- 1Biozentrum, Ludwig-Maximilians-Universität München, Munich, Germany
- 2Department of Agriculture, Food, Environment and Forestry (DAGRI), University of Florence, Florence, Italy
- 3Department of Biology, University of Florence, Florence, Italy
Fresh vegetables including baby greens, microgreens, and sprouts can host human pathogens without exhibiting any visible signs of spoilage. It is clear that the vast majority of foodborne disease outbreaks associated with vegetable produce are not simply a result of an oversight by a producer, as it was shown that zoonotic pathogens from Enterobacteriaceae can contaminate produce through various routes throughout the entire production cycle. In this context, phenotypic and genotypic signatures have been used since early ages in agriculture to obtain better produce, and can be used today as a strategy to reduce the risk of outbreaks through plant breeding. In this mini-review, we provide an updated view and perspectives on to what extent the selection of biological markers can be used to select safer cultivars of vegetable crops such as tomato (the most studied), leafy greens and cabbage. Once this knowledge will be better consolidated, these approaches should be integrated into the development of comprehensive farm-to-fork produce safety programs.
Human Pathogens in Crop Production Environment
Outbreaks linked to the consumption of fresh fruits, vegetables, and sprouts suggest that human pathogens can contaminate produce pre- and/or post-harvest (Bartz et al., 2014). Human enteric pathogens, such as non-typhoidal Salmonella and Shiga toxin-producing Escherichia coli can survive in the crop production environment, causing recurrent outbreaks (U.S. FDA, 2019). The majority of outbreaks of gastrointestinal illnesses have been associated with fruits, lettuce, alfalfa sprouts, spinach and tomatoes (Dewey-Mattia et al., 2018; CDC, 2019). Based on these observations, it is reasonable to hypothesize that enteric pathogens interact differently with various crops (although a number of other hypotheses can be offered and tested). Such pathogens also exhibit different contamination in greenhouses or in the field (Barak et al., 2011; Barak and Schroeder, 2012). These pathogens can survive in soil and water for extended periods of time and surface irrigation water improperly treated has been commonly identified as a source of contamination (Lapidot and Yaron, 2009; Moorman et al., 2014). Good Agricultural Practices and agronomical operations, as well as intervention technologies optimized to manage plant pathogens and safety have been put in place to minimize the risk of produce contamination and the spread of the pathogens through the supply chain (Bartz et al., 2014; Marvasi et al., 2015a; Murray et al., 2017). In addition to these tools, plant breeding has been recently suggested as another opportunity to enhance produce safety. This mini-review focuses on the feasibility of harnessing crop’s genetic potential to improve produce safety, with the emphasis on enteric pathogenic bacteria, primarily non-typhoidal Salmonella and Shiga toxin-producing E. coli, which have been the primary culprits of a large number of outbreaks linked to vegetable produce. In the first section of this mini-review we show the intra-species variability in the susceptibility to contamination and proliferation of enteric pathogens. Such variability among varieties and cultivars can be the result of specific genomic traits that can be transferred to the offspring. In the second section, we explore cases when potential biomarkers are identified and tentatively associated with the response to enteric pathogens (Figure 1).
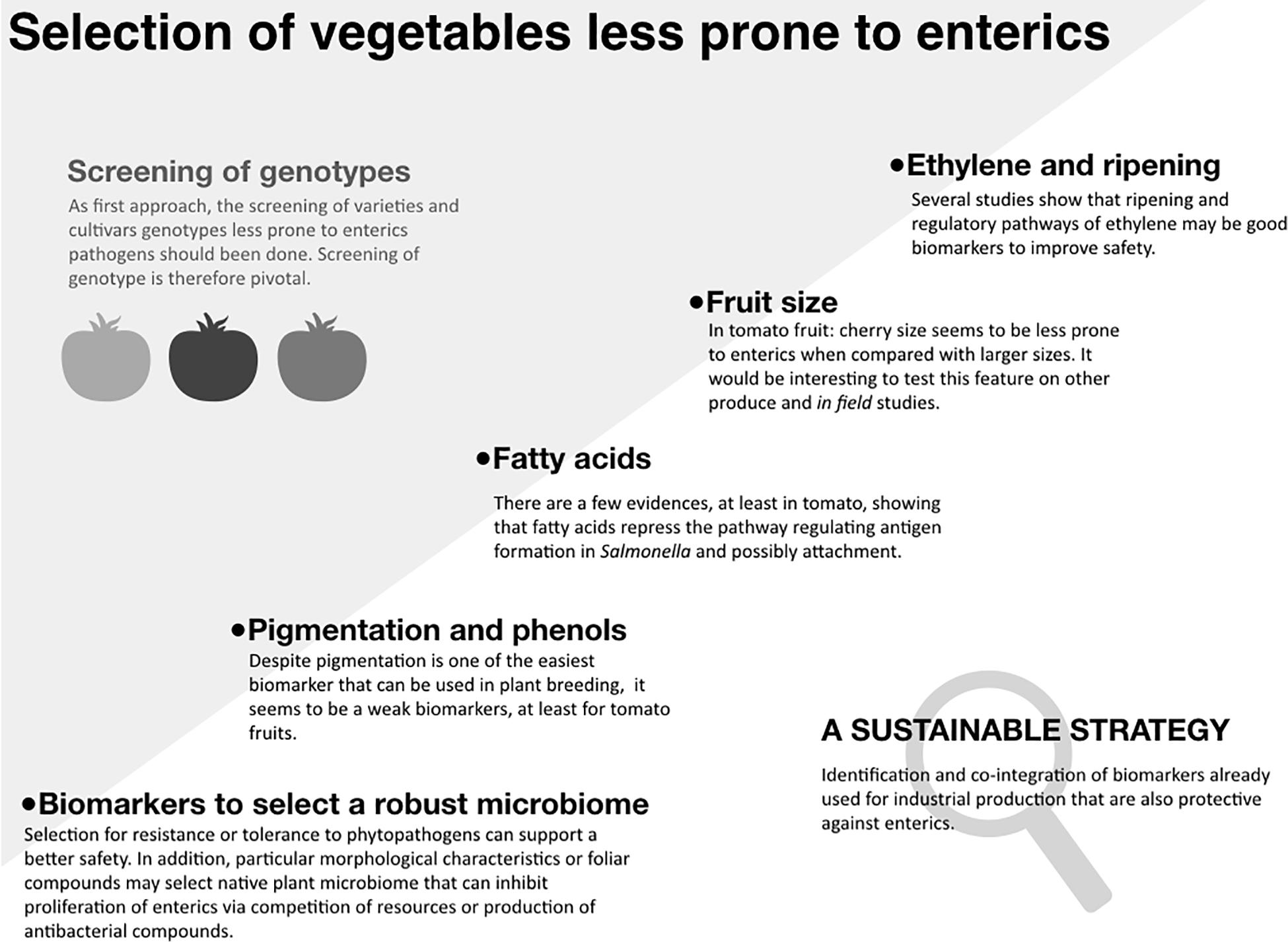
Figure 1. Selection of vegetables less prone to enteric pathogens. “Screening of genotypes”: The gray shadow panel shows that within same species there is a significant variability to the proliferation of enterics. Such different susceptibility can be the result of specific genetic traits that can be inherited by the offspring. The diagram shows the current advances in this direction, speculating which biomarkers may be the most promising. “Sustainable strategy” panel: To give an example, in tomato, some of the ripening related genes that are already used to increase shelf life could be used to develop vegetables less prone to enteric pathogens (Bai and Lindhout, 2007).
Cultivars of Vegetable Crops Differ in Their Susceptibility to Enteric Pathogens
Tomato
Tomato is by far the most studied species, probably due to the fact that it is the most important vegetable crop worldwide, and also often involved in enteric pathogens outbreaks. A greenhouse screening of 31 cultivars with characteristics that could conceivably affect how conducive tomatoes are to Salmonella proliferation has been carried out (Marvasi et al., 2014a). From this screening, tomato cultivar ‘Sun Gold’ and ‘Snow White’ were shown to be less prone to support Salmonella proliferation. This study also showed that cherry tomatoes (small size) were generally less conducive to proliferation of Salmonella when compared with tomato cultivars with a regular size (Marvasi et al., 2014a). A few studies were also carried out under field conditions. The post-harvest proliferation of Salmonella in the post-harvest tomato cultivars ‘Bonny Best,’ ‘Florida-47,’ and ‘Solar Fire’ grown under a variety of irrigation regimes showed that ‘Bonny Best’ was significantly less prone to Salmonella proliferation when compared with ‘Solar Fire’ but not when compared with ‘Florida-47’ (considering all maturity stages) (Marvasi et al., 2013a). Interestingly, in the greenhouse study, unripe ‘Bonny Best’ tomato fruits were less susceptible to Salmonella also when compared with ‘Florida-47’ (Marvasi et al., 2014a). This suggests that caution should be taken against directly translating the results of greenhouse studies to field conditions. The cultivar ‘Solar Fire’ was not significantly different from ‘Sebring’ when subjected to different fertilization regimes (Marvasi et al., 2013a, 2014b).
In another field study, 13 tomato cultivars selected based on a range of distinct fruit phenotypes, including morphology, pigmentation and resistance to phytopathogens were tested for susceptibility to Salmonella surface colonization (Han and Micallef, 2014). The study demonstrated that fruits and leaves of the same cultivar differed in their ability to suppress or support Salmonella growth arguing for the important role of tissue differentiation (Han and Micallef, 2014). Fruits of cultivar ‘Heinz-1706’ were the least colonized by Salmonella Newport, while the highest populations were observed on ‘Nyagous.’ By contrast, seedlings of the cultivar ‘Florida-91’ supported lowest populations of Salmonella Newport while the cherry variety ‘Virginia Sweets’ supported the highest. For seedling leaves infected with Salmonella Typhimurium the lowest susceptible was the cultivar ‘Nyagous’ and the highest were the cultivars ‘Heinz-1706’ and ‘Moneymaker’ (Han and Micallef, 2014). The comparative genomics of the pathogenic strains and in particular different serovars could help to determine why they behave differently during infection (e.g., Salmonella Typhimurium versus Newport) (Teplitski and de Moraes, 2018). Although tomato leaves are not edible, the information about Salmonella attachment and susceptibility to leaf colonization are relevant since Salmonella residing on leaves can be transmitted to fruits (Han and Micallef, 2014).
Cabbage
Response of cabbage cultivars to internalization or surface survival of Salmonella and Escherichia coli O157:H7 was studied by Erickson et al. (2019a;b). In a growth chamber study, internalized Salmonella was detected in cabbage within 24 h with prevalence ranging from 62% plants for the cultivar ‘Super Red 80’ to 92% for ‘Red Dynasty.’ The study showed that surface survival of both Salmonella and Escherichia coli O157:H7 on small cabbage plants over nine days was significantly affected by cultivars: pathogens survived the most on the cultivar “Farao” (Erickson et al., 2019b). In a field study which compared medium-sized cabbage heads, ‘Red Dinasty’ was more likely to be positive for Salmonella and E. coli O157:H7 (3.0 and 11.5 times, respectively) on day 5 post-inoculation when compared with the cultivar ‘Bravo F1’ (Erickson et al., 2019a).
Leafy Greens
When seeds of cilantro, parsley, radicchio, endive, lettuce and spinach were sown in the same pots containing contaminated soil, radicchio and endive had a significantly higher contamination index (CI) than lettuce (Barak et al., 2008). Since radicchio, endive, and lettuce all belong to the Asteraceae, these results revealed different response to enteric pathogens within the same Family, suggesting a possible effect of the genotype, at least under these greenhouse conditions (Barak et al., 2008). It must be mentioned that over the last decade, lettuce was instead associated with the highest number of outbreak investigations (U.S. FDA, 2019), but this could be due to the much higher acreage and consumption of lettuce versus other leafy crops (USDA - Statistics, 2019). The experiments described above were conducted in greenhouses and therefore may have missed some typical conditions of the field environment. Other field studies were carried out aiming to compare attachment of Salmonella on different lettuce cultivars. In this scenario prevalence of contamination was observed on leaves with the following order (from the less to the most): ‘Muir’ < ‘Gabriella’ < ‘Green Star’ = ‘New Red Fire’ < ‘Coastal Star’ (Erickson et al., 2019c).
Potential Biomarkers That Can Contribute to Produce Safety
We have previously seen that cultivars differ in their ability to support proliferation of enteric pathogens, therefore a number of potential biomarkers to be used in breeding programs can be obtained from studying such differences. Tomato is mainly used as a model plant for this purpose, but limited literature is also available on other raw eaten crops.
Flavonoids, Carotenoids, and Phenolics
Pigmentation, due to the presence of flavonoids and carotenoids, is the easiest biomarker that can be used in plant breeding programs. A recent study tested whether pigmentation can serve as an appropriate indicator of plant susceptibility to human pathogens. Thirty-one different tomato cultivars, including those that show different pigmentations have been screened. Despite color differences, pigmentation per se did not appear to account for the increased proliferation of Salmonella (Marvasi et al., 2014a). Follow-up studies demonstrated that tomato phenolics rutin, quercetin and kaempferol did not impact growth of Salmonella in rich laboratory media. Therefore, these three phenolics may not be useful as biomarkers for selection either (Marvasi et al., 2014a). The discovery of any association of pigmentation or other phenotypic characteristics (for example flavors) with enteric pathogens conductivity would be an easy phenotypic trait to be used. Less susceptible leafy greens or cabbage cultivars may be also potentially screened for pigments or phenolic compounds. To our knowledge no research has been carried out on these species.
Fatty Acids
The presence of fatty acids has been proposed as a possible indicator of susceptibility to enteric pathogens. Linolenic and linoleic acids are unsaturated fatty acids present in tomato and are precursors of hexanal which contributes to the fruity flavor (Jadhav et al., 1972). Linoleic acid seems to be involved in proliferation of Salmonella (Noel et al., 2010; Marvasi et al., 2013b). Linoleic acid is a polyunsaturated omega-6 fatty acid and its accumulation is progressively reduced in mature tomato fruits (Carrari et al., 2006). Linoleic acid has been demonstrated to play a role in the regulation of yihT gene which is involved in the synthesis of the O-antigen capsule in Salmonella. It is therefore reasonable to assume that capsule may be involved in attachment/persistence also outside the human host (Maruzani et al., 2019). Interestingly, deletion of yihT in S. enterica sv. Typhimurium reduced competitive fitness of the pathogen in immature but not in ripe tomatoes, regardless of their color at maturity. Linoleic acid is able to completely repress the yihT gene in Salmonella and the suppression of such gene could reduce the proliferation in immature tomatoes. Repression of the yihT reporter was stronger in tomato cultivars ‘Kumato,’ ‘Snow White,’ and ‘Mariana’ when compared with the variety ‘Bloody Butcher’ (Marvasi et al., 2013b). However, the response seemed to be generic to other fatty acids because decanoic and linolenic also affected the yihT gene reporter (Noel et al., 2010; Marvasi et al., 2013b). Other studies showed that linoleic acid was responsible for the upregulation of Salmonella fadH gene in green tomato fruit (Noel et al., 2010). The fadH gene encodes 2,4-dienoyl-CoA reductase, an iron-sulfur flavoenzyme required for metabolism of unsaturated fatty acids with double bonds at even carbon positions (Hubbard et al., 2003). More studies should be conducted to determine if fatty acids could serve as biomarkers due to their ability to repress key genes of enteric pathogens. As per phenolic, to our knowledge no additional literature is available to associate fatty acids and susceptibility to enteric pathogens in other crops, such as leafy greens or cabbage.
Ethylene
Ethylene is a plant hormone playing a key role in climacteric fruit ripening (Liu et al., 2015). Salmonella relies on a distinct set of metabolic and regulatory genes, which are differentially regulated in planta in response to the host genotype and during tomato fruit ripening (Noel et al., 2010). Because ethylene is involved in fruit ripening, it is clear that this metabolic pathway could be a good candidate to be further explored.
Fruits of three tomato cultivars, ‘Bonny Best,’ ‘Solar Fire,’ and ‘Florida-47,’ were harvested and infected with 100 CFU of Salmonella and incubated for a week at 22°C. In this experiment maturity stages of the fruits at the time of infection with Salmonella were assessed using the USDA tomato maturity chart. Maturity stages 5 and 6 at field harvest correspond to “ripe,” fruits that were harvested at 4 and 3 stages were considered “partially ripe,” and those that were harvested at 1 and 2 stages (and did not ripen beyond stage 5 during the experiment) were considered “unripe” (Marvasi et al., 2013a). The results showed that each ripening stage was significantly different in terms of Salmonella proliferation. The researchers found Salmonella counts to be at least 1 log higher in ripe tomatoes compared to the unripe tomatoes under the same growing conditions (Marvasi et al., 2013a). Salmonella ability to colonize bell peppers (cultivar ‘Aristotle’) at different maturity stages was also tested, showing that mature peppers were significantly more prone to support Salmonella when compared with the un-ripe ones (Marvasi et al., 2015b). The survival and/or proliferation of Escherichia coli O157:H7 and Salmonella on the surface of artificially bruised and unbruised tomatoes was tested at three ripeness stages (breaker, pink, and red) and two storage temperatures (10 and 20°C) (Tokarskyy et al., 2018). Tomatoes at the red ripeness stage showed a significant effect of bruising on Salmonella survival at both 10 and 20°C (Tokarskyy et al., 2018). Ripening is associated with tissue softening in tomato or bell peppers, therefore the proliferation of Salmonella during ripening may be associated with the ability of the bacterial cells to spread more easily into the softener tissue. In addition, a mature pericarp is also more prone to shallow wounds (Marvasi et al., 2014a). According with these results, it is reasonable to assume that testing a series of mutants in ethylene signaling could offer insights about the role of ethylene related genes and ripening during enteric pathogens proliferation. Results of the experiments with tomato mutants with specific defects in ethylene synthesis and perception suggested that ethylene signaling pathways mediated by RIN and NOR (MADS box and SPBP transcriptional factors, respectively) are more consequential that those that rely on the ethylene response sensor-like ethylene receptor NR (Fujisawa and Ito, 2013; Marvasi et al., 2014a). These mutants showed also changes in pigment production, being RIN and NOR not ripening (remaining green), while NR changing to dark red (Osorio et al., 2011; Liu et al., 2015). When post-harvest mature tomato fruits carrying RIN, NOR, and NR mutations were infected with Salmonella, reduced proliferation of Salmonella was observed when compared with the wild type parent ‘Ailsa Craig’ (Marvasi et al., 2014a). Some of these ripening related genes are already used in tomato breeding programs, therefore a systematic evaluation of the susceptibility of these cultivars to enteric pathogens can provide further information (Bai and Lindhout, 2007).
Another experiment to test the involvement of the ethylene cascade in the proliferations of enteric pathogens has been done by treating Medicago truncatula seedlings with the ethylene precursor 1-aminocyclopropane-1-carboxylic acid (ACC). The treatment strongly reduced endophytic populations of Salmonella (Iniguez et al., 2005). Are therefore ethylene mutants/variants good indicators? It is too early to advise, but the ethylene pathway may be a good candidate for further studies. In addition, the selection of plants that differently respond to the application of post-harvest ethylene can be interesting for produce safety (such as the ethylene receptor NR in tomato).
Breeding Plants to Support a Robust Microbiome
Microbiome has been shown to contribute to the proliferation of Salmonella in tomato fruits (Marvasi et al., 2013a; Devleesschauwer et al., 2017). Tomato fruits hosting the native microbiome were less prone to Salmonella infection when compared with surface cleaned fruits (Marvasi et al., 2013a; Devleesschauwer et al., 2017). Salmonella, for example, seems to harbor traits allowing for its interaction with plants and its microbiota (Brandl et al., 2013). It is tempting to speculate that the selection for leaf morphology, presence of exudates or waxes can stimulate the robust “healthy” microbiome and discourage establishment of enteric pathogens. There are no specific studies in this area, however, a few publications support the overall rationale. First, it is well established that leaf architecture, nutrients and secondary metabolites impact the composition of the plant epibiome (Singh et al., 2019). Second, recent studies demonstrated that certain members of the native plant microbiome can inhibit proliferation of Salmonella and pathogenic E. coli (Brandl et al., 2013).
For example, an antagonist epiphyte to Salmonella, Paenibacillus alvei TS-15, was isolated from different plants native to the Virginia Eastern Shore tomato-growing region (Allard et al., 2014). The strain TS-15 exhibited broad-range antimicrobial activity against both major foodborne pathogens and major bacterial phytopathogens of tomato. Survival of Salmonella after inoculation was measured for groups with and without the antagonist at days 0, 1, 2, and 3 and either day 5 on blossoms or day 6 for fruits and leaves. After P. alvei strain TS-15 was applied onto the fruits, leaves, and blossoms of tomato plants, the cell numbers of Salmonella Newport declined significantly compared with the controls. More than 90% of the plants tested had no detectable levels of Salmonella by day 5 for blossoms (Allard et al., 2014).
Bacterial phytopathogens also affect enteric pathogens proliferation in produce. Supermarket produce surveys showed that 60% of produce showing symptoms of soft rot also harbored presumptive Salmonella (Brandl et al., 2013). Salmonella cells inoculated in pre-existing aggregates of Pseudomonas syringae, Pseudomonas fluorescens, and Erwinia spp. had a greater probability of surviving dry conditions on lettuce and cilantro leaves than as solitary cells (Poza-Carrion et al., 2013). Within soft rots, Salmonella reached population densities 10- to 100-fold higher than within intact plants (Cox et al., 2013). The current view is that enteric pathogens appear to be successful secondary colonists in post-harvest produce, benefiting from the action of phytopathogens, e.g., suppression of plant defense and plant tissue damage (lesions, water soaking, and soft rots) (Brandl et al., 2013). Therefore, the selection of cultivars resistant to phytopathogens would be important not only to increase yield and quality but also to support produce safety (Brandl et al., 2013; Bettini et al., 2016).
Conclusion
The implementation of appropriate breeding practices could provide an additional important step toward produce safety. Currently the main picture is still in its infancy and it is difficult to provide directions on selecting safer cultivars or include them in risk assessments. Nevertheless, some data are useful: for example a study on tomato mutants showed that RIN gene, used to increase shelf life, has also showed some protection against enteric pathogens in ‘Ailsa Craig’ fruit. More studies must be done on different cultivars testing heterozygosity and alleles type (Marvasi et al., 2014a).
Excellent reviews also show that plants respond to Salmonella via defense pathways and that such genetic traits could also be used to achieve increase in produce safety (Teplitski et al., 2009; Brandl et al., 2013). At the same time, a better knowledge of the ecology of enteric pathogens in the environment, their interactions with plants and their persistence in wildlife are required to develop comprehensive solutions that build on the “One Health” concept.
Breeding practices have been used for a long time to reduce risk of plant pathogens. Can the same be done for pathogenic enteric pathogens? The question still remains extremely actual and open.
Author Contributions
TH and MM focused on the microbiological component. AL and AB on the agronomical part. All authors wrote the manuscript.
Funding
This work was supported by the BioMentoring Program, Faculty of Biology, LMU Munich, Germany 2018–2020.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Allard, S., Enurah, A., Strain, E., Millner, P., Rideout, S. L., Brown, E. W., et al. (2014). In situ evaluation of Paenibacillus alvei in reducing carriage of Salmonella enterica Serovar Newport on whole tomato plants. Appl. Environ. Microbiol. 80, 3842–3849. doi: 10.1128/AEM.00835-14
Bai, Y., and Lindhout, P. (2007). Domestication and breeding of tomatoes: what have we gained and what can we gain in the future? Ann. Bot. 100, 1085–1094. doi: 10.1093/aob/mcm150
Barak, J. D., Kramer, L. C., and Hao, L. Y. (2011). Colonization of tomato plants by Salmonella enterica is cultivar dependent, and type trichomes are preferred colonization sites. Appl. Environ. Microbiol. 77, 498–504. doi: 10.1128/AEM.01661-10
Barak, J. D., Liang, A., and Narm, K. E. (2008). Differential attachment to and subsequent contamination of agricultural crops by Salmonella enterica. Appl. Environ. Microbiol. 74, 5568–5570. doi: 10.1128/AEM.01077-08
Barak, J. D., and Schroeder, B. K. (2012). Interrelationships of food safety and plant pathology: the life cycle of human pathogens on plants. Annu. Rev. Phytopathol. 50, 241–266. doi: 10.1146/annurev-phyto-081211-172936
Bartz, J. A., Marvasi, M., and Teplitsi, M. (2014). “Salmonella and tomatoes,” in The Produce Contamination Problem: Causes and Solution, eds K. R. Matthews, G. M. Sapers, and C. P. Gerba (Cambridge, MA: Academic Press), 269–289.
Bettini, P. P., Marvasi, M., Fani, F., Lazzara, L., Cosi, E., Melani, L., et al. (2016). Agrobacterium rhizogenes rolB gene affects photosynthesis and chlorophyll content in transgenic tomato (Solanum lycopersicum L.) plants. J. Plant Physiol. 204, 27–35. doi: 10.1016/j.jplph.2016.07.010
Brandl, M. T., Cox, C. E., and Teplitski, M. (2013). Salmonella interactions with plants and their associated microbiota. Phytopathology 103, 316–325. doi: 10.1094/PHYTO-11-12-0295-RVW
Carrari, F., Baxter, C., Usadel, B., Urbanczyk-Wochniak, E., Zanor, M. I., Nunes-Nesi, A., et al. (2006). Integrated analysis of metabolite and transcript levels reveals the metabolic shifts that underlie tomato fruit development and highlight regulatory aspects of metabolic network behavior. Plant Physiol. 142, 1380–1396. doi: 10.1104/pp.106.088534
CDC (2019). Web site Centers for Disease Control. Available online at: https://www.cdc.gov/foodsafety/outbreaks/index.html (accessed December 20, 2019).
Cox, C. E., McClelland, M., and Teplitski, M. (2013). Consequences of disrupting Salmonella AI-2 signaling on interactions within soft rots. Phytopathology 103, 352–361. doi: 10.1094/PHYTO-09-12-0237-FI
Devleesschauwer, B., Marvasi, M., Giurcanu, M. C., Hochmuth, G. J., Speybroeck, N., Havelaar, A. H., et al. (2017). High relative humidity pre-harvest reduces post-harvest proliferation of Salmonella in tomatoes. Food Microbiol. 66, 55–63. doi: 10.1016/j.fm.2017.04.003
Dewey-Mattia, D., Manikonda, K., Hall, A. J., Wise, M. E., and Crowe, S. J. (2018). Surveillance for foodborne disease outbreaks – United States, 2009–2015. MMWR Surveill. Summ. 67:1. doi: 10.15585/mmwr.ss6710a1
Erickson, M. C., Liao, J. Y., Payton, A. S., Cook, P. W., Den Bakker, H. C., Bautista, J., et al. (2019a). Survival of Salmonella enterica and Escherichia coli O157:H7 sprayed onto the foliage of field-grown cabbage plants. J. Food Prot. 82, 479–485. doi: 10.4315/0362-028X.JFP-18-326
Erickson, M. C., Liao, J. Y., Payton, A. S., Cook, P. W., Den Bakker, H. C., Bautista, J., et al. (2019c). Pre-harvest internalization and surface survival of Salmonella and Escherichia coli O157:H7 sprayed onto different lettuce cultivars under field and growth chamber conditions. Int. J. Food Microbiol. 291, 197–204. doi: 10.1016/j.ijfoodmicro.2018.12.001
Erickson, M. C., Liao, J. Y., Payton, A. S., Cook, P. W., and Ortega, Y. R. (2019b). Survival and internalization of Salmonella and Escherichia coli O157:H7 sprayed onto different cabbage cultivars during cultivation in growth chambers. J. Sci. Food Agric. 99, 3530–3537. doi: 10.1002/jsfa.9573
Fujisawa, M., and Ito, Y. (2013). The regulatory mechanism of fruit ripening revealed by analyses of direct targets of the tomato MADS-box transcription factor RIPENING INHIBITOR. Plant Signal. Behav. 8:e24357. doi: 10.4161/psb.24357
Han, S., and Micallef, S. A. (2014). Salmonella Newport and Typhimurium colonization of fruit differs from leaves in various tomato cultivars. J. Food Prot. 77, 1844–1850. doi: 10.4315/0362-028X.JFP-13-562
Hubbard, P. A., Liang, X., Schulz, H., and Kim, J. J. P. (2003). The crystal structure and reaction mechanism of Escherichia coli 2,4-dienoyl-CoA reductase. J. Biol. Chem. 278, 37553–37560. doi: 10.1074/jbc.M304642200
Iniguez, A. L., Dong, Y., Carter, H. D., Ahmer, B. M. M., Stone, J. M., and Triplett, E. W. (2005). Regulation of enteric endophytic bacterial colonization by plant defenses. Mol. Plant Microbe Interact. 18, 169–178. doi: 10.1094/MPMI-18-0169
Jadhav, S., Singh, B., and Salunkhe, D. K. (1972). Metabolism of unsaturated fatty acids in tomato fruit: linoleic and linolenic acid as precursors of hexanal. Plant Cell Physiol. 13, 449–459. doi: 10.1093/oxfordjournals.pcp.a074758
Lapidot, A., and Yaron, S. (2009). Transfer of Salmonella enterica Serovar Typhimurium from contaminated irrigation water to parsley is dependent on curli and cellulose, the biofilm matrix components. J. Food Prot. 72, 618–623. doi: 10.4315/0362-028X-72.3.618
Liu, M., Pirrello, J., Chervin, C., Roustan, J. P., and Bouzayen, M. (2015). Ethylene control of fruit ripening: revisiting the complex network of transcriptional regulation. Plant Physiol. 169, 2380–2390. doi: 10.1104/pp.15.01361
Maruzani, R., Sutton, G., Nocerino, P., and Marvasi, M. (2019). Exopolymeric substances (EPS) from Salmonella enterica: polymers, proteins and their interactions with plants and abiotic surfaces. J. Microbiol. 57, 1–8. doi: 10.1007/s12275-019-8353-y
Marvasi, M., Cox, C. E., Xu, Y., Noel, J. T., Giovannoni, J. J., and Teplitski, M. (2013b). Differential regulation of Salmonella Typhimurium genes involved in O-antigen capsule production and their role in persistence within tomato fruit. Mol. Plant Microbe Interact. 26, 793–800. doi: 10.1094/MPMI-09-12-0208-R
Marvasi, M., George, A. S., Giurcanu, M., Hochmuth, G. J., Noel, J. T., Gause, E., et al. (2014b). Effects of nitrogen and potassium fertilization on the susceptibility of tomatoes to post-harvest proliferation of Salmonella enterica. Food Microbiol. 43, 20–27. doi: 10.1016/j.fm.2014.03.017
Marvasi, M., George, A. S., Giurcanu, M. C., Hochmuth, G. J., Noel, J. T., and Teplitski, M. (2015b). Effect of the irrigation regime on the susceptibility of pepper and tomato to post-harvest proliferation of Salmonella enterica. Food Microbiol. 46, 139–144. doi: 10.1016/j.fm.2014.07.014
Marvasi, M., Hochmuth, G., and Teplitski, M. (2015a). The role of crop production practices and weather conditions in microbiological safety of tomatoes and peppers. Food Microbiol. 46, 139–144. doi: 10.13140/RG.2.2.20981.01762
Marvasi, M., Hochmuth, G. J., Giurcanu, M. C., George, A. S., Noel, J. T., Bartz, J., et al. (2013a). Factors that affect proliferation of Salmonella in tomatoes post-harvest: the roles of seasonal effects, irrigation regime, crop and pathogen genotype. PLoS One 8:e80871. doi: 10.1371/journal.pone.0080871
Marvasi, M., Noel, J. T., George, A. S., Farias, M. A., Jenkins, K. T., Hochmuth, G., et al. (2014a). Ethylene signalling affects susceptibility of tomatoes to Salmonella. Microb. Biotechnol. 7, 545–555. doi: 10.1111/1751-7915.12130
Moorman, G. W., Gevens, A. J., Granke, L. H., Hausbeck, M. K., Hendricks, K., Roberts, P. D., et al. (2014). “Sources and distribution systems of irrigation water and their potential risks for crop health,” in Biology, Detection, and Management of Plant Pathogens in Irrigation Water, eds C. Hong, G. W. Moorman, W. Wohanka, and C. Buttner (St. Paul, MN: APS Press), 3–12. doi: 10.1094/9780890544914.002
Murray, K., Wu, F., Shi, J., Jun Xue, S., and Warriner, K. (2017). Challenges in the microbiological food safety of fresh produce: limitations of post-harvest washing and the need for alternative interventions. Food Qual. Saf. 1, 289–301. doi: 10.1093/fqsafe/fyx027
Noel, J. T., Arrach, N., Alagely, A., Mcclelland, M., and Teplitski, M. (2010). Specific responses of Salmonella enterica to tomato varieties and fruit ripeness identified by In Vivo expression technology. PLoS One 5:e12406. doi: 10.1371/journal.pone.0012406
Osorio, S., Alba, R., Damasceno, C. M. B., Lopez-Casado, G., Lohse, M., Zanor, M. I., et al. (2011). Systems biology of tomato fruit development: combined transcript, protein, and metabolite analysis of tomato transcription factor (nor, rin) and ethylene receptor (Nr) mutants reveals novel regulatory interactions. Plant Physiol. 157, 405–425. doi: 10.1104/pp.111.175463
Poza-Carrion, C., Suslow, T., and Lindow, S. (2013). Resident bacteria on leaves enhance survival of immigrant cells of Salmonella enterica. Phytopathology 103, 341–351. doi: 10.1094/PHYTO-09-12-0221-FI
Singh, P., Santoni, S., Weber, A., This, P., and Péros, J. P. (2019). Understanding the phyllosphere microbiome assemblage in grape species (Vitaceae) with amplicon sequence data structures. Sci. Rep. 9:14294. doi: 10.1038/s41598-019-50839-0
Teplitski, M., Barak, J. D., and Schneider, K. R. (2009). Human enteric pathogens in produce: un-answered ecological questions with direct implications for food safety. Curr. Opin. Biotechnol. 20, 166–171. doi: 10.1016/j.copbio.2009.03.002
Teplitski, M., and de Moraes, M. (2018). Of mice and men. and plants: comparative genomics of the dual lifestyles of enteric pathogens. Trends Microbiol. 26, 748–754. doi: 10.1016/j.tim.2018.02.008
Tokarskyy, O., De, J., Fatica, M. K., Brecht, J., and Schneider, K. R. (2018). Survival of Escherichia coli O157:H7 and Salmonella on bruised and unbruised tomatoes from three ripeness stages at two temperatures. J. Food Protect. 81, 2028–2033. doi: 10.4315/0362-028X.JFP-18-220
U.S. FDA (2019). Investigation Summary: Factors Potentially Contributing to the Contamination of Romaine Lettuce Implicated in the Fall 2018 Multi-State Outbreak of E. coli O157:H7. Silver Spring, ML: U.S. Food and Drug Administration.
USDA - Statistics (2019). Annual Reports: Crop Acreage, Yields, Areas Harvested, and Other Production Information. Available online at: https://www.usda.gov/topics/farming/crop-production (accessed December 21, 2019).
Keywords: plant breeding, enteric pathogens, biomarkers, cultivars, produce, food safety
Citation: Henriquez T, Lenzi A, Baldi A and Marvasi M (2020) Frontiers in Plant Breeding: Perspectives for the Selection of Vegetables Less Susceptible to Enteric Pathogens. Front. Microbiol. 11:1087. doi: 10.3389/fmicb.2020.01087
Received: 03 January 2020; Accepted: 30 April 2020;
Published: 28 May 2020.
Edited by:
Max Teplitski, University of Florida, United StatesReviewed by:
Abani Kumar Pradhan, University of Maryland, United StatesKalmia Kniel, University of Delaware, United States
Copyright © 2020 Henriquez, Lenzi, Baldi and Marvasi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Massimiliano Marvasi, massimiliano.marvasi@unifi.it
 Tania Henriquez
Tania Henriquez Anna Lenzi
Anna Lenzi Ada Baldi
Ada Baldi Massimiliano Marvasi
Massimiliano Marvasi