- Department of Biological Safety, German Federal Institute for Risk Assessment, Berlin, Germany
Methicillin-resistant Staphylococcus aureus (MRSA) have been found on German dairy farms and may be the cause of difficult-to-treat bovine mastitis. Considering the one health approach, MRSA might be transmitted from animals to humans raising the risk for severe infections. On 17 German dairy farms with a history of MRSA detection, MRSA strains were isolated from quarter milk, bulk tank milk, and swab samples of calves, heifers, pigs, and the environment. A selection of 33 isolates was analyzed using whole-genome sequencing and antimicrobial resistance testing. All detected MRSA strains were attributed to the livestock-associated sequence type 398. Methicillin-resistance was associated with the mecA gene in the staphylococcal cassette chromosome (SCC)mec types IVa (7/33) or V (26/33). The MRSA strains across the German federal states showed large allelic differences indicating independent development and distribution. On one farm, a clonal MRSA isolate was widely spread among different animals and the milking equipment. Moreover, MRSA transmission between two dairy farms in one federal state seems to be likely. In depth studies indicated that the resistance gene prediction and phenotypic resistance are in good agreement. Twenty eight strains were determined to exhibit a non-wildtype phenotype (resistant) against up to seven antimicrobial substances with an overall resistance to β-lactams and tetracycline. Ten different phenotypic antimicrobial resistance patterns were found among the MRSA strains. The strains harbored a wide virulence gene repertoire, of which some of them are related to bovine mastitis. However, the isolates lacked typical human infection associated factors such as the immune evasion cluster genes, staphylococcal enterotoxin genes, or Panton-Valentine leukocidin genes leading to the assumption for a low risk for severe human infections and foodborne diseases.
Introduction
Methicillin-resistant Staphylococcus (S.) aureus (MRSA) were repeatedly detected on German dairy farms (Tenhagen et al., 2018; Kadlec et al., 2019) and may be a cause of bovine mastitis (Holmes and Zadoks, 2011). MRSA infections are hard to cure since these bacteria are resistant against β-lactam antibiotics, which are widely used for S. aureus mastitis treatment. In addition to the animal health aspect, MRSA may be transmitted from animals to the farm personnel and sporadically cause severe infections in humans such as dermatitis, otitis, wound infection, pneumonia, endocarditis, or sepsis (Goerge et al., 2017). MRSA may carry resistance genes against several classes of antibiotics and even resistance against last resort antibiotics such as linezolid was found in isolates from various livestock (Cuny et al., 2017). Moreover, MRSA can be equipped with a wide arsenal of virulence factors such as immune evasion clusters (IECs), toxins, or leukocidins. Both, antibiotic resistance and virulence genes, are often encoded on mobile genetic elements (MGEs) giving the possibility to spread resistance or virulence between different strains. The most common MGE in MRSA with regard to antibiotic resistance is the staphylococcal cassette chromosome (SCC)mec element, in which the β-lactam antibiotic resistance gene mecA or its homolog mecC is located. The SCCmec is structurally divided into the types I–XIII (Lakhundi and Zhang, 2018). Moreover, antimicrobial resistance determinants may also be encoded on plasmids (Feßler et al., 2018). Likewise, also virulence factors are found in MGEs across the MRSA genome, e.g., in S. aureus pathogenicity islands (SaPIs) or phages. Livestock-associated MRSA (LA-MRSA) often lack the potential for causing severe human infections due to a lack of IEC genes or genes encoding the toxic shock syndrome toxin (TSST) or Panton-Valentine leucocidin (PVL; Cuny et al., 2015a). However, frequent monitoring of LA-MRSA strains from different livestock farms and the respective environment are necessary, since the genetic repertoire of MRSA strains might change spontaneously due to horizontal gene transfer leading to more harming strains with regard to animal and human health (Kraushaar et al., 2017).
The aim of the study was to compare the genotypes, antimicrobial resistance profiles, and virulence factors of MRSA strains from 17 dairy farms in eight German federal states. Whole-genome sequencing (WGS) of selected strains was conducted and the sequence data were analyzed regarding antimicrobial resistance genes and virulence factors to draw conclusions for a potential public health risk. Furthermore, the phylogenetic relationship between MRSA strains from various regions as well as within one farm was analyzed by core genome multi-locus sequence typing (cgMLST).
Materials and Methods
Sampling and MRSA Strain Selection
For this study, 17 dairy farms across eight German federal states were selected due to a previous positive MRSA detection. Samples from bovine mammary quarters (quarter milk samples, QMS), bulk tank milk (BTM), calves (nasal swabs), heifers (nasal swabs and udder cleft swabs), pigs (nasal swabs) on dairy farms as well as the milking equipment and environment, which were retrieved in a sampling campaign from September 2018 to December 2019, were examined for MRSA. Milk (1 ml) and swab samples were examined using a double selective enrichment method by incubation in Mueller Hinton broth (Thermo Fisher Scientific Oxoid Ltd., United Kingdom) supplemented with 6% of NaCl, tryptic soy broth (Merck, Germany) supplemented with 3.5 mg/l cefoxitin (Sigma-Aldrich, United States) and 50 mg/l aztreonam (Sigma-Aldrich, United States) and subsequent incubation on mannitol salt (Thermo Fisher Scientific Oxoid Ltd., United Kingdom) agar plates containing 4 mg/l cefoxitin (Sigma-Aldrich, United States). Each incubation step lasted for 24 ± 2 h at 37°C. With regard to a potential food intoxication or transmission to humans by the consumption of MRSA contaminated milk, in particular strains from QMS and BTM were chosen for sequencing if available. In total, 33 out of 184 MRSA isolates were selected as most interesting for comparison by WGS according to previous PCR results with regard to SCCmec type and spa type (Schnitt et al., 2020). In Table 1, all sequenced MRSA strains are listed. The strains originated from QMS, BTM, nasal swabs of calves, heifers, and a pig as well as a swab from a teatcup and a teat cleaning water sample. The data were anonymized due to the general data protection regulation. The code is a combination of the German federal state, the farm in the respective federal state and the sample number of the respective farm, e.g., AA1 means German federal state A, farm A from this federal state and sample number 1 from this farm. For studying the transmission of MRSA strains across one farm, isolated strains from various sample types were included from farm AA.
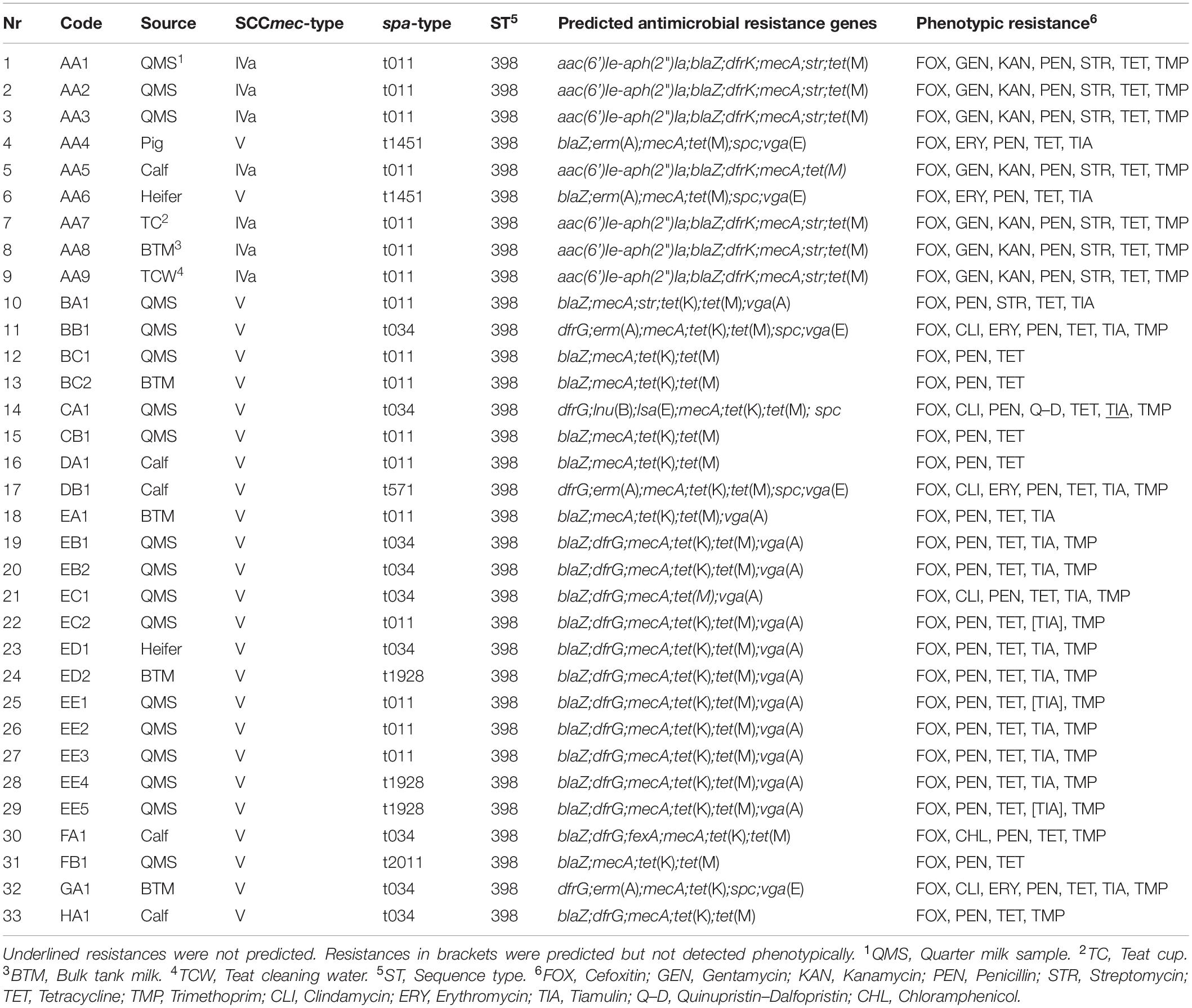
Table 1. Overview of sequenced MRSA strains, source, SCCmec-, spa-type, ST, prediction of antimicrobial resistance genes, and phenotypic resistance.
DNA Extraction and WGS
Methicillin-resistant Staphylococcus aureus isolates were cultured on sheep blood agar (Oxoid GmbH, 46483, Wesel, Germany) and DNA of one inoculation loop filled with MRSA colonies was extracted using the Qiagen DNeasy Blood and Tissue Kit (Qiagen, Germany) according to the manufacturer’s protocol modified by adding 10 μl lysostaphin to the lysis buffer. The DNA library was prepared using an Illumina Nextera DNA Flex kit (Illumina Inc., United States) and the 150 bp paired-end sequencing run was performed on an Illumina NextSeq 500 instrument.
Bioinformatic Analyses
Assembly and Quality Control
Raw Illumina reads were trimmed and de novo assembled with the in-house developed Aquamis pipeline1 which implements fastp (Chen et al., 2018) for trimming and shovill (based on SPAdes)2 for assembly. Furthermore, it performs mash v 2.1 for reference search (Ondov et al., 2016) as well as quast v 5.0.2 for assembly quality control (Mikheenko et al., 2018). The minimal coverage depth was >80. Quality of assemblies was checked by single-copy and duplicated orthologs analyses. The fraction majority species was >0.97. The total genome length was >2.7 Mbp.
Phylogenetic Analyses
The MLST sequence type was inferred using mlst3 with the pubmlst database (Jolley and Maiden, 2010). Moreover, SCCmec- and spa-types were predicted with respect to the software tools SCCmecFinder 1.2 and spaTyper 1.0 of the Centre for Genomic Epidemiology.4 In addition, the phylogenetic relationship of all sequenced MRSA strains was analyzed using cgMLST in Ridom SeqSphere + version 7.0.4. The default settings were kept so that clusters were defined at less than 24 allelic differences.
AMR Genes
Bacterial characterization was conducted with the in-house developed Bakcharak pipeline,5 which implements ABRicate6 for screening of antimicrobial resistance genes using the NCBI amrfinder database (Feldgarden et al., 2019).
Virulence Factor Genes
Virulence factor genes were predicted using the VFDB (Chen et al., 2005). Following the staphylococcal virulence factor classification of Naushad et al. (2019), the detected virulence factor genes were attributed to the functional categories adhesion, exoenzymes, hemolysis, immune evasion, iron uptake, and metabolism or secretion. A detailed sequence search for SaPIs and phages in the obtained sequences was performed using the NCBI blastn suite. Therefore, a collection of SaPIs and phages (phiNM3, phi80, phiPVL, phiETA, Saeq1, SaPI1-3, SaPIbov1-2, SaPIbov4-5, SaPIeq1, SaPIivm10, SaPIishikawa11, SaPIivm60, SaPIino10, SaPIhirosaki4, SaPIj11, SaPIhhms2, SaPINN54, SaPIPM1, SaPI68111, and SaPIj50) was chosen according to the publications of Walther et al. (2018) and Alibayov et al. (2014).
Antimicrobial Susceptibility Testing
Antimicrobial susceptibility testing (AST) was performed by broth microdilution according to the CLSI standard (ISO 20776-1:2006 or CLSI M31-A3) using a standardized antibiotic panel (EUVENC scheme) that is used in all member states of the European Union for resistance monitoring on staphylococci from livestock and food. For evaluation of minimal inhibitory concentrations (MIC) of the individual isolates the clinical breakpoints values of the CLSI were used. For quality control of resistance testing the S. aureus isolates ATCC 29213 and ATCC 25923 were used.
Results
MLST-, SCCmec-, and spa-Typing of MRSA Strains
Analyses of the sequence data showed that all strains belonged to the sequence type (ST) 398 (Table 1). MRSA strains with SCCmec type V dominated on the dairy farms. Solely on one farm AA, MRSA strains with SCCmec type IVa were detected in BTM, QMS, a nasal swab of a calf, a teatcup, and the teat cleaning water, whereas the MRSA strains AA4 (pig) and AA6 (heifer) from the animals placed in the pig barn carried SCCmec type V. Regarding the spa-types, t011, and t034 were mostly found. Beside, also spa-types t1451, t571, t1928, and t2011 were detected. On four farms (AA, EC, ED, and EE) various spa-types were found in different sample types. On farm AA and in accordance with the varying SCCmec types, spa-type t011 was found in BTM, QMS, the nasal swab of a calf, the teatcup and the teat cleaning water, whereas the MRSA strains from the nasal swabs of a pig and a heifer located in the pig barn carried spa-type t1451. On farm EC, two different spa-types (t011 and t034) were found in QMS. The spa-types t034 (nasal swab of heifer) and t1928 (BTM) were detected on farm ED. Moreover, on farm EE, different spa-types (t011 and t1928) were found in QMS.
Antimicrobial Resistance Profiles
A broad range of antimicrobial resistance genes was detected in the sequences of the different MRSA strains. Resistance to the antibiotic classes aminoglycoside, β-lactam, trimethoprim, tetracycline, macrolide, streptogramin, lincosamide, and phenicol were predicted showing differences between the dairy farms and sample types on farm AA (Table 1). All MRSA strains (33/33) carried the mecA gene, whereas the β-lactamase encoding blaZ gene was missing in four strains. Resistance to the other antibiotic classes was encoded by the following genes; aminoglycoside [aac(6’)Ie-aph(2”)Ia, str], aminocyclitol (spc), macrolide-lincosamide-streptogramin B [erm(A)], trimethoprim (dfrG, dfrK), tetracycline [tet(K), tet(M)], pleuromutilin–lincosamide–streptogramin A [lsa(E), vga(A), vga(E)], lincosamides [lnu(B)], and phenicol (fexA). All MRSA strains (33/33) harbored tetracycline resistance genes. Resistance to aminoglycosides (14/33), trimethoprim (24/33), and pleuromutilin–lincosamide–streptogramin A (19/33) was also commonly predicted. In contrast, resistance to macrolides–lincosamide–streptogramin B (5/33) and phenicol (1/33) was less frequently predicted.
The phenotypic resistance according to the MIC values was in good agreement with the predicted antimicrobial resistance genes (Table 1). The genotypic differences between the strains throughout the farms were also shown in the phenotypic resistance pattern. All MRSA strains were resistant to cefoxitin, penicillin, and tetracycline. Resistance to trimethoprim and tiamulin was also widespread. Only a few strains showed resistance to gentamicin (7/33), kanamycin (7/33), streptomycin (8/33), clindamycin (5/33), or erythromycin (5/33). Only the strain CA1 was phenotypically resistant to quinupristin–dalfopristin and strain FA1 was phenotypically resistant to chloramphenicol. In sum, ten different phenotypic resistance patterns occurred within the various MRSA strains.
Virulence Factors
The MRSA strains showed a diverse repertoire of virulence factor genes. Most of the analyzed genes (45/63) were present in every MRSA strain (Table 2). The cna (6/33), sdrE (32/33), and von Willebrand factor binding protein (32/33) genes were not found in all strains. Typical human MRSA IEC genes such as scn, sak, and chp or exfoliative toxin genes (eta/etb) were not detected in any of the sequenced isolates. Likewise, genes encoding toxins such as staphylococcal enterotoxins (SE; sea, seb, sec, sed, see, seh, selk, sell, selq) or TSST (tsst) were not found. Regarding PVL, the leukocidin subunit lukF-PV gene was only detected in seven MRSA strains, whereas the lukS-PV gene, which encodes the other PVL subunit, was not found. The pathogenicity island SaPIbov5 was detected in eight MRSA strains. Seven of these were isolates from the same farm AA. SaPIbov4 was found in eleven strains; with a sequence coverage regarding the reference of 89–90%. Other SaPIs or phages were not detected in the assembled sequences.

Table 2. Number of MRSA strains harboring various predicted virulence associated genes and corresponding functional categories.
Phylogenetic Relationship
The sequences were analyzed by cgMLST regarding the phylogenetic relationship of the MRSA strains from the dairy farms across the eight German federal states. Four different clusters were retrieved in the minimum spanning tree (MST) analysis (Figure 1). The MST analysis showed large allelic differences between the MRSA strains from different German federal states and also between farms from the same region. Moreover, MRSA strains from the same farm (EB, EC, and EE) sometimes differed significantly in the core genome. The MRSA strains EB1, ED1/2, and EE1/2/4/5 from three farms from German federal state E (cluster 2) clustered with 13–23 alleles differences closer together in comparison to the farms from other federal states. On farm AA, MRSA strains of the sample types QMS (AA1–3), BTM (AA8), the nasal swab of a calf (AA5) as well as the samples of the teatcup (AA7) and teat cleaning water (AA9) clustered closely together with a maximum difference of one core genome allele. Contrary to this, the strains AA4 (nasal swab of pig) and AA6 (nasal swab of heifer), with a pig barn origin, formed an own cluster, which is in accordance with the different SCCmec- and spa-types.
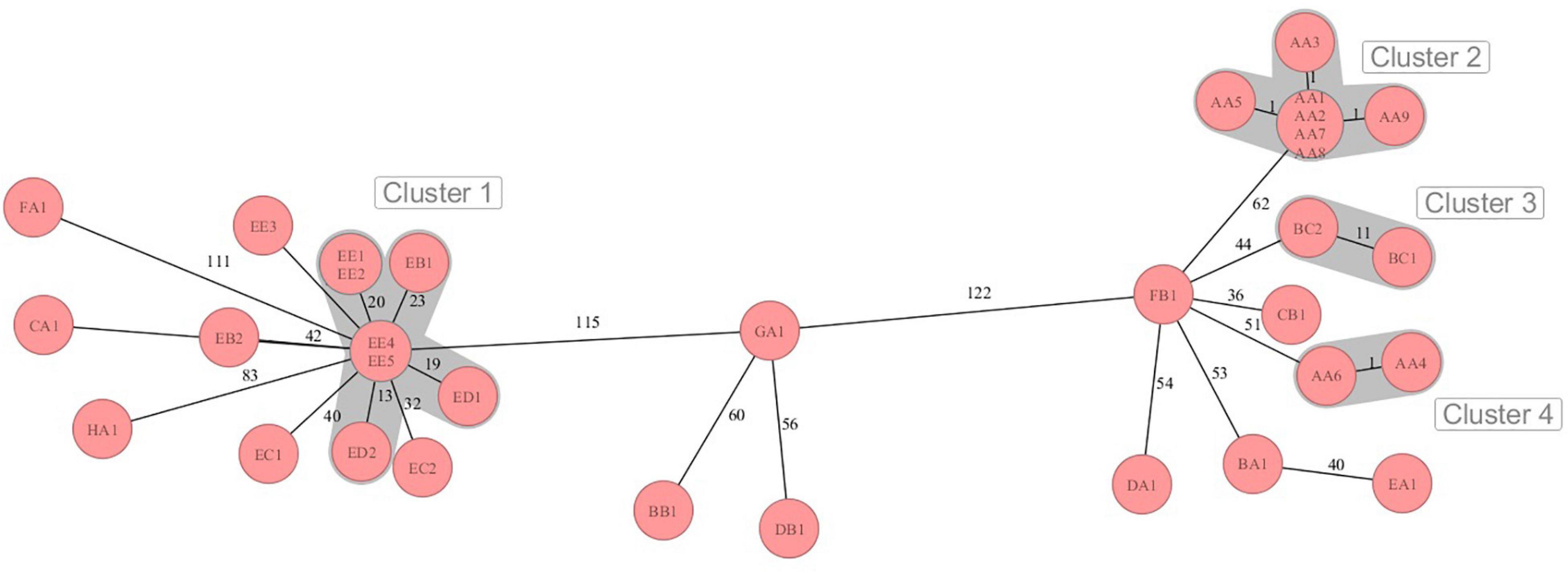
Figure 1. Minimum spanning tree visualization of cgMLST analysis of the MRSA strains on dairy farms from eight German federal states A-H with four clusters of close phylogenetic relationship. Numbers represent the allelic differences between the MRSA strains.
Discussion
Methicillin-resistant Staphylococcus aureus may be widespread on German dairy farms potentially causing infections such as bovine mastitis (Schnitt and Tenhagen, 2019). In addition to animal health and with regard to a transmission from animals to humans, monitoring of MRSA abundance and genotyping is also important in the public health context.
Genomic Distinctions Between MRSA Isolates Across Germany
In our study, 33 MRSA strains from 17 dairy farms across eight German federal states were phenotypically and genotypically analyzed. All strains were characterized as ST398 LA-MRSA, a sequence type, which is often found on dairy farms (Feßler et al., 2012; Kadlec et al., 2019) and which is the most widely disseminated LA-MRSA sequence type (Cuny et al., 2015b). SCCmec-types IV and V as well as the spa-type t011 dominated in LA-MRSA on Dutch dairy farms as reported by Feßler et al. (2012). This is in accordance with the results of our study, however, in contrast, SCCmec-type IVa was only found in 1/17 farms and the spa-type t034 co-dominated to the spa-type t011. Tenhagen et al. (2018) also detected the spa-types t011 and t034 as most frequent in MRSA of BTM from German dairy farms. Moreover, a dominance of MRSA carrying SCCmec type V on German dairy farms was shown in the study of Kadlec et al. (2019).
The phylogenetic analysis of the MRSA strains done by cgMLST showed mainly distinct allelic differences and only a few clusters of close relationship. Although the strains were similar according to SCCmec- or spa-typing, the core genome differed in some cases by more than 120 alleles. This illustrates the high genomic plasticity and strong evolution in terms of genetic recombination in MRSA. Although exhibiting differences of 56–60 alleles, the strains GA1, BB1, and DB1 clustered together in the MST. The phylogenetic relation of these strains was also indicated by the similar antimicrobial resistance genes profile and the identical phenotypic resistance pattern. Only some MRSA strains from federal state E showed lower genetic divergence, in particular the strains ED2 and EE4/5. These strains also shared the same spa-type t1928. It can be speculated that a transmission of these MRSA strains between the farms took place. Reasons for this might be humans, e.g., farm personnel or veterinarians, or animal trade transmitting MRSA from one farm to another. In line with that, the introduction and transmission of MRSA on pig farms in Norway by farm workers or livestock trade was illustrated in the studies of Grontvedt et al. (2016) and Elstrøm et al. (2019). The potential role of “humans” as MRSA vectors across dairy farms was also shown in a recently published review of Schnitt and Tenhagen (2019).
Since the MRSA genotype might also vary within farms, strains of different sample types from selected farms were investigated. MRSA strains with a maximum of one allele difference in cgMLST were found in BTM, QMS of different cows, a nasal swab of a calf, the teatcup of the milking equipment and water for teat cleaning prior to milking on farm AA. Moreover, the strains harbored the same antimicrobial resistance genes and showed the same phenotypic resistance pattern. Therefore, the spread of one clonal LA-MRSA strain between cows, calves, and the milking equipment seems likely. In contrast, the MRSA isolates of the pig and heifer placed in the pig barn showed a MRSA strain, which differed genotypically from the strain in the dairy barn. A spillover of MRSA from the pig production to veal calves or dairy cows was suggested by several studies (Locatelli et al., 2017; Hansen et al., 2019). However, in our study, the spillover from pigs to other animals was only found in close proximity in the pig barn. Various MRSA genotypes also co-existed on farms EB, EC, and EE. Accordingly, Feßler et al. (2012) showed a co-dominance of several MRSA strains on one dairy farm. A reason for this might be the purchase of new animals and thus MRSA genotypes in the farm or transmission of additional strains by humans or other vectors. On the other side, genetic diversification might have appeared on the farm leading to altered MRSA genotypes.
Widespread Multi-Resistant MRSA
The MRSA strains in our study carried a broad repertoire of antimicrobial resistance genes and 28 strains were multi-resistant to at least three classes of antibiotics. This is contrary to the recently published study of Kadlec et al. (2019), in which less than half of the MRSA strains from German dairy farms were multi-resistant with the constraint that only ten MRSA isolates were investigated by the authors. The prediction of the antimicrobial resistance genes in our study was mostly in agreement with the phenotypic antibiotic class resistance pattern. Resistance to β-lactam antibiotics such as cefoxitin or penicillin was mediated by the mecA gene in all strains, whereas the variant mecC gene was not found. In contrast, Schlotter et al. (2014) found MRSA harboring the mecC gene in 16 of 56 milk samples in a German dairy herd, probably due to a spread of this strain within the farm. However, in accordance with our study, MRSA strains in dairy cattle from Germany and Greece only harbored the mecA gene (Kadlec et al., 2019; Papadopoulos et al., 2019). The resistance to several classes of antibiotics was mediated by two to three different genes in our study. Feßler et al. (2018) summarized in their review that small plasmids play a pivotal role in the dissemination of certain antimicrobial resistance genes. Although not analyzed in detail, the transmission of resistance genes such as vga(A) or dfrK through plasmids likely also played a role in the strains of our study. In general, the antimicrobial resistance patterns differed between the farms indicating an independent development of the strains across Germany. Resistance to macrolides was rare in the MRSA strains. Accordingly, macrolide resistance in S. aureus retrieved from bovine mastitis was reported to be low (Pyorala et al., 2014; El Garch et al., 2020). In contrast, in a study of Tenhagen et al. (2018) macrolide resistance was detected in 17/41 MRSA isolates of BTM from German dairy farms. In our study, resistance to chloramphenicol was only found in one MRSA strain harboring the fexA gene. Accordingly, the fexA gene was only rarely found in a study analyzing ST398 MRSA isolates from bovine mastitis (Feßler et al., 2010). Resistance to tetracyclines was found in every strain from all farms in our study mediated by the tet(K) or tet(M) genes. In agreement with this, in the studies of Feßler et al. (2012) and Kadlec et al. (2019) all MRSA strains from dairy farms were resistant to tetracycline. Moreover, tetracycline resistance was detected in 99.4% of the MRSA isolates received from the cattle food chain (Tenhagen et al., 2014) and in 95.1% of the MRSA in BTM from German dairy farms (Tenhagen et al., 2018). Tetracyclines have been extensively used on animal farms, thus promoting the survival of tetracycline resistant strains (Granados-Chinchilla and Rodriguez, 2017). Furthermore, resistance to trimethoprim and tiamulin, a pleuromutilin, was detected in more than half of the MRSA strains in our study. Staphylococci are non-target bacteria with respect to pleuromutilins, however, their use in especially pig farming selects for multi-resistant MRSA (van Duijkeren et al., 2014). In this study, pleuromutilin resistance was transmitted by the vga(A) or vga(E) genes. In particular in the German federal state E, the vga(A) gene was present in MRSA strains from several farms. The vga(A) gene was shown to be transferred by plasmids (Feßler et al., 2018), whereas the vga(E) gene is located on a transferable transposon (Schwendener and Perreten, 2011). Hauschild et al. (2012) originally detected the vga(E) gene in dairy cattle. Although the vga(A) gene was reported to be most widespread among the vga genes (Feßler et al., 2018), in our study, also the vga(E) gene was equally distributed across the MRSA strains from the dairy farms. The resistance to trimethoprim was mediated by the dfrG or dfrK genes. The dfrK gene was only detected on farm AA in the MRSA strains with SCCmec type IVa. This is contrary to the finding of a 85.7% dissemination of the dfrK gene in MRSA isolates of bovine mastitis (Feßler et al., 2010). Moreover, the physical linkage of the dfrK and tet(L) genes, as described in Kadlec et al. (2012), was not found in our study. Aminoglycosides are widely used in veterinary medicine (EMA, 2018). Only the SCCmec IVa MRSA strains of farm AA showed phenotypical aminoglycoside resistance to streptomycin, gentamicin, and kanamycin. This can be explained by the different repertoire of antimicrobial resistance genes to aminoglycosides in the respective strains encoded by the aac(6’)Ie-aph(2”)Ia, and str genes. Moreover, the spc gene was detected in five strains, which mediates resistance to spectinomycin, an aminocyclitol, only (Zarate et al., 2018). The MRSA strain CA1 was phenotypically resistant to the streptogramin A and B quinupristin–dalfopristin. This is in agreement with the detection of the lsa(E) gene, since an eight-fold increased quinupristin–dalfopristin MIC was also previously detected for S. aureus harboring the lsa(E) gene (Wendlandt et al., 2013). Moreover, also resistance to clindamycin as detected for strain CA1 is associated with the lsa(E) gene (Wendlandt et al., 2013). Furthermore, erm genes confer inducible or constitutive resistance to macrolides, lincosamides, and streptogramin B (Leclercq, 2002). Therefore, in our study, the strains BB1 and GA1, which harbored the resistance gene erm(A), on the one hand showed phenotypical resistance to the macrolide erythromycin, but on the other side these strains were also resistant to the lincosamide clindamycin.
Large Repertoire of Virulence Factor Genes
The LA-MRSA strains from the dairy farms in our study harbored multiple virulence associated genes, most of them present in all strains. Piccinini et al. (2010) postulated that a specific virulence gene combination is related to the development of subclinical mastitis and the prevalence of S. aureus in dairy herds. Moreover, Magro et al. (2017) related some of the virulence factors to more contagious S. aureus strains with regard to mastitis. In accordance with the prediction of the hlb gene, β-hemolysis on sheep blood agar was found for every detected MRSA strain in our study (data not shown). The presence of hemolysins as a factor for bovine mastitis in Russian dairy herds was described in the study of Fursova et al. (2020). In our study, the clumping factor encoding genes clfA and clfB were detected in all strains. This is in contrast to the study of virulence factor genes in dairy cattle from Brazil conducted by Klein et al. (2012), in which the prevalence of the clfB gene was higher (91.8%) than the clfA gene prevalence (50.6%). In particular, the ClfB protein is associated with S. aureus nasal colonization and skin infections in humans (Wertheim et al., 2008; Lacey et al., 2019). With regard to the animal health aspect, it was related to S. aureus prevalence in bovine mastitis (Magro et al., 2017). The cna gene, which encodes a collagen adhesion protein, was found in six MRSA strains in our study. Accordingly, Klein et al. (2012) detected a cna gene prevalence of 22.4% in 85 MRSA isolates of dairy cattle from Brazil. Cna might play a pivotal role in binding collagen in wounded, injured, or inflamed tissue, e.g., in mastitis (Madani et al., 2017). In addition, in our study, the fibronectin-binding protein encoding fnbA gene was present in all MRSA strains. This protein was shown to be connected to mastitis in a mouse model (Brouillette et al., 2003) and it seems to be related to more contagious S. aureus strains in bovine mastitis (Magro et al., 2017). The sdrC, sdrD, and sdrE genes, which encode the serine-aspartate repeat proteins, were found in nearly all strains in our study. In particular, the presence of the sdrD gene was associated with bone infections (Trad et al., 2004) and more contagious S. aureus strains during mastitis (Magro et al., 2017). Moreover, biofilm formation plays a crucial role in virulence of S. aureus (Costerton et al., 1999). The genes icaA, icaB, icaC, icaD, and icaR, which are related to biofilm formation in several staphylococcal species, were present in all MRSA isolates in our study. Since biofilm formation is of high clinical impact (Martín-López et al., 2002), this might have also been an important issue regarding animal health in the MRSA isolates in our study.
Low Risk for Public Health
Virulence factor genes encoding the TSST, PVL, or elements of the IEC, which are associated with severe human infections, were not detected in our MRSA isolates. This is in agreement with the absence of most SaPIs and phages in the genomes, since in LA-MRSA these pathogenicity factors are often encoded in SaPIs (Ballhausen et al., 2017). SaPIbov5 and SaPIbov4, with a sequence coverage of 89–90%, were detected in 24 and 33% of our strains. Most likely, the von Willebrand factor-binding protein, a clotting factor encoded by the von Willebrand factor binding protein gene, was located on the SaPIs of the respective strains as shown by Viana et al. (2010). In particular, the SCCmec IVa MRSA strains from farm AA were equipped with both SaPIs. Cuny et al. (2016) associated some SCCmec IV and spa-type t011 LA-MRSA strains to human infections and Walther et al. (2018) found that 72% of ST398 and spa-type t011 MRSA strains in horse clinics harbored the human IEC encoded in a phiSa3 phage. Anyhow, the lack of TSST, PVL, and IEC genes in the strains from the farms investigated in our study indicates a low risk for severe human infections.
Furthermore, it has to be considered that food intoxication might appear by the consumption of raw milk or raw milk products, if SE producing MRSA strains are present in high numbers (Sergelidis and Angelidis, 2017). Yang et al. (2020) found high frequencies of SE genes in MRSA isolates of bovine mastitis cases from China. SE genes were also detected in MRSA strains of raw milk from Italy (Riva et al., 2015) and Egyptian dairy herds (El-Ashker et al., 2020). In contrast, Kadlec et al. (2019) and Kreausukon et al. (2012) did not detect any SE genes in LA-MRSA strains from German dairy farms. Accordingly, all strains investigated in our study were lacking genes for SEs, thus also lowering the possibility of a food poisoning. Moreover, the mastitis-related S. aureus genotype GTB, which is associated with the presence of the sea, sed, and sej genes (Graber et al., 2009), was not found in our study.
Our study is limited by the number of strains that were investigated with respect to the phylogenetic dynamics of MRSA strains across German dairy farms. Therefore, as a future perspective, sampling on dairy farms across Germany should be expanded and as a consequence, longitudinal core genome analysis of larger numbers of MRSA strains should be performed to better resolve the transmission pathways and evolutionary mechanisms of MRSA.
Conclusion
The results of our study show that MRSA on German dairy farms harbor a broad repertoire of antimicrobial resistance and virulence factor genes. Some of the virulence genes are associated to mastitis, but none of them are connected to human infections. Phylogenetic analyses indicate more than 24 allelic differences of the strains across Germany with some regional spots of minor allelic diversity. Transmission of MRSA between farms may occur and MRSA strains may also be expansively transmitted within the farm environment. The prediction of antimicrobial resistance through bioinformatics tools was in agreement with the phenotypic resistance profiles. MRSA monitoring on animal farms is of high significance, since the genetic repertoire might spontaneously change due to horizontal gene transfer of MGEs and transmission pathways need to be resolved for containment of MRSA on animal farms.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, BioProject PRJNA634452.
Ethics Statement
Ethical review and approval was not required for the animal study because sampling of milk and nasal swabs from calves and heifers was carried out in accordance with German legislation. No ethical approval from the Institutional Ethics Committee or the National Animal Experimentation Council was required. Samples were collected by a trained veterinarian with consent from the owners of the animals. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author Contributions
TL, AS, and B-AT: study design. TL, AS, and JAH: laboratory work. TL and AS: data analysis. All authors: manuscript preparation and review.
Funding
Our study was carried out in the framework of the project #1HealthPREVENT, funded by the German Federal Ministry of Education and Research, Grant No. 01KI1727C.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank all farmers, who voluntarily participated in the study. Many thanks to Thomas Peters, Ulrike Sorge, Karsten Donat, Sabine Reinhold, Oliver Claushues, Karin Eulenberger and Natalie Morgenstern for supporting the identification of study farms. Special thanks to Daniel Leeser-Boek, Pascal Witt and Ylanna Kelner-Burgos for NGS preparation and Heidi Wichmann-Schauer for help with study conception. We also thank Carlus Deneke and Simon Tausch for bioinformatical analysis.
Footnotes
- ^ https://gitlab.com/bfr_bioinformatics/AQUAMIS/
- ^ https://github.com/tseemann/shovill
- ^ https://github.com/tseemann/mlst
- ^ https://cge.cbs.dtu.dk/services/
- ^ https://gitlab.com/bfr_bioinformatics/bakcharak
- ^ https://github.com/tseemann/abricate
References
Alibayov, B., Baba-Moussa, L., Sina, H., Zdenkova, K., and Demnerova, K. (2014). Staphylococcus aureus mobile genetic elements. Mol. Biol. Rep. 41, 5005–5018. doi: 10.1007/s11033-014-3367-3
Ballhausen, B., Kriegeskorte, A., Van Alen, S., Jung, P., Kock, R., Peters, G., et al. (2017). The pathogenicity and host adaptation of livestock-associated MRSA CC398. Vet. Microbiol. 200, 39–45. doi: 10.1016/j.vetmic.2016.05.006
Brouillette, E., Talbot, B. G., and Malouin, F. (2003). The fibronectin-binding proteins of Staphylococcus aureus may promote mammary gland colonization in a lactating mouse model of mastitis. Infect. Immun. 71, 2292–2295. doi: 10.1128/iai.71.4.2292-2295.2003
Chen, L., Yang, J., Yu, J., Yao, Z., Sun, L., Shen, Y., et al. (2005). VFDB: a reference database for bacterial virulence factors. Nucleic Acids Res. 33, D325–D328.
Chen, S., Zhou, Y., Chen, Y., and Gu, J. (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890.
Costerton, J. W., Stewart, P. S., and Greenberg, E. P. (1999). Bacterial biofilms: a common cause of persistent infections. Science 284:1318. doi: 10.1126/science.284.5418.1318
Cuny, C., Abdelbary, M., Layer, F., Werner, G., and Witte, W. (2015a). Prevalence of the immune evasion gene cluster in Staphylococcus aureus CC398. Vet. Microbiol. 177, 219–223. doi: 10.1016/j.vetmic.2015.02.031
Cuny, C., Abdelbary, M. M. H., Kock, R., Layer, F., Scheidemann, W., Werner, G., et al. (2016). Methicillin-resistant Staphylococcus aureus from infections in horses in Germany are frequent colonizers of veterinarians but rare among MRSA from infections in humans. One Health 2, 11–17. doi: 10.1016/j.onehlt.2015.11.004
Cuny, C., Arnold, P., Hermes, J., Eckmanns, T., Mehraj, J., Schoenfelder, S., et al. (2017). Occurrence of cfr-mediated multiresistance in staphylococci from veal calves and pigs, from humans at the corresponding farms, and from veterinarians and their family members. Vet. Microbiol. 200, 88–94. doi: 10.1016/j.vetmic.2016.04.002
Cuny, C., Wieler, L. H., and Witte, W. (2015b). Livestock-associated MRSA: the impact on humans. Antibiotics (Basel) 4, 521–543. doi: 10.3390/antibiotics4040521
El Garch, F., Youala, M., Simjee, S., Moyaert, H., Klee, R., Truszkowska, B., et al. (2020). Antimicrobial susceptibility of nine udder pathogens recovered from bovine clinical mastitis milk in Europe 2015–2016: VetPath results. Vet. Microbiol. 245:108644. doi: 10.1016/j.vetmic.2020.108644
El-Ashker, M., Gwida, M., Monecke, S., El-Gohary, F., Ehricht, R., Elsayed, M., et al. (2020). Antimicrobial resistance pattern and virulence profile of S. aureus isolated from household cattle and buffalo with mastitis in Egypt. Vet. Microbiol. 240:108535. doi: 10.1016/j.vetmic.2019.108535
Elstrøm, P., Grøntvedt, C. A., Gabrielsen, C., Stegger, M., Angen, Ø, Åmdal, S., et al. (2019). Livestock-associated MRSA CC1 in Norway; introduction to pig farms, zoonotic transmission, and eradication. Front. Microbiol. 10:139. doi: 10.3389/fmicb.2019.00139
EMA (2018). Reflection Paper on Use of Aminoglycosides in Animals in the European Union: Development of Resistance and Impact on Human and Animal Health. EMA/CVMP/AWP/721118/2014. Amsterdam: European Medicines Agency.
Feldgarden, M., Brover, V., Haft, D. H., Prasad, A. B., Slotta, D. J., Tolstoy, I., et al. (2019). Validating the AMRFinder tool and resistance gene database by using antimicrobial resistance genotype-phenotype correlations in a collection of isolates. Antimicrob. Agents Chemother. 63:e00483-19.
Feßler, A., Kadlec, K., Wang, Y., Zhang, W. J., Wu, C., Shen, J., et al. (2018). Small antimicrobial resistance plasmids in livestock-associated methicillin-resistant Staphylococcus aureus CC398. Front. Microbiol. 9:2063. doi: 10.3389/fmicb.2018.02063
Feßler, A., Scott, C., Kadlec, K., Ehricht, R., Monecke, S., and Schwarz, S. (2010). Characterization of methicillin-resistant Staphylococcus aureus ST398 from cases of bovine mastitis. J. Antimicrob. Chemother. 65, 619–625. doi: 10.1093/jac/dkq021
Feßler, A. T., Olde Riekerink, R. G., Rothkamp, A., Kadlec, K., Sampimon, O. C., Lam, T. J., et al. (2012). Characterization of methicillin-resistant Staphylococcus aureus CC398 obtained from humans and animals on dairy farms. Vet. Microbiol. 160, 77–84. doi: 10.1016/j.vetmic.2012.05.005
Fursova, K., Sorokin, A., Sokolov, S., Dzhelyadin, T., Shulcheva, I., Shchannikova, M., et al. (2020). Virulence factors and phylogeny of Staphylococcus aureus associated with bovine mastitis in Russia based on genome sequences. Front. Vet. Sci. 7:135. doi: 10.3389/fvets.2020.00135
Goerge, T., Lorenz, M. B., Van Alen, S., Hubner, N. O., Becker, K., and Kock, R. (2017). MRSA colonization and infection among persons with occupational livestock exposure in Europe: prevalence, preventive options and evidence. Vet. Microbiol. 200, 6–12. doi: 10.1016/j.vetmic.2015.10.027
Graber, H. U., Naskova, J., Studer, E., Kaufmann, T., Kirchhofer, M., Brechbuhl, M., et al. (2009). Mastitis-related subtypes of bovine Staphylococcus aureus are characterized by different clinical properties. J. Dairy Sci. 92, 1442–1451. doi: 10.3168/jds.2008-1430
Granados-Chinchilla, F., and Rodriguez, C. (2017). Tetracyclines in food and feedingstuffs: from regulation to analytical methods, bacterial resistance, and environmental and health implications. J. Anal. Methods Chem. 2017:1315497.
Grontvedt, C. A., Elstrom, P., Stegger, M., Skov, R. L., Skytt Andersen, P., Larssen, K. W., et al. (2016). Methicillin-resistant Staphylococcus aureus CC398 in humans and pigs in Norway: a “One Health” perspective on introduction and transmission. Clin. Infect. Dis. 63, 1431–1438.
Hansen, J. E., Ronco, T., Stegger, M., Sieber, R. N., Fertner, M. E., Martin, H. L., et al. (2019). LA-MRSA CC398 in dairy cattle and veal calf farms indicates spillover from pig production. Front. Microbiol. 10:2733. doi: 10.3389/fmicb.2019.02733
Hauschild, T., Fessler, A. T., Kadlec, K., Billerbeck, C., and Schwarz, S. (2012). Detection of the novel vga(E) gene in methicillin-resistant Staphylococcus aureus CC398 isolates from cattle and poultry. J. Antimicrob. Chemother. 67, 503–504. doi: 10.1093/jac/dkr446
Holmes, M. A., and Zadoks, R. N. (2011). Methicillin resistant S. aureus in human and bovine mastitis. J. Mammary Gland Biol. Neoplasia 16, 373–382. doi: 10.1007/s10911-011-9237-x
Jolley, K. A., and Maiden, M. C. J. (2010). BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 11:595. doi: 10.1186/1471-2105-11-595
Kadlec, K., Entorf, M., and Peters, T. (2019). Occurrence and characteristics of livestock-associated methicillin-resistant Staphylococcus aureus in quarter milk samples from dairy cows in Germany. Front. Microbiol. 10:1295. doi: 10.3389/fmicb.2019.01295
Kadlec, K., Feßler, A. T., Hauschild, T., and Schwarz, S. (2012). Novel and uncommon antimicrobial resistance genes in livestock-associated methicillin-resistant Staphylococcus aureus. Clin. Microbiol. Infect. 18, 745–755. doi: 10.1111/j.1469-0691.2012.03842.x
Klein, R. C., Fabres-Klein, M. H., Brito, M. A., Fietto, L. G., and Ribon Ade, O. (2012). Staphylococcus aureus of bovine origin: genetic diversity, prevalence and the expression of adhesin-encoding genes. Vet. Microbiol. 160, 183–188. doi: 10.1016/j.vetmic.2012.05.025
Kraushaar, B., Hammerl, J. A., Kienol, M., Heinig, M. L., Sperling, N., Dinh Thanh, M., et al. (2017). Acquisition of virulence factors in livestock-associated MRSA: lysogenic conversion of CC398 strains by virulence gene-containing phages. Sci. Rep. 7:2004.
Kreausukon, K., Fetsch, A., Kraushaar, B., Alt, K., Muller, K., Kromker, V., et al. (2012). Prevalence, antimicrobial resistance, and molecular characterization of methicillin-resistant Staphylococcus aureus from bulk tank milk of dairy herds. J. Dairy Sci. 95, 4382–4388. doi: 10.3168/jds.2011-5198
Lacey, K. A., Mulcahy, M. E., Towell, A. M., Geoghegan, J. A., and Mcloughlin, R. M. (2019). Clumping factor B is an important virulence factor during Staphylococcus aureus skin infection and a promising vaccine target. PLoS Pathog. 15:e1007713. doi: 10.1371/journal.ppat.1007713
Lakhundi, S., and Zhang, K. (2018). Methicillin-resistant Staphylococcus aureus: molecular characterization, evolution, and epidemiology. Clin. Microbiol. Rev. 31:e00020-18.
Leclercq, R. (2002). Mechanisms of resistance to macrolides and lincosamides: nature of the resistance elements and their clinical implications. Clin. Infect. Dis. 34, 482–492. doi: 10.1086/324626
Locatelli, C., Cremonesi, P., Caprioli, A., Carfora, V., Ianzano, A., Barberio, A., et al. (2017). Occurrence of methicillin-resistant Staphylococcus aureus in dairy cattle herds, related swine farms, and humans in contact with herds. J. Dairy Sci. 100, 608–619. doi: 10.3168/jds.2016-11797
Madani, A., Garakani, K., and Mofrad, M. R. K. (2017). Molecular mechanics of Staphylococcus aureus adhesin, CNA, and the inhibition of bacterial adhesion by stretching collagen. PLoS One 12:e0179601. doi: 10.1371/journal.pone.0179601
Magro, G., Biffani, S., Minozzi, G., Ehricht, R., Monecke, S., Luini, M., et al. (2017). Virulence genes of S. aureus from dairy cow mastitis and contagiousness risk. Toxins (Basel) 9:195. doi: 10.3390/toxins9060195
Martín-López, J. V., Pérez-Roth, E., Claverie-Martín, F., Díez Gil, O., Batista, N., Morales, M., et al. (2002). Detection of Staphylococcus aureus clinical isolates harboring the gene cluster needed for biofilm establishment. J. Clin. Microbiol. 40, 1569–1570. doi: 10.1128/jcm.40.4.1569-1570.2002
Mikheenko, A., Prjibelski, A., Saveliev, V., Antipov, D., and Gurevich, A. (2018). Versatile genome assembly evaluation with QUAST-LG. Bioinformatics 34, i142–i150.
Naushad, S., Naqvi, S. A., Nobrega, D., Luby, C., Kastelic, J. P., Barkema, H. W., et al. (2019). Comprehensive virulence gene profiling of bovine non-aureus Staphylococci based on whole-genome sequencing data. mSystems 4:e0009818.
Ondov, B. D., Treangen, T. J., Melsted, P., Mallonee, A. B., Bergman, N. H., Koren, S., et al. (2016). Mash: fast genome and metagenome distance estimation using MinHash. Genome Biol. 17:132.
Papadopoulos, P., Angelidis, A. S., Papadopoulos, T., Kotzamanidis, C., Zdragas, A., Papa, A., et al. (2019). Staphylococcus Aureus and methicillin-resistant S. Aureus (MRSA) in bulk tank milk, livestock and dairy-Farm personnel in North-Central and North-Eastern Greece: prevalence, characterization and genetic relatedness. Food Microbiol. 84:103249. doi: 10.1016/j.fm.2019.103249
Piccinini, R., Borromeo, V., and Zecconi, A. (2010). Relationship between S. aureus gene pattern and dairy herd mastitis prevalence. Vet. Microbiol. 145, 100–105. doi: 10.1016/j.vetmic.2010.03.005
Pyorala, S., Baptiste, K. E., Catry, B., Van Duijkeren, E., Greko, C., Moreno, M. A., et al. (2014). Macrolides and lincosamides in cattle and pigs: use and development of antimicrobial resistance. Vet. J. 200, 230–239. doi: 10.1016/j.tvjl.2014.02.028
Riva, A., Borghi, E., Cirasola, D., Colmegna, S., Borgo, F., Amato, E., et al. (2015). Methicillin-resistant Staphylococcus aureus in raw milk: prevalence, SCCmec typing, enterotoxin characterization, and antimicrobial resistance patterns. J. Food Prot. 78, 1142–1146. doi: 10.4315/0362-028x.jfp-14-531
Schlotter, K., Huber-Schlenstedt, R., Gangl, A., Hotzel, H., Monecke, S., Muller, E., et al. (2014). Multiple cases of methicillin-resistant CC130 Staphylococcus aureus harboring mecC in milk and swab samples from a Bavarian dairy herd. J. Dairy Sci. 97, 2782–2788. doi: 10.3168/jds.2013-7378
Schnitt, A., Lienen, T., Wichmann-Schauer, H., Cuny, C., and Tenhagen, B. A. (2020). The occurrence and distribution of livestock-associated methicillin-resistant Staphylococcus aureus ST398 on German dairy farms. J. Dairy Sci. 103, 11806–11819. doi: 10.3168/jds.2020-18958
Schnitt, A., and Tenhagen, B. A. (2019). Risk factors for the occurrence of methicillin-resistant Staphylococcus aureus in dairy herds: an update. Foodborne Pathog. Dis. 17, 585–596. doi: 10.1089/fpd.2019.2638
Schwendener, S., and Perreten, V. (2011). New transposon Tn6133 in methicillin-resistant Staphylococcus aureus ST398 contains vga(E), a novel streptogramin A, pleuromutilin, and lincosamide resistance gene. Antimicrob. Agents Chemother. 55, 4900–4904. doi: 10.1128/aac.00528-11
Sergelidis, D., and Angelidis, A. S. (2017). Methicillin-resistant Staphylococcus aureus: a controversial food-borne pathogen. Lett. Appl. Microbiol. 64, 409–418. doi: 10.1111/lam.12735
Tenhagen, B. A., Alt, K., Pfefferkorn, B., Wiehle, L., Kasbohrer, A., and Fetsch, A. (2018). Short communication: methicillin-resistant Staphylococcus aureus in conventional and organic dairy herds in Germany. J. Dairy Sci. 101, 3380–3386. doi: 10.3168/jds.2017-12939
Tenhagen, B. A., Vossenkuhl, B., Käsbohrer, A., Alt, K., Kraushaar, B., Guerra, B., et al. (2014). Methicillin-resistant Staphylococcus aureus in cattle food chains – prevalence, diversity, and antimicrobial resistance in Germany. J. Anim. Sci. 92, 2741–2751. doi: 10.2527/jas.2014-7665
Trad, S., Allignet, J., Frangeul, L., Davi, M., Vergassola, M., Couve, E., et al. (2004). DNA macroarray for identification and typing of Staphylococcus aureus isolates. J. Clin. Microbiol. 42, 2054–2064. doi: 10.1128/jcm.42.5.2054-2064.2004
van Duijkeren, E., Greko, C., Pringle, M., Baptiste, K. E., Catry, B., Jukes, H., et al. (2014). Pleuromutilins: use in food-producing animals in the European Union, development of resistance and impact on human and animal health. J. Antimicrob. Chemother. 69, 2022–2031. doi: 10.1093/jac/dku123
Viana, D., Blanco, J., Tormo-Mas, M. A., Selva, L., Guinane, C. M., Baselga, R., et al. (2010). Adaptation of Staphylococcus aureus to ruminant and equine hosts involves SaPI-carried variants of von Willebrand factor-binding protein. Mol. Microbiol. 77, 1583–1594. doi: 10.1111/j.1365-2958.2010.07312.x
Walther, B., Klein, K. S., Barton, A. K., Semmler, T., Huber, C., Merle, R., et al. (2018). Equine methicillin-resistant sequence type 398 Staphylococcus aureus (MRSA) harbor mobile genetic elements promoting host adaptation. Front. Microbiol. 9:2516. doi: 10.3389/fmicb.2018.02516
Wendlandt, S., Lozano, C., Kadlec, K., Gomez-Sanz, E., Zarazaga, M., Torres, C., et al. (2013). The enterococcal ABC transporter gene lsa(E) confers combined resistance to lincosamides, pleuromutilins and streptogramin A antibiotics in methicillin-susceptible and methicillin-resistant Staphylococcus aureus. J. Antimicrob. Chemother. 68, 473–475. doi: 10.1093/jac/dks398
Wertheim, H. F., Walsh, E., Choudhurry, R., Melles, D. C., Boelens, H. A., Miajlovic, H., et al. (2008). Key role for clumping factor B in Staphylococcus aureus nasal colonization of humans. PLoS Med. 5:e17. doi: 10.1371/journal.pmed.0050017
Yang, F., Zhang, S., Shang, X., Li, H., Zhang, H., Cui, D., et al. (2020). Short communication: detection and molecular characterization of methicillin-resistant Staphylococcus aureus isolated from subclinical bovine mastitis cases in China. J. Dairy Sci. 103, 840–845. doi: 10.3168/jds.2019-16317
Keywords: LA-MRSA, dairy farms, phylogenetic relationship, antimicrobial resistance, one health
Citation: Lienen T, Schnitt A, Hammerl JA, Maurischat S and Tenhagen B-A (2021) Genomic Distinctions of LA-MRSA ST398 on Dairy Farms From Different German Federal States With a Low Risk of Severe Human Infections. Front. Microbiol. 11:575321. doi: 10.3389/fmicb.2020.575321
Received: 23 June 2020; Accepted: 08 December 2020;
Published: 08 January 2021.
Edited by:
Kristina Kadlec, Independent Researcher, Wunstorf, GermanyReviewed by:
Phil Giffard, Menzies School of Health Research, Charles Darwin University, AustraliaRenáta Karpíšková, Veterinary Research Institute (VRI), Czechia
Copyright © 2021 Lienen, Schnitt, Hammerl, Maurischat and Tenhagen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tobias Lienen, Tobias.Lienen@bfr.bund.de; Bernd-Alois Tenhagen, Bernd-Alois.Tenhagen@bfr.bund.de
 Tobias Lienen
Tobias Lienen Arne Schnitt
Arne Schnitt Jens Andre Hammerl
Jens Andre Hammerl Bernd-Alois Tenhagen
Bernd-Alois Tenhagen