- 1Institute of Applied Microbiology – iAMB, Aachen Biology and Biotechnology – ABBt, RWTH Aachen University, Aachen, Germany
- 2Leibniz Institute for Natural Product Research and Infection Biology – Hans Knöll Institute, Jena, Germany
- 3Faculty of Biological Sciences, Friedrich Schiller University Jena, Jena, Germany
Phenazine-1-carboxylic acid (PCA) and its derivative pyocyanin (PYO) are natural redox mediators in bioelectrochemical systems and have the potential to enable new bioelectrochemical production strategies. The native producer Pseudomonas aeruginosa harbors two identically structured operons in its genome, which encode the enzymes responsible for PCA synthesis [phzA1-G1 (operon 1), phzA2-G2 (operon 2)]. To optimize heterologous phenazines production in the biotech host Pseudomonas putida KT2440, we compared PCA production from both operons originating from P. aeruginosa strain PAO1 (O1.phz1 and O1.phz2) as well as from P. aeruginosa strain PA14 (14.phz1 and 14.phz2). Comparisons of phenazine synthesis and bioelectrochemical activity were performed between heterologous constructs with and without the combination with the genes phzM and phzS required to convert PCA to PYO. Despite a high amino acid homology of all enzymes of more than 97%, P. putida harboring 14.phz2 produced 4-times higher PCA concentrations (80 μg/mL), which resulted in 3-times higher current densities (12 μA/cm2) compared to P. putida 14.phz1. The respective PCA/PYO producer containing the 14.phz2 operon was the best strain with 80 μg/mL PCA, 11 μg/mL PYO, and 22 μA/cm2 current density. Tailoring phenazine production also resulted in improved oxygen-limited metabolic activity of the bacterium through enhanced anodic electron discharge. To elucidate the reason for this superior performance, a detailed structure comparison of the PCA-synthesizing proteins has been performed. The here presented characterization and optimization of these new strains will be useful to improve electroactivity in P. putida for oxygen-limited biocatalysis.
Introduction
Bioelectrochemical systems or BES can be utilized to generate bioelectricity from renewable (often waste) carbon sources or to sustainably produce platform chemicals through the interaction of microorganisms with a polarized electrode (Bajracharya et al., 2016). As a bioprocessing system, BES involve a microbial whole-cell biocatalysts to drive oxidation and reduction (redox) reactions at solid electrodes (Rosenbaum and Henrich, 2014). The electron exchange can thereby be either performed through a direct electron transfer (DET) mechanism, involving a physical contact between the cell and the electrode and typically requiring specific redox proteins, or a mediated electron transfer (MET), involving a soluble redox shuttling compound as the main electron transfer pathway. These two fundamental electron transfer modes will have a strong strategic consequence for the development of future bioprocesses, enabled or enhanced through microbial electroactivity. From a biotechnological standpoint, connecting a solution-based MET process to a target microbial production hosts has the great advantage to be easily compatible with classical bioprocess technology of stirred tank reactors. The optimization goal then would be to insert electrodes into the bioreactor infrastructure at as low as possible disturbance of the reactor fluid dynamics (Krieg et al., 2019). In contrast, a DET process would demand for completely new reactor concepts since the biochemical production would be limited to a two dimensional biofilm and a maximum packing density of biofilm and electrode material would be required for the bioreactor to reach high titers and productivities.
For MET processes, the electron mediator can be exogenous (artificially added in the system) or endogenous (naturally synthesized by the microorganism) (Harnisch et al., 2011). Advantages of endogenous mediators over exogenous mediators, like methylene blue (Rahimnejad et al., 2011), neutral red (Popov et al., 2012) or ferricyanide (Hu et al., 2015; Lai et al., 2016), are that they might be more biocompatible, do not require costly addition of chemical compounds, are likely produced throughout the process, and the production is linked to the activity of the producing microorganism. A disadvantage is that only certain microbial species are naturally equipped to produce endogenous redox mediators for extracellular electron transfer.
One group of these redox compounds studied for natural MET are microbial phenazines, which are produced by Pseudomonas species, especially by Pseudomonas aeruginosa. Among other functions, phenazines like phenazine-1-carboxylic acid (PCA) and pyocyanin (PYO) play an important role in anaerobic survival of P. aeruginosa through discharging surplus metabolic electrons to not directly accessible terminal electron acceptors, such as far away oxygen, iron oxides, or an anode (Rabaey et al., 2005; Pierson and Pierson, 2010; Glasser et al., 2014). The complex regulatory network of phenazine synthesis and the pathogenic nature of P. aeruginosa limit the application of phenazine redox mediation with this host for specific electrobiotechnological applications (Venkataraman et al., 2011). Thus, to avoid P. aeruginosa as microbial host, the utilization of a non-pathogenic species for heterologous phenazines production would be an advantage for future biotechnological utilization. However, because of their redox activity, phenazines also have strong antimicrobial activity through the generation of reactive oxygen species for those microorganisms, which do not possess a strong defense system against oxygen radicals. For example the top biotech host Escherichia coli is unable to tolerate the reactive phenazine PYO. On the other hand, tolerant microorganisms can make energetic use of provided phenazines by using them as a vehicle to allow for an anaerobic respiration under anaerobic conditions as it has been shown for the natural habitat partner Enterobacter aerogenes (Venkataraman et al., 2011; Schmitz and Rosenbaum, 2018).
One non-pathogenic Pseudomonas species closely related to the native phenazine producer and tolerant to phenazines, which has emerged as a versatile recombinant biotech host, is Pseudomonas putida KT2440. This host stands out because of the availability of extensive genetic and intrinsic metabolism information, combined with its tolerance to many relatively unpolar or even toxic chemical substances. Therefore, it has become very useful as a production host of various valuable compounds (Loeschcke and Thies, 2015). However, one current drawback of P. putida as production host is its obligate aerobic metabolism, which cannot always be sustained even in well mixed stirred tank reactors. So far, attempts to engineer alternative respirative or fermentative pathways showed only limited success (Nikel and de Lorenzo, 2013; Steen et al., 2013).
In a successful initial proof-of-principle study, we introduced an engineered strain of P. putida KT2440, in which heterologous phenazine synthesis enabled electron discharge to an extracellular anode under oxygen-limited conditions in BES (Schmitz et al., 2015). The heterologous expression of the nine phenazine synthesis genes from P. aeruginosa PAO1 (phzA1-G1, phzM, and phzS) in P. putida KT2440 resulted in the production of PCA and PYO, whereby the derivative PYO was only fully synthesized when active aeration (AA) was applied during the initial growth phase since the final enzyme PhzS is dependent on molecular oxygen. During subsequent passive aeration (PA) via opened vent filter (i.e., strong oxygen limitation of the culture), metabolic activity sustained over 10 days and PCA was further synthesized and accumulated. Thereby, the total produced phenazine concentrations in BES were in the upper range of phenazine concentrations produced by the native producer P. aeruginosa [30–40 μg/mL for P. putida pPhz (Schmitz et al., 2015) vs. 22–25 μg/mL for P. aeruginosa PA14 (Bosire et al., 2016)].
In order to develop an efficient P. putida biocatalyst for oxygen-limited bioelectrochemical production, such as the production of rhamnolipids (Wittgens et al., 2011) or aromatics (Wierckx et al., 2005), it is necessary to further explore and optimize the heterologous production of phenazine. The work conducted by Bosire et al. (2016) showed for several carbon sources that P. aeruginosa PA14 produced more phenazines and higher resulting anodic currents than P. aeruginosa PAO1 in oxygen-limited BES. This indicated that the phenazine production in P. aeruginosa is strain dependent.
Furthermore, P. aeruginosa encodes for two homologous operons; phzA1-G1 (operon 1) and phzA2-G2 (operon 2), which are responsible for the synthesis of PCA under different regulatory conditions. Although the two phenazine operons of P. aeruginosa are 98% identical on DNA level (97% in amino acid level), they differ distinctively in their promoter regions. A las box is present in the promoter region of the phz1 operon 390 bp upstream of the phzA1 translational region. With this regulatory genetic element, the quorum sensing regulators LasR and/or RhlR recognize their target genes to initiate quorum sensing controlled gene expression. In contrast, for phzA2, the las box is not present in the promoter region (Recinos et al., 2012). A more detailed analysis by Mavrodi et al. (2001) revealed that the two operons contain most dissimilarities at DNA sequence level within phzA1B1 and phzA2B2, while the regions from phzC to phzG of both operons are highly identical (Mavrodi et al., 2001). Hence, the heterologous expression of those two operons might be markedly different.
Therefore, in this study, we generated new strains of P. putida expressing phenazine genes from three different gene sources as follows; P. aeruginosa PAO1 (operon 1) and P. aeruginosa PA14 (operons 1 and 2) to produce PCA. Those three new constructs were compared with the performance of the already existing constructs originating from P. aeruginosa PAO1 operon 1 (Schmitz et al., 2015). The PCA producing- P. putida strains were combined further with the phzM+S plasmid to produce PYO. All phenazine producer strains were characterized for two different oxygen-limited conditions in BES; (i) 48 h active aeration (AA) during initial growth then subsequently switched to passive headspace aeration, and (ii) fully passive aeration (PA) throughout the experiment. With this approach, we aim to obtain the best heterologous P. putida phenazines producer for efficient biocatalysis under oxygen-limited condition BES.
Materials and Methods
Bacterial Strains, Media Preparation, and Strain Cultivation
Pseudomonas putida strain KT2440 (DSM 6125) was used for heterologous expression of phenazine genes from P. aeruginosa PAO1 (DSMZ 19880) and PA14 (DSMZ 19882). Escherichia coli strain DH5α (New England Biolabs) has been used as intermediary cloning recipient. Standard strain cultivations were performed in 250 mL shake flasks using LB medium (Carl Roth®) with or without antibiotics as required and were incubated at 30°C (P. putida) or 37°C (E. coli). For P. putida characterization in well plates, shake flasks and BES experiment, the strains have been cultivated at 30°C in Delft mineral salt medium (Hartmans et al., 1989), with a final composition (per L) of 3.88 g K2HPO4 (22 mM), 1.63 g NaH2PO4 (14 mM), 2.00 g (NH4)2SO4, 0.1 g MgCl2.6H2O, 10 mg EDTA, 2 mg ZnSO4.7H2O, 1 mg CaCl2.2H2O, 5 mg FeSO4.7H2O, 0.2 mg Na2MoO4.2H2O, 0.2 mg CuSO4.5H2O, 0.4 mg CoCl2.6H2O, 1 mg MnCl2.2H2O, with 20 mM glucose (PA) or 40 mM glucose (AA) and antibiotics as required (kanamycin at 50 μg/mL concentration or/and gentamycin at 30 μg/mL concentration).
Genetic Engineering of P. putida for Phenazine Synthesis
Table 1 gives an overview on the strains and plasmids used for construct generation. The pBNT plasmid harboring phzA1-G1 amplified from P. aeruginosa PAO1 genomic DNA (O1.phz1) was generated by Schmitz et al. (2015). The other three plasmids to synthesize PCA were newly constructed in this study; plasmid pBNT.O1.phz2 harboring phzA2-G2 amplified from P. aeruginosa PAO1; the plasmid pBNT14.phz1 and pBNT14.phz2 harboring phzA1-G1 and phzA2-G2, respectively, amplified from P. aeruginosa PA14. For plasmids assembly, the Gibson assembly method was used and performed according to the manufacturer’s instruction (New England Biolabs-Gibson Assembly) (Gibson et al., 2009).

Table 1. Bacterial strains and plasmids used for genetic engineering of P. putida for phenazine synthesis.
Subsequently, all constructed plasmids have been transformed into P. putida KT2440 via electroporation (Choi et al., 2006). For combined PCA and PYO synthesis, the best pre-selected strain producing PCA from each operon origin was transformed further with pJNN.phzM+S generated by Schmitz et al. (2015). Transformed cells were selected on LB agar plates with kanamycin (K, 50 μg/mL) for PCA, while for PCA and PYO producers LB agar plates with kanamycin (same concentration) and gentamycin (G, 30 μg/mL) were used. The constructs were verified using colony PCR, restriction digest analysis and DNA sequencing with targeted primers (Supplementary Table S1). DNA plasmid sequencing analysis was performed using the Sanger sequencing method (GATC-Eurofins Genomics). The DNA sequencing products of the promoter and phenazine gene regions were compared and verified with the reference plasmid map using Clone Manager Software. The successful expression of the phzA-G genes will show the PCA production by exhibiting yellow color in Delft medium, while in combination with phzM+S genes, the bacterial culture produced PCA and PYO and appears in a greenish blue color.
Aerobic Strain Characterization and Evaluation
Eight clones of each constructed strain were tested in triplicates for phenazine production in the multiplexed cultivation platform growth profiler (CR1424d, EnzyScreen, Heemstede, Netherlands), using 24 square-well plates filled with 5 mL Delft medium, 20 mM glucose, and supplemented with antibiotic/s (K, 50 μg/mL or/and G, 30 μg/mL). While the aerobic flask experiments were performed in 250 mL flasks; filled with 25 mL Delft medium. Both of the cultivations were starting at a uniform inoculation OD600: 0.1. After 3 h of incubation, sodium salicylate was added as an inducer of the phenazine gene expression. The incubation temperature was 30°C and the shaking frequency was 224 rpm (multiplexed cultivation) and 200 rpm (flasks experiment). The clone with the best growth as well as phenazine production obtained from each constructed strain was selected to be characterized further in the aerobic flasks as well as BES experiments.
Oxygen-Limited Bioelectrochemical System Experiments
Single chamber glass BES reactors (500 mL working volume) equipped with a water jacket for the temperature control was used (Supplementary Figure S1). The integrated three electrode set-up included: a 156.32 cm2 graphite comb of high-grade graphite (EDM-3, Novotec) as working electrode (anode), a 49.22 cm2 graphite block as counter electrode (cathode), and an Ag/AgCl, saturated KCl reference electrode (192 mV vs. SHE at 30°C, pH 7) as reference electrode. The BES reactor experiments were performed at 30°C at 200 rpm stirring with a magnetic stirrer, and potentiostatically controlled by a potentiostat (VMP3, Biologic) at 0.2 V vs. reference electrode. The electrical current signal was recorded continuously including 24 h of blank media measurement. Sampling for measurements of optical density at 600 nm, pH and HPLC analysis were performed daily after the bacterial culture inoculation. The duplicate BES reactor experiments were performed under two types of oxygen-limited condition: AA, in which the active aeration was applied via a sparger at 30 mL/min flow rate for 48 h aeration followed by PA of the headspace via two opened vent filters, and PA, in which only passive aeration via opened vent filters was applied for the entire experiment.
Phenazine Analysis
The quantification of phenazines was performed using high-performance liquid chromatography (HPLC) with a C18 column (Waters Corporation) equipped with a photodiode array UV/VIS detector (LC-168, Beckmann), which detected and quantified the PCA at 366 nm and PYO at 280 nm. Separation was achieved with a gradient of 0.1% trifluoroacetic acid (TFA) in acetonitrile (solution A) and 0.1% TFA in water (solution B) as mobile phase (with solution A at 15% for 2 min, 100% for 15 min, and 5% for 3 min) at a flow rate of 0.8 mL/min and a column temperature of 20°C (Kern and Newman, 2014). Analytical grade PCA (Princeton BioMolecular) and PYO (Cayman Chemical) were used as standard solutions of phenazines.
Analysis of Sugar Metabolites
Analysis of glucose and secreted metabolites was performed using HPLC equipped with a 300 × 8.0 mm polystyroldivinylbenzol copolymer separation column (CS-Chromatographie), a UV/VIS detector at 210 nm and a refractive index (RI) detector at 35°C. The separation was achieved at 60°C with 5 mM sulphuric acid mobile phase at 0.8 mL/min isocratic flow rate. Analytical grade glucose, 2-ketogluconate, gluconate, and acetate (Carl Roth®) were used as standard solutions.
Amino Acid Sequence Comparison of the Phenazine Operons
Sequences and annotations of P. aeruginosa PAO1 (assembly: GCA_000006765.1) and P. aeruginosa PA14 (assembly: GCA_000014625.1) were obtained from GenBank (Benson et al., 2005). Each strain contains two main clusters of seven genes organized in an operon for PCA production. All these genes were compared individually by multiple protein alignment and the entire operons (excluding the promoter) were compared by multiple nucleotide alignment. Clustal Omega 1.2.4 was used to perform the alignments (Sievers et al., 2011; Sievers and Higgins, 2018). In order to detect changes between the codon usage among genes in the four operons, codon adaptation indices/CAI (Sharp and Li, 1987) were calculated for all genes using an index previously generated for P. putida KT2440 (assembly: GCA_000007565.2). Plots for CAI comparison and operon analysis were created using RStudio (R Studio Team, 2015). All calculations were performed using Biopython 1.72 (Cock et al., 2009). For all proteins involved in phenazines production (except PhzF and PhzM due to full sequence identity), a 3D-model has been created by homology modeling using modeler (Webb and Sali, 2014). All structures were predicted in the same way, including those with an experimentally determined structure, to avoid bias. Models were built as homodimers, except PhzS that was built as a monomer. The predicted protein structures were superimposed and displayed using UCSF Chimera (Pettersen et al., 2004). RMSD (root-mean-square deviation of atomic positions) was calculated to measure the differences between structures.
Results and Discussion
Phenazine Gene Origin Determines the Phenazine Synthesis Capacity in P. putida
In an initial study, we transferred the capacity to synthesize the phenazines PCA and PYO from the pathogen P. aeruginosa into the GRAS (“generally recognized as safe”) organism P. putida KT2440 (Schmitz et al., 2015). The gene origins in that work came from P. aeruginosa strain PAO1 (i.e., O1.phz1 and phzM+S). To test the hypothesis that the genetic origin of the encoding genes impacts phenazine synthesis, the two strongly homologous copies of the 7-gene operon responsible for PCA synthesis from two P. aeruginosa strains (O1.phz1 and O1.phz2 vs. 14.phz1 and 14.phz2) were inserted into the inducible plasmid pBNT without their native promoter regions. Since the PhzM protein shares 100% amino acid identity, while the PhzS protein shares 99% amino acid identity in the two strains (without impact on the active site of the protein) (Supplementary Figure S13), the original plasmid construct for PYO synthesis (pJNN.phzM+S) with the genes originating from strain PAO1 was not changed in this study.
A comparison of phenazine synthesis of a strain version carrying only the PCA-production genes and the combination with the phzM+S genes to also synthesize PYO was undertaken (indicated by “ + ” right after the construct name, e.g., P. putida O1.phz1 makes only PCA and P. putida O1.phz1 + makes PCA and PYO). For each construct, eight colonies were picked from agar plates after transformation and compared in parallelized growth experiments on a micro-cultivation platform regarding the growth and yellow (PCA) or blue (PYO) pigment production (Supplementary Figures S2, S3).
For a more quantitative evaluation of phenazine synthesis, the best clone of each construct in the micro-cultivations was further cultivated in fully aerobic shake flask experiments (Supplementary Figures S4, S5). The strain constructs P. putida 14.phz2 and P. putida 14.phz2+ showed the most intense color formation compared to other strains (Figure 1A) with the phenazine quantification results shown in Figure 1B. The P. putida O1.phz1 strain was found to be the lowest PCA producer (23.34 ± 1.06 μg/mL) and P. putida 14.phz1+ was identified to be the lowest PCA and PYO producer (5.86 ± 0.30 μg/mL PCA and 0.60 ± 0.15 μg/mL PYO, respectively). In comparison to the P. putida O1.phz1 strain, the expression of the 14.phz1 operon alone (without phzM+S) boosted PCA synthesis to 79.79 ± 9.09 μg/mL. Thus, PCA production of P. putida expressing the 14.phz1 operon was higher than with the O1.phz1 operon. However, the combination of both plasmids in P. putida to produce PCA and PYO worked better with P. putida O1.phz1 + than with P. putida 14.phz1 + where very little of both phenazines was produced (Figure 1B), although the growth curve of P. putida 14.phz1 + was similar compared to P. putida O1.phz1 + (Supplementary Figure S5). In P. putida 14phz.1+, the presence of the phzM+S genes obviously negatively impacted the PCA synthesis, which resulted in low PYO production.
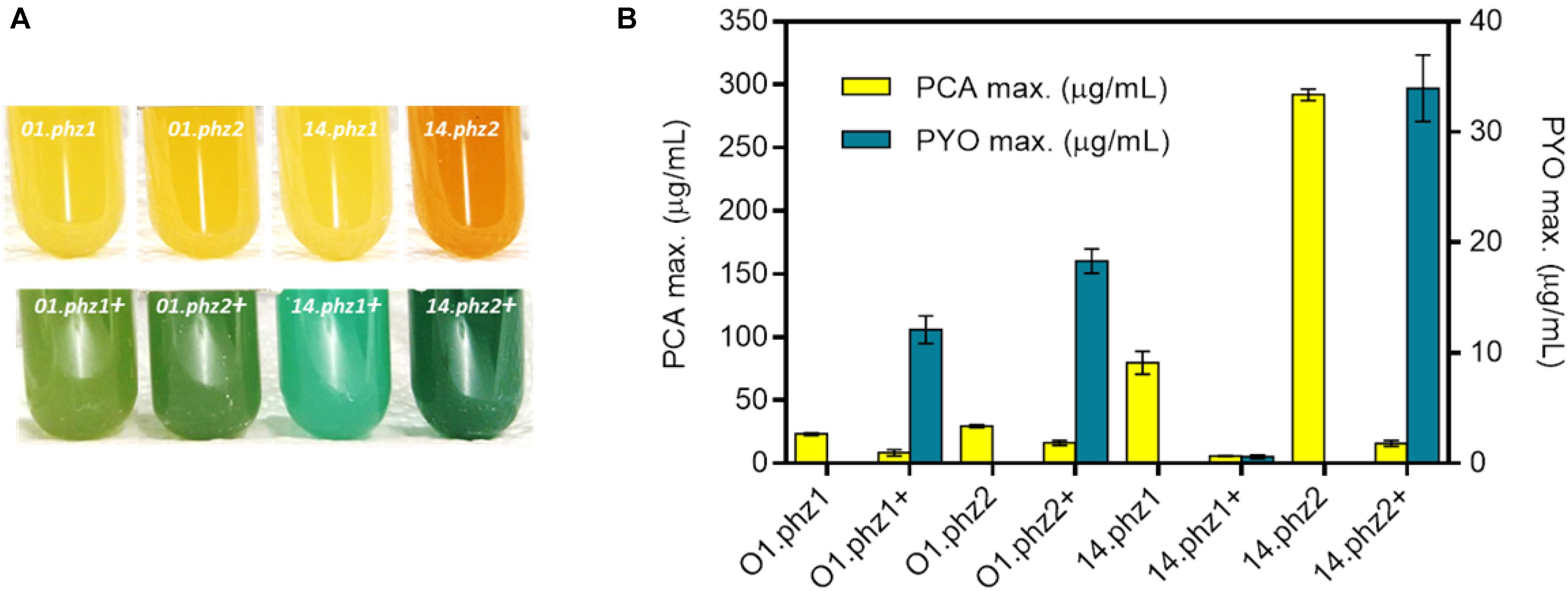
Figure 1. Heterologous expression of phz genes in P. putida KT2440. (A) P. putida cultures (from left to right) expressing O1.phz1, O1.phz2, 14.phz1, and 14.phz2 gene for PCA synthesis (top) and with combination of phzM+S genes for PCA and PYO synthesis PCA and PYO (bottom). (B) Phenazine production of engineered P. putida strains in triplicate shake flask experiments (fully aerobic), whereby the “+” in the strain designation indicates presence of phzM+S to convert PCA to PYO.
By far the best PCA producer was P. putida 14.phz2 (292 ± 4.58 μg/mL) and P. putida 14.phz2+ was the strongest PCA and PYO producer (16 ± 2.31 and 33.96 ± 3 μg/mL, respectively). The growth curve of P. putida 14.phz2 was similar with P. putida O1.phz1 and P. putida 14.phz1. Thus, the phzA-G operon 2 from P. aeruginosa PA14 allowed higher PCA production compared to operon 1. In P. putida 14.phz2+, although PYO production was highest compared to the other strains, PCA production was low. This was surprising given the high PCA production of P. putida 14.phz2. However, the two plasmid strategy requires two antibiotics that might add up to a significant metabolic burden, causing the low phenazines synthesis. But it is also possible that the presence of PYO has a toxic effect on the cells, reduces the biosynthetic activity, and thereby hinders further PCA synthesis, since it is known that PYO generates toxic oxygen radicals.
Overall, the experiments conducted under fully aerobic conditions showed that the PCA synthesis of P. putida expressing the phenazine genes (same strain origin) from operon 2 of P. aeruginosa was higher than from operon 1. Furthermore, PCA production of P. putida expressing the phenazine genes from P. aeruginosa PA14 was higher compared to PAO1, while for combined PCA and PYO production, the phenazine production was affected by either the metabolic burden of the two antibiotics applied and/or a restricted total phenazine production with increased levels of PYO, which shows toxic behavior at higher concentration through the formation of reactive oxygen species (Mentel et al., 2009). In general, we can conclude that phenazine synthesis in P. putida is strain origin- and operon-dependent under fully aerobic condition.
Phenazine Gene Origin Determines Electron Discharge Capacity in BES
The motivation for engineering P. putida to produce phenazines is driven by the idea to capacitate this obligate aerobic bacterium for metabolic activity and in future for biocatalysis in oxygen-limited bioreactors (Schmitz et al., 2015). P. putida will be able to utilize phenazines as reversible redox mediators for the anodic discharge of metabolic electrons. We have shown with the native producer P. aeruginosa that the effectivity of anodic electron discharge with the two produced phenazines PCA and PYO can be fine-tuned by electrochemical potential control (Bosire and Rosenbaum, 2017). In future, this oxygen-limited metabolism will be coupled with biotechnological productions in the BES, such as the production of rhamnolipids, which currently suffers from technical challenges because of extensive foam formation during high level aeration (Beuker et al., 2016). In order to characterize the capacity of the different P. putida strains for electron discharge, the BES performance of each engineered strain variant has been evaluated with just initial aeration for the first 48 h (Figures 2A–D). In accordance with the shake flask experiments, the lowest PCA production and current generation were observed from P. putida O1.phz1, which only produced 9.38 μg/mL PCA, corresponding to 1.68 μA/cm2 current density (Figure 2A). A control experiment, where ∼17 μg/mL synthetic PCA was added to the P. putida KT2440 wildtype in an oxygen-limited BES, confirmed that the PCA concentration stayed constant in the reactor until the end of the experiment, i.e., was not degraded or absorbed to the electrode and a current density of 8.57 μA/cm2 was observed (Supplementary Figure S6A). Furthermore, by far the lowest PCA and PYO producer in actively aerated BES was P. putida 14.phz1+, which only produced 2.71 μg/mL PCA, 1.63 μg/mL PYO, and 4.74 μA/cm2 maximum current density (Figure 2D). This result also mirrored the very low activity of the phenazine production of P. putida 14.phz1+ in the flask experiments, where the combination of the 14.phz1 gene with the phzM+S genes led to an almost complete suspension of phenazine production.
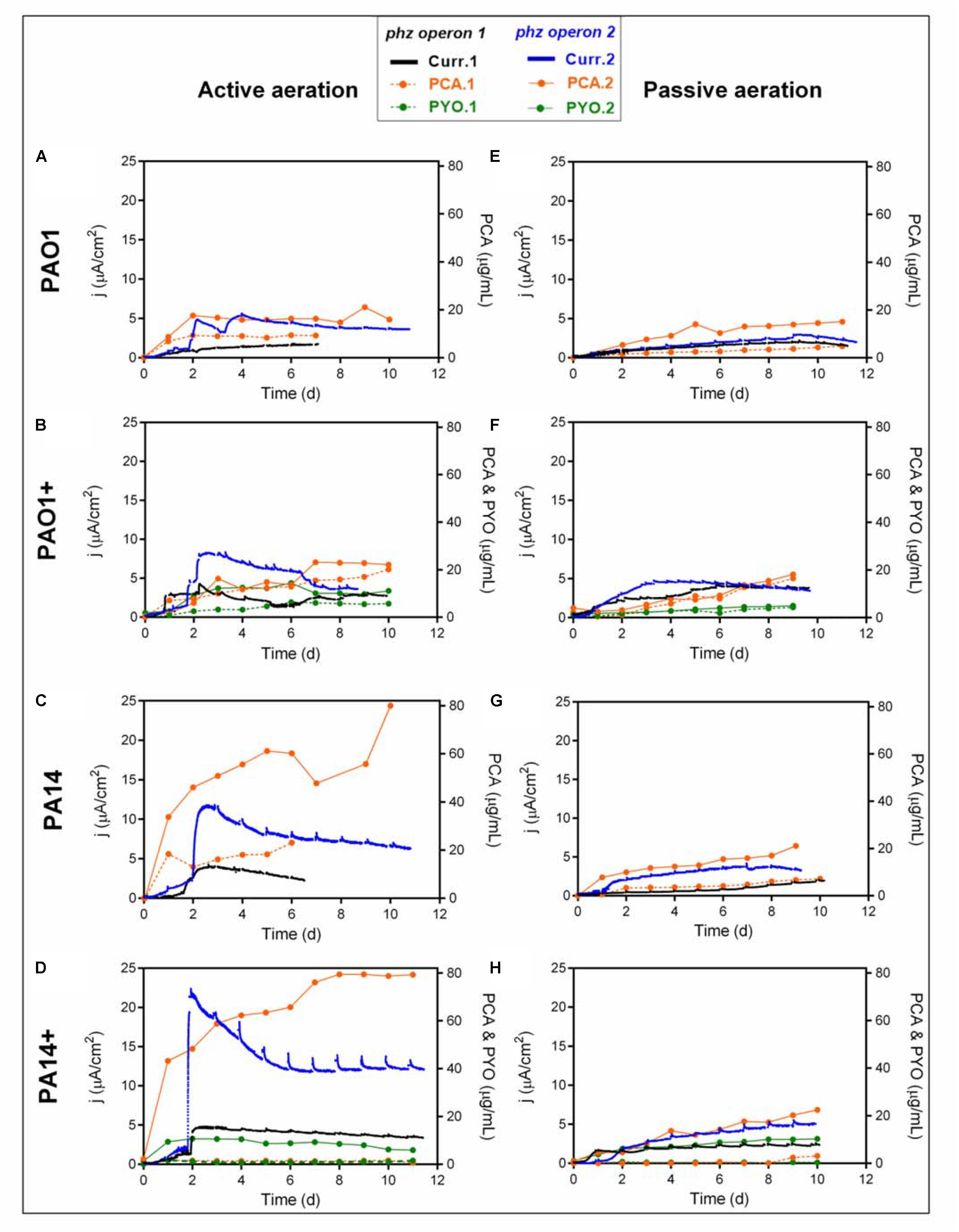
Figure 2. Heterologous phenazine production with P. putida under oxygen-limited conditions in a BES. Current densities generation and phenazine production of P. putida KT2440 harboring either phz operon 1 or operon 2 originating from P. aeruginosa strains PAO1 (A,E,B,F) or PA14 (C,G,D,H) under active (A–D) and passive (E–H) aeration. (B,F,D,H) indicated as PAO1+ and PA14+ are in combination with the phzM+S genes to synthesize both PCA and PYO. In the active aeration experiments, liquid phase aeration for the first 48 h was followed by passive headspace aeration. The highest currents generation and phenazine production were obtained from the engineered P. putida carrying operon 14.phz2+ under active aeration BES (Figure 2D). The BES data are means from biological duplicates.
The highest phenazine production and current generation was observed with P. putida 14.phz2, which produced 4-times higher PCA concentration, i.e., 80 μg/mL, resulting in 3-times higher maximum current density of 11.77 μA/cm2 compared to the other PCA gene clusters (Figure 2C). Also, the combination of PCA and PYO produced with P. putida 14.phz2+ outperformed the other strains with 80 μg/mL PCA, 11 μg/mL PYO, and 22.33 μA/cm2 maximum current density (Figure 2D). Again, the performance in oxygen-limited BES of this strain was in accordance with the aerobic flasks experiment. The presence of PYO as a soluble redox mediator in BES is markedly increasing electron shuttling to the anode. From control experiments with the P. putida KT2440 wildtype with ∼17 μg/mL exogenous PYO added to an oxygen-limited BES, we know that the PYO concentration is not sustained over time, likely because of adsorption to the electrode (Supplementary Figure S6B). Here, we see a fairly stable concentration of PYO over the course of the experiment with a continuously increasing concentration of PCA, suggesting continuous re-synthesis of PYO from PCA.
We also operated the BES under stringent oxygen limitation by only passively aerating the headspace (Figures 2E–H). Here, the lowest activities were observed for P. putida 14.phz1 (7.21 μg/mL of PCA, 2 μA/cm2 of current density) (Figure 2G) and P. putida 14.phz1 + (3.11 μg/mL of PCA, 0.65 μg/mL of PYO, 2.58 μA/cm2 current density) (Figure 2H). P. putida 14.phz2 was confirmed as the best PCA producer also under PA conditions with 21.09 μg/mL of PCA and 4.12 μA/cm2 current density (Figure 2G). Together with the phzM+S genes, P. putida 14.phz2+ was able to produce 22.48 μg/mL PCA and 10.29 μg/mL PYO, resulting in 5.53 μA/cm2 of current density (Figure 2H).
Overall, the performance of the P. putida phenazine producing strains in oxygen-limited BES is well in accordance with the performance in flasks experiment. Also, it is confirmed for these cultivation conditions that the phenazine synthesis in P. putida is strain origin- and operon-dependent. In direct comparison, the maximum produced amounts of PCA, however, are lower under oxygen-limited conditions compared to fully aerobic shake flasks because of the restricted energy availability. Notably, for the AA BES scenario, where the reactors were aerated for the first 48 h of growth to enable faster growth and cellular energy generation followed by completely PA with vent filters, a remarkable activity for PCA production resulting in roughly 1/3 of the concentrations of the aerobic cultivation was observed. This PCA production continued throughout the PA phase, confirming results from Schmitz et al. (2015) that stringently oxygen-limited conditions enable biosynthesis in P. putida if an anode is made accessible for metabolic electron discharge. The concentration of PYO did not further increase after switching from active to PA because of the requirement of molecular oxygen for PYO synthesis.
Evaluation of Metabolic Effects of Phenazine Usage in BES
To further evaluate the metabolic activity of the phenazine producing P. putida strains under oxygen-limited conditions, we analyzed biomass formation, glucose consumption, metabolite production, and the efficiency of donating metabolic electrons derived from the substrate glucose to the anode, which is measured as coulombic efficiency. Table 2 summarizes all results. We did not observe any strong biofilm formation with P. putida in our BES, and the evaluations at OD600 are well suited to compare the growth performance of the strains. This is different from BES experiments with P. aeruginosa as described by Bosire et al. (2016), where we found very strong and variable biofilm formation in the experiments, making a time-resolved estimation of growth difficult. The best growth, with the highest OD600 of 2.45, was observed for P. putida 14.phz2+ under AA, which sustained throughout the stationary phase until day 11 after inoculation. The average OD600 for AA was 1.95 compared to 1.57 for PA, reflecting the very stringent oxygen availability during PA. The glucose consumption is shown as the percentage of the provided glucose, which were 40 mM in AA BES and 20 mM in PA BES. Most cultures showed a glucose consumption of around 80% for active and around 70% for PA. Notably, the lowest glucose consumption of only ∼40% for both active and PA was found for strain P. putida 14.phz1. It has been reported before that P. putida produces several organic acids, such as 2-ketogluconate, gluconate, and acetate as side products from glucose degradation under oxygen-limited conditions (Nikel et al., 2015; Schmitz et al., 2015). For most strains, side product formation in these experiments stayed fairly low in the range of 0.1–2.9 mM. However, the best phenazine and most active current producer P. putida 14.phz2+ reached up to 4.80 mM gluconate. This finding is in line with bioelectrochemical studies of P. putida F1 with exogenous ferricyanide as an electron mediator, which showed a high efficiency of glucose to sugar acid conversion under oxygen-limited BES conditions (Lai et al., 2016).
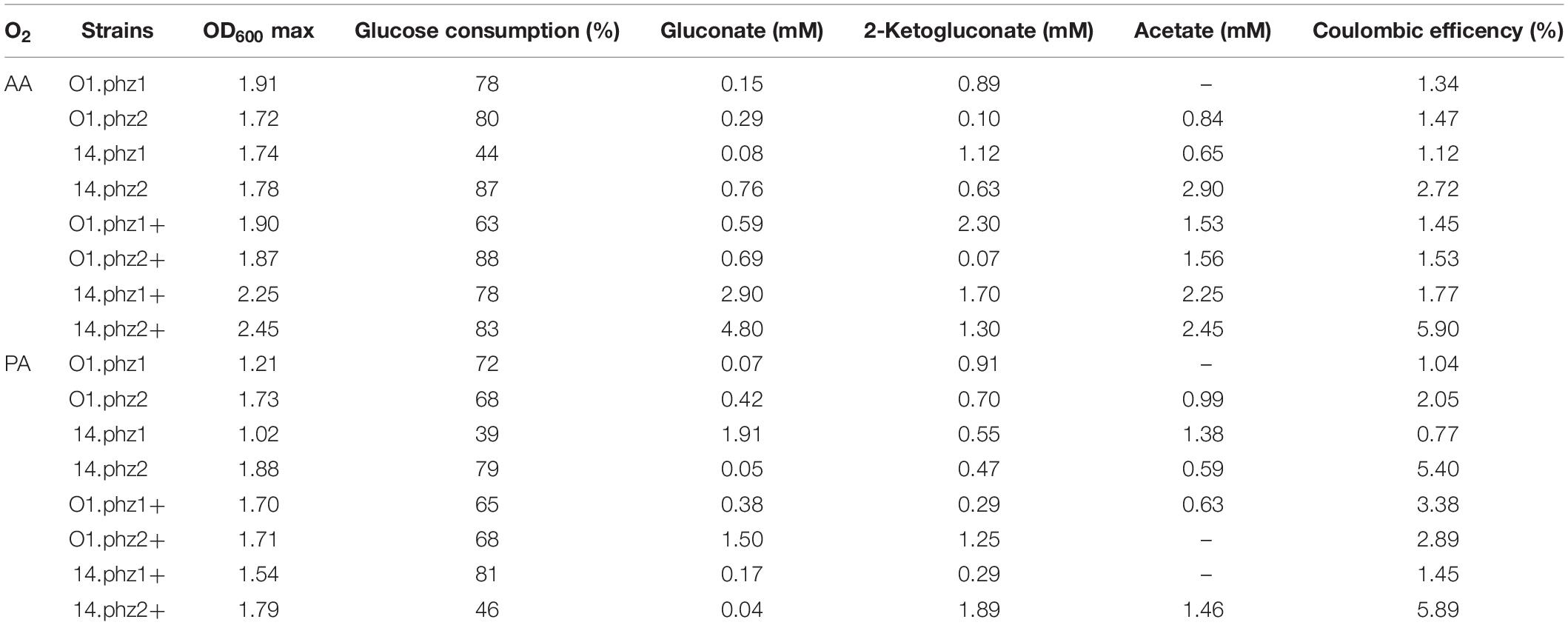
Table 2. Summary of glucose and metabolite evaluation of oxygen-limited BES experiments (Data are means of biological duplicates).
The estimation of energy retrieved from the carbon source in BES can be calculated as coulombic efficiency (Sleutels et al., 2011). Overall the coulombic efficiency observed from all strains was low and most of the consumed charge was used for slow respiration with oxygen (Table 2). However, the most active strains P. putida 14.phz2 and P. putida 14.phz2+ showed a significantly increased coulombic efficiency of up to 6% for both active and PA. This result shows that already small changes in the strain design for phenazine synthesis can not only improve phenazine production but also the efficiency of bioelectrochemical interaction of this strain. For a further improvement of phenazine based electron discharge to the anode, specific knowledge about the involved biochemical and energetic reaction pathways is required. However, this information is not available yet, neither for P. aeruginosa nor for P. putida. Work is on-going in our group to elucidate the specific phenazine electron transfer pathways and their integration into the energetic pathways of the microorganisms to remove electron discharge bottlenecks.
Deciphering the Effects of Operon and Strain Origin for Heterologous Production of Phenazine in P. putida
To elucidate possible reasons for the difference in phenazine production and resulting BES performance, we took a deeper look at the gene origin for PCA synthesis at DNA and protein level (Supplementary Data S1). In all constructs, we omitted the native promoter region of the operons and cloned the genes in a uniform salicylic acid inducible vector. Thus, any difference in performance and activity should originate from the sequence information of the genes. All plasmids have been re-sequenced to confirm the correctness of the original sequence and exclude mutations. In order to obtain a deeper understanding of the gene characteristic and its impact to dictate the phenazine production, a comparative in silico analysis of all genes used in this study has been conducted. Figures 3A,B shows the nucleotide alignment corresponding to two potential regulatory intergenic regions. These regions are long enough to allow for the attachment of DNA-binding proteins and they carry their own ribosome binding sites (consensus sequence: 5′-AGGAGG-3′). The first regulatory element (Figure 3A) is located between the genes phzA and phzB. The RBS of phzB (ribosome binding sites) is slightly interrupted by a deletion in the operons 1 compared with operons 2, where the RBS are complete and more conserved to the consensus sequence. The PhzB protein plays an important role to accelerate the condensation of two molecules of the intermediate product called 6-amino-5-oxocyclohex-2-ene-1-carboxylic acid (AOCHC) in the PCA synthesis (Blankenfeldt and Parsons, 2014). Thus, changes in PhzB numbers will significantly influence overall phenazine synthesis. Thus, a more functional RBS for phzB in operons 2 could be reflected in the overall higher phenazine synthesis from these operons compared to operons 1. Furthermore, the second regulatory element (Figure 3B) is located between the genes phzB and phzC. There is an insertion observed in operon 1 from strain PA14. The RBS in this operon seems to be slightly longer compared to all other operon. Other intergenic regions presenting RBS (Supplementary Data S2) were also detected but no differences observed among operons. Those regions are located between phzE and phzF as well as between phzF and phzG.
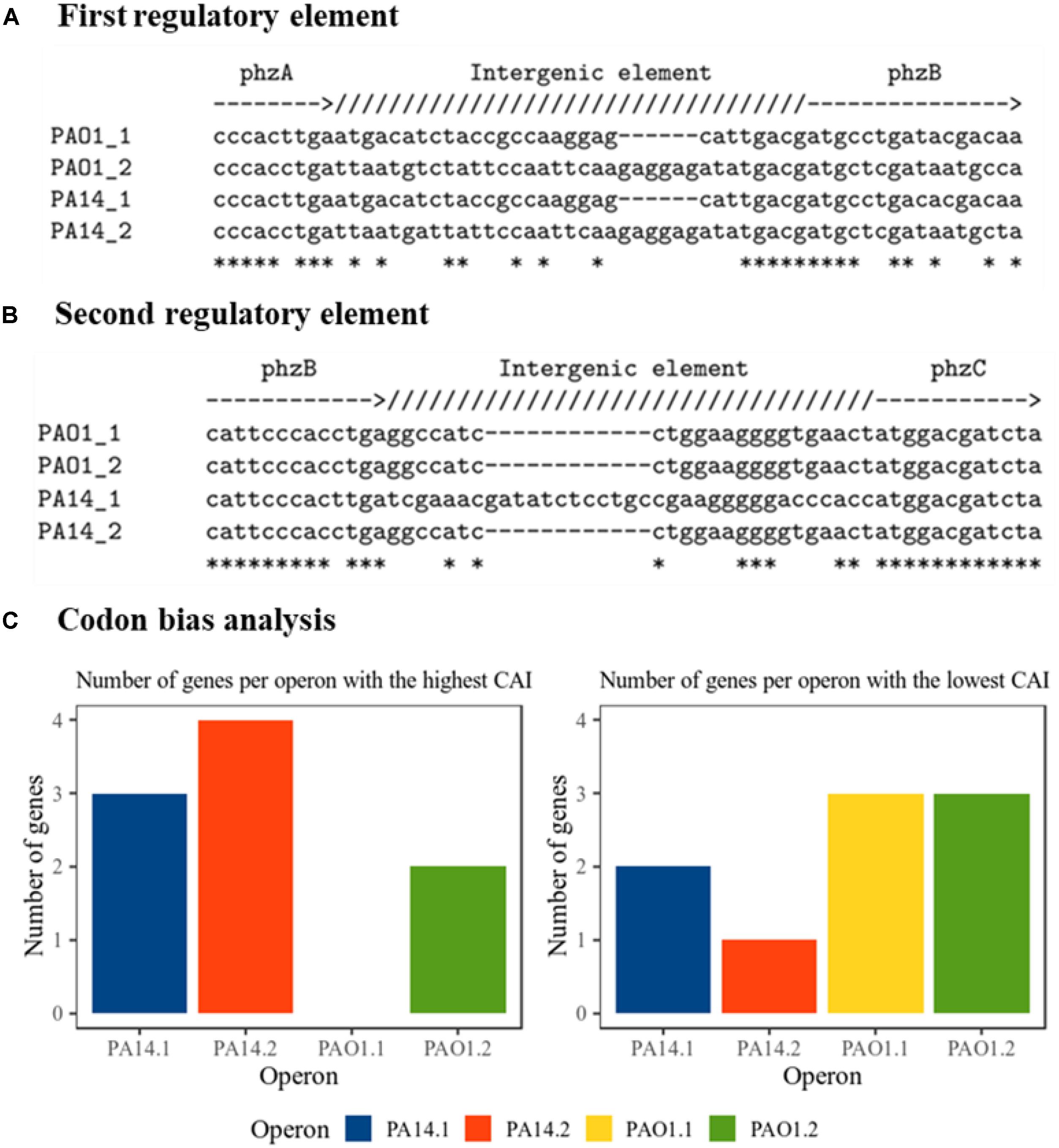
Figure 3. Nucleotide-based analysis of the phenazine clusters. (A) First intergenic regulatory element alignment; between phzA and phzB. Deleted nucleotides are marked as hyphen (-). Identical bases are marked as asterisk (∗). (B) Second intergenic regulatory element alignment; between phzB and phzC. Deleted nucleotides are marked as hyphen (-). Identical bases are marked as asterisk (∗). (C) Prediction of the expression tendency. Bars represent the number of genes in each operon detected with the highest or the lowest codon adaptation indexes, CAI, as detailed in Supplementary Table S2. Genes with equally high or low values are counted double.
An important aspect of heterologous gene expression is the fact that every organism is likely to prefer distinctive codons. In the genes of different organisms, a non-random distribution of synonymous codons has been explicated by comparative sequence analysis, the so-called codon bias (Quax et al., 2015). Besides influencing the expression levels of proteins, the codon bias is also affecting protein folding and regulation of protein expression (Pechmann and Frydman, 2013; Gingold et al., 2014). It has been revealed that even among genes in one genome, the codon usage might vary. In order to detect changes between the codon usages among genes in the four operons, codon adaptation indices CAI have been calculated for all genes using an index previously generated for P. putida KT2440 (Sharp et al., 2005). Counting the genes in each operon for which the highest (best fit of codon usage) or lowest (worst fit) CAI has been calculated (Figure 3C and Supplementary Table S2) gives an overall idea about the potential expression success of these operons. According to the results, operon 2 of PA14 would tend to have a better expression compared with the other constructs, which corresponds well to the experimental results.
A phylogenetic tree describing the evolutionary relation between the four operons has been generated (Supplementary Figure S7). The differences between operons within one strain are more prominent compared to the differences between strains. The detailed amino acid comparison between operon 1 and 2, either in PAO1 or PA14, gave the same result (Supplementary Data S1). The overall amino acid alignment analysis showed the biggest difference between operons 1 and 2 in the PhzB protein (91% identical), followed by the PhzA protein (97% identical). However, our protein structure analysis showed that these differences are in amino acids, which are located at the N- and C-terminus on the outer sphere of the protein, and are likely not affecting the protein function. In general, the structural analysis detected mostly small conformation changes in most of the enzymes involved in PCA synthesis (Supplementary Figures S8–S12). These changes are mainly located in the N-terminus, C-terminus and nearby loop regions, which could be the target of post-translational modifications and therefore these regions may not be present in the mature model protein. An exception is the protein PhzE in operon 2 of strain PA14 for which we observed relevant variations, which may be important in the activity of this protein (Figure 4). Structure differences are probably due to conformational changes of this protein resulting from two unique amino acid exchanges located close to each other and close to the enzyme catalytic site where the substrate chorismic acid binds (i.e., position 231 is a phenylalanine in PA14-phzE2 and a leucine for the other three PhzE versions; and position 266 is an isoleucine in PA14-phzE2 and a leucine in the other three PhzE versions). The PhzE protein plays an important role in the phenazine synthesis since it is the first protein of the synthesis pathway. It catalyzes the formation of 2-amino-2-desoxyisochorismic acid (ADIC) from the central metabolic intermediate chorismic acid (Spraggon et al., 2001). Therefore, the specific changes in the active site of this enzyme in the case of PA14-PhzE2 will likely have an impact on the overall phenazine synthesis and might be responsible for the superior performance of the P. putida 14.phz2 strain for PCA synthesis observed in this study.
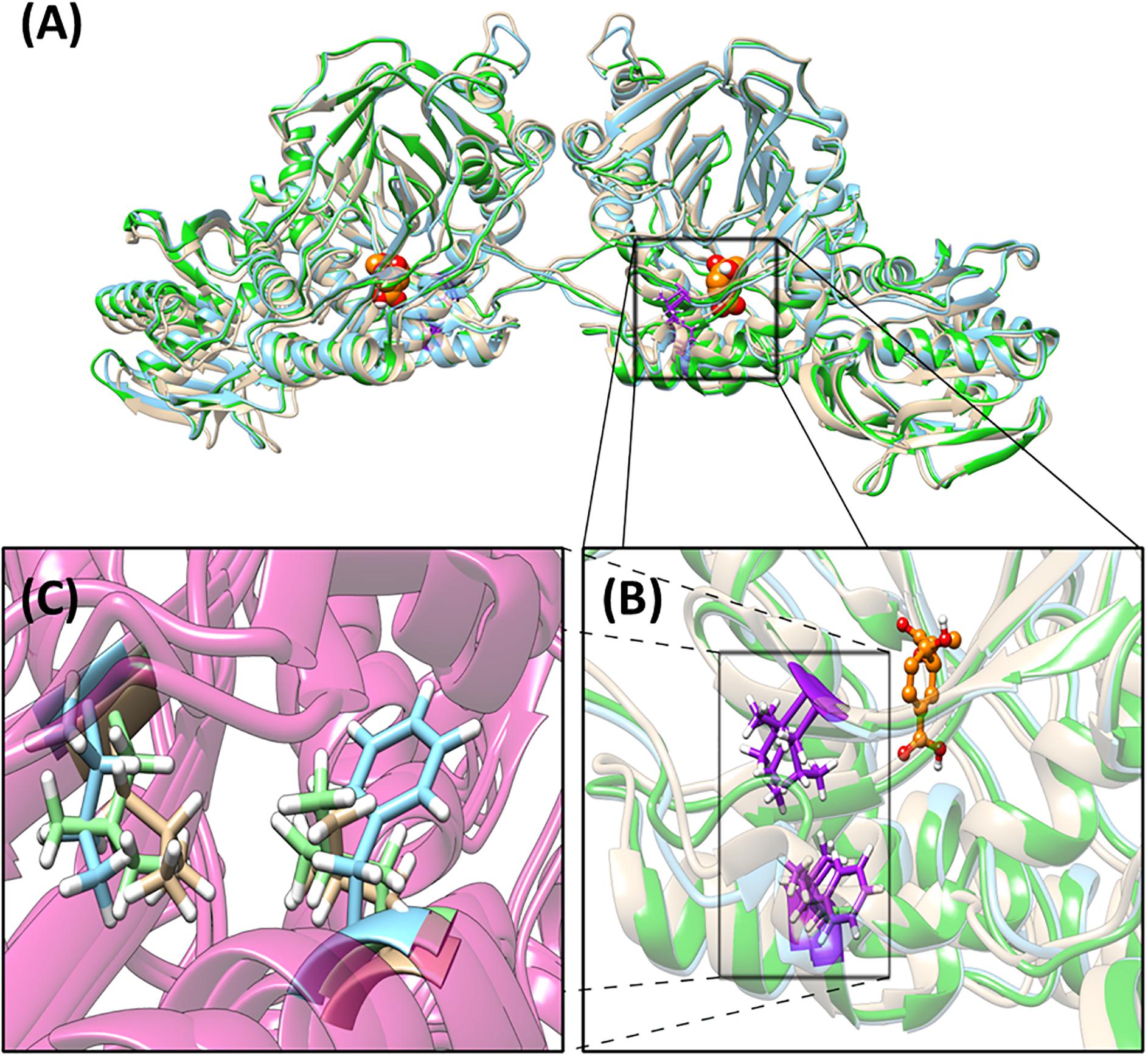
Figure 4. Structural analysis of PhzE. (A) PA14-PhzE2 is used as the reference for the alignment (blue). RMSD for PA14-PhzE1 (yellow): 1.056 Å. RMSD for PAO1-PhzE1 (green): 0.836 Å. PAO1-PhzE2 is identical to PAO1-PhzE1. Unique amino acid changes regarding in PA14-PhzE2 [p.(F231L;I266L)] are marked in purple. Substrate (chorismic acid) molecules are marked in yellow. (B) Detail of the substrate binding site vicinity. (C) Detail of the unique amino acid changes in PA14-PhzE2 and location inside the structure showing the phenylalanine in positon 231 and the isoleucine in position 266 for PA14-PhzE2 (both blue) compared to the leucine substitutions for both positions in the other PhzE proteins (green and yellow).
Conclusion
The activity of heterologous phenazine synthesis in P. putida is distinctively influenced by the origin of the genetic information. The gene origin dictates phenazine synthesis rate and yield under fully aerobic shake flask experiments and in oxygen-limited BES conditions. In this study, the phzA-G operon 2 from P. aeruginosa PA14 (14.phz2) was found to result in the best phenazine producer. P. putida 14.phz2 stunningly produced 4-times higher PCA concentrations (PCA only producer strain) as well as the respective PCA/PYO amounts (PCA and PYO producer strain) compared to the strains incorporating the other PCA synthesis operons. This elevated phenazine production also resulted in 3-times higher maximum current density and 5-times higher coulombic efficiency than for the other strains. Through deciphering potential sequence based reasons for this superior function, it has been revealed that the 14.phz2 operon contains distinctive regulatory elements for some of the contained genes and a codon usage prediction suggested a best compatibility of operon 14.phz2 with P. putida, potentially supporting protein synthesis. Most importantly, for the PhzE2 enzyme of strain PA14, the 3D-protein structure analysis showed two unique amino acid changes located close to the enzyme catalytic site where the substrate chorismic acid binds. This molecular difference might promote higher PCA synthesis with P. putida 14.phz2. With these results, P. putida 14.phz2 is a promising candidate to be employed as a biocatalyst in an oxygen-limited BES. The anodic electron discharge can now be coupled with the bioelectrochemical production of valuable compounds, such as rhamnolipids. Overall, the presented research findings will be valuable to establish an efficient, oxygen-limited biocatalysis of P. putida in BES.
Data Availability
All datasets generated for this study are included in the manuscript and/or the Supplementary Files.
Author Contributions
TA planned and conducted the experiments, analyzed the data and wrote the manuscript. SB performed the in silico sequence and structure analysis of the phenazine synthesis genes and proteins. LB co-supervised the work, discussed the results and revised the manuscript. MR conceived the work, planned the experiments, discussed the results and revised the manuscript.
Funding
The presented research was part of the DFG -funded project entitled “Mechanistic investigations of the syntrophy between Pseudomonas aeruginosa and 2,3-butanediol fermenters within the context of optimized phenazine-based current generation in bioelectrochemical systems” (Grant No. AG156/1-1) led by MR. TA and this work were also supported by University of Surabaya and BPP-LN Scholarship (RISTEK DIKTI) Indonesia. LB is partially funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany’s Excellence Strategy within the Clusters of Excellence TMFB 236 and FSC 2186.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors acknowledge the technical advice by Simone Schmitz and Christoph Lenzen.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.01990/full#supplementary-material
References
Bajracharya, S., Sharma, M., Mohanakrishna, G., Dominguez Benneton, X., Strik, D. P. B. T. B., Sarma, P. M., et al. (2016). An overview on emerging bioelectrochemical systems (BESs): technology for sustainable electricity, waste remediation, resource recovery, chemical production and beyond. Renew. Energ. 98, 153–170. doi: 10.1016/j.renene.2016.03.002
Benson, D. A., Karsch-Mizrachi, I., Lipman, D. J., Ostell, J., and Wheeler, D. L. (2005). GenBank. Nucleic Acids Res. 33, D34–D38. doi: 10.1093/nar/gki063
Beuker, J., Steier, A., Wittgens, A., Rosenau, F., Henkel, M., and Hausmann, R. (2016). Integrated foam fractionation for heterologous rhamnolipid production with recombinant Pseudomonas putida in a bioreactor. AMB Express 6:11. doi: 10.1186/s13568-016-0183-182
Blankenfeldt, W., and Parsons, J. F. (2014). The structural biology of phenazine biosynthesis. Curr. Opin. Struct. Biol. 29, 26–33. doi: 10.1016/j.sbi.2014.08.013
Bosire, E. M., Blank, L. M., and Rosenbaum, M. A. (2016). Strain- and substrate-dependent redox mediator and electricity production by Pseudomonas aeruginosa. Appl. Environ. Microbiol. 82, 5026–5038. doi: 10.1128/aem.01342-1316
Bosire, E. M., and Rosenbaum, M. A. (2017). Electrochemical potential influences phenazine production, electron transfer and consequently electric current generation by Pseudomonas aeruginosa. Front. Microbiol. 8:892. doi: 10.3389/fmicb.2017.00892
Choi, K. H., Kumar, A., and Schweizer, H. P. (2006). A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J. Microbiol. Methods 64, 391–397. doi: 10.1016/j.mimet.2005.06.001
Cock, P. J., Antao, T., Chang, J. T., Chapman, B. A., Cox, C. J., Dalke, A., et al. (2009). Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 25, 1422–1423. doi: 10.1093/bioinformatics/btp163
Gibson, D. G., Young, L., Chuang, R. Y., Venter, J. C., Hutchison, C. A. III, and Smith, H. O. (2009). Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345. doi: 10.1038/nmeth.1318
Gingold, H., Tehler, D., Christoffersen, N. R., Nielsen, M. M., Asmar, F., Kooistra, S. M., et al. (2014). A dual program for translation regulation in cellular proliferation and differentiation. Cell 158, 1281–1292. doi: 10.1016/j.cell.2014.08.011
Glasser, N. R., Kern, S. E., and Newman, D. K. (2014). Phenazine redox cycling enhances anaerobic survival in Pseudomonas aeruginosa by facilitating generation of ATP and a proton-motive force. Mol. Microbiol. 92, 399–412. doi: 10.1111/mmi.12566
Harnisch, F., Aulenta, F., and Schröder, U. (2011). “Microbial fuel cells and bioelectrochemical systems: industrial and environmental biotechnologies based on extracellular electron transfer,” in Comprehensive Biotechnology 2nd Edn., ed. M. Moo-Young. (Elsevier: Amsterdam), 643–659. doi: 10.1016/b978-0-08-088504-9.00462-1
Hartmans, S., Smits, J. P., van der Werf, M. J., Volkering, F., and de Bont, J. A. M. (1989). Metabolism of styrene oxide and 2-phenylethanol in the styrene-degrading Xanthobacter strain 124X. Appl. Environ. Microbiol. 55, 2850–2855.
Hu, J., Gao, G., and Xia, S. (2015). Development of a mediator-type bioelectrochemical sensor based on polypyrrole immobilized ferricyanide and microorganisms for biochemical oxygen demand fast detection. Int. J. Electroche 10, 9695–9705.
Kern, S. E., and Newman, D. K. (2014). Measurement of phenazines in bacterial cultures. Methods Mol. Biol. 1149, 303–310. doi: 10.1007/978-1-4939-0473-0-25
Krieg, T., Madjarov, J., Rosa, L. F. M., Enzmann, F., Harnisch, F., Holtmann, D., et al. (2019). “Reactors for microbial electrobiotechnology,” in Bioelectrosynthesis, eds F. Harnisch and D. Holtmann (Berlin: Springer International Publishing), 231–271. doi: 10.1007/10_2017_40
Lai, B., Yu, S., Bernhardt, P. V., Rabaey, K., Virdis, B., and Kromer, J. O. (2016). Anoxic metabolism and biochemical production in Pseudomonas putida F1 driven by a bioelectrochemical system. Biotechnol. Biofuels 9, 39–51. doi: 10.1186/s13068-016-0452-y
Loeschcke, A., and Thies, S. (2015). Pseudomonas putida-a versatile host for the production of natural products. Appl. Microbiol. Biotechnol. 99, 6197–6214. doi: 10.1007/s00253-015-6745-6744
Mavrodi, D. V., Bonsall, R. F., Delaney, S. M., Soule, M. J., Phillips, G., and Thomashow, L. S. (2001). Functional analysis of genes for biosynthesis of pyocyanin and phenazine-1-carboxamide from Pseudomonas aeruginosa PAO1. J. Bacteriol. 183, 6454–6465. doi: 10.1128/jb.183.21.6454-6465.2001
Mentel, M., Ahuja, E. G., Mavrodi, D. V., Breinbauer, R., Thomashow, L. S., and Blankenfeldt, W. (2009). Of two make one: the biosynthesis of phenazines. ChemBioChem 10, 2295–2304. doi: 10.1002/cbic.200900323
Nikel, P. I., Chavarria, M., Fuhrer, T., Sauer, U., and de Lorenzo, V. (2015). Pseudomonas putida KT2440 strain metabolizes glucose through a cycle formed by enzymes of the entner-doudoroff, embden-meyerhof-parnas, and pentose phosphate pathways. J. Biol. Chem. 290, 25920–25932. doi: 10.1074/jbc.M115.687749
Nikel, P. I., and de Lorenzo, V. (2013). Engineering an anaerobic metabolic regime in Pseudomonas putida KT2440 for the anoxic biodegradation of 1,3-dichloroprop-1-ene. Metab. Eng. 15, 98–112. doi: 10.1016/j.ymben.2012.09.006
Pechmann, S., and Frydman, J. (2013). Evolutionary conservation of codon optimality reveals hidden signatures of cotranslational folding. Nat. Struct. Mol. Biol. 20, 237–243. doi: 10.1038/nsmb.2466
Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C., et al. (2004). UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612. doi: 10.1002/jcc.20084
Pierson, L. S. III, and Pierson, E. A. (2010). Metabolism and function of phenazines in bacteria: impacts on the behavior of bacteria in the environment and biotechnological processes. Appl. Microbiol. Biotechnol. 86, 1659–1670. doi: 10.1007/s00253-010-2509-2503
Popov, A. L., Kim, J. R., Dinsdale, R. M., Esteves, S. R., Guwy, A. J., and Premier, G. C. (2012). The effect of physico-chemically immobilized methylene blue and neutral red on the anode of microbial fuel cell. Biotechnol. Bioproc. Eng. 17, 361–370. doi: 10.1007/s12257-011-0493-499
Quax, T. E., Claassens, N. J., Soll, D., and van der Oost, J. (2015). Codon bias as a means to fine-tune gene expression. Mol. Cell 59, 149–161. doi: 10.1016/j.molcel.2015.05.035
Rabaey, K., Boon, N., Höfte, M., and Verstraete, W. (2005). Microbial phenazine production enhances electron transfer in biofuel cells. Environ. Sci. Technol. 39, 3401–3408. doi: 10.1021/es048563o
Rahimnejad, M., Najafpour, G. D., Ghoreyshi, A. A., Shakeri, M., and Zare, H. (2011). Methylene blue as electron promoters in microbial fuel cell. Int. J. Hydrogen Energy 36, 13335–13341. doi: 10.1016/j.ijhydene.2011.07.059
Recinos, D. A., Sekedat, M. D., Hernandez, A., Cohen, T. S., Sakhtah, H., Prince, A. S., et al. (2012). Redundant phenazine operons in Pseudomonas aeruginosa exhibit environment-dependent expression and differential roles in pathogenicity. Proc. Natl. Acad. Sci. U.S.A. 109, 19420–19425. doi: 10.1073/pnas.1213901109
Rosenbaum, M. A., and Henrich, A. W. (2014). Engineering microbial electrocatalysis for chemical and fuel production. Curr. Opin. Biotechnol. 29, 93–98. doi: 10.1016/j.copbio.2014.03.003
Schmitz, S., Nies, S., Wierckx, N., Blank, L. M., and Rosenbaum, M. A. (2015). Engineering mediator-based electroactivity in the obligate aerobic bacterium Pseudomonas putida KT2440. Front. Microbiol. 6:284. doi: 10.3389/fmicb.2015.00284
Schmitz, S., and Rosenbaum, M. A. (2018). Boosting mediated electron transfer in bioelectrochemical systems with tailored defined microbial cocultures. Biotechnol. Bioeng. 115, 2183–2193. doi: 10.1002/bit.26732
Sharp, P. M., Bailes, E., Grocock, R. J., Peden, J. F., and Sockett, R. E. (2005). Variation in the strength of selected codon usage bias among bacteria. Nucleic Acids Res. 33, 1141–1153. doi: 10.1093/nar/gki242
Sharp, P. M., and Li, W. H. (1987). The codon adaptation index–a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 15, 1281–1295. doi: 10.1093/nar/15.3.1281
Sievers, F., and Higgins, D. G. (2018). Clustal omega for making accurate alignments of many protein sequences. Protein Sci. 27, 135–145. doi: 10.1002/pro.3290
Sievers, F., Wilm, A., Dineen, D., Gibson, T. J., Karplus, K., Li, W., et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using clustal omega. Mol. Syst. Biol. 7:539. doi: 10.1038/msb.2011.75
Sleutels, T. H., Darus, L., Hamelers, H. V., and Buisman, C. J. (2011). Effect of operational parameters on Coulombic efficiency in bioelectrochemical systems. Bioresour. Technol. 102, 11172–11176. doi: 10.1016/j.biortech.2011.09.078
Spraggon, G., Kim, C., Nguyen-Huu, X., Yee, M. C., Yanofsky, C., and Mills, S. E. (2001). The structures of anthranilate synthase of Serratia marcescens crystallized in the presence of (i) its substrates, chorismate and glutamine, and a product, glutamate, and (ii) its end-product inhibitor, L-tryptophan. Proc. Natl. Acad. Sci. U.S.A. 98, 6021–6026. doi: 10.1073/pnas.111150298
Steen, A., Utkür, F. Ö., Borrero-de Acuña, J. M., Bunk, B., Roselius, L., Bühler, B., et al. (2013). Construction and characterization of nitrate and nitrite respiring Pseudomonas putida KT2440 strains for anoxic biotechnical applications. J. Biotechnol. 163, 155–165. doi: 10.1016/j.jbiotec.2012.09.015
Venkataraman, A., Rosenbaum, M. A., Perkins, S. D., Werner, J. J., and Angenent, L. T. (2011). Metabolite-based mutualism between Pseudomonas aeruginosa PA14 and Enterobacter aerogenes enhances current generation in bioelectrochemical systems. Energy Environ. Sci. 4:4550. doi: 10.1039/c1ee01377g
Webb, B., and Sali, A. (2014). Comparative protein structure modeling using MODELLER. Curr. Protoc. Bioinformatics 47, 1–32. doi: 10.1002/0471250953.bi0506s47
Wierckx, N. J. P., Ballerstedt, H., de Bont, J. A. M., and Wery, J. (2005). Engineering of solvent-tolerant Pseudomonas putida S12 for bioproduction of phenol from glucose. Appl. Environ. Microbiol. 71, 8221–8227. doi: 10.1128/AEM.71.12.8221-8227.2005
Keywords: Pseudomonas putida, phenazine, PCA, pyocyanin, heterologous production, bioelectrochemical systems
Citation: Askitosari TD, Boto ST, Blank LM and Rosenbaum MA (2019) Boosting Heterologous Phenazine Production in Pseudomonas putida KT2440 Through the Exploration of the Natural Sequence Space. Front. Microbiol. 10:1990. doi: 10.3389/fmicb.2019.01990
Received: 20 June 2019; Accepted: 13 August 2019;
Published: 28 August 2019.
Edited by:
Amelia-Elena Rotaru, University of Southern Denmark, DenmarkReviewed by:
Alessandro Carmona, Independent Researcher, Logroño, SpainSatoshi Kawaichi, The University of Akron, United States
Copyright © 2019 Askitosari, Boto, Blank and Rosenbaum. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Miriam A. Rosenbaum, miriam.rosenbaum@leibniz-hki.de
 Theresia D. Askitosari
Theresia D. Askitosari Santiago T. Boto
Santiago T. Boto Lars M. Blank
Lars M. Blank Miriam A. Rosenbaum
Miriam A. Rosenbaum