- 1Department of Plant and Soil Sciences, University of Pretoria, Pretoria, South Africa
- 2Department of Science and Innovation, National Research Foundation Centre of Excellence in Food Security, Pretoria, South Africa
- 3Sequencing Core Facility, National Institute for Communicable Diseases, National Health Laboratory Service, Johannesburg, South Africa
The increasing occurrence of multidrug-resistant (MDR) extended-spectrum β-lactamase- (ESBL) and/or AmpC β-lactamase- (AmpC) producing Enterobacterales in irrigation water and associated irrigated fresh produce represents risks related to the environment, food safety, and public health. In South Africa, information about the presence of ESBL/AmpC-producing Enterobacterales from non-clinical sources is limited, particularly in the water–plant-food interface. This study aimed to characterize 19 selected MDR ESBL/AmpC-producing Escherichia coli (n=3), Klebsiella pneumoniae (n=5), Serratia fonticola (n=10), and Salmonella enterica (n=1) isolates from spinach and associated irrigation water samples from two commercial spinach production systems within South Africa, using whole genome sequencing (WGS). Antibiotic resistance genes potentially encoding resistance to eight different classes were present, with blaCTX-M-15 being the dominant ESBL encoding gene and blaACT-types being the dominant AmpC encoding gene detected. A greater number of resistance genes across more antibiotic classes were seen in all the K. pneumoniae strains, compared to the other genera tested. From one farm, blaCTX-M-15-positive K. pneumoniae strains of the same sequence type 985 (ST 985) were present in spinach at harvest and retail samples after processing, suggesting successful persistence of these MDR strains. In addition, ESBL-producing K. pneumoniae ST15, an emerging high-risk clone causing nosocomical outbreaks worldwide, was isolated from irrigation water. Known resistance plasmid replicon types of Enterobacterales including IncFIB, IncFIA, IncFII, IncB/O, and IncHI1B were observed in all strains following analysis with PlasmidFinder. However, blaCTX-M-15 was the only β-lactamase resistance gene associated with plasmids (IncFII and IncFIB) in K. pneumoniae (n=4) strains. In one E. coli and five K. pneumoniae strains, integron In191 was observed. Relevant similarities to human pathogens were predicted with PathogenFinder for all 19 strains, with a confidence of 0.635–0.721 in S. fonticola, 0.852–0.931 in E. coli, 0.796–0.899 in K. pneumoniae, and 0.939 in the S. enterica strain. The presence of MDR ESBL/AmpC-producing E. coli, K. pneumoniae, S. fonticola, and S. enterica with similarities to human pathogens in the agricultural production systems reflects environmental and food contamination mediated by anthropogenic activities, contributing to the spread of antibiotic resistance genes.
Introduction
The discovery of antibiotics in the 1940s led to a new age in medical care. However, the global increase in antimicrobial resistance (AMR) is reducing the effectiveness of clinically important antibiotics (Lobanovska and Pilla, 2017; Dandachi et al., 2019). Examples of shifting resistance profiles in bacteria within the β-lactam class of antibiotics, including penicillins and third generation cephalosporins, which are the most widely used in human and veterinary medicine and widely expressed AMR are being reported (Finton et al., 2020). Persistent exposure to these antibiotics has resulted in bacteria becoming resistant by evolving extended-spectrum β-lactamases (ESBLs), which hydrolyze the β-lactam ring within the antibiotic, thus rendering it inactive (Bush and Jacoby, 2010). Consequently, production of ESBLs is regarded as one of the most clinically significant resistance mechanisms (Bush and Jacoby, 2010), with ESBL-producing Enterobacterales (Escherichia coli, Klebsiella pneumoniae, and Serratia spp., among others) listed as priority pathogens for research and development in the new frontier of antibiotics (WHO, 2017).
Classified into several groups according to their amino acid sequence homology, the CTX-M, TEM, and SHV ESBL variants are the most common β-lactamases identified in Enterobacterales (van Duin and Doi, 2017). In addition, AmpC β-lactamases (AmpCs) are chromosomally encoded by several Enterobacterales species and play a key role in resistance development (van Duin and Doi, 2017). Plasmid encoded AmpC genes have been known since 1989 (Jacoby, 2009) and are now regularly reported in clinical and environmental strains (Khari et al., 2016; Colosi et al., 2020; Tekele et al., 2020). Both chromosomally encoded and plasmid-mediated AmpC β-lactamases confer resistance to a broad spectrum of β-lactams such as penicillins, oxyimino-cephalosporins (including cefotaxime and ceftazidime), cephamycins, and aztreonam at variable levels (Jacoby, 2009; Palzkill, 2018; Furlan and Stehling, 2021; Lopes et al., 2021b).
The increase in antimicrobial resistant strains and effective resistance mechanisms among Enterobacterales has led to numerous global reports of ESBLs, AmpC-, and more recently carbapenemase-producing Enterobacterales not only in clinical settings, but also in the agricultural environment (Ye et al., 2017; Al-Kharousi et al., 2019; Dandachi et al., 2019; Hassen et al., 2020; Richter et al., 2020). Although members of the Enterobacterales family occur naturally in human and animals’ gastrointestinal tracts as well as in the environment (water, soil, and plants; Blaak et al., 2014; Ye et al., 2017), the occurrence of multidrug-resistant (MDR) strains in the different habitats is concerning. Inadequately treated or untreated effluents from industries, households, and zootechnical farms are reported as one of the main contamination causes of South African surface- and ground water resources (Verlicchi and Grillini, 2020). It is also well-documented that the three principal antibiotic contamination channels in the environment are animal-, human-, and manufacturing waste (O’neill, 2016). Consequently, contamination of soil, irrigation-, and drinking water as well as crops can occur, adding additional exposure routes to humans (Finton et al., 2020; Lopes et al., 2021a).
Previous surveillance studies have shown prevalence of MDR ESBL/AmpC-producing Enterobacterales in fresh vegetables sold in South Africa (Richter et al., 2019) and in other countries e.g., the Netherlands, Switzerland, and Germany (Reuland et al., 2014; Zurfluh et al., 2015; Reid et al., 2020). Occurrence of ESBL-producing Enterobacterales has also been reported in corresponding irrigation water sources and cultivated crops (Blaak et al., 2014; Njage and Buys, 2014; Ye et al., 2017). Furthermore, Richter et al. (2020) reported the occurrence of ESBL/AmpC-producing Enterobacterales in different spinach supply chains from irrigation water and produce at harvest, throughout processing and at retail in the Gauteng Province of South Africa.
The high discriminatory power of whole genome sequencing (WGS) has led to an increase in use of this method for detecting points of contamination, source tracking, pathogen surveillance, and outbreak investigations (Oniciuc et al., 2018; CDC, 2019). WGS provides information regarding multiple AMR genes, genomic mutations, mobile genetic elements, and association with resistance genes, as well as molecular typing like multi-locus sequence typing (MLST; Oniciuc et al., 2018; CDC, 2019; Kim et al., 2020). Consequently, the WGS results can aid in elucidating the genetic relationship among isolates from different environments and along the food chain (Adator et al., 2020). Surveillance of antimicrobial resistant strains through WGS is increasingly being used due to increasing accessibility and affordability (Adator et al., 2020). In South Africa, WGS has been used for characterization of clinical ESBL-producing K. pneumoniae strains among others (Founou et al., 2019), as well as typing of Listeria monocytogenes from environmental and clinical settings during the 2017 listeriosis outbreak (Thomas et al., 2020). However, the use of WGS for surveillance of antimicrobial resistant potential pathogenic Enterobacterales in retailed fresh produce and the production environment has not been reported locally.
The World Health Organization (WHO) developed the Global AMR Surveillance System (GLASS) in 2015 supporting research and surveillance as well as a global data sharing through a standardized analysis approach (WHO, 2020). Initially, the GLASS focus was mainly on surveillance of human priority pathogens, but has since shifted to include AMR in foodborne pathogens (WHO, 2020). Moreover, the One Health framework for understanding AMR in pathogenic gram-negative bacteria is increasingly attracting attention (Collignon and McEwen, 2019). In South Africa, information regarding AMR in fresh produce production systems and specifically focusing on the Enterobacterales is lacking. The aim of this study was thus to use WGS for analysis of AMR genes, associated mobile genetic elements, virulence factors, serotypes, multi-locus sequence types, and pathogenicity of selected, partially characterized, ESBL/AmpC-producing environmental Enterobacterales from commercial spinach production systems (Richter et al., 2020). These isolates included four different species (E. coli, K. pneumoniae, S. fonticola, and S. enterica) listed by the WHO as a particular threat for gram-negative bacteria that are resistant to multiple antibiotics (WHO, 2017), while isolates harbouring integrons as described in Richter et al. (2020) were preferentially selected. The results of this study will contribute to address the problem of antimicrobial drug resistance at the water–plant-food interface and how it might impact human health and disease.
Materials and Methods
Isolation and DNA Extraction of ESBL/AmpC-Producing Enterobacterales
Irrigation water and fresh produce samples from spinach production systems were collected and ESBL-producing Enterobacterales were isolated as described (Richter et al., 2020). A selection of 19 isolates were further characterized (Table 1). The genomic DNA of each isolate was extracted with the DNeasy PowerSoil kit (Qiagen, South Africa) according to the manufacturer’s instructions. Following gDNA extraction, the concentrations were determined using the Qubit dsDNA Broad Range Assay and a Qubit 2.0 fluorometer (Life Technologies, Johannesburg) and quantification was determined on a Nanodrop 2000 (ThermoScientific, Johannesburg).
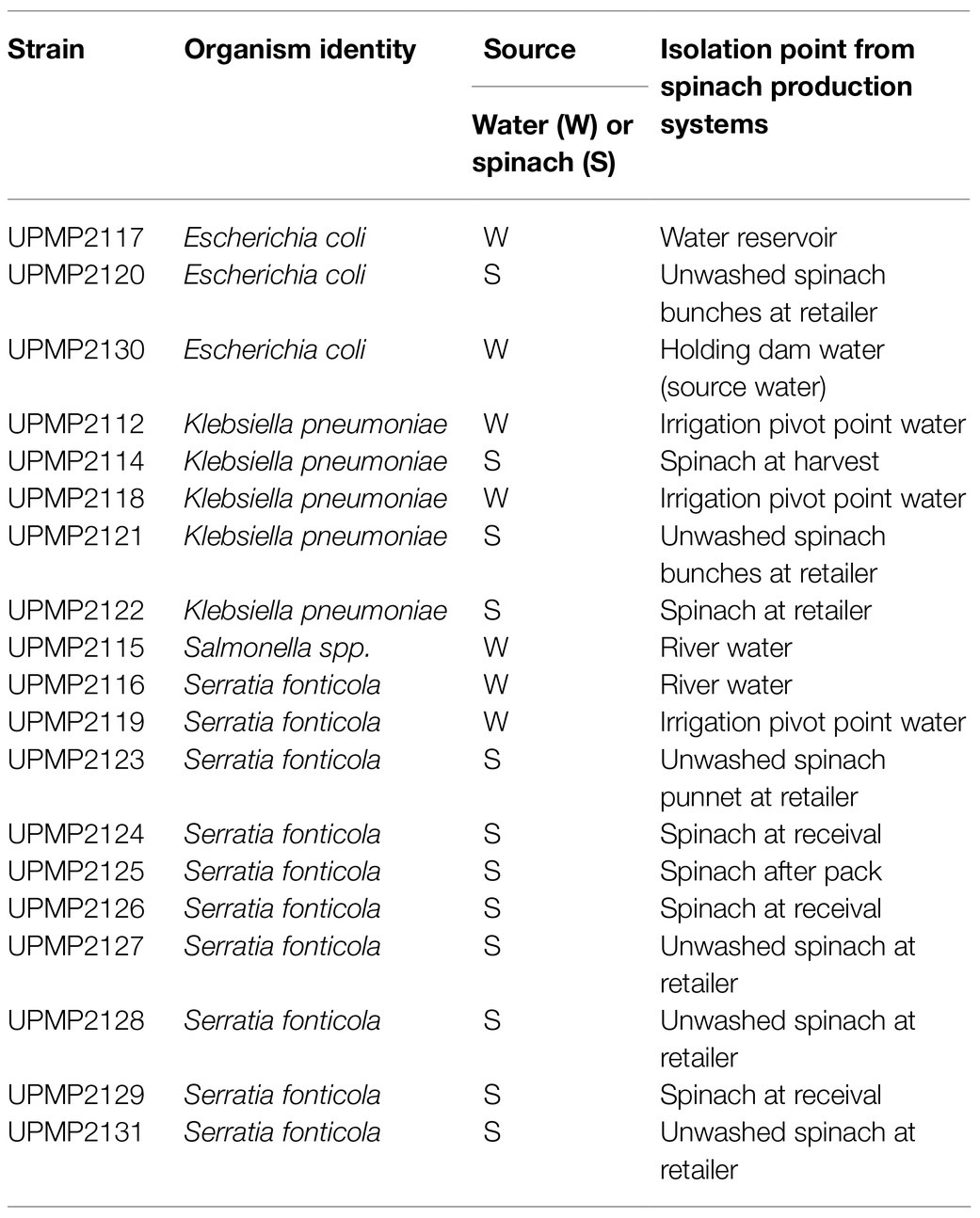
Table 1. Isolates selected for whole genome sequence analysis from the agricultural environment in spinach supply chains, Gauteng Province, South Africa.
DNA Sequencing and Whole Genome Analysis
Sequencing was performed on an Illumina MiSeq instrument (2×300bp) with 100× coverage by the National Institute for Communicable Diseases Sequencing Core Facility, South Africa, following preparation of multiplexed paired-end libraries with the Nextera XT DNA sample preparation kit (Illumina, San Diego, CA, United States). The resultant reads were quality trimmed using CLC version 201 and de novo assembled. The contiguous sequences were then submitted to the National Centre for Biotechnology Information (NCBI) Prokaryotic Genome Annotation Pipeline.2 The AMR gene presence was corroborated using ABRicate3 that included the Comprehensive Antibiotic Resistance Database (CARD), ARG-ANNOT, ResFinder, NCBI AMRFinder Plus, and MEGARes databases (Zankari et al., 2012; Gupta et al., 2014; Jia et al., 2017; Feldgarden et al., 2019; Doster et al., 2020). Plasmid replicon types were determined with PlasmidFinder (version 2.1; Carattoli et al., 2014). Using the Centre for Genomic Epidemiology (CGE) platform4, mobile genetic elements for all four species, sequence types of E. coli, K. pneumoniae, and S. enterica as well as the E. coli serotypes based on lipopolysaccharide (O-antigen) and capsular flagella (protein; H-antigen), and virulence genes of E. coli were determined with MGEFinder, Multilocus Sequence Typing (MLST; version 2.2), SeroTypeFinder (version 2.0), and VirulenceFinder (version 2.0), respectively (Larsen et al., 2012; Joensen et al., 2014, 2015; Johansson et al., 2021). The following parameters were used in the Serotype Finder Web-based tool: 85% threshold for %ID and 60% minimum length (the number of nucleotides in a sequence of interest that must overlap a serotype gene to count as a hit for that gene; Joensen et al., 2015). The in silico serotyping based on the capsule polysaccharide (K-antigen) of K. pneumoniae strains was conducted using Kaptive Web (Wick et al., 2018), while the presence of virulence genes for K. pneumoniae was identified by using the Institut Pasteur’s Klebsiella database.5 Additionally, paired reads of the WGS raw data files for the S. enterica strain were uploaded to the online SeroSeq tool version 1.0 which predicted the Salmonella serotype of the requested isolate (Zhang et al., 2015; Thompson et al., 2018). The Salmonella Pathogenicity Islands (SPI) were identified with SPIFinder 2.0 (Roer et al., 2016). Next, the existence of virulence factors in each SPI was analyzed by performing BLAST analysis on the predicted SPIs against the virulence factor database (VFDB; Chen et al., 2016; Ashari et al., 2019). The virulence factors of S. fonticola were determined using the VFDB with ABRicate (Chen et al., 2016). All sequences were submitted to the INTEGRALL database6 for annotation and integron number assignment. Using PathogenFinder (version 1.1) on the CGE platform, the strains’ pathogenicity towards humans was predicted (Cosentino et al., 2013).
Data Availability
The nucleotide sequences of the 19 Enterobacterales strains described in this paper were deposited in the National Center for Biotechnology Information GenBank database in the BioProject number: PRJNA642017, accession numbers NZ_JACAAL010000000, NZ_JACBIV000000000-NZ_JACBJE000000000, and NZ_JACNYM000000000-NZ_JACNYT000000000 (Table 2).
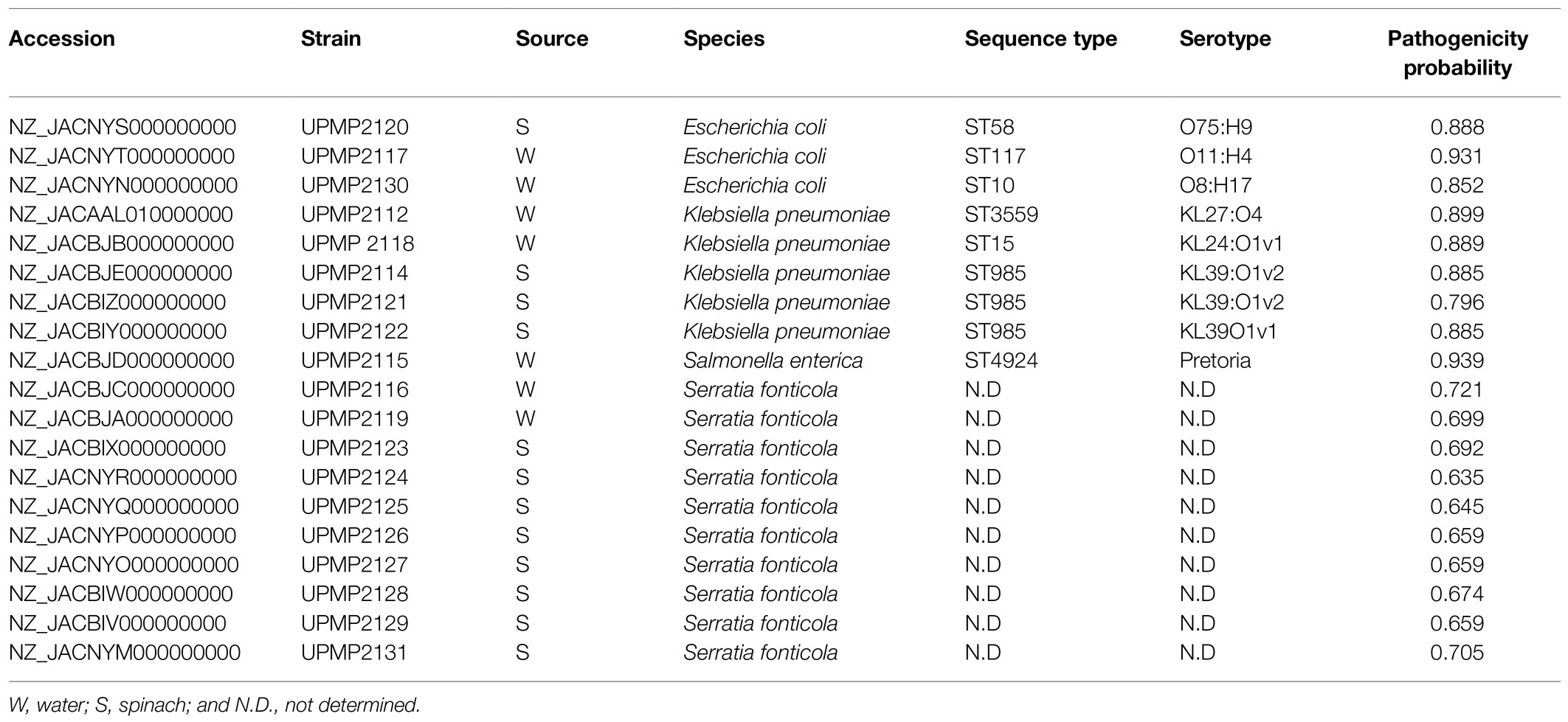
Table 2. In silico multi-locus sequence typing (MLST) analysis, predicted serotypes, and pathogenicity probability of Enterobacterales isolated from irrigation water and spinach throughout production from farm to retail.
Results
Detection of Antimicrobial Resistance Genes
The selected 19 ESBL/AmpC producing Enterobacterales isolates all harboured at least one β-lactamase encoding gene in addition to the ESBL/AmpC genetic determinants, accompanied by resistance genes from different antibiotic classes including fluoroquinolone, sulfonomide, fosfomycin, aminoglycoside, trimethroprim, phenicol, and/or tetracycline (Figure 1). The β-lactamase resistance genes included chromosomally encoded AmpC in the S. enterica strain as well as all three E. coli strains. Plasmid-mediated AmpC genes (blaCMY-113 and blaCMY-101) were present in two E. coli strains from irrigation water and blaACT-13, blaACT-38, blaACT-6, and/or blaACT-58 were present in 10 S. fonticola strains from irrigation water (n=2) and spinach (n=8) samples (Figure 1). Additionally, blaFONA-5 (n=8) from irrigation water and spinach and blaFONA-6 (n=2) from spinach were present in S. fonticola strains. The ESBL genes included blaSFO-1 in all 10 S. fonticola strains, blaCTX-M-15 in five K. pneumoniae strains from irrigation water and spinach, and one E. coli strain from spinach. It also included blaCTX-M-14 in an E. coli strain from irrigation water, while blaSHV-187 (n=3), blaSHV-106 (n=1), and blaSHV-178 (n=1) were present in K. pneumoniae strains (Figure 1).
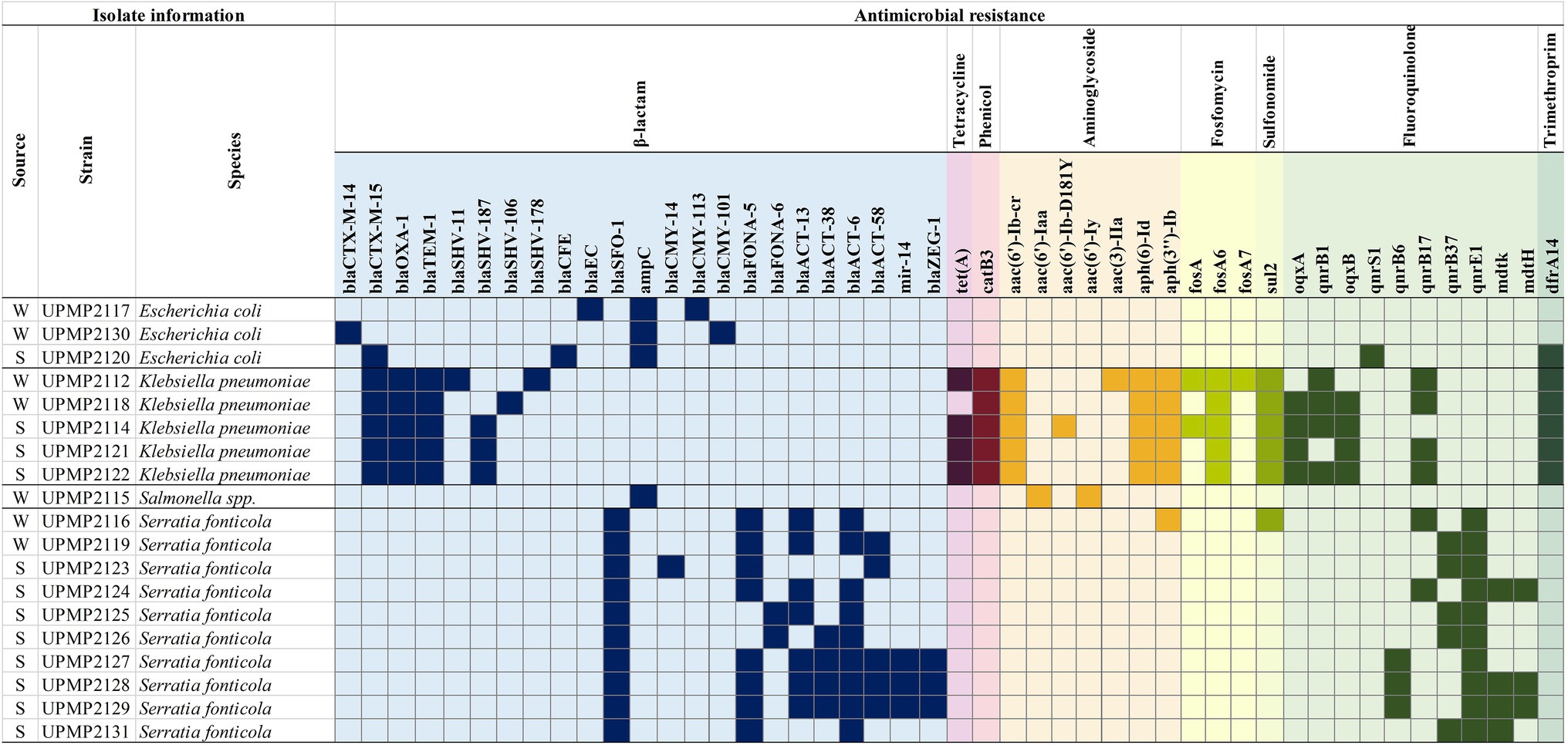
Figure 1. Antimicrobial resistance (AMR) genes present in Enterobacterales isolated from water and spinach from farm to retail. W, water; S, spinach.
Interestingly, a greater number of resistance genes across more classes were seen in all the K. pneumoniae strains (n=5), compared to the other genera tested. All five K. pneumoniae strains had chloramphenicol (catB3), aminoglycosides [aac(6')-Ib-cr, aph(6)-Id and aph(3")-Ib], fosfomycin (fosA6), and sulfonomide (sul2) resistance genes present (Figure 1). Other resistance genes included fluoroquinolone oqxA (n=4), oqxB (n=4), and qnrB1 (n=4) in K. pneumoniae from spinach and water, qnrS1 (n=1) in E. coli from spinach and qnrB6 (n=3), qnrB37 (n=5), qnrE1 (n=10) in S. fonticola from spinach and water, while mdtK (n=4), and mdtH (n=3) were present in S. fonticola from water only. The qnrB17 resistance gene was present in K. pneumoniae (n=4) and S. fonticola (n=2) strains from spinach and water (Figure 1). The S. enterica strain isolated from irrigation water also harboured aac(6')-Iaa and aac(6')-Iy aminoglycoside resistance genes (Figure 1) and a S. fonticola strain from irrigation water harboured an aminoglycoside [aph(3")-Ib] and sulfonomide (sul2) resistance gene (Figure 1).
Detection of Mobile Genetic Elements and Association to Antimicrobial Resistance Genes
Known resistance plasmid replicon types of Enterobacterales including IncFIB, IncFIA, IncFII, IncB/O, and IncHI1B were observed in all strains following analysis with PlasmidFinder (data not shown). The β-lactamase gene, blaCTX-M-15, was the only resistance gene associated with plasmids (IncFII_pKP91 and/or IncFIB(K)_1_Kpn3) in four K. pneumoniae strains upon further analysis (Table 3). The IS6 family elements (IS6100) have been reported to play a pivotal role in the dissemination of resistance determinants in gram-negative bacteria (Partridge et al., 2018), and were observed in relation to the dfrA14b resistance gene in all five K. pneumoniae strains (Table 3). The blaCTX-M-14 and sul2 resistance genes were related to the ISEcp1 element within the IS1380 family in one E. coli and three K. pneumoniae strains, respectively, while one S. fonticola strain carried a sul2 gene that was related to IS110 (Table 3). One E. coli strain carried the qnrS1 resistance gene that was related to ISKra4. Other insertion sequences detected belonged predominantly to the IS3 and IS110 families (data not shown), with one K. pneumoniae strain carrying the blaSHV-80 broad spectrum β-lactamase that was related to IS3 (Table 3). In all K. pneumoniae strains (n=5) where the qnrB1 resistance gene was present, association to Tn5403 was seen (Table 1). In one E. coli and five K. pneumoniae strains, integron In191 was observed, with dfrA14 in the cassette array (Table 3).
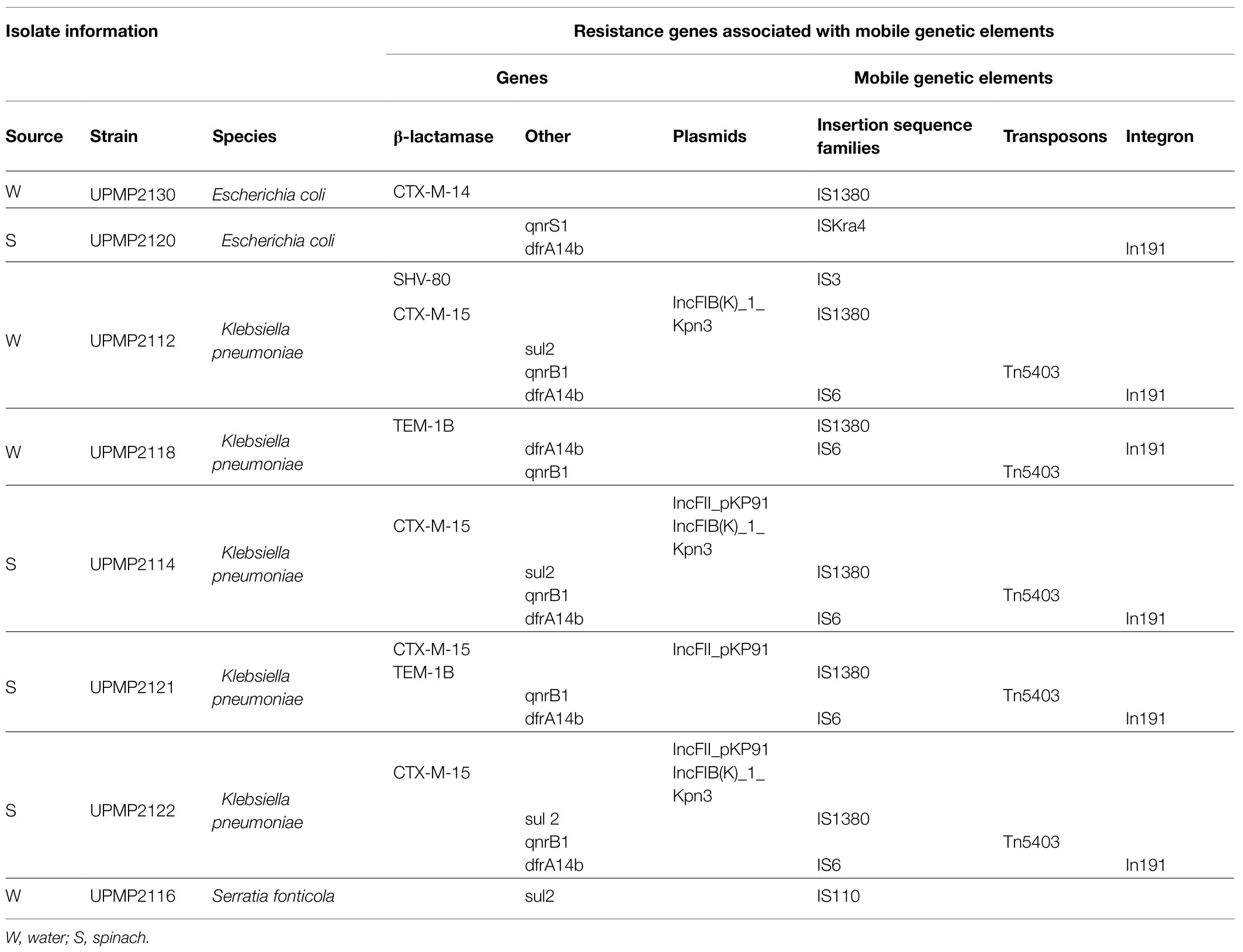
Table 3. Extended-spectrum β-lactamase (ESBL)/AmpC β-lactamase (AmpC)-producing Enterobacterales with resistance genes related to mobile genetic elements.
In silico Analysis of Serotypes, Multi-Locus Sequence Types, and Virulence Factors
The in silico MLST analysis, predicted serotypes, and pathogenicity probability of all 19 strains are shown in Table 2. Three different sequence types (ST58, ST117, and ST10) and three different serotypes (O75:H9, O11:H4, and O8:H17) were observed in the three E. coli strains. The five K. pneumoniae strains belonged to three different sequence types and three different serotypes (KL27, KL24, and KL39) which were observed based on the K-antigen, while the O-serotype included O4 and O1 (Table 2). The predicted antigenic profile of the S. enterica strain was O11:k:1,2. Furthermore, the S. enterica strain contained 11 Salmonella SPI, namely SPI-1, SPI-2, SPI-3, SPI-4, SPI-5, SPI-9, SPI-13, SPI-14, one unnamed, as well as the centisome 63 (C63PI) and 54 (CS54) pathogenicity islands, each harbouring between 20 and 60 virulence factors (Supplementary Table S1). A total of 42 virulence genes were identified in the E. coli and K. pneumoniae strains (Supplementary Tables S2, S3). Of these, 20 were detected in E. coli strains only and 20 in K. pneumoniae strains only, while fyuA (iron uptake associated with siderophores) and irp2 (iron uptake) virulence factors were detected in two E. coli strains from irrigation water as well as three K. pneumoniae strains from spinach samples. All three E. coli strains carried the terC (tellurite resistance) virulence gene (Supplementary Table S2) and in all five K. pneumoniae strains, the mrkA, mrkB, mrkC, mrkD, mrkE, (main structural subunit and assembly machinery for type 3 fimbriae) mrkH (regulatory protein), and mrkI (DNA binding protein) virulence factors were present (Supplementary Table S3). No shiga-toxin producing genes were present in the E. coli strains. A total of 89 virulence factors were identified in the S. fonticola strains (Supplementary Table S4). This included 25, 18, 16, and 6 of the virulence factors present in 100% (n=10), 90, 80, and 70% of the selected S. fonticola strains, respectively, while the remaining 24 virulence factors were present in varying numbers in 1–6 of the strains (Supplementary Table S4). The iroN salmochelin siderophore receptor which plays a role in disease establishment was present in three S. fonticola strains (two from unwashed baby spinach samples at the retailer and one from the irrigation pivot point water), one E. coli strain from the ground water, as well as in the SPI-13 in the S. enterica strain from river irrigation water. Relevant similarities to human pathogens were predicted for all 19 strains with a confidence of 0.635–0.721 in the S. fonticola strains (n=10), 0.852–0.931 in the E. coli strains (n=3), 0.796–0.899 in the K. pneumoniae strains (n=5), and 0.939 in the S. enterica strain (Table 2).
Discussion
To the authors knowledge, this is the first study to use WGS for in-depth molecular characterization of ESBL/AmpC-producing E. coli, K. pneumoniae, S. enterica, and S. fonticola isolates, previously identified and partially characterized, from spinach and irrigation water samples in commercial production chains (Richter et al., 2020). Characterization included AMR, mobile genetic elements (e.g., insertion sequences, plasmids, and integrons), serotypes, and determining the pathogenicity. All these factors are crucial in defining and attributing infection sources of food-related outbreaks caused by resistant microorganisms (Oniciuc et al., 2018). Overall, the results corresponded with main global findings where AMR genes and associated mobile genetic elements have been reported in Enterobacterales from fresh produce and irrigation water, with the potential to pose a health risk to humans upon exposure (Jones-Dias et al., 2016; Finton et al., 2020). Previously, the presence of intI3 was reported in a high percentage of isolates from the current study following conventional PCR and sequencing (Richter et al., 2020). However, in-depth WGS analysis showed that no attI fragment preceded the IntI3 genes; consequently, the IntI3 genes detected did not form part of complete integrons, which typically include an integrase intI gene encoding a site-specific recombinase, a recombination site attI as well as a promoter (Pc; Kaushik et al., 2018). Overall, six isolates in the current study were positive for Class 1 integrons (In191), similar to In191 positive clinical ESBL-producing Enterobacterales from an academic teaching hospital in Pretoria, SA (Sekyere et al., 2020). Additionally, these MDR environmental isolates harbored various virulence factors central to pathogenicity, including genes associated with urinary tract infections and iron sequestering systems crucial for disease establishment. All isolates had relevant similarity to human pathogens and form part of the WHO 3rd generation cephalosporin resistant critical priority pathogens (WHO, 2017).
Two of the E. coli strains from the current study harboured plasmid-mediated AmpC blaCMY-2-like genes (blaCMY-113 and blaCMY-101), which correspond to the phenotypic profile of resistance to expanded-spectrum cephalosporins previously reported for these isolates using traditional PCR analysis (Richter et al., 2020). The blaCMY-2 pAmpC genes are the most commonly reported in E. coli and other Enterobacterales species and have clinical relevance, as it inactivates third generation cephalosporins and mediates resistance to carbapenems (Jacoby, 2009; Bortolaia et al., 2014). Three different multi-locus sequence types, namely ST58, ST10, and ST117, were identified in the E. coli isolates. Isolated from the retailed unwashed spinach samples in the current study, ST58 E. coli have previously also been associated with human extra-intestinal infections including sepsis, and have emerged worldwide in wild and food-production animals (Reid et al., 2020). As an example, ST58 E. coli with serotype O75:H9 corresponded to an E. coli strain of bovine origin from Pakistan and also carried the IncFIB plasmid (Ali et al., 2020). Although the strain from the current study had less AMR genes than reported in ST58 E. coli with serotype O75:H9 by Ali et al. (2020), the trimethoprim (dfrA14), fluoroquinolone (qnrS1), and β-lactam (blaCTX-M-15) genes corresponded. Similarly, uropathogenic ST58 E. coli with resistance to fluoroquinolone and trimethoprim have previously been isolated from hospital patients in Australia (McKinnon et al., 2018). The blaCTX-M-15 gene identified in the ST58 E. coli strain from the current study was associated with the ISKra4 insertion sequence, previously identified in K. pneumoniae harbouring blaCTX-M-15, and was responsible for the movement to different parts of the genome through a replicative transposition mechanism (Razavi et al., 2020). In contrast to Hauser et al. (2013) who identified food-associated shiga-toxin producing E. coli ST58, no stx genes were present in the strains. The E. coli ST58 from the current study harboured the gad (glutamate decarboxylase) virulence gene, similar to E. coli ST58 strains isolated from aragula (rocket; Reid et al., 2020). However, the presence of lpfA (long polar fimbriae) and terC (tellurium ion resistance protein) virulence factors in the strain from the current study contrasted the virulence gene profiles reported by Reid et al. (2020). Escherichia coli ST10 have previously been associated with human clinical infections and has been isolated from different sources including recreational and/or wastewater samples (Falgenhauer et al., 2019). From the current study, the E. coli ST10 with serotype O8:H17 was isolated from borehole water used for irrigation. Although this sequence type has previously been associated with shiga-toxin-producing E. coli (STEC; Gonzalez-Escalona and Kase, 2018), no stx genes were detected in the current study. The virulence factors present were terC (tellurium ion resistance protein), astA (EAST-1 heat-stable toxin), fyuA (ferric yersiniabactin uptake receptor), irp2 (nonribosomal peptide synthetases), iss (increased serum survival), and sitA (iron transport protein). Previously, E. coli ST10 with similar virulence gene profiles were isolated from human blood cultures and reported as extra-intestinal pathogenic E. coli (ExPEC; Maluta et al., 2017). Additionally, ESBL-producing E. coli ST10 of the same serotype have been isolated from wastewater and are depicted as a probable environmental reservoir of blaCTX-M genetic determinants (Tanaka et al., 2019). In the current study, the ST58 E. coli strain harboured the blaCTX-M-15 genetic determinant, while blaCTX-M-14 was present in the ST10 E. coli strain. Globally, the CTX-M type ESBLs (especially blaCTX-M-14 and blaCTX-M-15) have become the dominant genotype and the most widely distributed (Cantón et al., 2012; Adamski et al., 2015). Escherichia coli blaCTX-M-14 positive strains have previously been isolated from store-bought produce in Germany and South Africa (Richter et al., 2019; Reid et al., 2020), food producing animals in China (Liao et al., 2015) and clinical settings in Brazil and South Africa (Cergole-Novella et al., 2010; Peirano et al., 2011).
The third E. coli sequence type (ST117) detected from irrigation source water in the current study has previously been reported as part of a group of multi-serotype extra-intestinal pathogenic E. coli (ExPEC) and avian pathogenic E. coli (APEC) strains (Kim et al., 2017). The E. coli ST117 strain from the current study harboured 20 virulence factors including the ExPEC hlyF (Hemolysin F) virulence gene. In previous studies, stx genes were identified in E. coli strains with the same STs detected in the current study, yet the virulence gene content and serotypes differ from the strains in the current study (Gonzalez-Escalona and Kase, 2018). However, the three non-STEC E. coli strains (ST58, ST10, and ST117) from the current study had a 93, 89, and 85% probability of being human pathogens, based on the pathogenic protein families.
In addition to E. coli, other Enterobacterales isolates harbouring blaCTX-M-15 have also been detected in different environments. In the current study, all five K. pneumoniae strains harboured the blaCTX-M-15 genetic determinant. The prevalence and dissemination of blaCTX-M throughout various environments globally underlines the different contamination routes through which fresh produce may also become contaminated with these MDR organisms. For instance, Gekenidis et al. (2020) have demonstrated the long-term persistence of E. coli harbouring blaCTX-M-15 in soil and lettuce after its introduction via irrigation water. Similarly, blaCTX-M-15 positive ST985 K. pneumoniae strains were present in spinach at harvest on the farm as well as retail samples after processing in the current study, suggesting successful persistence of these MDR strains. In four K. pneumoniae strains (ST3559, n=1 and ST985, n=3), the blaCTX-M-15 genes were associated with IncF replicons (IncFIIK and IncFIB) which have previously been linked to diverse K. pneumoniae outbreak strains (Dolejska et al., 2012, 2013; Löhr et al., 2015). Moreover, in K. pneumoniae ST3559, blaCTX-M-15 was also associated with ISEcp1 (also called ISEc9), a member of the widely reported IS1380 family, and can enable the independent transposition with insertion mutation and genetic relocations (Partridge, 2011). The K. pneumoniae strains in the current study also harboured blaSHV ESBL encoding genes (blaSHV-187, blaSHV-106, and blaSHV-178). Previously, SHV genetic determinants were reported in K. pneumoniae from hospitals and receiving wastewater treatment plants in Romania (Surleac et al., 2020) as well as irrigation water and agricultural soil in South Africa (Iwu et al., 2020; Richter et al., 2020). Interestingly, the K. pneumoniae ST15 strain isolated from water in the current study harboured blaSHV-106 which Liakopoulos et al. (2016) previously reported to be geographically constrained and have only been described in K. pneumoniae isolates from Portugal together with blaTEM-1. Similarly, the K. pneumoniae ST15 strain from the current study also harboured blaSHV-106 together with blaTEM-1. Klebsiella pneumoniae ST15 is regarded as an emerging international high-risk clone causing nosocomial outbreaks worldwide with high-levels of antibiotic resistance including production of ESBLs, mainly CTX-M-15 (Han et al., 2021).
The K. pneumoniae ST3559 strain isolated from irrigation water in the current study was capsular type 27 and serotype O4, which is similar to an O4 serotype MDR K. pneumoniae outbreak strain from a neonatal care unit in sub-Saharan Africa (Cornick et al., 2020). In addition, K. pneumoniae ST3559 harboured the blaSHV-178 gene which, to the best of our knowledge, have previously only been reported in clinical Enterobacter hormaechei strains from the First Affiliated Hospital of Zhejiang University in Hangzhou (Gou et al., 2020). Apart from β-lactamase genes, the K. pneumoniae strains also harboured aminoglycoside, fosfomycin, fluoroquinolone, tetracyline, phenicol, trimethoprim, and sulfonomide resistance genes, which is a greater diversity of resistance genes than previously reported in Enterobacterales isolates from German surface waters (Falgenhauer et al., 2019). Similar to the results of clinical K. pneumoniae strains reported by Mbelle et al. (2020), In191, harbouring dfrA14 was identified in the three different K. pneumoniae sequence types of the current study, reiterating that it is not a narrow spectrum integron. In addition, dfrA14b was associated with IS6 that has previously been reported as having a vital role in the rearrangement and dissemination of antibiotic resistance (Varani et al., 2021). The presence of fosA and sul2 in all the K. pneumoniae strains of the current study also corresponds to the results reported by Mbelle et al. (2020) from clinical K. pneumoniae strains in Pretoria. The high-level of trimethoprim resistance globally has however led to trimethoprim-sulfamethoxazole no longer being recommended for outpatient treatment of urinary tract infections and similarly, the use of fosfomycin might not be efficacious anymore (Mbelle et al., 2020). Four MDR K. pneumoniae isolates from irrigation water (ST15, n=1) and spinach (ST985, n=3) had O1 serotypes, previously reported as the most commonly isolated serotypes from human hosts and dominant in human disease (Follador et al., 2016). However, it is noteworthy that no genes encoding carbapenamases nor resistance to colistin were identified in the current study. All five characterized K. pneumoniae strains also harbored several virulence factors including those that coded for an iron uptake system (kfu) and type 3 fimbrial adhesins (mrk) that play an important role in adhesion to medical devices such as catheters (Albasha et al., 2020; Finton et al., 2020).
Serratia spp. are opportunistic pathogens that may pose a health threat to immunocompromised and hospitalized patients (Petersen and Tisa, 2013). The S. marcescens species is most often associated with nosocomial infections; however, S. fonticola has been reported to function as a human pathogen when detected alone or may be a bystander and act as a carrier of resistance genes when discovered with other organisms (Petersen and Tisa, 2013; Aljorayid et al., 2016). Characterizing virulence genes of the MDR environmental strains therefore becomes important within the plant-food producing environment. In the current study, all S. fonticola strains harboured blaSFO-1 and numerous plasmid incompatibility (Inc) groups were identified in these S. fonticola strains (data not shown). However more in-depth plasmid typing and analysis will be required to fully understand the risk/probability of blaSFO-1 dissemination in the environment where S. fonticola naturally occurs. In certain Enterobacterales species, ESBL genes are inherently carried on chromosomes (Naas et al., 2008). This includes the blaSFO-1 ESBL gene from S. fonticola that differs from most class A ESBLs, as the β-lactamases’ production can be induced by a high level of imipenem (Naas et al., 2008). The blaSFO-1 ESBL does not form part of the most clinically relevant ESBLs and are therefore rarely reported. Zhou et al. (2020) reported in contrast an increasing trend of the co-existence of plasmid-borne blaSFO-1 and carbapenemase genes in clinical Enterobacter spp. in China. All the S. fonticola strains also harboured numerous fluoroquinolone resistance genes, raising a health concern for treatment options, as fluoroquinolones are often used for management of conditions including typhoid fever and MDR tuberculosis (Richards et al., 2019). Interestingly, one S. fonticola strain harboured an acquired trimethoprim (sul2) resistance gene associated with IS110, corresponding to K. pneumoniae from a German university hospital (Schwanbeck et al., 2021). The Serratia genus naturally lacks resistance genes for trimethoprim and sulfonamides (Sandner-Miranda et al., 2018). Previous reports of potential pathogenic S. fonticola primarily focused on the antibiotic resistance profiles (Tasić et al., 2013; Aljorayid et al., 2016; Hai et al., 2020). The strains from the current study additionally harboured various virulence factors. This included flagellar biosynthesis- and chemotaxis-related genes as well as genes encoding iron uptake systems corresponding to those previously reported in important MDR nosocomial pathogenic S. marcescens (Iguchi et al., 2014).
Only one S. enterica strain isolated from river irrigation water was characterized in the current study. Irrigation water is well documented as a source for fresh produce contamination of foodborne pathogens including Salmonella spp. (Liu et al., 2018). The strain harboured an AmpC resistance gene, similar to S. enterica characterized from surface water in the United States (Li et al., 2014). In addition, the S. enterica from the current study carried aminoglycoside resistance genes [aac(6')-Iaa and aac(6')-Iy], similar to results reported by Nair et al. (2016) for non-typhoidal Salmonella spp. isolated from a United Kingdom population. Of the 23 known Salmonella SPIs previously described (Mansour et al., 2020), the isolate from the current study carried 11 SPIs. This included SPIs that are commonly reported in S. enterica and encode genes responsible for enabling invasion of epithelial cells (SPI1), facilitating the replication of intracellular bacteria (SPI2), adhesion to epithelial cells (SPI3, 4, 5, and 9; Waterman and Holden, 2003; Velásquez et al., 2016; Mansour et al., 2020), as well as SPI13 and 14 which corresponds to being part of the core genome of invasive non-typhoidal Salmonella spp. (Suez et al., 2013). Additionally, pathogenicity islands C63PI and CS54 were present in the S. enterica strain in this study, which has previously been found in the S. Typhimurium and S. Typhi genomes (Sabbagh et al., 2010; Jibril et al., 2021). Since no phenotypic indication of virulence was investigated, the prediction of virulence genes using in silico tools should be regarded with care; however, using PathogenFinder, the S. enterica strain from the current study showed 94% probability of being a human pathogen.
Conclusion
This is the first WGS analysis study of MDR ESBL/AmpC-producing E. coli, K. pneumoniae, S. fonticola, and S. enterica isolates from spinach production systems within South Africa. The selected isolates represent potential pathogenic genera listed by the WHO as a priority for surveillance of AMR screening. Numerous clinically relevant resistance genes were detected in the screened samples. This study showed the potential of using WGS in metadata studies for detailed molecular characterization of potential pathogenic Enterobacterales. Furthermore, the study highlighted the importance of the agricultural production environment as a source of antibiotic resistance genes within Enterobacterales at the water-plant-food interface. A more in-depth and controlled analysis, with a greater number of sequenced isolates from the farm-to-retail supply chain is required to better understand the prevalence and resistance gene transmission through the supply chain. The results from this study further highlight the need for expanded surveillance in agricultural systems.
Data Availability Statement
The nucleotide sequences of the 19 Enterobacteriaceae strains described in this paper were deposited in the National Center for Biotechnology Information GenBank database in the BioProject number: PRJNA642017, accession numbers NZ_JACAAL010000000, NZ_JACBIV000000000-NZ_JACBJE000000000, and NZ_JACNYM000000000-NZ_JACNYT000000000 (Table 2).
Author Contributions
EP, SD, LR, and LK contributed to the conception and design of the study. LR performed the experiments. LR, SD, MA, and AI analyzed the data. LR, EP, and SD contributed to interpretation and presentation. SD, EP, and LK were involved in funding acquisition. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by the Water Research Commission (WRC) funded project “Measurement of water pollution determining the sources and changes of microbial contamination and impact on food safety from farming to retail level for fresh vegetables” (WRC Project No K5/2706/4 and WRC Knowledge Review 2017/18) and the Partnerships for Enhanced Engagement in Research (PEER) a USAID/DST funded project “Characterizing and tracking of antimicrobial resistance in the water-plant-food public health interface” (Grant no. 48). Furthermore, the financial assistance of the Department of Science and Innovation (DSI) – National Research Foundation (NRF) Centre of Excellence in Food Security funded this research under the Food Safety Program’s “Safe Food for the Food Insecure” project (Project 160301 and 160302). This work is based on the research supported in part by the NRF of South Africa [Grant specific unique reference number (UID): 74426 and Grant Number: 120319]. Opinions expressed and conclusions arrived at, are those of the authors and are not necessarily to be attributed to the NRF.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors acknowledge the University of Pretoria, DSI-NRF Centre of Excellence in Food Security, PEER, and the WRC for funding and bursary support. The opinions, findings, conclusions, and/or recommendations expressed in this publication are the author(s) alone.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.734649/full#supplementary-material
Footnotes
1. ^https://digitalinsights.qiagen.com
2. ^https://pubmed.ncbi.nlm.nih.gov/27342282/
3. ^https://github.com/tseemann/abricate
4. ^https://cge.cbs.dtu.dk/services/
References
Adamski, C. J., Cardenas, A. M., Brown, N. G., Horton, L. B., Sankaran, B., Prasad, B. V. V., et al. (2015). Molecular basis for the catalytic specificity of the CTX-M extended-spectrum β-lactamases. Biochemistry 54, 447–457. doi: 10.1021/bi501195g
Adator, E. H., Walker, M., Narvaez-Bravo, C., Zaheer, R., Goji, N., Cook, S. R., et al. (2020). Whole genome sequencing differentiates presumptive extended spectrum beta-lactamase producing Escherichia coli along segments of the one health continuum. Microorganisms 8:448. doi: 10.3390/microorganisms8030448
Albasha, A. M., Osman, E. H., Abd-Alhalim, S., Alshaib, E. F., Al-Hassan, L., and Altayb, H. N. (2020). Detection of several carbapenems resistant and virulence genes in classical and hyper-virulent strains of Klebsiella pneumoniae isolated from hospitalized neonates and adults in Khartoum. BMC. Res. Notes 13:312. doi: 10.1186/s13104-020-05157-4
Ali, A., Ali, Q., Ali, R., and Mohsin, M. (2020). Draft genome sequence of an extended-spectrum β-lactamase-producing Escherichia coli ST58 isolate from cattle in Pakistan. J. Glob. Antimicrob. Resist. 21, 303–305. doi: 10.1016/j.jgar.2020.04.020
Aljorayid, A., Viau, R., Castellino, L., and Jump, R. L. P. (2016). Serratia fonticola, pathogen or bystander? A case series and review of the literature. IDCases 5, 6–8. doi: 10.1016/j.idcr.2016.05.003
Al-Kharousi, Z. S., Guizani, N., Al-Sadi, A. M., and Al-Bulushi, I. M. (2019). Antibiotic resistance of Enterobacteriaceae isolated from fresh fruits and vegetables and characterization of their AmpC b-lactamases. J. Food Prot. 82, 1857–1863. doi: 10.4315/0362-028X.JFP-19-089
Ashari, K. S., Roslan, N. S., Omar, A. R., Bejo, M. H., Ideris, A., and Isa, N. M. (2019). Genome sequencing and analysis of Salmonella enterica subsp. enterica serovar Stanley UPM 517: insights on its virulence-associated elements and their potentials as vaccine candidates. PeerJ 7:e6948. doi: 10.7717/peerj.6948
Blaak, H., van Hoek, A. H. A. M., Veenman, C., Docters van Leeuwen, A. E., Lynch, G., van Overbeek, W. M., et al. (2014). Extended spectrum ß-lactamase- and constitutively AmpC-producing Enterobacteriaceae on fresh produce and in the agricultural environment. Int. J. Food Microbiol. 168–169, 8–16. doi: 10.1016/j.ijfoodmicro.2013.10.006
Bortolaia, V., Hansen, K. H., Nielsen, C. A., Fritsche, T. R., and Guardabassi, L. (2014). High diversity of plasmids harbouring blaCMY-2 among clinical Escherichia coli isolates from humans and companion animals in the upper Midwestern USA. J. Antimicrob. Chemother. 69, 1492–1496. doi: 10.1093/jac/dku011
Bush, K., and Jacoby, G. A. (2010). Updated functional classification of β-lactamases. Antimicrob. Agents Chemother. 54, 969–976. doi: 10.1128/AAC.01009-09
Cantón, R., González-Alba, J. M., and Galán, J. C. (2012). CTX-M enzymes: origin and diffusion. Front. Microbiol. 3:110. doi: 10.3389/fmicb.2012.00110
Carattoli, A., Zankari, E., Garciá-Fernández, A., Larsen, M. V., Lund, O., Villa, L., et al. (2014). In silico detection and typing of plasmids using plasmidfinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903. doi: 10.1128/AAC.02412-14
CDC (2019). Antibiotic resistance threats in the United States. Atlanta. GA. Available at: https://www.cdc.gov/drugresistance/biggest_threats.html (Accessed June 18, 2021).
Cergole-Novella, M. C., Guth, B. E. C., Castanheira, M., Carmo, M. S., and Pignatari, A. C. C. (2010). First description of blaCTX-M-14-and blaCTX-M-15- producing Escherichia coli isolates in Brazil. Microb. Drug Resist. 16, 177–184. doi: 10.1089/mdr.2010.0008
Chen, L., Zheng, D., Liu, B., Yang, J., and Jin, Q. (2016). VFDB 2016: hierarchical and refined dataset for big data analysis—10 years on. Nucleic Acids Res. 44, D694–D697. doi: 10.1093/nar/gkv1239
Collignon, P. J., and McEwen, S. A. (2019). One health-its importance in helping to better control antimicrobial resistance. Trop. Med. Infect. Dis. 4:22. doi: 10.3390/tropicalmed4010022
Colosi, I. A., Baciu, A. M., Opriș, R. V., Peca, L., Gudat, T., Simon, L. M., et al. (2020). Prevalence of ESBL, AmpC and carbapenemase-producing enterobacterales isolated from raw vegetables retailed in Romania. Foods 9:1726. doi: 10.3390/foods9121726
Cornick, J., Musicha, P., Peno, C., Saeger, E., Iroh Toh, P. Y., Bennett, A., et al. (2020). Genomic investigation of a suspected multi-drug resistant Klebsiella pneumoniae outbreak in a neonatal care unit in sub-Saharan Africa. bioRxiv [Preprint]. doi: 10.1101/2020.08.06.236117
Cosentino, S., Voldby Larsen, M., Møller Aarestrup, F., and Lund, O. (2013). PathogenFinder—distinguishing friend from foe using bacterial whole genome sequence data. PLoS One 8:e77302. doi: 10.1371/journal.pone.0077302
Dandachi, I., Chaddad, A., Hanna, J., Matta, J., and Daoud, Z. (2019). Understanding the epidemiology of multi-drug resistant gram-negative bacilli in the middle east using a one health approach. Front. Microbiol. 10:1941. doi: 10.3389/fmicb.2019.01941
Dolejska, M., Brhelova, E., Dobiasova, H., Krivdova, J., Jurankova, J., Sevcikova, A., et al. (2012). Dissemination of IncFIIK-type plasmids in multiresistant CTX-M-15-producing Enterobacteriaceae isolates from children in hospital paediatric oncology wards. Int. J. Antimicrob. Agents 40, 510–515. doi: 10.1016/j.ijantimicag.2012.07.016
Dolejska, M., Vill, L., Dobiasova, H., Fortini, D., Feudi, C., and Carattoli, A. (2013). Plasmid content of a clinically relevant Klebsiella pneumoniae clone from the Czech Republic producing CTX-M-15 and QnrB1. Antimicrob. Agents Chemother. 57, 1073–1076. doi: 10.1128/AAC.01886-12
Doster, E., Lakin, S. M., Dean, C. J., Wolfe, C., Young, J. G., Boucher, C., et al. (2020). MEGARes 2.0: A database for classification of antimicrobial drug, biocide and metal resistance determinants in metagenomic sequence data. Nucleic Acids Res. 48, D561–D569. doi: 10.1093/nar/gkz1010
Falgenhauer, L., Schwengers, O., Schmiedel, J., Baars, C., Lambrecht, O., Heß, S., et al. (2019). Multidrug-resistant and clinically relevant gram-negative bacteria are present in german surface waters. Front. Microbiol. 10:2779. doi: 10.3389/fmicb.2019.02779
Feldgarden, M., Brover, V., Haft, D. H., Prasad, A. B., Slotta, D. J., Tolstoy, I., et al. (2019). Validating the AMRFINder tool and resistance gene database by using antimicrobial resistance genotype-phenotype correlations in a collection of isolates. Antimicrob. Agents Chemother. 63, e00483–e00419. doi: 10.1128/AAC.00483-19
Finton, M. D., Meisal, R., Porcellato, D., Brandal, L. T., and Lindstedt, B. A. (2020). Whole genome sequencing and characterization of multidrug-resistant (MDR) bacterial strains isolated from a Norwegian university campus pond. Front. Microbiol. 11:1273. doi: 10.3389/fmicb.2020.01273
Follador, R., Heinz, E., Wyres, K. L., Ellington, M. J., Kowarik, M., Holt, K. E., et al. (2016). The diversity of Klebsiella pneumoniae surface polysaccharides. Microb. Genomics 2:e000073. doi: 10.1099/mgen.0.000073
Founou, R. C., Founou, L. L., Allam, M., Ismail, A., and Essack, S. Y. (2019). Whole genome sequencing of extended spectrum β-lactamase (ESBL)-producing Klebsiella pneumoniae isolated from hospitalized patients in KwaZulu-Natal, South Africa. Sci. Rep. 9:6266. doi: 10.1038/s41598-019-42672-2
Furlan, J. P. R., and Stehling, E. G. (2021). Multiple sequence types, virulence determinants and antimicrobial resistance genes in multidrug- and colistin-resistant Escherichia coli from agricultural and non-agricultural soils. Environ. Pollut. 288:117804. doi: 10.1016/j.envpol.2021.117804
Gekenidis, M. T., Rigotti, S., Hummerjohann, J., Walsh, F., and Drissner, D. (2020). Long-term persistence of blactx-m-15 in soil and lettuce after introducing extended-spectrum β-lactamase (Esbl)-producing Escherichia coli via manure or water. Microorganisms 8:1646. doi: 10.3390/microorganisms8111646
Gonzalez-Escalona, N., and Kase, J. A. (2018). Virulence gene profiles and phylogeny of Shiga toxin-positive Escherichia coli strains isolated from FDA regulated foods during 2010-2017. bioRxiv [Preprint]. 1–26. doi: 10.1101/461327
Gou, J. J., Liu, N., Guo, L. H., Xu, H., Lv, T., Yu, X., et al. (2020). Carbapenem-resistant Enterobacter hormaechei ST1103 with IMP-26 carbapenemase and ESBL gene blashv-178. Infect. Drug Resist. 13, 597–605. doi: 10.2147/IDR.S232514
Gupta, S. K., Padmanabhan, B. R., Diene, S. M., Lopez-Rojas, R., Kempf, M., Landraud, L., et al. (2014). ARG-annot, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob. Agents Chemother. 58, 212–220. doi: 10.1128/AAC.01310-13
Hai, P. D., Hoa, L. T. V., Tot, N. H., Phuong, L. L., Quang, V. V., Thuyet, B. T., et al. (2020). First report of biliary tract infection caused by multidrug-resistant Serratia fonticola. New Microb. New Infect. 36:100692. doi: 10.1016/j.nmni.2020.100692
Han, Y., Huang, L., Liu, C., Huang, X., Zheng, R., Lu, Y., et al. (2021). Characterization of carbapenem-resistant Klebsiella pneumoniae st15 clone coproducing kpc-2, ctx-m-15 and shv-28 spread in an intensive care unit of a tertiary hospital. Infect. Drug Resist. 14, 767–773. doi: 10.2147/IDR.S298515
Hassen, B., Abbassi, M. S., Benlabidi, S., Ruiz-Ripa, L., Mama, O. M., Ibrahim, C., et al. (2020). Genetic characterization of ESBL-producing Escherichia coli and Klebsiella pneumoniae isolated from wastewater and river water in Tunisia: predominance of CTX-M-15 and high genetic diversity. Environ. Sci. Pollut. Res. 27, 44368–44377. doi: 10.1007/s11356-020-10326-w
Hauser, E., Mellmann, A., Semmler, T., Stoeber, H., Wieler, L. H., Karch, H., et al. (2013). Phylogenetic and molecular analysis of food-borne Shiga toxin-producing Escherichia coli. Appl. Environ. Microbiol. 79, 2731–2740. doi: 10.1128/AEM.03552-12
Iguchi, A., Nagaya, Y., Pradel, E., Ooka, T., Ogura, Y., Katsura, K., et al. (2014). Genome evolution and plasticity of serratia marcescens, an important multidrug-resistant nosocomial pathogen. Genome Biol. Evol. 6, 2096–2110. doi: 10.1093/gbe/evu160
Iwu, C. D., de Plessis, E. M., Korsten, L., Nontongana, N., and Okoh, A. I. (2020). Antibiogram signatures of some enterobacteria recovered from irrigation water and agricultural soil in two district municipalities of South Africa. Microorganisms 8:1206. doi: 10.3390/microorganisms8081206
Jacoby, G. A. (2009). AmpC Beta-Lactamases. Clin. Microbiol. Rev. 22, 161–182. doi: 10.1128/CMR.00036-08
Jia, B., Raphenya, A. R., Alcock, B., Waglechner, N., Guo, P., Tsang, K. K., et al. (2017). CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 45, 566–573. doi: 10.1093/nar/gkw1004
Jibril, A. H., Okeke, I. N., Dalsgaard, A., Menéndez, V. G., and Olsen, J. E. (2021). Genomic analysis of antimicrobial resistance and resistance plasmids in salmonella serovars from poultry in Nigeria. Antibiotics 10:99. doi: 10.3390/antibiotics10020099
Joensen, K. G., Scheutz, F., Lund, O., Hasman, H., Kaas, R. S., Nielsen, E. M., et al. (2014). Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J. Clin. Microbiol. 52, 1501–1510. doi: 10.1128/JCM.03617-13
Joensen, K. G., Tetzschner, A. M. M., Iguchi, A., Aarestrup, F. M., and Scheutz, F. (2015). Rapid and easy in silico serotyping of Escherichia coli isolates by use of whole-genome sequencing data. J. Clin. Microbiol. 53, 2410–2426. doi: 10.1128/JCM.00008-15
Johansson, M. H. K., Bortolaia, V., Tansirichaiya, S., Aarestrup, F. M., Roberts, A. P., and Petersen, T. N. (2021). Detection of mobile genetic elements associated with antibiotic resistance in Salmonella enterica using a newly developed web tool: MobileElementFinder. J. Antimicrob. Chemother. 76, 101–109. doi: 10.1093/jac/dkaa390
Jones-Dias, D., Manageiro, V., Ferreira, E., Barreiro, P., Vieira, L., Moura, I. B., et al. (2016). Architecture of class 1, 2, and 3 integrons from gram negative bacteria recovered among fruits and vegetables. Front. Microbiol. 7:1400. doi: 10.3389/fmicb.2016.01400
Kaushik, M., Kumar, S., Kapoor, R. K., Virdi, J. S., and Gulati, P. (2018). Integrons in Enterobacteriaceae: diversity, distribution and epidemiology. Int. J. Antimicrob. Agents 51, 167–176. doi: 10.1016/j.ijantimicag.2017.10.004
Khari, F. I. M., Karunakaran, R., Rosli, R., and Tay, S. T. (2016). Genotypic and phenotypic detection of AmpC β-lactamases in Enterobacter spp. isolated from a teaching hospital in Malaysia. PLoS One 11:e0150643. doi: 10.1371/journal.pone.0150643
Kim, S., Karns, J. S., Van Kessel, J. A. S., and Haley, B. J. (2017). Genome sequences of five multidrug-resistant Escherichia coli sequence type 117 isolates recovered from dairy calves. Genome Announc. 5, e00732–e00717. doi: 10.1128/genomeA.00732-17
Kim, S., Kim, H., Kim, Y., Kim, M., Kwak, H., and Ryu, S. (2020). Whole-genome sequencing-based characteristics in extended-spectrum beta-lactamase-producing Escherichia coli isolated from retail meats in Korea. Microorganisms 8:508. doi: 10.3390/microorganisms8040508
Larsen, M. V., Cosentino, S., Rasmussen, S., Friis, C., Hasman, H., Marvig, R. L., et al. (2012). Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 50, 1355–1361. doi: 10.1128/JCM.06094-11
Li, B., Vellidis, G., Liu, H., Jay-Russell, M., Zhao, S., Hu, Z., et al. (2014). Diversity and antimicrobial resistance of salmonella enterica isolates from surface water in southeastern United States. Appl. Environ. Microbiol. 80, 6355–6365. doi: 10.1128/AEM.02063-14
Liakopoulos, A., Mevius, D., and Ceccarelli, D. (2016). A review of SHV extended-spectrum β-lactamases: neglected yet ubiquitous. Front. Microbiol. 7:1374. doi: 10.3389/fmicb.2016.01374
Liao, X. P., Xia, J., Yang, L., Li, L., Sun, J., Liu, Y. H., et al. (2015). Characterization of CTX-M-14-producing Escherichia coli from food-producing animals. Front. Microbiol. 6:1136. doi: 10.3389/fmicb.2015.01136
Liu, H., Whitehouse, C. A., and Li, B. (2018). Presence and persistence of salmonella in water: the impact on microbial quality of water and food safety. Front. Public Health 6:159. doi: 10.3389/fpubh.2018.00159
Lobanovska, M., and Pilla, G. (2017). Penicillin’s discovery and antibiotic resistance: lessons for the future? Yale J. Biol. Med. 90, 135–145. doi: 10.1103/PhysRevA.32.435
Löhr, I. H., Hülter, N., Bernhoff, E., Johnsen, P. J., Sundsfjord, A., and Naseer, U. (2015). Persistence of a pKPN3-like CTX-M-15-encoding IncFIIK plasmid in a Klebsiella pneumonia ST17 host during two years of intestinal colonization. PLoS One 10:e0116516. doi: 10.1371/journal.pone.0116516
Lopes, R., Fuentes-Castillo, D., Fontana, H., Rodrigues, L., Dantas, K., Cerdeira, L., et al. (2021a). Endophytic lifestyle of global clones of extended-spectrum beta-lactamase-producing priority pathogens in fresh vegetables: a trojan horse strategy favoring human colonization? mSystems 6, e01125–e01120. doi: 10.1128/mSystems.01125-20
Lopes, R., Furlan, J. P. R., Dos Santos, L. D. R., Gallo, I. F. L., and Stehling, E. G. (2021b). Colistin-resistant mcr-1-positive Escherichia coli ST131-H22 carrying Bla CTX-M-15 and qnrB19 in agricultural soil. Front. Microbiol. 12:659900. doi: 10.3389/fmicb.2021.659900
Maluta, R. P., Leite, J. L., Rojas, T. C. G., Scaletsky, I. C. A., Guastalli, E. A. L., de Carvalho Ramos, M., et al. (2017). Variants of astA gene among extra-intestinal Escherichia coli of human and avian origin. FEMS Microbiol. Lett. 364, 1–5. doi: 10.1093/femsle/fnw285
Mansour, M. N., Yaghi, J., El Khoury, A., Felten, A., Mistou, M. Y., Atoui, A., et al. (2020). Prediction of salmonella serovars isolated from clinical and food matrices in Lebanon and genomic-based investigation focusing on Enteritidis serovar. Int. J. Food Microbiol. 333:108831. doi: 10.1016/j.ijfoodmicro.2020.108831
Mbelle, N. M., Feldman, C., Sekyere, J. O., Maningi, N. E., Modipane, L., and Essack, S. Y. (2020). Pathogenomics and evolutionary epidemiology of multi-drug resistant clinical Klebsiella pneumoniae isolated from Pretoria, South Africa. Sci. Rep. 10:1232. doi: 10.1038/s41598-020-58012-8
McKinnon, J., Roy Chowdhury, P., and Djordjevic, S. P. (2018). Genomic analysis of multidrug-resistant Escherichia coli ST58 causing urosepsis. Int. J. Antimicrob. Agents 52, 430–435. doi: 10.1016/j.ijantimicag.2018.06.017
Naas, T., Poirel, L., and Nordmann, P. (2008). Minor extended-spectrum β-lactamases. Clin. Microbiol. Infect. 14, 42–52. doi: 10.1111/j.1469-0691.2007.01861.x
Nair, S., Ashton, P., Doumith, M., Connell, S., Painset, A., Mwaigwisya, S., et al. (2016). WGS for surveillance of antimicrobial resistance: A pilot study to detect the prevalence and mechanism of resistance to azithromycin in a UK population of non-typhoidal salmonella. J. Antimicrob. Chemother. 71, 3400–3408. doi: 10.1093/jac/dkw318
Njage, P. M. K., and Buys, E. M. (2014). Pathogenic and commensal Escherichia coli from irrigation water show potential in transmission of extended spectrum and AmpC β-lactamases determinants to isolates from lettuce. Microb. Biotechnol. 8, 462–473. doi: 10.1111/1751-7915.12234
O’neill, J. (2016). Tackling Drug-Resistant Infections Globally: Final Report and Recommendations the Review on Antimicrobial Resistance.
Oniciuc, E. A., Likotrafiti, E., Alvarez-Molina, A., Prieto, M., Santos, J. A., and Alvarez-Ordóñez, A. (2018). The present and future of whole genome sequencing (WGS) and whole metagenome sequencing (WMS) for surveillance of antimicrobial resistant microorganisms and antimicrobial resistance genes across the food chain. Gene 9:268. doi: 10.3390/genes9050268
Palzkill, T. (2018). Structural and mechanistic basis for extended-spectrum drug-resistance mutations in altering the specificity of TEM, CTX-M, and KPC β-lactamases. Front. Mol. Biosci. 5:16. doi: 10.3389/fmolb.2018.00016
Partridge, S. R. (2011). Analysis of antibiotic resistance regions in gram-negative bacteria. FEMS Microbiol. Rev. 35, 820–855. doi: 10.1111/j.1574-6976.2011.00277.x
Partridge, S. R., Kwong, S. M., Firth, N., and Jensen, S. O. (2018). Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 31, e00088–e00017. doi: 10.1128/CMR.00088-17
Peirano, G., van Greune, C. H. J., and Pitout, J. D. D. (2011). Characteristics of infections caused by extended-spectrum β-lactamase-producing Escherichia coli from community hospitals in South Africa. Diagn. Microbiol. Infect. Dis. 69, 449–453. doi: 10.1016/j.diagmicrobio.2010.11.011
Petersen, L. M., and Tisa, L. S. (2013). Friend or foe? A review of the mechanisms that drive serratia towards diverse lifestyles. Can. J. Microbiol. 59, 627–640. doi: 10.1139/cjm-2013-0343
Razavi, M., Kristiansson, E., Flach, C.-F., and Larsson, D. G. J. (2020). The association between insertion sequences and antibiotic resistance genes. mSphere 5, 418–420. doi: 10.1128/mSphere.00418-20
Reid, C. J., Blau, K., Jechalke, S., Smalla, K., Djordjevic, S. P., and Campo, R.Del (2020). Whole genome sequencing of Escherichia coli from store-bought produce. Front. Microbiol. 10: 3050. doi: 10.3389/fmicb.2019.03050
Reuland, E. A., Al Naiemi, N., Raadsen, S. A., Savelkoul, P. H. M., Kluytmans, J. A. J. W., and Vandenbroucke-Grauls, C. M. J. E. (2014). Prevalence of ESBL-producing Enterobacteriaceae in raw vegetables. Eur. J. Clin. Microbiol. Infect. Dis. 33, 1843–1846. doi: 10.1007/s10096-014-2142-7
Richards, G. A., Brink, A. J., and Feldman, C. (2019). Rational use of the fluoroquinolones. South Afrn. Med. J. 109, 378–381. doi: 10.7196/SAMJ.2019.v109i6.14002
Richter, L., Du Plessis, E. M., Duvenage, S., and Korsten, L. (2019). Occurrence, identification, and antimicrobial resistance profiles of extended-spectrum and AmpC β-lactamase-producing enterobacteriaceae from fresh vegetables retailed in Gauteng province. South Afr. Foodborne Pathog. Dis. 16, 421–427. doi: 10.1089/fpd.2018.2558
Richter, L., du Plessis, E. M., Duvenage, S., and Korsten, L. (2020). Occurrence, phenotypic and molecular characterization of extended-spectrum- and AmpC- β-lactamase producing enterobacteriaceae isolated from selected commercial spinach supply chains in South Africa. Front. Microbiol. 11:638. doi: 10.3389/fmicb.2020.00638
Roer, L., Hendriksen, R. S., Leekitcharoenphon, P., Lukjancenko, O., Kaas, R. S., Hasman, H., et al. (2016). Is the evolution of Salmonella enterica subsp. enterica linked to restriction-modification systems? mSystems 1, e00009–e00016. doi: 10.1128/mSystems.00009-16
Sabbagh, S. C., Forest, C. G., Lepage, C., Leclerc, J. M., and Daigle, F. (2010). So similar, yet so different: uncovering distinctive features in the genomes of Salmonella enterica serovars typhimurium and Typhi. FEMS Microbiol. Lett. 305, 1–13. doi: 10.1111/j.1574-6968.2010.01904.x
Sandner-Miranda, L., Vinuesa, P., Cravioto, A., and Morales-Espinosa, R. (2018). The genomic basis of intrinsic and acquired antibiotic resistance in the genus Serratia. Front. Microbiol. 9:828. doi: 10.3389/fmicb.2018.00828
Schwanbeck, J., Bohne, W., Hasdemir, U., Groß, U., Pfeifer, Y., Bunk, B., et al. (2021). Detection of a new resistance-mediating plasmid chimera in a blaOXA-48-positive Klebsiella pneumoniae strain at a German university hospital. Microorganisms 9:720. doi: 10.3390/microorganisms9040720
Sekyere, J. O., Maningi, N. E., Modipane, L., and Mbelle, N. M. (2020). Emergence of mcr-9.1 in extended-spectrum-beta-lactamse-producing clinical Enterobacteriaceae in Pretoria, South Africa: global evolutionary Phylogenomics, Resistome, and Mobilome. mSystems 5, e00148–e00120. doi: 10.1128/mSystems.00148-20
Suez, J., Porwollik, S., Dagan, A., Marzel, A., Schorr, Y. I., Desai, P. T., et al. (2013). Virulence gene profiling and pathogenicity characterization of non-typhoidal salmonella accounted for invasive disease in humans. PLoS One 8:e58449. doi: 10.1371/journal.pone.0058449
Surleac, M., Barbu, I. C., Paraschiv, S., Popa, L. I., Gheorghe, I., Marutescu, L., et al. (2020). Whole genome sequencing snapshot of multidrug resistant Klebsiella pneumoniae strains from hospitals and receiving wastewater treatment plants in southern Romania. PLoS One 15:e0228079. doi: 10.1371/journal.pone.0228079
Tanaka, H., Hayashi, W., Iimura, M., Taniguchi, Y., Soga, E., Matsuo, N., et al. (2019). Wastewater as a probable environmental reservoir of extended-spectrum-beta-lactamase genes: detection of chimeric beta-lactamases CTX-M-64 and CTX-M-123. Appl. Environ. Microbiol. 85, e01740–e01719. doi: 10.1128/AEM.01740-19
Tasić, S., Obradović, D., and Tasić, I. (2013). Characterization of Serratia fonticola, an opportunistic pathogen isolated from drinking water. Arch. Biol. Sci. 65, 899–904. doi: 10.2298/ABS1303899T
Tekele, S. G., Teklu, D. S., Tullu, K. D., Birru, S. K., and Legese, M. H. (2020). Extended-spectrum Beta-lactamase and AmpC beta-lactamases producing gram negative bacilli isolated from clinical specimens at international clinical laboratories, Addis Ababa, Ethiopia. PLoS One 15:e0241984. doi: 10.1371/journal.pone.0241984
Thomas, J., Govender, N., McCarthy, K. M., Erasmus, L. K., Doyle, T. J., Allam, M., et al. (2020). Outbreak of listeriosis in South Africa associated with processed meat. N. Engl. J. Med. 382, 632–643. doi: 10.1056/NEJMoa1907462
Thompson, C. P., Doak, A. N., Amirani, N., Schroeder, E. A., Wright, J., Kariyawasam, S., et al. (2018). High-resolution identification of multiple salmonella serovars in a single sample by using CRISPRSeroSeq. Appl. Environ. Microbiol. 84, e01859–e01818. doi: 10.1128/AEM.01859-18
van Duin, D., and Doi, Y. (2017). The global epidemiology of carbapenemase-producing Enterobacteriaceae. Virulence 8, 460–469. doi: 10.1080/21505594.2016.1222343
Varani, A., He, S., Siguier, P., Ross, K., and Chandler, M. (2021). The IS6 family, a clinically important group of insertion sequences including IS26. Mob. DNA 12:11. doi: 10.1186/s13100-021-00239-x
Velásquez, J. C., Hidalgo, A. A., Villagra, N., Santiviago, C. A., Mora, G. C., and Fuentes, J. A. (2016). SPI-9 of Salmonella enterica serovar typhi is constituted by an operon positively regulated by RpoS and contributes to adherence to epithelial cells in culture. Microbiology 162, 1367–1378. doi: 10.1099/mic.0.000319
Verlicchi, P., and Grillini, V. (2020). Surface water and ground water quality in South Africa and Mozambique-analysis of the most critical pollutants for drinking purposes and challenges in water treatment selection. WaterSA 12:305. doi: 10.3390/w12010305
Waterman, S. R., and Holden, D. W. (2003). Functions of the salmonella pathogenicity island 2 (SPI-2) type III secretion system. Cell. Microbiol. 5, 501–511. doi: 10.1046/j.1462-5822.2003.00294.x
WHO (2017). Global Priority list of antibiotic-resistant bacteria to guide research, discovery, and develpment of new antibiotics.
WHO (2020). Global Antimicrobial Resistance and Use Surveillance System (GLASS) Report. Available at: https://www.who.int/glass/resources/publications/early-implementation-report-2020/en/ (Accessed June 15, 2021).
Wick, R. R., Heinz, E., Holt, K. E., and Wyres, K. L. (2018). Kaptive web: user-friendly capsule and lipopolysaccharide serotype prediction for Klebsiella genomes. bioRxiv [Preprint]. doi: 10.1101/260125
Ye, Q., Wu, Q., Zhang, S., Zhang, J., Yang, G., Wang, J., et al. (2017). Characterization of extended-spectrum β-lactamase-producing enterobacteriaceae from retail food in China. Front. Microbiol. 9:1709. doi: 10.3389/fmicb.2018.01709
Zankari, E., Hasman, H., Cosentino, S., Vestergaard, M., Rasmussen, S., Lund, O., et al. (2012). Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67, 2640–2644. doi: 10.1093/jac/dks261
Zhang, S., Yin, Y., Jones, M. B., Zhang, Z., Kaiser, B. L. D., Dinsmore, B. A., et al. (2015). Salmonella serotype determination utilizing high-throughput genome sequencing data. J. Clin. Microbiol. 53, 1685–1692. doi: 10.1128/JCM.00323-15
Zhou, K., Zhou, Y., Zhang, C., Song, J., Cao, X., Yu, X., et al. (2020). Dissemination of a ‘rare’ extended-spectrum β-lactamase gene blaSFO-1 mediated by epidemic clones of carbapenemase-producing Enterobacter hormaechei in China. Int. J. Antimicrob. Agents 56:106079. doi: 10.1016/j.ijantimicag.2020.106079
Zurfluh, K., Nuesch-Inderbinen, M., Morach, M., Berner, A. Z., Hachler, H., and Stephan, R. (2015). Extended-spectrum-beta-lactamase-producing Enterobacteriaceae isolated from vegetables imported from the Dominican Republic, India, Thailand, and Vietnam. Appl. Environ. Microbiol. 81, 3115–3120. doi: 10.1128/AEM.00258-15
Keywords: WGS, food safety, leafy greens, multidrug resistance, foodborne bacterial pathogens
Citation: Richter L, du Plessis EM, Duvenage S, Allam M, Ismail A and Korsten L (2021) Whole Genome Sequencing of Extended-Spectrum- and AmpC- β-Lactamase-Positive Enterobacterales Isolated From Spinach Production in Gauteng Province, South Africa. Front. Microbiol. 12:734649. doi: 10.3389/fmicb.2021.734649
Edited by:
Aloysius Wong, Kean University-Wenzhou, ChinaReviewed by:
Ralf Lopes, University of São Paulo, BrazilMasaki Yamamoto, Kyoto University Hospital, Japan
Copyright © 2021 Richter, du Plessis, Duvenage, Allam, Ismail and Korsten. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Erika M. du Plessis, Erika.duplessis@up.ac.za
 Loandi Richter
Loandi Richter Erika M. du Plessis
Erika M. du Plessis Stacey Duvenage
Stacey Duvenage Mushal Allam3
Mushal Allam3 Arshad Ismail
Arshad Ismail Lise Korsten
Lise Korsten