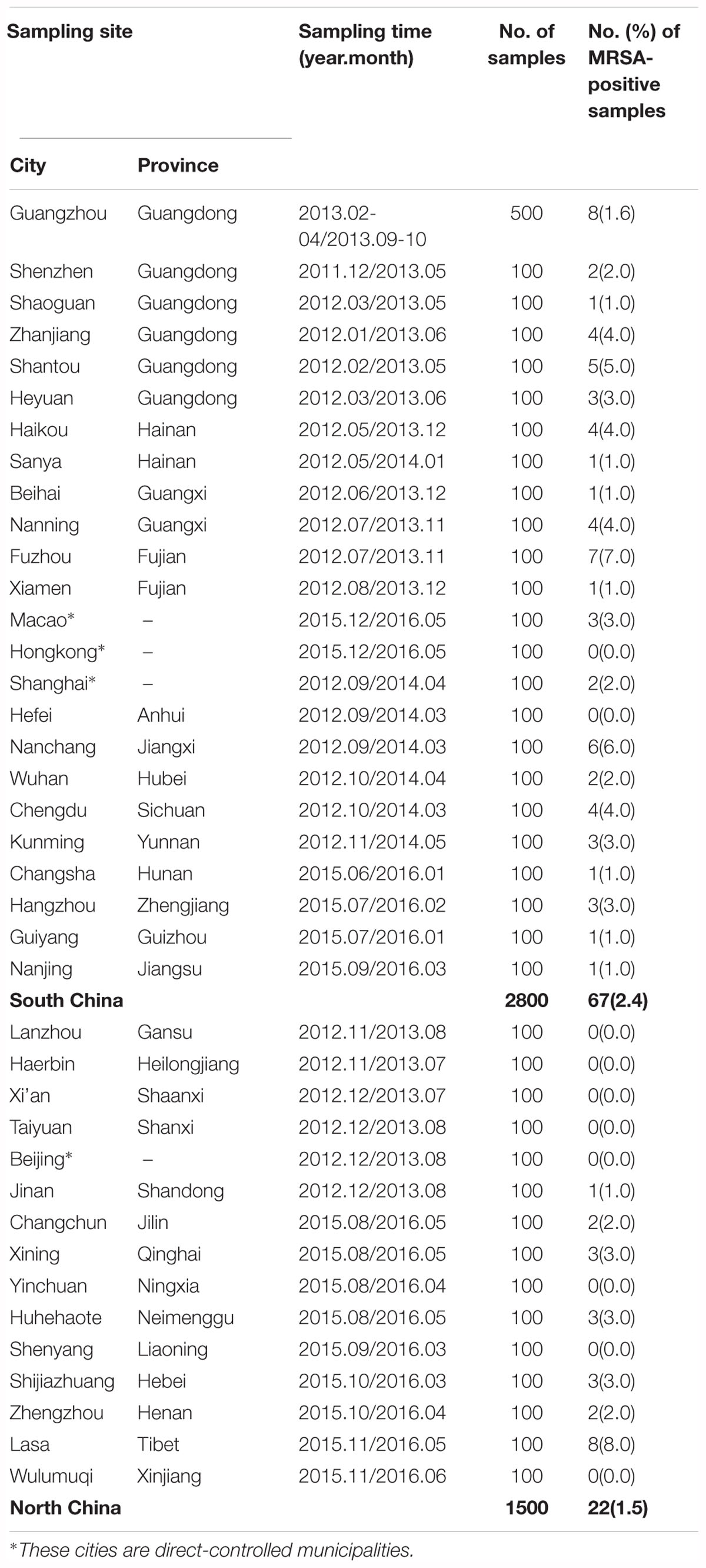
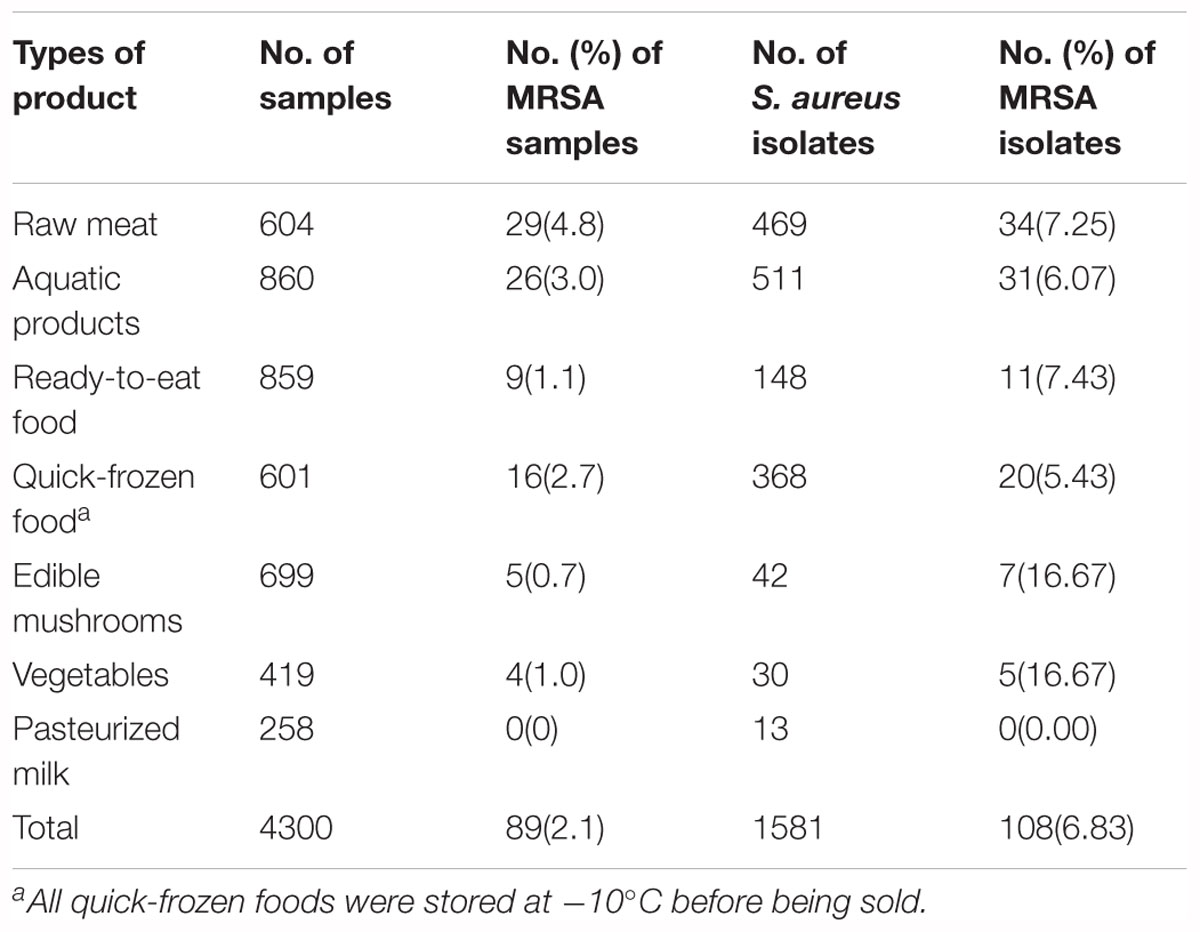
Overall, of the 1581 S. aureus isolates from retail food in China, 108 (6.83%) isolates from 89 positive samples (2.1%, 89/4300) were confirmed as MRSA, which exhibited cefoxitin resistance, 99.1% of isolates (107/108) were positive for mecA genes, and none were positive for the mecC gene. The distribution of MRSA among different sampling sites is shown in Supplementary Figure 2. In total, 29 of the 39 sampling cities (74.4%) had MRSA-positive samples, including 22 of the 24 southern cities and 7 of 15 the northern cities. Based on these results, there were 67 (2.4%) and 22 MRSA-positive samples (1.5%) in south China and north China, respectively. The most severe contamination level among the 29 cities was observed in Lasa (8%, 8/100), followed by Fuzhou (7%, 7/100), Nanchang (6%, 6/100), Shantou (5%, 5/100), Zhanjiang (4%, 4/100), Haikou (4%, 4/100), Nanning (4%, 4/100), Chengdu (4%, 4/100), Heyuan (3%, 3/100), Macao (3%, 3/100), Hangzhou (3%, 3/100), Xining (3%, 3/100), Huhehaote (3%, 3/100), Shijiazhuang (3%, 3/100), Shenzhen (2%, 2/100), Shanghai (2%, 2/100), Wuhan (2%, 2/100), Changchun (2%, 2/100), Zhengzhou (2%, 2/100), and Guangzhou (1.6%, 8/500); other cities were represented by one positive sample (Table 1). The analyzed food products were classified into seven categories, and the values of MRSA contamination in each sample were determined (Table 2). Among the analyzed categories, MRSA was detected in 4.8% (29/604) of raw meat, 3.0% (26/860) of aquatic products, 2.7% (16/601) of quick-frozen food, 1.1% (9/859) of ready-to-eat food, 1.0% (4/419) of vegetables and 0.7% (5/699) of edible mushrooms, whereas pasteurized milk was free of MRSA isolates. Of the 108 MRSA isolates, 34 (7.25%) of the 469 isolates were from raw meat, 31 (6.07%) of the 511 isolates were from aquatic products, 20 (5.43%) of the 368 isolates were from quick-frozen products, 11 (7.43%) of the 148 isolates were from ready-to-eat food, 7 (16.67%) of the 42 isolates were from edible mushrooms, and 5 (16.67%) of the 30 isolates were from vegetables, whereas pasteurized milk was free of MRSA isolates (Table 2).
Antibiotic Resistance Profiles of MRSA Isolates
The antibiotic susceptibility results of 108 MRSA isolates are shown in Table 3. All MRSA isolates were resistant to more than three antibiotics, including 33.3% of the isolates that were resistant to 4–10 antibiotics, 46.3% of the isolates that were resistant to 11–15 antibiotics and 20.4% of the isolates that were resistant to 16–26 antibiotics. The isolates were susceptible to linezolid, vancomycin and daptomycin, and the frequencies of resistance to individual agents were 100% for ampicillin and penicillin G, followed by 97.2% for ceftazidime, 87.0% for amoxicillin/clavulanic acid, 83.3% for erythromycin, 79.6% for clindamycin, 75.9% for kanamycin, 74.1% for telithromycin, 67.6% for streptomycin, 65.7% for cefepime and tetracycline, 38.0% for chloramphenicol, 27.8% for gentamycin, ciprofloxacin and fusidic acid, 25.9% for norfloxacin, 22.2% for amikacin, 13.95% for quinupristin/dalfopristin, 13.0% for trimethoprim/sulphamethoxazole 1:19, 7.4% for rifampicin, 2.8% for nitrofurantoin and 0.9% for teicoplanin. In different types of food products, the resistance to most antibiotics were equally distributed. Norfloxacin, clindamycin, quinupristin/dalfopristin, gentamycin, and trimethoprim/sulphamethoxazole 1:19 resistance was rare in MRSA isolated from non-animal sources (i.e., edible mushrooms and vegetables). It is worth noting that amikacin was the most commonly observed resistance antibiotic, whereas quinupristin/dalfopristin, gentamycin, ciprofloxacin and norfloxacin resistance were observed in low frequencies in aquatic product-related MRSA isolates compared to other types of food isolates (Supplementary Figure 2). However, AMC-AMP-FOX-P-CAZ-K-S-DA-TEL was the most common antibiotic resistance profile (55.6%, 60/108) in this study. All selected antibiotics were grouped into 15 classes of agents. Five MRSA isolates exhibited resistance to only β-lactam antibiotics, whereas 97 isolates exhibited a multidrug resistance phenotype, with resistance to ≥3 classes of antimicrobial agents.
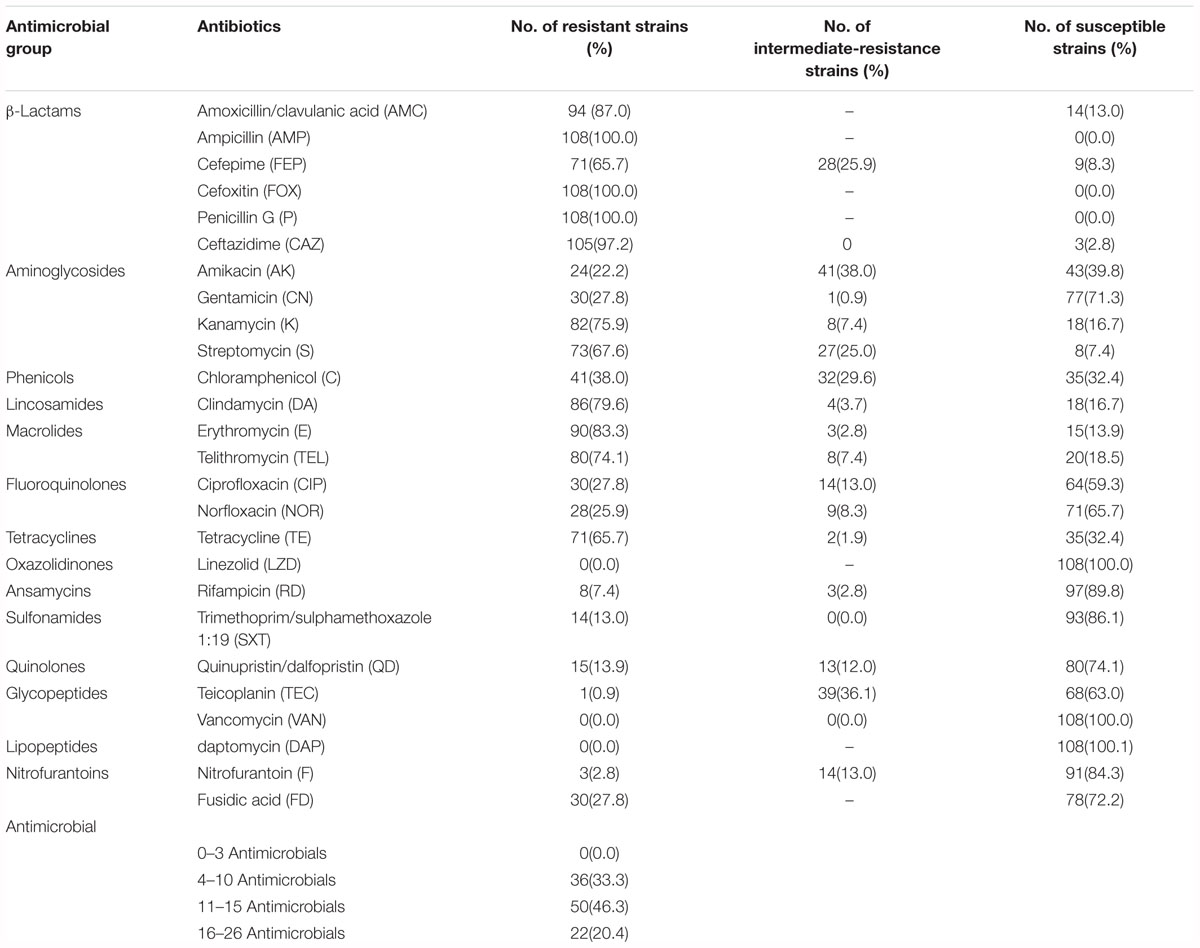
Table 3. Results of antimicrobial susceptibility tests of 108 methicillin-resistant Staphylococcus aureus isolates obtained from retail food in China.
Distribution of Virulence Genes
A total of 108 isolates of MRSA from retail food were detected for the presence of virulence genes. As shown in Figure 1, each isolate harboured at least one of the virulence genes, including 51 isolates carrying more than 10 genes. In total, 24.07% of isolates (26/108) were positive for the PVL gene lukSF-PV, whereas only 4 isolates (3.70%) were positive for the toxic shock syndrome toxin tsst alleles. Of the 18 investigated enterotoxin genes, the gene seg (82.41%, 88/108) was the most frequently detected, followed by the sei (80.56%, 86/108), seq (79.63%, 85/108), sek (77.78%, 82/108), sem (75.93%, 81/108), sec (75.00%, 79/108), sea (63.89%, 68/108), sep (63.89%, 67/108), sel (56.48%, 59/108), seb (50.93%, 54/108), ser (35.19%, 38/108), sej (34.26%, 37/108), seh (32.41%, 35/108), sen (27.78%, 30/108), seo (14.81%, 16/108), see (12.96%, 14/108), sed (12.96%, 14/108), and seu (4.63%, 5/108). In this study, 107 of 108 MRSA (99.07%) harboured one or more genes for classic SEs (sea, seb, sec, sed, and see), whereas 94 of 108 MRSA isolates (87.04%) harboured the genes of the egc cluster (seg, sei, sem, sen, seo, and seu). The classic SE genes showed 23.4% (195/833) of the detected genes, whereas the egc cluster accounted for 29.4% (245/833). Furthermore, some reported combinations of virulence genes of S. aureus were observed. The sec-sel gene combination, typical of the SaPIbov pathogenicity island, was harboured by 49.07% (53/108) of the isolates. The sea-sek-seq genes, which have been reported on ΦSa3ms and ΦSa3mw, were associated in 43.52% (47/108) of the isolates. In addition, seb-sek-seqwas observed on SaIP3, andsed-sej-serwas observed on pIB485, which also showed 47.22% (51/108) and 11.11% (12/108) of the isolates, respectively.
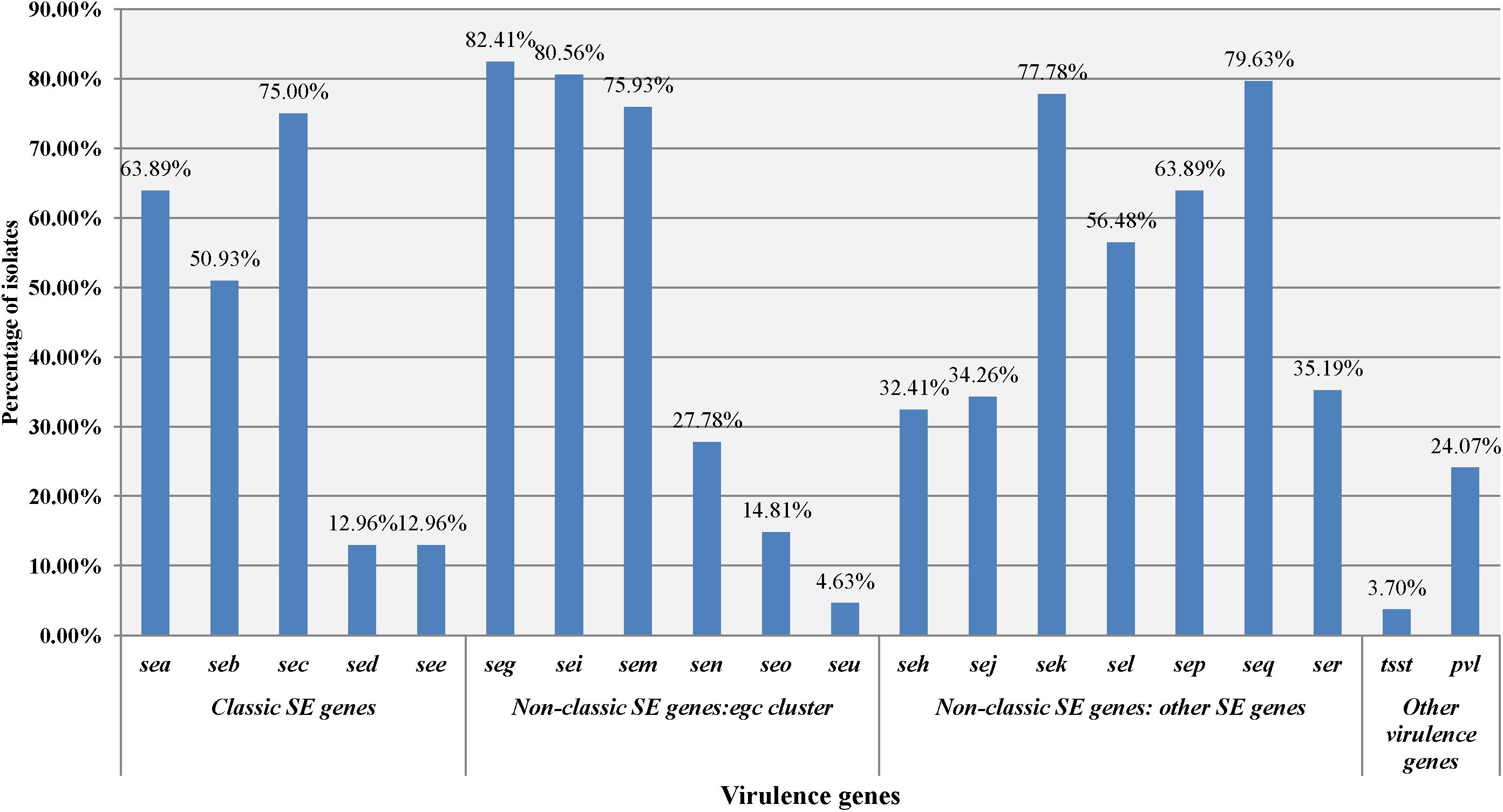
Figure 1. Distribution of the staphylococcal toxin gene profiles of MRSA isolates in different food types.
Molecular Types of MRSA Isolates
The molecular typing results are summarized in Table 4. For MLST, the STs of one strain (Sta2529-1) could not be determined. A total of 107 food-related MRSA isolates were assigned to 16 different STs, including ST1, ST5, ST6, ST7, ST9, ST10, ST45, ST59, ST88, ST188, ST338, ST398, ST630, ST943, ST3304, and ST3355. ST59 was the predominant clone and was observed in 47.7% of MRSA isolates (51/107). The remaining strains belonged to ST9 (15.9%, 17/107), ST1 (8.4%, 9/107), ST398 (6.5%, 7/107), ST7 (4.7%, 5/107), ST338 (4.7%, 5/107), ST630 (2.8%, 3/107), ST188 (1.9%, 2/107), and other STs. Based on eBURST analysis, three CCs were identified, including CC59 (ST59, ST338 and ST3355), CC7 (ST7, ST943), and CC1 (ST1, ST3304). spa-typing of all MRSA isolates yielded 24 spa types. t437, t899, t127, and t091 were the most predominant spa types, constituting 71.3% (78/108) of all of the isolates in this study. Other spa types, including t002, t034, t085, t114, t116, t163, t189, t377, t441, t528, t543, t571, t1751, t1764, t2874, t3485, t4549, t4792, t5554, and t9472, were distributed in fewer isolates (27.8%, 30/108). The MRSA isolates were also subjected to identified SCCmec types, and the types of 20 isolates could not be detected. The majority of MRSA isolates possessed SCCmec type IV, which was observed in 63.9% of isolates (69/108), including 50 for SCCmec type IVa, 17 for SCCmec type IVb and 2 for SCCmec type IVd. In addition, 9 isolates belonged to SCCmec III, and 10 isolates belonged to SCCmec V.
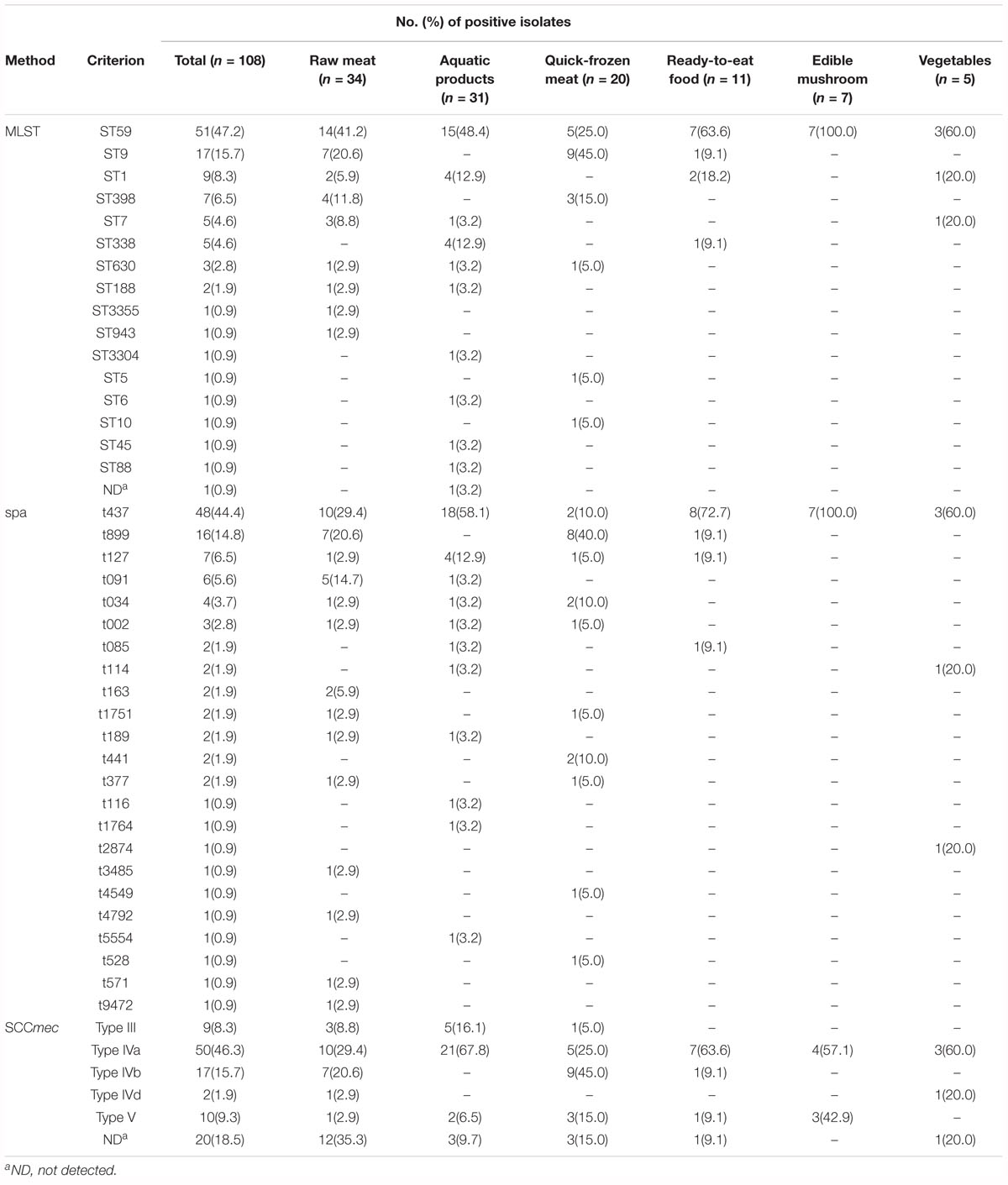
Table 4. The STs, spa types and SCCmec types of the MRSA strains isolated from retail food in China.
A phylogenetic tree based on the 7 concatenated MLST sequences (Figure 2) shows the relatedness between the MRSA strains. Two different clusters were observed in this study (designated as A and B). Cluster A included ST1, ST3304, ST188, ST9, ST15, ST6, ST5, ST7, ST943, ST630, and ST88, and cluster B included ST398, ST10, ST338, ST3355, and ST59, which showed distant genetic relationships. STs correlated well with spa types, such as ST1-t127, ST188-t189, ST9-t899, ST7-t091, ST59-t437, and ST338-t437. Overall, the genetic diversity among MRSA isolates was higher based on different cities and different food sources. For different food products, more than three subtypes were found in each type of food, except edible mushrooms and vegetables. CC59-t437 (45.4%, 49/108) was the predominant clone in this study, but ST9-t899-SCCmec IVb was the predominant clone in quick-frozen meat and was found only in animal-derived food (raw meat, quick-frozen meat and ready-to-eat meat). In addition, 80% (4/5) of SCCmec III-ST338-t437 isolates were found in aquatic products from the city of Kunming. However, some strains showed correlations among geographical locations, such as Sta223-2 (isolated from Shenzhen), Sta251 (isolated from Guangzhou), Sta403 (isolated from Shantou), Sta487 (isolated from Heyuan), and Sta1753 (isolated from Zhanjiang), which were isolated from neighboring cities in this study and clustered into one subtype (ST9-t899-SCCmec IVb).

Figure 2. The UPGMA (unweighted pair group method with arithmetic mean) tree of the 7 multilocus sequence typing loci of MRSA isolates. This tree was generated using S.T.A.R.T (version 2).
Discussion
MRSA is a significant public health concern in humans and animals. The rate of mortality due to MRSA infections has remained high in recent years. In hospitals, the prevalence rates of MRSA in some Asian countries, such as Taiwan, China, Japan, and South Korea, can reach 70–80% (Chuang and Huang, 2013). For CA-MRSA, the prevalence varies substantially worldwide and ranges from less than 1% to more than 50% in different countries (Tristan et al., 2007; Deurenberg and Stobberingh, 2008). MRSA has also been isolated from animals, as reported in many previous studies (Leonard and Markey, 2008). It is important to identify the origin of food-related MRSA and to evaluate the potential pathogenicity of these MRSA isolates. From July 2011 to June 2016, 4300 retail food samples were collected from supermarkets, fairs and farmers’ markets, covering most of the provincial capitals of China. This wide-scale and systematic investigation of S. aureus from retail food in China supplements nationwide qualitative and quantitative data on the prevalence and levels of S. aureus. In this study, we determined the MRSA isolates from these food-related S. aureus isolates and found 108 MRSA isolates (6.83%) in various types of food products (raw meat, aquatic products, quick-frozen products, ready-to-eat food, edible mushrooms and vegetables) from most of the sampling cities (29/39, 74.4%) in China, which suggested that retail food in China could be contaminated with MRSA.
Many studies have evaluated the presence of MRSA in retail food. In China, among studies that sampled retail food, 6.07% of MRSA isolates were found in quick-frozen dumpling samples of Shaanxi province (Hao et al., 2015) and in 1.7% of chicken samples (Wang et al., 2013), whereas MRSA was present in 29.5% of grain products, meat products and dairy products in southwest China. Overall, these rates varied from our results in this study. This incidence may be attributed to a number of factors, such as the sample size, sampling site, types of samples or isolation methods. In 2015, (Wang et al., 2017) collected 1150 S. aureus isolates from retail markets from 203 cities in 24 provinces in China and found 91 isolates (7.9%) that were identified as MRSA by PCR. The MRSA isolates were distributed in raw meat, rice and flour products, vegetable salads, sandwiches, meat products, and eggs. Compared with other countries’ studies, the prevalence of MRSA from retail foods in China was not low. For instance, Ge et al. (2017) conducted a 1-year survey in 2010-2011 from 3520 retail meats in eight U.S. states and found that 1.9% of samples were positive for MRSA. An Italian survey found that 6 of 160 (3.75%) foods of animal origin harbored MRSA (Normanno et al., 2007). MRSA was present in 3.6% (15/421) of retail meat in Korea (Lee, 2003), 1.6% (5/318) of food animals in Spain (Lozano et al., 2009), 0.75% (20/2662) in Switzerland (Huber et al., 2010), 3% (11/367) in Greece (Papadopoulos et al., 2018), and 1.9% (2/103) in the United Kingdom (Hadjirin et al., 2015). Therefore, Chinese food safety regulators should improve hygiene and supervision efforts.
Currently, MRSA isolated from food-producing animals has been recognized as LA-MRSA. The worldwide emergence of LA-MRSA since 2005 has prompted many of the surveys to focus on retail meat as a potential vehicle for this new MRSA clone (Ge et al., 2017). In general, ST398 was recognized as the most typical type in LA-MRSA. ST398 in swine, cattle and other animal species has been analyzed in several publications (Walther et al., 2009; Fessler et al., 2010; Graveland et al., 2011). It reported that 24–100% of pig farmers, 37% of poultry farmers, 30–38% of cattle farmers and up to 45% of veterinarians are colonized with MRSA CC398 in the nares (Köck et al., 2014). In this study, most MRSA isolates (88.9%, 96/108) were isolated from animal-derived food (raw meat, aquatic products, quick-frozen products and ready-to-eat food), but only 7 MRSA isolates belonged to this ST type. STs of MRSA isolated from retail food focused on CC59 (ST59, ST338, and ST3355). This showed significant genetic uniformity with the predominant Asian CA-MRSA lineage, which can reach 35.8–76.7% with CA-MRSA in China (Yang et al., 2017). In general, the CC59 clone always carried SCCmec IV/V and was concentrated in spa t437 and t441. In 2013, Li et al. (2013) analyzed the 110 CC59 isolates from invasive and noninvasive diseases in China by MLST, SCCmec typing and spa-typing and found that 65.5% of the clones were ST59-t437-IVa. From February 2016 to January 2017 Yang et al. (2017), collected S. aureus strains in Beijing Children’s hospital from the respiratory tract, skin and soft tissue, and sterile sites in 104 child cases. Of these, 54.8% were categorized as community-associated SA (CA-SA) infections, and ST59-SCCmec IV-t437 (61.7%) was the most prevalent MRSA genotype. In addition to China, ST59 has also been reported in Vietnam, Japan, Australia and other countries with CA-MRSA infection (Tang et al., 2007; Coombs et al., 2010; Higuchi et al., 2010). In contrast, ST239 and ST5 were found in a nationwide study to be two major MRSA clones with unique geographic distributions in Chinese hospitals (Liu et al., 2009). Accordingly, these results implied that the major food-related MRSA in China was closer to CA-MRSA, a finding that should be brought to public attention.
In the present study, 15.7% (17/108) of isolates belonged to ST9, the secondary clones of which were found only in animal-derived food (raw meat, quick-frozen meat and ready-to-eat meat). According to previous studies, ST9 was the predominant S. aureus and MRSA genotype in pigs and related workers in Asia (Chuang and Huang, 2015). In 2008, ST9 MRSA was first found in Chinese pigs, and farm workers carried ST9-t899-SCCmec III-PVL-negative (Cui et al., 2009). Studies in Taiwan, Hong Kong, Malaysia and Thailand have since reported the prevalence of this type of LA-MRSA (Neela et al., 2009; Graveland et al., 2011; Larsen et al., 2012; Lo et al., 2012). ST9 is the most prevalent LA-MRSA in most Asian countries and differs from the European pig-associated clone (ST398) with regard to clonal type, SCCmec content and resistance profile (Ye et al., 2016). In China, ST9 strains always showed SCCmec III with spa t899. These characteristics were in accordance with our results, which showed that the ST9 MRSA isolated from our study showed ST9-t899-SCCmec III. Therefore, this portion of the food-related MRSA isolates was supposed to be LA-MRSA. However, it is worth noting that in this study, all ST9 MRSA isolates were resistant to more than 15 antibiotics and showed a broader range of antimicrobial resistance than ST59 MRSA (Figure 2). More than 80% of ST9 MRSA was resistant to erythromycin, ciprofloxacin, gentamicin, tetracycline and clindamycin. Currently, more evidence has implicated animals as reservoirs of antimicrobial-resistant bacteria and has indicated that animals can potentially transmit resistance genes to humans (Liu et al., 2018). Thus, more attention should be focused on this type of strain among food chains.
Except for CC59 and ST9, the remaining strains belonged to ST1 (8.4%, 9/107), ST398 (6.5%, 7/107), ST7 (4.7%, 5/107), ST630 (2.8%, 3/107), and ST188 (1.9%, 2/107). Of these strains, most STs correlated well with spa types, but seven ST398 isolates distributed in four different spa types. In addition, SCCmec types were not detected in most (Figure 2). This finding is consistent with the results of a previous study suggesting that divergent SCCmec elements were inserted into the (clonal) ST398 MSSA (van Duijkeren et al., 2008; Smith and Pearson, 2011). Therefore, further studies evaluating MSSA ST398 in retail food are needed to determine the reason for the correlation. In this study, ST1-t127 and ST7-t091 were also detected. These STs showed high genetic diversity among MRSA isolates based on different cities and different food sources, which is a common finding in isolates of human and animal origin (Franco et al., 2011; Hummerjohann et al., 2014). Thus, these types of S. aureus isolates have been relevant to a variety of clinical infections and have theoretical pathogenic potential.
As is usually observed with CA-MRSA strains, CA-MRSA ST59 isolates had significantly more pronounced virulence in various animal infection models than the geographically matched HA-MRSA clones ST5 and ST239 (Li et al., 2009, 2016). For the strains of ST59, the evolutionary acquisition of PVL, the higher expression of α-toxin and, possibly, the loss of a large portion of the β-haemolysin-converting prophage probably contribute to its higher pathogenic potential (Chen et al., 2013; Chen and Huang, 2014). In this study, we also investigated the PVL gene and found 24.07% PVL-positive MRSA isolates. All PVL-positive MRSA isolates belonged to CC59, including 22 ST59 isolates and four ST338 isolates. For LA-MRSA ST9 isolates, certain important virulence factors, such as PVL, are absent. Despite a lack of virulence factors, ST9 strains have been found to cause disease in humans (van Loo et al., 2007; Liu et al., 2009; Chuang and Huang, 2015). Therefore, the hazards of these strains for consumers cannot be ignored.
As an important foodborne pathogen, S. aureus is involved in most staphylococcal food poisoning (SFP) events due to staphylococcal enterotoxins, the virulence factors that are heat stable and proteolytic or demonstrate emetic activity (Grumann et al., 2014). In this study, we also investigated most of the enterotoxin genes of S. aureus. All food-related MRSA isolates harbored at least one of the SE genes. The percentages of MRSA isolates containing sea, seb, and sec all exceeded 50%, whereas sed and see were detected in only fourteen MRSA isolates (12.96%). Except for the egc cluster, which accounted for 29.4% (245/833) of the detected genes in the present study, other SE genes showed 48.4% (403/833) of the detected genes. Generally, SEA, followed by SED, is the enterotoxin most frequently associated with SFP, although outbreaks caused by SEB, SEC, and SEE have also been reported (Argudín et al., 2012). In contrast to classical SEs, the relationship between the novel SEs and SFP is not fully understood, but most of them (SEG, SEH and SEI, SER, SES, and SET) have been shown to be emetic after oral administration in a primate model (Argudín et al., 2010). In this study, 94 of 108 MRSA isolates (87.04%) harbored egc cluster genes (seg, sei, sem, sen, seo, and seu), and 94.4% (102/108) of isolates harbored one or more genes for other novel SEs or tsst-1 genes. It is suggested that attention should not only be paid to classical enterotoxins but also to novel ones since an increasing number of foodborne outbreaks have been associated with novel enterotoxins.
Nowadays, multiple drug resistance is the most important characteristic of MRSA isolates. For the 26 clinically relevant antibiotics investigated, all MRSA isolates were resistant to more than three antibiotics. It seems higher than many previous studies in food-related MRSA isolates (Hanson et al., 2011; Shahraz et al., 2012; Ge et al., 2017; Tang et al., 2017). Interestingly, norfloxacin, clindamycin, trimethoprim/sulphamethoxazole 1:19, gentamycin and quinupristin/dalfopristin resistance were rare in MRSA isolated from non-animal sources (edible mushrooms and vegetables) (Supplementary Figure 2). The reason for this finding may be attributed to the food source. In general, most animal-derived food-related S. aureus came from animal farms that used these antibiotics as food supplements in animal feed (Wang et al., 2014), whereas S. aureus isolated from edible mushrooms or vegetables most likely originated in the environment. Furthermore, amikacin was the most commonly observed resistance antibiotic in aquatic product-related MRSA isolates, whereas gentamycin, quinupristin/dalfopristin, ciprofloxacin, and norfloxacin resistances were observed less frequently than other types of food isolates (Supplementary Figure 2). As we know, amikacin, gentamycin, kanamycin and streptomycin all belong to the aminoglycosides, which exert their bactericidal effects by irreversibly binding to the 30S ribosomal subunits of susceptible bacteria, inhibiting protein synthesis (Hammerberg et al., 1986). The most common clinical resistance mechanism to aminoglycosides is the structural modification of aminoglycosides by aminoglycoside-modifying enzymes (AMEs). Amikacin is a broad-spectrum semi-synthetic derivative of kanamycin and a poor substrate of many AMEs. Furthermore, even if amikacin is modified by AMEs, the modified amikacin can still bind to the 30S ribosomal subunit (Yuan et al., 2013). Therefore, amikacin is one of the most potent classes of antibiotics in S. aureus infection. In fact, the mechanism of MRSA resistance to amikacin is poorly understood. Thus, why amikacin resistance was higher in MRSA isolates from aquatic products should be further studied.
Conclusion
Essential for human survival, food is the one of most basic necessities of life. In this study, we investigated food-related MRSA and determined its genetic background in China. MRSA isolates were found in most investigated cities and were observed in different types of food samples. ST59 was the predominant clone in food-related MRSA in China, which indicated that the major food-related MRSA isolates in China were closer to CA-MRSA. Moreover, as the major LA-MRSA in Asian populations, ST9 MRSA was the secondary clone and showed a broader range of antimicrobial resistance. Determination of staphylococcal toxin genes presented their virulence potential, and antimicrobial susceptibility testing further confirmed the severe situation of MRSA isolated in retail food in China. In addition, some antibiotics were also found to be higher in some types of food. However, further studies are required to determine the reason for this correlation and to elucidate the transmission routes of MRSA in relation to retail foods in order to provide the tools for preventing the spread of MRSA.
Author Contributions
QW, JZ, SW, and TL conceived and designed the experiments. JH, FZ, and JD performed the experiments. SW, RP, and HZ analyzed the data. XW, LX, MC, and XY contributed reagents, materials, and analysis tools. SW and JW contributed to the writing of the manuscript.
Funding
We would like to acknowledge the financing support of National Natural Science Foundation of China (No. 31801657), China Postdoctoral Science Foundation (2017M612623), and GDAS’ Special Project of Science and Technology Development (2017GDASCX-0817).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.00304/full#supplementary-material
Footnotes
References
An, B., and Otto, M. (2008). The role of virulence determinants in community-associated MRSA pathogenesis. Trends Microbiol. 16, 361–369. doi: 10.1016/j.tim.2008.05.002
Argudín, M. A., Mendoza, M. C., González-Hevia, M. A., Bances, M., Guerra, B., and Rodicio, M. R. (2012). Genotypes, exotoxin gene content, and antimicrobial resistance of Staphylococcus aureus strains recovered from foods and food handlers. Appl. Environ. Microbiol. 78, 2930–2935. doi: 10.1128/AEM.07487-11
Argudín, M. Á, Mendoza, M. C., and Rodicio, M. R. (2010). Food poisoning and Staphylococcus aureus enterotoxins. Toxins 2:1751. doi: 10.3390/toxins2071751
Banerjee, R., Gretes, M., Li, B., Strynadka, N., and Chambers, F. H. (2008). In vitro selection and characterization of ceftobiprole-resistant methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 52, 2089–2096. doi: 10.1128/AAC.01403-07
Bens, C. C., Voss, A., and Klaassen, C. H. (2006). Presence of a novel DNA methylation enzyme in methicillin-resistant Staphylococcus aureus isolates associated with pig farming leads to uninterpretable results in standard pulsed-field gel electrophoresis analysis. J. Clin. Microbiol. 44, 1875–1876. doi: 10.1128/JCM.44.5.1875-1876.2006
Chen, C.-J., and Huang, Y.-C. (2014). New epidemiology of Staphylococcus aureus infection in Asia. Clin. Microbiol. Infect. 20, 605–623. doi: 10.1111/1469-0691.12705
Chen, C. J., Unger, C., Hoffmann, W., Lindsay, J. A., Huang, Y. C., and Götz, F. (2013). Characterization and comparison of 2 distinct epidemic community-associated methicillin-resistant Staphylococcus aureus clones of ST59 lineage. PLoS One 8:e63210. doi: 10.1371/journal.pone.0063210
Chuang, Y. Y., and Huang, Y. C. (2013). Molecular epidemiology of community-associated meticillin-resistant Staphylococcus aureus in Asia. Lancet Infect. Dis. 13:698. doi: 10.1016/S1473-3099(13)70136-1
Chuang, Y. Y., and Huang, Y. C. (2015). Livestock-associated meticillin-resistant Staphylococcus aureus in Asia: an emerging issue? Int. J. Antimicrob. Agents 45, 334–340. doi: 10.1016/j.ijantimicag.2014.12.007
Coombs, G. W., Monecke, S., Ehricht, R., Slickers, P., Pearson, J. C., Tan, H.-L., et al. (2010). Differentiation of clonal complex 59 community-associated methicillin-resistant Staphylococcus aureus in Western Australia. Antimicrob. Agents Chemother. 54, 1914–1921. doi: 10.1128/aac.01287-09
Cui, S., Li, J., Hu, C., Jin, S., Li, F., Guo, Y., et al. (2009). Isolation and characterization of methicillin-resistant Staphylococcus aureus from swine and workers in China. J. Antimicrob. Chemother. 64, 680–683. doi: 10.1093/jac/dkp275
Cui, S., and Li, J. C. (2009). Isolation and characterization of methicillin-resistant Staphylococcus aureus from swine and workers in China. J. Antimicrob. Chemother. 64, 680–683. doi: 10.1093/jac/dkp275
Deleo, F. R., Otto, M., Kreiswirth, B. N., and Chambers, H. F. (2010). Community-associated meticillin-resistant Staphylococcus aureus. Lancet 375, 767–768. doi: 10.1016/S0140-6736(10)61370-0
Deurenberg, R. H., and Stobberingh, E. E. (2008). The evolution of Staphylococcus aureus. Infect. Genet. Evol. 8, 747–763. doi: 10.1016/j.meegid.2008.07.007
Enright, M. C., Day, N. P. J., Davies, C. E., Peacock, S. J., and Spratt, B. G. (2013). Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 51, 306–310. doi: 10.1128/JCM.02421-12
Feil, E. J., Li, B. C., Aanensen, D. M., Hanage, W. P., and Spratt, B. G. (2004). eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 186, 1518–1530. doi: 10.1128/JB.186.5.1518-1530.2004
Fessler, A., Scott, C., Kadlec, K., Ehricht, R., Monecke, S., and Schwarz, S. (2010). Characterization of methicillin-resistant Staphylococcus aureus ST398 from cases of bovine mastitis. J. Antimicrob. Chemother. 65:619. doi: 10.1093/jac/dkq021
Franco, A., Hasman, H., Iurescia, M., Lorenzetti, R., Stegger, M., Pantosti, A., et al. (2011). Molecular characterization of spa type t127, sequence type 1 methicillin-resistant Staphylococcus aureus from pigs. J. Antimicrob. Chemother. 66, 1231–1235. doi: 10.1093/jac/dkr115
Garcíaálvarez, L., Holden, M. T., Lindsay, H., Webb, C. R., Brown, D. F., Curran, M. D., et al. (2011). Meticillin-resistant Staphylococcus aureus with a novel mecA homologue in human and bovine populations in the UK and Denmark: a descriptive study. Lancet Infect. Dis. 11, 595–603. doi: 10.1016/S1473-3099(11)70126-8
Ge, B., Mukherjee, S., Hsu, C. H., Davis, J. A., Tran, T. T. T., Yang, Q., et al. (2017). MRSA and multidrug-resistant Staphylococcus aureus in U.S. retail meats, 2010–2011. Food Microbiol. 62, 289–297. doi: 10.1016/j.fm.2016.10.029
Gombas, D. E., Chen, Y., Clavero, R. S., and Scott, V. N. (2003). Survey of listeria monocytogenes in ready-to-eat foods. J. Food Prot. 66, 559–569. doi: 10.4315/0362-028X-66.4.559
Graveland, H., Duim, B., Van, D. E., Heederik, D., and Wagenaar, J. A. (2011). Livestock-associated methicillin-resistant Staphylococcus aureus in animals and humans. Int. J. Med. Microbiol. 301, 630–634. doi: 10.1016/j.ijmm.2011.09.004
Grumann, D., Nübel, U., and Bröker, B. M. (2014). Staphylococcus aureus toxins–their functions and genetics. Infect. Genet. Evol. 21, 583–592. doi: 10.1016/j.meegid.2013.03.013
Hadjirin, N. F., Lay, E. M., Paterson, G. K., Harrison, E. M., Peacock, S. J., Parkhill, J., et al. (2015). Detection of livestock-associated meticillin-resistant Staphylococcus aureus CC398 in retail pork, United Kingdom, February 2015. Euro Surveill. 20:21156. doi: 10.2807/1560-7917.ES2015.20.24.21156
Hallin, M., De Mendonça, R., Denis, O., Lefort, A., El Garch, F., Butaye, P., et al. (2011). Diversity of accessory genome of human and livestock-associated ST398 methicillin resistant Staphylococcus aureus strains. Infect. Genet. Evol. 11, 290–299. doi: 10.1016/j.meegid.2010.10.021
Hammerberg, O., Elder, D., Richardson, H., and Landis, S. (1986). Staphylococcal resistance to aminoglycosides before and after introduction of amikacin in two teaching hospitals. J. Clin. Microbiol. 24, 629–632.
Hanson, B. M., Dressler, A. E., Harper, A. L., Scheibel, R. P., Wardyn, S. E., Roberts, L. K., et al. (2011). Prevalence of Staphylococcus aureus and methicillin-resistant Staphylococcus aureus (MRSA) on retail meat in Iowa. J. Infect. Public Health 4, 169–174. doi: 10.1016/j.jiph.2011.06.001
Hao, D., Xing, X., Li, G., Wang, X., Zhang, M., Zhang, W., et al. (2015). Prevalence, toxin gene profiles, and antimicrobial resistance of Staphylococcus aureus isolated from quick-frozen dumplings. J. Food Prot. 78, 218–223. doi: 10.4315/0362-028X.JFP-14-100
Higuchi, W., Hung, W. C., Takano, T., Iwao, Y., Ozaki, K., Isobe, H., et al. (2010). Molecular characteristics of the taiwanese multiple drug-resistant ST59 clone of panton-valentine leucocidin-positive community-acquired methicillin-resistant Staphylococcus aureus from pediatric cellulitis. J. Infect. Chemother. 16, 144–149. doi: 10.1007/s10156-010-0029-9
Huber, H., Koller, S., Giezendanner, N., Stephan, R., and Zweifel, C. (2010). Prevalence and characteristics of meticillin-resistant Staphylococcus aureus in humans in contact with farm animals, in livestock, and in food of animal origin, Switzerland, 2009. Euro Surveill. 15:19541.
Hummerjohann, J., Naskova, J., Baumgartner, A., and Graber, H. U. (2014). Enterotoxin-producing Staphylococcus aureus genotype B as a major contaminant in Swiss raw milk cheese. J. Dairy Sci. 97, 1305–1312. doi: 10.3168/jds.2013-7643
Jarraud, S., Mougel, C., Thioulouse, J., Lina, G., Meugnier, H., Forey, F., et al. (2002). Relationships between Staphylococcus aureus genetic background, virulence factors, agr groups (Alleles), and human disease. Infect. Immun. 70, 631–641. doi: 10.1128/IAI.70.2.631-641.2002
Kobayashi, S. D., Malachowa, N., and Deleo, F. R. (2015). Pathogenesis of Staphylococcus aureus abscesses. Am. J. Pathol. 185, 1518–1527. doi: 10.1016/j.ajpath.2014.11.030
Köck, R., Ballhausen, B., Bischoff, M., Cuny, C., Eckmanns, T., Fetsch, A., et al. (2014). The impact of zoonotic MRSA colonization and infection in Germany. Berl Munch. Tierarztl. Wochenschr. 127, 384–398.
Larsen, J., Imanishi, M., Hinjoy, S., Tharavichitkul, P., Duangsong, K., Davis, M. F., et al. (2012). Methicillin-resistant Staphylococcus aureus ST9 in pigs in Thailand. PLoS One 7:e31245. doi: 10.1371/journal.pone.0031245
Lee, J. H. (2003). Methicillin (Oxacillin)-resistant Staphylococcus aureus strains isolated from major food animals and their potential transmission to humans. Appl. Environ. Microbiol. 69, 6489–6494. doi: 10.1128/AEM.69.11.6489-6494.2003
Leonard, F. C., and Markey, B. K. (2008). Meticillin-resistant Staphylococcus aureus in animals: a review. Vet. J. 175, 27–36. doi: 10.1016/j.tvjl.2006.11.008
Li, J., Wang, L., Ip, M., Sun, M., Sun, J., Huang, G., et al. (2013). Molecular and clinical characteristics of clonal complex 59 methicillin-resistant Staphylococcus aureus infections in mainland China. PLoS One 8:e70602. doi: 10.1371/journal.pone.0070602
Li, M., Dai, Y., Zhu, Y., Fu, C. L., Tan, V. Y., Wang, Y., et al. (2016). Virulence determinants associated with the Asian community-associated methicillin-resistantStaphylococcus aureus lineage ST59. Sci. Rep. 6:27899. doi: 10.1038/srep27899
Li, M., Diep, B. A., Villaruz, A. E., Braughton, K. R., Jiang, X., Deleo, F. R., et al. (2009). Evolution of virulence in epidemic community-associated methicillin-resistant Staphylococcus aureus. Proc. Natl. Acad. Sci. U.S.A. 106, 5883–5888. doi: 10.1073/pnas.0900743106
Li, S., Skov, R. L., Han, X., Larsen, A. R., Larsen, J., Sørum, M., et al. (2011). Novel types of staphylococcal cassette chromosome mec elements identified in clonal complex 398 methicillin-resistant Staphylococcus aureus strains. Antimicrob. Agents Chemother. 55, 3046–3050. doi: 10.1128/AAC.01475-10
Liu, C. M., Stegger, M., Aziz, M., Johnson, T. J., Waits, K., Nordstrom, L., et al. (2018). Escherichia coli ST131-H22 as a foodborne uropathogen. mBio 9:e00470-18 doi: 10.1128/mBio.00470-18
Liu, Y., Wang, H., Du, N., Shen, E., Chen, H., Niu, J., et al. (2009). Molecular evidence for spread of two major methicillin-resistant staphylococcus aureus clones with a unique geographic distribution in chinese hospitals. Antimicrob. Agents Chemother. 53, 512–518. doi: 10.1128/aac.00804-08
Lo, Y. P., Wan, M. T., Chen, M. M., Su, H. Y., Lauderdale, T. L., and Chou, C. C. (2012). Molecular characterization and clonal genetic diversity of methicillin-resistant Staphylococcus aureus of pig origin in Taiwan. Comp. Immun. Microbiol. Infect. Dis. 35, 513–521. doi: 10.1016/j.cimid.2012.05.001
Lozano, C., López, M., Gómezsanz, E., Ruizlarrea, F., Torres, C., and Zarazaga, M. (2009). Detection of methicillin-resistant Staphylococcus aureus ST398 in food samples of animal origin in Spain. J. Antimicrob. Chemother. 64, 1325–1326. doi: 10.1093/jac/dkp378
Neela, V., Zafrul, A. M., Mariana, N. S., Belkum, A. V., Yun, K. L., and Rad, E. G. (2009). Prevalence of ST9 methicillin-resistant Staphylococcus aureus among pigs and pig handlers in Malaysia. J. Clin. Microbiol. 47, 4138–4140. doi: 10.1128/JCM.01363-09
Normanno, G., Corrente, M., Salandra, G. L., Dambrosio, A., Quaglia, N. C., Parisi, A., et al. (2007). Methicillin-resistant Staphylococcus aureus (MRSA) in foods of animal origin product in Italy. Int. J. Food Microbiol. 117, 219–222. doi: 10.1016/j.ijfoodmicro.2007.04.006
Papadopoulos, P., Papadopoulos, T., Angelidis, A. S., Boukouvala, E., Zdragas, A., Papa, A., et al. (2018). Prevalence of, Staphylococcus aureus, and of methicillin-resistant, S.?aureus, (MRSA) along the production chain of dairy products in north-western Greece. Food Microbiol. 69, 43–50. doi: 10.1016/j.fm.2017.07.016
Perez-Roth, E., Claverie-Martın, F., Villar, J., and Mendez-Alvarez, S. (2001). Multiplex PCR for simultaneous identification of Staphylococcus aureus and detection of methicillin and mupirocin resistance. J. Clin. Microbiol. 39, 4037–4041. doi: 10.1128/JCM.39.11.4037-4041.2001
Petinaki, E., and Spiliopoulou, I. (2012). Methicillin-resistant Staphylococcus aureus among companion and food-chain animals: impact of human contacts. Clin. Microbiol. Infect. 18, 626–634. doi: 10.1111/j.1469-0691.2012.03881.x
Shahraz, F., Dadkhah, H., Khaksar, R., Mahmoudzadeh, M., Hosseini, H., Kamran, M., et al. (2012). Analysis of antibiotic resistance patterns and detection of mecA gene in Staphylococcus aureus isolated from packaged hamburger. Meat Sci. 90, 759–763. doi: 10.1016/j.meatsci.2011.11.009
Shopsin, B., Gomez, M., Montgomery, S. O., Smith, D. H., Waddington, M., Dodge, D. E., et al. (1999). Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J. Clin. Microbiol. 37, 3556–3563.
Skov, R. L. (2009). Community-associated meticillin-resistant Staphylococcus aureus as a cause of hospital-acquired infections. J. Hosp. Infect. 73, 364–370. doi: 10.1016/j.jhin.2009.07.004
Smith, T. C., and Pearson, N. (2011). The emergence of Staphylococcus aureus ST398. Vector Borne Zoonotic Dis. 11, 327–339. doi: 10.1089/vbz.2010.0072
Stegger, M., Andersen, P. S., Kearns, A., Pichon, B., Holmes, M. A., Edwards, G., et al. (2012). Rapid detection, differentiation and typing of methicillin-resistant Staphylococcus aureus harbouring either mecA or the new mecA homologue mecA LGA251. Clin. Microbiol. Infect. 18, 395–400. doi: 10.1111/j.1469-0691.2011.03715.x
Tang, C. T., Tho, N. D., Hoa, N. T., Phuong, N. T. M., Le, V. T., Diep, T. S., et al. (2007). An outbreak of severe infections with community-acquired MRSA carrying the panton-valentine leukocidin following vaccination. PLoS One 2:e822. doi: 10.1371/journal.pone.0000822
Tang, Y., Larsen, J., Kjeldgaard, J., Andersen, P. S., Skov, R., and Ingmer, H. (2017). Methicillin-resistant and -susceptible Staphylococcus aureus from retail meat in Denmark. Int. J. Food Microbiol. 249, 72–76. doi: 10.1016/j.ijfoodmicro.2017.03.001
The Clinical, and Laboratory Standards Institute [Clsi]. (2015). Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fifth Informational Supplement. Approved Standard-M02-A12. Wayne, PA: The Clinical and Laboratory Standards Institute.
Tristan, A., Bes, M., Meugnier, H., Lina, G., Bozdogan, B., Courvalin, P., et al. (2007). Global distribution of panton-valentine leukocidin–positive methicillin-resistant Staphylococcus aureus, 2006. Emerg. Infect. Dis. 13, 594–600. doi: 10.3201/eid1304.061316
van Duijkeren, E. V., Ikawaty, R., Broekhuizen-Stins, M. J., Jansen, M. D., Spalburg, E. C., de Neeling, A. J., et al. (2008). Transmission of methicillin-resistant Staphylococcus aureus strains between different kinds of pig farms. Vet. Microbiol. 126, 383–389. doi: 10.1016/j.vetmic.2007.07.021
van Loo, I. H., Diederen, B. M., Savelkoul, P. H., Woudenberg, J. H., Roosendaal, R., Van, B. A., et al. (2007). Methicillin-resistant Staphylococcus aureus in meat products, the Netherlands. Emerg. Infect. Dis. 13, 1753–1755. doi: 10.3201/eid1311.070358
Varshney, A. K., Mediavilla, J. R., Robiou, N., Guh, A., Wang, X., Gialanella, P., et al. (2009). Diverse enterotoxin gene profiles among clonal complexes of Staphylococcus aureus isolates from the Bronx, New York. Appl. Environ. Microbiol. 75, 6839–6849. doi: 10.1128/AEM.00272-09
Walther, B., Monecke, S., Ruscher, C., Friedrich, A. W., Ehricht, R., Slickers, P., et al. (2009). Comparative molecular analysis substantiates zoonotic potential of equine methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 47, 704–710. doi: 10.1128/JCM.01626-08
Wang, W., Baloch, Z., Jiang, T., Zhang, C., Peng, Z., Li, F., et al. (2017). Enterotoxigenicity and antimicrobial resistance of Staphylococcus aureus isolated from retail food in China. Front. Microbiol. 8:2256. doi: 10.3389/fmicb.2017.02256
Wang, X., Li, G., Xia, X., Yang, B., Xi, M., and Meng, J. (2014). Antimicrobial susceptibility and molecular typing of methicillin-resistant Staphylococcus aureus in retail foods in Shaanxi, China. Foodborne Pathog. Dis. 11, 281–286. doi: 10.1089/fpd.2013.1643
Wang, X., Tao, X., Xia, X., Yang, B., Xi, M., Meng, J., et al. (2013). Staphylococcus aureus and methicillin-resistant Staphylococcus aureus in retail raw chicken in China. Food Control 29, 103–106. doi: 10.1016/j.foodcont.2012.06.002
Wu, S., Huang, J., Wu, Q., Zhang, F., Zhang, J., Lei, T., et al. (2018). Prevalence and characterization of Staphylococcus aureus isolated from retail vegetables in China. Front. Microbiol. 9:1263. doi: 10.3389/fmicb.2018.01263
Yang, X., Qian, S., Yao, K., Wang, L., Liu, Y., Dong, F., et al. (2017). Multiresistant ST59-SCCmecIV-t437 clone with strong biofilm-forming capacity was identified predominantly in MRSA isolated from Chinese children. BMC Infect. Dis. 17:733. doi: 10.1186/s12879-017-2833-7
Ye, X., Wang, X., Fan, Y., Peng, Y., Li, L., Li, S., et al. (2016). Genotypic and phenotypic markers of livestock-associated methicillin-resistant Staphylococcus aureus CC9 in humans. Appl. Environ. Microbiol. 82, 3892–3899. doi: 10.1128/AEM.00091-16
Yuan, W., Hu, Q., Cheng, H., Shang, W., Liu, N., Hua, Z., et al. (2013). Cell wall thickening is associated with adaptive resistance to amikacin in methicillin-resistant Staphylococcus aureus clinical isolates. J. Antimicrob. Chemother. 68, 1089–1096. doi: 10.1093/jac/dks522
Zhang, K., Mcclure, J. A., Elsayed, S., Louie, T., and Conly, J. M. (2005). Novel multiplex PCR assay for characterization and concomitant subtyping of staphylococcal cassette chromosome mec types I to V in methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 43, 5026–5033. doi: 10.1128/JCM.43.10.5026-5033.2005
Keywords: MRSA, antibiotic resistance, retail food, spa-typing, MLST
Citation: Wu S, Huang J, Zhang F, Wu Q, Zhang J, Pang R, Zeng H, Yang X, Chen M, Wang J, Dai J, Xue L, Lei T and Wei X (2019) Prevalence and Characterization of Food-Related Methicillin-Resistant Staphylococcus aureus (MRSA) in China. Front. Microbiol. 10:304. doi: 10.3389/fmicb.2019.00304
Received: 12 November 2018; Accepted: 05 February 2019;
Published: 20 February 2019.
Edited by:
Learn-Han Lee, Monash University Malaysia, MalaysiaReviewed by:
Santiago Castillo Ramírez, National Autonomous University of Mexico, MexicoLiang Li, Los Angeles Biomedical Research Institute, United States
Copyright © 2019 Wu, Huang, Zhang, Wu, Zhang, Pang, Zeng, Yang, Chen, Wang, Dai, Xue, Lei and Wei. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qingping Wu, wuqp203@163.com
†These authors have contributed equally to this work
 Shi Wu
Shi Wu Jiahui Huang1,2†
Jiahui Huang1,2† Qingping Wu
Qingping Wu Jumei Zhang
Jumei Zhang Rui Pang
Rui Pang Haiyan Zeng
Haiyan Zeng Moutong Chen
Moutong Chen Juan Wang
Juan Wang Liang Xue
Liang Xue