- 1Research Center of Horticultural Science and Engineering, Huaqiao University, Xiamen, China
- 2College of Plant Protection, Nanjing Agricultural University, Nanjing, China
- 3Institute of Fruit Tree Research, Guangdong Academy of Agricultural Sciences, Guangzhou, China
- 4UWA Institute of Agriculture, UWA School of Agriculture and Environment, The University of Western Australia, Perth, WA, Australia
Fusarium wilt is a devastating soil-borne fungal disease caused by Fusarium oxysporum f.sp. cubense (Foc). In recent years, some antifungal bacteria have been applied for the prevention and biocontrol of pathogenic fungi. In our study, a bacterial strain HQB-1, isolated from banana rhizosphere soil, was cultured for investigation. It showed broad-spectrum antifungal activities against representative phytopathogenic fungi including Fusarium oxysporum, Colletotrichum gloeosporioides, Botrytis cinerea, and Curvularia fallax. The strain HQB-1 was identified as Burkholderia sp. by morphological, physiological, and biochemical examinations, confirmed by 16S rRNA gene sequence analysis. Among the metabolites produced by the strain, we identified an antifungal compound which was identified phenazine-1-carboxylic acid (PCA) (C13H8N2O2) through ultraviolet, liquid chromatography quadrupole-time of flight mass spectrometer, and nuclear magnetic response. Furthermore, PCA exhibited the lowest minimum inhibitory concentration (MIC) against F. oxysporum (1.56 μg/ml) and yielded the highest MIC against C. gloeosporioides. Pot experiments showed that application of 5 μg/ml or more of PCA efficiently controlled banana wilt and promoted the growth of banana plants. These results suggested that Burkholderia sp. HQB-1, as an important microbial resource of PCA, could be a promising biological agent against wilt diseases and promoting banana growth.
Introduction
Banana (Musa spp.), is one of the most important fruit crops, widely cultivated in tropical and subtropical areas, providing staple food for over 400 million people (Dale et al., 2017). With the increase in exportation, banana becomes an important source for foreign exchange in China (Wang B. et al., 2013). However, in recent years, Fusarium wilt, also known as Panama disease, has become one of the most destructive soil-borne diseases threatening banana production worldwide (Davis et al., 1996; Ploetz, 2005; Fourie et al., 2011). Fusarium wilt is caused by Fusarium oxysporum f. sp. cubense (Foc) with the Tropical Race 4 (Foc TR4) causing the most severe damage among the four races of this pathogen. It has been demonstrated that the pathogen Foc TR4 can infect banana roots, penetrate into the root xylem, spread into the rhizome and pseudostem xylem within several days after infection (Warman and Aitken, 2018), and eventually cause the occlusion of vascular bundle and the blocking of water and nutrients transportation (Li et al., 2017). Once Foc TR4 was introduced to or detected in banana orchard, it is quite difficult to eradicate. Because the chlamydospores of Foc TR4 can survive for a long period of time in soil (Viljoen, 2002), it is becoming one of the most crucial limiting factors in banana production (O’Donnell et al., 1998; Lin et al., 2009). Therefore, it is necessary and imperative to efficiently control the occurrence and spread of Foc TR4 in banana.
Traditionally, chemical pesticide is considered to be a simple and direct approach to control Fusarium wilt (Fuchs et al., 1999; Getha et al., 2005). However, chemical residues not only provided unsatisfactory control levels but also lead to severe environmental pollutions and induce the development of resistant strains (Kanini et al., 2013). Though resistant breeding cultivars have been studied and widely used for a long time, polyploidy, low reproductive fertility and complete sterility of banana are the main reasons for the limited success of the breeding program (Shepherd, 1987). Crop rotation slowly reduces pathogen density in the soil but its effectiveness is restricted once the disease outbreak occurred (Chellemi et al., 2016). Recently, biological control, especially the application of biocontrol microorganisms, has been widely studied due to its advantage as an economically and environmentally friendly method (Leeman et al., 1996). However, the available microorganisms for biocontrol of plant diseases may also lead to inconsistent performance and poor activity in the soil environment (Saravanan et al., 2003). Thus, the isolation and screening of highly efficient and broad-spectrum antagonistic microorganisms are the key for the development of biocontrol agents. New bioformulation technologies like nanotechnologies, formulation of microbial consortia and/or their metabolites need to be developed to improve their efficacy and deepen the knowledge of biocontrol microorganisms (Bubici et al., 2019).
The genus Burkholderia has become increasingly important during the past several decades because of its capability in producing abundant secondary metabolites with antimicrobial, insecticidal, herbicidal, or growth-promotion traits (Jeong et al., 2003; Singh et al., 2013; Kunakom and Eustáquio, 2019). Several Burkholderia strains like B. vietnamiensis, B. ambifaria, and B. pyrrocinia have been reported to be potential and efficient biological control agents (Mahenthiralingam et al., 2008). In fact, the key mechanism for the antagonistic effects of Burkholderia against plant pathogens is the production of antimicrobial secondary metabolites (Schmidt et al., 2010; Eberl and Vandamme, 2016). Among them, cepacin, phenazine, and pyrrolnitrin have been shown to play roles as antibiotics in controlling plant diseases (Parker et al., 1984; Cartwright et al., 1995; Xu et al., 2013). Siderophores produced by Burkholderia not only exhibit survival competition with pathogens but also promote the growth of the plant (Jiang et al., 2008). However, a large number of new natural compounds produced by Burkholderia is still unknown, and the isolation, purification, and identification of these bio-functional natural products, especially as antimicrobials, need to be further investigated.
In this study, we screened and isolated a broad-spectrum antagonistic bacterial strain HQB-1 from the rhizosphere soil of banana plants. The taxonomic status of HQB-1 was determined as Burkholderia sp. according to the 16S rRNA gene sequence analysis combined with morphological, physiological, and biochemical characteristics. Moreover, the antifungal component phenazine-1-carboxylic acid (PCA) produced by Burkholderia sp. HQB-1 was separated and identified by column chromatography, ultraviolet (UV), liquid chromatography quadrupole-time of flight mass spectrometer (LC-Q-TOF-MS), and nuclear magnetic response (NMR). The bio-controlling effect of PCA was further investigated via pot experiments. The aim of this study was to analyze the antifungal metabolites from potential beneficial microbial resources and to identify the main efficient compound that could be used to prevent Fusarium wilt in banana.
Materials and Methods
Soil Sample Collection, Isolation of Fusarium Wilt Antagonistic Bacteria and Antifungal Activity
Twelve rhizosphere soil samples including clay and silt loam and were collected from healthy plants found in a pathogen-infested banana orchard, which has been planted banana for 15 years, at Changtai County, Zhangzhou City, Fujian Province, China (24.65°N, 117.72°E) as described by Chen et al. (2013). The plants were gently removed from the farmland, the loose soil was removed by shaking, and the soil attached to the roots was collected by a brush. The soil samples were placed into clean, dry, and sterile polythene bags, transported to the laboratory, and stored at 4°C until further analysis.
The samples were suspended in 0.85% of NaCl and spread on Luria-Bertani (LB) agar. After incubation at 30°C for 2 days, bacterial colonies with different morphological characteristics were selected and re-streaked on fresh LB agar until the homogeneous colonies appeared. Fifty-six isolated strains were further sub-cultured in LB broth on a rotary shaker. For long-term storage of bacterial strains, 50% (v/v) glycerol solution was added to the liquid culture for freezing at −80°C.
Several important phytopathogenic fungi were chosen and used in the present study, included Fusarium oxysporum f. sp. cubense Tropical Race 4 (Foc TR4, ATCC 76255), Colletotrichum gloeosporioides (ATCC 16330), Botrytis cinerea (ATCC 11542), and Curvularia fallax (ATCC 38579). These strains were provided by the Institute of Tropical Bioscience and Biotechnology, China Academy of Tropical Agricultural Sciences, Haikou, China. The antifungal activity of isolated strains was determined according to Reyes-Chilpa et al. (1997) and Chen et al. (2018) with slight modifications. Briefly, a fungal plug (Φ = 6 mm) was inoculated in the center of a plate filled with potato dextrose agar (PDA) medium; Each isolated bacterium was seeded with a sterile stick at a distance of 2.5 cm from the fungi. A fungal disk in the center of a plate without seeding isolated bacterium was served as a control. After incubation at 28°C for 5 days, the inhibition zone was recorded by measuring the distance between the edge of the fungal mycelium and the bacterial colony. The percentage inhibition of mycelial radial growth was calculated by the following formula:
Percent mycelial inhibition = , where A is the mean colony diameter for the control, and B is the mean colony for the treatment (Nimaichand et al., 2015).
Morphology, Physiological, and Biochemical Tests of Antagonistic Bacteria
The morphological, physiological, and biochemical characteristics tests of the strain HQB-1 were further analyzed including: Gram-staining, temperature and pH range for growth, presence of pigment, catalase activity, oxidative or fermentative reaction, nitrate reduction, hydrolysis of casein, starch, Tween 80 and gelatin, activity of urease and pyruvate deaminase, tolerance to sodium chloride, production of indole, reaction of H2S, Methyl Red-staining, and Voges-Proskauer tests. All these characteristics were then compared with the standard description of Bergey’s Manual of Determinative Bacteriology (Buchanan and Gibbons, 1984).
To obtain scanning electron microscope (SEM) image of HQB-1, a 1 cm2 transverse section of the bacterial strain on LB agar plate was obtained and fixed in 2.5% glutaraldehyde buffer at 4°C for 3 h. Then the bacterial cells were washed by phosphate-buffered saline (PBS) for three times and fixed in 1% osmic acid at 25°C for 5 h. After washing, the cells were dehydrated with 50, 70, 95, and 100% ethanol, respectively, for critical point drying and platinum-coating using an ion-sputter coater. The morphology of the specimen was observed at an opening voltage of 5 kV using a field-emission scanning electron microscopy (S-4800IIESEM, Hitachi, Japan).
16S rRNA Gene Sequencing and Phylogenetic Analysis
The strain HQB-1 was cultured in LB broth at 28°C in an orbital shaker for 12 h. The genomic DNA of the strain was extracted using the bacterial genomic DNA extraction kit (Takara, Beijing, China). The 16S rRNA sequences were amplified by PCR with the universal primers 27F (5′-GAGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-CTACGGCTACCTTGTTACGA-3′) (Gupta et al., 2014). The PCR conditions were as follows: initial denaturation at 94°C for 5 min, 30 cycles of 94°C for 45 s, 58°C for 60 s, and 72°C for 90 s, and a final extension at 72°C for 10 min. The PCR products were purified by Quick Gel Extraction Kit (TransGen Biotech, Beijing, China) and sequenced by Sangon Biotech (Shanghai, China). The 16S rRNA gene sequence was submitted to the GenBank database. The sequences were analyzed by a global alignment algorithm implemented in the EzBioCloud database1 (Kim et al., 2012; Yoon et al., 2017) and blasted with the sequences of known species in NCBI database2. After multiple alignments of sequences via CLUSTAL_X (Thompson et al., 1997), the phylogenetic analysis was performed using the neighbor-joining method (Saitou and Nei, 1987) with MEGA version 6.0 (Tamura et al., 2007).
Fermentation of HQB-1 and Separation of Antifungal Components
Forty liters of LB broth with HQB-1 were grown in a 200 L fermentation tank.at 30°C for 4 days. At the end of fermentation, the cultural broth with HQB-1 was centrifuged at 10,000 × g for 20 min, and an equal volume of ethyl acetate (EA) was added into the cultural supernatant. This procedure was repeated three times. The EA layers were combined and the solvent was evaporated under vacuum at 40°C using a rotary evaporator for obtaining the final crude extract with approximately 36 g. The crude EA extract was then subjected to macroporous resin (XAD-16, Amberlite, United States) column chromatography and eluted with different (30, 60, and 100%) aqueous MeOH to yield three fractions (Fractions 1–3). The fractions were tested in plates for their antifungal activity, where a Foc TR4 mycelial plug (6 mm in diameter) was inoculated into 10 ml potato dextrose broth (PDB) per dish with 25 μg/ml of each fraction or DMSO (0.1% in sterile water, negative control). The most active fraction (Fraction 3) with visible inhibition was then passed through a silica gel (200 meshes) column (Sigma-Aldrich, Shanghai, China) and eluted with different ratios of methanol and chloroform (subfractions 1–3 were eluted with methanol and chloroform at the ratio of 1:3, 1:6, and 1:9, respectively).
Purification and Chemical Analysis of the Main Antifungal Component
The subfractions were also tested in the bioassays as described above, and the most active fraction was subjected to the analysis of the UV absorption spectrum (Shimadzu UV-2450, Japan). Liquid chromatographic separation was conducted in a reverse-phased C18 analytical column (Zorbax RRHD Eclipse Plus, 2.1 × 50 mm, 1.8 μm, Agilent, United States). A syringe pump delivering 5 μl was adjusted by the direct loop injection method (Hu et al., 2005). The mobile phase A was water with 0.1% acetic acid (v/v) and the mobile phase B was methanol. The chromatographic separation was held at the initial mobile phase composition (5% methanol) constant for 0.1 min, followed by a linear gradient to 95% methanol for 6 min, then decreased to 5% in 6–8 min, and followed by a one min post-run time after each analysis. All samples were performed on high performance liquid chromatography (HPLC) using a solvent containing 30% mobile phase A and 70% mobile phase B for 2 min, operated at a flow rate of 0.4 ml/min and the UV absorption at 365 nm. The HPLC-separated samples were then analyzed by quadrupole-time of flight mass spectrometer (TOF-MS) (Agilent1290-6545, United States). Electrospray ionization (ESI) was operated in negative ionization mode with other parameters used as following: capillary voltage was 4.0 kV; the temperature of dry gas (N2) was 200°C; the flow rate of dry gas was 9 L/h at 35 psig spray pressure; the temperature of sheath was 350°C; the flow rate of sheath was 10 L/h, and the ion scanning range was m/z 100∼1100. The collision energy was set at 10∼40 eV for the MS/MS analysis. The mass data of the molecular ions were processed through the Mass Hunter Qualitative Analysis (version B.07.00) software.
Meanwhile, the bioactive subfraction was solved in deuterated chloroform (CDCl3, δ = 77.0 ppm) at the concentration of 10 mg/ml for the 1H-NMR and 13C-NMR experiments with a 5 mm probe by a Bruker spectrometer (500 MHz/Advance III, Advance Digital, Bruker, Germany). The spectra were obtained at 30°C using tetramethylsilane (TMS) as the internal standard. The NMR spectra were analyzed by MestReNova (version 6.1.1-6384) software.
Determination of the Minimum Inhibitory Concentration (MIC) of PCA
A standardized 96-well microdilution broth assay (Wedge and Kuhajek, 1998) was used to evaluate the antifungal activity of the pure compound from Burkholderia sp. HQB-1.
The MICs of phenazine-1-carboxylic acid (PCA) from the cultural broth of the strain HQB-1 against the fungal pathogens including F. oxysporum, C. gloeosporioides, B. cinerea, and C. fallax. Different concentrations of PCA (50.0, 25.0, 12.5, 6.25, 3.13, 1.56, 0.78, 0.39 and 0.20 μg/ml) were prepared by using 2-fold serial dilutions for minimum inhibitory concentration (MIC) test. The lowest concentration of PCA that inhibited fungal growth was recorded as the MIC. The concentration of the inoculated fungal pathogens was 1 × 106 CFU/ml. Carbendazim was used as a positive control and 0.1% DMSO as a negative control. The 96-well plates were covered with a plastic lid and incubated at 25°C for 72 h. The absorbance was measured at 620 nm by a microplate photometer (BioTek, BioTek Instrument Co., United States). Mean absorbance and standard errors were used to evaluate fungal growth after 72 h incubation (Wang X. et al., 2013).
Biological Assays
Biological assays were carried out in July–September 2019 at the greenhouse in Huaqiao University, Xiamen. Pathogen suspensions were prepared as follows: a 6 mm diameters agar disk containing 7-day-old pathogen was added to a flask containing PDB. The flask was incubated at 28°C, 170 rpm for 7 days, and then the suspension was filtered through four layers of sterilized gauze. The conidial suspension was determined using a biological microscope (Nikon, Japan) and adjusted with distilled water to give a final concentration of 1.0 × 106 CFU/ml for further use.
The soil used in this study was a mixture of peat and perlite (2:1, v/v) and sterilized at 120°C for 2 h. Three to four true leaves old banana (Musa acuminata L.) AAA group, cv. Cavendish seedlings (Shishengyuan Biological Technology Co., Ltd., Gaozhou, China) were transplanted in plastic pots (8 cm × 8 cm, with one plant standing in one cup) with 80 g soil for pot experiment. The fungal pathogen F. oxysporum Tropical Race 4 with a concentration of 1.0 × 106 CFU/ml was inoculated into the potting substrates 5 days before PCA treatments. Six treatment group were conducted as follows: (1) CK1 (negative control, non-inoculated Foc TR4 and application of 0.1% DMSO); (2) CK2 (positive control, inoculated Foc TR4 and application with sterile water); (3) PCA 1 μg/ml; (4) PCA 5 μg/ml; (5) PCA 25 μg/ml; (6) PCA 50 μg/ml. Each PCA treatment group was watered weekly with 5 ml PCA at the concentration of 1, 5, 25, or 50 μg/ml mixed in sterile water. Seven plants were included for each treatment, and the experiment was repeated three times. The banana seedlings were placed on a greenhouse bench with natural light for 60 days and watered regularly.
The physiological indexes of banana seedlings treated for 60 days were determined, including root length, plant height, stem diameter, leaf width and dry weight. The disease symptoms on each plant were recorded and rated in five classes from 0 to 4 according to Mak et al. (2004). The disease index (DI) was calculated as DI (%) = Σ (Class × Number of plants in that class)/(4 × Total number of assessed plants) × 100. Biocontrol efficiency (BE) was calculated using the following formula: BE (%) = (DI in sterile water controlled - DI in treatments)/DI in sterile water controlled × 100.
Statistical Analysis
Statistical analysis was performed using SPSS 19. The difference among treatments was determined based on one-way analysis of variance (ANOVA), and means were subjected to Duncan’s multiple range test with significance set at P < 0.05.
Results
Isolation of the Strain HQB-1
A total of 11 bacterial isolates with 33.3 to 56.7% antagonistic effect was isolated from the soil samples in banana orchard. Among them, the isolate HQB-1 showed a more significant and stronger antagonistic activity against Foc TR4 on the plate (Figure 1) and exhibited a broad-spectrum antifungal activity against other plant pathogens. The growth inhibition of F. oxysporum, C. gloeosporioides, B. cinerea, and C. fallax was 56.7, 36.7, 52.4, and 52.2% respectively, after 5-day cultivation with HQB-1 (Table 1).
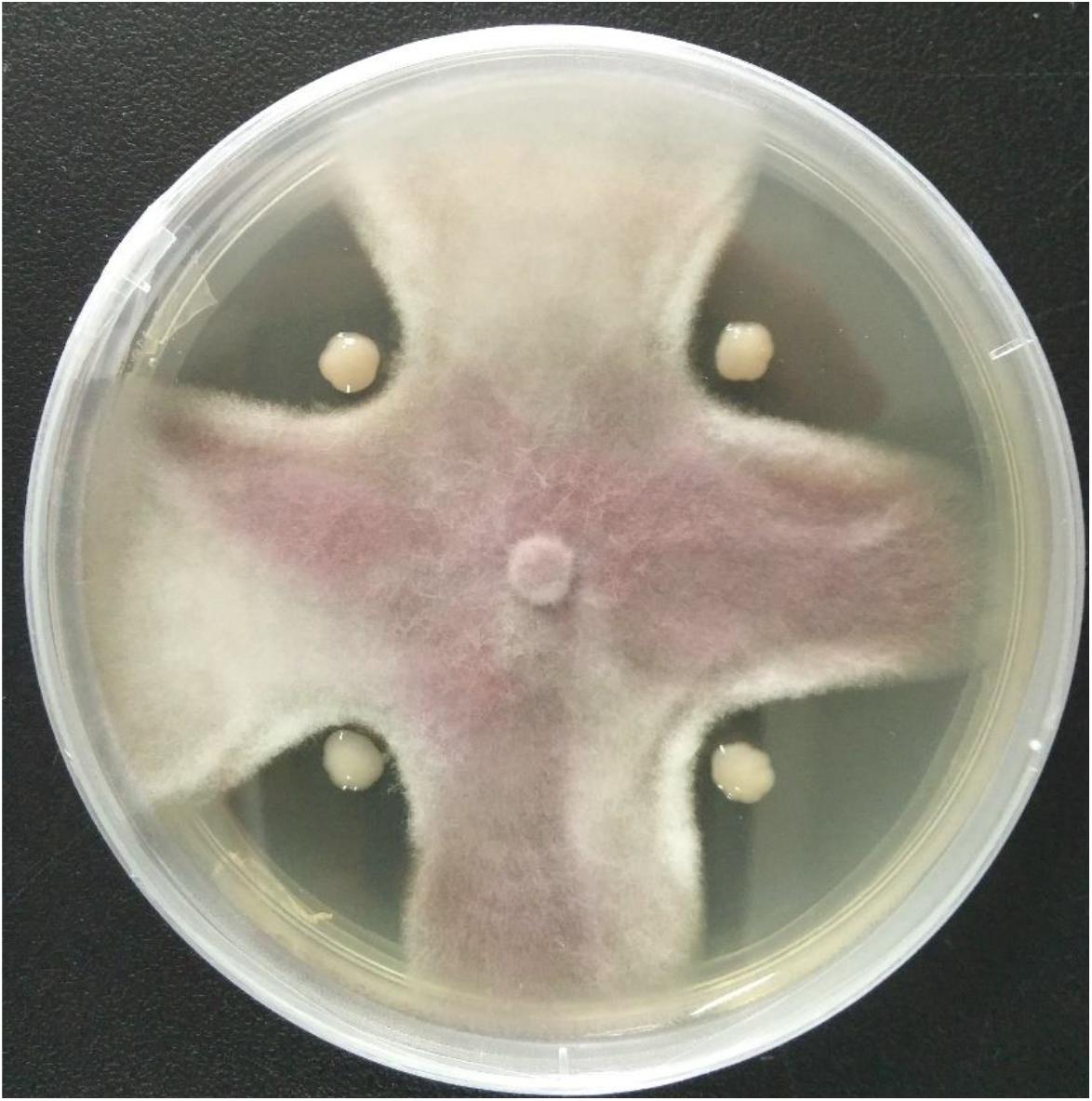
Figure 1. Antagonistic activity of the strain HQB-1 against Fusarium oxysporum Tropical Race 4 (Foc TR4). The co-cultivation HQB-1 and Foc TR4 grown on PDA medium at 28°C for 5 days.
Morphological, Physiological, and Biochemical Identification of the Strain HQB-1
The strain HQB-1 was slightly yellow, shining, and smooth in colony appearance on the nutrient agar (NA) plate. The biochemical tests indicated that it is a gram-negative and rod-shaped bacterium. The shape of the strain HQB-1 was straight rod, and the size was (1.2–2.5) μm × (0.5–1.0) μm (Figure 2) under scanning electron microscope (SEM). The tests of catalase, methyl red, oxidative or fermentative acid production from carbohydrates, liquefaction of gelatin, phenylalanine deaminase and indole were positive; and the Voges-Proskauer tests, tests for the presence of pigment, hydrolysis of starch, urease, Tween 80, H2S production, and creatine were negative. Besides, the strain HQB-1 can grow at a temperature range of 18–39°C.
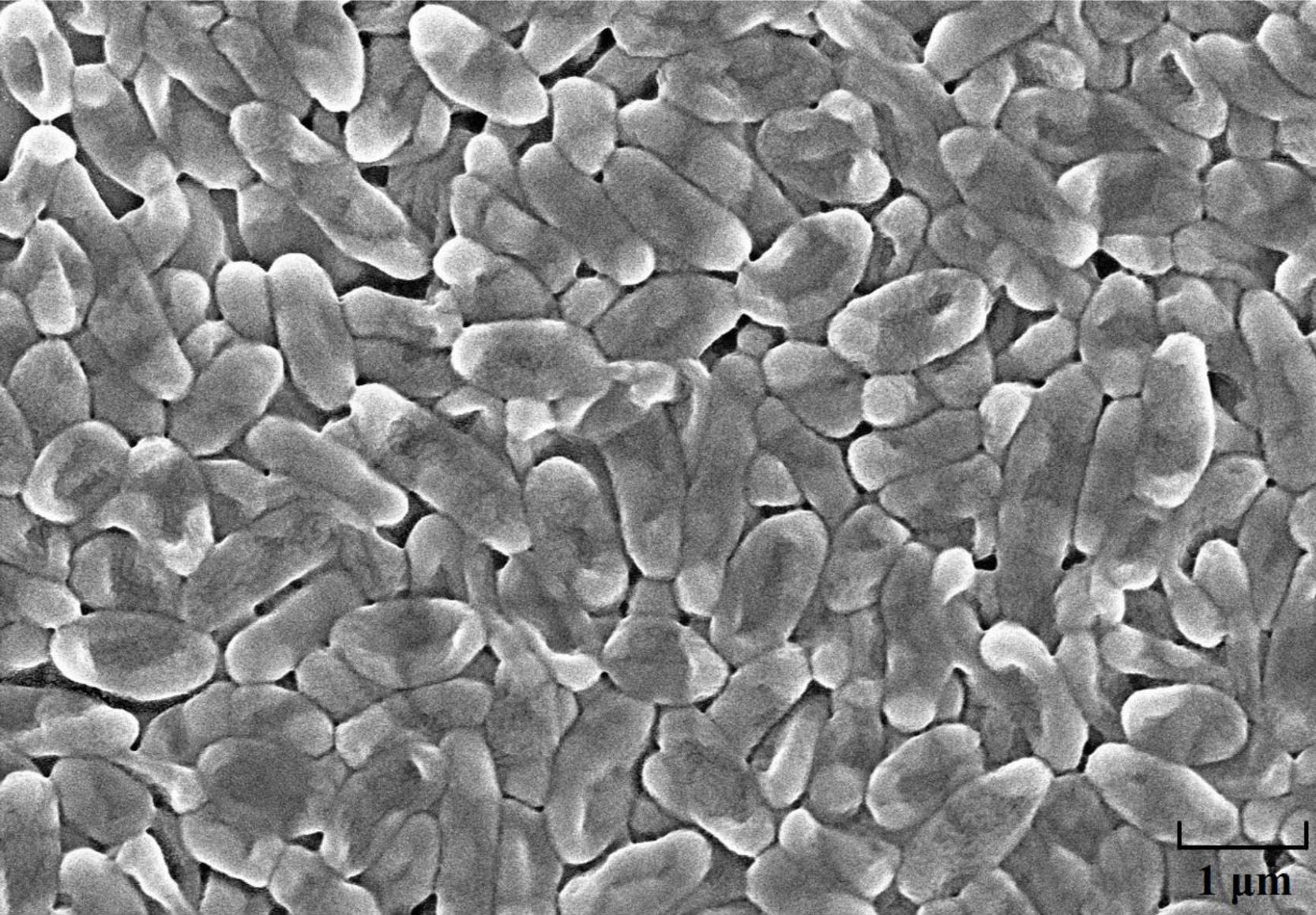
Figure 2. Scanning electron microscope image of HQB-1. Morphological characters of Burkholderia sp. HQB-1 viewed using SEM. Scale bar means 1 μm in the image.
16S rRNA Gene Sequencing and Phylogenetic Analysis of the Strain HQB-1
The 16S rRNA gene sequence (1471 bp) was submitted to GenBank with accession number MK612762. The sequence exhibited 99.7% similarity with Burkholderia stagnalis LMG 28156T (NR136495) by comparison against the EzBioCloud database and the BLASTn database in GenBank. After multiple alignments of sequences via CLUSTAL_X, a phylogenetic tree based on the neighbor-joining method using MEGA version 6.0 software demonstrated that HQB-1 fell into the cluster comprised of Burkholderia species (Figure 3). Therefore, based on morphological, physiological, and biochemical characteristics as well as the 16S rRNA gene sequence analysis, the strain HQB-1 is supposed to be affiliated to the genus Burkholderia.
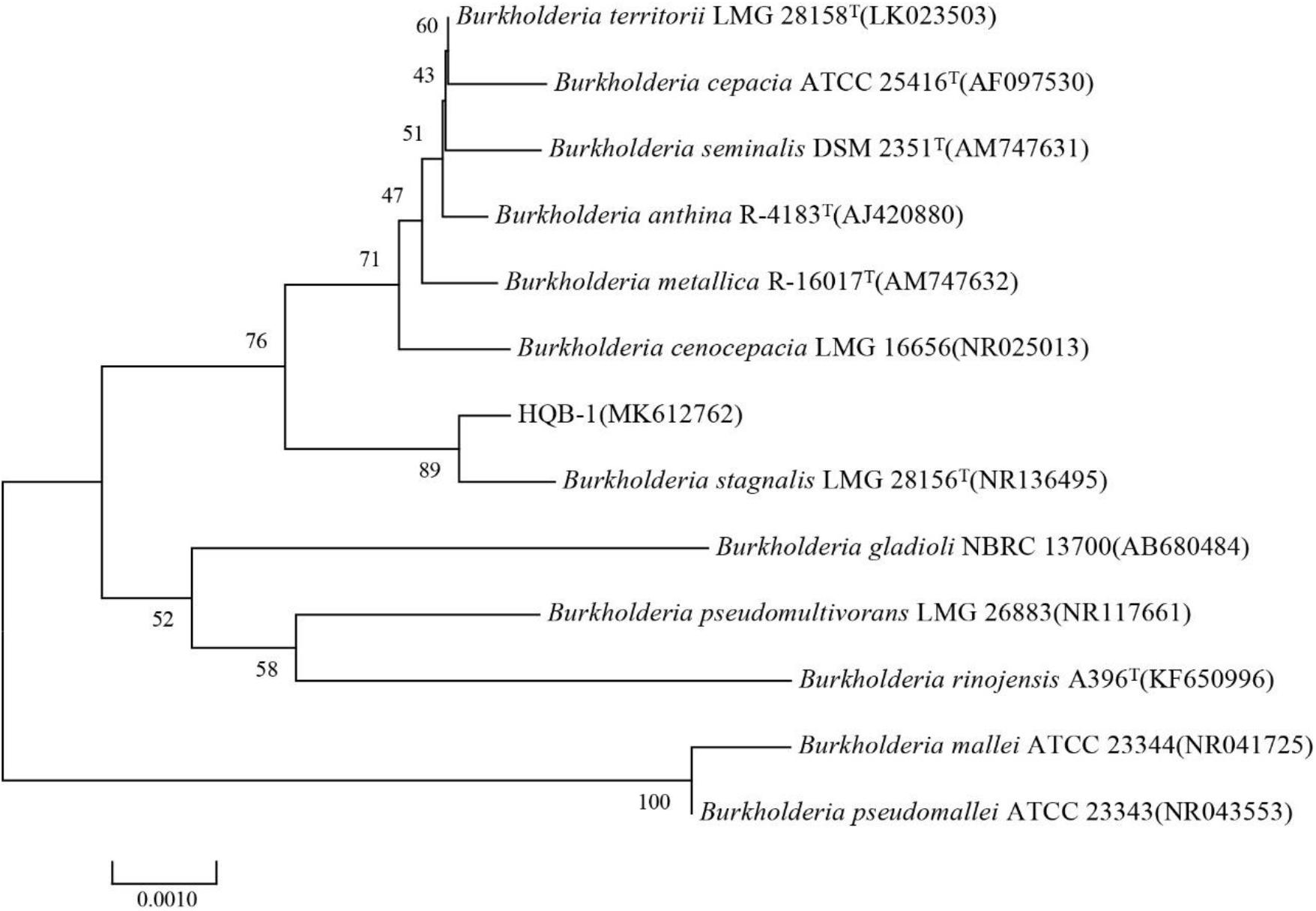
Figure 3. Phylogenetic tree of the strain HQB-1 based on 16S rRNA gene sequence analysis. The bootstrap values (%) presented at the branches were calculated from 1,000 replications. The numbers in parentheses are the GenBank accession numbers. The scale bar 0.001 represents 2 nucleotide substitutions per 1,000 nucleotides. T means type strain.
Separation and Purification of the Main Active Antifungal Component
Antifungal activity assays showed that the most active antifungal compound was eluted with 100% MeOH by macroporous resin column (Fraction 3) (Supplementary Figure 1). The subfraction with the strongest antifungal activity eluted in methanol and chloroform (1:9, v/v) was discovered using silica gel column chromatography (Supplementary Table 1). The solvent was slowly evaporated at room temperature to obtain greenish-yellow crystals. The chemical analysis of this active compound was performed by using UV and HPLC. The UV spectrum data (Figure 4A) showed a strong absorption and a broad peak at 365 nm, which indicated the presence of a phenazine moiety in the molecule. Then the active compound was subjected to HPLC with a C18 column, and a single peak with a retention time of 1.617 min under the 365 nm was detected (Figure 4B).
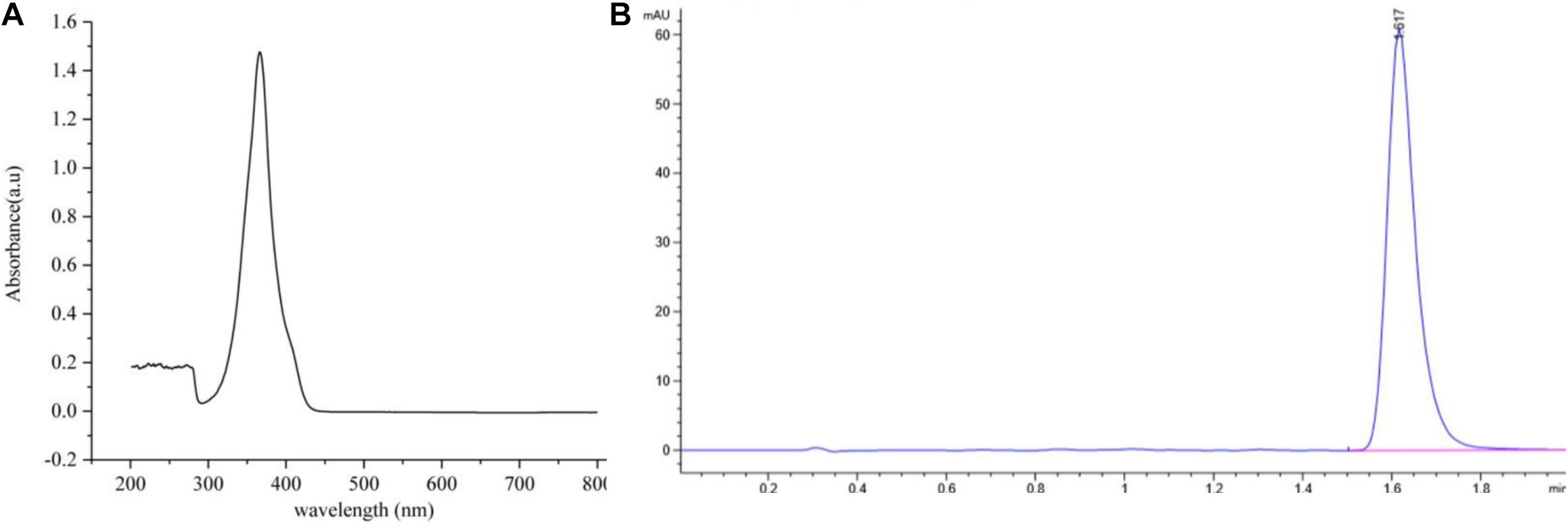
Figure 4. The ultraviolet and HPLC data of the main active antifungal component produced by Burkholderia sp. HQB-1. (A) The UV absorption of the main active subfraction at a wavelength of 0-800 nm, the peak means the maximum absorption at the wavelength of 365 nm; (B) Chromatogram of the main active component conducted in C18 column with detection at 365 nm. The flow rate was set at 0.4 ml⋅min–1 and the presence of a single peak at 1.617 min indicated that the high purity compound. Absorbance × Retention Time.
Structure Identification of the Main Active Antifungal Component
The main active antifungal component was then subjected to Q-TOF-MS and NMR. The MS spectra showed a molecular ion peak at m/z 223.0458 and the molecular formula as C13H8N2O2 was deduced (Figure 5A). The theoretical MS (Q-TOF) value of C13H8N2O2 ([M-H]–) was calculated for mass 223.0513 and in this study mass 223.0512 was found. The MS/MS spectra of the component displayed a fragment ion peak at m/z 179.0574, which may be due to the loss of COOH. The major fragment ion at m/z 179 was close to the relative molecular mass of a phenazine ring.
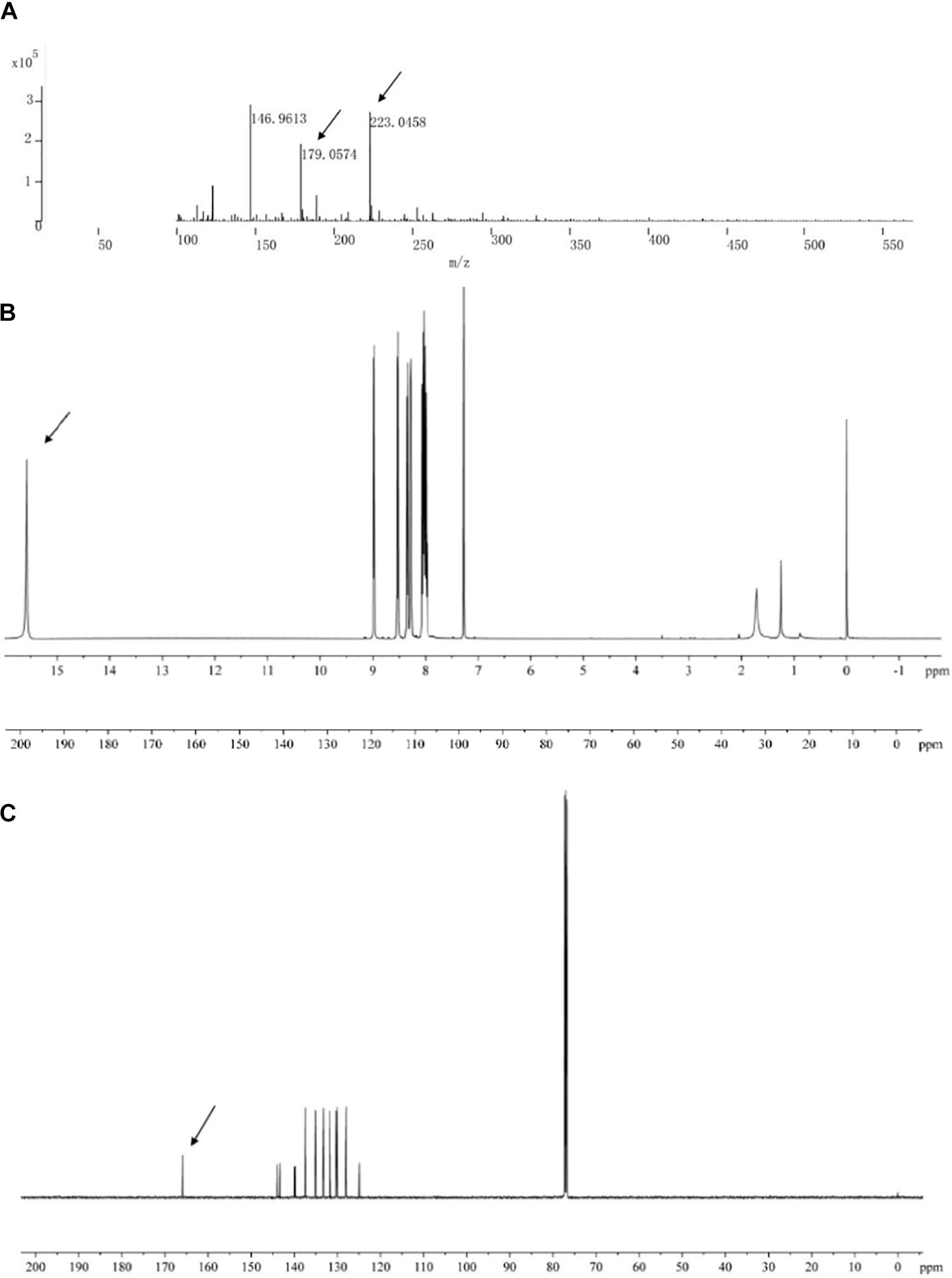
Figure 5. Chemical analysis of the main active antifungal component produced by Burkholderia sp. HQB-1. (A) liquid chromatography quadrupole-time of flight mass spectrometer (LC-Q-TOF-MS). The ion peaks pointed by the arrows indicate the presence of carbon and proton MS data of m/z 179.0574 and m/z 223.0458, respectively. (B,C) 1H-NMR and 13C-NMR spectrum. The spectral data (δ in ppm, J in Hz) were measured at 30°C with CD3Cl as the solvent. The tetramethylsilane (TMS) was used as internal standard.
The structure of the active compound was further identified by 1H-NMR and 13C-NMR spectroscopy. There were eight and thirteen well-resolved signals available in the 1H-NMR and 13C-NMR spectrum, respectively. Seven peaks in the 7.30–9.00 ppm region were assumed to be aromatic protons and the peak at 15.58 ppm was assigned to the carboxylic acid proton (Figure 5B). In the 13C-NMR spectrum, twelve peaks in 124.91–144.07 ppm were assigned to be aromatic carbons, which indicated a symmetrical structure for the molecule. The peak at 165.93 ppm clearly indicated the presence of carbonyl carbon (Figure 5C). Meanwhile, the NMR spectra were compared with Scifinder3 and SDBS information database4, which indicated the target molecule as phenazine-1-carboxylic acid (PCA) (C13H8N2O2) (Figure 6).
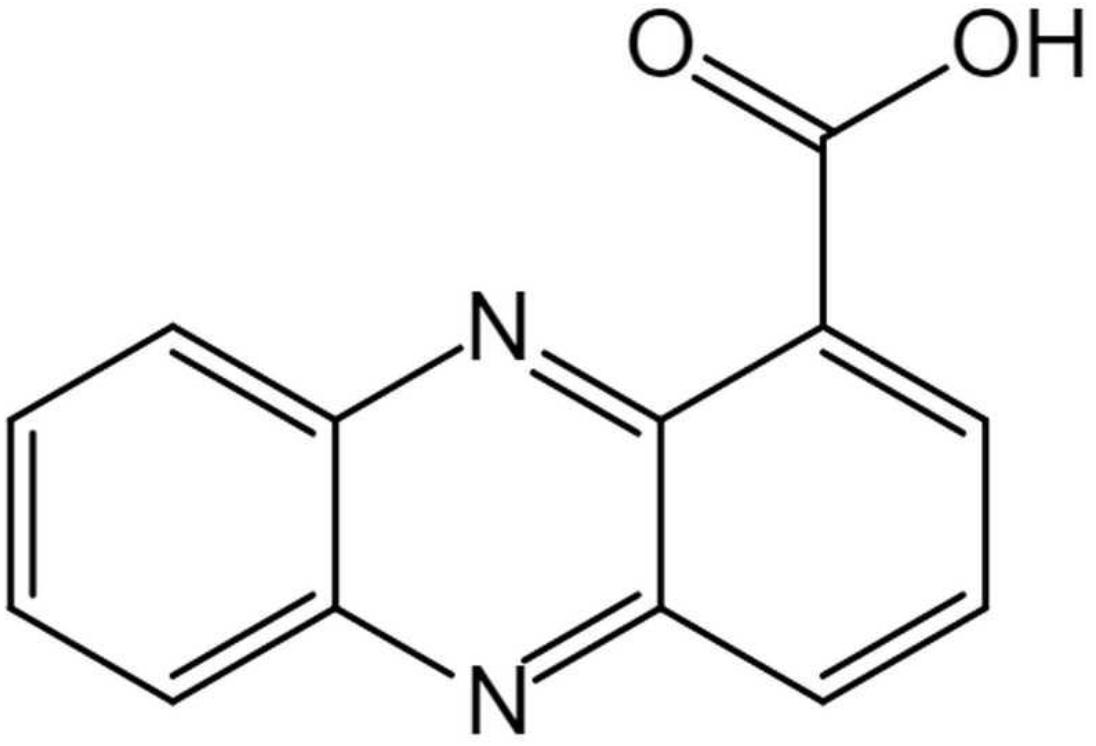
Figure 6. Chemical structure of (phenazine-1-carboxylic acid) PCA isolated produced by Burkholderia sp. HQB-1.
MIC of PCA
According to the 96-well microtiter assay, the MIC values of purified PCA against F. oxysporum, C. gloeosporioides, B. cinerea, and C. fallax were 1.56, 6.13, 1.56, and 3.13, respectively (Table 2), which were lower than carbendazim treatments used as positive control. The lowest MIC of PCA was 1.56 μg/ml against F. oxysporum and B. cinerea. Whereas, the highest MIC of PCA was 6.13 μg/ml against C. gloeosporioides. The negative control (0.1% DMSO treated) had no inhibitory effect on the tested pathogenic fungi.

Table 2. Values of minimum inhibitory concentration (MIC) of (phenazine-1-carboxylic acid) PCA against pathogenic fungi.
Effect of the Active Antifungal Component in Pot Experiments
The disease incidence measured for various treatments with different PCA concentrations (1, 5, and 25 μg/ml) were 40.8 ± 1.55, 19.1 ± 2.08, and 14.29% ± 1.84, respectively (Table 3). Meanwhile, the biocontrol efficiency was 51.2 ± 1.68, 77.2 ± 1.75, and 82.9% ± 1.43 respectively. PCA treatment at the concentration of 50 μg/ml completely inhibited Fusarium wilt in banana plants demonstrating strong antifungal activity against F. oxysporum in vivo (Figure 7). Furthermore, PCA treatments significantly promoted the growth of banana plants including root length, plant height, stem girth, leaf width, and dry weight compared with the control (Table 4). When the concentration of PCA reached more than 5 μg/ml, there was no significant difference in root length of banana plants between treatments. However, when the concentration of PCA reached more than 25 μg/ml, the difference in the growth of plant height was insignificant compared with DMSO treatment.

Figure 7. Biocontrol by treatments with different concentrations of phenazine-1-carboxylic acid (PCA) on banana plants. Plants were grown for 60 days and watered weekly using water mixed with 5 ml of PCA at different concentrations. From left to right: banana plants were treated by 50, 25, 5, and 1 μg/ml PCA, respectively; CK1, negative control, non-inoculated Foc TR4 and application of 0.1% DMSO; CK2, positive control, inoculated Foc TR4 and application with sterile water. Scale bar means 5 cm in the image.
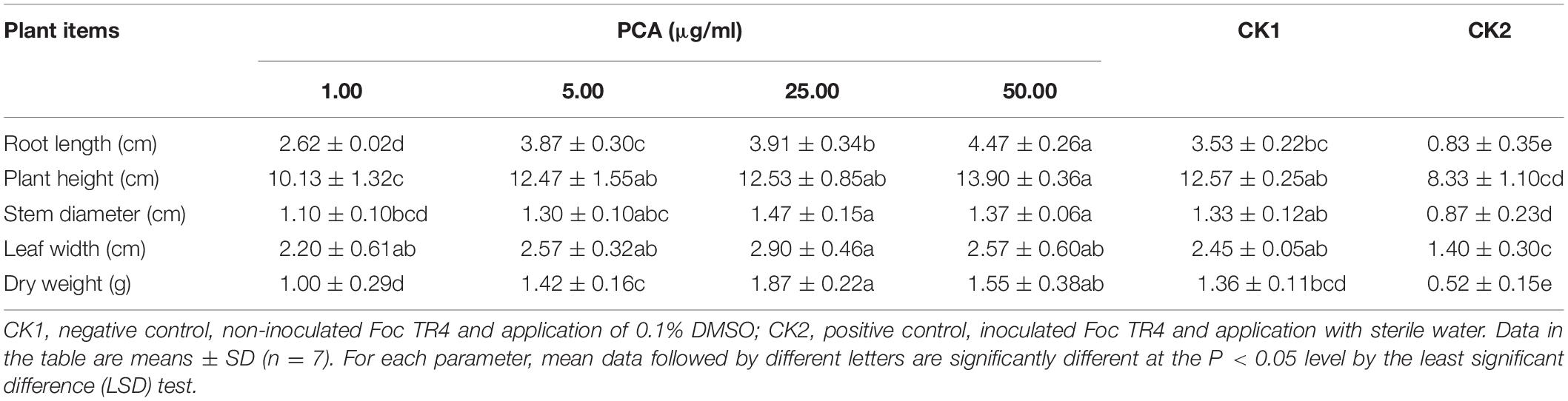
Table 4. Effects of different concentrations of (phenazine-1-carboxylic acid) PCA treatments on the growth of banana plants.
Discussion
Soil-borne plant diseases, especially Fusarium wilt, cause a significant decline of the production in banana field (Ploetz, 2005). Biological control is considered to be a novel and efficient way to prevent and inhibit banana Fusarium wilt. The screening of antagonistic bacteria from soil microorganisms in fields with banana Fusarium wilt is of great practical significance in controlling this disease (Duan et al., 2020). In the soil bacteria community of healthy banana plants, the Burkholderia may be one of the most abundant genera that contribute to suppressing Fusarium wilt (Shen et al., 2014). It was well demonstrated that most of the Burkholderia sp. strains isolated from the rhizosphere of healthy plants have been used for controlling such plant disease and promoting the growth of the plant (Parke and Gurian-Sherman, 2001; Compant et al., 2008; Gu et al., 2009). In our study, the Burkholderia sp. strain HQB-1 was isolated and screened from banana rhizosphere soil. It exhibited an excellent antagonistic effect (56.7 ± 0.64%) against F. oxysporum and a broad-spectrum antifungal activity, which indicated that Burkholderia sp. HQB-1 has the potential to be a promising candidate against banana Fusarium wilt.
The genus Burkholderia was used as a biological agent against phytopathogenic fungi mainly due to its production of plentiful metabolites with antimicrobial activity (Hu and Young, 1998). In this study, to identify the potential antagonistic mechanisms of Burkholderia sp. HQB-1, we investigated the effect of the secondary metabolites secreted by this strain on inhibiting the mycelial growth of Foc TR4. The HQB-1 cultural broth was filtered through the 0.22 μm microfiltration membranes for salting-out and dialysis, whereas, the antimicrobial tests indicated that neither the proteins nor the enzymes in the HQB-1 metabolites possessed antifungal activity. The siderophore produced by the strain HQB-1 was determined by the chrome azurol S (CAS) method (Bultreys et al., 2001), but the quantitative tests showed that HQB-1 was not high-yield in siderophore (data not shown). However, the crude HQB-1 ethyl acetate extract possessed antifungal activity against F. oxysporum, which suggested that the secondary metabolites secreted from HQB-1 might contain antimicrobials. A greenish-yellow crystal was further purified by column macroporous resin and silica gel column chromatography, which exhibited a strong antifungal activity against Foc TR4 mycelial plug in plates. The antifungal component showed a strong absorbance peak at 365 nm (Figure 4A) with UV spectral analysis, which indicated that the antifungal component might be a molecule bearing phenazine moiety (Hu et al., 2005). Furthermore, the 1H-NMR spectrum (Figure 5B) revealed the presence of 8 aromatic protons and an ABCD pattern in the aromatic ring, the characteristics of which are typically found in phenazine derivatives. The 13C-NMR spectrum (Figure 5C) revealed the presence 13 aromatic carbons that indicated a highly symmetrical structure for the target molecule (Gurusiddaiah et al., 1986). Both of the peaks at 15.58 ppm in 1H-NMR and at 165.93 ppm in the 13C-NMR spectrum demonstrated the presence of a carboxyl. Additionally, by compared with the NMR spectrum data in the SDBS information database, the chemical structure of the antifungal component was confirmed as phenazine-1-carboxylic acid (PCA) (Figure 6; Abraham et al., 2015).
Gurusiddaiah et al. (1986) found that the MIC values of PCA isolated from Pseudomonas fluorescens strain 2-79 against F. oxysporum was 25–30 μg/ml using the agar dilution method. PCA at 3.12 μg/ml reduced the mycelial growth of B. cinerea by 50%, and inhibited the formation of exopolysaccharide (Simionato et al., 2017). Zheng et al. (2019) reported that the MIC value of crude extracts of Streptomyces sp. FJAT-31547 against F. oxysporum was 6.250 μg/ml. In this study, the MIC values of PCA against plant pathogenic fungi were 1.563–6.13 μg/ml according to the results from 96-well microtiter assay (Table 2). The lowest MIC was 1.56 μg/ml against F. oxysporum, which indicated that PCA was more efficient than carbendazim. Thus, we proposed that PCA produced by Burkholderia sp. HQB-1 might be a key factor for the bacterial broad-spectrum antifungal activity against plant phytopathogenic fungi.
Phenazine-1-carboxylic acid has been reported to play an important role in inhibiting F. oxysporum in diverse crops such as chickpea, tomato, and wheat (Anjaiah et al., 1998; Chin-A-Woeng et al., 1998; Pastor et al., 2016). It seemed to be a more contributor to biocontrol Fusarium oxysporum than pyrrolnitrin in Pseudomonas fluorescens (Upadhyay and Srivastava, 2011). PCA is the main active component of the newly registered green biopesticide “Shenqinmycin,” and its broad-spectrum inhibition of plant fungal pathogens of has been applied (Jin et al., 2015). The Shenqinmycin suspension with a concentration as low as 1% can prevent rice sheath blight, pepper blight, and cucumber seedling damping-off (Xu, 2013). However, the biocontrol effect of PCA against banana Fusarium wilt has not been well studied. In the present study, we investigated not only the antifungal activity of the PCA produced by HQB-1 against several phytopathogenic fungi was effective both in vitro and in vivo. Most of the banana plants in the current study could be efficiently protected from F. oxysporum infection, thus preventing Fusarium wilt, by 5 μg/ml PCA (BE, 77.2 ± 1.75%). Treatment with PCA at 500 μg/ml could protect most of the pepper plants from Phytophthora infection, and it also showed significant protective effect against anthracnose development on cucumber leaves (Lee et al., 2003). However, the mechanism of the antifungal effect of PCA remains unclear. Previous research reported that phenazine compounds are involved in inhibiting RNA synthesis and DNA binding (Morrison and Marley, 1976; Palchykovska et al., 2008), thereby interfering with redox balance and inducing the generation of reactive oxygen species (ROS) which eventually causes fungi death (Morales et al., 2010). Moreover, the phenazines may also have important physiological functions within phenazine-producing bacteria themselves. For example, it has been reported that the phenazine derivative pyocyanin leads to decreased levels of NADH in Pseudomonas aeruginosa grown under anoxic condition (Xu et al., 2013). We also found that proper concentrations (5–25 μg/ml) of PCA could increase the height and biomass, especially the root length of the banana plants compare with CK2 group (Table 4). This indicated the potential of PCA in facilitating water uptake, nutrient mobilization and growth promotion in banana plants. Thus, we propose that PCA might be critical for the broad-spectrum antifungal activity of Burkholderia sp. HQB-1 and it possesses a strong biocontrol effect on banana plants. The study of the antagonistic mechanism of PCA against F. oxysporum in banana and its practical application needs to be further investigation.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
Author Contributions
ZX and MW conceived the study and prepared the manuscript. ZX, JD, and TH performed the experiments and analyzed the data. MW, JL, TD, and YC supervised and provided the suggestion of the research work. YC helped to revise the manuscript. All authors have read and approved the manuscript before submission.
Funding
This project was funded by the Promotion Program for Young and Middle-aged Teacher in Science and Technology Research of Huaqiao University (ZQN-YX507), the Special Project on Education and Research of Fujian Province (2017N5009), and the Subsidized Project for Postgraduates’ Innovative Fund in Scientific Research of Huaqiao University (17013087004).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We greatly thank Dr. Dengbo Zhou of the Institute of Tropical Bioscience and Biotechnology, China Academy of Tropical Agricultural Sciences, Haikou, China for providing the phytopathogens. Additionally, we thank colleagues of the Instrumental Analysis Center of Huaqiao University for their support and providing access to the facilities needed for the chemical analysis of the microbial metabolites.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.605152/full#supplementary-material
Footnotes
- ^ http://eztaxon-e.ezbiocloud.net/
- ^ https://blast.ncbi.nlm.nih.gov/Blast.cgi
- ^ http://scifinder.cas.org
- ^ https://sdbs.db.aist.go.jp/
References
Abraham, A., Philip, S., Jacob, M. K., Narayanan, S. P., Jacob, C. K., and Kochupurackal, J. (2015). Phenazine-1-carboxylic acid mediated anti-oomycete activity of the endophytic Alcaligenes sp. eil-2 against Phytophthora meadii. Microbiol. Res. 170, 229–234. doi: 10.1016/j.micres.2014.06.002
Anjaiah, V., Koedam, N., Nowak-Thompson, B., Loper, J. E., Höfte, M., Tambong, J. T., et al. (1998). Involvement of phenazines and anthranilate in the antagonism of Pseudomonas aeruginosa PNA1 and Tn5 derivatives toward Fusarium spp. and Pythium spp. Mol. Plant Microbe Interact. 11, 847–854. doi: 10.1094/MPMI.1998.11.9.847
Bubici, G., Kaushal, M., Prigigallo, M. I., Gómez-Lama Cabanás, C., and Mercado-Blanco, J. (2019). Biological control agents against Fusarium wilt of banana. Front. Microbiol. 10:616. doi: 10.3389/fmicb.2019.00616
Buchanan, R. E., and Gibbons, N. E. (1984). Bergey’s Manual of Determinative Bacteriology, 8th Edn. Baltimore, MD: Williams & Wilkins Co.
Bultreys, A., Gheysen, I., Maraite, H., and de Hoffmann, E. (2001). Characterization of fluorescent and non-fluorescent peptide siderophores produced by Pseudomonas syringae strains and their potential use in strain identification. Appl. Environ. Microb. 67, 1718–1727. doi: 10.1128/AEM.67.4.1718-1727.2001
Cartwright, D. K., Chilton, W. S., and Benson, D. M. (1995). Pyrrolnitrin and phenazine production by Pseudomonas cepacia, strain 5.5B, a biocontrol agent of Rhizoctonia solani. Appl. Microbiol. Biotechnol. 43, 211–216. doi: 10.1007/BF00172814
Chellemi, D. O., Gamliel, A., Katan, J., and Subbarao, K. V. (2016). Development and deployment of systems-based approaches for the management of soilborne plant pathogens. Phytopathology 106, 216–225. doi: 10.1094/PHYTO-09-15-0204-RVW
Chen, B., Zhang, X. Y., Huang, X., Liu, X. Y., Zhou, D. B., Tan, X., et al. (2013). Diversity analysis of soil fungi in Fusarium wilt of banana disease area. Jiangsu Agric. Sci. 41, 354–357.
Chen, Y., Zhou, D., Qi, D., Gao, Z., Xie, J., and Luo, Y. (2018). Growth promotion and disease suppression ability of a Streptomyces sp. CB-75 from banana rhizosphere soil. Front. Microbiol. 8:2704. doi: 10.3389/fmicb.2017.02704
Chin-A-Woeng, T. F. C., Bloemberg, G. V., van der Bij, A. J., van der Drift, K. M. G. M., Schripsema, J., Kroon, B., et al. (1998). Biocontrol by Phenazine-1-carboxamide-Producing Pseudomonas chlororaphis PCL1391 of tomato root rot caused by Fusarium oxysporum f. sp. radicis-lycopersici. Mol. Plant Microbe Interact. 11, 1069–1077. doi: 10.1094/MPMI.1998.11.11.1069
Compant, S., Nowak, J., Coenye, T., Clément, C., and Ait Barka, E. (2008). Diversity and occurrence of Burkholderia spp. in the natural environment. FEMS Microbiol. Rev. 32, 607–626. doi: 10.1111/j.1574-6976.2008.00113.x
Dale, J., James, A., Paul, J., Khanna, H., Smith, M., Peraza-Echeverria, S., et al. (2017). Transgenic cavendish bananas with resistance to Fusarium wilt tropical race 4. Nat. Commun. 8:1496. doi: 10.1038/s41467-017-01670-6
Davis, R. D., Moore, N. Y., and Kochman, J. K. (1996). Characterisation of a population of Fusarium oxysporum f. sp. vasinfectum causing wilt of cotton in Australia. Aust. J. Agric. Res. 47, 1143–1156. doi: 10.1071/AR9961143
Duan, Y., Chen, J., He, W., Chen, J., Pang, Z., Hu, H., et al. (2020). Fermentation optimization and disease suppression ability of a Streptomyces ma. FS-4 from banana rhizosphere soil. BMC Microbiol. 20:24. doi: 10.1186/s12866-019-1688-z
Eberl, L., and Vandamme, P. (2016). Members of the genus Burkholderia: good and bad guys. F1000res. 5:1007. doi: 10.12688/f1000research.8221.1
Fourie, G., Steenkamp, E. T., Ploetz, R. C., Gordon, T. R., and Viljoen, A. (2011). Current status of the taxonomic position of Fusarium oxysporum formae specialis cubense within the Fusarium oxysporum complex. Infect. Genet. Evol. 11, 533–542. doi: 10.1016/j.meegid.2011.01.012
Fuchs, J. G., Moënne-Loccoz, Y., and Défago, G. (1999). Ability of nonpathogenic Fusarium oxysporum Fo47 to protect tomato against Fusarium wilt. Biol. Control 14, 105–110. doi: 10.1006/bcon.1998.0664
Getha, K., Vikineswary, S., Wong, W. H., Seki, T., Ward, A., and Goodfellow, M. (2005). Evaluation of Streptomyces sp. strain g10 for suppression of Fusarium wilt and rhizosphere colonization in pot-grown banana plantlets. J. Ind. Microbiol. Biotechnol. 32, 24–32. doi: 10.1007/s10295-004-0199-5
Gu, G. Y., Wang, N., Noel, C., Leif, S., and Lu, E. S. (2009). Ambr1 is a key transcriptional regulator for production of antifungal activity of Burkholderia contaminans strain MS14. FEMS Microbiol. Lett. 1, 54–60. doi: 10.1111/j.1574-6968.2009.01653.x
Gupta, V. K., Shivasharanappa, N., Kumar, V., and Kumar, A. (2014). Diagnostic evaluation of serological assays and different gene based PCR for detection of Brucella melitensis in goat. Small Rumin. Res. 117, 94–102. doi: 10.1016/j.smallrumres.2013.11.022
Gurusiddaiah, S., Weller, D. M., Sarkar, A., and Cook, R. J. (1986). Characterization of an antibiotic produced by a strain of Pseudomonas fluorescens inhibitory to Gaeumannomyces graminis var. tritici and Pythium spp. Antimicrob. Agents Chemother. 29, 488–495. doi: 10.1128/AAC.29.3.488
Hu, F. P., and Young, J. M. (1998). Biocidal activity in plant pathogenic Acidovorax, Burkholderia, Herbaspirillum, Ralstonia and Xanthomonas spp. J. Appl. Microbiol. 84, 263–271. doi: 10.1046/j.1365-2672.1998.00340.x
Hu, H. B., Yu, Q. X., Feng, C., Xue, H. Z., and Hur, B. K. (2005). Isolation and characterization of a new fluorescent Pseudomonas strain that produces both phenazine 1-carboxylic acid and pyoluteorin. J. Microbiol. Biotechnol. 15, 86–90.
Jeong, Y., Kim, J., Kim, S., Kang, Y., Nagamatsu, T., and Hwang, I. (2003). Toxoflavin produced by Burkholderia glumae causing rice grain rot is responsible for inducing bacterial wilt in many field crops. Plant Dis. 87, 890–895. doi: 10.1094/PDIS.2003.87.8.890
Jiang, C., Sheng, X., Qian, M., and Wang, Q. (2008). Isolation and characterization of a heavy metal-resistant Burkholderia sp. from heavy metal-contaminated paddy field soil and its potential in promoting plant growth and heavy metal accumulation in metal-polluted soil. Chemosphere 72, 157–164. doi: 10.1016/j.chemosphere.2008.02.006
Jin, K., Zhou, L., Jiang, H., Sun, S., Fang, Y., Liu, J., et al. (2015). Engineering the central biosynthetic and secondary metabolic pathways of Pseudomonas aeruginosa strain PA1201 to improve phenazine-1-carboxylic acid production. Metab. Eng. 32, 30–38. doi: 10.1016/j.ymben.2015.09.003
Kanini, G. S., Katsifas, E. A., Savvides, A. L., and Karagouni, A. D. (2013). Streptomyces rochei ACTA1551, an indigenous Greek isolate studied as a potential biocontrol agent against Fusarium oxysporum f. sp. lycopersici. BioMed Res. Int. 12:387230. doi: 10.1155/2013/387230
Kim, O. S., Cho, Y. J., Lee, K., Yoon, S. H., Kim, M., Na, H., et al. (2012). Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int. J. Syst. Evol. Microbiol. 62, 716–721. doi: 10.1099/ijs.0.038075-0
Kunakom, S., and Eustáquio, A. S. (2019). Burkholderia as a source of natural products. J. Nat. Prod. 82, 2018–2037. doi: 10.1021/acs.jnatprod.8b01068
Lee, J. Y., Moon, S. S., and Hwang, B. K. (2003). Isolation and in vitro and in vivo activity against Phytophthora capsici and Colletotrichum orbiculare of phenazine-1-carboxylic acid from Pseudomonas aeruginosa strain GC-B26. Pest Manag. Sci. 59, 872–882. doi: 10.1002/ps.688
Leeman, M., den Ouden, F. M., Dirkx, F. P. M., Steijl, H., Bakker, P. A. H. M., and Schippers, B. (1996). Iron availability affects induction of systemic resistance to Fusarium wilt of radish by Pseudomonas fluorescens. Phytopathology 86, 149–155. doi: 10.1094/Phyto-86-149
Li, C., Yang, J., Li, W., Sun, J., and Peng, M. (2017). Direct root penetration and rhizome vascular colonization by Fusarium oxysporum f. sp. cubense are the key steps in the successful infection of Brazil Cavendish. Plant Dis. 101, 2073–2078. doi: 10.1094/PDIS-04-17-0467-RE
Lin, Y., Chang, J., Liu, E., Chao, C., Huang, J., and Chang, P. L. (2009). Development of a molecular marker for specific detection of Fusarium oxysporum f. sp. cubense race 4. Eur. J. Plant Pathol. 123, 353–365. doi: 10.1007/s10658-008-9372-4
Mahenthiralingam, E., Baldwin, A., and Dowson, C. G. (2008). Burkholderia cepacia complex bacteria: opportunistic pathogens with important natural biology. J. Appl. Microbiol. 104, 1539–1551. doi: 10.1111/j.1365-2672.2007.03706.x
Mak, C., Mohamed, A. A., Liew, K. W., and Ho, Y. W. (2004). “Early screening technique for Fusarium wilt resistance in banana micropropagated plants,” in Banana Improvement: Cellular, Molecular, Biology, and Induced Mutations, eds S. M. Jain R. Swennen (Enfield, NH: Science Publishers, Inc), 219–227.
Morales, D. K., Jacobs, N. J., Rajamani, S., Krishnamurthy, M., Cubillos-Ruiz, J. R., and Hogan, D. A. (2010). Antifungal mechanisms by which a novel Pseudomonas aeruginosa phenazine toxin kills Candida albicans in biofilms. Mol. Microbiol. 78, 1379–1392. doi: 10.1111/j.1365-2958.2010.07414.x
Morrison, N. E., and Marley, G. M. (1976). The mode of action of clofazimine DNA binding studies. Int. J. Lepr. Other. Mycobact. Dis. 44, 133–134.
Nimaichand, S., Devi, A. M., Tamreihao, K., Ningthoujam, D. S., and Li, W. J. (2015). Actinobacterial diversity in limestonedeposit sites in Hundung, Manipur (India) and their antimicrobial activities. Front. Microbiol. 6:413. doi: 10.3389/fmicb.2015.00413
O’Donnell, K., Kistler, H. C., Cigelnik, E., and Ploetz, R. C. (1998). Multiple evolutionary origins of the fungus causing Panama disease of banana: concordant evidence from nuclear and mitochondrial gene genealogies. P. Natl. Acad. Sci. U.S.A. 95, 2044–2049. doi: 10.1073/pnas.95.5.2044
Palchykovska, L. G., Alexeeva, I. V., Kostina, V. G., Platonov, M. O., and Shved, A. D. (2008). New amides of phenazine-1-carboxylic acid: antimicrobial activity and structure-activity relationship. Ukr. Biokhim. Zh. 3, 140–147.
Parke, J. L., and Gurian-Sherman, D. (2001). Diversity of the Burkholderia Cepacia complex and implications for risk assessment of biological control strains. Annu. Rev. Phytopathol. 39, 225–258. doi: 10.1146/annurev.phyto.39.1.225
Parker, W. L., Rathnum, M. L., Seiner, V., Trejo, W. H., Principe, P. A., and Sykes, R. B. (1984). Cepacin A and cepacin B, two new antibiotics produced by Pseudomonas cepacia. J. Antibiot. 37, 431–440. doi: 10.7164/antibiotics.37.431
Pastor, N., Masciarelli, O., Fischer, S., Luna, V., and Rovera, M. (2016). Potential of Pseudomonas putida PCI2 for the protection of tomato plants against fungal pathogens. Curr. Microbiol. 73, 346–353. doi: 10.1007/s00284-016-1068-y
Ploetz, R. C. (2005). Fusarium-induced diseases of tropical, perennial crops. Phytopathology 96, 648–652. doi: 10.1094/PHYTO-96-0648
Reyes-Chilpa, R., Jimenez-Estrada, M., and Estrada-Muniz, E. (1997). Antifungal Xanthones from Calophyllum brasiliensis heartwood. J. Chem. Ecol. 23, 1901–1911. doi: 10.1023/B:JOEC.0000006459.88330.61
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. doi: 10.1093/oxfordjournals.molbev.a040454
Saravanan, T., Muthusamy, M., and Marimuthu, T. (2003). Development of integrated approach to manage the fusarial wilt of banana. Crop Prot. 9, 1117–1123. doi: 10.1016/S0261-2194(03)00146-7
Schmidt, S., Blom, J. F., Pernthaler, J., Berg, G., and Eberl, L. (2010). Production of the antifungal compound pyrrolnitrin is quorum sensing-regulated in members of the Burkholderia cepacia complex. Environ. Microbiol. 11, 1422–1437. doi: 10.1111/j.1462-2920.2009.01870.x
Shen, Z. Z., Wang, D. S., Ruan, Y. Z., Xue, C., Zhang, J., Li, R., et al. (2014). Deep 16S rRNA pyrosequencing reveals a bacterial community associated with banana Fusarium wilt disease suppression induced by bio-organic fertilizer application. PLoS One 9:e98420. doi: 10.1371/journal.pone.0098420
Shepherd, K. (1987). Banana breeding—past and present. Acta Hortic. 196, 37–43. doi: 10.17660/ActaHortic.1987.196.3
Simionato, A. S., Navarro, M. O. P., de Jesus, M. L. A., Barazetti, A. R., da Silva, C. S., Simões, G. C., et al. (2017). The effect of phenazine-1-carboxylic acid on mycelial growth of Botrytis cinerea produced by Pseudomonas aeruginosa LV strain. Front. Microbiol. 8:1102. doi: 10.3389/fmicb.2017.01102
Singh, A., Goel, Y., Rai, A. K., and Banerjee, U. C. (2013). Lipase catalyzed kinetic resolution for the production of (S)-3-[5-(4-fluoro-phenyl)-5-hydroxy-pentanoyl]-4-phenyl-oxazolidin-2-one: an intermediate for the synthesis of ezetimibe. J. Mol. Catal. B Enzym. 85-86, 99–104. doi: 10.1016/j.molcatb.2012.08.014
Tamura, K., Dudley, J., Nei, M., and Kumar, S. (2007). MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24, 1596–1599. doi: 10.1093/molbev/msm092
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F., and Higgins, D. G. (1997). The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882. doi: 10.1093/nar/25.24.4876
Upadhyay, A., and Srivastava, S. (2011). Phenazine-1-carboxylic acid is a more important contributor to biocontrol Fusarium oxysporum than pyrrolnitrin in Pseudomonas fluorescens strain Psd. Microbiol. Res. 4, 323–335. doi: 10.1016/j.micres.2010.06.001
Viljoen, A. (2002). The status of Fusarium wilt (Panama disease) of banana in South Africa. S. Afr. J. Sci. 98, 341–344.
Wang, B., Yuan, J., Zhang, J., Shen, Z., Zhang, M., Li, R., et al. (2013). Effects of novel bioorganic fertilizer produced by Bacillus amyloliquefaciens W19 on antagonism of Fusarium wilt of banana. Biol. Fertil. Soils 49, 435–446. doi: 10.1007/s00374-012-0739-5
Wang, X., Radwan, M. M., Taráwneh, A. H., Gao, J., Wedge, D. E., Rosa, L. H., et al. (2013). Antifungal activity against plant pathogens of metabolites from the endophytic fungus Cladosporium cladosporioides. J. Agric. Food Chem. 61, 4551–4555. doi: 10.1021/jf400212y
Warman, N. M., and Aitken, E. A. B. (2018). The movement of Fusarium oxysporum f. sp. cubense (Sub-Tropical Race 4) in susceptible cultivars of banana. Front. Plant Sci. 9:1748. doi: 10.3389/fpls.2018.01748
Wedge, D. E., and Kuhajek, J. M. (1998). A microbioassay for fungicide discovery. SAAS Bull. Biochem. Biotechnol. 11, 1–7.
Xu, N., Ahuja, E. G., Janning, P., Mavrodi, D. V., Thomashow, L. S., and Blankenfeldt, W. (2013). Trapped intermediates in crystals of the FMN-dependent oxidase PhzG provide insight into the final steps of phenazine biosynthesis. Acta Crystallogr. D 69, 1403–1413. doi: 10.1107/S0907444913008354
Xu, Y. (2013). “Genomic features and regulation of phenazine biosynthesis in the rhizosphere strain Pseudomonas aeruginosa M18,” in Microbial Phenazines, eds S. Chincholkar and L. Thomashow (Berlin: Springer), 177–198. doi: 10.1007/978-3-642-40573-0_9
Yoon, S., Ha, S., Kwon, S., Lim, J., Kim, Y., Seo, H., et al. (2017). Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 67, 1613–1617. doi: 10.1099/ijsem.0.001755
Keywords: antifungal activity, Burkholderia, banana Fusarium wilt, LC-Q-TOF-MS, phenazine-1-carboxylic acid
Citation: Xu Z, Wang M, Du J, Huang T, Liu J, Dong T and Chen Y (2020) Isolation of Burkholderia sp. HQB-1, A Promising Biocontrol Bacteria to Protect Banana Against Fusarium Wilt Through Phenazine-1-Carboxylic Acid Secretion. Front. Microbiol. 11:605152. doi: 10.3389/fmicb.2020.605152
Received: 11 September 2020; Accepted: 18 November 2020;
Published: 10 December 2020.
Edited by:
Yong Wang, Guizhou University, ChinaReviewed by:
Orlando Borras-Hidalgo, Qilu University of Technology, ChinaSonia Rippa, University of Technology Compiegne, France
Copyright © 2020 Xu, Wang, Du, Huang, Liu, Dong and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mingyuan Wang, w_mingyuan@163.com; Yinglong Chen, yinglong.chen@uwa.edu.au
 Zhizhou Xu
Zhizhou Xu Mingyuan Wang
Mingyuan Wang Jinpeng Du1
Jinpeng Du1