- 1Department of Life Sciences, University of Modena and Reggio Emilia, Modena, Italy
- 2Nanyang Technological University Food Technology Centre (NAFTEC), Singapore, Singapore
- 3School of Chemical and Biomedical Engineering, Nanyang Technological University, Singapore, Singapore
- 4Biogest-Siteia, University of Modena and Reggio Emilia, Modena, Italy
Leuconostoc carnosum is a known colonizer of meat-related food matrices. It reaches remarkably high loads during the shelf life in packaged meat products and plays a role in spoilage, although preservative effects have been proposed for some strains. In this study, the draft genomes of 17 strains of L. carnosum (i.e., all the strains that have been sequenced so far) were compared to decipher their metabolic and functional potential and to determine their role in food transformations. Genome comparison and pathway reconstruction indicated that L. carnosum is a compact group of closely related heterofermentative bacteria sharing most of the metabolic features. Adaptation to a nitrogen-rich environment, such as meat, is evidenced by 23 peptidase genes identified in the core genome and by the autotrophy for nitrogen compounds including several amino acids, vitamins, and cofactors. Genes encoding the decarboxylases yielding biogenic amines were not present. All the strains harbored 1–4 of 32 different plasmids, bearing functions associated to proteins hydrolysis, transport of amino acids and oligopeptides, exopolysaccharides, and various resistances (e.g., to environmental stresses, bacteriophages, and heavy metals). Functions associated to bacteriocin synthesis, secretion, and immunity were also found in plasmids. While genes for lactococcin were found in most plasmids, only three harbored the genes for leucocin B, a class IIa antilisterial bacteriocin. Determinants of antibiotic resistances were absent in both plasmids and chromosomes.
Introduction
Leuconostoc is a genus of heterofermentative Lactobacillaceae (Zheng et al., 2020). The general traits of Leuconostoc species include facultative anaerobiosis, intrinsic vancomycin resistance, catalase negativity, ovococcoid morphology, and dextran production. At the time of writing, the genus Leuconostoc encompassed at least 20 recognized species or subspecies, mostly isolated from plant materials, dairy products, meats, and fermented vegetables (Jeon et al., 2017; Chen et al., 2020). Among them, Leuconostoc carnosum is a known colonizer of food matrices, particularly the meat-related ones (Shaw and Harding, 1989; van Laack et al., 1992; Parente et al., 1996; Budde et al., 2003; Geeraerts et al., 2018; Raimondi et al., 2018, 2019).
Leuconostoc carnosum is reported to grow during the first days of the shelf-life in packaged meat products, which may range up to some weeks. The charge of contaminant bacteria, often dominated by L. carnosum, tends to increase and peaks up to 108 cells/g in some foods (Raimondi et al., 2018, 2019). These remarkably high levels of L. carnosum contamination were observed in modified atmosphere packaged (MAP) cooked ham, where it prevailed over the microbiota well before the expiry date (Björkroth et al., 1998; Vasilopoulos et al., 2010; Raimondi et al., 2019); in other meat products, such as sausages, vacuum-packaged smoked bacon, and sliced cooked poultry (Geeraerts et al., 2018; Li et al., 2019); and in processed vegetables (Hong et al., 2014; Jung et al., 2014). As a result, a high load of living L. carnosum may be ingested with certain foods, including many popular ready-to-eat products for which the consumption is steadily increasing all over in the world. Nonetheless, any potential impact of L. carnosum on human health has never been assessed.
The high moisture and the low salt content, the near-neutral pH, and the availability of nutrients of meat-related products encourage microbial growth, eventually leading to spoilage. With respect to food wholesomeness, the role of L. carnosum is controversial. Many studies describe it as a spoilage bacterium that causes meat deterioration and affects sensorial properties by souring, discoloration, gas production, and slime formation (Shaw and Harding, 1989; Björkroth et al., 1998; Samelis et al., 2006; Raimondi et al., 2019). On the other hand, likewise other Leuconostoc species, L. carnosum yields organic acid and hydrogen peroxide, which exert an intrinsic antimicrobial effect, and can produce class II heat-stable bacteriocins (van Laack et al., 1992; Felix et al., 1994; Hastings et al., 1994; Stiles, 1994). Typical bacteriocins produced by L. carnosum are leucocins, consisting of small peptides of 30–50 residues with differences due to the length of the prepeptide sequence. Their genetic information generally resides in plasmids but can occur also in the chromosome. Most of leucocins are active against both Listeria monocytogenes and Listeria innocua, but can also inhibit some lactic acid bacteria, especially at low pH (Felix et al., 1994; Parente et al., 1996; Wan et al., 2015).
Contamination of meat products with L. carnosum may restrict the growth of spoiling and pathogenic bacteria (Jacobsen et al., 2003; Lawton et al., 2020). As a matter of fact, the capability of L. carnosum to deteriorate meat products seems to be a strain-specific feature. A survey of MAP cooked ham samples from different producers revealed that, at the end of the shelf life, L. carnosum dominated the microbiota of both the good and the spoiled ones (Raimondi et al., 2019). These evidences opened the perspective to develop protective starters with specifically selected strains of L. carnosum that may preserve meat without impacting on the sensorial properties (Budde et al., 2003).
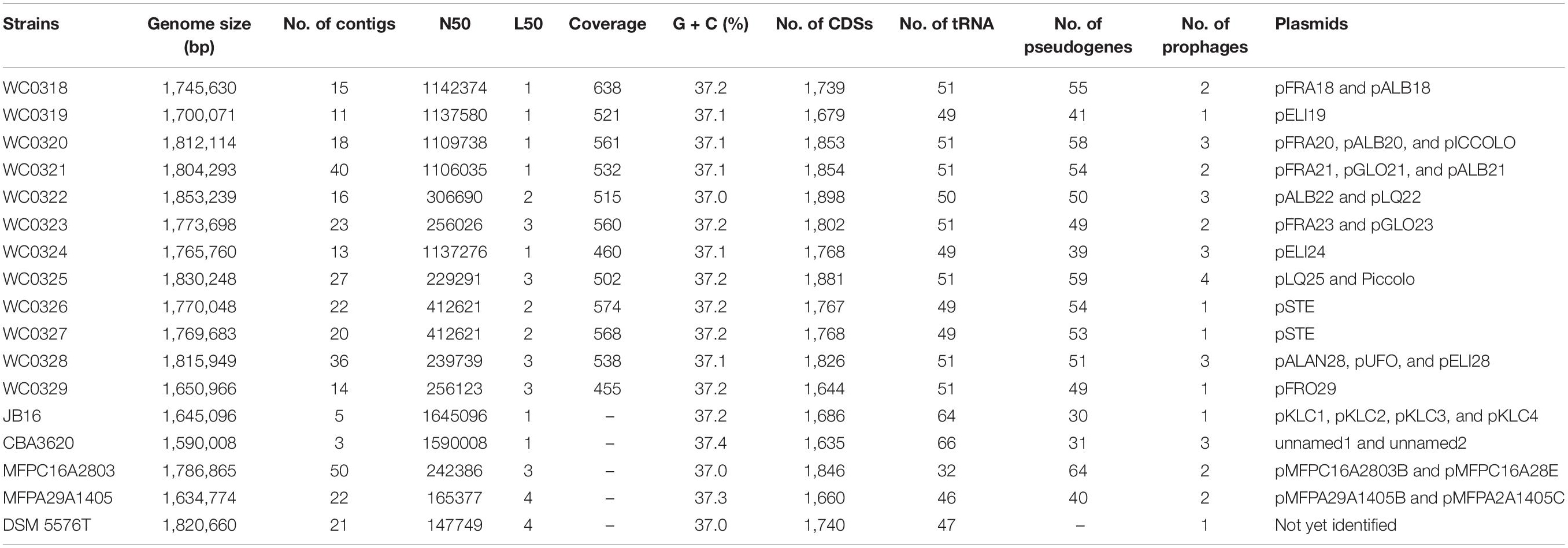
The dual role of L. carnosum in spoilage and preservation, the high loads of these bacteria entering the human diet and possibly affecting health, and the lack of studies aimed to highlight strain to strain differences prompted us to explore more deeply in this species. In the present study, comparative genomics was performed to explore the main genetic determinants of all the 17 strains of L. carnosum that have been sequenced so far. Twelve strains were recently isolated from meat matrices and newly sequenced (Candeliere et al., 2020), whereas five had a genome already publicly available (Table 1; Jung et al., 2012). Using the dataset of the 17 chromosomal sequences, a comparative genome analysis of the L. carnosum taxon was undertaken through the assessment of the phylogeny and of pan- and core genome. Genetic diversity, plasmids, phages, Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR-Cas) systems, bacteriocin, antibiotic resistances, and metabolic capabilities were also investigated.
Materials and Methods
Genomes and Genomes Analysis
The 12 genomes of L. carnosum strains published by Candeliere et al. (2020) are available with the following GenBank accessions: SAMN11618753, SAMN11618754, SAMN11618755, SAMN11 618756, SAMN11618757, SAMN11618758, SAMN11618759, SAMN11618761, SAMN11618763, SAMN11618764, SAMN11618767, and SAMN11618768. The genomes of the other L. carnosum strains, i.e., JB16, CBA3620, MFPC16A2803, MFPA29A1405, and DSM 5576T, are available with the accessions SAMN02603179, SAMN11843679, SAMEA104699786, SAMEA104699785, and SAMN14908560, respectively. The strains were isolated from different sources, mostly meat-related products: MAP cooked ham (WC0318, WC0319, WC0320, WC0321, WC0322, WC0323, WC0324, WC0325, and WC0329), MAP sausages (WC0326, WC0327, and WC0328), vacuum-packed beef carpaccio (MFPC16A2803), MAP beef carpaccio (MFPA29A1405), vacuum-packaged beef (DSM 5576T), and kimchi (JB16 and CB3620).
Plasmids contigs were identified using plasmidSPAdes v 3.12 (Nurk et al., 2017). Average Nucleotide Identity (ANI) and digital-DNA/DNA hybridization (dDDH) were calculated utilizing the web tools ANI Matrix1 and Genome-to-Genome Distance Calculator GGDC 2.12, by all-against-all approach (Meier-Kolthoff et al., 2013; Rodriguez-R and Konstantinidis, 2016). The thresholds for species demarcation were 95 and 70% for ANI and dDDH, respectively (Richter and Rosselló-Móra, 2009).
Gene clusters, such as prophages, CRISPR/Cas systems, insertion sequences (ISs), bacteriocins genes, and antimicrobial resistances, were identified with specific web tools. Prophage sequences were searched with PHASTER3 (Arndt et al., 2016). CRISPRs and Cas genes were searched utilizing CRISPRCasFinder4 (Couvin et al., 2018), with default settings and subtype clustering of Cas genes. Bacteriocins genes were searched with BAGEL 4 server5 (van Heel et al., 2018). ISs were identified with ISfinder6) (Siguier et al., 2006). Antimicrobial resistance was assessed by Resistance Gene Identifier (RGI) tool of Comprehensive Antibiotic Resistance Database (CARD), processing the contigs file for “Perfect, Strict, and Loose hits7” (Alcock et al., 2020).
Genomes Annotation and Functional Characterization
Genomes were annotated with Prokka, utilizing default parameters (Seemann, 2014). Roary was utilized to calculate the pangenome and to delineate core, soft core, accessory, shell, and cloud genes, utilizing Prokka annotation files (Page et al., 2015). To differentiate plasmids and chromosomal accessory genes and highlight the plasmid contribution, Roary was used to calculate the pangenome with and without the plasmid contigs. COG annotation was conducted utilizing WebMGA server8 (Wu et al., 2011), using as input the protein files predicted by Prokka. A phylogenetic tree based on core genome alignment was constructed with FastTree (Price et al., 2009) utilizing the alignment file produced by Roary and was visualized with iTOL (Letunic and Bork, 2019).
To investigate the similarity among genomes and among plasmids, Jaccard’s distance was computed based on the presence/absence of predicted genes and subjected to Principal Coordinate Analysis (PCoA).
The functional prediction of the genomes was carried out with the KEGG tools BlastKOALA9 and Mapper10 with the aim to predict the metabolic functions, such as transporters, sugars catabolism, and biosynthetic pathways of amino acids, vitamins, and bases (Kanehisa et al., 2016; Kanehisa and Sato, 2020).
The presence of specific genes that are not included in KEGG annotation was investigated with a BLASTp search of cognate proteins recognized in a closely related taxon (i.e., in L. carnosum, in the genus Leuconostoc, in other Lactobacillales, or in other bacteria). The following enzymes for citrate metabolism and acetoin pathway were searched (García-Quintáns et al., 2008): citrate permease (AAA60396) of Leuconostoc mesenteroides subsp. mesenteroides, citrate lyase alpha and beta subunits (CAA71633 and CAA71632) of L. mesenteroides subsp. cremoris, oxaloacetate decarboxylase (AFS39629) of Leuconostoc gelidum, alpha-acetolactate synthase (SPJ44178) of L. carnosum, alpha-acetolactate decarboxylase (AFT82058) of L. carnosum, diacetyl reductase (SPJ42929) of L. carnosum, and 2,3-butanediol dehydrogenase (WP_135197409) of L. carnosum. The genes encoding the carboxylases yielding biogenic amines were searched (Li et al., 2018; Rodrigo-Torres et al., 2019): tyrosine decarboxylase (AAN77279) and agmatine deaminase (ABS19476 and ABS19477) of Lactobacillus brevis (proposed Levilactobacillus brevis), ornithine decarboxylase (ANJ65946) of Lactobacillus rossiae (proposed Furfurilactobacillus rossiae), histidine decarboxylase (N877767) of Lactobacillus reuteri (proposed Limosilactobacillus reuteri), and lysine decarboxylase (NP_414728) of Escherichia coli. For menaquinone biosynthesis, the presence of 1,4-dihydroxy-2-naphthoyl-CoA hydrolase from L. mesenteroides subsp. mesenteroides ATCC 8293 (ABJ61187) was searched.
Results
General Genome Features
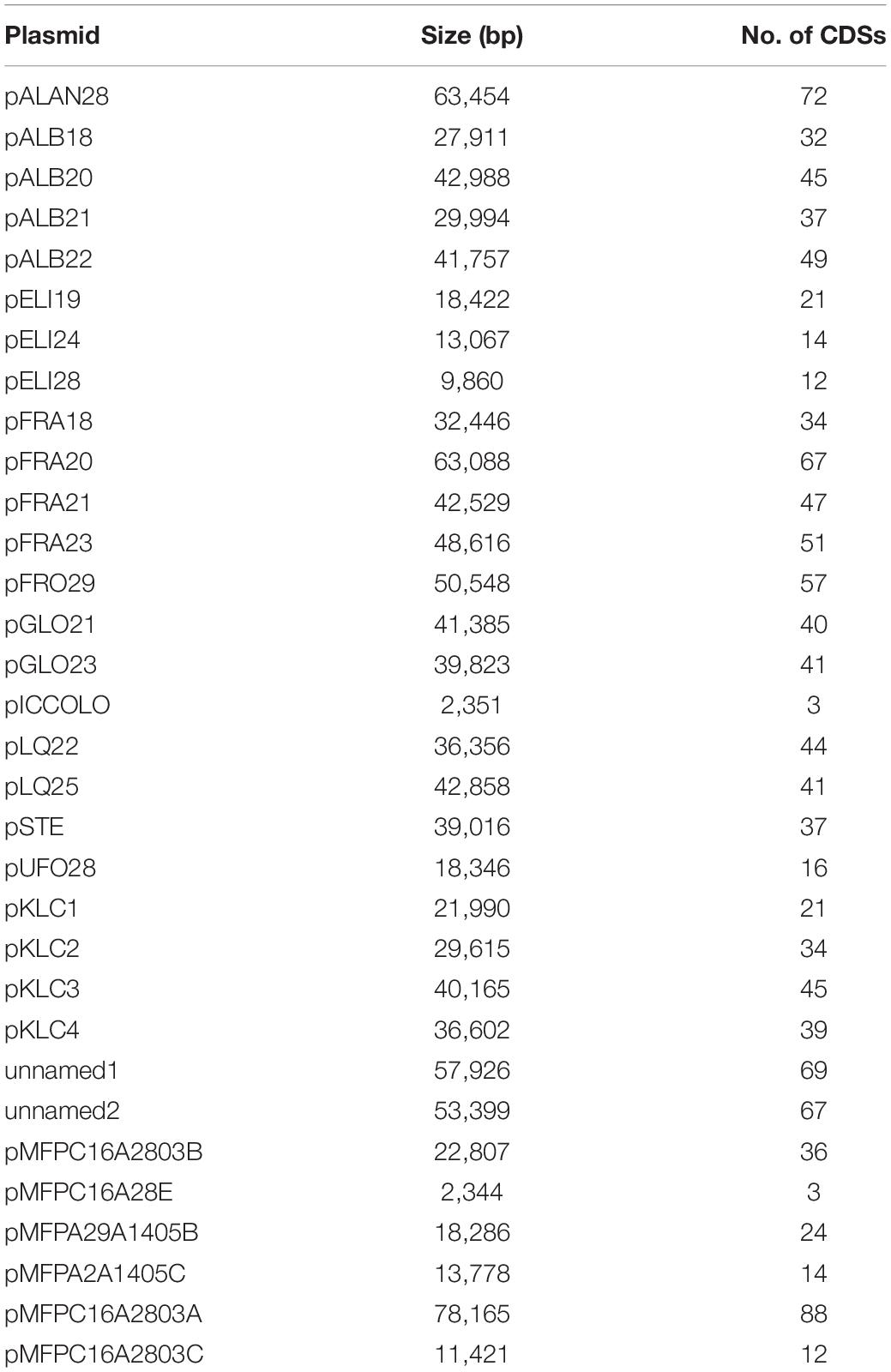
The 12 newly assembled genomes of L. carnosum had a mean size of 1.77 Mbp, laying in the range between 1.65 and 1.85 Mbp of strains L. carnosum WC0329 and L. carnosum WC0322, respectively, and a GC content of 37.0–37.2% (Table 1). The genomes of the five already published strains had a size of 1.59–1.79 Mbp and a GC content of 37.0–37.4% (Table 1). A > 455-fold coverage was obtained for all the 12 genomes, with a mean of 535.3-fold. The draft genomes encompassed from 11 to 40 contigs, on average 21 per genome. For all the assemblies, 1–3 contigs together constituted more than 50% of the genome size (L50 value, mean = 1.92; N50 value, mean = 645,510 bp) (Table 1). A total of 20 natural plasmids, ranging in size from 2.4 to 63.5 kbp, were identified in the new 12 draft genomes, 1–3 per strain (Tables 1, 2). Twelve other plasmids occurred in four already available genomes (Table 2), whereas the information was not available for L. carnosum DSM 5576T. For all the pairwise comparisons between 17 genomes, both ANI and dDDH were higher than the corresponding threshold for species demarcation (95 and 70%, respectively); thus, all the strains were confirmed to belong to the same species with only minor intra-species differences (Supplementary Table 1). L. carnosum WC326 and WC327 resulted in the same strain according to both ANI and dDDH.
The number of coding sequences (CDSs) in the 12 new genomes ranged between 1,644 and 1,898, with a mean of 1,790. Single plasmids harbored from 3 to 67 genes; thus, each draft genome included 14–124 plasmid genes, accounting for 0.8–6.7% of the CDS content. The number of CDSs in the five genomes already available ranged from 1,635 and 1,846, with plasmids of four strains harboring from 3 to 69 genes. All the CDSs in the 17 genomes were compared by a blast all-against-all approach to identify orthologous gene groups and construct pan- and core genome matrices. The pangenome of L. carnosum encompassed 3,221 orthologous genes, whereas the core genome included 1,383 chromosomal genes (on average 43% of the total genes). The accessory genome was composed of 689 shell genes (548 of which in the chromosome and 141 in plasmids) and 1,149 cloud genes (868 of which in the chromosome and 281 in plasmids) (Figure 1A). As per the Heap’s law, the value of γ was 0.35 for 17 genomes, and thus L. carnosum pangenome was considered open (Figure 1B; Tettelin et al., 2008).
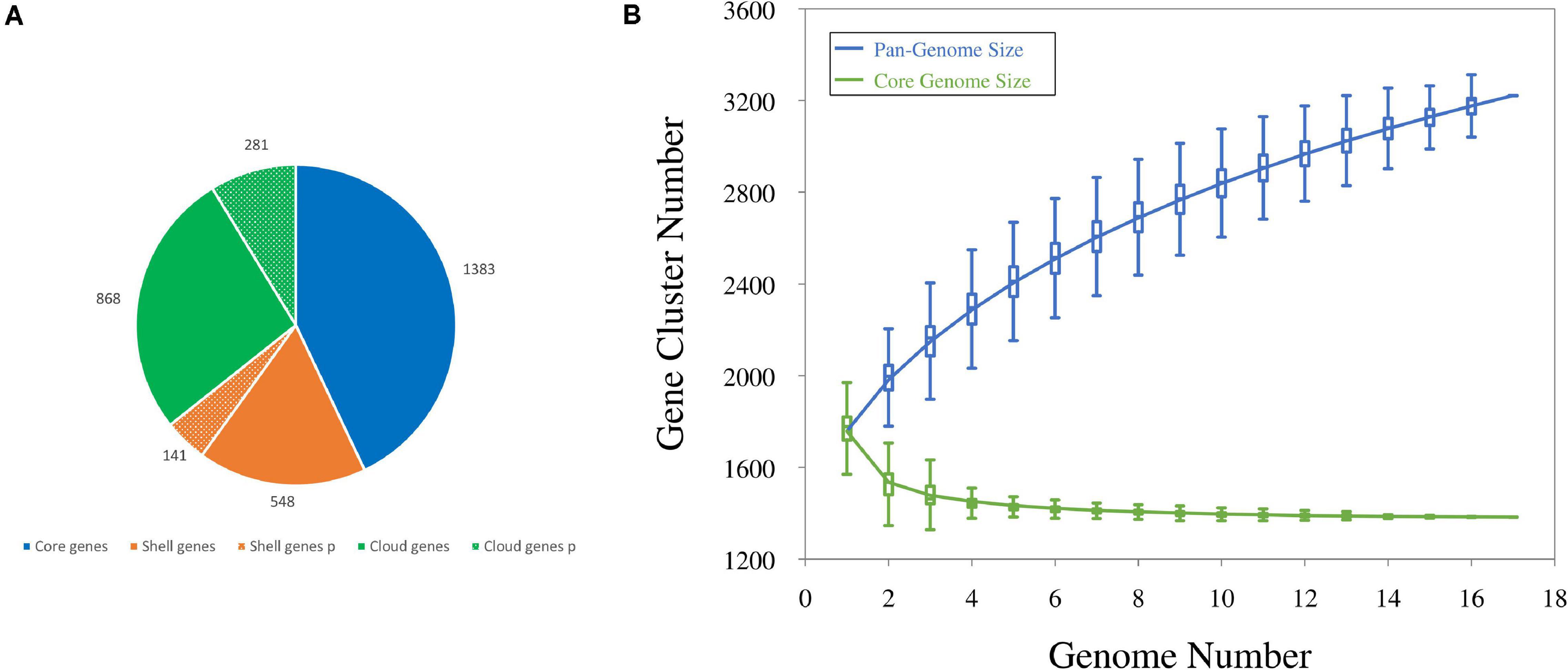
Figure 1. (A) Distributions of CDSs found in the pangenome of L. carnosum: core genes (blue), shell genes (orange), and cloud genes (green) in chromosome (plain colors) and plasmids (dotted colors). (B) Estimation of the pangenome (blue) and the core genome (green) by including genomes one by one.
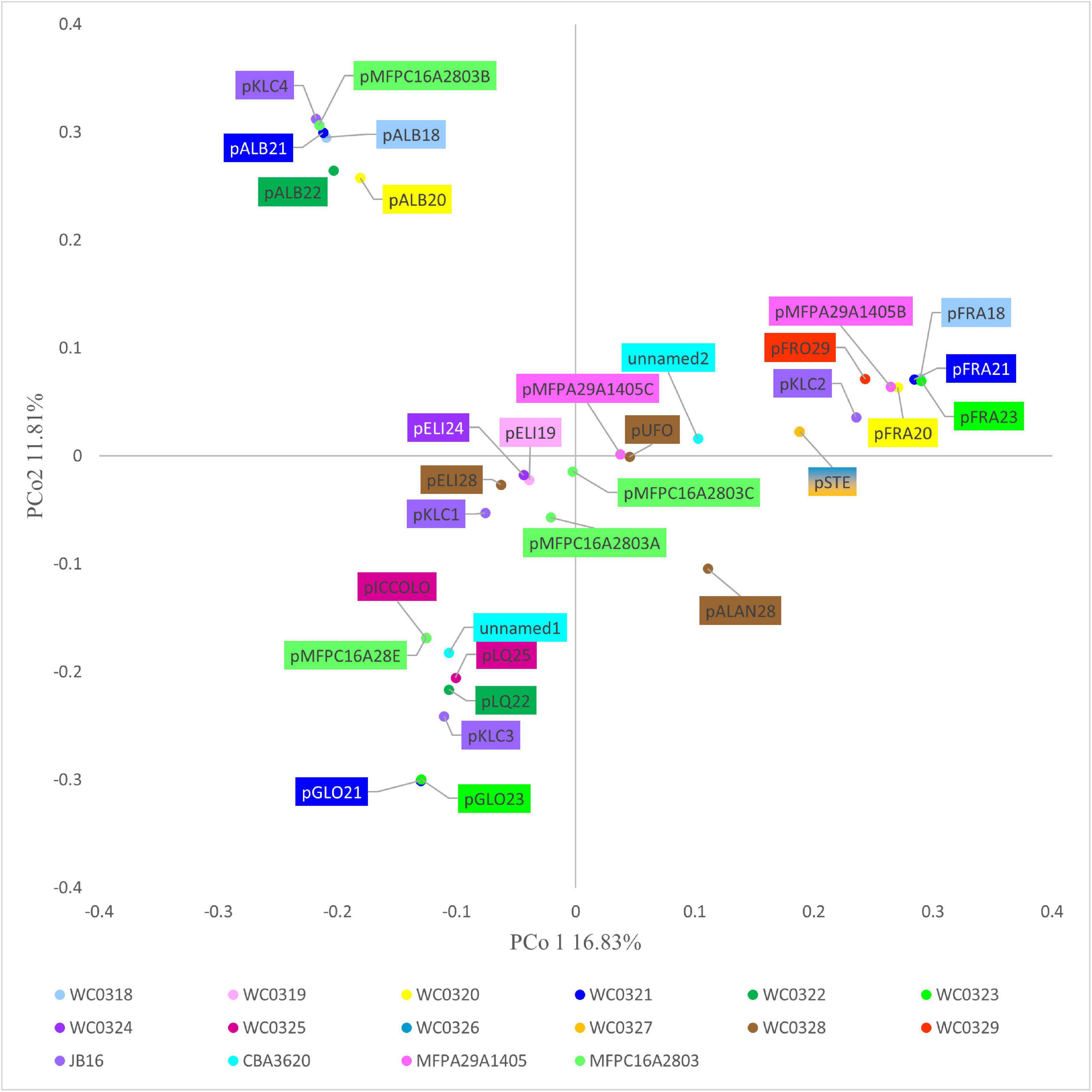
The Jaccard distance matrix among genomes, due to the presence/absence of the genes, is displayed in the PCoA plot of Figure 2A. Five genomes laying at negative values of PCo1 (i.e., WC0319, WC0324, WC0328, JB16, and CBA3620) were separated from the others dispersed at positive values of PCoA1, among which three lay at positive values of PCo2 (WC0318, WC0326, and WC0327), and the others grouped together at lower values. The phylogenetic relationship between the 17 strains was constructed using relative hierarchical clustering based on core genome alignment (Figure 2B). The low phylogenetic distances confirmed a close relatedness among the L. carnosum strains, even though sequence differences within the core genome revealed three major clades. All the genomes that lay at positive PCo1 values were in the same phylogenetic clade, whereas the five genomes that lay at negative PCo1 values were clearly separated in two different clades.
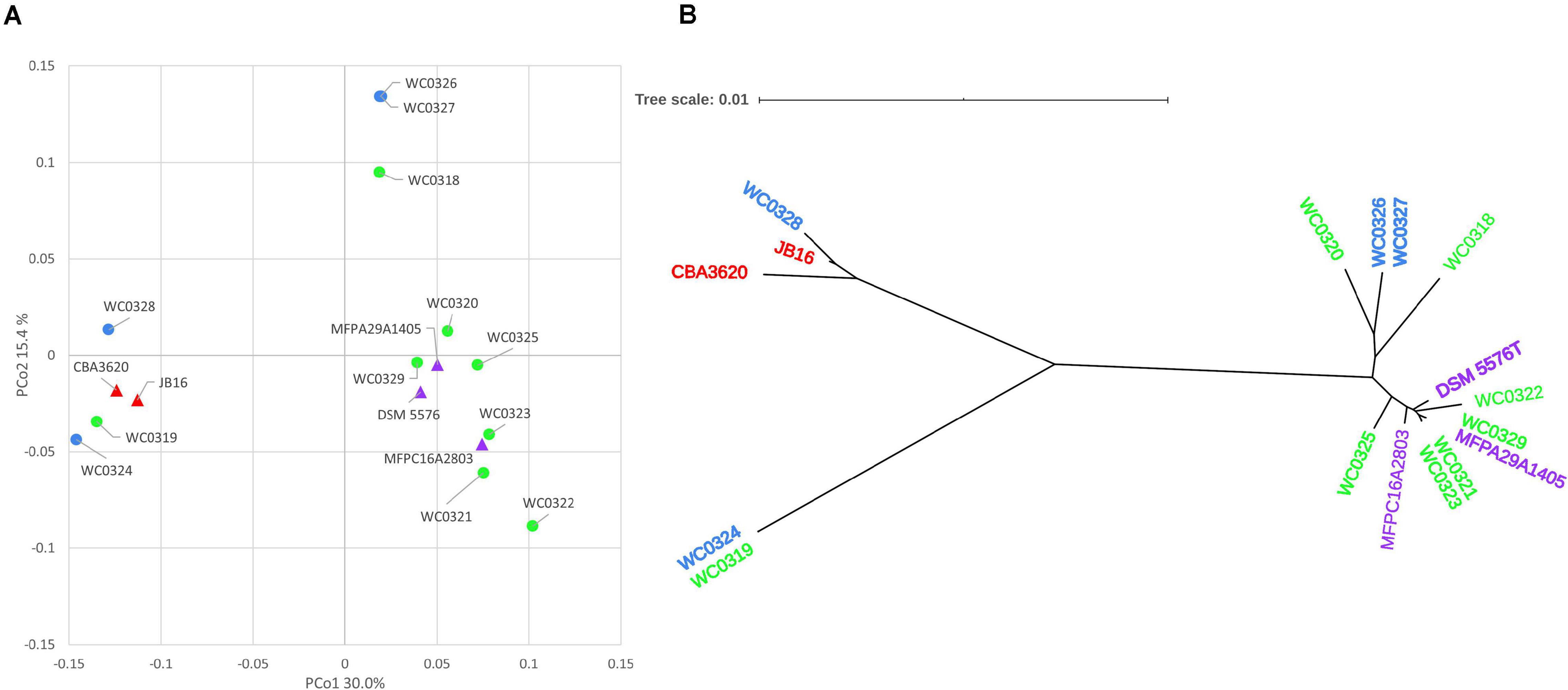
Figure 2. (A) PCoA visualization of Jaccard distances among genomes, based on shared genes. Circles are the genomes published by Candeliere et al. (2020); triangles are the genomes already published. (B) Neighbor joining unrooted phylogenetic tree based on core genome alignment. In both panels, colors indicate the source of the strains: MAP cooked ham, green; MAP sausage, blue; packaged beef products, purple; and kimchi, red.
A comparative analysis of chromosomal sequences was carried out for all the genomes. The 1,383 genes of the core genome were ascribed to 24 COG families (Figure 3). Functional distribution of the COGs of the core showed that the majority encoded components of the information processing systems associated to translation, ribosomal structure, and biogenesis (13.1%) and of amino acid transport and metabolism (10.1%). The core also included 87 genes of function unknown (6.1%) and 114 genes with only a general prediction of biochemical activity (8.0%).
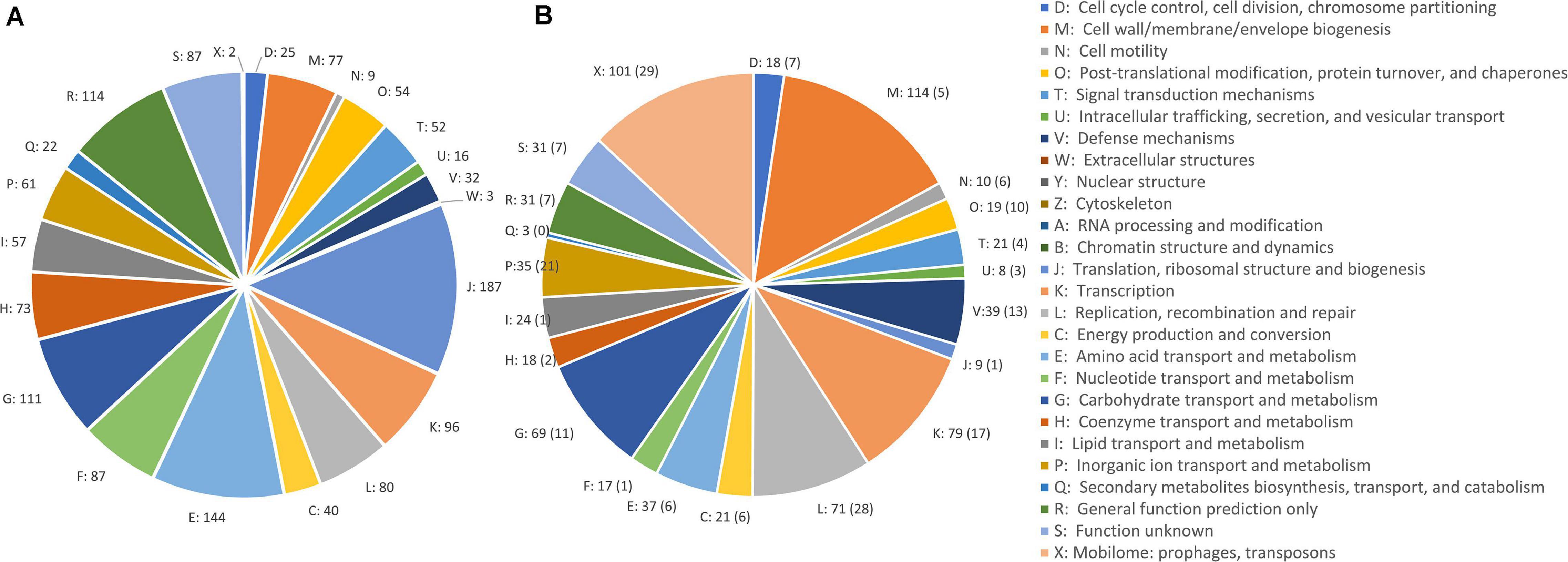
Figure 3. COG class abundances in core genome (A) and in the accessory genome (B). The number of CDSs predicted within each class is reported. The CDSs in plasmids are in brackets.
Mobilome
The chromosome sequences were analyzed for the presence of mobile elements, such as genes putatively encoding ISs and transposases. Twenty-nine genes encoding transposases were identified in the accessory genome, 20 of them as part of IS. Except for L. carnosum MFPA29A1405 that did not harbor any, all the strains harbored at least one IS. The ISs belonged to the families IS3, IS5, IS6, IS30, IS256, and IS1182. The IS presented similarities with counterparts of other Lactobacillales: IS3, Lactobacillus sakei (proposed Latilactobacillus sakei by Zheng et al., 2020) and Weissella cibaria; IS5 and IS1182, Lactobacillus plantarum (proposed Lactiplantibacillus plantarum by Zheng et al., 2020); IS6, Enterococcus faecium and L. mesenteroides; IS30, L. plantarum, Leuconostoc lactis, and Pediococcus pentosaceus; and IS256, Enterococcus hirae and Lactobacillus helveticus. The most represented were IS3 and IS30 transposases (seven and six genes, respectively).
Plasmids
The 32 plasmids were annotated (Supplementary Datasheet 1). The Jaccard distance matrix among plasmids, due to the presence of shared genes and predicted functions, is displayed in the PCoA plot of Figure 4.
pALB plasmids, pKLC4, and pMFPC16A2803B harbored genes encoding all the functions for conjugative transfer, including mating pair formation and DNA replication, mobilization, and transfer (Supplementary Datasheet 1). The series pGLO and pLQ and the plasmids pMFPA29A1405B, pMFPC16A28E, and pKLC1 had some conjugation genes, but lacked the genes encoding mobilization proteins (relaxases), resulting not mobilizable. The series pELI and pSTE, pFRO29, pALAN28, pUFO, pKLC2, pKLC3, pMFPC16A2803A, pMFPC16A2803C, pMFPA2A1405C, unnamed1, and unnamed2 were mobilizable but not conjugative plasmids, for the presence of 2–4 mobility-related genes.
pICCOLO and pMFPC16A28E were likely the same small plasmid of approximately predicted 2,350 bp, occurring in the strains L. carnosum WC0320, WC0325, and MFPC16A2803. They showed a very high identity (>97.8%) with the small plasmid pM411 (2,303 bp) of L. plantarum, replicating via the rolling circle mechanism. They encoded an N-acetyltransferase and the rolling circle initiator protein RepB. Genes encoding RepB proteins occurred in 16 of the 32 replicons, such as in the pELI series (Supplementary Datasheet 1). IS3 transposase was found in the series pFRA and in pSTE, pFRO29, pKLC2, pKLC3, pKLC4, pMFPC16A2803A, pMFPC16A2803C, pMFPA29A1405B, unnamed1, and unnamed2. pSTE and pKLC2 also shared IS30 transposase, whereas pFRO29, pMFPA29A1405B, and the series pFRA shared IS110 that contributed to their clustering. The series of pGLO plasmids harbored the IS6 transposase and other common recombinases and transposases, whereas the pSTE plasmids and pKLC2 encoded both IS3 and IS30 transposases. Peculiar was the scarce presence or absence of genes associated to transposases in pALB plasmids, albeit the large size of these replicons, likely due to their conjugative feature. The plasmid unnamed2 and series pALB and pELI were also characterized by genes encoding DNA-methyl transferase and/or restriction endonucleases.
Known antibiotic resistance genes were not identified in the new plasmids. Genes encoding ImmA/IrrE and RelE/ParE toxin–antitoxin systems were present in the series pSTE, pLQ25, and pGLO and in pKLC3, pMFPC16A2803A, and unnamed1. The latter also harbored RelB/DinJ system. ImmA/IrrE, RelE/ParE, and RelB/DinJ thus added to HicB family and PemK/MazF toxin–antitoxin systems located in the core genome. The plasmids often harbored genes involved in stress responses and participating in arsenic and heavy metal detoxification. Genetic determinants of thioredoxin were identified in pALB and pGLO series and in pKLC1, pKLC3, pKLC4, pMFPC16A2803A, pMFPC16A2803B, unnamed1, and unnamed2 plasmids. Interestingly, only pKLC1, pMFPC16A2803A, and pGLO series also encoded a glutathione reductase gene, the product of which can contribute for regulating cellular redox homeostasis, in concert with the glutathione peroxidase present in the core genome.
Genes associated to bacteriocin production were detected in four plasmids. pFRA21, pFRA23, and pFRO29 held the genes encoding the leucocin B and an immunity protein. Moreover, the genes encoding an additional immunity protein and an accessory secretion protein for the bacteriocin were predicted in pFRA21 and pFRA23. Conversely, pFRA18 did not harbor any gene associated to bacteriocin production. pALAN28 had three other genes linked to antibacterial activity: a lactococcin, a mesentericin, and another immunity protein.
Reconstructed Metabolic Pathways
Pathway reconstruction based on KEGG gene annotation of the core genome predicted at least five complete phosphotransferase system (PTS) transporters, i.e., one for sucrose, two belonging to mannose–fructose–sorbose family, and one each to glucose–β-glucoside and cellobiose–diacetyl chitobiose families. On the other hand, a complete PTS transporter for fructose and two belonging to ascorbate and galactitol families were accessory. Genes encoding other PTS components were also recognized as part of incomplete transporters in the core or in the accessory genome, such as EIIA components (or proteins with EIIA domains) with similarity to agaF, crr, and ptsN that may play a role in the uptake of substrates if they could interact with unidentified EIIB and EIIC components (Figure 5 and Supplementary Datasheet 2).
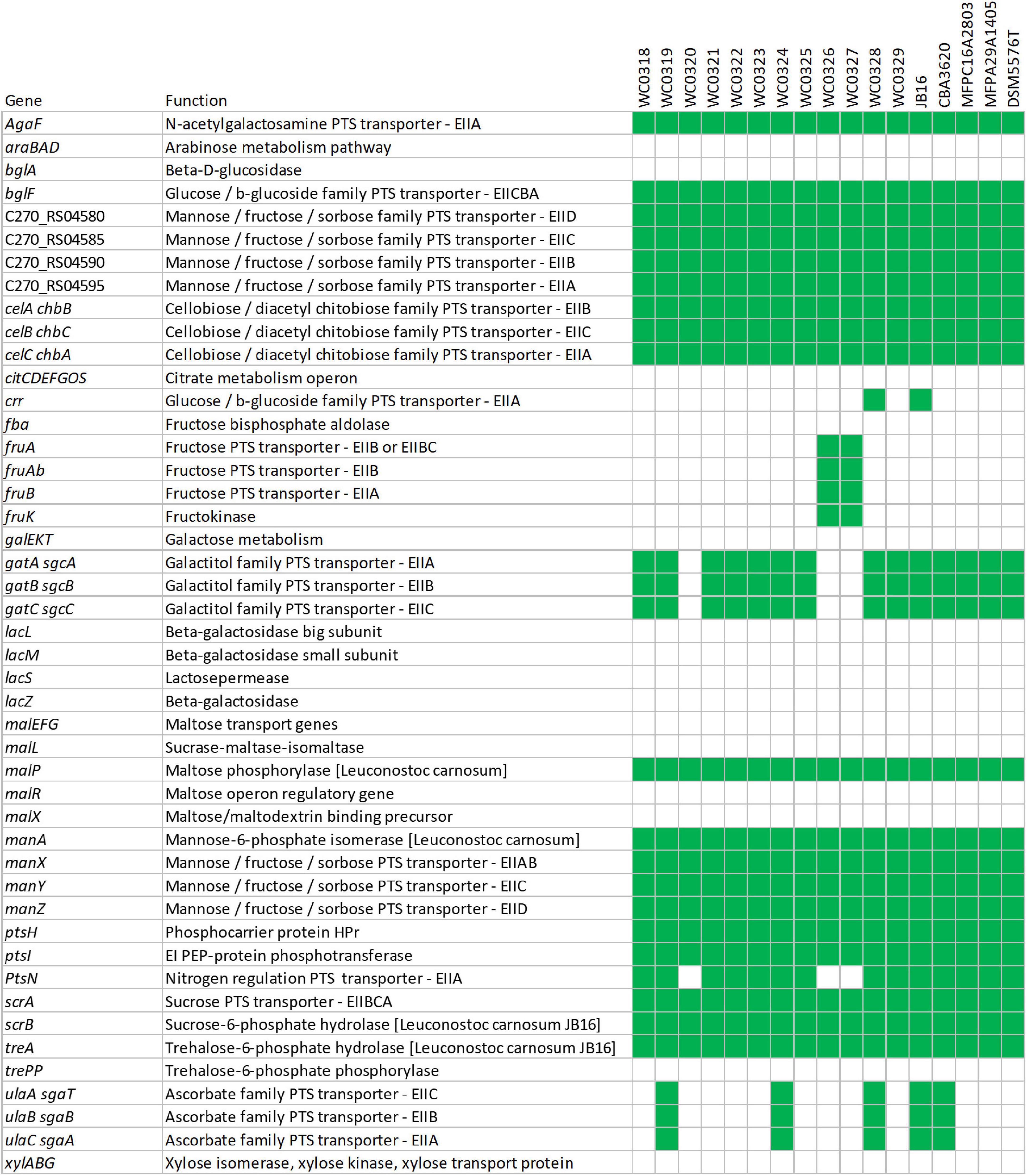
Figure 5. Genetic potential for metabolism of carbohydrates based on the presence (green) or absence (white) of predicted transporters or enzymes.
Complete ABC transporters for the uptake of ribose and xylose, oligopeptides, many amino acids, and other compounds or nutrients, such as nucleosides, amines, biotin, phosphate, and manganese, were predicted in the core genome (Supplementary Datasheet 2). The accessory genome included a complete ABC transporter for the uptake of choline and two exporters presenting similarity to those encoded by the operons TagGH and NodIJ for the export of teichoic acid and capsular polysaccharides, respectively. Three eukaryotic-type ABC exporters were also recognized in the core genome. One is involved in cytochrome export and assembly, whereas two have similarity to the multidrug efflux systems mdlAB and lmrCD. The accessory genome included a further eukaryotic-type ABC transporter, similar to blpA that is involved in bacteriocin export (Supplementary Datasheet 2).
Leuconostoc carnosum is heterofermentative, with the sugars being metabolized via the pentose phosphate and phosphoketolase pathway (PKP), yielding lactate, acetate, or ethanol and CO2 (Supplementary Datasheet 3). The pivot enzyme of PKP, D-xylulose-5-phosphate phosphoketolase (EC 4.1.2.9), was encoded in the core genome. Consistently, the main bacterial glycolytic pathways were incomplete, the Embden–Meyerhof’s one lacking 6-phosphofructokinase and fructose-bisphosphate aldolase, and the Entner–Doudoroff’s one lacking phosphogluconate dehydratase and 2-dehydro-3-deoxy-phosphogluconate aldolase. Leloir’s pathway for galactose utilization was incomplete in all the strains, even though some strains harbored the genes encoding galactose epimerase (EC 5.1.3.3) and UDP-glucose 4-epimerase (EC 5.1.3.2). The whole route for ascorbate transformation into xylulose 5-phosphate and channeling into the pentose phosphate pathway was accessory, the seven enzymes being encoded in some strains, or all missing in the others (Supplementary Datasheet 3). The citrate pathway was absent in all the strains, due to the lack of the Cit genes encoding citrate transporter, citrate lyase, and oxaloacetate decarboxylase. Nonetheless, all the strains harbored the genes encoding α-acetolactate synthase, α-acetolactate decarboxylases, and diacetyl acetoin reductases and may yield acetoin and 2,3-butanediol from pyruvate.
A total of 25 peptidase genes were identified (Supplementary Figure 1). The vast majority was encoded by core genes, whereas a minority were accessory. Reconstruction of amino acid biosynthesis pathways suggested that the strains did not differ regarding the ability to produce amino acids, except for tryptophan (Supplementary Datasheet 3). Some metabolic routes involved in the addition of an amino group to carbon backbones seemed interrupted, thus hampering the synthesis of some amino acids. In particular, the genes encoding glutamate dehydrogenase and glutamate synthase for the biosynthesis of glutamate from 2-oxoglutarate were not identified. The lack of alanine dehydrogenase and aspartate aminotransferase interrupted the synthesis of alanine and aspartate, respectively, whereas the pathway of serine production from glycolysis intermediates seemed ineffective due to the lack of phosphoserine phosphatase.
The pathways depending on pre-formed serine to yield glycine and cysteine, on glutamate to yield glutamine, arginine, and proline, and on aspartate to yield asparagine, threonine, and methionine were complete in the core genome (Supplementary Datasheet 3). Many genes encoding enzymes of lysine biosynthesis from aspartate were predicted in the core genome. The biosynthetic pathway seemed complete from aspartate to 2,3,4,5-tetrahydrodipicolinate. Some enzymes ascribable to the bacterial routes transforming this intermediate into lysine (i.e., the succinylase, acetylase, and dehydrogenase pathways) could be predicted, yet none of the routes seemed complete due to the lack of one or two enzymes. Interestingly, the carboxy-lyase catalyzing lysine formation from the last intermediate onto which the three routes converge (meso-2,6-diaminoheptanedioate) was predicted.
The anabolic pathways leading to branched chain amino acids were all complete in the core genome. With regard to aromatic amino acids, the route from 5-phosphoribosyl diphosphate to histidine and the shikimate pathway from erythrose 4-phosphate to chorismate were complete. The pathway leading to tyrosine seemed interrupted due to the lack of chorismate mutase, whereas the pathway leading to phenylalanine also lacked prephenate dehydratase. Nonetheless, the other enzymes necessary for the synthesis of tyrosine and phenylalanine were predicted. The metabolic pathway for tryptophan was complete in most of the strains, whereas it was absent in L. carnosum WC0318, WC0319, and WC0324.
All the strains harbored the whole set of genes and operons for the synthesis of adenine and guanine ribonucleotides from inosine monophosphate and of pyrimidine ribonucleotides from UMP. On the other hand, the pathway for uridine monophosphate biosynthesis from glutamine missed the gene encoding the aspartate carbamoyltransferase regulatory subunit, whereas the catalytic subunit was present. For pyrimidine deoxyribonucleotide biosynthesis from CDP/CTP, all the strains lacked the gene encoding the dCTP deaminase.
All the strains seemed unable to synthetize any biogenic amine, based on the absence of the genes encoding histidine, lysine, tyrosine, and ornithine decarboxylases (hdcA, ldc, tyrDC, and odc, respectively) and agmatine deiminase (aguD and aguA).
The metabolic pathways for the biosynthesis of most vitamins and cofactors were missing or largely incomplete in all the strains that seemed unable to synthetize thiamine, riboflavin, pyridoxal, NAD, pantothenate, biotin, folate, heme, cobalamin, and ubiquinone. Only the pathway for menaquinone biosynthesis was complete in all the strains. In this pathway, the module encoding 1,4-dihydroxy-2-naphthoyl-CoA hydrolase, a thioesterase of PaaI family, was not predicted using KEGG but was found utilizing the sequence of the cognate protein of L. mesenteroides ATCC 8293 as query for a BLASTp search against the proteins of L. carnosum. A PaaI family thioesterase presenting 68% of identity was detected in all the strains, suggesting that they all have the whole set of genes for the biosynthesis of menaquinone.
Stress Responses and Two-Component Signal Transduction Systems
The pangenome of L. carnosum lacked genetic determinants associated to superoxide dismutase, catalase, and de novo biosynthesis of glutathione. On the other hand, the core genome harbored other genes for coping with oxidative stress and reactive oxygen species, such as those encoding thioredoxin and thioredoxin reductase, a peroxidase, and two peroxiredoxins. Moreover, genes encoding thioredoxin-related proteins were harbored by 14 out of 32 plasmids. The core genome harbored the gene encoding a glutathione peroxidase (EC 1.11.1.9) that relies on pre-formed glutathione for H2O2 detoxification, whereas two plasmids encoded the counterpart encoding the glutathione reductase.
The genes of the stringent response mediated by the alarmone nucleotides (P)PPGPP were present in the core genome. Putative members of the two-component system were identified: six histidine kinases (HKs) and seven response regulators (RRs) were located in the core genome, and single HK and RR in the accessory portion. Four complete two-component systems presented similarity to known regulators, such as PhoR/PhoP, VicK/VicR, AgrC/AgrA, and CiaH/CiaR.
Antibiotic Resistance
The search for antibiotic resistance genes with RGI tool did not return any hit marked as “Perfect” or “Strict.” Many lactic acid bacteria possess intrinsic resistance to vancomycin due to D-alanine–D-alanine ligase encoded by the gene ddl. This resistance is correlated to the presence of a conserved phenylalanine residue (F) in the active site of the enzyme, whereas the sensitive strains possess a tyrosine residue (Y) in this position (Campedelli et al., 2019). The multiple sequence alignment of the ddl enzyme highlighted that all the 17 strains of L. carnosum possess the F-type enzyme, resulting in intrinsic vancomycin resistance.
Bacteriophages and Phage Defense
The genome sequences were investigated for prophages using PHASTER. For each strain, the tool identified 1–4 prophage gene clusters. Several intact prophages and a few incomplete ones were detected (Supplementary Table 2). Complete sequences of putatively active prophages were found in 9 out of 17 genomes (WC0319, WC0320, WC0321, WC0322, WC0323, WC0324, WC0325, WC0328, and MFPC16A2803). In all the strains without active prophages, sequences of prophages likely inactive were identified. In three strains, WC0321, WC0328, and CBA3620, questionable regions have also been found.
A type II CRISPR-Cas locus was found in the chromosomes of L. carnosum WC0319 and WC0324. The CRISPR array of WC0319 and WC0324 contained 16 and 11 spacers, respectively, the 11 of WC0324 identified as a first string of WC319 array, followed by other five unshared spacers. This arrangement suggested an iterative acquisition of the spacers and a common origin of the two strains, in agreement with the negligible phylogenetic distance, with the close position in the PCoA plot, with the hold of single similar plasmids (pELI19 and pELI24).
Bacteriocin Production
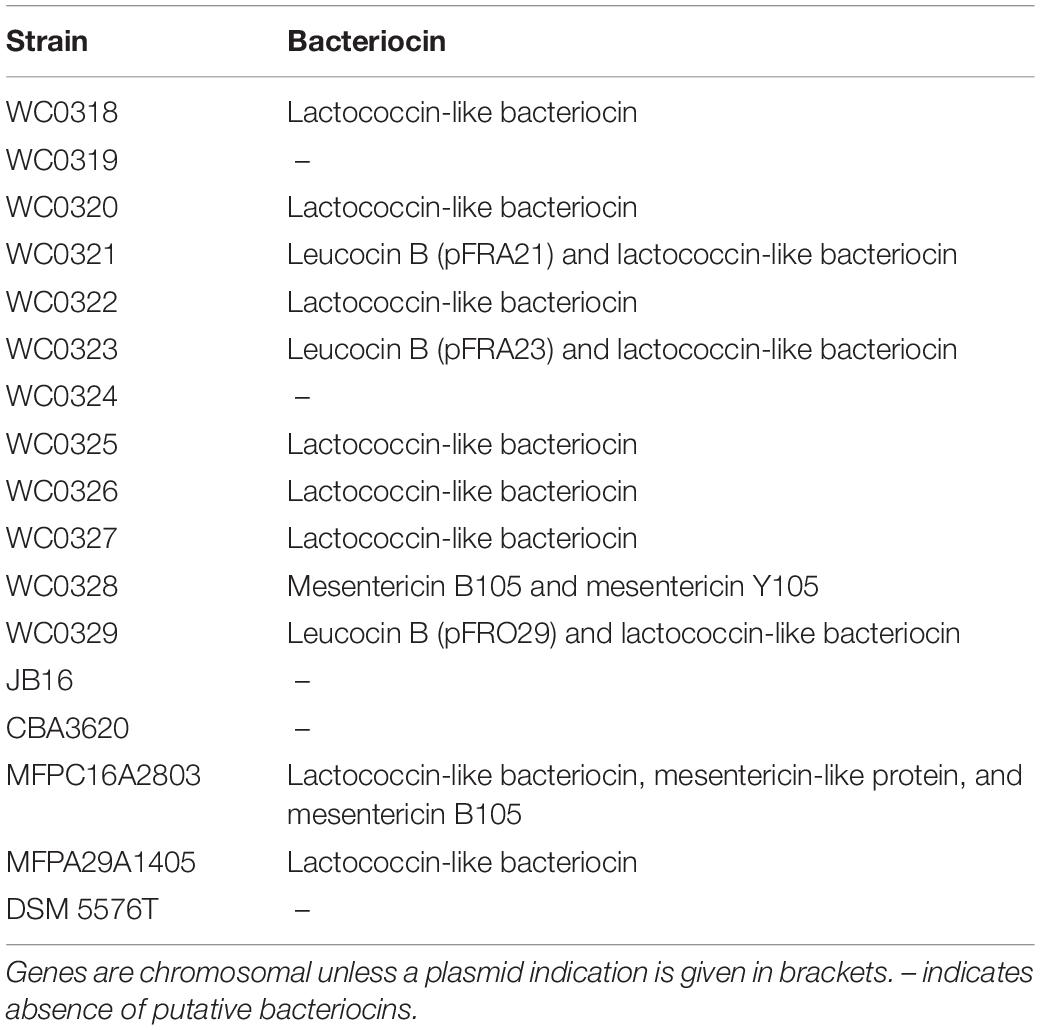
Screening of the entire genomes of L. carnosum using the bacteriocin database BAGEL 4 revealed the presence of five types of putative bacteriocin encoding loci (Table 3). Plasmids of L. carnosum WC0321, WC0323, and WC0329 harbored the genes for the synthesis of leucocin B. The genome of L. carnosum WC0321 and WC0323 encompassed also the gene encoding the transporter LanT, missing in WC0329. Most of the strains encoded a chromosomal lactococcin-like protein with double glycine leader peptide. No further information on the type of lactococcin was yielded by the prediction tool. Genes encoding mesentericin B105, mesentericin Y105, and/or a mesentericin-like protein were found in the genome of L. carnosum WC0328 and L. carnosum MFPC16A2803.
Discussion
In this study, all the genomes available for L. carnosum were compared to decipher their metabolic and functional potential and to determine the ecological role in food matrices (mainly meat) and the impact on food transformations. Most strains (15) were isolated from packed meat products of various origin (cooked ham, sausages, and beef), whereas a minority were isolated from kimchi (Jung et al., 2012; Raimondi et al., 2018, 2019). Genome comparison indicated that the 17 strains of L. carnosum are a compact group of bacteria, with values of sequence similarity much higher than the threshold required for species demarcation (ddDH > 90.5%; ANI > 98.9%), albeit their pangenome remained open. Differences within the core genome, revealed by sequence alignment, indicated three clades of strains. The clade encompassing most of the strains, all isolated from meat, was also separated in the PCoA plot computed from the matrix of the presence/absence of genes. The two strains isolated from kimchi were closely related, laying in the same clade and PCoA cluster, together with meat strains. Differences of location in PCoA plot with respect to phylogenetic distances can be attributed to accessory genes, with some contribution of genes encoded by plasmids. For instance, L. carnosum WC0328 is in a different clade than L. carnosum WC0324 and WC0319, but they all cluster together in the PCoA plot and harbor a plasmid of the pELI series. L. carnosum WC326 and WC327, which resulted to be the same strain, were isolated from the same factory, in fresh sausages in diverse lots sampled 2 months apart (Raimondi et al., 2018), suggesting the persistence of this species in the environment of production plants.
Leuconostoc carnosum harbored the genes for heterolactic fermentation sugars, yielding L-lactate, ethanol, acetate, and CO2 (Posthuma et al., 2002). The availability in meat of a small range of carbohydrates, mainly glucose deriving from glycogen hydrolysis (Pereira and Vicente, 2013), is consistent with the small potential to ferment sugars, restricted to glucose and few others. With a sole exception, carbohydrate metabolic capabilities were shared by all the strains and were not strain-dependent. The genes for ribose assimilation and fermentation were present only in L. carnosum WC0322, encoded by plasmid pLQ22. Unlike other lactic acid bacteria, including Leuconostoc species, L. carnosum seemed unable to uptake and metabolize citrate via citrate lyase and oxaloacetate decarboxylase (Bekal et al., 1998; García-Quintáns et al., 2008). However, all the strains may yield flavored four carbon metabolites, such as acetoin and butanediol from pyruvate, even though the role of L. carnosum in food ripening and aroma development remains to be clarified.
Meat is a rich substrate mainly composed of proteins, albeit an abundant fraction of amino acids or small peptides is also available (Cobos and Díaz, 2015). Despite the ability to synthetize some amino acids, all the strains seemed unable to grow utilizing ammonium as the sole nitrogen source and must rely on a pool of few pre-formed amino acids that should include at least glutamate, aspartate, serine, and alanine. Since the biosynthetic pathway of other amino acids (e.g., tyrosine, phenylalanine, and lysine) is present but incomplete, it remains to be clarified whether some genes are not correctly annotated, and prediction could make up for the interruption. Adaptation of L. carnosum to a nitrogen-rich environment explains the autotrophy not only for several amino acids but also for some ribonucleotide and deoxyribonucleotide and for several vitamins and cofactors. On the other hand, L. carnosum clearly took advantage of the 23 peptidase genes identified in the core genome and the other two encoded by accessory genes.
The presence of a choline transporter indicates that L. carnosum may use this compound that is abundant in meat to cope with salt and other osmotic stress. However, while in other bacteria (e.g., Bacillus subtilis) choline uptake is functional to the biosynthesis of the osmoprotectant betaine (Kappes et al., 1999), any gene involved in choline transformation to betaine is missing.
All the strains herein compared harbored 1–4 plasmids that presented main functions associated to: hydrolysis of proteins; transport and metabolism of amino acids, oligopeptides, and carbohydrates; production of bacteriocin and exopolysaccharide; resistance to bacteriophage, heavy metal, and other stress responses; and DNA restriction–modification systems. A peculiar trait of these replicons was the absence of genetic determinants for antibiotic resistance that were not detected either in the chromosomes. However, the genes mdlAB and lmrCD, putatively encoding the multidrug efflux systems, were located in the genome. There is no information on whether mdlAB confers any antibiotic resistance to bacteria, whereas lmrCD confers to Lactococcus lactis the resistance to some toxic compounds, including daunomycin (Kobayashi et al., 2001; Lubelski et al., 2006). The lack of genes encoding antibiotic resistance may be due to the specificity of L. carnosum for meat matrices that seclude the species in an environment where the selective pressure is limited and the interaction with gut bacteria, the most exposed to the antibiotics utilized in animal production, is low. The safety of L. carnosum is a key point, considering the high load of bacteria belonging to this species that are ingested mostly by consuming products, such as MAP sliced meat. Safety of this species was associated to the absence not only of any antibiotic resistance genetic determinants but also of decarboxylase genes for biogenic amine production, assessing the incapability of these strains to produce biogenic amine.
In most of the strains, the potential of bacteriocin synthesis, secretion, and immunity was identified, including the production of leucocin B, a class IIa bacteriocin effective against L. monocytogenes (Felix et al., 1994). It is quite interesting the fact that, at the end of the shelf-life, MAP cooked ham generally presented an abundant population of L. carnosum, regardless of the sensorial evolution of the product (Raimondi et al., 2019). Indeed, L. carnosum dominated both in seriously spoiled products and in those that maintained good sensorial properties. However, studies of biochemistry and physiology of L. carnosum, also focused on molecular mechanisms affecting meat spoilage, are still lacking. The utilization of L. carnosum strains as bioprotective starters could be encouraged by the ability to produce bacteriocins; thus, a deeper investigation is required to determine their activity spectrum against spoilage and pathogen bacteria. Production and activity of the predicted lactococcin-like proteins need to be analyzed further.
As a whole, this study provided a deep insight into the genomic and metabolic features of this important bacterium ubiquitous in meat products, revealing that it is safe and opening new possibilities of exploitation in the food conservation industry.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
SR, JS, and MR conceived and designed the experiments. FC carried out the DNA extraction and the bioinformatics for genome assembly and annotation. MT supervised genome sequencing, assembly, and annotation. FC, GS, and AA performed data analysis. SR, AA, and MR wrote the manuscript with contributions from all other authors. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.605127/full#supplementary-material
Footnotes
- ^ http://enve-omics.ce.gatech.edu/g-matrix/
- ^ https://www.dsmz.de/services/online-tools/genome-to-genome-distance-calculator-ggdc
- ^ http://phaster.ca/
- ^ https://crisprcas.i2bc.paris-saclay.fr/CrisprCasFinder/Index
- ^ http://bagel4.molgenrug.nl/index.php
- ^ https://isfinder.biotoul.fr/
- ^ https://card.mcmaster.ca/analyze/rgi
- ^ http://weizhong-lab.ucsd.edu/webMGA/server/
- ^ https://www.kegg.jp/blastkoala
- ^ https://www.genome.jp/kegg/tool/map_pathway.html
References
Alcock, B. P., Raphenya, A. R., Lau, T. T. Y., Tsang, K. K., Bouchard, M., Edalatmand, A., et al. (2020). CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 48, D517–D525. doi: 10.1093/nar/gkz935
Arndt, D., Grant, J. R., Marcu, A., Sajed, T., Pon, A., Liang, Y., et al. (2016). PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 44, W16–W21. doi: 10.1093/nar/gkw387
Bekal, S., Van Beeumen, J., Samyn, B., Garmyn, D., Henini, S., Diviès, C., et al. (1998). Purification of Leuconostoc mesenteroides citrate lyase and cloning and characterization of the citCDEFG gene cluster. J. Bacteriol. 180, 647–654. doi: 10.1128/JB.180.3.647-654.1998
Björkroth, K. J., Vandamme, P., and Korkeala, H. J. (1998). Identification and characterization of Leuconostoc carnosum, associated with production and spoilage of vacuum-packaged, sliced, cooked ham. Appl. Environ. Microbiol. 64, 3313–3319. doi: 10.1128/AEM.64.9.3313-3319.1998
Budde, B. B., Hornbaek, T., Jacobsen, T., Barkholt, V., and Koch, A. G. (2003). Leuconostoc carnosum 4010 has the potential for use as a protective culture for vacuum-packed meats: culture isolation, bacteriocin identification, and meat application experiments. Int. J. Food Microbiol. 83, 171–184. doi: 10.1016/S0168-1605(02)00364-1
Campedelli, I., Mathur, H., Salvetti, E., Clarke, S., Rea, M. C., Torriani, S., et al. (2019). Genus-wide assessment of antibiotic resistance in Lactobacillus spp. Appl. Environ. Microbiol. 85, e1738–e1718. doi: 10.1128/AEM.01738-18
Candeliere, F., Raimondi, S., Spampinato, G., Tay, M. Y. F., Amaretti, A., Schlundt, J., et al. (2020). Draft genome sequences of 12 Leuconostoc carnosum strains isolated from cooked ham packaged in a modified atmosphere and from fresh sausages. Microbiol Resour Announc. 9, e1247–e1219. doi: 10.1128/MRA.01247-19
Chen, Y. S., Wang, L. T., Wu, Y. C., Mori, K., Tamura, T., Chang, C. H., et al. (2020). Leuconostoc litchii sp. nov., a novel lactic acid bacterium isolated from lychee. Int. J. Syst. Evol. Microbiol. 70, 1585–1590. doi: 10.1099/ijsem.0.003938
Cobos, Á, and Díaz, O. (2015). “Chemical composition of meat and meat products,” in Handbook of Food Chemistry, eds P. Cheung and B. Mehta (Berlin: Springer), doi: 10.1007/978-3-642-36605-5_6
Couvin, D., Bernheim, A., Toffano-Nioche, C., Touchon, M., Michalik, J., Néron, B., et al. (2018). CRISPRCasFinder, an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for Cas proteins. Nucleic Acids Res. 46, W246–W251. doi: 10.1093/nar/gky425
Felix, J. V., Papathanasopoulos, M. A., Smith, A. A., von Holy, A., and Hastings, J. W. (1994). Characterization of leucocin B-talla: a bacteriocin from Leuconostoc carnosum talla isolated from meat. Curr. Microbiol. 29, 207–212. doi: 10.1007/BF01570155
García-Quintáns, N., Blancato, V. S., Repizo, G. D., Magni, C., and López, P. (2008). “Citrate metabolism and aroma compound production in lactic acid bacteria,” in Molecular Aspects of Lactic Acid Bacteria for Traditional and New Applications, eds B. Mayo, P. López, and G. Pérez-Martínez (Thiruvananthapuram: Research Signpost), 65–88.
Geeraerts, W., Pothakos, V., De Vuyst, L., and Leroy, F. (2018). Variability within the dominant microbiota of sliced cooked poultry products at expiration date in the Belgian retail. Food Microbiol. 73, 209–215. doi: 10.1016/j.fm.2018.01.019
Hastings, J. W., Stiles, M. E., and von Holy, A. (1994). Bacteriocins of leuconostocs isolated from meat. Int. J. Food Microbiol. 24, 75–81. doi: 10.1016/0168-1605(94)90107-4
Hong, Y., Yang, H. S., Li, J., Han, S. K., Chang, H. C., and Kim, H. Y. (2014). Identification of lactic acid bacteria in salted Chinese cabbage by SDS-PAGE and PCR-DGGE. J Sci Food Agric. 94, 296–300. doi: 10.1002/jsfa.6257
Jacobsen, T., Budde, B. B., and Koch, A. G. (2003). Application of Leuconostoc carnosum for biopreservation of cooked meat products. J. Appl. Microbiol. 95, 242–249. doi: 10.1046/j.1365-2672.2003.01970.x
Jeon, H. H., Kim, K. H., Chun, B. H., Ryu, B. H., Han, N. S., and Jeon, C. O. (2017). A proposal of Leuconostoc mesenteroides subsp. jonggajibkimchii subsp. nov. and reclassification of Leuconostoc mesenteroides subsp. suionicum (GU et al., 2012) as Leuconostoc suionicum sp. nov. based on complete genome sequences. Int. J. Syst. Evol. Microbiol. 2017, 2225–2230. doi: 10.1099/ijsem.0.001930
Jung, J. Y., Lee, S. H., and Jeon, C. O. (2012). Complete genome sequence of Leuconostoc carnosum strain JB16, isolated from kimchi. J. Bacteriol. 194, 6672–6673. doi: 10.1128/JB.01805-12
Jung, J. Y., Lee, S. H., and Jeon, C. O. (2014). Kimchi microflora: history, current status, and perspectives for industrial kimchi production. Appl. Microbiol. Biotechnol. 98, 2385–2393. doi: 10.1007/s00253-014-5513-1
Kanehisa, M., and Sato, Y. (2020). KEGG Mapper for inferring cellular functions from protein sequences. Protein Sci. 29, 28–35. doi: 10.1002/pro.3711
Kanehisa, M., Sato, Y., and Morishima, K. (2016). BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J. Mol. Biol. 428, 726–731. doi: 10.1016/j.jmb.2015.11.006
Kappes, R. M., Kempf, B., Kneip, S., Boch, J., Gade, J., Meier-Wagner, J., et al. (1999). Two evolutionarily closely related ABC transporters mediate the uptake of choline for synthesis of the osmoprotectant glycine betaine in Bacillus subtilis. Mol. Microbiol. 32, 203–216. doi: 10.1046/j.1365-2958.1999.01354.x
Kobayashi, N., Nishino, K., and Yamaguchi, A. (2001). Novel macrolide-specific ABC-type efflux transporter in Escherichia coli. J. Bacteriol. 183, 5639–5644. doi: 10.1128/JB.183.19.5639-5644.2001
Lawton, M. R., Jencarelli, K. G., Kozak, S. M., and Alcaine, S. D. (2020). Short communication: evaluation of commercial meat cultures to inhibit Listeria monocytogenes in a fresh cheese laboratory model. J. Dairy Sci. 103, 1269–1275. doi: 10.3168/jds.2019-17203
Letunic, I., and Bork, P. (2019). Interactive tree of life (iTOL) v4: recent updates and new developments. Nucleic Acids Res 47, W256–W259. doi: 10.1093/nar/gkz239
Li, L., Wen, X., Wen, Z., Chen, S., Wang, L., and Wei, X. (2018). Evaluation of the biogenic amines formation and degradation abilities of Lactobacillus curvatus from chinese bacon. Front. Microbiol. 9:1015. doi: 10.3389/fmicb.2018.01015
Li, X., Li, C., Ye, H., Wang, Z., Wu, X., Han, Y., et al. (2019). Changes in the microbial communities in vacuum-packaged smoked bacon during storage. Food Microbiol. 77, 26–37. doi: 10.1016/j.fm.2018.08.007
Lubelski, J., de Jong, A., van Merkerk, R., Agustiandari, H., Kuipers, O. P., Kok, J., et al. (2006). LmrCD is a major multidrug resistance transporter in Lactococcus lactis. Mol. Microbiol. 61, 771–781. doi: 10.1111/j.1365-2958.2006.05267.x
Meier-Kolthoff, J. P., Auch, A. F., Klenk, H. P., and Göker, M. (2013). Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics 14:60. doi: 10.1186/1471-2105-14-60
Nurk, S., Meleshko, D., Korobeynikov, A., and Pevzner, P. A. (2017). metaSPAdes: a new versatile metagenomic assembler. Genome Res. 27, 824–834. doi: 10.1101/gr.213959.116
Page, A. J., Cummins, C. A., Hunt, M., Wong, V. K., Reuter, S., Holden, M. T., et al. (2015). Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31, 3691–3693. doi: 10.1093/bioinformatics/btv421
Parente, E., Moles, M., and Ricciardi, A. (1996). Leucocin F10, a bacteriocin from Leuconostoc carnosum. Int. J. Food Microbiol. 33, 231–243. doi: 10.1016/0168-1605(96)01159-2
Pereira, P. M., and Vicente, A. F. (2013). Meat nutritional composition and nutritive role in the human diet. 2013. Meat Sci. 93, 586–592. doi: 10.1016/j.meatsci.2012.09.018
Posthuma, C. C., Bader, R., Engelmann, R., Postma, P. W., Hengstenberg, W., and Pouwels, P. H. (2002). Expression of the xylulose 5-phosphate phosphoketolase gene, xpkA, from Lactobacillus pentosus MD363 is induced by sugars that are fermented via the phosphoketolase pathway and is repressed by glucose mediated by CcpA and the mannose phosphoenolpyruvate phosphotransferase system. Appl. Environ. Microbiol. 68, 831–837. doi: 10.1128/aem.68.2.831-837.2002
Price, M. N., Dehal, P. S., and Arkin, A. P. (2009). FastTree: computing large minimum-evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26, 1641–1650. doi: 10.1093/molbev/msp077
Raimondi, S., Luciani, R., Sirangelo, T. M., Amaretti, A., Leonardi, A., Ulrici, A., et al. (2019). Microbiota of sliced cooked ham packaged in modified atmosphere throughout the shelf life: microbiota of sliced cooked ham in MAP. Int. J. Food Microbiol. 289, 200–208. doi: 10.1016/j.ijfoodmicro.2018.09.017
Raimondi, S., Nappi, M. R., Sirangelo, T. M., Leonardi, A., Amaretti, A., Ulrici, A., et al. (2018). Bacterial community of industrial raw sausage packaged in modified atmosphere throughout the shelf life. Int. J. Food Microbiol. 280, 78–86. doi: 10.1016/j.ijfoodmicro.2018.04.041
Richter, M., and Rosselló-Móra, R. (2009). Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. U.S.A. 106, 19126–19131. doi: 10.1073/pnas.0906412106
Rodrigo-Torres, L., Yépez, A., Aznar, R., and Arahal, D. R. (2019). Genomic insights into five strains of lactobacillus plantarum with biotechnological potential isolated from chicha, a traditional maize-based fermented beverage from northwestern argentina. Front. Microbiol. 10:2232. doi: 10.3389/fmicb.2019.02232
Rodriguez-R, L. M., and Konstantinidis, K. T. (2016). The enveomics collection: a toolbox for specialized analyses of microbial genomes and metagenomes. PeerJ 4:e1900v1. doi: 10.7287/peerj.preprints.1900v1
Samelis, J., Björkroth, J., Kakouri, A., and Rementzis, J. (2006). Leuconostoc carnosum associated with spoilage of refrigerated whole cooked hams in Greece. J. Food Prot. 69, 2268–2273.
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Shaw, B. G., and Harding, C. D. (1989). Leuconostoc gelidum sp. nov. and Leuconostoc carnosum sp. nov. from chill-stored meats. Int. J. Syst. Bacteriol. 39, 217–223. doi: 10.1099/00207713-39-3-217
Siguier, P., Perochon, J., Lestrad, E. L., Mahillon, J., and Chandler, M. (2006). ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 34, D32–D36. doi: 10.1093/nar/gkj014
Stiles, M. E. (1994). Bacteriocins produced by Leuconostoc species. J. Dairy Sci. 77, 2718–2724. doi: 10.3168/jds.S0022-0302(94)77214-3
Tettelin, H., Riley, D., Cattuto, C., and Medini, D. (2008). Comparative genomics: the bacterial pan-genome. Curr. Opin. Microbiol. 11, 472–477. doi: 10.1016/j.mib.2008.09.006
van Heel, A. J., de Jong, A., Song, C., Viel, J. H., Kok, J., and Kuipers, O. P. (2018). BAGEL4: a user-friendly web server to thoroughly mine RiPPs and bacteriocins. Nucleic Acids Res. 46, W278–W281. doi: 10.1093/nar/gky383
van Laack, R. L. J. M., Schillinger, U., and Holzapfel, W. H. (1992). Characterization and partial purification of a bacteriocin produced by Leuconostoc carnosum LA44A. Int. J. Food Microbiol. 16, 183–195.
Vasilopoulos, C., De Mey, E., Dewulf, L., Paelinck, H., De Smedt, A., Vandendriessche, F., et al. (2010). Interactions between bacterial isolates from modified-atmosphere-packaged artisan-type cooked ham in view of the development of a bioprotective culture. Food Microbiol. 27, 1086–1094. doi: 10.1016/j.fm.2010.07.013
Wan, X., Saris, P. E. J., and Takala, T. M. (2015). Genetic characterization and expression of leucocin B, a class IId bacteriocin from Leuconostoc carnosum 4010. Res. Microbiol. 166, 494–503. doi: 10.1016/j.resmic.2015.04.003
Wu, S., Zhu, Z., Fu, L., Niu, B., and Li, W. (2011). WebMGA: a customizable web server for fast metagenomic sequence analysis. BMC Genomics 12:444. doi: 10.1186/1471-2164-12-444
Zheng, J., Wittouck, S., Salvetti, E., Franz, C. M. A. P., Harris, H. M. B., Mattarelli, P., et al. (2020). A taxonomic note on the genus Lactobacillus: description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 70, 2782–2858. doi: 10.1099/ijsem.0.004107
Keywords: Leuconostoc carnosum, genomics, pangenome analysis, bacteriocin, metabolism
Citation: Candeliere F, Raimondi S, Spampinato G, Tay MYF, Amaretti A, Schlundt J and Rossi M (2021) Comparative Genomics of Leuconostoc carnosum. Front. Microbiol. 11:605127. doi: 10.3389/fmicb.2020.605127
Received: 11 September 2020; Accepted: 04 December 2020;
Published: 11 January 2021.
Edited by:
Victor Ladero, Consejo Superior de Investigaciones Científicas (CSIC), SpainReviewed by:
Raul Raya, CONICET Centro de Referencia para Lactobacilos (CERELA), ArgentinaOtávio Guilherme Gonçalves De Almeida, University of São Paulo, Brazil
Takeshi Zendo, Kyushu University, Japan
Copyright © 2021 Candeliere, Raimondi, Spampinato, Tay, Amaretti, Schlundt and Rossi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maddalena Rossi, maddalena.rossi@unimore.it
 Francesco Candeliere
Francesco Candeliere Stefano Raimondi
Stefano Raimondi Gloria Spampinato
Gloria Spampinato Moon Yue Feng Tay2,3
Moon Yue Feng Tay2,3 Alberto Amaretti
Alberto Amaretti Joergen Schlundt
Joergen Schlundt Maddalena Rossi
Maddalena Rossi