- State Key Laboratory of Developmental Biology of Freshwater Fish, College of Life Science, Hunan Normal University, Changsha, China
The food-borne pathogen Salmonella enterica serovar Typhimurium (S. Typhimurium) causes self-limiting gastroenteritis in humans and is not easily eradicated because it often attaches to suitable surfaces to form biofilms that have high resistance to disinfectants and antimicrobials. To develop an alternative strategy for the treatment of biofilms, it is necessary to further explore the effects of flagellar motility on the development process of Salmonella biofilms. Here, we constructed flagella mutants (ΔflgE and ΔfliC) to systematically study this process. By comparing them with wild-type strains, we found that these mutants lacking flagellar motility form fewer biofilms in the early stage, and the formed mature biofilms contain more cells and extracellular polymeric substances (EPS). In addition, fewer mutant cells adhered to glass plates compared with wild-type cells even after 6 h of incubation, suggesting that flagellar motility plays a significant role in preliminary cell-surface interactions. More importantly, the motility of wild-type strain was greatly decreased when they were treated with carbonyl cyanide m-chlorophenylhydrazone, which inhibited flagellar motility and reduced biofilm formation, as in the case of the ΔflgE mutant. Overall, these findings suggest that flagellar motility plays an important role in Salmonella biofilm initiation and maturation, which can help us to counteract the mechanisms involved in biofilm formation and to develop more rational control strategies.
Introduction
Contamination of fresh produce with food-borne pathogens is a potential threat to public health. Salmonella enterica serovar Typhimurium (S. Typhimurium) is a food-borne pathogen that causes high morbidity worldwide, including life-threatening infections in fetuses, newborns, and immunocompromised individuals (Achinas et al., 2019). Its natural habitat is the gut of animals and humans, and infection occurs through fecal contamination of soil, water, plants and medical equipment, which spreads the bacteria to other hosts (Semenov et al., 2010; Jacobsen and Bech, 2012; MacKenzie et al., 2017). Once attached to one of these surfaces, the bacteria begin to form biofilms. Bacteria inside biofilms are more resistant to external antibiotics, disinfectants, and environmental stress, so they can be extremely difficult to eradicate, in contrast to planktonic bacteria (Pande et al., 2016). The biofilms cause food safety issues and can lead to enormous economic losses.
Biofilms are the preferred lifestyle for most microorganisms in nature (Stoodley et al., 2002). Biofilms begin to develop when bacteria adhere reversibly to a surface. The adhered cells then form microcolonies, which synthesize extracellular matrices to support mature three-dimensional biofilms. Bacteria within the biofilms can undergo controlled dissociation, resulting in biofilm dispersal (Watnick and Kolter, 2000; Koo et al., 2017). Biofilm formation is influenced by the structural composition (curli and other fimbriae, flagella, BapA et al.), the bacterial genome, environmental signals, and stress factors (Watnick and Kolter, 2000; Simm et al., 2014; MacKenzie et al., 2017). A variety of biofilms inhibitors have been developed that target these factors, including some that target the flagella. Previous research has shown that furanones reduce biofilm formation through interfering with Salmonella flagellar synthesis (Janssens et al., 2008). Moreover, zinc is required for flagellar expression, and the absence of flagella impairs the ability of S. Typhimurium to produce biofilms (Ammendola et al., 2016). However, in order to make more effective strategies for the treatment of biofilms targeting flagella, it is necessary to further clarify the effect of flagellar motility on the development process and composition of Salmonella biofilms.
Flagella, as main motility organs of bacteria, play important roles in the formation biofilm of several gram-negative bacteria (O’Toole et al., 2000; Haiko and Westerlund-Wikström, 2013). Some studies have proposed that flagella might act in biofilm formation both as providers of motility and as surface adhesins, however, in Escherichia coli and Listeria monocytogenes it is motility itself that is critical (O’Toole and Kolter, 1998; Pratt and Kolter, 1998; Lemon et al., 2007). In S. Typhimurium, previous studies performed with motA and fliA mutants have suggested that motility is necessary for biofilm development on the glass (Huber et al., 2002; Prouty and Gunn, 2003). Moreover, it has been described that Salmonella mutants with defective flagella (flhC or flgE) are unable to develop complete biofilms in the presence of bile (Crawford et al., 2010; Tsai et al., 2019). Previously, work regarding QseBC two-component system (TCS) function in S. Typhimurium revealed that QseBC functions as a global regulator of flagella, biofilm formation, and virulence (Merighi et al., 2009; Bearson et al., 2010; Ji et al., 2017). Recent studies have shown that the cyclic di-guanylate monophosphate (c-di-GMP) receptor YcgR and the phosphodiesterase YhjH can distinctively inhibit flagellar motility in S. Typhimurium (Le Guyon et al., 2015; Han and Yoon, 2019). Taken together, the role of flagellar motility in the development of Salmonella biofilm is very important. By observing the number and distribution of polysaccharides and cells, we can more intuitively judge the effect of flagellar motility on biofilm initiation and maturation. Therefore, this study aims to deepen the understanding of the effect of flagellar motility on biofilm development and to provide theoretical support for the development of biofilms inhibitors that interfere with flagella motility.
In many motile bacteria, the synthesis and assembly of flagella involve at least 50 genes that are divided into early, middle, and late genes, which are hierarchically and temporally synchronized (Aizawa, 1996). flgE is a middle gene that encodes the major component of the hook-basal body structure, which is necessary for flagellar filament elongation (Bonifield et al., 2000). fliC is a late gene that encodes the major component of the flagellin structure, which is necessary for the formation of the helical filament (Chilcott and Hughes, 2000). Mutations in these two genes may impair flagellar motility, therefore, our primary approach involved deleting flgE and fliC and observing the effect on flagellar motility. Our results confirmed that a lack of flagellar motility affects bacterial contact with abiotic surfaces, and reduces cell colonization and early biofilm formation. Moreover, the mature biofilms formed by these flagella mutants had denser matrices and contained more aggregated cells.
Experimental
Bacterial Strains, Plasmids, and Growth Media, and Chemicals
S. Typhimurium CMCC 50115 is a S. Typhimurium LT2 derivative. All strains and plasmids used in this work are described in Supplementary Table S1. All strains were grown on Luria-Bertani (LB) broth agar plates or in LB broth liquid medium (Tryptone 10 g/L; Yeast Extract 5 g/L; NaCl 10 g/L) at 37°C. Chloramphenicol (Cm, 25 μg/mL), ampicillin (100 μg/mL), L − (+)− Arabinose (5 mmol/L) were added when required. SYTO® 9 (Invitrogen, S 34854); Alexa Fluor® 647 conjugate of Con A (Invitrogen, C 21421); Carbonyl cyanide m-chlorophenylhydrazone (CCCP, Sigma) acted as an inhibitor of proton-driven force of the flagella (Kosaka et al., 2019).
Rdar Morphotype and Pellicle Formation
After one colony of the bacteria from LB plates was transferred into a tube containing 3 mL of LB medium and incubated for 18 h at 37°C, the amount of inoculation was adjusted to optical density OD600 = 1.0, and confirmed by plate counts of 10 fold dilutions of the bacteria. These bacterial cultures contained approximately 108CFU/mL. The Salmonella morphotype was judged visually on Congo red agar plates, 2 μL of bacterial suspension (OD600 of 4.0) was plated onto LB agar plates without NaCl and complemented with Congo red (40 mg/L, Aladdin) and Coomassie brilliant blue G-250 (20 mg/L, Sangon). The inoculated plates were incubated at 28°C for 48 h and colonies were visualized macroscopically.
The method measures biofilm production on the air-liquid interface in a modified method as previously reported (Ramachandran et al., 2016). Briefly, overnight Salmonella strains were transferred into 3 mL of LB medium without NaCl (1:100 dilutions) for 5 days at 28°C without shaking. To observe the biofilms that form at the liquid-air interface, the culture was gently poured out and further stained with 1% crystal violet.
Biofilm Formation and Quantification Assay
Biofilm formation was observed as described previously with some modifications (Baugh et al., 2012). Experiments in 96-well polystyrene microtiter plates (Sangon) were performed. Overnight cultures were diluted to 106 CFU/mL in LB medium without NaCl. A total volume of 0.2 mL was added per well, followed by 24 h incubation at 28°C in static conditions. The total amount of biofilm biomass was quantified by crystal violet. After careful removal of the medium from wells, the biofilms were rinsed three with deionized water. 1% crystal violet was used to stain biofilms for 15 min, and then the excess of stain was removed by gently washing with deionized water. Residual crystal violet was solubilized with 200 μL of 33% glacial acetic acid per well and the OD595 was measured using an ELISA reader (Thermo Scientific Labsystems 354). LB medium without NaCl was used as a negative control in all biofilm assays. All experiments were repeated three times at different time points.
Construction of Mutants and the Complemented Strains
The flagella-deficient strains were constructed by allelic replacement via homologous recombination (Datsenko and Wanner, 2000). Homologous regions and a chloramphenicol cassette with two FRT sites were PCR-amplified from pKD3, and the purified PCR product was digested with DpnI (Invitrogen) and electroporated into bacteria carrying pKD46. Antibiotic selection and the λRed recombineering system led to homologous recombination between the fragments and the host strain genome, and the recombinants were selected for on agar plates containing chloramphenicol. The pCP20 plasmid was introduced to the recombinant strains to remove the DNA fragment containing the chloramphenicol resistance gene, resulting in a single FRT site within the targeted genomic segment. The markerless mutant strains were verified by genomic DNA PCR using primers that annealed to sequences flanking the target gene and further confirmed by sequencing analysis (Supplementary Table S2 and Supplementary Figures S1, S2).
To generate the ΔflgE complemented strain, the full-length flgE gene was PCR-amplified from S. Typhimurium wild-type strain CMCC50115 genomic DNA using the primers flgE–F3 and flgE–F4 (Supplementary Table S2). The PCR product was ligated into pBad/gIIIA, and the resulting plasmid was transformed into the ΔflgE strain by electroporation. The ΔfliC complemented strain was constructed by the same method.
Motility Assays
Swimming motility was performed as described previously (Duan et al., 2012). Briefly, 30 μL cultured overnight of bacterial was re-inoculated into 3 mL of LB and incubated at 37°C with a shaker at 200 rpm until a density value of approximately 1.0 at OD600. The culture samples were inoculated onto 0.4% tryptone agar plates as 1 μL aliquots (Tryptone 1%, NaCl 0.5%, Agar 0.4%). The bacteria grew for 12 h at 37°C and the swimming diameter was measured. Photographs were taken by using the SONY Alpha 7 camera.
Transmission Electron Microscopy (TEM) Analysis
Flagellar morphology was visualized by TEM with negative staining. A single colony was picked and inoculated into fresh LB medium, and the OD600 value of the bacteria was adjusted to about 0.5. The cells were washed once with deionized water and resuspended in sterile water. A sample of 5 μL of the bacterial solution was added dropwise to a copper mesh having a carbon film and dried at room temperature. Using 1% phosphotungstic acid (pH 7.4) for negative staining 2 min, excess solution was blotted with filter paper and observed via electron microscopy (TecnaiG2 F20, FEI, America).
Western Blot Analysis
Flagellar expression using whole-cell lysates was detected by western blot analysis (Shao et al., 2018). Overnight cultures of the tested bacteria were transformed into the fresh LB medium and cultivated an optimal density value of 1.0 at OD600. 2 mL cultures were centrifuged at 9000 × g for 10 min at 4°C to pellet the bacteria. The bacterial cells from whole-cell lysates were loaded and separated by 12% SDS-PAGE, followed by transfer onto a PVDF membrane (GE Healthcare, Life Science). The blot was incubated with mouse polyclonal antiserum to H1 flagella (1: 2000 dilutions, TBC, China), followed by HRP-conjugated Goat anti-mouse IgG (1: 200 dilutions, Sangon, China) orderly. The signal was detected by Luminata Forte Western HRP Substrate (Millipore). Photographs were taken by using the Tanon 5500 imaging system.
Confocal Laser Scanning Microscopy (CLSM) Imaging of Biofilms
After being cultured overnight in LB medium in a shaking 37°C incubator, the cultures were taken to dilute to 107 CFU/mL in 50 mL LB medium without NaCl in a 100 mL Conical flask. A sterile glass slide was gently added into the diluted culture and was taken to statically incubate at 28°C (Supplementary Figure S4). After the culture time was terminated, the glass slide with biofilms was carefully taken out with tweezers and placed in a clean culture dish, and the residual culture medium on them was skimmed with deionized water. The glass slide was stained with 100 μL of SYTO 9 (1 μg/mL, green fluorescence, labeled bacterial cells) and Alexa Fluor 647 (50 μg/mL, red fluorescence, labeled EPS) solution for 30 min, in the dark at room temperature. Confocal microscopy images were obtained on a Leica DMi8 microscope by using a 40 × objective. The settings of the confocal microscope were as follows: the excitation/emission of Alexa Fluor 647 and SYTO 9 were 650/668 and 480/500 nm, respectively. Stack images were obtained by scanning the biofilms along the Z-axis at 0.5 μm intervals. The confocal images were analyzed using COMSTAT software for simultaneous visualization and quantification of EPS and bacterial cells within intact biofilms (Heydorn et al., 2000). For each image stack, cell biomass (μm3/μm2) is defined as the percentage of area occupied by cells labeled by SYTO 9 (green fluorescence); EPS biomass (μm3/μm2) is defined as the percentage of area occupied by EPS labeled by concanavalin A-Alexa Fluor 647 (red fluorescence); Total biomass (μm3/μm2) is cell biomass plus EPS biomass.
Statistical Analysis
The statistical significance of numerical data was compared using variance analysis technology. Significant levels of p values were reported by the following symbols: ∗p < 0.05, ∗∗p < 0.005, and ns, with no difference.
Results
S. Typhimurium CMCC 50115 Biofilm Production
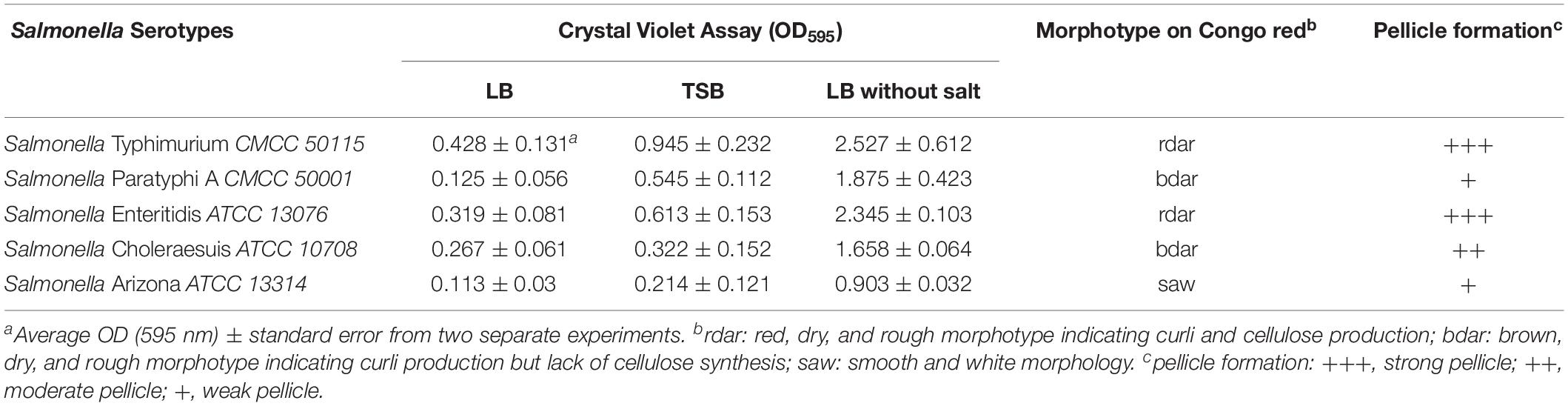
The best-studied Salmonella biofilm phenotype is the rdar morphotype (Römling et al., 1998). After culturing on media containing Congo red at 28°C for 2 days, both S. Typhimurium CMCC50115 and S. Enteritidis ATCC 13076 formed strong rdar morphotypes (Figure 1). In liquid culture, biofilms form a matrix comprising curli and cellulose called pellicles, which appear as films of cell growth at the air-liquid interface (Solano et al., 2002). S. Typhimurium and S. Enteritidis, but not the other strains, formed obvious pellicles at the liquid-air interface (Figure 1). Since crystal violet staining showed that S. Typhimurium CMCC50115 formed more biofilms compared with the other strains (Table 1), we selected it for use as our wild-type control.
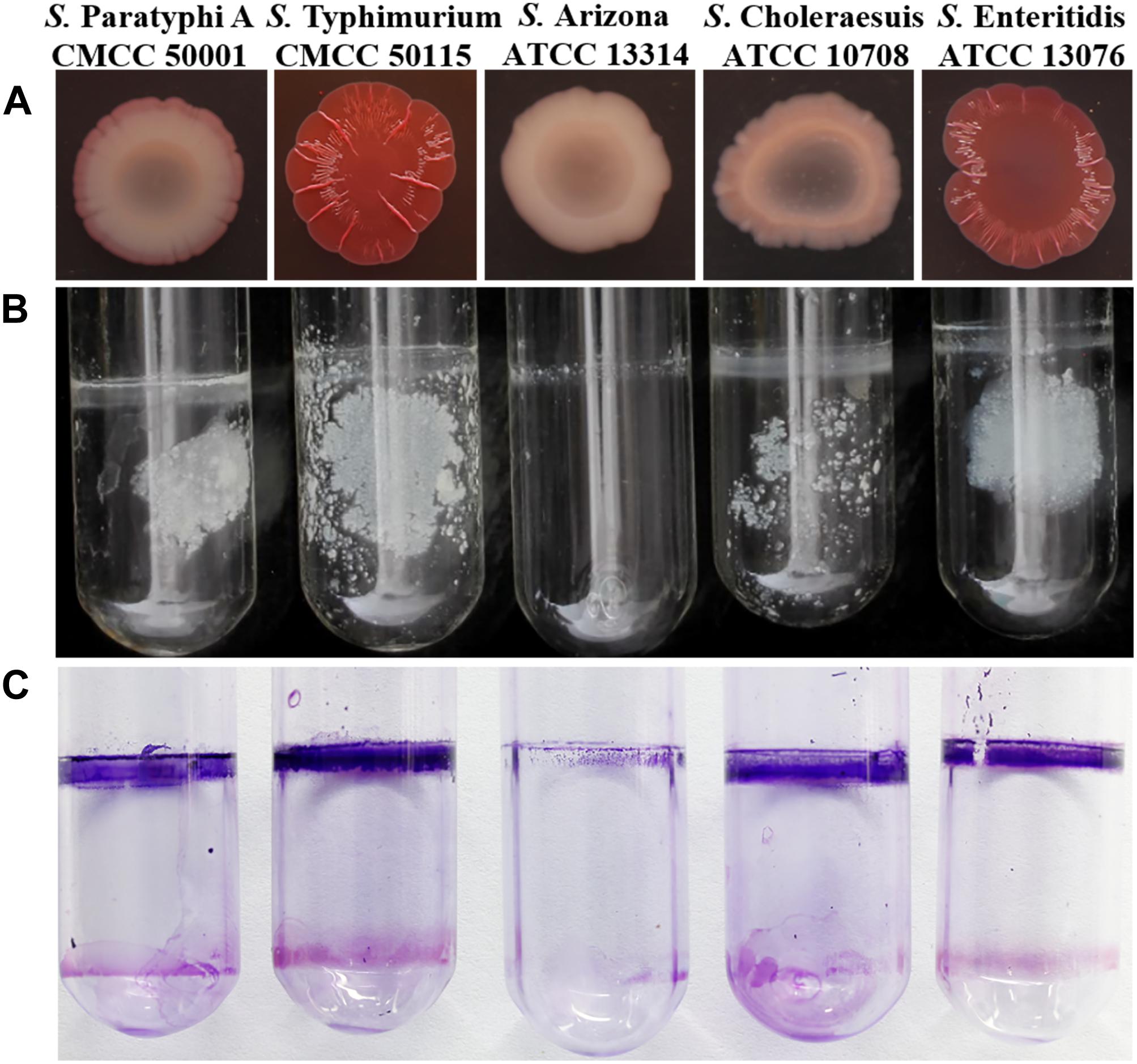
Figure 1. Phenotype of Salmonella assay. (A) Congo red morphotype assay five Salmonella strains; (B) Pellicle formation at the liquid-air interface of five Salmonella strains; (C) Crystal violet stained and observed the formed pellicle of five Salmonella strains.
Construction and Characterization of Flagella Mutants
The wild-type strain was used to construct the ΔflgE and ΔfliC isogenic mutants via homologous recombination. The resulting ΔflgE and ΔfliC mutants exhibited impaired motility on 0.4% tryptone agar plates (Figure 2A). In the case of the ΔflgE strain, this may have been because of the loss of flagella. Confirming this, the flagellin protein was not detected by western blot analysis of whole-cell lysates (Figure 2G), and flagella were not observed by TEM in this mutant strain (Figures 2B,D). In contrast, flagellar protein expression (Figure 2G) and flagella production (Figure 2E) was restored in the complemented strain (ΔflgE/pflgE), which contained a recombinant plasmid expressing FlgE. In the ΔfliC strain, the impaired motility phenotype may be related to the absence of the FliC subunit, which plays a significant role in flagellar motility. As S. Typhimurium expresses two filament proteins, FliC (H1 flagellin) and FljB (H2 flagellin), the ΔfliC mutant could be flagellated under some circumstances. Corroborating this, flagellin protein was not detected by western blot analysis of whole-cell lysates (Figure 2G), but the ΔfliC strain had flagella (presumably composed of FljB), as observed by TEM (Figure 2C). In contrast, flagellar protein expression (Figure 2G) and flagella production (Figure 2F) was restored in the complemented strain (ΔfliC/pfliC), which contained a recombinant plasmid expressing FliC. In addition, it was verified that the fljB sequence of the ΔfliC strain did not mutate (Supplementary Figure S7). Therefore, the ΔfliC strain lacking motility through FljB probably depends on some characteristics of the used strain. Taken together these data show that flgE and fliC are indispensable for flagellar motility.
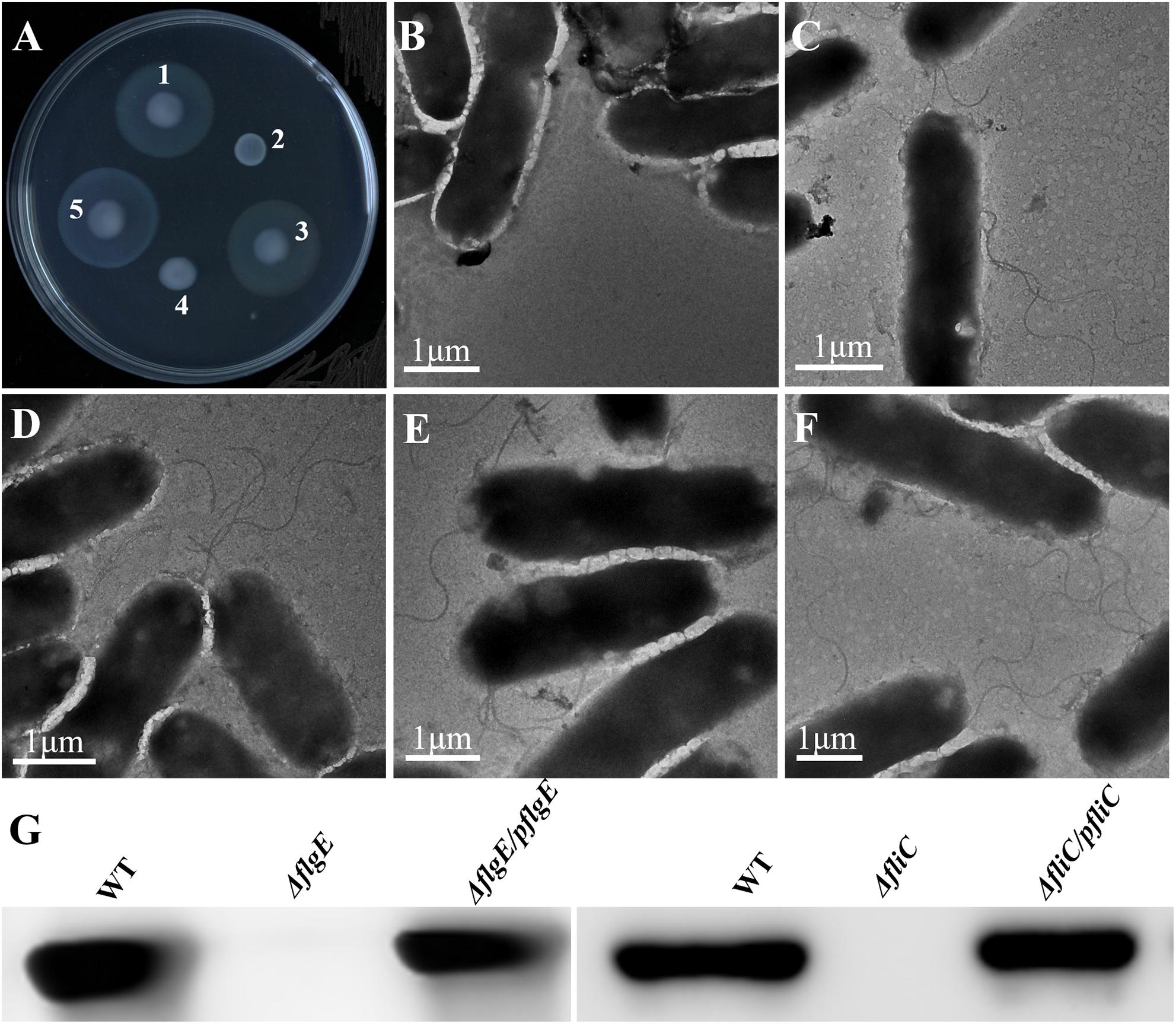
Figure 2. Phenotypes of wild-type S. Typhimurium and isogenic mutants. (A) Motility of (1) S. Typhimurium; (2) ΔflgE (S. Typhimurium flgE–); (3) ΔflgE/pflgE (ΔflgE complemented with pflgE); (4) ΔfliC (S. Typhimurium fliC–); (5) ΔfliC/pfliC (ΔfliC complemented with pfliC) incubated for 12 h at 37°C. (B–F) Expression of flagella by the isogenic mutants was assessed by TEM, (B): ΔflgE, (C): ΔfliC, (D): WT, (E): ΔflgE/pflgE, (F): ΔfliC/pfliC; (G): Flagellin deficiency was confirmed by immunoblot analysis with mouse polyclonal antiserum to H1 flagella. WT, wild-type.
Flagella Mutants Are Defective in the Early Stage of Biofilm Formation
The initial attachment of bacteria to a surface is a key step in their ability to form a biofilm. Compared with the wild-type strain, the two flagellar mutants were defective in the early stage of biofilm formation (Figure 3A). We measured biofilm formation by crystal violet staining and found that the two mutants formed fewer biofilms than the wild-type strain, with an average absorbance at 595 nm (OD595) of 0.93 ± 0.13 and 1.61 ± 0.21, respectively, compared with an OD595 of 2.47 ± 0.16 for the wild-type strain (Figures 3A,B). There was no difference in viability between the flagella mutants and the wild-type strain (Figure 3C). Importantly, biofilm formation was restored to wild-type levels when the mutants complemented (Figures 3A,B). Overall, these data demonstrate that flagella-mediated motility plays an important role in the early stage of biofilm formation.
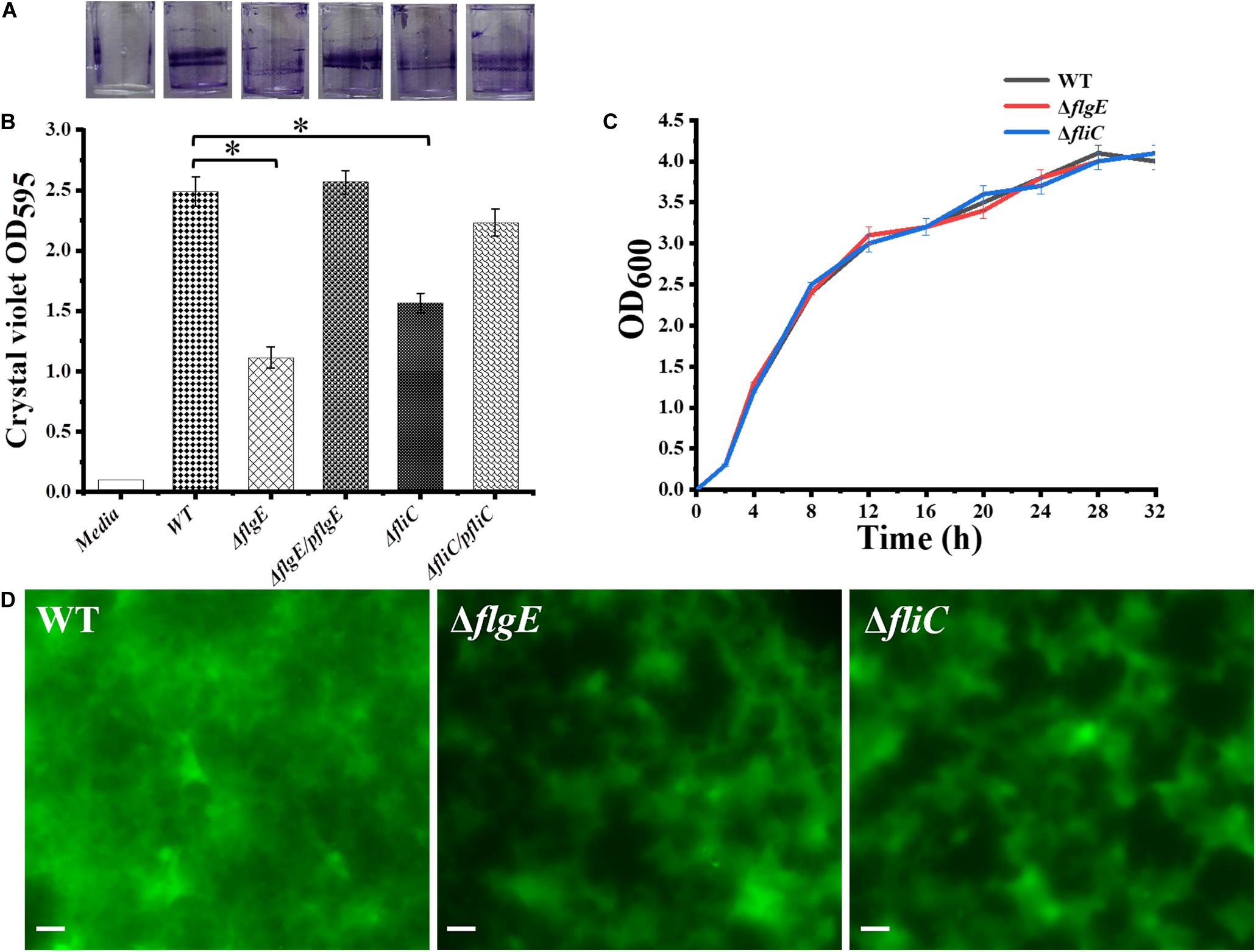
Figure 3. Deletion of the gene encoding flagella was defective in biofilm formation. (A) Crystal violet stained S. Typhimurium biofilms formed on wells of a 96-well polystyrene plate. The first well has medium alone (Media), followed by the wild-type (WT), flgE mutants (ΔflgE), ΔflgE complemented with pflgE (ΔflgE/pflgE), fliC mutants (ΔfliC), ΔfliC complemented with pfliC (ΔfliC/pfliC). (B) Graph of surface-adhered biofilms 24 h biomass of controls (Media and WT), mutants (ΔflgE, ΔfliC) and complemented strains (ΔflgE/pflgE, ΔfliC/pfliC) were quantified by staining with cystal violet. The data are averages of four independent experiments and were analyzed using the Student’s two-tailed t-test. Error bars represent the standard deviation. (C) Growth curves for WT, ΔfliC, ΔflgE. Bacteria were grown in LB broth at 37°C for 28 h with agitation, and the OD600 was determined at hourly intervals. The data are the means of three independent experiments and error bars represent the standard deviation. (D) Top-down view of 24 h biofilms stained with SYTO 9 and propidium iodide. Scale bar: 20 μm. ∗p < 0.05.
Biofilm architecture can be assessed by staining with SYTO 9 and propidium iodide (Vermilyea et al., 2019). Fluorescence microscopy confirmed that biofilms formed by the ΔflgE and ΔfliC mutants comprised less biomass than those formed by the wild-type strain (Figures 3D, 4G). Biofilm composition was also evaluated by staining with SYTO 9 (which detects all bacteria) and concanavalin A-Alexa Fluor 647 (which detects EPS). There were significantly fewer bacteria in biofilms formed by the flagella mutants compared with those formed by the wild-type strain (Figure 4E). However, there was no significant difference in EPS content between the biofilms formed by the mutant and wild-type strains (Figure 4F). In addition, there was no significant difference in biofilm thickness between the flagellar mutants and the wild-type strain (Figures 4A–D). Therefore, the reduced biofilm formation exhibited by the flagellar mutants could be due to reduced adhesion of planktonic cells at the initial stage compared with the wild-type strain.
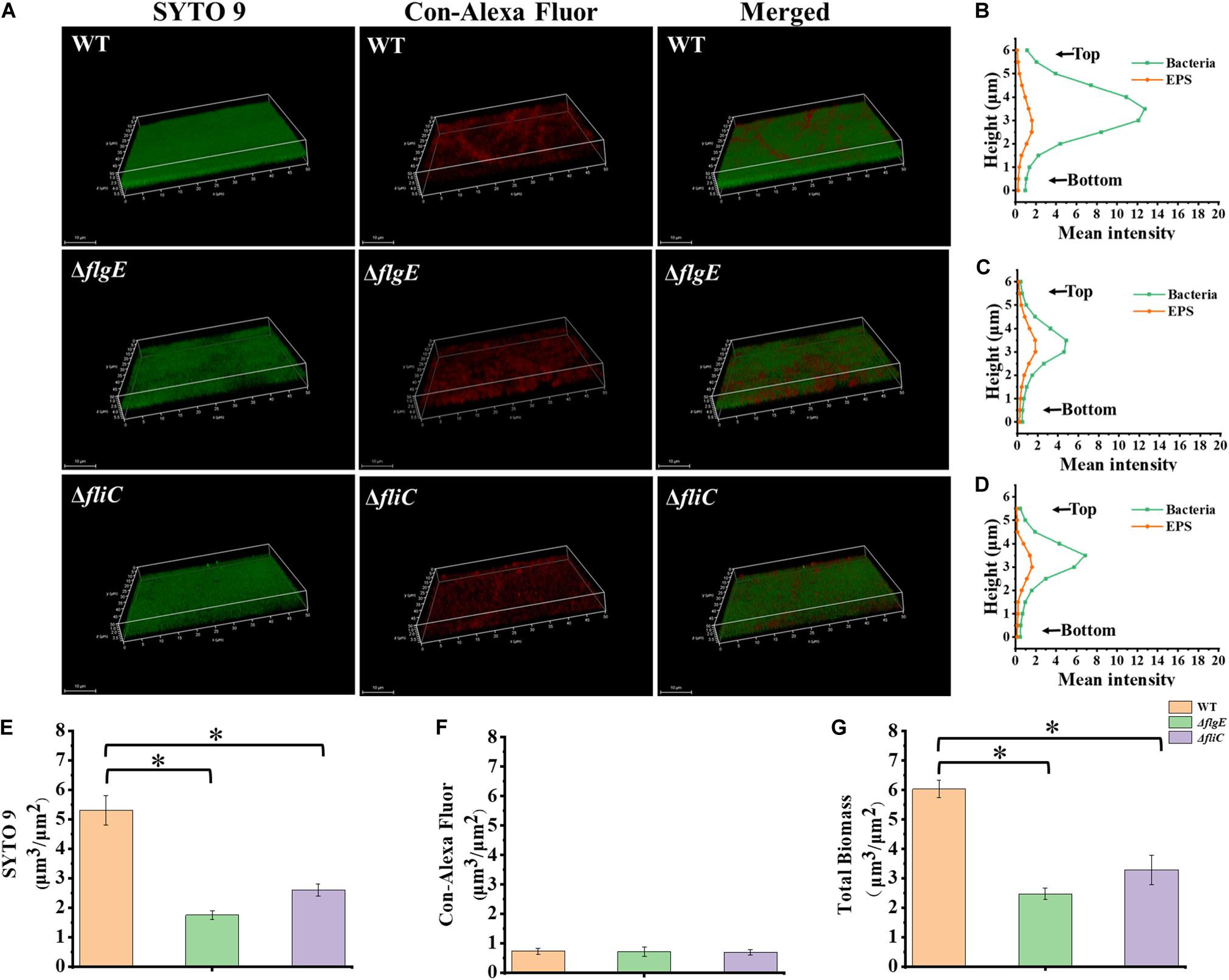
Figure 4. ΔflgE and ΔfliC biofilms were comprised of less bacterial cells than S. Typhimurium wild-type biofilms. (A) ΔflgE,ΔfliC, and wild-type were grown for 24 h on the glass, stained with SYTO 9 (green) and Con-Alexa Fluor (red), and 3D images were acquired by CLSM, Scale bar: 10 μm. (B–D), the bacteria and EPS of biofilm thickness in strains WT, ΔflgE, and ΔfliC are, respectively, indicated. (E) SYTO 9, (F) Con-Alexa Fluor, and (G) Total biomass were quantified using Comstat 2. Data are averages of three replicates (n = 3). Error bars represent the standard deviation. The data were analyzed using the Student’s two-tailed t-test. *p < 0.05.
Flagella Mutants Produce More Thick and Dense Biofilms
To explore the role of flagella on biofilm maturation, CLSM was used to directly observe biofilms that had been allowed to form for 48 h. The wild-type biofilms were twice as thick as the ΔflgE biofilms and had a looser inner structure than the mutant biofilms (Figures 5A–D). Additionally, the ΔflgE biofilms contained more cells and EPS than the wild-type biofilms (Figures 5E–G). The ΔfliC and ΔflgE biofilms exhibited similar phenotypes, and flagellar motility was impaired in both strains, suggesting that flagellar motility plays an important role in forming the structure of biofilms. Taken together, these findings indicate that the thick phenotype of biofilms formed by the flagella mutants is due to the accumulation of cells and EPS.
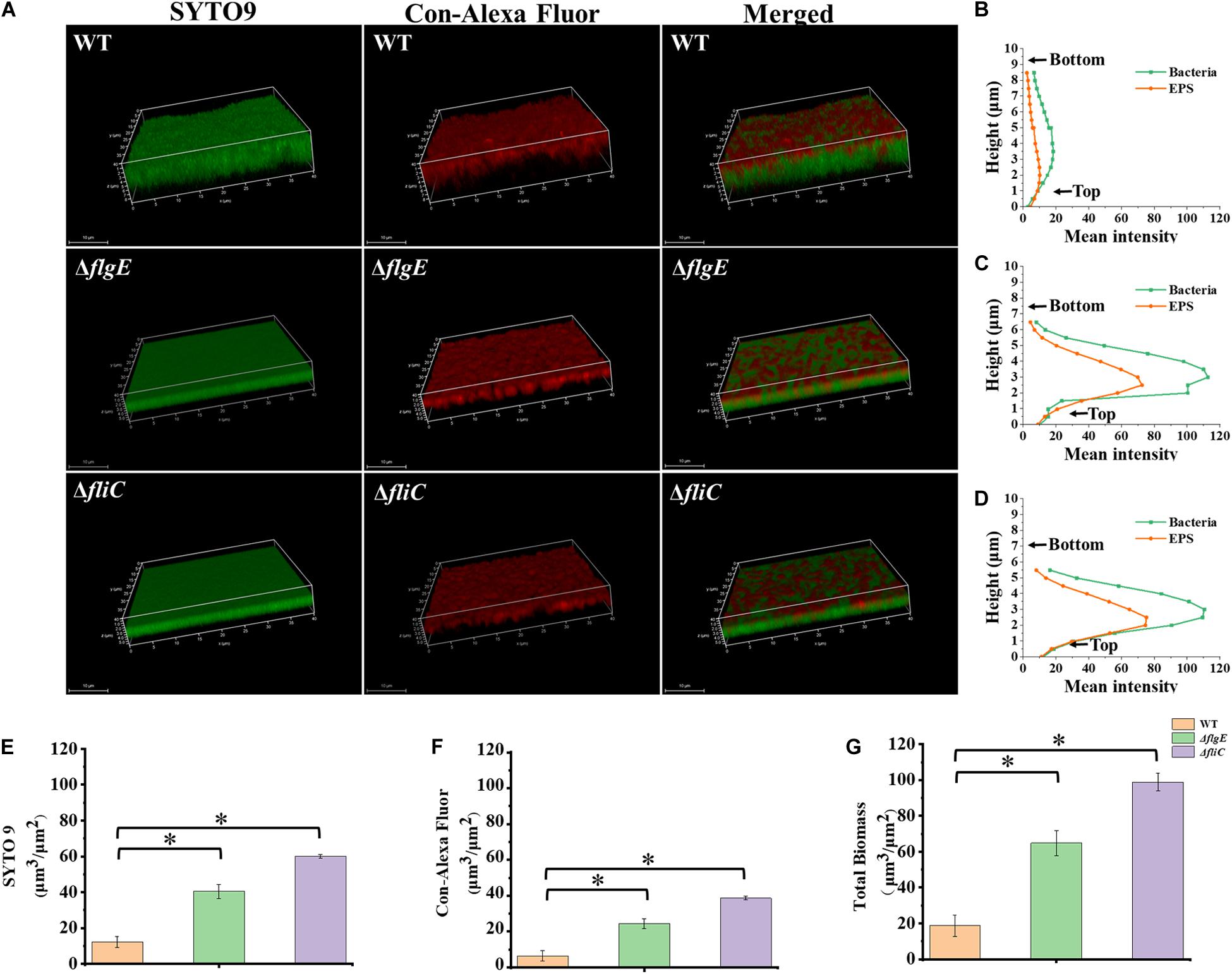
Figure 5. ΔflgE and ΔfliC biofilms were comprised of more bacterial cells and EPS than S. Typhimurium wild-type biofilms. (A) ΔflgE, ΔfliC, and wild-type were grown for 48 h on the glass, stained with SYTO 9 and Con-Alexa Fluor, and 3D images were acquired by CLSM, Scale bar: 10 μm. (B–D), the bacteria and EPS of biofilm thickness in strains WT, ΔflgE, and ΔfliC are, respectively, indicated. (E) SYTO 9, (F) Con-Alexa Fluor, and (G) Total biomass were quantified using Comstat 2. Data are averages of three replicates (n = 3). Error bars represent the standard deviation. The data were analyzed using the Student’s two-tailed t-test. *p < 0.05.
Flagella Mutants Exhibit Poor Adhesion, Which Inhibits the Initiation of Biofilm Formation
Next, we investigated the role of flagellar motility in the early steps in biofilm formation. To assess the initiation of biofilm formation by the flagella mutants and the wild-type strain, we directly visualized biofilm formation on sterile glass slides using a confocal fluorescence microscope. Figure 6A shows a time-course of the development of wild-type biofilms on a sterile glass slide over 20 h at 28°C. Six hours after inoculation, the bacteria had aggregated into small clusters that gradually cross-linked together over the next 6–9 h. By 20 h, the clusters had developed into microcolonies comprising multiple layers of cells.
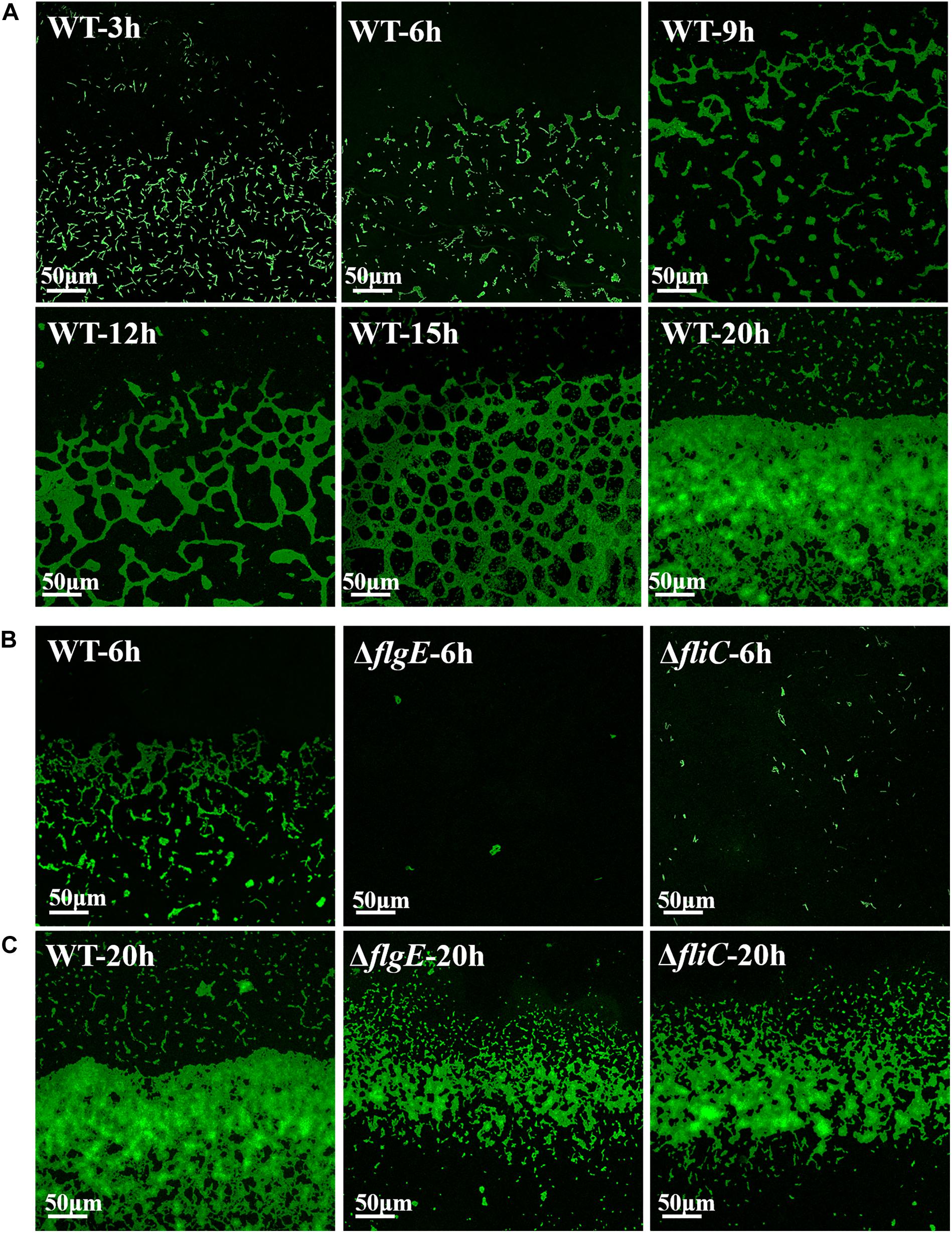
Figure 6. Direct observation of biofilm formation on a sterile glass slide using a confocal fluorescence microscope. (A) Observed the adhesion of wild-type biofilms on the sterile glass slide over 20 h. (B) Shown are fluorescence micrographs of the wild-type strain, ΔflgE, and ΔfliC after incubation for 6 h at 28°C. (C) The biofilms of the wild-type, ΔflgE, and ΔfliC at 20 h were displayed. The micrographs were taken at 400 × magnification and the representative fields are shown and the experiments were repeated three times.
In contrast, 6 h after inoculation, the ΔflgE mutant had only formed a few scattered cell clusters (Figure 6B). By 20 h, the ΔflgE strain exhibited fewer colonies and a smaller total attachment area compared with the wild-type strain (Figure 6C). Further statistics on them continually supported the results (Supplementary Figures S5, S6). The ΔfliC mutants exhibited similar phenotypes. These observations are consistent with our earlier results showing the importance of flagella-mediated motility in biofilm formation.
Effect of the Protonophore CCCP on Biofilm Formation
The motility of most bacterial flagella is driven by a proton gradient across the cytoplasmic membrane, so the protonophore CCCP was applied to explore the role of flagellar motility on biofilm formation (Kosaka et al., 2019). First, we treated the wild-type strain with CCCP to determine the optimal concentration for inhibiting cell spreading on semisolid agar. In the presence of 10 μM of the protonophore CCCP on 0.4% tryptone agar plates, the growth of the wild-type strain was not affected (Figure 7A), but its motility was dramatically reduced (Figures 7B,C). Crystal violet staining revealed that the phenotype of the CCCP-treated wild-type strain was consistent with the phenotype of the ΔflgE strain (Figure 7D). These results clearly show that flagellar motility plays a key role in S. Typhimurium biofilm formation. Treatment with CCCP inhibited the motility of S. Typhimurium flagella and thereby reduced the formation of biofilms. Similar results were obtained when Erwinia carotovora subsp. carotovora was treated with CCCP, indicating that the role of flagellar motility in biofilm formation is relatively conserved (Hossain and Tsuyumu, 2006).
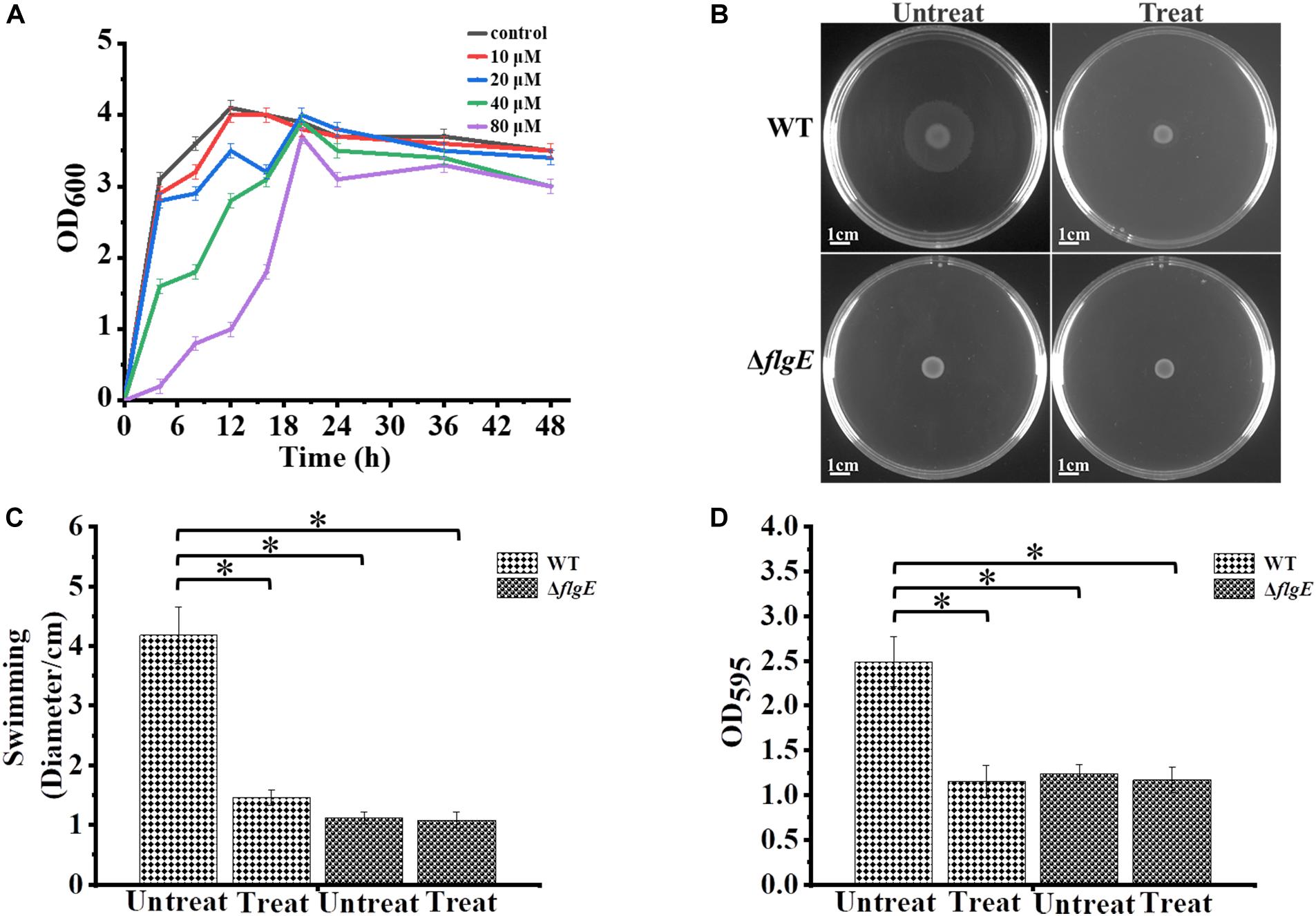
Figure 7. Biofilm formation by S. Typhimurium after treatment with CCCP. (A) Analysis of the growth curve of wild-type strains treated with different concentrations of CCCP. (B) Swimming plates of different groups captured after incubation at 37°C for 12 h. Treat: add 1 μL of 100 mM CCCP to a final concentration of 10 μM CCCP per plate, Untreat: add 1 μL DMSO according to the above treatment. (C) The quantitative analysis diameter of swimming. (D) The bacteria were cultured with or without 10 μM protonophore CCCP, a quantitative analysis of the formation of biofilms cultured for 24 h by crystal violet staining. Data in (B–D) are representative of three independent experiments. Error bars represent the standard deviation. ∗p < 0.05.
Discussion
Salmonella is an important food-borne pathogen found all over the world. It has been implicated in the etiology of life-threatening diarrheal diseases, partially due to its ability to colonize and form biofilms on various equipment used in the food production industry (Waldner et al., 2012). Our findings show that the impaired flagellar motility mutant exhibits poor adhesion and smaller colonies in initial biofilms. However, the mutant has a denser and flatter inner structure in mature biofilms. In E. coli, flagellar motility is essential for overcoming electrostatic repulsion and tethering cells together during biofilm formation (Pratt and Kolter, 1998; Walker et al., 2004; Serra et al., 2013). Furthermore, high motility leads to vertical structures and low motility leads to flatter microcolonies in mature biofilm architecture (Wood et al., 2006). Our results also confirm that the flagellar motility and biofilm machinery of S. Typhimurium and E. coli are very similar.
Previous studies have shown that the behavior of a bacterial community at an air-liquid, surface-liquid, or cell-liquid interface changes in the absence of flagella (Römling and Rohde, 1999). Additionally, the flagellar filament (containing the FliC and FljB subunits), and specifically the FliC subunit, of serovar Typhimurium is necessary and specific for cholesterol binding during biofilm initiation (Crawford et al., 2010). Our results show that flagellar motility is impaired in the ΔfliC strain, which exhibits a similar phenotype to the ΔflgE strain (which is aflagellate). Therefore, it was speculated that the FliC-type filament may play a key role in biofilm formation on abiotic surfaces. The ΔfliC also showed a somewhat intermediate biofilm phenotype between the WT and ΔflgE, which may be the compensation effect of the fljB subunit. Detailed observation by confocal fluorescence microscopy revealed that the flagella mutants (ΔflgE and ΔfliC) formed mature biofilms with increased content of aggregated cells and EPS compared with biofilms formed by the wild-type strain. It appears that motility and extracellular matrix production are mutually exclusive processes, as many motile bacteria only begin to produce matrix once they have made contact with a surface (Kolter and Greenberg, 2006). We speculate that this may also be true for Salmonella, although the specific mechanism regulating this switch requires further study.
Biofilm formation is a complex process that is affected by environmental conditions, gene expression profiles, and physiological shifts within the cell (Mireles et al., 2001). Modulating some of these factors could help prevent biofilm formation. Our results confirmed that flagella-mediated motility is very important in early biofilm formation by S. Typhimurium. Our observations regarding the early stages of biofilm development help clarify the mechanisms involved in biofilm resistance and hold promise for developing more effective control strategies for use in the food industry. For example, aptamers or zinc targeting flagella motility (Nielubowicz et al., 2010; Ning et al., 2015; Ammendola et al., 2016), anti-adhesive materials, and the combination of some anti-adhesion materials with antibiotics could be used to control biofilm formation. The results regarding the composition of mature biofilms help us make methods to clear them. Finally, the methods might be directed toward the degradation of the EPS and kill the bacteria in biofilms, which can achieve better therapeutic effects.
Conclusion
In summary, the formation of Salmonella biofilms is a threat to food safety and public health. Understanding the mechanisms underlying biofilm formation will help identify more effective means of treating and preventing the problems it causes. In this study, we constructed ΔflgE and ΔfliC mutants to further explore the development process of Salmonella biofilms. It has been well proved by a series of experiments that impaired flagellar motility mutants exhibit poor adhesion and small colonies in initial biofilms, conversely, they have more dense and complex structures in their mature biofilms. In the near future, flagellar motility could be inhibited to help eliminate biofilms, but it is necessary to select the appropriate treatment time to achieve the desired therapeutic effect.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
Author Contributions
FW designed, supervised the experiments, analyzed the results, revised the first draft, and prepared the last draft of the manuscript. LD performed part of the experiments, contributed significantly to analysis and manuscript preparation, and revised the last draft of the manuscript. FH performed part of the experiments, analyzed the data, and participated in the first draft of the manuscript. ZW and QL analyzed the data and helped perform the analysis with constructive discussions. CX collaborated in the design of the experiments and revised different versions of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (31770109) and the Opening Fund of Key Laboratory of Chemical Biology and Traditional Chinese Medicine Research (Hunan Normal University), Ministry of Education (KLCBTCMR18-03).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01695/full#supplementary-material
References
Achinas, S., Charalampogiannis, N., and Euverink, G.-J. (2019). A brief recap of microbial adhesion and biofilms. Appl, Sci. 9:2801. doi: 10.3390/app9142801
Aizawa, S.-I. (1996). Flagellar assembly in Salmonella typhimurium. Mol. Microbiol. 19, 1–5. doi: 10.1046/j.1365-2958.1996.344874.x
Ammendola, S., D’Amico, Y., Chirullo, B., Drumo, R., Civardelli, D., Pasquali, P., et al. (2016). Zinc is required to ensure the expression of flagella and the ability to form biofilms in Salmonella enterica sv Typhimurium. Metallomics 8, 1131–1140. doi: 10.1039/C6MT00108D
Baugh, S., Ekanayaka, A. S., Piddock, L. J. V., and Webber, M. A. (2012). Loss of or inhibition of all multidrug resistance efflux pumps of Salmonella enterica serovar Typhimurium results in impaired ability to form a biofilm. J. Antimicrob. Chemother. 67, 2409–2417. doi: 10.1093/jac/dks228
Bearson, B. L., Bearson, S. M. D., Lee, I. S., and Brunelle, B. W. (2010). The Salmonella enterica serovar Typhimurium QseB response regulator negatively regulates bacterial motility and swine colonization in the absence of the QseC sensor kinase. Microb. Pathog. 48, 214–219. doi: 10.1016/j.micpath.2010.03.005
Bonifield, H. R., Yamaguchi, S., and Hughes, K. T. (2000). The flagellar hook protein, FlgE, of Salmonella enterica serovar typhimurium is posttranscriptionally regulated in response to the stage of flagellar assembly. J. Bacteriol. 182, 4044–4050. doi: 10.1128/jb.182.14.4044-4050.2000
Chilcott, G. S., and Hughes, K. T. (2000). Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar typhimurium and Escherichia coli. Microbiol. Mol. Biol. Rev. 64, 694–708. doi: 10.1128/mmbr.64.4.694-708.2000
Crawford, R. W., Reeve, K. E., and Gunn, J. S. (2010). Flagellated but not hyperfimbriated Salmonella enterica serovar typhimurium attaches to and forms biofilms on cholesterol-coated surfaces. J. Bacteriol. 192, 2981–2990. doi: 10.1128/jb.01620-09
Datsenko, K. A., and Wanner, B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645. doi: 10.1073/pnas.120163297
Duan, Q., Zhou, M., Zhu, X., Bao, W., Wu, S., Ruan, X., et al. (2012). The flagella of F18ab Escherichia coli is a virulence factor that contributes to infection in a IPEC-J2 cell model in vitro. Vet. Microbiol. 160, 132–140. doi: 10.1016/j.vetmic.2012.05.015
Haiko, J., and Westerlund-Wikström, B. (2013). The role of the bacterial flagellum in adhesion and virulence. Biology 2, 1242–1267. doi: 10.3390/biology2041242
Han, D., and Yoon, J. W. (2019). Cyclic di-guanosine monophosphate signaling regulates bacterial life cycle and pathogenicity. J. Prev. Vet. Med. 43, 38–46. doi: 10.13041/jpvm.2019.43.1.38
Heydorn, A., Nielsen, A. T., Hentzer, M., Sternberg, C., Givskov, M., Ersbøll, B. K., et al. (2000). Quantification of biofilm structures by the novel computer program comstat. Microbiology 146, 2395–2407. doi: 10.1099/00221287-146-10-2395
Hossain, M. M., and Tsuyumu, S. (2006). Flagella-mediated motility is required for biofilm formation by Erwinia carotovora subsp. carotovora. J. Gen. Plant Pathol. 72, 34–39. doi: 10.1007/s10327-005-0246-8
Huber, B., Riedel, K., Köthe, M., Givskov, M., Molin, S., and Eberl, L. (2002). Genetic analysis of functions involved in the late stages of biofilm development in Burkholderia cepacia H111. Mol. Microbiol. 46, 411–426. doi: 10.1046/j.1365-2958.2002.03182.x
Jacobsen, C. S., and Bech, T. B. (2012). Soil survival of Salmonella and transfer to freshwater and fresh produce. Food Res. Int. 45, 557–566. doi: 10.1016/j.foodres.2011.07.026
Janssens, J. C. A., Steenackers, H., Robijns, S., Gellens, E., Levin, J., Zhao, H., et al. (2008). Brominated furanones inhibit biofilm formation by Salmonella enterica serovar typhimurium. Appl. Environ. Microbiol. 74, 6639–6648. doi: 10.1128/AEM.01262-08
Ji, Y., Li, W., Zhang, Y., Chen, L., Zhang, Y., Zheng, X., et al. (2017). QseB mediates biofilm formation and invasion in Salmonella enterica serovar Typhi. Microb. Pathog. 104, 6–11. doi: 10.1016/j.micpath.2017.01.010
Kolter, R., and Greenberg, E. P. (2006). The superficial life of microbes. Nature 441, 300–302. doi: 10.1038/441300a
Koo, H., Allan, R. N., Howlin, R. P., Stoodley, P., and Hall-Stoodley, L. (2017). Targeting microbial biofilms: current and prospective therapeutic strategies. Nat. Rev. Microbiol. 15, 740–755. doi: 10.1038/nrmicro.2017.99
Kosaka, T., Goda, M., Inoue, M., Yakushi, T., and Yamada, M. (2019). Flagellum-mediated motility in Pelotomaculum thermopropionicum SI. Biosci. Biotechnol. Biochem. 83, 1362–1371. doi: 10.1080/09168451.2019.1597618
Le Guyon, S., Simm, R., Rehn, M., and Romling, U. (2015). Dissecting the cyclic di-guanylate monophosphate signalling network regulating motility in Salmonella enterica serovar Typhimurium. Environ. Microbiol. 17, 1310–1320. doi: 10.1111/1462-2920.12580
Lemon, K. P., Higgins, D. E., and Kolter, R. (2007). Flagellar motility is critical for listeria monocytogenes biofilm formation. J. Bacteriol. 189, 4418–4424. doi: 10.1128/jb.01967-06
MacKenzie, K. D., Palmer, M. B., Köster, W. L., and White, A. P. (2017). Examining the link between biofilm formation and the ability of pathogenic Salmonella strains to colonize multiple host species. Front. Vet. Sci. 4:138. doi: 10.3389/fvets.2017.00138
Merighi, M., Septer, A. N., Carroll-Portillo, A., Bhatiya, A., Porwollik, S., McClelland, M., et al. (2009). Genome-wide analysis of the PreA/PreB (QseB/QseC) regulon of Salmonella enterica serovar Typhimurium. BMC Microbiol. 9:42. doi: 10.1186/1471-2180-9-42
Mireles, J. R., Toguchi, A., and Harshey, R. M. (2001). Salmonella enterica serovar typhimurium swarming mutants with altered biofilm-forming abilities: surfactin inhibits biofilm formation. J. Bacteriol. 183, 5848–5854. doi: 10.1128/JB.183.20.5848-5854.2001
Nielubowicz, G. R., Smith, S. N., and Mobley, H. L. T. (2010). Zinc uptake contributes to motility and provides a competitive advantage to Proteus mirabilis during experimental urinary tract infection. Infect. Immun. 78, 2823–2833. doi: 10.1128/iai.01220-09
Ning, Y., Cheng, L., Ling, M., Feng, X., Chen, L., Wu, M., et al. (2015). Efficient suppression of biofilm formation by a nucleic acid aptamer. Pathog. Dis. 73:ftv034. doi: 10.1093/femspd/ftv034
O’Toole, G., Kaplan, H. B., and Kolter, R. (2000). Biofilm formation as microbial development. Annu. Rev. Microbiol. 54, 49–79. doi: 10.1146/annurev.micro.54.1.49
O’Toole, G. A., and Kolter, R. (1998). Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30, 295–304. doi: 10.1046/j.1365-2958.1998.01062.x
Pande, V. V., McWhorter, A. R., and Chousalkar, K. K. (2016). Salmonella enterica isolates from layer farm environments are able to form biofilm on eggshell surfaces. Biofouling 32, 699–710. doi: 10.1080/08927014.2016.1191068
Pratt, L. A., and Kolter, R. (1998). Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol. Microbiol. 30, 285–293. doi: 10.1046/j.1365-2958.1998.01061.x
Prouty, A. M., and Gunn, J. S. (2003). Comparative analysis of Salmonella enterica serovar typhimurium biofilm formation on gallstones and on glass. Infect. Immun. 71, 7154–7158. doi: 10.1128/iai.71.12.7154-7158.2003
Ramachandran, G., Aheto, K., Shirtliff, M. E., and Tennant, S. M. (2016). Poor biofilm-forming ability and long-term survival of invasive Salmonella Typhimurium ST313. Pathog. Dis. 74:ftw049. doi: 10.1093/femspd/ftw049
Römling, U., Sierralta, W. D., Eriksson, K., and Normark, S. (1998). Multicellular and aggregative behaviour of Salmonella typhimurium strains is controlled by mutations in the agfD promoter. Mol. Microbiol. 28, 249–264. doi: 10.1046/j.1365-2958.1998.00791.x
Römling, U., and Rohde, M. (1999). Flagella modulate the multicellular behavior of Salmonella typhimurium on the community level. FEMS Microbiol. Lett. 180, 91–102. doi: 10.1111/j.1574-6968.1999.tb08782.x
Semenov, A. M., Kuprianov, A. A., and van Bruggen, A. H. C. (2010). Transfer of enteric pathogens to successive habitats as part of microbial cycles. Microb. Ecol. 60, 239–249. doi: 10.1007/s00248-010-9663-0
Serra, D. O., Richter, A. M., Klauck, G., Mika, F., and Hengge, R. (2013). Microanatomy at cellular resolution and spatial order of physiological differentiation in a bacterial biofilm. mBio 4:e00103-13. doi: 10.1128/mBio.00103-13
Shao, X., Zhang, X., Zhang, Y., Zhu, M., Yang, P., Yuan, J., et al. (2018). RpoN-dependent direct regulation of quorum sensing and the type VI secretion system in Pseudomonas aeruginosa PAO1. J. Bacteriol. 200:e00205-18. doi: 10.1128/jb.00205-18
Simm, R., Ahmad, I., Rhen, M., Le Guyon, S., and Römling, U. (2014). Regulation of biofilm formation in Salmonella enterica serovar Typhimurium. Future Microbiol. 9, 1261–1282. doi: 10.2217/fmb.14.88
Solano, C., García, B., Valle, J., Berasain, C., Ghigo, J.-M., Gamazo, C., et al. (2002). Genetic analysis of Salmonella enteritidis biofilm formation: critical role of cellulose. Mol. Microbiol. 43, 793–808. doi: 10.1046/j.1365-2958.2002.02802.x
Stoodley, P., Sauer, K., Davies, D. G., and Costerton, J. W. (2002). Biofilms as complex differentiated communities. Annu. Rev. Microbiol. 56, 187–209. doi: 10.1146/annurev.micro.56.012302.160705
Tsai, M.-H., Liang, Y.-H., Chen, C.-L., and Chiu, C.-H. (2019). Characterization of Salmonella resistance to bile during biofilm formation. J. Microbiol. Immunol. Infect. 53, 518–524. doi: 10.1016/j.jmii.2019.06.003
Vermilyea, D. M., Ottenberg, G. K., and Davey, M. E. (2019). Citrullination mediated by PPAD constrains biofilm formation in P. gingivalis strain 381. npj Biofilms Microb. 5:7. doi: 10.1038/s41522-019-0081-x
Waldner, L. L., MacKenzie, K. D., Köster, W., and White, A. P. (2012). From exit to entry: long-term survival and transmission of Salmonella. Pathogens 1, 128–155.
Walker, S. L., Redman, J. A., and Elimelech, M. (2004). Role of cell surface lipopolysaccharides in Escherichia coli K12 adhesion and transport. Langmuir 20, 7736–7746. doi: 10.1021/la049511f
Watnick, P., and Kolter, R. (2000). Biofilm, city of microbes. J. Bacteriol. 182, 2675–2679. doi: 10.1128/JB.182.10.2675-2679.2000
Keywords: flagellum, biofilms, motility, S. Typhimurium, confocal laser scanning microscopy
Citation: Wang F, Deng L, Huang F, Wang Z, Lu Q and Xu C (2020) Flagellar Motility Is Critical for Salmonella enterica Serovar Typhimurium Biofilm Development. Front. Microbiol. 11:1695. doi: 10.3389/fmicb.2020.01695
Received: 19 September 2019; Accepted: 29 June 2020;
Published: 09 September 2020.
Edited by:
Yang Deng, Qingdao Agricultural University, ChinaReviewed by:
Jose Antonio Ibarra, National Polytechnic Institute, MexicoIrfan Ahmad, University of Health Sciences, Pakistan
Andrea Battistoni, University of Rome Tor Vergata, Italy
Copyright © 2020 Wang, Deng, Huang, Wang, Lu and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Le Deng, Dengle@hunnu.edu.cn
 Feiying Wang
Feiying Wang Le Deng
Le Deng