- 1Institute of Postharvest and Food Sciences, Agricultural Research Organization, Volcani Center, Rishon LeZion, Israel
- 2Institute of Biochemistry, Food Science and Nutrition, The Robert H. Smith Faculty of Agriculture, Food and Environment, The Hebrew University of Jerusalem, Rehovot, Israel
- 3College of Food Science and Engineering, Gansu Agricultural University, Lanzhou, China
Pathogenic fungi must respond effectively to changes in environmental pH for successful host colonization, virulence and toxin production. Aspergillus carbonarius is a mycotoxigenic pathogen with the ability to colonize many plant hosts and secrete ochratoxin A (OTA). In this study, we characterized the functions and addressed the role of PacC-mediated pH signaling in A. carbonarius pathogenicity using designed pacC gene knockout mutant. ΔAcpacC mutant displayed an acidity-mimicking phenotype, which resulted in impaired fungal growth at neutral/alkaline pH, accompanied by reduced sporulation and conidial germination compared to the wild type (WT) strain. The ΔAcpacC mutant was unable to efficiently acidify the growth media as a direct result of diminished gluconic and citric acid production. Furthermore, loss of AcpacC resulted in a complete inhibition of OTA production at pH 7.0. Additionally, ΔAcpacC mutant exhibited attenuated virulence compared to the WT toward grapes and nectarine fruits. Reintroduction of pacC gene into ΔAcpacC mutant restored the WT phenotype. Our results demonstrate important roles of PacC of A. carbonarius in OTA biosynthesis and in pathogenicity by controlling transcription of genes important for fungal secondary metabolism and infection.
Introduction
In order to survive and proliferate under diverse conditions, fungal microorganisms must be able to sense and respond to rapidly changing environmental stresses (Prusky et al., 2013). Fungal pathogens are not only able to survive in a diverse range of environmental conditions, but also have evolved abilities to recognize, penetrate and attack their hosts, while responding to chemical and physical signals from the host. Ambient pH is one of these environmental conditions and acts as an important signal for fungal growth, development, secondary metabolism and host infection (Drori et al., 2003; Li et al., 2010; Vylkova et al., 2011; Barad et al., 2014, 2016). Aspergillus carbonarius is frequently responsible for post-harvest decay of various fresh fruits, including grapes, peaches, pears, citrus and nectarines (JECFA, 2001). In addition to its pathogenicity, A. carbonarius is also considered as the main producer of ochratoxin A (OTA) – a potent nephrotoxin which may exhibit carcinogenic, teratogenic and immunotoxic properties in animals and possibly in humans (IARC, 1993; Pfohl-Leszkowicz and Manderville, 2007). Our recent study demonstrated that ambient pH plays an important role in A. carbonarius pathogenicity and OTA biosynthesis (Maor et al., 2017). Secretion of gluconic acid (GLA) by A. carbonarius caused direct fruit tissue acidification and induced accumulation of OTA in colonized grapes. Previous findings indicated that acidification of the apple fruit host environment through secretion of organic acids enhanced maceration and colonization of the fruit by Penicillium expansum (Prusky et al., 2004; Barad et al., 2012). Barad et al. (2014) pointed out the importance of the acidification process driven by GLA production in the activation of patulin biosynthesis and its contribution to the enhanced pathogenicity of P. expansum in apples.
Ambient pH signaling in filamentous fungi was first discovered in Aspergillus nidulans; it’s mediated by transcription factor PacC and six Pal proteins (PalA, PalB, PalC, PalF, PalH, and PalI), which may regulate both acid- and alkaline-expressed genes in several fungal species (Caddick et al., 1986; Tilburn et al., 1995; Penalva et al., 2008; Ment et al., 2015). The external pH signal is transmitted by fungal signaling pathway from the extracellular environment to the nucleus, where it regulates the expressions of PacC dependent genes, which are involved in secondary metabolism, cell wall biosynthesis and fungal pathogenesis (Penalva and Arst, 2002; You et al., 2007; Luo et al., 2017). PacC has been previously shown to differentially regulate virulence as well as biosynthesis and secretion of secondary metabolites in several fungal species. Decreased fungal virulence as well as reduction in biosynthesis of secondary metabolites, as a result of pacC knockout, have been observed in several Aspergillus and Penicillium species (Suarez and Penalva, 1996; Keller et al., 1997; Bergh and Brakhage, 1998; Zhang et al., 2013; Chen et al., 2018; Wang et al., 2018). In contrast, disruption of pacC resulted in increased pathogenicity of Fusarium oxysporum (Caracuel et al., 2003) and higher trichothecene and fumonisin production in F. graminearum and Fusarium verticillioides, respectively, compared to the wild type strains (Flaherty et al., 2003; Merhej et al., 2011), suggesting that this transcription factor acts as a negative regulator of virulence and secondary metabolism in Fusarium spp.
It has been already reported that OTA biosynthesis is regulated by PacC in Aspergillus ochraceus (Wang et al., 2018). Although the growth of the ΔAopacC mutant and its ability to produce OTA were compromised to some degree, an increase in conidia formation has been observed compared to that of the wild type strain, suggesting that PacC in A. ochraceus positively regulates growth and OTA biosynthesis, but has a negative regulatory role in sporulation. In the present study we investigated the role of the pH regulatory factor PacC in virulence and OTA production in A. carbonarius, which is one of the most important mycotoxigenic pathogens. AcpacC gene deletion showed the significance of this transcription factor in germination, sporulation, mycelial growth, OTA biosynthesis and virulence in fruits. Our results suggest that AcPacC is a positive regulator of virulence in A. carbonarius by mediating expression of the glucose oxidase encoding gene (gox) and the genes encoding for cell wall degrading enzymes.
Materials and Methods
Fungal Strain and Growth Conditions
The wild type (WT) strain A. carbonarius NRRL 368 was obtained from USDA Agricultural Research Service Culture Collection (Northern Regional Research Laboratory, Peoria, IL, United States). The WT strain and mutants generated in this study were grown at 28°C and maintained on Potato Dextrose Agar (PDA) plates (BD, Franklin Lakes, NJ, United States). Conidia were harvested and adjusted using a haemocytometer to the indicated concentrations.
Gene Knockout and Complementation
Construction of the AcpacC gene replacement plasmid was achieved by PCR-amplifying genomic flanking regions using specific primer pairs that incorporated a single 2-deoxyuridine nucleoside near the 5′ ends (primers U-f1 × U-r1 for the promoter region and primers D-f1 × D-r1 for the terminator region). Both DNA fragments and the pre-digested pRFHU2 binary vector (Frandsen et al., 2008) were mixed together and were treated with the USER enzyme (New England Biolabs, Ipswich, MA, United States) to obtain the plasmid pRFHU2-AcpacC. An aliquot of the mixture was used directly in chemical transformation of high-efficiency Escherichia coli DH5α cells (New England Biolabs, Ipswich, MA, United States) without prior ligation. Kanamycin resistant transformants were screened by PCR for validation of proper fusion events. Then, the plasmid was introduced into electro-competent Agrobacterium tumefaciens AGL-1 cells.
A single colony of A. tumefaciens AGL-1 carrying plasmid pRFHU2-AcpacC was used to inoculate a starter culture and incubated for 24 h. Bacterial cells were centrifuged, washed with induction medium (10 mM K2HPO4, 10 mM KH2PO4, 2.5 mM NaCl, 2 mM MgSO4, 0.6 mM CaCl2, 9 μM FeSO4, 4 mM (NH4)2SO4, 10 mM glucose, 40 mM MES pH 5.3, 0.5% glycerol) and diluted in the same medium amended with 200 μM acetosyringone (IMAS) to an OD600 = 0.15. Cells were grown at 28°C and 200 rpm until they reached an OD600 = 0.75. Equal volumes of IMAS induced bacterial culture and conidial suspension of A. carbonarius (106 conidia/ml) were mixed and spread onto Whatman filter papers, which were placed on agar plates containing the co-cultivation medium (same as IMAS, but containing 5 mM instead of 10 mM of glucose). After co-cultivation at 28°C for 48 h, the membranes were transferred to PDA plates containing hygromycin B (100 μg/mL) as the selection agent for fungal transformants, and cefotaxime (200 μg/mL) to inhibit growth of A. tumefaciens cells. Hygromycin resistant colonies appeared after 3–4 days of incubation at 28°C. Disruption of AcpacC was confirmed by PCR analyses of the transformants.
A restriction free cloning method (van den Ent and Lowe, 2006) was employed in order to replace the hygromycin resistance gene hph of the pRFHU2 vector with the phleomycin resistance gene ble amplified from the pBC-Phleo plasmid (Silar, 1995) using primer pair RFC-f1 x RFC-r1. Proper substitution of the resistance genes was confirmed by DNA sequencing and the new plasmid was termed pRFPU2. For the construction of the complementation vector, two genomic fragments consisting of the entire AcpacC cassette (primer pair U-f1 × U-r2) and the gene’s terminator region (primer pair D-f1 × D-r1), were USER cloned to the pre-digested pRFPU2 vector to generate pRFPU2-AcpacC-c as described before. Conidia of the ΔAcpacC knockout strain were transformed using A. tumefaciens AGL-1 cells carrying the plasmid pRFPU2-AcpacC-c as described before. For selecting phleomycin resistant complementation transformants, phleomycin (50 μg/mL) was enough to prevent growth of untransformed conidia. Analysis of transformants for reintroduction of the endogenous AcpacC cassette was done by PCR. All primers used to create and confirm the mutant and complement strains are listed in Supplementary Table S1.
Physiological Analysis
Radial growth and sporulation (conidial production) were assessed on solid YES media (20 g bacto yeast extract, 150 g sucrose and 15 g bacto agar per liter) adjusted with 10 M HCl or 10 M NaOH to pH 4.0, 7.0 and 8.0, while conidial germination was evaluated in YES broth media (20 g bacto yeast extract and 150 g sucrose per liter) adjusted to pH 4.0 and pH 7.0.
For radial growth assessment, 90 mm agar plates were point inoculated with 102 conidia of either the WT, knockout or complement strain and incubated at 28°C. Growth was monitored by diameter measurements on a daily basis for 10 days using three replicate plates per strain.
For conidial production quantification, 55 mm agar plates containing 105 conidia of each strain were incubated at 28°C for 7 days. To accurately count conidia, two 1 cm plugs from each plate were homogenized in 3 ml water containing 0.01% Tween 20, diluted and counted with a haemocytometer. Conidial production was quantified starting by the 3rd day post-inoculation using three replicate plates per strain.
For germination evaluation, the conidia concentration of all strains was adjusted to 104 conidia/ml in the medium. 0.5 ml of each conidial suspension was distributed into three replicate wells of a 24-well sterile culture plate (SPL Life Sciences, Pocheon, South Korea). Time-course microscopy was carried out over 24 h at 28°C using a Nikon Eclipse Ti inverted microscope (Nikon, Tokyo, Japan) equipped with a ProScan motorized XY stage (Prior Scientific, Cambridge, United Kingdom) with a LAUDA ECO RE 415 temperature-controlled incubator (LAUDA-Brinkmann, Delran, NJ, United States). Images were captured at 1-h intervals, beginning 2 h post-incubation using an ANDOR zyla 5.5 MP sCMOS camera (Andor Technology, Belfast, Northern Ireland) and processed using the NIS elements AR 4.6 (64 bit) software package. Conidia were considered to have germinated when germ tubes arose from the swollen conidial base. Number of conidia germlings were counted for each strain and the percent of germinated conidia was plotted against time.
pH Measurements, Organic Acids and OTA Analysis
A 106 fungal conidia/ml solution (100 μl) was inoculated onto 55 mm petri dishes containing 10 ml of solid YES media adjusted to pH 4.0 or pH 7.0. The plates were incubated at 28°C in the dark for 2–13 days as needed for sample collection.
pH was measured directly in the agar cultures with a double pore slim electrode connected to a Sartorius PB-11 Basic Meter (Sartorius, Göttingen, Germany).
For assessment of organic acids production, five 1-cm diameter discs of agar were placed in 5 ml of sterilized water and crushed to homogeneity. A 1 ml aliquot of the solution was sampled in a 1.5 ml microcentrifuge tube and centrifuged for 10 min at 20,800 g. The supernatant was taken for GLA and citric acid analysis using test kits applying enzymatic methods for the specific measurement of total D-Gluconic acid and citric acid contents (Megazyme, Wicklow, Ireland) according to the manufacturer’s instructions.
To evaluate OTA levels, five 1-cm diameter discs of agar were added to 1.7 ml of HPLC grade methanol (Bio-Lab, Jerusalem, Israel) and crushed to homogeneity. OTA was extracted by shaking for 30 min at 150 RPM on an orbital shaking platform and centrifuged for 10 min at 20,800 g. The supernatant was filtered through a 0.22 μm PTFE syringe filter (Agela Technologies, Tianjin, China) and kept at −20°C prior to HPLC analysis. OTA was quantitatively analyzed by injection of 20 μl into a reverse phase UHPLC system (Waters ACQUITY Arc, FTN-R, Milford, MA, United States). The mobile phase consisted of acetonitrile:water:acetic acid (99:99:2, v/v/v) at 0.5 ml/min through a Kinetex 2.6 μm XB-C18 (100 × 2.1 mm) with a security guard column C18 (4 × 2 mm) (Phenomenex, Torrance, CA, United States). The column temperature was maintained at 30°C. The OTA peak was detected with a fluorescence detector (excitation at 330 nm and emission at 450 nm) and quantified by comparing with a calibration curve of the standard mycotoxin (Fermentek, Jerusalem, Israel).
Colonization and Pathogenicity Experiments
“Zani” seedless grapes and “Sun Snow” nectarines were obtained from a local supermarket. Fruits were subjected to surface sterilization using 1% sodium hypochlorite solution for 1 min, and immediately rinsed in sterile distilled water. A 10 μl conidial suspension containing 106 conidia/ml of either the WT, the ΔAcpacC mutant strain or the AcpacC-c complement strain was injected directly into the sterilized fruits at 2 mm depth. Following inoculation, the fruits were incubated in covered plastic containers at 28°C for 2–9 days as needed for symptom monitoring and sample collection, and the diameters of the rotten spots were recorded daily.
The pH of nectarine tissues was measured by inserting a double pore slim electrode directly into the tested area. To analyze GLA content in inoculated nectarine fruits, 1.7 gr of the macerated necrotic area were taken, 5 ml of sterilized water were added and the tissues were homogenized. A 1 ml aliquot of the solution was sampled in a 1.5 ml microcentrifuge tube, centrifuged for 10 min at 20,800 g, and the amounts of GLA produced were measured as described above.
For OTA analysis in colonized grapes and nectarines, 1.7 gr of the macerated necrotic area were taken, 1.7 ml of HPLC grade methanol (Bio-Lab, Jerusalem, Israel) were added and the tissues were homogenized. Then, OTA was quantitatively analyzed as described above.
RNA Extraction and qRT-PCR Analysis of Gene Transcription Profile
Mycelia from the in vitro experiments were harvested at the appropriate time, weighed, frozen in liquid nitrogen, lyophilized for 24 h and kept at −80°C until use. In colonized nectarines, mycelia containing exocarp (peel) was removed, frozen in liquid nitrogen and lyophilized prior to RNA extraction. Total RNA was extracted from 100 mg of lyophilized tissue of the selected samples using the Hybrid-R RNA isolation kit (GeneAll, Seoul, South Korea) according to the manufacturer’s protocol. The DNase and reverse-transcription reactions were performed on 1 μg of total RNA with the Maxima First-Strand cDNA Synthesis Kit (Thermo Scientific, Waltham, MA, United States) according to the manufacturer’s instructions. The cDNA samples were diluted 1:10 (v/v) with ultrapure water. The quantitative real-time PCR was performed using Fast SYBR green Master Mix (Applied Biosystems, Waltham, MA, United States) in a StepOnePlus Real-Time PCR System (Applied Biosystems, Waltham, MA, United States). The PCR conditions were as follows: 95°C for 20 s, followed by 40 cycles of 95°C for 3 s and 60°C for 20 s. The samples were normalized using β-tubulin as endogenous control and the relative expression levels were measured using the 2(–ΔΔCt) analysis method. Results were analyzed with StepOne software v2.3. Primer sequences used for qRT-PCR analysis are listed in Supplementary Table S2.
Statistical Analysis
Student’s t-test was performed when data was normally distributed and the sample variances were equal. For multiple comparisons, one-way ANOVA was performed when the equal variance test was passed. Significance was accepted at p < 0.05. All experiments described here are representative of at least three independent experiments with the same pattern of results.
Results and Discussion
Creation and Validation of pacC Deletion and Complementation Strains of A. carbonarius
In order to explore the functional roles of PacC in the physiology and pathogenicity of A. carbonarius, AcpacC deletion and complementation strains were generated. For AcpacC deletion, a targeted gene deletion strategy was employed using A. tumefaciens mediated transformation of A. carbonarius NRRL 368 WT strain (Supplementary Figure S1). Gene replacement plasmid, pRFHU2-AcpacC, was obtained by a USER Friendly cloning system (Frandsen et al., 2008). Co-cultivation of A. tumefaciens cells carrying pRFHU2-AcpacC with the conidia of A. carbonarius led to the appearance of hygromycin B-resistant colonies approximately 4 days after transfer to the selective PDA plates. Disruption of AcpacC was confirmed by several PCR analyses for the introduction of the hygromycin resistance gene coding sequence, correct genomic placement of the 5′ and 3′ flanking sequences and the absence of the AcpacC sequence. With respect to the AcpacC complement strain, twelve single transformants were initially selected on PDA medium supplemented with phleomycin (50 μg/ml). The phleomycin resistant strains were diagnosed by PCR to confirm the integration of the WT allele in the ΔAcpacC strain using the same set of primers that amplify the AcpacC ORF. As expected, the AcpacC-c strain revealed the expected band of 472 bp (Supplementary Figure S2). One of the correct AcpacC-c strains was used for the following experiments.
AcPacC Is Required for Fungal Growth, Conidial Formation and Germination
Physiological analysis revealed that compared to the WT strain the growth of the ΔAcpacC mutant was reduced on YES synthetic media under acidic condition at pH 4.0, significantly impaired at pH 7.0 and completely inhibited under alkaline condition at pH 8.0 (Figures 1A,B). Due to the inability of the mutant strain to grow in alkaline conditions, the following experiments were performed under acidic and neutral pH conditions. Conidia production in ΔAcpacC mutant strain was severely inhibited at all examined time points under both acidic and neutral pH conditions (Figure 1C). Our study demonstrated that pH, as an environmental factor, may affect conidial germination of A. carbonarius. Germination rate of A. carbonarius was inhibited by 29% under ambient pH 7.0 (Figure 1D). It has been previously reported that germination of P. expansum conidia was inhibited significantly under ambient pH 2.0 and 8.0, probably through impairing protein synthesis and folding (Li et al., 2010). Conidial germination in the ΔAcpacC mutant strain was delayed compared to the WT at both pH conditions, although 100% germination of these conidia has been observed following 17 h of incubation (Figure 1D). By this time point, the ability of the mutant strain to develop hyphae in liquid culture at acidic pH was normal as was the ability to maintain hyphal growth (Figure 2A). In contrast, at pH 7.0 hyphal formation and growth of ΔAcpacC strain was significantly stunted and characterized by severe hyper-branching compared to the WT (Figure 2B). Normal growth, sporulation, conidial germination and hyphal formation were recovered in the AcpacC-c strain, indicating that AcPacC is required for morphogenetic development of A. carbonarius, especially in alkaline conditions.
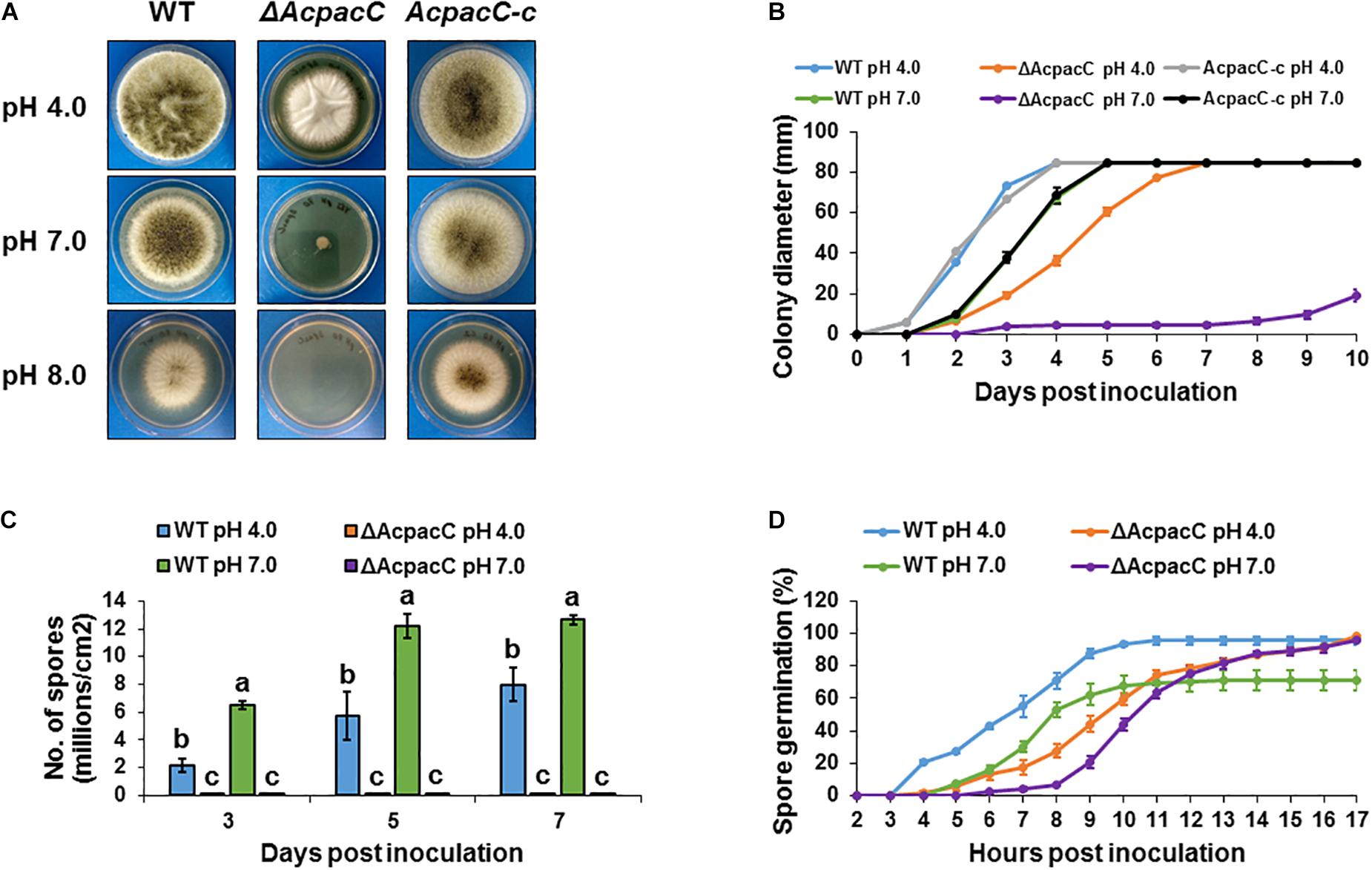
Figure 1. Physiological analyses of the WT and mutant strains of A. carbonarius at different pH conditions. (A) Growth phenotype and (B) radial growth of the WT, ΔAcpacC and AcpacC-c strains on solid YES media at 28°C under pH 4.0, 7.0, and 8.0. (C) Conidiation of the WT and ΔAcpacC strains on solid YES media at pH 4.0 and 7.0. (D) Germination rates in the WT and ΔAcpacC strains were assessed in static YES broth media at 28°C under pH 4.0 and 7.0. Error bars represent the standard error of the mean (SEM) across three independent replicates. Different letters above the columns indicate statistically significant differences at p < 0.05, as determined using the Tukey’s honest significant difference test.
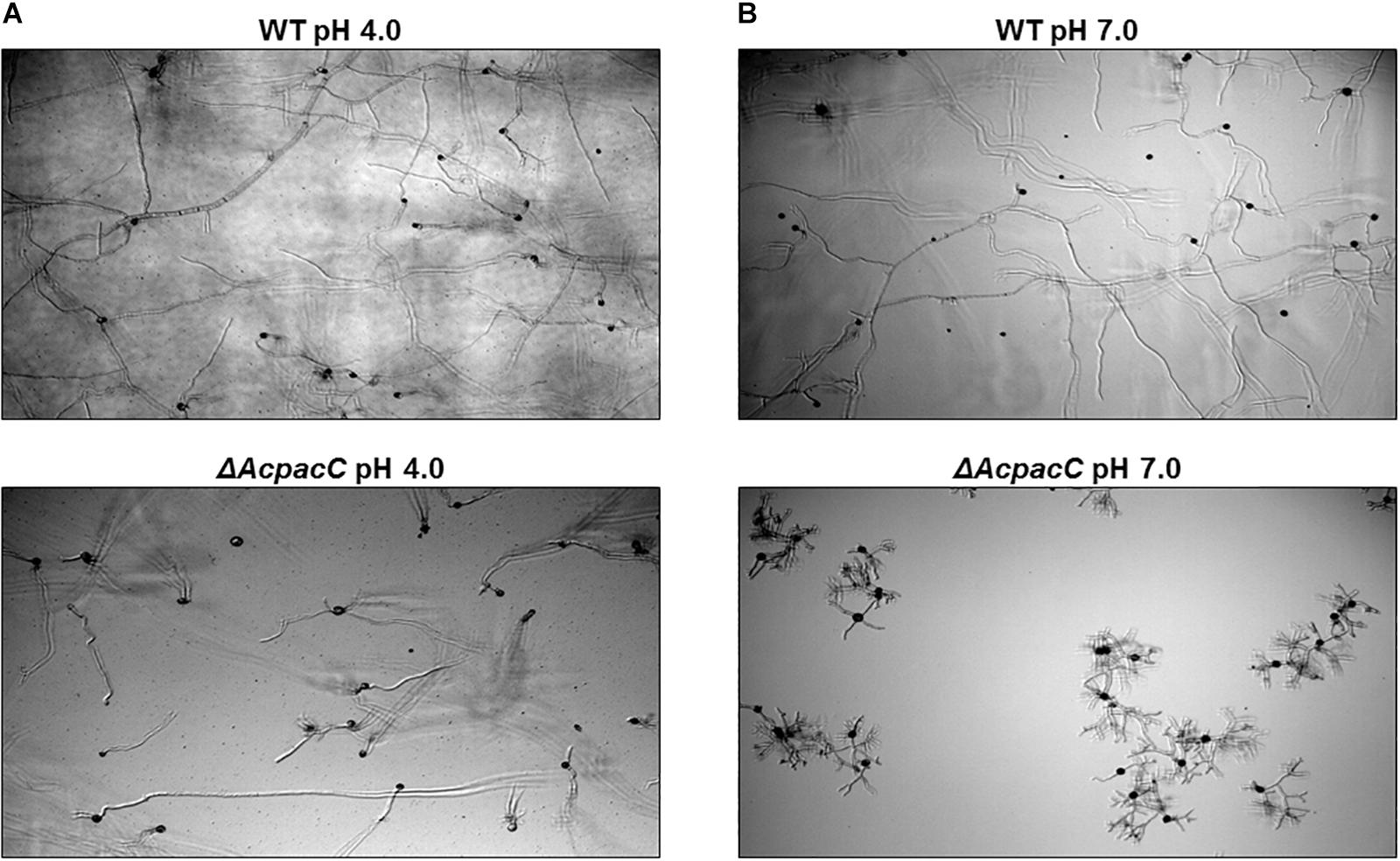
Figure 2. Microscopic observation of hyphal morphology of WT and ΔAcpacC strains. Images of time-course microscopy were captured 17 h following incubation of WT and ΔAcpacC conidia suspensions in YES broth media adjusted to pH 4.0 (A) and pH 7.0 (B).
Similarly to ΔAcpacC, growth of the pacC disrupted mutants in Sclerotinia sclerotiorum, F. oxysporum, Colletotrichum acutatum, F. graminearum and Penicillium digitatum was slightly reduced under acidic pH but severely impaired under alkaline conditions (Caracuel et al., 2003; Rollins, 2003; You et al., 2007; Merhej et al., 2011; Zhang et al., 2013). However, deletion of pacC showed poor growth phenotype under both acidic and alkaline pH conditions in P. expansum, Metarhizium robertsii and Ganoderma lucidum (Huang et al., 2015; Wu et al., 2016; Chen et al., 2018). Interestingly, A. ochraceus ΔpacC mutant strain had slightly impaired growth under alkaline conditions, but similar growth rate to the WT in acidic pH. On the contrary, unlike ΔAcpacC strain, an increase in conidia formation was observed in A. ochraceus ΔpacC mutant compared to that of the WT strain under all the pH conditions (Wang et al., 2018). Therefore, it is likely that PacC plays different roles in mycelial growth and sporulation of different fungal pathogens.
AcPacC Regulates Production of Organic Acids in A. carbonarius
Post-harvest fungal pathogens were reported to enhance their virulence by locally modulating the host’s ambient pH (Prusky et al., 2014). Previous studies have shown that Penicillium spp. (P. expansum, P. digitatum) and A. carbonarius acidify the ambient environments of deciduous fruits during decay development by secretion of significant amounts of organic acids, mainly citric and gluconic acids (Prusky et al., 2004; Barad et al., 2012, 2014; Maor et al., 2017). Gluconic acid accumulation by P. expansum and A. carbonarius is pH-dependent and is mainly regulated by glucose oxidase (GOX) that catalyzes the oxidation of glucose to gluconic acid. In the current study, a significant decrease in the formation of both citric and gluconic acids was observed in ΔAcpacC knockout mutant compared with the WT under acidic and neutral pH conditions at all tested time points (Figures 3A,B). The kinetics of the gene transcript level shows that Acgox gene expression was markedly down-regulated in ΔAcpacC in both acidic and neutral pH conditions, compared to the WT strain (Figure 3C), indicating that AcPacC directly regulates gluconic acid production by positive modulation of A. carbonarius glucose oxidase-encoding gene. In P. expansum, two pacC-RNAi mutants with downregulation of PacC (silenced by RNAi technology) resulted in a 63 and 27% reduction in gluconic acid production, respectively (Barad et al., 2014). This relatively moderate reduction could be attributed to residual PacC expressions in pacC-RNAi mutants. A recent study reported that GOX, which was identified by proteome analysis as an alkaline-expressed protein, is directly regulated by P. expansum transcription factor PacC (Chen et al., 2018). At acidic pH, the PacC protein is inactive and therefore unable to bind to the promoter sites of the target genes, however, under alkaline conditions PacC acts as an activator of alkaline-expressed genes and as a repressor of acid-expressed genes (Penalva et al., 2008).
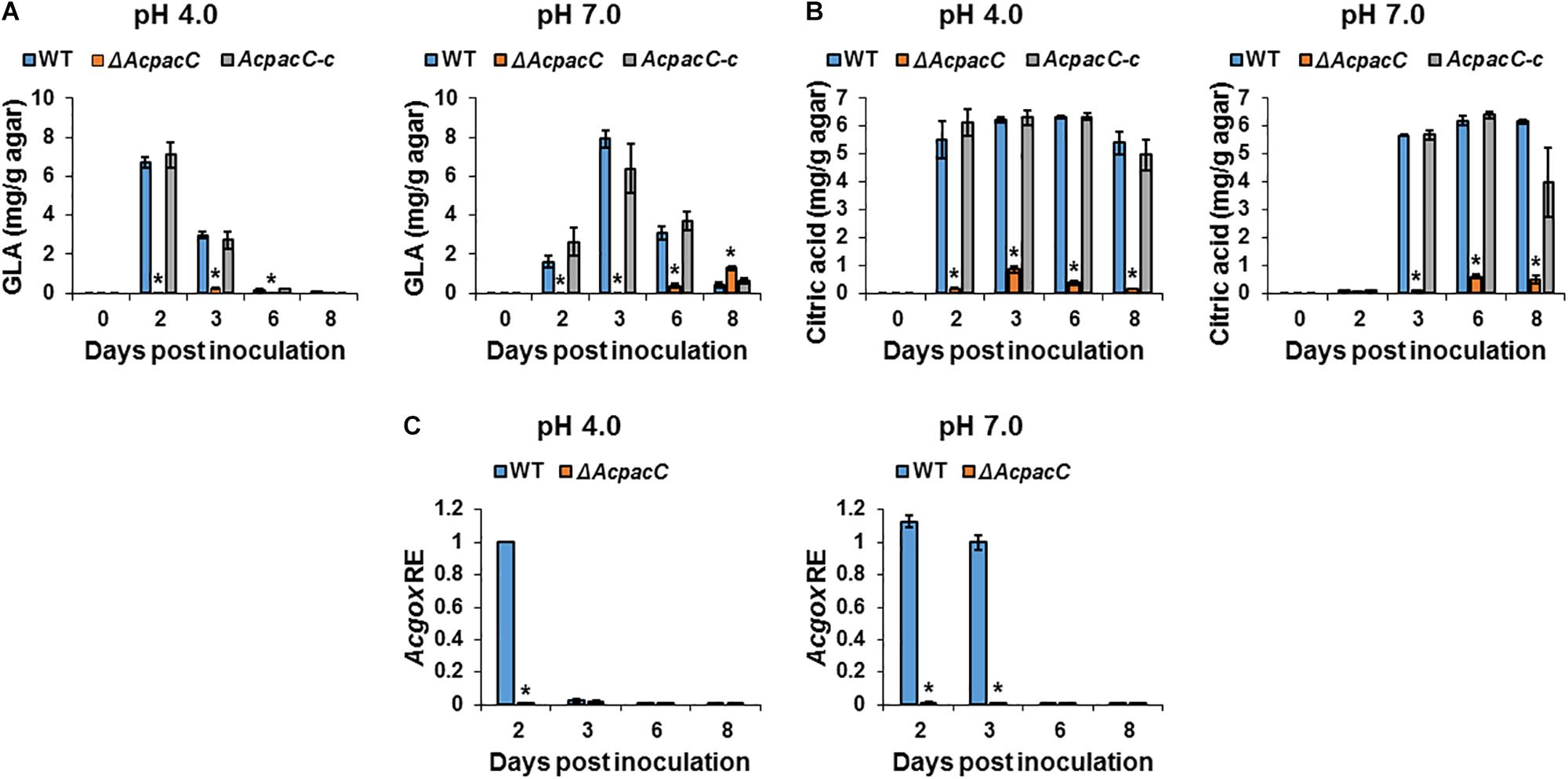
Figure 3. Effects of AcPacC on organic acids production in A. carbonarius. Gluconic acid (GLA) (A) and citric acid (B) accumulation by the WT, ΔAcpacC and AcpacC-c strains under different pH conditions. (C) Differential expression of the Acgox gene between WT and ΔAcpacC at pH 4.0 and 7.0. Average values of three replicates (± standard error) are reported. Experiments were repeated three times and results of a single representative experiment are shown. Asterisks denote significant differences between strains at p < 0.05.
OTA Biosynthesis in A. carbonarius Is Regulated by AcPacC
Among black aspergilli, A. carbonarius has shown a consistent ability to produce OTA and is the most probable source of this mycotoxin in a wide range of foods. Our previous study showed a clear pattern of pH modulation through secretion of organic acids by A. carbonarius, which acidify the ambient environment and induce OTA production in culture (Maor et al., 2017). In addition, Barad et al. (2014) demonstrated that down-regulation of gox gene in P. expansum (using RNAi technology) resulted in impaired ability to produce gluconic acid, which was accompanied by down-regulation in the relative expression of idh gene (encodes the isoepoxydon dehydrogenase enzyme, a key enzyme in the patulin biosynthesis pathway) and reduction in patulin accumulation. As shown in Figure 4A, OTA production by WT strain was significantly higher throughout the experiment under acidic condition, at pH 4.0, compared to the accumulation at pH 7.0. Deletion of AcpacC resulted in complete inhibition of OTA production at both pH 4.0 and 7.0 during the first 6 days after inoculation (Figure 4A). Nevertheless, under acidic condition a small amount of OTA was secreted by ΔAcpacC at day 8 of the experiment (Figure 4A). These results indicated that AcPacC is an important regulator in OTA biosynthesis in A. carbonarius under different pH conditions. Our findings suggest that not only the organic acids production could influence the accumulation of OTA, but also low pH itself might stimulate the mycotoxin biosynthesis. Moreover, we investigated the differential expression of all the five OTA biosynthetic cluster genes in ΔAcpacC and WT strains at ambient pH 4.0 and 7.0. As shown in Figure 4B, expressions of all five genes at day 4 of the experiment were down-regulated in ΔAcpacC at acidic pH. Under this condition, the relative expression of bZIP transcription factor, halogenase (HAL) and polyketide synthase (PKS) encoding genes in ΔAcpacC was similar to that of the WT strain at day 7 post-inoculation (Figure 4B); apparently, this is reflected in OTA production by the mutant at day 8 of the experiment. Although ΔAcpacC lost the ability of OTA production under neutral condition, the expression levels of several genes in the mutant strain, encoding bZIP transcription factor, PKS or HAL, were either unaffected or upregulated at pH 7.0 compared to the WT strain (Figure 4B), suggesting that these genes might be regulated by other transcription factors. PacC may act either as a positive or negative regulator of secondary metabolites biosynthesis (Brakhage, 2013). Similar to our findings, PacC was found to serve as a positive regulator of penicillin synthesis in A. nidulans and patulin biosynthesis in P. expansum (Bergh and Brakhage, 1998; Barad et al., 2014; Chen et al., 2018). On the contrary, PacC negatively regulates fumonisin biosynthesis in F. verticillioides and trichothecene biosynthesis in F. graminearum (Flaherty et al., 2003; Merhej et al., 2011). In A. ochraceus, another OTA producing pathogen, PacC played a positive role in regulating OTA biosynthesis, which was slightly impaired in AopacC loss-of-function mutant (Wang et al., 2018). Thus, PacC appears to function differently in regulating secondary metabolites in different fungal pathogens; yet, many unanswered questions remain on the mechanism of this regulation.
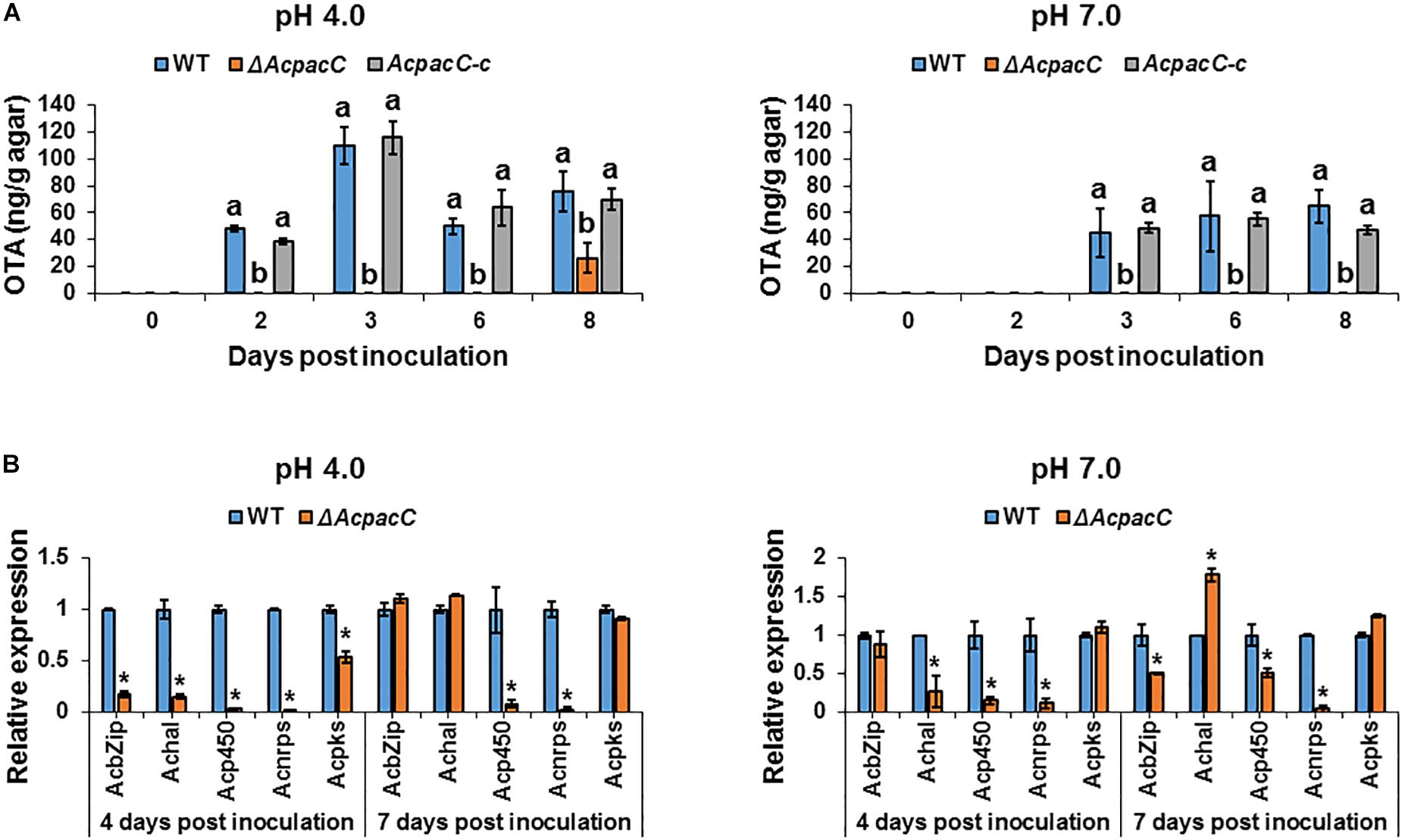
Figure 4. Effects of AcPacC on OTA biosynthesis in A. carbonarius at different pH conditions. (A) OTA production by the WT, ΔAcpacC and AcpacC-c strains at pH 4.0 and pH 7.0. (B) Relative expression of OTA cluster genes in WT and ΔAcpacC at days 4 and 7 post-inoculation. Relative expression was normalized using β-tubulin as an internal control. Average values of three replicates (± standard error) are presented. Experiments were repeated three times and results of a single representative experiment are shown. Different letters above the columns indicate statistically significant differences at p < 0.05, as determined using the Tukey’s honest significant difference test. Asterisks denote significant differences between strains at p < 0.05.
PacC Is Required for Pathogenicity in A. carbonarius and OTA Contamination of Deciduous Fruits
Colonization of “Sun Snow” nectarines and white “Zani” grape berries by ΔAcpacC strain showed a significant reduction in the rotten colonized area relative to that of the WT strain (Figures 5A,B and Supplementary Figures S3A,B). Four days after inoculation, ΔAcpacC strain showed an inhibition of the rotten area in nectarines and grape berries by up to 47 and 26%, respectively, compared with the WT strain (Figure 5B and Supplementary Figure S3B). The virulence was reverted by the complemented AcpacC-c strain (Figure 5A). The analysis of OTA accumulation in the infected nectarine tissue 5 days after inoculation revealed a three-fold reduction in OTA synthesis by the mutant strain, compared with the WT strain (Figure 5C); at the same time point, no OTA accumulation was detected by the ΔAcpacC in the inoculated grape berries (Supplementary Figure S3C). Nectarines infected with the ΔAcpacC mutant showed a 2–10 fold down-regulation of the transcript levels of all the five genes involved in OTA biosynthesis, compared with infections with the WT strain (Figure 5D), suggesting that AcPacC is essential for OTA production and probably directly involved in regulating transcription of the genes in OTA biosynthetic pathway. One would expect that reduction in OTA production by ΔAcpacC mutant may contribute to a reduction in virulence in this strain, however, in this regard, it should be noted that pathogenicity of the A. carbonarius pks mutant, which is unable to produce OTA, remained very similar to that of the WT (data not shown).
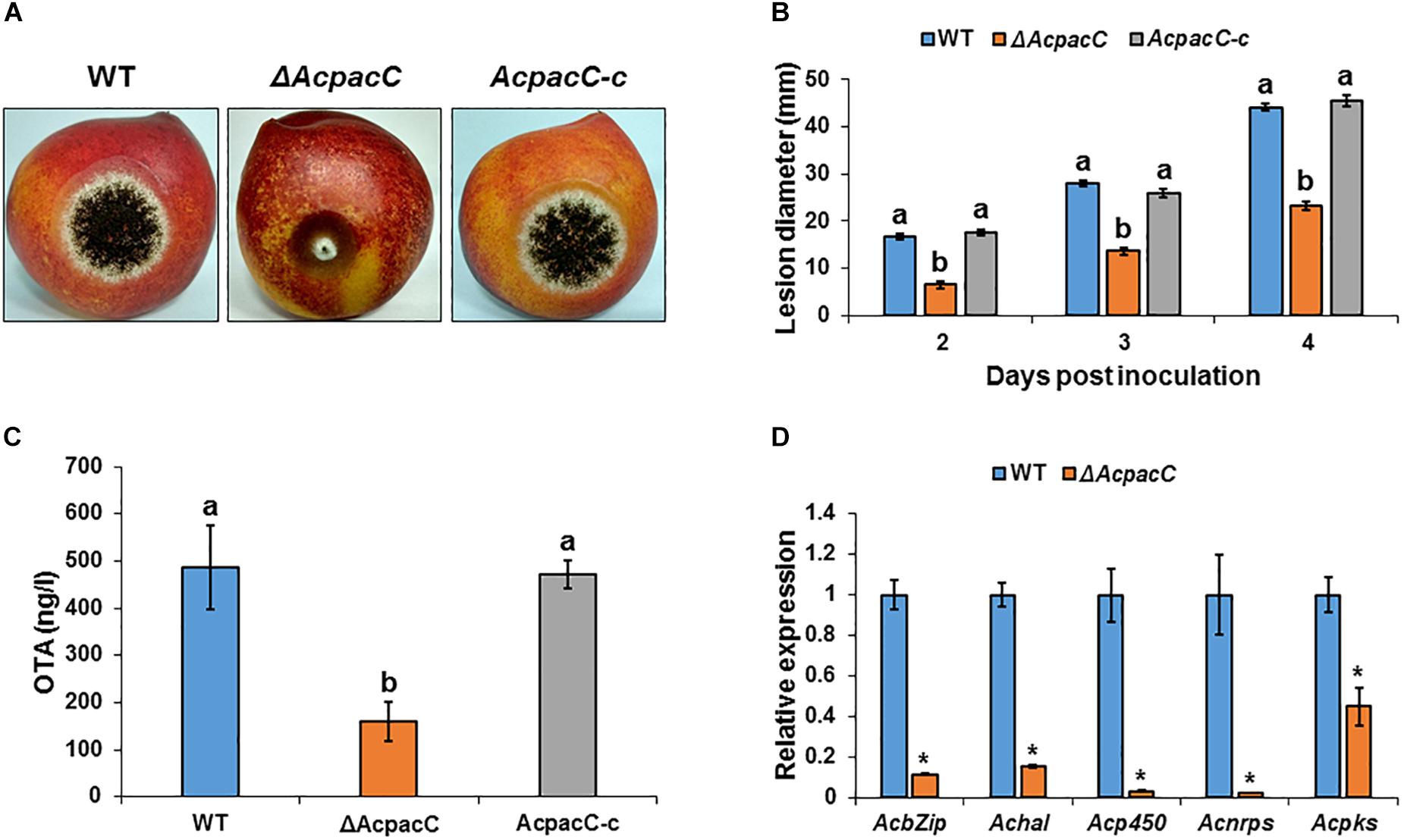
Figure 5. Effects of AcPacC on pathogenicity of A. carbonarius and OTA production in nectarines. (A) Disease symptoms on nectarine fruits inoculated with conidia of WT, ΔAcpacC and AcpacC-c strains at 3 days after inoculation. (B) Histogram showing the diameters of the rotten spots on infected nectarines. (C) OTA accumulation in infected nectarines, and (D) relative expression of OTA cluster genes in WT and ΔAcpacC strains. RNA was extracted from infected nectarines at day 4 post-inoculation. Relative expression was normalized using β-tubulin as an internal control. Error bars represent standard error of three independent biological replicates. Different letters above the columns indicate statistically significant differences at p < 0.05, as determined using the Tukey’s honest significant difference test. Asterisks denote significant differences between strains at p < 0.05.
To gain an understanding of the potential mechanism underlying the reduced pathogenicity of the ΔAcpacC strain, the mutant was assessed for several physiological characteristics that have previously been associated with virulence in this pathogen. It has been proposed that one of the factors that contribute to pathogenicity of A. carbonarius is its ability to reduce the pH of infected grape tissue through the production of gluconic acid (Maor et al., 2017). Indeed, 4 days after inoculation, colonization of nectarine tissue by A. carbonarius WT strain reduced pH from 4.3 in the healthy part of the fruit to 3.5 in the decayed tissue (Figure 6A). This further acidification of the rotten tissue was accompanied by an accumulation of 9 mg/g of gluconic acid (Figure 6B). In contrast, the ΔAcpacC strain showed a smaller reduction in pH and resulted in the accumulation of minimum amount of the gluconic acid (0.24 mg/g; Figures 6A,B). Gene expression analysis in the tissue inoculated with the ΔAcpacC mutant showed a 10-fold down-regulation of Acgox expression, which may explain poor gluconic acid formation in vivo (Figure 6C). Thus, our data indicate that AcPacC is required for A. carbonarius virulence in fruits, most likely by the regulation of the expression of the gox gene.
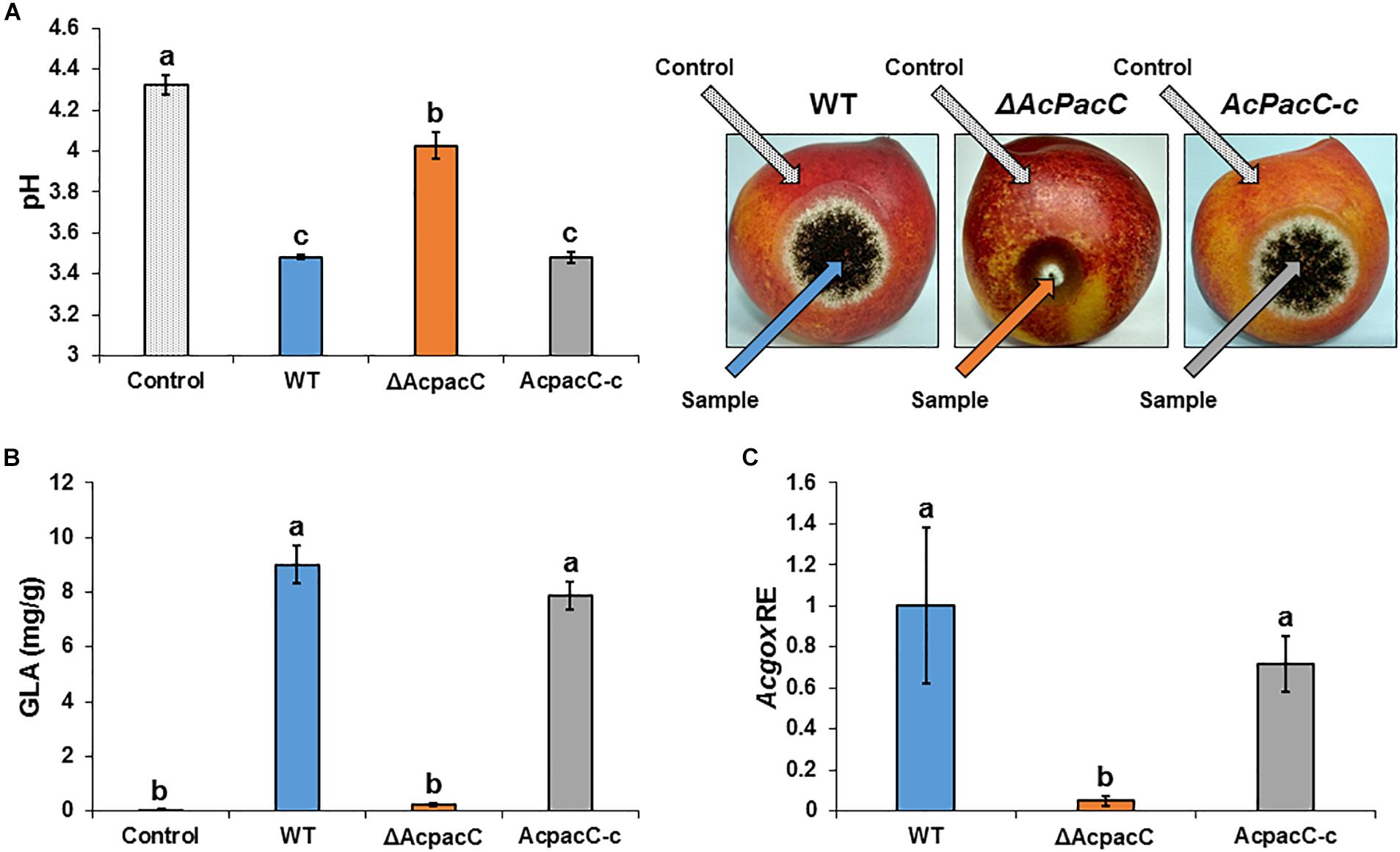
Figure 6. Effects of AcPacC on GLA production in colonized nectarines. pH of nectarine tissues (A), GLA accumulation (B), and Acgox relative expression (C) were measured in fruits infected with the WT, ΔAcpacC and AcpacC-c strains at day 4 post-inoculation. Error bars represent standard error of three independent biological replicates. Different letters above the columns indicate statistically significant differences at p < 0.05, as determined using the Tukey’s honest significant difference test.
Disruption of pacC resulted in reduced pathogenicity of P. expansum in pear and apple fruits through mediating a virulence factor glucose oxidase (Chen et al., 2018). In that study, glucose oxidase was identified as alkaline-expressed protein by proteome analysis and proved to be involved in the virulence of P. expansum. These results confirmed the findings of an earlier study of Barad et al. (2014), where P. expansum pacC-RNAi mutants reduced gluconic acid (which is regulated by gox expression) and patulin accumulation in apples and showed a 45% reduction in fungal pathogenicity, compared to the WT.
Furthermore, since Aspergillus enzymes are involved in degradation of plant cell wall polysaccharides, the expression levels of four genes encoding polygalacturonase, pectate lyase, cellulase and hemicellulase were also analyzed during fruit colonization, and all of them were down-regulated in the ΔAcpacC strain compared with the WT (Supplementary Figure S4). This down-regulation suggests possible involvement of AcPacC in the regulation of the cell wall-degrading enzymes during fruit colonization. Zhang et al. (2013) have reported similar findings, where PdPacC has been shown to be important for the pathogenicity of P. digitatum in citrus fruits via regulation of polygalacturonase and the pectin lyase genes. In Colletotrichum gloeosporioides, pac1 mutants showed reduction of pelb gene expression level, with consequent delayed pectate lyase secretion and dramatically reduced virulence in avocado fruits (Miyara et al., 2008). Overall, our results suggest that AcPacC may contribute to the pathogenesis of A. carbonarius through regulating different PacC-dependent genes or pathways involved in virulence.
Conclusion
In this study we demonstrated that disruption of the pH signaling transcription factor PacC significantly decreased the virulence of A. carbonarius on deciduous fruits. This phenotype is associated with an impairment in fungal growth, decreased accumulation of gluconic acid and reduced synthesis of pectolytic enzymes. We showed that glucose oxidase-encoding gene, which is essential for gluconic acid production and acidification during fruit colonization, was significantly down-regulated in the ΔAcpacC mutant, suggesting that gox is PacC-responsive gene. Recently we have provided evidence that deletion of gox gene in A. carbonarius led to a reduction in virulence toward nectarine and grape fruits (data not shown), further indicating that GOX is a virulence factor of A. carbonarius, and its expression is regulated by PacC. The deletion of AcpacC may also affect the pathogenesis of A. carbonarius through the down-regulation of the cell wall-degrading enzymes such as polygalacturonase, pectate lyase, cellulase and hemicellulose. It is also clear from the present data that PacC in A. carbonarius is a key factor for the biosynthesis of secondary metabolites, such as OTA. Additional work is needed in order to gain a genomic perspective of the function of PacC during pathogenesis. Therefore, comparison of the transcriptomes of the WT and the ΔAcpacC mutant during fruit infection would contribute for the better understanding of the molecular regulatory network in pathogenicity and OTA biosynthesis of A. carbonarius.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Author Contributions
OB, DP, YB, and ES conceived and designed the experiments. OB, UM, SS, and VZ performed the experiments. OB, SS, DP, YB, and ES analyzed the data. OB, DP, and ES wrote the manuscript. All authors read and approved the final manuscript.
Funding
This study was supported by a grant (No. 2432/18) from the Joint Israel Science Foundation (ISF) and the National Natural Science Foundation of China (NSFC) Research Program.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00210/full#supplementary-material
References
Barad, S., Espeso, E. A., Sherman, A., and Prusky, D. (2016). Ammonia activates pacC and patulin accumulation in an acidic environment during apple colonization by Penicillium expansum. Mol. Plant. Pathol. 17, 727–740. doi: 10.1111/mpp.12327
Barad, S., Horowitz, S. B., Kobiler, I., Sherman, A., and Prusky, D. (2014). Accumulation of the mycotoxin patulin in the presence of gluconic acid contributes to pathogenicity of Penicillium expansum. Mol. Plant Microbe Interact. 27, 66–77. doi: 10.1094/MPMI-05-13-0138-R
Barad, S., Horowitz, S. B., Moskovitch, O., Lichter, A., Sherman, A., and Prusky, D. (2012). A Penicillium expansum glucose oxidase-encoding gene, GOX2, is essential for gluconic acid production and acidification during colonization of deciduous fruit. Mol. Plant Microbe Interact. 25, 779–788. doi: 10.1094/MPMI-01-12-0002
Bergh, K. T., and Brakhage, A. A. (1998). Regulation of the Aspergillus nidulans penicillin biosynthesis gene acvA (pcbAB) by amino acids: implication for involvement of transcription factor PACC. Appl. Environ. Microbiol. 64, 843–849. doi: 10.1128/aem.64.3.843-849.1998
Brakhage, A. A. (2013). Regulation of fungal secondary metabolism. Nat. Rev. Microbiol. 11, 21–32. doi: 10.1038/nrmicro2916
Caddick, M. X., Brownlee, A. G., and Arst, H. N. (1986). Regulation of gene expression by pH of the growth medium in Aspergillus nidulans. Mol. Gen. Genet. 203, 346–353. doi: 10.1007/bf00333978
Caracuel, Z., Roncero, M. I. G., Espeso, E. A., Gonzalez-Verdejo, C. I., Garcia-Maceira, F. I., and Di Pietro, A. (2003). The pH signalling transcription factor PacC controls virulence in the plant pathogen Fusarium oxysporum. Mol. Microbiol. 48, 765–779. doi: 10.1046/j.1365-2958.2003.03465.x
Chen, Y., Li, B. Q., Xu, X. D., Zhang, Z. Q., and Tian, S. P. (2018). The pH-responsive PacC transcription factor plays pivotal roles in virulence and patulin biosynthesis in Penicillium expansum. Environ. Microbiol. 20, 4063–4078. doi: 10.1111/1462-2920.14453
Drori, N., Kramer-Haimovich, H., Rollins, J., Dinoor, A., Okon, Y., Pines, O., et al. (2003). External pH and nitrogen source affect secretion of pectate lyase by Colletotrichum gloeosporioides. Appl. Environ. Microbiol. 69, 3258–3262. doi: 10.1128/aem.69.6.3258-3262.2003
Flaherty, J. E., Pirttila, A. M., Bluhm, B. H., and Woloshuk, C. P. (2003). PAC1, a pH-regulatory gene from Fusarium verticillioides. Appl. Environ. Microbiol. 69, 5222–5227. doi: 10.1128/aem.69.9.5222-5227.2003
Frandsen, R. J., Andersson, J. A., Kristensen, M. B., and Giese, H. (2008). Efficient four fragment cloning for the construction of vectors for targeted gene replacement in filamentous fungi. BMC Mol. Biol. 9:70. doi: 10.1186/1471-2199-9-70
Huang, W., Shang, Y., Chen, P., Gao, Q., and Wang, C. (2015). MrpacC regulates sporulation, insect cuticle penetration and immune evasion in Metarhizium robertsii. Environ. Microbiol. 17, 994–1008. doi: 10.1111/1462-2920.12451
IARC (1993). Monographs on the Evaluation of Carcinogenic Risks to Humans. Ochratoxin A. In: Some Naturally Occurring Substances: Food Items and Constituents, Heterocyclic Aromatic Amines and Mycotoxins. Lyon: IARC.
JECFA (2001). “Fifty-sixth meeting of the joint FAO/WHO expert commiatee on food additives Ochratoxin A” in Safety evaluation of certain mycotoxins in food. WHO Additives Series 47 and FAO Food and Nutrition Paper. WHO: Geneva 74, 281–416.
Keller, N. P., Nesbitt, C., Sarr, B., Phillips, T. D., and Burow, G. B. (1997). pH regulation of sterigmatocystin and aflatoxin biosynthesis in Aspergillus spp. Phytopathology 87, 643–648. doi: 10.1094/phyto.1997.87.6.643
Li, B., Lai, T., Qin, G., and Tian, S. (2010). Ambient pH stress inhibits spore germination of Penicillium expansum by impairing protein synthesis and folding: a proteomic-based study. J. Proteome Res. 9, 298–307. doi: 10.1021/pr900622j
Luo, Z. B., Ren, H., Mousa, J. J., Rangel, D. E. N., Zhang, Y. J., Bruner, S. D., et al. (2017). The PacC transcription factor regulates secondary metabolite production and stress response, but has only minor effects on virulence in the insect pathogenic fungus Beauveria bassiana. Environ. Microbiol. 19, 788–802. doi: 10.1111/1462-2920.13648
Maor, U., Sadhasivam, S., Zakin, V., Prusky, D., and Sionov, E. (2017). The effect of ambient pH modulation on ochratoxin A accumulation by Aspergillus carbonarius. World Mycotoxin J. 10, 339–348. doi: 10.3920/wmj2017.2200
Ment, D., Alkan, N., Luria, N., Bi, F. C., Reuveni, E., Fluhr, R., et al. (2015). A Role of AREB in the regulation of PACC-dependent acid-expressed-genes and pathogenicity of Colletotrichum gloeosporioides. Mol. Plant Microbe Interact. 28, 154–166. doi: 10.1094/MPMI-09-14-0252-R
Merhej, J., Richard-Forget, F., and Barreau, C. (2011). The pH regulatory factor Pac1 regulates Tri gene expression and trichothecene production in Fusarium graminearum. Fungal Genet. Biol. 48, 275–284. doi: 10.1016/j.fgb.2010.11.008
Miyara, I., Shafran, H., Kramer Haimovich, H., Rollins, J., Sherman, A., and Prusky, D. (2008). Multi-factor regulation of pectate lyase secretion by Colletotrichum gloeosporioides pathogenic on avocado fruits. Mol. Plant Pathol. 9, 281–291. doi: 10.1111/j.1364-3703.2007.00462.x
Penalva, M. A., and Arst, H. N. (2002). Regulation of gene expression by ambient pH in filamentous fungi and yeasts. Microbiol. Mol. Biol. R. 66, 426–446. doi: 10.1128/mmbr.66.3.426-446.2002
Penalva, M. A., Tilburn, J., Bignell, E., and Arst, H. N. (2008). Ambient pH gene regulation in fungi: making connections. Trends Microbiol. 16, 291–300. doi: 10.1016/j.tim.2008.03.006
Pfohl-Leszkowicz, A., and Manderville, R. A. (2007). Ochratoxin A: an overview on toxicity and carcinogenicity in animals and humans. Mol. Nutr. Food Res. 51, 61–99. doi: 10.1002/mnfr.200600137
Prusky, D., Alkan, N., Mengiste, T., and Fluhr, R. (2013). Quiescent and necrotrophic lifestyle choice during postharvest disease development. Annu. Rev. Phytopathol. 51, 155–176. doi: 10.1146/annurev-phyto-082712-102349
Prusky, D., McEvoy, J. L., Saftner, R., Conway, W. S., and Jones, R. (2004). Relationship between host acidification and virulence of Penicillium spp. on apple and citrus fruit. Phytopathology 94, 44–51. doi: 10.1094/PHYTO.2004.94.1.44
Prusky, D., Barad, S., Luria, N., and Ment, D. (2014). “Post-harvest Pathology” in Plant Pathology in the 21st Century (Contributions to the 9th International Congress), eds D. Prusky, and M. Gullino, (Cham: Springer), 11–25.
Rollins, J. A. (2003). The Sclerotinia sclerotiorum pac1 gene is required for sclerotial development and virulence. Mol. Plant Microbe Interact. 16, 785–795. doi: 10.1094/mpmi.2003.16.9.785
Suarez, T., and Penalva, M. A. (1996). Characterization of a Penicillium chrysogenum gene encoding a PacC transcription factor and its binding sites in the divergent pcbAB-pcbC promoter of the penicillin biosynthetic cluster. Mol. Microbiol. 20, 529–540. doi: 10.1046/j.1365-2958.1996.5421065.x
Tilburn, J., Sarkar, S., Widdick, D. A., Espeso, E. A., Orejas, M., Mungroo, J., et al. (1995). The Aspergillus PacC zinc-finger transcription factor mediates regulation of both acid- and alkaline-expressed genes by ambient pH. EMBO J. 14, 779–790. doi: 10.1002/j.1460-2075.1995.tb07056.x
van den Ent, F., and Lowe, J. (2006). RF cloning: a restriction-free method for inserting target genes into plasmids. J. Biochem. Biophys. Methods 67, 67–74. doi: 10.1016/j.jbbm.2005.12.008
Vylkova, S., Carman, A. J., Danhof, H. A., Collette, J. R., Zhou, H., and Lorenz, M. C. (2011). The fungal pathogen Candida albicans autoinduces hyphal morphogenesis by raising extracellular pH. MBio 2, e00055–11. doi: 10.1128/mBio.00055-11
Wang, Y., Liu, F., Wang, L. Q., Wang, Q., Selvaraj, J. N., Zhao, Y. J., et al. (2018). pH-signaling transcription factor AopacC regulates ochratoxin A biosynthesis in Aspergillus ochraceus. J. Agric. Food Chem. 66, 4394–4401. doi: 10.1021/acs.jafc.8b00790
Wu, F. L., Zhang, G., Ren, A., Dang, Z. H., Shi, L., Jiang, A. L., et al. (2016). The pH-responsive transcription factor PacC regulates mycelial growth, fruiting body development, and ganoderic acid biosynthesis in Ganoderma lucidum. Mycologia 108, 1104–1113. doi: 10.3852/16-079
You, B. J., Choquer, M., and Chung, K. R. (2007). The Colletotrichum acutatum gene encoding a putative pH-responsive transcription regulator is a key virulence determinant during fungal pathogenesis on citrus. Mol. Plant Microbe Interact. 20, 1149–1160. doi: 10.1094/mpmi-20-9-1149
Keywords: Aspergillus carbonarius, PacC, pH regulation, OTA biosynthesis, post-harvest disease
Citation: Barda O, Maor U, Sadhasivam S, Bi Y, Zakin V, Prusky D and Sionov E (2020) The pH-Responsive Transcription Factor PacC Governs Pathogenicity and Ochratoxin A Biosynthesis in Aspergillus carbonarius. Front. Microbiol. 11:210. doi: 10.3389/fmicb.2020.00210
Received: 02 December 2019; Accepted: 29 January 2020;
Published: 13 February 2020.
Edited by:
István Pócsi, University of Debrecen, HungaryReviewed by:
Giancarlo Perrone, Italian National Research Council, ItalyFuguo Xing, Institute of Food Science and Technology (CAAS), China
Copyright © 2020 Barda, Maor, Sadhasivam, Bi, Zakin, Prusky and Sionov. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Edward Sionov, edwardsio@volcani.agri.gov.il
 Omer Barda
Omer Barda Uriel Maor1,2
Uriel Maor1,2 Sudharsan Sadhasivam
Sudharsan Sadhasivam Edward Sionov
Edward Sionov