- 1Facultad de Ciencias Exactas, Centro de Investigación y Desarrollo en Criotecnología de Alimentos, Universidad Nacional de La Plata – Consejo Nacional de Investigaciones Científicas y Técnicas Centro Científico-Tecnológico La Plata – Comisión de Investigaciones Científicas de la Provincia de Buenos Aires, La Plata, Argentina
- 2Área Bioquímica y Control de Alimentos – Facultad de Ciencias Exactas, Universidad Nacional de La Plata, La Plata, Argentina
Microbiota coexists in true symbiosis with the host playing pivotal roles as a key element for well-being and health. Exopolysaccharides from lactic acid bacteria are an alternative as novel potential prebiotics that increase microbiota diversity. Considering this, the aim of the present work was to evaluate the capacity of the EPS produced by two L. paracasei strains isolated from kefir grains, to be metabolized in vitro by fecal microbiota producing short chain fatty acids. For this purpose, fecal samples from healthy children were inoculated in a basal medium with EPS and incubated in anaerobiosis at 37°C for 24, 48, and 72 h. DGGE profiles and the production of SCFA after fermentation were analyzed. Additionally, three selected samples were sequenced by mass sequencing analysis using Ion Torrent PGM. EPS produced by L. paracasei CIDCA 8339 (EPS8339) and CIDCA 83124 (EPS83124) are metabolized by fecal microbiota producing a significant increase in SCFA. EPS8339 fermentation led to an increment of propionate and butyrate, while fermentation of EPS83124 increased butyrate levels. Both EPS led to a profile of SCFA different from the ones obtained with inulin or glucose fermentation. DGGE profiles of 72 h fermentation demonstrated that both EPS showed a different band profile when compared to the controls; EPS profiles grouped in a cluster that have only 65% similarity with glucose or inulin profiles. Mass sequencing analysis demonstrated that the fermentation of EPS8339 leads to an increase in the proportion of the genera Victivallis, Acidaminococcus and Comamonas and a significant drop in the proportion of enterobacteria. In the same direction, the fermentation of EPS83124 also resulted in a marked reduction of Enterobacteriaceae with a significant increase in the genus Comamonas. It was observed that the changes in fecal microbiota and SCFA profile exerted by both polymers are different probably due to differences in their structural characteristics. It can be concluded that EPS synthesized by both L. paracasei strains, could be potentially used as bioactive compound that modify the microbiota increasing the production of propionic and butyric acid, two metabolites highly associated with beneficial effects both at the gastrointestinal and extra-intestinal level.
Introduction
Microbiota coexists in true symbiosis with the host playing pivotal roles as a key element for well-being and health (Weiss and Hennet, 2017; Cani, 2018). The relevance of microbial ecology at the intestinal level on health status led the “International Scientific Association for Probiotics and Prebiotics” (ISAPP) to propose the concept of “normobiosis” to characterize a healthy microbiota where microorganisms with potential benefits for health predominate in number compared to potentially harmful ones, in contrast to “dysbiosis” in which one or a few potentially harmful microorganisms are dominant creating a disease situation (Roberfroid et al., 2010; Thursby and Juge, 2017). The intestinal microbiota responds to multiple stress factors such as diet, antibiotic use, inflammation of the intestinal tract and/or infection of the host with enteric pathogens (Conlon and Bird, 2015; Shen and Wong, 2016; Durack and Lynch, 2019). A stable microbiota with an adequate balance is necessary to maintain the integrity of the epithelial barrier, the immune balance and the physiological control of inflammatory processes. In turn, a “dysbiotic” microbial composition leads to an intestine with loss of integrity of the epithelial barrier which favors bacterial translocation and inflammation (Round and Mazmanian, 2009; Tsai et al., 2019). In this context, the use of diet as a basis for modifying the microbiota has re-emerged in recent years, validating the ancient concepts of the relevant role of nutrition in health (Requena et al., 2018). Therefore, the interest for probiotics or novel sources of prebiotic compounds is increasing all over the world (Ewaschuk and Dieleman, 2006; Gareau et al., 2010; Alagón Fernández del Campo et al., 2019; Venema et al., 2020).
Prebiotics has recently been defined as “a substrate that is selectively utilized by host microorganisms conferring a health benefit,” expanding the concept of prebiotics to include non-carbohydrate substances with healthy effects even at distal sites (Gibson et al., 2017). Prebiotics has been studied for the modulation of infant microbiota on account of their long-lasting effects, extended even after the administration period. Otherwise, they have low risk of serious adverse effects and are easy to administrate in infant foods (Miqdady et al., 2020). The administration of prebiotics in children is associated with a number of beneficial health outcomes, such as reduced risk of some allergic reactions, reduced inflammation and risk of infections (Miqdady et al., 2020). Additionally, they may contribute to reduce the risk of development of dysbiosis associated chronic diseases like intestinal bowel disease, irritable bowel syndrome, and type 1 diabetes (Milani et al., 2017). Oligosaccharides present in human milk (HMO) as well as galactooligosacharides (GOS) and fructooligosaccharides (FOS) usually included in infant milk formulae are the most studied prebiotics used for children, which have been proved to induce specific changes in the composition and metabolic activity of the intestinal microbiota (Braegger et al., 2011; Parker et al., 2020). The end products of prebiotics fermentation are acetic, propionic and butyric acids, lactic acid, hydrogen, methane and carbon dioxide (Louis et al., 2014). Lactate and short chain fatty acids (SCFA) are used by host cells as an energy source. They participate as mediators of the host response since they are able to interact with G protein-coupled receptors (GPR43, GPR41, GPR81, and GPR109A) modulating positively or negatively the activity of enzymes that originate second messengers. Besides, some SCFA act as epigenetic regulators by inhibition of histone deacetylases (HDAC) (Kasubuchi et al., 2015). In addition, they can promote the integrity of the epithelial barrier function by reinforcing tight junctions (Morrison and Preston, 2016) and play a role in the regulation of inflammatory response mediated by inflammasome (Offermanns, 2014). They participate in the absorption of water and electrolytes (Vinolo et al., 2011; Koh et al., 2016) and are relevant not only in the context of gastrointestinal pathologies (Louis and Flint, 2009, 2017; Thorburn et al., 2014) but also of extraintestinal diseases (Durack and Lynch, 2019).
Fermented foods containing lactic acid bacteria, whether probiotic or not, are the main source of microorganisms that temporarily complete the microbial community of the gastrointestinal tract, constituting what is known as the transient microbiome (Koh et al., 2016) that can reach 1010–1011 viable bacteria ingested per day, depending on the eating habits of each individual (Plé et al., 2015; Rezac et al., 2018). Some lactic acid bacteria produce exopolysaccharides (EPS) during fermentation, which, when ingested with the fermented product, can serve as a substrate for commensal bacteria (Ryan et al., 2015), stimulating the development of beneficial microorganisms at the intestinal level and the production of bioactive metabolites (Salazar et al., 2011; Hamet et al., 2016). Being selectively fermented by the microbiota, exopolysaccharides from lactic acid bacteria constitute an alternative as novel potential prebiotic compounds (Balzaretti et al., 2017; Lynch et al., 2018).
Kefir is an artisanal fermented food obtained by milk fermentation with the complex microbiota present in kefir grains. This fermented milk has a long tradition of offering health benefits such as antimicrobial activity, stimulation of immune system, anti-inflammatory, anti-obesity, cholesterol lowering and antioxidant effects, improvement of lactose tolerance, and enhancement of intestinal bacterial microbiota, among others. Lactic acid bacteria, yeast and acetic acid bacteria of different genera, species and even strains coexist in this product and they and/or the metabolites synthesized by them during fermentation could contribute to beneficial health properties attributed to its consumption (Garrote et al., 2010; Bengoa et al., 2019b). Lactobacillus paracasei CIDCA 8339 and CIDCA 83124 are EPS-producing strains isolated from Argentine kefir grains (Hamet et al., 2013; Bengoa et al., 2018a) that have good technological properties and fulfill safe requirement for food application of Argentine and European regulation (Bengoa et al., 2019a). L. paracasei strains produce EPS both in milk (Hamet et al., 2015) and culture media (Bengoa et al., 2018a). Additionally, it has been demonstrated that these strains present potential probiotic properties like the adhesion ability to intestinal epithelial cells which is increased after passage through the gastrointestinal tract (Bengoa et al., 2018b) and the protective effect against Salmonella infection in vitro (Zavala et al., 2016).
Considering that microorganisms’ metabolites may contribute to health properties of the fermented product, the aim of the present work was to evaluate the capacity of the EPS produced by L. paracasei CIDCA 8339 and CIDCA 83124 in milk to be metabolized in vitro by fecal microbiota producing short chain fatty acids.
Materials and Methods
Microorganisms, Growth Conditions, and Fermented Milks Production
L. paracasei CIDCA 8339 and CIDCA 83124 were grown in MRS broth (Difco Laboratories, Detroit, MI, United States) under aerobic conditions at 30°C for 24 h. For fermented milks production, 10 mL of an active culture of the corresponding L. paracasei strain containing ≈ 1 × 109 CFU/mL were inoculated in 1,000 mL of UHT low-fat milk (La Serenísima, Mastellone Hnos S.A, Argentina) and then incubated in aerobic conditions at 30°C for 24 h.
Exopolysaccharide Obtainment
EPS extraction from the fermented milk was performed according to Rimada and Abraham (2003). Fermented milks (500 mL) were heated for 30 min at 100°C to promote the detachment and dissolution of the polysaccharide bound to the cells and the inactivation of enzymes that could hydrolyze EPS. Trichloroacetic acid 8% (Ciccarelli, Santa Fe, Argentina) was added to precipitate proteins and the samples were then centrifuged at 10,000 × g for 20 min at 20°C in an Avanti J25 centrifuge (Beckman Coulter Inc., Carlsbad, CA, United States). The EPS suspended in the supernatant was precipitated by adding two volumes of ethanol per volume of supernatant. Finally, the samples were dialyzed for 48 h at 4°C with stirring through a 1 kDa cut-off dialysis membrane (Spectra/Por, Spectrum laboratories, CA, United States) to remove lactose residues. In order to evaluate samples purity, the protein content was determined qualitatively by the Bradford method (Bradford, 1976). Thin layer chromatography (TLC) was used to determine the absence of lactose and other simple sugars in the EPS samples. EPS were finally lyophilized and preserved at room temperature until use. EPS extraction was performed from two independent cultures.
EPS Molecular Mass Determination
Average molecular weight (Mw) was determined by high-performance size exclusion chromatography using a OH-PAK SB-805HQ gel filtration column (SHODEX, Kawasaki, Japan) with refractive index (RI) detection system according to Piermaria et al. (2008). Samples were filtered through a 0.45 μm membrane (Millipore Corporation, Milford, MA, United States) and 50 μL of polysaccharide solutions (0.5 g/L in NaNO3 0.1 M) were injected for each run. Samples elution was performed at room temperature using NaNO3 0.1 M as mobile phase with a flow rate of 0.95 mL/min (pressure 120–130 psi). Dextrans with Mw ranging from 97,000 to 3,800,000 Da (ALO-2770, Phenomenex, Torrance, CA, United States) were used as standard.
In vitro EPS Fermentation by Human Fecal Microbiota
Fermentation assay was carried out using fecal samples from five healthy children aged between 8 months and 3 years old. The donors were selected taking into account that they had an optimal health state (normal anthropometric values, without overweight, and without previous pathologies), an omnivorous diet and that they had not consumed antibiotics in the last 6 months prior to the assay. In addition, a survey was conducted to the parents in relation to the aforementioned aspects (Supplementary Figure S1). Samples were collected by the parents according to the protocol and with the sterile materials provided and sent to the laboratory the same day, together with a note of informed consent in obedience to the protocol approved by the Central Bioethical Committee, National University of La Plata (May 2017). All of them were kept at 4°C and processed within 24 h after deposition to guarantee the viability of the microorganisms present.
For the fecal homogenate, equal amounts of the five samples (5 g) were suspended into sterile phosphate buffer saline (225 mL) and mixed to obtain a 1/10 diluted pool (Aguirre et al., 2014). Homogenates were inoculated to a carbohydrate-free basal medium (1/10) with the EPS under study as the only sugar source at a final concentration of 0.3% w/v. The carbohydrate-free basal medium was formulated according to Salazar et al. (2008): peptone water 2 g/L, yeast extract 2 g/L, NaCl 0.1 g/L, K2HPO4 0.04 g/L, KH2PO4 0.04 g/L, MgSO4 0.01 g/L, CaCl2 6H2O 0.01 g/L, NaHCO3 2 g/L, cysteine HCl 2.5 g/L, bile salts 0.5 g/L, and tween 80 2 g/L. The medium was autoclaved and then 1 mL/L of hemin solution (50 mg/ml) and 10 μL/L of vitamin K previously sterilized by filtration were added.
Samples were incubated for 24, 48, or 72 h at 37°C in anaerobiosis using jars (AnaeroPack, Mitsubishi Gas Chemical Company, Japan) according to Salazar et al. (2008). Controls without sugar (BM), with glucose (GLU) (Britania, Buenos Aires, Argentina) and with inulin (INU) (Saporiti, Buenos Aires, Argentina) in the same concentration (0.3% w/v) were included in the experiment. Each fermentation condition was performed in triplicate. After fermentation, the samples were centrifuged for 10 min at 10,000 xg, the supernatant was filtered through a 0.45 μm pore membrane and stored at −20°C for SCFA quantification by gas chromatography. The pellets were stored at −80°C for the subsequent characterization of microbial populations.
Determination of Organic Acids by Gas Chromatography
Chromatographic analysis was carried out using an Agilent 7890a GC system with a DB23 column (Agilent Technologies, Santa Clara, CA, United States) coupled to a flame detector (FID). The temperature at the injection port and at the FID was 250°C. Helium was used as carrier gas at a flow rate of 1.6 mL/min. For the run, 1 μL of the sample was injected with a 1:25 split and a temperature program that consisted of a ramp from 100 to 200°C at a speed of 8°C/min, keeping constant at 200°C for 3 min was used. Calibration curves with standards of glacial acetic acid (1–50 mM), propionic acid (0.5–20 mM), butyric acid (0.5–20 mM), iso-butyric acid (0.5–20 mM), and iso-valeric acid (0.5–20 mM) were prepared (Sigma-Aldrich, San Luis, MO, United States). Organic acids were identified by comparison to standard retention times and quantified with the corresponding peak area using the calibration curve. Differences were statistically tested using One-way analysis of variance (ANOVA) with Tukey’s multiple comparison test (p < 0.05) conducted by the GraphPad Prism® software.
DNA Isolation
DNA extractions were performed by using a commercial QIAamp PowerFecal DNA kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions. DNA concentration and quality were determined using a NanoDrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA, United States). The DNA samples were used for DGGE analysis and mass sequencing. DNA extracted from pure cultures of Lactobacillus casei, L. plantarum, L. kefiri, and Bifidobacterium adolescentis were used as reference strains for DGGE analysis.
Microbiota Evolution Analysis by DGGE
The fecal microbiota evolution during batch fermentations was analyzed by partial amplification of the 16S rRNA gene using universal primers 518R and 338F-GC (Table 1). PCR amplification was performed using Taq polymerase Pegasus (PB-L Biological Products, Argentina) following the manufactures instruction and using 1 ng/μL of DNA template. The reaction was carried out in a T100 thermal cycler (Bio-rad laboratories, Irvine, CA, United States) with the following amplification program: 94°C for 5 min; 35 cycles of 94°C for 30 s, 60°C for 45 s and 72°C for 20 s; and a final extension step at 72°C for 1 min. The amplification products were analyzed by electrophoresis in 1% w/v agarose gels with ethidium bromide and revealed under UV light.
Denaturing-gradient-gel electrophoresis (DGGE) was performed in a DGGE-2401 analyzer (C.B.S. Scientific Co., Del Mar, CA, United States). The PCR products (15 μL) were seeded in 8 g/100 mL polyacrylamide gels (15 × 20 × 0.075 cm) in TAE buffer [50X TAE is 2 M Tris, 1 M acetic acid, and 50 mM EDTA (pH 8.0)]. A denaturing gradient of Urea-Formamide 40–60% (100% corresponds to Urea 7 M and Formamide 40% v/v) was used to achieve the optimal separation of the bands corresponding to Eubacteria. Electrophoresis was carried out at 90 V for 16 h at 60°C. Gels were then stained by immersion for 30 min in a 0.1 μL/mL Sybr-Gold solution (Invitrogen, United States) in TAE buffer and observed under UV light. Band patterns obtained for each sample were compered using the Bionumerics 6 program (Applied maths NV, Sint-Martens-Latem, Belgium). The percentage of similarity between the samples was calculated using the Dice Similarity Coefficient and the corresponding UPGMA dendrograms were constructed.
Mass Sequencing Analysis Using Ion Torrent PGM
The mass sequencing analysis was carried out in the MR DNA molecular research laboratory (TX, United States1), based on established and validated protocols2. Primers 515 and 806 that amplify the V4 variable region of the gene that codes for 16S rRNA were used (Table 1). For PCR, the HotStarTaq Plus Master Mix kit (Qiagen, Hilden, Germany) was used with an amplification program that consisted of 94°C 3 min, 30 cycles of 94°C 30 s, 53°C 40 s, 72°C 1 min and finally 72°C 5 min.
The mass sequencing analysis was performed using the Ion Torrent Personal Genome machine (PGM) system (Thermo Fisher Scientific, Waltham, MA, United States) following the manufacturer’s guidelines. The generated data was demultiplexed and analyzed using a pipeline developed in the MR DNA molecular research laboratory. Raw data sequencing reads were quality trimmed using the QIIME suite of tools. Sequences were depleted of barcodes and primers, followed by removal of short sequences (<150 bp), sequences with ambiguous base calls and with homopolymer runs exceeding 6 bp. Noise from sequences and chimeras were also removed. Sequencing data were grouped into 3% divergence operating taxonomic units (OTUs) and taxonomically classified using the BLASTn.NET algorithm with the database derived from RDPII3 and NCBI4.
Results
EPS Production by L. paracasei Strains in Milk
EPS production during milk fermentation at 30°C by L. paracasei CIDCA 8339 and CIDCA 83124 were about 130–145 and 140–160 mg of EPS per liter of fermented milk respectively. These values are within the expected range, since EPS yield by LAB is normally very low (Ruas-Madiedo et al., 2008; Llamas-Arriba et al., 2019). Crude EPS isolated from fermented milk with L. paracasei CIDCA 8339 (EPS8339) and CIDCA 83124 (EPS83124) were partially characterized by analyzing their molecular weight distribution (Mw) by gel permeation chromatography. EPS8339 consists of two fractions, a high Mw fraction of about 4 105 Da and a low Mw fraction of about 1 104 Da. On the other hand, EPS83124 presents four fractions: a low Mw fraction of 1 104 Da, an intermediate Mw fraction of 7 104 Da and a high Mw fraction constituted by two Mw distributions of 7 105 and 6 106 Da.
Evaluation of Fecal Microbiota Evolution During EPS Fermentation
PCR DGGE profiles were employed to monitor major qualitative changes in the compositions of microbial groups of fecal homogenates with and without sugar source added after 24, 48, and 72 h fermentation. DGGE profiles obtained were both time and sugar source dependent (Figure 1A). After 24 h of incubation, DGGE profiles of the homogenates fermented in the presence of EPS8339 and EPS83124 had a high similarity percentage (77–81%) with respect to the profile obtained for those grown in the carbohydrate-free basal medium (Figure 1B). In contrast, profiles of homogenates fermented with glucose or inulin presented less similarity when compared to the basal medium. However, after 72 h fermentation, the electrophoretic profile of the homogenates fermented in the presence of glucose and inulin resembled more the profile obtained in the basal medium, while those fermented in the presence of EPS8339 and EPS83124 showed a different band pattern, locating in a separated cluster and showing only 60% similarity with the two controls (GLU and INU). In the presence of glucose or inulin, changes in microbiota were observed after short fermentation times indicating the rapid use or assimilation of these sugars by fecal microorganisms. On the other hand, when EPS8339 and EPS83124 were added to basal medium, the changes in fecal microbiota occurred after long fermentation times, probably because the microorganisms need to adapt to this new carbon source.
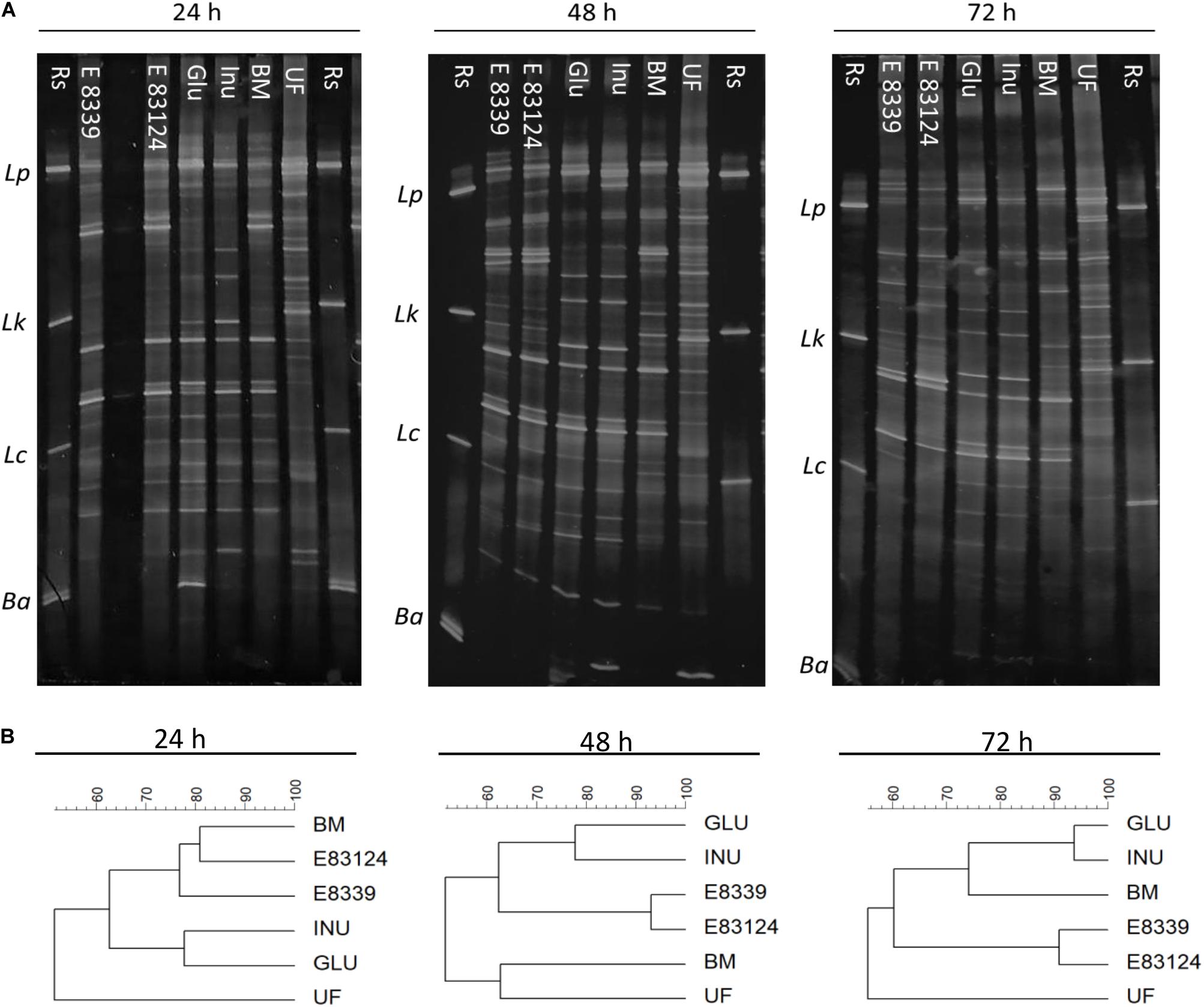
Figure 1. PCR-based DGGE profiles (A) and the respective dendrograms (B) of fecal homogenates after 24, 48, and 72 h fermentation with EPS8339 (E 8339), EPS83124 (E 83124), glucose (Glu), inulin (Inu) or without extra sugar added (BM). UF profile corresponds to the one obtained with the unfermented fecal homogenate and Rs corresponds to reference strains. Lp, Lactobacillus. plantarum; Lk, Lactobacillus kefiri; Lc, Lactobacillus casei; Ba, Bifidobacterium adolescentis. Dendrograms were obtained by similarity analysis of DGGE profiles using Dice similarity coefficient and UPGMA cluster analysis.
Dendrogram comparing V3 DGGE profiles obtained with all growth media assayed at different fermentation times is shown in Figure 2. Two main clusters that have a similarity value lower than 48% were observed. One of them joined the samples corresponding to the 72 h of fermentation in the media containing EPS8339 and EPS83124 and the other cluster joined the rest samples grouped in two subclusters. It can be observed that all the samples obtained after 24 h fermentation joined in the same subcluster with 71% similarity. The other subcluster grouped DGGE profiles of the rest of the samples obtained after 48 and 72 h fermentation (61% of similarity). Within this second subcluster inulin and glucose fermentation after 48 and 72 h grouped together with 72% of similarity.
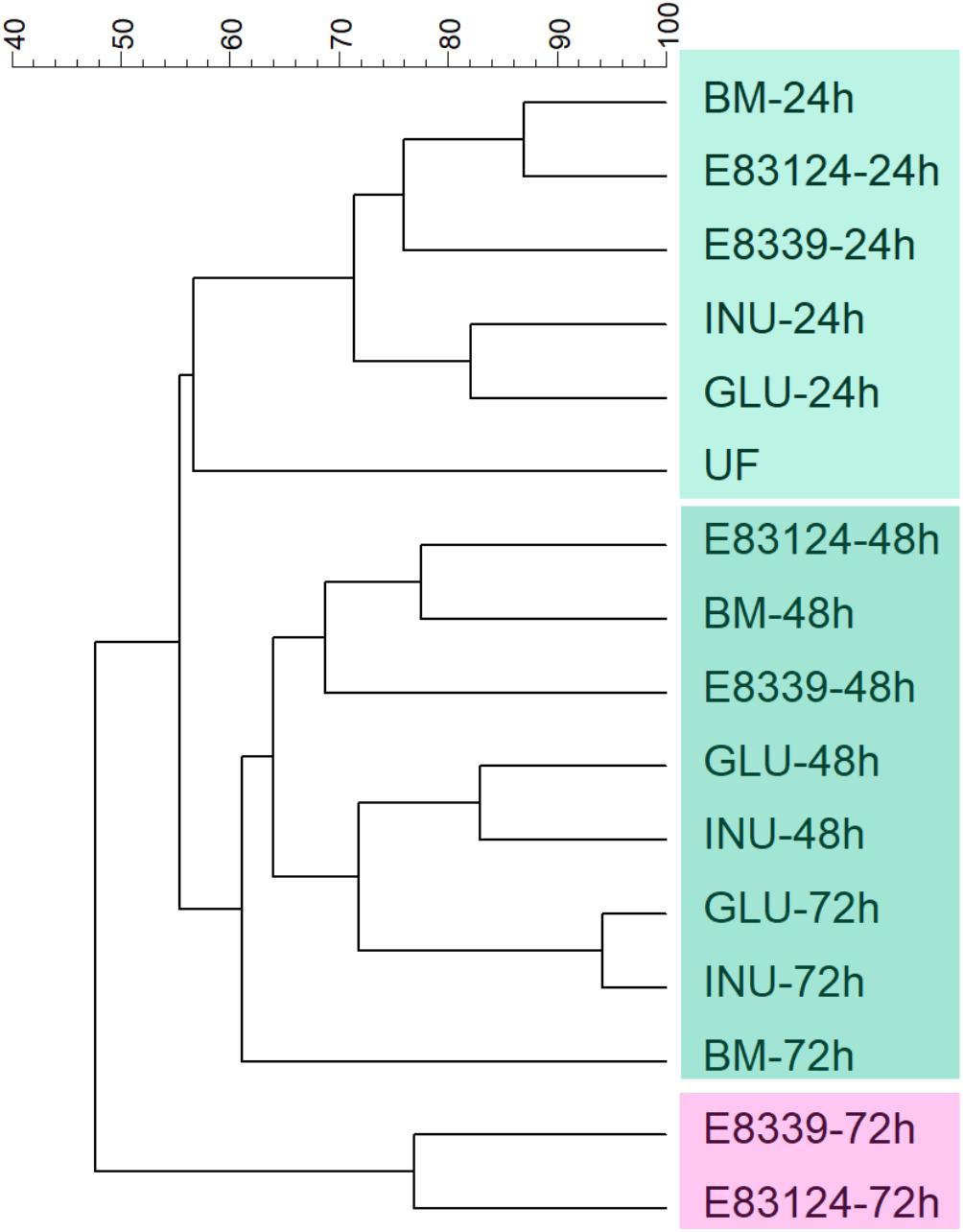
Figure 2. UPGMA dendrogram of DGGE profiles of fecal homogenates microbiota after different fermentation times with EPS8339 (E 8339), EPS83124 (E 83124), glucose (GLU), inulin (INU) or without sugar source added (BM). UF corresponds to the unfermented fecal homogenate. Dendrogram was obtained by similarity analysis of DGGE profiles using Dice similarity coefficient and UPGMA cluster analysis.
Since fecal homogenates fermented during 72 h with EPS8339 and EPS83124 presented the higher differences in the DGGE band pattern compared to basal medium control, we proceed to perform a mass sequencing analysis of these samples using Ion Torrent PGM. Sequence depth performed in the analysis was adequate since rarefaction curves reached a plateau for all samples (Supplementary Figure S2). The analysis of alpha diversity across the samples gave Shannon diversity indices of 3.79, 3.23, and 3.30 for fermentation in carbohydrate-free basal medium, basal medium with EPS8339 and basal medium with EP83124, respectively, indicating high diversity in the three samples.
Figure 3A shows the distribution of the populations present in homogenates samples after 72 h fermentation at the phylum level. The main phyla present in the sample from basal medium (control) were Proteobacteria (58%), Bacteroidetes (19%), Firmicutes (17%), and Actinobacteria (2%). Fermentation of EPS8339 led to an increase in the relative proportion of Firmicutes (29%) and Lentisphaerae (32%), accompanied by the decrease in Actinobacteria (0.5%), Proteobacteria (27%) and Bacteroidetes (8%). On the other hand, fermentation of EPS83124 caused a reduction of the Actinobacteria (0.7%) and Bacteroidetes (9%) phyla with an increase in the proportion of Proteobacteria (73%).
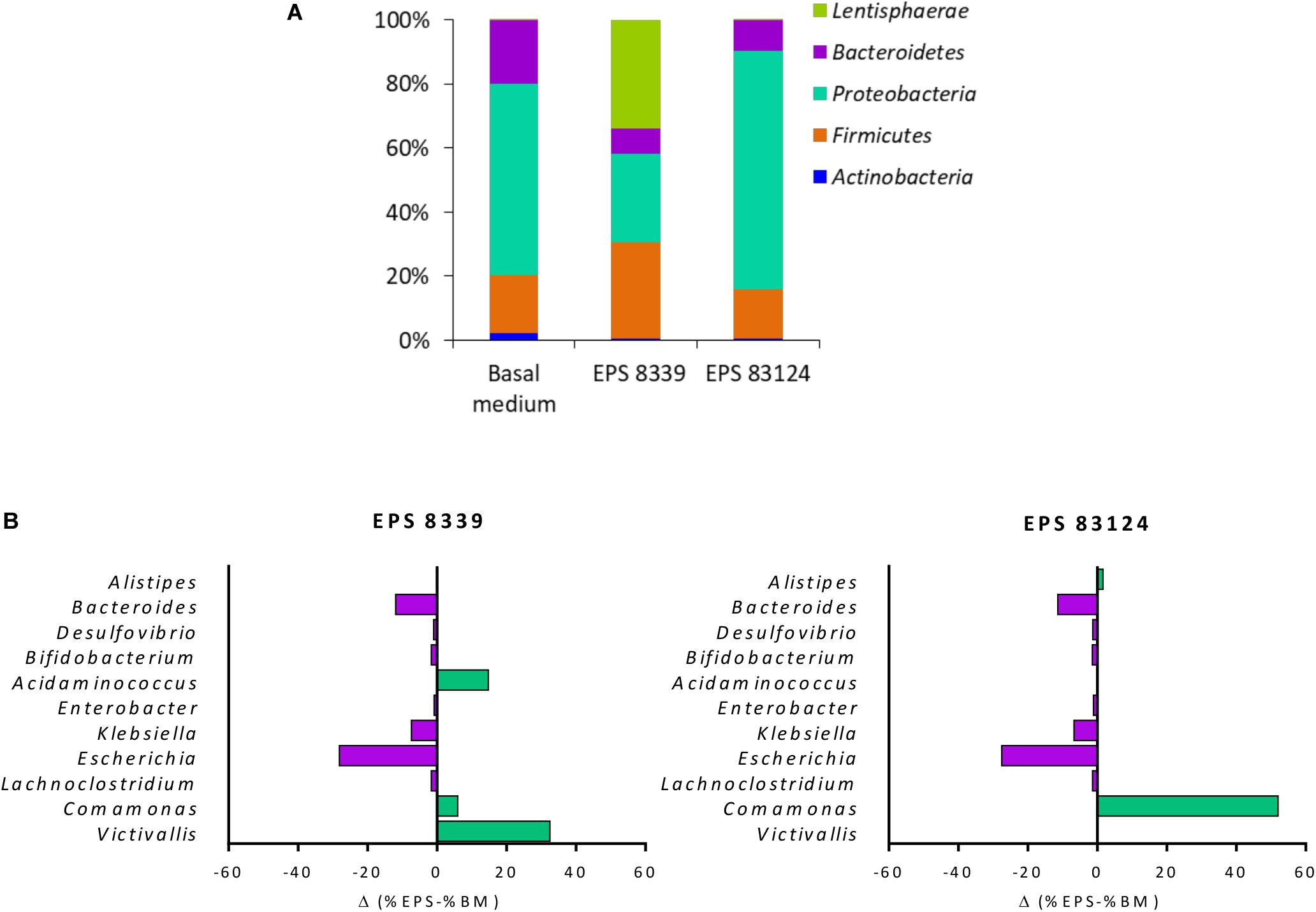
Figure 3. Relative abundance at the phylum level of fecal homogenates fermented for 72 h in the presence EPS8339, EPS83124 or without extra sugar added (BM). Only phyla with relative abundance higher than 1% in at least one sample were included (A). Differences in bacterial relative abundance at the genus level in fecal homogenates fermented for 72 h with EPS8339 and EPS83124 in comparison with homogenates fermented 72 h without extra sugar added (BM). Only genera that showed differences higher than 1% in at least one sample were included (B).
At the genus level, it was observed that, regardless of the EPS used during the fermentation, the proportion of Klebsiella and Escherichia (γ-Proteobacteria) were reduced in 6 and 28% respectively while Bacteroides (Bacteroidetes) dropped in about 11–12% (Figure 3B). Nonetheless, the genera that showed an increment in their relative abundance after fermentation were different in both EPS samples, indicating that EPS8339 and EPS83124 are selectively used by different microorganisms present in fecal microbiota. The proportion of Comamonas (β-Proteobacteria) increased 6% for EPS8339 and 52% for EPS83124. This substantial rise of Comamonas genera explains the difference in the proportion of Proteobacteria phylum previously mentioned. Furthermore, fermentation of EPS8339 increased the proportion of the genera Victivallis (33%) and Acidaminococcus (15%) which correspond to almost the total rise in Lentisphaerae and Firmicutes phyla evidenced. However, fermentation of both EPS did not induce major changes in the population of Lactobacillus and Bifidobacterium.
SCFA Production During EPS Fermentation by Fecal Microbiota in Batch Cultures
When analyzing the SCFA levels in the supernatant of fermented samples, it was evidenced that EPS8339 and EPS83124 are metabolized by the fecal microbiota producing, consequently, a significant increase in organic acids with recognized biological activity (propionate and butyrate) compared to basal medium. As an example, chromatograms obtained with fecal homogenates with EPS83124 after different fermentation times are shown in Supplementary Figure S3. Total SCFA levels increased during the first 24 and 48 h of incubation with and without external carbon sources while no changes or a decrease in total SCFA were observed after 72 h depending on the sugar source. Fermentation of EPS8339 showed a pattern similar to glucose, where the greatest increase in SCFA was evidenced at 48 and 72 h. The fermentation of EPS83124 showed a significant increase after 24 h followed by a drop after 72 h of fermentation. This reduction could be due to the fact that some SCFA produced can be then consumed by some microbial population whose activity was selectively stimulated by this EPS (Figure 4A).
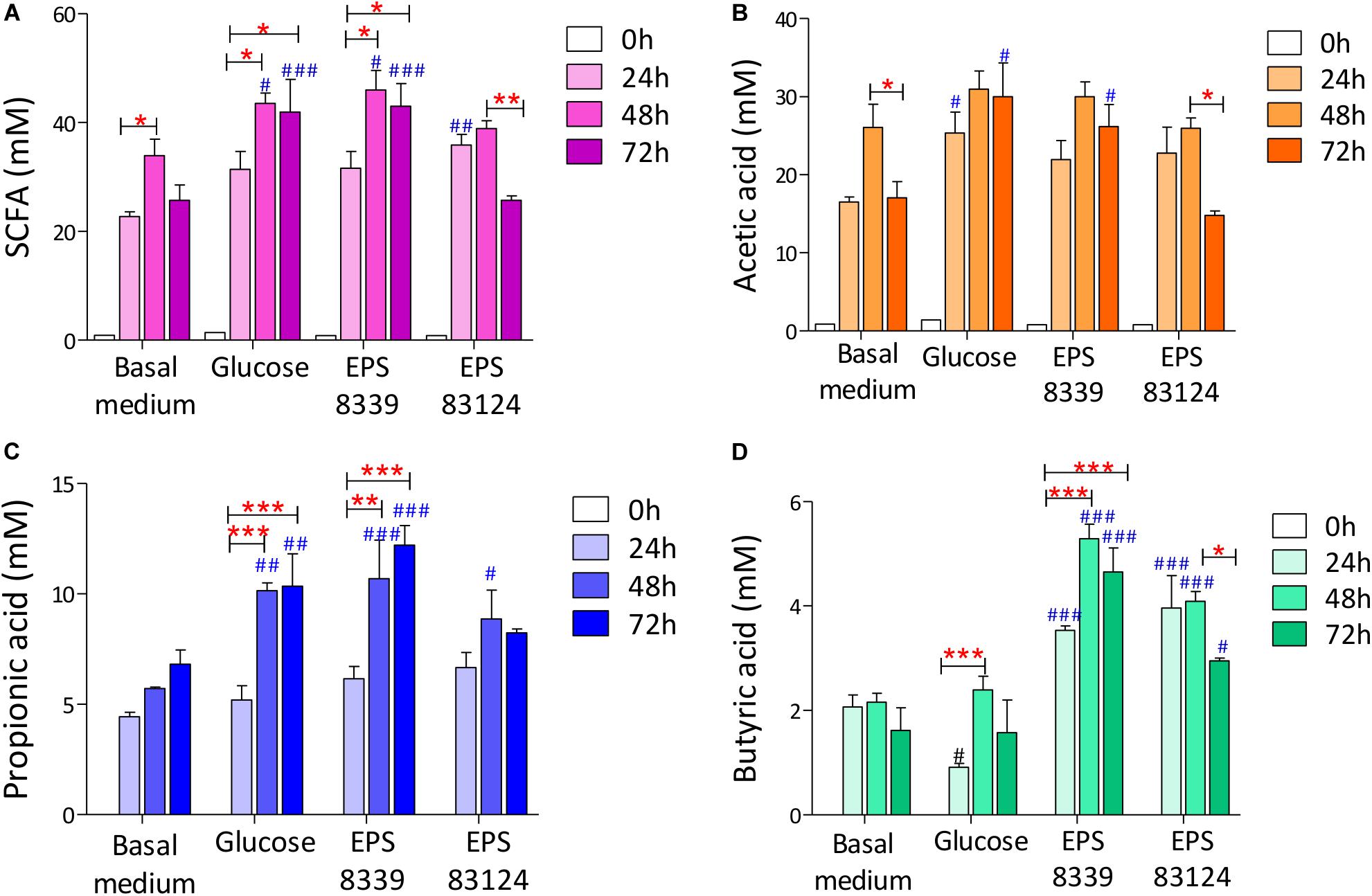
Figure 4. Total SCFA (A) and individual acetate (B), propionate (C) and butyrate (D) levels of fecal homogenates fermented during 24, 48, and 72 h with and without sugar source added. # indicates significant differences against carbohydrate-free basal medium at the same fermentation time. *indicates significant differences between samples with a specific sugar at different fermentation time. (*/#p < 0.05, **/##p < 0.01, ***/###p < 0.001, Tukey’s multiple comparison test).
On account of the levels of each individual SCFA with biological activity, it was observed that neither EPS8339 nor EPS83124 significantly increased acetate levels. However, in the case of EPS83124 a reduction in acetate levels was observed after 72 h fermentation similar to the results obtained with carbohydrate-free basal medium (Figure 4B). Regarding propionate, EPS8339 and glucose fermentation led to a significant increase of this organic acid at 48 and 72 h, while fermentation of EPS83124 produced a significant increase in propionate only at 48 h (Figure 4C). However, the levels of propionate achieved in the presence of EPS are low compared to inulin fermentation, since this prebiotic was metabolized by fecal microbiota producing propionate in concentrations of about 100 mM (Table 2), 10 times higher than those obtained with EPS8339, EPS83124 or glucose. Furthermore, it is noteworthy that fermentation of both EPS led to a significant increase in butyrate at all fermentation times, unlike the results observed with glucose and inulin where no increment in butyrate was observed (Figure 4D and Table 2).
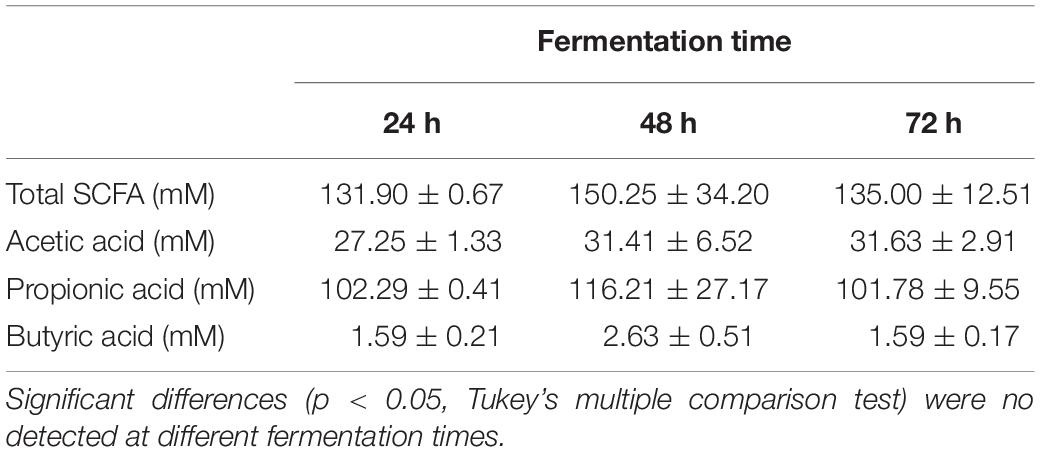
Table 2. Short chain fatty acids (SCFA) concentration of the homogenates fermented in basal medium with inulin at different times.
These results show that EPS8339 fermentation led to an increment of propionate and butyrate, while fermentation of EPS83124 increased butyrate levels. Noteworthy, both EPS generated a different pattern of SCFA than inulin fermentation, which significantly increases propionate but does not modify butyrate levels.
In addition, two peaks that elute at 2.63 and 3.25 min and that were identified as isobutyric and isovaleric acid respectively, progressively increased with fermentation time when both bacterial EPS were used as sugar sources (Table 3). In contrast, these organic acids were not detected when inulin or glucose were present.
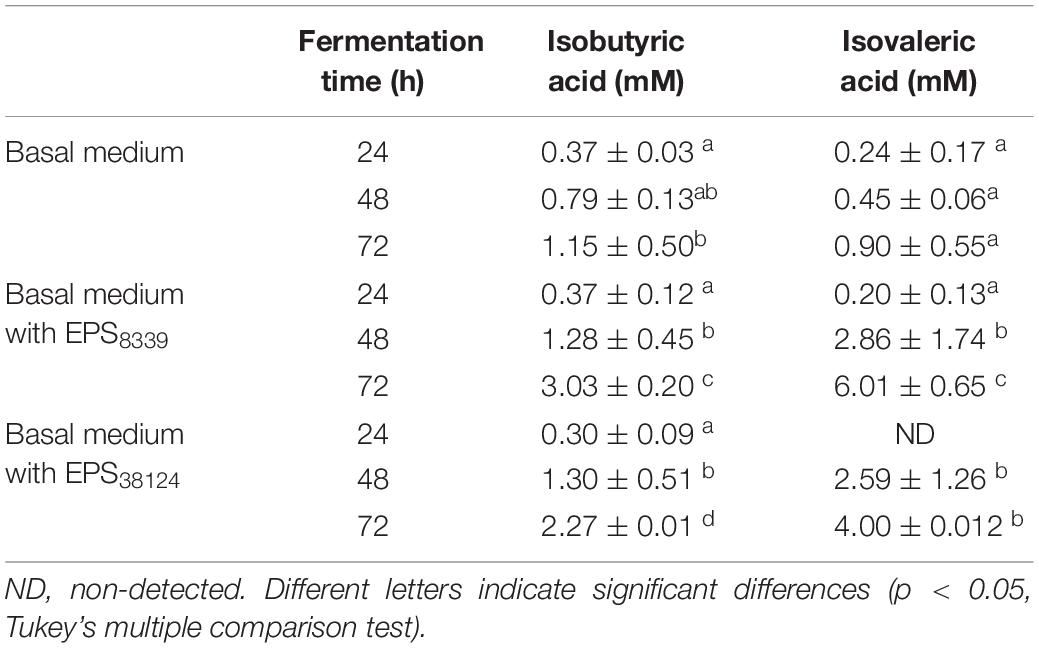
Table 3. Isobutyric and isovaleric levels from homogenates fermented in basal medium with or without EPS at different incubation times.
Discussion
The understanding of the mechanisms by which a balanced microbiome contributes to health has been considerably expanded in the last years, being the role of the metabolites one of the focus of research. In particular, SCFA results of great interest because of their wide variety of health benefit effects at both intestinal and extra-intestinal level. The homeostasis of the intestinal microbiota and its corresponding metabolome depends on the characteristics of the host (age, sex, genetic background) and on environmental conditions (stress, medications, gastrointestinal surgery, infectious, and toxic agents) (Conlon and Bird, 2015). Although microbiota composition is diverse between individuals in terms of genus and species, it is dominated by four main phyla: Firmicutes, Bacteroidetes, Actinobacteria, and Proteobacteria (Hollister et al., 2014; Hugon et al., 2015) as it was described in the present work. It is worth to note that the percentage of each phylum described herein was in concordance to those described previously (Milani et al., 2017). The early colonization process is crucial for long-term health benefits having the infant gut microbiota a main role in modulating risk factors related to adult health conditions (Milani et al., 2017). It was observed that gut bacterial microbiome rapidly diversifies over the first years of life in healthy children while is less diverse in those who develop allergy or asthma or who are malnourished (Durack and Lynch, 2019). As the composition of the infant’s microbiome can have a profound effect on adult life, the search of new compounds that contribute to microbiota modulation for their inclusion in infant diet results of relevance. In the present work, the changes in infant fecal microbiota and SCFA levels induced by EPS8339 and EPS83124 fermentation were studied. For that purpose, fecal homogenates prepared in a basal medium were added with different sugars including glucose and inulin that were used as controls.
Lactic acid bacteria exopolysaccharides are widely studied for its contribution to food texture. Still, they can also act as bioactive compounds that are able to exert their effect by direct interaction to the epithelial cells or indirectly by inducing specific changes in the composition and metabolic activity of the intestinal microbiota (Gibson et al., 2017). They are highly diverse in structure and sugar composition and it has been evidenced that the biological activity attributed to each biopolymer, such as the prebiotic potential, is mostly dependent on its molecular characteristics (Salazar et al., 2016). The fermentation of EPS synthetized by L. paracasei CIDCA 8339 and CIDCA 83124 led to changes in fecal microbiota as well as in SCFA profile. These results indicate in first place, that both EPS are fermentable by fecal microbiota. However, EPS synthetized by L. paracasei CIDCA 8339 and CIDCA 83124 in milk showed different molecular weight distribution. Thus, it is not surprising that the changes in the fecal microbiota and in SCFA profile exerted by both polymers are different.
When analyzing the mass sequencing data from 72 h fermentation samples, it was evidenced that neither EPS8339 nor EPS83124 favored the growth of the genera commonly consider as beneficial such as Lactobacillus and Bifidobacterium. Moreover, despite the production of propionate and butyrate evidenced in these samples, none of the microorganisms generally associated to the production of those SCFA including Faecalibacterium prausnitzii, Eubacterium rectale, Eubacterium hallii, Ruminococcus bromii, Akkermansia municiphilla, and Roseburia intestinalis (Morrison and Preston, 2016) were increased after 72 h fermentation when compared to basal medium. It has been reported that species from the genera Victivallis, Acidaminococcus and Comamonas, the most favored by EPS8339 and EPS83124 fermentation, are part of the human gastrointestinal tract (Jumas-Bilak et al., 2007; Segata et al., 2012; Samb-Ba et al., 2014; Ricaboni et al., 2017). However, they have not been widely studied and their role in the intestinal microbiota is not clearly yet. Although they do not correspond to any of the genera commonly associated with the production of SCFA, it has been reported in the literature that Acidaminococcus species are capable of producing acetate, propionate and butyrate (Jumas-Bilak et al., 2007). Furthermore, the genus Victivallis also contributes to acetate production (Zoetendal et al., 2003). Despite no significant increase in acetate levels were observed in the samples fermented in the presence of EPS, it must be considered that acetate can be used by many gut commensals to produce propionate and butyrate in a growth-promoting cross-feeding process (Verkhnyatskaya et al., 2019). Considering this, the production of acetate by Acidaminococcus and Victivallis could indirectly promote the production of other SCFA, such as butyrate through the butyryl-CoA: acetate-CoA transferase pathway present in some microorganisms of the microbiota (Louis and Flint, 2017). In this way, these populations could be directly or indirectly responsible for the significant increase in butyrate observed during fermentation of EPS8339. The fermentation of EPS83124 results in an increase of species of the genera Comamonas such as C. aquatica and C. kerstersii (data not shown) that do not produce SCFA but instead are able to consume them (Wauters et al., 2003). Therefore, the pronounced increase in Comamonas due to EPS83124 fermentation could explain the significant drop in acetate and butyrate evidenced after 72 h fermentation. It has been evidenced that the presence of this genus, that is normally located in the Lieberkühn crypts, is beneficial since it participates in the maintenance of local homeostasis that is essential for epithelial regeneration (Pédron et al., 2012). Similarly, the consumption of milk containing L. casei BL23 in Balb/c mice also generated a significant increase in Comamonas (Yin et al., 2014). Moreover, the significant decrease in the Enterobacteriaceae family, that include genera usually associated with pathogens (Klebsiella and Escherichia), evidenced with both EPS also contributes to a more anti-inflammatory and healthy gut state.
It can also be highlighted that results obtained with these EPS differ from the results obtained with inulin that was included as a positive prebiotic control. Fermentation of both EPS conducted to a different SCFA profile and led to greater changes in the microbial population after 72 h fermentation. In the DGGE profiles obtained for glucose (a sugar easily fermentable by microorganisms that was included as control of bacterial growth and activity) and inulin it can be observed a high percentage of similarity between both sugars at 48 and 72 h fermentation with practically identical band patterns. These similar patterns obtained with inulin and glucose could be attributed to the fact that the donors of fecal samples regularly consumed formula milk with inulin, FOS or GOS, so their microbiota is probably adapted to inulin-type prebiotics. These results highlight the relevance of finding new compounds that can exert a beneficial effect on the intestinal microbiota different from those observed with the prebiotics commonly used in food in order to contribute to establish a widely diverse intestinal microbiota usually associated with a healthy state (Durack and Lynch, 2019). In this context, the EPS produced by L. paracasei CIDCA 8339 and CIDCA 83124 emerge as alternative potential prebiotics that can be fermented by the fecal microbiota in vitro, producing modifications in the DGGE microbiological profile after 72 h that differs from the changes induced by inulin. Additionally, in contrast to inulin that induced mainly the production of propionate, the fermentation of EPS8339 and EPS83124 led to a significant increase in butyrate levels, a bioactive metabolite with several beneficial effects at the intestinal level. It is remarkable that, even though bacterial EPS8339 and EPS83124 required 72 h fermentation to induce changes in microbial population, they rapidly modified microbiota activity as can be evidenced by the increase in SCFA after 24 h. Considering this, both EPS could be used as complementary prebiotics that contribute to consumers’ health inducing favorable changes at the intestinal microbiota that are different from the ones induced by inulin.
The increase of butyrate as a consequence of EPS8339 and EPS83124 fermentation may bring a wide range of health benefits, particularly at the intestinal level. Butyrate can be used as an energy source by enterocytes (van der Beek et al., 2015). Moreover, it contributes to strengthen the intestinal epithelial barrier through a mechanism that involves the induction of tight junctions’ proteins expression such as Claudin 1 and ZO-1 and their redistribution in the membrane (Morrison and Preston, 2016). The loss of integrity of the intestinal barrier and the consequent increase in its permeability is generally associated with an increase in bacterial translocation and/or its wall components, which results in a mild chronic inflammatory state that has been associated with pathologies such as obesity, insulin resistance and diabetes type 2 (Cani et al., 2008; Qin et al., 2012). Furthermore, propionate production would also be beneficial in people that suffer of obesity since it inhibits cholesterol synthesis at the liver, regulates lipogenesis in adipose tissue (Ríos-Covián et al., 2016; Tsai et al., 2019) and regulates appetite through the expression of leptin, PYY and GLP-1 (Chambers et al., 2015). Thus, the use of prebiotics that leads to the production of butyrate and/or propionate at the intestinal level results interesting in individuals with these kinds of metabolic disorders. On the other hand, SCFA regulate the immune response at the intestinal level, contributing to the healthy state in patients suffering from inflammatory bowel diseases. Butyrate, for instance, exerts an anti-inflammatory effect by inhibiting the activation of the transcription factor NFκB in macrophages and the expression of proinflammatory cytokines (IL-6 and IL-12) in dendritic cells. Moreover, butyrate and propionate are able to regulate the production and function of regulatory T cells by inhibiting histone deacetylases (Morrison and Preston, 2016; Requena et al., 2018). Dysbiosis observed in inflammatory bowel diseases, including ulcerative colitis and Crohn’s disease, is generally associated with a reduction in SCFA levels (Alagón Fernández del Campo et al., 2019) and an increase in species of the Enterobacteriaceae family and other opportunistic pathogens (Gonçalves et al., 2018; Uchiyama et al., 2019). The microbiota modulation favoring the production of SCFA during infancy is relevant to reduce the risk of disease development in the future. In this context, Roduit et al. (2019) studied the role of SCFA in the prevention of allergy and asthma by analyzing SCFA levels in 1-year-old children fecal samples and correlating them with the development of disease when those children were 6 years old. The authors evidenced that children that presented the highest levels of propionate and/or butyrate when they were 1 year old, were less likely to develop asthma, food allergy and allergic rhinitis when they grew up. In the same way, it has been suggested that butyrate-producing bacteria play a key role in reducing the risk of developing type 1 diabetes in children between 1 and 5 years old (de Goffau et al., 2014).
This study revealed that the EPS produced by L. paracasei CIDCA 8339 and CIDCA 83124 isolated from kefir induce substantial distinct effects on fecal microbiota activity and composition of healthy children leading to selective enrichments of those microorganisms that possess the ability to adapt their growth to the respective substrates. Mass sequencing analysis demonstrated that the fermentation of EPS8339 leads to an increase in the proportion of the genera Victivallis, Acidaminococcus, and Comamonas and a significant drop in the proportion of enterobacteria. In the same direction, the fermentation of the EPS83124 also resulted in a marked reduction in the population of Enterobacteriaceae with a significant increase in the genus Comamonas. These responses were linked to directed changes in SCFA toward butyrate production in higher concentration than controls. It was observed that both EPS presented a different fermentation profile probably due to differences in their structural characteristics. EPS8339 fermentation led to an increment of propionate and butyrate, while fermentation of EPS83124 increased mainly butyrate levels. These increase in the production of propionate and/or butyrate, accompanied by a decrease in the population of Enterobacteriaceae allowed us to hypothesize that the consumption of both EPS could contribute to reduce the inflammation at the intestinal level.
Although fecal microbiota composition partially correlates with gut microbiota, these results are a first step in the knowledge of the ability of two EPS from L. paracasei strains isolated from kefir to be fermented by human microbiota. It can be concluded that the EPS synthesized by L. paracasei CIDCA8339 and CIDCA83124 in milk can be considered bioactive compounds that modify the microbiota increasing the production of propionic and/or butyric acid, two metabolites highly associated with beneficial effects both at the gastrointestinal and extra-intestinal level.
Data Availability Statement
SRA data generated in this study was uploaded in NCBI database. Project number PRJNA665182. BioSample accessions SAMN16245014, SAMN16245015, and SAMN16245016.
Author Contributions
AB contributed in study design and conception, and performed experimental work, data interpretation, and manuscript writing. CD contributed with DGGE experiments, and contributed to microbiota data interpretation and writing the manuscript. NG contributed to acid organic determination and DNA extraction and revised the manuscript. GG participated in study design and conception, funding, and manuscript revising. AA participated in study design and conception, funding, and manuscript revising. All authors contributed to the article and approved the submitted version.
Funding
The present work was supported by CONICET, Universidad Nacional de La Plata (UNLP 18/X813, 2018-2021) and ANPCyT (PICT 2014-3137 and PICT 2016-0639).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
AB and NG are fellows of Consejo Nacional de Ciencia y Tecnologia (CONICET). CD is fellow of Agencia Nacional de Promoción Científica y Tecnológica (ANPCyT). GG and AA are members of Scientific Career of CONICET. We thank M. S. Sinkec and C. Reyes Cantera for chromatographic technical assistance.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.583254/full#supplementary-material
Footnotes
- ^ www.mrdnalab.com, Shallowater,
- ^ http://www.mrdnalab.com/
- ^ http://rdp.cme.msu.edu
- ^ http://www.ncbi.nlm.nih.gov
References
Aguirre, M., Ramiro-Garcia, J., Koenen, M. E., and Venema, K. (2014). To pool or not to pool? Impact of the use of individual and pooled fecal samples for in vitro fermentation studies. J. Microbiol. Methods 107, 1–7. doi: 10.1016/j.mimet.2014.08.022
Alagón Fernández del Campo, P., De Orta Pando, A., Straface, J. I., López Vega, J. R., Toledo Plata, D., Niezen Lugo, S. F., et al. (2019). The use of probiotic therapy to modulate the gut microbiota and dendritic cell responses in inflammatory bowel diseases. Med. Sci. 7, 1–17. doi: 10.3390/medsci7020033
Bakke, I., De Schryver, P., Boon, N., and Vadstein, O. (2011). PCR-based community structure studies of Bacteria associated with eukaryotic organisms: a simple PCR strategy to avoid co-amplification of eukaryotic DNA. J. Microbiol. Methods 84, 349–351. doi: 10.1016/j.mimet.2010.12.015
Balzaretti, S., Taverniti, V., Guglielmetti, S., Fiore, W., Minuzzo, M., Ngo, H. N., et al. (2017). A novel rhamnose-rich hetero-exopolysaccharide isolated from Lactobacillus paracasei DG activates THP-1 human monocytic cells. Appl. Environ. Microbiol. 83, e2702–e2716. doi: 10.1128/aem.02702-16
Bengoa, A. A., Iraporda, C., Acurcio, L. B., de Cicco Sandes, S. H., Costa, K., Moreira Guimarães, G., et al. (2019a). Physicochemical, immunomodulatory and safety aspects of milks fermented with Lactobacillus paracasei isolated from kefir. Food Res. Int. 123, 48–55. doi: 10.1016/j.foodres.2019.04.041
Bengoa, A. A., Iraporda, C., Garrote, G. L., and Abraham, A. G. (2019b). Kefir micro-organisms: their role in grain assembly and health properties of fermented milk. J. Appl. Microbiol. 126, 686–700. doi: 10.1111/jam.14107
Bengoa, A. A., Llamas, M. G., Iraporda, C., Dueñas, M. T., Abraham, A. G., and Garrote, G. L. (2018a). Impact of growth temperature on exopolysaccharide production and probiotic properties of Lactobacillus paracasei strains isolated from kefir grains. Food Microbiol. 69, 212–218. doi: 10.1016/j.fm.2017.08.012
Bengoa, A. A., Zavala, L., Carasi, P., Trejo, S. A., Bronsoms, S., Serradell, M., et al. (2018b). Simulated gastrointestinal conditions increase adhesion ability of Lactobacillus paracasei strains isolated from kefir to Caco-2 cells and mucin. Food Res. Int. 103, 462–467. doi: 10.1016/j.foodres.2017.09.093
Bradford, M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. doi: 10.1006/abio.1976.9999
Braegger, C., Chmielewska, A., Decsi, T., Kolacek, S., Mihatsch, W., Moreno, L., et al. (2011). Supplementation of infant formula with probiotics and/or prebiotics: a systematic review and comment by the ESPGHAN committee on nutrition. J. Pediatr. Gastroenterol. Nutr. 52, 238–250. doi: 10.1097/MPG.0b013e3181fb9e80
Cani, P. D. (2018). Human gut microbiome: hopes, threats and promises. Gut 67, 1716–1725. doi: 10.1136/gutjnl-2018-316723
Cani, P. D., Bibiloni, R., Knauf, C., Neyrinck, A. M., and Delzenne, N. M. (2008). Changes in gut microbiota control metabolic diet-induced obesity and diabetes in mice. Diabetes Metab. Res. Rev. 57, 1470–1481. doi: 10.2337/db07-1403
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Lozupone, C. A., Turnbaugh, P. J., et al. (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. U.S.A. 108, 4516–4522. doi: 10.1073/pnas.1000080107
Chambers, E. S., Viardot, A., Psichas, A., Morrison, D. J., Murphy, K. G., Zac-Varghese, S. E. K., et al. (2015). Effects of targeted delivery of propionate to the human colon on appetite regulation, body weight maintenance and adiposity in overweight adults. Gut 64, 1744–1754. doi: 10.1136/gutjnl-2014-307913
Conlon, M., and Bird, A. (2015). The impact of diet and lifestyle on gut microbiota and human health. Nutrients 7, 17–44. doi: 10.3390/nu7010017
de Goffau, M. C., Fuentes, S., van den Bogert, B., Honkanen, H., de Vos, W. M., Welling, G. W., et al. (2014). Aberrant gut microbiota composition at the onset of type 1 diabetes in young children. Diabetologia 57, 1569–1577. doi: 10.1007/s00125-014-3274-0
Durack, J., and Lynch, S. V. (2019). The gut microbiome: relationships with disease and opportunities for therapy. J. Exp. Med. 216, 20–40. doi: 10.1084/jem.20180448
Ewaschuk, J. B., and Dieleman, L. A. (2006). Probiotics and prebiotics in chronic inflammatory bowel diseases. World J. Gastroenterol. 12, 5941–5950. doi: 10.3748/wjg.v12.i37.5941
Gareau, M. G., Sherman, P. M., and Walker, W. A. (2010). Probiotics and the gut microbiota in intestinal health and disease. Nat. Rev. Gastroenterol. Hepatol. 7, 503–514. doi: 10.1038/nrgastro.2010.117
Garrote, G. L., Abraham, A. G., and De Antoni, G. L. (2010). “Microbial interactions in kefir: a natural probiotic drink,” in Biotechnology of Lactic Acid Bacteria, eds F. Mozzi, R. R. Raya, and G. M. Vignolo (Ames, IO: Wiley-Blackwell), 327–340.
Gibson, G. R., Hutkins, R., Sanders, M. E., Prescott, S. L., Reimer, R. A., Salminen, S. J., et al. (2017). Expert consensus document: the international scientific association for probiotics and prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat. Rev. Gastroenterol. Hepatol. 14, 491–502. doi: 10.1038/nrgastro.2017.75
Gonçalves, P., Araújo, J. R., and Di Santo, J. P. (2018). A cross-talk between microbiota-derived short-chain fatty acids and the host mucosal immune system regulates intestinal homeostasis and inflammatory bowel disease. Inflamm. Bowel Dis. 24, 558–572. doi: 10.1093/ibd/izx029
Hamet, M. F., Londero, A., Medrano, M., Vercammen, E., Van Hoorde, K., Garrote, G. L., et al. (2013). Application of culture-dependent and culture-independent methods for the identification of Lactobacillus kefiranofaciens in microbial consortia present in kefir grains. Food Microbiol. 36, 327–334. doi: 10.1016/j.fm.2013.06.022
Hamet, M. F., Medrano, M., Pérez, P. F., and Abraham, A. G. (2016). Oral administration of kefiran exerts a bifidogenic effect on BALB/c mice intestinal microbiota. Benef. Microb. 7, 237–246. doi: 10.3920/BM2015.0103
Hamet, M. F., Piermaria, J. A., and Abraham, A. G. (2015). Selection of EPS-producing Lactobacillus strains isolated from kefir grains and rheological characterization of the fermented milks. LWT Food Sci. Technol. 63, 129–135. doi: 10.1016/j.lwt.2015.03.097
Hollister, E. B., Gao, C., and Versalovic, J. (2014). Compositional and functional features of the gastrointestinal microbiome and their effects on human health. Gastroenterology 146, 1449–1458. doi: 10.1053/j.gastro.2014.01.052
Hugon, P., Dufour, J.-C., Colson, P., Fournier, P.-E., Sallah, K., and Raoult, D. (2015). A comprehensive repertoire of prokaryotic species identifi ed in human beings. Lancet Infect. Dis. 15, 1211–1230. doi: 10.1016/S1473-3099(15)00293-5
Jumas-Bilak, E., Carlier, J. P., Jean-Pierre, H., Mory, F., Teyssier, C., Gay, B., et al. (2007). Acidaminococcus intestini sp. nov., isolated from human clinical samples. Int. J. Syst. Evol. Microbiol. 57, 2314–2319. doi: 10.1099/ijs.0.64883-0
Kasubuchi, M., Hasegawa, S., Hiramatsu, T., Ichimura, A., and Kimura, I. (2015). Dietary gut microbial metabolites, short-chain fatty acids, and host metabolic regulation. Nutrients 7, 2839–2849. doi: 10.3390/nu7042839
Koh, A., De Vadder, F., Kovatcheva-Datchary, P., and Bäckhed, F. (2016). From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell 165, 1332–1345. doi: 10.1016/j.cell.2016.05.041
Llamas-Arriba, M. G., Hernández-Alcántara, A. M., Yépez, A., Aznar, R., Dueñas, M. T., and López, P. (2019). Functional and nutritious beverages produced by lactic acid bacteria. Nutr. Beverag. 12, 419–465. doi: 10.1016/b978-0-12-816842-4.00012-5
Louis, P., and Flint, H. J. (2009). Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol. Lett. 294, 1–8. doi: 10.1111/j.1574-6968.2009.01514.x
Louis, P., and Flint, H. J. (2017). Formation of propionate and butyrate by the human colonic microbiota. Environ. Microbiol. 19, 29–41. doi: 10.1111/1462-2920.13589
Louis, P., Hold, G. L., and Flint, H. J. (2014). The gut microbiota, bacterial metabolites and colorectal cancer. Nat. Rev. Microbiol. 12, 661–672. doi: 10.1038/nrmicro3344
Lynch, K. M., Zannini, E., Coffey, A., and Arendt, E. K. (2018). Lactic acid bacteria exopolysaccharides in foods and beverages: isolation, properties, characterization, and health benefits. Annu. Rev. Food Sci. Technol. 9, 155–176.
Milani, C., Duranti, S., Bottacini, F., Casey, E., Turroni, F., Mahony, J., et al. (2017). The first microbial colonizers of the human gut: composition, activities, and health implications of the infant gut microbiota. Microbiol. Mol. Biol. Rev. 81:e0036-17. doi: 10.1128/mmbr.00036-17
Miqdady, M., Al Mistarihi, J., Azaz, A., and Rawat, D. (2020). Prebiotics in the infant microbiome: the past, present, and future. Pediatr. Gastroenterol. Hepatol. Nutr. 23, 1–14. doi: 10.5223/pghn.2020.23.1.1
Morrison, D. J., and Preston, T. (2016). Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microb. 7, 189–200. doi: 10.1080/19490976.2015.1134082
Offermanns, S. (2014). Free fatty acid (FFA) and hydroxy carboxylic acid (HCA) receptors. Annu. Rev. Pharmacol. Toxicol. 54, 407–434. doi: 10.1146/annurev-pharmtox-011613-135945
Parker, A., Fonseca, S., and Carding, S. R. (2020). Gut microbes and metabolites as modulators of blood-brain barrier integrity and brain health. Gut Microb. 11, 135–157. doi: 10.1080/19490976.2019.1638722
Pédron, T., Mulet, C., Dauga, C., Frangeul, L., Chervaux, C., Grompone, G., et al. (2012). A crypt-specific core microbiota resides in the mouse colon. mBio 3:e00116-12. doi: 10.1128/mBio.00116-12
Piermaria, J. A., de la Canal, M. L., and Abraham, A. G. (2008). Gelling properties of kefiran, a food-grade polysaccharide obtained from kefir grain. Food Hydrocoll. 22, 1520–1527. doi: 10.1016/j.foodhyd.2007.10.005
Plé, C., Breton, J., Daniel, C., and Foligné, B. (2015). Maintaining gut ecosystems for health: are transitory food bugs stowaways or part of the crew? Int. J. Food Microbiol. 213, 139–143. doi: 10.1016/j.ijfoodmicro.2015.03.015
Qin, J., Li, Y., Cai, Z., Li, S., Zhu, J., Zhang, F., et al. (2012). A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 490, 55–60. doi: 10.1038/nature11450
Requena, T., Martínez-Cuesta, M. C., and Peláez, C. (2018). Diet and microbiota linked in health and disease. Food Funct. 9, 688–704.
Rezac, S., Kok, C. R., Heermann, M., and Hutkins, R. (2018). Fermented foods as a dietary source of live organisms. Front. Microbiol. 9:1785. doi: 10.3389/fmicb.2018.01785
Ricaboni, D., Mailhe, M., Benezech, A., Cadoret, F., Fournier, P. E., and Raoult, D. (2017). ‘Acidaminococcus timonensis’ sp. nov. and ‘Acidaminococcus massiliensis’ sp. nov. isolated from human gut. New Microb. New Infect. 15, 46–48. doi: 10.1016/j.nmni.2016.11.010
Rimada, P. S., and Abraham, A. G. (2003). Comparative study of different methodologies to determine the exopolysaccharide produced by kefir grains in milk and whey. Lait 83, 79–87. doi: 10.1051/lait:2002051
Ríos-Covián, D., Ruas-Madiedo, P., Margolles, A., Gueimonde, M., De los Reyes-Gavilán, C. G., and Salazar, N. (2016). Intestinal short chain fatty acids and their link with diet and human health. Front. Microbiol. 7:185. doi: 10.3389/fmicb.2016.00185
Roberfroid, M., Gibson, G. R., Hoyles, L., McCartney, A. L., Rastall, R., Rowland, I., et al. (2010). Prebiotic effects: metabolic and health benefits. Br. J. Nutr. 104, S1–S63. doi: 10.1017/S0007114510003363
Roduit, C., Frei, R., Ferstl, R., Loeliger, S., Westermann, P., Rhyner, C., et al. (2019). High levels of butyrate and propionate in early life are associated with protection against atopy. Allergy 74, 799–809. doi: 10.1111/all.13660
Round, J. L., and Mazmanian, S. K. (2009). The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 9, 313–323. doi: 10.1038/nri2515
Ruas-Madiedo, P., Abraham, A. G., Mozzi, F., and de los Reyes-Gavilán, C. G. (2008). “Functionality of exopolysaccharides produced by lactic acid bacteria,” in Molecular Aspects of Lactic Acid Bacteria for Traditional and New Applications, eds B. Mayo, P. López, and G. Pérez-Martínez (Kerala: Research Signpost), 137–166.
Ryan, P. M., Ross, R. P., Fitzgerald, G. F., Caplice, N. M., and Stanton, C. (2015). Sugar-coated: Exopolysaccharide producing lactic acid bacteria for food and human health applications. Food Funct. 6, 679–693. doi: 10.1039/c4fo00529e
Salazar, N., Binetti, A., Gueimonde, M., Alonso, A., Garrido, P., González del Rey, C., et al. (2011). Safety and intestinal microbiota modulation by the exopolysaccharide-producing strains Bifidobacterium animalis IPLA R1 and Bifidobacterium longum IPLA E44 orally administered to Wistar rats. Int. J. Food Microbiol. 144, 342–351. doi: 10.1016/j.ijfoodmicro.2010.10.016
Salazar, N., Gueimonde, M., Gonzalez de los Reyes-Gavilán, C., and Ruas-Madiedo, P. (2016). Exopolysaccharides produced by lactic acid bacteria and Bifidobacteria as fermentable substrates by the intestinal microbiota. Crit. Rev. Food Sci. Nutr. 56, 1440–1453. doi: 10.1080/10408398.2013.770728
Salazar, N., Gueimonde, M., Hernández-Barranco, A. M., Ruas-Madiedo, P., and Gonzalez de los Reyes-Gavilán, C. (2008). Exopolysaccharides produced by intestinal Bifidobacterium strains act as fermentable substrates for human intestinal bacteria. Appl. Environ. Microbiol. 74, 4737–4745. doi: 10.1128/AEM.00325-08
Samb-Ba, B., Mazenot, C., Gassama-Sow, A., Gory Dubourg, G., Richet, H., Hugon, P., et al. (2014). MALDI-TOF identification of the human gut microbiome in people with and without diarrhea in senegal. PLoS One 9:e87419. doi: 10.1371/journal.pone.0087419
Segata, N., Haake, S. K., Mannon, P., Lemon, K. P., Waldron, L., Gevers, D., et al. (2012). Composition of the adult digestive tract bacterial microbiome based on seven mouth surfaces, tonsils, throat and stool samples. Genome Biol. 13, 1–18. doi: 10.1186/gb-2012-13-6-r42
Shen, S., and Wong, C. H. (2016). Bugging inflammation: role of the gut microbiota. Clin. Transl. Immunol. 5:e72. doi: 10.1038/cti.2016.12
Thorburn, A. N., Macia, L., and Mackay, C. R. (2014). Diet, metabolites, and “Western-Lifestyle” inflammatory diseases. Immunity 40, 833–842. doi: 10.1016/j.immuni.2014.05.014
Thursby, E., and Juge, N. (2017). Introduction to the human gut microbiota. Biochem. J. 474, 1823–1836. doi: 10.1042/BCJ20160510
Tsai, Y.-L., Lin, T.-L., Chang, C.-J., Wu, T.-R., Lai, W.-F., Lu, C.-C., et al. (2019). Probiotics, prebiotics and amelioration of diseases. J. Biomed. Sci. 26:3. doi: 10.1186/s12929-018-0493-6
Uchiyama, K., Naito, Y., and Takagi, T. (2019). Intestinal microbiome as a novel therapeutic target for local and systemic inflammation. Pharmacol. Ther. 199, 164–172. doi: 10.1016/j.pharmthera.2019.03.006
van der Beek, C. M., Bloemen, J. G., van den Broek, M. A., Lenaerts, K., Venema, K., Buurman, W. A., et al. (2015). Hepatic uptake of rectally administered butyrate prevents an increase in systemic butyrate concentrations in humans. J. Nutr. 145, 2019–2024. doi: 10.3945/jn.115.211193
Venema, K., Verhoeven, J., Verbruggen, S., and Keller, D. (2020). Xylo-oligosaccharides from sugarcane show prebiotic potential in a dynamic computer-controlled in vitro model of the adult human large intestine. Benef. Microb. 11, 191–200. doi: 10.3920/BM2019.0159
Verkhnyatskaya, S., Ferrari, M., De Vos, P., and Walvoort, M. T. C. (2019). Shaping the infant microbiome with non-digestible carbohydrates. Front. Microbiol. 10:343. doi: 10.3389/fmicb.2019.00343
Vinolo, M. A. R., Rodrigues, H. G., Hatanaka, E., Sato, F. T., Sampaio, S. C., and Curi, R. (2011). Suppressive effect of short-chain fatty acids on production of proinflammatory mediators by neutrophils. J. Nutr. Biochem. 22, 849–855. doi: 10.1016/j.jnutbio.2010.07.009
Wauters, G., De Baere, T., Willems, A., Falsen, E., and Vaneechoutte, M. (2003). Description of Comamonas aquatica comb. nov. and Comamonas kerstersii sp. nov. for two subgroups of Comamonas terrigena and emended description of Comamonas terrigena. Int. J. Syst. Evol. Microbiol. 53, 859–862. doi: 10.1099/ijs.0.02450-0
Weiss, G. A., and Hennet, T. (2017). Mechanisms and consequences of intestinal dysbiosis. Cell. Mol. Life Sci. 74, 2959–2977. doi: 10.1007/s00018-017-2509-x
Yin, X., Yan, Y., Kim, E. B., Lee, B., and Marco, M. L. (2014). Short communication: effect of milk and milk containing Lactobacillus casei on the intestinal microbiota of mice. J. Dairy Sci. 97, 2049–2055. doi: 10.3168/jds.2013-7477
Zavala, L., Golowczyc, M. A., Van Hoorde, K., Medrano, M., Huys, G., Vandamme, P., et al. (2016). Selected Lactobacillus strains isolated from sugary and milk kefir reduce Salmonella infection of epithelial cells in vitro. Benef. Microb. 7, 585–595. doi: 10.3920/BM2015.0196
Keywords: prebiotics, probiotics, microbiota, short chain fatty acids, exopolysaccharide, lactic acid bacteria
Citation: Bengoa AA, Dardis C, Gagliarini N, Garrote GL and Abraham AG (2020) Exopolysaccharides From Lactobacillus paracasei Isolated From Kefir as Potential Bioactive Compounds for Microbiota Modulation. Front. Microbiol. 11:583254. doi: 10.3389/fmicb.2020.583254
Received: 14 July 2020; Accepted: 23 September 2020;
Published: 16 October 2020.
Edited by:
Katia Sivieri, São Paulo State University, BrazilReviewed by:
Alex Galanis, Democritus University of Thrace, GreeceFabien Magne, University of Chile, Chile
Copyright © 2020 Bengoa, Dardis, Gagliarini, Garrote and Abraham. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Analía G. Abraham, aga@biol.unlp.edu.ar
 Ana Agustina Bengoa
Ana Agustina Bengoa Carolina Dardis
Carolina Dardis Nina Gagliarini
Nina Gagliarini Graciela L. Garrote
Graciela L. Garrote Analía G. Abraham
Analía G. Abraham