- 1College of Food Science, Northeast Agricultural University, Harbin, China
- 2Green Food Research Institute of Heilongjiang (National Research Center of Dairy Engineering and Technology), Northeast Agricultural University, Harbin, China
- 3Food Department, Heilongjiang Administration for Market Regulation, Harbin, China
- 4Department of Food Science, Rutgers, The State University of New Jersey, New Brunswick, NJ, United States
The aim of this study was to investigate the prevalence and genotypic characteristics of Cronobacter isolated from powdered infant formula (PIF) manufacturing facilities and to identify a potential source of contamination. A total of 42 Cronobacter isolates (5%) were detected in 835 environmental samples collected during the surveillance study. These isolates included C. sakazakii (n = 37), C. malonaticus (n = 3), and C. turicensis (n = 2). The isolates were divided into 14 sequence types (STs) by multi-locus sequence typing (MLST) and 21 pulsotypes (PTs) using pulsed-field gel electrophoresis (PFGE). The dominant C. sakazakii sequence types were ST3 (n = 12) and ST21 (n = 10), followed by ST136 (n = 6). The major PTs were PT22 (n = 12) and PT17 (n = 4) based on 100% similarity. Strains isolated from samples collected at the same production facility showed closer phylogenetic relation than those collected from distinct facilities. The result of extensive traceback sampling showed that PIF residues (PIF dust in production areas), fluid beds, drying areas, floors, and soil samples collected adjacent to the production facilities were the primary positive areas for Cronobacter. The present study outlines an effective approach to determine prevalence and genetic diversity of Cronobacter isolates associated with contamination of PIF.
Introduction
Cronobacter spp. belongs to the family Enterobacteriaceae, which were previously classified as Enterobacter sakazakii, consisting of C. sakazakii, C. malonaticus, C. turicensis, C. muytjensii, C. condimenti, C. universalis, and C. dublinensis (Stephan et al., 2014). Cronobacter infections in infants can lead to necrotizing enterocolitis, septicemia, meningitis, and death (FAO/WHO 2004, 2006, 2008). Powdered infant formula (PIF) is a major vehicle for Cronobacter infections in neonates (Stephan et al., 2011; Hunter and Bean, 2013). Building on information related to Cronobacter prevalence and genetic diversity will prove valuable in conducting risk analyses to identify the potential sources of Cronobacter during production of PIF.
A plethora of studies have shown that Cronobacter spp. can be isolated from PIF and PIF manufacturing facilities (Iversen et al., 2009; Craven et al., 2010; Fei et al., 2018b). Mullane et al. (2007) profiled Cronobacter spp. isolates by pulsed-field gel electrophoresis (PFGE) from a PIF production site and demonstrated the persistence of specific bacterial clones in the production facility. Proudy et al. (2008) highlighted that the environment is the major source of Cronobacter contamination of PIF and PIF production facilities. However, the contamination source and transmission routes of Cronobacter may differ depending on the production process and country of production. In depth studies focused on sources of Cronobacter within production environments are limited; most sampling is conducted on finished products.
In the present study, traceback sampling of the PIF production facility environment was conducted to determine: (1) areas within a production facility and area(s) immediately outside the facility that may harbor Cronobacter, (2) identify processing step(s) that represent a greatest risk for introduction of Cronobacter into PIF, and (3) determine prevalence and genotypic characterization of Cronobacter. A total of 835 environmental samples were collected and 42 Cronobacter strains were identified. Assessment of the genetic diversity of Cronobacter isolates was conducted using multi-locus sequence typing (MLST) and pulsed-field gel electrophoresis (PFGE) to determine whether single or multiple strains were dominant in a given PIF production facility and to establish potential sources of contamination.
Materials and Methods
Sample Collection
A sampling scheme was developed to cover all the potential source sites of contamination within a PIF production facility collectively referred to as environmental samples. A total of 835 samples were collected from four PIF production facilities in China (Table 1). The environmental samples were collected from clean work areas and quasi work areas (including the spray-drying areas along the entire production line), PIF residues (PIF dust that typically settles on the environmental surfaces) on the equipment surfaces and the floor. In addition to samples collected from the indoor production environment, areas outside the production facility were sampled including air conditioning units, water, and soil. The uniforms, hands, and gloves of technicians conducting the sampling were also sampled.
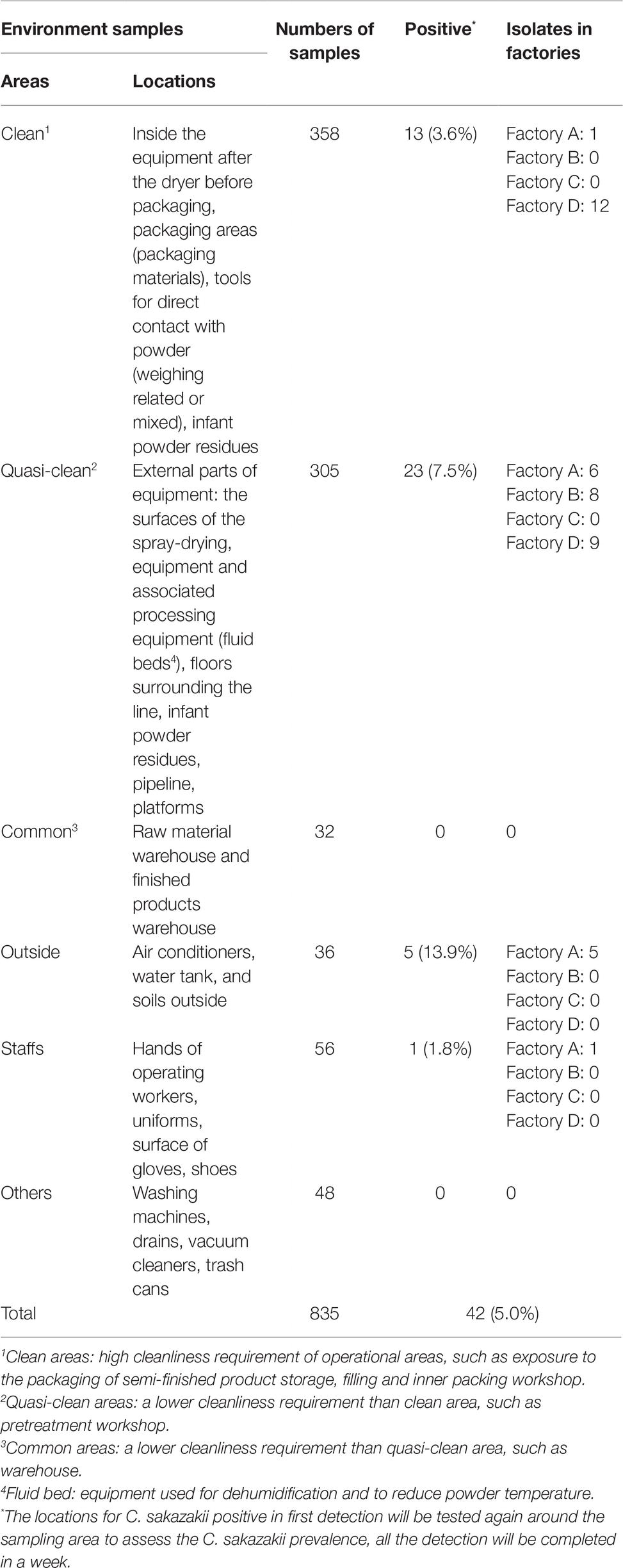
Table 1. Occurrence of Cronobacter strains of environmental samples in four infant formula production factories in China.
A sterile sponge-stick was used to swab the surface of a defined area (100 cm2 or 1 m2) of the floor, the equipment, and the packaging materials. A sterile swab was used to sample a defined area (20 cm2) of irregular surfaces and the entire area of a hand, or inner wall of a PIF can. Sponges and swabs were returned to the laboratory and processed within 4 h. Surfaces were disinfected with ethanol and dried after sampling.
Isolation and Identification of Cronobacter Strains
Processing of samples was conducted as follows: (1) sponge samples were added into 100 ml of sterile buffered peptone water (BPW, LandBridge, China), mixed well, then incubated for 18 ± 2 h at 36°C; (2) swab samples containing 10 ml of sterile BPW were incubated directly for 18 ± 2 h at 36°C; and (3) PIF residue samples were mixed with BPW to yield a tenfold dilution for pre-enrichment (36°C for 18 ± 2 h). After incubation, a 1 ml aliquot of the pre-enrichment culture was transferred into 10 ml of modified lauryl sulfate tryptose (mLST, LandBridge, China)/vancomycin medium and then incubated at 44 ± 0.5°C for 24 ± 2 h. All samples were streaked onto the CHROMagar™ Cronobacter spp. agar (CHROMagar, France) and incubated at 37°C for 24 ± 2 h. Presumptive colonies of Cronobacter were selected, and streaked onto tryptone soy agar (TSA, LandBridge, China), followed by incubation at 25°C for 44–48 h. After incubation, presumptive Cronobacter colonies were selected to confirm their identity using biochemical identification and 16S rRNA gene sequencing (Chen et al., 2010; ISO 22964, 2017).
All the confirmed Cronobacter isolates were cryopreserved at −80°C in 40% (v/v) glycerol in water. Prior to analysis of MLST and PFGE, isolates were cultured in BPW at 36°C for 18 ± 2 h, streaked onto TSA and incubated at 36°C for 24 h to isolate a single colony.
MLST Subtyping of Cronobacter
Genomic DNA was extracted using a Tsingke Bacteria DNA kit (TsingKe Biotech Co., Ltd., Beijing, China). Primers for the seven housekeeping genes (atpD, fusA, glnS, gltB, gyrB, infB, and ppsA) and PCR was performed according to Joseph et al. (2012) and Cui et al. (2015). The PCR products were sequenced by TsingKe Biotech Co., Ltd. (Beijing, China). The assignments of each allele and sequence types (STs) profile were obtained from the Cronobacter MLST open-access database (http://pubmlst.org/Cronobacter/). The phylogenetic relationship of the concatenated sequences (3,036 bp) of the seven housekeeping genes was analyzed using the maximum-likelihood algorithm in MEGA (version 6) with 1,000 bootstrap replicates. A minimum spanning tree was constructed to analyze the relatedness of all isolates using BioNumerics (version 5).
PFGE Subtyping of Cronobacter
Cronobacter isolates were analyzed as described in the PulseNet standardized PFGE Cronobacter protocol using XbaI (TaKaRa, Japan) as the restriction enzyme (Brengi et al., 2012; Yan and Fanning, 2015). DNA fragments were separated by electrophoresis (CHEF Mapper, Bio-Rad Laboratories, Hercules, California, US) through a 1% (w/v) agarose gel (Seakem Gold, Rockland, Maine, US) in 0.56 TBE buffer at 6 V/cm with an initial switch time of 1.8 s and a final switch time of 25 s. Gels were stained in deionized water containing GelRed Nucleic Acid Gel Stain (Biotium, CA, US), and visualized under UV light using a GelDoc XR+ system (Bio-Rad laboratories, Hercules, California, US). XbaI-digested Salmonella Braenderup H 9812 was used as the molecular weight standard. Dendrograms were constructed using Bionumerics software (Version 5.1, Applied-Maths, Belgium), and the cluster analysis was conducted using the DICE coefficient and unweighted pair group method (UPGMA) with arithmetic means, with a 1.2% band position tolerance. When comparing the DNA fingerprint patterns, a cutoff value of 100% similarity was applied.
Results
Prevalence of Cronobacter
In total, 835 samples were screened for the presence of Cronobacter spp., resulting in 42 isolates, 13 isolates from Factory A, eight from Factory B, zero in Factory C, and 21 from Factory D, respectively. In Factory A, 10 C. sakazakii isolates were associated with samples collected from inside and immediately outside the factory, two C. turicensis and one C. sakazakii were recovered from samples collected from a water tank and air conditioner. In Factory B, eight Cronobacter isolates were recovered from “quasi-clean” work areas, adjacent to fluid beds and air intake equipment. Twenty-one C. sakazakii isolates; 12 isolated associated with “clean work” areas and nine associated with “quasi-clean” work areas (PIF residues, powder control room, processing line, and the drying tower) in Factory D (Table 1 and Figure 1).
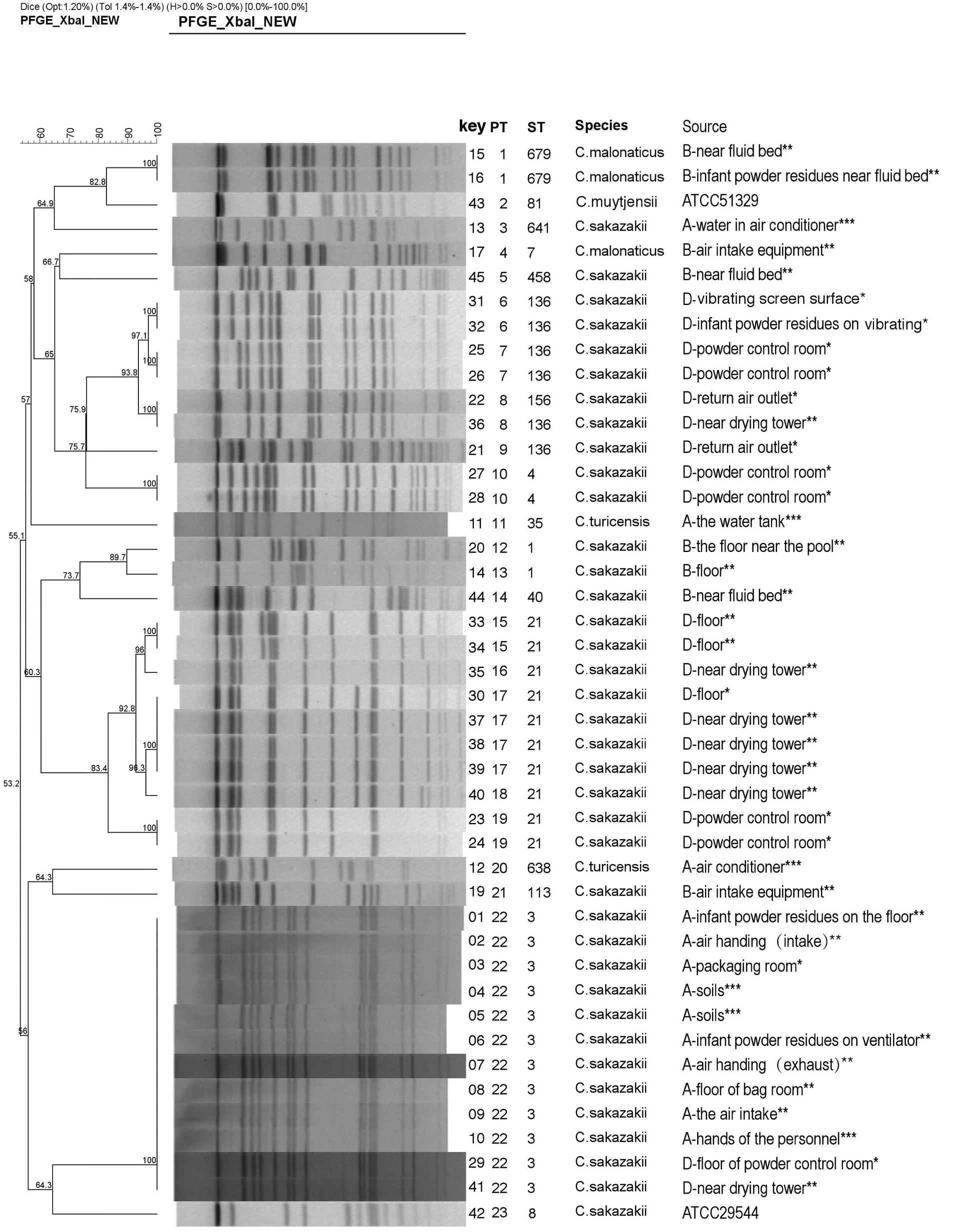
Figure 1. Dendrogram based on XbaI-mediated PFGE profiles of 44 Cronobacter spp. The tree was drawn using UPGMA and the Dice coefficient with 1.2% tolerance. Forty-four Cronobacter spp. include 42 Cronobacter isolates and two reference strains (ATCC 29544 and ATCC 51329). PT, pulsotype; ST, sequence type. (A, B, and D) indicate the three PIF factories, while no Cronobacter was not isolated in Factory C. *The strains from the clean work areas. **The strains from the quasi-clean work areas. ***The external environment of the factory.
The majority of Cronobacter strains were recovered from outside areas in this study (13.9%, 5/36). The “quasi-clean” work areas including the spray-drying areas, and the floor (23 strains) also accounted for large number of strains. In “clean work” areas, positive samples were associated with the PIF control room, return air outlet, and PIF residues (associated with various surfaces) (3.6%, 13/358) (Table 1). In Factories A, B, and D, four positive sites for Cronobacter were associated with PIF residues. The remaining positive samples were recovered from outside the production facility and staff. No positive samples were collected from common areas (Figure 1).
MLST Profiling
Sequence trace files were used to generate a minimum spanning tree (Figure 2). MLST subtyping was performed with all 42 isolates which were clustered into 14 STs, of which 37 isolates were identified as C. sakazakii, three isolates as C. malonaticus, and two isolates of C. turicensis. C. sakazakii was the dominant species identified in this study, which included 10 STs, with ST3 (n = 12) as the main sequence type, followed by ST21 (n = 10) and ST136 (n = 6). C. malonaticus included two STs (ST679 and ST7), and ST679 was assigned as the new ST in the MLST database. C. turicensi included two STs (ST35 and ST638).
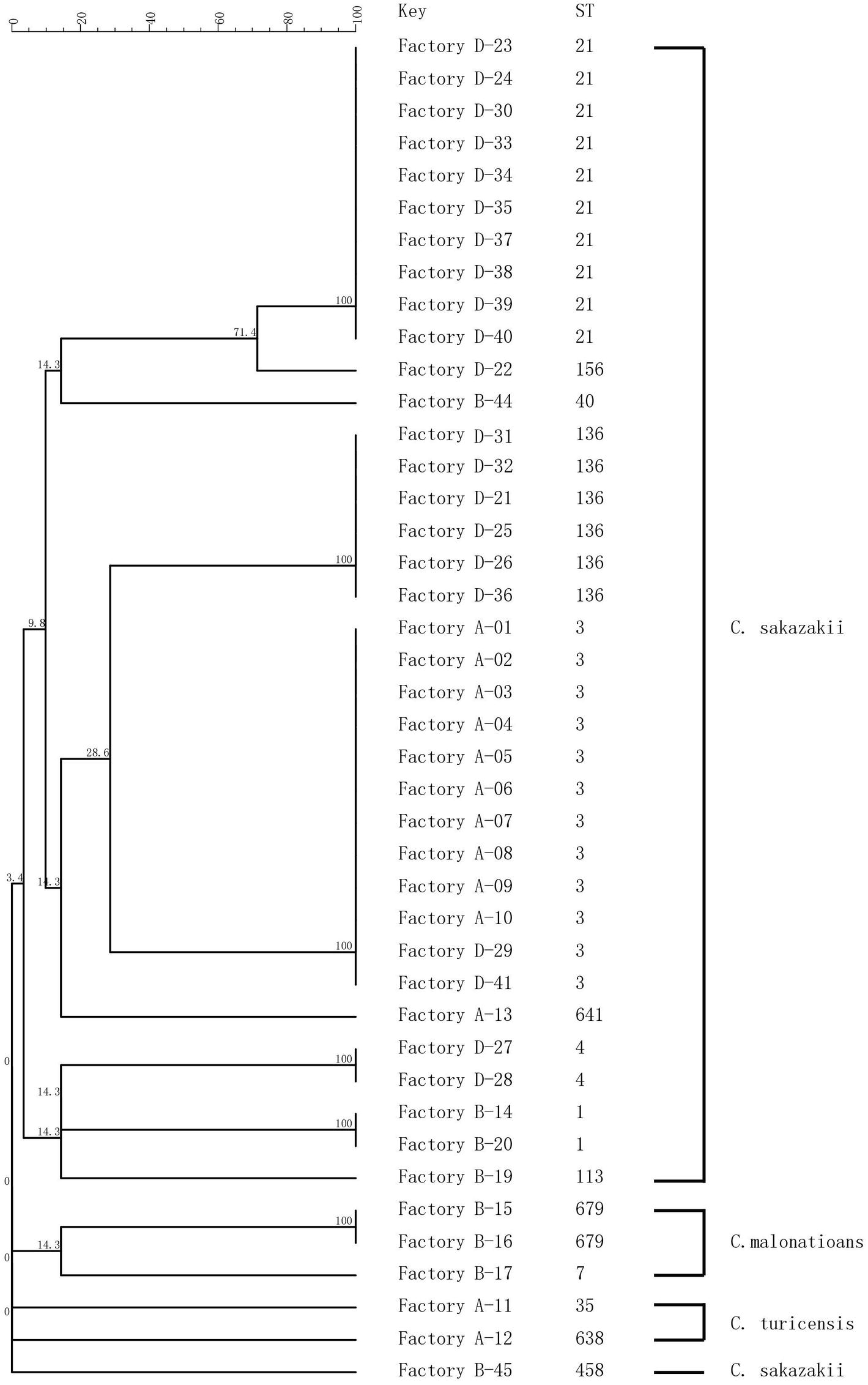
Figure 2. MLST dendrogram of 42 Cronobacter strains. Comparison is constructed from concatenated sequences of the seven MLST loci.
PFGE Analysis
Forty-two Cronobacter isolates were divided into 21 pulsotypes (PTs) by using 100% similarity type as the critical threshold. The major PTs were PT22 (n = 12) and PT17 (n = 4). The isolates identified as C. sakazakii (n = 37) were typed into 17 PTs. C. malonaticus (n = 3) and C. turicensis (n = 2) were typed into two PTs, respectively (Figure 1).
The Tracing of Cronobacter spp. in PIF Factories
In Factory A, 10 C. sakazakii isolates exhibited the same type (ST3 and PT22), and they were collected from the hands of technicians as well as the environment inside and outside the factory. In Factory B, five C. sakazakii isolates and three C. malonaticus isolates were recovered and exhibited six STs and seven PTs. In Factory D, 21 C. sakazakii isolates were recovered and included five STs and 11 PTs. Collectively, Cronobacter isolates with different sequence types and pulsotypes from the environment exhibited a high diversity. Although the geographic separation of PIF manufacturing facilities included in the study was minimal, each factory harbored strains unique to that factory (Figures 1, 2).
Discussion
Cronobacter spp. are opportunistic foodborne pathogens that can cause severe infections in neonates and infants through the ingestion of contaminated PIF; therefore, they are of global interest to human health (Forsythe et al., 2014; Fei et al., 2018a; Li et al., 2019). Results of this study suggest that should Cronobacter be detected in clean area or quasi-clean areas, measures must be immediately implemented to prevent contamination of the end products’ subsequent greater contamination of the processing environment (Pei et al., 2019). In manufacturing, Cronobacter may contaminate the processing line and product, since the current technology cannot completely eliminate the pathogen from the manufacturing environment (Codex, 2008; Fei et al., 2015). The study of multiple PIF plants is necessary and justified since surveillance of a single PIF plant would likely fail to provide sufficient evidence for the occurrence of Cronobacter. Therefore, in the present study, understanding areas within and external to a PIF production facility that may harbor Cronobacter in four PIF factories will aid in developing risk assessments and provide guidance to manufacturers when reviewing environmental monitoring plans.
Focusing on Factory A, ST3 was the predominant strain associated with samples collected from inside and outside of the Factory. C. sakazakii PT22 showed that ST3 isolates were identical. Collectively, these results demonstrate that C. sakazakii ST3 persistently and extensively contaminated Factory A. Moreover, ST3 exhibited the same type as soil associated isolates suggesting that soil may be the source of the Cronobacter contamination in Factory A. Bioaerosols are easily translocated by wind and air currents from one ecosystem to another, facilitating the spread of potentially pathogenic organisms (Wijnand et al., 2012). The bacteria may be brought into the factory through dust particles or condensation droplets, on worker shoes and clothes (Kucerova et al., 2011; Brandl et al., 2014). Dust particles in the air of a manufacturing plant can be a vector of Cronobacter dispersal, and the highest particle loads were observed in areas where filling, bagging, and the final packaging of PIF occurs (Brandl et al., 2005; Mullane et al., 2008). To reduce Cronobacter contamination in PIF manufacturing facilities, methods that reduce the number of dust particles in the air and treatment of PIF residues may be considered.
In Factory B, one C. sakazakii and one C. malonaticus were isolated from the air intake equipment, which is likely an essential portal for the transfer of bacteria from the outside environment to the inside of a facility. However, neither was isolated from the inside of Factory B. Two identical C. malonaticus (ST679/ PT1) and two C. sakazakii were isolated from near the fluid bed, but the two C. sakazakii exhibited different sequence types and pulsotypes. C. malonaticus strains were not persistent inside the plant, and only isolated in Factory B. Collectively, this suggests that there were multiple sources by which Cronobacter was introduced into Factory B. Results also indicated inadequate cleaning and sanitizing of the fluid bed and the floor in Factory B.
In Factory D, the isolate from the return air outlet was identical to the dominant isolates (ST136), suggesting that C. sakazakii inside the production facility contaminated the air return outlet by virtue of air flow and PIF transmission (Figure 1). During PIF production, particularly in the spray-drying and packing areas, once the air is contaminated with Cronobacter spp., the risk of final product contamination increases (Sonbol et al., 2013).
The best environmental controls for Cronobacter of all the factories were perhaps associated with Factory C in which no samples were positive for the pathogen. Factory C used a drying processing method which differs from the wet-mix processing method used by Factories A, B, and D. Perhaps implementation of more effective manufacturing procedures and sanitation programs in conjunction with the reduction of water in the dry-mix process may have contributed to the control of Cronobacter by Factory C. Based on the results of this study, more research is needed on the effects of processing technologies to control contamination of PIF by Cronobacter spp. PIF processors should implement practices that control the spread of Cronobacter, including keeping the air humidity low, reducing formation and spread of dust particles, timely removal of PIF residues, and regular cleaning and sanitizing of production equipment (Codex, 2008; Brandl et al., 2014).
Importantly, sites of contamination in the processing environment and routes of transmission were necessary to aid in risk analysis. Craven et al. (2010) investigated Cronobacter contamination in five Australian milk powder plants using PFGE. They suggest that Cronobacter strains are spread by air movement, milk powder dust particles, and personnel moving through the production environment. Pei et al. (2019) identified 10 isolates from six sample areas including conveyer belts, floor, walls, water powder around the product line, and vacuum cleaners. Cronobacter prevalence was the highest in samples collected from vacuum cleaners (28.0% positive samples). However, in the present study, no isolations were recovered from the vacuum cleaners. Rather, the extensive traceback sampling showed that PIF residues, fluid beds, drying areas, floors, and soil samples were the major contamination sites, which represent the greatest risk for introduction of Cronobacter during the production of PIF.
The isolates from samples collected at the same production facility showed closer phylogenetic relation than those collected from distinct facilities. The only exception was ST3, which was isolated from Factories A and D (Figure 2). The main purpose of the present study was to investigate the prevalence and potential contamination source of Cronobacter linked to PIF. The study was not designed to address how given strains may contaminate several factories. Consequently, a potential perspective future study could focus on the gene sequence of ST3 strains and the potential source of multiple factory contaminations.
Infant powder formula residues recovered from the production environment were more likely to harbor C. sakazakii. Although PIF is a low water activity product, Cronobacter spp. can survive in PIF for extended periods of time. Therefore, PIF should be included in environmental monitoring programs for Cronobacter. Pei et al. (2019) reported that the occurrence of Cronobacter is mainly associated with powder clumps on the vibrating screen in the processing environment. Those results are consistent with the present study where four positive sites for Cronobacter were associated with PIF residues.
In the present study, multiple locations within a PIF manufacturing facility were positive for the pathogen. Notably, C. sakazakii ST3, ST21, and ST136 were the dominant ST in this study. They are widely isolated from food (mainly PIF) and the environment according to PubMLST database, but no report was associated with human infection in China. Some pathogenic STs (e.g., ST1, ST7 from Factory B and ST4 from Factory D) were also identified in this study. They are commonly related to clinical cases based on MLST analysis. For example, C. sakazakii ST4 is linked to cases of meningitis (Hariri et al., 2013) and C. malonaticus ST7 is associated with adult infections, though the source was not identified (Joseph et al., 2012). Therefore, it is critical to investigate the Cronobacter distribution in the environment, which will help in understanding how the final product was contaminated and where the pathogens came from. Comparison of MLST and PFGE, as performed in this study, are useful for PIF manufactures to understand the diversity and characteristics of Cronobacter.
This study has improved our understanding of the prevalence and genetic diversity of Cronobacter spp. isolated from the production environment of PIF manufacturing facilities in China. Prevention efforts at manufacturing facilities must be multi-faceted and should include environmental sampling, and best practices supported by government regulatory agencies. Public education programs supported by health-care providers that emphasize safe handling practices of PIF will contribute to ensuring consumer safety. PIF manufacturers can utilize the results as a basis to review their existing monitoring program as part of a food safety plan and to make appropriate revisions to control PIF contamination.
Data Availability
The raw data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.
Author Contributions
XX and YL conceived and designed the experiments. YL, PL, MS, and JF performed the experiments. YL, XX, and CL generated and analyzed the data. YL, JG, and KM wrote the paper.
Funding
This study was supported by the Food department in Heilongjiang Administration for Market Regulation (HSRP2018001), the postdoctoral grant from Heilongjiang province government used for the study on the analysis of microorganisms in the environment of food processing enterprises (HBBH2017-2).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Brandl, H., Bachofen, R., and Bischoff, M. (2005). Generation of bioaerosols during manual mail unpacking and sorting. J. Appl. Microbiol. 99, 1099–1107. doi: 10.1111/j.1365-2672.2005.02700.x
Brandl, H., Fricker-Feer, C., Ziegler, D., Mandal, J., Stephan, R., and Lehner, A. (2014). Distribution and identification of culturable airborne microorganisms in a Swiss milk processing facility. J. Dairy Sci. 97, 240–246. doi: 10.3168/jds.2013-7028
Brengi, S. P., O’Brien, S. B., Pichel, M., Iversen, C., Arduino, M., Binsztein, N., et al. (2012). Development and validation of a PulseNet standardized protocol for subtyping isolates of Cronobacter species. Foodborne Pathog. Dis. 9, 861–867. doi: 10.1089/fpd.2012.1161
Chen, Y., Song, K. Y., Brown, E. W., and Lampel, K. A. (2010). Development of an improved protocol for the isolation and detection of Enterobacter sakazakii (Cronobacter) from powdered infant formula. J. Food Prot. 73, 1016–1022. doi: 10.4315/0362-028X-73.6.1016
Codex (2008). CAC/RCP 66-2008 code of hygienic practice for powdered infant formulae for infants and young children.
Craven, H. M., McAuley, C. M., Duffy, L. L., and Fegan, N. (2010). Distribution, prevalence and persistence of Cronobacter (Enterobacter sakazakii) in the non-processing and processing environments of five milk powder factories. J. Appl. Microbiol. 109, 1044–1052. doi: 10.1111/j.1365-2672.2010.04733.x
Cui, J. H., Du, X. L., Wei, R. J., Zhou, H. J., Li, W., Forsythe, S., et al. (2015). Multilocus sequence typing analysis of Cronobacter spp. isolated from China. Arch. Microbiol. 197, 665–672. doi: 10.1007/s00203-015-1097-0
FAO/WHO (2004). Workshop on Enterobacter sakazakii and other microorganisms in powdered infant formula, Geneva, 2–5 February 2004. Available at: http://www.who.int/foodsafety/micro/jemra/meetings/feb2004/en/index.html
FAO/WHO (2006). Expert meeting on Enterobacter sakazakii and Salmonella in powdered infant formula, Rome, 16–20 January 2006. Available at: http://www.who.int/foodsafety/micro/jemra/meetings/jan2006/en/index.html
FAO/WHO (2008). Enterobacter sakazakii (Cronobacter spp.) in powdered follow-up formulae. Microbiological Risk Assessment Series no. 15. Available at: http://www.who.int/foodsafety/publications/micro/mra_followup/en/
Fei, P., Ali, M. A., Gong, S., Sun, Q., Bi, X., Liu, S., et al. (2018a). Antimicrobial activity and mechanism of action of olive oil polyphenols extract against Cronobacter sakazakii. Food Control 94, 289–294. doi: 10.1016/j.foodcont.2018.07.022
Fei, P., Jiang, Y., Gong, S., Li, R., Jiang, Y., Yuan, X., et al. (2018b). Occurrence, genotyping, and antibiotic susceptibility of Cronobacter spp. in drinking water and food samples from Northeast China. J. Food Prot. 81, 456–460. doi: 10.1007/978-1-4939-2599-5_6
Fei, P., Man, C., Lou, B., Forsythe, S. J., Chai, Y., Li, R., et al. (2015). Genotyping and source tracking of Cronobacter sakazakii and C. malonaticus isolates from powdered infant formula and an infant formula production factory in China. Appl. Environ. Microbiol. 81, 5430–5439. doi: 10.1128/AEM.01390-15
Forsythe, S. J., Dickins, B., and Jolley, K. A. (2014). Cronobacter, the emergent bacterial pathogen Enterobacter sakazakii comes of age; MLST and whole genome sequence analysis. BMC Genomics 15:1121. doi: 10.1186/1471-2164-15-1121
Hariri, S., Joseph, S., and Forsythe, S. J. (2013). Cronobacter sakazakii ST4 strains and neonatal meningitis, United States. Emerg. Infect. Dis. 19, 175–177. doi: 10.3201/eid1901.120649
Hunter, C. J., and Bean, J. F. (2013). Cronobacter: an emerging opportunistic pathogen associated with neonatal meningitis, sepsis and necrotizing enterocolitis. J. Perinatol. 33, 581–585. doi: 10.1038/jp.2013.26
ISO 22964 (2017). Microbiology of the food chain-horizontal method for the detection of Cronobacter spp. Geneva: International Organization for Standardization.
Iversen, C., Lehner, A., Fricker-Feer, C., Gschwend, K., and Stephan, R. (2009). Genotyping of Cronobacter (Enterobacter sakazakii) strains isolated from an infant formula processing plant. Arch. Food Hyg. 60, 66–72. doi: 10.5851/kosfa.2009.29.6.702
Joseph, S., Sonbol, H., Hariri, S., Desai, P., McClelland, M., and Forsythe, S. J. (2012). Diversity of the Cronobacter genus as revealed by multilocus sequence typing. J. Clin. Microbiol. 50, 3031–3039. doi: 10.1128/JCM.00905-12
Kucerova, E., Joseph, S., and Forsythe, S. (2011). The Cronobacter genus: ubiquity and diversity. Qual. Assur. Saf. Crops Foods 3, 104–122. doi: 10.1111/j.1757-837X.2011.00104.x
Li, C., Zeng, H., Zhang, J., He, W., Ling, N., Chen, M., et al. (2019). Prevalence, antibiotic susceptibility, and molecular characterization of Cronobacter spp. isolated from edible mushrooms in China. Front. Microbiol. 10:283. doi: 10.3389/fmicb.2019.00283
Mullane, N., Healy, B., Meade, J., Whyte, P., Wall, P. G., and Fanning, S. (2008). Dissemination of Cronobacter spp. (Enterobacter sakazakii) in a powdered milk protein manufacturing facility. Appl. Environ. Microbiol. 74, 5913–5917. doi: 10.1128/AEM.00745-08
Mullane, N. R., Whyte, P., Wall, P. G., Quinn, T., and Fanning, S. (2007). Application of pulsed-field gel electrophoresis to characterize and trace the prevalence of Enterobacter sakazakii in an infant formula processing facility. Int. J. Food Microbiol. 116, 73–81. doi: 10.1016/j.ijfoodmicro.2006.12.036
Pei, X., Li, Y., Zhang, H., Zhan, L., Yu, X., Lan, G., et al. (2019). Surveillance and characterisation of Cronobacter in powdered infant formula processing factories. Food Control 96, 318–323. doi: 10.1016/j.foodcont.2018.09.009
Proudy, I., Bouglé, D., Leclercq, R., and Vergnaud, M. (2008). Tracing of Enterobacter sakazakii isolates in infant milk formula processing by BOX-PCR genotyping. J. Appl. Microbiol. 105, 550–558. doi: 10.1111/j.1365-2672.2008.03775.x
Sonbol, H., Joseph, S., McAuley, C. M., Craven, H. M., and Forsythe, S. J. (2013). Multilocus sequence typing of Cronobacter spp. from powdered infant formula and milk powder production factories. Int. Dairy J. 30, 1–7. doi: 10.1016/j.idairyj.2012.11.004
Stephan, R., Grim, C. J., Gopinath, G. R., Mammel, M. K., Sathyamoorthy, V., Trach, L. H., et al. (2014). Re-examination of the taxonomic status of Enterobacter helveticus, Enterobacter pulveris, and Enterobacter turicensis as members of the genus Cronobacter and their reclassification in the genera Franconibacter gen. nov. and Siccibacter gen. nov. as Franconibacter helveticus comb. nov., Franconibacter pulveris comb. nov. and Siccibacter turicensis comb. nov., respectively. Int. J. Syst. Evol. Microbiol. 64, 3402–3410. doi: 10.1099/ijs.0.059832-0
Stephan, R., Lehner, A., Tischler, P., and Rattei, T. (2011). Complete genome sequence of Cronobacter turicensis LMG 23827, a food-borne pathogen causing deaths in neonates. J. Bacteriol. 193, 309–310. doi: 10.1128/JB.01162-10
Wijnand, E., Dick, H., Duchaine, C., and Green, B. J. (2012). Bioaerosol exposure assessment in the workplace: the past, present and recent advances. J. Environ. Monit. 14, 334–339. doi: 10.1039/c2em10717a
Keywords: Cronobacter spp., genetic diversity, infant formula production facilities, multi-locus sequence typing, pulsed-field gel electrophoresis
Citation: Lu Y, Liu P, Li C, Sha M, Fang J, Gao J, Xu X and Matthews KR (2019) Prevalence and Genetic Diversity of Cronobacter Species Isolated From Four Infant Formula Production Factories in China. Front. Microbiol. 10:1938. doi: 10.3389/fmicb.2019.01938
Edited by:
Eugenia Bezirtzoglou, Democritus University of Thrace, GreeceReviewed by:
Hana Drahovska, Comenius University, SlovakiaYuanhong Li, Xuzhou Medical University, China
Amin N. Olaimat, Hashemite University, Jordan
Copyright © 2019 Lu, Liu, Li, Sha, Fang, Gao, Xu and Matthews. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoxi Xu, xiaoxi_xu01@163.com; Karl R. Matthews, bijan@sebs.rutgers.edu
 Yan Lu
Yan Lu Peng Liu2
Peng Liu2 Karl R. Matthews
Karl R. Matthews