- 1Department of Bacteriology, Osaka Dental University, Hirakata, Japan
- 2National Institute of Molecular Biology and Biotechnology, College of Science, University of the Philippines Diliman, Quezon City, Philippines
- 3Department of Fixed Prosthodontics, Osaka Dental University, Hirakata, Japan
- 4Department of Synthetic Chemistry and Biological Chemistry, Graduate School of Engineering, Kyoto University, Kyoto, Japan
- 5Graduate School of Biostudies, Kyoto University, Kyoto, Japan
Archaeal species encode a variety of distinct lineage-specific chromosomal proteins. We have previously shown that in Thermococcus kodakarensis, histone, Alba, and TrmBL2 play distinct roles in chromosome organization. Although our understanding of individual archaeal chromosomal proteins has been advancing, how archaeal chromosomes are folded into higher-order structures and how they are regulated are largely unknown. Here, we investigated the primary and higher-order structures of archaeal chromosomes from different archaeal lineages. Atomic force microscopy of chromosome spreads out of Thermoplasma acidophilum and Pyrobaculum calidifontis cells revealed 10-nm fibers and 30–40-nm globular structures, suggesting the occurrence of higher-order chromosomal folding. Our results also indicated that chromosome compaction occurs toward the stationary phase. Micrococcal nuclease digestion indicated that fundamental structural units of the chromosome exist in T. acidophilum and T. kodakarensis but not in P. calidifontis or Sulfolobus solfataricus. In vitro reconstitution showed that, in T. acidophilum, the bacterial HU protein homolog HTa formed a 6-nm fiber by wrapping DNA, and that Alba was responsible for the formation of the 10-nm fiber by binding along the DNA without wrapping. Remarkably, Alba could form different higher-order complexes with histone or HTa on DNA in vitro. Mass spectrometry detected HTa and Rad50 in the T. acidophilum chromosome but not in other species. A putative transcriptional regulator of the AsnC/Lrp family (Pcal_1183) was detected on the P. calidifontis chromosome, but not on that of other species studied. Putative membrane-associated proteins were detected in the chromosomes of the three archaeal species studied, including T. acidophilum, P. calidifontis, and T. kodakarensis. Collectively, our data show that Archaea use different combinations of proteins to achieve chromosomal architecture and functional regulation.
Introduction
Genomic DNA needs to be folded properly in the cell to simultaneously accomplish compaction of the genetic material and accessibility to the transcription and replication machinery in all three domains of life. In eukaryotic cells, genomic DNA folding is primarily achieved through the nucleosome, a structure composed of histone octamers with approximately 150 bp of DNA wrapped around them (Luger et al., 1997; Davey et al., 2002). Additional proteins, including linker histone H1 and DNA topoisomerases, have been proposed to contribute to the higher-order folding of the eukaryotic genome (Thomas, 1984; Woodcock et al., 2006; Hizume et al., 2007; Fyodorov et al., 2018). Structural regulation of the chromosome is also pivotal for cell cycle progression. For example, condensin, a structural maintenance of chromosomes (SMC)-superfamily protein, is required for mitotic chromosome condensation in eukaryotes (Hirano et al., 1997; Ono et al., 2003). Epigenetic modifications also reportedly regulate gene expression (Grewal and Moazed, 2003; Li and Reinberg, 2011). Moreover, recent technological advances in chromosome conformation capture (3C) have shown that the eukaryotic genome is hierarchically folded into chromosomal domains, such as topologically associating domains (TADs) (Gibcus and Dekker, 2013; Barutcu et al., 2017).
In bacteria, genome compaction is achieved through the functions of nucleoid-associated proteins (NAPs) including HU, Fis, H-NS, and IHF (Skoko et al., 2006; Sarkar et al., 2009; Wang et al., 2013; Badrinarayanan et al., 2015; Hammel et al., 2016; Japaridze et al., 2017). Bacterial SMC condensin complexes, such as mukBEF in Escherichia coli and Smc-ScpAB in Bacillus subtilis, have also been implicated in higher-order organization and segregation of the nucleoid (Nolivos and Sherratt, 2014; Lioy et al., 2018). The B. subtilis Smc-ScpAB complex is targeted to parS sites on the nucleoid via ParB-SMC interaction (Marbouty et al., 2015; Wang et al., 2015). A highly transcribed ribosomal DNA region has also been proposed as a target for the SMC loading (Yano and Niki, 2017). Non-protein factors, such as macromolecular crowding, also contribute to the higher-order folding and regulation of the bacterial nucleoid (De Vries, 2010). It has also become evident that the bacterial nucleoid is segmented into many highly self-interacting regions called chromosomal interaction domains (CIDs), which are equivalent to the eukaryotic TADs, and the CIDs are further organized into large spatially distinct domains called macrodomains (Dame and Tark-Dame, 2016; Brocken et al., 2018; Verma et al., 2019).
Archaea constitute one of the three domains of life, along with Eukarya and Bacteria. Although Archaea are considered extremophiles that live in extreme environments, including high temperature, high salinity, or low pH (Forterre, 2016), they have also been found in moderate environments, including marine environments (Lloyd et al., 2013), soil (Bates et al., 2011), and even the human body (Probst et al., 2013; Koskinen et al., 2017; Nkamga et al., 2017; Pausan et al., 2019). Moreover, although no archaeal pathogens have been identified, Archaea have been associated with human diseases, including periodontal disease (Lepp et al., 2004) and inflammatory bowel disease (Blais Lecours et al., 2014). Thus, understanding how Archaea respond to and influence their environment has become crucial. Whereas control of gene expression in Archaea is achieved in part through transcription factors (TFs) resembling bacterial TFs, recent studies have suggested that chromosome structure and the interplay between chromosomal proteins and basal transcription machinery might also play important roles in gene expression (Peeters et al., 2015; Lemmens et al., 2019).
Archaeal chromosomal proteins are diverse (Sandman and Reeve, 2005; Luijsterburg et al., 2008; Driessen and Dame, 2011). For example, most species in Euryarchaeota, one of the major archaeal phyla, encode proteins homologous to eukaryotic histone (Nishida and Oshima, 2017; Henneman et al., 2018). Species in newly proposed phyla such as Nanoarchaeota, Thaumarchaeota, and Lokiarchaeota also encode histone (Henneman and Dame, 2015; Henneman et al., 2018). Exceptions in Euryarchaeota are the members of the order Thermoplasmatales, which lack histones and instead encode HTa (H istone-like protein of Thermoplasma acidophilum), a protein homologous to bacterial HU (DeLange et al., 1981; Hocher et al., 2019). Whether HTa has a role similar to that of bacterial HU or an Archaea-specific function remains elusive. Proteins such as Cren7, CC1, and Sul7 are specific to species in Crenarchaeota, another major archaeal phylum (Driessen and Dame, 2011). Alba, a 10-kDa DNA/RNA-binding protein, is found in both Euryarchaeota and Crenarchaeota (Guo et al., 2003; Laurens et al., 2012), as well as in newly proposed phyla including Nanoarchaeota, Korarchaeota, Thaumarchaeota, and Lokiarchaeota (Henneman and Dame, 2015; Sanders et al., 2019). Alba has been shown to undergo post-translational modifications including methylation and acetylation (Bell et al., 2002; Cao et al., 2018). Alba has also been shown to have several different modes of interaction with DNA, such as DNA stiffening or bridging (Laurens et al., 2012). However, how Alba cooperates with other chromosomal proteins in higher-order chromosome folding and whether it plays different roles in Euryarchaeota and Crenarchaeota remain to be elucidated.
Besides these DNA-binding proteins that are usually smaller than 10 kDa, some larger TF-like proteins also behave as chromosomal proteins in Archaea. TrmB-like 2 (TrmBL2), a protein homologous to the sugar-responsive transcriptional regulator TrmB, is an abundant chromosomal protein that is completely conserved among Thermococcales (Gindner et al., 2014; Kim et al., 2016). Lines of experimental evidence suggest that TrmBL2 is a chromosome-associated protein resembling bacterial H-NS, with functions that include filament formation on DNA and the global suppression of gene expression (Maruyama et al., 2011; Efremov et al., 2015). More recently, Hi-C chromosome conformation capture experiments on Sulfolobus species showed that coalescin, a new class of SMC protein, mediates the crenarchaeal chromosome into a two-domain organization resembling the eukaryotic A/B compartments (Takemata et al., 2019).
In the present study, we analyzed the protein composition and structure of chromosomes from diverse archaeal lineages, aiming to understand the general principles of genome folding. Species used in this study belong to either Euryarchaeota or Crenarchaeota, two major phyla in Archaea, and encode different combinations of chromosomal proteins. Thermococcus kodakarensis is a euryarchaeon that encodes histone, Alba, and TrmBL2 as major chromosomal proteins (Maruyama et al., 2011). Thermoplasma acidophilum is a euryarchaeon that exceptionally lacks histones, but encodes HTa, a protein homologous to bacterial HU, together with Alba (Ruepp et al., 2000). Since it is unclear whether HTa plays roles similar to that of bacterial HU, we investigated how HTa contributes to genome folding. Pyrobaculum calidifontis is a hyperthermophilic crenarchaeon that encodes Alba, Cren7, CC1, and Sso10a (Amo et al., 2002; Peeters et al., 2015). Sulfolobus solfataricus is a crenarchaeon that encodes Alba, Cren7, Sul7, and Sso10a (Peeters et al., 2015). Both the primary and higher-order chromosome structures were analyzed with atomic force microscopy (AFM). Chromosomal proteins in each species were identified through a combination of chromosome isolation and mass spectrometry. The minimal structural unit of each chromosome was investigated with micrococcal nuclease (MNase) digestion analysis and the in vitro reconstitution of protein–DNA complexes.
Results
The Archaeal Chromosome Is Commonly Composed of 10-nm Fibers and 30–40-nm Globular Structures
We previously showed that, in T. kodakarensis, the chromosome is more compact in the stationary phase than in the log-phase (Maruyama et al., 2011). Here, the extent of chromosome compaction in two archaeal species from different phyla, T. acidophilum and P. calidifontis, were analyzed by using the on-substrate lysis method (see section “Materials and Methods”). Cells were grown and applied on coverslips, then mildly lysed with detergent. Staining of the lysed cells with 4′,6-diamidino-2-phenylindole (DAPI) showed stretched and decondensed chromosome released from log-phase cells for both archaeal species (Figure 1A). In contrast, compact chromosome structures were dominant after the lysis of stationary phase cells based on DAPI staining (Figure 1B). These results suggest the difference in chromosome organization of archaeal cells between the growth phases.
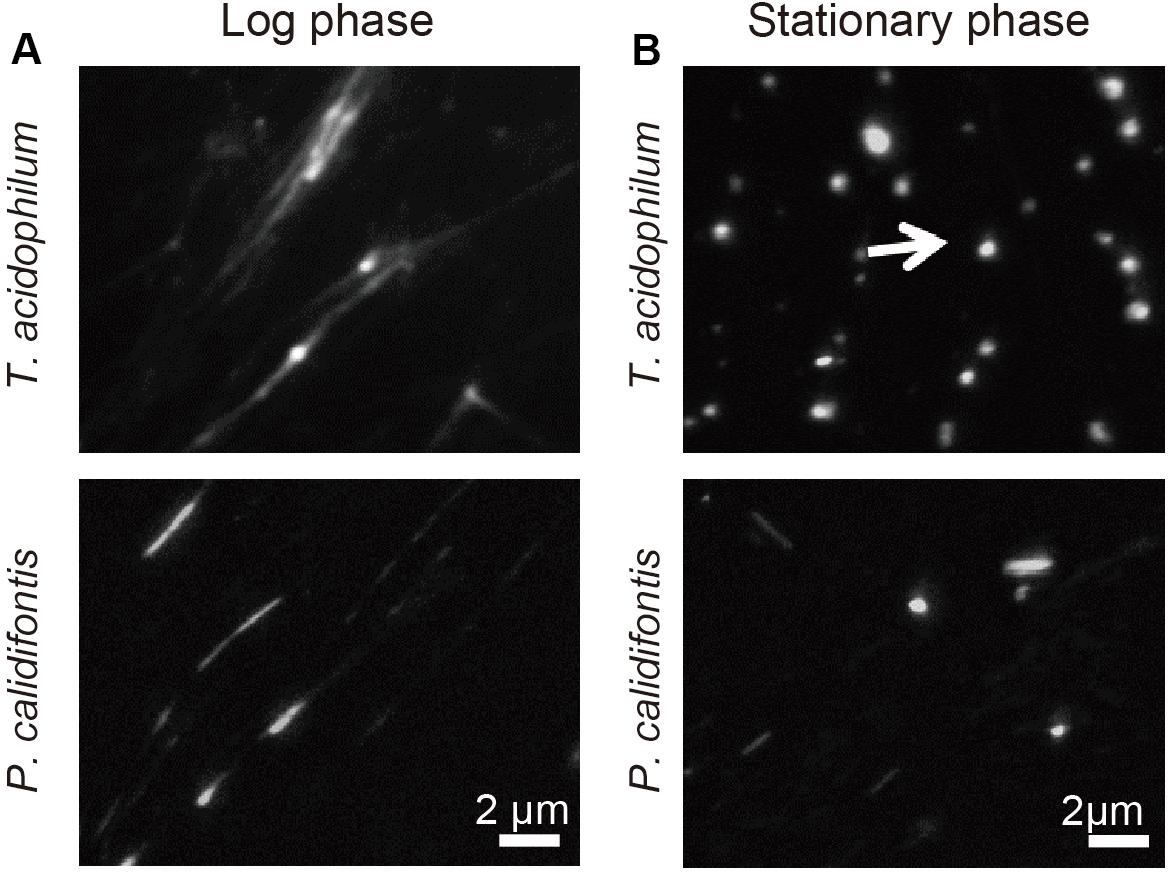
Figure 1. Genomic material released from T. acidophilum and P. calidifontis cells. DAPI staining was performed after on-substrate lysis of the cells during (A) log and (B) stationary phases. Fluorescence microscopy images are shown. (A) Chromosomes were decondensed and elongated in the log-phase for both T. acidophilum (upper panel) and P. calidifontis (lower panel). (B) The stationary phase chromosome was more compact than the log-phase chromosome (arrow). Scale bars: 2 μm.
Detailed AFM analysis of chromosomes released from the lysed cells identified two distinct chromosomal structures common to both species, 10-nm fiber and 30–40-nm globular structures. The 10-nm fiber structure was prevalent on the well-spread chromosome in the log-phase in both species (Figure 2A). The diameter of the fibers was 10.1 ± 4.2 nm (mean ± SD, n = 119) in T. acidophilum and 11.5 ± 4.2 nm (n = 110) in P. calidifontis (Figure 2B). Larger globular structures were found where chromosomes were less spread in both species in the stationary phase, wherein most cells showed limited chromosome release (Figure 2C). The diameters of the globular structures differed between the two species and were specifically 38.6 ± 10.1 nm (n = 217) nm in T. acidophilum and 29.1 ± 5.1 nm (n = 99) nm in P. calidifontis (Figure 2D), suggesting differences in the fundamental structure or the higher-order architecture of the chromosome. Although the fiber and globular structures are seen in both growth phases, these results show that the extent of archaeal chromosome folding is greater in the stationary phase.
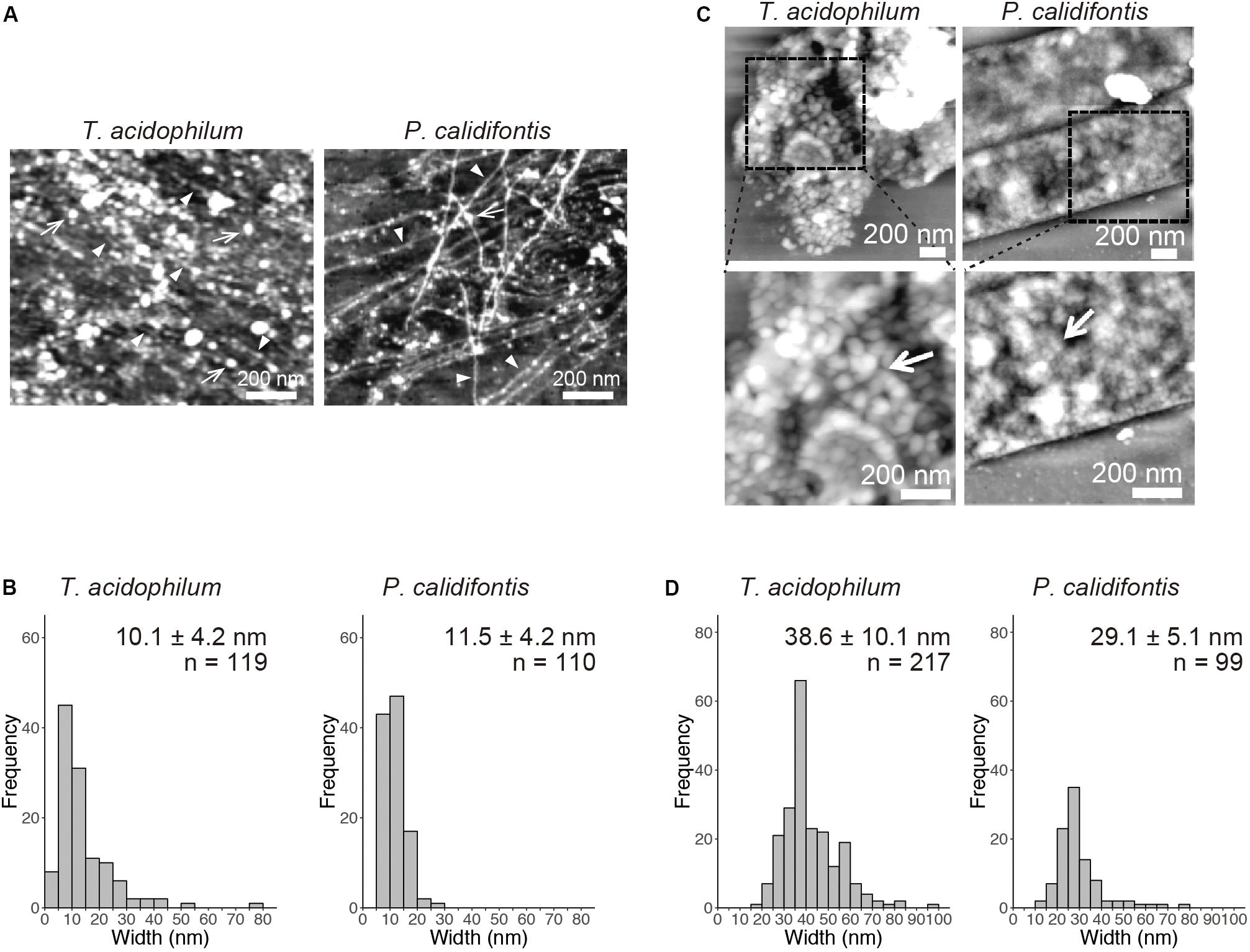
Figure 2. Structural details of genomic materials from T. acidophilum and P. calidifontis after on-substrate lysis visualized with atomic force microscopy (AFM). (A,B) Extensively released chromosome fibers of log-phase cells. (A) AFM images of the released genomic materials show relatively thin fibers (arrow heads) and globular structures (arrows). (B) Histograms indicate the diameters of the released fiber structures indicated by arrow heads in (A). (C,D) Compact chromosome of stationary phase cells. (C) AFM images of the lysed cells. Boxed areas in the upper panels were shown in detail in the lower panels. Globular structures (arrows) were observed more frequently than in the log phase. (D) Histograms indicate the diameters of the globular structures. Scale bars: 200 nm.
Minimal Unit of the Archaeal Chromosome Revealed With MNase Digestion
On-substrate lysis results revealed that the size of the primary structure of the archaeal chromosome is approximately 10 nm in diameter (Figures 2A,B). We next investigated the minimal structural unit through MNase digestion of purified chromosomes from four archaeal species: T. kodakarensis and T. acidophilum from Euryarchaeota, and P. calidifontis and S. solfataricus from Crenarchaeota. The MNase digestion was performed without chemical fixation. The analysis revealed a variation in minimal structural units in each species (Figure 3). As we have previously shown, MNase digestion of the T. kodakarensis chromosome resulted in a 30-bp laddered pattern with a minimum of 60 bp of DNA (Figure 3A). The 30-bp intervals detected are typical of some histone-containing Archaea and reflect the flexible histone multimeric structure, which was shown previously (Maruyama et al., 2013; Mattiroli et al., 2017; Henneman et al., 2018). Interestingly, MNase digestion of the T. acidophilum chromosome resulted in the accumulation of 40∼50 or 80∼90 bp of DNA depending on the enzyme concentration (Figure 3B), corresponding with the results of the previous reports (Searcy and Stein, 1980; Hocher et al., 2019). The 40∼50 bp fragment suggests the size of DNA involved in the primary unit. The larger DNA fragment of 80∼90 bp identified with a lower MNase concentration indicated that this unit can be positioned adjacent to each other (Hocher et al., 2019). In contrast, the MNase digestion of the chromosomes of the two crenarchaeal species, P. calidifontis and S. solfataricus, did not yield the accumulation of DNA fragments of a particular size (Figures 3C,D). These results demonstrate that in crenarchaeal species, which encode neither histone nor HTa, the chromosomes do not have a defined-size structural unit that protects the genomic DNA from MNase digestion. Whereas on-substrate cell lysis revealed similar fibrous chromosome structures among T. acidophilum and P. calidifontis (Figure 2), as well as in T. kodakarensis, as previously reported (Maruyama et al., 2011), the MNase digestion patterns indicated that the fine details in the organization of these structures vary, probably due to differences in the chromosomal proteins involved.
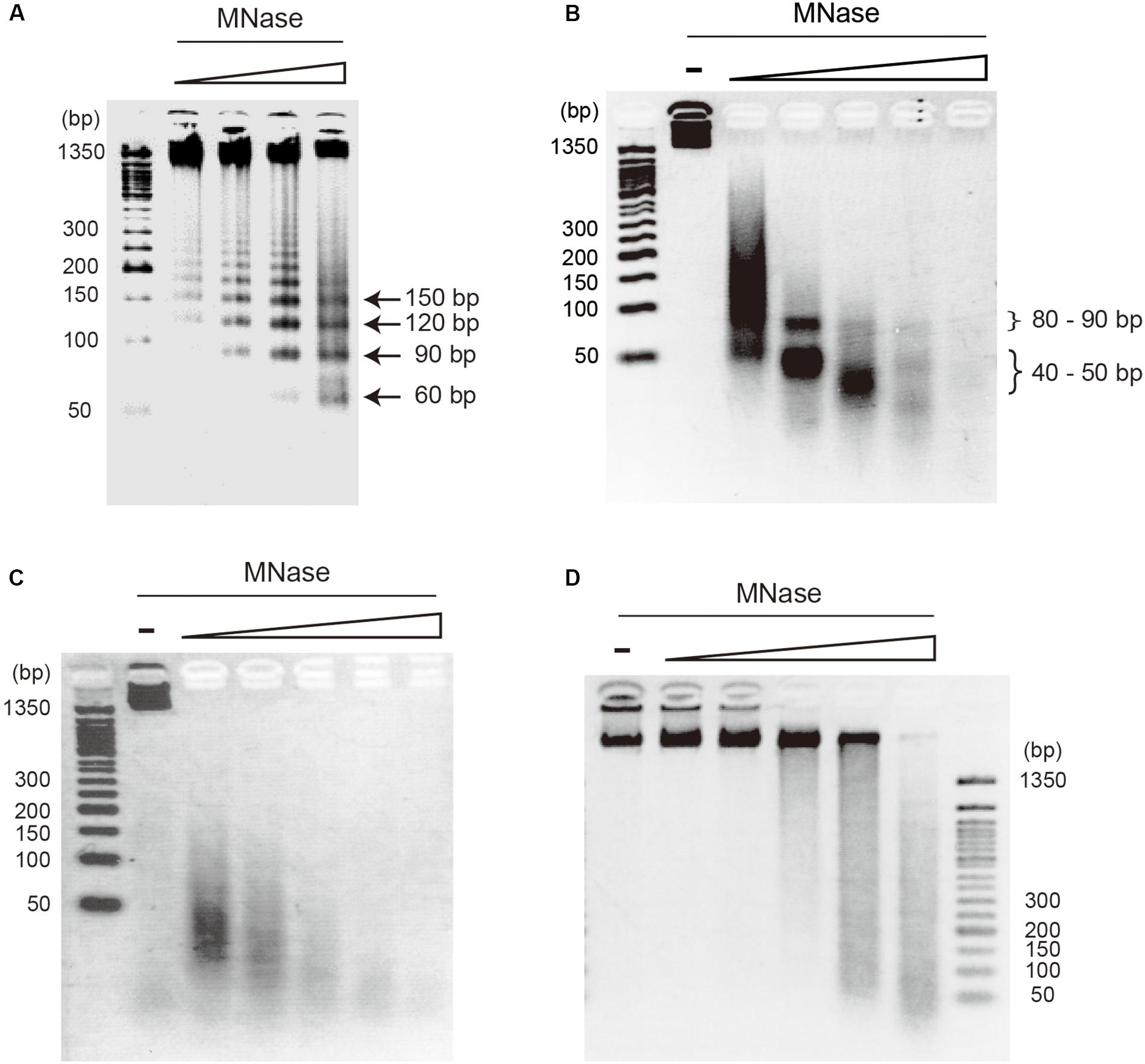
Figure 3. Digestion pattern of archaeal chromosomes treated with micrococcal nuclease (MNase). Purified chromosomes of (A) T. kodakarensis, (B) T. acidophilum, (C) P. calidifontis, and (D) S. solfataricus were digested with increasing concentrations of MNase and separated on 2.5% agarose gels in 1X TBE. The accumulation of DNA of particular sizes was observed with T. kodakarensis (arrows) and T. acidophilum (curly brackets) but not with P. calidifontis and S. solfataricus chromosomes. MNase concentration was 0.3, 1, 3, 10 U MNase in 100 μl reaction (A) or 0, 0.3, 1, 3, 10, and 30 U MNase in 100 μl reaction (B–D).
Mass Spectrometry Identifies Chromosome-Associated Proteins in Archaeal Species
Protein components of isolated chromosomes of T. acidophilum, P. calidifontis, and T. kodakarensis were analyzed using mass spectrometry (Figure 4). Tables 1–3 provide an overview of the distribution of chromosomal proteins in each archaeal species. Proteins predicted to be involved in replication, transcription, and translation were detected in the three species. These include the DNA primase DnaG detected in T. acidophilum and P. calidifontis and DNA-directed RNA polymerase subunits detected in T. acidophilum and T. kodakarensis. Ribosome subunits were commonly found in all species and constituted a large portion of the identified proteins, which is reasonable considering that transcription and translation are coupled in Archaea (French et al., 2007).
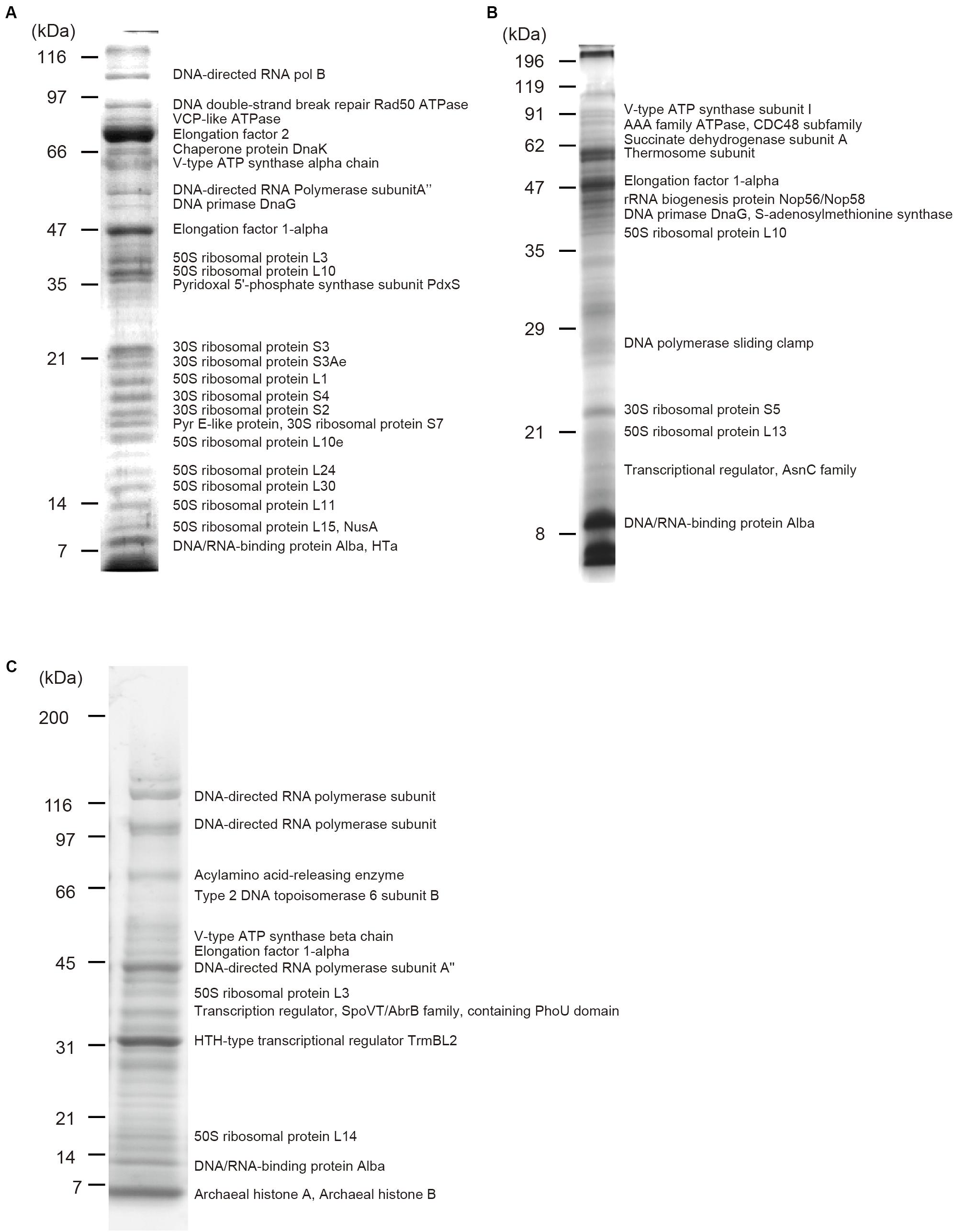
Figure 4. Protein components of isolated chromosomes. Chromosomes isolated from (A) T. acidophilum (B) P. calidifontis (C) T. kodakarensis were digested with MNase, separated through SDS-PAGE and stained with Comassie Brilliant Blue. Representative proteins detected with mass spectrometry are indicated next to the corresponding bands. See Tables 1–3 for details.
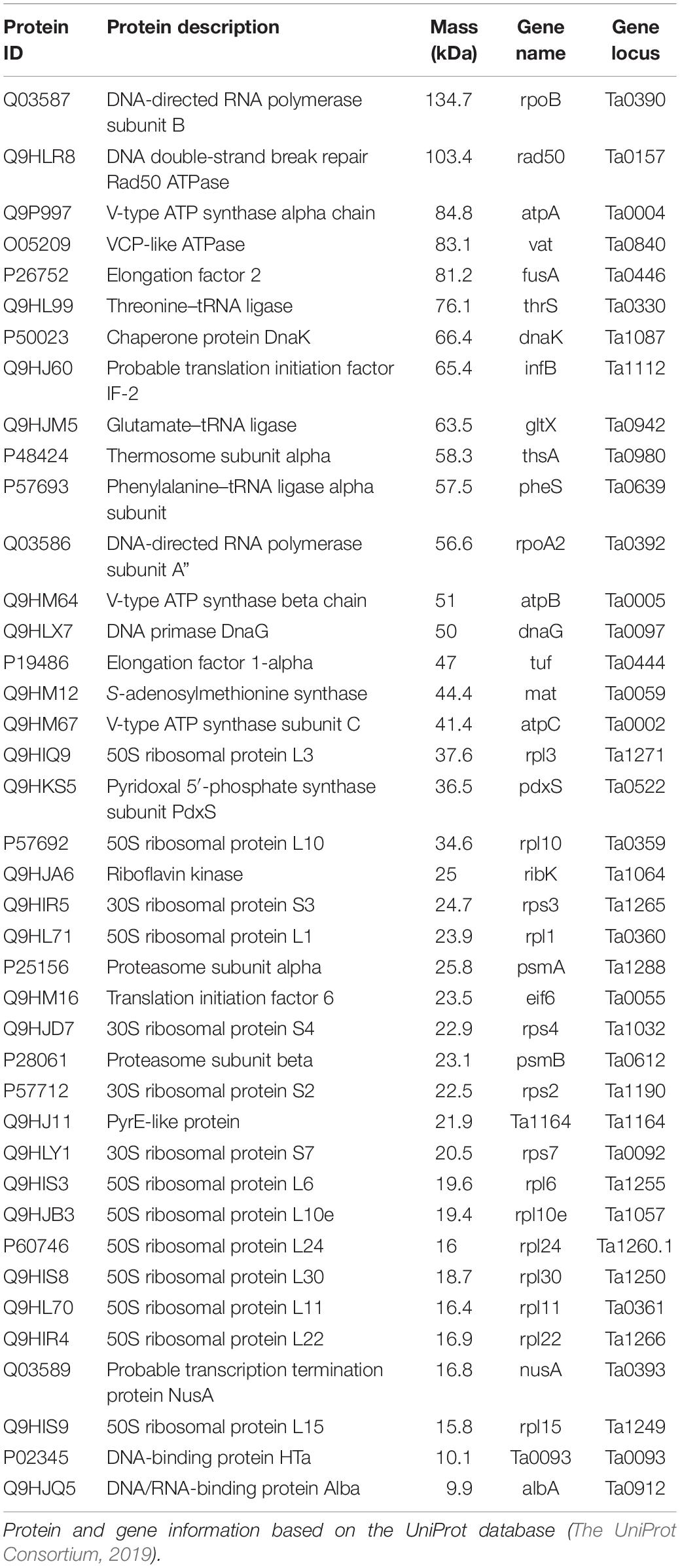
Table 1. Proteins identified by mass spectrometry from isolated chromosomes of Thermoplasma acidophilum.
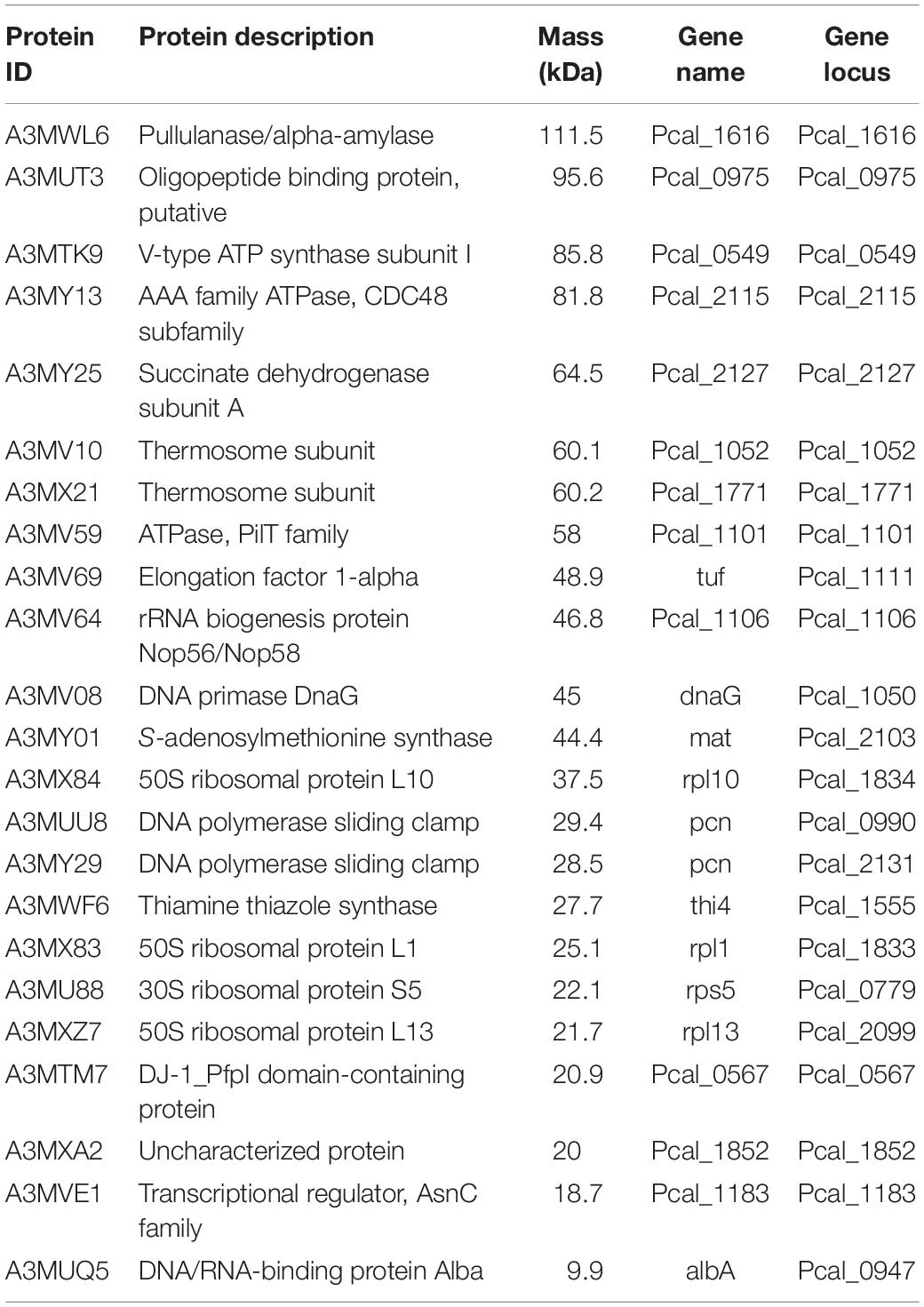
Table 2. Proteins identified by mass spectrometry from isolated chromosomes of Pyrobaculum calidifontis.
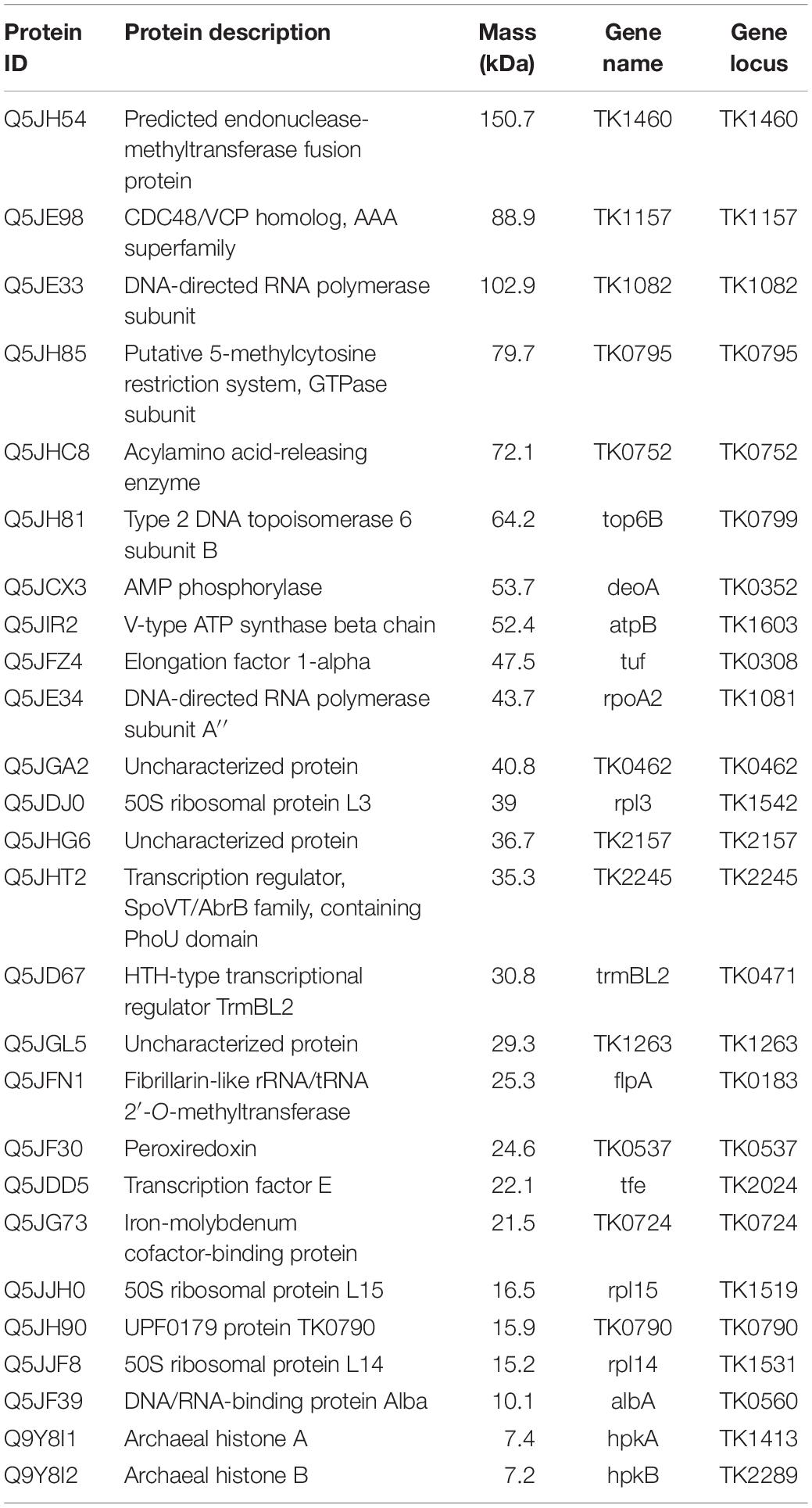
Table 3. Proteins identified by mass spectrometry from isolated chromosomes of Thermococcus kodakarensis.
Apart from these general information processing proteins, DNA-binding proteins that might contribute to chromosome folding and regulation were also detected. Alba was detected in all three species studied (Figure 4 and Tables 1–3). In the T. acidophilum chromosome, HTa and Alba were detected together with Rad50 ATPase, an SMC-family protein (Figure 4A and Table 1). In the P. calidifontis chromosome, a protein annotated as ‘Transcriptional regulator, AsnC family’ (UniProtKB/TrEMBL ID: A3MVE1, gene ID: Pcal_1183) was detected in addition to Alba (Figure 4B and Table 2). It has been suggested that some AsnC/Lrp family transcriptional regulators have global gene regulatory roles as well as an architectural function, instead of being a specific transcriptional regulator (Peeters et al., 2015). The identification of Pcal_1183 in P. calidifontis, which has not been reported as a chromosomal protein, supports the view that TF-like proteins act as chromosome proteins in Archaea, as is the case for TrmBL2 in Thermococcales (Maruyama et al., 2011). Cren7, although encoded in the P. calidifontis genome, was not detected in the present mass spectrometry analysis, suggesting that the amount of Cren7 on the P. calidifontis chromosome is lower than the detection limit of our method. In T. kodakarensis, histones, Alba, and TrmBL2 were detected as previously reported (Figure 4C and Table 3) (Maruyama et al., 2011).
Interestingly, in addition to proteins with apparent DNA-binding ability, a few putative membrane-associated proteins were detected in the chromosome. For example, V-type ATPases were found in all three species (Figure 4 and Tables 1–3), suggesting a general role of these proteins in archaeal chromosome folding and regulation. Because the chromosome isolation procedure included cell lysis with a detergent and sucrose density gradient sedimentation, we assume that the isolated chromosome was not contaminated with cell membrane or membrane-bound proteins, as shown previously for T. kodakaresis (Maruyama et al., 2011).
Distinct Fundamental Structures Formed With Alba and HTa
Alba was common among the three species studied (Figure 4 and Tables 1–3). When recombinant Alba proteins from T. acidophilum or P. calidifontis genes expressed in E. coli were mixed with linear 3-kb DNA in vitro, a fiber structure was formed at Alba:DNA weight ratio higher than 6:1 (Figure 5A). The diameter of the fiber was 10.0 ± 2.8 nm (n = 80) in T. acidophilum and 12.1 ± 2.2 nm (n = 119) in P. calidifontis (Figure 5B). These correlate well with the structures visualized in the on-substrate lysis experiment (Figures 2A,B), suggesting that Alba is responsible for the formation and maintenance of the 10-nm fiber structures. The contour length of the filamentous structures was comparable to the theoretical length of a 3-kb DNA, which is ∼1000 nm (Maruyama et al., 2011), indicating that DNA does not wrap around Alba molecules (Figure 5C). At relatively higher Alba concentrations, these filaments seemed to be capable of folding onto themselves and forming more complex structures. The results strongly support a role of Alba protein contributing to primary order folding of the DNA into 10-nm fibers. In addition, the data suggest the role of Alba in forming more complex, higher-order structures.
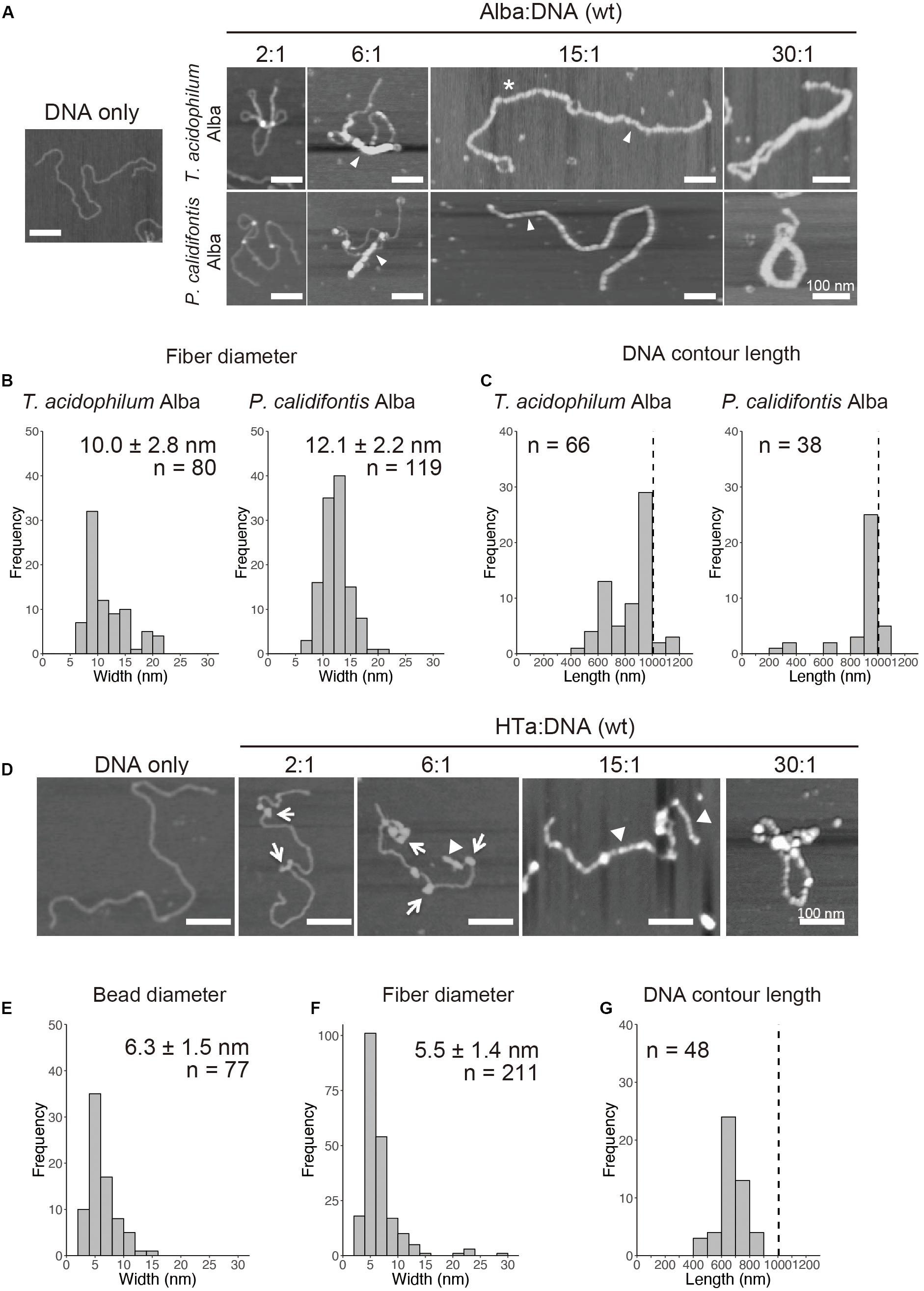
Figure 5. In vitro reconstitution of chromosome structures with recombinant Alba and HTa. (A–C) Reconstitution on a linear 3-kb plasmid using recombinant Alba from T. acidophilum and P. calidifontis at different Alba to DNA weight ratios. (A) AFM images showing various structures formed with Alba and DNA. Fibers are indicated with arrow heads. Alba:DNA weight ratios are indicated. The asterisk indicates a structure in which two 3-kb DNA molecules are joined together by Alba binding. (B) Histograms show the diameters of the fibrous structures formed (indicated with arrow heads in A). (C) Histograms show the respective contour lengths of DNA at a 15:1 Alba:DNA ratio. The theoretical length of a linear 3-kb DNA (∼1000 nm) is indicated with a dashed line. See Figure 6A for a histogram of unbound DNA length. (D–G) Reconstitution with histidine-tagged HTa from T. acidophilum at varying protein to DNA ratio. (D) AFM images showing beaded (arrows) and filamentous (arrow heads) structures. (E) Diameter of the beads formed at a relatively lower HTa concentration. (F) Width of the filaments formed at relatively higher HTa concentration. (G) Histogram shows the contour DNA lengths of the structures formed at a 15:1 HTa:DNA ratio. Dashed line indicates the theoretical length of a 3-kb unbound DNA. Scale bars: 100 nm.
Whereas most species in Euryarchaeota commonly encode histones, species belonging to Thermoplasmatales lack histones and encode HTa, a homolog of bacterial HU. It has been proposed that Thermoplasmatales acquired the HU gene from bacteria through horizontal gene transfer (HGT) and lost the histone gene and that HTa plays a role similar to that of archaeal histone (Hocher et al., 2019). We investigated how HTa interacts with DNA at the primary level. The reconstitution using recombinant histidine-tagged HTa from T. acidophilum showed a beaded structure (Figure 5D), similar to the beads-on-a-string structure formed with archaeal histone (Maruyama et al., 2011, 2013) or bacterial HU (Rouvière-Yaniv et al., 1979). The diameter of the HTa beads was 6.3 ± 1.5 nm (n = 77) (Figure 5E), which is thinner than the 10-nm Alba filaments (Figures 5A,B). At higher concentrations, HTa formed a filament of 5.5 ± 1.4 nm in width (n = 211) on the DNA (Figures 5D,F). The contour DNA length of reconstituted nucleoprotein structure was shorter than that of naked DNA (Figure 5G), reflecting DNA wrapping around HTa particles. These results support the notion that archaeal HTa is functionally more closely related to histones than bacterial HU (Hocher et al., 2019). The ∼6-nm particle might be a fundamental unit of the Thermococcales chromosome, together with the 10-nm fiber formed with Alba (Figures 5A,B).
Relationship Between Alba and Other Chromosomal Proteins in Forming Structural Complexes With DNA
Alba can form various structures with DNA, depending on its amount (Figure 5A). This flexibility might facilitate its interaction with other structural proteins, forming DNA-based complexes. In the cell, Alba coexists with other DNA-binding proteins. For example, T. kodakarensis encodes histone and Alba, whereas T. acidophilum encodes HTa and Alba. We therefore combined Alba with the chromosomal proteins, histone and HTa, to determine how they cooperate in compacting DNA.
In vitro reconstitution using histone alone at a 1:1 histone:DNA weight ratio resulted in compact beaded fiber structures displaying decreased contour length of the DNA-protein complex (Figure 6A). Next, histone and Alba were mixed together with DNA. Alba was added at the concentration range at which it started to form the 10-mn fiber structure when mixed with DNA by itself (Figure 5A). Reconstitution using both histone and Alba from T. kodakarensis at a 1:3 or 1:7 histone:Alba ratio resulted in the formation of beaded structures somewhat similar to those formed with DNA and histone alone (Figure 6A), but with a larger proportion of DNA molecules having a longer contour length (>500 nm) (Figure 6A). As Alba does not wrap DNA (Figures 5A,C), this result suggests that Alba binding decreases the extent of histone-mediated DNA folding or that Alba competes with histone for DNA-binding, leading to less compact structures. Although the amount of Alba relative to histone in T. kodakarensis cell (3:1 histone:Alba in weight) (Maruyama et al., 2011) is estimated to be lower than that used in this experiment, these results indicate that Alba can compete with histone at least at the local level.
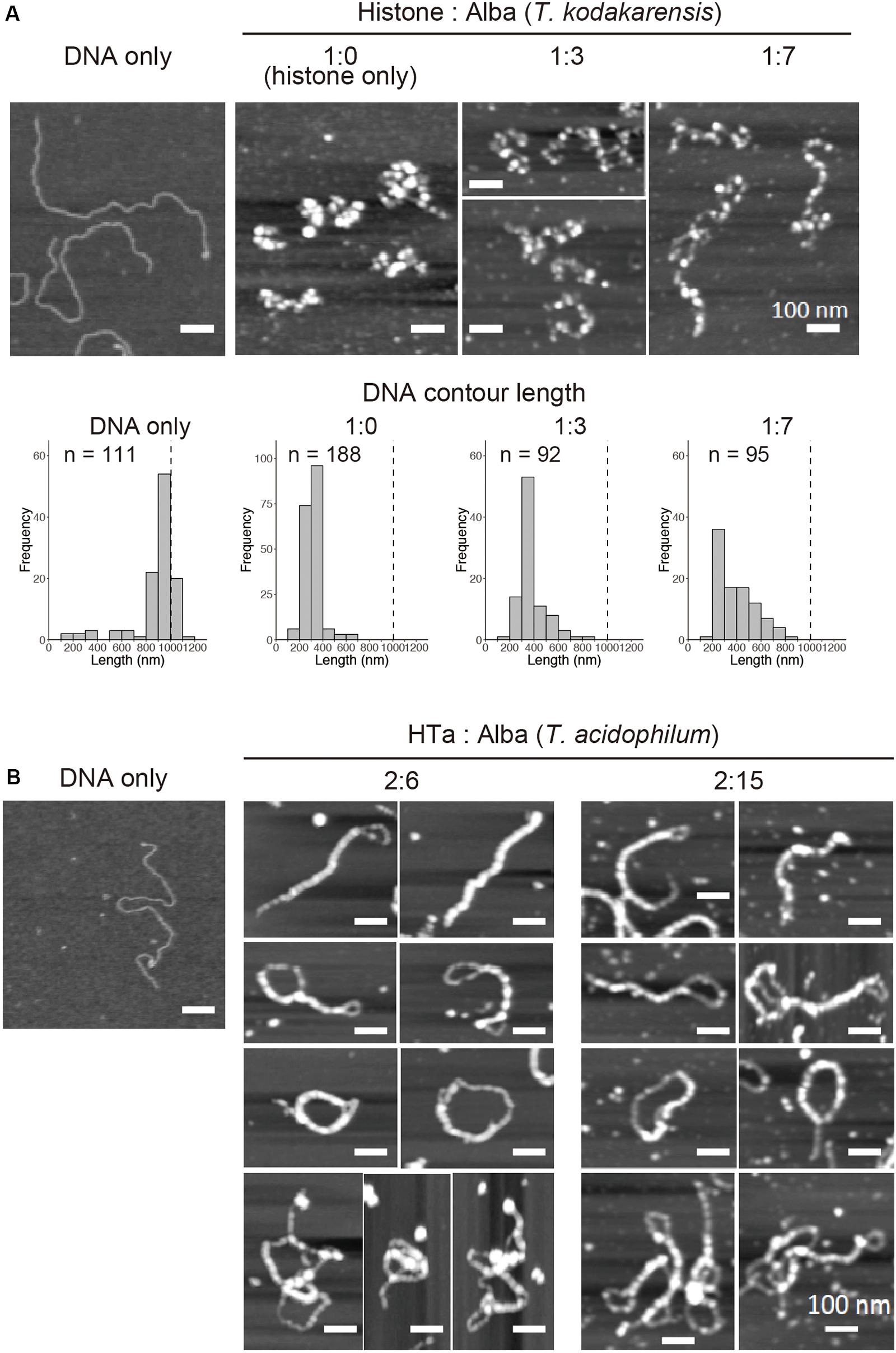
Figure 6. In vitro reconstitution using a combination of archaeal proteins. Chromatin structures were reconstituted on a linear 3-kb plasmid using recombinant proteins at varying Alba concentrations. (A) AFM images showing reconstitution using histone and Alba from T. kodakarensis. Histograms below the AFM images show the contour DNA length in each condition. Dashed line indicates the theoretical length of a 3-kb unbound DNA. (B) AFM images showing reconstitution using HTa and Alba from T. acidophilum. Scale bars: 100 nm. Note that histograms are not shown in (B) because of the inability to accurately measure the DNA contour length due to a high degree of folding or joining of the fiber structures.
Next, HTa and Alba from T. acidophilum were mixed together with DNA (Figure 6B). The concentration of HTa was fixed to 1:2 DNA:HTa, at which it had formed separate beaded structure when mixed with DNA by itself (Figure 5D). Alba was mixed at the concentration range at which it formed fiber structure on DNA (1:6 or 1:15 DNA:Alba) when it was mixed alone with DNA (Figure 5A). Reconstitution using HTa and Alba from T. acidophilum at 2:6 and 2:15 weight ratios (Figure 6B) resulted in structures similar to those formed by Alba alone (Figure 5A). Structures with DNA strands joined together were dominant, and DNA molecules were considerably folded (Figure 6B). Moreover, some complexes were able to fold upon themselves, providing additional compaction (Figure 6B). These data indicate that unlike with histone (Figure 6A), Alba with HTa facilitates DNA compaction through DNA strand bridging (Figure 6B).
Thus, Alba encoded in each species might play different roles depending on the interaction or synergy with other proteins associated with the chromosome in each archaeal lineage. Binding of HTa might increase the efficacy of Alba-mediated DNA bridging, leading to complexes that are more compact than those with either HTa or Alba alone. Whether this compaction is achieved through local structural change in chromatin due to the binding of individual protein or through direct protein–protein interaction between HTa and Alba needs to be elucidated.
Discussion
In the present study, we investigated the fundamental and higher-order structures of archaeal chromosomes from different lineages. We show that archaeal chromosomes comprise step-wise structures that consist of primary fibrous structures of 10 nm in diameter and higher-order globular structures of 30–40 nm. The minimal unit of the chromosome varies among species. Archaeal chromosomes undergo compaction toward the stationary phase (Figure 7A). We also show that archaeal HTa can wrap DNA and form a 6-nm beaded fiber, which might be folded into the higher-order structures in association with Alba in T. acidophilum. Interestingly, TF-like proteins were found in T. kodakarensis and P. calidifontis. Membrane-associated proteins were detected in the chromosomes of all three species. Finally, we demonstrate that Alba-mediated chromosomal structures vary depending on the presence and identity of additional architectural proteins.
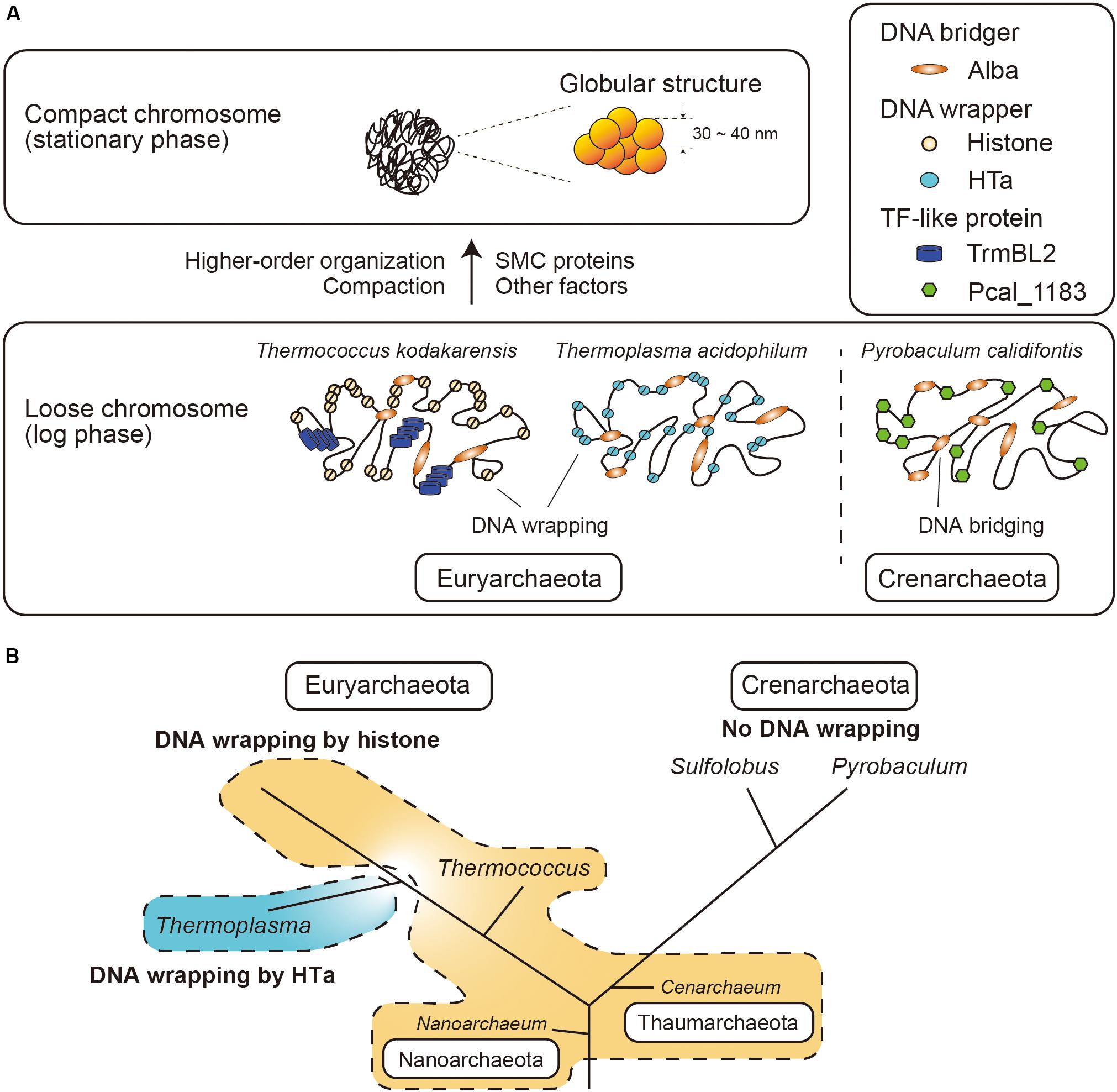
Figure 7. Model of archaeal chromosome folding. (A) In Euryarchaeota, histone or HTa wraps genomic DNA, whereas there is no DNA wrapper in Crenarchaeota. Alba can bridge distant genomic DNA. Chromosome compaction occurs toward the stationary phase, when 30∼40-nm globular structures are formed. SMC proteins might be involved in higher-order organization of the chromosome. (B) Schematic phylogenetic tree of selected Archaea (Sandman and Reeve, 2005) together with the distributions of histone and HTa. Species in Euryarchaeota utilize histone or HTa (Thermoplasmatales) to wrap DNA at the primary level. Members of Nanoarchaeota and Thaumarchaeota also encode histone that might wrap DNA. Species in Crenarchaeota neither encode histone nor wrap DNA. The branch lengths do not correspond to evolutionary distances. Note that Cenarchaeum used to be classified as a member of Crenarchaeota, but is now considered a member of Thaumarchaeota (Spang et al., 2010).
Primary and Higher-Order Folding of Archaeal Chromosomes
Higher-order chromosome architecture and its regulation have been extensively studied in bacteria and eukaryotes. In bacteria, the coordination of various NAPs is required for proper folding and regulation of the nucleoid (Verma et al., 2019). For example, the E. coli nucleoid is composed of 10-, 40-, and 80-nm fibers and undergoes compaction in the stationary phase, in which Dps is highly expressed (Ali Azam et al., 1999; Kim et al., 2004). Oxidative stress also causes Dps-dependent nucleoid compaction in Staphylococcus aureus (Morikawa et al., 2019). In eukaryotes, it has been viewed that beads-on-a-string chromatin is folded into a 30-nm fiber with the linker histone H1 (Hizume et al., 2005; Prieto and Maeshima, 2019). Our results indicate that archaeal chromosomes also comprise step-wise structures, sharing features with both bacterial and eukaryotic chromosomes. A hierarchical mode of archaeal genome packing can be inferred wherein the 6- or 10-nm fiber structures form the 30–40-nm globular structures. The 6- or 10-nm fibers might represent the first order of folding in the archaeal chromosome, wherein DNA is bound and constrained by one or more abundant chromosomal protein(s). The abundant chromosomal proteins present in each species and their interactions with each other might contribute to species-specific primary and higher-order chromosome structures.
We show that the archaeal chromosome undergoes compaction toward the stationary phase, similarly to what happens in the bacterial nucleoid. However, how the compaction is achieved remains elusive. SMC-superfamily proteins are known to be involved in higher-order architecture and the regulation of chromosomes in eukaryotes and bacteria (Uhlmann, 2016; Lioy et al., 2018). In archaea, the Hi-C experiments on Sulfolobus species showed that the SMC-family protein coalescin contributes to the formation of distinct chromosome domains that undergo discrete and specific higher-order interactions (Takemata et al., 2019). Although SMC homologs are present in archaeal genomes based on genomic sequences, they were not commonly detected using mass spectrometry in the present study, except for Rad50 ATPase detected in T. acidophilum chromosome (Figure 4A and Table 1). Low amounts of SMC proteins might be sufficient for their function in archaeal cells, probably cooperating with other chromosomal proteins to achieve higher-order folding of the chromosome (Figure 7A). An indirect association of SMC protein with the chromosomes might also be a reason why they were not detected in our mass spectrometry analysis. Indeed, bacterial SMC proteins are loaded onto the chromosome by ParB proteins (Marbouty et al., 2015; Wang et al., 2015).
Post-translational modifications of archaeal DNA-binding proteins such as Alba and Cren7 have been reported (Bell et al., 2002; Niu et al., 2013; Cao et al., 2018). Detailed analyses of the influence of such post-translational modifications on protein–protein and protein–DNA interactions might also provide insights into mechanisms of chromosome folding and regulation.
Role of HTa in T. acidophilum
HTa is an abundant protein in members of Thermococcales, which lack histones. Our present study shows that T. acidophilum HTa can form a beads-on-a-string structure by wrapping DNA and that the wrapping protects DNA from MNase digestion (Figures 3B, 5D). Taken together with a recently reported analysis of HTa (Hocher et al., 2019), these results indicate that HTa has characteristics more similar to those of archaeal histone than to those of bacterial HU. Moreover, HTa was recently reported to form oligomers (Hocher et al., 2019) resembling the flexible “hypernucleosome” formed by multiple archaeal histone dimers stacked onto each other (Maruyama et al., 2013; Mattiroli et al., 2017; Henneman et al., 2018). Thus, it seems that for species in Euryarchaeota, a protein that fulfills the DNA wrapping function (i.e., histone or HTa) seems to be indispensable for the proper folding and regulation of the chromosome. Based on the results of the MNase assay (Figures 3C,D), we assume that most crenarchaeal species do not utilize DNA wrapping as the primary level of DNA folding (Figure 7B). Given the efficiency of genome compaction by DNA wrapping, why most crenarchaeal chromosomes do not require DNA wrapping is an interesting question that needs to be answered in the future.
The phylogenetic reconstitution of the HTa/HU gene family has suggested that HTa was acquired through the horizontal transfer of bacterial HU at the root of the clade which includes Thermoplasmatales (Hocher et al., 2019). From an evolutionary aspect, an intriguing question is how histone could be replaced by a protein with apparently different characteristics; for example, archaeal histone wraps DNA, whereas bacterial HU bends it (Henneman et al., 2018; Verma et al., 2019). Although some species have been shown to tolerate the complete deletion of histone gene(s) from their genome (Weidenbach et al., 2008), complete deletion of histone genes seems to be impossible in most species (Čuboňová et al., 2012). Besides, since all species in Euryarchaeota (except for Thermoplasmatales) encode histone, loss of histone does not seem to be favorable in the living environments. Thus, it seems unlikely that the loss of histone occurred prior to the acquisition of HU. Interestingly, the expression of archaeal histone in E. coli does not cause a severe growth defect (Rojec et al., 2019), indicating that histone and HU can coexist in a single organism. These facts are consistent with the idea that the horizontal transfer of bacterial HU to Archaea and its adaptation might have occurred before the loss of the histone gene. It is important to note here that histone is assumed to have existed prior to the acquisition of HU. Without this assumption, other scenarios are possible.
Role of Transcription Factor-Like Proteins in P. calidifontis and T. kodakarensis
Our present results showed the abundance of a putative TF-like protein in the P. calidifontis chromosome. Several archaeal TFs and TF-like proteins have been found to combine a global gene regulatory role with an architectural role (Maruyama et al., 2011; Peeters et al., 2015). For example, TrmBL2 (TK0471) in T. kodakarensis has a helix-turn-helix motif with non-specific DNA-binding ability (Maruyama et al., 2011; Ahmad et al., 2015). Single-molecule studies have shown that TrmBL2 forms a stiff nucleoprotein filament and that histone competes with TrmBL2, modulating the stiffness of the filament (Efremov et al., 2015). The functions of TrmBL2 resemble those of bacterial H-NS, a NAP that suppresses the transcription of horizontally acquired genetic elements by forming a nucleoprotein filament on genomic DNA (Navarre et al., 2006; Oshima et al., 2006; Higashi et al., 2016). Considering the functional similarity between H-NS and TrmBL2 and the fact that HGT occurs between bacteria and Archaea (López-García et al., 2015; Wagner et al., 2017), TrmBL2 might also function in suppressing the expression of genes acquired by horizontal transfer in Archaea (Maruyama et al., 2019). Proteins homologous to TrmBL2 have been found in Archaea other than members of the Thermococcales, as well as in bacteria (Maruyama et al., 2011). The discovery of Pcal_1183, a TF that belongs to the AsnC/Lrp family, as an abundant chromosomal protein in P. calidifontis, adds another candidate TF-like chromosomal protein to Archaea. Searching for abundant chromosomal proteins with the method used in the present study would facilitate the identification of novel TF-like nucleoid proteins possibly involved in chromosomal folding or the suppression of horizontally transferred genes both in bacteria and Archaea in the future.
Conclusion
Although the chromosomes of Euryarchaeota and Crenarchaeota are commonly folded into higher-order structures, their structural units are fundamentally different. Specifically, DNA wrappers such as histones or HTa are necessary for Euryarchaeota, whereas DNA wrapping as a primary DNA folding is missing in Crenarchaeota (Figures 7A,B). Both Euryarchaeota and Crenarchaeota use Alba, which basically functions as a DNA bridger. The function of Alba might be different depending on the interaction with other lineage-specific proteins such as histone or HTa in Euryarchaeota, or Sul7 or Cren7 in Crenarchaeota. TF-like proteins are found in both Euryarchaeota and Crenarchaeota. Possible functions of such TF-like proteins might include architectural roles as well as the suppression of horizontally transferred genetic elements (Maruyama et al., 2019). Further, the interplay between the chromosomal proteins, as well as differential expression and post-translational modifications, might contribute to higher-order folding and regulation of the archaeal chromosomes (Figure 7A).
We also propose that membrane-associated proteins might play roles in chromosome folding and regulation. In the present study, membrane-associated proteins, together with proteins involved in translation, transcription, and replication, were detected by mass spectrometry of proteins associated with isolated archaeal chromosomes. The association between membrane-bound proteins and chromosomes has been proposed in both eukaryotes and bacteria (Maruyama et al., 2014; Chang et al., 2015; Monterroso et al., 2019). In eukaryotes, the linker of nucleoskeleton and cytoskeleton (LINC) complex has been shown to interconnect the chromosome, nuclear membrane, and cytoskeletal filaments, performing diverse functions, including mechanotransduction and meiotic chromosome movements in mice, yeast, and nematodes (Hiraoka and Dernburg, 2009; Sato et al., 2009; Chang et al., 2015). In E. coli, the actin-like protein MreB, a member of a membrane-bound complex (MreBCD), is associated with the nucleoid, playing a role in bacterial chromosome segregation (Thanbichler et al., 2005). It can be inferred that archaeal membrane-associated proteins are similarly involved in chromosome segregation. The present findings suggest that membrane-associated proteins might be involved in genome architecture and dynamics in all three domains of life.
Materials and Methods
Strain and Growth Conditions
Thermoplasma acidophilum strain DSM 1728 was grown at 58°C under aerobic conditions in modified Allan’s basal salt medium [in 1 L: 0.2 g (NH4)2SO4, 3.0 g KH2PO4, 0.5 g MgSO4⋅7H2O, 0.25 g CaCl2⋅2 H2O] adjusted to pH 1.5 with H2SO4 and supplemented with 0.1% yeast extract and 1% glucose (Searcy, 1975). Pyrobaculum calidifontis strain VA1 was cultivated in the TY medium containing 0.3% sodium thiosulfate pentahydrate at 90°C (Pelve et al., 2012). Thermococcus kodakarensis strain KOD1 was cultured as previously described (Maruyama et al., 2011). Sulfolobus solfataricus strain P1 was cultured at 80°C in S. solfataricus medium 171 specified by the Japan Collection of Mircoorganisms (https://jcm.brc.riken.jp/en/).
On-Substrate Lysis
On-substrate lysis of archaeal cells was performed as previously described (Maruyama et al., 2011; Ohniwa et al., 2018) with slight modifications. T. acidophilum and P. calidifontis cells at the log and stationary phases were harvested, washed, and resuspended to a final density of approximately 0.5 OD600. The cell suspension was then applied to a round coverslip (15-mm round; Matsunami Glass, Ind., Kishiwada, Japan) and dried with nitrogen gas. The cells were lysed with the addition of lysis buffer (10 mM tris pH 6.8, 10 mM EDTA, 0.5% Triton X-100) onto the coverslip and then dried again with nitrogen gas. To achieve sufficient cell lysis, the detergent Triton X-100 was added at the same concentration used for chromosome isolation (see below). To assess the extent of chromosome spreading, the samples were stained with DAPI and imaged using fluorescence microscopy. AFM was used to analyze the detailed structure of the chromosomes.
Atomic Force Microscopy
Samples were imaged in the air with the tapping mode of Nanoscope IIIa or IV (Digital Instruments, Santa Barbara, CA, United States) using an OMCL AC160TS probe (Olympus, Tokyo, Japan). Images were captured in a 512 × 512-pixel format, plane-fitted, and flattened using the computer program supplied in the imaging module. Measurements were made, and correction for the “tip-effect” was performed accordingly (Maruyama et al., 2011). Diameters of the chromosomal structures (fibers or particles) were calculated based on the measured bottom-to-bottom distance (apparent measured distance) using the previously described equation S = 0.75W - 16.4, where S is the corrected width of the sample and W is the apparent measured distance (Ohniwa et al., 2007). The contour DNA lengths were measured manually using Nanoscope software; DNA paths were divided into short straight lines and their lengths were summed. When fibers or beads-like structures were encountered, the center of the fiber or beads was used as the DNA path. In this way, the apparent length of DNA wrapped around a protein particle is shortened. When the DNA pathway was not clear, it was excluded from the measurement.
Isolation of Archaeal Chromosome
Chromosomes from T. acidophilum, P. calidifontis, T. kodakarensis, and S. solfataricus were obtained as described previously with some modifications (Maruyama et al., 2011). Cells were harvested at log or stationary phase and lysed for 10 min at 4°C with extraction buffer (25 mM HEPES, pH 7.0, 15 mM MgCl2, 100 mM NaCl, 0.4M sorbitol, 0.5% Triton X-100). The lysate was then layered on top of a pre-cooled linear sucrose gradient (15–45% sucrose) in 10 mM tris pH 6.8 and 3 mM Mg(CH3COO)2. The samples were centrifuged in a swinging-bucket MLS-50 rotor (Beckman Coulter, Brea, CA, United States) at 10,000 × g and 4°C for 20 min. The DNA-enriched material, visible as an opalescent band, was collected, washed with MNase buffer (20 mM Tris pH 8.0, 5 mM NaCl, 2.5 mM CaCl2), and stored at −20°C until further use.
MNase Digestion of Archaeal Chromosome
T. acidophilum, P. calidifontis, T. kodakarensis, and S. solfataricus genomic DNA was incubated with MNase (Worthington Biochemical, Lakewood, NJ, United States) at various concentrations (0.3, 1, 3, 10 and 30 U MNase/100 μl) for 20 min at 37°C. The digestion reaction was stopped by adding stop solution (100 mM EDTA, 100 mM EGTA, 2% SDS). Chromosomal fragments were then purified with phenol/chloroform extraction and ethanol precipitation. The DNA pellet was dissolved in TE buffer (10 mM tris pH 7.5, 1 mM EDTA), resolved with a 2.5% agarose gel, and visualized with ethidium bromide staining.
Mass Spectrometry
Chromosomes were isolated from cells in the stationary phase as described previously herein, digested with MNase (Worthington Biochemical, Corp., Lakewood, NJ, United States), and run on a 15% SDS-PAGE gel to separate proteins associated with archaeal chromosomes. After staining with Coomassie Brilliant Blue, visible protein bands were excised from the gel and processed for liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS) as described previously (Maruyama et al., 2011). Top hits of each band are summarized in Tables 1–3.
Expression and Purification of Recombinant Proteins
Genes encoding the protein Alba from T. acidophilum and P. calidifontis and HTa from T. acidophilum were PCR-amplified from their respective genomic DNA template and inserted into a pET16b vector containing 10X histidine tag at the N-terminus (Novagen, Madison, WI, United States). The following primers were used: for T. acidophilum Alba, forward 5′-ATCGCCATG GCAGAGGAGAACATAATCTTTG-3′ and reverse 5′-ATCG GGATCCTCAACGTGACAGC-3′; for P. calidifontis Alba, forward 5′-ATCGCCATGGCGACAGAACAGACAATAC-3′ and reverse 5′-ATCGGATCCTTAGGCCATTTCGAGCACT-3′; for T. acidophilum HTa, forward 5′-ATCGCATATGTAGGAATC AGTGAGCTAT-3′ and reverse 5′-ATCGGGATCCTTACTGC TGGTATTTTATCTTGC-3′. The Alba gene was inserted using the NcoI and BamHI restriction sites, resulting in the removal of the his-tag. HTa gene was inserted via the NdeI and BamHI sites and thus tagged with histidine. Competent E. coli BL21-CodonPlus (DE3)-RIL (Agilent Technologies, Santa Clara, CA, United States) was transformed with the resulting constructs. Protein expression was induced via the addition of IPTG. Recombinant proteins were then extracted and purified. Alba proteins were run on an ion-exchange column (HiTrap SP column, 1 ml, GE Healthcare UK, Ltd., Buckinghamshire, United Kingdom) following the manufacturer’s protocol and as described previously (Maruyama et al., 2011). The His-tagged HTa was purified with Ni-NTA agarose beads (Qiagen Hilden, Germany) following the manufacturer’s protocol. Histones from T. kodakarensis were expressed and purified as previously described (Maruyama et al., 2011).
In vitro Reconstitution
The plasmid pBlueScriptII (Agilent Technologies, Inc., Santa Clara, CA, United States) was digested with Hind III restriction enzyme digestion, resulting in a 2961-bp linearized double-stranded DNA. Each recombinant protein was mixed with the 3-kb linearized DNA at various protein-to-DNA weight ratios in 10 mM tris pH 6.8 and 200 mM NaCl. In each experiment, DNA and the protein were mixed in a 20 μl volume. The DNA concentration was fixed to 5 ng/μl (100 ng total). Protein was included at a concentration that achieves the desired protein:DNA weight ratio. The mixture was heated for 10 min at appropriate temperatures, 58°C for T. acidophilum and 90°C for P. calidifontis, incubated for 10 min at 25°C, and then processed for AFM imaging. For AFM observation, the samples were diluted 1:20 with AFM fixation buffer (10 mM tris pH 8.0, 5 mM NaCl, 0.3% glutaraldehyde) and incubated for 30 min at 25°C. The protein–DNA complexes were then deposited onto a freshly cleaved mica surface pretreated with 10 mM spermidine. After 10 min, the mica was washed with 1 ml of pure water, dried with nitrogen gas, and observed with AFM. Note that the theoretical length of a 2961-bp dsDNA in B-form is ∼1007 nm (0.34 nm/bp). The theoretical length is indicated in the histograms as a dashed line. Reconstitution experiments using a combination of different chromosomal proteins were also performed by mixing the 3-kb linearized DNA with the protein pair of interest at specified amounts. In this case, the protein weight ratio are indicated relative to DNA (e.g., 2:6 HTa:Alba means 1:2:6 DNA:HTa:Alba weight ratio).
Data Availability Statement
All datasets generated for this study are included in the article/supplementary material.
Author Contributions
HM and KT conceived and designed the study. EP performed experiments, analyzed data, and wrote the first draft of the manuscript. TN, CM, and TO wrote sections of the manuscript and gave critical comments on the manuscript. HM, EP, HA, and KT revised the manuscript. All authors read and approved the final version of the manuscript.
Funding
This work was supported by a JSPS KAKENHI Grant-in-Aid for Young Scientist (B), Grant numbers 26850057 and 17K15254 to HM.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Mr. Yuzo Watanabe (Proteomics Facility, Graduate School of Biostudies, Kyoto University) and Dr. Ryosuke Ohniwa (Faculty of Medicine, University of Tsukuba) for mass spectrometry assistance and Editage (www.editage.com) for English editing.
References
Ahmad, M. U. D., Waege, I., Hausner, W., Thomm, M., Boos, W., Diederichs, K., et al. (2015). Structural insights into nonspecific binding of DNA by TrmBL2, an archaeal chromatin protein. J. Mol. Biol. 427, 3216–3229. doi: 10.1016/j.jmb.2015.08.012
Ali Azam, T., Iwata, A., Nishimura, A., Ueda, S., and Ishihama, A. (1999). Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J. Bacteriol. 181, 6361–6370. doi: 10.1128/jb.181.20.6361-6370.1999
Amo, T., Paje, M. L. F., Inagaki, A., Ezaki, S., Atomi, H., and Imanaka, T. (2002). Pyrobaculum calidifontis sp. nov., a novel hyperthermophilic archaeon that grows in atmospheric air. Archaea 1, 113–121. doi: 10.1155/2002/616075
Badrinarayanan, A., Le, T. B., and Laub, M. T. (2015). Bacterial chromosome organization and segregation. Annu. Rev. Cell Dev. Biol. 31, 171–199.
Barutcu, A. R., Lian, J. B., Stein, J. L., Stein, G. S., and Imbalzano, A. N. (2017). The connection between BRG1, CTCF and topoisomerases at TAD boundaries. Nucleus 8, 150–155. doi: 10.1080/19491034.2016.1276145
Bates, S. T., Berg-Lyons, D., Caporaso, J. G., Walters, W. A., Knight, R., and Fierer, N. (2011). Examining the global distribution of dominant archaeal populations in soil. ISME J. 5, 908–917. doi: 10.1038/ismej.2010.171
Bell, S. D., Botting, C. H., Wardleworth, B. N., Jackson, S. P., and White, M. F. (2002). The interaction of Alba, a conserved archaeal chromatin protein, with Sir2 and its regulation by acetylation. Science 296, 148–151. doi: 10.1126/science.1070506
Blais Lecours, P., Marsolais, D., Cormier, Y., Berberi, M., Haché, C., Bourdages, R., et al. (2014). Increased prevalence of methanosphaera stadtmanae in inflammatory bowel diseases. PLoS One 9:e87734. doi: 10.1371/journal.pone.0087734
Brocken, D. J. W., Tark-Dame, M., and Dame, R. T. (2018). The organization of bacterial genomes: towards understanding the interplay between structure and function. Curr. Opin. Syst. Biol. 8, 137–143. doi: 10.1016/j.coisb.2018.02.007
Cao, J., Wang, Q., Liu, T., Peng, N., and Huang, L. (2018). Insights into the post-translational modifications of archaeal Sis10b (Alba): lysine-16 is methylated, not acetylated, and this does not regulate transcription or growth. Mol. Microbiol. 109, 192–208. doi: 10.1111/mmi.13973
Chang, W., Worman, H. J., and Gundersen, G. G. (2015). Accessorizing and anchoring the LINC complex for multifunctionality. J. Cell Biol. 208, 11–22. doi: 10.1083/jcb.201409047
Čuboňová, L., Katano, M., Kanai, T., Atomi, H., Reeve, J. N., and Santangelo, T. J. (2012). An archaeal histone is required for transformation of Thermococcus kodakarensis. J. Bacteriol. 194, 6864–6874. doi: 10.1128/jb.01523-12
Dame, R. T., and Tark-Dame, M. (2016). Bacterial chromatin: converging views at different scales. Curr. Opin. Cell Biol. 40, 60–65. doi: 10.1016/j.ceb.2016.02.015
Davey, C. A., Sargent, D. F., Luger, K., Maeder, A. W., and Richmond, T. J. (2002). Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 Å resolution. J. Mol. Biol. 319, 1097–1113. doi: 10.1016/s0022-2836(02)00386-8
De Vries, R. (2010). DNA condensation in bacteria: Interplay between macromolecular crowding and nucleoid proteins. Biochimie 92, 1715–1721. doi: 10.1016/j.biochi.2010.06.024
DeLange, R. J., Green, G., and Searcy, D. (1981). A histone-like protein (HTa) from Thermoplasma acidophilum. I. purification and properties. J. Biol. Chem. 256, 900–904.
Driessen, R. P. C., and Dame, R. T. (2011). Nucleoid-associated proteins in Crenarchaea. Biochem. Soc. Trans. 39, 116–121. doi: 10.1042/bst0390116
Efremov, A. K., Qu, Y., Maruyama, H., Lim, C. J., Takeyasu, K., and Yan, J. (2015). Transcriptional repressor TrmBL2 from Thermococcus kodakarensis forms filamentous nucleoprotein structures and competes with histones for DNA binding in a salt- and DNA supercoiling-dependent manner. J. Biol. Chem. 290, 15770–15784. doi: 10.1074/jbc.M114.626705
French, S. L., Santangelo, T. J., Beyer, A. L., and Reeve, J. N. (2007). Transcription and translation are coupled in Archaea. Mol. Biol. Evol. 24, 893–895. doi: 10.1093/molbev/msm007
Fyodorov, D. V., Zhou, B. R., Skoultchi, A. I., and Bai, Y. (2018). Emerging roles of linker histones in regulating chromatin structure and function. Nat. Rev. Mol. Cell Biol. 19, 192–206. doi: 10.1038/nrm.2017.94
Gibcus, J. H., and Dekker, J. (2013). The hierarchy of the 3D genome. Mol. Cell 49, 773–782. doi: 10.1016/j.molcel.2013.02.011
Gindner, A., Hausner, W., and Thomm, M. (2014). The TrmB family: a versatile group of transcriptional regulators in Archaea. Extremophiles 18, 925–936. doi: 10.1007/s00792-014-0677-672
Grewal, S. I., and Moazed, D. (2003). Heterochromatin and epigenetic control of gene expression. Science 301, 798–802. doi: 10.1126/science.1086887
Guo, R., Xue, H., and Huang, L. (2003). Ssh10b, a conserved thermophilic archaeal protein, binds RNA in vivo. Mol. Microbiol. 50, 1605–1615. doi: 10.1046/j.1365-2958.2003.03793.x
Hammel, M., Amlanjyoti, D., Reyes, F. E., Chen, J.-H., Parpana, R., Tang, H. Y., et al. (2016). HU multimerization shift controls nucleoid compaction. Sci. Adv. 2:e1600650. doi: 10.1126/sciadv.1600650
Henneman, B., and Dame, R. T. (2015). Archaeal histones: dynamic and versatile genome architects. AIMS Microbiol. 1, 72–81. doi: 10.3934/microbiol.2015.1.72
Henneman, B., van Emmerik, C., van Ingen, H., and Dame, R. T. (2018). Structure and function of archaeal histones. PLoS Genet. 14:e1007582. doi: 10.1371/journal.pgen.1007582
Higashi, K., Tobe, T., Kanai, A., Uyar, E., Ishikawa, S., Suzuki, Y., et al. (2016). H-NS facilitates sequence diversification of horizontally transferred DNAs during their integration in host chromosomes. PLoS Genet. 12:e1005796. doi: 10.1371/journal.pgen.1005796
Hirano, T., Kobayashi, R., and Hirano, M. (1997). Condensins, chromosome condensation protein complexes containing XCAP-C, XCAP-E and a Xenopus homolog of the Drosophila Barren protein. Cell 89, 511–521. doi: 10.1016/s0092-8674(00)80233-0
Hiraoka, Y., and Dernburg, A. F. (2009). The SUN rises on meiotic chromosome dynamics. Dev. Cell 17, 598–605. doi: 10.1016/j.devcel.2009.10.014
Hizume, K., Araki, S., Yoshikawa, K., and Takeyasu, K. (2007). Topoisomerase II, scaffold component, promotes chromatin compaction in vitro in a linker-histone H1-dependent manner. Nucleic Acids Res. 35, 2787–2799. doi: 10.1093/nar/gkm116
Hizume, K., Yoshimura, S. H., and Takeyasu, K. (2005). Linker histone H1 per se can induce three-dimensional folding of chromatin fiber. Biochemistry 44, 12978–12989. doi: 10.1021/bi050623v
Hocher, A., Rojec, M., Swadling, J. B., Esin, A., and Warnecke, T. (2019). The DNA-binding protein HTa from Thermoplasma acidophilum is an archaeal histone analog. eLife 8:e052542. doi: 10.7554/eLife.52542
Japaridze, A., Renevey, S., Sobetzko, P., Stoliar, L., Nasser, W., Dietler, G., et al. (2017). Spatial organization of DNA sequences directs the assembly of bacterial chromatin by a nucleoid-associated protein. J. Biol. Chem. 292, 7607–7618. doi: 10.1074/jbc.m117.780239
Kim, J., Yoshimura, S. H., Hizume, K., Ohniwa, R. L., Ishihama, A., and Takeyasu, K. (2004). Fundamental structural units of the Escherichia coli nucleoid revealed by atomic force microscopy. Nucleic Acids Res. 32, 1982–1992. doi: 10.1093/nar/gkh512
Kim, M., Park, S., and Lee, S.-J. (2016). Global transcriptional regulator TrmB family members in prokaryotes. J. Microbiol. 54, 639–645. doi: 10.1007/s12275-016-6362-7
Koskinen, K., Pausan, M. R., Perras, A. K., Beck, M., Bang, C., Mora, M., et al. (2017). First insights into the diverse human archaeome: specific detection of archaea in the gastrointestinal tract, lung, and nose and on skin. mBio 8:e824-17. doi: 10.1128/mBio.00824-817
Laurens, N., Driessen, R. P., Heller, I., Vorselen, D., Noom, M. C., Hol, F. J., et al. (2012). Alba shapes the archaeal genome using a delicate balance of bridging and stiffening the DNA. Nat. Commun. 3:1328.
Lemmens, L., Maklad, H. R., Bervoets, I., and Peeters, E. (2019). Transcription regulators in archaea: homologies and differences with bacterial regulators. J. Mol. Biol. 431, 4132–4146. doi: 10.1016/j.jmb.2019.05.045
Lepp, P. W., Brinig, M. M., Ouverney, C. C., Palm, K., Armitage, G. C., and Relman, D. A. (2004). Methanogenic Archaea and human periodontal disease. Proc. Natl. Acad. Sci. U.S.A. 101, 6176–6181. doi: 10.1073/pnas.0308766101
Li, G., and Reinberg, D. (2011). Chromatin higher-order structures and gene regulation. Curr. Opin. Genet. Dev. 21, 175–186. doi: 10.1016/j.gde.2011.01.022
Lioy, V. S., Cournac, A., Marbouty, M., Duigou, S., Mozziconacci, J., Espéli, O., et al. (2018). Multiscale structuring of the E. coli chromosome by nucleoid-associated and condensin proteins. Cell 172, 771–783. doi: 10.1016/j.cell.2017.12.027
Lloyd, K. G., Schreiber, L., Petersen, D. G., Kjeldsen, K. U., Lever, M. A., Steen, A. D., et al. (2013). Predominant archaea in marine sediments degrade detrital proteins. Nature 496, 215–218. doi: 10.1038/nature12033
López-García, P., Zivanovic, Y., Deschamps, P., and Moreira, D. (2015). Bacterial gene import and mesophilic adaptation in archaea. Nat. Rev. Microbiol. 13, 447–456. doi: 10.1038/nrmicro3485
Luger, K., Mader, A. W., Richmond, R. K., Sargent, D. F., and Richmond, T. J. (1997). Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389, 251–260. doi: 10.1038/38444
Luijsterburg, M. S., White, M. F., van Driel, R., and Dame, R. T. (2008). The major architects of chromatin: architectural proteins in bacteria, archaea and eukaryotes. Crit. Rev. Biochem. Mol. Biol. 43, 393–418. doi: 10.1080/10409230802528488
Marbouty, M., Le Gall, A., Cattoni, D. I., Cournac, A., Koh, A., Fiche, J.-B., et al. (2015). Condensin-and replication-mediated bacterial chromosome folding and origin condensation revealed by Hi-C and super-resolution imaging. Mol. Cell 59, 588–602. doi: 10.1016/j.molcel.2015.07.020
Maruyama, H., Harwood, J. C., Moore, K. M., Paszkiewicz, K., Durley, S. C., Fukushima, H., et al. (2013). An alternative beads-on-a-string chromatin architecture in Thermococcus kodakarensis. EMBO Rep. 14, 711–717. doi: 10.1038/embor.2013.94
Maruyama, H., Kent, N. A., Nishida, H., and Oshima, T. (2019). “Functions of archaeal nucleoid proteins: archaeal silencers are still missing,” in DNA Traffic in the Environment, eds H. Nishida and T. Oshima (Singapore: Springer), 29–45. doi: 10.1007/978-981-13-3411-5_2
Maruyama, H., Ohniwa, R. L., Prieto, E., Hejna, J., and Takeyasu, K. (2014). “Genome-folding mechanisms in the three domains of life revealed by AFM,” in Atomic Force Microscopy in Nanobaiology, ed. K. Takeyasu (New York, NY: Pan Stanford Publishing), 275–310. doi: 10.1201/b15671-15
Maruyama, H., Shin, M., Oda, T., Matsumi, R., Ohniwa, R. L., Itoh, T., et al. (2011). Histone and TK0471/TrmBL2 form a novel heterogeneous genome architecture in the hyperthermophilic archaeon Thermococcus kodakarensis. Mol. Biol. Cell 22, 386–398. doi: 10.1091/mbc.E10-08-0668
Mattiroli, F., Bhattacharyya, S., Dyer, P. N., White, A. E., Sandman, K., Burkhart, B. W., et al. (2017). Structure of histone-based chromatin in Archaea. Science 357, 609–612. doi: 10.1126/science.aaj1849
Monterroso, B., Zorrilla, S., Sobrinos-Sanguino, M., Robles-Ramos, M. Á, Alfonso, C., Soderstrom, B., et al. (2019). The bacterial DNA binding protein MatP involved in linking the nucleoid terminal domain to the divisome at midcell interacts with lipid membranes. mBio 10, e376–e319.
Morikawa, K., Ushijima, Y., Ohniwa, R. L., Miyakoshi, M., and Takeyasu, K. (2019). What happens in the staphylococcal nucleoid under oxidative stress? Microorganisms 7:631. doi: 10.3390/microorganisms7120631
Navarre, W. W., Porwollik, S., Wang, Y., McClelland, M., Rosen, H., Libby, S. J., et al. (2006). Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science 313, 236–238. doi: 10.1126/science.1128794
Nishida, H., and Oshima, T. (2017). Archaeal histone distribution is associated with archaeal genome base composition. J. Gen. Appl. Microbiol. 63, 28–35. doi: 10.2323/jgam.2016.07.003
Niu, Y., Xia, Y., Wang, S., Li, J., Niu, C., Li, X., et al. (2013). A prototypic lysine methyltransferase 4 from archaea with degenerate sequence specificity methylates chromatin proteins Sul7d and Cren7 in different patterns. J. Biol. Chem. 288, 13728–13740. doi: 10.1074/jbc.m113.452979
Nkamga, V. D., Henrissat, B., and Drancourt, M. (2017). Archaea: essential inhabitants of the human digestive microbiota. Hum. Microb. J. 3, 1–8. doi: 10.1016/j.humic.2016.11.005
Nolivos, S., and Sherratt, D. (2014). The bacterial chromosome: architecture and action of bacterial SMC and SMC-like complexes. FEMS Microbiol. Rev. 38, 380–392. doi: 10.1111/1574-6976.12045
Ohniwa, R. L., Maruyama, H., Morikawa, K., and Takeyasu, K. (2018). Atomic force microscopy imaging and analysis of prokaryotic genome organization. Methods Mol. Biol. 1837, 147–160. doi: 10.1007/978-1-4939-8675-0_9
Ohniwa, R. L., Morikawa, K., Takeshita, S. L., Kim, J., Ohta, T., Wada, C., et al. (2007). Transcription-coupled nucleoid architecture in bacteria. Genes Cells 12, 1141–1152. doi: 10.1111/j.1365-2443.2007.01125.x
Ono, T., Losada, A., Hirano, M., Myers, M. P., Neuwald, A. F., and Hirano, T. (2003). Differential contributions of condensin I and condensin II to mitotic chromosome architecture in vertebrate cells. Cell 115, 109–121. doi: 10.1016/s0092-8674(03)00724-4
Oshima, T., Ishikawa, S., Kurokawa, K., Aiba, H., and Ogasawara, N. (2006). Escherichia coli histone-like protein H-NS preferentially binds to horizontally acquired DNA in association with RNA polymerase. DNA Res. 13, 141–153. doi: 10.1093/dnares/dsl009
Pausan, M. R., Csorba, C., Singer, G., Till, H., Schöpf, V., Santigli, E., et al. (2019). Exploring the archaeome: detection of archaeal signatures in the human body. Front. Microbiol. 10:2796. doi: 10.3389/fmicb.2019.02796
Peeters, E., Driessen, R. P., Werner, F., and Dame, R. T. (2015). The interplay between nucleoid organization and transcription in archaeal genomes. Nat. Rev. Microbiol. 13, 333–341. doi: 10.1038/nrmicro3467
Pelve, E. A., Lindås, A. C., Knöppel, A., Mira, A., and Bernander, R. (2012). Four chromosome replication origins in the archaeon Pyrobaculum calidifontis. Mol. Microb. 85, 986–995. doi: 10.1111/j.1365-2958.2012.08155.x
Prieto, E. I., and Maeshima, K. (2019). Dynamic chromatin organization in the cell. Essays Biochem. 63, 133–145. doi: 10.1042/ebc20180054
Probst, A. J., Auerbach, A. K., and Moissl-Eichinger, C. (2013). Archaea on human skin. PLoS One 8:e65388. doi: 10.1371/journal.pone.0065388
Rojec, M., Hocher, A., Stevens, K. M., Merkenschlager, M., and Warnecke, T. (2019). Chromatinization of Escherichia coli with archaeal histones. eLife 8:e049038. doi: 10.7554/eLife.49038
Rouvière-Yaniv, J., Yaniv, M., and Germond, J.-E. (1979). E. coli DNA binding protein HU forms nucleosome-like structure with circular double-stranded DNA. Cell 17, 265–274. doi: 10.1016/0092-8674(79)90152-1
Ruepp, A., Graml, W., Santos-Martinez, M.-L., Koretke, K. K., Volker, C., Mewes, H. W., et al. (2000). The genome sequence of the thermoacidophilic scavenger Thermoplasma acidophilum. Nature 407, 508–513. doi: 10.1038/35035069
Sanders, T. J., Marshall, C. J., and Santangelo, T. J. (2019). The role of archaeal chromatin in transcription. J. Mol. Biol. 431, 4103–4115. doi: 10.1016/j.jmb.2019.05.006
Sandman, K., and Reeve, J. N. (2005). Archaeal chromatin proteins: different structures but common function? Curr. Opin. Microbiol. 8, 656–661. doi: 10.1016/j.mib.2005.10.007
Sarkar, T., Petrov, A. S., Vitko, J. R., Santai, C. T., Harvey, S. C., Mukerji, I., et al. (2009). Integration host factor (IHF) dictates the structure of polyamine-DNA condensates: implications for the role of IHF in the compaction of bacterial chromatin. Biochemistry 48, 667–675. doi: 10.1021/bi8019965
Sato, A., Isaac, B., Phillips, C. M., Rillo, R., Carlton, P. M., Wynne, D. J., et al. (2009). Cytoskeletal forces span the nuclear envelope to coordinate meiotic chromosome pairing and synapsis. Cell 139, 907–919. doi: 10.1016/j.cell.2009.10.039
Searcy, D. G. (1975). Histone-like protein in the prokaryote Thermoplasma acidophilum. Biochim. Biophys. Acta 395, 535–547. doi: 10.1016/0005-2787(75)90076-3
Searcy, D. G., and Stein, D. B. (1980). Nucleoprotein subunit structure in an unusual prokaryotic organism: Thermoplasma acidophilum. Biochim. Biophys. Acta 609, 180–195. doi: 10.1016/0005-2787(80)90211-7
Skoko, D., Yoo, D., Bai, H., Schnurr, B., Yan, J., McLeod, S. M., et al. (2006). Mechanism of chromosome compaction and looping by the Escherichia coli nucleoid protein Fis. J. Mol. Biol. 364, 777–798. doi: 10.1016/j.jmb.2006.09.043
Spang, A., Hatzenpichler, R., Brochier-Armanet, C., Rattei, T., Tischler, P., Spieck, E., et al. (2010). Distinct gene set in two different lineages of ammonia-oxidizing archaea supports the phylum Thaumarchaeota. Trends Microbiol. 18, 331–340. doi: 10.1016/j.tim.2010.06.003
Takemata, N., Samson, R. Y., and Bell, S. D. (2019). Physical and functional compartmentalization of archaeal chromosomes. Cell 179, 165–179. doi: 10.1016/j.cell.2019.08.036
Thanbichler, M., Wang, S. C., and Shapiro, L. (2005). The bacterial nucleoid: a highly organized and dynamic structure. J. Cell. Biochem. 96, 506–521. doi: 10.1002/jcb.20519
The UniProt Consortium (2019). UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 47, D506–D515. doi: 10.1093/nar/gky1049
Thomas, J. O. (1984). The higher order structure of chromatin and histone H1. J. Cell Sci. Suppl. 1(Suppl. 1), 1–20. doi: 10.1242/jcs.1984.supplement_1.1
Uhlmann, F. (2016). SMC complexes: from DNA to chromosomes. Nat. Rev. Mol. Cell Biol. 17, 399–412. doi: 10.1038/nrm.2016.30
Verma, S. C., Qian, Z., and Adhya, S. L. (2019). Architecture of the Escherichia coli nucleoid. PLoS Genet. 15:e1008456. doi: 10.1371/journal.pgen.1008456
Wagner, A., Whitaker, R. J., Krause, D. J., Heilers, J.-H., Van Wolferen, M., Van Der Does, C., et al. (2017). Mechanisms of gene flow in archaea. Nat. Rev. Microbiol. 15:492.
Wang, X., Le, T. B., Lajoie, B. R., Dekker, J., Laub, M. T., and Rudner, D. Z. (2015). Condensin promotes the juxtaposition of DNA flanking its loading site in Bacillus subtilis. Genes Dev. 29, 1661–1675. doi: 10.1101/gad.265876.115
Wang, X., Llopis, P. M., and Rudner, D. Z. (2013). Organization and segregation of bacterial chromosomes. Nat. Rev. Genet. 14:191. doi: 10.1038/nrg3375
Weidenbach, K., Gloer, J., Ehlers, C., Sandman, K., Reeve, J. N., and Schmitz, R. A. (2008). Deletion of the archaeal histone in Methanosarcina mazei Go1 results in reduced growth and genomic transcription. Mol. Microbiol. 67, 662–671. doi: 10.1111/j.1365-2958.2007.06076.x
Woodcock, C. L., Skoultchi, A. I., and Fan, Y. (2006). Role of linker histone in chromatin structure and function: H1 stoichiometry and nucleosome repeat length. Chromosom. Res. 14, 17–25. doi: 10.1007/s10577-005-1024-3
Keywords: archaea, higher-order chromosome structure, nucleoid associated proteins (NAPs), chromatin, histone, structural maintenance of chromosomes (SMC) proteins, horizontal gene transfer (HGT), atomic force microscopy (AFM)
Citation: Maruyama H, Prieto EI, Nambu T, Mashimo C, Kashiwagi K, Okinaga T, Atomi H and Takeyasu K (2020) Different Proteins Mediate Step-Wise Chromosome Architectures in Thermoplasma acidophilum and Pyrobaculum calidifontis. Front. Microbiol. 11:1247. doi: 10.3389/fmicb.2020.01247
Received: 13 March 2020; Accepted: 15 May 2020;
Published: 12 June 2020.
Edited by:
Eveline Peeters, Vrije University Brussel, BelgiumReviewed by:
Cynthia Haseltine, Washington State University, United StatesRemus T. Dame, Leiden University, Netherlands
Daniela Barilla‘, University of York, United Kingdom
Copyright © 2020 Maruyama, Prieto, Nambu, Mashimo, Kashiwagi, Okinaga, Atomi and Takeyasu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hugo Maruyama, maruyama@cc.osaka-dent.ac.jp; maruyama.hugo@gmail.com
†These authors have contributed equally to this work
 Hugo Maruyama
Hugo Maruyama Eloise I. Prieto
Eloise I. Prieto Takayuki Nambu1
Takayuki Nambu1 Haruyuki Atomi
Haruyuki Atomi Kunio Takeyasu
Kunio Takeyasu