- 1Genetics Graduate Program, Michigan State University, East Lansing, MI, United States
- 2Department of Plant, Soil, and Microbial Sciences, Michigan State University, East Lansing, MI, United States
Erwinia amylovora, causative agent of fire blight disease of apple and pear trees, has evolved to use small RNAs for post-transcriptional regulation of virulence traits important for disease development. The sRNA ArcZ regulates several virulence traits, and to better understand its roles, we conducted a transcriptomic comparison of wild-type and ΔarcZ mutant E. amylovora. We found that ArcZ regulates multiple cellular processes including genes encoding enzymes involved in mitigating the threat of reactive oxygen species (katA, tpx, osmC), and that the ΔarcZ mutant has reduced catalase activity and is more susceptible to exogenous hydrogen peroxide. We quantified hydrogen peroxide production by apple leaves inoculated with E. amylovora and found that the while wild-type E. amylovora cells produce enough catalase to cope with defense peroxide, the ΔarcZ mutant is likely limited in virulence because of inability to cope with peroxide levels in host leaves. We further found that the ArcZ regulon overlaps significantly with the regulons of transcription factors involved in oxidative sensing including Fnr and ArcA. In addition, we show that ArcZ regulates arcA at the post-transcriptional level suggesting a role for this system in mediating adaptations to oxidative state, especially during disease development.
Introduction
When pathogenic microbes arrive on a host plant, the plant perceives the arrival of a threat through recognition of pathogen associated molecular patterns (PAMPs) (Jones and Dangl, 2006). The recognized patterns include conserved molecules associated with pathogenic microbes, such as chitin (Miya et al., 2007), flagellin (Chinchilla et al., 2007), and translation elongation factor Tu (Kunze et al., 2004). The binding of these PAMPs to surface receptors triggers a complex signaling cascade that activates defense responses (Jones and Dangl, 2006). Host plant defense responses are diverse and include actions such as stomatal closure (Melotto et al., 2006), hormone signaling (Heese et al., 2007), callose deposition (Thilmony et al., 2006), and production of reactive oxygen species (Bolwell and Wojtaszek, 1997). Plant pathogenic microbes have responded to these host defenses through the evolution of effector proteins that act to suppress and subvert host defense signaling and activity (Guo et al., 2009). In the case of bacterial pathogens, the effectors are often translocated directly into the host cytoplasm via the type III secretion system, a needle-like protein structure (He, 1998). In an ongoing biochemical arms race, hosts and pathogens alike have evolved numerous effector-target relationships that affect disease outcomes (Jones and Dangl, 2006; Asai and Shirasu, 2015; Khan et al., 2016). For many bacterial pathogens, this has resulted in a number of effector proteins that are essential for full virulence (Jamir et al., 2004; Toruño et al., 2016). In addition to effector proteins, bacterial pathogens have evolved additional virulence strategies that allow them to flourish in the environment of a host plant and avoid host defenses. For example, Erwinia amylovora, causative agent of fire blight disease of apple and pear trees, utilizes several virulence strategies to avoid, suppress, and cope with host defenses (Geier and Geider, 1993; Bogdanove et al., 1998; Berry et al., 2009; Koczan et al., 2009, 2011; Zhao et al., 2010). For pathogenesis, E. amylovora requires effective translocation of the type III effectors DspE and AvrRpt2Ea into host cells to suppress host defenses and induce necrosis (Bogdanove et al., 1998; Zhao et al., 2006). Additional virulence traits that play a key role for E. amylovora include exopolysaccharide production and biofilm formation (Bellemann et al., 1994; Nimtz et al., 1996; Koczan et al., 2009; Castiblanco and Sundin, 2018), motility (Raymundo and Ries, 1980; Bayot and Ries, 1986), ability to mitigate the threat of reactive oxygen species (Santander et al., 2018), and ability to acquire and utilize essential nutrients (Aldridge et al., 1997; Dellagi et al., 1998).
Production of the exopolysaccharides amylovoran (Bellemann et al., 1994; Nimtz et al., 1996), levan (Gross et al., 1992; Koczan et al., 2009), and cellulose (Castiblanco and Sundin, 2018) along with proteinaceous attachment structures (Koczan et al., 2011) contribute to biofilm formation. Biofilm formation provides protective layers that can serve to both prevent host defense molecules, like reactive oxygen species, from reaching the bacteria (Danhorn and Fuqua, 2007), and to conceal the bacteria from host detection, reducing the degree of host defense response (D’Haeze and Holsters, 2004). Motility enables bacteria to use flagella or pili to migrate and move to more favorable locations where host defenses may be reduced or nutrient availability may be more favorable (Bayot and Ries, 1986; Berry et al., 2009). Although E. amylovora can be concealed through some virulence traits, move away from host defenses, and even directly reduce the host defense response through type III effectors, the bacteria will still have to cope with host defense compounds and responses as well as acquire sufficient nutrients to maintain growth during infection (Kamber et al., 2016). Thus, the ability to face host defenses and mitigate the threat of reactive oxygen species is also critical for full virulence (Santander et al., 2018). To coordinately express each virulence-associated trait under the precise conditions, E. amylovora has evolved elaborate environmental sensing and signal transduction cascades (Zhao et al., 2009). Efforts to characterize these regulatory pathways have successfully linked several regulatory systems with virulence associated traits.
Recent work has revealed the importance of small non-coding RNAs (sRNAs) in the regulation of virulence and virulence-associated traits in E. amylovora (Zeng et al., 2013; Zeng and Sundin, 2014). sRNAs are typically involved in post-transcriptional regulation. One class of sRNAs that affects virulence in E. amylovora includes those that are dependent on the chaperone protein Hfq (Zeng et al., 2013). The Hfq chaperone stabilizes a family of trans-acting sRNAs that regulate targets by RNA-RNA base-pairing (Sun et al., 2002; Vogel and Luisi, 2011; Sauer et al., 2012). In E. amylovora, 42 Hfq-dependent sRNAs have been identified, and the Hfq-dependent sRNA ArcZ in particular is critical for virulence and several virulence-associated traits including production of the exopolysaccharides levan and amylovoran, normal biofilm formation, flagellar motility and translocation of type III effectors to plant cells (Zeng and Sundin, 2014). We have recently shown that ArcZ regulates flagellar motility in E. amylovora through a direct interaction with the flagellar master regulator FlhD (Schachterle et al., 2019) and that ArcZ impacts exopolysaccharide production and biofilm formation through the leucine responsive regulator protein Lrp (Schachterle and Sundin, 2019). However, it is not known if there are further virulence-associated traits being regulated by ArcZ, nor is it known how ArcZ regulates type III secretion.
Because of the breadth of phenotypes ArcZ regulates, we conducted a transcriptomic comparison of the ΔarcZ mutant relative to wild-type to gain additional insights into the mechanisms of ArcZ regulation of virulence-associated traits. In addition to previously known interactions between ArcZ and Lrp, we found that ArcZ regulates several genes involved in mitigating the threat of reactive oxygen species, and present evidence that this regulation is critical for in planta survival. We also found a significant amount of overlap between the ArcZ regulon and regulons of global transcription factors associated with oxidative state signaling, including the ArcBA (anoxic redox control) two-component system. We further present evidence that ArcZ regulates arcA post-transcriptionally, indicating that ArcZ plays a major role in the oxidative status responsive regulatory pathways.
Materials and Methods
Strain Growth and Culture Conditions
Bacterial strains were routinely grown using LB culture media. E. amylovora strains were cultured at 28°C and Escherichia coli strains were cultured at 37°C. When appropriate, antibiotics were used in the following final concentrations: ampicillin 100 μg mL–1, kanamycin 30 μg mL–1, chloramphenicol 20 μg mL–1. Bacterial strains and oligonucleotides used in this study are found in Table 1 and Supplementary Table S1, respectively.
RNA Extraction and Sequencing
RNA was isolated from cells induced in hrp-inducing minimal medium (HIMM, Huynh et al., 1989). For induction, cells were grown overnight in one volume of LB medium, collected by centrifugation, washed with HIMM, and then resuspended in an equal volume of HIMM and incubated at 28°C with shaking for the time specified. RNA was extracted using the approach of Rivas et al. (2001), with documented specific modifications (Schachterle and Sundin, 2019). RNA was quantified using the Qubit fluorescence method (Thermo Fisher Scientific, Waltham, MA, United States). RNA quality was ensured by visualization of ribosomal RNA bands in agarose gel and by LabChipGX HS RNA analysis (Caliper Life Sciences, Waltham, MA, United States). Total RNA was depleted of ribosomal RNA using bacterial Ribo-Zero kits (Illumina, San Diego, CA, United States) and remaining RNA was used for library preparation with the Illumina TruSeq Stranded Total RNA Library Preparation Kit on a Perkin Elmer Sciclone G3 robot using manufacturer’s recommendations (Perkin Elmer, Waltham, MA, United States). Completed libraries were quality checked and quantified using a combination of Qubit RNA HS (Thermo Fisher Scientific, Waltham, MA, United States) and Caliper LabChipGX HS RNA assays. All libraries were combined in equimolar amounts and pools were quantified using the Kapa Biosystems Illumina Library Quantification qPCR kit. Sequencing was performed in a single-end 50 bp read format using HiSeq 4000 SBS reagents and base calling was done by Illumina Real Time Analysis (RTA) v.2.7.6. Output of RTA was demultiplexed and converted to FastQ format with Illumina Bcl2fastq v2.19.0.
Differential Gene Expression Analysis
Reads obtained from RNA sequencing were trimmed of adapter sequences and filtered to remove low-quality reads using Trimmomatic SE (Bolger et al., 2014) with the following parameters: -phred33 ILLUMINACLIP:2:30:10 LEADING:10 TRAILING:10 SLIDINGWINDOW:4:15 MINLEN:30. Trimmed and filtered reads were mapped to the E. amylovora ATCC49946 genome (Sebaihia et al., 2010) using bowtie2 (Langmead and Salzberg, 2012) with parameters: -q –phred33 -N 1. The resulting SAM file of mapped reads was sorted for downstream applications using SAMTools (Li et al., 2009). The E. amylovora ATCC49946 genome annotation file was used in conjunction with HTSeq (Anders et al., 2015) to count the number of reads mapping to each annotated feature. Read counts by feature across all samples were analyzed using the R package DESeq (Anders and Huber, 2012) to determine statistically differentially expressed genes between wild-type and ΔarcZ mutant samples with a false-detection rate of 0.05.
Quantitative Real-Time PCR
For qRT-PCR validation of select differentially expressed genes, RNA samples were collected in the same manner as for RNA sequencing, from E. amylovora cultures grown in LB medium and then induced for 18 h in HIMM. Three replicate RNA isolations were made from independent bacterial cultures. 500 ng of total RNA was used as template for reverse transcriptase reactions using the High-Capacity Reverse Transcriptase kit (Applied Biosystems, Foster City, CA, United States) following prescribed protocols. Resulting cDNA was utilized as template in qRT-PCR reactions set up using SYBR green 2X master mix (Applied Biosystems, Foster City, CA, United States) according to manufacturer’s protocols and run on an Applied Biosystems StepOnePlus instrument. The housekeeping gene recA was included as an endogenous control, and relative mRNA abundance was calculated using the 2–ddCt method (Livak and Schmittgen, 2001).
Catalase Activity, Zone of Inhibition, and Minimum Inhibitory Concentration Assays
Catalase activity assays were conducted as described (Iwase et al., 2013), using cells grown overnight in liquid LB and adjusted to an OD600 nm of 0.5. Briefly, cells were mixed in a 1:1:1 volumetric ratio with 8 M hydrogen peroxide and 1% (v/v) Triton X-100, allowed to incubate for 10 min and height of stabilized bubbles measured. These assays were carried out with 100 μL volumes in 15 mm tubes, and measurements were normalized to those of wild-type cells. Zone of inhibition was assayed by spread-plating 100 μL of bacteria cultures with an OD600 nm of 0.2 onto agar plates and then placing a filter paper disk in the center of the plate. A total of 10 μL of 8 M H2O2 was dripped onto the filter paper, and plates were incubated for 24 h at 28°C, after which the plate was imaged and the area of the zone of clearing around the filter paper disk was quantified using ImageJ image analysis software (Abràmoff et al., 2004). For determination of the minimum inhibitory concentration (MIC) of H2O2, LB or minimal media (4 g L–1 L-asparagine, 2 g L–1 K2HPO4, 0.2 g L–1 MgSO4 7H2O, 3 g L–1 NaCl, 0.2 g L–1 nicotinic acid, 0.2 g L–1 thiamin hydrochloride, 10 g L–1 sorbitol) were prepared with concentrations of hydrogen peroxide ranging from 0 to 20 mM. Cells were inoculated into these media at an initial density of 1 × 107 cfu mL–1 and incubated with shaking at 28°C. The MIC was determined to be the concentration of hydrogen peroxide at which bacterial growth was completely inhibited by visual assessment after 24 h. Each of these assays was repeated at least four times on separate days with independent bacterial cultures.
Survival in Tobacco Apoplast
The ability of bacterial cells to survive in the apoplast of Nicotiana tabacum leaves was assessed as described (Santander et al., 2018), with the modification that surviving bacterial populations were enumerated at 5 days post-inoculation by dilution plating, rather than across a time-course. Each strain was tested in at least three experiments with a total of at least six replicates.
Quantitation of Hydrogen Peroxide in Apple Leaves
Hydrogen peroxide levels in apple leaves were determined using a potassium iodide method (Junglee et al., 2014). For the assay, apple leaves were inoculated as described (Koczan et al., 2009) with a cell suspension of wild-type E. amylovora cells at a density of 5 × 108 cfu mL–1. Inoculated leaves were sampled at indicated time points and 1 cm diameter disks were punched from the leaves, homogenized in 1 mL reaction buffer (0.5 M potassium iodide, 0.025% trichloroacetic acid, 2.5 mM potassium phosphate, pH 7), and supernatants from homogenates were transferred to wells of a 96-well microtiter plate and incubated in the dark for 30 min. Following incubation, 345 nm absorbance was measured, and background color from leaf tissue was subtracted by using leaf disks punched from the same leaf, homogenized in reaction buffer in lacking potassium iodide. Absorbance values were converted to concentrations of hydrogen peroxide using a standard curve as described (Junglee et al., 2014). This experiment was repeated twice, and in each experiment at least two leaves were sampled at each time point and two pairs of punches were made from each leaf. In total, at least 12 replicate measurements were made at each time point.
Swimming Motility
Swimming motility assays were conducted in soft-agar as described (Schachterle et al., 2019). Briefly, cells were grown overnight in LB, normalized to an OD600 nm of 0.2, and stab inoculated in swimming motility medium (10 g L–1 tryptone, 5 g L–1 sodium chloride, 2.5 g L–1 agar). After 24 h of incubation at 28°C, plates were photographed and the area covered by swimming cells was quantified using ImageJ (Abràmoff et al., 2004). Four biological replicates were assessed.
Immature Pear Virulence Assay
Virulence of strains was assessed in immature pears as described (Zhao et al., 2005). Briefly, immature pears were washed and sterilized using 10% bleach, after which they were wounded and inoculated with 103–104 cfu in a 1 μL droplet and incubated at 28°C under high humidity conditions. Inoculated pears were assessed every 24 h for water soaking or necrotic symptom development. Each strain was tested with eight biological replicates.
Reporter Fusion Generation and Testing
For translational fusions, the 5’ untranslated region (UTR) of each gene of interest was amplified from the transcriptional start site through 20 amino acids into the coding region and cloned in-frame with gfp in plasmid pXG20 (Urban and Vogel, 2007) using an in vivo assembly approach (García-Nafría et al., 2016). For the katA promoter fusion, the 500 bases upstream from the katA start codon were amplified and cloned into plasmid pPROBE-NT (Miller et al., 2000). Strains harboring the reporter fusions were grown overnight in LB medium and assessed for GFP fluorescent output using a Tecan Spark plate reader (Tecan, Männedorf, Switzerland) with excitation wavelength of 488 nm and emission wavelength of 535 nm. Relative fluorescence was determined by normalizing arbitrary fluorescence units to cell density, and relative to the wild-type strain. Each strain was tested in at least four experiments.
Regulon Analysis
Known E. coli transcription factor regulons were obtained from RegulonDB (Gama-Castro et al., 2010) and corresponding gene sequences were extracted from the E. coli K-12 genome (Blattner et al., 1997). E. coli gene sequences were used as queries to search for presence in E. amylovora using tblastx from BLAST+ (Camacho et al., 2009). If a BLAST hit had an e-value of less than 0.001, that gene from E. coli was considered present in E. amylovora. Using the assumption that if a transcription factor and its regulated genes are conserved across E. coli and E. amylovora, regulatory relationships are likely to be similar, we used this assessment to generate putative E. amylovora regulons for several transcription factors. Putative E. amylovora regulons were tested for significant overlap with the ArcZ regulon determined herein using Fisher’s exact test with adjustment for multiple hypothesis testing.
Results
Transcriptomic Characterization of the E. amylovora ΔarcZ Mutant Relative to Wild-Type
We sequenced the E. amylovora Ea1189 transcriptome using RNA from wild-type and ΔarcZ mutant cells induced for six or eighteen hrs in HIMM (Huynh et al., 1989). Our sequencing resulted in a total of 128.4 million reads generated, of which 96.9% had per-base quality scores greater than 30. Of these reads, 97.2 percent mapped to the E. amylovora ATCC49946 genome. Following normalization and statistical analysis, we found a total of 342 differentially expressed genes between wild-type and ΔarcZ mutant cells. Of these, 62 genes were differentially regulated after 6 h of induction (27 up-regulated, 35 down-regulated) and 302 were differentially expressed after 18 h of induction (176 up-regulated, 126 down-regulated) with 22 genes differentially expressed at both time points (19 down-regulated, 3 up-regulated). Principal component analysis, based on differentially expressed genes, showed that samples clustered by strain and time point (Supplementary Figure S1).
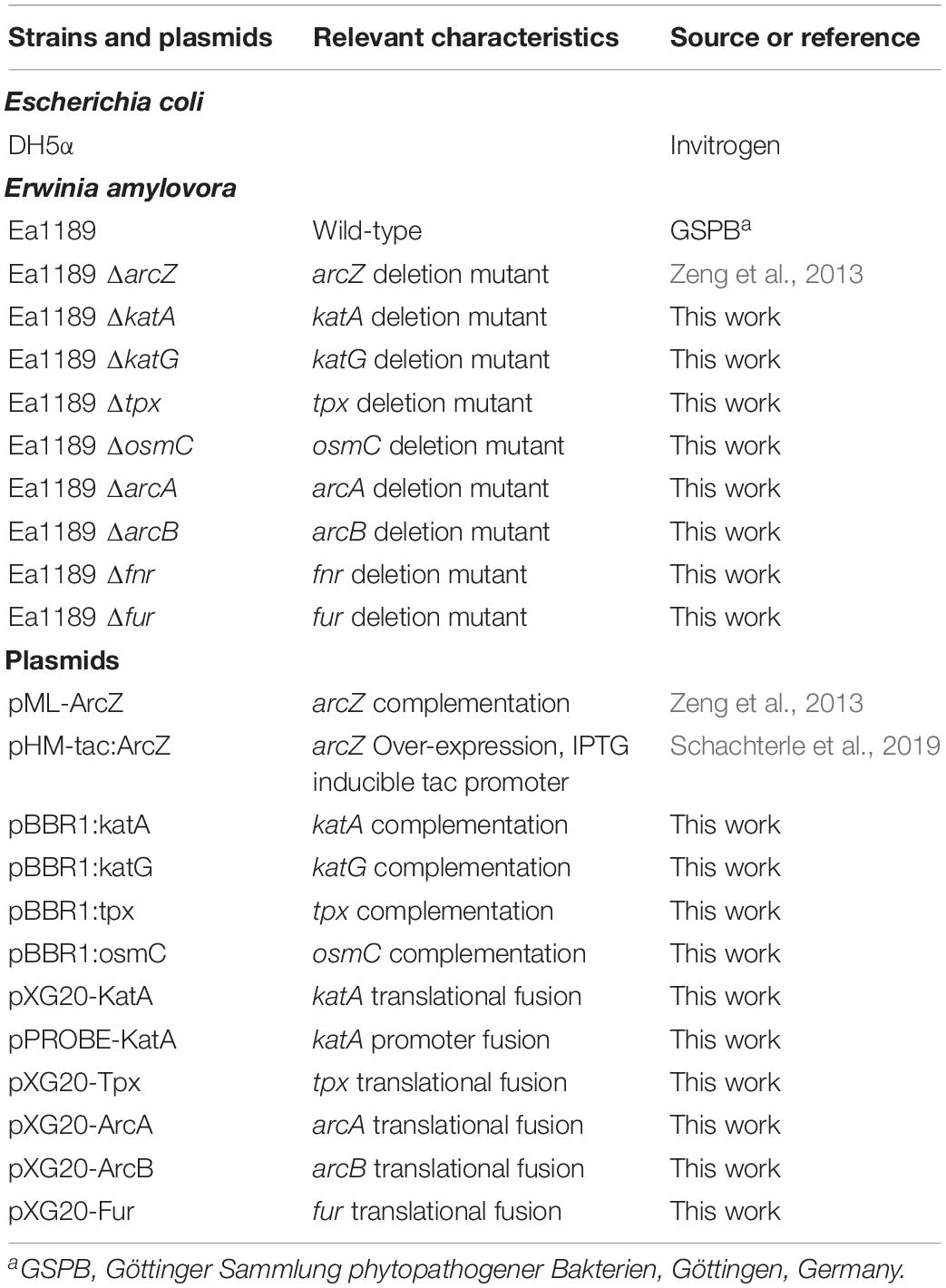
Visualization of differentially expressed genes across samples is provided as a heatmap in Figure 1. Genes clustered into four main groups by strain and time point differences, designated groups I, II, III, and IV. Group I genes are characterized by higher expression in the ΔarcZ mutant after 6 h of induction in HIMM, but no dramatic differences between wild-type and ΔarcZ at 18 h. Genes of interest in group I include the aerotaxis receptor, aer, and the leucine responsive regulatory protein, lrp, which we recently demonstrated is destabilized post-transcriptionally by ArcZ (Schachterle and Sundin, 2019). Group II genes are characterized by higher expression in wild-type samples at 6 h of induction relative to 18 h of induction in HIMM and reduced expression in general in the ΔarcZ mutant at both time points. This is the largest cluster of differentially expressed genes and includes genes involved in several metabolic and virulence processes. Examples of virulence associated genes include flagellar motility genes (flhC, motB, and flgE) and type III secretion genes (hrpA, hrpW, and hrpJ). Examples of metabolic genes include crp encoding the global regulator catabolite repressor protein, and other genes involved in metabolism such as argD, cysD, gcvP, livM, and metB. Group III genes are characterized by higher expression in wild-type at 18 h in HIMM compared to wild-type after 6 h of induction in HIMM, but not elevated in the ΔarcZ samples after 18 h of induction in HIMM. Many of these genes are also general metabolism genes and include tktA and rpsS. Group IV genes have elevated expression in the ΔarcZ mutant cells after 18 h of induction in HIMM. Most of these genes are uncharacterized, but multiple genes in this group are likely involved in reactions with phospho-sugars, such as pgsA and EAM_1622. For a complete list of all differentially expressed genes, see Supplementary Tables S2, S3. Because of the long duration of our time-course sampling, it is likely that many of the observed differentially expressed genes may be affected only indirectly by ArcZ. Because our approach did not distinguish between direct and indirect regulation by ArcZ, for the purposes of this study, we consider all of the observed differentially expressed genes to be a part of the ArcZ regulon.
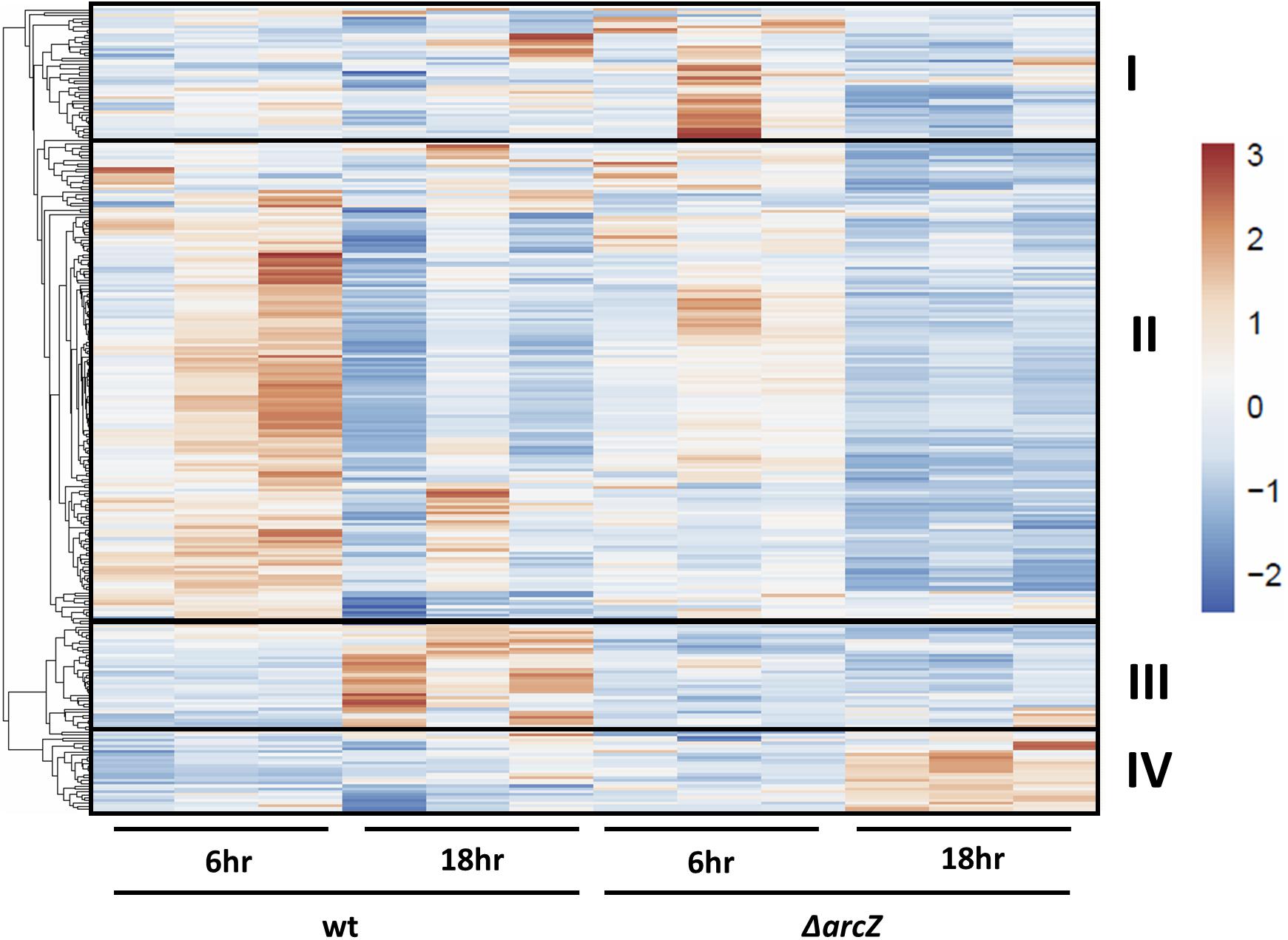
Figure 1. RNAseq heatmap comparing expression of differentially expressed genes across all biological replicate samples. Rows are centered and unit variance scaling is applied to rows. Rows are clustered using correlation distance and average linkage. Four main clusters of differentially expressed genes have been designated as groups I, II, III, and IV.
Pathway Enrichment in ArcZ Regulon
We tested for enrichment of specific cell pathways as annotated by the Kyoto Encyclopedia of Genes and Genomes (KEGG) (Kanehisa and Goto, 2000). We found no pathways significantly enriched in the set of genes differentially expressed in the 6 h time point, however, at the 18 h time point we found several pathways that were significantly enriched in differentially expressed genes (Figure 2). Several pathways that were enriched were involved in carbon metabolism and amino acid biosynthesis and metabolism. Because we observed that crp mRNA was affected by deletion of arcZ, it is possible that the carbon metabolism related pathway effects are due to this regulation, but it remains unknown if these are direct or indirect effects. The several genes and pathways involved in amino acid biosynthesis and metabolism are likely targets of the transcription factor Lrp, which is known to be regulated by ArcZ (Schachterle and Sundin, 2019) and which we found to be differentially regulated in the ΔarcZ mutant in our transcriptomic analysis. The type III secretion system was also significantly enriched for differentially expressed genes, the function of which is known to be affected by deletion of arcZ (Zeng and Sundin, 2014). Other affected KEGG pathways included sulfur metabolism, selenocompound metabolism, monobactam biosynthesis, RNA polymerase, and quorum sensing. Some of these pathways, although annotated in the KEGG database, may not be functional in E. amylovora as experimental evidence is lacking.
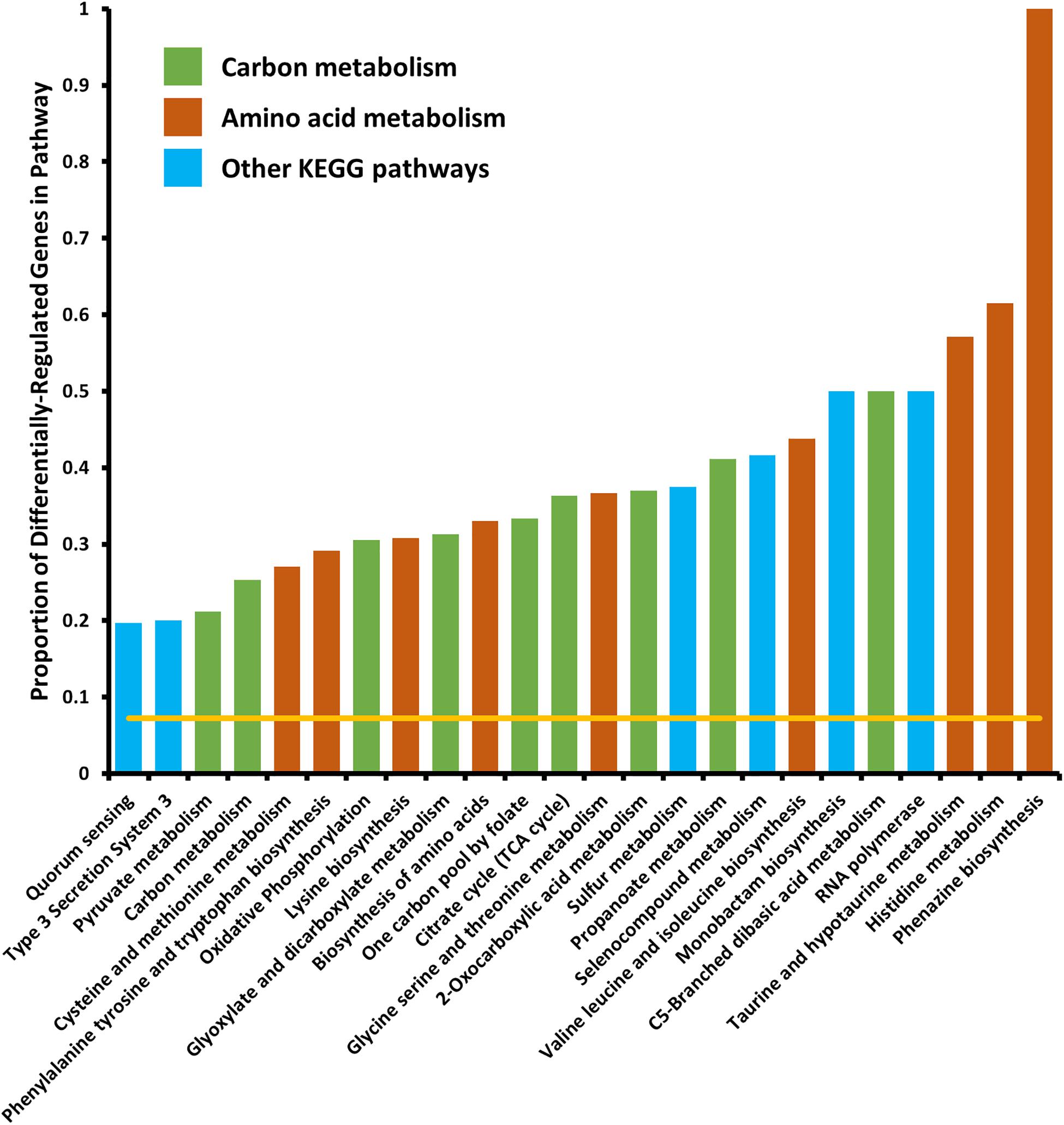
Figure 2. Kyoto encyclopedia of genes and genomes pathways significantly enriched in differentially expressed genes. Yellow line indicates expected proportion of differentially expressed genes if randomly distributed. Bars correspond to observed proportions of differentially expressed genes in KEGG pathways. Green bars indicate KEGG pathways related to carbon and central metabolism, burnt-orange bars indicate KEGG pathways relating to amino acid biosynthesis and metabolism and blue bars indicate other KEGG pathways. All bars shown are significantly enriched (Padj < 0.05) by Fisher’s exact test.
When analyzing the KEGG pathway for glycolate/glyoxylate metabolism, we found that E. amylovora has neither genes coding for enzymes that generate glycolate nor glyoxylate. In other organisms, glycolate oxidase, which converts glycolate to glyoxylate, generates hydrogen peroxide as a byproduct of this enzymatic reaction (Fahnenstich et al., 2008), and catalase is considered to be a part of this pathway for the detoxification of the peroxide. Although E. amylovora does not code for a glycolate oxidase enzyme, plants do, and have been shown to use this enzyme for generating hydrogen peroxide as a pathogen defense mechanism (Rojas and Mysore, 2012; Rojas et al., 2012). This led us to search for other genes differentially regulated by ArcZ that may play a role in coping with oxidative stress.
ArcZ Regulates Oxidative Stress Response Genes
In our search of differentially expressed genes that have links to the oxidative stress response, we found katA, encoding a catalase, tpx, encoding a thiol-peroxidase, and osmC, encoding an osmotically inducible peroxiredoxin. katA and osmC were both down-regulated in the ΔarcZ mutant, and tpx mRNA was more abundant (Figure 3A). Although recent work has indicated that another catalase, KatG, plays a role in E. amylovora mitigation of oxidative stress (Santander et al., 2018), katG was not differentially expressed in the ΔarcZ mutant relative to wild-type. Nonetheless, as an additional oxidative stress mitigation enzyme, we have included katG in several of our experiments to better understand its role with the other ArcZ-regulated oxidative stress mitigation enzymes. We independently verified by quantitative real-time PCR that katA and osmC are down-regulated in the ΔarcZ mutant, and that tpx is up-regulated (Figure 3B). Consistent with our RNAseq data, there was no difference in relative abundance of katG mRNA between wild-type and the ΔarcZ mutant.
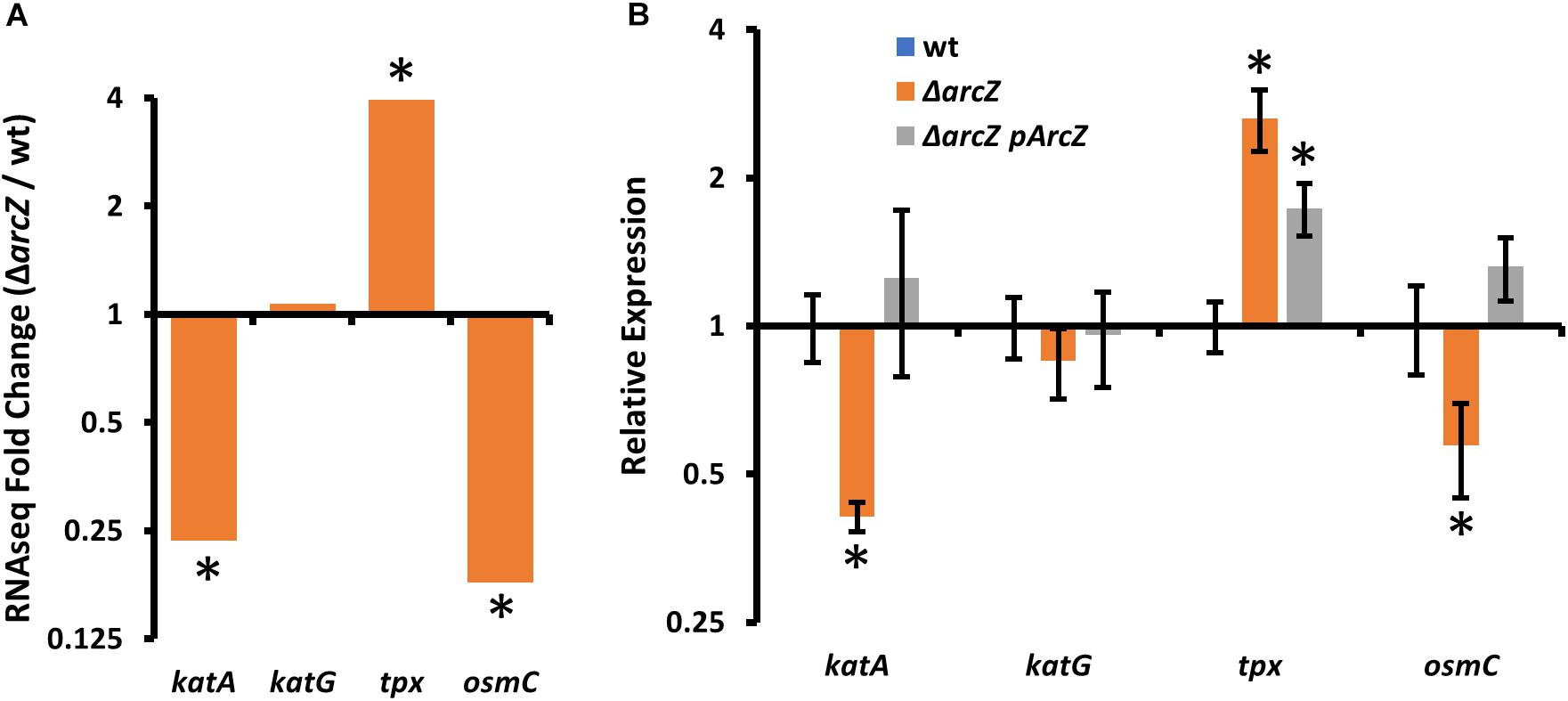
Figure 3. Oxidative stress mitigation enzymes are differentially expressed in E. amylovora Ea1189 ΔarcZ mutant relative to wild-type. Relative expression of katA, katG, tpx, and osmC based on (A) RNAseq (ΔarcZ/wt) or (B) quantitative real-time PCR. Asterisks indicate significant difference (P < 0.05) from wild-type by negative binomial distribution (RNAseq; through DESeq) or Student’s t-test (qPCR data). qPCR experiments used cDNA synthesized from three replicate RNA samples from independent bacterial cultures.
ArcZ Regulated Oxidative Stress Response Genes Are Critical for Survival of Exogenous Hydrogen Peroxide
Because KatA and KatG have been shown to play a role in E. amylovora response to exogenous hydrogen peroxide (Santander et al., 2018), we tested the ΔarcZ mutant, along with the ΔkatA, ΔkatG, Δtpx, and ΔosmC mutants for their catalase activity and survival after treatment with excess hydrogen peroxide. We found that the ΔkatA mutant had no detectable catalase activity (Figure 4A) and exhibited increased susceptibility to hydrogen peroxide in a disk diffusion assay (Figure 4B). The catalase activity of the ΔarcZ mutant was reduced nearly 10-fold relative to wild-type and the mutant was also increased in sensitivity to hydrogen peroxide in a disk-diffusion assay. The Δtpx mutant had a reduction in catalase activity of about 3-fold and increased sensitivity to hydrogen peroxide in the disk diffusion assay. The ΔkatG and ΔosmC mutants had only a slight decrease in overall catalase activity, and the ΔkatG mutant had increased susceptibility in the disk diffusion assay. It is likely that the ΔkatG mutant did not show decreased catalase activity in the catalase activity assay but does have increased susceptibility in the disk-diffusion assay because of the differences in growth in liquid culture for the catalase activity assay and growth on solid media for the disk-diffusion assay, as it is known that katG expression is growth phase dependent (Santander et al., 2018). The growth of the ΔosmC mutant was not different from wild-type in the disk-diffusion assay.
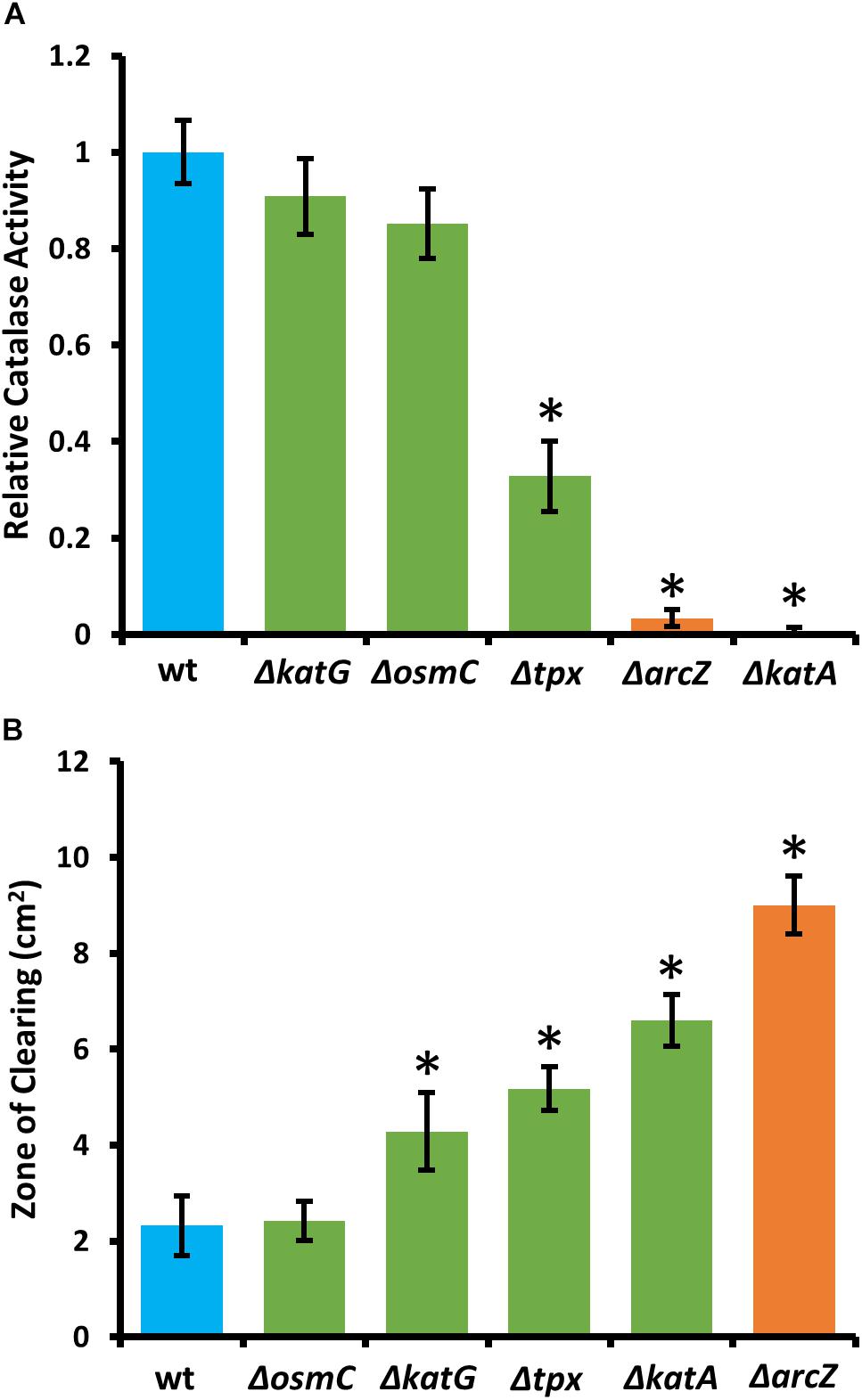
Figure 4. E. amylovora Ea1189 ΔarcZ mutant has reduced catalase activity and increased susceptibility to exogenous hydrogen peroxide. Relative catalase activity (A), determined from cells grown overnight in LB broth. Zone of clearing (B) around a filter paper disk treated with 10 μL of 8 M hydrogen peroxide on LB solid media. Assays were repeated at least four times. Asterisks indicate significant (P < 0.05) difference from wild-type by Student’s t-test.
During our determination of catalase activity in E. amylovora, we observed that a small amount of catalase activity is released into the culture medium. To determine whether the catalase activity in culture supernatants is from KatA or KatG, we concentrated culture supernatants from overnight cultures of the ΔkatA and ΔkatG mutants. Concentrated supernatants were mixed with hydrogen peroxide and monitored for evolution of gas through formation of bubbles. Catalase activity was observed in the ΔkatG culture supernatants, but not in the ΔkatA culture supernatants, indicating that KatA is responsible for the catalase activity present in culture supernatants. Because secreted catalase activity has not been reported in other Enterobacterales, we conducted a multiple sequence alignment of KatA and KatE protein sequences from phylogenetically diverse bacteria. This analysis revealed that E. amylovora KatA is more similar to KatA from Bacillus subtilis and Pseudomonas aeruginosa than to KatE from E. coli (Supplementary Figure S2). Protein BLAST (Camacho et al., 2009) further showed that the most similar hits for a search with E. amylovora KatA as query came from the genera Erwinia, Pantoea, and Pseudomonas.
Mutation of ArcZ Can Be Complemented by katA
Because ΔarcZ has reduced catalase activity relative to wild-type and is more susceptible than wild-type to exogenous hydrogen peroxide both on solid media and in liquid media, we wanted to determine if any of the oxidative stress mitigation enzymes would be able to restore wild-type phenotypes in these tests. To test this, we complemented the ΔarcZ mutant with katA, katG, tpx, or osmC, each with the respective native promoter in a medium-copy-number plasmid, where we hypothesized that increased gene copy number would compensate for moderate repressive regulatory effects. When tested for catalase activity, we found that introduction of any of these genes in this manner led to increased catalase activity relative to the ΔarcZ mutant (Figure 5A). However, providing katG, tpx, or osmC in the ΔarcZ mutant still resulted in catalase activity well below that of wild-type cells. Only providing katA on a plasmid restored catalase activity to greater than wild-type levels. When we tested the ΔarcZ mutant complemented with katA in the disk-diffusion assay for susceptibility to exogenous hydrogen peroxide, we found that katA restored wild-type levels of growth in the ΔarcZ mutant (Figure 5B).
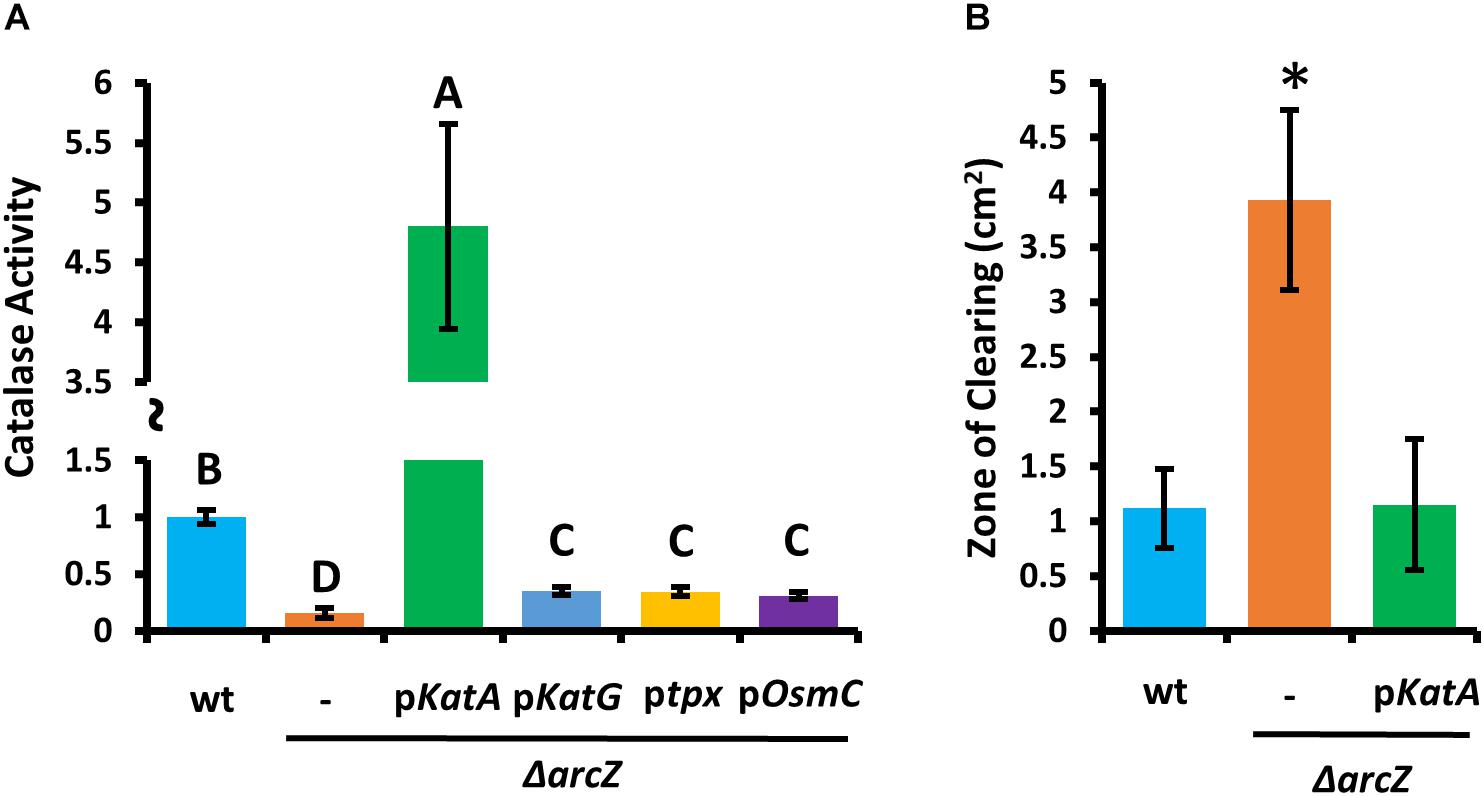
Figure 5. Providing katA on a plasmid recovers catalase activity and susceptibility to exogenous hydrogen peroxide in the E. amylovora Ea1189 ΔarcZ mutant. Relative catalase activity (A) of wild-type or ΔarcZ mutant carrying empty plasmid (-) or the indicated gene with its corresponding native promoter. Groups with shared letter designation do not differ from each other significantly (P < 0.05) by Tukey’s HSD test. Zone of clearing (B) around a filter paper disk treated with 10 μL of 1 M hydrogen peroxide on LB solid media. Tests were conducted at least four times and asterisks denote significant differences (P < 0.05) from wild-type by Student’s t-test.
Hydrogen Peroxide Produced by Inoculated Apple Shoots
In order to relate the difference in hydrogen peroxide susceptibility of our various strains to the interactions between E. amylovora and host apple shoots, we quantified hydrogen peroxide levels in apple leaves over the course of infection with wild-type E. amylovora cells. We detected a baseline of approximately 1 mM hydrogen peroxide in uninfected apple leaves (Figure 6A). One day post-inoculation, before visual disease symptoms developed, hydrogen peroxide levels doubled to nearly 2 mM. After 2 days post-inoculation, when visual symptoms had developed in the main vein of the leaf, hydrogen peroxide levels had doubled again, to over 4 mM. After three- and 4-days post-inoculation, as visual fire blight symptoms spread from the main vein to the rest of the leaf, hydrogen peroxide levels decreased again to below 2 mM (Figure 6A).
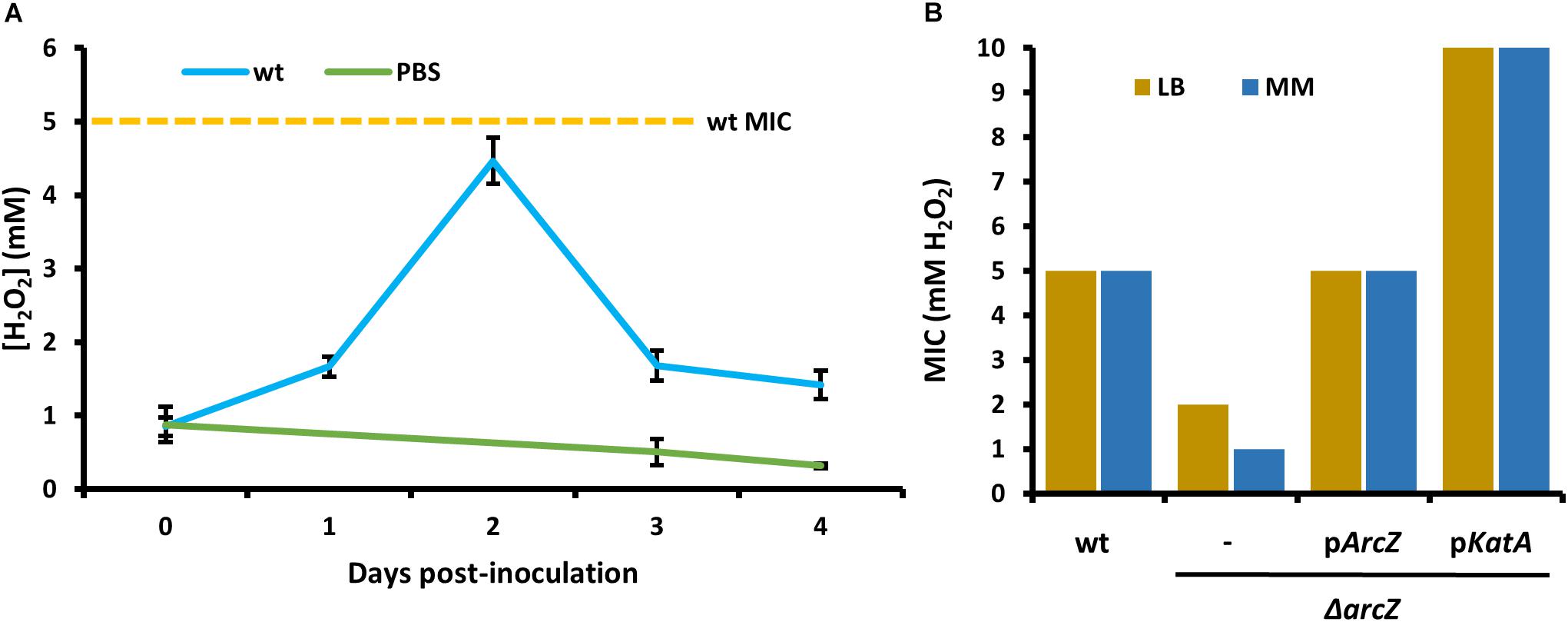
Figure 6. Erwinia amylovora Ea1189 elicits hydrogen peroxide production response from apple leaves and has evolved to cope with high levels of exogenous hydrogen peroxide. (A) Levels of hydrogen peroxide in apple leaves inoculated with wild-type E. amylovora or mock-inoculated with phosphate buffered saline (PBS), across time following inoculation. Averages come from two independent experiments with a total of at least 12 replicate measurements per time point, and error bars represent standard error of the mean. Dotted line indicates the H2O2 minimum inhibitory concentration (MIC) of wild-type E. amylovora Ea1189. (B) The MIC of hydrogen peroxide for wild-type or ΔarcZ mutant carrying empty plasmid (–) or the indicated gene with its native promoter. MIC was tested in LB and minimal media with sucrose as the carbon source (MM).
In order to determine the specific concentration to which E. amylovora wild-type and ΔarcZ mutant cells are susceptible to exogenous hydrogen peroxide, we tested for the MIC of hydrogen peroxide. We found that the MIC of hydrogen peroxide for wild-type cells is 5 mM whether tested in minimal medium or rich LB medium (Figure 6B). The MIC of hydrogen peroxide for ΔarcZ mutant cells was found to be 1 mM in minimal medium and 2 mM when tested in LB medium. This is consistent with the finding that metabolism of specific amino acids available in rich media can help to mitigate oxidative threats (Carlioz and Touati, 1986) The hydrogen peroxide MIC of the ΔarcZ mutant was complemented back to wild-type levels by providing arcZ on a plasmid under control of its native promoter. The ΔarcZ mutant with katA on a plasmid grew uninhibited at concentrations of hydrogen peroxide up to 10 mM. It is noteworthy that the hydrogen peroxide MIC for wild-type cells was determined to be 5 mM, but in planta hydrogen peroxide levels peaked at just over 4 mM.
ArcZ and KatA Are Critical for Survival of E. amylovora During the Hypersensitive Response in Tobacco
Because the hydrogen peroxide MIC for wild-type and ΔarcZ mutant cells and our quantification of hydrogen peroxide levels in apple leaves suggested that the inability of the ΔarcZ mutant to cope with oxidative stress may play an important role in ability of the bacteria to survive and successfully infect host plants, we wanted to test the impact of catalase activity on bacterial survival in planta. Because loss of arcZ leads to decreases in several virulence-associated traits, we also wanted to uncouple survival during the in planta oxidative burst from other virulence defects. To accomplish this, we assessed survival in non-host Nicotiana tabacum (tobacco) which will undergo a hypersensitive response, including an oxidative burst (Montillet et al., 2005), in response to type III effector translocation when E. amylovora cells are infiltrated into the tobacco apoplast (Wei et al., 1992). We infiltrated tobacco leaves with E. amylovora Ea1189 wild-type and ΔarcZ mutant cells at a density of 109 CFU mL–1 and assessed survival 5 days post-infiltration by sampling a 1 cm2 leaf disk. We found that on average 107 CFU/cm2 wild-type cells survived but only 105 CFU/cm2 of ΔarcZ mutant cells survived (Figure 7). The survival defect in the ΔarcZ mutant could be rescued by providing katA on a plasmid, suggesting that the survival defect in the ΔarcZ mutant is due to increased susceptibility to reactive oxygen species, and not just to other pleiotropic effects of ArcZ. To verify whether provision of katA on a plasmid in the ΔarcZ mutant would be sufficient to complement the ΔarcZ virulence defect, we inoculated immature pears and monitored symptom development. We found that providing katA on a plasmid did not increase virulence of the ΔarcZ mutant in immature pears (data not shown).
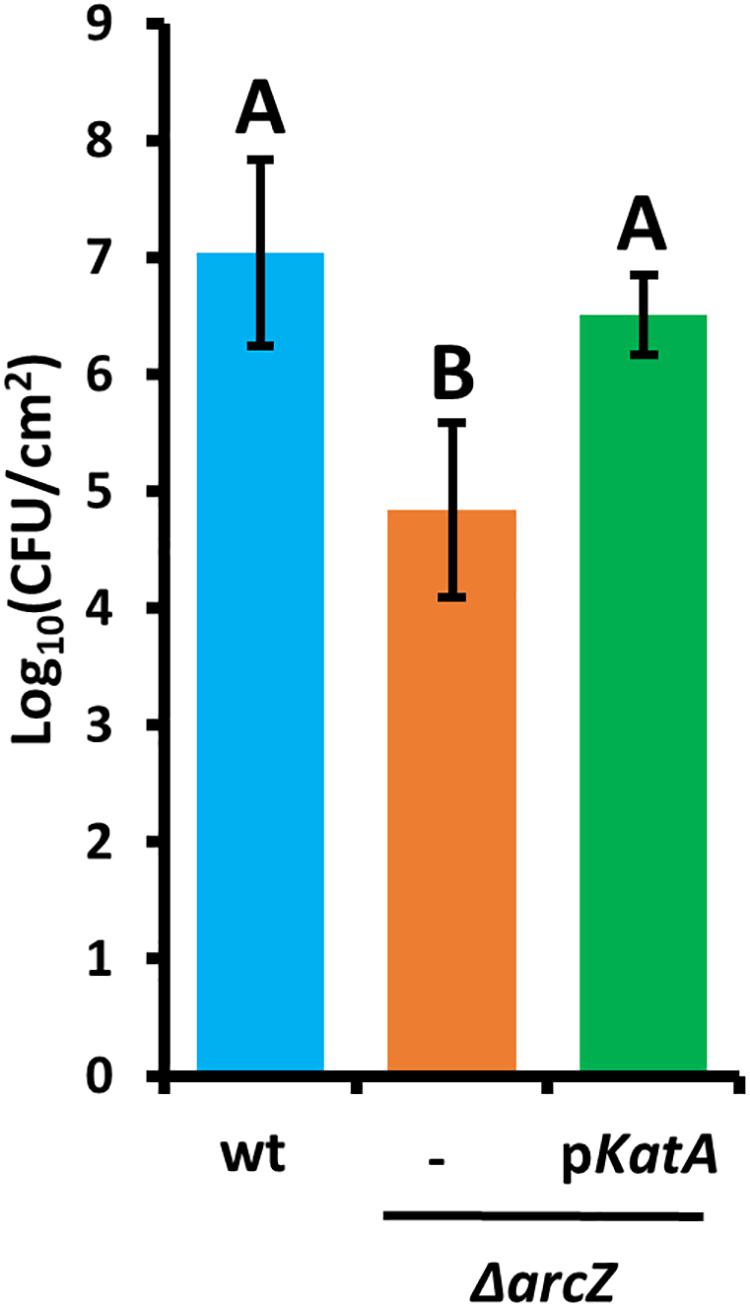
Figure 7. Survival of E. amylovora Ea1189 cells in tobacco leaves following elicitation of the hypersensitive response. Leaf disks were sampled 5 days post-infiltration. Each strain was tested at least three times with a total of at least six replicates. Error bars represent standard deviation and groups with shared letter designation do not differ significantly from each other (P < 0.05) by Tukey’s HSD test.
ArcZ Regulates katA Transcriptionally and tpx Post-transcriptionally
Because ArcZ is a post-transcriptional regulator and modulates katA transcript abundance, we assessed whether ArcZ regulates katA at the transcriptional or post-transcriptional level. To do so, we constructed a promoter fusion with the katA promoter upstream of a promoter-less gfp in plasmid pPROBE-NT (Miller et al., 2000), and a translational fusion with the 5’ UTR of katA and first 18 amino acids in-frame with gfp in plasmid pXG20 (Urban and Vogel, 2007). We observed reduced katA promoter activity in the ΔarcZ mutant relative to wild-type but no difference on the katA translational fusion between wild-type and ΔarcZ (Figure 8).
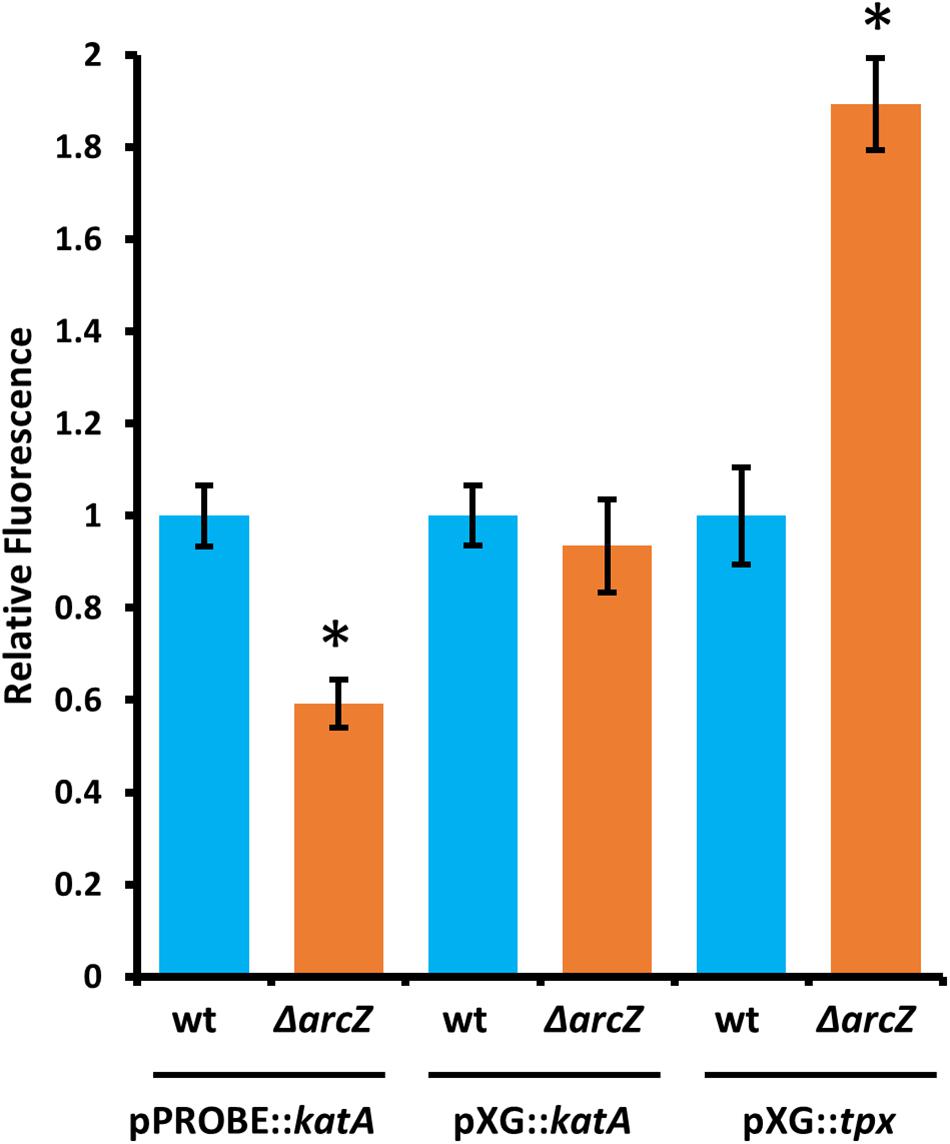
Figure 8. ArcZ of E. amylovora Ea1189 regulates katA promoter activity and regulates tpx post-transcriptionally. Relative fluorescence of indicated reporter fusions tested in ΔarcZ mutant cells and relative to fluorescence of wild-type cells carrying the same construct. Asterisks indicate significant difference (P < 0.05) by Student’s t-test between ΔarcZ and wild-type carrying the same construct. Error bars represent standard deviation, and each strain was tested at least four times.
Because ArcZ is known to post-transcriptionally repress tpx mRNA in Salmonella enterica serovar Typhimurium through a direct interaction (Papenfort et al., 2009), we tested whether ArcZ repression of tpx also occurs through post-transcriptional regulation in E. amylovora. We generated a translational fusion with the 5’ UTR of tpx and first 33 amino acids in-frame with gfp in plasmid pXG20 (Urban and Vogel, 2007), and compared relative fluorescence between wild-type and ΔarcZ mutant cells. We found increased GFP fluorescence in the ΔarcZ mutant relative to wild-type (Figure 8), suggesting that the ArcZ-tpx interaction is likely conserved between Salmonella Typhimurium and E. amylovora. To determine if this interaction is likely to occur between the same bases in these two organisms, we predicted the interaction between ArcZ and tpx using RNAhybrid (Krüger and Rehmsmeier, 2006), and found that the same region is predicted to interact in E. amylovora as in Salmonella Typhimurium (Supplementary Figure S3). Because the same interaction is predicted, and the fact that ArcZ has a high degree of conservation in the interacting region (Schachterle et al., 2019), it is likely that the post-transcriptional repression of tpx mRNA in E. amylovora occurs through the same interaction as in Salmonella Typhimurium. Because our qPCR analysis showed that arcZ complementation did not fully restore wild-type levels of tpx transcript, it is possible that in addition to post-transcriptional interactions, ArcZ may affect tpx indirectly at the transcriptional level.
ArcZ Regulon Overlaps With Known Transcription Factor Regulons
Because ArcZ regulates katA at the transcriptional level, we utilized our RNAseq data to search for candidate regulators that could explain the ArcZ regulation of katA. We analyzed the ArcZ regulon for overlap with known transcription factors with known regulons. We inferred E. amylovora transcription factor regulons on the assumption that if a transcription factor and its target gene are conserved between E. coli and E. amylovora then the target is also a part of the regulon in E. amylovora. We acquired E. coli regulon information from regulondb.com and utilized BLAST+ to search for transcription factor and target homologs in E. amylovora. Using this approach, we found 38 conserved regulators with conserved targets in those regulons, with an average of 48.5% of targets conserved in each regulon. When we tested these putative regulons for overlap with our determined E. amylovora ArcZ regulon, we found six regulons with a significant (Padj < 0.05; Fisher’s exact test) amount of overlap (Figure 9). The six transcription factors with overlapping regulons are ArcA, Fnr, IHF, Lrp, NarL, and PurR. We note also that the overlap between the ArcZ and Fur regulons was nearly significant (Padj = 0.069). Of these regulons, ArcA, Fnr and Fur all form a network of interactions and are known to have impacts on oxidative sensing and response (including catalase) in E. coli (Niederhoffer et al., 1990; Hassan and Sun, 1992; Compan and Touati, 1994; Benov and Sequeira, 2003). Furthermore, this core set ArcA, Fnr, and Fur also has known interactions with IHF (Mettert and Kiley, 2007), NarL (Tyson et al., 1993), and PurR (Stojiljkovic et al., 1994), three of the remaining regulators with ArcZ regulon overlap. Additionally, we recently reported that Lrp is regulated post-transcriptionally by ArcZ (Schachterle and Sundin, 2019).
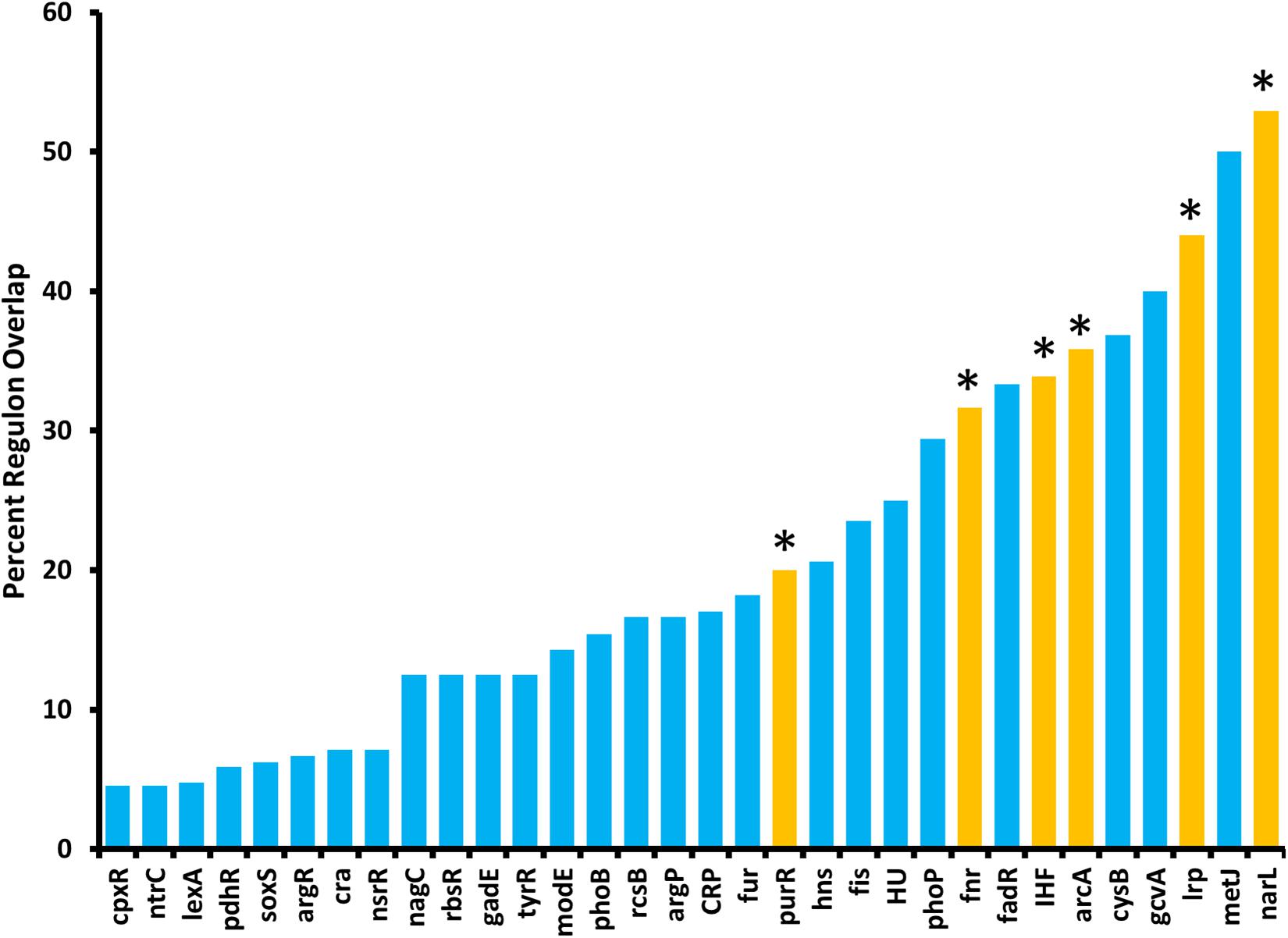
Figure 9. The E. amylovora Ea1189 ArcZ regulon overlaps with several putative transcription factor regulons. Transcription factor regulons were inferred in E. amylovora based on documented regulons in Escherichia coli and tested for significant (Padj < 0.05; indicated by asterisks above bars) overlap with the ArcZ regulon by Fisher’s exact test. Transcription factor regulons with no overlap with the ArcZ regulon are not shown.
arcZ Mutant Phenotypes Are Recapitulated by arcA and arcB Mutants
To determine the regulatory roles that the ArcBA two-component system, along with Fnr and Fur may share with ArcZ, we generated single-gene deletion mutants for each of the genes encoding these transcriptional regulators. We determined the effect of these mutations on swimming motility and susceptibility to exogenous hydrogen peroxide, two phenotypic traits affected by deletion of arcZ. We found that the ΔarcA mutant had reduced swimming motility compared to wild-type, but that deletion of arcB, fnr, or fur had no effect (Figure 10A). Similarly, we found increased susceptibility to exogenous hydrogen peroxide in the ΔarcA and ΔarcB mutants compared to wild-type, but no difference in susceptibility to hydrogen peroxide in the Δfnr and Δfur mutants (Figure 10B).
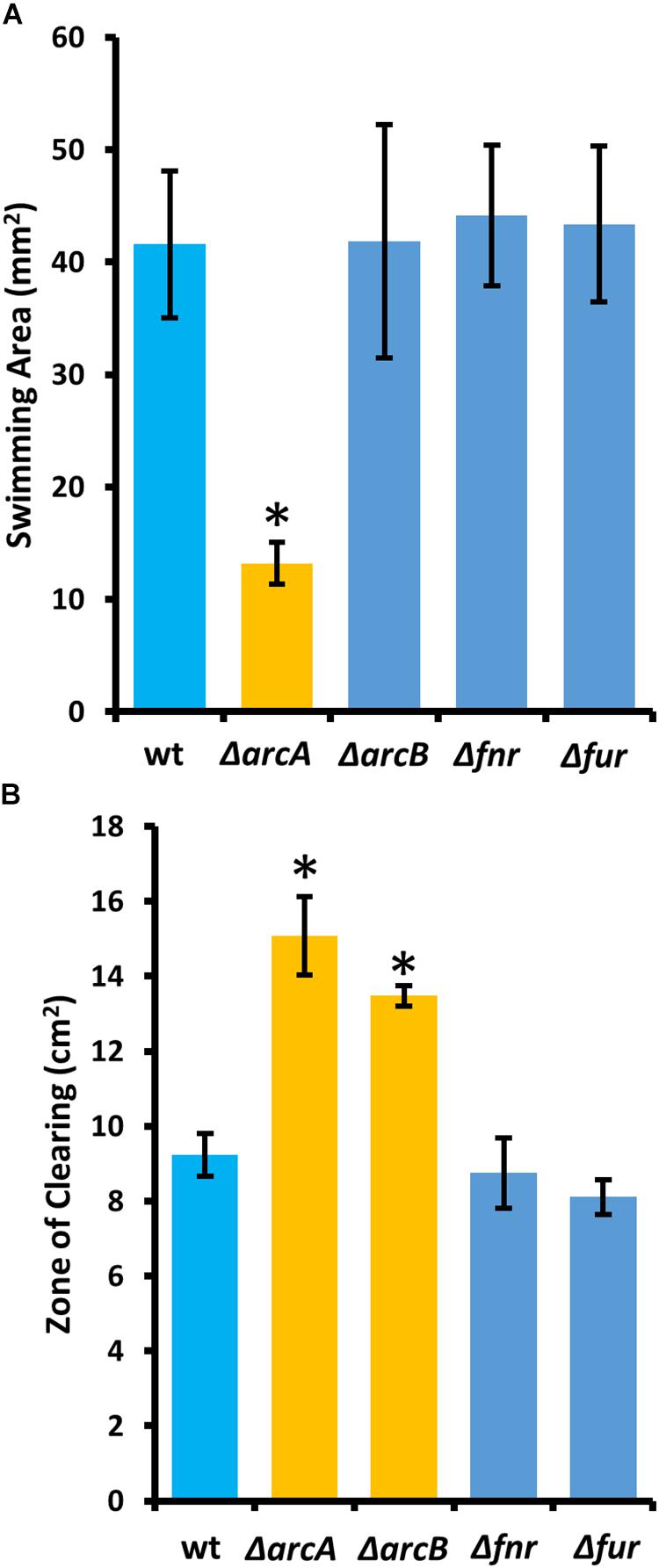
Figure 10. The E. amylovora Ea1189 ArcBA two-component system affects swimming motility and hydrogen peroxide susceptibility. (A) Swimming motility area of indicated strains 24 h after stab inoculation into soft agar (0.25% w/v) plates. (B) Zone of clearing around a filter paper disk treated with 8 M hydrogen peroxide on LB solid media. Asterisks indicate significant difference (P < 0.05) from wild-type strain by Student’s t-test. Error bars represent standard deviations from four biological replicates for swimming motility and from three biological replicates for hydrogen peroxide susceptibility.
ArcZ Regulates ArcA Post-transcriptionally
Because deletions in arcA or arcB of the ArcBA two-component system had similar effects to ΔarcZ on the motility and susceptibility to hydrogen peroxide phenotypes, we generated translational fusions for arcA and arcB to test whether ArcZ regulates these genes post-transcriptionally. We additionally generated a fur translational fusion to determine if ArcZ regulates fur post-transcriptionally because Fur is a transcriptional regulator of the catalase katE in E. coli (Benov and Sequeira, 2003). The arcA, arcB, and fur translational fusions with gfp reporter were tested in an E. coli strain carrying arcZ under control of an IPTG-inducible tac promoter. Upon induction of arcZ expression, we found no difference in the strain carrying the arcB or fur translational fusion constructs but did find increased fluorescence in the strain carrying the arcA construct (Figure 11A). To confirm this result, we tested the arcA translational fusion in E. amylovora wild-type and ΔarcZ mutant cells and found a 20 percent reduction in fluorescence in the ΔarcZ mutant compared to wild-type (Figure 11B). Together these results indicate that ArcZ regulates arcA post-transcriptionally in E. amylovora. We predicted candidate interactions between ArcZ and arcA mRNA using RNAHybrid (Krüger and Rehmsmeier, 2006) and found a strong candidate interaction 50 bases upstream of the ArcA start codon (Supplementary Figure S4). Consistent with the idea that ArcZ is affecting katA at the transcriptional level through post-transcriptional regulation of arcA, we found three direct repeats of the ArcA binding motif upstream of katA in the E. amylovora genome (Supplementary Figure S5). These three direct repeats of the ArcA binding motif represent a common arrangement of binding motifs in ArcA regulated genes (Park et al., 2013).
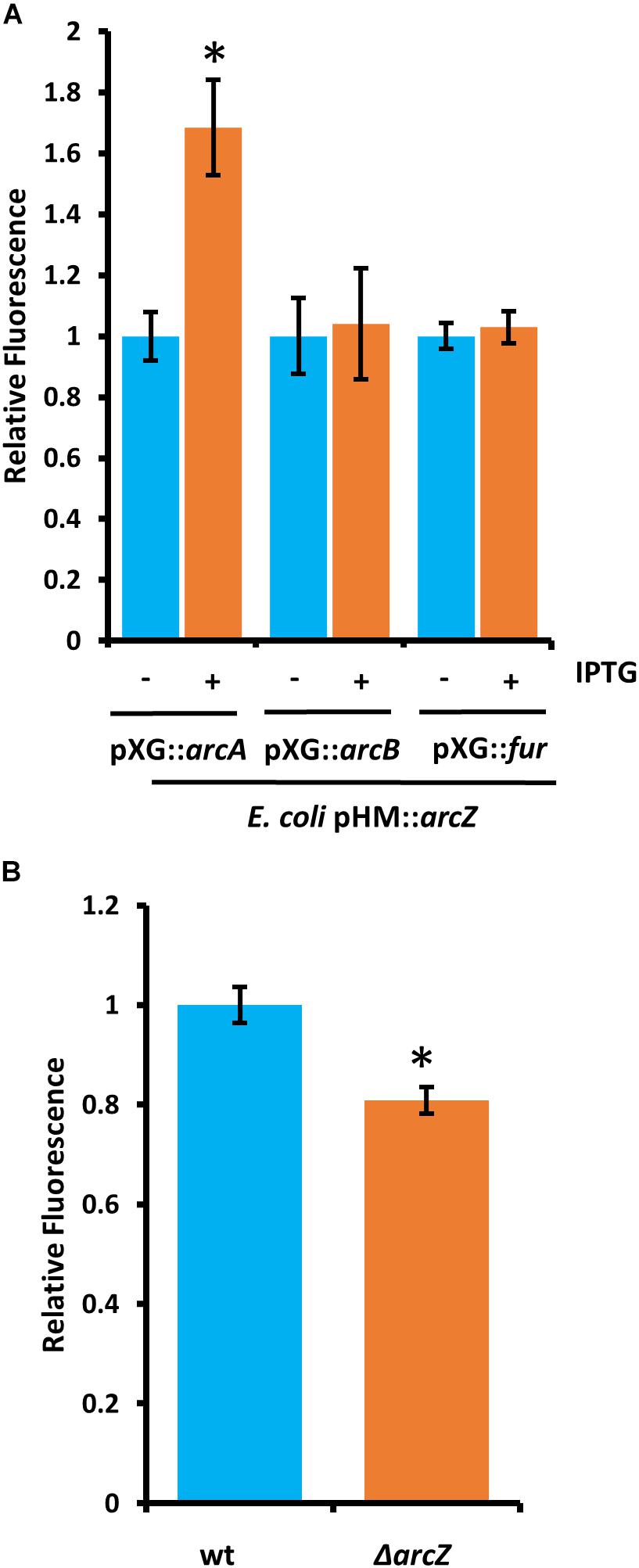
Figure 11. ArcZ of E. amylovora Ea1189 regulates arcA post-transcriptionally. (A) Escherichia coli carrying E. amylovora arcZ on a plasmid under control of the IPTG-inducible tac promoter, with the indicated translational fusions comparing relative fluorescence between un-induced and induced cells. (B) Relative fluorescence of E. amylovora wild-type or ΔarcZ mutant cells carrying the arcA translational fusion. Asterisks indicate significant difference (P < 0.05) from wild-type or un-induced cells carrying the same reporter plasmid by Student’s t-test. Error bars represent standard deviation from at least four biological replicates.
Discussion
Here, we present transcriptomic analysis of the sRNA ArcZ regulon, providing evidence that in E. amylovora, ArcZ is a global regulator with a regulon of 342 genes, or 9.8% of the genome, based on the culture conditions used in our study. Furthermore, analysis of the ArcZ regulon identified an important role for ArcZ in regulation of genes involved in coping with oxidative stress. We found that ArcZ regulates katA at the transcriptional level and while it affects tpx transcript abundance, ArcZ represses tpx post-transcriptionally.
In addition to transcriptional regulation of katA and post-transcriptional regulation of tpx, we found that ArcZ regulates arcA post-transcriptionally. ArcA is the response regulator of the ArcBA (anoxic redox control) two-component system, which is responsive to oxidative status of the cell (Iuchi et al., 1990). This two-component system is activated in a sigmoidal response pattern in response to oxidative state of quinones (Bekker et al., 2010) and plays a major role in modulating expression of several genes encoding redox-active enzymes (Morales et al., 2013). The sRNA ArcZ has received this Arc acronym for its position adjacent to arcB in the genome as an arc-associated sRNA (Papenfort et al., 2009). Although arcB and arcZ are distal to arcA in the genome, it has been found in E. coli that arcZ is transcriptionally regulated in response to oxygen levels in an ArcA dependent manner (Mandin and Gottesman, 2010). Because we are reporting that ArcZ regulates arcA post-transcriptionally in E. amylovora, this suggests that if these regulatory relationships are conserved between E. amylovora and E. coli, ArcZ and ArcA may form a feedback loop to reinforce cellular responses in response to oxygen availability and oxidative status.
Given our findings that ArcZ regulates katA at the transcriptional level and arcA and tpx post-transcriptionally, we propose a regulatory model in which the ArcBA two-component system acts as an oxygen sensor to transcriptionally regulate arcZ and katA, and that ArcZ in turn activates arcA post-transcriptionally, providing positive feedback on catalase activity. ArcA regulates transcription of arcZ in E. coli in an oxygen dependent manner (Mandin and Gottesman, 2010), but further work is necessary to confirm that this same regulation occurs in E. amylovora. We hypothesize that this proposed regulatory loop is significant during infection of host tissue, because of variations in oxygen accessibility across tissues. For example, in tissues with high oxygen availability such as leaves and flowers, E. amylovora cells are interacting with living host cells that are the most prone to mount defense responses including production of reactive oxygen species. It has been shown previously that E. amylovora cells trigger host defense mechanisms including generation of an oxidative burst during compatible interactions (i.e., successful infection) (Venisse et al., 2002, 2003; Iakimova et al., 2013; Abdollahi et al., 2015). Indeed, we demonstrate here that concentrations of hydrogen peroxide in infected apple leaves peak at levels of 4–5 mM at 2 days post-inoculation (Figure 6A). In contrast, host cells are dead in mature xylem vessels, and host-produced reactive oxygen species are likely to be scarce. Furthermore, in woody xylem, it has been shown that oxygen levels are typically reduced to half of atmospheric oxygen with ample water flow, but that when xylem flow is restricted, oxygen levels can drop to anaerobic levels (Gansert, 2003). The oxygen-responsive nature of the proposed ArcZ-ArcA-KatA feedback loop suggests that oxygen and oxidative state may play an essential role in proper expression of genes for coping with reactive oxygen species during disease progression. Future work to determine the specific roles of oxygen availability as an environmental signal modulating virulence gene expression shows great promise to provide novel insights into how E. amylovora integrates environmental signals to determine virulence behaviors. Such insights are of great importance in understanding the basic biology of this pathogen to guide development of strategies that can limit its devastating effects.
In support of the importance of ability to cope with reactive oxygen species during infection, we found that provision of katA on a plasmid in the ΔarcZ mutant background not only restored catalase activity and wild-type susceptibility to exogenous hydrogen peroxide in in vitro tests, but also restored survival in non-host tobacco during hypersensitive response. This suggests that although ΔarcZ mutant cells are deficient in several virulence factors (Zeng and Sundin, 2014), coping with reactive oxygen species is a major limiting factor for this mutant in planta independent of other virulence-associated traits. This also matches the recent finding that the activity of the catalases KatA and KatG plays an important role in E. amylovora survival in the plant environment (Santander et al., 2018).
We found that ArcZ regulation of katA occurs at the transcriptional level and not at the post-transcriptional level. Previous research found little change in katA expression between exponential and stationary phase cultures (Santander et al., 2018), suggesting that ArcZ expression may not vary greatly either between these growth stages and that this regulatory system may respond to other environmental cues. We found that ArcZ regulates arcA and tpx post-transcriptionally and that interaction predictions between ArcZ and the arcA 5′ UTR indicate a likely interaction that could explain the effect of ArcZ on the arcA 5′ UTR, but further work is needed to provide experimental confirmation that these bases participate in direct interactions. The presence of three sequential ArcA binding sites upstream of katA suggests that the ArcZ regulation of katA is through the observed post-transcriptional effects on arcA. Again, future experimentation is necessary to confirm that ArcA directly regulates katA transcription.
The determined ArcZ regulon had significant overlap with the inferred regulons of ArcA, Fnr, PurR, Lrp, IHF, and NarL. Our previous work indicated that ArcZ regulates lrp (Schachterle and Sundin, 2019), and that finding was confirmed in this work in the significant amount of overlap between the ArcZ and Lrp regulons. In E. coli, the remaining transcription factors with regulon overlap with ArcZ form a complex web of inter-regulation, which is also involved in transcriptional regulation of catalases and thiol peroxidase (Niederhoffer et al., 1990; Hassan and Sun, 1992; Compan and Touati, 1994; Kim et al., 1999; Benov and Sequeira, 2003). The finding that ArcZ regulates arcA post-transcriptionally provides a connection between this sRNA and this transcription factor network, although additional links may exist. Although ArcZ affected abundance of osmC transcripts, deletion of osmC had little effect on the oxidative stress phenotypes we tested. Because osmC is a part of the lrp regulon (Bouvier et al., 1998), it seems likely that ArcZ is regulating osmC through its post-transcriptional regulation of lrp (Schachterle and Sundin, 2019). Because we found weak effects in the ΔosmC mutant when testing with hydrogen peroxide, it is possible that in E. amylovora as a peroxiredoxin OsmC functions to reduce the threat of organic peroxides but has little activity against inorganic hydrogen peroxide. Future work to understand the role of osmC and additional interactions between ArcZ and these transcription factors will help to uncover the contributions of these regulatory networks to E. amylovora physiology and virulence.
In this study, we observed catalase activity present in culture supernatants, and determined that katA is responsible for this activity. This suggests that during infection E. amylovora may be releasing or actively secreting catalase to reduce damage done to cellular structures when peroxide production is elicited as a part of host-defense responses. In support of the hypothesis that early protection may be important, production of hydrogen peroxide in infected apple and pear shoots occurs early during disease development, as elevated production of hydrogen peroxide occurred ahead of symptom development (Abdollahi et al., 2015). Additionally, because the protein sequence of E. amylovora KatA is more similar to catalases from gram-positive Bacillus subtilis than it is to KatE from E. coli, E. amylovora may have acquired this gene during its evolution as a plant pathogen. Indeed, KatA from E. amylovora is most similar to catalases from Pantoea and Pseudomonas species, suggesting it may have been horizontally acquired from one of these during evolution as bacteria from these genera all colonize apple flowers (Shade et al., 2013). Because katA does not encode a secretion signal peptide, further work will be needed to determine if KatA is being actively secreted or released through some other means such as simply as a result of cell lysis. Because we observed that KatA catalase activity is stable in the extracellular milieu, we hypothesize that extracellular KatA may play an important role during disease development.
In E. amylovora, ArcZ has been shown to directly interact with flhDC mRNA (Schachterle et al., 2019) and to post-transcriptionally regulate lrp (Schachterle and Sundin, 2019). In Salmonella Typhimurium, ArcZ is known to regulate and interact with sdaCB, tpx, and a gene encoding a horizontally acquired methyl-accepting chemotaxis protein (Papenfort et al., 2009). In E. coli, ArcZ is also known to interact with and post-transcriptionally regulate rpoS (Mandin and Gottesman, 2010). Herein we provide additional evidence that in E. amylovora ArcZ post-transcriptionally represses tpx similar to S. Typhimurium, and also acts as a post-transcriptional regulator of arcA. These interactions explain several of the phenotypes observed in the E. amylovora ΔarcZ mutant, however, additional phenotypes remain unexplained, such as the effects of arcZ on type III secretion. This transcriptomic and molecular analysis of the ArcZ regulon will serve to guide and inform future studies to more fully understand the mechanisms and specific roles that ArcZ plays as a global regulator in coordinating virulence-associated traits in E. amylovora.
Data Availability Statement
The sequencing datasets generated in this study can be found in the NCBI Sequence Read Archive (SRA) under BioProject accession number: PRJNA543432 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA543432).
Author Contributions
JS and GS conceived and designed the experiments. JS and DO conducted the experiments. JS, DO, and GS analyzed the data, and wrote and edited the manuscript.
Funding
This project was supported by the Agriculture and Food Research Initiative Competitive Grant No. 2015-67013-23068 of the US Department of Agriculture National Institute of Food and Agriculture, Michigan State University AgBioResearch, and Project GREEEN, a Michigan plant agriculture initiative at Michigan State University. This material is also based upon work supported by the National Science Foundation Graduate Research Fellowship under Grant No. DGE1424871 to JS.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02775/full#supplementary-material
References
Abdollahi, H., Zahra, G., Kobra, E., and Mehrabi, R. (2015). Role of electron transport chain of chloroplasts in oxidative burst of interaction between Erwinia amylovora and host cells. Photosynth. Res. 124, 231–242. doi: 10.1007/s11120-015-0127-8
Abràmoff, M. D., Magalhães, P. J., and Ram, S. J. (2004). Image processing with ImageJ. Biophotonics Int. 11, 36–42.
Aldridge, P., Metzger, M., and Geider, K. (1997). Genetics of sorbitol metabolism in Erwinia amylovora and its influence on bacterial virulence. Mol. Gen. Genet. 256, 611–619. doi: 10.1007/s004380050609
Anders, S., and Huber, W. (2012). Differential Expression of RNA-Seq Data at The Gene Level–the DESeq Package. Heidelberg: European Molecular Biology Laboratory (EMBL).
Anders, S., Pyl, P. T., and Huber, W. (2015). HTSeq-a python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. doi: 10.1093/bioinformatics/btu638
Asai, S., and Shirasu, K. (2015). Plant cells under siege: plant immune system versus pathogen effectors. Curr. Opin. Plant Biol. 28, 1–8. doi: 10.1016/j.pbi.2015.08.008
Bayot, R. G., and Ries, S. M. (1986). Role of motility in apple blossom infection by Erwinia amylovora and studies of fire blight control with attractant and repellent compounds. Phytopathology 76, 441–445.
Bekker, M., Alexeeva, S., Laan, W., Sawers, G., Teixeira De Mattos, J., and Hellingwerf, K. (2010). The ArcBA two-component system of Escherichia coli is regulated by the redox state of both the ubiquinone and the menaquinone pool. J. Bacteriol. 192, 746–754. doi: 10.1128/JB.01156-09
Bellemann, P., Bereswill, S., Berger, S., and Geider, K. (1994). Visualization of capsule formation by Erwinia amylovora and assays to determine amylovoran synthesis. Int. J. Biol. Macromol. 16, 290–296. doi: 10.1016/0141-8130(94)90058-2
Benov, L., and Sequeira, F. (2003). Escherichia coli Δ fur mutant displays low HPII catalase activity in stationary phase. Redox Rep. 8, 379–383. doi: 10.1179/135100003225003357
Berry, M. C., McGhee, G. C., Zhao, Y., and Sundin, G. W. (2009). Effect of a waaL mutation on lipopolysaccharide composition, oxidative stress survival, and virulence in Erwinia amylovora. FEMS Microbiol. Lett. 291, 80–87. doi: 10.1111/j.1574-6968.2008.01438.x
Blattner, F. R., Plunkett, G., Bloch, C. A., Perna, N. T., Burland, V., Riley, M., et al. (1997). The complete genome sequence of Escherichia coli K-12. Science 277, 1453–1462. doi: 10.1126/science.277.5331.1453
Bogdanove, A. J., Bauer, D. W., and Beer, S. V. (1998). Erwinia amylovora secretes DspE, a pathogenicity factor and functional AvrE homolog, through the Hrp (type III secretion) pathway. J. Bacteriol. 180, 2244–2247.
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bolwell, G. P., and Wojtaszek, P. (1997). Mechanisms for the generation of reactive oxygen species in plant defence–a broad perspective. Physiol. Mol. Plant Pathol. 51, 347–366. doi: 10.1006/pmpp.1997.0129
Bouvier, J., Gordia, S., Kampmann, G., Lange, R., Hengge-Aronis, R., and Gutierrez, C. (1998). Interplay between global regulators of Escherichia coli: effect of RpoS, Lrp and H-NS on transcription of the gene osmC. Mol. Microbiol. 28, 971–980. doi: 10.1046/j.1365-2958.1998.00855.x
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421
Carlioz, A., and Touati, D. (1986). Isolation of superoxide dismutase mutants in Escherichia coli: is superoxide dismutase necessary for aerobic life? EMBO J. 5, 623–630. doi: 10.1002/j.1460-2075.1986.tb04256.x
Castiblanco, L. F., and Sundin, G. W. (2018). Cellulose production, activated by cyclic di-GMP through BcsA and BcsZ, is a virulence factor and an essential determinant of the three-dimensional architectures of biofilms formed by Erwinia amylovora Ea1189. Mol. Plant Pathol. 19, 90–103. doi: 10.1111/mpp.12501
Chinchilla, D., Zipfel, C., Robatzek, S., Kemmerling, B., Nürnberger, T., Jones, J. D. G., et al. (2007). A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448:497. doi: 10.1038/nature05999
Compan, I., and Touati, D. (1994). Anaerobic activation of arcA transcription in Escherichia coli: roles of Fnr and ArcA. Mol. Microbiol. 11, 955–964. doi: 10.1111/j.1365-2958.1994.tb00374.x
Danhorn, T., and Fuqua, C. (2007). Biofilm formation by plant-associated bacteria. Annu. Rev. Microbiol. 61, 401–422. doi: 10.1146/annurev.micro.61.080706.093316
Dellagi, A., Brisset, M. N., Paulin, J. P., and Expert, D. (1998). Dual role of desferrioxamine in Erwinia amylovora pathogenicity. Mol. Plant Microb. Interact. 11, 734–742.
D’Haeze, W., and Holsters, M. (2004). Surface polysaccharides enable bacteria to evade plant immunity. Trends Microbiol. 12, 555–561. doi: 10.1016/j.tim.2004.10.009
Fahnenstich, H., Scarpeci, T. E., Valle, E. M., Flügge, U. I., and Maurino, V. G. (2008). Generation of hydrogen peroxide in chloroplasts of Arabidopsis overexpressing glycolate oxidase as an inducible system to study oxidative stress. Plant Physiol. 148, 719–729. doi: 10.1104/pp.108.126789
Gama-Castro, S., Salgado, H., Peralta-Gil, M., Santos-Zavaleta, A., Muniz-Rascado, L., Solano-Lira, H., et al. (2010). RegulonDB version 7.0: transcriptional regulation of Escherichia coli K-12 integrated within genetic sensory response units (Gensor Units). Nucleic Acids Res. 39(Suppl._1), D98–D105. doi: 10.1093/nar/gkq1110
Gansert, D. (2003). Xylem sap flow as a major pathway for oxygen supply to the sapwood of birch (Betula pubescens Ehr.). Plant Cell Environ. 26, 1803–1814. doi: 10.1046/j.1365-3040.2003.01097.x
García-Nafría, J., Watson, J. F., and Greger, I. H. (2016). IVA cloning: a single-tube universal cloning system exploiting bacterial in vivo assembly. Sci. Rep. 6:27459. doi: 10.1038/srep27459
Geier, G., and Geider, K. (1993). Characterization and influence on virulence of the levansucrase gene from the fireblight pathogen Erwinia amylovora. Physiol. Mol. Plant Pathol. 42, 387–404. doi: 10.1006/pmpp.1993.1029
Gross, M., Geier, G., Rudolph, K., and Geider, K. (1992). Levan and levansucrase synthesized by the fireblight pathogen Erwinia amylovora. Physiol. Mol. Plant Pathol. 40, 371–381. doi: 10.1016/0885-5765(92)90029-u
Guo, M., Tian, F., Wamboldt, Y., and Alfano, J. R. (2009). The majority of the type III effector inventory of Pseudomonas syringae pv. tomato DC3000 can suppress plant immunity. Mol. Plant Microb. Interact. 22, 1069–1080. doi: 10.1094/MPMI-22-9-1069
Hassan, H. M., and Sun, H. C. (1992). Regulatory roles of Fnr, Fur, and Arc in expression of manganese-containing superoxide dismutase in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 89, 3217–3221. doi: 10.1073/pnas.89.8.3217
He, S. Y. (1998). Type III protein secretion systems in plant and animal pathogenic bacteria. Annu. Rev. Phytopathol. 36, 363–392. doi: 10.1146/annurev.phyto.36.1.363
Heese, A., Hann, D. R., Gimenez-Ibanez, S., Jones, A. M. E., He, K., Li, J., et al. (2007). The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc. Natl. Acad. Sci. U.S.A. 104, 12217–12222. doi: 10.1073/pnas.0705306104
Huynh, T. V., Dahlbeck, D., and Staskawicz, B. J. (1989). Bacterial blight of soybean: regulation of a pathogen gene determining host cultivar specificity. Science 245, 1374–1377. doi: 10.1126/science.2781284
Iakimova, E. T., Sobiczewski, P., Michalczuk, L., Wêgrzynowicz-Lesiak, E., Mikiciñski, A., and Woltering, E. J. (2013). Morphological and biochemical characterization of Erwinia amylovora-induced hypersensitive cell death in apple leaves. Plant Physiol. Biochem. 63, 292–305. doi: 10.1016/j.plaphy.2012.12.006
Iuchi, S., Matsuda, Z., Fujiwara, T., and Lin, E. C. C. (1990). The arcB gene of Escherichia coli encodes a sensor-regulator protein for anaerobic repression of the arc modulon. Mol. Microbiol. 4, 715–727. doi: 10.1111/j.1365-2958.1990.tb00642.x
Iwase, T., Tajima, A., Sugimoto, S., Okuda, K., Hironaka, I., Kamata, Y., et al. (2013). A simple assay for measuring catalase activity: a visual approach. Sci. Rep. 3:3081. doi: 10.1038/srep03081
Jamir, Y., Guo, M., Oh, H. S., Petnicki-Ocwieja, T., Chen, S., Tang, X., et al. (2004). Identification of Pseudomonas syringae type III effectors that can suppress programmed cell death in plants and yeast. Plant J. 37, 554–565. doi: 10.1046/j.1365-313x.2003.01982.x
Junglee, S., Urban, L., Sallanon, H., and Lopez-Lauri, F. (2014). Optimized assay for hydrogen peroxide determination in plant tissue using potassium iodide. Am. J. Anal. Chem. 5:730. doi: 10.4236/ajac.2014.511081
Kamber, T., Buchmann, J. P., Pothier, J. F., Smits, T. H. M., Wicker, T., and Duffy, B. (2016). Fire blight disease reactome: RNA-seq transcriptional profile of apple host plant defense responses to -pathogen infection. Sci. Rep. 6:21600. doi: 10.1038/srep21600
Kanehisa, M., and Goto, S. (2000). KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30.
Khan, M., Subramaniam, R., and Desveaux, D. (2016). Of guards, decoys, baits and traps: pathogen perception in plants by type III effector sensors. Curr. Opin. Microbiol. 29, 49–55. doi: 10.1016/j.mib.2015.10.006
Kim, S. J., Han, Y. H., Kim, I. H., and Kim, H. K. (1999). Involvement of ArcA and Fnr in expression of Escherichia coli thiol peroxidase gene. IUBMB Life 48, 215–218. doi: 10.1080/713803496
Koczan, J. M., Lenneman, B. M., McGrath, M. J., and Sundin, G. W. (2011). Cell surface attachment structures contribute to biofilm formation and xylem colonization of Erwinia amylovora. Appl. Environ. Microbiol. 77, 7031–7039. doi: 10.1128/AEM.05138-11
Koczan, J. M., McGrath, M. J., Zhao, Y., and Sundin, G. W. (2009). Contribution of Erwinia amylovora exopolysaccharides amylovoran and levan to biofilm formation: implications in pathogenicity. Phytopathology 99, 1237–1244. doi: 10.1094/PHYTO-99-11-1237
Krüger, J., and Rehmsmeier, M. (2006). RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 34(Suppl._2), W451–W454.
Kunze, G., Zipfel, C., Robatzek, S., Niehaus, K., Boller, T., and Felix, G. (2004). The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. Plant Cell 16, 3496–3507. doi: 10.1105/tpc.104.026765
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Meth. 9:357. doi: 10.1038/nmeth.1923
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2- ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Mandin, P., and Gottesman, S. (2010). Integrating anaerobic/aerobic sensing and the general stress response through the ArcZ small RNA. EMBO J. 29, 3094–3107. doi: 10.1038/emboj.2010.179
Melotto, M., Underwood, W., Koczan, J., Nomura, K., and He, S. Y. (2006). Plant stomata function in innate immunity against bacterial invasion. Cell 126, 969–980. doi: 10.1016/j.cell.2006.06.054
Mettert, E. L., and Kiley, P. J. (2007). Contributions of [4Fe-4S]-FNR and integration host factor to fnr transcriptional regulation. J. Bacteriol. 189, 3036–3043. doi: 10.1128/jb.00052-07
Miller, W. G., Leveau, J. H. J., and Lindow, S. E. (2000). Improved gfp and inaZ broad-host-range promoter-probe vectors. Mol. Plant Microb. Interact. 13, 1243–1250.
Miya, A., Albert, P., Shinya, T., Desaki, Y., Ichimura, K., Shirasu, K., et al. (2007). CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 104, 19613–19618. doi: 10.1073/pnas.0705147104
Montillet, J. L., Chamnongpol, S., Rustérucci, C., Dat, J., Van De Cotte, B., Agnel, J. P., et al. (2005). Fatty acid hydroperoxides and H2O2 in the execution of hypersensitive cell death in tobacco leaves. Plant Physiol. 138, 1516–1526. doi: 10.1104/pp.105.059907
Morales, E. H., Collao, B., Desai, P. T., Calderón, I. L., Gil, F., Luraschi, R., et al. (2013). Probing the ArcA regulon under aerobic/ROS conditions in Salmonella enterica serovar Typhimurium. BMC Genom. 14:626. doi: 10.1186/1471-2164-14-626
Niederhoffer, E. C., Naranjo, C. M., Bradley, K. L., and Fee, J. A. (1990). Control of Escherichia coli superoxide dismutase (sodA and sodB) genes by the ferric uptake regulation (fur) locus. J. Bacteriol. 172, 1930–1938. doi: 10.1128/jb.172.4.1930-1938.1990
Nimtz, M., Mort, A., Domke, T., Wray, V., Zhang, Y., Qiu, F., et al. (1996). Structure of amylovoran, the capsular exopolysaccharide from the fire blight pathogen Erwinia amylovora. Carbohydr. Res. 287, 59–76. doi: 10.1016/0008-6215(96)00070-5
Papenfort, K., Said, N., Welsink, T., Lucchini, S., Hinton, J. C. D., and Vogel, J. (2009). Specific and pleiotropic patterns of mRNA regulation by ArcZ, a conserved, Hfq-dependent small RNA. Mol. Microbiol. 74, 139–158. doi: 10.1111/j.1365-2958.2009.06857.x
Park, D. M., Akhtar, M. S., Ansari, A. Z., Landick, R., and Kiley, P. J. (2013). The bacterial response regulator ArcA uses a diverse binding site architecture to regulate carbon oxidation globally. PLoS Genet. 9:e1003839. doi: 10.1371/journal.pgen.1003839
Raymundo, A. K., and Ries, S. M. (1980). Motility of Erwinia amylovora. Phytopathology 70, 1062–1065.
Rivas, R., Vizcaino, N., Buey, R. M., Mateos, P. F., Martinez-Molina, E., and Velazquez, E. (2001). An effective, rapid and simple method for total RNA extraction from bacteria and yeast. J. Microbiol. Meth. 47, 59–63. doi: 10.1016/s0167-7012(01)00292-5
Rojas, C. M., and Mysore, K. S. (2012). Glycolate oxidase is an alternative source for H2O2 production during plant defense responses and functions independently from NADPH oxidase. Plant Signal. Behav. 7, 752–755. doi: 10.4161/psb.20429
Rojas, C. M., Senthil-Kumar, M., Wang, K., Ryu, C. M., Kaundal, A., and Mysore, K. S. (2012). Glycolate oxidase modulates reactive oxygen species–mediated signal transduction during nonhost resistance in Nicotiana benthamiana and Arabidopsis. Plant Cell 24, 336–352. doi: 10.1105/tpc.111.093245
Santander, R. D., Figàs-Segura, A., and Biosca, E. G. (2018). Erwinia amylovora catalases KatA and KatG are virulence factors and delay the starvation-induced viable but non-culturable (VBNC) response. Mol. Plant Pathol. 19, 922–934. doi: 10.1111/mpp.12577
Sauer, E., Schmidt, S., and Weichenrieder, O. (2012). Small RNA binding to the lateral surface of Hfq hexamers and structural rearrangements upon mRNA target recognition. Proc. Natl. Acad. Sci. U.S.A. 109, 9396–9401. doi: 10.1073/pnas.1202521109
Schachterle, J. K., and Sundin, G. W. (2019). The Leucine-responsive regulatory protein Lrp participates in virulence regulation downstream of small RNA ArcZ in Erwinia amylovora. mBio 10:e00757-19. doi: 10.1128/mBio.00757-19
Schachterle, J. K., Zeng, Q., and Sundin, G. W. (2019). Three Hfq-dependent small RNA s regulate flagellar motility in the fire blight pathogen Erwinia amylovora. Mol. Microbiol. 111, 1476–1492. doi: 10.1111/mmi.14232
Sebaihia, M., Bocsanczy, A. M., Biehl, B. S., Quail, M. A., Perna, N. T., Glasner, J. D., et al. (2010). Complete genome sequence of the plant pathogen Erwinia amylovora strain ATCC 49946. J. Bacteriol. 192, 2020–2021. doi: 10.1128/JB.00022-10
Shade, A., McManus, P. S., and Handelsman, J. (2013). Unexpected diversity during community succession in the apple flower microbiome. mBio 4, e602–e612. doi: 10.1128/mBio.00602-12
Stojiljkovic, I., Bäumler, A. J., and Hantke, K. (1994). Fur regulon in gram-negative bacteria: identification and characterization of new iron-regulated Escherichia coli genes by a Fur titration assay. J. Mol. Biol. 236, 531–545. doi: 10.1006/jmbi.1994.1163
Sun, X., Zhulin, I., and Wartell, R. M. (2002). Predicted structure and phyletic distribution of the RNA-binding protein Hfq. Nucleic Acids Res. 30, 3662–3671. doi: 10.1093/nar/gkf508
Thilmony, R., Underwood, W., and He, S. Y. (2006). Genome-wide transcriptional analysis of the Arabidopsis thaliana interaction with the plant pathogen Pseudomonas syringae pv. tomato DC3000 and the human pathogen Escherichia coli O157: H7. Plant J. 46, 34–53.
Toruño, T. Y., Stergiopoulos, I., and Coaker, G. (2016). Plant-pathogen effectors: cellular probes interfering with plant defenses in spatial and temporal manners. Annu. Rev. Phytopathol. 54, 419–441. doi: 10.1146/annurev-phyto-080615-100204
Tyson, K. L., Bell, A. I., Cole, J. A., and Busby, S. J. W. (1993). Definition of nitrite and nitrate response elements at the anaerobically inducible Escherichia coli nirB promoter: interactions between FNR and NarL. Mol. Microbiol. 7, 151–157. doi: 10.1111/j.1365-2958.1993.tb01106.x
Urban, J. H., and Vogel, J. (2007). Translational control and target recognition by Escherichia coli small RNAs in vivo. Nucleic Acids Res. 35, 1018–1037. doi: 10.1093/nar/gkl1040
Venisse, J. S., Barny, M. A., Paulin, J. P., and Brisset, M. N. (2003). Involvement of three pathogenicity factors of Erwinia amylovora in the oxidative stress associated with compatible interaction in pear. FEBS Lett. 537, 198–202. doi: 10.1016/s0014-5793(03)00123-6
Venisse, J. S., Malnoy, M., Faize, M., Paulin, J. P., and Brisset, M. N. (2002). Modulation of defense responses of Malus spp. during compatible and incompatible interactions with Erwinia amylovora. Mol. Plant Microb. Interact. 15, 1204–1212. doi: 10.1094/mpmi.2002.15.12.1204
Vogel, J., and Luisi, B. F. (2011). Hfq and its constellation of RNA. Nat. Rev. Microbiol. 9:578. doi: 10.1038/nrmicro2615
Wei, Z. M., Laby, R. J., Zumoff, C. H., Bauer, D. W., He, S. Y., Collmer, A., et al. (1992). Harpin, elicitor of the hypersensitive response produced by the plant pathogen Erwinia amylovora. Science 257, 85–88. doi: 10.1126/science.1621099
Zeng, Q., McNally, R. R., and Sundin, G. W. (2013). Global small RNA chaperone Hfq and regulatory small RNAs control virulence in the fire blight pathogen Erwinia amylovora. J. Bacteriol. 195, 1706–1717. doi: 10.1128/jb.02056-12
Zeng, Q., and Sundin, G. W. (2014). Genome-wide identification of Hfq-regulated small RNAs in the fire blight pathogen Erwinia amylovora discovered small RNAs with virulence regulatory function. BMC Genom. 15:414. doi: 10.1186/1471-2164-15-414
Zhao, Y., Blumer, S. E., and Sundin, G. W. (2005). Identification of Erwinia amylovora genes induced during infection of immature pear tissue. J. Bacteriol. 187, 8088–8103. doi: 10.1128/jb.187.23.8088-8103.2005
Zhao, Y., He, S. Y., and Sundin, G. W. (2006). The Erwinia amylovora avrRpt2EA gene contributes to virulence on pear and AvrRpt2EA is recognized by Arabidopsis RPS2 when expressed in Pseudomonas syringae. Mol. Plant Microb. Interact. 19, 644–654. doi: 10.1094/mpmi-19-0644
Zhao, Y., Wang, D. P., Nakka, S., Sundin, G. W., and Korban, S. S. (2009). Systems level analysis of two-component signal transduction systems in Erwinia amylovora: role in virulence, regulation of amylovoran biosynthesis and swarming motility. BMC Genom. 10:245. doi: 10.1186/1471-2164-10-245
Keywords: sRNA, ArcZ, catalase, fire blight, Erwinia amylovora, peroxide
Citation: Schachterle JK, Onsay DM and Sundin GW (2019) Small RNA ArcZ Regulates Oxidative Stress Response Genes and Regulons in Erwinia amylovora. Front. Microbiol. 10:2775. doi: 10.3389/fmicb.2019.02775
Received: 31 July 2019; Accepted: 14 November 2019;
Published: 29 November 2019.
Edited by:
Catherine Ayn Brissette, University of North Dakota, United StatesReviewed by:
Tim McNellis, Pennsylvania State University (PSU), United StatesElena G. Biosca, University of Valencia, Spain
Copyright © 2019 Schachterle, Onsay and Sundin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: George W. Sundin, sundin@msu.edu
 Jeffrey K. Schachterle
Jeffrey K. Schachterle Daphne M. Onsay
Daphne M. Onsay George W. Sundin
George W. Sundin