- 1National Research Laboratory of Molecular Microbiology and Toxicology, Seoul National University, Seoul, South Korea
- 2Department of Agricultural Biotechnology, and Center for Food Safety and Toxicology, Seoul National University, Seoul, South Korea
- 3Department of Microbiology and Immunology, Chosun University School of Dentistry, Gwangju, South Korea
- 4Department of Veterinary Physiology, College of Veterinary Medicine, Research Institute for Veterinary Medicine, BK21 PLUS Creative Veterinary Research Center, Seoul National University, Seoul, South Korea
- 5College of Pharmacy, Pusan National University, Busan, South Korea
Nitric oxide (NO) and its derivatives are important effectors of host innate immunity, disrupting cellular function of infecting pathogens. Transcriptome analysis of Vibrio vulnificus, an opportunistic human pathogen, identified a set of genes induced upon exposure to NO. Among them, VvhmpA (V. vulnificus hmpA), encoding a multidomain NO dioxygenase, was the most greatly induced upon exposure to NO and was thus further characterized. Absorption spectra demonstrated that VvHmpA is a heme protein in which the heme iron can exist in either reduced, NO-bound, or oxidized state. Biochemical studies revealed that VvHmpA is a flavohemoglobin containing equimolar amounts of heme and FAD as cofactors. The KM and kcat values of VvHmpA for NO at 37°C, the temperature encountered by V. vulnificus in the host, were greater than those at 30°C, indicating that VvHmpA detoxifies high levels of NO effectively during infection. Compared with the wild type, the VvhmpA mutant exhibited a lower NO-decomposition activity and impaired growth in the presence of NO in vitro. Also, the cytotoxicity and survival of the VvhmpA mutant infecting the NO-producing murine macrophage cells were lower than those of the wild type. Furthermore, the mouse lethality of the VvhmpA mutant was reduced compared to that of the parental wild type. The combined results revealed that VvHmpA is a potent virulence factor that is induced upon exposure to NO and important for the survival and pathogenesis of V. vulnificus during infection.
Introduction
Nitric oxide (NO) and its derivatives, collectively called reactive nitrogen species (RNS), are among the most important components of the host innate immune system, the first line of defense against infecting pathogens (Fang, 2004). Under infectious conditions, NO is produced by inducible NO synthase (iNOS) which is expressed in phagocytes, particularly in macrophages (Fang, 2004). iNOS catalyzes the formation of NO and citrulline from L-arginine and oxygen (Stuehr, 1999). NO produced by iNOS can subsequently be converted into derivatives such as nitrogen dioxide (NO2), peroxynitrite (ONOO–), and dinitrogen trioxide (N2O3) (Fang, 2004; Stern and Zhu, 2014). Furthermore, nitrate in the diet can be reduced by commensals to nitrite, which interacts with gastric acid to result in RNS (Sobko et al., 2005; Davies et al., 2011; Tiso and Schechter, 2015) that act as antimicrobial barriers against ingested enteric pathogens (Fang, 2004). RNS can lead to the damage of cellular components, including metal centers of proteins, membrane lipids and nucleotide bases, and thereby inhibit respiration and interfere with DNA replication of pathogens (Fang, 1999). Therefore, pathogens have evolved sophisticated mechanisms to overcome nitrosative stress caused by the increased level of RNS, and the mechanisms are closely linked to their virulence (Bang et al., 2006; Richardson et al., 2006; Stern et al., 2012).
To defend against the nitrosative stress, pathogens rely on a variety of detoxifying enzymes such as NO dioxygenase, flavorubredoxin and associated oxidoreductase, cytochrome c nitrite reductase, S-nitrosoglutathione reductase, and peroxynitrite reductase (Bryk et al., 2000; Liu et al., 2001; Fang, 2004; Karlinsey et al., 2012). Among these, multidomain NO dioxygenases are a family of flavohemoglobins (Hmp) composed of the N-terminal globin domain carrying the heme-binding site and the C-terminal oxidoreductase domain containing NAD- and FAD-binding sites (Bonamore and Boffi, 2008). The NO dioxygenases typically detoxify the potentially harmful NO by oxidizing it to a less toxic metabolite NO3– under aerobic conditions (Hausladen et al., 2001), although some of the NO dioxygenases have an additional activity to reduce NO to N2O in the absence of oxygen (Mills et al., 2008; Forrester and Foster, 2012). Under conditions where O2 is not limiting, the N-terminal ferrous-oxy (Fe2+-O2) heme reacts with NO to yield NO3– and ferric-deoxy (Fe3+) heme, in which both atoms of O2 are incorporated into the NO3– (Gardner A. M. et al., 2000; Forrester and Foster, 2012). The C-terminal oxidoreductase domain transfers electrons from FADH2 to the ferric heme to regenerate the ferrous heme and the resulting FAD+ is then reduced back to FADH2 by utilizing the reducing power of cellular NAD(P)H (Forrester and Foster, 2012).
The opportunistic human pathogen, V. vulnificus, is a causative agent of life-threatening septicemia and necrotizing fasciitis in individuals with predisposing conditions, such as liver damage and kidney failure (Gulig et al., 2005; Oliver, 2015). It is reasonable to assume that V. vulnificus has to cope with the nitrosative stress imposed by the immune system in order to survive in the host and in turn ensure developing illness. Nevertheless, no definitive analysis on the mechanisms of V. vulnificus in surviving under nitrosative stress and thereby exhibiting virulence has been undertaken at a molecular level until now. Accordingly, we initiated a transcriptome analysis and identified hmpA, a homolog of Escherichia coli hmp (Echmp), which is the most preferentially expressed in V. vulnificus cells exposed to NO. The biochemical and kinetic properties of V. vulnificus HmpA (VvHmpA), the product of VvhmpA, were verified experimentally. Construction of the isogenic VvhmpA mutant and evaluation of its phenotypes provided evidence that VvHmpA could contribute to the survival and thereby the pathogenesis of V. vulnificus during infection.
Materials and Methods
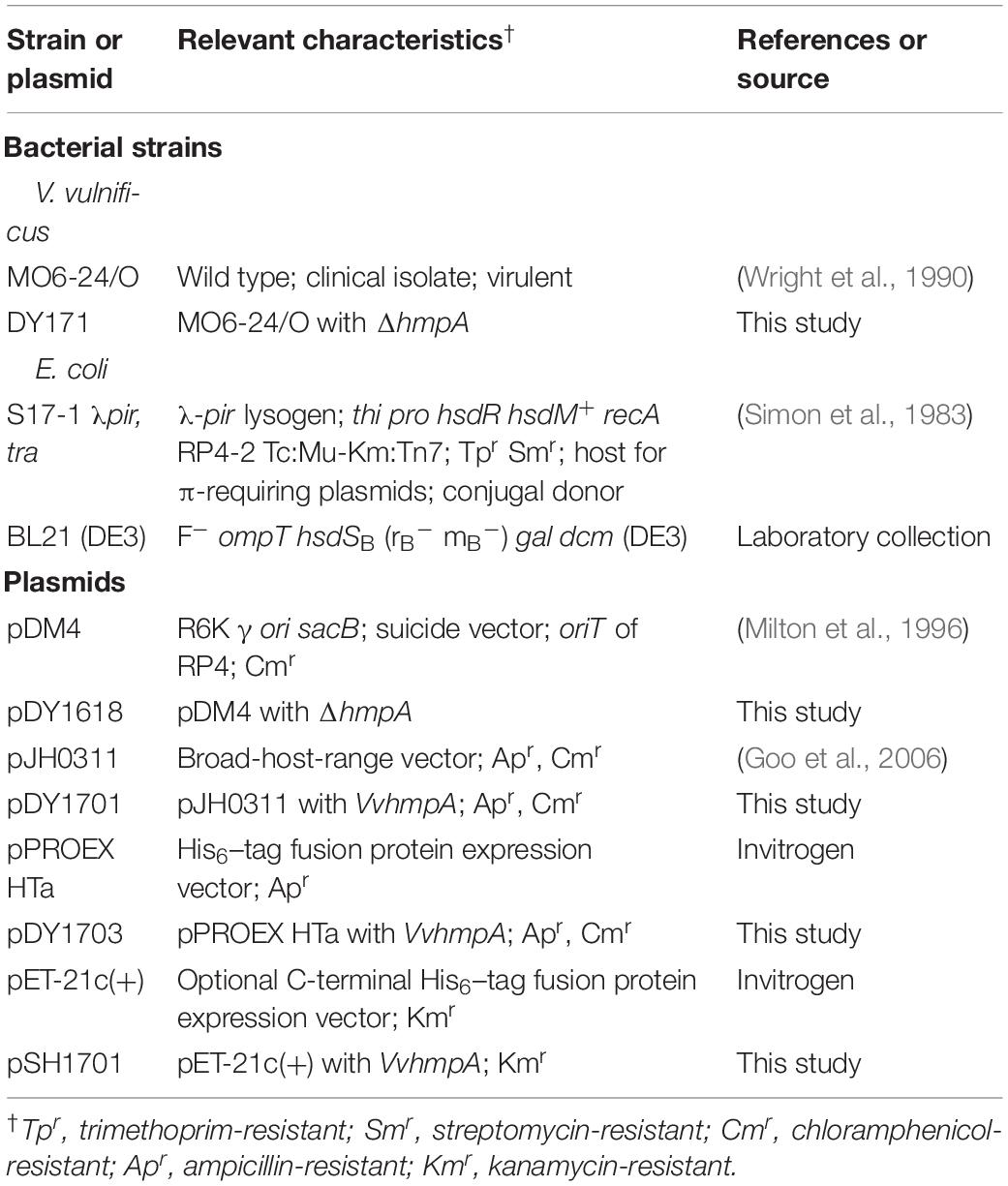
Strains, Plasmids, and Culture Conditions
The strains and plasmids used in this study are listed in Table 1. Unless otherwise noted, the V. vulnificus MO6-24/O (wild type), VvhmpA mutant, and VvhmpA-complemented strain were grown aerobically in Luria-Bertani (LB) medium supplemented with 2.0% (w/v) NaCl (LBS) at 30°C, and their growth was monitored spectrophotometrically at 600 nm (A600). The RAW 264.7 murine macrophage cells were obtained from Korean Cell Line Bank (Seoul, South Korea) and grown in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (FBS) and appropriate antibiotics [100 units ml–1 penicillin G and 100 μg ml–1 streptomycin (Gibco-BRL, Gaithersburg, MD)] in air supplemented with 5% CO2 at 37°C.
RNA Purification and Transcriptome Analysis
For transcriptome analysis, total RNAs were isolated from biological duplicates of V. vulnificus MO6-24/O, grown aerobically to A600 of 0.5 in M9 minimal media supplemented with 0.4% (w/v) glucose (M9G) and exposed to different types of NO donors for 10 min. As NO donors, excess amounts of either NO-releasing poly(lactic-co-glycolic acid)-polyethylenimine nanoparticles (NO/PPNPs) (releasing NO with t1/2 = 24 h at 37°C) or Spermine NONOate (releasing NO with t1/2 = 39 min at 37°C, Cayman Chemical, Ann Arbor, MI) were used (Bang et al., 2006; Nurhasni et al., 2015). The RNAs were further purified by removing DNA using TURBO DNase (Ambion, Austin, TX), and mRNA was selectively enriched by depleting rRNA using a Ribo-Zero rRNA removal kit (Epicentre, Madison, WI) according to the manufacturer’s procedure.
The cDNA libraries were constructed using a TruSeq Stranded mRNA Sample Prep kit (Illumina, San Diego, CA). Strand-specific paired-end 100-bp sequences were read from each cDNA library using HiSeq 2500 (Illumina) as described previously (Kim et al., 2018). The raw sequencing reads were analyzed using CLC Genomics Workbench 5.5.1 (CLC Bio, Aarhus, Denmark) and mapped on to the V. vulnificus MO6-24/O genome sequence (GenBankTM accession numbers: CP002469 and CP002470). The expression level of each gene was defined using the number of fragments per kilobase of transcript per million mapped reads (FPKM) (Conesa et al., 2016). Quantile-normalized FPKM values were then statistically analyzed by t-tests to identify the genes expressed differentially (fold change ≥2; p value of <0.05) in the V. vulnificus exposed to NO/PPNPs.
qRT-PCR
One microgram of the total RNA was used to synthesize cDNA with the iScriptTM cDNA synthesis kit (Bio-Rad, Hercules, CA), and real-time PCR amplification of the cDNA was performed by using the Chromo 4 real-time PCR detection system (Bio-Rad) with pairs of specific primers (Supplementary Table S1) as described previously (Jang et al., 2017). Relative expression levels of the VvhmpA mRNA in the same amounts of total RNA were calculated by using the 16S rRNA expression level as the internal reference for normalization (Jang et al., 2017). All qRT-PCR was conducted in biological triplicates.
Generation and Complementation of the VvhmpA Mutant
The VvhmpA gene was inactivated in vitro by deletion of the VvhmpA ORF (660-bp of 1,185-bp) using the PCR-mediated linker-scanning mutation method as described previously (Kim et al., 2014). Briefly, two pairs of primers HMPA01-F and -R (for amplification of the 5′ amplicon) and HMPA02-F and -R (for amplification of the 3′ amplicon) were designed and used (Supplementary Table S1). The VvhmpA gene with the 660-bp deletion was amplified by PCR using the mixture of both amplicons as the template and HMPA01-F and HMPA02-R as primers. The resulting ΔhmpA was ligated into SpeI-SphI-digested pDM4 to form pDY1618 (Table 1). E. coli S17-1 λpir, tra (Simon et al., 1983) strain containing pDY1618 was used as a conjugal donor to V. vulnificus MO6-24/O to generate the VvhmpA mutant, DY171 (Table 1). The conjugation and isolation of the transconjugant were conducted using the method described previously (Lim and Choi, 2014).
To complement the VvhmpA mutation, the upstream region and ORF of VvhmpA were amplified by PCR using HMPA03-F and HMPA03-R as primers (Supplementary Table S1). The amplified VvhmpA was cloned into the broad-host-range vector pJH0311 (Goo et al., 2006) to create pDY1701 (Table 1). Either pJH0311 or pDY1701 was transferred into DY171 by conjugation as described above.
Western Blot Analysis
The ORF of VvhmpA was amplified by PCR using a pair of primers, HMPA04-F and -R (Supplementary Table S1), digested with NcoI and SalI, and then ligated into pPROEX HTa (Invitrogen, Carlsbad, CA) to result in pDY1703 (Table 1). The His6-tagged VvHmpA was expressed in E. coli BL21 (DE3) containing pDY1703 and purified by using affinity chromatography (Qiagen) according to the manufacturer’s procedure, and used to raise rabbit anti-VvHmpA polyclonal antibody (AbFrontier, Seoul, South Korea) (Jang et al., 2017).
For Western blot analysis, V. vulnificus MO6-24/O grown to A600 of 0.5 in M9G was exposed either to Spermine NONOate as described above or to M9G (negative control) and then harvested to isolate total cellular proteins. To detect VvHmpA, the total cellular proteins (20 μg) were resolved on SDS-PAGE under reducing conditions and immunoblotted using the rabbit anti-VvHmpA antibody as described previously (Jang et al., 2017). A mouse antibody to E. coli DnaK was purchased (Enzo Life Sciences, Farmingdale, NY) and used to detect V. vulnificus DnaK (VvDnaK, a loading control) (Jang et al., 2017).
Purification of VvHmpA and Absorption Spectra of the Reduced, NO-Bound, and Oxidized VvHmpA
The VvhmpA ORF was amplified by PCR using a pair of primers, HMPA05-F and -R (Supplementary Table S1), digested with NdeI and XhoI, and then ligated into pET-21c(+) (Invitrogen) to result in pSH1701 (Table 1). The non-His6-tagged VvHmpA was expressed in E. coli BL21 (DE3) containing pSH1701 and purified by using a HiTrap Q anion-exchange column (GE Healthcare, Chicago, IL) and then a HiLoad Superdex 16/60 200 column (GE Healthcare) as described previously (Ahn et al., 2018). The protein was dissolved in the buffer with 20 mM Tris–HCl (pH 8.0) and 0.25 M NaCl at 4°C until use. The purified VvHmpA was quantitated using the Bradford method (Bradford, 1976).
To obtain the reduced VvHmpA, the purified VvHmpA (10 μM) was mixed with sodium hydrosulfite powder (1 μM, Sigma, St. Louis, MO) under anaerobic conditions (Ioannidis et al., 1992). Immediately, the residual sodium hydrosulfite was removed by using Ultracell-10K centricon (Millipore, Burlington, MA) in the anaerobic chamber with an atmosphere of 90% N2, 5% CO2, and 5% H2 (Coy Laboratory Products, Grass Lake, MI) (Kim et al., 2014). The reduced VvHmpA (10 μM) was incubated under anaerobic conditions with 40 μM Diethylamine NONOate sodium salt hydrate (Sigma) (releasing NO with t1/2 = 2 min at 37°C) for 10 min to obtain the NO-bound VvHmpA, or under aerobic conditions for 20 min to obtain the oxidized VvHmpA. The UV–vis absorption spectra of the VvHmpA proteins were recorded by using Shimadzu UV-1800 UV/VIS spectrophotometer (Shimadzu, Kyoto, Japan) at room temperature.
Reconstitution of VvHmpA With Heme and FAD and Quantitation of the Cofactors
To reconstitute VvHmpA with heme in vitro, 10 μM of the purified VvHmpA was gradually mixed with stoichiometric excess of hemin (1 mM in 0.01 M NaOH), and then incubated for 40 min at 4°C (Gardner A. M. et al., 2000). After the residual hemin aggregates were removed by centrifugation, the VvHmpA saturated with heme was further purified using a HiPrepTM 26/10 column (GE Healthcare). Similarly, 10 μM of the purified VvHmpA was incubated with 80 μM of FAD for an hour at room temperature, and then residual FAD was removed from the resulting reconstituted VvHmpA using Ultracell-10K centricon (Fang et al., 2017). The contents of the heme and FAD in VvHmpA both before and after the reconstitution with each of the cofactor were determined using the pyridine hemochromagen assay (Barr and Guo, 2015) and FAD fluorometric assay (Hirano and Namihira, 2017), respectively, as described previously.
Kinetic Analysis
Fresh NO stock solution was prepared daily as described previously (Poole et al., 1996) with modifications. Briefly, NO gas was formed by reacting 0.25 g sodium nitrite with 5 ml acidic ferrous sulfate solution, and purified by passing through 1 M NaOH solution and then degassed distilled water. The purified NO gas was collected and then dissolved in degassed distilled water to form the NO stock solution of approximately 500 μM, which was serially diluted to appropriate concentrations immediately before use.
To determine the kinetic properties of VvHmpA, 10 μl of the distilled water containing varying concentrations of NO and 10 μl of the purified and heme-reconstituted VvHmpA (1 μM) were delivered via separate syringes to 980 μl of the reaction buffer (50 mM potassium phosphate (pH 7.8) with 100 μM EDTA, 1 μM FAD and 100 μM NADH), that is pre-incubated at either 37 or 30°C (Gardner A. M. et al., 2000). The initial rates of NO decomposition were obtained by measuring the residual NO in the reaction mixture amperometrically using an ISO-NOP electrode (World Precision Instruments, Sarasota, FL) and plotted against the concentrations of NO. KM was determined as the NO concentration at which VvHmpA decomposes NO at one half rate of its Vmax (Smagghe et al., 2008) and the kcat was determined through the Lineweaver–Burk plotting (Chapman and Reid, 1999). Presuming only VvHmpA molecules containing heme are active, the kcat was expressed relative to heme.
NO-Decomposition Activity and Survival of V. vulnificus
The V. vulnificus strains grown to A600 of 0.5 in M9G were pre-incubated with 50 μM Spermine NONOate for 30 min to induce VvHmpA. The V. vulnificus cells were harvested with centrifugation, washed twice using PBS, and resuspended with 10 ml PBS. The PROLI NONOate (releasing NO with t1/2 = 1.8 s at 37°C, Cayman Chemical) was administered to the resuspended V. vulnificus cells to achieve 2 μM NO at a final concentration, and their NO-decomposition activities were determined by measuring the residual NO using the ISO-NOP electrode.
To examine the effect of VvHmpA on the survival of V. vulnificus under nitrosative stress, equal numbers (approximately 107 cells ml–1) of the V. vulnificus strains were used to inoculate the M9G containing 0.15 mg ml–1 NO/PPNPs at a final concentration. The resulting cultures were further incubated aerobically with shaking and the viable cells were counted at time intervals.
Survival and Cytotoxicity of V. vulnificus Infecting Immune Cells
The macrophage RAW 264.7 cells were resuspended in fresh DMEM containing 500 ng ml–1 E. coli O111:B4 lipopolysaccharide (Sigma) and 1 mM L-arginine (Sigma) to induce NO production (Walker et al., 1997) either with or without 500 μM L-NG-monomethyl arginine citrate (L-NMMA, Sigma), which is a known NO synthase inhibitor (Nathan and Hibbs, 1991). The RAW 264.7 cells were seeded into 24-well culture dishes at a concentration of 5 × 105 cells per well, and infected with the V. vulnificus strains at a multiplicity of infection (MOI) of 1. To determine the survival of the V. vulnificus strains directly affected by the NO from the RAW 264.7 cells, the culture dishes were washed two times to remove bacteria non-adherent to the macrophages as described previously (Lim and Choi, 2014). Following the last wash, the RAW 264.7 cells were broken with 0.1% Triton X-100 treatment for 20 min, and the recovered bacterial cells were enumerated as cfu per well (Lim and Choi, 2014). The numbers of live RAW 264.7 cells were determined at each time point using the LIVE/DEAD Viability/Cytotoxicity Kit for mammalian cells (Invitrogen) following manufacturer’s procedure, and used to result in numbers of bacteria per macrophage at each time point. The cytotoxicity of the V. vulnificus strains was determined by measuring the lactate dehydrogenase (LDH) activity released into the supernatant as described previously (Lim and Choi, 2014), and expressed using the LDH activity released from the RAW 264.7 cells completely lysed by 1.5% Triton X-100 (Sigma) as 100%. Statistical significance was determined by the Student’s t test.
Mouse Lethality Assay
The V. vulnificus strains grown to A600 of 0.5 were harvested and resuspended in PBS to 1.0 × 107 cfu ml–1. To determine mouse lethality, 100 μl of the suspension of either the wild type or VvhmpA mutant was used for intraperitoneal infection of the 7-week-old Institute of Cancer Research (ICR) female mice (specific-pathogen-free, Seoul National University) (n = 5 for each infection). The infection experiments were performed three times with the mice (total n = 15) to ensure reproducibility and percentages of the mice survival were recorded for 24 h. Statistical significance was determined by the log-rank test. All manipulations for mouse lethality assay were approved by the Animal Care and Use Committee at Seoul National University.
Data Analyses and Transcriptome Data Accession Number
Averages and standard deviations (S.D.) were calculated from at least three independent experiments. Statistical analyses were performed using GraphPad Prism 7.0 (GraphPad Software). All raw transcriptome data were deposited in Sequence Read Archive (SRA)1 under accession number PRJNA5134632.
Results
Effects of NO on V. vulnificus Transcriptome
The V. vulnificus MO6-24/O cells exposed to NO/PPNPs (Nurhasni et al., 2015) were harvested and their transcriptomes were analyzed. Transcriptome analysis revealed 551 genes that were differentially expressed (fold change ≥2; p value <0.05) upon exposure to NO; 320 genes were upregulated and 231 genes were downregulated (data were deposited in SRA, see text footnote 1, under accession number PRJNA513463, Supplementary Tables S2, S3). Among the genes upregulated upon exposure to NO, 8 genes potentially involved in nitrosative stress defense were selected (Figure 1). Six of the genes are predicted to encode proteins involved in the defense against nitrosative stress: NO dioxygenase, nitrite reductase large subunit, nitrite reductase small subunit, NrfA (cytochrome c nitrite reductase subunit c552), NnrS (putative heme- and copper-containing transmembrane protein), and NrfD (cytochrome c nitrite reductase subunit). The other two genes are predicted to encode transcriptional regulators that control the expression of the genes involved in nitrosative stress defense: NorR (NO reductase transcription regulator) and NsrR (nitrite-sensitive transcriptional repressor).
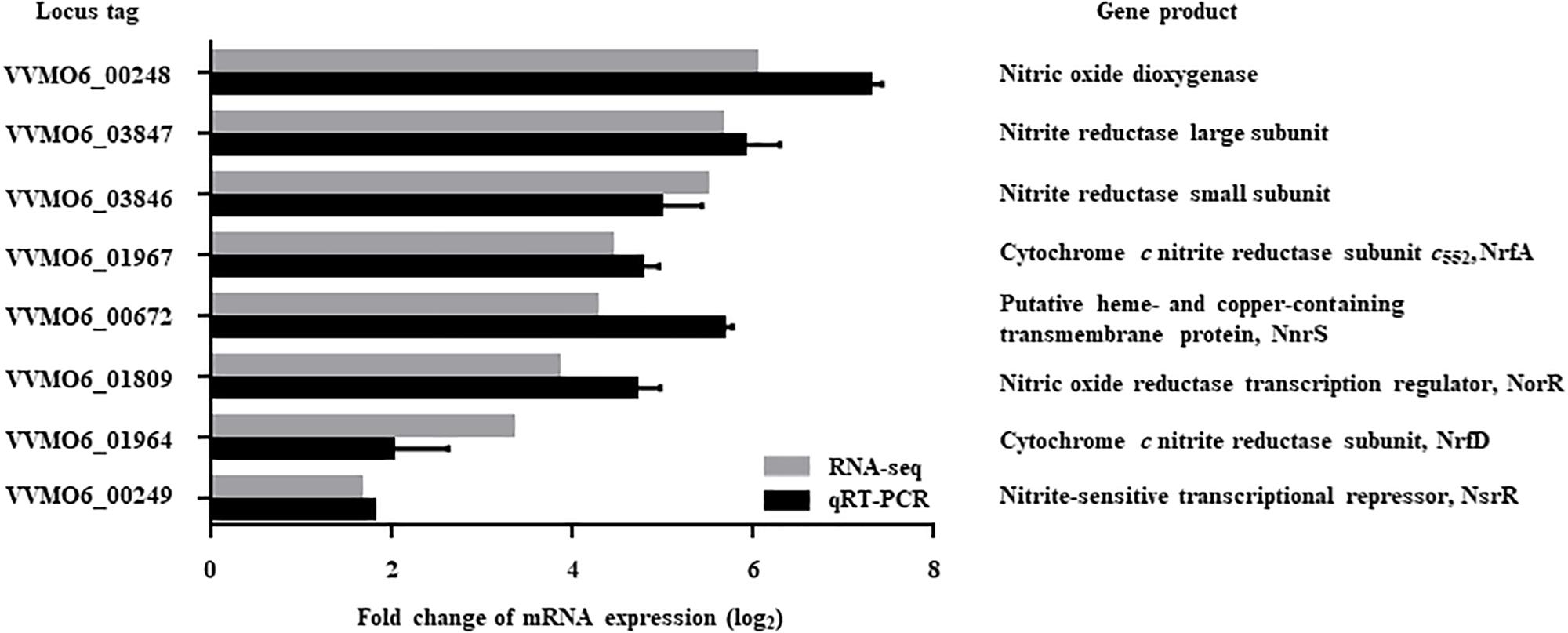
Figure 1. Genes upregulated by NO exposure and possibly involved in nitrosative stress defense. Among the NO-upregulated genes (fold change ≥2; p value of <0.05) identified by transcriptome analysis, 8 genes potentially involved in nitrosative stress defense were selected and their upregulation was confirmed by qRT-PCR. Each column represents the mRNA expression level of V. vulnificus MO6-24/O exposed to NO/PPNPs relative to that exposed to PPNPs (negative control). Error bars represent the S.D. Locus tags are based on the V. vulnificus MO6-24/O genome sequence (GenBankTM accession numbers: CP002469 and CP002470) and the products of the genes are presented on the right. NO/PPNPs, NO-releasing poly(lactic-co-glycolic acid)-polyethylenimine nanoparticles; PPNPs, poly(lactic-co-glycolic acid)-polyethylenimine nanoparticles.
Expression of the 8 genes in V. vulnificus exposed to NO/PPNPs was reevaluated by using quantitative real-time PCR (qRT-PCR) analyses, further confirming that NO exposure induced transcription of the genes (Figure 1). Since the expression of the VVMO6_00248 gene, predicted to encode an NO dioxygenase, increased the most upon exposure to NO (Figure 1), the gene was selected for further study. To verify that the induction of the VVMO6_00248 gene is not confined to a specific NO donor, expression of the gene upon exposure of V. vulnificus to Spermine NONOate as an alternate NO donor was determined. As shown in Figure 2, exposure to the Spermine NONOate increased the levels of the gene product as well as the transcript of VVMO6_00248 as determined by Western blot analysis and qRT-PCR, respectively. The combined results confirmed that the expression of the VVMO6_00248 gene in V. vulnificus is induced upon exposure to NO.
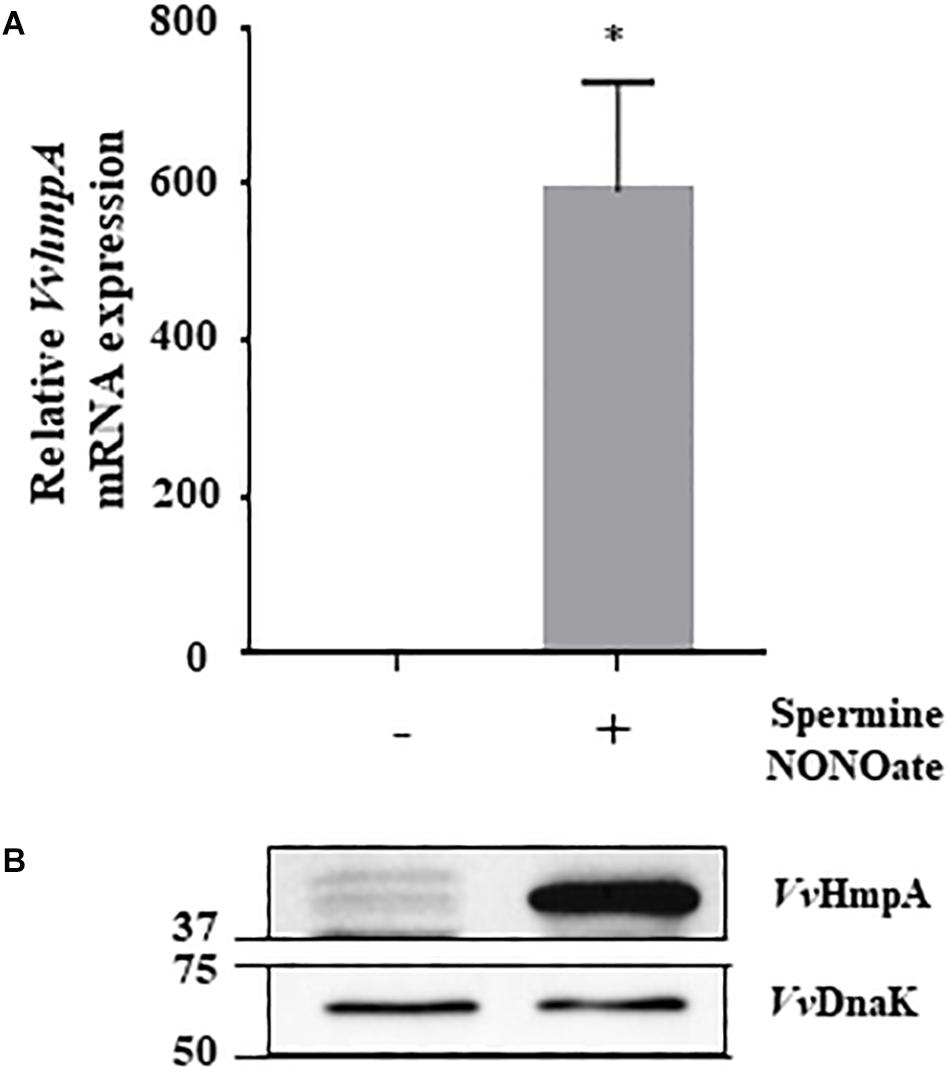
Figure 2. Expression of VvhmpA under nitrosative stress. RNAs and proteins were extracted from V. vulnificus MO6-24/O exposed either to Spermine NONOate or M9G (negative control). (A) The VvhmpA mRNA levels were determined by qRT-PCR analyses, and expressed using the VvhmpA mRNA level of the culture unexposed to NO as 1. Error bars represent the S.D. ∗p < 0.05 relative to the culture unexposed to NO. (B) Total proteins of the cultures were resolved on reducing SDS-PAGE, and VvHmpA and VvDnaK were immunoblotted using the rabbit anti-VvHmpA antibody and the mouse anti-DnaK antibody, respectively. The protein size markers (Bio-Rad) on the left are in kilodaltons.
Identification and Sequence Analysis of VvHmpA
The amino acid sequence deduced from the nucleotide sequence of the VVMO6_00248 gene revealed a putative protein, composed of 394 amino acids with a theoretical molecular mass of 44.3 kDa and a pI of 5.21. The deduced amino acid sequence of VVMO6_00248 was 62%, 61%, and 51% identical to known NO dioxygenases such as E. coli Hmp (EcHmp), Salmonella enterica serovar Typhimurium Hmp (StHmp), and Vibrio cholerae HmpA (VcHmpA) (Figure 3), respectively. These findings led us to assume that the protein is an NO dioxygenase of V. vulnificus and thus named it VvHmpA. Amino acid sequence analysis of VvHmpA further revealed that the protein possesses the highly conserved heme-binding domain of known NO dioxygenases (Bonamore and Boffi, 2008) (Figure 3). Moreover, VvHmpA contains the putative NAD- and FAD-binding domains, which are also conserved in NO dioxygenases (Bonamore and Boffi, 2008) (Figure 3). The combined results proposed that VvHmpA is a multidomain NO dioxygenase that possibly contains heme and FAD as its cofactors.

Figure 3. Sequence analysis of V. vulnificus HmpA (VvHmpA), E. coli Hmp (EcHmp), S. Typhimurium Hmp (StHmp), and V. cholerae HmpA (VcHmpA). The amino acid sequences retrieved from the NCBI protein database (accession numbers: WP_013570994.1 for VvHmpA, NP_417047.1 for EcHmp, WP_000883146.1 for StHmp, and WP_000957477.1 for VcHmpA) were aligned using the Clustal Omega program. Identical (black boxes), conserved (gray boxes), and missing (dashes) sequences are indicated. The conserved amino acid residues potentially involved in the binding of heme and FAD are indicated above the amino acid sequences by asterisks and dots, respectively. The putative NAD-binding domain is boxed by a black line.
VvHmpA Is a Flavohemoglobin
Absorption spectra of VvHmpA in solution were characterized to reveal physical properties of the protein. A narrow intense band centered at approximately 433 nm, referred to as the Soret band and characteristic of heme proteins (Anderson and Robertson, 1995), was observed in the absorption spectrum of the reduced VvHmpA (Figure 4A). The 433 nm Soret peak of the reduced VvHmpA was altered to the 420 and 404 nm Soret peaks of the NO-bound and oxidized VvHmpAs, respectively (Figure 4A). These Soret peak alterations most probably resulted from the changes in the physical state of iron in the heme protein, as demonstrated previously with a heme protein EcHmp which showed Soret peaks at 431.5, 419, and 403.5 nm in its reduced, NO-bound, and oxidized states, respectively (Ioannidis et al., 1992; Anderson and Robertson, 1995; Gardner A. M. et al., 2000). Therefore, it is reasonable to propose that VvHmpA is also a heme protein, in which iron can exist in either reduced, NO-bound, or oxidized state.
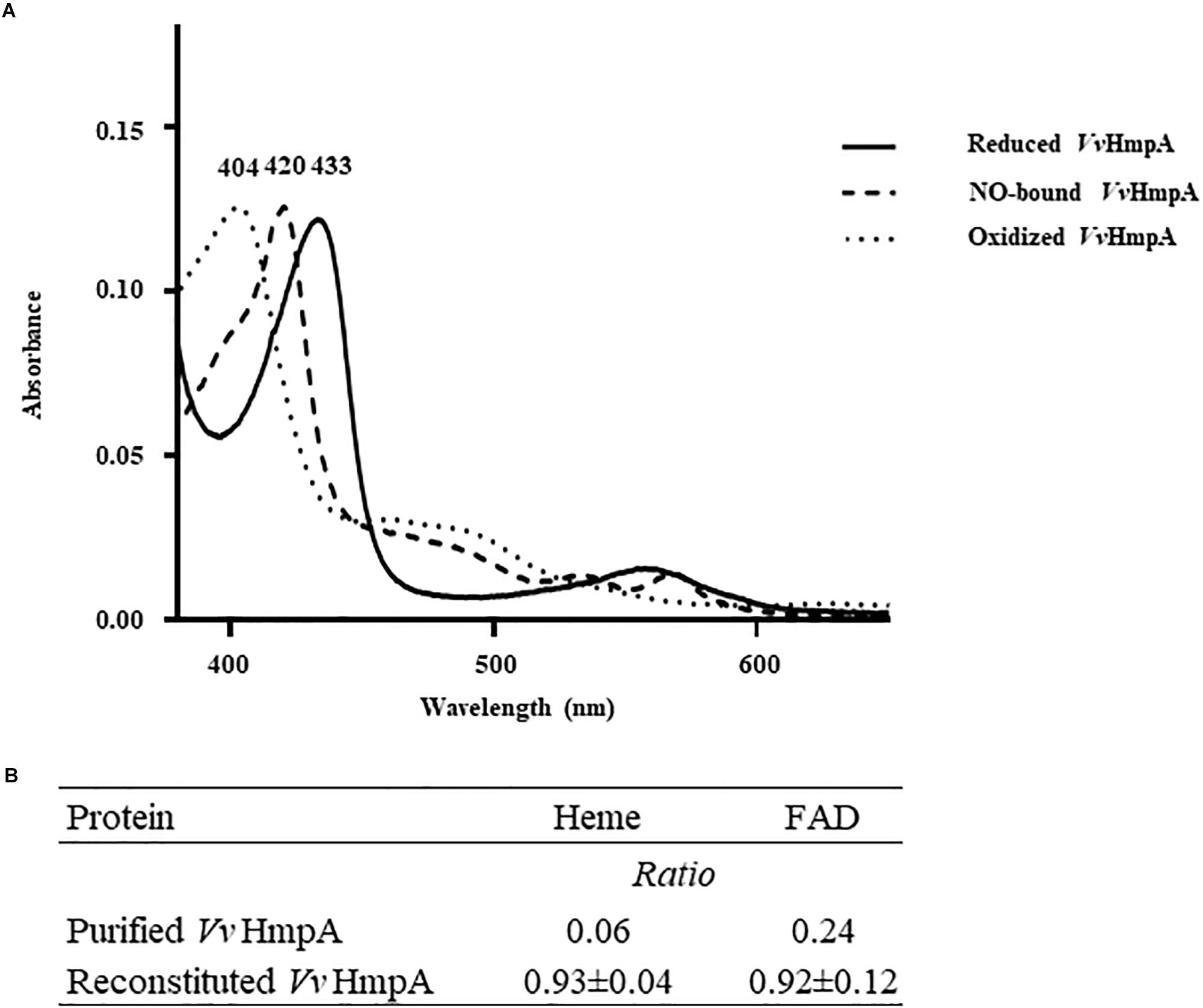
Figure 4. Absorption spectra and cofactors of VvHmpA. (A) The absorption spectra of the purified VvHmpA in the reduced (solid line), NO-bound (dashed line), and oxidized (spotted line) states were measured using a UV–vis spectrophotometer. The numbers above the peaks indicate the wavelengths of the observed Soret peaks. (B) The ratios of heme and FAD per one molecule of purified and reconstituted VvHmpA.
Measurement of heme content revealed that the overexpressed and purified VvHmpA contains 0.06 molecules of heme per VvHmpA monomer (Figure 4B). It was possible that the amount of heme provided in the condition used for the overexpression and purification of the VvHmpA protein is not sufficient to saturate the protein. To examine the possibility, the purified VvHmpA was saturated with excess hemin in vitro (Gardner A. M. et al., 2000), and then the content of heme in the reconstituted VvHmpA was determined. Following the saturation, the content of heme in the VvHmpA increased to 0.93 ± 0.04 molecules of heme per VvHmpA monomer (Figure 4B). These results suggested that holo-VvHmpA contains approximately one molecule of heme per protein monomer.
FAD fluorometric assay was carried out in order to determine the FAD content of the overexpressed and purified VvHmpA. Prior to saturation with excess FAD in vitro, the purified VvHmpA contained 0.24 molecules of FAD per VvHmpA monomer (Figure 4B). After saturation with excess FAD in vitro, the content of FAD in the reconstituted VvHmpA increased to 0.92 ± 0.12 molecules of FAD per VvHmpA monomer (Figure 4B), suggesting that the holo-VvHmpA contains approximately one molecule of FAD per protein monomer. The combined results proposed that the holo-VvHmpA protein is a flavohemoglobin that contains equimolar amounts of heme and FAD as cofactors.
Kinetic Properties of VvHmpA for NO Decomposition
To determine the kinetic properties of VvHmpA, the initial rates of NO decomposition were measured at different concentrations of NO. At the concentrations of NO exceeding 0.15 μM, which are most probably encountered in the human immune system (Toledo and Augusto, 2012), the NO-decomposition rate of VvHmpA was higher at 37°C rather than at 30°C (Figure 5A). The Vmax of VvHmpA for NO, obtained from the Michaelis–Menten plot, was approximately 110.1 ± 6.1 nM s–1 at 37°C and the NO-decomposition rate at the concentration of 1 μM NO was approximately 80% of the Vmax value (Figure 5). In contrast, the Vmax of VvHmpA for NO at 30°C was about 47 ± 1.5 nM s–1 and the NO-decomposition rate at the concentration of 1 μM NO was approximately 90% of the Vmax value (Figure 5). The KM values of VvHmpA for NO were 0.3 ± 0.04 μM and 0.1 ± 0.01 μM at 37 and 30°C, respectively (Figure 5B). The kcat values of VvHmpA for NO were 21.4 ± 1.2 s–1 and 9.1 ± 0.2 s–1 at 37 and 30°C, respectively (Figure 5B). Considering that V. vulnificus is a pathogen infecting humans with the body temperature of 37°C, the results indicated that VvHmpA is more efficient at decomposing high levels of toxic NO in the host than in the natural environment.
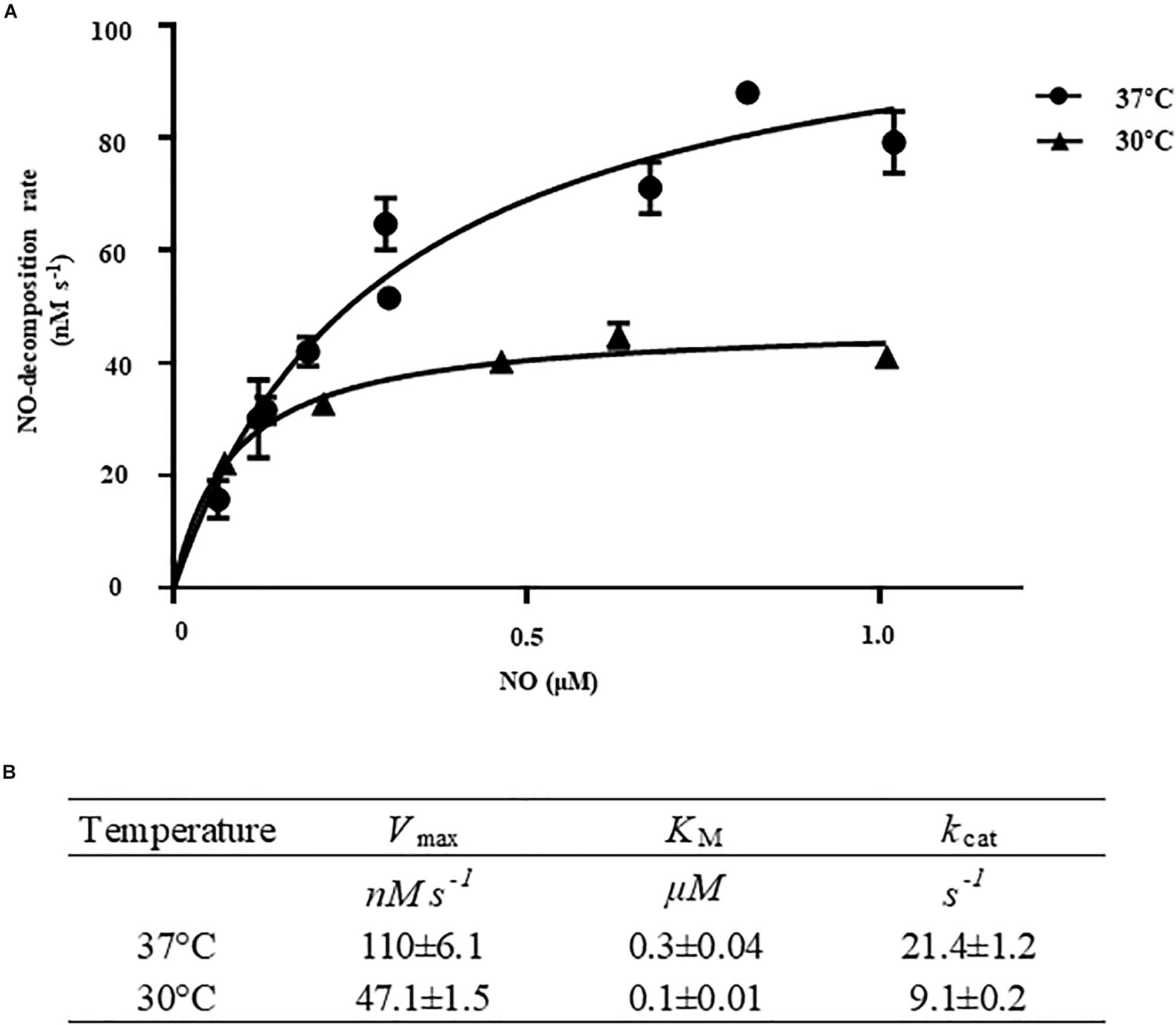
Figure 5. Kinetic analysis of VvHmpA. (A) The VvHmpA protein and various concentrations of NO were delivered into the reaction buffer. The initial rates of NO decomposition were determined by measuring the residual NO in the reaction mixture and plotted against the corresponding initial concentrations of NO. Error bars represent the S.D. (B) Vmax, KM, and kcat values for NO were determined by fitting the curve (A) to a classical Michaelis–Menten enzyme kinetic equation. The turnover rate (kcat) is expressed relative to heme.
VvHmpA Is Essential for the Survival of V. vulnificus Under Nitrosative Stress in vitro
To evaluate the role of VvHmpA in V. vulnificus encountering nitrosative stress, the NO-decomposition activities of the V. vulnificus strains were compared (Figure 6A). When NO was administered to the V. vulnificus wild-type culture, the NO concentration decreased rapidly in the culture and the residual NO was not detectable after 100 s, indicating that the V. vulnificus wild type effectively decomposes NO in vitro. In contrast, the rate at which NO levels decrease in the VvhmpA mutant culture was much slower than that in the wild-type culture (Figure 6A). The rate at which NO levels decrease in the VvhmpA mutant culture was close to that in the medium without bacterial inoculation (control, PBS), indicating that the NO-decomposition activity of the VvhmpA mutant was significantly impaired (Figure 6A). By complementation of the VvhmpA gene, the impaired NO-decomposition activity of the VvhmpA mutant was restored to a level even higher than that of the wild type (Figure 6A). The results suggested that the NO-decomposition activity of V. vulnificus is mostly dependent on VvHmpA in vitro.
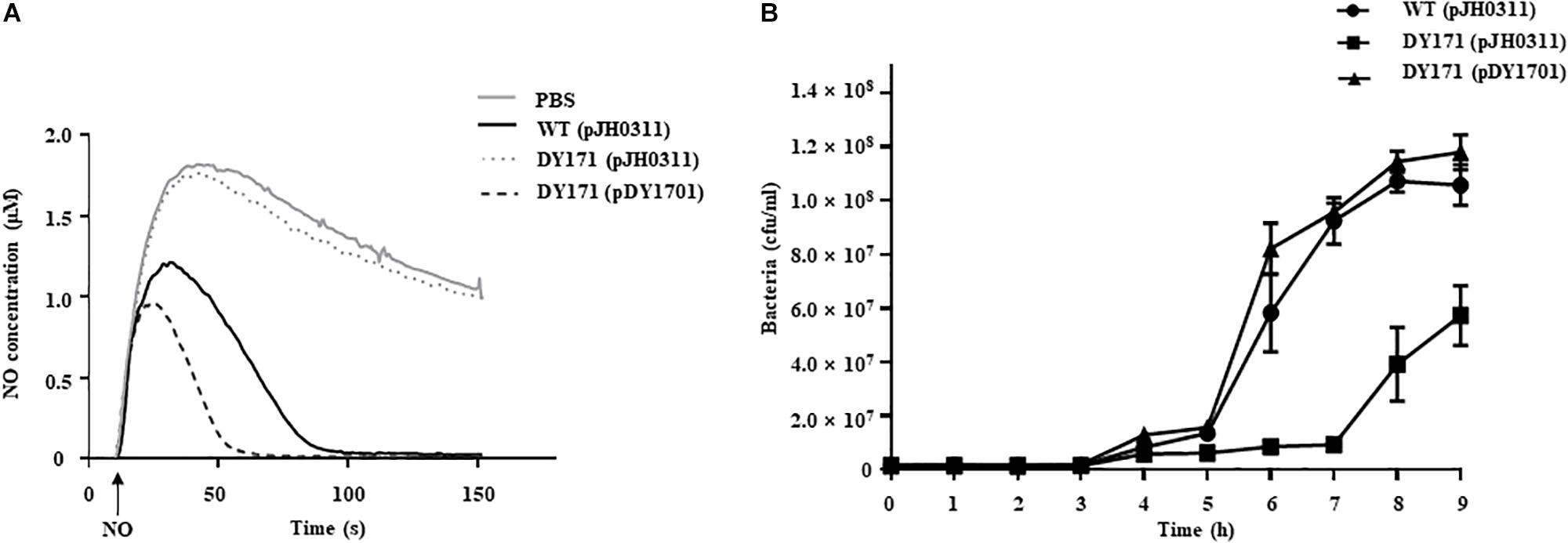
Figure 6. NO decomposition and survival of the V. vulnificus strains under nitrosative stress. (A) The V. vulnificus strains were pre-exposed to Spermine NONOate to induce VvHmpA, and then 2 μM of NO was administered to the strains at the time designated by an arrow. The residual NO in the mixtures was measured to determine the NO decomposition. (B) Survival of the V. vulnificus strains exposed to excess NO was monitored by counting viable cells at time intervals. Error bars represent the S.D. PBS, control; WT (pJH0311), wild type; DY171 (pJH0311), VvhmpA mutant; DY171 (pDY1701), VvhmpA-complemented strain.
To examine the effects of VvHmpA on the survival of V. vulnificus under nitrosative stress in vitro, the growth of V. vulnificus strains in M9G was compared in the presence of NO (Figure 6B). The growth of the VvhmpA mutant was significantly delayed compared with that of the wild type. That is, the VvhmpA mutant did not enter the exponential phase until 7 h post-inoculation, which is delayed for approximately 2 h compared with the wild type. The delayed growth of the VvhmpA mutant was restored in the VvhmpA-complemented strain (Figure 6B), indicating that VvHmpA is able to effectively decompose toxic NO to the level at which V. vulnificus can manage to grow. The combined results led us to conclude that VvHmpA detoxifies NO effectively in vitro, which in turn ensures survival of the pathogen under nitrosative stress.
VvHmpA Is Essential for the Virulence of V. vulnificus ex vivo
To examine the survival of V. vulnificus strains in the presence of NO-producing murine macrophage RAW 264.7 cells, the numbers of the V. vulnificus cells adherent to the RAW 264.7 cells were measured. The numbers of the VvhmpA mutant per macrophage were significantly lower than those of the wild type and VvhmpA-complemented strain (Figure 7A), indicating that VvHmpA is required for V. vulnificus to survive in proximity to the NO-producing macrophages. Accordingly, when the NO production from the RAW 264.7 cells was inhibited by the NO synthase inhibitor L-NMMA, the numbers of the wild type, VvhmpA mutant, and VvhmpA-complemented strain adherent to the RAW 264.7 cells were comparable (Figure 7B). The results indicated that VvHmpA is crucial for V. vulnificus to overcome nitrosative stress imposed by the host immune cells and thereby survive during infection.
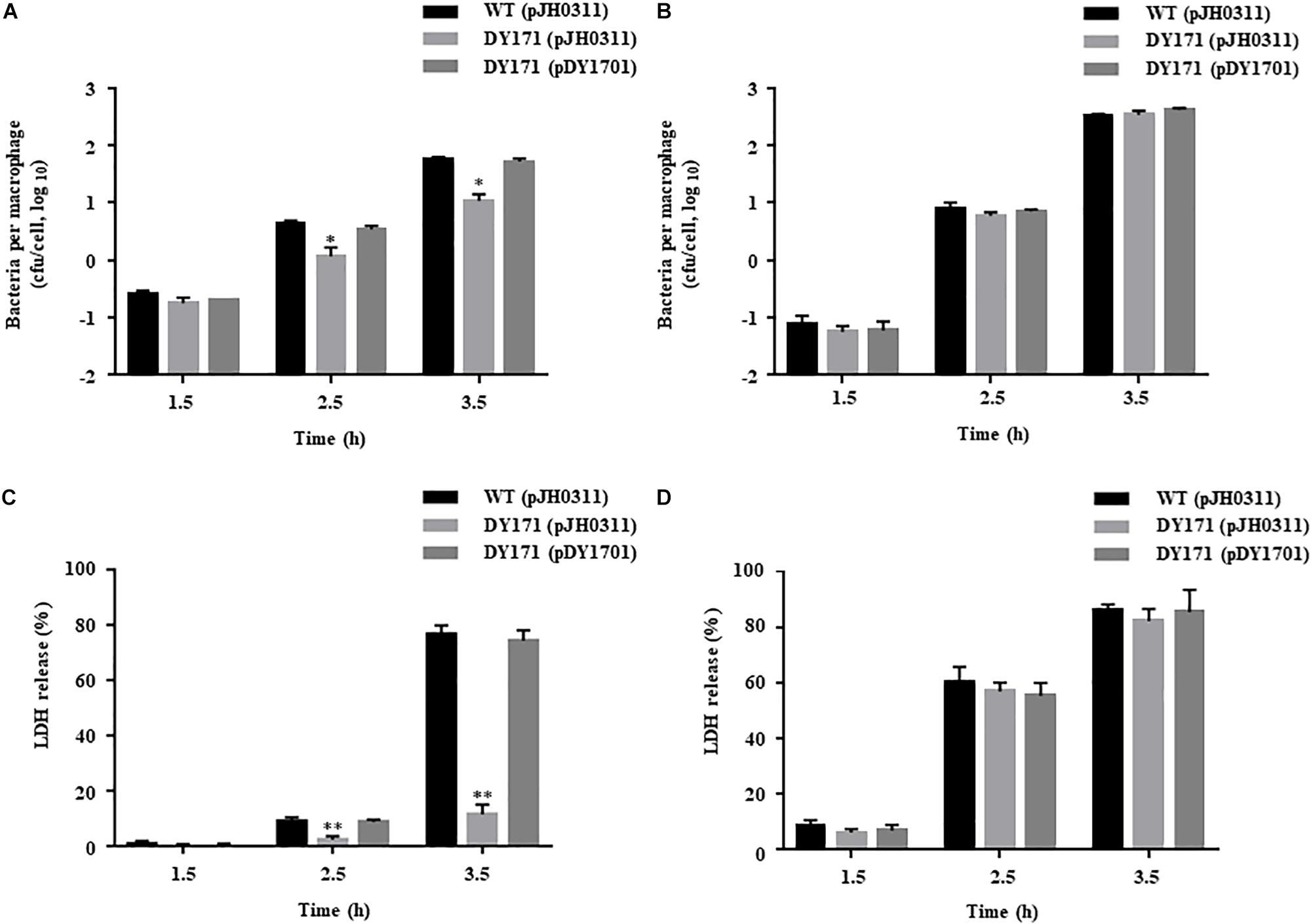
Figure 7. Survival and cytotoxicity of the V. vulnificus strains infecting the RAW 264.7 cells. The NO-producing RAW 264.7 cells were infected with the V. vulnificus strains at the MOI of 1 for various incubation times in the absence (A,C) or presence (B,D) of the NO synthase inhibitor, L-NMMA. (A,B) The V. vulnificus cells adherent to the RAW 264.7 cells were enumerated in cfu per macrophage at each time point after infection. (C,D) The cytotoxicity was expressed using the total LDH activity of the RAW 264.7 cells completely lysed by 1.5% Triton X-100 as 100%. Error bars represent the S.D. ∗p < 0.05 and ∗∗p < 0.005 relative to groups infected with the wild type at each incubation time. WT (pJH0311), wild type; DY171 (pJH0311), VvhmpA mutant; DY171 (pDY1701), VvhmpA-complemented strain.
To examine the role of VvHmpA in the virulence of V. vulnificus ex vivo, the activities of LDH released from RAW 264.7 cells infected with the V. vulnificus strains were determined. As shown in Figure 7C, the VvhmpA mutant exhibited significantly lower LDH-releasing activity compared with those of the wild type or the VvhmpA-complemented strain. In contrast, when the NO production from the RAW 264.7 cells was inhibited by L-NMMA, the wild type, VvhmpA mutant, and VvhmpA-complemented strain exhibited comparable levels of LDH-releasing activity (Figure 7D). Notably, the expression levels of the well-known cytotoxic virulence factors RtxA and VvhA in the wild type, VvhmpA mutant, and VvhmpA-complemented strain were not significantly different (Supplementary Figure S1). These results combined indicated that VvHmpA contributes to the virulence of V. vulnificus by coping with NO released from the host immune cells.
VvHmpA Is Important for the Pathogenesis of V. vulnificus in vivo
The importance of VvHmpA in the V. vulnificus pathogenesis was further investigated in a mouse model. As shown in Figure 8, the survival time of mice infected with the VvhmpA mutant was consistently prolonged (p value of 0.0420, log-rank test) compared with that of mice infected with the wild type. At 24 h post-infection, percentages of mice that survived after challenge with the VvhmpA mutant or the wild type were 66.6 and 33.3%, respectively, indicating that the deletion of VvhmpA attenuated the virulence of V. vulnificus in mice. Taken together, the combined results suggest that VvHmpA is important for NO decomposition, survival adjacent to NO-producing immune cells, and thereby the pathogenesis of V. vulnificus.
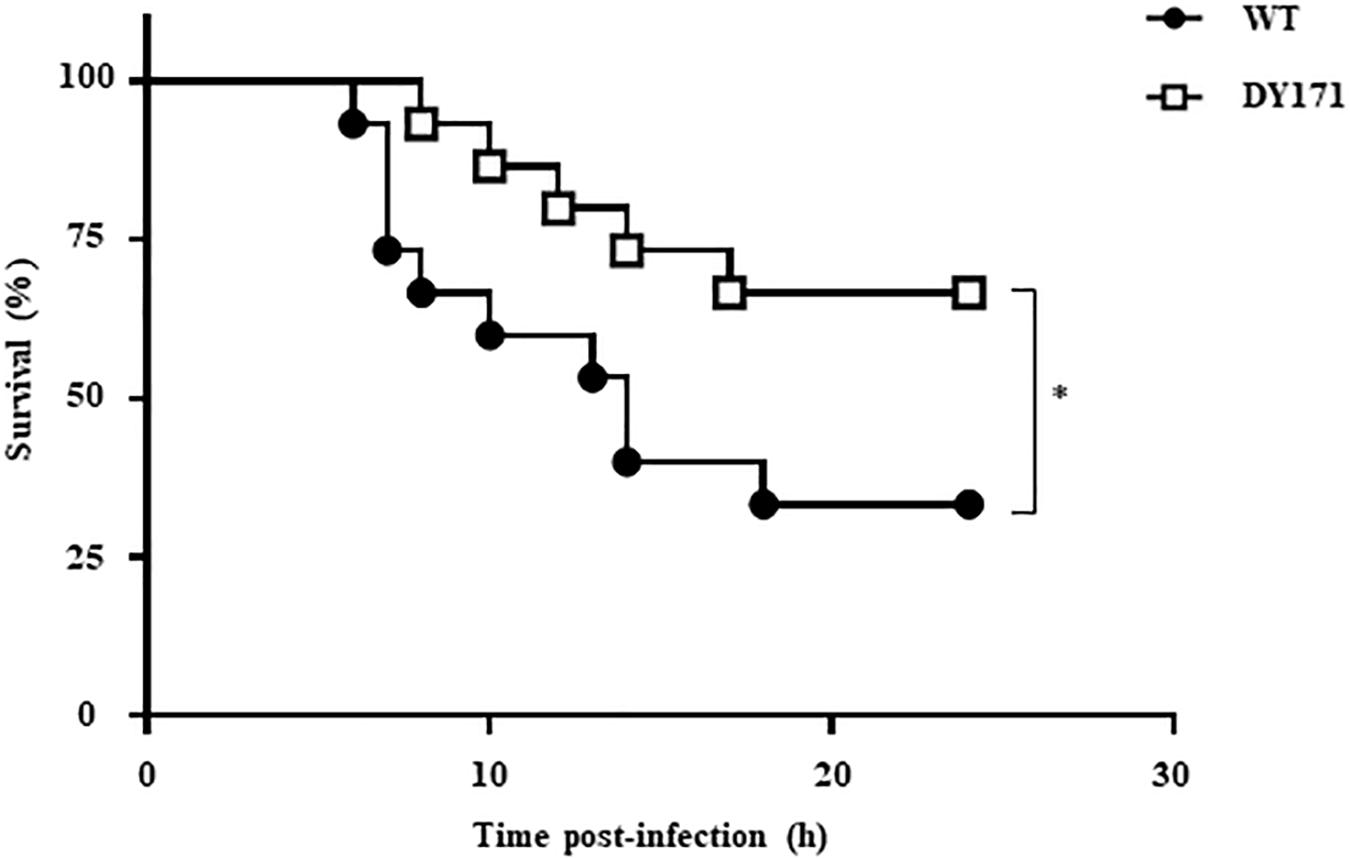
Figure 8. Mouse lethality of the V. vulnificus strains. Groups (n = 15) of 7-week-old specific pathogen-free female ICR mice were intraperitoneally infected with either the wild type or the VvhmpA mutant of V. vulnificus at doses of 1.0 × 106 cfu. Mouse survival percentage was monitored for 24 h. ∗p < 0.05; WT, wild type; DY171, VvhmpA mutant.
Discussion
In order to establish infection successfully, pathogens need to overcome nitrosative stresses that originate from the host immune system (Fang, 2004). In addition to the 8 genes, whose expression was further confirmed by qRT-PCR analysis in the present study (Figures 1, 2), transcriptome analysis of V. vulnificus identified 15 more genes that are induced upon exposure to NO and potentially involved in nitrosative stress defense (Supplementary Table S2). Among the 23 genes, VvhmpA was the most greatly induced and thus its gene product VvHmpA was selected and further analyzed at amino acid sequence levels. The deduced amino acid sequence revealed that VvHmpA is a multidomain NO dioxygenase consisting of the N-terminal globin domain with the heme-binding site and the C-terminal oxidoreductase domain with NAD- and FAD-binding sites (Figure 3). As well appreciated in a multidomain EcHmp, the N-terminal ferrous-oxy heme reacts with NO to yield NO3– and ferric-deoxy heme, and then the C-terminal domain transfers electrons from NAD(P)H to the ferric heme via FAD (Gardner A. M. et al., 2000). This endogenous electron transfer could allow the efficient regeneration of the ferrous heme, which is ready for another catalytic cycle of NO decomposition. In contrast, single-domain NO dioxygenases found in some pathogens such as Campylobacter jejuni lack the C-terminal domain and rely on an exogenous redox partner(s) to regenerate ferrous heme, resulting in less efficient NO decomposition (Shepherd et al., 2011). Therefore, V. vulnificus seems to adopt the multidomain VvHmpA to efficiently overcome nitrosative stress during infection.
Biochemical and kinetic analyses of EcHmp proposed two possible reaction mechanisms to initiate NO decomposition, namely dioxygenation and nitrosylation (Gardner A. M. et al., 2000; Hausladen et al., 2001; Forrester and Foster, 2012). Dioxygenation begins with O2 binding to the ferrous heme to result in ferrous-oxy heme, most probably under conditions where O2 is not limiting. The ferrous-oxy heme reacts with NO to form a transient Fe-ONOO– intermediate which releases NO3– as described above. In contrast, nitrosylation involves initial reaction of the ferrous heme with NO to form a ferrous-nitrosyl heme, converting transiently to a ferric-nitrosyl heme which in turn reacts with O2 to release NO3– under O2 limiting conditions. Regardless of which mechanistic pathway predominates, they both result in rapid enzymatic turnover of NO to NO3–. Although the exact mechanisms of VvHmpA detoxifying NO have not been yet clarified, the similarities found in the amino acid sequences and absorption spectra of EcHmp and VvHmpA (Figures 3, 4A) led us to postulate that VvHmpA also could convert NO to NO3– through one of the two pathways depending on O2 availability. Supporting this postulation, the kinetic values of VvHmpA for NO decomposition under aerobic conditions (Figure 5) are also comparable to those of EcHmp, in which the kcat and KM values are 10-670 s–1 and 0.25 μM for NO, respectively (Gardner P. R. et al., 2000; Gardner, 2005).
After infection, pathogenic bacteria probably encounter increased levels of NO due to the expression of iNOS in cells of the immune system and must overcome the nitrosative stress for successful pathogenesis (Bang et al., 2006; Richardson et al., 2006; Stern et al., 2012). The NO concentration in humans and experimental animals increases to the micromolar range under infectious and inflammatory conditions (Toledo and Augusto, 2012). It is noteworthy that the KM and kcat values of VvHmpA for NO at 37°C, the temperature to which V. vulnificus is inevitably exposed in the host, are greater than those at 30°C (Figure 5). This kinetic property indicates that VvHmpA expressed in the host may be more optimized for acting on the large amounts of NO and decomposing them rapidly to a safe level during infection, before the increased nitrosative stress impairs cellular components of V. vulnificus. It should be noted, however, that VvHmpA may not be the sole enzyme that protects V. vulnificus from NO as the VvhmpA mutation reduced the growth and survival of the pathogen significantly but not completely (Figures 6B, 7A). Indeed, analysis of the V. vulnificus transcriptome upon exposure to NO showed upregulation of the genes encoding a number of NO detoxifying enzymes in addition to NO dioxygenase (Figure 1 and Supplementary Table S2) (Poole, 2005). Nevertheless, it is reasonable to hypothesize that VvHmpA could provide an evolutionary advantage for V. vulnificus to survive in the host rather than in nature, where lower concentration of NO may be occasionally encountered. Supporting this hypothesis, loss of VvHmpA led V. vulnificus to show significantly reduced survival adjacent to the NO-producing macrophage cells (Figures 7A,B), cytotoxicity to the immune cells (Figures 7C,D), and possibly virulence in mice (Figure 8).
It has been reported that the Hmp proteins of miscellaneous pathogenic bacteria including V. cholerae, Salmonella Typhimurium and Staphylococcus aureus were responsible for NO detoxification and thus required for their survival under nitrosative stress and successful pathogenesis (Bang et al., 2006; Richardson et al., 2006; Stern et al., 2012). However, little has been known about the biochemical and kinetic properties of the Hmp proteins of the pathogens until now. In the present study, absorption spectral and biochemical analyses revealed that VvHmpA is a flavohemoglobin containing equimolar amounts of heme and FAD as cofactors. Kinetic properties of VvHmpA with greater KM and kcat values at 37°C than 30°C indicated that VvHmpA is optimized to act on and decompose large amounts of NO more effectively in the host. Reduced NO-decomposition activity and growth rate of the VvhmpA mutant in the presence of NO, along with the attenuated virulence observed both in the cell culture and mouse models, indicated that VvHmpA contributes to the survival of V. vulnificus under nitrosative stress, and might play an important role in the pathogenesis of the pathogen. Therefore, VvHmpA could be a promising target for development of new antibacterial agents.
Data Availability Statement
The datasets generated for this study can be found in the Sequence Read Archive – PRJNA513463; https://www.ncbi.nlm.nih.gov/sra/?term=PRJNA513463.
Ethics Statement
The animal study was reviewed and approved by Animal Care and Use Committee at Seoul National University.
Author Contributions
DK, EN, and SC designed the research. DK, EN, SK, JK, YJ, and JC performed the research. DK, EN, IB, N-CH, and SC wrote the manuscript. All authors made major contributions to the acquisition, analysis, and interpretation of the data.
Funding
This work was supported by the National Research Foundation of Korea, funded by the Ministry of Science, ICT, and Future Planning (2017R1E1A1A01074639) and the Institute of Planning and Evaluation for Technology (IPET) of the Ministry of Agriculture, Food, and Rural Affairs (710012-03-3-SB110) to SC. This work was also supported by the BK21 Plus Program of the Department of Agricultural Biotechnology, Seoul National University, Seoul, South Korea.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank all members of the Choi laboratory for valuable discussion and technical support.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02208/full#supplementary-material
Footnotes
References
Ahn, J., Jang, K. K., Jo, I., Nurhasni, H., Lim, J. G., Yoo, J. W., et al. (2018). Crystal structure of peroxiredoxin 3 from Vibrio vulnificus and its implications for scavenging peroxides and nitric oxide. IUCrJ 5(Pt 1), 82–92. doi: 10.1107/S205225251701750X
Anderson, A. B., and Robertson, C. R. (1995). Absorption-spectra indicate conformational alteration of myoglobin adsorbed on polydimethylsiloxane. Biophys. J. 68, 2091–2097. doi: 10.1016/S0006-3495(95)80388-7
Bang, I. S., Liu, L., Vazquez-Torres, A., Crouch, M. L., Stamler, J. S., and Fang, F. C. (2006). Maintenance of nitric oxide and redox homeostasis by the Salmonella flavohemoglobin hmp. J. Biol. Chem. 281, 28039–28047. doi: 10.1074/jbc.M605174200
Barr, I., and Guo, F. (2015). Pyridine hemochromagen assay for determining the concentration of heme in purified protein solutions. Bio Protoc. 5:e1594. doi: 10.21769/BioProtoc.1594
Bonamore, A., and Boffi, A. (2008). Flavohemoglobin: structure and reactivity. Iubmb Life 60, 19–28. doi: 10.1002/iub.9
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. doi: 10.1006/abio.1976.9999
Bryk, R., Griffin, P., and Nathan, C. (2000). Peroxynitrite reductase activity of bacterial peroxiredoxins. Nature 407, 211–215. doi: 10.1038/35025109
Conesa, A., Madrigal, P., Tarazona, S., Gomez-Cabrero, D., Cervera, A., McPherson, A., et al. (2016). A survey of best practices for RNA-seq data analysis. Genome Biol. 17:13. doi: 10.1186/s13059-016-0881-8
Davies, B. W., Bogard, R. W., Dupes, N. M., Gerstenfeld, T. A., Simmons, L. A., and Mekalanos, J. J. (2011). DNA damage and reactive nitrogen species are barriers to Vibrio cholerae colonization of the infant mouse intestine. PLoS Pathog. 7:e1001295. doi: 10.1371/journal.ppat.1001295
Fang, F. C. (2004). Antimicrobial reactive oxygen and nitrogen species: concepts and controversies. Nat. Rev. Microbiol. 2, 820–832. doi: 10.1038/nrmicro1004
Fang, X., Liang, P., Raba, D. A., Rosas-Lemus, M., Chakravarthy, S., Tuz, K., et al. (2017). Kinetic characterization of Vibrio cholerae ApbE: substrate specificity and regulatory mechanisms. PLoS One 12:e0186805. doi: 10.1371/journal.pone.0186805
Forrester, M. T., and Foster, M. W. (2012). Protection from nitrosative stress: a central role for microbial flavohemoglobin. Free Radic. Biol. Med. 52, 1620–1633. doi: 10.1016/j.freeradbiomed.2012.01.028
Gardner, A. M., Martin, L. A., Gardner, P. R., Dou, Y., and Olson, J. S. (2000). Steady-state and transient kinetics of Escherichia coli nitric-oxide dioxygenase (flavohemoglobin) - The B10 tyrosine hydroxyl is essential for dioxygen binding and catalysis. J. Biol. Chem. 275, 12581–12589. doi: 10.1074/jbc.275.17.12581
Gardner, P. R. (2005). Nitric oxide dioxygenase function and mechanism of flavohemoglobin, hemoglobin, myoglobin and their associated reductases. J. Inorg. Biochem. 99, 247–266. doi: 10.1016/j.jinorgbio.2004.10.003
Gardner, P. R., Gardner, A. M., Martin, L. A., Dou, Y., Li, T. S., Olson, J. S., et al. (2000). Nitric-oxide dioxygenase activity and function of flavohemoglobins - sensitivity to nitric oxide and carbon monoxide inhibition. J. Biol. Chem. 275, 31581–31587. doi: 10.1074/jbc.M004141200
Goo, S. Y., Lee, H. J., Kim, W. H., Han, K. L., Park, D. K., Lee, H. J., et al. (2006). Identification of OmpU of Vibrio vulnificus as a fibronectin-binding protein and its role in bacterial pathogenesis. Infect. Immun. 74, 5586–5594. doi: 10.1128/Iai.00171-06
Gulig, P. A., Bourdage, K. L., and Starks, A. M. (2005). Molecular pathogenesis of Vibrio vulnificus. J. Microbiol. 43, 118–131.
Hausladen, A., Gow, A., and Stamler, J. S. (2001). Flavohemoglobin denitrosylase catalyzes the reaction of a nitroxyl equivalent with molecular oxygen. Proc. Natl. Acad. Sci. U.S.A. 98, 10108–10112. doi: 10.1073/pnas.181199698
Hirano, K., and Namihira, M. (2017). FAD influx enhances neuronal differentiation of human neural stem cells by facilitating nuclear localization of LSD1. FEBS Open Bio. 7, 1932–1942. doi: 10.1002/2211-5463.12331
Ioannidis, N., Cooper, C. E., and Poole, R. K. (1992). Spectroscopic studies on an oxygen-binding hemoglobin-like flavohaemoprotein from Escherichia coli. Biochem. J. 288, 649–655. doi: 10.1042/bj2880649
Jang, K. K., Lee, Z. W., Kim, B., Jung, Y. H., Han, H. J., Kim, M. H., et al. (2017). Identification and characterization of Vibrio vulnificus plpA encoding a phospholipase A(2) essential for pathogenesis. J. Biol. Chem. 292, 17129–17143. doi: 10.1074/jbc.M117.791657
Karlinsey, J. E., Bang, I. S., Becker, L. A., Frawley, E. R., Porwollik, S., Robbins, H. F., et al. (2012). The NsrR regulon in nitrosative stress resistance of Salmonella enterica serovar Typhimurium. Mol. Microbiol. 85, 1179–1193. doi: 10.1111/j.1365-2958.2012.08167.x
Kim, B. S., Jang, S. Y., Bang, Y. J., Hwang, J., Koo, Y., Jang, K. K., et al. (2018). QStatin, a selective inhibitor of quorum sensing in Vibrio Species. MBio 9:e2262-17. doi: 10.1128/mBio.02262-17
Kim, S., Bang, Y. J., Kim, D., Lim, J. G., Oh, M. H., and Choi, S. H. (2014). Distinct characteristics of OxyR2, a new OxyR-type regulator, ensuring expression of peroxiredoxin 2 detoxifying low levels of hydrogen peroxide in Vibrio vulnificus. Mol. Microbiol. 93, 992–1009. doi: 10.1111/mmi.12712
Lim, J. G., and Choi, S. H. (2014). IscR is a global regulator essential for pathogenesis of Vibrio vulnificus and induced by host cells. Infect. Immun. 82, 569–578. doi: 10.1128/Iai.01141-13
Liu, L., Hausladen, A., Zeng, M., Que, L., Heitman, J., and Stamler, J. S. (2001). A metabolic enzyme for S-nitrosothiol conserved from bacteria to humans. Nature 410, 490–494. doi: 10.1038/35068596
Mills, P. C., Rowley, G., Spiro, S., Hinton, J. C., and Richardson, D. J. (2008). A combination of cytochrome c nitrite reductase (NrfA) and flavorubredoxin (NorV) protects Salmonella enterica serovar Typhimurium against killing by NO in anoxic environments. Microbiol 154(Pt 4), 1218–1228. doi: 10.1099/mic.0.2007/014290-0
Milton, D. L., OToole, R., Horstedt, P., and WolfWatz, H. (1996). Flagellin A is essential for the virulence of Vibrio anguillarum. J. Bacteriol. 178, 1310–1319. doi: 10.1128/jb.178.5.1310-1319.1996
Nathan, C. F., and Hibbs, J. B. (1991). Role of nitric-oxide synthesis in macrophage antimicrobial activity. Curr. Opin. Immunol. 3, 65–70. doi: 10.1016/0952-7915(91)90079-G
Nurhasni, H., Cao, J., Choi, M., Kim, I., Lee, B. L., Jung, Y., et al. (2015). Nitric oxide-releasing poly(lactic-co-glycolic acid)-polyethylenimine nanoparticles for prolonged nitric oxide release, antibacterial efficacy, and in vivo wound healing activity. Int. J. Nanomed. 10, 3065–3080. doi: 10.2147/IJN.S82199
Oliver, J. D. (2015). The biology of Vibrio vulnificus. Microbiol. Spect. 3:VE-0001-2014. doi: 10.1128/microbiolspec.VE-0001-2014
Poole, R. K., Anjum, M. F., Membrillo-Hernandez, J., Kim, S. O., Hughes, M. N., and Stewart, V. (1996). Nitric oxide, nitrite, and Fnr regulation of hmp (flavohemoglobin) gene expression in Escherichia coli K-12. J. Bacteriol. 178, 5487–5492. doi: 10.1128/jb.178.18.5487-5492.1996
Poole, R. K. (2005). Nitric oxide and nitrosative stress tolerance in bacteria. Biochem. Soc. Trans. 33, 176–180. doi: 10.1042/BST0330176
Richardson, A. R., Dunman, P. M., and Fang, F. C. (2006). The nitrosative stress response of Staphylococcus aureus is required for resistance to innate immunity. Mol. Microbiol. 61, 927–939. doi: 10.1111/j.1365-2958.2006.05290.x
Shepherd, M., Bernhardt, P. V., and Poole, R. K. (2011). Globin-mediated nitric oxide detoxification in the foodborne pathogenic bacterium Campylobacter jejuni proceeds via a dioxygenase or denitrosylase mechanism. Nitric Oxide 25, 229–233. doi: 10.1016/j.niox.2010.12.006
Simon, R., Priefer, U., and Puhler, A. (1983). A broad host range mobilization system for in vivo genetic engineering - transposon mutagenesis in gram negative bacteria. Biotechnology 1, 784–791. doi: 10.1038/nbt1183-784
Smagghe, B. J., Trent, J. T. III, and Hargrove, M. S. (2008). NO dioxygenase activity in hemoglobins is ubiquitous in vitro, but limited by reduction in vivo. PLoS One 3:e0002039. doi: 10.1371/journal.pone.0002039
Sobko, T., Reinders, C. I., Jansson, E., Norin, E., Midtvedt, T., and Lundberg, J. O. (2005). Gastrointestinal bacteria generate nitric oxide from nitrate and nitrite. Nitric Oxide 13, 272–278. doi: 10.1016/j.niox.2005.08.002
Stern, A. M., Hay, A. J., Liu, Z., Desland, F. A., Zhang, J., Zhong, Z. T., et al. (2012). The NorR regulon is critical for Vibrio cholerae resistance to nitric oxide and sustained colonization of the intestines. Mbio 3:e00013-12. doi: 10.1128/mBio.00013-12
Stern, A. M., and Zhu, J. (2014). An introduction to nitric oxide sensing and response in bacteria. Adv. Appl. Microbiol. 87, 187–220. doi: 10.1016/B978-0-12-800261-2.00005-0
Stuehr, D. J. (1999). Mammalian nitric oxide synthases. Biochim. Biophys. Acta. 1411, 217–230. doi: 10.1016/s0005-2728(99)00016-x
Tiso, M., and Schechter, A. N. (2015). Nitrate reduction to nitrite, nitric oxide and ammonia by gut bacteria under physiological conditions. PLoS One 10:e0119712. doi: 10.1371/journal.pone.0119712
Toledo, J. C. Jr., and Augusto, O. (2012). Connecting the chemical and biological properties of nitric oxide. Chem. Res. Toxicol. 25, 975–989. doi: 10.1021/tx300042g
Walker, G., Pfeilschifter, J., and Kunz, D. (1997). Mechanisms of suppression of inducible nitric-oxide synthase (iNOS) expression in interferon (IFN)-gamma-stimulated RAW 264.7 cells by dexamethasone - Evidence for glucocorticoid-induced degradation of iNOS protein by calpain as a key step in post-transcriptional regulation. J. Biol. Chem. 272, 16679–16687. doi: 10.1074/jbc.272.26.16679
Keywords: Vibrio vulnificus, microbiology, nitric oxide dioxygenase, flavohemoglobins, virulence factors, gene expression profiling
Citation: Kim D, Na EJ, Kim S, Kim JS, Jung YH, Cao J, Han HJ, Bang IS, Yoo J-W, Ha N-C and Choi SH (2019) Transcriptomic Identification and Biochemical Characterization of HmpA, a Nitric Oxide Dioxygenase, Essential for Pathogenesis of Vibrio vulnificus. Front. Microbiol. 10:2208. doi: 10.3389/fmicb.2019.02208
Received: 15 July 2019; Accepted: 09 September 2019;
Published: 24 September 2019.
Edited by:
Ulrike Kappler, University of Queensland, AustraliaReviewed by:
Carmen Amaro, University of Valencia, SpainTakashige Kashimoto, Kitasato University, Japan
Copyright © 2019 Kim, Na, Kim, Kim, Jung, Cao, Han, Bang, Yoo, Ha and Choi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nam-Chul Ha, hanc210@snu.ac.kr; Sang Ho Choi, choish@snu.ac.kr
†These authors have contributed equally to this work
 Dukyun Kim1,2†
Dukyun Kim1,2† Jung Sung Kim
Jung Sung Kim Young Hyun Jung
Young Hyun Jung Ho Jae Han
Ho Jae Han Nam-Chul Ha
Nam-Chul Ha Sang Ho Choi
Sang Ho Choi