- Wageningen University and Research (WUR), Wageningen Research (WR), Wageningen, Netherlands
Escherichia coli strains carrying Shiga toxins 1 and 2 (stx1 and stx2), intimin (eae), and hemolysin (ehxA) production genes were found in grass shoot, rhizosphere soil, and stable manure samples from a small-scale cattle farm located at the center of Netherlands, using cultivation-dependent and -independent microbiological detection techniques. Pasture land with grazing heifers in the first year of sampling in 2014 and without grazing cattle in 2015 was physically separated from the stable that housed rose calves during both years. Manure from the stable was applied to pasture via injection into soil once per year in early spring. Among a variety of 35 phylogenetic distinctly related E. coli strains, one large group consisting of 21 closely resembling E. coli O150:H2 (18), O98:H21 (2), and O84:H2 (1) strains, all belonging to phylogenetic group B1 and carrying all screened virulence traits, was found present on grass shoots (10), rhizosphere soil (3), and stable manure (8) in 2014, but not anymore in 2015 when grazing heifers were absent. Presence and absence of these strains, obtained via enrichments, were confirmed via molecular detection using PCR-NALFIA in all ecosystems in both years. We propose that this group of Shiga toxin-producing E. coli phylogenetic group B1 strains was originally introduced via stable manure injection into the pasture. Upon grazing, these potential pathogens proliferated in the intestinal track systems of the heifers resulting in defecation with higher loads of the STEC strain onto the grass cover. The STEC strain was further smeared over the field via the hooves of the heifers resulting in augmentation of the potential pathogen in the pasture in 2014, whereas in 2015, in the absence of heifers, no augmentation occurred and only a more diverse group of potentially mild virulent E. coli phylogenetic group A and B1 strains, indigenous to pasture plants, remained present. Via this model, it was postulated that human pathogens can circulate between plants and farm animals, using the plant as an alternative ecosystem. These data indicate that grazed pasture must be considered as a potential carrier of human pathogenic E. coli strains and possibly also of other pathogens.
Introduction
Human pathogenic bacteria such as Shiga toxin-producing Escherichia coli (STEC) strains can circulate across the different ecosystems relevant in plant production (van Overbeek et al., 2014). From there, these pathogens can enter the human food production chain, either directly via colonization of freshly consumable plants (Franz et al., 2008; Deering et al., 2012; van der Linden et al., 2013) or indirectly via colonization of plants that are used as animal feed in livestock production (Dodd et al., 2003; Williams et al., 2007). It seems likely that these pathogens circulate between plant and animal ecosystems via grazing, intestinal track colonization, excretion, and plant colonization.
Human pathogens often are transmitted to plant production systems via animal excrements, and the transmission route via manure application to soil may be the best studied contamination route of these pathogens to crop plants (van Overbeek et al., 2014). However, excrements of wild animals and (migratory) birds also may be held responsible for contamination of crop plants, either via droppings in production fields or via contamination of open surface water bodies that are used for irrigation (Atwill et al., 2015). Plants must be considered as alternative environments for human pathogens that are used as vectors for digestive track colonization of animals and humans via ingestion of plant-derived feed and food (Teplitski and de Moreas, 2018). Considered from the perspective of the pathogen, persistence in alternative environments such as plants is advantageous for eventual later transmission to intestinal track systems of other warm-blooded animals (van Elsas et al., 2011; Teplitski and de Moreas, 2018).
Plant contamination with human pathogens has considerable implications on human health as was demonstrated at the outbreak of the food-borne disease in Hamburg and surrounding area in 2011. The outbreak was caused by the enterohemorrhagic E. coli O104:H4 strain carrying Shiga toxin2a (stx2a) genes (Mellmann et al., 2011). Although, this strain may be considered as an exceptional agent in food-borne outbreaks, it became clear that this highly virulent strain emerged from a less virulent variant of E. coli O104:H4 that was involved in a previous outbreak (Mellmann et al., 2011; Ahmed et al., 2012). Horizontal transmission of virulence genes played an important role in the rapid evolution of the outbreak strain, and it may be questioned whether horizontal transmission of virulence genes takes place in plant production systems, especially in those that are practiced under intensive management systems (Guttierrez-Rodriguez and Adhikari, 2018). In plant production systems, microbial communities from different ecosystems, such as from soil, manure, water, and plants amalgamate, and interactions between these communities in typical microbial hotspots such as the rhizospheres of crop plants were proposed to result in higher gene exchanges between microbes originating from the different habitats in van Overbeek et al. (2014). The fact that different plasmid-borne extended beta-lactamase resistances (ESBL) were found in different enteric species from herbal plants grown in Southeast Asia (Veldman et al., 2014) can be an indication for the fact that horizontal gene transmission to E. coli does occur in the phytobiome.
Shiga toxin-producing Escherichia coli (STEC) and Salmonella enterica can colonize plants from emerging seeds (Liu et al., 2018). Later during plant growth, contamination with these pathogens can occur via agricultural handlings such as at irrigation of plants (van der Linden et al., 2013) and at manure application to soil (Franz et al., 2008). Internalization of plants has been described for both enteric pathogens (Ávila-Quezada et al., 2010; Deering et al., 2012; Scott et al., 2017). Many commonalities in gene distribution exist among endophytic enterobacterial strains highlighting the conservation of functions that overlap with enteric human pathogens (Lòpez-Fernàndez et al., 2015). The enterohemorrhagic E. coli O157:H7 strain was found to be present in the apoplast of edible plants, and binding to the plant surface via curli and biofilm formation appeared to play an important role in the initial steps of colonization of internal compartments of plants (Wright et al., 2016). From studies on plant-colonizing Salmonella enterica cells, it appeared that the mechanisms used for infection of plants and animals are conserved (Schikora et al., 2011; Wiedemann et al., 2015; de Moraes et al., 2017; Cox et al., 2018). Inside plants, the invading enteric pathogens must protect themselves from the plant immune system, and genes were shown to be differentially expressed in E. coli O157:H7 cells exposed to spinach and lettuce extracts, and genes typically involved in stress responses were higher expressed (Crozier et al., 2016). Residence of E. coli K12 cells in lettuce plants also resulted in higher resistance to oxidative stress, but also to increased chemo-attraction to lettuce leaf extracts indicating that these are important features for E. coli cells residing in plants (de los Angeles Dublan et al., 2014).
In livestock animals, the intestinal track system must be considered as an important hot spot for microbial colonization and horizontal gene transmission (van Overbeek et al., 2014). Many different E. coli serotypes were found in pigs, and after pig slaughtering, some serotypes persisted throughout the entire pork food production chain (Colello et al., 2016). Different E. coli serotypes and biotypes were also found present in cattle feces (Bettelheim et al., 2005), and especially, the diet that the animals received played an important role in a selection of different E. coli types, including the ones that pose serious threats to humans upon oral ingestion such as E. coli O157:H7 (Hovde et al., 1999; Bettelheim et al., 2005; Callaway et al., 2009; Cote et al., 2015; Berry et al., 2017). From livestock animals, gut microorganisms enter arable soils upon manure applications. Survival of E. coli was higher in cattle manure-amended soil than in cattle manure itself (Oliver et al., 2006), and manure-to-soil ratio, temperature, and competition with soil-indigenous microbes were found to be important factors influencing the survival time of E. coli O157:H7 in manure-amended soil (Jiang et al., 2002). Mutations in the stress response-controlling gene rpoS in E. coli O157:H7 appeared to be a determinant factor in survival of this species in manure-amended soil (van Hoek et al., 2013), resulting in attenuated survival. Higher rpoS mutation rates were found in human isolates of E. coli O157:H7 than in the ones originating from food and from cattle, suggesting that larger variation in survival time will exist in human than in environmental isolates. No relationship was found to exist between possession of virulence genes in E. coli O157:H7 and survival time in manure-amended soil (Franz et al., 2011). However, from the same study, it became apparent that increased oxidizing capacities of particular metabolites such propionic acid, α-ketobutyric acid, and α-hydroxybutyric acid were important factors in survival of this pathogen in manure-amended soil (Franz et al., 2011). Many factors, thus, can influence the selection of E. coli strains in plant and animal production systems, and survival time of the enteric pathogens in arable soil is an important factor for eventual later plant colonization. For over almost two decades, agroproduction systems are considered to be important risk factors in the dissemination of potentially life-threatening pathogens such as E. coli O157:H7, whereas no information is available on the roles that pastures play in the accumulation of this type of pathogens (Jones, 1999).
The aim of this study was to demonstrate that circulation of enteric human pathogens between plants and animals is realistic, using E. coli as model. For that purpose, a small-scale cattle farm located in the center of Netherlands was chosen, where in the previous year E. coli O157:H7 was found present in feces of two rose calves. In our study, we investigated the presence of STEC in grass from the pastureland with and without grazing heifers in two successive years, by making use of cultivation-independent and dependent microbiological detection techniques. Via isolation upon enrichment, STECs can be isolated from different environments (grass shoots, rhizosphere soil, and manure), and genomic comparisons can be made between obtained isolates and E. coli strains from culture collections representative of different phylogenetic groups.
Materials and Methods
Nijkerk Farm Location
A small-scale cattle farm located in the neighborhood of the village of Nijkerk, Netherlands, was selected for screening for eventual presence of E. coli O157:H7 and other Shiga toxin-producing E. coli (STEC) bacteria in pasture land and stable manure. The reason for selection of this farm was a previous finding made by the faculty of animal health sciences of the University of Utrecht, Netherlands, that E. coli O157:H7 was present in feces of two out of 50 rose calves during a survey over Dutch farms in April 2013. The farmer used pasture land for grazing by 15 heifers until 2014, whereas in a separate stable, 50 rose calves were housed (Figure 1). Heifers and calves never had direct contact with each other before and during the study. The distance between stable and pasture land was 242 me (in a direct line from the stable to the middle of the field), and there was no direct contact between stable and pasture land with the exception of one single yearly application of manure, via injection into soil, in March 2014 and March 2015.
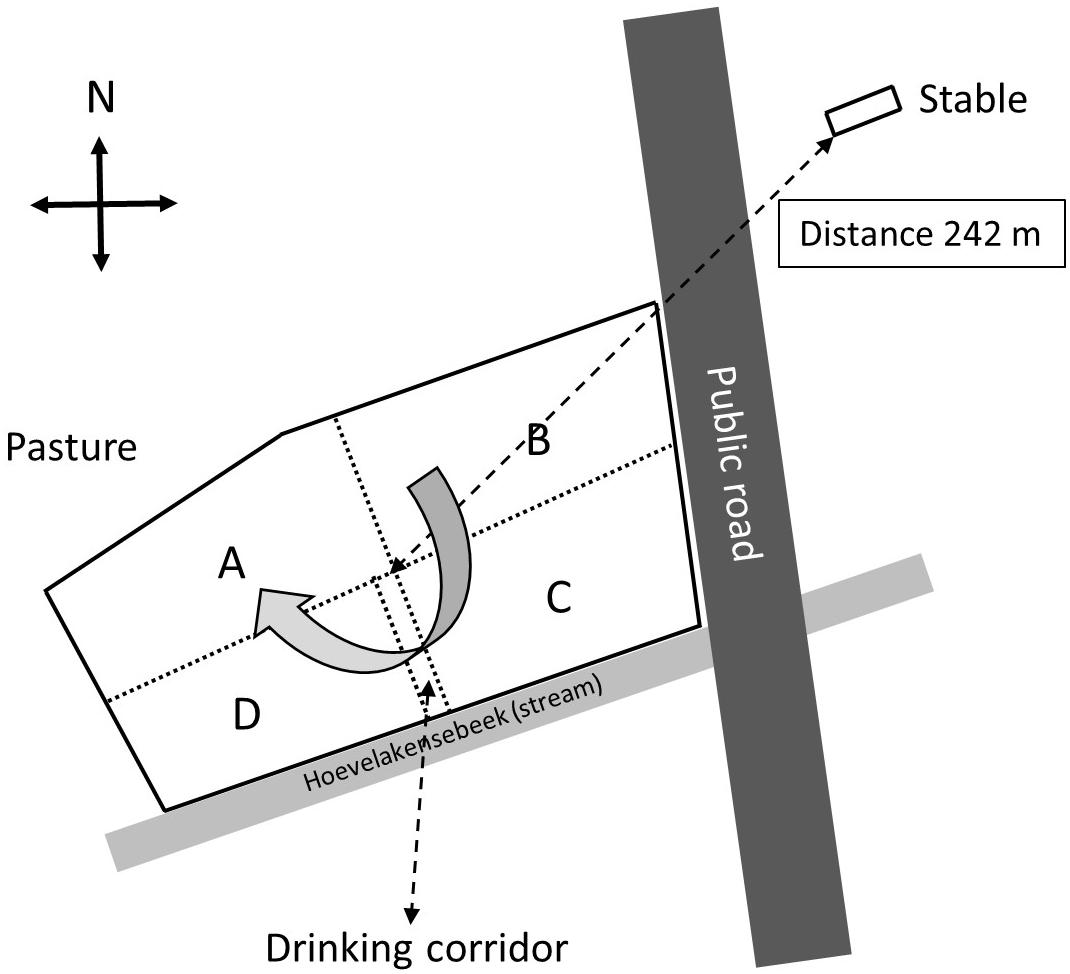
Figure 1. Schematic overview of the surveyed farm near Nijkerk including the stable with 50 calves and the pasture land with 15 heifers (grazing heifers were only present in 2014). The pasture was separated by electric wiring into four sectors (A through D) and a drinking corridor to allow the heifers to drink from a water reservoir near the stream. The heifers were weekly, clockwise, rotated to another sector with freshly grown grass, and at the moment of the first sampling (August 2014), the heifers were located in sector A.
Sampling of the Nijkerk Farm
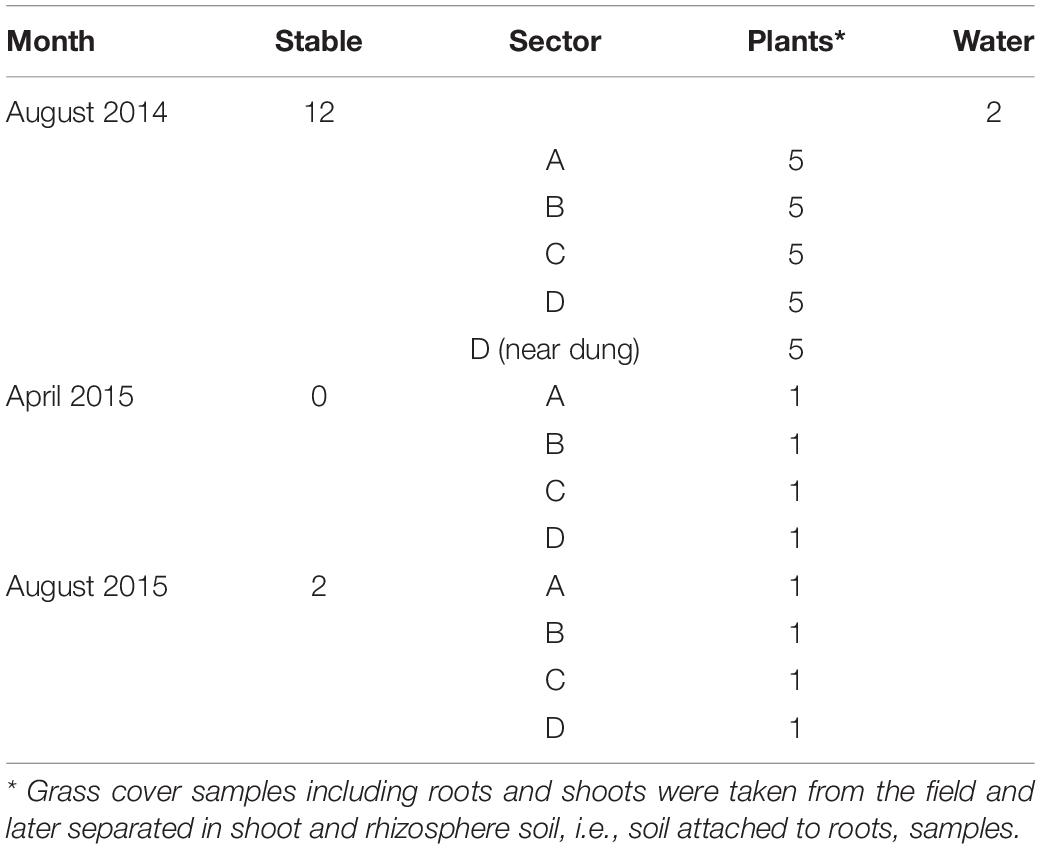
The farm was visited for sampling at three occasions in August 2014 and in April and August 2015. In 2014 and 2015, there were 50 rose calves at the age of between 6 and 7 months present in the stable. The calves were distributed over 12 compartments and per compartment samples, composed of five subsamples of 10 g of fresh manure, taken in August 2014 and stored in plastic bags (Table 1). In August 2015, 10 g of fresh manure samples were taken from two randomly selected stable compartments. The size of the grazed pasture land was 2.42 ha, and the land was separated into four sectors of about equal sizes (A through D) by electric fences (Figure 1) to allow grass plants to recover and grow for 3 weeks in the absence of grazers. In 2014, a herd of 15 heifers was present on the pasture land, whereas in 2015, the pasture land was not used for grazing anymore by heifers or any other grazers. At the moment of sampling in 2014, the heifers were located in a sector that was denoted by “A.” Based on information provided by the farmer, the presence of the heifers in the weeks before sampling was known, which was 1 week before sampling for sector D, 2 weeks before sampling for C, and 3 weeks before sampling for B. Five replicate samples were taken from each sector, and in addition, five samples were taken adjacent to separate cowpats in sector D only, resulting in a total of 25 samples in August 2014 (Table 1). The cowpats in sector D were not older than 2 weeks, and samples were taken at distances of not more than 5 cm from the cowpat where grass was still alive and green and not decolorized by ammonia emission from the cowpat. In April and August 2015, one composite sample per sector, composed of four subsamples from different sites in each sector, was taken from the pasture at both occasions. At all field sampling dates, grass cover was sampled using a soil bore (diameter of 6 cm) to a maximal depth of 8 cm. Grass shoots were separated from the roots using sterilized scissors, and roots with adhering soil were shaken by hand to remove excessive soil from the roots. Soil that remained attached to roots was considered as “rhizosphere soil.” Grass shoots and roots with rhizosphere soil were placed in plastic bags for transport to the laboratory. A corridor was created with electric fence to allow the heifers to move from their sector to a drinking place, which was adjacent to a small stream (Figure 1). The location nearby the drinking place appeared muddy because of excessive water spill and grass cover disturbance by the heifers. Because of its proximity to the drinking place, the stream was sampled for eventual presence of STECs because of the observed run-off of muddy water mixed with cow feces. Water from the stream flanking the grazed pasture land was sampled at one occasion in August 2014. At this occasion, two 50-ml water samples were taken at different locations at a distance of 10 m from each other (Table 1). All samples were kept at 4°C during transport and sample processing, and the period between sampling and sample processing never exceeded more than 4 h. Short before and during samplings in 2014 and 2015, the weather conditions were normal for the time of the year in Netherlands, and the mixed grass-clover coverage appeared normal, devoid of any yellow decolorization due to nutrient limitation or drought stress.
Sample Processing
For rhizosphere soil sample analyses, 10-g roots with adhering soil were suspended in 30 ml of 1/4 strength Ringers (Merck, Darmstadt, Germany) solution (further denoted as “Ringers solution”) in sterile 100-ml Erlenmeyer flasks with 1 g of gravel. Suspensions were shaken at 20°C for 20 min at 150 rpm. Volumes of 100 μl of the thus obtained rhizosphere soil suspensions were directly plated onto fresh CHROMagarTM O157 plates (Becton Dickinson GmbH, Heidelberg, Germany) for eventual STECcolony formation. CHROMagarTM O157 is a semi-selective medium for a broader group of E. coli strains than just E. coli O157:H7, however, most likely not supporting growth of all non-O157:H7 STECs present in complex microbial habitats (Fan et al., 2018). Only colony types resembling the color (mauve) and morphology of E. coli O157:H7 strain EDL933 (ATCC® 43895TM) on this medium were counted and eventually selected for isolation. Mauve-stained colonies from all studied habitats were presumed to be STECs, although their identities still need to be confirmed by molecular serotyping and genomic analysis for presence of stx, and other STEC-related virulence genes such as eae and ehxA genes. Plates were incubated at 37°C for 16 h, including one control plate with E. coli O157:H7 strain EDL933 streaked to purity as reference. From the same rhizosphere soil suspensions, enrichment cultures were prepared using the ISO16654: 2001 “Horizontal method for the detection of Escherichia coli O157” protocol1. For that purpose, 1 ml of rhizosphere soil suspension was added to 9 ml of modified tryptone soya broth amended with 20 μg of Novobiocin (mTSB-N) and incubated for 18 h at 41.5°C, all in accordance with the ISO 16654 protocol. After incubation, between 2 and 10 loop-full subsamples (10 μl) of the pre-enriched cultures were streaked to purity on fresh CHROMagarTM O157 plates, and plates were incubated at 37°C for 16 h after which colonies were inspected for mauve colorization. For DNA extraction, 2 ml of rhizosphere suspension from each sample were centrifuged at 7,000 × g for 1 min in an Eppendorf centrifuge and resulting pellets were immediately frozen and stored at −70°C. DNA was extracted from the frozen pellets using the PowerMag® Soil DNA isolation kit, optimized for KingFisher® application (Qiagen, Hilden, GE), according to the protocol provided by the manufacturer.
For grass shoot sample analyses, 5 g of fresh grass shoots were transferred to 12 × 14-cm size Bioreba (BIOREBA AG, Reinach, Switzerland) extraction bags containing 3 ml of Ringers solution amended with 60 μg of sodium diethyldithiocarbamate trihydrate (DIECA; Merck, Darmstadt, Germany), to prevent oxidation of the grass samples during and after homogenization. Grass shoots were homogenized by hammering for 1 min using a plastic hammer and a volume of 100 μl of filtered grass shoot homogenate per sample was used for direct plating onto CHROMagarTM O157 plates, for STEC colony recovery, whereas a volume of 1 ml per sample was added to 9 ml of mTSB-N for STEC enrichment. Further incubation and processing steps for STEC colony growth were identical as described for rhizosphere soil sample analysis. For DNA extraction, 15 g of grass shoot samples were first freeze dried, resulting in dry weight samples of approximately 0.75 g, followed by grinding. Subsamples of 0.25 g of dried and ground shoots were used for DNA extraction using the same PowerMag® Soil DNA isolation kit as used for rhizosphere soil DNA extraction.
For stable manure sample analyses, 10 g of stable manure per sample was suspended in 30 ml of Ringer solution, shaken at 150 rpm in 100-ml Erlenmeyer flasks at 20°C for 20 min, and 1-ml subsamples of the resulting suspensions were used for STEC enrichment according to the same procedure as described for grass shoot and rhizosphere soil sample analyses. DNA was extracted from frozen pellets (−70°C) from 2 ml of centrifuged (7,000 rpm for 1 min) manure suspensions, using the Fast DNA Stool Mini Kit (Qiagen), according to the protocol provided by the manufacturer.
For STEC enrichment from stream water samples, 1-ml volumes were added to 9 ml of mTSB-N, and mixtures were further processed as previously described for STEC enrichment from grass shoot, rhizosphere soil, and stable manure samples. Colonies, presumed to be STECs, from all enrichments from grass shoot, rhizosphere soil, stable manure, and stream water samples on CHROMagarTM O157 plates were streaked to purity on fresh CHROMagarTM O157 plates, and single colonies were grown in Luria Bertani [LB; peptone (Oxoid), 10 g; yeast extract (Oxoid), 5 g; sodium chloride, 5 g; water, 1 L] broth under shaking (180 rpm) at 37°C for 16 h. Thus, the obtained end-logarithmic phase cell cultures were stored frozen in LB broth with 20% glycerol at −70°C.
DNA Extraction From Presumptive STECs and Downstream Molecular Analyses of Genomic and Environmental DNA Extracts
DNA was extracted from end-logarithmic phase cell cultures of all presumptive STECs using a “chelex DNA purification” method. For that purpose, 300 μl of cell culture was thoroughly mixed with chelex-100 resin, 50–100 mesh (BioRad) and heated to 100°C for 11 min and then cooled down to 4°C. After precipitation of the chelex resin, the cell lysate, containing between 10 and 25 ng per μl of DNA, was used for downstream analysis using PCR-nucleic acid lateral flow immunoassay (NALFIA; see below). For whole genome DNA sequencing, DNA was extracted from end-logarithmic cell cultures using the “Wizard magnetic DNA purification kit for food” (Promega), resulting in DNA yields of between 1 and 2 μg. DNA solutions (50 μl) containing between 1 and 1.5 μg of DNA were used for whole genome DNA sequencing performed by the Wageningen Plant Research Bioscience group using the Illumina MiSeq platform.
PCR-NALFIA, designed and evaluated for detection of STECs from cattle feces (Noguera et al., 2011), was applied. The following genes were PCR amplified: E. coli 16S rRNA (covering a broader group of species including E. coli, Escherichia vulneris, and Shigella species) (Huijsdens et al., 2002), intimin (eae), hemolysin (ehxA), and Shiga toxins 1 and 2 (respectively, stx1 and stx2). Positive PCR reactions were visualized by NALFIA using nitrocellulose strips containing colloidal carbon nanoparticles attached to neutravidin. Polyclonal antibodies against digoxigenin and the different fluorescent tags then immobilized at different positions specific for each fluorescent tag–primer system combination. Double-labeled amplicons were sandwiched between immobilized antibodies and the carbon–neutravidin conjugate, resulting in visualization of positive reactions by appearance of gray/black stripes at positions where amplicons were sandwiched (Noguera et al., 2011).
Draft genomes of 35 presumptive STECs were analyzed using the CLC genomics workbench 10 (Qiagen). First, phylogenetic relatedness between the 35 presumptive STECs obtained in this study with 15 E. coli and one E. fergusonii strains was determined based on DNA sequences, often cellular household genes, i.e., adenylate kinase (adk, 645 bp), class II fumarate hydratase (fumC, 1,404 bp), glycerol kinase (glpK, 1,509 bp), DNA gyrase subunit B (gyrB, 2,415 bp), 3-isopropylmalate dehydrogenase (icd, 1,092 bp), diaminopimelate decarboxylase (lysA, 1,263 bp), malate/lactate/ureidoglycolate dehydrogenase (mdh, 1,086 bp), methionine-tRNA ligase (metG, 2,034 bp), adeylosuccinate synthetase (purA, 1,299 bp), and DNA recombination/repair protein (recA, 1,062 bp). A maximum likelihood phylogenetic tree, with 1,000 bootstrap iterations, was constructed in MEGA 6 using concatenated and aligned loci. Reference genomes belonging to E. coli phylogenetic groups A (O9:H4 strain HS, K12 strain MG1655), B1 (O104:H4 strain 2011C-3493, O104:H4 strain 55989, O103:H2strain 12009, O152:H28 strain SE11, O26:H11 strain 11368, O111:H8strain 11128, O8:H9 strain IAI1), B2 (O150:H5 strain SE15, and O18:H7 strain UT189), D (O17:K52:H18 strain UMN026), E (O157:H7 Sakai strain), F (O7:H45 strain IAI39), and E. fergusonii ATCC35469 (as outgroup), all obtained from public databases, were used for assignment of the presumptive STECs to phylogenetic groups. Then, these presumptive STECs were in silico serotyped for identification of the outer membrane (O) antigen and flagellin protein (H) types using the SerotypeFinder tool from the Centre for Genomic Epidemiology (CGE) website2 (Joensen et al., 2015). For identification of virulence genes, six loci from the E. coli O157:H7 Sakai strain complete genome, i.e., stx1a (948 bp), stx1b (270 bp), stx2a (960 bp), stx2b (270 bp), ehxA (998 bp), and gamma intimin gene eae (2,805 bp) were selected and mapped against the genome sequences of the presumptive STECs. This Whole Genome Shotgun project consisting of genomes of 35 strains was deposited at the NCBI database under reference of PRJNA6242293.
Results
Grass, Manure, and Water Analysis by Application of Cultivation-Independent and -Dependent Microbial Approaches
Grassland sampling in August 2014 resulted in DNA extracts of high quality (high molecular weight DNA, low in DNA polymerase-inhibiting impurities) from 25 grass rhizosphere soil and corresponding grass shoot, and of 12 stable manure samples (Table 1). In April and August 2015, high-quality DNA extracts from four rhizosphere soil and corresponding grass shoot samples were obtained at both occasions and from two manure samples in August 2015 only. All 47 community DNA extracts were used for downstream molecular analyses in the presence of E. coli and virulence genes relevant for E. coli O157:H7 and other STECs.
Direct plating of grass shoot and rhizosphere soil samples taken in August 2014 on CHROMagarTM O157 resulted in absence of any mauve-stained colonies (identical to E. coli O157:H7 on CHROMagarTM and presumed to be STEC) on samples from grass shoots, with the exception of grass shoots near cowpats in sector D. In these samples, between 0 and 200 mauve colonies were found on plates that had received undiluted extract, resulting in presumptive STEC numbers of between log [10Log (n + 1)] 0 and 4.78 CFU (average of 3.11) per g of fresh grass near cowpats in sector D. In the rhizosphere soil, numbers of mauve colonies on plates that had received undiluted rhizosphere soil suspensions were between 0 and 96, including samples taken near cowpats, resulting in log CFU numbers of between 0 and 2.55 CFU per g of dry soil in all rhizosphere soils. Prevalence of detectable mauve colony forms in rhizosphere soil was low; five samples of 25 were found positive, of which two came from samples near cowpats. Isolation of mauve colony types (three from each positive sample) by streaking to purity on fresh CHROMagarTM O157 plates never resulted in single isolated mauve colonies. Either colonies initially colorizing mauve turned blue after purification (other E. coli and non-E. coli strains different from the reference E. coli O157:H7 strain EDL933), or mauve colony types were overgrown by blue-stained colonies. Repeated purification steps from these mauve colony types on fresh CHROMagarTM O157 again remained futile. It was, therefore, concluded that STEC quantification in grass shoot and rhizosphere soils via direct plating might give an indication on the number of presumptive STECs in these environments, but confirmation of these numbers cannot be provided because of failure in subsequent purification steps required for further identification.
Therefore, enrichments for presumptive STECs from these samples, using the ISO 16654 protocol including colony to purity streaking on CHROMagarTM O157, was performed, which resulted, after purification steps, in single isolated mauve-stained colony types. A total of 85 presumptive STECs were obtained from grass shoots, rhizosphere soil, and manure over both years (Table 2). Three presumptive STECs were from rhizosphere soil sample 90 (sector B) by three independent enrichments from the same sample resulting in strains N116–N118. Four presumptive STECs were obtained from grass shoot sample 65 (sector A), and all four came from the same enrichment of the same sample, and these strains were denoted as N102–N105. In two other grass shoot samples, 70 (sector B) and 78 (sector D), respectively, six (N106–N111) and four (N112–N115) strains were obtained, all by independent enrichments. Further, two presumptive STECs were obtained from two independent stream water samples (denoted as N119 and N120), and 60 presumptive STECs were obtained from 60 independent enrichments from stable manure samples. The reason that multiple strains were isolated from the same enrichment (N102–N105) or from different enrichments of the same sample (N106–N118) was to check whether phenotypically and genotypically different variants might arise from the same enrichment or from different enrichments of the same sample. In April 2015, no presumptive STECs were obtained via enrichment from all eight rhizosphere soil and corresponding grass shoot samples. In August 2015, presumptive STECs were found in four independent enrichments from rhizosphere soil samples from sector A (N164), sector B (N165 and N166), and sector C (N163) and in two grass shoot samples from sector A (N167) and sector B (N168).
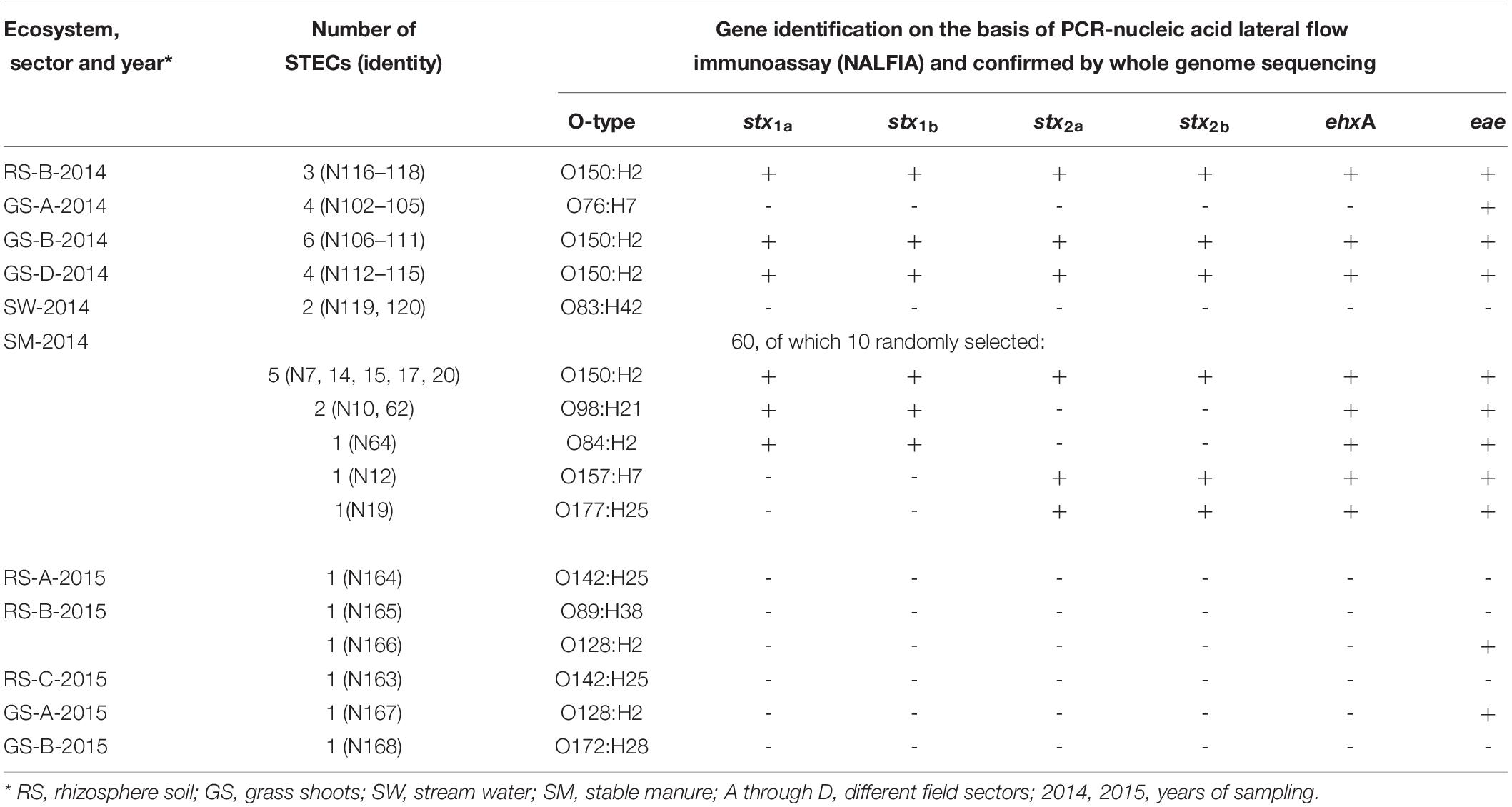
Table 2. Identities and presence of virulence genes in Shiga toxin-producing Escherichia coli (STECs) isolated from different ecosystems over 2 years from the Nijkerk cattle farm.
Prevalence of E. coli and STEC Virulence Genes in Grass, Manure, and Water Samples
The presence of E. coli and closely related E. vulneris and Shigella species (16S rRNA gene) and STEC virulence genes stx1 and stx2 (Shiga toxins 1 and 2 genes, respectively), eae (intimin gene), and ehxA (hemolysin gene) was demonstrated using the PCR-NALFIA diagnostic platform, on community DNA samples from grass shoots, rhizosphere soil, and stable manure (Figures 2, 3). Over all samples taken in 2014 (n = 37), E. coli was found present in all grass shoot (n = 25) and corresponding rhizosphere soil samples and in all stable manure samples (n = 12) (Figure 2). The Shiga toxin stx1 gene was found in one grass shoot and in none of the rhizosphere and stable manure samples, whereas stx2 was found present in 19 grass shoot, two rhizosphere soil, and in one stable manure samples. The hemolysin gene ehxA was found to be present in eight grass shoot, five rhizosphere soil, and 11 stable manure samples, the intimin gene eae in zero grass shoot, one rhizosphere soil, and four stable manure samples. Over the 18 grass shoot, rhizosphere soil, and manure samples from April and August 2015, E. coli was also found present in all samples, whereas the ehxA gene was present in only one grass shoot sample, in all eight rhizosphere soils and in both manure samples. Further, the eae gene was only present in one grass shoot sample and not found in any of the other samples, whereas the stx1 and stx2 genes were not found in any of the 18 samples taken in 2015. Based on these data, it is obvious that E. coli was always detectably present in all community DNA samples, whereas most virulence genes and especially the stx2 gene were only present in 2014 samples, when heifers were grazing on the pasture land and absent in 2015 samples when no heifers were grazing anymore. Over all 2014 samples, the highest prevalence for stx2 was found in the grass shoots, whereas the highest prevalence for ehxA was found in stable manure.
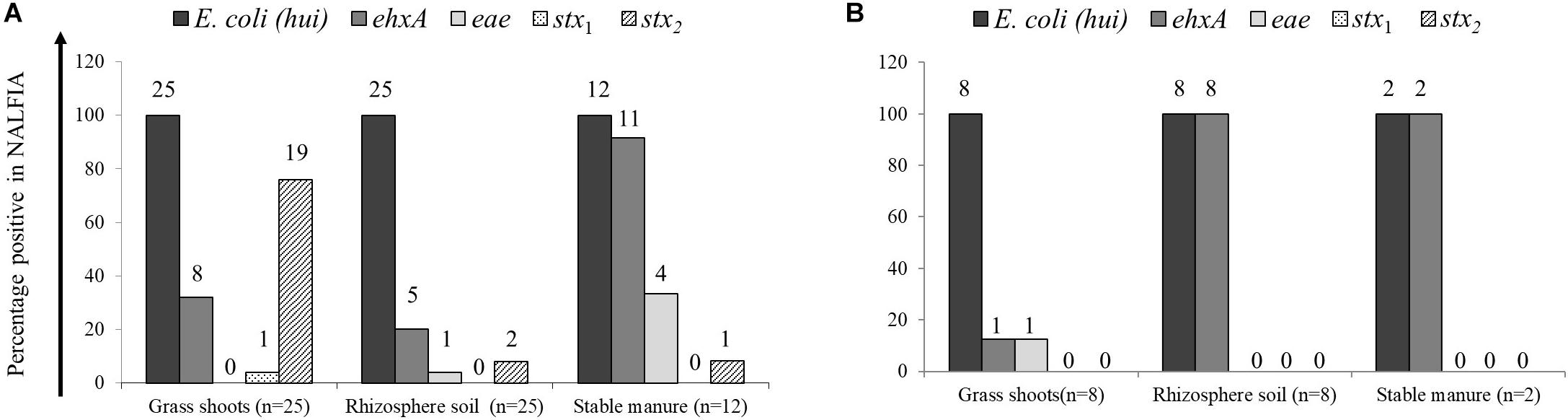
Figure 2. Prevalence of Escherichia coli 16S rRNA and virulence genes, as determined by PCR-nucleic acid lateral flow immunoassay (NALFIA), in DNA extracts from grass shoots, rhizosphere soil, and stable manure taken in August 2014 (A) and August 2015 (B) and expressed as fractions of total sample numbers from each ecosystem. Number above bars represents the absolute numbers of positive reactions per target gene for each ecosystem.
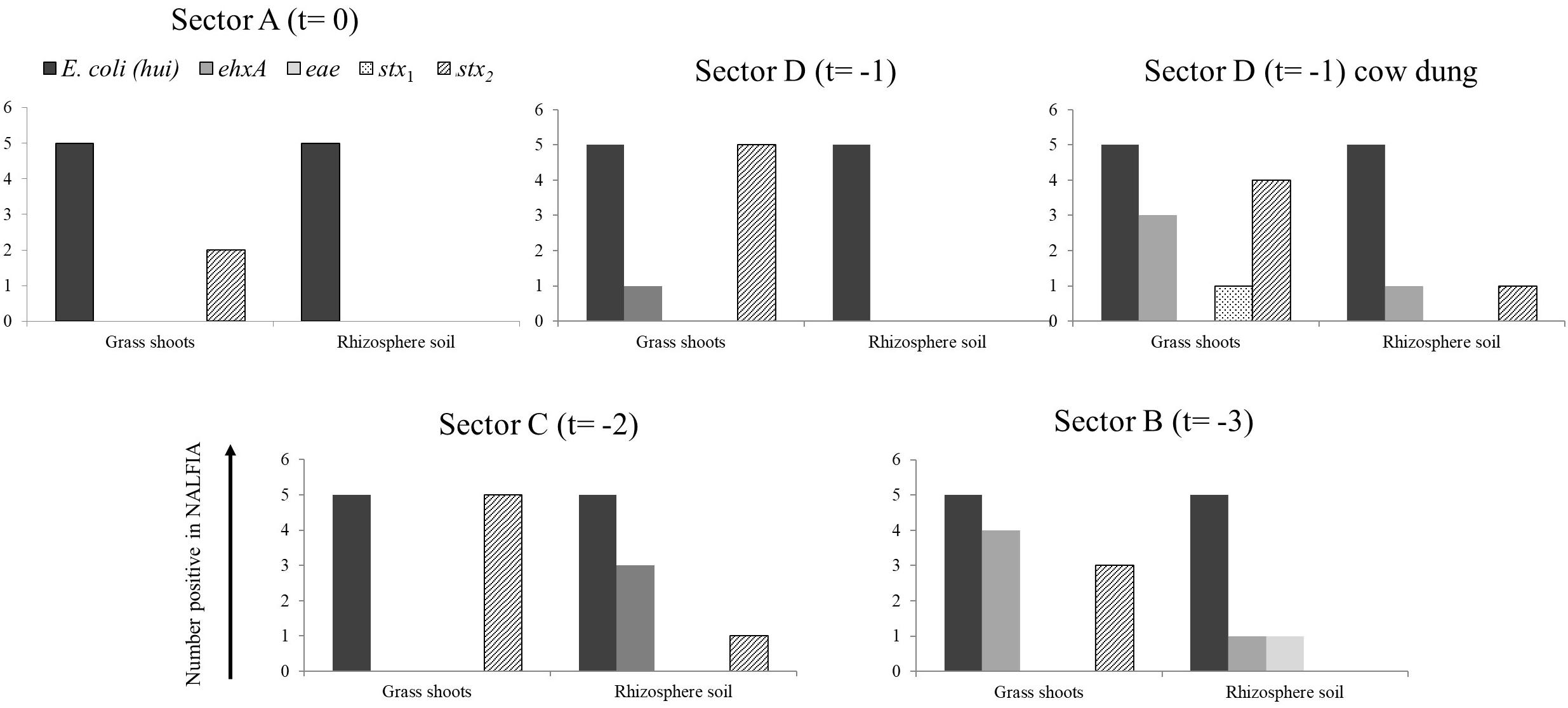
Figure 3. Number of positive PCR-NALFIA reactions to E. coli 16S rRNA and virulence genes in DNA extracts from grass shoots, rhizosphere soil, and stable manure taken in different sectors of the pasture in August 2014 (A). Numbers on vertical axis represent the number of positive NALFIA reactions for each target gene in grass shoots and rhizosphere soil DNA extracts for each sector (n = 5).
Divided over the four sectors of the pasture land in 2014 (Figure 3), the stx2 gene was found present in one grass shoot sample in sector A, the sector where heifers were present at the moment of sampling. However, the stx2 gene was present in all five grass shoot samples from sectors D and C; the sectors where heifers were grazing, respectively, 1 and 2 weeks before sampling and in three samples from sector B (3 weeks before sampling). Of the other virulence genes, only the ehxA gene was found present in grass shoots from sector D (one sample) and B (four samples). This indicates that the stx2 gene, and occasionally the ehxA gene, is present in the microbial community associated with grass shoots, even in the absence of grazers for 3 weeks. In rhizosphere soil, the picture was different from that of the corresponding grass shoots, namely the stx2 gene was only found in one sample from sector C, whereas the ehxA and the eae genes were both found at one occasion in samples from sector B. Rhizosphere soil and corresponding grass shoot sampling most proximate to a cowpat in sector D also revealed consistent presence of E. coli in all 10 samples, and presence of the stx2 gene in four grass shoot and one rhizosphere soil sample. Shiga toxin 1 gene was only found once in a grass shoot sample and the ehxA gene three times in grass shoot and one time in a rhizosphere soil sample. Overall a higher prevalence of the stx2 gene was found in grass shoots in comparison with rhizosphere soil. A cultivation-based approach was applied to demonstrate the presence of stx2 genes in STECs.
Full genome analysis was, therefore, performed on DNA extracts made from all 25 presumptive STECs from grass shoots, rhizosphere soil, and stream water (over both years) and from a randomly selected set of 10 STECs from stable manure samples taken in 2014 and denoted as N7, N10, N12, N14, N15, N17, N19, N20, N62, and N64 for further downstream molecular typing.
Identity of Presumptive STECs From the Nijkerk Farm
Identification of the 35 STECs was performed on the basis of full genome DNA sequence analysis by: (1) multilocus sequence typing of 10 E. coli household genes, namely, adk, fumC, glpK, gyrB, icd, lysA, mdh, metG, purA, and recA, (2) in silico serotyping using the CGE website, and (3) gene mapping for stx1a, stx1b, stx2a, stx2b, intimin gene eae, and hemolysin gene ehxA from the E. coli O157:H7 Sakai strain genome. The presence of the four virulence genes in draft genomes of 16 strains from grass shoots (N102, N106, N112), rhizosphere soil (N116), stream water (N119 and N120) and manure (N7, N10, N12, N14, N15, N17, N19, N20, N62, and N64) corresponded to observations made by PCR-NALFIA on DNA extracts made from these strains (Table 2).
Phylogenetic clustering on the basis of multilocus DNA sequence similarities revealed one cluster of 21 strains that closely resembled each other on the basis of MLST (Figure 4). These 21 strains showed phylogenetically closest relationship with two E. coli O104:H4 strains and more distinct relationships with five other E. coli strains, namely, O103:H2, SE11, O26:H11, O111:H8, and IAI1, all from phylogenetic group B1. Ten of these strains originated from grass shoots, eight from stable manure samples, and three from rhizosphere soil, all from 2014. Of the 10 grass shoot strains, six were from sector B and four from sector D. Altogether, these 21 strains originated from 11 independent samples. The fact that closely resembling strains originated both from the stable manure (eight) and from pasture field (three) samples indicates that interactions between stable and pasture land must have taken place in spite of the fact that both were distantly located from each other. These 21 strains also resembled each other on the basis of presence of virulence genes, i.e., all six tested virulence genes were present in the genomes of 18 strains, whereas in three strains, both stx2 genes were shown to be absent (Table 2). The 18 strains containing all virulence genes, including stx2 genes were identified to belong to the same O150:H2 E. coli serotype, whereas the three other strains (all originating from manure) were typed to belong to O98:H21 (two) and O84:H2 (one) E. coli serotypes. Further, only one strain, N12, only containing stx2 and no stx1 genes, from stable manure, was serotyped as E. coli O157:H7 (phylogenetic group E), and this was a remarkable fact because it was anticipated on forehand that the E. coli O157:H7 serotype would prevail among the manure isolates. One other strain from manure was shown to possess stx2, and no stx1 genes in its genome and this strain was identified to belong to E. coli serotype O177:H25 (N19). Altogether, 23 strains, of which 22 belonged to phylogenetic group B1 and one to E, carried one or more stx genes in their genomes, and therefore, these 23 strains were confirmed to be STEC positive. Twelve strains did not possess any stx gene in their genomes, and thus, these strains are further denoted as “non-Shiga toxin-producing E. coli strains,” i.e., non-STECs. These non-STECs did not cluster together in the phylogenetic tree, and most (8) belonged to phylogenetic group B1, two to A and two to D. Two strains, denoted as N166 and N167 and containing the intimin eae gene, were closely related to E. coli O103:H2. Both strains originated, respectively, from grass shoots and rhizosphere soil taken in August 2015 and were identified to belong to E. coli serotype O128:H2. All other 10 non-STECs were shown to belong to different serogroups (O142:H25, O76:H7, O83:H42, O89:H38, O172:H28) and must be considered as “commensal” E. coli isolates. Of these 10, two, denoted as N163 and N164 (O142:H25), and showing closest relationship with E. coli K12, were from rhizosphere soil sampled in August 2015. Four strains, denoted as N102–N104 (O76:H7), and showing closest relationship with E. coli strain IAI1, were from grass shoots sampled in 2014, and their resemblance was expected on forehand because these four strains originated from the same enrichment culture. Two non-STECs, originating from stream water adjacent to the pasture land (N119 and N120 and serotyped as O83:H42), showed closest relationship with E. coli strain IAI39 and most likely belong to phylogenetic group D. These two strains were most distinct from the other 33 strains, indicative of the fact that stream water may carry other E. coli strains than present in pasture land and stable manure. Two other non-STECs, N165 (O89:H38) from rhizosphere soil from August 2015 and N168 (O172:H28) from grass shoots from August 2015, showed close relationship with each other and with strains N102–N104, and belonged to phylogenetic group B1.
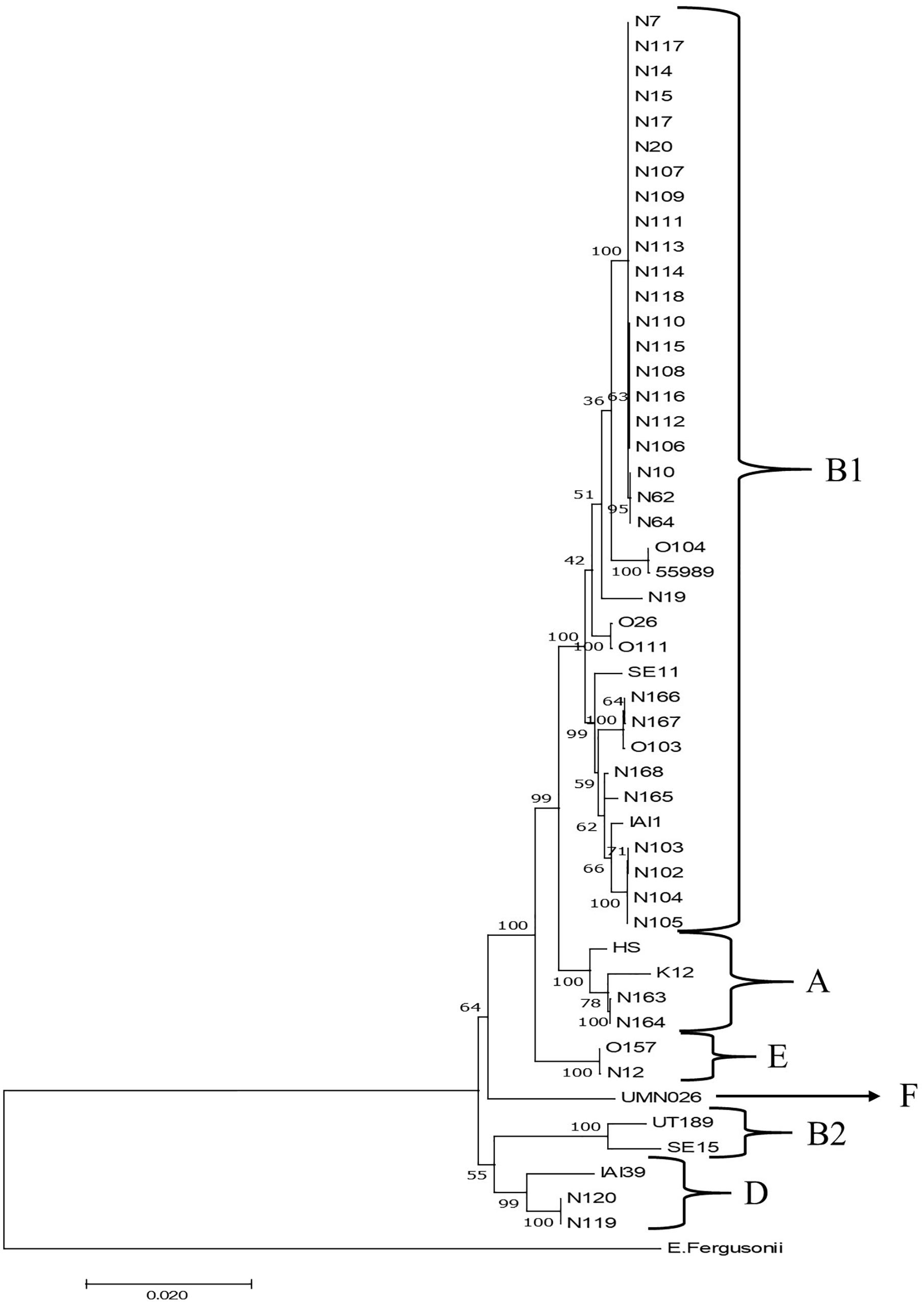
Figure 4. Maximum likelihood phylogenetic comparison of 35 STECs from different ecosystems on the Nijkerk cattle farm and 15 E. coli/Escherichia fergusonii strains. The tree was rooted using E. fergusonii (ATCC35469) as outgroup. Phylogenetic relationships between E. coli strains were inferred by making use of 10 concatenated household genes (adk, fumC, glpK, gyrB, icd, lysA, mdh, metG, purA, and recA) with 1,000 bootstrap iterations. Relevant phylogenetic groups (A, B1, B2, D, E, and F) are indicated in the tree. Reference strain abbreviations: HS, O9:H4 strain HS; K12, O16:H48 K12 strain MG1655; O104, O104:H4 strain 2011C-3493; 55989, O104:H4 strain 55989; O103, O103:H2 strain 12009; SE11, O173:H4 strain SE11; O26, O26:H11 strain 11368; O111, O111:H8 strain 11128; IAI1, O8:H9 strain IAI1; SE15, O150:H5 strain SE15; UT189, O18:H7 strain UT189; UMNO26, O17:H18 strain UMN026; O157, O157:H7 Sakai strain; IAI39, O7:H45 strain IAI39; E. fergusonii, E. fergusonii ATCC35469.
In summary, it is obvious that most of the STEC and non-STEC (commensal E. coli) strains obtained from the pasture land in 2014 and 2015 belonged to phylogenetic group B1. The majority of these 2014 strains were STECs and closely resembling strains isolated from stable manure. The non-STECs from this phylogenetic group from 2014 (strains N102–N104) were closely related with four strains from pasture land from 2015.
Discussion
The presence of STECs and stx2 genes on grass shoots and in rhizosphere soil at the first sampling, when heifers were grazing on the pastureland, but not in the second and third samplings, in the absence of grazing animals, indicate a positive relationship between grazers and STECs/stx2-phages in pastureland. This is a main conclusion that can be drawn from this study. The fact that the farmer would withhold his grazing animals from the pastureland in the second year was not anticipated on forehand in the design of our study. His decision was fortunate to us because now we were able to investigate the same pastureland in the absence of grazing animals during the summertime of the succeeding year. Stable manure was applied to the pastureland in the same way in both years, which indicates that manure application, itself, may not be enough to cause field contamination with STECs or stx2-phages. Potential risks, indeed, exist with respect to persistence of human pathogens in agricultural environments as already stated before in Jones (1999), but now, with the implication of grazing animals on pasture land, that can lead to contamination of grass plants with non-typical STECs.
It has been shown before that E. coli O157:H7 and S. enterica can survive for over a month in bulk and rhizosphere soil and that both species can percolate to deeper soil layers, depending on manure type and type of manure application to soil (Semenov et al., 2009). It is, thus, realistic to presume that E. coli strains present in stable manure were able to survive long enough in deeper soil layers upon manure injection into soil to make contact with growing grass roots. The E. coli strains could establish themselves in the rhizosphere soil by consumption of root-released nutrients. Further spread to above-soil parts of the grass plants might be possible either actively via internal plant colonization, or passively, e.g., at rainfall via splashing of water droplets with contaminated soil to the above-soil parts of the grass plants. However, it was active grazing by the heifers instead of stable manure application to soil that was the main driving factor behind STEC contamination of the grass plants. Grazing animals sometimes pull out grass from the soil that, itself, may lead to smearing of soil with contaminating agents over the pasture. However, excrements from grazers may be held responsible for most of the contamination of the pasture. E. coli was shown to persist in decomposing cow pats and neighboring soil for 6 months and over in Muirhead (2010). In our study, the presence of stx2 and other virulence genes near and on grass plants in the neighborhood of heifer pats did not seriously deviate from the other investigated locations in the pasture. This would indicate that contaminating agents may be spread from the pats over the pasture, and most likely smearing occurred via the hooves of the grazers when walking over the land.
The near clonal group of 21 STECs, belonging to phylogenetic group B1 and typed as E. coli O150:H2, O98:H21, and O84:H2 and carrying three (O98:H21 and O84:H2) or four (O150:H2) of the virulence genes screened for, was found in two physically separated locations in this study, namely, in stable manure and near and on grass plants at the first sampling event. The fact that stable manure applications into the field in early springtime of both years were the only clear links between stable and pasture may indicate that a single origin of this particular group of E. coli strains was present. The fact that E. coli O150:H2 was only found at the first sampling may be a consequence of grazers on the pasture. A possible explanation can be that stable manure injection into the soil led to field contaminations with this E. coli line. When the heifers were present, E. coli O150:H2-contaminated grass with roots were ingested by the grazers. Uptake of E. coli O150:H2 resulted in outgrowth of the E. coli line in the intestinal track system of the animals, and excretion of larger quantities of E. coli O150:H2 were returned to the pasture via their feces, from where E. coli O150:H2 was further smeared over the field. Without grazers, no outgrowth, and further smearing of E. coli O150:H2 occurred, explaining the absence, or below the limits of detection, of this particular bacterial line in the second and third sampling events in 2015. This explanation is in favor with our hypothesis that human pathogens likely circulate between plants as feed and grazing herbivores such as (young) cattle. The plant must be considered as an alternative environment for this particular group of potential human pathogens waiting for the opportunity to colonize intestinal track systems of other grazing animals and from an ecological perspective creating the opportunity to disseminate over other herds of grazing animals feeding from the same land.
Escherichia coli O150:H2 is not often found as contaminating agent in foods and environments, although it has been described as an emerging serogroup for over several years (Monaghan et al., 2012; Onyeka et al., 2020). The group of 21 closely resembling STECs from our study showed closest relationship with strains belonging to phylogroup B1, the group to which E. coli O104:H4 strains belong to. Escherichia coli phylogenetic group B1 strains are known to be environmental strains, commonly occurring in different household animal species (Carlos et al., 2010) and freshwater beaches (Walk et al., 2007), whereas E. coli O150:H2 strains are commonly associated with beef products (Monaghan et al., 2012). The E. coli O150:H2 strains obtained from our study are not related to E. coli O150:H5, which belong to phylogroup B2; a group consisting of extra-intestinal pathogenic and uropathogenic E. coli strains, including the commensal strain SE15 and the multidrug-resistant sequence type ST131 (Toh et al., 2010; Petty et al., 2014). Three other strains from the same group belonged to serogroups O98:H21 and O84:H2, and all three strains carried stx1, eae, and ehxA, but no stx2 genes, and all three were derived from manure. Although closely related with the other 18 O150:H2 strains, these three strains were different on the basis of their sero- and virulotypes. Strains belonging to O98 and O84:H2 serotypes were found before in, respectively, diseased monkeys in China (Qi et al., 2017) and in humans, cattle, and sheep in New Zealand (Cookson et al., 2006). That STEC virulence genes are located on genomes of serotypes different from the most commonly observed ones can be an indication for the fact that new STEC types still arise (Hao et al., 2012). For example, it was suggested that adaptation to the plant environment could lead to more virulent E. coli O157:H7 types (Kulasekara et al., 2009).
Besides the closely resembling STECs found in our study belonging to serogroups O150:H2, O98:H21, O84:H2, O177:H25, and O157:H7, other less virulent E. coli types were found at the first (O76:H7; four strains) and third sampling events (O128:H2; two strains), respectively, in 2014 and 2015. E. coli O76 and O128 strains carrying stx genes were isolated before from humans in, respectively, Spain (Sánchez et al., 2014) and Germany (Beutin et al., 2004). In our study, strains of both serotypes did not carry stx and ehxA genes, but contained eae genes. The fact that these strains were found over 2 years in different habitats (grass shoots and rhizosphere soil) may imply that mild virulent E. coli strains are continuously present in pasture, also in the absence of grazers. Further, six E. coli strains with different serotypes were detected in water samples (2014) and in grass shoot and rhizosphere soil samples from 2015, and all strains did not carry any of the tested virulence genes. This indicates that a second, more diverse, group of non-virulent and commensal E. coli strains exist in pasture land and adjacent stream water. Escherichia coli has been found before in plants (Whitman et al., 2005; NandaKafle et al., 2017), and niche partition among different E. coli strains was observed in pasture land (NandaKafle et al., 2017). In the last study, it was proposed that certain E. coli types could survive winter conditions, including freezing temperatures. It is possible that these commensal E. coli types from our study may be representatives of E. coli populations that naturally reside in or on grass plants or on/in other plants (clover) that grow in pasture. Upon manure application, the more threatening E. coli types, among which the STECs serotyped as O150:H2, were introduced into pasture. In the absence of grazing animals in 2015, no augmentation of these STECs occurred, and only the naturally residing E. coli strains remained present in pasture in 2015. This would be the best explanatory model on the presence of the different E. coli strains in the Nijkerk farm. The fact that no STECs were found in the stream near the drinking place indicates that STECs from feces at least not vastly contaminated the water of the stream. This observation corresponds to a previous observation made in surface water bodies nearby agricultural fields in Netherlands where E. coli O157 was detected although without carriage of Shiga toxin production genes (Heijnen and Medema, 2006).
The presence of STECs in pastureland was confirmed by molecular analysis using PCR-NALFIA, although by following this approach, it was not possible to make a physical link between E. coli and the detected stx2 gene. It was obvious that stx2 genes were more prevalent in pasture in 2014 than all of the other virulence genes screened for, whereas derived E. coli O150:H2 strains possessed all virulence genes. The observed inconsistence in “stoichiometry” between the screened virulence genes was obvious in grass shoot and rhizosphere soil DNA extracts, and this may be related to the relatively higher selectivity of the CHROMagar TM O157 medium for serotype O157:H7 over non-O157 STECs (Fan et al., 2018), but also may be related to the fact that phage DNA containing stx2 genes were co-extracted with bacterial DNA from soil and plant samples. This corresponds to the observation made by Grau-Leal et al. (2015), where numbers of stx2 bacteriophages were found to be higher than stx1 bacteriophages in many different extraintestinal environments, including soils.
Both cultivation-dependent and independent approaches did not allow us to make any quantification of STECs or stx2 carrying microorganisms in these environments. However, based on direct CFU counts from mauve-stained colonies from the grass shoots nearby pats and in rhizosphere soil, the order of magnitude may lay between log 2 and log 3 per gram of soil or gram of fresh plant weight. These numbers occasionally may be too low for molecular detection in environmental DNA extracts, but possibly still high enough for cultivation by enrichment. Considering the fact that further purification attempts of the mauve-stained colonies on CHROMagarTM O157 plates remained futile, it may be that stx2 genes were present in other STECs that were not able to grow on CHROMagarTM O157 medium, or that STEC cells directly recovered from grass shoot and rhizosphere soil were seriously stressed and that a pre-enrichment step was needed to overcome cellular damage (Hepburn et al., 2002; Ishii et al., 2006).
In conclusion, this study makes clear that the combination of plants and grazing animals is the most important driver behind the establishment and augmentation of a potentially dangerous non-typical STEC population in pasture land. The circulation of human pathogens between plants as feed for grazing animals in the pasture land would be indicative for the fact that circulations of human pathogens exist at larger and more complex scale levels between plant (as feed) and animal production systems. Possibly, these circulations remained unnoticed in studies that were only focused on typical STECs, such as O157:H7. This is an important message especially with respect to safe production of healthy foods such as fresh produce (Guttierrez-Rodriguez and Adhikari, 2018). Circulation of human pathogens between different plant and animal production systems is a topic that would require more attention in safe food production programs.
Data Availability Statement
The datasets generated for this study are available on request to the corresponding author.
Ethics Statement
A microbiological survey was carried out on a private property. For that purpose, permission was granted by the owner of the farm. The farmer fully collaborated in the research performed on his property, but indicated to remain anonymous in scientific communications.
Author Contributions
LO, AA, HR, and PW conceived the project and designed the experiments together with JW and PZ. PW, PZ, and LO were the main responsible persons for field surveying and microbiological analyses. JW was responsible for PCR-NALFIA analysis and data analysis. All authors contributed to the article and approved the submitted version.
Funding
This work was carried out under the knowledge basis program 12 on Agro-production chains financed by the Dutch Ministry of Economic Affairs. Publication of the manuscript was supported by EU COST Action HUPLANTcontrol (16110).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to the owner of the Nijkerk farm for his support and for providing us information about farming practices and Gerrit Hooijer and Aniek Lotterman of the Department of Veterinary Sciences of Utrecht University for providing us information about the E. coli O157 livestock farm survey done in Netherlands in 2013. Further, we would like to thank Khan Pham, Carin Lombaers, and Carin van Tongeren for practical assistance and Marjon Krijger for CLC analysis.
Footnotes
- ^ https://www.iso.org/standard/29821.html
- ^ http://www.genomicepidemiology.org/
- ^ https://www.ncbi.nlm.nih.gov/Traces/study/?acc=PRJNA624229
References
Ahmed, S. A., Awosika, J., Baldwin, C., Bishop-Lilly, K. A., Biswas, B., Broomall, S., et al. (2012). Genomic comparison of Escherichia coli O104:H4 isolates from 2009 and 2011 reveals plasmid, and prophage heterogeneity, including Shiga toxin encoding phage stx2. PLoS One 7:e48228. doi: 10.1371/journal.pone.0048228
Atwill, E. R., Chase, J. A., Oryang, D., Bond, R. F., Koike, S. T., Cahn, M. D., et al. (2015). Transfer of Escherichia coli O157:H7 from simulated wildlife scat onto romaine lettuce during foliar irrigation. J. Food Prot. 78, 240–247. doi: 10.4315/0362-028X.JFP-14-277
Ávila-Quezada, G., Sánchez, E., Gardea-Béjar, A. A., and Acedo-Félix, E. (2010). Salmonella spp. and Escherichia coli: survival and growth in plant tissue. N. Z. J. Crop Horti. Sci. 38, 47–55. doi: 10.1080/0114067100367834
Berry, E. D., Wells, J. E., Varel, V. H., Hales, K. E., and Kalchayanand, N. (2017). Persistence of Escherichia coli O157:H7 and total Escherichia coli in feces and feedlot surface manure from cattle fed diets with and without corn or sorghum wet distillers grains with solubles. J. Food Prot. 80, 1317–1327. doi: 10.4315/0362-028X.JFP-17-018
Bettelheim, K. A., Kuzevski, A., Gilbert, R. A., Krause, D. O., and McSweeney, C. S. (2005). The diversity of Escherichia coli serotypes and biotypes in cattle faeces. J. Appl. Microbiol. 98, 699–709. doi: 10.1111/J.1365-2672.2004.02501.x
Beutin, L., Krause, G., Zimmermann, S., Kaulfuss, S., and Gleier, K. (2004). Characterization of Shiga toxin-producing Escherichia coli strains isolated from human patients in Germany over a 3-year period. J. Clin. Microbiol. 42, 1099–1108. doi: 10.1128/JCM.42.3.1099-1108.2004
Callaway, T. R., Carr, M. A., Edrington, T. S., Anderson, R. C., and Nisbet, D. J. (2009). Diet, Escherichia coli O157:H7, and cattle: a review after 10 years. Curr. Issues Mol. Biol. 11, 67–80.
Carlos, C., Pires, M. M., Stoppe, N. C., Hachich, E. M., and Sato, M. I. (2010). Escherichia coli phylogenetic group determination and its application in the identification of the major animal source of fecal contamination. BMC Microbiol. 10:161. doi: 10.1186/1471-2180-10-161
Colello, R., Cáceres, M. E., Ruiz, M. J., Sanz, M., Etcheverria, A. I., and Padola, N. L. (2016). From farm to table: follow-up of Shiga toxin-producing Escherichia coli throughout the porc production chain in Argentina. Front. Microbiol. 7:93. doi: 10.3389/fmicb.2016.00093
Cookson, A. L., Croucher, D., Pope, C., Bennett, J., Thomson-Carter, F., and Attwood, G. T. (2006). Isolation, characterization, and epidemiological assessment of Shiga toxin-producing Escherichia coli O84 isolates from New Zealand. J. Clin. Microbiol. 44, 1863–1866. doi: 10.1128/JCM.44.5.1863-1866.2006
Cote, R., Katani, R., Moreau, M. R., Kudva, I. T., Arthur, T. M., DebRoy, C., et al. (2015). Comparative analysis of super-shedder strains of Escherichia coli O157:H7 reveals distinctive genomic features and a strongly aggregative adherent phenotype on bovine rectoanal junction squamous epithelial cells. PLoS One 10:e0116743. doi: 10.1371/journal.pone.0116743
Cox, C. E., Brandl, M. T., de Moraes, M. H., Gunasekera, S., and Teplitski, M. (2018). Production of the plant hormone auxin by Salmonella and its role in the interactions with plants and animals. Front. Microbiol. 8:2668. doi: 10.3389/fmicb.2017.02668
Crozier, L., Hedley, P. E., Morris, J., Wagstaff, C., Andrews, S. C., Toth, I., et al. (2016). Whole-transcriptome analysis of verocytotoxigenic Escherichia coli O157:H7 (Sakai) suggests plant-species-specific metabolic response on exposure to spinach and lettuce extracts. Front. Microbiol. 7:1088. doi: 10.3389/fmicb.2016.01088
de los Angeles Dublan, M., Ortiz-Marquez, J. C. F., Lett, L., and Curatti, L. (2014). Plant-adapted Escherichia coli show increased lettuce colonizing ability, resistance to oxidative stress and chemotactic response. PLoS One 9:e110416. doi: 10.1371/journal.pone.0110416
de Moraes, M. H., Desai, P., Porwollik, S., Canals, R., Perez, D. R., Chu, W., et al. (2017*). Salmonella persistence in tomatoes requires a distinct set of metabolic functions identified by transposon insertion sequencing. App. Environ. Microbiol. 83:e03028-16. doi: 10.1128/AEM.03028-16
Deering, A. J., Mauer, L. J., and Pruitt, R. E. (2012). Internalization of E. coli O157:H7 and Salmonella spp. in plants: a review. Food Res. Int. 45, 567–575. doi: 10.1016/j.foodres.2011.06.058
Dodd, C. C., Sanderson, M. W., Sargeant, J. M., Nagaraja, T. G., Oberst, R. D., Smith, R. A., et al. (2003). Prevalence of Escherichia coli O157 in cattle feed is Midwestern feedlots. Appl. Environ. Microbiol. 69, 5243–5247. doi: 10.1128/AEM.69.9.5243-5247.2003
Fan, R., Bai, X., Fu, S., Xu, Y., Sun, H., Wang, H., et al. (2018). Tellurite resistance profiles and performance of different chromogenic agars for detection of non-O157 Shiga toxin-producing Escherichia coli. Int. J. Food Microbiol. 266, 295–300. doi: 10.1016/j.ijfoodmicro.2017.12.016
Franz, E., Semenov, A. V., and van Bruggen, A. H. C. (2008). Modelling the contamination of lettuce with Escherichia coli O157:H7 from manure-amended soil and the effect of intervention strategies. J. Appl. Microbiol. 105, 1569–1584. doi: 10.1111/j.1365-2672.2008.03915.x
Franz, E., van Hoek, H. A. M., Bouw, E., and Aarts, H. J. M. (2011). Variability of Escherichia coli O157 strain survival in manure-amended soil in relation to strain origin, virulence profile, and carbon nutrition profile. Appl. Environ. Microbiol. 77, 8088–8096. doi: 10.1128/AEM.00745-11
Grau-Leal, F., Quirós, P., Martínez-Castillo, A., and Muniesa, M. (2015). Free Shiga toxin 1-encoding bacteriophages are less prevalent than Shiga toxin 2 phages in extraintestinal environments. Environ. Microbiol. 17, 4790–4801. doi: 10.1111/1462-2920.13053
Guttierrez-Rodriguez, E., and Adhikari, A. (2018). Preharvest farming practices impacting fresh produce safety. Microbiol. Spectrum 6:FS-0022-2018. doi: 10.1128/microbiolspec.PFS-0022-2018
Hao, W., Allen, V. G., Jamieson, F. B., Low, D. E., and Alexander, D. C. (2012). Phylogenetic incongruence in E. coli O104: understanding the evolutionary relationships of emerging pathogens in the face of homologous recombination. PLoS One 7:e33971. doi: 10.1371/journal.pone.0033971
Heijnen, L., and Medema, G. (2006). Quantitative detection of E. coli, E. coli O157 and other shiga toxin producing E. coli in water samples using a culture method combined with real-time PCR. J. Water Health 04. 4, 487–497. doi: 10.2166/wh.2006.026
Hepburn, N. F., MacRae, M., Johnston, M., Mooney, J., and Ogden, I. D. (2002). Optimizing enrichment conditions for the isolation of Escherichia coli O157 in soils by immunomagnetic separation. Lett. App. Microbiol. 34, 365–369. doi: 10.1046/J.1472-765X.2002.01098.x
Hovde, C. J., Austin, P. R., Cloud, K. A., Williams, C. J., and Hunt, C. W. (1999). Effect of cattle diet on Escherichia coli O157:H7 acid resistance. Appl. Environ. Microbiol. 65, 3233–3235. doi: 10.1128/aem.65.7.3233-3235.1999
Huijsdens, X. W., Linskens, R. K., Mak, M., Meuwissen, S. G. M., and Vandenbroucke-Grauls, C. M. J. E. (2002). Quantification of bacteria adherent to gastrointestinal mucosa by real-time PCR. J. Clin. Microbiol. 40, 4423–4427. doi: 10.1128/JCM.40.12.4423-4427.2002
Ishii, S., Ksoll, W. B., Hicks, R. E., and Sadowsky, M. J. (2006). Presence and growth of naturalized Escherichia coli in temperate soils from lake superior watersheds. Appl. Environ. Microbiol. 72, 612–621. doi: 10.1128/AEM.72.1.612-621.2006
Jiang, X., Morgan, J., and Doyle, M. P. (2002). Fate of Escherichia coli O157:H7 in manure-amended soil. Appl. Environ. Microbiol. 68, 2605–2609. doi: 10.1128/AEM.68.5.2605-2609.2002
Joensen, K. G., Tetzschner, A. M. M., Iguchi, A., Aarestrup, F. M., and Scheutz, F. (2015). Rapid and easy In Silico serotyping of Escherichia coli isolates by use of whole-genome sequencing data. J. Clin. Microbiol. 53, 2410–2426. doi: 10.1128/JCM.00008-15
Jones, D. L. (1999). Potential health risks associated with the persistence of Escherichia coli O157 in agricultural environments. Soil Manag. 15, 76–83. doi: 10.1111/J.1475-2743.1999.tb00069.x
Kulasekara, B. R., Jacobs, M., Zhou, Y., Wu, Z., Sims, E., Saenphimmachak, C., et al. (2009). Analysis of the genome of the Escherichia coli O157:H7 2006 spinach-associated outbreak isolate indicates candidate genes that may enhance virulence. Infect. Immun. 77, 3713–3721. doi: 10.1128/IAI.00198-09
Liu, D., Cui, Y., Walcott, R., and Chen, J. (2018*). Fate of Salmonella enterica and enterohemorrhagic Escherichia coli cells artificially internalized into vegetable seeds during germination. Appl. Environ. Microbiol. 84:e01888-17. doi: 10.1128/AEM.01888-17
Lòpez-Fernàndez, S., Sonego, P., Moretto, M., Pancher, M., Engelen, K., and Campisano, A. (2015). Whole-genome comparative analysis of virulence genes unveils similarities and differences between endophytes and other symbiotic bacteria. Front. Microbiol. 6:419. doi: 10.3389/fmicb.2015.00419
Mellmann, A., Harmsen, D., Cummings, C., Zentz, E. B., Leopold, S. R., Rico, A., et al. (2011). Prospective genomic characterization of the German Enterohemorrhagic Escherichia coli O104:H4 outbreak by rapid next generation sequencing technology. PLoS One 6:e22751. doi: 10.1371/journal.pone.0022751
Monaghan, Á, Byrne, B., Fanning, S., Sweeney, T., McDowell, D., and Bolton, D. J. (2012). Serotypes and virulotypes of non-O157 shiga-toxin producing Escherichia coli (STEC) on bovine hides and carcasses. Food Microbiol. 32, 223–229. doi: 10.1016/j.fm.2012.06.002
Muirhead, R. W. (2010). Soil and faecal material reservoirs of Escherichia coli in a grazed pasture. N. Z. J. Agri. Res. 52, 1–8. doi: 10.1080/00288230909510483
NandaKafle, G., Seale, T., Flint, T., Nepal, M., Venter, S. N., and Brözel, V. S. (2017). Distribution of diverse Escherichia coli between cattle and pasture. Microbes Environ. 32, 226–233. doi: 10.1264/jsme2.ME17030
Noguera, P., Posthuma-Trumpie, G. A., van Tuil, M., van der Wal, F. J., and de Boer, A. (2011). Carbon nanoparticles in lateral flow methods to detect genes encoding virulence factors of Shiga toxin-producingEscherichia coli. Anal. Bioanal. Chem. 399, 831–838. doi: 10.1007/s00216-010-4334-z
Oliver, D. M., Haygarth, P. M., Clegg, C. D., and Heathwaite, A. L. (2006). Differential E. coli die-off patterns associated with agricultural matrices. Environ. Sci. Technol. 40, 5710–5716. doi: 10.1021/es0603249
Onyeka, L. O., Adesiyun, A. A., Keddy, K. H., Madoroba, E., Manqele, A., and Thompson, P. N. (2020). Shiga toxin–producing Escherichia coli contamination of raw beef and beef-based ready-to-eat products at retail outlets in Pretoria, South Africa. J. Food Prot. 83, 476–484. doi: 10.4315/0362-028X.JFP-19-372
Petty, N. K., Ben Zakour, N. L., Stanton-Cook, M., Skippington, E., Totsika, M., Forde, B. M., et al. (2014). Global dissemination of a multidrug resistant Escherichia coli clone. Proc. Natl. Acad. Sci. U.S.A. 11, 5694–5699. doi: 10.1073/pnas.1322678111
Qi, M., Wang, Q., Tong, S., Zhao, G., Hu, C., Chen, Y., et al. (2017). Identification of atypical enteropathogenic Escherichia coli O98 from golden snub-nosed monkeys with diarrhea in China. Front. Vet. Sci. 4:217. doi: 10.3389/fvets.2017.00217
Sánchez, S., García Cenoz, M., Martin, C., Beristain, X., Llorente, M. T., and Herrera-Leon, S. (2014). Cluster investigation of mixed O76:H19 Shiga toxin-producing Escherichia coli and atypical enteropathogenic E. coli infection in a Spanish household. Epidemiol. Infect. 142, 1029–1033. doi: 10.1017/S0950268813001842
Schikora, A., Virlogeux-Payant, I., Bueso, E., Garcia, A. V., Nilau, T., Charrier, A., et al. (2011). Conservation of Salmonella infection mechanisms in plants and animals. PLoS One 6:e24112. doi: 10.1371/journal.pone.0024112
Scott, R. A., Thilmony, R., Harden, L. A., Zhou, Y., and Brandl, M. T. (2017). Escherichia coli O157:H7 converts plant-derived choline to glycine betaine for osmoprotection during pre- and post-harvest colonization of injured lettuce leaves. Front. Microbiol. 8:2436. doi: 10.3389/fmicb.2017.02436
Semenov, A. V., van Overbeek, L. S., and van Bruggen, A. H. C. (2009). Percolation and survival of Escherichia coli O157:H7 and Salmonella enterica serovar Typhimurium in soil amended with contaminated dairy manure or slurry. Appl. Environ. Microbiol. 75, 3206–3215. doi: 10.1128/AEM.01791-08
Teplitski, M., and de Moreas, M. (2018). Of mice and men. and plants: comparative genomics of the dual lifestyles of enteric pathogens. Trends Microbiol. 26, 748–754. doi: 10.1016/j.tim.2018.02.008
Toh, H., Oshima, K., Toyoda, A., Ogura, Y., Ooka, T., Sasamoto, H., et al. (2010). Complete genome sequence of the wild-type commensal Escherichia coli strain SE15, belonging to phylogenetic group B2. J. Bacteriol. 192, 1165–1166. doi: 10.1128/JB.01543-09
van der Linden, I., Cottyn, B., Uyttendaele, M., Vlaemynck, G., Heyndrickx, M., and Maes, M. (2013). Survival of enteric pathogens during butterhead lettuce growth: crop stage, leaf age, and irrigation. Foodborne Pathogens Dis. 10:1386. doi: 10.1089/fpd.2012.1386
van Elsas, J. D., Semenov, A. V., Costa, R., and Trevors, J. T. (2011). Survival of Escherichia coli in the environment: fundamental and public health aspects. ISME J. 5, 173–183. doi: 10.1038/ismej.2010.80
van Hoek, A. H. A. M., Aarts, H. J. M., Bouw, E., van Overbeek, W. M., and Franz, E. (2013). The role of rpoS in Escherichia coli O157 manure-amended soil survival and distribution of allelic variations among bovine, food and clinical isolates. FEMS Microbiol. Lett. 383, 18–23. doi: 10.1111/1574-6968.12024
van Overbeek, L. S., van Doorn, J., Wichers, J., van Amerongen, A., van Roermund, H. J. W., and Willemsen, P. T. J. (2014). The arable ecosystem as battleground for emergence of new pathogens. Front. Microbiol. 5:104. doi: 10.3389/fmicb.2014.00104
Veldman, K., Kant, A., Dierikx, C., van Essen-Zandbergen, A., Wit, B., and Mevius, D. (2014). Enterobacteriaceae resistant to third-generation cephalosporins and quinolones in fresh culinary herbs imported from Southeast Asia. Int. J. Food Microbiol. 177, 72–77. doi: 10.1016/j.ijfoodmicro.2014.02.014
Walk, S. T., Calhoun, L. M., Mladoncky, J. M., and Whittam, T. S. (2007). Genetic diversity and population structure of Escherichia coli isolated from freshwater beaches. Env. Microbiol. 9, 2274–2288. doi: 10.1111/j.1462-2920.2007.01341.x
Whitman, R. L., Byers, S. E., Shively, D. A., Ferguson, D. M., and Byappanahalli, M. (2005). Occurrence and growth characteristics of Escherichia coli and enterococci within the accumulated fluid of the northern pitcher plant (Sarracenia purpurea L.). Can. J. Microbiol. 51, 1027–1037. doi: 10.1139/W05-091
Wiedemann, A., Virlogeux-Payant, I., Chaussé, A., Schikora, A., and Velge, P. (2015). Interaction of Salmonella with animals and plants. Front. Microbiol. 5:791. doi: 10.3389/fmicb.2014.00791
Williams, A. P., Avery, L. M., Killham, K., and Jones, D. L. (2007). Survival of Escherichia coli O157:H7 in the rhizosphere of maize grown in waste-amended soil. J. Appl. Microbiol. 102, 319–326. doi: 10.1111/j.1365-2672.2006.03104.x
Wright, K. M., Crozier, L., Marshall, J., Merget, B., Holmes, A., and Holden, N. J. (2016). Differences in internalization and growth of Escherichia coli O157:H7 within the apoplast of edible plants, spinach and lettuce, compared with the model species Nicotiana benthamiana. Microb. Biotechnol. 10, 555–569. doi: 10.1111/1751-7915.12596
Keywords: Escherichia coli, animal production systems, calves, human pathogen, pasture, Shiga toxin, stable manure, Shiga toxin-producing Escherichia coli (STEC)
Citation: van Overbeek LS, Wichers JH, van Amerongen A, van Roermund HJW, van der Zouwen P and Willemsen PTJ (2020) Circulation of Shiga Toxin-Producing Escherichia coli Phylogenetic Group B1 Strains Between Calve Stable Manure and Pasture Land With Grazing Heifers. Front. Microbiol. 11:1355. doi: 10.3389/fmicb.2020.01355
Received: 16 January 2020; Accepted: 27 May 2020;
Published: 30 June 2020.
Edited by:
Jorge Blanco, University of Santiago de Compostela, SpainReviewed by:
Sabine Delannoy, Agence Nationale de Sécurité Sanitaire de l’Alimentation, de l’Environnement et du Travail (ANSES), FranceMaite Muniesa, University of Barcelona, Spain
Copyright © 2020 van Overbeek, Wichers, van Amerongen, van Roermund, van der Zouwen and Willemsen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Leonard S. van Overbeek, leo.vanoverbeek@wur.nl
 Leonard S. van Overbeek
Leonard S. van Overbeek Jan H. Wichers
Jan H. Wichers Aart van Amerongen
Aart van Amerongen Herman J. W. van Roermund
Herman J. W. van Roermund Patricia van der Zouwen
Patricia van der Zouwen Peter T. J. Willemsen
Peter T. J. Willemsen